Abstract
Domesticated plants and animals played crucial roles as models for evolutionary change by means of natural selection and for establishing the rules of inheritance, originally proposed by Charles Darwin and Gregor Mendel, respectively. Here, we review progress that has been made during the last 35 y in unraveling the molecular genetic variation underlying the stunning phenotypic diversity in crops and domesticated animals that inspired Mendel and Darwin. We notice that numerous domestication genes, crucial for the domestication process, have been identified in plants, whereas animal domestication appears to have a polygenic background with no obvious “domestication genes” involved. Although model organisms, such as Drosophila and Arabidopsis, have replaced domesticated species as models for basic research, the latter are still outstanding models for evolutionary research because phenotypic change in these species represents an evolutionary process over thousands of years. A consequence of this is that some alleles contributing to phenotypic diversity have evolved by accumulating multiple changes in the same gene. The continued molecular characterization of crops and farm animals with ever sharper tools is essential for future food security.
Keywords: Mendel, crops, domestic animals, genetics, domestication
Domestication is a coevolutionary process that arises from a mutualistic interaction, most prominently between humans, crop plants, and domesticated animals (1). Domestication began ∼30,000 y ago with the dog, Canis familiaris (2), and occurred in earnest during the Holocene ∼11,000 y ago (3), when several domesticated species and populations evolved to provide humans with food, material, and various services. As the evolution of domesticated plant and animal species proceeded, it led not only to genetic differentiation between the domesticated species and their wild ancestors (i.e., domestication) but also, to increased phenotypic variation within species as new traits appeared and were selected (i.e., diversification). This selection pressure was primarily related to adapting the species to the farm environment and increasing productivity, but it is clear that there has also been selection for visual phenotypic diversity in morphology and color (4). As a result of these different types of selection pressures, domesticated plants and animals constituted a rich source of phenotypic variation that inspired both Gregor Mendel and Charles Darwin.
In 1859, Charles Darwin (5) published his seminal book On the Origins of Species by Means of Natural Selection or the Preservation of Favoured Races in the Struggle for Life. In the introduction, he noted how the study of domesticated plants and animals provides “the best chance of making out this obscure problem”—that of the mechanisms of evolutionary change (5). He devoted the first chapter of the book to the variation of domesticated species and followed this up 9 y later with the two-volume exposition The Variation of Animals and Plants under Domestication (6). In fact, Darwin developed his theory of evolution by natural selection long before the publication of On the Origins of Species by Means of Natural Selection or the Preservation of Favoured Races in the Struggle for Life and spent decades of research to gather support for his theory, an important component of which was the study of domesticated plants and animals, including his breeding experiments with pigeons.
The rediscovery of Mendel’s work at the dawn of the twentieth century led to a proliferation of studies as biologists sought to advance genetic investigation across a number of different species. Just as Mendel had done, these initial studies relied largely on domesticated varieties. Early geneticists, such as Hugo de Vries, Carl Correns, and William Bateson, continued work using domesticated plants, not only in peas but also, in maize and snapdragons. In maize, studies focused on various kernel characteristics, including sugar/starch variation and color, propelling this crop species as a major model system for plant genetic studies (7). In the first studies on Mendelian inheritance in animals in 1902, Bateson (8) and Bateson and Saunders (9) reported to the Royal Society in England on the inheritance of five traits in chickens (Gallus gallus): rose comb, pea comb, polydactyly, yellow skin, and dominant white color. A few years later, Bateson and Punnett (10) described one of the first examples of epistatic interaction between loci, namely that the walnut comb is caused by the combined effect of Rose-comb and Pea-comb.
The study of the genetics of domesticated plants and animals continued throughout the last century, when the foundations for modern genetics were established. A major breakthrough was the advent of molecular-based genetic mapping and cloning techniques during the 1980s. It then became possible to isolate and study genes underlying phenotypic diversity—probing the molecular details of the variation in domesticated plants and animals that fascinated Darwin and Mendel.
Molecular Genetic Variation of Plants under Domestication
Numerous plant loci have been isolated and linked to specific crop domestication or diversification phenotypes, uncovering trends on the nature of genetic variation. In this context, domestication genes are those that lead to phenotypic differentiation between the crop and its wild ancestor, while diversification genes are those that result in phenotypic variation within the crop species, possibly between varieties (11).
Differences within crop species were the focus of genetic experimentation in the early twentieth century after the rediscovery of Mendel’s work. Maize was particularly interesting, and Mendel himself noted in an 1867 letter to the botanist Carl Nageli that he had done experiments with maize, although he did not report on the results (12). De Vries and Correns, who rediscovered Mendel’s laws in 1900, also worked on maize and studied the naturally variable starchy/sugary phenotype of kernels (7, 13). Soon after, R. A. Emerson and E. M. East developed maize as one of the first plant genetic model systems (7), and geneticists started focusing on various naturally occurring phenotypes found in this species, including different kernel properties, pericarp and aleurone color, and pod corn (Fig. 1). The study of maize genetics allowed for the assignment of genetic linkage groups to specific chromosomes (7, 13), showed the relationship between genetic and chromosomal crossing over (14), led to the discovery of the first genetically characterized transposable elements (TEs) (15), and provided early evidence that quantitative traits could be explained by numerous Mendelian genes (16).
Fig. 1.
Some key Mendelian traits in domesticated plants. (A) Early genetic work in plants focused on maize, including various kernel traits, such as aleurone and pericarp color. Image credit: David Spender, United Kingdom (CC Commons Attribution 2.0 Generic license). (B) Glutinous phenotype caused by mutations at the Wx gene leads to sticky rice valued by cultures in Northeast and Southeast Asia. (C) The round vs. (D) wrinkled pea phenotype maps to the r allele first described by Mendel, with the latter caused by a 0.8-kb TE insertion into a starch branching enzyme gene. Image credit: Claire Domoney (John Innes Center, Norwich, United Kingdom). (E) Tall vs. dwarf pea plants are controlled by Mendel’s Le gene. Image credit: Julie Hofer (John Innes Center, Norwich, United Kingdom).
The molecular isolation of genes associated with maize diversification traits began in the 1980s. The first isolated gene linked to a specific phenotype was the Waxy (Wx) locus (17), which encodes a starch biosynthetic enzyme responsible for amylose formation and whose mutation resulted in a distinctive kernel phenotype. It was shown that numerous naturally occurring alleles of this gene contained deletions and retrotransposon insertions (18–20). Interestingly, the isolation of Wx in maize led to the identification of the homologous gene in rice, Oryza sativa (21), where selection for a naturally occurring splice donor mutation in the gene’s first intron leads to the sticky or glutinous rice phenotypes prized by several East and Southeast Asian cultures (22) (Fig. 1).
Other genes associated with distinct maize varieties were similarly isolated at the molecular level. For example, the sugary1 locus, first studied by Correns (23), is responsible for lower starch and greater sugar content in kernels; mutations at this locus result in kernels that appear glassy, translucent, and partially wrinkled when dried. The gene was cloned in 1995 and shown to encode a starch debranching enzyme that hydrolyzes α-(l-6) glucosyl linkages (24).
Genes associated with kernel color were also identified, including the Y1 locus responsible for the yellow/white kernel polymorphism, which was shown to encode a phytoene synthase enzyme as part of the carotenoid biosynthetic pathway (25). Interestingly, the wild teosinte ancestor of maize only possesses white kernels, and the yellow kernels in some varieties of the domesticate suggest an expansion of Y1 gene expression into the seed endosperm (25). Moreover, population genetic analysis indicates a selective sweep of ∼0.85 to 1 Mb surrounding the gene in yellow kernel maize lines (26). Another seed color gene, the C1 locus responsible for red pigmentation in some traditional maize varieties, was shown to encode an myb-like transcription factor (27) (Fig. 1).
Soon, genes that displayed a clear relationship of molecular genotype to crop phenotypes were also isolated in other domesticated species, and today, hundreds of such loci across more than 20 crop species have been studied. Among the most notable ones are those that led to shorter crop plants and were the basis for the Green Revolution in agriculture 60 y ago. The reduced height (Rht) gene in wheat (Triticum aestivum) has been shown through molecular genetic analysis to encode a DELLA-containing member of the GRAS family of transcription activators that regulates the gibberellic acid (GA) hormone biosynthetic pathway (28). The allele that led to Green Revolution wheat varieties contains a base substitution that results in a premature stop codon in the gene. Interestingly, the semidwarfing1 (sd1) gene used to develop the rice Green Revolution variety IR8 encodes a GA 20-oxidase enzyme (29). The sd1 allele first identified in the traditional Chinese cultivar Dee-geo-woo-gen and subsequently introduced into IR8 contains a 383-bp deletion that also leads to a premature stop codon (29).
Molecular Genetics of Mendel’s Traits in Peas.
Coming full circle, four of the seven loci that Mendel studied in peas have now been identified at the molecular level (Fig. 1 and SI Appendix, Table S1). Mendel’s experiments used green pea (Pisum sativum) varieties and focused on genes for seed shape (R; round vs. wrinkled), stem length (Le; tall vs. dwarf), cotyledon color (I; green vs. yellow), seed coat/flower color (A; purple vs. white), pod color (green vs. yellow), pod form (inflated vs. constricted), and flower position (axial vs. terminal) (30). The seed shape gene was the first to be cloned in 1990, when it was shown that the R locus cosegregated with the starch-branching enzyme gene SBE1. Molecular analysis showed that the wrinkled r allele contained a 0.8-kb insertion of a TE in the Ac/Ds family and that the loss of function resulted in metabolic changes in seed metabolism (31).
Three others of Mendel’s pea genes were subsequently isolated. The Le gene for plant height was shown to encode a GA 3β-hydroxylase, and the dwarf allele has an alanine to threonine mutation near the enzyme’s active site (32, 33). The I gene for green cotyledons was demonstrated to be homologous to the rice Stay-green gene that appears to be involved in chlorophyll catabolism. The recessive i allele in pea contains a 6-bp insertion that leads to the addition of two amino acids in the encoded protein and prevents degradation of chlorophyll b (34, 35). Finally, the A gene for purple vs. white seed coat, which is also associated with purple/white flowers, encodes a basic helix–loop–helix transcription factor that regulates anthocyanin biosynthesis (36). Two naturally occurring white (a) alleles are known, one of which was thought to be widespread in European pea varieties in the nineteenth century; this allele had a G to A splice donor mutation in intron 6 of the gene, which results in the use of a cryptic splice donor site 8 bp downstream and produces a transcript with a premature stop codon (36).
Molecular Genetic Variation of Animals under Domestication
In the early part of the twentieth century, the mode of inheritance for many phenotypes in domestic animals was established; over the last 35 y, a long list of disorders and traits has been characterized at the molecular level (Online Mendelian Inheritance in Animals; https://omia.org/home/) (37). The first molecular genetic characterization of a Mendelian phenotype in a domesticated animal concerned hereditary goiter caused by a nonsense mutation in the thyroglobulin gene, a gene identification guided by a similar disease association in humans (38). An early discovery of major economic importance in livestock was the identification of a missense mutation Arg615Cys in ryanodine receptor 1 causing malignant hyperthermia in pigs (39). This mutation had increased to high frequency in several pig breeds intensively selected for lean meat because the mutation was associated with lean muscle growth, most likely because the calcium channel encoded by this gene affects muscle contractions by controlling calcium flow. However, in the homozygous condition, the mutation is predisposing to malignant hyperthermia that may be induced by exposure to stress: for instance, during transport. This mutation was, therefore, a major economic problem in the pig industry because of an increasing incidence of malignant hyperthermia as the frequency of the mutation increased in pig populations, which was solved by widespread diagnostic testing for the causal mutation. This important finding in pigs led to the discovery that exactly the same missense mutation predisposes to malignant hyperthermia in humans (40).
The First Mendelian Traits in Animals Reported by Bateson and Saunders ( 9 ) in 1902.
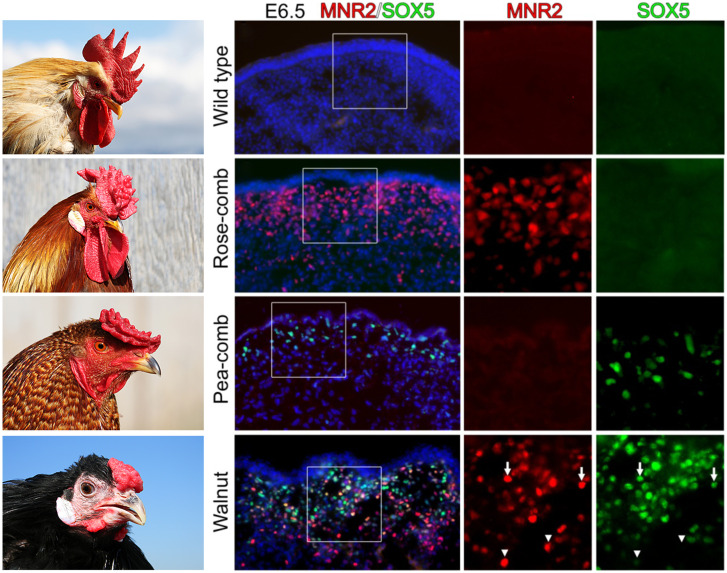
As molecular genetic analyses of domesticated animals progressed, the mutations causing the five phenotypes first studied by Bateson (8) were characterized at the molecular level. The dominant mutations Pea-comb and Rose-comb (Fig. 2) are both caused by structural changes affecting cis-acting regulatory elements controlling expression of the transcription factors encoded by SOX5 (41) and MNR2 (42), respectively. Pea-comb is due to a copy number expansion in a SOX5 intron, whereas Rose-comb is caused by a 7.4-Mb inversion that translocates the MNR2 gene to the vicinity of the proximal inversion break point. Immunohistochemistry revealed that these two mutations lead to ectopic expression of SOX5 and MNR2 in a layer of mesenchymal cells during a few days of development in the area where the comb develops (Fig. 2). Birds that carry both mutations show the walnut phenotype and accompanying ectopic expression of SOX5 and MNR2 in the same cells (Fig. 2), providing a molecular explanation for the epistatic interaction causing the walnut phenotype as described by Bateson and Punnett (10) more than 100 y ago. A third locus affecting comb morphology, Duplex comb, is also caused by a structural rearrangement leading to ectopic expression of an important transcription factor (43). In this case, a 20-kb tandem duplication located in an intron of CMC1 leads to ectopic expression of eomesdermin (EOMES) during comb development, although the duplication is located 200 kb upstream of EOMES.
Fig. 2.
Comb morphology in domesticated chickens and its molecular basis. Four comb phenotypes in chickens, wild type (or single comb), Rose-comb, Pea-comb, and Walnut-comb, and immunohistochemical labeling of MNR2 and SOX5 in comb tissue sections from embryonic day (E) 6.5. Nuclei are visualized by DAPI (4′,6-diamidino-2-phenylindole). Boxed regions are shown magnified as a single color. Arrows in the Walnut-comb tissue sections indicate double-labeled cells, whereas arrowheads indicate single-labeled cells. Reproduced from ref. 42, which is licensed under CC BY-NC-ND.
The three other phenotypes studied by Bateson, polydactyly, yellow skin, and dominant white, are caused by mutations in an intron of LMBR1 affecting the expression of sonic hedgehog in the posterior limb (44, 45), in the BCO2 gene (46), and in PMEL (47), respectively.
Gene Variants Underlying Phenotypic Variation in Domestic Animals.
Mutations with major effects on phenotypic variation in domestic animals are often noncoding, especially if an altered or disrupted coding sequence has pleiotropic effects. The allelic series in the microphthalmia-associated transcription factor (MITF) gene (48), which causes white spotting patterns in dogs (including the variant alleles Irish spotting, piebald, and extreme white) (49), is an illustrative example. MITF is a master regulator of gene expression in pigment cells as well as other cell types. MITF alleles in dogs have mutations in the noncoding regions, and these alleles are all fully viable, with the only negative pleiotropic effect being hearing loss in some dogs homozygous for the extreme white phenotype (e.g., some Dalmatians and white boxers; https://omia.org/OMIA000214/9615/). This is in contrast to mice, where more than 30 MITF alleles have been described (Mouse Genome Informatics [MGI]; www.informatics.jax.org), the majority of which disrupt the coding sequence and cause severe negative pleiotropic effects in homozygotes: defects in coat and eye pigmentation, microphthalmia, hearing loss, mast cell deficiency, bone resorption anomalies, and lethality. The importance of noncoding mutations is also illustrated by the comb phenotypes in chickens (Fig. 2), which are all caused by cis-acting regulatory mutations affecting expression of pleiotropic transcription factors (41–43).
Indeed, unlike the causal mutations for phenotypic diversity, Mendelian genetic disorders and disease in domestic animals are usually caused by mutations in coding sequences (https://omia.org/home/), similar to the situation in humans (https://omim.org). Changes in coding sequences contributing to phenotypic diversity in domesticated animals are more common in genes with tissue-specific expression, which limits their pleiotropic effects. The best examples are those contributing to pigmentation variation. Mutations in the genes encoding melanocortin receptor-1 (MC1R) and its antagonist agouti-signaling protein occur in almost all domestic species, including complete loss-of-function mutations without obvious negative pleiotropic effects (MC1R: https://omia.org/gene427562/;ASIP and https://omia.org/gene492296/)
Numerous structural changes (duplications, deletions, inversions, and complex rearrangements) contributing to phenotypic diversity in domestic animals have been characterized (SI Appendix, Table S2), some of which may alter promoter–enhancer interactions and act as regulatory mutations. The three comb phenotypes in chicken are striking examples in which the consequences of structural changes have been characterized in detail by immunohistochemistry (Fig. 2). Another example is the 4.5-kb duplication in an intron of syntaxin 17 underlying the iconic greying with age phenotype in horses, common in Lippizaners and Arabians, that leads to premature hair greying and predisposing to melanoma (50). Functional studies revealed that the duplication transforms a weak melanocyte-specific enhancer to a strong enhancer, and the duplicated sequence contains two MITF binding sites critical for up-regulated expression (51). The majority of domestic horses show the nondun phenotype characterized by more intense pigmentation than the wild-type dun phenotype, most likely because intense pigmentation is considered more attractive. The most common nondun allele has a 1.6-kb deletion of a region containing an enhancer controlling melanocyte-specific expression of the TBX3 transcription factor (52). Interestingly, the horse reference genome assembly is missing this sequence because the reference horse is homozygous for the deletion.
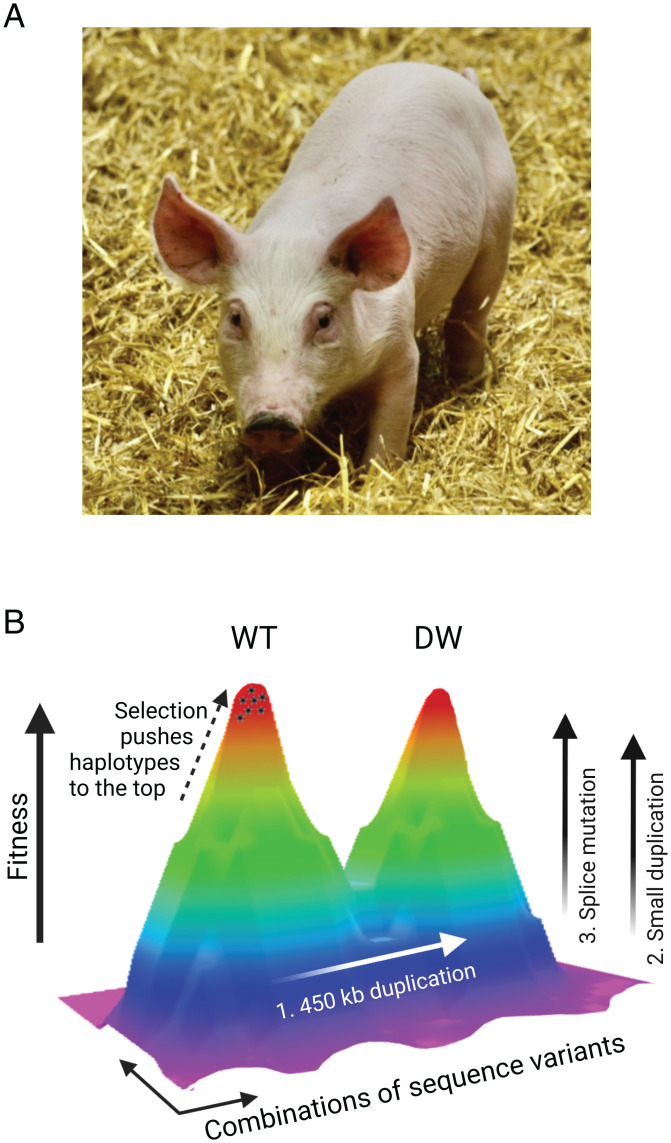
Mendel and Darwin used domesticated plants and animals as models to study inheritance and evolution. Since then, model organisms, like yeast, Caenorhabditis elegans, Drosophila, zebrafish, zebra finch, Arabidopsis, and mouse, have taken over as model organisms for basic biological research. Nevertheless, domesticated species are still excellent models for evolutionary research as they have gone through an evolutionary process over thousands of years while adapting to farm and/or home environments. One interesting consequence is the evolution of alleles carrying multiple functionally important mutations affecting the same gene (53). The first reported example of this is the evolution of dominant white color in domestic pigs (Fig. 3A). The majority of pigs used for meat production in the western world are white, a consequence of a strong tradition in consuming pig meat lacking skin pigmentation. The difference between the wild-type and the Dominant white allele is at least three consecutive mutations affecting KIT (52–54), which encodes a tyrosine kinase receptor of critical importance for migration of embryonic stem cell populations, including melanocyte precursor cells (MGI; www.informatics.jax.org).
Fig. 3.
Evolution of the Dominant white (DW) allele at the KIT locus in pigs. (A) Large white piglet with the DW phenotype. Image credit: Per Jensen (Linköping University, Linköping, Sweden). (B) Illustration of the evolution of the DW allele from the wild-type (WT) allele involving three steps: 1) a 450-kb duplication encompassing the entire KIT gene and flanking regions, 2) one or more smaller duplications of noncoding sequence (SI Appendix, Table S2), and 3) splice mutation resulting in exon skipping of exon 17 that encodes the tyrosine kinase domain in one of the two KIT copies.
The seed for this evolutionary process was the occurrence of a 450-kb duplication encompassing the entire KIT gene and part of flanking regions harboring long-range regulatory elements controlling KIT expression (Fig. 3B). Presence of the duplication on its own causes the patch phenotype (partial white spotting) (54). A splice mutation in one of the copies and at least one smaller duplication of flanking noncoding sequence (55, 56) were subsequently added, resulting in the Dominant white allele (Fig. 3B). The duplications most likely act as regulatory mutations altering KIT expression, while the splice mutation leads to skipping of the exon encoding the tyrosine kinase domain, resulting in a KIT receptor with normal ligand binding but no kinase activity. Such a splice mutation would be a recessive lethal in a wild-type mammal carrying a single KIT copy because this gene is crucial for normal hematopoiesis. In pigs, the splice mutation is fully viable because the second copy provides sufficient KIT function, and billions of pigs worldwide carry the Dominant white allele. This KIT allele in pigs has a stronger effect on pigmentation than any of the >90 Kit alleles described in mice (MGI; www.informatics.jax.org) and is still fully viable, whereas mouse mutations disrupting the coding sequences have negative pleiotropic effects on hematopoiesis and fertility. The Dominant white allele in pigs is not a single-hit mutant but a product of an evolutionary process with multiple gene alterations.
Another example of allelic evolution is in domesticated chickens, where the Rose1 allele has an inversion that translocates the MNR2 gene. This causes the comb phenotype, but one of the inversion break points disrupts the testis CCDC108 gene, causing reduced sperm motility in male homozygotes (42). A second allele, Rose2, arose by recombination between Rose1 and the wild-type allele, which restored the wild-type configuration but left an extra MNR2 copy in a translocated 91-kb fragment, a remnant of the inversion. This allele causes an unaltered Rose-comb phenotype but with normal male fertility as the CCDC108 gene is intact again. Other examples of the evolution of alleles in domestic animals are summarized in SI Appendix, Table S2. The evolutionary processes leading to the Dominant white allele in pigs and Rose-comb2 in chicken were seeded by the occurrence of a large duplication and an inversion, respectively. These two examples also illustrate another advantage of using domesticated species as models for evolutionary change, namely their recent history increasing the chance that the intermediate alleles, in these cases Patch in pig and Rose1 in chicken, are still present and their associated phenotypes are known. These are models for evolution of alleles and haplotypes with increasing fitness by means of natural selection, which are most likely of crucial importance for adaptability and evolvability as suggested for ecological adaptation in Atlantic herring (57) and for the adaptive radiation of Darwin’s finches (58). This model for evolutionary change suggests that it may not be possible to go from one adaptive haplotype/allele to another by a single mutation, as is the case for the Dominant white allele in pigs (Fig. 3B). The ongoing evolution of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mimics the evolution of adaptive haplotypes/alleles as it accumulates multiple mutations, increasing its ability to propagate in the human population.
Despite the successes in identifying Mendelian genes in domesticated animals, the majority of production traits in livestock have a polygenic background (59), and it has been generally challenging to identify causal mutations affecting polygenic traits. However, it is worth noticing that the first identification of a quantitative trait nucleotide (QTN) affecting a polygenic trait and residing in a noncoding sequence was not identified in fruit flies, mice, or humans but in pigs. Van Laere et al. (60) reported in 2003, well before there was a genome sequence in pigs, that a single base change in an intron of insulin-like growth factor 2 (IGF2) is the causal mutation for a major quantitative trait locus (QTL) affecting muscle growth and size of the heart. The mutation is in a highly conserved CpG island and leads to up-regulated expression of IGF2 messenger RNA (mRNA) in postnatal skeletal muscle and heart. This mutation has gone through a massive selective sweep, and most populations used for meat production are fixed for the mutation. Its identification was possible because the mutation arose on an Asian IGF2 haplotype still common in Asian pigs, and it is the only sequence difference between the Asian haplotype and the derived haplotype causing increased muscle growth (60). It was later demonstrated that a previously unknown transcription factor named ZBED6, which evolved from a domesticated DNA transposon, binds to the QTN region in pigs and most likely, the corresponding regulatory region in all placental mammals (61).
The access to data from whole-genome sequencing and high-density single-nucleotide polymorphism (SNP) panels combined with extensive pedigree records now provide powerful tools to explore genotype–phenotype relationships in domesticated animals. This makes it possible to identify even rare mutations causing serious disorders and establish diagnostic tests for an efficient elimination of such rare variants from breeding populations (62–64).
Genetics of Domestication
An important aspect of Darwin’s studies of domesticated plants and animals was how domesticated varieties differ from their wild ancestral forms (6). The tools of modern genetics have revealed the genetic basis for many of the striking differences between domesticated plants and animals and their wild ancestors.
Domestication of Plants.
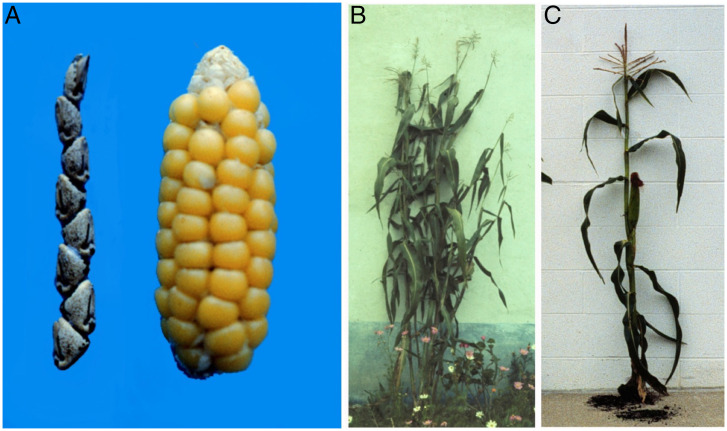
The most comprehensive analysis of genes underlying domestication is in maize, Zea mays ssp. mays (65), which is unsurprising given the advanced state of maize genetic analyses. The origin of maize had been a subject of speculation, but it became clear that teosinte, Z. mays ssp. parviglumis, is the ancestor of maize (66); domestication of ssp. mays from teosinte began ∼9,000 y ago, possibly in southwestern Mexico. The domestication of maize was accompanied by several substantial phenotypic changes in plant and inflorescence architecture, including increasing apical dominance, enlargement of seed-bearing ears, and kernel properties (65, 67) (Fig. 4).
Fig. 4.
Domestication traits in maize. (A, Left) An example of a Z. mays ssp. parviglumis (teosinte) ear vs. (A, Right) a domesticated Z. mays ssp. mays ear. Shoot architecture of (B) teosinte and (C) maize, which is controlled by the tb1 gene. Image credit: John Doebley (University of Wisconsin, Madison, WI).
The ability to cross maize and teosinte provided the basis for genetic studies of domestication in maize (68, 69). Attention was focused on the ear and whole-plant architecture as indicative of the transition from wild teosinte to maize. In the ears, four features differentiate wild vs. domesticated plants. In maize, the cupules and glumes are reduced to form the cob, the ears do not disarticulate upon maturity, there is only a single spikelet in each rachis segment, and the ears are four ranked or more (70). As to plant architecture, maize has short lateral branches with ears at the tip, while teosinte has long branches with tassels as well as secondary lateral branches.
Doebley et al. (69) provided the clearest genetic analysis of differences between maize vs. teosinte using QTL analysis. Their analysis indicated that QTLs for domestication could be found in all 10 maize chromosomes, although five to six genomic regions had markedly strong effects. They suggested that this may indicate that maize domestication was governed by just a few major genes (or linkage blocks of several genes) plus a greater number of loci with smaller effects (69).
With this QTL analysis, it became possible to identify several of the key maize domestication genes. The first gene to be isolated at the molecular level was teosinte-branched1 (tb1), which was first identified as a maize mutant that reduced apical dominance and transformed the maize plant to a teosinte architecture (70), and it was in a domestication QTL region on chromosome 1. Molecular isolation showed that tb1 encoded a class II TCP transcription factor that regulated apical dominance and bud dormancy in maize. It was shown that the maize tb1 allele contained a Hopscotch retrotransposon ∼60 kb upstream of the coding sequence, which appears to enhance tb1 expression in the domesticated plant (71, 72). In support of this, chromosomal contact data indicate physical interaction between this retrotransposon and the tb1 coding region via chromosome looping (73).
Another major Mendelian locus associated with a domestication trait, the formation of the naked maize kernels, was mapped near the centromere of the short arm of chromosome 4 (69). This locus was designated as teosinte glume architecture1 (tga1), and developmental analysis shows that the teosinte allele makes the glumes longer and thicker (74). Molecular analysis indicates that tga1 encodes a member of the SBP domain family of transcription factors (74). There are seven nucleotide differences between the maize and teosinte allele, including a lysine to asparagine amino acid change in position 6 that appears to affect protein dimer formation (75). Other genes have also been identified as involved in maize domestication, including zfl2 (76), zag1 (77), and gt1 (78), the latter two also encoding transcription factors.
Similarly, domestication genes have also been identified in other crop species. In rice, the sh4 gene (79) controls nonshattering, prog1 controls upright growth (80, 81), rc leads to white pericarp in the seed (82), and laba1 reduces seed awns (83). Interestingly, the first three genes also encode transcription factors, while laba1 produces a cytokinin-activating enzyme. Domestication genes in wheat (84), barley (85), beans (86), and tomato (87), among others, have also been identified.
While these genetic analyses suggest that only a few genes underlie domestication in crop plants, it is likely that a greater number of loci may be involved. A study in maize estimates that ∼1,200 genes may have been affected by natural selection; this suggests that the number of loci underlying crop domestication and diversification could indeed be large (88). It should also be noted that this may be a biased view of the genetic architecture of plant domestication, as the genetics of this process in root crop, vegetable and perennial tree species are less well understood.
Domestication of Animals.
In contrast to plants, no obvious “domestication gene” has been identified in domestic animals if we define this as a gene that was crucial for the domestication process. There are, however, many gene variants that are fixed in certain breeds and responsible for a characteristic feature of that breed. These include loss-of-function mutations in myostatin causing muscular hypertrophy in some breeds of meat-producing cattle (89), a premature stop codon in DMRT3 in horses that perform various types of ambling gaits (90), and polledness (lack of horn) in cattle caused by noncoding changes on cattle chromosome 1 (91). However, none of these gene variants have been crucial for domestication as many individuals of the respective species do not carry them. The genetic basis for domestication has also been studied by cross-breeding experiments between domestic pigs and European wild boars (92) and between red junglefowl and domestic chicken (93). These studies revealed several genes explaining pigmentation differences between wild and domestic species (47, 55) and some major QTLs with large effects on phenotype (58), but they did not reveal any candidate domestication genes.
An alternative approach for finding domestication genes in animals has been to carry out whole-genome sequencing of domestic species and their wild ancestors, first pioneered in chicken (94) and subsequently done in many species, including pig (56), dog (95), sheep (96), and rabbit (97). These studies have all revealed numerous loci under strong selection during animal domestication but still no obvious domestication genes. For instance, a study in chickens revealed a missense mutation in the thyroid-stimulating hormone receptor gene that is widespread among domestic chicken, which was later reported to affect photoperiodic response and reproduction (98), but the variant is not fixed in domestic chicken. Another example is the dog/wolf genome comparison, which revealed a copy number expansion of the amylase gene that occurs at high frequency in dogs, most likely a response to a more starch-rich diet subsequent to domestication (95).
The comparison of wild and domesticated rabbits is particularly informative (97). Rabbit domestication is relatively recent and occurred about 1,500 y before present. Domestication was initiated in southern France, where large populations of wild rabbits are still present. Whole-genome sequencing revealed that differences between wild and domestic rabbits may be brought about by shifts in allele frequencies at many loci rather than strong selection at a limited number of domestication genes (97). Allele frequency differences between wild and domestic rabbits were strikingly enriched at noncoding sites in the vicinity of genes affecting brain and neuronal development, in line with the fact that the most consistent difference between wild and domesticated animals is a change in behavior that allows them to tolerate close interaction with humans. The only way such a strong gene enrichment can be obtained is if rabbit domestication has a highly polygenic background.
Genetic Architecture of Crop and Domesticated Animal Evolution
The molecular analyses that have advanced over the last few decades allow us to compare the genetic basis of domestication and variation in major crop plants and domesticated animals. A key difference noted in the previous section is the difference in the genetic architecture of domestication genes between crops and domesticated animals. In the former, there are clear examples of a few genes of large effect associated with domestication, while animal domestication appears to have a more polygenic basis. The initial stages of animal domestication most likely took place by a gradual change at many loci affecting tameness rather than by disruption of a few critical genes, and this probably explains why extensive studies on this topic have not revealed any obvious domestication genes. Moreover, the modular nature of plant development (99) may also partly explain the greater frequency of major effect genes, as the phenotypic consequences of mutations at these loci may be less pleiotropic.
What types of mutations underlie variation in domesticated phenotypes? In a compilation of validated or putative causative mutations in 60 known crop genes (11), 41% are SNPs, 38% are insertion/deletions, 15% are TE insertions, and 5% are duplications or chromosomal rearrangements. This contrasts with the mutations observed in domesticated animal systems, in which there are numerous examples of large-scale deletions, inversions, or translocations associated with key alleles with major phenotypic effects (SI Appendix, Tables S2 and S3).
Of interest are the cases of TE insertions associated with the evolution of domesticated species. For example, the wrinkled R phenotype in Mendel’s peas (31) and the color polymorphism of date palm (Phoenix dactylifera) fruits controlled by the Virescens gene are the result of coding region transposon insertions (100). The role of TEs in providing for allelic variation in domesticated plants may reflect greater activity of these mobile sequences in plant genomes than in animal genomes. In maize, for example, where the original Ac/Ds and Spm/dSpm TEs were first described by Barbara McClintock (15, 101), analyses of the Wx locus indicate that ∼40% of spontaneous mutations that lead to phenotypes are the result of TE insertions ranging in size from ∼150 bp to >6 kb (18–20). Nevertheless, there are clear examples of mobile element insertions associated with domesticated animal phenotypes, including an endogenous retrovirus insertion in CYP19A1 associated with henny feathering in chicken (102, 103) and a retrogene insertion encoding fibroblast growth factor 4 that is associated with chondrodysplasia, a short-legged phenotype found in several dog breeds (104). These examples suggest that TE activity may be a significant factor in diversifying domesticated species phenotypes.
Another key element is the extent to which mutations are associated with regulatory vs. coding mutations. In 60 previously analyzed crop genes (11), we observed mutations both in the regulatory regions and in the coding sequence. Less than half of these genes have putative or validated cis-regulatory mutations, while most had mutations that affected the coding region, including missense mutations, frameshifts, or premature stop codons. Moreover, there is a preponderance of coding region mutations and loss-of-function alleles in domesticated plant genes; indeed, 20% of previously analyzed crop genes had loss-of-function alleles (11). This is not as common in domesticated animals except in those causing inherited disorders (37), and this difference may again arise from reduced pleiotropy of plant loci.
There is a wide range of proteins encoded by genes that underlie domesticated plant phenotypes. In an analysis of genes associated with crop evolution (11), there is a preponderance of regulatory genes, with 65% of isolated genes encoding transcriptional regulators. This suggests that regulatory evolution may play a key role in the genetics of plant domestication and diversification. Among the other genes, 25% encode enzymes, and 3% are transporter protein genes (11).
Finally, the wide range of domesticated plant and animal species that have been analyzed provides ways to compare similar phenotypes across multiple taxa. Interestingly, in some cases, the same gene underlies parallel evolution of similar phenotypic variation in different species. For example, glutinous rice is caused by a splice donor defect in the Wx gene, and mutations in this gene also underlie the sticky cereal phenotypes in barley, corn, and Job’s tears (105). Mutations in an myb-like transcription factor gene lead to color variation in fruits in date palms, oil palms, grapes, apples, cacao, and citrus (100, 106, 107). A similar pattern is observed in domesticated animal phenotypes, where allelic variation at the KIT and/or MC1R loci are causing variation in color in goats, pigs, horses, cattle, and chickens (SI Appendix, Tables S2 and S3). These results suggest that, in some cases, mutations in homologous genes are responsible for similar phenotypes in distinct domesticated species, which provide the molecular basis for Vavilov’s law of homologous series in variation (108) across domesticated plants and animals.
Implications for Food Security
The study of the genetics of domesticated species has been one of the key drivers of progress in food security over the last century. The development of efficient breeding strategies in crops and animals based on quantitative genetics theory has been crucial; it would not have been possible to feed 8 billion people with the crop varieties and farm animals that were available 100 y ago. The improved breeding strategies were initially based on the efficient use of phenotypic records, later complemented with DNA marker–assisted selection, and more recently, further developed using so-called genomic selection (109). Nevertheless, food security remains a key challenge for the future because of increasing world populations, greater urbanization, and global climate change. Approximately 9% of the world’s population is currently undernourished, and this number is projected to grow to 9.8% by 2030; at this point, more than 850 million people are predicted to face hunger (110). Moreover, agricultural operations continue to have a large footprint on the planet, taking up 38% of the Earth’s land surface, taking up ∼70% of the world’s fresh water, and consuming 1.2% of global energy. The ability to genetically improve crop varieties and livestock breeds to provide food in a sustainable manner has benefited from the genetic approaches that were launched by Mendel ∼150 y ago.
Conclusion
The seminal experiments by Gregor Mendel on peas in the middle of the nineteenth century laid the foundations for modern genetics. His genetic studies relied on domesticated crop species and paralleled Charles Darwin’s interest in domesticated taxa in formulating his own ideas of variation, selection, and evolutionary change. Since then, geneticists have consistently relied on domesticated species as systems to understand both genetics and evolution, and the advent of molecular tools has allowed for the dissection of the genetics of domestication and diversification. Continuing studies on the genetics of crops and livestock species will provide greater insights into the molecular basis of trait variation and the mechanisms of evolutionary change as well as help ensure continued global food security.
Supplementary Material
Acknowledgments
We thank Noel Ellis, Claire Domoney, Julie Hofer, and John Doebley for providing photographs; Mårten Larsson for assistance in preparing Fig. 3; and Frank Nicholas for a careful review of an earlier version of the paper.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122150119/-/DCSupplemental.
Data Availability
There are no data underlying this work.
References
- 1.Purugganan M., What is domestication? Trends Ecol. Evol. 37, in press (2022). [DOI] [PubMed] [Google Scholar]
- 2.Botigué L. R., et al. , Ancient European dog genomes reveal continuity since the Early Neolithic. Nat. Commun. 8, 16082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larson G., et al. , Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. U.S.A. 111, 6139–6146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang M., Larson G., Ribeiro H. S., Li N., Andersson L., Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 5, e1000341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darwin C., On the Origins of Species by Means of Natural Selection or the Preservation of Favoured Races in the Struggle for Life (John Murray, London, United Kingdom, 1859). [Google Scholar]
- 6.Darwin C., The Variation of Animals and Plants under Domestication (John Murray, London, United Kingdom, 1868). [Google Scholar]
- 7.Rhoades M. M., The early years of maize genetics. Annu. Rev. Genet. 18, 1–29 (1984). [DOI] [PubMed] [Google Scholar]
- 8.Bateson W., Experimental studies in the physiology of heredity. Part II. Poultry. Rep. Evol. Comm. Roy. Soc 1, 87–124 (1902). [Google Scholar]
- 9.Bateson W., Saunders E. R., The facts of heredity in the light of Mendel’s discovery. Rep. Evol. Comm. Roy. Soc. 1, 125–160 (1902). [Google Scholar]
- 10.Bateson W., Punnett R. C., Experimental studies in the physiology of heredity. Poultry. Rep. Evol. Comm. Roy. Soc 4, 18–35 (1908). [Google Scholar]
- 11.Meyer R. S., Purugganan M. D., Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852 (2013). [DOI] [PubMed] [Google Scholar]
- 12.van Dijk P. J., Ellis T. H. N., The full breadth of Mendel’s genetics. Genetics 204, 1327–1336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coe E. H. Jr., The origins of maize genetics. Nat. Rev. Genet. 2, 898–905 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Creighton H. B., McClintock B., A correlation of cytological and genetical crossing over in Zea mays. Proc. Natl. Acad. Sci. U.S.A. 17, 492–497 (1931). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClintock B., The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. U.S.A. 36, 344–355 (1950). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emerson R. A., East E. M., The inheritance of quantitative characters in maize. Bull. Agric. Exp. Stn. Nebr. 2, 1–120 (1913). [Google Scholar]
- 17.Shure M., Wessler S., Fedoroff N., Molecular identification and isolation of the Waxy locus in maize. Cell 35, 225–233 (1983). [DOI] [PubMed] [Google Scholar]
- 18.Wessler S. R., Varagona M. J., Molecular basis of mutations at the waxy locus of maize: Correlation with the fine structure genetic map. Proc. Natl. Acad. Sci. U.S.A. 82, 4177–4181 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varagona M. J., Purugganan M., Wessler S. R., Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell 4, 811–820 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purugganan M. D., Wessler S. R., Molecular evolution of magellan, a maize Ty3/gypsy-like retrotransposon. Proc. Natl. Acad. Sci. U.S.A. 91, 11674–11678 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai X.-L., Wang Z. Y., Xing Y. Y., Zhang J. L., Hong M. M., Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J. 14, 459–465 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Olsen K. M., et al. , Selection under domestication: Evidence for a sweep in the rice waxy genomic region. Genetics 173, 975–983 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Correns C., Bastarde zwischen maisrassen, mit besonder Berucksichtung der Xenien. Bibl. Bot. 53, 1–161 (1901). [Google Scholar]
- 24.James M. G., Robertson D. S., Myers A. M., Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7, 417–429 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buckner B., San Miguel P., Janick-Buckner D., Bennetzen J. L., The yl gene of maize codes for phytoene synthase. Genetics 143, 479–488 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palaisa K., Morgante M., Tingey S., Rafalski A., Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc. Natl. Acad. Sci. U.S.A. 101, 9885–9890 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paz-Ares J., Ghosal D., Wienand U., Peterson P. A., Saedler H., The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6, 3553–3558 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng J., et al. , ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Spielmeyer W., Ellis M. H., Chandler P. M., Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. U.S.A. 99, 9043–9048 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid J. B., Ross J. J., Mendel’s genes: Toward a full molecular characterization. Genetics 189, 3–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharyya M. K., Smith A. M., Ellis T. H. N., Hedley C., Martin C., The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell 60, 115–122 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Lester D. R., Ross J. J., Davies P. J., Reid J. B., Mendel’s stem length gene (Le) encodes a gibberellin 3 beta-hydroxylase. Plant Cell 9, 1435–1443 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin D. N., Proebsting W. M., Hedden P., Mendel’s dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 8907–8911 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato Y., Morita R., Nishimura M., Yamaguchi H., Kusaba M., Mendel’s green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 14169–14174 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aubry S., Mani J., Hörtensteiner S., Stay-green protein, defective in Mendel’s green cotyledon mutant, acts independent and upstream of pheophorbide a oxygenase in the chlorophyll catabolic pathway. Plant Mol. Biol. 67, 243–256 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Hellens R. P., et al. , Identification of Mendel’s white flower character. PLoS One 5, e13230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholas F. W., Online Mendelian Inheritance in Animals (OMIA): A record of advances in animal genetics, freely available on the Internet for 25 years. Anim. Genet. 52, 3–9 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Ricketts M. H., et al. , A nonsense mutation causes hereditary goitre in the Afrikander cattle and unmasks alternative splicing of thyroglobulin transcripts. Proc. Natl. Acad. Sci. U.S.A. 84, 3181–3184 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujii J., et al. , Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science 253, 448–451 (1991). [DOI] [PubMed] [Google Scholar]
- 40.Gillard E. F., et al. , A substitution of cysteine for arginine 614 in the ryanodine receptor is potentially causative of human malignant hyperthermia. Genomics 11, 751–755 (1991). [DOI] [PubMed] [Google Scholar]
- 41.Wright D., et al. , Copy number variation in intron 1 of SOX5 causes the Pea-comb phenotype in chickens. PLoS Genet. 5, e1000512 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imsland F., et al. , The Rose-comb mutation in chickens constitutes a structural rearrangement causing both altered comb morphology and defective sperm motility. PLoS Genet. 8, e1002775 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorshorst B., et al. , A genomic duplication is associated with ectopic eomesodermin expression in the embryonic chicken comb and two duplex-comb phenotypes. PLoS Genet. 11, e1004947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorshorst B., Okimoto R., Ashwell C., Genomic regions associated with dermal hyperpigmentation, polydactyly and other morphological traits in the Silkie chicken. J. Hered. 101, 339–350 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Maas S. A., Suzuki T., Fallon J. F., Identification of spontaneous mutations within the long-range limb-specific Sonic hedgehog enhancer (ZRS) that alter Sonic hedgehog expression in the chicken limb mutants oligozeugodactyly and silkie breed. Dev. Dyn. 240, 1212–1222 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eriksson J., et al. , Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 4, e1000010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerje S., et al. , The Dominant white, Dun and Smoky color variants in chicken are associated with insertion/deletion polymorphisms in the PMEL17 gene. Genetics 168, 1507–1518 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karlsson E. K., et al. , Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet. 39, 1321–1328 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Little C. C., The Inheritance of Coat Colour in the Dogs (Cornell University Press, Ithaca, NY, 1957). [Google Scholar]
- 50.Rosengren Pielberg G., et al. , A cis-acting regulatory mutation causes premature hair graying and susceptibility to melanoma in the horse. Nat. Genet. 40, 1004–1009 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Sundström E., et al. , Identification of a melanocyte-specific, MITF-dependent regulatory element in the intronic duplication causing hair greying and melanoma in horses. Pigment Cell Melanoma Res. 25, 28–36 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Imsland F., et al. , Regulatory mutations in TBX3 disrupt asymmetric hair pigmentation that underlies Dun camouflage color in horses. Nat. Genet. 48, 152–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andersson L., Molecular consequences of animal breeding. Curr. Opin. Genet. Dev. 23, 295–301 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Johansson Moller M., et al. , Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mamm. Genome 7, 822–830 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Marklund S., et al. , Molecular basis for the dominant white phenotype in the domestic pig. Genome Res. 8, 826–833 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rubin C.-J., et al. , Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. U.S.A. 109, 19529–19536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han F., et al. , Ecological adaptation in Atlantic herring is associated with large shifts in allele frequencies at hundreds of loci. eLife 9, e61076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rubin C.-J., et al. Darwin’s finches—an adaptive radiation constructed from ancestral genetic modules. bioRxiv [Preprint] (2021). 10.1101/2021.09.17.460815 (Accessed 3 January 2022). [DOI]
- 59.Goddard M. E., Hayes B. J., Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 10, 381–391 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Van Laere A. S., et al. , A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425, 832–836 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Markljung E., et al. , ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 7, e1000256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Charlier C., et al. , NGS-based reverse genetic screen for common embryonic lethal mutations compromising fertility in livestock. Genome Res. 26, 1333–1341 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bourneuf E., et al. , Rapid discovery of de novo deleterious mutations in cattle enhances the value of livestock as model species. Sci. Rep. 7, 11466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reynolds E. G. M., et al. , Non-additive association analysis using proxy phenotypes identifies novel cattle syndromes. Nat. Genet. 53, 949–954 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Doebley J., The genetics of maize evolution. Annu. Rev. Genet. 38, 37–59 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Matsuoka Y., et al. , A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. U.S.A. 99, 6080–6084 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stitzer M. C., Ross-Ibarra J., Maize domestication and gene interaction. New Phytol. 220, 395–408 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Doebley J., Stec A., Inheritance of the morphological differences between maize and teosinte: Comparison of results for two F2 populations. Genetics 134, 559–570 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doebley J., Stec A., Wendel J., Edwards M., Genetic and morphological analysis of a maize-teosinte F2 population: Implications for the origin of maize. Proc. Natl. Acad. Sci. U.S.A. 87, 9888–9892 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doebley J., Stec A., Hubbard L., The evolution of apical dominance in maize. Nature 386, 485–488 (1997). [DOI] [PubMed] [Google Scholar]
- 71.Studer A., Zhao Q., Ross-Ibarra J., Doebley J., Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43, 1160–1163 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clark R. M., Wagler T. N., Quijada P., Doebley J., A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 38, 594–597 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Xu G., et al. , Evolutionary and functional genomics of DNA methylation in maize domestication and improvement. Nat. Commun. 11, 5539 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang H., et al. , The origin of the naked grains of maize. Nature 436, 714–719 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang H., Studer A. J., Zhao Q., Meeley R., Doebley J. F., Evidence that the origin of naked kernels during maize domestication was caused by a single amino acid substitution in tga1. Genetics 200, 965–974 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bomblies K., Doebley J. F., Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics 172, 519–531 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wills D. M., Fang Z., York A. M., Holland J. B., Doebley J. F., Defining the role of the Mads-box gene, Zea agamous-like1, a target of selection during maize domestication. J. Hered. 109, 333–338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whipple C. J., et al. , grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. U.S.A. 108, E506–E512 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li C., Zhou A., Sang T., Rice domestication by reducing shattering. Science 311, 1936–1939 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Jin J., et al. , Genetic control of rice plant architecture under domestication. Nat. Genet. 40, 1365–1369 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Tan L., et al. , Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364 (2008). [DOI] [PubMed] [Google Scholar]
- 82.Sweeney M. T., Thomson M. J., Pfeil B. E., McCouch S., Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18, 283–294 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hua L., et al. , LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 27, 1875–1888 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simons K. J., et al. , Molecular characterization of the major wheat domestication gene Q. Genetics 172, 547–555 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pourkheirandish M., et al. , Evolution of the grain dispersal system in barley. Cell 162, 527–539 (2015). [DOI] [PubMed] [Google Scholar]
- 86.Repinski S. L., Kwak M., Gepts P., The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theor. Appl. Genet. 124, 1539–1547 (2012). [DOI] [PubMed] [Google Scholar]
- 87.Tanksley S. D., The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16 (suppl.), S181–S189 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wright S. I., et al. , The effects of artificial selection on the maize genome. Science 308, 1310–1314 (2005). [DOI] [PubMed] [Google Scholar]
- 89.Grobet L., et al. , Molecular definition of an allelic series of mutations disrupting the myostatin function and causing double-muscling in cattle. Mamm. Genome 9, 210–213 (1998). [DOI] [PubMed] [Google Scholar]
- 90.Andersson L. S., et al. , Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 488, 642–646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Medugorac I., et al. , Bovine polledness—an autosomal dominant trait with allelic heterogeneity. PLoS One 7, e39477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Andersson L., et al. , Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science 263, 1771–1774 (1994). [DOI] [PubMed] [Google Scholar]
- 93.Kerje S., et al. , The twofold difference in adult size between the red junglefowl and White Leghorn chickens is largely explained by a limited number of QTLs. Anim. Genet. 34, 264–274 (2003). [DOI] [PubMed] [Google Scholar]
- 94.Rubin C.-J., et al. , Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464, 587–591 (2010). [DOI] [PubMed] [Google Scholar]
- 95.Axelsson E., et al. , The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495, 360–364 (2013). [DOI] [PubMed] [Google Scholar]
- 96.Kijas J. W., et al. ; International Sheep Genomics Consortium Members, Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 10, e1001258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carneiro M., et al. , Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science 345, 1074–1079 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Karlsson A. C., et al. , A domestication related mutation in the thyroid stimulating hormone receptor gene (TSHR) modulates photoperiodic response and reproduction in chickens. Gen. Comp. Endocrinol. 228, 69–78 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Hallé F., Modular growth in seed plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 313, 77–87 (1986). [Google Scholar]
- 100.Hazzouri K. M., et al. , Whole genome re-sequencing of date palms yields insights into diversification of a fruit tree crop. Nat. Commun. 6, 8824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feschotte C., Jiang N., Wessler S. R., Plant transposable elements: Where genetics meets genomics. Nat. Rev. Genet. 3, 329–341 (2002). [DOI] [PubMed] [Google Scholar]
- 102.Matsumine H., Herbst M. A., Ou S. H., Wilson J. D., McPhaul M. J., Aromatase mRNA in the extragonadal tissues of chickens with the henny-feathering trait is derived from a distinctive promoter structure that contains a segment of a retroviral long terminal repeat. Functional organization of the Sebright, Leghorn, and Campine aromatase genes. J. Biol. Chem. 266, 19900–19907 (1991). [PubMed] [Google Scholar]
- 103.Li J., et al. , Characterization of the endogenous retrovirus insertion in CYP19A1 associated with henny feathering in chicken. Mob. DNA 10, 38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Parker H. G., et al. , An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 325, 995–998 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Purugganan M. D., Evolutionary insights into the nature of plant domestication. Curr. Biol. 29, R705–R714 (2019). [DOI] [PubMed] [Google Scholar]
- 106.Singh R., et al. , The oil palm VIRESCENS gene controls fruit colour and encodes a R2R3-MYB. Nat. Commun. 5, 4106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Allan A. C., Hellens R. P., Laing W. A., MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102 (2008). [DOI] [PubMed] [Google Scholar]
- 108.Vavilov N. I., The law of homologous series in variation. J. Genet. 12, 48–89 (1922). [Google Scholar]
- 109.T. Meuwissen, B. Hayes, M. Goddard, Accelerating improvement of livestock with genomic selection. Ann. Rev. Anim. Biosci. 1, 221–237 (2013) [DOI] [PubMed]
- 110.Food and Agriculture Organization, The State of Food Security and Nutrition in the World 2020 (FAO, Rome, Italy, 2019). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are no data underlying this work.