Abstract
The ability to coat scaffolds and wound dressings with therapeutic short interfering RNA (siRNA) holds much potential for applications in wound healing, cancer treatment, and regenerative medicine. Layer-by-layer (LbL) technology is an effective method to formulate polyelectrolyte thin films for local delivery of siRNA; however, the formation and efficacy of LbL coatings as drug delivery systems are highly contingent on the assembly conditions. Here, we investigate the effects of LbL assembly parameters on film composition and consequent siRNA-mediated gene knockdown efficiency in vitro. Films comprising poly(β-amino ester) (PBAE) and siRNA were built on polyglactin 910 (Vicryl) sutures consisting of poly(10% l-lactide, 90% glycolide). A fractional factorial design was employed, varying the following LbL assembly conditions: pH, ionic strength, PBAE concentration, and siRNA concentration. Effects of these parameters on PBAE loading, siRNA loading, their respective weight ratios, and in vitro siRNA-mediated knockdown were elucidated. The parameter effects were leveraged to create a rationally designed set of solution conditions predicted to give effective siRNA-mediated knockdown (47%), but not included in any of the original experimental conditions. This level of knockdown with our rationally designed loading conditions is comparable to previous formulations from our lab while being simpler in construction and requiring fewer film layers, which could save time and cost in manufacturing. This study highlights the importance of LbL solution conditions in the preparation of surface-mediated siRNA delivery systems and presents an adaptable methodology for extending these electrostatically-assembled coatings to the delivery of other therapeutic nucleic acids.
Keywords: Layer-by-layer, siRNA, delivery, PBAE, design of experiment
Graphical Abstract
1. Introduction:
Small interfering RNAs (siRNA) have great therapeutic potential for modulating protein expression at the post-transcriptional level. By associating with the endogenous RNA-induced silencing complex (RISC), siRNA cleaves messenger RNA (mRNA) of complementary sequence [1,2]. Thus, siRNA may be synthesized to theoretically target and knock down expression of any protein of choice, without concerns of integration and modification to host DNA. FDA approvals have recently generated much excitement around siRNA therapies [3], and potential applications are vast, ranging from inflammation management within a wound to personalized treatment of cancer [4,5].
Nonetheless, siRNA therapy is not without its challenges; naked siRNA administered intravenously is prone to nuclease degradation, rapid clearance, and repulsion by the cellular membrane due to its negative charge [6]. Once in the cell, siRNA faces barriers of endosomal entrapment and lysosomal degradation. These hurdles must be overcome for effective siRNA treatment. Efforts to address these challenges in siRNA delivery have largely focused on nanoparticle delivery [7]. While there are undoubtedly benefits to intravenous delivery of siRNA nanoparticles, localized administration of siRNA, when possible, may reduce both off-target effects and therapeutic load requirements. Moreover, localized delivery approaches hold promise across a variety of applications, such as treatment for bone repair, muscle regeneration, wound healing promotion, and fibrosis mitigation [5,8].
Research groups, including our own, have investigated using layer-by-layer (LbL) technology, which leverages electrostatic interactions to create conformal thin films with enhanced therapeutic loading and stability [9]. LbL has enabled coating of orthopedic implants, wound dressings, and other medically relevant substrates with proteins and nucleic acids for local therapeutic delivery [10-15]. Additionally, recent works from our lab and others have used the LbL approach to coat nanoparticles for the systemic delivery of siRNA therapies [16-22]. In general, LbL thin films can enable in vitro, ex vivo, and in vivo delivery of gene therapeutic approaches [5,24,25]. For example, we have demonstrated the coating of wound dressings and sutures for local delivery of siRNA to promote wound healing in a diabetic mouse model and to reduce scarring in a rat burn model, respectively [11,12]. These studies, among others, highlight the potential for clinical translation of LbL delivery systems for siRNA upon further system optimization.
While reports have shown varied success in delivery of siRNA via LbL thin films, the effects of assembly conditions on efficacy remain unclear. A large body of literature investigated the effects of the solution conditions, such as pH [26-28] and salt concentration [29,30], for the layering of weak polyelectrolytes into thin films [31]; however, the rigidity of siRNA means that it may not behave in a similar manner to more flexible polymer chains [32-34] which are greatly affected by changes in pH and ionic strength. A recent study in our lab investigated the effects of some of these parameters on siRNA loading in colloidal LbL systems, but differences exist between building LbL constructs on colloids versus planar surfaces, as colloids must stay stable in solution [35]. Additionally, since the parameter space of solution conditions that may influence siRNA thin film composition is so large and interdependent, a systematic approach is needed to generate rules that enable the optimized loading of siRNA into LbL thin films and promote its therapeutic efficacy.
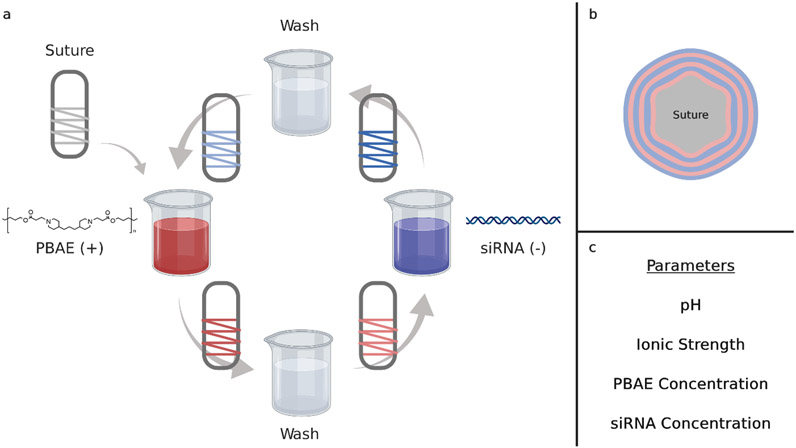
In this work, we elucidate LbL thin film fabrication parameters for optimizing efficacy of film-delivered siRNA. To complement the negatively charged siRNA in LbL assembly, we chose to use a poly(β-amino ester) (PBAE) polymer, as these polycations are known for their biocompatibility and efficacy in gene delivery [36]. Studies have shown that for nanoparticle delivery, therapeutic efficiency varies with the respective weight ratio (w/w ratio) of PBAE to siRNA [37]. Under physiological conditions, a PBAE-containing film degrades due to the hydrolyzable ester bonds in the polymer backbone and releases its components for therapeutic activity [38,39]. A schematic for assembling our LbL films is shown in Figure 1a. For these studies, LbL films were assembled on mechanically robust polyglactin 910 (Vicryl®) sutures. By alternately dipping the suture in solutions of PBAE and siRNA, with wash steps in between each deposition step, bilayer films [PBAE/siRNA]15 were conformally assembled on the suture surface (Figure 1b). Here, we investigate the effects of LbL assembly parameters on PBAE incorporation, siRNA incorporation, and the resultant ability to induce specific silencing of gene expression in vitro.
Figure 1.
Layer-by-layer coating of suture and the parameters varied. (a) Schematic representation of dip LbL assembly for coating Vicryl 3-0 suture with [PBAE/siRNA] bilayers. (b) Cross-section depiction of a suture coated with 3 bilayers. Films assembled in this study consisted of 15 bilayers. (c) List of parameters varied in the fractional factorial design. The pH (pH 4.5 – 6.0) of the entire process, ionic strength (150– 250 mM) and PBAE concentration (0.5 – 2.0 mg/mL) of the PBAE bath, and the siRNA concentration (20 – 30 μg/mL) of the siRNA bath were varied. Wash baths and the siRNA bath were buffered to 10 mM sodium acetate.
The assembly parameters that were studied consist of: pH, ionic strength, PBAE concentration, and siRNA concentration (Figure 1c). Experimental ranges were determined based on parameter values previously published by our lab [11,12,40]. Guided by Design of Experiment (DOE) principles, we conducted a fractional factorial design to reduce the number of experiments required to determine statistically significant trends. Through this approach, we systematically elucidated the effects of assembly conditions on the loading of polymer and siRNA into the film; careful tuning of these parameters is critical to enable siRNA to be taken up by cells and maintain biological activity. We also demonstrated how fundamental knowledge on the effects of solution conditions can be leveraged to rationally design solution conditions for an siRNA-releasing thin film formulation. This highlights the need for a DOE setup to identify solution conditions that yield optimal siRNA-mediated gene knockdown and how fractional factorial design can limit the number of conditions to be tested in the multifactorial solution condition design space. Optimization of siRNA incorporation in LbL thin films to maximize its efficacy is particularly important given the costs associated with therapeutic nucleic acids and the need to maximize the target biological effect with a given amount. Ultimately, this work describes vital considerations for improving gene silencing from LbL-constructed siRNA films and presents a critical, systematic evaluation of LbL film formulations. We believe this work will enhance the development of clinically translatable nucleic acid therapeutic delivery systems.
2. Experimental Section:
2.1. Materials
siRNA targeting green fluorescent protein (GFP) was purchased from Dharmacon (Lafayette, CO); AllStars Neg. siRNA AF 647 Alexa Fluor 647-labeled siRNA was purchased from Qiagen (Germantown, MD). The GFP-targeting siRNA has sequence 5’-GCA AGC TGA CCC TGA AGT TC–3’. For fluorescence microscopy and scanning electron microscopy (SEM) of sutures, a dsDNA molecule with 21 base pairs and Alexa Fluor 647 (AF647) label served as model of siRNA molecules and was acquired from IDT (Coralville, IA). The sequence was 5’-AF647-GT CAG AAA TAG AAA CTG GTC ATC-3’ (sense) and 5’-GAT GAC CAG TTT CTA TTT CTG AC-3’ (anti-sense), per the literature [41]. Lipofectamine RNAiMAX transfection reagent was obtained from ThermoFisher Scientific (Waltham, MA). Undyed, braided Vicryl 3-0 sutures were purchased from Ethicon Inc. (Somerville, NJ). RNase free UltraPure water was purchased from Life Technologies (Carlsbad, CA). Sodium acetate buffer and 99% 1,6-hexanediol diacrylate stabilized with 90 ppm hydroquinone were purchased from Alfa Aesar (Haverhill, MA). 97% 4,4’-trimethylene dipiperidine and 99.9% inhibitor-free anhydrous tetrahydrofuran (THF) were obtained from Sigma-Aldrich (St. Louis, MO). Hexanes were purchased from Fisher Scientific (Waltham, MA). Chemicals were stored per manufacturer's instructions.
HeLa cervical cancer cells stably expressing a destabilized GFP (HeLa d2eGFP) were a gift from Professor Piyush Jain’s Lab at the University of Florida. The plasmid construct used to express GFP in HeLa cells was CMV-d2eGFP-empty, which was a gift from Phil Sharp’s Lab (Addgene plasmid # 26164). Media was Corning Dulbecco’s Modified Eagle Medium (DMEM, Corning, NY) supplemented with Gibco 10% Fetal Bovine Serum (FBS, Waltham, MA) and Corning 1% Penicillin-Streptomycin (Corning, NY). The cells tested negative for mycoplasma on arrival, after thawing from storage, and periodically during culture, using a Lonza MycoAlert kit (Morristown, NJ).
2.2. Polymer synthesis
Synthesis of the PBAE polymer proceeded according to the literature [42]. Briefly, a 50 mL round bottom (RB) flask and stir bar were cleaned and dried in a drying oven. An oil bath was set to warm to 50°C with the stir rate set to 500 rpm. Meanwhile, 2.167 g (10.30 mmol, 1.02 equivalents) of 4,4’-trimethylene dipiperidine and 2.285 g (10.10 mmol, 1.0 equivalents.) of 1,6-hexanediol diacrylate were measured to the nearest half milligram and added to the RB flask. Using the Schlenk line and dry nitrogen gas to maintain the solvent as anhydrous, a needle and syringe were used to draw up about 17 mL of THF, which was added to the RB flask. A rubber stopper was used to cap the flask. Dry nitrogen gas was flushed into the reaction pot to purge any air, and the synthesis proceeded for 48 hours. The polymer was re-precipitated three times in ice-cold hexanes through a filter funnel, re-dissolving in THF between precipitations. The polymer (Mn ~ 7 kDa, Mw ~13 kDa per gel permeation chromatography against polystyrene standards) was dried and stored under inert gas in a vacuum desiccator. This synthesis has been successfully scaled up to three times the present synthesis, changing only the RB flask size to 250 mL.
2.3. Layer-by-layer film preparation
The PBAE was dissolved fresh daily using a rotating mixer due to its hydrolyzable nature and then filtered through a 0.2 μm cellulose acetate filter. Dissolution took a few hours and varied in duration based on solution conditions. The concentration of PBAE, buffer pH, and ionic strength were defined by the design of experiments setup for each run. LbL films were deposited on plasma treated Vicryl sutures. Sutures were cleaned with a 70% ethanol/30% water mixture, rinsed with water, and consequently dried. Sutures were then wrapped around a stainless steel wire frame for ease in handling. Air plasma treatment was performed for 10 minutes on high setting in a plasma cleaner (PDC-32G, Harrick, USA). Sutures were then immediately immersed in a solution of PBAE for a duration of 1 hour in order to create a well-adsorbed layer of polycation on the surface of the suture, as performed in previous work [11,12].
A Carl Zeiss HMS-DS50 slide stainer (Oberkochen, Germany) was used to automate assembly of the LbL films. Bilayer films were constructed through alternating adsorption steps. The adsorption time was determined based on previous studies in our lab using LbL to coat siRNA onto surfaces [11,12]. PBAE was adsorbed for 10 minutes, and siRNA was adsorbed for 15 minutes. Between the adsorption steps, the sutures were dipped in two wash baths of buffered RNase free water for 30 seconds each. The fractional factorial design setup determined the pH of the entire process, the ionic strength of the PBAE bath, the PBAE concentration of the PBAE adsorption bath, and the siRNA concentration of the siRNA adsorption bath. All solutions were prepared in RNase free water, adjusted to the predetermined pH with sodium acetate buffer. The wash baths and the siRNA baths were buffered to an ionic strength of 10 mM of sodium acetate. The baths consisted of the wells of 24-well plates, which can hold about 3.5 mL of solution.
Sutures were stored at −20°C until prepared for characterization and transfection studies. For consistency, coated sutures were divided into sections. Sutures from the top half were used for transfection studies, and sutures from the bottom half were used for characterization studies.
2.4. LbL film characterization
Total PBAE and siRNA incorporation were measured as follows: sutures coated with the LbL films were immersed in a 3 M NaCl solution. Sutures were incubated for 1 hour at 37°C then subjected to vigorous agitation for complete dissolution of the film. Releasate was split for quantification of PBAE and siRNA. The Pierce Micro BCA Protein Assay Kit (ThermoFisher Scientific, Waltham, MA), typically used to measure protein concentration via detection of copper ion reduction [43], was repurposed to quantify the PBAE polycation, which also contains amide groups that chelate and reduce copper, in the releasate. To quantify siRNA, 100 μL releasate was first incubated with 50 μL 0.1 M NaOH for 1 hour at room temperature to hydrolyze the PBAE. Hydrolysis of the polymer was necessary, as we found that polymer complexation with siRNA interfered with RNA quantification. The releasate was then neutralized with an equal volume of 0.1 M HCl (50 μL), and the Quant-iT RiboGreen RNA Assay Kit (Invitrogen, Waltham, MA) was used to quantify the siRNA, following manufacturer’s instructions. See Figure S1 and Figure S2 in Supporting Information for standard curves and further rationale for these quantitation methods.
2.5. Polyplex Transfection Evaluation
HeLa d2eGFP cells in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin were grown to confluence in a T75 flask at 37°C and 5% CO2. The cells were incubated for 10 minutes with 5 mL Trypsin-EDTA solution to dissociate them. The dissociation was quenched with 5 mL of warm media, and the cells were pelleted at 1000 rpm in a centrifuge for 5 minutes at room temperature. The cells were resuspended in warm media and seeded at 5,000 per well in a 96-well plate. The next day, transfection was performed. 17 pmol siRNA (approx. 274 ng of labeled siRNA or 240 ng of GFP-targeting non-labeled siRNA) was mixed at varying w/w ratios with PBAE (dissolved at 1 mg/mL in 50 mM pH 5.2 sodium acetate buffer). 50 mM pH 5.2 sodium acetate buffer was added to a total volume of 85 μL. Additionally, 17 pmol siRNA was added to 85 μL 50 mM pH 5.2 sodium acetate buffer with 0.68 μL of RNAiMax to serve as a positive control. The cell media was replaced with 75 μL of OPTI-MEM, and 25 uL of the polyplexes was added to a well in the 96-well plate. This correlates to 50 nM siRNA in the final solution. Note that each stock solution therefore was plated in triplicate, with 10 μL to spare. Three wells with just 100 μL of OPTI-MEM served as negative controls. The cells were returned to the incubator. After 6 hours, the media was replaced with warm DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. The cells were incubated for three days after treatment at 37°C and 5% CO2 and then prepared for flow cytometry.
2.6. LbL Suture Transfection Evaluation
HeLa d2eGFP cells in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin were grown to confluence in a T75 flask at 37°C and 5% CO2. The cells were incubated for 10 minutes with 5 mL Trypsin-EDTA solution to dissociate them. The dissociation was quenched with 5 mL of warm media, and the cells were pelleted at 1000 rpm in a centrifuge for 5 minutes at room temperature. The cells were resuspended in warm media and seeded at 7,000 per well in 48-well plates. The next day, transfection was performed. Three 1 cm pieces of coated suture were added per well by placing directly in the well on top of the cells. The cells were kept in DMEM. The cells were incubated for three days after treatment at 37°C and 5% CO2 and then prepared for flow cytometry.
2.7. Flow Cytometry
Three days after treatment, cells were prepared for flow cytometry. The cells were dissociated with Trypsin-EDTA, as before, and the reaction was neutralized with warm media. The cells were spun down at 1000 rpm for 5 minutes at room temperature and the old media removed. The cells were resuspended in 50 μL of cold PBS with NucBlue Live Cell Stain (2 drops/mL, per manufacturer's instructions) and incubated on ice, shielded from light, for 30 minutes. The cells were centrifuged once again with the same conditions as before, and the PBS replaced with 150 μL of OPTI-MEM. Fluorescence was measured using a BD LSR II flow cytometer with a high throughput sampler attachment in the Swanson Biotechnology Center Flow Cytometry Facility at the Koch Institute for Integrative Cancer Research. NucBlue stain fluorescence was read with a 355 nm laser and 450/50 filter set, GFP fluorescence was read with a 488 nm laser and 530/30 filter set, and AlexaFluor647 labeled scrambled siRNA was read with a 640 nm laser and 660/20 filter set. 50 μL or up to 10000 live single cells, whichever came first, were read for each sample. Analysis was performed in BD FlowJo software (Ashland, OR). Live single-cell populations were analyzed for siRNA fluorescence, signaling uptake, and GFP expression, signaling siRNA-mediated knockdown for the polyplex data. For the suture data, only the NucBlue and GFP channels were read. The gating strategy is shown in Figure S3. Histograms of the GFP fluorescence by run is shown in Figure S4.
2.8. Fluorescence microscopy
HeLa cells that express GFP were used to perform live imaging of siRNA uptake from sutures. HeLa d2eGFP cells were seeded at 5,000 cells per well in Nunc Lab-Tek II Chambered Coverglass w/ cover #1.5 with 8 wells (Rochester, NY) in 500 μL DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were allowed to adhere overnight in the same incubator conditions as in section 2.6. The next day, 2 pieces of 1 cm suture from each run were placed directly in their respective well of the chambered slide. After one day, the media was changed to OPTI-MEM supplemented with 10% FBS. This enabled imaging without fluorescence signal from the phenol in DMEM. The chamber was then sealed with parafilm and transferred to the heated 37°C environmental chamber of the microscope for live cell imaging.
An Applied Precision DeltaVision Ultimate Focus Microscope (Issaquah, WA) with inverted Olympus X71 microscope, TIRF Module, and Photometrics CoolSNAP HQ camera was used to image the live cells. This microscope, housed in the Koch Institute Microscopy Core, excites GFP at 475 nm and reads emission at 528 nm. It excites AlexaFluor647 labeled scrambled siRNA at 632 nm and reads emission at 685 nm. The 100x/1.40 objective was used with oil. Cells near the sutures were located. Care was taken not to find clusters of cells that were too close to one another, which limits ability to co-locate siRNA signal within the GFP-expressing cell. The transmittance was set to 100%, and the exposure time was set using the automated system (0.2 seconds for the GFP channel and 0.08-0.15 seconds for AlexaFluor647 channel, depending on the sample). The top and bottom of the cells in the z direction were found, and image slices of 0.2 μm were taken throughout the sample. Images were deconvolved, and a fast projection was obtained. The Softworx software was used to acquire and process images. Raw image files were loaded into ImageJ for further processing and creation of a composite image. The brightness/contrast was adjusted starting with auto and then fine tuning the minimum and maximum levels so that individual clusters of LbL siRNA material could be clearly seen, while minimizing noise. The composite image from the red and green channel was then created.
2.9. Scanning electron microscopy
To evaluate the morphology of the sutures and the overlaying coating, LBL sutures were analyzed by SEM (Zeiss Crossbeam 540, Oberkochen, Germany) and compared to an uncoated, plasma-treated specimen. Samples were prepared by freezing in liquid nitrogen and using a sharp razor blade to halve the sample perpendicular to the plane of the fibers. Representative samples for each group of study were coated with a thin layer of gold and observed under an operating voltage of 3 kV.
2.10. Statistics
Each LbL film was assembled in triplicate (n = 3) and measurements were done in duplicate. Design of Experiment (DOE) was conducted with the assistance of JMP Pro 14 statistical software (SAS, Cary, NC). JMP was used to generate Runs 1-8 of the fractional factorial design. Runs 9-12 were added as center points for pH and PBAE. Assembly parameters pH, ionic strength, PBAE concentration, and siRNA concentration served as quantitative factors. Parameter ranges: pH (4.5 - 6.0), ionic strength (150 mM - 250 mM), PBAE concentration (0.5 mg/mL - 2 mg/mL), siRNA concentration (20 μg/mL - 30 μg/mL), were determined based on parameters previously published by our lab [11,12,40]. pH 5.2 buffer and 1 mg/mL PBAE served as center points for their respective factors. JMP Pro software was used to fit the standard least squares linear regression models, determine fitting parameters for these models, calculate p values, and generate plots. Statistical significance was pre-specified at the ɑ = 0.05 level.
3. Results and Discussion
3.1. Characterization of Films
LbL films were assembled on Polyglactin 910 (Vicryl®) sutures according to the fractional factorial design (Table 1). The siRNA used was designed to target the reporter gene, green fluorescent protein (GFP). Once sutures were coated, loading was characterized, and in vitro knockdown studies were conducted as fully described in the methods section. The effects of changing the pH, ionic strength, PBAE concentration, and siRNA concentration on PBAE loading, siRNA loading, w/w ratio, and knockdown were measured. The response variables are reported as a mean with standard deviation in Table 1 and ranged from 0.57 to 1.55 μg/cm, 0.09 to 2.17 μg/cm, 0.41 to 8.62, and 0 to 37%, respectively. Thus, simply modulating the solution conditions for LbL with siRNA can improve the loading of PBAE and siRNA by a factor of approximately 3 and 24, respectively. Of note, we highlight 2 formulations. The formulation with the highest knockdown was run 6 (pH 6.0, 150 mM ionic strength, 0.5 mg/mL PBAE, 30 μg/mL siRNA), which demonstrated about 37% knockdown of GFP expression. Run 6 also had the highest siRNA loading on the suture, at 2.17 μg/cm. Only Run 4 had near this amount of siRNA loaded, while all other runs had about 75% or less this amount of siRNA. In contrast, run 7 (pH 6.0, 250 mM ionic strength, 2.0 mg/mL PBAE, 20 μg/mL siRNA) demonstrated no knockdown of GFP-expression, had the lowest siRNA loading (0.09 μg/cm), and had the second to least PBAE loading (0.67 μg/cm), suggesting poor film formation. Note that this formulation did demonstrate the highest w/w ratio, but the lack of siRNA drove this result rather than a large excess of polymer to compensate for the incorporated siRNA. No significant cytotoxicity was observed in cells treated with coated sutures (Table S1).
Table 1.
Fractional factorial design parameters and response variable data. The parameters tested include pH, ionic strength, PBAE concentration, and siRNA concentration. Ranges were determined based on parameters previously published by our lab. Response variables of PBAE loading, siRNA loading, w/w ratio, and knockdown are reported as the mean and the standard deviation (SD). Cells are colored to depict differences in magnitude. JMP was used to determine Runs 1-8 for the fractional factorial design. Runs 9 – 12 were added as midpoints for pH and polymer concentration to test for non-linearity.
| Run | pH | Ionic Strength (mM) |
PBAE Conc. (mg/mL) |
siRNA Conc. (μg/mL) |
PBAE Loading (μg/cm) |
siRNA Loading (μg/cm) |
w/w ratio | Knockdown (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||||||||
| 1 | 4.5 | 150 | 2.0 | 20 | 1.18 | 0.14 | 0.66 | 0.08 | 1.80 | 0.20 | 17.88 | 6.49 | |||
| 2 | 4.5 | 150 | 2.0 | 30 | 0.90 | 0.03 | 1.30 | 0.21 | 0.71 | 0.11 | 8.16 | 3.44 | |||
| 3 | 4.5 | 250 | 0.5 | 20 | 0.57 | 0.05 | 1.08 | 0.06 | 0.53 | 0.05 | 10.82 | 10.19 | Response Variables | High | |
| 4 | 4.5 | 250 | 0.5 | 30 | 0.80 | 0.02 | 1.95 | 0.07 | 0.41 | 0.02 | 9.23 | 2.39 | |||
| 5 | 6.0 | 150 | 0.5 | 20 | 1.55 | 0.09 | 1.42 | 0.23 | 1.13 | 0.27 | 7.54 | 3.81 | |||
| 6 | 6.0 | 150 | 0.5 | 30 | 1.30 | 0.10 | 2.17 | 0.08 | 0.60 | 0.05 | 36.96 | 17.98 | |||
| 7 | 6.0 | 250 | 2.0 | 20 | 0.67 | 0.10 | 0.09 | 0.02 | 8.62 | 3.24 | 0.00 | 0.00 | Low | ||
| 8 | 6.0 | 250 | 2.0 | 30 | 0.89 | 0.11 | 0.29 | 0.01 | 3.07 | 0.31 | 1.04 | 1.13 | |||
| 9 | 5.2 | 150 | 1.0 | 20 | 1.07 | 0.01 | 0.89 | 0.09 | 1.22 | 0.13 | 2.26 | 1.60 | |||
| 10 | 5.2 | 150 | 1.0 | 30 | 1.18 | 0.11 | 1.41 | 0.15 | 0.85 | 0.09 | 2.10 | 1.49 | |||
| 11 | 5.2 | 250 | 1.0 | 20 | 1.41 | 0.08 | 0.72 | 0.11 | 2.02 | 0.45 | 0.34 | 0.48 | |||
| 12 | 5.2 | 250 | 1.0 | 30 | 1.16 | 0.07 | 1.34 | 0.08 | 0.87 | 0.05 | 3.61 | 2.47 | |||
3.2. Least Squares Linear Regression Model
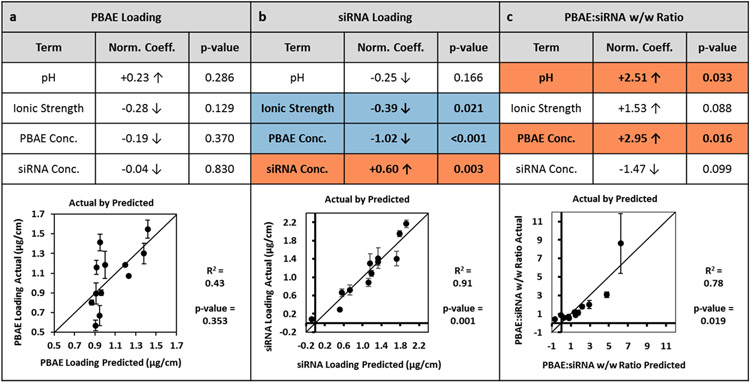
A standard least squares linear regression model was applied to PBAE and siRNA loadings, as well as their w/w ratio using the assembly parameters as the explanatory variables (Figure 2). Predicted values are plotted against actual values to visualize goodness-of-fit for each regression. PBAE loading is not well-predicted by the least squares linear regression model (R2 = 0.43). In contrast, the siRNA loading is well-predicted (R2 = 0.91). This leads to moderate prediction of the PBAE to siRNA w/w ratio (R2 = 0.78). Normalized coefficients, representing the correlation between the explanatory variable and the response, are reported for each parameter along with their p-values. While no significant trends (p < 0.05) were found for PBAE loading, several parameters were found to be statistically significant predictors of the siRNA loading and the w/w ratio. The ionic strength and PBAE concentration were negative predictors of siRNA loading while the siRNA concentration was a positive predictor of siRNA loading. pH and PBAE concentration were both positive predictors of w/w ratio. The way in which pH, ionic strength, PBAE concentration, and siRNA concentration impact the polymers both in solution and upon adsorption to the film underlies the observed trends. These trends and their implications for the observed results are explored in the subsections below.
Figure 2.
Standard Least Squares Fit of Parameters on Response Variables. A standard least squares fit was performed on the average (mean) values from each run for (a) PBAE loading, (b) siRNA loading, and (c) PBAE to siRNA w/w ratio. “Actual by Predicted” plots are shown with R2 and p-values reported. The coefficients for each parameter were normalized by the range tested and p-values are reported. Parameter terms with correlation p-values < 0.05 are highlighted; significant positive coefficients are highlighted in orange and negative coefficients are highlighted in blue. Error bars represent standard deviation.
3.2.1. Effects of pH
The assembly pH governs the percent ionization of the polymers in solution. The assembly pH was not found to have a statistically significant effect on either the PBAE loading or the siRNA loading. Nonetheless, the slight positive correlation to PBAE loading compounded with the slight negative correlation to the siRNA resulted in a significant positive coefficient for the w/w ratio. This phenomenon is supported by previous research on the effect of pH in constructing LbL films [27]. As pH increases towards the pKa, the weak electrolyte PBAE becomes less positively charged. The decrease in charge leads to a loopier conformation of the bulk polyelectrolyte in the solution bath [34]. For the less charged PBAE, more PBAE molecules are needed to compensate charge of an equivalent amount of polyanion than a more charged PBAE, explaining the increase in loading of PBAE. Since the phosphate groups on the RNA backbone have a pKa near 0, siRNA is completely ionized with a negative charge across the pH range in this study. Furthermore, the rigid conformation of siRNA remains unchanged due to its short double stranded structure. Nevertheless, the decrease in charge of the adsorbed PBAE results in a lower amount of siRNA required to neutralize the charge, and thus decreased adsorption. Overall, increasing assembly pH towards pKa increases polymer loading while decreasing siRNA loading, leading to increased polymer to siRNA w/w ratio in the thin films.
3.2.2. Effects of ionic strength
Ionic strength of the buffer can change the amount of charge shielding along the polyelectrolyte backbone during film assembly and can mediate changes in protonation of the polymers in different pH buffers. In the present study, ionic strength of the PBAE deposition bath was not found to have a significant effect on PBAE loading within the experimental range. Previous studies have reported that the thickness of a film has a parabolic dependence on salt concentration, with the maximum thickness co-dependent on pH [44]. As the ionic strength within the PBAE deposition bath increases, charge shielding occurs to extrinsically compensate the PBAE charge with acetate ions while decreasing intrinsic charge compensation by the siRNA. Thus, loopier structures of PBAE would be formed, leading to greater incorporation of the polymer. However, if the salt concentration exceeds a certain point, the charge shielding of PBAE chains becomes so extensive that adsorption to the LbL film decreases. The ionic strength at which the maximum PBAE adsorption occurs is thus related to the ionization of the PBAE and therefore the solution pH. We have observed separately that at pH 4.5, ionic strength has a positive correlation with PBAE loading while at pH 6.0, the ionic strength has a negative correlation (data not shown). As the pH shifts from 4.5 to 6.0, the solution approaches the measured PBAE pKa of about 7 (Figure S5), which represents the titratable moiety responsible for our pH-responsive polymer. Thus, the degree of ionization decreases. This supports the observed phenomenon that the ionic strength at which maximum PBAE loading occurs decreases as the pH is increased towards the pKa. Due to this interaction between pH and ionic strength, the linear regression does not comprehensively capture the effects of ionic strength on PBAE loading.
The ionic strength of the PBAE deposition bath was found to be a significant term in only the siRNA loading. As the ionic strength increased, siRNA loading decreased. This suggests that across the pH levels tested, the increased ionic strength in the PBAE deposition bath may induce swelling and partial decomposition of the film. At higher salt concentrations, extrinsic compensation occurs within the film, prompting ejection of some siRNA from the film into the PBAE bath. Thus, in the range tested, ionic strength of the PBAE deposition buffer exhibits pH-dependent effects on PBAE loading, and increased ionic strength may lead to partial decomposition of the film to decrease siRNA loading.
3.2.3. Effects of PBAE deposition bath concentration
Changing the concentration of the PBAE deposition bath could influence equilibrium adsorption of polymers in the thin film. In our experiments, the PBAE concentration did not have a significant effect on the PBAE loading in the film. This suggests that adsorption of PBAE is not diffusion limited in the range of concentrations tested. At the lowest concentration of 0.5 mg/mL PBAE, deposition equilibrium is already reached in the allotted dipping time of 10 minutes. Increase of PBAE concentration does not affect its loading.
Interestingly, the concentration of the PBAE deposition bath has a negative effect on siRNA loading. In separate experiments using fluorescently tagged siRNA, the fluorescent dye is seen in the PBAE bath at the conclusion of film assembly when high PBAE concentrations are used (data not shown), indicating that siRNA is removed from the film and solubilized in the dipping bath. This concentration-dependent stripping phenomenon has been previously reported in LbL coating of nylon fibers [45]. Within the polycation bath during film assembly, polycation is typically deposited onto the growing thin film, but this process is in equilibrium with stripping of the polyanion from the surface by positively charged polyelectrolytes in solution [46]. As polyplexes are known to have greater stability in solution in the presence of excess of one of the polyelectrolyte species [47], a greater PBAE concentration within the deposition bath would promote stability of PBAE/siRNA complexes in solution. Coupling this effect with translational and configurational entropic gains of polyplex formation over film deposition [46], increasing the PBAE concentration within our experimental range leads to increased stripping of siRNA from the film. The negative correlation PBAE concentration has on siRNA loading contributes to the significant positive correlation with the w/w ratio. Overall, we demonstrate that in the ranges tested, increasing PBAE concentration during adsorption does not affect PBAE loading, but does inversely correlate with siRNA loading.
3.2.4. Effects of siRNA deposition bath concentration
Changes to the siRNA concentration in the deposition bath could also change equilibrium adsorption. In the range tested, the PBAE loading does not seem to be affected by the siRNA concentration. The lack of the aforementioned stripping effect may be due to the relatively low concentration of siRNA within the deposition bath and the smaller size of the polycation compared to the siRNA.
The siRNA concentration exhibited a significant positive correlation to the siRNA loading. This suggests deposition of siRNA is diffusion limited in this concentration regime. As siRNA is the most expensive component of the assembly process, the concentrations used are much lower than those of the complementary PBAE by a factor of approximately 17-100, depending on the exact concentrations used as part of the fractional factorial design experimental setup. While the siRNA concentration of 20 μg/mL appears to be sufficient for adsorption and subsequent surface charge reversal for film formation, the greater concentration of 30 μg/mL results in greater loading for all formulations, holding pH, ionic strength, and PBAE concentration constant. Overall, increasing siRNA concentration during adsorption does not affect PBAE loading but increases siRNA loading.
3.3. Fluorescence microscopy of siRNA interaction with cells
To corroborate the GFP-reporter expression knockdown data, we also investigated four of the runs for their uptake of siRNA upon elution from the LbL suture. The four chosen runs were 1 (pH 4.5, 150 mM ionic strength, 2.0 mg/mL PBAE, 20 μg/mL siRNA), 6 (pH 6.0, 150 mM ionic strength, 0.5 mg/mL PBAE, 30 μg/mL siRNA), 7 (pH 6.0, 250 mM ionic strength, 2.0 mg/mL PBAE, 20 μg/mL siRNA), and 11 (pH 5.2, 250 mM ionic strength, 1.0 mg/mL PBAE, 20 μg/mL siRNA). These runs were chosen since the sutures coated by these solution conditions generated the two runs with the most and the two runs with the least knockdown, respectively. The cells are distinguished by their GFP fluorescence, and it is notable that runs 1 and 6 seem to have the greatest uptake of the fluorescently labeled control siRNA (Figure S6). In contrast, run 7, which exhibited no knockdown and very poor loading of siRNA, demonstrates almost no uptake of fluorescent siRNA. Interestingly, even though run 11 showed very poor knockdown, there is some uptake of siRNA into the cell. Without any membrane staining, it is hard to determine whether the siRNA that has been taken up is trapped in endosomes and or has escaped to the cytosol; however, qualitatively, most of the siRNA is punctate in appearance for all runs, which suggests the prior is true. It is necessary for the siRNA to escape endosomes for its biological activity, but it is believed that endosomal escape is a very rare event that occurs for only 1-2% of endosomes when using an effective lipid nanoparticle carrier [48]. Our LbL delivery system likely suffers from similar inefficiencies in endosomal escape. Use of Gal8 intracellular tracking in future experiments could help to elucidate endosomal escape of our siRNA [49]. Overall, the fluorescence microscopy data of siRNA uptake into the HeLa cells corroborates the observed knockdown in Table 1.
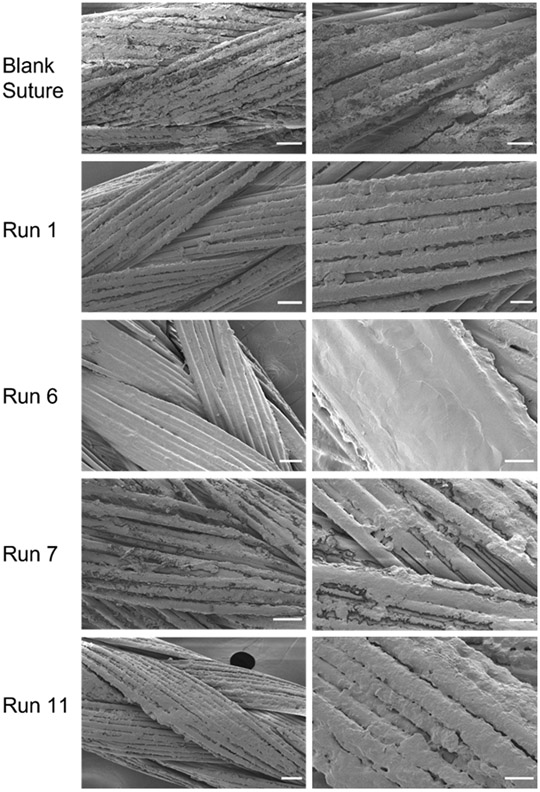
3.4. Scanning electron microscopy of LbL film surface morphology
In addition to characterizing the composition of the formed LbL films as a function of the solution conditions, it was necessary to assess how the solution conditions affect the morphology of the LbL surface formed. The same four runs as in 3.3 were studied. The fluorescently labeled siRNA has a blue hue and its qualitative coating of the surface was first evaluated with digital imaging (Figure S7), suggesting high correlation with the quantitative loading seen in Table 1.
Scanning electron microscopy was used to image the surface of the LbL sutures (Figure 3). The surface of the control sutures that received plasma treatment but no LbL coating show the braided nature of the sutures. The calcium stearate coating that is applied to all Vicryl® sutures to enhance handling properties is visualized as regions of rough coating on the surface of the smooth PLGA suture filaments [50]. Run 6 shows a heavily coated suture with a smooth morphology with thicknesses that, in some regions, appear to obfuscate the underlying braided texture of the underlying suture fibers. Runs 1 and 11 depict a moderate coating of the sutures such that the features of the underlying braided suture are apparent, but the rough original calcium stearate coating of the untreated suture appears to be totally covered. The surfaces of the LbL films in runs 1 and 11 appear rougher than that of run 6. Finally, run 7 shows some aspects of splotchy coating on the suture with areas of the rough calcium stearate on the suture surface still visible. The SEM images confirm poor coating of the suture with the solution conditions in run 7. Overall, the qualitative interrogation of the LbL thin film coating on the suture by solution conditions aligns with the compositional analysis in section 3.1.
Figure 3.
Morphological analysis of LbL thin film surface on sutures. SEM was acquired for the uncoated, plasma-treated suture and 4 different solution conditions (runs 1, 6, 7, and 11). Representative low (left) and high (right) magnification images are shown. Scale bars for low magnification images (left) represent 50 μm and those for high magnification images (right) represent 20 μm.
3.5. Optimization of Assembly Parameters for siRNA-mediated gene silencing
When the standard least squares linear regression was applied to the knockdown with the assembly parameters as the explanatory variables, significant terms were not identified (Figure S8a). Additionally, the ability to predict knockdown using these parameters was poor (R2 = 0.29). A regression using PBAE loading, siRNA loading, and w/w ratio as the explanatory variable produced a slightly better prediction (Figure S8b), albeit still not very correlative (R2 = 0.37). Despite the lack of a strong fit across variables, the analysis does indicate parameters that dominate in mediating transfection, such as siRNA loading and polymer to siRNA w/w ratio. The importance of siRNA loading on knockdown follows a dose-response relationship where higher dose leads to increased knockdown. This dose-response relationship between dosed siRNA and mRNA/resultant protein levels was similarly shown in vivo during the phase I clinical trials of patisiran, the first clinically-approved siRNA therapeutic [51]. Thus, these results are consistent with the literature and known dose-response relationships of siRNA therapies.
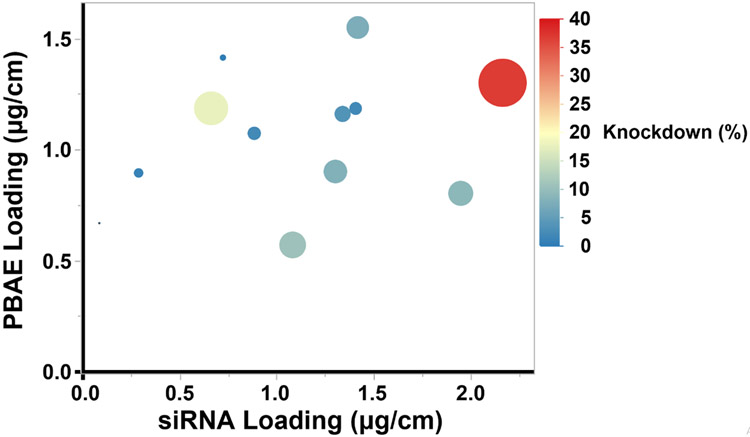
Studies have shown that nanoparticle transfection of nucleic acids is dependent on the w/w ratio of its components [37,52,53]. In vitro transfection experiments with PBAE/siRNA polyplexes confirm that siRNA uptake and knockdown increased as w/w ratio increased (Figure S9). As the N:P ratio increases in the polyplexes, uptake of fluorescent siRNA increases. Accordingly, GFP knockdown also increases with N:P ratio. To further investigate how these effects seen in the polyplexes translate to self-assembled thin films on sutures, we plotted the knockdown efficacy by the siRNA and PBAE loadings of each individual suture in a bubble plot (Figure 4). In this visualization, we find that the suture condition that produces the greatest knockdown have the greatest siRNA loading. Furthermore, the sutures of high siRNA loading show greater knockdown if they also have high PBAE loading.
Figure 4.
Bubble plot of in vitro knockdown by PBAE loading and siRNA loading. Resulting GFP knockdown data of LbL films are represented with siRNA loading on the x-axis and PBAE loading on the y-axis. Knockdown of biological replicates was averaged. LbL coated sutures that effected greater knockdown are represented with larger bubbles. The knockdown is also color coded with high knockdown in red and low knockdown in blue.
Informed by these experiments, we aimed to maximize knockdown by tuning assembly parameters to maximize siRNA loading while also maintaining an adequate w/w ratio. From the significant terms in the standard least squares fit, a low ionic strength and high siRNA concentration in our experimental range both contribute to greater siRNA loading. A high pH contributes to a greater w/w ratio. Since the polymer concentration is negatively correlated to siRNA loading and positively correlated to the w/w ratio, a moderate concentration was chosen.
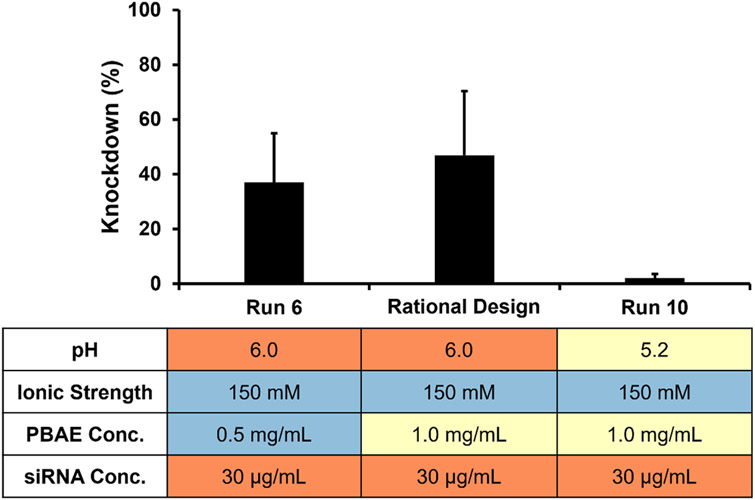
Through this study, we elucidated rules on how solution conditions effect PBAE loading, siRNA loading, PBAE to siRNA w/w ratio, and knockdown. Using rational design and knowledge of these design rules, we selected solution conditions for another formulation that was not included in the original design of experiments but that was expected to give high siRNA-mediated knockdown. Within our parameter space, the assembly conditions were thus optimized as follows: pH 6.0, ionic strength of 150 mM, PBAE concentration of 1.0 mg/mL, and siRNA concentration of 30 μg/mL. An LbL film was constructed on sutures with these conditions and in vitro knockdown was evaluated (Figure 5). The rational design conditions resulted in roughly equivalent knockdown to run 6 (pH 6.0, 150 mM buffer, 0.5 mg/mL PBAE, 30 μg/mL siRNA), but enhanced knockdown compared to Run 10 (pH 5.2, 150 mM buffer, 1 mg/mL PBAE, 30 μg/mL siRNA), which has similar solution conditions to the rational design formulation except pH. This suggests that the design of experiments selected runs that optimized the siRNA knockdown and elucidated rules that can enable rational selection of other solution conditions that produce effective knockdown.
Figure 5.
Knockdown comparison of rationally designed formulation vs. runs with neighboring conditions. The rationally selected LbL parameters of pH 6, ionic strength of 150 mM, PBAE concentration of 1.0 mg/mL, and siRNA concentration of 30 μg/mL were found to achieve in vitro knockdown of 47%. Run 6 (37% knockdown) and Run 10 (2% knockdown) from the fractional factorial design both neighbor these conditions with a single solution condition parameter varied. Colors denote high (red), moderate (yellow), and low (blue) values of parameters within the experimental range of the design. Error bars represent standard deviation.
Previous efforts from our lab in incorporating siRNA on sutures involve a hierarchical LbL structure with a [PBAE/Dextran Sulfate]20 degradable layer deposited first, followed by a [Chitosan/siRNA]25 bilayer. 37% knockdown was achieved in HeLa cells after three days of transfection [12]. Here we demonstrate that the optimized conditions result in a film that is simpler in construction (bilayer formulation vs. hierarchical structure of two bilayers) and requires fewer film layers (15 bilayers vs. a total of 45 bilayers) yet produces comparable knockdown.
In further investigation of these optimized conditions, films of 30 bilayers were constructed on the Vicryl sutures and in vitro efficacy was evaluated (Figure S10). The increased GFP knockdown with 30 bilayers compared to 15 bilayers reveals the dose-responsive nature of our LbL-coated sutures. With 30 bilayers, the optimal assembly conditions resulted in nearly 70% knockdown. This enhanced knockdown at double the number of bilayers could be essential to increasing the efficacy of localized siRNA therapies and highlights the modularity of our approach.
One notable limitation of this study is that the fractional factorial design was not set up for interaction terms. Thus, the study was not powered correctly to detect the effects of interactions between pH, ionic strength, PBAE concentration, and siRNA concentration. We were additionally limited by the need to find pH, buffer strengths, and PBAE concentrations that were compatible with complete aqueous dissolution of PBAE. Future studies could look at an expanded design of experiments that is powered to account for interactions. We also note that some of the runs demonstrate knockdown at levels that seem counter to the observed trends, particularly that increased siRNA loading is needed for effective knockdown. Further studies are needed to clarify what aspects of the solution conditions for these runs enables effective knockdown despite lower loading of siRNA. Finally, this study focuses on composition rather than structural morphology characteristics of the film itself. Further investigation of the thin film morphological characteristics could further elucidate how pH, ionic strength, PBAE concentration, and siRNA concentration impact the assembly of thin films containing siRNA on degradable sutures.
4. Conclusions
In this study we have investigated the incorporation of siRNA in LbL thin films for local delivery. By conducting a fractional factorial design, we determined the effects of assembly parameters: pH, ionic strength, PBAE concentration, and siRNA concentration on the loadings of PBAE, siRNA, and their w/w ratio in an LbL film assembled on commercially available polyglactin 910 (Vicryl®) sutures. The significant effects drawn from a standard least squares linear regression model are corroborated from theory and findings from past studies on films assembled with weak polyelectrolytes. Using the elucidated design rules of how assembly solution conditions effect siRNA-mediated knockdown from LbL thin films on sutures, we demonstrate that a rationally designed set of solution conditions that were not investigated in the original fractional factorial design can produce knockdown at levels seen in the best run from the original fractional factorial design experiment. Compared to previously published siRNA LbL films from our lab [12], films identified in this study are simpler in construction and require fewer film layers to achieve similar or even improved siRNA efficacy in vitro. The reduction of required film layers could translate to savings in time and materials in therapeutic LbL film production. The discussed effects of assembly conditions on LbL formulation suggests that investigation of values outside the parameter ranges of this study may yield even more efficient films. We believe that this work demonstrates the applicability of design of experiment principles to optimize LbL assembly conditions for modulating the loading of PBAE and siRNA into the film while also suggesting how aspects of film composition may modulate knockdown. Furthermore, this study highlights that a full factorial design is not needed to elucidate the effects of solution conditions on thin film composition. This DOE approach has the potential to aid in the rational design of process conditions for other polyelectrolyte-based self-assembled thin film delivery systems, particularly those for the delivery of other short therapeutic nucleic acids.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Short interfering RNA (siRNA) therapeutics are powerful tools to silence aberrant gene expression in the diseased state; however, the clinical utility of these therapies relies on effective controlled delivery approaches. Electrostatic self-assembly through the Layer-by-layer (LbL) process enables direct release from surfaces, but this method is highly dependent upon the specific solution conditions used. Here, we use a fractional factorial design to illustrate how these assembly conditions impact composition of siRNA-eluting LbL thin films. We then elucidate how these properties mediate in vitro transfection efficacy. Ultimately, this work presents a significant step towards understanding how optimization of assembly conditions for surface-mediated LbL delivery can promote transfection efficacy while reducing the processing and material required.
ACKNOWLEDGMENTS
The authors thank the MIT Koch Institute Swanson Biotechnology Center, which is supported by the Koch Institute Core Grant P30CA14051 from the NCI, for the use of the Flow Cytometry Core Facility, Peterson (1957) Nanotechnology Materials Core Facility, and Microscopy Core. The authors would like to thank John Martin, MayLin Howard, Sheryl Wang, and Natalie Boehnke for helpful advice on the work and manuscript. A donation to the Henrietta Lacks Foundation was made in honor of the use of HeLa cells in this work.
Funding Sources
This work was supported by the Army Research Office (grant number W911NF-18-2-0048), the National Institutes of Health (grant numbers 1R01CA235375, 1R01DE024747), and the Koch Institute/Dana Farber Cancer Institute Bridge and FootBridge funds.
ABBREVIATIONS
- siRNA
short interfering RNA
- LbL
Layer-by-Layer
- PBAE
poly(β-amino ester)
- mRNA
messenger RNA
- FDA
Food and Drug Administration
- DOE
Design of Experiments
- GFP
green fluorescent protein
Footnotes
Supporting Information
The following files are available free of charge.
Quantitation methods for PBAE and siRNA, gating strategy for flow cytometry, complete data for fractional factorial design, averaged histograms of flow cytometry data, PBAE titration curve, fluorescence microscopy, digital imaging of fluorescent sutures, standard least squares fit on knockdown, polyplex transfection, and knockdown data by number of bilayers. (PDF)
Conflicts of Interest
PTH has funding to her lab in conjunction with SomaGenics, a company focused on developing therapeutic nucleic acids for wound healing applications. PTH is on the Scientific Advisory Board of Moderna Therapeutics and the Boards of Alector and LayerBio. The authors have no other relevant disclosures.
REFERENCES
- [1].Ameres SL, Martinez J, Schroeder R, Molecular Basis for Target RNA Recognition and Cleavage by Human RISC, Cell. 130 (2007) 101–112. 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- [2].Rand TA, Petersen S, Du F, Wang X, Argonaute2 Cleaves the Anti-Guide Strand of siRNA during RISC Activation, Cell. 123 (2005) 621–629. 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- [3].O. of the Commissioner, FDA approves first-of-its kind targeted RNA-based therapy to treat a rare disease, FDA. (2020). https://www.fda.gov/news-events/press-announcements/fda-approves-first-its-kind-targeted-rna-based-therapy-treat-rare-disease (accessed May 25, 2020). [Google Scholar]
- [4].Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee S-S, Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine, Molecular Therapy - Nucleic Acids. 8 (2017) 132–143. 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Berger AG, Chou JJ, Hammond PT, Approaches to modulate the chronic wound environment using localized nucleic acid delivery, Advances in Wound Care. (2020). 10.1089/wound.2020.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Whitehead KA, Langer R, Anderson DG, Knocking down barriers: advances in siRNA delivery, Nat Rev Drug Discov. 8 (2009) 129–138. 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tatiparti K, Sau S, Kashaw SK, Iyer AK, siRNA Delivery Strategies: A Comprehensive Review of Recent Developments, Nanomaterials. 7 (2017) 77. 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sarett SM, Nelson CE, Duvall CL, Technologies for controlled, local delivery of siRNA, Journal of Controlled Release. 218 (2015) 94–113. 10.1016/j.jconrel.2015.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Alkekhia D, Hammond PT, Shukla A, Layer-by-Layer Biomaterials for Drug Delivery, Annu. Rev. Biomed. Eng (2020). 10.1146/annurev-bioeng-060418-052350. [DOI] [PubMed] [Google Scholar]
- [10].Song W, Song X, Yang C, Gao S, Klausen LH, Zhang Y, Dong M, Kjems J, Chitosan/siRNA functionalized titanium surface via a layer-by-layer approach for in vitro sustained gene silencing and osteogenic promotion, Int J Nanomedicine. 10 (2015) 2335–2346. 10.2147/IJN.S76513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Castleberry SA, Almquist BD, Li W, Reis T, Chow J, Mayner S, Hammond PT, Self-Assembled Wound Dressings Silence MMP-9 and Improve Diabetic Wound Healing In Vivo, Advanced Materials. 28 (2016) 1809–1817. 10.1002/adma.201503565. [DOI] [PubMed] [Google Scholar]
- [12].Castleberry SA, Golberg A, Sharkh MA, Khan S, Almquist BD, Austen WG, Yarmush ML, Hammond PT, Nanolayered siRNA delivery platforms for local silencing of CTGF reduce cutaneous scar contraction in third-degree burns, Biomaterials. 95 (2016) 22–34. 10.1016/j.biomaterials.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hossfeld S, Nolte A, Hartmann H, Recke M, Schaller M, Walker T, Kjems J, Schlosshauer B, Stoll D, Wendel H-P, Krastev R, Bioactive coronary stent coating based on layer-by-layer technology for siRNA release, Acta Biomaterialia. 9 (2013) 6741–6752. 10.1016/j.actbio.2013.01.013. [DOI] [PubMed] [Google Scholar]
- [14].Song W, Song X, Yang C, Gao S, Klausen LH, Zhang Y, Dong M, Kjems J, Chitosan/siRNA functionalized titanium surface via a layer-by-layer approach for in vitro sustained gene silencing and osteogenic promotion, Int J Nanomedicine. 10 (2015) 2335–2346. 10.2147/IJN.S76513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Koenig O, Neumann B, Schlensak C, Wendel HP, Nolte A, Hyaluronic acid/poly(ethylenimine) polyelectrolyte multilayer coatings for siRNA-mediated local gene silencing, PLOS ONE. 14 (2019) e0212584. 10.1371/journal.pone.0212584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Choi KY, Correa S, Min J, Li J, Roy S, Laccetti KH, Dreaden E, Kong S, Heo R, Roh YH, Lawson EC, Palmer PA, Hammond PT, Binary Targeting of siRNA to Hematologic Cancer Cells In Vivo Using Layer-by-Layer Nanoparticles, Advanced Functional Materials. 29 (2019) 1900018. 10.1002/adfm.201900018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Deng ZJ, Morton SW, Ben-Akiva E, Dreaden EC, Shopsowitz KE, Hammond PT, Layer-by-Layer Nanoparticles for Systemic Codelivery of an Anticancer Drug and siRNA for Potential Triple-Negative Breast Cancer Treatment, ACS Nano. 7 (2013) 9571–9584. 10.1021/nn4047925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Boehnke N, Correa S, Hao L, Wang W, Straehla JP, Bhatia SN, Hammond PT, Theranostic Layer-by-Layer Nanoparticles for Simultaneous Tumor Detection and Gene Silencing, Angewandte Chemie International Edition. 59 (2020) 2776–2783. 10.1002/anie.201911762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gu L, Deng ZJ, Roy S, Hammond PT, A Combination RNAi-Chemotherapy Layer-by-Layer Nanoparticle for Systemic Targeting of KRAS/P53 with Cisplatin to Treat Non–Small Cell Lung Cancer, Clin Cancer Res. 23 (2017) 7312–7323. 10.1158/1078-0432.CCR-16-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Elbakry A, Zaky A, Liebl R, Rachel R, Goepferich A, Breunig M, Layer-by-Layer Assembled Gold Nanoparticles for siRNA Delivery, Nano Lett. 9 (2009) 2059–2064. 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- [21].Tan YF, Mundargi RC, Chen MHA, Lessig J, Neu B, Venkatraman SS, Wong TT, Layer-by-Layer Nanoparticles as an Efficient siRNA Delivery Vehicle for SPARC Silencing, Small. 10 (2014) 1790–1798. 10.1002/smll.201303201. [DOI] [PubMed] [Google Scholar]
- [22].Labala S, Jose A, Venuganti VVK, Transcutaneous iontophoretic delivery of STAT3 siRNA using layer-by-layer chitosan coated gold nanoparticles to treat melanoma, Colloids and Surfaces B: Biointerfaces. 146 (2016) 188–197. 10.1016/j.colsurfb.2016.05.076. [DOI] [PubMed] [Google Scholar]
- [23].Alkekhia D, Hammond PT, Shukla A, Layer-by-Layer Biomaterials for Drug Delivery, Annual Review of Biomedical Engineering. 22 (2020) null. 10.1146/annurev-bioeng-060418-052350. [DOI] [PubMed] [Google Scholar]
- [24].Linnik DS, Tarakanchikova YV, Zyuzin MV, Lepik KV, Aerts JL, Sukhorukov G, Timin AS, Layer-by-Layer technique as a versatile tool for gene delivery applications, Expert Opinion on Drug Delivery. 0 (2021) 1–19. 10.1080/17425247.2021.1879790. [DOI] [PubMed] [Google Scholar]
- [25].Tarakanchikova YV, Linnik DS, Mashel T, Muslimov AR, Pavlov S, Lepik KV, Zyuzin MV, Sukhorukov GB, Timin AS, Boosting transfection efficiency: A systematic study using layer-by-layer based gene delivery platform, Materials Science and Engineering: C. 126 (2021) 112161. 10.1016/j.msec.2021.112161. [DOI] [PubMed] [Google Scholar]
- [26].Yoo D, Shiratori SS, Rubner MF, Controlling Bilayer Composition and Surface Wettability of Sequentially Adsorbed Multilayers of Weak Polyelectrolytes, Macromolecules. 31 (1998) 4309–4318. 10.1021/ma9800360. [DOI] [Google Scholar]
- [27].Shiratori SS, Rubner MF, pH-Dependent Thickness Behavior of Sequentially Adsorbed Layers of Weak Polyelectrolytes, Macromolecules. 33 (2000) 4213–4219. 10.1021/ma991645q. [DOI] [Google Scholar]
- [28].Choi J, Rubner MF, Influence of the Degree of Ionization on Weak Polyelectrolyte Multilayer Assembly, Macromolecules. 38 (2005) 116–124. 10.1021/ma048596o. [DOI] [Google Scholar]
- [29].Kovačević D, van der Burgh S, de Keizer A, Cohen Stuart MA, Specific Ionic Effects on Weak Polyelectrolyte Multilayer Formation, J. Phys. Chem. B 107 (2003) 7998–8002. 10.1021/jp0273777. [DOI] [Google Scholar]
- [30].Fery A, Schöler B, Cassagneau T, Caruso F, Nanoporous Thin Films Formed by Salt-Induced Structural Changes in Multilayers of Poly(acrylic acid) and Poly(allylamine), Langmuir. 17 (2001) 3779–3783. 10.1021/la0102612. [DOI] [Google Scholar]
- [31].Yuan W, Weng G-M, Lipton J, Li CM, Van Tassel PR, Taylor AD, Weak polyelectrolyte-based multilayers via layer-by-layer assembly: Approaches, properties, and applications, Advances in Colloid and Interface Science. 282 (2020) 102200. 10.1016/j.cis.2020.102200. [DOI] [PubMed] [Google Scholar]
- [32].Rana TM, Illuminating the silence: understanding the structure and function of small RNAs, Nature Reviews Molecular Cell Biology. 8 (2007) 23–36. 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- [33].Hayashi K, Chaya H, Fukushima S, Watanabe S, Takemoto H, Osada K, Nishiyama N, Miyata K, Kataoka K, Influence of RNA Strand Rigidity on Polyion Complex Formation with Block Catiomers, Macromolecular Rapid Communications. 37 (2016) 486–493. 10.1002/marc.201500661. [DOI] [PubMed] [Google Scholar]
- [34].Netz RR, Andelman D, Neutral and charged polymers at interfaces, Physics Reports. 380 (2003) 1–95. 10.1016/S0370-1573(03)00118-2. [DOI] [Google Scholar]
- [35].Correa S, Boehnke N, Deiss-Yehiely E, Hammond PT, Solution Conditions Tune and Optimize Loading of Therapeutic Polyelectrolytes into Layer-by-Layer Functionalized Liposomes, ACS Nano. 13 (2019) 5623–5634. 10.1021/acsnano.9b00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Akinc A, Anderson DG, Lynn DM, Langer R, Synthesis of poly(beta-amino ester)s optimized for highly effective gene delivery, Bioconjug. Chem 14 (2003) 979–988. 10.1021/bc034067y. [DOI] [PubMed] [Google Scholar]
- [37].Vandenbroucke RE, Geest BGD, Bonné S, Vinken M, Haecke TV, Heimberg H, Wagner E, Rogiers V, Smedt SCD, Demeester J, Sanders NN, Prolonged gene silencing in hepatoma cells and primary hepatocytes after small interfering RNA delivery with biodegradable poly(β-amino esters), The Journal of Gene Medicine. 10 (2008) 783–794. 10.1002/jgm.1202. [DOI] [PubMed] [Google Scholar]
- [38].Wood KC, Boedicker JQ, Lynn DM, Hammond PT, Tunable Drug Release from Hydrolytically Degradable Layer-by-Layer Thin Films, Langmuir. 21 (2005) 1603–1609. 10.1021/la0476480. [DOI] [PubMed] [Google Scholar]
- [39].Chuang HF, Smith RC, Hammond PT, Polyelectrolyte Multilayers for Tunable Release of Antibiotics, Biomacromolecules. 9 (2008) 1660–1668. 10.1021/bm800185h. [DOI] [PubMed] [Google Scholar]
- [40].Castleberry S, Wang M, Hammond PT, Nanolayered siRNA Dressing for Sustained Localized Knockdown, ACS Nano. 7 (2013) 5251–5261. 10.1021/nn401011n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL, Balancing Cationic and Hydrophobic Content of PEGylated siRNA Polyplexes Enhances Endosome Escape, Stability, Blood Circulation Time, and Bioactivity in Vivo, ACS Nano. 7 (2013) 8870–8880. 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lynn DM, Langer R, Degradable Poly(β-amino esters): Synthesis, Characterization, and Self-Assembly with Plasmid DNA, J. Am. Chem. Soc 122 (2000) 10761–10768. 10.1021/ja0015388. [DOI] [Google Scholar]
- [43].Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC, Measurement of protein using bicinchoninic acid, Anal. Biochem 150 (1985) 76–85. 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- [44].Dubas ST, Schlenoff JB, Polyelectrolyte Multilayers Containing a Weak Polyacid: Construction and Deconstruction, Macromolecules. 34 (2001) 3736–3740. 10.1021/ma001720t. [DOI] [Google Scholar]
- [45].Dubas ST, Limsavarn L, Iamsamai C, Potiyaraj P, Assembly of polyelectrolyte multilayers on nylon fibers, Journal of Applied Polymer Science. 101 (2006) 3286–3290. 10.1002/app.23826. [DOI] [Google Scholar]
- [46].Sui Z, Salloum D, Schlenoff JB, Effect of Molecular Weight on the Construction of Polyelectrolyte Multilayers: Stripping versus Sticking, Langmuir. 19 (2003) 2491–2495. 10.1021/la026531d. [DOI] [Google Scholar]
- [47].Dautzenberg H, Kriz J, Response of Polyelectrolyte Complexes to Subsequent Addition of Salts with Different Cations, Langmuir. 19 (2003) 5204–5211. 10.1021/la0209482. [DOI] [Google Scholar]
- [48].Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stöter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M, Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape, Nature Biotechnology. 31 (2013) 638–646. 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- [49].Kilchrist KV, Dimobi SC, Jackson MA, Evans BC, Werfel TA, Dailing EA, Bedingfield SK, Kelly IB, Duvall CL, Gal8 Visualization of Endosome Disruption Predicts Carrier-Mediated Biologic Drug Intracellular Bioavailability, ACS Nano. 13 (2019) 1136–1152. 10.1021/acsnano.8b05482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tomihata K, Suzuki M, Sasaki I, Surgical suture, US6616687B1, 2003. https://patents.google.com/patent/US6616687B1/en (accessed July 23, 2021).
- [51].Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, Perez J, Chiesa J, Warrington S, Tranter E, Munisamy M, Falzone R, Harrop J, Cehelsky J, Bettencourt BR, Geissler M, Butler JS, Sehgal A, Meyers RE, Chen Q, Borland T, Hutabarat RM, Clausen VA, Alvarez R, Fitzgerald K, Gamba-Vitalo C, Nochur SV, Vaishnaw AK, Sah DWY, Gollob JA, Suhr OB, Safety and Efficacy of RNAi Therapy for Transthyretin Amyloidosis, New England Journal of Medicine. 369 (2013) 819–829. 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- [52].Lavertu M, Méthot S, Tran-Khanh N, Buschmann MD, High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation, Biomaterials. 27 (2006) 4815–4824. 10.1016/j.biomaterials.2006.04.029. [DOI] [PubMed] [Google Scholar]
- [53].Mao S, Sun W, Kissel T, Chitosan-based formulations for delivery of DNA and siRNA, Advanced Drug Delivery Reviews. 62 (2010) 12–27. 10.1016/j.addr.2009.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.