Abstract
Solar evaporation ponds are commonly used to reduce the volume of seleniferous agricultural drainage water in the San Joaquin Valley, Calif. These hypersaline ponds pose an environmental health hazard because they are heavily contaminated with selenium (Se), mainly in the form of selenate. Se in the ponds may be removed by microbial Se volatilization, a bioremediation process whereby toxic, bioavailable selenate is converted to relatively nontoxic dimethylselenide gas. In order to identify microbes that may be used for Se bioremediation, a 16S ribosomal DNA phylogenetic analysis of an aerobic hypersaline pond in the San Joaquin Valley showed that a previously unaffiliated group of uncultured bacteria (belonging to the order Cytophagales) was dominant, followed by a group of cultured γ-Proteobacteria which was closely related to Halomonas species. Se K-edge X-ray absorption spectroscopy of selenate-treated bacterial isolates showed that they accumulated a mixture of predominantly selenate and a selenomethionine-like species, consistent with the idea that selenate was assimilated via the S assimilation pathway. One of these bacterial isolates (Halomonas-like strain MPD-51) was the best candidate for the bioremediation of hypersaline evaporation ponds contaminated with high Se concentrations because it tolerated 2 M selenate and 32.5% NaCl, grew rapidly in media containing selenate, and accumulated and volatilized Se at high rates (1.65 μg of Se g of protein−1 h−1), compared to other cultured bacterial isolates.
The soils in the Central Valley of California and other western states are derived from shale rocks that naturally contain high levels of Se, B, As, V, Sr, and other potentially toxic elements (12). When the fields are irrigated, large volumes of agricultural drainage water are produced that contain high levels of Se, mainly in the form of selenate (25). Solar evaporation ponds are commonly used to reduce the volume of selenate-contaminated agricultural drainage water (29). Agricultural drainage water is pumped into the ponds and allowed to evaporate, leaving patches of salt and brine containing high concentrations of Se, which pose a threat to wildlife via bioaccumulation and biomagnification in the food chain (30). For example, when Se-contaminated agricultural drainage water was released to the Kesterson reservoir, fish and birds accumulated very high concentrations of Se that resulted in reproductive and developmental deformities and even death (28, 31).
In spite of the ecotoxicological effects of Se and other elements that accumulate to high levels, solar evaporation ponds are still being used in California and other western states to reduce the large volumes of Se-contaminated drainage water (29). An alternative approach to reducing the volume of Se-contaminated drainage water is to use integrated on-farm drainage management (IFDM); i.e., drainage water is reduced in volume by being recycled through a successive series of crops, each of which is more salt tolerant than the preceding crop. The small amount of drainage water that remains after recycling through the different crops is ultimately collected in a plastic-lined solar evaporation pond. The IFDM approach has been investigated at Red Rock Ranch in Five Points, Calif. (6).
The microbial composition of this terminal solar evaporation pond is of considerable interest with respect to Se bioremediation. This is because the microbes are able to survive the extremely high levels of salt, Se, and other potentially toxic elements (substrates that totally exclude the growth of higher plants) and because the microbes have the ability to remove the Se through volatilization to the atmosphere (11). There are several ways in which microbes from the solar evaporation pond are of interest with respect to the remediation of Se-contaminated drainage water. First, they could be used for the bioaugmentation of evaporation ponds to increase Se removal by increasing the rate of Se volatilization from the ponds. Second, they represent a reservoir of microbes that could be used in a bioreactor especially developed for the remediation of agricultural drainage water. Third, they represent a reservoir of genes encoding enzymes that are able to facilitate tolerance to extremely high concentrations of Se and salt, possibly by allowing the uptake and assimilation of Se, for example, to volatile forms, so that Se may be dissipated harmlessly. Such genes could be used, for example, for incorporation into plants for the phytoremediation of Se.
Biological volatilization of inorganic forms of Se (selenate and selenite) to gaseous forms can be carried out by plants and microbes (see references 11 and 35 for reviews) and is an especially important pathway of Se removal. Selenium-volatilizing microalgae and bacteria have been isolated from Se-contaminated ponds containing agricultural drainage water (10, 26). The overwhelming advantage of Se volatilization is that it physically removes Se from the site, minimizing the ecotoxic effects of Se. Furthermore, dimethylselenide, the predominant form of volatile Se produced by microbes in solar evaporation ponds (36), is 500 to 700 times less toxic than selenate (38). Terry and Lin (34) made field measurements of the rate of Se volatilization by different components of the IFDM system at Red Rock Ranch. From their measurements of Se volatilization from the solar evaporation pond, they estimated that 6% of the Se entering the pond was removed via volatilization in 1 year (34). It would be tremendously beneficial to find ways to significantly enhance Se removal above this 6% level.
A major factor influencing Se volatilization in evaporation ponds is microbial community composition (in addition to other factors, such as the levels of nutrients and sulfate, oxygen tension, temperature, pH, Se forms, and concentration) (11). The goal of the present work was to determine the role of microbial community composition as a prime factor controlling Se volatilization in a solar evaporation pond. This was achieved by means of a 16S ribosomal DNA (rDNA) phylogenetic analysis that was carried out on salt from the Red Rock Ranch evaporation pond. The specific objectives were to (i) identify the microbial populations present in the solar evaporation pond at Red Rock Ranch and (ii) determine which culturable microbes have superior capacities for Se assimilation and volatilization and therefore bioremediation. To this end, the culturable microbes were tested with respect to their efficiency in accumulating, assimilating, and volatilizing Se and for their tolerance to high levels of Se and salt. Using Se K-edge X-ray absorption spectroscopy (XAS), the chemical forms of Se accumulated by the bacteria were identified so as to ascertain their biochemical pathway of Se assimilation.
MATERIALS AND METHODS
Site description.
The solar evaporation pond which was used as a sampling site in the present study was the terminal step of an IFDM system at Red Rock Ranch (6, 34). The IFDM system consists of several cells in series. The drainage water from 192 ha of salt-sensitive crops (Alfalfa) is used to irrigate 52 ha of salt-tolerant crops (Brassica napus), whose drainage water is supplied to 5 ha of trees (Eucalyptus). The saline drainage water from the trees is used to irrigate 1.85 ha of halophytes (Salicornia bigelovii, Atriplex patula, Distichilis spicata, and Spartina gracilis). The hypersaline drainage water from the halophyte cell is sprayed into an unvegetated, plastic-lined 0.73-ha solar evaporation pond. A damp salt sample was collected from the solar evaporation pond in March 1997. At the time of sample collection, the electrical conductivity (EC) of the brine in the pond was 75 mS cm−1, the pH was 8.5, and the Se concentration was 5 mg liter−1 (34).
Isolation of bacteria from the solar evaporation pond.
The sample of solar evaporation pond salt was maintained at 4°C until it was brought back to the laboratory. The sample was stirred to mix it, and 0.5 g of the sample was immediately frozen at −80°C for DNA extraction. Another 15 g of the mixed salt sample was added to 100 ml of an enrichment medium and maintained for 1 week in the dark on a shaker (100 rpm) at room temperature; the enrichment medium contained (per liter) 500 ml of Hoagland's solution (15), 1 ml of vitamin solution (7), 0.1 g of yeast extract, and 5 mM sodium acetate. The pH of the medium was 8.5. Aliquots (50 μl) of the enrichment culture were spread on plates of isolation medium, which was similar to enrichment medium but also contained 70 μM (5.6 mg of Se liter−1) sodium selenate (Sigma), 5 g of NaCl liter−1, and 15 g of Difco agar liter−1.
DNA isolation and cloning of 16S rDNA.
In order to determine the identities of both unculturable and culturable microbes in the solar evaporation pond, DNA was isolated by bead beating of a frozen salt sample and then by using the phenol-chloroform procedure described in detail for the extraction of DNA from soil (9). A smear of DNA was observed after electrophoresis on a 1% polyacrylamide gel. This smear was cut out from the gel, and the DNA was cleaned using a Geneclean II kit (Bio 101, Inc., La Jolla, Calif.). Seven 10-fold dilutions of the DNA were made with water and used as templates for PCRs with small-subunit (SSU) rDNA primers 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′) and 533F (5′-GTG CCA GCM GCC GCG GTA A-3′) and the PCR protocol described earlier (9). A PCR product was obtained for the DNA at a 1:100 dilution. This product was cloned using a TA cloning kit as described in the kit instructions (Invitrogen). Clones were checked for the presence of the insert by preparing plasmids with a Qiagen kit, digesting the plasmids with EcoRI, and checking for the presence of the insert by electrophoresis on a 1% polyacrylamide gel.
Restriction fragment length polymorphism (RFLP) analysis of the 16S rDNA clones was carried out to rapidly estimate the microbial diversity in the solar evaporation pond. rDNA inserts from recombinant clones were reamplified by PCR, after which 25 μl of PCR product was digested overnight at 37°C with 1.5 U of each of the 4-base-specific restriction endonucleases MspI and HinPII in NEB2 buffer (New England Biolabs) at a final volume of 30 μL. Digested fragments were separated by electrophoresis on a 2% agarose gel and visualized by staining with ethidium bromide and UV illumination. RFLP patterns were grouped visually, and representatives were selected for DNA sequencing.
Sequencing of rDNA clones, phylogenetic analyses, and chimera detection.
Plasmid templates from representative clones were sequenced using an ABI 373 Stretch DNA sequencer (Dye-Terminator Cycle Sequencing Ready Reaction FS kit; PE Applied Biosystems) according to the manufacturer's instructions. Primers used for sequencing included vector primers and the 1492R and 533F SSU rDNA primers. Sequences were compared to available databases by use of the BLAST (Basic Local Alignment Search Tool) network service (1) to determine their approximate phylogenetic affiliations. Partial sequences were compiled with AutoAssembler 2.1 (PE Applied Biosystems); compiled sequences were aligned by use of the ARB database (http://www.mikro.biologie.tu-muenchen.de). Chimeric sequences were identified with the CHECK_CHIMERA program (20) and by secondary-structure anomalies. Sequence alignments used for phylogenetic inference were minimized by use of the Lane mask (17) for bacterial data. The dendrogram was constructed by use of the ARB database with evolutionary distance (neighbor-joining algorithms with Olsen correction). The robustness of inferred topologies was tested by bootstrap resampling of trees calculated with evolutionary distance (version 4.0b2 of PAUP*; neighbor-joining algorithm with either Kimura two-parameter correction or maximum-likelihood correction with an empirically determined gamma distribution model of site-to-site rate variation and empirically determined base frequencies) (33), parsimony (version 4.0b2 of PAUP*; heuristic search) (33), and maximum likelihood (fast DNAml in the ARB database) (http://www.mikro.biologie.tu-muenchen.de).
Measurements of bacterial growth rates, Se accumulation, and volatilization.
Phylogenetically distinct bacterial strains were characterized with regard to their growth in the presence of high Se concentrations and their ability to take up and volatilize Se. Cells were grown in sterile Nalgene beakers that contained 600 ml of sterile isolation medium (minus the agar) and that were placed in airtight Se volatilization chambers (8). Three replicate chambers were used for each bacterial strain. The chambers were sterilized by treating them with 20% bleach and washing them with sterile distilled water. Volatile Se that was produced by the cultures was trapped in gas washing bottles (Fisher), which contained 200 ml of alkaline peroxide (40 ml of 30% H2O2, 160 ml of 0.05 M NaOH). Volatile Se was trapped by pulling a vacuum on the chamber outlet, which resulted in sterile air being bubbled into the culture via a 0.2-μm filter (Gelman) on the inlet to the chamber. Samples (5 ml) of the alkaline peroxide trap solution were collected at 12 different time points during the course of bacterial growth. The samples were heated at 92°C for 30 min in a water bath, 5 ml of concentrated HCl was added, and the samples were heated again at 92°C for 30 min. The Se content in these samples was determined by atomic absorption hydride-generation spectroscopy (22). The rates of Se volatilization (micrograms of Se gram of protein−1 hour−1) were calculated from the amount of Se volatilized during the last two time points in the exponential phase of growth and normalized to the protein content at the last time point in the exponential phase.
A 1-ml sample of culture solution was withdrawn from each of the chambers during the first five time points to measure bacterial growth. After the first five time points, there was enough bacterial growth to measure Se bioaccumulation. Twenty-five milliliters of culture solution was collected at each of the remaining seven time points to measure Se bioaccumulation in addition to growth. Strain MPD-72 was the only exception; for this strain, 25 ml of culture solution was collected for Se analysis at the fourth time point as well as the remaining eight time points. This exception was made because preliminary growth curves showed that MPD-72 had a shorter lag phase than the other strains. Growth was monitored by measuring the optical density at 600 nm and by measuring the protein content of the culture. The bacterial cells in 0.5 ml of each culture were lysed using alkaline hydrolysis (7), and their protein contents were measured using the Bradford assay (Bio-Rad) in accordance with the manufacturer's instructions.
Se bioaccumulation in the bacterial cultures was measured as follows. The 25-ml sample of each bacterial culture was centrifuged at 8,000 × g for 10 min. The pellets were washed with 10 ml of sterile saline (0.85% NaCl) and recentrifuged at 8,000 × g for 10 min. The washed pellets were digested overnight at room temperature in 50-ml Pyrex digestion tubes with 1 ml of concentrated nitric acid. Glass funnels were placed on the digestion tubes, which were then heated in a digestion block (Tecator model 2040) at 130°C for 5 h. The volume of the acid digests was brought to 10 ml with deionized water. One milliliter of these digests was heated with 1 ml of 30% hydrogen peroxide at 92°C for 30 min in a water bath, after which 5 ml of concentrated HCl was added and the samples were heated again at 92°C for 30 min. Atomic absorption spectroscopy was used to measure total Se in the acid digests (22). At the end of the growth experiment, the remaining culture (∼300 ml) was centrifuged and washed as described above. Glycerol (100 μl) was added to each of the washed pellets, which were then frozen at −80°C for XAS analysis of the chemical forms of Se that accumulated in the bacterial cells.
In a separate experiment, the bacterial strains were tested for halotolerance and Se tolerance, based on their ability to grow in media containing different concentrations of NaCl and selenate, respectively. The cultures were grown aseptically in flasks containing 250 ml of isolation medium (minus the agar). The flasks were placed on a shaker at 150 rpm for 3 days at room temperature. The cultures were centrifuged at 8,000 × g and washed with sterile saline, and the pellets were resuspended in 5 ml of saline. These cultures (25 μl) were used to inoculate culture tubes containing 5-ml quantities of different batches of liquid medium to determine the effect of 6 NaCl concentrations and 8 selenate concentrations on bacterial growth. The medium for the NaCl experiment was the same as isolation medium except that it contained 0, 0.1, 1, 5, 10, 15, 20, or 32.5% NaCl but did not contain any selenate or agar. Culture solutions containing 5, 10, 15, and 20% NaCl culture solutions gave EC values of 79, 148, 296, and 422 mS cm−1, respectively, when measured with a Checkmate EC meter (Corning). These values were obtained after the solutions were diluted 10- or 100-fold with deionized water. Solutions containing 25 or 32.5% NaCl gave the same EC value as the 20% NaCl solution. The medium for the selenate experiment was the same as isolation medium except that it contained 5 g of NaCl liter−1; no sodium selenate or 0.2 μM, 2 μM, 20 μM, 200 μM, 2 mM, 20 mM, 200 mM, or 2 M sodium selenate; and no agar. For conversion of these molar quantities into mass concentrations, 0.2 μM selenate is approximately equivalent to 16 μg of Se liter−1. The bacterial cultures were inoculated into triplicate tubes containing each type of medium. All tubes were placed in a shaker maintained at 250 rpm for 1 week at room temperature. The absorbance of each culture tube was measured at 600 nm as an indicator of growth.
Identification of bioaccumulated Se.
XAS was used to determine the form of bioaccumulated Se in the frozen pellets of each bacterial culture as described previously (37). XAS analyses of all frozen samples were performed at the Stanford Synchrotron Radiation Laboratory (SSRL) with Beam Line 4-3. A Si(220) double-crystal monochromator was used with an upstream vertical aperture of 1 mm, and harmonic rejection was achieved by detuning one crystal by 50%. The electron energy was 3.0 GeV, with a current of 50 to 100 mA. Frozen samples were positioned at a 45° angle to the X-ray beam and were maintained at 15 K in a liquid He cryostat. Se K-edge X-ray absorption spectra were collected by monitoring the Se Kα fluorescence using a Canberra 13-element Ge detector in a series of replicate scans dependent on trace element concentrations. Spectra were also collected for Se references, i.e., 10 mM aqueous solutions of sodium selenate, sodium selenite, and selenomethionine (SeMet) and solid red elemental selenium (23), the latter being collected in transmission. All samples were calibrated against a hexagonal elemental Se reference foil, the spectrum of which was collected simultaneously with the data in transmission and the first energy inflection of which was assumed to be 12,658.0 eV. Data were collected using the XAS-Collect program (13) and analyzed using the EXAFSPAK suite of programs (http://ssrl.slac.stanford.edu/exafspak.html). Quantitative analysis using an edge-fitting method was carried out according to the methods of Pickering and coworkers (23) and Van Fleet-Stalder et al. (37). The advantage of using XAS is that it determines the form of Se in vivo, without any need for chemical extraction of Se, which may alter the form of Se in the cell. The production of elemental Se was also evaluated by visually examining the cultures for orange particles, which are characteristic of elemental Se produced via bacterial dissimilatory selenate reduction (32).
Nucleotide sequence accession numbers.
The sequences of the rDNA clones have GenBank accession numbers AF348707 to AF348733.
RESULTS
Based on the composition of the salt crystals from the Red Rock Ranch evaporation pond, it seems clear that any microbes surviving in the salt could withstand high concentrations of Se and salt (Table 1). The salt crystals contained a Se concentration of 200 mg kg of dry weight−1 and very high concentrations of other elements, especially sulfur, which made up almost 25% of the salt. The Se concentration in the salt was 40-fold higher than that in the brine in the solar evaporation pond, which had a Se concentration of ∼5 mg liter−1 in March 1997, when salt samples were collected (34).
TABLE 1.
Elemental analysis of the salt from the solar evaporation pond at Red Rock Rancha
| Element | Element content (mg kg−1)
|
|
|---|---|---|
| Mean | SD | |
| Cd | 0.2 | 0.1 |
| Mo | 1.5 | 0.1 |
| Pb | 3.7 | 0.4 |
| As | 5.7 | 0.1 |
| Ni | 31 | 0.8 |
| V | 42 | 0.6 |
| Cu | 43 | 1.0 |
| Zn | 46 | 1.7 |
| Cr | 88 | 3.7 |
| Ba | 120 | 2.8 |
| Mn | 160 | 4.2 |
| Se | 200 | 4.4 |
| Sr | 800 | 41 |
| B | 5,100 | 16 |
| Fe | 10,000 | 430 |
| Al | 13,000 | 230 |
| S | 260,000 | 40,000 |
This salt sample was used for the phylogenetic study and to isolate the bacterial strains. Means and standard deviations for three replicates are shown.
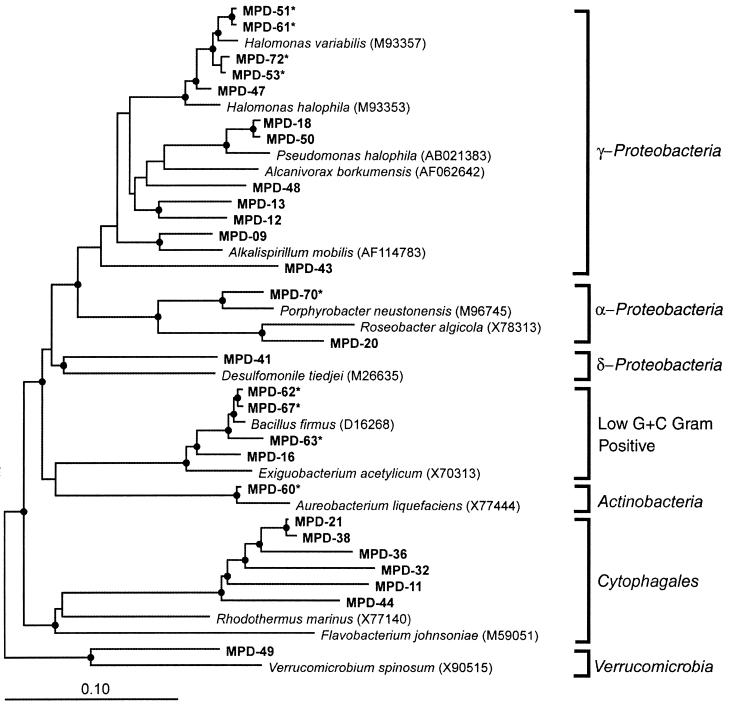
In order to identify evaporation pond microbes that would be superior for Se bioremediation, the microbial community in the salt from the solar evaporation pond was identified by sequencing clones of the community rDNA. These clones were obtained from the total DNA extracted from the salt. A phylogenetic analysis of 72 clones of 16S rDNA identified members of the Bacteria; no members of the Archaea were found (Fig. 1). The most dominant group was a previously unaffiliated group of bacteria placed in the order Cytophagales. This group had 30 clones with similar sequence identities or RFLP patterns (Table 2) and 88% sequence identity to Rhodothermus marinus. However, none of these bacteria could be cultured. The second most dominant group consisted of Halomonas species placed in the class γ-Proteobacteria; some of these bacteria could be cultured (Table 2). Several additional clones were identified as members of different bacterial groups, i.e., α-Proteobacteria, δ-Proteobacteria, low G+C-content gram-positive bacteria, Actinobacteria, and Verrucomicrobiae.
FIG. 1.
Evolutionary distance dendrogram of bacterial 16S rDNA sequence types obtained from the selenium-contaminated solar evaporation pond. Bacterial groups are listed outside the brackets. Reference sequences were chosen with the ARB parsimony insertion tool and database (http://www.mikro.biologie.tu-muenchen.de) and the BLAST program (1). Branch points supported (bootstrap values of >74%) by rate-corrected maximum-likelihood, parsimony, and distance analyses are indicated by closed circles. Cultivated organisms are indicated with asterisks.
TABLE 2.
16S rDNA sequences identified in the Se-contaminated solar evaporation pond
| Straina | Other strains in group (MDP-) | No. of clonesb | Group | Strain match | % Identity |
|---|---|---|---|---|---|
| MPD-21 | 1, 3, 4, 7, 8, 10, 15, 21, 22, 23, 24, 25, 26, 28, 34, 35, 37, 40, 42, 44, 45 | 30 | Cytophagales | R. marinus | 88 |
| MPD-32 | 1 | Cytophagales | Sphingobacterium multivorum | 89 | |
| MPD-36 | 38 | 2 | Cytophagales | Cytophaga sp. strain BD7-14 | 89, 86 |
| MPD-11 | 1 | Cytophagales | WCHA2-47 | 87 | |
| MPD-60 | 1 | Actinobacteria | M. saperdae | 99 | |
| MPD-20 | 1 | α-Proteobacteria | Agrobacterium gelatinovorum | 95 | |
| MPD-70 | 1 | α-Proteobacteria | “A. sanguineum” | 99.8 | |
| MPD-41 | 1 | δ-Proteobacteria | Geobacter metallireducens | 88 | |
| MPD-51 | 52, 53, 54, 55, 56, 58 | 14 | γ-Proteobacteria | HTB069 (Halomonas) | 98 |
| MPD-61 | 1 | γ-Proteobacteria | Halomonas subglaciescola | 95 | |
| MPD-72 | 1 | γ-Proteobacteria | Unidentified strain HTB143 | 97 | |
| MPD-09 | 1 | γ-Proteobacteria | Alkalispirillum mobilis | 95 | |
| MPD-12 | 30 | 2 | γ-Proteobacteria | Alcanivorax borkumensis | 92, 91 |
| MPD-13 | 1 | γ-Proteobacteria | Pseudomonas jessenii | 91 | |
| MPD-14 | 1 | γ-Proteobacteria | Obligately oligotrophic strain | 93 | |
| MPD-18 | 50, 27 | 3 | γ-Proteobacteria | Pseudomonas halophila | 98, 95, 95 |
| MPD-43 | 47 | 3 | γ-Proteobacteria | Deleya salina | 96, 99 |
| MPD-48 | 1 | γ-Proteobacteria | Marinobacter sp. strain PCOB-2 | 98 | |
| MPD-16 | 1 | Low G+C content | WFeA1-16 | 94 | |
| MPD-62 | 67 | 2 | Low G+C content | B. firmus | 99 |
| MPD-63 | 65 | 2 | Low G+C content | Bacillus simplex | 98 |
| MPD-49 | 1 | Verrucomicrobiae | Verrucomicrobium spinosum | 88 | |
| Total | 72 |
The strains shown in bold are the ones that were cultured. Most of the type sequences were used as representatives in the phylogenetic analysis (Fig. 1) and were obtained by sequencing both strands.
Number of clones which had ≥98% identity to the type sequence, based on sequence comparison or inferred from RFLP patterns.
The physiology of Se assimilation and volatilization was studied with six bacterial strains cultured from the hypersaline pond: three strains closely related to Halomonas species (MPD-51, MPD-61, and MPD-72) and strains related to Microbacterium saperdae (MPD-60), Bacillus firmus (MPD-67), and “Agrobacterium sanguineum” (MPD-70). These six cultured strains of bacteria were grown in a medium containing 5 mg of selenate liter−1, a concentration similar to that of the water in the solar evaporation pond at the time of sample collection (34). The bacterial strains were studied with respect to their growth, salt and Se tolerance, rate of Se volatilization, and amount and chemical form of Se that they accumulated.
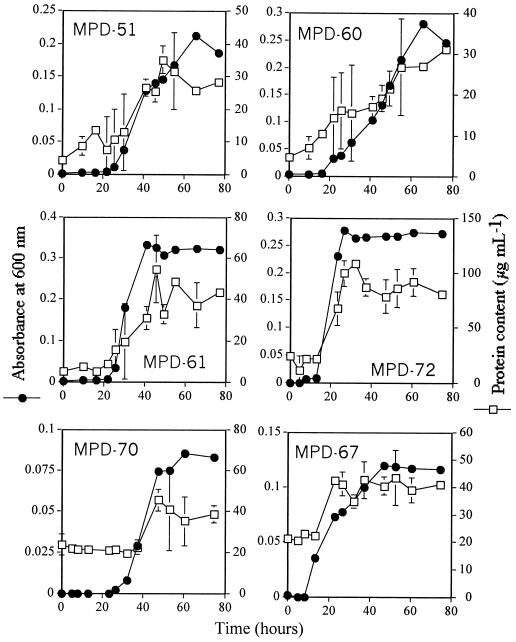
Two measures of growth, i.e., the turbidity of the culture and the protein content of the bacteria, were used to determine the rate of bacterial growth. Both indicators of growth showed that the strains related to Halomonas (MPD-51, MPD-61, and MPD-72) and M. saperdae (MPD-60) grew best in the liquid medium containing selenate, whereas the strains related to “A. sanguineum ” (MPD-70) and B. firmus (MPD-67) grew relatively poorly (Fig. 2). The growth curve based on the protein content of the bacterial cells paralleled the growth curve obtained from the turbidity measurements.
FIG. 2.
Growth curves and protein contents of bacterial isolates from the selenium-contaminated solar evaporation pond. Means and standard deviations for three replicates are shown.
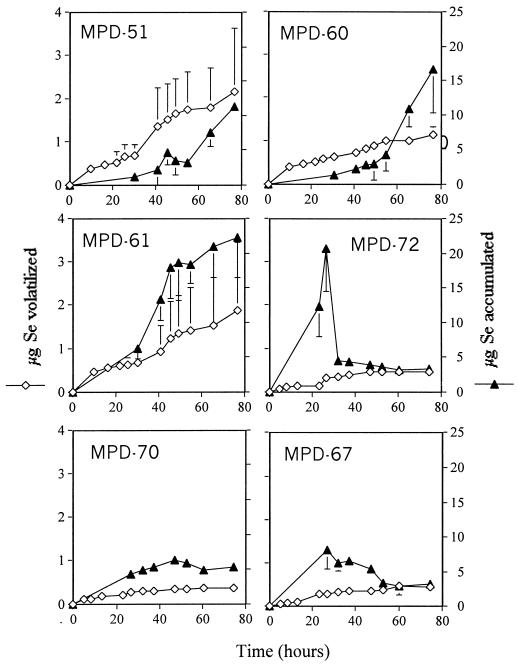
The amount of Se volatilized by the cultures increased with the growth of the cultures over time (Fig. 2 and 3). Two of the Halomonas-like strains, MPD-51 and MPD-61, and the M. saperdae-like strain (MPD-60), which grew well in the selenate-supplied liquid medium, produced volatile Se at the highest rates, 1.65, 1.09, and 0.88 μg of Se g of protein−1 h−1, respectively. The slow-growing bacterial isolates, i.e., those related to “A. sanguineum ” and B. firmus, exhibited ∼5-fold lower rates of Se volatilization, 0.29 and 0.35 μg of Se g of protein−1 h−1, respectively, than the best Se volatilizer—Halomonas strain HTB0639-like MPD-51. Strain MPD-72, which is related to Halomonas, grew well in the selenate-supplied medium, but its rate of Se volatilization (0.16 μg of Se g of protein−1 h−1) was only 10 to 15% that measured for its relatives, strains MPD-51 and MPD-61, even though it accumulated a large amount of Se in the exponential phase. However, the Se content of strain MPD-72 decreased once the culture entered the stationary phase. The amount of Se accumulated by strains MPD-51, MPD-60, and MPD-61 increased with the growth of the cells over time (Fig. 2 and 3). These bacteria accumulated the most Se by the end of the growth curve, whereas strains MPD-70 and MPD-67 did not accumulate much Se over time. The total amounts of Se removed (bioaccumulation plus volatilization) by strains MPD-51, MPD-60, MPD-61, MPD-67, MPD-70, and MPD-72 at the end of the growth curve were 13.5, 17.68, 24.09, 3.72, 5.58, and 3.72 μg, respectively. Thus, two strains closely related to Halomonas (MPD-51 and MPD-61) and the strain related to M. saperdae (MPD-60) removed the largest amount of Se from solution because they volatilized Se at the highest rates and accumulated the most Se compared to the other cultured bacterial strains.
FIG. 3.
Amounts of Se volatilized or accumulated by cultures of bacteria that were isolated from the selenium-contaminated solar evaporation pond. Means and standard deviations for three replicates are shown.
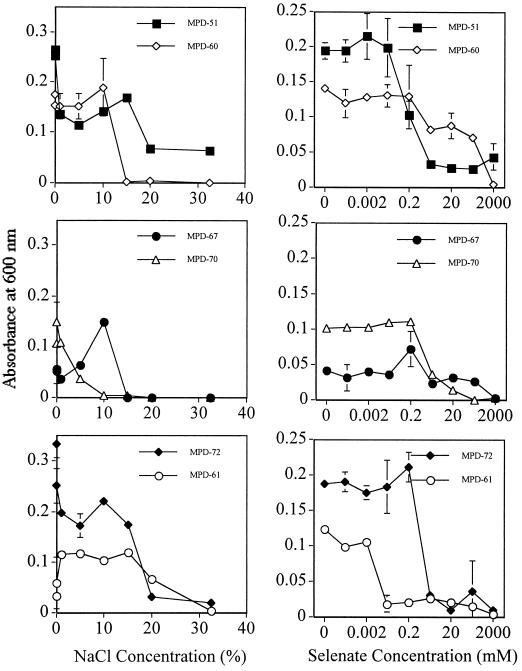
In order to use the cultured bacterial strains for the bioremediation of Se-contaminated drainage water, e.g., by bioaugmentation into evaporation ponds, the bacteria need to tolerate the high concentrations of Se and salt which are often present in the ponds due to fluctuating water levels. The ability of the bacteria to withstand extremely high Se and salt concentrations was tested under control conditions. The Halomonas-like strains (MPD-51, MPD-61, and MPD-72) could tolerate 20% NaCl (a salinity of 422 mS cm−1) and 0.2 mM selenate (16 mg of Se liter−1), concentrations which are higher than those measured in the evaporation pond brine at Red Rock Ranch in March 1997, when the samples were collected (34). These results suggest that the Halomonas-like bacterial strains will tolerate and survive at the very high Se and salt concentrations found in evaporation ponds. Two of the three strains that were related to Halomonas, strains MPD-51 and MPD-72, even grew at the highest concentration of NaCl tested (Fig. 4). The B. firmus-like strain MPD-67 and the M. saperdae-like strain MPD-60 grew at NaCl concentrations of up to 10%, while the strain related to “A. sanguineum,” MPD-70, grew at NaCl concentrations of up to 5%. With regard to selenate tolerance, most strains tolerated selenate concentrations of up to 0.2 mM very well. There was still significant growth for most strains at higher Se concentrations, and strain MPD-51 grew even at 2 M Se, a result which demonstrates its superior tolerance to Se.
FIG. 4.
Effects of different concentrations of NaCl (left) and selenate (right) on the growth of bacterial isolates from the selenium-contaminated solar evaporation pond. Means and standard deviations for three replicates are shown. Note that the different selenate concentrations are plotted on a log scale. Growth was estimated by measuring the absorbance of the cultures at 600 nm. The absorbance of the culture immediately after inoculation was 0.005.
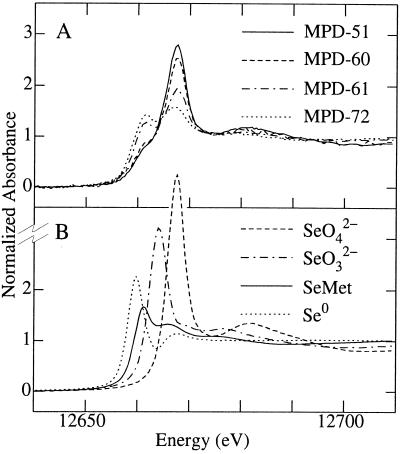
XAS was used to determine the form of Se accumulated in the washed bacterial cells. The near-edge spectra of Se in the bacterial pellets of strains MPD-51 and MPD-60 were similar to those of selenate, with a low-energy shoulder on the selenate edge corresponding to a component of organic Se (Fig. 5). In comparison, strains MPD-61 and MPD-72 showed proportionally lower levels of selenate and higher levels of organic species (Fig. 5). The spectra of MPD-70 and MPD-67 were considerably more noisy due to their lower selenium contents but appeared similar to those of MPD-51 and MPD-60 in having higher proportions of selenate. A quantitative analysis of the different forms of bioaccumulated Se was carried out using an edge-fitting method with a mixture of selenate, selenite, SeMet, and elemental selenium (Table 3). The edge fit showed that the major components of bioaccumulated Se were similar to those of organic Se and selenate, with only small contributions from selenite or elemental Se. The small fraction of bioaccumulated Se identified as elemental Se in the cultures was supported by the fact that none of the cultures contained the orange color indicative of elemental Se at the end of the growth curve. Thus, it is very likely that the bacterial strains did not produce significant amounts of elemental Se from selenate.
FIG. 5.
Se K-edge X-ray absorption near-edge spectra for bioaccumulated Se in four of the bacterial strains that were isolated from the selenium-contaminated solar evaporation pond (A) compared to aqueous selenate, selenite, and l-SeMet and solid red elemental Se standards (B).
TABLE 3.
Percent contributions to the fit for Se K-edge X-ray absorption near-edge spectra for bacterial strains isolated from a selenate-treated hypersaline evaporation pond (Fig. 5)a
| Strain | % Contribution of:
|
|||
|---|---|---|---|---|
| SeO42− | SeO32− | SeMet | Se0 | |
| MPD-51 | 55 (1) | 16 (2) | 22 (4) | 7 (2) |
| MPD-60 | 48 (1) | 10 (3) | 34 (7) | 8 (4) |
| MPD-61 | 24 (1) | 4 (2) | 67 (5) | 5 (3) |
| MPD-72 | 11 (1) | 9 (1) | 66 (3) | 14 (2) |
| MPD-70 | 52 (6) | 19 (9) | 29 (12) | NS |
| MPD-67 | 42 (4) | NS | 58 (6) | NS |
Values in parentheses are three times the estimated standard deviation and indicate the 95% confidence limit of the parameter in the fit. The higher values of the confidence limits for the last two strains are reflective of the poorer data quality due to the low Se concentration. NS, not significant.
DISCUSSION
Despite the extreme conditions of the solar evaporation pond at Red Rock Ranch (i.e., the high concentrations of Se and other elements as well as various osmotic and heat stresses), the phylogenetic analysis showed that the pond supported a large diversity of Bacteria. The dominant group in the pond was a previously unaffiliated group of bacteria in the order Cytophagales. The microbial community composition found in the present study was strikingly similar to that found for two out of three limnologically distinct hypersaline lakes in Antarctica; two of the lakes had clones that grouped within Proteobacteria and Cytophagales, while the microbial community of the third lake was made up almost exclusively of Archaea (5). Our analysis showed that there were no Archaea in the 72 clones screened, a result which is surprising because Archaea have been commonly found in hypersaline environments (3, 4, 14). Results similar to those of the present study were also found for a solar saltern, where Bacteria constituted up to 25% of the microbial community (2). These results, along with those for hyperthermophilic communities containing Bacteria (16, 27), further support the view that Bacteria can exist in and dominate some extreme environments that were previously assumed to contain only Archaea.
Phylogenetic studies of extreme environments have revealed a wealth of previously undiscovered microbial lineages (2, 16, 27). The fact that the dominant group in the pond is a previously unaffiliated group of bacteria (Cytophagales) is an exciting result because these bacteria may well have superior abilities for Se assimilation and volatilization and could therefore represent a source of novel genes for Se bioremediation. However, none of the bacteria belonging to this unaffiliated group could be cultured. Bacterial strains that could be cultured mostly belonged to the γ-Proteobacteria. This result is similar to that found for a study of bacterial strains from a saltern in Spain, where most of the strains grouped in the γ-Proteobacteria and belonged to marine or halotolerant members of the genera Halomonas, Deleya, Pseudomonas, Alteromonas, Acinetobacter, and Vibrio (21).
There are two possible pathways for selenate metabolism by microbes, dissimilatory selenate reduction (32), which immobilizes Se as elemental Se, and assimilation of selenate, which leads to the formation of volatile Se. Dissimilatory selenate reduction takes place mostly under anaerobic conditions (32), although there are a few bacterial strains that carry out selenate reduction to elemental Se under microaerophilic conditions (19). Because the pond at Red Rock Ranch is largely aerobic in nature, it is unlikely that selenate is metabolized by dissimilatory selenate reduction. Furthermore, the data from the XAS analysis support the idea that the bacterial strains mainly carry out assimilatory Se reduction to SeMet, which could be methylated to Se-methylselenomethionine, which then could be cleaved to form dimethylselenide. The aerobically grown selenate-treated bacterial strains accumulated a mixture of predominantly selenate and a SeMet-like species (Fig. 5 and Table 3), analogous to the amino acid intermediates of the S assimilation pathway. Elemental Se, the product of dissimilatory reduction of selenate or selenite, did not make a substantial contribution to the fits of the Se K-edge spectra (Table 3), in contrast with the results of previous studies of anaerobically grown Rhodobacter sphaeroides treated with selenite (37). Since dissimilatory selenate reduction can be ruled out as a significant metabolic pathway for the bacterial strains in the present study, the culturable bacteria very likely use enzymes of the S assimilation pathway to assimilate and volatilize Se, in a manner similar to that of plants (35).
The EC in the Red Rock Ranch evaporation pond fluctuates from 15 mS cm−1 in the winter, when the pond is flooded by rain, to 400 mS cm−1 in the summer (34). Total Se levels in the pond can fluctuate from 3 to 45 mg liter−1. All the bacterial strains in the present study tolerated NaCl concentrations of 5 to 10% (ECs of 79 to 148 mS cm−1) very well (Fig. 4). The salt tolerance of all these strains was considerably higher than that of a previously isolated Aeromonas strain, whose growth was severely affected by an EC of 40 mS cm−1 (26). In fact, three of the cultured bacterial strains, MPD-51, MPD-61, and MPD-72, all of which were related to Halomonas species, were halotolerant, tolerating NaCl concentrations of greater than 20% (Fig. 4).
The bacterial strains in the present study also tolerated Se concentrations of up to 0.2 mM (∼16 mg liter−1), with significant growth at up to 20 mM (∼1,600 mg liter−1) (Fig. 4), a concentration higher than the Se concentration in the salt from the pond (200 mg liter−1) (Table 1). Strains MPD-51, MPD-60, and MPD-67 grew at 200 mM selenate, and strain MPD-51 even grew at 2 M selenate (∼160 g liter−1) (Fig. 4). Thus, the culturable bacteria isolated in the present study tolerated very high concentrations of Se and NaCl, characteristics which make them good candidates for Se bioremediation by bioaugmentation of evaporation ponds or by treatment of hypersaline pond water in bioreactors.
The best strain for the bioremediation of hypersaline selenate-contaminated solar evaporation ponds is most likely MPD-51, a member of the Halomonas group that was the second most dominant group in the pond. This is because it was the only cultured bacterial strain that was halotolerant and tolerant of Se at concentrations up to 2 M; it also accumulated, assimilated, and volatilized Se at very high rates compared to the other cultured bacterial strains. The rates of Se volatilization by Halomonas-like strain MPD-51 (1.65 μg of Se g of protein−1 h−1) supplied with 70 μM selenate were ∼300 to 3,000-fold higher than those measured for Brassica juncea supplied with 100 μM selenate (8) and various wetland plants supplied with 20 μM selenate (24). These rates were 100-fold higher than those measured for an Aeromonas strain isolated from evaporation pond water and cultured with 0.125 to 6.25 μM selenate to an optical density similar to that of strain MPD-51 (26). Since Halomonas-like strain MPD-51 grew under conditions of various salinities and Se concentrations, the fluctuations in the salt and Se levels caused by various water levels in solar evaporation ponds should not affect its ability to survive or carry out Se bioremediation processes under these conditions.
Solar evaporation ponds contain very high concentrations of S (Table 1), which is present mainly as sodium sulfate in agricultural drainage water and in solar evaporation pond salt (6). Since the physiological experiments (Fig. 3, 4, and 5) showed that the bacterial isolates took up, assimilated, and volatilized Se (supplied as 70 μM selenate or 5 mg of Se liter−1) at very high rates in the presence of high concentrations of sulfate in the medium (1 mM or 32 mg of sulfate liter−1), it is clear that the culturable bacteria have a transporter(s) that can compete effectively for selenate in the presence of sulfate. The gene for such a transporter could prove useful in the genetic engineering of plants for enhanced selenate uptake in the presence of sulfate. The bioaccumulation of Se is of little practical significance for Se bioremediation in that the microbial biomass is small and could not be harvested.
As stated earlier, volatilization is of particular interest for Se bioremediation because it allows Se to be removed from the local food chain into the atmosphere, where it is dispersed to other areas (18); in California, this is not a problem because many areas (e.g., the east side of the San Joaquin Valley) are known to be deficient in Se and farm animals require Se supplementation. Volatilization may be enhanced by bioaugmentation with the rapidly volatilizing microbes (e.g., Halomonas-like strains MPD-51 and MPD-61) identified in the present study and by environmental manipulation. The primary environmental factors that promote bacterial Se volatilization are the addition of nutrients (especially C and N), highly aerobic conditions, high temperatures (∼35°C), high pHs (∼8), adequate moisture, Se forms and concentrations, and microbial community composition (11). The water in the evaporation pond at Red Rock Ranch is aerobic and has a pH of 8.5, a temperature of >30°C for 8 months of the year, and high levels of nitrate (6, 34).
One factor that could be easily manipulated to increase Se volatilization rates above those measured in the field is the addition of a carbon source. The availability of a carbon source is very likely to affect Se volatilization because the production of volatile Se in the present physiological study paralleled the growth curve of the bacterial isolates (Fig. 2 and 3). The addition of organic carbon sources has been shown to significantly enhance the rates of microbial Se volatilization in situ, sometimes up to 10-fold (11, 36). If the addition of an economical carbon source to the Red Rock Ranch evaporation pond sustains the rates of Se volatilization that were measured in the physiological experiments, it may be possible to remove most of the Se from the pond in an environmentally friendly, relatively nontoxic manner.
ACKNOWLEDGMENTS
This work was supported by grants W08021-30 and W04163 from the Electric Power Research Institute. The XAS analysis was performed at SSRL, which is funded by the Department of Energy, Offices of Basic Energy Sciences and Biological and Environmental Research, by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and by the National Institute of General Medical Sciences.
We thank Zhiqing Lin for collecting samples of solar evaporation pond salt and Marina Ma for technical assistance.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anton J, Rossello-Mora R, Rodriguez-Valera F, Amann R. Extremely halophilic bacteria in crystallizer ponds from solar salterns. Appl Environ Microbiol. 2000;66:3052–3057. doi: 10.1128/aem.66.7.3052-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anton J, Llobet-Brossa E, Rodriguez-Valera F, Amann R. Fluorescence in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ Microbiol. 1999;1:517–523. doi: 10.1046/j.1462-2920.1999.00065.x. [DOI] [PubMed] [Google Scholar]
- 4.Benlloch S, Martinez-Murcia A J, Rodriguez-Valera F. Sequencing of bacterial and archaeal 16S rRNA genes directly amplified from a hypersaline environment. Syst Appl Microbiol. 1995;18:574–581. [Google Scholar]
- 5.Bowman J P, McCammon S A, Rea S M, McMeekin T A. The microbial composition of three limnologically disparate hypersaline Antarctic lakes. FEMS Microbiol Lett. 2000;183:81–88. doi: 10.1111/j.1574-6968.2000.tb08937.x. [DOI] [PubMed] [Google Scholar]
- 6.Cervinka V. Agroforestry farming system for the management of selenium and salt on irrigated farmland. In: Frankenberger W T, Benson S, editors. Selenium in the environment. New York, N.Y: Marcel Dekker, Inc; 1994. pp. 237–250. [Google Scholar]
- 7.de Souza M P, Yoch D C. Purification and characterization of dimethyl-sulfoniopropionate (DMSP) lyase from an Alcaligenes-like dimethyl sulfide-producing marine isolate. Appl Environ Microbiol. 1995;61:21–26. doi: 10.1128/aem.61.1.21-26.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Souza M P, Pilon-Smits E A H, Lytle C M, Hwang S, Tai J, Honma T S U, Yeh L, Terry N. Rate limiting steps in Se assimilation and volatilization by Brassica juncea. Plant Physiol. 1998;117:1487–1494. doi: 10.1104/pp.117.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan T W-M, Higashi R M, Lane A N. Biotransformations of selenium oxyanion by filamentous cyanophyte-dominated mat cultured from agricultural drainage waters. Environ Sci Technol. 1998;32:3185–3193. [Google Scholar]
- 11.Frankenberger W T, Jr, Karlson U. Microbial volatilization of selenium from soils and sediments. In: Frankenberger W T, Benson S, editors. Selenium in the environment. New York, N.Y: Marcel Dekker, Inc; 1994. pp. 369–387. [Google Scholar]
- 12.Fujii R S, Deverel J, Hatfield D B. Distribution of selenium in soils of agricultural fields, western San Joaquin Valley, California. Soil Sci Soc Am J. 1988;52:1274–1283. [Google Scholar]
- 13.George M J. XAS-Collect: a computer program for X-ray absorption spectroscopic data acquisition. J Synchrotron Rad. 2000;7:283–286. doi: 10.1107/S090904950000683X. [DOI] [PubMed] [Google Scholar]
- 14.Guixa-Boixereu N, Calderon-Paz J I, Heldal M, Bratbak G, Pedros-Alio C. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat Microb Ecol. 1996;11:215–227. [Google Scholar]
- 15.Hoagland, D., and D. I. Arnon. 1938. The water culture method for growing plants without soil. Bull. Calif. Agric. Stat. 346.
- 16.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division-level bacterial diversity in a Yellowstone hot spring. Appl Environ Microbiol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Z-Q, Schemenauer R S, Cervinka V, Zayed A, Lee A, Terry N. Selenium volatilization from a soil-plant system for the remediation of contaminated water and soil in the San Joaquin Valley. J Environ Qual. 2000;29:1048–1056. [Google Scholar]
- 19.Losi M E, Frankenberger W T., Jr Reduction of selenium by Enterobacter cloacae SLD1a-1: isolation and growth of the bacterium and its expulsion of selenium particles. Appl Environ Microbiol. 1997;63:3079–3084. doi: 10.1128/aem.63.8.3079-3084.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marquez M C, Ventosa A, Ruiz-Berraquero F. A taxonomic study of heterotrophic halophilic and non-halophilic bacteria from a solar saltern. J Gen Microbiol. 1987;133:45–46. [Google Scholar]
- 22.Martin T D. Determining selenium in wastewater sediment and sludge by flameless atomic absorption. Atomic Abs Newsl. 1975;14:109–116. [Google Scholar]
- 23.Pickering I J, Brown G E, Jr, Tokunaga T K. Quantitative speciation of selenium in soils using X-ray absorption spectroscopy. Environ Sci Technol. 1995;29:2456–2459. doi: 10.1021/es00009a043. [DOI] [PubMed] [Google Scholar]
- 24.Pilon-Smits E A H, Hwang S, Lytle C M, Zhu Y, Tai J C, Bravo R C, Chen Y, Leustek T, Terry N. Overexpression of ATP sulfurylase in Indian mustard leads to increased selenate uptake, reduction and tolerance. Plant Physiol. 1999;119:123–132. doi: 10.1104/pp.119.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Presser T S, Ohlendorf H M. Biogeochemical cycling of selenium in the San Joaquin Valley, California. Environ Manag. 1987;11:805–821. [Google Scholar]
- 26.Rael R M, Frankenberger W T., Jr Influence of pH, salinity, and selenium on the growth of Aeromonas veronii in evaporation agricultural drainage water. Water Res. 1996;30:422–430. [Google Scholar]
- 27.Reysenbach A-L, Wickham G S, Pace N R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saiki M K, Lowe T P. Selenium in aquatic organisms from subsurface agricultural drainage water, San Joaquin Valley, California. Arch Environ Contam Toxicol. 1987;16:657–670. doi: 10.1007/BF01055416. [DOI] [PubMed] [Google Scholar]
- 29.Seiler R L. Prediction of lands susceptible to irrigation-induced selenium contamination of water. In: Frankenberger W T Jr, Engberg R A, editors. Environmental chemistry of selenium. New York, N.Y: Marcel Dekker, Inc; 1998. pp. 397–418. [Google Scholar]
- 30.Skorupa J. Selenium poisoning of fish and wildlife in nature: lessons from twelve real-world examples. In: Frankenberger W T Jr, Engberg R A, editors. Environmental chemistry of selenium. New York, N.Y: Marcel Dekker, Inc; 1998. pp. 315–354. [Google Scholar]
- 31.Skorupa J P, Ohlendorf H M. Contaminants in drainage water and avian risk thresholds. In: Dinar A, Zilberman D, editors. The economy and management of water and drainage. Boston, Mass: Kluwer Academic Publishers; 1991. pp. 345–368. [Google Scholar]
- 32.Stolz J F, Oremland R S. Bacterial respiration of arsenic and selenium. FEMS Microbiol Rev. 1999;23:615–627. doi: 10.1111/j.1574-6976.1999.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 33.Swofford D L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 34.Terry N, Lin Z-Q. Managing high selenium in agricultural drainage water by agroforestry system: role of selenium volatilization. Technical Report for Project DWR B-80665. Sacramento: California Department of Water Resources; 1999. [Google Scholar]
- 35.Terry N T, Zayed A M, de Souza M P, Tarun A S. Selenium in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:401–432. doi: 10.1146/annurev.arplant.51.1.401. [DOI] [PubMed] [Google Scholar]
- 36.Thompson-Eagle E T, Frankenberger W T., Jr Protein-mediated selenium biomethylation in evaporation pond water. Environ Toxicol Chem. 1990;9:1453–1462. [Google Scholar]
- 37.Van Fleet-Stalder V, Chasteen T G, Pickering I J, George G N, Prince R C. Fate of selenate and selenite metabolized by Rhodobacter sphaeroides. Appl Environ Microbiol. 2000;66:4849–4853. doi: 10.1128/aem.66.11.4849-4853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilber C G. Toxicology of selenium: a review. Clin Toxicol. 1980;17:171–230. doi: 10.3109/15563658008985076. [DOI] [PubMed] [Google Scholar]