Abstract
Bottom‐up synthetic biology is the science of building systems that mimic the structure and function of living cells from scratch. To do this, researchers combine tools from chemistry, materials science, and biochemistry to develop functional and structural building blocks to construct synthetic cell‐like systems. The many strategies and materials that have been developed in recent decades have enabled scientists to engineer synthetic cells and organelles that mimic the essential functions and behaviors of natural cells. Examples include synthetic cells that can synthesize their own ATP using light, maintain metabolic reactions through enzymatic networks, perform gene replication, and even grow and divide. In this Review, we discuss recent developments in the design and construction of synthetic cells and organelles using the bottom‐up approach. Our goal is to present representative synthetic cells of increasing complexity as well as strategies for solving distinct challenges in bottom‐up synthetic biology.
Keywords: artificial cells, artificial organelles, cell mimics, microreactors, synthetic biology
This Review highlights strategies used to build synthetic cells and organelles from scratch. These cell‐like materials, which mimic the structure and function of natural cells, represent the next generation of biologically inspired smart systems. The discussion begins by describing basic functions and components, such as energy supply and compartmentalization, and evolves into systems with an increasing degree of sophistication.
1. Introduction
Synthetic cells are supramolecular chemical systems designed to mimic the behavior, function, and structure of living cells. [1] Depending on how they are produced, synthetic cells can have properties and components that are not present in living systems, which significantly increases their technological value. [2] Designing and building synthetic cells from scratch gives researchers full control over the functional and structural properties of these materials. In addition, the process of making synthetic cells advances our knowledge of how biological cells work and how they might have evolved from nonbiological components.
The focus of this Review is the bottom‐up approach, which consists of building synthetic cells by combining functional and structural parts, biological or synthetic, into integrated systems with cell‐like properties. [3] The building blocks or modules must be engineered to improve their mutual compatibility, thereby paving the way for synthetic cells that are structurally and functionally adaptable. Thus, the bottom‐up approach is fundamentally different from the top‐down approach, where the goal is to create synthetic cells by adding or removing cell components from existing organisms, such as a bacterium.
The top‐down approach allows researchers to probe the effects caused by changing the original constitution of cells, and can provide answers to questions such as “what is the smallest number of components a cell needs to stay alive or to sustain certain properties essential for life? [4] Researchers in the field of bottom‐up synthetic biology explore questions such as ”How could primitive cells have arisen from nonbiological components?“ or ”Can we discover the fundamental design principles of life and apply them to create synthetic cells for technological applications? [5]
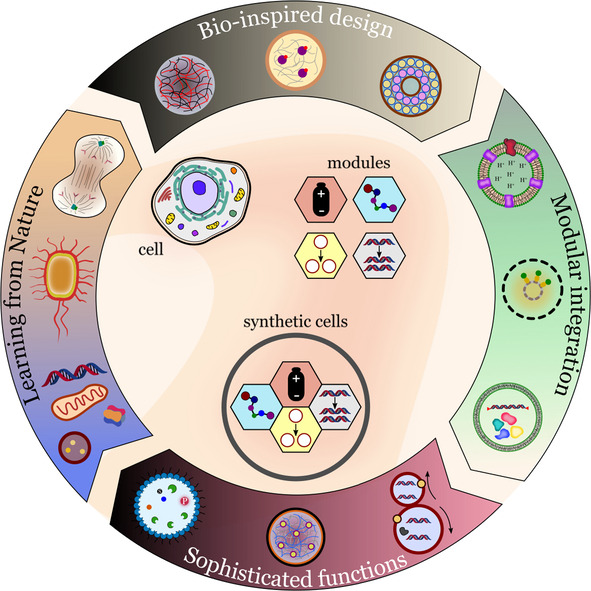
Researchers in the field of bottom‐up synthetic biology have focused their attention on solving specific engineering problems, such as compartmentalization, energy supply, metabolism, gene replication, and biosynthesis.[ 2b , 6 ] A feature of current research in the field is the focus on developing robust and versatile functional modules that can be combined to create synthetic cells with desired properties (Figure 1). [7]
Figure 1.
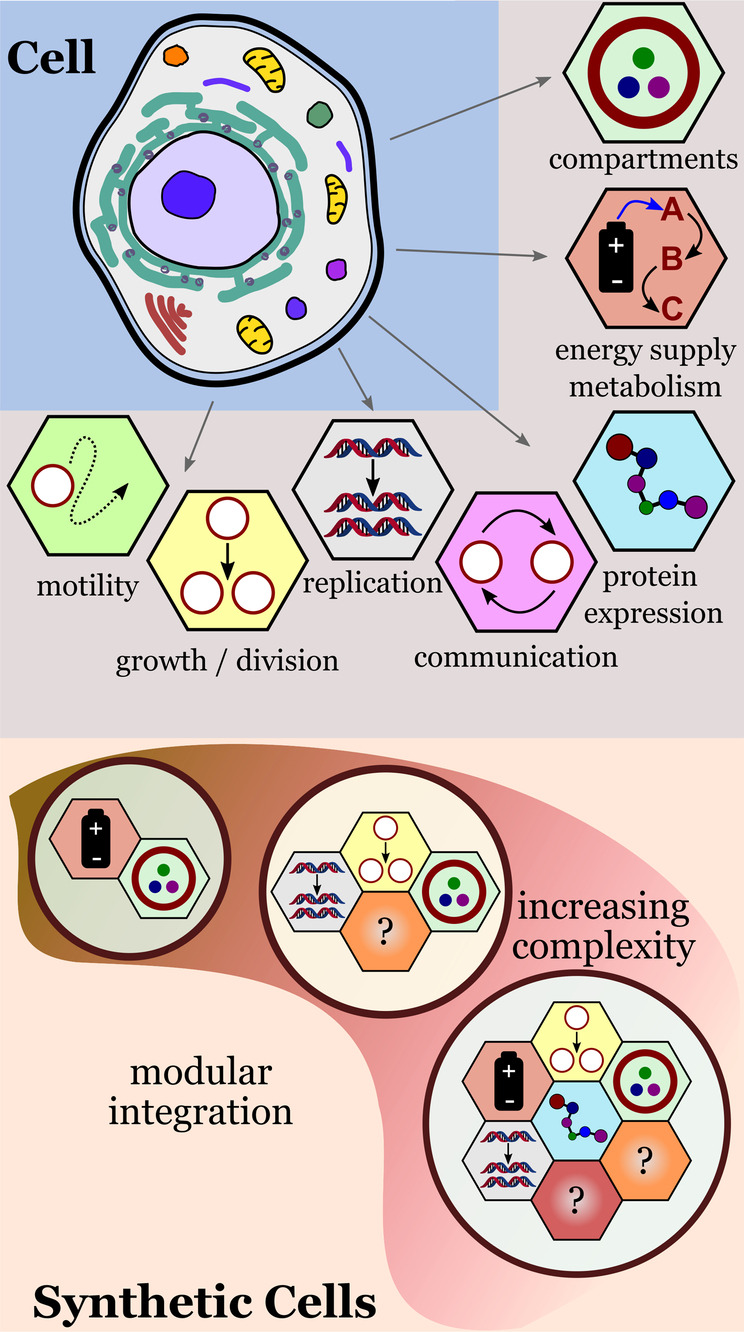
Modular approach for building synthetic cells with cell‐like properties. The integration of functional modules creates synthetic cells with increasing complexity.
For example, energy supply and biosynthesis can be considered as two independent functional modules that can be combined to create synthetic cells capable of producing proteins using their own, self‐made ATP. [8] Other properties that can be programmed in synthetic cells through modular integration include triggered division events, gene replication, biosynthesis, and communication (signaling) between different synthetic cells. [9]
In this Review, we will discuss representative examples of strategies, materials, and methods found in the ever‐growing toolbox of bottom‐up synthetic biology. First, we will discuss recent advances in the development of individual functional modules: compartmentalization, energy supply and metabolism, protein expression, communication, gene replication, growth and division, and motility. We then discuss recent studies describing more sophisticated synthetic cells that are generated by integrating different functional modules. Finally, we present and discuss new strategies and applications of synthetic cells designed to exhibit adaptive behavior and how researchers are now facing the challenge of implementing evolvability by combining the functional modules and strategies discussed in this Review. For simplicity, we use the term synthetic cells to describe protocells, cell mimics, bioreactors, and artificial cells. The distinctions between the different types of synthetic cells will become evident from the discussion.
2. Basic Biomimetic Functions
2.1. Compartmentalization
The spatial organization of functional components is a vital feature of eukaryotic cells, which is made possible by the compartmentalization provided by the cell membrane and cell organelles. Cells need compartments to store, protect, and process sensitive molecular materials (e.g. genes, enzymes, proteins) that are necessary for cell integrity, maintenance, and reproduction. Therefore, compartments not only serve as structural components of cells, but play an important role in cellular functionality and behavior.
Synthetic compartments can be divided into two main classes: membrane‐containing and membraneless compartments. Liposomes, polymersomes, proteinosomes, and colloidosomes belong to the first type. Coacervates and simple liquid droplets, hydrogels, and aqueous two‐phase systems (ATPS) are examples of membraneless compartments. For details on membraneless compartments, proteinosomes, and colloidosomes, we refer the reader to excellent publications on the subject. [10]
Liposomes and polymersomes remain the most widely employed types in bottom‐up synthetic cells because of their close structural similarity to cell membranes. As in cells, synthetic polymers and liposomes allow functional macromolecules (e.g. proteins, electron carriers) to be incorporated into their self‐assembled membranes. [11] The hydrophobic layer of these cell‐like compartments may contain functional biomolecules, such as pores, or be synthetically tuned to be endowed with the desired permeability properties. [12]
A common feature of liposomes and polymers is that they can produce vesicles with a wide range of sizes (10 nm–100 μm), depending on how they are prepared. [13] As a result of their wide size range, polymers and liposomes can be used to create synthetic nanocompartments with sizes comparable to that of cellular organelles, as well as microcompartments with sizes comparable to whole biological cells. [14] Liposomes are generally unstable, have a high sensitivity to the chemical composition of the medium, and a limited range of chemical functionality. [15] Polymers, on the other hand, are made of amphiphilic polymers with molecular weights that are significantly higher (>1000 g mol−1) than those of ordinary lipids, thereby resulting in generally stronger and more robust compartments. [16]
Polymersomes can be designed to exhibit different chemical functionalities and a wide range of molecular structures. [17] For example, poly(butadiene)‐b‐poly(ethylene oxide) block copolymers with end groups such as hydroxy, alkyne, or acrylate allow the chemical functionalization of polymersome surfaces through covalent integration. [18] This approach has been used to create polybutadiene‐poly(ethylene oxide) polymers with a spiropyran end group that allows the polymer to self‐assemble into giant vesicles when exposed to light. [19] Surface functionalization of polymer vesicles is also possible through noncovalent integration of functional molecules. Luo et al. showed that cholesterol‐tagged DNA molecules could be anchored to the surface of polymer vesicles through the hydrophobic interactions between the polymers and the cholesterol tag. [20] By using complementary DNA strands, the researchers were able to induce the sequence‐specific assembly of polymer vesicles (Figure 2).
Figure 2.
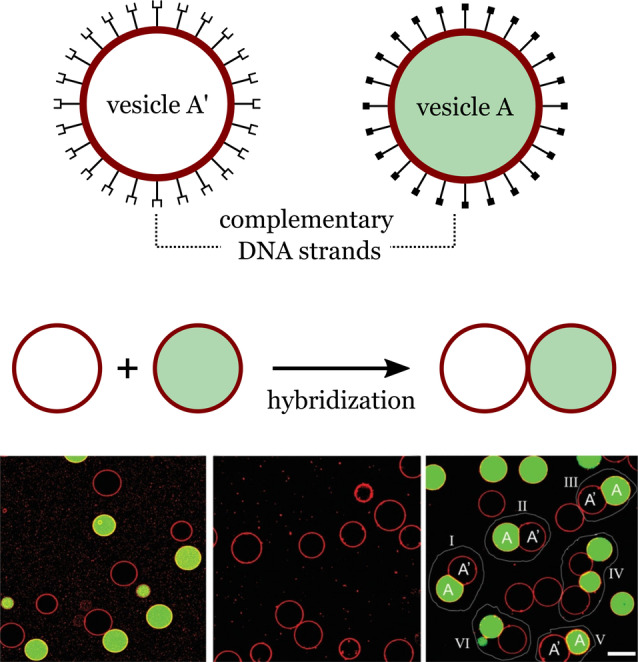
Formation of a polymer network. Hybridization of complementary DNA strands present on the surface of the polymer vesicles promotes directed adhesion. Scale bar: 100 μm. Adapted from Ref. [20]. Creative Commons CC BY license 2020.
Hybrid lipid–polymer vesicles are a very promising compartment type that combines the best properties of polymersomes and liposomes. [15] Peyret et al. have described the composition‐dependent properties of lipid–polymer hybrid vesicles with a complete membrane asymmetry. The researchers demonstrated that the transverse and translational diffusion of hybrid membranes containing polymers and lipids can be similar to that of cell membranes. [21] Rottet et al. have demonstrated the versatility of hybrid vesicles, using them to gain control over the transport of molecules across compartment walls. [22] The researchers reconstituted two distinct ATP binding cassette (ABC) membrane transporters into nanosized hybrid vesicles obtained from a blend of phospholipids and the block copolymer poly(butadiene)‐poly(ethylene oxide). The lipid fraction promoted an environment that preserved the protein function (transporter), while the block copolymer part offered high structural stability and low passive permeability.
As the complexity of synthetic cells increases, so does the demand for multi‐compartmentalized architectures that provide more sophisticated organizational and functional properties. These structures are achieved by the encapsulation of subcompartments into larger compartments. Examples include liposomes‐in‐liposome systems, [23] multiple emulsions, [24] liposomes in layer‐by‐layer (LbL) capsules, [25] polymersomes‐in‐polymersome,[ 20 , 26 ] and capsosomes. [27] In 2017, Peyret et al. described a liposome‐in‐polymersome system with a temperature‐release mechanism based on the thermosensitivity of the internal liposomes. [28] Cargoes inside the liposomes could be released in a sequence‐controlled manner at specific temperatures. Recently, Mason et al. developed robust multi‐compartmentalized synthetic cells through the encapsulation of small semipermeable enzyme‐loaded polymersomes (160 nm diameter) into cell‐sized coacervate microdroplets (ca. 20 μm diameter; Figure 3 a). [29] The small enzyme‐loaded vesicles behaved like synthetic organelles inside the main coacervate compartment.
Figure 3.
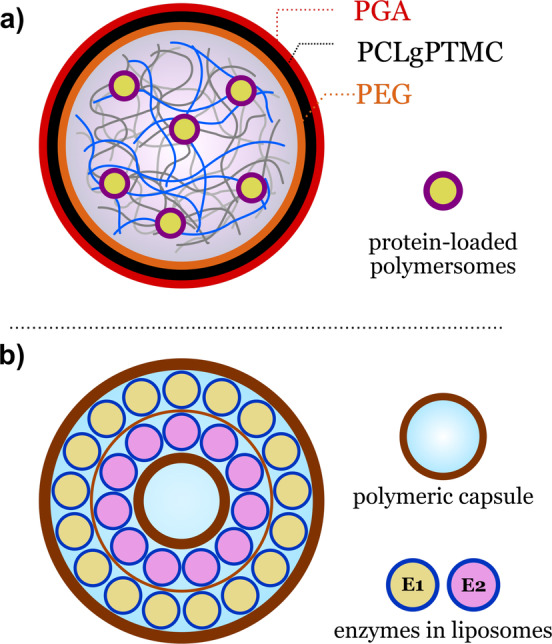
Multicompartmentalized hybrid systems. a) Synthetic cells obtained from nanopolymersomes in membranized coacervates. b) Synthetic organelles obtained from nanoliposomes arranged in layers within a polymeric capsule. Redrawn from Ref. [29] and Ref. [27b].
The synthetic organelles were spontaneously recruited by amylose‐based coacervate microdroplets, carrying the enzymatic cargo inside the synthetic cell. The multi‐compartmentalized coacervate droplets were formed into a membrane using a tailor‐made terpolymer (PEG‐PCLgPTMC‐PGA) that interacts electrostatically with the amylose‐based coacervate droplets. The spatial organization of the synthetic organelles within the synthetic cells, together with the semipermeable characteristics of the engineered polymer membrane, allowed internal enzymatic cascade reactions to occur efficiently on addition of substrates into the surrounding aqueous medium.
Another example of a multi‐compartmentalized system that combines polymers and lipids was given by Godoy‐Gallardo et al. [27b] The researchers designed synthetic organelle capsosomes: polymeric capsules that contain liposomes arranged internally in layers (Figure 3 b). The different layers inside the capsosomes were composed of gold nanoclusters (AuNCs) and liposomes used as enzymatic nanoreactors carrying trypsin and horseradish peroxidase. Macrophages could internalize the synthetic organelles, allowing two distinct enzymatic reactions to take place simultaneously inside the phagocytes. The protection provided by the compartments allowed the enzymatic reactions to occur even after being taken up by macrophages. Free unprotected liposomes, on the other hand, showed no enzymatic activity under similar conditions, thus highlighting the importance of compartment design in the stability and applicability of synthetic organelles.
Permeability and structural adaptability remain a challenge for most polymeric and phospholipid membrane based systems. This is not the case for coacervate microdroplets. Coacervates consist of liquid–liquid phase‐separated droplets that can concentrate biomolecules inside, thereby acting as a permeable and highly dynamic compartment. Donau et al. designed RNA coacervate droplets that were formed when a fuel was available and decomposed if the fuel was depleted. Functional RNA molecules could be transiently sequestered by the droplets during this process, which is reminiscent of the action of biological condensates in cells. [30] The lack of a membrane can be a drawback for coacervates, as they have a high tendency to fuse with other droplets or wet surfaces. In a recent study, Zhang et al. have demonstrated that robust long‐lived coacervate droplets can be obtained by coating the droplets with a phospholipid membrane. This produces stable coacervate vesicles with desirable properties such as a molecularly crowded interior, enhanced molecule sequestration power, and selective molecular transport. [11c]
2.2. Energy Supply and Metabolism
Energy can be supplied externally or internally to synthetic cells. The energy can then be used to power enzymatic cascade reactions and other biochemical processes needed to maintain the function and integrity of the synthetic cells. For example, synthetic cells can be powered by feeding energy‐rich molecules such as carbohydrates or enzyme‐specific substrates. More energy‐intensive tasks, such as protein expression, can be powered by ATP produced by synthetic cells equipped with an ATP‐synthesis module. Redox chemistry, such as in enzymatic reactions controlled by oxidoreductases, can also be regulated by synthetic cells if they are equipped with the corresponding functional modules designed to recycle NADH/NAD coenzymes.
In the early 2000s, Noireaux and Libchaber reported a synthetic cell consisting of a cell‐free protein expression system from Escherichia coli encapsulated in a phospholipid vesicle. [31] This simple bioreactor allowed protein expression to proceed for 100 h if nutrients were continuously added to system. The process eventually came to a halt because of the accumulation of toxic by‐products, a problem that typically limits the long‐term performance of synthetic cells that depend on continuous feeding. If not remediated, by‐products can poison catalysts, create unfavorable chemical gradients, and cause harmful osmotic pressure changes. Recent progress in the rational design of oscillatory reaction networks has indicated that the chemical accumulation problem could be partially addressed in systems equipped with self‐regulatory mechanisms, where the activation and deactivation of a process would depend on the concentration of transient species in the system. [32]
An alternative to continuous feeding of external high energy chemicals is the internal production of ATP by the synthetic cells. ATP‐producing synthetic cells were previously reported by Pitard et al. [33] in 1996 and by Steinberg‐Yfrach et al. [34] in 1998. These two research groups conducted important studies on light‐induced ATP synthesis inside proteoliposomes and liposomes based on the reconstitution of the transmembrane enzyme F0F1‐type ATP synthase. Kleineberg et al. demonstrated the successful integration of ATP‐synthase and bacteriorhodopsin in polymer and polymer–lipid hybrid vesicles. [35] The composition and structure of the membranes directly affected the performance of the ATP synthesis process. For example, protein stability in hybrid vesicles was significantly enhanced compared to pure liposomes. [35]
In 2018, Lee et al. took an important step toward the development of energy‐self‐sufficient synthetic cells. [36] Photosynthetic liposomes were constructed with two types of membrane‐embedded photoconverters (Figure 4): a plant‐derived photosystem II (PSII) and a bacteria‐derived proteorhodopsin (PR). Encapsulation of the photosynthetic liposomes into larger liposomes resulted in light‐responsive synthetic cells able to produce their own ATP using light as an energy source. The photosynthetic liposomes behaved as synthetic organelles for the corresponding synthetic cells, a level of structural organization and functionality that shows similarities to chloroplasts in cells.
Figure 4.
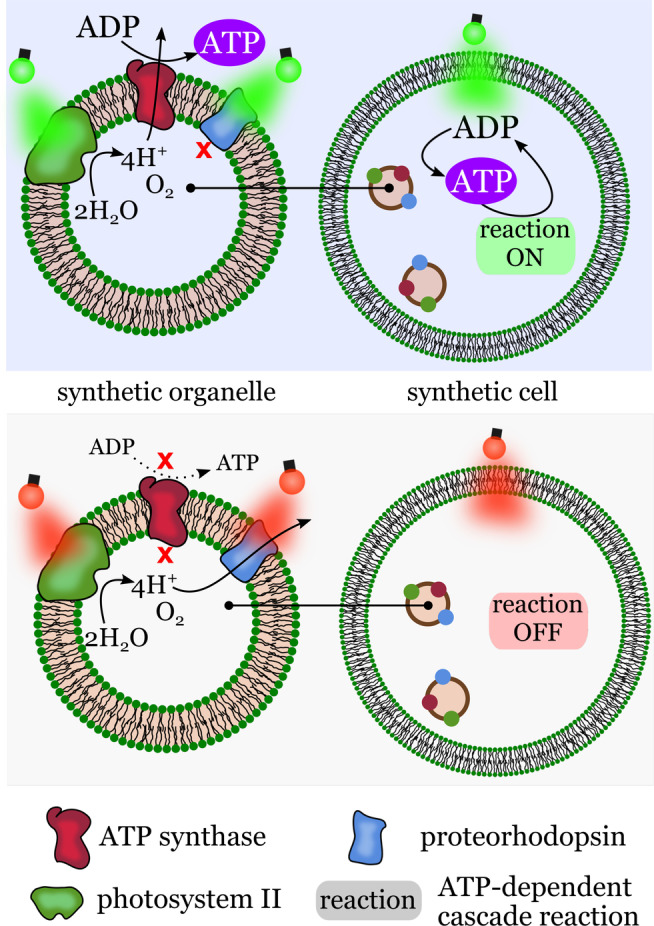
Energy self‐sufficient synthetic cells. Top left: Synthetic organelles featuring transmembrane enzymes for light‐controlled proton generation and ATP synthesis. Top right: Synthetic cells with encapsulated synthetic organelles capable of ATP generation when exposed to green light. The internal ATP drives reactions inside the synthetic cells. Bottom: synthetic organelles and cells do not operate if illuminated with red light. Redrawn from Ref. [36].
Exposing the synthetic cells to red light activated the electron transport chain, thereby establishing a transmembrane proton gradient between the interior of the synthetic organelle and the interior of the giant vesicle. Similar to the situation in cells, the proton gradient was used to generate ATP from ADP and Pi by ATP‐synthase. Green light, on the other hand, suspended proton generation by PSII and activated proton pumping by PR, which stopped ATP synthesis through elimination of the proton gradient. This engineered photoactivated system allowed the synthetic cells to perform two ATP‐dependent enzymatic cascade reactions: carbon fixation and actin polymerization.
NAD is an essential electron carrier involved in redox reactions carried out by oxidoreductases. In synthetic cells, the redox transformation of NAD to NADH, or vice versa (regeneration), offers a way to dynamically select the direction of redox reactions. NAD regeneration strategies include photocalysis, chemical and electrochemical regeneration, and enzymatic regeneration. [37]
In 2019, Ma et al. demonstrated the light‐driven regeneration of NAD using metal‐free nanoparticles made of a conjugated polymer (Figure 5 a). [38]
Figure 5.
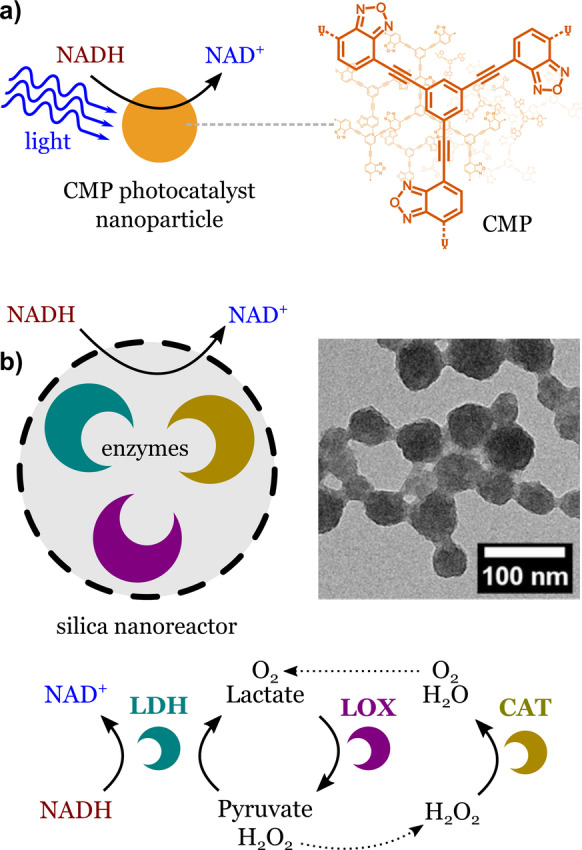
Regeneration of NAD with nanoparticles. a) Photocatalysis: a conjugated microporous polymer nanoparticle exposed to visible light oxidizes NADH to NAD. Adapted from Ref. [38]. b) Enzymatic cascade reaction: enzymes encapsulated in porous silica nanoparticles recycle NAD in a continuous manner. The nanoreactor contains three enzymes: lactate dehydrogenase (LDH), lactate oxidase (LOX), and catalase (CAT). Adapted from Ref. [39]. Creative Commons CC BY license 2021.
The nanoparticle photocatalyst behaves as a synthetic organelle that regenerates NAD if properly illuminated. Photocatalysis usually creates by‐products that can be harmful to biological molecules. A common problem is the deactivation of enzymes. An alternative is to develop NAD regeneration systems that are fully enzymatic. Jo et al. built enzyme‐loaded nanoreactors that can regenerate NAD in a continuous manner through enzymatic reactions only (Figure 5 b). [39] The nanoreactors were made of porous silica nanoparticles carrying lactate dehydrogenase, lactate oxidase, and catalase. The system features a pyruvate‐recycling cycle and a by‐product‐elimination cycle that ensured the continuous regeneration of NAD. The silica matrix protected the enzymes from proteolysis and heat, and allowed the enzymes to maintain a high activity after encapsulation. [40] Other robust materials, such as metal organic frameworks (MOFs), have also been used to create synthetic cells based on the encapsulation of enzymes. [41]
An interesting approach to create synthetic cells with ATP and NAD regeneration systems is to use biological parts to do the job. For example, Miller et al. used thykaloid membranes (TMs) from chloroplasts to energize enzymatic reactions in metabolically active microcompartments. [42] The system used light to regenerate ATP and NADPH through the biochemical activity of the TMs. These molecules were used by enzymes to power metabolic processes such as the fixation of CO2.
It is evident that most strategies to manipulate ATP and NAD(H) in synthetic cells still require sensitive biological components as main functional parts. The generation of ATP, for example, relies on a very sensitive, complex transmembrane enzyme that is hard to manipulate. Research in this area will certainly evolve towards nonbiological ways to recreate energy‐supply systems that can be integrated with biological parts, but that are not entirely biological.
2.3. Protein Expression
Encapsulation of transcription/translation systems inside a synthetic vesicle is the first step towards building systems that can carry out protein expression.[ 31 , 43 ] Elani et al. designed synthetic cells capable of synthesizing two fluorescent proteins (GFP and RFP) located in distinct internal compartments of the same synthetic cell (Figure 6). [44]
Figure 6.
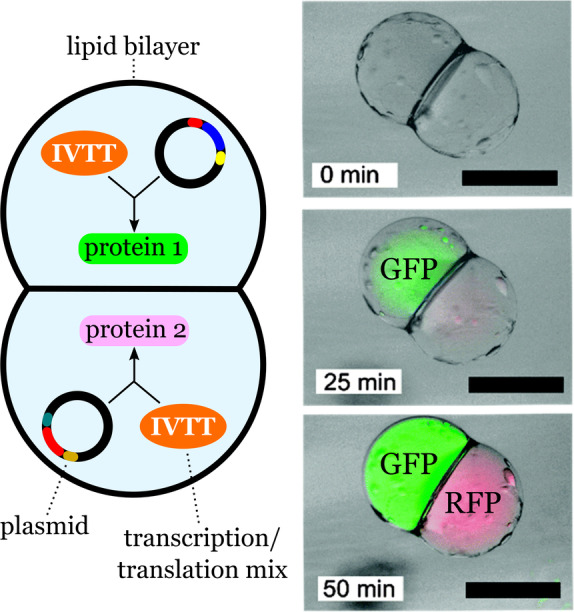
Compartmentalized protein synthesis in a synthetic cell with two fused lipid compartments. Each compartment carries the components necessary to synthesize two distinct proteins (GFP and RFP). The bright‐field fluorescent composite images show the synthesis of the compartmentalized cell‐free protein. Scale bar: 200 μm. Adapted from Ref. [44] with permission from the PCCP Owner Societies.
The two‐compartment vesicles carrying the machinery and components for in vitro transcription and translation (IVTT) were obtained by a phase‐transfer method. Each droplet, one carrying the genetic information for protein 1 and the other for protein 2, was created independently before the formation of the two‐vesicle system. Therefore, each sub‐compartment contained the biochemical components necessary for protein expression in the presence of the corresponding plasmids. The biochemical synthesis of green fluorescent protein (GFP) and red fluorescent protein (RFP) was spatially separated by a lipid bilayer. These results represent a new level of cell‐like organization that allows the simultaneous expression of distinct proteins in different compartments. Since each sub‐compartment could, in principle, be designed independently, the number and types of processes in the final multicompartment system could be adjusted.
In addition to spatial organization, natural cells can control gene expression in response to chemical or physical signals, which is an essential feature that allows living cells to adapt to new environments. Therefore, to create adaptable synthetic cells, different processes should not only be compartmentalized, but the possibility should exist to switch different process on and off in response to external cues. One example of a synthetic cell with switchable gene expression was reported by Dwidar et al. [43b] The researchers were able to modulate gene expression inside synthetic cells using riboswitches. To create synthetic cells with a switchable gene expression capability, the researchers used an RNA aptamer for the switch. The aptamers were engineered to function as a histamine‐specific riboswitch for the reconstituted transcription/translation system. The riboswitch was fused to a reporter gene and other genes and encapsulated in phospholipid vesicles, thereby resulting in synthetic cells with an intrinsic histamine‐activated control mechanism for protein synthesis. Although many features of such responsive system still have to be improved, such as the long‐term stability, immunogenicity of the expressed protein, and the permeability of the lipid bilayers to the signaling molecules, the strategy demonstrated by Dwidar et al. represents a significant advance in the engineering of synthetic cells with DNA‐programmable protein‐expression capabilities.
2.4. Communication
All living organisms, unicellular or multicellular, have to respond to changes in the environment in order to remain alive. These responses are coordinated by the cells through their ability to sense and react to internal and external physical and chemical changes. In synthetic cells, communication occurs through the exchange of chemical signals between different cells (synthetic–synthetic, or synthetic–natural) or between the cells and their environment.
An early report of a synthetic cell capable of communicating with natural cells comes from 2009 by Gardner et al. [45] The researchers developed a lipid‐based synthetic cell capable of initiating a quorum‐sensing response in a unicellular organism, the marine bacterium Vibrio harveyi (Figure 7). The signal generated by the synthetic cell consisted of low‐molecular‐weight carbohydrates. These were synthesized inside the synthetic cells by the autocatalytic formose reaction involving encapsulated precursors. The resulting sugars produced by this primitive carbohydrate metabolism were transported to the surrounding aqueous medium and could interact with transduction proteins present on the surface of the bacteria. This initiated a cascade of transformations inside the bacteria mediated by its autoinducer‐2 quorum‐sensing system, thereby resulting in the production of a bioluminescent protein.
Figure 7.
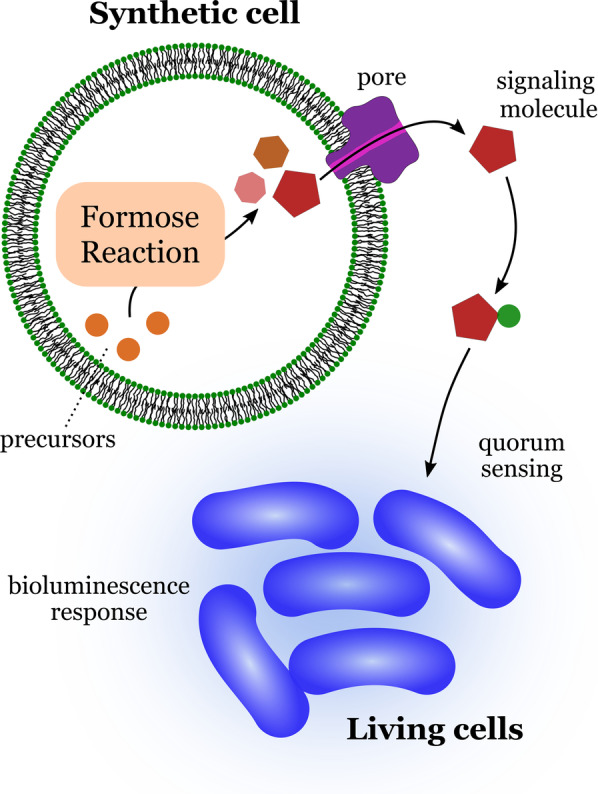
Communication between a synthetic cell and bacteria (Vibrio harveyi). Products from the encapsulated formose reaction escape from the interior of the synthetic cells through a transmembrane protein pore. The bacteria sense the low molecular weight signaling molecules, which triggers a bioluminescence response. Redrawn from Ref. [45].
The results obtained by Gardner et al. demonstrated that communication between a synthetic cell and a living cell is possible if they share a common “chemical language” and if they can coexist in the same environment. This approach was expanded by Lentini et al. in 2017. [46] The researchers created lipid‐based synthetic cells containing a biochemical machinery that allowed synthetic cells to sense and send chemical signals to different bacteria (V. fischeri, V. harveyi, E. coli, and P. aeruginosa). In a recent report, researchers from the Mansy Group demonstrated that artificial cells could induce neural differentiation. [47] The artificial cells consisted of liposomes containing the machinery for the transcription and translation of DNA templates coding for brain‐derived neurotropic factor (BDNF) and other components, which were necessary for regulating BDNF release from the artificial cells. The artificial cells were stable and functional under physiological conditions and successfully communicated with neurons.
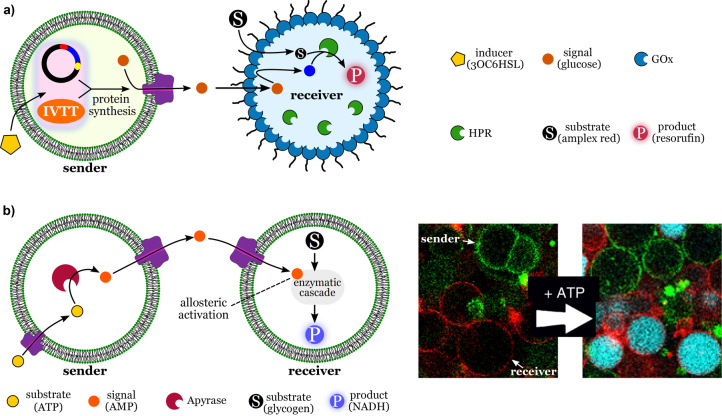
Considerable progress has been made towards developing new strategies to achieve communication between communities of synthetic cell mimics.[ 9a , 9b , 48 ] Tang et al. engineered lipid‐based synthetic cells (transmitters) that could start an enzymatic cascade reaction in a distinct population of synthetic cells (receivers) consisting of proteinosomes carrying glucose oxidase (GOx) and horseradish peroxidase (HRP; Figure 8 a). [48a]
Figure 8.
Communication between synthetic cells. a) Signaling mediated by cell‐free expression of a pore protein. The sender cell contains the components necessary for the synthesis of the membrane porin α‐hemolysin. The porin inserts itself into the membrane of the sender. The sender releases a signal molecule (glucose) through the pore, which reaches the receiver cells. There, the signal initiates an internal cascade reaction that produces the fluorescent product (resorufin). Redrawn from Ref. [48a]. b) Signaling mediated by allosteric triggers. The sender cell produces an internal signal molecule (AMP) that diffuses to the receiver cell. Inside the receiver, AMP triggers a response cascade reaction through the allosteric activation of the first enzyme in the cascade. Adapted from Ref. [9b]. Scale bar: 30 μm. Creative Commons CC BY license 2020.
The communication between transmitters and receivers was mediated by a unidirectional signaling pathway involving two chemical signals: N‐(3‐oxohexanoyl)‐l‐homoserine lactone (3OC6HSL) and glucose. The first signal, 3OC6HSL, induced the cell‐free expression of a transmembrane‐pore‐forming protein (α‐hemolysin), which allowed glucose (second signal) to be released by the transmitters and processed by the receivers. The glucose signal was oxidized by the interfacial GOx of the receivers, thereby producing hydrogen peroxide, which was then consumed by the internal HRP. This cascade reaction was detected using Amplex red, which gives a fluorescent signal upon oxidation in the presence of HRP and peroxide. The results illustrate a robust design principle for unidirectional communication, where receivers only respond if sender cells are activated. In communities of synthetic cells, the ability to control the behavior of different cell populations is of great interest, as it will offer researchers the option to design multifunctional high‐order systems. A drawback of communication based on molecular diffusion is that chemical signals released by sender cells will become diluted in the surrounding medium. The more diluted the signal becomes, the weaker is the response produced by the receivers. A solution to this problem has recently been offered by Buddingh′ et al. In their approach, a signal molecule is used to trigger a cascade reaction inside a receiver cell based on the allosteric activation of enzymes (Figure 8 b). [9b] The signal, adenosine monophosphate (AMP), is generated by a fuel (ATP) through the action of apyrase encapsulated in the sender cells. AMP is released through a porin (α‐hemolysin) and diffuses into the receiver cell through another porin. The signal is not consumed by the response reaction, but is only needed in small concentrations to act as an allosteric trigger that “switches on” the first enzyme of a NADH‐producing cascade reaction involving three enzymes present inside the receivers. Therefore, the receiver cells amplify the signal effect through the cascade reaction. This strategy is quite powerful, and one could explore the use of specific allosteric activation of enzymes to selectively control the activity of distinct populations in communities of synthetic cells. However, dilution is still a disadvantage, although to a lesser degree, compared to simple chemical signaling where the signal is a reactant. An alternative to chemical communication is to use light as a signal. Researchers in the Wegner group created synthetic sender cells that emit a bioluminescence signal that triggers adhesion between the sender and receiver cells. [49] The adhesion was controlled by photoswitchable proteins on the surface of the synthetic cells. Adhesion was followed by the unblocking of membrane porins and the contact‐dependent transfer of Ca2+ ions from the senders to the receivers. This activated the Ca2+‐responsive apo‐PLA2 inside the receivers, triggering the cleavage of the phospholipid membrane of the receivers.
Research on the collective behavior of synthetic cells has also seen considerable success. The Mann group has developed systems that exhibit complex behavior, such as artificial phagocytosis, self‐induced reconfiguration of internal compartments, and even a rudimentary predatory activity between synthetic cells. [50] For example, the researchers were able to induce high‐order behavior in synthetic cells through the exchange of “chemical packages” between microcontainers. [50a] These systems offer excellent examples of how clever designs and strategies can result in biomimetic materials featuring complex dynamic behavior.
2.5. Replication of Genetic Material
Synthetic cells capable of replication represent a crucial step, and a current challenge, towards the development of advanced biomimetic systems that can process genetic information. Replication introduces a new level of complexity to synthetic cells, bringing researchers closer to the ambitious goal of understanding how evolvable and self‐maintaining chemical systems can create life. [51]
In 2008, Kita et al. described a simple lipid‐based synthetic cell that could replicate its encapsulated genetic material through a self‐encoding system. [52] The self‐encoding system consisted of an in vitro translation system and a template RNA encoding the catalytic subunit of Qβ replicase, the protein responsible for RNA replication. In comparison to studies published previously, where emulsion droplets were used for the compartmentalization of the genetic material, the system proposed by Kita et al. was more analogous to natural cells since liposomes were used as compartments. [53] The results also indicated a more fundamental outcome: that the encapsulation of a functional self‐encoding system into liposomes opens up the possibility of evolution occurring. The self‐replicating synthetic cell provided the two basic elements needed for evolution: 1) genetic variability (mutations), which was promoted by the high error rate of the self‐encoded Qβ replicase; and 2) a linkage between the genotype and phenotype, as a result of the space limitation inside the liposomes.
More recently, in 2018, van Nies et al. demonstrated the self‐replication of DNA in liposomes based on self‐encoded viral proteins (Figure 9). [54] The genetic information was contained in a synthetic linear DNA which encoded the viral proteins required for DNA replication. The proteins were part of the protein‐primed replication machinery of the Φ29 virus, which can amplify a linear synthetic genome. A PURE transcription‐translation system was used to translate the replication proteins encoded in the synthetic DNA. Once combined inside lipid vesicles, the activity of both systems (replication and protein expression) resulted in the autocalytic replication of the DNA present in the synthetic cells. Figure 9 b shows confocal micrographs of the synthetic cells during the DNA amplification in the presence of deoxyribonucleoside triphosphates (dNTPs). The membrane of the synthetic cells remained intact, as shown by the images obtained with a membrane‐dye (in red). The signals from acridine orange, a DNA dye (green), revealed the areas containing amplified DNA inside the liposomes. Undesired cross‐reactions usually become a problem in such highly complex systems. For example, the replication machinery (Φ29) can be inhibited by components of the PURE system, which then limits the performance of the synthetic cells. Therefore, fine‐tuning becomes essential to guarantee the sustained synthesis of DNA, mRNA, and protein in synthetic cells. Other factors such as temperature and concentrations of metabolites and enzymes must also be taken into account and must be optimized.
Figure 9.
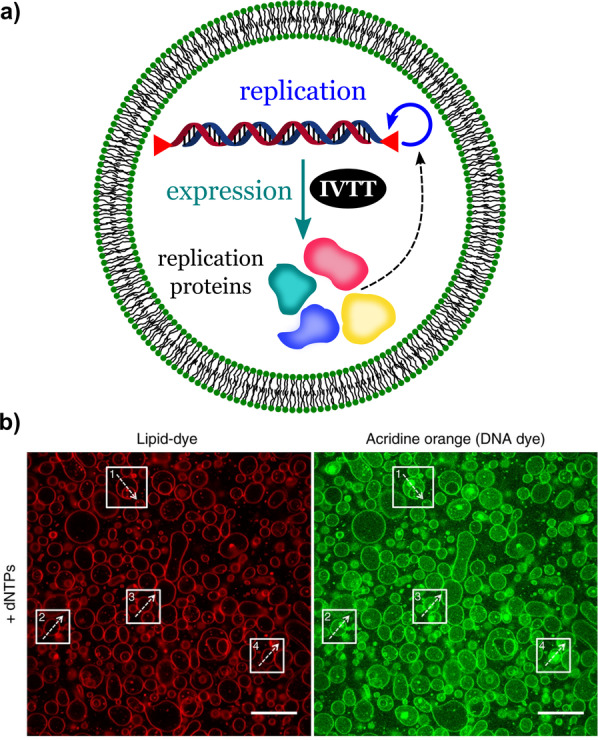
DNA replication, transcription, and translation in a synthetic cell. DNA‐replication module plus gene‐expression module. a) Scheme showing the internal composition and synthetic processes (replication and expression) of the synthetic cell. b) Fluorescent confocal micrographs of liposomes labeled with a membrane dye (red) or acridine orange (DNA dye). Scale bar: 20 μm. Adapted from Ref. [54]. Creative Commons CC BY license 2018.
Synthetic cells carrying cell‐free replication systems (DNA or RNA) provide a powerful tool for the study and development of compartmentalized systems that can evolve in a similar way as living organisms. AS the ability to adapt to changes in the environment is essential for life, studies of synthetic adaptive systems are fundamental to our understanding of how life arose from nonliving chemical systems. [55]
2.6. Growth and Division
Every cell originates from a pre‐existing one. The ability to grow and divide is an important property that allows cells to dynamically adapt to their environment and search for nutrients. In synthetic cells, growth and division are processes that require designs and materials that can elicit considerable molecular reorganization of the membranes and interfaces, coordinated by internal or external signals. Several research groups have dedicated their efforts to creating synthetic cells capable of growing and dividing. [56]
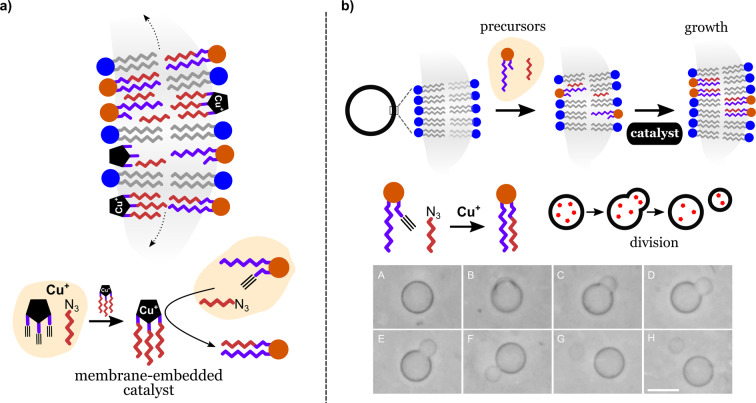
One effective strategy is to design synthetic cells that can produce the additional membrane molecules needed for growth and division. Hardy et al. have developed a synthetic lipid membrane that contains an embedded self‐reproducing oligotriazole‐Cu+ catalyst. [56d] The Cu complex catalyses the synthesis of new phospholipids that become incorporated in the vesicle membrane. Additionally, the Cu complex can catalyze its own assembly, producing new Cu complexes (self‐reproduction) that become incorporated into the expanding vesicle membrane (Figure 10 a). This dual role of the Cu complex confers on the lipid membranes the ability to continuously self‐synthesize from simpler precursors without being limited by the dilution of the catalyst as a result of membrane expansion. Lipid precursors typically achieved 95 % conversion in less than 35 h and the process could be repeated several times. A similar mechanism has been used by Castro et al. to induce division and payload partitioning in giant phospholipid vesicles (Figure 10 b). [57] In this case, division was induced by a copper(I)‐catalyzed elongation of acyl chains from membrane precursors and free catalyst present in solution. The elongation produced amphiphilic membrane molecules that increased the membrane area of the original vesicles, thereby resulting in the budding and splitting of daughter vesicles. The examples discussed represent systems in which the availability of “nutrients” and a suitable membrane catalyst are the only triggers for growth and division events. The simplicity of such systems suggests that simple reactions involving catalytic components and amphiphilic precursors may have preceded more complex membrane biochemistry in the early development of life on Earth.
Figure 10.
Synthetic cell growth promoted by catalytic self‐synthesis and self‐assembly of phospholipids. a) Growth based on a self‐reproducing oligotriazole‐Cu+ catalyst. Redrawn from Ref. [56d]. b) Growth based on Cu+‐mediated catalysis. Scale bar: 10 μm. Adapted from Ref. [57]. Creative Commons CC BY license 2019.
Division does not always require the synthesis and integration of additional membrane molecules. In 2020, Miele et al. described self‐dividing giant unilamellar vesicles (GUVs) that do the opposite. [58] Division was induced by the pH‐driven depletion of oleic acid molecules from the GUV membrane. This caused the inner membrane area of the vesicles to decrease asymmetrically. The process was driven by the pH increase produced by an internal urea–urease reaction inside the GUVs. The resulting membrane and osmotic stresses caused by the depletion of membrane molecules forced the GUVs to divide, thereby producing two smaller daughter vesicles.
One of the theories about the origin of life states that small liquid droplets, such as coacervates, could have provided the first type of compartmentalization in primitive cells before the transition to membrane‐bound vesicles. [59] These droplets, like vesicles, could foster the molecular segregation needed to sustain evolution in living systems. [60] Zwicker et al. have studied the physics involved in the growth and division of active droplets using numerical calculation. [56a] The study focused on droplets maintained away from thermodynamic equilibrium by adding droplet material created by chemical reactions inside the droplets. The results showed that droplets can grow and divide in response to the internal accumulation of droplet material. Growth happens first, with droplets eventually reaching a stationary size. This is followed by shape instabilities that lead to droplet splitting, thereby resulting in new droplets that are able to repeat the cycle.
The simple droplet growth model proposed by Zwicker et al. [56a] supports the idea that prebiotic protocells with the ability to grow and divide could have been simple active chemical droplets before they acquired biological membranes.
In 2020, Steinkühler et al. demonstrated that systematic morphological changes and vesicle division can be induced by the binding of proteins on the surface of giant lipid vesicles (GUVs). [9c] The effect was caused by the asymmetric adsorption of a bulky His‐tagged fluorescent protein (GFP) on the external leaflet of the vesicle membrane (Figure 11). The asymmetric coverage induced a preferred curvature that forced the vesicles to adopt different shapes. Vesicle division was induced by adjusting the protein coverage in the dilute regime, where the protein–protein interactions were negligible. According to fluorescence measurements, the protein coverage on the vesicle membranes increased linearly with the protein concentration. Furthermore, the relationship between the GFP coverage and the spontaneous curvature obtained by both theoretical calculations and experimental data also revealed a linear trend. Control of asymmetric spontaneous curvature by the adsorption of low‐density protein offers a simple way to manipulate shape and division of GUV membranes with minimum changes in the chemical composition of the system. Since the effect requires a low protein coverage (<1000 μm−2), other components such as channels and ion pumps could be accommodated in the GUVs without limitations caused by protein crowding.
Figure 11.
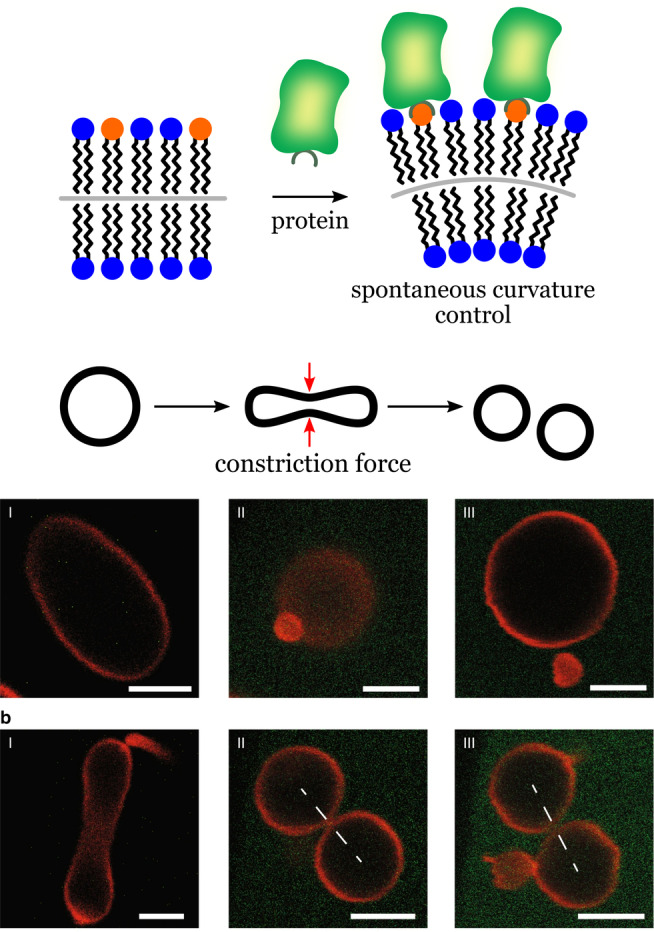
Control of vesicle shape by protein adsorption. The scheme shows His‐tagged GFP molecules binding to lipid anchors (orange) in the membrane. The degree of coverage by GPF controls the membrane curvature and morphology. Scale bars: 5 μm. Adapted from Ref. [9c]. Creative Commons CC BY license 2020.
In general, a current challenge concerning synthetic cells is the development of systems capable of self‐regulated growth/division. Can synthetic cells create building blocks when and where they are needed? Or must they always be provided externally? Another characteristic of current systems is the composition simplicity of the vesicle membranes. Growth and division are typically demonstrated using relatively simple membranes. Therefore, it is not yet clear how nanopores and other membrane‐localized components will influence growth and division in synthetic cells. To answer these questions, it is necessary to learn more about the underlying forces and complex dynamics of biomimetic membranes of higher complexity.
2.7. Motility
Cell motility gives rise to important biological processes such as embryonic development, wound healing, and immune response. Motility is also critical for the cell's ability to adapt and interact with its habitat. Motility in synthetic cells can give rise to complex behavior, such as programmable spatial migration and collective interaction involving groups of synthetic cells. One strategy to provide synthetic cells with motility is to encapsulate enzymes that will produce a propellant in the presence of a fuel. Jang et al. described protocells consisting of catalase encapsulated in micrometer‐sized polymersomes. [61] The impulse force was generated by the enzymatic disproportionation of hydrogen peroxide into water and O2. Motility could be sustained for up to 10 hours. The motion, however, was random because of the high symmetry of the system. As noted by the authors, a break in symmetry could lead to a more directional motion. Motility has also been induced in liposomes using other enzymes, such as ATPase, acid phosphatase, and urease, that were attached to the membrane of the vesicles. [62] A main difficulty for systems that depend on hydrogen peroxide is the high reactivity of the fuel, which can become a problem in the presence of biological molecules and other sensitive components. However, the generation of oxygen by catalase remains a convenient way to control the motion of synthetic cells. A different use for O2 was developed in the Mann group. The researchers used the oxygen bubbles from the disproportionation of H2O2 to regulate the buoyancy of microcapsules, which behaved like tiny submarines. [63] The system consisted of organoclay/DNA microcapsules with co‐encapsulated catalase and glucose oxidase. Buoyancy was reversibly controlled through the formation or consumption of the O2 bubbles by the coordinated action of catalase (O2 production) and glucose oxidase (O2 consumption). By controlling the vertical position of the microcapsules, the authors demonstrated sophisticated behavior, such as position‐dependent chemical reactions and vertical transportation of macroscopic objects.
Apart from enzymes, motion can also be induced by controlling the adhesion state of synthetic cells to a substrate. To do that, researchers in the Wegner group used photoswitchable proteins that bind to each other under irradiation with blue light, but dissociate in the dark. [64] GUVs decorated with one of the proteins (Micro) were able to move on a substrate coated with the other protein of the pair (iLID). Asymmetric illumination provided the driving force to induce motion as a result of the stronger adhesion of the illuminated areas relative to the weak adhesion in darker regions. As a result, the GUVs could be guided by light using an elegant strategy that is much simpler than the highly complex mechanisms found in cells.
3. Integration of Biomimetic Functions
In the previous section, we described strategies and materials that introduce basic individual biological functions into synthetic cells. Each of these basic functions and strategies can be considered as a module, with each module being responsible for a task. If modules are compatible with one another, they can be combined to create synthetic cells of higher complexity. We will now discuss examples of multimodular and more sophisticated synthetic cell systems designed to solve important engineering problems in bottom‐up synthetic biology.
3.1. Energy Supply and Protein Expression
Biosynthesis in cells is powered by high‐energy molecules such as ATP. This can develop in synthetic cells with functional modules for ATP synthesis and the corresponding ATP‐dependent synthetic machinery. In 2018, Lee et al. described a photoactivated synthetic cell capable of inducing and controlling two ATP‐dependent cascade reactions: carbon fixation and actin polymerization. [36] Inspired by these results, Berhanu et al. were able to create a synthetic cell that coupled a light‐driven synthetic organelle capable of ATP synthesis with a cell‐free protein synthesis system (PURE system; Figure 12). [65]
Figure 12.
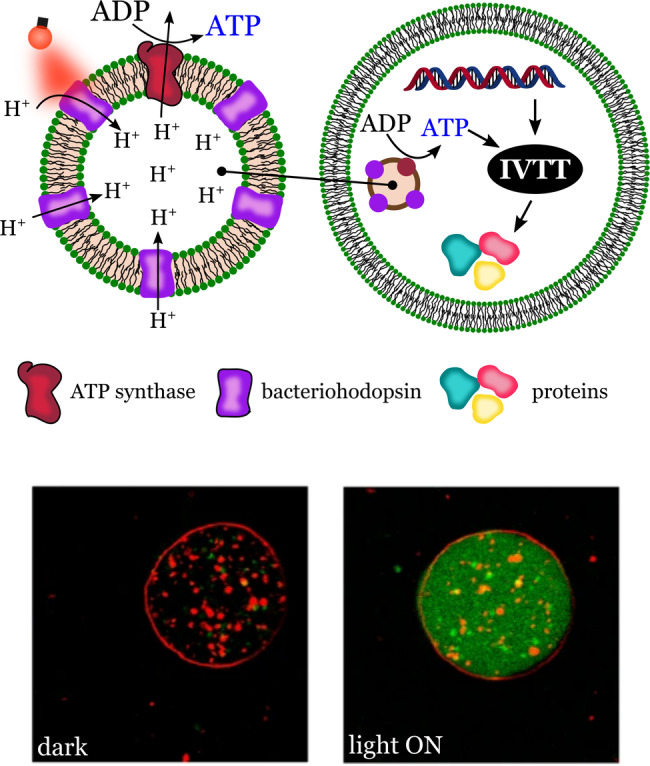
Synthetic cell showing protein synthesis powered by self‐produced ATP. Vesicle diameter ca. 10 μm. Internal synthetic organelles synthesize ATP on the surface of lipid compartments carrying light‐driven proton pumps (bacteriorhodopsin) and ATP synthase embedded in the lipid membrane. Adapted from Ref. [65]. Creative Commons CC BY license 2019.
The synthetic organelles were capable of producing ATP from ADP under irradiation with light, a transformation reminiscent of photosynthesis. ATP was synthesized by the photosynthetic organelles consisting of lipid vesicles carrying a light‐driven proton pump (bacteriorhodopsin, bR) and ATP synthase embedded in the vesicle membrane. ATPase was powered by the proton gradient maintained by the light‐driven proton‐pumping activity of bacteriorhodopsin. To form energy‐sufficient synthetic cells, the photosynthetic organelles (100–200 nm, diameter) were encapsulated in GUVs (10 μm). The ATP obtained from the photosynthetic organelles was used to drive the synthesis of bR and parts of the ATP synthase by the co‐encapsulated in vitro transcription translation system. These results are impressive, as they show that the synthetic cells could synthesize functional parts of their own photosynthetic organelles.
Multicomponent systems, such as the one shown in Figure 12, can be limited by the uncertainties in the composition caused by the low encapsulation efficiency and low membrane integration efficiency of the functional components in the lumen and membranes of the synthetic cells. Furthermore, it is still unclear how a synthetic cell could function continuously, and not be dependent on the external replenishment of essential nutrients.
3.2. Protein Expression and Signaling
The signaling processes during cell‐to‐cell communication in natural multicellular organisms mainly involve the secretion of proteins that act as growth factors or morphogens. This strategy has been explored to grant communication capabilities to synthetic cells by engineering synthetic systems that can produce, release, and sense diffusive protein signals. Niederholtmeyer et al. have described porous synthetic cells capable of gene expression and intercellular communication through diffusive protein signals. [9a] The synthetic cells were assembled using a microfluidic device and were composed of a porous polymeric shell (about 40 μm diameter) containing an encapsulated hydrogel sub‐compartment (about 20 μm) that carried the genetic machinery for protein synthesis (Figure 13).
Figure 13.
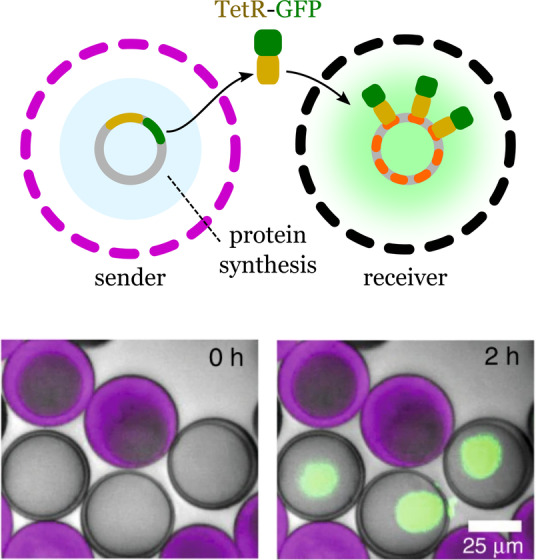
Gene expression and intercellular communication between synthetic cells. Sender lipid‐based cells synthesize fusion proteins (TetR‐sfGFP) that reach the receiver cells by molecular diffusion. The protein accumulates only in the receiver cells by interaction between the protein and tetO array plasmids, which bind to TetR. Adapted from Ref. [9a]. Creative Commons CC BY license 2018.
Protein synthesis occurred inside the cell mimics upon addition of cell‐free transcription and translation reagents. The authors chose a fusion protein of the tetracycline repressor TetR and sfGFP (TetR‐sfGFP), used as a fluorescent reporter, to be expressed by the hydrogel nuclei. The signal from the reporter protein was localized at the hydrogel nucleus with a co‐enpsulated tetO array plasmid, which binds to TetR. The authors reported that the cell mimics showed long‐term functional stability, with protein synthesis being detected even after 2 years of storage. The proteins synthesized by the hydrogel nuclei and transcription factors could freely diffuse, thus allowing communication to be stablished. To demonstrate that the synthetic cells could exchange proteins, the authors created sender cells carrying the tetR‐sfGFP expression plasmid, and receiver cells containing the tetO array plasmid to capture the reporter protein produced by the sender. After mixing together (1:1 ratio) the sender and receiver cells for 2 h, only the nuclei of the receivers showed an increase in fluorescence. Communication between the senders and receivers was also demonstrated using T3 RNA polymerase as a signal that could activate the synthesis of tetR‐sfGFP in receiver cells. Successful communication between the two populations was confirmed by the increased fluorescence in the receiver cells. Moreover, the response of the receivers depended on the density of the synthetic cells, thus allowing them to demonstrate quorum‐sensing properties. The system reported by Niederholtmeyer et al. can be programmed to collectively sense and respond to the environment, thereby showing potential applications as sensors and self‐organizing materials.
3.3. Gene Replication and Division
Gene replication and division are intrinsically connected during the cellular reproduction of living organisms. Gene replication and cell division guarantees the exchange of genetic information between parent and daughter cells. Combining gene replication and division in synthetic cells remains a challenging problem.
In 2011, Kurihara et al. described the construction of an advanced synthetic cell that combines DNA self‐replication with the self‐reproduction of giant vesicles (GV). [6c] To create responsive GVs, the researchers used a mixture of different components. Two phospholipids, POPC and POPG, were used as the main components of the lipid membrane. The GV membrane contained an amphiphilic acid catalyst capable of hydrolyzing an externally supplied membrane precursor (V*). Once hydrolyzed, the new amphiphilic membrane molecule (V) was incorporated by the GV, thereby promoting growth and division (Figure 14).
Figure 14.
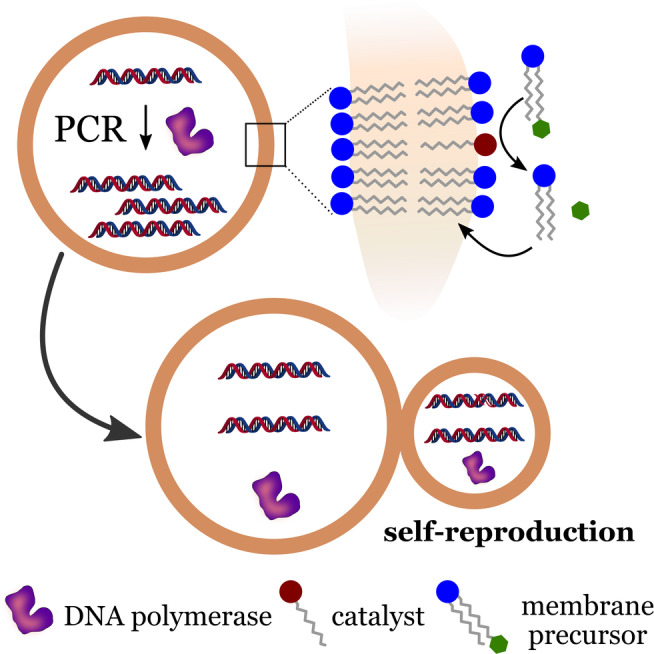
DNA replication and self‐reproduction in synthetic cells. Self‐reproduction is characterized by the partition of internal content between parent and daughter cells during the process. Redrawn from Ref. [6c].
Additionally, the vesicles contained a cationic membrane molecule, which was used as a ligand to assist the portioning of DNA during vesicle division. A PCR solution containing a 1229‐base pair (bp) template DNA was used for gene amplification. The solution was encapsulated into the GVs, which resulted in about 30 DNA templates per GV. The amplification of the DNA was carried out within the GVs using a two‐step thermal cycling. Amplification was monitored through the fluorescence of the product, the DNA‐SYBR Green I complex. After the amplification process, the authors observed the emission of intense fluorescence, and gel electrophoresis confirmed that the amplified DNA and the template DNA had the same the length.
Self‐reproduction was initiated by the addition of the bolaamphiphilic membrane precursor (V*), which resulted in the production of amphiphilic membrane molecules (V). Morphological changes and GV growth could be observed within 4 min of the addition of V*. GV division was observed within 15 min, which resulted in daughter GVs with sizes similar to the mother GVs. Daughter GVs inherited the amplified DNA from the mother GVs, with the rate of division in the presence of DNA being higher than in cases where DNA amplification did not occur. This effect was attributed to beneficial morphological changes caused by the interaction between DNA and the GV membrane. Therefore, the amplified DNA assisted in the GV division process, thus indicating that vesicles containing the DNA had an advantage in terms of their propagation compared to vesicles that did not contain DNA. Such interactions must have been essential in primitive cells. One can imagine that interactions of primitive cell membranes with charged or surface‐active materials could have had an important regulatory role for such systems. The main drawback reported by Kurihara et al. was that the self‐reproduction could be limited by gradual changes in the composition of the membrane, which happened after every thermal cycle. These limitations were addressed in a subsequent study published in 2015 by the same research group. [56c] The main challenge was to guarantee that the composition of the daughter vesicles were similar to that of the parent vesicles. Kurihara et al. showed that the composition of the daughter GVs could be restored by a continual replacement of the depleted substrates involved in the DNA amplification. To do that, the researchers developed conveyer GVs filled with the depleted substrates. The daughter GVs were able to fuse with conveyer GVs when they were mixed at a low pH value and incubated overnight. This process produced daughter GVs with a replenished supply of substrates that were able to undergo a new amplification/division cycle. The simple cell model developed by Kurihara et al. was capable of maintaining reproduction cycles over at least three generations, which represents a significant advance in the field.
However, the system cannot be classified as self‐maintained. A great amount of external intervention by the experimenter is required to keep the synthetic cells active. Gradual changes in membrane composition cannot be avoided, which would eventually lead the system to functional failure.
3.4. Growth‐Driven Metabolism
Biological systems can resist changes in their environment through internal regulatory mechanisms that sense and respond to external physical and chemical signals. This resistance is associated with homeostasis, that is, the tendency of living organisms to reach a chemical and mechanical steady state that optimizes their performance in different situations. Variables associated with homeostasis (e.g. body temperature, pH value of fluids, ions concentration, sugar levels) are controlled by regulators or complex homeostatic mechanisms, which together maintain life. Since homeostasis is essential for life, a question remains as to whether homeostatic processes were present at all in the ancestors of the cells we know today. Did protocells feature intrinsic physicochemical processes able to confer some degree of homeostatic response? For example, if one considers a simple lipid‐based protocell carrying active components, any growth and eventual splitting of this protocell would cause an irreversible dilution of the active components, thus limiting the performance of further generations. This limitation could be partially avoided if one considers Oparin's hypothesis that primitive cells were contained within membraneless microdroplets (coacervates), as opposed to lipid‐based membranes. [59] In this case, the concentration of biomolecules inside the primitive cells would not be determined by the selective permeability of a membrane, but rather by the relative affinity of the compounds to the liquid phase in the coacervates.
Considering this matter, Engelhart et al. have demonstrated a simple physical process that compensates for volume changes resulting from cell growth. [66] Using fatty acid vesicles containing split ribozymes as model synthetic cells, the researchers were able to show that the inhibition of the ribozymes by short oligonucleotides could be regulated by dilution through a growth‐driven mechanism. This was achieved by fusion of the synthetic cells and small empty phospholipid vesicles, causing dilution of vesicle content. Upon dilution, the inhibitors dissociated, allowing reconstitution of the ribozyme, and thus increasing its catalytic activity after vesicle growth. The authors pointed out that short oligonucleotides were probably abundant in primitive cells and could have offered a simple homeostatic regulation mechanism similar to the one observed in their model protocells. Therefore, a growth‐driven activation of catalysts could have provided a simple form of homeostatic regulation in primitive cells, a crucial step towards cellular evolution.
3.5. Biohybrid Synthetic Cells
Synthetic cells can be constructed using whole cells or entire biological systems as functional modules. This strategy creates hybrid systems that bring together the richness of cellular biochemistry and the robustness and chemical versatility offered by synthetic chemistry.
In 2018, Elani et al. presented their results on the assembly of biohydrid synthetic cells obtained by the encapsulation of living cells in synthetic lipid vesicles (Figure 15). [67] The researchers used a droplet microfluidic technology which allowed the cells and other components to be efficiently encapsulated in the lipid vesicles. The encapsulation method was quite robust, allowing a bacterium and several eukaryotic cell lines to be encapsulated without being damaged by the process. The encapsulated cells became the internal organelles for synthetic cells with the task of producing glucose. Glucose was used as the first substrate in a model cascade reaction that produced an intense fluorescence signal (Figure 15). Another important feature of the biohybrid synthetic cells was the protection of the biological cells from damaging chemicals present in the surrounding medium. This ability was tested through exposing the encapsulated cells to solutions containing Cu2+ ions. The results revealed that the tested system prevented the inflow of harmful materials. This extra layer of protection, however, also caused the accumulation of waste products inside the synthetic cells.
Figure 15.
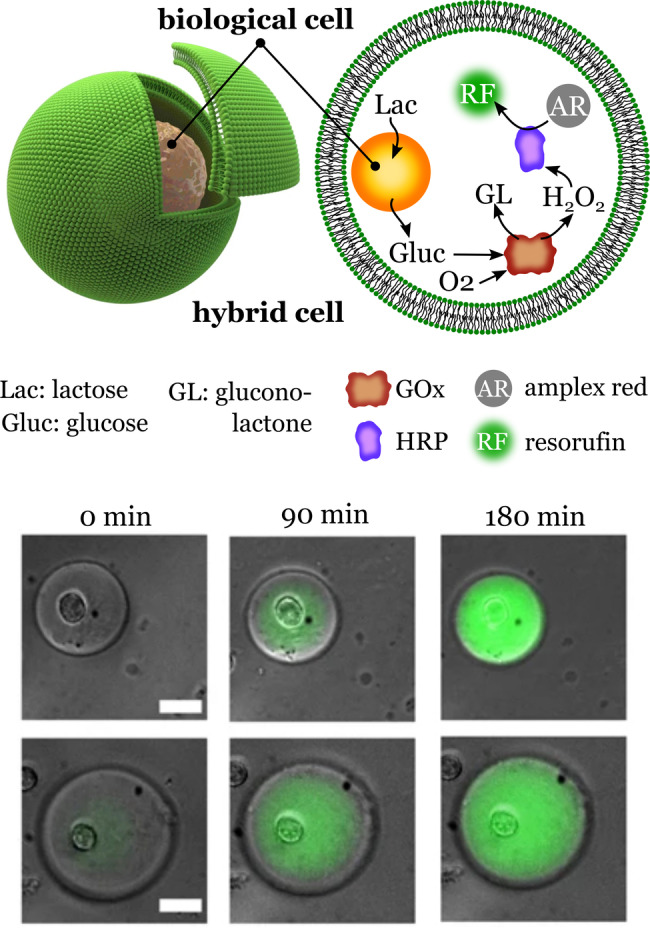
Synthetic hybrid cells. Encapsulation of biological cells into lipid vesicles creates hybrid synthetic cells. The micrographs show the synthesis of resorufin, the final product of an enzymatic cascade reaction that uses the glucose produced by the internalized biological cell. Scale bar: 25 μm. Adapted from Ref. [67]. Creative Commons CC BY license 2018.
Hybrid systems can also be obtained by coating a synthetic substrate with biological membranes. This process is also known as “membranization”, and has been used to create compartmentalized structures surrounded by membranes obtained from natural cells. [68] The biological composition of the membranes imparts cell‐like functionality and compatibility to the synthetic compartments, while the interior of the cells can be designed to perform a desired task. A recent example was described by researchers in the Mann group. Synthetic cells consisting of coacervate microdroplets were coated with hemoglobin‐containing membrane fragments obtained from erythrocytes. [69] Glucose oxidase was encapsulated in the coacervate droplets, thereby creating a two‐enzyme system that was able to produce nitric oxide (NO) in the presence of glucose and hydroxyurea. The researchers showed that the NO produced by the synthetic cells could induce vasodilation in a model system. As a result of the biological origin of the membrane, the system showed a considerable higher stability in blood. The system must be improved to account for the short lifetime of NO and the dilution effect in the surrounding medium. For example, local administration of NO could be improved by surface functionalization of the synthetic cells with targeting groups or the immobilization of the synthetic cells in devices.
In 2020, Einfalt et al. published a ground‐breaking study on the development of hybrid systems. [70] They introduced a general strategy for creating microscale bioinspired molecular factories (MFs) through the incorporation of synthetic organelles into giant plasma membrane vesicles (GPMVs; Figure 16).
Figure 16.
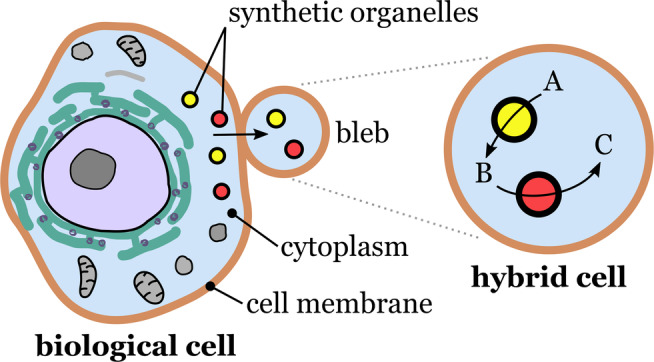
Fabrication of molecular factories. Cell blebbing releases giant plasma membrane vesicles that become independent hybrid cells. Synthetic organelles initially internalized by the biological cells become part of the hybrid cells along with other cytosolic components. Redrawn from Ref. [70].
GPMVs are an interesting class of vesicular microcompartments that can be isolated from living cells. Under certain conditions, cells can release GPMVs through blebbing. A major advantage of GMPVs over synthetic microcompartments is that the membrane of the GMPVs and their cytosolic composition are closely related to that of the donor cells. As a result, GMPVs retain the high biological complexity of the donor cells. The researchers used HepG2 liver carcinoma, a human‐derived model cell line, as donor cells to produce GMPVs. To fabricate the MFs, the researchers introduced nanosized synthetic organelles (polymersomes and liposomes) into the donor cells, and harvested the resulting micrometer‐sized GMPVs, which carried the synthetic organelles internalized by the donor cells. By this approach, MFs were obtained which mimicked the functionality and architecture of eukaryotic cells. A highlight of this study was the in vivo evaluation of the functionality of the molecular factories. This was demonstrated with zebrafish embryos which were injected with MFs. The embryos did not show signs of loss of integrity or functionality even 24 h after the injection of the MFs. Additionally, the MFs survived the shear stress caused by blood circulation in the embryo, thus indicating a high structural stability. The results obtained by Einfalt et al. open new possibilities for the development of sophisticated in vitro diagnostics and biologically compatible in vivo therapeutics. A drawback of using living cells as donors is that they must be resistant to the activity of the components that will be encapsulated. In addition, some materials may not be easily taken up by the cells without additional surface functionalization.
4. Evolution and Adaptability
The synthetic cells and strategies discussed so far can mimic complex cellular behavior such as communication, division, motility, and protein expression. In the last part of this Review, we will focus on recent efforts towards mimicking and understanding evolvability and adaptability in synthetic cells.
4.1. Rudimentary Evolvability
A mechanism that links genotype and phenotype is a basic requirement for Darwinian evolution. Here, the genetic information stored by the cells (genotype) contain the instructions for the synthesis of the biomolecules responsible for the structural, functional, and behavioral properties developed by the cells (phenotype). Compartmentalization has been postulated as one requirement for Darwinian evolution, and it has been demonstrated experimentally using simple cell mimics.
In 1998, Tawfik and Griffiths experimentally demonstrated the link between genotype and phenotype using a model compartmentalized system. [71] For this, the researchers constructed a simple cell mimic using water‐in‐oil droplets as compartments. The cell mimics contained only one of two possible genes (HaeIII, folA) encoding DNA methyltransferase III (DNMT) or dihydrofolate reductase (DHFR), respectively. The genes were linked to a substrate (HaeIII/M sequence) that was methylated only in the presence of DNMT but not by DHFR. This allowed the methylated genes to be selectively enriched in further rounds. Compartmentalization was crucial, as selection could not have occurred if the two genes were present in the medium.
In a more recent study, Ichihashi et al. demonstrated that genetic information can be replicated in a synthetic cell and undergo the same replication‐error events that form the basis of evolution in living organisms. [72] The synthetic cell, a water‐in‐oil compartment containing RNA and a purified translation system, was able to synthesize its own RNA replicase. Inside the synthetic cells, the self‐encoded RNA replicase was able to replicate either the genomic RNA or parasitic, small RNAs generated during the process. Compared to genomic RNA, the replication of parasitic RNA occurred rapidly, but it did not encode RNA replicase. Compartmentalization was essential as it allowed the system to evolve without being overwhelmed by parasitic sequences. The compartment spatially restricted the parasitic replicators and enhanced the effect of positive mutations that increased the replication efficiency of the genomic RNA. Another example of directed evolution made possible by the encapsulation of genetic material in compartments was reported by Fujii et al. [73] They demonstrated that α‐hemolysin (AH), a membrane protein that forms pores, could be artificially evolved in synthetic cells comprising a DNA template and an in vitro translation system (IVT) encapsulated in giant unilamellar vesicles (GUVs). The DNA template was encapsulated at the single‐molecule level, with each resulting vesicle containing only a single DNA molecule obtained from a library of mutagenized DNA. By measuring the pore‐forming activity of AH synthesized inside the GUVs, the researchers were able to select the fittest DNAs, which became the subsequent DNA library used in further selection rounds (Figure 17).
Figure 17.
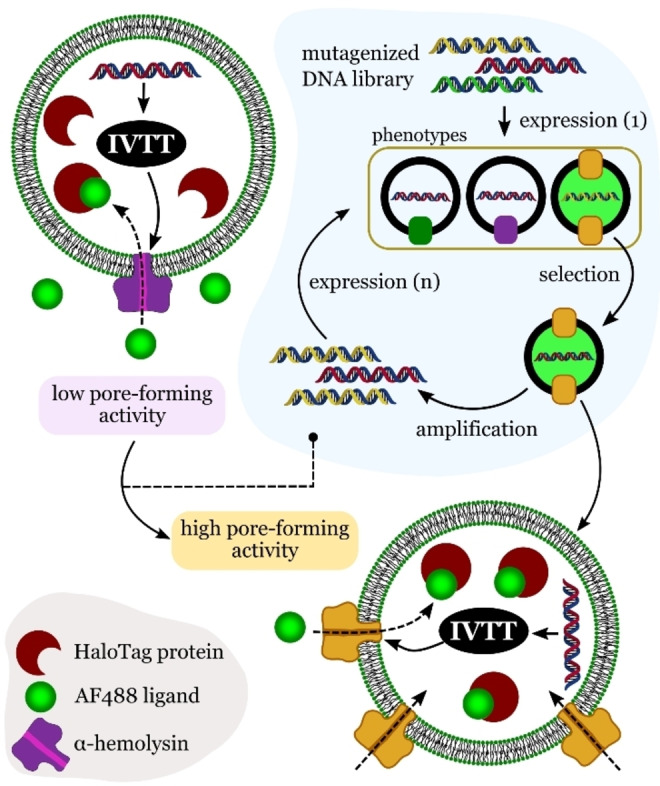
Evolution of a pore‐membrane protein in synthetic cells. The method involves the cell‐free synthesis of α‐hemolysin in liposomes. The performance of the pores was evaluated by the diffusion of a tracer (AF488) present inside the liposomes. Redrawn from Ref. [73].
After 20 rounds of selection, the evolved AH exhibited a 30‐fold increase in pore‐formation activity relative to the wild‐type AH. This method, referred to as liposome display, can be considered a rapid and efficient strategy to evolve a wide range of membrane proteins.
Adamala and Szostak developed liposomes capable of competitive growth, which indicated clear evidence of their capability for evolution. [74] In their study, the authors assumed that evolvability may have emerged in primitive cellular systems through competition between protocells for common resources. [51] They developed a simple model (Figure 18 a) consisting of fatty acid vesicles containing a dipeptide catalyst (seryl‐histidine, Ser‐His), which catalyzed the formation of a hydrophobic dipeptide (N‐acetyl‐l‐phenylalanine leucinamide, AcPheLeuNH2).
Figure 18.
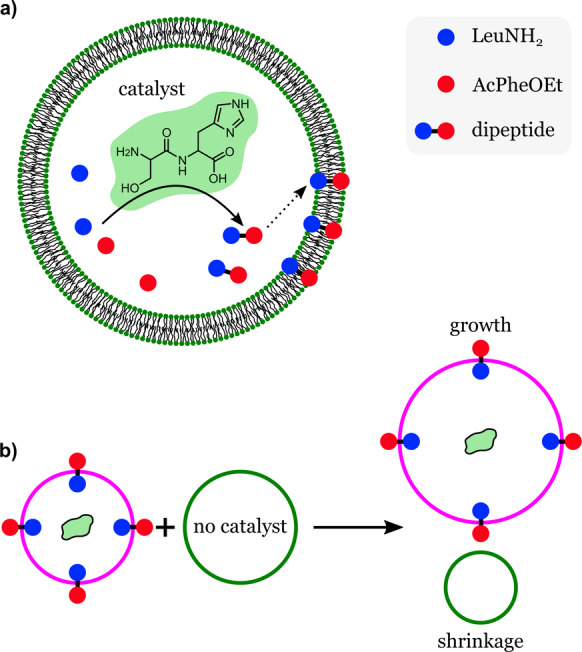
Competitive growth in synthetic cells. Lipid vesicles carrying a dipeptide catalyst produce a hydrophobic dipeptide from precursors inside the vesicles. The hydrophobic dipeptides accumulate in the vesicle membrane. b) Membranes with embedded dipeptides show an enhanced uptake of lipids and can grow by consuming lipids from vesicles that do not carry the dipeptide catalyst. Redrawn from Ref. [74].
The hydrophobic dipeptide, formed inside the vesicles, was taken up by the vesicle membranes, increasing their affinity for fatty acids and consequently promoting vesicle growth if a source of fatty acids was present. Vesicles that were able to make AcPheLeuNH2 grew when mixed with vesicles that did not contain the catalyst (Figure 18 b). Therefore, the presence of the catalyst enhanced the fitness of the synthetic cells in terms of their ability to grow at the expense of vesicles that lacked the capability of regulating the composition of their membranes. In another experiment, the authors investigated the competition between vesicles containing catalytic peptides with different catalytic efficiencies. Vesicles carrying the dipeptide catalyst (Ser‐His) had an advantage over vesicles carrying a modified catalyst (Ser‐His‐Gly) which had a lower catalytic efficiency. The addition of substrates to the mixed system caused the vesicles carrying Ser‐His to grow by consuming the fatty acids of the Ser‐His‐Gly‐containing vesicles, which shrank. This study demonstrated that competitive behavior could arise between populations of protocells, which according to the authors is analogous to a predator–prey interaction. If the system developed by the authors was capable of self‐replication (heredity), it could be considered as a protocell capable of growing and capable of Darwinian evolution.
4.2. Adaptability
Coacervate microdroplets have been proposed as an ideal medium to explore cell‐like adaptability in synthetic systems. [75] Coacervation results from the phase separation of oppositely charged polyelectrolytes (such as polymers, proteins, nucleic acids) in solution, which results in concentrated condensate droplets similar to biological condensates present in cells. [76] Biological condensates, such as P granules and Cajal bodies, have important regulatory roles, including RNA metabolism and gene replication. [60] In contrast to polymer or lipid‐based vesicles, coacervates show a high degree of molecular crowding, plasticity, and a relatively free molecular transport between the membraneless condensates and the surrounding medium. [77] Consequently, coacervates are highly dynamic, showing dramatic changes in their shape and internal composition in response to internal or external physicochemical changes. [78]
An example of adaptable coacervate‐based synthetic cells was described by Samanta et al. [79] The synthetic cells consisted of coacervate DNA microdroplets with DNA strands bound to artificial catalysts (metalloenzymes) by attractive biotin–streptavidin noncovalent interactions (Figure 19).
Figure 19.
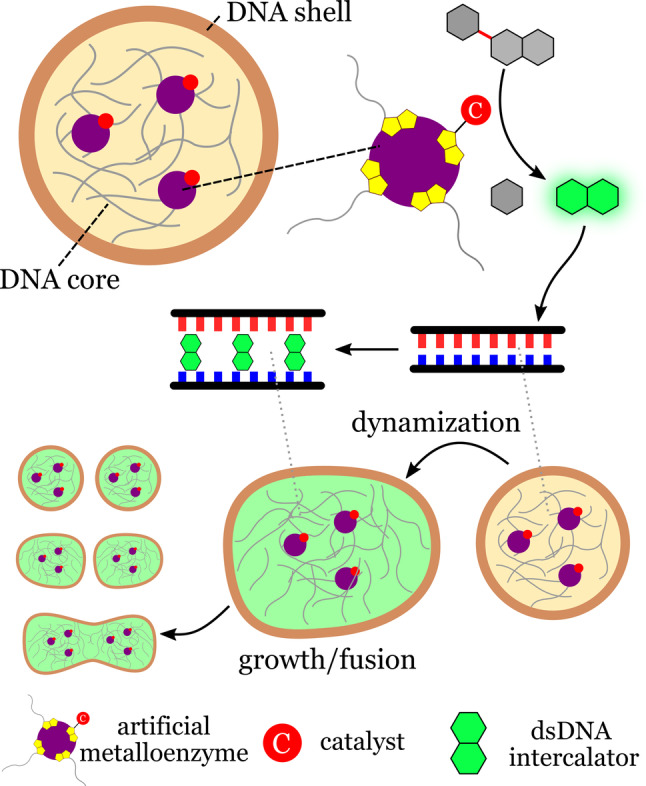
Morphogenesis in coacervate‐based synthetic cells. Artificial metalloenzymes within coacervate microdroplets catalyze the internal production of a DNA intercalator from an inactive precursor. The intercalator dynamizes the coacervates by weakening the DNA interactions, thereby resulting in morphological responses such as fusion and growth. Redrawn from Ref. [79].
Within the synthetic cells, the artificial enzymes produced a DNA intercalator from an inactive precursor present in the medium. The intercalator caused a weakening of the DNA duplex interaction, thereby resulting in dramatic morphological transformations as a consequence of coacervate dynamization. The observed dynamic responses included the division and fusion of synthetic cells. The system illustrates how chemically transduced processes and morphological adaptation can be induced in a cell mimic using abiotic components. Examples such as the those reported by Samanta and co‐workers shows that complex cell‐like behavior can be induced in soft materials without the need for complex biological machinery.
5. Summary and Outlook
Synthetic cells obtained through the bottom‐up approach have come a long way in terms of sophistication and overall performance. This success has resulted from the development of new techniques, materials, and strategies that have simplified the challenging task of building cell‐like materials from basic components. We have seen synthetic cells that can mimic a variety of cell functions and structural organization. They can divide, move, synthesize ATP, and communicate with each other. However, these systems are still very simple compared to what a simple bacterium can do. There are still many challenges ahead and improvements that need to be made before synthetic cells can develop to their full potential.
The development of hybrid systems will be essential for the future technological application of synthetic cells. Although biological molecules are relatively easy to use as building blocks in synthetic cells, they typically suffer from low stability and low compatibility in nonbiological environments. One solution to this problem would be to replace sensitive biological components with artificial substitutes. For example, robust artificial enzymes, such as artificial metalloenzymes and other nanozymes, could replace sensitive enzymes. This would allow synthetic cells to be used in applications where conditions are not ideal for biomolecules.
Many interesting applications of synthetic cells, such as tissue formation, drug delivery, and biosynthesis, will depend on the versatility, multifunctionality, and robustness of the membranes or interfaces of their compartments. However, the current compartments used to create synthetic cells are still too simple in terms of structure and function. The development of artificial membrane pores and channels (e.g. DNA origami and supramolecular channels) as well as artificial receptors will expand the range of tools available to design suitable synthetic biomimetic membranes.
An important milestone in bottom‐up synthetic biology is the engineering of synthetic cells and organelles capable of establishing a symbiotic relationship with living cells. The premise is that synthetic cells could be used to control the behavior of biological cells. A clear demonstration of the potential of this approach was provided by the Mann and Mansy research groups, who showed that relatively simple synthetic cells could influence the behavior of muscle cells and neurons.[ 47 , 69 ] Communication between synthetic and natural cells represents an exciting way to modulate cellular behavior and function, or to use cells as regulators in communities of synthetic and natural cells to create hybrid tissues.
Bottom‐up synthetic biology has produced synthetic cells with an ever‐increasing degree of complexity. The scientific benefits of this research are twofold. On the one hand, it has introduced exciting new technologies that have applications in biomedicine, tissue engineering, and materials chemistry. On the other hand, building synthetic cells from scratch improves our understanding of how cells work, which leads to broader questions. This completes a conceptual cycle in bottom‐up synthetic biology: more advanced questions lead to more sophisticated systems, which in turn lead to more advanced questions. How many turns of this conceptual cycle will it take before a synthetic cell becomes indistinguishable from a simple bacterium? Only time will tell. Or perhaps we will find that a piece of the puzzle is still missing. Either way, the answers will have profound implications on how we define life.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Camila Guindani obtained her PhD in chemical engineering from the Federal University of Santa Catarina in 2018 with Prof. Débora de Oliveira, with part of her thesis carried out at the Max Planck Institute for Polymer Research, Germany, with Prof. Katharina Landfester. She is currently a postdoctoral researcher with Prof. Márcio Nele at the Chemical Engineering Program of the Federal University of Rio de Janeiro, Brazil, where her research focuses on the synthesis and modification of polymers for biological purposes, functionalized nanostructures for cell targeting, and applying polymersomes to cell mimicking.

Biographical Information
Lucas Caire da Silva studied chemistry at the University of Campinas in Brazil and obtained his PhD in polymer chemistry with Prof. Kenneth Wagener at the University of Florida. In 2016, he joined Prof. Katharina Landfester's group at the Max Planck Institute for Polymer Research as a postdoctoral fellow to work on bottom‐up synthetic cell engineering. He is currently a research group leader at the same institute, where he develops adaptive synthetic cell mimics and compartmentalized catalytic bioreactors.

Biographical Information
Shoupeng Cao obtained his PhD in 2020 at the Eindhoven University Technology under the guidance of Prof. Jan C. M. van Hest. His thesis focused on the engineering of well‐defined biodegradable polymeric nanoparticles for tailored biomedical applications. Currently, he is a postdoctoral researcher at the Max Planck Institute for Polymer Research in the group of Prof. Katharina Landfester, where he engineers biomimetic microcompartments with adaptive and cell‐like features.

Biographical Information
Tsvetomir Ivanov is currently a PhD student in the group of Prof. Katharina Landfester at the Max Planck Institute for Polymer Research. He obtained his double master's degree in chemical engineering through a cooperation between the University of Chemical Technology and Metallurgy Sofia and Hamburg University of Technology in 2019. His thesis focused on the solute‐dependent stability of GUVs. He is currently working on engineering vesicle‐based artificial cells that mimic the structure and functions of living cells.

Biographical Information
Katharina Landfester received her PhD in 1995 with Prof. Hans‐Wolfgang Spiess. She then worked at the Max Planck Institute of Colloids and Interfaces in Potsdam (Germany) leading the miniemulsion group. From 2003 to 2008, she was full professor at the University of Ulm, and in 2008 she was appointed as one of the directors of the Max Planck Institute for Polymer Research. Her research focuses on creating functional colloids for technological and biomedical applications.

Acknowledgements
This work was supported by the Max Planck Consortium for Synthetic Biology (MaxSynBio) jointly funded by the Federal Ministry of Education and Research of Germany (BMBF) and the Max Planck Society. C.G. thanks FAPERJ (Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro) for financial support (process number E‐26/201.911/2020). Open Access funding enabled and organized by Projekt DEAL.
C. Guindani, L. C. da Silva, S. Cao, T. Ivanov, K. Landfester, Angew. Chem. Int. Ed. 2022, 61, e202110855; Angew. Chem. 2022, 134, e202110855.
Contributor Information
Dr. Lucas Caire da Silva, Email: silva@mpip-mainz.mpg.de.
Prof. Dr. Katharina Landfester, Email: landfester@mpip-mainz.mpg.de.
References
- 1.
- 1a. Noireaux V., Maeda Y. T., Libchaber A., Proc. Natl. Acad. Sci. USA 2011, 108, 3473–3480; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b. Xu C., Hu S., Chen X. Y., Mater. Today 2016, 19, 516–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.
- 2a. Elani Y., Angew. Chem. Int. Ed. 2021, 60, 5602–5611; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 5662–5671; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2b. Jeong S., Nguyen H. T., Kim C. H., Ly M. N., Shin K., Adv. Funct. Mater. 2020, 30, 1907182. [Google Scholar]
- 3. Schwille P., Science 2011, 333, 1252–1254. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a. Koonin E. V., Nat. Rev. Microbiol. 2003, 1, 127–136; [DOI] [PubMed] [Google Scholar]
- 4b. Wu C. Y., Rupp L. J., Roybal K. T., Lim W. A., Curr. Opin. Immunol. 2015, 35, 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blain J. C., Szostak J. W., Annu. Rev. Biochem. 2014, 83, 615–640. [DOI] [PubMed] [Google Scholar]
- 6.
- 6a. Schoonen L., van Hest J. C. M., Adv. Mater. 2016, 28, 1109–1128; [DOI] [PubMed] [Google Scholar]
- 6b. Poudyal R. R., Guth-Metzler R. M., Veenis A. J., Frankel E. A., Keating C. D., Bevilacqua P. C., Nat. Commun. 2019, 10, 490; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6c. Kurihara K., Tamura M., Shohda K., Toyota T., Suzuki K., Sugawara T., Nat. Chem. 2011, 3, 775–781. [DOI] [PubMed] [Google Scholar]
- 7. Schwille P., Spatz J., Landfester K., Bodenschatz E., Herminghaus S., Sourjik V., Erb T. J., Bastiaens P., Lipowsky R., Hyman A., Dabrock P., Baret J. C., Vidakovic-Koch T., Bieling P., Dimova R., Mutschler H., Robinson T., Tang T. Y. D., Wegner S., Sundmacher K., Angew. Chem. Int. Ed. 2018, 57, 13382–13392; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 13566–13577. [Google Scholar]
- 8. Jia Y., Li J. B., Nat. Rev. Chem. 2019, 3, 361–374. [Google Scholar]
- 9.
- 9a. Niederholtmeyer H., Chaggan C., Devaraj N. K., Nat. Commun. 2018, 9, 5027; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b. Buddingh′ B. C., Elzinga J., van Hest J. C. M., Nat. Commun. 2020, 11, 1652; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9c. Steinkuhler J., Knorr R. L., Zhao Z. L., Bhatia T., Bartelt S. M., Wegner S., Dimova R., Lipowsky R., Nat. Commun. 2020, 11, 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.
- 10a. Mason A. F., van Hest J. C. M., Emerging Top. Life Sci. 2019, 3, 567–571; [DOI] [PubMed] [Google Scholar]
- 10b. Deshpande S., Brandenburg F., Lau A., Last M. G. F., Spoelstra W. K., Reese L., Wunnava S., Dogterom M., Dekker C., Nat. Commun. 2019, 10, 1800; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10c. Douliez J. P., Martin N., Beneyton T., Eloi J. C., Chapel J. P., Navailles L., Baret J. C., Mann S., Beven L., Angew. Chem. Int. Ed. 2018, 57, 7780–7784; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 7906–7910; [Google Scholar]
- 10d. Huang X., Patil A. J., Li M., Mann S., J. Am. Chem. Soc. 2014, 136, 9225–9234; [DOI] [PubMed] [Google Scholar]
- 10e. Li M., Harbron R. L., Weaver J. V. M., Binks B. P., Mann S., Nat. Chem. 2013, 5, 529–536. [DOI] [PubMed] [Google Scholar]
- 11.
- 11a. Itel F., Najer A., Palivan C. G., Meier W., Nano Lett. 2015, 15, 3871–3878; [DOI] [PubMed] [Google Scholar]
- 11b. Wang M., Weber A., Hartig R., Zheng Y., Krafft D., Vidakovic-Koch T., Zuschratter W., Ivanov I., Sundmacher K., Bioconjugate Chem. 2021, 32, 897–903; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11c. Zhang Y. W., Chen Y. F., Yang X. H., He X. X., Li M., Liu S. Y., Wang K. M., Liu J. B., Mann S., J. Am. Chem. Soc. 2021, 143, 2866–2874. [DOI] [PubMed] [Google Scholar]
- 12.
- 12a. Garni M., Thamboo S., Schoenenberger C. A., Palivan C. G., BBA-Biomembranes 2017, 1859, 619–638; [DOI] [PubMed] [Google Scholar]
- 12b. Wang X. R., Yao C. Z., Zhang G. Y., Liu S. Y., Nat. Commun. 2020, 11, 1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jesorka A., Orwar O., Annu. Rev. Anal. Chem. 2008, 1, 801–832. [DOI] [PubMed] [Google Scholar]
- 14.
- 14a. Jo S. M., Wurm F. R., Landfester K., Nano Lett. 2020, 20, 526–533; [DOI] [PubMed] [Google Scholar]
- 14b. Otrin L., Kleineberg C., da Silva L. C., Landfester K., Ivanov I., Wang M. H., Bednarz C., Sundmacher K., Vidakovic-Koch T., Adv. Biosyst. 2019, 3, 1800323; [DOI] [PubMed] [Google Scholar]
- 14c. Bhatia T., Robinson T., Dimova R., Soft Matter 2020, 16, 7359–7369; [DOI] [PubMed] [Google Scholar]
- 14d. Houbrechts M., da Silva L. C., Ethirajan A., Landfester K., Soft Matter 2021, 17, 4942–4948. [DOI] [PubMed] [Google Scholar]
- 15. Le Meins J. F., Schatz C., Lecommandoux S., Sandre O., Mater. Today 2013, 16, 397–402. [Google Scholar]
- 16.
- 16a. Lensen D., Vriezema D. M., van Hest J. C. M., Macromol. Biosci. 2008, 8, 991–1005; [DOI] [PubMed] [Google Scholar]
- 16b. Discher B. M., Won Y. Y., Ege D. S., Lee J. C. M., Bates F. S., Discher D. E., Hammer D. A., Science 1999, 284, 1143–1146. [DOI] [PubMed] [Google Scholar]
- 17. Rideau E., Dimova R., Schwille P., Wurm F. R., Landfester K., Chem. Soc. Rev. 2018, 47, 8572–8610. [DOI] [PubMed] [Google Scholar]
- 18. Petit J., Thomi L., Schultze J., Makowski M., Negwer I., Koynov K., Herminghaus S., Wurm F. R., Baumchen O., Landfester K., Soft Matter 2018, 14, 894–900. [DOI] [PubMed] [Google Scholar]
- 19. da Silva L. C., Rideau E., Landfester K., Macromol. Rapid Commun. 2019, 40, 1900027. [DOI] [PubMed] [Google Scholar]
- 20. Luo R. C., Gopfrich K., Platzman I., Spatz J. P., Adv. Funct. Mater. 2020, 30, 2003480. [Google Scholar]
- 21. Peyret A., Ibarboure E., Le Meins J. F., Lecommandoux S., Adv. Sci. 2018, 5, 1700453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rottet S., Iqbal S., Beales P. A., Lin A. R., Lee J., Rug M., Scott C., Callaghan R., Polymers-Basel 2020, 12, 1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bolinger P. Y., Stamou D., Vogel H., Angew. Chem. Int. Ed. 2008, 47, 5544–5549; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2008, 120, 5626–5631. [Google Scholar]
- 24. Chu L. Y., Utada A. S., Shah R. K., Kim J. W., Weitz D. A., Angew. Chem. Int. Ed. 2007, 46, 8970–8974; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2007, 119, 9128–9132. [Google Scholar]
- 25. Kreft O., Prevot M., Mohwald H., Sukhorukov G. B., Angew. Chem. Int. Ed. 2007, 46, 5605–5608; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2007, 119, 5702–5705. [Google Scholar]
- 26.
- 26a. Peters R. J. R. W., Marguet M., Marais S., Fraaije M. W., van Hest J. C. M., Lecommandoux S., Angew. Chem. Int. Ed. 2014, 53, 146–150; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 150–154; [Google Scholar]
- 26b. Mason A. F., Thordarson P., J. Polym. Sci. Part A 2017, 55, 3817–3825. [Google Scholar]
- 27.
- 27a. Städler B., Chandrawati R., Price A. D., Chong S. F., Breheney K., Postma A., Connal L. A., Zelikin A. N., Caruso F., Angew. Chem. Int. Ed. 2009, 48, 4359–4362; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2009, 121, 4423–4426; [Google Scholar]
- 27b. Godoy-Gallardo M., Labay C., Jansman M. M. T., Ek P. K., Hosta-Rigau L., Adv. Healthcare Mater. 2017, 6, 1601190. [DOI] [PubMed] [Google Scholar]
- 28. Peyret A., Ibarboure E., Pippa N., Lecommandoux S., Langmuir 2017, 33, 7079–7085. [DOI] [PubMed] [Google Scholar]
- 29. Mason A. F., Yewdall N. A., Welzen P. L. W., Shao J. X., van Stevendaal M., van Hest J. C. M., Williams D. S., Abdelmohsen L. K. E. A., ACS Cent. Sci. 2019, 5, 1360–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Donau C., Spath F., Sosson M., Kriebisch B. A. K., Schnitter F., Tena-Solsona M., Kang H. S., Salibi E., Sattler M., Mutschler H., Boekhoven J., Nat. Commun. 2020, 11, 5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Noireaux V., Libchaber A., Proc. Natl. Acad. Sci. USA 2004, 101, 17669–17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Semenov S. N., Wong A. S. Y., van der Made R. M., Postma S. G. J., Groen J., van Roekel H. W. H., de Greef T. F. A., Huck W. T. S., Nat. Chem. 2015, 7, 160–165. [DOI] [PubMed] [Google Scholar]
- 33. Pitard B., Richard P., Dunach M., Rigaud J. L., Eur. J. Biochem. 1996, 235, 779–788. [DOI] [PubMed] [Google Scholar]
- 34. Steinberg-Yfrach G., Rigaud J. L., Durantini E. N., Moore A. L., Gust D., Moore T. A., Nature 1998, 392, 479–482. [DOI] [PubMed] [Google Scholar]
- 35. Kleineberg C., Wolfer C., Abbasnia A., Pischel D., Bednarz C., Ivanov D. I., Heitkamp D. T., Borsch M., Sundmacher K., Vidakovic-Koch T., ChemBiochem 2020, 21, 2149–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee K. Y., Park S. J., Lee K. A., Kim S. H., Kim H., Meroz Y., Mahadevan L., Jung K. H., Ahn T. K., Parker K. K., Shin K., Nat. Biotechnol. 2018, 36, 530–535. [DOI] [PubMed] [Google Scholar]
- 37.
- 37a. Yuan M. W., Kummer M. J., Milton R. D., Quah T., Minteer S. D., ACS Catal. 2019, 9, 5486–5495; [Google Scholar]
- 37b. Wang M. H., Woelfer C., Otrin L., Ivanov I., Vidakovic-Koch T., Sundmacher K., Langmuir 2018, 34, 5435–5443; [DOI] [PubMed] [Google Scholar]
- 37c. Beneyton T., Krafft D., Bednarz C., Kleineberg C., Woelfer C., Ivanov I., Vidakovic-Koch T., Sundmacher K., Baret J. C., Nat. Commun. 2018, 9, 2391; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37d. Lee S. H., Choi D. S., Kuk S. K., Park C. B., Angew. Chem. Int. Ed. 2018, 57, 7958–7985; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 8086–8116. [Google Scholar]
- 38. Ma B. C., da Silva L. C., Jo S. M., Wurm F. R., Bannwarth M. B., Zhang K. A. I., Sundmacher K., Landfester K., ChemBiochem 2019, 20, 2593–2596. [DOI] [PubMed] [Google Scholar]
- 39. Jo S. M., Wurm F. R., Landfester K., Angew. Chem. Int. Ed. 2021, 60, 7728–7734; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 7807–7813. [Google Scholar]
- 40. Jo S. M., Wurm F. R., Landfester K., ACS Appl. Mater. Interfaces 2018, 10, 34230–34237. [DOI] [PubMed] [Google Scholar]
- 41. Liu J., Guo Z. Y., Liang K., Adv. Funct. Mater. 2019, 29, 1905321. [Google Scholar]
- 42. Miller T. E., Beneyton T., Schwander T., Diehl C., Girault M., McLean R., Chotel T., Claus P., Cortina N. S., Baret J. C., Erb T. J., Science 2020, 368, 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.
- 43a. Nomura S., Tsumoto K., Hamada T., Akiyoshi K., Nakatani Y., Yoshikawa K., ChemBiochem 2003, 4, 1172–1175; [DOI] [PubMed] [Google Scholar]
- 43b. Dwidar M., Seike Y., Kobori S., Whitaker C., Matsuura T., Yokobayashi Y., J. Am. Chem. Soc. 2019, 141, 11103–11114. [DOI] [PubMed] [Google Scholar]
- 44. Elani Y., Law R. V., Ces O., Phys. Chem. Chem. Phys. 2015, 17, 15534–15537. [DOI] [PubMed] [Google Scholar]
- 45. Gardner P. M., Winzer K., Davis B. G., Nat. Chem. 2009, 1, 377–383. [DOI] [PubMed] [Google Scholar]
- 46. Lentini R., Martin N. Y., Forlin M., Belmonte L., Fontana J., Cornella M., Martini L., Tamburini S., Bentley W. E., Jousson O., Mansy S. S., ACS Cent. Sci. 2017, 3, 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Toparlak O. D., Zasso J., Bridi S., Dalla Serra M., Macchi P., Conti L., Baudet M. L., Mansy S. S., Sci. Adv. 2020, 6, eabb4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.
- 48a. Tang T. Y. D., Cecchi D., Fracasso G., Accardi D., Coutable-Pennarun A., Mansy S. S., Perriman A. W., Anderson J. L. R., Mann S., ACS Synth. Biol. 2018, 7, 339–346; [DOI] [PubMed] [Google Scholar]
- 48b. Qiao Y., Li M., Booth R., Mann S., Nat. Chem. 2017, 9, 110–119; [DOI] [PubMed] [Google Scholar]
- 48c. Joesaar A., Yang S., Bogels B., van der Linden A., Pieters P., Kumar B. V. V. S. P., Dalchau N., Phillips A., Mann S., de Greef T. F. A., Nat. Nanotechnol. 2019, 14, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chakraborty T., Wegner S. V., ASC Nano 2021, 15, 9434–9444. [DOI] [PubMed] [Google Scholar]
- 50.
- 50a. Rodríguez-Arco L., Kumar B. V. V. S. P., Li M., Patil A. J., Mann S., Angew. Chem. Int. Ed. 2019, 58, 6333–6337; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 6399–6403; [Google Scholar]
- 50b. Martin N., Douliez J. P., Qiao Y., Booth R., Li M., Mann S., Nat. Commun. 2018, 9, 3652; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50c. Rodríguez-Arco L., Li M., Mann S., Nat. Mater. 2017, 16, 857–863. [DOI] [PubMed] [Google Scholar]
- 51. Szostak J. W., Bartel D. P., Luisi P. L., Nature 2001, 409, 387–390. [DOI] [PubMed] [Google Scholar]
- 52. Kita H., Matsuura T., Sunami T., Hosoda K., Ichihashi N., Tsukada K., Urabe I., Yomo T., ChemBiochem 2008, 9, 2403–2410. [DOI] [PubMed] [Google Scholar]
- 53.
- 53a. Miller O. J., Bernath K., Agresti J. J., Amitai G., Kelly B. T., Mastrobattista E., Taly V., Magdassi S., Tawfik D. S., Griffiths A. D., Nat. Methods 2006, 3, 561–570; [DOI] [PubMed] [Google Scholar]
- 53b. Griffiths A. D., Tawfik D. S., Trends Biotechnol. 2006, 24, 395–402. [DOI] [PubMed] [Google Scholar]
- 54. van Nies P., Westerlaken I., Blanken D., Salas M., Mencia M., Danelon C., Nat. Commun. 2018, 9, 1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Szostak J. W., Angew. Chem. Int. Ed. 2017, 56, 11037–11043; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 11182–11189. [Google Scholar]
- 56.
- 56a. Zwicker D., Seyboldt R., Weber C. A., Hyman A. A., Julicher F., Nat. Phys. 2017, 13, 408–413; [Google Scholar]
- 56b. Zhu T. F., Szostak J. W., J. Am. Chem. Soc. 2009, 131, 5705–5713; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56c. Kurihara K., Okura Y., Matsuo M., Toyota T., Suzuki K., Sugawara T., Nat. Commun. 2015, 6, 8352; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56d. Hardy M. D., Yang J., Selimkhanov J., Cole C. M., Tsimring L. S., Devaraj N. K., Proc. Natl. Acad. Sci. USA 2015, 112, 8187–8192; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56e. Dervaux J., Noireaux V., Libchaber A. J., Eur. Phys. J. Plus 2017, 132, 284; [Google Scholar]
- 56f. Bhattacharya A., Brea R. J., Devaraj N. K., Chem. Sci. 2017, 8, 7912–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Castro J. M., Sugiyama H., Toyota T., Sci. Rep. 2019, 9, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miele Y., Medveczky Z., Holl G., Tegze B., Derenyi I., Horvolgyi Z., Altamura E., Lagzi I., Rossi F., Chem. Sci. 2020, 11, 3228–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Oparin A. I., The Origin of Life, Dover Publications, 1953. [Google Scholar]
- 60. Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jang W. S., Kim H. J., Gao C., Lee D., Hammer D. A., Small 2018, 14, 1801715. [DOI] [PubMed] [Google Scholar]
- 62. Ghosh S., Mohajerani F., Son S., Velegol D., Butler P. J., Sen A., Nano Lett. 2019, 19, 6019–6026. [DOI] [PubMed] [Google Scholar]
- 63. Kumar B. V. V. S. P., Patil A. J., Mann S., Nat. Chem. 2018, 10, 1154–1163. [DOI] [PubMed] [Google Scholar]
- 64. Bartelt S. M., Steinkuhler J., Dimova R., Wegner S. V., Nano Lett. 2018, 18, 7268–7274. [DOI] [PubMed] [Google Scholar]
- 65. Berhanu S., Ueda T., Kuruma Y., Nat. Commun. 2019, 10, 1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Engelhart A. E., Adamala K. P., Szostak J. W., Nat. Chem. 2016, 8, 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Elani Y., Trantidou T., Wylie D., Dekker L., Polizzi K., Law R. V., Ces O., Sci. Rep. 2018, 8, 4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhao C. Y., Li J. B., Wang S. L., Xu Z. J., Wang X. L., Liu X. M., Wang L., Huang X., ACS Nano 2021, 15, 10048–10057. [DOI] [PubMed] [Google Scholar]
- 69. Liu S. Y., Zhang Y. W., Li M., Xiong L., Zhang Z. J., Yang X. H., He X. X., Wang K. M., Liu J. B., Mann S., Nat. Chem. 2020, 12, 1165–1173. [DOI] [PubMed] [Google Scholar]
- 70. Einfalt T., Garni M., Witzigmann D., Sieber S., Baltisberger N., Huwyler J., Meier W., Palivan C. G., Adv. Sci. 2020, 7, 1901923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tawfik D. S., Griffiths A. D., Nat. Biotechnol. 1998, 16, 652–656. [DOI] [PubMed] [Google Scholar]
- 72. Ichihashi N., Usui K., Kazuta Y., Sunami T., Matsuura T., Yomo T., Nat. Commun. 2013, 4, 2494. [DOI] [PubMed] [Google Scholar]
- 73. Fujii S., Matsuura T., Sunami T., Kazuta Y., Yomo T., Proc. Natl. Acad. Sci. USA 2013, 110, 16796–16801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Adamala K., Szostak J. W., Nat. Chem. 2013, 5, 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.
- 75a. Mason A. F., Buddingh′ B. C., Williams D. S., van Hest J. C. M., J. Am. Chem. Soc. 2017, 139, 17309–17312; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75b. Koga S., Williams D. S., Perriman A. W., Mann S., Nat. Chem. 2011, 3, 720–724; [DOI] [PubMed] [Google Scholar]
- 75c. Buddingh′ B. C., van Hest J. C. M., Acc. Chem. Res. 2017, 50, 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shin Y., Brangwynne C. P., Science 2017, 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- 77.
- 77a. Abbas M., Lipin W. P., Wang J., Spruijt E., Chem. Soc. Rev. 2021, 50, 3690–3705; [DOI] [PubMed] [Google Scholar]
- 77b. Yewdall N. A., Andre A. A. M., Lu T. M., Spruijt E., Curr. Opin. Colloid Interface Sci. 2021, 52, 101416. [Google Scholar]
- 78. Yin Y. D., Niu L., Zhu X. C., Zhao M. P., Zhang Z. X., Mann S., Liang D. H., Nat. Commun. 2016, 7, 10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Samanta A., Sabatino V., Ward T. R., Walther A., Nat. Nanotechnol. 2020, 15, 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]