Abstract
Sexual reproduction is commonly assumed to occur in the vast majority of diatoms due to the intimate association of this process with cell size control. Surprisingly, however, little is known about the impact of sexual events on diatom population dynamics. The Sig1 gene is strongly upregulated during sexual reproduction in the centric diatom Thalassiosira weissflogii and has been hypothesized to encode a protein involved in gamete recognition. In the present study, degenerate PCR primers were designed and used to amplify a portion of Sig1 from three closely related species in the cosmopolitan genus Thalassiosira, Thalassiosira oceanica, Thalassiosira guillardii, and Thalassiosira pseudonana. Identification of Sig1 in these three additional species facilitated development of this gene as a molecular marker for diatom sexual events. Examination of the new sequences indicated that multiple copies of Sig1 are probably present in the genome. Moreover, compared to the housekeeping gene β-tubulin, the Sig1 genes of isolates of T. weissflogii collected from different regions of the Atlantic and Pacific oceans displayed high levels of divergence. The Sig1 genes of the four closely related Thalassiosira species also displayed high levels of sequence divergence compared to the levels observed with a second gene, Fcp, probably explaining why Sig1 could not be amplified from more distantly related species. The high levels of sequence divergence both within and between species suggest that Sig1 is rapidly evolving in a manner reminiscent of the manner observed in other genes that encode gamete recognition proteins. A simple model is presented for Sig1 evolution and the implications of such a rapidly evolving sexual reproduction gene for diatom speciation and population dynamics.
Diatoms are the most species-rich group of phytoplankton known. Conservative estimates suggest that tens of thousands of different species of diatoms are distributed throughout marine and freshwater ecosystems (27). Explosive diversification of diatom species has therefore occurred over the last 200 million years (27, 30). Typically, tens to perhaps hundreds of species of diatoms comprise the phytoplankton community of any given body of water. Under most circumstances, diatoms are likely to be essentially indifferent to whether neighboring cells are the same or different species. During sexual events, however, the ability of a given species to distinguish between itself and all other species becomes critical.
The onset of sexual reproduction in diatoms is commonly coupled to control of cell size. Due to physical and developmental constraints associated with generation of the silica frustule, each mitotic division results in the formation of two daughter cells of different sizes, one that is the same size as the parent and one that is slightly smaller. Thus, over successive generations the mean cell size of a diatom population decreases (26, 37). Interestingly, only relatively small cells within a population respond to environmental signals and undergo sexual reproduction, an event that ultimately restores cell size (10). Consequently, multiple species of diatoms may undergo sexual reproduction simultaneously in a single body of water (reviewed in reference 11). In centric diatoms, flagellated sperm formed during sexual events must distinguish not only between vegetative cells and egg cells still encased within their frustule but also between vegetative and egg cells of different species.
The molecular basis of species-specific gamete recognition during external fertilization has been examined in marine invertebrates such as abalone (42, 45), sea urchin (31), and teguline gastropods (17). A common feature of sexual recognition proteins appears to be rapid diversification of amino acid sequences in closely related species (33, 43). In many instances, strong selection for sequence variation, known as positive Darwinian selection, appears to occur. The unicellular freshwater algal genus Chlamydomonas is the only phytoplankton genus in which evolution of a sex-related protein has been examined (12). The C. reinhardtii protein, MID, is required for gamete differentiation, and the sequences from two closely related species display dramatic differences, although positive selection does not appear to underlie the evolution of this protein. Ultimately, rapid diversification of sexual recognition proteins is expected to lead to speciation, although it remains unclear what forces underlie the evolution of new species (12, 45).
We recently identified in the centric diatom Thalassiosira weissflogii a gene family, composed of Sig1, -2, and -3, whose transcription is highly upregulated during the onset of sexual reproduction. The proteins encoded by these genes appear to be part of the extracellular matrix and have been hypothesized to play a role in mediating sperm-egg recognition (1). Our initial goal in the present study was to determine whether Sig homologues could be identified in other species of centric diatoms and whether upregulation of these genes could serve as a molecular marker for the occurrence of sexual reproduction in field populations. In this work, we found that the gene on which we focused, Sig1, is a multicopy gene that appears to be undergoing rapid divergence both within and between species, a feature that has come to be expected for genes encoding proteins involved in sexual recognition.
MATERIALS AND METHODS
Culture conditions.
Seven T. weissflogii isolates (CCMP1336 clone Actin from Long Island Sound, New York; CCMP1049 clone 4C from Long Island Sound, New York; CCMP1050 clone WTFLU from Del Mar Slough, California; CCMP1051 clone THALA7 from King Kalakaua's Fishpond, Hawaii; CCMP1052 clone TTW1 from Segerrak Sea, Norway; CCMP1053 clone SA from Portugal; CCMP1587 clone JA92I from Jakarta Harbor, Indonesia) and isolates of four additional Thalassiosira species, Thalassiosira guillardii (CCMP988 clone 7-15 from the North Atlantic Ocean), Thalassiosira oceanica (CCMP1005 clone 13-1 from the Sargasso Sea), Thalassiosira rotula (CCMP1647 from the Bay of Naples, Italy), and Thalassiosira pseudonana (CCMP1335 clone3H from Moriches Bay, New York) were purchased from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton (CCMP), Bigelow Laboratory for Ocean Sciences. A Thalassiosira antarctica isolate was obtained from R. J. Olson of Woods Hole Oceanographic Institution (Table 1). All cultures were maintained in f/2-enriched seawater (14). All species except T. guillardii and T. antarctica were maintained at 20°C with continuous illumination at 120 μmol of photons · m−2 · s−1. The T. guillardii culture was maintained at 14°C with a cycle consisting of 16 h of light (66 μmol of photons · m−2 · s−1) and 8 h of darkness. The T. antarctica culture was maintained at 2°C with constant illumination at 20 μmol of photons · m−2 · s−1.
TABLE 1.
Isolates and species used in this study
| Species | Clone | CCMP no. | Isolation site | Year isolated |
|---|---|---|---|---|
| T. weissflogii | Actin | 1336 | Long Island Sound, New York | 1958 |
| 4C | 1049 | Long Island Sound, New York | 1968 | |
| TTW1 | 1052 | Segerrak Sea, Norway | 1978 | |
| SA | 1053 | Portugal, North Atlantic Ocean | 1973 | |
| WTFLU | 1050 | Del Mar Slough, California | 1959 | |
| THALA7 | 1051 | King Kalakaua's Fishpond, Hawaii | 1985 | |
| JA921 | 1587 | Jakarta Harbor, Indonesia | 1992 | |
| T. guillardii | 7-15 | 988 | North Atlantic Ocean | 1958 |
| T. oceanica | 13-1 | 1005 | Sargasso Sea | 1958 |
| T. rotula | 1647 | Bay of Naples, Italy | 1993 | |
| T. pseudonana | 3H | 1335 | Moriches Bay, New York | 1958 |
| T. antarctica | NAa | NA | NA | NA |
NA, not available.
Clonal isolates of T. weissflogii clone Actin were obtained by plating cells on f/2-enriched seawater solidified with 1.5% agar (Difco). Individual colonies were then transferred to and maintained in liquid f/2 media. The T. weissflogii clone Actin isolates were induced to undergo sexual reproduction by interrupting exponential growth in continuous light with 12 h of darkness (1, 3).
Nucleic acid isolation and generation of cDNAs.
Genomic DNA was isolated with a DNeasy Plant Mini Kit (Qiagen). Total RNA was isolated with an RNeasy Plant Mini Kit (Qiagen). First-strand cDNAs were generated from 500 ng of total RNA with a 1st Strand cDNA synthesis kit (Clontech).
Detection of DNA polymorphisms.
DNA polymorphisms were examined in two gene fragments, β-tubulin (2) and Sig1 (1). An internal fragment of the β-tubulin gene spanning a single intron was amplified by using two gene-specific PCR primers, 5′-TTCGACCGGATAACTTTG-3′ (forward) and 5′-CGACTAGTCAAAGGAGC-3′ (reverse). PCR amplifications in reaction mixtures (final volume, 20 μl) containing each deoxynucleoside triphosphate (dNTP) at a concentration of 0.1 mM, 2.5 mM MgCl2, 10 pmol of each primer, and 0.75 U of Taq DNA polymerase (Display) began with a 2-min denaturation step at 94°C, which was followed by 35 cycles of 94°C for 10 s, 50°C for 30 s, and 72°C for 90 s and then by a final extension at 72°C for 10 min. The genomic fragment was eluted from a low-melting-point agarose gel (40), cloned into pCR2.1-TOPO, and transformed into TOP10 Escherichia coli cells with a TOPO TA cloning kit (Invitrogen). Positive transformants containing inserts of the correct size were identified by PCR using the vector-specific M13F and M13R primers. Plasmid DNAs were isolated from the transformants with a Mini Prep kit (Qiagen), were sequenced with a DYEnamic ET dye terminator kit (Amersham Pharmacia Biotech Inc.), and were analyzed with a MegaBACE 1000 (Molecular Dynamics).
Sig1 DNA fragments were obtained in the following manner. Blockmaker (http://blocks.fhcrc.org/blockmkr/make_blocks.html) was used to identify conserved amino acid domains within the SIG1, SIG2, and SIG3 proteins (1). Based on the amino acid sequences of the conserved domains, degenerate PCR primers were designed by using the CODEHOP algorithm (36) to amplify a fragment from the Sig1 and Sig3 genes. The forward Sig primer was 5′-AACGCTGCTCTGGCCACGGNWCTTGYGG-3′, and the reverse primer was 5′-GGGCCGGTATATCCAGGATCRCAYTTRCAWCC-3′. PCR amplifications in reaction mixtures (final volume, 20 μl) containing each dNTP at a concentration of 0.1 mM, 2.5 mM MgCl2, 10 pmol of each primer, and 0.75 U of Taq DNA polymerase (Display) began with a 2-min denaturation step at 94°C, which was followed by 30 cycles of 94°C for 10 s, 62°C for 30 s, and 72°C for 90 s and then by a final extension at 72°C for 10 min. Genomic or cDNA fragments corresponding to Sig1 were cloned, and the resulting transformants were screened as described above for inserts of the correct size.
Positive Sig1 transformants were PCR amplified a second time with the degenerate Sig-specific primers as described above, but this time the forward primer was labeled with FAM (Operon). The resulting fluorescent PCR products were analyzed by using single-strand conformational polymorphism (SSCP) (23). PCR products were diluted 1:1 with deionized formamide, denatured for 5 min at 95°C, and immediately placed on ice before they were loaded onto a 10% acrylamide (ratio of acrylamide to bisacrylamide, 99:1) gel (20 by 20 cm). The gel was electrophoresed in a water-cooled apparatus (Owl Scientific Inc.) at 6 V for 17 h. Fluorescent products were detected with a FluorImager 595 (Molecular Dynamics).
Clones whose PCR products displayed unique SSCP patterns were chosen for DNA sequencing. Plasmid DNAs were isolated from the original transformants as described above. Either the DNAs were sequenced with a Thermosequenase II dye terminator cycle sequencing kit (Amersham Pharmacia Biotech Inc.) and analyzed with a 373A DNA sequencer (Applied Biosystems), or they were sequenced with a DYEnamic ET dye terminator kit (Amersham Pharmacia Biotech Inc.) and analyzed with a MegaBACE 1000 (Molecular Dynamics).
All sequence data were compiled and analyzed by using a combination of the Wisconsin Package (version 10.0) of the Genetics Computer Group, Madison, Wis.; Sequencher 4.0.5 (Gene Codes); and SeqApp (http://ftp.bio.indiana.edu/soft/molbio/seqapp).
Phylogenetic analysis.
The 18S rRNA genes from each species were isolated by using the universal 18sA and 18sB primers lacking the 5′ restriction sites (28). PCR amplifications in reaction mixtures (final volume, 10 μl) containing each dNTP at a concentration of 0.1 mM, 3.125 mM MgCl2, 10 pmol of each primer, and 0.75 U of Taq DNA polymerase (Promega) began with a 2-min denaturation step at 94°C, which was followed by 35 cycles of 94°C for 10 s, 50°C for 30 s, and 72°C for 60 s and then by a final extension at 72°C for 10 min. PCR products of the correct size were cloned into pCR2.1-TOPO as described above and were sequenced by using a combination of vector-specific and gene-specific primers. The 18S rRNA gene-specific forward primers were 5′-CTGCCCTATCAGCTTTGG-3′ (primer C) and 5′-TTGACTCAACACGGGAAAAC-3′ (primer E); the 18S rRNA gene-specific reverse primers were 5′-CGGCCATGCACCACC-3′ (primer D) and 5′-ATCCAAAGCTGATAGGGCAG-3′ (primer F). Phylogenetic analyses were performed by using the default settings of the PAUP program (Smithsonian Institution, 1997) accessed through the Genetics Computer Group. Consensus (50% majority rule) trees were constructed by using neighbor-joining distances with 1,000 bootstrap replicates and were viewed by using TREEVIEW (32). Complete coding and intron sequences of Sig1 and β-tubulin gene fragments were used for phylogenetic analyses. For phylogenetic analysis of the 18S rRNA gene, 1,635 nucleotides of an informative sequence were used.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the GenBank database under the following accession numbers: 18S rRNA genes, AF374477 to AF374482; β-tubulin genes, AF374483 to AF374489; Sig1 genomic DNAs, AF374490 to AF374539; and Sig1 cDNAs, AF374540 to AF374552.
RESULTS
Multiple copies of Sig1 are transcribed shortly after T. weissflogii cells undergo sexual reproduction.
Identification of five highly conserved amino acid domains in SIG1, SIG2, and SIG3 proteins of T. weissflogii (1) suggested that these regions could represent functional domains that might also be conserved in SIG homologues in different species of Thalassiosira. The CODEHOP algorithm (36) was used to design degenerate PCR primers to amplify DNA encoding the region spanning two of the domains, domains I and IV (1), which display the greatest amino acid identity for SIG1 and SIG3. Although the SIG2 protein also displayed significant amino acid identity with the other SIG proteins in these two domains, the level of degeneracy needed to recognize Sig2 as well was quite high, and no attempts were made to amplify this gene.
The utility of the newly designed degenerate Sig primers was tested by using genomic DNA isolated from a culture that had originated from a single cell of T. weissflogii clone Actin, the clone in which the Sig genes were originally identified (1). As expected, two fragments, which were 706 and 483 bp long, were amplified; the sizes corresponded to the sizes predicted for genomic fragments (each with a single intron) of Sig1 and Sig3, respectively. To ensure that the degenerate Sig primers were Sig specific, the 706-bp fragment was cloned, and DNA inserts from three transformants were sequenced. Pairwise comparisons of the new sequences and the previously published Sig1 sequence indicated that the four sequences differed from one another by anywhere from 1 to 5 bp (data not shown). Assuming that the diploid clone from which the DNA was isolated was a heterozygote, the presence of four distinct Sig1 sequences implied that at least two Sig1 loci might be present in an individual.
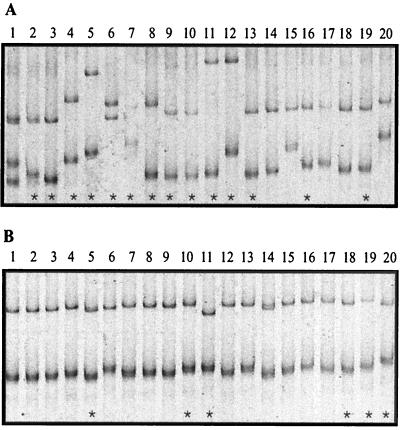
SSCP was used to provide an estimate of the number of unique copies of Sig1 in an individual. Analysis of SSCP patterns can be used to detect, relatively quickly, sequence differences between DNA fragments of the same length due to sequence-dependent rates of migration of denatured, single-stranded DNA fragments in a nondenaturing gel (23). When SSCP was used with the Sig1 clones, at least 19 variants that migrated differently were detected (Fig. 1A). Assuming that the clone from which the genomic DNA was isolated was heterozygous at every Sig1 locus, the SSCP results suggested that Sig1 was composed of at least 10 different loci.
FIG. 1.
Representative SSCP patterns of genomic (A) and cDNA (B) versions of Sig1, each amplified from 20 E. coli transformants. The asterisks indicate a subset of the original clones that were sequenced to completion.
This potentially high number of Sig1 loci in a single individual was unexpected. To confirm the predicted sequence diversity, the original plasmids corresponding to 15 of the 19 Sig1 inserts that migrated differently were sequenced. SSCP analysis proved to be an extremely sensitive predictor of DNA sequence variation as each clone with a different DNA sequence migrated differently in the SSCP gel (although two clones differed only in the primer sequence), indicating that the high level of SSCP variation was not simply an artifact of a second round of PCR amplification. One Sig1 genomic fragment displayed a 1-bp insertion, and two fragments displayed 1-bp deletions in the coding sequence (data not shown). These copies were not analyzed further since they were assumed to represent nonfunctional pseudogenes.
The DNA sequences of the remaining 11 unique copies differed at 30 positions scattered throughout the 645-bp fragment (not including the two primer sites). Surprisingly, only four variable sites were located within the 87-bp intron (Table 2). At six positions an identical sequence change was found in two copies, and at one position an identical change was present in three copies. Pairwise comparisons of all sequences indicated that the greatest number of substitutions for any two copies was seven, corresponding to a maximum sequence difference of about 1.1%. Relative to the reference sequence, two fragments displayed seven substitutions, five displayed four substitutions, two displayed two substitutions, and only one displayed one substitution (Table 2). Assuming that the potential Taq replication error rate is 0.07% per base (24), then about one substitution in 1,400 bp is expected to occur simply due to a PCR artifact (background). The substitution rate observed with Sig1 sequences was about sixfold greater than this, which indicates that multiple copies of Sig1 are present in an individual. Interestingly, there was no obvious grouping of the different sequences on the basis of shared similarities, as has been seen with other multicopy genes (4).
TABLE 2.
Variable nucleotides in genomic or cDNA copies of Sig1 amplified from T. weissflogii clone Actin
| DNA | Clone | Nucleotides at the following positionsa:
|
||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coding
|
Intron
|
Coding
|
||||||||||||||||||||||||||||||||||||||||||||||
| 1 | 25 | 26 | 27 | 29 | 69 | 84 | 122 | 134 | 149 | 175 | 202 | 203 | 212 | 239 | 251 | 264 | 272 | 281 | 284 | 288 | 319 | 369 | 384 | 388 | 441 | 443 | 476 | 495 | 497 | 501 | 502 | 516 | 523 | 532 | 561 | 588 | 590 | 597 | 601 | 607 | 614 | 627 | 630 | 638 | 641 | 642 | ||
| Nuclear | Reference | A | T | A | G | A | A | T | T | A | C | T | T | A | G | T | A | C | T | A | T | A | T | G | T | T | C | A | A | T | A | C | A | A | T | A | A | T | G | T | T | C | T | A | A | A | A | T |
| 1 | C | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | A | . | . | . | . | . | G | . | . | C | . | . | . | . | . | A | . | . | . | . | . | . | |
| 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | G | . | . | T | C | |
| 3 | . | . | . | . | . | G | . | . | . | . | A | . | C | . | . | C | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | C | G | . | . | . | . | . | . | . | . | . | . | ||||
| 4 | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | ||
| 5 | . | . | . | . | . | . | C | . | G | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | |
| 6 | . | . | . | A | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 7 | . | . | G | . | . | . | . | . | . | . | . | C | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 8 | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | ||
| 9 | . | . | . | . | . | . | C | . | . | . | . | . | . | A | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | |
| 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| cDNA | 1 | C | . | . | . | . | — | — | — | — | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2 | . | . | . | . | . | — | — | — | — | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | |
| 3 | . | . | . | . | . | — | — | — | — | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | ||||
| 4 | . | . | . | . | . | — | — | — | — | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 5 | . | . | . | . | . | — | — | — | — | . | . | . | G | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 6 | . | . | . | . | . | — | — | — | — | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | |
| 7 | C | . | . | . | . | — | — | — | — | . | . | . | . | . | . | . | . | . | G | . | . | G | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | |
| 8 | . | . | . | . | . | — | — | — | — | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | |
| 9 | . | . | . | . | . | — | — | — | — | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 10 | C | . | . | . | . | — | — | — | — | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 11 | . | . | . | . | . | — | — | — | — | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | |
| 12 | . | . | . | . | . | — | — | — | — | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 13 | . | . | . | . | . | — | — | — | — | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
The positions are numbered relative to the first nucleotide immediately following the forward primer. A dot indicates that the nucleotide is identical to the nucleotide in the reference clone; dashes indicate gaps.
When the intron sequence was excluded from the 11 unique Sig1 genomic sequences, 558-bp open reading frames predicted to encode 186 amino acids were identified, suggesting that the different gene copies might be transcribed. To determine whether multiple copies of the Sig1 gene were in fact transcribed, the T. weissflogii clone used for genomic DNA analysis was induced to undergo sexual reproduction. RNA was isolated 5 h into sexual reproduction and reverse transcribed into cDNAs. When first-strand cDNAs were used as templates for PCR with the degenerate Sig primers, two bands, at 558 and 400 bp corresponding to Sig1 and Sig3 mRNAs, respectively, were obtained.
The cDNA fragment that was the size of Sig1 was cloned, and 34 different transformants were analyzed with SSCP. The differences in the migration rates of cDNA fragments were not as great as the differences in the migration rates of genomic copies, which made it more difficult to unambiguously identify cDNA variants using SSCP alone (Fig. 1B). Based on apparent differences in SSCP migration patterns, plasmids corresponding to 15 cDNA fragments were chosen for sequencing. Thirteen of the 15 cDNA sequences were unique, and the same sequence change was observed at three positions in both genomic and cDNA clones (Table 2). The same sequence change was found at a single nucleotide position in two copies of the cDNA fragment and at a single position in three copies of the fragment. One cDNA sequence was identical to the genomic coding sequence. There were fewer substitutions per cDNA copy than per genomic copy: four cDNA sequences displayed a single substitution, seven displayed two substitutions, and only one displayed five substitutions. Regardless, the substitution rate was about four times greater than the background rate, suggesting that multiple copies of Sig1 were also transcribed. As with the genomic copies, there was no obvious grouping of the different cDNA sequences.
A total of 56 nucleotide substitutions were present in the coding sequence of the cDNA and genomic clones. Nineteen of these were first-position substitutions, 19 were second-position substitutions, and 18 were third-position substitutions. Only 15 third-position substitutions were silent and resulted in no change in the predicted amino acid sequence. All other nucleotide substitutions resulted in amino acid changes. Thus, there appeared to be no bias towards silent substitutions, as would be expected under conditions of stabilizing selection in which amino acid divergence is selected against (19).
The 15 cDNAs were predicted to encode 11 unique proteins with different amino acid sequences. When genomic sequences with the intron excluded were included in this analysis, an additional nine unique amino acid sequences were observed; thus, a total of 20 potentially unique proteins were encoded by Sig1 loci (Table 3). Thirty-seven amino acid changes were scattered throughout the predicted protein sequences. Amino acid substitutions can be categorized as conservative, moderate, radical, or very radical depending on the predicted change in composition, polarity, and molecular volume (13). Eighteen of the amino acid changes were conservative, 12 were moderate, and 7 were radical (Table 3). Different protein variants were expected to display slightly different characteristics, suggesting that individuals might express multiple SIG1 proteins. Furthermore, this potential variation in SIG1 within an individual suggested that different individuals likely possessed different combinations of Sig1 copies; this was particularly true of individuals isolated from different locations.
TABLE 3.
Variable amino acids in predicted open reading frames of either genomic or cDNA copies of Sig1 amplified from T. weissflogii clone Actin
| Template | Clone | Amino acids at the following positionsa:
|
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 | 10 | 22 | 43 | 52 | 60 | 63 | 66 | 67 | 68 | 95 | 100 | 119 | 131 | 137 | 138 | 139 | 144 | 148 | 159 | 168 | 169 | 171 | 177 | 181 | 182 | 185 | 186 | ||
| Genomic | Reference | R | N | P | D | S | T | F | M | S | D | G | L | S | N | F | N | A | H | E | H | L | A | L | Y | D | K | T | M |
| 1 | . | . | . | . | . | . | L | . | . | . | . | . | Y | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | S | |
| 3 | . | . | . | . | P | . | L | . | . | . | . | . | Y | . | . | . | . | . | D | R | . | . | . | . | . | . | . | . | |
| 4 | . | . | . | . | . | . | . | . | . | . | V | P | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 5 | . | . | . | . | P | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | R | . | . | |
| 6 | K | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 7 | G | . | . | N | . | . | . | . | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . | . | . | . | |
| 8 | . | D | . | . | . | . | . | . | . | . | . | . | . | . | Y | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 9 | . | . | . | . | P | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | R | . | . | |
| 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | R | . | . | . | . | . | . | . | . | . | . | |
| cDNA | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | P | . | . | . | . | . | . | . | |
| 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | P | . | |
| 4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . | . | . | . | . | . | . | . | . | . | . | . | |
| 5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 6 | . | . | . | . | . | . | . | . | . | . | . | . | Y | . | . | . | . | . | D | . | . | . | . | . | . | . | . | . | |
| 7 | . | . | . | . | . | . | . | V | . | . | . | . | . | H | . | . | . | . | . | . | . | . | S | . | . | . | . | . | |
| 8 | . | . | S | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | H | . | . | . | . | |
| 9 | . | . | . | . | . | I | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 10 | . | . | . | . | . | . | . | . | P | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | |
| 12 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 13 | . | . | . | . | . | . | . | . | . | G | . | . | Y | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
The positions are numbered relative to the first predicted amino acid of the open reading frame. Roman type indicates conservative changes, italic type indicates moderate changes, and underlining indicates radical changes. A dot indicates that the amino acid is identical to the amino acid in the reference clone.
Sig1 displays relatively high levels of sequence divergence in T. weissflogii isolates collected from different ocean regions.
Intraspecific DNA sequence divergence is expected to mirror the extent to which populations are geographically isolated from one another (33). Intraspecific sequence divergence was compared for two gene fragments, β-tubulin, which is required for cellular housekeeping (8), and Sig1. Genomic fragments corresponding to the two genes were amplified from a number of T. weissflogii isolates that had been collected from different oceanic regions over the course of 34 years (Table 1).
When β-tubulin-specific primers (2) were used with genomic DNAs isolated from the seven T. weissflogii isolates, a single 671-bp fragment was obtained, and it was subsequently cloned. The gene encoding β-tubulin appears to be a single-copy gene in T. weissflogii (2), and only one clone was sequenced for each isolate. The DNA sequences of the four Atlantic isolates were identical except for a single transition in the intron of the Norwegian isolate. The DNA sequences of the three Pacific isolates showed slightly more variation; two nucleotide positions in the intron and one position in the coding sequence were variable (Table 4). In contrast, the DNA sequences of the β-tubulin fragment from the four Atlantic isolates differed from the DNA sequences of the β-tubulin fragment from the three Pacific isolates at 32 positions; 17 of the variable positions were localized to the intron, and 15 were localized to the coding sequence (Table 4). The β-tubulin fragment from T. weissflogii clone Actin, the isolate for which the primers were originally developed (2), has an 87-bp intron and 582 bp of coding sequence. This means that 19.5% of the sites in the intron and 2.6% of the sites in the coding sequence varied for β-tubulin sequences from different isolates.
TABLE 4.
Variable nucleotides in the β-tubulin gene fragment amplified from seven isolates of T. weissflogii
| Isolatea | Nucleotides at the following positionsb:
|
|||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intron
|
Coding
|
|||||||||||||||||||||||||||||||
| 42 | 46 | 55 | 56 | 57 | 68 | 76 | 77 | 79 | 81 | 85 | 86 | 87 | 90 | 91 | 109 | 127 | 174 | 180 | 204 | 231 | 333 | 408 | 411 | 417 | 444 | 483 | 510 | 579 | 585 | 621 | 627 | |
| Long Island Sound clone Actin | A | C | — | — | G | G | C | C | G | A | T | C | G | A | T | C | A | C | A | C | T | A | T | C | G | T | T | C | G | C | A | G |
| Long Island Sound clone 4C | . | . | — | — | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Norway | . | . | — | — | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Portugal | . | . | — | — | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| South Pacific | C | — | T | A | A | C | G | G | T | T | . | G | A | G | . | T | G | A | T | T | C | T | C | T | A | C | C | T | A | T | T | C |
| California | C | — | T | A | A | C | G | G | T | T | . | G | A | G | . | T | G | A | T | T | C | T | C | T | A | C | C | T | A | T | T | C |
| Hawaii | C | — | T | A | . | C | G | G | T | T | C | G | A | G | . | T | G | . | T | T | C | T | C | T | A | C | C | T | A | T | T | C |
Long Island Sound clones Actin and 4C and the Norway and Portugal isolates were obtained from the Atlantic Ocean, and the South Pacific, California, and Hawaii isolates were obtained from the Pacific Ocean basin.
The positions are numbered relative to the first nucleotide immediately following the forward primer. A dot indicates that the nucleotide is identical to the nucleotide in the Long Island Sound clone Actin reference clone; dashes indicate gaps.
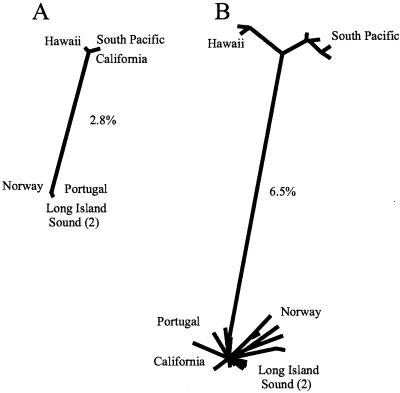
Evolutionary distances between β-tubulin sequences were calculated by using the Jukes-Cantor model of DNA sequence divergence, in which substitutions among nucleotides are assumed to occur at the same rate at the different nucleotide positions and each nucleotide can change with equal probability to the other three nucleotides (for a discussion, see reference 15). Based on this model, the β-tubulin DNA coding sequences from the Atlantic and Pacific isolates displayed a maximum divergence of 2.8%. A distance-based analysis grouped the three Pacific strains together and the four Atlantic strains together (Fig. 2A), which suggested that the two groups of isolates had been physically separated for a long time. All nucleotide changes in the coding sequences of different isolates were silent, which resulted in identical amino acid sequences for all β-tubulin molecules regardless of the ocean from which the organisms originated. Thus, although DNA sequences distinguished the Atlantic and Pacific isolates, stabilizing selection appears to have maintained the same amino acid sequence for each predicted protein.
FIG. 2.
Unrooted, neighbor-joining trees for β-tubulin gene fragments (A) and Sig1 gene fragments isolated from seven T. weissflogii isolates from Long Island Sound, New York (CCMP1336, CCMP1049); Norway (CCMP1052); Portugal (CCMP1053); California (CCMP1050); Hawaii (CCMP1051); and the Java Sea (CCMP1587). The greatest estimated distances are the distances for the Atlantic and Pacific isolates (A) and the distances for the Long Island and Hawaii-South Pacific isolates (B).
The Sig1 genomic fragment was amplified, cloned, and sequenced from the same seven T. weissflogii isolates. DNA sequence polymorphisms were observed for sequences from each T. weissflogii isolate, confirming the multicopy nature of this gene. Five different Sig1 genomic DNA sequences of each isolate were chosen randomly for comparison with clone Actin genomic sequences (Table 5). Even greater overall sequence variation was observed in T. weissflogii Sig1 sequences when the comparison included genomic clones from all the isolates; 86 nucleotide positions displayed variation. Due to this high number of polymorphisms, only data for substitutions present in two or more clones are summarized in Table 5. More variable positions were observed if cDNA clones were included in the comparison (Tables 2 and 5). Only 16 of the 86 variable positions in Sig1 were localized to the intron. For example, the intron from the South Pacific isolate had a 2-bp insertion relative to other isolates, and the 3′ splice site was TAG rather than CAG. In contrast to what was observed with β-tubulin, more than 80% of sequence variation between isolates occurred within the coding sequence of Sig1. Similar to the β-tubulin intron sequence variation, about 19% of the sites in the 84-bp Sig1 intron were variable. In contrast, about 12.5% of the sites in the Sig1 coding region were variable; this value was nearly five times higher than the value observed for the coding sequence of β-tubulin.
TABLE 5.
Variable nucleotides present in two or more copies of the Sig1 gene fragment amplified from seven isolates of T. weissflogii
| Isolatea | Clone | Nucleotides present in two or more copies at the following positionsb:
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coding
|
Intron
|
Coding Coding
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | 19 | 25 | 28 | 31 | 53 | 64 | 65 | 66 | 85 | 86 | 91 | 98 | 102 | 114 | 119 | 121 | 132 | 140 | 161 | 189 | 196 | 198 | 211 | 212 | 228 | 229 | 234 | 237 | 241 | 267 | 274 | 297 | 315 | 318 | 335 | 345 | 387 | 405 | 417 | 435 | 443 | 483 | 531 | 534 | 558 | 579 | 596 | 605 | 618 | 630 | 632 | 638 | 639 | ||
| Long Island Sound clone Actin | Reference | A | C | T | G | C | T | T | — | — | A | T | A | G | T | A | C | C | T | C | A | T | G | G | G | G | A | A | A | T | T | C | T | T | C | G | A | T | T | A | T | A | C | G | C | A | A | C | T | A | C | T | A | T | C |
| 1 | C | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | A | . | . | C | . | . | . | . | . | . | . | . | . | |
| 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | ||
| 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | C | . | . | . | . | . | . | . | . | . | A | . | . | C | . | . | . | . | . | . | . | . | ||
| 4 | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||
| 5 | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | |
| 6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 7 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 8 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 9 | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | |
| 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| Long Island Sound clone 4C | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | |
| 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | |
| 5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | |
| Norway | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | C | . | . | T | A | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | T | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 5 | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| Portugal | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||
| 5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| California | 1 | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| South Pacific | 1 | . | T | . | A | T | C | C | G | C | T | . | T | A | G | . | T | . | C | T | . | . | . | A | . | . | T | G | G | C | . | G | . | . | T | A | . | C | G | C | C | G | C | A | T | . | . | A | A | G | T | C | . | C | A |
| 2 | . | T | . | A | T | C | C | G | C | C | . | T | A | G | . | T | . | C | T | . | . | . | A | . | . | T | G | G | C | . | G | . | . | T | . | . | C | G | C | C | G | . | A | T | . | . | A | A | G | T | C | . | C | A | |
| 3 | . | T | . | A | T | C | C | G | C | T | . | T | A | G | . | T | . | C | T | . | . | . | A | . | . | T | G | G | C | . | G | . | . | T | A | . | C | G | C | C | G | . | A | T | . | . | A | A | G | T | C | . | C | A | |
| 4 | . | T | . | A | T | C | C | G | C | T | . | T | A | G | . | T | G | C | T | G | . | . | A | . | . | T | G | G | C | . | G | . | . | T | A | . | C | G | C | C | G | . | A | T | . | . | A | A | G | T | C | . | C | A | |
| 5 | . | T | . | A | T | C | C | G | C | T | . | T | A | G | . | T | G | C | T | G | . | . | A | . | . | T | G | G | C | . | G | . | . | T | A | . | C | G | C | C | G | . | A | T | . | . | A | A | G | T | C | . | C | A | |
| Hawaii | 1 | . | T | . | A | T | C | C | . | . | T | . | T | A | G | T | T | . | C | . | . | . | . | . | . | . | T | G | G | C | . | G | . | C | T | A | . | C | G | C | C | G | . | A | T | . | G | A | A | . | T | C | . | C | A |
| 2 | . | T | . | A | T | C | C | . | . | T | . | T | A | G | T | T | . | C | . | . | . | . | . | . | . | T | G | G | C | . | G | . | C | T | A | . | C | G | C | C | G | . | A | T | . | G | A | A | . | T | C | . | C | A | |
| 3 | . | T | . | A | T | C | C | . | . | T | . | T | A | G | T | T | . | C | . | . | . | . | . | . | . | T | G | G | C | . | G | . | C | T | A | . | C | G | C | C | G | . | A | T | . | G | A | A | . | T | C | . | C | A | |
| 4 | . | T | . | A | T | C | C | . | . | T | . | T | A | G | T | T | . | C | . | . | . | . | . | . | . | T | G | G | C | . | G | . | C | T | A | . | C | G | C | C | G | . | A | T | . | G | A | A | . | T | C | . | C | A | |
| 5 | . | T | . | A | T | C | C | . | . | T | . | T | A | G | T | T | . | C | . | . | . | . | . | . | . | T | G | G | C | . | G | . | C | T | A | . | C | G | C | C | G | . | A | T | . | G | A | A | . | T | C | . | C | A | |
Long Island Sound Clones Actin and 4C and the Norway and Portugal isolates were obtained from the Atlantic Ocean, and the California, South Pacific, and Hawaii isolates were obtained from the Pacific Ocean basin.
The positions are numbered relative to the first nucleotide immediately following the forward primer. A dot indicates that the nucleotide is identical to the nucleotide in the Long Island Sound Clone Actin reference clone; dashes indicate gaps.
Despite the frequent occurrence of within-individual nucleotide polymorphisms (Table 2), the same DNA substitution was observed at 30 different positions in all Sig1 copies examined from Hawaiian and South Pacific isolates. Five positions had a substitution found only in South Pacific copies; three positions had a substitution found only in Hawaiian copies; one position had a substitution found only in Portuguese copies; and one position had a substitution found in all South Pacific copies and four of the five Hawaiian copies (the fifth Hawaiian copy had a C rather than a T at this position) (Table 5). The greatest divergence between isolates was the divergence between the Long Island clone Actin isolate and the Hawaiian and South Pacific isolates; the estimated distance was about 6.5% (Fig. 2B), almost 2.5-fold greater than the distance found with β-tubulin (Fig. 2A).
Remarkably, the isolate from California did not display variation at the same positions as other Pacific isolates (Table 5). In fact, the divergence between the Sig1 sequences of the California isolate and the Long Island clone Actin isolate was only 1.4%, which was comparable to the divergence observed within individual isolates. The California isolate clustered with and was essentially indistinguishable from other Atlantic isolates (Fig. 2B). This result directly contrasts with what was observed with the β-tubulin phylogeny (Fig. 2A) and suggests that perhaps with Sig1 the groups are not determined by ocean basin but instead are determined by a division between tropical or subtropical regions and temperate regions, with the South Pacific and Hawaiian isolates belonging to the tropical-subtropical group.
In contrast to the uniformity of β-tubulin amino acid sequences, pairwise comparisons of different SIG1 amino acid sequences identified five changes that were isolate specific (Table 6). The Portuguese isolate was characterized by a moderate amino acid change of alanine to serine at position 37 (the position is the amino acid position in the fragment). The South Pacific isolate was characterized by a conservative amino acid change of histidine to arginine at position 173. Both the South Pacific and Hawaiian isolates were distinguished from other isolates by three amino acid changes, a conservative change of leucine to valine at position 48, a conservative change of phenylalanine to tyrosine at position 169, and a moderate change of valine to alanine at position 184. Stabilizing selection appears to have maintained the same amino acid sequence in β-tubulin, whereas SIG1 appears to be less constrained. The high level of within-species polymorphism suggested that the levels of divergence of Sig1 between species might also be high.
TABLE 6.
Variable amino acids in predicted open reading frames of Sig1 genomic copies amplified from seven isolates of T. weissflogii
| Isolate | Clone | Amino acids at the following positionsa:
|
|||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 9 | 16 | 25 | 37 | 42 | 43 | 48 | 52 | 60 | 63 | 68 | 83 | 84 | 95 | 98 | 108 | 118 | 119 | 126 | 149 | 159 | 170 | 173 | 178 | 181 | 182 | 184 | 186 | ||
| Long Island Sound clone Actin | Reference | M | C | R | N | K | A | G | D | L | S | T | F | D | E | C | G | E | C | N | S | Q | E | H | F | H | D | D | K | V | M |
| 1 | . | . | . | . | . | . | . | . | . | . | . | L | . | . | . | . | . | . | . | Y | . | . | . | . | . | . | . | . | . | . | |
| 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | S | |
| 3 | . | . | . | . | . | . | . | . | . | P | . | L | . | . | . | . | . | . | . | Y | . | D | R | . | . | . | . | . | . | . | |
| 4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . | . | . | . | . | . | . | ||
| 5 | . | . | . | . | . | . | . | . | . | P | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | R | . | . | |
| 6 | . | . | K | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 7 | . | . | G | . | . | . | . | N | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 8 | . | . | . | D | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 9 | . | . | . | . | . | . | . | . | . | P | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | R | . | . | |
| 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| Long Island Sound Clone 4C | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . | . | . | . | . | . | . | . |
| 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | D | . | . | . | . | . | . | . | . | |
| 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | R | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | D | . | . | . | G | . | . | . | . | |
| 5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| Norway | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2 | . | . | . | . | . | . | Y | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 3 | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 4 | . | . | . | . | . | . | Y | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 5 | . | . | . | . | . | . | Y | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | E | . | . | . | . | |
| Portugal | 1 | . | . | . | . | . | S | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2 | . | . | . | . | . | S | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 3 | . | . | . | . | . | S | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 4 | . | . | . | . | . | S | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 5 | . | . | . | . | . | S | . | . | . | . | . | . | . | . | . | . | . | . | . | . | R | . | . | . | . | . | . | . | . | . | |
| California | 1 | L | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |
| South Pacific | 1 | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . | . | . | . | . | . | . | Y | R | . | . | . | A | . |
| 2 | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . | . | . | . | . | . | . | Y | R | . | . | . | A | . | |
| 3 | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . | . | D | . | . | . | . | Y | R | . | . | . | A | . | |
| 4 | . | R | . | . | R | . | . | . | V | . | . | . | . | . | . | . | . | . | . | . | . | . | . | Y | R | . | . | . | A | . | |
| 5 | . | . | . | . | R | . | . | . | V | . | . | . | . | . | . | . | . | R | . | . | . | . | . | Y | R | . | . | . | A | . | |
| Hawaii | 1 | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . | . | . | . | . | . | . | Y | . | . | . | . | A | . |
| 2 | T | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . | . | . | . | . | . | . | Y | . | . | . | . | A | . | |
| 3 | . | . | . | . | . | . | . | . | V | . | . | . | H | . | . | . | . | . | . | . | . | . | . | Y | . | . | . | . | A | . | |
| 4 | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . | . | . | . | . | . | . | Y | . | . | . | . | A | . | |
| 5 | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . | . | . | . | . | . | . | Y | . | . | . | . | A | . | |
The positions are numbered relative to the first predicted amino acid of the open reading frame. A dot indicates that the amino acid is identical to the amino acid in the reference sequence. Lightface roman type indicates conservative changes, italic type indicates moderate changes, underlining indicates radical changes, and boldface roman type indicates very radical changes.
Sig1 homologues display high levels of divergence in Thalassiosira species.
Five species of Thalassiosira were chosen for comparison with T. weissflogii. Three non-chain-forming species, T. pseudonana, T. oceanica, and T. guillardii, were chosen based on their presumed close relationships to one another and to T. weissflogii (6, 16). The chain-forming species T. rotula was chosen because it is a common member of diatom blooms, and T. antarctica was chosen because of its presumed distant relationship to the temperate species T. weissflogii.
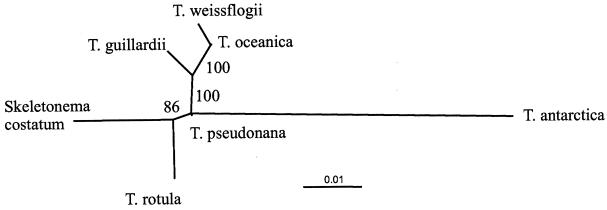
The nucleus-encoded 18S rRNA gene of each species was sequenced to confirm these predicted relationships. A well-supported distance-based phylogeny revealed that T. oceanica, T. guillardii, and T. pseudonana formed a tight cluster closely related to T. weissflogii (Fig. 3). In fact, the T. oceanica sequence differed by only 0.4% from the T. weissflogii sequence. As expected, T. antarctica was only distantly related to T. weissflogii.
FIG. 3.
Unrooted, neighbor-joining tree for the 18S ribosomal gene from T. weissflogii CCMP1336, T. oceanica CCMP1005, T. guillardii CCMP988, T. pseudonana CCMP1335, T. rotula CCMP1647, T. antarctica, and S. costatum (GenBank accession no. X85395). Bootstrap values are indicated at the nodes.
When total genomic DNAs of the five Thalassiosira species other than T. weissflogii were used as templates for PCR with the degenerate Sig primers, two fragments, at 706 and 483 bp, were readily amplified from T. guillardii and T. oceanica; only the 706-bp fragment was amplified from T. pseudonana. Neither fragment was amplified from T. antarctica or T. rotula despite numerous modifications to the amplification protocol and redesign of the PCR primers (data not shown).
The Sig1-sized fragment from T. guillardii, T. pseudonana, and T. oceanica was cloned, and SSCP was used to determine that multiple copies of the Sig1 homologue were likely to be present in each species (data not shown). Three copies of Sig1 were sequenced for each species. The Sig1 DNA sequences fell into three distinct groups, one composed of sequences from T. weissflogii and T. oceanica, one composed of sequences from T. pseudonana, and one composed of sequences from T. guillardii. The Sig1 DNA sequences from T. oceanica and the Atlantic and California isolates of T. weissflogii were very similar to one another, displaying no more variation than that observed within single isolates. In contrast, the Sig1 DNA sequences from members of different groups differed from one another at more than 225 positions in the 538-bp coding sequence.
The intron occurred at the same site in each Sig1 fragment and had very similar 5′ and 3′ splice sites. The T. oceanica intron sequences were readily recognized as they were essentially identical to those of T. weissflogii clone Actin. In contrast, the intron sequences from the other species had diverged so dramatically from one another that it was not possible to align them (data not shown). The T. guillardii intron was 91 bp long, and the T. pseudonana intron was 95 bp long; both of these introns were larger than the T. weissflogii and T. oceanica introns, which were 87 bp long. The 5′ splice site for the T. guillardii and T. pseudonana introns was GTGAG rather than GTAAG, as found in T. weissflogii. The 3′ splice site in T. pseudonana, T. oceanica, and each T. weissflogii isolate except the South Pacific isolate was CAG. The South Pacific isolate and T. guillardii both had a TAG 3′ splice site.
Due to the high level of DNA divergence among Sig1 genes from members of the different groups, the best DNA alignment was achieved by using two steps. The intron was excluded to determine the predicted amino sequence of the resulting open reading frames. The DNA sequences were then aligned based on amino acid alignment, although there were two regions consisting of four to five amino acids at positions 126 to 129 and 146 to 150 where alignment was ambiguous. In this manner, one 3-bp gap was introduced at the same position into the T. guillardii, T. oceanica, and T. weissflogii DNA sequences, and one 3-bp gap was introduced into the T. pseudonana DNA sequences at a different position. As expected, the smallest evolutionary distance was the distance between T. weissflogii and T. oceanica; this distance was only 1.3%, which was comparable to the intraspecific variation in T. weissflogii. The greatest evolutionary distance was the distance between the Sig1 sequences from T. guillardii and T. weissflogii clone Actin and the distance between the T. guillardii and T. pseudonana Sig1 sequences. Both of these comparisons resulted in a maximum evolutionary distance of 37% (the evolutionary distance between T. weissflogii and T. pseudonana was only slightly less, around 35%), a remarkably high value given the extremely close relationship of these three species (Fig. 3). Even this value probably underestimates the true divergence since the Jukes-Cantor model assumes that the probabilities of nucleotide change are equal and does not take into account the possibility of saturation.
The only other nuclear gene fragment that has been isolated from different Thalassiosira species is the multicopy gene Fcp, which encodes fucoxanthin chlorophyll a/c binding protein. Sequences of Fcp cDNA fragments have been reported previously for T. weissflogii clone Actin (22) and T. pseudonana (GenBank accession number U66184). An Fcp cDNA (41) has also been isolated from the relatively closely related centric diatom Skeletonema costatum (29). The estimated distance for the Fcp genes of T. weissflogii and T. pseudonana was 15%, about 2.5 times less than the distance observed for the Sig1 fragments from the same two species. Even more striking is the fact that the evolutionary distance between Fcp copies is only 20% in T. weissflogii and S. costatum, and only 20% in T. pseudonana and S. costatum.
SIG1 and FCP amino acid sequences from the different species were compared to determine if low levels of DNA sequence identity between Sig1 groups reflected a high number of synonymous substitutions at third positions of codons. The percentages of identical codons in SIG1 in members of the three groups of Thalassiosira species ranged from 38 to 43%. In contrast, 64 to 71% of the codons were identical in FCP regardless of whether T. weissflogii and T. pseudonana or T. weissflogii and S. costatum were compared. Furthermore, 24% (45 of 186) of the amino acids in SIG1 differed when T. weissflogii and T. pseudonana were compared (Table 7). In contrast, only 7% (8 of 110) of the amino acids in FCP differed in the two species. Moreover, pairwise comparisons of the three Thalassiosira SIG1 protein groups indicated that a relatively high percentage of the amino acid differences were radical or very radical substitutions (Table 7). Finally, the SIG1 amino acid sequences reflected higher numbers of synonymous changes between T. weissflogii and T. pseudonana (36% for SIG1 and 24% for FCP).
TABLE 7.
Amino acids that are fixed in all SIG1 copies in a given species or isolate but differ between T. guillardii and T. pseudonana and T. weissflogii isolates from Long Island Sound, Portugal, the South Pacific, and Hawaii.
| Isolate | Amino acids at the following positionsa
|
||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 11 | 14 | 25 | 32 | 37 | 40 | 44 | 47 | 48 | 50 | 54 | 59 | 63 | 67 | 72 | 74 | 75 | 78 | 93 | 94 | 96 | 106 | 116 | 120 | 126 | 127 | 128 | 129 | 133 | 137 | 139 | 140 | 144 | 146 | 147 | 148 | 149 | 150 | 151 | 153 | 154 | 169 | 170 | 173 | 174 | 176 | 184 | ||
| Long Island Sound clone Actin | S | F | A | K | R | A | T | H | I | I | A | L | I | F | S | T | I | A | A | I | N | L | A | K | S | Q | G | E | R | N | F | A | N | H | R | A | — | A | E | V | V | R | D | A | F | H | G | T | V |
| Portugal | . | . | . | . | . | S | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| South Pacific | . | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | Y | R | . | . | A |
| Hawaii | . | . | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | Y | . | . | . | A |
| T. guillardii | . | W | . | M | K | . | S | . | . | V | T | V | T | Y | E | N | . | . | . | A | V | . | V | . | N | . | . | D | . | S | . | . | Q | N | . | . | . | S | T | . | P | . | . | . | . | Y | . | V | S |
| T. pseudonana | P | W | S | . | K | F | A | S | V | V | T | V | T | Y | A | N | L | D | G | G | S | Q | . | N | H | T | — | . | G | . | Y | G | K | L | A | G | R | L | S | I | T | T | N | S | Y | Y | N | V | M |
The positions are numbered relative to the first predicted amino acid of the open reading frame. Lightface roman type indicates conservative changes, italic type indicates moderate changes, and boldface roman type indicates radical changes relative to the reference Long Island Sound clone Actin isolate. A dot indicates that the amino acid is identical to the amino acid in the reference sequence; dashes indicate introduced alignment gaps.
Similar to the within-species comparison of β-tubulin and Sig1, a between-species comparison of Fcp and Sig1 also suggested that different selective forces shaped Sig1 diversity. A commonly used means of determining divergence between genes from different species is to compare the number of synonymous changes per synonymous site (Ks) and the number of nonsynonymous changes per nonsynonymous site (Ka) (19). When T. weissflogii and T. pseudonana were compared, Ks was 1.46 ± 0.40 for Sig1 and 0.43 ± 0.11 for Fcp. The number of synonymous changes per site was almost 3.5-fold higher in Sig1 than in Fcp. The Ka values for the same comparison were 0.17 ± 0.04 for Sig1 and 0.06 ± 0.03 for Fcp. The number of nonsynonymous changes per nonsynonymous site was about threefold higher for Sig1 than for Fcp. Both results suggested that Sig1 genes are diverging more rapidly than Fcp genes. Positive selection, a potential driving force for divergence, can be invoked when the Ka/Ks ratio is greater than 1 (19). The Ka/Ks ratio for Sig1 was 0.12, and the ratio for Fcp was 0.13, indicating that in neither case is positive selection at work.
DISCUSSION
Sexual reproduction in diatoms is intimately connected to the control of cell size (38). Consequently, the vast majority of species are assumed to undergo sexual reproduction, even if they do so only infrequently. In reality, however, the sexual cycles of only about 200 of the more than 10,000 species of diatoms have been described. Furthermore, sexual events in natural populations of marine species have only rarely been documented (9, 46), due in large part to inherent difficulties in identifying sexual stages in mixed diatom communities. Thus, our understanding of the frequency of sexual events and the potential generation of genetic diversity within diatom populations remains limited (however, see reference 39).
The genes in the gene family composed of Sig1, -2, and -3 in T. weissflogii were recently identified as potential molecular markers for sexual reproduction in centric diatoms since transcription of these genes is strongly upregulated during the sexual cycle (1). Detection of transcription of genes encoding key enzymes, such as ribulose bisphosphate carboxylase (34) or nitrogenase (47), has proven to be an extremely sensitive means of determining when and where key processes occur in natural populations. In the present study, degenerate PCR primers were designed that amplified Sig1 homologues from four closely related species of Thalassiosira, T. weissflogii, T. oceanica, T. guillardii, and T. pseudonana. Identification of Sig1 in different species of this cosmopolitan genus should facilitate determination of when and where sexual reproduction occurs in natural populations of these target species.
Examination of Sig1 DNA sequences suggests that the likely reason that this gene could be isolated only from closely related Thalassiosira species is that the gene is undergoing rapid sequence divergence. For example, 24% of the amino acids in the predicted SIG1 proteins from T. weissflogii and T. pseudonana differed; in contrast, a comparison of FCP proteins from the same two species indicated that only 7% of the amino acids differed. High levels of sequence divergence between closely related species appear to characterize proteins involved in sexual recognition (43), the hypothesized role of SIG1 (1). In animals, positive Darwinian selection is believed to underlie extreme sequence divergence and is commonly assumed to be a molecular signature for recognition proteins (7). No evidence of positive selection was observed with Sig1. However, a recent study with abalone indicated that an egg receptor protein also does not display positive selection (42). Interestingly, no evidence of positive selection was found with a full-length gamete differentiation gene that nonetheless displayed high levels of divergence in two species of the unicellular algal genus Chlamydomonas (12). Thus, evidence of positive selection does not appear to be required for all recognition and/or differentiation genes.
The most striking feature of Sig1 is the presence of high levels of intraspecific variation and intraindividual variation in addition to high levels of interspecific divergence. The marine invertebrate recognition genes described thus far are all present as single-copy genes, and only the sea urchin gene bindin displays evidence of intraspecific polymorphisms (31). Sig1, on the other hand, is repeated multiple times in the genomes of individual T. weissflogii cells and multiple times in the genomes of all Sig1-positive species. Significantly, multiple copies of different Sig1 genes are transcribed in T. weissflogii, and presumably the same is true for other Sig1-positive species. The multicopy nature of Sig1 suggests that different variants of the protein are expressed during sexual reproduction. Assuming that only a subset of SIG1 proteins are required for proper function, selective pressure on any individual gene copy would be reduced, presumably permitting the observed high levels of intraindividual polymorphisms to persist (25).
The comparison of between-individual polymorphisms in Sig1 and the housekeeping gene β-tubulin is particularly intriguing. Divergence in β-tubulin is relatively easy to explain. Isolates from the Atlantic and Pacific oceans have apparently been physically separated from one another long enough (about 2 million years since North America and South America joined) to display slight DNA sequence divergence (about 2.8%) in their β-tubulin genes, but amino acid sequence divergence is not evident due to purifying selection (19). In contrast, the patterns of divergence in Sig1 are more complicated. As expected, Atlantic isolates have similar Sig1 sequences. What is surprising, however, is the fact that the Sig1 sequence from the California isolate is indistinguishable from Atlantic sequences but is very different (6.5% divergence) from Hawaiian and South Pacific sequences.
How could this difference in Sig1 sequences between California and other Pacific isolates be maintained? Based on average surface circulation patterns, for example, one would expect Hawaiian and California populations to mix, with genetic recombination during sexual reproduction homogenizing any molecular divergence. The apparent differences between these two populations could simply reflect a sampling error; if Sig1 from a second isolate from California, for example, was analyzed, perhaps less divergence would be observed. Unfortunately, no other California isolates of T. weissflogii are currently available to test this hypothesis. However, Sig1 sequences from two Long Island Sound isolates collected 10 years apart were analyzed. These two sets of Sig1 sequences were very similar to one another (Table 5). The maintenance of sequence similarity for 10 years suggests that the largest source of Sig1 diversity in a given population may be diversity within individuals rather than diversity between individuals. This suggests that the differences in the Sig1 DNA sequences from California and Hawaiian isolates may reflect true divergence rather than a sampling artifact. Regardless of the explanation, some form of regional selection pressure associated with the Hawaiian and South Pacific environment, rather than drift, appears to drive the high level of intraspecific Sig1 divergence.
Potential selective pressures that underlie rapid protein evolution remain unclear. The most intuitive hypotheses have been developed for proteins displayed on cell surfaces. For example, the surface proteins of pathogens appear to evolve rapidly, presumably to avoid recognition by the immune system (18). Similarly, surface proteins of externally fertilized gametes have been hypothesized to evolve rapidly to avoid recognition by pathogens (44, 45). If Sig1 does encode a protein displayed in the extracellular matrix, similar pathogen-induced evolution could occur. Two dominant types of diatom pathogens are viruses (35) and parasitoids (21), both of which appear to possess species-specific recognition mechanisms. Sexually reproducing diatoms could be particularly vulnerable to infection since sperm entirely, and auxospores temporarily, lack the silica frustule. SIG1 variants with amino acid substitutions that prevent infection by a pathogen could presumably sweep through a population to fixation. Region-specific differences in pathogen communities could then generate region-specific differences in SIG1 sequences, such as the apparent temperate and tropical divergence. Thus, there is the intriguing possibility that avoidance of infection by pathogens in the ocean might actually lead to speciation and further division of niche space (20).
An alternate, more recent hypothesis is that no external forces drive divergence of surface molecules; the process simply results from the repetitive structure common to these kinds of proteins. In abalone, for example, the egg surface receptor protein is composed of multiple, tandemly repeated amino acid domains, each of which binds the cognate sperm protein to some degree (42). Selective pressures on individual repeat units are relaxed, and concerted evolution is predicted to propagate identical nucleotide changes throughout the molecule. A rapidly changing amino acid sequence of the egg receptor is believed to drive rapid evolution of the cognate sperm protein (42). An analogous mechanism could drive Sig1 evolution. Instead of multiple tandemly repeated sequences in a single gene, the entire Sig1 sequence appears to be repeated multiple times as distinct genes. It is interesting that multicellular organisms commonly combine into a single protein domains that are present as individual proteins in unicellular organisms (5). If Sig1 is tandemly repeated, then concerted evolution could propagate particular nucleotide changes to the multiple gene copies.
The simplest explanation for the observed intraspecific and interspecific divergence is that different forces, acting on different times scales, combine to shape Sig1 evolution. First, selective pressure on individual copies of Sig1 has apparently been relaxed, allowing high levels of intraindividual polymorphisms to arise, either because Sig1 duplicated relatively recently (25) or because Sig1 has somehow escaped selection pressures. Superimposed on this high rate of divergence appears to be a lower rate of convergent evolution, in which specific nucleotide changes are spread throughout all gene copies. The effect of these two processes in combination with stochastic factors could lead to large-scale region-specific differences in Sig1 sequences without any need for external driving forces. Small-scale regional differences would be expected to persist if particular SIG1 variants conveyed a region-specific selective advantage, such as resistance to infection by pathogens. If, for example, the selective forces in the tropics differ from those in more temperate environments, collection of T. weissflogii isolates from tropical regions in the Atlantic Ocean could provide insight into these processes. A true understanding of the evolutionary pressures acting on SIG1, however, awaits determination of the exact role played by this protein.
The potential scenarios described here, although speculative, should not be considered specific to diatoms. Similar mechanisms could explain speciation in any group of phytoplankton in which multiple, closely related species simultaneously undergo sexual reproduction in a body of water.
ACKNOWLEDGMENTS
We thank Paul Bentzen, Mike Canino, and Tatiana Rynearson for many helpful discussions.
This work was supported in part by National Science Foundation grant OCE 9702158 (to E.V.A.), by Office of Naval Research DURIP award N000140010597 (to E.V.A.), and by a Mary Gates Endowment for Students undergraduate research training grant (to H.M.G.).
REFERENCES
- 1.Armbrust E V. Identification of a new gene family expressed during the onset of sexual reproduction in the centric diatom Thalassiosira weissflogii. Appl Environ Microbiol. 1999;65:3121–3128. doi: 10.1128/aem.65.7.3121-3128.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armbrust E V. Structural features of nuclear genes in the centric diatom Thalassiosira weissflogii (Bacillariophyceae) J Phycol. 2000;36:942–946. [Google Scholar]
- 3.Armbrust E V, Olson R J, Chisholm S W. Role of light and the cell cycle on the induction of spermatogenesis in a centric diatom. J Phycol. 1990;26:470–478. [Google Scholar]
- 4.Bhaya D, Grossman A R. Characterization of gene clusters encoding the fucoxanthin chlorophyll proteins of the diatom Phaeodactylum tricornutum. Nucleic Acids Res. 1993;21:4458–4466. doi: 10.1093/nar/21.19.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bork P, Downing A K, Kieffer B, Campbell I D. Structure and distribution of modules in extracellular proteins. Q Rev Biophys. 1996;29:119–167. doi: 10.1017/s0033583500005783. [DOI] [PubMed] [Google Scholar]
- 6.Brand L E. Genetic variability in reproduction rates in marine phytoplankton populations. Evolution. 1981;35:1117–1127. doi: 10.1111/j.1558-5646.1981.tb04981.x. [DOI] [PubMed] [Google Scholar]
- 7.Civetta A, Singh R S. Sex-related genes, directional sexual selection, and speciation. Mol Biol Evol. 1998;15:901–909. doi: 10.1093/oxfordjournals.molbev.a025994. [DOI] [PubMed] [Google Scholar]
- 8.Cleveland D W, Sullivan K F. Molecular biology and genetics of tubulin. Annu Rev Biochem. 1985;54:331–365. doi: 10.1146/annurev.bi.54.070185.001555. [DOI] [PubMed] [Google Scholar]
- 9.Crawford R M. The role of sex in the sedimentation of a marine diatom bloom. Limnol Oceanogr. 1995;40:200–204. [Google Scholar]
- 10.Drebes G. Sexuality. In: Werner D, editor. The biology of diatoms. Berkeley: University of California Press; 1977. pp. 250–283. [Google Scholar]
- 11.Edlund M B, Stoermer E F. Ecological, evolutionary, and systematic significance of diatom life histories. J Phycol. 1997;33:897–918. [Google Scholar]
- 12.Ferris P J, Pavlovic C, Fabry S, Goodenough U W. Rapid evolution of sex-related genes in Chlamydomonas. Proc Natl Acad Sci USA. 1997;94:8634–8639. doi: 10.1073/pnas.94.16.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 14.Guillard R R L. Culture of phytoplankton for feeding marine invertebrates. In: Smith W L, Chaney M H, editors. Culture of marine invertebrate animals. New York, N.Y: Plenum Press; 1975. pp. 29–60. [Google Scholar]
- 15.Hartl D L, Clark A G. Principles of population genetics. Sunderland, Mass: Sinauer Associates, Inc.; 1997. [Google Scholar]
- 16.Hasle G R. Some freshwater and brackish water species of the diatom genus Thalassiosira Cleve. Phycologia. 1978;17:263–292. [Google Scholar]
- 17.Hellberg M E, Moy G W, Vacquier V D. Positive selection and propeptide repeats promote rapid interspecific divergence of a gastropod sperm protein. Mol Biol Evol. 2000;17:458–466. doi: 10.1093/oxfordjournals.molbev.a026325. [DOI] [PubMed] [Google Scholar]
- 18.Hughes A L. Positive selection and interallelic recombination at the merozoite surface antigen-1 (MSA-1) locus of Plasmodium falciparum. Mol Biol Evol. 1992;9:381–393. doi: 10.1093/oxfordjournals.molbev.a040730. [DOI] [PubMed] [Google Scholar]
- 19.Hughes A L. Adaptive evolution of genes and genomes. Oxford, United Kingdom: Oxford University Press; 1999. p. 270. [Google Scholar]
- 20.Hutchinson G E. The paradox of the plankton. Am Nat. 1961;95:137–145. [Google Scholar]
- 21.Kuehn S F. Infection of Coscinodiscus spp. by the parasitoid nanoflagellate Pirsonia diadema. 1. Behavioural studies on the infection process. J Plankton Res. 1997;19:791–804. [Google Scholar]
- 22.Leblanc C, Falciatore A, Watanabe M, Bowler C. Semi-quantitative RT-PCR analysis of photoregulated gene expression in marine diatoms. Plant Mol Biol. 1999;40:1031–1044. doi: 10.1023/a:1006256300969. [DOI] [PubMed] [Google Scholar]
- 23.Lessa E P, Applebaum G. Screening techniques for detecting allelic variation in DNA sequences. Mol Ecol. 1993;2:119–129. doi: 10.1111/j.1365-294x.1993.tb00006.x. [DOI] [PubMed] [Google Scholar]
- 24.Lundberg K S, Shoemaker D D, Adams M W W, Short J M, Sorge J A, Mathur E J. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene. 1991;108:1–6. doi: 10.1016/0378-1119(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 25.Lynch M, Conery J S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald J D. On the structure of the diatomaceous frustule, and its genetic cycle. Ann Mag Nat Hist Ser. 1869;4:1–8. [Google Scholar]
- 27.Mann D G. The species concept in diatoms. Phycol Rev. 1999;38:437–495. [Google Scholar]
- 28.Medlin L, Elwood H J, Stickel S, Sogin M L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 29.Medlin L K, Barker G L A, Campbell L, Green J C, Hayes P K, Marie D, Wrieden S, Vaulot D. Genetic characterisation of Emiliania huxleyi (Haptophyta) J Mar Syst. 1996;9:13–31. [Google Scholar]
- 30.Medlin L K, Kooistra W H C F, Gersonde R, Sims P A, Wellbrock U. Is the origin of the diatoms related to the end-Permian mass extinction? Nova Hedwigia. 1997;65:1–11. [Google Scholar]
- 31.Metz E C, Palumbi S R. Positive selection and sequence rearrangements generate extensive polymorphism in the gamete recognition protein bindin. Mol Biol Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- 32.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Applic Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 33.Palumbi S R. Genetic divergence, reproductive isolation, and marine speciation. Annu Rev Ecol Syst. 1994;25:547–572. [Google Scholar]
- 34.Paul J H, Pichard S L, Kang J B, Watson G M F, Tabita F R. Evidence for a clade-specific temporal and spatial separation in ribulose bisphosphate carboxylase gene expression in phytoplankton populations off Cape Hatteras and Bermuda. Limnol Oceanogr. 1999;44:12–23. [Google Scholar]
- 35.Proctor L M. Advances in the study of marine viruses. Microsc Res Tech. 1997;37:136–161. doi: 10.1002/(SICI)1097-0029(19970415)37:2<136::AID-JEMT3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 36.Rose T M, Schultz E R, Henikoff J G, Pietrokovski S, McCallum C M, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly-related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Round F E. The problem of cell size during diatom cell division. Nova Hedwigia. 1972;31:485–493. [Google Scholar]
- 38.Round F E, Crawford R M, Mann D G. The diatoms. Cambridge, United Kingdom: Cambridge University Press; 1990. p. 747. [Google Scholar]
- 39.Rynearson T A, Armbrust E V. DNA fingerprinting reveals extensive genetic diversity in a field population of the centric diatom Ditylum brightwellii. Limnol Oceanogr. 2000;45:1329–1340. [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Smith G J, Gao Y, Alberte R S. The fucoxanthin-chlorophyll A/C proteins comprise a large family of coexpressed genes in the marine diatom Skeletonema costatum (Greve): characterization of eight unique cDNAs. Plant Physiol. 1997;114:1136. [Google Scholar]
- 42.Swanson W J, Vacquier V D. Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science. 1998;281:710–712. doi: 10.1126/science.281.5377.710. [DOI] [PubMed] [Google Scholar]
- 43.Vacquier V D. Evolution of gamete recognition proteins. Science. 1998;281:1995–1998. doi: 10.1126/science.281.5385.1995. [DOI] [PubMed] [Google Scholar]
- 44.Vacquier V D, Lee Y-H. Abalone sperm lysin: unusual mode of evolution of a gamete recognition protein. Zygote. 1993;1:181–196. doi: 10.1017/s0967199400001465. [DOI] [PubMed] [Google Scholar]
- 45.Vacquier V D, Swanson W J, Lee Y-H. Positive Darwinian selection of two homologous fertilization proteins: what is the selective pressure driving their divergence? J Mol Evol. 1997;44(Suppl. 1):S15–S22. doi: 10.1007/pl00000049. [DOI] [PubMed] [Google Scholar]
- 46.Waite A, Harrison P J. Role of sinking and ascent during sexual reproduction in the marine diatom Ditylum brightwellii. Mar Ecol Prog Ser. 1992;87:113–122. [Google Scholar]
- 47.Wyman M, Zehr J P, Capone D G. Temporal variability in nitrogenase gene expression in natural populations of the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1996;62:1073–1075. doi: 10.1128/aem.62.3.1073-1075.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]