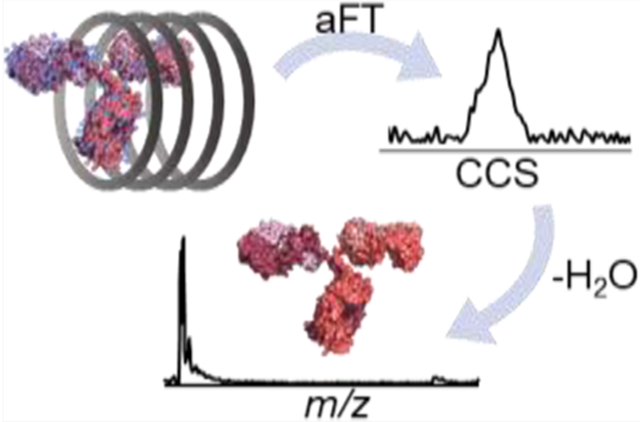
Abstract
New developments in analytical technologies and biophysical methods have advanced the characterization of increasingly complex biomolecular assemblies using native mass spectrometry (MS). Ion mobility methods, in particular, have enabled a new dimension of structural information and analysis of proteins, allowing separation of conformations and providing size and shape insights based on collision cross sections (CCSs). Based on the concepts of absorption-mode Fourier transform (aFT) multiplexing ion mobility spectrometry (IMS), here, a modular drift tube design proves capable of separating native-like proteins up to 148 kDa with resolution up to 45. Coupled with high-resolution Orbitrap MS, binding of small ligands and cofactors can be resolved in the mass domain and correlated to changes in structural heterogeneity observed in the ion-neutral CCS distributions. We also demonstrate the ability to rapidly determine accurate CCSs for proteins with 1-min aFT-IMS-MS sweeps without the need for calibrants or correction factors.
Graphical Abstract
INTRODUCTION
The success of native mass spectrometry (nMS) for the structural characterization of intact proteins and protein complexes in the gas phase has inspired the development of new methods and instrumentation for analyzing even larger macromolecules and uncovering more comprehensive information about their conformations, architectures, and interfaces.1-3 The majority of nMS applications have entailed the analysis of proteins in low charge states generated with nano-electrospray ionization (nESI). The gentle nature of nESI has long been shown to allow retention of noncovalent interactions and thus observation of multimeric species.4-7 Ion mobility spectrometry (IMS) has proven to be a landmark tool for studying the structures of gas-phase ions and more recently extended for determining stabilities and folding states of both native-like protein complexes and ones activated to induce unfolding and disassembly.8,9 Coupling nESI with IMS has provided key evidence that the structure and topology of complexes are maintained in the gas phase.10,11 The measured ion-neutral collision cross sections (CCSs) calculated from the observed mobilities of protein ions have also been used to assist with the molecular modeling of protein structures in solution.12,13
Several ion mobility methods have been commercialized and used for applications involving native-like proteins, including drift tube IMS,14-16 traveling-wave IMS (TWIMS),10,11,17-19 trapped IMS (TIMS),20-22 and structures for lossless ion manipulations (SLIM).23-25 A traveling-wave SLIM device has recently been commercialized to allow high-resolution separations at low pressures using long, serpentine pathlengths (≥13 m) prior to MS analysis;26 however, the suitability of this RF-confining system with respect to native protein application is an ongoing area of research. These IMS devices are most commonly implemented on time-of-flight mass spectrometers that offer the microsecond sampling speeds necessary for monitoring drift-tube IMS separations that occur on the millisecond timescale in the gas phase. Despite the robust performance of IMS-enabled Q-ToF systems, practical limits constrain the mass resolution of time-of-flight analyzers and inhibit applications combining drift tube IMS and Q-ToF platforms for large, complex proteins. Stated differently, as the resolving power requirements in the IMS domain increase (i.e., narrower peak widths), the increased flight times needed in a ToF platform to capture high m/z species are concurrently increased, which establishes a functional mismatch between sampling peaks in the IM domain and measurement of m/z based upon flight time. This limitation has motivated significant interest in extending IM strategies to FT-based mass analyzers (FT-ICR and Orbitraps) that offer unprecedented mass resolution allowing confident determination of small m/z differences of proteins from binding molecules such as drugs,27,28 lipids,29-31 and biological cofactors.32 Moreover, Orbitrap mass spectrometers with extended mass ranges have been developed,33,34 thus offering opportunities to expand the scope of IMS applications.3
Modular drift-tube systems have recently been developed and coupled to various ion trap and Orbitrap instruments.35-38 One of the first IMS-enabled Orbitrap systems utilized ambient-pressure drift tubes to separate small isomers (<2 kDa), as demonstrated for a series of peptides, saccharides, lipids, and metabolites.35,39 However, the incompatibility of the timescales of the IMS separation (milliseconds) and the Orbitrap mass analysis (tens of milliseconds to seconds) results in a poor duty cycle of <1%, thus wasting more than 99% of ions that are generated using an ESI source. The design of Fourier transform (FT)-based multiplexing strategies, originally implemented on quadrupole ion trap and linear ion trap systems, involves controlled frequency modulation of a dualgate pulsing scheme of a drift tube, increasing the duty cycle to 25%.37,38,40-43 This type of FT multiplexing offers an innovative approach to overcome the duty-cycle mismatch and establishes the framework for the development of FT-IMS-Orbitrap platforms. Advancements have also been made in data collection and processing to improve the throughput, resolution, and sensitivity of FT methods, including the adoption of basis pursuit denoising43 and absorption-mode FT.44 The latter method was implemented in the workflow described herein. Improvements in hardware have also been developed, such as the implementation of a tristate ion shutter to increase ion throughput and minimize gate depletion of larger ions.45,46
The first FT multiplexing-IMS-Orbitrap workflow was implemented using a 58 cm home-built, periodic-focusing drift tube adapted to the back-end of an Orbitrap mass spectrometer, ultimately achieving an IMS resolving power of up to 40 for analysis of streptavidin, transthyretin, and GlnK bound to their respective biological cofactors.42 Extension of the same drift tube design to 1.5 m improved resolution to ~60.47 It was also demonstrated that CCS calculations from ion mobilities through the periodic-focusing IMS system were possible using a dampening term, α, without the need for calibration.47 Conversely, TWIMS- and TIMS-based separations rely on calibration standards for accurate CCS determination and discovery of robust calibrants (particularly for native-like proteins) remains a critical quest for these methods.48,49
Our group has adopted an ambient drift tube module constructed from printed circuit boards (PCB) based on an open-source design50 and integrated it with Orbitrap mass spectrometers modified with an excimer laser for 193 nm ultraviolet photodissociation (UVPD).44,51 Conformer-specific fragments generated from UVPD of lipid double-bond isomers allowed postmobility separation of isobaric ions that were not fully resolved using IMS alone.51 UVPD has also been shown to generate conformer-dependent fragmentation of proteins and peptides,52,53 underscoring the capabilities of exploring conformational landscapes of gas-phase ions by coupling IMS with UVPD.
Here, the PCB-IMS coupled to an Ultra High Mass Range Orbitrap mass spectrometer proves capable of transferring and separating large, native-like protein ions for first-principles determination of CCSs without the need for correction factors or calibrants. Resolving powers up to 45 were obtained for native protein complexes ranging from a 14 kDa metal-loprotein, azurin, to a 148 kDa monoclonal antibody, benralizumab. High resolution in the m/z (Rm/z) and IMS (RIMS) dimensions are both crucial for resolving small but significant structural changes resulting from changes in the environment. In this case, proteins streptavidin and transferrin exhibited increases in structural homogeneity upon coordinating to their native ligands and cofactors, biotin and iron(III), respectively. Calmodulin also displayed conformational shifts as a function of calcium binding. IMS afforded separation of ions with overlapping m/z as demonstrated for monomers and dimers of azurin, offering the possibility of individual characterization of each. The ability to further characterize protein structures using collisional activation dissociation and UVPD are demonstrated for streptavidin homotetramers.
EXPERIMENTAL SECTION
Reagents.
Streptavidin (SA) was obtained from Proteo-Chem (Hurricane, UT). Hemoglobin (Hb) and apo- and holo-transferrin (TF) were prepared from separate lyophilized stocks, each from Sigma-Aldrich (St. Louis, MO). Azurin (Az) was expressed and purified as described previously.32 Benralizumab (Ben) was a gift from Dr. Yury Tsybin (Spectroswiss Sèrl), and apo- and holo-calmodulin (CaM) were donated by the Aldrich group (The University of Texas at Austin, Department of Neuroscience). Stocks of each protein were exchanged into ammonium acetate solutions using BioSpin size exclusion spin columns (BioRad, Hercules, CA) per manufacturer’s instructions and diluted to final protein concentrations of 5 μM unless otherwise stated. Biotin was added to the streptavidin solution at a 4:1 concentration with the tetramer before analysis.
Data Acquisition.
The PCB-DT with a 10 cm desolvation region and a 10 cm drift region was assembled as previously described.44,50,51 A schematic representation of the instrument coupled to a prototype Thermo Scientific Q Exactive Plus UHMR Orbitrap mass spectrometer (Bremen, Germany) is shown in Figure S1a, and FT-IMS parameters are listed in Table S1. Further details about the modular drift tube can be found in the Supporting Information.
Orbitrap resolution settings were kept at 12,500 at m/z 400 for FT-IMS-MS experiments and 200,000 for FT-IMS-MS/MS experiments for both higher-energy collisional dissociation (HCD) and UVPD. Supplemental in-source trapping (IST), a collisional method employed for the removal of adducts, was utilized to aid in desolvation following IMS separation and optimized for each protein as follows: −40 V for SA, Az, and CaM; −125 V for TF; and −200 V for Ben. No desolvation was applied to hemoglobin to prevent disassembly and maximize detection of the intact tetramer. IST occurs after drift tube separation, and thus has no effect (i.e., unfolding) on the observed mobilities of the ions nor their CCS distributions.
Data Analysis.
Thermo.raw files were converted to .mzXML using MSConvert54 and analyzed using a custom Matlab (R2020a) program as previously described.44,51 Briefly, extracted ion chromatograms (XICs) were generated for protein ions (Figure S1b) and averaged across five replicates then zero-padded and apodized (Figure S1c). Resultant XICs were then transformed to arrival time distributions (ATDs) using aFT and baseline-corrected (Figure S1d).44 Peak locations and FWHM values were determined using Matlab functions and used to calculate resolution RIMS (). Graphical representations of these results were generated using the Multipeak Fitting package embedded in Igor Pro 8 (Lake Oswego, OR) and applied to both arrival time and CCS distribution datasets. Arrival times were converted to CCS values using the Mason-Schamp equation as shown in the Supporting Information.
RESULTS AND DISCUSSION
The PCB-DT was implemented on a UHMR Orbitrap mass spectrometer and initially evaluated with streptavidin (SA). SA is a 52 kDa homotetramer that has been extensively studied by IMS and other MS methods.42,48,55 It binds biotin with a Kd of 10−13 M, the strongest noncovalent interaction known in nature.56 Mass spectra of unbound (apo) and biotin-bound (holo) SA using the PCB-DT source are presented in Figure S3a. Approximately 2 orders of magnitude of ion signal is lost relative to a standard nESI source, which is not unexpected for a front-end drift tube module operated at atmospheric pressure. This significant loss of signal has inspired methods to improve the transmission of ions, particularly those with low mobilities, through improved gating techniques.45,46 The mass shift for holo-SA observed in Figure S3a is consistent with the addition of four biotin molecules (~977 Da total). ATDs resulting from replicate FT-IMS sweeps are shown in Figure S3b. The drift times for each ion m/z were normalized and converted to a collision cross section distribution using equations 1 and 2. The resulting CCS distributions for SA are shown in Figure S4a and the maximum CCS values, representing the most abundant conformer, are summarized for SA and all other proteins in Table S2. The CCS values for apo-SA (12+ to 14+ charge states) of ~38 nm2 are in good agreement with a previous report (<2% difference).48
The addition of four biotins to the SA tetramer results in a slight but reproducible shift to larger CCS, believed to be caused by the added mass of the ligands as previously described.42,57 The peak width of holo-SA also narrows by an average of 11% for the three charge states (Table S2), suggesting increased homogeneity and decreased flexibility. These observations are consistent with the reported increased structural stability of biotin-bound streptavidin58 and were also noted by Poltash et al. in their initial studies using a periodic-focusing drift tube.42 Interestingly, a component with a shorter drift time (smaller CCS) is observed for the 12+ charge state of holo-SA (Figure S4a). This feature, with a CCS of 35.4 nm2, may result from a population of a more compact tetrameric structure that becomes pronounced upon binding biotin. Alternatively, it may indicate that proton loss from the 13+ tetramer is occurring after IM separation and entry into the vacuum of the mass spectrometer as evidenced by the similarity in drift times of 33.4 ms and 33.1 ms from the ATDs of the tetramers in the 12+ and 13+ charge states, respectively (Figure S3b). In this case, one would expect that an analogous proton loss from the 14+ tetramer to produce the 13+ tetramer might also occur and, indeed, a similar feature is seen for the 13+ SA tetramer (Figures S4a and S3b), albeit at relatively lower abundance than that observed for 12+ SA. However, proton loss following IMS separation is not observed for the other proteins analyzed in this study and is thus not believed to be a general confounding factor. Overall, the results for SA demonstrate the ability to accurately determine CCS values for native-like proteins and observe changes in the dynamic structures of entire protein populations through both their arrival time distributions and CCS profiles.
Increased structural homogeneity upon ligand binding is also demonstrated by the iron-transport protein, transferrin (TF). Mass spectra of the 79 kDa monomeric protein (apo and holo forms) are shown in Figure S5, and the expanded insets illustrate the addition of one or two iron atoms that are well resolved even at the relatively low-resolution setting (12,500) of the Orbitrap analyzer. The apo-TF sample, however, contained some residual iron as apparent by the distribution of TF bound to 0, 1, and 2 Fe(III), which adds complexity and heterogeneity to the spectra and ATDs. Shown in Figure S4b are the CCS distributions of apo- and holo-TF for the 15+ through 17+ charge states, each centered around ~50 nm2. The addition of Fe(III) atoms is not sufficient to result in a measurable increase in CCS due to mass alone. Narrowing of the CCS distributions is again observed for holo-TF compared to apo-TF by an average of more than 20% for the three charge states (Table S2). However, the heterogeneity of apo-TF resulting from residual iron may contribute to the broader CCS distributions. A greater uniformity in structure within the entire protein population is thus reached upon complete binding of transferrin to its cofactor (i.e., 2:1 Fe(III):TF).
Calmodulin (CaM) is another, much smaller metalloprotein that is involved in calcium transport to hundreds of targets in eukaryotes.59 The overall structure of CaM has been observed to be either globular, with the two terminal lobes in relatively close proximity, or dumbbell-like, with the lobes separated by a central helix, which are predominantly modulated by ligand binding.59 The CCSs of the globular and dumbbell structures of CaM are proposed to exist in equilibrium in solution, with and without calcium bound,59 and are estimated to be 16.4 and 20.5 nm2 based on a trajectory method approximation60 using the relevant crystal structures (PDB: 1PRM and 3CLN), respectively. Here, the structures of metal-free and metal-bound CaM are investigated using aFT-IMS-MS. Figure S6a,b displays representative mass spectra of apo- and holo-CaM with the progressive mass shift of each additional Ca2+ ion illustrated in Figure S6c. The CCS distributions of the 6+ charge state of CaM bound to 0, 2, or 4 Ca2+ ions are displayed in Figure S6d with peaks centered around 17 nm2 (Table S2). The size measured here suggests that the 6+ charge state represents the predominantly globular structure present in solution for both apo- and holo-CaM, which supports a previous observation for the 6+ charge state.61 A slight shift to larger CCS is observed upon binding calcium, which was also reported in the previous study.61 No significant changes in peak width suggestive of a change in structural homogeneity of the protein population are observed.
Hemoglobin (Hb), an oxygen transport protein found in blood, forms a 64 kDa heterotetramer with two α and two β subunits that each noncovalently bind a prosthetic heme group.62 In solution, the heterodimer has been observed in equilibrium with the heterotetramer,63,64 which is reflected in the nESI mass spectrum in Figure S7. IMS separation of Hb dimers and tetramers resulted in the ATDs in Figure S8a with drift times between 29 and 35 ms. The m/z information provided by MS facilitates the conversion of the observed drift times to CCS, as shown in Figure S8b. The Hb tetramers exhibit CCS values between 41 and 43 nm2 and are within 1% of previous reports.63,65 The population of heterotetramers appears to be quite homogeneous with narrow peak widths (~1 nm2) equivalent to an RIMS up to 45 for the 16+ charge state. This contrasts the broad distributions of the heterodimers with CCS values between 26 and 29 nm2 (RIMS of ~22), with peak values within 3% of reported CCSs of the canonical α1β1 dimers.63,65 The heterogeneity observed for the dimers may be due to differences in subunit orientation and be composed of a mixture of α1β1 and α1β2 dimers as described previously.63
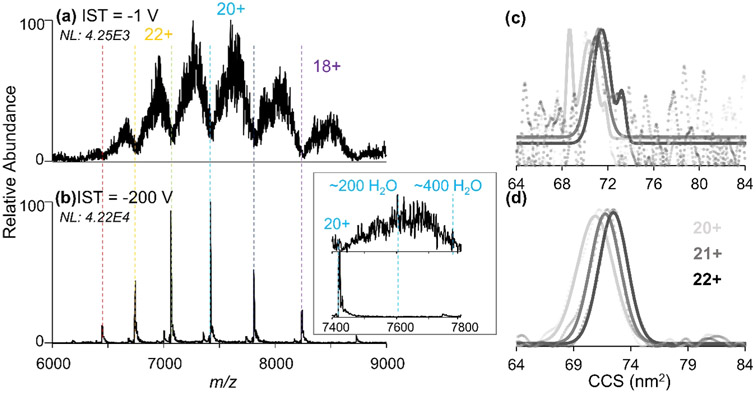
The max RIMS of 45 obtained for the Hb heterotetramer using this 10 cm drift tube is notably different from previous reports for tetraalkylammonium ions with RIMS values of up to 75 using an identical IMS-MS system44 and up to 92 using a similar drift tube with a Faraday plate detector.50 This difference in resolving power is attributed to the increased heterogeneity of large proteins that result in a broad CCS landscapes compared to the uniform structural populations afforded by small salts and metabolites. Moreover, the transfer of ions into the ambient drift tube does not result in full desolvation of the proteins, thus resulting in additional heterogeneity arising from adducted water and/or salt molecules. The impact of neutral molecule adducts is more apparent for larger proteins, as demonstrated by the examination of mass spectra acquired for the 148 kDa monoclonal antibody asthma therapeutic, benralizumab (Ben)66 in Figure 1. Without IST (Figure 1a), each charge state of Ben spans several hundred m/z, which is attributed to the presence of hundreds of adducts. Application of −200 V IST energy to cause desolvation via collisional heating (Figure 1b) results in the release of nearly all adducts and a 10-fold increase in signal. The CCS distributions of Ben following IMS separation with or without desolvation are shown in Figure 1c,d. The signal-to-noise ratio of the CCS distributions for the desolvated ions are greatly improved, but the observed CCS values for both cases are equivalent and fall between 72 and 73 nm2, reflecting the size of the water-adducted protein ions. Because IST occurs only after the proteins exit the drift tube, the collision energy has no effect on the mobilities of drift-separated ions. Thus, the ambient drift tube provides a means by which to observe the heterogeneous, solvated populations generated from ESI of native solutions prior to their transmission to vacuum. The CCS values for Ben are in good agreement with past measurements of the NIST monoclonal antibody (72.2 nm2)67 that possesses a similar mass and structure to Ben. These earlier studies were conducted using TWIMS following collisional heating to desolvate the mAb ions.67 Agreement in the CCS values reported previously and herein suggest that, while the incomplete desolvation may influence ATD peak broadness (i.e., heterogeneity), it has a negligible impact on CCS. Additionally, the agreement in the DTCCS values collected at ambient pressures and the TWCCS collected in vacuum67 suggests that any structural rearrangement of the native-like protein upon transmission into the vacuum is minor. This observation is encouraging for native MS analyses as preservation of native-like structures is a key goal.
Figure 1.
nESI mass spectra of benralizumab transferred through the drift tube to the UHMR with gates open with either (a) −1 V or (b) −200 V of applied in-source trapping (IST) desolvation energy for gentle collisional heating. The inset shows the effect of desolvation on the 20+ charge state. CCS distributions of Ben (20+ through 22+) obtained using (c) −25 V and (d) −200 V IST desolvation energy with the raw data represented as circles and the fitted data overlaid as lines. The increased signal abundance of Ben following desolvation results in an improved signal-to-noise ratio of subsequent ATDs.
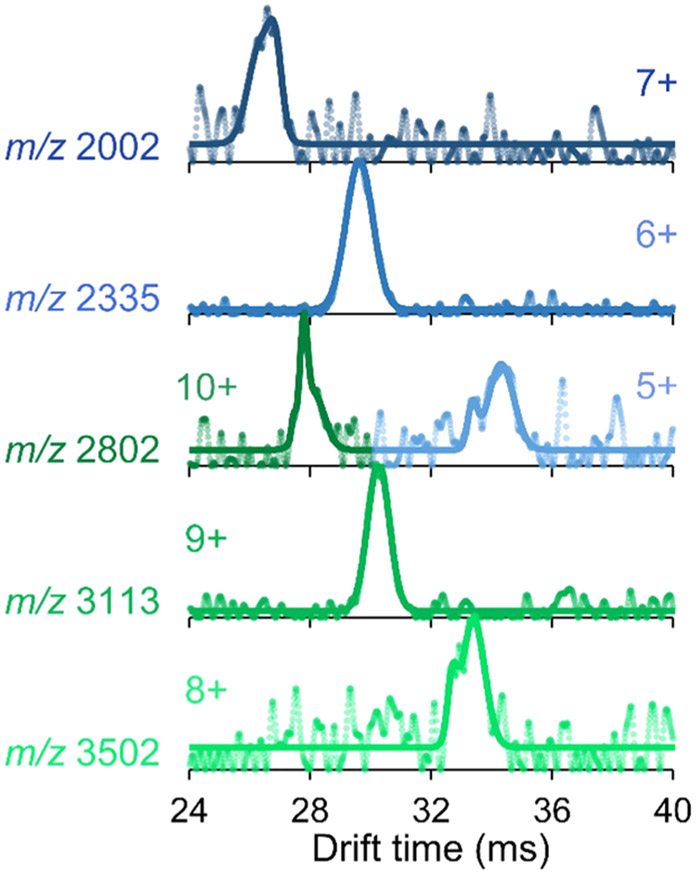
The ability of the drift tube to separate ions by size and shape provides an additional dimension of analysis to the mass and charge information from the mass spectrometer. That is, ions that cannot be separated with a mass filter owing to overlapping m/z can be size-separated by the drift tube. This capability is demonstrated with the electron transfer protein, azurin (Az). Az is known to oligomerize depending on solution conditions including protein and salt concentration.68 Figure S9a displays the mass spectrum of Az, illustrating an overlapping distribution of monomers and dimers from a solution containing 20 μM protein in 20 mM ammonium acetate. The m/z overlap of the 5+ monomer with the 10+ dimer (both m/z 2802) is resolved using FT-IMS-MS, as seen in the resulting ATDs in Figure 2. Conversion of the drift times to CCS shows two distinct populations centered at 16 and 24 nm2 (Figure S9b) corresponding to the monomers and dimers.
Figure 2.
Arrival time distributions or azurin monomers (blue; ana dimers (green) obtained showing separation of the overlapping 5+ monomer and 10+ dimer at m/z 2802. The raw data are represented as circles, and the fitted data are overlaid as solid lines.
By applying a fixed gate acquisition waveform on the two ion shutters of the drift tube (Figure S2b), the Az monomer (5+) and dimer (10+) pair can be isolated prior to mass analysis. This fixed gating acquisition method isolates all ions with a select drift time (td) within a designated pulse width (tw) of the two gates, allowing for transmission of only a few, designated species. Figure S10 demonstrates the effectiveness of this isolation mode for Az. Using a 2 ms window centered at 26 ms, only the 7+ monomer is transmitted past the second ion gate into the mass spectrometer, as shown in Figure S10b. Shifting the window across the drift time domain allows isolation of the 10+ dimer (Figure S10c) from the 5+ monomer (Figure S10f). Shorter gate pulse widths would further increase the selectivity of individual ions at the expense of duty cycle and sensitivity. This approach is necessary for the independent characterization of the two overlapping m/z species.
For enhanced characterization of proteins, tandem mass spectrometry (MS/MS) is employed to activate and dissociate selected precursors into informative fragment ions. Traditional collisional methods are available on most commercial platforms and can provide adequate sequence coverage for protein identification. Other ion activation methods have been implemented on the UHMR platform to generate more structurally informative fragmentation, including surface-induced dissociation,69,70 electron capture dissociation,71 and ultraviolet photodissociation (UVPD).72 Because the PCB-DT is adapted to the front of the Orbitrap mass spectrometer, it does not impede the collection of MS/MS data. To demonstrate this functionality, the 12+ through 14+ charge states of the apo-SA tetramer were isolated using the quadrupole mass filter for an FT-IMS-MS/MS experiment using either higher-energy collisional dissociation (HCD) or UVPD. Due to the reduced sensitivity of the ambient IMS method, longer injection times and thus longer sweep times (up to 20 min) were utilized for this approach. Representative MS/MS spectra are displayed in Figure S11a,b. At the collision and laser energies utilized, both methods predominantly produce monomers and trimers from the tetrameric precursors. Interferograms can be extracted for each of the fragment ions as done for the 6+ and 7+ monomers shown in Figures S11d,e. Because activation occurs following FT-IMS, the fragments are encoded with the frequency of the precursor from which they were generated. The frequency-encoded monomers exhibit multimodal distributions with peaks corresponding to the drift times of 13+ and 14+ tetramers at 32.5 and 30.5 ms, respectively (Figure S11c). Monomers (6+ and 7+) resulting from HCD of the 12+ tetramer (35 ms) are not observed in the ATDs in Figure S11d owing to the increased stability of low-charged precursors and thus decreased dissociation efficiency using fixed activation conditions. Typically, HCD collision energy is normalized to account for the charge of the precursor, but because all tetramer charge states were co-isolated and a single HCD collision energy was used, the highest charge state dissociated most readily as evident by the extensive precursor depletion of the 14+ charge state in Figure S11a. UVPD of protein complexes has been shown to be less dependent on precursor charge as evidenced here by the similar charge state distributions of the surviving 12+ through 14+ tetramers in Figure S11b compared to the MS1 spectrum in Figure S3a. Due to signal-to-noise limitations, however, the 6+ and 7+ monomers generated from UVPD do not appear to have appreciable contributions evolving from the 12+ tetramer (Figure S11e). A hallmark of UVPD is the ability to generate primary sequence information in addition to stoichiometry for protein complexes. Here, the reduced sensitivity of the ambient drift tube impairs the ability to achieve adequate signal-to-noise ratio of MS/MS spectra for the detection of low-abundance fragments such as ones that originate from covalent cleavage of the protein backbone. Work is ongoing to incorporate UVPD with a low-pressure drift tube that offers significantly greater transmission efficiency to identify and characterize protein conformers with higher sensitivity.
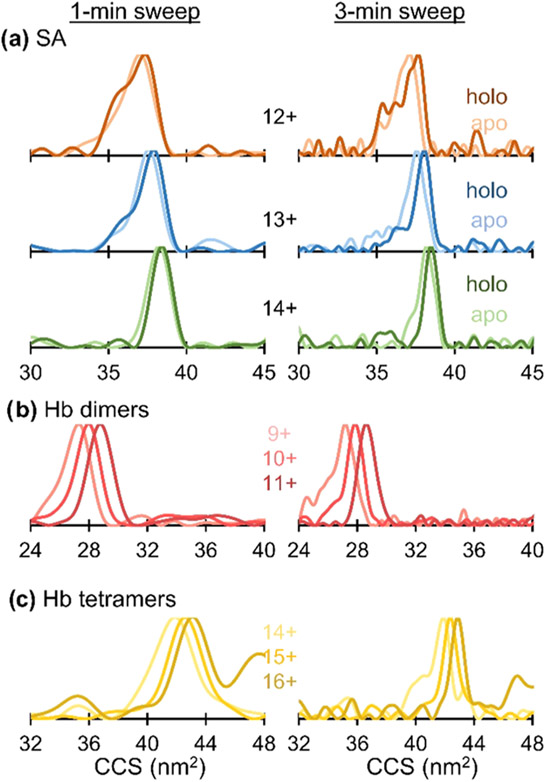
FT-IMS acquisitions commonly require ≥5 minutes sweeps to adequately sample CCS distributions.42,44,47,51,73 Shorter acquisitions down to 1 min were recently shown to be possible by artificially extending the XICs (Figure S1b) using zero-padding and apodization (Figure S1c). Here, the 8–10 min sweeps collected for all proteins listed in Table S1 were truncated to 1 min and 3 min and reprocessed using the aFT method as described previously. The “new” end frequencies for the 1- and 3-min sweeps are listed in Table S3. The resulting CCS distributions for select proteins are shown in Figure 3 with the remaining proteins shown in Figure S12. CCS distributions for the 3-min truncation exhibit lower resolution compared to the full-length sweeps by only 11% overall and still retain the features highlighted previously. That is, shifts in peak CCS and slight changes in peak width are still observed in the 3-min sweeps in Figures 3a and S12a,b. Further shortening the sweeps to 1 min decreases the resolution by an average of 42% compared to the full-length acquisitions, obscuring subtle changes in the distributions. However, accurate CCS values are still afforded by the 1-min sweeps, allowing for the rapid determination of protein size.
Figure 3.
CCS distributions for (a) apo- and holo-streptavidin, (b) hemoglobin heterodimers, and (c) hemoglobin heterotetramers using truncated FT-IMS acquisition times.
CONCLUSIONS
A compact, modular drift tube constructed from printed circuit boards demonstrates the ability to provide accurate CCS measurements for proteins up to 148 kDa in as little as 1 min. More detailed structural information can be obtained using extended aFT-IMS-MS sweep parameters, which offers insights into population dynamics. Increased homogeneity is achieved for streptavidin and transferrin when they bind their native ligands, and broad distributions from hemoglobin heterodimers reflect the heterogeneous arrangements they may adopt in solution. Resolution is limited by structural heterogeneity and incomplete desolvation, but RIMS values up to 45 were achieved, which was sufficient to resolve a monomer–dimer pair with overlapping m/z. The ensemble changes observed for protein complexes are still resolved with truncated acquisition times and accurate CCS values are obtained in just 1 min. The modular characteristic of this drift tube allows retention of MS/MS capabilities, and IMS multiplexing allows simultaneous activation of multiple charge states of protein complexes while tracking distinctive product ions via frequency-encoding in the ion mobility dimension. To our knowledge, this is the first report on the structural characterization of native-like protein ions using an ambient, modular IMS-Orbitrap system. Because the design of this low-cost drift tube is freely available and easily assembled50 (or available as a commercial product with minor modification),35 we anticipate greater adoption of these native IMS-MS methods across the field of structural biology. The short timescales of aFT multiplexing experiments also pave the way for high-throughput, automated methods, such as online buffer exchange,74 for the rapid determination of protein CCSs. The greater transmission efficiency of a low-pressure drift tube, now under development, will enable higher MS/MS sensitivity than achieved using the current atmospheric pressure device. The exploration of IM technologies remains an active frontier, and the exciting advances in methods like SLIM and TIMS, as well as drift tube strategies like the one presented here, continue to extend the limits of sensitivity and mobility resolution in addition to expanding the range of applications on many different mass spectrometer platforms.
Supplementary Material
ACKNOWLEDGMENTS
Funding from NSF (CHE-2203602) and the Robert A. Welch Foundation (F-1155) is gratefully acknowledged. Researchers at WSU were supported by NIGMS (R01GM138863).
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.2c01653.
Experimental FT-IMS parameters, representative nESI and MS/MS spectra, and arrival time and CCS distributions of all proteins discussed herein (PDF)
Contributor Information
Sarah N. Sipe, Department of Chemistry, The University of Texas at Austin, Austin, Texas 78712, United States
James D. Sanders, Department of Chemistry, The University of Texas at Austin, Austin, Texas 78712, United States
Tobias Reinecke, Department of Chemistry, Washington State University, Pullman, Washington 99164, United States.
Brian H. Clowers, Department of Chemistry, Washington State University, Pullman, Washington 99164, United States.
Jennifer S. Brodbelt, Department of Chemistry, The University of Texas at Austin, Austin, Texas 78712, United States.
REFERENCES
- (1).Tamara S; den Boer MA; Heck AJR Chem. Rev 2021, DOI: 10.1021/acs.chemrev.1c00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Zhou M; Lantz C; Brown KA; Ge Y; Paša-Tolić L; Loo JA; Lermyte F Chem. Sci 2020, 11, 12918–12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Poltash ML; McCabe JW; Shirzadeh M; Laganowsky A; Russell DH TrAC, Trends Anal. Chem 2020, 124, No. 115533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ganem B; Li YT; Henion JD J. Am. Chem. Soc 1991, 113, 6294–6296. [Google Scholar]
- (5).Light-Wahl KJ; Schwartz BL; Smith RD J. Am. Chem. Soc 1994, 116, 5271–5278. [Google Scholar]
- (6).Loo JA Mass Spectrom. Rev 1997, 16, 1–23. [DOI] [PubMed] [Google Scholar]
- (7).Rostom AA; Robinson CV J. Am. Chem. Soc 1999, 121, 4718–4719. [Google Scholar]
- (8).Allen SJ; Eaton RM; Bush MF Anal. Chem 2017, 89, 7527–7534. [DOI] [PubMed] [Google Scholar]
- (9).Dixit SM; Polasky DA; Ruotolo BT Curr. Opin. Chem. Biol 2018, 42, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ruotolo BT; Giles K; Campuzano I; Sandercock AM; Bateman RH; Robinson CV Science 2005, 310, 1658–1661. [DOI] [PubMed] [Google Scholar]
- (11).Zhou M; Wysocki VH Acc. Chem. Res 2014, 47, 1010–1018. [DOI] [PubMed] [Google Scholar]
- (12).Hall Z; Politis A; Robinson CV Structure 2012, 20, 1596–1609. [DOI] [PubMed] [Google Scholar]
- (13).Lee JW; L Davidson K; F Bush M; I Kim H Analyst 2017, 142, 4289–4298. [DOI] [PubMed] [Google Scholar]
- (14).May JC; Goodwin CR; Lareau NM; Leaptrot KL; Morris CB; Kurulugama RT; Mordehai A; Klein C; Barry W; Darland E; Overney G; Imatani K; Stafford GC; Fjeldsted JC; McLean JA Anal. Chem 2014, 86, 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).May JC; Jurneczko E; Stow SM; Kratochvil I; Kalkhof S; McLean JA Int. J. Mass Spectrom 2018, 427, 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Vallejo DD; Polasky DA; Kurulugama RT; Eschweiler JD; Fjeldsted JC; Ruotolo BT Anal. Chem 2019, 91, 8137–8146. [DOI] [PubMed] [Google Scholar]
- (17).Giles K; Pringle SD; Worthington KR; Little D; Wildgoose JL; Bateman RH Rapid Commun. Mass Spectrom 2004, 18, 2401–2414. [DOI] [PubMed] [Google Scholar]
- (18).Uetrecht C; Barbu IM; Shoemaker GK; van Duijn E; Heck AJR Nat. Chem 2011, 3, 126–132. [DOI] [PubMed] [Google Scholar]
- (19).Giles K; Ujma J; Wildgoose J; Pringle S; Richardson K; Langridge D; Green M Anal. Chem 2019, 91, 8564–8573. [DOI] [PubMed] [Google Scholar]
- (20).Fernandez-Lima F; Kaplan DA; Suetering J; Park MA Int. J. Ion Mobility Spectrom 2011, 14, 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Benigni P; Marin R; Molano-Arevalo JC; Garabedian A; Wolff JJ; Ridgeway ME; Park MA; Fernandez-Lima F Int. J. Ion Mobility Spectrom 2016, 19, 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Fouque KJD; Fernandez-Lima F TrAC, Trends Anal. Chem 2019, 116, 308–315. [Google Scholar]
- (23).Webb IK; Garimella SVB; Tolmachev AV; Chen T-C; Zhang X; Norheim RV; Prost SA; LaMarche B; Anderson GA; Ibrahim YM; Smith RD Anal. Chem 2014, 86, 9169–9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Deng L; Ibrahim YM; Hamid AM; Garimella SVB; Webb IK; Zheng X; Prost SA; Sandoval JA; Norheim RV; Anderson GA; Tolmachev AV; Baker ES; Smith RD Anal. Chem 2016, 88, 8957–8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Nagy G; Attah IK; Conant CR; Liu W; Garimella SVB; Gunawardena HP; Shaw JB; Smith RD; Ibrahim YM Anal. Chem 2020, 92, 5004–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).May JC; Leaptrot KL; Rose BS; Moser KLW; Deng L; Maxon L; DeBord D; McLean JA J. Am. Soc. Mass Spectrom 2021, 32, 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tian Y; Lippens JL; Netirojjanakul C; Campuzano IDG; Ruotolo BT Protein Sci 2019, 28, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Marcoux J; Champion T; Colas O; Wagner-Rousset E; Corvaïa N; Dorsselaer AV; Beck A; Cianférani S Protein Sci. 2015, 24, 1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Cong X; Liu Y; Liu W; Liang X; Russell DH; Laganowsky AJ Am. Chem. Soc 2016, 138, 4346–4349. [DOI] [PubMed] [Google Scholar]
- (30).Allison TM; Reading E; Liko I; Baldwin AJ; Laganowsky A; Robinson CV Nat. Commun 2015, 6, No. 8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Patrick JW; Boone CD; Liu W; Conover GM; Liu Y; Cong X; Laganowsky A Proc. Natl. Acad. Sci. U.S.A 2018, 115, 2976–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Crittenden CM; Novelli ET; Mehaffey MR; Xu GN; Giles DH; Fies WA; Dalby KN; Webb LJ; Brodbelt JS J. Am. Soc. Mass Spectrom 2020, 31, 1140–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fort KL; Waterbeemd M; van de Boll D; Reinhardt-Szyba M; Belov ME; Sasaki E; Zschoche R; Hilvert D; Makarov AA; Heck AJR Analyst 2018, 143, 100–105. [DOI] [PubMed] [Google Scholar]
- (34).van de Waterbeemd M; Fort KL; Boll D; Reinhardt-Szyba M; Routh A; Makarov A; Heck AJR Nat. Methods 2017, 14, 283–286. [DOI] [PubMed] [Google Scholar]
- (35).Keelor JD; Zambrzycki S; Li A; Clowers BH; Fernández FM Anal. Chem 2017, 89, 11301–11309. [DOI] [PubMed] [Google Scholar]
- (36).Hollerbach A; Fedick PW; Cooks RG Anal. Chem 2018, 90, 13265–13272. [DOI] [PubMed] [Google Scholar]
- (37).Morrison KA; Siems WF; Clowers BH Anal. Chem 2016, 88, 3121–3129. [DOI] [PubMed] [Google Scholar]
- (38).Schrader RL; Marsh BM; Cooks RG Anal. Chem 2020, 92, 5107–5115. [DOI] [PubMed] [Google Scholar]
- (39).Kaszycki JL; Rotta AL; Colsch B; Fenaille F; Dauly C; Kamleh A; Wu C Rapid Commun. Mass Spectrom 2019, 33, 3–10. [DOI] [PubMed] [Google Scholar]
- (40).Ibrahim YM; Garimella SVB; Prost SA; Wojcik R; Norheim RV; Baker ES; Rusyn I; Smith RD Anal. Chem 2016, 88, 12152–12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Knorr FJ; Eatherton RL; Siems WF; Hill HH Anal. Chem 1985, 57, 402–406. [DOI] [PubMed] [Google Scholar]
- (42).Poltash ML; McCabe JW; Shirzadeh M; Laganowsky A; Clowers BH; Russell DH Anal. Chem 2018, 90, 10472–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Davis AL; Reinecke T; Morrison KA; Clowers BH Anal. Chem 2019, 91, 1432–1440. [DOI] [PubMed] [Google Scholar]
- (44).Sanders JD; Butalewicz JP; Clowers BH; Brodbelt JS Anal. Chem 2021, 93, 9513–9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Kwantwi-Barima P; Reinecke T; H Clowers B Analyst 2019, 144, 6660–6670. [DOI] [PubMed] [Google Scholar]
- (46).Kirk AT; Grube D; Kobelt T; Wendt C; Zimmermann S Anal. Chem 2018, 90, 5603–5611. [DOI] [PubMed] [Google Scholar]
- (47).McCabe JW; Mallis CS; Kocurek KI; Poltash ML; Shirzadeh M; Hebert MJ; Fan L; Walker TE; Zheng X; Jiang T; Dong S; Lin C-W; Laganowsky A; Russell DH Anal. Chem 2020,92, 11155–11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Stiving AQ; Jones BJ; Ujma J; Giles K; Wysocki VH Anal. Chem 2020, 92, 4475–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Bush MF; Hall Z; Giles K; Hoyes J; Robinson CV; Ruotolo BT Anal. Chem 2010, 82, 9557–9565. [DOI] [PubMed] [Google Scholar]
- (50).Reinecke T; Clowers BH HardwareX 2018, 4, No. e00030. [Google Scholar]
- (51).Sanders JD; Shields SW; Escobar EE; Lanzillotti MB; Butalewicz JP; James VK; Blevins MS; Sipe SN; Brodbelt JS Anal. Chem 2022, 94, 4252–4259. [DOI] [PubMed] [Google Scholar]
- (52).Warnke S; Helden G; von Pagel K PROTEOMICS 2015, 15, 2804–2812. [DOI] [PubMed] [Google Scholar]
- (53).Theisen A; Black R; Corinti D; Brown JM; Bellina B; Barran PE J. Am. Soc. Mass Spectrom 2019, 30, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Chambers MC; Maclean B; Burke R; Amodei D; Ruderman DL; Neumann S; Gatto L; Fischer B; Pratt B; Egertson J; Hoff K; Kessner D; Tasman N; Shulman N; Frewen B; Baker TA; Brusniak M-Y; Paulse C; Creasy D; Flashner L; Kani K; Moulding C; Seymour SL; Nuwaysir LM; Lefebvre B; Kuhlmann F; Roark J; Rainer P; Detlev S; Hemenway T; Huhmer A; Langridge J; Connolly B; Chadick T; Holly K; Eckels J; Deutsch EW; Moritz RL; Katz JE; Agus DB; MacCoss M; Tabb DL; Mallick P Nat. Biotechnol 2012, 30, 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Morrison LJ; Brodbelt JS J. Am. Chem. Soc 2016, 138, 10849–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Loosli A; Rusbandi UE; Gradinaru J; Bernauer K; Schlaepfer CW; Meyer M; Mazurek S; Novic M; Ward TR Inorg. Chem 2006, 45, 660–668. [DOI] [PubMed] [Google Scholar]
- (57).Quintyn RS; Yan J; Wysocki VH Chem. Biol 2015, 22, 583–592. [DOI] [PubMed] [Google Scholar]
- (58).González M; Bagatolli LA; Echabe I; Arrondo JLR; Argaraña CE; Cantor CR; Fidelio GD J. Biol. Chem 1997, 272, 11288–11294. [DOI] [PubMed] [Google Scholar]
- (59).Kursula P Amino Acids 2014, 46, 2295–2304. [DOI] [PubMed] [Google Scholar]
- (60).Marklund EG; Degiacomi MT; Robinson CV; Baldwin AJ; Benesch JLP Structure 2015, 23, 791–799. [DOI] [PubMed] [Google Scholar]
- (61).Wyttenbach T; Grabenauer M; Thalassinos K; Scrivens JH; Bowers MT J. Phys. Chem. B 2010, 114, 437–447. [DOI] [PubMed] [Google Scholar]
- (62).Park S-Y; Yokoyama T; Shibayama N; Shiro Y; Tame JRH J. Mol. Biol 2006, 360, 690–701. [DOI] [PubMed] [Google Scholar]
- (63).Woodall DW; Brown CJ; Raab SA; El-Baba TJ; Laganowsky A; Russell DH; Clemmer DE Anal. Chem 2020, 92, 3440–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Herzik MA; Wu M; Lander GC Nat. Commun 2019, 10, No. 1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Scarff CA; Patel VJ; Thalassinos K; Scrivens JH J. Am. Soc. Mass Spectrom 2009, 20, 625–631. [DOI] [PubMed] [Google Scholar]
- (66).Castro M; Wenzel SE; Bleecker ER; Pizzichini E; Kuna P; Busse WW; Gossage DL; Ward CK; Wu Y; Wang B; Khatry DB; van der Merwe R; Kolbeck R; Molfino NA; Raible DG Lancet Respir. Med 2014, 2, 879–890. [DOI] [PubMed] [Google Scholar]
- (67).Campuzano IDG; Larriba C; Bagal D; Schnier PD Ion Mobility and Mass Spectrometry Measurements of the Humanized IgGk NIST Monoclonal Antibody. In State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 3. Defining the Next Generation of Analytical and Biophysical Techniques; Schiel JE; Davis DL; Borisov OV, Eds.; American Chemical Society: Washington, DC, 2015; Vol. 1202, pp 75–112, DOI: 10.1021/bk-2015-1202.ch004. [DOI] [Google Scholar]
- (68).Sokolová L; Williamson H; Sýkora J; Hof M; Gray HB; Brutschy B; Vlček A J. Phys. Chem. B 2011, 115, 4790–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).VanAernum ZL; Gilbert JD; Belov ME; Makarov AA; Horning SR; Wysocki VH Anal. Chem 2019, 91, 3611–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Snyder DT; Panczyk EM; Somogyi A; Kaplan DA; Wysocki V Anal. Chem 2020, 92, 11195–11203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Shaw JB; Malhan N; Vasil’ev YV; Lopez NI; Makarov A; Beckman JS; Voinov VG Anal. Chem 2018, 90, 10819–10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Mehaffey MR; Sanders JD; Holden DD; Nilsson CL; Brodbelt JS Anal. Chem 2018, 90, 9904–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Kwantwi-Barima P; Ouyang H; Hogan CJ; Clowers BH Anal. Chem 2017, 89, 12416–12424. [DOI] [PubMed] [Google Scholar]
- (74).VanAernum ZL; Busch F; Jones BJ; Jia M; Chen Z; Boyken SE; Sahasrabuddhe A; Baker D; Wysocki VH Nat. Protoc 2020, 15, 1132–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.