Significance
Mammalian complex IV (CIV), the final complex of the mitochondrial electron transport chain, uses electrons from cytochrome c to reduce oxygen to water, driving aerobic life. Although CIV functions as the main site for respiratory regulation, there is little structural or biochemical information on how this regulation occurs. Previous studies provided evidence of CIV regulation by steroids, but the steroid binding site and regulatory mechanism remain unclear. Using single particle cryogenic electron microscopy (cryo-EM), we discover the binding site of the steroid-derived detergent, glyco-diosgenin. Results from flash photolysis kinetic experiments with CIV in the presence of glyco-diosgenin and cholesteryl hemisuccinate are combined with cryo-EM and molecular simulations to elucidate how steroid binding limits proton uptake by the complex.
Keywords: electron transport chain, complex IV, cryo-EM, kinetics, molecular simulations
Abstract
The mitochondrial electron transport chain maintains the proton motive force that powers adenosine triphosphate (ATP) synthesis. The energy for this process comes from oxidation of reduced nicotinamide adenine dinucleotide (NADH) and succinate, with the electrons from this oxidation passed via intermediate carriers to oxygen. Complex IV (CIV), the terminal oxidase, transfers electrons from the intermediate electron carrier cytochrome c to oxygen, contributing to the proton motive force in the process. Within CIV, protons move through the K and D pathways during turnover. The former is responsible for transferring two protons to the enzyme’s catalytic site upon its reduction, where they eventually combine with oxygen and electrons to form water. CIV is the main site for respiratory regulation, and although previous studies showed that steroid binding can regulate CIV activity, little is known about how this regulation occurs. Here, we characterize the interaction between CIV and steroids using a combination of kinetic experiments, structure determination, and molecular simulations. We show that molecules with a sterol moiety, such as glyco-diosgenin and cholesteryl hemisuccinate, reversibly inhibit CIV. Flash photolysis experiments probing the rapid equilibration of electrons within CIV demonstrate that binding of these molecules inhibits proton uptake through the K pathway. Single particle cryogenic electron microscopy (cryo-EM) of CIV with glyco-diosgenin reveals a previously undescribed steroid binding site adjacent to the K pathway, and molecular simulations suggest that the steroid binding modulates the conformational dynamics of key residues and proton transfer kinetics within this pathway. The binding pose of the sterol group sheds light on possible structural gating mechanisms in the CIV catalytic cycle.
The transmembrane protein complexes of the electron transport chain power the production of adenosine triphosphate (ATP) by maintaining an electrochemical proton motive force (PMF) across the mitochondrial inner membrane or the plasma membrane of bacteria. The first component of the mitochondrial chain, complex I, initiates electron transport by oxidizing reduced nicotinamide adenine dinucleotide (NADH) produced by the Krebs cycle, glycolysis, and fatty acid oxidation. Complex I uses electrons from NADH to reduce membrane-bound quinone to quinol while pumping protons from the negatively charged matrix side (N side) of the mitochondrial inner membrane to the positively charged intermembrane space side (P side). Complex II oxidizes succinate to fumarate and reduces quinone, contributing to the quinol pool. Complex III oxidizes quinol to quinone through a mechanism known as the Q cycle, adding to the PMF and reducing soluble cytochrome c in the intermembrane space. Reduced cytochrome c then binds to complex IV (CIV), the terminal enzyme of the electron transport chain. CIV contributes to the PMF while transferring electrons from cytochrome c to the final electron acceptor, O2, which is reduced to H2O. Each electron transfer from cytochrome c to O2 is linked to proton uptake from the N side and pumping of one proton from the N to the P side of the membrane. Thus, overall, the eukaryotic electron transport chain couples proton translocation across the mitochondrial inner membrane with electron transfer from NADH and succinate to O2.
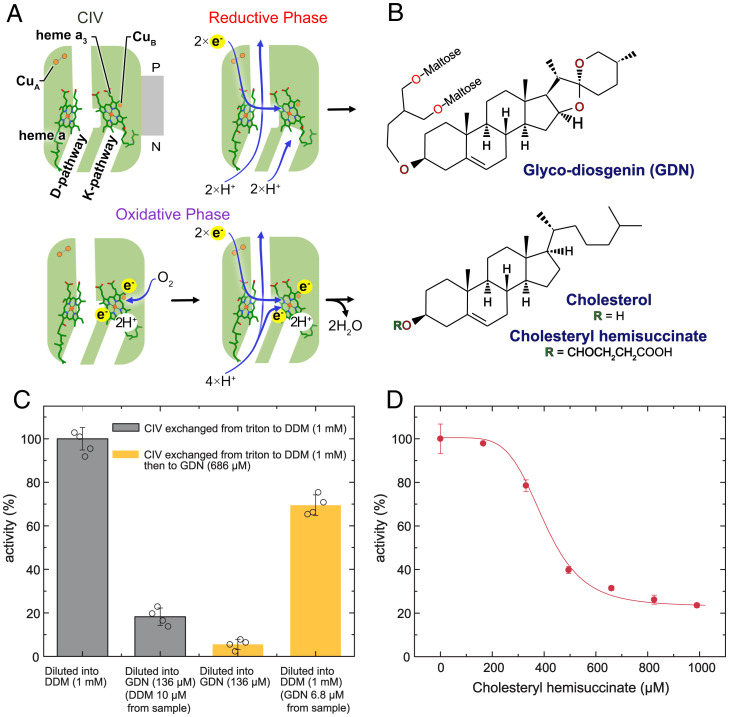
Reduced cytochrome c binds to CIV and is oxidized by its primary electron acceptor, CuA. The electron is then transferred to heme a and onwards to the enzyme’s catalytic site, which includes heme a3 and CuB (Fig. 1A, Top Left) (for reviews see refs. 1, 2). Overall, CIV catalyzes the oxidation of four cytochrome c molecules, uptake of eight protons from the N side, and reduction of one dioxygen molecule to produce two water molecules and add four protons to the P side. Turnover of CIV can be divided into a reductive phase and an oxidative phase. In the reductive phase, two electrons are transferred from cytochrome c molecules to the oxidized catalytic site (Fig. 1A, Top Right). In the oxidative phase, dioxygen binds to the reduced heme a3 and CIV receives two more electrons from cytochrome c molecules to complete the reaction cycle (Fig. 1A, Bottom). Reduction of the oxidized catalytic site is coupled to the transfer of two protons from the N side to the catalytic site through the K pathway (3, 4), which comprises conserved residues including the Lys319 residue (Bos taurus CIV amino acid residue numbering) that gives the pathway its name (5). After binding of dioxygen to the catalytic site and transfer of the next two electrons from cytochrome c molecules, proton transfer from the N side to the catalytic site occurs through the D pathway. This pathway is composed of water molecules and conserved polar and protonatable residues, including Glu242 and Asp91, the latter giving the pathway its name (6). During each electron transfer from cytochrome c to the catalytic site, protons are pumped across the membrane through the D pathway (for review, see refs. 2, 7–11). A third pathway (the H pathway) has also been suggested for the mammalian CIV (8, 12), but remains controversial because it is not present in the mitochondrial CIV from Saccharomyces cerevisiae (13) or bacterial CIVs (14).
Fig. 1.
Inhibition of CIV by cholesterol analogs. (A) Cartoon of CIV structure, with electron carriers and channels labeled (Top Left structure). In the reductive phase, CIV pumps protons across the inner membrane through the D pathway and transfers protons to the catalytic site through the K pathway (Top Right structure). In the oxidative phase, oxygen binds to the catalytic site (Bottom Left structure) followed by transfer of protons across the inner membrane and to the catalytic site through the D pathway (Bottom Right structure). (B) Structure of GDN, cholesterol, and CHS. (C) Cytochrome c oxidase activity measured with a Clark-type oxygen electrode. CIV incubated in DDM then diluted into DDM (first gray bar) or GDN (second gray bar), incubated in GDN and diluted into DDM (first orange bar) or GDN (second orange bar). (D) CIV activity in LMNG with varying concentrations of CHS (red points, mean ± SD, n = 3 independent measurements), and fit to data with an empirical Hill equation (red line).
Complex IV is thought to be the central regulatory site for oxidative phosphorylation in mitochondria (15). This regulation is of paramount importance as mitochondrial dysfunction can lead to the progression of numerous human diseases, including neurodegeneration, ischemia reperfusion injury, cancer, and diabetes (15). Mammalian CIV is regulated by phosphorylation of subunits (16) and differential expression of subunit isoforms (17). CIV also possesses conserved lipid binding sites (18) that allow the enzyme’s lipid environment to modulate its activity (19, 20). Lipophilic small molecules, including detergents, steroids, and bile salts, have a complex effect on the activity of CIV from Rhodobacter sphaeroides (18, 21). Steroids as well as thyroid hormones have been shown to inhibit mammalian CIV (22, 23), possibly by interfering with K pathway proton translocation (24), and thyroid hormones also appear to suppress superoxide formation by CIV (23). This effect has also been seen when mitochondria are exposed to steroids and glucocorticoids, which causes decreased oxygen consumption, an effect that is being explored in relation to cancer therapies (25, 26). Further, accumulation of bilirubin inhibits CIV, possibly by blocking K pathway proton translocation, which was suggested to be linked with development of neonatal encephalopathy. This inhibition can be reversed by the bile salt glycoursodeoxycholic acid (10, 27).
Here, we explore the interaction between mammalian CIV and the steroids cholesteryl hemisuccinate (CHS) and glyco-diosgenin (GDN). CIV activity measurements show that both GDN, a detergent with a steroid-based lipophilic group (28), as well as CHS, reversibly inhibit CIV. Flash photolysis reveals that binding of each of these compounds results in impaired K pathway proton translocation, similar to K pathway mutants in R. sphaeroides. TheA cryogenic electron microscopy (cryo-EM) structure of CIV with GDN defines the sterol binding site and pose, which is different from a site previously proposed from crystal structures of proteins purified and crystallized with sodium cholate and deoxycholate (29–32). Importantly, the binding site is adjacent to helix VII of subunit I, which is in close contact with Lys319 and may have a direct role in proton gating. Molecular simulations explain the inhibitory effect of sterol binding on proton transfer through the K pathway at a molecular level.
Results and Discussion
Inhibition of Mammalian CIV by Cholesterol Derivatives.
A number of bile acids and other sterol-containing compounds, including detergents, have been shown to inhibit mammalian and bacterial CIV activity in vitro (21, 22). To explore this effect, CIV was purified from bovine heart mitochondria (33) and its activity was assayed with a Clark electrode, which measures the oxygen concentration as a function of time. Concentrated CIV was diluted into an aerobic buffer containing a pool of cytochrome c, which is kept reduced by an excess of ascorbate and the electron mediator N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD). Under these conditions, CIV uses the electrons from cytochrome c to reduce dissolved oxygen until it is depleted. Clark electrode measurements with CIV purified in Triton X-100 and exchanged into n-dodecyl-β-D-maltoside (DDM) showed an electron transfer rate of 220 ± 10 e−/s (mean ± SD, n = 4 measurements from two separate purifications), similar to previous studies in DDM (33). However, upon dilution of concentrated DDM-solubilized CIV into buffer containing GDN, which includes a steroid-derived lipophilic group (Fig. 1B, Top) (28), CIV activity decreased by ∼80% (Fig. 1C, second bar). CIV exchanged into GDN buffer during ion exchange chromatography and assayed in buffer containing GDN displays only ∼5% of the activity of the DDM-solubilized CIV assayed in DDM-containing buffer (Fig. 1C, third bar), but upon dilution into DDM buffer it regains ∼60% of the activity of DDM-solubilized CIV assayed in DDM-containing buffer (Fig. 1C, fourth bar). These data show that GDN inhibits CIV, and that DDM can reverse the inhibition. This observation is consistent with previous data showing reversible inhibition of mammalian CIV by sterol-containing compounds (22). The ability of DDM to reverse inhibition of CIV by small molecules was demonstrated with CIV from R. sphaeroides (21, 34) and is likely related to DDM’s ability to strip hydrophobic molecules from proteins (35).
To confirm that the observed inhibition was caused by the steroid moiety of GDN, CIV was titrated with CHS, a cholesterol derivative that is more soluble than cholesterol due to the addition of a succinate group (Fig. 1B, Bottom). CIV in DDM was exchanged to the lower critical micelle concentration (CMC) detergent, lauryl maltose neopentyl glycol (LMNG), before titration with CHS. Detergents with lower CMCs are often considered more “mild” than higher CMC detergents and exchange from DDM to LMNG was performed to minimize the risk of removal of CHS from CIV by the detergent. In the absence of CHS, the activity of CIV in LMNG was 200 ± 15 e−/s (mean ± SD, n = 4 measurements from two separate purifications), comparable to its activity in DDM. As CHS was gradually increased to 990 µM, the activity of CIV decreased to ∼20% of its initial activity (45 ± 5 e−/s) (Fig. 1D). Together, these data show that the sterol-containing molecules GDN and CHS can inhibit CIV, likely due to the sterol moieties of the molecules.
Steroids Inhibit Proton Uptake through the K Pathway.
The absorbance spectra of the redox cofactors in CIV change with redox state, making it possible to determine how electrons are distributed among the internal electron carriers. Results from earlier stopped-flow measurements with R. sphaeroides CIV revealed that bile salts caused accumulation of electrons at heme a, suggesting that the electron transfer from heme a to the catalytic site is rate limiting (21, 23, 36).
To identify the internal electron transfers that are inhibited by GDN and CHS, we investigated intramolecular electron transfer and proton-coupled electron transfer reactions in CIV with the so-called flow-flash technique (see, e.g., ref. 4). To study the oxidative phase with this technique, CIV is reduced in the absence of O2 and the catalytic site is blocked by binding of carbon monoxide (CO). After addition of O2, the CO ligand is removed by a short laser flash. Removal of CO allows O2 to bind to the catalytic site and initiate oxidation of the enzyme, which can be followed spectrophotometrically with microseconds (µs) time resolution. As indicated above, in the oxidative phase all protons are transferred through the D pathway and any effects from slowed proton uptake are typically manifested in slowing electron transfers that are linked to D pathway proton uptake (37). The time constant of these components (SI Appendix, Fig. S1) were the same in GDN and DDM, indicating that the impaired activity of CIV in GDN is not due to slowed intramolecular electron transfer or proton uptake through the D pathway.
The reductive phase is typically investigated by mixing oxidized CIV with reduced cytochrome c. However, in this reaction, electron transfer from cytochrome c to CIV is rate limiting, which precludes identification of specific intramolecular reactions that may be slowed. To overcome this limitation, we measured internal electron transfer and proton-coupled electron transfer in flash photolysis experiments (38, 39). In these experiments, CIV is incubated with CO in an anaerobic environment creating a mixed valence state where CuA and heme a are oxidized, while CuB and heme a3 at the catalytic site are reduced, and CO is bound to heme a3 (Fig. 2A, Top Left) (4). The heme a3-CO bond is photolabile and upon a short laser flash the CO ligand dissociates and equilibrates with the gas dissolved in solution (Fig. 2A, pink arrow). Upon dissociation of CO, the apparent midpoint potential of heme a3 decreases, which results in electron transfer from heme a3 to heme a and then to CuA over time scales of ∼3 and 30 µs, respectively (4). Because this electron transfer occurs in the opposite direction of the electron transfer during turnover, the reaction is referred to as “electron backflow.” However, the rate constants of the kinetic components reflect the sum of the forward and backward rate constants and therefore provide information on intramolecular electron transfer in the reductive phase. With the mammalian CIV, after CO dissociation the difference in heme a/a3 midpoint potentials yields ∼10 to 30% (depending on the source of CIV) oxidation of heme a3 (38, 40), with the remaining population retaining the electron at heme a3 (Fig. 2A, Bottom). In this remaining population, the electron equilibrium shifts further toward heme a once a proton is transferred through the K pathway (4, 38) (Fig. 2A, purple arrow). Both the fraction of this component and its time constant increase with increasing pH (pKa ≅ 8 to 9, depending on the source of CIV) (38, 40), and therefore the reaction is monitored at pH >7. This electron transfer event can be monitored by measuring the change in absorbance at 598 nm (38).
Fig. 2.
Flash photolysis experiments. (A) CIV in the mixed valence state (Top Left structure) is excited by a laser flash (pink arrow). In a population where heme a3 remains reduced, electron transfer from heme a3 to heme a is coupled with proton translocation through the K pathway (purple arrow), producing a change in absorbance at 598 nm. (B) Absorbance changes at 598 nm. The data within the purple box show a kinetic phase attributed to proton-coupled electron transfer from heme a3 to heme a for LMNG only (black curve), LMNG + CHS (red curve), and GDN (orange curve). Inset shows zoom out of absorbance changes over 400 ms.
To investigate the effect of sterol binding on the reductive phase, CIV in LMNG was first prepared in the mixed-valence state for flash photolysis experiments. Upon laser-induced photolysis, CO disassociates and an electron is transferred from heme a3 to heme a in a subpopulation of CIV complexes, causing a large increase in the absorbance at 598 nm in the time range 0 to 50 µs (38) (Fig. 2B, black curve, first 0.05 ms). In the remaining subpopulation of CIV complexes a proton must leave through the K pathway for the electron transfer to occur with a time constant of 1.6 ± 0.2 ms (average of ∼200 traces from each of two separate purifications) (Fig. 2A, purple arrow and Fig. 2B, black curve in purple box). Over a longer time scale the signal at 598 nm slowly decreases back to baseline as CO recombines with CIV (Fig. 2B, black curve, Inset) bringing the system to equilibrium with a time constant of ∼58 ms and allowing for repetition of the experiment. CIV with LMNG and CHS, as well as CIV exchanged to GDN, were also prepared in the mixed valence state. Upon flash photolysis, both samples showed a rapid increase in absorbance in the 0 to 50-µs time range, indicating that the photolysis and electron transfer occurs in a portion of the population. This observation shows that internal electron transfer is not affected by binding of GDN or CHS (Fig. 2B, blue and red curves, first 50 μs). However, after this rapid increase in absorbance there is no further absorbance increase, so that the 1.6-ms component is not observed (Fig. 2B, blue and red curves in purple box). The absence of this component demonstrates that the proton-coupled electron transfer does not occur. These results show that proton transfer through the K pathway is blocked in the presence of GDN or CHS. Furthermore, this loss of the 1.6-ms component is identical to data from flash photolysis studies in which the K pathway of CIV from R. sphaeroides was deactivated by site-directed mutagenesis (4). Therefore, these data also support the suggestion that slowed proton uptake through the K pathway is the reason for accumulation of electrons at heme a when R. sphaeroides CIV is treated with bile salts (21, 36). As shown in the Inset in Fig. 2B, recombination of the CO with the LMNG sample was a factor of ∼2 (∼58 ms) slower than with the LMNG + CHS and GDN samples (∼28 ms). As shown previously (38), the CO ligand binds only to the fraction of CIV in which heme a3 is reduced, yielding a CO-recombination time constant that is dependent on the heme a3/heme a electron equilibrium. Because GDN and CHS block proton-coupled electron transfer from heme a3 to heme a, the fraction heme a3 that is reduced increases and CO recombination with CO can occur faster.
Structure of GDN-Inhibited CIV Reveals Steroid Proximity to K Pathway.
To gain insight into how steroids limit proton translocation along the K pathway, CIV in GDN was subjected to structural analysis by cryo-EM. GDN was selected over LMNG with CHS because of the possibility that LMNG could compete with CHS for interaction with the sterol binding site, while with CIV in GDN there is no possibility for this competition. To detect conformational changes induced by reduction of the complex not seen in crystal structures, CIV was reduced and inhibited with CO prior to freezing cryo-EM specimens. Subsequent imaging and image analysis led to a 3.1-Å resolution map of the complex (Fig. 3A, Top and SI Appendix, Fig. S2A–D and Table S1). The map of CIV showed density for three core and 10 peripheral subunits of the complex, but is missing the NDUFA4 subunit originally thought to be part of complex I (41). Previous atomic models from X-ray crystallography (42) fit well into the cryo-EM map (Fig. 3A and SI Appendix, Fig. S2E and Table S1) indicating no conformational changes in the present structure. The previous crystal structure was higher resolution than the cryo-EM structure and consequently no further refinement was performed prior to validation. In addition, three-dimensional (3D) variability analysis (43) did not detect any motions, suggesting a rigid protein structure. Clear density was apparent for the lipids phosphatidylethanolamine (PE) and phosphatidylglycerol (PG) between subunits I and III (Fig. 3A, yellow models and SI Appendix, Fig. S2E), which were seen in earlier crystal structures (18, 42). These lipids have been suggested to be important for CIV function (31).
Fig. 3.
Cryo-EM structure of GDN-inhibited CIV. (A) Cryo-EM map (Top), and atomic model for CIV (Bottom) with lipids PE and PG modeled and GDN shown in orange. (B) Closeup of K pathway with map and model for the GDN (orange map and model).
The proton translocation pathways in CIV are formed by the three core subunits (subunits I to III) of CIV. The entrance to the K pathway is found on the N side of the inner membrane at the interface of helix VII from subunit I and helix II of subunit II (Fig. 3B), the latter bearing Glu62, the first amino acid in the K pathway proton transfer chain. This region of the map is at a lower resolution than surrounding regions (SI Appendix, Fig. S2D), consistent with the higher B factors for residues at the entrance of the K pathway in earlier crystal structures (21). These higher B factors, which correspond to uncertainty in the positions of atoms, have been attributed to movements at the K pathway opening that are necessary for proton uptake (24). The remainder of the pathway is formed by helices VI and VII of subunit I, bearing the CuB ligands His240 (helix VI), His290 (helix VII), and His291 (helix VII), as well as helix VIII of subunit I, which contains the eponymous Lys319 residue. Subunit I also contains Thr316 and Tyr244, the latter being part of the catalytic site, which participates in the reduction of oxygen to water (1). Helix VII lies between the K pathway and the outside of the protein and is in close contact with Lys319 (Fig. 3B). The cryo-EM map reveals clear density for the sterol portion of a GDN molecule adjacent to helix VII (Fig. 3A and B). With the SD of the cryo-EM map normalized to σ = 1, the sterol is apparent at 8σ, similar to the density from helix VII, suggesting high occupancy of GDN in this site. However, no clear density corresponding to the maltose groups of GDN could be detected, suggesting that these portions of the detergent molecule are more dynamic. The sterol from GDN sits in a hydrophobic cleft formed by helices V, VI, and VII of subunit I. This position puts GDN in close contact (∼4.6 Å) with Met278 on helix VII (Fig. 3B, cyan dashed line) where it is positioned to affect dynamics of the helix and proton translocation as described below.
Cryo-EM shows that the CIV preparation used in the present study was monomeric. Both dimers and monomers of CIV have been observed in solution previously (44–47), with monomers reported to show more activity than dimers (46). Bile salts such as sodium cholate, which include a sterol moiety, promote formation of CIV dimers (47), with both oligomeric states having been resolved by X-ray crystallography (12, 46). X-ray crystallography of bovine CIV dimers in the presence of cholate showed cholate in the crystals at the dimer interface (21, 48). Diffraction analysis of R. sphaeroides CIV crystals grown in the presence of cholate showed cholate between CIV complexes at crystal contacts in the lattice (32). These studies led to the proposal that the dimer interface forms a sterol binding site near the entrance to the K pathway for both species. More recently, cholate was modeled far from the K pathway in a X-ray crystal structure of monomeric bovine CIV, indicating that cholate remains in the sample after purification with cholate but is not bound at the dimerization interface (46). Further, the CIV NDUFA4 subunit seen in a recent cryo-EM structure of human CIV (41) is bound where the monomer–monomer interface forms in dimers, blocking the proposed cholate binding site. Together, the inconsistent location of cholate in X-ray crystal structures suggests that the presence of cholate in CIV crystals is due to the crystallization process and does not indicate a specific sterol binding site. The present results show that sterols can indeed bind monomeric CIV. Dimers of bovine CIV appear to exclude steroids from the binding site seen here because the N-terminal tail of subunit VIa in each monomer occupies the sterol binding pocket of the opposite monomer (SI Appendix, Fig. S2F) (42). In the simpler four subunit CIV from R. sphaeroides, the sterol binding site seen here with bovine CIV monomers is blocked by subunit IV (49), which is not present in the mammalian enzyme.
Role of Helix VII in K pathway Gating.
Proton transfer along the K and D pathways requires continuous hydrogen-bonded chains of water molecules as well as polar and protonatable amino acid side chains. While no water molecules could be resolved in the current map, crystal structures of CIV show that the D pathway is highly hydrated (50). The K pathway, on the other hand, shows gaps in the water networks (42, 50, 51). This lack of a continuous proton-conducting pathway implies that reorientation of side chains and hydration must occur to accommodate proton transfer through the K pathway (50, 52, 53). Prior molecular dynamics (MD) simulations showed that Lys319 can adopt multiple conformations, with the deprotonated side chain adopting a “down” conformation far from Tyr244 and the protonated side chain adopting an “up” conformation near to Tyr244, which allows for water-mediated proton transfer to the catalytic site (53–55).
To understand how GDN binding affects proton transfer through the K pathway, we performed MD simulations of the bovine CIV with and without GDN modeled next to helices VI and VII in the orientation observed by cryo-EM (Fig. 4A). Similar to previous simulations, Lys319 adopts both an up and down conformation (Fig. 4A). During the MD simulations without GDN, up to ∼10 water molecules diffuse into the K pathway from the N side of the membrane (Fig. 4B and SI Appendix, Fig. S3A, dark green curve). The Lys319 side chain flips from its down conformation (Lys319-Tyr244 distance >16 Å) to its up conformation (Lys319-Tyr244 distance <12 Å), which results in formation of a hydrogen-bonded water wire with three to four water molecules leading from Lys319 to Tyr244 via Thr316 (Fig. 4B, circled in blue). These three to four water molecules allow for proton transfer between Lys319 and Tyr244. Reorientation of Lys319 into its up conformation is observed only in simulations where the CuB is reduced (53), whereas with an oxidized catalytic site Lys319 samples the down conformation far from Y244, consistent with previous results (53, 56, 57). In contrast to the up conformation of Lys319 in the absence of GDN (Fig. 4C, blue curve), when the MD simulations are performed with GDN modeled between helices VI and VII, Lys319 samples only the down orientation, independent of the modeled redox state at CuB (Fig. 4C, orange curve). This finding, in addition to previous simulations (57), are also consistent with recent cryo-EM structures of CIV from Paracoccus denitrificans showing that Lys319 (Lys354 in P.denitrificans) moves toward Tyr244 (Tyr280 in P. denitrificans) when the enzyme is in the reduced state (58).
Fig. 4.
Molecular simulations of CIV K pathway dynamics and proton uptake. (A) Snapshot of MD trajectories. Phe251 and Lys319 are displayed in the down conformation (dark gray) and the up conformation (light gray), with the Lys319-Y244 distance labeled. (B) Residues involved in the proton transfer reaction, showing the structure of the water wire connecting Lys319 and CuB, with the water molecules between Lys319 and Tyr244 circled in blue. (C) Simulation trajectories in the presence (orange curve) and absence (blue curve) of GDN. In the presence of GDN, Lys319 samples the down conformation (d > 16 Å), in the absence of GDN, Lys319 can flip to its up conformation (d < 12 Å). (D) Distribution of the Phe251 side chain dihedral angle (χ1) from simulations 1 through 6 (SI Appendix, Table S2). Simulations with GDN show an enhanced sampling of the down conformation of Phe251 (χ1 = 60°) as compared to the simulations without a sterol. The down conformation of Phe251 could prevent Lys319 from flipping upwards (see B) and suppresses proton uptake through the K pathway. (E) When Phe251 and Lys319 are in the down conformation Lys319 can extract a proton form the N side of the inner membrane (Left structure). The correlated movement of Phe251 and Lys319 to the up conformation allows proton transfer to the catalytic site (Middle structure). Steroids block movement of Phe251, which in turn blocks Lys319, preventing proton transfer through the K pathway.
The simulation trajectories suggest that GDN binding affects the conformation of aliphatic residues in helices VI and VII, which in turn affect the conformation of Lys319. In particular, Phe251 on helix VI samples a down orientation (χ1 of Phe251 ∼170°) in the presence of GDN (Fig. 4D, orange and SI Appendix, Fig. S3B–D), which appears to block rotation of Lys319 into its up conformation. In contrast, in the absence of GDN the aromatic side chain is only found in the up conformation (Fig. 4D, blue). Simulations with cholesterol in the sterol binding cavity show similar results to those with GDN (SI Appendix, Fig. S3C and D, red). As described above, X-ray crystallography of dimeric CIV shows that the N-terminal tail of subunit VIa in each monomer binds the opposing monomer of the dimer (SI Appendix, Fig. S3E, red ribbon). The binding site for the N-terminal tail overlaps with the sterol binding pocket observed in cryo-EM of the monomeric protein (SI Appendix, Fig. S3E, cyan model). Occupancy of this site would prevent sterol binding to the site identified here by cryo-EM of the monomeric CIV. To investigate this hypothesis we performed MD simulations of the dimeric bovine CIV. During these simulations, the N-terminal tail of subunit VIa appears dynamic but remains associated with its binding site (SI Appendix, Fig. S3E), consistent with competition between the two interactions. Despite the persistence of the N-terminal tail in the sterol binding pockets of the dimer, Lys319 can still sample the up conformation (SI Appendix, Fig. S3F). This observation suggests that binding of the N-terminal tail from subunit IVa is not equivalent to sterol binding and is not the reason for the reported difference in activity between CIV monomers and dimers (46).
To further probe how the up conformation of Lys319 is connected to the proton transfer along the K pathway, we performed hybrid quantum/classical (QM/MM) calculations. These calculations suggest that protons are transferred from Lys319 to Tyr244 along water molecules in Grotthuss-type reaction steps, where excess protons form hydronium and Zundel ion intermediates (SI Appendix, Fig. S4A). The reaction is energetically feasible with a barrier of ∼11 kcal mol−1 (SI Appendix, Fig. S4B and C) suggesting that the reaction can take place on microsecond to millisecond time scales. In contrast, the down state of Lys319 sampled in the GDN simulations leads to loss of the water connectivity between Lys319 and Tyr244, effectively blocking the water-mediated proton transfer reaction to the catalytic site heme a3/CuB binuclear center. Together the classical MD simulations and the hybrid QM/MM simulations suggest that the up conformation of Phe251 is required for Lys319 to adopt its up conformation, which in turn is needed for water wire formation and proton translocation (Fig. 4E, Left two structures). GDN binding alters the orientation of Phe251, which in turn blocks Lys319 and prevents proton translocation (Fig. 4E, Right structure). Together, these results show how GDN binding to the site observed by cryo-EM induces the changes in internal electron transfers seen in kinetic experiments.
Materials and Methods
Preparation of Bovine Heart Mitochondria.
Bovine hearts were kept on ice and all subsequent steps were carried out at 4 °C. Fat, blood vessels, and connective tissues were removed from the hearts and the remaining material was cut into ∼2-cm3 pieces before being ground with a meat grinder. Portions of 200 g of ground heart were mixed with 600 mL of buffer (250 mM sucrose and 10 mM Tris pH 7.8) and 3 mL of 2 M Tris (pH unadjusted) in a blender for 45 s. A total of 4 mL of 2 M Tris was then added and mixed for another 45 s and pH was adjusted to 7.8 if necessary. The homogenate was centrifuged at 1,600 × g for 15 min to remove unbroken cells and nuclei. The supernatant was filtered through two layers of cheesecloth to remove lipid granules and centrifuged at 17,000 × g for 30 min. The pellet was then resuspended in 250 mM sucrose and 10 mM Tris pH 7.8 and centrifuged at 17,000 × g for 30 min. The resulting pellet consisted of two layers, one soft and light in color, and one darker and firmer. The lighter fraction (broken mitochondrial fragments) was removed by decantation. The procedure was then repeated until only the heavy fraction of mitochondria remained after centrifugation. The heavy fraction of mitochondria was frozen in liquid nitrogen and stored at −80 °C until use.
Purification of Mammalian CIV.
Purification of bovine CIV was performed as described in ref. 33 with some modifications (changes refer to the procedure described in section “Purification of cytochrome c oxidase” in ref. 33). In the original protocol, separation of CIII and CIV involves centrifugation for 45 min at 100,000 × g. Here, the centrifugation time was 1 h at 126,000 × g. In the original protocol, after elution of CIV from the hydroxyapatite column and pooling of fractions containing the enzyme, the buffer is exchanged on a Sepharose CL-6B column. Here, this step was excluded and instead the pH was adjusted to 8.5 by titration with a 0.5 M Tris solution. In the next step, Triton X-100 was removed using Bio-Beads (Bio-Rad) instead of dialysis: Bio-Beads were added to the CIV fraction at 0.3 g/mL and stirred for 3 h at 4 °C. The solution was then filtered to remove the Bio-Beads and the filtrate was centrifuged at 30,000 × g for 20 min at 4 °C. The pellet was resuspended in 2% DDM (39 mM), 20 mM Tris⋅HCl pH 8.5, 200 mM NaCl, and centrifuged at 2,000 × g for 20 min at 4 °C. The supernatant containing CIV was then collected.
A HiTrap Q ion exchange column (GE Healthcare) preequilibrated with buffer (50 mM 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES) pH 7.4 and 0.08% [wt/vol] GDN, 686 µM) was used to exchange CIV into GDN using an Äkta Pure M25 chromatography system (GE Healthcare) operated at 4 °C with UV detection at 280 nm and 420 nm. CIV was eluted with a gradient of increasing KCl (elution ∼400 mM KCl). Fractions containing CIV were concentrated with a 100-kDa molecular weight cutoff concentrator (Merck Millipore) and into 50 mM KCl, 50 mM HEPES pH 7.4 and 0.016% (wt/vol) GDN (136 µM) before being concentrated again. For cryo-EM samples, CIV was applied to a Superdex 200 Increase 10/300 GL size exclusion column to remove CIV aggregates. To make CHS solutions, 0.1% (wt/vol) CHS (2 mM) was mixed in 50 mM KCl, 50 mM HEPES pH 7.4, and 0.004% (wt/vol) LMNG (40 µM) or 50 mM KCl, 50 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES) pH 9.0, and 0.004% (wt/vol) LMNG (40 µM) and sonicated for 4 min at 60% of the maximum power (Vibra-Cell VCX 130, Sonics & Materials, Inc.) until it was clear, followed by centrifugation to remove large particles.
Activity Measurements.
The activity of CIV was measured by monitoring the oxygen-reduction rate with an Oxygraph Clark-type oxygen electrode (Hansatech) operated at 25 °C. A baseline oxygen concentration was first recorded with 50 μM oxidized cytochrome c from bovine heart, 10 mM ascorbate, 100 μM TMPD in 50 mM KCl, 50 mM Hepes pH 7.4, and either 0.016% (wt/vol) GDN (136 µM) or 0.05% (wt/vol) DDM (1 mM). For the CHS titration, 0.004% (wt/vol) LMNG (40 µM) was used in the buffer instead of GDN or DDM with CHS concentrations in the range of 165 μM to 990 μM, and CIV was incubated in the buffer containing CHS prior to measurement. The reaction was initiated by addition of 15 nM CIV. The CHS titration was fit with an empirical four-parameter Hill equation, using SigmaPlot (Alfasoft).
Preparation of the Two-Electron Reduced State and Flash Photolysis.
CIV was diluted to 5 μM in 50 mM KCl, 50 mM CHES pH 9.0, and either 0.016% (wt/vol) GDN (136 µM) or 0.004% (wt/vol) LMNG (40 µM) with or without 0.06% (wt/vol) CHS (1.2 mM). The samples were then transferred to a modified anaerobic Thunberg cuvette. The atmosphere was exchanged consecutively for N2 and CO on a vacuum line, after which the sample was incubated for 1 to 2 h. During that time, CO donates two electrons to each CIV, which results in formation of the “mixed-valence state” in which the catalytic site heme a3/CuB is reduced while heme a and CuA remain oxidized. Another CO molecule then binds to the reduced heme a3, resulting in formation of the CIV-CO complex (as monitored spectrophotometrically) (59): 2CO + H2O + CuB2+Fea33+ → CO2 + 2H+ + CuB+ Fea32+-CO. The CO ligand was dissociated by a 10-ns laser flash (Nd-YAG laser at 532 nm, Quantel) and absorbance changes were monitored using a flash photolysis setup (Applied Photophysics). The CO dissociation and recombination, and the following electron transfer from heme a3 to heme a, were monitored at 445 nm. The proton-coupled electron transfer at a millisecond time scale was monitored at 598 nm, which is an isosbestic point for the CO-recombination reaction. The 598-nm traces were normalized to the absorbance changes associated with CO dissociation, measured at 445 nm.
Preparation of the Four-Electron Reduced CIV and Flow Flash.
CIV was diluted to 5 μM in 50 mM KCl, 50 mM HEPES pH 8.0, and either 0.016% (wt/vol) GDN (136 µM) or 0.05% (wt/vol) DDM (1 mM). The samples were transferred to Thunberg cuvettes, and the atmosphere was exchanged to nitrogen before reduction by 2 mM ascorbate and 1 μM of the electron mediator phenazine methosulfate (PMS). After the samples were fully reduced, nitrogen was exchanged for CO. The flow-flash measurements were performed using a locally modified laser flash-photolysis system combined with a stopped-flow apparatus (39). Briefly, the reduced CO-bound enzyme was mixed at a ratio of 1:1 with an oxygen-saturated solution containing 50 mM KCl, 50 mM HEPES pH 8.0, and either 0.016% (wt/vol) GDN (136 µM) or 0.05% (wt/vol) DDM (1 mM). The CO ligand was dissociated by a short laser flash (Nd-YAG laser at 532 nm, Quantel) at 0.2 s after mixing, which initiated the reaction. The reaction with oxygen was monitored at 445 nm.
Grid Preparation and Cryo-Electron Microscopy.
Purified CIV (35 µL) at a concentration of ∼8 mg/mL, was bubbled with carbon monoxide (Praxair) in a sealed container for ∼10 min to ensure an oxygen-free environment. The sample was reduced by adding 3.5 µL of a mixture of 1.4 mM TMPD (Millipore Sigma) and 112 mM ascorbate (Chem-Impex International) then bubbled with carbon monoxide for another ∼10 min. The reduced CO-bound CIV was applied to homemade nanofabricated holey gold grids (60, 61) that had been glow discharged in air (120 s, 20 mA using PELCO easiGlow). Cryo-EM specimen preparation was performed in a low-light environment to prevent dissociation of carbon monoxide. Grids were blotted for 28 s at 4 °C and 100% humidity before rapid freezing in liquid ethane-propane mixture with a Vitrobot Mark III (Thermo Fisher Scientific). Cryo-EM data were collected with a Titan Krios G3 electron microscope (Thermo Fisher Scientific) operated at 300 kV equipped with a prototype Falcon 4 direct detector device camera. Automated data collection was done with the EPU software package. An initial dataset of 4,222 movies, each consisting of 30 exposure fractions, was collected at a nominal magnification of 75,000×, corresponding to a calibrated pixel size of 1.03 Å. The camera exposure rate and the total exposure of the specimen were ∼4.7 e−/pixel/s and ∼42.7 e−/Å2, respectively (SI Appendix, Table S1).
Cryo-EM Image Processing.
All image analysis was performed with cryoSPARC v2 (62). Movies were aligned with MotionCor2 (63) and contrast transfer function (CTF) parameters were estimated in patches with a 7 × 7 grid. The dataset was manually curated to remove movies with devitrification, large cracks in the ice, or poor CTF fit parameters, reducing the dataset size to 4,161 movies. Templates for particle selection were generated by two-dimensional classification of manually selected particle images leading to 911,268 selected particles. After particle selection, images were corrected for local motion (64) and extracted in 256 × 256 pixel boxes (SI Appendix, Table S1). Extracted particle images were cleaned with four rounds of ab initio 3D classification and heterogeneous refinement, taking only the classes that corresponded to CIV after each round. This procedure further reduced the size of the dataset to 112,880 particle images, which were subject to nonuniform refinement to produce the final map. Chimera and Coot were used to perform a ridged body fit of the CIV molecular model from a previous crystal structure (PDB ID:1V54) (48) and GDN into the final map. A validation report for the rigid body fit was produced with Phenix (SI Appendix, Table S1).
Classical Molecular Dynamics Simulations.
Classical atomistic MD simulations were performed on the 13 subunit bovine CIV embedded in a lipid membrane comprising a 2:2:1 mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and cardiolipin constructed using CHARMM-GUI (65), The system was solvated in TIP3P water molecules, and 150 mM NaCl was added to neutralize the system. Water molecules in the D pathway were modeled based on a prior X-ray structure (PDB ID:1V54). The total system comprised ∼250,000 atoms. To probe the effect of the sterol on the K pathway dynamics, CIV was modeled either without a bound steroid, with an entire GDN molecule comprising the steroid moiety and the hydrophilic maltose moiety, or with cholesterol. The catalytic site was modeled in the OH state (FeIII-OH−/CuII-H2O Y244-O−), with heme a reduced or in the OH,R state (FeIII-OH−/CuI-H2O Y244-O−) with heme a oxidized. The protein, membrane, and water molecules were modeled using the CHARMM36 force field, whereas the redox cofactors were modeled using in-house parameters based on density functional theory calculations (66), with force field parameters (67) derived also for GDN. MD simulations were performed at T = 310 K and P = 1 atm with a 2 fs timestep, and treating long-range electrostatics with the particle mesh Ewald approach. All MD simulations were performed with NAMD2.14 and NAMD3.0 (68). See SI Appendix, Table S2 for the list of MD simulations. To probe the effect of the N-terminal tail of subunit VIa that occupies the same position as the steroid ring moiety in the dimer, we constructed a model of the CIV dimer based on the X-ray crystal structure (PDB ID: 1V54). The dimer model was built and simulated in a manner analogous to the monomer system with a total system size of ∼ 460,000 atoms.
QM/MM Calculations.
The proton transfer energetics along the K pathway were studied with hybrid quantum/classical (QM/MM) simulations. A structure extracted from MD simulation S5 (without bound GDN) was selected as a starting point for the QM/MM models. The QM region contained Lys319, Thr316, Tyr244, His240, His290, and His291 of subunit I, CuB, and six water molecules that had diffused between Lys319 and Tyr244/CuB during the MD simulations. Link atoms were introduced between Cβ and Cα atoms. The QM region, comprising ∼100 atoms, was modeled at the B3LYP-D3/def2-SVP level of theory (with def2-TZVP for Cu), and the surrounding MM region was described at the CHARMM36 level or with our in-house force field for the cofactors (66). During the initial QM/MM geometry optimization, the MM atoms within 10 Å from the QM region were allowed to relax, while the surrounding protein environment was kept fixed. The total QM/MM system was trimmed to include 50,000 atoms around the K pathway region. Subsequent reaction coordinate optimizations were performed by allowing the MM hydrogen atoms to relax during minimization. The reaction coordinate was described as a linear combination of bond breaking and bond forming distances along the proton wire from Lys319 to Tyr244 (SI Appendix, Fig. S4C). The reaction coordinate was optimized along subsequent forward and backward cycles until convergence was reached. All QM/MM calculations were performed with CHARMM c38b1 (69), TURBOMOLE v. 7.5 (70), coupled together with a Python interface (71).
Supplementary Material
Acknowledgments
This work was supported by the Knut and Alice Wallenberg Foundation via Grants 2019.0043 (P.B. and V.R.I.K) and 2019.0251 (V.R.I.K), the Swedish Research Council Grant 2018-04619 (P.B.), and the Canadian Institutes of Health Research Grant PJT162186 (J.L.R.). This work was also supported by the Swedish National Infrastructure for Computing (2021/1-40) at Center for High-Performance Computing, partially funded by the Swedish Research Council through Grant Agreement 016-07213. J.D.T. was supported by a Canadian Institutes of Health Research postdoctoral fellowship and J.L.R. was supported by the Canada Research Chairs program. Titan Krios cryo-EM data were collected at the Toronto High-Resolution High-Throughput Cryo-EM facility supported by the Canada Foundation for Innovation and Ontario Research Fund. Molecular graphics and analyses were performed with University of California, San Francisco (UCSF) Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at UCSF, with support from NIH P41-GM103311.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205228119/-/DCSupplemental.
Data Availability
The electron cryomicroscopy map and associated model described in this article have been deposited in the Electron Microscopy Data Bank (EMDB) (accession no. EMD-27196) (72) and the Protein Data Bank (PDB ID: 8D4T) (73).
References
- 1.Rich P. R., Mitochondrial cytochrome c oxidase: Catalysis, coupling and controversies. Biochem. Soc. Trans. 45, 813–829 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Wikström M., Sharma V., Kaila V. R. I., Hosler J. P., Hummer G., New perspectives on proton pumping in cellular respiration. Chem. Rev. 115, 2196–2221 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Konstantinov A. A., Siletsky S., Mitchell D., Kaulen A., Gennis R. B., The roles of the two proton input channels in cytochrome c oxidase from Rhodobacter sphaeroides probed by the effects of site-directed mutations on time-resolved electrogenic intraprotein proton transfer. Proc. Natl. Acad. Sci. U.S.A. 94, 9085–9090 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brzezinski P., Ädelroth P., Pathways of proton transfer in cytochrome c oxidase. J. Bioenerg. Biomembr. 30, 99–107 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Hosler J. P., et al. , Polar residues in helix VIII of subunit I of cytochrome c oxidase influence the activity and the structure of the active site. Biochemistry 35, 10776–10783 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Fetter J. R., et al. , Possible proton relay pathways in cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 92, 1604–1608 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adelroth P., Gennis R. B., Brzezinski P., Role of the pathway through K(I-362) in proton transfer in cytochrome c oxidase from R. sphaeroides. Biochemistry 37, 2470–2476 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Yoshikawa S., Shimada A., Reaction mechanism of cytochrome c oxidase. Chem. Rev. 115, 1936–1989 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Hosler J. P., Ferguson-Miller S., Mills D. A., Energy transduction: Proton transfer through the respiratory complexes. Annu. Rev. Biochem. 75, 165–187 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson-Miller S., Hiser C., Liu J., Gating and regulation of the cytochrome c oxidase proton pump. Biochim. Biophys. Acta 1817, 489–494 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brzezinski P., Moe A., Ädelroth P., Structure and mechanism of respiratory III-IV supercomplexes in bioenergetic membranes. Chem. Rev. 121, 9644–9673 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukihara T., et al. , The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272, 1136–1144 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Maréchal A., et al. , A common coupling mechanism for A-type heme-copper oxidases from bacteria to mitochondria. Proc. Natl. Acad. Sci. U.S.A. 117, 9349–9355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H. M., et al. , Mutations in the putative H-channel in the cytochrome c oxidase from Rhodobacter sphaeroides show that this channel is not important for proton conduction but reveal modulation of the properties of heme a. Biochemistry 39, 2989–2996 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Arnold S., The power of life—Cytochrome c oxidase takes center stage in metabolic control, cell signalling and survival. Mitochondrion 12, 46–56 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Hüttemann M., Lee I., Grossman L. I., Doan J. W., Sanderson T. H., Phosphorylation of mammalian cytochrome c and cytochrome c oxidase in the regulation of cell destiny: Respiration, apoptosis, and human disease. Adv. Exp. Med. Biol. 748, 237–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hüttemann M., Kadenbach B., Grossman L. I., Mammalian subunit IV isoforms of cytochrome c oxidase. Gene 267, 111–123 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Qin L., Hiser C., Mulichak A., Garavito R. M., Ferguson-Miller S., Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 103, 16117–16122 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musatov A., Robinson N. C., Bound cardiolipin is essential for cytochrome c oxidase proton translocation. Biochimie 18, 1199–1216 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson N. C., Zborowski J., Talbert L. H., Cardiolipin-depleted bovine heart cytochrome c oxidase: Binding stoichiometry and affinity for cardiolipin derivatives. Biochemistry 29, 8962–8969 (1990). [DOI] [PubMed] [Google Scholar]
- 21.Hiser C., Buhrow L., Liu J., Kuhn L., Ferguson-Miller S., A conserved amphipathic ligand binding region influences k-path-dependent activity of cytochrome C oxidase. Biochemistry 52, 1385–1396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Buuren K. J. H., Van Gelder B. F., Biochemical and biophysical studies on cytochrome c oxidase. XIII. Effect of cholate on the enzymic activity. Biochim. Biophys. Acta 333, 209–217 (1974). [DOI] [PubMed] [Google Scholar]
- 23.Oleynikov I., Azarkina N., Vygodina T., Direct interaction of mitochondrial cytochrome c oxidase with thyroid hormones: Evidence for two binding sites. Cells 11, 908 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiser C., Liu J., Ferguson-Miller S., The K-path entrance in cytochrome c oxidase is defined by mutation of E101 and controlled by an adjacent ligand binding domain. Biochim. Biophys. Acta Bioenerg. 1859, 725–733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashton T. M., McKenna W. G., Kunz-Schughart L. A., Higgins G. S., Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. 24, 2482–2490 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Simon N., et al. , Glucocorticoids decrease cytochrome c oxidase activity of isolated rat kidney mitochondria. FEBS Lett. 435, 25–28 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Vaz A. R., et al. , Bilirubin selectively inhibits cytochrome c oxidase activity and induces apoptosis in immature cortical neurons: Assessment of the protective effects of glycoursodeoxycholic acid. J. Neurochem. 112, 56–65 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Chae P. S., et al. , A new class of amphiphiles bearing rigid hydrophobic groups for solubilization and stabilization of membrane proteins. Chemistry 18, 9485–9490 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimada S., et al. , Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode. EMBO J. 36, 291–300 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yano N., et al. , X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state at 2.0 Å resolution. Acta Crystallogr. F Struct. Biol. Commun. 71, 726–730 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinzawa-Itoh K., et al. , Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 26, 1713–1725 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin L., Mills D. A., Buhrow L., Hiser C., Ferguson-Miller S., A conserved steroid binding site in cytochrome C oxidase. Biochemistry 47, 9931–9933 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandt U., Schägger H., von Jagow G., Purification of cytochrome-c oxidase retaining its pulsed form. Eur. J. Biochem. 182, 705–711 (1989). [DOI] [PubMed] [Google Scholar]
- 34.Hiser C., Liu J., Ferguson-Miller S., The K-path entrance in cytochrome c oxidase is defined by mutation of E101 and controlled by an adjacent ligand binding domain. Biochim. Biophys. Acta Bioenerg. 1859, 725–733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ilgü H., et al. , Variation of the detergent-binding capacity and phospholipid content of membrane proteins when purified in different detergents. Biophys. J. 106, 1660–1670 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J., Hiser C., Ferguson-Miller S., Role of conformational change and K-path ligands in controlling cytochrome c oxidase activity. Biochem. Soc. Trans. 45, 1087–1095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ädelroth P., Ek M. S., Mitchell D. M., Gennis R. B., Brzezinski P., Glutamate 286 in cytochrome aa3 from Rhodobacter sphaeroides is involved in proton uptake during the reaction of the fully-reduced enzyme with dioxygen. Biochemistry 36, 13824–13829 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Ädelroth P., Brzezinski P., Malmström B. G., Internal electron transfer in cytochrome c oxidase from Rhodobacter sphaeroides. Biochemistry 34, 2844–2849 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Brändén M., et al. , On the role of the K-proton transfer pathway in cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 98, 5013–5018 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallén S., Brzezinski P., Light-induced structural changes in cytochrome c oxidase: Implication for the mechanism of electron and proton gating. Biochim. Biophys. Acta 1184, 207–218 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Zong S., et al. , Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 28, 1026–1034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aoyama H., et al. , The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272, 1136–1144 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Punjani A., Fleet D. J., 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Liko I., et al. , Dimer interface of bovine cytochrome c oxidase is influenced by local posttranslational modifications and lipid binding. Proc. Natl. Acad. Sci. U.S.A. 113, 8230–8235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolli R., Nałecz K. A., Azzi A., The interconversion between monomeric and dimeric bovine heart cytochrome c oxidase. Biochimie 67, 119–128 (1985). [DOI] [PubMed] [Google Scholar]
- 46.Shinzawa-Itoh K., et al. , Monomeric structure of an active form of bovine cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 116, 19945–19951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Musatov A., Robinson N. C., Cholate-induced dimerization of detergent- or phospholipid-solubilized bovine cytochrome C oxidase. Biochemistry 41, 4371–4376 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Tsukihara T., et al. , The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc. Natl. Acad. Sci. U.S.A. 100, 15304–15309 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svensson-Ek M., et al. , The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J. Mol. Biol. 321, 329–339 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Koepke J., et al. , High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: New insights into the active site and the proton transfer pathways. Biochim. Biophys. Acta 1787, 635–645 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Rich P. R., Maréchal A., Functions of the hydrophilic channels in protonmotive cytochrome c oxidase. J. R. Soc. Interface 10, 20130183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wikström M., Krab K., Sharma V., Oxygen activation and energy conservation by cytochrome c oxidase. Chem. Rev. 118, 2469–2490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Supekar S., Kaila V. R. I., Dewetting transitions coupled to K-channel activation in cytochrome c oxidase. Chem. Sci. (Camb.) 9, 6703–6710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woelke A. L., Wagner A., Galstyan G., Meyer T., Knapp E. W., Proton transfer in the K-channel analog of B-type cytochrome c oxidase from Thermus thermophilus. Biophys. J. 107, 2177–2184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hofacker I., Schulten K., Oxygen and proton pathways in cytochrome c oxidase. Proteins 30, 100–107 (1998). [PubMed] [Google Scholar]
- 56.Lepp H., Svahn E., Faxén K., Brzezinski P., Charge transfer in the K proton pathway linked to electron transfer to the catalytic site in cytochrome c oxidase. Biochemistry 47, 4929–4935 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Woelke A. L., Galstyan G., Knapp E. W., Lysine 362 in cytochrome c oxidase regulates opening of the K-channel via changes in pKA and conformation. Biochim. Biophys. Acta 1837, 1998–2003 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Kolbe F., et al. , Cryo-EM structures of intermediates suggest an alternative catalytic reaction cycle for cytochrome c oxidase. Nat. Commun. 12, 6903 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brzezinski P., Malmström B. G., The reduction of cytochrome c oxidase by carbon monoxide. FEBS Lett. 187, 111–114 (1985). [DOI] [PubMed] [Google Scholar]
- 60.Marr C. R., Benlekbir S., Rubinstein J. L., Fabrication of carbon films with ∼ 500nm holes for cryo-EM with a direct detector device. J. Struct. Biol. 185, 42–47 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Russo C. J., Passmore L. A., Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rubinstein J. L., Brubaker M. A., Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 192, 188–195 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Johansson M. P., Kaila V. R. I., Laakkonen L., Charge parameterization of the metal centers in cytochrome c oxidase. J. Comput. Chem. 29, 753–767 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Best R. B., et al. , Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Phillips J. C., et al. , Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 153, 044130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brooks B. R., et al. , CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Balasubramani S. G., et al. , TURBOMOLE: Modular program suite for ab initio quantum-chemical and condensed-matter simulations. J. Chem. Phys. 152, 184107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riahi S., Rowley C. N., The CHARMM-TURBOMOLE interface for efficient and accurate QM/MM molecular dynamics, free energies, and excited state properties. J. Comput. Chem. 35, 2076–2086 (2014). [DOI] [PubMed] [Google Scholar]
- 72.J. M. Di Trani et al., Structural basis of mammalian complex IV inhibition by steroids. EMD-27196. https://www.ebi.ac.uk/emdb/EMD-27196. Deposited 2 June 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.J. M. Di Trani et al., Structural basis of mammalian complex IV inhibition by steroids. PDB ID: 8D4T. https://www.rcsb.org/structure/8D4T. Deposited 2 June 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The electron cryomicroscopy map and associated model described in this article have been deposited in the Electron Microscopy Data Bank (EMDB) (accession no. EMD-27196) (72) and the Protein Data Bank (PDB ID: 8D4T) (73).