ABSTRACT
Melioidosis is an underreported human disease caused by the Gram-negative intracellular pathogen Burkholderia pseudomallei (Bpm). Both the treatment and the clearance of the pathogen are challenging, with high relapse rates leading to latent infections. This has been linked to the bacterial persistence phenomenon, a growth arrest strategy that allows bacteria to survive under stressful conditions, as in the case of antibiotic treatment, within a susceptible clonal population. At a molecular level, this phenomenon has been associated with the presence of toxin–antitoxin (TA) systems. We annotated the Bpm K96243 genome and selected 11 pairs of genes encoding for these TA systems, and their expression was evaluated under different conditions (supralethal antibiotic conditions; intracellular survival bacteria). The predicted HigB toxin (BPSL3343) and its predicted antitoxin HigA (BPS_RS18025) were further studied using mutant construction. The phenotypes of two mutants (ΔhigB and ΔhigB ΔhigA) were evaluated under different conditions compared to the wild-type (WT) strain. The ΔhigB toxin mutant showed a defect in intracellular survival on macrophages, a phenotype that was eliminated after levofloxacin treatment. We found that the absence of the toxin provides an advantage over the WT strain, in both in vitro and in vivo models, during persister conditions induced by levofloxacin. The lack of the antitoxin also resulted in differential responses to the conditions evaluated, and under some conditions, it restored the WT phenotype, overall suggesting that both toxin and antitoxin components play a role in the persister-induced phenotype in Bpm.
KEYWORDS: Burkholderia pseudomallei, toxin-antitoxin, HigBA, persistence, antibiotics
INTRODUCTION
Burkholderia pseudomallei (Bpm) is a Gram-negative, facultative intracellular pathogen, and the causative agent of melioidosis. Melioidosis is a multifaceted disease affecting humans and some mammals that reaches up to 50% mortality with an incidence of approximately 165,000 cases per year, although this prediction is considered underreported (1–3). Melioidosis disease includes a myriad of symptoms, while the most common ones are pneumonia and skin localized abscesses, usually leading to sepsis during the acute stage of the infection; the chronic stage of the infection can stay asymptomatic for long periods of time (1). Clearance of the pathogen is challenging due to its endogenous multidrug resistance, but also results in relapse rates ranging between 15 and 23%. This relapse is associated with the bacterial intracellular lifestyle, which causes latent infections similar to the ones observed during tuberculosis caused by Mycobacterium tuberculosis (2). The current recommended treatment for melioidosis consists of 2–8 weeks of intravenous antimicrobial therapy followed by 3–6 months of oral antimicrobial therapy (1). For Bpm, both treatment failure and chronic infection have been linked to persistence (4).
Bacterial persistence is a cell cycle arrest phenomenon in which phenotypic variants that are highly tolerant to antimicrobial treatment, among other environmental stresses, exist within bacterial communities (5, 6). This phenomenon differs from antibiotic resistance because persister bacteria do not express gene products that enable the organism to grow in the presence of the antibiotic, so they do not proliferate. Persisters’ biological relevance lies in the fact that this phenomenon has been associated with latent and chronic infections, as well as implicated in nearly all bacterial infections that result in treatment failure (4). This cellular phenotype has already been linked to the ability of the bacterium to generate latent infections in tuberculosis, skin infections by Staphylococcus aureus, and Escherichia coli urinary tract infections (4, 7).
At a molecular level, the switching mechanism between growth and persistence phenotypes has been associated with toxin–antitoxin (TA) systems, which were first described in E. coli with the hipQ gene (8). These TA modules are small genetic elements broadly distributed in bacteria, which encode a two-component system constituted by a toxin and its associated antitoxin. A typical TA pair comprises a stable toxin protein that interferes with multiple cellular processes and an unstable protein or RNA antitoxin that neutralizes the toxic effect of the active form of the toxin (6, 9). According to the genetic structures and their regulation, these systems are assigned to four main classes, although eight different classes have been described. Differences among the classes rely on how the antitoxin interferes and neutralizes the toxin effect (6, 9–11).
Among all of them, the type II is still the largest and best-studied class of TA systems. In them, the antitoxin (protein) neutralizes the toxic effect through two main mechanisms: directly forming a complex toxin–antitoxin, or as a transcriptional regulator that represses the expression of the operon (10). Under normal growth conditions, the antitoxin forms a protein–protein complex with the toxin that counteracts its toxic effect and regulates the transcription of the operon. However, under different stressful stimuli, the antitoxin could be rapidly degraded by proteases while the toxin is released, changing into its active form (9). This regulation, especially in type II TA systems, could be flexible or promiscuous, because a toxin belonging to a superfamily can also form a complex with the antitoxin from another system, or an antitoxin can counteract the toxicity from different noncognate toxins from same or different superfamilies (6, 12). Emerging evidence has shown that some type II systems are involved in pathogen adaptation to host tissues during chronic or recurrent infections, which is linked to the persister phenotype (13–15). For example, the HigBA system in Pseudomonas aeruginosa has been shown to participate in biofilm formation, iron uptake, carbon metabolism, and virulence involving Type 3 and Type 6 secretion systems (16, 17). More recently, a transcriptional regulatory role of virulence genes has been attributed to the antitoxin HigA (18).
Through bioinformatics prediction approaches, between 62 and 106 toxin–antitoxin systems have been identified in B. pseudomallei genomes. Using transcriptional analysis data, many of these were associated with responses to different stresses or host conditions (19, 20). Some isogenic mutants lacking these toxins demonstrated an attenuation in a murine model of chronic melioidosis infection, with a lower bacterial burden in target organs (19, 21).
In this study, we have analyzed the expression of 11 of those predicted TA systems. While several pairs were differentially expressed under the persister-induced conditions evaluated, we focused on one predicted system encoded by BPSL3343 (toxin) and BPS_RS18025 (antitoxin) genes that encode a putative type II HigB-HigA system. The toxin mutant (ΔBPSL3343, named ICG001) showed a defect on intracellular survival in macrophages, a phenotype which was eliminated after levofloxacin treatment. The absence of the toxin provides an advantage over the wild-type (WT) strain, both in vitro and in vivo models, during persister-induced conditions. In most of these conditions, the differential responses after levofloxacin treatment disappeared when both toxin and antitoxin genes were deleted, which might suggest a role of the antitoxin in the regulatory process.
RESULTS
Genomic annotation and gene expression.
The genome sequence from the reference Bpm strain K96243 was obtained from the NCBI database. Both Prokka (22) and RAST (23) databases were used to annotate the genome and search for toxin–antitoxin predicted modules. Both databases were used to compare predictions between them, because RAST mainly annotates described genes with Enterobacteriaceae names, while Prokka utilizes specific assigned names when they are available (24). The presence of genomic islands (GIs) was also checked with Prokka software, because the toxin–antitoxin (TA) systems are usually predicted to be located in relationship with those genomic structures, due to their first-described role as plasmid maintenance systems (25, 26). The predicted/annotated systems were compared with the TA-encoding genes that we have previously assigned into three clusters, based on their transcriptomic profile and their association with host–environment conditions and antibiotic treatment (19). Those three clusters were manually established according to specific expression patterns when Bpm was exposed to different host-like conditions or to antibiotics. Both independent approach results were evaluated, and results compared. Finally, orthologues of the chosen systems were identified in a second Bpm reference strain (1026b) to determine whether they were present and conserved across different strains. The genes, the identified systems, their association to a previously described cluster, and their link to any genomic island are shown in Table 1.
TABLE 1.
List of toxin–antitoxin predicted systems in Bpm K96243a
| Toxin | Antitoxin | System predicted | GIs | Cluster | 1026b orthologue | Chromosome |
|---|---|---|---|---|---|---|
| BPSS0390 | BPSS0391 | HicA/HicB | 13 | 3 | No | 2 |
| BPSS0395 | BPSS0394 | BrnT/BrnA | 3 | Yes | 2 | |
| BPSS1584 | BPSS1583 | HipA/HipB | 3 | Yes | 2 | |
| BPSL0175 | BPSL0174 | RelE/ParE | 2 | 3 | Yes | 1 |
| BPSL0559 | BPSL0558 | HipA/XRE regulator | 3 | No | 1 | |
| BPSL2333 | BPSL2334 | RelE/ParE | 3 | Yes | 1 | |
| BPSL3261 | BPSL3260 | PtaRNA1/HigA | 11 | No | 1 | |
| BPSS1060 | BPSS1061 | RelE/ParE | 3 | Yes | 2 | |
| BPSL3343 | BPS_RS18025 | HigB/HigA | 12 | 3 | Yes | 1 |
| BPSL0034 | BPSL0033 | SAM-methyltransferase/MerR regulator | 2 | Yes | 1 | |
| BPSL0952 | BPSL0953 | Replication protein/AlpA regulator | 5 | 3 | Yes | 1 |
The list shows the chosen systems evaluated in this work, their relationship with genomic islands (GIs), if they belong to a described cluster (19), if they are present in the Bpm 1026 strain, and in which K96243 chromosome they are located.
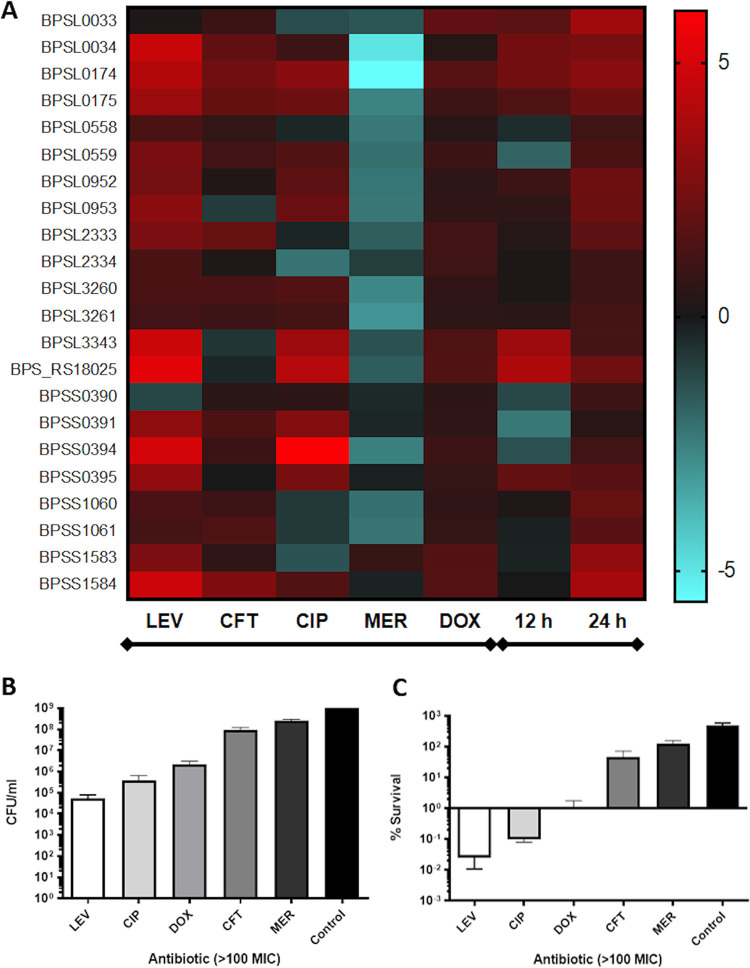
The results for gene expression are shown as a heatmap in Fig. 1A. Antibiotic treatment upregulated most TA systems, while meropenem resulted in a general downregulation across most of those modules, where we hypothesized that Bpm may exhibit resistance to the high concentration of this antibiotic (Fig. 1B and C) and that hence the toxin–antitoxin systems are not needed. For most of them, both toxin and antitoxin predicted genes were upregulated because they are usually cotranscribed (Fig. 1A). The most upregulated TA systems were observed with levofloxacin, especially the pairs encoded by BPSL3343/BPS_RS18025 (annotated as HigB/HigA), BPSS0395/BPSS0394 (annotated as BrnT/BrnA), and BPSS1584/BPSS1583 (annotated as HipA/HipB), the three of them belonging to the type II TA class (Table 1). The expression of those predicted TA systems genes, summarized in Table 1, was evaluated in the surviving bacteria after supralethal concentration of five different clinically relevant antibiotics (e.g., levofloxacin, ciprofloxacin, doxycycline, ceftazidime, and meropenem) (Fig. 1B and C) (21). The HigB/HigA (BPSL3343/BPS_RS18025) and BrnT/BrnA (BPSS0395/BPSS0394) predicted systems also showed higher overexpression after ciprofloxacin treatment (Fig. 1A). In Fig. 1B, the total bacteria after 24 h of antibiotic treatment or nonsupplemented LB (control) is shown. Using the input numbers (2 × 108 CFU/mL) and the CFU enumeration after 24 h, the survival rates were established as % survival (Fig. 1C). The antibiotics impacted bacterial CFU (Fig. 1B) compared with the nontreated group, particularly those exposed to fluoroquinolones (levofloxacin, ciprofloxacin), which showed the lower survival rates (Fig. 1C).
FIG 1.
Expression of predicted toxin–antitoxin systems under different conditions and Bpm K96243 survival after antibiotic treatment. (A) Heatmap with the predicted toxin and antitoxin genes (left) from Bpm K96243 and the conditions evaluated on the bottom (left group, antibiotics exposition; right group, intracellular survival bacteria at 12 h and 24 h). As control of expression, bacteria were grown in media (LB or DMEM) without supplements. Each condition was measured at least in triplicate from three independent replicates. (B) CFU enumeration and (C) survival based on the input after 24 h of supralethal antibiotic concentration exposure of Bpm K96243 to LEV, levofloxacin; CIP, ciprofloxacin; DOX, doxycycline; CFT, ceftazidime; MER, meropenem; Control, LB medium.
Because host-associated conditions and the intracellular life cycle of the pathogen are interconnected, intracellular survival bacteria from macrophages were also used to analyze gene expression at 12 h and 24 h (Fig. 1A). In both cases, antibiotic-induced and intracellular conditions, bacteria grown in nonsupplemented media (LB or Dulbecco's modified Eagle medium [DMEM]) were used as baseline expression to establish the fold change (Fig. 1A, Fig. 2A and B). During intracellular conditions, all the systems are upregulated at 24 h postinfection compared with bacteria in DMEM, which was used as baseline. However, to understand the activation dynamics among the systems, we only focused on the 12 h time point. Among all of them, the system annotated as BPSL3343 (toxin) and BPS_RS18025 (antitoxin) demonstrated the strongest upregulation (Fig. 1A). These two genes were predicted to be a HigBA type II toxin–antitoxin system (as shown in Table 1).
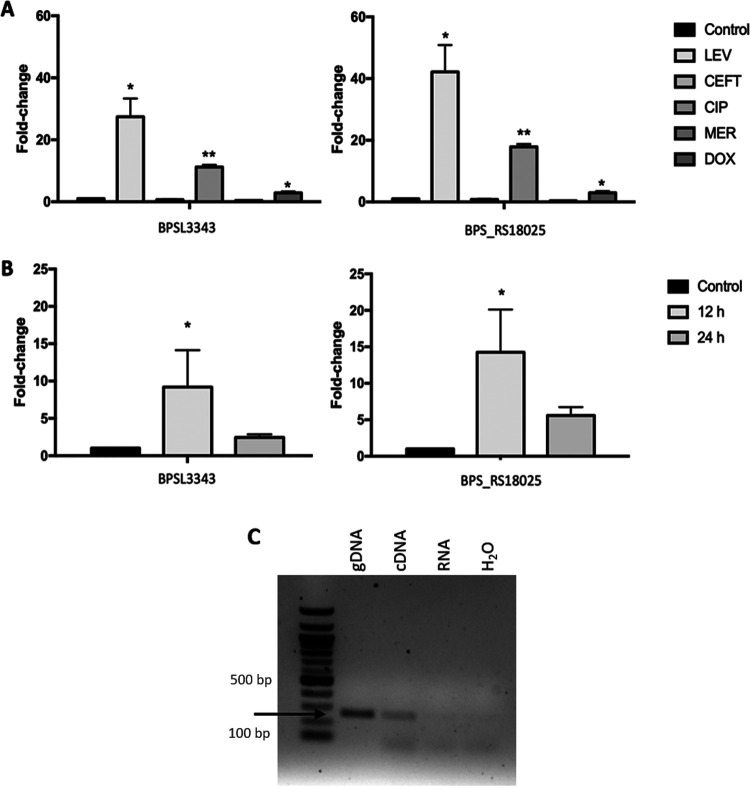
FIG 2.
BPSL3343 and BPS_RS18025 TA system expression. (A) BPSL3343 (toxin) and BPS_RS18025 (antitoxin) expression under different antibiotic exposure for 24 h in LB media, using bacteria kept in nonsupplemented LB as control for expression. LEV, levofloxacin; CIP, ciprofloxacin; DOX, doxycycline; CEFT, ceftazidime; MER, meropenem. (B) BPSL3343 (toxin) and BPS_RS18025 (antitoxin) expression during intracellular survival in murine macrophages after 12 h or 24 h, using bacteria maintained in DMEM as control for expression. (C) Coexpression of both genes. The gDNA (second lane) was used as template, and cDNA (third lane) and RNA (fourth lane) from Bpm K96243 were used for the same purpose; black arrow shows the PCR product with same size between samples. 100 bp DNA ladder (NEB) was loaded in the first lane. *, P < 0.05; **, P < 0.01.
In Fig. 2 (A–B), the gene expression of BPSL3343 and BPS_RS18025 genes is shown in all conditions as well as the ones in which they are significantly overexpressed compared with the controls. The most potent activation of expression appears to occur with antibiotics of the fluoroquinolone class, with levofloxacin and ciprofloxacin treatment resulting in the highest levels of expression among all the antibiotics evaluated (Fig. 1A, 2A). The intracellular environment also triggers elevated levels of expression, suggesting that redox and nutrient stress might play a role in activation (Fig. 1A, 2B). Because both genes showed the same overexpression pattern, cotranscription between them was assessed. The PCR was performed using either genomic DNA or cDNA as a template with primers designed to only amplify the genes while cotranscribed. As shown in Fig. 2C, the same amplicon size was obtained for both template sources, confirming the fact that they are cotranscribed.
Mutant construction and phenotypic characterization.
To identify the role of the predicted HigBA toxin–antitoxin system in the persister and virulence features of Bpm, we constructed two isogenic mutants in the Bpm K96243 background. The isogenic mutant carrying a deletion in the predicted toxin encoded by the annotated higB gene (ICG001) was obtained through allelic exchange using K96243 as donor strain. Construction of the predicted antitoxin higA deletion mutant could not be accomplished, suggesting that the lack of this antitoxin could be deleterious for the bacteria, which has been previously described in other TA systems (27). For the double mutant (ICG002) construction, the ICG001 mutant strain was used as donor strain, with a high selection efficiency, highlighting the fact that the antitoxin deletion could not be achieved due to the unhindered activity of the toxin.
Once all strains were obtained, a phenotypic characterization was performed to rule out the differences in growth among the strains was dependent on a fitness loss in the different media evaluated (Fig. S1A–C in the supplemental material). The growth in any of the media (LB, M9, or DMEM) was not different among the strains, suggesting that the lack of these genes does not affect its fitness in nutrient-rich, minimal, or host-like media, when no other stresses are present. There were no differences in colony morphology while growing in Ashdown’s selective medium (Fig. S1D).
Persister-induction after supralethal antibiotic concentrations.
One of the main problems of melioidosis disease is the failure of antibiotic treatment, which is linked to the survival population that persists after antimicrobial exposure via growth arrest. The ability of Bpm to survive after exposure to supralethal concentrations of clinically relevant antibiotics has been previously demonstrated (21). Following that premise, five antibiotics were chosen, and the survival bacteria was quantified after 24 h in the presence of 100× MIC treatment with the WT and each isogenic mutant strain (Fig. 3).
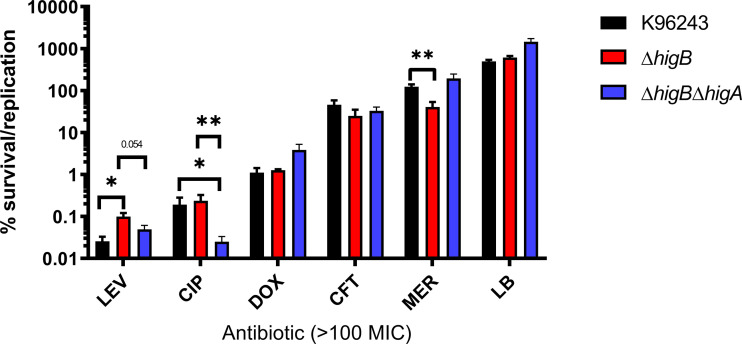
FIG 3.
Supralethal antibiotic concentration induced persister survival. Percentage of survival after 24 h of antibiotic exposure compared with the input bacteria of Bpm K96243, ΔhigB, and ΔhigB ΔhigA. LEV, levofloxacin; CIP, ciprofloxacin; DOX, doxycycline; CFT, ceftazidime; MER, meropenem. Bars represent the average of three independent experiments in triplicate ± standard deviation (SD). *, P < 0.05; **, P < 0.01.
The ΔhigB mutant demonstrated increased fitness under levofloxacin stress, with significantly higher survival compared to wild type K96243 and higher trend of survival versus ΔhigB ΔhigA. The presence of the toxin did not seem to affect survival under ciprofloxacin treatment because the ΔhigB and wild-type strains showed similar survival rates when exposed to this antibiotic; both survived at higher levels compared to ΔhigB ΔhigA. This suggests that the lack of the antitoxin or the entire system impacts the survival phenotype. This was surprising considering the remarkably similar effect both fluoroquinolone compounds had on gene expression (Fig. 1A and 2A). Conversely, meropenem treatment resulted in decreased survival for the ΔhigB mutant compared to wild type and double mutant ΔhigB ΔhigA. This antibiotic treatment caused a global downregulation of tested TA systems (Fig. 1A), suggesting that activation of TA systems during meropenem stress is not needed, which could suggest a resistant more than a persister phenotype. However, the decreased survival of ΔhigB and equitable survival between wild-type K96243 and ΔhigB ΔhigA demonstrates that the activation of this HigBA system and other redundant TA systems might be deleterious under meropenem stress. Nevertheless, considering that persister cells do not proliferate during antibiotic exposure, meropenem survivals seem to be due to a resistance phenotype (Fig. 1C). Doxycycline and ceftazidime showed no significant differences or trends, reinforcing the idea that TA activation and persister formation is specific to different stresses.
Both ceftazidime and meropenem are intravenous antimicrobials used in the treatment of this bacterium (28); however, they showed to be the less effective for in vitro killing. Both fluoroquinolones were the most effective compounds in vitro, exhibiting a different survival capacity among the strains with the TA HigBA system disrupted, and the different antimicrobials have different impacts on the Bpm K96243 survival rates.
Biofilm formation in antibiotic- or macrophage-induced persister conditions.
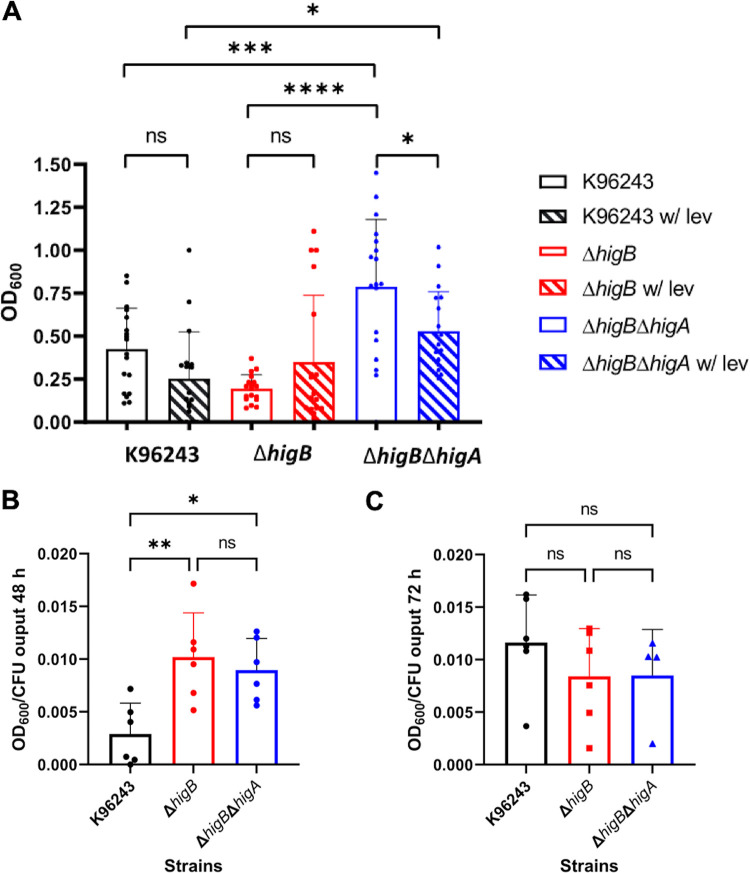
Biofilm formation is usually considered a virulence-associated phenotype because many chronic infections are linked to the capacity of the pathogens to form these 3D structures (29). It is also known that biofilm creates an environment for the survival of the persister cells (29). For Bpm, it is suggested that biofilm may play a role in the pathogen adherence to epithelial cells and participate in relapses of melioidosis disease (30, 31). As shown in Fig. 4A, the biofilm formation capacity is significantly reduced in the isogenic ΔhigB mutant strain as compared with the wild type, while biofilm is increased in the double mutant when compared to both the toxin mutant and the WT strains (Fig. 4A). When the wild-type biofilm was treated with levofloxacin, the ability to form this 3D structure was reduced as well as in the double mutant, but the biofilm formed by the toxin mutant was slightly increased. While those differences, in general, were not significant, the result highlights the differential behavior of these strains in the presence of fluoroquinolone treatment.
FIG 4.
Biofilm formation in the presence or absence of levofloxacin or from intracellular bacteria. (A) White bars represent the biofilm formed in nonsupplemented LB and the pattern labeled bars the biofilm formed in the presence of levofloxacin. Among strains, only significant differences are shown. Bars represent the average of six independent experiments in triplicate ± SD. (B–C) Intracellular bacteria recovered from RAW 264.7 cells and treatment with levofloxacin for 24 h were further incubated in LB for biofilm formation at (B) 48 h or (C) 72 h. The biofilm formed was normalized with bacterial enumeration at the respective time points. Each bar represents six independent replicates ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Because biofilm formation is linked with the persister phenotype, the competency of the intracellular Bpm to form this 3D structure was also evaluated. The intracellular survival Bpm obtained from phagocytic RAW 264.7 cells treated with the permeable antibiotic levofloxacin for 24 h, was used to evaluate the capacity to form biofilm when transferred into fresh media. Because of the different input used, the quantified biofilm was normalized with the CFU output bacteria, as previously described (32, 33). Because standard biofilm formation assay from overnight cultures can be quantified at 24 h (Fig. 4A), we hypothesized that the persister bacteria might take longer to resuscitate, so the incubation time was extended to 48 and 72 h (we could not detect any growth at 24 h). A significant reduction on the biofilm formed by the wild-type strain compared with both mutant strains was found at 48 h (Fig. 4B), but not at 72 h (Fig. 4C), highlighting the transient state of the persister phenotype and the switch between cell growth arrest and normal growing bacteria.
Overexpression of plasmid-encoded HigB.
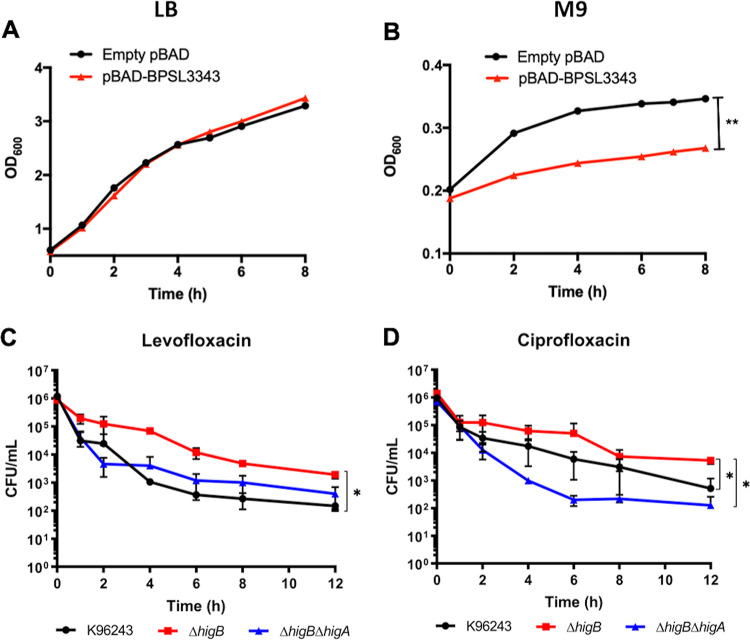
To confirm the predicted function of BPSL3343, we performed an overexpression assay in E. coli using an inducible pBAD plasmid carrying the toxin gene. When the bacteria were grown in nutrient rich LB media, there was no difference between the empty vector and the toxin expressing plasmid after arabinose induction (Fig. 5A). However, cell growth arrest was observed in the strain expressing the toxin when the strains were grown in minimal media M9 (Fig. 5B). These media differences rely on the nutrient content, with the lack of nutrients from the minimal medium generating a stress for the bacteria (34). The significant different growth rate suggests the existence of a growth arrest when the toxin gene is induced and expressed, as previously described for other toxins in Bpm (19), and for the HigBA system in other bacterial species (35, 36).
FIG 5.
Overexpression of toxin induced system in nutrient rich or minimal media and killing curves in the presence of quinolones. Growth curves of empty pBAD vector and the one carrying the toxin BPSL3343 (predicted HigB) after induced with arabinose in (A) LB or (B) minimal M9 media. Killing curves of Bpm K96243, ΔhigB, and ΔhigB ΔhigA in presence or supralethal concentrations of (C) levofloxacin or (D) ciprofloxacin. *, P < 0.05; **, P < 0.01.
Killing curves in the presence of fluoroquinolones.
Presence of a fluoroquinolone in the cultured media seems to be the major trigger for the expression of the HigBA system in Bpm, among all the stress conditions assessed. Because both mutants showed differential survival in the presence of levofloxacin and ciprofloxacin as compared with the wild type K96243 (Fig. 3), the response of the three strains to both antibiotics in a time course analysis was performed (Fig. 5C and D). Like the results found at 24 h (Fig. 3), the ΔhigB strain showed a significant better survival in the presence of levofloxacin than the wild-type strain, and although the ΔhigB ΔhigA mutant also exhibited higher survival tendency, this was not significant (Fig. 5C). In the presence of ciprofloxacin, the double mutant strain showed the lowest survival rate compared to the other strains, while the toxin mutant also survived better than the wild type (Fig. 5D).
Murine macrophage uptake, survival, and persistence.
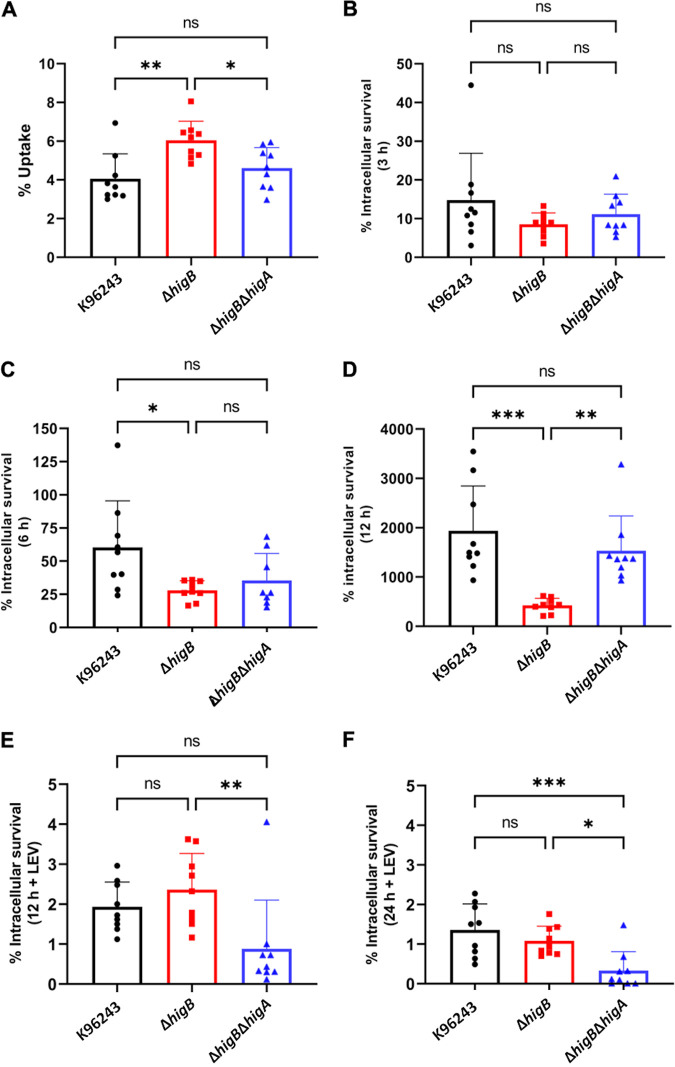
It is well known that Bpm can survive and replicate inside macrophages (37). Due to the intracellular lifestyle of this pathogen, its survival inside these cells is one of the most studied features. However, the bacterial gene products that participate in this process are poorly understood, but many of those genes have been found differentially regulated during this stage (19, 38, 39). As shown in Fig. 1A, many of the predicted TA systems are upregulated during intracellular conditions, including BPSL3343 (higB) and BPS_RS18025 (higA), which showed the highest level of upregulation. For that reason, we analyzed both the uptake and the intracellular survival of the wild type and the isogenic mutant strains in murine RAW 264.7 macrophages, under different conditions and time points (Fig. 6).
FIG 6.
Uptake and survival under macrophage induced-persistence conditions. Murine macrophages were infected (MOI 10) with the different Bpm strains for 30 min. The uptake percentage was calculated from the input bacteria (A). After 30 min, media were removed and replaced with fresh media supplemented with kanamycin for (B) 3 h, (C) 6 h, or (D) 12 h, or with the cell permeable levofloxacin for (E) 12 h and (F) 24 h. Bars represent the average of three independent experiments in triplicate ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
The bacteria uptake rate was obtained through the input data, while the survival at the different time points was established with the uptake numbers. The bacteria uptake by the phagocytic cells was significantly increased in the toxin-deleted strain (ΔhigB), while the phenotype of the double mutant (ΔhigB ΔhigA) was like the wild-type strain (Fig. 6A). However, the intracellular survival of the ΔhigB strain was significantly reduced compared to both the wild type and the double mutant after 12 h (Fig. 6D), and it was reduced compared with the wild type at 6 h after infection (Fig. 6C). These results demonstrated that the higB mutant had a defect on intracellular survival at least after 6 h, but no defect at entry or at the first stage of the infection. Moreover, our results also suggested that the antitoxin HigA might affect intracellular survival.
The ΔhigB strain had shown a differential phenotype after levofloxacin exposure, confirming that quinolones are the antimicrobials that trigger a persister state in association with the HigBA system (17, 27). For that reason, the intracellular survival of the bacteria was also evaluated after addition of the cell-permeable antibiotic levofloxacin after 12 h (Fig. 6E) or 24 h (Fig. 6F). The reduction in intracellular survival that the ΔhigB strain displayed in Fig. 6C and D when compared with the wild type disappeared when cells were treated with levofloxacin, showing a similar survival than the Bpm K96243 strain. In contrast, the double mutant strain displayed a survival defect after the fluoroquinolone treatment at both time points.
Plaque formation.
Another important feature of Bpm pathogenesis is the ability to disseminate from cell to cell, which under in vitro tested conditions could lead to the formation of plaques on monolayers of epithelial cells (40). Due to the different phenotypes of the tested strains infecting and surviving in macrophages, we tested their ability to form plaques in HeLa epithelial cells (Fig. S2). Both ΔhigB and ΔhigB ΔhigA strains exhibited a similar number of plaques formed compared to wild-type K96243, which indicates that there was no difference in cell-to-cell spread among the tested strains at the used MOIs (Fig. S2). The MOI 10 was also evaluated, but the monolayers were destroyed by the intracellular bacteria. There were no significant differences between both mutants’ ability to form plaques; however, ΔhigB seems to form slightly more plaques than ΔhigB ΔhigA, especially at the lowest MOI.
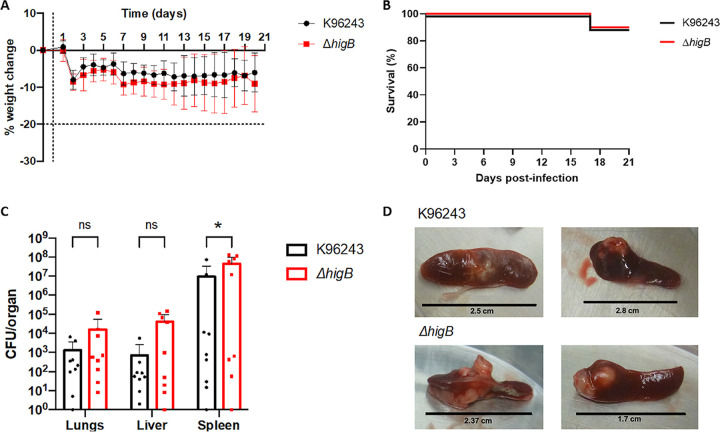
In vivo bacterial infection model.
To examine the role of the HigBA system during chronic melioidosis infection, we induced chronic-persistent melioidosis in a murine model (19, 21). BALB/c mice were intranasally infected with wild type or ΔhigB mutant followed by intraperitoneal administration of levofloxacin on days 1–5 postinfection to favor the transition of the infection into a chronic-persistent stage (relapse rates on the disease are associated with this dormant state) (4). BALB/c mice were used because of their susceptibility to melioidosis infection (41). There were no significant differences in weight changes (Fig. 7A) or the survival rate (Fig. 7B) of the mice after infection with the wild-type and mutant strains. After 21 days postinfection, surviving animals were euthanized and lungs, livers, and spleens (these are target organs for the bacterial infection) were collected and processed to quantify the bacterial burden. There were no significant differences in the organs’ weights between both groups (data not shown). However, animals infected with the mutant showed higher bacterial loads in all three collected organs as compared with the wild type, but significant difference was only observed in spleen colonization (Fig. 7C). Examples of the gross pathology and abscesses observed in the spleens are shown in Fig. 7D. Most of the infected animals with both strains carried mild to large abscesses on the spleens, while the ones infected with the mutant strain were more compact, with some of them comprising almost half of the organ size.
FIG 7.
Persistence of Bpm in vivo after levofloxacin treatment. (A) Weight change during the 21 days of infection. Vertical dotted line represents the challenge day; horizontal dotted line shows the endpoint established in the protocol. (B) Survival graph of mice infected with wild type K96243 or mutant ΔhigB for 21 days postchallenge. (C) Bacterial burden in lungs, liver, and spleen of n = 9 mice challenged with each strain ± SD. (D) Images of gross pathology of infected spleens of mice infected with wild type of ΔhigB after treatment. Two-way ANOVA was used to establish statistical differences (C). *, P < 0.05.
DISCUSSION
Melioidosis is a neglected and re-emerging, underreported, human disease that has a mortality rate of up to 50%, even with treatment (1–3). Besides the problem of proper and reliable diagnosis, which could lead to untreated or delayed treatment, the other concerning issues are treatment failure and prevalence of latent infections (1). This disease is caused by the Gram-negative bacteria B. pseudomallei, which is a pathogen categorized by the Centers for Disease Control and Prevention (CDC) as a Tier 1 Selected Agent due to its possible use as a biothreat agent (42). The treatment of the disease is challenging due to the inherent multidrug resistance profile of the clinical isolates of Bpm, as well as the high relapse rates associated with the latent and chronic infections (2, 5, 43, 44).
Previous work has determined the presence of many toxin–antitoxin systems in these bacterial genomes, as well as the role of some of them in triggering the persister state (19, 21, 45, 46). However, while dozens of systems have been predicted by bioinformatics tools, only a few have been experimentally studied (19, 21, 45, 46). Both the identification and the characterization of new TA modules in human pathogens could improve our knowledge of pathogenic bacterial virulence, leading to more efficient or successful treatments. In the case of B. pseudomallei, that would represent reducing the relapse rates caused by latent or chronic infections, reducing the current treatment duration, and improving the outcomes.
Evaluating the in vitro efficiency of clinically used antibiotics has let us to show that three of the main treatment options for melioidosis (ceftazidime, meropenem, and doxycycline) were the least efficient in killing the bacteria (Fig. 1B and C). However, exposure to both quinolones levofloxacin and ciprofloxacin showed the highest reduction in bacterial survival rates, while a subpopulation remained alive even after supralethal concentrations were used (Fig. 1B and C). That subpopulation of survivors is what we define as persister cells. When we checked the expression of the 11 predicted toxin–antitoxin systems under these treatments, we found a higher number of overexpressed systems in levofloxacin-survival cells (Fig. 1A), which might suggest redundant or complementary functions of these systems. Among all of them, the system formed by BPSL3343 (toxin) and BPS_RS18025 (antitoxin) had the higher fold change values, being significantly overexpressed not only after the quinolones treatment but also during intracellular survival of the bacteria on macrophages (Fig. 2A and B). Based on those results, we proposed that this system might affect bacterial behavior under these stresses and/or host-like conditions.
These genes (BPSL3343/BPS_RS18025), located in the chromosome 1 of Bpm K96243 in the genomic island 12, encode for a predicted type II TA system annotated as HigB/HigA. The HigBA system has been found in other human pathogens such as M. tuberculosis (47) and P. aeruginosa (16, 48), among others. HigB is a ribosome-dependent endoribonuclease from the RelE toxin family, enzymes that specifically cleave prokaryotic mRNA rich in A bases (27, 49, 50). HigA is a transcriptional regulator that under normal growing conditions sequesters its cognate toxin, and whose regulation relies on being rapidly degraded in response to stress signals (antibiotic treatment, host-like environment, phagocytosis) (18, 27). The genetic pair is one of the examples from type II systems with a reverse gene organization, in which the toxin gene is upstream of the antitoxin gene, allowing the antitoxin HigA to repress the toxin transcription binding to its promoter (6, 18, 27). The HigBA complex was recently resolved in E. coli, showing that both components form an heterotetrameric structure, exhibiting high affinity for the promoter sequence, as the antitoxin homodimer does (35). The properties of the antitoxin, as well as its role as virulence transcriptional regulator (18), might explain the differences found with the tested strains, because the HigA antitoxin may be interfering in other processes when its cognate toxin is not present.
To characterize the role of each gene as well as the system in those survival strategies of Bpm, we generated isogenic mutants in the wild-type K96243, a clinical isolate widely used as reference strain (51). While we easily achieved the toxin mutant and the toxin–antitoxin double deletion strains, we were unable to obtain the antitoxin mutant, which suggests that the lack of this gene product could be deleterious to the strain because the free toxin could be lethal for the bacteria (27).
The deletion of the toxin leads to a significant increase survival after levofloxacin treatment since the first moment of exposure (Fig. 3 and 5C). This phenomenon of differential survival in toxin or antitoxin mutants has been previously described in other bacteria (52, 53). In accordance with our data, other work has recently showed the dichotomous role of these systems in which the toxin gene is induced in response to high-lethal concentrations of antibiotic, but at the same time, lacking of the system allows the bacteria to better survive when exposed to it (54). In contrast, meropenem-treated bacteria showed a reduction in survival rates of the ΔhigB ICG001 mutant, even though there was a reduction in the expression of the system, although not significant (Fig. 1A and 2A). The double mutant ICG002 that lacks both functional components showed a high reduction in survival after supralethal concentrations of the other quinolone assessed, ciprofloxacin, compared to not only the wild type but also the toxin mutant (Fig. 3 and 5D).
The type II TA system is considered the most adaptable because while the proteins that belong to that class are organized in superfamilies based on their sequence, one toxin from one superfamily can also be sequestered by the antitoxin from another (6). Moreover, the hyperpromiscuity of some of those components have been already described, which indicates that under specific conditions, a cross talk between the redundant systems is occurring (12). Following that hypothesis, the different phenotypes observed between ΔhigB and ΔhigB ΔhigA strains under the different conditions tested might be due, at least in part, to the interaction of the antitoxin with another target, which could be influencing the observed phenotype. In the case of the antitoxin HigA, it has been found to play a role as a virulence regulator by itself in P. aeruginosa (18), and HigA recognizes the promoter region with same affinity as when is part of the intact TA complex (35), suggesting that not only the toxins from these systems participate in the survival strategies of the bacteria.
Similarly, biofilm formation is shown to be reduced in the toxin mutant compared with the wild-type strain, which disappears when levofloxacin is added to the culture (Fig. 4A). After an antibiotic treatment, susceptible bacteria die, leaving only the persister cells, which might not be cleared by the immune system if they are embedded in the biofilm structure (29). For both wild type and double mutant, the biofilm formation is reduced after levofloxacin treatment, while only the double mutant structure is significantly modified when the antibiotic is present compared with nonsupplemented LB. However, the ΔhigB mutant tends to form more biofilm in the presence than in the absence of levofloxacin (Fig. 4A). A different behavior was also found between the wild type and the isogenic mutant strains when biofilm formation was evaluated in the antibiotic-induced persister ex vivo model, using the intracellular bacteria obtained from phagocytic cells treated with levofloxacin (Fig. 4B and C). A defect in biofilm formation with the wild-type strain as compared with both mutants was found (Fig. 4B), a phenotype that disappeared with time (Fig. 4C). This experiment highlighted the transient state of the persister phenotype when the stressful condition is removed. Those phenotypes also indicated the that different “resuscitation” times of the persisters are associated with a functional TA system.
Also, when the toxin expression was induced in E. coli, a differential growing rate was found if the bacteria was maintained in minimal media (Fig. 5B), suggesting that its presence in stressful conditions triggers some types of growth arrest. This bacteriostatic but nonlethal effect of the toxin has been also seen with other predicted HigB protein as well as other toxins (19, 35, 36).
When murine macrophages were infected with the three strains, we found differences among all of them. The uptake rate is higher in ΔhigB compared with the other isolates, which might indicate that this mutant is more easily taken by the macrophages (Fig. 6A). However, the intracellular survival of this mutant was significantly reduced after 6 h as compared with the wild type and after 12 h as compared with both the wild type and double mutant strain, which highlights a defect in intracellular survival within the macrophage (Fig. 6C and D). In the case of ΔhigB ΔhigA, the phenotype obtained was like the one observed with the wild-type strain. On the other hand, due to the different responses of these two isolates to levofloxacin, we replaced the treatment with kanamycin to test the role of this compound on their intracellular survival properties. Under these conditions, the survival differences between the ΔhigB and wild-type strains were eliminated (Fig. 6E and F), indicating that these bacteria react differently to the presence of the broad-spectrum quinolone. However, the loss of both TA genes resulted in a survival defect under this condition, as it occurred when it was treated in vitro compared with the single mutant strain (Fig. 3 and 5C). During the evaluation of the cell-to-cell dissemination capacity of these strains in epithelial cells, both mutants showed a similar ability as the wild type in forming plaques (Fig. S2). Also, no major differences were observed between the two mutants, except for a relative reduced number of plaques in the double mutant at MOI 0.1 compared with the toxin mutant strain.
The in vivo chronic melioidosis infection model with levofloxacin treatment demonstrated that both wild type K96243 and toxin mutant ΔhigB showed the same lethality as well as the same bacterial burden in lungs and livers. In contrast, the CFU burden in the spleen was significantly higher in the animals infected with the toxin mutant (Fig. 7C), which suggested a better capacity of the mutant strain to disseminate and persist in the spleen. Additionally, the gross pathology showed higher spleen abscesses in those mice infected with the ΔhigB ICG001 mutant (Fig. 7D). This phenotype was previously described with other toxin mutant strains when animals were not treated with antibiotics (19).
Overall, these data suggest that both components of the type II toxin–antitoxin system formed by BPSL3343 and BPS_RS18025, annotated as HigBA, play a role in B. pseudomallei survival under specific conditions, being the presence of the quinolones, a major trigger of the observed phenotypes. The toxin mutant showed a clear phenotype when the antibiotic levofloxacin participates in all the tested assays, while the double mutant showed less distinct phenotypes. The ΔhigB strain showed better survival than the wild-type strain when exposed in vitro to supralethal concentration of levofloxacin (Fig. 3 and 5C); the biofilm formed is apparently enhanced (not significant) after levofloxacin treatment (Fig. 4A); the reduced intracellular survival disappears when levofloxacin is present in the media (Fig. 6); and the bacteria colonized better in the mice spleen when animals were treated with the quinolone (Fig. 7C). However, the double mutant did not show different survival after levofloxacin at a supralethal concentration, but was reduced after ciprofloxacin (Fig. 3 and 5D); the biofilm formed was significantly reduced in the presence of levofloxacin (Fig. 4A); and the intracellular survival in macrophages was the same than wild type in normal conditions but significantly reduced after levofloxacin supplementation (Fig. 6). Overall, there is still a need to increase knowledge regarding how Bpm differentially responds to the different treatments, especially during an infection course, and while there are so many TA systems encoded in their genomes, which can easily complement the mutation and provide just partial information about the role of HigBA in pathogenesis and survival.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 2. B. pseudomallei strain K96243 was obtained from BEI Resources, Manassas, VA, USA. B. pseudomallei and E. coli strains were routinely cultured on LB agar plates at 37°C, and antibiotics were supplemented as needed for selection. Bacterial culture stocks were preserved in 20% glycerol (vol/vol) and stored at −80°C.
TABLE 2.
Strains and plasmids used in the study
| Strains | Characteristics | Reference |
|---|---|---|
| B. pseudomallei K96243 | BEI Resources (Manassas, VA, USA) | 57 |
| B. pseudomallei ICG001 | Bpm K96243 ΔhigB | This publication |
| B. pseudomallei ICG002 | Bpm K96243 ΔhigB ΔhigA | This publication |
| E. coli S17 λpir pMo130higB | higB cloned into pMo130 | This publication |
| E. coli S17 λpir pMo130higA | higA cloned into pMo130 | This publication |
| E. coli DH10 pBAD | Inducible vector | 58 |
| E. coli DH10 pBAD-BPSL3343 | BPSL3343 in pBAD | This publication |
Genomic annotation.
The genomic sequence from the Bpm strain K96243 was obtained from the public deposit in the NCBI website (accession numbers, NC_006350.1 [chromosome 1] and NC_006351.1 [chromosome 2]). Annotation of the genome was performed with the Rapid Annotations using Subsystems Technology (RAST) server, as well as Prokka (version 1.13) to identify the coding sequences (CDS) predicted as toxin or antitoxin that could belong to the TA systems category, as well as genomic islands (22, 23). When only the toxin was predicted, antitoxin was manually annotated.
RNA extraction and cDNA synthesis.
RNA from intracellular bacteria or antibiotic-treatment survival bacteria was extracted using the Direct-zol RNA Miniprep Kit (Zymo Research) according to the manufacturer’s instructions. RNA concentration and purity were measured using an Epoch microplate spectrophotometer (Biotek) and samples stored at −80°C for future use. The cDNA was synthesized using the different RNA samples as templates employing the iScript cDNA Synthesis Kit (Bio-Rad), following manufacturer’s instructions, with the following conditions: 25°C for 5 min, 42°C for 30 min, 85°C for 5 min, 25°C for 5 min, 12°C for 5 min, 4°C store. The cDNA concentration and purity were measured, and samples were stored at −20°C for further use. For intracellular bacteria, after cell permeabilization, two steps of differential centrifugation were used, to enhance the ratio of bacterial RNA over the eukaryotic RNA (38).
qPCR.
Quantitative PCR was used to analyze the expression of the predicted pair of toxin-antitoxin genes from the survival bacteria after antibiotic treatment or intracellular bacterial survival. Gene expression quantification was performed using QuantiNova SYBR green (Qiagen) following the manufacturer’s instructions, after cDNA synthesis and normalization at 100 ng/μL. Bacteria grown in LB without antibiotics (persister) or grew in Dulbecco's Modified Eagle Medium (DMEM) (intracellular survival) were used as control for expression. All primers used for expression are summarized in Table 3, and their specificity was evaluated in a PCR gradient (56–76°C) using Q5 High-Fidelity DNA polymerase (NEB). Expression was normalized with the housekeeping genes rpoB and 16S for each condition, following the ΔΔCt method (55). The PCR cycling program was set as follows: initial heat activation step 95°C 2 min; 2-step 40 cycles of 5 s 95°C and 30 s 60°C. The threshold cycle (CT) and melting curve of each gene were automatically established and recorded by the software CFX Maestro Software (version 4.0). Multiple t test analysis was conducted to establish significant differences among strains and conditions.
TABLE 3.
List of primers designed and used in this study
| Primer | Sequence | Experiment |
|---|---|---|
| F1F_3343 | GAGCTGATATCAGGGCCCCGTAATTCTTCTTGCTTCTACTCCATC | Mutant construction |
| F1R_3343 | TGCGGACTACAGAGGTGCGTAATGGCTAAG | |
| F2F_3343 | ACGCACCTCTGTAGTCCGCAAGAATCCCAG | |
| F2R_3343 | GATTAATTGTCAACAGCTCATGAATAGTGCGGCGCTGTC | |
| F1F_RS | GAGCTGATATCAGGGCCCCGATCTCGATCCCATCCGGC | |
| F1R_RS | TCTGCTCGCATTGTGATTTCGTTCGCTTGAC | |
| F2F_RS | GAAATCACAATGCGAGCAGACGAGCTGTG | |
| F2R_RS | GATTAATTGTCAACAGCTCAAAATTTGCTAACATGAAATTCGATTGGACG | |
| BPSL3343-BPS_RS18025 F | CCGCCTTGACCTTACTATCTG | Coexpression |
| BPSL3343-BPS_RS18025 R | GTTTGAGTTGCGTCCAAAGG | |
| BPSL3343 F | CTGGGATTCTTGCGGACTAC | qPCR |
| BPSL3343 R | GCAACTCAAACAACCCGTTAC | |
| BPS_RS18025 F | GCTCGCAGATAGTAAGGTCAAG | |
| BPS_RS18025 R | CTTGAGTCAATCCAGCCTCTT | |
| BPSS0390 F | GATGGCTGGAGGTTGGTT | |
| BPSS0390 R | CCAATCGGTAGGTCCTTCTTC | |
| BPSS0391 F | TGATCGAGCTTGGAGAAGATG | |
| BPSS0391 R | GCTTGGAATCAAGCTGAGAAAG | |
| BPSS0394 F | GACATTGTCGACGCCTTCA | |
| BPSS0394 R | CTACGCAGGCTGATGTTCC | |
| BPSS0395 F | CCTCTATTGCGTGGTGTTCA | |
| BPSS0395 R | GCTTGCTCGACATAGCTCTT | |
| BPSL0175 F | TCGGATCGAACTCATCATGC | |
| BPSL0175 R | CCTTCCATGTCGACACCTG | |
| BPSL0174 F | GAATGTCACCGCGAGTCTT | |
| BPSL0174 R | CTGGAGTCGATGTGTGATCTG | |
| BPSL0559 F | CCTGAGCACCGACTTTCAATA | |
| BPSL0559 R | CAGCCATCAGGCTCAGTATC | |
| BPSL0558 F | GGGCTTTCTACGTCATCCAT | |
| BPSL0558 R | GGAGGAGATTCAGGATGTTCAG | |
| BPSL2333 F | TTAGCCCTGGTGCAAATGA | |
| BPSL2333 R | TTGTGATGCACCACCATCT | |
| BPSL2334 F | ACCGGATGAGGTCGAGAT | |
| BPSL2334 R | CGCGCAACCATTTGTCATAG | |
| BPSL3261 F | CCTCTTTGTGGTCGTCTTCTATC | |
| BPSL3261 R | ACTTCAACTGTTCCGCATACT | |
| BPSL3260 F | GACCTTGCAGAGCAACTATGA | |
| BPSL3260 R | ATCCACCTCACCATGCAAAT | |
| BPSS1060 F | CATGGACGAGATACCGAGATG | |
| BPSS1060 R | GAAACGTGAGGAATACGCAATAG | |
| BPSS1061 F | TTCGACGGTGTTTCTGATCG | |
| BPSS1061 R | GCGGCACACGTCGTATT | |
| BPSS1583 F | AACGGCAAGACCGACAA | |
| BPSS1583 R | CATACGGCTTGATGAAGTTGAC | |
| BPSS1584 F | CGTCGAGGAACGCATCAC | |
| BPSS1584 R | TTGTCGCGCATCGTATCG | |
| BPSL0033 F | CGAGGTCGCCTCGATGTT | |
| BPSL0033 R | GAGGTCAGCCTGCGGAT | |
| BPSL0034 F | GCGAGCTTCGATACGATCATT | |
| BPSL0034 R | CGTCATGAACGGGTTCTCC | |
| BPSL0952 F | TCGCCTTGACAAGCATACAG | |
| BPSL0952 R | GTCCATGTTCAGCCTCAAGTAG | |
| BPSL0953 F | TGCCCGAAGTGCTCAAA | |
| BPSL0953 R | TCAGGCGGTCAATGTCAC | |
| 16S F | GCGTAGAGATGTGGAGGAATAC | |
| 16S R | ACCAGGGTATCTAATCCTGTTTG | |
| rpoB F | CCGAAGGACGTGCTGTATTT | |
| rpoB R | GTGAAGTTGTCGAAGACGAAGA |
Mutant construction.
The single (ΔBPSL3343, ΔhigB, named ICG001) and double mutant (ΔBPSL3343 ΔBPS_RS18025, ΔhigB ΔhigA, named ICG002) strains were constructed using a biparental matting approach for allelic exchange using the plasmid pMo130, as previously described (56). All the primers used are shown in Table 3. As described (21), pMo130 was linearized with HindIII-HF (NEB) and NheI-HF (NEB) following the established protocol. Both upstream and downstream fragments by 400–600 bp from the target gene were amplified with Q5 High-Fidelity DNA polymerase (NEB), harboring the beginning and the end of each gene. Assembly of the three fragments was constructed using the Gibson Assembly kit (NEB) and transformed into E. coli S17-1 λpir. The plasmids carrying intragenic in-frame deletions were confirmed by PCR sequencing in the University of Texas Medical Branch Core Service. The E. coli S17-1 λpir strain was used as donor for allelic exchange with recipient Bpm through biparental mating. For the single mutant strain construction, Bpm K96243 wild type (WT) was used for the matting, while for the double mutant construction, the ΔhigB strain ICG001, was used. Bpm K96243 wild type was also used as donor for the ΔBPS_RS18025 construction (ΔhigA), but construction of that strain was not successful after multiple trials. Merodiploids were selected by supplementing kanamycin (500 μg/mL) and polymyxin B (30 μg/mL) into LB agar plates. Colonies that turned yellow after pyrocatechol exposure were selected as positive for the deletions, and they were counterselected on YT agar supplemented with 15% sucrose. The intragenic deletions from the selected clones were confirmed by PCR sequencing. Because Bpm is a Tier 1 Selected Agent, complementation of the mutants was not performed, to avoid the gain of function or enhance virulence phenotypes.
Bacterial growth curves.
Growth curves were conducted to confirm that differences among the strains were not present. Isolates from overnight (O/N) cultures were inoculated in 30 mL of LB, minimal medium M9, or DMEM, and incubated at 37°C for 24 h with shaking at 200 rpm. The optical density (600 nm with a BioSpec-mini spectrophotometer, Shimadzu) and CFU enumeration were measured and validated at each time point.
Antibiotic-induced persistence assays.
To quantify the persister frequency or the survival population after antibiotic treatment, wild-type and mutant strains were grown for 12 h in LB with shaking at 200 rpm at 37°C, and the overnight cultures were inoculated in LB at 2 × 108 CFU/mL containing 100× MIC of levofloxacin (LEV), ciprofloxacin (CIP), ceftazidime (CFT), meropenem (MER), or doxycycline (DOX) and further incubated in static conditions at 37°C for 24 h. Bacteria inoculated in LB without antibiotics were used as control. Bacteria were serially diluted for CFU enumeration after 24 h, and the survival rates were quantified with the input numbers. All assays were performed in triplicate.
Biofilms.
Biofilm formation capacity was evaluated in Corning 96-well PP 1.2-mL cluster tubes. The strains were grown overnight in LB for 12 h and diluted 1:100 in 1 mL LB in a 96-well plate, with at least 6 wells per replicate. Cultures were statically incubated at 37°C for 24 h. In parallel, levofloxacin at a final 80 μg/mL concentration was added to each well after 12 h of incubation then incubated for an additional 12 h. After that, the wells were washed three times with water and stain with 1% crystal violet (vol/vol) for 30 min. Then, the stain was discarded and the remaining was rinsed with water. The stained biofilms were solubilized with 1 mL of 33% acetic acid. Optical density was measured at 600 nm in a BioSpec-mini spectrophotometer (Shimadzu), using wells with LB only as blank. One-way ANOVA was used to establish statistical differences.
Overexpression assays.
The inducible vector pBAD (Thermo Fisher Scientific) maintained in E. coli DH10 (NEB) was used for the toxin overexpression. The toxin gene was amplified with Q5 High-fidelity DNA polymerase (NEB) using the primers (forward) att aac cat gga tcc gag ctA TGA AAT TCG ATT GGA CGG, and (reverse) ttc gaa ttc cca tat ggt acT TAG CCA TTA CGC ACC TC. The PCR product was cleaned using the QIAquick PCR purification kit (Qiagen), and the vector was digested with KpnI-HF and SacI-HF (NEB), following the manufacturer’s instructions. Vector and insert fragments were assembled using the Gibson Assembly kit (NEB). Selected plasmid sequences were confirmed by sequencing at the UTMB Core Sequencing Service.
The overexpression assays were conducted in LB and minimal M9 media supplemented with ampicillin for plasmid maintenance. Both the empty vector (pBAD) and the vector with the toxin inserted (pBAD-BPSL3343) were cultured overnight for 12 h and diluted in 100 mL of fresh media. Toxin expression was induced with 1% arabinose when the culture reached OD600 0.2 in M9 and 0.5 in LB, and optical density was measured every hour for 8 h. Statistical t test analysis was conducted to establish significant differences between growth.
Killing curves.
Killing growth curves were performed in the Bpm K96243 wild type and both isogenic mutant (ICG001 and ICG002) strains, in the presence of supralethal concentrations of both fluoroquinolones (levofloxacin and ciprofloxacin). Isolates from O/N cultures were used to inoculate 20 mL of LB broth with 1 × 106 CFU/mL, and then supplemented with the compounds. Different time points up to 12 h were established, and CFU enumeration was measured to establish the killing rates of the different strains. Cultures were incubated at 37°C in shaking, and the assay performed with 4 independent replicates. One-way ANOVA was used to establish statistical differences.
Macrophage survival assays.
The intracellular survival of wild-type and mutant strains were evaluated using the murine macrophage cell line RAW 264.7 (TIB-71). Cells were routinely grown in DMEM supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 1 mM nonessential amino acids, and 1% penicillin-streptomycin. B. pseudomallei strains were cultured overnight for 12 h in LB at 37°C with 200 rpm shaking. The murine macrophages were seeded at 2 × 105 cells/mL in Costar 24-well plates (Corning), and they were infected with the different strains at an MOI of 10, for 30 min (uptake step). After that, the supernatants were collected, cells were washed 2× with PBS and incubated with DMEM supplemented with 250 μg/mL kanamycin for extracellular bacterial killing. For bacterial enumeration, cells were washed with PBS before lysis with 0.1% Triton X-100. The lysed cells were serially diluted and plated on LB agar. The uptake percentage was determined with the input bacterial number, while the intracellular bacteria were normalized using the uptake concentration for each strain. All the assays were performed in triplicate.
Alternatively, to test the antibiotic-induced persister ex vivo phenotype, DMEM media were supplemented with 80 μg/mL of the cell-permeable antibiotic levofloxacin (instead of kanamycin) then incubated for 30 min for the uptake step, as previously described (21). The intracellular bacteria were collected at 12 h and 24 h postinfection and plated for CFU enumeration. The intracellular bacteria were normalized using the uptake concentration for each strain. All the assays were performed in triplicate. One-way ANOVA was used to establish statistical differences.
Biofilms from intracellular bacteria.
To evaluate the biofilm formation properties of the macrophage-induced persisters, intracellular bacteria were used as input as follows: bacteria from RAW 264.7 cells treated with levofloxacin for 24 h were collected and after cell lysis with 500 μL, 100 μL were used to inoculate 1 mL of LB per well in 24-well plates. Each strain and condition were repeated for a total of 6 replicates. Plates were incubated for 24, 48, and 72 h, and the output at each time point was plated for biofilm normalization (32, 33). Staining and solubilization of the biofilm with 1% crystal violet and 33% acetic acid were performed as described above. For normalization, the OD was divided by the output of each well, as the input cannot be standardized because the persister cells “resuscitate” at different rates. One-way ANOVA was used to establish statistical differences.
Plaque assay.
To assess the differential plaque formation among the different strains, the human epithelial cell line HeLa (CRM-CCL-2) was used. Cells were routinely grown in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Before infection, cells were seeded at 5 × 105 cells/mL in a Costar 12-well plate (Corning) and B. pseudomallei isolates were cultured overnight (12 h) in 20 mL LB with shaking at 200 rpm at 37°C. HeLa cells were infected with Bpm strains at MOI of 0.1 and 1 for 1 h, and media supplemented kanamycin was replaced and incubated for 24 h. After the infection, cells were fixed with 4% paraformaldehyde (PFA) and stained with Giemsa for 30 min.
In vivo bacterial infection model.
Female, 6- to 8-week-old BALB/cJ mice, were purchased from Jackson Laboratories (Bar Harbor, ME). Mice were housed in microisolator cages under pathogen‐free conditions, provided with rodent feed and water ad libitum, and maintained on a 12 h light cycle. Before experiments, mice were acclimated for 7 days. Anesthetized BALB/cJ mice (n = 10 per group) were intranasally (i.n.) inoculated with 50 μL (25 μL/nare) of B. pseudomallei K96243 wild type or ΔhigB (ICG001), equivalent to 3.5 LD50 (1 LD50 = 312 CFU). Mice were daily treated with intraperitoneal (i.p.) injections of levofloxacin (25 mg/kg per day in PBS), starting at 24 h postinfection for 5 days, using a previously established model for persister Bpm infections (18, 34). Animals were monitored and weighed daily up to 21 days postinfection, and the organs of survival mice were collected for bacterial burden determination. Human endpoints were established and strictly monitored every day, as described in the animal protocol IACUC #0503014D approved by the Animal Care and Use Committee of the University of Texas Medical Branch.
ACKNOWLEDGMENTS
We thank the seminal published contributions of Brittany N. Ross that ignited the advancement of this project. This work was supported by NIH NIAID grant AI148913 awarded to A.G.T. J.L.S. received an USDA NBAF Scientist Training Program fellowship.
The contents are solely the responsibility of the authors and do not necessarily represent the official views of NIH or the USDA.
Footnotes
Supplemental material is available online only.
Contributor Information
Alfredo G. Torres, Email: altorres@utmb.edu.
Igor E. Brodsky, University of Pennsylvania
REFERENCES
- 1.Wiersinga WJ, Virk HS, Torres AG, Currie BJ, Peacock SJ, Dance DAB, Limmathurotsakul D. 2018. Melioidosis. Nat Rev Dis Primers 4:17107. 10.1038/nrdp.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Limmathurotsakul D, Golding N, Dance DAB, Messina JP, Pigott DM, Moyes CL, Rolim DB, Bertherat E, Day NPJ, Peacock SJ, Hay SI. 2016. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol 1:15008. 10.1038/nmicrobiol.2015.8. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous A. 2021. Melioidosis. J Spec Oper Med 21:104–105. 10.55460/WEJ5-A5CA. [DOI] [PubMed] [Google Scholar]
- 4.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 5.Amato SM, Fazen CH, Henry TC, Mok WWK, Orman MA, Sandvik EL, Volzing KG, Brynildsen MP. 2014. The role of metabolism in bacterial persistence. Front Microbiol 5:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srivastava A, Pati S, Kaushik H, Singh S, Garg LC. 2021. Toxin-antitoxin systems and their medical applications: current status and future perspective. Appl Microbiol Biotechnol 105:1803–1821. 10.1007/s00253-021-11134-z. [DOI] [PubMed] [Google Scholar]
- 7.Maisonneuve E, Gerdes K. 2014. Molecular mechanisms underlying bacterial persisters. Cell 157:539–548. 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 8.Kussell E, Kishony R, Balaban NQ, Leibler S. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169:1807–1814. 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harms A, Brodersen DE, Mitarai N, Gerdes K. 2018. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol Cell 70:768–784. 10.1016/j.molcel.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Yao J, Sun Y-C, Wood TK. 2021. Type VII toxin/antitoxin classification system for antitoxins that enzymatically neutralize toxins. Trends Microbiol 29:388–393. 10.1016/j.tim.2020.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Choi JS, Kim W, Suk S, Park H, Bak G, Yoon J, Lee Y. 2018. The small RNA, SdsR, acts as a novel type of toxin in Escherichia coli. RNA Biol 15:1319–1335. 10.1080/15476286.2018.1532252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tatsuaki K, Kumar SC, Bj A, Toomas M, Tetiana B, Tk J, Ao F, Alves OSR, Steffi J, Karin E, Maxence D, Karina P, Tanel T, Henrik S, Vasili H, Ag C. 2022. A hyperpromiscuous antitoxin protein domain for the neutralization of diverse toxin domains. Proc Natl Acad Sci USA 119:e2102212119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agarwal S, Sharma A, Bouzeyen R, Deep A, Sharma H, Mangalaparthi KK, Datta KK, Kidwai S, Gowda H, Varadarajan R, Sharma RD, Thakur KG, Singh R. 2020. VapBC22 toxin-antitoxin system from Mycobacterium tuberculosis is required for pathogenesis and modulation of host immune response. Sci Adv 6:eaba6944. 10.1126/sciadv.aba6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma D, Mandell JB, Donegan NP, Cheung AL, Ma W, Rothenberger S, Shanks RMQ, Richardson AR, Urish KL. 2019. The toxin-antitoxin MazEF drives Staphylococcus aureus biofilm formation, antibiotic tolerance, and chronic infection. mBio 10:e01658. 10.1128/mBio.01658-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul P, Sahu BR, Suar M. 2019. Plausible role of bacterial toxin-antitoxin system in persister cell formation and elimination. Mol Oral Microbiol 34:97–107. 10.1111/omi.12258. [DOI] [PubMed] [Google Scholar]
- 16.Wood TL, Wood TK. 2016. The HigB/HigA toxin/antitoxin system of Pseudomonas aeruginosa influences the virulence factors pyochelin, pyocyanin, and biofilm formation. Microbiologyopen 5:499–511. 10.1002/mbo3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Long Y, Liu Y, Liu Y, Chen R, Shi J, Zhang L, Jin Y, Yang L, Bai F, Jin S, Cheng Z, Wu W. 2016. HigB of Pseudomonas aeruginosa enhances killing of phagocytes by up-regulating the type III secretion system in ciprofloxacin induced persister cells. Front Cell Infect Microbiol 6:125. 10.3389/fcimb.2016.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Y, Zhang S, Luo G, Shen Y, Li C, Zhu Y, Huang Q, Mou X, Tang X, Liu T, Wu S, Tong A, He Y, Bao R. 2021. Type II antitoxin HigA is a key virulence regulator in Pseudomonas aeruginosa. ACS Infect Dis 7:2930–2940. 10.1021/acsinfecdis.1c00401. [DOI] [PubMed] [Google Scholar]
- 19.Ross BN, Thiriot JD, Wilson SM, Torres AG. 2020. Predicting toxins found in toxin–antitoxin systems with a role in host-induced Burkholderia pseudomallei persistence. Sci Rep 10:16923. 10.1038/s41598-020-73887-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood TK, Knabel SJ, Kwan BW. 2013. Bacterial persister cell formation and dormancy. Appl Environ Microbiol 79:7116–7121. 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross BN, Micheva-Viteva S, Hong-Geller E, Torres AG. 2019. Evaluating the role of Burkholderia pseudomallei K96243 toxins BPSS0390, BPSS0395, and BPSS1584 in persistent infection. Cell Microbiol 21:e13096. 10.1111/cmi.13096. [DOI] [PubMed] [Google Scholar]
- 22.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 23.Aziz RK, Bartels D, Best A, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapartegui-González I, Fernández-Martínez M, Rodríguez-Fernández A, Rocha DJP, Aguiar ERGR, Pacheco LGC, Ramos-Vivas J, Calvo J, Martínez-Martínez L, Navas J. 2020. Antimicrobial susceptibility and characterization of resistance mechanisms of Corynebacterium urealyticum clinical isolates. Antibiotics 9:404–414. 10.3390/antibiotics9070404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamruzzaman M, Wu AY, Iredell JR. 2021. Biological functions of type II toxin-antitoxin systems in bacteria. Microorganisms 9:1276. 10.3390/microorganisms9061276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerdes K, Rasmussen PB, Molin S. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci USA 83:3116–3120. 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi Q, Kamruzzaman M, Iredell JR. 2021. The higBA-type toxin-antitoxin system in IncC plasmids is a mobilizable ciprofloxacin-inducible system. mSphere 6:e0042421. 10.1128/mSphere.00424-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Currie BJ. 2015. Melioidosis: evolving concepts in epidemiology, pathogenesis, and treatment. Semin Respir Crit Care Med 36:111–125. 10.1055/s-0034-1398389. [DOI] [PubMed] [Google Scholar]
- 29.Wen Y, Behiels E, Devreese B. 2014. Toxin-antitoxin systems: their role in persistence, biofilm formation, and pathogenicity. Pathog Dis 70:240–249. 10.1111/2049-632X.12145. [DOI] [PubMed] [Google Scholar]
- 30.Kunyanee C, Kamjumphol W, Taweechaisupapong S, Kanthawong S, Wongwajana S, Wongratanacheewin S, Hahnvajanawong C, Chareonsudjai S. 2016. Burkholderia pseudomallei biofilm promotes adhesion, internalization and stimulates proinflammatory cytokines in human epithelial A549 cells. PLoS One 11:e0160741. 10.1371/journal.pone.0160741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pakkulnan R, Anutrakunchai C, Kanthawong S, Taweechaisupapong S, Chareonsudjai P, Chareonsudjai S. 2019. Extracellular DNA facilitates bacterial adhesion during Burkholderia pseudomallei biofilm formation. PLoS One 14:e0213288. 10.1371/journal.pone.0213288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bravo Z, Chapartegui-Gonzalez I, Lazaro-Diez M, Ramos-Vivas J. 2018. Acinetobacter pittii biofilm formation on inanimate surfaces after long-term desiccation. J Hosp Infect 98:74–82. 10.1016/j.jhin.2017.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Chapartegui-González I, Lázaro-Díez M, Bravo Z, Navas J, Icardo JM, Ramos-Vivas J. 2018. Acinetobacter baumannii maintains its virulence after long-time starvation. PLoS One 13:e0201961. 10.1371/journal.pone.0201961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaiswal S, Paul P, Padhi C, Ray S, Ryan D, Dash S, Suar M. 2016. The Hha-TomB toxin-antitoxin system shows conditional toxicity and promotes persister cell formation by inhibiting apoptosis-like death in S. typhimurium. Sci Rep 6:38204. 10.1038/srep38204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jadhav PV, Sinha VK, Chugh S, Kotyada C, Bachhav D, Singh R, Rothweiler U, Singh M. 2020. 2.09 Å resolution structure of E. coli HigBA toxin–antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin. Biochem J 477:4001–4019. 10.1042/BCJ20200363. [DOI] [PubMed] [Google Scholar]
- 36.Choi E, Huh A, Oh C, Oh J-I, Kang HY, Hwang J. 2022. Functional characterization of HigBA toxin-antitoxin system in an Arctic bacterium, Bosea sp. PAMC 26642. J Microbiol 60:192–206. 10.1007/s12275-022-1619-9. [DOI] [PubMed] [Google Scholar]
- 37.Thiriot JD, Martinez-Martinez YB, Endsley JJ, Torres AG. 2020. Hacking the host: exploitation of macrophage polarization by intracellular bacterial pathogens. Pathog Dis 78:ftaa009. 10.1093/femspd/ftaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jitprasutwit S, Jitprasutwit N, Hemsley CM, Onlamoon N, Withatanung P, Muangsombut V, Vattanaviboon P, Stevens JM, Ong C, Stevens MP, Titball RW, Korbsrisate S. 2020. Identification of Burkholderia pseudomallei genes induced during infection of macrophages by differential fluorescence induction Front Microbiol 11:72. 10.3389/fmicb.2020.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chieng S, Carreto L, Nathan S. 2012. Burkholderia pseudomallei transcriptional adaptation in macrophages. BMC Genomics 13:328. 10.1186/1471-2164-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stockton JL, Torres AG. 2020. Multinucleated giant cell formation as a portal to chronic bacterial infections. Microorganisms 8:1637. 10.3390/microorganisms8111637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bearss JJ, Hunter M, Dankmeyer JL, Fritts KA, Klimko CP, Weaver CH, Shoe JL, Quirk AV, Toothman RG, Webster WM, Fetterer DP, Bozue JA, Worsham PL, Welkos SL, Amemiya K, Cote CK. 2017. Characterization of pathogenesis of and immune response to Burkholderia pseudomallei K96243 using both inhalational and intraperitoneal infection models in BALB/c and C57BL/6 mice. PLoS One 12:e0172627. 10.1371/journal.pone.0172627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng AC, Dance DAB, Currie BJ. 2005. Bioterrorism, glanders and melioidosis. Euro Surveill 10:E1–2. [PubMed] [Google Scholar]
- 43.Rhodes KA, Schweizer HP. 2016. Antibiotic resistance in Burkholderia species. Drug Resist Updat 28:82–90. 10.1016/j.drup.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweizer HP. 2012. Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis. Future Microbiol 7:1389–1399. 10.2217/fmb.12.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butt A, Müller C, Harmer N, Titball RW. 2013. Identification of type II toxin–antitoxin modules in Burkholderia pseudomallei. FEMS Microbiol Lett 338:86–94. 10.1111/1574-6968.12032. [DOI] [PubMed] [Google Scholar]
- 46.Butt A, Higman VA, Williams C, Crump MP, Hemsley CM, Harmer N, Titball RW. 2014. The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation. Biochem J 459:333–344. 10.1042/BJ20140073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuessler DL, Cortes T, Fivian-Hughes AS, Lougheed KEA, Harvey E, Buxton RS, Davis EO, Young DB. 2013. Induced ectopic expression of HigB toxin in Mycobacterium tuberculosis results in growth inhibition, reduced abundance of a subset of mRNAs and cleavage of tmRNA. Mol Microbiol 90:195–207. 10.1111/mmi.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Gao Z, Liu G, Geng Z, Dong Y, Zhang H. 2019. Structural insights into the transcriptional regulation of HigBA toxin–antitoxin system by antitoxin HigA in Pseudomonas aeruginosa. Front Microbiol 10:3158. 10.3389/fmicb.2019.03158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goeders N, Drèze P-L, Van Melderen L. 2013. Relaxed cleavage specificity within the RelE toxin family. J Bacteriol 195:2541–2549. 10.1128/JB.02266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lalaouna D, Massé E. 2017. Cut in translation: ribosome-dependent mRNA decay. EMBO J 36:1120–1122. 10.15252/embj.201797019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Limmathurotsakul D, Funnell SGP, Torres AG, Morici LA, Brett PJ, Dunachie S, Atkins T, Altmann DM, Bancroft G, Peacock SJ. 2015. Consensus on the development of vaccines against naturally acquired melioidosis. Emerg Infect Dis 21:e1–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 53.Rosendahl S, Tamman H, Brauer A, Remm M, Hõrak R. 2020. Chromosomal toxin-antitoxin systems in Pseudomonas putida are rather selfish than beneficial. Sci Rep 10:9230. 10.1038/s41598-020-65504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su W, Bredèche M, Dion S, Dauverd J, Condamine B, Gutierrez A. 2022. TisB protein protects Escherichia coli cells suffering massive DNA damage from environmental toxic compounds. mBio 23:e0038522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Hamad MA, Zajdowicz SL, Holmes RK, Voskuil MI. 2009. An allelic exchange system for compliant genetic manipulation of the select agents Burkholderia pseudomallei and Burkholderia mallei. Gene 430:123–131. 10.1016/j.gene.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holden MTG, Titball RW, Peacock SJ, Cerdeño-Tárraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason M, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PCF, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA 101:14240–14245. 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2. Download iai.00035-22-s0001.pdf, PDF file, 0.3 MB (282.8KB, pdf)