Abstract
The increasing emergence of resistances against established antibiotics is a substantial threat to human health. The discovery of new compounds with potent antibiotic activity is thus of utmost importance. Within this work, we identify strong antibiotic activity of the natural product myxocoumarin B from Stigmatella aurantiaca MYX‐030 against a range of clinically relevant bacterial pathogens, including clinical isolates of MRSA. A focused library of structural analogs was synthesized to explore initial structure‐activity relationships and to identify equipotent myxocoumarin derivatives devoid of the natural nitro substituent to significantly streamline synthetic access. The cytotoxicity of the myxocoumarins as well as their potential to cure bacterial infections in vivo was established using a zebrafish model system. Our results reveal the exceptional antibiotic activity of the myxocoumarin scaffold and hence its potential for the development of novel antibiotics.
Keywords: antibiotics, myxocoumarins, natural products, Staphylococcus aureus, structure-activity relationships
Myxocoumarin B from Stigmatella aurantiaca MYX‐030 is shown to possess strong antibiotic activity against a range of clinically relevant pathogens. Synthesis of a diverse compound library of structural analogs enabled investigations into initial structure‐activity relationships and revealed the potential of the myxocoumarin scaffold as lead structure for the development of new antibiotics. Myxocoumarin B and analogs show low toxicity in vivo in a zebrafish infection model and were superior in clearing bacterial infections when compared to established antibiotics such as vancomycin.
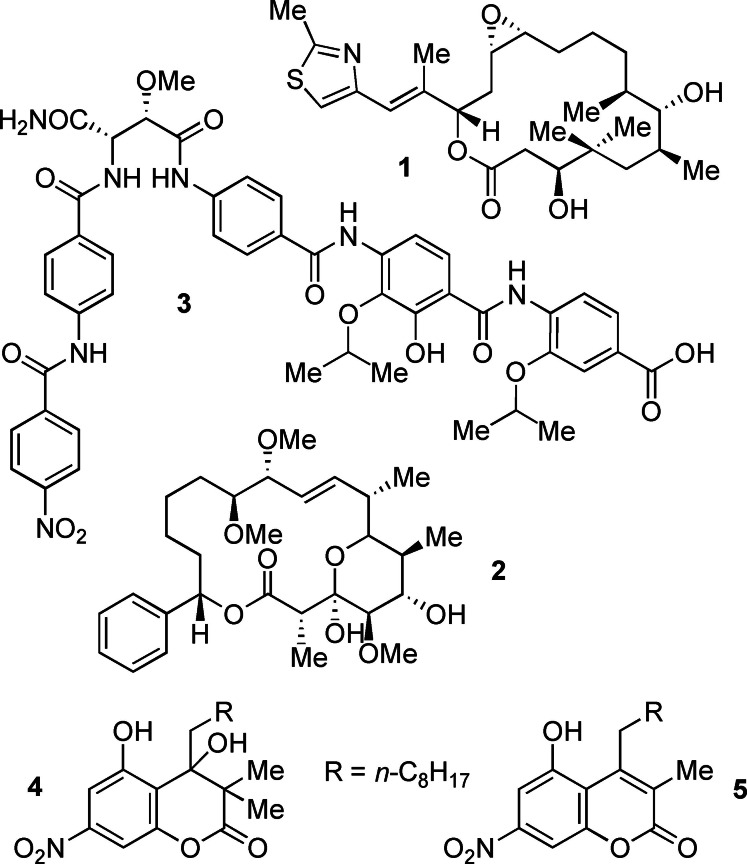
Myxobacteria are talented producers of complex, biomedically interesting specialized metabolites from most diverse natural product classes. [1] Prominent examples include epothilone A (1) and analogs as anti‐cancer chemotherapeutics, [2] the antifungal soraphen A1α (2), [3] or the antibacterial cystobactamids, [4] such as 3 (Figure 1). These organisms have thus proven to be a promising resource for the discovery of new small molecules with potent biological functions, particularly for applications with increasing demand due to emerging resistance against currently known agents, both in the agrochemical and medical sectors. In 2013, we have described the discovery of two natural products from Stigmatella aurantiaca MYX‐030, myxocoumarins A (4) and B (5), both of which being equipped with a long alkyl side chain and a rare aromatic nitro group. [5] Myxocoumarin A (4) displayed potent inhibitory effects against a range of agrochemically relevant pathogenic fungi, while 5 could not be biologically assessed due to the low production titers and the loss of a viable producing strain. To allow for antifungal evaluation of 5, a concise total synthesis of this compound was developed, revealing a complete lack of the anticipated antifungal properties. [6] A possible natural function as well as prospective applications of myxocoumarin B (5) thus remained elusive. This raised our interest in exploring potential other biological functions of 5. We therefore set out to more broadly explore potential antimicrobial properties of myxocoumarin‐type scaffolds within this work.
Figure 1.
Chemical structures of the myxobacterial natural products epothilone A (1), soraphen A1α (2), cystobactamide 3 and the myxocoumarins A (4) and B (5).
Initial screening of synthetic 5 against Candida albicans, Pseudomonas aeruginosa, Escherichia coli, Enterococcus faecalis and Staphylococcus aureus using classical disk‐diffusion assays revealed a strong activity of 5 against the latter two, Gram‐positive bacteria, comparable to the employed standard kanamycin. Owing to these promising preliminary results, a closer inspection of the antibiotic potential of the myxocoumarins was targeted. Given the straightforward and flexible synthetic access developed for 5, [6] the preparation of a focused compound library to give first insights into structure‐activity relationships across the myxocoumarin core was conducted. Of particular interest were variations at the prototypical myxocoumarin alkyl side chain to assess its role in granting antibacterial activity. Furthermore, the evaluation of analogs without the nitro substituent was of high interest, to exclude potential formation of non‐specific toxic nitroso side products in vivo. [7] In addition, the synthesis of myxocoumarin derivatives lacking the nitro function is significantly simplified, giving very fast access to the target molecules.
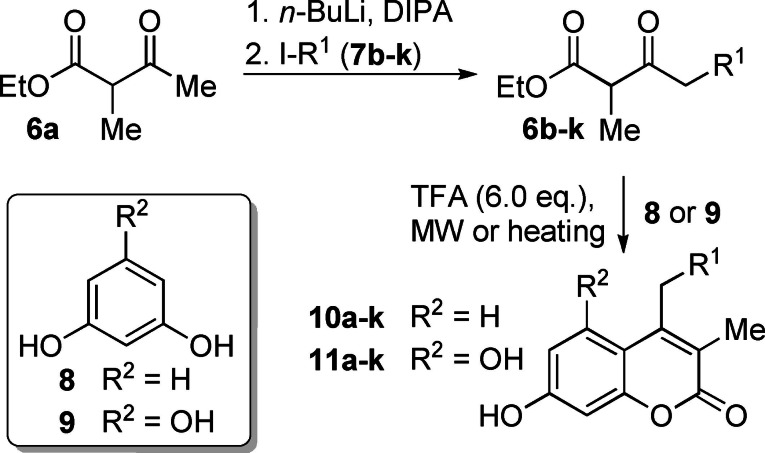
The method of choice for the synthesis of myxocoumarin analogs was the condensation of β‐keto esters with diverse alkyl substitution with phenol building blocks in a TFA‐mediated Pechmann condensation. [8] Ethyl 2‐methylacetoacetate (6 a) was deprotonated with n‐BuLi/DIPA and selectively alkylated with iodoalkanes 7 b–k to deliver the desired β‐keto esters 6 b–k (Scheme 1). The analogs bearing linear side chains from C4 to C11 (6 b–6 i) and C13 (6 j) were obtained in yields ranging from 43 % to 62 %. The terminally branched 6 k gave a lower yield of 38 %. The β‐keto esters were subsequently condensed with resorcinol (8) and phloroglucinol (9) to give the corresponding mono‐ (10 a–k) and dihydroxy (11 a–k) myxocoumarin analogs (for yields, see Table 1).
Scheme 1.
Synthesis of hydroxy‐substituted myxocoumarin analogs 10 a–k and 11 a–k by Pechmann condensation with β‐keto esters 6 a–k with different side‐chains R1.
Table 1.
Substitution pattern and synthetic yields of analogs 10 and 11.
|
Compound |
R1 |
Yield [%] of 10 (R2=H) |
Yield [%] of 11 (R2=OH) |
|---|---|---|---|
|
a |
H |
69 |
73 |
|
b |
n‐C3 |
42 |
30 |
|
c |
n‐C4 |
42 |
33 |
|
d |
n‐C5 |
26 |
27 |
|
e |
n‐C6 |
36 |
32 |
|
f |
n‐C7 |
35 |
25 |
|
g |
n‐C8 |
33 |
42 |
|
h |
n‐C9 |
30 |
25 |
|
i |
n‐C10 |
30 |
26 |
|
j |
n‐C12 |
25 |
28 |
|
k |
iso‐C5 |
37 |
19 |
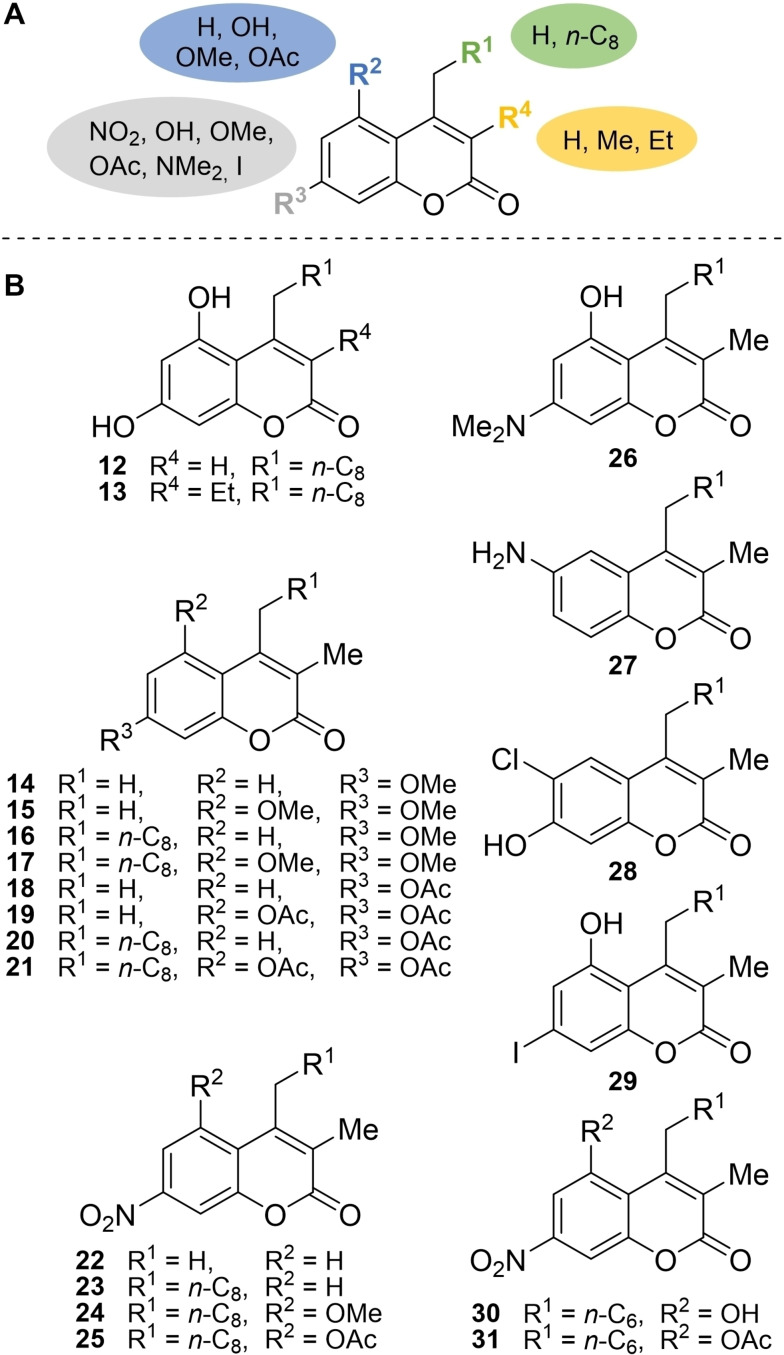
To test the role of the methyl group at the position neighboring the ester functionality, derivatives bearing a proton (12) or an ethyl group (13) at R4 were prepared by alkylation of ethyl acetoacetate or ethyl 2‐ethylacetoacetate with 7 g followed by Pechmann condensation with phloroglucinol (9).
To further increase the structural diversity of the myxocoumarin chemical library beyond hydroxyl analogs of 5, a selection of O‐methylated (14–17) and O‐acetylated (18–21) derivatives was additionally prepared (Figure 2), either bearing a proton at position R1 or an n‐C8 alkyl chain as in the original myxocoumarin scaffold. While compounds 14 and 18–21 were obtained by either O‐methylation or O‐acetylation of the corresponding phenol analogs 10 a and 10 a, 11 a, 10 g, 11 g using MeI or Ac2O, respectively, 15–17 were generated by Pechmann condensation of 3,5‐dimethoxyphenol or 3‐methoxyphenol with either 6 a or 6 g. Furthermore, four myxocoumarin derivatives 22–25 bearing the natural nitro function were prepared. Compounds 22–24 were accessed by triflation and a subsequent nitration reaction of the corresponding hydroxy coumarins, [6] while 25 was synthesized by O‐acetylation of myxocoumarin B (5). In addition, a dimethylamino‐ (26), amino‐ (27), chloro‐ (28) and iodo‐ (29) analog were accessed by Pechmann condensation of the correspondingly functionalized phenols with 6 g (see Supporting Information for experimental details on the syntheses of 12–29).
Figure 2.
A. Structural changes introduced into the myxocoumarin scaffold in analogs 12–25. B. Chemical structures of phenolic (12, 13), O‐methylated‐ (14–17) and O‐acetylated‐ (18–21) myxocoumarin analogs, and further nitro myxocoumarins (22–25) as well as dimethylamino‐ (26), amino‐ (27), chloro‐ (28) and iodo‐ (29) substituted myxocoumarin analogs (for 26–29, R1=n‐C8) and additional short‐chain analogs 30 and 31.
Having this myxocoumarin library composed of the natural product 5 and 40 synthetic structural analogs in hands, we set out to evaluate their antimicrobial potential. All compounds were inactive against the opportunistic pathogenic yeast C. albicans, the gram‐negative bacterium Pseudomonas aeruginosa, as well as the gram‐positive Micrococcus luteus and Listeria monocytogenes at the maximum tested concentrations (250 μg/mL). However, strong antibacterial potential against the gram‐positive bacteria Bacillus subtilis NCTC5398 and S. aureus NCTC6571 was observed for a number of analogs (Table 2, see Table S1). The natural product 5 exhibited very strong antibacterial activity against these test strains, with MIC values as low as 0.3 μg/mL against S. aureus NCTC6571 combined with an up to 400‐fold higher LC50 value in a zebrafish model (119.6 μg/mL). For the mono‐hydroxylated 10 and dihydroxylated 11 compound series, a clear structure‐activity relationship became obvious, with the activity correlating with the length of the alkyl side chain at R1. Considering the activity of analogs 10 against S. aureus NCTC6571, strong effects were only observed for 10 e–g with C7 to C9 alkyl substitution, with lowest MICs for the natural C9 substituent. Inhibition of B. subtilis followed a similar trend, but activity extended to C10 and to a smaller extent to C11 substitution. The series of dihydroxylated analogs generally had a broader activity profile in terms of alkyl chain length, with significant inhibition already starting from C4 up to C9 substitution.
Table 2.
Heat map of the antibacterial activity profile of myxocoumarin B (5) and its structural analogs 10–31 against B. subtilis NCTC5398, S. aureus NCTC6571, S. aureaus MRSA compared to toxicity in zebrafish embryos. NCTC=National Collection of Type Cultures (NCTC, Culture Collection of Public Health, Salisbury, UK). Green color indicates antibiotic activity (darker green equals higher activity), organge color toxicity (darker orange equals higher toxicity).
|
Compound |
B. subtilis NCTC5398 [μg/mL] |
S. aureus NCT6571 [μg/mL] |
S. aureus MRSA [μg/mL] |
Zebrafish LC50 [μg/mL] |
|---|---|---|---|---|
|
5 |
8 |
0.3 |
0.6 |
119.6 |
|
10 a |
>250 |
>250 |
>250 |
8.9 |
|
10 b |
31.2 |
62.5 |
62.5 |
2.1 |
|
10 c |
200 |
200 |
200 |
2.5 |
|
10 d |
200 |
200 |
200 |
1.5 |
|
10 e |
2 |
2 |
2 |
2.7 |
|
10 f |
15.6 |
7.8 |
7.8 |
21.5 |
|
10 g |
2 |
3.9 |
7.8 |
>151.2 |
|
10 h |
7.8 |
>250 |
250 |
30.5 |
|
10 i |
62.5 |
>250 |
>250 |
>82.6 |
|
10 j |
>250 |
>250 |
>250 |
>89.6 |
|
10 k |
15.6 |
>250 |
>250 |
5.2 |
|
11 a |
125 |
125 |
62.5 |
14.7 |
|
11 b |
15.6 |
15.6 |
15.6 |
9.0 |
|
11 c |
7.8 |
7.8 |
7.8 |
5.2 |
|
11 d |
7.8 |
4 |
2 |
4 |
|
11 e |
7.8 |
2.5 |
4 |
24.3 |
|
11 f |
4 |
15.6 |
7.8 |
>152.2 |
|
11 g |
31.2 |
7.8 |
31.2 |
>159.2 |
|
11 h |
>250 |
250 |
250 |
>158.2 |
|
11 i |
>250 |
>250 |
>250 |
>173.2 |
|
11 j |
>250 |
>250 |
>250 |
>187.3 |
|
11 k |
250 |
10 |
10 |
10.9 |
|
12 |
250 |
100 |
100 |
>250 |
|
13 |
5 |
12.5 |
50 |
>250 |
|
14 |
>250 |
>250 |
>250 |
12.7 |
|
15 |
>250 |
>250 |
>250 |
1.3 |
|
16 |
250 |
>250 |
>250 |
>158.2 |
|
17 |
62.5 |
>250 |
>250 |
>166.2 |
|
18 |
250 |
>250 |
>250 |
18.4 |
|
19 |
125 |
>250 |
>250 |
11.3 |
|
20 |
15.6 |
31.2 |
15.6 |
118.9 |
|
21 |
31.2 |
31.2 |
62.5 |
180.2 |
|
22 |
>250 |
>250 |
>250 |
25.8 |
|
23 |
>250 |
>250 |
>250 |
>165.7 |
|
24 |
>250 |
>250 |
>250 |
>180.7 |
|
25 |
0.3 |
0.15 |
0.15 |
27.8 |
|
26 |
>250 |
>250 |
>250 |
>172.7 |
|
27 |
>250 |
>250 |
>250 |
21.5 |
|
28 |
125 |
250 |
>250 |
>168.4 |
|
29 |
62.5 |
>250 |
250 |
67.6 |
|
30 |
2 |
0.3 |
0.3 |
20.2 |
|
31 |
1 |
0.15 |
1.25 |
18.6 |
|
Vancomycin |
0.4 |
0.8 |
0.5 |
>250 |
|
Linezolid |
0.8 |
3.1 |
3.1 |
>250 |
Alkyl chain lengths beyond the natural C9 substituent lead to a fast drop in antibacterial activity for both compound series, 10 and 11. When correlating antibacterial structure‐activity relationships with toxicity in the zebrafish model, it became evident that longer side chains also lead to drastically increased LC50 values and thus less toxicity. Overall, besides the natural product 5, 10 f (C8), 10 g (C9) and 11 e–11 g (C7‐C9) exhibited the most promising activity profiles.
When considering R4 substitution, analog 12 bearing a proton at this position had dramatically reduced activity when compared to its methylated congener 11 g, while activity in ethyl derivative 13 was retained. Among all other tested compounds 14–29, the O‐acetylated analogs 20 and 21 and to an even lesser extent O‐methylated derivative 17 and chlorinated 28 all bearing the natural C9 alkyl substitution, retained weak antibacterial effects. Interestingly, the antibacterial activity of the O‐acetylated analog 25 of myxocoumarin B (5) was approx. 25‐fold higher against B. subtilis NCTC5398 and 2–4‐fold higher against S. aureus when compared to the natural product. However, this was accompanied with an approx. 4‐fold increase in toxicity in the zebrafish model, overall leading to similar activity versus toxicity ratios.
Overall, these generated structure‐activity data indicated that substitution at R2 is required for strong antibiotic activity, preferably with OH or OAc, that analogs with nitro substitution at R3 generally show higher activity (roughly 10‐fold when compared to the respective phenolic derivatives), and that alkyl side‐chain lengths from C7 to C9 seem to be optimal for activity, with increased side‐chain lengths correlating with decreased toxicity in the zebrafish model. This structure‐activity correlation was corroborated by synthesis of the C7 analogs of 5, without (30) and with (31) O‐acetylation of the phenol at R2. Both compounds indeed showed activity comparable to that of 5, yet with approx. 6‐fold increased toxicity. Most importantly, the antibacterial potential of all active myxocoumarin analogs was also retained against S. aureus MRSA, showing the high potential of these compounds to combat this clinically relevant, highly resistant bacterial pathogen.
We next evaluated the cytotoxicity of all myxocoumarin analogs with significant antimicrobial activity (at least one MIC value at or below 15.6 μg/mL) by screening against healthy human MRC‐5 fibroblast cells (see Table S2). This revealed low selectivity indices (SI) for some derivatives (e.g., below 1 for 11 g and 17), with up to an SI of 87.5 for the natural product 5. For further in‐depth studies, we selected three analogs with high antibacterial activity combined with a range of selectivity indices, namely myxocoumarin B (5) (SI=87.5), its O‐acetyl analog 25 (SI=25), and the most potent dihydroxylated congener 11 e equipped with a C7 alkyl chain (SI=3.8). These compounds were evaluated against a larger panel of S. aureus strains, including clinical isolates (Table 3). The strains were collected from veterinary specimens (dog urine, ear swab and mouth swab) and showed resistance to one or more commonly used antibiotics in veterinary practice. Enterococcus faecium, as another Gram‐positive opportunistic pathogen, was also included in the assessment. To our delight, all compounds showed pronounced antibacterial effects against all tested strains, with 25 being the most active antibacterial across all evaluated organisms (Table 3).
Table 3.
Antibiotic activity profile of 5, 25 and 11 e against a selection of S. aureus strains and Enterococcus sp. (top), including clinical isolates (bottom, strains isolated and identified from the clinical specimens delivered to veterinary Laboratory “VetLab”, Belgrade, Serbia including dog urine, mouth and ear swab).
|
Compound |
S. aureus MRSA ATCC43300 [μg/mL] |
S. aureus ATCC9144 [μg/mL] |
Enterococcus faecium ATCC6057 [μg/mL] |
|---|---|---|---|
|
5 |
0.6 |
0.3 |
2 |
|
25 |
0.15 |
0.15 |
0.6 |
|
11 e |
4 |
2.5 |
2.5 |
|
Compound |
Staphylococcus sp. 80103770 |
Staphylococcus sp. 80100861 |
Staphylococcus sp. 80100865 |
|---|---|---|---|
|
5 |
0.3 |
0.3 |
2 |
|
25 |
0.15 |
0.15 |
0.6 |
|
11 e |
2.5 |
5 |
2.5 |
The likelihood of resistance development against 5, 25 and 11 e was assessed by multistep resistance selection of S. aureus ATCC9144 by propagating cultures with subinhibitory concentrations of the respective individual compound for 20 rounds, followed by one round of growth without selective pressure. [9] Over the course of this treatment, S. aureus developed a 5‐fold increase in MIC against 5, a 4‐fold increase against 25 and only a 2‐fold increase for 11 e. Therefore, high antibiotic potential was retained for all compounds. This indicates a low likelihood of S. aureus for developing rapid resistance to the tested myxocoumarins.
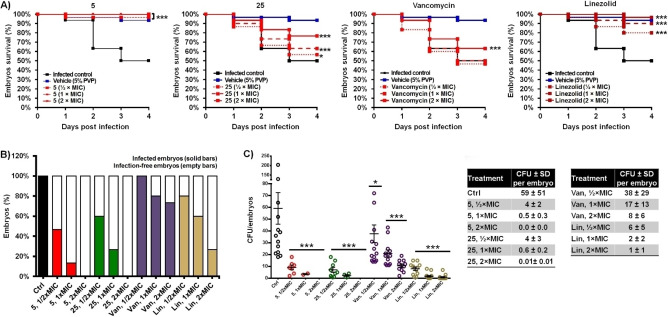
Based on above resistance development assays and SI values, 5 and 25 were selected for evaluation of the antibiotic potential in vivo in a S. aureus‐ zebrafish infection model (Figure 3). The zebrafish infection model is a well‐recognized platform for studying host‐pathogen interactions, as well as for the development of therapeutic strategies. [10] In this experiment, wild type zebrafish embryos were challenged with the lethal systemic infection with S. aureus MRSA 43300, and followed for the survival and bacterial burden after 3‐days treatments with 5 and 25. Antibacterial efficacy of the selected myxocoumarins was evaluated in relation to the efficacy of vancomycin and linezolid, two antibiotics of clinical relevance. Analyses of zebrafish embryo survival coupled with bacterial proliferation confirmed that both compounds are highly efficient in rescuing zebrafish from infection, with the effect being dose dependent (Figure 3 A, B). Remarkably, both 5 and 25 proved approx. 10‐fold more efficient at 0.5×MIC, with even approx. 35‐fold increased activity at 1×MIC when compared to vancomycin, and approx. 2‐fold higher activity at these concentrations when compared against linezolid.
Figure 3.
Myxocoumarin B (5) and derivative 25 rescued zebrafish embryos of S. aureus MRSA 43300 infection (A−B) and significantly decreased the bacterial burden (C). The Kaplan‐Meier curves of the infected embryos survival upon different doses of vancomycin (Van), linezolid (Lin) and myxocoumarins 5 and 25 are shown. Survival of the treated infected fish was compared to those in the group without treatment (infected control) and the uninfected group (injected with 5 % PVP [polyvinylpyrrolidone], which was used as a vehicle for the S. aureus suspension for infection). Embryos were monitored daily for survival. Data are compilations of two independent experiments using two replicates (n=20 embryos/replicates) for each group. Bacterial burden was determined at 4 dpi by plating of the crushed embryos for colony forming units (CFUs). Data are compilations of two independent experiments using ten embryos for each group. Each dot represents an individual fish (square – untreated embryos, circle – treated embryos). The mean CFUs±SEM are shown.
Using 2×MIC of either 5 or 25 completely eliminated bacterial infection in vivo, as reflected in the efficiency of reduction of the bacterial burden (Figure 3C). Most importantly, treatment of infected zebrafish with 5 restored survival rates at all tested concentrations to the level of uninfected specimen (Figure 3A), thus proving both, high antibiotic efficiency and low toxicity of this compound upon application in vivo.
Overall, our work provides first structure‐activity relationship data on a new class of antibiotics based on the myxocoumarin scaffold. It is interesting to note that the natural nitro substitution seems to be crucial for strong antibiotic activity. The most potent congeners provide outstanding sub‐nanomolar antibacterial potency, even against drug‐resistant S. aureus strains accompanied with low toxicity, both, in vitro and in vivo. Particularly encouraging are the obtained results on eliminating bacterial S. aureus infections in zebrafish by 5, clearly outcompeting the established antibiotics vancomycin and linezolid, not only in terms of clearing infections, but most importantly also in terms of restoring survival of infected zebrafish to levels of healthy specimen. The myxocoumarins thus represent a valuable scaffold for the development of new antibiotics against clinically relevant bacterial pathogens, particularly for desperately needed new treatment option against MRSA. [11]
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This work was generously funded by the German Research Foundation (DFG GU 1233/1‐1) and by the Ministry of Education, Science and Technological Development of the Republic of Serbia (agreement no. 451‐03‐68/2020‐14/200042) and DAAD (Deutscher Akademischer Austauschdienst, Bilateral Project of Germany with the Republic of Serbia to SV and TAMG – 2020/2021). Open Access funding enabled and organized by Projekt DEAL.
G. Hertrampf, K. Kusserow, S. Vojnovic, A. Pavic, J. I. Müller, J. Nikodinovic-Runic, T. A. M. Gulder, Chem. Eur. J. 2022, 28, e202200394.
Contributor Information
Dr. Jasmina Nikodinovic‐Runic, Email: jasmina.nikodinovic@gmail.com, Email: jasmina.nikodinovic@imgge.bg.ac.rs.
Prof. Dr. Tobias A. M. Gulder, Email: tobias.gulder@tu-dresden.de.
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.
References
- 1.
- 1a. Schäberle T. F., Lohr F., Schmitz A., König G. M., Nat. Prod. Rep. 2014, 31, 953–972; [DOI] [PubMed] [Google Scholar]
- 1b. Herrmann J., Abou Fayad A., Müller R., Nat. Prod. Rep. 2017, 34, 135–160; [DOI] [PubMed] [Google Scholar]
- 1c. Gemperlein K., Zaburannyi N., Garcia R., La Clair J. J., Müller R., Mar. Drugs 2018, 16, 314–329; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1d. Bader C. D., Panter F., Müller R., Biotechnol. Adv. 2020, 39, 107480. [DOI] [PubMed] [Google Scholar]
- 2.
- 2a. Bollag D. M., McQueney P. A., Zhu J., Hensens O., Koupal L., Liesch J., Götz M., Lazarides E., Woods C. M., Cancer Res. 1995, 55, 2325–2333; [PubMed] [Google Scholar]
- 2b. Gerth K., Bedorf N., Höfle G., Irschik H., Reichenbach H., J. Antibiot. 1996, 49, 560–563; [DOI] [PubMed] [Google Scholar]
- 2c. Reichenbach H., Höfle G., Drugs 2008, 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.
- 3a. Gerth K., Bedorf N., Irschik H., Höfle G., Reichenbach H., J. Antibiot. 1994, 47, 23–31; [DOI] [PubMed] [Google Scholar]
- 3b. Naini A., Sasse F., Brönstrup M., Nat. Prod. Rep. 2019, 36, 1394–1411. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a. Baumann S., Herrmann J., Raju R., Steinmetz H., Mohr K. I., Hüttel S., Harmrolfs K., Stadler M., Müller R., Angew. Chem. Int. Ed. 2014, 53, 14605–14609; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 14835–14839; [Google Scholar]
- 4b. Hüttel S., Testolin G., Herrmann J., Planke T., Gille F., Moreno M., Stadler M., Brönstrup M., Kirsching A., Müller R., Angew. Chem. Int. Ed. 2017, 56, 12760–12764; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 12934–12938. [Google Scholar]
- 5. Gulder T. A. M., Neff S., Schütz T., Winkler T., Gees R., Böhlendorf B., Beilstein J. Org. Chem. 2013, 9, 2579–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Müller J. I., Kusserow K., Hertrampf G., Pavic A., Nikodinovic-Runic J., Gulder T. A. M., Org. Biomol. Chem. 2019, 17, 1966–1969. [DOI] [PubMed] [Google Scholar]
- 7.
- 7a. Nepali K., Lee H.-Y., Liou J.-P., J. Med. Chem. 2019, 62, 2851–2893; [DOI] [PubMed] [Google Scholar]
- 7b. Purohit V., Basu A. K., Chem. Res. Toxicol. 2000, 13, 673–692. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. v. Pechmann H., Ber. Dtsch. Chem. Ges. 1884, 17, 929–936; [Google Scholar]
- 8b. Katkevičs M., Kontijevskis A., Mutule I., Sūna E., Chem. Heterocycl. Compd. 2007, 43, 151–159. [Google Scholar]
- 9. Farrell D. J., Robbins M., Rhys-Williams W., Love W. G., Antimicrob. Agents Chemother. 2011, 55, 1177–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.
- 10a. Torraca V., Mostowy S., Trends Cell Biol. 2018, 28, 143–156; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10b. Medina C., Royo J. L., Methods 2013, 62, 241–245; [DOI] [PubMed] [Google Scholar]
- 10c. Li Y.-J., Hu B., J. Genet. Genomics 2012, 39, 521–534. [DOI] [PubMed] [Google Scholar]
- 11. Cascioferro S., Carbone D., Parrino B., Pecoraro C., Giovannetti E., Cirrincione G., Diana P., ChemMedChem 2020, 16, 65–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.