Abstract
Short sequence repeats (SSRs) with a potential variable number of tandem repeat (VNTR) loci were identified in the genome of the citrus pathogen Xylella fastidiosa and used for typing studies. Although mono- and dinucleotide repeats were absent, we found several intermediate-length 7-, 8-, and 9-nucleotide repeats, which we examined for allelic polymorphisms using PCR. Five genuine VNTR loci were highly polymorphic within a set of 27 X. fastidiosa strains from different hosts. The highest average Nei's measure of genetic diversity (H) estimated for VNTR loci was 0.51, compared to 0.17 derived from randomly amplified polymorphic DNA (RAPD) analysis. For citrus X. fastidiosa strains, some specific VNTR loci had a H value of 0.83, while the maximum value given by specific RAPD loci was 0.12. Our approach using VNTR markers provides a high-resolution tool for epidemiological, genetic, and ecological analysis of citrus-specific X. fastidiosa strains.
Xylella fastidiosa has been associated with diseases in economically important crops such as grapevine, plum, almond, peach, citrus, and more recently, coffee (13, 17) as well as some diseases in ornamental plants (3). Reciprocal transmission tests including X. fastidiosa from several hosts evidenced the occurrence of host infection groups (for details, see reference 10). In Brazil, X. fastidiosa has been responsible for causing diseases in important crops such as citrus and coffee (5, 13), although it has also been observed in plum (7) and Hibiscus schizopetalus (E. W. Kitajima, H. D. Coletta-Filho, M. A. Machado, and Q. S. Novaes, 33th Congr. Brazil Phytopathol. Soc., abstr. 323, 2000). The major economic damage is in the sweet orange crop (Citrus sinensis Osb.) which has suffered an annual loss of about $100 million. However, the total cost to Brazilian agriculture is probably higher because the economic damage caused to the coffee crop by X. fastidiosa has not been estimated.
Methods for distinguishing between bacterial strains are important for detecting disease outbreaks for epidemiological analysis, and for understanding the genetic structure of microbial populations. Molecular techniques of DNA profiling based on PCR of randomly amplified polymorphic DNA (RAPD) and repetitive element polymorphism PCR (rep-PCR) have been used with great success for studies of genetic variation and the relationships between X. fastidiosa strains (2, 6, 8, 16). However, the data produced by these methods are biallelic, which limits the amount of genetic information per locus and thus the use of these methods in estimating genetic diversity. Interlaboratory reproducibility is also a weakness of RAPD and rep-PCR analysis (20). Short sequence repeats (SSRs) with a potential variable number of repeats (VNTR) within prokaryotic DNA have been used as markers for discrimination between strains (1, 9, 12, 23). Some of these repetitive regions are located within or near DNA coding regions and so could have the potential to affect gene expression (22).
The complete sequencing of the genome of the pathogenic X. fastidiosa strain 9a5c (19) has allowed the identification of repetitive DNA motifs. In the present study, we conducted a search for SSR size variation in different strains of X. fastidiosa, comparing the results with the frequently used RAPD method.
MATERIALS AND METHODS
Strains and DNA isolation.
Table 1 shows the X. fastidiosa strains used, all of which were cultured on BCYE agar medium (24) for 7 to 10 days at 28°C. The hexadecyltrimethylammonium bromide (CTAB) miniprep method (25) was used for DNA preparation from each strain. Briefly, cells were harvested, washed twice in washing buffer (pH 8.0) containing 20 mM Tris HCl and 10 mM EDTA, and treated with proteinase K and sodium dodecyl sulfate. Proteins and other cellular components were removed by using CTAB and by two chloroform extractions. The DNA present in the aqueous phase was precipitated with absolute ethanol followed by washing with 70% ethanol. The pellet obtained was dissolved in TE buffer (10 mM Tris HCl, 1 mM EDTA) (pH 8.0) containing 20 mM RNase. The DNA was subjected to electrophoresis in a 0.8% agarose gel. After staining with ethidium bromide, the samples were visualized under ultraviolet light and the DNA concentration was estimated. The DNA solution was diluted to 5 ng/μl and stored at −20°C.
TABLE 1.
X. fastidiosa strains used in this study
| Strain | Host | Geographic origin | Source |
|---|---|---|---|
| 9a5c | Citrus sinensis | São Paulo, Brazil | INRAa |
| B14 | Citrus sinensis | São Paulo, Brazil | CCSMb |
| 11779 | Citrus sinensis | Santa Catarina, Brazil | IAPARc |
| M2-1 | Citrus sinensis | São Paulo, Brazil | CCSM |
| ITAa | Citrus sinensis | São Paulo, Brazil | CCSM |
| 11834 | Citrus sinensis | Paraná, Brazil | IAPAR |
| NP3.2 | Citrus sinensis | São Paulo, Brazil | CCSM |
| 11775 | Citrus sinensis | Rio Grande do Sul, Brazil | IAPAR |
| 11066 | Citrus sinensis | Paraná, Brazil | IAPAR |
| 11067 | Citrus sinensis | Paraná, Brazil | IAPAR |
| NP2.3 | Citrus sinensis | São Paulo, Brazil | CCSM |
| 10438 | Citrus sinensis | Paraná, Brazil | IAPAR |
| 11038 | Citrus sinensis | São Paulo, Brazil | IAPAR |
| 11347 | Citrus sinensis | Paraná, Brazil | IAPAR |
| SR9.2 | Citrus sinensis | São Paulo, Brazil | CCSM |
| 11037 | Citrus sinensis | São Paulo, Brazil | IAPAR |
| 11399 | Citrus sinensis | Sergipe, Brazil | IAPAR |
| 11380 | Citrus sinensis | Santa Catarina, Brazil | IAPAR |
| SL4 | Citrus sinensis | Minas Gerais, Brazil | CCSM |
| C3 | Coffea arabica | São Paulo, Brazil | CCSM |
| C6 | Coffea arabica | São Paulo, Brazil | CCSM |
| C13 | Coffea arabica | São Paulo, Brazil | CCSM |
| 12288 | Coffea arabica | Paraná, Brazil | IAPAR |
| 8935 | Vitis vinifera | United States | ATCC 35879d |
| Hib. 5 | Hibiscus schizopetalus | Brasilia, Brazil | CCSM |
| 9746 | Prunus salicina | Paraná, Brazil | IAPAR |
| 12319 | Catharantus sp. | São Paulo, Brazil | IAPAR |
INRA, Institut National de la Recherche Agronomique et Université Victor Ségale, Bordeaux, France.
CCSM, Centro de Citricultura Sylvio Moreira, Instituto Agronômico, Cordeiropolis, São Paulo, Brazil.
IAPAR, Instituto Agronômico do Paraná, Paraná, Brazil.
ATCC, American Type Culture Collection, Manassas, Va.
Computer analysis of repetitive DNA.
The genomic DNA sequence of X. fastidiosa strain 9a5c (available on the UNICAMP website at http://onsona.lbi.ic.unicamp.br/xf) was screened for repetitive DNA with the Tandem Repeat Finder version 2.02 software (4), freely available at http://c3.biomath.mssm.edu /trf/. This software scores all DNA motif categories, although in our study, only perfect repeats were selected. BLASTN searches of the regions upstream and downstream of the repeat motif were conducted on the complete X. fastidiosa 9a5c genome to localize the repeats and related genes.
Repeat analysis.
For some 7-, 8-, and 9-nucleotide repeats, sets of primers (Table 2) with the potential for locus-specific amplification were designed using Lasergene 99 software (DNASTAR, Inc). Most of the primers were deduced from sequences bordering the repeat, 5 nucleotides upstream and downstream of the locus. The other primers (SSR26, SSR30, and SSR32) were designed up to 30 nucleotides distant from the repeat locus. Amplifications were conducted in a volume of 25 μl containing 10% of 10× PCR buffer (200 mM Tris-HCl, pH 8.4, 500 mM KCl), 2.5 mM MgCl2, 0.25 mM (each) deoxynucleoside triphosphate (GIBCO BRL), 50 ng (each) primer, 50 ng of template DNA, and 1.5 U of Taq DNA polymerase (GIBCO BRL). An initial denaturation step at 94°C for 3 min was followed by a touchdown amplification program. DNA was denatured at 94°C for 1 min, and primers were annealed for 30 s and extended at 72°C for 1 min. The initial annealing temperature was 64°C for 1 cycle. The temperature was subsequently dropped 1°C every cycle until a final annealing temperature of 55°C was reached. For the remaining 22 cycles, the annealing temperature was 55°C. A final extension step of 4 min at 72°C was followed by 10°C soak. PCR products were separated by gel electrophoresis in 3% agarose gels and stained with ethidium bromide, with the fragment size estimated based on migration relative to that of a 100-bp size marker (GIBCO BRL) loaded together with the samples in the gel.
TABLE 2.
SSR regions monitored for polymorphism in 27 strains of X. fastidiosa isolated from different hostsa
| Potential VNTR name | Repeat motif | Repeat position in genome | Primer sequence for VNTR (5′ → 3′ amplification) |
|---|---|---|---|
| SSR20 | (ATTGCTG)13 | 18527–18619 | ATGAAGAAGCCAGGATACAT |
| GCTACACGTGCAACAAC | |||
| SSR21 | (TGTTATC)21 | 52902–53049 | AACACGGATCAAGCTCATG |
| GGAACACGCAATAGTAAGAC | |||
| SSR26 | (GTGTGTGA)37 | 49439–49733 | CTGTGATCGGTGAATTGA |
| TCAAGCACACTTCCTACG | |||
| SSR28 | (GTGTGCCT)11 | 1046415–1046503 | GCAACGCTGTTATCTCAAT |
| ATTACGCTTCTTATCGCTGT | |||
| SSR30 | (TGATCCTG)15 | 1512303–1512422 | TACGCTGCACCTGTCTG |
| CTGTGAACTTCCATCAATCC | |||
| SSR32 | (CTGATGTG)9 | 2050614–2050688 | AGATGAACCTCGCCAC |
| GTACTCATCTGCGATGG | |||
| SSR34 | (TTGGGTAG)22/(TTGGGTAA)35 | 2289266–2289727 | TGATAGAACTGTTTGACGCATTTG |
| TCGGGAAGTTTGGGGTGAC | |||
| SSR36 | (TGTTGGGG)10 | 2639987–2640070 | ATGTCACTCAGGTCAGG |
| CAGAACCACCGACTG | |||
| SSR40 | (GAAGGCGTA)27 | 1220096–1220340 | ACCTTGACGACGGATG |
| TAGGAACTGCTGCTACTGAT |
All data are based on the genome sequence of X. fastidiosa strain 9a5c.
RAPD analysis.
RAPD analysis was performed in a volume of 13 μl containing 10% 10× PCR buffer, 2.0 mM MgCl2, 0.25 mM (each) deoxynucleoside triphosphate (GIBCO BRL), 15 ng of template DNA, 1.5 U of Taq DNA polymerase (GIBCO BRL), and 15 ng of primer (Operon kit of 10-mer primers; OPG10, OPG17, OPG19, OPH03, OPH07, OPH12, OPN04, OPQ05, and OPW07). Amplification was performed using a temperature profile, with initial denaturation at 94°C for 3 min followed by 36 cycles of denaturation (1 min at 94°C), annealing (1 min at 36°C), and extension (2 min at 72°C) with a final 10-min extension step at 72°C. The RAPD reaction products were separated by electrophoresis in 1.3% agarose gels and stained with ethidium bromide, with the RAPD band size estimated by comparison with a 1-kb DNA ladder marker (GIBCO BRL).
Data analysis.
All genotypes used for SSR and RAPD were included in the analysis. For the RAPD methodology, only high-intensity markers of unambiguous interpretation and good reproducibility were scored. Nei's measure of genetic diversity was calculated as H = [1 − Σpi2], where pi is the frequency of allele i at the locus (15).
RESULTS
Database searches for SSRs.
There were no 1- or 2-base SSRs, but there were 67 perfect SSRs with 3 to 33 nucleotides per unit (Table 3). Most common were 8-nucleotide SSRs, which were present at twelve loci at copy numbers varying from 3 to 37 copies per repeat. In one of these loci, we encountered mixed repeats harboring the (TTGGGTAG)22/(TTGGGTAA)35 motif.
TABLE 3.
Perfect SSR regions encountered in the genome of X. fastidiosa strain 9a5c by computerized screening
| Repeat unit length | No. of copiesa |
|---|---|
| 3 | 8, 10 |
| 4 | 7, 8, 13 |
| 5 | 11, 12, 21, 22 |
| 6 | (5)b 4, 7, 8, 10, 15, 16 |
| 7 | 4, 5, (2) 7, 13, 21 |
| 8 | (3) 3, 4, 5, (2) 9, 10, 11, 15, 22, 35, 37 |
| 9 | (2) 3, 27 |
| 10 | 3, 4 |
| 11 | 2 |
| 12 | (7) 2, 3 |
| 13 | (2) 2 |
| 14 | 2 |
| 15 | (6) 2 |
| 17 | 2 |
| 18 | 4 |
| 21 | 2 |
| 23 | 2 |
| 29 | 2 |
| 33 | 2 |
The number of copies in different positions in the genome is shown.
Number of times that the number of copies of repeat were encountered through the genome.
SSR amplification by PCR.
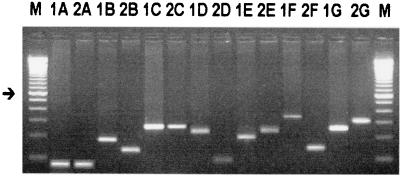
For DNA polymorphism analysis of the SSR regions, we selected two 7-nucleotide SSRs, six 8-nucleotide SSRs, and one 9-nucleotide SSR (Table 2). These SSRs were selected as potential VNTRs because they were of intermediate size, uncommon for prokaryotic SSRs (22), and presented a large number of copies per repeat unit. Amplifications using all these potential VNTR primers were performed in X. fastidiosa strains 9a5c and B14 (Fig. 1), although for two SSR primers (SSR32 and SSR34), the amplification step failed even though variations in the amplification program and reaction mixtures were tried. Although the SSR26 and SSR36 primers generated the same product for both strains 9a5c and B14, they were still used for typing studies together with primers SSR20, -21, -28, -30, and -40.
FIG. 1.
Gel electrophoresis of seven SSR primers for two X. fastidiosa strains. X. fastidiosa strains 9a5c (lanes 1) and B14 (lanes 2) and SSR primers SSR36 (A lanes) SSR20 (B lanes), SSR26 (C lanes), SSR21 (D lanes), SSR28 (E lanes), SSR40 (F lanes), and SSR30 (G lanes) were used. The intensely stained band represents a 600-bp fragment (arrow). Lanes M, 100-bp ladder marker (GIBCO BRL).
VNTR analysis.
Table 4 shows the SSR and RAPD primers used and the genetic diversity values obtained. For the SSR26 and SSR36 primers, no variation in the number of repeats was observed in the strains. Five pairs of primers (SSR20, -21, -28, -30, and -40) showed definite length polymorphism in their respective SSR regions and thus appear to represent genuine VNTRs. Even though the reactions were repeated, doublet bands were sometimes observed when the SSR20 and SSR40 primers were used with strain 12319 and when the SSR30 primer was used in some other strains. This indicates the presence of two alleles for the same SSR locus in some strains, although no doublet bands were observed in strain 9a5c.
TABLE 4.
Mean H values for the SSR and RAPD loci in 27 strains of X. fastidiosa isolated from different hosts
| Locusa |
H value for X. fastidiosa strain isolated from:
|
|||
|---|---|---|---|---|
| All hosts | Citrus | Coffee | Other hostsb | |
| SSR | ||||
| SSR20 | 0.77 | 0.70 | 0.50 | 0.72 |
| SSR21 | 0.44 | 0.28 | 0.63 | 0.38 |
| SSR26 | 0.00 | 0.00 | 0.00 | 0.00 |
| SSR28 | 0.81 | 0.80 | 0.75 | 0.63 |
| SSR30 | 0.69 | 0.64 | 0.63 | 0.48 |
| SSR36 | 0.00 | 0.00 | 0.00 | 0.00 |
| SSR40 | 0.83 | 0.83 | 0.63 | 0.56 |
| Avg | 0.51 | 0.46 | 0.45 | 0.39 |
| RAPD | ||||
| OPG10 | 0.23 | 0.12 | 0.09 | 0.36 |
| OPG17 | 0.16 | 0.02 | 0.15 | 0.33 |
| OPG19 | 0.22 | 0.16 | 0.05 | 0.27 |
| OPH03 | 0.14 | 0.07 | 0.06 | 0.39 |
| OPH07 | 0.16 | 0.04 | 0.08 | 0.43 |
| OPH12 | 0.13 | 0.09 | 0.06 | 0.21 |
| OPN04 | 0.19 | 0.11 | 0.06 | 0.35 |
| OPQ05 | 0.07 | 0.00 | 0.00 | 0.28 |
| OPW07 | 0.20 | 0.00 | 0.13 | 0.48 |
| Avg | 0.17 | 0.07 | 0.07 | 0.34 |
SSR and RAPD primers.
Data from strains isolated from plum, grapevine, hibiscus, and periwinkle.
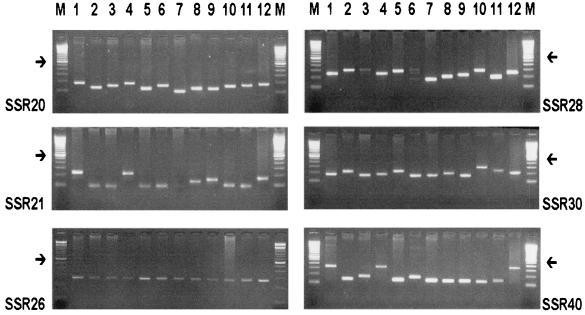
A minimum of six and a maximum of eight alleles per locus were observed with the VNTR primers, generating a high level of polymorphism. The discriminatory power of each VNTR locus and RAPD analysis was estimated from the genetic diversity (H) values based on the number of alleles and their frequency. In the analysis of citrus and coffee X. fastidiosa strains, the average H value was higher for VNTR markers than for RAPD markers (Table 4), with VNTR primers SSR28 and SSR40 showing the highest H values. For citrus strains, these values reached 0.80 (SSR28) and 0.83 (SSR40). It has previously been shown that VNTR typing of Bacillus anthracis and Yersinia pestis has produced H values of 0.80 and 0.82, respectively (1, 11). Table 4 also shows that in the analysis of X. fastidiosa strains isolated from other hosts (plum, grapevine, hibiscus, and periwinkle), the average VNTR marker H value (0.39) was only slightly higher than the average RAPD marker H value (0.34).
DISCUSSION
The presence of SSRs and their usefulness for typing have previously been shown for both human and animal pathogens (22), but this is the first time that these molecular markers have been used to type phytopathogenic bacteria.
The characteristics of the SSR regions found in the X. fastidiosa genome were quite different from the SSRs already known in other bacteria. No mono-or dinucleotide SSRs were detected in X. fastidiosa, but there was a large number of 7- and 8-nucleotide repeats (Table 3), a fact which led us to use intermediate-sized repeats in our typing studies. Shorter, 1- or 2-nucleotide repeats are abundant in other species of pathogenic bacteria such as Helicobacter, Neisseria, Mycobacterium, and Escherichia coli, while intermediate-sized repeats have rarely been found (22). This phenomenon appears to be independent of genome size, because in the 4.2-Mb chromosome of Bacillus subtilis, there are only repeats with short sequences, while in the 0.58-Mb chromosome of Mycoplasma genitalium, there are other classes of repeats (21).
The VNTR typing method presented in this paper is a powerful method for the genetic characterization of X. fastidiosa isolates. The potential VNTR regions selected for typing were conserved in all X. fastidiosa strains tested, irrespective of host (Table 4 and Fig. 2). The genetic diversity (H) values based on VNTR primers were extremely high, especially for citrus- and coffee-specific X. fastidiosa strains. This contrasts with the RAPD technique, which although it has allowed the construction of well-defined X. fastidiosa host groups (5, 16) has shown limited discriminatory capacity between X. fastidiosa strains isolated from citrus species (18). Although the RAPD technique has the potential for genome-wide analysis, SSRs have the advantage that they can link to the genomic hypervariable regions produced as a result of variation in sequence composition and DNA polymerase activity. It seems that the high discriminatory power of VNTR locus analysis is a result of the hypervariability of this molecular marker rather than the occurrence of artifacts due to serial subculture. The VNTR assay is also fairly reproducible (1, 14), and VNTR mutation rates, as seen in Bacillus anthracis, are less than 10−5 (11).
FIG. 2.
Gel electrophoresis of SSR20, SSR21, SSR26, SSR28, SSR30, and SSR40 primers from 12 X. fastidiosa strains. X. fastidiosa strains 9a5c, B14, 11779, M2.1, ITAa, 11834, 9713, C13, C3, Hib 5, 9746, and NP3.2 are shown in lanes 1 to 12, respectively (see Table 1 for host identification). The intensely stained bands represent a 600-bp fragment (arrows). Lanes M, 100-bp ladder marker (GIBCO BRL).
The growing numbers of prokaryotic DNA sequences (including those from plant pathogens) in databases and computer programs available for the detection of SSR loci have facilitated the evaluation of SSR within DNA sequences. Not only are the hypervariability and reproducibility of SSRs markers useful, but if the primers are X. fastidiosa specific, there is also the possibility of in situ analysis of a known gene without isolating the bacteria. This approach would greatly facilitate epidemiological, genetic, and ecological studies of fastidious bacteria, such as X. fastidiosa, which are normally difficult to isolate and grow.
ACKNOWLEDGMENTS
This work was supported by FAPESP, CNPq, and FUNDECITRUS.
REFERENCES
- 1.Adair D M, Worsham P L, Hill K K, Klevytska A M, Jackson P J, Friedlander A M, Keim P. Diversity in a variable-number tandem repeat from Yersinia pestis. J Clin Microbiol. 2000;38:1516–1519. doi: 10.1128/jcm.38.4.1516-1519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albibi R, Chen J, Lamikanra O, Banks D, Jarret R L, Smith B J. RAPD fingerprinting Xylella fastidiosa Pierce's disease strains isolated from a vineyard in North Florida. FEMS Microbiol Lett. 1998;165:347–352. [Google Scholar]
- 3.Barnard E L, Ash E C, Hopkins D L, McGovern R J. Distribution of Xylella fastidiosa in oaks in Florida and its association with growth decline in Quercus laevis. Plant Dis. 1998;82:569–572. doi: 10.1094/PDIS.1998.82.5.569. [DOI] [PubMed] [Google Scholar]
- 4.Benson G. Tandem repeats finder: a program to analyzed DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang C J, Garnier M, Zreik L, Rossetti V, Bové M L. Culture and serological detection of the xylem-limited bacterium causing citrus variegated chlorosis and its identification as a strain of Xylella fastidiosa. Curr Microbiol. 1993;27:137–142. doi: 10.1007/BF01576010. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Lamikanra O, Chang C J, Hopkins D L. Randomly amplified polymorphism DNA analysis of Xylella fastidiosa Pierce's disease and oak leaf scorch pathotypes. Appl Environ Microbiol. 1995;61:1688–1690. doi: 10.1128/aem.61.5.1688-1690.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.French W J, Kitajima E W. Occurrence of plum leaf scald in Brazil and Paraguay. Plant Dis Rep. 1978;62:1035–1038. [Google Scholar]
- 8.Hendson M, Purcell A H, Chen D, Smart C, Guilhabert M, Kirkpatrick B. Genetic diversity of Pierce's disease and other pathotypes of Xylella fastidiosa. Appl Environ Microbiol. 2001;67:895–903. doi: 10.1128/AEM.67.2.895-903.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hood D W, Deadman M E, Jennings M P, Bisercic M, Fleischmann R D, Venter J C, Moxon E R. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:1121–1125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopkins D L. Xylella fastidiosa. In: Singh U S, Singh K P, Kohmoto K, editors. Pathogenesis and host specificity in plant diseases: histopathological, biochemical, genetic and molecular bases. I. Prokaryotes. Oxford, United Kingdom: Elsevier Science; 1995. pp. 185–197. [Google Scholar]
- 11.Keim P, Price L B, Klevytska A M, Smith K L, Chupp J M, Okinaka R, Jackson P J, Hugh-Jones M E. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol. 2000;182:2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kremer K, van Soolingen D, Frothingham R, Haas W H, Hermans P W M, Martín C, Palittapongarnpim P, Plikaytis B B, Riley L W, Yakrus M A, Musser J M, Embden J D A. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lima J E O, Miranda V S, Hartung J S, Brlansky R H, Coutinho A, Roberto S R, Carlos E F. Coffee leaf scorch bacterium: axenic culture, pathogenicity, and comparison with Xylella fastidiosa of citrus. Plant Dis. 1998;82:94–97. doi: 10.1094/PDIS.1998.82.1.94. [DOI] [PubMed] [Google Scholar]
- 14.Lunel F V, Licciardello L, Stefani S, Verbrugh H A, Melchers W J G, Meis J F G M, Scherer S, van Belkum A. Lack of consistent short sequence repeat polymorphisms in genetically homologous colonizing and invasive Candida albicans strains. J Bacteriol. 1998;180:3771–3778. doi: 10.1128/jb.180.15.3771-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pooler M R, Hartung J S. Genetic relationships among strains of Xylella fastidiosa from RAPD-PCR data. Curr Microbiol. 1995;31:134–137. doi: 10.1007/BF00294290. [DOI] [PubMed] [Google Scholar]
- 17.Purcell A H, Hopkins D L. Fastidious xylem-limited bacterial plant pathogens. Annu Rev Phytopathol. 1996;34:131–151. doi: 10.1146/annurev.phyto.34.1.131. [DOI] [PubMed] [Google Scholar]
- 18.Rosato Y B, Neto J R, Miranda V S, Carlos E F, Manfio G P. Diversity of Xylella fastidiosa population isolated from Citrus sinensis affected by citrus variegated chlorosis in Brazil. Syst Appl Microbiol. 1998;21:593–598. [Google Scholar]
- 19.Simpson A J G, Reinach F C, Arruda P, et al. The genome sequence of plant pathogen Xylella fastidiosa. Nature. 2000;406:151–159. doi: 10.1038/35018003. [DOI] [PubMed] [Google Scholar]
- 20.van Belkum A, Kluytmans J, van Leeuwen W, Bax R, Quint W, Peters E, Fluit A, Vandenbroucke-Grauls C, van den Brule A, Koeleman H, Melchers W, Meis J, Elaichouni A, Vaneechoutte M, Moonens F, Maes N, Struelens M, Tenover F, Verbrugh H A. Multicenter evaluation of arbitrarily primed PCR for typing of Staphylococcus aureus strains. J Clin Microbiol. 1995;33:1537–1547. doi: 10.1128/jcm.33.6.1537-1547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Belkum A, van Leeuwen W, Scherer S, Verbrugh H. Occurrence and structure-function relationship of pentameric short sequence repeats in microbial genomes. Res Microbiol. 1999;150:617–626. doi: 10.1016/s0923-2508(99)00129-1. [DOI] [PubMed] [Google Scholar]
- 22.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viana-Niero C, Gutierrez C, Sola C, Filliol I, Boulahbal F, Vincent V, Rastogi N. Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J Clin Microbiol. 2001;39:57–65. doi: 10.1128/JCM.39.1.57-65.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells J M, Raju B C, Thonson J M, Lowe S K. Evidence of the common etiology of phony peach and plum leaf scald diseases. Phytopathology. 1981;71:1156–1161. [Google Scholar]
- 25.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1987. pp. 2.4.1–2.4.2. [Google Scholar]