Abstract
Background
Although the majority of human papillomavirus (HPV) infections are cleared by the immune system, a small percentage of them progress to develop HPV‐driven cancers. Cervical cancer studies highlight that HPV persistence and cancer risk are associated with genetic factors, especially at the human leukocyte antigen (HLA) genes. This study was conducted to investigate such associations in head and neck cancer (HNC).
Methods
In all, 192 patients with HNC and 384 controls were genotyped with the Infinium Global Screening Array (Illumina, Inc). HLA variants were imputed with SNP2HLA, and an association analysis was performed by logistic regression.
Results
HPV‐positive HNCs were significantly associated with single‐nucleotide polymorphisms (SNPs) at DRB1_32660090 (P = 1.728 × 10–6) and DRB1_32660116 (P = 1.728 × 10–6) and with the amino acid variant DRB1_11_32660115 (P = 1.728 × 10–6). None of these associations were observed in the HPV‐negative cohort, and this suggested their specificity to convey risk for HPV‐associated HNCs. In general, associations observed for HPV‐negative HNC were relatively weak, and variants in the HLA‐DPA1 region were the strongest among them (P = 4.531 × 10–4). Several lead signals reported by previous HNC genome‐wide association studies, including SNPs rs3135001 (P = .012), rs1049055 (P = .012), and rs34518860 (P = .029) and allele HLA‐DQB1*06 (P = .009), were replicated in the current study. However, these associations were limited to the HPV‐positive HNC group. Several cervical cancer–associated HLA variants, including SNPs rs9272143 (P = .002) and rs9271858 (P = .002) and alleles HLA‐B‐1501 (P = .009) and HLA‐B‐15 (P = .015), were also exclusively associated with HPV‐positive HNC.
Conclusions
HPV‐positive HNC risk is associated with distinct HLA variants, and some of them are shared by both cervical cancer and HPV‐positive HNC.
Human papillomavirus (HPV)–positive head and neck cancer (HNC) risk is associated with distinct human leukocyte antigen variants, and some of them are shared by both cervical cancer and HPV‐positive HNC.
Lay Summary
Cervical cancer studies highlight that human papillomavirus (HPV)–driven cancer risk is linked with human leukocyte antigen (HLA) polymorphism.
Hence, the current study was designed to investigate the HLA associations in HPV‐positive and HPV‐negative head and neck cancer (HNC) and compare these associations with cervical cancer.
Several lead signals reported by previous HNC and cervical genome‐wide association studies were replicated in the current study.
However, these associations were limited to the HPV‐positive HNC group, and this suggests that HPV‐positive HNC risk is associated with distinct HLA variants, and some of them are shared by both cervical cancer and HPV‐positive HNC.
Keywords: cervical cancer, head and neck cancer, human leukocyte antigen, human papillomavirus
Introduction
Oncogenic strains of human papillomavirus (HPV) account for a considerable number of human cancers originating in different anatomical sites of the human body. These cancers include almost all cervical cancers, a considerable fraction of other anogenital cancers, and head and neck cancer (HNC). 1 Despite the differences in the pathology of these cancers, they appear to share common HPV‐driven oncogenic pathways. 2 , 3 HPV infection in any of these sites is not necessarily followed by cancer development, the vast majority of the infections being eliminated by the immune system over a period of months to a few years. 4 , 5 , 6 However, certain individuals fail to clear the infection; this leads to persistent HPV infection and, consequently, to HPV‐driven cancers. 7
Epidemiological studies have reported many factors, including older age, infections such as HIV, and smoking, to be associated with HPV persistence and HPV‐driven cancer risk. 8 , 9 , 10 , 11 , 12 , 13 However, heritable genetic factors associated with HPV persistence and HPV‐driven cancers in parallel to these acquired risks have been demonstrated in many studies. The early evidence suggesting the heritability of HPV‐driven cancer risk arose from familial studies and twin studies of cervical cancer. 14 , 15 These observations were further supported by candidate gene studies and confirmed by subsequent genome‐wide association studies (GWASs). 16 , 17 , 18 , 19 , 20
In GWASs of cervical cancer, the strongest associations were seen within the human leukocyte antigen (HLA) genes located on the short arm of chromosome 6 (6p21.3). 18 , 20 , 21 HLA genes encode proteins that construct major histocompatibility complex (MHC) molecules involved in antigen presentation. These are key components of the immune system and thus play a major role in modulating the immune response against viruses such as HPV. As such, these associations appear to have a plausible functional relationship.
HNC studies have also reported associations between HLA variations and HNC risk. 22 , 23 , 24 However, unlike cervical cancer, which is almost always caused by HPV infection, only a fraction of HNCs are associated with HPV. As such, it is unclear whether the observed associations in HNC are general or are confined only to the HPV‐positive cohort. Intriguingly, the only study that stratified HNC by HPV status reported stronger associations with HLA variants in the HPV‐positive cohort compared with the HPV‐negative cohort. 22
This study was designed to further evaluate HLA associations with HPV‐positive and HPV‐negative HNC by replicating previous GWAS findings in an independent cohort and to compare the HLA associations with those reported for cervical cancer.
Materials and Methods
Ethical Consideration and Patient Recruitment
Ethical approval for the study was obtained from the Metro South Human Research Ethics Committee (reference numbers HREC/12/QPAH/381 and HREC/16/QPAH/125). Samples were collected at Princess Alexandra Hospital and Royal Brisbane and Women’s Hospital from 2012 to 2019.
Salivary Sample Collection and Processing
Unstimulated saliva samples were collected according to a previously published procedure. 25 , 26
Determination of the HPV Status of HNC
The tumor p16 status, determined with the CINtec p16INK4a histology kit (E6H4 clone; Roche MTM Laboratories), was used as a surrogate marker for the HPV status of the tumor. Strong, diffused nuclear and cytoplasmic staining over 75% of the tumor tissue was considered p16 overexpression and considered p16 positive. Salivary HPV DNA detection was used to validate the tumor HPV status. Salivary DNA was tested for high‐risk HPV types with quantitative polymerase chain reaction and the iPLEX MassARRAY (Agena Bioscience, San Diego, California) according to a protocol described previously. 26 , 27
Genotyping
All samples were genotyped with the Infinium Global Screening Array (chip GSAMD‐24v2‐0‐20024620_A1; Illumina, Inc, San Diego, California) according to the manufacturer's protocol at the Australian Translational Genetic Centre (Woolloongabba, Queensland, Australia).
Statistical Methods
Quality control (QC) was performed with PLINK. 28 Samples with an excess genotyping missing rate (>10%) or an outlying heterozygosity rate (beyond mean ± 3 SD) were excluded; single‐nucleotide polymorphisms (SNPs) with an excess genotyping rate or individual missing rate (>5% and >5% separately), a minor allele frequency < 1%, or a Hardy‐Weinberg equilibrium P value < 1 × 10–6 were identified and excluded. Then, duplicated or related individuals with identity by descent > 0.185 were excluded, and this was followed by standard population stratification analysis using FlashPCA2 (https://github.com/gabraham/flashpca) and the HapMap3 data set (https://www.sanger.ac.uk/data/hapmap‐3/) as a reference. Ten principal components (PCs) with a gradually diminishing weight for each individual were generated. The top 2 components, PC1 and PC2, were used to exclude ancestry outliers (PC1 and/or PC2 beyond mean ± 3 SD). PCs were recalculated after the removal of ancestry outliers, and the top 4 PCs, which could capture most of the information, were used as covariates in the following association analysis to control for population stratification because additional components did not further reduce inflation in the test statistics.
A detailed investigation in the HLA region was performed with SNP2HLA, which performs HLA allele and amino acid imputation from passed‐QC SNP data and association analysis. The imputation was performed with the Type 1 Diabetes Genetics Consortium reference panel and was followed by QC, which included the exclusion of imputed loci with r 2 < 0.3 and samples with the allele dosage at any HLA type exceeding 2.5. An association analysis was then conducted via logistic regression with PC1 to PC4 as covariates.
To apply the polygenic risk score (PRS) derived from our previous study 20 for persistent HPV infections in cervical cancer, SNP imputation on passed‐QC SNP data was performed first to predict unobserved SNPs. The Michigan imputation server with the Haplotype Reference Consortium reference panel was used where imputed loci with r 2 > 0.3 were kept. After that, the genetic risk score for each individual was calculated with PLINK, and the predictivity was evaluated with the area under the receiver operator curve.
Results
With consideration of their tumor p16 status and salivary HPV status, 192 patients with HNC, including 96 with HPV‐positive HNC (salivary HPV positive and tumor p16 positive) and 96 with HPV‐negative HNC (salivary HPV negative and tumor p16 negative) who had sufficient DNA in their salivary samples, were selected for genotyping. Demographic and clinical information for the patients with HNC is listed in Supporting Table 1. The HPV‐positive HNC cohort was mainly composed of patients with oropharyngeal cancer (OPC; 93.75%), and the HPV‐negative HNC cohort mainly consisted of patients with oral cancer (55.21%). For an unselected control, a cohort of 384 samples previously described was used. 29 , 30 Both the HNC cohort and the control cohort were mostly of European descent. After QC, a total of 91 HPV‐positive patients with HNC, 88 HPV‐negative patients with HNC, and 364 controls with 458,881 common SNPs remained (Fig. 1).
Figure 1.
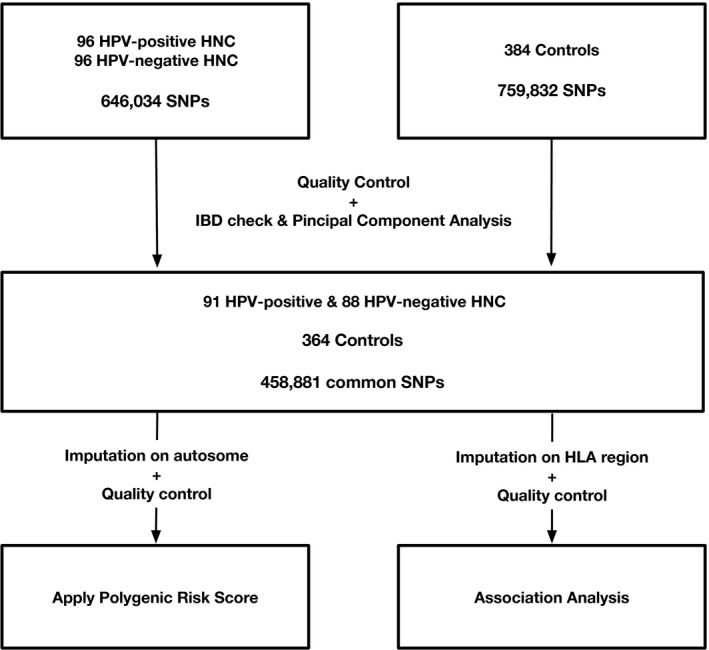
Workflow of the study. HLA indicates human leukocyte antigen; HNC, head and neck cancer; HPV, human papillomavirus; IBD, identity by descent; SNP, single‐nucleotide polymorphism.
Associations Observed in the Current Study
Although this study was designed for replication, we investigated the overall structure of the 8445 variants in the HLA region, including 256 HLA alleles, 1060 amino acids, and 7129 SNPs that were imputed with SNP2HLA. An association analysis of the HLA region was performed on HPV‐positive HNC against controls (HPV‐positive HNC group) and on HPV‐negative HNC against controls (HPV‐negative HNC group) via logistic regression with 4 PCs as covariates (Supporting Table 2).
In the HPV‐positive HNC group, the strongest associations with suggestive genome‐wide significance were seen with SNP_DRB1_32660090_G (odds ratio [OR], 2.406 [1.679‐3.448]; P = 1.728 × 10–6), SNP_DRB1_32660116_C ([OR], 2.406 [1.679‐3.448]; P = 1.728 × 10–6), and AA_DRB1_11_32660115_SPL (presence of serine, proline, or leucine in HLA‐DRB1 amino acid position 11; [OR], 2.406 [1.679‐3.448]; P = 1.728 × 10–6; Fig. 2). The targets were in strong linkage disequilibrium with each other (r 2 = 1), and conditioning on any of these targets masked most of the signals leaving only minor residual associations in the region (P > .0001; Fig. 2). Notably, conditioning AA_DRB1_11_32660115_SPL of these variants also controlled for the associations HLA‐DRB1*13, HLA‐DRB1*1301, HLA‐DQA1*0103, and HLA‐DQB1*0603 previously reported in HNC GWASs. The linkage disequilibrium structure with these amino acids and the classical HLA type is shown in Figure 2.
Figure 2.

Zoom plot indicating HLA associations in HNC: (A) HLA associations in the HPV‐positive HNC group, (B) associations with HLA classical alleles in the HPV‐positive HNC group, (C) HLA associations in the HPV‐negative HNC group indicating that the associations are quite different from the HPV‐positive HNC group, and (D) associations with HLA classical alleles in the HPV‐negative HNC group. SNP associations are reported as filled‐in dots, and HLA amino acid associations are reported as hollow diamonds. Colors represent the extent of linkage disequilibrium with amino acid position 11 of HLA‐DRB1 (the top signal for the HPV‐positive HNC group). A: top left, B: top right, C: bottom left, D: bottom right. HLA indicates human leukocyte antigen; HNC, head and neck cancer; HPV, human papillomavirus; SNP, single‐nucleotide polymorphism.
In the HPV‐negative HNC group, no signal reached suggestive genome‐wide significance. Relatively weak associations were observed with a group of variants with r 2 = 1, including SNPs, amino acids in the HLA‐DPA1 region, and HLA‐DPA1*2 ([OR], 2.085 [1.383‐3.144]; P = 4.531 × 10–4). No associations were observed with the top signals observed to be associated with the HPV‐positive HNC group. This provided further evidence for the role of AA_DRB1_11_32660115_SPL/SNP_DRB1_32660090_G/SNP_DRB1_32660116_C in HPV‐positive HNC and confidence in the replication study.
Replication of HLA Associations Reported by Other HNC GWASs
Replication of lead signals
The observations of 2 previous large‐scale GWASs were compared with the current study to identify replicated targets (Table 1). When lead signals reported by these GWASs were considered, a number of HLA variants reported by Shete et al 24 were identified to be associated with the HPV‐positive HNC group but not with the HPV‐negative HNC group in the current study. rs3135001, located in HLA‐DQB1, which was previously reported to be associated with overall HNC ([OR], 0.77 [0.73‐0.82]; P = 1.44 × 10–16), was found to significantly correlate with HPV‐positive HNC risk ([OR], 0.56 [0.36‐0.88]; P = .012). Similarly, HLA‐DQB1 rs1049055, which was previously reported to be associated with oral cavity cancer ([OR], 0.78 [0.72‐0.85]; P = 2.96 × 10–9), and SNP HLA‐DQA1 rs34518860, which was reported to be linked with OPC ([OR], 0.61 [0.54‐0.68]; P = 2.61 × 10–17), were among the targets replicated in the current study to be significantly correlated with HPV‐positive HNC risk (rs1049055: [OR], 0.56 [0.36‐0.88]; P = .012; rs34518860: [OR], 0.39 [0.16‐0.91]; P = .029). Furthermore, HLA‐DQB1*06, which had been reported to be associated with overall HNC ([OR], 0.77 [0.7‐0.84]; P = 4.34 × 10–8) and OPC ([OR], 0.77 [0.7‐0.84]; P = 4.34 × 10–8), was observed to be associated with HPV‐positive HNC ([OR], 0.55 [0.35‐0.86]; P = .009).
TABLE 1.
Replication of Previous HNC Genome‐Wide Association Studies
| Variant (Minor Allele) | Gene | Chromosome | Previous Studies | Current Study | |||
|---|---|---|---|---|---|---|---|
| Shete et al 24 (2020) | Lesseur et al 22 (2016) | HPV‐Positive HNC | HPV‐Negative HNC | ||||
| Sample size |
Discovery: 2171 cases 4493 controls Validation: 5205 cases 3232 controls |
6034 cases 6585 controls |
91 cases 364 controls |
88 cases 364 controls |
|||
| Overall HNC | |||||||
| rs1229984 (A) | ADH1B | Chr4 |
P = 2.29 × 10–15 OR = 0.56 |
N/A | N/A | ||
| rs259919 (A) | ZNRD1ASP | Chr6 |
P = 2.96 × 10–9 OR = 1.17 (1.11‐1.24) |
P = .977 OR = 1.01 (0.71‐1.44) |
P = .976 OR = 0.99 (0.69‐1.43) |
||
| rs3135001 (T) | HLA‐DQB1 | Chr6 |
P = 1.44 × 10–16 OR = 0.77 (0.73‐0.82) |
P = .012 OR = 0.56 (0.36‐0.88) |
P = .439 OR = 0.85 (0.56‐1.29) |
||
| rs1265081 (C) | CCHCR1 | Chr6 |
P = 3.75 × 10–10 OR = 0.85 (0.81‐0.9) |
P = .323 OR = 0.84 (0.60‐1.19) |
P = .609 OR = 0.92 (0.65‐1.29) |
||
| rs3828805 (C) | HLA‐DQB1 | Chr6 |
P = 3.35 × 10–13 OR = 1.28 |
P = .407 OR = 1.16 (0.81‐1.67) |
P = .650 OR = 1.08 (0.76‐1.54) |
||
| rs201982221 | LHPP | Chr10 |
P = 1.58 × 10–9 OR = 1.67 |
NA | NA | ||
| rs1453414 (C) | OR52N2/TRIM5 | Chr11 |
P = 4.78 × 10–8 OR = 1.19 |
P = .857 OR = 1.03 (0.73‐1.44) |
P = .503 OR = 1.12 (0.8‐1.56) |
||
| HLA‐B*37/3701 |
P = 2.35 × 10–25 OR = 2.77 (2.29‐3.35) |
P = .088 OR = 2.49 (0.87‐7.11) |
P = .369 OR = 1.73 (0.52‐5.67) |
||||
| HLA‐DQB1*06 |
P = 4.20 × 10–8 OR = 0.71 (0.63‐0.8) |
P = .009 0.55 (0.35‐0.86) |
P = .537 0.88 (0.59 ‐1.32) |
||||
| Oral cavity cancer | |||||||
| rs6547741 (A) | GPN1 | Chr2 |
P = 3.97 × 10–8 OR = 0.83 |
P = .107 OR = 1.31 (0.93‐1.85) |
P = .975 OR = 1.00 (0.71‐1.40) |
||
| rs1229984 (A) | ADH1B | Chr4 |
P = 1.09 × 10–9 OR = 0.57 |
N/A | N/A | ||
| rs10462706 (T) | CLPTM1L | Chr5 |
P = 7.87 × 10–11 OR = 0.73 (0.66‐0.80) |
P = 5.54 × 10–10 OR = 0.74 |
P = .533 OR = 1.16 (0.72‐1.87) |
P = .239 OR = 1.31 (0.84‐2.05) |
|
| rs1049055 (C) | HLA‐DQB1 | Chr6 |
P = 2.96 × 10–9 OR = 0.78 (0.72‐0.85) |
P = .012 OR = 0.56 (0.36‐0.88) |
P = .419 OR = 0.84 (0.56‐1.27) |
||
| rs8181047 (A) | CDKN2B‐AS1 | Chr9 |
P = 3.80 × 10–9 OR = 1.24 |
P = .350 OR = 1.19 (0.83‐1.71) |
P = .407 OR = 1.17 (0.81‐1.70) |
||
| rs928674 (G) | LAMC3 | Chr9 |
P = 2.09 × 10–8 OR = 1.33 |
P = .130 OR = 0.66 (0.39 ‐1.13) |
P = .718 OR = 0.92 (0.57‐1.48) |
||
| HLA‐B*37/3701 |
P = 1.07 × 10–12 OR = 2.63 (2.02‐3.44) |
P = .088 OR = 2.49 (0.87‐7.11) |
P = .369 OR = 1.73 (0.52‐5.67) |
||||
| Oropharyngeal cancer | |||||||
| rs4318431 (T) | GALNT14 | Chr2 |
P = 3.13 × 10–9 OR = 1.39 (1.25‐1.55) |
P = .589 OR = 0.84 (0.46‐1.56) |
P = .496 OR = 0.79 (0.41‐1.54) |
||
| rs1229984 (A) | ADH1B | Chr4 |
P = 8.53 × 10–9 OR = 0.55 |
N/A | N/A | ||
| rs13211972 (A) | MUC21 | Chr6 |
P = 1.04 × 10–10 OR = 1.55 (1.36‐1.77) |
P = .142 OR = 1.60 (0.86‐2.99) |
P = .868 OR = 1.07 (0.53‐2.17) |
||
| rs34518860 (A) | HLA‐DQA1 | Chr6 |
P = 2.61 × 10–17 OR = 0.61 (0.54‐0.68) |
P = .029 OR = 0.39 (0.16‐0.91) |
P = .893 OR = 1.04 (0.58‐1.88) |
||
| rs3828805 (C/T) | HLA‐DQB1 | Chr6 |
P = 2.21 × 10–12 OR = 1.37 |
P = .407 OR = 1.16 (0.81‐1.67) |
P = .65 OR = 1.08 (0.76‐1.53) |
||
| HLA‐B*37/3701 |
P = 3.52 × 10–22 OR = 2.94 (2.36‐3.66) |
P = .088 OR = 2.49 (0.87‐7.11) |
P = .369 OR = 1.73 (0.52‐5.67) |
||||
| HLA‐DQB1*06 |
P = 4.2 × 10–8 OR = 0.71 (0.63‐0.8) |
P = .009 OR = 0.55 (0.35‐0.86) |
P = .537 OR = 0.88 (0.59 ‐1.32) |
||||
| HLA‐DRB1*13 |
P = 2.69 × 10–8 OR = 0.6 (0.51‐0.72) |
P = .107 OR = 0.57 (0.29‐1.13) |
P = .8083 OR = 1.07 (0.61‐1.87) |
||||
| HLA‐DRB1*1301/HLA‐DQA1*0103/HLA‐DQB1*0603 | Haplotype |
P = 2.7 × 10–9 OR = 0.59 |
P = .122 OR = 0.44 (0.15‐1.25) P = .088 OR = 0.40 (0.14‐1.15) P = .068 OR = 0.38 (0.13‐1.07) |
P = .739 OR = 0.87 (0.38 ‐1.97) P = .801 OR = 0.91 (0.43‐1.93) P = .958 OR = 0.98 (0.47‐2.05) |
|||
| Hypopharyngeal and laryngeal cancer | |||||||
| rs142021700 (C) | RTTN | Chr18 |
P = 2.54 × 10–9 OR = 3.95 (2.51‐6.21) |
N/A | N/A | ||
Abbreviations: N/A, not available; HLA, human leukocyte antigen; HNC, head and neck cancer; HPV, human papillomavirus; OR, odds ratio.
Replication of Other Validated Signals
Several stronger associations were observed when other validated HNC‐associated genetic variants reported by these 2 GWASs were considered (Supporting Table 3). A number of SNPs located at class II HLA variants, including DQA1, DRB1, and DRB5, that were reported to be associated with OPC risk by Shete et al 24 were observed to reach suggestive genome‐wide significance (P < 1 × 10–5) in the HPV‐positive HNC group. The strongest signal among them was observed for rs9271776, an SNP located at HLA‐DQA1 ([OR], 2.42 [1.68‐3.48]; P = 2.06 × 10–6), which was reported by Shete et al to be linked with OPC risk ([OR], 1.23 [1.15‐1.32]; P = 5.65 × 10–10). Strong associations with HPV‐positive HNC were also observed for several SNPs located at HLA‐DQA1, including rs17612562 ([OR], 2.08 [1.451‐2.98]; P = 6.617 × 10–5); this confirmed associations with OPC previously reported by Lesseur et al 22 ([OR], 1.29; P = 1.6 × 10–9). Among the allelic variations, HLA‐DQA1*03 ([OR], 2.153 [1.483‐3.125]; P = 5.5 × 10–5) and HLA‐DQA1*0301 ([OR], 2.153 [1.483‐3.125]; P = 5.5 × 10–5), which have been previously reported to promote OPC risk (HLA‐DQA1*03: [OR], 1.29 [1.15‐1.46]; P = 2.43 × 10–5; HLA‐DQA1*0301: [OR], 1.29 [1.15‐1.46]; P = 2.43 × 10–5), had the strongest associations in the current study. 24
In contrast, only a few HLA variants were replicated with the HPV‐negative HNC cohort. These associations were limited to SNPs located at HLA‐DQB1 and HLA‐DQA1 and were relatively weak in comparison with HLA associations observed with HPV‐positive HNC. Among them, rs9273415 at HLA‐DQB1 ([OR], 0.64 [0.45‐0.90]; P = .011), which had been previously reported to be associated with OPC by Lesseur et al 22 ([OR], 1.28; P = 2.8 × 10–9), was the strongest signal.
Replication of HLA Associations Reported by Cervical Cancer Studies
A number of HLA targets that had been identified to be associated with cervical cancer were also found to be significantly associated with HPV‐positive HNC in the current study (Table 2 and Supporting Table 4). Among the associations shared by both cervical cancer and HPV‐positive HNC, there were several lead signals previously reported by cervical cancer GWASs. Both rs9272143 ([OR], 0.67 [0.62‐0.72]; P = 9.3 × 10−24), the top signal reported by Chen et al for cervical cancer, 21 and rs9271858 ([OR], 7.44; P = 5.20 × 10−15), the top signal reported by Leo et al 20 for cervical cancer, also significantly correlated with HPV‐positive HNC risk (rs9272143: [OR], 0.58 [0.41‐0.82]; P = 2 × 10–3; rs9271858: [OR], 1.72 [1.22‐2.44]; P = 2 × 10–3). 1
TABLE 2.
Replication of Previous Cervical Cancer Genome‐Wide Association Studies
| Variant ID (Minor Allele) | Location | Chromosome | Previous Studies | Current Study | ||||
|---|---|---|---|---|---|---|---|---|
| Chen et al 21 (2013) | Chen et al 19 (2016) | Leo et al 20 (2017) | HPV‐Positive HNC | HPV‐Negative HNC | ||||
| Sample size |
Discovery:
Validation:
|
Discovery:
Validation:
|
2866 CIN2‐3/cervical cancer 6481 controls |
91 cases 364 controls |
88 cases 364 controls |
|||
| SNPs | rs9272143 (C) | Between HLA‐DRB1 and HLA‐DQA1 | 6 |
P = 9.3 × 10−24 OR = 0.67 (0.62‐0.72) |
P = 2.04 × 10–3 OR = 0.58 (0.41‐0.82) |
P = .228 OR = 1.23 (0.88‐1.71) |
||
| rs2516448 (T) | Adjacent to MICA | 6 |
P = 1.6 × 10−18 OR = 1.42 (1.31‐1.54) |
P = 1.10 × 10–15 OR = 1.39 (1.28‐1.52) |
P = .297 OR = 1.19 (0.85‐1.6) |
P = .974 OR = 1.00 (0.71‐1.38) |
||
| rs3117027 (A) | Pseudogene HLA‐DPB2 | 6 |
P = 4.9 × 10−8 OR = 1.25 (1.15‐1.35) |
P = .716 OR = 1.07 (0.74‐1.54) |
P = .769 OR = 1.06 (0.74‐1.51) |
|||
| rs9271898 (A) | Between HLA‐DRB1 and HLA‐DQA1 | 6 |
P = 1.2 × 10−24 OR = 0.64 (0.59‐0.7) |
— | — | |||
| rs3130196 (C) | Between HLA‐DPB1 and HLA‐DPA1 | 6 |
P = 2.3 × 10−9 OR = 1.4 (1.26‐1.57) |
— | — | |||
| rs115625939 (G) | Between HLA‐DRB1 and HLA‐DQA1 | 6 |
P = 1.4 × 10−15 OR = 0.58 (0.51‐0.67) |
— | — | |||
| rs9271858 (A) | Near HLA‐DQA1‐0102 | 6 |
P = 5.2 × 10−15 OR = 7.44 |
P = 2.0 × 10–3 OR = 1.72 (1.22‐2.44) |
P = .228 OR = 0.81 (0.58‐1.14) |
|||
| Alleles | HLA‐B‐0702 | 6 |
P = 7.9 × 10–8 OR = 1.42 (1.25‐1.61) |
P = 1.4 × 10−10 OR = 1.41 (1.27‐1.57) |
P = 3.9 × 10−9 OR = 1.31 |
P = .864 OR = 1.04 (0.66‐1.63) |
P = .227 OR = 0.73 (0.44‐1.21) |
|
| HLA‐DRB1‐1301 | 6 |
P = 3.9 × 10–10 OR = 0.47 (0.37‐0.59) |
P = 1.8 × 10–13 OR = 0.49 (0.40‐0.59) |
P = 2.9 × 10–8 OR = 0.62 |
P = .122 OR = 1.44 (0.15‐1.25) |
P = .739 OR = 0.87 (0.38‐1.97) |
||
| HLA‐DRB1‐1501 | 6 |
P = 2.7 × 10–7 OR = 1.39 (1.23‐1.57) |
P = 9.7 × 10−9 OR = 1.36 (1.22‐1.51) |
P = 5.55 × 10–12 OR = 1.43 |
P = .151 OR = 0.69 (0.42‐1.14) |
P = .394 OR = 0.81 (0.51‐1.31) |
||
| HLA‐DQA1‐0103 | 6 |
P = 1.9 × 10–9 OR = 0.49 (0.39‐0.62) |
P = 5.6 × 10−14 OR = 0.49 (0.40‐0.59) |
P = 3.36 × 10–8 OR = 0.63 |
P = .088 OR = 0.4 (0.14‐1.14) |
P = .801 OR = 0.91 (0.43‐1.93) |
||
| HLA‐DQB1‐0603 | 6 |
P = 5.7 × 10–10 OR = 0.48 (0.38‐0.60) |
P = 1.5 × 10−11 OR = 0.54 (0.45‐0.64) |
P = 4.17 × 10–8 OR = 0.63 |
P = .068 OR = 0.38 (0.13‐1.07) |
P = .957 OR = 0.98 (0.47‐2.05) |
||
| HLA‐DQB1‐0602 | 6 |
P = 3.5 × 10–7 OR = 1.39 (1.22‐1.58) |
P = 2.5 × 10−7 OR = 1.32 (1.19‐1.47) |
P = 4.46 × 10–12 OR = 1.44 |
P = .295 OR = 0.77 (0.47‐1.26) |
P = .219 OR = 0.73 (0.44‐1.21) |
||
| HLA‐B‐0702 | 6 |
P = 1.4 × 10−10 OR = 1.41 (1.27‐1.57) |
P = 3.86 × 10–9 OR = 1.31 |
P = .864 OR = 1.04 (0.66‐1.63) |
P = .227 OR = 0.73 (0.44‐1.21) |
|||
| HLA‐B‐1501 | 6 |
P = 2.6 × 10−8 OR = 0.67 (0.58‐0.77) |
P = 4.44 × 10–9 OR = 0.63 |
P = .009 OR = 0.21 (0.06‐0.67) |
P = .595 OR = 0.83 (0.43‐1.63) |
|||
| HLA‐C‐0702 | 6 |
P = 2.6 × 10−9 OR = 1.37 (1.24‐1.52) |
P = 2.40 × 10–7 OR = 1.26 |
P = .934 OR = 0.98 (0.63‐1.53) |
P = .509 OR = 0.85 (0.53‐1.36) |
|||
| HLA‐B‐15 | 6 |
P = 1.56 × 10–9 OR = 0.64 |
P = .015 OR = 0.31 (0.12‐0.80) |
P = .564 OR = 0.83 (0.43‐1.58) |
||||
| HLA‐DRB1‐0401 |
P = 7.13 × 10–5 OR = 1.24 |
P = .140 OR = 1.47 (0.88‐2.43) |
P = .869 OR = 1.05 (0.60‐1.82) |
|||||
Abbreviations: CIN, cervical intraepithelial neoplasia; HLA, human leukocyte antigen; HNC, head and neck cancer; HPV, human papillomavirus; OR, odds ratio; SNP, single‐nucleotide polymorphism.
Among the allelic associations reported by cervical cancer studies, HLA‐B‐15, reported by Leo et al 20 to be associated with cervical cancer ([OR], 0.64; P = 1.56× 10−9), was observed to be associated with HPV‐positive HNC ([OR], 0.31 [0.12‐0.80]; P = .015). Similarly, HLA‐B‐1501, a suballele of the HLA‐B‐15 family, which was reported to be associated with cervical cancer by both Chen et al 19 ([OR], 0.67 [0.58‐0.77]; P = 2.6 × 10−8) and Leo et al 20 ([OR], 0.63; P = 4.44 × 10–9), was observed to be associated with HPV‐positive HNC ([OR], 0.21 [0.06‐0.67]; P = .009). This HLA type has not been previously reported in HNC GWASs; however, it is also a relatively rare allele with a population frequency of 8% in controls and 2% in cases. Importantly, none of these associations were observed to be linked with HPV‐negative HNC.
Considering that there are some similarities between cervical cancer and HPV‐positive HNC, we evaluated the efficacy of the PRS algorithm developed for cervical cancer to predict HNC risk under the hypothesis that genomic variants for persistent HPV infection might be shared. This PRS had an average area under the receiver operator curve of 0.678, including 35 regions and 692 predictors (including 234 on MHC regions), and it showed that women with the highest 10% and 5% risk scores had approximately greater than 7.1% and 21.6% risks of developing cervical cancer, respectively. 20 However, the cervical cancer PRS model was unable to stratify the HPV‐positive and HPV‐negative cases or separate the HPV‐positive cases from the unselected controls (Table 3). We conclude that the underlying risks for HPV‐driven diseases are not similar.
TABLE 3.
Performance of the Cervical Cancer Polygenic Risk Score Algorithm for Predicting HNC Risk
| Comparison | Mean AUC of 10‐Fold Cross‐Validation |
|---|---|
| HPV‐positive HNC vs HPV‐negative HNC | 0.515 |
| HPV‐positive HNC vs controls | 0.530 |
| HPV‐negative HNC vs controls | 0.528 |
Abbreviations: AUC, area under the receiver operator curve; HNC, head and neck cancer; HPV, human papillomavirus.
Discussion
Mounting a timely and effective immune response relies on prompt antigen recognition, and thus cells have evolved efficient mechanisms to present antigens to immune cells. Among them, the MHC‐based antigen presentation pathway is the key pathway for peptide antigen presentation. 31 Except for the β2‐microglobulin fragment of MHC class I molecules, protein molecules that construct MHC class I and class II are encoded by an MHC gene complex located at the short arm of chromosome 6 (6p21). This is one of the most polymorphic regions of the human genome, where certain genes have more than 4000 known allelic variants. 32 Changes in MHC peptide interactions due to these allelic variations allow MHC molecules to efficiently present a broad range of antigens. This diversification of immunity can be considered an evolutionary adaptation to ensure that at least certain individuals survive against an emerging infection.
However, this survival advantage comes with a price as some of these variants have a lower affinity toward certain antigens affecting the efficacy of antigen presentation. This results in an inability to competently respond to certain infections or mount immune reactions against certain types of cancers in individuals having these alleles. Consequently, HLA allelic variations have been indicated to be associated with a range of diseases. 33 Cervical cancer is one such disease where the link between HLA variations and the risk for acquiring cervical cancer is well established. 34 Because almost all cervical cancers are triggered by HPV infection, these HLA variations are assumed to be responsible for either defective viral clearance or a defective immune response against cellular transformation.
Similarly, the current study together with previous HNC GWASs establishes the associations between HLA changes and HNC risk. 22 , 24 However, these associations were apparent in HPV‐positive HNC in contrast to HPV‐negative HNC, where only a limited number of associations were identified. When HPV‐positive HNC was taken into consideration, significant associations were observed with both class I and class II MHC variants. The strongest associations were observed with class II MHC variants, with several SNPs and amino acid positions located at HLA‐DRB1 and HLA‐DQA1 reaching P values < 10−5. Furthermore, we have been able to replicate several HLA targets that have been reported by previous HNC GWASs in the comparison between HPV‐positive HNC and controls. The majority of the replicated targets have been reported to be associated with OPC (Supporting Table 3). Because a significant number of OPCs are caused by HPV, it can be reasoned that the majority of these associations are likely to be due to the higher number of HPV‐positive cases. The only GWAS that has considered the HPV status in a subgroup of samples also suggests that there are stronger associations when HPV‐positive OPC is considered. 22
Strengthening the notion that there are common genetic factors associated with HPV‐driven cancer risk, several cervical cancer–associated HLA variants were also observed to be associated with HPV‐positive HNC. Among these targets, there were several lead signals observed by cervical cancer GWASs, including the leading SNPs reported by Chen et al 21 and Leo et al 20 and component alleles of the major risk and protective haplotypes reported by Leo et al. Confirming the specific role played by these HLA variants in HPV‐driven oncogenesis, none of them were observed to be associated with HPV‐negative HNC. Despite these associations, the risk score prediction model developed for cervical cancer by Leo et al was unable to predict HPV‐driven HNC risk. This could be due either to the limited number of samples considered in this study or to characteristic HLA associations in HPV‐positive HNC in addition to HLA variants shared with cervical cancer.
In conclusion, HPV‐positive HNC risk is associated with HLA variations, and at least some of these associations are shared by both cervical cancer and HPV‐positive HNC.
Funding Support
Chamindie Punyadeera reports TRI Clinical Collaboration Seed Grants (2019/2020) as well as funding from Cancer Australia, the National Institutes of Health, and the National Health and Medical Research Council (Ideas Grant). Chameera Ekanayake Weeramange reports a joint scholarship from the University Grants Commission (Sri Lanka) and the Queensland University of Technology (Australia).
Conflict of Interest Disclosures
The authors made no disclosures.
Author Contributions
Chameera Ekanayake Weeramange: Formal analysis, writing–original draft, data curation, investigation, methodology, resources, visualization, and writing–review and editing. Danhua Shu: Formal analysis, writing–original draft, data curation, investigation, methodology, resources, visualization, and writing–review and editing. Kai Dun Tang: Data curation, investigation, methodology, resources, visualization, and writing–review and editing. Jyotsna Batra: Data curation, investigation, methodology, resources, visualization, and writing–review and editing. Rahul Ladwa: Data curation, investigation, methodology, resources, visualization, and writing–review and editing. Lizbeth Kenny: Data curation, investigation, methodology, resources, visualization, and writing–review and editing. Sarju Vasani: Data curation, investigation, methodology, resources, visualization, and writing–review and editing. Ian H. Frazer: Data curation, investigation, methodology, resources, visualization, and writing–review and editing. Riccardo Dolcetti: Data curation, investigation, methodology, resources, visualization, and writing–review and editing. Jonathan J. Ellis: Data curation, investigation, methodology, resources, visualization, and writing–review and editing. Richard A. Sturm: Data curation, investigation, methodology, resources, visualization, and writing–review and editing. Paul Leo: Conceptualization, project administration, formal analysis, validation, software, supervision, data curation, investigation, methodology, resources, visualization, and writing–review and editing. Chamindie Punyadeera: Conceptualization, funding acquisition, project administration, supervision, validation, data curation, investigation, methodology, resources, visualization, and writing–review and editing.
Supporting information
Table S1
Table S2
Table S3
Table S4
Ekanayake Weeramange C, Shu D, Tang KD, Batra J, Ladwa R, Kenny L, Vasani S, Frazer IH, Dolcetti R, Ellis JJ, Sturm RA, Leo P, Punyadeera C. Analysis of human leukocyte antigen associations in human papillomavirus–positive and –negative head and neck cancer: Comparison with cervical cancer. Cancer. 2022. 10.1002/cncr.34148
The first 2 authors contributed equally to this article.
We acknowledge all the members of the Centre for Genomics and Personalized Health, including Sahana Manoli; all the clinical research coordinators, including Trang Le, Jennifer Edmunds, Charmaine Micklewright, Jacqui Keller, and Dana Middleton; and Darryl Irwin and Vandhana Bharti from Agena Bioscience (Australia).
References
- 1. de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141:664‐670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eckhardt M, Zhang W, Gross AM, et al. Multiple routes to oncogenesis are promoted by the human papillomavirus–host protein network. Cancer Discov. 2018;8:1474‐1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Otter S, Whitaker S, Chatterjee J, Stewart A. The human papillomavirus as a common pathogen in oropharyngeal, anal and cervical cancers. Clin Oncol. 2019;31:81‐90. [DOI] [PubMed] [Google Scholar]
- 4. Taylor S, Bunge E, Bakker M, Castellsagué X. The incidence, clearance and persistence of non‐cervical human papillomavirus infections: a systematic review of the literature. BMC Infect Dis. 2016;16:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wood ZC, Bain CJ, Smith DD, Whiteman DC, Antonsson A. Oral human papillomavirus infection incidence and clearance: a systematic review of the literature. J Gen Virol. 2017;98:519‐526. [DOI] [PubMed] [Google Scholar]
- 6. Bulkmans NWJ, Berkhof J, Bulk S, et al. High‐risk HPV type–specific clearance rates in cervical screening. Br J Cancer. 2007;96:1419‐1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. D’Souza G, McNeel TS, Fakhry C. Understanding personal risk of oropharyngeal cancer: risk‐groups for oncogenic oral HPV infection and oropharyngeal cancer. Ann Oncol. 2017;28:3065‐3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beachler DC, Sugar EA, Margolick JB, et al. Risk factors for acquisition and clearance of oral human papillomavirus infection among HIV‐infected and HIV‐uninfected adults. Am J Epidemiol. 2014;181:40‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kero K, Rautava J, Syrjanen K, Willberg J, Grenman S, Syrjanen S. Smoking increases oral HPV persistence among men: 7‐year follow‐up study. Eur J Clin Microbiol Infect Dis. 2014;33:123‐133. [DOI] [PubMed] [Google Scholar]
- 10. Nyitray AG, Carvalho da Silva RJ, Chang M, et al. Incidence, duration, persistence, and factors associated with high‐risk anal human papillomavirus persistence among HIV‐negative men who have sex with men: a multinational study. Clin Infect Dis. 2016;62:1367‐1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li W, Meng Y, Wang Y, et al. Association of age and viral factors with high‐risk HPV persistence: a retrospective follow‐up study. Gynecol Oncol. 2019;154:345‐353. [DOI] [PubMed] [Google Scholar]
- 12. Obiri‐Yeboah D, Akakpo PK, Mutocheluh M, et al. Epidemiology of cervical human papillomavirus (HPV) infection and squamous intraepithelial lesions (SIL) among a cohort of HIV‐infected and uninfected Ghanaian women. BMC Cancer. 2017;17:688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Louie KS, Castellsague X, de Sanjose S, et al. Smoking and passive smoking in cervical cancer risk: pooled analysis of couples from the IARC multicentric case‐control studies. Cancer Epidemiol Biomarkers Prev. 2011;20:1379‐1390. [DOI] [PubMed] [Google Scholar]
- 14. Magnusson PK, Lichtenstein P, Gyllensten UB. Heritability of cervical tumours. Int J Cancer. 2000;88:698‐701. [DOI] [PubMed] [Google Scholar]
- 15. Moore EE, Wark JD, Hopper JL, Erbas B, Garland SM; CeCaGeEn Study Group . The roles of genetic and environmental factors on risk of cervical cancer: a review of classical twin studies. Twin Res Hum Genet. 2012;15:79‐86. [DOI] [PubMed] [Google Scholar]
- 16. Miura K, Mishima H, Yasunami M, et al. A significant association between rs8067378 at 17q12 and invasive cervical cancer originally identified by a genome‐wide association study in Han Chinese is replicated in a Japanese population. J Hum Genet. 2016;61:793‐796. [DOI] [PubMed] [Google Scholar]
- 17. Ivansson EL, Juko‐Pecirep I, Erlich HA, Gyllensten UB. Pathway‐based analysis of genetic susceptibility to cervical cancer in situ: HLA‐DPB1 affects risk in Swedish women. Genes Immun. 2011;12:605‐614. [DOI] [PubMed] [Google Scholar]
- 18. Shi Y, Li L, Hu Z, et al. A genome‐wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat Genet. 2013;45:918‐922. [DOI] [PubMed] [Google Scholar]
- 19. Chen D, Enroth S, Liu H, et al. Pooled analysis of genome‐wide association studies of cervical intraepithelial neoplasia 3 (CIN3) identifies a new susceptibility locus. Oncotarget. 2016;7:42216‐42224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leo PJ, Madeleine MM, Wang S, et al. Defining the genetic susceptibility to cervical neoplasia—a genome‐wide association study. PLoS Genet. 2017;13:e1006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen D, Juko‐Pecirep I, Hammer J, et al. Genome‐wide association study of susceptibility loci for cervical cancer. J Natl Cancer Inst. 2013;105:624‐633. [DOI] [PubMed] [Google Scholar]
- 22. Lesseur C, Diergaarde B, Olshan AF, et al. Genome‐wide association analyses identify new susceptibility loci for oral cavity and pharyngeal cancer. Nat Genet. 2016;48:1544‐1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei Q, Yu D, Liu M, et al. Genome‐wide association study identifies three susceptibility loci for laryngeal squamous cell carcinoma in the Chinese population. Nat Genet. 2014;46:1110‐1114. [DOI] [PubMed] [Google Scholar]
- 24. Shete S, Liu H, Wang J, et al. A genome‐wide association study identifies two novel susceptible regions for squamous cell carcinoma of the head and neck. Cancer Res. 2020;80:2451‐2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tang KD, Kenny L, Frazer IH, Punyadeera C. High‐risk human papillomavirus detection in oropharyngeal cancers: comparison of saliva sampling methods. Head Neck. 2019;41:1484‐1489. [DOI] [PubMed] [Google Scholar]
- 26. Ekanayake Weeramange C, Liu Z, Hartel G, et al. Salivary high‐risk human papillomavirus (HPV) DNA as a biomarker for HPV‐driven head and neck cancers. J Mol Diagn. 2021;23:1334‐1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang KD, Menezes L, Baeten K, et al. Oral HPV16 prevalence in oral potentially malignant disorders and oral cavity cancers. Biomolecules. 2020;10:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Purcell S, Neale B, Todd‐Brown K, et al. PLINK: a tool set for whole‐genome association and population‐based linkage analyses. Am J Hum Genet. 2007;81:559‐575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Duffy DL, Jagirdar K, Lee KJ, et al. Genes determining nevus count and dermoscopic appearance in Australian melanoma cases and controls. J Invest Dermatol. 2020;140:498‐501.e417. [DOI] [PubMed] [Google Scholar]
- 30. Koh U, Janda M, Aitken JF, et al. ‘Mind Your Moles’ study: protocol of a prospective cohort study of melanocytic naevi. BMJ Open. 2018;8:e025857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neefjes J, Jongsma MLM, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823‐836. [DOI] [PubMed] [Google Scholar]
- 32. Robinson J, Soormally AR, Hayhurst JD, Marsh SGE. The IPD‐IMGT/HLA database—new developments in reporting HLA variation. Hum Immunol. 2016;77:233‐237. [DOI] [PubMed] [Google Scholar]
- 33. Dendrou CA, Petersen J, Rossjohn J, Fugger L. HLA variation and disease. Nat Rev Immunol. 2018;18:325‐339. [DOI] [PubMed] [Google Scholar]
- 34. Brown MA, Leo PJ. Genetic susceptibility to cervical neoplasia. Papillomavirus Res. 2019;7:132‐134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3
Table S4


