Abstract
In Comamonas testosteroni BR60 (formerly Alcaligenes sp. strain BR60), catabolism of the pollutant 3-chlorobenzoate (3CBA) is initiated by enzymes encoded by cbaABC, an operon found on composite transposon Tn5271 of plasmid pBRC60. The cbaABC gene product CbaABC converts 3CBA to protocatechuate (PCA) and 5-Cl-PCA, which are then metabolized by the chromosomal PCA meta (extradiol) ring fission pathway. In this study, cbaA was found to possess a ς70 type promoter. O2 uptake experiments with whole cells and expression studies with cbaA-lacZ constructs showed that cbaABC was induced by 3CBA. Benzoate, which is not a substrate of the 3CBA pathway, was a gratuitous inducer, and CbaR, a MarR family repressor coded for by a divergently transcribed gene upstream of cbaABC, could modulate induction mediated by benzoate. Purified CbaR bound specifically to two regions of the cbaA promoter (PcbaA); site I, a high-affinity site, is between the transcriptional start point (position +1) and the start codon of cbaA, while site II, a lower-affinity site, overlaps position +1. 3CBA at concentrations as low as 40 μM interfered with binding to PcbaA. PCA also interfered with binding, while benzoate only weakly disrupted binding. Unexpectedly, benzoate with a hydroxyl or carboxyl at position 3 improved CbaR binding. Data are also presented that suggest that an unidentified regulator is encoded on the chromosome that induces cbaABC in response to benzoate and 3CBA.
The chlorinated benzoic acids (CBA) are a common class of pollutants that occur in the environment as a result of intentional introduction (e.g., in the form of herbicides) or incomplete bacterial metabolism of some accidentally released chemicals (e.g., polychlorinated biphenyls) (46). Bacteria possess a remarkable assortment of metabolic pathways for biodegradation of CBA, and the innate ability of bacteria to degrade CBA has been exploited for bioremediation of contaminated sites (47). Several aerobic degradation pathways have been characterized at the biochemical and genetic levels. The most intensively studied pathway is encoded by the clc genes of Pseudomonas putida that specify intradiol ring fission of 3-chlorocatechol, a metabolite generated by nonspecific activity of benzoate or toluate dioxygenases with 3-chlorobenzoate (3CBA) (19). In contrast, the cba-encoded pathway involves a dioxygenase and a dehydrogenase that convert 3CBA, 4CBA, or 3,4-dichlorobenzoate to the vicinal diol intermediates protocatechuate (PCA) and 5-Cl-PCA (Fig. 1A) (40, 41). Other CBA degradation operons include the cbd-encoded pathway of Burkholderia cepacia (22) and the ohb-encoded pathway of Pseudomonas aeruginosa (60), both of which specify dioxygenase-mediated conversion of 2CBA to catechol; the fcb pathway of Arthrobacter globiformis for conversion of 4CBA to 4-hydroxybenzoate by a coenzyme A ligase and a hydrolase (61); and an Alcaligenes sp. pathway that converts 3CBA to 3-hydroxybenzoate (31, 32). Proven or putative regulatory factors for these pathways are encoded by genes closely linked to the catabolic operons. These factors include ClcR, a LysR-like regulator for clc (9); CbdR, an AraC-like regulator for cbd (22); and OhbR, an IclR-like regulator for ohb (60).
FIG. 1.
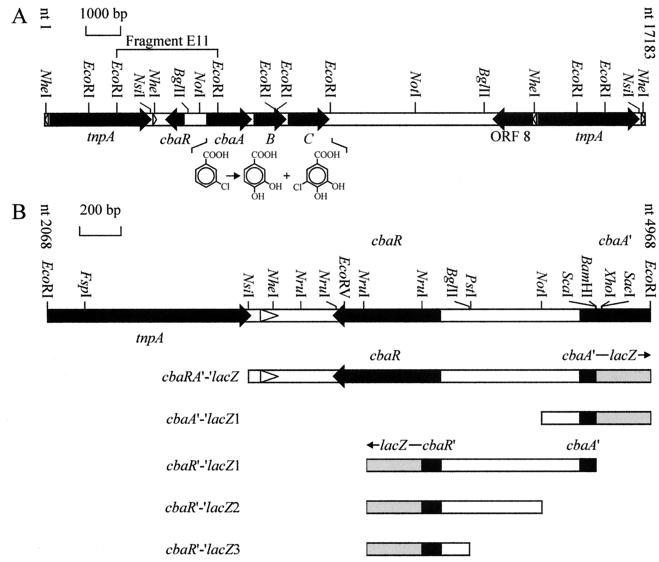
(A) Physical and restriction map of 3CBA catabolism transposon Tn5271 found on plasmid pBRC60 of C. testosteroni BR60. The solid arrows indicate various genes, including cbaABC, which encode the dioxygenase (CbaAB) and dehydrogenase (CbaC) that convert 3CBA to PCA or 5-Cl-PCA; cbaR, which codes for a regulatory protein (this study); the transposase tnpA gene of the two copies of IS1071; and an ORF that codes for a truncated aryl coenzyme A ligase-like product (ORF 8). Flanking tnpA are the left and right inverted repeats of IS1071 (open triangles). (B) Restriction map of EcoRI fragment 11 (E11) from pBRC60 and schematic diagrams of lacZ constructs made to study expression of cbaA and cbaR. Note that lacZ (grey boxes) is not drawn to scale.
The cbaABC operon is located in composite transposon Tn5271 (Fig. 1A) found on the self-transmissible IncPβ plasmid pBRC60 (38, 67). Despite the potential for being mobilized to members of various subclasses of the class Proteobacteria (14, 20, 21, 47, 65), the natural host range of this plasmid includes primarily members of the β subclass of the Proteobacteria, particularly strains that degrade PCA, the product of CbaABC-mediated catabolism of CBA, by an extradiol pathway (39). From an ecological point of view, little is known about factors that limit the horizontal spread of this mobile pathway. We therefore sought to better understand regulation of cbaABC expression, not only because horizontal transfer in contaminated environments brings this operon into a variety of genetic backgrounds and we are curious about whether the capacity to effectively regulate cbaABC plays a role in determining the host range of these genes, but also because cbaABC encodes an additional pathway by which bacteria metabolize CBA and thus has potential applications in bioremediation of contaminated sites. We therefore characterized induction of cbaABC in the original host, and in this report we show that a cis-encoded MarR-like regulator, CbaR, plays a role in modulating expression of cbaABC. We also present evidence that a chromosomally encoded protein may be involved in regulating this operon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Escherichia coli was routinely grown at 37°C in Luria-Bertani medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl) containing ampicillin (250 mg liter−1), kanamycin (40 mg liter−1), or chloramphenicol (50 mg liter−1), as required. Comamonas testosteroni strains (formerly Alcaligenes sp. strains [see below]) were grown as described below in minimal medium A (64) containing 10 mM succinate, aromatic substrate at a concentration of 4 mM, and chloramphenicol (50 to 100 mg liter−1), as required. All chemicals and antibiotics were purchased from Sigma-Aldrich (Oakville, Ontario, Canada). When necessary, growth media were solidified by adding agar to a final concentration of 1.6% (wt/vol).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant phenotypic and/or genotypic characteristicsa | Reference or source |
|---|---|---|
| C. testosteroni strains | ||
| BR60(pBRC60) | Wild type; Tn5271 proficient | 66 |
| BR6020b | BR60 cured of pBRC60; Tn5271 deficient | 66 |
| BR6024(pBRC60) | Trp− and Cmr strain derived from BR6020; Tn5271 proficient; used to conjugate pBRC60 into BR6020 strains harboring lacZ expression constructs | 42 |
| E. coli strains | ||
| XL-1 Blue | Used during cloning and DNA manipulation; α-lacZ complementing strain | Stratagene |
| CC118::λpir | Host for pUTCm and pUTCm-based suicide delivery vectors | 12 |
| M15 | Host for pQE30, pQE30-based expression vectors, and pREP4 | Qiagen |
| Source plasmids | ||
| pCRScript SK+ | Ampr; α-lacZ MCS; general-purpose cloning vector | Stratagene |
| pUC18 | Ampr; α-lacZ MCS; general-purpose cloning vector | 68 |
| pUC18Not | Ampr; α-lacZ MCS; general-purpose cloning vector | 13 |
| pUC128 | Ampr; α-lacZ MCS; general-purpose cloning vector | 27 |
| pUTCm | Ampr Cmr; Tn5-based suicide delivery vector | 12 |
| pUTCmMCS.3 | Ampr Cmr; pUTCm with oligonucleotide containing additional restriction sites cloned into NotI sitec | This study |
| pRK2013 | Kmr RK2 tra; used in triparental mating to mobilize pUTCm-based plasmids | 18 |
| pBW30::TnphoA′-4 | Ampr Kmr; vector used as source of lacZ gene; removed either as a BamHI-HindIII fragment or a BamHI-BglII fragment, which releases a promoterless lacZ gene lacking DNA encoding the first nine amino acids | 63 |
| pMP10.1 | Ampr; lacZ cloned as BamHI-HindIII fragment into pUC18Not | This study |
| pMP28.1 | Ampr; lacZ cloned as a BamHI-BglII fragment into pUC18Not digested with BamHI | This study |
| pBRE11 | Ampr; EcoRI fragment 11 from pBRC60 cloned into pUC18 | 38 |
| pQE30 | Ampr, IPTG-inducible expression vector for adding a six-histidine affinity tag to the N terminus of proteins cloned into the MCS | Qiagen |
| pREP4 | Kmr; codes for Laclq, which represses expression from pQE30 promoter | Qiagen |
| LacZ expression constructs | ||
| pBRCW34 | Ampr; NruI-ScaI fragment from pBRE11 cloned into SrfI site of pCR-Script SK(+) so that the ScaI end (i.e., cbaA′) is adjacent to BamHI | This study |
| pBRCW34L | Ampr; lacZ cloned as a BamHI-HindIII fragment into pBRCW34, placing it in frame with cbaA′ | This study |
| pMP68.3 | Ampr; FspI-NotI fragment from pBRE11 cloned into pBRCW34L digested with SacII (blunt ended) and NotI; DNA coding for cbaR, a divergently transcribed ORF between cbaA and IS1071L, is placed upstream of cbaA′ | This study |
| pMP70.5 | Ampr Cmr; blunt-ended NsiI-HindIII fragment from pMP68.3 containing construct cbaRA′-′lacZ cloned into pUTCm digested with NotI and also blunt ended | This study |
| pMP14.10 | Ampr; BglII fragment from pBRCW34L cloned into pUC18Not digested with BamHI | This study |
| pMP17.6 | Ampr Cmr; NotI fragment from pMP14.10 containing construct cbaA′-′lacZ1 cloned into pUTCm | This study |
| pBRCW27 | Ampr; NruI-ScaI fragment from pBRE11 cloned into SrfI site of pCRScript SK (+) so that the NruI end (i.e., cbaR′) is adjacent to BamHI | This study |
| pBRCW27L | Ampr; lacZ cloned as a BamHI-HindIII fragment into pBRCW27, placing it in frame with cbaR′ | This study |
| pMP105.1 | AmprSacI fragment from pBRCW27L cloned into pMP28.1 | This study |
| pMP123.2 | Ampr; SacII-XbaI fragment from pMP105.1 cloned into pUC128 | This study |
| pMP125.1 | Ampr Cmr; NsiI-NotI fragment from pMP123.2 containing construct cbaR′-′lacZl cloned into pUTCmMCS.3 | This study |
| pMP106.1 | Ampr Cmr; NotI fragment from pMP105.1 containing construct cbaR′-′lacZ2 cloned into pUTCm | This study |
| pMP50.5 | Ampr; BglII fragment from pBRCW27L cloned into pUC18Not digested with BamHI | This study |
| pMP54.9 | Ampr Cmr; NotI fragment from pMP50.5 containing construct cbaR′-′lacZ3 cloned into pUTCm | This study |
| pMP19.9 | Ampr Cmr; NotI fragment from pMP10.1 containing control expression construct (promoterless lacZ) cloned into pUTCm | This study |
| Affinity-tagged CbaR plasmids | ||
| pMP130.4 | Ampr; PCR-amplified cbaR cloned into SmaI site of pCRScript SK (+) | This study |
| pQE30cbaR | Ampr; cbaR from pMP130.4 subcloned into pQE30 as a BamHI-SacI fragment | This study |
Abbreviations: MCS, multiple cloning site; Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance.
BR6020 strains harboring chromosomal inserts of the various lacZ expression constructs (see Materials and Methods) are not listed because of space considerations.
The sequence of the modifed cloning site in pUTCmMCS.3 is 5′-GCGGCCGCAA GCTTGCATGC CTGCAGGTCG ACTCTAGAGG ATCCCCCGGG GGATTTAAAT ATGCATCGGC CGC-3′. This sequence contains restriction sites for NotI, HindIII, SphI, PstI, SalI, XbaI, BamHI, SmaI, SwaI, NsiI, and EagI. Only the restriction sites for NotI, SwaI, and NsiI are unique to this vector. The 5′ end is proximal to the Cmr gene.
DNA sequencing and sequence analysis.
To add to the phenotypic characteristics previously determined for 3CBA-degrading strain BR60(pBRC60) (67), both strands of the first 523 bases of the 16S rRNA gene were sequenced by MIDI Labs (Newark, Del.) by using primers that anneal beginning at positions 5 and 531 in the 16S rRNA gene of E. coli. The sequences of both strands of Tn5271 DNA between IS1071L and cbaA (Fig. 1A) were determined by the chain-terminating dideoxy method with an ABI Prism automated sequencer (Biotechnology Research Institute, University of Ottawa, Ottawa, Ontario, Canada) using appropriate primers and pBRE11 (Table 1) as the template. This DNA was analyzed for similarities to entries in the GenBank nonredundant database by using the BLAST (5) network service of the National Center for Biotechnology Information, Bethesda, Md. (http://www.ncbi.nlm.nih.gov). A divergently transcribed open reading frame (ORF) upstream of cbaA was identified and designated cbaR. Because the putative product of this ORF showed similarity to MarR and various other transcriptional regulators that are known to respond to aromatic compounds or control aromatic catabolism genes (see below), this region was taken into consideration when we designed the various lacZ expression constructs described below. DNA between cbaC and IS1071R (Fig. 1A) had been sequenced previously and was not taken into consideration as it contains ORFs whose products show similarity to proteins involved in uptake of aromatic compounds (unpublished results) and a truncated aryl coenzyme A ligase (14).
Determination of cbaA transcriptional start site and reverse transcription (RT)-PCR assay.
The transcriptional start of cbaA was determined by primer extension analysis as described elsewhere (53). Briefly, C. testosteroni BR60(pBRC60) was grown to the mid-log phase in minimal medium A containing 3CBA, which maximally induced 3CBA degradation activity (see below). Total RNA was isolated, and primer CBAAR (5′-CATGAGGCCGCCCATCG-3′; complementary to the N-terminal coding region of CbaA) was annealed to it and extended with Moloney murine leukemia virus reverse transcriptase (New England Biolabs). During extension, [α-32P]dCTP (Amersham Canada, Oakville, Ontario, Canada) was included in the reaction mixture to label the product. The extension product and the product of a sequencing reaction conducted with the same primer (using pMP14.10 as the template) were resolved by electrophoresis on a denaturing 8% polyacrylamide sequencing gel, and DNA was detected by autoradiography; both of these analyses were performed by using standard methods (55).
Levels of cbaA RNA in induced cultures and uninduced cultures were qualitatively compared by an RT-PCR assay, as follows. BR60(pBRC60) was grown to the mid-log phase on minimal medium A containing 3CBA or succinate (a noninducing substrate as determined by O2 uptake analysis [see below]), and DNA-free total RNA was extracted with an E.Z.N.A bacterial RNA kit (PeqLab, Erlangen, Germany) according to the manufacturer's instructions. CBAAR was annealed to equal amounts of RNA from each source and reverse transcribed by using a RevertAid First Strand cDNA synthesis kit (MBI Fermentas, St. Leon-Rot, Germany) as recommended by the manufacturer. Aliquots from the RT step were then analyzed by PCR performed with forward primer PCBAF (5′-ACCAACTACATGGATCGAA-3′; the sequence corresponds to the extreme 5′ end of the cbaA transcript) and primer CBAAR as the reverse primer. The intensity of the signal (a 167-bp fragment) directly reflected the initial level of the transcript. Two independent RT-PCRs were conducted, and the PCR products were analyzed by agarose gel electrophoresis. As a control to ensure that no contaminating DNA was present in the RNA samples, RNAs from both sources were treated as recommended for first-strand synthesis, except that no reverse transcriptase was added. Aliquots were then analyzed by PCR as described above, and no signal was observed.
Regulation of 3CBA degradation activity.
Control of 3CBA catabolism in C. testosteroni BR60(pBRC60) was studied by first determining which growth substrates other than 3CBA were capable of inducing 3CBA degradation activity. As discussed above, CbaABC-mediated metabolism of 3CBA generates PCA and 5-Cl-PCA, which are then degraded by an extradiol (meta) ring fission pathway (38). Because benzoate, 3-hydroxybenzoate, 4-hydroxybenzoate, and PCA also induce this pathway (41, 62), we tested whether 3CBA degradation activity was induced by growth on these four aromatic substrates. Control cells were grown on succinate, a nonaromatic tricarboxylic acid cycle intermediate that presumably does not induce activity. O2 uptake assays were conducted as described previously (41). Briefly, cultures were grown to the mid-log phase on one of the substrates mentioned above, harvested, and washed, and the respiration rates of cells with the various test compounds were determined polarographically. For each growth substrate, two or three independent trials were performed, and O2 consumption was measured with replicate samples of the culture from each trial. Spontaneous deletion of Tn5271 occurs at a high frequency (66), and we therefore assessed the loss of the 3CBA degradation genes for cultures grown on carbon sources other than 3CBA by diluting and plating samples on minimal medium A agar containing 3CBA (Tn5271-proficient cells) or succinate (total count). During the incubation period used in these experiments, there was no noticeable loss of Tn5271.
Construction and insertion of cbaA and cbaR expression constructs and measurement of LacZ activity.
Additional studies were conducted to investigate the effect of CbaR on cbaA expression. Two translational fusion constructs were used: cbaRA′-′lacZ, which included the gene cbaR, and cbaA′-′lacZ1, which lacked it (Fig. 1B). Expression of cbaR was studied by using three translational fusion constructs which contained different amounts of DNA upstream of the cbaR start codon (Fig. 1B). Finally, a control expression construct (promoterless lacZ) was also designed. Table 1 provides information on the structures and derivations of the relevant plasmids used in these studies. The nucleotide sequences of the junctions between cbaA′ and ′lacZ in pBRCW34L and between cbaR′ and ′lacZ in pBRCW27L were determined to confirm that both genes were in frame with lacZ. The various expression constructs were then cloned into suicide delivery vector pUTCm or pUTCmMCS.3 (Table 1) so that lacZ was transcribed in the direction opposite the direction of transcription of the chloramphenicol resistance gene, and they were mobilized from E. coli CC118λpir and inserted into the chromosome of C. testosteroni BR6020 (Table 1) by triparental mating (41). BR6020 is a pBRC60/Tn5271-deficient derivative of BR60 and thus is not able to grow on 3CBA. This strain allowed expression to be measured in a genetic background lacking plasmid/Tn5271-encoded functions. Transconjugants were recovered by plating cells on minimal medium A agar containing 4-hydroxybenzoate, chloramphenicol, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Because insertion of the expression constructs into the chromosome was random, we tested whether the location of transposition affected LacZ levels by isolating four independent transconjugants for each construct and measuring the LacZ activity of succinate-grown cells, as described below. Levels could vary by as much as twofold; therefore, the transconjugant that had the lowest LacZ activity was used as the sole representative in subsequent studies in which expression with that construct was measured. Transposition of the expression constructs into the chromosome as single copies was confirmed by Southern blotting. A Tn5271-proficient background, which restored the ability to grow on 3CBA, was created for the representative strains harboring an expression construct by transferring pBRC60 from C. testosteroni BR6024(pBRC60) and recovering transconjugants on minimal medium A agar containing 3CBA, chloramphenicol, and X-Gal (41). The LacZ levels in cells harboring the expression constructs were measured as described elsewhere (55). Cultures were grown on succinate (a noninducing substrate) and 3CBA or benzoate (inducing substrates). The averages ± standard errors based on three independent trials are reported below for each carbon source, and the statistical significance of observed effects was determined by a paired t test (P = 0.05).
Cloning and overexpression of cbaR
Affinity-purified CbaR for gel shift studies and DNase I protection assays (described below) was obtained as follows. The gene coding for CbaR was PCR amplified by using forward primer CBAR3958 (5′-cgggatccCTTGCAAGAGATCCTCGA-3′) and reverse primer CBAR3440 (5′-ccgagctcCTACTCCTGAGGAGATTC-3′) (lowercase letters indicates nucleotides that are not native to cbaR, and the underlined nucleotides in the two sequences are restriction sites for BamHI and SacI, respectively). CBAR3958 is complementary to the portion of cbaR coding for the N terminus but lacks the native start codon, while CBAR3440 is complementary to the C-terminal coding portion of cbaR up to and including the native stop codon. The amplification product was initially cloned into pCRScript SK(+) digested with SmaI (resulting in pMP132.4), and the insert was sequenced to confirm that PCR amplification had not introduced mutations. cbaR was then placed downstream from and in frame with DNA coding for the N-terminal affinity tag MRGSH6GS by digesting pMP132.4 with BamHI and SacI and cloning the ∼550-bp fragment that was generated into pQE30 digested with the same enzymes, which resulted in pQE30cbaR. CbaR was then overexpressed and affinity purified with an Ni-nitriloacetate column as recommended by the manufacturer (Qiagen, Mississauga, Ontario, Canada), dialyzed twice for ∼18 h against glycerol storage buffer (300 mM NaCl, 50 mM NaH2PO4, 50% [vol/vol] glycerol; pH 8.0) by using a Pierce Slyde-A-Lyzer cassette (10-kDa molecular mass cutoff; MJS BioLynx, Brockville, Ontario, Canada), and stored at −20°C. The protein concentration was determined by using the Bradford reagent (Bio-Rad Laboratories, Mississauga, Ontario, Canada) with bovine serum albumin as the standard. Based on the results of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the purified extract contained traces of contaminating proteins or degradation products (see Results). 3A). Therefore, as a control to confirm that CbaR caused the effects observed in gel shift assays (see below), an extract was prepared as described above from cells harboring the expression construct with no insert.
Gel shift assays.
Binding of CbaR to DNA containing the cbaA or cbaR transcriptional start site was characterized by gel shift assays using three DNA templates, shown schematically in Fig. 3B. In the initial assays we tested interactions with two templates, templates 1 and 2. Template 1 putatively contained the transcriptional start site of cbaR, which was roughly mapped in expression studies (see below), and spanned nucleotides (nt) 4098 (BglII site) to 4446 (NotI site). Template 2 contained the transcriptional start site of cbaA (determined by primer extension analysis [see below]), as well as sufficient upstream DNA so that cbaA expression was normal (as determined in expression studies with construct cbaA′-′lacZ1 [see below]). The latter sequence spanned nt 4446 (NotI site) to 4705 within cbaA (ScaI site). Templates 1 and 2 were obtained by digesting pMP14.10 with NotI and BamHI and gel purifying the ∼400- and ∼260-bp fragments, respectively. For more detailed gel shift studies we used a 186-bp fragment (molecular weight, 114,000) that was similar to template 2 but slightly shorter at both ends (Fig. 3B). The sequence of this third template, template 3, is shown in Fig. 2B, and it was generated by PCR with primers PCBA4480B (5′-CTCGGGTAAACACCTAGA-3′) and CBAA4665 (5′-TCGCACCAAATCTTCGT-3′). The amplification product was agarose gel purified and quantified by UV spectroscopy. All templates were end labeled with digoxigenin-11-ddUTP by using terminal transferase as recommended in the instructions for a DIG Gel Shift kit (Roche Molecular Biochemicals, Laval, Québec, Canada). Templates were separated from unincorporated label by spin column chromatography (Qiagen) and treated as described below with respect to binding.
FIG. 3.
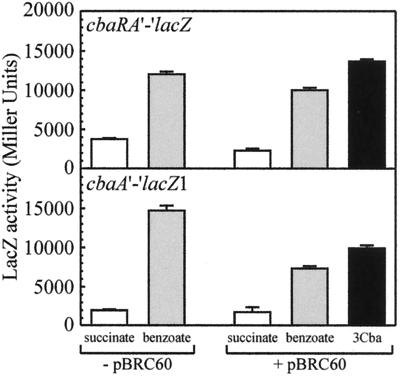
Levels of cbaA expression in C. testosteroni BR6020 harboring different lacZ constructs. Expression was measured with a growth substrate that did not induce 3CBA degradation activity (succinate) or did induce this activity (benzoate or 3CBA) in Tn5271-deficient cells (−pBRC60) and Tn5271-proficient cells (+pBRC60). The data are averages based on three independent trials; the error bars indicate standard errors. 3Cba, 3-chlorobenzoate.
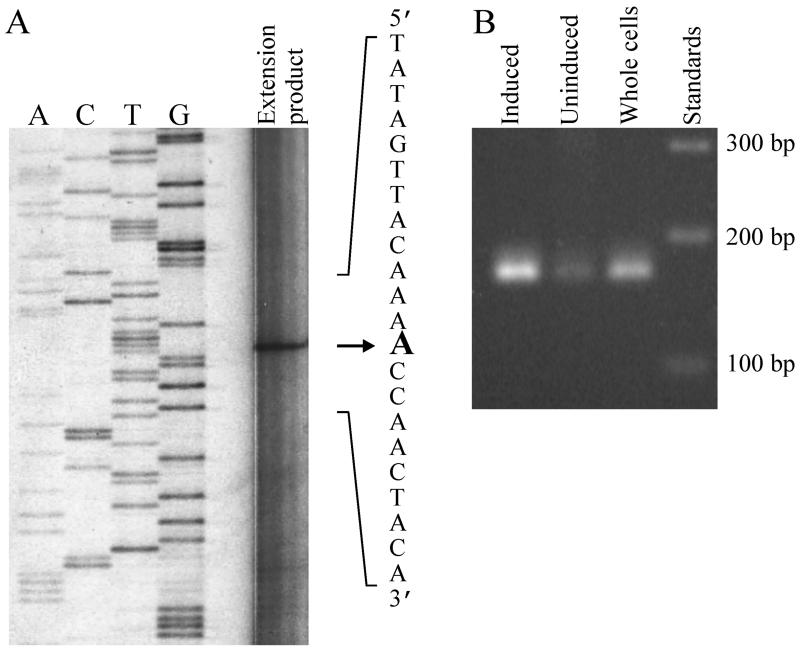
FIG. 2.
(A) Transcriptional initiation site of cbaA, as determined by primer extension analysis of total RNA isolated from 3CBA-grown C. testosteroni BR60(pBRC60). The extension product was loaded onto a sequencing gel next to four sequencing reaction mixtures (lanes A, C, T, and G) from reactions conducted with the same primer. The primer extension lane was digitally manipulated to enhance image quality. (B) RT-PCR assay to compare levels of cbaA mRNA in succinate-grown (uninduced) and 3CBA-grown (induced) BR60(pBRC60). The RT step was performed with the primer used for primer extension analysis (see Materials and Methods). For reference purposes, the PCR signal obtained with whole cells is also shown. The lengths of DNA size standards are indicated on the right.
DNA binding assays were performed in a total volume of 10 μl, and the reaction mixtures contained 2 μl of 5× binding buffer [100 mM HEPES (pH 7.6), 5 mM EDTA, 50 mM (NH4)2SO4, 5 mM dithiothreiotol, 1% (wt/vol) Tween 20, 150 mM KCl], 0.5 μl of poly(dI-dC) (1 μg/μl), 0.5 μl of poly-l-lysine (0.1 μg/μl; Roche Molecular Biochemicals), 4 ng of digoxigenin-labeled template DNA, various amounts of amendments depending on the experiment (see below), and 1 to 3 μl of CbaR in glycerol storage buffer. Glycerol storage buffer containing no CbaR was added as required so that the final concentration of glycerol was 15% (vol/vol), and sterile distilled H2O was added as required to adjust the volume of the reaction mixture to the desired value. The final concentration of glycerol in the binding reaction mixtures made loading buffer for subsequent electrophoresis unnecessary. Samples were kept at room temperature for 15 to 30 min and placed on ice for at least 5 min, and then a 4- to 5-μl aliquot of each binding reaction mixture was applied to a nondenaturing polyacrylamide gel (10 by 10 by 0.08 cm) at 4°C that had been prerun for 1 to 2 h at a constant current in a running buffer containing Tris base, boric acid, and EDTA (TBE) (final concentrations, 22.25, 22.25, and 0.5 mM, respectively) (pH 8.0). The first one-third of the gel (stacking gel) contained 3.5% (wt/vol) polyacrylamide, and the last two-thirds (separating gel) contained 12% (wt/vol) polyacrylamide; both parts of the gel contained TBE at the same concentration as the running buffer. Samples were resolved by electrophoresis at 10 V/cm for 3 to 4 h and then transferred to a positively charged nylon membrane (Roche Molecular Biochemicals) by electroblotting with TBE for 60 to 90 min at 300 mA in a Panther Semi-Dry electroblotter (Owl Separation Systems, Portsmouth, N.H.). DNA was detected by chemiluminescence with an antidigoxigenin system used as recommended by the manufacturer (Roche Molecular Biochemicals).
DNase I protection assays.
The specific areas bound by CbaR were determined by DNase I protection assays. The two templates ([32P]PCBA and [32P]CBAA) were identical to the 186-bp gel shift template described above, except that one of the primers used during amplification had been previously radioactively end labeled using [γ-32P]ATP (Amersham Canada Ltd.) and T4 polynucleotide kinase so that only the noncoding strand ([32P]PCBA) or the coding strand ([32P]CBAA) was phosphorylated. Following amplification, PCR products were separated from unincorporated label by spin column chromatography (Qiagen). 32P-labeled DNA templates (∼6 ng) were bound to various amounts of CbaR with and without 4 mM 3CBA or 4 mM 3-carboxybenzoate (see below) as described above for the gel shift assays; the only differences were that the binding buffer contained 5 mM MgCl2 and 2.5 mM CaCl2 and the reaction volume was 12 μl. After binding, 2.5 × 10−3 U of DNase I (Sigma-Aldrich) was added, and template DNA was digested at 22°C as follows. For [32P]PCBA, 4-μl samples were removed from the reaction mixture at 30, 60, and 120 s and combined into one tube containing 1.5 μl of stop solution (500 mM EDTA); for [32P]CBAA, 1.5 μl of stop solution was added after 60 s of digestion. Three microliters of a glycerol loading buffer was then added, and samples were placed in a boiling water bath for 3 min. A 3-μl aliquot was applied to a denaturing sequencing gel (8% [wt/vol] polyacrylamide, 8 M urea), and sequencing ladders (generated with the appropriate primer using pMP14.10 as the template) were included in adjacent lanes. Fragments were resolved electrophoretically and visualized by autoradiography.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in the GenBank database under accession numbers U18133 (for CbaR) and AF345907 (for 16S ribosomal DNA).
RESULTS
Classification of strain BR60(pBRC60).
The identity of 3CBA-degrading strain BR60(pBRC60), originally described as an Alcaligenes sp. strain (67), was not firmly established, and we therefore sequenced a 523-nt portion of the 16S rRNA gene. This portion of the 16S rRNA gene was identical to the corresponding portion of the 16S rRNA gene of the C. testosteroni type strain, and because BR60(pBRC60) has several genetic and metabolic similarities to other C. testosteroni strains (see below), Alcaligenes sp. strain BR60 is reclassified as C. testosteroni BR60.
Identification of cbaR and determination of cbaA transcriptional start.
BLAST analysis of DNA upstream of Tn5271 revealed the presence of a divergently oriented ORF beginning 667 bp upstream of cbaA (Fig. 1). This ORF was designated cbaR and codes for a 19.4-kDa protein that exhibits identity to various members of the MarR family of transcriptional regulators (Table 2). Because many proteins belonging to this family respond to specific aromatic compounds or are involved in regulating genes for metabolism of aromatic compounds, we focused on the effect of CbaR on expression of cbaA (see below).
TABLE 2.
Proteins in the GenBank database showing identity to CbaRa
| Protein | Accession no. | % Identity to CbaR | Organism | Function and effectors, if known | Reference(s) |
|---|---|---|---|---|---|
| BadR | U75363 | NAb | Rhodopseudomonas palustris CGA009 | Activates genes required for anaerobic growth on benzoate; may respond to benzoyl coenzyme A | 15, 16 |
| CinR | U64802 | NA | Butyrivibrio fibrisolvens E14 | Represses genes for cinnamoyl ester hydrolase; responds to feruloyl-containing sugars | 11 |
| EmrR | U19993 | 20 | Escherichia coli | Represses multidrug resistance pump genes; responds to 2,4-dinitrophenol | 7, 34 |
| HpcR | S56952 | 19 | Escherichia coli C | Represses homoprotocatechuate catabolic genes | 25, 52 |
| MarR | M96235 | 28 | Escherichia coli | Represses multiple antibiotic resistance genes; responds to 2-hydroxybenzoate and 2,4-dinitrophenol | 2, 6, 56 |
| MexR | U23763 | 28 | Pseudomonas aeruginosa | Represses multiple antibiotic resistance genes; may respond to chloramphenicol | 17, 48, 57 |
| NhhD | D67027 | NA | Rhodococcus rhodochrous JI | Activates nitrile hydratase genes | 30 |
| PecS | X74409 | 22 | Erwinia chrysanthemi | Represses pectinase and cellulase genes | 51 |
| SlyA | U03842 | 25 | Salmonella enterica serovar Typhimurium | Activates genes required for hemolysis and infection of and survival in mouse macrophages | 8, 33, 43 |
Various uncharacterized ORFs showing identity to CbaR were also identified, but they were not included as they had no known function.
NA, not applicable. A BLASTP search with CbaR as the query did not identify this protein as a CbaR homologue. However, the protein is included because it is a MarR family member based on sequence similarity determined by using homologues of CbaR as the query in a second round of BLASTP analysis.
The transcriptional start site of cbaA was determined in order to better characterize the promoter of this gene. As determined by primer extension analysis with RNA from 3CBA-grown cells, this transcriptional start site is an adenine 99 nt upstream of the putative start codon (Fig. 2A). A hexamer starting at position −12, TATAGT, is similar to the consensus enterobacterial −10 hexamer for ς70-dependent genes (TATAAT), indicating that cbaA possesses a ς70-like promoter. However, no sequence similar to TTGACA, the consensus enterobacterial −35 hexamer, was detected. An RT-PCR assay was conducted to compare levels of cbaA mRNA in uninduced cells (succinate grown) and induced cells (3CBA grown), and the intensity of the signal obtained with the former cells was much lower than the intensity obtained with the latter cells (Fig. 2B), indicating that growth on 3CBA leads to an increase in cbaA transcription. This experiment also showed that CBAAR, the primer used in the RT step of these two assays, was specific for cbaA mRNA.
Induction of 3CBA degradation activity and expression of cbaA
In order to determine which growth substrates induced 3CBA degradation activity, we initially investigated control of 3CBA catabolism in C. testosteroni BR60(pBRC60) by performing O2 uptake assays with whole cells, and the results are shown in Table 3. As expected, maximal 3CBA degradation activity was induced by 3CBA. Benzoate, which is not a substrate of this pathway and is not channeled via the PCA extradiol pathway (50), gratuitously induced 3CBA degradation activity (∼29% of the maximum activity). Similarly, benzoate degradation activity was gratuitously induced by 3CBA (∼22% of the maximum activity observed following growth on benzoate). Succinate, a nonaromatic carbon source, did not induce 3CBA activity, nor did 3-hydroxybenzoate, 4-hydroxybenzoate, and PCA, three aromatic carbon sources that are channeled through the PCA extradiol ring fission pathway, suggesting that the cba pathway, at least with respect to 3CBA, responds to the parent substrate but not to downstream metabolites that are common to these degradative pathways (i.e., PCA or the ring fission products). We observed good reproducibility between trials for all cultures except those grown on benzoate. For unknown reasons, cells exhibited variable lag phases (18 to 36 h) in the three trials before the onset of logarithmic growth. This apparently affected O2 uptake rates with benzoate but not with the other carbon sources (data not shown). The results presented here are the results for the trial in which the log phase was achieved within 18 h, the time necessary for the other growth substrates.
TABLE 3.
Rates of oxygen consumption by C. testosteroni BR60(pBRC60) grown on one substrate and exposed to various other carbon sources
| Growth substrate | O2 consumption rate (mg of O2 liter−1 min−1) when exposed to a:
|
|||||
|---|---|---|---|---|---|---|
| 3CBA | Benzoate | 3-Hydroxybenzoate | 4-Hydroxybenzoate | PCA | Succinate | |
| 3CBA | 1.10 (0.05) | 0.69 (0.02) | 0.06 (0.01) | 0.40 (0.03) | 1.36 (0.05) | 0.13 (0.01) |
| Benzoate | 0.32 (0.04) | 3.08 (0.31) | 0.13 (0.00) | 0.38 (0.06) | NDb | 0.08 (0.03) |
| 3-Hydroxybenzoate | <0.05 | <0.05 | 3.07 (0.20) | <0.05 to 0.60 (0.01)c | 1.36 (0.04) | 0.35 (0.02) |
| 4-Hydroxybenzoate | <0.05 | <0.05 | <0.05 to 0.40 (0.01)d | 3.51 (0.13) | 1.47 (0.04) | 0.22 (0.02) |
| PCA | <0.05 | <0.05 | <0.05 | <0.05 | 1.15 (0.04) | 0.22 (0.01) |
| Succinate | <0.05 | <0.05 | <0.05 | <0.05 | ND | 0.74 (0.03) |
Rates were corrected for endogenous oxygen uptake. Averages (standard errors) based on the results of independent trials are shown.
ND, not determined. However, in benzoate-grown cultures the levels of PCA degradation activity were comparable to those in 3CBA-grown cultures (41).
In two of the three trials, levels of activity greater than 0.05 mg of O2 liter−1 min−1 were observed.
In one of the three trials, a level of activity greater than 0.05 mg of O2 liter−1 min−1 was observed; the value in parentheses is the standard deviation for the replicate samples of the culture in that trial.
Control of cbaABC-mediated catabolism of 3CBA was further characterized by studying cbaA expression in the presence and absence of CbaR. Expression levels were measured with strain BR6020 (Table 1) harboring the constructs cbaA′-′lacZ1 and cbaRA′-′lacZ (Fig. 1B) and grown on 3CBA or the gratuitous inducer benzoate (see above). The levels of expression were compared to those observed following growth on the noninducing substrate succinate (see above). We initially hypothesized that CbaR was a repressor and that in its absence we would observe constitutive (i.e., maximal) cbaA expression. The Tn5271-deficient genetic background provided by BR6020 allowed us to specifically test its role by eliminating trans-encoded CbaR, while reintroduction of pBRC60, which created a Tn5271-proficient background, restored all functions necessary for growth on 3CBA and created a strain that was essentially identical to the parent strain, BR60(pBRC60). The results are summarized in Fig. 3. In Tn5271-deficient cells, LacZ activity under noninducing conditions was roughly the same with both constructs. For benzoate-grown cells, the induction ratios compared to succinate-grown cells were 3.2- and 7.4-fold in BR6020 harboring cbaRA′-′lacZ and BR6020 harboring cbaA′-′lacZ1, respectively. From these data, it appeared that control of cbaA was not due to simple repression by CbaR and that CbaR might be a modulator of benzoate-induced activity. In addition, because cbaA was induced by benzoate in the absence of any pBRC60/Tn5271-encoded functions, our data suggested that there is a benzoate-responsive chromosomally encoded regulator.
In a Tn5271-proficient background, the LacZ activities under noninducing conditions were roughly the same with both constructs, and compared to succinate-grown cells, the induction ratios for benzoate-grown cells (4.1- to 4.2-fold) and 3CBA-grown cells (5.5- to 5.8-fold) were virtually the same with both constructs (Fig. 3). After pBRC60 was introduced to create a Tn5271-proficient background, the levels of cbaA expression compared to those in Tn5271-deficient cells tended to decrease with both constructs (Fig. 3). For succinate-grown cells the effect was insignificant to moderate (1.1- to 1.6-fold decreases), but for benzoate-grown cells harboring cbaA′-′lacZ1 the effect was relatively strong (2-fold decrease), probably because of trans-encoded CbaR from pBRC60. For benzoate-grown cells harboring cbaRA′-′lacZ, the effect was slight (1.2-fold), presumably because a cis-encoded CbaR was already provided by the construct. In BR6020 harboring the control construct (promoterless lacZ) and grown as described above in a Tn5271-deficient or Tn5271-proficient background, LacZ activity was negligible (∼1 Miller unit).
Expression of cbaR and gel shift and DNase I footprinting assays with affinity-purified CbaR.
In order to better understand the gene coding for CbaR, we investigated expression of cbaR with various LacZ constructs, not only to determine whether the levels of cbaR expression were affected by growth conditions and genetic backgrounds that affected cbaA expression but also to roughly map the transcription starting point. LacZ activity in BR6020 harboring cbaR′-′lacZ1 and cbaR′-′lacZ2 was very low (∼20 Miller Units), but compared to the control (see above) the activity was significant. These levels of expression were observed in Tn5271-deficient cells grown on succinate or benzoate and in Tn5271-proficient cells grown on succinate or 3CBA, implying that Tn5271-encoded functions, particularly CbaR, did not have an effect on cbaR expression. With BR6020 harboring cbaR′-′lacZ3 in both genetic backgrounds and grown under the same conditions, the activity was negligible (∼1 Miller unit). This implied that cbaR transcription began before nt 4098 (i.e., the BglII site defining the end of construct cbaR′-′lacZ3) but after nt 4446 (i.e., the NotI site defining the end of construct cbaR′-′lacZ2, in which expression was observed) (Fig. 1B).
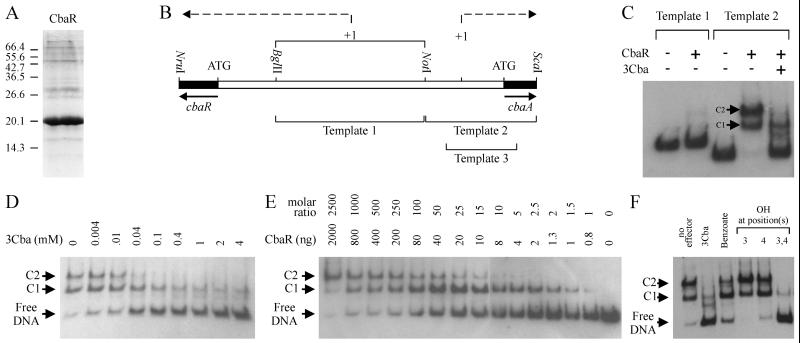
In an effort to determine if cbaR coded for a functional DNA binding protein, CbaR was tagged at the N terminus, overexpressed in E. coli, and affinity purified. As determined by SDS-PAGE analysis, affinity-purified extracts of isopropyl-β-d-thiogalactopyranoside (IPTG)-induced cultures of M15(pQE30cbaR, pREP4) contained large amounts of a protein that comigrated with the 20.1-kDa size standard (Fig. 4A), which is very similar to the predicted molecular mass of CbaR with the six-histidine tag (20.7 kDa). Initial gel shift assays with purified CbaR were then conducted to determine if CbaR bound DNA containing either the transcriptional start site of cbaR (template 1) or cbaA (template 2), and the results are shown in Fig. 4C. CbaR did not affect migration of template 1 but retarded migration of template 2.With template 2, two CbaR-DNA complexes that moved more slowly were observed; these complexes were designated C1 and C2, and addition of 4 mM 3CBA strongly disrupted their formation. If unlabeled template 2 was added as competitor DNA to a binding reaction mixture containing template 2, a shift towards less retarded DNA was observed (data not shown). As a control, the effect of an affinity-purified extract from E. coli containing the expression vector with no insert on migration of the two DNA templates was tested, and no change was observed (data not shown), indicating that the effects elicited by the extract were caused by CbaR.
FIG. 4.
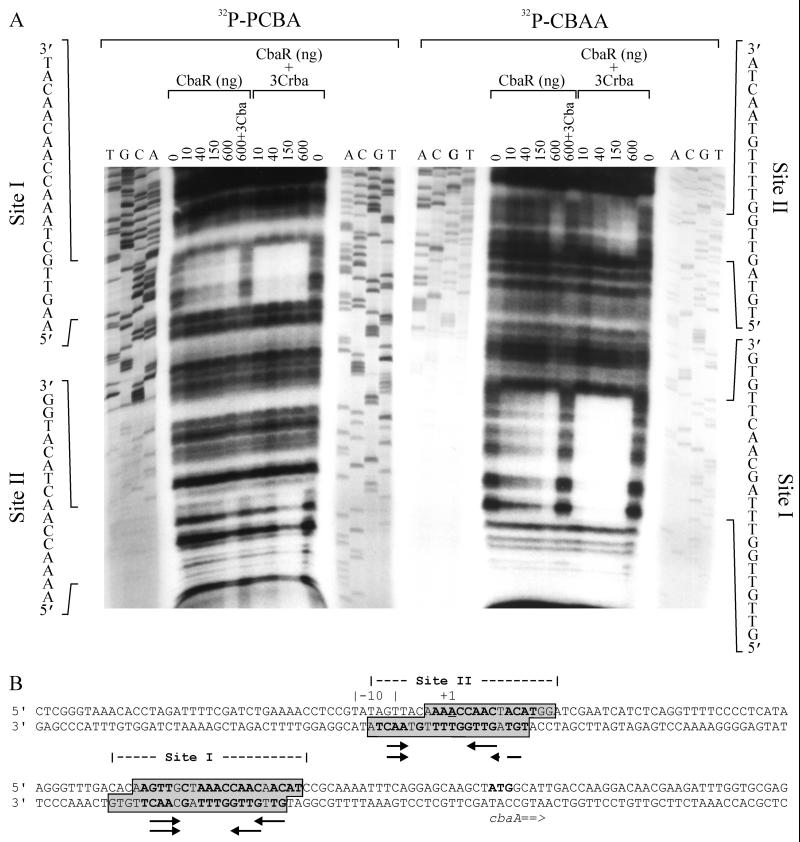
(A) Analysis of affinity-purified CbaR by SDS-PAGE. Ten micrograms of protein was loaded onto the gel. The positions of protein size standards (in kilodaltons) are indicated on the left. (B) Schematic diagram of the three DNA templates used in gel shift assays. Template 1 putatively contains the transcriptional start site of cbaR, while templates 2 and 3 harbor the transcriptional start site and promoter of cbaA (PcbaA). (C to F) Unless indicated otherwise, each binding reaction mixture contained 0.4 μg of CbaR, 4 ng of DNA, and an aromatic compound at a concentration of 4 mM. The positions of unbound template (Free DNA), CbaR-PcbaA DNA complex 1 (C1), and CbaR-PcbaA DNA complex 2 (C2) are indicated. Template 3 was used for all assays except those whose results are shown in panel C, which tested the effect of CbaR on the mobility of templates 1 and 2. For template 2, the effect of including 3CBA in the binding reaction mixture was also tested. (D) Assay to determine the minimum concentration at which 3CBA interfered with CbaR binding to PcbaA DNA. The concentration of 3CBA was varied, while the amounts of CbaR and DNA were kept constant. (E) Stoichiometry of CbaR-PcbaA DNA complex formation. The amount of CbaR per reaction mixture was varied, while the amount of DNA was kept constant. For reference purposes, the approximate molar ratios of CbaR to DNA are also shown. (F) Effects of benzoate and various hydroxylated benzoates on CbaR binding to PcbaA DNA. 3Cba, 3-chlorobenzoate.
More detailed gel shift assays to study interactions between CbaR and the cbaA promoter (PcbaA) were then performed by using template 3 (Fig. 4B). Experiments with a range of 3CBA concentrations revealed that significant disruption occurred with 40 μM 3CBA (Fig. 4D). The stoichiometry of C1 and C2 formation was also studied, and the results are shown in Fig. 4E. Small amounts of C1 appeared when CbaR and PcbaA DNA were present at approximately equimolar concentrations, and equal amounts of unbound DNA and C1 resulted when there was a 10-fold molar excess of CbaR. Small amounts of C2 were already apparent by this point, but equal amounts of C1 and C2 did not appear until there was a 250-fold molar excess of CbaR. C2 did not predominate until there was at least a 2,500-fold molar excess of CbaR. Because the template used in these assays contained two CbaR binding sites (see below), we were not able to unambiguously determine binding constants for PcbaA DNA. A final set of gel shift assays was conducted to determine whether other aromatic compounds disrupted the CbaR-PcbaA DNA complexes. We tested the effect of benzoate or one of the three hydroxylated benzoates used in the O2 uptake experiments (see above), and the results are presented in Fig. 4F. Benzoate was a weak disrupter (i.e., it caused a small shift of DNA towards less retarded forms); 4-hydroxybenzoate had no effect; PCA was a strong disrupter (i.e., it interfered with binding at levels comparable to the 3CBA levels); and unexpectedly, 3-hydroxybenzoate improved CbaR binding (i.e., it caused DNA to shift towards more retarded forms). The latter effect was also elicited by 3-carboxybenzoate (see below).
DNase I protection assays were then performed to determine the specific binding sites of CbaR on PcbaA, and the results are presented in Fig. 5A. Approximately 6 ng of DNA was used per reaction mixture, and according to gel shift data presented in Fig. 4E, the smallest amount of CbaR tested (10 ng or a ∼9-fold molar excess compared to the amount of PcbaA DNA) resulted mostly in C1 and very little C2, while the largest amount (600 ng or an ∼800-fold molar excess) resulted in approximately equal amounts of C1 and C2. Two protected regions were identified: site I (a higher-affinity site), which was clearly evident with 10 ng of CbaR, and site II (a lower-affinity site), which became apparent only with larger amounts of CbaR. If 3CBA was included in the binding reaction mixture, neither site was protected from DNase I digestion. If the binding enhancer 3-carboxybenzoate was included, protection of both sites was improved ∼60-fold (i.e., 10 ng of CbaR in the presence of 3-carboxybenzoate protected sites I and II as strongly as 600 ng of CbaR in the absence of 3-carboxybenzoate). Furthermore, it allowed us to more clearly define nucleotides protected within site II.
FIG. 5.
(A) DNase I protection assay to determine specific regions of PcbaA DNA protected by CbaR. Template DNA was end labeled with 32PO4 on the noncoding strand (32P-PCBA) or the coding strand (32P-CBAA). Various amounts of CbaR were added, and the effect of 4 mM 3CBA (3Cba) or 4 mM 3-carboxybenzoate (3Crba) on CbaR protection of two regions (sites I and II) was determined. A sequencing reaction conducted with the appropriate primers was included during electrophoretic resolution of DNase I-digested samples. (B) Sequences of both strands of the template DNA used for the assay, indicating the positions of sites I and II relative to the transcriptional start site of cbaA (position +1); the putative −10 hexamer; and the start codon of cbaA (ATG). Inverted repeats found in site I and modified versions of these repeats in site II are indicated by arrows, and bases present in both sites are indicated by boldface type.
Site II overlaps the transcription starting point (position +1) of cbaA, and site I is approximately 40 nt downstream (Fig. 5B). Site I possesses a 4-nt inverted repeat (IR) separated by 6 or 9 nt (5′-GTTG[N]6/9CAAC-3′). Modified forms of the two IRs are present in the lower strand of site II (5′-GTTG[N]6TAAC-3′ or 5′-GTAG[N]9TAAC-3′).
DISCUSSION
Classification of strain BR60(pBRC60).
We reclassified Alcaligenes sp. strain BR60(pBRC60) as C. testosteroni BR60(pBRC60) based on several metabolic and genetic similarities between this bacterium and various other strains of the β-proteobacterium C. testosteroni (59, 62). The metabolic similarities include an inability to grow on various carbohydrates (67); induction of the PCA meta ring fission enzymes by growth on 3-hydroxybenzoate, 4-hydroxybenzoate, benzoate, or PCA (41); and a lower growth rate with benzoate than with hydroxylated benzoates (62; Providenti, unpublished data). The genetic similarities are the presence of the same partial 16S ribosomal DNA sequence (see above) and highly homologous genes for PCA meta ring fission pathway enzymes (J. Mampel, M. A. Providenti, R. C. Wyndham, and A. M. Cook, unpublished data).
Control of 3CBA degradation activity.
We initially hypothesized that cbaABC was negatively regulated by CbaR, which was consistent with the predominant role of the MarR family of proteins (Table 2) (see below), but studies with the cbaA expression constructs implied that the major role of CbaR may be to modulate gratuitous induction by compounds like benzoate (see above). Gel shift experiments showing that benzoate only weakly disrupted CbaR binding to PcbaA supported this model (Fig. 4F). Furthermore, previous work that showed that the cbaR region is not required for cbaABC-mediated growth on 3CBA (41) suggested that CbaR is not essential for cbaA expression. Because studies with Tn5271-deficient cells harboring construct cbaA′-′lacZ1 showed that growth on benzoate (Fig. 3) or growth on succinate in the presence of 3CBA (data not shown) induced cbaA expression, we propose that there is a chromosomally encoded regulator that, along with CbaR, controls cbaABC. 3CBA and benzoate gratuitously activate each other's pathways and exhibit similar induction spectra with respect to 3-hydroxybenzoate, 4-hydroxybenzoate, and PCA (Table 3), and we therefore hypothesize that there is a shared regulator for the two pathways. The cbaABC regulatory system may thus have similarities to CatR activation of both the chromosomally encoded catBCA operon for catechol metabolism and the plasmid-encoded pheBA operon for phenol metabolism (26, 44). Alternatively, we may have observed a cross-activation phenomenon, like induction of the lower pathway of the plasmid-encoded xyl genes in response to benzoate despite the absence of XylS, the normal activator (10, 24, 28), or CatR- and ClcR-mediated cross-activation of the clc- and cat-encoded catabolic pathways (37, 45).
CbaR: expression of the gene and functional characterization.
The gene that encodes CbaR appears to be expressed at a very low level (see above), and neither benzoate nor 3CBA, two growth substrates that increased cbaA expression, altered the level of cbaR expression. In addition, CbaR itself does not appear to exert control over expression of its own gene. This conclusion is suggested by the observation that the levels of cbaR expression were the same regardless of the presence of Tn5271, which provided a trans-encoded CbaR (see above), and the observation that CbaR did not bind to DNA spanning the region putatively containing the cbaR transcription start site (see above) (Fig. 4C). Furthermore, no sequences similar to the two binding sites of CbaR were detected when the complete region upstream of cbaA was analyzed.
CbaR bound the cbaA promoter at two sites and exhibited different affinities for these sites, protecting site I strongly and site II less strongly (Fig. 5B). Sites I and II have many identical bases, and IR structures were detected in site I (Fig. 5B). Although a direct role for the IR structures has not been determined, it is noteworthy that many of the sequence differences between sites I and II occur in the possible modified forms of the IR structures in site II (Fig. 5B), which may explain the different binding affinities. By correlating gel shift stoichiometry data (Fig. 4F) with DNase I protection results (Fig. 5A), it was shown that C1 represents CbaR bound to site I while C2 represents CbaR bound to both site I and site II. Presumably, independent CbaR proteins bind to each site, but this remains to be shown. When binding inhibitors are present, binding to site II is affected first, as disappearance of C2 always precedes disappearance of C1 (Fig. 4D), presumably because the lower affinity of CbaR for site II makes it more susceptible to binding disrupters. This may allow CbaR-mediated repression at site II, which overlaps the transcription start site of cbaA (Fig. 5A), to be overcome more easily and is probably an adaptation that increases the sensitivity of the cba pathway to potential substrates and allows it to respond to the concentrations of CBA (which are expressed in parts per million) typically encountered in contaminated environments (67). Our gel shift assays showing that 40 μM 3CBA (∼6 ppm) disrupted binding (Fig. 4D) support this view.
A brief survey of the effects of other aromatic compounds on CbaR binding identified PCA as a strong disrupter of the CbaR-PcbaA complex (Fig. 4F), suggesting that CbaR evolved so that it responds to both 3CBA, the substrate of the cba pathway, and at least one downstream metabolite. This may be a positive feedback mechanism that ensures derepression of cbaABC under conditions in which PCA is being produced because of CBA metabolism. Unexpectedly, two meta-substituted benzoates (3-hydroxybenzoate and 3-carboxybenzoate) improved CbaR binding to PcbaA DNA (Fig. 4F and 5A). To our knowledge, this is the first time that this phenomenon has been reported for a MarR-like protein, and we are currently exploring the effect in greater detail and studying its physiological significance. Conceivably, improved binding by CbaR could lead to increased repression of cbaA, a phenomenon observed with MarR mutant proteins that bind more strongly to their cognate operator and superrepress marRAB (3).
As a member of the MarR family of regulators, CbaR belongs to a diverse group of proteins that control a variety of microbial functions, including antibiotic resistance, catabolism of various substrates, plant pathogenicity, and animal virulence (Table 3). On a functional level, CbaR exhibits some similarities to MarR, PecS, and MexR, which form two or three protein-DNA complexes in a concentration-dependent fashion when they are combined with DNA containing the appropriate cognate promoter; the number of complexes reflects the number of binding sites (17, 35, 36, 49, 56). Furthermore, the binding sites of MexR (17) contain an inverted repeat structure (5′-GTTGA[N]6TCAAC-3′) that is extremely similar to one of the possible inverted repeats detected in the higher-affinity binding site of CbaR (see above). Otherwise, the binding sites for CbaR, MarR (36), PecS (49), and MexR (17) differ with respect to sequence, size, spacing relative to each other, and position relative to the transcriptional start site and start codons for target genes. For the most part, regulators belonging to the MarR family are repressors, but BadR, NhdR, and SlyA are activators of the operons that they control (15, 30, 43). The organizations of genes that encode MarR family proteins relative to the genes that they control and the methods by which expression of the gene that encodes a MarR family protein is regulated vary substantially. Interesting contrasts include genes that encode MarR-like proteins that are divergently transcribed from their target genes, including hpcR, mexR, nhhD, and cbaR (29, 48, 52; this study); that are part of the same operon, including emrR and marR (4, 34); or that are part of a different operon but are transcribed in the same direction, including badR (15). Expression of the genes that encode some MarR-like regulators is subject to induction by other regulators; these genes include nhhD, which is induced by NhhC (29), and marR, which is induced by MarA (4). In addition, autorepression has been demonstrated for emrR, mexR, and marR (4, 34, 48). For the most part, MarR-like proteins appear to act alone on target genes, but in some cases they act in concert with other regulators. An example of this is BadR, which works together with AadR to fully activate the bad genes of Rhodopseudomonas palustris (15). Similarly, PecS is part of an intricate regulatory network which, in concert with a variety of other regulators (both repressors and activators), controls or attenuates expression of a host of physically unlinked operons spread over the Erwinia chrysanthemi chromosome (23, 54). Despite the diversity of the MarR-like proteins, shared features of these proteins include an ability to respond to aromatic compounds (Table 3) and a conserved motif in the center of the protein (58) whose function may be to bind DNA (1).
In conclusion, we found that the cba catabolic pathway of C. testosteroni BR60(pBRC60) is controlled in part by CbaR, a cis-encoded 3CBA-responsive MarR-like regulator whose major role may be to modulate gratuitous induction by benzoate or compounds like benzoate. There also appears to be an unidentified, chromosomally encoded benzoate-3CBA-responsive regulator that induces cbaABC, and we are currently attempting to isolate this protein in order to better understand factors that control cbaA expression.
ACKNOWLEDGMENTS
We thank Suzanne Paterson for invaluable assistance with primer extension and DNase I footprinting assays, Barb Holland for construction of several clones, Christina Matula for generation of some transconjugants, and Alasdair M. Cook and Tewes Tralau for assistance with RT-PCR.
M.A.P. was the recipient of an Ontario Graduate Scholarship and an Alexander von Humboldt Fellowship. This work was supported by a Natural Sciences and Engineering Research Council of Canada grant to R.C.W.
REFERENCES
- 1.Alekshun M N, Kim Y S, Levy S B. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol Microbiol. 2000;35:1394–1404. doi: 10.1046/j.1365-2958.2000.01802.x. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun M N, Levy S B. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol. 1999;181:4669–4672. doi: 10.1128/jb.181.15.4669-4672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alekshun M N, Levy S B. Characterization of MarR superrepressor mutants. J Bacteriol. 1999;181:3303–3306. doi: 10.1128/jb.181.10.3303-3306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alekshun M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ariza R R, Cohen S P, Bachhawat N, Levy S B, Demple B. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1994;176:143–148. doi: 10.1128/jb.176.1.143-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooun A, Tomashek J J, Lewis K. Purification and ligand binding of EmrR, a regulator of a multidrug transporter. J Bacteriol. 1999;181:5131–5133. doi: 10.1128/jb.181.16.5131-5133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchmeier N, Bossie S, Chen C Y, Fang F C, Guiney D G, Libby S J. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect Immun. 1997;65:3725–3730. doi: 10.1128/iai.65.9.3725-3730.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coco W M, Rothmel R K, Henikoff S, Chakrabarty A M. Nucleotide sequence and initial functional characterization of the clcR gene encoding a LysR family activator of the clcABD chlorocatechol operon in Pseudomonas putida. J Bacteriol. 1993;175:417–427. doi: 10.1128/jb.175.2.417-427.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuskey S M, Sprenkle A B. Benzoate-dependent induction from the OP2 operator-promoter region of the TOL plasmid pWWO in the absence of known plasmid regulatory genes. J Bacteriol. 1988;170:3742–3746. doi: 10.1128/jb.170.8.3742-3746.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalrymple B P, Swadling Y. Expression of a Butyrivibrio fibrisolvens E14 gene (cinB) encoding an enzyme with cinnamoyl ester hydrolase activity is negatively regulated by the product of an adjacent gene (cinR) Microbiology. 1997;143:1203–1210. doi: 10.1099/00221287-143-4-1203. [DOI] [PubMed] [Google Scholar]
- 12.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 14.Di Gioia D, Peel M, Fava F, Wyndham R C. Structures of homologous composite transposons carrying cbaABC genes from Europe and North America. Appl Environ Microbiol. 1998;64:1940–1946. doi: 10.1128/aem.64.5.1940-1946.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egland P G, Harwood C S. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, an Fnr family member. J Bacteriol. 1999;181:2102–2109. doi: 10.1128/jb.181.7.2102-2109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egland P G, Pelletier D A, Dispensa M, Gibson J, Harwood C S. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc Natl Acad Sci USA. 1997;94:6484–6489. doi: 10.1073/pnas.94.12.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans K, Adewoye L, Poole K. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J Bacteriol. 2001;183:807–812. doi: 10.1128/JB.183.3.807-812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frantz B, Chakrabarty A M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci USA. 1987;84:4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulthorpe R R, Wyndham R C. Involvement of a chlorobenzoate-catabolic transposon, Tn5271, in community adaptation to chlorobiphenyl, chloroaniline, and 2,4-dichlorophenoxyacetic acid in a freshwater ecosystem. Appl Environ Microbiol. 1992;58:314–325. doi: 10.1128/aem.58.1.314-325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fulthorpe R R, Wyndham R C. Transfer and expression of the catabolic plasmid pBRC60 in wild bacterial recipients in a freshwater ecosystem. Appl Environ Microbiol. 1991;57:1546–1553. doi: 10.1128/aem.57.5.1546-1553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haak B, Fetzner S, Lingens F. Cloning, nucleotide sequence, and expression of the plasmid-encoded genes for the two-component 2-halobenzoate 1,2-dioxygenase from Pseudomonas cepacia 2CBS. J Bacteriol. 1995;177:667–675. doi: 10.1128/jb.177.3.667-675.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Regulation of pectinolysis in Erwinia chrysanthemi. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 24.Jeffrey W H, Cuskey S M, Chapman P J, Resnick S, Olsen R H. Characterization of Pseudomonas putida mutants unable to catabolize benzoate: cloning and characterization of Pseudomonas genes involved in benzoate catabolism and isolation of a chromosomal DNA fragment able to substitute for xylS in activation of the TOL lower-pathway promoter. J Bacteriol. 1992;174:4986–4996. doi: 10.1128/jb.174.15.4986-4996.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins J R, Cooper R A. Molecular cloning, expression, and analysis of the genes of the homoprotocatechuate catabolic pathway of Escherichia coli C. J Bacteriol. 1988;170:5317–5324. doi: 10.1128/jb.170.11.5317-5324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasak L, Horak R, Nurk A, Talvik K, Kivisaar M. Regulation of the catechol 1,2-dioxygenase- and phenol monooxygenase-encoding pheBA operon in Pseudomonas putida PaW85. J Bacteriol. 1993;175:8038–8042. doi: 10.1128/jb.175.24.8038-8042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 28.Kessler B, Marques S, Kohler T, Ramos J L, Timmis K N, de Lorenzo V. Cross talk between catabolic pathways in Pseudomonas putida: XylS-dependent and -independent activation of the TOL meta operon requires the same cis-acting sequences within the Pm promoter. J Bacteriol. 1994;176:5578–5582. doi: 10.1128/jb.176.17.5578-5582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi M, Shimizu S. Metalloenzyme nitrile hydratase: structure, regulation, and application to biotechnology. Nat Biotechnol. 1998;16:733–736. doi: 10.1038/nbt0898-733. [DOI] [PubMed] [Google Scholar]
- 30.Komeda H, Kobayashi M, Shimizu S. Characterization of the gene cluster of high-molecular-mass nitrile hydratase (H-NHase) induced by its reaction product in Rhodococcus rhodochrous J1. Proc Natl Acad Sci USA. 1996;93:4267–4272. doi: 10.1073/pnas.93.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krooneman J, Moore E R B, Van Velzen J C L, Prins R A, Forney L J, Gottschal J C. Competition for oxygen and 3-chlorobenzoate between two aerobic bacteria using different degradation pathways. FEMS Microbiol Ecol. 1998;26:171–179. [Google Scholar]
- 32.Krooneman J, Wieringa E B, Moore E R, Gerritse J, Prins R A, Gottschal J C. Isolation of Alcaligenes sp. strain L6 at low oxygen concentrations and degradation of 3-chlorobenzoate via a pathway not involving (chloro)catechols. Appl Environ Microbiol. 1996;62:2427–2434. doi: 10.1128/aem.62.7.2427-2434.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Libby S J, Goebel W, Ludwig A, Buchmeier N, Bowe F, Fang F C, Guiney D G, Songer J G, Heffron F. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc Natl Acad Sci USA. 1994;91:489–493. doi: 10.1073/pnas.91.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lomovskaya O, Lewis K, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin R G, Jair K W, Wolf R E, Jr, Rosner J L. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J Bacteriol. 1996;178:2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin R G, Rosner J L. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McFall S M, Klem T J, Fujita N, Ishihama A, Chakrabarty A M. DNase I footprinting, DNA bending and in vitro transcription analyses of ClcR and CatR interactions with the clcABD promoter: evidence of a conserved transcriptional activation mechanism. Mol Microbiol. 1997;24:965–976. doi: 10.1046/j.1365-2958.1997.4041763.x. [DOI] [PubMed] [Google Scholar]
- 38.Nakatsu C, Ng J, Singh R, Straus N, Wyndham C. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc Natl Acad Sci USA. 1991;88:8312–8316. doi: 10.1073/pnas.88.19.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakatsu C H, Fulthorpe R R, Holland B A, Peel M C, Wyndham R C. The phylogenetic distribution of a transposable dioxygenase from the Niagara River watershed. Mol Ecol. 1995;4:593–603. doi: 10.1111/j.1365-294x.1995.tb00259.x. [DOI] [PubMed] [Google Scholar]
- 40.Nakatsu C H, Providenti M, Wyndham R C. The cis-diol dehydrogenase cbaC gene of Tn5271 is required for growth on 3-chlorobenzoate but not 3,4-dichlorobenzoate. Gene. 1997;196:209–218. doi: 10.1016/s0378-1119(97)00229-1. [DOI] [PubMed] [Google Scholar]
- 41.Nakatsu C H, Wyndham R C. Cloning and expression of the transposable chlorobenzoate-3,4-dioxygenase genes of Alcaligenes sp. strain BR60. Appl Environ Microbiol. 1993;59:3625–3633. doi: 10.1128/aem.59.11.3625-3633.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng J, Wyndham R C. IS1071-mediated recombinational equilibrium in Alcaligenes sp. BR60 carrying the 3-chlorobenzoate catabolic transposon Tn5271. Can J Microbiol. 1993;39:92–100. [Google Scholar]
- 43.Oscarsson J, Mizunoe Y, Uhlin B E, Haydon D J. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol Microbiol. 1996;20:191–199. doi: 10.1111/j.1365-2958.1996.tb02500.x. [DOI] [PubMed] [Google Scholar]
- 44.Parsek M R, Kivisaar M, Chakrabarty A M. Differential DNA bending introduced by the Pseudomonas putida LysR-type regulator, CatR, at the plasmid-borne pheBA and chromosomal catBC promoters. Mol Microbiol. 1995;15:819–828. doi: 10.1111/j.1365-2958.1995.tb02352.x. [DOI] [PubMed] [Google Scholar]
- 45.Parsek M R, McFall S M, Shinabarger D L, Chakrabarty A M. Interaction of two LysR-type regulatory proteins CatR and ClcR with heterologous promoters: functional and evolutionary implications. Proc Natl Acad Sci USA. 1994;91:12393–12397. doi: 10.1073/pnas.91.26.12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peel M C, Wyndham R C. The impact of industrial contamination on microbial chlorobenzoate degradation in the Niagara watershed. Microb Ecol. 1997;33:59–68. doi: 10.1007/s002489900008. [DOI] [PubMed] [Google Scholar]
- 47.Peel M C, Wyndham R C. Selection of clc, cba, and fcb chlorobenzoate-catabolic genotypes from groundwater and surface waters adjacent to the Hyde Park, Niagara Falls, chemical landfill. Appl Environ Microbiol. 1999;65:1627–1635. doi: 10.1128/aem.65.4.1627-1635.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D E, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Praillet T, Nasser W, Robert-Baudouy J, Reverchon S. Purification and functional characterization of PecS, a regulator of virulence-factor synthesis in Erwinia chrysanthemi. Mol Microbiol. 1996;20:391–402. doi: 10.1111/j.1365-2958.1996.tb02626.x. [DOI] [PubMed] [Google Scholar]
- 50.Providenti M A, Mampel J, MacSween S, Cook A M, Wyndham R C. Comamonas testosteroni BR6020 possesses a single genetic locus for extradiol cleavage of protocatechuate. Microbiology. 2001;147:2157–2167. doi: 10.1099/00221287-147-8-2157. [DOI] [PubMed] [Google Scholar]
- 51.Reverchon S, Nasser W, Robert-Baudouy J. pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol Microbiol. 1994;11:1127–1139. doi: 10.1111/j.1365-2958.1994.tb00389.x. [DOI] [PubMed] [Google Scholar]
- 52.Roper D I, Fawcett T, Cooper R A. The Escherichia coli C homoprotocatechuate degradative operon: hpc gene order, direction of transcription and control of expression. Mol Gen Genet. 1993;237:241–250. doi: 10.1007/BF00282806. [DOI] [PubMed] [Google Scholar]
- 53.Rothmel R K, Chakrabarty A M, Berry A, Darzins A. Genetic systems in Pseudomonas. Methods Enzymol. 1991;204:485–514. doi: 10.1016/0076-6879(91)04025-j. [DOI] [PubMed] [Google Scholar]
- 54.Rouanet C, Nomura K, Tsuyumu S, Nasser W. Regulation of pelD and pelE, encoding major alkaline pectate lyases in Erwinia chrysanthemi: involvement of the main transcriptional factors. J Bacteriol. 1999;181:5948–5957. doi: 10.1128/jb.181.19.5948-5957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 56.Seoane A S, Levy S B. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J Bacteriol. 1995;177:3414–3419. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srikumar R, Paul C J, Poole K. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol. 2000;182:1410–1414. doi: 10.1128/jb.182.5.1410-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sulavik M C, Gambino L F, Miller P F. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995;1:436–446. [PMC free article] [PubMed] [Google Scholar]
- 59.Tamaoka J, Ha D M, Komagata K. Reclassification of Pseudomonas acidovorans den Dooren de Jong 1926 and Pseudomonas testosteroni Marcus and Talalay 1956 as Comamonas acidovorans comb. nov. and Comamonas testosteroni comb. nov., with an emended description of the genus Comamonas. Int J Syst Bacteriol. 1987;37:52–59. [Google Scholar]
- 60.Tsoi T V, Plotnikova E G, Cole J R, Guerin W F, Bagdasarian M, Tiedje J M. Cloning, expression, and nucleotide sequence of the Pseudomonas aeruginosa 142 ohb genes coding for oxygenolytic ortho dehalogenation of halobenzoates. Appl Environ Microbiol. 1999;65:2151–2162. doi: 10.1128/aem.65.5.2151-2162.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsoi T V, Zaitsev G M, Plotnikova E G, Kosheleva I A, Boronin A M. Cloning and expression of the Arthrobacter globiformis KZT1 fcbA gene encoding dehalogenase (4-chlorobenzoate-4-hydroxylase) in Escherichia coli. FEMS Microbiol Lett. 1991;65:165–169. doi: 10.1016/0378-1097(91)90298-o. [DOI] [PubMed] [Google Scholar]
- 62.Wheelis M L, Palleroni N J, Stanier R Y. The metabolism of aromatic acids by Pseudomonas testosteroni and P. acidovorans. Arch Microbiol. 1967;59:302–314. doi: 10.1007/BF00406344. [DOI] [PubMed] [Google Scholar]
- 63.Wilmes-Riesenberg M R, Wanner B L. TnphoA and TnphoA′ elements for making and switching fusions for study of transcription, translation, and cell surface localization. J Bacteriol. 1992;174:4558–4575. doi: 10.1128/jb.174.14.4558-4575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyndham R C. Evolved aniline catabolism in Acinetobacter calcoaceticus during continuous culture of river water. Appl Environ Microbiol. 1986;51:781–789. doi: 10.1128/aem.51.4.781-789.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wyndham R C, Nakatsu C, Peel M, Cashore A, Ng J, Szilagyi F. Distribution of the catabolic transposon Tn5271 in a groundwater bioremediation system. Appl Environ Microbiol. 1994;60:86–93. doi: 10.1128/aem.60.1.86-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wyndham R C, Singh R K, Straus N A. Catabolic instability, plasmid gene deletion and recombination in Alcaligenes sp. BR60. Arch Microbiol. 1988;150:237–243. doi: 10.1007/BF00407786. [DOI] [PubMed] [Google Scholar]
- 67.Wyndham R C, Straus N A. Chlorobenzoate catabolism and interactions between Alcaligenes and Pseudomonas species from Bloody Run Creek. Arch Microbiol. 1988;150:230–236. doi: 10.1007/BF00407785. [DOI] [PubMed] [Google Scholar]
- 68.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]