Abstract
The aryl hydrocarbon receptor (AHR) is required for vertebrate development and is also activated by exogenous chemicals, including polycyclic aromatic hydrocarbons (PAHs) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). AHR activation is well-understood, but roles of downstream molecular signaling events are largely unknown. From previous transcriptomics in 48 h postfertilization (hpf) zebrafish exposed to several PAHs and TCDD, we found wfikkn1 was highly coexpressed with cyp1a (marker for AHR activation). Thus, we hypothesized wfikkn1’s role in AHR signaling, and showed that wfikkn1 expression was Ahr2 (zebrafish ortholog of human AHR)-dependent in developing zebrafish exposed to TCDD. To functionally characterize wfikkn1, we made a CRISPR-Cas9 mutant line with a 16-bp deletion in wfikkn1’s exon, and exposed wildtype and mutants to dimethyl sulfoxide or TCDD. 48-hpf mRNA sequencing revealed over 700 genes that were differentially expressed (p < .05, log2FC > 1) between each pair of treatment combinations, suggesting an important role for wfikkn1 in altering both the 48-hpf transcriptome and TCDD-induced expression changes. Mass spectrometry-based proteomics of 48-hpf wildtype and mutants revealed 325 significant differentially expressed proteins. Functional enrichment demonstrated wfikkn1 was involved in skeletal muscle development and played a role in neurological pathways after TCDD exposure. Mutant zebrafish appeared morphologically normal but had significant behavior deficiencies at all life stages, and absence of Wfikkn1 did not significantly alter TCDD-induced behavior effects at all life stages. In conclusion, wfikkn1 did not appear to be significantly involved in TCDD’s overt toxicity but is likely a necessary functional member of the AHR signaling cascade.
Keywords: aryl hydrocarbon receptor (AHR), zebrafish, TCDD, wfikkn1, transcriptomics, behavior
Aryl hydrocarbon receptors (AHRs) are ligand-dependent transcription factors (TFs) of the basic-helix-loop-helix superfamily of TFs (Kewley et al., 2004; Schmidt and Bradfield, 1996). AHRs are well-known for their ability to interact with several xenobiotics, particularly polycyclic aromatic hydrocarbons (PAHs; Billiard et al., 2006), polychlorinated biphenyls (Kafafi et al., 1993), and dioxins, such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD; Mimura and Fujii-Kuriyama, 2003). Both AHR-knockout mice and rats (Fernandez-Salguero et al., 1996; Harrill et al., 2016), as well as mice that have mutations in either the nuclear localization or DNA binding domains of the AHR (Bunger et al., 2003, 2008) are resistant to TCDD-induced toxicity. In addition to the AHR’s role in mediating xenobiotic effects, this receptor has been implicated in endogenous roles (Barouki et al., 2007). In rodents, the AHR is crucial during development and plays a physiological role in overall growth rate, blood pressure regulation, and immune, hepatic, and urinary systems (Larigot et al., 2018).
Upon ligand activation and translocation to the nucleus, the AHR forms a heterodimer with the AHR nuclear translocator (ARNT; Hoffman et al., 1991), which recognizes aryl hydrocarbon response elements (AHREs) in the promoters of downstream target genes, such as the cytochrome P450 family of genes (CYPs) and the AHR repressor (AHRR; Fujii-Kuriyama and Mimura, 2005; Mimura et al., 1999; Watson and Hankinson, 1992). CYP1A is critical for either metabolic activation or deactivation of chemicals (Ma and Lu, 2007), and the AHRR has important role in inflammation and tumorigenesis (Vogel and Haarmann-Stemmann, 2017). Evidence demonstrates that the AHR’s role as a master transcriptional regulator is critical during both development and upon xenobiotic exposure; however, the specific functional roles of many downstream genes within the AHR signaling cascade remain unknown.
The zebrafish (Danio rerio) model is an invaluable used in the fields of molecular toxicology, developmental biology, and neurobehavior (Bailey et al., 2013; Bambino and Chu, 2017; Bugel et al., 2014). Zebrafish have high fecundity, rapid ex-utero development, and high sensitivity to chemical exposures, which are ideal traits for high-throughput chemical toxicity screening in a whole animal (Hill et al., 2005). Zebrafish and mammals have highly conserved physiological and genetic similarities, with 71.4% of human genes having at least 1 zebrafish ortholog (Howe et al., 2013). Thus, zebrafish are routinely used to unravel complex molecular pathways and have become an important platform to investigate the AHR signaling pathway. Zebrafish have 3 AHR orthologs: Ahr1a, Ahr1b, and Ahr2, and while developmental toxicity of PAHs is differentially dependent on the zebrafish AHRs (Incardona et al., 2006, 2011), Ahr2 is most responsive to TCDD (Shankar et al., 2020). Similar to rodents, ahr2-knockout zebrafish have demonstrated the receptor’s requirement for TCDD toxicity (Garcia et al., 2018a; Goodale et al., 2012). Ahr2 is also required for normal reproductive, neurobehavioral, and skeletal development in zebrafish (Garcia et al., 2018a; Goodale et al., 2012).
Zebrafish have been utilized to elucidate transcriptional events downstream of AHR activation. For example, the ahr2-dependent gene, cox2b, was discovered to influence pericardiac edema caused by TCDD exposure in developing zebrafish (Teraoka et al., 2014). Similarly, sox9b is one of the most downregulated transcripts upon TCDD exposure and is associated with TCDD-induced cardiac toxicity and craniofacial malformations (Garcia et al., 2017; Hofsteen et al., 2013; Xiong et al., 2008). Declining cost and widening availability of high-throughput sequencing have made RNA sequencing a popular discovery tool for transcriptomic changes from chemical exposure. Developing zebrafish exposed to Ahr2 activators exhibit several ligand-dependent transcriptional changes (Goodale et al., 2015; Shankar et al., 2019), and multiple transcriptional factors are predicted to regulate expression of genes associated with Ahr2 activation (Garland et al., 2020; Goodale et al., 2015), emphasizing the intricate crosstalk between pathways upon chemical exposure.
Our group has published multiple transcriptomic studies in developmental zebrafish exposed to various chemicals. The studies identified wfikkn1—whey acidic protein (WAP), follistatin/Kazal, immunoglobulin (IG), Kunitz/bovine pancreatic trypsin inhibitor (BPTI), and netrin (NTR) domain containing 1—as highly induced in 48 h postfertilization (hpf) zebrafish exposed to individual PAHs and TCDD (Garcia et al., 2018b; Goodale et al., 2013, 2015; Shankar et al., 2019). Additionally, a comparison of several of these gene expression studies revealed wfikkn1’s high coexpression with cyp1a, suggesting wfikkn1’s role in Ahr2 signaling (Shankar et al., 2021). Morpholino knockdown of ahr2 led to a significant decrease in wfikkn1 expression when zebrafish were developmentally exposed to the oxyPAH, benz[a]anthracene-7,12-dione (Goodale et al., 2015). High induction of wfikkn1 in developing zebrafish by structurally diverse PAHs and TCDD suggested a role for wfikkn1 in their toxicity, a hypothesis further supported by the repression of wfikkn1 upon ahr2 knockdown. In this study, we leveraged a CRISPR-Cas9-generated wfikkn1 mutant zebrafish to investigate the functional role of wfikkn1 in both normal and TCDD-exposed zebrafish. We hypothesized that the lack of wfikkn1 would significantly alter whole-animal transcriptomics and proteomics, and sensitive neurobehavioral endpoints.
MATERIALS AND METHODS
Fish Husbandry
Tropical 5D zebrafish were housed at the Sinnhuber Aquatic Research Laboratory at Oregon State University (Corvallis, Oregon), and maintained according to Institutional Animal Care and Use Committee protocols (ACUP 5143). Adult fish were raised in recirculating filtered water supplemented with Instant Ocean salts in densities of approximately 500 fish/50-gallon tank at 28°C under a 14/10-h light/dark cycle. Adult zebrafish were fed GEMMA Micro 300 or 500 (Skretting, Inc., Fontaine les Vervins, France) twice a day, and larval and juvenile zebrafish were fed GEMMA Micro 75 and 150, respectively, 3 times a day (Barton et al., 2016). Spawning funnels were placed in the tanks the night prior, and the following morning, embryos were collected, staged, and maintained in embryo medium (EM) in a 28°C incubator (Westerfield, 2007). EM consisted of 15 mM NaCl, 0.5 mM KCl, 1 mM MgSO4, 0.15 mM KH2PO4, 0.05 mM Na2HPO4, and 0.7 mM NaHCO3 (Mandrell et al., 2012).
Waterborne TCDD Exposure
Depending on the experiment, tropical 5D wild-type (WT) zebrafish, wfikkn1 mutant zebrafish, or ahr2 mutant zebrafish (ahr2osu1; Garcia et al., 2018a) were exposed to TCDD. Shield-stage embryos (6 hpf) were exposed to 0.1% dimethyl sulfoxide (DMSO; vehicle control) and TCDD at 50 pg/ml (juvenile and adult studies) or 1 ng/ml (for all other analyses) with gentle rocking for 1 h. 250-ml (juvenile and adult studies) or 20-ml (all other analyses) glass vials with 10 embryos/ml were used for the exposures. The 50 pg/ml TCDD concentration was previously shown to have no significant effect on mortality, and skeletal growth and deformity of zebrafish exposed at 4 hpf (Baker et al., 2013). The 1 ng/ml TCDD concentration has been used in previous studies, as it results in 99–100% of 120-hpf zebrafish developing overt TCDD-induced malformations (Garcia et al., 2018b; Henry et al., 1997). During the exposures, vials were inverted every 15 min to ensure proper mixing and even exposure. After 1 h, embryos were rinsed 3 times with EM and raised in 100-mm Petri dishes at 28°C with up to 100 embryos in 50 ml of EM until collection at 48 hpf (quantitative reverse transcriptase polymerase chain reaction [RT-qPCR] and RNA sequencing) or 120 hpf (juvenile and adult studies). From the exposures for juvenile and adult studies, a subset of the embryos (n = 96/treatment) was placed in round bottom 96-well plates (Falcon, product number: 353227) with 1 embryo per well prefilled with 100 µl EM for assessment of larval photomotor response (LPR; described below).
Creation and Generation of a wfikkn1 Mutant Zebrafish Line
Design, Assembly, and Preparation of Single-Guide RNA
Two single-guide RNA (sgRNA) targets for wfikkn1 were generated using the http://www.crisprscan.org website (Moreno-Mateos et al., 2015) (last accessed April 6, 2022). This program finds 23-bp target sequences, including the protospacer adjacent motif (PAM) site, with predicted high activity scores, number of off targets, and 5′ end of sequence beginning with GG that is required for efficient T7 transcription. The wfikkn1 gene has a single exon, and we selected 2 sgRNAs targeting regions as close as possible to the 5′ end of the gene to maximize the potential for frameshift mutations resulting in an early premature stop codon.
To assemble the sgRNAs, we used the cloning-free method described previously (Varshney et al., 2016), with slight modifications. Briefly, gene-specific oligo templates consisted of an 18-bp T7 promoter sequence, a 20-bp gene-specific sequence (without the PAM), and a 15-bp overlap sequence to the constant tracrRNA (trans-activating CRISPR RNA) which also is an 80-bp sequence required for CAS9 recognition. Additionally, we used universal forward and reverse primers matching the 5′ ends of the gene-specific and constant oligos, respectively (Garcia et al., 2018a). sgRNA templates and primers for guide assembly were purchased from IDT (Coralville, Iowa), and their sequences are in Supplementary Table 1. Amplification of the template was performed using the KOD Hot Start DNA Polymerase Kit (Millipore Sigma) according to the manufacturer’s recommendation. 0.2 μM each of the gene-specific oligo, forward, and reverse universal primers, and 0.008 μM of the constant oligo were added to the reaction, and PCR was performed under the following conditions: denaturation at 95°C for 2 min, followed by 35 cycles of amplification (95°C for 20 s, 62°C for 10 s, and 72°C for 10 s), and a final extension at 72°C for 5 min. 2 µl of PCR template was used as input for in vitro transcription of the sgRNAs with the HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs) according to manufacturer’s instructions. We included the optional DNase treatment. The sgRNAs were precipitated using ethanol/sodium acetate (pH 5.2), quality was checked on a 1.2% agarose gel, and RNA quantity was determined using a BioTek SynergyMix microplate reader with the Gen5 Take3 module.
Embryo Injections
The Tropical 5D WT strain was used to create the wfikkn1 mutant line. Microinjections of pooled sgRNAs of 6 different genes were conducted, with each gene target located on a different chromosome to avoid large genomic deletion events (Shah et al., 2016). Briefly, a multiplex injection mix of pooled guides and CAS9 protein was calculated to deliver approximately 150–200 pg of each sgRNA (diluted to 1000 ng/µl) and 225–300 pg CAS9 protein in a 1.5–2 nl injection volume into the yolk of a 1-cell embryo. Purified Cas9 protein with a nuclear localization signal derived from SV40 Large T Ag was purchased (Clonetech; Mountain View, California).
Founder Screening
Injected embryos were raised to maturity and screened for heritable mutations by pair crossing potential founder zebrafish to WT 5D fish. Screening was performed using fluorescent PCR and melt curve analysis (see below). DNA was extracted from 8 individual embryos per mating pair, and 8 WT zebrafish embryos were used as controls. Embryos (4–5 days postfertilization [dpf]) were plated in 96-well PCR plates with 1 embryo per well and euthanized on ice, and excess water was removed from the wells. Embryos were then lysed by incubating in 20 μl of 50 mM NaOH, heated at 95°C for 10 min, and vortexed for 10–15 s to dissociate remaining tissue. 4 μl of 1 M Tris-HCl (pH 8.0) neutralization solution was added followed by the addition of 100 μl of ultrapure water to dilute out PCR inhibitors. The mixture was then spun down at 4680 rpm for 10 min to pellet debris.
RT-qPCR and Melt Curve Analysis/Sequencing
The 20-μl PCR reactions for melt curve analysis consisted of 10 μl 2× SYBR Green Master Mix (Applied Biosystems, Foster City, California), 0.4 μl each of 10 μM forward and reverse primers, and 2 μl of diluted DNA. The primers were designed to be at least 15 bp away from the predicted Cas9 cut site (−3 bp from PAM site) and to produce 70–90 bp products. “High-melt primer” sequences are in Supplementary Table 1. The reaction was performed using a StepOnePlus Real-Time PCR System (Applied Biosystems) under the following conditions: denaturation and activation of SYBR polymerase at 95°C for 5 min, followed by 40 cycles of amplification (95°C for 15 s, 58°C for 1 min). Melt curve analysis was conducted with a ramp rate of +0.2 (95°C for 2 min, 70°C for 2 min, and 95°C for 2 min). Founders were identified by a shift in the melt-curve peak in their offspring compared with the WT controls. To determine if the identified founders produce frameshift mutations, we amplified a region 200–400 bp up- and downstream of the sgRNA target site and sequenced the amplicon. The 25 μl PCR reaction had a 0.2 μM final concentration of forward and reverse primers (see Supplementary Table 1 for “sequencing primer” sequences), 2 μl of diluted DNA, and KOD Hot Start DNA Polymerase (Millipore Sigma) according to the manufacturer’s instructions. PCR conditions were as follows: denaturation at 95°C for 2 min, 35 cycles of amplification (95°C for 20 s, 62°C for 10 s, and 72°C for 10 s), followed by an extension at 72°C for 5 min. The Core Facilities of the Center for Genome Research and Biocomputing at Oregon State University performed Sanger sequencing using an ABI 3730 capillary sequence machine. Sequencing results for mixed base reads in heterozygous offspring of potential founders at the mutation site were interpreted manually and confirmed using online tools for characterizing indel mutations (https://tide.nki.nl/ and http://yosttools.genetics.utah.edu/PolyPeakParser/; last accessed December 1, 2021).
RNA Sequencing Analyses
RNA Extraction
For developmental expression of wfikkn1, total RNA was extracted from pooled groups of 15 WT embryos at 2.5, 4, 12, and 24 hpf, and 11 WT larvae at 48, 72, 96, and 120 hpf. For ahr2-dependence, total RNA was extracted from pooled groups of four 48- and 120-hpf WT and ahr2 mutant (ahr2osu1) larvae exposed to 0.1% DMSO or 1 ng/ml TCDD, as described above. The lower number of embryos was a result of impaired fertility of the ahr2 mutant zebrafish, as described previously (Garcia et al., 2018a). RNA extraction was conducted using RNAzol (Molecular Research Center, Inc.) and a bullet blender with 0.5 mm zirconium oxide beads (Next Advance, Averill Park, New York) for 3 min at speed 8, as recommended by the manufacturer. The RNA was purified using the Direct-zol MiniPrep kit (Zymo Research, Irvine, California) as recommended by the manufacturer. The optional in-column DNase 1 digestion step was performed for 15 min. RNA quality and quantity were determined using a BioTek Synergy Mx microplate reader (BioTek Instruments, Inc., Winooski, Vermont) with the Gen5 Take3 module.
mRNA Quantification using RT-qPCR
A total of 10 µl one-step RT-qPCR reactions were set up consisting of 5 µl SYBR Green master Mix and 0.08 µl reverse transcriptase enzyme mix (Power SYBR Green RNA-to-CT 1-Step Kit; Applied Biosystems), 0.2 µl each of 10 µM forward and reverse primers, and 20 ng RNA per reaction. The QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific, Waltham, Massachusetts) was used under the following cycling conditions: reverse transcription at 48°C for 30 min, denaturation and activation of SYBR polymerase at 95°C for 10 min, followed by 40 cycles of amplification (95°C for 15 s, 60°C for 1 min). Expression values were normalized to β-actin and analyzed using the 2−ΔΔCT method (Livak and Schmittgen, 2001). For the developmental expression of wfikkn1, the 2.5 hpf time point served as the calibrator. For all other experiments, the DMSO treatment of WT zebrafish served as the calibrator.
Each treatment consisted of 2–4 replicates. Results were statistically analyzed using R (version 3.6.3) within RStudio (version 1.1.456), and graphed using the ggplot2 package (version 3.3.3; Wickham, 2016). The data were log2 transformed and assessed for normality and equal variance using the Shapiro-Wilk test and Levene’s test, respectively. Statistical significance was determined using a Type I 1-way ANOVA with a post hoc Tukey (TCDD concentration response) for data that were normal, or a Kruskal-Wallis test followed by Dunn’s test (developmental expression of cyp1a and wfikkn1) for data that failed normality testing. For the 2-factor experiments (ahr2-dependence), a Type I 2-way ANOVA with a post hoc Tukey’s test was used.
Sample Preparation and RNA Sequencing
WT and wfikkn1 mutant zebrafish embryos were exposed to 0.1% DMSO or 1 ng/ml TCDD as described above. Total RNA was isolated from pooled groups of 9 zebrafish embryos at 48 hpf with 4 replicates per treatment group. The 48-hpf time point was chosen to acquire a snapshot of the transcriptome in the absence of wfikkn1 that has been deleted throughout development, before the manifestation of behavioral phenotypes beginning at 120 hpf. RNA integrity was confirmed (RIN score > 9) using a Bioanalyzer 2100 (Agilent, Santa Clara, California). Samples were submitted to Oregon State University’s Center for Genome Research and Biocomputing (Corvallis, Oregon) for library preparation and sequencing. mRNA was polyA-selected using the Robotic PolyA Enrichment Library Prep and libraries were prepared with the Robotic Stranded RNA Library Prep kit (WaferGen Biosystems, Fremont, California). Single-end 100-bp sequencing was conducted with an Illumina HiSeq 3000 sequencer. Raw sequence reads were filtered based on Illumina quality scores and their quality was checked prior to processing using FastQC (version 0.11.7). Reads were trimmed using Cutadapt version 1.8.1 to remove the adapter (AGATCGGAAGAGCA) with no other options (Martin, 2011). All sequencing data and processing details have been deposited in the NCBI Gene Expression Omnibus (GSE172418).
Sequence Mapping, Transcriptome Assembly, and Statistical Analyses
The trimmed files were verified and aligned using HISAT2 (version 2.2.0; Siren et al., 2014) with default options against the Zebrafish build 11 genome (D. rerio Ensembl GRCz11 release 97; http://ftp://ftp.ensembl.org/pub/release-97/fasta/danio_rerio/dna/; last accessed December 1, 2021). The number of reads mapped to the reference genome was sorted and converted to binary alignment files using Samtools with default options (version 1.10; Li et al., 2009). Read counts per gene were estimated using HTSeq with default options (version 0.12.4; Anders et al., 2015) with the GRCz11 Ensembl GTF annotation (release 97; http://ftp://ftp.ensembl.org/pub/release-97/gtf/danio_rerio/Danio_rerio.GRCz11.97.gtf.gz; last accessed December 1, 2021). Normalization of counts and differential gene expression analysis was conducted using Bioconductor’s edgeR (Robinson et al., 2010), following a previously developed workflow (Chen et al., 2016) using RStudio (version 1.1.456). The custom R script (version 4.0.3) was modified from a previously published study (Garcia et al., 2018b). Briefly, edgeR (version 3.22.5) was used to determine differentially expressed genes (DEGs), and multidimensional scaling was leveraged to identify outliers. One outlier each from the WT_DMSO and WT_TCDD treatments was removed, resulting in 3 replicates each for these 2 treatments, and 4 replicates each for the mut_DMSO and mut_TCDD treatments for a total of 14 samples (n = 3–4). Genes were filtered to exclude those with low counts across all 14 libraries (20–27 million reads), only keeping genes that were expressed in a minimum of 3 samples with a minimum read count of 15 (Chen et al., 2016). This corresponded to a cutoff of 0.64296 for the average counts per million reads per sample. Filtered genes were normalized using the trimmed mean of M values (TMM) method to eliminate composition biases between libraries. Principle components analysis (PCA) was performed in RStudio (version 1.1.456) and visualized using ggplot2 (version 3.3.3). Differential expression of WT vs. wfikkn1 mutants (mut) exposed to DMSO was calculated using edgeR which uses the negative binomial generalized linear model extended with quasi-likelihood estimates to fit the gene count data accounting for gene-specific variability, the Cox-Reid profile-adjusted likelihood method for estimating dispersions, and an empirical Bayes F-test (with the option, “robust = TRUE”) for differential expression. All calculated p values were adjusted for false discovery using the Benjamini-Hochberg (BH) method. We enlisted a log2 (fold change) (log2FC) cutoff of 1 for all genes, and those with a BH-adjusted p ≤ .5 were considered significantly DEGs.
Pathway and Interaction Analyses
To evaluate the functional role of wfikkn1 on the 48-hpf zebrafish transcriptome, we identified the significant expression changes in the mut_DMSO samples compared with WT_DMSO. We conducted biological process network enrichment using the MetaCore GeneGo software (version 19.3, build 69800; Clarivate Analytics), as previously described (Garcia et al., 2018b; Haggard et al., 2016). To make the data compatible with MetaCore, the biomaRt package (version 2.40.4) was used to assign the human orthologs of the zebrafish genes (Durinck et al., 2005, 2009), which were used for all downstream analyses. 74.99% of all zebrafish genes were converted to their human orthologs. Processes with a false discovery rate (FDR)-adjusted p < .05 were considered significant. The Enrichment Ratio was manually calculated as the intersection size divided by the expected number of terms in each network. Interaction analysis was conducted using a MetaCore network building algorithm, as described previously (Haggard et al., 2016) with some modifications. MetaCore connects genes that have been shown to interact with each other based on the manually curated MetaCore knowledgebase. The network was generated using the Shortest Paths algorithm allowing for 2-step path through all supplied genes. We included AHR and WFIKKN1 in the algorithm, and the network was visualized using Cytoscape (Shannon et al., 2003).
To understand how the lack of Wfikkn1 alters TCDD-induced gene expression changes in 48-hpf zebrafish, the union of the DEGs between DMSO and TCDD in the WT and wfikkn1 mutants was determined. The TMM-normalized, regular log-transformed gene values for each replicate of all 4 treatment groups were normalized to the mean of the WT_DMSO (control) group. A heatmap with hierarchical clustering was created using the R package gplots (version 3.1.1; Warnes et al., 2016) to identify the changes in gene expression between DMSO and TCDD in the 2 zebrafish genotypes. Biological process network enrichment analyses were conducted on the human orthologs of the DEGs, as described above. To explore interactions of genes that were differentially expressed in opposite directions in WT zebrafish compared with the wfikkn1 mutants upon TCDD exposure, we used the MetaCore “Transcriptional Regulation” network building algorithm that allows for only 1 step between the genes in the input list and potential TFs. We also included AHR and WFIKKN1 into the network building algorithm since none of the zebrafish ahrs or wfikkn1 were expressed in opposite directions in the WT and mutant zebrafish. From the top 30 enriched TF networks identified by the algorithm, we identified the DEGs in our input gene list that was shown to directly interact with AHR and extracted their predicted TFs. The network was visualized using Cytoscape (Shannon et al., 2003).
Proteomic Analysis
Sample Collection and Deyolking
At 48 hpf, WT and wfikkn1 mutant morphologically normal larvae (n = 6) were transferred to 1.5-ml Eppendorf tubes in EM and euthanized by placing on ice for 15 min. EM was carefully removed, and the individual zebrafish were deyolked with mechanical disruption, as described previously (Link et al., 2006) with slight modifications. Briefly, 200 µl of ice-cold ½ Ginzburg Fish Ringer with Calcium and PMSF was added to each tube. Yolk sacs were disrupted by pipetting up and down with a 20-µl pipette tip until the yolk sac was disrupted. Tubes were vortexed briefly to dissolve the yolk and centrifuged at 1000 × g for 30 s. After supernatant was removed and discarded, embryos were washed with HEPES buffer and PMSF (vortexed and centrifuged). Supernatant was discarded, and tubes were placed on ice.
Extraction and Digestion
To extract embryos for proteomic analysis, solvent (100 µl 90:10 v/v methanol:water) with protease inhibitors (cOmplete protease inhibitor cocktail with EDTA, Millipore Sigma) was added to each tube. Embryos were homogenized with 0.5 mm zirconium oxide beads using a counter-top bullet blender (Next Advance) for 3 min at speed 8 and placed on ice for 3 min. This was repeated for a total of 3 times. Solution was transferred to new 1.5-ml tubes and combined with 100 µl bead wash solution: 90:10 v/v methanol:water with ProteaseMax Trypsin Enhancer detergent (Promega). Tubes were placed on ice. Protein was quantified using the BCA Protein assay (ThermoFisher).
To reduce the disulfide bonds of the proteins, the samples were incubated at 56°C for 1 h with 5 mM dithiothreitol (ThermoFisher). Then, samples were incubated with 10 mM iodoacetamide (MilliPore Sigma) for 1 h at room temperature in the dark in order to carbamidomethylate cysteine residues. Samples were digested overnight at 37°C using Trypsin Gold (Mass Spectrometry Grade, Promega). After digestion, samples were spun down at 12 000 × g for 30 s to collect condensate, and digestion was stopped by addition of 0.5% (v/v) trifluoroacetic acid (37°C incubation for 45 min). We cleaned the samples using Oasis HLB cartridges (Waters). After solid-phase activation by pure methanol, approximately 6 µg of protein per sample was loaded, washed with 5% methanol, eluted with pure acetonitrile, and transferred to LC vials.
Mass-Spectrometry and Data Analysis
A Waters nanoAcquity UPLC system (Waters, Milford, Massachusetts) was coupled online to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Peptides were loaded onto a trap 2G nanoAcquity UPLC Trap Column (100 mm, 50 mm, and 5 µm) at a flow rate of 5 μl/min for 5 min. The separation was performed on a commercially available Acquity UPLC Peptide BEH C18 column (100 µm, 100 mm, and 1.7 µm). Column temperature was maintained at 37°C using the integrated column heater. Solvent A was 0.1% formic acid in liquid chromatography-mass spectrometry (LC-MS) grade water, and solvent B was 0.1% formic acid in LC-MS grade acetonitrile. The separation was performed at a flow rate of 0.5 μl/min and using linear gradients of 3–10% B for 3 min, 10–30% B for 2 min, 30–90% B for 3 min, 90% B for 1 min, 95–3% B for 3 min, and 3% B for 8 min. Total method length was 120 min. The outlet of the column was connected to the Thermo Nanospray Flex ion source and +2300 V were applied to the needle.
MS1 spectra were acquired at a resolution of 120 000 (at m/z 200) in the Orbitrap using a maximum IT of 50 ms and an automatic gain control target value of 2e5. For MS2 spectra, up to 10 peptide precursors were isolated for fragmentation (isolation width of 1.6 Th, maximum IT of 10 ms, AGC value of 2e4). Precursors were fragmented by HCD using 30% normalized collision energy and analyzed in the Orbitrap at a resolution of 30 000. The dynamic exclusion duration of fragmented precursor ions was set to 30 s.
All proteomics raw data have been uploaded to ProteomeXchange with accession number: PXD032309. Raw files were processed by Thermo Proteome Discoverer 2.3. Precursor ion mass tolerance was set to 5 ppm, whereas fragment ion mass tolerance was 0.1 Da. The SequestHT search engine was used to search against the zebrafish proteome (downloaded January 2021, 46 897 proteins) from Uniprot (Swissprot) database. b and y ions only were considered for peptide spectrum matching. Minimum peptide length was set to 6 amino acids, and peptide spectrum matches were grouped together into peptide groups. Peptide groups were then assembled into corresponding proteins. MS1 precursor quantification was used for label-free quantitation of the peptides. Protein abundances were calculated as a sum of abundances of unique peptides detected. p values adjusted by a BH FDR correction engine were calculated for evaluation of statistical significance of the differences between WT samples and wfikkn1 mutants.
Significant proteins were further filtered by considering only the ones that were expressed in at least 3 of the 6 biological replicates of either the WT or wfikkn1 mutant zebrafish and absent in all replicates of the other genotype. Proteins were converted to their corresponding gene IDs and human orthologs using the biomaRt package (version 2.40.4) on R (47.8% conversion), and functional enrichment was carried out using MetaCore’s (version 19.3, build 69800) Process Networks analysis.
Behavior Analyses
Larval Photomotor Response
The Viewpoint Zebrabox (Viewpoint Behavior Technology) was used to evaluate photoinduced larval locomotor activity at 120 hpf in 96-well plates. Each plate consisted of 24 larvae/treatment, and there was a total of 4 plates (n = 96). Each plate was placed in a Zebrabox, and the video tracking protocol on the Viewpoint Zebrabox software was used to track total larval movement over 3 alternating light/dark cycles. Each cycle consisted of 3 min in visible light (1000 lux) and 3 min in the dark (infrared light). The integration time was set to 6 s bins, and raw data files were processed using custom R scripts (version 4.0.3) to average the total distance traveled for each integration. Larvae that exhibited mortality or any morphological malformations at 120 hpf were excluded from the LPR analysis.
Juvenile Assays
WT and wfikkn1 mutant embryos were exposed to 0.1% DMSO or 50 pg/ml TCDD and raised to 120 hpf as described above. At 120 hpf, the larvae were placed on the recirculating water system and raised at a density of 1 larva/28 ml until juvenile behavior studies. Once an assay was completed, the zebrafish were returned to their tanks.
Mirror response assay (28 dpf)
The 28-dpf mirror response assay was conducted as described previously (Shen et al., 2020). Briefly, individual zebrafish (n = 48) were transferred from their tanks to individual chambers of a custom, 3D printed, 24-chamber plate with 2 ml laboratory fish water. Each chamber had a 2 mm thick acrylic mirror on 1 side, with the other 3 sides made of white polyacetic acid. Each chamber was placed in a Zebrabox, and the video tracking protocol on the Viewpoint Zebrabox software was used to track zebrafish location integrated into 30 s bins over the 10 min duration of the assay. A 3 mm wide arena adjacent and along the mirror was designated as the “mirror zone.” The average percent time spent in the mirror zone by the fish was calculated, and data collected from minute 7.0 to 9.5 (2.5-min duration) were statistically analyzed.
Shoaling assay (28 dpf)
The 28-dpf shoaling assay was conducted as described previously (Shen et al., 2020). Four zebrafish per treatment (n = 12) were placed in 50 ml laboratory fish water in a custom 3D-printed chamber. Each chamber was placed in a Zebrabox, and the video tracking protocol on the Viewpoint Zebrabox software was used to record the swimming behavior of the zebrafish in the light every 30 s for 7.5 min. Data were statistically analyzed from minute 3.5 to 5 (1.5-min duration). The nearest neighbor distance (NND) and interindividual distance (IID) were calculated to investigate shoaling behavior or tightness of the shoaling group. NND is the average distance between the nearest zebrafish to each of the 4 zebrafish in the shoal, and IID is the average distance between each of the 4 zebrafish to the other 3 zebrafish in the shoal (Miller and Gerlai, 2012). Average swim speed was recorded to assess motor function.
Adult Assays
The adult behavior assays were conducted over a 2-week period when zebrafish were approximately 6 months old. Zebrafish were raised in groups of 16 (8 males and 8 females) in 2.8-l tanks until each assay was conducted and returned to the tanks after the completion of the assay.
Shoaling assay
Groups of 4 adult zebrafish (2 males and 2 females; n = 16) were tested for their shoaling behavior. The assay was conducted in 1.8-l shoaling tanks, as described previously (Knecht et al., 2017). Briefly, zebrafish were allowed to swim for 30 min, and movement was captured using Q-See cameras and Noldus Media Recorder software (Leesburg, Virginia). The Viewpoint Zebrabox software was used to analyze movement of individual zebrafish. Similar to the juvenile shoaling assay (described above), NND, IID, and speed were computed every minute.
Free swim assay
After 30 min of the shoaling assay, each individual zebrafish was separated into a new 1.8-l tank and recorded for an additional 30 min. Total swim distance was calculated for each zebrafish, measured every minute.
Statistical Analyses for Behavior Assays
The normality of all neurobehavior data was checked using normality q-q and residual plots. Data from all assays were normal apart from the 28-dpf mirror assay. However, the sample size (n ≥ 42) was high enough to tolerate deviance from normality. Prior to analysis, any zebrafish that showed no movement throughout the duration of any assay was removed from the analysis.
For the larval assay (LPR), all 3 light/dark cycles were analyzed. Statistical significance (p < .05) was quantified using a Type III ANOVA test to determine the presence of an interaction between the wfikkn1 mutation and TCDD exposure.
For juvenile and adult zebrafish assays, the acclimation period for the control (WT_DMSO) was determined using the Friedman test (p < .05). Data were analyzed using the Type III ANOVA test (p < .05) to determine the effect of the mutation, the chemical exposure, and any interactions between the 2. Custom R scripts were used for data analyses of all assays.
RESULTS
wfikkn1 Is Highly Induced in Parallel With cyp1a Expression
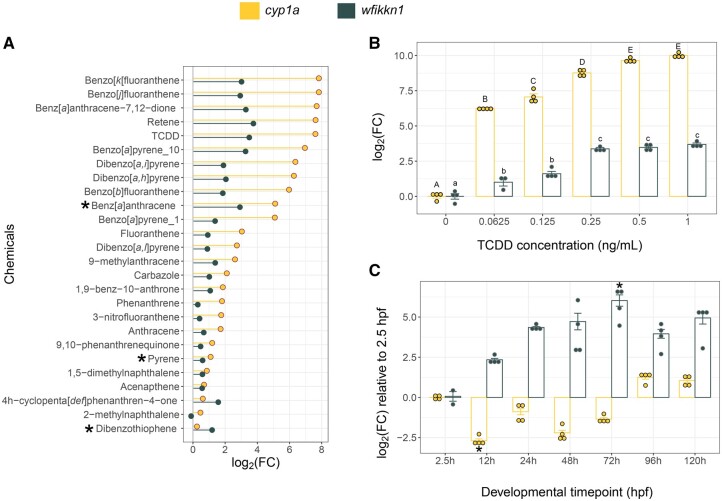
wfikkn1 was first identified as being highly induced in 48-hpf zebrafish exposed to the PAHs benz[a]anthracene, dibenzothiophene, and pyrene (Goodale et al., 2013). The same study showed that expression of the transcript had a similar temporal pattern to the induction of cyp1a, and similar to cyp1a, wfikkn1 was also not significantly induced at 24 hpf. When we compared multiple microarray and RNA sequencing datasets from 48-hpf zebrafish exposed to a variety of chemicals (Goodale et al., 2013; Shankar et al., 2021), we discovered that both wfikkn1 and cyp1a were highly induced by similar PAHs and TCDD (Figure 1A). Additionally, while cyp1a expression was generally higher than wfikkn1, exceptions included dibenzothiophene and 4 h-cyclopenta[d, e, f]phenanthren-4-one. Of the datasets that we analyzed for cyp1a and wfikkn1 expression at 48 hpf, only benzo[a]pyrene data came from 2 concentrations, 1 and 10 µM. Both genes had higher benzo[a]pyrene-associated expression at 10 µM. This suggested a concentration-dependent expression profile for wfikkn1. To test this hypothesis, we exposed WT zebrafish to a broad concentration range of TCDD (0, 0.0625, 0.125, 0.25, 0.5, and 1 ng/ml) and found that expression of wfikkn1 increased with concentration (Figure 1B). Although wfikkn1 expression was consistently lower than cyp1a, the levels of the 2 transcripts significantly increased in parallel. Expression of cyp1a plateaued from 0.5 ng/ml TCDD, and expression of wfikkn1 plateaued from 0.25 ng/ml TCDD.
Figure 1.
Comparison between cyp1a and wfikkn1 expression in developing zebrafish. A, Comparison of cyp1a and wfikkn1 gene expression from previously published microarray (*) and RNA sequencing datasets collected from 48-h postfertilization (hpf) zebrafish. wfikkn1 expression increases in parallel with cyp1a induction, with chemicals that strongly induce cyp1a, also strongly inducing wfikkn1 expression. All microarray* (benz[a]anthracene, dibenzothiophene, and pyrene) data are previously published (Goodale et al., 2013). All RNA sequencing datasets (remaining chemicals) were reanalyzed with the same data analysis pipeline (Shankar et al., in progress). B, Quantitative reverse transcriptase polymerase chain reaction (RT-qPCR) concentration response of cyp1a and wfikkn1 expression in 48-hpf wild-type (WT) zebrafish embryos (n = 3–4, represented by the individual dots) exposed to 0.0625, 0.125, 0.25, 0.5, and 1 ng/ml 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Expression of both genes increased in a concentration-dependent manner. The control was 0.1% dimethyl sulfoxide, which is listed as 0 ng/ml TCDD. Expression values were determined using the 2−ΔΔCT method, and the normalization control was β-actin. The data were log2-transformed, and data are represented as the mean ± standard error of mean (error bars). Normality was tested using a Shapiro test, and statistical significance for each gene was determined using a 1-way ANOVA followed by a post hoc Tukey test. Uppercase (cyp1a) and lowercase (wfikkn1) letters indicate expression values statistically different (p < .01) from each other. C, RT-qPCR expression of cyp1a and wfikkn1 in developing WT zebrafish (n = 2–4, represented by the individual dots) at multiple time points from 2.5 to 120 hpf in the absence of xenobiotic ahr2 activation. Data analysis was similar to (B), and statistical significance was determined compared with 2.5 hpf for each gene using the Kruskal-Wallis followed by Dunn’s test. * indicates p < .05 compared with respective 2.5-h log2FC.
We investigated the developmental expression of the 2 genes in zebrafish from 2.5 hpf to 120 hpf using RT-qPCR. wfikkn1 expression at 72 hpf was significantly higher than at 2.5 hpf and showed a trend of being higher at all other time points than at 2.5 hpf (Figure 1C). On the contrary, cyp1a expression at 12 hpf was significantly lower than at 2.5 hpf and showed an increasing trend to be higher at 96 and 120 hpf, which is consistent with previous work (Andreasen et al., 2002). In order to capture where the wfikkn1 gene is being expressed, we designed several in situ hybridization probes for the transcript (Thisse and Thisse, 2008) but were unable to capture the location(s) of expression in whole-animal developing zebrafish (data not shown). We hypothesize that either the low abundance of wfikkn1 and/or the highly specific nature of its expression might be contributing to challenges around localizing its gene expression.
Induction of wfikkn1 Expression Is Ahr2-Dependent
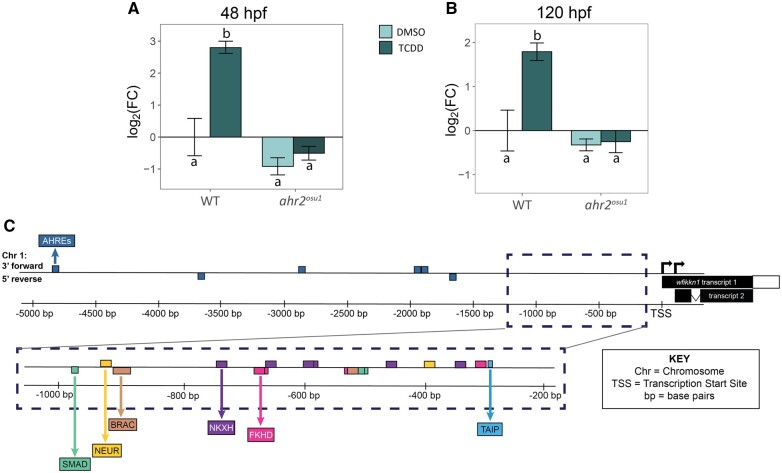
Since wfikkn1 expression increased upon exposure to those PAHs that also induced cyp1a, and in a concentration-dependent manner with TCDD exposure, we hypothesized that activation of Ahr2 was necessary for induction of wfikkn1 expression. We tested this hypothesis in ahr2 mutant and WT zebrafish exposed to 0.1% DMSO or 1 ng/ml TCDD and measured wfikkn1 expression at 48 and 120 hpf. At both time points, TCDD exposure significantly induced wfikkn1 expression in WT but not in ahr2 mutant zebrafish (Figs. 2A and 2B) demonstrating that wfikkn1 induction is Ahr2-dependent for xenobiotic ligands like TCDD. Of note, the background wfikkn1 expression levels in ahr2 mutant fish at both 48 and 120 hpf trended towards being lower than WT background levels at each time point, suggestive of Ahr2’s control on endogenous wfikkn1 expression in developing zebrafish, which should be investigated further in a future study.
Figure 2.
ahr2-dependence of wfikkn1 developmental expression. wfikkn1 expression in 48 h postfertilization (hpf) (A) and 120 hpf (B) wild-type 5D (WT) and ahr2-null (ahr2osu1) whole embryos developmentally exposed to 0.1% dimethyl sulfoxide or 1 ng/ml 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD; n = 3–4 reps/treatment). wfikkn1 expression was significantly increased upon exposure to TCDD only in WT zebrafish, and there was no significant increase in ahr2-null zebrafish at both timepoints. Expression values were analyzed using the 2−ΔΔT method, and β-actin was used as the normalization control. Different letters indicate statistical difference (p < .001, Type I 2-way ANOVA with a post hoc Tukey test). C, Schematic diagram of the wfikkn1 gene locus (not to scale) on the reverse strand of Chromosome 1 of the zebrafish genome. Zebrafish have 2 predicted wfikkn1 transcripts (TSS, transcription start site). The 6 identified aryl hydrocarbon response element consensus sequences, “GCGTC” (Chang et al., 2013; Karchner et al., 2002) in the 5-kb region upstream of the TSS are highlighted in the top panel. The bottom panel shows the top 10 predicted transcription factor binding sites from Genomatix (Cartharius et al., 2005). Abbreviations: NEUR, NeuroD; Beta2, HLH domain; BRAC, Brachyury gene, mesoderm developmental factor; NKXH, NKX homeodomain factors; FKHD, fork head domain factors; TAIP, TGF-β-induced apoptosis proteins).
Upon activation and translocation of the AHR, the AHR-ARNT heterodimer recognizes and binds to AHREs (also known as DREs or XREs). The AHRE, 5′-GCGTG-3′, is a conserved sequence and has been identified in upstream promoter sequences of several downstream target genes of zebrafish and other species (Chang et al., 2013; Karchner et al., 2002). To this end, we conducted a search for putative TF binding sites in the upstream region of the predicted wfikkn1 sequence in zebrafish using MatInspector (version 3.14; Cartharius et al., 2005). MatInspector did not identify putative AHRE sites in the upstream predicted promoter region of the gene; however, we manually detected 6 potential AHREs in the 5 kb bp upstream from the gene (Figure 2C). The top 10 results from MatInspector had a matrix similarity of 1.000, which refers to a 100% match for every nucleotide between the most conserved nucleotides of the binding sequence and the sequence detected upstream of the wfikkn1 gene locus (Figure 2C). These sequences included 3 putative SMAD binding sites: 2 at approximately 500 bp and 1 at approximately 1000 bp from the transcription start site (TSS). There were multiple fork-head box domain factor sequences and a putative binding site for the TGF-β-induced apoptosis proteins at approximately 275 bp upstream from the wfikkn1 TSS.
Creation, Generation, and Validation of the wfikkn1 Mutant Line
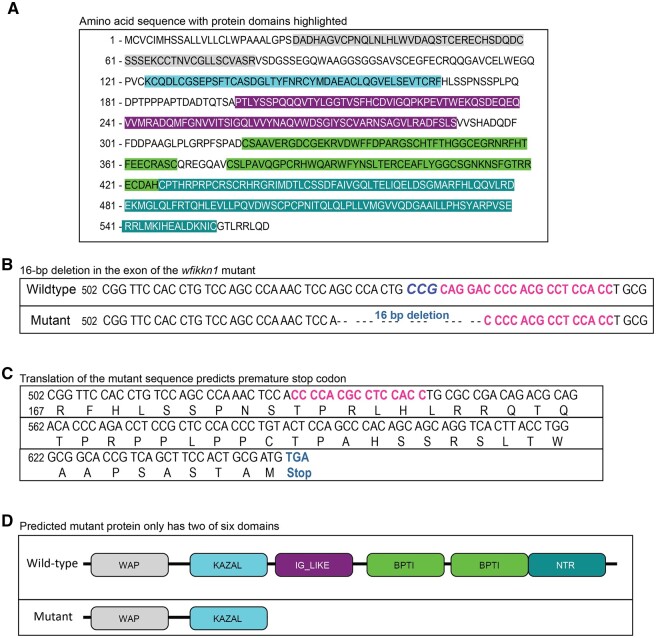
We first confirmed the wfikkn1 RNA sequence in our tropical 5D WT zebrafish. The zebrafish wfikkn1 gene (ENSDARG00000101364) is located on chromosome 1 and has a single exon with 2 predicted transcripts, ENSDART00000167087.2 (transcript 1) and ENSDART00000193546.1 (transcript 2; Figure 2C). There was lack of consensus between the predicted transcripts on Ensembl (Release 102, GRCz11; Yates et al., 2020) and the National Center for Biotechnology Information’s (NCBI) GenBank (Gene ID: 406570; Sayers et al., 2020). The NCBI mRNA sequence (https://www.ncbi.nlm.nih.gov/nuccore/47550770; last accessed April 6, 2022) consisted of an additional 175-bp section upstream of the predicted sequence of transcript 2, which was distinct from the 27-bp addition in transcript 1. We chose the longest of the 3 predicted sequences (NCBI) as the best chance of capturing the protein coding region of the wfikkn1 transcript (see Supplementary Figure 1 for details on amplification methods). The amplified PCR product from our tropical 5D WT zebrafish had >99% sequence similarity to the NCBI-predicted mRNA sequence (Supplementary Figure 1). The deduced Wfikkn1 protein has 564 amino acids (Figure 3A) with a predicted molecular mass of 61.72 kDa. The predicted-protein domains of the Wfikkn1 protein are highlighted: WAP-type “4-disulfide core” (gray), Kazal serine protease inhibitors (blue), IG-like domain (purple), 2 pancreatic trypsin inhibitor (Kunitz or BPTI) domains (light green), and the NTR domain (dark green).
Figure 3.
Schematic diagrams of predicted Wfikkn1 protein in wild-type (WT) zebrafish, and characterization of wfikkn1 mutant CRISPR-Cas9 line. A, Predicted amino acid sequence of Wfikkn1 with protein domains highlighted in different colors (gray: whey acidic protein [WAP]-type “4-disulfide core,” blue: Kazal serine protease inhibitors, purple: immunoglobulin-like domain, light green: 2 pancreatic trypsin inhibitor (Kunitz or BPTI) domains, and dark green: NTR domain). (B) The DNA sequence of the wfikkn1 exon in wild-type (top) and wfikkn1 mutant (bottom) zebrafish. The protospacer adjacent motif site is bolded and in italics (CCG), and the single-guide RNA sequence is bolded and in pink. C, The translated mutant sequence results in a frameshift mutation and is predicted to result in a premature stop codon at amino acid residue 216. D, The truncated protein in the mutant line only contains the WAP and KAZAL domains. A color version of this figure appears in the online version of this article.
In order to investigate the functional role of wfikkn1 in zebrafish, we generated a zebrafish wfikkn1 mutant line (wfikkn1osu2/osu2) using the CRISPR/Cas9 system in WT 5D Tropical zebrafish. Of the 2 sgRNAs we designed, we identified 1 founder female injected with our second sgRNA. She had a 16-bp deletion in the wfikkn1 gene (Figure 3B) in her germ cells that were transferred to her offspring. The 16-bp deletion was predicted to cause a frameshift mutation resulting in a premature stop codon (Figure 3C). The expected truncated mutant protein should only contain the WAP and KAZAL domains (Figure 3D).
For the generation of homozygous wfikkn1 mutant zebrafish, we first identified 5 F1 adult heterozygous zebrafish by sequencing. The fish were incrossed to produce a 25% homozygous population confirmed with sequencing the region around the 16-bp nucleotide deletion. The established homozygous wfikkn1 mutant zebrafish line was screened for possible gene mutations carried over from initial multiplex injections, of which none were found. Additionally, the mutant line appeared to be morphologically normal compared with WT zebrafish, hatched at the normal timing between 48 and 72 hpf, and did not present any reproductive deficiencies (data not shown). Mutant zebrafish also did not have overt deficiencies in larval cartilage structure or adult fin regeneration (data not shown), 2 phenotypic endpoints previously associated with activation of the Ahr2 signaling pathway (Burns et al., 2015; Zodrow and Tanguay, 2003). It is possible that wfikkn1 plays a major role in an organ- or tissue-specific manner that we were unable to observe via the experiments we conducted.
Unbiased Transcriptomics in TCDD-Exposed WT and wfikkn1 Mutant Zebrafish
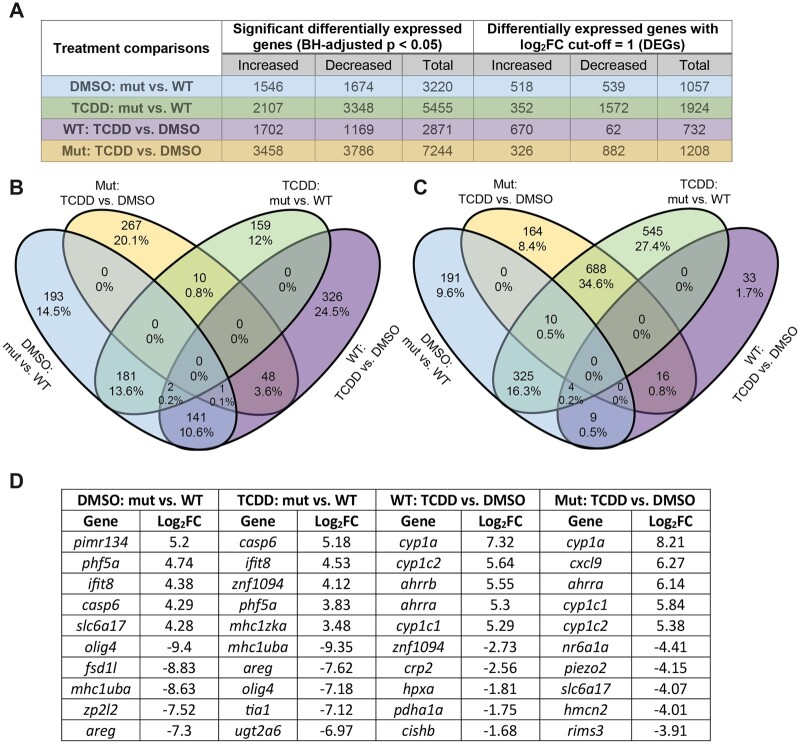
To elucidate the functional role of wfikkn1 in influencing normal and TCDD-induced gene expression changes, we conducted whole embryo transcriptomic profiling of 48-hpf WT and wfikkn1 mutant (mut) zebrafish exposed to 0.1% DMSO or 1 ng/ml TCDD. This led to 4 different treatment comparisons: mut_DMSO versus WT_DMSO, mut_TCDD versus mut_DMSO, mut_TCDD versus WT_TCDD, and WT_TCDD versus WT_DMSO. PCA indicated that the replicate samples of each treatment clustered together, and there was a strong separation between WT and wfikkn1 mutant zebrafish along PC2 (y-axis, 30%; Supplementary Figure 2). Separation between chemicals (DMSO and TCDD) occurred along the PC1 (x-axis, 50%). A total of 15 754 unique significant differentially expressed (BH-adjusted p < .05) genes were identified across all treatments, and there were no significant genes that were common to all treatments. In total, we identified >2500 significant genes between each of the treatment comparisons (Figure 4A). Due to the high number of significant genes, we also enlisted a log2FC = 1 cutoff (Figure 4A) for downstream analyses of DEGs.
Figure 4.
Overview of RNA sequencing data from 48-h postfertilization wild-type (WT) and wfikkn1 mutants (mut) exposed to 0.1% dimethyl sulfoxide or 1 ng/ml 2,3,7,8-tetrachlorodibenzo-p-dioxin. A, Total number of significant differentially expressed genes (DEGs) with a log2FC = 1 cutoff (DEGs) across the 4 treatment comparisons. Venn diagrams showing all DEGs (Benjamini-Hochberg-adjusted p < .05) that had increased (B) or decreased (C) expression in each of the treatment comparisons. D, The top 5 annotated DEGs with most increased and most decreased expressions between each of the treatment comparisons with their log2FC values.
We identified some potentially important trends when increased (Figure 4B) or decreased (Figure 4C) DEGs were considered separately. In any 2 comparisons, at most 13.6% of DEGS had increased expression (between mut_TCDD vs WT_TCDD and mut_DMSO vs WT_DMSO), suggestive of the wfikkn1 mutation having a significant role on the 48-hpf zebrafish transcriptome. A high number of increased DEGs (159 or 12%) and decreased DEGs (545 or 27.4%) were unique to the mut_TCDD versus mut_DMSO comparison, suggestive of a unique transcriptomic response to TCDD in the absence of Wfikkn1. Thus, our overall observations were: (1) Lack of Wfikkn1 caused significant expression alterations in 48-hpf zebrafish, and (2) TCDD exposure affected the 48-hpf zebrafish transcriptome of wfikkn1 mutants differently compared with WT zebrafish. The top 5 increased and decreased DEGs for each treatment comparison are listed in Figure 4D. We made an important observation that the canonical AHR signaling pathway genes including cyp1a, cyp1c1, cyp1c2, and ahrra were present at similar log2FC levels in both WT and wfikkn1 mutant zebrafish when they were exposed to TCDD.
Impact of Lack of Basal wfikkn1 Expression on the 48-Hpf Zebrafish Transcriptome
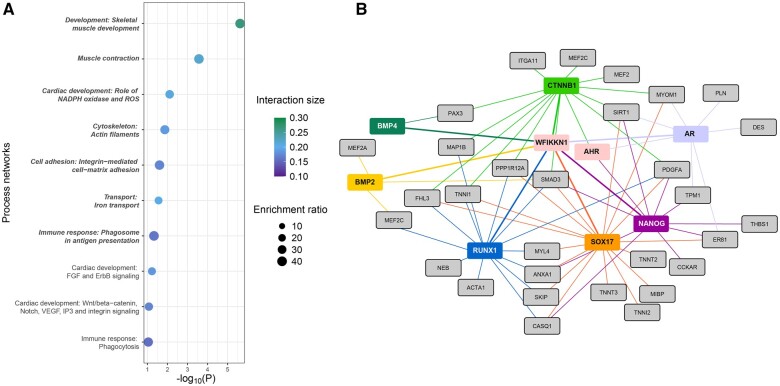
We identified 3220 significant genes of which 1057 were DEGs in mut_DMSO compared with WT_DMSO zebrafish (Figure 4A). Functional enrichment of all significant genes led to 7 significantly altered process networks in the wfikkn1 mutant zebrafish (Figure 5A). The top 20 process networks with their corresponding human gene IDs are in Supplementary Table 3. We observed processes related to skeletal muscle development and muscle contraction as the most significantly enriched in the dataset. To explore how the absence of Wfikkn1 could have a role in the identified muscle-related processes, we generated a network in MetaCore (see Materials and Methods: RNA Sequencing Analyses—Pathway and Interaction analyses) investigating the interactions between the genes in the 2 most enriched pathways. We added both AHR and WFIKKN1 as additional inputs. MetaCore identified 7 proteins that have been predicted to interact with WFIKKN1 and at least one of the genes in the input dataset (Figure 5B). SOX17, RUNX1, CTNNB1 (β-catenin), and NANOG had the greatest number of connections with the genes involved in the muscle-related processes. Importantly, AHR also had interactions with CTNNB1 and NANOG indicating that these 2 TFs may be particularly important hubs mediating the normal function of WFIKKN1 on skeletal muscle development and contraction processes.
Figure 5.
Impact of the lack of wfikkn1 on the 48-h postfertilization zebrafish transcriptome. A, Top 10 MetaCore process network enrichments of human orthologs of differentially expressed genes between WT_DMSO and mut_DMSO. Significant processes (FDR-adjusted p < .05) are bolded and italicized. The interaction size is the number of observed genes from each cluster that belongs to the process network, and enrichment ratio is the ratio of the number of observed genes to the expected genes from each network. The top 2 processes are skeletal muscle development and muscle contraction. B, MetaCore interactome of genes enriched in the top 2 biological process networks (gray). AHR and WFIKKN1 were manually added to the network. Proteins predicted by MetaCore that interact with AHR or WFIKKN1, and the gray genes are depicted in other colors (BMP4, CTNNB1, AR, NANOG, SOX17, RUNX1, BMP2, and BMP4). Thicker colored lines show the direct interactions with WFIKKN1. Abbreviations: WT, wild type; DMSO, dimethyl sulfoxide; FDR, false discovery rate. AHR, aryl hydrocarbon receptor. A color version of this figure appears in the online version of this article.
Role of wfikkn1 in TCDD-Induced Gene Expression Alterations
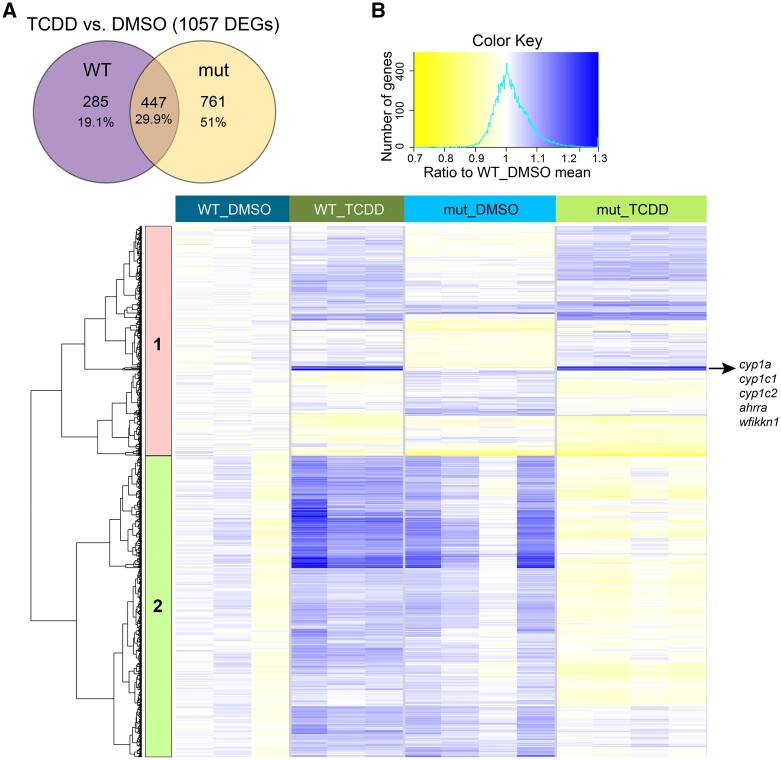
To understand the functional role of wfikkn1 in TCDD-induced gene expression changes, we analyzed the 48-hpf transcriptome of WT and mutant zebrafish exposed to TCDD. WT zebrafish exposed to TCDD resulted in 732 DEGs (Figure 4A). The mut_TCDD compared with mut_DMSO zebrafish had 1208 DEGs. When comparing the gene expression changes in WT and wfikkn1 mutant zebrafish, each exposed to TCDD, 285 DEGs (19.1%) were unique to WT, 761 DEGs (54%) were unique to wfikkn1 mutants, and 447 DEGs (29.9%) were common to both zebrafish strains (Figure 6A). Our data demonstrate that most of the variability was driven by baseline differences in the WT_DMSO and mut_DMSO samples (Supplementary Figure 2 and Figure 4A). Thus, we normalized each of the treatments to a common baseline, WT_DMSO (see Materials and Methods: RNA Sequencing Analyses—Pathway and Interaction analyses) followed by unidirectional hierarchical clustering on all the DEGs significantly altered by TCDD in both the WT and mut samples (Figure 6A). Based on the responses of the genes, we identified 2 major clusters (Figure 6B).
Figure 6.
Effect of the lack of wfikkn1 on the 48-h postfertilization transcriptome when zebrafish are exposed to 1 ng/ml 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). A, Venn diagram of differentially expressed genes in 1 ng/ml TCDD-exposed wild-type (WT) or mut zebrafish compared with the respective dimethyl sulfoxide (DMSO) controls. B, Hierarchical clustering of the union of the genes in (A) whose trimmed mean of M-normalized, regular log-transformed counts have been normalized to the control WT_DMSO. Blue indicates genes that have increased expression and yellow indicates genes that have decreased expression compared with WT_DMSO. We identified two unique clusters, with Cluster 2 consisting of genes that had distinct gene expression changes in the WT zebrafish line compared with the wfikkn1 mutants upon exposure to TCDD. A color version of this figure appears in the online version of this article.
In general, genes in Cluster 1 consisted of genes that mostly responded in the same direction (increased) in the WT_TCDD and mut_TCDD samples relative to their individual normalized DMSO controls. Of note, multiple known Ahr2-regulated genes, including cyp1a, cyp1c1, cyp1c2, and ahrra, were found in this cluster, seen as a bright blue band (Figure 6B). In addition, wfikkn1 was in this cluster of genes and was significantly induced in both WT (log2FC = 4.81, BH-adjusted p = 4.47 × 10−6) and mutant (log2FC = 3.25, BH-adjusted p = 9.92 × 10−4) zebrafish exposed to TCDD, with mutants having nearly a 40% lower log2FC. We found that while ahrrb was highly induced in TCDD-exposed WT zebrafish (log2FC = 5.55, BH-adjusted p = 1.28 × 10−3), its expression was reduced in the wfikkn1 mutants (log2FC = 2.93, BH-adjusted p = 1.47 × 10−3; Supplementary Tables 2A–D), which suggested a potential direct or indirect wfikkn1-ahrrb interaction.
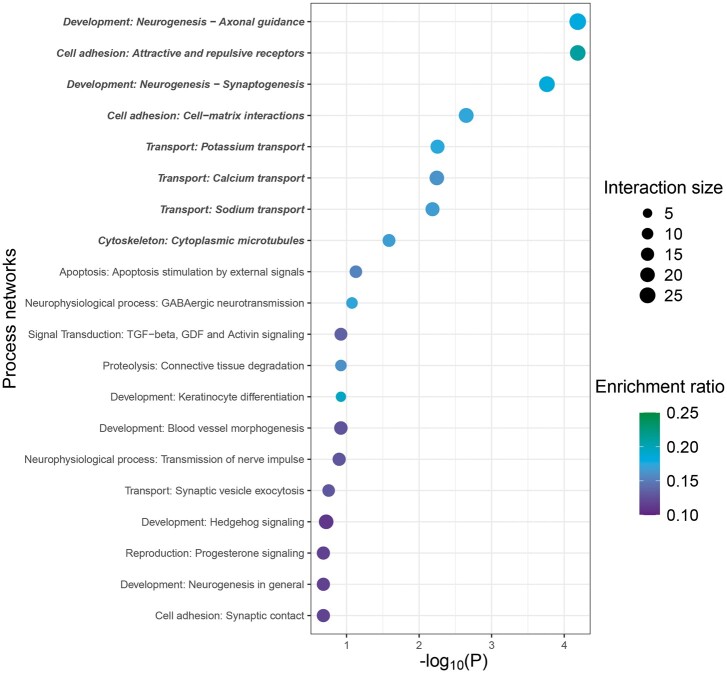
After normalization to respective DMSO controls, genes in Cluster 2 (1208) had opposite patterns of change in expression in WT compared with the wfikkn1 mutants—gene expression increased in WT_TCDD and decreased in mut_TCDD compared with WT_DMSO and mut_DMSO, respectively. Dissimilar expression profiles of these genes were indicative of wfikkn1-dependent TCDD effects on the 48-hpf zebrafish transcriptome. We conducted biological process network enrichment analysis of the human orthologs of the genes in this cluster using the Metacore GeneGo software. Of the top 20 MetaCore process network enrichments, there were 8 significant processes that were related to developmental neurogenesis, cell adhesion, ion transport, and cytoskeleton structure (Figure 7). The remaining pathways were associated with apoptosis, signal transduction, proteolysis, development (keratinocyte differentiation and blood vessel morphogenesis), and reproduction (progesterone signaling; Figure 7).
Figure 7.
Functional enrichment of genes differently differentially expressed in wild-type and wfikkn1 mutant zebrafish upon exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Top 20 MetaCore process network enrichments of genes in Cluster 2. Significant processes (false discovery rate-adjusted p < .05) bolded and italicized. The interaction size is the number of observed genes from each cluster that belongs to the process network, and enrichment ratio is the ratio of the number of observed genes to the expected genes from each network.
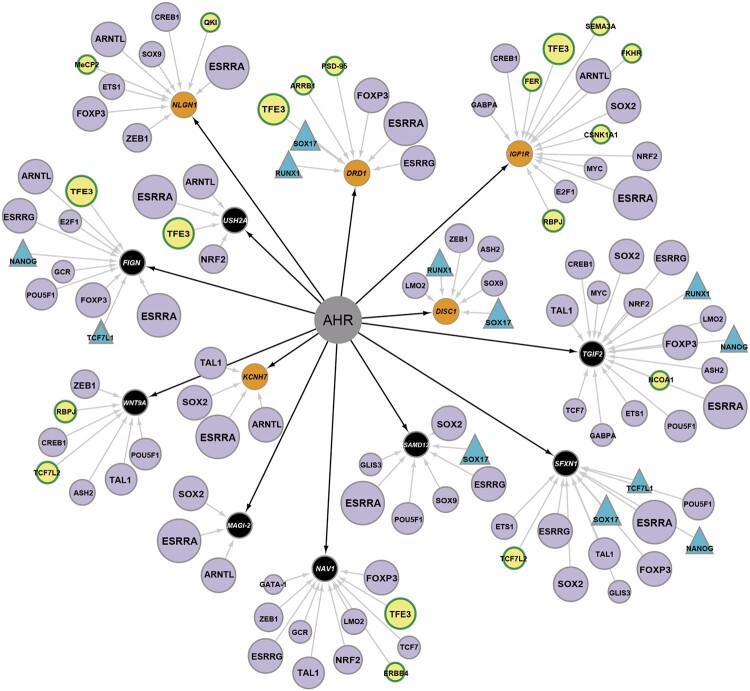
We sought to explore how genes that behaved differently upon exposure to TCDD in WT compared with wfikkn1 mutant zebrafish (Cluster 2) are associated with the AHR signaling pathway. As a first step, we used the Transcriptional Regulation algorithm to identify the top 30 significantly enriched transcriptional factors that directly interact with the genes in our dataset. In this analysis, we added both AHR and WFIKKN1 as additional nodes, since neither were present in Cluster 2. In the second step, we filtered for the genes that have direct interactions with AHR, a well-known known TF. This analysis allowed us to build a network of enriched TFs (purple) that potentially interact with the AHR signaling pathway via the genes altered by TCDD in wfikkn1 mutant zebrafish (orange and black; Figure 8).
Figure 8.
Transcriptional regulation of aryl hydrocarbon receptor (AHR)-associated differentially expressed genes in wild-type and wfikkn1 mutant zebrafish exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Orange circles (NLGN1, DRD1, IGF1R, DISC1, KCNH7) depict the genes from Cluster 2 (Figure 6) and belong to at least one of the enriched biological process networks (Figure 7) that are predicted to have direct interactions with the AHR. Black circles (TGIF2, SFXN1, SAMD12, NAV1, MAGI-2, WNT9A, FIGN, USH2A) are genes in Cluster 2 that do not belong to a biological process network but have a direct interaction with the AHR. The transcription factors (TFs; purple) either belong to the top 30 predicted enriched TFs for the dataset or are found in the dataset (yellow). TF circle sizes refer to the number of predicted targets here, with the largest circles having 10 targets (eg, ESRRA), and the smallest circles having 1 target (eg, QK1 and FER). TFs are shown as triangles interact with wfikkn1 in MetaCore’s manually curated database. A color version of this figure appears in the online version of this article.
We identified 13 unique genes that had direct interactions with AHR, with 5 (orange) belonging to at least one of the significant enriched process networks. Of these genes, NLGN1, DISC1, and DRD1 belonged to one of the nervous system process networks (Supplementary Table 4). KCHN7 was predicted to be involved in the potassium transport network, and IGF1R has a role in signal transduction and reproduction (progesterone signaling). ESRRA, ESRRG, FOXP3, BMAL1, and SOX2 were predicted to interact with several genes in this AHR network, including the genes in the highly enriched biological process networks. In addition, several TFs that interacted with the AHR-associated genes were found in our dataset (yellow). For example, TFE3 was predicted to regulate 5 of the 13 AHR-associated genes. MetaCore also predicted that SOX17, NANOG, TCF7L1, and RUNX1 regulate WFIKKN1 in addition to regulating many of the AHR-associated genes.
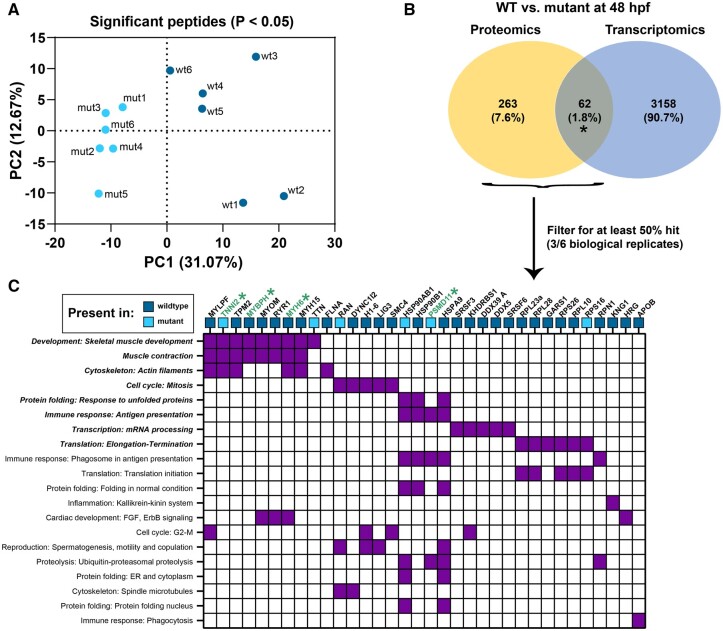
Impact of Lack of Wfikkn1 on Basal Protein Expression in 48-Hpf Zebrafish
The predicted protein sequence of Wfikkn1 in zebrafish, similar to its human ortholog (Trexler et al., 2001), has multiple serine protease inhibitor domains. Thus, we hypothesized that the lack of Wfikkn1’s protease inhibitor activity might significantly alter the zebrafish proteome consequently leading to the significant transcriptomic differences that we identified in our RNA sequencing analyses. To evaluate our hypothesis, we conducted whole-animal mass spectrometry-based proteomics on whole-animal 48-hpf WT and wfikkn1 mutant individual zebrafish and identified 481 significant peptides that differed significantly in abundance and were associated with 326 unique protein isoforms. PCA revealed a clear separation between the WT and mutants (PC1 = 31.07%) when only the significant peptides were considered (Figure 9A). Further, there was only a 1.8% concordance between proteomic and transcriptomic profiles (Figure 9B). To have the most confidence in our proteomic data, we further filtered the proteins for downstream analysis (see Materials and Methods: Proteomic Analysis—Mass-Spectrometry and Data Analysis), and functional enrichment identified significant skeletal muscle and muscle contraction processes. The absence of Wfikkn1 was associated with the absence of several proteins in these categories, including MYLPF, TPM2, MYOM, RYR1, MYH6, and MYH15. Although TNNI2 was detected in mutants but not in the WT, TNNI2, MYBH, and MYH6 were all identified as genes significantly altered in their mRNA expression between the WT and zebrafish mutants (Supplementary Tables 2A–D).
Figure 9.
Whole-animal proteomic comparison between wild-type (WT) and wfikkn1 mutant zebrafish at 48 hpf. A, PCA plot of 6 biological replicates of the wild-type (wt) and wfikkn1 (mut) significant peptides (false discovery rate [FDR] p < .05). B, Comparison between genes of proteins predicted from significant peptides (FDR p < .05; proteomics) and significant differentially expressed genes (FDR p < .05; transcriptomics). C, Heatmap showing MetaCore process networks (significant processes are bolded) on the y-axis, and proteins from proteomic and transcriptomic (*) datasets on the x-axis. Blue boxes represent proteins expressed in at least 3 biological replicates of WT or mutants and absent in all replicates of the other genotype.
wfikkn1 Mutant Zebrafish Do Not Have Altered Neurobehaviors in Response to TCDD at All Life Stages
To the best of our knowledge, the wfikkn1 CRISPR-Cas9 mutant line that we generated is the first of its kind in any species. To determine the functionality of the Wifikkn1 protein in our mutant line, we first exposed WT and wfikkn1 mutant zebrafish to 0.1% DMSO or a concentration range of TCDD (Garcia et al., 2018b) at 6 hpf for 1 h. We did not detect a shift in the concentration response curve in the wfikkn1 mutant zebrafish compared with the WT controls, and mutants exposed to 1 ng/ml TCDD had the expected TCDD-induced toxicity phenotype at 120 hpf, including binary yes or no responses for cardiac edema, blood pooling, and a bent axis (data not shown). We also exposed the 2 zebrafish lines to 0.1% DMSO or a low concentration of TCDD (50 pg/ml) and raised the fish until adulthood. We did not expect to see any morphological defects in WT zebrafish at this 50 pg/ml TCDD exposure concentration (Baker et al., 2013), and in general, neither zebrafish line had any visible malformations.
We evaluated the effect of lack of Wfikkn1 on zebrafish neurobehavior responses to TCDD (Figure 10). These assays have not been used previously to measure the effect of TCDD. We were interested in 2 main questions: (1) Does the lack of a functional Wfikkn1 protein alter behavioral responses in the mutants compared with WT zebrafish? (2) Does the lack of Wfikkn1 alter how the zebrafish respond to TCDD exposure? We conducted 5 assays at multiple time points in larval, juvenile, and adult zebrafish.
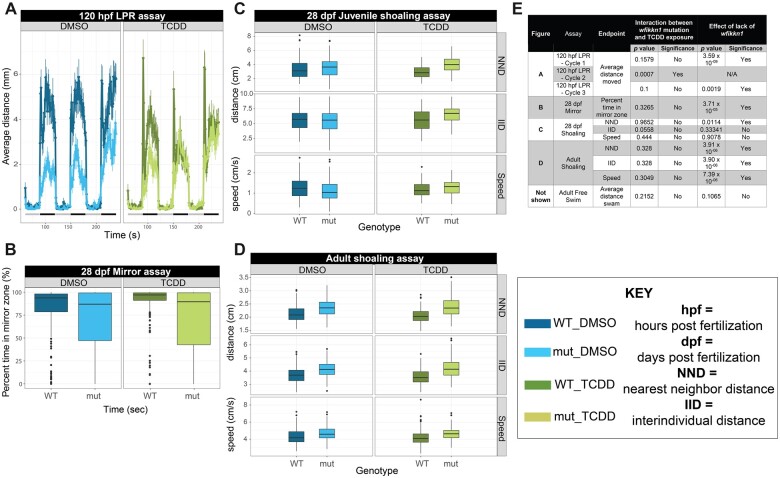
Figure 10.
Behavior analyses of wfikkn1 mutant zebrafish exposed to 50 pg/ml 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). A, 120-h postfertilization larval photomotor response (LPR) showing wfikkn1 mutant zebrafish significantly hypoactive compared with wild-type (WT) zebrafish in light (quiescent) and dark (active) periods of the assay indicated by the gray-black bar at the bottom, respectively. Significant interaction between lack of wfikkn1 and TCDD exposure was detected in Cycle 2. Error bars are a representation of the standard deviation of the mean every 6 seconds. B, 28-dpf mirror assay showing mutant zebrafish spending significantly less time in the “mirror zone” compared with WT zebrafish. Boxplots (for B–D) show spread of the data, with the horizontal line representing each median, and dots representing outliers. 28-dpf juvenile (C) and adult (D) shoaling measured using the nearest neighbor distance (NND), interindividual distance, and speed of zebrafish within shoals of 4 zebrafish. NND was significantly different between mutants and WT in juvenile zebrafish, and all 3 endpoints were significant in adult zebrafish. E, Summary table showing assay details, including p values and corresponding significance (Type III ANOVA).
In general, we concluded that there was no significant interactive effect between the lack of Wfikkn1 and TCDD in our assays conducted at all lifestages of zebrafish (Figure 10E). Reaction norm plots for the assays are presented in Supplementary Figure 3. At 120 hpf, zebrafish were subjected to 3 alternating light-dark cycles in the LPR assay. During all 3 cycles of the assay with DMSO exposure, wfikkn1 mutant zebrafish were significantly hypoactive compared with WT zebrafish (p < .01; Figure 10A). There was a significant interaction between the mutation and TCDD exposure only in 1 of the 3 cycles of the assay, cycle 2 (p = .0007), and thus we concluded a lack of an interactive effect. At 28 dpf, the percent time spent in the “mirror zone” (see Materials and Methods: Behavior Analyses—Juvenile assays—Mirror response assay [28 dpf]) was calculated, and we found that the wfikkn1 mutant zebrafish spent significantly less time (p = 3.71 × 10−5) in the mirror zone compared with WT zebrafish, irrespective of the chemical treatment (Figure 10B).
Shoaling behavior was assessed at 28 dpf and at adulthood. At both time points, there was no significant interactive effect between the wfikkn1 mutation and TCDD exposure for IID, NND, and shoaling (Figures 10C). However, at adulthood, a significant effect from lack of Wfikkn1 (p < .05) on shoaling behavior for all 3 measured endpoints was associated with both DMSO and TCDD treatments (Figure 10D), while at 28 dpf, a significant effect from lack of Wfikkn1 (p < .05) was observed only for NND (Figure 10C). We also evaluated the free swim behavior of individual zebrafish at adulthood and found no effects from lack of Wfikkn1 or from TCDD exposure on the total distance moved by the zebrafish (data not shown). Behaviors of the wfikkn1 mutant zebrafish were also generally more variable than the WT.
DISCUSSION
Characterization of wfikkn1 Expression in Zebrafish
WFIKKN (human chromosome 16) was first discovered in human studies being expressed in several adult organs, including the pancreas, liver, and thymus, but not in the brain and ovary (Trexler et al., 2001), and in fetal lung, skeletal muscle, and the liver (Trexler et al., 2002). Two orthologs of this gene are annotated in humans, rodents, and zebrafish; WFIKKN2 is more easily detected in both mouse and human serum samples (Hill et al., 2003b). To date, the protease inhibitory activity of these multidomain proteins remains unclear, as their functions have been studied only in the context of their interactions with myostatin (growth and differentiation factor-8, and GDF-8) and other TGFβ proteins (Szlama et al., 2010). In zebrafish, wfikkn1 was first identified as being highly induced in 48-hpf zebrafish exposed to multiple PAHs (Goodale et al., 2013). Here, for the first time, we definitively demonstrate wfikkn1’s presence in the Ahr2 signaling pathway and investigate its potential functional role in zebrafish. While wfikkn1 is highly induced by several Ahr2 ligands, its paralog, wfikkn2, was not induced by Ahr2 activation (data not shown). This study used a combination of CRISPR-Cas9-mediated gene editing, transcriptomics and proteomics, and multiple sensitive neurobehavioral assays to functionally characterize this novel AHR signaling gene.
The high correlation between cyp1a and wfikkn1 expression following exposure to environmentally relevant PAHs as well as across multiple concentrations of TCDD strongly suggested that the 2 genes are likely regulated by AHR signaling. Previous work has shown similar concentration-dependent expression of cyp1a and other AHR-regulated genes, including ahrra and ahrrb (Evans et al., 2005). Our lab’s previous study demonstrated via transcriptomics that, although certain PAHs predominantly activate Ahr2 and consequently induce cyp1a expression, each chemical can elicit unique gene expression signatures (Shankar et al., 2019). We found that despite chemical-specific gene expression changes, wfikkn1 was always induced when cyp1a expression was significantly increased. Thus, we hypothesized that wfikkn1, just like cyp1a, is a downstream gene in the Ahr2 signaling pathway. To validate this, we measured wfikkn1 levels in absence of ahr2 and showed a significant reduction of expression compared with WT zebrafish. This was in agreement with our lab’s previous study that showed wfikkn1 expression at 48 hpf was reduced in Ahr2 morphants treated with benz[a]anthracene-7,12-dione (Goodale et al., 2015). Our study has definitively placed wfikkn1 in the Ahr2 signaling pathway. We also identified 6 putative core AHREs (5′-GCGTG-3′) between 1500 and 5000 bp upstream of the predicted wfikkn1 transcriptional start site. One previous study in MCF-7 cells identified that while the AHR/ARNT heterodimer’s peak density was within 1000 bp of the TSS, binding was detected as far as 100 kb from the TSS (Lo and Matthews, 2012) suggesting that wfikkn1 could be a direct target gene of Ahr2. However, future work investigating the functionality of the identified AHREs with either in vitro (Mimura et al., 1999) or zebrafish (Chang et al., 2013) studies will help provide evidence for wfikkn1 being a direct AHR target, since it is possible that wfikkn1 induction is in fact a secondary response to AHR activation.
Lack of Basal wfikkn1 Expression Influences Zebrafish Transcriptome, Proteome, and Behavior
Even though we detected wfikkn1 mRNA expression during development in WT zebrafish, we were surprised by the large number of DEGs (1057) induced at 48 hpf by the lack of Wfikkn1 as we did not see any obvious morphological effects associated with the loss of wfikkn1. It is possible that the 16 bp deletion we generated in the wfikkn1 mutant led to the loss of functional Wfikkn1 protein with a compensatory response (Buglo et al., 2020), or the mutant line was in fact a hypomorph, especially since the premature stop codon that we introduced only deleted 4 of the 6 protein domains. We did, however, identify abnormal behavioral responses in wfikkn1 mutant zebrafish at all stages of development (Figure 10). wfikkn1 mutants were significantly hypoactive in both the light and dark phases of our LPR assay compared with WT zebrafish, an endpoint seen previously due to neurotoxicants (Sun et al., 2016; Truong et al., 2016). LPR has also been used as a direct measurement for visual function (Ahmad et al., 2012). It is possible that wfikkn1 mutants have impaired visual development, since the juveniles also had significant behavior abnormalities in the mirror assay. Iterations of the mirror assay have traditionally been used as a measure of aggression or social interaction (Shen et al., 2020; Way et al., 2015). Although it is too premature to conclude how wfikkn1 alters LPR behavior and mirror response, future research including assays such as the optomotor response assay (Xie et al., 2019), which specifically interrogates vision, might further illuminate wfikkn1’s functional roles in zebrafish. Both xenobiotic Ahr2 activation by TCDD and other Ahr2 agonists (Hill et al., 2003a; Knecht et al., 2017), as well as ahr2-knockout (Garcia et al., 2018a) have all been associated with neurodevelopmental abnormalities in larval and adult zebrafish. These studies highlight the importance of maintaining normal AHR signaling for proper neurodevelopment and sensory system development, but do not identify specific molecular alterations that cause the observed phenotypes. It is plausible that wfikkn1 is a player in some aspect of baseline Ahr2 signaling required for normal zebrafish behavior. We also observed a robust signature of transcripts involved in skeletal muscle development, particularly muscle contraction in our wfikkn1 mutants. While we did not conduct in-depth analyses of muscle structure or function, a reasonable hypothesis is that the associated hypoactive zebrafish behavior is due to impaired skeletal muscle development (Babin et al., 2014). Results from our transcriptomic and behavior analyses were corroborated by the proteomics that also showed significantly enriched muscle-related processes. Several of the proteins that were identified to be differentially abundant between the WT and mutants, including TNNI2 (Ferrante et al., 2011), TPM2 (Dube et al., 2017), and RYR (ryanodine receptor; Hirata et al., 2007), are also important for zebrafish skeletal muscle structure, development, and contraction. A previous study demonstrated the ortholog of wfikkn2 in rodents significantly inhibits myostatin activity, which is a negative regulator of skeletal muscle mass (Hill et al., 2003b). Thus, it is conceivable that Wfikkn1 has a role in muscle developmental processes, evidenced by both our transcriptomic and proteomic data. It should, however, be noted that although our proteomics data suggested a response specific to muscle contraction, a research gap remains regarding this mechanism, specifically, if Wfikkn1’s protease inhibitor domains are involved in biological activity. In addition, there was high variability for most of the peptides identified; therefore, future work should consider experiments with more biological and technical replicates or pooled animals to yield more robust data. Our efforts at conducting single-animal proteomics for the first time in developing zebrafish show us with reasonable confidence that Wfikkn1 may be involved in skeletal muscle regulation.
Our network analysis highlighted different players, such as BMP2 and BMP4 that could mediate wfikkn1’s role in muscle development. The 2 WFIKKN orthologs in humans were found to bind members of the TGFβ family of proteins, including GDF8 and GDF11 without altering their activity (Kondas et al., 2008; Szlama et al., 2010). It is possible that WFIKKN1 is a reservoir for these growth factor proteins which can alter their gradients and consequently their downstream effects. smad3 mRNA expression was significantly altered in wfikkn1 mutants compared with WT zebrafish, and its role as a mediator of TGFβ signaling (Nakao et al., 1997), especially via β-catenin (CTNNB1; Jian et al., 2006; Zhang et al., 2010) might suggest that Wfikkn1 in zebrafish influences muscle development pathways via any of these important signaling mechanisms. Our identification of multiple Smad binding sites in the zebrafish wfikkn1 gene promoter should be functionally investigated in future studies as a potential way by which Wfikkn1 interacts with Smad3 and its associated pathways. Additionally, we observed that 4 h-cyclopenta[def]phenanthrene-4-one and dibenzothiophene, 2 PAHs that did not significantly induce cyp1a, but induced an approximately 2 log2FC, increase of wfikkn1 mRNA. This was different from the remaining PAHs and TCDD that we investigated (Figure 1A), suggesting that ahr2 might not be the only driver of wfikkn1 expression, especially when ahr2 is not activated by a xenobiotic chemical.
Role of wfikkn1 in TCDD-Induced Gene Expression and Behavior Alterations
Our neurobehavioral assays demonstrated that wfikkn1 does not appear to play a role in TCDD-induced behavioral responses with statistical interactions absent for the majority of the endpoints measured. We noted that wfikkn1 mutants at 120 hpf had reduced distance moved in the dark cycles of the LPR assay, which was also true for cycle 1 of the TCDD exposure (Figure 10A), suggesting that wfikkn1 mutants are deficient in the ability to move in response to a sudden dark cycle, independent of AHR2. This conclusion is corroborated by the adult shoaling assay (Figure 10D), where wfikkn1 mutants moved more and faster than WT zebrafish, irrespective of chemical exposure. Thus, while it remains possible that wfkkn1 could alter baseline neurobehavioral responses via AHR2, the lack of interactive effects upon TCDD exposure points to wfikkn1 regulating some aspect of circadian rhythms, muscle movement, ability to coordinate stress response, or sensory system development, in a manner independent of AHR2.
In contrast to the absence of neurobehavioral interactive effects, the lack of wfikkn1 altered TCDD-induced transcriptomics. Of the gene expression changes that were inversely altered in the WT and wfikkn1 mutants with TCDD, the most significantly enriched pathways were associated with neurogenesis, cell adhesion, and ion transport, indicative that lack of Wfikkn1 targeted these functions. Interestingly, NLGN1, DISC1, and DRD1, genes associated with neurogenesis, were predicted by MetaCore to be regulated by AHR. To the best of our knowledge, there is no evidence that NLGN1 and DISC1 are strongly induced by AHR activation, but DRD1 had nearly a 3× increase in its expression in mammalian breast cancer cell culture with TCDD exposure (DuSell et al., 2010). Additionally, all 3 genes were predicted to be associated with ESRRA (estrogen-related receptor alpha), and there is some evidence from previous work suggesting AHR regulation of ESRRA (Tijet et al., 2006). In zebrafish too, estrogen receptor-AHR signaling crosstalk has been demonstrated (Bugel et al., 2013; Shaya et al., 2019) with neurobehavioral signals, making it conceivable that wfikkn1 could be involved in this potential estrogen receptor-AHR interaction upon TCDD exposure during neurodevelopment. Although we detected several signatures for altered neurodevelopment upon TCDD exposure of wfikkn1 mutant zebrafish, the absence of a neurobehavioral interactive phenotype in any of the assays we conducted suggests that if in fact wfikkn1 is involved in TCDD-induced neurobehavior, just the lack of its expression is not enough to produce a neurobehavioral phenotype. Other AHR-regulated targets will need to be investigated in the future to identify the combination of molecular events that collaborate to influence behavioral outcomes. The transcriptional regulation network analysis provides some candidate genes and signaling pathways that can be investigated in future studies to untangle the relationships between Ahr2, Wfikkn1, and neurobehavior in developing zebrafish.
CONCLUSIONS AND FUTURE DIRECTIONS
Overall, we have begun to characterize a novel AHR-regulated gene, wfikkn1, in developing zebrafish using a multiomic approach paired with CRISPR-Cas9 gene editing and neurobehavioral assays in larval, juvenile, and adult zebrafish. We have multiple lines of evidence from our transcriptomics and proteomics studies to believe that Wfikkn1 is involved in skeletal muscle processes in developing zebrafish. Upon xenobiotic AHR activation by TCDD, we did not identify significant statistical interaction between TCDD and Wfikkn1 for neurobehavioral outcomes at all zebrafish lifestages; however, we discovered several significant transcriptomic events that point to wfikkn1’s potential role in neurodevelopment. Several questions remain, the most important being the mechanism by which Wfikkn1 is inducing its significant effects in developing zebrafish and the extent to which the AHR is involved. Additionally, future work should investigate the functions of wfikkn1 specifically upon AHR activation by other AHR ligands that might induce an apparent phenotype. Also, investigation of tissue-specific effects, with a focus on skeletal muscle structures, and in-depth studies on the various molecular signatures identified via our transcriptomics and proteomics experiments could functionally confirm the role of wfikkn1 in zebrafish and provide more resolution on its interactions as they relate to zebrafish development and Ahr2 signaling.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank members of the Sinnhuber Aquatic Research Laboratory (SARL), especially Carrie Barton, Kimberly Hayward, Dante Perone, and Connor Leong for all their help regarding fish husbandry and behavior phenotyping. We would also like to thank Drs Michael Simonich and Subham Dasgupta for careful review of the article. PNNL is operated by Battelle Memorial Institute for the DOE under contract DE-AC05-76RL01830.
FUNDING
The National Institute of Environmental Health Sciences (NIEHS) grants (T32 ES007060, P42 ES016465, and R01 ES030017), and the National Institutes of Health (NIH) grant (S10OD020111).
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflict of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Ahmad F., Noldus L. P. J. J., Tegelenbosch R. A. J., Richardson M. K. (2012). Zebrafish embryos and larvae in behavioural assays. Behaviour 149, 1241–1281. [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq – A python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen E. A., Spitsbergen J. M., Tanguay R. L., Stegeman J. J., Heideman W., Peterson R. E. (2002). Tissue-specific expression of AHR2, ARNT2, and CYP1A in zebrafish embryos and larvae: Effects of developmental stage and 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure. Toxicol. Sci. 68, 403–419. [DOI] [PubMed] [Google Scholar]
- Babin P. J., Goizet C., Raldua D. (2014). Zebrafish models of human motor neuron diseases: Advantages and limitations. Prog. Neurobiol. 118, 36–58. [DOI] [PubMed] [Google Scholar]
- Bailey J., Oliveri A., Levin E. D. (2013). Zebrafish model systems for developmental neurobehavioral toxicology. Birth Defects Res. C Embryo Today 99, 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. R., Peterson R. E., Heideman W. (2013). Early dioxin exposure causes toxic effects in adult zebrafish. Toxicol. Sci. 135, 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambino K., Chu J. (2017). Zebrafish in toxicology and environmental health. Curr. Top. Dev. Biol. 124, 331–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouki R., Coumoul X., Fernandez-Salguero P. M. (2007). The aryl hydrocarbon receptor, more than a xenobiotic-interacting protein. FEBS Lett. 581, 3608–3615. [DOI] [PubMed] [Google Scholar]
- Barton C. L., Johnson E. W., Tanguay R. L. (2016). Facility design and health management program at the Sinnhuber aquatic research laboratory. Zebrafish 13(Suppl 1), S39–S43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiard S., Timme-Laragy A. R., Wassenberg D. M., Cockman C., Di Giulio R. T. (2006). The role of the aryl hydrocarbon receptor pathway in mediating synergistic developmental toxicity of polycyclic aromatic hydrocarbons to zebrafish. Toxicol. Sci. 92, 526–536. [DOI] [PubMed] [Google Scholar]
- Bugel S. M., Tanguay R. L., Planchart A. (2014). Zebrafish: A marvel of high-throughput biology for 21(st) century toxicology. Curr. Environ. Health Rep. 1, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugel S. M., White L. A., Cooper K. R. (2013). Inhibition of vitellogenin gene induction by 2,3,7,8-tetrachlorodibenzo-p-dioxin is mediated by aryl hydrocarbon receptor 2 (AHR2) in zebrafish (Danio rerio). Aquat. Toxicol. 126, 1–8. [DOI] [PubMed] [Google Scholar]
- Buglo E., Sarmiento E., Martuscelli N. B., Sant D. W., Danzi M. C., Abrams A. J., Dallman J. E., Zuchner S. (2020). Genetic compensation in a stable slc25a46 mutant zebrafish: A case for using F0 CRISPR mutagenesis to study phenotypes caused by inherited disease. PLoS One 15, e0230566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger M. K., Glover E., Moran S. M., Walisser J. A., Lahvis G. P., Hsu E. L., Bradfield C. A. (2008). Abnormal liver development and resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA-binding domain of the aryl hydrocarbon receptor. Toxicol. Sci. 106, 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger M. K., Moran S. M., Glover E., Thomae T. L., Lahvis G. P., Lin B. C., Bradfield C. A. (2003). Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J. Biol. Chem. 278, 17767–17774. [DOI] [PubMed] [Google Scholar]
- Burns F. R., Peterson R. E., Heideman W. (2015). Dioxin disrupts cranial cartilage and dermal bone development in zebrafish larvae. Aquat. Toxicol. 164, 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005). Matinspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942. [DOI] [PubMed] [Google Scholar]
- Chang C. T., Chung H. Y., Su H. T., Tseng H. P., Tzou W. S., Hu C. H. (2013). Regulation of zebrafish CYP3A65 transcription by AHR2. Toxicol. Appl. Pharmacol. 270, 174–184. [DOI] [PubMed] [Google Scholar]
- Chen Y., Lun A. T., Smyth G. K. (2016). From reads to genes to pathways: Differential expression analysis of RNA-seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 5, 1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube D. K., Dube S., Abbott L., Wang J., Fan Y., Alshiekh-Nasany R., Shah K. K., Rudloff A. P., Poiesz B. J., Sanger J. M., et al. (2017). Identification, characterization, and expression of sarcomeric tropomyosin isoforms in zebrafish. Cytoskeleton (Hoboken) 74, 125–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. (2005). Biomart and bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. [DOI] [PubMed] [Google Scholar]
- Durinck S., Spellman P. T., Birney E., Huber W. (2009). Mapping identifiers for the integration of genomic datasets with the R/bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuSell C. D., Nelson E. R., Wittmann B. M., Fretz J. A., Kazmin D., Thomas R. S., Pike J. W., McDonnell D. P. (2010). Regulation of aryl hydrocarbon receptor function by selective estrogen receptor modulators. Mol. Endocrinol. 24, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B. R., Karchner S. I., Franks D. G., Hahn M. E. (2005). Duplicate aryl hydrocarbon receptor repressor genes (ahrr1 and ahrr2) in the zebrafish Danio rerio: Structure, function, evolution, and AHR-dependent regulation in vivo. Arch. Biochem. Biophys. 441, 151–167. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero P. M., Hilbert D. M., Rudikoff S., Ward J. M., Gonzalez F. J. (1996). Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 140, 173–179. [DOI] [PubMed] [Google Scholar]
- Ferrante M. I., Kiff R. M., Goulding D. A., Stemple D. L. (2011). Troponin T is essential for sarcomere assembly in zebrafish skeletal muscle. J. Cell Sci. 124, 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii-Kuriyama Y., Mimura J. (2005). Molecular mechanisms of AHR functions in the regulation of cytochrome P450 genes. Biochem. Biophys. Res. Commun. 338, 311–317. [DOI] [PubMed] [Google Scholar]
- Garcia G. R., Bugel S. M., Truong L., Spagnoli S., Tanguay R. L. (2018a). AHR2 required for normal behavioral responses and proper development of the skeletal and reproductive systems in zebrafish. PLoS One 13, e0193484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G. R., Goodale B. C., Wiley M. W., La Du J. K., Hendrix D. A., Tanguay R. L. (2017). In vivo characterization of an AHR-dependent long noncoding RNA required for proper Sox9b expression. Mol. Pharmacol. 91, 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G. R., Shankar P., Dunham C. L., Garcia A., La Du J. K., Truong L., Tilton S. C., Tanguay R. L. (2018b). Signaling events downstream of AHR activation that contribute to toxic responses: The functional role of an AHR-dependent long noncoding RNA (slincR) using the zebrafish model. Environ. Health Persp. 126, 117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland M. A., Geier M. C., Bugel S. M., Shankar P., Dunham C. L., Brown J. M., Tilton S. C., Tanguay R. L. (2020). Aryl hydrocarbon receptor mediates larval zebrafish fin duplication following exposure to benzofluoranthenes. Toxicol. Sci. 176, 46–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodale B. C., La Du J. K., Bisson W. H., Janszen D. B., Waters K. M., Tanguay R. L. (2012). AHR2 mutant reveals functional diversity of aryl hydrocarbon receptors in zebrafish. PLoS One 7, e29346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodale B. C., La Du J., Tilton S. C., Sullivan C. M., Bisson W. H., Waters K. M., Tanguay R. L. (2015). Ligand-specific transcriptional mechanisms underlie aryl hydrocarbon receptor-mediated developmental toxicity of oxygenated PAHs. Toxicol. Sci. 147, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodale B. C., Tilton S. C., Corvi M. M., Wilson G. R., Janszen D. B., Anderson K. A., Waters K. M., Tanguay R. L. (2013). Structurally distinct polycyclic aromatic hydrocarbons induce differential transcriptional responses in developing zebrafish. Toxicol. Appl. Pharm. 272, 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggard D. E., Noyes P. D., Waters K. M., Tanguay R. L. (2016). Phenotypically anchored transcriptome profiling of developmental exposure to the antimicrobial agent, triclosan, reveals hepatotoxicity in embryonic zebrafish. Toxicol. Appl. Pharmacol. 308, 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J. A., Layko D., Nyska A., Hukkanen R. R., Manno R. A., Grassetti A., Lawson M., Martin G., Budinsky R. A., Rowlands J. C., et al. (2016). Aryl hydrocarbon receptor knockout rats are insensitive to the pathological effects of repeated oral exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Appl. Toxicol. 36, 802–814. [DOI] [PubMed] [Google Scholar]
- Henry T. R., Spitsbergen J. M., Hornung M. W., Abnet C. C., Peterson R. E. (1997). Early life stage toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in zebrafish (Danio rerio). Toxicol. Appl. Pharmacol. 142, 56–68. [DOI] [PubMed] [Google Scholar]
- Hill A., Howard C. V., Strahle U., Cossins A. (2003a). Neurodevelopmental defects in zebrafish (Danio rerio) at environmentally relevant dioxin (TCDD) concentrations. Toxicol. Sci. 76, 392–399. [DOI] [PubMed] [Google Scholar]
- Hill A. J., Teraoka H., Heideman W., Peterson R. E. (2005). Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicol. Sci. 86, 6–19. [DOI] [PubMed] [Google Scholar]
- Hill J. J., Qiu Y., Hewick R. M., Wolfman N. M. (2003b). Regulation of myostatin in vivo by growth and differentiation factor-associated serum protein-1: A novel protein with protease inhibitor and follistatin domains. Mol. Endocrinol. 17, 1144–1154. [DOI] [PubMed] [Google Scholar]
- Hirata H., Watanabe T., Hatakeyama J., Sprague S. M., Saint-Amant L., Nagashima A., Cui W. W., Zhou W., Kuwada J. Y. (2007). Zebrafish relatively relaxed mutants have a ryanodine receptor defect, show slow swimming and provide a model of multi-minicore disease. Development 134, 2771–2781. [DOI] [PubMed] [Google Scholar]
- Hoffman E. C., Reyes H., Chu F. F., Sander F., Conley L. H., Brooks B. A., Hankinson O. (1991). Cloning of a factor required for activity of the Ah (dioxin) receptor. Science 252, 954–958. [DOI] [PubMed] [Google Scholar]
- Hofsteen P., Plavicki J., Johnson S. D., Peterson R. E., Heideman W. (2013). Sox9b is required for epicardium formation and plays a role in TCDD-induced heart malformation in zebrafish. Mol. Pharmacol. 84, 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., Muffato M., Collins J. E., Humphray S., McLaren K., Matthews L., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona J. P., Day H. L., Collier T. K., Scholz N. L. (2006). Developmental toxicity of 4-ring polycyclic aromatic hydrocarbons in zebrafish is differentially dependent on ah receptor isoforms and hepatic cytochrome p4501a metabolism. Toxicol. Appl. Pharmacol. 217, 308–321. [DOI] [PubMed] [Google Scholar]
- Incardona J. P., Linbo T. L., Scholz N. L. (2011). Cardiac toxicity of 5-ring polycyclic aromatic hydrocarbons is differentially dependent on the aryl hydrocarbon receptor 2 isoform during zebrafish development. Toxicol. Appl. Pharmacol. 257, 242–249. [DOI] [PubMed] [Google Scholar]
- Jian H., Shen X., Liu I., Semenov M., He X., Wang X. F. (2006). Smad3-dependent nuclear translocation of beta-catenin is required for TGF-beta1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 20, 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafafi S. A., Afeefy H. Y., Ali A. H., Said H. K., Kafafi A. G. (1993). Binding of polychlorinated biphenyls to the aryl hydrocarbon receptor. Environ. Health Perspect. 101, 422–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchner S. I., Franks D. G., Powell W. H., Hahn M. E. (2002). Regulatory interactions among three members of the vertebrate aryl hydrocarbon receptor family: AHR repressor, AHR1, and AHR2. J. Biol. Chem. 277, 6949–6959. [DOI] [PubMed] [Google Scholar]
- Kewley R. J., Whitelaw M. L., Chapman-Smith A. (2004). The mammalian basic helix–loop–helix/pas family of transcriptional regulators. Int. J. Biochem. Cell Biol. 36, 189–204. [DOI] [PubMed] [Google Scholar]
- Knecht A. L., Truong L., Simonich M. T., Tanguay R. L. (2017). Developmental benzo[a]pyrene (b[a]p) exposure impacts larval behavior and impairs adult learning in zebrafish. Neurotoxicol. Teratol. 59, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondas K., Szlama G., Trexler M., Patthy L. (2008). Both WFIKKN1 and WFIKKN2 have high affinity for growth and differentiation factors 8 and 11. J. Biol. Chem. 283, 23677–23684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larigot L., Juricek L., Dairou J., Coumoul X. (2018). AhR signaling pathways and regulatory functions. Biochim. Open 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup. (2009). The sequence alignment/map format and samtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link V., Shevchenko A., Heisenberg C. P. (2006). Proteomics of early zebrafish embryos. BMC Dev. Biol. 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lo R., Matthews J. (2012). High-resolution genome-wide mapping of AHR and ARNT binding sites by Chip-Seq. Toxicol. Sci. 130, 349–361. [DOI] [PubMed] [Google Scholar]
- Ma Q., Lu A. Y. (2007). Cyp1a induction and human risk assessment: An evolving tale of in vitro and in vivo studies. Drug Metab. Dispos. 35, 1009–1016. [DOI] [PubMed] [Google Scholar]
- Mandrell D., Truong L., Jephson C., Sarker M. R., Moore A., Lang C., Simonich M. T., Tanguay R. L. (2012). Automated zebrafish chorion removal and single embryo placement: Optimizing throughput of zebrafish developmental toxicity screens. J. Lab. Autom. 17, 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. [Google Scholar]
- Miller N., Gerlai R. (2012). From schooling to shoaling: Patterns of collective motion in zebrafish (Danio rerio). PLoS One 7, e48865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura J., Fujii-Kuriyama Y. (2003). Functional role of AHR in the expression of toxic effects by TCDD. Biochim. Biophys. Acta 1619, 263–268. [DOI] [PubMed] [Google Scholar]
- Mimura J., Ema M., Sogawa K., Fujii-Kuriyama Y. (1999). Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev. 13, 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos M. A., Vejnar C. E., Beaudoin J. D., Fernandez J. P., Mis E. K., Khokha M. K., Giraldez A. J. (2015). CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao A., Imamura T., Souchelnytskyi S., Kawabata M., Ishisaki A., Oeda E., Tamaki K., Hanai J., Heldin C. H., Miyazono K., et al. (1997). TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 16, 5353–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W., Cavanaugh M., Clark K., Ostell J., Pruitt K. D., Karsch-Mizrachi I. (2020). Genbank. Nucleic Acids Res. 48, D84–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. V., Bradfield C. A. (1996). Ah receptor signaling pathways. Annu. Rev. Cell Dev. Biol. 12, 55–89. [DOI] [PubMed] [Google Scholar]
- Shah A. N., Davey C. F., Whitebirch A. C., Miller A. C., Moens C. B. (2016). Rapid reverse genetic screening using CRISPR in zebrafish. Zebrafish 13, 152–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar P., Dasgupta S., Hahn M. E., Tanguay RL, (2020). A review of the functional roles of the zebrafish aryl hydrocarbon receptors. Toxicol. Sci. 178, 215–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar P., Geier M. C., Truong L., McClure R. S., Pande P., Waters K. M., Tanguay R. L. (2019). Coupling genome-wide transcriptomics and developmental toxicity profiles in zebrafish to characterize polycyclic aromatic hydrocarbon (PAH) hazard. Int. J. Mol. Sci. 20, 2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar P., McClure R. S., Waters K. M., Tanguay R. L. (2021). Gene co-expression network analysis in zebrafish reveals chemical class specific modules. BMC Genomics 22, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaya L., Jones D. E., Wilson J. Y. (2019). CYP3C gene regulation by the aryl hydrocarbon and estrogen receptors in zebrafish. Toxicol. Appl. Pharmacol. 362, 77–85. [DOI] [PubMed] [Google Scholar]
- Shen Q., Truong L., Simonich M. T., Huang C., Tanguay R. L., Dong Q. (2020). Rapid well-plate assays for motor and social behaviors in larval zebrafish. Behav. Brain Res. 391, 112625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siren J., Valimaki N., Makinen V. (2014). Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans. Comput. Biol. Bioinform. 11, 375–388. [DOI] [PubMed] [Google Scholar]
- Sun L., Xu W., Peng T., Chen H., Ren L., Tan H., Xiao D., Qian H., Fu Z. (2016). Developmental exposure of zebrafish larvae to organophosphate flame retardants causes neurotoxicity. Neurotoxicol. Teratol. 55, 16–22. [DOI] [PubMed] [Google Scholar]
- Szlama G., Kondas K., Trexler M., Patthy L. (2010). WFIKKN1 and WFIKKN2 bind growth factors TGFβ1, BMP2 and BMP4 but do not inhibit their signalling activity. FEBS J. 277, 5040–5050. [DOI] [PubMed] [Google Scholar]
- Teraoka H., Okuno Y., Nijoukubo D., Yamakoshi A., Peterson R. E., Stegeman J. J., Kitazawa T., Hiraga T., Kubota A. (2014). Involvement of COX2-thromboxane pathway in TCDD-induced precardiac edema in developing zebrafish. Aquat. Toxicol. 154, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. [DOI] [PubMed] [Google Scholar]
- Tijet N., Boutros P. C., Moffat I. D., Okey A. B., Tuomisto J., Pohjanvirta R. (2006). Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol. Pharmacol. 69, 140–153. [DOI] [PubMed] [Google Scholar]
- Trexler M., Banyai L., Patthy L. (2001). A human protein containing multiple types of protease-inhibitory modules. Proc. Natl. Acad. Sci. U.S.A. 98, 3705–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trexler M., Banyai L., Patthy L. (2002). Distinct expression pattern of two related human proteins containing multiple types of protease-inhibitory modules. Biol. Chem. 383, 223–228. [DOI] [PubMed] [Google Scholar]
- Truong L., Gonnerman G., Simonich M. T., Tanguay R. L. (2016). Assessment of the developmental and neurotoxicity of the mosquito control larvicide, pyriproxyfen, using embryonic zebrafish. Environ. Pollut. 218, 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney G. K., Carrington B., Pei W., Bishop K., Chen Z., Fan C., Xu L., Jones M., LaFave M. C., Ledin J., et al. (2016). A high-throughput functional genomics workflow based on CRISPR/CAS9-mediated targeted mutagenesis in zebrafish. Nat. Protoc. 11, 2357–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C. F. A., Haarmann-Stemmann T. (2017). The aryl hydrocarbon receptor repressor - More than a simple feedback inhibitor of AhR signaling: Clues for its role in inflammation and cancer. Curr. Opin. Toxicol. 2, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes G. R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S., et al. (2016). Gplots: Various r programming tools for plotting data. R package version 3.0.1 ed. Available at: https://cran.r-project.org/web/packages/gplots/gplots.pdf. Accessed December 1, 2021.
- Watson A. J., Hankinson O. (1992). Dioxin- and Ah receptor-dependent protein binding to xenobiotic responsive elements and G-rich DNA studied by in vivo footprinting. J. Biol. Chem. 267, 6874–6878. [PubMed] [Google Scholar]
- Way G. P., Ruhl N., Snekser J. L., Kiesel A. L., McRobert S. P. (2015). A comparison of methodologies to test aggression in zebrafish. Zebrafish 12, 144–151. [DOI] [PubMed] [Google Scholar]
- Westerfield M. 2007. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene, OR: University of Oregon Press. [Google Scholar]
- Wickham H. 2016. Ggplot2: Elegant Graphics for Data Analysis. New York: Springer. [Google Scholar]
- Xie J., Jusuf P. R., Bui B. V., Goodbourn P. T. (2019). Experience-dependent development of visual sensitivity in larval zebrafish. Sci. Rep. 9, 18931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong K. M., Peterson R. E., Heideman W. (2008). Aryl hydrocarbon receptor-mediated down-regulation of sox9b causes jaw malformation in zebrafish embryos. Mol. Pharmacol. 74, 1544–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A. D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., Amode M. R., Armean I. M., Azov A. G., Bennett R., et al. (2020). Ensembl 2020. Nucleic Acids Res. 48, D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Wang M., Tan X., Li T. F., Zhang Y. E., Chen D. (2010). Smad3 prevents beta-catenin degradation and facilitates beta-catenin nuclear translocation in chondrocytes. J. Biol. Chem. 285, 8703–8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zodrow J. M., Tanguay R. L. (2003). 2,3,7,8-tetrachlorodibenzo-p-dioxin inhibits zebrafish caudal fin regeneration. Toxicol. Sci. 76, 151–161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.