SUMMARY
Scent bouquets produced by the flowers of Petunia spp. (petunia) are composed of a complex mixture of floral volatile benzenoid and phenylpropanoid compounds (FVBPs), which are specialized metabolites derived from phenylalanine (Phe) through an interconnected network of enzymes. The biosynthesis and emission of high levels of these volatiles requires coordinated transcriptional activation of both primary and specialized metabolic networks. The petunia R2R3‐MYB transcription factor ODORANT 1 (ODO1) was identified as a master regulator of FVBP production and emission; however, our knowledge of the direct regulatory targets of ODO1 has remained limited. Using chromatin immunoprecipitation followed by sequencing (ChIP‐seq) in petunia flowers, we identify genome‐wide ODO1‐bound genes that are enriched not only in genes involved in the biosynthesis of the Phe precursor, as previously reported, but also genes associated with the specialized metabolic pathways involved in generating phenylpropanoid intermediates for FVBPs. ODO1‐bound genes are also involved in methionine and S‐adenosylmethionine metabolism, which could modulate methyl group supplies for certain FVBPs. Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) and RNA‐seq analysis in an ODO1 RNAi knockdown line revealed that ODO1‐bound targets are expressed at lower levels when ODO1 is suppressed. A cis‐regulatory motif, CACCAACCCC, was identified as a potential binding site for ODO1 in the promoters of genes that are both bound and activated by ODO1, which was validated by in planta promoter reporter assays with wild‐type and mutated promoters. Overall, our work presents a mechanistic model for ODO1 controlling an extensive gene regulatory network that contributes to FVBP production to give rise to floral scent.
Keywords: floral scent, volatiles, plant metabolism, ChIP‐seq, promoter motif, transcription factor, Petunia hybrida
Significance Statement
Petunia floral scent production and emission is highly regulated, with a major role for the transcription factor ODORANT 1 (ODO1). This paper describes the ODO1‐regulated gene network of petunia, which extends to branches involved in the production of phenylpropanoid intermediates and S‐adenosylmethionine biosynthesis, along with the identification of an ODO1‐binding motif.
INTRODUCTION
Volatile organic compounds emitted as constituents of floral scent bouquets play an important role in the reproductive success of many flowering plants by attracting pollinators. The volatile metabolite profiles and the timing of their emission are attuned to specific pollinators, and can be a driver of speciation through reproductive isolation (Byers et al., 2014a, 2014b; Whitehead and Peakall, 2014). In Petunia spp. (petunia), the scent bouquet is comprised of a mixture of floral volatile benzenoids and phenylpropanoids (FVBPs), which are derived from phenylalanine (Phe) (Boatright et al., 2004; Colquhoun et al., 2010; Hahlbrock and Scheel, 1989; Herrmann, 1995; Verdonk et al., 2003). The FVBPs are synthesized in and emitted from the corolla. Their biosynthesis and emission are developmentally regulated with sharp induction at anthesis and follow a diurnal rhythm, with the maximum occurring around midnight (Hoballah et al., 2005; Verdonk et al., 2003). The production of FVBPs relies on primary metabolic pathways, which provide precursors and energy, as well as on a large network of enzymes generating numerous intermediates for their formation (Muhlemann et al., 2014a). Thus, the biosynthesis and release of FVBPs requires coordinated regulation of gene expression of multiple enzymatic steps among and within biosynthetic pathways to attain the production of high levels of Phe and FVBPs in the corolla. The R2R3‐MYB transcription factor ODORANT 1 (ODO1) has been identified as a key master regulator of FVBP biosynthesis and emission in petunia (Verdonk et al., 2005). ODO1 is highly expressed in corolla of fragrant cultivars of the popular domesticated bedding plant, Petunia hybrida, whereas in most non‐fragrant cultivars there is little to no ODO1 expression (Moerkercke et al., 2011; Verdonk et al., 2005). Similarly, in wild petunia species, ODO1 transcript levels are high in fragrant but low in non‐fragrant plants. A comparative study of two wild petunia species with distinct pollination syndromes revealed that ODO1 is one of the two major quantitative trait loci that determines pollinator choice by controlling FVBP emission (Klahre et al., 2011).
The 1.2‐kbp promoter of ODO1 is sufficient to drive its developmental and diurnal expression specifically in corolla (Moerkercke et al., 2011). The circadian clock transcription factor LATE ELONGATED HYPOCOTYL (LHY) negatively regulates ODO1 expression by binding to the two evening elements in the ODO1 promoter and thus regulating diurnal changes in FVBP emission (Fenske et al., 2015). Two other R2R3‐MYB transcription factors, EMISSION OF BENZENOIDS (EOB) I and II, also bind the promoter region, but, in contrast to LHY, activate ODO1 expression and positively regulate FVBP production and emission (Moerkercke et al., 2011; Spitzer‐Rimon et al., 2012). Indeed, EOBII recognizes two MYB binding sites in the ODO1 promoter, which are required for its strong activity (Moerkercke et al., 2011). In addition, EOBI and EOBII appear to have overlapping roles in regulating petunia FVBP biosynthesis: they can both activate the promoters of ISOEUGENOL SYNTHASE (IGS) and l‐PHENYLALANINE AMMONIA LYASE (PAL) involved in the formation of FVBPs (Spitzer‐Rimon et al., 2010, 2012). In Arabidopsis an MYB transcription factor belonging to the same clade as ODO1, MYB99, forms a similar transcriptional network with MYB21 and MYB24, orthologs of petunia EOBI and EOBII, respectively (Battat et al., 2019). AtMYB99 binds to promoters of primary metabolic and phenylpropanoid biosynthetic genes to direct the synthesis of pollen wall components. This suggests that in other plants, orthologous transcriptional networks to ODO1, EOBI and EOBII can regulate diverse branches of phenylpropanoid biosynthesis.
Although ODO1 was among the first transcription factors recognized to regulate biosynthesis and the emission of FVBPs, the exact nature and range of its regulatory targets remains largely unknown. To date, the genes regulated by ODO1 have primarily been inferred from changes in their expression measured in transgenic lines. Suppression of ODO1 by RNAi approach (odo1i lines) in fragrant P. hybrida cv. W115 led to a strong reduction in the emission of FVBPs, including the C6‐C2 compounds phenylacetaldehyde and phenylethylbenzoate, the C6‐C1 compounds benzylbenzoate, methylbenzoate and benzylacetate and their intermediate precursor benzoic acid, and the C6‐C3 compounds vanillin and isoeugenol (Verdonk et al., 2005). This reduction in emission in ODO1 RNAi lines was accompanied by the differential expression of several genes encoding enzymes contributing to Phe biosynthesis and phenylpropanoid metabolism. Transcript levels of genes encoding 3‐DEOXY‐d‐ARABINO‐HEPTULOSONATE‐7‐PHOSPHATE SYNTHASE (DAHPS), 5‐ENOLPYRUVYLSHIKIMATE‐3‐PHOSPHATE SYNTHASE (EPSPS) and CHORISMATE MUTASE (CM), members of the shikimate and Phe biosynthetic pathways, were lower when ODO1 expression was suppressed. Moreover, the contribution of ODO1 to transcriptional regulation of EPSPS was supported by the activation of the EPSPS promoter in the presence of transiently expressed ODO1 (Verdonk et al., 2005). The expression of two PAL genes, which encode enzymes directing the precursor flux into the formation of FVBPs, was also reduced in odo1i lines. In contrast, transcript levels of S‐ADENOSYL‐l‐METHIONINE:BENZOIC ACID/SALICYLIC ACID CARBOXYL METHYLTRANSFERASE (BSMT) and BENZOYL‐COA:BENZYLALCOHOL/2‐PHENYLETHANOL BENZOYLTRANSFERASE (BPBT), which encode enzymes acting at the final steps of volatile compound formation, were higher in odo1i lines. Based on these results, it was proposed that the primary function of ODO1 is to potentiate FVBP production by increasing the supply of Phe precursor through transcriptional activation of the shikimate pathway genes. However, the far‐reaching effects of ODO1 suppression on FVBP emission in petunia suggest that ODO1 directly regulates a larger range of target genes, including the specialized metabolic pathway genes. This was also supported by the observation that expression of PhODO1 in Solanum lycopersicum (tomato) led to increased transcript levels of not only the shikimate pathway genes but also downstream phenylpropanoid pathway biosynthetic genes (Cin et al., 2011). In addition, orthologs of ODO1 in other species have been found to be involved in regulating both primary and specialized metabolism (Battat et al., 2019).
Here, we show that the role of ODO1 extends well beyond regulating Phe precursor supply to FVBP biosynthesis by identifying the direct genome‐wide targets of ODO1 using chromatin immunoprecipitation followed by sequencing (ChIP‐seq). We found that in addition to genes from the shikimate pathway, ODO1 binds to promoters of the specialized metabolic pathway genes from several phenylpropanoid branches. Transcriptional regulation of these ODO1 targets was confirmed with qRT‐PCR and RNA‐seq analysis in an odo1i line. A cis‐regulatory motif was identified as enriched in the promoters of ODO1‐bound genes that exhibit reduced expression in the odo1i plants. Mutating this motif in the promoters of EPSPS1 and PALc fully ablated their transcriptional activation by ODO1 in planta. Overall, we propose that ODO1 binds and activates a wide range of FVBP biosynthesis genes through the recognition of a specific promoter motif to activate floral volatile synthesis and emission in the corolla.
RESULTS
ODO1 protein shows evening accumulation in the nucleus
It was previously shown that ODO1 transcript levels exhibited a diurnal oscillation pattern post‐anthesis, peaking at dusk and coinciding with the expression of FVBP biosynthetic genes and maximum volatile emission (Verdonk et al., 2005). qRT‐PCR analysis of ODO1 expression in petunia flowers grown under controlled light conditions confirmed that the ODO1 transcript level was very low early in the day before reaching a maximum at 5:00 pm, an hour before dark (Figure S1). To test whether this diurnal oscillation of mRNA level is at least partially driven by the promoter activity, the day–night oscillations of ODO1 promoter activity were analyzed using a transgenic petunia line carrying a pODO1:LUC construct (Video Clip S1). Hourly measurements of luciferase intensity display a pattern similar to that of qRT‐PCR expression, with a peak at 5:00 pm (Figure S1), thus supporting the proposal that both the promoter of ODO1 and its transcript level are subject to diurnal regulation.
However, whether ODO1 protein levels show similar fluctuations during the day–night cycle was unknown. Thus, P. hybrida cv. W115 was stably transformed with a construct containing GFP‐tagged ODO1 protein driven by its native promoter, pODO1:GFP‐ODO1. The rhythmic cycling of GFP‐ODO1 was then investigated by confocal microscopy (Figure 1). For visualization of the nucleus, corollas were additionally transiently transformed with 35S:NLS‐mCherry (nuclear localization signal fused to mCherry) (Moerkercke et al., 2012). As expected, except for auto‐fluorescence of the cell wall, no fluorescent GFP signal was observed in corollas of wild‐type plants (Figure 1). In pODO1:GFP‐ODO1 plants, no nuclear GFP signal was detected in corollas at 9:00 am. Conversely, at 5:00 pm a strong nuclear GFP signal was observed at both the adaxial and abaxial sides of the corollas (Figure 1). These results support the localization of ODO1 protein in the nucleus, and show that nuclear ODO1 protein levels fluctuate in a diurnal pattern with an evening peak similar to the ODO1 transcript levels (Figure S1) and concomitant with ODO1 promoter activity, as measured using the luciferase reporter (Figure S1; Video Clip S1). The evening accumulation of ODO1 within the nucleus is concordant with the prominent role of ODO1 in activating the evening expression of FVBP biosynthesis genes.
Figure 1.
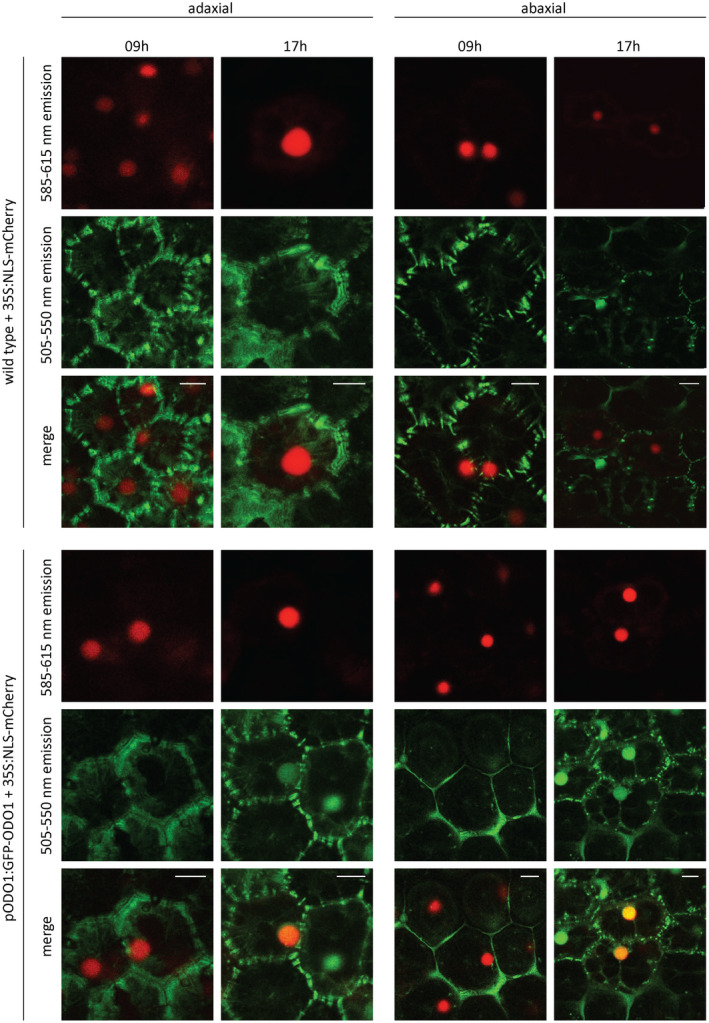
Nuclear localization and rhythmic cycling of GFP‐ODO1 protein. Corollas of pODO1:GFP‐ODO1 line 10 (T2) and WT petals were transiently transformed with 35S:NLS‐mCherry. At 9:00 h in the morning, seven hours after end of dark, and at 17:00 h, one hour before dark, pictures were taken from the adaxial and abaxial site of corollas with a confocal microscope. mCherry signal (585‐615nm emission band) is in red, GFP signal and autofluorescence in the 505‐550nm band pass emission filter is in green, while overlapping mCherry and GFP signal appears yellow/orange. Abbreviations: GFP: GREEN FLUORESCENT PROTEIN, ODO1: ODORANT1; pODO1, 1.9 kbp promoter of ODO1; NLS, nuclear localization signal; WT, wild type.
ODO1 binds a wide range of genes throughout the FVBP metabolic network
To better understand the regulatory regime of ODO1 in governing Phe and FVBP metabolic networks, ChIP‐seq was performed to identify the directly bound targets of ODO1. Petunia corollas were harvested 1 hour before dark (5:00 pm) from two transgenic petunia lines, either stably transformed with pODO1:GFP‐ODO1 or from flowers transiently expressing 35S:GFP‐ODO1. Chromatin was extracted from corollas, and after shearing into nucleosomes, it was immunoprecipitated with an anti‐GFP antibody to pull down GFP‐tagged ODO1. Subsequently, the co‐precipitated genomic DNA was sequenced by Illumina sequencing, and ChIP‐seq reads were aligned with the P. hybrida parental genomes, Petunia axillaris and Petunia inflata (Bombarely et al., 2016). Binding peaks were identified genome wide by comparing mapped ChIP‐seq reads with unimmunoprecipated chromatin background using the macs2 algorithm (false discovery rate (FDR)‐corrected P < 0.05) (Zhang et al., 2008). Genes that were bound by ODO1 were determined using P. axillaris genome annotation, as 86% of the mapped ChIP‐seq reads align with the P. axillaris genome, in agreement with our previous study (Patrick et al., 2021). To determine the direct gene targets of ODO1 with high confidence, only binding peaks identified as present in both the transient 35S:GFP‐ODO1 and the stable pODO1:GFP‐ODO1 corollas were used to generate the set of ODO1‐bound genes.
This analysis revealed 1713 ODO1‐bound genes (Supplementary information, with two genes shown as examples in Figures 2a and S3). Interestingly, the ODO1 ChIP‐seq signal spans from the promoter to the gene body (Figure 2a), peaking around the transcription start site (TSS) and the first exon. A similar genic positional distribution of ChIP‐seq signals has been reported for other transcription factors (TFs) in plants (Liu et al., 2018; Martin et al., 2018; Shamimuzzaman and Vodkin, 2013), which could be the result of regulatory binding in both the promoter and the gene body (Meng et al., 2021) or of cross‐linking with other proteins in the transcriptional machinery. To determine the function of the ODO1‐bound genes, Gene Ontology (GO) term enrichment analysis was performed, as previously described (Patrick et al., 2021). Overall, 29 GO terms were found to be significantly enriched (Table S1), which primarily encompassed terms related to stress responses and specific metabolic pathways (Figure 2b). Given the association of ODO1 with the transcriptional regulation of FVBP metabolic genes, the 11 metabolism‐related significant GO terms were examined further (Figure 2c). Genes bound by ODO1 were associated with ‘chorismate biosynthesis’ (FDR‐corrected P < 0.00178), ‘aromatic amino acid family biosynthetic process’ (P < 0.0113) and ‘phenylpropanoid metabolic process’ (P < 0.0241), consistent with previous observations that ODO1 regulates genes in the shikimate pathway in order to increase Phe biosynthesis (Verdonk et al., 2005). In addition, it was also found that ODO1‐bound genes were involved in the ‘lignin biosynthetic process’ (P < 0.0138), indicating that ODO1 may also play a role in directly regulating genes that are responsible for generating the monolignol precursor coniferyl alcohol for the synthesis of the FVBPs eugenol and isoeugenol (Muhlemann et al., 2014b; Shaipulah et al., 2016). Finally, GO terms ‘methionine biosynthetic process’ (P < 1.01e–04) and ‘S‐adenosylmethionine (SAM) biosynthetic process’ (P < 8.28e–06) were found to be significantly enriched. This is in line with our previous report that the expression of SAM SYNTHETASE was affected by ODO1 knockdown (Verdonk et al., 2005), and our new findings now expand this to the genome‐wide level and reveal that ODO1 directly interacts with multiple genes involved in the methionine biosynthetic pathway. Such regulation by ODO1 is likely to increase the level of SAM for the production of the methylated‐FVBP methyl benzoate, and the intermediate feruloyl‐CoA for the synthesis of eugenol and isoeugenol (Negre et al., 2003).
Figure 2.
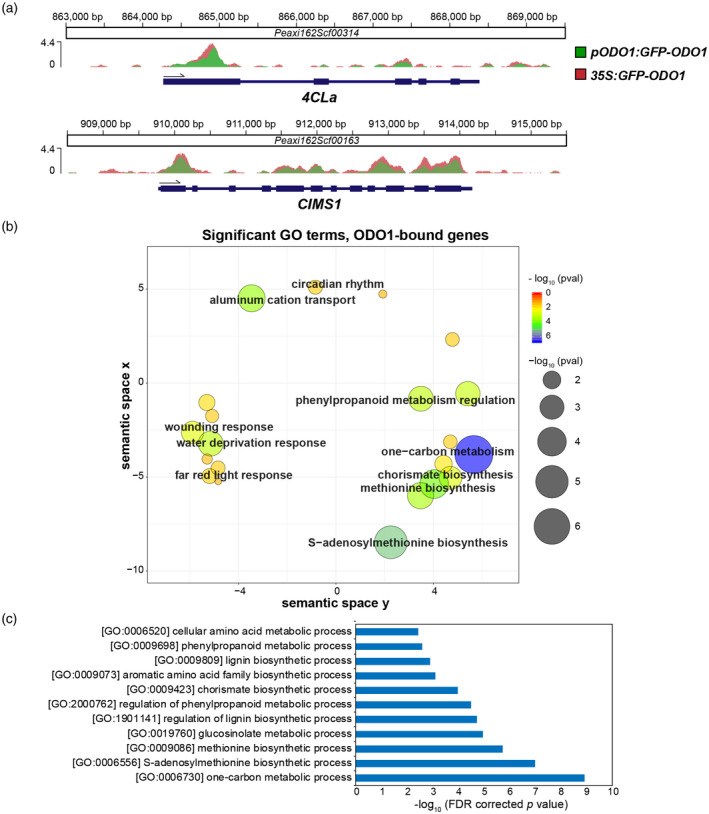
ODO1‐bound genes determined by ChIP‐seq. (a) ChIP‐seq signals at two exemplary gene loci with detected ODO1 binding. The Y axis represents the sequencing depth covered by ChIP‐seq in fragments per million, after normalization to the total number of unique aligned fragments in each library. Ruler markings indicate location of ChIP signal along the corresponding petunia scaffolds, with annotated gene features below (thicker bar sections for coding regions, thinner sections for introns and untranslated regions). (b) ReviGO plot showing enriched GO terms among ODO1‐bound genes. Significantly enriched GO terms were determined among ODO1‐bound genes against P. axillaris background (FDR < 0.05), and then visualized by ReviGO to reduce the redundancy within the list of identified GO terms. In the ReviGO plot, significant GO terms are shown as circles in a two‐dimensional space, which was derived by multidimensional scaling of a semantic similarity matrix of the GO terms. Therefore, similar GO terms are close to each other in the RevoGO plot. Size and color of the circles represent the significance of enrichment measured as ‐log10(FDR corrected p value). (c) Metabolic GO terms significantly enriched among ODO1‐bound genes are shown and sorted in order of significance. Abbreviations: 4CL, 4‐coumaryl‐CoA ligase; CIMS, cobalamin‐independent methionine synthase; ODO1, ODORANT1.
We further examined the overlap between the ODO1‐bound genes and genes upregulated during flower opening, when ODO1 is induced (Verdonk et al., 2005), and the majority of FVBP‐related genes are strongly upregulated (Patrick et al., 2021). There was a significant overlap between the ODO1‐bound genes and the genes that were upregulated on day 2 post‐anthesis in the corolla (enrichment P < 2.04e–27, determined by the hypergeometric method), but not with those that were downregulated (Figure S4), consistent with the expected role of ODO1 in activating genes during flower opening to induce FVBP production and emission.
Next, the specific points of transcriptional regulation by ODO1 within the FVBP pathways were identified based on extensive knowledge of these pathways in petunia. A total of 68 genes involved in producing phenylalanine‐derived FVBPs have been annotated in the P. axillaris genome (Patrick et al., 2021), including the shikimate/phenylalanine pathway (Dewick, 1994), the general phenylpropanoid reactions (that convert Phe to coumaroyl‐CoA; Vogt, 2010), volatile benzenoid and phenylpropanoid (C6‐C1 and C6‐C2) biosynthesis, and eugenol and isoeugenol (C6‐C3) synthesis through the monolignol branch (Figure 3a). Interestingly, we observed a broad distribution of ODO1‐bound genes throughout the FVBP metabolic network (Figure 3a), with a significant enrichment of ODO1‐bound genes among the entirety of the FVBP gene networks (32 out of 68, enrichment P < 2.02e–24 determined by the hypergeometric method; Figure 3b). Specifically, the binding of ODO1 was concentrated among genes involved in synthesis of FVBP precursors and intermediate metabolites (30 of 56, enrichment P < 3.83e–25) (Figure 3b). In contrast, ODO1 binding was not significantly enriched among gene loci encoding end‐step enzymes that synthesize volatile products for emission, with binding only detected at two gene loci, which encode copies of BPBT and PHENYLACETALDEHYDE SYNTHASE (PAAS) enzymes. There was significant enrichment of ODO1‐bound genes within all of the individual pathways examined (Figure 3b), including eight out of 24 shikimate and phenylalanine biosynthetic genes (P < 9.96e–06), seven out of 11 general phenylpropanoid pathway genes (P < 1.58e–07), eight out of 19 volatile benzenoid biosynthetic genes (P < 9.96e–06), and nine out of 15 monolignol pathway and eugenol/isoeugenol biosynthetic genes (P < 4.95e–09). These results suggest that ODO1 binding is distributed widely throughout the branches of the FVBP synthesis network and is not only limited to the formation of primary metabolic precursors, as had been proposed previously (Verdonk et al., 2005).
Figure 3.
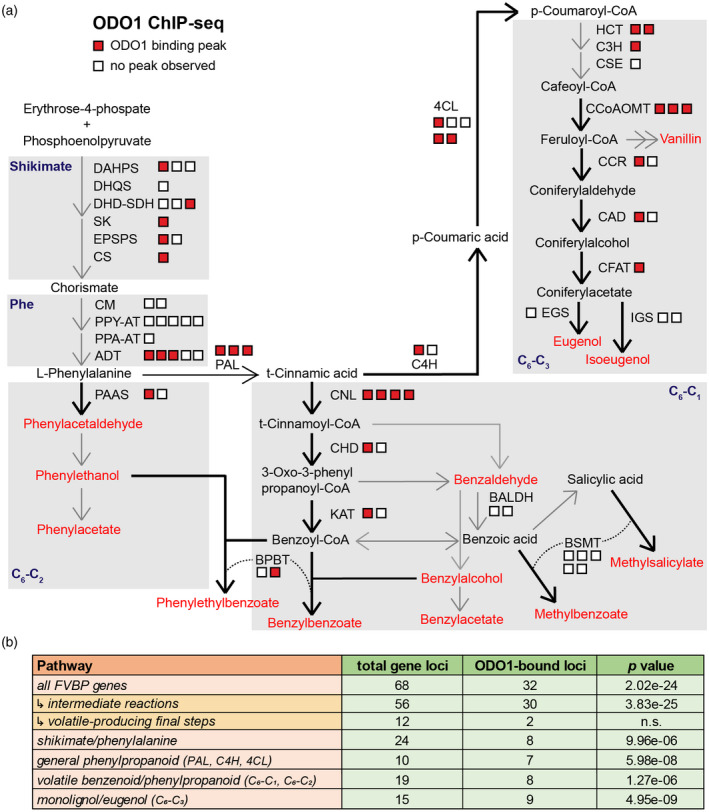
Pathways enriched for ODO1 ChIP‐seq binding. (a) Gene loci encoding enzymes contributing to phenylalanine biosynthesis and phenylpropanoid pathways leading to production of volatiles (emitted FVBPs in red text). Squares represent individual loci for a gene encoding a protein that is active in a particular pathway or that catalyzes the conversion of depicted metabolites, with red squares representing ODO1‐bound loci. (b) Hypergeometric p value for enrichment of ODO1‐bound genes among those belonging to FVBP and phenylpropanoid pathways in petunia (Patrick et al., 2021). Genes were assigned to the shikimate/phenylalanine pathways, the general phenylpropanoid reactions (PAL, C4H, and 4CL genes), the volatile benzenoid and phenylpropanoid branches (C6‐C1 and C6‐C2), and monolignol/eugenol branch (C6‐C3). Abbreviations: 4CL, 4‐coumaryl‐CoA ligase; ADT, arogenate dehydratase; BALDH, benzaldehyde dehydrogenase; BPBT, benzoyl‐CoA:benzylalcohol/phenylethanol benzoyltransferase; BSMT, benzoic acid/salicylic acid carboxyl methyltransferase; C3H, coumarate 3‐hydroxylase; C4H, cinnamate 4‐hydroxylase; CAD, cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl‐CoA O‐methyltransferase; CCR, cinnamoyl‐CoA reductase; CFAT, coniferyl alcohol acetyltransferase; CHD, cinnamoyl‐CoA hydratase‐dehydrogenase; CM, chorismate mutase; CNL, cinnamoyl‐CoA ligase; CS, chorismate synthase; CSE, caffeoyl shikimate esterase; DAHPS, 3‐deoxy‐D‐arabino‐heptulosonate 7‐phosphate synthase; DHD‐SDH, 3‐dehydroquinate dehydratase and shikimate dehydrogenase; DHQS, 3‐dehydroquinate synthase; EGS, eugenol synthase; EPSPS, 5‐enolpyruvylshikimate 3‐phosphate; HCT, hydroxycinnamoyl‐CoA:shikimate/quinate hydroxycinnamoyl transferase; IGS, isoeugenol synthase; KAT, 3‐ketoacyl‐CoA thiolase; PAAS, phenylacetaldehyde synthase; ODO1, ODORANT1; PAL, phenylalanine ammonia lyase; PPA‐AT, prephenate aminotransferase; PPY‐AT, phenylpyruvate aminotransferase; SK, shikimate kinase.
As GO terms associated with methionine and SAM biosynthesis were enriched among ODO1‐bound genes (Figures 2b,c), we investigated this in further detail. Based on homology with Arabidopsis thaliana genes, we identified petunia genes involved in the SAM cycle and the folate cycle (Hanson and Roje, 2001; Sauter et al., 2013) (Table S2). The folate cycle regenerates 5‐methyltetrahydrofolate, which is a source of single‐carbon units necessary for methionine biosynthesis from homocysteine (Figure S5). There was strong enrichment of ODO1‐bound genes among genes involved in generating methionine and SAM (14 out of 22, with enrichment P < 6.95e–14) (Figure S5). As the biosynthesis of major FVBPs, including methylbenzoate, methylsalicylate, eugenol, isoeugenol and vanillin, requires methyltransferases that use SAM as the source of methyl groups, our finding further supports a broader role for ODO1 in modulating the expression of methionine and SAM genes to potentiate FVBP production.
The ODO1‐bound genes are downregulated by RNAi knockdown of ODO1
To determine whether ODO1 indeed regulates the transcription of ODO1‐bound genes in primary and specialized metabolic pathways, qRT‐PCR was performed using RNA isolated from 2‐day‐old flowers of the wild type (WT, P. hybrida cv. W115) and a previously characterized RNAi suppression line, odo1i (Verdonk et al., 2005). In this line, the ODO1 expression level was moderately suppressed, to about 58% of wild‐type expression (Figure 4a). Concordant with this, most ODO1‐bound genes tested showed a statistically significant decreases in expression, including shikimate pathway genes (EPSPS1 and DHD‐SDH3), two PAL genes, monolignol pathway genes (HCT2 and CAD1) and methionine/SAM genes (CIMS1 and SAHH1) (Figure 4a). In contrast, the expression of several FVBP genes, which based on ChIP‐seq analysis are not ODO1 targets, was unaffected in odo1i (Figure 4b). Therefore, ODO1 binding determined by ChIP‐seq appears to be associated with transcriptional activation by ODO1, including genes from metabolic pathways outside of shikimate/phenylalanine biosynthesis that contribute to FVBP production.
Figure 4.
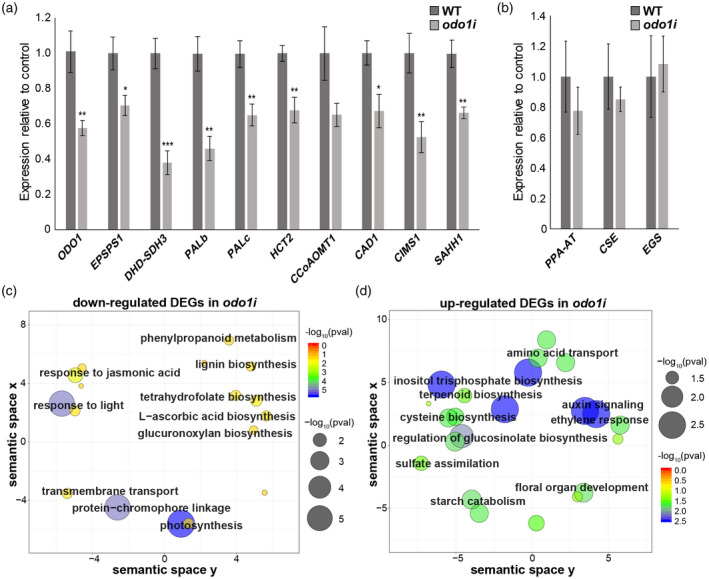
ODO1‐regulated genes. (a) Expression levels of selected ODO1‐bound genes in WT and ODO1 RNAi (odo1i) flowers measured by qRT‐PCR. Expression was normalized to the petunia housekeeping reference gene EF1a (Mallona et al., 2010). Error bars represent standard error of the mean for biological replicates (n = 6). Asterisks denote significance of difference in measurement between WT and odo1i as determined by Student’s t test: (*), P < 0.05, (**), P < 0.01, (***), P < 0.001. (b) Expression levels of volatile biosynthesis genes in WT and odo1i which are not predicted ODO1 binding targets, normalized to EF1a. Error bars represent standard error of the mean for biological replicates (n = 3). (c–d) ReviGO plots showing GO terms significantly enriched among differentially expressed genes (DEGs) downregulated (c) or upregulated (d) in odo1i compared to wild‐type determined by RNA‐seq. In the ReviGO plot, significant GO terms are shown as circles in a two‐dimensional space, which was derived by multidimensional scaling of a semantic similarity matrix of the GO terms. Size and color of the circles represent the significance of enrichment measured as ‐log10(FDR corrected p value). Abbreviations: CAD, cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl‐CoA O‐methyltransferase; CIMS, cobalamin‐independent methionine synthase; CSE, caffeoyl shikimate esterase; DHD‐SDH, 3‐dehydroquinate dehydratase and shikimate dehydrogenase; EGS, eugenol synthase; EPSPS, 5‐enolpyruvylshikimate 3‐phosphate; HCT, hydroxycinnamoyl‐CoA:shikimate/quinate hydroxycinnamoyl transferase; ODO1, ODORANT1; PAL, phenylalanine ammonia lyase; PPA‐AT, prephenate aminotransferase; SAHH, S‐adenosyl‐L‐homocysteine hydrolase.
To gain a comprehensive insight into the transcriptional regulation of ODO1‐bound targets at a genome‐wide level, RNA‐seq was performed to compare wild‐type (cv. W115) and odo1i transgenic flowers using corolla tissue collected 2 days post‐anthesis, 1 hour before dark. Differentially expressed genes in the odo1i line relative to the wild‐type background were determined from the RNA‐seq data using deseq2 (Love et al., 2014). As shown by the qRT‐PCR results, the expression of ODO1 was only mildly reduced in the odo1i line, to 56% compared with the wild type. In the odo1i line, transcript levels of 634 genes were downregulated and 519 genes were upregulated relative to the wild type (FDR < 0.05) (Data S2). Subsequently, enriched GO terms were determined for upregulated and downregulated genes (Figure 4c,d; Tables S3 and S4). Consistent with our results for ODO1‐bound genes, downregulated genes were enriched with GO terms (Figure 4c; Table S3), including ‘phenylpropanoid metabolism’ (P < 0.029) and ‘lignin biosynthesis’ (P < 0.029) that are potentially related to FVBP biosynthesis (Figure 3), as well as ‘tetrahydrofolate biosynthesis’ (P < 0.019), contributing to the SAM cycle (Figure S5). Upregulated genes were associated with several GO terms for sucrose and starch metabolism and signaling pathways, and interestingly for ‘cysteine biosynthesis’ (P < 0.0025), which provides the precursor to methionine biosynthesis (Figure 4d; Table S4). As expected for the role of ODO1 as a positive transcriptional regulator (Verdonk et al., 2005), ODO1‐bound genes show a significant overlap (hypergeometric P < 7.83e–09) with genes that are downregulated in the odo1i line (Figure 5a). Unexpectedly, there is also a significant overlap (P < 4.88e–06) between genes upregulated in the odo1i line and ODO1‐bound genes (Figure 5b), indicating a previously unknown role of ODO1 in repressing the expression of certain genes.
Figure 5.
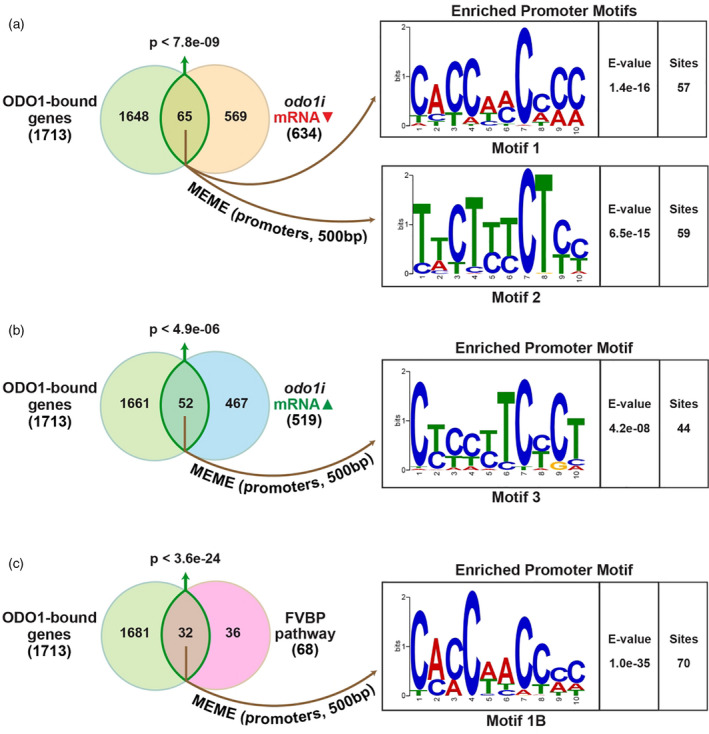
Promoter motifs enriched in ODO1‐bound genes. Overlaps between ODO1‐bound genes determined by ChIP‐seq with (a‐b) genes differentially regulated in odo1i flowers detected by RNA‐seq or (c) FVBP pathway genes were shown, with significance of the overlap calculated using hypergeometric distribution. Enriched cis‐regulatory motifs in the 500bp promoters of the intersecting gene sets were identified by the MEME toolkit with corresponding E‐value for significance of identified motif occurrence relative to random background and number of sites matched within supplied promoter regions.
An MYB binding motif is associated with ODO1 binding
To identify the cis‐regulatory binding motif for ODO1, the meme toolkit (Bailey et al., 2009) was applied to analyze the promoter sequence of genes that are bound by ODO1 as well as differentially expressed in the odo1i line. A cis motif, CACCAACCCC (motif 1; Figure 5a), was enriched among the promoters of genes bound by ODO1 and activated by ODO1 (i.e. downregulated in odo1i), but was not detected among the promoters of ODO1‐bound genes that are upregulated in odo1i (Figure 5b). This motif was therefore considered as a candidate binding site of ODO1 in the promoters of target genes for their transcriptional activation.
The motif 1 identified is similar to the ACC(T/A)ACC motif bound by ZmMYB31, a regulator of the lignin biosynthetic pathway in Zea mays (maize) (Fornalé et al., 2010). It is also similar to the AC element sequence AC‐II (ACCAACC) involved in the regulation of phenylpropanoid and lignin biosynthetic genes in Arabidopsis (Raes et al., 2003; Zhong and Ye, 2012). A comparison between motif 1 and the motifs present in the DAP‐SEQ database (O’Malley et al., 2016) using tomtom (Gupta et al., 2007) revealed that the binding motifs for MYB63, MYB83 and MYB58 were the best matches (Figure S6). MYB58 and MYB63 are involved in regulating lignin biosynthetic pathways in Arabidopsis by transcriptionally activating general phenylpropanoid and monolignol biosynthetic pathways (Zhou et al., 2009). MYB83 also plays a role in lignin biosynthesis, at least in part through the activation of MYB63 (McCarthy et al., 2009). All three Arabidopsis transcription factors have been experimentally verified to bind target promoters through a core AC element sequence identical to that predicted for ODO1 in our study (Zhong and Ye, 2012; Zhou et al., 2009). Motifs 2 and 3, on the other hand, do not resemble the canonical MYB binding sequence and lack significant similarity to any known binding motifs present in the DAP‐SEQ database. They may therefore represent targets bound via ODO1 interaction with another unknown transcriptional regulator.
To gain mechanistic insights into the regulation of FVBP pathways, the meme toolkit was applied to detect significant cis‐regulatory motifs in the promoters of FVBP pathway genes that are bound by ODO1 (Figure 5c). A significantly over‐represented cis‐regulatory motif almost identical to motif 1 was discovered by this approach (motif 1B, Figure 5c). Motif 1B was found in promoters of the shikimate (DAHP1, DHD‐SDH3, SK, EPSPS1 and CS), phenylalanine (ADT1 and ADT3) and monolignol (CCoAOMT1, CCoAOMT3, HCT1, HCT2, C3H and CAD1) biosynthetic genes, as well as in genes involved in the general phenylpropanoid pathway (PAL, C4Ha and three isoforms of 4CL) (Figure S7). However, few occurrences of motif 1B were observed in genes encoding enzymes catalyzing the formation of volatile benzenoid products, with the exception of benzyl benzoate (BPBT2) and its precursor benzoyl‐CoA (CNL3 and CHD1) (Figure S7). Overall, motif 1/1B (CACCAACCCC) is a highly promising candidate for the cis‐regulatory motif recognized by ODO1 to activate its target genes.
The ODO1‐binding motif identified is required for the transcriptional activation of FVBP promoters by ODO1
To test whether the putative ODO1‐binding motif (motif 1/1B, CACCAACCCC) in the promoter of FVBP genes is required for their transcriptional activation by ODO1, promoter activity assays were performed using Nicotiana benthamiana leaves transiently co‐expressing 35S:ODO1 and a GUS reporter driven by either native or mutated promoters of EPSPS1 and PALc. Promoter regions immediately upstream of the predicted TSS, containing all the predicted binding motifs (Figure S7), were cloned based on the P. hybrida cv. W115 sequence. Specifically, the ODO1‐binding motif was identified at two locations within the 262‐bp EPSPS1 promoter (Figure 6a) and three locations within the 270‐bp PALc promoter (Figure 6b) using meme (see Figure S7). Mutant promoter constructs were generated by changing the third, fourth, seventh and eighth nucleotides in each motif (Figure 6a,b) to ablate the binding sites. The unrelated tomato transcription factor MYC1 (Spyropoulou et al., 2014) was used as a control for baseline promoter activity, in addition to an empty vector (EV) control. Furthermore, we also tested whether the GFP‐ODO1 fusion protein could activate the native promoters. Although native promoters for EPSPS1 (Figure 6c) and PALc (Figure 6d) were able to activate the transcription of GUS in the presence of ODO1, promoters with mutated ODO1‐binding motifs exhibited only the baseline level of GUS transcription, similar to that observed with the native promoter in the presence of MYC1 or EV (Figure 6c,d). The native promoters were also activated in the presence of GFP‐ODO1, confirming its functionality. These results provide in planta evidence that motif 1/1B, as predicted from genome‐wide analysis of ODO1 binding and regulatory targets, is indeed a bona fide ODO1‐binding sequence necessary for full transcriptional activity of the target genes of ODO1.
Figure 6.
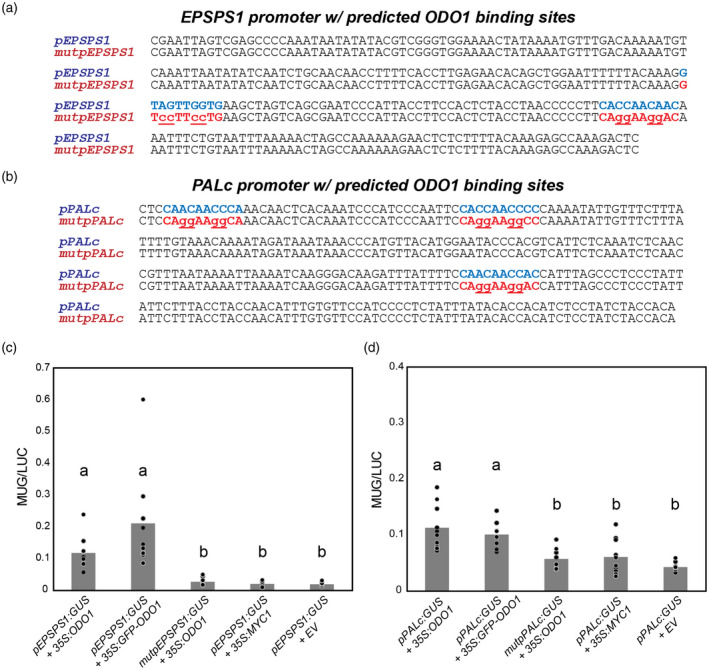
Activation of EPSPS1 and PALc promoters by ODO1 in N. benthamiana. Promoter regions immediately preceding the predicted transcription start site and containing predicted ODO1 binding sites from MEME (see also Figure S7) in blue for (a) EPSPS1 and (b) PALc. Binding sites in pEPSPS1 and pPALC were mutated to form mutpEPSPS1 and mutpPALc constructs as shown (mutated bases lowercase, underlined). N. benthamiana leaves were infiltrated with A. tumefaciens harboring the effector constructs 35S:ODO1, 35S:MYC1, 35S:GFP‐ODO1, or empty vector (pBINPLUS, EV) and the reporter constructs (c) pEPSPS1:GUS or mutpEPSPS1:GUS or (d) pPALc:GUS or mutpPALc:GUS. GUS activity was normalized with 35S:LUC activity. For both promoter constructs, in the presence of ODO1, mutation of predicted ODO1 binding sites led to reduction of promoter GUS activity to background levels in the range observed for MYC1 or empty vector. Tagged GFP‐ODO1 as used in ChIP‐Seq showed similar activity to untagged protein. Bars that do not share a letter have a significant difference determined by Kruskal‐Wallis test and pairwise comparisons adjusted by Bonferroni correction for multiple tests (P < 0.05). Individual data points are depicted with dots. Abbreviations: EPSPS, 5‐enolpyruvylshikimate 3‐phosphate; GUS, β‐glucuronidase; LUC, luciferase; ODO1, ODORANT1; PAL, phenylalanine ammonia lyase.
DISCUSSION
Using ChIP‐seq, we identified an extensive genome‐wide network of targets bound by ODO1, which includes not only the genes involved in the biosynthesis of the Phe precursor but also metabolic genes responsible for the formation of FVBPs and intermediates in their biosynthesis (Figure 3). Interestingly, the ODO1 ChIP‐seq signal is present at the TSS as well as further downstream within the gene body (Figure 2a), similar to what has been reported for other TFs in plants (Liu et al., 2018; Martin et al., 2018; Shamimuzzaman and Vodkin, 2013). Although such observations could be caused by cross‐linking ODO1 with other proteins in transcriptional machinery (e.g. RNA polymerase II, mediator complex proteins and histone‐modifying proteins) during ChIP, they could also indicate that there are indeed regulatory sites in the introns or other parts of the gene body.
Our results showed a statistically significant 10% overlap between ODO1‐bound genes determined by ChIP‐seq and ODO1‐regulated genes determined by RNA‐Seq (Figures 5a,b). This is in agreement with the 1–40% overlap between TF‐bound targets and genes that are misregulated under TF perturbation, observed across plants, animals and yeast (Swift and Coruzzi, 2017). The bound but unregulated targets are possibly poised for transcriptional regulation until the desired developmental or environmental cues are perceived. On the other hand, the regulated but unbound genes could be indirect targets or could be direct targets in which the interaction with ODO1 was not captured as a result of the transient nature of such interactions or the limitations of the ChIP‐seq technique. For example, the 2‐kbp promoter of the FVBP transporter PhABCG1 has previously been shown to be transcriptionally activated by ODO1 (Van Moerkercke et al., 2012), but did not meet the stringency requirements to be termed an ODO1‐bound gene in this study as it only had binding peaks in the 35S:GFP‐ODO1 library and not in the pODO1:GFP‐ODO1 ChIP‐seq library. It is therefore likely to represent a false negative because of the difficulty in capturing the transient interaction.
Unexpectedly, we also found that a portion of ODO1‐bound genes are upregulated in the odo1i line, indicating that, in addition to activating many target genes, ODO1 also functions to repress the expression of other target genes (Figure 5b). It is possible that ODO1 interacts with other transcription factors, which recognize a binding motif (Figure 5b) different from the canonical MYB binding motif (Figure 5a), to downregulate specific gene targets. On the other hand, we do not exclude the possibility of the existence of an incoherent feedforward loop (Alon, 2007) in the ODO1 regulatory network, where ODO1 activates a repressor X that downregulates gene Y, and at the same time binds the promoter of gene Y to prime it for future activation under adequate developmental or environmental conditions. For instance, some MYB proteins in Arabidopsis upregulate repressive transcription factors, as mutants of these MYBs led to their downregulation and activation of the metabolic pathways that they repress (Geng et al., 2020). In our study, ODO1 binds and activates MYB4 (Table S5), a negative regulator of the phenylpropanoid pathway in petunia (Colquhoun et al., 2011). Overall, the exact mechanism of how ODO1 controls gene regulation either directly or through downstream targets requires further investigation.
ODO1 is most closely related to a clade of MYB proteins that includes Arabidopsis MYB20, MYB42, MYB43, MYB85, MYB40 and MYB99 (Battat et al., 2019; Verdonk et al., 2005; Geng et al., 2020, Chen et al., 2021, Guanqun et al., 2021). Among them, MYB20, MYB42, MYB43 and MYB85 have recently been shown to regulate phenylalanine synthesis, the general phenylpropanoid pathway and monolignol biosynthetic genes (Geng et al., 2020). In addition, Arabidopsis MYB99 was found to play a specialized role in phenylpropanoid metabolism in pollen (Battat et al., 2019). Other evolutionarily related members of this clade have been identified as regulators of phenylpropanoid and monolignol biosynthesis genes, including PtoMYB92 in Populus (poplar) xylem (Li et al., 2015), EjODO1 in Eriobotrya japonica (loquat) fruits (Zhang et al., 2016) and CsMYB85 in Citrus × sinensis (orange) fruits (Jia et al., 2019). Consistent with these recent findings that this MYB clade regulates phenylpropanoid gene expression across multiple species, our genome‐wide study has uncovered an expansive network for specialized metabolism regulated by ODO1 in petunia that includes shikimate/phenylalanine biosynthetic genes and phenylpropanoid/monolignol pathway genes, as well as methionine biosynthetic genes, to provide a source of methyl groups for FVBP biosynthesis.
The ODO1 binding motif shares the core sequence of the AC‐II (ACCAACC) element, which is recognized by transcription factors in Arabidopsis that regulate lignification and cell wall biosynthesis (Raes et al., 2003; Zhong and Ye, 2012; Zhou et al., 2009). Indeed, at least one member of the PAL, 4CL, HCT, C3H, CCoAOMT, CCR and CAD gene families were shown to be regulated via the AC element in their promoters (Raes et al., 2003). Interestingly, their homologous genes in petunia have the ODO1‐binding motif in their promoters, except for CCR (Figure S7), and are bound by ODO1 (Figure 3a). ODO1 therefore appears to use the cis‐regulatory motifs similar to those essential for general lignin biosynthesis, but in a flower‐specific manner to produce monolignol precursors for eugenol and isoeugenol in the corolla, where laccase and peroxidase genes that polymerize monolignols into lignin are strongly downregulated (Patrick et al., 2021). In addition, similar AC elements have also been identified in the promoters of shikimate biosynthetic genes in Arabidopsis, including DAHP SYNTHASE, DHD‐SDH, SK, EPSPS and CS (Chen et al., 2006), the petunia homologs of which were also found to be bound by ODO1 (Figures 3a and S7). Probably by recognizing the conserved motifs, petunia ODO1 was able to activate both the shikimate pathway genes and the phenylpropanoid/monolignol biosynthetic genes, including PAL, C4H and 4CL, via HCT, CCoAOMT and CCR, in transgenic tomato fruits (Cin et al., 2011). Similarly, fruit‐specific expression of AtMYB12 in tomato led to the upregulation of DAHPS and several PAL5 genes via the binding of an AC‐I (ACCTACC) element‐containing motif (Zhang et al., 2015). Tomato SlMYB12 binds the same motif (Zhang et al., 2015) and regulates genes in both primary and secondary metabolic pathways (Fernandez‐Moreno et al, 2016). It has also been found that the expression of an ODO1 ortholog from scented lily species in petunia was sufficient to activate shikimate pathway genes and PAL expression (Yoshida et al., 2018), indicating that the ODO1 regulatory network is at least partly conserved between monocots and dicots. Therefore, the regulatory regime used by ODO1 to attain the wide‐ranging activation of genes involved in Phe biosynthesis and phenylpropanoid pathways for FVBP production and emission could be widely present in flowering plants to produce high levels of a diverse range of FVBPs.
As repressive MYBs also target phenylpropanoid gene promoters through the AC element in diverse plant species, ODO1 is likely to compete with MYB repressors for the same binding motif (Ma and Constabel, 2019). This has been suggested for AtMYB4, which may act as a direct repressor and compete with activators at certain MYB binding motifs in promoters of phenylpropanoid‐related genes (Jin et al., 2000). Another example is ZmMYB31, which binds to a motif highly similar to that observed for ODO1, ACC[T/A]ACC, and when expressed in Arabidopsis represses the expression of genes involved in the monolignol biosynthetic pathway (Fornalé et al., 2010). In petunia, PhMYB4 has been identified as a negative regulator of phenylpropene emission, at least in part through repressing C4H transcription (Colquhoun et al., 2011). As ODO1 protein levels fluctuate during the day–night cycle, reaching a maximum in the evening, at its binding site ODO1 may compete with transcriptional repressors that lower the transcriptional activity of these genes in the morning, resulting in diurnal fluctuation of the shikimate and phenylpropanoid pathway gene expression for FVBP biosynthesis and emission.
Remarkably, despite the enormous importance of SAM for many processes in the cell, including ethylene and polyamine biosynthesis and DNA methylation (Lindermayr et al., 2020; Sauter et al., 2013), relatively little is known about the transcriptional regulation of SAM SYNTHETASE (SAMS). In tomato fruit, the auxin response factor 6A binds to the SAMS1 promoter to repress its activity (Yuan et al., 2019). In Arabidopsis, more is known about the post‐translational regulation of SAMS, including its nitrosylation, phosphorylation and interaction with FERONIA, a receptor‐like kinase (for a review see Pattyn et al., 2021). Our data indicate that in petunia flowers, the SAM cycle genes are transcriptionally activated by ODO1. Methyl groups leave the flower via the emission of methylated FVBPs in large quantities, a situation quite different from tissues where only small quantities of ethylene are produced or methylated compounds are recycled (e.g. such as in modifications of histone, DNA, and RNA). SAMS transcription is highly induced in Nicotiana suaveolens petals where methyl benzoate is produced, and SAM levels fluctuate within the day–night cycle as they are consumed for FVBP production (Roeder et al., 2009). Flowers that emit methylated volatiles might thus have adapted this type of transcriptional activation, whereas post‐translational regulatory mechanisms are involved to regulate SAMS activity and SAM homeostasis when essential (Pattyn et al., 2021).
In summary, we discovered that ODO1 binds and regulates an extensive set of genes involved in multiple networks contributing to floral volatile metabolism, including the biosynthesis of the Phe precursor, the formation of the specialized metabolites and the biosynthesis of methionine and S‐adenosylmethionine, which provides methyl groups. Although fine‐tuning individual enzymes to increase the metabolic flux is likely to be constrained by substrate availability and/or feedback inhibition, the systems‐level regulation of primary and secondary metabolic pathways, simultaneously, through tuning master regulators such as ODO1 could provide a desirable method to increase the metabolic outputs of high‐value specialized metabolites. Our findings therefore have practical implications in the metabolic engineering of secondary metabolism, although is relevant to economically important food, industrial and pharmaceutical applications.
EXPERIMENTAL PROCEDURES
Plant material, growth conditions and transformations
Petunia hybrida cv. W115/Mitchell and N. benthamiana were grown in a glasshouse with a 16‐h photoperiod of 300 μmol m–2 sec–1 light, day/night temperatures of 22°C/17°C and 60–65% humidity. Transgenic pODO1:GFP‐ODO1 plants were generated as described by Moerkercke et al. (2009). Transgene integration was confirmed in T0 lines by PCR amplification of GFP‐ODO1 (the primers are listed in Table S6). Experiments have been performed with T0 pODO1:GFP‐ODO1 lines, unless stated otherwise. A transgenic P. hybrida cv. W115 line silenced for ODO1 (odo1i) was generated previously by Verdonk et al. (2005). For the transient Agrobacterium‐mediated transformation, plants were moved to a glasshouse compartment without artificial light. For qRT‐PCR, RNA‐seq and ChIP‐seq of stable transformed lines, plants were moved at least 2 days before sample collection to a climate chamber with a 16‐h photoperiod of 300 μE m–2 sec–1 light, constant temperature of 21°C and 70% humidity (kept dark from 6:00 pm to 2:00 am). Flowers harvested 2 days after anthesis were used for all experiments in this study, unless otherwise specified.
Construct design
The 35S:GFP‐ODO1, pODO1:GFP‐ODO1 with 1.9‐kbp ODO1 promoter, 35S:ODO1, 35S:NLS‐mCherry, 35S:MYC1 and 35S:LUC constructs have been described previously (Moerkercke et al., 2011, 2012; Quattrocchio et al., 1999; Spyropoulou et al., 2014a, 2014b). Mutated and non‐mutated PALc and EPSPS1 promoter fragments, 271 and 262 bp, respectively (Figure S8), with flanking SacI, XbaI or NotI restriction sites, were synthesized by eurofins (https://eurofinsgenomics.eu) and cloned into pEX‐A128 (eurofins). The promoter fragments were subsequently released with NotI and XbaI and cloned into a 35S:GUS:tNOS shuttle vector replacing the 35S promoter (Moerkercke et al., 2011). Next, the promoter:GUS:tNos cassettes were AscI and SfoI ligated in the multiple cloning site (MCS) of the binary vector pBINPLUS between the AscI and SmaI sites (van Engelen et al., 1995).
To create the ODO1 promoter‐luciferase (pODO1:LUC) construct, 1880 bp of the ODO1 promoter from P. hybrida cv. W115 was PCR‐amplified using a forward primer containing a HindIII site and reverse primer containing a KpnI site (Table S6). The promoter fragment was cloned into a pMON999 vector, with the luciferase CDS followed by the Nos terminator (tNos), and sequenced (Verdonk et al., 2005). The pODO1:LUC:tNos cassette was digested with HindIII/SmaI and ligated in the binary vector pBinPLUS (van Engelen et al., 1995).
Confocal microscopy
Transient Agrobacterium‐mediated transformations of corollas of wild type (WT) and pODO1:GFP‐ODO1 line 10 (T2) with 35S:NLS‐mCherry were performed as described previously (Moerkercke et al., 2012; Verweij et al., 2008). After 2 days, GFP (505–550 nm bandpass emission filter, 488 nm excitation) and mCherry (585–615 nm bandpass emission filter, 561 nm excitation) were imaged with a Zeiss LSM 510 inverted microscope (Zeiss, https://www.zeiss.com). Images were analyzed with imagej (https://fiji.sc).
ChIP‐seq sample preparation
Corollas of P. hybrida cv. W115 lines stably transformed with pODO1:GFP‐ODO1 were harvested in the climate chamber 1 h before dark (5:00 pm). Petunia hybrida cv. W115 corollas transiently transformed with 35S:GFP‐ODO1 were harvested in a glasshouse compartment without artificial light 1 h before dark (with the natural dark period spanning from 5:00 pm to 8:00 am). Agrobacterium‐mediated transient transformation assays were performed as described by Moerkercke et al. (2011). For each ChIP sample, 5 g of corollas were collected on ice, divided over two 50‐ml Greiner tubes and directly cross‐linked (Kaufmann et al., 2010). Nuclei were isolated according to the method described by Kaufmann et al. (2010) and samples were pooled again by resuspending the nuclei in 330 μl of lysis buffer (Myltenyi Biotec, 130‐091‐125, https://www.miltenyibiotec.com/). DNA was sheared with a Branson probe sonicator for 45 cycles of 5‐sec pulses, with 20‐sec cooling in between at 10% strength. The samples were centrifuged for 10 min at 15 000 g . The supernatant was incubated for 1 h at 20–25 °C with 100 μl of paramagnetic anti‐GFP MicroBeads (Myltenyi Biotec, https://www.miltenyibiotec.com). The mixture was transferred to a calibrated μ‐column on a μMACS Separator (Myltenyi Biotec). Subsequently the column was washed twice with 200 μl of lysis buffer (Myltenyi Biotec), high‐salt buffer (500 mM NaCl, 0.1% SDS, 1% Triton X‐100, 2 mM EDTA, 20 mm Tris‐HCl, pH 8.0), LiCL buffer (10 mM Tris‐HcL, pH 8.0, 1 mM EDTA, 1% NP‐40, 1% sodium deoxycholate, 0.25 m LiCl) and TE buffer (pH 8.0). Anti‐GFP bound proteins were eluted by incubating the column with 20 μl of boiling elution buffer (1% SDS, 50 mM Tris, pH 8.0, 1 mm EDTA, 50 mM DTT) for 5 min, and adding three volumes of 50 μl of boiling elution buffer to the column. The cross‐linking was reversed by adding 100 μl TE (pH 8.0) and 0.5 mg ml–1 proteinase K to the eluate. After at least 12 h of incubation at 37°C, fresh proteinase K (0.5 mg ml–1) was added and samples were incubated for another 6 h at 65°C. DNA was precipitated with 2.5 volumes of 100% ethanol, 0.1 volumes of 3 M NaAc (pH 5.4) and 1 μl of glycogen (20 μg μl–1) for at least 12 h at −20°C. DNA was centrifuged for 30 min at 15 000 g at 4°C, resuspended in 100 μl of Milli‐Q water and purified with QIAquick PCR Purification Kit (QIAGEN, https://www.qiagen.com) in 35 μl of elution buffer.
ChIP‐seq Illumina library preparation
A 15‐μl volume of ChIP DNA was end‐repaired with Fast DNA End Repair Kit (K0771; ThermoFisher Scientific, https://www.thermofisher.com) and purified with QIAquick PCR Purification Kit (QIAGEN). For A‐tailing, end‐repaired DNA was incubated with 10 U of Klenow fragment (OP0051; ThermoFisher Scientific) and 0.2 mM dATP for 30 min at 37°C, and then purified with MinElute PCR Purification Kit (QIAGEN). End‐repaired DNA was ligated with 1 μl of 1.5 μM NEBNext Multiplex Oligos for Illumina (Index Primers Set 1) (E7335; New England BioLabs, https://www.neb.uk.com) by 1 U of T4 ligase (EL0016; ThermoFisher Scientific) at 16°C for 20 h. Adapters were excised by adding 3 μl of USER enzyme (E7335; New England BioLabs) for 15 min at 37°C. DNA was purified with the QIAquick PCR Purification Kit (QIAGEN) before amplification. DNA was amplified with 2 U Phusion™ Hot Start II High‐Fidelity DNA Polymerase, HF buffer (F‐537S; ThermoFisher Scientific), 0.2 mM dNTPs, 2.5 μl of universal and index primers (E7335S; New England BioLabs) for 18 cycles with an annealing temperature of 65°C and extension time of 30 sec. Free adapters were removed following the manufacturer’s instructions for adapter removal of the MagJET NGS Cleanup and Size Selection Kit (K2821; ThermoFisher Scientific). The quality and quantity of the libraries were verified with tapestation 2200 (Agilent Technologies, https://www.agilent.com). Libraries were diluted to 10 nM and pooled. Pooled libraries were sequenced with HiSeq 2000 (1 × 100 base) (Illumina, https://www.illumina.com; DNA Core, https://dnacore.missouri.edu).
ChIP sequencing analysis
ChIP sequencing reads were trimmed using cutadapt (Martin, 2011) and aligned with the P. axillaris and P. inflata genomes (Bombarely et al., 2016) using bowtie2 (Langmead and Salzberg, 2012), as previously described (Patrick et al., 2021). To identify the ODO1 binding peaks in the pODO1:GFP‐ODO1 and 35S:GFP‐ODO1 libraries, the macs2 ‘callpeak’ function was used (Zhang et al., 2008). Pooled input chromatin from mature (day‐2 open flower) P. hybrida (National Center for Biotechnology Information (NCBI) Sequence Read Archive BioProject PRJNA650505) was used as a background control. ODO1 binding peaks were then determined by intersecting those present in both the native promoter and overexpression libraries with bedtools (Quinlan and Hall, 2010). ODO1‐bound genes were determined by the intersection of ODO1 peaks with P. axillaris genes. Enriched GO terms among ODO1‐bound genes were determined using an in‐house pipeline developed for petunia, as previously described (Patrick et al., 2021). GO term enrichment results were visualized using revigo (Supek et al., 2011). The enrichment of ODO1‐bound genes among lists of developmentally regulated genes or curated metabolic pathway genes were determined by hypergeometric test, as previously described (Patrick et al., 2021). Genes involved in SAM and the folate cycle (Table S2) were determined from gene loci expressed in the corolla with BLAST homology to A. thaliana, as previously described (Patrick et al., 2021).
RNA‐seq sample preparation and sequencing
For qRT‐PCR and RNA‐seq corollas of P. hybrida cv. W115, wild‐type and odo1i plants were harvested 1 h before dark in a climate chamber. RNA was isolated with the RNeasy plant mini kit (QIAGEN) following the manufacturer’s intructions. gDNA was removed with the Ambion DNA‐free kit (Invitrogen, now ThermoFisher Scientific, https://www.thermofisher.com/nl/en/home.html). Following gDNA removal, RNA was purified by RNeasy plant mini kit (QIAGEN). The integrity and concentration of the RNA was determined by gel electrophoresis, Nanodrop (ThermoFisher Scientific) and TapeStation (Agilent Technologies). cDNA synthesis and qRT‐PCR were performed to confirm the downregulation of ODO1 in the odo1i line compared with the WT plants. RNA sequencing, mRNA isolation and library preparation were performed using standard Illumina protocols by Novogene (https://en.novogene.com). Paired‐end (2 × 150 bp) sequencing was performed on the NovaSeq 6000 platform (Illumina) with three biological replicates for WT and odo1i.
RNA‐seq analysis
Adaptors were trimmed from RNA sequencing reads using cutadapt (Martin, 2011). Trimmed reads were then aligned with the petunia genomes (Bombarely et al., 2016) using tophat2 (Kim et al., 2013). htseq (Anders et al., 2015) was used to assign aligned reads to petunia genes and differentially expressed genes were determined by deseq2 (Love et al., 2014) using P. axillaris genome features, as previously described (Patrick et al., 2021), and with a significance cut‐off FDR of <0.05.
ODO1 binding motif discovery
To determine the ODO1 binding motif, a 500‐bp promoter sequence upstream of the TSS was extracted for gene sets of interest, generated by intersecting ODO1‐bound genes and genes transcriptionally regulated by ODO1 or previously known FVBP pathway genes. For motif discovery, meme (Bailey et al., 2009) was run using ‘zoops’ mode (zero or one occurrence per sequence), with a width of eight to 10 bases, for genes that are ODO1‐bound and transcriptionally regulated by ODO1. ANR mode (any number of repeats) mode was used for motif discovery among ODO1‐bound FVBP genes, to determine all potential binding sites for mutational analysis. tomtom (Gupta et al., 2007) was used to find similar motifs in Arabidopsis.
cDNA synthesis and qRT‐PCR
Fermentas RevertAid H MinusReverse transcriptase kit and 5 mM oligo(dT)18 were used to synthesize cDNA from 1 μg of total RNA. Quantitative RT‐PCR was performed on the ABI‐7500 (7500 Real‐Time PCR Systems; Applied Biosystems, now ThermoFisher Scientific) with an RNA equivalent of 10 ng of cDNA and HOT FIREPol EvaGreen qPCR mix with ROX (Solis BioDyne, https://solisbiodyne.com). Normalization was performed with EF1a (Mallona et al., 2010) or FBP1 (Shaipulah et al., 2016) as a housekeeping gene. Primers are listed in Table S6.
Transient Agrobacterium‐mediated transformation
Transient Agrobacterium‐mediated transformations and trans‐activation assays were performed as described by Moerkercke et al. (2011). Nicotiana benthamiana leaves were co‐infiltrated with Agrobacterium tumefaciens GV3101 (pMP90) harboring different reporter constructs and incubated for 3 days (n = 10). LUC and GUS activity (465 nm emission filter, 355 nm excitation) were measured with a Synergy H1 plate reader (Biotek, https://www.biotek.com).
AUTHOR CONTRIBUTIONS
MRB carried out the ChIP‐seq experiments, protein localization and the qRT‐PCR. RMP analyzed ChIP‐seq and RNA‐seq data. SJ performed RNA isolations. SJ and PS conducted Agrobacterium‐mediated transient assays. NFS performed the luciferase experiments. RS, MH, YL and ND designed the research. MRB, RMP, YL, ND and RS wrote and edited the article.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest associated with this work.
Supporting information
Figure S1. Diurnal fluctuation of ODO1 transcriptional activity. (a) Corolla tissue from two‐day‐old W115 wild‐type flowers was collected every three hours for RNA extraction and expression of ODO1 was measured by qRT‐PCR. Petunia housekeeping reference gene FLORAL BINDING PROTEIN 1 (FBP1) was used for normalization (Angenent et al., 1992; Shaipulah et al., 2016). White and black bars represent periods of light and dark, respectively. Error bars represent standard error of the mean (n = 3). Transcript levels were determined to have significantly rhythmic pattern at P < 0.001 (ANOVA followed by Tukey’s post hoc analysis). (b) Measurement of ODO1 promoter luciferase activity in pODO1:LUC petunia flowers (Video Clip 1). Pixel intensity was measured using the MetaVueTM Imaging System (Universal Imaging Corporation, USA). Luciferase activity was determined by collection at hourly intervals of luminescence counts per flower to determine intensity in relative light units per pixel (rlu/pixel). Image collection was performed in constant dark; grey and black bars represent light and dark periods respectively to which plants were acclimated. Diamond markers represent the mean intensity from pODO1:LUC flowers with error bars representing standard error of the mean (n = 4), while square markers show background levels measured from a wild‐type flower (n = 1). Luciferase activity in pODO1:LUC flowers was determined to have a significantly rhythmic pattern at P < 0.05 (ANOVA followed by Tukey’s post hoc analysis). The experiment was repeated twice with similar results.
Figure S2. Setup of detached flowers for luciferase imaging. Depiction of the arrangement of four pODO1:LUC flowers (bottom) with one wild‐type flower (top) for imaging in Video Clip 1.
Figure S3. ChIP‐Seq signal in gene space surrounding example ODO1 targets. ChIP‐seq signals at 4CLa (Peaxi162Scf00314g00086) and CIMS1 (Peaxi162Scf00163g00910) with surrounding upstream and downstream regions. The Y axis represents the sequencing depth covered by ChIP‐seq in fragments per million, after normalization to the total number of unique aligned fragments in each library. Ruler markings indicate location of ChIP signal along the corresponding petunia scaffolds, with annotated gene features below (thicker bar sections for coding regions, thinner sections for introns and untranslated regions).
Figure S4. Developmental regulation of ODO1‐bound genes. ODO1‐bound genes are significantly overlapped with genes that are upregulated (a) at day 2 relative to day 0 post‐anthesis (Patrick et al., 2021, but not with genes that are downregulated (b) at day 2 relative to day 0 post‐anthesis (Patrick et al., 2021. The hypergeometric P value for the overlap between gene sets are shown. n.s. = not significant.
Figure S5. ODO1 binding of SAM cycle genes determined by ChIP‐seq. (a) Methionine pathway with SAM and folate cycle genes identified in petunia (see also Table S2). The genes bound by ODO1 are indicated by red squares. (b) Overlap between ODO1‐bound genes and SAM/THF genes with a p value representing the significance of the observed overlap determined by hypergeometric distribution against the whole genome as background. Abbreviations: CIMS, cobalamin‐independent methionine synthase; MTHFR, methylenetetrahydrofolate reductase; MTs, methyltransferases; ODO1, ODORANT1; SAHH, S‐adenosyl‐L‐homocysteine hydrolase; SAMS, S‐adenosylmethionine synthetase; SHMT, serine hydroxymethyltransferase; THF, tetrahydrofolate.
Figure S6. Comparison between the putative ODO1 binding motif with previously reported binding motifs of transcription factors in Arabidopsis. The putative binding motif of ODO1 (Motif 1, Figure 5a) was queried against the Arabidopsis DAP‐Seq database (O’Malley et al., 2016) using Tomtom (Gupta et al., 2007). Top three matches by lowest E‐value shown, all relating to MYB transcription factors.
Figure S7. Motif 1B occurrences within promoters of ODO1‐bound FVBP genes. Sequence matches to the predicted ODO1 binding motif within 500 bp promoter of FVBP genes (Figure 5c) with P value < 1.0e‐04 as determined by MEME (Bailey et al., 2009) are shown. Abbreviations: 4CL, 4‐coumaryl‐CoA ligase; ADT, arogenate dehydratase; BPBT, benzoyl‐CoA:benzylalcohol/phenylethanol benzoyltransferase; C3H, coumarate 3‐hydroxylase; C4H, cinnamate 4‐hydroxylase; CAD, cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl‐CoA O‐methyltransferase; CHD, cinnamoyl‐CoA hydratase‐dehydrogenase; CNL, cinnamoyl‐CoA ligase; CS, chorismate synthase; DAHP, 3‐deoxy‐D‐arabino‐heptulosonate 7‐phosphate synthase; DHD‐SDH, 3‐dehydroquinate dehydratase and shikimate dehydrogenase; EPSPS, 5‐enolpyruvylshikimate 3‐phosphate; HCT, hydroxycinnamoyl‐CoA:shikimate/quinate hydroxycinnamoyl transferase; ODO1, ODORANT1; PAL, phenylalanine ammonia lyase; SK, shikimate kinase.
Figure S8. Synthetic promoter constructs. Restriction sites are underlined and ODO1 binding sites are bold. Mutated nucleotides are displayed in lower case and bold.
Table S1. Significantly enriched GO terms among ODO1‐bound genes determined by ChIP‐seq.
Table S2. P. axillaris genes involved in SAM and folate cycle determined by BLAST against Arabidopsis whole gene set. E‐Value: Expect value for sequence match relative to chance.
Table S3. Significantly enriched GO terms for genes downregulated in odo1i.
Table S4. Significantly enriched GO terms for genes upregulated in odo1i.
Table S5. Transcription factor regulatory targets of ODO1.
Table S6. Primer sequences.
Video Clip 1. Rhythmic luciferase activity driven by the ODO1 promoter. Four transgenic (T2) flowers of P. hybrida cv. W115 plants stably transformed with pODO1:LUC construct are shown, with one wild‐type flower at top of frame as a background control (see Figure S2). Flowers from plants acclimated to a 16 h light, 8 h dark period were sprayed with luciferin solution (1 mm D‐Luciferin and 0.01% Silwet) one day prior to imaging, subsequently detached and placed upright in a glass beaker containing water and then sprayed once again with luciferin solution prior to being placed in constant dark for imaging. Bioluminescence images were captured using a CCD camera (Pixis 1024, https://www.princetoninstruments.com) every hour for 48 h with a 1‐minute exposure. Quantification of the bioluminescence images is shown in Figure S1b.
ACKNOWLEDGEMENT
Michel de Vries is kindly acknowledged for generating the transgenic W115 pODO1:GFP‐ODO1 lines. This research was supported by a seed grant to YL and ND from the Purdue Center for Plant Biology, a National Science Foundation (NSF) grant (no. 10001896) to YL and ND from the Division of Molecular and Cellular Biosciences, United States Department of Agriculture (USDA) National Institute of Food and Agriculture Hatch project numbers 1013620, to YL, and 177845, to ND, and an Agriculture and Food Research Initiative Postdoctoral Fellowship (grant no. 2019‐67012‐29660) to RMP from the USDA National Institute of Food and Agriculture. MRB was supported by a Dutch Research Council (NWO) NWO‐talent grant 022.001.018. NFS was supported by a grant from the Malaysian ministry of higher education. SJ was supported by a grant from the University of Amsterdam (UvA).
DATA AVAILABILITY STATEMENT
The ChIP and RNA sequencing data used in this study are available at the NCBI Sequence Read Archive under BioProject ID PRJNA729780 and additional data are available at NCBI‐GEO GSE189280.
REFERENCES
- Alon, U. (2007) Network motifs: theory and experimental approaches. Nature Reviews Genetics, 8, 450–461. 10.1038/nrg2102 [DOI] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P.T. & Huber, W. (2015) HTSeq–a Python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent, G.C. , Busscher, M. , Franken, J. , Mol, J.N. & van Tunen, A.J. (1992) Differential expression of two MADS box genes in wild‐type and mutant petunia flowers. The Plant Cell, 4, 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L. , Boden, M. , Buske, F.A. , Frith, M. , Grant, C.E. , Clementi, L. et al. (2009) MEME Suite: tools for motif discovery and searching. Nucleic Acids Research, 37, W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battat, M. , Eitan, A. , Rogachev, I. , Hanhineva, K. , Fernie, A. , Tohge, T. et al. (2019) A MYB triad controls primary and phenylpropanoid metabolites for pollen coat patterning. Plant Physiology, 180, 87–108. 10.1104/pp.19.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright, J. , Negre, F. , Chen, X. , Kish, C.M. , Wood, B. , Peel, G. et al. (2004) Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiology, 135, 1993–2011. 10.1104/pp.104.045468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely, A. , Moser, M. , Amrad, A. , Bao, M. , Bapaume, L. , Barry, C.S. et al. (2016) Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nature Plants, 2, 1–9. 10.1038/nplants.2016.74 [DOI] [PubMed] [Google Scholar]
- Byers, K.J.R.P. , Bradshaw, H.D. & Riffell, J.A. (2014a) Three floral volatiles contribute to differential pollinator attraction in monkeyflowers (Mimulus). Journal of Experimental Biology, 217, 614–623. 10.1242/jeb.092213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, K.J.R.P. , Vela, J.P. , Peng, F. , Riffell, J.A. & Bradshaw, H.D. (2014b) Floral volatile alleles can contribute to pollinator‐mediated reproductive isolation in monkeyflowers (Mimulus). The Plant Journal, 80, 1031–1042. 10.1111/tpj.12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. , He, W. , Guo, X. & Pan, J. (2021) Genome‐wide identification, classification and expression analysis of the MYB transcription factor family in petunia. International Journal of Molecular Sciences, 22, 4838. 10.3390/ijms22094838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Zhang, X. , Wu, W. , Chen, Z. , Gu, H. & Qu, L.‐J. (2006) Overexpression of the wounding‐responsive gene AtMYB15 activates the shikimate pathway in Arabidopsis. Journal of Integrative Plant Biology, 48, 1084–1095. 10.1111/j.1744-7909.2006.00311.x [DOI] [Google Scholar]
- Cin, V.D. , Tieman, D.M. , Tohge, T. , McQuinn, R. , de Vos, R.C.H. , Osorio, S. et al. (2011) Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit. The Plant Cell, 23, 2738–2753. 10.1105/tpc.111.086975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun, T.A. , Kim, J.Y. , Wedde, A.E. , Levin, L.A. , Schmitt, K.C. , Schuurink, R.C. et al. (2011) PhMYB4 fine‐tunes the floral volatile signature of Petunia×hybrida through PhC4H. Journal of Experimental Botany, 62, 1133–1143. 10.1093/jxb/erq342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun, T.A. , Verdonk, J.C. , Schimmel, B.C.J. , Tieman, D.M. , Underwood, B.A. & Clark, D.G. (2010) Petunia floral volatile benzenoid/phenylpropanoid genes are regulated in a similar manner. Phytochemistry, 71, 158–167. 10.1016/j.phytochem.2009.09.036 [DOI] [PubMed] [Google Scholar]
- Dewick, P.M. (1994) The biosynthesis of shikimate metabolites. Natural Products Reports, 11, 173–203. 10.1039/NP9941100173 [DOI] [PubMed] [Google Scholar]
- Fenske, M.P. , Hazelton, K.D.H. , Hempton, A.K. , Shim, J.S. , Yamamoto, B.M. , Riffell, J.A. et al. (2015) Circadian clock gene LATE ELONGATED HYPOCOTYL directly regulates the timing of floral scent emission in Petunia. PNAS, 112, 9775–9780. 10.1073/pnas.1422875112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Moreno, J.P. , Tzfadia, O. , Forment, J. , Presa, S. , Rogachev, I. , Meir, S. et al. (2016) Characterization of a new pink‐fruited tomato mutant results in the identification of a null allele of the SlMYB12 transcription factor. Plant Physiology, 171(3), 1821–1836. 10.1104/pp.16.00282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornalé, S. , Shi, X. , Chai, C. , Encina, A. , Irar, S. , Capellades, M. et al. (2010) ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. The Plant Journal, 64, 633–644. 10.1111/j.1365-313X.2010.04363.x [DOI] [PubMed] [Google Scholar]
- Geng, P. , Zhang, S. , Liu, J. , Zhao, C. , Wu, J. , Cao, Y. et al. (2020) MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiology, 182, 1272–1283. 10.1104/pp.19.01070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guanqun, C. , Weizhi, H. , Xiangxin, G. & Junsong, P. (2021) Genome‐wide identification, classification and expression analysis of the MYB transcription factor family in petunia. International Journal of Molecular Sciences, 22(9), 4838. 10.3390/ijms22094838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S. , Stamatoyannopoulos, J.A. , Bailey, T.L. & Noble, W.S. (2007) Quantifying similarity between motifs. Genome Biology, 8, R24. 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock, K. & Scheel, D. (1989) Physiology and molecular biology of phenylpropanoid metabolism. Annual Review of Plant Physiology and Plant Molecular Biology, 40, 347–369. [Google Scholar]
- Hanson, A.D. & Roje, S. (2001) One‐carbon metabolism in higher plants. Annual Review of Plant Physiology and Plant Molecular Biology, 52, 119–137. 10.1146/annurev.arplant.52.1.119 [DOI] [PubMed] [Google Scholar]
- Herrmann, K. (1995) The shikimate pathway as an entry to aromatic secondary metabolism. Plant Physiology, 107, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoballah, M.E. , Stuurman, J. , Turlings, T.C.J. , Guerin, P.M. , Connétable, S. & Kuhlemeier, C. (2005) The composition and timing of flower odour emission by wild Petunia axillaris coincide with the antennal perception and nocturnal activity of the pollinator Manduca sexta. Planta, 222, 141–150. 10.1007/s00425-005-1506-8 [DOI] [PubMed] [Google Scholar]
- Jia, N. , Liu, J. , Tan, P. , Sun, Y. , Lv, Y. , Liu, J. et al. (2019) Citrus sinensis MYB transcription factor CsMYB85 induce fruit juice sac lignification through interaction with other CsMYB transcription factors. Frontiers in Plant Science, 10, 10.3389/fpls.2019.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H. , Cominelli, E. , Bailey, P. , Parr, A. , Mehrtens, F. , Jones, J. et al. (2000) Transcriptional repression by AtMYB4 controls production of UV‐protecting sunscreens in Arabidopsis. EMBO Journal, 19(22), 6150–6161. 10.1093/emboj/19.22.6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, K. , Muiño, J.M. , Østerås, M. , Farinelli, L. , Krajewski, P. & Angenent, G.C. (2010) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP‐SEQ) or hybridization to whole genome arrays (ChIP‐CHIP). Nature Protocols, 5, 457–472. 10.1038/nprot.2009.244 [DOI] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. & Salzberg, S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 14, R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre, U. , Gurba, A. , Hermann, K. , Saxenhofer, M. , Bossolini, E. , Guerin, P.M. et al. (2011) Pollinator choice in petunia depends on two major genetic loci for floral scent production. Current Biology, 21, 730–739. 10.1016/j.cub.2011.03.059 [DOI] [PubMed] [Google Scholar]
- Langmead, B. & Salzberg, S.L. (2012) Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Wang, X. , Ran, L. , Tian, Q. , Fan, D. & Luo, K. (2015) PtoMYB92 is a transcriptional activator of the lignin biosynthetic pathway during secondary cell wall formation in populus tomentosa. Plant and Cell Physiology, 56, 2436–2446. 10.1093/pcp/pcv157 [DOI] [PubMed] [Google Scholar]
- Lindermayr, C. , Rudolf, E.E. , Durner, J. & Groth, M. (2020) Interactions between metabolism and chromatin in plant models. Molecular Metabolism, You are What You Eat, 38, 100951. 10.1016/j.molmet.2020.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Wang, B. , Li, Z. , Peng, Z. & Zhang, J. (2018) TsNAC1 Is a key transcription factor in abiotic stress resistance and growth. Plant Physiology, 176, 742–756. 10.1104/pp.17.01089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. & Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, D. & Constabel, C.P. (2019) MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends in Plant Science, 24, 275–289. 10.1016/j.tplants.2018.12.003 [DOI] [PubMed] [Google Scholar]
- Mallona, I. , Lischewski, S. , Weiss, J. , Hause, B. & Egea‐Cortines, M. (2010) Validation of reference genes for quantitative real‐time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biology, 10, 4. 10.1186/1471-2229-10-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. (2011) Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet.journal, 17, 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martin, R.C. , Vining, K. & Dombrowski, J.E. (2018) Genome‐wide (ChIP‐seq) identification of target genes regulated by BdbZIP10 during paraquat‐induced oxidative stress. BMC Plant Biology, 18, 58. 10.1186/s12870-018-1275-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, R.L. , Zhong, R. & Ye, Z.‐H. (2009) MYB83 Is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant and Cell Physiology, 50, 1950–1964. 10.1093/pcp/pcp139 [DOI] [PubMed] [Google Scholar]
- Meng, F. , Zhao, H. , Zhu, B. , Zhang, T. , Yang, M. , Li, Y. et al. (2021) Genomic editing of intronic enhancers unveils their role in fine‐tuning tissue‐specific gene expression in Arabidopsis thaliana. The Plant Cell, 33(6), 1997–2014. 10.1093/plcell/koab093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerkercke, A.V. , Haring, M.A. & Schuurink, R.C. (2011) The transcription factor EMISSION OF BENZENOIDS II activates the MYB ODORANT1 promoter at a MYB binding site specific for fragrant petunias. The Plant Journal, 67, 917–928. 10.1111/j.1365-313X.2011.04644.x [DOI] [PubMed] [Google Scholar]
- Moerkercke, A.V. , Schauvinhold, I. , Pichersky, E. , Haring, M.A. & Schuurink, R.C. (2009) A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. The Plant Journal, 60, 292–302. 10.1111/j.1365-313X.2009.03953.x [DOI] [PubMed] [Google Scholar]
- Muhlemann, J.K. , Klempien, A. & Dudareva, N. (2014a) Floral volatiles: from biosynthesis to function. Plant, Cell & Environment, 37, 1936–1949. 10.1111/pce.12314 [DOI] [PubMed] [Google Scholar]
- Muhlemann, J.K. , Woodworth, B.D. , Morgan, J.A. & Dudareva, N. (2014b) The monolignol pathway contributes to the biosynthesis of volatile phenylpropenes in flowers. New Phytologist, 204, 661–670. 10.1111/nph.12913 [DOI] [PubMed] [Google Scholar]
- Negre, F. , Kish, C.M. , Boatright, J. , Underwood, B. , Shibuya, K. , Wagner, C. et al. (2003) Regulation of methylbenzoate emission after pollination in snapdragon and petunia flowers. The Plant Cell, 15, 2992–3006. 10.1105/tpc.016766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley, R.C. , Huang, S.C. , Song, L. , Lewsey, M.G. , Bartlett, A. , Nery, J.R. et al. (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell, 165, 1280–1292. 10.1016/j.cell.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, R.M. , Huang, X.‐Q. , Dudareva, N. & Li, Y. (2021) Dynamic histone acetylation in floral volatile synthesis and emission in petunia flowers. Journal of Experimental Botany, 72(10), 3704–3722. 10.1093/jxb/erab072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn, J. , Vaughan‐Hirsch, J. & Van de Poel, B. (2021) The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytologist, 229, 770–782. 10.1111/nph.16873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F. , Wing, J. , van der Woude, K. , Souer, E. , de Vetten, N. , Mol, J. et al. (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. The Plant Cell, 11, 1433–1444. 10.1105/tpc.11.8.1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, A.R. & Hall, I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes, J. , Rohde, A. , Christensen, J.H. , Van de Peer, Y. & Boerjan, W. (2003) Genome‐wide characterization of the lignification toolbox in Arabidopsis. Plant Physiology, 133, 1051–1071. 10.1104/pp.103.026484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, S. , Dreschler, K. , Wirtz, M. , Cristescu, S.M. , van Harren, F.J.M. , Hell, R. et al. (2009) SAM levels, gene expression of SAM synthetase, methionine synthase and ACC oxidase, and ethylene emission from N. suaveolens flowers. Plant Molecular Biology, 70, 535–546. 10.1007/s11103-009-9490-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter, M. , Moffatt, B. , Saechao, M.C. , Hell, R. & Wirtz, M. (2013) Methionine salvage and S‐adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochemical Journal, 451, 145–154. 10.1042/BJ20121744 [DOI] [PubMed] [Google Scholar]
- Shaipulah, N.F.M. , Muhlemann, J.K. , Woodworth, B.D. , Moerkercke, A.V. , Verdonk, J.C. , Ramirez, A.A. et al. (2016) CCoAOMT down‐regulation activates anthocyanin biosynthesis in petunia. Plant Physiology, 170, 717–731. 10.1104/pp.15.01646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamimuzzaman, M. & Vodkin, L. (2013) Genome‐wide identification of binding sites for NAC and YABBY transcription factors and co‐regulated genes during soybean seedling development by ChIP‐Seq and RNA‐Seq. BMC Genomics, 14, 477. 10.1186/1471-2164-14-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer‐Rimon, B. , Farhi, M. , Albo, B. & Cna’ani, A. , Zvi, M.M.B. , Masci, T. , et al. (2012) The R2R3‐MYB–like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent‐related genes in petunia. The Plant Cell, 24, 5089–5105. 10.1105/tpc.112.105247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer‐Rimon, B. , Marhevka, E. , Barkai, O. , Marton, I. , Edelbaum, O. , Masci, T. et al. (2010) EOBII, a gene encoding a flower‐specific regulator of phenylpropanoid volatiles’ biosynthesis in petunia. The Plant Cell, 22, 1961–1976. 10.1105/tpc.109.067280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulou, E.A. , Haring, M.A. & Schuurink, R.C. (2014a) Expression of Terpenoids 1, a glandular trichome‐specific transcription factor from tomato that activates the terpene synthase 5 promoter. Plant Molecular Biology, 84, 345–357. 10.1007/s11103-013-0142-0 [DOI] [PubMed] [Google Scholar]
- Spyropoulou, E.A. , Haring, M.A. & Schuurink, R.C. (2014b) RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genomics, 15, 402. 10.1186/1471-2164-15-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek, F. , Bošnjak, M. , Škunca, N. & Šmuc, T. (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One, 6, e21800. 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift, J. & Coruzzi, G.M. (2017) A matter of time — How transient transcription factor interactions create dynamic gene regulatory networks. Biochimica et Biophysica Acta (BBA) ‐ Gene Regulatory Mechanisms. Plant Gene Regulatory Mechanisms and Networks, 1860, 75–83. 10.1016/j.bbagrm.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelen, F.A. , Molthoff, J.W. , Conner, A.J. , Nap, J.‐P. , Pereira, A. & Stiekema, W.J. (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Research, 4, 288–290. 10.1007/BF01969123 [DOI] [PubMed] [Google Scholar]
- Van Moerkercke, A. , Galvan‐Ampudia, C.S. , Verdonk, J.C. , Haring, M.A. & Schuurink, R.C. (2012) Regulators of floral fragrance production and their target genes in petunia are not exclusively active in the epidermal cells of petals. Journal of Experimental Botany, 63, 3157–3171. 10.1093/jxb/ers034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdonk, J.C. , Haring, M.A. , van Tunen, A.J. & Schuurink, R.C. (2005) ODORANT1 regulates fragrance biosynthesis in petunia flowers. The Plant Cell, 17, 1612–1624. 10.1105/tpc.104.028837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdonk, J.C. , Ric de Vos, C.H. , Verhoeven, H.A. , Haring, M.A. , van Tunen, A.J. & Schuurink, R.C. (2003) Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry, Plant Metabolomics, 62, 997–1008. 10.1016/S0031-9422(02)00707-0 [DOI] [PubMed] [Google Scholar]
- Verweij, W. , Di Sansebastiano, G.‐P. , Quattrocchio, F. & Dalessandro, G. (2008) Agrobacterium ‐mediated transient expression of vacuolar GFPs in Petunia leaves and petals. Plant Biosystems ‐ an International Journal Dealing with All Aspects of Plant Biology, 142, 343–347. 10.1080/11263500802150779 [DOI] [Google Scholar]
- Vogt, T. (2010) Phenylpropanoid biosynthesis. Molecular Plant, 3, 2–20. [DOI] [PubMed] [Google Scholar]
- Whitehead, M.R. & Peakall, R. (2014) Pollinator specificity drives strong prepollination reproductive isolation in sympatric sexually deceptive orchids. Evolution, 68, 1561–1575. 10.1111/evo.12382 [DOI] [PubMed] [Google Scholar]
- Yoshida, K. , Oyama‐Okubo, N. & Yamagishi, M. (2018) An R2R3‐MYB transcription factor ODORANT1 regulates fragrance biosynthesis in lilies (Lilium spp.). Molecular Breeding, 38, 144. 10.1007/s11032-018-0902-2 [DOI] [Google Scholar]
- Yuan, Y. , Xu, X. , Gong, Z. , Tang, Y. , Wu, M. , Yan, F. et al. (2019) Auxin response factor 6A regulates photosynthesis, sugar accumulation, and fruit development in tomato. Horticulture Research, 6, 85. 10.1038/s41438-019-0167-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Ge, H. , Zang, C. , Li, X. , Grierson, D. , Chen, K. et al. (2016) EjODO1, a MYB transcription factor, regulating lignin biosynthesis in developing loquat (Eriobotrya japonica) fruit. Frontiers in Plant Science, 7, 10.3389/fpls.2016.01360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Butelli, E. , Alseekh, S. , Tohge, T. , Rallapalli, G. , Luo, J. et al. (2015) Multi‐level engineering facilitates the production of phenylpropanoid compounds in tomato. Nature Communications, 6, 8635. 10.1038/ncomms9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Liu, T. , Meyer, C.A. , Eeckhoute, J. , Johnson, D.S. , Bernstein, B.E. et al. (2008) Model‐based analysis of ChIP‐Seq (MACS). Genome Biology, 9, R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R. & Ye, Z.‐H. (2012) MYB46 and MYB83 Bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant and Cell Physiology, 53, 368–380. 10.1093/pcp/pcr185 [DOI] [PubMed] [Google Scholar]
- Zhou, J. , Lee, C. , Zhong, R. & Ye, Z.‐H. (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis . The Plant Cell, 21, 248–266. 10.1105/tpc.108.063321 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Diurnal fluctuation of ODO1 transcriptional activity. (a) Corolla tissue from two‐day‐old W115 wild‐type flowers was collected every three hours for RNA extraction and expression of ODO1 was measured by qRT‐PCR. Petunia housekeeping reference gene FLORAL BINDING PROTEIN 1 (FBP1) was used for normalization (Angenent et al., 1992; Shaipulah et al., 2016). White and black bars represent periods of light and dark, respectively. Error bars represent standard error of the mean (n = 3). Transcript levels were determined to have significantly rhythmic pattern at P < 0.001 (ANOVA followed by Tukey’s post hoc analysis). (b) Measurement of ODO1 promoter luciferase activity in pODO1:LUC petunia flowers (Video Clip 1). Pixel intensity was measured using the MetaVueTM Imaging System (Universal Imaging Corporation, USA). Luciferase activity was determined by collection at hourly intervals of luminescence counts per flower to determine intensity in relative light units per pixel (rlu/pixel). Image collection was performed in constant dark; grey and black bars represent light and dark periods respectively to which plants were acclimated. Diamond markers represent the mean intensity from pODO1:LUC flowers with error bars representing standard error of the mean (n = 4), while square markers show background levels measured from a wild‐type flower (n = 1). Luciferase activity in pODO1:LUC flowers was determined to have a significantly rhythmic pattern at P < 0.05 (ANOVA followed by Tukey’s post hoc analysis). The experiment was repeated twice with similar results.
Figure S2. Setup of detached flowers for luciferase imaging. Depiction of the arrangement of four pODO1:LUC flowers (bottom) with one wild‐type flower (top) for imaging in Video Clip 1.
Figure S3. ChIP‐Seq signal in gene space surrounding example ODO1 targets. ChIP‐seq signals at 4CLa (Peaxi162Scf00314g00086) and CIMS1 (Peaxi162Scf00163g00910) with surrounding upstream and downstream regions. The Y axis represents the sequencing depth covered by ChIP‐seq in fragments per million, after normalization to the total number of unique aligned fragments in each library. Ruler markings indicate location of ChIP signal along the corresponding petunia scaffolds, with annotated gene features below (thicker bar sections for coding regions, thinner sections for introns and untranslated regions).
Figure S4. Developmental regulation of ODO1‐bound genes. ODO1‐bound genes are significantly overlapped with genes that are upregulated (a) at day 2 relative to day 0 post‐anthesis (Patrick et al., 2021, but not with genes that are downregulated (b) at day 2 relative to day 0 post‐anthesis (Patrick et al., 2021. The hypergeometric P value for the overlap between gene sets are shown. n.s. = not significant.
Figure S5. ODO1 binding of SAM cycle genes determined by ChIP‐seq. (a) Methionine pathway with SAM and folate cycle genes identified in petunia (see also Table S2). The genes bound by ODO1 are indicated by red squares. (b) Overlap between ODO1‐bound genes and SAM/THF genes with a p value representing the significance of the observed overlap determined by hypergeometric distribution against the whole genome as background. Abbreviations: CIMS, cobalamin‐independent methionine synthase; MTHFR, methylenetetrahydrofolate reductase; MTs, methyltransferases; ODO1, ODORANT1; SAHH, S‐adenosyl‐L‐homocysteine hydrolase; SAMS, S‐adenosylmethionine synthetase; SHMT, serine hydroxymethyltransferase; THF, tetrahydrofolate.
Figure S6. Comparison between the putative ODO1 binding motif with previously reported binding motifs of transcription factors in Arabidopsis. The putative binding motif of ODO1 (Motif 1, Figure 5a) was queried against the Arabidopsis DAP‐Seq database (O’Malley et al., 2016) using Tomtom (Gupta et al., 2007). Top three matches by lowest E‐value shown, all relating to MYB transcription factors.
Figure S7. Motif 1B occurrences within promoters of ODO1‐bound FVBP genes. Sequence matches to the predicted ODO1 binding motif within 500 bp promoter of FVBP genes (Figure 5c) with P value < 1.0e‐04 as determined by MEME (Bailey et al., 2009) are shown. Abbreviations: 4CL, 4‐coumaryl‐CoA ligase; ADT, arogenate dehydratase; BPBT, benzoyl‐CoA:benzylalcohol/phenylethanol benzoyltransferase; C3H, coumarate 3‐hydroxylase; C4H, cinnamate 4‐hydroxylase; CAD, cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl‐CoA O‐methyltransferase; CHD, cinnamoyl‐CoA hydratase‐dehydrogenase; CNL, cinnamoyl‐CoA ligase; CS, chorismate synthase; DAHP, 3‐deoxy‐D‐arabino‐heptulosonate 7‐phosphate synthase; DHD‐SDH, 3‐dehydroquinate dehydratase and shikimate dehydrogenase; EPSPS, 5‐enolpyruvylshikimate 3‐phosphate; HCT, hydroxycinnamoyl‐CoA:shikimate/quinate hydroxycinnamoyl transferase; ODO1, ODORANT1; PAL, phenylalanine ammonia lyase; SK, shikimate kinase.
Figure S8. Synthetic promoter constructs. Restriction sites are underlined and ODO1 binding sites are bold. Mutated nucleotides are displayed in lower case and bold.
Table S1. Significantly enriched GO terms among ODO1‐bound genes determined by ChIP‐seq.
Table S2. P. axillaris genes involved in SAM and folate cycle determined by BLAST against Arabidopsis whole gene set. E‐Value: Expect value for sequence match relative to chance.
Table S3. Significantly enriched GO terms for genes downregulated in odo1i.
Table S4. Significantly enriched GO terms for genes upregulated in odo1i.
Table S5. Transcription factor regulatory targets of ODO1.
Table S6. Primer sequences.
Video Clip 1. Rhythmic luciferase activity driven by the ODO1 promoter. Four transgenic (T2) flowers of P. hybrida cv. W115 plants stably transformed with pODO1:LUC construct are shown, with one wild‐type flower at top of frame as a background control (see Figure S2). Flowers from plants acclimated to a 16 h light, 8 h dark period were sprayed with luciferin solution (1 mm D‐Luciferin and 0.01% Silwet) one day prior to imaging, subsequently detached and placed upright in a glass beaker containing water and then sprayed once again with luciferin solution prior to being placed in constant dark for imaging. Bioluminescence images were captured using a CCD camera (Pixis 1024, https://www.princetoninstruments.com) every hour for 48 h with a 1‐minute exposure. Quantification of the bioluminescence images is shown in Figure S1b.
Data Availability Statement
The ChIP and RNA sequencing data used in this study are available at the NCBI Sequence Read Archive under BioProject ID PRJNA729780 and additional data are available at NCBI‐GEO GSE189280.


