Abstract
Recent molecular characterizations of microbial communities from deep-sea hydrothermal sites indicate the predominance of bacteria belonging to the epsilon subdivision of Proteobacteria (epsilon Proteobacteria). Here, we report the first enrichments and characterizations of four epsilon Proteobacteria that are directly associated with Alvinella pompejana, a deep sea hydrothermal vent polychete, or with hydrothermal vent chimney samples. These novel bacteria were moderately thermophilic sulfur-reducing heterotrophs growing on formate as the energy and carbon source. In addition, two of them (Am-H and Ex-18.2) could grow on sulfur lithoautrotrophically using hydrogen as the electron donor. Optimal growth temperatures of the bacteria ranged from 41 to 45°C. Phylogenetic analysis of the small-subunit ribosomal gene of the two heterotrophic bacteria demonstrated 95% similarity to Sulfurospirillum arcachonense, an epsilon Proteobacteria isolated from an oxidized marine surface sediment. The autotrophic bacteria grouped within a deeply branching clade of the epsilon Proteobacteria, to date composed only of uncultured bacteria detected in a sample from a hydrothermal vent along the mid-Atlantic ridge. A molecular survey of various hydrothermal vent environments demonstrated the presence of two of these bacteria (Am-N and Am-H) in more than one geographic location and habitat. These results suggest that certain epsilon Proteobacteria likely fill important niches in the environmental habitats of deep-sea hydrothermal vents, where they contribute to overall carbon and sulfur cycling at moderate thermophilic temperatures.
Several recent molecular studies have demonstrated the presence and dominance of bacteria belonging to the epsilon subdivision of Proteobacteria (epsilon Proteobacteria) that are both free-living and found in association with metazoans at deep-sea hydrothermal vents (4, 5, 14, 25, 29, 35; Campbell et al., unpublished data). Epsilon Proteobacteria have also been detected and/or isolated from deep subsurface sediments, oil fields, activated sludge, and marine snow (13, 20, 34, 40, 45). Until now, however, epsilon Proteobacteria have not been cultured from hydrothermal vent environments. All epsilon Proteobacteria isolated to date are involved in the sulfur cycle by either reducing elemental sulfur to sulfide or oxidizing sulfide to sulfur. In many cases, a single bacterium is able to do both (24, 37). A hallmark of the epsilon Proteobacteria is their ability to utilize a variety of electron acceptors, including oxygen (under microaerophilic conditions), nitrate, several sulfur species, and, in some cases, arsenate, selenate, manganese, and Fe(III) (12, 18, 27, 28, 42). Because of these capabilities, it is not surprising that they flourish at hydrothermal vents, where there are high levels of many sulfur species as well as an abundance of heavy metals (10, 17, 22, 23, 36).
Although inferences can be drawn about the biochemistry of hydrothermal vent epsilon Proteobacteria, little is actually known about the chemical and thermal conditions needed for the growth of this dominant bacterial group. There is one report that suggests that an epsilon Proteobacteria belonging to the Arcobacter group is involved in filamentous sulfur production at hydrothermal vents, although it has not been isolated or phylogenetically characterized (44). Similar filamentous production of sulfur occurred in continuous-flow H2S reactors with an Arcobacter sp. isolated from shallow coastal marine waters (43).
Our laboratory has been investigating the symbiotic relationship between Alvinella pompejana, a deep-sea hydrothermal vent polychete, and the morphologically and phylogenetically diverse episymbiont community that is integrated into its dorsal epithelium (4, 5, 7, 14). We have demonstrated through a variety of molecular techniques that members of a single clade of the epsilon Proteobacteria dominate the microbial community (4, 5, 14). Several attempts have been made in the past to isolate A. pompejana epibionts under mesophilic, aerobic, and heterotrophic conditions (16, 31–33). In a recent study, we confirmed that these attempts did not isolate any epsilon Proteobacteria and that members of the A. pompejana episymbiont community identified previously by molecular studies (14) are not present in these extensive culture collections (Campbell et al., unpublished data).
The goal in this study was to isolate epsilon Proteobacteria from the dorsal epithelium of A. pompejana by enrichment culture techniques. Positive enrichments would likely further our understanding of the biochemical conditions necessary for both epibiont and free-living bacterial growth. In addition, we extended our epsilon Proteobacteria isolation attempts to sulfidic chimney samples from hydrothermal vents at 13°N latitude along the East Pacific Rise (EPR) and at the Guaymas basin. A molecular survey of various hydrothermal vent sites was also performed to investigate the ecology of two novel epsilon Proteobacteria that were enriched from the 13°N A. pomejana epibiont community.
MATERIALS AND METHODS
Sampling and enrichment conditions.
Initial enrichments were from A. pompejana worms sampled from various hydrothermal vents along the EPR during the Amistad cruise, May to June 1999, to 13°N (12°49′N, 103°56′W) at a depth of approximately 2,500 m (Table 1). A. pompejana specimens (in their associated tubes) were collected and transported to the surface via the deep-submergence vehicle (DSV) Nautile in an enclosed container. Once on board, A. pompejana worms were removed from their tubes and associated chimneys, and washed three times in sterile 0.22-μm-filtered seawater, and the epibiont community (found mainly on the hair-like projections) were scraped into a sterile 50-ml tube. The hairs were slightly homogenized with a 20-gauges needle in a final volume of 10 ml containing sterile seawater and aseptically transferred to anaerobic medium (described below). A portion of the homogenate was saved for DNA extraction and preservation in glycerol at −80°C.
TABLE 1.
Details of samples used for enrichments and PCR analysis
| Site | Vent (latitude, longitude) | Dive | Specimen type | Designation | Growth pattern
|
|
|---|---|---|---|---|---|---|
| Am-H-like | Am-N-like | |||||
| 13°N | Grandbonum (12°48′7"N, 103°56′4"W) | AM01, AM13, AM17 | Hairs | A, N, X | − | + |
| PP55 (12°49′84"N′, 103°56′8"W) | AM08 | Hairs | H | + | − | |
| PP57 (12°50′32"N, 103°56′80"W) | AM14 | Hairs | P | − | − | |
| PPHot14 (12°48′18"N, 103°56′39"W) | AM12 | Chimney | M | ND | − | |
| Grandbonum (12°48′7"N, 103°56′4"W) | AM04 | Hairs | E | − | − | |
| PPHot14 (12°48′18"N, 103°56′39"W) | AM04 | Hairs | F, G | − | − | |
| PP57 (12°50′32"N, 103°56′80"W) | AM16 | Hairs | R | − | − | |
| 9°N | Q vent (9°50′727"N, 104°17′58"W) | 3395 | Chimney | 97 chim. | ND | ND |
| M vent (9°50′83"N, 104°17′58"W) | 3399 | Chimney | 215 chim. | ND | ND | |
| Guaymas | K2 (27°00′84"N, 111°24′48"W) | 3518 | Flange | 18.1 | − | + |
| Robin's Roost (27°00′88"N, 111°24′63"W) | 3518 | Flange | 18.2 | + | − | |
| Robin's Roost (27°00′88"N, 111°24′63"W) | 3519 | Chimney | 19.1 | − | + | |
| K2 (27°00′84"N, 111°24′48"W) | 3523 | Flange | 23.2 | + | − | |
| Kristin's Summit (27°00′83"N, 111°24′68"W) | 3523 | Flange | 23.3 | + | − | |
| Rebecca's Roost (27°00′66"N, 111°24′42"W) | 3517, 3521 | Flange | 17.1, 21.1 | − | − | |
Growth with acetate, pyruvate, formate, and sulfur at 50°C for the 13°N samples and acetate, formate and sulfur at 45°C for the Guaymas samples. Based on morphologic and identical migrations on DGGE. ND, not done.
For the enrichments from chimney samples, sulfides were collected from the Guaymas basin, a sediment- and hydrocarbon-rich hydrothermal site in the Gulf of California (27°00′N, 111°24′W) at a depth of approximately 2,000 m, during the Extreme 2000 cruise, January 2000 (Table 1). The outsides of chimneys were scraped aseptically into sterile tubes, and anaerobic sterile seawater was added (10×, vol/vol).
Approximately 1 ml of diluted sample (hairs or chimney) was used for each enrichment.
Medium used for enrichments was modified from that of Widdel and Bak (47) and contained (per liter): 20 g of NaCl, 3 g of MgCl2 · 6H2O, 0.15 g of CaCl2 · 2H2O, 0.5 g of KCl, 0.25 g of NH4Cl, 0.2 g of KH2PO4, 1 ml of trace element solution (46), 1 ml of selenite-tungstate solution, 0.015 g of resazurin, 30 ml of 1 M NaHCO3, 1 ml of vitamin mixture solution, 1 ml of vitamin B12 solution, 1 ml of thiamine solution, and 5 ml of 0.2 M Na2S as a reductant (47). Elemental sulfur (approximately 5 g/liter) was sterilized by heating to 100°C three times and added aseptically after the medium was autoclaved. The final pH was adjusted to approximately 7.0, and the headspace consisted of N2-CO2 (80:20; 150 kPa).
A combination of three potential electron donors and carbon sources (formate [20 mM final concentration], acetate [2 mM final concentration], and pyruvate [20 mM final concentration]) was added separately from sterile, anoxic stocks to individual tubes before inoculation for the 13°N enrichments. Enrichments from the chimneys collected at Guaymas basin were performed in the sulfur medium with added formate and acetate. The A. pompejana enrichments were incubated at 30, 50, and 65°C, while the chimney enrichments were incubated at 45 and 60°C. Growth was monitored microscopically.
Positive enrichments were subcultured five times on board ship and/or in the laboratory until stable cultures (cultures that did not change, based on microscopic or molecular analysis) were obtained. These subcultures were considered pure when microscopic and molecular evidence indicated only one type of bacterium per culture. DNA was extracted from all the positive enrichments and stable cultures and subjected to DNA fingerprinting analysis (denaturing gradient gel electrophoresis [DGGE]) as described previously (4, 38). Universal primers for both bacteria and archaea were used in the DGGE analysis to assess the purity of the cultures. In addition purity of the cultures was assessed by microscopic analysis.
Growth characterizations.
Two of the epsilon Proteobacteria cultures that were considered pure (Am-H and Am-N) were subjected to limited physiological assessment. Initial cultures were grown in the sulfur medium described above with the addition of formate and acetate as potential electron donor and carbon source, respectively.
The temperature range tested was from 30 to 65°C. Other carbon sources (CO2, pyruvate [20 mM], fumarate [0.2%], 0.2% peptone, formate [10 mM], and acetate [10 mM]), electron donors (H2 and formate), and electron acceptors [sulfite (5 mM), thiosulfate (10 mM), and Fe(III)] were evaluated. In addition, growth with various gas mixtures (H2-CO2, 90:10, 150 kPa, and N2, 100%, 150 kPa) was tested. Growth using Fe(III)-oxyhydroxide was evaluated on media prepared and manipulated as above (47).
We modified the medium for Fe reducers by adding 2 mM ferrous chloride as a reductant in place of dissolved sulfide. Amorphous Fe(III)-oxyhydroxide was added to the autoclaved medium as the sole electron acceptor to a final concentration of 50 mM. The Fe(III) oxide was prepared by neutralizing a solution of FeCl3, rinsing, and autoclaving as described previously (21). All medium preparation and manipulations were carried out under strictly anoxic conditions unless otherwise specified. Sterile medium components for the Fe(III) medium were combined, and the medium was dispensed into serum bottles which were sealed with butyl rubber stoppers under a gas stream of 90% N2 and 10% CO2 (100 kPa). Inocula for all the metabolic characterizations were 1/20th volume. Positive cultures were subcultured an additional time to confirm growth in the tested medium (and not growth from the original inoculum). Negative cultures were tested from the source inoculum at least twice. Additional negative controls included growth with no added substrates (other than the basal minimal medium with and without added sulfur). Cells were counted after 3 days by epifluorescent microscopy after fixation with 3.7% formaldehyde, staining with DAPI (4′,6′-diamidino-2-phenylindole) (2 μg/ml), and filtration onto a 0.22-μm polycarbonate filter (30). Growth was scored as positive if there was a greater than fivefold increase in cells compared to control tubes with no added substrates.
Growth curves of the stable subcultures were performed four times at their optimal temperatures in minimal enrichment medium with added sulfur and formate (20 mM) under an N2-CO2 gas headspace. Growth was also measured in sulfur medium without formate. Cells were counted by epifluorescent microscopy as described above.
Hydrogen sulfide production was measured using the Cline method (6). Light photomicrographs were obtained after staining with DAPI (2 μg/ml) as described above. The lengths and widths of the bacteria were measured on an Olympus Provis AX70 microscope using a 100× objective with a Chroma 31000 band pass filter set. Lengths and widths of the bacteria were estimated from a frequency plot of the values for approximately 100 individual bacteria.
Phylogenetic assessment.
The 16S ribosomal DNAs (rDNAs) of the bacteria were amplified from extracted DNA using the 21F and 1518R primers as described previously (14) and cloned into a Topo-TA vector (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. The resulting 16S rDNA clones were bidirectionally sequenced on an ABI 310 sequencer (Applied Biosystems, Inc. [ABI], Foster City, Calif.) using the TA vector-specific primers M13F and M13R (Invitrogen) as well as 519F, 519R, 1100F, and 1100R (1). DNA sequences were assembled using the ABI Autoassembler program (ABI) and aligned to other 16S rDNA sequences using Genetic Data Environment (GDE) (39) as described previously (14). DNA distance similarities were determined by the method of Olsen (26). Neighbor-joining and parsimony trees were obtained in GDE as previously described (14).
Presence of bacteria in the environment.
DNA was extracted from the A. pompejana epibiont samples listed in Table 1 using an Isoquick DNA extraction kit (ORCA Research, Bothwell, Wash.) as described previously (4). Several DNA extraction protocols were used on various chimney-flange samples (Table 1) to evaluate extraction efficiencies and potential PCR inhibition effects.
DNA was initially extracted from approximately 500 μl of ground chimney samples (slurries) from 9°N and 13°N with acetyltrimethylammonium bromide–polyvinylpyrrolidone–β-mercaptoethanol (CTAB/PVP/β-ME) method as described previously (8) and resuspended in 50 μl of sterile H2O. We found better yields and less inhibition when extracting from an equal amount of chimney slurry with the QIAamp DNA stool mini kit (Qiagen, Valencia, Calif.). DNA was extracted from the Guaymas chimney-flange samples using this kit and resuspended in 50 μl of sterile H2O. From 1 to 10 ng of DNA was used in PCR for DGGE analysis as described previously (4, 38). The universal forward primer 338F (with a GC clamp) was also used in combination with two strain-specific 16S rDNA primers (for Am-H, H607R [5′- CTCCCGAACTCTAGTCTGA], and for Am-N, N601R [5′- CTAGATAAACAGTTTCAAGA], based on Escherichia coli numbering [3]) in PCR amplifications for DGGE to determine the presence of strain Am-H or Am-N in the indicated samples.
Nucleotide sequence accession numbers.
The 16S rDNA sequences for Am-H, Am-N, Ex-18.1, and Ex-18.2 were deposited in GenBank and assigned accession numbers AF357197, AF357198, AF357199, and AF357196, respectively.
RESULTS
Enrichments.
Enrichments from A. pompejana samples collected from 13°N that were grown at 30°C yielded bacteria that varied widely in their morphologies, while little to no growth occurred at 65°C. Successful enrichments of two morphologically different populations of bacteria that were grown at 50°C were obtained from A. pompejana samples collected from two separate hydrothermal vent sites (Table 1). Initially, both enrichments contained a dominant bacterial morphotype, with several minor morphotypes. After subculturing, only the dominant bacterium in each enrichment was detected by DGGE and microscopy. The first bacterial morphotype recovered from three separate A. pompejana specimens (A, N, and X) grew on acetate-formate-sulfur medium (Table 1). These cultures consisted of slow-growing (doubling time, >24 h at 50°C) motile vibrioid cells (Table 2). Stable subcultures of the dominant morphotypes were obtained after decreasing the incubation temperature to 45°C. DGGE analysis of the three subcultures demonstrated three identically migrating bands that were indistinguishable by sequence analysis of 110 bp (data not shown). These strains were considered similar. Therefore, a subculture from the N enrichment was chosen to be characterized; it was designated Am-N.
TABLE 2.
Characteristics of Am-N and Am-H enriched from A. pompejana episymbiont biomass
| Characteristic | Am-N | Am-H |
|---|---|---|
| Morphology | Curved rods | Slightly curved rods |
| Motility | + | + |
| Length (μm) | 0.8 | 0.4 |
| Width (μm) | 0.3 | 0.3 |
| Temp range (°C) | 30–50 | 30–55 |
| Temp optimum (°C) | 41 | 45 |
| Growtha on carbon source (in presence of S0) | ||
| Acetate | − | − |
| Formate | +b | + |
| Pyruvate | − | + |
| Autotrophic growth (with H2 in presence of S0) | − | + |
| Electron donor (with S0) | ||
| H2 | − | + |
| Acetate | − | − |
| Formate | + | + |
| Pyruvate | − | + |
| Fermentation | ||
| Fumarate | + | − |
| Pyruvate, peptone | − | − |
| Electron acceptorc | ||
| Thiosulfate, sulfite | − | − |
| Fe(III)-oxyhydroxide | − | − |
As measured by turbidity, microscopic counts, and H2S production.
Positive growth on sulfur granules only (microscopic evaluation and H2S production; cultures were not turbid).
In the presence of formate and acetate.
The second bacterial morphotype (Am-H) was enriched from a single A. pompejana specimen collected from PP55, also located along the EPR at 13°N (Table 1). After several subcultures in the same medium as above, it was also confirmed to be a single bacterium by both microscopy and DGGE analysis (data not shown). Compared to the first bacterium, it grew faster at 50°C, was smaller in size, and was also motile (Table 2).
Two other epsilon Proteobacteria, designated Ex-18.1 and Ex-18.2, were enriched from several chimney samples collected from hydrothermal vents in the Guaymas basin using similar conditions as above, except the initial incubation temperature was reduced to 45°C and the sulfur medium contained only formate and acetate. The first bacterium, Ex-18.1, was morphologically and phylogenetically similar to Am-N. Morphologic and DGGE analysis of enrichments from K2 and Robin's Roost indicated that bacteria identical to Ex-18.1 were also found at these vent sites (Table 1 and data not shown). The types of samples used in the enrichments were somewhat different; the sample from K2 was a flange outcropping, while the sample from Robin's Roost was a sulfidic chimney. The second chimney bacterium, Ex-18.2, was enriched from three other samples collected from Guaymas: Robin's Roost flange, another K2 flange, and a flange collected from Kristin's Summit (Table 1 and data not shown). According to morphologic and DGGE analysis, it was morphologically and phylogenetically similar to Am-H.
Preliminary characterization of bacteria and growth rates.
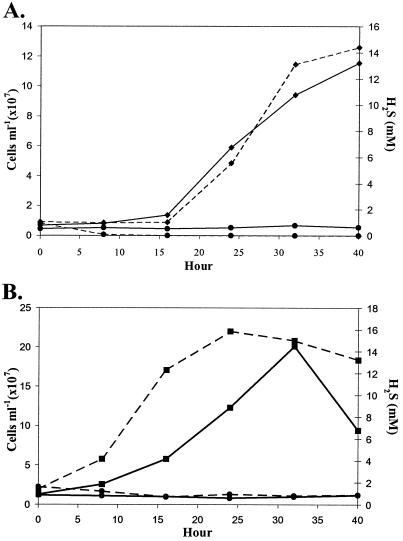
Two of the isolates, Am-H and Am-N, were chosen for further characterization. Am-H and Am-N are slightly curved rods with widths of 0.3 and 0.3 μm and lengths of 0.4 and 0.8 μm, respectively (Table 2). Am-H is highly motile, while Am-N is less motile. As shown in Table 2, the temperature growth ranges of these organisms varied slightly; Am-H generally grew at higher temperatures (up to 55°C but not above) and had a higher temperature optimum (45°C) than Am-N (50 and 41°C, respectively). Growth curves for the two bacteria were performed at least four separate times in a basal minimal medium with added elemental sulfur and formate. Representative curves are illustrated in Fig. 1. Am-N had a slightly longer doubling time than Am-H (9 h versus 6 h, respectively), as calculated by the slope of the growth curves during the linear phase of growth (Fig. 1). Based on the graphic comparison of the number of cells per mole of H2S produced, growth yields of Am-H were approximately twice that of Am-N (data not shown).
FIG. 1.
Representative growth curves of strains Am-H (A, ⧫) and Am-N (B, ■). Cell densities (solid lines) and hydrogen sulfide production (dashed lines) were measured with formate as the electron donor and elemental sulfur as the electron acceptor at their individual temperature optimums under anaerobic conditions (N2-CO2 gas phase). Controls were inoculated tubes without added formate (●).
Growth of Am-N and Am-H was tested with a limited series of gas mixtures, electron acceptors, and carbon sources (Table 2). Both Am-N and Am-H grew heterotrophically using formate as a carbon source and sulfur as the electron acceptor. They were not able to use thiosulfate, sulfite, and Fe(III)-oxyhydroxide as alternative electron acceptors in the presence of formate. None used acetate as an energy and carbon source. With sulfur, Am-H was able to grow lithoautotrophcally using hydrogen and formate as electron donors and heterotrophically in the presence of pyruvate. Fermentation of fumarate was performed by Am-N. The ability of Am-N and Am-H to grow under low levels of oxygen and with nitrate as electron acceptors was not tested.
Phylogenetic affiliations.
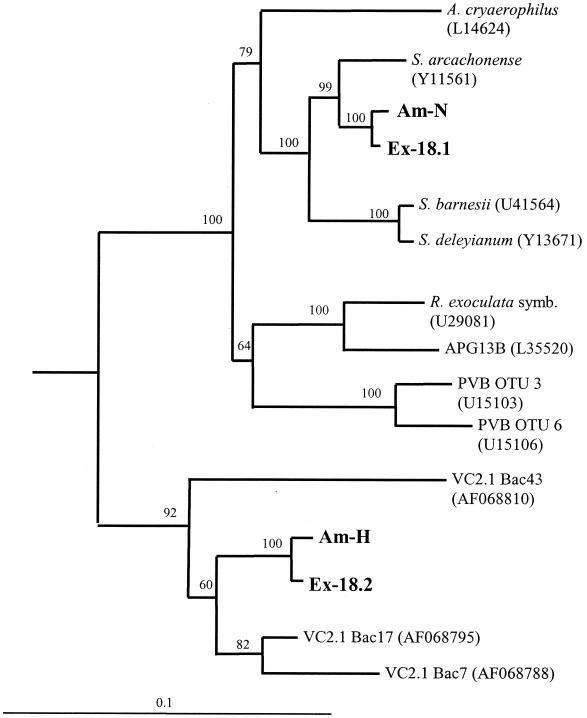
According to their 16S rDNA sequences, Am-N and Am-H and their close relatives from the Guaymas basin (Ex-18.1 and Ex-18.2) grouped into the epsilon subdivision of the Proteobacteria (Fig. 2). Am-N showed 99.2% identity with Ex-18.1 and 96.5 and 94.5% identity with Sulfurospirillum arcachonense and Sulfurospirillum barnesii, respectively, by DNA distance analysis. Am-H and Ex-18.2 (Fig. 2) were much more distantly related to the Sulfurospirillum, grouping into a previously described deeply branching epsilon clade that contains uncultured 16S rDNA clones from a hydrothermal vent cap deployed at the Snake Pit vent along the mid-Atlantic ridge (35). Am-H was 99% identical to Ex-18.2 and showed 95.4 and 87.5% identity to VC2.1 Bac43 and VC2.1 Bac30, respectively.
FIG. 2.
Phylogenetic tree showing the relationships between isolated strains with other members of the epsilon subdivision of the Proteobacteria. The trees are based on alignments of approximately 1,500 bp from the 16S rDNA gene minus insertions, deletions, and ambiguous bases. E. coli was used as the outgroup. Bootstrap values from 100 resamplings are indicated prior to the branch points of the tree. Sequences from the isolates are marked in boldface type. The scale bar represents the calculated number of changes per nucleotide position.
Ecological significance of isolates.
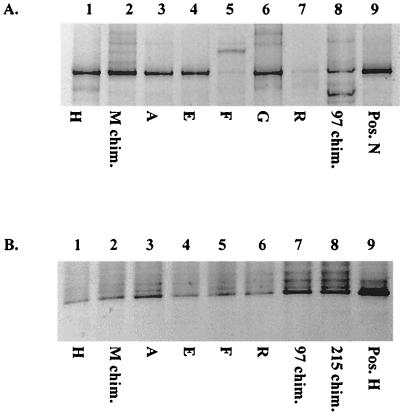
Nine A. pompejana samples and nine chimney or flange samples (samples designated by letters and numbers in Table 1) were tested by PCR with strain-specific primers, followed by DGGE analysis for the presence of bacteria with migration patterns identical to those of either Am-N or Am-H (Fig. 3). Positive PCRs which migrated identically to Am-N on a DGGE gel were obtained from all A. pompejana specimens tested from the 13°N latitude with the primer designed to specifically amplify Am-N (representative amplifications are shown in Fig. 3A). Bands migrating identically to Am-N were also amplified from DNA extracted from two chimney samples, one from 13°N (M chim.) and one from 9°N (97 chim.). No positive PCRs were obtained with the Am-N-specific primer with DNA extracted from one chimney sample from 9°N (215 chim.) or from any of the samples collected from the Guaymas basin. Similar results were obtained with an Am-H-specific primer. However, two A. pompejana specimens were negative (Am-G and Am-N), while all the chimney samples from 9°N and 13°N were positive (Fig. 3B). Very weak amplification products were obtained with an Am-H-specific primer on two samples collected from the Guaymas basin, and three others were negative.
FIG. 3.
DGGE of positive PCR products obtained after amplification using strain-specific primers for Am-N (A) and Am-H (B). Lanes 1 to 8, separated amplification products obtained from samples of various A. pompejana epibiont biomass and hydrothermal chimney samples as listed in Table 1. Lane 9, positive controls (Am-N in A, Am-H in B). A slight frown occurred in the gel shown in panel B.
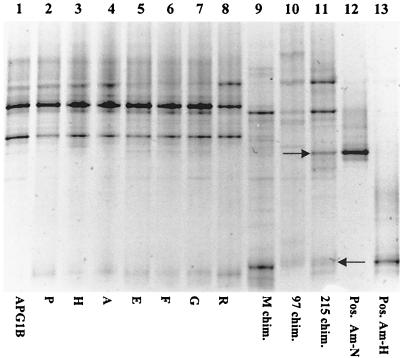
The A. pompejana worms and the three chimney samples from 9°N and 13°N were also tested for the presence of the isolates Am-H and Am-N by DGGE analysis with universal primers to detect all bacteria present in the samples (Fig. 4). The A. pompejana bacterial communities from 13°N contained very similar members, as indicated by the number of identically migrating bands. However, no bands corresponding to Am-H or Am-N were observed on the gel, suggesting that these bacteria were minor members of the communities tested. This was also the case for two of the chimney samples (M and 97). Chimney 215 did have observable bands migrating similarly to PCR amplicons from the bacterial cultures Am-H and Am-N (indicated by the arrows in Fig. 4). After sequencing these highly visible bands, it was determined that they were not identical to either Am-H or Am-N, confirming the previous negative PCR results with the Am-N-specific primers on the sample from chimney 215.
FIG. 4.
DGGE of positive PCR products obtained after amplification using universal primers. Lanes 1 to 11, separated amplification products obtained from samples of various A. pompejana epibiont biomass and hydrothermal chimney samples as listed in Table 1. Lanes 12 and 13, positive controls. Arrows indicate bands that were reamplified for DNA sequencing.
DISCUSSION
This is the first report of the isolation of Proteobacteria belonging to the epsilon subdivision from deep-sea hydrothermal vents. Our and previous PCR-based experiments demonstrated that in hydrothermal vent environments, epsilon Proteobacteria dominate the free-living organisms on the outer surfaces of chimneys and/or are closely associated with invertebrate hosts (4, 5, 14, 25, 29). Large percentages of diverse epsilon Proteobacteria have also been detected from a vent cap deployment along the mid-Atlantic ridge (35). These reports suggest that the epsilon subdivision of Proteobacteria plays a major role in the bacterial communities at hydrothermal vents. One of the strains described here (Am-H) has some properties similar to other cultured epsilon Proteobacteria, such as growth with fumarate. However, both strains are novel in that they grow at moderate thermophilic temperatures. While we have no direct evidence for carbon fixation by Am-H, the deeply branching epsilon Proteobacteria are able to autotrophically use elemental sulfur, hydrogen, and CO2 for growth under anaerobic conditions.
Autotrophy involving the reduction of elemental sulfur is not unique to the Proteobacteria, but has only been described in one other previously identified epsilon Proteobacteria (an Arcobacter sp.) isolated from oil field brine (13). Until this report, all anaerobic elemental sulfur-reducing chemolithoautotrophic bacteria described from hydrothermal vents were thermophiles and hyperthermophiles (2, 19, 41). Many sulfur-oxidizing chemolithoautotrophs have been described from marine environments, including hydrothermal vents, but these microorganisms oxidize sulfides in the presence of O2 (9, 15). Alternate chemolithoautotrophic metabolisms involving the disproportionation of elemental sulfur have been described in marine environments (11), but have not been described from bacteria isolated at deep-sea hydrothermal vents or by epsilon Proteobacteria. It seems likely that the deeply branching epsilon Proteobacteria described here (Am-H), and possibly other phylogenetically similar bacteria (35), fill an important niche in the environmental habitat of deep-sea hydrothermal vents, where they may contribute both to an increase in biomass and to overall carbon production.
Two of the epsilons (Am-N and Ex-18.1) described in this report phylogenetically group with the Sulfurospirillum spp., a distinct clade within the epsilon subdivision of Proteobacteria (12, 37, 42). Other members of the Sulfurospirillum group are not able to grow at 42°C but have pH requirements similar to that of Am-N. Sulfurospirillum spp. also use a variety of electron donors and are able to ferment fumarate (42). S. arcachonense, the closest phylogenetic representative to Am-N, also seems very close metabolically since, like Am-N, it does not reduce thiosulfate, sulfite, and Fe(III). However, another species of this genus, S. barnesii, is able to use a diverse spectrum of electron acceptors, including arsenate, selenate, and Fe(III) (18, 27, 42), indicating the potential of diverse physiological abilities of these bacteria, an adaptation certainly appropriate for organisms thriving in deep-sea hydrothermal vent environments.
The epsilon Proteobacteria described in this paper were enriched from both A. pompejana samples and geographically distinct chimney samples from deep-sea hydrothermal vents from 13°N and the Guaymas basin. Because of the presumed and measured chemical differences in the samples from the sediment-starved EPR and the hydrocarbon and sediment-rich Guaymas basin (10, 22, 23; Luther et al., unpublished data), we were initially surprised by our ability to enrich for such phylogenetically similar bacteria (99%) from these two areas. We therefore believe that the physiological abilities of the cultured epsilon Proteobacteria reported in this paper are possibly far more diverse than we have described. The molecular survey that demonstrated these isolates at geographically and chemically distinct hydrothermal vent sites (9°N, 13°N, and the Guaymas basin) supports the hypothesis that these epsilon Proteobacteria potentially have wide physiological abilities.
While we were able to cultivate hydrothermal vent epsilon Proteobacteria from the A. pompejana episymbiont community as well as from chimney samples, we were unsuccessful in enriching for the dominant filamentous epsilon Proteobacteria phylotypes found integrated into the hair-like projections on the worm's dorsal epithelium (5, 14). We found, during the course of this investigation, that the medium designed for cultivation of epsilons was limited and selected for specific growth of two types of epsilon Proteobacteria. The culturing conditions were restricted by temperature range, carbon source used, electron donor-acceptor pairs tested, and pH. Any one or a combination of these factors will need to be tested further for potential growth of the free-living counterparts of the dominant episymbionts that were detected in chimney samples during our previous investigation (5). Furthermore, as determined by their relative band intensities by DGGE analysis, neither Am-N nor Am-H was numerically dominant in any of the 13°N A. pompejana samples or chimney samples from 9°N. However, a bacterium phylogenetically identical to Am-H was detected by DGGE analysis in a 10−7 dilution of a hydrothermal vent chimney enrichment for Fe(III) reducers from the same cruise at 13°N (38). Additionally, phylogenetically similar deeply branching bacteria have been observed at other hydrothermal vent sites devoid of A. pompejana specimens (35). It seems likely, then, that Am-H (or phylogenetically similar bacteria) exists in higher numbers in the chimney samples than on A. pompejana specimens from 13°N EPR.
Bacteria belonging to the epsilon subdivision of the Proteobacteria are clearly important in the ecology of hydrothermal vents, as indicated by their dominance in several molecular surveys (4, 14, 25, 35). The enrichment of autotrophic and heterotrophic epsilon Proteobacteria contributes to our understanding of carbon and sulfur cycling in hydrothermal vent environments. Our successful culturing of four phylogenetically distinct epsilon Proteobacteria from different hydrothermal vent environments paves the way for more biochemical testing of these isolates and further attempts to culture additional epsilons from these extreme environments.
ACKNOWLEDGMENTS
This research was supported by grants to S.C.C. from the LEXEN initiative (OCE-9907666) and the Delaware Sea Grant Program (R/B37) as well as a LEXEN initiative grant to G. Luther and S.C.C. (OCE-9729784). The Amistad cruise was organized by the Centre National de la Recherche Scientifique.
We gratefully acknowledge the following people for their technical assistance: L. Waidner, S. L'Haridon, D. Dalton, and M. Cottrell. We thank K. Coyne, C. DiMeo, and two anonymous reviewers for critically reviewing the manuscript. We thank the captains and crews of the R/Vs Atlantis and L'Atalante and especially the pilots of the DSVs Alvin and Nautile for their essential roles in the collection of specimens.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blochl E, Rachel R, Burggraf S, Hafenbradl D, Jannasch H W, Stetter K O. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113 degrees C. Extremophiles. 1997;1:14–21. doi: 10.1007/s007920050010. [DOI] [PubMed] [Google Scholar]
- 3.Brosius J, Dull T J, Sleeter D D, Noller H. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 4.Campbell B J, Cary S C. Characterization of a novel spirochete associated with the hydrothermal vent polychaete annelid Alvinella pompejana. Appl Environ Microbiol. 2001;67:110–117. doi: 10.1128/AEM.67.1.110-117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cary S C, Cottrell M T, Stein J L, Camacho F, Desbruyères D. Molecular identification and localization of a filamentous symbiotic bacteria associated with the hydrothermal vent annelid Alvinella pompejana. Appl Environ Microbiol. 1997;63:1124–1130. doi: 10.1128/aem.63.3.1124-1130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cline J D. Spectrophotometric determination of hydrogen sulfide in naturals waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 7.Cottrell M T, Cary S C. Diversity of dissimilatory bisulfite reductase genes of bacteria associated with the deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl Environ Microbiol. 1999;65:1127–1132. doi: 10.1128/aem.65.3.1127-1132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dempster E L, Pryor K V, Francis D, Young J E, Rogers H J. Rapid DNA extraction from ferns for PCR-based analyses. Biotechniques. 1999;27:66–68. doi: 10.2144/99271bm13. [DOI] [PubMed] [Google Scholar]
- 9.Durand P, Reysenbach A L, Prieur D, Pace N. Isolation and characterization of Thiobacillus hydrothermalis sp. nov., a mesophilic obligately chemolithotrophic bacterium isolated from a deep-sea hydrothermal vent in Fiji Basin. Arch Microbiol. 1993;159:39–44. [Google Scholar]
- 10.Edmond J M, Von Damm K L. Chemisty of ridge crest hot springs. Biol Soc Wash Bull. 1985;6:43–47. [Google Scholar]
- 11.Finster K, Liesack W, Thamdrup B. Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment. Appl Environ Microbiol. 1998;64:119–125. doi: 10.1128/aem.64.1.119-125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finster K, Liesack W, Tindall B J. Sulfurospirillum arcachonense sp. nov., a new-microaerophilic sulfur-reducing bacterium. Int J Syst Bacteriol. 1997;47:1212–1217. doi: 10.1099/00207713-47-4-1212. [DOI] [PubMed] [Google Scholar]
- 13.Gevertz D, Telang A J, Voordouw G, Jenneman G E. Isolation and characterization of strains CVO and FWKOB, two novel nitrate-reducing, sulfide-oxidizing bacteria isolated from oil field brine. Appl Environ Microbiol. 2000;66:2491–2501. doi: 10.1128/aem.66.6.2491-2501.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haddad M A, Camacho F, Durand P, Cary S C. Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaete Alvinella pompejana. Appl Environ Microbiol. 1995;61:1679–1687. doi: 10.1128/aem.61.5.1679-1687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jannasch H W, Wirsen C O, Nelson D C, Robertson L A. Thiomicrospira crunogena sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1985;35:422–424. [Google Scholar]
- 16.Jeanthon C, Prieur D. Susceptibility to heavy metals and characterization of heterotrophic bacteria isolated from two hydrothermal vent polychaetes, Alvinella pompejana and Alvinella caudata. Appl Environ Microbiol. 1990;56:3308–3314. doi: 10.1128/aem.56.11.3308-3314.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juniper S K, Sarrizan J. Interaction of vent biota and hydrothermal deposits: present evidence and future experimentation. In: Humpris S E, Zierenberg R A, Mullineaux L S, Thomson R E, editors. Seafloor hydrothermal systems: physical, chemical, biological, and geological interactions. Washington, D.C.: American Geophysical Union; 1995. pp. 178–193. [Google Scholar]
- 18.Laverman A M, Blum J S, Schaefer J K, Phillips E J P, Lovley D R, Oremland R S. Growth of strain SES-3 with arsenate and other diverse electron acceptors. Appl Environ Microbiol. 1995;61:3556–3561. doi: 10.1128/aem.61.10.3556-3561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L'Haridon S, Cilia V, Messner P, Raguenes G, Gambacorta A, Sleytr U B, Prieur D, Jeanthon C. Desulfurobacterium thermolithotrophum gen. nov., sp. nov., a novel autotrophic, sulpfur-reducing bacterium isolated from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1998;48:701–711. doi: 10.1099/00207713-48-3-701. [DOI] [PubMed] [Google Scholar]
- 20.Li L N, Kato C, Horikoshi K. Bacterial diversity in deep-sea sediments from different depths. Biodivers Conserv. 1999;8:659–677. [Google Scholar]
- 21.Lovley D R, Phillips E J P. Organic-matter mineralization with reduction of ferric iron in anaerobic sediments. Appl Environ Microbiol. 1986;51:683–689. doi: 10.1128/aem.51.4.683-689.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luther G W, Glazer B T, Hohmann L, Popp J I, Taillefert M, Rozan T F, Brendel P J, Theberge S M, Nuzzio D B. Sulfur speciation monitored in situ with solid state gold amalgam voltammetric microelectrodes: polysulfides as a special case in sediments, microbial mats and hydrothermal vent waters. J Environ Manage. 2001;62:61–66. doi: 10.1039/b006499h. [DOI] [PubMed] [Google Scholar]
- 23.Luther G W, Rozan T F, Taillefert M, Nuzzio D B, Di Meo C, Shank T M, Lutz R A, Cary S C. Chemical speciation drives hydrothermal vent ecology. Nature. 2001;410:813–816. doi: 10.1038/35071069. [DOI] [PubMed] [Google Scholar]
- 24.Macy J M, Schroder I, Thauer R K, Kroger A. Growth the Wolinella succinogenes on H2S plus fumarate and on formate plus sulfur as energy-sources. Arch Microbiol. 1986;144:147–150. [Google Scholar]
- 25.Moyer C L, Dobbs F C, Karl D M. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen G J. Phylogenetic analysis using ribosomal RNA. Methods Enzymol. 1988;164:793–812. doi: 10.1016/s0076-6879(88)64084-5. [DOI] [PubMed] [Google Scholar]
- 27.Oremland R S, Blum J S, Culbertson C W, Visscher P T, Miller L G, Dowdle P, Strohmaier F E. Isolation, growth, and metabolism of an obligately anaerobic, selenate-respiring bacterium, strain SES-3. Appl Environ Microbiol. 1994;60:3011–3019. doi: 10.1128/aem.60.8.3011-3019.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfenning N, Biebl H. The dissimilatory sulfur-reducing bacteria. In: Starr M P, Stolp H, Truper H G, Balows A, Schlegel H G, editors. The prokaryotes. Vol. 1. New York, N.Y: Springer-Verlag; 1981. pp. 941–947. [Google Scholar]
- 29.Polz M F, Cavanaugh C M. Dominance of one bacterial phylotype at a mid-Atlantic Ridge hydrothermal vent site. Proc Natl Acad Sci USA. 1995;92:7232–7236. doi: 10.1073/pnas.92.16.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceangr. 1980;25:943–948. [Google Scholar]
- 31.Prieur D, Jeanthon C. Preliminary study of heterotrophic bacteria isolated from deep sea hydrothermal vent invertebrates: Alvinella pompejana (Polychaete) and Bathymodiolus thermophilus (Bivalve) Symbiosis. 1987;4:87–98. [Google Scholar]
- 32.Prieur D, Chamroux S, Durand P, Erauso G, Fera P, Jeanthon C, Le borgne L, Mével G, Vincent P. Metabolic diversity in epibiotic flora associated with the pompeii worms, Alvinella pompejana and Alvinella caudata (Polychaeta: Annelida) from deep-sea hydrothermal vents. Mar Biol. 1990;106:361–367. [Google Scholar]
- 33.Raguenes G, Pignet P, Gauthier G, Peres A, Christen R, Rougeaux H, Barbier G, Guezennec J. Description of a new polymer-secreting bacterium from a deep-sea hydrothermal vent, Alteromonas macleodii subsp. fijiensis, and preliminary characterization of the polymer. Appl Environ Microbiol. 1996;62:67–73. doi: 10.1128/aem.62.1.67-73.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rath J, Wu K Y, Herndl G J, DeLong E F. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol. 1998;14:261–269. [Google Scholar]
- 35.Reysenbach A L, Longnecker K, Kirshtein J. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a mid-Atlantic Ridge hydrothermal vent. Appl Environ Microbiol. 2000;66:3798–3806. doi: 10.1128/aem.66.9.3798-3806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozan T F, Theberge S M, Luther G. Quantifying elemental sulfur (S0), bisulfide (HS−) and polysulfides (Sx2−) using a voltammetric method. Anal Chim Acta. 2000;415:175–184. [Google Scholar]
- 37.Schumacher W, Kroneck P M H, Pfennig N. Comparative systematic study on “Spirillum” 5175, Campylobacter, and Wolinella species—description of “Spirillum” 5175 as Sulfurospirillum deleyianum gen., nov. spec. nov. Arch Microbiol. 1992;158:287–293. [Google Scholar]
- 38.Slobodkin A, Campbell B J, Cary S C, Bonch-Osmolovskaya E, Jeanthon C. Thermophilic Fe(III)-reducing microorganisms inhabit deep-sea hydrothermal vents on the east Pacific rise. FEMS Microbiol Ecol. 2001;36:235–243. doi: 10.1111/j.1574-6941.2001.tb00844.x. [DOI] [PubMed] [Google Scholar]
- 39.Smith S W, Overbeek R, Olsen G, Woese C, Gillevet P M, Gilbert W. Genetic data environment and the Harvard genome database: genome mapping and sequencing. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 40.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stetter K O. Hyperthermophilic procaryotes. FEMS Microbiol Rev. 1996;18:149–158. [Google Scholar]
- 42.Stolz J F, Ellis D J, Blum J S, Ahmann D, Lovley D R, Oremland R S. Sulfurospirillum barnesii sp. nov. and Sulfurospirillum arsenophilum sp. nov., new members of the Sulfurospirillum clade of the epsilon Proteobacteria. Int J Syst Bacteriol. 1999;49:1177–1180. doi: 10.1099/00207713-49-3-1177. [DOI] [PubMed] [Google Scholar]
- 43.Taylor C D, Wirsen C O. Microbiology and ecology of filamentous sulfur formation. Science. 1997;277:1483–1485. [Google Scholar]
- 44.Taylor C D, Wirsen C O, Gaill F. Rapid microbial production of filamentous sulfur mats at hydrothermal vents. Appl Environ Microbiol. 1999;65:2253–2255. doi: 10.1128/aem.65.5.2253-2255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe K, Kodama Y, Syutsubo K, Harayama S. Molecular characterization of bacterial populations in petroleum-contaminated groundwater discharged from underground crude oil storage cavities. Appl Environ Microbiol. 2000;66:4803–4809. doi: 10.1128/aem.66.11.4803-4809.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Widdel F. Methods for enrichment and pure culture isolation of filamentous gliding sulfate-reducing bacteria. Arch Microbiol. 1983;134:282–285. [Google Scholar]
- 47.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. IV. New York, N.Y: Springer-Verlag; 1992. pp. 3352–3378. [Google Scholar]