Significance
Methanol-based biomanufacturing is considered a promising way to achieve carbon neutrality. However, efficient methanol biotransformation toward chemical production is challenging. To address this challenge, we enhanced the precursor supply and coenzyme regeneration in Pichia pastoris, combined with reinforcing methanol assimilation, which resulted in significant improvements in fatty acid production. Furthermore, we offered an effective approach for rapidly constructing versatile cell factories that share common precursors. This study represents an important step forward in the rewiring of P. pastoris as industrial microbial cell factories for chemical production.
Keywords: metabolic engineering, methylotrophic yeast, synthetic biotechnology, oleochemicals, biofuels
Abstract
Methanol-based biorefinery is a promising strategy to achieve carbon neutrality goals by linking CO2 capture and solar energy storage. As a typical methylotroph, Pichia pastoris shows great potential in methanol biotransformation. However, challenges still remain in engineering methanol metabolism for chemical overproduction. Here, we present the global rewiring of the central metabolism for efficient production of free fatty acids (FFAs; 23.4 g/L) from methanol, with an enhanced supply of precursors and cofactors, as well as decreased accumulation of formaldehyde. Finally, metabolic transforming of the fatty acid cell factory enabled overproduction of fatty alcohols (2.0 g/L) from methanol. This study demonstrated that global metabolic rewiring released the great potential of P. pastoris for methanol biotransformation toward chemical overproduction.
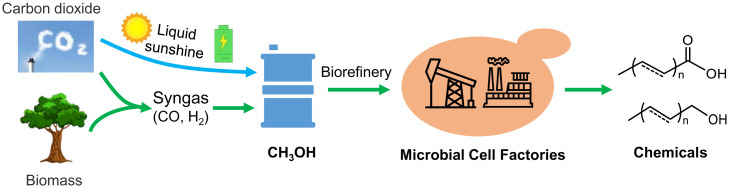
The rapid growth of the global energy demand has exacerbated environmental and climate issues and enhanced the need for sustainable feedstocks. Methanol is considered as an ideal one-carbon (C1) resource for the chemical industry and biomanufacturing (1) since it can be produced from CO2 and hydrogen (electrolysis of water) by photocatalysis (2) or electrocatalysis (3). This strategy is considered one of the most promising ways to achieve carbon neutrality, and cheap sources and flexible production processes have lowered the cost of methanol and increased the competitiveness of methanol biomanufacturing (Fig. 1). Furthermore, methanol is easy to store and transport and is thus considered an excellent energy carrier and a promising building block for synthesizing complex chemicals, and its high reduction potential is beneficial for achieving higher product yields (4, 5).
Fig. 1.
The concept of methanol-based biomanufacturing from renewable resources. Methanol can be produced economically and in a large scale from renewable resources and by using green energy, representing an ideal feedstock for future biomanufacturing, and will contribute to the carbon neutrality goal by capturing CO2.
The core consideration in methanol-based biomanufacturing is the construction of efficient microbial cell factories for synthesis of target products from methanol. However, the complex metabolism of methanol and the fatal toxicity of formaldehyde, the intermediate of methanol metabolism, bring challenges in driving metabolic flux toward target chemicals (6). Recently, great efforts have been made in engineering synthetic methylotrophs such as Escherichia coli (7–9) and Corynebacterium glutamicum (10) for methanol utilization by constructing heterologous and artificial utilization pathways (11, 12). However, the methanol utilization efficiency is extremely low for the synthesis of target chemicals (13–16). Alternatively, native methylotrophs might be promising hosts for methanol-based biomanufacturing given their capacity for methanol metabolism. Among them, methylotrophic yeasts show great potential in methanol biotransformation because of their natural efficiency in methanol assimilation and available methanol induction system (17). In particular, Pichia pastoris (syn. Komagataella phaffii) is an important industrial yeast that has been recognized as a GRAS (generally regarded as safe) microbe and is widely used for the production of recombinant proteins (18), such as human serum albumin (19) and plectasin (20). In addition, the Crabtree-negative phenotype facilitates the use of P. pastoris for high-cell-density fermentation in industrial processes (6). Although some chemicals have been synthesized from methanol in P. pastoris (21–26), the efficiency needs to be largely improved. Previous studies have elucidated the basic features of the methanol metabolism in P. pastoris (27–31), but more efforts are still needed to accurately regulate the methanol metabolism to promote chemicals production.
Fatty acids and their derivatives are promising raw materials for manufacturing advanced biofuels, detergents, lubricants, surfactants, etc. (32). Microbes have been extensively engineered for overproduction of fatty acid derivatives from glucose (33–36). For example, we previously extensively reprogrammed the metabolism of Saccharomyces cerevisiae from alcoholic fermentation to lipogenesis, which enabled the production of 33.4 g/L free fatty acids (FFAs). However, there has been no report of the production of fatty acid derivatives from methanol thus far, which might be attributed to the complex regulation of methanol metabolism. Recently, we and others developed efficient genetic tools for P. pastoris with high homologous recombination, various promoters, and genome integration sites (37–43), which should facilitate precise and global metabolic rewiring of P. pastoris for methanol biotransformation.
Here, we comprehensively engineered the central metabolism of P. pastoris for overproduction of FFAs (23.4 g/L) and fatty alcohols (2.0 g/L) from methanol by enhancing the supply of precursors and cofactors. A metabolic transforming strategy was developed for the rapid transformation of an FFA cell factory toward fatty alcohols overproduction. The engineering endeavors described here provide insight into methanol metabolism, which should be beneficial for the production of other chemicals from methanol.
Results
Engineering a P. pastoris Chassis for FFA Production.
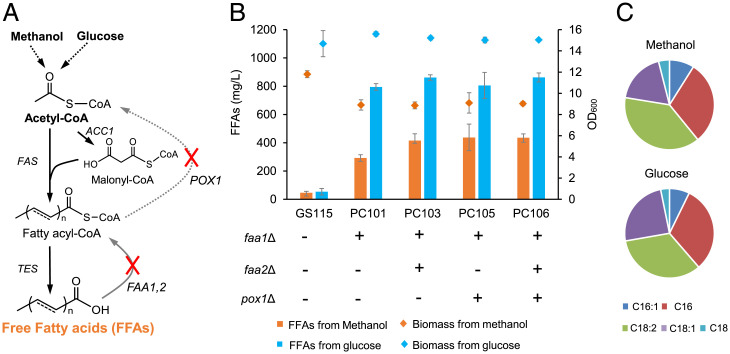
We first tried to establish a platform P. pastoris strain for FFA overproduction. In yeast, fatty acids are de novo synthesized by cytosolic type I fatty acid synthase (Fas) as fatty acyl-CoAs by using acetyl-CoA as a primer and malonyl-CoA as an extension unit. Trace amounts of FFAs can be synthesized from fatty acyl-CoA through a nonspecific thioesterase (Tes), and excess amounts of FFAs can be rapidly converted back to fatty acyl-CoA under the catalysis of fatty acyl-CoA synthetases (36) (Fig. 2A). Consistent with this, the wild-type strain GS115 accumulated only ∼50 mg/L fatty acids using either glucose or methanol as the carbon source (Fig. 2B). A search of the genome of P. pastoris identified two fatty acyl-CoA synthetase genes, namely, FAA1 and FAA2. Deletion of FAA1 (strain PC101) significantly improved FFA accumulation and further deletion of FAA2 (strain PC103) slightly improved FFA production (Fig. 2B), which suggested that FAA1 played the dominant role in FFA activation. Knocking out the fatty acyl-CoA oxidase gene POX1 to reduce the β-oxidation of fatty acyl-CoA did not increase FFA production. We thus retained POX1 in subsequent experiments since it may play an important role in peroxisome function. The engineered strain PC103 produced 860 and 415 mg/L FFAs from 20 g/L glucose and 20 g/L methanol with cell densities (OD600) of 15.2 and 8.9, respectively (Fig. 2B). This result demonstrated that methanol metabolism was much more rigid for FFA synthesis and needed to be carefully rewired. The percentage of C18:2 fatty acids from cells cultured with methanol was slightly higher than that of cells cultured with glucose, although the unsaturated fatty acids content was more than 65% (Fig. 2C).
Fig. 2.
Blocking FFA activation and consumption in P. pastoris. (A) Schematic view of the interruption of fatty acids consumption. (B) FFA titers of engineered strains in shake flasks after 96 h of cultivation in 20 g/L glucose medium or 120 h of cultivation in 20 g/L methanol medium at 220 rpm and 30 °C. The histogram shows the FFA titers, and the rhombus scatter points show the biomass of engineered strains. (C) FFA profiles of the strain PC103 in methanol- and glucose medium. Error bars correspond to the SD of the mean ± SD (n = 3, corresponding to three biological replicates).
Rewiring Carbon Metabolism to Improve FFA Production from Methanol.
The lower production of FFAs from methanol than from glucose suggested that the metabolic flux needed to be enhanced. We thus used a bottom-up rational strategy to improve the biosynthesis of fatty acids from methanol in P. pastoris (Fig. 3A). Knocking out FAA1 and FAA2 in PC111B, a homologous recombination-enhanced strain PC110 with a genome-integrated CAS9 gene at PNSI-3 site (37), exhibited similar FFA production as strain PC103 (Figs. 2B and 3B).
Fig. 3.
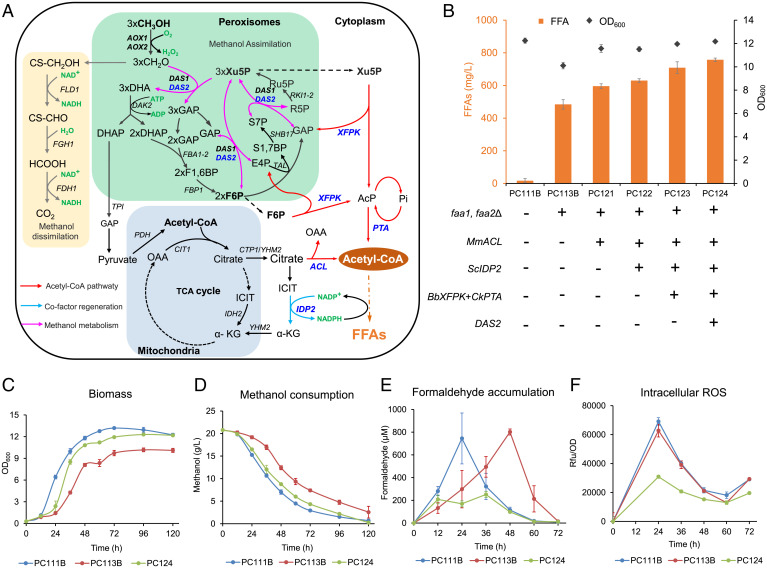
Metabolic rewiring for FFA production in P. pastoris. (A) Schematic illustration of rewired central metabolism for improved FFA production from methanol. Overexpressed genes are shown in bright blue. The Mus musculus ATP:citrate lyase gene MmACL was integrated into the genomic PNSI-2 site; the Bifidobacterium breve phosphoketolase gene BbXFPK and Clostridium kluyveri phosphotransacetylase gene CkPTA were integrated into the genomic PNSIII-5 site. For NADPH regeneration, the S. cerevisiae NADP+-dependent isocitrate dehydrogenase gene ScIDP2 was integrated at the PNSI-4 site. Another copy of endogenous DAS2 was overexpressed at the PNSII-4 site to improve formaldehyde assimilation. (B) FFA production and cell density of strains in shake flasks after 120 h of cultivation at 220 rpm and 30 °C with 20 g/L methanol minimal medium. (C–F) Growth curves, methanol consumption, formaldehyde accumulation and intracellular ROS levels. Error bars represent SD of triplicate samples.
Acetyl-CoA and malonyl-CoA are the direct precursors of fatty acid synthesis, and one molecule of acetyl-CoA is required for each two-carbon unit extension of the fatty acid carbon chain. Therefore, sufficient metabolic flux of acetyl-CoA and malonyl-CoA is crucial for fatty acid synthesis. We first tried to enhance the supply of acetyl-CoA. Overexpressing ATP:citrate lyase gene (MmACL) from Mus musculus, catalyzing the decomposition of citric acid to form acetyl-CoA and oxaloacetate (36), improved FFA production by 23% (596 mg/L in strain PC121) compared to PC113B (Fig. 3B). The phosphoketolase-phosphotransacetylase (PK-PTA) pathway was hypothesized to be beneficial for enhancing acetyl-CoA supply by diverting carbon flux from glycolysis and pentose phosphate pathway via acetyl phosphate (AcP) (44–46). The heterologous PK-PTA pathway was constructed by expressing BbPK from Bifidobacterium breve and CkPTA from Clostridium kluyveri, which significantly increased FFA production to 709 mg/L (strain PC123). The intracellular malonyl-CoA content is usually very low (47). Here, the promoter replacement method was used for malonyl-CoA engineering. Unfortunately, no transformants were obtained either by PGAP or PTEF1, which was speculated to be caused by the cytotoxicity of malonyl-CoA.
Since FFA biosynthesis requires a large amount of NADPH, we also tried several strategies to increase the NADPH supply. The NADP+-dependent isocitrate dehydrogenase (Idp2) is responsible for converting isocitrate to α-ketoglutaric acid and producing NADPH in the cytoplasm (33). Overexpressing ScIDP2 from S. cerevisiae in strain PC122 slightly increased FFA production (Fig. 3B). We also constructed an artificial malate cycle in the cytoplasm (36) to improve NADPH supply (SI Appendix, Fig. S1A), which resulted in a lower FFA production regardless of whether the strong or moderate promoters were used (SI Appendix, Fig. S1B). Cofactor quantification showed that the P. pastoris wild-type strain PC111B had much higher NADPH/NADP+ levels (0.62 at 18 h and 0.29 at 42 h) than S. cerevisiae (0.39 at 18 h and 0.04 at 42 h) when cultured in glucose media (SI Appendix, Fig. S2). More importantly, methanol-cultured P. pastoris PC111B had significantly higher NADPH/NADP+ levels (1.00 at 24 h, and 1.07 at 48 h) than S. cerevisiae. Although FFA biosynthesis consumed a large amount of NADPH in strains PC113B and PC124, their NADPH/NADP+ levels were still much higher than that of S. cerevisiae. These results suggested that methanol-cultured P. pastoris cells had sufficient NADPH supply for FFA biosynthesis, which might explain why the NADPH engineering strategies were successful in S. cerevisiae (33) but failed in P. pastoris.
We then overexpressed the mitochondrial acid transporter Yhm2 from S. cerevisiae (33) to drive metabolic flux through cytoplasmic ATP:citrate lyation and isocitrate dehydrogenation, which, however, dramatically lowered FFA production and biomass yield (SI Appendix, Fig. S1B). This observation suggested that the metabolic flux of tricarboxylic acid (TCA) cycle in P. pastoris was not strong enough when using methanol as carbon source compared to that of the glucose-based TCA cycle in S. cerevisiae (33). As xylulose-5-phosphate (Xu5P) is a critical precursor for methanol assimilation in methylotrophic yeast (48), we constructed a compensation pathway for Xu5P supply by expressing the fructose-1,6-bisphosphatase gene (FBP1) and D-ribulose-5-phosphate 3-epimerase gene (RPE1), as well as the glucose-6-phosphate dehydrogenase gene (ZWF1) and 6-phosphogluconate dehydrogenase gene (GND2) for NADPH generation (SI Appendix, Fig. S3A). However, these engineering endeavors resulted in reduced FFA production (SI Appendix, Fig. S3B).
Formaldehyde is the first intermediate of methanol utilization and confers the main toxicity that impairs cellular robustness (16). Indeed, we found that the FFA-overproducing strain PC113B exhibited decreased cell density (Fig. 3C) and methanol utilization (Fig. 3D) with higher formaldehyde accumulation (Fig. 3E) than the control strain PC111B. This observation suggested that FFA biosynthesis caused cellular stress and drove enhanced methanol oxidation toward a high level of formaldehyde accumulation. Interestingly, engineering the central metabolism to improve the supply of acetyl-CoA and NADPH (strain PC123) resulted in reduced formaldehyde accumulation (SI Appendix, Fig. S4), which suggested that a smoother metabolic flux of methanol utilization was driven toward FFA production. To further drive formaldehyde assimilation, we expressed an additional copy of the endogenous dihydroxyacetone synthase gene DAS2 (strain PC124), which further decreased formaldehyde accumulation with higher biomass yield and methanol utilization (Fig. 3C–E). In particular, the accumulation of formaldehyde in strain PC124 was much lower than that in the non-FFA producing strain PC111B (Fig. 3E). Furthermore, DAS2 overexpression together with central metabolism rewiring (strain PC124) significantly reduced the reactive oxygen species (ROS) stress (Fig. 3F) and improved FFA production (Fig. 3B) compared with the parent strain PC113B, which suggested that enhancing formaldehyde assimilation was beneficial for detoxification and improving cellular robustness. These results showed that global rewiring of the central metabolism was essential for driving the metabolic flux toward product biosynthesis with relieving the toxicity caused by methanol oxidation.
Fed-Batch Fermentation for FFA Production.
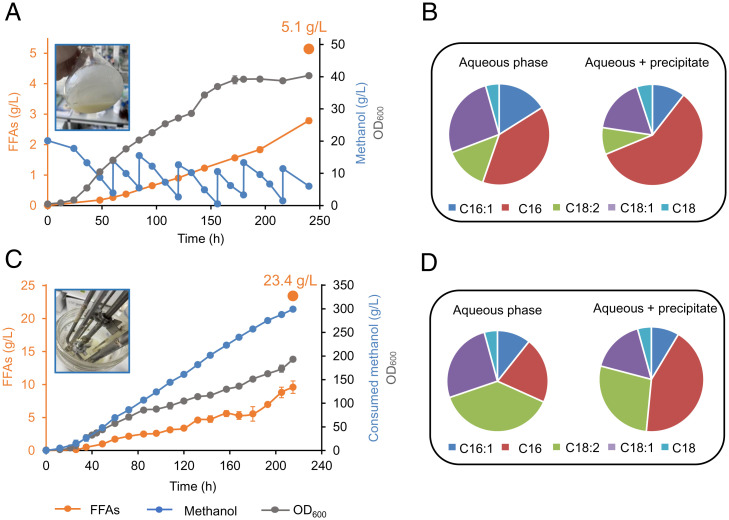
To test the potential of our engineered strain for FFA production from methanol, we carried out fed-batch fermentation in shake flasks and bioreactors by using the prototrophic strain PC124H (in situ restoration of the HIS4 gene in PC124). In shake-flask fed-batch fermentation, the engineered strain produced 5.1 g/L FFAs with 2.8 g/L in the aqueous phase (Fig. 4A and B). In the bioreactor process, the FFA titer reached 23.4 g/L (9.6 g/L in the aqueous phase) with a methanol consumption rate of 1.4 g/L/h (Fig. 4C and D). The FFA yield (0.078 g/g methanol, 24.4% of the theoretical yield) was comparable with the current maximal FFA yield from glucose in S. cerevisiae (33), indicating the outstanding potential of our engineered strain for industrial application. Interestingly, the percentage of unsaturated fatty acids (C18:1, C18:2, and C16:1) from bioreactor-cultured cells (54%) was much higher than that of shake flask-cultured cells (Fig. 4B and D), which might be attributed to the more abundant supply of oxygen in the bioreactor, since oxygen acts as an electron acceptor in the desaturation procedure (49).
Fig. 4.
Fed-batch fermentation for FFA production. Fed-batch fermentation of PC124H in shake flasks (A) or 1 L bioreactor (C). PC124H is a prototrophic strain with complementation of the HIS4 marker in PC124. Time courses of fatty acid titers (in orange), growth curves (in gray) and methanol consumption (in blue) are shown. Fatty acids production at the end of fermentation are also shown as filled orange circles. FFA profile of the strain PC124H in shake flasks (B) and bioreactors (D). Due to the precipitation, ethyl acetate was used for the dissolution of all fatty acids. Error bars correspond to the SD of the mean (n = 3 for shake flask fed-batch, corresponding to three biological replicates; n = 2 for bioreactor fed-batch, corresponding to two independent biological samples).
Metabolic Transforming for the Production of Fatty Alcohols.
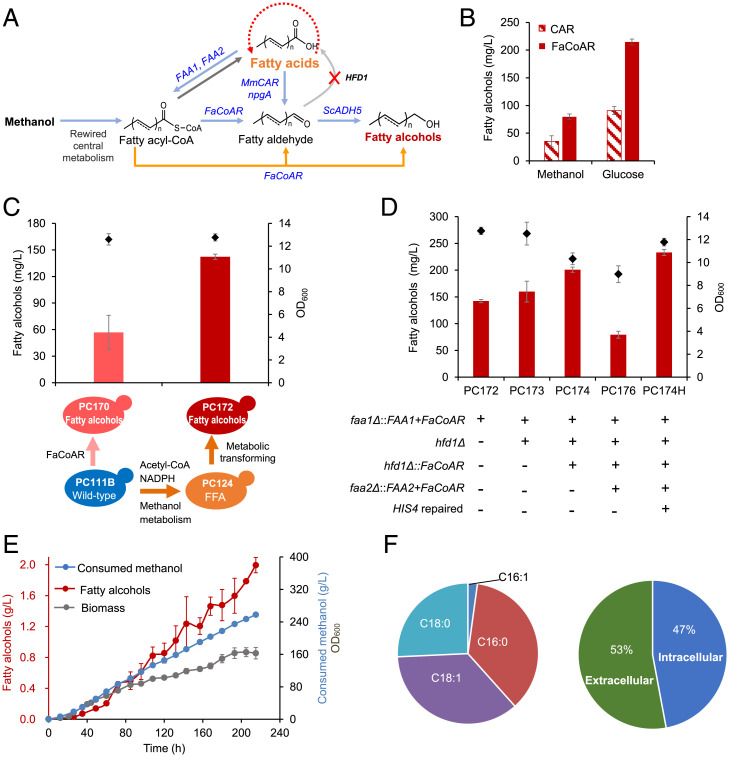
Fatty alcohols are widely used as detergents, cosmetic ingredients, pharmaceutical ingredients, and potential substitutes for fossil fuels. We thus explored the de novo synthesis of fatty alcohols from methanol in P. pastoris. Two main fatty alcohol biosynthetic pathways have been constructed in S. cerevisiae (36, 50): the FFA reduction pathway and the fatty acyl-CoA reduction pathway (Fig. 5A). Construction of the FFA reduction pathway in the FFA-overproducing background by expressing the codon-optimized carboxylic acid reductase gene MmCAR from Mycobacterium marinum, the cofactor 4′-phosphopantetheinyl transferase gene NpgA from Aspergillus nidulans and alcohol dehydrogenase ScADH5 from S. cerevisiae, produced 35.2 and 90.8 mg/L fatty alcohols from methanol and glucose, respectively. Expressing the fatty acyl-CoA reductase gene FaCoAR resulted in much higher fatty alcohol production than that obtained from FFA reduction pathway (Fig. 5B), which was consistent with our previous observation (37). The low titer of fatty alcohols from FFA reduction pathway might be attributed to FFAs secretion, which makes it difficult in access and utilization of the substrate FFAs.
Fig. 5.
Metabolic transforming for fatty alcohol production. (A) Schematic illustration of metabolic transforming of an FFAs-overproducing strain for overproduction of fatty alcohols. (B) Comparison of FFA reduction (CAR) and fatty acyl-CoA reduction (FaCoAR) pathways for fatty alcohol production from methanol. (C) Comparing fatty alcohol production between the wild-type and metabolic rewired strain. One copy of the FaCoAR gene was expressed in PC111B (generating strain PC170) and PC124 (generating strain PC172 by simultaneously reintroducing FAA1). (D) Optimization of fatty acyl-CoA reduction pathway. The engineered strains were cultivated in shake flasks containing 20 g/L methanol for 120 h at 220 rpm and 30 °C. (E) Fed-batch fermentation of the prototrophic strain PC174H. Time courses of fatty alcohol titers (deep red), growth curve (gray-purple) and consumption of methanol (blue) are shown. (F) The composition (Left) and distribution (Right) of fatty alcohols from fed-batch fermentation. Error bars correspond to the SD of the mean (n = 3, corresponding to three biological replicates).
We next tried to enhance the supply of fatty acyl-CoA to improve fatty alcohol biosynthesis by using the fatty acyl-CoA reduction pathway. It is easy to get the idea to use metabolic engineering strategies as described for FFA production such as enhancing the supply of acetyl-CoA, NADPH, and methanol assimilation (Fig. 3B), which, however, is time consuming when conducting multiple rounds of genetic manipulation. We thus transformed the FFA-overproducing strain toward fatty alcohol production by restoring FAA1 and FAA2 together with genomic integration of FaCoAR to their original genomic locus. Consistent with this, integration of one copy of FaCoAR with restoration of FAA1 in the FFA-overproducing strain PC124 resulted in strain PC172 and enabled the production of 142 mg/L fatty alcohols, which was 2.5 times higher than the titer of the wild-type–derived strain PC170 without engineered central metabolism (Fig. 5C). Reintroducing FAA1 diminished FFA accumulation (SI Appendix, Fig. S5), again suggesting that FAA1 plays a major role in activating FFAs in P. pastoris. We speculated that FaCoAR expression might be a bottleneck in the background of enhanced acetyl-CoA and NADPH levels, and our previous studies showed that deletion of the aldehyde dehydrogenase gene HFD1 was beneficial for fatty alcohol production by blocking the consumption of fatty aldehydes in S. cerevisiae (36, 50). We therefore knocked out HFD1 by integrating another copy of the FaCoAR gene at the HFD1 site (strain PC174), which improved fatty alcohol production by 41% (201 mg/L), while HFD1 deletion alone (strain PC173) resulted in only 10% improvement of fatty alcohol production (Fig. 5D). Further integration of another copy of the FaCoAR gene at the genomic site FAA2 (strain PC176) resulted in a significant decrease in fatty alcohol production and cell growth. This result suggested that two copies of FaCoAR were optimal for transforming cellular fatty acyl-CoA toward fatty alcohol production and that excessive FaCoAR expression placed a metabolic burden by draining cellular fatty acyl-CoA.
For fed-batch fermentation, a prototrophic strain PC174H was constructed by in situ restoring the auxotrophic marker gene HIS4 in PC174, which produced 233 mg/L fatty alcohols in shake-flask cultivation (Fig. 5D). Fed-batch cultivation of PC174H in bioreactor resulted in high-level production of fatty alcohols of 2.0 g/L with a methanol consumption rate of 1.2 g/L/h and cell density (OD600) of 163 (Fig. 5E). The main components of fatty alcohols (Fig. 5F) were hexadecanol (C16:0, 36%), octadecanal (C18:0, 26%), and 9-octadecenal (C18:1, 36%). It is worth mentioning that 53% of the fatty alcohols were secreted (Fig. 5F), while previous studies showed that fatty alcohols mainly accumulated in the cells when glucose was used as the carbon source (51–55).
Discussion
Methanol is a promising feedstock for biomanufacturing by linking clean energy storage and CO2 capture (1). Engineering efficient microbial cell factories is one of the key steps for economical methanol biotransformation. Here, we harnessed the industrially important yeast P. pastoris for high-level production of fatty acids and fatty alcohols from sole methanol as the carbon source in minimal media (Table 1) by rewiring the central metabolism and metabolic transforming. We rationally designed and engineered the cellular metabolism using a bottom-up approach: 1) engineering FFA accumulation by blocking FFA consumption; 2) enhancing the supply of precursor acetyl-CoA and cofactor NADPH; and 3) enhancing methanol assimilation. This engineering endeavor resulted in the comparable production of FFAs and fatty alcohols with those from glucose in S. cerevisiae (33) and much higher than several other chemicals biosynthesized from methanol (Table 1). Furthermore, the secretion of FFAs and fatty alcohols should be convenient for subsequent separation and purification.
Table 1.
Chemical production from methanol in P. pastoris
| Chemicals | Titer (g/L) | Yield (g/g) | Cultivation condition | Ref. |
|---|---|---|---|---|
| D-lactic acid | 3.48 | N.C.* | YPM†, Test tube | (21) |
| Dammarenediol-II | 4.8 × 10−3 | N.C.* | YPM, squalene | (67) |
| Citrinin | 6 × 10−4 | 1.2 × 10−4 | Methanol (MM‡) | (22) |
| 6-Methylsalicylic acid | 2.2 | 9 × 10−3 | Glycerol, methanol (MM) Fed-batch |
(23) |
| Chondroitin sulfate | 2.1 | N.C.* | Glycerol, methanol (MM) Fed-batch |
(24) |
| Lovastatin | 0.25 | 1 × 10−3 | Glycerol, methanol (MM) Fed-batch |
(25) |
| Monacolin J | 0.59 | 2 × 10−3 | ||
| Malic acid | 2.8 | N.C.* | Methanol, Yeast extract Fed-batch |
(26) |
| (+)-Nootkatone | 0.21 | 5 × 10−4 | Glucose, methanol (MM) Fed-batch |
(68) |
| Fatty alcohols | 2.0 | 8 × 10−3 | Methanol (MM), Fed-batch | This study |
| Fatty acids | 23.4 | 7.8 × 10−2 | Methanol (MM), Fed-batch | This study |
*N.C., not calculated due to the lack of methanol consumption data or containing complex media component such as yeast extract.
†YPM, a nutrient-rich medium containing yeast extract, peptone, and methanol.
‡MM, minimal medium.
P. pastoris is considered an ideal host for methanol-based biomanufacturing due to its natural ability of methanol utilization. Long-term evolution results in efficient methanol utilization with high-level expression of alcohol oxidase 1 (Aox1) (30). Thus, the promoter of AOX1 (PAOX1) can drive high-level production of proteins at the gram-per-liter level with methanol induction (56). However, economical production of small molecules requires much higher titers with relatively low costs, which can be realized by global rewiring of the central metabolism and balancing of the biosynthetic pathways. The established efficient genetic engineering tools (18, 56–58) can facilitate comprehensive metabolic engineering of P. pastoris as described in this study. For example, we used a combination of methanol-induced and constitutive promoters for pathway construction and optimization (SI Appendix, Table S2), which showed good performance in enhancing the production of FFAs and fatty alcohols with marginal effects on cellular fitness.
Currently, it is still challenging in engineering methanol biotransformation toward efficient chemical biosynthesis and the milligram-per-liter level of production is far from the requirement for industrial application (59). As the precursor of fatty acid biosynthesis, acetyl-CoA is a key central intermediate involving several cellular processes with tight regulation (60). Acetyl-CoA is mainly formed from pyruvate by mitochondrial pyruvate dehydrogenase (Pdh) and then enters the TCA cycle, or PDH bypass in the cytoplasm. During glucose metabolism, the TCA cycle is the main process for providing cellular energy, which indicates the relative abundance of acetyl-CoA flux. In methanol metabolism, the main energy source is methanol dissimilation rather than the TCA cycle (48), which suggests the relatively limited availability of acetyl-CoA and more challenges in enhancing the acetyl-CoA supply by using methanol as the carbon source. Indeed, most acetyl-CoA engineering strategies that were successful in improving FFA production from glucose in S. cerevisiae (33) failed to improve FFA production from methanol in P. pastoris. Only expressing MmACL and a PK-PTA pathway significantly improved FFAs biosynthesis, which suggested that the central metabolism was relatively rigid and required exploration of alternative engineering targets.
The cofactor NADPH is also essential for fatty acid biosynthesis, driving two reduction steps (reduction of β-ketoacyl-ACP and enoyl-ACP) during one round of extension. The NADP+-dependent isocitrate dehydrogenase (Idp2) slightly promoted FFA production by enhancing NADPH regeneration. This reaction might compete with citrate that is also used for acetyl-CoA supply, and this carbon-energy trade-off might offset the positive effect on FFA biosynthesis. Here, we observed that P. pastoris had much higher cellular NADPH/NADP+ levels than S. cerevisiae, which suggested P. pastoris is an ideal host for NADPH-dependent chemical biosynthesis.
As described previously, high level of AOX1 expression is essential for efficient methanol utilization by oxidizing methanol toward the active intermediate formaldehyde (30). However, the accumulation of formaldehyde causes toxicity stress (48), and we found that FFA biosynthesis further exacerbated formaldehyde accumulation. Overexpression of the dihydroxyacetone synthase gene DAS2, for accelerating formaldehyde assimilation, significantly improved FFA production and decreased formaldehyde accumulation to a much lower level than that of the wild-type strain. This observation showed that accelerating formaldehyde utilization is a feasible approach to drive methanol biotransformation.
The construction of microbial cell factories is time-consuming and laborious since it requires multiple rounds of genetic manipulation. Here, we developed a metabolic transforming strategy for rapidly transforming an FFA-overproducing strain toward fatty alcohol overproduction by taking advantage of the enhanced supply of acetyl-CoA and NADPH. Two rounds of genetic manipulation realized the construction of a fatty alcohol overproducer, while building from scratch required six rounds of genetic engineering. Thus, this concept can be easily applied to construct versatile cell factories for production of a variety of chemicals, particularly those with common precursors such as acetyl-CoA, as described here.
In summary, we successfully reprogrammed the metabolism of P. pastoris for high-level production of FFAs and fatty alcohols by using methanol as the sole carbon source. The strategies can be used for other methylotrophic yeasts and even other nonconventional microbes, with the rapid expansion of the genetic engineering toolbox (61, 62).
Materials and Methods
Plasmids and Strains.
E. coli DH5α was used for plasmid propagation, and P. pastoris GS115 was used as the host for yeast strains construction. All the plasmids and strains used in this study are shown in SI Appendix, Tables S1 and S2, respectively. PC111B derived from PC110 with a Cas9 expression cassette at the neutral site PNSI-3 (37). The engineering strategy and the flowcharts of yeast strain construction are shown in SI Appendix, Fig. S6.
Genetic Manipulation.
For construction of gRNA expression plasmids, 20 bp target sequences of gRNAs for genome targeting were designed by using the web toolbox (http://chopchop.cbu.uib.no). pPICZ-Cas9-gPOX1 for targeting POX1 was constructed as follows. The promoter part and gRNA part were obtained by PCR amplification with primers AOX1t-Kpn-ARS-R/gPOX1-F and HTX1-Cas9-F/gPOX1-R, respectively. The two parts were fused by overlap-extension PCR, and then ligated to the linearized pPICZ-Cas9-gRNA backbone (digested by Kpn I and Spe I) by Gibson cloning. Transformations were screened on LB (with 5 g/L NaCl) agar plate with 25 μg/L Zeocin. pCAI-gFAA1t and pCAI-gFAA2t were constructed as follows. Primers gFAA1t-F/gFAA1t-R and gFAA2t-F/gFAA2t-R were used to amplify the vector skeleton of pCAI-gRNA, and 20 bp target sequences of TFAA1 and TFAA2 were introduced. PCR products after digested by DpnI were directly transformed into DH5α competent cells. Transformations were screened on LB agar plate with 100 μg/L kanamycin. Other plasmids used in this study have been described in our previous studies (37).
For seamless deletion of POX1, 1 kb homologous arms flanking the ORF were amplified from genomic DNA by using the primers POX1-HRUP-F/POX1-HRUP-R1 and POX1-HRDN-F1/POX1-HRDN-R, which were used for constructing the seamless-knockout donor through overlap-extension PCR. The expression cassettes were also constructed by overlap-extension PCR. All the endogenous genes were amplified from the genomic DNA of P. pastoris GS115. The gene fragments of ScPYC1 and Sc'MDH3 (without mitochondrial localization signal) were amplified from S. cerevisiae S288C genomic DNA. OpRPE1 was amplified from Ogataea polymorpha NCYC495 genomic DNA. All the other heterologous genes were codon optimized and chemical synthesized at Genewiz. Custom DNA oligonucleotide primers were synthesized by Sangon Biotech, and the sequences are listed in SI Appendix, Table S3. Information of integration sites has been given in our previous study (37).
Strain Cultivation.
Shake-flask batch fermentations for production of fatty acids and fatty alcohols were carried out in 100 mL shaking flasks with a working volume of 20 mL minimal medium, which contained 2.5 g/L (NH4)2SO4, 14.4 g/L KH2PO4, 0.5 g/L MgSO4·7H2O, 20 g/L glucose or 20 g/L methanol, trace metal and vitamin solutions supplemented with 40 mg/L histidine if needed. The yeast cells were cultivated at 30 °C, 220 rpm (Zhichu Shaker ZQZY-CS8) for 5 d with initial OD600 of 0.3 when using methanol as the carbon source, or 4 d with initial OD600 of 0.1 when using glucose as the carbon source. The shake-flask fed-batch fermentations in 250 mL shake conical flasks with an initial working volume of 50 mL and an initial OD600 of 0.5. The temperature was set as 30 °C, and agitation was set as 220 rpm. The pH was adjusted to 5.6 by adding 4 M KOH every 24 h. During the fed-batch process, 632 μL of methanol was supplemented each time when the methanol concentration in the medium was lower than 10 g/L. The cells were fed with 0.5 mL 10× supplementary components containing 25 g/L (NH4)2SO4, 144 g/L KH2PO4, 5 g/L MgSO4·7H2O, 10× trace metal, and 10× vitamin solutions at the same time.
The fed-batch fermentation was conducted in 1.0 L DasGip Parallel Bioreactors System (DasGip) bioreactors with an initial working volume of 300 mL The initial batch stage was conducted with an inoculation OD600 of 1.0 in minimal medium containing 2.5 g/L (NH4)2SO4, 14.4 g/L KH2PO4, 0.5 g/L MgSO4·7H2O, 10 g/L methanol, trace metal, and vitamin solutions. The temperature, agitation, aeration, and pH were monitored and controlled using a DasGip Control 4.0 System. The temperature was kept at 30 °C, the initial agitation was set to 400 rpm and increased to maximally 1,200 rpm depending on the dissolved oxygen (DO) level. Aeration was initially set as 36 sL/h and increased to maximally 48 sL/h depending on the DO level. The dissolved oxygen level was maintained above 30%, the pH was kept at pH 5.6 by automatic addition of 4 M KOH and 2 M HCl. During the fed-batch cultivation, the cells were fed with 600 g/L methanol and 10× supplementary components [25 g/L (NH4)2SO4, 144 g/L KH2PO4, 5 g/L MgSO4·7H2O, 10× trace metal, and 10× vitamin solutions]. The methanol replenishment rate was 1.0 to 1.5 mL/h to keep the methanol concentration below 20 g/L. The feeding speed of supplementary component solution was set as half of feeding speed of methanol solution.
Extraction and Quantification of Products and Metabolites.
The extraction and quantification of FFAs were performed as our previous studies (33, 36) with slight modification. For aqueous FFA analysis, 100 μL cell cultures from shake flask were diluted 2-fold with water and those from bioreactor were diluted 10-fold. To analyze total fatty acids including those in precipitation, 5 mL and 100 mL ethyl acetate were added into the shake flask and bioreactor, respectively, and stirred for 30 min to dissolve fatty acid precipitation. The solution was stratified after standing for 10 min. The ethyl acetate phase in the supernatant samples was removed by rotary evaporation after 50-fold dilution. The followed extracted step was manipulated according to the reported methods (36). The extracted methyl esters were resuspended in hexane and analyzed by gas chromatography (Focus GC, Thermo Fisher Scientific) equipped with a Zebron ZB-5MS GUARDIAN capillary column (30 m * 0.25 mm * 0.25 μm; Phenomenex). The GC program was as follows: initial temperature of 40 °C, held for 2 min; ramp to 130 °C at a rate of 30 °C per minute, then raised to 280 °C at a rate of 10 °C per min and held for 3 min. Fatty alcohols were extracted and quantified as a previous report (36, 50) by using 2 mL cell culture from shake flasks and 0.2 mL cell culture from fed-batch fermentation.
The methanol concentration was determined by high-performance liquid chromatography analysis. One milliliter broth sample was centrifuged at 12,000 × g for 10 min and then filtered through a 0.2 μm syringe filter and analyzed with an Aminex HPX-87G column (Bio-Rad) on a 1260 infinity II HPLC (Agilent) equipped with a differential refractive index detector. The samples were eluted with 5 mM H2SO4 at a flow rate of 0.6 mL/min at 50 °C for 26 min. Formaldehyde was quantified as described in reference (63) with slight modification. Briefly, 0.2 mL samples were mixed with 0.2 mL 10% trichloroacetic acid, 0.1 mL 2,4-dinitrophenylhydrazine (DNPH, 0.1 g/L) and 0.5 mL acetonitrile via vigorous vortexing at 1,600 rpm for 1 min, then incubated in a 60 °C water bath for 30 min. After incubation, samples were centrifuged at 12,000 × g for 10 min and filtered through a 0.2 μm syringe filter. Formaldehyde-DNPH was then quantified by using an HPLC (LC-2030, Shimadzu) equipped with an UV detector and a C18 reversed-phase column, with 65% acetonitrile as the mobile phase.
Quantification of Cellular NADP(H).
The quantification method was modified from the previous references (64, 65). About 4 × 108 cells of each sample were collected and washed twice with cold PBS. Cells were centrifuged (4,000 × g at 4 °C for 5 min) and immediately resuspended in 0.3 mL of 0.2 M HCl (for NADP+) or 0.2 M NaOH (for NADPH). The cells were vigorously shaken with appropriate amount of glass beads to crush the cells at low temperature, and then incubated at 60 °C for 30 min. Next, 0.3 mL of 0.2 M NaOH (for NADP+) and 0.2 M HCl (for NADPH) were added for the neutralization reaction. Samples were centrifuged at 12,000 × g for 5 min.
The cycling assay (SI Appendix, Fig. S2A) was performed by using a reagent mixture consisting of equal volumes of 1.0 M Bicine buffer (pH 8.0), 30 mM glucose-6-phosphate, 40 mM EDTA (pH 8.0), 4.2 mM 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), twice the volume of 16.6 mM phenazine ethosulfate, and three volumes of water, previously incubated at 25 °C. The following volumes were added to 96-well plates: 10 μL neutralized extract, 180 μL reagent mixture. At last, the reaction was started by adding 10 μL of yeast glucose-6-phosphate dehydrogenase (200 U/mL, A003981, Sangon Biotech). The absorbance at 570 nm was recorded for 2 min at 25 °C by microplate reader (Tecan Spark 3). All fluorescence measurements were normalized to the protein concentration which was calculated by the BCA assay kit (Sangon Biotech).
ROS Measurement.
The cellular ROS level was estimated by using the oxidant-sensitive probe 2’,7’-dichlorofluorescin diacetate (DCFH-DA) as described previously (66). Samples were collected every 12 h during the first 3 d of cultivation and washed twice with distilled water. The cells were then resuspended in 0.5 mL 10 mM PBS (pH 7.0) containing 10 μM DCFH-DA, and incubated at 37 °C for 30 min. Fluorescence was measured with λEX 485 nm and λEM 525 nm using a SPARK multifunctional microplate platform (Tecan) with a gain value of 50.
Supplementary Material
Acknowledgments
This research was supported by National Natural Science Foundation of China (21922812 and 22161142008), National Key Research and Development Program of China (2021YFC2103500), DICP innovation grant (DICP I201920, and I202111) from Dalian Institute of Chemicals Physics, CAS. Funding for open access charge: National Natural Science Foundation of China.
Footnotes
Competing interest statement: P.C., and Y.J.Z. have one patent (202010516436.1) for protecting part of the work described herein. All other authors declare no competing financial interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201711119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
Change History
July 22, 2022: The editor’s affiliation has been updated to correct an error. Jens Nielsen’s affiliation is: BioInnovation Institute, DK2200 Copenhagen, Denmark.
References
- 1.Zhou Y. J., Kerkhoven E. J., Nielsen J., Barriers and opportunities in bio-based production of hydrocarbons. Nat. Energy 3, 925–935 (2018). [Google Scholar]
- 2.Lais A., Gondal M. A., Dastageer M. A., Al-Adel F. F., Experimental parameters affecting the photocatalytic reduction performance of CO2 to methanol: A review. Int. J. Energy Res. 42, 2031–2049 (2018). [Google Scholar]
- 3.Al-Rowaili F. N., Jamal A., Ba Shammakh M. S., Rana A., A review on recent advances for electrochemical reduction of carbon dioxide to methanol using metal–organic framework (MOF) and non-MOF catalysts: Challenges and future prospects. ACS Sustain. Chem. Eng. 6, 15895–15914 (2018). [Google Scholar]
- 4.Whitaker W. B., Sandoval N. R., Bennett R. K., Fast A. G., Papoutsakis E. T., Synthetic methylotrophy: Engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization. Curr. Opin. Biotechnol. 33, 165–175 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Fan L., Tuyishime P., Zheng P., Sun J., Synthetic methylotrophy: A practical solution for methanol-based biomanufacturing. Trends Biotechnol. 38, 650–666 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Fabarius J. T., Wegat V., Roth A., Sieber V., Synthetic methylotrophy in yeasts: Towards a circular bioeconomy. Trends Biotechnol. 39, 348–358 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Müller J. E. N., et al. , Engineering Escherichia coli for methanol conversion. Metab. Eng. 28, 190–201 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez J. E., Bennett R. K., Papoutsakis E. T., Antoniewicz M. R., Methanol assimilation in Escherichia coli is improved by co-utilization of threonine and deletion of leucine-responsive regulatory protein. Metab. Eng. 45, 67–74 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Chen C. T., et al. , Synthetic methanol auxotrophy of Escherichia coli for methanol-dependent growth and production. Metab. Eng. 49, 257–266 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Tuyishime P., et al. , Engineering Corynebacterium glutamicum for methanol-dependent growth and glutamate production. Metab. Eng. 49, 220–231 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Lu X., et al. , Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design. Nat. Commun. 10, 1378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou A., Lee S. H., Zhu F., Clomburg J. M., Gonzalez R., An orthogonal metabolic framework for one-carbon utilization. Nat. Metab. 3, 1385–1399 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Wang Y., et al. , Adaptive laboratory evolution enhances methanol tolerance and conversion in engineered Corynebacterium glutamicum. Commun. Biol. 3, 217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett R. K., et al. , Improving the methanol tolerance of an Escherichia coli methylotroph via adaptive laboratory evolution enhances synthetic methanol utilization. Front. Microbiol. 12, 638426 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espinosa M. I., et al. , Adaptive laboratory evolution of native methanol assimilation in Saccharomyces cerevisiae. Nat. Commun. 11, 5564 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen F. Y., Jung H. W., Tsuei C. Y., Liao J. C., Converting Escherichia coli to a synthetic methylotroph growing solely on methanol. Cell 182, 933–946.e14 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Vogl T., et al. , A toolbox of diverse promoters related to methanol utilization: Functionally verified parts for heterologous pathway expression in Pichia pastoris. ACS Synth. Biol. 5, 172–186 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Schwarzhans J. P., Luttermann T., Geier M., Kalinowski J., Friehs K., Towards systems metabolic engineering in Pichia pastoris. Biotechnol. Adv. 35, 681–710 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Zhu W., et al. , High level expression and purification of recombinant human serum albumin in Pichia pastoris. Protein Expr. Purif. 147, 61–68 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Zhang J., et al. , Expression of plectasin in Pichia pastoris and its characterization as a new antimicrobial peptide against Staphyloccocus and Streptococcus. Protein Expr. Purif. 78, 189–196 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Yamada R., Ogura K., Kimoto Y., Ogino H., Toward the construction of a technology platform for chemicals production from methanol: D-lactic acid production from methanol by an engineered yeast Pichia pastoris. World J. Microbiol. Biotechnol. 35, 37 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Xue Y., et al. , Methylotrophic yeast Pichia pastoris as a chassis organism for polyketide synthesis via the full citrinin biosynthetic pathway. J. Biotechnol. 242, 64–72 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Gao L. M., et al. , Engineered fungal polyketide biosynthesis in Pichia pastoris: A potential excellent host for polyketide production. Microb. Cell Fact. 12, 77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin X., et al. , Biosynthesis of non-animal chondroitin sulfate from methanol using genetically engineered Pichia pastoris. Green Chem. 23, 4365–4374 (2021). [Google Scholar]
- 25.Liu Y., et al. , Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol. Metab. Eng. 45, 189–199 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Guo F., et al. , Metabolic engineering of Pichia pastoris for malic acid production from methanol. Biotechnol. Bioeng. 118, 357–371 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Lin-Cereghino G. P., et al. , Mxr1p, a key regulator of the methanol utilization pathway and peroxisomal genes in Pichia pastoris. Mol. Cell. Biol. 26, 883–897 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prielhofer R., et al. , Pichia pastoris regulates its gene-specific response to different carbon sources at the transcriptional, rather than the translational, level. BMC Genomics 16, 167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moser J. W., et al. , Implications of evolutionary engineering for growth and recombinant protein production in methanol-based growth media in the yeast Pichia pastoris. Microb. Cell Fact. 16, 49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartner F. S., Glieder A., Regulation of methanol utilisation pathway genes in yeasts. Microb. Cell Fact. 5, 39 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou R., et al. , Comparative proteomics analysis of Pichia pastoris cultivating in glucose and methanol. Synth. Syst. Biotechnol. 7, 862–868 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fillet S., Adrio J. L., Microbial production of fatty alcohols. World J. Microbiol. Biotechnol. 32, 152 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Yu T., et al. , Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell 174, 1549–1558.e14 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Kim H. M., Chae T. U., Choi S. Y., Kim W. J., Lee S. Y., Engineering of an oleaginous bacterium for the production of fatty acids and fuels. Nat. Chem. Biol. 15, 721–729 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Ledesma-Amaro R., Dulermo R., Niehus X., Nicaud J. M., Combining metabolic engineering and process optimization to improve production and secretion of fatty acids. Metab. Eng. 38, 38–46 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Zhou Y. J., et al. , Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun. 7, 11709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai P., et al. , Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res. 49, 7791–7805 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weninger A., Hatzl A. M., Schmid C., Vogl T., Glieder A., Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J. Biotechnol. 235, 139–149 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Liu Q., et al. , CRISPR-Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb. Cell Fact. 18, 144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weninger A., et al. , Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers. J. Cell. Biochem. 119, 3183–3198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prielhofer R., et al. , GoldenPiCS: A Golden Gate-derived modular cloning system for applied synthetic biology in the yeast Pichia pastoris. BMC Syst. Biol. 11, 123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gassler T., Heistinger L., Mattanovich D., Gasser B., Prielhofer R., CRISPR/Cas9-mediated homology-directed genome editing in Pichia pastoris. Methods Mol. Biol. 1923, 211–225 (2019). [DOI] [PubMed] [Google Scholar]
- 43.De S., Mattanovich D., Ferrer P., Gasser B., Established tools and emerging trends for the production of recombinant proteins and metabolites in Pichia pastoris. Essays Biochem. 65, 293–307 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Chuang D. S., Liao J. C., Role of cyanobacterial phosphoketolase in energy regulation and glucose secretion under dark anaerobic and osmotic stress conditions. Metab. Eng. 65, 255–262 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Liu Q., et al. , Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 10, 4976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiao K., Wasylenko T. M., Zhou K., Xu P., Stephanopoulos G., Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat. Biotechnol. 35, 173–177 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Wen J., et al. , A synthetic malonyl-CoA metabolic oscillator in Komagataella phaffii. ACS Synth. Biol. 9, 1059–1068 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Rußmayer H., et al. , Systems-level organization of yeast methylotrophic lifestyle. BMC Biol. 13, 80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagao K., Murakami A., Umeda M., Structure and function of Δ9-fatty acid desaturase. Chem. Pharm. Bull. (Tokyo) 67, 327–332 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y. J., et al. , Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition. J. Am. Chem. Soc. 138, 15368–15377 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Cordova L. T., Butler J., Alper H. S., Direct production of fatty alcohols from glucose using engineered strains of Yarrowia lipolytica. Metab. Eng. Commun. 10, e00105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu Y., et al. , Engineering carboxylic acid reductase for selective synthesis of medium-chain fatty alcohols in yeast. Proc. Natl. Acad. Sci. U.S.A. 117, 22974–22983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu Y., Zhu Z., Nielsen J., Siewers V., Heterologous transporter expression for improved fatty alcohol secretion in yeast. Metab. Eng. 45, 51–58 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Tang X., Chen W. N., Enhanced production of fatty alcohols by engineering the TAGs synthesis pathway in Saccharomyces cerevisiae. Biotechnol. Bioeng. 112, 386–392 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Wang G., et al. , Exploring fatty alcohol-producing capability of Yarrowia lipolytica. Biotechnol. Biofuels 9, 107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Z., Zhang Z., Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: A review. Biotechnol. Adv. 36, 182–195 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Vogl T., Hartner F. S., Glieder A., New opportunities by synthetic biology for biopharmaceutical production in Pichia pastoris. Curr. Opin. Biotechnol. 24, 1094–1101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischer J. E., Glieder A., Current advances in engineering tools for Pichia pastoris. Curr. Opin. Biotechnol. 59, 175–181 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Peña D. A., Gasser B., Zanghellini J., Steiger M. G., Mattanovich D., Metabolic engineering of Pichia pastoris. Metab. Eng. 50, 2–15 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Nielsen J., Keasling J. D., Engineering cellular metabolism. Cell 164, 1185–1197 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Gao J., Gao N., Zhai X., Zhou Y. J., Recombination machinery engineering for precise genome editing in methylotrophic yeast Ogataea polymorpha. iScience 24, 102168 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu W., Gao J., Zhai X., Zhou Y. J., Screening neutral sites for metabolic engineering of methylotrophic yeast Ogataea polymorpha. Synth. Syst. Biotechnol. 6, 63–68 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu J., Miao J., Su T., Liu Y., He R., Formaldehyde induces hyperphosphorylation and polymerization of Tau protein both in vitro and in vivo. Biochim. Biophys. Acta 1830, 4102–4116 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Vemuri G. N., Eiteman M. A., McEwen J. E., Olsson L., Nielsen J., Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 104, 2402–2407 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Y. J., et al. , Determining the extremes of the cellular NAD(H) level by using an Escherichia coli NAD(+)-auxotrophic mutant. Appl. Environ. Microbiol. 77, 6133–6140 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang M. M., Zhao X. Q., Cheng C., Bai F. W., Improved growth and ethanol fermentation of Saccharomyces cerevisiae in the presence of acetic acid by overexpression of SET5 and PPR1. Biotechnol. J. 10, 1903–1911 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Liu X. B., et al. , Metabolic engineering of Pichia pastoris for the production of dammarenediol-II. J. Biotechnol. 216, 47–55 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Wriessnegger T., et al. , Production of the sesquiterpenoid (+)-nootkatone by metabolic engineering of Pichia pastoris. Metab. Eng. 24, 18–29 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.