Abstract
Regeneration of the mammalian adult skeletal muscle is a well-orchestrated process regulated by multiple proteins and signaling pathways. Cytokines constitute a major class of regulators of skeletal myogenesis. It is well established that infiltrating immune cells at the site of muscle injury secrete cytokines, which play critical roles in the myofiber repair and regeneration process. In the past 10–15 years, skeletal muscle itself has emerged as a prolific producer of cytokines. Much attention in the field has been focused on the endocrine effects of muscle-secreted cytokines (myokines) on metabolic regulation. However, ample evidence suggests that muscle-derived cytokines also regulate myogenic differentiation and muscle regeneration in an autocrine manner. In this review, we survey cytokines that meet two criteria: (a) evidence of expression by muscle cells; (b) evidence demonstrating a myogenic function. Dozens of cytokines representing several major classes make up this group, and together they regulate all steps of the myogenic process. How such a large array of cytokines coordinate their signaling to form a regulatory network is a fascinating, pressing question. Functional studies that can distinguish the source of the cytokines in vivo are also much needed in order to facilitate exploration of their full therapeutic potential.
Keywords: Cytokine, skeletal muscle, regeneration, myogenesis, myogenic differentiation
Introduction
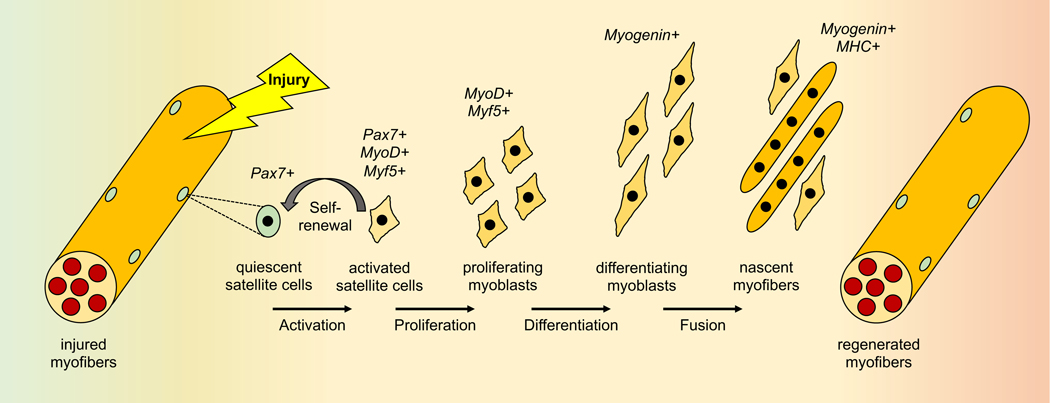
Skeletal muscle is one of the few adult mammalian tissues that can undergo robust regeneration after injury, owing at least partly to a resident population of stem cells that imparts a great regenerative capacity. These myogenic progenitor cells, called satellite cells, exist in a quiescent state under the basal lamina of myofibers until stimulated to divide by muscle injury as a result of acute trauma, toxin, exercise, or diseases. A subset of the activated satellite cells returns to quiescence, allowing for self-renewal of the muscle stem cell pool, while the remaining activated cells proliferate at the site of injury to allow for new myofiber formation or repairing of injured myofibers [1, 2] (Figure 1). Effective regeneration depends on the new myoblasts successfully completing a number of different processes, including expression of many of the same myogenic genes seen in embryonic development, such as the MEF2 and MyoD families of transcription factors [3, 4]. The differentiating myoblasts must also undergo massive cytoskeletal rearrangement, migration, cell-cell adhesion and alignment, and finally membrane fusion in order to form a multinucleated myofiber. Myocyte fusion occurs in two stages that are regulated by distinct molecular pathways [5, 6]. The first stage entails fusion between differentiated myoblasts, leading to small, nascent myotubes. The second stage of fusion occurs between myoblasts and these nascent myotubes, resulting in mature myofibers. This well-orchestrated myogenic process is regulated by numerous intracellular signaling pathways, including p38 [7], Jak/STAT [8], mTOR [9] and PI3K/AKT [10], to name just a few. This review will discuss the roles of cytokines and their signaling in regulating myogenesis, with a special focus on cytokines of muscle cell origin.
Figure 1. The sequential events of injury-induced muscle regeneration.
The expression of Pax7, MyoD, Myf5 and myogenin marks myocytes at different stages. MHC: myosin heavy chain.
The exact definition of what constitutes a cytokine has been evolving since their discovery in purulent exudates in the 1940s [11]. The earliest identified cytokines were of hematopoietic origin and thought to be dedicated to modulating inflammatory responses. Today, it is well established that nearly every cell type can produce secreted factors to some degree, and that even the original cytokines have other sources and pleiotropic functions beyond the immune system. Biochemical, structural, and functional understanding of these secreted factors have led to various, sometimes overlapping, forms of nomenclature. For instance, most of the interleukins and all interferons signal through cytokine receptors associated with the Jak kinases and they are still considered by some to be the only bona fide “cytokines”. Most growth factors (e.g., EGF, IGF, FGF, VEGF, etc.) act through receptor tyrosine kinases. Chemokines, so named for their chemoattractant properties, are ligands for G protein coupled receptors. TGFβ’s and TNFs bind distinct families of receptors known as TGFβ receptors and TNF receptors, respectively. For all practical purposes, the term “cytokine” has been extended to include all small (<30 kDa) secreted proteins that function via cell surface receptors, although there are certainly exceptions to this already broad definition. For instance, almost all TNF family members exist in a transmembrane form in addition to the extracellular soluble form resulting from proteolytic cleavage, both biologically active; some cytokines are larger than the 30 kDa molecular weight cutoff (e.g. IL-12, VEGF).
A large number of infiltrating immune cells at the site of skeletal muscle injury play a critical role in not only clearing the damaged tissue but also promoting muscle regeneration [12, 13]. It is well accepted that these immune cells produce a milieu of secreted factors that modulate every stage of injury-induced muscle regeneration. However, in the past two decades the skeletal muscle has also gained increasing attention as a secretory organ. Originally coined by Pedersen et al. [14], the term “myokine” refers to a cytokine that is released by skeletal muscle cells (often as a result of exercise) into the circulation to exert endocrine effects and metabolic regulation [15, 16]. IL-6, for which the term myokine was first proposed [14], is secreted by exercising muscles and may exert a multitude of beneficial effects on metabolism [15]. At an ever-growing number, myokines have attracted intense interest and been the subject of many excellent reviews (e.g., [15–17]). The autocrine and paracrine functions of muscle- or myogenic cell-secreted cytokines on modulating myogenesis have received less attention. With the advent of single-cell RNA sequencing (scRNAseq), which enables deep interrogation of the transcriptomics of the injured muscle, it has become evident that vast heterogeneity of gene expression exists in the many types and subpopulations of cells contributing to the regeneration process [18–21]. For instance, Oprescu et al. uncovered a subpopulation of muscle satellite cells enriched for immune gene expression in the regenerating muscle, which they termed “immunomyoblasts” [19]. It is reasonable to assume that these cells contribute to cytokine secretion during regeneration. scRNAseq data from other groups have also hinted at the existence of such a cytokine-expressing satellite cell subpopulation [18, 21]. The exact source of, and cell-specific responses to, each of the cytokines released during specific time points in regeneration are fascinating questions we are only just beginning to address. To help clarify the specific contribution of the myocyte lineage to the myogenic process, the current review focuses on muscle cell-derived cytokines that act in an autocrine manner to regulate skeletal muscle differentiation and regeneration.
Skeletal muscle cells are prolific producers of cytokines during differentiation
It has long been known that conditioned medium of differentiating skeletal myoblasts or myotubes in culture contain factors that influence myogenic differentiation, implicating muscle-derived cytokines in the autocrine/paracrine regulation of myogenesis. Whereas muscle tissue homogenates can contain proteins from non-muscle sources, analyses of in vitro muscle cell cultures have provided definitive evidence for muscle cells as prolific secretors of cytokines. About 10 years ago, significant efforts by several research groups led to the first proteome-wide glimpse of the skeletal muscle secretome. Two groups independently performed stable isotope labeling by amino acids in cell culture (SILAC) followed by proteomic analysis of the conditioned media of mouse C2C12 myoblasts at various stages of differentiation [22–24]. While Henningsen et al. identified 635 secreted proteins among which 75 were cytokines/growth factors [24], Chan et al. reported 34 secreted proteins [22]. Both studies concluded that a large portion of the proteins were secreted differentially during the course of differentiation, providing potential support for the idea that those secreted factors may have regulatory roles in the myogenic process. Another commonly used myogenic cell line, the rat L6 myoblast, was studied by Yoon et al., and they identified 254 proteins in the conditioned medium of fully differentiated myotubes, a small subset of which were up- or down-regulated by short-term (5 hours) insulin treatment [25]. Myotubes differentiated from human myoblasts were also analyzed by Norheim et al. and 18 classically annotated secreted proteins were reported [26]. Around the same time as the cluster of proteomic studies, Griffin et al. examined the mRNA expression of 84 chemokines and genes encoding proteins involved in chemokine signaling during differentiation of mouse primary myoblasts. They found 80 of those genes to be expressed, with the majority peaking at the time of myocyte fusion, consistent with a potential role of these chemokines in cell migration necessary for fusion [27]. It is worth noting that Henningsen et al. did not observe a marked correlation between the levels of cytokine proteins in their secretome and the levels of the corresponding mRNAs [24], suggesting that a significant part of regulation of cytokine secretion by muscle cells may be at a post-transcriptional level.
Many of those identified muscle-secreted cytokines remain to be characterized for their potential functions in myogenesis. Nevertheless, the collective evidence raised the intriguing possibility that muscle cell-secreted proteins might have a previously under-appreciated role in modulating myogenic differentiation and muscle regeneration. Our lab conducted a functional screen of 134 mouse cytokines for their impact on myogenic differentiation in C2C12 myoblasts using RNAi, which led us to identify 29 potential regulators of myogenesis in distinct functional groups [28]. Several of those candidates have since been characterized for their myogenic functions and mechanisms (discussed later).
Early discoveries of cytokines with autocrine functions in myogenic differentiation
The concept of autocrine regulation of myogenesis by muscle-secreted factors is not new. Below we briefly describe a few of such cytokines first reported. Where comprehensive reviews have been published, we will refer the readers to those reviews in lieu of citing primary literature.
IGFs
The insulin-like growth factors IGF1 and IGF2 were among the earliest characterized and most extensively studied cytokines that promote myogenesis in an autocrine fashion [29–31]. IGFs are critically involved in skeletal muscle development, hypertrophy, and regeneration [29, 32, 33]. The autocrine function of IGF was first reported 30 years ago in cultured myoblasts found to secrete IGF2 that was indispensable for the initiation of differentiation [34]. Indeed, C2C12 myoblasts secrete high levels of both IGF1 and IGF2 [35, 36], and both stimulate the expression of myogenin, an early marker of myogenic differentiation [29]. Autocrine IGF2 is also a survival factor for muscle cells [37].
FGF, PDGF, HGF
Around the time when IGFs emerged as positive regulators of myogenesis, it became clear that fibroblast growth factors (FGFs) were autocrine inhibitors of myogenic differentiation in vitro [30]. FGF2 (basic FGF) is more potent than FGF1 (acidic FGF) in this regard, both possibly acting as mitogens to prevent cell cycle withdrawal and consequently suppress differentiation, although direct inhibition of myogenic gene expression is also proposed [30]. Several FGFs (at least FGF1, 2, 4 and 6) are expressed in satellite cells and, not surprisingly, they can also stimulate cultured satellite cell proliferation in an autocrine manner [38]. It would therefore appear that FGFs could have dual roles in muscle regeneration – promoting satellite cell expansion and suppressing myogenic differentiation. However, in vivo evidence for FGF function in myogenesis is limited (to be discussed later).
Another growth factor, platelet derived growth factor (PDGF), is expressed by myoblasts and exerts inhibitory effects on myogenic differentiation by promoting cell proliferation and inhibiting cell cycle exit [39–41]. Likewise, hepatic growth factor (HGF) is found to be expressed in satellite cells during rat muscle regeneration [42], stimulate satellite cell proliferation, and inhibit differentiation [43, 44]. Although IGFs are also growth factors that stimulate myoblast proliferation, unlike FGF, PDGF and HGF, the mitogenic activity of IGFs – especially IGF2 [45] – does not interfere with their myogenic activity at the time of differentiation [29].
TGFβ
In 1986 transforming growth factor β (TGFβ) was reported independently by three groups to be an inhibitor of myogenic differentiation in vitro [46–48]. Those initial reports concluded that the effect of TGFβ was independent of cell proliferation. However, it is now confirmed in at least C2C12 cultures that TGFβ stimulates myoblast proliferation through Smad2 activation and consequently suppresses cell cycle exit [49, 50]. On the other hand, TGFβ activation of Smad3 leads to direct suppression of the transcriptional activity of MyoD and possibly other myogenic regulatory factors [51]. Consistent with an autocrine function, TGFβ is found to be secreted by myoblasts in vitro and its mRNA is expressed in mouse skeletal muscles [52].
Myostatin
A member of the TGFβ family, myostatin (also known as growth/differentiation factor-8 or GDF8) was reported in 1997 to be an autocrine inhibitor of muscle mass in mice [53]. Deletion of the myostatin gene leads to drastic muscle hypertrophy, manifesting the double-muscled phenotype in mice, cattle, and humans [54]. The mechanism by which myostatin inhibits muscle growth and myogenesis is multifold and will be discussed later. Two other TGFβ family members closely related to myostatin, activin A and GDF11, are also secreted by muscle cells and inhibit myogenic differentiation [55, 56]. All three ligands signal through the same activin receptors, and the receptor binding is antagonized by these ligands’ physical interaction with muscle-secreted follistatin [55, 57]. Consistent with its anti-myostatin function, follistatin is a potent inducer of muscle hypertrophy [58, 59], although a myostatin-independent mechanism has also been suggested for follistatin’s hypertrophic effects [60].
Regulation of myogenic processes by muscle-derived cytokines
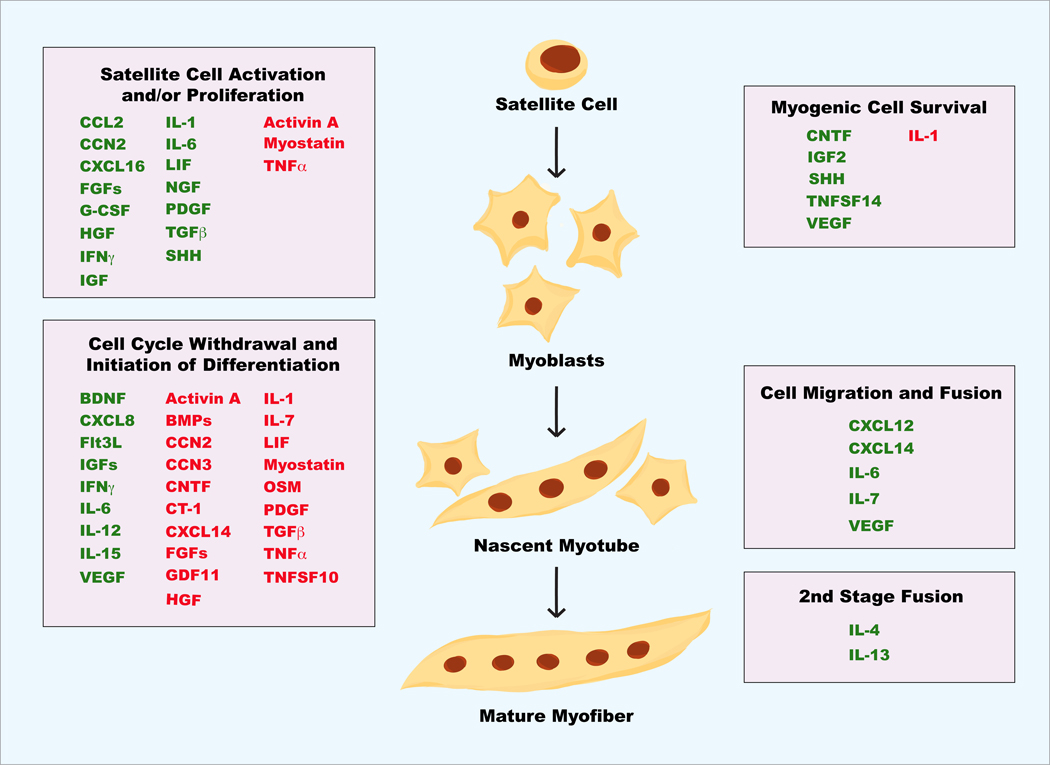
In our literature survey we have set two criteria to look for cytokines that are autocrine regulators of muscle differentiation/regeneration: (a) evidence of expression by muscle cells in culture or in vivo and (b) evidence of myogenic regulation by the cytokine – either endogenous or recombinant. To date, dozens of cytokines have met these criteria as summarized in Table 1. These cytokines encompass several major classes of extracellular ligands based on the types of their cognate receptors, including members of TGFβ family, members of TNF family, chemokines, ligands for receptor tyrosine kinases, and interleukins/interferons that signal through cytokine receptors. Although not all these cytokines have been defined for their mechanisms of myogenic action, there is a sizable number of reports on the cellular mechanisms of autocrine regulation in myogenesis by many of the cytokines. Remarkably, every stage of the myogenic process involves regulation by muscle-derived cytokines, and many cytokines contribute to myogenic regulation at more than one stage (Figure 2).
Table 1. Muscle-derived cytokines and their reported functions in myogenic regulation.
Cytokines reported to be expressed in muscle cells and have a myogenic function are listed. “+” indicates positive regulation of the process whereas “−” indicates negative regulation. Differential expression of these cytokines in muscles or muscle progenitor cells in vivo under the following conditions are indicated: Dev – developing muscle; Reg – regenerating muscle after acute injury; Hyp – satellite cell-dependent compensatory hypertrophy; Dys – DMD or mdx.
| Cytokine | Function in myogenic differentiation | Expression in vivo |
|---|---|---|
| Ligands for RTKs: | ||
| BDNF | + cell cycle withdrawal; + differentiation [81, 82] | Dev [183, 184]; Reg [81] |
| FGF1,2,4,6 | + proliferation; − cell cycle withdrawal; − differentiation [30, 38] | Dev [133, 134, 185]; Reg [135, 136]; Hyp [188]; Dys [186, 187] |
| Flt3L | + cell cycle withdrawal; + differentiation [78] | |
| HGF (Scatter factor) [191] | + proliferation; − cell cycle withdrawal; − differentiation [43, 44] | Dev [189]; Reg [190]; Hyp [188]; Dys |
| IGF1, IGF2 [193]; | + proliferation; + initiation of differentiation; + survival [29–31, 37] | Dev [192]; Reg [138, 139, 190]; Hyp Dys [191] |
| NGF | + satellite cell activation/proliferation [68] | Dev [194]; Dys [195] |
| PDGF | + proliferation; − cell cycle withdrawal; − differentiation [39–41] | Dev [196]; Reg [196]; Dys [197] |
| VEGF | + cell cycle withdrawal; + migration; + differentiation; + survival [84, 112] | |
| Reg [198]; Hyp [199]; Dys [187] | ||
| Ligands for cytokine receptors: | ||
| CNTF | + survival; − differentiation; + dedifferentiation [122, 200] | |
| CT-1 | − initiation of differentiation [201, 202] | Dev [203]; Reg [204]; Hyp [204] |
| G-CSF | + proliferation [205–208] | Dev [209]; Reg [209]; Hyp [210] |
| IFNγ | + proliferation; +/− differentiation [64] [87, 155, 156, 158] | Reg [64] |
| IL-4 | + second-stage fusion [118] | Hyp [162] |
| IL-6 | + proliferation; + migration; + differentiation [66, 110, 111, 173] | Hyp [66]; Reg [164]; Dys [187] |
| IL-7 | + migration; − differentiation [113] | |
| IL-12 | + differentiation [211, 212] | |
| IL-13 | + second-stage fusion [118, 213, 214] | |
| IL-15 | + differentiation [215, 216] | |
| LIF | + proliferation; − cell cycle withdrawal; − differentiation [67, 98, 99] | Hyp [148]; Reg [148, 164]; Dys [98, 187] |
| OSM | + satellite cell quiescence; − differentiation [217, 218] | Dys [187] |
| Ligands for GPCRs: | ||
| CCL2 (MCP1) | + proliferation [23, 61, 219] | Reg [219, 221]; Dys [187, 220] |
| CXCL8 (IL-8) | + differentiation [222] | Dys [223] |
| CXCL12 (SDF1) | + migration; + fusion [27, 224, 225] | Dev [224]; Reg [226]; Dys [227] |
| CXCL14 (BRAK) | − cell cycle withdrawal; + migration [92, 114] | Reg [92]; Dys [220, 227] |
| CXCL16 | + proliferation; + modulating immune cells [62] | Reg [62] |
| TGFβ family: | ||
| Activin A | − proliferation; − differentiation [55, 58, 75] | Dev [55]; Reg [75] |
| BMP4, 7 | − cell cycle withdrawal; − differentiation [88–91] | Dev [228, 229] |
| GDF11 | − differentiation [56, 109] | Dev [230] |
| Myostatin (GDF8) [187] | − proliferation; − initiation of differentiation [73, 74, 108, 109] | Dev [231]; Reg [148]; Hyp [148]; Dys |
| TGFβ | + proliferation; − cell cycle withdrawal; − differentiation [46–50] | Dev [232]; Reg [235]; Dys [233, 234] |
| TNF family: | ||
| TNFα (Cachectin) | − satellite cell activation; − cell cycle withdrawal; −/+ differentiation [87, 155, 156, 158–161] | |
| Dev [236]; Reg [159]; Dys [187, 237] | ||
| TNFSF10 (TRAIL) | − cell cycle withdrawal; − differentiation [93] | |
| TNFSF14 (LIGHT) | + survival; + differentiation [123] | Reg [123] |
| Others: | ||
| CCN2 (CTGF) | + proliferation; − differentiation; + dedifferentiation [238, 239] | Dev [240]; Dys [241, 242] |
| CCN3 (NOV) | − myogenic cell lineage; − cell cycle withdrawal; − differentiation [243] | |
| Dev [244] | ||
| IL-1 | + proliferation; − differentiation; − survival [65, 87, 126] | Reg [235] |
| SHH | + satellite cell activation and proliferation; +/− differentiation; + survival [69, 70, 72] | |
| Dev [245]; Reg [72] | ||
Figure 2. Muscle-derived cytokines involved in various stages of myogenesis.
Green: positive regulators; red: negative regulators.
Satellite cell activation and proliferation
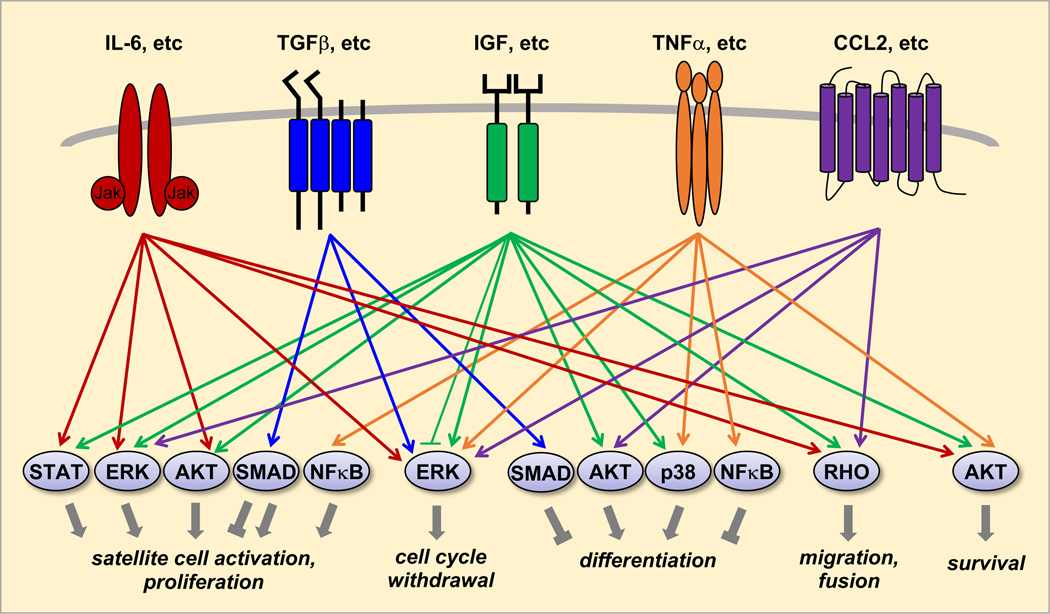
Cytokines that have been reported to positively regulate satellite cell activation and myoblast proliferation in a muscle-autonomous manner include CCL2 [61], CXCL16 [62], FGFs (1, 2, 4, 6 and possibly others) [30, 38], G-CSF [63], HGF [43, 44], IFNγ [64], IGFs [29–31], IL-1 [65], IL-6 [66], LIF [67], NGF [68], PDGF [39–41], TGFβ [49, 50], and Sonic hedgehog (SHH) [69, 70]. These cytokines signal through their cognate receptors and intracellular signaling pathways well established to regulate proliferation of various cell types, including ERK, AKT, SMADs and STATs (Figure 3). SHH plays a critical role in embryonic myogenesis [71] and its expression is absent in adult muscles. Muscle injury reactivates SHH signaling, and pharmacological inhibition of the signaling is reported to impair regeneration [72]. While its receptor Ptch1 and downstream target Gli1 are expressed in mouse primary myoblasts and C2C12 cells, SHH itself is not produced by those cells [70]. Instead, injured myofibers appear to be the source of SHH production in vivo [72], making SHH a bona fide muscle-derived cytokine.
Figure 3. Schematic diagram of muscle-derived cytokine signaling regulating myogenic processes.
Five classes of cytokines and receptors are represented, and only the most downstream components of signaling pathways are shown. A major omission is SHH, which signals through its receptor Ptch1 and regulates satellite cell activation and proliferation, differentiation, and survival.
Myostatin, among its many reported inhibitory functions in muscle, negatively regulates satellite cell self-renewal and proliferation by impinging on the levels of the G1 cell cycle regulators p21CIP and Cdk2 [73, 74]. Similarly, activin A is also reported to negatively regulate satellite cell activation and proliferation [75]. TNFα inhibits regeneration of mdx muscles by suppressing satellite cell activation via NF-κB signaling and epigenetic silencing of Notch1 [76, 77].
Cell cycle withdrawal
Two events during the earliest stage of myogenic differentiation – cell cycle withdrawal and initiation of differentiation – are regulated by a large number of cytokines (Figure 2). This is perhaps not surprising, as the commitment to differentiation is a point of no return for the cell fate. Muscle-derived Flt3L promotes C2C12 myoblast exit from the cell cycle, and knockdown of Flt3L impairs myoblast differentiation in vitro and muscle regeneration in vivo [78]. As an example of a ligand-receptor pair utilizing distinct signaling mechanisms in different biological contexts, Flt3L via its receptor Flt3 (an RTK) stimulates hematopoietic cell proliferation by activating ERK signaling [79], whereas in myoblasts Flt3L-Flt3 signaling activates cell cycle withdrawal by suppressing ERK through a non-canonical p120RasGAP pathway [78]. Brain derived neurotrophic factor (BDNF) is reported to be expressed in L6 [80], C2C12 [68], and both mouse [81] and human [82] primary myoblasts. Depletion or deletion of BDNF in primary myoblasts results in impaired cell cycle withdrawal and differentiation in vitro [81, 82], suggesting a positive role of BDNF in facilitating cell cycle exit of myoblasts. Similar to Flt3L, BDNF has an opposite effect in non-myogenic cells – it promotes cell proliferation in the subventricular zone and hippocampal dentate gyrus [83]. VEGF’s effect on suppressing cell proliferation at the onset of differentiation also makes it a likely candidate as a positive regulator of cell cycle withdrawal [84].
Interestingly, a large number of muscle-derived cytokines have been found to block cell cycle withdrawal and negatively regulate myogenic differentiation including, not surprisingly, many of the growth factors that promote myoblast proliferation – FGFs [38, 85, 86], PDGF [39–41], HGF [43, 44] and TGFβ [49, 50], as well as TNFα [87], BMP4/7 [88–91], CXCL14 [92], and TNFSF10/TRAIL [93]. These cytokines belong to different classes and signal through different types of receptors, and yet, ERK signaling has emerged as a common downstream pathway responsible for modulating the cell cycle (Figure 3). It has been well established that the ubiquitous mitogenic signaling by ERK suppresses myogenic differentiation by preventing cell cycle withdrawal [94, 95], which requires the expression of cyclin dependent kinase inhibitor p21CIP [96, 97]. In principle, it is also possible that distinct downstream pathways can transduce the signals from different cytokines and converge on the cell cycle machinery.
Not all published studies reporting effects of cytokines on the early stage of differentiation directly examined their involvement in the cell cycle, but some of those cytokines may well be regulating cell cycle withdrawal. For instance, LIF is expressed in mouse primary myoblasts [98]; recombinant LIF inhibits myoblast differentiation at an early stage accompanied by reduced p21CIP expression and increased ERK activation, and inhibition of ERK signaling rescues differentiation in LIF-treated cells [99]. An earlier study in fact had reported LIF stimulating mouse primary myoblast proliferation [67]. These observations taken together strongly suggest that LIF negatively regulates myogenic differentiation through suppressing cell cycle withdrawal. Our RNAi screen [28] has led to the identification of additional 10 or so cytokines (belonging to several distinct families) that may negatively regulate myogenic differentiation by blocking cell cycle withdrawal (D. Kim & J. Chen, unpublished observations). It is not clear why myocytes secrete such a large number of distinct cytokines to modulate a single step of myogenesis. Do these cytokines support myoblast/satellite cell proliferation and simply need to be removed at the time of differentiation? The results of our unpublished preliminary studies do not seem to support this notion (R. Waldemer-Streyer, D. Kim, & J. Chen). Or is this arsenal of inhibitors of differentiation necessary for the maintenance of satellite cell pool? The knockdown of any one of those cytokines in myoblasts results in a differentiation phenotype, suggesting that these regulators are not redundant. How do they functionally interact? Do their signaling pathways interact and if so how?
Initiation of differentiation
Whereas the initiation of differentiation can be a direct consequence of cell cycle withdrawal regulated by the cytokines discussed above, some cytokines may regulate this process independently of cell cycle withdrawal. IGF1 and IGF2, through their receptor IGF1R, signal through the PI3K-AKT pathway to robustly activate the myogenic differentiation program [10, 100]. Mammalian target of rapamycin (mTOR) plays a multi-faceted role in the myogenic regulation by IGF2 [9]: (a) a rapamycin-sensitive and kinase-independent function of mTOR governs IGF2 expression in muscle cells [101–103]; (b) mTOR complex 2 (mTORC2) is a kinase for AKT [104] and hence a positive regulator of IGF2 signaling; (c) mTOR complex 1 (mTORC1) dampens PI3K-AKT myogenic signaling through serine phosphorylation of IRS1 [105, 106].
SHH was also reported to promote C2 myoblast differentiation through the PI3K pathway [69], although another study found recombinant SHH to inhibit C2C12 differentiation [70]. The marked differences between C2 and its subclone line C2C12 [107] could have contributed to this discrepancy. Alternatively, since SHH also stimulates myoblast/satellite cell proliferation [69, 70], the timing of SHH addition to cell culture could determine the outcome of proliferation versus differentiation.
Myostatin, acting through the activin type II receptor, has been shown to reduce MyoD expression and activity via Smad3, hence inhibiting the initiation of differentiation [108]. Signaling from mTORC1 and mTORC2 may also be involved in mediating myostatin inhibition of differentiation of human myoblasts [109]. Several other TGFβ family members may have a similar role as myostatin, including TGFβ1, GDF11, and activin A [55, 56, 109].
In addition to regulating satellite cell proliferation as discussed earlier, the autocrine function of IL-6 is necessary for myogenin expression and myotube fusion, which is impaired in C2C12 cells with IL-6 knockdown and mouse primary myoblasts with IL-6 KO [110, 111].
Myoblast migration and fusion
Upon cell cycle withdrawal and activation of the myogenic gene expression program, mononucleated myocytes fuse to form multi-nucleated myotubes/myofibers. Proper positioning or alignment of the cells is necessary for this fusion, which likely requires myocyte migration. Known to induce cell migration in a number of cell types, VEGF is reported to stimulate myoblast migration and differentiation [84, 112]. Both IL-6 [66] and IL-7 [113] also play a positive role in myoblast migration. The chemokine CXCL12 and its receptor CXCR4 are demonstrated to be critical regulators of myocyte migration and fusion [27]. Another chemokine, CXCL14, is reported to stimulate migration of C2C12 cells as well [114]. In addition, Griffin et al. found a larger number of chemokines to be expressed at the mRNA levels during myoblast fusion, raising the possibility that a network of chemokines may regulate the fusion process [27]. Intracellular signaling pathways involving the Rho small GTPases are known to regulate actin cytoskeleton dynamics and cell migration [115, 116] and they can be downstream targets of muscle-derived cytokine signaling. Furthermore, it is possible that cytokine signaling can be linked to many of the regulators identified to control myoblast migration and myocyte fusion [5, 6].
There are two distinct phases of skeletal myocyte fusion during both embryonic and adult myogenesis in mammals – myoblast-myoblast fusion to form nascent myotubes and myoblast-myotube fusion to form mature myotubes/myofibers [6, 117]. Remarkably, the two fusion processes are separable at the molecular level, with a distinct set of molecules and signaling pathways regulating each [6, 117]. IL-4 was the first example of a muscle-derived cytokine that specifically regulates the second stage of myocytes fusion, reported by Horsley et al. [118]. However, the origin of this cytokine in vivo has been disputed [119]. Heredia et al., detected no IL-4 expression in regenerating muscles in vivo; instead, they demonstrated that eosinophils recruited to the muscle injury site produce IL-4 to regulate muscle resident fibro/adipocyte progenitor cells essential for myofiber regeneration [119]. mTOR is found to govern multiple stages of myogenic differentiation via distinct pathways, one of which is the regulation of second-stage myocyte fusion via a secreted factor [120]. Follistatin partially fulfills the criteria for this secreted factor as its expression in muscle cells is regulated by an mTOR pathway involving microRNA-1 and HDAC4, although follistatin contributes to both stages of myocyte fusion [121].
Cell survival
Although not a direct step in the myogenic process, maintenance of myocyte survival is obviously necessary for the process. Among the muscle-derived cytokines, CNTF [122], IGF2 [37], SHH [70], TNFSF14 [123], and VEGF [84, 112] have been reported to support the survival of myoblasts. Signaling by the Ser/Thr kinase AKT is a major survival pathway in various cell types [124, 125] and, indeed, at least CNTF and TNFSF14 have been shown to signal through AKT to regulate myoblast survival [122, 123]. Other muscle-derived cytokines may exert pro-apoptotic effects. One example is IL-1, which has been reported to be expressed by human primary myoblasts; exogenous IL-1β induces myoblast apoptosis without affecting cell proliferation or fusion [126]. It is possible that optimal cell number/density is controlled by counter-acting cytokine signaling to regulate muscle size during myogenesis.
Muscle-derived cytokines in myogenesis in vivo
The critical role of cytokines in muscle regeneration is well established, but immune cells at the site of muscle injury have been traditionally considered the main source of those cytokines [12, 13]. Although cytokine involvement in muscle diseases, such as muscular dystrophy, cachexia and sarcopenia, has been widely known, the emphasis is again on inflammation and immune cell-produced cytokines [127, 128]. Much of what we know to date about muscle-derived cytokines in regulating myogenesis came from in vitro studies. In the literature there is surprisingly a paucity of in vivo studies of myogenesis that entail modulating cytokine expression in a muscle-specific manner. Nevertheless, many of the cytokines covered in this review (i.e., expressed by muscle cells and have demonstrated myogenic functions) are reported to be expressed in myogenic cells or myofibers during acute injury-induced muscle regeneration, or in muscles undergoing satellite cell-dependent compensatory hypertrophy upon mechanical overload. In addition, some of the cytokines are expressed in dystrophic muscles, in Duchenne muscular dystrophy (DMD) for example, which undergo spontaneous cycles of degeneration and regeneration [129]. In Table 1 we include information of cytokine expression in the regenerating muscle, hypertrophic muscle, dystrophic muscle, and developing muscle. Below we discuss selected cytokines with evidence of functional involvement in the regeneration of normal or dystrophic muscles.
BDNF
Initially identified as a myokine produced in human skeletal muscles in response to exercise and found to confer autocrine/paracrine regulation of muscle metabolism [130, 131], the expression of BDNF is also upregulated in mouse regenerating muscles after injury [81]. BDNF is expressed in normal human skeletal muscles as well as by immune cells located near regenerating myofibers from patients of inflammatory myopathies [82]. A myogenic role of BDNF produced by human satellite cells post-exercise has been implicated by an increase in the number of BDNF+/myogenin+ cells [132]. Muscle regeneration is impaired in mice with targeted BDNF gene disruption driven by satellite cell-specific Myf5-Cre [81], providing definitive evidence for a physiological role of muscle-derived BDNF in myogenesis.
FGF6
The expression of FGF6 is restricted to developing skeletal muscle during embryogenesis [133, 134] and also found in regenerating muscles post-injury [135, 136]. It is reported that FGF6-null mice have impaired muscle regeneration upon injury, most likely due to diminished satellite cell expansion [135], although another study found no defect in injury-induced muscle regeneration with FGF6−/− mice [137]. The phenotype of impaired regeneration in FGF6-null mice is corroborated by accelerated regeneration of injured mouse soleus muscle upon intramuscular delivery of recombinant FGF6 [136]. While these observations combined provide support for a function of muscle-derived FGF6, definitive evidence is yet to come from muscle-specific ablation of the cytokine.
IGF
The expression of both IGF1 and IGF2 in regenerating muscles is associated with regenerative capacity [138, 139]. Muscle-specific transgene expression of IGF1 in mice has been well studied, the effects of which include hypertrophy, accelerated regeneration upon injury, prevention of muscle decline in mdx mice, and enhanced muscle maintenance and regenerative capacity during aging [140–143]. Muscle-specific deletion of IGFR1 (hence impairment of both IGF1 and IGF2 signaling) suppresses muscle growth [144, 145]. However, muscle-specific knockout of IGF1 or IGF2 has not been reported and hence definitive genetic evidence for the autocrine function of IGF in vivo is still lacking.
Myostatin
Systemic knockout of myostatin in mice leads to drastically enhanced muscle growth [53], and a mutation in the bovine myostatin gene is responsible for the double-muscled phenotype in cattle [146]. Since myostatin is almost exclusively expressed in skeletal muscles in mice [53], its systemic knockout provides sufficient evidence for a muscle-autonomous function. Muscle-specific expression of follistatin, which antagonizes myostatin, leads to enhanced muscle growth in mice to a similar degree as myostatin knockout [147]. The expression of myostatin is elevated in mechanically overloaded hypertrophic muscles but decreased in injury-induced regenerative muscles [148]. Nevertheless, myostatin KO may promote muscle regeneration and improve muscle strengths in the mdx mouse, a commonly used (albeit imperfect) model of DMD [149], making myostatin a potential therapeutic target for treating DMD [150, 151].
TNFα
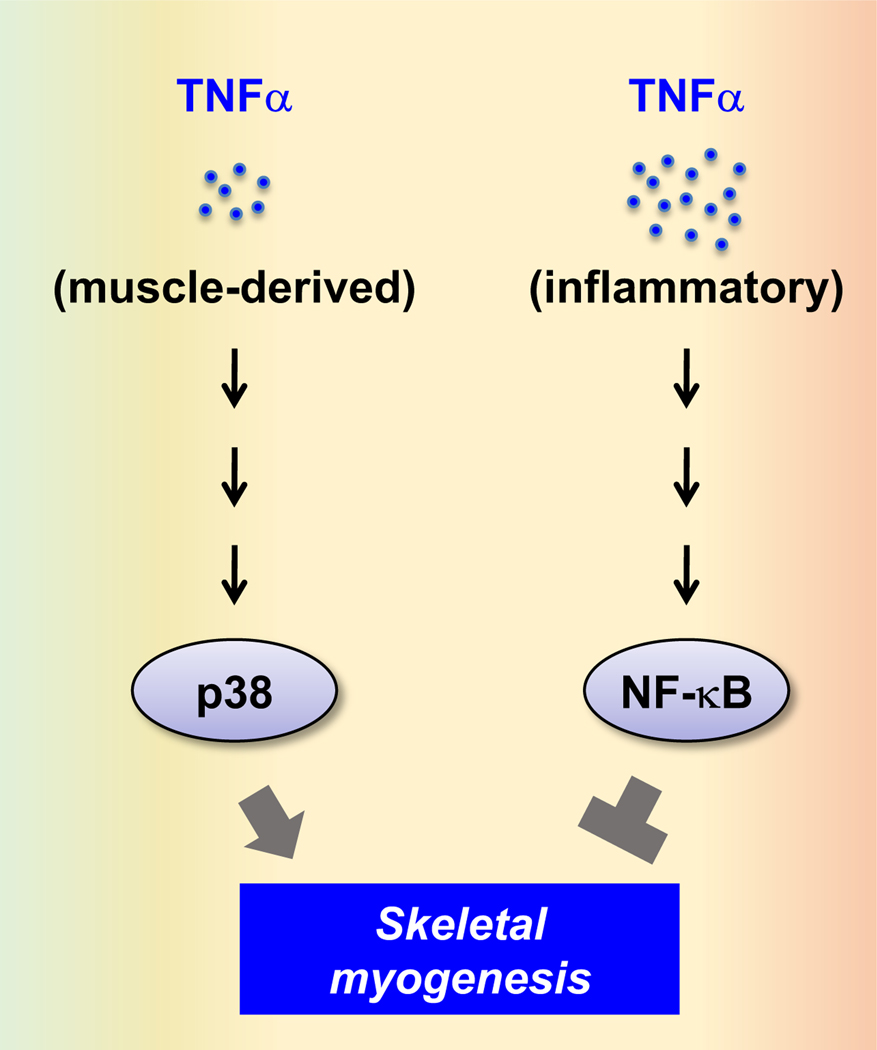
Implicated as a major inflammatory mediator of muscle wasting in aging or disease [152–154], recombinant TNFα inhibits myoblast differentiation in human and mouse cultures [87, 155, 156]. TNFα is reported to be expressed in regenerating myofibers of mdx mice, although the source of this cytokine in DMD can also be immune cells [76, 128, 157]. The elevated level of TNFα in the mdx dystrophic muscles may suppress regeneration by inhibiting satellite cell activation [76, 157]. However, interfering with endogenous TNFα expression or signaling leads to inhibition of myoblast differentiation in vitro and reduction of muscle regeneration in vivo [158–161]. A model to potentially reconcile these paradoxical observations is that normal muscle-derived TNFα has a positive role in myogenic differentiation whereas immune cell- or dystrophic muscle-derived TNFα has a negative role. Concentrations of the cytokine and/or activation of distinct downstream signaling pathways may determine the outcome. Indeed, the MAP kinase p38 is a relevant target of TNFα signaling that supports myogenic differentiation [160, 161], whereas NF-κB mediates the inhibitory function of TNFα [76, 77, 87] (Figure 4).
Figure 4. Context-dependent TNFα signaling in myogenesis.
The source and concentration of TNFα can determine its downstream signaling pathways and effects on myogenesis.
IL-6
A pro-myogenic function of IL-6 in vivo is supported by the observations that expression of this cytokine is induced in satellite cells and growing myofibers during load-induced compensatory hypertrophy [66, 162], and that satellite cell-dependent hypertrophy is impaired in IL-6 KO mice [66]. LIF, which is closely related to IL-6, also has a critical role in compensatory hypertrophy [148, 163]. In addition, IL-6 and LIF are expressed in regenerating muscles after acute injury [98, 148, 164, 165], and a positive role of endogenous LIF in muscle regeneration in mice has been reported [165, 166]. IL-6 expression is also upregulated in muscles of DMD [167] and young mdx mice [168], the latter manifest a disease phenotype similar to human patients. A decreased level of IL-6 accompanies lessened dystrophy in adult mdx mice [169], suggesting a detrimental role of IL-6 in DMD disease severity. Indeed, forced expression of recombinant IL-6 in adult mdx mice recapitulates the severe phenotype of DMD in humans [169]. Conversely, treatment of mdx mice with an IL-6 receptor-blocking antibody led to enhanced muscle regeneration and alleviated morphological and functional defects of the dystrophic muscle [168]. However, another study employing a similar strategy did not find statistically significant improvement in mdx muscles [170]. The discrepancy between the two studies may be attributed to differences in the antibody dosing regimen and/or functional analyses. Administration of the IL-6 receptor-neutralizing antibody also normalized gastrointestinal dysfunction in mdx mice [171]. Therefore, it appears that elevated levels of IL-6 may exacerbate the dystrophic phenotype in DMD and that IL-6 could be a therapeutic target. Currently it is believed that transient and local production of IL-6, such as during injury-induced regeneration or load-induced compensatory hypertrophy, is associated with pro-myogenic effects, whereas chronic and systemically increased IL-6 levels are coupled with muscle atrophy [172]. In vitro, IL-6 has been reported to play both positive and negative roles in myogenic differentiation. The autocrine function of IL-6 promotes satellite cell proliferation, myoblast migration and differentiation [66, 110, 111] as discussed earlier. However, exogenous IL-6 treatment of C2C12 cells suppresses myogenic differentiation, possibly through down-regulation of the kinases p90RSK and p70S6K [173]. This is reminiscent of the paradoxical roles of TNFα in differentiation discussed earlier (Figure 4).
IFNγ
Similar to TNFα and IL-6, paradoxical observations have been reported for the role of IFNγ in myogenesis. IFNγ is found to be expressed by both immune cells and muscle cells in regenerating muscles upon injury, and muscle regeneration is impaired in IFNγ KO mice or by the administering of an IFNγ receptor blocking antibody [64]. Clearly, IFNγ has a positive function in myogenesis. However, administration of exogenous IFNγ also inhibits muscle regeneration in mice, whereas an IFNγ neutralizing antibody rescues regeneration in mice with elevated circulating levels of IFNγ and impaired regeneration as a result of KO of immunity-related GTPase family M1 protein (lRGM1) [174]. Consistent with these in vivo observations, antibody blocking of IFNγ receptor inhibits proliferation and fusion of C2C12 cells [64], and exogenous IFNγ also suppresses differentiation of C2C12 [175] and human skeletal myoblasts [176]. Again, all these observations can be potentially reconciled by considering muscle-derived IFNγ pro-myogenic and pathological levels of IFNγ anti-myogenic. It should be noted, however, a role of muscle-derived IFNγ is yet to be demonstrated in vivo.
Concluding remarks and future directions
Released by infiltrating immune cells at skeletal muscle injury sites, cytokines play critical roles in facilitating muscle repair and regeneration. Although autocrine functions of a few muscle-secreted cytokines have been known for a long time, only in the last 10–15 years has the muscle emerged as a significant source of a large number of cytokines from all families. Many of these cytokines have now been assigned functions in the well-orchestrated process of myogenic differentiation, although many more remain to be functionally characterized. However, very few studies thus far have directly examined the source of cytokines (i.e., muscle versus immune cells) for their contribution to muscle regeneration in vivo. Filling this knowledge gap not only is of intellectual intrigue but can also lead to more effective designs of stem cell therapy in muscular dystrophy and other muscle diseases. To that end, the advent of CRISPR/Cas9 gene editing technology should enable more rapid generation of mice with muscle-specific ablation of cytokines. For instance, the recently reported transgenic mice with Cas9 expression driven by human skeletal actin (HSA)-Cre [177] and muscle creatine kinase (MCK)-Cre [178] would offer a convenient way to knock out or knock down cytokine genes of interest in muscle by delivering specific sgRNAs. Muscle-specific activation of cytokine gene expression would also be informative and potentially applicable in therapeutics, and the CRISPR activation technology (CRISPRa) provides a powerful tool [179]. For instance, the dCas9-SunTag system [180, 181] can be combined with appropriate promoters [182] to drive Cre in order to achieve muscle-specific gene activation.
Many cytokines are detected at low levels in non-myeloid cell types including skeletal muscle cells. A potential implication is that individual cytokines may primarily contribute to fine-tuning mechanisms during differentiation and regeneration. Alternatively, it is possible that the local concentrations of muscle-derived cytokines are sufficiently high to support autocrine functions. To make matters more complicated, it is likely that many of the same cytokines are secreted by both muscle cells and immune cells, and that the source of a cytokine could determine its specific function, as we have seen with TNFα, IL-6 and IFNγ as examples. How does the muscle cell distinguish cytokines coming from different sources – what are the biochemical and cellular mechanisms for the differential signaling? How are muscle-derived cytokines regulated at the expression level and is there any inter-connectivity or hierarchy among them? Do these cytokines interact with each other at the signaling level to form a network in the regulation of the multiple processes of myogenesis? What would be the heterogeneity, if any, within a muscle cell population in their production of and response to cytokines? Multi-pronged experimental approaches coupled with computational modelling may offer the best chance to delineate such a complex regulatory system.
Acknowledgements
We would like to thank the anonymous reviewers of this manuscript for their insightful comments that have helped improve this paper. The authors’ work was supported by grants from the National Institutes of Health to JC (AR048914 from NIAMS and GM089771 from NIGMS).
Abbreviations
- BDNF
brain-derived neurotrophic factor
- BMP
bone morphogenetic protein
- CCL
C-C motif chemokine ligand
- CCN
connective tissue growth factor (CTGF), cystein rich protein, and nephroblastoma overexpressed gene (NOV)
- CNTF
ciliary neurotrophic factor
- CT-1
cardiotrophin-1
- CXCL
C-X-C motif ligand
- DMD
Duchenne muscular dystrophy
- ERK
extracellular signal-regulated kinase
- FGF
fibroblast growth factor
- Flt3
fms like tyrosine kinase 3
- Flt3L
fms like tyrosine kinase 3 ligand
- G-CSF
granulocyte colony stimulating factor
- GDF
growth differentiation factor
- GPCR
G protein-coupled receptor
- HGF
hepatocyte growth factor
- IFN
interferon
- IGF
insulin-like growth factor
- IL
interleukin
- LIF
leukemia inhibitory factor
- mTOR
mammalian target of rapamycin
- NGF
nerve growth factor
- OSM
oncostatin M
- PDGF
platelet-derived growth factor
- PI3K
phosphoinositide 3-kinase
- RTK
receptor tyrosine kinase
- SHH
sonic hedgehog
- STAT
signal transducer and activator of transcription
- TGF
transforming growth factor
- TNF
tumor necrosis factor
- TNFSF
TNF superfamily
- VEGF
vascular endothelial growth factor
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Montarras D, L’honoré A, and Buckingham M, Lying low but ready for action: the quiescent muscle satellite cell. FEBS Journal, 2013. 280: p. 4036–4050. [DOI] [PubMed] [Google Scholar]
- 2.Tidball JG, Mechanisms of muscle injury, repair, and regeneration. Compr Physiol, 2011. 1(4): p. 2029–62. [DOI] [PubMed] [Google Scholar]
- 3.Buckingham M. and Rigby PW, Gene regulatory networks and transcriptional mechanisms that control myogenesis. Developmental Cell, 2014. 28(3): p. 225–238. [DOI] [PubMed] [Google Scholar]
- 4.Parker MH, Seale P, and Rudnicki MA, Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nature Reviews, Genetics, 2003. 4: p. 495–505. [DOI] [PubMed] [Google Scholar]
- 5.Pavlath GK, Spatial and functional restriction of regulatory molecules during mammalian myoblast fusion. Exp Cell Res, 2010. 316(18): p. 3067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simionescu A. and Pavlath GK, Molecular mechanisms of myoblast fusion across species. Advances in Experimental Medicine and Biology, 2011. 713: p. 113–135. [DOI] [PubMed] [Google Scholar]
- 7.Lluís F, Perdiguero E, Nebreda AR, and Muñoz-Cánoves P, Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends Cell Biol, 2006. 16(1): p. 36–44. [DOI] [PubMed] [Google Scholar]
- 8.Jang YN and Baik EJ, JAK-STAT pathway and myogenic differentiation. JAKSTAT, 2013. 2(2): p. e23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge Y. and Chen J, Mammalian Target of Rapamycin (mTOR) Signaling Network in Skeletal Myogenesis. J Biol Chem., 2012. 287(52): p. 43928–35. (PMC3527976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang BH, Aoki M, Zheng JZ, Li J, and Vogt PK, Myogenic signaling of phosphatidylinositol 3-kinase requires the serine-threonine kinase Akt/protein kinase B. Proc Natl Acad Sci U S A, 1999. 96(5): p. 2077–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinarello CA, Historical insights into cytokines. European journal of immunology, 2007. 37(S1): p. S34–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tidball JG, Regulation of muscle growth and regeneration by the immune system. Nat Rev Immunol, 2017. 17(3): p. 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tidball JG and Villalta SA, Regulatory interactions between muscle and the immune system during muscle regeneration. Am J Physiol Regul Integr Comp Physiol., 2010. 298(5): p. R1173–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen BK, Steensberg A, Fischer C, Keller C, Keller P, Plomgaard P, Febbraio M, and Saltin B, Searching for the exercise factor: is IL-6 a candidate? J Muscle Res Cell Motil, 2003. 24(2–3): p. 113–9. [DOI] [PubMed] [Google Scholar]
- 15.Pedersen BK and Febbraio MA, Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nature Reviews Endocrinology, 2012. 8(8): p. 457–465. [DOI] [PubMed] [Google Scholar]
- 16.Whitham M. and Febbraio MA, The ever-expanding myokinome: discovery challenges and therapeutic implications. Nat Rev Drug Discov, 2016. 15(10): p. 719–29. [DOI] [PubMed] [Google Scholar]
- 17.Peake JM, Della Gatta P, Suzuki K, and Nieman DC, Cytokine expression and secretion by skeletal muscle cells: regulatory mechanisms and exercise effects. Exerc Immunol Rev, 2015. 21: p. 8–25. [PubMed] [Google Scholar]
- 18.Yartseva V, Goldstein LD, Rodman J, Kates L, Chen MZ, Chen YJ, Foreman O, Siebel CW, Modrusan Z, Peterson AS, and Jovičić A, Heterogeneity of Satellite Cells Implicates DELTA1/NOTCH2 Signaling in Self-Renewal. Cell Rep, 2020. 30(5): p. 1491–1503.e6. [DOI] [PubMed] [Google Scholar]
- 19.Oprescu SN, Yue F, Qiu J, Brito LF, and Kuang S, Temporal Dynamics and Heterogeneity of Cell Populations during Skeletal Muscle Regeneration. iScience, 2020. 23(4): p. 100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dell’Orso SA-O, Juan AH, Ko KD, Naz F, Perovanovic J, Gutierrez-Cruz G, Feng X, and Sartorelli VA-O, Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. LID - 10.1242/dev.174177 [doi] LID - dev174177. Development, 2019. 146: p. dev174177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Micheli AJ, Laurilliard EJ, Heinke CL, Ravichandran H, Fraczek P, Soueid-Baumgarten S, De Vlaminck I, Elemento O, and Cosgrove BD, Single-Cell Analysis of the Muscle Stem Cell Hierarchy Identifies Heterotypic Communication Signals Involved in Skeletal Muscle Regeneration. Cell Rep, 2020. 30(10): p. 3583–3595 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan CY, Masui O, Krakovska O, Belozerov VE, Voisin S, Ghanny S, Chen J, Moyez D, Zhu P, Evans KR, McDermott JC, and Siu KW, Identification of differentially regulated secretome components during skeletal myogenesis. Mol Cell Proteomics, 2011. 10(5): p. M110.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henningsen J, Pedersen BK, and Kratchmarova I, Quantitative analysis of the secretion of the MCP family of chemokines by muscle cells. Mol Biosyst., 2011. 7(2): p. 311–21. [DOI] [PubMed] [Google Scholar]
- 24.Henningsen J, Rigbolt KT, Blagoev B, Pedersen BK, and Kratchmarova I, Dynamics of the skeletal muscle secretome during myoblast differentiation. Mol Cell Proteomics., 2010. 9(11): p. 2482–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon JH, Yea K, Kim J, Choi YS, Park S, Lee H, Lee CS, Suh P-G, and Ryu SH, Comparative proteomic analysis of the insulin-induced L6 myotube secretome. Proteomics, 2009. 9(1): p. 51–60. [DOI] [PubMed] [Google Scholar]
- 26.Norheim F, Raastad T, Thiede B, Rustan AC, Drevon CA, and Haugen F, Proteomic identification of secreted proteins from human skeletal muscle cells and expression in response to strength training. Am J Physiol Endocrinol Metab, 2011. 301(5): p. E1013–21. [DOI] [PubMed] [Google Scholar]
- 27.Griffin CA, Apponi LH, Long KK, and Pavlath G, Chemokine expression and control of muscle cell migration during myogenesis. Journal of Cell Science, 2010. 123: p. 3052–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge Y, Waldemer RJ, Nalluri R, Nuzzi PD, and Chen J, RNAi Screen Reveals Potentially Novel Roles of Cytokines in Myoblast Differentiation. PLoS ONE, 2013. 8(7): p. e68068 (PMC3699544). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Florini JR, Ewton DZ, and Coolican SA, Growth hormone and the insulin-like growth factor system in myogenesis. Endocr Rev., 1996. 17(5): p. 481–517. [DOI] [PubMed] [Google Scholar]
- 30.Florini JR, Ewton DZ, and Magri KA, Hormones, growth factors, and myogenic differentiation. Annu Rev Physiol, 1991. 53: p. 201–16. [DOI] [PubMed] [Google Scholar]
- 31.Stewart CE and Rotwein P, Growth, differentiation, and survival: multiple physiological functions for insulin-like growth factors. Physiol Rev, 1996. 76(4): p. 1005–26. [DOI] [PubMed] [Google Scholar]
- 32.Saini A, Al-Shanti N, and Stewart CE, Waste management - cytokines, growth factors and cachexia. Cytokine Growth Factor Rev, 2006. 17(6): p. 475–86. [DOI] [PubMed] [Google Scholar]
- 33.Glass DJ, Molecular mechanisms modulating muscle mass. Trends Mol Med., 2003. 9(8): p. 344–50. [DOI] [PubMed] [Google Scholar]
- 34.Florini JR, Magri KA, Ewton DZ, James PL, Grindstaff K, and Rotwein PS, “Spontaneous” differentiation of skeletal myoblasts is dependent upon autocrine secretion of insulin-like growth factor-II. J Biol Chem, 1991. 266(24): p. 15917–23. [PubMed] [Google Scholar]
- 35.Tollefsen SE, Lajara R, McCusker RH, Clemmons DR, and Rotwein P, Insulin-like growth factors (IGF) in muscle development. Expression of IGF-I, the IGF-I receptor, and an IGF binding protein during myoblast differentiation. J Biol Chem, 1989. 264(23): p. 13810–7. [PubMed] [Google Scholar]
- 36.Tollefsen SE, Sadow JL, and Rotwein P, Coordinate expression of insulin-like growth factor II and its receptor during muscle differentiation. Proc Natl Acad Sci U S A, 1989. 86(5): p. 1543–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart CE and Rotwein P, Insulin-like growth factor-II is an autocrine survival factor for differentiating myoblasts. J Biol Chem., 1996. 271(19): p. 11330–8. [DOI] [PubMed] [Google Scholar]
- 38.Pawlikowski B, Vogler TO, Gadek K, and Olwin BB, Regulation of skeletal muscle stem cells by fibroblast growth factors. Dev Dyn, 2017. 246(5): p. 359–367. [DOI] [PubMed] [Google Scholar]
- 39.Yablonka-Reuveni Z, Balestreri TM, and Bowen-Pope DF, Regulation of proliferation and differentiation of myoblasts derived from adult mouse skeletal muscle by specific isoforms of PDGF. J Cell Biol, 1990. 111(4): p. 1623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin P, Sejersen T, and Ringertz NR, Recombinant platelet-derived growth factor-BB stimulates growth and inhibits differentiation of rat L6 myoblasts. Journal of Biological Chemistry, 1991. 266(2): p. 1245–1249. [PubMed] [Google Scholar]
- 41.Sejersen T, Betsholtz C, Sjölund M, Heldin C-H, Westermark B, and Thyberg J, Rat skeletal myoblasts and arterial smooth muscle cells express the gene for the A chain but not the gene for the B chain (c-sis) of platelet-derived growth factor (PDGF) and produce a PDGF-like protein. Proceedings of the National Academy of Sciences, 1986. 83(18): p. 6844–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jennische E, Ekberg S, and Matejka GL, Expression of hepatocyte growth factor in growing and regenerating rat skeletal muscle. American Journal of Physiology-Cell Physiology, 1993. 265(1): p. C122–C128. [DOI] [PubMed] [Google Scholar]
- 43.Gal-Levi R, Leshem Y, Aoki S, Nakamura T, and Halevy O, Hepatocyte growth factor plays a dual role in regulating skeletal muscle satellite cell proliferation and differentiation. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1998. 1402(1): p. 39–51. [DOI] [PubMed] [Google Scholar]
- 44.Sheehan SM, Tatsumi R, Temm-Grove CJ, and Allen RE, HGF is an autocrine growth factor for skeletal muscle satellite cells in vitro. Muscle & Nerve, 2000. 23(2): p. 239–245. [DOI] [PubMed] [Google Scholar]
- 45.Ewton DZ, Roof SL, Magri KA, McWade FJ, and Florini JR, IGF-II is more active than IGF-I in stimulating L6A1 myogenesis: greater mitogenic actions of IGF-I delay differentiation. J Cell Physiol, 1994. 161(2): p. 277–84. [DOI] [PubMed] [Google Scholar]
- 46.Florini JR, Roberts AB, Ewton DZ, Falen SL, Flanders KC, and Sporn MB, Transforming growth factor-beta. A very potent inhibitor of myoblast differentiation, identical to the differentiation inhibitor secreted by Buffalo rat liver cells. J Biol Chem, 1986. 261(35): p. 16509–13. [PubMed] [Google Scholar]
- 47.Massagué J, Cheifetz S, Endo T, and Nadal-Ginard B, Type beta transforming growth factor is an inhibitor of myogenic differentiation. Proceedings of the National Academy of Sciences, 1986. 83(21): p. 8206–8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olson EN, Sternberg E, Hu JS, Spizz G, and Wilcox C, Regulation of myogenic differentiation by type beta transforming growth factor. J Cell Biol, 1986. 103(5): p. 1799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodgers BD, Wiedeback BD, Hoversten KE, Jackson MF, Walker RG, and Thompson TB, Myostatin stimulates, not inihibits, C2C12 myoblast proliferation. Endocrinology, 2014. 155(3): p. 670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schabort EJ, van der Merwe M, Loos B, Moore FP, and Niesler CU, TGF-beta’s delay skeletal muscle progenitor cell differentiation in an isoform-independent manner. Exp Cell Res, 2009. 315(3): p. 373–84. [DOI] [PubMed] [Google Scholar]
- 51.Liu D, Black BL, and Derynck R, TGF-β inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes & Development, 2001. 15(22): p. 2950–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lafyatis R, Lechleider R, Roberts AB, and Sporn MB, Secretion and transcriptional regulation of transforming growth factor-beta 3 during myogenesis. Molecular and cellular biology, 1991. 11(7): p. 3795–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McPherron AC, Lawler AM, and Lee SJ, Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature, 1997. 387(6628): p. 83–90. [DOI] [PubMed] [Google Scholar]
- 54.Rodgers BD and Garikipati DK, Clinical, agricultural, and evolutionary biology of myostatin: a comparative review. Endocr Rev, 2008. 29(5): p. 513–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Link BA and Nishi R, Opposing effects of activin A and follistatin on developing skeletal muscle cells. Experimental cell research, 1997. 233(2): p. 350–362. [DOI] [PubMed] [Google Scholar]
- 56.Egerman MA, Cadena SM, Gilbert JA, Meyer A, Nelson HN, Swalley SE, Mallozzi C, Jacobi C, Jennings LL, Clay I, Laurent G, Ma S, Brachat S, Lach-Trifilieff E, Shavlakadze T, Trendelenburg AU, Brack AS, and Glass DJ, GDF11 Increases with Age and Inhibits Skeletal Muscle Regeneration. Cell Metab, 2015. 22(1): p. 164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodino-Klapac LR, Haidet AM, Kota J, Handy C, Kaspar BK, and Mendell JR, Inhibition of myostatin with emphasis on follistatin as a therapy for muscle disease. Muscle Nerve, 2009. 39(3): p. 283–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilson H, Schakman O, Kalista S, Lause P, Tsuchida K, and Thissen J-P, Follistatin induces muscle hypertrophy through satellite cell proliferation and inhibition of both myostatin and activin. American Journal of Physiology-Endocrinology and Metabolism, 2009. 297(1): p. E157–E164. [DOI] [PubMed] [Google Scholar]
- 59.Kota J, Handy CR, Haidet AM, Montgomery CL, Eagle A, Rodino-Klapac LR, Tucker D, Shilling CJ, Therlfall WR, and Walker CM, Follistatin gene delivery enhances muscle growth and strength in nonhuman primates. Science translational medicine, 2009. 1(6): p. 6ra15–6ra15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winbanks CE, Weeks KL, Thomson RE, Sepulveda PV, Beyer C, Qian H, Chen JL, Allen JM, Lancaster GI, Febbraio MA, Harrison CA, McMullen JR, Chamberlain JS, and Gregorevic P, Follistatin-mediated skeletal muscle hypertrophy is regulated by Smad3 and mTOR independently of myostatin. J Cell Biol., 2012. 197(7): p. 997–1008. Epub 2012 Jun 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yahiaoui L, Gvozdic D, Danialou G, Mack M, and Petrof BJ, CC family chemokines directly regulate myoblast responses to skeletal muscle injury. The Journal of physiology, 2008. 586(16): p. 3991–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L, Ran L, Garcia GE, Wang XH, Han S, Du J, and Mitch WE, Chemokine CXCL16 regulates neutrophil and macrophage infiltration into injured muscle, promoting muscle regeneration. The American journal of pathology, 2009. 175(6): p. 2518–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hara M, Yuasa S, Shimoji K, Onizuka T, Hayashiji N, Ohno Y, Arai T, Hattori F, Kaneda R, and Kimura K, G-CSF influences mouse skeletal muscle development and regeneration by stimulating myoblast proliferation. The Journal of experimental medicine, 2011. 208(4): p. 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng M, Nguyen M-H, Fantuzzi G, and Koh TJ, Endogenous interferon-γ is required for efficient skeletal muscle regeneration. American Journal of Physiology-Cell Physiology, 2008. 294(5): p. C1183-C1191. [DOI] [PubMed] [Google Scholar]
- 65.Otis JS, Niccoli S, Hawdon N, Sarvas JL, Frye MA, Chicco AJ, and Lees SJ, Pro-inflammatory mediation of myoblast proliferation. PloS one, 2014. 9(3): p. e92363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Serrano AL, Baeza-Raja B, Perdiguero E, Jardi M, and Munoz-Canoves P, Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab., 2008. 7(1): p. 33–44. [DOI] [PubMed] [Google Scholar]
- 67.Austin L. and Burgess AW, Stimulation of myoblast proliferation in culture by leukaemia inhibitory factor and other cytokines. J Neurol Sci, 1991. 101(2): p. 193–7. [DOI] [PubMed] [Google Scholar]
- 68.Seidl K, Erck C, and Buchberger A, Evidence for the participation of nerve growth factor and its low-affinity receptor (p75NTR) in the regulation of the myogenic program. J Cell Physiol, 1998. 176(1): p. 10–21. [DOI] [PubMed] [Google Scholar]
- 69.Elia D, Madhala D, Ardon E, Reshef R, and Halevy O, Sonic hedgehog promotes proliferation and differentiation of adult muscle cells: Involvement of MAPK/ERK and PI3K/Akt pathways. Biochim Biophys Acta, 2007. 1773(9): p. 1438–46. [DOI] [PubMed] [Google Scholar]
- 70.Koleva M, Kappler R, Vogler M, Herwig A, Fulda S, and Hahn H, Pleiotropic effects of sonic hedgehog on muscle satellite cells. Cell Mol Life Sci, 2005. 62(16): p. 1863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borycki AG, Brunk B, Tajbakhsh S, Buckingham M, Chiang C, and Emerson CP Jr., Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development, 1999. 126(18): p. 4053–63. [DOI] [PubMed] [Google Scholar]
- 72.Straface G, Aprahamian T, Flex A, Gaetani E, Biscetti F, Smith RC, Pecorini G, Pola E, Angelini F, Stigliano E, Castellot JJ Jr., Losordo DW, and Pola R, Sonic hedgehog regulates angiogenesis and myogenesis during post-natal skeletal muscle regeneration. J Cell Mol Med, 2009. 13(8b): p. 2424–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas M, Langley B, Berry C, Sharma M, Kirk S, Bass J, and Kambadur R, Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. Journal of Biological Chemistry, 2000. 275(51): p. 40235–40243. [DOI] [PubMed] [Google Scholar]
- 74.McCroskery S, Thomas M, Maxwell L, Sharma M, and Kambadur R, Myostatin negatively regulates satellite cell activation and self-renewal. The Journal of cell biology, 2003. 162(6): p. 1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yaden BC, Wang YX, Wilson JM, Culver AE, Milner A, Datta-Mannan A, Shetler P, Croy JE, Dai GL, and Krishnan V, Inhibition of Activin A Ameliorates Skeletal Muscle Injury and Rescues Contractile Properties by Inducing Efficient Remodeling in Female Mice. American Journal of Pathology, 2014. 184(4): p. 1152–1166. [DOI] [PubMed] [Google Scholar]
- 76.Acharyya S, Sharma SM, Cheng AS, Ladner KJ, He W, Kline W, Wang H, Ostrowski MC, Huang TH, and Guttridge DC, TNF inhibits Notch-1 in skeletal muscle cells by Ezh2 and DNA methylation mediated repression: implications in duchenne muscular dystrophy. PLoS one, 2010. 5(8): p. e12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Acharyya S, Villalta SA, Bakkar N, Bupha-Intr T, Janssen PM, Carathers M, Li Z-W, Beg AA, Ghosh S, and Sahenk Z, Interplay of IKK/NF-κB signaling in macrophages and myofibers promotes muscle degeneration in Duchenne muscular dystrophy. The Journal of clinical investigation, 2007. 117(4): p. 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ge Y, Waldemer RJ, Nalluri R, Nuzzi PD, and Chen J, Flt3L is a novel regulator of skeletal myogenesis. J Cell Sci., 2013. 126(Pt 15): p. 3370–9 (PMC3730246). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Masson K. and Ronnstrand L, Oncogenic signaling from the hematopoietic growth factor receptors c-Kit and Flt3. Cell Signal., 2009. 21(12): p. 1717–26. Epub 2009 Jun 18. [DOI] [PubMed] [Google Scholar]
- 80.Mousavi K. and Jasmin BJ, BDNF is expressed in skeletal muscle satellite cells and inhibits myogenic differentiation. J Neurosci, 2006. 26(21): p. 5739–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clow C. and Jasmin BJ, Brain-derived neurotrophic factor regulates satellite cell differentiation and skeltal muscle regeneration. Mol Biol Cell, 2010. 21(13): p. 2182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Colombo E, Bedogni F, Lorenzetti I, Landsberger N, Previtali SC, and Farina C, Autocrine and immune cell-derived BDNF in human skeletal muscle: implications for myogenesis and tissue regeneration. The Journal of Pathology, 2013. 231(2): p. 190–198. [DOI] [PubMed] [Google Scholar]
- 83.Ferreira FF, Ribeiro FF, Rodrigues RS, Sebastião AM, and Xapelli S, Brain-Derived Neurotrophic Factor (BDNF) Role in Cannabinoid-Mediated Neurogenesis. Front Cell Neurosci, 2018. 12: p. 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arsic N, Zacchigna S, Zentilin L, Ramirez-Correa G, Pattarini L, Salvi A, Sinagra G, and Giacca M, Vascular endothelial growth factor stimulates skeletal muscle regeneration in vivo. Molecular Therapy, 2004. 10(5): p. 844–854. [DOI] [PubMed] [Google Scholar]
- 85.Sheehan SM and Allen RE, Skeletal muscle satellite cell proliferation in response to members of the fibroblast growth factor family and hepatocyte growth factor. J Cell Physiol, 1999. 181(3): p. 499–506. [DOI] [PubMed] [Google Scholar]
- 86.Hannon K, Kudla AJ, McAvoy MJ, Clase KL, and Olwin BB, Differentially expressed fibroblast growth factors regulate skeletal muscle development through autocrine and paracrine mechanisms. J Cell Biol, 1996. 132(6): p. 1151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langen RC, Schols AM, Kelders MC, Wouters EF, and Janssen-Heininger YM, Inflammatory cytokines inhibit myogenic differentiation through activation of nuclear factor-kappaB. Faseb J., 2001. 15(7): p. 1169–80. [DOI] [PubMed] [Google Scholar]
- 88.Ono Y, Calhabeu F, Morgan JE, Katagiri T, Amthor H, and Zammit PS, BMP signalling permits population expansion by preventing premature myogenic differentiation in muscle satellite cells. Cell Death Differ, 2011. 18(2): p. 222–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sterrenburg E, van der Wees CG, White SJ, Turk R, de Menezes RX, van Ommen G-JB, den Dunnen JT, and AC’t Hoen P, Gene expression profiling highlights defective myogenesis in DMD patients and a possible role for bone morphogenetic protein 4. Neurobiology of disease, 2006. 23(1): p. 228–236. [DOI] [PubMed] [Google Scholar]
- 90.Friedrichs M, Wirsdöerfer F, Flohé SB, Schneider S, Wuelling M, and Vortkamp A, BMP signaling balances proliferation and differentiation of muscle satellite cell descendants. BMC Cell Biol, 2011. 12: p. 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gozo M, Aspuria P, Cheon D, Walts A, Berel D, Miura N, Karlan B, and Orsulic S, Foxc2 induces Wnt4 and Bmp4 expression during muscle regeneration and osteogenesis. Cell Death & Differentiation, 2013. 20(8): p. 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Waldemer-Streyer RJ, Reyes-Ordonez A, Kim D, Zhang R, Singh N, and Chen J, Cxcl14 depletion accelerates skeletal myogenesis by promoting cell cycle withdrawal. NPJ Regen Med, 2017. 2: p. pii: 16017 (PMC553773). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim D, Singh N, Waldemer-Streyer RJ, Yoon MS, and Chen J, Muscle-derived TRAIL negatively regulates myogenic differentiation. Exp Cell Res, 2020. 394(1): p. 112165 (PMC7434709). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bennett AM and Tonks NK, Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science, 1997. 278(5341): p. 1288–1291. [DOI] [PubMed] [Google Scholar]
- 95.Coolican SA, Samuel DS, Ewton DZ, McWade FJ, and Florini JR, The mitogenic and myogenic actions of insulin-like growth factors utilize distinct signaling pathways. J Biol Chem, 1997. 272(10): p. 6653–62. [DOI] [PubMed] [Google Scholar]
- 96.Guo K, Wang J, Andres V, Smith RC, and Walsh K, MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol Cell Biol., 1995. 15(7): p. 3823–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, and Lassar AB, Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science., 1995. 267(5200): p. 1018–21. [DOI] [PubMed] [Google Scholar]
- 98.Kurek JB, Nouri S, Kannourakis G, Murphy M, and Austin L, Leukemia inhibitory factor and interleukin-6 are produced by diseased and regenerating skeletal muscle. Muscle & nerve, 1996. 19(10): p. 1291–1301. [DOI] [PubMed] [Google Scholar]
- 99.Jo C, Kim H, Jo I, Choi I, Jung SC, Kim J, Kim SS, and Jo SA, Leukemia inhibitory factor blocks early differentiation of skeletal muscle cells by activating ERK. Biochim Biophys Acta, 2005. 1743(3): p. 187–97. [DOI] [PubMed] [Google Scholar]
- 100.Kaliman P, Canicio J, Shepherd PR, Beeton CA, Testar X, Palacin M, and Zorzano A, Insulin-like growth factors require phosphatidylinositol 3-kinase to signal myogenesis: dominant negative p85 expression blocks differentiation of L6E9 muscle cells. Mol Endocrinol, 1998. 12(1): p. 66–77. [DOI] [PubMed] [Google Scholar]
- 101.Erbay E. and Chen J, The Mammalian Target of Rapamycin Regulates C2C12 Myogenesis via a Kinase-independent Mechanism. Journal of Biological Chemistry, 2001. 276(39): p. 36079–36082. [DOI] [PubMed] [Google Scholar]
- 102.Erbay E, Park IH, Nuzzi PD, Schoenherr CJ, and Chen J, IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J Cell Biol, 2003. 163(5): p. 931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ge Y, Wu AL, Warnes C, Liu J, Zhang C, Kawasome H, Terada N, Boppart MD, Schoenherr CJ, and Chen J, mTOR regulates skeletal muscle regeneration in vivo through kinase-dependent and kinase-independent mechanisms. Am J Physiol Cell Physiol., 2009. 297(6): p. C1434–44. (PMC2793064). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sarbassov DD, Guertin DA, Ali SM, and Sabatini DM, Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science, 2005. 307(5712): p. 1098–101. [DOI] [PubMed] [Google Scholar]
- 105.Ge Y, Yoon MS, and Chen J, Raptor and Rheb negatively regulate skeletal myogenesis through suppression of insulin receptor substrate 1 (IRS1). J Biol Chem., 2011. 286(41): p. 35675–82. (PMC3195566). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Son K, You JS, Yoon MS, Dai C, Kim JH, Khanna N, Banerjee A, Martinis SA, Han G, Han JM, Kim S, and Chen J, Nontranslational function of leucyl-tRNA synthetase regulates myogenic differentiation and skeletal muscle regeneration. J Clin Invest, 2019. 129(5): p. 2088–2093 (PMC6486340). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sharples AP, Al-Shanti N, and Stewart CE, C2 and C2C12 murine skeletal myoblast models of atrophic and hypertrophic potential: relevance to disease and ageing? J Cell Physiol, 2010. 225(1): p. 240–50. [DOI] [PubMed] [Google Scholar]
- 108.Langley B, Thomas M, Bishop A, Sharma M, Gilmour S, and Kambadur R, Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. Journal of Biological Chemistry, 2002. 277(51): p. 49831–49840. [DOI] [PubMed] [Google Scholar]
- 109.Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, and Glass DJ, Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol., 2009. 296(6): p. C1258–70. Epub 2009 Apr 8. [DOI] [PubMed] [Google Scholar]
- 110.Baeza-Raja B. and Munoz-Canoves P, p38 MAPK-induced nuclear factor-kappaB activity is required for skeletal muscle differentiation: role of interleukin-6. Mol Biol Cell., 2004. 15(4): p. 2013–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hoene M, Runge H, Häring HU, Schleicher ED, and Weigert C, Interleukin-6 promotes myogenic differentiation of mouse skeletal muscle cells: role of the STAT3 pathway. Am J Physiol Cell Physiol, 2013. 304(2): p. C128–36. [DOI] [PubMed] [Google Scholar]
- 112.Bryan BA, Walshe TE, Mitchell DC, Havumaki JS, Saint-Geniez M, Maharaj AS, Maldonado AE, and D’Amore PA, Coordinated vascular endothelial growth factor expression and signaling during skeletal myogenic differentiation. Mol Biol Cell., 2008. 19(3): p. 994–1006. Epub 2007 Dec 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haugen F, Norheim F, Lian H, Wensaas AJ, Dueland S, Berg O, Funderud A, Skålhegg BS, Raastad T, and Drevon CA, IL-7 is expressed and secreted by human skeletal muscle cells. American Journal of Physiology-Cell Physiology, 2010. 298(4): p. C807–C816. [DOI] [PubMed] [Google Scholar]
- 114.Hayashi Y, Murakami M, Kawamura R, Ishizaka R, Fukuta O, and Nakashima M, CXCL14 and MCP1 are potent trophic factors associated with cell migration and angiogenesis leading to higher regenerative potential of dental pulp side population cells. Stem Cell Research & Therapy, 2015. 6(1): p. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hall A, Rho family GTPases. Biochem Soc Trans, 2012. 40(6): p. 1378–82. [DOI] [PubMed] [Google Scholar]
- 116.Hodge RG and Ridley AJ, Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol, 2016. 17(8): p. 496–510. [DOI] [PubMed] [Google Scholar]
- 117.Jansen KM and Pavlath GK, Molecular Control of Mammalian Myoblast Fusion. Methods in Molecular Biology, 2008. 475: p. 115–133. [DOI] [PubMed] [Google Scholar]
- 118.Horsley V, Jansen KM, Mills ST, and Pavlath GK, IL-4 Acts as a Myoblast Recruitment Factor during Mammalian Muscle Growth. Cell, 2003. 113(4): p. 483–94. [DOI] [PubMed] [Google Scholar]
- 119.Heredia JE, Mukundan L, Chen FM, Mueller AA, Deo RC, Locksley RM, Rando TA, and Chawla A, Type 2 Innate Signals Stimulate Fibro/Adipogenic Progenitors to Facilitate Muscle Regeneration. Cell, 2013. 153(2): p. 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Park I-H and Chen J, Mammalian Target of Rapamycin (mTOR) Signaling Is Required for a Late-stage Fusion Process during Skeletal Myotube Maturation. Journal of Biological Chemistry, 2005. 280(36): p. 32009–32017. [DOI] [PubMed] [Google Scholar]
- 121.Sun Y, Ge Y, Drnevich J, Zhao Y, Band M, and Chen J, Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. Journal of Cell Biology, 2010. 189(7): p. 1157–1169 (PMC2894448). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hiatt K, Lewis D, Shew M, Bijangi-Vishehsaraei K, and Halum S, Ciliary neurotrophic factor (CNTF) promotes skeletal muscle progenitor cell (MPC) viability via the phosphatidylinositol 3-kinase–Akt pathway. Journal of tissue engineering and regenerative medicine, 2014. 8(12): p. 963–968. [DOI] [PubMed] [Google Scholar]
- 123.Waldemer-Streyer R. and Chen J, Myocyte-derived Tnfsf14 is a survival factor necessary for myoblast differentiation and skeletal muscle regeneration. Cell Death & Disease, 2015. 6(12): p. e2026 (PMC4720906). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Manning BD and Toker A, AKT/PKB Signaling: Navigating the Network. Cell, 2017. 169(3): p. 381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Manning BD and Cantley LC, AKT/PKB signaling: navigating downstream. Cell., 2007. 129(7): p. 1261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Plonquet A, Eliezer-Vanerot M-C, Poron F, Belec L, Barlovatz-Meimon G, and Gherardi RK, Differential expression of the IL-1 system components during in vitro myogenesis: Implication of IL-1b in induction of myogenic cell apoptosis. Cell death and differentiation, 1999. 6: p. 1012–1021. [DOI] [PubMed] [Google Scholar]
- 127.Zhou J, Liu B, Liang C, Li Y, and Song YH, Cytokine Signaling in Skeletal Muscle Wasting. Trends Endocrinol Metab, 2016. 27(5): p. 335–347. [DOI] [PubMed] [Google Scholar]
- 128.De Paepe B. and De Bleecker JL, Cytokines and chemokines as regulators of skeletal muscle inflammation: presenting the case of Duchenne muscular dystrophy. Mediators Inflamm, 2013. 2013: p. 540370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guiraud S, Aartsma-Rus A, Vieira NM, Davies KE, van Ommen GJ, and Kunkel LM, The Pathogenesis and Therapy of Muscular Dystrophies. Annu Rev Genomics Hum Genet, 2015. 16: p. 281–308. [DOI] [PubMed] [Google Scholar]
- 130.Matthews VB, Aström MB, Chan MH, Bruce CR, Krabbe KS, Prelovsek O, Akerström T, Yfanti C, Broholm C, Mortensen OH, Penkowa M, Hojman P, Zankari A, Watt MJ, Bruunsgaard H, Pedersen BK, and Febbraio MA, Brain-derived neurotrophic factor is produced by skeletal muscle cells in response to contraction and enhances fat oxidation via activation of AMP-activated protein kinase. Diabetologia, 2009. 52(7): p. 1409–18. [DOI] [PubMed] [Google Scholar]
- 131.Pedersen BK, Pedersen M, Krabbe KS, Bruunsgaard H, Matthews VB, and Febbraio MA, Role of exercise-induced brain-derived neurotrophic factor production in the regulation of energy homeostasis in mammals. Exp Physiol, 2009. 94(12): p. 1153–60. [DOI] [PubMed] [Google Scholar]
- 132.McKay BR, Nederveen JP, Fortino SA, Snijders T, Joanisse S, Kumbhare DA, and Parise G, Brain-derived neurotrophic factor is associated with human muscle satellite cell differentiation in response to muscle-damaging exercise. Appl Physiol Nutr Metab, 2020. 45(6): p. 581–590. [DOI] [PubMed] [Google Scholar]
- 133.deLapeyrière O, Ollendorff V, Planche J, Ott MO, Pizette S, Coulier F, and Birnbaum D, Expression of the Fgf6 gene is restricted to developing skeletal muscle in the mouse embryo. Development, 1993. 118(2): p. 601–11. [DOI] [PubMed] [Google Scholar]
- 134.Han JK and Martin GR, Embryonic expression of Fgf-6 is restricted to the skeletal muscle lineage. Dev Biol, 1993. 158(2): p. 549–54. [DOI] [PubMed] [Google Scholar]
- 135.Floss T, Arnold H-H, and Braun T, A role for FGF-6 in skeletal muscle regeneration. Genes & development, 1997. 11(16): p. 2040–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Armand A-S, Launay T, Pariset C, Della Gaspera B, Charbonnier F, and Chanoine C, Injection of FGF6 accelerates regeneration of the soleus muscle in adult mice. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 2003. 1642(1): p. 97–105. [DOI] [PubMed] [Google Scholar]
- 137.Fiore F, Sébille A, and Birnbaum D, Skeletal muscle regeneration is not impaired in Fgf6 −/− mutant mice. Biochem Biophys Res Commun, 2000. 272(1): p. 138–43. [DOI] [PubMed] [Google Scholar]
- 138.Jennische E. and Matejka GL, IGF-I binding and IGF-I expression in regenerating muscle of normal and hypophysectomized rats. Acta Physiol Scand, 1992. 146(1): p. 79–86. [DOI] [PubMed] [Google Scholar]
- 139.Levinovitz A, Jennische E, Oldfors A, Edwall D, and Norstedt G, Activation of insulin-like growth factor II expression during skeletal muscle regeneration in the rat: correlation with myotube formation. Mol Endocrinol, 1992. 6(8): p. 1227–34. [DOI] [PubMed] [Google Scholar]
- 140.Barton ER, Morris L, Musaro A, Rosenthal N, and Sweeney HL, Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J Cell Biol, 2002. 157(1): p. 137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Barton-Davis ER, Shoturma DI, Musaro A, Rosenthal N, and Sweeney HL, Viral mediated expression of insulin-like growth factor I blocks the aging-related loss of skeletal muscle function. Proc Natl Acad Sci U S A, 1998. 95(26): p. 15603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Musaro A, McCullagh K, Paul A, Houghton L, Dobrowolny G, Molinaro M, Barton ER, Sweeney HL, and Rosenthal N, Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat Genet, 2001. 27(2): p. 195–200. [DOI] [PubMed] [Google Scholar]
- 143.Coleman ME, DeMayo F, Yin KC, Lee HM, Geske R, Montgomery C, and Schwartz RJ, Myogenic vector expression of insulin-like growth factor I stimulates muscle cell differentiation and myofiber hypertrophy in transgenic mice. J Biol Chem, 1995. 270(20): p. 12109–16. [DOI] [PubMed] [Google Scholar]
- 144.Mavalli MD, DiGirolamo DJ, Fan Y, Riddle RC, Campbell KS, van Groen T, Frank SJ, Sperling MA, Esser KA, Bamman MM, and Clemens TL, Distinct growth hormone receptor signaling modes regulate skeletal muscle development and insulin sensitivity in mice. J Clin Invest, 2010. 120(11): p. 4007–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.O’Neill BT, Lauritzen HP, Hirshman MF, Smyth G, Goodyear LJ, and Kahn CR, Differential Role of Insulin/IGF-1 Receptor Signaling in Muscle Growth and Glucose Homeostasis. Cell Rep, 2015. 11(8): p. 1220–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Grobet L, Martin LR, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Ménissier F, and Massabanda J, A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nature genetics, 1997. 17(1): p. 71–74. [DOI] [PubMed] [Google Scholar]
- 147.Lee SJ and McPherron AC, Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci U S A, 2001. 98(16): p. 9306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sakuma K, Watanabe K, Sano M, Uramoto I, and Totsuka T, Differential adaptation of growth and differentiation factor 8/myostatin, fibroblast growth factor 6 and leukemia inhibitory factor in overloaded, regenerating and denervated rat muscles. Biochim Biophys Acta, 2000. 1497(1): p. 77–88. [DOI] [PubMed] [Google Scholar]
- 149.Wagner KR, McPherron AC, Winik N, and Lee SJ, Loss of myostatin attenuates severity of muscular dystrophy in mdx mice. Ann Neurol, 2002. 52(6): p. 832–6. [DOI] [PubMed] [Google Scholar]
- 150.Crone M. and Mah JK, Current and Emerging Therapies for Duchenne Muscular Dystrophy. Curr Treat Options Neurol, 2018. 20(1092–8480 (Print)). [DOI] [PubMed] [Google Scholar]
- 151.Shieh PB, Emerging Strategies in the Treatment of Duchenne Muscular Dystrophy. Neurotherapeutics, 2018. 15(4): p. 840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cohen S, Nathan JA, and Goldberg AL, Muscle wasting in disease: molecular mechanisms and promising therapies. Nature Reviews Drug Discovery, 2015. 14(1): p. 58–74. [DOI] [PubMed] [Google Scholar]
- 153.Fearon KC, Glass DJ, and Guttridge DC, Cancer cachexia: mediators, signaling, and metabolic pathways. Cell Metabolism, 2012. 16(2): p. 153–166. [DOI] [PubMed] [Google Scholar]
- 154.Michaud M, Balardy L, Moulis G, Gaudin C, Peyrot C, Vellas B, Cesari M, and Nourhashemi F, Proinflammatory cytokines, aging, and age-related diseases. Journal of the American Medical Directors Association, 2013. 14(12): p. 877–882. [DOI] [PubMed] [Google Scholar]
- 155.Miller S, Ito H, Blau H, and Torti F, Tumor necrosis factor inhibits human myogenesis in vitro. Molecular and cellular biology, 1988. 8(6): p. 2295–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Langen RC, Schols AM, Kelders MC, Van Der Velden JL, Wouters EF, and Janssen-Heininger YM, Tumor necrosis factor-α inhibits myogenesis through redox-dependent and-independent pathways. American Journal of Physiology-Cell Physiology, 2002. 283(3): p. C714–C721. [DOI] [PubMed] [Google Scholar]
- 157.Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, Leahy P, Li J, Guo W, and Andrade FH, A chronic inflammatory response dominates the skeletal muscle molecular signature in dystrophin-deficient mdx mice. Hum Mol Genet, 2002. 11(3): p. 263–72. [DOI] [PubMed] [Google Scholar]
- 158.Li YP and Schwartz RJ, TNF-alpha regulates early differentiation of C2C12 myoblasts in an autocrine fashion. Faseb j, 2001. 15(8): p. 1413–5. [DOI] [PubMed] [Google Scholar]
- 159.Warren GL, Hulderman T, Jensen N, McKinstry M, Mishra M, Luster MI, and Simeonova PP, Physiological role of tumor necrosis factor alpha in traumatic muscle injury. Faseb j, 2002. 16(12): p. 1630–2. [DOI] [PubMed] [Google Scholar]
- 160.Chen S-E, Gerken E, Zhang Y, Zhan M, Mohan RK, Li AS, Reid MB, and Li Y-P, Role of TNF-α signaling in regeneration of cardiotoxin-injured muscle. American Journal of Physiology-Cell Physiology, 2005. 289(5): p. C1179-C1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen S-E, Jin B, and Li Y-P, TNF-α regulates myogenesis and muscle regeneration by activating p38 MAPK. American Journal of Physiology-Cell Physiology, 2007. 292(5): p. C1660-C1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Guerci A, Lahoute C, Hébrard S, Collard L, Graindorge D, Favier M, Cagnard N, Batonnet-Pichon S, Précigout G, Garcia L, Tuil D, Daegelen D, and Sotiropoulos A, Srf-dependent paracrine signals produced by myofibers control satellite cell-mediated skeletal muscle hypertrophy. Cell Metab, 2012. 15(1): p. 25–37. [DOI] [PubMed] [Google Scholar]
- 163.Spangenburg EE and Booth FW, Leukemia inhibitory factor restores the hypertrophic response to increased loading in the LIF(−/−) mouse. Cytokine, 2006. 34(3–4): p. 125–30. [DOI] [PubMed] [Google Scholar]
- 164.Kami K. and Senba E, Localization of leukemia inhibitory factor and interleukin-6 messenger ribonucleic acids in regenerating rat skeletal muscle. Muscle Nerve, 1998. 21(6): p. 819–22. [DOI] [PubMed] [Google Scholar]
- 165.Barnard W, Bower J, Brown M, Murphy M, and Austin L, Leukemia inhibitory factor (LIF) infusion stimulates skeletal muscle regeneration after injury: injured muscle expresses lif mRNA. Journal of the neurological sciences, 1994. 123(1): p. 108–113. [DOI] [PubMed] [Google Scholar]
- 166.Kurek JB, Bower JJ, Romanella M, Koentgen F, Murphy M, and Austin L, The role of leukemia inhibitory factor in skeletal muscle regeneration. Muscle Nerve, 1997. 20(7): p. 815–22. [DOI] [PubMed] [Google Scholar]
- 167.Messina S, Vita GL, Aguennouz M, Sframeli M, Romeo S, Rodolico C, and Vita G, Activation of NF-kappaB pathway in Duchenne muscular dystrophy: relation to age. Acta Myol, 2011. 30(1): p. 16–23. [PMC free article] [PubMed] [Google Scholar]
- 168.Pelosi L, Berardinelli MG, De Pasquale L, Nicoletti C, D’Amico A, Carvello F, Moneta GM, Catizone A, Bertini E, De Benedetti F, and Musarò A, Functional and Morphological Improvement of Dystrophic Muscle by Interleukin 6 Receptor Blockade. EBioMedicine, 2015. 2(4): p. 285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pelosi L, Berardinelli MG, Forcina L, Spelta E, Rizzuto E, Nicoletti C, Camilli C, Testa E, Catizone A, De Benedetti F, and Musarò A, Increased levels of interleukin-6 exacerbate the dystrophic phenotype in mdx mice. Hum Mol Genet, 2015. 24(21): p. 6041–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kostek MC, Nagaraju K, Pistilli E, Sali A, Lai SH, Gordon B, and Chen YW, IL-6 signaling blockade increases inflammation but does not affect muscle function in the mdx mouse. BMC Musculoskelet Disord, 2012. 13: p. 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Manning J, Buckley MM, O’Halloran KD, and O’Malley D, In vivo neutralization of IL-6 receptors ameliorates gastrointestinal dysfunction in dystrophin-deficient mdx mice. Neurogastroenterol Motil, 2016. 28(7): p. 1016–26. [DOI] [PubMed] [Google Scholar]
- 172.Muñoz-Cánoves P, Scheele C, Pedersen BK, and Serrano AL, Interleukin-6 myokine signaling in skeletal muscle: a double-edged sword? Febs j, 2013. 280(17): p. 4131–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pelosi M, De Rossi M, Barberi L, and Musarò A, IL-6 impairs myogenic differentiation by downmodulation of p90RSK/eEF2 and mTOR/p70S6K axes, without affecting AKT activity. Biomed Res Int, 2014. 2014: p. 206026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang L, Wang G, Chen X, Zhang C, Jiang Y, Zhao W, Li H, Sun J, Li X, Xu H, Weng Y, Zhang X, Hou L, Kong Q, Liu Y, Xu H, Mu L, and Wang J, Irgm1 knockout indirectly inhibits regeneration after skeletal muscle injury in mice. Int Immunopharmacol, 2020. 84: p. 106515. [DOI] [PubMed] [Google Scholar]
- 175.Grzelkowska-Kowalczyk K, Wicik Z, Majewska A, Tokarska J, Grabiec K, Kozłowski M, Milewska M, and Błaszczyk M, Transcriptional regulation of important cellular processes in skeletal myogenesis through interferon-γ. J Interferon Cytokine Res, 2015. 35(2): p. 89–99. [DOI] [PubMed] [Google Scholar]
- 176.Kalovidouris AE, Plotkin Z, and Graesser D, Interferon-gamma inhibits proliferation, differentiation, and creatine kinase activity of cultured human muscle cells. II. A possible role in myositis. J Rheumatol, 1993. 20(10): p. 1718–23. [PubMed] [Google Scholar]
- 177.Bond ST, Zhuang A, Yang C, Gould EAM, Sikora T, Liu Y, Fu Y, Watt KI, Tan Y, Kiriazis H, Lancaster GI, Gregorevic P, Henstridge DC, McMullen JR, Meikle PJ, Calkin AC, and Drew BG, Tissue-specific expression of Cas9 has no impact on whole-body metabolism in four transgenic mouse lines. (2212–8778 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Li F, Yin C, Ma Z, Yang K, Sun L, Duan C, Wang T, Hussein A, Wang L, Zhu X, Gao P, Xi Q, Zhang Y, Shu GA-O, Wang S, and Jiang Q, PHD3 mediates denervation skeletal muscle atrophy through Nf-κB signal pathway. (1530–6860 (Electronic)). [DOI] [PubMed] [Google Scholar]
- 179.Dominguez AA, Lim WA, and Qi LS, Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol, 2016. 17(1): p. 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, and Vale RD, A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell, 2014. 159(3): p. 635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhou H, Liu J, Zhou C, Gao N, Rao Z, Li H, Hu X, Li C, Yao X, Shen X, Sun Y, Wei Y, Liu F, Ying W, Zhang J, Tang C, Zhang X, Xu H, Shi L, Cheng L, Huang P, and Yang H, In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat Neurosci, 2018. 21(3): p. 440–446. [DOI] [PubMed] [Google Scholar]
- 182.Skopenkova VV, Egorova TV, and Bardina MV, Muscle-Specific Promoters for Gene Therapy. Acta Naturae, 2021. 13(1): p. 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Griesbeck O, Parsadanian AS, Sendtner M, and Thoenen H, Expression of neurotrophins in skeletal muscle: quantitative comparison and significance for motoneuron survival and maintenance of function. J Neurosci Res, 1995. 42(1): p. 21–33. [DOI] [PubMed] [Google Scholar]
- 184.Sakuma K. and Yamaguchi A, The Recent Understanding of the Neurotrophin’s Role in Skeletal Muscle Adaptation. Journal of Biomedicine and Biotechnology, 2011. 2011: p. 201696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Olwin BB, Hannon K, and Kudla AJ, Are fibroblast growth factors regulators of myogenesis in vivo? Prog Growth Factor Res, 1994. 5(2): p. 145–58. [DOI] [PubMed] [Google Scholar]
- 186.Anderson JE, Liu E. Fau - Kardami L, and Kardami E, Distinctive patterns of basic fibroblast growth factor (bFGF) distribution in degenerating and regenerating areas of dystrophic (mdx) striated muscles. Developmental Biology, 1991. 147: p. 96–109. [DOI] [PubMed] [Google Scholar]
- 187.Zhou S, Qian B, Wang L, Zhang C, Hogan MV, and Li H, Altered bone-regulating myokine expression in skeletal muscle Of Duchenne muscular dystrophy mouse models. Muscle Nerve, 2018. 58(4): p. 573–582. [DOI] [PubMed] [Google Scholar]
- 188.Yamaguchi A, Ishii H, Morita I, Oota I, and Takeda H, mRNA expression of fibroblast growth factors and hepatocyte growth factor in rat plantaris muscle following denervation and compensatory overload. Pflugers Arch, 2004. 448(5): p. 539–46. [DOI] [PubMed] [Google Scholar]
- 189.Sonnenberg E, Meyer D, Weidner KM, and Birchmeier C, Scatter factor/hepatocyte growth factor and its receptor, the c-met tyrosine kinase, can mediate a signal exchange between mesenchyme and epithelia during mouse development. J Cell Biol, 1993. 123(1): p. 223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hayashi S, Aso H, Watanabe K, Nara H, Rose MT, Ohwada S, and Yamaguchi T, Sequence of IGF-I, IGF-II, and HGF expression in regenerating skeletal muscle. Histochem Cell Biol, 2004. 122(5): p. 427–34. [DOI] [PubMed] [Google Scholar]
- 191.Honda H, Abe S, Ishida R, Watanabe Y, Iwanuma O, Sakiyama K, and Ide Y, Expression of HGF and IGF-1 during regeneration of masseter muscle in mdx mice. J Muscle Res Cell Motil, 2010. 31(1): p. 71–7. [DOI] [PubMed] [Google Scholar]
- 192.Gerrard DE, Okamura CS, Ranalletta MA, and Grant AL, Developmental expression and location of IGF-I and IGF-II mRNA and protein in skeletal muscle. J Anim Sci, 1998. 76(4): p. 1004–11. [DOI] [PubMed] [Google Scholar]
- 193.DeVol DL, Rotwein P, Sadow JL, Novakofski J, and Bechtel PJ, Activation of insulin-like growth factor gene expression during work-induced skeletal muscle growth. Am J Physiol, 1990. 259(1 Pt 1): p. E89–95. [DOI] [PubMed] [Google Scholar]
- 194.Wheeler EF and Bothwell M, Spatiotemporal patterns of expression of NGF and the low-affinity NGF receptor in rat embryos suggest functional roles in tissue morphogenesis and myogenesis. J Neurosci, 1992. 12(3): p. 930–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Toti P, Villanova M, Vatti R, Schuerfeld K, Stumpo M, Barbagli L, Malandrini A, and Costantini M, Nerve growth factor expression in human dystrophic muscles. Muscle & nerve, 2003. 27(3): p. 370–373. [DOI] [PubMed] [Google Scholar]
- 196.Contreras O, Córdova-Casanova A, and Brandan E, PDGF-PDGFR network differentially regulates the fate, migration, proliferation, and cell cycle progression of myogenic cells. Cell Signal, 2021. 84: p. 110036. [DOI] [PubMed] [Google Scholar]
- 197.Piñol-Jurado P, Gallardo E, de Luna N, Suárez-Calvet X, Sánchez-Riera C, Fernández-Simón E, Gomis C, Illa I, and Díaz-Manera J, Platelet-Derived Growth Factor BB Influences Muscle Regeneration in Duchenne Muscle Dystrophy. Am J Pathol, 2017. 187(8): p. 1814–1827. [DOI] [PubMed] [Google Scholar]
- 198.Wagatsuma A, Tamaki H, and Ogita F, Sequential expression of vascular endothelial growth factor, Flt-1, and KDR/Flk-1 in regenerating mouse skeletal muscle. Physiol Res, 2006. 55(6): p. 633–40. [DOI] [PubMed] [Google Scholar]
- 199.Huey KA, Smith SA, Sulaeman A, and Breen EC, Skeletal myofiber VEGF is necessary for myogenic and contractile adaptations to functional overload of the plantaris in adult mice. J Appl Physiol (1985), 2016. 120(2): p. 188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Johnson RW, White JD, Walker EC, Martin TJ, and Sims NA, Myokines (muscle-derived cytokines and chemokines) including ciliary neurotrophic factor (CNTF) inhibit osteoblast differentiation. Bone, 2014. 64: p. 47–56. [DOI] [PubMed] [Google Scholar]
- 201.Miyake T, Alli NS, Aziz A, Knudson J, Fernando P, Megeney LA, and McDermott JC, Cardiotrophin-1 maintains the undifferentiated state in skeletal myoblasts. J Biol Chem., 2009. 284(29): p. 19679–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pennica D, Arce V, Swanson T, Vejsada R, Pollock R, Armanini M, Dudley K, Phillips H, Rosenthal A, and Kato A, Cardiotrophin-1, a cytokine present in embryonic muscle, supports long-term survival of spinal motoneurons. Neuron, 1996. 17(1): p. 63–74. [DOI] [PubMed] [Google Scholar]
- 203.Sheng Z, Pennica D, Wood WI, and Chien KR, Cardiotrophin-1 displays early expression in the murine heart tube and promotes cardiac myocyte survival. Development, 1996. 122(2): p. 419–28. [DOI] [PubMed] [Google Scholar]
- 204.Nishikawa J, Sakuma K, Sorimachi Y, Yoshimoto K, and Yasuhara M, Increase of Cardiotrophin-1 immunoreactivity in regenerating and overloaded but not denervated muscles of rats. Neuropathology, 2005. 25(1): p. 54–65. [DOI] [PubMed] [Google Scholar]
- 205.Hayashiji N, Yuasa S, Miyagoe-Suzuki Y, Hara M, Ito N, Hashimoto H, Kusumoto D, Seki T, Tohyama S, Kodaira M, Kunitomi A, Kashimura S, Takei M, Saito Y, Okata S, Egashira T, Endo J, Sasaoka T, Takeda S, and Fukuda K, G-CSF supports long-term muscle regeneration in mouse models of muscular dystrophy. Nat Commun, 2015. 6: p. 6745. [DOI] [PubMed] [Google Scholar]
- 206.Naito T, Goto K, Morioka S, Matsuba Y, Akema T, Sugiura T, Ohira Y, Beppu M, and Yoshioka T, Administration of granulocyte colony-stimulating factor facilitates the regenerative process of injured mice skeletal muscle via the activation of Akt/GSK3αβ signals. European journal of applied physiology, 2009. 105(4): p. 643–651. [DOI] [PubMed] [Google Scholar]
- 207.Hara M, Yuasa S, Shimoji K, Onizuka T, Hayashiji N, Ohno Y, Arai T, Hattori F, Kaneda R, Kimura K, Makino S, Sano M, and Fukuda K, G-CSF influences mouse skeletal muscle development and regeneration by stimulating myoblast proliferation. J Exp Med., 2011. 208(4): p. 715–27. Epub 2011 Mar 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wright CR, Brown EL, Della-Gatta PA, Ward AC, Lynch GS, and Russell AP, G-CSF does not influence C2C12 myogenesis despite receptor expression in healthy and dystrophic skeletal muscle. Front Physiol, 2014. 5: p. 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hara M, Yuasa S, Shimoji K, Onizuka T, Hayashiji N, Ohno Y, Arai T, Hattori F, Kaneda R, Kimura K, Makino S, Sano M, and Fukuda K, G-CSF influences mouse skeletal muscle development and regeneration by stimulating myoblast proliferation. J Exp Med, 2011. 208(4): p. 715–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ohashi M, Okubo K, Mizuno S, Yoda M, Shirasawa H, Chiba K, Horiuchi K, Matsumoto M, and Nakamura M, Granulocyte-colony stimulating factor enhances load-induced muscle hypertrophy in mice. Biochem Biophys Res Commun, 2018. 506(4): p. 944–949. [DOI] [PubMed] [Google Scholar]
- 211.Gomez-Merino D, Drogou C, Guezennec C, and Chennaoui M, Effects of chronic exercise on cytokine production in white adipose tissue and skeletal muscle of rats. Cytokine, 2007. 40(1): p. 23–29. [DOI] [PubMed] [Google Scholar]
- 212.Romanazzo S, Forte G, Morishima K, and Taniguchi A, IL-12 involvement in myogenic differentiation of C2C12 in vitro. Biomaterials science, 2015. 3(3): p. 469–479. [DOI] [PubMed] [Google Scholar]
- 213.Jiang LQ, Franck N, Egan B, Sjögren RJ, Katayama M, Duque-Guimaraes D, Arner P, Zierath JR, and Krook A, Autocrine role of interleukin-13 on skeletal muscle glucose metabolism in type 2 diabetic patients involves microRNA let-7. Am J Physiol Endocrinol Metab, 2013. 305(11): p. E1359–66. [DOI] [PubMed] [Google Scholar]
- 214.Jacquemin V, Butler-Browne GS, Furling D, and Mouly V, IL-13 mediates the recruitment of reserve cells for fusion during IGF-1-induced hypertrophy of human myotubes. J Cell Sci., 2007. 120(Pt 4): p. 670–81. Epub 2007 Jan 30. [DOI] [PubMed] [Google Scholar]
- 215.Grabstein KH, Eisenman J, Shanebeck K, Rauch C, Srinivasan S, Fung V, Beers C, Richardson J, Schoenborn MA, Ahdieh M, and et al. , Cloning of a T cell growth factor that interacts with the beta chain of the interleukin-2 receptor. Science, 1994. 264(5161): p. 965–8. [DOI] [PubMed] [Google Scholar]
- 216.Quinn LS, Haugk KL, and Grabstein KH, Interleukin-15: a novel anabolic cytokine for skeletal muscle. Endocrinology, 1995. 136(8): p. 3669–72. [DOI] [PubMed] [Google Scholar]
- 217.Xiao F, Wang H, Fu X, Li Y, Ma K, Sun L, Gao X, and Wu Z, Oncostatin M inhibits myoblast differentiation and regulates muscle regeneration. Cell Res., 2011. 21(2): p. 350–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sampath SC, Sampath SC, Ho ATV, Corbel SY, Millstone JD, Lamb J, Walker J, Kinzel B, Schmedt C, and Blau HM, Induction of muscle stem cell quiescence by the secreted niche factor Oncostatin M. Nat Commun, 2018. 9(1): p. 1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lu H, Huang D, Ransohoff RM, and Zhou L, Acute skeletal muscle injury: CCL2 expression by both monocytes and injured muscle is required for repair. Faseb J., 2011. 25(10): p. 3344–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Porter JD, Guo W, Merriam AP, Khanna S, Cheng G, Zhou X, Andrade FH, Richmonds C, and Kaminski HJ, Persistent over-expression of specific CC class chemokines correlates with macrophage and T-cell recruitment in mdx skeletal muscle. Neuromuscular Disorders, 2003. 13(3): p. 223–235. [DOI] [PubMed] [Google Scholar]
- 221.Iwasaki S, Miyake M, Hayashi S, Watanabe H, Nagasawa Y, Terada S, Watanabe K, Ohwada S, Kitazawa H, Rose MT, and Aso H, Effect of myostatin on chemokine expression in regenerating skeletal muscle cells. Cells Tissues Organs, 2013. 198(1): p. 66–74. [DOI] [PubMed] [Google Scholar]
- 222.Polesskaya A, Pinna G, Sassi Y, Vandamme M, Bigot A, Mouly V, Morozova N, Harel-Bellan A, and Degerny C, Post-transcriptional modulation of interleukin 8 by CNOT6L regulates skeletal muscle differentiation. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 2016. 1863(2): p. 263–270. [DOI] [PubMed] [Google Scholar]
- 223.De Paepe B, Creus KK, Martin JJ, and De Bleecker JL, Upregulation of chemokines and their receptors in Duchenne muscular dystrophy: potential for attenuation of myofiber necrosis. Muscle Nerve, 2012. 46(6): p. 917–25. [DOI] [PubMed] [Google Scholar]
- 224.Vasyutina E, Stebler J, Brand-Saberi B, Schulz S, Raz E, and Birchmeier C, CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes & Development, 2005. 19(18): p. 2187–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bae GU, Gaio U, Yang YJ, Lee HJ, Kang JS, and Krauss RS, Regulation of myoblast motility and fusion by the CXCR4-associated sialomucin, CD164. J Biol Chem., 2008. 283(13): p. 8301–9. Epub 2008 Jan 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Bobadilla M, Sainz N, Abizanda G, Orbe J, Rodriguez JA, Páramo JA, Prósper F, and Pérez-Ruiz A, The CXCR4/SDF1 axis improves muscle regeneration through MMP-10 activity. Stem cells and development, 2014. 23(12): p. 1417–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Pescatori M, Broccolini A, Minetti C, Bertini E, Bruno C, D’Amico A, Bernardini C, Mirabella M, Silvestri G, Giglio V, Modoni A, Pedemonte M, Tasca G, Galluzzi G, Mercuri E, Tonali PA, and Ricci E, Gene expression profiling in the early phases of DMD: a constant molecular signature characterizes DMD muscle from early postnatal life throughout disease progression. Faseb j, 2007. 21(4): p. 1210–26. [DOI] [PubMed] [Google Scholar]
- 228.Selever J, Liu W, Lu MF, Behringer RR, and Martin JF, Bmp4 in limb bud mesoderm regulates digit pattern by controlling AER development. Dev Biol, 2004. 276(2): p. 268–79. [DOI] [PubMed] [Google Scholar]
- 229.Pizette S, Abate-Shen C, and Niswander L, BMP controls proximodistal outgrowth, via induction of the apical ectodermal ridge, and dorsoventral patterning in the vertebrate limb. Development, 2001. 128(22): p. 4463–74. [DOI] [PubMed] [Google Scholar]
- 230.Nakashima M, Toyono T, Akamine A, and Joyner A, Expression of growth/differentiation factor 11, a new member of the BMP/TGFbeta superfamily during mouse embryogenesis. Mech Dev, 1999. 80(2): p. 185–9. [DOI] [PubMed] [Google Scholar]
- 231.Manceau M, Gros J, Savage K, Thome V, McPherron A, Paterson B, and Marcelle C, Myostatin promotes the terminal differentiation of embryonic muscle progenitors. Genes & Development, 2008. 22(5): p. 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.McLennan IS, Localisation of transforming growth factor beta 1 in developing muscles: implications for connective tissue and fiber type pattern formation. Dev Dyn, 1993. 197(4): p. 281–90. [DOI] [PubMed] [Google Scholar]
- 233.Bernasconi P, Torchiana E, Confalonieri P, Brugnoni R, Barresi R, Mora M, Cornelio F, Morandi L, and Mantegazza R, Expression of transforming growth factor-beta 1 in dystrophic patient muscles correlates with fibrosis. Pathogenetic role of a fibrogenic cytokine. Journal of Clinical Investigation, 1995. 96(2): p. 1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cohn RD, van Erp C, Habashi JP, Soleimani AA, Klein EC, Lisi MT, Gamradt M, ap Rhys CM, Holm TM, and Loeys BL, Angiotensin II type 1 receptor blockade attenuates TGF-β–induced failure of muscle regeneration in multiple myopathic states. Nature medicine, 2007. 13(2): p. 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Goetsch SC, Hawke TJ, Gallardo TD, Richardson JA, and Garry DJ, Transcriptional profiling and regulation of the extracellular matrix during muscle regeneration. Physiol Genomics, 2003. 14(3): p. 261–71. [DOI] [PubMed] [Google Scholar]
- 236.Wride MA and Sanders EJ, Expression of tumor necrosis factor-alpha (TNF alpha)-cross-reactive proteins during early chick embryo development. Dev Dyn, 1993. 198(3): p. 225–39. [DOI] [PubMed] [Google Scholar]
- 237.Kuru S, Inukai A, Kato T, Liang Y, Kimura S, and Sobue G, Expression of tumor necrosis factor-alpha in regenerating muscle fibers in inflammatory and non-inflammatory myopathies. Acta Neuropathol, 2003. 105(3): p. 217–24. [DOI] [PubMed] [Google Scholar]
- 238.Vial C, Zúñiga LM, Cabello-Verrugio C, Cañón P, Fadic R, and Brandan E, Skeletal muscle cells express the profibrotic cytokine connective tissue growth factor (CTGF/CCN2), which induces their dedifferentiation. Journal of cellular physiology, 2008. 215(2): p. 410–421. [DOI] [PubMed] [Google Scholar]
- 239.Nishida T, Kubota S, Aoyama E, Janune D, Lyons KM, and Takigawa M, CCN family protein 2 (CCN2) promotes the early differentiation, but inhibits the terminal differentiation of skeletal myoblasts. Journal of biochemistry, 2014: p. mvu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ohkawara B, Kobayakawa A, Kanbara S, Hattori T, Kubota S, Ito M, Masuda A, Takigawa M, Lyons KM, Ishiguro N, and Ohno K, CTGF/CCN2 facilitates LRP4-mediated formation of the embryonic neuromuscular junction. EMBO Rep, 2020. 21(8): p. e48462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Morales MG, Gutierrez J, Cabello-Verrugio C, Cabrera D, Lipson KE, Goldschmeding R, and Brandan E, Reducing CTGF/CCN2 slows down mdx muscle dystrophy and improves cell therapy. Hum Mol Genet, 2013. 22(24): p. 4938–51. [DOI] [PubMed] [Google Scholar]
- 242.Sun G, Haginoya K, Wu Y, Chiba Y, Nakanishi T, Onuma A, Sato Y, Takigawa M, Iinuma K, and Tsuchiya S, Connective tissue growth factor is overexpressed in muscles of human muscular dystrophy. J Neurol Sci, 2008. 267(1–2): p. 48–56. [DOI] [PubMed] [Google Scholar]
- 243.Calhabeu F, Lafont J, Le Dreau G, Laurent M, Kazazian C, Schaeffer L, Martinerie C, and Dubois C, NOV/CCN3 impairs muscle cell commitment and differentiation. Experimental cell research, 2006. 312(10): p. 1876–1889. [DOI] [PubMed] [Google Scholar]
- 244.Kocialkowski S, Yeger H, Kingdom J, Perbal B, and Schofield PN, Expression of the human NOV gene in first trimester fetal tissues. Anat Embryol (Berl), 2001. 203(6): p. 417–27. [DOI] [PubMed] [Google Scholar]
- 245.Krüger M, Mennerich D, Fees S, Schäfer R, Mundlos S, and Braun T, Sonic hedgehog is a survival factor for hypaxial muscles during mouse development. Development, 2001. 128(5): p. 743–52. [DOI] [PubMed] [Google Scholar]