Abstract
Re‐establishment of desiccation tolerance is essential for the survival of germinated seeds facing water deficit in the soil. The molecular and ultrastructural features of desiccation tolerance maintenance and loss within the nuclear compartment are not fully resolved. In the present study, the impact of desiccation‐induced genotoxic stress on nucleolar ultrastructure and ribogenesis was explored along the rehydration−dehydration cycle applied in standard seed vigorization protocols. Primed and overprimed Medicago truncatula seeds, obtained through hydropriming followed by desiccation (dry‐back), were analysed. In contrast to desiccation‐tolerant primed seeds, overprimed seeds enter irreversible germination and do not survive dry‐back. Reactive oxygen species, DNA damage and expression profiles of antioxidant/DNA Damage Response genes were measured, as main hallmarks of the seed response to desiccation stress. Nuclear ultrastructural features were also investigated. Overprimed seeds subjected to dry‐back revealed altered rRNA accumulation profiles and up‐regulation of genes involved in ribogenesis control. The signal molecule PAP (3′‐phosphoadenosine 5′‐phosphate) accumulated during dry‐back only in primed seeds, as a distinctive feature of desiccation tolerance. The presented results show the molecular and ultrastructural landscapes of the seed desiccation response, including substantial changes in nuclear organization.
Keywords: 5.8S RNA, hydropriming/dry‐back, nucleolus and ribogenesis, overpriming, seed germination
Summary Statement
Novelty on desiccation tolerance in overprimed seeds is brought by distinctive changes in the genotoxic stress response, profiles of precursor and mature rRNAs, and accumulation of the signal molecule PAP.
1. INTRODUCTION
Severe drought conditions caused by climate change threaten sensitive crop species, highlighting their physiological limitations and the need for new varieties more adapted to adverse environments (Howden et al., 2007). Desiccation tolerance (DT), or the ability to survive extreme water losses (Bewley, 1979; Leprince et al., 2017; Oliver et al., 2020), is a key trait for the design of effective strategies to fight drought and limit the progressive expansion of global dryland while targeting fundamental issues, such as food security (Anderson et al., 2020). DT, a typical feature of orthodox seeds, includes a plethora of complex molecular events maintained as long as the embryo radicle is kept inside the seed envelope (Buitink et al., 2006; González‐Moralesa et al., 2016; Smolikova et al., 2020; Terrasson et al., 2013). DT is progressively lost during germination; however, it can be rescued within a short developmental window between germination and seedling establishment, by exposing seeds to polyethylene glycol (PEG)‐induced osmotic stress (Bruggink & van der Toorn, 1995; Peng et al., 2017). Such a developmental window corresponds to the stage where the length of the protruding radicle ranges between 1 and 3 mm in Medicago truncatula (Buitink et al., 2003; Maia et al., 2011). Abscisic acid (ABA) mediates the PEG‐induced re‐establishment of DT, and the phytohormone alone can also trigger the process (Costa et al., 2015; Peng et al., 2017). Proteomic analysis of seeds collected during the PEG‐ and ABA‐induced re‐establishment of DT has provided further insights into the complex mechanisms underlying this trait (Peng et al., 2018; Wang et al., 2021). DT re‐establishment is essential for the survival of germinated seeds facing a transient water deficit condition. It has been hypothesized that this physiological trait has a relevant biological and ecological significance since it might contribute to optimizing seedling establishment (Dekkers et al., 2015). Resumption of DT can help seeds that encounter unpredictable natural drying conditions in the soil, during and after germination. By exploiting this post‐germination window for DT, early seedlings can survive until a favourable environment is restored (Dekkers et al., 2015).
In the context of seed technology, loss and gain of DT are critical issues in seed priming (Fabrissin et al., 2021; Farooq et al., 2019; Paparella et al., 2015). Priming starts with seed imbibition under controlled conditions with water (hydropriming) or specific priming agents to boost seed metabolism and to allow germination to proceed without reaching the final stage of radicle protrusion (Soeda et al., 2005). Although treatments must be tailored, based on the plant species, seed morphology and physiology, they share common cellular and molecular dynamics associated with the activation of the pre‐germinative metabolism (Bailly, 2004; Macovei et al., 2017). Excessive reactive oxygen species (ROS) generated during imbibition are detrimental to nucleic acids; thus the pre‐germinative metabolism features the combined effect of enhanced antioxidant activity and DNA repair to preserve genome integrity (Bailly, 2004; Forti et al., 2020a, 2020b, 2021; Macovei et al., 2010, 2011; Pagano et al., 2017, 2019; Waterworth et al., 2019). Increasing priming duration brings seeds to exceed a critical threshold and triggers the irreversible phase of germination, a process known as overpriming. At this point, DT is lost and the dry‐back of seeds with radicle protrusion results in the death of early seedlings. Indeed, acceleration imposed by prolonged priming shortens the lag time between imbibition and radicle emergence (Bradford et al., 1990). Based on these premises, the rehydration−dehydration cycle routinely applied in standard seed vigorization protocols represents an ideal biological context to explore some critical issues of DT, including mechanisms underlying the maintenance of genome integrity.
There is emerging evidence that the plant nucleolus participates in stress signalling pathways, and the maintenance of genome integrity (Kalinina et al., 2018). Recent findings suggest the involvement of plant‐specific transcription factors in the nucleolar stress response. Ohbayashi et al. (2017) proposed that the ANAC082 transcription factor might have roles similar to those played by the animal p53 protein. In addition, some studies implicate the plant nucleolus in tyrosyl‐DNA phosphodiesterases (TDPs)‐dependent DNA damage response (DDR) processes (Donà et al., 2013; Macovei et al., 2018). The nucleolus harbours several proteins involved in both DNA repair and ribosome assembly, suggesting a crosstalk between these two main cellular mechanisms (Orgawa & Baserga, 2017). Such noncanonical roles expand the interface connecting ribogenesis and DDR, bringing additional layers of complexity still unexplored in plants. In the present work, we applied the rehydration−dehydration cycle (hydropriming followed by dry‐back) to M. truncatula seeds, as a model to gain insights into the molecular events associated with the progressive loss of DT. The focus on DDR and ribogenesis allowed us to observe molecular players and ultrastructural features linked to dehydration‐induced genotoxic stress. Shedding light on these basic processes associated with DT will help determine novel molecular hallmarks for a more rational design of seed vigorization protocols, as urgently needed to preserve crop productivity in the current global climate emergency.
2. MATERIALS AND METHODS
2.1. Plant material, priming and germination tests
M. truncatula seeds (commercial seed lot, kindly provided by Fertiprado L.d.a.) were treated as follows. For hydropriming, seeds were transferred to Petri dishes (diameter 90 mm) containing two filter papers moistened with 2.5 ml of H2O, sealed and kept in a growth chamber at 22°C under light conditions with a photon flux density of 150 μmol m−2 s−1, photoperiod of 16/8 h and 70%−80% relative humidity. Hydropriming treatments were performed by imbibing seeds for increasing time (from 12 h up to 32 h), to induce overpriming, namely radicle protrusion with consequent loss of DT. For each treatment, three independent replications with 20 seeds per replication were analysed. For dehydration (dry‐back, DB), primed (P) and overprimed (OP) seeds were transferred into glass tubes, placed between two cotton disks, covered with silica beads (disidry® Orange Silica Gel, The Aerodyne) with a seed: silica ratio of 1:10, and kept at 24−25°C. Seed fresh weight was monitored every 15 min for 6 h when the weight of dry seed was reached. For germination tests, seeds were transferred to Petri dishes (diameter 90 mm) containing two filter papers moistened with 2.5 ml of H2O, sealed, and kept in a growth chamber as previously described. Seeds with protrusion of the primary radicle were considered germinated. Medicago sativa seeds (commercial genotype) underwent hydropriming for 24 h, dry‐back for 6 h, and germination tests as described above. Primed seeds and overprimed seeds (showing radicle protrusion) were harvested at the indicated time points, and the fresh weight was measured. Samples were frozen using liquid N2 and stored at −80°C for subsequent molecular analyses.
2.2. Moisture content
Relative water content (RWC) was calculated for M. truncatula dry and imbibed (primed and overprimed) seeds. Fresh weight (F w) was measured at the indicated timepoints. Dry weight (D w) was measured after 24 h dehydration at 80°C. RWC was calculated and expressed as percentage relative to F w according to the following formula: RWC [%] = [(F w − D w)/F w] × 100 (Romero‐Rodríguez et al., 2018). Measures were performed with an analytical scale (Mettler AJ100; Mettler‐Toledo). Water content was also assessed by thermogravimetric analysis (TGA), using a Mettler Toledo TGA 1 instrument with a fixed heating rate of 20°C min−1. The temperature range was from 25°C to 800°C with a gas flow in the oven (air or nitrogen) of 4 l h−1. The samples were grounded into powder and sieved to a size of 100 μm. About 5 mg of sample was used in each test.
2.3. ROS detection
ROS levels were quantified in M. truncatula dry and imbibed (primed and overprimed) seeds, using the fluorogenic dye 2′,7′‐dichlorofluorescein diacetate (DCFH‐DA; Sigma‐Aldrich). The dye is converted to a nonfluorescent molecule following deacetylation mediated by cellular esterases, and it is subsequently oxidized by ROS into the fluorescent compound 2′,7′‐dichlorofluorescein. DFC can be detected by fluorescence spectroscopy with maximum excitation and emission spectra of 495 and 529 nm, respectively. The assay was carried out as described (Forti et al., 2020a). Samples (three seeds per time point) were incubated for 15 min with 50 μl of 10 μM DCFH‐DA and subsequently, fluorescence was determined at 517 nm using a Rotor‐Gene 6000 PCR apparatus (Corbett Robotics), setting the program for one cycle of 30 s at 25°C. As a negative control, a sample containing only DCFH‐DA was used to subtract the baseline fluorescence. Relative fluorescence was calculated by normalizing samples to controls and expressed as relative fluorescence units (R.F.U.).
2.4. Alkaline comet assay and detection of γH2AX foci
The alkaline version of the comet assay allows quantifying single‐strand breaks (SSBs) formed from alkali‐labile sites as well as DNA−DNA or DNA−protein crosslinks (Collins, 2004). Seeds were harvested at the indicated time points, embryo axes (primed seeds) and embryos with radicle protrusion (overprimed seeds) were isolated from the cotyledons and seed coat using a razor blade. Nuclei were extracted as previously described (Ventura et al., 2014). The resulting suspension containing purified nuclei was mixed in equal volume with a solution containing 1% low melting point (Sigma‐Aldrich) agarose in phosphate‐buffered saline buffer (PBS: 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) maintained at 38°C. Two drops of the resulting suspension were then pipetted onto agarose‐precoated slides and solidified on ice. Slides were incubated for 30 min at 4°C in alkaline buffer (1 mM Na2 EDTA, 300 mM NaOH, pH 13) and then electrophoresed in the same buffer for 25 min at 0.72 V cm−1 in a cold chamber under dark conditions. After electrophoresis, slides were washed in 0.4 M Tris HCl pH 7.5 three times for 5 min, rinsed in 70% ethanol (v/v) three times for 5 min at 4°C, and dried overnight at room temperature. Slides were stained with 20 μl 4′,6‐diamidino‐2‐phenylindole (1 mg ml−1, Sigma‐Aldrich). For each slide, 100 nucleoids were scored, visualized using an Olympus BX51 fluorescence microscope with an excitation filter of 340−380 nm and a barrier filter of 400 nm. Images were captured using an Olympus MagnaFire camera equipped with Olympus Cell‐F software. Nucleoids were classified as previously described by Collins (2004), where each type of nuclei morphology belongs to a class from 0 to 4. The results were expressed in arbitrary units (a.u.), calculated using the following formula: a.u. = [Σ(N c × c) × 100]/N tot, where Nc is the number of nuclei of each class, c is the class number (e.g., 0, 1, 2, 3, 4), and N tot is the total number of counted nuclei (Collins, 2004). The occurrence and distribution of γH2AX foci in the nucleus of embryo axes and embryos with radicle protrusion, isolated from primed (P) and overprimed (OP) seeds was monitored as follows. Thin sections of 70−80 nm were cut with an ultramicrotome and collected on formvar‐carbon‐coated nickel grids (300 mesh). Sections were blocked in 2% NGS (normal goat serum, Sigma‐Aldrich) for 5 min, and then incubated with the primary antibody Phospho‐Histone H2A.X (Ser139) Polyclonal Antibody from rabbit (ThermoFisher Scientific) diluted 1:30 in 0.05% PBS‐TWEEN®20 (PBT) overnight at 4°C in a humidified chamber. Subsequently, grids were blotted with a filter paper to remove excess solution and rinse twice with PBT and twice with 1× PBS, 5 min each. Specimens were blocked again in 2% NGS at room temperature for 5 min and incubated with a secondary antibody coupled with 12 nm colloidal gold grain, diluted 1:20 in 1× PBS, and finally rinsed twice with the same buffer and twice with dH2O (5 min each), at room temperature. Before imaging with transmission electron microscopy (TEM), the tissue sections were stained according to EDTA regressive technique (Bernhard, 1969) by incubating them in 4% uranyl acetate aqueous solution for 10 min, followed by incubation in 40 mM EDTA for 17 s, to remove uranyl from DNA, and finally in lead citrate for 2 min. After each incubation step, grids were washed thoroughly. Images were acquired as previously described, 10 nuclei for each treatment/sample were scored for the presence of γH2AX foci. The density of foci was calculated as follows: 100 squares (each one with an area of 200 nm2) were identified and the number of foci per single area was counted. The measurement was performed considering 10 cells for each sample and 10 squares per single cell. The results are expressed as mean values ± SD. Statistical significance was evaluated by unpaired Student's t test.
2.5. RNA extraction, cDNA synthesis and qRT‐PCR
RNA isolation was carried out as described by Oñate‐Sanchez and Vicente‐Carbajosa (2008). cDNAs were obtained using the RevertAid First Strand cDNA Synthesis Kit (ThermoFisher Scientific) according to the manufacturer's suggestions. qRT‐PCR was performed with the Maxima SYBR Green qPCR Master Mix (2X) (ThermoFisher Scientific) according to the supplier's indications, using a Rotor‐Gene 6000 PCR apparatus (Corbett Robotics Pty Ltd). Amplification conditions were as follows: denaturation at 95°C for 10 min, and 45 cycles of 95°C for 15 s and 60°C for 30 s and 72°C for 30 s. Oligonucleotide primers were designed using the Real‐Time PCR Primer Design program Primer3Plus (https://primer3plus.com) from GenScript and further validated through the online software Oligo Analyser (https://eu.idtdna.com/calc/analyzer) (Table S1). Quantification was carried out using as reference genes MtELF1a (ELONGATION FACTOR 1a) (Medtr6g021805) and MtACT (ACTIN) (Medtr2g096840) for the experimental conditions (primed vs. overprimed) used in this study (Macovei et al., 2019). The raw, background‐subtracted fluorescence data provided by the Rotor‐Gene 6000 Series Software 1.7 (Corbett Robotics) were used to estimate PCR efficiency (E) and threshold cycle number (C t) for each transcript quantification. Relative quantification of transcript accumulation was carried out as described by Pfaffl (2001) and statistical analysis was performed with REST2009 Software V2.0.13 (Qiagen GmbH). The following genes were tested: MtSOD (SUPEROXIDE DISMUTASE) (Medtr7g114240), MtAPX (ASCORBATE PEROXIDASE) (Medtr4g061140), MtTRH1 (THIOREDOXIN 1) (Medtr3g112410), MtSPMS (SPERMINE/SPERMIDINE SYNTHASE) (MTR_3g091090), MtSPDS (SPERMIDINE SYNTHASE) (MTR_5g034470), MtMRE11 (MEIOTIC RECOMBINATION) (Medtr2g081100), MtRAD50 (RADIATION SENSITIVE) (Medtr3g084300), MtNBS1 (NIJMEGEN BREAKAGE SYNDROME) (Medtr5g076180), MtSOG1‐1 (SUPPRESSOR OF GAMMA RESPONSE) (Medtr5g053430), MtOGG1 (8‐OXOGUANINE GLYCOSYLASE/LYASE) (Medtr3g088510), MtTDP1α (TYROSYL‐DNA PHOSPHODIESTERASE) (Medtr7g050860), MtLAS1 (LETHAL IN THE ABSENCE OF SSD1) (Medtr5g089190), MtNOL9 (NUCLEAR PROTEIN 9) (Medtr8g080140), MtXRN3 (5′ TO 3′ EXORIBONUCLEASE 3) (Medtr7g114570), MtSAL1/FRY1 (FIERY) (Medtr2g012510), MtRPS3 (RIBOSOMAL PROTEIN S3) (Medtr1g023000), MtRAB28 (RESPONSIVE TO ABSCISIC ACID 28) (Medtr3g034660), MtANAC082 (ARABIDOPSIS NAC 82) (Medtr3g096140), MtFANCJB (FANCONI ANEMIA) (Medtr1g026490), MtRTEL1 (REGULATOR OF TELOMERE LENGTH) (Medtr2g102150). To monitor rRNA synthesis and processing, five M. truncatula rDNA regions hereby named amplicon 1 (18S rRNA, LOC112418413), amplicon 2 (ITS1‐18S rRNA), amplicon 3 (5.8S rRNA, LOC112418314), amplicon 4 (ITS1‐5.8S rRNA, LOC112418314), amplicon 5 (25S rRNA, LOC112418308) and amplicon 6 (ITS2‐25S rRNA, LOC112418308) were designed. For M. sativa the following amplicons were designed: 5.8S rRNA and ITS1‐5.8S rRNA (M. sativa locus: MH712681).
2.6. Nuclear staining with toluidine blue and Hoechst 33258
M. truncatula embryo axes and embryos with radicle protrusion isolated from primed (P) and overprimed (OP) seeds, respectively, were collected at the indicated time points during the rehydration−dehydration cycle and fixed with 2% paraformaldehyde/0.2% glutaraldehyde (Sigma‐Aldrich) in 1× PBS pH 7.2−7.4 for 3 h at room temperature. Embryos were rinsed in the same buffer overnight, incubated in 0.5 M NH4Cl for 30 min, to prevent aspecific signals due to free aldehyde groups which may react with any tested antibody, dehydrated in progressively concentrated ethanol solution (starting from 30% to 100%), and finally embedded in LR‐White acrylic resin (Agar Scientific Ltd) for 24 h at 60°C. Semi‐thin sections of 500 nm in thickness were cut using an ultramicrotome from seed tissues embedded in LR‐White acrylic resin, and used for nuclear staining with toluidine blue and Hoechst 33258. The sections were covered with a drop of 0.3% toluidine blue solution and incubated for 5 min at 100°C. Sections were then washed thoroughly with dH2O to remove excess dye, air‐dried, mounted in Mowiol® 4‐88 (Sigma‐Aldrich), and finally imaged using a Zeiss Axioskop 2 Plus microscope. For Hoechst 33258 staining, sections were initially hydrated by four washes in 1× PBS pH 7.2−7.4 (2 min each) and then incubated for 5 min in Hoechst 33258 (1 μg ml−1, Sigma‐Aldrich) in the dark. Subsequently, four washes, 2 min each, followed by one wash for 5 min in 1× PBS pH 7.2−7.4, were performed. Glass slides were finally mounted using 90% glycerol in 1× PBS pH 7.2−7.4. Seed tissue sections were imaged using an Olympus BX51 fluorescence microscope.
2.7. Staining with terbium citrate and osmium ammine staining
The seed tissue sections prepared as previously described were used for TEM. Thin sections of 70−80 nm were cut with an ultramicrotome and collected on nickel grids without coating for terbium staining and on gold grids without coating for the osmium ammine staining, respectively. The staining with terbium citrate, which selectively contrasts RNA (Biggiogera & Fakan, 1998), was performed as follows. The specimen was floated on a 50 μl drop of terbium citrate for 30 min at room temperature. Without drying, the grid is washed in 100−150 μl drops of dH2O for 10 s and immediately afterward for 5 s at room temperature and air‐dried for at least 30 min. Half of the specimen stained by terbium citrate was previously treated with RNase I (1 mg ml−1) as follows. Each sample was hydrated for 5 min in 1× PBS pH 7.2−7.4 and incubated in the presence of 10 µl RNase I overnight at room temperature, in a humidified chamber. The grids were then washed twice with the same buffer and twice in dH2O, 5 min each, stained air‐dried, and imaged using a Jeol JEM‐1200EXIII electron microscope equipped with a 30 mm objective aperture and operating at 80 kV. The osmium ammine staining allows the detection of nuclear domains characterized by high DNA concentration, like heterochromatin (Vazquez‐Nin et al., 1995). It is accomplished by a Feulgen‐type reaction, consisting of acid hydrolysis to obtain free aldehyde groups on DNA followed by their binding to the Schiff‐type reagent osmium ammine. The grids were transferred in 5 N HCl for 30 min and subsequently, seven quick washes in dH2O followed by three washes (2 min each) in dH2O were performed. Afterward, the grids were placed in osmium amine solution for 60 min and finally washed in dH2O to reduce precipitates formation as follows: seven quick washes, three washes (2 min each), three washes (5 min each), a single wash (20 min). At the end of each wash, the grids were rinsed on an absorbent paper dish. M. truncatula embryo axes and embryos with radicle protrusion isolated from P and OP, respectively, were also fixed with 1.5% glutaraldehyde (Sigma‐Aldrich) for 3 h at room temperature, rinsed in 1× PBS (pH 7.2), post‐fixed in 1% aqueous OsO4 (Sigma‐Aldrich) for 2 h at room temperature, and finally embedded in epoxy resin (Sigma‐Aldrich). These sections were routinely stained with uranyl acetate and lead citrate. For morphometric analysis, all specimens were observed with a Jeol JEM‐1200EXIII electron microscope equipped with a 30 mm objective aperture and operating at 80 kV. Morphometric analyses were carried out using the software ImageJ (https://imagej.nih.gov/ij).
2.8. Determination of PAP content
The phosphonucleotide PAP (3′‐phosphoadenosine 5′‐phosphate) was quantified in M. truncatula dry and imbibed (primed and overprimed) seeds collected at the indicated time points and ground in liquid N2. PAP was extracted as described by Estavillo et al. (2011), with the following modifications: 100 mg powder per sample were dissolved into 1 ml of 0.1 M HCl and incubated on ice for 30 min, then centrifuged twice at 16 000g for 5 min at 4°C. Subsequently, 150 µl of the supernatant were mixed with 770 µl of CP buffer (620 mM citric acid‐1‐hydrate and 760 mM Na2HPO4.2H2O, pH 4). Samples were centrifuged at 16 000g for 45 min at room temperature and filtered before injection into the HPLC system (Kontron Instrument 420, equipped with C18 reverse‐phase Zorbax ODS column [250 mm, 4.6 mm, 5 μm, Agilent Technologies] and with a UV‐Vis detector [Kontron]). The analyte was eluted with a mobile phase consisting of methanol: water (20:80, v/v) containing 75 mM KH2PO4, 100 mM NH4Cl, 1 mM 1‐octylamine (pH 4.55) at a flow rate of 1.0 ml min−1 (Xu et al., 2012). The injection volume was 20 µl, the column temperature was set at 30°C and the detection wavelength was monitored at 254 nm. Standard solutions containing PAP (Sigma‐Aldrich) in the range 0−1.20 mg ml−1 were used to obtain a calibration curve (Figure S1).
2.9. Statistical analysis
Statistical analysis of germination tests, radicle length and fresh/dry biomass was performed using two‐tailed heteroscedastic Student's t test with asterisks indicating statistically significant differences (*p < 0.05; **p < 0.01, ***p < 0.001). Statistical analysis of ROS, comet assays, γH2AX foci, PAP, and qRT‐PCR data was performed using two‐way ANOVA and Tukey's test. Means without a common superscript letter differ (p < 0.05) as analysed by two‐way ANOVA and TUKEY test.
3. RESULTS
3.1. Impact of overpriming on germination performance and seedling phenotype
To set an experimental system allowing selection of primed and overprimed seeds during rehydration and monitoring their response during dry‐back, M. truncatula seeds underwent imbibition with water during controlled periods (12, 16, 20, 24, 28 and 32 h; hydropriming, HP) followed by 6 h of dehydration (dry‐back, DB) (Figure 1a). These treatments were named HP12H6D, HP16H6D, HP20H6D, HP24H6D, HP28H6D and HP32H6D. Germination tests showed that HP16H6D, HP20H6D and HP24H6D could anticipate germination (Figure 1b) while HP28H6D and HP32H6D resulted in extensive overpriming, as evidenced by the occurrence of radicle protrusion. In these treatments, no germination tests were performed. Abnormal M. truncatula seedlings, characterized by a rootless phenotype, were produced by OP seeds (Figure 1c). UP and HP12H6D seeds showed 91 ± 2% and 95 ± 7% germination, respectively, and produced only seedlings with normal phenotypes (100%). For the HP16H6D treatment, the percentage of seedlings with normal phenotype was significantly (p = 0.037) reduced (75 ± 4%), compared with HP12H6D, and the aberrant phenotype appeared (18 ± 2%) (Figure 1d). The occurrence of aberrant seedlings further increased, accounting for 68 ± 8% (HP20H6D), 61 ± 2% (HP24H6D) and 60 ± 10% (HP28H6D) (p < 0.016) of the total seedlings. The HP32H6D treatment resulted in the highest percentage of aberrant seedlings (81 ± 8%). Based on these results, the HP24H6D treatment was selected to produce primed and overprimed seeds used in this study.
Figure 1.
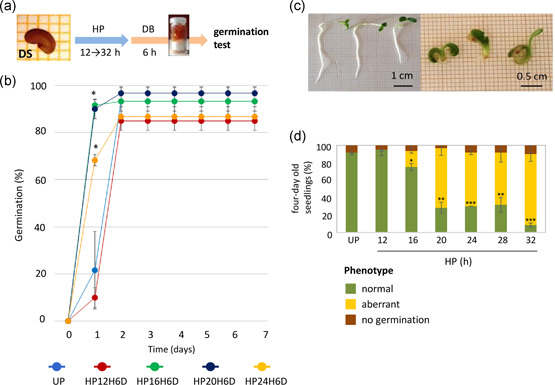
(a) Schematic representation of the hydropriming (HP) treatment (from 12 to 32 h) and dry‐back (DB) procedure (6 h) applied to Medicago truncatula seeds in this study. (b) Results of germination tests performed with unprimed (UP) and primed (HP) M. truncatula seeds. Seeds underwent HP for increasing time (from 12 up to 28 h) followed by 6 h of dry‐back (treatments hereby named HP12H6D, HP16H6D, HP20H6D, HP24H6D, HP28H6D). HP28H6D and HP32H6D treatments resulted in extensive overpriming, as evidenced by the occurrence of radicle protrusion during treatment; thus, no germination tests were performed. (c) Four‐day‐old M. truncatula seedlings showing the normal phenotype (left) and aberrant (rootless) phenotype (right) developed from primed and overprimed seeds, respectively. (d) Percentage of normal and aberrant phenotypes in M. truncatula seedlings developed from unprimed (UP) seeds and seeds subjected to hydropriming for increasing time. Values are expressed as mean ± SD of three independent replications with 20 seeds for each replication. Asterisks indicate statistically significant differences in the percentage of normal seedlings, compared with UP, determined using Student's t test (*p < 0.05, **p < 0.01, ***p < 0.001)
3.2. DNA damage and dynamics of genotoxic stress response in overprimed versus primed M. truncatula seeds
The experimental design included dry seeds (DS), imbibed primed (P) and overprimed seeds (OP), and seeds collected during dry‐back (PDB, OPDB) (Figure 2a). Water content ranged from 6 ± 0% in the DS up to 58 ± 5% and 68 ± 2% in imbibed P and OP seeds, respectively (Figure S2). Along the dry‐back, water content progressively decreased with estimated values of 18 ± 1% and 17 ± 0% (PDB2 and OPDB2), 8 ± 1% and 9 ± 1% (PDB4 and OPDB4). At the end of the treatment, water content was 6 ± 0% and 7 ± 3% (PDB6 and OPDB6) (Figure S2). ROS amounts increased in OP seeds during dry‐back, compared with P seeds (Figure 2b), triggering changes in transcript levels of the antioxidant genes MtAPX and MtTRH1, as well as the MtSPMS and MtSPDS genes involved in polyamine biosynthesis (Figure 2c). These genes were selected for assessing the antioxidant response along the rehydration−dehydration cycle due to their documented role in the context of the seed pre‐germinative metabolism (Bailly et al., 2008; Balestrazzi et al., 2011; Forti et al., 2020a; Pagano et al., 2017, 2019). The h‐type thioredoxin TRH1, abundant in M. truncatula seeds, has been reported to act as an effective ROS scavenger able to prevent oxidative damage upon imbibition (Renard et al., 2011). Polyamines protect the cellular components against toxic radicals (Liu et al., 2015) and their role in the seed antioxidant response has been demonstrated (Kim et al., 2019). MtSPDS and MtSPMS genes encoding spermidine synthase and spermine/spermidine synthase, respectively, were significantly up‐regulated during imbibition in M. truncatula seeds facing antioxidant stress (Pagano et al., 2019).
Figure 2.
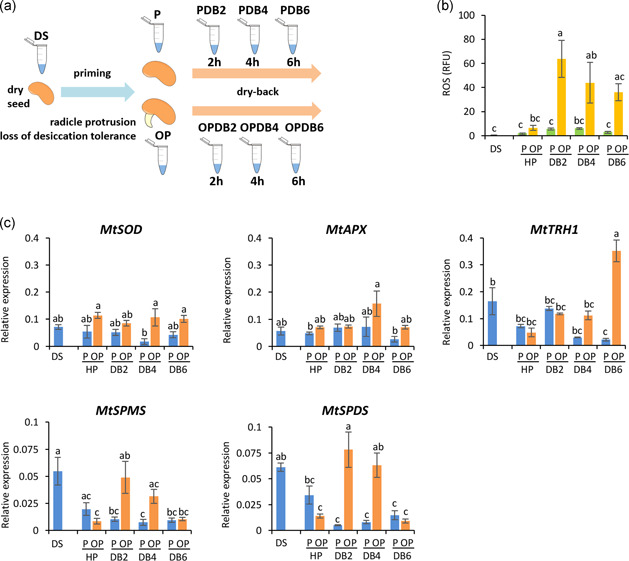
Distinctive responses of primed and overprimed Medicago truncatula seeds coping with oxidative damage along the rehydration−dehydration cycle. (a) Experimental design. Dry seeds (DS), primed (P), and overprimed (OP; occurrence of radicle protrusion during treatment) seeds were collected at 24 h of hydropriming, and subsequently during dry‐back (DB) at 2 h (PDB2, OPDB2), 4 h (PDB4, OPDB4), and 6 h (PDB6, OPDB6). (b) ROS (reactive oxygen species) levels, measured in whole seeds at the selected time points using DCFH‐DA (2′,7′‐dichlorofluorescein diacetate). (c) Expression profiles of MtSOD, MtAPX, MtTRH1, MtSPMS, and MtSPDS genes in isolated embryos. Data were analysed with two‐way ANOVA and TUKEY test. Means without common letters are significantly different (p < 0.05). R.F.U., relative fluorescence unit
The genotoxic impact generated during the rehydration−dehydration cycle was assessed in isolated embryos, using the alkaline comet assay (Figure 3a). When comparing P and OP embryos, no significant differences were observed; however, at 2 h of dry‐back, DNA damage was significantly increased in OP embryos, as well as at the subsequent time points (Figure 3a). The DNA damage profile of OP embryos correlated with ROS patterns. Upon DNA damage, ATM phosphorylates the histone variant H2AX on Ser139 (Burma et al., 2001) and such modification (γH2AX) can spread for up to 1 Mbp away from the break site, acting as a platform to recruit the DNA repair enzymes (Iacovoni et al., 2010). The spatial distribution of γH2AX foci was mapped in P and OP embryos, along the rehydration−dehydration cycle (Figure S3) whereas the estimated frequency is reported in Figure 3b. Samples were obtained from a pool of 10 embryo axes. For each sample, 10 nuclei were scored for the presence of γH2AX foci. In P and OP embryos there were no significant differences in the frequency of γH2AX foci. After 2 h of dry‐back, a significant decrease in the average number of γH2AX foci was measured in both P and OP embryos (Figure 3b, PDB2: 0.8 ± 0.1, OPDB2: 0.9 ± 0.1). At 4 h of dry‐back, the average number of γH2AX foci in P embryos did not change (Figure 3b, PDB4: 0.9 ± 0.1) whereas a significant increase was noticed in OP embryos (Figure 3b, OPDB4: 1.2 ± 0.1). The density of γH2AX foci was further enhanced in OP embryos at the end of dry‐back (Figure 3b, OPDB6: 2.4 ± 0.2), differently from what was recorded in P embryos (Figure 3b, PDB6: 1.3 ± 0.1).
Figure 3.
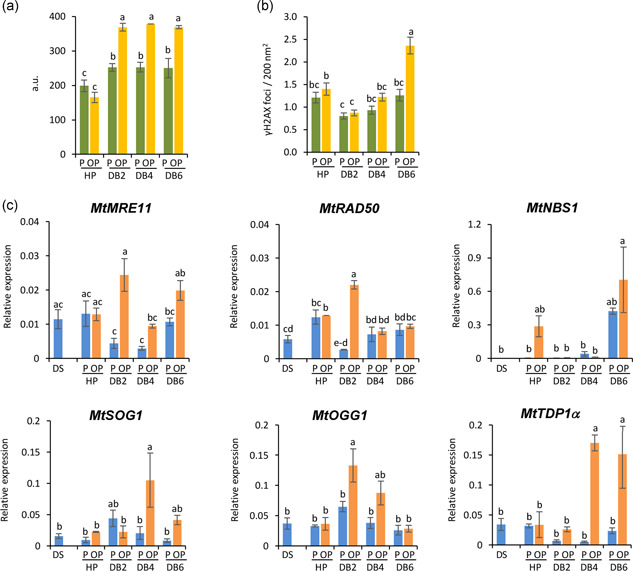
Distinctive responses of primed and overprimed Medicago truncatula seeds coping with genotoxic damage along the rehydration−dehydration cycle. (a) Alkaline comet assay was used to measure DNA strand breaks accumulation in M. truncatula embryos, along the rehydration−dehydration cycle. Values are expressed as mean ± SD of three independent replications with 20 seeds for each replication. (b) Distribution of γH2AX foci in the nucleolus and perinucleolar region of M. truncatula embryos isolated along the rehydration−dehydration cycle. Detection of γH2AX foci was performed by immunocytochemical and TEM analyses. Samples were obtained from a pool of 10 embryo axes. For each sample, 10 nuclei were scored for the presence of γH2AX foci. Density of γH2AX foci is expressed as average number of foci per 200 nm2 ± standard error. (c) Expression profiles of MtMRE11, MtRAD50, MtNBS1, MtSOG1, MtOGG1, and MtTDP1α genes in isolated embryos. Data were analysed with two‐way ANOVA and TUKEY test. Means without common letters are significantly different (p < 0.05). a.u., arbitrary unit; TEM, transmission electron microscopy
To provide evidence of the seed response to DNA damage, the expression profiles of DDR genes involved in DNA damage sensing and signalling as well as in DNA repair were evaluated (Figure 3c). The MtMRE11, MtRAD50 and MtNBS1 genes encode the components of the MRN complex, able to activate the DSB‐induced cell cycle checkpoints and recognize DSB‐repair effectors (Nikitaki et al., 2018). Downstream the MRN complex, the transcription activator SOG1 (SUPPRESSOR OF GAMMA RESPONSE) acts as master regulator of plant DDR (Yoshiyama, 2016). The bifunctional DNA glycosylase/lyase OGG1 (8‐oxoguanine glycosylase/lyase; EC: 3.2.2−4.2.99.18) catalyses the release of 7,8‐dihydro‐8‐oxoguanine (8‐oxodG) and the cleavage of DNA at the resulting abasic site. The role of OGG1 in counteracting oxidative DNA damage accumulated during both seed desiccation and imbibition has been documented (Balestrazzi et al., 2011; Chen et al., 2012; Cordoba‐Canero et al., 2014; Forti et al., 2020a; Macovei et al., 2010, 2011; Pagano et al., 2017). The multitask TDP1 enzyme removes topoisomerase I‐mediated DNA lesions and contributes to the repair of oxidized DNA lesions (Pommier et al., 2014). The plant TDP1α gene is up‐regulated during seed imbibition under physiological and oxidative stress conditions (Balestrazzi et al., 2011; Macovei et al., 2010; Pagano et al., 2017, 2019). The MtMRE11, MtRAD50 and MtNBS1 genes showed up‐regulation in OP seeds during dry‐back (Figure 3c). Up‐regulation of the MtSOG1 gene observed in OP embryos at 4 h of dry‐back suggests active DDR signalling under dehydration and for the activation of downstream components such as the MtOGG1 and MtTDP1α genes (Figure 3c).
Preliminary analyses carried out using Hoechst 33258 staining revealed nuclear shrinkage with an expansion of heterochromatin domains in both P and OP nuclei of desiccated embryos (Figures S4 and S5). When osmium ammine staining was used to detect ultrastructural changes in DNA distribution, the nuclei of both P and OP rehydrated embryos showed osmiophilic and widely dispersed chromatin, with few heterochromatic domains located at the nuclear periphery (Figure 4a, (i) and (iii), arrows). Upon dehydration, the dispersed chromatin exhibited clear segregation and condensation patterns; however, heterochromatin regions appeared to be expanded in OP nuclei (Figure 4a, (ii) and (iv), arrows).
Figure 4.
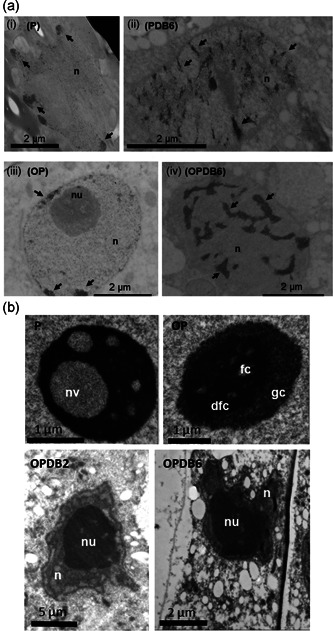
Nuclear and nucleolar ultrastructure of primed and overprimed Medicago truncatula embryos. (a) Changes in chromatin distribution revealed by osmium ammine staining in the nucleus of M. truncatula embryos along the rehydration−dehydration cycle. In P and OP embryo axes, the nuclei of hydrated cells showed dispersed chromatin, with few heterochromatic domains at the nuclear periphery (i and iii, arrows). Chromatin condensation patterns were enhanced at the end of the dry‐back treatment (DB6) in both P and OP embryos (ii and iv, arrows). (b) Ultrastructural analysis reveals distinctive nucleolar architectures in imbibed P and OP seeds. For each sample, pools of 10 embryo axes were collected and 10−20 nuclei per sample were analysed. dfc, dense fibrillar centre; fc, fibrillar centre; gc, granular centre; n, nucleus; nu, nucleolus; nv, nucleolar vacuole
3.3. Nucleolar architecture of primed and overprimed M. truncatula embryos
Substantial differences in the nucleolar architecture of rehydrated embryos were observed by TEM analysis. For each sample, pools of 10 embryo axes were collected and 10−20 nuclei per sample were analysed. Nucleolar vacuolization, typical of actively transcribing nucleoli (Macovei et al., 2018; Stepinski, 2014), was recurrent in P embryos (Figure 4b, P) and absent from OP embryos where a tripartite nucleolar organization was observed (Figure 4b, OP). OP nucleoli showed an expanded dense fibrillar component (dfc) surrounding the fibrillar centres (fc). The granular component (gc) was visible in the peripheral region (Figure 4b, OP). These morphologies might reflect the two metabolic conditions of primed and overprimed seeds. During the dry‐back, the nuclear area was progressively reduced due to water loss but there was no evidence of changes in the size of nucleolus which accounted for most of the nuclear compartment in desiccated embryos (Figure 4b, OPDB2 and OPDB6). Such profile was observed in all the scored nucleoli of both PDB and OPDB embryos.
3.4. Changes in rRNA profiles of overprimed embryos challenged with dry‐back
qRT‐PCR was used to measure the 18S, 5.8S and 25S rRNA unspliced precursors and mature forms. The rDNA region and target amplicons are reported in Figure 5a (see also Figure S6 for the nucleotide sequence) whereas rRNA profiles are shown in Figure 5b. Both 18S rRNA precursors and form increased at 4 h of dry‐back in OP embryos whereas the 5.8S rRNA peaked at 2 h of dry‐back. In addition, the 25S rRNA precursors and mature form were enhanced at 4 h of dry‐back. In contrast, only slight changes in rRNA levels occurred in P embryos (Figure 5b). The early 5.8S rRNA peak observed in OP embryos suggested a stress hallmark potentially useful for monitoring the response to dry‐back. To better assess whether the 5.8S rRNA accumulation during early dry‐back might represent a conserved legume stress hallmark, useful for diagnostic applications in seed technology, the same experimental design was applied in the closely related legume alfalfa (Medicago sativa L.). The tailored hydropriming treatments were able to speed up seed germination in alfalfa (Figures 6a and S7a). As previously observed for M. truncatula, alfalfa seedlings developed from OP seeds showed an abnormal phenotype (Figure S7b), with a significant reduction in radicle length (Figure 6b) and fresh weight (Figure S7c) compared with seedlings developed from P seeds. The levels of 5.8S rRNA unspliced precursors were consistently increased in M. sativa OP seeds, compared with P seeds, along the rehydration−dehydration cycle, with a peak at the end of dry‐back (Figure 6c, ITS1‐5.8S, DB6). In contrast to the response observed in M. truncatula, the 5.8S rRNA mature form accumulated only in imbibed P seeds (Figure 6c, 5.8S, HP).
Figure 5.
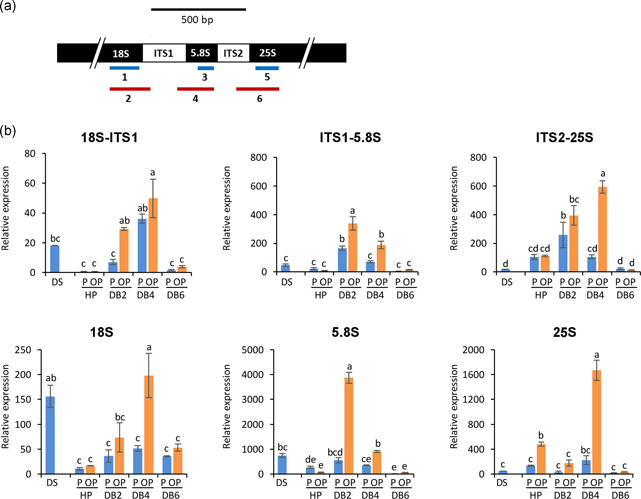
(a) Schematic representation of the Medicago truncatula rDNA region. Amplicon 1 (18S rRNA), amplicon 2 (ITS1‐18S rRNA), amplicon 3 (5.8S rRNA), amplicon 4 (ITS1‐5.8S rRNA), amplicon 5 (25S rRNA) and amplicon 6 (ITS2‐25S rRNA) are highlighted. (b) Levels of precursor and mature forms of 18S, 5.8S, and 25S rRNA measured by qRT‐PCR along the rehydration−dehydration cycle in M. truncatula embryos excised from P and OP seeds. qRT‐PCR data were analysed with two‐way ANOVA and TUKEY test. Means without common letters are significantly different (p < 0.05)
Figure 6.
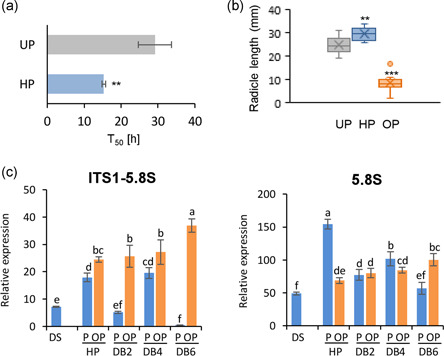
Proof of concept carried out in alfalfa to assess the role of 5.8S rRNA as early hallmark of desiccation stress in overprimed seeds. (a) Hydropriming applied for 24 h (HP) accelerates alfalfa seed germination, compared with unprimed control (UP). T50, time required to reach 50% germination. (b) Radicle length of 4‐day‐old alfalfa seedlings developed from P and OP seeds. Ten seedlings for each treatment were analysed. Asterisks indicate statistically significant differences determined using two‐tailed heteroscedastic Student's t test (*p < 0.05; **p < 0.01, ***p < 0.001). (c) Levels of precursor and mature forms of 5.8S rRNA measured by qRT‐PCR along the rehydration−dehydration cycle in alfalfa P and OP seeds. qRT‐PCR data were analysed with two‐way ANOVA and TUKEY test. Means without common letters are significantly different (p < 0.05)
3.5. Ribogenesis‐associated MtLAS1, MtXRN3 and MtRPS3 genes are up‐regulated in overprimed embryos under desiccation stress
There is increasing evidence of a possible connection between ribosome assembly and DNA repair (Goffová & Fajkus, 2021; Yang et al., 2019). Although the molecular basis of such interaction is still poorly explored, it has been suggested that ITS2 RNA processing factors might be involved (Pillon et al., 2019). ITS2 removal, required for the maturation of the 60S ribosome subunit, is initiated by a multienzyme complex, composed of the ribonuclease LAS1 and the RNA polynucleotide kinase NOL9 (Pillon et al., 2019). In the context of the rehydration−dehydration cycle, MtLAS1 and MtNOL9 genes showed different expression profiles, and only MtLAS1 gene was up‐regulated in response to dehydration in OP embryos (Figure 7b). XRN3 is part of a highly conserved 5′−3′‐exoribonuclease family responsible for 5′processing or turnover of uncapped RNAs, thus participating in a surveillance mechanism for aberrant transcripts (Crisp et al., 2018). Furthermore, the MtXRN3 gene was up‐regulated during dehydration in OP seeds (Figure 7b). The highly conserved plant RPS3 (ribosomal protein S3) has key roles in DDR in animals, including the ability to bind the BER enzymes OGG1 and APE/Ref‐1 (Hegde et al., 2004) and to inhibit the NHEJ (nonhomologous end joining) pathway (Park et al., 2020). Up‐regulation of the MtRPS3 gene was detected at 4 h of dry‐back in OP embryos (Figure 7b). Although the reported data do not provide direct evidence of a role played by the nucleolus in the desiccation stress response, ribogenesis‐associated genes might represent helpful targets in future studies focused on this issue.
Figure 7.
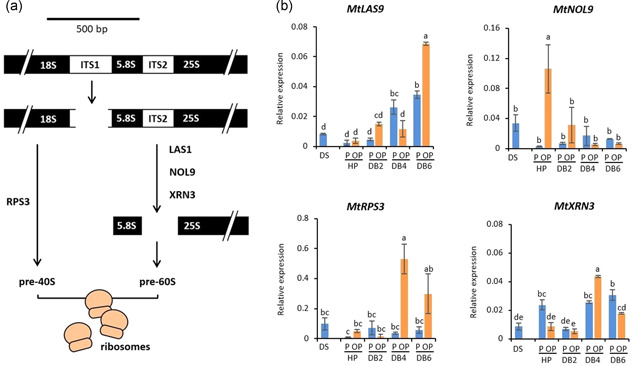
(a) Schematic representation of ribogenesis. (b) Expression profiles of MtLAS1, MtNOL9, MtXRN3, MtRPS3 genes, qRT‐PCR data were analysed with two‐way ANOVA and TUKEY test. Means without common letters are significantly different (p < 0.05)
3.6. Primed M. truncatula seeds accumulate PAP during dry‐back
The MtXRN gene encodes a nuclear 5′−3′‐exoribonucleases that participates in the SAL1‐PAP‐XRN retrograde signalling pathway (Crisp et al., 2018). The redox‐sensitive SAL1 protein modulates the signalling molecule PAP. PAP migrates from chloroplasts and mitochondria to the nucleus where it inhibits the activity of XRN exoribonucleases, thereby altering RNA metabolism (Estavillo et al., 2011). PAP also contributes to the drought stress response by intersecting ABA‐mediated signalling pathways (Pornsiriwong et al., 2017). Based on these premises, the expression profile of MtSAL1 gene was investigated in primed and overprimed embryos. The MtSAL1 gene encodes an inositol polyphosphate 1‐phosphatase able to dephosphorylate PAP. The SAL1 protein is also regarded as an oxidative stress sensor in plant organelles (Chan et al., 2016). MtSAL1 transcript levels changed in both P and OP embryos at along dry‐back (Figure 8a). PAP was not detected in dry (DS) and imbibed seeds (P and OP) (Figure 8b) but it accumulated in response to dehydration. The highest content was observed in P seeds at 2 h of dry‐back (PDBH2, 212 ± 14 pmol mg−1 FW) followed by a progressive decrease (Figure 8b). In OP seeds, during dry‐back, PAP levels were in the 15−32 pmol mg−1 FW range, from 4‐fold up to 13‐fold lower than those reported for P seeds (Figure 8b).
Figure 8.
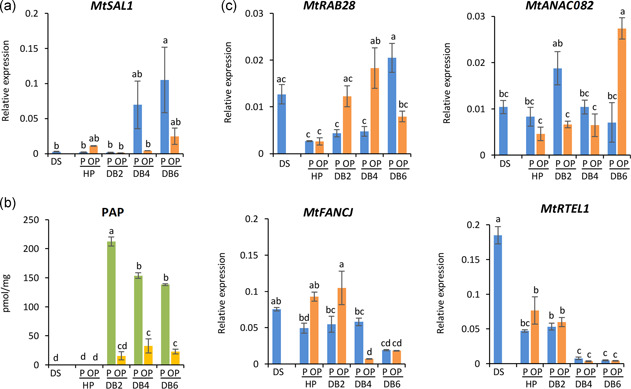
Evidence for the role of the SAL1‐PAP retrograde signalling in primed Medicago truncatula seeds coping with dry‐back. (a) Expression profiles of the MtSAL1 gene in isolated primed (P) and overprimed (OP) embryos. (b) PAP (3'‐phosphoadenosine 5'‐phosphate) levels in P and OP seeds along the rehydration−dehydration cycle. (c) Expression profiles of the MtRAB28, MtANAC082, MtFANC1, and MtRTEL1 genes in P and OP embryos. Data were analysed with two‐way ANOVA and TUKEY test. Means without common letters are significantly (p < 0.05) different
On the other hand, the response of P and OP embryos to dehydration shared some common features such as the up‐regulation of MtRAB28 and MtANAC082 genes (Figure 8c). MtRAB28 encodes a group 5 LEA protein able to improve drought stress tolerance in maize embryos. RAB28 showed nucleolar localization and its overexpression resulted in enhanced seed germination under water deficit (Amara et al., 2013). ANAC082, the plant counterpart of the p53 protein (master regulator of DDR in animals), is involved in the ribosome stress signalling pathways and possibly linked to nucleolar DDR (Ohbayashi et al., 2017). Nucleolar DDR also features the ATM‐dependent recruitment of the MRN complex at damaged sites and the subsequent translocation of DSBs‐containing rDNA from the interior to the periphery of the nucleolus, possibly to avoid interchromosomal recombination with lethal consequences in mitosis and facilitate repair (Korsholm et al., 2019). In this context, the Fe‐S cluster helicases FANCJB and RTEL1 have been recently characterized for their specific involvement in rDNA maintenance (Dorn et al., 2019). P and OP embryos also shared up‐regulation of MtFANCJB and MtRTEL1 genes (Figure 8c).
4. DISCUSSION
Our results with M. truncatula seeds revealed that, when hydropriming was prolonged up to 24 h, germination was accelerated but, at the same time, an increase in the number of aberrant seedlings resulting from OP seeds was observed. The aberrant phenotype was because only cotyledons of OP seeds were able to withstand dehydration as reported by Reisdorph and Koster (1999). Based on the work by Buitink et al. (2003), OP seeds with radicle protrusion ≤ 3 mm were collected for this study, as representative of a physiological stage resulting from the priming‐imposed metabolic acceleration, in which DT was lacking. The underpinning idea was to develop a model allowing the comparison between the molecular response displayed by the two subpopulations (P and OP seeds) to highlight differences in terms of genotoxic impact and seed repair ability at the nuclear and nucleolar level, useful to improve our understanding of DT dynamics.
OP seeds were challenged by oxidative stress at 2 h of dry‐back and the up‐regulation of MtSPMS and MtSPDS genes suggests a protective role of polyamines. In OP embryos, higher ROS and DNA damage levels measured during dehydration correlated with the attempt to preserve genome integrity through the up‐regulation of DDR genes, for example, the MtSOG1 gene involved in embryo growth restoration following DNA damage accumulation (Johnson et al., 2017). Phosphorylation of histone H2AX DNA at the DSBs sites represents the earliest, highly conserved, DDR hallmark which triggers DSB repair (Branzei & Foiani, 2008; Waterworth et al., 2011) whereas chromatin condensation protects DNA from damage and blocks the expansion of H2AX phosphorylation (Nair et al., 2017). The decrease in density of γH2AX foci observed in P and OP embryos after 2 h of dry‐back might reflect the effects of progressive chromatin compaction induced by dehydration. On the other hand, chromatin condensation did not impair transcription since changes in gene expression profiles were observed in OP embryos during desiccation, similarly to what was reported during the desiccation phase of seed maturation (Verdier et al., 2013).
This comparative study provides for the first time a picture of the events resulting from loss of DT in the nuclear compartment of OP embryos versus the ability to cope with damage found in primed seeds. The first hallmark of nucleolar perturbation was the boost in the precursor and mature rRNA production observed only in OP embryos with the progression of dehydration. Both the precursor and mature 5.8S rRNA showed an early peak at 2 h of dry‐back only in OP embryos. The role played by 5.8S rRNA processing in the context of DT has been so far documented only in yeast where mutants selected for increased DT were defective in genes involved in 60S ribosome biogenesis, particularly in 5.8S rRNA maturation (Welch et al., 2013). Accumulation of 5.8S rRNA in nucleoli of human myoblasts upon exposure to sodium arsenite reported by Zhu et al. (2008) suggests a general role of 5.8S rRNA processing in the oxidative stress response. When looking at dehydrated OP seeds in alfalfa, only the 5.8S rRNA unspliced precursors showed a profile similar to that observed in M. truncatula, highlighting the heterogeneity of the response in closely related species. Validation of 5.8S rRNA as a stress hallmark in the context of seed priming will require extensive testing carried out across different species. The search for molecular hallmarks is complicated by the complexity of seed responses. Such complexity has been recently highlighted in eggplant (Solanum melongena L.) when looking at the dynamics of the pre‐germinative metabolism stimulated by controlled rehydration (Forti et al., 2020a, 2020b). Even more challenging is translating the acquired knowledge from a domesticated plant to its wild crop relatives (Forti et al., 2021). In addition, the conditions and timing of the dry‐back step should be carefully assessed to design a successful priming protocol (Chen & Arora, 2013).
It has been reported that slow drying of primed seeds induces accumulation of LEA (late embryogenesis abundant) proteins (Gurusinghe et al., 2002) whereas drying at different rates results in the differential expression of DNA repair and stress tolerance genes (Soeda et al., 2005). Such findings show the need for deeper studies of the molecular processes underlying seed responses to dry‐back, as a strategy to disclose novel seed quality hallmarks.
ITS2 processing factors, such as the LAS1‐GRC3/NOL9 complex, acting upstream of the 5.8S rRNA maturation, have been implicated in genome maintenance pathways in animal cells (Pillon et al., 2019). Although changes in MtLAS1 and MtNOL9 gene expression were reported in OP embryos along the rehydration−dehydration cycle, there was no obvious correlation with altered ribogenesis.
Up‐regulation of MtXRN3 gene involved in surveillance mechanisms for the removal of aberrant transcripts (Crisp et al., 2018) was another distinctive nucleolar feature of OP embryos. According to Pornsiriwong et al. (2017), PAP plays a role in Arabidopsis mature seeds as a signal molecule migrating to the nucleus from small, non‐photosynthetic immature plastids derived from chloroplast dedifferentiation during seed desiccation. Our results suggest novel roles of the PAP molecule in stress adaptation. PAP‐mediated responses are still in part unknown (Chen et al., 2011) and plastid retrograde signals have been recently associated with genome instability and DDR (Duan et al., 2020). Based on this, the rehydration−dehydration system hereby investigated might represent an informative source in future studies. Furthermore, M. truncatula is also an attractive model to study DT re‐establishment (Buitink et al., 2003, 2006). Water content and the drying rate could also influence DT re‐establishment whereas up‐regulation of the MtDHN (DEHYDRIN) gene and accumulation of protective metabolites were associated with DT resumption under osmotic stress conditions (Buitink et al., 2003). Notably, DT can be induced only when radicle growth has been stopped. Indeed, the re‐establishment of DT is highly demanding in terms of metabolic energy (Leprince et al., 2000). The comparison between primed and overprimed M. truncatula embryos and their distinctive molecular and ultrastructural landscapes related to genotoxic damage provide novel, although indirect, insights on DT. The potential of ribogenesis as a source of novel hallmarks of the seed response to priming and dry‐back has been explored (Figure 9). Future work focused on OP embryos responding to desiccation stress will expand this emerging picture, providing direct evidence and further molecular interpretations of the link between DDR and nucleolar functions in the context of seed priming. Responses of OP embryos reflect the need of the seed to maintain genome integrity and adapt to environmental change and thus reveal promising targets for basic and applied research.
Figure 9.
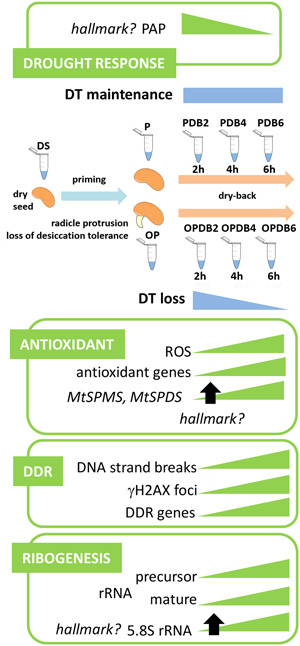
Schematic representation of the main distinctive features highlighted in this study, through the comparison between P and OP Medicago truncatula embryos coping differently with desiccation‐induced genotoxic damage
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supporting information.
ACKNOWLEDGEMENTS
This study was supported by the University of Pavia Crowdfunding (UNIVERSITIAMO) campaign, 'The other side of the seed' (https://universitiamo.eu/en/campaigns/the-other-side-of-theseed/), and by the Italian Ministry of Education, University and Research (MIUR): Dipartimenti di Eccellenza Program (2018‐2022)—Department of Biology and Biotechnology 'L. Spallanzani', University of Pavia (to M. B., A. M., and A. B.). A. P. is supported by a Departmental Research Fellowship (type A—COD. DBB2020‐A01). S. S. A. acknowledges the financial support from Fundação para a Ciência e Tecnologia throughout the Centre BIO R&D Unit (UIDB/05083/2020), NORTE 2020 throughout the I‐CERES project (NORTE‐01‐0145‐FEDER‐000082), funded by the Fundo Europeu de Desenvolvimento Regional (FEDER) and project NORTE‐06‐3559‐FSE‐000103 funded by the Fundo Social Europeu (FSE). Open access funding provided by Universita degli Studi di Pavia within the CRUI‐CARE Agreement.
Pagano, A. , Zannino, L. , Pagano, P. , Doria, E. , Dondi, D. , Macovei, A. , et al. (2022) Changes in genotoxic stress response, ribogenesis and PAP (3′‐phosphoadenosine 5′‐phosphate) levels are associated with loss of desiccation tolerance in overprimed Medicago truncatula seeds. Plant, Cell & Environment, 45, 1457–1473. 10.1111/pce.14295
Andrea Pagano and Lorena Zannino equally contributed to this study.
DATA AVAILABILITY STATEMENT
Data are available upon contact with the corresponding author.
REFERENCES
- Amara, I. , Capellades, M. , Ludevid, M.M.D. , Pagés, M. & Goday, A. (2013) Water stress tolerance of transgenic maize plants over‐expressing LEA Rab28 gene. Journal of Plant Physiology, 170(9), 864–873. Available from: 10.1016/j.jplph.2013.01.004 [DOI] [PubMed] [Google Scholar]
- Anderson, R. , Bayer, P.E. & Edwards, D. (2020) Climate change and the need for agricultural adaptation. Current Opinion in Plant Biology, 56, 197–202. Available from: 10.1016/j.pbi.2019.12.006 [DOI] [PubMed] [Google Scholar]
- Bailly, C. (2004) Active oxygen species and antioxidants in seed biology. Seed Science Research, 14(2), 93–107. Available from: 10.1079/SSR2004159 [DOI] [Google Scholar]
- Bailly, C. , El‐Maarouf‐Bouteau, H. & Corbineau, F. (2008) From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. Comptes Rendus Biologies, 331(10), 806–814. Available from: 10.1016/j.crvi.2008.07.022 [DOI] [PubMed] [Google Scholar]
- Balestrazzi, A. , Confalonieri, M. , Macovei, A. & Carbonera, D. (2011) Seed imbibition in Medicago truncatula Gaertn.: expression profiles of DNA repair genes in relation to PEG‐mediated stress. Journal of Plant Physiology, 168(7), 706–713. Available from: 10.1016/j.jplph.2010.10.008 [DOI] [PubMed] [Google Scholar]
- Bernhard, W. (1969) A new staining procedure for electron microscopical cytology. Journal of Ultrastructural Research, 27(3−4), 250–265. Available from: 10.1016/S0022-5320(69)80016-X [DOI] [PubMed] [Google Scholar]
- Bewley, J.D. (1979) Physiological aspects of desiccation tolerance. Annual Review of Plant Physiology, 30, 195–238. Available from: 10.1146/annurev.pp.30.060179.001211 [DOI] [Google Scholar]
- Biggiogera, M. & Fakan, S. (1998) Fine structural specific visualization of RNA on ultrathin sections. Journal of Histochemistry and Cytochemistry, 46(3), 389–395. Available from: 10.1177/002215549804600313 [DOI] [PubMed] [Google Scholar]
- Bradford, K.J. , Steiner, J.J. & Trawatha, S.E. (1990) Seed priming influence on germination and emergence of pepper seed lots. Crop Science, 30(3), 718–721. Available from: 10.2135/cropsci1990.0011183X003000030049x [DOI] [Google Scholar]
- Branzei, D. & Foiani, M. (2008) Regulation of DNA repair throughout the cell cycle. Nature Reviews Molecular Cell Biology, 9(4), 297–308. Available from: 10.1038/nrm2351 [DOI] [PubMed] [Google Scholar]
- Bruggink, T. & van der Toorn, P. (1995) Induction of desiccation tolerance in germinated seeds. Seed Science Research, 5(1), 1–4. Available from: 10.1017/S096025850000252X [DOI] [Google Scholar]
- Buitink, J. , Leger, J.J. , Guisle, I. , Ly Vu, B. , Wuilleme, S. & Leprince, O. (2006) Transcriptome profiling uncovers metabolic and regulatory processes occurring during the transition from desiccation‐sensitive to desiccation‐tolerant stages in Medicago truncatula seeds. The Plant Journal, 47(5), 735–750. Available from: 10.1111/j.1365-313X.2006.02822.x [DOI] [PubMed] [Google Scholar]
- Buitink, J. , Ly Vu, B. , Satour, P. & Leprince, O. (2003) The re‐establishment of desiccation tolerance in germinated radicles of Medicago truncatula Gaertn. seeds. Seed Science Research, 13(4), 273–286. Available from: 10.1079/SSR2003145 [DOI] [Google Scholar]
- Burma, S. , Chen, B.P. , Murphy, M. , Kurimasa, A. & Chen, D.J. (2001) ATM phosphorylates histone H2AX in response to DNA double‐strand breaks. Journal of Biological Chemistry, 276(45), 42462–42467. Available from: 10.1074/jbc.C100466200 [DOI] [PubMed] [Google Scholar]
- Chan, K.X. , Mabbitt, P.D. , Phua, S.Y. , Mueller, J.W. , Nisar, N. , Gigolashvili, T. et al. (2016) Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase. Proceedings of the National Academy of Sciences of the United States of America, 113(31), E4567–E4576. Available from: 10.1073/pnas.1604936113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Chu, P. , Zhou, Y. , Li, Y. , Liu, J. & Huang, S. (2012) Overexpression of AtOGG1, a DNA glycosylase/AP lyase, enhances seed longevity and abiotic stress tolerance in Arabidopsis. Journal of Experimental Botany, 63(11), 4107–4121. Available from: 10.1093/jxb/ers093 [DOI] [PubMed] [Google Scholar]
- Chen, H. , Zhang, B. , Hicks, L.M. & Xiong, L.A. (2011) Nucleotide metabolite controls stress‐responsive gene expression and plant development. PLoS ONE, 6, e26661. Available from: 10.1371/journal.pone.0026661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. & Arora, R. (2013) Priming memory invokes seed stress‐tolerance. Environmental and Experimental Botany, 94, 33–45. Available from: 10.1016/j.envexpbot.2012.03.005 [DOI] [Google Scholar]
- Collins, A.R. (2004) The comet assay for DNA damage and repair. Molecular Biotechnology, 26(3), 249–261. Available from: 10.1385/MB:26:3:249 [DOI] [PubMed] [Google Scholar]
- Cordoba‐Canero, D. , Roldan‐Arjona, T. & Ariza, R.R. (2014) Arabidopsis ZDP DNA 30‐phosphatase and ARP endonuclease function in 8‐oxoG repair initiated by FPG and OGG1 DNA glycosylases. The Plant Journal, 79(5), 824–834. Available from: 10.1111/tpj.12588 [DOI] [PubMed] [Google Scholar]
- Costa, M.C.D. , Righetti, K. , Nijveen, H. , Yazdanpanah, F. , Ligterink, W. , Buitink, J. et al. (2015) A gene co‐expression network predicts functional genes controlling the re‐establishment of desiccation tolerance in germinated Arabidopsis thaliana seeds. Planta, 242, 435–449. Available from: 10.1007/s00425-015-2283-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp, P.A. , Smith, A.B. , Gaunguly, D.R. , Murray, K.D. , Eichten, S.R. , Millar, A.A. et al. (2018) RNA polymerase II read‐through promotes expression of neighboring genes in SAL1‐PAP‐XRN retrograde signaling. Plant Physiology, 178(4), 1614–1630. Available from: 10.1104/pp.18.00758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers, B.J.W. , Costa, M.C.D. , Maia, J. , Bentsink, L. , Ligterink, W. & Hilhorst, H.M.W. (2015) Acquisition and loss of desiccation tolerance in seeds: from experimental model to biological relevance. Planta, 241(3), 563–577. http://www.jstor.org/stable/43565862 [DOI] [PubMed] [Google Scholar]
- Donà, M. , Confalonieri, M. , Minio, A. , Biggiogera, M. , Buttafava, A. & Balestrazzi, A. (2013) RNA‐Seq analysis discloses early senescence and nucleolar dysfunction triggered by Tdp1a depletion in Medicago truncatula . Journal of Experimental Botany, 64(7), 1941–1951. Available from: 10.1093/jxb/ert063 [DOI] [PubMed] [Google Scholar]
- Dorn, A. , Feller, L. , Castri, D. , Röhrig, S. , Enderle, J. , Herrmann, N.J. et al. (2019) An Arabidopsis FANCJ helicase homologue is required for DNA crosslink repair and rDNA repeat stability. PLoS Genetics, 15(5), e1008174. Available from: 10.1371/journal.pgen.1008174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, S., Hu, H., Dong, B., Jin, H‐L., Wang, H‐B. (2020) Signaling from plastid genome stability modulates endoreplication and cell cycle during plant development. Cell Reports, 32, 108019. 10.1016/j.celrep.2020.108019 [DOI] [PubMed] [Google Scholar]
- Estavillo, G.M. , Crisp, P.A. , Pornsiriwong, W. , Wirtz, M. , Collinge, D. , Carrie, C. et al. (2011) Evidence for a SAL1‐PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. The Plant Cell, 23(11), 3992–4012. Available from: 10.1105/tpc.111.091033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrissin, I. , Sano, N. , Seo, M. & North, H.M. (2021) Ageing beautifully: can the benefits of seed priming be separated from a reduced lifespan trade‐off? Journal of Experimental Botany, 72(7), 2312–2333. Available from: 10.1093/jxb/erab004 [DOI] [PubMed] [Google Scholar]
- Farooq, M. , Usman, M. , Nadeem, F. , Rehman, H. , Wahid, A. , Basra, S.M.A. et al. (2019) Seed priming in field crops: potential benefits, adoption and challenges. Crop Pasture Science, 70(9), 731–771. Available from: 10.1071/CP18604 [DOI] [Google Scholar]
- Forti, C. , Ottobrino, V. , Bassolino, L. , Toppino, L. , Rotino, G.L. , Pagano, A. et al. (2020a) Molecular dynamics of pre‐germinative metabolism in primed eggplant (Solanum melongena L.) seeds. Horticulture Research, 7, 87. Available from: 10.1038/s41438-020-0310-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forti, C. , Ottobrino, V. , Doria, E. , Bassolino, L. , Toppino, L. , Rotino, G.L. et al. (2021) Hydropriming applied on fast germinating Solanum villosum Miller seeds: impact on pre‐germinative metabolism. Frontiers in Plant Sciences, 12, 639336. Available from: 10.3389/fpls.2021.639336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forti, C. , Shankar, A. , Singh, A. , Balestrazzi, A. , Prasad, V. & Macovei, A. (2020b) Seed priming improves germination on heavy‐metal contaminated soil by inducing upregulation of genes involved in DNA repair and antioxidant response. Genes, 11(3), 242. Available from: 10.3390/genes11030242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffová, I. & Fajkus, J. (2021) The rDNA Loci‐Intersections of replication, transcription, and repair pathways. International Journal of Molecular Sciences, 22(3), 1302. Available from: 10.3390/ijms22031302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Moralesa, S.I. , Chávez‐Montesa, R.A. , Hayano‐Kanashiroa, C. , Alejo‐Jacuindea, G. , Rico‐Cambrona, T.Y. , de Foltera, S. et al. (2016) Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana . Proceedings of the National Academy of Science of the United States of America, 113(35), E5232–E5241. Available from: 10.1073/pnas.1610985113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurusinghe, S. , Powell, A.L.T. & Bradford, K.J. (2002) Enhanced expression of BiP is associated with treatments that extend storage longevity of primed tomato seeds. Journal of the American Society for Horticultural Science, 127(4), 528–534. [Google Scholar]
- Hegde, V. , Wang, M. & Deutsch, W.A. (2004) Human ribosomal protein S3 interacts with DNA base excision repair proteins hAPE/Ref‐1 and hOGG1. Biochemistry, 43(44), 14211–14217. Available from: 10.1021/bi049234b [DOI] [PubMed] [Google Scholar]
- Howden, S.M. , Soussana, J.‐F. , Tubiello, F.N. , Chhetri, N. , Dunlop, M. & Meinke, H. (2007) Adapting agriculture to climate change. Proceedings of the National Academy of Sciences of the United States of America, 104(50), 19691–19696. Available from: 10.1073/pnas.0701890104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni, J.S. , Caron, P. , Lassadi, I. , Nicolas, E. , Massip, L. , Trouche, D. et al. (2010) High‐resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO Journal, 29(8), 1446–1457. Available from: 10.1038/emboj.2010.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.A. , Conklin, P.A. , Tjahjadi, M. , Missirian, V. , Toal, T. , Brady, S.M. et al. (2017) SUPPRESSOR OF GAMMA RESPONSE 1 links DNA damage response to organ regeneration. Plant Physiology, 176(2), 1665–1675. Available from: 10.1104/pp.17.01274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina, N.O. , Makarova, S. , Makhotenko, A. , Love, A.J. & Taliansky, M. (2018) The multiple functions of the nucleolus in plant development, disease and stress responses. Frontiers in Plant Science, 9, 132. Available from: 10.3389/fpls.2018.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, W. , Zeljković, S.Ć. , Piskurewicz, U. , Megies, C. , Tarkowski, P. & Lopez‐Molina, L. (2019) Polyamine uptake transporter 2 (put2) and decaying seeds enhance phyA‐mediated germination by overcoming PIF1 repression of germination. PLoS Genetics, 15(7), e1008292. Available from: 10.1371/journal.pgen.1008292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsholm, L.M. , Gal, Z. , Lin, L. , Quevedo, O. , Ahmad, D.A. , Dulina, E. et al. (2019) Double‐strand breaks in ribosomal RNA genes activate a distinct signaling and chromatin response to facilitate nucleolar restructuring and repair. Nucleic Acids Research, 47(15), 8019–8035. Available from: 10.1093/nar/gkz518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince, O. , Harren, F.J.M. , Buitink, J. , Alberda, M. & Hoekstra, F.A. (2000) Metabolic dysfunction and unabated respiration precede the loss of membrane integrity during dehydration of germinating radicles. Plant Physiology, 122(2), 597–608. Available from: 10.1104/pp.122.2.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince, O. , Pellizzaro, A. , Berriri, S. & Buitink, J. (2017) Late seed maturation: drying without dying. Journal of Experimental Botany, 68(4), 827–841. Available from: 10.1093/jxb/erw363 [DOI] [PubMed] [Google Scholar]
- Liu, J.‐H. , Wang, W. , Wu, H. , Gong, X. & Moriguchi, T. (2015) Polyamines function in stress tolerance: from synthesis to regulation. Frontiers in Plant Science, 6, 827. Available from: 10.3389/fpls.2015.00827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei, A. , Balestrazzi, A. , Confalonieri, M. & Carbonera, D. (2010) The Tdp1 (Tyrosyl‐DNA phosphodiesterase) gene family in barrel medic (Medicago truncatula Gaertn.): bioinformatic investigation and expression profiles in response to copper‐ and PEG‐mediated stress. Planta, 232(2), 393–407. Available from: 10.1007/s00425-010-1179-9 [DOI] [PubMed] [Google Scholar]
- Macovei, A. , Balestrazzi, A. , Confalonieri, M. , Faè, M. & Carbonera, D. (2011) New insights on the barrel medic MtOGG1 and MtFPG functions in relation to oxidative stress response in planta and during seed imbibition. Plant Physiology and Biochemistry, 49(9), 1040–1050. Available from: 10.1016/j.plaphy.2011.05.007 [DOI] [PubMed] [Google Scholar]
- Macovei, A. , Faè, M. , Biggiogera, M. , de Sousa Araujo, S. , Carbonera, D. & Balestrazzi, A. (2018) Ultrastructural and molecular analyses reveal enhanced nucleolar activity in Medicago truncatula cells overexpressing the MtTdp2alpha gene. Frontiers in Plant Science, 9, 596. Available from: 10.3389/fpls.2018.00596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei, A. , Pagano, A. , Cappuccio, M. , Gallotti, L. , Dondi, D. , De Sousa Araujo, S. et al. (2019) A snapshot of the trehalose pathway during seed imbibition in Medicago truncatula reveals temporal‐ and stress‐dependent shifts in gene expression patterns associated with metabolite changes. Frontiers in Plant Science, 10, 1590. Available from: 10.3389/fpls.2019.01590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei, A. , Pagano, A. , Leonetti, P. , Carbonera, D. , Balestrazzi, A. & Araújo, S. (2017) Systems biology approaches to unveil the molecular players involved in the pre‐germinative metabolism: implications on seed technology traits. Plant Cell Reports, 36(5), 669–688. Available from: 10.1007/s00299-016-2060-5 [DOI] [PubMed] [Google Scholar]
- Maia, J. , Dekkers, B.J.W. , Provart, N.J. , Ligterink, W. & Hilhorst, H.W.M. (2011) The re‐establishment of desiccation tolerance in germinated Arabidopsis thaliana seeds and its associated transcriptome. PLoS ONE, 6, e29123. Available fom: 10.1371/journal.pone.0029123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, N. , Shoaib, M. & Storgaard Sorensen, C. (2017) Chromatin dynamics in genome stability: roles in suppressing endogenous DNA damage and facilitating DNA repair. International Journal of Molecular Sciences, 18(7), 1486. Available from: 10.3390/ijms18071486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitaki, Z. , Holá, M. , Donà, M. , Pavlopoulou, A. , Michalopoulos, J. , Angelis, K.J. et al. (2018) Integrating plant and animal biology for the search of novel DNA damage biomarkers. Mutation Research—Reviews in Mutation Research, 775, 21–38. Available from: 10.1016/j.mrrev.2018.01.001 [DOI] [PubMed] [Google Scholar]
- Ohbayashi, I. , Lin, C.‐Y. , Shinohara, N. , Matsumura, Y. , Machids, Y. , Horiguchi, G. et al. (2017) Evidence for a role of ANAC082 as a ribosomal stress response mediator leading to growth defects and developmental alterations in Arabidopsis. The Plant Cell, 29(10), 2644–2660. Available from: 10.1105/tpc.17.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, M.J. , Farrant, J.M. , Hilhorst, H.W.M. , Mundree, S. , Williams, B. & Bewley, J.D. (2020) Desiccation tolerance: avoiding cellular damage during drying and rehydration. Annual Review of Plant Biology, 71, 435–460. Available from: 10.1146/annurev-arplant-071219-105542 [DOI] [PubMed] [Google Scholar]
- Oñate‐Sanchez, L. & Vicente‐Carbajosa, J. (2008) DNA‐free RNA isolation protocols from Arabidopsis thaliana, including seeds and siliques. BMC Research Notes, 1, 93. Available from: 10.1186/1756-0500-1-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgawa, L.M. & Baserga, S.J. (2017) Crosstalk between the nucleolus and the DNA damage response. Molecular BioSystems, 13, 443. Available from: 10.1039/C6MB00740F [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano, A. , Araújo, S.S. , Macovei, A. , Leonetti, P. & Balestrazzi, A. (2017) The seed repair response during germination: disclosing correlations between DNA repair, antioxidant response, and chromatin remodeling in Medicago truncatula . Frontiers in Plant Science, 8, 1972. Available from: 10.3389/fpls.2017.01972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano, A. , De Sousa Araújo, S. , Macovei, A. , Dondi, D. , Lazzaroni, S. & Balestrazzi, A. (2019) Metabolic and gene expression hallmarks of seed germination uncovered by sodium butyrate in Medicago truncatula . Plant Cell & Environment, 42(1), 259–269. Available from: 10.1111/pce.13342 [DOI] [PubMed] [Google Scholar]
- Paparella, S. , Araujo, S.S. , Rossi, G. , Wijayasinghe, M. , Carbonera, D. & Balestrazzi, A. (2015) Seed priming: state of the art and new perspectives. Plant Cell Reports, 34(8), 1281–1293. Available from: 10.1007/s00299-015-1784-y [DOI] [PubMed] [Google Scholar]
- Park, Y.J. , Kim, T.‐S. , Kim, E.‐H. , Kim, H.D. & Kim, J. (2020) Ribosomal protein S3 is a novel negative regulator of nonhomologous end joining repair of DNA double‐strand breaks. FASEB Journal, 34(6), 8102–8113. Available from: 10.1096/fj.201903245R [DOI] [PubMed] [Google Scholar]
- Peng, L. , Lang, S. , Wang, Y. , Pritchard, H.W. & Wang, X.F. (2017) Modulating role of ROS in reestablishing desiccation tolerance in germinating seeds of Caragana korshinskii Kom. Journal of Experimental Botany, 68(13), 3585–3601. Available from: 10.1093/jxb/erx172 [DOI] [PubMed] [Google Scholar]
- Peng, L. , Sun, Q. , Xue, H. & Wang, X. (2018) iTRAQ‐based quantitative proteomic analysis reveals pathways associated with re‐establishing desiccation tolerance in germinating seeds of Caragana korshinskii Kom. Journal of Proteomics, 179(2018), 1–16. Available from: 10.1016/j.jprot.2018.01.009 [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Research, 29(9), e45. Available from: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon, M.C. , Lo, Y.‐H. & Stanley, R.E. (2019) IT's 2 for the price of 1: multifaceted ITS2 processing machines in RNA and DNA maintenance. DNA Repair, 81, 102653. Available from: 10.1016/j.dnarep.2019.102653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier, Y. , Huang, S.Y. , Gao, R. , Das, B.B. , Murai, J. & Marchand, C. (2014) Tyrosyl‐DNA‐phosphodiesterases (TDP1 and TDP2). DNA Repair, 19, 114–129. Available from: 10.1016/j.dnarep.2014.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornsiriwong, W. , Estavillo, G.M. , Chan, K.X. , Tee, E.E. , Ganguly, D. , Grisp, P.A. et al. (2017) A chloroplast retrograde signal, 3′‐ phosphoadenosine 5′‐phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. eLife, 6, e23361. Available from: 10.7554/eLife.23361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisdorph, N.A. & Koster, K. (1999) Progressive loss of desiccation tolerance in germinating pea (Pisum sativum) seeds. Physiologia Plantarum, 105(2), 266–271. Available from: 10.1034/j.1399-3054.1999.105211.x [DOI] [Google Scholar]
- Renard, M. , Alkhalfioui, F. , Schmitt‐Keichinger, C. , Ritzenthaler, C. & Montrichard, F. (2011) Identification and characterization of thioredoxin h isoforms differentially expressed in germinating seeds of the model legume Medicago truncatula . Plant Physiology, 155(3), 1113–1126. Available from: 10.1104/pp.110.170712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero‐Rodríguez, M.C. , Archidona‐Yuste, A. , Abril, N. , Gil‐Serrano, A.M. , Meijón, M. & Jorrín‐Novo, J.V. (2018) Germination and early seedling development in Quercus ilex recalcitrant and non‐dormant seeds: targeted transcriptional, hormonal, and sugar analysis. Frontiers in Plant Science, 9, 1508. Available from: 10.3389/fpls.2018.01508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikova, G. , Leonova, T. , Vashurina, N. , Frolov, A. & Medvedev, S. (2020) Desiccation tolerance as the basis of long‐term seed viability. International Journal of Molecular Science, 22(1), 101. Available from: 10.3390/ijms22010101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda, Y. , Konings, M.C.J.M. , Vorst, O. , van Houwelingen, A.M.M.L. , Stoopen, G.M. , Maliepaard, C.A. et al. (2005) Gene expression programs during Brassica oleracea seed maturation, osmopriming, and germination are indicators of progression of the germination process and the stress tolerance level. Plant Physiology, 137(1), 354–368. Available from: 10.1104/pp.104.051664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepinski, D. (2014) Functional structure of the plant nucleolus. Protoplasma, 251(6), 1285–1306. Available from: 10.1007/s00709-014-0648-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrasson, E. , Buitink, J. , Righetti, K. , Ly Vu, B. , Pelletier, S. , Zinsmeister, J. et al. (2013) An emerging picture of the seed desiccome: confirmed regulators and newcomers identified using transcriptome comparison. Frontiers in Plant Science, 4, 497. Available from: 10.3389/fpls.2013.00497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez‐Nin, G.H. , Biggiogera, M. & Echeverria, O.M. (1995) Activation of osmium ammine by SO2‐generating chemicals for EM Feulgen‐type staining of DNA. European Journal of Histochemistry, 39(2), 101–106. [PubMed] [Google Scholar]
- Ventura, L. , Macovei, A. , Donà, M. , Paparella, S. , Buttafava, A. , Giovannini, A. et al. (2014) Genotoxic effects due to in vitro culture and H2O2 treatments in Petunia x hybrida cells monitored through DNA diffusion assay, FPG‐SCGE and gene expression profile analyses. Acta Physiologiae Plantarum, 36(2), 331–341. Available from: 10.1007/s11738-013-1415-6 [DOI] [Google Scholar]
- Verdier, J. , Lalanne, D. , Pelletier, S. , Torres‐Jerez, I. , Righetti, K.A. , Bandyopadhyay, K. et al. (2013) Regulatory network‐based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiology, 163(2), 757–774. Available from: 10.1104/pp.113.222380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W.‐Q. , Wang, Y. , Song, X.‐J. , Zhang, Q. , Cheng, H.‐Y. , Liu, J. et al. (2021) Proteomic analysis of desiccation tolerance and its re‐establishment in different embryo axis tissues of germinated pea seeds. Journal of Proteome Research, 20, 2352–2363. Available from: 10.1021/acs.jproteome.0c00860 [DOI] [PubMed] [Google Scholar]
- Waterworth, W.M. , Bray, C.M. & West, C.E. (2019) Seeds and the art of genome maintenance. Frontiers in Plant Science, 10, 706. Available from: 10.3389/fpls.2019.00706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth, W.M. , Drury, G.E. , Bray, C.M. & West, C.E. (2011) Repairing breaks in the plant genome: the importance of keeping it together. New Phytologist, 192(4), 805–822. Available from: 10.1111/j.1469-8137.2011.03926.x [DOI] [PubMed] [Google Scholar]
- Welch, A.Z. , Gibney, P.A. , Botstein, D. & Koshland, D.E. (2013) TOR and RAS pathways regulate desiccation 885 tolerance in Saccharomyces cerevisiae . Molecular Biology of the Cell, 24(2), 115–128. Available from: 10.1091/mbc.E12-07-0524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Chen, Y. , Li, L. , Li, Z. , Wang, C. , Zhou, T. et al. (2012) An improved HPLC method for the quantitation of 3′‐phosphoadenosine 5′‐phosphate (PAP) to assay sulfotransferase enzyme activity in HepG2 cells. Journal of Pharmaceutical and Biomedical Analysis, 62, 182–186. Available from: 10.1016/j.jpba.2011.12.015 [DOI] [PubMed] [Google Scholar]
- Yang, C. , Zang, W. , Ji, Y. , Li, T. , Yang, Y. & Zheng, X. (2019) Ribosomal protein L6 (RPL6) is recruited to DNA damage sites in a poly(ADP‐ribose) polymerase‐dependent manner and regulates the DNA damage response. Journal of Biological Chemistry, 294, 2827–2830. Available from: 10.1074/jbc.RA118.007009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama, K. (2016) SOG1: a master regulator of the DNA damage response in plants. Genes Genetic Systems, 90(4), 209–216. Available from: 10.1266/ggs.15-00011 [DOI] [PubMed] [Google Scholar]
- Zhu, C.H. , Kim, J. , Shay, J.W. & Wright, W.E. (2008) SGNP: an essential Stress Granule/Nucleolar Protein potentially involved in 5.8s rRNA processing/transport. PLoS ONE, 3(11), e3716. Available from: 10.1371/journal.pone.0003716 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
Data are available upon contact with the corresponding author.


