Abstract
Clostridioides difficile is an anaerobic, Gram-positive pathogen that is responsible for C. difficile infection (CDI). To survive in the environment and spread to new hosts, C. difficile must form metabolically dormant spores. The formation of spores requires activation of the transcription factor Spo0A, which is the master regulator of sporulation in all endospore-forming bacteria. Though the sporulation initiation pathway has been delineated in the Bacilli, including the model spore-former Bacillus subtilis, the direct regulators of Spo0A in C. difficile remain undefined. C. difficile Spo0A shares highly conserved protein interaction regions with the B. subtilis sporulation proteins Spo0F and Spo0A, although many of the interacting factors present in B. subtilis are not encoded in C. difficile. To determine if comparable Spo0A residues are important for C. difficile sporulation initiation, site-directed mutagenesis was performed at conserved receiver domain residues and the effects on sporulation were examined. Mutation of residues important for homodimerization and interaction with positive and negative regulators of B. subtilis Spo0A and Spo0F impacted C. difficile Spo0A function. The data also demonstrated that mutation of many additional conserved residues altered C. difficile Spo0A activity, even when the corresponding Bacillus interacting proteins are not apparent in the C. difficile genome. Finally, the conserved aspartate residue at position 56 of C. difficile Spo0A was determined to be the phosphorylation site that is necessary for Spo0A activation. The finding that Spo0A interacting motifs maintain functionality suggests that C. difficile Spo0A interacts with yet unidentified proteins that regulate its activity and control spore formation.
Keywords: Clostridium, Bacillus subtilis, spore, Spo0A, Spo0F
INTRODUCTION
Sporulation initiation is a complex developmental process that allows for prolonged survival when environmental conditions become unfavorable. Some members of the Firmicutes phylum transition into metabolically dormant endospores (spores) that remain inert until environmental conditions are favorable again and the spore germinates to produce vegetative cells. Sporulation is energetically costly and, as such, highly regulated [1–5]. Sporulation initiation is controlled by the conserved transcription factor, Spo0A, the essential regulator of the sporulation gene expression program. Spo0A consists of a receiver domain and a DNA-binding domain and is encoded in all endospore-forming species (Figure S1) [6, 7]. Spo0A is a response regulator, and its DNA-bindng activity is regulated by phosphorylation of a conserved aspartate residue [8]. In the activated form, phosphorylated Spo0A undergoes a conformational change that facilitates self-dimerization. Activated Spo0A can then bind specific promoter regions, referred to as “0A boxes”, to regulate gene expression and trigger entry into the sporulation pathway [9, 10].
Sporulation initiation has been extensively studied in the model spore-former, Bacillus subtilis. In B. subtilis and other Bacilli, the phosphorylation status of Spo0A is controlled through a multicomponent phosphorelay, with the orphan sensor histidine kinases, KinA, KinB, KinC, KinD, and KinE, transferring phosphate to the intermediate response regulator, Spo0F. Spo0F in turn mediates the flow of phosphate to the phosphotransferase Spo0B [11]. Spo0B then directly phosphorylates Spo0A, which activates sporulation-specific gene expression [2]. The Rap phosphatases, such as RapA, RapB, and RapH, can dephosphorylate Spo0F, while the Spo0E family of proteins dephosphorylate Spo0A (Figure 1A). The ability of Spo0B to interact with both Spo0F and Spo0A at shared, highly conserved motifs suggests a critical role for these residues in the regulation of Spo0A activity [12, 13].
Figure 1. Evolutionarily divergent strategies for Spo0A activation.
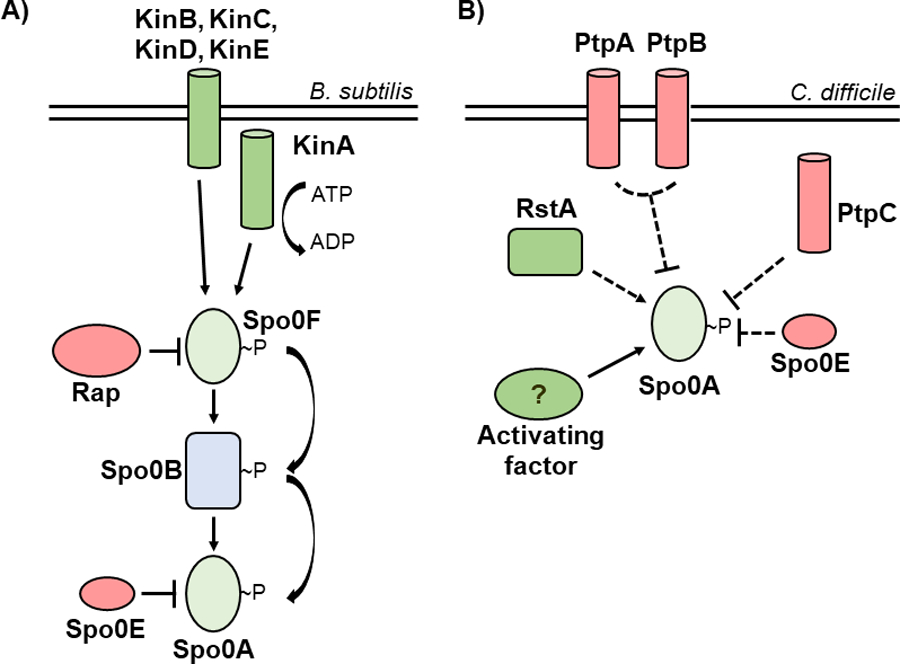
A) In Bacillus species, Spo0A is activated via the phosphorelay, with kinases KinA, KinB, KinC, KinD, and KinE transferring phosphate to Spo0A via Spo0F and Spo0B, while the Rap and Spo0E phosphatases repress Spo0A activation. B) In C. difficile, the phosphotransfer proteins PtpA and PtpB act in coordination to prevent Spo0A activation, with PtpC and Spo0E also acting to repress Spo0A activity. RstA promotes sporulation through an unknown mechanism, and a yet unidentified activating factor is hypothesized to phosphorylate Spo0A.
Like the Bacilli, all spore-forming members of the anaerobic Clostridia encode spo0A [14]. However, the mechanisms of Spo0A regulation in the Clostridia, including C. difficile, are poorly characterized. The Spo0F-Spo0B phosphorelay is not apparent in clostridial genomes, suggesting that there are divergent mechanisms of Spo0A activation [15]. In some clostridial species, phosphotransfer proteins interact directly with Spo0A to activate or inactivate sporulation in a manner consistent with a traditional two-component system [16–19]. C. difficile encodes five orphan putative histidine kinases, three of which resemble the B. subtilis Spo0A-associated kinases and negatively regulate sporulation (PtpA, PtpB, and PtpC), and two that are not involved in sporulation [20, 21]. While one orphan kinase, PtpC, was reported to phosphorylate Spo0A in vitro [22], it was recently shown that a ptpC null mutant exhibits variably increased sporulation, demonstrating that PtpC negatively impacts Spo0A activity in the conditions tested [20] (Figure 1B). As none of the C. difficile orphan kinases are verified activators of Spo0A, it is challenging to predict the specific strategy of C. difficile Spo0A regulation.
Although Spo0F and Spo0B are not found in C. difficile, the regions of the Spo0A receiver domain that interact with these and other Bacillus regulators appear to be conserved in C. difficile. We hypothesized that conserved Bacillus Spo0A and Spo0F residues are also functionally important for C. difficile Spo0A regulation. To better understand how C. difficile Spo0A activity is regulated, we performed site-directed mutagenesis of conserved regions of the receiver domain that are functionally important for B. subtilis Spo0A and Spo0F, or are in areas likely to be important for functional interactions, and examined the effects on sporulation. Here we report on the residues and potential interaction surfaces that are important for regulation of C. difficile Spo0A activity.
RESULTS
The Spo0A B. subtilis and C. difficile N-terminal receiver domains are highly conserved.
In B. subtilis, Spo0A and Spo0F share similar response regulator receiver domains (residues 6 – 116 in Spo0F and 6 – 120 in Spo0A), and both proteins interact with the phosphotransfer protein Spo0B using conserved secondary structure [12, 23–26]. The residues of B. subtilis Spo0F and Spo0A that are important for signal transduction were previously identified and characterized [4, 12, 13, 27–30]. We aligned the amino acid sequence of the B. subtilis Spo0A (Figure 2) and Spo0F receiver domains (Figure S2) to C. difficile Spo0A to predict orthologous functional residues. After identifying corresponding functional residues in the C. difficile Spo0A amino acid sequence, including many residues not previously investigated in Spo0A proteins and solely in Spo0F, we performed site-directed mutagenesis of 30 C. difficile Spo0A residues to alanine, with the exception of native alanine residues, which were mutated to serine (Figure 2B). Mutated spo0A alleles driven by the C. difficile spo0A native promoter were expressed in a C. difficile spo0A mutant [31]. To assess the stability of mutant Spo0A proteins, we performed western blotting using an anti-Spo0A antibody [32] and found that all mutant Spo0A proteins, except those containing the Q17A, V18A, and P60A mutations, were stable under sporulating conditions (Figure S3).
Figure 2. Conservation of the Spo0A receiver domains in B. subtilis and C. difficile.
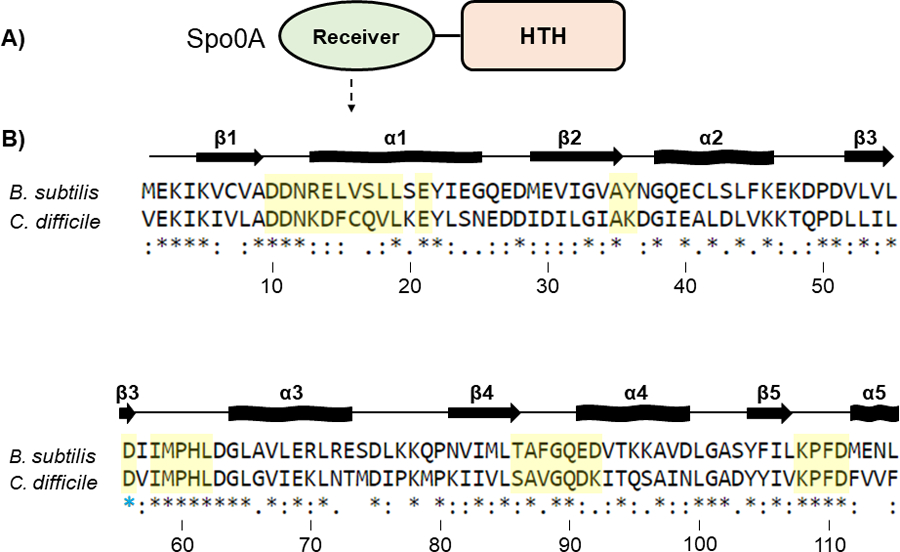
A) Graphic representation of Spo0A domain structure. Functional residues responsible for protein - protein interaction and Spo0A activation are located in the N-terminal receiver domain. The C-terminal region of Spo0A is defined by a helix-turn-helix (HTH) DNA-binding domain. B) Amino acid sequences of the Spo0A receiver domains for B. subtilis str. 168 (BSU_24220, top) and C. difficile 630 (CD630_12140, bottom). Residues important for activity that were chosen for mutation in C. difficile are highlighted in yellow. The blue star (*) is the conserved site of phosphorylation. Alignment performed using Clustal Omega. Arrows represent beta sheets, and waved rectangles represent alpha helices.
Conserved amino acid residues impact Spo0A function in C. difficile
To determine the functional significance of the individual mutant Spo0A proteins, the ability for these proteins to restore sporulation when expressed in a C. difficile spo0A mutant was assessed. The sporulation frequencies for the mutant C. difficile spo0A alleles tested are displayed in Table 1. The corresponding B. subtilis Spo0A amino acid residue location and the functional significance of each site-directed mutant are also included for reference (Table 1).
Table 1.
Sporulation frequencies of C. difficile Spo0A site-directed mutants
| C. difficile Spo0A allele | Spo0A region |
Corresponding B. subtilis Spo0A residue | Corresponding B. subtilis Spo0F residue | Spo0A function in B. subtilis | Spo0F function in B. subtilis | Average sporulation frequency (%)a,c | Mutant phenotype in B. subtilisb |
|---|---|---|---|---|---|---|---|
| Wildtype | - | - | - | Interaction with Spo0B, Spo0E | Interaction with KinA, KinB, KinC, KinD, and KinE, interaction with Spo0B | 12.1±1.0 | - |
| spo0A::erm | - | - | - | - | - | 0.00 | |
| D10A | β1-α1 | D10 | D10 | Forms aspartyl pocket, Spo0A dimerization, divalent cation binding, predicted Spo0E interaction [4, 25, 36, 68] | Forms aspartyl pocket, divalent cation binding [57] | 0.0002±0.0001** | n.d. |
| D11A | β1-α1 | D11 | D11 | Forms aspartyl pocket, Spo0A dimerization, divalent cation binding, predicted Spo0E interaction [4, 25, 36, 68] | Forms aspartyl pocket, divalent cation binding [57] | 0.00055±0.0004** | n.d. |
| N12A | β1-α1 | N12 | Q12 | Interaction with Spo0E, N12K is resistant to hyperactive Spo0E, predicted Spo0B interaction [12, 28, 33–35, 68] | Interaction with Spo0B, inferred interaction with KinA [12, 13, 39, 69] | 14.8±1.86 | Spo0A gain-of-function |
| K13A | α1 | R13 | Y13 | n.d. | Interaction with RapA and RapB, Y13S is resistant to RapB [70–73] | 19.1±3 | Spo0F gain-of-function |
| D14A | α1 | E14 | G14 | E14A allows direct interaction with KinC, resistant to hyperactive Spo0E [28, 33] | Interaction with Spo0B, and RapB [12, 71–73] | 49.7±6.5* | Spo0A gain-of-function |
| F15A | α1 | L15 | I15 | n.d. | Interaction with KinA, Spo0B, and RapB [12, 13, 71–73] | 23.1±1.7 | Spo0F decreased sporulation |
| C16A | α1 | V16 | R16 | n.d. | Interaction with KinA, Spo0B, and RapB [12, 13, 71] | 0.7±0.3* | Spo0F decreased sporulation |
| Q17A | α1 | S17 | I17 | n.d. | Interaction with RapB [13, 69, 71–73] | <LOD ** | Spo0F gain-of-function |
| V18A | α1 | L18 | L18 | n.d. | Interaction with KinA, Spo0B, RapB [12, 13, 71–73] | <LOD ** | Spo0F decreased sporulation |
| L19A | α1 | L19 | L19 | n.d. | Inferred structural importance [13] | 6.4±1.5 | Spo0F decreased sporulation |
| E21A | α1 | E21 | E21 | Inferred to interact with Spo0B [12] | Interaction with Spo0B, inferred interaction with KinA [12, 39] | 0.01±0.01** | n.d. |
| A35S | β2 | A35 | A33 | n.d. | n.d. | 0.15±0.1* | n.d. |
| K36A | β2-α2 | Y36 | A34 | Interaction with Spo0B [12] | 26.4±5.7 | n.d. | |
| D56A | β3 | D56 | D54 | Site of phosphorylation by Spo0B, forms aspartyl pocket, Spo0A dimerization, predicted to interact with Spo0E [4, 25, 36, 68] | Site of phosphorylation by KinA, KinB, KinC, KinD, and KinE,[27], forms aspartyl pocket [4, 25, 36, 57] | 0.008±0.001** | Spo0A |
| I58A | β3-α3 | I58 | K56 | Stabilizes aspartyl pocket, Spo0A dimerization [25] | Interaction with KinA, Spo0B, and RapB, stabilizes aspartyl pocket [12, 13, 57, 71] | 2±0.9 | Spo0F decreased sporulation |
| M59A | β3-α3 | M59 | I57 | n.d. | Interaction with KinA [13] | 0.006±0.001** | Spo0F decreased sporulation |
| P60A | β3-α3 | P60 | P58 | Interaction with Spo0E, P60S is resistant to hyperactive Spo0E; active without phosphorelay [28, 33] | Interaction with Spo0B [12] | <LOD ** | Spo0A gain-of-function |
| H61A | β3-α3 | H61 | G59 | n.d. | Interaction with Spo0B [12] | 0.6±0.2* | n.d. |
| L62A | β3-α3 | L62 | M60 | Interaction with Spo0E, L62P is resistant to hyperactive Spo0E [28] | M60A results in reduced spoIIG transcription [12] | 11.5±1.8 | Spo0A gain-of-function |
| S86A | β4 | T86 | T82 | Stabilizes phosphorylation of active site [47] | Interaction with KinA, interaction with RapH, interaction with Spo0B [13, 38, 39, 74] | 0.1±0.1* | Spo0F decreased sporulation |
| A87S | β4 | A87 | A83 | n.d. | Interaction with Spo0B [12] | 0.01±0.01** | n.d. |
| V88A | β4-α4 | F88 | Y84 | Interaction with Spo0E; F88L is resistant to hyperactive Spo0E [28] | Interaction with Spo0B, KinA, RapH, Y84A is resistant to RapB, RapH [13, 38, 71] | 17.9±1.0 | Spo0F reduced sporulation |
| G89A | β4-α4 | G89 | G85 | Inferred to interact with Spo0B [12] | Interaction with Spo0B [12] | 5.6±2.1 | n.d. |
| Q90A | β4-α4 | Q90 | E86 | Q90R allows for direct interaction with KinC, and resistant to hyperactive Spo0E [34, 35] | Interaction with Spo0B, interaction with KinA [12, 13] | 59.3±10.7* | Spo0F reduced sporulation, Spo0A gain-of-function |
| D91A | α4 | E91 | L87 | n.d. | Interaction with Spo0B, interaction with KinA [12, 13] | 37.8±12.5 | Spo0F reduced sporulation |
| K92A | α4 | D92 | D88 | D92Y is resistant to hyperactive Spo0E and is functional without phosphorelay [27, 33] | n.d. | 33±2.7* | Spo0A gain-of-function |
| K108A | β5 | K108 | K104 | Stabilizes aspartyl pocket, Spo0A dimerization, predicted Spo0E and Spo0B interaction [12, 25, 68] | Interaction with Spo0B, stabilizes aspartyl pocket, inferred interaction with KinA [12, 39, 57] | 0.6±0.2* | n.d. |
| P109A | β5-α5 | P109 | P105 | Spo0E and Spo0B interaction [12, 68] | Interaction with Spo0B [12] | 76.9±9.3* | n.d. |
| F110A | β5-α5 | F110 | F106 | Predicted Spo0E interaction [68] | Interaction with Spo0B [12] | 17.9±2.1 | Spo0F reduced sporulation |
| D111A | β5-α5 | D111 | D107 | Predicted Spo0E interaction [68] | Interaction with Spo0B, inferred interaction with KinA [12, 39] | 7.6±0.9 | n.d. |
, P = > .05
, P = > .01
B. subtilis phenotype that differs from C. difficile phenotype noted in bold
Spo0A site-directed mutant sporulation frequency where protein was undetectable by western blot is underlined
n.d., not determined
The strains containing the D14A, Q90A, K92A, and P109A Spo0A site-directed mutants exhibited significantly increased sporulation compared to the control strain expressing wildtype spo0A allele (Figure 3A, 3C). The strains containing the F15A, K36A, and D91A Spo0A site-directed mutants also displayed increased sporulation but were not statistically significant (Table 1). The C. difficile Spo0A Q90A gain-of-function sporulation phenotype was similar to the increased sporulation phenotype observed with the B. subtilis Spo0A Q90R mutant, which facilitates interaction with the activating protein KinC [33–35]. The increased sporulation phenotype displayed by the C. difficile Spo0A D14A mutant was similar to the sporulation phenotype observed when the orthologous B. subtilis Spo0A residue E14 is mutated [28, 33]. The B. subtilis Spo0A E14A mutant confers resistance to hyperactive Spo0E, resulting in increased Spo0A phosphorylation and activity [28, 33]. The gain of function phenotype of the C. difficile D14A mutant suggests that this residue may also be important for recognition by Spo0E in C. difficile [28, 33]. The B. subtilis Spo0F residues G14, L87, and P105 are all important for positively influencing sporulation through interaction with Spo0B (Table 1), yet the corresponding C. difficile site-directed mutants (Spo0A D14A, D91A, and P109A) all exhibited increased sporulation, suggesting that these residues serve a divergent role in C. difficile Spo0A activation [12].
Figure 3. Mutagenesis of conserved Spo0A residues results in both increased and decreased C. difficile sporulation frequency.
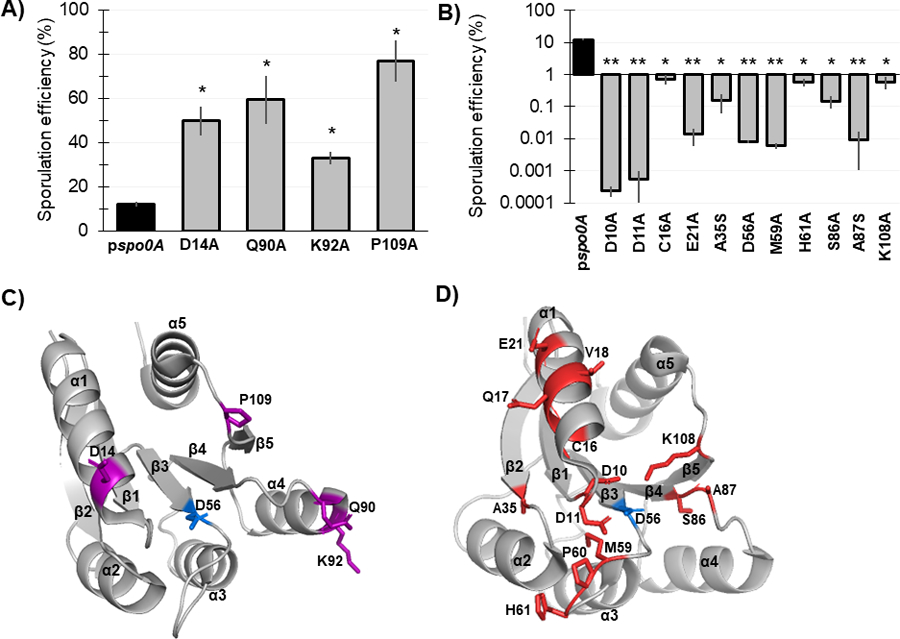
A) Ethanol-resistant spore formation of 630Δerm spo0A pspo0A (MC848) expressed on a plasmid compared to the Spo0A site-directed mutants D14A (MC1671), Q90A (MC1712), K92A (MC1185), and P109A (MC1621) with increased sporulation frequency. B) Ethanol-resistant spore formation of 630Δerm spo0A pspo0A (MC848) expressed on a plasmid compared to the Spo0A site-directed mutants D10A (MC1618), D11A (MC1703), C16A (MC1057), E21A (MC1058), A35S (MC1059), D56A (MC849), M59A (MC1184), H61A (MC1036), S86A (MC1846), A87S (MC1061), and K108A (MC1064) with decreased sporulation frequency, displayed on log10 scale. Sporulation assays were performed independently at least four times. Statistical significance was determined using Kruskal-Wallis test and uncorrected Dunn’s test (*, P = > 0.05; **, P = > 0.01). C) 3D structure of Spo0A with residues (highlighted purple) that cause increased sporulation when mutated, orientated around the activation site (D56, highlighted blue). D) 3D structure of Spo0A with residues (highlighted red) that reduce sporulation when mutated, orientated around the active site (D56, highlighted blue). Spo0A PDB code 5WQ0, edited in PyMOL (The PyMOL Molecular Graphics System, Version 2.4.0 Schrödinger, LLC).
Conversely, 15 of the 30 Spo0A site-directed mutants had reduced sporulation compared to expression the wildtype spo0A allele, representing a much larger proportion of the mutants assessed (Table 1, Figure 3B, 3D). Expression of eleven of the mutant spo0A alleles resulted in significantly reduced sporulation: D10A, D11A, C16A, E21A, A35S, D56A, M59A, H61A, S86A, A92S, and K108A. Spo0A Q17A, V18A, and P60A demonstrated sporulation frequencies below the limit of detection (>0.0002%); however, through western blotting we found these Spo0A site-directed mutants were not stably produced (Table 1, Figure S3).
The C. difficile Spo0A mutants that demonstrate a loss-of-function sporulation phenotype may represent amino acid residues that facilitate direct interactions with positive regulators of sporulation. The C. difficile Spo0A C16A and E21A mutations are located within the α1 region (Figure 3D). The B. subtilis Spo0F equivalents, R16 and E21, promote direct interactions with KinA and Spo0B [13], while B. subtilis Spo0A E21 is also expected to interact with Spo0B [12]. Altogether, these data suggest that C. difficile C16 and E21 coordinate the interaction with a positive regulator of Spo0A activity. The B. subtilis Spo0A residues D10, D11, I58, and K108 form the aspartyl pocket and are important for Spo0A homodimerization, which is necessary for DNA-binding activity [25, 36]. Mutation of the C. difficile Spo0A equivalent residues D10, D11, and K108 all produced severe sporulation defects. The C. difficile Spo0A I58A mutant had decreased sporulation, although these results were not statistically significant (Table 1).
The aspartate residue at position 56 of B. subtilis Spo0A serves as the phosphorylation site and is critical for sporulation, consistent with findings for the conserved aspartate residue in other species’ Spo0A orthologs [4, 16–19, 37]. As expected, mutation of the predicted C. difficile Spo0A phosphorylation site (D56A) resulted in dramatically reduced sporulation (>1000-fold decrease, Table 1), suggesting that this aspartate residue is required for C. difficile Spo0A phosphorylation and activation. C. difficile Spo0A I58, M59, and H61 are located immediately adjacent to the phosphorylation site in the open face between β3-α3 (Figure 3D). Mutation of the B. subtilis Spo0F K56 residue results in a loss-of-function phenotype, and several residues in this region facilitate B. subtilis Spo0A and Spo0F interactions with kinases or phosphatases [12, 13]. These data correspond with the low sporulation frequencies of the orthologous C. difficile Spo0A I58A, M59A, and H61A mutants (Figure 3B), suggesting that this region functions similarly in C. difficile. The β4 region of B. subtilis Spo0A is important for phosphotransfer between Spo0F or Spo0B. The C. difficile Spo0A S86 and A87 residues are located at the C-terminal end of β4 (Figure 3D), and site-directed mutagenesis of these residues significantly reduced sporulation frequency (Table 1), suggesting that the β4 region is likewise important for phosphotransfer to C. difficile Spo0A [12, 38]. Additionally, the B. subtilis Spo0F residue T82 is equivalent to C. difficile Spo0A S86, and is involved in stabilizing the phosphorylation of Spo0F [39]. Since both threonine and serine have polar side chains, C. difficile S86 may also facilitate phosphorylation of the Spo0A active site. Finally, the C. difficile Spo0A A35 residue is conserved in both B. subtilis Spo0A (A35) and Spo0F (A33), though the function of these residues in Bacilli have not been determined.
The Spo0A mutants N12A, K13A, L19A, L62A, F110A, and D111A had sporulation frequencies that were comparable to the wildtype Spo0A allele. N12, K13, and L19 appear to be dispensable for sporulation, even though residues located in this region are important for interaction of B. subtilis Spo0A and Spo0F with both positive and negative regulators (Table 1) [13, 33]. However, the Spo0A L19A mutant exhibited a translucent plate morphology on sporulation agar, suggesting some functional importance in other physiological processes outside of sporulation (Table 2). This result is not surprising given the pleiotropic effects Spo0A displays in C. difficile and other species [10, 19, 40–46]. Mutation of the Bacillus Spo0A and Spo0F residues that are comparable to C. difficile Spo0A L62 result in gain-of-function phenotypes but was not important for C. difficile Spo0A activity (Table 1). The C. difficile Spo0A residues F110A and D111A are located at the open face of β5-α5 in a motif (KPFD) that is highly conserved in the CheY superfamily of response regulators [12, 47]. Our data indicate that this region is also important for C. difficile Spo0A regulation, as the K108A mutant had decreased sporulation and the P109A mutant had increased sporulation (Table 1). While the F110A or D111A mutants did not affect sporulation, mutation of these residues produced a translucent and crushed plate morphologies, respectively (Table 2), suggesting they impact Spo0A function. Lastly, we used RoseTTAFold to model Spo0A site-directed mutants with the greatest changes in sporulation relative to wildtype, and did not observe major predicted changes to Spo0A structure in the site-directed mutants relative to wildtype Spo0A, suggesting that the changes in sporulation in the site-directed mutants are not likely to be due to major structural differences (Figure S4) [48].
Table 2.
Spo0A site-directed mutant morphology and growth phenotypes
| Spo0A mutant | Morphology phenotype |
|---|---|
| D14A | Mucoidal |
| F15A | Mucoidal |
| C16A | Mucoidal |
| Q17A | Poor growth |
| L19A | Translucent |
| E21A | Mucoidal, poor growth |
| A35S | Mucoidal, poor growth |
| H61A | Mucoidal, poor growth |
| A87S | Mucoidal, poor growth |
| Q90A | Mucoidal |
| D91A | Mucoidal |
| P109A | Mucoidal |
| F110A | Translucent |
| D111A | Crushed |
Altered growth and morphology of Spo0A mutants
Fourteen of the mutated spo0A alleles produced phenotypes that impacted growth in BHIS broth and growth and morphology on sporulation agar (Table 2). The most commonly observed phenotype was a stringy, mucoidal morphology that was observed for ten of the mutants after 24 h growth on sporulation plates (Spo0A D14A, F15A, C16A, E21A, A35S, H61A, A87S, Q90A, D91A, and P109A). The mucoidal phenotype was observed in both hyper- and hyposporulating strains, indicating that mucoidy is not directly correlated with the sporulation outcome. The Spo0A L19A and F110A mutants produced flat, translucent lawns, but sporulation was not affected in either mutant background. Similarly, the Spo0A D111A mutant did not affect sporulation, but produced a rigid, crushed lawn morphology. Strains expressing spo0A Q17A, E21A, A35S, H61A, and A87S exhibited poor growth in BHIS liquid compared to expression of the wildtype spo0A (data not shown). The Spo0A mutants with poor growth all had reduced sporulation (Table 1). However, only 5 of the 14 hyposporulating mutants grew slowly, indicating that defects in Spo0A that reduce sporulation do not necessarily retard growth.
C. difficile Spo0A requires phosphorylation of the conserved aspartate for activation.
In B. subtilis, Spo0A is phosphorylated at the conserved aspartate residue D56, which is required for activation [36, 49]. In the activated state, Spo0A homodimerizes and binds to specific DNA sequences, or “0A boxes”, to regulate Spo0A-dependent gene expression [50, 51]. Sequence comparison to B. subtilis Spo0A and other response regulators implicated D56 as the conserved site of C. difficile Spo0A phosphorylation and activation. The C. difficile Spo0A D56A site-directed mutation also dramatically reduced sporulation, further supporting the necessity of this residue for activity (Figure 3B). To determine if C. difficile Spo0A is also phosphorylated at the conserved aspartate residue, we isolated total protein from strains expressing either pspo0A-3XFLAG 3x-FLAG-Spo0A or pspo0A-D56A-3XFLAG and separated phosphorylated and unphosphorylated Spo0A species using phos-tag SDS-polyacrylamide gel electrophoresis followed by western blotting with an α-FLAG antibody [52–54] (Figure 4A). In the phos-tag assay, higher molecular weight bands that are present in the unheated sample but absent in the heated sample represent phosphorylated protein, as phosphoryl groups are heat-labile. In the strain expressing wildtype spo0A, two bands were observed in the unheated sample, with the upper band denoting phosphorylated Spo0A and the lower band corresponding to unphosphorylated Spo0A. In contrast, the Spo0A D56A mutant displayed only the lower, unphosphorylated band in both the unheated and heated samples, indicating that D56 is the primary site of phosphorylation. The ratio of phosphorylated Spo0A to total Spo0A is significantly greater in the wildtype compared to the Spo0A D56A mutant (Figure 4B). Altogether, the sporulation defect and the absence of Spo0A phosphorylation of the D56A mutant demonstrate that residue D56 is the primary site of Spo0A phosphorylation.
Figure 4. The conserved aspartate residue of C. difficile Spo0A is phosphorylated.
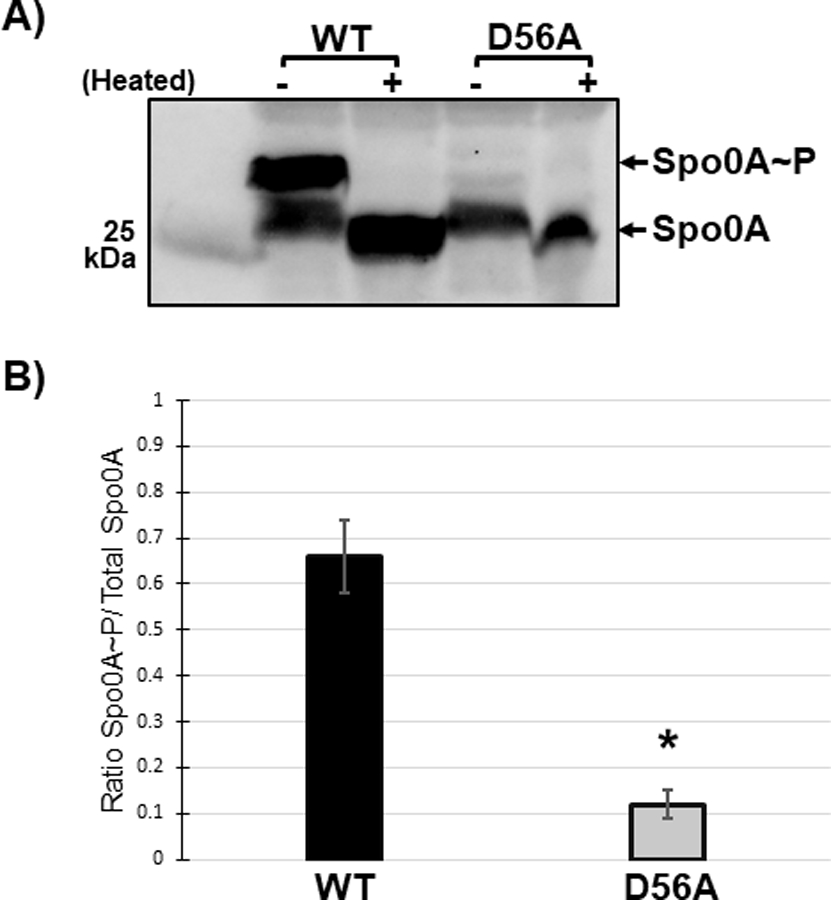
A) Anti-FLAG western blot after phos-tag gel separation of unphosphorylated and phosphorylated Spo0A (Spo0A~P) species in 630∆erm spo0A pspo0A-3XFLAG (MC1003) and 630∆erm spo0A pspo0A D56A-3XFLAG (MC1690) grown on sporulation agar. Phos-tag SDS-PAGE was performed on protein extracts (10 ug) and visualized using an anti-FLAG antibody. The molecular weight marker (25 kDa) is indicated on the left of the panel and experiments were performed 3 independent times. B) Ratio of phosphorylated Spo0A to total Spo0A. Densitometry calculations were performed using ImageJ 1.53a. (*, P = < 0.01) as determined by unpaired two-tailed Student’s t-test.
Residues necessary for Spo0A dimerization in other species have conserved functions in C. difficile.
Residues that are important for Spo0A homodimerization were previously identified in Bacilli [25, 36, 47]. The residues of C. difficile Spo0A that facilitate dimerization have not been characterized; however, C. difficile Spo0A contains five residues that are identical to those involved in dimerization in B. subtilis and other aerobic spore-formers: D10, D11, D56, I58, and K108. The alanine mutants of these five residues all produced defects in sporulation, indicating that they are important for Spo0A function. To test if these residues are involved in Spo0A dimerization in vivo, we performed split-luciferase reporter assays. Here, luciferase enzyme is fragmented into either a SmBit or LgBit subunit and fused to a gene(s) of interest to test for protein-protein interaction [55, 56]. We constructed C-terminal fusions of the SmBit and LgBit luciferase subunits to the wildtype, D10A, D11A, D56A, I58A, and K108A mutant spo0A alleles. All five site-directed mutants had less activity than the wildtype Spo0A fusions, with the D10A and D56A alleles exhibiting significantly less output (Figure 5A, Table S1), indicating that these Spo0A site-directed mutations reduce the ability for these mutant proteins to form homodimers. The D10, D11, I58, and K108 residues are all oriented around the D56 activation site (Figure 5B), further supporting the importance of these residues for Spo0A homodimerization [25]. These results demonstrate that the functional residues that are involved in Bacilli Spo0A dimerization are also important for C. difficile Spo0A dimerization.
Figure 5. Residues necessary for Spo0A dimerization in other Firmicutes are functionally conserved in C. difficile.
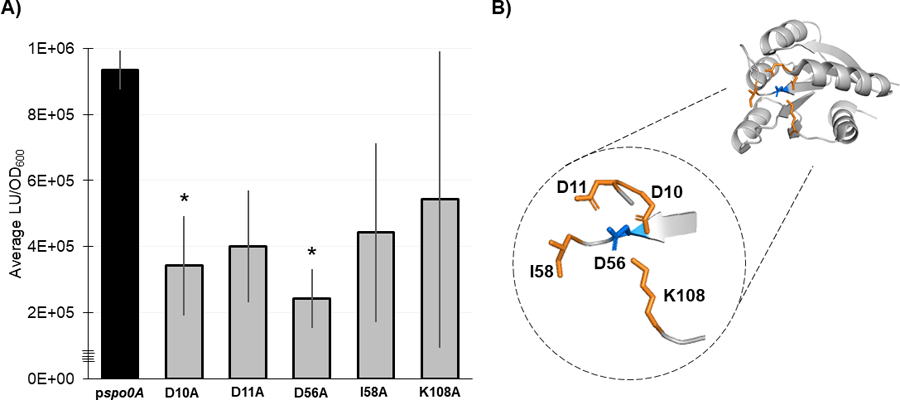
A) Split-luciferase activity in strains 630Δerm spo0A pspo0A (MC1906) and the Spo0A site-directed mutants D10A (MC2001), D11A (MC2002), D56A (MC2003), I58A (MC2004), and K108A (MC2005) fused to SmBit and LgBit fragments after cultures were grown in 70:30 sporulation broth to OD600 = 0.8 – 0.9 and induced with anhydrous tetracycline (ATc) for 1 h. Average luminescence outputs are normalized to optical densities (LU/OD600). Error bars represent the standard deviation of three independent experiments (*, P = < 0.05) as determined by a one-way ANOVA with Dunnett’s multiple comparisons test. B) 3D structure of Spo0A with the residues that form the aspartyl pocket and facilitate dimerization highlighted orange near the site of activation (D56, highlighted blue). Spo0A PDB code 5WQ0, edited in PyMOL (The PyMOL Molecular Graphics System, Version 2.4.0 Schrödinger, LLC).
DISCUSSION
In this study, we employed alanine-scanning mutagenesis to define the regions of the C. difficile Spo0A receiver domain that are important for regulation of sporulation. Altogether, we examined the ability of C. difficile Spo0A to initiate sporulation through mutational analysis of 30 residues located within 10 different regions of the Spo0A receiver domain secondary structure (Figure 2). The results demonstrated that mutation of many residues that influence B. subtilis Spo0A and Spo0F activation also have profound effects on C. difficile Spo0A function, even though few of the interacting partner proteins are conserved between these species. We also established Spo0A residues that are important for homodimerization and found altered growth and morphology phenotypes by mutating the receiver domain of Spo0A.
The receiver domain of the C. difficile Spo0A shares 47% identity with the B. subtilis Spo0A and 30% identify with B. subtilis Spo0F, but the protein architectures of the receiver motifs are highly conserved. By probing the function of conserved regions and residues that have been implicated in protein interaction in Bacilli, we demonstrate conservation of sporulation phenotypes in many C. difficile Spo0A residues relative to Bacillus Spo0F and Spo0A (Table 1) [12, 13, 25, 27, 30, 34]. As in Bacilli, we found that the receiver domain α-helices and the β1-α1, β3-α3, β4-α4, and β5-α5 open faces are all important for C. difficile Spo0A activity (Table 1) [12, 13, 25]. The majority of residues that were mutated in this study that produced major changes in sporulation are orientated on the same face as the site of activation, an effect observed for other response regulator receiver domains (Figure 3) [13, 47, 57].
C. difficile Spo0A and B. subtilis Spo0A perform the same sporulation function, but there are gaps in knowledge about the contribution of specific residues to Spo0A activity in both species. Our data demonstrate that most residues within the receiver domains of B. subtilis and C. difficile Spo0A proteins have similar impacts on sporulation. However, many of the characterized B. subtilis Spo0A site-directed mutants are gain-of-function suppressor mutations that are not alanine substitutions or were characterized in strain backgrounds lacking elements of the phosphorelay [27, 33]. Additionally, many of the described residues that are important for sporulation in B. subtilis Spo0F have not been characterized in Spo0A. Our sporulation results in C. difficile suggest open questions remain about the function of the following B. subtilis Spo0A residues: L15, V16, S17, L18, E21, A35, I58, M59, P60, H61, T86, A87, Q90, E91, D92, K108, and P109. These residues may also be important for Spo0A function in B. subtilis and other spore-forming Firmicutes.
The receiver domain of B. subtilis Spo0F has been more extensively characterized than Spo0A and more is understood about the impact of specific Spo0F residues on the regulation of sporulation [12, 13, 39]. We found several differences in the sporulation outcomes for mutations in conserved residues of B. subtilis Spo0F and C. difficile Spo0A, which is not surprising, considering the differences in these species’ sporulation pathways. Mutation of C. difficile Spo0A residues K13, F15, Q17, L62, V88, Q90, D91, and F110 resulted in different impacts on sporulation relative to similar mutations in Spo0F (Table 1). Some of these residues are important for Spo0F interaction with factors that are not present in C. difficile, such as the Rap phosphatases, Spo0B, and the specific sporulation kinases of Bacillus (Figure 1, Table 1). In particular, the C. difficile Spo0A mutants F15A, Q90A, and D91A displayed higher sporulation, while corresponding residues in B. subtilis Spo0F (I15A, E86A, L87A) resulted in sporulation defects [13]. C. difficile Spo0A V88A and F110A maintained wildtype sporulation levels, while B. subtilis Spo0F Y84A and F106A resulted in sporulation defects [13]. These results suggest that the importance of these residues is maintained for both proteins, although the interacting partners and the resulting effects on sporulation differ.
Distinct effects on growth and colony morphology were observed for the 14 Spo0A mutants listed in Table 2. The growth and morphology phenotypes are likely due to altered Spo0A regulation or function, as we found that deletion of spo0A in C. difficile does not change growth or morphology under the conditions tested, as previously observed [22, 40, 41]. The mucoidal phenotype observed on sporulation agar was the most commonly observed effect and was found in 10 of the 30 characterized Spo0A mutants. Mucoidy was only observed when the mutants grew for at least 12 hours as a lawn on sporulation agar, suggesting this phenotype is linked to either a facet of sporulation or conditions that facilitate sporulation (data not shown). However, the mucoidal phenotype was present in both hyposporulating and hypersporulating mutants and did not have an obvious impact on the capacity to sporulate. Additional changes in morphology, but not sporulation, were observed in Spo0A L19A, F110A and D111A. The L19A and F110A mutants produced flat, translucent lawns on sporulation agar, and the D111A mutant had a crushed morphology on sporulation agar. While our findings were unexpected, changes in plate morphology in C. difficile spo0A mutants in various strain backgrounds, including 630Δerm, have been previously described [40]. However, altered morphology was described explicitly in spo0A null mutants, not specific site-directed mutants, and we did not observe changes in morphology in our spo0A null mutant. Further, the mutants Q17A, E21A, A35S, H61A, and A87S all had poor growth in BHIS broth relative to both the wildtype and spo0A mutant, and all had defects in sporulation. To our knowledge, this is the first report that specific Spo0A residues impact colony morphology or growth. While it is unclear why Spo0A mutant alleles would affect morphology or growth, the simplest explanation is that the altered Spo0A alleles can interact with additional partner proteins that control these cellular processes. Future experiments to determine the differences in binding partners between wildtype Spo0A and site-directed Spo0A mutants with altered morphology or growth may help explain the impact specific Spo0A site-directed mutants have in vivo in processes outside of sporulation. It remains unknown if the morphology and growth phenotypes of specific Spo0A site-directed mutants are unique to C. difficile or if these phenotypes are conserved for Spo0A of other spore-forming Firmicutes.
We found that Spo0A is phosphorylated at the conserved site of activation (D56), and that a D56A mutation results in loss of phosphorylation (Figure 4). Interestingly, the D56A mutant does exhibit reduced, but not total loss, of sporulation. This could be a result of low Spo0A DNA-binding activity present in unphosphorylated Spo0A. Although all studied Spo0A are regulated by phosphorylation and dephosphorylation, the proteins that directly interact with Spo0A vary considerably within the Clostridia and the Spo0A proteins in these species have diverged (Figure S5A, S5B). In other Clostridia in which Spo0A regulation has been studied, Spo0A is directly phosphorylated by orphan histidine kinases or phosphatases to regulate Spo0A activity, and all encode at least one kinase that induces sporulation [16–20, 58, 59]. While a Spo0A-activating kinase has not yet been identified in C. difficile, our data confirm that phosphorylation of Spo0A at the conserved site of activation is critical for Spo0A activity. Despite the lack of evidence of an activating kinase to date, we expect that Spo0A is directly phosphorylated by at least one histidine kinase to positively regulate sporulation.
To our knowledge, this represents the first report on residues important for Spo0A dimerization in C. difficile (Figure 5) [25, 36]. The fact that the mechanism of dimerization is maintained in C. difficile is likely due to the conserved architecture of response regulator receiver domains, defined by (β/α)5 folding and functional residues that are orientated near the site of activation [25, 36, 47, 57].
The Bacilli and Clostridia diverged roughly 2.4 billion years ago during the Great Oxidation Event [60]. While the mechanism(s) of C. difficile Spo0A regulation remains unclear, we have identified conserved regions of Spo0A that are important for activity. Because the phosphorelay interactions are not retained in C. difficile, our results suggest that C. difficile Spo0A uses functionally conserved regions for interaction with both positive and negative regulators that are not part of the Bacilli mechanism for Spo0A regulation. Elucidation of the factors that regulate Spo0A in C. difficile will provide greater insight on the biology and lifestyle of this clinically important pathogen.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in (Table 3). C. difficile strains were routinely grown in BHIS broth or on BHIS agar supplemented with 2–5 μg ml−1 thiamphenicol (Sigma-Aldrich) as needed [61]. C. difficile cultures were supplemented with 0.2% fructose and 0.1% taurocholate (Sigma-Aldrich) to prevent sporulation and induce germination as indicated [32, 61]. C. difficile was grown on 70:30 agar to assess sporulation frequency as previously described [32]. C. difficile strains were grown in a 37°C anaerobic chamber (Coy) with an atmosphere consisting of 10% H2, 5% CO2, and 85% N2, as previously described [62]. Strains of Escherichia coli were cultured in LB at 37°C [63] and supplemented with 100 μg ml−1 ampicillin or 20 μg ml−1 chloramphenicol as needed. Kanamycin 100 μg ml−1 was used for counterselection of E. coli HB101 pRK24 after conjugation with C. difficile [64].
Table 3.
Bacterial Strains and plasmids
| Plasmid or Strain | Relevant genotype or features | Source, construction or reference | |
|---|---|---|---|
| Strains | |||
| E. coli | |||
| HB101 | F− mcrB mrr hsdS20(rB− mB−) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 | B. Dupuy | |
| C. difficile | |||
| 630Δerm | ErmS derivative of strain 630 | N. Minton [75] | |
| MC310 | 630Δerm spo0A::erm |
[31] | |
| MC324 | 630Δerm pMC123 | [31] | |
| MC848 | 630Δerm spo0A::erm pMC566 | This study | |
| MC849 | 630Δerm spo0A::erm pMC567 | This study | |
| MC855 | 630Δerm spo0A::erm pMC123 | This study | |
| MC961 | 630Δerm spo0A::erm pMC656 | This study | |
| MC962 | 630Δerm spo0A::erm pMC657 | This study | |
| MC981 | 630Δerm spo0A::erm pMC663 | This study | |
| MC1003 | 630Δerm spo0A::erm pMC674 | This study | |
| MC1033 | 630Δerm spo0A::erm pMC684 | This study | |
| MC1036 | 630Δerm spo0A::erm pMC685 | This study | |
| MC1057 | 630Δerm spo0A::erm pMC697 | This study | |
| MC1058 | 630Δerm spo0A::erm pMC698 | This study | |
| MC1059 | 630Δerm spo0A::erm pMC699 | This study | |
| MC1060 | 630Δerm spo0A::erm pMC700 | This study | |
| MC1061 | 630Δerm spo0A::erm pMC701 | This study | |
| MC1062 | 630Δerm spo0A::erm pMC702 | This study | |
| MC1063 | 630Δerm spo0A::erm pMC703 | This study | |
| MC1064 | 630Δerm spo0A::erm pMC704 | This study | |
| MC1184 | 630Δerm spo0A::erm pMC768 | This study | |
| MC1185 | 630Δerm spo0A::erm pMC770 | This study | |
| MC1527 | 630Δerm pMC917 | This study | |
| MC1529 | 630Δerm pMC930 | This study | |
| MC1618 | 630Δerm spo0A::erm pMC732 | This study | |
| MC1619 | 630Δerm spo0A::erm pMC742 | This study | |
| MC1620 | 630Δerm spo0A::erm pMC769 | This study | |
| MC1621 | 630Δerm spo0A::erm pMC771 | This study | |
| MC1664 | 630Δerm spo0A::erm pMC967 | This study | |
| MC1665 | 630Δerm spo0A::erm pMC969 | This study | |
| MC1666 | 630Δerm spo0A::erm pMC970 | This study | |
| MC1670 | 630Δerm spo0A::erm pMC965 | This study | |
| MC1671 | 630Δerm spo0A::erm pMC966 | This study | |
| MC1690 | 630Δerm spo0A::erm pMC971 | This study | |
| MC1711 | 630Δerm spo0A::erm pMC975 | This study | |
| MC1712 | 630Δerm spo0A::erm pMC976 | This study | |
| MC1713 | 630Δerm spo0A::erm pMC986 | This study | |
| MC1778 | 630Δerm spo0A::erm pMC968 | This study | |
| MC1846 | 630Δerm spo0A::erm pMC1055 | This study | |
| MC1904 | 630Δerm spo0A::erm pMC922 | This study | |
| MC1905 | 630Δerm spo0A::erm pMC924 | This study | |
| MC1906 | 630Δerm spo0A::erm pMC944 | This study | |
| MC1991 | 630Δerm spo0A::erm pMC1097 | This study | |
| MC1992 | 630Δerm spo0A::erm pMC1098 | This study | |
| MC1993 | 630Δerm spo0A::erm pMC1099 | This study | |
| MC1994 | 630Δerm spo0A::erm pMC1100 | This study | |
| MC1995 | 630Δerm spo0A::erm pMC1101 | This study | |
| MC1996 | 630Δerm spo0A::erm pMC1102 | This study | |
| MC1997 | 630Δerm spo0A::erm pMC1103 | This study | |
| MC1998 | 630Δerm spo0A::erm pMC1104 | This study | |
| MC1999 | 630Δerm spo0A::erm pMC1105 | This study | |
| MC2000 | 630Δerm spo0A::erm pMC1106 | This study | |
| MC2001 | 630Δerm spo0A::erm pMC1107 | This study | |
| MC2002 | 630Δerm spo0A::erm pMC1108 | This study | |
| MC2003 | 630Δerm spo0A::erm pMC1109 | This study | |
| MC2004 | 630Δerm spo0A::erm pMC1110 | This study | |
| MC2005 | 630Δerm spo0A::erm pMC1111 | This study | |
| Plasmids | |||
| pRK24 | Tra+, Mob+; bla, tet | [76] | |
| pUC19 | Cloning vector; bla | [77] | |
| pMC123 | E. coli-C. difficile shuttle vector; bla, catP | [59] | |
| pMC566 | pMC123 WT Spo0A | This study | |
| pMC567 | pMC123 Spo0A D56A | This study | |
| pMC656 | pMC123 Spo0A N12A | This study | |
| pMC657 | pMC123 Spo0A K13A | This study | |
| pMC663 | pMC123 Spo0A I58A | This study | |
| pMC674 | pMC123 Spo0A 3xFLAG | This study | |
| pMC684 | pMC123 Spo0A V18A | This study | |
| pMC685 | pMC123 Spo0A H61A | This study | |
| pMC697 | pMC123 Spo0A C16A | This study | |
| pMC698 | pMC123 Spo0A E21A | This study | |
| pMC699 | pMC123 Spo0A A35S | This study | |
| pMC700 | pMC123 Spo0A P60A | This study | |
| pMC701 | pMC123 Spo0A A87S | This study | |
| pMC702 | pMC123 Spo0A V88A | This study | |
| pMC703 | pMC123 Spo0A G89A | This study | |
| pMC704 | pMC123 Spo0A K108A | This study | |
| pMC742 | pMC123 Spo0A D91A | This study | |
| pMC768 | pMC123 Spo0A M59A | This study | |
| pMC769 | pMC123 Spo0A L62A | This study | |
| pMC770 | pMC123 Spo0A K92A | This study | |
| pMC771 | pMC123 Spo0A P109A | This study | |
| pMC915 | pAF 256 HupA-SmBit-LgBit | Wiep Klaas Smits [56] | |
| pMC916 | pAF257 SmBit-HupA-LgBit | Wiep Klaas Smits [56] | |
| pMC917 | pAF 259 BitLuc | Wiep Klaas Smits [56] | |
| pMC918 | pAP118 HupA-SmBit-HupA-LgBit | Wiep Klaas Smits [56] | |
| pMC922 | pAP118 Spo0A-SmBit-LgBit | This study | |
| pMC924 | pAP118 SmBit-Spo0A-LgBit | This study | |
| pMC930 | pAF256 SmBit-LgBit | This study | |
| pMC932 | pAF257 Spo0A-SmBit-LgBit | This study | |
| pMC944 | pMC932 Spo0A-SmBit-Spo0A-LgBit | This study | |
| pMC965 | pMC123 Spo0A D11A | This study | |
| pMC966 | pMC123 Spo0A D14A | This study | |
| pMC967 | pMC123 Spo0A F15A | This study | |
| pMC968 | pMC123 Spo0A Q17A | This study | |
| pMC969 | pMC123 Spo0A L19A | This study | |
| pMC970 | pMC123 Spo0A D111A | This study | |
| pMC971 | pMC123 Spo0A D56A 3xFLAG | This study | |
| pMC975 | pMC123 Spo0A K36A | This study | |
| pMC976 | pMC123 Spo0A Q90A | This study | |
| pMC986 | pMC123 Spo0A F110A | This study | |
| pMC1055 | pMC123 Spo0A S86A | This study | |
| pMC1088 | pMC123 Spo0A D10A | This study | |
| pMC1097 | pAF256 Spo0A D10A-SmBit | This study | |
| pMC1098 | pAF256 Spo0A D11A-SmBit | This study | |
| pMC1099 | pAF256 Spo0A D56A-SmBit | This study | |
| pMC1100 | pAF256 Spo0A I58A-SmBit | This study | |
| pMC1101 | pAF256 Spo0A K108A-SmBit | This study | |
| pMC1102 | pAF257 Spo0A D10A-LgBit | This study | |
| pMC1103 | pAF257 Spo0A D11A-LgBit | This study | |
| pMC1104 | pAF257 Spo0A D56A-LgBit | This study | |
| pMC1105 | pAF257 Spo0A I58A-LgBit | This study | |
| pMC1106 | pAF257 Spo0A K108A-LgBit | This study | |
| pMC1107 | pAP118 D10A-SmBit-D10A-LgBit | This study | |
| pMC1108 | pAP118 D11A-SmBit-D11A-LgBit | This study | |
| pMC1109 | pAP118 D56A-SmBit-D56A-LgBit | This study | |
| pMC1110 | pAP118 I58A-SmBit-I58A-LgBit | This study | |
| pMC1111 | pAP118 K108A-SmBit-K108A-LgBit | This study | |
Strain and plasmid construction
Table 4 contains oligonucleotides used in this study. C. difficile 630 strain (GenBank accession number AJP10906.1) was used as a template for primer design and C. difficile 630Δerm genomic DNA was used for PCR amplification. C. difficile 630Δerm has a known 18 nucleotide duplication outside of the Spo0A receiver domain and was used for strain creation [10]. Strain construction is described in Table S2.
Table 4.
Oligonucleotides
| Primer | Sequence (5’3’)a, b | Use/locus tag/reference |
|---|---|---|
| oMC305 | CACAGGAGGTATCGTACAG | Forward primer for sequencing Spo0A |
| oMC306 | GCGAAACGGTATAACCCTAG | Reverse for sequencing Spo0A |
| oMC1249 | GTCGAGGATCCGATGACAAGTTATTGGAATACACAG | Forward primer for Spo0A expression from pMC123 |
| oMC1250 | GACTCGAATTCCCCTAGTGGTTATACCGTTTCG | Reverse primer for Spo0A expression from pMC123 |
| oMC1251 | ATTAATACTAGCTGTAATAATGCCACATC | Forward SOEing primer for Spo0A D56A |
| oMC1252 | GAT GTGGCATTATTACAGCTAGTATTAAT | Reverse SOEing primer for Spo0A D56A |
| oMC1513 | GTTTTAGCAGATGACGCTAAGGATTTTTGTCAG | Forward SOEing primer for Spo0A N12A |
| oMC1514 | CTGACAAAAATCCTTAGCGTCATCTGCTAAAAC | Reverse SOEing primer for Spo0A N12A |
| oMC1515 | TTAGCAGATGACAATGCAGATTTTTGTCAGGTA | Forward SOEing primer for Spo0A K13A |
| oMC1516 | TACCTGACAAAAATCTGCATTGTCATCTGCTAA | Reverse SOEing primer for Spo0A K13A |
| oMC1517 | AAGGATTTTTGTCAGGCATTAAAAGAGTATTTG | Forward SOEing primer for Spo0A V18A |
| oMC1518 | CAAATACTCTTTTAATGCCTGACAAAAATCCTT | Reverse SOEing primer for Spo0A V18A |
| oMC1519 | TTAATACTAGATGTAGCAATGCCACATCTAGAT | Forward SOEing primer for Spo0A I58A |
| oMC1520 | ATCTAGATGTGGCATTGCTACATCTAGTATTAA | Reverse SOEing primer for Spo0A I58A |
| oMC1547 | GATGCGAATTCTCACTTGTCATCGTCATCCTTGTAA TCTATGTCATGATCTTTATAATCACCGTCATGGT CTTTGTAGTCACCTCCTTTAACCATACTATGTTC TAGTCTTAA |
Reverse primer for Spo0A with 3x FLAG tag and homology to pMC123 |
| oMC1583 | GATGTAATAATGCCAGCACTAGATGGATTAGGT | Forward SOEing primer for Spo0A H61A |
| oMC1584 | ACCTAATCCATCTAGTGCTGGCATTATTACATC | Reverse SOEing primer for Spo0A H61A |
| oMC2354 | AGGTTATAGACTTTTTGAAGAAATTCTATAGCT CGATCGGTGTAAAAAGTTTAGTTTTCTGTAATA AGAAGATGT |
Forward primer to amplify Spo0A fused to LgBit fragment |
| oMC2437 | CTTGATCGTAGCGTTAACAGATCTGAGCTCGTG TAAAAAGTTTAGTTTTCTGTAATAAGAAGATGT |
Forward primer to amplify Spo0A fused to SmBit fragment |
| oMC2439 | CACCACCACTAGAACCCCCTCGAGATTTAACCAT ACTATGTTCTAGTCTTAATTTATCAGC |
Reverse primer to amplify Spo0A fused to SmBit fragment |
| oMC2447 | ACCACCACCACTAGAACCTGCGGCCGCTCCTTTAA CCATACTATGTTCTAGTCTTAATTTATCAGC |
Reverse primer to amplify Spo0A fused to LgBit fragment |
Restriction sites underlined
Nucleotides for site-directed mutagenesis are noted in bold
Dendrogram
The Spo0A dendrogram rooted to B. subtilis Spo0A was created using the MUSCLE Multiple Alignment plugin and Geneious Tree Builder in Geneious Prime v2020.2.2. Spo0A amino acid sequences from C. difficile 630 (GenBank accession AJP10906.1), C. perfringens SM101 (GenBank CP000312.1), C. acetobutylicum ATCC 824 (GenBank NC_003030.1), A. thermocellus DSM 1313 (GenBank NC_017304.1), C. botulinum A str. ATCC 3502 (GenBank NC_009495.1), and B. subtilis str. 168 (GenBank NC_000964.3) were retrieved, aligned, and assembled into a dendrogram. The percentage of identity for each Spo0A protein relative to B. subtilis and the heatmap comparing Spo0A percent identities to each species was generated using Geneious Tree Builder in Geneious Prime v2020.2.2. (https://www.geneious.com).
Sporulation assays
C. difficile ethanol resistance sporulation assays were performed on 70:30 sporulation agar supplemented with 2 μg ml−1 thiamphenicol for plasmid maintenance, as previously described [65–67]. Following growth on sporulation agar for 24 h, cells were resuspended in BHIS broth to an OD600 of 1.0. To determine total vegetative cell counts ml−1, cultures were serially diluted in BHIS and plated on BHIS agar with 2 μg ml−1 thiamphenicol. Concurrently, 0.5 ml of resuspended cells were treated with a mixture of 0.3 ml 95% ethanol and 0.2 ml dH2O for 15 min to kill all vegetative cells, then serially diluted in a mixture of 1X PBS and 0.1% taurocholate and plated onto BHIS agar with 2 μg ml−1 thiamphenicol and 0.1% taurocholate to enumerate the total number of spores per ml. After 48 h growth, CFU were calculated and the sporulation frequency was determined as the number of spores that germinated following ethanol treatment divided by the total number of spores and vegetative cells [65]. A spo0A mutant complemented with wildtype spo0A driven from its native promoter on a plasmid was used as a positive control (MC848), and a spo0A null mutant containing the empty vector was used as the negative control (MC855). Statistical analyses were performed using the Welch’s ANOVA with Dunnett’s multiple comparisons test to compare spo0A site-directed mutants to the wildtype control (MC848) using GraphPad Prism v8.0.
Western blotting
C. difficile strains were grown in BHIS supplemented with 5 µg ml−1 thiamphenicol, 0.2% fructose, and 0.1% taurocholate. Cultures were then diluted, grown to an OD600 of 0.5, and 250 µL of culture was plated on 70:30 agar. After 12 h, 5 ml of cells were scraped from agar, pelleted, and then washed with 1x PBS. Cells were resuspended in 1X sample buffer (10% glycerol, 5% 2-mercaptoethanol, 62.5 mM upper tris, 3% SDS, 5 mM PMSF) and lysed using a Biospec BeadBeater. Total protein concentration was then measured using a BCA protein assay kit (Pierce), and 2.5 µg of protein was separated by SDS-PAGE using pre-cast TGX 4–15% gradient gels (BioRad) and performed in triplicate. Stain-free imaging using BioRad ChemiDoc MP System was performed for densitometric analysis, and protein was then transferred to a 0.45 µm nitrocellulose membrane. Spo0A was detected using anti-Spo0A antibody [32]. Goat anti-mouse IgG Alexa fluor 488 (Invitrogen) was used as a secondary antibody, and western blots were visualized using a BioRad ChemiDoc MP System. Densitometry calculations were performed using Image Lab 6.0.1 (BioRad). Detected Spo0A protein was normalized to a major band on the stain-free image located at ~40 kDa as a loading control, and then each site-directed mutant was normalized to the Spo0A detected in the parental control strain.
Spo0A modeling for structural changes
RoseTTAFold was used for 3D predictive modeling of wildtype Spo0A and the following Spo0A site-directed mutants: D10A, D11A, D14A, Q17A, V18A, D56A, P60A, A87S, Q90A, K92A, and P109A using default settings and full-length Spo0A amino acid sequences [48]. The angstroms error estimate values for wildtype and the corresponding Spo0A site-directed mutants were derived from the first generated model (Model 1), and the confidence values of the accuracy of the predicted structures were recorded to demonstrate the similarities of the wildtype and mutant Spo0A predicted structures.
Phos-tag blotting
C. difficile strains were cultured as described for western blotting. Cells from two plates for each strain were collected and pelleted. Cell pellets were suspended in 1 ml of 1X sample buffer (5% SDS, 93 mM Tris, 10% glycerol, 100 mM DTT). Protease Inhibitor Cocktail II (Sigma-Aldrich) was included in the sample buffer to inhibit protein degradation. Cells were lysed using a bead beater as described above. Total protein was measured using a BCA protein assay kit (Pierce). 10 μg protein aliquots were kept at 4°C or heated to 99°C for 10 min to dephosphorylate Spo0A prior to loading onto a 12.5% SuperSep Phos-tag gel (Fujifilm Wako)[53, 54]. Total protein was electrophoresed at 125 V for two hours at 4°C. The gel was rinsed three times in transfer buffer with 10% methanol and 10 mM EDTA to remove zinc present within the gel, and subsequently transferred to a low-fluorescence PVDF membrane (Thermo Scientific) in transfer buffer containing 10% methanol and 0.5% SDS overnight at 4°C. Western blot analysis was conducted with anti-FLAG M2 antibody (Sigma-Aldrich), followed by goat anti-mouse Alexa Fluor 488-conjugated antibody (Invitrogen) as the secondary. Experiments were performed in triplicate, and imaging was performed using the BioRad ChemiDoc MP system. Densitometry calculations were performed using ImageJ 1.53a.
Two-hybrid luciferase assays
Two-hybrid assays were performed using a C. difficile codon-optimized split luciferase system previously described [55, 56]. C. difficile strains were grown in 70:30 broth supplemented with 2 µg ml−1 thiamphenicol. Cultures were grown to an OD600 of 0.8 – 0.9, then induced with 50 ng ml−1 anhydrous tetracycline for 1 hour. After induction, the OD600 were recorded, and 100 µL of each culture was added in technical duplicate to a chimney-style 96 well plate. Split-luciferase assay was then performed per manufacturer’s instructions (Promega). Luminescence output was immediately recorded at 135 nm using a BioTek plate reader. Output was normalized to cell density (OD600). A one-way ANOVA with Dunnett’s multiple comparisons test was performed to determine the statistical significance of luminescence outputs of the site-directed mutants relative to the wildtype using GraphPad Prism v8.0.
Supplementary Material
ACKNOWLEDGEMENTS
We are appreciative for the gift of the anti-Spo0A antibody from Aimee Shen. We give special thanks to members of the McBride lab for suggestions during the completion of this work and preparation of this manuscript. This research was supported by the U.S. National Institutes of Health through research grants AI116933 and AI156052 to S.M.M. and GM008490 to M.A.D. The content of this manuscript is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Footnotes
Declaration of Interest: all authors were supported by funding from the National Institutes of Health (NIH). The content of this manuscript is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
REFERENCES
- [1].Ferrari FA, Trach K, LeCoq D, Spence J, Ferrari E, Hoch JA. Characterization of the spo0A locus and its deduced product. Proceedings of the National Academy of Sciences of the United States of America 1985;82:2647–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Perego M, Cole SP, Burbulys D, Trach K, Hoch JA. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. Journal of bacteriology 1989;171:6187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Perego M, Spiegelman GB, Hoch JA. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Molecular microbiology 1988;2:689–99. [DOI] [PubMed] [Google Scholar]
- [4].Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 1991;64:545–52. [DOI] [PubMed] [Google Scholar]
- [5].Strauch MA, Wu JJ, Jonas RH, Hoch JA. A positive feedback loop controls transcription of the spo0F gene, a component of the sporulation phosphorelay in Bacillus subtilis. Molecular microbiology 1993;7:967–74. [DOI] [PubMed] [Google Scholar]
- [6].Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic acids research 2022;50:D439–D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Strauch MA, Hoch JA. Signal transduction in Bacillus subtilis sporulation. Current opinion in genetics & development 1993;3:203–12. [DOI] [PubMed] [Google Scholar]
- [9].Zhao H, Msadek T, Zapf J, Madhusudan, Hoch JA, Varughese KI. DNA complexed structure of the key transcription factor initiating development in sporulating bacteria. Structure 2002;10:1041–50. [DOI] [PubMed] [Google Scholar]
- [10].Rosenbusch KE, Bakker D, Kuijper EJ, Smits WK. C. difficile 630Deltaerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PloS one 2012;7:e48608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Trach K, Burbulys D, Strauch M, Wu JJ, Dhillon N, Jonas R, et al. Control of the initiation of sporulation in Bacillus subtilis by a phosphorelay. Research in microbiology 1991;142:815–23. [DOI] [PubMed] [Google Scholar]
- [12].Zapf J, Sen U, Madhusudan, Hoch JA, Varughese KI. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure 2000;8:851–62. [DOI] [PubMed] [Google Scholar]
- [13].Tzeng YL, Hoch JA. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. Journal of molecular biology 1997;272:200–12. [DOI] [PubMed] [Google Scholar]
- [14].Brown DP, Ganova-Raeva L, Green BD, Wilkinson SR, Young M, Youngman P. Characterization of spo0A homologues in diverse Bacillus and Clostridium species identifies a probable DNA-binding domain. Molecular microbiology 1994;14:411–26. [DOI] [PubMed] [Google Scholar]
- [15].Stephenson K, Hoch JA. Evolution of signalling in the sporulation phosphorelay. Molecular microbiology 2002;46:297–304. [DOI] [PubMed] [Google Scholar]
- [16].Worner K, Szurmant H, Chiang C, Hoch JA. Phosphorylation and functional analysis of the sporulation initiation factor Spo0A from Clostridium botulinum. Molecular microbiology 2006;59:1000–12. [DOI] [PubMed] [Google Scholar]
- [17].Mearls EB, Lynd LR. The identification of four histidine kinases that influence sporulation in Clostridium thermocellum. Anaerobe 2014;28:109–19. [DOI] [PubMed] [Google Scholar]
- [18].Steiner E, Dago AE, Young DI, Heap JT, Minton NP, Hoch JA, et al. Multiple orphan histidine kinases interact directly with Spo0A to control the initiation of endospore formation in Clostridium acetobutylicum. Molecular microbiology 2011;80:641–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Freedman JC, Li J, Mi E, McClane BA. Identification of an Important Orphan Histidine Kinase for the Initiation of Sporulation and Enterotoxin Production by Clostridium perfringens Type F Strain SM101. mBio 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Edwards AN, Wetzel D, DiCandia MA, McBride SM. Three orphan histidine kinases inhibit Clostridioides difficile sporulation. Journal of Bacteriology 2022. e0010622. doi: 10.1128/jb.00106-22. [DOI] [PMC free article] [PubMed]
- [21].Suarez JM, Edwards AN, McBride SM. The Clostridium difficile cpr locus is regulated by a noncontiguous two-component system in response to type A and B lantibiotics. Journal of bacteriology 2013;195:2621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Underwood S, Guan S, Vijayasubhash V, Baines SD, Graham L, Lewis RJ, et al. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. Journal of bacteriology 2009;191:7296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bourret RB, Borkovich KA, Simon MI. Signal Transduction Pathways Involving Protein-Phosphorylation in Prokaryotes. Annual review of biochemistry 1991;60:401–41. [DOI] [PubMed] [Google Scholar]
- [24].Stock JB, Ninfa AJ, Stock AM. Protein-Phosphorylation and Regulation of Adaptive Responses in Bacteria. Microbiological reviews 1989;53:450–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee CW, Park SH, Lee SG, Shin SC, Han SJ, Kim HW, et al. Crystal structure of the inactive state of the receiver domain of Spo0A from Paenisporosarcina sp TG-14, a psychrophilic bacterium isolated from an Antarctic glacier. J Microbiol 2017;55:464–74. [DOI] [PubMed] [Google Scholar]
- [26].Feher VA, Zapf JW, Hoch JA, Whiteley JM, McIntosh LP, Rance M, et al. High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, SpoOF: Implications for phosphorylation and molecular recognition. Biochemistry 1997;36:10015–25. [DOI] [PubMed] [Google Scholar]
- [27].Quisel JD, Burkholder WF, Grossman AD. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. Journal of bacteriology 2001;183:6573–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stephenson SJ, Perego M. Interaction surface of the Spo0A response regulator with the Spo0E phosphatase. Molecular microbiology 2002;44:1455–67. [DOI] [PubMed] [Google Scholar]
- [29].Strauch M, Webb V, Spiegelman G, Hoch JA. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proceedings of the National Academy of Sciences of the United States of America 1990;87:1801–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jiang M, Tzeng YL, Feher VA, Perego M, Hoch JA. Alanine mutants of the Spo0F response regulator modifying specificity for sensor kinases in sporulation initiation. Molecular microbiology 1999;33:389–95. [DOI] [PubMed] [Google Scholar]
- [31].Edwards AN, Nawrocki KL, McBride SM. Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infection and immunity 2014;82:4276–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Putnam EE, Nock AM, Lawley TD, Shen A. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. Journal of bacteriology 2013;195:1214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Spiegelman G, Van Hoy B, Perego M, Day J, Trach K, Hoch JA. Structural alterations in the Bacillus subtilis Spo0A regulatory protein which suppress mutations at several spo0 loci. Journal of bacteriology 1990;172:5011–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kobayashi K, Shoji K, Shimizu T, Nakano K, Sato T, Kobayashi Y. Analysis of a suppressor mutation ssb (kinC) of sur0B20 (spo0A) mutation in Bacillus subtilis reveals that kinC encodes a histidine protein kinase. Journal of bacteriology 1995;177:176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].LeDeaux JR, Grossman AD. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. Journal of bacteriology 1995;177:166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lewis RJ, Brannigan JA, Muchova K, Barak I, Wilkinson AJ. Phosphorylated aspartate in the structure of a response regulator protein. J Mol Biol 1999;294:9–15. [DOI] [PubMed] [Google Scholar]
- [37].Bird TH, Grimsley JK, Hoch JA, Spiegelman GB. Phosphorylation of Spo0A activates its stimulation of in vitro transcription from the Bacillus subtilis spoIIG operon. Molecular microbiology 1993;9:741–9. [DOI] [PubMed] [Google Scholar]
- [38].Parashar V, Mirouze N, Dubnau DA, Neiditch MB. Structural basis of response regulator dephosphorylation by Rap phosphatases. PLoS biology 2011;9:e1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Varughese KI, Tsigelny I, Zhao HY. The crystal structure of beryllofluoride Spo0F in complex with the phosphotransferase Spo0B represents a phosphotransfer pretransition state (vol 188, pg 4970, 2006). Journal of bacteriology 2007;189:3328-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mackin KE, Carter GP, Howarth P, Rood JI, Lyras D. Spo0A differentially regulates toxin production in evolutionarily diverse strains of Clostridium difficile. PloS one 2013;8:e79666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infection and immunity 2012;80:2704–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pettit LJ, Browne HP, Yu L, Smits WK, Fagan RP, Barquist L, et al. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC genomics 2014;15:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dawson LF, Valiente E, Faulds-Pain A, Donahue EH, Wren BW. Characterisation of Clostridium difficile biofilm formation, a role for Spo0A. PloS one 2012;7:e50527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Molle V, Fujita M, Jensen ST, Eichenberger P, González‐Pastor JE, Liu JS, et al. The Spo0A regulon of Bacillus subtilis. Molecular microbiology 2003;50:1683–701. [DOI] [PubMed] [Google Scholar]
- [45].Huang IH, Waters M, Grau RR, Sarker MR. Disruption of the gene (spo0A) encoding sporulation transcription factor blocks endospore formation and enterotoxin production in enterotoxigenic Clostridium perfringens type A. FEMS microbiology letters 2004;233:233–40. [DOI] [PubMed] [Google Scholar]
- [46].Mirouze N, Desai Y, Raj A, Dubnau D. Spo0A similar to P Imposes a Temporal Gate for the Bimodal Expression of Competence in Bacillus subtilis. PLoS genetics 2012;8. [DOI] [PMC free article] [PubMed]
- [47].Volz K Structural conservation in the CheY superfamily. Biochemistry 1993;32:11741–53. [DOI] [PubMed] [Google Scholar]
- [48].Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021;373:871-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fujita M, Sadaie Y. Feedback loops involving Spo0A and AbrB in in vitro transcription of the genes involved in the initiation of sporulation in Bacillus subtilis. J Biochem 1998;124:98–104. [DOI] [PubMed] [Google Scholar]
- [50].Asayama M, Yamamoto A, Kobayashi Y. Dimer form of phosphorylated Spo0A, a transcriptional regulator, stimulates the spo0F transcription at the initiation of sporulation in Bacillus subtilis. Journal of molecular biology 1995;250:11–23. [DOI] [PubMed] [Google Scholar]
- [51].Lewis RJ, Scott DJ, Brannigan JA, Ladds JC, Cervin MA, Spiegelman GB, et al. Dimer formation and transcription activation in the sporulation response regulator Spo0A. Journal of molecular biology 2002;316:235–45. [DOI] [PubMed] [Google Scholar]
- [52].Barbieri CM, Stock AM. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Analytical biochemistry 2008;376:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kinoshita E, Kinoshita-Kikuta E. Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics 2011;11:319–23. [DOI] [PubMed] [Google Scholar]
- [54].Kinoshita-Kikuta E, Kusamoto H, Ono S, Akayama K, Eguchi Y, Igarashi M, et al. Quantitative monitoring of His and Asp phosphorylation in a bacterial signaling system by using Phos-tag Magenta/Cyan fluorescent dyes. Electrophoresis 2019;40:3005–13. [DOI] [PubMed] [Google Scholar]
- [55].Oliveira Paiva AM, Friggen AH, Hossein-Javaheri S, Smits WK. The Signal Sequence of the Abundant Extracellular Metalloprotease PPEP-1 Can Be Used to Secrete Synthetic Reporter Proteins in Clostridium difficile. ACS synthetic biology 2016;5:1376–82. [DOI] [PubMed] [Google Scholar]
- [56].Oliveira Paiva AM, Friggen AH, Qin L, Douwes R, Dame RT, Smits WK. The Bacterial Chromatin Protein HupA Can Remodel DNA and Associates with the Nucleoid in Clostridium difficile. Journal of molecular biology 2019;431:653–72. [DOI] [PubMed] [Google Scholar]
- [57].Madhusudan, Zapf J, Whiteley JM, Hoch JA, Xuong NH, Varughese KI. Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis (vol 4, pg 679, 1996). Structure 1996;4:999-. [DOI] [PubMed] [Google Scholar]
- [58].Obana N, Nakao R, Nagayama K, Nakamura K, Senpuku H, Nomura N. Immunoactive Clostridial Membrane Vesicle Production Is Regulated by a Sporulation Factor. Infection and immunity 2017;85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].McBride SM, Sonenshein AL. Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infection and immunity 2011;79:167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Stephenson K, Lewis RJ. Molecular insights into the initiation of sporulation in Gram-positive bacteria: new technologies for an old phenomenon. FEMS microbiology reviews 2005;29:281–301. [DOI] [PubMed] [Google Scholar]
- [61].Sorg JA, Dineen SS. Laboratory maintenance of Clostridium difficile. Current protocols in microbiology 2009;Chapter 9:Unit9A 1. [DOI] [PubMed]
- [62].Edwards AN, Suarez JM, McBride SM. Culturing and maintaining Clostridium difficile in an anaerobic environment. Journal of visualized experiments : JoVE 2013:e50787. [DOI] [PMC free article] [PubMed]
- [63].Luria SE, Burrous JW. Hybridization between Escherichia coli and Shigella. Journal of bacteriology 1957;74:461–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. Journal of bacteriology 2012;194:3307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Edwards AN, Tamayo R, McBride SM. A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Molecular microbiology 2016;100:954–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Edwards AN, McBride SM. Determination of the in vitro Sporulation Frequency of Clostridium difficile. Bio Protoc 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Childress KO, Edwards AN, Nawrocki KL, Woods EC, Anderson SE, McBride SM. The Phosphotransfer Protein CD1492 Represses Sporulation Initiation in Clostridium difficile. Infection and immunity 2016. [DOI] [PMC free article] [PubMed]
- [68].Diaz AR, Stephenson S, Green JM, Levdikov VM, Wilkinson AJ, Perego M. Functional role for a conserved aspartate in the Spo0E signature motif involved in the dephosphorylation of the Bacillus subtilis sporulation regulator Spo0A. The Journal of biological chemistry 2008;283:2962–72. [DOI] [PubMed] [Google Scholar]
- [69].Cheng RR, Morcos F, Levine H, Onuchic JN. Toward rationally redesigning bacterial two-component signaling systems using coevolutionary information. Proceedings of the National Academy of Sciences of the United States of America 2014;111:E563–E71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Perego M, Hanstein C, Welsh KM, Djavakhishvili T, Glaser P, Hoch JA. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell 1994;79:1047–55. [DOI] [PubMed] [Google Scholar]
- [71].Tzeng YL, Feher VA, Cavanagh J, Perego M, Hoch JA. Characterization of interactions between a two-component response regulator, Spo0F, and its phosphatase, RapB. Biochemistry 1998;37:16538–45. [DOI] [PubMed] [Google Scholar]
- [72].McLaughlin PD, Bobaya BG, Regel EJ, Thompson RJ, Hoch JA, Cavanagh J. Predominantly buried residues in the response regulator Spo0F influence specific sensor kinase recognition. FEBS letters 2007;581:1425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Perego M, Hoch JA. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proceedings of the National Academy of Sciences of the United States of America 1996;93:1549–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Feher VA, Tzeng YL, Hoch JA, Cavanagh J. Identification of communication networks in SpoOF: a model for phosphorylation-induced conformational change and implications for activation of multiple domain bacterial response regulators. FEBS letters 1998;425:1–6. [DOI] [PubMed] [Google Scholar]
- [75].Hussain HA, Roberts AP, Mullany P. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Δerm) and demonstration that the conjugative transposon Tn916ΔE enters the genome of this strain at multiple sites. Journal of medical microbiology 2005;54:137–41. [DOI] [PubMed] [Google Scholar]
- [76].Thomas CM, Smith CA. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol 1987;41:77–101. [DOI] [PubMed] [Google Scholar]
- [77].Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 1985;33:103–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


