Abstract
Purpose
Checkpoint therapy is now the cornerstone of treatment for patients with renal cell carcinoma (RCC) with advanced disease, but biomarkers are lacking to predict which patients will benefit. This study proposes potential immunological biomarkers that could developed for predicting therapeutic response in patients with RCC.
Methods
Using flow cytometry, RNA sequencing, and T-cell receptor (TCR) sequencing, we investigated changes in T cells in the peripheral blood of patients with advanced RCC after receiving immunotherapy. We used immunofluorescence (IF) imaging and flow cytometry to investigate how intratumoral T cells in patients’ tumors (resected months/years prior to receiving checkpoint therapy) predicted patient outcomes after immunotherapy.
Results
We found that a small proportion of CD4 and CD8 T cells in the blood activate following checkpoint therapy, expressing the proliferation marker Ki67 and activation markers HLA-DR and CD38. Patients who had the highest increase in these HLA-DR +CD38+CD8 T cells after treatment had the best antitumor immune response and experienced clinical benefit. Using RNA sequencing, we found that while these cells expanded in most patients, their phenotype did not drastically change during treatment. However, when we analyzed the TCR repertoire of these HLA-DR +CD38+CD8+T cells, we found that only patients who clinically benefitted had a burst of new clonotypes enter this pool of activated cells. Finally, we found that abundant T cells in the untreated tumors predicted clinical benefit to checkpoint therapy on disease progression.
Conclusions
Together, these data suggest that having a strong pre-existing immune response and immediate peripheral T-cell activation after checkpoint therapy is a predictor of clinical benefit in patients with RCC.
Keywords: Biomarkers, Tumor; CD8-Positive T-Lymphocytes; Immunotherapy; Kidney Neoplasms; Lymphocytes, Tumor-Infiltrating
Key messages.
-
WHAT IS ALREADY KNOWN ON THIS TOPIC: Checkpoint therapy is a key treatment for patients with advanced renal cell carcinoma (RCC), but current biomarkers unable to predict which patients will respond to this therapy.
WHAT THIS STUDY ADDS: This study offers potential immunological biomarkers for predicting therapeutic response in patients with RCC.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE AND/OR POLICY: The data presented here suggests that patients with RCC with a strong pre-existing immune response and immediate peripheral T-cell activation after therapy may be more likely to clinically benefit from checkpoint blockade. Future investigation should validate these biomarkers for prospective use.
Introduction
Renal cell carcinoma (RCC) has been identified as an immunogenic tumor, and patients with RCC have been treated with immunotherapy since the mid-1980s.1 High dose interleukin (IL)-2 remains an option, although for only select patients given substantial toxicity. Objective responses are seen in approximately 20% of patients, of whom a subset experience complete response.2 Currently, most patients with advanced clear cell RCC are treated with checkpoint blockade,3–5 and the latest clinical trials in clear cell RCC have led to Food and Drug Administration approval or breakthrough therapy designations for various tyrosine kinase inhibitor +checkpoint inhibitor combinations as frontline therapy.6–9 Most recently, anti-programmed cell death protein-1 (PD-1) therapy was approved in the adjuvant setting for patients with intermediate and high-risk disease, and data from several ongoing neo-adjuvant trials are pending.10 While early data show both safety and efficacy signals, checkpoint inhibition is not without risk of immune mediated adverse events, and many patients still fail to respond. Unfortunately, common biomarkers of response to immunotherapy in other solid tumors, such as programmed death-ligand 1 expression and tumor mutation burden, have not shown a clear role in predicting clinical benefit to immunotherapy in RCC,11 despite usefulness in other tumor types.12 Thus, it is crucial to better understand the immunologic mechanisms that underlie an effective immune response in order to advance treatment options for patients with high-risk localized or advanced RCC.
Much has been learned about what immune factors correlate with therapeutic outcome since the advent of immunotherapy. Many studies have found that features of CD8 T cells predict how a patient will respond to therapy, such as increased expression of Ki67 by CD8 cells in the blood,13 and this marker has been reported to be associated with improved clinical response in patients with melanoma and lung cancer.14–22 Several studies investigating the T-cell receptor (TCR) repertoire of peripheral CD8 T cells have found that new clonotypes enter the blood after treatment and that this increase in clonotypes is associated with better survival in many cancers.23–25 These studies suggest that these proliferating CD8 T cells may be newly activated or reactivated, previously dormant clones.26 In addition to these dynamic peripheral immune changes, CD8 T-cell infiltration into the tumor is an important correlate of patient survival and response to therapy.27–32 This was first shown in melanoma, where patients with higher CD8 T-cell infiltration into their tumor at the time of therapy were more likely to benefit from treatment, and similar findings have since been reported in other cancers.29–31 33–35 More recent studies have found that a subset of tumor infiltrating T cells, TCF-1 +CD8 T cells, are mechanistically important for the clinical efficacy of PD-1 blockade and that TCF-1 +CD8 T cell numbers in tumors correlated with therapeutic efficacy in melanoma.33 34 36–38 Together, these studies highlight a common feature of the immune response to cancer—some patients generate a strong T-cell response to their cancer either before or during checkpoint therapy, while others do not. Based on these studies, we examined how the T-cell response in patients with RCC was affected by immunotherapy, and how the pre-existing immune response to that cancer might correlate with a later capacity to clinically benefit from checkpoint therapy.
Methods
Sample collection, preparation, and storage
Patients with stage IV RCC, either treated with nivolumab, nivolumab +ipilimumab, or on clinical trial (nivolumab +bempegaldesleukin (IL-2) formulation) were recruited.39 Patients consented for blood collection. Blood collection time points were cycle dependent, coinciding with scheduled phlebotomy for standard laboratory analysis. This practical approach, as well as approval of extended nivolumab dosing,40 led to collection intervals that ranged from 2 to 4 weeks. Peripheral blood was obtained in cell preparation tubes and processed to cryopreserve peripheral blood mononuclear cells (PBMCs) and plasma.
Patient tumor samples were collected immediately after undergoing partial or radical nephrectomy. Patients received immune checkpoint blockade, either alone or in combination with targeted therapy (tables 1 and 2). Tumor samples for flow cytometric analysis were harvested in Hank’s Balanced Salt Solution, cut into small pieces, digested using Liberase enzyme cocktail (Roche), and homogenized using an MACS Dissociator. Single cell suspensions were obtained, RBC ACK lysed, and stored at –80°C in freezing media. Samples for IF analysis were formaldehyde fixed and embedded in paraffin blocks by Emory Pathology. Unstained and H&E-stained sections of formalin fixed parafin embedded (FFPE) blocks were obtained from Emory Pathology.
Table 1.
Flow cytometry antibodies
| Target | Clone | Fluorochrome | Source |
| CD3 | UCHT1 | FITC | BioLegend |
| CD4 | OKT4 | PerCP | BioLegend |
| CD28 | CD28.2 | PE/Cy7 | eBioscience |
| CD39 | A1 | BV421 | BioLegend |
| CD38 | HIT2 | BV510 | BioLegend |
| CD38 | HIT2 | BV711 | BioLegend |
| CD8 | RPA-T8 | BV605 | BioLegend |
| HLA-DR | L243 | BV711 | BioLegend |
| HLA-DR | L243 | APC/Cy7 | BioLegend |
| PD-1 | 29F.1A12 | BV786 | BioLegend |
| CD45RA | HI100 | BV785 | BioLegend |
| CD25 | M-A251 | APC | BioLegend |
| CCR7 | G043HI | PE-TR | BioLegend |
| CD19 | HIB19 | 700 | BioLegend |
| CD14 | HCD14 | 700 | BioLegend |
| Tbet | 4B10 | 421 | BioLegend |
| FOXP3 | PCH101 | PE | Invitrogen |
| Ki67 | B56 | APC | BD Biosciences |
| GZMB | GB11 | 700 | BD Biosciences |
Table 2.
Immunofluorescence antibodies
| Target | Antibody type | Clone | Concentration | Secondary | Concentration |
| MHC-II (HLA-DR, DP, DQ) | Mouse IgG2a | Tu39 | 1:100 | Goat anti-mouse IgG2a A488 | 1:250 |
| TCF-1 | Rabbit | C63D9 | 1:150 | Goat anti-rabbit A568 | 1:250 |
| CD8 | Mouse IgG1 | C8/144B | 1:150 | Goat anti-mouse IgG1 A647 | 1:500 |
Assessment of therapeutic response
Clinical benefit was defined as a best response of complete response (CR), partial response (PR), or stable disease (SD) and a progression-free survival (PFS) of 3 months or greater. Objective response was determined by using Response Evaluation Criteria in Solid Tumor V.1.1 (RECIST V.1.1)41 by a board-certified radiologist. Restaging radiograph interval varied among patients on standard of care and clinical trial treatments, and modalities may have switched between CT and MRI depending on other clinical factors.
Flow cytometry and fluorescence activated cell sorting
Single cell suspensions from human tumors and peripheral blood were stained with antibodies listed in table 1. Live/dead discrimination was performed using fixable Aqua or Near-IR Dead Cell Stain Kit (Invitrogen). Samples were acquired with a Becton Dickinson LSRII or sorted with a Becton Dickinson FACSAria II and analyzed using FlowJo software. For intracellular staining, cells were fixed and permeabilized using the FOXP3 Transcription Factor Staining Buffer Set (eBioscience). For cell sorting, cryopreserved samples were thawed, stained, and sorted. For RNA and TCR sequencing experiments, total and naïve CD8 T cells were sorted. For TCR sequencing experiments, CD8 T cells were gated as shown, and CD38 +HLA-DR+ were sorted.
RNA and TCR sequencing
RNA was isolated from sorted CD8 populations (circulating HLA-DR +CD38+and PD-1 +CD45RA− TILs) (online supplemental figure 5G) using a Qiagen AllPrep DNA/RNA Micro Isolation Kit. RNA was sequenced using the Clontech SMARTSeq kit following manufacturers instruction. RNA was sequenced on an Illumina HiSeq 3000 at the Yerkes Genomic. For some samples, RNA processing to obtain complete TCR V(D)J clonotypes of TCR transcripts was done using the SMARTer Human TCR a/b Profiling Kit (Takara Biosciences) following manufacturer’s user manual. SMARTer Human TCR a/b Profiling Kit (Takara Bio) following manufacturers’ instructions. Briefly, first strand complementary DNA synthesis (from RNA) is dT-primed and performed by the SMARTScribe Reverse Transcriptase, adding non-templated nucleotides at the 5’end of each messenger RNA template, followed by two rounds of PCR to uniquely amplify complete V(D)J regions of TCR-a and TCR-b. TCR products were purified with a final product with a range of 600–800 bp which was confirmed by a bioanalyzer. Raw sequences were aligned for full CDR3 sequences, clonotypes and full-length variable sequences using MiXCR. Overall TCR sequencing analysis was performed on TCR-b regions using the immunarch R package (V.0.6.5) and custom R scripts. The diversity was calculated using three methods: Shannon Entropy, Morisita Horn Index, and the number of clones to account for 50% of the total sampled repertoire. Clonotype distribution was defined as the proportion of each clonotype within the sampled TCRs, with the most dominant clonotype accounting for the top TCR-b CDR3 region shared within each sample. TCR rankings (figure 3D) was performed according to the clonotype dominance for the top 20 clonotypes within each time point. Time points within each patient were then matched for clonotype overlap within the top rankings to determine whether clonotype dominance was maintained post-treatment in each patient.
jitc-2022-004803supp005.pdf (773.8KB, pdf)
IF
Sections were deparaffinized in successive incubations with xylene and decreasing concentrations (100, 95, 75, 50, 0%) of EtOH in ddH2O. Antigen retrieval utilized Abcam 100× TrisEDTA Antigen Retrieval Buffer (pH=9) heated under high pressure and washed in phosphate buffered saline (PBS)+0.1% Tween 20. Sections were blocked for 30 min with 10% goat serum in 1×PBS+0.1% Tween 20 before staining. Primary antibodies were used at a concentration of 1:100 (MHC-II) or 1:150 (CD8, TCF-1) and incubated for 1 hour at room temperature. Secondary antibodies were used at a concentration of 1:250 (A488, A568) or 1:500 (A647) and incubated for 30 min at room temperature. Details about antibodies used are listed in table 2. Sections were counterstained with 4’,6-diamidino-2-pehnylindole (DAPI) according to manufacturer instructions (Thermo Fisher). IF images were collected using a Zeiss Z.1 Slide Scanner equipped with a Colibri 7 Flexible Light Source, and Zeiss ZenBlue software was used for post-acquisition image processing. CellProfiler42 43 and custom R and python scripts were used for image analysis, as previously described,44 to determine the xy coordinates of cells within tissue slices, measure fluorescence intensity within each cell, calculate cellular density, and create spatial maps of features within the tissue.
Patient survival analysis
Survival analysis of patients was performed using the log-rank test from the R package, Survminer.
Results
Activation of CD8 T cells in blood after the first treatment predicts response to checkpoint therapy
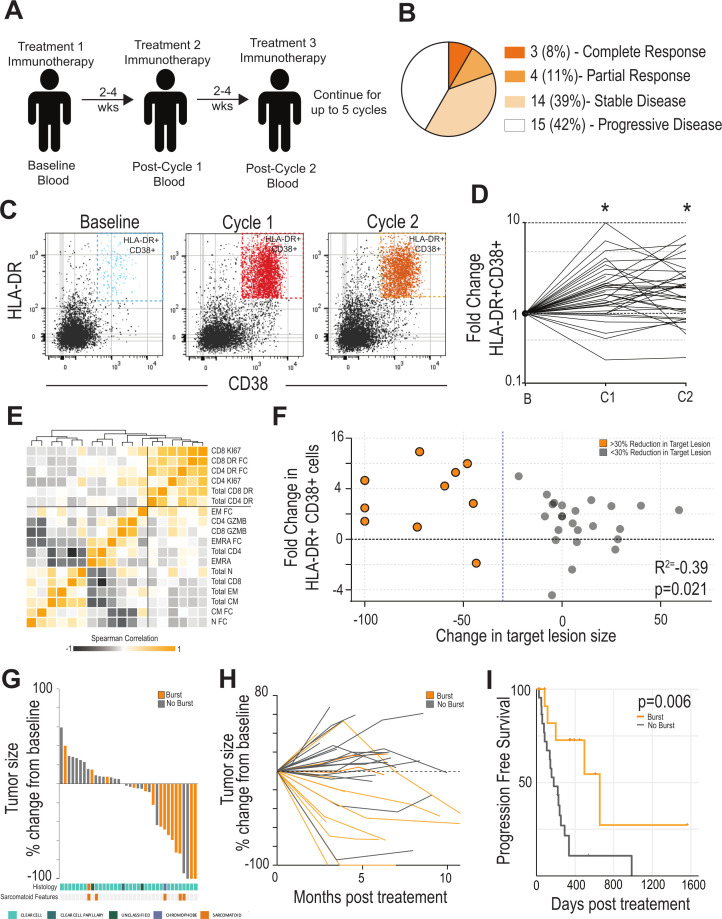
We enrolled 36 patients with metastatic RCC who were to receive checkpoint therapy (figure 1A, online supplemental figure 1A). These patients received either nivolumab, ipilimumab +nivolumab, bempegaldesleukin (IL-2)+nivolumab.39 Twenty-seven patients had clear cell RCC, and the remainder had other, non-clear cell RCC histologies (online supplemental figure 1B). Of these patients, 3 had a CR to therapy, 4 had a PR, 14 had SD, and 15 had progressive disease (PD). Patients in the SD group had an average PFS of 326 days after starting immunotherapy. A total of 21 of 36 (58.3%) patients derived clinical benefit (either SD, CR, or PR) from this treatment and 7/36 (PR or CR) (19.4%) achieved an objective response (figure 1B). Blood was collected from patients immediately before initiating immunotherapy (ie, baseline) and following each treatment cycle (ie, C1, C2) in order to examine how changes in the circulating T cell immune response might correlate with clinical benefit after therapy. CD4 and CD8 T cells (as a per cent of total PBMCs) were unchanged over the course of the study (online supplemental figure 2A, B). Similarly, the breakdown of naïve, Tem, Tcm, and Temra CD8 +T cell subsets (as defined by CD45RA and CCR7 expression) were not significantly different at any time point during treatment (online supplemental figure 2C). The proportion of CD8 T cells expressing granzyme B (GZMB) did not differ following treatment (online supplemental figure 2D).
Figure 1.
Expansion of HLA-DR +CD38+CD8 T cells after checkpoint immunotherapy (IO) is predictive of response in patients with RCC. (A) Study Design. Patients with metastatic RCC receiving checkpoint immunotherapy were recruited. Blood was collected at baseline and at each treatment interval. Blood was assessed for various immune parameters over the course of treatment. (B) Clinical response. Patients were assessed for clinical outcome using RECIST criteria. Data shown indicates outcomes for the cohort of 36 patients. (C) HLA-DR and CD38 expression by CD8 T cells during treatment. Blood was analyzed by flow cytometry. Flow plots shown are gated on CD8 +T cells and show the expression of HLA-DR and CD38 by these cells. (D) Fold change in HLA-DR +CD38+CD8 T cells after treatment. Baseline was set as the untreated level for each patient, and fold change in these cells expressed versus this time point. * Indicates paired t-test <0.05 versus baseline. (E) Correlation of various T-cell parameters in blood. Heatmap shows pairwise correlation of each parameter measured. Squares with darker orange have similar fold changes in this parameter from baseline to cycle 1. Order of parameters are based on similarity clustering by Euclidean distance. (F) Correlation with burst of HLA-DR +CD38+T cells and change in tumor size. Patients in orange are those with at least a 30% reduction in the target lesion. Patients in gray are those with less than a 30% reduction in the target lesion. (G) Waterfall plot. Target lesions were assessed by a pathologist and waterfall plots show the greatest change at any time point during the treatment. Patients with a >2.7-fold increase in HLA-DR +CD38+ cells are highlighted in orange. Patients with a <2.7-fold increase in HLA-DR +CD38+ cells are shown in gray. Histologic subtypes identified in tumor tissue by board-certified pathologists highlighted at the bottom (cyan: clear cell, dark tea: clear cell papillary, green: unclassified, chromophobe: purple). Patients’ tumors exhibiting sarcomatoid features are highlighted in orange beneath histologic subtype. (H) Spider plot showing change in major lesion size over treatment course. Patients underwent scans at 3-month to 6-month intervals. Target lesion was assessed by a pathologist for changes in size during the treatment course. Patients with a >2.7-fold increase in HLA-DR +CD38+ cells (‘burst’) are highlighted in orange, while those with <2.7-fold increase in HLA-DR +CD38+ cells (‘no burst’) are shown in gray. (I) Progression-free survival in patients with or without an increase in HLA-DR +CD38+ cells: Patients were stratified into above (n=14) or below (n=22) the 2.7-fold increase in HLA-DR +CD38+ cells after treatment. Initiation of IO was set as starting point for survival analysis. RCC, renal cell carcinoma; RECIST, Response Evaluation Criteria in Solid Tumor.
jitc-2022-004803supp001.pdf (56.7KB, pdf)
jitc-2022-004803supp002.pdf (1.2MB, pdf)
In comparison to these general T-cell parameters, we found increased expression of several markers of T-cell activation. The most striking change following therapy was an expansion of HLA-DR +CD38+CD8 T cells (recently activated CD8 T cells) in the blood with at least a 2.7-fold increase compared with baseline after cycle 1 (figure 1C, D, online supplemental figure 2F). This increase over baseline was also found at cycle 2, but to a somewhat lesser extent (2.2-fold). A similar pattern was found in HLA-DR +CD38+CD4 T cells—there was a large increase in these cells after one cycle of treatment that somewhat diminished in later treatment cycles (online supplemental figure 2G, H). There was also a significant increase in the proportion of both CD4 and CD8 cells expressing Ki67 +after treatment (online supplemental figure 2E). Interestingly, patients with the largest increase in HLA-DR +CD38+CD8+T cells also had expansion of HLA-DR +CD38+CD4 T cells (online supplemental figure 2I). Importantly, most of the CD8 or CD4 cells expressing Ki67 + cells in the blood were captured in the HLA-DR +CD38+ populations (online supplemental figure 2J, K). Generally, there was a strong correlation between proliferation of CD4 and CD8 in blood and cells that express HLA-DR and CD38, while other blood markers like the number of memory subsets, or cells expression GZMB did not have strong correlations with any parameter (figure 1E).
We were next interested in how the magnitude of this T-cell activation might correlate with a change in tumor size. Each patient’s change in target tumor lesions were assessed using RECIST V.1.1 criteria,41 and we correlated the change in the target lesion size with the magnitude of HLA-DR +CD38+T cell expansion. Patients with the largest fold increase in HLA-DR +CD38+CD8 T cells in their blood had the most significant reductions in tumor size (figure 1F, rho=−0.39, p<0.05). Importantly, patients with >2.7-fold increase in HLA-DR +CD38+CD8 cells were far more likely to clinically benefit from therapy than those with smaller expansion of these cells (figure 1G). Patients with a large burst of new HLA-DR +CD38+CD8+T cells generally had an antitumor effect seen on the first scan on treatment that continued for several treatment cycles (figure 1H). Most importantly, patients with a burst of HLA-DR +CD38+ cells had a longer PFS of 655 days compared 178 days in patients without the burst (p=0.006, figure 1I). Together, these data show that a strong T-cell response measured in the blood immediately after the first cycle of checkpoint therapy is a strong predictor of antitumor effect and thus clinical benefit.
HLA-DR+CD38+CD8 T cells are phenotypically and transcriptionally stable throughout the treatment course
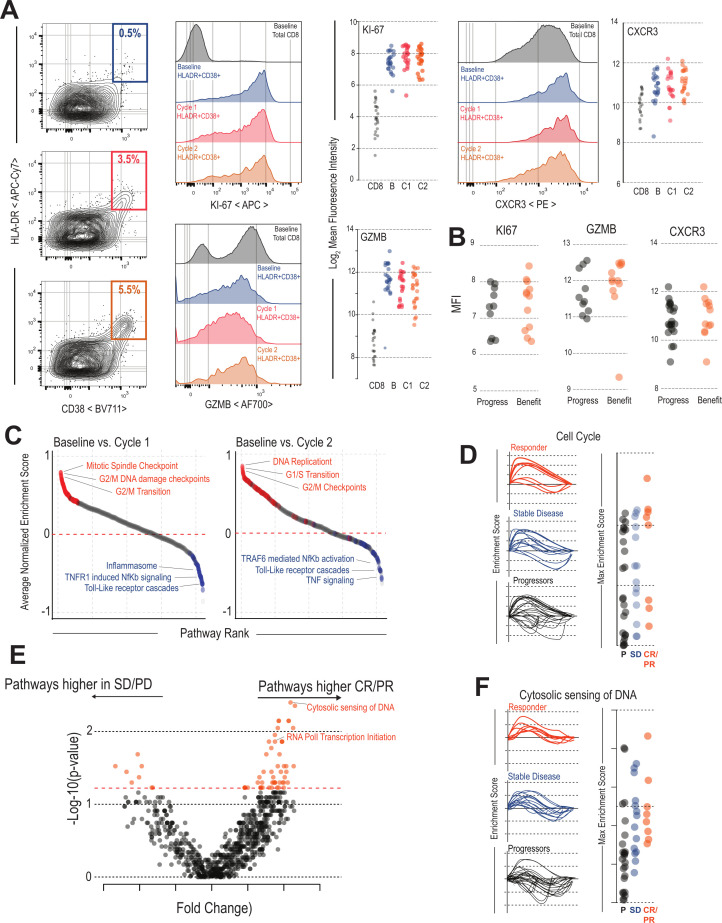
Given that expansion of peripheral HLA-DR +CD38+CD8+T cells had such a strong correlation with clinical benefit after therapy, we wondered how the phenotype of these cells might change throughout treatment. Compared with the total CD8 T-cell population, HLA-DR +CD38+ cells had the highest levels of proliferation marker Ki67, cytotoxic molecule GZMB, and migration marker CXCR3, indicating that these cells had acquired three key traits of effector CD8 T cells: (1) ongoing division, (2) cytotoxicity, and (3) the ability to migrate to areas of inflammation (figure 2A). Expression of these key functional markers remained stable in HLA-DR +CD38+ cells at all time points during treatment (figure 2A). Of note, there was no significant difference in the phenotype of activated CD8 T cells between patients who had clinical benefit compared with those with PD (figure 2B).
Figure 2.
Transcriptional and phenotypic characteristics of HLA-DR +CD38+CD8 T cells during immunotherapy: (A) Flow cytometry analysis of molecules associated with T cell function. HLA-DR +CD38+ cells expression of Ki67, Gzmb, and CXCR3 at baseline, cycle 1, and cycle 2 of immunotherapy. Total CD8 T cells are shown as a control. Summary plots show MFI of these markers for each patient. (B) HLA-DR +CD38+CD8 T cells are phenotypically similar in patients with and without clinical benefit: HLA-DR +CD38+ cells expression of Ki67, Gzmb, and CXCR3 were measured at cycle 1 in with or without clinical benefit. Plots show MFI of these parameters (n=21). (C) Pathways altered in HLA-DR +CD38+CD8 T cells after checkpoint blockade. Pathway analysis comparing baseline to either cycle 1 or cycle 2 cells was performed for each patient. The average of all patients for each pathway is plotted in order of the pathways most upregulated post-treatment. Highlighted in red are pathways within the top 100 related to cell cycle and blue pathways related to innate inflammation. Selected pathways are listed in plots. (D) Cell cycle response after checkpoint blockade. GSEA plots show the enrichment scores of the cell cycle pathway comparing baseline to either cycle 1 or cycle 2 after checkpoint blockade. Summary plots show most patients had increased cell-cycle activity compared with baseline and there was no significant difference between patients with different immunologic outcomes. (E) Pathway analysis comparing patients with and without objective response. GSEA for all REACTOME pathways was performed on all samples comparing HLA-DR +CD38+ cells from baseline to either cycle 1 or cycle 2 cells. The enrichment scores for each pathway were then compared between patients with and without objective response as defined by RECIST criteria. Volcano plot shows the pathways most significantly activated or deactivated in patients with objective responses. (F) Cytosolic sensing of DNA after checkpoint blockade. GSEA plots show the enrichment scores of the Cytosolic sending of DNA pathway comparing baseline to either cycle 1 or cycle 2 after checkpoint blockade. This pathway is highly related to interferon signaling and includes many molecules related to this process. This pathway is significantly upregulated in the HLA-DR +CD38+T cells of patients with clinical benefit after checkpoint blockade. CR, complete response; PR, partial response; PD, progressive disease; RECIST, Response Evaluation Criteria in Solid Tumor; SD, stable disease; GSEA, gene set enrichment analysis; MFI, mean fluorescence intensity.
We were also interested if more subtle changes might occur in this population that might not be detected by measuring these limited markers of T-cell function by flow cytometry, so we performed RNA sequencing on the HLA-DR +CD38+CD8 T cells from 28 patients at baseline, cycle 1, and cycle 2 after therapy. We first compared HLA-DR +CD38+ cells from baseline samples to naïve CD8 T cells collected in previous studies.45 HLA-DR +CD38+CD8 cells expressed high levels of many genes associated with CD8 effector programming, such as perforin-1 (PRF1), GZMB, Tbe, and interferon gamma (IFN-g) (online supplemental figure 3A). We performed gene set enrichment analysis against canonical effector CD8 T cell gene sets from yellow fever vaccine recipients and lymphocytic choriomeningitis virus (LCMV) infected mice.45 46 The HLA-DR +CD38+ cells were highly enriched for these effector signatures suggesting the HLA-DR +CD38+ cells in these patients are similar to newly activated CD8 T cells in antiviral immune responses (online supplemental figure 3B). In addition to enrichment with these effector signatures, HLA-DR +CD38+ cells had enrichment in pathways associated with T-cell activation like TCR signaling and IFN a/b signaling (online supplemental figure 3C). Similar to our findings by flow cytometry, the HLA-DR +CD38+T cells expressed genes associated with T-cell activation, showed high expression of effector molecules (GZMB, PRF1, IFN-g), and showed reduced expression of genes typically expressed by naïve cells, such as CCR7 or IL-7R that did not appreciably change over the treatment course (online supplemental figure 3D).
jitc-2022-004803supp003.pdf (504.3KB, pdf)
We were next interested in how this population of HLA-DR +CD38+ cells changed over the course of treatment and performed gene set enrichment analysis comparing baseline samples to cycle 1 or cycle 2. Of the top 100 pathways turned on after the first treatment, 77 were related to the cell cycle (figure 2C). The pathways most significantly downregulated after treatment were pathways such as tumor necrosis factor signaling, TLR signaling, and the inflammasome. Similar results were found when comparing gene expression at cycle 2 to expression at baseline (figure 2C).
We next compared pathway enrichment between patients who had CR or PR after treatment compared with those with PD or SD. Cell cycle pathways were upregulated in most patients and were not significantly different between patients who progressed, with SD, or PR or CRs (figure 2D). We then aimed to specifically identify pathways that were up or downregulated in these cells in patients who had a clinical response to treatment (figure 2E). Of these, the most significantly upregulated pathway in patients with CR or PR was ‘cytosolic sensing of DNA’, a pathway related to type I IFN signaling (figure 2F). We also compared individual genes that were significantly upregulated between patients with an objective response (CR or PR) (online supplemental figure 4A). Genes that were upregulated in patients without an objective response included checkpoint molecules like PD-1 and TIGIT, in addition to transcription factors such as TOX and EOMES. In comparison, genes upregulated in T cells from patients with an objective response included IRF7 and IRF9, as well as many canonical IFN-stimulated genes like MX1, MX2, OAS1, and OAS2 (online supplemental figure 4B). Similarly, when we compared the size of the HLA-DR +CD38+burst following treatment, patients with the largest increase in these cells had the largest increase in many of the type I IFN related genes like IRF-9 and MX2 (online supplemental figure 4C, D). Finally, we compared how these pathways were altered by the different treatments that patients received. Patients receiving either nivolumab or nivolumab +bempegaldesleukin (IL-2) had no significant differences in pathways turned on by these treatments (online supplemental figure 4E). Patients receiving ipilimumab +nivolumab had a small number of pathways upregulated including the citric acid cycle and antigen cross presentation (online supplemental figure 4E, F). Together, these data indicate that there are some differences in the HLA-DR +CD38+ cells between patients with or without a clinical benefit, mostly related to IFN signaling. However, the most consistent and clear pattern in T-cell activation after therapy is the induction of a proliferative transcriptional program.
jitc-2022-004803supp004.pdf (729KB, pdf)
Responding patients have an influx of new TCR clonotypes to the HLA-DR+CD38+pool after checkpoint blockade
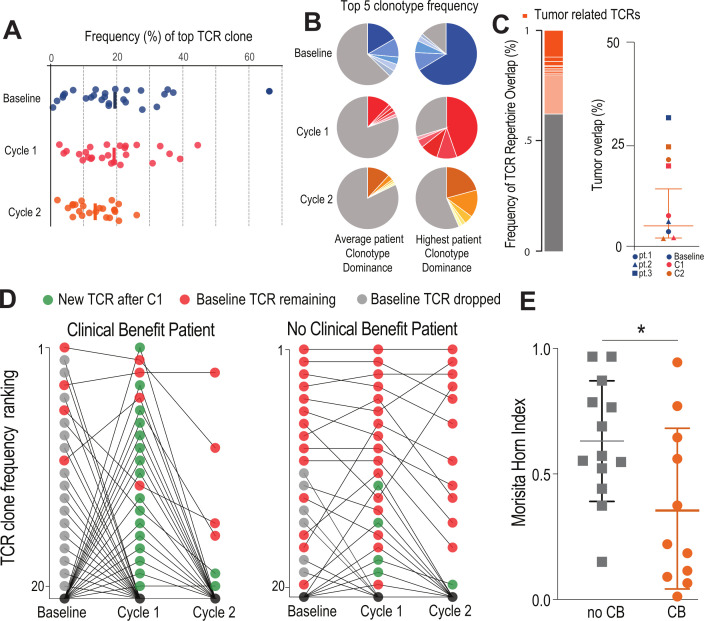
We were next interested in how the TCR repertoire of the HLA-DR +CD38+population might change during treatment. The top clonotype accounted for around 20% of the cells at baseline, cycle 1, and cycle 2, and it took on average 34 TCRs to account for 50% of the total repertoire suggesting this is a clonally expanded population (figure 3, online supplemental figure 5A). There was no difference in this level of immunodominance across the treatment course for any of the different treatment regimens (online supplemental figure 5B). Likewise, there was no change in TCR diversity over the treatment time course (online supplemental figure 5C). For most patients, the top five clonotypes made up around 30% of the repertoire, but for some patients this number was as high as 80%, suggesting that this population of newly activated effector cells was expanded against a small range of antigens (figure 3A, B).
Figure 3.
Patients with clinical benefit have a burst of new TCR clonotypes: (A) HLA-DR CD38+CD8 T cells are clonally expanded. Plot shows the frequency distribution of the dominant TCR clone of circulating activated CD8 T cells (HLA-DR+CD38+) at baseline and after each cycle of therapy (n=26). (B) Clonotype distribution of HLA-DR+CD38+CD8 T cells. Example circle graphs for patients with average (left,~20%) and highest (right) TCR clonotype dominance among their activated circulating CD8 T cells. Shown in different shades of each color are the top five TCR clonotypes in the same patient at each time point. (C) Frequency of TCR repertoire overlap between circulating CD8 T cells before/after therapy and tumor infiltrating CD8 T cells at the time of surgery. Representative proportion of the detected TCR repertoire in circulating CD8 T cells that is unique (gray) or overlapping (orange) with tumor infiltrating CD8 T cells at the time of surgery. Summary of TCR repertoire overlap with primary tumors at the time of surgery for three patients at three time points. Shapes denote a single patient while colors represent the time point at which the samples were collected. (D) Dynamic clonotype rank in responding patients after treatment. Plots show how the top 20 TCR clones present at baseline change over the treatment time course in a representative patient with and without clinical benefit. In the representative patient with clinical benefit, 16 of the top 20 clonotypes are replaced by new TCR clones after the first treatment (green). (E) Similarity of TCR clonotypes from baseline to treatment is significantly different in patients with clinical benefit. Morisita-Horn index was used to quantify the similarity between the different therapy cycles within each individual patient. Mean and SD are shown. *P<0.05 determined by unpaired Welch’s t-test. CB, clinical benefit; TCR, T-cell receptor.
For three patients, we were able to sort infiltrating PD-1 +CD8 T cells from the resected tumor prior to undergoing immunotherapy. In all three of these patients, we found TCR overlap (1%–30%) between circulating HLA-DR +CD38+CD8 T cells at the time of therapy and PD-1 +CD8 TILs at the time of surgery (figure 3C, online supplemental figure 5D), suggesting that HLA-DR +CD38+effector T cells are related to the antitumor immune response prior to and throughout treatment.
We investigated if there were any changes in the TCR repertoire during checkpoint blockade therapy that correlated with response. Interestingly, only in patients who had clinical benefit did we find significant changes in the top clones of the TCR repertoire. Figure 3D shows the top 20 clonotypes ordered by TCR clone frequency for an example patient who had a significant reduction in tumor size. In this patient, 16 out of the top 20 clonotypes lose their dominance in the repertoire after one cycle of treatment (gray dots) and 16 new TCRs now occupy these top spots (green dots). In comparison, in a patient who had no clinical benefit, only 4 new clonotypes enter the top 20 dominant clones after treatment, while most TCR clones kept exactly the same rank as baseline (figure 3D). This change was not associated with an overall change in TCR diversity among responding or non-responding patients (online supplemental figure 5E). Given the TCR dominance within this expanded population, the top 20 clones account for the majority of the responding TCRs in the repertoire. Thus, the changes in the top 20 TCR clonotypes are reflective of changes in the overall TCR repertoire, suggesting that only in patients with clinical benefit is there a dynamic shift in TCR dominance after immunotherapy. We used the Morisita-Horn (MH) index, a measurement of the similarity of TCR clonotypes, to quantify this change in TCR repertoire between baseline and post-treatment samples. We found a significantly lower MH index in patients with clinical benefit (MH=0.36) when compared with patients with worsening disease (MH=0.63), supporting the idea that clinical benefit is associated with a dynamic TCR repertoire in the HLA-DR +CD38+T cell population (figure 3E). This effect was not associated with any particular treatment the patients received (online supplemental figure 5F). Together, these data indicate that a feature of the HLA-DR +CD38+population in responding patients is an influx of new clonotypes to the immune response, and patients who do not have this occur are much less likely to respond to treatment.
Pre-existing tumor immunity is an important predictor of later immunologic response to checkpoint blockade
Activation of new TCR clonotypes and a large burst of HLA-DR +CD38+T cells following checkpoint blockade raises the question of why some patients have this response to treatment while others do not. Prior studies have found that a TCF-1 +CD8+T cell is the cell that proliferates following checkpoint blockade, and that having these cells present in tumors is an important predictor objective clinical response to checkpoint therapy.33 34 Our previous work found that kidney tumors harbor TCF-1 +CD8 T cells in regions of dense antigen presenting cells, and that having these regions in a tumor predicts both the magnitude of total T-cell infiltration and patient survival.44 Based on these data, and in keeping with previous reports in other tumor types,29 35 47 we reasoned that having a strong CD8 T-cell response in the tumor at the time of surgery might predict features of a patient’s later clinical response to immunotherapy.
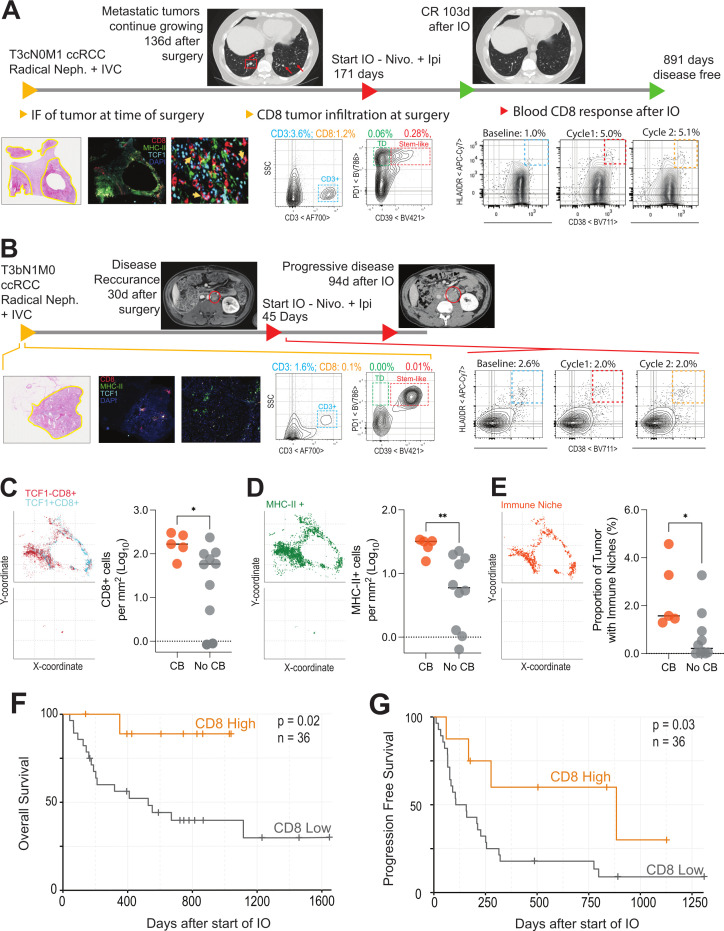
We analyzed surgically resected tumors from 15 patients of the original 36 analyzed in figures 1–3 by IF (online supplemental figure 6A-C). In patients with clinical benefit, there were significantly more total CD8 T cells, more TCF-1 +CD8 T cells, and more MHC-II + cells in the tumor at the time of surgery (figure 4C–E). Figure 4A, B highlight examples of patients with or without clinical benefit following checkpoint therapy. The patient in figure 4A was diagnosed with stage IV (pT3N0M1) clear cell RCC. The patient underwent a radical nephroureterectomy with adrenalectomy, inferior vena cava (IVC) tumor thrombectomy, partial IVC resection, resection of psoas and diaphragm crus, retroperitoneal lymph node dissection, and omentoplasty. The tumor invaded the perinephric and renal sinus fat and angiolymphatics, and showed eosinophilic features, WHO/ISUP grade 3, and a multifocal growth pattern. By IF, we found extensive CD8 T cell and MHC-II + T-cell infiltration and that TCF-1 +CD8 T cells were predominantly resident in dense antigen presenting niches (figure 4A and C–E). By flow cytometry, this patient had 1.2% CD8 T cell infiltration, which is near the mean for all patients with RCC we have previously analyzed (online supplemental figure 6K). Importantly, we also found many TCF-1 +stem like CD8 T cells in the tumor by flow cytometry, and we observed a sizeable population of CD39 + terminally differentiated effector T cells in the tumor. Five months after surgery, the patient’s previously suspicious but non-diagnostic pulmonary nodules increased in size, indicating growing metastases. This patient then received checkpoint immunotherapy, and measurement of peripheral blood T-cell activation revealed a large expansion of HLA-DR +CD38+ cells in the blood when compared with baseline levels (figure 4A). The patient had a CR to therapy, with resolution of all lung metastasis at the 3-month follow-up scan and has remained disease free at our censoring date more than 800 days after starting immunotherapy.
Figure 4.
Long-term immunological status is correlated with clinical benefit following immunotherapy: (A and B) Examples of longitudinal immune analysis in a responding and non-responding patient: Plots show the timeline of a patient and immunologic status at each point. This timeline includes when they were diagnosed and received surgery, details of disease progression, time point where immunotherapy was started, subsequent scans after immunotherapy. Below the timeline are the immunological parameters collected showing H&E with tumor areas outlined in yellow, immunofluorescence of tumor at the time of surgery, flow cytometry of the tumor at time of surgery showing total CD3 infiltration and the phenotype of CD8 cells, and the CD8 response in the blood after receiving IO. (C and D) Quantitative analysis of CD8 T cells and MHC-II+ cells in patients with and without clinical benefit from immunotherapy. Immunofluorescence images for 15 patients, stratified into those with and without clinical benefit, were analyzed for infiltration by total, TCF-1+and TCF-1–CD8 T cells (C), and MHC-II+ cells (D). Spatial plots show where each of these subsets are found in tumor tissue and summary plots show the proportion of these cells in tumors of representative patients who had clinical benefit (CB) versus those who did not (no CB). * Indicates p<0.05, n=15. (E) Patients who clinically benefit from immunotherapy have a higher proportion of immunologic niches in their tumors. Immune niches were defined as regions with ≥1 MHC-II+ cells and ≥1 TCF-1+CD8 T cells in the same local neighborhood (100μm×100μm) and proportions were calculated out of the total number of 100μm×100μm neighborhoods contained within the entire tissue slice. Spatial plots demonstrate these immune niches in representative patients with clinical benefit (top) and without clinical (bottom), and a summary plot demonstrates the proportion of immune niches in tumors of patients who had CB versus those who did not (no CB). * Indicates p<0.05. (F) Overall survival after initiation of immunotherapy for patients with high or low CD8 infiltration into tumors measured by flow cytometry. Patients were stratified into CD8 high (>2.2% CD8 infiltration into tumor, n=8) or CD8 low (<2.2% CD8 infiltration into tumor, n=28) groups. Initiation of immunotherapy (IO) was set as starting point for survival analysis. (G) Progression-free survival after initiation of immunotherapy for patients with high or low CD8 infiltration into tumors. Patients were stratified in the same manner as ((F) and the time until progression as defined by Response Evaluation Criteria in Solid Tumor criteria was measured. IVC, inferior vena cava; TCF-1, T cell factor 1; ccRCC, clear cell renal cell carcinoma.
jitc-2022-004803supp006.pdf (3.1MB, pdf)
In comparison, figure 4B shows a patient with similarly advanced disease who had no clinical benefit after receiving checkpoint immunotherapy. This patient was also diagnosed with stage IV disease (pT3N1M1) clear cell RCC and underwent radical nephrectomy, adrenalectomy, psoas resection, lymph node dissection, and IVC resection with tumor thrombectomy. The tumor invaded the perinephric and renal sinus fat, angiolymphatic invasion was present, several para-aortic and retroperitoneal lymph nodes were positive, and extracapsular extension was noted. The tumor showed eosinophilic and rhabdoid features and WHO/ISUP grade 4. Extremely few immune cells were found in this patient’s tumor at the time of surgery by IF (figure 4B–E). By flow cytometry this patient had less than 0.1% of the tumor as CD8 +T cells, which places this patient in the bottom 10% of all patients. When they received immunotherapy approximately 1 month later, there was no appreciable T-cell activation in the blood and their disease progressed rapidly.
This trend of strong immune infiltration in the tumor and later systemic response to checkpoint therapy was found in 15 total patients (table 3) analyzed by IF. A representative patient, demonstrating an intermediate amount of immune infiltration, is shown, highlighting the location of the tumor and immune infiltrate (online supplemental figure 6A, B). Whole slide IF data were quantified to discern the xy location of each cell, as well as the intensity of staining for each marker in each cell. The immuno maps shown in online supplemental figure 6C demonstrate the xy location of all CD8 T cells (red, top) and of TCF-1 +CD8 T cells (cyan, top), as well as the xy location of each MHC-II + cell (green, middle). Importantly, we are also able to define areas that contain both TCF-1 +CD8+T cells and MHC-II + cells, which we term ‘immune niches’ (orange, bottom, defined as 100μm×100μm areas with ≥1 TCF-1 +CD8 T cells and ≥1 MHC-II + cells). In line with our previous studies,44 we found the number of T cells correlates strongly with the amount of MHC-II+ (online supplemental figure 6D) and TCF-1 +CD8 T cells in the tissue (online supplemental figure 6E). Similarly, the proportion of the tumor that was made up with immune niche strongly correlated with the proportion of MHC-II +and TCF + cells in the tumor (online supplemental figure 6F, G). We then examined if these immunologic features of the tumor microenvironment correlated with patients’ later response to immunotherapy. In this group of 15 patients, those with clinical benefit to checkpoint blockade had significantly higher numbers of total CD8 T cells in their tumor (figure 4C), and a trend towards more TCF-1 +CD8 T cells (p=0.055, online supplemental figure 6H). These patients also had more areas of MHC-II + cell density (where TCF-1 + cells are usually found) (figure 4D), and more areas defined as immune niches. Having both more MHC-II dense regions and immunological niches was strongly correlated with the size of the HLA-DR +CD38+burst in CD8 T cells after later checkpoint therapy (online supplemental figure 6I, J). Among these 15 patients, those with an above median increase in HLA-DR +CD38+ cells had significantly more of these immune niches in their tumors at the time of surgery (figure 4E).
Table 3.
| N | % | ||
| Total | 15 | 100.00 | |
| Age at time of surgery | Median (range) | 62 (33–75) | |
| Sex | Male | 9 | 60.00 |
| Female | 6 | 40.00 | |
| Race | Black/African American | 2 | 13.33 |
| White/Caucasian | 13 | 86.67 | |
| Histologic subtype | Clear cell RCC | 11 | 73.33 |
| Other RCC | 4 | 26.67 | |
| Treatment | Nivolumab | 3 | 20.00 |
| Ipilimumab+nivolumab | 9 | 60.00 | |
| Study drug+ipilimumab+nivolumab | 3 | 20.00 | |
| Histologic subtype | Clear cell RCC | 11 | 73.33 |
| Other RCC | 3 | 20.00 | |
| Stage at diagnosis | I | 1 | 6.67 |
| II | 0 | 0.00 | |
| III | 6 | 40.00 | |
| IV | 8 | 53.33 | |
RCC, renal cell carcinoma.
To extend this analysis, we also had data available from 36 additional patients for whom we had flow cytometry analysis quantitating immune infiltration in their primary tumors that later went on to receive checkpoint blockade treatments. In these patients, the overall T-cell infiltration spans the range we have typically seen and reported in RCC (online supplemental figure 6K), and importantly, we were able to identify terminally differentiated and stem-like CD8 T cells, as we have previously extensively characterized (as in online supplemental figure 6L).44 In this cohort of 36 patients (see table 4), those that had >2.2% of the total cells in the tumor as CD8 +T cells had significantly longer overall survival (figure 4F), and significantly longer time to tumor progression after initiating checkpoint blockade therapy (figure 4G). Together, these two cohorts highlight that a patient’s ability to mount an immune response against their tumor is a feature that extends across the course of their disease and impacts patient outcomes following systemic immunotherapy. When patients have strong intratumoral T-cell activity at the time of surgery, characterized by infiltration of TCF-1 +CD8 T cells and dense regions of antigen presenting cells, the immunological response to later therapy is stronger and patients have improved outcomes.
Table 4.
| N | % | ||
| Total | 36 | 100.00 | |
| Age at time of surgery | Median (range) | 63.3 (32.5–76.6) | |
| Sex | Male | 23 | 63.89 |
| Female | 13 | 36.11 | |
| Race | Black/African American | 9 | 25.00 |
| White/Caucasian | 27 | 75.00 | |
| Histologic subtype | Clear cell RCC | 27 | 75.00 |
| Other RCC | 9 | 25.00 | |
| Treatment | Nivolumab | 14 | 38.89 |
| Ipilimumab+nivolumab | 12 | 33.33 | |
| Cabozantinib+nivolumab | 6 | 16.67 | |
| Levatinib+nivolumab | 1 | 2.78 | |
| Atezolizumab | 1 | 2.78 | |
| Axitinib+avelumab | 1 | 2.78 | |
| Study drug+ipilimumab+nivolumab | 1 | 2.78 | |
| Histologic subtype | Clear cell RCC | 27 | 75.00 |
| Other RCC | 9 | 25.00 | |
| Stage at diagnosis | I | 2 | 5.56 |
| II | 2 | 5.56 | |
| III | 13 | 36.11 | |
| IV | 19 | 52.78 | |
| %CD8 strata | High | 10 | 27.78 |
| Low | 26 | 72.22 | |
| %CD8 | Median (range) | 1.1 (0.0–24.5) | |
RCC, renal cell carcinoma.
Discussion
In this study, we investigated how T-cell activation in the blood of patients with RCC was altered after checkpoint blockade. We found a rapid expansion in HLA-DR +CD38+CD4 and CD8 T cells immediately after the first cycle of immunotherapy. These cells expressed the proliferation marker Ki67 and important effector molecules like GZMB and CXCR3 and were strongly enriched for cell cycle genes. Patients with clinical benefit typically had a much larger increase in these cells, in line with previous reports,14–16 19 such as Kim et al, where a higher fold percentage in Ki67 + cells among PD-1 +CD8 T cells after immunotherapy predicted durable clinical benefit in non-small cell lung cancer and thymic epithelial tumors.19 Interestingly, in this work by Kim et al, they found a fold change of Ki67 of 2.8 from baseline was predicative of survival, very much in line with our cut-off of 2.7-fold change. We also found that in patients who had clinical benefit had a TCR repertoire that was replaced by new clonotypes. In comparison, patients with no clinical benefit had a stable TCR repertoire where the most frequent clones before treatment maintained their dominance after therapy. Several studies have previously reported that outcomes in patients treated with checkpoint therapy are associated with new clones entering the total CD8 or the activated PD-1 +CD8 T cell pool.24 48 49 Importantly, recent work reported that there is clonal replacement in the tumor of patients receiving anti-PD-1.49 Our data indicates that only responding patients had an influx of new clones to the blood and this may be the cells that replace TCRs in the tumors of patients responding to treatment. Interestingly, despite this new influx of clones, the RNA sequencing profiles of these cells is mostly similar between responding and non-responding patients, suggesting that the phenotype of the effector is less important than the overall number and repertoire of T cells responding to the treatment. Finally, we find that the T-cell response in a patient’s originally resected tumor (with surgery occurring months to years before receiving immunotherapy) predicts later outcomes following checkpoint therapy, where patients with more CD8 T cells in their tumor exhibit clinical benefit. Many of these individual observations have been made alone in other cancers, but here we have been able to show how immunobiology at the time of surgery has an enduring impact on subsequent immune response to checkpoint inhibitor therapy.
In the past few years, details of the cellular mechanisms that control activation of T cells after PD-1 blockade have been identified. Most notably, the TCF-1 +CD8 T cells we have previously described in these patient’s tumors are likely the same cells that proliferate after PD-1 blockade.36 50 51 On treatment with anti-PD-1, these stem-like CD8 T cells proliferate, but more importantly, give rise to cytotoxic daughter cells that are responsible for clearing virally infected cells36 or tumor cells. Based on these studies and our data here, we hypothesize that there is a pool of tumor specific TCF-1 +stem like CD8 T cells that are not proliferating or generating antitumor effector cells but that are the biologic reservoir that is unleashed by checkpoint blockade. Currently it is unclear if these dormant TCF-1 + cells are the ones in the tumor we identified here or are in other locations outside the tumor such as the tumor draining lymph nodes, and this is important to identify in future studies. Based on this hypothesis, we propose that patients who have this dormant TCF-1 + stem like T-cell pool are those who can generate a large new repertoire of T cells after checkpoint blockade. Evidence of this process occurring is given by the large burst of HLA-DR +CD38+CD8 T cells seen in the blood following immune checkpoint blockade and is likely why measuring this burst in activated cells is promising indicator of clinical benefit.
This study has a few key real-world limitations. To minimize impact on patients, the blood collection time points were cycle dependent, coinciding with scheduled phlebotomy, thus intervals for the standard of care and clinical trial immunotherapy regimens ranged from 2 to 4 weeks. Previous studies have shown that the initial burst in activated CD8 T cells may be seen as early as 1 week post-treatment,19 therefore sample collection for patients on the longer dosing interval may have missed the greatest magnitude of the burst. Similarly, the timing and type of re-staging radiographs varied among patients on standard of care and clinical trial treatments. The evaluation of baseline tumor T-cell infiltration was limited to those patients who underwent resection at Emory University Hospital, which introduces bias in clinical presentation as the surgically resected patients may be different than those who underwent biopsy elsewhere. Despite these practical variances, among patients with predominantly clear cell RCC treated with different immunotherapy regimens, we show that an early burst in activated CD8 T cells in the peripheral blood and presence of TCF-1 +stem like CD8 T cells in the tumor as seen in archival samples are associated with clinical benefit. In future work, larger prospective and randomized studies are needed to confirm the utility of these accessible predictors of response to immunotherapy, with the hope of informing selection of optimal treatment.
jitc-2022-004803supp007.pdf (157.4KB, pdf)
Footnotes
Twitter: @careyjans, @sowalsky, @bilenma, @haydnkissick
Contributors: Conception and design: JWC, CJ, MAB, HK. Acquisition of data: JWC, CJ, MAC, AMR, RG, LDB, NP, DB, FH, RL. Analysis and interpretation of data: JWC, CJ, MAC, ES, AMR, RG, NP, PCM, AO. Technical resources and expertise: SW, RL, AS. Clinical samples: OK, BCC, PCM, AO, VAM, MAB. Statistical analysis: YL, HK. Manuscript drafting: JWC, CJ, MAC, HK. Manuscript revision: JWC, CJ, MAC, HK. Guarantor: HK. Final approval: all authors.
Funding: CJ is supported by a National Cancer Institute grant (1-F30-CA-243250).
Competing interests: MAB has acted as a paid consultant for and/or as a member of the advisory boards of Exelixis, Bayer, BMS, Eisai, Pfizer, AstraZeneca, Janssen, Calithera Biosciences, Genomic Health, Nektar, and Sanofi and has received grants to his institution from Xencor, Bayer, Bristol Myers Squibb, Genentech/Roche, Seattle Genetics, Incyte, Nektar, AstraZeneca, Tricon Pharmaceuticals, Genome & Company, AAA, Peloton Therapeutics, and Pfizer for work performed as outside of the current study. HK acted as a paid consultant for Nektar and received grants to his institution from Nektar.
Provenance and peer review: Not commissioned; externally peer reviewed.
Author note: JWC and CJ are co-first authors. VAM, MAB, and HK are co-last authors.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Data is available under GSE181465.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved under the Emory University Urological Satellite Specimen Bank in accordance with the Institutional Review Board (IRB00055316). Participants gave informed consent to participate in the study before taking part.
References
- 1.Klapper JA, Downey SG, Smith FO, et al. High‐dose interleukin‐2 for the treatment of metastatic renal cell carcinoma. Cancer 2008;113:293–301. 10.1002/cncr.23552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang JC, Sherry RM, Steinberg SM, et al. Randomized study of high-dose and low-dose interleukin-2 in patients with metastatic renal cancer. J Clin Oncol 2003;21:3127–32. 10.1200/JCO.2003.02.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motzer RJ, Jonasch E, Boyle S, et al. NCCN guidelines insights: kidney cancer, version 1.2021. J Natl Compr Canc Netw 2020;18:1160–70. 10.6004/jnccn.2020.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motzer RJ, Tannir NM, McDermott DF, et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med 2018;378:1277–90. 10.1056/NEJMoa1712126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med 2015;373:1803–13. 10.1056/NEJMoa1510665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2019;380:1116–27. 10.1056/NEJMoa1816714 [DOI] [PubMed] [Google Scholar]
- 7.Motzer RJ, Penkov K, Haanen J, et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2019;380:1103–15. 10.1056/NEJMoa1816047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motzer R, Alekseev B, Rha S-Y, et al. Lenvatinib plus pembrolizumab or everolimus for advanced renal cell carcinoma. N Engl J Med 2021;384:1289–300. 10.1056/NEJMoa2035716 [DOI] [PubMed] [Google Scholar]
- 9.Choueiri TK, Powles T, Burotto M, et al. Nivolumab plus cabozantinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2021;384:829–41. 10.1056/NEJMoa2026982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choueiri TK, Tomczak P, Park SH, et al. Adjuvant pembrolizumab after nephrectomy in renal-cell carcinoma. N Engl J Med 2021;385:683–94. 10.1056/NEJMoa2106391 [DOI] [PubMed] [Google Scholar]
- 11.Xu W, Atkins MB, McDermott DF. Checkpoint inhibitor immunotherapy in kidney cancer. Nat Rev Urol 2020;17:137–50. 10.1038/s41585-020-0282-3 [DOI] [PubMed] [Google Scholar]
- 12.Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US food and drug administration (FDA) approvals of immune checkpoint inhibitors. J Immunother Cancer 2019;7:278. 10.1186/s40425-019-0768-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim KH, Cho J, Ku BM, et al. The First-week Proliferative Response of Peripheral Blood PD-1+CD8+ T Cells Predicts the Response to Anti-PD-1 Therapy in Solid Tumors. Clin Cancer Res 2019;25:2144–54. 10.1158/1078-0432.CCR-18-1449 [DOI] [PubMed] [Google Scholar]
- 14.Huang AC, Postow MA, Orlowski RJ, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017;545:60–5. 10.1038/nature22079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang AC, Orlowski RJ, Xu X, et al. A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat Med 2019;25:454–61. 10.1038/s41591-019-0357-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamphorst AO, Pillai RN, Yang S, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci U S A 2017;114:4993–8. 10.1073/pnas.1705327114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dronca RS, Liu X, Harrington SM, et al. T cell Bim levels reflect responses to anti-PD-1 cancer therapy. JCI Insight 2016;1. 10.1172/jci.insight.86014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairfax BP, Taylor CA, Watson RA, et al. Peripheral CD8+ T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat Med 2020;26:193–9. 10.1038/s41591-019-0734-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim KH, Cho J, Ku BM, et al. The First-week Proliferative Response of Peripheral Blood PD-1+CD8+ T Cells Predicts the Response to Anti-PD-1 Therapy in Solid Tumors. Clin Cancer Res 2019;25:2144. 10.1158/1078-0432.CCR-18-1449 [DOI] [PubMed] [Google Scholar]
- 20.Valpione S, Galvani E, Tweedy J, et al. Immune-awakening revealed by peripheral T cell dynamics after one cycle of immunotherapy. Nat Cancer 2020;1:210–21. 10.1038/s43018-019-0022-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boland GM, Flaherty KT. Tracking early response to immunotherapy. Nat Cancer 2020;1:160–2. 10.1038/s43018-020-0032-8 [DOI] [PubMed] [Google Scholar]
- 22.Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu TD, Madireddi S, de Almeida PE, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 2020;579:274–8. 10.1038/s41586-020-2056-8 [DOI] [PubMed] [Google Scholar]
- 24.Han J, Duan J, Bai H, et al. TCR Repertoire Diversity of Peripheral PD-1+CD8+ T Cells Predicts Clinical Outcomes after Immunotherapy in Patients with Non-Small Cell Lung Cancer. Cancer Immunol Res 2020;8:146. 10.1158/2326-6066.CIR-19-0398 [DOI] [PubMed] [Google Scholar]
- 25.Cha E, et al. Improved survival with T cell clonotype stability after Anti–CTLA-4 treatment in cancer patients. Science Translational Medicine 2014;6:ra270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hui E, Cheung J, Zhu J, et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science 2017;355:1428. 10.1126/science.aaf1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagès F, Berger A, Camus M, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 2005;353:2654–66. 10.1056/NEJMoa051424 [DOI] [PubMed] [Google Scholar]
- 28.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960. 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 29.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peranzoni E, Lemoine J, Vimeux L, et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti–PD-1 treatment. Proc Natl Acad Sci U S A 2018;115:E4041. 10.1073/pnas.1720948115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herbst RS, Soria J-C, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563–7. 10.1038/nature14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-Infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol 2012;30:2678–83. 10.1200/JCO.2011.37.8539 [DOI] [PubMed] [Google Scholar]
- 33.Sade-Feldman M, Yizhak K, Bjorgaard SL, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 2018;175:998–1013. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller BC, Sen DR, Al Abosy R, et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20:326–36. 10.1038/s41590-019-0312-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen P-L, Roh W, Reuben A, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov 2016;6:827. 10.1158/2159-8290.CD-15-1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Im SJ, Hashimoto M, Gerner MY, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 2016;537:417–21. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurtulus S, Madi A, Escobar G, et al. Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1-CD8+ Tumor-Infiltrating T Cells. Immunity 2019;50:181–94. 10.1016/j.immuni.2018.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siddiqui I, Schaeuble K, Chennupati V, et al. Intratumoral Tcf1+PD-1+CD8+ T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 2019;50:195–211. 10.1016/j.immuni.2018.12.021 [DOI] [PubMed] [Google Scholar]
- 39.Diab A, Tannir NM, Bentebibel S-E, et al. Bempegaldesleukin (NKTR-214) plus nivolumab in patients with advanced solid tumors: phase I dose-escalation study of safety, efficacy, and immune activation (PIVOT-02). Cancer Discov 2020;10:1158. 10.1158/2159-8290.CD-19-1510 [DOI] [PubMed] [Google Scholar]
- 40.Long GV, Tykodi SS, Schneider JG, et al. Assessment of nivolumab exposure and clinical safety of 480 mg every 4 weeks flat-dosing schedule in patients with cancer. Ann Oncol 2018;29:2208–13. 10.1093/annonc/mdy408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 42.Kamentsky L, Jones TR, Fraser A, et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 2011;27:1179–80. 10.1093/bioinformatics/btr095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carpenter AE, Jones TR, Lamprecht MR, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006;7:R100. 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jansen CS, Prokhnevska N, Master VA, et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 2019;576:465–70. 10.1038/s41586-019-1836-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akondy RS, Fitch M, Edupuganti S, et al. Origin and differentiation of human memory CD8 T cells after vaccination. Nature 2017;552:362–7. 10.1038/nature24633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wherry EJ, Ha S-J, Kaech SM, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 2007;27:670–84. 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- 47.Daud AI, Loo K, Pauli ML, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest 2016;126:3447–52. 10.1172/JCI87324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li T, Zhao L, Yang Y, et al. T Cells Expanded from PD-1+ Peripheral Blood Lymphocytes Share More Clones with Paired Tumor-Infiltrating Lymphocytes. Cancer Res 2021;81:2184. 10.1158/0008-5472.CAN-20-2300 [DOI] [PubMed] [Google Scholar]
- 49.Yost KE, Satpathy AT, Wells DK, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 2019;25:1251–9. 10.1038/s41591-019-0522-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Utzschneider DT, Charmoy M, Chennupati V, et al. T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 2016;45:415–27. 10.1016/j.immuni.2016.07.021 [DOI] [PubMed] [Google Scholar]
- 51.He R, Hou S, Liu C, et al. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature 2016;537:412–6. 10.1038/nature19317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-004803supp005.pdf (773.8KB, pdf)
jitc-2022-004803supp001.pdf (56.7KB, pdf)
jitc-2022-004803supp002.pdf (1.2MB, pdf)
jitc-2022-004803supp003.pdf (504.3KB, pdf)
jitc-2022-004803supp004.pdf (729KB, pdf)
jitc-2022-004803supp006.pdf (3.1MB, pdf)
jitc-2022-004803supp007.pdf (157.4KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Data is available under GSE181465.