Abstract
Eukaryotic cells partition enzymes and other cellular components into distinct subcellular compartments to generate specialized biochemical niches. A subclass of these compartments form in the absence of lipid membranes, via liquid-liquid phase separation of proteins to form biomolecular condensates or “membraneless organelles” such as nucleoli, stress granules, and P-bodies. Because of their ability to form compartments from simple starting materials, membraneless organelles are an attractive target for engineering new functionalities in both living cells and protocells. In this work, we demonstrate incorporation of novel enzymatic activity in protein coacervates with a light-generating enzyme, NanoLuc, to produce bioluminescence. Using condensates comprised of the disordered RGG domain of C. elegans LAF-1, we show functionalization of condensates with enzymatic activity in vitro and that localization to protein coacervates enhances the assembly and activity of split enzymes. To build condensates that function as light emitting reactors, we designed a NanoLuc enzyme flanked by RGG domains. The resulting condensates concentrated NanoLuc by 10-fold over bulk solution and display significantly increased net reaction rates. We further show that condensate viscosity impacts light emission due to diffusion-limited behavior. By splitting NanoLuc enzyme into its constituent components, we demonstrate that NanoLuc activity can be reconstituted via co-condensation. Further, we demonstrate control of the spatial localization of enzyme within condensates by targettng NanoLuc to the surface of in vitro condensates. Collectively, this work demonstrates that membraneless organelles can be endowed with localized enzymatic activity, and that this activity can be spatially and temporally controlled via enzyme reconstitution and design of protein surfactants.
Introduction
Cells regulate biological function by controlling subcellular compartmentalization. These compartments, or organelles, allow highly specific partitioning of proteins and enzymes and spatiotemporal control over a variety of cellular functions. Membraneless organelles are proteinaceous compartments and not bound by a lipid membrane and instead form from liquid-liquid phase separation (LLPS). Previous studies proposed that liquid-like coacervates could increase biochemical reaction rates by sequestering enzymes at locally high concentrations (1–4). Such phenomena have also been suggested to have played a potential role in the origins of life, perhaps before development of membrane-encapsulated organelles, to allow for protein or nucleic acid self-assembly and function (3, 5–7). Conversely, protein condensates can also isolate client molecules from the surrounding cellular milieu by macromolecular coacervation, utilizing a range of primarily weak interactions including electrostatic, dipole-dipole, pi-pi, cation-pi, hydrophobic, and hydrogen bonding (8–11). This allows a liquid-like environment and permits diffusion of substrates or other small molecules (3, 12). Examples of naturally occurring liquid-like membraneless organelles include P granules observed during C. elegans embryonic development (13), the nucleoli in germinal vesicles of Xenopus laevis oocytes (14), Cajal bodies (15), and stress granules (16). Functions of liquid biomolecular condensates have been linked to a growing number of activities including intracellular signaling (16–18), cell cycle control (19), endocytosis and autophagy (20–22), and gene transcription and regulation (23–25). Further, dysfunction of some phase separating proteins may disrupt cellular processes and has been linked with neurodegenerative diseases and some cancers (1, 3, 4, 8, 10, 11, 26, 27).
Engineering condensates with tailored functionalities is an attractive route for constructing functional membraneless organelles in protocell engineering. If appropriately designed, protein coacervates could serve as biochemical hubs for catalyzing enzymatic activity. Previously, biochemical reactions have been carried out in vitro using membrane-free polysaccharide/polypeptide (28, 29), polypeptide/mononucleotide (30), and polyelectrolyte/mononucleotide (31) coacervates. Recent work also demonstrated biochemical reactions in FUS- and SPD5-based membrane-less organelles in vivo (32, 33). Here, we utilize the intrinsically disordered region (IDR) of C. elegans P granule protein LAF-1 as the scaffold for our study. The disordered N-terminal RGG domain of LAF-1 is comprised of 168 aa, enriched with arginines (R) and glycines (G) with frequent repeats of the ‘RGG’ motif, which is observed in other RNA binding proteins. The RGG domain alone is sufficient for LLPS and formation of liquid-like coacervates (34). In previous work, we demonstrated that valency of the RGG domain could be used to tune LLPS behavior (35). Multivalent RGG polypeptides display more potent coacervation than the single domain and allow for the incorporation of additional domains as tools for engineering coacervates (36, 37). For example, we were able to add protease sites or photocleavable domains to control the magnitude of protein condensation and could localize cargos into coacervates using recruitment tags in vitro (36, 37). Further, multivalent IDR scaffolds with similar recruitment motifs were recently demonstrated to effectively sequester endogenous enzymes in vivo, leading to control over cell behavior (38).
Using an IDR scaffold, we demonstrate that biochemical reactions can be catalyzed within our condensate system. We incorporated a light-emitting NanoLuc enzyme (39) into coacervates by flanking it with RGG domains (RGG-NanoLuc-RGG). Incorporation of enzymes into coacervates can lead to enhancement of reaction rates, as shown for examples such as β-galactosidase (40). NanoLuc is an ultrabright variant of luciferase, and we reasoned that this chemical feature combined with the increased concentration as a result of protein condensation would generate bright light emitted from condensates. Further, RGG-NanoLuc-RGG self-assembly into liquid droplets can be controlled in vitro by temperature, ionic strength and protein concentration. Remarkably, protein condensation increased NanoLuc concentration by approximately 10-fold over bulk solution and increased overall reaction rates. Interestingly, changes in condensate viscosity result in changes in light emission, providing a potential strategy for high-throughput screening of condensate targeting drugs. Incorporation of split enzyme subunits fused to RGG domains also enabled reconstitution of enzyme activity within co-condensates. We further demonstrate fine control over the dynamics and degree of coacervation, and thus split enzyme activity, using solubility tags and proteolytic cleavage as well as spatial control over enzyme localization in condensates, driving NanoLuc to the coacervate-water interface. Taken together, our results demonstrate the potential for IDR-based protein condensates as hubs for driving or regulating biochemical reactions.
Experimental
Molecular cloning.
NanoLuc and NanoBit were engineered by Promega (Madison, WI, USA). The plasmid encoding NanoLuc (41), was purchased from Addgene (Watertown, MA, USA). DNA fragments encoding LgBit and SmBit were ordered from Integrated DNA Technologies (Coralville, IA, USA). Plasmids encoding RGG-RGG and MBP have been previously described (36). Oligonucleotide primers for cloning work were ordered from Integrated DNA Technologies (Coralville, IA, USA). PCR products of NanoLuc and DNA fragments above were cloned into bacterial expression vectors by In-Fusion cloning (Takara Bio, Kusatsu, Shiga, Japan) and verified by Sanger sequencing (GENEWIZ, South Plainfield, NJ, USA). All constructs contained a C-terminal 6-His tag for affinity chromatography.
Protein purification and storage.
Plasmids were transformed into BL21 (DE3) competent cells (New England Biolabs, Ipswich, MA, USA). Starter culture was grown overnight at 37 °C in 5 mL LB (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 50μg/mL kanamycin and then inoculated into 500 mL Terrific Broth (TB) Auto-induction Media supplemented with 50 μg/mL kanamycin. The culture was grown at 37 °C to an optical density (OD600) of 0.4 – 0.8, and then was grown at 18 °C overnight. E. coli pellets were obtained by centrifugation at 4500 RCF for 10min. Pellets were resuspended in buffer containing 500 mM NaCl, 20 mM Tris, 20 mM imidazole, 1 mM DTT, and protease inhibitor cocktail tablets (EDTA-free, Roche, Basel, Switzerland) followed by lysis via sonication. Lysate was centrifuged at 15000 RCF for 20 min and the supernatant was incubated with HisPur Ni-NTA resin (Thermo Scientific, Waltham, MA, USA), washed with 500 mM NaCl, 20 mM Tris, 20 mM imidazole, 1 mM DTT, and eluted with 500 mM NaCl, 20 mM Tris, 500 mM imidazole, 1 mM DTT on a disposable 5 mL polypropylene column (Thermo Scientific, Waltham, MA, USA). Eluted RGG-RGG protein was dialyzed into 500 mM NaCl, 20 mM Tris buffer, PH 7.5, at 42 °C. Other eluted proteins were dialyzed into 500 mM NaCl, 20 mM Tris, 10 v/v% glycerol, 1 mM DTT. RGG-NanoLuc-RGG was dialyzed overnight at 42 °C. To purify a fluorescent RGG construct (RGG-GFP-RGG), pellets were resuspended in a lysis buffer (Tris-HCl, pH 7.5, 1 M NaCl, 50 mM Imidazole, 1 mM β-mercaptoethanol) with a dissolved tablet of protease inhibitor cocktail (Roche), lysed, cleared by centrifugation, and incubated with Ni-NTA beads as above. Beads were then washed in five column volumes of lysis buffer and eluted with lysis buffer containing 500 mM imidazole and 1 mM DTT. Elutions were dialyzed overnight into buffer containing 1M NaCl, 20 mM Tris-HCl, pH 7.5, 1 mM DTT at room temperature. Proteins were concentrated by centrifugation in filter concentrators with 10kDa cutoff (Amgen; Thousand Oaks, CA, USA). Protein concentrations were determined by measuring absorbance at 280 nm on a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) after preclearing by centrifugation (9800 RCF, 10 min) and mixing 1:1 with 8M urea dissolved in PBS. RGG-GFP-RGG concentrations were determined by Bradford assay (BioRad; Hercules, CA, USA). Protein aliquots were snap frozen in liquid nitrogen and stored at −80°C.
Sample preparation for microscopic assays.
Protein solutions were precleared by centrifugation as above. RGG-NanoLuc-RGG and RGG-RGG were mixed and diluted with salt-free buffer (0 mM NaCl, 20 mM Tris, 1 mM DTT) and 150 mM NaCl buffer (150 mM NaCl, 20 mM Tris, 1 mM DTT) to induce phase separation in a mixture containing 150 mM NaCl, 20 mM Tris, 1 mM DTT. Phase separation proceeded at room temperature. MBP-RGG-LgBit-RGG and MBP-RGG-SmBit-RGG were mixed and diluted as above to reach a mixture containing 150 mM NaCl, 20 mM Tris, 1 mM DTT. Phase separation was then induced by proteolytic cleavage of MBP from the rest of the polypeptide with addition of HRV3C to final concentration of 1 μM. In all cases, samples were in a final volume of 25 to 50 μl and were placed in wells of a CultureWell chambered coverglass with 0.17 mm bottom thickness (Invitrogen, Carlsbad, CA, USA), passivated with Pluronic F-127 (Sigma-Aldrich, St. Louis, MO, USA) overnight and rinsed before use. In all cases, condensates were allowed to form for 1hr prior to experiments.
Microscopy and bioluminescence imaging.
Samples were imaged on a Leica (Wetzlar, Germany) DMi8 inverted microscope using a Leica 63x / 1.4 Harmonic Compound (HC) plan-apochromatic (PL-APO) oil-immersion objective (Article No. 11506349). Imaging was performed in a dark room, with all shutters closed, and a 460 nm emission filter with 50nm bandwidth loaded. Differential interference contrast (DIC) and bioluminescence images were captured by a Hamamatsu (Shizuoka, Japan) ORCA-Flash 4.0 V2 digital sCMOS camera (C11440-22CU). with a quantum efficiency (QE) of ~70 % at 460 nm and a conversion factor of 0.48 electrons/ADU. Bioluminescence images were taken immediately after addition of 50x Nano-Glo luciferase assay substrate (Promega, Madison, WI, USA).
Image processing and analysis.
Bioluminescence images were background subtracted by the built-in function of ImageJ for overlaying. Images were analyzed by a custom written MATLAB algorithm modified from Reed and coworkers (37). A previously described algorithm was used to create circular masks in an automated fashion (42). To estimate NanoLuc concentrations in the protein coacervates, analogue-to-digital units (ADU) in each pixel were converted to photons per pixel per second on the camera sensor: Photon flux = C · (I−D)/(QE · t), where C is the conversion factor, I is the light intensity in ADU, D is the average intensity value from 3 dark images taken under the same exposure, and t is the exposure time. We simulated collection efficiency of photons traveling from protein coacervates of different radii onto the camera sensor and used these values for NanoLuc concentration measurements (43). Briefly, we calculated the fraction of emitted light collected by the objective and which contributes to the image of the droplet. We calculated approximate solution by geometrical ray tracing similar to Velesco and Schweiger (44) with forward propagation of rays from the source onwards. Figure S1 illustrates a droplet on a cover glass and exemplifies the propagation of light from a luminescent source towards the objective. We assumed 52 % transmission efficiency of the filter set according to Leica (Wetzlar, Germany) including ~90 % transmission through the dichroic mirror and ~96 % transmission through the emission filter with ~60 % coverage of the bioluminescence spectrum. Images for fluorescence recovery after photobleaching (FRAP) were analyzed using ImageJ plugins written by Jay Unruh at Stowers Institute for Medical Research in Kansas City, MO.
Bioluminescence measurements by plate reader assay.
Bioluminescence was measured in 96-well plates by a Tecan infinite 200 Pro plate reader (Tecan, Männedorf, Switzerland) at room temperature. The wells were passivated with Pluronic F-127 (Sigma-Aldrich, St. Louis, MO, USA) overnight and rinsed before addition of 25μL samples. LLPS was induced by dilution of protein into a low salt condition (150 mM NaCl). All samples were in a final buffer of 150 mM NaCl, 20 mM Tris, 1 mM DTT. The samples were incubated at room temperature for 1hr before the experiment. For kinetics measurement, Nano-Glo assay substrate (Promega, Madison, WI, USA) was diluted into different concentrations in buffer containing 150 mM Nacl, 20 mM Tris, 1 mM DTT and 5 μL was pipetted into each well. The bioluminescence signals were read with 20 s waiting time and 1 s integration time. For all other experiments, Nano-Glo assay substrate (Promega, Madison, WI, USA) was diluted 200x in buffer containing 150 mM Nacl, 20 mM Tris, 1 mM DTT and 5 μL was injected into each well by a Te-inject reagent injector (Tecan, Männedorf, Switzerland). The bioluminescence signals were then read with 2 s waiting time and 10 s integration time.
Fluorescence recovery after photobleaching (FRAP).
FRAP experiments were performed on an Olympus IX81 inverted confocal microscope (Olympus Life Science; Tokyo, Japan) equipped with a Yokogawa CSU-X1 spinning disk, Mercury lamp, 488 and 561 nm laser launches, iLas targeted laser system for photobleaching, and an iXon3 EMCCD camera (Andor; Belfast, UK). Image acquisition was controlled by MetaMorph software (Molecular Devices; Downingtown, PA). Samples were prepared as above in 50μl and incubated at room temperature for 1hr prior to imaging. RGG-GFP-RGG at a final concentration of 200nM was used as a fluorescent tracer for FRAP experiments. Samples were illuminated using a 488 nm laser and imaged through a 100x/1.4 NA oil-immersion objective. Photobleaching was performed using a 405 nm laser from the iLas laser system. For photobleaching of internal regions of droplets, ROIs of similar sizes were selected and bleached. For photobleaching of whole droplets, an ROI encompassing an entire droplet was selected and photobleached as above.
Results and Discussion
Recruitment of NanoLuc Enzyme into protein condensates.
We previously showed that LLPS of RGG-based protein coacervates can be tuned by control over RGG domain valency and multivalent IDR scaffolds can be further engineered to localize or concentrate other biomolecules (36, 38). First, the assembly and disassembly of the RGG-based coacervates can be reversibly controlled by temperature, protein concentration, ionic strength, and light (37). Second, the platform is capable of selectively recruiting multiple protein cargos (36). Third, the permeability of RGG-based protein coacervates to small molecules enables substrates and products to diffuse in and out of the coacervates. Given these findings, we reasoned that this platform may be further functionalized to concentrate enzymes and catalyze biochemical activities within protein coacervates. We chose to test a light-emitting NanoLuc enzyme as a cargo such that enzymatic activity inside the coacervates can be directly visualized and measured by microscopy. Bioluminescence microscopy has been thought as challenging because light emission can be dim (45–47) and specialized microscopy may be needed for the detection of bioluminescent activity (46–49). NanoLuc is small (19 kDa), more stable, and ~150-fold brighter than its traditional counterpart (39). We reasoned that local concentration of NanoLuc luciferase in protein coacervates would catalyze high enzymatic activity and lead to bright luminescence signal (Figure 1a).
Figure 1.
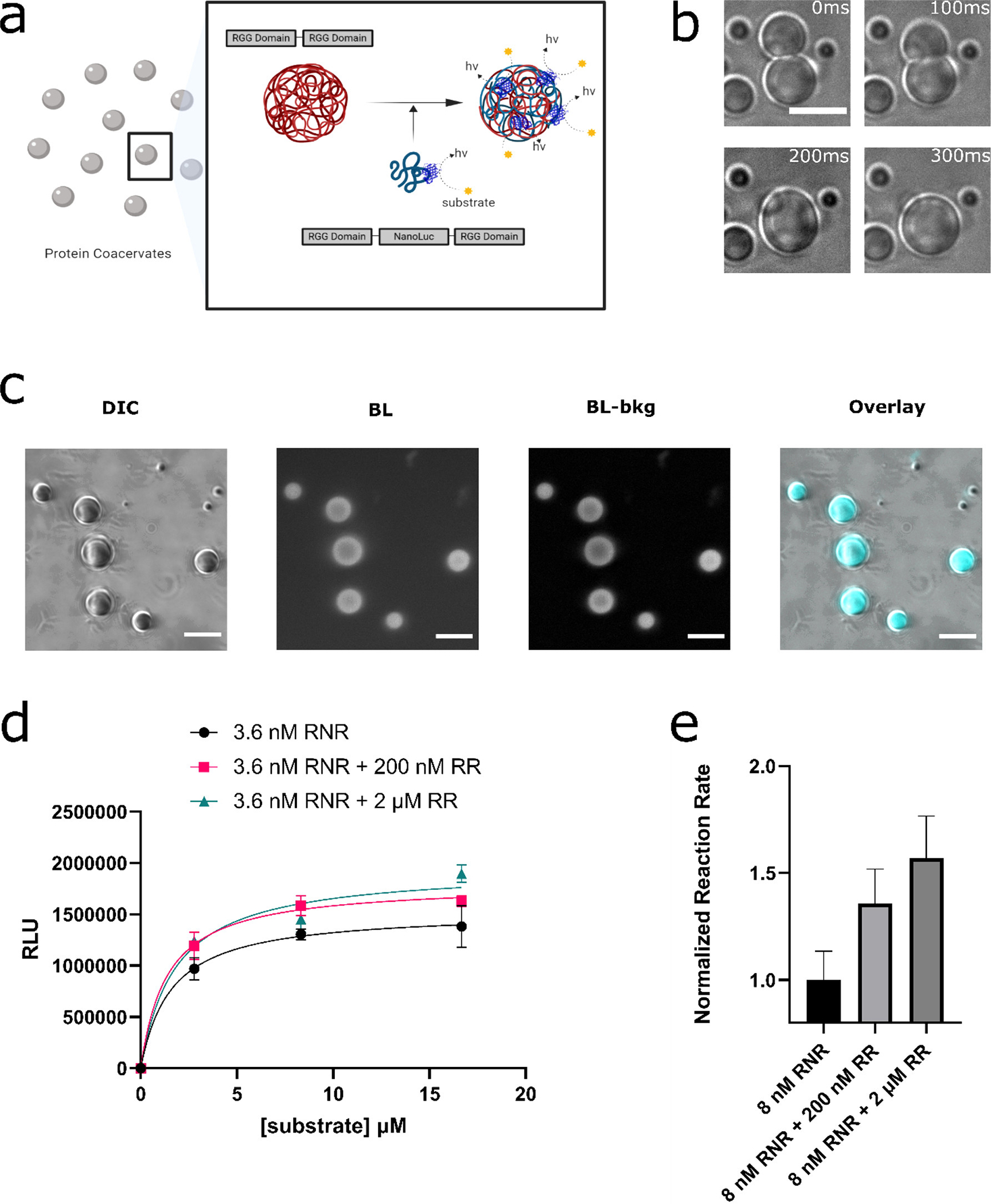
Incorporation of NanoLuc enzyme into RGG based protein coacervates. (a) Schematic of design goal: (i) localization of NanoLuc enzyme molecules in RGG-based protein coacervates and (ii) studying the enzymatic reaction via bioluminescence imaging under a microscope or quantification by a luminometer. (b) RGG-NanoLuc-RGG coacervates displayed fusion, demonstrating their liquid-like properties, and ultimately attained a spherical shape. The coacervates can also be seen as transparent in this figure. Scale bar: 10 μm. (c) DIC and bioluminescence images of 6μM RGG-NanoLuc-RGG, imaged by an inverted microscope, confirming the enzymatic activity of NanoLuc inside the liquid coacervates. From left to right: DIC image; Bioluminescence image after 5 s exposure at a substrate concentration of ~833 μM (6x dilution of Nano-Glo assay substrate) (50); Bioluminescence image – background subtracted; Overlay, with pseudo-color. Scale bars: 10 μm. (d-e) Reaction kinetics of NanoLuc in different coacervation states measured by a microplate reader. Low concentration of RGG-NanoLuc-RGG (abbreviated as ‘RNR’) was expected to have a low tendency for LLPS. In contrast, with the presence with elevated concentration of scaffold RGG-RGG (abbreviated as ‘RR’), RGG-NanoLuc-RGG was expected to partition into the RGG-RGG condensates. (d) Comparison of reaction kinetics between 3.6 nM RGG-NanoLuc-RGG, 3.6 nM RGG-NanoLuc-RGG with 200 nM RGG-RGG, and 3.6 nM RGG-NanoLuc-RGG with 2 μM RGG-RGG, in a final solution of 150 mM NaCl, 20 mM Tris, 1 mM DTT. Sample volume: 25 μl. Data were fit into Michaelis–Menten equation in GraphPad Prism 9. (e) Comparison of normalized reaction rates. Nano-Glo assay substrate was diluted 100x and 5 μl was added into each well containing 25 μl sample in 150 mM NaCl, 20 mM Tris, 1 mM DTT.
We purified a construct of NanoLuc flanked by RGG domains (RGG-NanoLuc-RGG) and confirmed it is capable of phase separation into spherical protein coacervates in physiological salt concentrations. We also observed rapid fusion events of RGG-NanoLuc-RGG coacervates, a hallmark for liquid-like behavior (Figure 1b). Strikingly, after addition of Nano-Glo substrate into the solution, we observed clear generation of bioluminescence emanating from the condensates by bioluminescence microscopy (Figure 1c). To quantify reaction kinetics with respect to the extent of coacervation, we utilized a microplate reader to measure luminescence. We compared the reaction kinetics of nanomolar levels RGG-NanoLuc-RGG well below its saturation concentration (thus remaining in the aqueous phase) or mixed this with a higher concentrations of an RGG-RGG scaffold to drive coacervation and incorporation of RGG-NanoLuc-RGG into condensates (Figure 1d–e). Our measurements indicated an elevated reaction rates corresponding with the amount of condensate formation. Specifically, the reaction rate increased by ~1.3 fold in the presence of 200 nM RGG-RGG (a molar ratio of RGG-NanoLuc-RGG: RGG-RGG of 1:25), and by ~1.5 fold in the presence of 2 μM RGG-RGG (a molar ratio of 1:250) (Figure 1e). This suggests that concentrating enzymes within condensates improves reaction efficiency.
By microscopy, we observed an exponential decay (y = 3850e−0.025t, R2 = 0.98) of bioluminescence from condensates as the substrate in solution is consumed (Figure 2a–b). This indicates a signal lifetime on the order of 40 s. Because of high reaction rates resulting from local concentration of NanoLuc inside the condensates, maintaining a high substrate to NanoLuc concentration ratio may improve the signal lifetime. Nevertheless, NanoLuc concentration should be high enough to allow photon collection by the camera. Practically, for our setting with a normal inverted microscope with 63x/1.4 NA objective, a ~1 μM NanoLuc concentration with 10 μl substrate (or roughly 800 μM furimazine) coupled with 5 s exposure by a sCOMS camera is sufficient to capture bioluminescence from a condensate. As brightness is proportional to the square of the numerical aperture (NA) and inversely proportional to the square of the objective magnification (M) (46, 47), a high NA objective (1.4 NA) was used for imaging at high magnification. Although we did not use them, in future work we recommend the use of a light-proof box (49) or a specialized bioluminescence microscope (47 for improved light harvesting. Imaging of NanoBit (see section ‘Reconstitution of fragmented NanoLuc inside the RGG coacervates’) or NanoBRET (51) inside the condensates may have the advantage of ultra-low background, considering the elevated protein-protein interactions inside the condensed phase, although we have not tested the latter in the current study.
Figure 2.
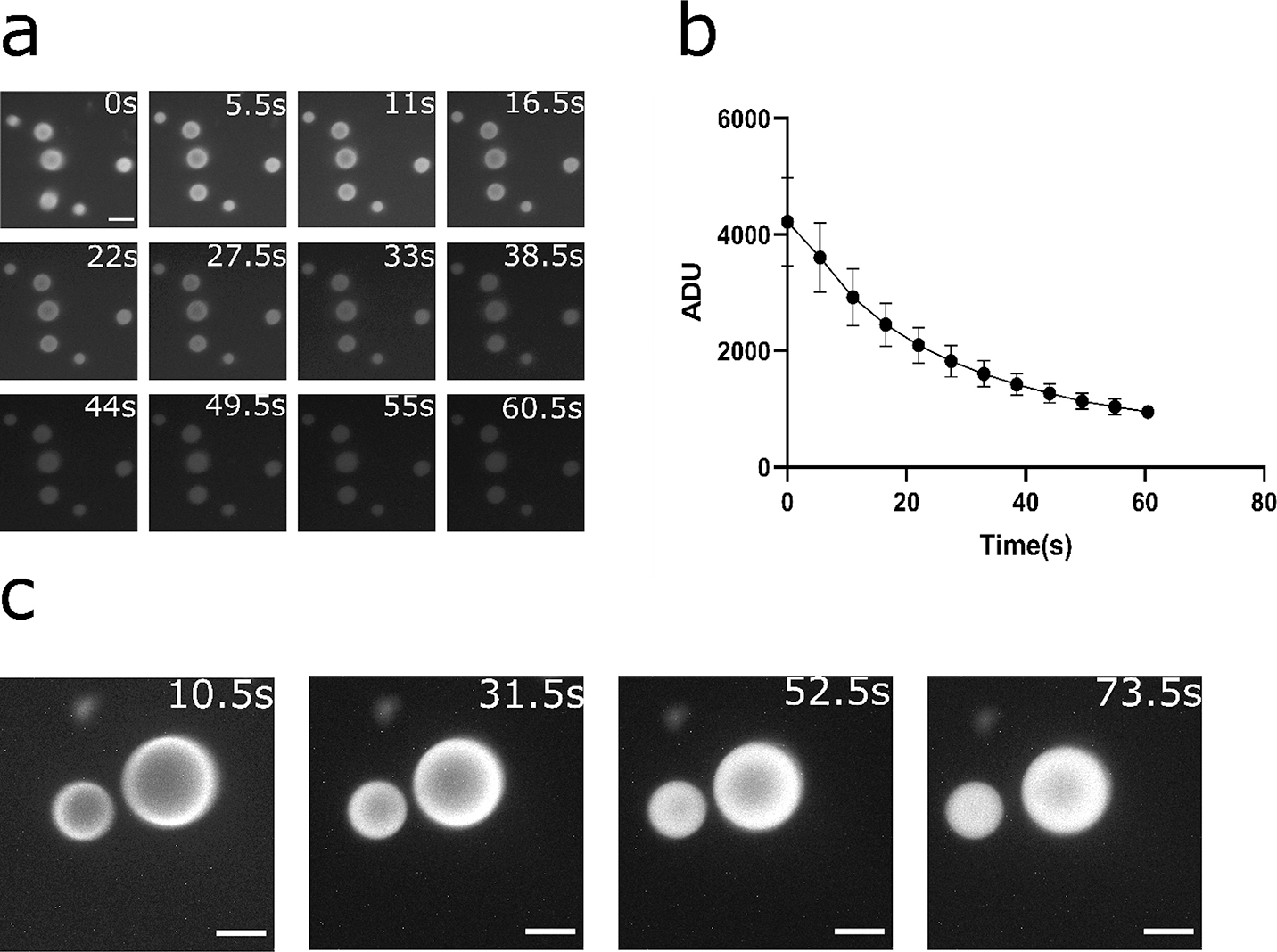
Rapid turnover of substrates and diffusion-limited behavior in the RGG protein coacervates containing NanoLuc. (a-b) Kinetics of bioluminescence from the coacervates for 6μM RGG-NanoLuc-RGG, with substrate concentration of ~833 μM (6x dilution of Nano-Glo assay substrate). (a) Bioluminescence images at different time points. Contrast at default for comparison between time points. Scale bar: 10 μm. (b) Plot of mean pixel intensity inside coacervates over time. Exposure: 5 s. Substrate: 10 μl (~833 μM). Data presented as mean ± SD. (c) Bioluminescence images of RGG-NanoLuc-RGG coacervates over time indicating evidence of diffusion-limited reaction. Reaction was initially predominantly occurring at the coacervate-water interface and gradually reached inside corresponding to substrate diffusion into the coacervates. 20 μl substrate (~1430 μM final substrate concentration) was added into a mixture of 2.25 μM RGG-NanoLuc-RGG with 13.5 μM RGG-RGG. Exposure: 10 s. Scale bars: 10 μm.
It has been reported that NanoLuc has potentially high turnover rates (39). Indeed, we detected diffusion-limited behavior of the reaction inside a portion of the RGG-based coacervates (Figure 2c). Specifically, a ring of bioluminescence can be seen from the initial frames of the bioluminescence image series, indicating that the reaction generated significantly more photons at short time scales at the coacervate-water interface than the interior of these condensates. Over time, bioluminescence intensity gradually encompassed the entirety of individual condensates, indicating diffusion of substrate into the coacervates may be rate limiting. We use several seconds of exposure in these assays and thus assume that diffusion of the substrate into these coacervates were on a similar time-scale. The diffusion time scale (τ) can be estimated by the Einstein-Smoluchowski equation: τ = L2/D, where L is the characteristic length and D is the diffusion coefficient. The Stokes-Einstein equation gives an estimate of the diffusion coefficient: D = kT/(6πηa), where k is the Boltzmann constant, T is the absolute temperature, η is the viscosity of the diffusion medium, and a is the radius of the diffusing molecule. Using the reported viscosity value of RGG-RGG condensates (1.6 Pa·s) (52) and an estimated simple radius of furimazine calculated from the topological polar surface area (0.2 nm), we estimate a diffusion coefficient of furimazine in RGG-based condensates D ≈ 0.7 μm2/s. This indicates that for a condensate of 1 μm radius, the diffusion time scale of the substrate is on the order of 1 to 2 s; for a condensate of 5 μm radius, however, the diffusion time scale of the substrate is on the order of 35 s. We believe this diffusion-limited behavior is potentially useful, as condensate viscosity might affect the light output when screened by a microplate reader. We propose that this system may aid in high-throughput screening (HTS) for discovery of condensate targeting molecules or drugs for industrial or therapeutic applications (see section ‘Condensate viscosity affects light output’).
Unexpectedly, NanoLuc activity resulted in the formation of fibril-like structures around the coacervates or in the surrounding aqueous phase (Figure 3a). Negative controls, which included the addition of Nano-Glo substrate itself into pure RGG-RGG coacervates or addition of only 150 mM NaCl, 20 mM Tris buffer into coacervates containing RGG-NanoLuc-RGG, did not cause fibril formation (Figure 3a). We observed formation of similar structures after adding 10μl 50x Nano-Glo luciferase assay substrate into 10 μl 0.4 mg/mL pure NanoLuc enzyme solution (Figure 3a), suggesting that these fibril-like structures are an enzyme-specific event. Nevertheless, the RGG-NanoLuc-RGG coacervates remained active even after fibril formation, as bioluminescence images showed that further additions of substrate after the exhaustion of initial reaction could reinitiate the reaction and led to the generation of luminescence. Subsequent reaction rates appeared lower than that of the initial reaction, as the bioluminescence images showed a 65 % reduction in light intensity from the coacervates for the second reaction (Figure 3b–c).
Figure 3.
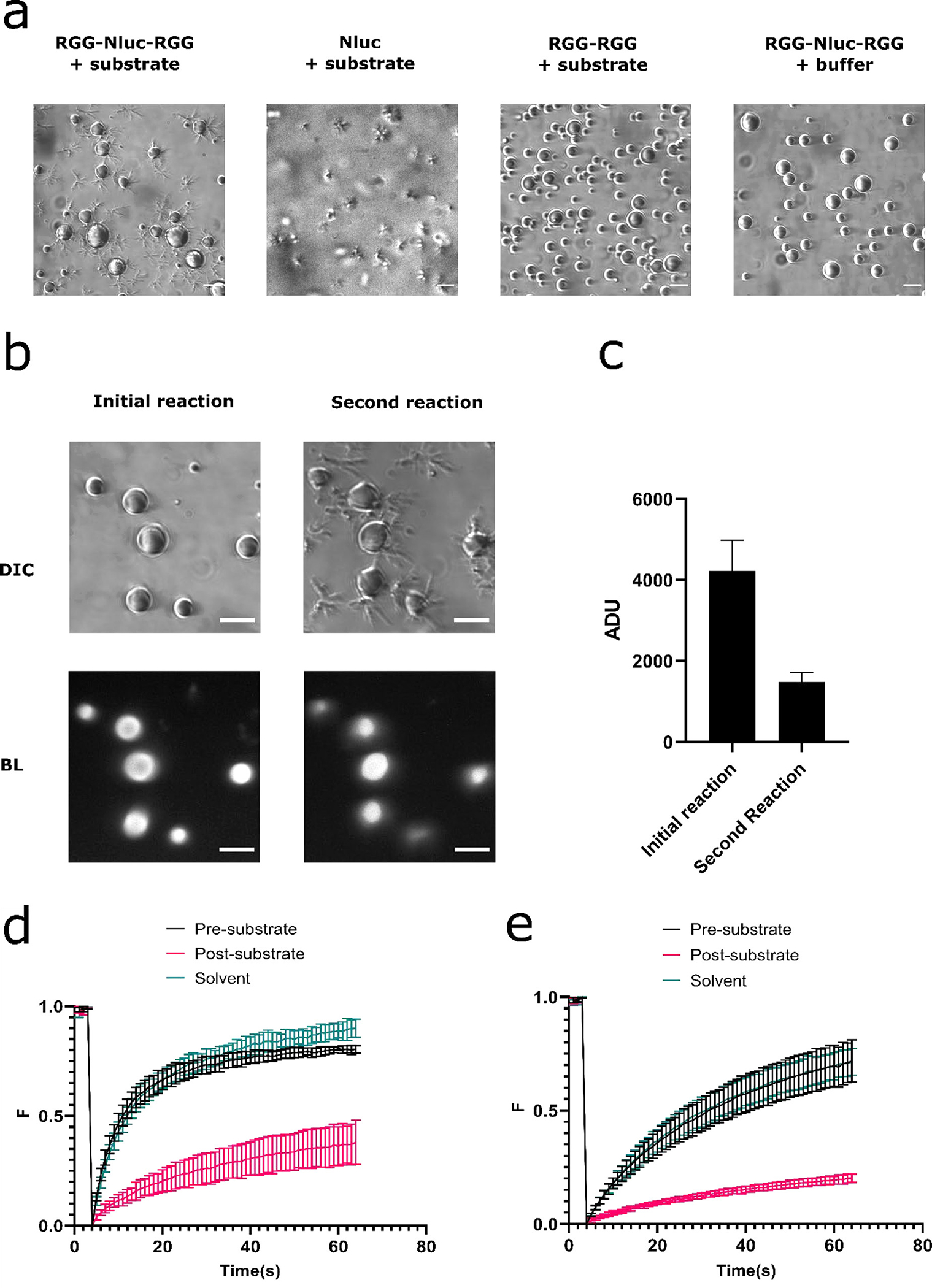
Morphology and structural changes of the coacervates after the Nanoluc enzymatic reaction. (a) Formation of fibril-like structures after NanoLuc reaction. RGG-NanoLuc-RGG coacervates generated fibril-like structures after reaction. Substrate: 10 μl. Similar structure was also observed after reaction of pure NanoLuc enzyme. 10 μl substrate was added into 10 μl of 0.4 mg/mL pure NanoLuc enzyme. We did not observe formation of such structures after adding 10 μl substrate into 6 μM RGG-RGG, or adding 10 μl buffer containing 150mM NaCl, 20 mM Tris into a mixture of 6 μM RGG-NanoLuc-RGG with 2 μM RGG-RGG. Scale bars: 10 μm. (b) NanoLuc enzyme remained active in RGG-based coacervates despite formation of fibril-like structures. Top left: DIC image of the coacervates before initial reaction. Bottom left: Bioluminescence image of the coacervates during the initial reaction. Substrate: 10 μl (~833 μM final concentration). Top right: DIC image of the coacervates after the initial reaction and before the second reaction. Bottom right: Bioluminescence images during the second reaction. 10μl substrate (~714 μM final concentration) was re-added into the well after the initial reaction exhausted. Exposure: 5 s. Scale bars: 10 μm. (c) Comparison of mean pixel intensity in the coacervates between initial and second reaction. Data presented as mean ± SD. (d) FRAP curves on internal spots of the coacervates before and 20 minutes after addition of 10 μl substrate. Sample contains 6 μM RGG-NanoLuc-RGG with 200 nM RGG-GFP-RGG in 150 mM NaCl, 20 mM Tris, 1 mM DTT buffer. Total volume was 50 μL. A control group was measured 20 minutes after adding the same volume of substrate solvent (85% ethanol and 15% glycerol) into the well. (e) FRAP curves on the entire coacervates before and 20 minutes after addition of 10 μl substrate. Sample contains 6 μM RGG-NanoLuc-RGG with 200 nM RGG-GFP-RGG in 150 mM NaCl, 20 mM Tris, 1 mM DTT buffer. Total volume was 50 μL. A control group was measured 20 minutes after addition of the same volume of substrate solvent (85% ethanol and 15% glycerol).
To test whether NanoLuc enzymatic activity caused changes to physical properties of condensates, we performed fluorescence recovery after photobleaching (FRAP) experiments with in vitro condensates as described above, supplemented with 200 nM of a fluorescent RGG construct (RGG-GFP-RGG) to function as a tracer. We photobleached internal regions of condensates (Figure 3d, Figure S7b) or entire condensates (Figure 3e, Figure S7c) to measure internal diffusion and diffusion of fluorescent molecules from solution into the condensates, respectively. RGG-GFP-RGG partitioned into the RGG-NanoLuc-RGG coacervates, but not in the fibrils (Figure S7a). FRAP was performed before and after enzymatic reactions (starting 20 min after substrate addition). These experiments revealed that condensate dynamics were significantly reduced after NanoLuc activity. Before substrate addition, the amplitude of fluorescence was 0.72 ± 0.03 after 60 s and half recovery time was 8.3 ± 1.7 s for internally photobleached condensates. After addition, the amplitude dropped to 0.38 ± 0.11 and half time increased to 27.7 ± 3.8 s. Addition of the same volume of substrate solvent (85 % ethanol and 15 % glycerol) without furimazine showed a similar amplitude (0.79 ± 0.03) and half time (11.6 ± 0.9 s) to the non-catalyzed control, indicating the drop in fluorescence recovery is not due to the solvent effects on the condensates (Figure 3d). We observed similar results when analyzing condensates that were photobleached in their entirety (Figure 3e). Overall, our FRAP data suggest that the liquidity of the coacervates decreased after addition of the substrate and enzymatic activity, and this was not an effect caused by the solvent component of the substrate. Therefore, we conclude that the enzymatic reaction of NanoLuc inside the RGG protein coacervates caused a transition of the coacervates from a liquid to a more gel-like state.
Condensate viscosity affects light output.
The physical properties of the condensates have been associated with various pathologies. For example, multiple IDR-containing RNP granule proteins associated with amyotrophic lateral sclerosis (ALS) transit into a more solid-like state over time (10, 11). This liquid-to-solid transition is accelerated by ALS-causing mutations (11, 27, 53). Both liquid-to-gel or liquid-to-solid transitions can potentially be associated with disease (11, 27, 54, 55). This suggests that, in some cases, molecules that fluidize disease-associated condensates could alleviate or reverse pathological transitions. In addition, a recent study showed drugs that cause condensate hardening can also bring benefits such as blocking viral replication in the condensate viral factories (56).
Condensate targeting drugs represent a new therapeutic avenue that has gained considerable momentum in drug discovery in recent years (57, 58). However, screening for molecules that target condensates remains a daunting task and new techniques are in demand (57). Inspired by our finding that the NanoLuc reaction is diffusion-limited inside our model condensate system and the resulting luminescence can be easily detected by a microplate reader, we asked whether our system can provide a new strategy for high-throughput screening (HTS) of the condensate targeting molecules by detecting changes in light signal resulting from changes in the physical properties of condensates, namely their viscosity.
Although we are unaware of a drug molecule that could lead to hardening or solidification of our RGG condensates, we show in figure 3d–e that RGG-NanoLuc-RGG coacervates exhibit a liquid-to-gel-like transition following addition of furimazine, associated with a dampened recovery in FRAP experiments. We used a microplate reader to determine whether this property change of the condensates could lead to a reduction in light signal in the following reactions (Figure 4). We added furimazine-containing Nano-Glo substrate into wells containing 2 μM RGG-NanoLuc-RGG and let the reaction proceed, then measured luminescence after 3.5 hr, following the second addition of substrate. Compared to condensates not treated with furimazine, RGG-NanoLuc-RGG coacervates treated with furimazine showed a 43% reduction in light signal. This reduction is in line with our observation by bioluminescence microscopy shown in figure 3c. In comparison, we did not observe a significant change in light signal for Nanoluc in free solution. Combined with our observation that NanoLuc showed diffusion-limited behavior inside condensates, we demonstrate that more viscous condensates result in decreased luminescence signal, likely by hindering the diffusion of substrate through the coacervates.
Figure 4.
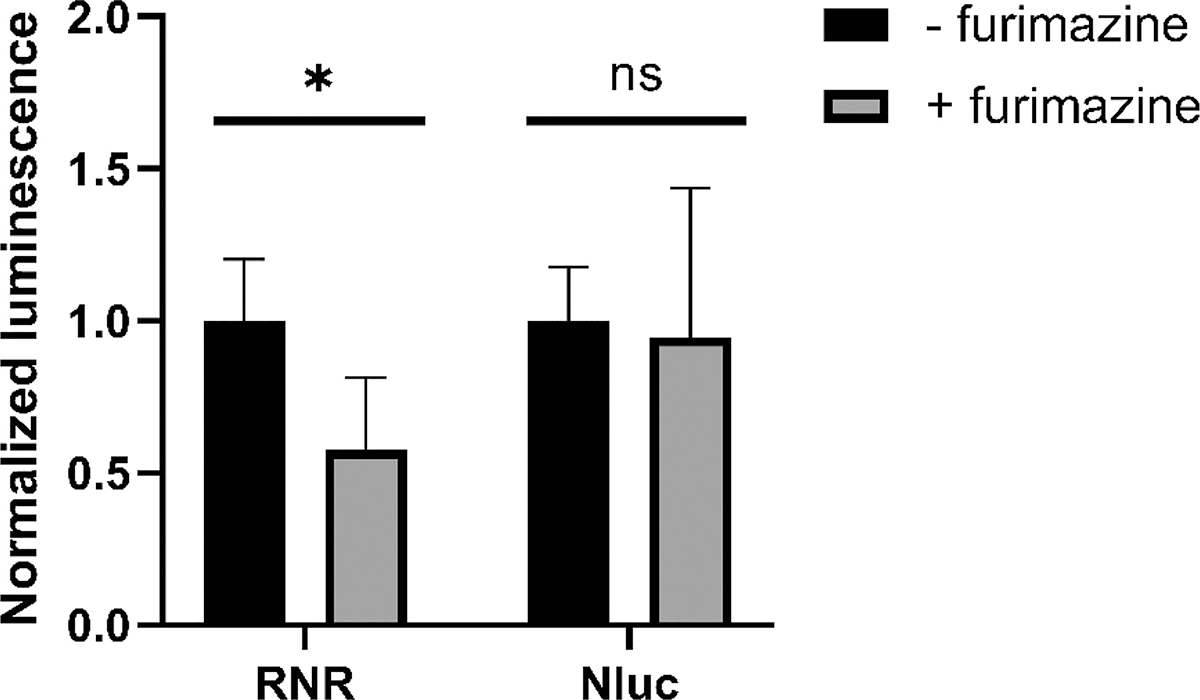
Hardened RGG-NanoLuc-RGG coacervates caused a change in light signal detected by a plate reader. 2 μL furimazine-containing Nano-Glo substrate was added into wells containing 2 μM RGG-NanoLuc-RGG (abbreviated as ‘RNR’) or 2 μM NanoLuc (‘Nluc’) in 150 mM NaCl, 20 mM Tris, 1 mM DTT buffer. In control groups, no furimazine was added. After 3.5 hr, light signal was measured following injection of 5 μL 200x diluted Nano-Glo substrate. Data presented as mean ± SD (n = 6). *, p < 0.05; ns, not significant; multiple unpaired t-test.
Considering that RGG-RGG condensates have lower viscosity (1.6 Pa·s) than most other IDP condensates (52), we expect the NanoLuc reaction to be diffusion-limited in most condensates that have a comparable or higher viscosity. We believe our model system can be easily translated to other condensate platforms by replacing the RGG domain with the pathological IDR. A condensate-Nanoluc system combined with simple plate reader assays can be used to screen for molecules that cause changes in physical properties or phase behavior. Theoretically, a condensate fluidizing drug would lead to an increase in light signal in hardened or solidified condensates, and a condensate hardening drug would cause a decrease of the light signal. Future investigations are needed to define on the effectiveness of this strategy.
Dose dependence of NanoLuc enzyme concentration and light intensity inside condensates.
We hypothesized that NanoLuc concentration and therefore its enzymatic activity in the coacervates can be regulated by adjusting the composition of RGG-NanoLuc-RGG in a mixture with non-reactive RGG-RGG. We blended RGG-NanoLuc-RGG with RGG-RGG at different ratios and imaged bioluminescence from mixtures containing 25 %, 50 %, 75 %, and 100 % RGG-NanoLuc-RGG after addition of Nano-Glo luciferase substrate (Figure 5a). The average intensities in coacervates were analyzed and plotted with respect to NanoLuc composition (Figure 5b). We observed a linear relationship between the light intensity from the coacervates with respect to percentage of RGG-NanoLuc-RGG in the mixture, with mean pixel intensity (in ADU) of 2246, 3762, 6861, and 8159 at 25%, 50%, 75%, and 100% NanoLuc, respectively.
Figure 5.
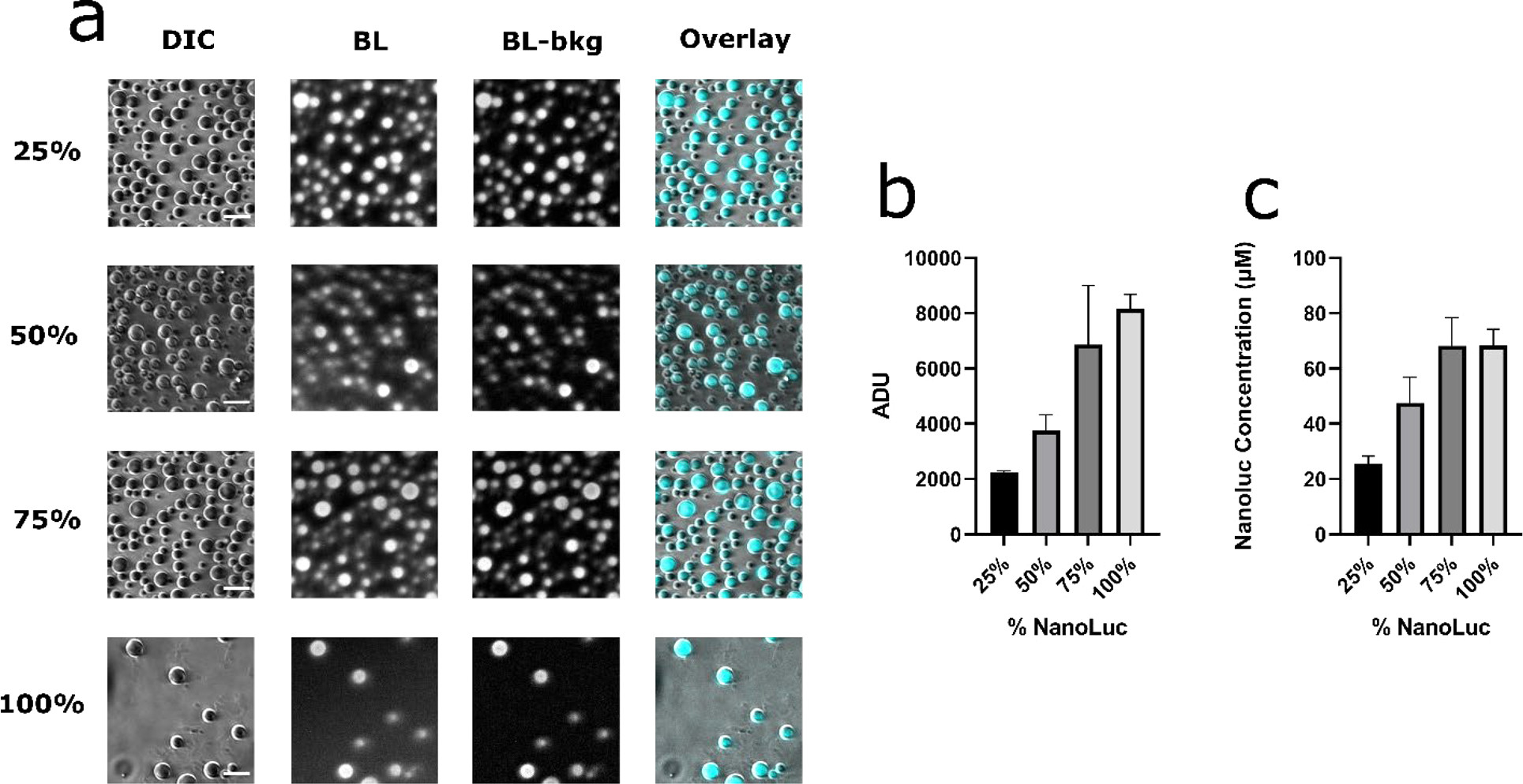
Dose dependence of light intensity as a function of NanoLuc concentration in the protein coacervates. (a) DIC and bioluminescence images of RGG-NanoLuc-RGG combined with RGG-RGG mixed at different ratios. From left to right: DIC image, Bioluminescence image, Bioluminescence image – background subtracted, Overlay. From top to bottom: RGG-NanoLuc-RGG at 25 %, 50 %, 75 %, and 100 % total composition. Scale bars: 10 μm. Total concentration of RGG-NanoLuc-RGG and RGG-RGG upon mixing: 9 μM. Exposure: 10 s. Substrate: 10 μl (~833 μM final concentration). (b) Average pixel intensity in the protein coacervates at different mixing ratios. (c) Estimate of NanoLuc concentration in the protein coacervates at different mixing ratios. Data presented as mean ± SEM.
To estimate NanoLuc enzyme concentration in condensates, bioluminescence intensity values in condensates were converted to photon flux on each camera pixel using parameters of the camera and exposure time. Photon collection rate on the camera sensor from each coacervate was obtained by summing up the pixels in each protein coacervate. We divided this value by a photon collection efficiency simulated for coacervates of different radius to obtain the photon emission rate from each coacervate. Assuming maximum reaction rate at a reported value of 1.9 photon · s−1 · molecule−1 for NanoLuc enzyme (59), we obtained molar amount of NanoLuc enzyme in the coacervates, and by dividing the coacervate volume, we obtained estimates of NanoLuc concentration and plotted against NanoLuc composition (Figure 5c). Although we obtained the images immediately after addition of substantial amount of substrate, we believe the NanoLuc enzyme concentrations are still underestimated, especially at high NanoLuc composition, as Figure 2a shows a decay of light signal during the time scale of exposure due to rapid enzyme turnover. In addition, coacervate movement during exposure and substrate diffusion-limited behavior in a portion of the coacervates are also causes for this underestimation. Nevertheless, we observed that NanoLuc is concentrated in the coacervates.
At 25 % composition of NanoLuc (corresponding to 2.25 μM in the mixture), we estimate a NanoLuc concentration of 25.6 μM inside the coacervates, or roughly a 10-fold increase in concentration. At 50 % (4.5 μM in the mixture), we estimate this concentration to be 47.5 μM. At 75% (6.75 μM in the mixture) and 100 % (9 μM in the mixture) we estimate NanoLuc concentrations of 68.2 and 68.5 μM, respectively, in the coacervate phase. In each case, we consistently observed a ~10-fold increase in local concentration of NanoLuc enzyme inside coacervates, compared to the bulk concentration prior to condensate formation. Altogether, experimental data upheld our hypothesis that enzymes could be incorporated and concentrated within protein condensates and that enzyme activity can be tuned by mixing RGG-NanoLuc-RGG and RGG-RGG at different ratios. NanoLuc enzyme was highly concentrated in the protein coacervate phase, enabling the capture of high-quality bioluminescence images by conventional light microscopy. This concentrating effect of enzymes by protein coacervation supports the importance of LLPS and biomolecular condensates as hubs for biochemical activity.
Reconstitution of fragmented NanoLuc inside the RGG condensates.
Having demonstrated that enzymes can be incorporated and concentrated in protein coacervates, we wondered whether such concentration could reconstitute enzymatic activity from split enzymes inside coacervates. This hypothesis is based on early observations that fragmented peptides of the enzyme ribonuclease A spontaneously self-assembled into a native and functional folded structure (60, 61). Fusing the ‘split’ parts of a fluorescent or bioluminescent protein with two interacting proteins may promote reconstitution of function by increasing the effective concentration of the split parts, and thus has become an approach for probing protein-protein interactions (61, 62). Considering the high concentration and the fluidity of the phase-separated protein condensates, we hypothesize that they would be especially powerful for the assembly of such split proteins.
To test this hypothesis directly, we incorporated the split components of the NanoLuc enzyme inside the RGG protein coacervates. We used NanoBiT, an engineered split NanoLuc system for detecting protein-protein interactions, consisting of a small 1.3 kDa peptide (SmBiT) and a larger 17.6 kDa peptide (LgBiT) (63). The pair has a dissociation constant of 190 μM, making them less likely to assemble in aqueous phase but sequestration into the coacervate would theoretically increase the effective concentration of the components inside and thus promote increased associations (Figure 6a). We flanked SmBit and LgBit with RGG domains and fused an MBP domain at the N-termini of the recombinant proteins along with Human Rhinovirus 3C (HRV3C) protease cleavage site (MBP-x-RGG-LgBit-RGG and MBP-x-RGG-SmBit-RGG, where x denotes for HRV3C cut site). We confirmed that HRV3C effectively cleaved MBP domains from both of these constructs. We observe nearly complete cleavage within 20 minutes (Figure S8). A mixture of 3 μM MBP-RGG-LgBit-RGG and 3 μM MBP-RGG-SmBit-RGG in physiological salt concentrations does not phase separate into observable protein condensates in microscopic assays (Figure 6b). Addition of HRV3C resulted in micron-scale protein coacervates within 60 minutes (Figure 6b). Bioluminescence images show that LgBit and SmBit were reconstituted inside the RGG protein coacervates and generate a detectable light signal after addition of Nano-Glo substrate (Figure 6c). Again, we observe the formation of fibril-like structures as described above following the reaction. In comparison, 6 μM MBP-RGG-LgBit-RGG or 6 μM MBP-RGG-SmBit-RGG alone did not generate a signal after addition of HRV3C and Nano-Glo substrate. This confirms that bioluminescence is a result of reconstitution and assembly of LgBit and SmBit and not from either split part of the enzyme. To test how phase separation induced by HRV3C cleavage contributed to the reconstitution of the split NanoLuc, we measured bioluminescence from samples containing single or both split parts, with or without the presence of HRV3C protease, using a plate reader (Figure 6d). Nano-Glo substrate was injected in each well containing protein solution; samples containing only a single NanoBit component did not generate a signal, regardless of HRV3C addition. In contrast, we observed a 1.5-fold increase in luminescence signal in samples containing 3 μM of both components and 2 μM HRV3C compared to samples containing 3 μM of both components but without HRV3C; we noticed although the latter group did not generate a signal detectable by microscopy, plate reader assays were able to detect measurable signal, indicating LgBit and SmBit at a concentration far below their dissociation constant reconstituted even without protease-induced LLPS. We believe it is likely that LgBit and SmBit reconstituted in aqueous phase via homotypic interactions of the RGG domains, or even formed coacervates at a small size scale. Nevertheless, reconstitution in the aqueous phase is significantly lower than in the condensed phase, and does not generate detectable signal in our microscopic assays. Taken together, we conclude that the increase in bioluminescence signal is resulted from LLPS and coacervate formation which promoted the reconstitution of LgBit and SmBit into a functional enzyme. Overall, our results demonstrate that formation of protein coacervates could promote reconstitution of an enzyme inside the coacervates by increasing the local concentration of the split parts.
Figure 6.
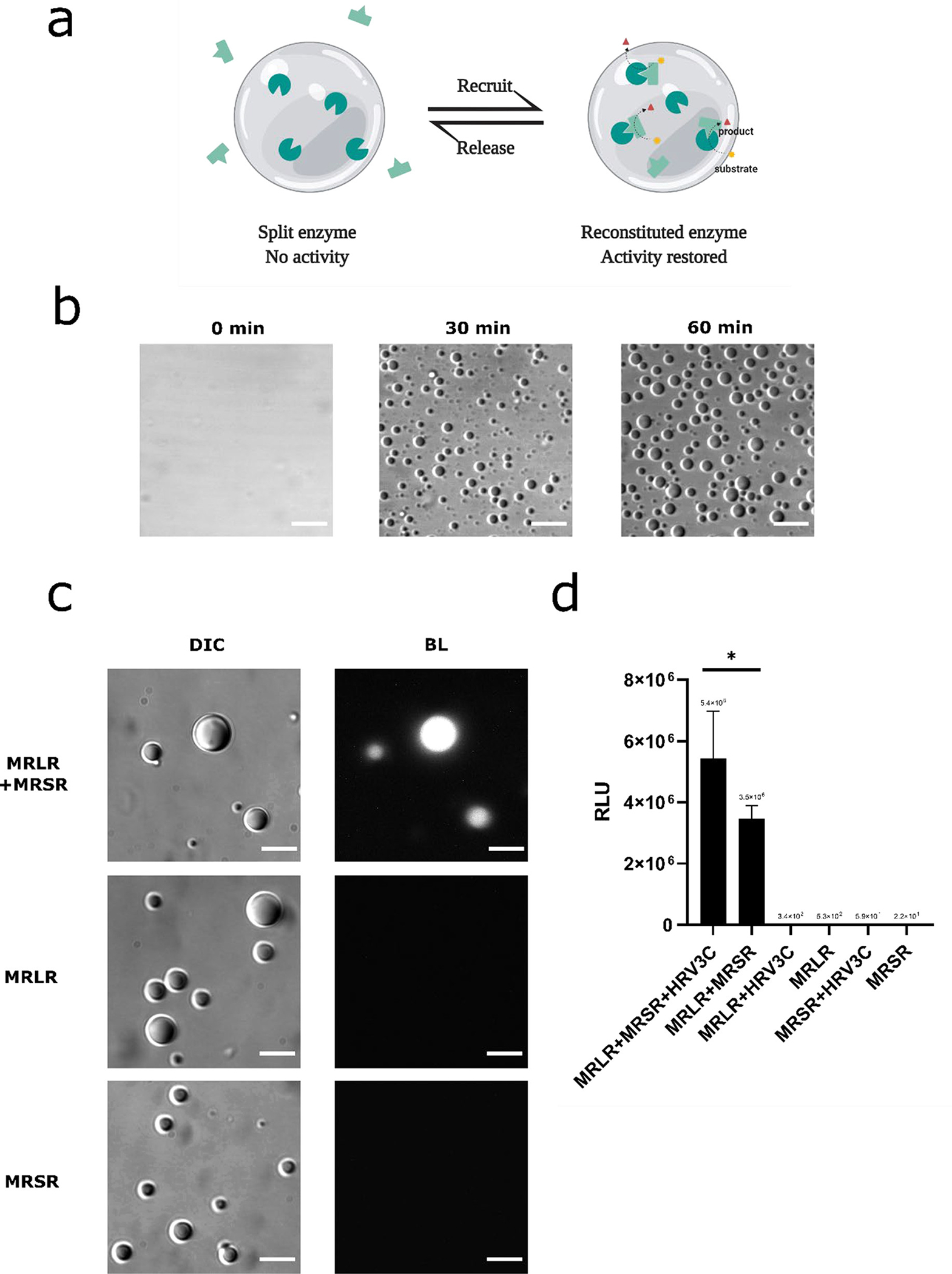
Incorporation of fragmented Nanoluc enzyme into the RGG protein coacervates. (a) design schematic: Formation of RGG protein coacervates promotes reconstitution of the split enzyme and turns on biochemical reaction inside the coacervates. (b) RGG-LgBit-RGG and RGG-SmBit-RGG form coacervates. HRV3C protease (1μM final concentration) was added into a mixture of 3 μM MBP-x-RGG-LgBit-RGG and 3 μM MBP-x-RGG-SmBit-RGG (‘x’ denotes for HRV3C cleavage site) to cleave off the MBP domain and promote phase separation. Images of droplets were taken at different time points after protease addition. Scale bars: 10 μm. (c) Bioluminescence images showing reconstitution of LgBit and SmBit inside RGG protein coacervates. Top row: DIC and Bioluminescence images of protein droplets taken 1hr after adding HRV3C protease (1 μM final concentration) into a solution containing 3 μM MBP-x-RGG-LgBiT-RGG (abbreviated as ‘MRLR’) with 3 μM MBP-x-RGG-SmBiT-RGG (abbreviated as ‘MRSR’, ‘x’ denotes for HRV3C cleavage site), confirming reconstitution of NanoLuc activity from LgBit and SmBit inside the RGG-based coacervates. Middle row: DIC and Bioluminescence images of protein droplets taken 1 hr after adding HRV3C protease (1 μM final concentration) into a solution containing 6 μM MBP-x-RGG-LgBiT-RGG, indicating LgBit alone does not generate a light signal inside RGG-based droplets. Bottom row: DIC and Bioluminescence images of protein droplets taken 1hr after adding HRV3C protease (1 μM final concentration) into a solution containing 6 μM MBP-x-RGG-SmBiT-RGG, indicating SmBit alone does not generate a light signal inside RGG-based droplets. Scale bars: 10 μm. Substrate: 10 μl. Exposure: 5 s. (d) Light intensity readings from a plate reader for samples containing single or both components of NanoBit. From left to right: MBP-RGG-LgBit-RGG (abbreviated as ‘MRLR’) (3 μM) + MBP-RGG-SmBit-RGG (abbreviated as ‘MRSR’) (3 μM) + HRV3C (2 μM); MBP-RGG-LgBit-RGG (3 μM) + MBP-RGG-SmBit-RGG (3 μM); MBP-RGG-LgBit-RGG (6 μM) + HRV3C (2 μM); MBP-RGG-LgBit-RGG (6 μM); MBP-RGG-SmBit-RGG (6 μM) + HRV3C (2 μM); MBP-RGG-SmBit-RGG (6 μM). 5 μL of 200x diluted Nano-Glo assay substrate was injected into each well. Data presented as mean ± SD. *, p < 0.05; 2-tailed t-test.
Recruitment of NanoLuc enzyme at the condensate-water interface.
Microarchitectures have been observed in naturally occurring protein coacervates, with functions including vectoral organization of biochemical processes (64, 65). One typical example is the nucleolus, which has a core-shell architecture that produces ribosomes in a successive and vectoral manner through different nucleolar subcompartments (66). The multiphase substructures result from distinct surface tensions of the multiple phases of the membrane-less organelle (10). Such hierarchical IDP assemblies have been made in synthetic models by programming intermolecular interactions governing these interfacial tensions (67, 68). We previously engineered a surfactant protein molecule (GST-mCherry-RGG) that at low concentrations would localize at the interface of the RGG protein coacervates _ENREF_59 (69). The surfactant consists of an N-terminal hydrophilic protein domain Glutathione S-transferase (GST) which is often used as a water solubility enhancer during protein purification (70), a C-terminal RGG domain which tends to partition into the RGG coacervates, and a fluorescent reporter mCherry. We hypothesized we could recruit a cargo NanoLuc enzyme at the coacervate-water interface of the RGG coacervates by linking it with a surfactant using protein interaction domains like SynZips (SZ) (Figure 7a). To demonstrate this strategy, we fused the surfactant GST-mCherry-RGG with an N-terminal SZ1 domain (SZ1-GST-mCherry-RGG) and mixed it with a GFP cargo linked to SZ2 (GFP-SZ2), and then titrated a small amount into the RGG-RGG protein solution. SZ1 and SZ2 form a coiled-coil heterodimer with parallel orientation (71). Confocal microscopy imaging shows that both the surfactant and the GFP cargo are recruited at coacervate-water interfaces (Figure 7b).
Figure 7.
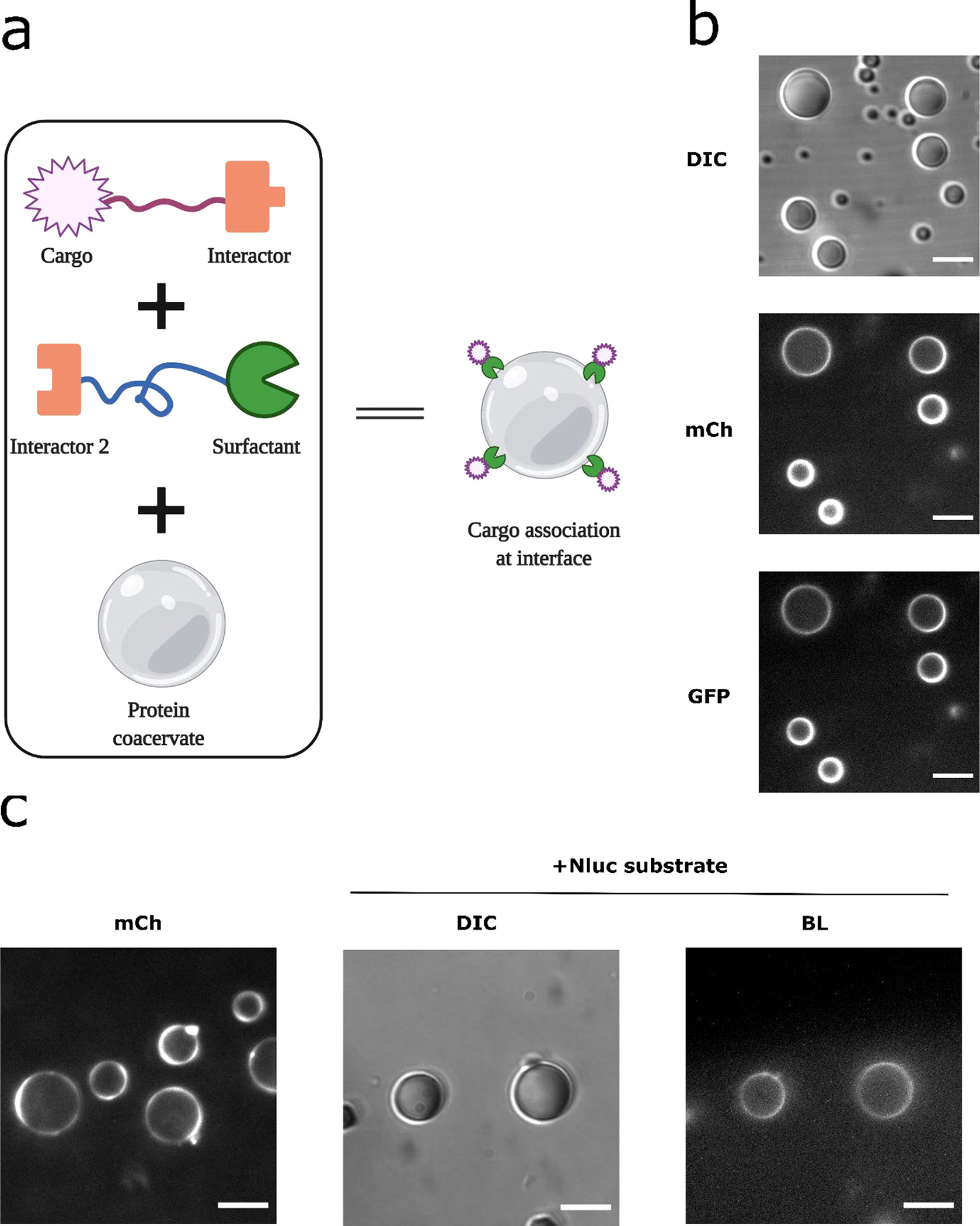
Recruitment of NanoLuc enzyme at coacervate-water interface. (a) Schematic of design strategy: linking the cargo via interactor modules onto a surfactant molecule that localizes at the coacervate-water interface. (b) Recruitment of a sample cargo protein. SZ1-GST-mCherry-RGG and GFP-SZ2 were mixed at a ratio of 2:1 and incubated for 1 hr to optimize binding, then 2 μl of the mixture was titrated into 50 ul of protein solution containing 6 μM RGG-RGG, 150 mM NaCl. Final concentrations for SZ1-GST-mCherry-RGG, GFP-SZ2, and RGG-RGG were about 200 nM, 100 nM and 6 μM, respectively. From top to bottom: DIC image (exposure: 100 ms) on the droplets, mCherry channel (exposure: 100 ms) showing surfactant localization at the droplet interface, GFP channel (exposure: 1000 ms) showing localization of the GFP cargo at the coacervate interface. Scale bars: 10 μm. (c) Recruitment of NanoLuc enzyme at the coacervate interface. SZ1-GST-mCherry-RGG and NanoLuc-SZ2 were mixed and incubated, then titrated into protein solution containing 6 μM RGG-RGG, 150 mM NaCl. Final concentrations for SZ1-GST-mCherry-RGG, NanoLuc-SZ2, and RGG-RGG were about 800 nM, 400 nM and 6 μM, respectively. Left: mCherry image showing surfactant localization at the droplet interface, imaged using a spinning disk confocal to minimize out-of-focus light. Middle to right: DIC and Bioluminescence images showing NanoLuc recruitment at the coacervate-water interface, after addition of 10 μL Nano-Glo substrate. Exposure for bioluminescence imaging: 10 s. Scale bars: 10 μm.
We then tested whether a NanoLuc enzyme cargo can be recruited at the coacervate-water interface. We fused NanoLuc with a SZ2 motif (NanoLuc-SZ2) and mixed it with SZ1-GST-mCherry-RGG and RGG-RGG. Confocal microscopy confirmed that the surfactant molecule localized at the coacervate-water interface (Figure 7c, left). We then added the Nano-Glo substrate and imaged the bioluminescence signal from the coacervates. A striking ring pattern of bioluminescence was clearly visible indicating peripheral localization of the NanoLuc enzyme at coacervate-water interfaces (Figure 7c, right). Combined with the fluorescence imaging showing surface localization of the surfactant, we demonstrate altered spatial patterns of cargo localization and localized function of the NanoLuc cargo at the coacervate-water interface.
Conclusions
In this work, NanoLuc enzymatic activity was incorporated into RGG-based protein coacervates to produce bioluminescence. We show that NanoLuc concentrated ten-fold by protein coacervation in RGG-based condensates resulting in acceleration of biochemical reactions. Component composition and viscosity could affect the resulting light signal, which is easily detectable by plate reader or microscopy. We demonstrate the diffusion limited reaction behavior of NanoLuc inside condensates could be utilized as a powerful tool to aid in drug discovery. We also demonstrated that the NanoLuc biochemical activity could be reconstituted by concentrating individual non-functional split parts of the NanoLuc enzyme within coacervates. Split enzymes could be useful as a reporter for condensate fusion in protocells or living systems. Reactions of NanoLuc resulted in fibrillar formation within the droplets; however, the droplets retained partially liquid-like character, as assessed by fluorescence recovery after photobleaching. Additionally, we demonstrate spatial patterning of enzymatic activity, localizing this function to the condensate interface via modular surfactant protein anchors. Overall, the results of this study provide paradigms for regulating and patterning biochemical reactions via self-assembling membrane-less organelles and suggests new avenues for using these designer condensates in cellular and protocell engineering.
Supplementary Material
Figure S1. Ray tracing inside the protein droplet
Figure S2. Collection efficiency simulation
Figure S3. Amino acid sequence for RGG-NanoLuc-RGG
Figure S4. SDS-PAGE gel of purified RGG-NanoLuc-RGG
Figure S5. Amino acid sequence for MBP-RGG-LgBit-RGG
Figure S6. Amino acid sequence for MBP-RGG-SmBit-RGG
Figure S7. FRAP on RGG-NanoLuc-RGG droplets
Figure S8. Gel on MBP cleavage by HRV3C
Acknowledgements
This work was supported by a grant from the DOE Basic Energy Sciences, Biomolecular Materials Program (DE-SC0007063) (DAH, MCG) and National Institutes of Biomedical Imaging and Bioengineering (R01 EB028320) (MCG, DAH). We gratefully thank Drs. Ellen H. Reed and Rajarshi Chattaraj from the Hammer lab and the journal reviewers for valuable discussion, Ken T. Anderson from BioVision and Ciprian Almonte from Hamamatsu Photonics for technical assistance, Marylin C. Huff from the department of Chemical and Biomolecular Engineering, University of Pennsylvania for access to the plate reader. M. Leutenegger acknowledges funding by the German Ministry of Education and Research (BMBF), grant no. 13N14122 to the Max Planck Institute for Biophysical Chemistry, Department of NanoBiophotonics.
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
Protein Accession IDs
References
- 1.Buchan JR. mRNP granules: assembly, function, and connections with disease. RNA biology. 2014;11(8):1019–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strulson CA, Molden RC, Keating CD, Bevilacqua PC. RNA catalysis through compartmentalization. Nature chemistry. 2012;4(11):941. [DOI] [PubMed] [Google Scholar]
- 3.Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annual review of cell and developmental biology. 2014;30:39–58. [DOI] [PubMed] [Google Scholar]
- 4.Protter DSW, Parker R. Principles and properties of stress granules. Trends in cell biology. 2016;26(9):668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazcano A Historical development of origins research. Cold Spring Harbor perspectives in biology. 2010;2(11):a002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oparin AI. The origin of life and the origin of enzymes. Adv Enzymol Relat Areas Mol Biol. 1965;27:347–80. [DOI] [PubMed] [Google Scholar]
- 7.Hyman T, Brangwynne C. In retrospect: the origin of life. Nature. 2012;491(7425):524. [Google Scholar]
- 8.Brangwynne CP, Tompa P, Pappu RV. Polymer physics of intracellular phase transitions. Nature Physics. 2015;11(11):899–904. [Google Scholar]
- 9.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nature reviews Molecular cell biology. 2017;18(5):285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357(6357):eaaf4382. [DOI] [PubMed] [Google Scholar]
- 11.Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L. Protein phase separation: a new phase in cell biology. Trends in cell biology. 2018;28(6):420–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei M-T, Elbaum-Garfinkle S, Holehouse AS, Chen CC-H, Feric M, Arnold CB, Priestley RD, Pappu RV, Brangwynne CP. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nature Chemistry. 2017;9(11):1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324(5935):1729–32. [DOI] [PubMed] [Google Scholar]
- 14.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proceedings of the National Academy of Sciences. 2011;108(11):4334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strzelecka M, Trowitzsch S, Weber G, Lührmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nature structural & molecular biology. 2010;17(4):403. [DOI] [PubMed] [Google Scholar]
- 16.Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152(4):791–805. [DOI] [PubMed] [Google Scholar]
- 17.Takahara T, Maeda T. Transient sequestration of TORC1 into stress granules during heat stress. Molecular cell. 2012;47(2):242–52. [DOI] [PubMed] [Google Scholar]
- 18.Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352(6285):595–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boisvert F-M, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nature reviews Molecular cell biology. 2007;8(7):574. [DOI] [PubMed] [Google Scholar]
- 20.Bergeron-Sandoval L-P, Heris HK, Hendricks AG, Ehrlicher AJ, Francois P, Pappu RV, Michnick SW. Endocytosis caused by liquid-liquid phase separation of proteins. bioRxiv. 2017:145664. [Google Scholar]
- 21.Zhang G, Wang Z, Du Z, Zhang H. mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell. 2018;174(6):1492–506. [DOI] [PubMed] [Google Scholar]
- 22.Sun D, Wu R, Zheng J, Li P, Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell research. 2018;28(4):405–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547(7662):236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. Phase separation drives heterochromatin domain formation. Nature. 2017;547(7662):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361(6400):eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HJ, Kim NC, Wang Y-D, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162(5):1066–77. [DOI] [PubMed] [Google Scholar]
- 28.Drobot B, Iglesias-Artola JM, Le Vay K, Mayr V, Kar M, Kreysing M, Mutschler H, Tang TYD. Compartmentalised RNA catalysis in membrane-free coacervate protocells. Nature communications. 2018;9(1):3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang TYD, van Swaay D, deMello A, Anderson JLR, Mann S. In vitro gene expression within membrane-free coacervate protocells. Chemical Communications. 2015;51(57):11429–32. [DOI] [PubMed] [Google Scholar]
- 30.Deshpande S, Brandenburg F, Lau A, Last MGF, Spoelstra WK, Reese L, Wunnava S, Dogterom M, Dekker C. Spatiotemporal control of coacervate formation within liposomes. Nature communications. 2019;10(1):1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crosby J, Treadwell T, Hammerton M, Vasilakis K, Crump MP, Williams DS, Mann S. Stabilization and enhanced reactivity of actinorhodin polyketide synthase minimal complex in polymer–nucleotide coacervate droplets. Chemical Communications. 2012;48(97):11832–4. [DOI] [PubMed] [Google Scholar]
- 32.Zhao EM, Suek N, Wilson MZ, Dine E, Pannucci NL, Gitai Z, Avalos JL, Toettcher JE. Light-based control of metabolic flux through assembly of synthetic organelles. Nature chemical biology. 2019;15(6):589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinkemeier CD, Girona GE, Lemke EA. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science. 2019;363(6434):eaaw2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC-H, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proceedings of the National Academy of Sciences. 2015;112(23):7189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuster BS, Dignon GL, Tang WS, Kelley FM, Ranganath AK, Jahnke CN, Simpkins AG, Regy RM, Hammer DA, Good MC. Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proceedings of the National Academy of Sciences. 2020;117(21):11421–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuster BS, Reed EH, Parthasarathy R, Jahnke CN, Caldwell RM, Bermudez JG, Ramage H, Good MC, Hammer DA. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nature communications. 2018;9(1):2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed EH, Schuster BS, Good MC, Hammer DA. SPLIT: Stable Protein Coacervation Using a Light Induced Transition. ACS Synthetic Biology. 2020;9(3):500–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garabedian MV, Wang W, Dabdoub JB, Tong M, Caldwell RM, Benman W, Schuster BS, Deiters A, Good MC. Designer membraneless organelles sequester native factors for control of cell behavior. Nature Chemical Biology. 2021:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS chemical biology. 2012;7(11):1848–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dzuricky M, Rogers BA, Shahid A, Cremer PS, Chilkoti A. De novo engineering of intracellular condensates using artificial disordered proteins. Nature chemistry. 2020;12(9):814–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim B, Zimmermann M, Barry NA, Goodman AL. Engineered regulatory systems modulate gene expression of human commensals in the gut. Cell. 2017;169(3):547–58. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoelson B. createCirclesMask.m. (https://www.mathworks.com/matlabcentral/fileexchange/47905-createcirclesmask-m), MATLAB Central File Exchange. [Google Scholar]
- 43.Leutenegger M, Lasser T. Detection efficiency in total internal reflection fluorescence microscopy. Optics Express. 2008;16(12):8519–31. [DOI] [PubMed] [Google Scholar]
- 44.Velesco N, Schweiger G. Geometrical optics calculation of inelastic scattering on large particles. Applied optics. 1999;38(6):1046–52. [DOI] [PubMed] [Google Scholar]
- 45.Welsh DK, Noguchi T. Cellular bioluminescence imaging. Cold Spring Harbor Protocols. 2012;2012(8):pdb-top070607. [DOI] [PubMed] [Google Scholar]
- 46.Kim TJ, Tuerkcan S, Ceballos A, Pratx G. Modular platform for low-light microscopy. Biomedical optics express. 2015;6(11):4585–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogoh K, Akiyoshi R, Sugiyama T, Dosaka S, Hatta-Ohashi Y, Suzuki H. Bioluminescence microscopy using a short focal-length imaging lens. Journal of microscopy. 2014;253(3):191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goda K, Hatta-Ohashi Y, Akiyoshi R, Sugiyama T, Sakai I, Takahashi T, Suzuki H. Combining fluorescence and bioluminescence microscopy. Microscopy research and technique. 2015;78(8):715–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki T, Kondo C, Kanamori T, Inouye S. Video rate bioluminescence imaging of secretory proteins in living cells: localization, secretory frequency, and quantification. Analytical biochemistry. 2011;415(2):182–9. [DOI] [PubMed] [Google Scholar]
- 50.Stacer AC, Nyati S, Moudgil P, Iyengar R, Luker KE, Rehemtulla A, Luker GD. NanoLuc reporter for dual luciferase imaging in living animals. Molecular imaging. 2013;12(7):7290–2013. [PMC free article] [PubMed] [Google Scholar]
- 51.Machleidt T, Woodroofe CC, Schwinn MK, Méndez J, Robers MB, Zimmerman K, Otto P, Daniels DL, Kirkland TA, Wood KV. NanoBRET A Novel BRET Platform for the Analysis of Protein–Protein Interactions. ACS chemical biology. 2015;10(8):1797–804. [DOI] [PubMed] [Google Scholar]
- 52.Wang H, Kelley FM, Milovanovic D, Schuster BS, Shi Z. More than just oil droplets in water: surface tension and viscosity of protein condensates quantified by micropipette aspiration. bioRxiv. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163(1):123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murakami T, Qamar S, Lin JQ, Schierle GSK, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FTS, Michel CH. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88(4):678–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu-Yesucevitz L, Bilgutay A, Zhang Y-J, Vanderwyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PloS one. 2010;5(10):e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Risso-Ballester J, Galloux M, Cao J, Le Goffic R, Hontonnou F, Jobart-Malfait A, Desquesnes A, Sake SM, Haid S, Du M. A condensate-hardening drug blocks RSV replication in vivo. Nature. 2021:1–4. [DOI] [PubMed] [Google Scholar]
- 57.Mullard A Biomolecular condensates pique drug discovery curiosity. Nature Reviews Drug Discovery. 2019:NA–NA. [DOI] [PubMed] [Google Scholar]
- 58.Strzyz P Drugs enter a liquid phase. Nature Reviews Molecular Cell Biology. 2020;21(8):419-. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki K, Kimura T, Shinoda H, Bai G, Daniels MJ, Arai Y, Nakano M, Nagai T. Five colour variants of bright luminescent protein for real-time multicolour bioimaging. Nature communications. 2016;7:13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richards FM. On the enzymic activity of subtilisin-modified ribonuclease. Proceedings of the National Academy of Sciences of the United States of America. 1958;44(2):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lowder MA, Appelbaum JS, Hobert EM, Schepartz A. Visualizing protein partnerships in living cells and organisms. Current opinion in chemical biology. 2011;15(6):781–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luker KE, Smith MCP, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein–protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proceedings of the National Academy of Sciences. 2004;101(33):12288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS chemical biology. 2016;11(2):400–8. [DOI] [PubMed] [Google Scholar]
- 64.Lyon AS, Peeples WB, Rosen MK. A framework for understanding the functions of biomolecular condensates across scales. Nature Reviews Molecular Cell Biology. 2020:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schuster BS, Regy RM, Dolan EM, Kanchi Ranganath A, Jovic N, Khare SD, Shi Z, Mittal J. Biomolecular Condensates: Sequence Determinants of Phase Separation, Microstructural Organization, Enzymatic Activity, and Material Properties. The Journal of Physical Chemistry B. 2021;125(14):3441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165(7):1686–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simon JR, Carroll NJ, Rubinstein M, Chilkoti A, López GP. Programming molecular self-assembly of intrinsically disordered proteins containing sequences of low complexity. Nature chemistry. 2017;9(6):509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roberts S, Miao V, Costa S, Simon J, Kelly G, Shah T, Zauscher S, Chilkoti A. Complex microparticle architectures from stimuli-responsive intrinsically disordered proteins. Nature communications. 2020;11(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kelley FM, Favetta B, Regy RM, Mittal J, Schuster BS. Amphiphilic proteins coassemble into multiphasic condensates and act as biomolecular surfactants. bioRxiv. 2021:2021.05.28.446223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Current opinion in biotechnology. 2006;17(4):353–8. [DOI] [PubMed] [Google Scholar]
- 71.Thompson KE, Bashor CJ, Lim WA, Keating AE. SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains. ACS synthetic biology. 2012;1(4):118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Ray tracing inside the protein droplet
Figure S2. Collection efficiency simulation
Figure S3. Amino acid sequence for RGG-NanoLuc-RGG
Figure S4. SDS-PAGE gel of purified RGG-NanoLuc-RGG
Figure S5. Amino acid sequence for MBP-RGG-LgBit-RGG
Figure S6. Amino acid sequence for MBP-RGG-SmBit-RGG
Figure S7. FRAP on RGG-NanoLuc-RGG droplets
Figure S8. Gel on MBP cleavage by HRV3C


