Abstract
Variants in multiple lysosomal enzymes increase Parkinson’s disease (PD) risk, including the genes encoding glucocerebrosidase (GCase), acid sphingomyelinase (ASMase) and galactosylceramidase. Each of these enzymes generates ceramide by hydrolysis of sphingolipids in lysosomes, but the role of this common pathway in PD pathogenesis has not yet been explored. Variations in GBA1, the gene encoding GCase, are the most common genetic risk factor for PD. The lysosomal enzyme cathepsin B has recently been implicated as an important genetic modifier of disease penetrance in individuals harboring GBA1 variants, suggesting a mechanistic link between these enzymes. Here, we found that ceramide activates cathepsin B, and identified a novel role for cathepsin B in mediating prosaposin cleavage to form saposin C, the lysosomal coactivator of GCase. Interestingly, this pathway was disrupted in Parkin-linked PD models, and upon treatment with inhibitor of ASMase which resulted in decreased ceramide production. Conversely, increasing ceramide production by inhibiting acid ceramidase activity was sufficient to upregulate cathepsin B- and saposin C-mediated activation of GCase. These results highlight a mechanistic link between ceramide and cathepsin B in regulating GCase activity and suggest that targeting lysosomal ceramide or cathepsin B represents an important therapeutic strategy for activating GCase in PD and related disorders.
Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disease that leads to α-synuclein accumulation in neurons and dopaminergic ‘neuronal death in the substantia nigra’ pars compacta. PD is clinically characterized by bradykinesia, tremor, postural instability and non-motor symptoms including depression, cognitive impairment and sleep disturbances (1,2). Multiple cellular pathways involving impaired mitochondrial quality control, endosomal-lysosomal dysfunction, immune dysfunction and impaired lysosomal sphingolipid metabolism have been associated with PD pathogenesis (3). These pathways have been informed by the emergence of genetic forms of PD, including variations in GBA1, which is now recognized as one of the most important risk factors for PD, with approximately 5–10% of PD patients carrying a GBA1 variant. GBA1 encodes for the lysosomal enzyme β-glucocerebrosidase (GCase) which hydrolyzes glucosylceramide (GluCer) into glucose and ceramide in lysosomes (4,5). Importantly, multiple studies have found reduced GCase activity in idiopathic PD cases in individuals who do not carry GBA1 variants (6–10), suggesting a key role of GCase activity in PD pathogenesis across multiple patient cohorts.
GCase activity is regulated by saposin C, an important endogenous activator of GCase activity in the lysosome. Proteolytic cleavage of prosaposin generates four saposins (A, B, C and D) in the lysosome. The saposins are 80 amino acid-long heat-stable proteins that play specific roles in the lysosomal sphingolipid degradation pathway by coactivating distinct lysosomal enzymes (11). Saposin C deficiency results in a disease phenotype that is similar to Gaucher disease, a condition that results from homozygous GBA1 variants. This therefore suggests a key role for saposin C in regulating GCase activity in disease states (12). However, the mechanisms regulating prosaposin cleavage into saposin C are still not well understood.
Interestingly, lysosomal ceramide which is generated by GCase (GBA1) can also be generated by other lysosomal enzymes including acid sphingomyelinase (ASMase) (SMPD1) and galactosylceramidase (GALC). Importantly, loss-of-function variants of GBA1, SMPD1 and GALC all increase PD risk (5,13,14) and are predicted to result in decreased ceramide in the lysosome (15,16), suggesting that ceramide levels may play an important role in regulating PD pathogenesis, but how ceramide modulates pathways implicated in PD has not been well studied. Cathepsin B has recently been identified as an important modifier of PD risk in individuals with GBA1 variants (17), suggesting a potential mechanistic link between cathepsin B and GCase-mediated enzymatic pathways, including ceramide production.
Genetic forms of PD have also been linked to mutations in PARK2 which encodes Parkin, an E3 ubiquitin ligase. Parkin is the most common cause of autosomal recessive juvenile-onset and young-onset PD (18) and plays an important role in mitochondrial quality control as well as the ubiquitin proteasome system. Impaired mitophagy and mitochondrial dysfunction are the main mechanisms proposed for the development of PD due to mutations of PARK2 (19). However, Parkin has also been shown to have roles in inflammation (20) and vesicle trafficking, including exosome secretion, endocytosis and retromer trafficking (21–23). The mechanisms by which cathepsin B, GCase activity and prosaposin cleavage may be modulated in genetic models of PD, including Parkin-deficient cells, remain unclear.
In the current study, we found a novel role for cathepsin B in mediating prosaposin to saposin C cleavage. Importantly, we further found that ceramide was able to activate cathepsin B maturation and activity which increased prosaposin processing, resulting in more saposin C that coactivates GCase activity. This pathway was disrupted in both Parkin-deficient cells and via treatment with an ASMase inhibitor which results in decreased ceramide production. Furthermore, this pathway could be rescued upon treatment with carmofur, an acid ceramidase inhibitor which increases ceramide production, thereby identifying lysosomal ceramide as a potential therapeutic target.
Results
Lysosomal GCase activity, but not GCase protein, is decreased in Parkin KO cells
Multiple studies have suggested that abnormal sphingolipid metabolism in lysosomes may contribute to the pathogenesis of PD (5,9,13,14,24,25). In addition, it has been reported that GCase activity is reduced in various forms of PD (6–8,10,26). Using Parkin-linked PD as a model, we explored possible roles for Parkin in the regulation of GCase activity. To this end, we generated a Parkin-deficient HEK293-FT cell line (Parkin knockout (KO)) using CRISPR/Cas9-based genome editing. Parkin KO cells showed complete loss of Parkin protein by immunoblot analysis with Parkin antibody (Fig. 1B). Using a live cell GCase assay (27), we found that Parkin KO cells displayed a significant reduction in fluorescence intensity, compared with wild-type (WT) cells (Fig. 1A), suggesting that lysosomal GCase activity was significantly reduced in Parkin KO cells.
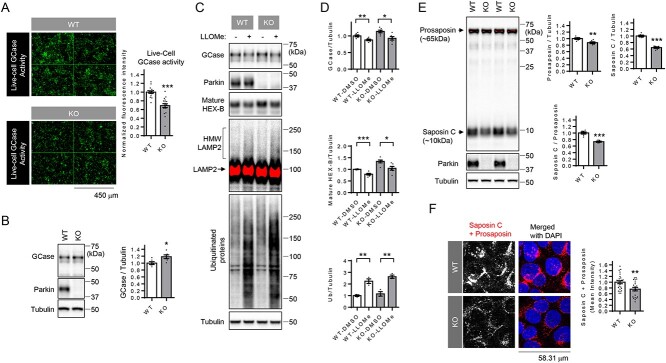
Figure 1.
Live-cell GCase activity and saposin C levels are reduced in Parkin knockout (KO) cells. (A) Representative fluorescent images of live-cell GCase activity in WT and Parkin KO cells. Two-tailed unpaired t-test ***P < 0.001, n = 16 microscopic fields from three independent dishes and 18 microscopic fields from three independent dishes from left to right. (B) Representative immunoblot data and quantification of cellular GCase protein levels in WT and Parkin KO cells. Band intensities were normalized to tubulin, and compared with WT cells. Two-tailed unpaired t-test, *P < 0.05. n = 6. (C) Representative immunoblot data of indicated proteins in WT and Parkin KO cells upon LLOMe (lysophagy inducer) treatment. Saturated pixels are shown in red. Ubiquitinated high-molecular-weight (HMW) LAMP2 upon LLOME treatment is indicated. (D) Quantification of indicated proteins in WT and Parkin KO cells upon LLOMe treatment. Two-way ANOVA followed by Tukey’s multiple comparisons test, *P < 0.05; **P < 0.01; ***P < 0.001, n = 6 for GCase and mature Hex-B, n = 3 for ubiquitin. (E) Representative immunoblot data using saposin C-specific prosaposin antibody and quantification of prosaposin and saposin C in WT and Parkin KO cells. Saturated pixels are shown in red. Band intensities were normalized to tubulin and compared with WT cells. For the quantitation of saposin C to prosaposin ratio, band intensity of saposin C was divided with prosaposin intensity and normalized with average of WT cells. Two-tailed unpaired t-test, **P < 0.01; ***P < 0.001, n = 6. (F) Representative fluorescent image of prosaposin staining using saposin C-specific prosaposin antibody in WT and Parkin KO cells. Quantitation of mean fluorescence intensity is shown. n = 26 microscopic fields from two coverslips for WT, n = 21 microscopic fields from two coverslips for KO cells, **P < 0.01, two-tailed unpaired t-test.
Interestingly, GCase activity was not reduced when analyzing GCase using whole-cell lysates and 4-methylumbelliferyl-β-D-glucopyranoside (4-MUG) as a fluorometric substrate (Supplementary Material, Fig. S1A). Moreover, GCase protein levels were not decreased in Parkin KO cells compared with WT cells (Fig. 1B), suggesting that reduced lysosomal GCase activity was not due to a reduction in total GCase protein. To further explain the discrepancy between live-cell GCase activity and the whole-cell GCase activity, we first examined whether GCase was mis-localized to other cellular compartments instead of lysosomes. Using double immunostaining with antibodies against GCase and LIMP2, a receptor for lysosomal targeting of GCase (28), we found that WT and Parkin KO cells showed comparable levels of LIMP-2 protein. Co-localization of GCase and LIMP-2 was largely unaffected in Parkin KO cells (Supplementary Material, Fig. S1B). Although LIMP-2 protein is considered a reliable marker of lysosomes, the persistent co-localization of GCase and LIMP-2 in Parkin KO cells did not rule out a possibility that GCase may be localized to non-functional lysosomal compartments.
To further address this issue, we used a lysosomotropic reagent, l-leucyl-l-leucine methyl ester (LLOMe), that accumulates within lysosomes. Damaged lysosomes are tagged by ubiquitination of LAMP2 and cleared by lysophagy (29,30) that can be indirectly assayed by measuring the degree of reduction of lysosomal enzymes after LLOMe treatment. Consistent with previous reports, we found that LLOME treatment induced the appearance of high-molecular-weight (HMW) LAMP2 and ubiquitinated protein species (Fig. 1C). The degree of reduction of GCase, as well as mature forms of another enzyme HEXB, in response to LLOMe treatment was similar in WT and Parkin KO cells (Fig. 1C and D), indicating that GCase was located to LLOMe-responsive functional lysosomal compartments. Collectively, these results suggested that decreased live-cell GCase lysosomal activity in Parkin KO cells was not due to changes in GCase protein levels or alterations in GCase localization.
Saposin C level is reduced in Parkin KO cells
Lysosomal GCase activity is known to be regulated by saposin C, the lysosomal coactivator of GCase (12,31,32). Saposins are generated from prosaposin which undergoes proteolytic processing to generate four saposin fragments (saposin A, saposin B, saposin C and saposin D) in lysosomes (33) (Supplementary Material, Fig. S2). To examine whether reduced saposin C levels might contribute to decreased GCase activity in Parkin KO cells, we performed immunoblot analysis of whole lysates from WT and KO cells using a prosaposin antibody directed against the internal saposin C domain of prosaposin (Supplementary Material, Fig. S2). Protein levels of prosaposin were reduced by ~10% in Parkin KO cells, while Saposin C levels were reduced by ~40% (Fig. 1E). Importantly, the ratio of saposin C to prosaposin was also reduced by ~30% in Parkin KO cells, compared with WT cells (Fig. 1E), suggesting an impairment of prosaposin processing into saposin C in Parkin KO cells. Immunocytochemistry analysis using the prosaposin antibody directed to the internal saposin C domain also confirmed that there was a significant reduction in total protein levels of prosaposin and saposin C in Parkin KO cells, compared with WT cells (Fig. 1F). In addition, both Saposin C levels and the ratio of saposin C to prosaposin in an independent Parkin KO cell line (Parkin KO #2), which was generated with a different CRISPR/Cas9 gRNA, were reduced by ~40%, compared with WT cells (Supplementary Material, Fig. S3A and B). These results suggest that decreased saposin C may be responsible for the reduced GCase activity observed in Parkin KO cells.
Cathepsin B promotes cleavage of prosaposin to saposin C
We next investigated the mechanism responsible for decreased prosaposin cleavage to saposin C in Parkin KO cells. As cathepsin D has been previously suggested to mediate prosaposin processing to saposin C (33), we first sought to confirm these findings. To this end, we treated cells (WT and KO) with a specific and membrane-permeant cathepsin D inhibitor (Pepstatin A-Methyl Ester, PepA-ME) (34,35). We found that treatment with PepA-ME significantly increased levels of mature forms of cathepsin D, presumably due to the stabilization of inactive mature cathepsin D, complexed with PepA-ME (Fig. 2A). However, neither prosaposin levels nor saposin C levels were affected by PepA-ME treatment in either WT or KO cells (Fig. 2A), suggesting that cathepsin D may not regulate prosaposin cleavage.
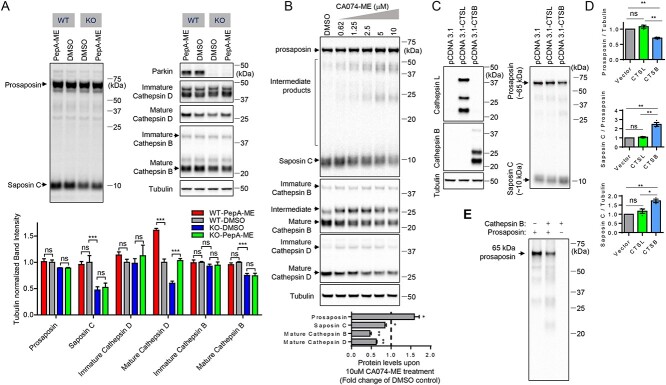
Figure 2.
Cathepsin B promotes proteolytic processing of prosaposin to saposin C. (A) Representative immunoblots of prosaposin, saposin c, Parkin, Cathepsin B, cathepsin D and tubulin upon PepstainA-ME (PepA-ME) treatment (4 μM, ~ 24 h in WT and Parkin KO cells). Quantitation of prosaposin, saposin c, Parkin, Cathepsin B, cathepsin D and tubulin upon PepA-ME treatment (4 μM, ~ 24 h) in WT and KO cells. Band intensities were normalized to tubulin, and compared with DMSO-treated WT cells. n = 3, Two-way ANOVA with Tukey’s multiple comparisons test, ns, not significant, ***P < 0.001. (B) Representative immunoblot of prosaposin, saposin C, cathepsin B and cathepsin D upon CA074-ME treatment (0.625, 1.25, 2.5, 5 and 10 μM, ~24 h) in WT HEK293-FT cells. The Graph shows immunoblot quantitation of prosaposin, saposin C, mature cathepsin B and mature cathepsin D upon 10 μM CA074-ME treatment. Protein band intensities are normalized with the level of tubulin. Data from the sample treated with 10 μM CA074-ME were compared with the DMSO control. Two-tailed paired t-test. n = 3, *P < 0.05; **P < 0.01. (C) Representative immunoblot data of indicated proteins upon empty vector, cathepsin B or cathepsin L overexpression in WT cells. (D) Quantitation of prosaposin, saposin C and saposin C to prosaposin ratio upon overexpression of cathepsin B or cathepsin L. Band intensities are normalized with tubulin levels and compared with vector transfected cells. Data are means ± SEM. Repeated-measures ANOVA with Tukey’s post hoc analysis. n = 3. *P < 0.05; **P < 0.01; ns, not significant. (E) In vitro prosaposin cleavage with recombinant cathepsin B and recombinant prosaposin. Representative immunoblot data from three independent experiments with similar results are shown.
To investigate if other lysosomal proteases, rather than cathepsin D, might be responsible for prosaposin processing to saposin C, we tested the effect of other lysosomal cathepsin inhibitors. E64D ((2S,3S)-trans-epoxysuccinyl-L-leucylamido-3-methylbutane ethyl ester) is a cell-permeable cysteine cathepsin inhibitor, known to inhibit cathepsin B and cathepsin L, but not cathepsin D (36). Strikingly, treatment with E64D strongly increased levels of ~65 kDa prosaposin and intermediate prosaposin cleavage products in both WT and KO cells. Moreover, E64D treatment in WT cells, but not in KO cells, significantly reduced saposin C levels (Supplementary Material, Fig. S4A and B), suggesting that E64D-sensitive lysosomal proteases promote the proteolytic processing of prosaposin to saposin C. We further found that cells treated with E64D exhibited reduced levels of mature cathepsin B, whereas levels of intermediate and immature forms of cathepsin B were increased (Supplementary Material, Fig. S4A and B), indicating that E64D may inhibit cathepsin B maturation, and that cathepsin B may be mediating prosaposin cleavage to saposin C. Interestingly, mature cathepsin B levels were reduced in Parkin KO, compared with WT cells (Fig. 2A, Supplementary Material, Fig. S4A and B). Moreover, an independent Parkin KO cell line (Parkin KO #2) also showed significant reduction of mature cathepsin B levels, compared with WT cells (Supplementary Material, Fig. S3A and B). Consistent with our results, Okarmus et al. (2020) (37) recently reported that cathepsin B levels were reduced in Parkin KO iPS-derived dopaminergic neurons, compared with isogenic controls.
To directly test the involvement of cathepsin B in prosaposin processing to Saposin C, we utilized a cathepsin B inhibitor, CA074-ME, which is a cell-permeable covalent inhibitor of cathepsin B (38). We further validated that CA074-ME indeed inhibited cathepsin B by demonstrating reduction in mature forms of cathepsin B and an increase in its intermediate forms and immature forms (Fig. 2B). Treatment of WT cells with increasing doses of CA074-ME resulted in a gradual increase of ~65 kDa prosaposin and intermediate prosaposin cleavage products. In addition, CA074-ME treatment decreased ~10 kDa saposin C levels in a dose-dependent manner (Fig. 2B), suggesting that cathepsin B activity is involved in saposin C generation through proteolytic processing of prosaposin.
Similar to the data from PepA-ME (cathepsin D inhibitor)-treated cells, cathepsin D-RNAi in WT cells did not increase levels of ~65 kDa prosaposin (Supplementary Material, Fig. S5A and B). Knockdown of endogenous cathepsin D with three independent RNAi constructs did not change the ratio of saposin C to prosaposin (Supplementary Material, Fig. S5A and B). Three independent heterogeneous cathepsin D KO pools generated with CRISPR/Cas9 also did not show any significant changes in prosaposin or saposin C levels (Supplementary Material, Fig. S6A and B).
Moreover, we demonstrated that cathepsin D is not responsible for prosaposin cleavage by ectopically overexpressing cathepsin D to test its effects on prosaposin processing. Both mature and immature forms of cathepsin D were greatly increased upon transient transfection of cathepsin D, but we could not detect any increase in saposin C levels in either WT or Parkin KO cells. In particular, the ratio of saposin to prosaposin did not change upon cathepsin D overexpression (Supplementary Material, Fig. S5C and D), and mature forms of cathepsin B levels were not affected by cathepsin D overexpression (Supplementary Material, Fig. S5C and D), suggesting that cathepsin D does not play a major role in the proteolytic processing of prosaposin to saposin C. In contrast, transiently overexpressing cathepsin B, but not cathepsin L which is also a target of E64D, generated more saposin C with concomitant reduction of prosaposin levels (Fig. 2C and D). Moreover, RNAi knockdown of endogenous cathepsin L in WT cells with two independent RNAi constructs did not affect prosaposin processing (Supplementary Material, Fig. S7A and B). Neither prosaposin nor saposin levels were affected significantly by the knockdown of endogenous cathepsin L (Supplementary Material, Fig. S7A and B), suggesting that cathepsin L does not play a major role in prosaposin processing either. Consistent with the previous reports that cathepsin L degrades cathepsin D (39), protein levels of both mature and immature cathepsin D levels were increased significantly upon cathepsin L knockdown (Supplementary Material, Fig. S7A and B). Both immature and mature cathepsin B were increased upon cathepsin L knockdown (Supplementary Material, Fig. S7A and B).
Interestingly, co-immunoprecipitation assay revealed that prosaposin interacts with immature forms of Cathepsin B, but not cathepsin D (Supplementary Material, Fig. S8), raising a possibility that cathepsin B and prosaposin may traffic to lysosomes together, facilitating cathepsin B-mediated prosaposin processing in the lysosome.
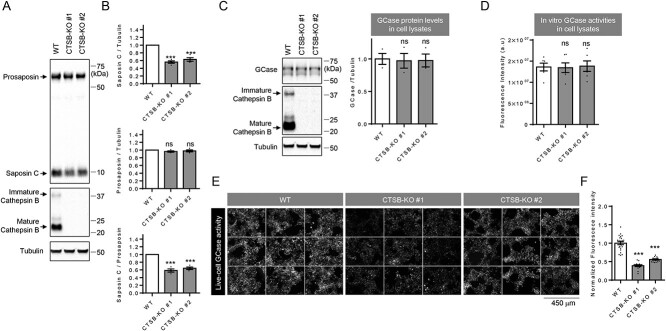
Having shown that cathepsin B can promote prosaposin processing to saposin C, and that prosaposin forms a complex with cathepsin B, we tested whether cathepsin B can cleave prosaposin directly in vitro using prosaposin cleavage assays with purified recombinant human prosaposin and recombinant human cathepsin B. Indeed, the in vitro cleavage assay showed a robust prosaposin cleavage by cathepsin B (Fig. 2E). We additionally established two independent cathepsin B KO cell lines using CRISPR/Cas9 genome editing, and both saposin C levels and the ratio of saposin C to prosaposin C were significantly reduced in these lines (Fig. 3A and B). Importantly, two independent cathepsin B KO cell lines showed decreased live-cell GCase activity (Fig. 3E and F), whereas whole-cell GCase activity or protein levels were not changed in these cells, further supporting the critical role of saposin C in promoting lysosomal GCase activity (Fig. 3C and D).
Figure 3.
Effects of cathepsin B on saposin C, GCase protein and GCase activity. (A, B) Effects of cathepsin B knockout (KO) on prosaposin processing to saposin C. Total cell lysates from WT and two cathepsin B KO cell lines (CTSB-KO #1 and CTSB-KO #2) were analyzed with immunoblot using indicated antibodies. Representative immunoblot data are shown in A. (B) Quantitation of immunoblot data. Band intensities are normalized with tubulin levels. Data are means ± SEM. Repeated-measures analysis of variance (ANOVA) with Dunnett’s multiple comparison test. n = 4. ***P < 0.001; ns, not significant. (C, D) Effects of cathepsin B KO on GCase protein levels, in vitro GCase activity and live-cell GCase activity. (C) GCase protein levels in WT cells and cathepsin B KO cell lines (CTSB-KO #1 or CTSB-KO #2). Representative immunoblot data and quantitation of immunoblot data. Band intensities are normalized with tubulin levels. Data are means ± SEM. One-way ANOVA followed by Dunnett’s test. n = 3, ns = P > 0.05. (D) In vitro GCase activity using 4-MUG (4-methylumbelliferyl-β-D-glucopyranoside) substrate in WT or two CTSB-KO cell lines (CTSB-KO #1 or CTSB-KO #2). Data are means ± SEM. One-way ANOVA followed by Dunnett’s test. n = 6, ns, not significant. (E, F) Live-cell GCase activities are reduced in cathepsin B KO cell lines, compared with WT cells. (E) Representative fluorescent images of live-cell GCase assay from WT and two CTSB-KO cell lines (CTSB-KO #1 and CTSB-KO#2). (F) Normalized mean fluorescence intensities. One-way ANOVA followed by Dunnett’s test. n = 24 microscopic fields from three independent dishes for WT, 16 microscopic fields from three independent dishes for CTSB-KO #1 cells and 16 microscopic fields from three independent dishes for CTSB-KO #2 cells.; ***P < 0.001.
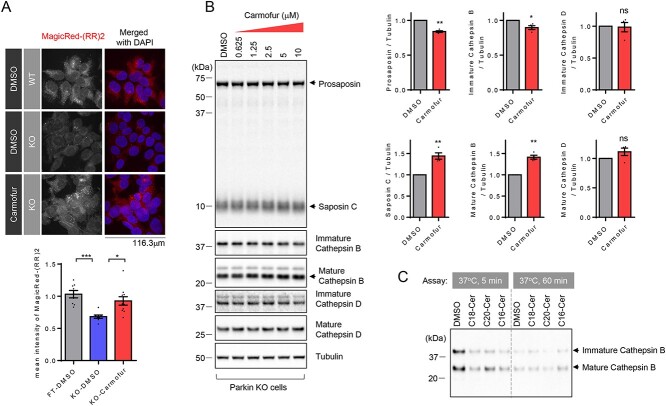
As we identified cathepsin B as a key regulator of prosaposin to saposin C cleavage, we next investigated whether cathepsin B activity was altered in Parkin KO cells to account for the observed decrease in prosaposin processing. These experiments revealed that mature forms of cathepsin B (Supplementary Material, Fig. S4A and B), as well as its activity, as assayed with Cathepsin B substrate MagicRed-RR2, (Fig. 4A) were reduced in Parkin KO cells compared with WT cells.
Figure 4.
Acid ceramidase inhibition leads to increased cathepsin B activity and cleavage of prosaposin into saposin C. (A) Representative fluorescence images showing intracellular cathepsin B activity in WT and Parkin KO cells. And effect of acid ceramidase inhibitor (7.5 μM, carmofur) on intracellular cathepsin B activity in KO cells. Intracellular cathepsin B activity was assayed using cathepsin B substrate (MagicRed-RR2). Graph shows mean fluorescence intensity of cleaved cathepsin B substrate (MagicRed-RR2). n = 8 microscopic fields from two dishes, 9 microscopic fields from two dishes and 10 microscopic fields from two dishes from left to right. One-way ANOVA followed by Tukey’s multiple comparisons test, *P < 0.05, ***P < 0.001. (B) Representative immunoblot data showing effects of acid ceramidase inhibitor (carmofur) treatment (0.625, 1.25, 2.5, 5 and 10 μM, ~6 h) on cathepsin B, cathepsin D and prosaposin cleavage to saposin C in Parkin KO cells. The Graph shows immunoblot quantitation for prosaposin, saposin C, mature cathepsin B and mature cathepsin D upon 10 μM carmofur treatment, compared with DMSO control. Protein band intensities are normalized with the level of tubulin. Two-tailed paired t-test. n = 3 *P < 0.05; **P < 0.01. (C) Effect of ceramide on in vitro cathepsin B activity, based on auto-degradation of recombinant cathepsin B in vitro. Representative immunoblot data from three independent experiments with similar results is shown.
Ceramide increases cathepsin B-mediated prosaposin cleavage into saposin C in Parkin KO cells
A ceramide is composed of sphingosine and a fatty acid, which can be generated via de novo synthesis in the endoplasmic reticulum (40,41). In addition, ceramides can also be produced in lysosomes via the catabolic salvage pathway by three lysosomal enzymes including GCase, ASMase or galactosylceramidase (15,16). Because ceramide has been reported to activate cathepsin B activity (42), we wondered whether defective cathepsin B activity could be corrected by elevating lysosomal ceramide levels with carmofur, a lysosomal acid ceramidase inhibitor (43). Indeed, treatment of Parkin KO cells with carmofur was able to increase cathepsin B activity (Fig. 4A). Moreover, increasing concentrations of carmofur in Parkin KO cells resulted in a gradual increase in cathepsin B maturation (Fig. 4B), as well as a gradual reduction of ~65 kDa prosaposin with concomitant increase of ~10 kDa saposin C (Fig. 4B). Similarly, treatments of WT cells (Supplementary Material, Fig. S9) or an independent Parkin KO line (Supplementary Material, Fig. S10) with carmofur were able to increase cathepsin B maturation and prosaposin processing to saposin C.
Next, we wondered whether lysosomal ceramide–cathepsin B–prosaposin mechanism also operates in microglia since dysfunction of microglia has been implicated in neurodegenerative diseases. In particular, expression of prosaposin and cathepsin B is highest in microglia among many brain cell types (www.brainrnaseq.org). Consistent with our data in HEK293-FT cells, treatment of Human Microglial Cell line 3 (HMC3) with cathepsin B inhibitor (CA074-ME) impaired prosaposin processing (Supplementary Material, Fig. S11), whereas acid ceramidase inhibitor (carmofur) promoted cathepsin B maturation and prosaposin processing (Supplementary Material, Fig. S12). Importantly, these results were also confirmed in primary cortical neurons where we found that inhibition of acid ceramidase with carmofur promoted cathepsin B maturation with concomitant reduction of prosaposin (Supplementary Material, Fig. S13).
To further examine whether ceramide could directly activate cathepsin B, we used the auto-degradation of recombinant cathepsin B as a readout of its activity. Recombinant cathepsin B in the presence of DMSO showed a time-dependent auto-degradation (compare 5 min incubation with DMSO and 60 min incubation with DMSO) (Fig. 4C) that was accelerated in the presence of multiple different species of ceramide (C18-Cer; C20-Cer; C16-Cer). The remaining cathepsin B levels after 5 min incubation of cathepsin B in the presence of ceramides was comparable to those after 60 min incubation, suggesting that ceramide can directly activate cathepsin B.
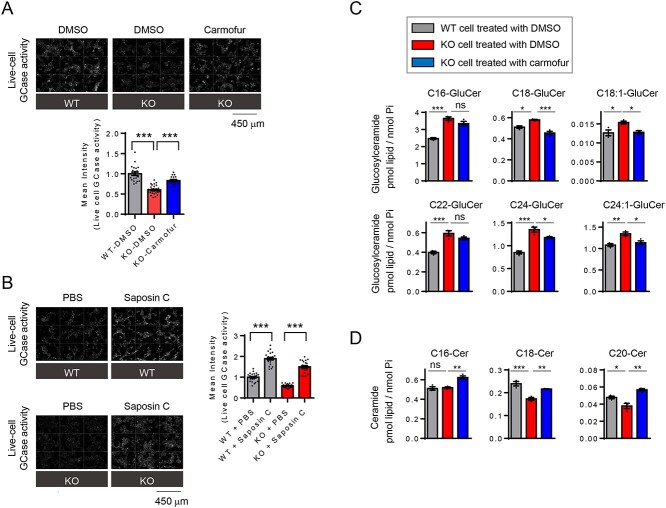
Since carmofur treatment in Parkin KO cells increased saposin C levels (Fig. 4B), we asked whether carmofur treatment could also rescue reduced GCase activity. Indeed, treatment of Parkin KO cells with carmofur rescued reduced GCase activity (Fig. 5A). Importantly, exogenous recombinant saposin C also significantly increased lysosomal GCase activity both in WT and KO cells (Fig. 5B), further demonstrating the importance of Saposin C in modulating GCase activity. This also suggests that increasing saposin C levels may represent a therapeutic strategy to restore reduced GCase activity.
Figure 5.
Acid ceramidase inhibition or recombinant saposin C increase lysosomal GCase activity. (A) Effect of acid ceramidase inhibitor (7.5 μM, carmofur) on live-cell GCase activity. Representative fluorescence images from live-cell GCase assay are shown. Graph shows mean fluorescence intensity from a live-cell GCase assay. Data were compared with DMSO-treated WT cells. For the comparison pair, two-tailed unpaired t-test, ***P < 0.001. n = 21, 29 and 17 microscopic fields from left to right. Images were taken from minimum two independent dishes for each condition. (B) Effect of exogenous recombinant saposin C on live-cell GCase activity in WT and Parkin KO cells. Representative fluorescence images from live-cell GCase assay are shown. Graph shows mean fluorescence intensity from live-cell GCase assay. One-way ANOVA followed by Tukey’s multiple comparisons test, ***P < 0.001. n = 25, 22, 28 and 22 microscopic fields from left to right. Images were taken from minimum two independent dishes for each condition. (C) Lipidomic analysis of glucosylceramide (GluCer) species in WT cell and Parkin KO cells and effects of carmofur (7.5 μM) on GluCer species in KO cells. GluCer were quantified and expressed as p mol/n mole inorganic phosphate (Pi). One-way ANOVA followed by Tukey’s multiple comparisons test, *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant, n = 3. Full data set is in Supplementary Material, Figure S11A. (D) Lipidomic analysis of ceramide species (Cer) in WT cell and Parkin KO cells and effects of carmofur (7.5 μM) on ceramide species in KO cells. Ceramide species were quantified and expressed as p mol/n mole Pi. One-way ANOVA followed by Tukey’s multiple comparisons test, *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant, n = 3. Full data set is in Supplementary Material, Figure S11B.
Mass spectrometry-based lipid analysis further revealed that Parkin KO cells contained higher levels of GluCer species with various acyl chain length, compared with WT cells (Fig. 5C and Supplementary Material, Fig. S14A), in agreement with decreased live-cell GCase activity in Parkin KO cells (Fig. 1A). Similar to the previous results from GCase-deficient cells (44), specific ceramide species with longer acyl chain (C18-, C18:1-, C20-, C20:1-ceramide) were selectively reduced in Parkin KO cells (Fig. 5D and Supplementary Material, Fig. S14B). In contrast, the levels of ceramide species with shorter acyl chain (C14- and C16-ceramide) were not altered in Parkin KO cells (Fig. 5D and Supplementary Material, Fig. S14B). Importantly, carmofur treatment was sufficient to reduce GluCer levels (Fig. 5C and Supplementary Material, Fig. S14A) and correct reduced levels of specific ceramide species (C18- and C20-ceramide) in Parkin KO cells (Fig. 5D and Supplementary Material, Fig. S14B).
Reducing ceramide by inhibition of ASMase impairs cathepsin B-mediated prosaposin cleavage
Loss-of-function variants of GBA1, SMPD1 and GALC all increase PD risk and are predicted to result in decreased ceramide in the lysosome. For example, ASMase catalyzes the breakdown of sphingomyelin to generate ceramide in lysosomes, and loss-of-function ASMase variant is causally linked to increased PD risk (14,24).
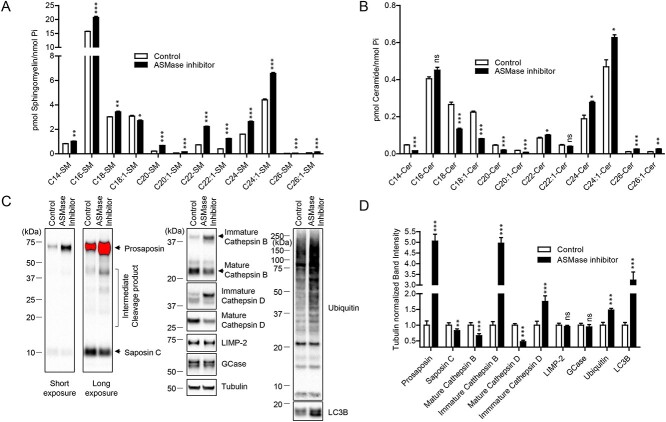
To further test our hypothesis that lysosomal ceramide regulates cathepsin B-mediated prosaposin cleavage into saposin C, we thus examined whether decreasing ceramide production by ASMase inhibition was sufficient to downregulate this pathway. Using a highly potent and selective chemical ASMase inhibitor (1-aminodecylidene bis-phosphonic acid) (45,46), we first showed that this was effective in increasing levels of most sphingomyelin species as expected (Fig. 6A) while reducing specific ceramide species including C14-, C18-, C18:1-, C20-, C20:1-ceramide, suggesting that these might be the species normally generated by the hydrolysis of sphingomyelin by ASMase (Fig. 6B).
Figure 6.
Inhibition of acid sphingomyelinase (ASMase) leads to alterations in sphingomyelin/ceramide profiles, cathepsin B maturation and processing of prosaposin. (A, B) HEK-293-FT cells were treated either PBS or 5 μM of ASMase inhibitor (1-aminodecylidene bis-phosphonic acid) for ~17 h. (A) Lipidomic analysis of sphingomyelin species upon ASMase inhibitor treatment. Sphingomyelin species were quantified and expressed as p mol/n mole inorganic phosphate (Pi), and compared with control (PBS-treated) samples. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant, two-tailed unpaired t-test. n = 3. (B) Lipidomic analysis of ceramide species upon ASMase inhibitor. Ceramide species were quantified and expressed as p mol/n mole Pi, and compared with control (PBS-treated) samples. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant, two-tailed unpaired t-test. n = 3. (C, D) Cells were treated either DMSO or 5 μM of ASMase inhibitor (1-aminodecylidene bis-phosphonic acid) for 2 days. (C) Representative immunoblot data of indicated proteins upon ASMase inhibitor. (D) Quantitation of indicated proteins ASMase. Band intensities were normalized to tubulin, and compared with control (PBS-treated) cells. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant, two-tailed unpaired t-test. n = 4.
Next, we investigated the functional consequences of ASMase inhibition on cathepsin B maturation and prosaposin processing to saposin C. Supporting the notion that reduced lysosomal ceramide results in reduced cathepsin B maturation and impaired cathepsin B-mediated prosaposin processing to saposin C, cells treated with ASMase inhibitor showed reduced mature cathepsin B level by ~50%, and strongly increased immature cathepsin B levels by 5-fold (Fig. 6C and D). Importantly, ASMase inhibition also resulted in a large ~5-fold increase in ~65 kDa prosaposin levels with a concomitant reduction of ~10 kDa saposin C levels, demonstrating that ASMase inhibitor treatment results in impaired cathepsin B maturation and defective prosaposin processing to saposin C.
Furthermore, we found that ASMase inhibition resulted in the accumulation of ubiquitinated protein species and autophagic substrates LC3B, a protein involved in formation of autophagosomes, levels, suggesting that lysosomal degradation of autophagic substrates were also inhibited by ASMase inhibition, potentially due to the reduction of cathepsin B and cathepsin D activities. Interestingly, inhibition of ASMase did not affect the protein levels of GCase or LIMP-2, indicating that the observed cellular phenotypes are mainly driven by the chemical inhibition of ASMase activity (Fig. 6C and D). Together, these data further support our findings that lysosomal ceramide activates cathepsin B activity which mediates prosaposin cleavage, and that disrupting ceramide production by inhibiting ASMase activity blocks this pathway.
Reduced cathepsin B levels and impaired prosaposin to saposin processing in Parkin KO mice
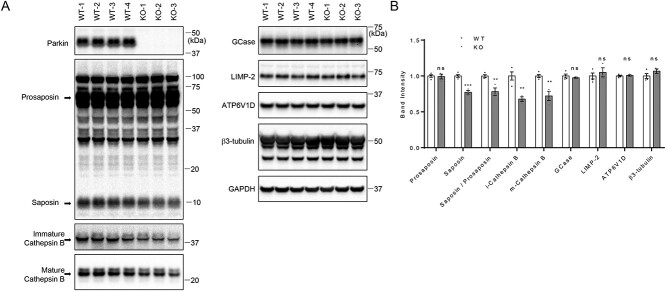
To confirm our findings in vivo, we analyzed 12-week-old Parkin KO mouse brains. Consistent with the data from Parkin KO cell lines, both mature and immature cathepsin B levels were reduced in Parkin KO brains, compared with WT mice. Furthermore, we found decreased saposin and saposin to prosaposin ratios in Parkin KO brain, compared with WT mice, whereas prosaposin levels were unchanged (Fig. 7A and B). However, levels of GCase, LIMP-2, ATP6V1D (lysosomal ATPase H+ transporting V1 subunit D) or neuronal marker β3-tubulin were not different in Parkin KO compared with WT mice (Fig. 7A and B).
Figure 7.
Saposin and cathepsin B levels are reduced in Parkin knockout (KO) mouse brain; (A) 20 μg of tissue lysates isolated form whole brains from 12-week-old WT and Parkin KO mice were analyzed with immunoblot analysis using indicated antibodies. Representative immunoblot data are shown. (B) Quantification of immunoblots. Protein band intensities were normalized with GAPDH and compared with WT. Two-tailed unpaired t-test was used to compare KO to WT mice (n = 4 for WT; n = 3 for KO) (**P < 0.01; ***P < 0.001; ns, not significant).
Discussion
Multiple genetic studies have demonstrated that variations in GBA1, SMPD1 or GALC increase PD risk (5,13,14). Importantly, while these variants cause accumulation of different lipid substrates in lysosomes (GluCer for GBA1, sphingomyelin for SMPD1 and galactosylceramide for GALC), they all contribute to ceramide deficiency, as these lysosomal hydrolases normally generate ceramide in the lysosome.
Here, we found that ceramide activates cathepsin B, which subsequently mediates the cleavage of prosaposin into saposin C. As saposin C is the lysosomal coactivator of GCase activity, this has important implications for understanding the upstream regulation of GCase activity in PD and its potential to be modulated by ceramide levels. Our study also demonstrated that measuring total cellular GCase activity with artificial 4-MUG substrates does not always match lysosomal activity, suggesting that the presence of physiological modulators such as saposin C plays an important role in regulating lysosomal GCase activity. The use of in vitro GCase activity assays using 4-MUG has had a significant impact on GCase research and therapeutic development. However, a major limitation of this assay is that it does not account for endogenous factors such as lysosomal PH, or the presence of cofactor such as saposin C that could influence GCase activity in cells (47).
Notably, a recent GWAS study identified genetic modifiers of GBA1-associated PD and dementia with Lewy bodies (DLB) (17). In this study, the authors identified variants in close proximity to SNCA and CTSB as the most significant contributors. Risk variants in the CTSB gene locus were reported to decrease mRNA expression of cathepsin B. Additionally, they showed decreased mature cathepsin B protein in multiple lines of induced pluripotent stem cell-derived neurons carrying GBA1 N370S variant. Our data provide a possible mechanistic explanation of this modifying effect of cathepsin B through saposin C-mediated regulation of GCase activity, but further studies will be required to specifically examine the identified variants of CTSB in regulating Saposin C levels. Interestingly, recent genetic screening identified pathogenic prosaposin heterozygous variant from three families with autosomal dominant PD, further implicating this pathway in the pathogenesis of PD (48). It will be of interest to examine whether variants in prosaposin affect GCase activity through deficient production of saposin C in these families.
Several studies have previously reported reduced ceramide species in idiopathic PD brains (10,49). In particular, one study by Abbot et al. (49) showed that total ceramide was reduced by ~53% in the anterior cingulate cortex, an affected brain area in PD. Our study suggests that ceramide deficiency causes reduced cathepsin B activity, whereas increasing lysosomal ceramide levels by inhibiting acid ceramidase leads to activation of cathepsin B. Previously, we found that specific ceramide species including C18- and C20-ceramide were reduced in GBA1-KO cells (44). In the current study, we used an independent strategy to lower lysosomal ceramide with chemical inhibitors of ASMase (encoded by SMPD1), whose variant is also a risk factor for PD. Interestingly, inhibition of ASMase also caused a reduction of C18-ceramide and C20-ceramide levels, suggesting that C18- and C20-ceramide species were generated by catabolic hydrolysis of GluCer or sphingomyelin in lysosomes. Notably, our lipidomic analysis from Parkin KO cells also showed that Parkin KO cells have reduced levels of C18-ceramide and C20-ceramide. Importantly treatment with carmofur, a lysosomal acid ceramidase inhibitor, corrected the reduced levels of C18- and C20-ceramide, and further rescued decreased cathepsin activity by promoting cathepsin B maturation. These data showed that lowering lysosomal ceramide by multiple independent approaches resulted in reduction of mature cathepsin B levels suggesting a direct link between lysosomal ceramides and cathepsin B.
Together, our data suggest that loss of function of GBA1, SMPD1 or GALC in PD causes lysosomal ceramide deficiency, and that reduced ceramide-mediated cathepsin B activation in lysosomes causes subsequent impairment of prosaposin processing to saposin C, ultimately resulting in impaired GCase activity (Supplementary Material, Fig. S15). As a therapeutic strategy, we propose that acid ceramidase inhibition will increase ceramide levels and consequently promote activation of cathepsin B and GCase activity. Thus, development of novel acid ceramidase inhibitors may serve as a potential therapeutic strategy for PD and related disorders (Supplementary Material, Fig. S15).
Materials and Methods
Antibodies and chemicals
The following antibodies were used: mouse anti-Parkin (Santacruz, sc-32 282); mouse anti-GCase (SantaCruz, sc-365 745 and sc-166 407 for immunoblotting); mouse anti-GBA (Abnova, Taipei, Taiwan, H00026229 for immunostaining); mouse anti-tubulin (Sigma, St. Louis, MO, T5168); mouse anti-LAMP2 (Developmental Studies Hybridoma Bank, Iowa City, IA, H4B4-s); rabbit anti-prosaposin (Proteintech, Rosemont, IL, 18 398-1-AP); rabbit anti-mouse prosaposin (ThermoFisher, Waltham, MA, PA581461); rabbit anti-LIMP-2 (a gift from Dr Michael Schake); rabbit anti-HA antibody (Y-11, Santa Cruz); goat anti-cathepsin B (R&D systems, AF953); rabbit cathepsin B (cell signaling, 31718S); goat anti-cathepsin D (Clone R20, SantaCruz, sc-6478); mouse anti-cathepsin L (Sigma, C4618); mouse anti-HEXB (clone D9, SantaCruz, sc-376 781); mouse anti-ubiquitin (clone P4D1, SantaCruz, sc-8017); mouse anti-progranulin (ThermoFisher, MA1-187); mouse anti-ATP6V1D (Santa Cruz, sc-166 218); mouse anti-GAPDH (Millipore, Burlington, MA, MAB374); rabbit β3-tubulin (Covance, Princeton, NJ, PRB-435P); CA-074 methyl ester (CA074-ME) (SantaCruz, sc-214 647), E64D (Enzo Life Sciences, Farmingdale, New York, BML-PI107-0001); carmofur (Cayman Chemicals, Ann Arbor, MI, Item 14 416), ASMase inhibitor (1-aminodecylidene bis-phosphonic acid, Cayman Chemicals, Item 13 583); LLOMe (hydrochloride) (Cayman Chemicals, Item 16 008); C18-ceramide (d18:1/18:0) (Avanti Polar Lipid, Birmingham, AL, 860518P); C16-ceramide (d18:1/16:0) (Avanti Polar Lipid, 860516P) and C20-ceramide (d18:1/20:0) (Avanti Polar Lipid, 860520P). Other chemicals were purchased from Sigma, unless otherwise stated.
Recombinant proteins
Saposin C, recombinant (Enzo, ALX-201-262-C050); recombinant prosaposin (Abnova, H00005660-P01); recombinant human cathepsin B (R&D Systems, Minneapolis, MN, 953-CY-010).
Plasmid, cell culture and transfection
The following plasmids were obtained from Addgene: pcDNA3.1-human cathepsin B and pcDNA3.1-human cathepsin L were gifts from Hyeryun Choe (Addgene plasmid #11249, #11250). The scramble shRNA control vector (Addgene plasmid #1864) was a gift from David Sabatini. The CRISPR/Cas9 construct targeting human PARK2 gene, CTSB gene or CTSD gene were constructed into pSpCas9(BB)-2A-Puro (PX459) V2.0 (a gift from Feng Zhang, Addgene plasmid, #62988). Target sequences of human PARK2-CRISPR/Cas-9 construct are 5′- GTG TCA GAA TCG ACC TCC AC-3′ for the Parkin KO cell and 5′-AGT GCC GTA TTT GAA GCC TC-3′ for the Parkin KO #2 cell. Target sequence of human CTSB-CRISPR/Cas-9 construct is 5′-ATG CTG TGG CAG CAT GTG TG-3′. Target sequence of human CTSD-CRISPR/Cas-9 constructs are 5’-TCT CCT TCT ACC TGA GCA GG-3′ for pX459-CTSD KO#1, 5’-CGT TGT TGA CGG AGA TGC GG-3′ for pX459-CTSD KO#2 and 5′-TGG GCG GTG TCA AAG TGG AG-3′ for pX459-CTSD KO#3. CTSL-RNAi #1 (TRCN0000318682) and CTSL-RNAi #2 (TRCN0000318611) were purchased from Millipore Sigma. CTSD-RNAi #1 (TRCN0000003660), CTSD-RNAi #2 (TRCN0000003661) and CTSD-RNAi #3 (TRCN0000003663) were obtained from Dharmacon, Inc. WT human embryonic kidney 293-FT (HEK293-FT) cells and KO cell lines were maintained in Dulbecco’s modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum, 10 U/ml penicillin and 10 μg/ml streptomycin at 37°C with 5% CO2 incubation. Cells were transfected with Lipofectamine® 2000 Transfection Reagent (Invitrogen, Waltham, MA, 11 668-019), according to the manufacture’s instruction.
Generation of KO cell lines
HEK293-FT cells grown in 12-well plates were transfected with the CRISPR/Cas9 construct targeting the human PARK2 gene or CTSB gene. One day after transfection, cells were trypsinized and re-seeded on a new 10 cm dish with low density (~50 cells in 10 cm dish), and cultured until single cells formed into cell colonies. Clonal colonies were picked from the 10 cm culture dish and expanded in new culture dishes. KO clones were identified by immunoblot analysis.
Generation of heterogeneous KO cell pools
HEK293-FT cells were transfected with either empty CRISPR/Cas9 vector (pX459) or pX459 vectors harboring gRNA targeting human CTSD gene. One day after transfection, transfected cells were selected with puromycin (2.5 μg/ml) for two days. Puromycin-resistant cells were harvested and re-plated onto 12-well plate with cell density of 100 000 cells per well, and cultured for additional three days without puromycin. Cells were then harvested and analyzed with immunoblot.
Live-cell GCase assay
Live-cell GCase assay was performed by using LysoLive™ Lysosomal β-Glucosidase Assay Kit (M2775) from MarkerGene. In brief, cells were seeded onto poly-D-lysine-coated 35 mm glass bottom dish with cell density of ~200 000 cell per dish. One day later, cells were washed with PBS, and incubated with LysoLive™-GlucGreen with 1:2500 dilution in 2 ml of OPTI-MEM medium (Invitrogen) for ~16 h. For imaging, cells were washed twice with PBS, prior to the addition of 2 ml of MakerGene-Opti-Klear Live Cell imaging buffer. For the experiment testing the effect of recombinant saposin C on live-cell GCase activity, 10 μg recombinant saposin C were included during the incubation with LysoLive™-GlucGreen for ~16 h. For the experiment testing the effect of carmofur on live-cell GCase activity, ~14 h before live-cell imaging, either final 7.5 μM of carmofur or equivalent volume of DMSO was added in the LysoLive™-GlucGreen labeling media. Images were taken by widefield epifluorescence microscope system equipped with bandpass filters with EX/EM: 490/520. The acquisition settings were kept the same for all imaging when fluorescence intensity was compared.
In vitro GCase activity assay
Cells were washed twice with PBS, harvested and lysed with sonication in ice-cold GCase lysis buffer (0.25% triton X100, 0.25% taurocholic acid, 1 mM EDTA in citrate/phosphate buffer, pH 5.4). Protein concentrations were measured by BCA method. GCase activity in 10 μg of cleared cell lysates was assayed in 50 μl of GCase assay buffer (1 mM 4-methylumbelliferyl-β-D-glucopyranoside (4-MUG, Sigma-Aldrich) 0.25% Triton X100, 1% BSA, 0.25% (w/v) Taurocholic acid, 1 mM EDTA, in citrate/phosphate buffer, pH 5.4). Enzyme reaction mixtures were incubated for 30 min at 37°C, and the enzyme reaction was stopped by the addition of equal volumes of 1 M glycine, pH 12.5. In endpoint mode, 4MU fluorescence (excitation wavelength = 355 nm, emission wavelength = 460 nm) was measured with SpectraMax i3 (Molecular Devices, San Jose, CA). The fluorescence value of control no-cell lysates were subtracted from 4MU fluorescence values.
Cell lysis and immunoblotting
Cells were seeded ~180 000 cells per well in 12-well plates. Two days later, cells were harvested for biochemical assays, unless otherwise stated. Cells were washed with PBS and lysed with 2× SDS sample buffer (100 mM Tris-Cl (pH 6.8), 4% (w/v) SDS, 0.05% (w/v) bromophenol blue, 20% (v/v) glycerol, 200 mM dithiothreitol). Protein samples from total cell lysates were subjected to SDS-PAGE. Proteins were then transferred to nitrocellulose membranes using Trans-Blot Turbo Transfer system (Bio-Rad, Hercules, CA). The membranes were incubated overnight with the indicated primary antibodies. All primary antibodies were diluted in an antibody dilution buffer (25 mM Tris, 0.15 M NaCl, 0.05% Tween-20, 5% BSA, 0.05% sodium azide). Primary antibodies were visualized using the appropriate horseradish peroxidase-conjugated secondary antibody (anti-mouse [Jackson Immuno Research Labs, Philadelphia, PA, #115-035-146], anti-rabbit [Jackson Immuno Research Labs, #111-035-144], anti-goat [Jackson Immuno Research Labs, #805-035-180] and SuperSignal West Femto Maximum Sensitivity Substrate [Thermo Scientific, #34096]). Chemiluminescence signals from blot were imaged on the ChemiDoc MP System (Bio-Rad) with a 16-bit CCD camera. Signal accumulation mode was used to acquire images at progressively longer exposure times. This allowed for acquisition of immunoblot images with band intensities within the linear range of the system. Quantification of protein levels was done using Bio-Rad ImageLab software and ImageJ (NIH) using non-saturated raw image files. Data were normalized to tubulin and expressed relative to control levels as indicated.
Immunostaining and imaging
WT HEK293-FT cells or Parkin KO cells were seeded onto poly-D-lysine-coated glass coverslips with a density of ~60 000 cells per coverslip in 12-well plates. Two days later, cells were washed once with PBS and fixed with 4% formaldehyde in PBS for 15 min at room temperature, and subsequently formaldehyde-fixed cells were fixed again with cold methanol for 5 min. After PBS washing, fixed cells were incubated in blocking solution (0.1% gelatin, 5% goat serum and 0.1% Triton X-100 in PBS) for 30 min. Cells were then incubated with indicated antibodies in GDB buffer (30 mM phosphate buffer, pH 7.4, containing 0.1% gelatin, 0.3% Triton X-100 and 0.45 M NaCl) overnight at 4°C. Primary antibodies were visualized using goat Alexa dye-conjugated secondary antibodies against rabbit or mouse. Immunostaining images were acquired with a Leica confocal microscope with a 63X oil objective. The confocal microscope settings were kept the same for all scans when fluorescence intensity was compared. Fluorescence intensity measurements were performed using ImageJ (NIH) using non-saturated raw image files. Data were expressed relative to WT levels as indicated. Pearson’s correlation coefficient of GCase and LIMP-2 was analyzed using Coloc2 plugin (Fiji).
Mass spectrometry-based lipid analysis
Cells were grown on 10 cm dishes at a cell density of ~ 2.5 million cells per dish (three 10 cm dishes for WT cells; six 10 cm dishes for Parkin KO cells). The next day, cells were treated with either DMSO or 7.5 μM carmofur for ~20 h. Cells were washed with cold PBS, and were then counted. Roughly three million cells from each 10 cm dish were pelleted with spin at 300xg for 5 min, and frozen at −80°C. At the University of South Carolina Medical Lipidomics Center, lipids were extracted from frozen cell pellets as a service provided by their core facilities. Levels of GluCer and ceramide were measured by a high-performance liquid chromatography/mass spectrometer (LC–MS/MS) method. Lipid analysis results were normalized with inorganic phosphate (Pi). For each group, three independent samples were analyzed. For the experiment testing the effect of ASMase inhibitor on sphingomyelin and ceramide species, cells were treated either with 5 μM ASMase inhibitor or equivalent volume of PBS for ~20 h. Lipid analysis results were normalized with Pi. For each group, three independent samples were analyzed.
In vitro prosaposin cleavage with recombinant human cathepsin B
800 ng of affinity purified recombinant GST-prosaposin (Abnova) was mixed with 150 ng activated recombinant human cathepsin B (R&D systems) in 100 μl assay buffer (25 mM MES, pH 5.0) for 2 h at 37°C. As controls, prosaposin only or cathepsin B only samples were assayed in parallel. The reaction was stopped by boiling in 2X SDS sample buffer. After the reaction, 100 μl of 4x SDS sample buffer was added into reaction mixtures to stop the reaction. Samples were then analyzed with immunoblot using saposin C-specific prosaposin antibody. For the cathepsin B activation, recombinant human cathepsin B was diluted to 10 μg/ml in activation buffer (25 mM MES, 5 mM DTT, pH 5.0), and subsequently incubated at room temperature for 15 min.
Ceramide effect on cathepsin B activity, based on auto-degradation of cathepsin B in vitro
Ceramides (C16-, C18-, C20-ceramide) were purchased from Avanti Polar Lipids, and were dissolved in DMSO with 0.5 mg/ml concentration. For the auto-cleavage assay of cathepsin B, 500 ng of each ceramide or same volumes of DMSO were added into separate tubes containing 100 μl cathepsin B assay buffer (25 mM MES, pH 5.0), and sonicated for 3 min in a sonic water bath. Auto-degradation reactions of cathepsin B were initiated by adding 25 μl of activated cathepsin B (3 μg/ml in ice-cold assay buffer) into ceramide dilutions in assay buffer. Reaction mixtures were incubated for 5 and 60 min at 37°C. After reactions, samples were boiled in 2x SDS sample buffer with DTT. Samples were analyzed with immunoblot using goat anti-cathepsin B antibody.
Co-immunoprecipitation assay
HEK293-FT cells in two 10 cm culture dishes were lysed with 2 ml of ice-cold Triton X-100 lysis buffer (1% Triton X-100, 150 mM NaCl, 20 mM Tris pH 7.4, 1 mM EDTA, 1 mM EGTA) containing cOmplete Protease inhibitor cocktail (Roche). After incubating the cells for 15 min in the lysis buffer on ice, the cell debris was pelleted at 14000 r.p.m. for 20 min. The cleared cell lysates were immunoprecipitated either with control rabbit antibody (rabbit HA antibody [Y-11, Santa Cruz] or rabbit prosaposin antibody [Proteintech] for 2 h at 4°C). 40 μL of protein G-coupled bead (Santa Cruz) was added and further incubated for 2 h at 4°C. Resulting immune complexes were washed four times with ice-cold Triton X-100 lysis buffer and boiled in 2X SDS sample buffer for immunoblot analysis.
Cathepsin B activity with cathepsin B substrate (MagicRed-(RR)2)
The Magic Red-RR2™ Cathepsin B Kit was obtained from Bio-Rad (cat #, ICT937). For the assay, cells were seeded onto poly-D-lysine coated glass coverslips at a density of 60 000/well (12-well plate). Two days later, cells were pre-treated with DMSO or 7.5 μM carmofur for ~2 h before the cathepsin B substrates labeling. And then cathepsin B substrate (MagicRed-RR2), which was reconstituted according to the manufacture’s protocol, was added to the cells at 1:100 dilution, and incubated for 60 min in a cell culture incubator with occasional mixing. Cells were then washed twice with PBS and fixed in 4% formaldehyde in PBS for 8 min, and stained with DAPI. Fluorescence images were taken with confocal microscope.
Biochemical analysis of mouse brain tissue
Whole brains, including brain stem, from three 12-week-old homozygous Parkin KO mouse (B6.129S4-Prkntm1Shn/J, The Jackson Laboratory Stock No: 006582) and four WT control mouse (C57BL/6 J, The Jackson Laboratory Stock No: 000664) were collected from The Jackson Laboratory. Frozen whole brains were thawed on ice and homogenized with Dounce homogenizer in ice-cold T-PER lysis buffer (Pierce) containing cOmplete Protease inhibitor cocktail (Roche). Protein concentration of brain tissue lysates was measured with the BCA method. 30ug of tissue lysates were analyzed with immunoblot.
Statistical analysis
All values in figures and text refer to mean ± SEM unless otherwise stated. N refers to the number of independent experiments unless otherwise indicated. Statistics and graphing were performed using Prism (GraphPad) software. Statistical analysis of data was performed with two-tailed unpaired t-test unless otherwise indicated.
Supplementary Material
Contributor Information
Myung Jong Kim, The Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Hyunkyung Jeong, The Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Dimitri Krainc, The Ken and Ruth Davee Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Data availability
All data and/or analyses generated during the current study are available from the corresponding author upon request.
Authors’ contributions
M.J.K. and D.K. conceived and designed the experiments. M.J.K. and H.K. performed the experiments. M.J.K analyzed the data. M.J.K. and D.K. wrote the manuscript.
Funding
This work was supported by National Institute of Neurological Disorders and Stroke (NINDS) R01NS076054 and R37NS96241 (D.K.). Lipidomics were supported in part by the Lipidomics Shared Resource, Hollings Cancer Center, Medical University of South Carolina (P30 CA138313 and P30 GM103339).
Conflict of Interest statement: D.K. is the Founder and Scientific Advisory Board Chair of Lysosomal Therapeutics Inc. and Vanqua Bio; serves on the scientific advisory boards of The Silverstein Foundation, Intellia Therapeutics and Prevail Therapeutics; and is a Venture Partner at OrbiMed. The authors declare no other competing financial interests.
References
- 1. Poewe, W., Seppi, K., Tanner, C.M., Halliday, G.M., Brundin, P., Volkmann, J., Schrag, A.E. and Lang, A.E. (2017) Parkinson disease. Nat Rev Dis Primers, 3, 17013. [DOI] [PubMed] [Google Scholar]
- 2. Kalia, L.V. and Lang, A.E. (2015) Parkinson's disease. Lancet, 386, 896–912. [DOI] [PubMed] [Google Scholar]
- 3. Abeliovich, A. and Gitler, A.D. (2016) Defects in trafficking bridge Parkinson's disease pathology and genetics. Nature, 539, 207–216. [DOI] [PubMed] [Google Scholar]
- 4. Schapira, A.H. (2015) Glucocerebrosidase and Parkinson disease: recent advances. Mol. Cell. Neurosci., 66, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sidransky, E., Nalls, M.A., Aasly, J.O., Aharon-Peretz, J., Annesi, G., Barbosa, E.R., Bar-Shira, A., Berg, D., Bras, J., Brice, A.et al. (2009) Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N. Engl. J. Med., 361, 1651–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burbulla, L.F., Song, P., Mazzulli, J.R., Zampese, E., Wong, Y.C., Jeon, S., Santos, D.P., Blanz, J., Obermaier, C.D., Strojny, C.et al. (2017) Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson's disease. Science, 357, 1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiasserini, D., Paciotti, S., Eusebi, P., Persichetti, E., Tasegian, A., Kurzawa-Akanbi, M., Chinnery, P.F., Morris, C.M., Calabresi, P., Parnetti, L.et al. (2015) Selective loss of glucocerebrosidase activity in sporadic Parkinson's disease and dementia with Lewy bodies. Mol. Neurodegener., 10, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gegg, M.E., Burke, D., Heales, S.J., Cooper, J.M., Hardy, J., Wood, N.W. and Schapira, A.H. (2012) Glucocerebrosidase deficiency in substantia nigra of Parkinson disease brains. Ann. Neurol., 72, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mazzulli, J.R., Xu, Y.H., Sun, Y., Knight, A.L., McLean, P.J., Caldwell, G.A., Sidransky, E., Grabowski, G.A. and Krainc, D. (2011) Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell, 146, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Murphy, K.E., Gysbers, A.M., Abbott, S.K., Tayebi, N., Kim, W.S., Sidransky, E., Cooper, A., Garner, B. and Halliday, G.M. (2014) Reduced glucocerebrosidase is associated with increased alpha-synuclein in sporadic Parkinson's disease. Brain, 137, 834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tamargo, R.J., Velayati, A., Goldin, E. and Sidransky, E. (2012) The role of saposin C in Gaucher disease. Mol. Genet. Metab., 106, 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schnabel, D., Schroder, M. and Sandhoff, K. (1991) Mutation in the sphingolipid activator protein 2 in a patient with a variant of Gaucher disease. FEBS Lett., 284, 57–59. [DOI] [PubMed] [Google Scholar]
- 13. Chang, D., Nalls, M.A., Hallgrimsdottir, I.B., Hunkapiller, J., van derBrug, M., Cai, F., International Parkinson's Disease Genomics, C, Me Research, T., Kerchner, G.A., Ayalon, G.et al. (2017) A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat. Genet., 49, 1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gan-Or, Z., Ozelius, L.J., Bar-Shira, A., Saunders-Pullman, R., Mirelman, A., Kornreich, R., Gana-Weisz, M., Raymond, D., Rozenkrantz, L., Deik, A.et al. (2013) The p.L302P mutation in the lysosomal enzyme gene SMPD1 is a risk factor for Parkinson disease. Neurology, 80, 1606–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kitatani, K., Idkowiak-Baldys, J. and Hannun, Y.A. (2008) The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell. Signal., 20, 1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kitatani, K., Sheldon, K., Rajagopalan, V., Anelli, V., Jenkins, R.W., Sun, Y., Grabowski, G.A., Obeid, L.M. and Hannun, Y.A. (2009) Involvement of acid beta-glucosidase 1 in the salvage pathway of ceramide formation. J. Biol. Chem., 284, 12972–12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blauwendraat, C., Reed, X., Krohn, L., Heilbron, K., Bandres-Ciga, S., Tan, M., Gibbs, J.R., Hernandez, D.G., Kumaran, R., Langston, R.et al. (2020) Genetic modifiers of risk and age at onset in GBA associated Parkinson's disease and Lewy body dementia. Brain, 143, 234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kitada, T., Asakawa, S., Hattori, N., Matsumine, H., Yamamura, Y., Minoshima, S., Yokochi, M., Mizuno, Y. and Shimizu, N. (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature, 392, 605–608. [DOI] [PubMed] [Google Scholar]
- 19. Pickrell, A.M. and Youle, R.J. (2015) The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron, 85, 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sliter, D.A., Martinez, J., Hao, L., Chen, X., Sun, N., Fischer, T.D., Burman, J.L., Li, Y., Zhang, Z., Narendra, D.P.et al. (2018) Parkin and PINK1 mitigate STING-induced inflammation. Nature, 561, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Fallon, L., Belanger, C.M., Corera, A.T., Kontogiannea, M., Regan-Klapisz, E., Moreau, F., Voortman, J., Haber, M., Rouleau, G., Thorarinsdottir, T.et al. (2006) A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat. Cell Biol., 8, 834–842. [DOI] [PubMed] [Google Scholar]
- 22. Song, P., Trajkovic, K., Tsunemi, T. and Krainc, D. (2016) Parkin modulates endosomal organization and function of the Endo-lysosomal pathway. J. Neurosci., 36, 2425–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Williams, E.T., Glauser, L., Tsika, E., Jiang, H., Islam, S. and Moore, D.J. (2018) Parkin mediates the ubiquitination of VPS35 and modulates retromer-dependent endosomal sorting. Hum. Mol. Genet., 27, 3189–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alcalay, R.N., Mallett, V., Vanderperre, B., Tavassoly, O., Dauvilliers, Y., Wu, R.Y.J., Ruskey, J.A., Leblond, C.S., Ambalavanan, A., Laurent, S.B.et al. (2019) SMPD1 mutations, activity, and alpha-synuclein accumulation in Parkinson's disease. Mov. Disord., 34, 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith, B.R., Santos, M.B., Marshall, M.S., Cantuti-Castelvetri, L., Lopez-Rosas, A., Li, G., vanBreemen, R., Claycomb, K.I., Gallea, J.I., Celej, M.S.et al. (2014) Neuronal inclusions of alpha-synuclein contribute to the pathogenesis of Krabbe disease. J. Pathol., 232, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burbulla, L.F., Jeon, S., Zheng, J., Song, P., Silverman, R.B. and Krainc, D. (2019) A modulator of wild-type glucocerebrosidase improves pathogenic phenotypes in dopaminergic neuronal models of Parkinson's disease. Sci. Transl. Med., 11. 10.1126/scitranslmed.aau6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harlan, F.K., Lusk, J.S., Mohr, B.M., Guzikowski, A.P., Batchelor, R.H., Jiang, Y. and Naleway, J.J. (2016) Fluorogenic substrates for visualizing acidic organelle enzyme activities. PLoS One, 11, e0156312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reczek, D., Schwake, M., Schroder, J., Hughes, H., Blanz, J., Jin, X., Brondyk, W., Van Patten, S., Edmunds, T. and Saftig, P. (2007) LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of beta-glucocerebrosidase. Cell, 131, 770–783. [DOI] [PubMed] [Google Scholar]
- 29. Hasegawa, J., Maejima, I., Iwamoto, R. and Yoshimori, T. (2015) Selective autophagy: lysophagy. Methods, 75, 128–132. [DOI] [PubMed] [Google Scholar]
- 30. Yoshida, Y., Yasuda, S., Fujita, T., Hamasaki, M., Murakami, A., Kawawaki, J., Iwai, K., Saeki, Y., Yoshimori, T., Matsuda, N.et al. (2017) Ubiquitination of exposed glycoproteins by SCF(FBXO27) directs damaged lysosomes for autophagy. Proc. Natl. Acad. Sci. U. S. A., 114, 8574–8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ho, M.W. and O'Brien, J.S. (1971) Gaucher's disease: deficiency of 'acid' -glucosidase and reconstitution of enzyme activity in vitro. Proc. Natl. Acad. Sci. U. S. A., 68, 2810–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weiler, S., Kishimoto, Y., O'Brien, J.S., Barranger, J.A. and Tomich, J.M. (1995) Identification of the binding and activating sites of the sphingolipid activator protein, saposin C, with glucocerebrosidase. Protein Sci., 4, 756–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hiraiwa, M., Martin, B.M., Kishimoto, Y., Conner, G.E., Tsuji, S. and O'Brien, J.S. (1997) Lysosomal proteolysis of prosaposin, the precursor of saposins (sphingolipid activator proteins): its mechanism and inhibition by ganglioside. Arch. Biochem. Biophys., 341, 17–24. [DOI] [PubMed] [Google Scholar]
- 34. Knight, C.G. and Barrett, A.J. (1976) Interaction of human cathepsin D with the inhibitor pepstatin. Biochem. J., 155, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McAdoo, M.H., Dannenberg, A.M., Jr., Hayes, C.J., James, S.P. and Sanner, J.H. (1973) Inhibition of cathepsin D-type proteinase of macrophages by pepstatin, a specific pepsin inhibitor, and other substances. Infect. Immun., 7, 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wilcox, D. and Mason, R.W. (1992) Inhibition of cysteine proteinases in lysosomes and whole cells. Biochem. J., 285(Pt 2), 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Okarmus, J., Bogetofte, H., Schmidt, S.I., Ryding, M., Garcia-Lopez, S., Ryan, B.J., Martinez-Serrano, A., Hyttel, P. and Meyer, M. (2020) Lysosomal perturbations in human dopaminergic neurons derived from induced pluripotent stem cells with PARK2 mutation. Sci. Rep., 10, 10278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murata, M., Miyashita, S., Yokoo, C., Tamai, M., Hanada, K., Hatayama, K., Towatari, T., Nikawa, T. and Katunuma, N. (1991) Novel epoxysuccinyl peptides. Selective inhibitors of cathepsin B, in vitro. FEBS Lett., 280, 307–310. [DOI] [PubMed] [Google Scholar]
- 39. Zheng, X., Chu, F., Mirkin, B.L., Sudha, T., Mousa, S.A. and Rebbaa, A. (2008) Role of the proteolytic hierarchy between cathepsin L, cathepsin D and caspase-3 in regulation of cellular susceptibility to apoptosis and autophagy. Biochim. Biophys. Acta, 1783, 2294–2300. [DOI] [PubMed] [Google Scholar]
- 40. Grosch, S., Schiffmann, S. and Geisslinger, G. (2012) Chain length-specific properties of ceramides. Prog. Lipid Res., 51, 50–62. [DOI] [PubMed] [Google Scholar]
- 41. Wegner, M.S., Schiffmann, S., Parnham, M.J., Geisslinger, G. and Grosch, S. (2016) The enigma of ceramide synthase regulation in mammalian cells. Prog. Lipid Res., 63, 93–119. [DOI] [PubMed] [Google Scholar]
- 42. Taniguchi, M., Ogiso, H., Takeuchi, T., Kitatani, K., Umehara, H. and Okazaki, T. (2015) Lysosomal ceramide generated by acid sphingomyelinase triggers cytosolic cathepsin B-mediated degradation of X-linked inhibitor of apoptosis protein in natural killer/T lymphoma cell apoptosis. Cell Death Dis., 6, e1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Realini, N., Solorzano, C., Pagliuca, C., Pizzirani, D., Armirotti, A., Luciani, R., Costi, M.P., Bandiera, T. and Piomelli, D. (2013) Discovery of highly potent acid ceramidase inhibitors with in vitro tumor chemosensitizing activity. Sci. Rep., 3, 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim, M.J., Jeon, S., Burbulla, L.F. and Krainc, D. (2018) Acid ceramidase inhibition ameliorates alpha-synuclein accumulation upon loss of GBA1 function. Hum. Mol. Genet., 27, 1972–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gorelik, A., Illes, K., Heinz, L.X., Superti-Furga, G. and Nagar, B. (2016) Crystal structure of mammalian acid sphingomyelinase. Nat. Commun., 7, 12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roth, A.G., Drescher, D., Yang, Y., Redmer, S., Uhlig, S. and Arenz, C. (2009) Potent and selective inhibition of acid sphingomyelinase by bisphosphonates. Angew Chem Int Ed Engl, 48, 7560–7563. [DOI] [PubMed] [Google Scholar]
- 47. Ysselstein, D., Young, T.J., Nguyen, M., Padmanabhan, S., Hirst, W.D., Dzamko, N. and Krainc, D. (2021) Evaluation of strategies for measuring lysosomal glucocerebrosidase activity. Mov. Disord., 36, 2719–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oji, Y., Hatano, T., Ueno, S.I., Funayama, M., Ishikawa, K.I., Okuzumi, A., Noda, S., Sato, S., Satake, W., Toda, T.et al. (2020) Variants in saposin D domain of prosaposin gene linked to Parkinson's disease. Brain, 143, 1190–1205. [DOI] [PubMed] [Google Scholar]
- 49. Abbott, S.K., Li, H., Munoz, S.S., Knoch, B., Batterham, M., Murphy, K.E., Halliday, G.M. and Garner, B. (2014) Altered ceramide acyl chain length and ceramide synthase gene expression in Parkinson's disease. Mov. Disord., 29, 518–526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and/or analyses generated during the current study are available from the corresponding author upon request.