ABSTRACT
Modifications in gene regulation are driving forces in the evolution of organisms. Part of these changes involve cis-regulatory elements (CREs), which contact their target genes through higher-order chromatin structures. However, how such architectures and variations in CREs contribute to transcriptional evolvability remains elusive. We use Hoxd genes as a paradigm for the emergence of regulatory innovations, as many relevant enhancers are located in a regulatory landscape highly conserved in amniotes. Here, we analysed their regulation in murine vibrissae and chicken feather primordia, two skin appendages expressing different Hoxd gene subsets, and compared the regulation of these genes in these appendages with that in the elongation of the posterior trunk. In the two former structures, distinct subsets of Hoxd genes are contacted by different lineage-specific enhancers, probably as a result of using an ancestral chromatin topology as an evolutionary playground, whereas the gene regulation that occurs in the mouse and chicken embryonic trunk partially relies on conserved CREs. A high proportion of these non-coding sequences active in the trunk have functionally diverged between species, suggesting that transcriptional robustness is maintained, despite considerable divergence in enhancer sequences.
KEY WORDS: Chromatin topology, TADs, Enhancers, Evolution, Gene regulation, Development, Teguments, Placodes
Summary: Analyses of the relationships between chromatin architecture and regulatory activities at the HoxD locus show that ancestral transcription patterns can be maintained while new regulations evolve.
INTRODUCTION
Changes in the spatial and temporal regulation of genes crucial for developmental processes have greatly contributed to the evolution of animal morphologies (reviewed by Carroll, 2008; Rebeiz et al., 2015). The expression of such genes is generally modulated by combinations of cis-regulatory elements (CREs), short DNA sequences enriched for transcription factor binding sites, which tend to evolve more rapidly than the genes they control (reviewed by Edwards et al., 2013; Long et al., 2016; Spitz and Furlong, 2012). CREs can be spread over large distances around the gene(s) they regulate, which highlights the importance of the spatial organization of chromatin. Indeed, the existence of such ‘regulatory landscapes’ (Spitz et al., 2003) implies that enhancer-promoter interactions involve spatial proximity, which is generally thought to occur through large chromatin loops (Rao et al., 2014).
The extent of regulatory landscapes often correspond to topologically associating domains (TADs) (Andrey et al., 2013; Harmston et al., 2017), which were defined as chromatin domains in which the probabilities of interactions, as measured by chromosome conformation capture (Belton et al., 2012), are higher than those of the neighbouring regions (Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012). The distribution of TADs tends to be conserved in various vertebrate species, a phenomenon likely associated with regulatory constraints exerted by the complex and pleiotropic regulations found around many vertebrate developmental genes. Such domains, in which many interactions might occur, were proposed to form chromatin niches, rich in various DNA-binding proteins and in which the evolutionary emergence of novel regulatory sequences could be favoured due to the presence of both a pre-existing scaffold and appropriate co-factors (Darbellay and Duboule, 2016). Although genomic rearrangements that altered the structure of TADs were reported to impact proper gene regulation during development, sometimes leading to genetic syndromes (Bolt et al., 2021; Bompadre and Andrey, 2019; Lupiáñez et al., 2015), these rearrangements also affected enhancer-promoter spatial relationships in most cases, which makes a causal assessment difficult.
Genome-wide analyses carried out in different species, comparing DNA sequence conservation, TAD organization or some epigenetic modifications, have started to reveal some of the molecular mechanisms underlying both the evolution of gene regulation and, by extension, the modification of species-specific traits. However, integrated functional approaches are required to better understand how changes in chromatin architecture and modifications in CREs may influence the evolvability and the robustness of gene transcription. To address such questions, we used the HoxD locus as a paradigm of pleiotropic regulation of a multi-gene family, with a strong impact both on the control of important developmental steps and on the emergence of evolutionary novelties. The HoxD cluster contains a series of genes in cis, which encode transcription factors that are expressed in different combinations across several embryonic structures. The cluster is flanked by two large TADs, each one matching a gene desert highly enriched in CREs (Amândio et al., 2020; Darbellay and Duboule, 2016). These two TADs are separated by a boundary region which is localized within the cluster, close to its centromeric extremity (Andrey et al., 2013; Rodríguez-Carballo et al., 2017). The telomeric TAD comprises a large number of enhancers controlling transcription of various Hoxd genes in most of the tissues where HOX proteins operate (Andrey et al., 2013; Darbellay et al., 2019; Delpretti et al., 2013; Guerreiro et al., 2016; Schep et al., 2016). In contrast, the centromeric TAD contains enhancers specific for the transcription of genes involved in the development of terminal structures such as digits or external genitals (Amândio et al., 2020; Montavon et al., 2011).
Amongst the many tissues in which Hoxd genes exert their functions are the embryonic primordia of different skin derivatives, including those of mammary glands, vibrissae and pelage hairs of mammals, as well as those of chicken feathers (Kanzler et al., 1997; 1994; Reid and Gaunt, 2002; Reynolds et al., 1995; Schep et al., 2016). Despite the fact that these ectodermal structures share common developmental mechanisms and gene regulatory networks such as the Wnt, EDA and BMP signalling pathways (Biggs and Mikkola, 2014; Dhouailly et al., 2019; Di-Poï and Milinkovitch, 2016; Pantalacci et al., 2008; Wu et al., 2004), each type of primordium expresses different subsets of contiguous Hoxd genes. Because the specific topological organization of the HoxD locus can be observed in fishes (Acemel et al., 2016; Woltering et al., 2014), it likely predates the emergence of hairs and feathers during amniote evolution (Dhouailly, 2009; Wu et al., 2004). For this reason, these organs are valuable model systems to understand how CRE evolution within the pre-existing chromatin architecture could lead to the implementation of new gene-specific expression patterns. In contrast, the overall general organization of Hoxd gene expression along the main embryonic body axis is maintained across vertebrate species, despite low sequence conservation, raising the question as to how evolving Hox clusters could maintain globally similar transcriptional outputs.
Here, we addressed these issues by taking a comparative look at a well-defined regulatory landscape, positioned telomeric to the HoxD gene cluster, in mammals and in birds. These two syntenic regions contain CREs involved either in the same regulatory tasks or in controlling regulatory aspects specific to each taxon. In the latter context, we characterized Hoxd gene regulation in the mouse-specific vibrissae primordia (VPs) and in the chicken-specific feather primordia (FPs). We show that different subsets of contiguous Hoxd genes are expressed in the mouse VP mesoderm and in the chicken FP ectoderm, indicating an independent co-option of Hoxd paralogues in these structures. Although the transcription of these genes in VPs and FPs relies on lineage-specific CREs located within different segments of the regulatory landscape, these CREs exploit a largely conserved three-dimensional (3D) chromatin architecture.
Our comparative analysis of the gene regulation during trunk extension also revealed that Hoxd1 displays an expression pattern markedly different from that of its neighbouring paralogues, both in mouse and chick, and that this Hoxd1-specific expression is driven by an evolutionarily conserved CRE located in a region predominantly interacting with Hoxd1 in both species. However, the cis-regulatory code involved in Hoxd1 gene regulation differs between the two species, suggesting that inter-species conservation of both gene and chromatin architectures is compatible with high plasticity of the cis-regulatory sequences involved. Extrapolating this observation across the genome suggests that the divergence of regulatory activities at conserved CRE sequences is more frequent than their maintainance.
RESULTS
Hoxd gene expression differs between mammalian and avian skin appendages
We initially analysed the expression profiles of Hoxd genes in mouse VPs and chicken FPs (Fig. 1). Skin appendage development starts with the interaction of the skin ectoderm with its underlying mesoderm, resulting in the thickening of the epithelium into a placode and in the condensation of dermal cells into a papilla (Mikkola, 2007; Sawyer and Knapp, 2003; Widelitz and Chuong, 1999). VP development starts at early embryonic day (E) 12, forming a stereotyped array of eight columns and five rows of VPs (Wrenn and Wessells, 1984) (Fig. 1A). The epithelial placode subsequently invaginates and engulfs the dermal papilla, forming the follicle primordium (Fig. S1). Likewise, in birds, ectoderm-mesoderm interactions result in the evagination of feather buds around E7 [stage Hamburger–Hamilton (HH) 30-32 (Hamburger and Hamilton, 1951; Michon et al., 2007)] (Fig. S2).
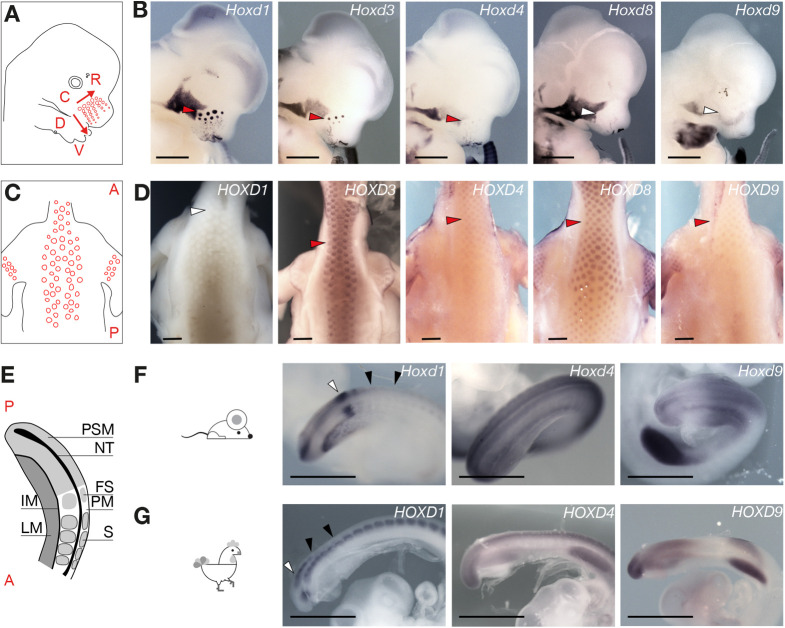
Fig. 1.
Transcription of Hoxd genes in mammalian and avian skin primordia. (A) Schematic of an E12.5 mouse embryo with vibrissae primordia (VPs) represented as red circles. The dorsoventral (D-V) and the caudorostral (C-R) axes are shown as red arrows, with the directions representing the timing of appearance of the VPs. (B) WISH on E12.5 faces. The arrowheads point to the most caudodorsal placode, which is the largest one and the first to appear. The red arrowheads indicate the detection of transcripts, white arrowheads indicate no detection of transcripts. Staining intensity progressively decreases from Hoxd1 to Hoxd8. (C) Schematic of the back of a HH35 chicken embryo with feather placodes indicated as red circles. A, anterior; P, posterior. (D) WISH on HH35 chickens showing the skin of the upper back. The arrowheads point to the neck at the level of the shoulders. HOXD1 is not expressed, whereas HOXD3 and HOXD8 signal intensities are stronger than for HOXD4 and HOXD9. The red arrowhead indicates the detection of transcripts, white arrowheads indicate no detection of transcripts. (E) Schematic representation of a E9.5 or HH20 posterior trunk and tailbud. A, anterior; P, posterior; FS, forming somites; IM, intermediate mesoderm; LM, lateral mesoderm; NT, neural tube; PM, paraxial mesoderm; PSM, presomitic mesoderm; S, somites. (F) WISH on E9.5 mouse posterior trunk and tail bud. Hoxd1 transcription pattern is different from that of Hoxd4 and Hoxd9. (G) WISH on HH20 chicken posterior trunk and tail bud. HOXD1 transcription pattern is different from HOXD4 and HOXD9. The white arrowheads in panels F and G point to the Hoxd1 stripe in the PSM. The black arrowheads show the difference between mouse (F) and chicken (G) in the persistence of Hoxd1 mRNAs in formed somites. Images are representative of at least three different embryos processed in at least two different WISH experiments. Scale bars: 1 mm.
Whole-mount in situ hybridization (WISH) revealed strong Hoxd1 expression in the dermal papillae of the VPs (Fig. 1B; Fig. S1). Furthermore, a comparative analysis of VPs from single E12.5 embryos revealed that Hoxd1 transcripts accumulate slightly after ectodermal placode formation, marked by the expression of Shh (Chiang et al., 1999) (Fig. S1). Hoxd3 and Hoxd4 are also expressed in VPs, although at much lower levels than Hoxd1 (Fig. 1B; Fig. S1). Faint levels of Hoxd8 mRNA were sporadically detected in the VPs, whereas Hoxd9 to Hoxd13 mRNAs were never observed in these structures (Fig. 1B; Fig. S1). In situ hybridization on cryostat sections confirmed that Hoxd1 transcripts accumulated in the dermal mesenchyme of VPs, but not in the ectoderm-derived region of the follicle (Fig. S1). Therefore, Hoxd1, Hoxd3 and Hoxd4 are transcriptionally active in the dermal papillae of developing VPs, with Hoxd1 being the main paralogue expressed in these structures.
WISH experiments in HH35 chicken embryos revealed expression of HOXD3, HOXD4, HOXD8 and HOXD9 in the FPs, whereas HOXD1 and HOXD11 mRNAs were not detected (Fig. 1C,D; Fig. S2). Cryostat sections indicated that HOXD gene expression in chicken FPs occurred in the follicle ectoderm (Fig. S2), in contrast with murine VPs. Different Hoxd genes thus operate in the dermal papillae of mouse VPs and in ectodermal placodes of chicken FPs, likely reflecting the implementation of different regulatory mechanisms in the two lineages.
Hoxd gene expression in the mouse and chicken embryonic body axis
Hox genes are expressed in the paraxial and lateral mesoderm of the main embryonic axis, as well as in the neural tube, and display progressively more restricted expression domains depending on their position within the cluster (Gaunt et al., 1988). Comparative WISH analysis of mouse and chicken embryos revealed that Hoxd1 expression patterns in the trunk were markedly similar between the two species, but were largely divergent from those of the 5′-located neighbouring Hoxd paralogues (Fig. 1E-G). Although Hoxd4 was strongly and uniformly transcribed in the mouse paraxial and lateral mesoderm as well as in the neural tube, Hoxd1 displayed a biphasic expression pattern. Hoxd1 transcripts accumulated in the embryonic tailbud; however, they rapidly disappeared from the presomitic mesoderm and remained undetected in lateral mesoderm progenitors or in the neural tube (Fig. 1F). Instead, Hoxd1 was specifically re-activated during somite condensation, possibly coupling the transcription of this gene to the segmentation clock (Dale and Pourquié, 2000; Zákány et al., 2001). Similar to the expression patterns in mice, chicken HOXD1 expression differed from its neighbouring paralogues, even though its transcripts remained detectable in the formed somites all along the main body axis (Fig. 1G). These observations suggested that Hoxd1 transcription in developing somites may depend on a specific set of CREs, which are different from the CREs controlling the neighbouring paralogues, and that this locus-specific transcription was evolutionarily conserved across amniotes.
An evolutionarily conserved topology at the HoxD locus
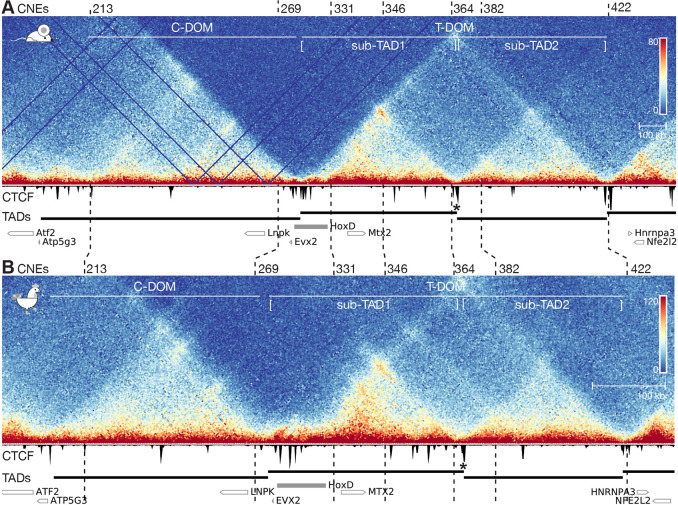
In order to see whether these differences and similarities in regulatory specificities could be associated to variations in the global chromatin architecture at the HoxD locus, we performed Capture Hi-C (CHi-C; high throughput chromosome conformation capture in which the library is enriched for regions of interest by RNA probe hybridization) experiments using dissected mouse and chicken posterior trunks (Fig. 2A,B). Even though the global size of TADs at this locus is approximately two times smaller in the chicken genome (i.e. 1.7 Mb in mouse versus 0.8 Mb in chicken), the chicken HOXD region closely resembled its mouse counterpart, with the telomeric domain (T-DOM) being further organized into two sub-TADs in both species (Fig. 2, sub-TAD1 and sub-TAD2) (Rodríguez-Carballo et al., 2020; Yakushiji-Kaminatsui et al., 2018). Of note, the presence and relative distribution of conserved non-coding elements (CNEs) were maintained within this similar chromatin architecture (Fig. 2; Fig. S2D).
Fig. 2.
Capture Hi-C-seq at the mouse and chicken HoxD loci. (A,B) Capture Hi-C-seq heatmaps using dissected posterior trunk cells. Below each heatmap, a CTCF ChIP-seq track is shown, produced from the same material. The similarities in the structural organization of the mouse and the chicken loci are underlined either by the positions of syntenic CNEs at key positions (numbered vertical dashed lines), the domains produced by TAD calling (black bars below) and the presence of a sub-TAD boundary (asterisk) within T-DOM. Genes are represented by empty rectangles, the filled grey rectangle indicates the HoxD cluster. In panel A, E9.5 mouse posterior trunk cells were used and the CHi-C heatmap was mapped on mm10 with a bin size of 5 kb (chr2:73,800,000-75,800,000). In panel B, HH20 chicken posterior trunk cells were used and the CHi-C heatmap was mapped on galGal6 with a bin size of 2.5 kb (chr7:15,790,000-16,700,000) and on an inverted x-axis. The positions of the two TADs (T-DOM and C-DOM) and of both sub-TADs are shown on top. The scales on the x-axes were adjusted to comparable sizes for ease of comparison, yet the chicken locus is more compacted (scale bars are shown on the right).
We corroborated these observations by assessing the interactions established by the Hoxd1, Hoxd4 and Hoxd9 promoters in mouse and chicken embryonic posterior trunk cells, using circularized chromosome conformation capture coupled with high-throughput sequencing (4C-seq) (Fig. S3). As an indicator for non-tissue-specific interactions, we used embryonic brain cells, in which Hoxd genes are not transcribed. In both species, the Hoxd1, Hoxd4 and Hoxd9 viewpoints established frequent contacts with the T-DOM, whereas interactions with the centromeric domain (C-DOM) were virtually absent. In addition, all viewpoints contacted T-DOM sub-TAD1 with a higher probability than T-DOM sub-TAD2, showing specific and non-overlapping regions where interactions were enriched. Hoxd1 contacted the first half of sub-TAD1 (D1-region; Fig. 3A; Fig. S3) more intensively than Hoxd4 or Hoxd9 (Fig. S3). In turn, Hoxd4 contacts within the 3′ half of sub-TAD1 (D4-region; Fig. 3A; Fig. S3) were increased to the sub-TAD boundary (Fig. S3). Finally, Hoxd9 interactions were mostly limited to the sub-TAD boundary region (D9-region; Fig. 3A; Fig. S3), where they were higher than for Hoxd1 and Hoxd4. This distribution of interacting regions within the 3′ TAD were found in both brain and posterior trunk cells, albeit with an overall lower contact frequency in brain cells, reflecting the transcription-independent default conformation of the region, in agreement with previous reports (Andrey et al., 2013; Dekker and Heard, 2015; Noordermeer et al., 2011).
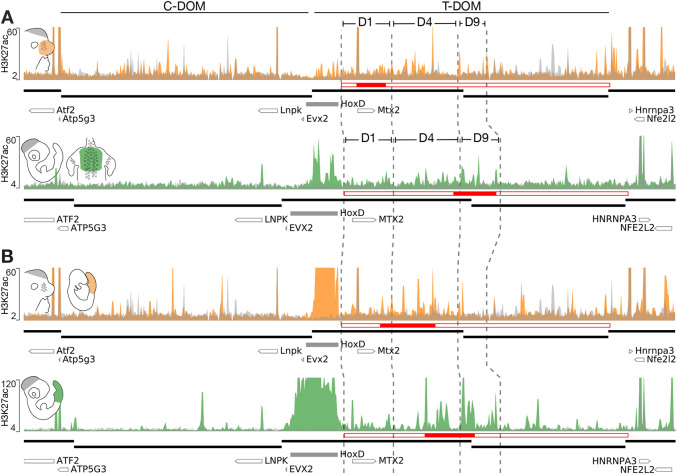
Fig. 3.
Tissue- and gene-specific interactions of dense H3K27ac regions. (A) The top panel shows a ChIP-seq profile of H3K27ac using mouse E12.5 VPs (orange) superimposed over the E12.5 forebrain cells (grey) (mm10, chr2:73,800,000-75,800,000). The lower panel shows a H3K27ac profile produced from dissected chicken HH35 dorsal skin (green), with HH18 brain cells (grey) (galGal6, chr7:15,790,000-16,700,000, inverted x-axis). The highest interacting regions are depicted as D1, D4 and D9 (see also Fig. S3B). The positions of conserved CNEs were used to delimit these regions in both species (vertical dashed lines) and the extents of TADs are shown below (thick black lines). The T-DOM (empty red rectangle) is split in overlapping genomic windows and the densest window (most acetylated region; MAR) is shown as a filled red rectangle. In the mouse VP, the MAR is within the D1 DNA segment, whereas the chicken MAR overlaps with the D4- and D9-regions. (B) The top panel shows a H3K27ac ChIP-seq using mouse E9.5 posterior trunk cells (orange) superimposed over forebrain cells (grey) (mm10, chr2:73,800,000-75,800,000). The lower panel shows the H3K27ac profile obtained from the same sample but dissected from a HH18 chicken embryo (green) with brain cells (grey) as a control (galGal6, chr7:15,790,000-16,700,000, inverted x-axis). Although the mouse posterior trunk MAR (red rectangle) coincides with the D4 segment, the chicken MAR counterpart also covers D4 and a small part of D9.
The regions of preferential interactions were annotated using a combination of CNE positions and 4C-seq coverage comparisons (Fig. S3C,D). The centromeric border of D1-region was set to CNE331, which is the CNE that is the most proximal from the HoxD cluster, but within the regulatory landscape. To define the boundary between the D1- and D4-regions and between the D4- and D9-regions, we used the coordinates for which the cumulative sum of the difference between the scores of two viewpoints was at its minimum, corresponding to the genomic location of the switch of preferential contacts from Hoxd1 to Hoxd4 and from Hoxd4 to Hoxd9, respectively. Finally, the telomeric border of the D9-region was set to CNE382, which is located just after a clear, tissue-specific peak of Hoxd9 contact frequencies.
Chromatin architectures partly rely upon the presence of the zinc-finger DNA-binding protein CTCF, which, together with the cohesin complex, can induce the formation and stabilization of large loops (Hansen et al., 2017; Nanni et al., 2020; Pugacheva et al., 2020). Chromatin immunoprecipitation sequencing (ChIP-seq) analysis of CTCF profiles revealed that both at the mouse and chicken HoxD loci, the telomeric end of the D1-region maps close to an occupied CTCF site (Fig. S3, blue arrowhead). Additionally, a cluster of CTCF-binding sites delimits the boundary between the mouse and the chicken sub-TAD1 and sub-TAD2 (Rodríguez-Carballo et al., 2017, 2020; Yakushiji-Kaminatsui et al., 2018), thus correlating with the limit between the D4- and D9-regions in both species (Fig. 2; Fig. S3, blue arrowheads in panels A,B).
Overall, these results show that Hoxd1, Hoxd4 and Hoxd9 preferentially interact with different regions of the T-DOM and that these segments are determined by the distribution of CTCF-binding sites in T-DOM subTAD-1. These distinct interaction patterns are constitutive and conserved between mouse and chicken. From these results, we concluded that the evolutionarily conserved contact topology of Hoxd genes with T-DOM may contribute to the implementation of their specific regulatory strategies in mouse and chicken embryonic structures.
Tissue-specific regulatory sequences are located in regions of preferential interactions
To evaluate whether the promoter-specific partition of contacts with T-DOM is associated with the presence of differential regulatory activities, we proceeded to identify the CREs controlling Hoxd gene expression in the VPs, FPs and the embryonic trunk of both mouse and chicken embryos. We carried out ChIP-seq analyses using an antibody against H3K27ac, a histone modification indicative of an active chromatin state (Creyghton et al., 2010; Rada-Iglesias et al., 2011), and annotated tissue-specific regions with increased H3K27ac enrichment as most acetylated regions (MARs, Fig. 3). MARs were identified as genomic regions accounting for the highest H3K27ac enrichment after subtraction of the corresponding signal in brain cells (see Materials and Methods).
In the mouse VPs, the MAR was mapped within the D1-region (Fig. 3A, top, filled red rectangle), in agreement with the stronger expression of Hoxd1 compared with its neighbouring paralogues (Fig. 1B; Fig. S1). However, in chicken dorsal skin samples in which HOXD1 is not active (Fig. 1D), the MAR largely overlapped the D4- and D9-regions (Fig. 3A, bottom, filled red rectangle). Therefore, VPs and FPs display different distributions of H3K27ac enrichment across T-DOM, in agreement with the various subsets of Hoxd genes expressed in these tissues. In contrast, the MARs of both mouse and chicken posterior trunk samples largely overlapped the D4-region (Fig. 3B, filled red rectangles) as expected from their similar expression patterns in the embryonic trunk of both species, with the expression domain of Hoxd4 being more extended than that of Hoxd9 (Fig. 1F). In summary, the different enrichments of H3K27ac marks across T-DOM were located within the genomic segments that preferentially interacted with the genes expressed in each tissue.
Hoxd1 regulation in mutant regulatory landscapes
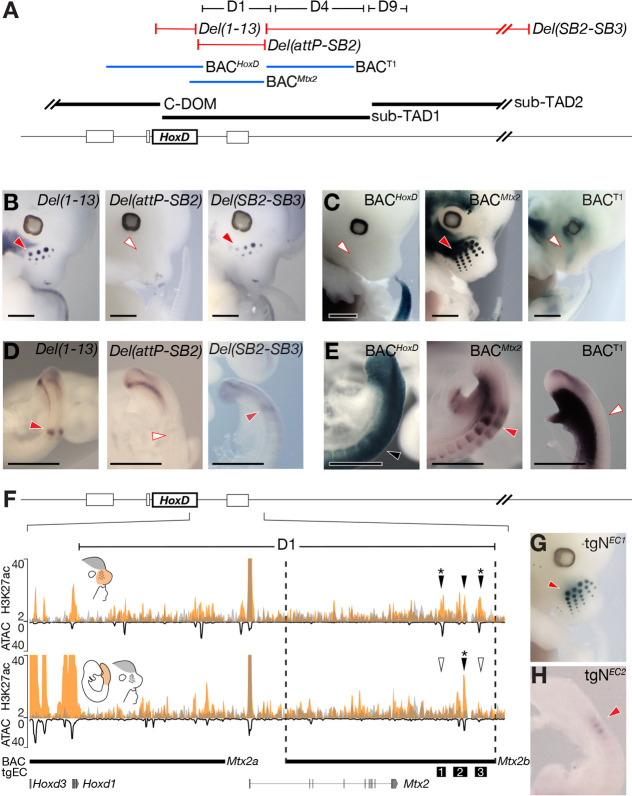
To evaluate the functional importance of the D1-region, we assessed Hoxd1 transcription in different mouse lines carrying mutations affecting this regulatory domain. We first analysed Hoxd1 expression in the VPs of mouse embryos carrying various deletions of T-DOM (Fig. 4A, red lines). In the HoxDDel(attP-TpSB2)lac allele, the D1-region was removed [hereafter referred to as Del(attP-SB2)]. As Del(attP-SB2) homozygous embryos are not viable due to the deletion of the essential Mtx2 gene (Andrey et al., 2013), we crossed this line with the mouse line HoxDDel(1-13)d9lac (Spitz et al., 2001) [hereafter referred to as Del(1-13)]. In the latter line, the entire HoxD cluster is deleted, in order to see the effect of the HoxDDel(attP-SB2)lac deletion on Hoxd gene regulation in cis. Hoxd1 expression in the VPs of trans-heterozygous HoxDDel(attP-TpSB2)lac/Del(1-13) mutant embryos was completely abolished, in contrast with Del(1-13) heterozygous control littermates (Fig. 4B). Conversely, the HoxDDel(TpSB2-TpSB3)lac allele [hereafter Del(SB2-SB3)] (Andrey et al., 2013) carries a large deletion that encompasses the whole T-DOM except the D1-region. We detected robust Hoxd1 expression in the VPs of E12.5 HoxDDel(TpSB2-TpSB3)lac/Del(1-13) trans-heterozygous mutant embryos (Fig. 4B).
Fig. 4.
Transcriptional regulation of Hoxd1. (A) The three deletion lines used are shown in red and the transgenic BACs in blue. The extent of the D1- to D9-regions are shown on top (black) as well as the positions of both sub-TADs below (thick black lines). (B) WISH using a Hoxd1 probe on E12.5 mouse embryos. Hoxd1 mRNAs were detected in both the control and the Del(SB2-SB3) lines (red arrowheads), but the signal was absent from Del(attP-SB2) mutant embryos (white arrowhead). (C) X-gal staining of E12.5 mouse embryos carrying randomly integrated BACs. A strong staining was detected in VPs when BACMtx2 was used (red arrowhead), whereas staining was not scored with the other two flanking BAC clones (white arrowheads). Therefore, a region directly downstream the HoxD cluster is necessary and sufficient to activate Hoxd1 transcription in VPs, referred to as D1-region as it coincides with this previously defined region. (D) WISH of Hoxd1 on E9.5 embryos focusing on tail buds. Hoxd1 was detected in forming somites in both the control and the Del(SB2-SB3) lines (red arrowheads), but was absent from Del(attP-SB2) mutant embryos (white arrowhead). (E) X-gal staining of E9.5 mouse embryos carrying various BAC transgenes. Staining was scored in the entire neural tube and paraxial mesoderm with the BACHoxD (black arrowhead). In contrast, embryos carrying BACMtx2 displayed staining in forming somites (red arrowhead), a staining that was absent when BACT1 was used (white arrowhead). (F) Enlargement of the D1-region along with a H3K27ac ChIP-seq using dissected E12.5 VPs (orange, top) and E9.5 posterior trunk cells (orange, bottom) superimposed over E12.5 forebrain (FB) cells (grey) (mm10; chr2:74,747,751-74,916,570). ATAC-seq using E12.5 VPs and E9.5 PTs are shown as a black line with an inverted y-axis to indicate levels of chromatin accessibility. Above the profiles are the three VP-acetylated elements (black arrowheads for H3K27ac-positive elements, asterisks for ATAC-positive elements) and below are MACS2 narrowPeaks for VPs and PTs (orange) and FB (grey). The position of the three EC sequences used as transgenes (tgEC) are shown as numbered black boxes. (G) X-gal staining of VPs from an E12.5 embryo transgenic (tgN) for EC1. (H) X-gal staining of forming somites in an E9.5 embryo transgenic for the EC2 sequence. Red arrowheads in F,G indicate the detection of transcripts in VPs. White arrowheads indicate no transcript detection in VPs. Images are representative of at least two different embryos processed in at least two different WISH experiments. Scale bars: 1 mm.
We complemented these results by producing mouse transgenic embryos carrying bacterial artificial chromosomes (BACs) spanning the HoxD cluster and/or parts of sub-TAD1 (blue lines, Fig. 4A). In agreement with the analysis of the T-DOM deletions, the BACs covering either the HoxD cluster (BACHoxD) (Schep et al., 2016) or mapping outside of the D1-region (BACT1) (Delpretti et al., 2013) did not display any lacZ reporter activity in E12.5 to E13.5 VPs (Fig. 4C). Instead, mouse embryos that were transgenic for BACMtx2 (Allais-Bonnet et al., 2021), which covers most of the D1-region, displayed strong reporter expression in the VPs (Fig. 4C). Notably, expression of Hoxd1 was also abolished from facial mesenchymal progenitors in Del(attP-SB2) mouse embryos, whereas it was reported by X-gal staining of BACMtx2 transgenic embryos, indicating that the D1-region regulates the transcription of Hoxd1 in several tissues.
To assess whether the D1-region is also necessary and sufficient for the specific Hoxd1 expression pattern in somites, we looked at Hoxd1 transcription in posterior trunk cells using the same set of mutant alleles. Hoxd1 expression was abolished in developing somites of Del(attP-SB2) E9.5 embryos compared with their control Del(1-13) littermates (Fig. 4A,D), whereas similar transcript levels were maintained in the tailbuds, suggesting that Hoxd1 regulation in both structures involves different regulatory sequences. Accordingly, embryos transgenic for BACMtx2 displayed strong lacZ expression in somites but not in the tailbud, with stronger activity in the last formed somites (Fig. 4A,E), thus closely mimicking native Hoxd1 transcription in these structures (Zákány et al., 2001). In contrast, lacZ expression was uniform along the paraxial mesoderm of BACHoxD transgenic embryos, without showing any Hoxd1-like specific reporter activity in the forming somites. Altogether, these results confirmed the regulatory potential of the mouse D1-region for Hoxd1 expression patterns in facial tissues and in forming somites.
Emergence of tissue-specific enhancers within conserved chromatin domains
Hoxd1 transcription in VPs could have appeared through the evolution of new CREs, in association with the emergence of these structures in mammals. An alternative possibility would be the re-deployment of a pre-existing element already at work in somites. The latter hypothesis comes from the fact that in both VPs and somites, transcription of Hoxd1 follows a cyclic dynamic, with a time course in phase with the production either of somites or of whisker pads. To gain insights into the evolution of Hoxd1 regulation, we isolated individual VP enhancers to see if they would be active in somites. We generated two smaller BAC transgenes through homologous recombination of a lacZ reporter construct, starting with BACMtx2. Although BACMtx2a displayed no signal in VPs, BACMtx2b reported stable β-galactosidase activity (Fig. S4A). Within the BACMtx2b region, three candidate enhancers (ECs) were selected based on H3K27ac enrichment, two of them also being positive by assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) (Fig. 4F, black stars on arrowheads), thus corresponding to open chromatin regions in VPs. Of note, EC2 was ATAC negative in VPs, whereas H3K27ac was ATAC positive in posterior trunk cells (Fig. 4F, black and white arrowheads). The regulatory potential of the enhancer candidates was assessed using transient lacZ reporter assays in transgenic mouse embryos, and we identified a strong and reproducible enhancer activity of transgenic EC1 (tgNEC1) in VPs, comparable with that observed in embryos transgenic for BACMtx2 (Fig. 4G). Instead, β-galactosidase activity was not detected either in tgNEC2 or in tgNEC3 transgenic embryos (Fig. S4B). We next tested these candidate enhancers at E9.5 to evaluate their regulatory potential in forming somites. EC2, which was the only candidate sequence with significant H3K27ac enrichment in posterior trunk cells, displayed reproducible activity in forming somites (Fig. 4H), unlike tgNEC1 or tgNEC3 (Fig. S4C). We concluded that the only VP enhancer sequence identified was not active during somite formation, unlike a somite enhancer located nearby (2.7 kb) within the D1-region.
The fact that two independent tissue-specific regulations rely on enhancers that are located in close proximity to a region of strong interactions suggests that a constrained topology may aid the emergence of novel regulatory sequences (Darbellay and Duboule, 2016). To investigate how enhancers might evolve within domains of preferential interactions and how their activities might relate to the observed transcriptional outputs, we compared our mouse and chicken H3K27ac and ATAC datasets with the levels of DNA sequence conservation. We analysed the number and distribution of putative enhancers (non-coding H3K27ac peaks overlapping with ATAC peaks) in the D1-, D4- and D9-regions of both mouse and chicken. CNEs were annotated by using BLASTZ pairwise alignments downloaded from the University of California, Santa Cruz (UCSC) Genome Browser and from which all elements overlapping with exons or promoters were discarded (Fig. S4D,E).
Overall, the number and distribution of putative enhancers in the different regions correlated with the transcriptional activity of Hoxd genes. A majority of putative enhancer sequences did not coincide with CNEs, suggesting a lineage-specific evolution of regulatory elements. Amongst those putative enhancers coinciding with CNEs, several displayed regulatory activities in one species only (Fig. S4, black arrows), indicating a divergence in the combinations of enhancers at work, even in posterior trunk cells, in which the transcriptional activity was expected to be similar in both species. This observation indicates that the sequence conservation of individual regulatory elements is not an obligatory requirement to maintain similar transcription patterns.
Divergent enhancer activities of conserved sequences
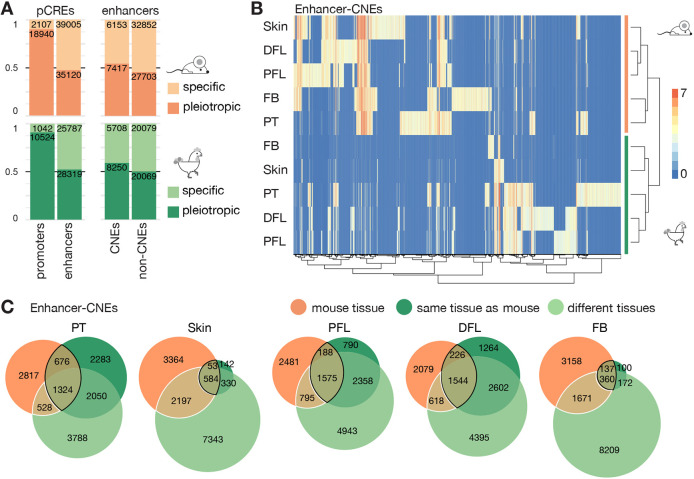
To take this analysis genome-wide, we compared the H3K27ac profiles across different mouse and chicken embryonic tissues for the presence of CREs, by adding available proximal and distal forelimb datasets (Beccari et al., 2016; Yakushiji-Kaminatsui et al., 2018) to our posterior trunk and skin H3K27ac ChIP-seq datasets. Putative CREs (pCREs) were separated into putative enhancers and promoters, hereafter referred to as ‘enhancers’ and ‘promoters’ for simplicity, with ‘enhancers’ mapping more than 2 kb away from a gene transcription start site and ‘promoters’ mapping less than 2 kb away. pCREs which were H3K27ac positive in more than one of the analysed tissues were categorized as ‘pleiotropic’, whereas those being active in only one tissue were classified as ‘specific’. In both mouse and chicken, most promoters were pleiotropic, whereas the majority of the enhancers were specific (Fig. 5A), a result in agreement with the usual pleiotropic expression of genes carrying important developmental functions. When enhancers were divided into ‘CNEs’ or ‘non-CNEs’, i.e. those that displayed sequence similarities between mouse and chicken and those that did not, respectively, the proportion of pleiotropic enhancers was higher amongst the former (Fig. 5A; P<10−15 in mouse and in chicken, Fisher test), suggesting an increased level of sequence conservation that may reflect the higher evolutionary pressure on enhancers endorsing multiple regulatory functions.
Fig. 5.
Conserved sequences with divergent functions. (A) Bar plots showing the proportion of putative cis-regulatory elements (pCREs) harbouring the H3K27ac mark in one (specific) versus more than one (pleiotropic) tissue, in either mouse (orange) or chicken (green). For simplification purposes, distal pCREs (>2 kb away from the transcription start site) are referred to as enhancers and proximal pCREs (<2 kb away from transcription start site) are referred to as promoters. The numbers of enhancers and promoters (left), and the numbers of conserved enhancers and non-conserved enhancers (right) are indicated. The proportion of specific versus pleiotropic elements is shown for CNE and non-CNE enhancers (right). In both mouse and chicken, enhancers are more specific than promoters and amongst enhancers, conserved elements are found to be acetylated in more tissues than non-conserved elements. (B) Hierarchical clustering obtained using the pheatmap R package. The x-axis indicates enhancer-CNEs, i.e. conserved sequences that are non-coding in mouse and in chicken and overlap with a H3K27ac mark. The y-axis shows the tissues used to obtain MACS2-processed peaks. The values on the heatmap correspond to the enrichment scores of the peaks. Enhancer-CNEs cluster by species and not by tissues. (C) Euler diagrams representing the numbers of enhancer-CNEs in the indicated tissues that are acetylated in mouse (orange), in the corresponding chicken tissue (dark green) or in unrelated chicken tissue (light green), showing that conserved sequences diverge in their regulatory activities. The intersection between the same tissue in mouse and in chicken is outlined in black, and the intersection between a chicken tissue different from the mouse tissue is outlined in white. DFL, distal forelimb; FB, forebrain; PFL, proximal forelimb; PT, posterior trunk.
Next, we used CNEs to compare the regulatory activities of murine and chicken orthologous sequences. In agreement with what was observed at the HoxD locus, the enhancer activities of CNEs, as inferred by H3K27ac peaks, more often diverged than they matched between the related mouse and chicken tissues (Fig. 5B,C). Hierarchical cluster analysis revealed that the enhancer profiles of CNEs were more similar between different tissues of the same species, than between the same tissues in mouse and chicken (Fig. 5B); a surprising observation as these CNEs were initially selected owing to their sequence conservation between the two species. We hypothesised that some sequences could have been conserved over time despite divergence in their regulatory activities because they would endorse other functions in other tissues. To evaluate this possibility, we used Euler diagrams to represent the relationships between mouse and chicken CNEs displaying enhancer features (Fig. 5C). Each mouse tissue was used as a reference in independent diagrams, in which chicken enhancer CNEs were represented as different sets, depending on whether they were active in the same or in different tissues compared with the mouse reference.
The intersection between the same tissue in mouse and in chicken represents elements that have both a conserved sequence and regulatory function (Fig. 5C, overlaps outlined in black). This intersection is smaller than any difference between two sets, which indicates that DNA sequence similarity between two species can be maintained amongst regulatory sequences despite divergence in their regulatory functions. We observed that CNEs often displayed enhancer activities in a chicken tissue that was different from the mouse tissue, supporting this idea (Fig. 5C, overlaps circled in white). Consequently, the regulatory activity of a given sequence conserved between these two species cannot be used to predict its function(s) in the two species.
To control for the technical specificity of our H3K27ac-based approach, we showed that chicken and mouse H3K27ac peaks reproducibly overlap ATAC-seq peaks. We first quantified the proportion of H3K27ac peaks that were also ATAC positive and found that most of them were indeed in an open chromatin state (Fig. S5A). We then repeated the enhancer-CNE quantifications to obtain ‘open enhancer CNEs’ by intersecting CNEs with open pCREs (Fig. S5). Again, open enhancer CNEs clustered by species (Fig. S5B), and the intersections on Euler diagrams were smaller than any difference between two sets (Fig. S5C). Therefore, the open pCRE quantifications recapitulated the results obtained using pCREs, thus supporting the validity of our H3K27ac approach.
Altogether, we show that both the global genomic organisation of the HoxD locus and specific domains of preferential enhancer-promoter interactions are highly conserved between mouse and chicken. However, the regulatory activities of individual CREs are largely species-specific and such sequences are systematically found within those segments of regulatory landscapes that preferentially interact with the promoters of the most highly transcribed genes. We also show that the lineage-specific evolution of regulatory activities could either generate new regulatory relationships or, conversely, maintain ancestral transcription patterns.
DISCUSSION
Versatile functions of Hoxd genes in skin appendages
Hox genes play a dual role in skin appendage development. Although their colinear expression is initially necessary to pattern the skin dermis (Kanzler et al., 1997), they are subsequently transcribed in the dermal papillae and/or ectoderm of embryonic and adult hair pelage follicles and vibrissae precursors (Godwin and Capecchi, 1998; Packer et al., 2000; Reynolds et al., 1995; Yu et al., 2018), as well as in the avian buds (Chuong et al., 1990; Kanzler et al., 1997). Here, we show that the dermal papillae of embryonic mouse vibrissae express Hoxd1, Hoxd3 and Hoxd4, with the mRNA of Hoxd1 being the first to be detected, followed by considerably lower levels of Hoxd3 and Hoxd4 mRNAs. The transcription in VPs is driven by CREs located in a region encompassing approximately 120 kb of DNA adjacent to the telomeric end of the mouse HoxD cluster, a region that is preferentially contacted by the Hoxd1 gene. Therefore, a proximity effect is observed in the building of the chromatin architecture, based on constitutive contacts between Hoxd1 and the neighbouring sub-TAD, whereby most of the Hoxd1 regulatory sequences are located in the adjacent region of the regulatory landscape.
Although a detailed functional characterization of Hoxd genes during the development of whiskers was not the aim of this work, we did not observe any major morphological alteration in the vibrissae of mice carrying genetic deletions of this precise region, despite the complete abrogation in the expression of Hoxd1, Hoxd3 and Hoxd4 in these structures. This lack of a visible abnormal phenotype may be due to functional redundancy with other Hox paralogy groups expressed there, such as Hoxc8 (Yu et al., 2018), a lack of analytical power or a lack of the appropriate behavioural paradigms that may have revealed functionally abnormal whiskers. The diverse expression of Hox paralogues in mammalian and avian skin appendages nonetheless illustrates the plasticity in the usage of Hox gene functions in these structures. This plasticity likely relies upon the independent acquisition of specific CREs for each gene cluster, as these skin derivatives emerged after the genome duplication events thought to be at the origin of the vertebrate lineage (Holland et al., 2008; Holland and Garcia-Fernàndez, 1996).
Although Hoxd9, Hoxd11 and Hoxd13 are expressed in the ectoderm-derived hair fibres (Godwin and Capecchi, 1998; Packer et al., 2000; Reynolds et al., 1995; Yu et al., 2018), we did not observe any Hoxd1 in the vibrissae shaft, nor any lacZ expression when the BACMtx2 (carrying the D1-region) transgenic animals were used, indicating that these expression patterns may depend on different sets of CREs. Our data also show that the expression of HOXD3, HOXD4, HOXD8 and HOXD9 in the ectoderm of the chicken FPs depends on CREs unrelated to those operating in the mouse VPs. These observations may indicate that different sets of Hoxd genes were independently co-opted during the evolution of skin derivatives, likely through de novo enhancer acquisition in the mammalian and avian lineages, in a way related to the regulation of Hoxd9 and Hoxc13 in the embryonic mammary buds and in the nails and hairs of mammals, respectively (Fernandez-Guerrero et al., 2020; Schep et al., 2016). Alternatively, the rapid pace of tegument evolution in tetrapods may have been accompanied by a divergence in enhancer sequences, which makes comparisons difficult. In this context, we cannot rule out the possibility that Hoxd gene regulation in skin appendages of both mammals and birds derives from an initial pan-Hoxd expression pattern both in the ectoderm and mesoderm of skin appendages, which then evolved differently in each lineage through modification of the CRE complement located within the adjacent TAD. The fact that the two CNEs acetylated in mouse VPs and chick FPs are also acetylated in mouse and chick posterior trunk cells may indicate that the former structures co-opted parts of the regulatory mechanisms at work in the latter. This indicates that DNA sequence conservation may be due to constraints imposed by the function in the trunk rather than by the function in the skin.
Enhancer acquisition in evolving tetrapod skin appendages
TADs containing pleiotropic genes of key importance for vertebrate development are frequently conserved across tetrapods, and topological changes in their organization often correlate with important modifications in gene expression (Eres et al., 2019; Laverré et al., 2022; Torosin et al., 2020; Yakushiji-Kaminatsui et al., 2018). Our results show that despite a considerable divergence in the non-coding elements localized in the mouse and chicken T-DOM, the global pattern of interactions is conserved, leading to similar chromatin topologies. Indeed, in both cases, different Hoxd genes interact preferentially with distinct regions of sub-TAD1. These contacts are for the most part constitutive, even though we observed an activity-dependent increase in interactions in the mouse VPs and in mouse and chicken posterior trunk cells. Of interest, the regions within T-DOM (D1 to D9), which are preferentially contacted by the Hoxd1, Hoxd4 or Hoxd9 genes, are organized in a linear sequence opposite to the order of the genes themselves, as if Hoxd1 was folding in towards its closest possible chromatin segment within the T-DOM, bringing along Hoxd4 towards the next chromatin segment and positioning the interactions involving Hoxd9 further away into the T-DOM. This sequential positioning of interactions in the reverse order of spatial colinearity may impose a generic contact pattern, whereby Hoxd9, for example, is unable to establish strong interactions with the D1-region and its enhancers, as this region is primarily contacting Hoxd1, thus imposing constraints upon the distributions of interactions (see below).
Furthermore, the distribution of H3K27ac-positive regions in the chicken FPs and mouse VPs across these interacting regions correlates well with the specific subsets of Hoxd genes expressed in each of these tissues. Finally, the regulation of Hoxd1 in developing somites relies on evolutionarily conserved elements located in the D1-region. This general regulatory topology is close to that observed in teleost fishes (Acemel et al., 2016; Woltering et al., 2014), suggesting that it was already established at the root of the vertebrate lineage and that its hijacking by nascent enhancer sequences may have favoured the co-option of specific Hoxd gene subsets as targets of the novel regulations. The regulation of Hoxd genes in mouse VPs and chicken FPs, as well in mouse mammary buds (Schep et al., 2016), may illustrate this process.
Plasticity of the cis-regulatory code versus conservation in TAD organization
Owing to their functional relevance in the regulation of gene expression, CREs tend to be evolutionarily conserved across species (Sandelin et al., 2004; Sanges et al., 2013; 2006). However, several studies have shown that enhancers controlling developmental gene expression can be functionally conserved even across distant phylogenetic lineages, despite a strong divergence in their DNA sequences (Ambrosino et al., 2019; Berthelot et al., 2018; Wong et al., 2020; Eichenlaub and Ettwiller, 2011; Schmidt et al., 2010; Villar et al., 2015). Our inter-species comparison of CNEs enriched in H3K27ac across different embryonic structures revealed that a considerable portion of evolutionarily conserved CREs diverged in their patterns of activity between the two species analysed, in agreement with previous observations (Dermitzakis and Clark, 2002; Schmidt et al., 2010; Vierstra et al., 2014; Villar et al., 2015). This dichotomy between the conservation of enhancer sequences and their function is illustrated by the CREs controlling Hoxd1 expression in mouse and chicken developing somites. Although some of the sequences active in the mouse posterior trunk are evolutionarily and functionally conserved in chicken, such as EC2, which can drive lacZ expression in the last formed somites of the murine embryo, other elements conserved at the nucleotide level displayed H3K27ac enrichment only in the mouse.
In fact, a large fraction of H3K27ac-positive sequences detected either in mouse or in chicken posterior trunk cells were not conserved between the two species, suggesting that they evolved in a species-specific manner. In contrast, the high proportion of chicken promoters that display similar H3K27ac enrichments in mice, together with the similar expression patterns observed for mouse and chicken Hoxd genes in embryonic trunk cells, demonstrates that conserved gene expression can occur despite a high evolutionary plasticity in the cis-regulatory code usage, in agreement with previous reports (Berthelot et al., 2018; Snetkova et al., 2021). This may point to a regulatory strategy in which multiple enhancers coordinate to regulate the same target gene(s), thus creating a quantitative effect in a way similar to the reported action of super-enhancers (Sabari et al., 2018), yet in which various subsets of CREs may display distinct tissue-specificities. This process may be implemented preferentially within large regulatory landscapes, e.g. in the case of Fgf8 (Marinic et al., 2013) or in the TAD controlling Hoxd gene transcription in external genitals (Amândio et al., 2020).
A regulatory playground with constraints
In those cases that were investigated, the respective positions of regulatory sequences within large syntenic regulatory landscapes are generally conserved, and the global regulatory architecture found at one particular developmental locus in one species can be extrapolated to its cognate locus in another amniote species (Kragesteen et al., 2018), implying that enhancer-promoter interactions are also evolutionarily well conserved, even for CREs acting over long distances (Irimia et al., 2012; Laverré et al., 2022), although some mechanistic elements may vary (Ushiki et al., 2021). In agreement with this, and contrasting with the apparent divergence in the cis-regulatory code controlling Hoxd gene expression in mouse and chicken embryos, our phylogenetic footprinting and epigenetic analysis revealed that non-coding elements conserved in the DNA sequence between the mouse and chicken genomes were distributed across the two species in a very similar manner. Therefore, the potential gain and/or loss of particular CREs in the two lineages did not significantly impact the internal organization of those conserved regulatory elements, nor did they impact the global chromatin topology of the locus. This confirms the observation that even the significant divergence in the CREs located at the HoxD locus, which accompanied the evolution of the snake body plan, did not result in any substantial alteration of the overall corn-snake Hoxd gene interaction map (Guerreiro et al., 2016). Altogether, these observations suggest that the internal organization of the TADs at the HoxD locus and the specific interaction profiles for various subsets of Hoxd target genes are resilient to considerable variation in their CREs.
These results also highlight the somewhat dual properties that large regulatory landscapes may have imposed in the course of regulatory evolution. On the one hand, a pre-established chromatin topology, as materialized by TADs, may have provided unique ‘structural niches’ to evolve new enhancer sequences, owing to the proximity of factors already at work. This might especially apply to landscapes in which a strong quantitative parameter may be instrumental, and a fortiori when the target gene(s) are acting in combination, coding for proteins of rather low specificities (Bolt and Duboule, 2020). This may be illustrated nowadays by large landscapes, in which all kinds of enhancer sequences are mixed within the same TAD. On the other hand, although this regulatory playground may stimulate the emergence of regulations, it will constrain the realms of action of the enhancer sequences, due to both the very architecture that favours their evolution and the accessibility to a single subset of contiguous target genes, as a result of the global distribution of interactions at this precise place in the landscape.
MATERIALS AND METHODS
Mouse strains and chicken eggs
All mutant mouse strains are listed in Table S1 and were backcrossed continuously to Bl6/CBA mixed animals and maintained as heterozygous stocks. Chick embryos from a White Leghorn strain were incubated at 37.5°C and staged according to Hamburger and Hamilton (1951).
Enhancer cloning and transgenic animals
The different CRE candidate sequences were amplified using PCR-specific primers (see Table S2) and the Expand High Fidelity PCR system (Roche). Amplified fragments were gel purified and cloned into the pSK-lacZ vector (GenBank X52326.1; Beccari et al., 2016). For the transgenesis assays, each lacZ reporter construct was digested with NotI and KpnI, and the restriction fragment encoding the potential enhancer sequence, the β-globin minimal promoter and the lacZ reporter gene were gel purified and injected into the male pronucleus of fertilized oocytes. F0 embryos were dissected either at E10 or at E12, fixed and stained for β-galactosidase activity according to standard protocols.
In situ hybridization
The probes used in this study for in situ hybridization were either previously reported or produced by PCR amplification of a fragment subsequently ligated in pGEMT Easy vector (Promega). Probe sequences and references are listed in Table S3. Digoxigenin (Dig)-labelled probes were synthesized in vitro by linearizing the respective plasmids using specific restriction enzymes and transcribed in vitro with T3 (Promega, P2083), T7 (Promega, P2078) or Sp6 (Promega, P1085) RNA polymerase (Table S3) and the Dig-RNA labelling mix (Roche). The probes were purified using the RNeasy Mini Kit (QIAGEN, NC9677589). WISH experiments were performed as described in Woltering et al. (2009). In situ experiments in sections were performed according to the protocol of Sockanathan (2015; https://protocolexchange.researchsquare.com/article/nprot-3781/v1). Cryostat sections were prepared after fixing embryos in 4% paraformaldehyde solution at 4°C overnight. Subsequently, embryos were washed three times with PBS and passed to increasing sucrose solutions at 4°C in agitation (15% sucrose solution for 3-4 h, followed by incubation in 30% sucrose solution overnight) and finally included in OCT blocks (TissueTek O.C.T Compound 4583). Sections of 20 µm were obtained with a Leica CM 1850 cryostat and stored at −80°C.
Capture Hi-C-seq experiments
The SureSelectXT RNA probe design and capture Hi-C experiments were performed as described by Bolt et al. (2021) for mouse and as by Yakushiji-Kaminatsui et al. (2018) for chicken. Dissected tissues were processed as in Yakushiji-Kaminatsui et al. (2018) with the following change: cells were crosslinked in 2% formaldehyde in PBS and washed three times instead of being quenched by glycine. Hi-C-libraries were prepared as in Yakushiji-Kaminatsui et al. (2018). The first part of the data analysis was performed on our local Galaxy server (Afgan et al., 2016). Raw reads were preprocessed with CutAdapt (v1.16) (-a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC -A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATCATT --minimum-length=15 --pair-filter=any --quality-cutoff=30) (Martin, 2011). Then, Hicup (v0.6.1) (Wingett et al., 2015) and Samtools 1.2 (Danecek et al., 2021) were used with default parameters. The pairs were then loaded to 5 kb (mouse) and 2.5 kb (chicken) resolution matrices with Cooler v0.7.4 (Abdennur and Mirny, 2020). These matrices were balanced with HiCExplorer hicCorrectMatrix (v3.7.2) using ICE as the correction method, the mad threshold from HiCExplorer hicCorrectMatrix diagnostic plot as the minimum threshold value and five as the maximum threshold value. The TAD separation scores were computed with HiCExplorer hicFindTADs (Ramírez et al., 2018; Wolff et al., 2020) with a fixed window of 240 kb in mouse and 120 kb in chicken. See CHiC_processRaws.sh https://gitlab.unige.ch/Aurelie.Hintermann/hintermannetal2022 for details.
ATAC-seq
Embryos were collected and placed into PBS with 10% fetal calf serum and 15 μl collagenase at 50 mg/ml (Sigma-Aldrich, C9697) at 37°C for approximately 10 to 15 min. Two replicates were done using one embryo each and 50,000 cells were isolated for processing with the Nextera Tn5 enzyme (Illumina, FC-131-1096) as previously described (Buenrostro et al., 2013). Tn5-treated DNA was amplified with Nextera Library primers using the NEBNext library amplification master mix (New England BioLabs, M0541) and sequenced on an Illumina NextSeq 500. Sequenced DNA fragments were processed as previously reported (Amândio et al., 2021) with the following minor modification on a local Galaxy server (Afgan et al., 2016): the BAM file was converted to BED prior to peak calling with bedtools version 2.30.0 (Quinlan and Hall, 2010). Peak calling was done using MACS2 (v2.1.1.20160309) callpeak (--no-model --shift −100 --extsize 200 --call-summits --keep-dup all). Peak regions for each condition were obtained from the union of the peak region of both replicates using bedtools merge (v2.30.0) and extended by 660 bp each side using bedtools slop. Each replicate was normalized per million reads mapped in their own peaks, and the displayed coverage is the mean of normalized replicates.
ChIP-seq
For genomics analyses, we used the mouse assembly mm10 and the chicken galGal6 and plotted genomic data using pyGenomeTracks 3.6 (Lopez-Delisle et al., 2021). ChIP-seq experiments were performed using the ChIP-IT High Sensitivity Kit according to the manufacturer's instructions with minor modifications (Active Motif). For the mouse samples, 12 pairs of E12.5 whisker pads (WPs) or 100 E8.75 posterior trunks were used, the latter dissected at the level of the second to fourth pair of somites. For the chicken samples, the dorsal skin of five HH35 embryos, the posterior trunk or 80 HH19-21 posterior trunk regions were isolated. In all cases, the samples were fixed for 10 min in 1% formaldehyde at room temperature and the crosslinking reaction was quenched with glycine. Subsequently, nuclei were extracted and sonicated in 50 mM Tris-HCl (pH 8.0), 10 mM EDTA and 1% SDS to an average fragment size of 200-500 bp using a Bioruptor sonicator (Diagenode). Approximately 10-20 µg of sonicated chromatin was diluted tenfold with the dilution buffer [20 mM HEPES (pH 7.3), 1 mM EDTA, 150 mM NaCl, 0.1% NP-40] and incubated with 2 µg of anti-H3K27ac antibody (Abcam, ab4729). All buffers contained 1× Complete Proteinase Inhibitor cocktail (EDTA-free; Roche) and 10 mM sodium butyrate to prevent protein degradation and histone deacetylation. Chromatin–antibody complexes were immunoprecipitated with protein G-coupled agarose beads (Active Motif), purified and de-crosslinked according to the manufacturer instructions. Approximately 5-10 ng of purified DNA was used for ChIP library preparation by the Geneva IGE3 Genomics Platform (University of Geneva) and sequenced to obtain 100 bp single-end reads on an Illumina HiSeq2500 or HiSeq4000 system. Published fastq files were downloaded from the Sequence Read Archive (SRA) for H3K27ac: mouse distal forelimb (DFL; GSM2713703, SRR5855214), mouse proximal forelimb (PFL; GSM2713704, SRR5855215), chicken DFL (GSM3182462, SRR7288104), chicken PFL (GSM3182459, SRR7288101); and for CTCF in mouse posterior trunk (PT; GSM5501395, SRR15338248). ChIP-seq reads processing was done as in Amândio et al. (2020) with some modifications, using the laboratory Galaxy server (Afgan et al., 2016). Adapters and bad-quality bases were removed with Cutadapt (v1.16.8) (Martin, 2011) (options -m 15 -q 30 -a GATCGGAAGAGCACACGTCTGAACTCCAGTCAC). Reads were mapped to the mouse genome (mm10) and to the chicken genome (galGla6) using Bowtie2 (v2.4.2) (Langmead and Salzberg, 2012) with standard settings. Only alignments with a mapping quality >30 were kept (Samtools v1.13) (Danecek et al., 2021). H3K27ac ChIP-mapped reads were downsampled with Samtools view (v1.13) according to the sample with the smallest number of reads after filtering and duplicate removal (mouse: 71,741,225 reads; chicken 25,872,853 reads). Mouse CTCF ChIP-mapped reads were downsampled to a quarter and chicken CTCF ChIP reads were not downsampled. When two replicates were available, alignment BAM files were merged using Samtools merge (v1.13) and subsampled to the same number as single replicates. The coverage and peaks were obtained as the output of MACS2 (v2.1.1.20160309.6) (Zhang et al., 2008) with a fixed extension of 200 bp (--call-summits --nomodel --extsize 200 -B) and, for the WP sample, the q-value cutoff for peak detection was set to 0.1 instead of the 0.05 default. CTCF motif orientation was assessed using peaks extended by 100 bp each side and the CTCFBSDB 2.0 database (Ziebarth et al., 2012) with MIT_LM7-identified motifs. Open H3K27ac peaks were obtained by the intersection of H3K27ac peaks with ATAC peak regions in matching tissues using betdtools intersect. The proportions of open H3K27ac peaks were obtained using R (v3.6.1) and the bar plots of Fig. S5A were made with the ggplot2 package (v3.2.1) (https://www.R-project.org/).
MAR annotation
MARs were defined as genomic segments of the T-DOM with the highest density of specific H3K27ac coverage and were annotated as follows: for mm10, chr2:74,872,605-75,696,338 and for galGal6, chr7:15,856,252-16,252,401; sliding windows were of 10 kb every 2 kb. For each window, the coverage of the ChIP was computed and the difference with the coverage in the corresponding H3K27ac ChIP in the brain was considered as the specific signal. The smallest sequence of consecutive windows cumulating 30% (for the dorsal skin and WP samples) or 50% (for the PT samples) of the total specific signal is the MAR. See ChIP_scanRegionForHighestDensity_2files.py at https://gitlab.unige.ch/Aurelie.Hintermann/hintermannetal2022 for details.
Annotation of open pCREs and pCREs
pCREs were defined as non-coding genomic intervals that overlap with H3K27ac MACS2 peaks in at least one of the samples analysed per species. To obtain the genomic coordinates of pCREs, MACS2 peaks of all samples were first pooled into one file, the size of each peak was increased by 660 bp each side to account for nucleosome occupancy using bedtools slop (v2.30.0) and overlapping MACS2 peaks were combined into a single interval using bedtools merge. Intervals were annotated according to their colocalization with tissue-specific H3K27ac peaks, exons, promoters and sequence conservation using bedtools intersect. Intervals overlapping with exons were discarded to obtain pCREs, which were then further annotated as ‘enhancers’ and ‘promoters’ depending on whether they mapped more or less than 2 kb away from a gene TSS, respectively. pCREs that were H3K27ac positive in more than one tissue were called ‘pleiotropic’, whereas those being active in one tissue were labelled ‘specific’. Open pCREs are H3K27ac peaks which overlapped with ATAC peaks using bedtools intersect. pCREs were quantified using R (v3.6.1) and the bar plots of Fig. 5A were made with the ggplot2 package (v3.2.1). See CNEs_01.sh at https://gitlab.unige.ch/Aurelie.Hintermann/hintermannetal2022 for details.
CNEs
Pairwise BLASTZ alignments were downloaded from http://hgdownload.soe.ucsc.edu/goldenPath/mm10/vsGalGal6/mm10.galGal6.synNet.maf.gz and transformed to intervals with maf to interval (Blankenberg et al., 2011). Conserved elements were considered to be non-coding if they did not overlap with annotated exons either in mouse or in chicken. The synteny plot on Fig. S3A was made using the R package ggplot2 (v3.2.1). H3K27ac peaks and open H3K27ac peaks were first extended by 660 bp on each side to account for nucleosome occupancy. Then, CNEs that were located 2 kb or more from the next TSS and which intersected with either H3K27ac peaks or with open H3K27ac peaks using bedtools intersect were annotated as enhancer CNEs and open enhancer CNEs, respectively. Enhancer-CNE heatmaps were made with the pheatmap R package (v1.0.12) using a matrix in which columns were enhancer CNEs (Fig. 5B) or open enhancer CNEs (Fig. S5B), rows were the tissues in which acetylation peaks were observed and values were the scores of acetylation peaks. If an enhancer CNE comprised several peaks of the same tissue, the average was calculated. In Fig. 5C and Fig. S5C, the relationships of the three sets formed by the conditions of being an enhancer CNE (or an open enhancer CNE) in the mouse tissue, in the chicken tissue or in a given mouse tissue, in the matching chicken tissue or in another chicken tissue were represented on Euler diagrams using the eulerr R package (v6.0.0), in which each tissue analysed was considered independently. See CNEs_01.sh at https://gitlab.unige.ch/Aurelie.Hintermann/hintermannetal2022 for details.
4C-seq experiments
4C-seq experiments were performed according to Noordermeer et al. (2011). Briefly, the tissues were dissected, dissociated with collagenase (Sigma-Aldrich/Fluka) and filtered through a 35 μm mesh. Cells were fixed with 2% formaldehyde (in PBS with 10% fetal bovine serum) for 10 min at room temperature and the reaction was quenched on ice with glycine. Nuclei were extracted using a cell lysis buffer [10 mM Tris-HCl (pH 7.5), 10 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA] and stored at −80°C. Approximately 200 E9.5 posterior trunk regions dissected at the level of the second to fourth pair of somites, as well as 120 chicken HH19-21 posterior trunk samples, were used. Nuclei were digested with NlaIII (New England BioLabs) and ligated with T4 DNA ligase HC (Promega) in diluted conditions to promote intramolecular ligation. Samples were digested again with DpnII (New England BioLabs) and re-ligated with T4 DNA ligase HC. These templates were amplified using the Expand Long Template (Roche) and PCR-barcoded primers flanked with adaptors. For each library, eight to ten independent PCR reactions were pooled together and purified using the PCR purification kit (QIAGEN). Multiplexed libraries were sequenced on Illumina HiSeq 2500 to obtain 100 bp single-end reads. Demultiplexing, mapping and 4C-seq analysis were performed using a local version of the pipeline described in David et al. (2014) on the mouse mm10 and chicken galGal6 genome assemblies. The profiles were smoothened using a window size of 11 fragments and normalized to the mean score in ±5 Mb around the viewpoint. When multiple independent biological replicates were available, average 4C-seq profiles were calculated. When viewpoints were subtracted, the score of each window of the normalized, smoothed profiles were subtracted (see 4C_subset_subtract at https://gitlab.unige.ch/Aurelie.Hintermann/hintermannetal2022) for details.
Annotation of the D-regions
The definition of the interacting regions was based on 4C-seq interaction profiles in mouse and sequence conservation. The regions are delimited by CNEs to be able to transpose them to the chicken genome. CNE331 was chosen for the 5′ border of the D1-region as it is the only CNE located in the region that spans from the 3′ of the HoxD cluster to the promoter of Mtx2. At the other extremity, the 3′ border of the D9-region was set to CNE382, where the Hoxd9 signal goes back to the same level as Hoxd4 (Fig. S3A). The limits between the Hoxd1- and D4-regions and between the D4- and D9-regions were established based on 4C scores normalized by the number of mapped reads. To obtain the relative distribution of contacts across our region of interest (i.e. from CNE331 to CNE382), we first normalized 4C scores by the total score of the whole region. We then computed the cumulative sum of the difference between the normalized scores of two viewpoints (Fig. S3C). Accordingly, the delimitation between Hoxd1-region and D4-region was set to CNE346, which is the closest conserved element to the minimum value of the cumulative sum using the score difference between Hoxd4 and Hoxd1, and thus corresponds to the end of the genomic segment where Hoxd1 values are higher than those of Hoxd4. Similarly, the D4/D9 border was set to CNE364, which corresponds to the genomic location for which the cumulative sum of the score difference between Hoxd9 and Hoxd4 is minimum and where Hoxd9 values become higher than those of Hoxd4. The limits of the D-regions were then transposed to the chicken genome using the corresponding CNEs. Finally, the normalized 4C scores for each viewpoint were quantified in each region and normalized by the region size in Mb (Fig. S3D). The D-regions were quantified using R (v3.6.1) and the bar plots of Fig. S3D were made with the ggplot2 package (v3.2.1).
Ethics approval
All experiments involving animals were performed in agreement with the Swiss Law on Animal Protection (LPA), under licence no. GE 81/14 (to D.D.).
Supplementary Material
Acknowledgements
We thank all members of the Duboule laboratories for comments and discussions, Nayuta Yakushiji-Kaminatsui for the chicken HOXD8 probe, Andrew P. MacMahon for the mouse Shh probe, Jozsef Zakany for his series of mouse RNA probes and Gregory Loichot for artwork. This work was supported in part by using the resources and services of both the Gene Expression Core Facility at the School of Life Sciences of the École polytechnique fédérale de Lausanne (EPFL) and the genomic platform from the Institute of Genetics and Genomics of Geneva (iGE3) centre at the University of Geneva. Transgene injections were performed at the transgenesis platform of the University of Geneva Medical School.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.H., I.G., D.D., L.B.; Methodology: A.H., I.G., L.L.-D., C.C.B., D.D.; Software: A.H., L.L.-D.; Validation: A.H., L.L.-D., D.D., L.B.; Formal analysis: A.H., D.D., L.L.-D., L.B.; Investigation: A.H., I.G., C.C.B., S.G., L.B.; Resources: L.L.-D., S.G.; Data curation: A.H., L.L.-D.; Writing - original draft: A.H., D.D., L.B.; Writing - review & editing: L.L.-D., C.C.B.; Supervision: D.D., L.B.; Project administration: A.H., D.D., L.B.; Funding acquisition: D.D.
Funding
This work was supported by funds from the Université de Genève, the École Polytechnique Fédérale de Lausanne (EPFL, Lausanne), the Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (no. 310030B_138662) and the European Research Council grants SystemHox (no. 232790) and RegulHox (no. 588029) to D.D. The funding bodies had no role in the design of the study, in the collection, analysis and interpretation of data and in the writing of the manuscript. Open Access funding provided by Université de Genève. Deposited in PMC for immediate release.
Data availability
All raw and processed datasets are available in the Gene Expression Omnibus (GEO) repository under accession number GSE195592. All scripts necessary to reproduce figures from raw data are available at https://gitlab.unige.ch/Aurelie.Hintermann/hintermannetal2022 (https://doi.org/10.5281/zenodo.6554916).
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.200594.
References
- Abdennur, N. and Mirny, L. A. (2020). Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311-316. 10.1093/bioinformatics/btz540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acemel, R. D., Tena, J. J., Irastorza-Azcarate, I., Marlétaz, F., Gómez-Marín, C., de la Calle-Mustienes, E., Bertrand, S., Diaz, S. G., Aldea, D., Aury, J.-M.et al. (2016). A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat. Genet 48, 336-341. 10.1038/ng.3497 [DOI] [PubMed] [Google Scholar]
- Afgan, E., Baker, D., van den Beek, M., Blankenberg, D., Bouvier, D., Čech, M., Chilton, J., Clements, D., Coraor, N., Eberhard, C.et al. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3-W10. 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allais-Bonnet, A., Hintermann, A., Deloche, M.-C., Cornette, R., Bardou, P., Naval-Sanchez, M., Pinton, A., Haruda, A., Grohs, C., Zakany, J.et al. (2021). Analysis of polycerate mutants reveals the evolutionary co-option of HOXD1 for horn patterning in bovidae. Mol. Biol. Evol. 38, 2260-2272. 10.1093/molbev/msab021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amândio, A. R., Lopez-Delisle, L., Bolt, C. C., Mascrez, B. and Duboule, D. (2020). A complex regulatory landscape involved in the development of mammalian external genitals. Elife 9, e52962. 10.7554/eLife.52962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amândio, A. R., Beccari, L., Lopez-Delisle, L., Mascrez, B., Zakany, J., Gitto, S. and Duboule, D. (2021). Sequential in cis mutagenesis in vivo reveals various functions for CTCF sites at the mouse HoxD cluster. Genes Dev. 35, 1490-1509. 10.1101/gad.348934.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosino, L., Vassalli, Q. A., D'Agostino, Y., Esposito, R., Cetrangolo, V., Caputi, L., Amoroso, A., Aniello, F., D'Aniello, S., Chatzigeorgiou, M.et al. (2019). Functional conserved non-coding elements among tunicates and chordates. Dev. Biol. 448, 101-110. 10.1016/j.ydbio.2018.12.012 [DOI] [PubMed] [Google Scholar]
- Andrey, G., Montavon, T., Mascrez, B., Gonzalez, F., Noordermeer, D., Leleu, M., Trono, D., Spitz, F. and Duboule, D. (2013). A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340, 1234167. 10.1126/science.1234167 [DOI] [PubMed] [Google Scholar]
- Beccari, L., Yakushiji-Kaminatsui, N., Woltering, J. M., Necsulea, A., Lonfat, N., Rodríguez-Carballo, E., Mascrez, B., Yamamoto, S., Kuroiwa, A. and Duboule, D. (2016). A role for HOX13 proteins in the regulatory switch between TADs at the HoxD locus. Genes Dev. 30, 1172-1186. 10.1101/gad.281055.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton, J.-M., McCord, R. P., Gibcus, J., Naumova, N., Zhan, Y. and Dekker, J. (2012). Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58, 268-276 10.1016/j.ymeth.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot, C., Villar, D., Horvath, J. E., Odom, D. T. and Flicek, P. (2018). Complexity and conservation of regulatory landscapes underlie evolutionary resilience of mammalian gene expression. Nat. Ecol. Evol. 2, 152-163. 10.1038/s41559-017-0377-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs, L. C. and Mikkola, M. L. (2014). Early inductive events in ectodermal appendage morphogenesis. Semin. Cell Dev. Biol. 25-26, 11-21. 10.1016/j.semcdb.2014.01.007 [DOI] [PubMed] [Google Scholar]
- Blankenberg, D., Taylor, J. and Nekrutenko, A. (2011). Making whole genome multiple alignments usable for biologists. Bioinformatics 27, 2426-2428. 10.1093/bioinformatics/btr398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt, C. C. and Duboule, D. (2020). The regulatory landscapes of developmental genes. Development 147, dev171736. 10.1242/dev.171736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt, C. C., Lopez-Delisle, L., Mascrez, B. and Duboule, D. (2021). Mesomelic dysplasias associated with the HOXD locus are caused by regulatory reallocations. Nat. Commun. 12, 5013. 10.1038/s41467-021-25330-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bompadre, O. and Andrey, G. (2019). Chromatin topology in development and disease. Curr. Opin. Genet. Dev. 55, 32-38. 10.1016/j.gde.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. and Greenleaf, W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213-1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, S. B. (2008). Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25-36. 10.1016/j.cell.2008.06.030 [DOI] [PubMed] [Google Scholar]
- Chiang, C., Swan, R. Z., Grachtchouk, M., Bolinger, M., Litingtung, Y., Robertson, E. K., Cooper, M. K., Gaffield, W., Westphal, H., Beachy, P. A.et al. (1999). Essential role for Sonic hedgehog during hair follicle morphogenesis. Dev. Biol. 205, 1-9. 10.1006/dbio.1998.9103 [DOI] [PubMed] [Google Scholar]
- Chuong, C. M., Oliver, G., Ting, S. A., Jegalian, B. G., Chen, H. M. and De Robertis, E. M. (1990). Gradients of homeoproteins in developing feather buds. Development 110, 1021-1030. 10.1242/dev.110.4.1021 [DOI] [PubMed] [Google Scholar]
- Creyghton, M. P., Cheng, A. W., Welstead, G. G., Kooistra, T., Carey, B. W., Steine, E. J., Hanna, J., Lodato, M. A., Frampton, G. M., Sharp, P. A.et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931-21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, K. J. and Pourquié, O. (2000). A clock-work somite. BioEssays 22, 72-83. [DOI] [PubMed] [Google Scholar]
- Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., Whitwham, A., Keane, T., McCarthy, S. A., Davies, R. M.et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10, giab008. 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbellay, F. and Duboule, D. (2016). Chapter sixteen - topological domains, metagenes, and the emergence of pleiotropic regulations at Hox Loci. In Current Topics in Developmental Biology, Essays on Developmental Biology, Part A, (ed. Wassarman P. M.), pp. 299-314. Academic Press. 10.1016/bs.ctdb.2015.11.022 [DOI] [PubMed] [Google Scholar]
- Darbellay, F., Bochaton, C., Lopez-Delisle, L., Mascrez, B., Tschopp, P., Delpretti, S., Zakany, J. and Duboule, D. (2019). The constrained architecture of mammalian Hox gene clusters. Proc. Natl. Acad. Sci. USA 116, 13424-13433. 10.1073/pnas.1904602116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, F. P. A., Delafontaine, J., Carat, S., Ross, F. J., Lefebvre, G., Jarosz, Y., Sinclair, L., Noordermeer, D., Rougemont, J. and Leleu, M. (2014). HTSstation: a web application and open-access libraries for high-throughput sequencing data analysis. PLoS One 9, e85879. 10.1371/journal.pone.0085879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker, J. and Heard, E. (2015). Structural and functional diversity of topologically associating domains. FEBS Lett. 589, 2877-2884. 10.1016/j.febslet.2015.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpretti, S., Montavon, T., Leleu, M., Joye, E., Tzika, A., Milinkovitch, M. and Duboule, D. (2013). Multiple enhancers regulate Hoxd genes and the Hotdog LncRNA during cecum budding. Cell Rep. 5, 137-150. 10.1016/j.celrep.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Dermitzakis, E. T. and Clark, A. G. (2002). Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol. Biol. Evol. 19, 1114-1121. 10.1093/oxfordjournals.molbev.a004169 [DOI] [PubMed] [Google Scholar]
- Dhouailly, D. (2009). A new scenario for the evolutionary origin of hair, feather, and avian scales. J. Anat. 214, 587-606. 10.1111/j.1469-7580.2008.01041.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhouailly, D., Godefroit, P., Martin, T., Nonchev, S., Caraguel, F. and Oftedal, O. (2019). Getting to the root of scales, feather and hair: as deep as odontodes? Exp. Dermatol. 28, 503-508. 10.1111/exd.13391 [DOI] [PubMed] [Google Scholar]
- Di-Poï, N. and Milinkovitch, M. C. (2016). The anatomical placode in reptile scale morphogenesis indicates shared ancestry among skin appendages in amniotes. Sci. Adv. 2, e1600708. 10.1126/sciadv.1600708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., Hu, M., Liu, J. S. and Ren, B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376-380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, S. L., Beesley, J., French, J. D. and Dunning, A. M. (2013). Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 93, 779-797. 10.1016/j.ajhg.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub, M. P. and Ettwiller, L. (2011). De Novo genesis of enhancers in vertebrates. PLoS Biol. 9, e1001188. 10.1371/journal.pbio.1001188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eres, I. E., Luo, K., Hsiao, C. J., Blake, L. E. and Gilad, Y. (2019). Reorganization of 3D genome structure may contribute to gene regulatory evolution in primates. PLoS Genet. 15, e1008278. 10.1371/journal.pgen.1008278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Guerrero, M., Yakushiji-Kaminatsui, N., Lopez-Delisle, L., Zdral, S., Darbellay, F., Perez-Gomez, R., Bolt, C. C., Sanchez-Martin, M. A., Duboule, D. and Ros, M. A. (2020). Mammalian-specific ectodermal enhancers control the expression of Hoxc genes in developing nails and hair follicles. Proc. Natl. Acad. Sci. USA 117, 30509-30519. 10.1073/pnas.2011078117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt, S., Sharpe, P. T. and Duboule, D. (1988). Spatially restricted domains of homeo-gene transcripts in mouse embryos: relation to a segmented body plan. Development 104, 169-179. 10.1242/dev.104.Supplement.169 [DOI] [Google Scholar]
- Godwin, A. R. and Capecchi, M. R. (1998). Hoxc13 mutant mice lack external hair. Genes Dev. 12, 11-20. 10.1101/gad.12.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro, I., Gitto, S., Novoa, A., Codourey, J., Nguyen Huynh, T. H., Gonzalez, F., Milinkovitch, M. C., Mallo, M. and Duboule, D. (2016). Reorganisation of Hoxd regulatory landscapes during the evolution of a snake-like body plan. Elife 5, e16087. 10.7554/eLife.16087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger, V. and Hamilton, H. L. (1951). A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49-92. 10.1002/jmor.1050880104 [DOI] [PubMed] [Google Scholar]
- Hansen, A. S., Pustova, I., Cattoglio, C., Tjian, R. and Darzacq, X. (2017). CTCF and Cohesin regulate chromatin loop stability with distinct dynamics. Elife 6, e25776. 10.7554/eLife.25776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmston, N., Ing-Simmons, E., Tan, G., Perry, M., Merkenschlager, M. and Lenhard, B. (2017). Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nat. Commun. 8, 441. 10.1038/s41467-017-00524-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, P. W. H. and Garcia-Fernàndez, J. (1996). HoxGenes and chordate evolution. Dev. Biol. 173, 382-395. 10.1006/dbio.1996.0034 [DOI] [PubMed] [Google Scholar]
- Holland, L. Z., Albalat, R., Azumi, K., Benito-Gutiérrez, È., Blow, M. J., Bronner-Fraser, M., Brunet, F., Butts, T., Candiani, S., Dishaw, L. J.et al. (2008). The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18, 1100-1111. 10.1101/gr.073676.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia, M., Tena, J. J., Alexis, M. S., Fernandez-Miñan, A., Maeso, I., Bogdanović, O., de la Calle-Mustienes, E., Roy, S. W., Gómez-Skarmeta, J. L. and Fraser, H. B. (2012). Extensive conservation of ancient microsynteny across metazoans due to cis-regulatory constraints. Genome Res. 22, 2356-2367. 10.1101/gr.139725.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzler, B., Viallet, J. P., Le Mouellic, H., Boncinelli, E., Duboule, D. and Dhouailly, D. (1994). Differential expression of two different homeobox gene families during mouse tegument morphogenesis. Int. J. Dev. Biol. 38, 633-640. [PubMed] [Google Scholar]
- Kanzler, B., Prin, F., Thelu, J. and Dhouailly, D. (1997). CHOXC-8 and CHOXD-13 expression in embryonic chick skin and cutaneous appendage specification. Dev. Dyn. 210, 274-287. [DOI] [PubMed] [Google Scholar]
- Kragesteen, B. K., Spielmann, M., Paliou, C., Heinrich, V., Schöpflin, R., Esposito, A., Annunziatella, C., Bianco, S., Chiariello, A. M., Jerković, I.et al. (2018). Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat. Genet. 50, 1463-1473. 10.1038/s41588-018-0221-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverré, A., Tannier, E. and Necsulea, A. (2022). Long-range promoter-enhancer contacts are conserved during evolution and contribute to gene expression robustness. Genome Res. 32, 280-296. 10.1101/gr.275901.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, H. K., Prescott, S. L. and Wysocka, J. (2016). Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167, 1170-1187. 10.1016/j.cell.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Delisle, L., Rabbani, L., Wolff, J., Bhardwaj, V., Backofen, R., Grüning, B., Ramírez, F. and Manke, T. (2021). pyGenomeTracks: reproducible plots for multivariate genomic datasets. Bioinformatics 37, 422-423. 10.1093/bioinformatics/btaa692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez, D. G., Kraft, K., Heinrich, V., Krawitz, P., Brancati, F., Klopocki, E., Horn, D., Kayserili, H., Opitz, J. M., Laxova, R.et al. (2015). Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012-1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinic, M., Aktas, T., Ruf, S. and Spitz, F. (2013). An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Dev. Cell 24, 530-542. 10.1016/j.devcel.2013.01.025 [DOI] [PubMed] [Google Scholar]
- Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J 17, 10. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Michon, F., Charveron, M. and Dhouailly, D. (2007). Dermal condensation formation in the chick embryo: requirement for integrin engagement and subsequent stabilization by a possible Notch/integrin interaction. Dev. Dyn. 236, 755-768. 10.1002/dvdy.21080 [DOI] [PubMed] [Google Scholar]
- Mikkola, M. L. (2007). Genetic basis of skin appendage development. Semin. Cell Dev. Biol. 18, 225-236. 10.1016/j.semcdb.2007.01.007 [DOI] [PubMed] [Google Scholar]
- Montavon, T., Soshnikova, N., Mascrez, B., Joye, E., Thevenet, L., Splinter, E., de Laat, W., Spitz, F. and Duboule, D. (2011). A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132-1145. 10.1016/j.cell.2011.10.023 [DOI] [PubMed] [Google Scholar]
- Nanni, L., Ceri, S. and Logie, C. (2020). Spatial patterns of CTCF sites define the anatomy of TADs and their boundaries. Genome Biol. 21, 197. 10.1186/s13059-020-02108-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer, D., Leleu, M., Splinter, E., Rougemont, J., De Laat, W. and Duboule, D. (2011). The dynamic architecture of Hox gene clusters. Science 334, 222-225. 10.1126/science.1207194 [DOI] [PubMed] [Google Scholar]
- Nora, E. P., Lajoie, B. R., Schulz, E. G., Giorgetti, L., Okamoto, I., Servant, N., Piolot, T., van Berkum, N. L., Meisig, J., Sedat, J.et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381-385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer, A. I., Jane-wit, D., McLean, L., Panteleyev, A. A., Christiano, A. M. and Wolgemuth, D. J. (2000). Hoxa4 expression in developing mouse hair follicles and skin. Mech. Dev. 99, 153-157. 10.1016/S0925-4773(00)00471-8 [DOI] [PubMed] [Google Scholar]
- Pantalacci, S., Chaumot, A., Benoît, G., Sadier, A., Delsuc, F., Douzery, E. J. P. and Laudet, V. (2008). Conserved features and evolutionary shifts of the EDA signaling pathway involved in vertebrate skin appendage development. Mol. Biol. Evol. 25, 912-928. 10.1093/molbev/msn038 [DOI] [PubMed] [Google Scholar]
- Pugacheva, E. M., Kubo, N., Loukinov, D., Tajmul, M., Kang, S., Kovalchuk, A. L., Strunnikov, A. V., Zentner, G. E., Ren, B. and Lobanenkov, V. V. (2020). CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc. Natl. Acad. Sci. USA 117, 2020-2031. 10.1073/pnas.1911708117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, A. R. and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias, A., Bajpai, R., Swigut, T., Brugmann, S. A., Flynn, R. A. and Wysocka, J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279-283. 10.1038/nature09692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, F., Bhardwaj, V., Arrigoni, L., Lam, K. C., Grüning, B. A., Villaveces, J., Habermann, B., Akhtar, A. and Manke, T. (2018). High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9, 189. 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, S. S. P., Huntley, M. H., Durand, N. C., Stamenova, E. K., Bochkov, I. D., Robinson, J. T., Sanborn, A. L., Machol, I., Omer, A. D., Lander, E. S.et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665-1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz, M., Patel, N. H. and Hinman, V. F. (2015). Unraveling the tangled skein: the evolution of transcriptional regulatory networks in development. Annu. Rev. Genomics Hum. Genet. 16, 103-131. 10.1146/annurev-genom-091212-153423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, A. I. and Gaunt, S. J. (2002). Colinearity and non-colinearity in the expression of Hox genes in developing chick skin. Int. J. Dev. Biol. 46, 209-215. 10.1387/ijdb.011495 [DOI] [PubMed] [Google Scholar]
- Reynolds, A. J., Lawrence, C. M. and Jahoda, C. A. (1995). Hair follicle and fiber reconstruction. J. Invest. Dermatol. 104, 39S-40S. 10.1038/jid.1995.57 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Carballo, E., Lopez-Delisle, L., Zhan, Y., Fabre, P. J., Beccari, L., El-Idrissi, I., Huynh, T. H. N., Ozadam, H., Dekker, J. and Duboule, D. (2017). The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev. 31, 2264-2281. 10.1101/gad.307769.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Carballo, E., Lopez-Delisle, L., Willemin, A., Beccari, L., Gitto, S., Mascrez, B. and Duboule, D. (2020). Chromatin topology and the timing of enhancer function at the HoxD locus. Proc. Natl. Acad. Sci. USA 117, 31231-31241. 10.1073/pnas.2015083117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari, B. R., Dall'Agnese, A., Boija, A., Klein, I. A., Coffey, E. L., Shrinivas, K., Abraham, B. J., Hannett, N. M., Zamudio, A. V., Manteiga, J. C.et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958. 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin, A., Bailey, P., Bruce, S., Engström, P. G., Klos, J. M., Wasserman, W. W., Ericson, J. and Lenhard, B. (2004). Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5, 99. 10.1186/1471-2164-5-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanges, R., Kalmar, E., Claudiani, P., D'Amato, M., Muller, F. and Stupka, E. (2006). Shuffling of cis-regulatory elements is a pervasive feature of the vertebrate lineage. Genome Biol. 7, R56. 10.1186/gb-2006-7-7-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanges, R., Hadzhiev, Y., Gueroult-Bellone, M., Roure, A., Ferg, M., Meola, N., Amore, G., Basu, S., Brown, E. R., De Simone, M.et al. (2013). Highly conserved elements discovered in vertebrates are present in non-syntenic loci of tunicates, act as enhancers and can be transcribed during development. Nucleic Acids Res. 41, 3600-3618. 10.1093/nar/gkt030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, R. H. and Knapp, L. W. (2003). Avian skin development and the evolutionary origin of feathers. J. Exp. Zool. 298B, 57-72. 10.1002/jez.b.26 [DOI] [PubMed] [Google Scholar]
- Schep, R., Necsulea, A., Rodríguez-Carballo, E., Guerreiro, I., Andrey, G., Nguyen Huynh, T. H., Marcet, V., Zákány, J., Duboule, D. and Beccari, L. (2016). Control of Hoxd gene transcription in the mammary bud by hijacking a preexisting regulatory landscape. Proc. Natl. Acad. Sci. USA 113, E7720-E7729. 10.1073/pnas.1617141113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, D., Wilson, M. D., Ballester, B., Schwalie, P. C., Brown, G. D., Marshall, A., Kutter, C., Watt, S., Martinez-Jimenez, C. P., Mackay, S.et al. (2010). Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328, 1036-1040. 10.1126/science.1186176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton, T., Yaffe, E., Kenigsberg, E., Bantignies, F., Leblanc, B., Hoichman, M., Parrinello, H., Tanay, A. and Cavalli, G. (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458-472. 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Snetkova, V., Ypsilanti, A. R., Akiyama, J. A., Mannion, B. J., Plajzer-Frick, I., Novak, C. S., Harrington, A. N., Pham, Q. T., Kato, M., Zhu, Y.et al. (2021). Ultraconserved enhancer function does not require perfect sequence conservation. Nat. Genet. 53, 521-528. 10.1038/s41588-021-00812-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz, F. and Furlong, E. E. M. (2012). Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613-626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- Spitz, F., Gonzalez, F., Peichel, C., Vogt, T. F., Duboule, D. and Zákány, J. (2001). Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes Dev. 15, 2209-2214. 10.1101/gad.205701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz, F., Gonzalez, F. and Duboule, D. (2003). A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113, 405-417. 10.1016/s0092-8674(03)00310-6 [DOI] [PubMed] [Google Scholar]
- Torosin, N. S., Anand, A., Golla, T. R., Cao, W. and Ellison, C. E. (2020). 3D genome evolution and reorganization in the Drosophila melanogaster species group. PLoS Genet. 16, e1009229. 10.1371/journal.pgen.1009229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushiki, A., Zhang, Y., Xiong, C., Zhao, J., Georgakopoulos-Soares, I., Kane, L., Jamieson, K., Bamshad, M. J., Nickerson, D. A., University of Washington Center for Mendelian Genomics, Shen, Y., et al. (2021). Deletion of CTCF sites in the SHH locus alters enhancer-promoter interactions and leads to acheiropodia. Nat. Commun. 12, 2282. 10.1038/s41467-021-22470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, J., Rynes, E., Sandstrom, R., Zhang, M., Canfield, T., Hansen, R. S., Stehling-Sun, S., Sabo, P. J., Byron, R., Humbert, R.et al. (2014). Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346, 1007-1012. 10.1126/science.1246426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar, D., Berthelot, C., Aldridge, S., Rayner, T. F., Lukk, M., Pignatelli, M., Park, T. J., Deaville, R., Erichsen, J. T., Jasinska, A. J.et al. (2015). Enhancer evolution across 20 mammalian species. Cell 160, 554-566. 10.1016/j.cell.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widelitz, R. B. and Chuong, C. M. (1999). Early events in skin appendage formation: induction of epithelial placodes and condensation of dermal mesenchyme. J. Investig. Dermatol. Symp. Proc. 4, 302-306. 10.1038/sj.jidsp.5640234 [DOI] [PubMed] [Google Scholar]
- Wingett, S., Ewels, P., Furlan-Magaril, M., Nagano, T., Schoenfelder, S., Fraser, P. and Andrews, S. (2015). HiCUP: pipeline for mapping and processing Hi-C data. F1000Res 4, 1310. 10.12688/f1000research.7334.1. eCollection 201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, J., Rabbani, L., Gilsbach, R., Richard, G., Manke, T., Backofen, R. and Grüning, B. A. (2020). Galaxy HiCExplorer 3: a web server for reproducible Hi-C, capture Hi-C and single-cell Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 48, W177-W184. 10.1093/nar/gkaa220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltering, J. M., Vonk, F. J., Müller, H., Bardine, N., Tuduce, I. L., de Bakker, M. A. G., Knöchel, W., Sirbu, I. O., Durston, A. J. and Richardson, M. K. (2009). Axial patterning in snakes and caecilians: evidence for an alternative interpretation of the Hox code. Dev. Biol. 332, 82-89. 10.1016/j.ydbio.2009.04.031 [DOI] [PubMed] [Google Scholar]
- Woltering, J. M., Noordermeer, D., Leleu, M. and Duboule, D. (2014). Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits. PLoS Biol. 12, e1001773. 10.1371/journal.pbio.1001773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, E. S., Zheng, D., Tan, S. Z., Bower, N. I., Garside, V., Vanwalleghem, G., Gaiti, F., Scott, E., Hogan, B. M., Kikuchi, K.et al. (2020). Deep conservation of the enhancer regulatory code in animals. Science 370, eaax8137. 10.1126/science.aax8137 [DOI] [PubMed] [Google Scholar]
- Wrenn, J. T. and Wessells, N. K. (1984). The early development of mystacial vibrissae in the mouse. J. Embryol. Exp. Morphol. 83, 137-156. [PubMed] [Google Scholar]
- Wu, P., Hou, L., Plikus, M., Hughes, M., Scehnet, J., Suksaweang, S., Widelitz, R., Jiang, T.-X. and Chuong, C.-M. (2004). Evo-Devo of amniote integuments and appendages. Int. J. Dev. Biol. 48, 249-270. 10.1387/ijdb.041825pw [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushiji-Kaminatsui, N., Lopez-Delisle, L., Bolt, C. C., Andrey, G., Beccari, L. and Duboule, D. (2018). Similarities and differences in the regulation of HoxD genes during chick and mouse limb development. PLoS Biol. 16, e3000004. 10.1371/journal.pbio.3000004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z., Jiang, K., Xu, Z., Huang, H., Qian, N., Lu, Z., Chen, D., Di, R., Yuan, T., Du, Z.et al. (2018). Hoxc-dependent mesenchymal niche heterogeneity drives regional hair follicle regeneration. Cell Stem Cell 23, 487-500.e6. 10.1016/j.stem.2018.07.016 [DOI] [PubMed] [Google Scholar]
- Zákány, J., Kmita, M., Alarcon, P., de la Pompa, J.-L. and Duboule, D. (2001). Localized and transient transcription of Hox Genes suggests a link between patterning and the segmentation clock. Cell 106, 207-217. 10.1016/S0092-8674(01)00436-6 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Liu, T., Meyer, C. A., Eeckhoute, J., Johnson, D. S., Bernstein, B. E., Nusbaum, C., Myers, R. M., Brown, M., Li, W.et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebarth, J. D., Bhattacharya, A. and Cui, Y. (2012). CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res. 41, D188-D194. 10.1093/nar/gks1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.