Abstract
Chitinolytic and glucanolytic fungal cell wall-degrading enzymes have been suggested to be primary determinants of biocontrol by Trichoderma spp. We examined the effects of ammonium, glucose, chitin, and chito-oligomers on transcription of specific genes and secretion of fungal cell wall-degrading enzymes. The genes ech42, nag1, and gluc78 were examined, as were the enzymes they encode (endochitinase CHIT42, N-acetylhexosaminidase CHIT73, and glucan exo-1,3-β-glucanase GLUC78, respectively). gluc78 could be induced by nitrogen starvation alone, while both ech42 and nag1 required nitrogen starvation and the presence of chitin for induction. Starvation for both ammonium and glucose resulted in very early expression and secretion of all cell wall-degrading enzymes examined. In the presence of low levels of ammonium (10 mM), both chito-oligomers and chitin triggered CHIT42 and CHIT40 (chitobiosidase) production. CHIT73 secretion occurred in the presence of N-acetylglucosamine and chito-oligomers, while chitin was less effective. The presence of different chito-oligomers resulted in secretion of specific N-acetylhexosaminidases, of which CHIT73 is one. Our results indicate that the expression and secretion of cell wall-degrading enzymes is nitrogen repressed, that effects of carbon and nitrogen nutrition are interactive, and that especially for chitinolytic enzymes, the inductive effect of chitin is altered by the level of ammonium or glucose in the medium.
Fungi in the genus Trichoderma are common soil inhabitants (16), and some strains are effective fungal parasites. Among the gene products that mediate these responses are various enzymes capable of degrading fungal cell wall components such as chitin, glucans, and proteins (3). Consequently, cell wall-degrading enzymes (CWDEs) have been the subject of considerable research interest, including isolation and characterization of several hydrolases and the genes that encode them (see references 1 and 17 for reviews).
Expression of these extracellular enzymes frequently has been reported to be induced by fungal cell wall components and repressed by carbon catabolite repressors, such as glucose (2, 9, 10, 22, 24). In some cases, starvation conditions alone could trigger CWDE production (23), while in others, cell walls or cell wall components were needed (10). The transcription factor Cre1 (15) can bind to the upstream regulatory region of the gene (ech42) encoding the endochitinase CHIT42 (2, 11, 14) from Trichoderma atroviride, suggesting that this factor may be involved in glucose repression in Trichoderma (19).
However, several lines of evidence indicate that CWDEs are regulated by factors other than carbon catabolite repression and induction by fungal cell wall components. Upstream regulatory regions of gluc78 (which encodes GLUC78, a 78-kDa exo-1,3-β-glucosidase [9]), ech42, and nag1 (which encodes CHIT73, a 73-kDa N-acetylhexosaminidase [22]) from T. atroviride contain clustered putative binding sites, such as GATA, stress response elements, BrlA response elements, and AbaA response elements (Donzelli and Harman, unpublished data), suggesting that they are regulated by a number of stimuli.
In addition, fungal cell walls and the cytoplasm of target fungi all are potential sources of both nitrogen and carbon for mycoparasitic Trichoderma spp. Therefore, it seems likely that these fungi have evolved mechanisms to respond not only to the scarcity of simple carbon sources but also to the lack of easily exploitable nitrogenous compounds. This scenario requires regulatory control of CWDEs that respond to a lack of carbon or nitrogen, or both, in order to yield these nutrients from target fungi. The primary objective of this study was to test the hypothesis that nitrogen nutrition regulates CWDE gene expression and enzyme secretion. We also tested the hypothesis that effects attributable to carbon and nitrogen nutrition and inducers such as chitin are interactive. Our results resolve discrepancies between previous studies that reported variable effects of inducing conditions for CWDEs and better define the conditions that elicit biocontrol enzymes. An abstract summary of these results has been published (B. Giuliano Garisto Donzelli and G. E. Harman, Abstr. Am. Phytopathol. Soc. Annu. Meet., Phytopathology 90[Suppl.]:S20, 2000).
MATERIALS AND METHODS
Strain and growth conditions.
We used Trichoderma atroviride (formerly Trichoderma harzianum [20]) strain P1 (ATCC 74058). Conidia were collected from colonies sporulating on potato dextrose agar (Difco, Detroit, Mich.) and used to inoculate 250-ml Erlenmeyer flasks containing 50 ml of potato dextrose broth (Difco) to give 2 × 107 spores/flask. Cultures were incubated at 25°C on a rotary shaker (150 rpm) for 18 to 24 h. The germlings were collected on Miracloth (Calbiochem, La Jolla, Calif.), washed three times with sterile deionized water, and used to inoculate 250-ml Erlenmeyer flasks containing 50 ml of enhanced minimal medium (EMM) that contained, per liter, the following: 23.4 g of 2-[N-morpholino]ethanesulfonic acid, 5 g of KH2PO4, 0.6 g of MgSO4, 0.6 g of CaCl2, 5 mg of FeSO4 · 7H2O, 10 mg of FeCl3, 1.6 mg of MnSO4 · H2O, 1 mg of ZnSO4 · 7H2O, 1 mg of CoCl2, 40 μg of CuSO4 · 5H2O and 13 μg of (NH4)6Mo7O24 · 4H2O. Glucose was added as the carbon source, and the nitrogen sources were either ammonium sulfate or ammonium acetate. Colloidal chitin (2 g/liter dry weight, wt/vol) or dried mycelia of Botrytis cinerea (0.2 g/liter dry weight, wt/vol) were added in some cases. Media were adjusted to pH 6.0. To avoid browning of the media that occurred during autoclaving, EMM salt base, glucose, and ammonium salt stock solutions were autoclaved separately and then blended under aseptic conditions.
In some experiments, chito-oligomers were used rather than chitin or B. cinerea mycelium as inducers. All experiments involving chito-oligomers were performed with 10-fold-smaller culture medium volumes than other experiments. One hour after adding biomass to EMM, four oligomers of N-acetylglucosamine (GlcNac1, GlcNac2, GlcNac3, or GlcNac4) (all from Sigma Chemical Co., St. Louis, Mo.) were added separately to give a final concentration of 1 mM each. All experiments were conducted at least twice with similar results.
Enzymes and genes examined.
We evaluated expression and secretion of an endochitinase (EC 3.2.1.14; CHIT42) (18) which is encoded by ech42 (2, 10, 14), an N-acetylhexosaminidase (nahase) (EC 3.2.1.52; CHIT73) (22) which is encoded by nag1, and a glucan 1,3-β-glucosidase (EC 3.2.1.58, GLUC78) which is encoded by gluc78 (9). We also monitored activity of chitin 1,4-β-chitobiosidase (CHIT40) (18), an additional 1,3-β-glucosidase, and several nahases in addition to CHIT73, since strain P1 produces up to five separate nahases, including CHIT73 (Donzelli and Harman, unpublished data).
Enzyme activity assays.
Chitinolytic activity was measured in 50 mM sodium acetate buffer (pH 5.0). For N-acetylhexosaminidase, the substrate was 4-methylumbelliferyl N-acetyl-β-d-glucosaminide, and for endochitinase, the substrate was 4-methylumbelliferyl β-d-N,N′-diacetyl-chitobioside (Sigma), both at a 0.1 mM final concentration. Release of 4-methylumbelliferone after 30 min at 25°C was measured using a fluorescence (360-nm excitation, 460-nm emission) microtiter plate reader (Cytofluor II; PerSeptive Biosystems, Framingham, Mass.).
Protein gel electrophoresis.
Polyacrylamide gel electrophoresis (PAGE) of culture filtrates was carried out under native conditions (13). Activity of chitinase on gels was assessed using 4-methylumbelliferyl N-acetyl-β-d-glucosaminide and β-d-N,N′-diacetyl-chitobioside as the substrates (25), while 1,3-β-glucanase activity was detected by the absence of fluorescence of laminarin in the presence of decolorized aniline blue (5).
RNA extraction and Northern analyses.
RNA was extracted from about 100 mg of mycelia (4). Total RNA was separated electrophoretically on a 1.2% agarose–6% formaldehyde gel and transferred onto a MagnaGraph nylon membrane (MSI, Westboro, Mass.). Probes prepared from the coding regions of ech42, nag1, and gluc78 (850, 1,400, and 1,100 nucleotides, respectively) were labeled with the PCR DIG Probe Synthesis Kit, following the manufacturer's protocols (Roche Molecular Biochemicals, Indianapolis, Ind.). Membrane prehybridization, hybridization development, and stripping were performed as described by Roche protocols. CDP-STAR (Roche) was used as the chemiluminescent substrate.
RESULTS
Nitrogen starvation and chitinase production in T. atroviride strain P1.
We used media (EMM) containing glucose at repressive levels (3%, wt/vol), dried mycelia of B. cinerea (0.02%, wt/vol) as an inducer, and nitrogen either as ammonium sulfate (5 or 50 mM) or ammonium acetate (10 or 100 mM). Both N-acetylhexosaminidase and endochitinase activities were high in culture filtrates when ammonium was present at 10 mM but low at 100 mM (data not shown). Similar results were obtained with either ammonium sulfate or ammonium acetate, but the pH was more stable in media containing ammonium acetate, and all subsequent experiments used ammonium acetate as the nitrogen source.
ech42, nag1, and gluc78 expression.
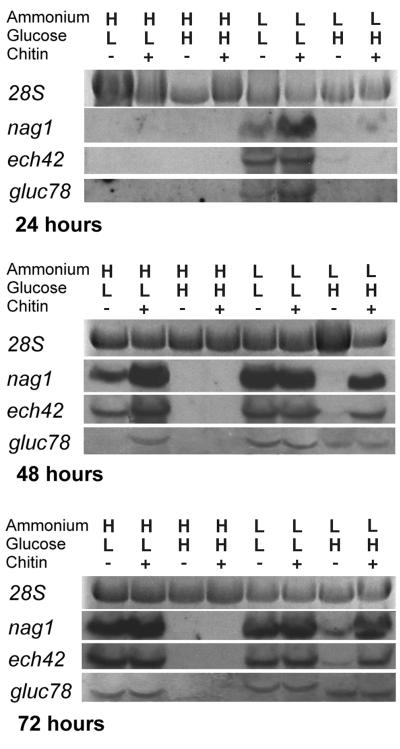
With low ammonium (10 mM) and high glucose (3%) levels, transcription of ech42 and nag1 occurred within 48 h (Fig. 1) when chitin was present. When chitin was absent, mRNA transcription was evident only 72 h after the induction. Under these conditions, low gluc78 expression levels were detected 48 h after induction. Expression of this gene was unaffected by chitin (Fig. 1).
FIG. 1.
Northern analyses of ech42, nag1, and gluc78 by T. atroviride after growth in EMM containing different levels of ammonium acetate (H = 100 mM; L = 10 mM) and glucose (H = 3%; L = 0.1%) in the presence or absence of colloidal chitin (1%). Expression levels at 24, 48, and 72 h after medium replacement are shown. All lanes were loaded with 7.5 μg of total RNA. Hybridizations were performed for 18 h at high stringency using PCR-generated DIG-labeled double-stranded-DNA-specific probes.
With high ammonium (100 mM) and low glucose (0.1%) levels, nag1 and ech42 transcription occurred within 48 h, regardless of the presence or absence of chitin (Fig. 1). Initial transcription levels were higher in the presence of chitin than in its absence, but by 72 h there were no detectable differences. gluc78 was expressed within 48 h in the presence of chitin but only after 72 h in its absence.
With low ammonium (10 mM) and low glucose (0.1%) levels, nag1, ech42, and gluc78 all were expressed at high levels within 24 h, and the presence of chitin had little additional effect (Fig. 1).
With high ammonium (100 mM) and high glucose (3%) levels, none of the genes were expressed, either in the presence or in the absence of chitin.
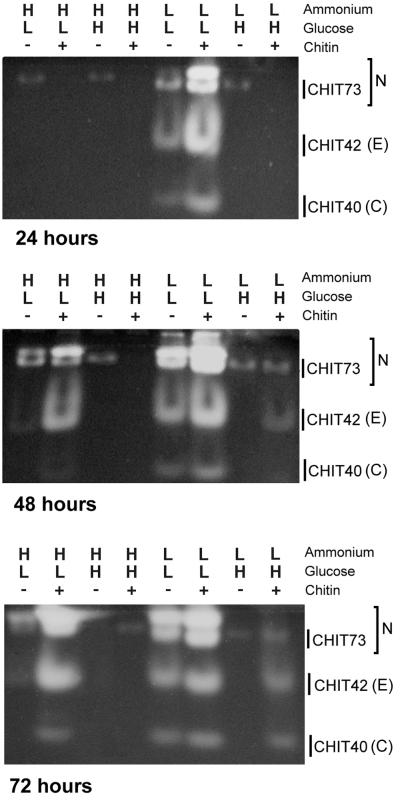
PAGE analyses of enzyme activity.
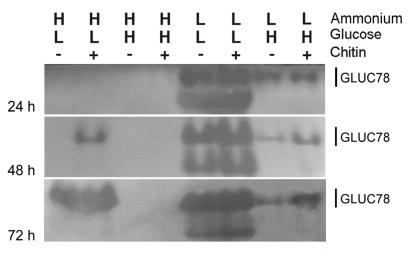
With chitin, low ammonium, and high glucose, both CHIT42 and CHIT73 activities were present in the culture filtrate by 48 h. By 72 h, CHIT40 also was evident (Fig. 2). In the absence of chitin, CHIT42 was not detected. Low constitutive levels of nahase were detected at all times (Fig. 2). GLUC78, or a band that migrates to the same gel location, was present at all times tested with and without chitin (Fig. 3). Although enzyme activity was detectable at 24 h, transcripts were not. Enzyme activity increased between 48 and 72 h (Fig. 3).
FIG. 2.
Expression of nahases (N), endochitinase (E), and chitobiosidase (C) by T. atroviride as assessed by separation of enzymes using native PAGE and visualization of activity from culture filtrates. Filtrates were harvested from cultures grown in EMM containing different levels of ammonium acetate (H = 100 mM; L = 10 mM) and glucose (H = 3%; L = 0.1%) in the presence or absence of colloidal chitin (1%). Activity was assessed by release of methylumbelliferone by digestion of the appropriate methylumbelliferyl substrate. Culture filtrates (15 μl) were loaded for each lane and separated at constant voltage (4 V/cm).
FIG. 3.
Expression of 1,3-β-glucosidase by T. atroviride as assessed by separation of enzymes using native PAGE and visualization of activity from culture filtrates. Filtrates were harvested from cultures grown in EMM containing different levels of ammonium acetate (H = 100 mM; L = 10 mM) and glucose (H = 3%; L = 0.1%) in the presence or absence of colloidal chitin (1%). Activity was detected by a decrease in fluorescence of laminarin conjugates with aniline blue decolorized at pH 9.0. Culture filtrates (15 μl) were loaded for each lane and separated at constant voltage (4 V/cm). Shown are the 1,3-β glucosidase GLUC78 and another glucosidase that has been heretofore unknown.
With high ammonium and low glucose levels, nahase and endochitinase activities were detectable 48 and 72 h after induction, with higher activity levels in the presence than in the absence of chitin (Fig. 2). At 72 h, CHIT40 also was present in chitin-supplemented cultures. We saw two nahase bands; the one with the greatest mobility corresponded to CHIT73 (Fig. 2). 1,3-β-glucanase activity was evident in chitin-containing cultures after 48 h (Fig. 3) and in both chitin-supplemented and chitin-free cultures after 72 h.
With low ammonium and low glucose levels, nahase, chitinase, chitobiosidase (Fig. 2), and 1,3-β-glucanase (Fig. 3) activities were strong in cultures at all times tested in both the presence and absence of chitin. In addition to CHIT73 and GLUC78, other bands with nahase and 1,3-β-glucanase activities were detected (Fig. 2 and 3). This was the only condition under which an additional 1,3-β-glucanase band was seen (Fig. 3).
With high ammonium and high glucose levels, neither chitinolytic nor glucanolytic enzyme activities were evident in these culture filtrates (Fig. 2 and 3) except for an extremely low level of nahase. This enzyme was detected at 24 and 48 h in the absence but not in the presence of chitin.
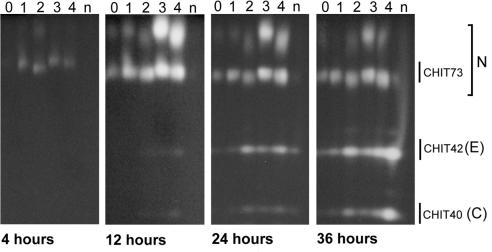
Induction of ech42 and nag1 by chito-oligomers under nitrogen-limiting conditions.
After 4 h of culture in the presence of chito-oligomers, at least two nahases were produced when GlcNac1 to GlcNac4 were present singly in the medium (Fig. 4). After 12 h, nahases were detected in the cultures without chitin, with the greatest levels in media containing GlcNac3 and GlcNac4. At 24 and 36 h, the induction patterns of the nahases were unchanged. By 36 h, activity levels of nahases with low electrophoretic mobilities decreased relative to enzymes at or near the mobility level of CHIT73 (Fig. 4). CHIT42 activity was detectable, at low levels, at 12 h in cultures containing GlcNac3 and GlcNac4 (Fig. 4). At 24 h, CHIT42 activity was strong in the media supplemented with GlcNac2–4, while lower levels of both CHIT42 and CHIT40 were detectable in chitin-supplemented media. Very low levels of these enzymes were present in media supplemented with N-acetylglucosamine or that lacked chitin derivatives. At 36 h CHIT42 was detectable in all cultures, with the strongest activity in media supplemented with chitin.
FIG. 4.
Induction of nahases (N), endochitinase (E), and chitobiosidase (C) from T. atroviride by chitin or chito-oligomers as assessed by separation of enzymes using native PAGE and visualization of activity from culture filtrates. Filtrates were harvested from cultures grown in EMM containing 10 mM ammonium acetate and 3% glucose. Activity was assessed by release of methylumbelliferone by digestion of the appropriate methylumbelliferyl substrate. Lanes: 0, no chito-oligomers; 1, N-acetylglucosamine; 2, N,NI-diacetylchitobiose; 3, N,NINII-triacetylchitotriose; 4, NI,NII,NIII,NIV-tetra-N-acetylchitotetraose; n, colloidal chitin. Culture filtrates (75 μl) were loaded in each lane and separated at constant voltage (4 V/cm).
DISCUSSION
Chitinolytic and glucanolytic enzymes may play crucial roles in biocontrol of fungi by Trichoderma spp. An important component in understanding their ecological role is defining factors affecting regulation of these genes (12, 19). Generally, CWDEs have been considered to be induced by the presence of chitin and repressed by catabolic repressors, such as glucose (2, 6, 7), although induction of enzyme expression by chitin has not been consistent (20).
We found that as with glucose, high levels of ammonium repress expression and secretion of extracellular chitinolytic (CHIT40, CHIT42, and CHIT78 and other nahases) and glucanolytic (e.g., GLUC78) enzymes. These results are consistent with a recent report that ammonium represses another chitinolytic gene, chit33, in a strain of T. harzianum (8).
At high levels of both glucose and ammonium, expression and secretion of all CWDEs measured were completely repressed, except for very low levels of a nahase. When both nutrients were at low levels, all enzymes were strongly expressed.
The levels of glucose and ammonium also affected whether chitin induced the expression of chitinolytic enzymes. When ammonium was low and glucose was high, enzyme expression occurred only in the presence of chitin or chito-oligomers. Thus, under this condition, chitin or chito-oligomers were essential for ech42 and nag1 transcription and for the secretion of CHIT42, CHIT40, and various nahases, including CHIT73. If glucose was low, irrespective of the ammonium concentration, chitin was not essential, but it enhanced and hastened expression or secretion of the chitinolytic enzymes.
We also found that chitinolytic enzymes are differentially induced by chito-oligomers under conditions of low ammonium and high glucose. Chito-oligomers of different chain lengths induce different nahases and are effective in inducing the production of CHIT40 and CHIT42.
These results may explain some of the discrepancies between different studies on the abilities of chitin to induce CWDEs. Mach et al. (20) reported that chitin or chito-oligomers did not induce transcription. Instead, ech42 was expressed only after prolonged carbon starvation or stresses, such as ethanol or cold shocks. However, they used a medium that contained high levels of nitrogen, analogous to our high-ammonium conditions. Thus, multiple repressive and inductive components of medium or environment should be considered when studying expression of CWDEs.
In some cases there were differences between patterns of gene expression and levels of enzymes secreted into the medium. For example, transcription levels of ech42 and nag1 were similar in media containing the following: low glucose and high ammonium plus chitin, high glucose and low ammonium plus chitin, and low glucose plus low ammonium. However, secreted levels of CHIT42 and CHIT73 were higher under the low-carbon plus low-ammonium condition than in the other two. These results suggest that posttranscriptional events affect enzyme production or secretion.
Under some circumstances, including repressive conditions, low levels of nahase activity were detected even though no transcription of nag1 occurred. The nahase produced under this condition has lower electrophoretic mobility than CHIT73, suggesting that this protein is likely to be encoded by a gene other than nag1.
We hypothesize that control of enzyme expression is of ecological importance. In soils the level of available nitrogen and organic carbon are below the lowest levels we used (21). Even in the rhizosphere, where root exudates increase the amount of simple sugars and organic acids, the amount of readily available nitrogen (e.g., ammonium salts and amino acids) is likely to be much less than 10 mM. Thus, soil conditions appear favorable for the expression of Trichoderma CWDEs, even in relatively nutrient-rich niches such as rhizodeposition sheaths (21).
The overall dynamics of CWDE expression are no doubt even more complex than described here, with other factors, e.g., light (2) or temperature (8), playing a role. Further, different genes respond differently to the presence of target fungi; ech42 is expressed prior to physical contact by the two fungi, while nag1 is expressed only after contact occurs (26). In the present study, we determined that nitrogen nutrition regulates CWDE expression; that effects of carbon and nitrogen are interactive upon CWDEs; that especially for chitinolytic enzymes, the inductive efficacy of chitin is dependent upon ammonium levels in the medium; and that chito-oligomers specifically induce particular nahases. Thus, regulation of CWDE expression in Trichoderma is complex and is regulated by multiple interactive factors.
ACKNOWLEDGMENTS
This work was supported by the U.S.-Israel Binational Agricultural Research and Development (BARD) Fund IS-2880-97.
We thank Kristen Ondik for her editorial work.
REFERENCES
- 1.Benitez T, Limon C, Delgado-Jarana J, Rey M. Glucanolytic and other enzymes and their genes. In: Harman G E, Kubicek C P, editors. Trichoderma and Gliocladium. Vol. 2. London, United Kingdom: Taylor and Francis; 1998. pp. 101–127. [Google Scholar]
- 2.Carsolio C, Guitierrez A, Jiminez B, Van Montagu M, Herrera-Estrella A. Characterization of ech-42, a Trichoderma harzianum endochitinase gene expressed during mycoparasitism. Proc Natl Acad Sci USA. 1994;91:10903–10907. doi: 10.1073/pnas.91.23.10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chet I, Benhamou N, Haran S. Mycoparasitism and lytic enzymes. In: Harman G E, Kubicek C P, editors. Trichoderma and Gliocladium. Vol. 2. London, United Kingdom: Taylor and Francis; 1998. pp. 153–172. [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Coté F, Letarte J, Grenier J, Trudel J, Asselin A. Detection of β-1,3-glucanase activity after native polyacrylamide gel electrophoresis: application to tobacco pathogenesis-related proteins. Electrophoresis. 1989;10:527–529. doi: 10.1002/elps.1150100714. [DOI] [PubMed] [Google Scholar]
- 6.de la Cruz J, Pintor-Toro J A, Benitez T, Llobell A, Romero L C. A novel endo-β-1,3-glucanase, BGN13.1, involved in the mycoparasitism of Trichoderma harzianum. J Bacteriol. 1995;177:6937–6945. doi: 10.1128/jb.177.23.6937-6945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Cruz J, Rey M, Lora J M, Hidalgo-Gallego A, Dominguez F, Pintor-Toro J A, Llobell A, Benitez T. Carbon source control on β-glucanases, chitobiase and chitinase from Trichoderma harzianum. Arch Microbiol. 1993;159:316–322. [Google Scholar]
- 8.de las Mercedes Dana M, Limon M C, Mejias R, Mach R L, Benitez T, Pintor-Toro J A, Kubicek C P. Regulation of chitinase 33 (chit33) gene expression in Trichoderma harzianum. Curr Genet. 2001;38:335–342. doi: 10.1007/s002940000169. [DOI] [PubMed] [Google Scholar]
- 9.Donzelli B G G, Lorito M, Scala F, Harman G E. Cloning, sequence and structure of a gene encoding an antifungal glucan 1,3-β-glucosidase from Trichoderma atroviride (T. harzianum) Gene. 2001;277:199–208. doi: 10.1016/s0378-1119(01)00681-3. [DOI] [PubMed] [Google Scholar]
- 10.Elad Y, Chet I, Henis Y. Degradation of plant pathogenic fungi by Trichoderma harzianum. Can J Microbiol. 1982;28:719–725. [Google Scholar]
- 11.Garcia I, Lora J M, de la Cruz J, Benitez T, Llobell A, Pintor-Toro J A. Cloning and characterization of a chitinase (CHIT42) cDNA from the mycoparasitic fungus Trichoderma harzianum. Curr Genet. 1994;27:83–89. doi: 10.1007/BF00326583. [DOI] [PubMed] [Google Scholar]
- 12.Harman G E. Myths and dogmas of biocontrol. Changes in perceptions derived from research on Trichoderma harzianum T-22. Plant Dis. 2000;84:377–393. doi: 10.1094/PDIS.2000.84.4.377. [DOI] [PubMed] [Google Scholar]
- 13.Harris E L V, Angal S. Protein purification methods: a practical approach. New York, N.Y: IRL Press; 1989. [Google Scholar]
- 14.Hayes C K, Klemsdal S, Lorito M, Di Pietro A, Peterbauer C, Nakas J P, Tronsmo A, Harman G E. Isolation and sequence of an endochitinase-encoding gene from a cDNA library of Trichoderma harzianum. Gene. 1994;138:143–148. doi: 10.1016/0378-1119(94)90797-8. [DOI] [PubMed] [Google Scholar]
- 15.Ilmen M, Thrane C, Penttilä M. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol Gen Genet. 1996;251:451–460. doi: 10.1007/BF02172374. [DOI] [PubMed] [Google Scholar]
- 16.Klein D, Eveleigh D E. Ecology of Trichoderma. In: Kubicek C P, Harman G E, editors. Trichoderma and Gliocladium. Vol. 1. London, United Kingdom: Taylor and Francis; 1998. pp. 57–74. [Google Scholar]
- 17.Lorito M. Chitinolytic enzymes and their genes. In: Harman G E, Kubicek C P, editors. Trichoderma and Gliocladium. Vol. 2. London, United Kingdom: Taylor and Francis; 1998. pp. 73–99. [Google Scholar]
- 18.Lorito M, Harman G E, Hayes C K, Broadway R M, Tronsmo A, Woo S L, Di Pietro A. Chitinolytic enzymes produced by Trichoderma harzianum: antifungal activity of endochitinase and chitobiosidase. Phytopathology. 1993;83:302–307. [Google Scholar]
- 19.Lorito M, Mach R L, Sposato P, Strauss J, Peterbauer C K, Kubicek C P. Mycoparasitic interaction relieves binding of the Cre1 carbon catabolite repressor protein to promoter sequences of the ech42 (endochitinase-encoding) gene in Trichoderma harzianum. Proc Natl Acad Sci USA. 1996;93:14868–14872. doi: 10.1073/pnas.93.25.14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mach R L, Peterbauer C K, Payer K, Jaksits S, Woo S L, Zeilinger S, Kullnig C M, Lorito M, Kubicek C P. Expression of two major chitinase genes of Trichoderma atroviride (T. harzianum P1) is triggered by different regulatory signals. Appl Environ Microbiol. 1999;65:1858–1863. doi: 10.1128/aem.65.5.1858-1863.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marschner H. Mineral nutrition of higher plants. 2nd ed. San Diego, Calif: Academic Press Ltd.; 1995. [Google Scholar]
- 22.Peterbauer C K, Lorito M, Hayes C K, Harman G E, Kubicek C P. Molecular cloning and expression of the nag1 gene (N-acetyl-β-D-glucosaminidase-encoding gene) from Trichoderma harzianum P1. Curr Genet. 1996;30:325–331. doi: 10.1007/s002940050140. [DOI] [PubMed] [Google Scholar]
- 23.Ramot O, Cohen-Kupiek R, Chet I. Regulation of β-1,3-glucanase by carbon starvation in the mycoparasite Trichoderma harzianum. Mycol Res. 2000;104:415–420. [Google Scholar]
- 24.Tronsmo A, Harman G E. Coproduction of chitinolytic enzymes and biomass for biological control by Trichoderma harzianum on media containing chitin. Biol Control. 1992;2:272–277. [Google Scholar]
- 25.Tronsmo A, Harman G E. Detection and quantification of N-acetyl-β-D-glucosaminidase, chitobiosidase and endochitinase in solutions and on gels. Anal Biochem. 1993;208:74–79. doi: 10.1006/abio.1993.1010. [DOI] [PubMed] [Google Scholar]
- 26.Zeilinger S, Galhaup C, Payer K, Woo S L, Mach R L, Fekete C, Lorito M, Kubicek C P. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet Biol. 1999;26:131–140. doi: 10.1006/fgbi.1998.1111. [DOI] [PubMed] [Google Scholar]