Abstract
Approximately 30% of elderly adults are cognitively unimpaired at time of death despite the presence of Alzheimer’s disease neuropathology at autopsy. Studying individuals who are resilient to the cognitive consequences of Alzheimer’s disease neuropathology may uncover novel therapeutic targets to treat Alzheimer’s disease. It is well established that there are sex differences in response to Alzheimer’s disease pathology, and growing evidence suggests that genetic factors may contribute to these differences. Taken together, we sought to elucidate sex-specific genetic drivers of resilience.
We extended our recent large scale genomic analysis of resilience in which we harmonized cognitive data across four cohorts of cognitive ageing, in vivo amyloid PET across two cohorts, and autopsy measures of amyloid neuritic plaque burden across two cohorts. These data were leveraged to build robust, continuous resilience phenotypes. With these phenotypes, we performed sex-stratified [n (males) = 2093, n (females) = 2931] and sex-interaction [n (both sexes) = 5024] genome-wide association studies (GWAS), gene and pathway-based tests, and genetic correlation analyses to clarify the variants, genes and molecular pathways that relate to resilience in a sex-specific manner.
Estimated among cognitively normal individuals of both sexes, resilience was 20–25% heritable, and when estimated in either sex among cognitively normal individuals, resilience was 15–44% heritable. In our GWAS, we identified a female-specific locus on chromosome 10 [rs827389, β (females) = 0.08, P (females) = 5.76 × 10−09, β (males) = −0.01, P(males) = 0.70, β (interaction) = 0.09, P (interaction) = 1.01 × 10−04] in which the minor allele was associated with higher resilience scores among females. This locus is located within chromatin loops that interact with promoters of genes involved in RNA processing, including GATA3. Finally, our genetic correlation analyses revealed shared genetic architecture between resilience phenotypes and other complex traits, including a female-specific association with frontotemporal dementia and male-specific associations with heart rate variability traits. We also observed opposing associations between sexes for multiple sclerosis, such that more resilient females had a lower genetic susceptibility to multiple sclerosis, and more resilient males had a higher genetic susceptibility to multiple sclerosis.
Overall, we identified sex differences in the genetic architecture of resilience, identified a female-specific resilience locus and highlighted numerous sex-specific molecular pathways that may underly resilience to Alzheimer’s disease pathology. This study illustrates the need to conduct sex-aware genomic analyses to identify novel targets that are unidentified in sex-agnostic models. Our findings support the theory that the most successful treatment for an individual with Alzheimer’s disease may be personalized based on their biological sex and genetic context.
Keywords: Alzheimer’s disease, sex differences, resilience, genetics, GWAS
By performing a series of sex-stratified and sex-interaction genetic analyses on continuous metrics of resilience to Alzheimer’s disease pathology, Eissman et al. identify a genome-wide significant resilience locus in females and numerous molecular pathways that may contribute to resilience in each sex.
See Groot et al. (https://doi.org/10.1093/brain/awac216) for a scientific commentary on this article.
See Groot et al. (https://doi.org/10.1093/brain/awac216) for a scientific commentary on this article.
Introduction
Alzheimer’s disease is a progressive, neurodegenerative disorder leading to cognitive impairment. Alzheimer’s disease is marked by two primary neuropathologies: amyloid plaques and neurofibrillary tangles. However, ∼30% of elderly adults are cognitively resilient to the downstream consequences of Alzheimer’s disease pathology, as they meet neuropathological criteria for Alzheimer’s disease at autopsy, yet remain cognitively unimpaired throughout life.1 Studying resilient individuals may uncover quintessential information about Alzheimer’s disease progression and enable the discovery of novel therapeutic targets. In a recent study from our group2, we conducted the largest genome-wide meta-analysis on cognitive resilience to date and demonstrated a unique genetic architecture of cognitive resilience that is distinct from that of Alzheimer’s disease.
Our original analysis did not investigate whether certain variants, genes, or molecular pathways relate to cognitive resilience in a sex-specific manner. An emerging body of evidence suggests there are sex differences in response to Alzheimer’s disease neuropathology. There are notable sex differences in both Alzheimer’s disease neuropathology burden and the association between neuropathology burden and longitudinal cognitive decline. Specifically, a one-unit increment in Alzheimer’s disease neuropathology at autopsy is associated with a 22-fold higher odds for clinical Alzheimer’s disease during life in females, but only a 3-fold higher odds in males.3,4 Similar sex differences are also observed in biomarker studies of Alzheimer’s disease, such that females with more pronounced Alzheimer’s disease neuropathology biomarkers show faster cognitive decline and faster hippocampal atrophy than males with comparable levels of Alzheimer’s disease biomarkers.5,6 Additionally, amyloid-positive females show both a faster rate of CSF tau7 and more pronounced tau accumulation in the medial temporal lobe as measured with tau PET6 compared to amyloid-positive males. Taken together, there is strong evidence that the occurrence and downstream consequences of Alzheimer’s disease neuropathology differ by biological sex.
In addition to the notable sex differences in Alzheimer’s disease biomarkers, there is similar evidence that sex-specific genetic factors contribute to sex differences in response to Alzheimer’s disease pathology. The most robust genetic risk factor of late-onset Alzheimer’s disease, the apolipoprotein E ε4 (APOE ε4) allele, has a stronger association with clinical Alzheimer’s disease among females compared to males, particularly between the ages of 55 and 70.8,9 Amyloid-positive females with the APOE ε4 allele have a faster rate of cognitive decline10,11 and show higher tau burden compared to male counterparts.12 Beyond APOE, work from our group has demonstrated sex-specific genome-wide associations with CSF amyloid and tau levels,13 and autopsy measures of neurofibrillary tangles,14 including a male-specific locus (e.g. TSPAN13) that was recently replicated in the UK Biobank dataset.15 Together, these findings highlight the importance of including sex-stratification in genomic models to better understand the genetic architecture of Alzheimer’s disease.
To this end, we took a precision medicine approach to elucidate sex differences in the genetic architecture of cognitive resilience to Alzheimer’s disease pathology. We harmonized cognitive data across four cohorts of cognitive ageing, leveraged a published model of cognitive resilience that implements latent variable modeling,2,16 and performed a series of sex-aware genetic analyses. We hypothesized that genetic drivers of cognitive resilience differ between males and females downstream of amyloidosis. By identifying sex-specific variants, candidate genes, and molecular pathways driving cognitive resilience to Alzheimer’s disease pathology, the results of this study will contribute to our understanding of Alzheimer’s disease progression in each biological sex and to the identification of novel therapeutic targets to treat Alzheimer’s disease.
Materials and methods
Participants
Our study included four cohorts of cognitive ageing [n (both sexes) = 5024, n (males) = 2093, n (females) = 2931]: Adult Changes in Thought (ACT), Religious Orders Study and Rush Memory and Aging Project (ROS/MAP), The Alzheimer’s Disease Neuroimaging Initiative (ADNI) and Anti-Amyloid Treatment in Asymptomatic Alzheimer’s Disease (A4). The A4 study began in 2014 as part of a clinical trial and recruited cognitively unimpaired individuals.17 ADNI launched in 2003 and is comprised of four phases of which ADNI1, ADNI2 and ADNI-GO were included in this study. There are now over 1800 individuals aged 55–90 who have participated in ADNI and are comprised of a mix of individuals who are cognitively unimpaired and individuals that have mild cognitive impairment, or Alzheimer’s disease dementia (http://adni.loni.usc.edu/). ACT began in 1994, recruiting cognitively unimpaired individuals from the Seattle area.18 ROS began in 1994 and recruited Catholic nuns, priests, and brothers living in orders. MAP launched in 1997 and recruited cognitively unimpaired individuals from the Chicago area.19 Written informed consent was obtained from all participants, and research was carried out in accordance with Institutional Review Board-approved protocols. Secondary analyses of all data were approved by the Vanderbilt University Medical Center Institutional Review Board.
Amyloid-PET acquisition
We leveraged in vivo amyloid PET data for two cohorts, ADNI and A4. ADNI’s methods and protocols for their in vivo amyloid PET imaging can be found on their website, http://adni.loni.usc.edu/. ADNI and A4 used a combination of GE, Philips, and Siemens technologies. Scans were conducted 50–70 min after tracer injection, and acquired frames were 5 min in length. Both cohorts utilized the 18F-florbetapir tracer, and a portion of ADNI’s study utilized the 11C-Pittsburgh compound B (PiB) tracer instead. For each brain region, standardized uptake value ratios (SUVRs) were calculated and scaled using the cerebellum as the reference brain region. For each participant, a SUVR composite score was calculated, comprised of cortical brain regions. For more extensive details regarding amyloid PET acquisition, see our recent paper.2
Amyloid-PET harmonization
As referenced in the above section, raw SUVR composite scores (comprised of cortical brain regions) were obtained from ADNI and A4. To normalize SUVR scores across cohorts and tracers, we performed Gaussian mixture modelling (GMM) within each cohort leveraging a previously published algorithm.20 Since ADNI scores were a mix of PiB and florbetapir, we performed separate GMM for each tracer within ADNI. Models were estimated among those that were cognitively normal and then were subsequently applied to all participants. Each GMM leveraged a two-component model fit, as this best fit the bimodal property of the amyloid distribution. The mean and the standard deviation of the amyloid negative distribution from each GMM was applied to standardize SUVR composites across all participants. The resulting harmonized SUVR composite scores were on a z-score scale, representative of individual amyloid burden. Our group recently published a paper testing different methods of amyloid PET harmonization, and more details can be found in Raghavan et al.21 Overall, we concluded that there are only minor differences between harmonization methods, and the minor differences have less import with amyloid as a linear predictor in our models.2,21
Post-mortem assessment of neuropathology
Post-mortem assessments were conducted for participants in the ACT and ROS/MAP cohorts. A well established measure of amyloid plaque burden, the Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) neuritic plaque staging scores, were determined for each participant using standard protocols.17,18,22 CERAD scores were standardized between ACT and ROS/MAP, such that higher CERAD scores were representative of higher individual amyloid burden in both cohorts. For more extensive details regarding post-mortem assessment of neuropathology, see our recent paper.2
Cognitive harmonization
Cognitive data were harmonized across all cohorts using published, modern psychometric techniques.23 Briefly, qualified neuropsychologists/behavioural neurologists categorized test items into memory or executive function domains (or neither). Overlapping test items across cohorts were set as anchor items. Test items were indicators in a confirmatory factor analysis, with the scaled anchor items allowing non-overlapping test items to be freely estimated. Memory was successfully harmonized across all four cohorts, as well as executive function across ACT, ROS/MAP and ADNI. A4 did not have sufficient anchor items for executive function harmonization, so a previously published composite, the Preclinical Alzheimer Cognitive Composite (PACC) was leveraged as an additional score across ADNI and A4. A four-item version was calculated in each, including logical memory, Mini-Mental State Exam along with Selective Reminding Test and digit symbol in A4 and ADAS-Cog and Trail Making Test B in ADNI. As detailed in the supplement of our first publication with this phenotype, the PACC behaves quite comparably in both ADNI and A4.2 The harmonized memory, executive function, and PACC composite scores were extracted and leveraged in building resilience phenotypes. For more extensive details regarding cognitive harmonization, see our recent paper.2
Latent variable modelling
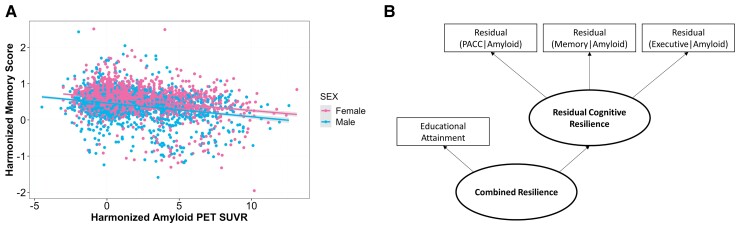
Cognitive resilience models were created using previously published protocols.2,16 Linear models were built in the combined autopsy dataset (ACT and ROS/MAP) and in the combined PET dataset (ADNI and A4). Memory and executive function harmonized scores were used as outcomes in the autopsy datasets covarying for age, sex, and CERAD staging scores. In the PET datasets, memory and PACC harmonized scores were used as outcomes, as well as harmonized executive function in ADNI, covarying for age, sex and harmonized amyloid PET SUVR. See Fig. 1A for an example of harmonized memory scores by harmonized amyloid PET scores, by sex.
Figure 1.
Cognitive and biomarker data harmonization and cognitive resilience model. Memory, executive function, and Preclinical Alzheimer Cognitive Composite (PACC) scores were harmonized across cohorts. Additionally, in vivo amyloid PET SUVRs were harmonized with Gaussian mixture modelling. (A) Harmonized in vivo amyloid PET SUVR by harmonized memory scores are plotted by sex. (B) Linear models leveraging harmonized cognitive and amyloid data (harmonized in vivo PET or autopsy measures of amyloid plaque burden, CERAD scores) were residualized and fed as indicators into a residual cognitive resilience latent variable model. The combined resilience latent variable model included educational attainment as an additional indicator.
Standardized residuals from all linear models were extracted and entered as indicators into latent variable models in Mplus (version 7.31).24 Two resilience models were built: residual cognitive resilience, with the standardized residuals as indicators, and a second-order latent variable, combined resilience, which included residual cognitive resilience and educational attainment as indicators (Fig. 1B). Inclusion criteria for the models required participants to have cognitive scores for at least two of the cognitive domains. Models were run in all individuals as well as in cognitively normal individuals only. Factor scores were extracted from all models. For average resilience scores by sex, see Table 1, and for more extensive details regarding latent variable modelling, see our recent paper.2
Table 1.
Cohort demographics by sex
| Males (n = 2093) | Females (n = 2931) | Both sexes (n = 5024) | |
|---|---|---|---|
| Age, years | 76.70 ± 8.80 | 76.84 ± 10.26 | 76.78 ± 9.68 |
| Educationa, years | 17.01 ± 3.07 | 16.05 ± 2.84 | 16.45 ± 2.98 |
| Residual Cognitive Resilience Score | 0.06 ± 0.89 | 0.06 ± 0.87 | 0.06 ± 0.88 |
| Combined Resilience Scorea | 0.08 ± 0.42 | −0.02 ± 0.39 | 0.02 ± 0.40 |
| Amyloid statusa | 857 (40.95%) | 1287 (43.91%) | 2144 (42.68%) |
| Alzheimer’s disease diagnosisa | 236 (11.28%) | 381 (13.00%) | 617 (12.28%) |
| APOE ε4 carrier status | 659 (31.49%) | 898 (30.64%) | 1557 (30.99%) |
Categorial values given in n (%); continuous values given in mean ± standard deviation.
Significant difference between sexes via a t-test (continuous variables) or via a chi-square test (categorical variables).
Genotyping, quality control and imputation
Participants included in this study were genotyped using DNA extracted from either brain or whole blood. Each cohort used the following genotype chips: A4 implemented the Illumina Global Screening Array and ACT implemented the Illumina Human660W-Quad Array. ADNI implemented three chips: Human610-Quad, HumanOmniExpress, and Omni 2.5 M. Finally, ROS/MAP implemented three chips: Affymetrix Genechip 6.0, Illumina Human1 M, and Illumina Global Screening Array.
All genetic data were processed with a standardized quality control and imputation pipeline. Raw genetic data were filtered to remove variants with >5% sample missingness and minor allele frequency (MAF) <1%. Then genetic data were filtered to remove individuals with >1% sample missingness, related individuals, and individuals with mismatched sex. Additionally, X-chromosome genetic data were compared between sexes, and variants with differential missingness (P < 1 × 10−07) were removed. Individuals who were non-Hispanic white were retained for analysis. Those who self-reported as non-Hispanic white but were deemed to be genetic ancestry outliers in a principal component analysis (including the 1000 Genomes reference dataset) were subsequently removed (based on a standard deviation ± five cut-point or by visual inspection).
Prior to imputation, variants were lifted over to genome build 38 (hg38). Then a Hardy-Weinberg equilibrium (HWE) exact test (P < 1 × 10−06) was performed in the whole sample for autosomal variants and a HWE exact test (P < 1 × 10−06) was performed in the female sample only for X-chromosome variants and the male sample was filtered accordingly. All variants were filtered to remove palindromic variants, same position variants, and variants with alleles mismatching with the reference panel. Genetic data were then imputed on the Trans-Omics for Precision Medicine (TOPMed) program server.25–27 Post-imputation, genetic data were filtered to remove variants with an imputed R2 < 0.8 and duplicated/multi-allelic variants were dropped. All genotyped variants were then dropped from the imputed data and the original genotypes were merged back in with the rest of the imputed data, and then variants with a MAF <1% were dropped. Finally, a HWE exact test (P < 1 × 10−06) was performed on the imputed data in the whole sample for autosomal variants and in the female sample only for X-chromosome variants and the male sample was filtered accordingly.
Genetic data requiring multiple datasets to be merged (ADNI, ACT and ROS/MAP) were then checked for overlapping samples across genotype chips. If sample overlap was present, the sample was dropped from the chip with the lower coverage. Cleaned, imputed, genetic data from each chip were compared and subsequently filtered to remove variants with mis-matching reference alleles and MAF differences of >10% and then the genetic datasets across chips were merged. The merged datasets were filtered for cryptic relatedness. Genetic ancestry was assessed in the merged dataset using a principal component analysis. Individuals who self-reported as non-Hispanic white but were deemed to be genetic ancestry outliers (based on a standard deviation ± five cut-point or by visual inspection) were subsequently removed.
Statistical analysis
Prior to performing genome-wide association studies (GWAS), cryptic relatedness across all four genetic datasets was assessed, removing 38 related individuals in total. In addition, for the combined ACT and ROS/MAP dataset and for the combined ADNI and A4 dataset, variants were filtered for reference allele mismatches and MAF differences >10%. Then ACT and ROS/MAP were merged to result in a combined autopsy dataset. Likewise, ADNI and A4 were merged to result in a combined PET dataset. Combined genetic datasets were subsequently used for all genetic analyses to facilitate joint analysis.
GWAS and genome-wide meta-analyses
GWAS were performed with PLINK linear association models (v1.90b5.2, https://www.cog-genomics.org/plink/1.9).28 All GWAS were run in the combined autopsy dataset and in the combined PET dataset for all resilience phenotypes. Sex-stratified GWAS covaried for age and the first three genetic principal components. The sex-interaction GWAS also covaried for sex and included a single nucleotide polymorphism (SNP) × sex interaction term. GWAS results were then meta-analysed across cohorts using a fixed-effects model with beta and standard error input (GWAMA v2.2.2).29 The above models were also run identically in the sample restricted to cognitively normal individuals, with the fixed effects meta-analyses implementing the minor allele frequencies calculated based on these individuals only. Additionally, an identical GWAS and meta-analysis pipeline as described above was implemented with the X-chromosome genetic data. All meta-analysis results were restricted to SNPs present in both the autopsy and the PET dataset, and these filtered results were leveraged for all post-GWAS steps discussed below.
SNP-heritability analysis
To determine the heritability of each resilience phenotype estimated in each sex and if estimates significantly differed between sexes, we performed a sex-aware heritability analysis that was outlined by Martin and colleagues.30,31 We first leveraged the Genome-Wide Complex Trait Analysis (GCTA) software tool to calculate genetic relatedness matrices in all individuals, in males only, and in females only. Then we implemented the GCTA restricted maximum likelihood statistical method with the genetic relatedness matrices to calculate SNP-based heritability estimates in all individuals, in males only, and in females only.32 Next, we performed a test to determine if the heritability estimates for each resilience phenotype significantly differed between sexes. To perform this test, we calculated z-scores with the following formula: z-score = (h2females−h2males)/√[h2females(SE)2 + h2males(SE)2]. Then, we obtained P-values for each z-score from the normal distribution based on a one-tailed test.
Variant annotation
Functional annotation was performed with Functional Mapping and Annotation (FUMA, v1.3.6a)33 on genome-wide significant loci from the meta-analyses. All variants in linkage disequilibrium with top variants were also considered in annotation. In brief, FUMA performs three types of mapping: expression quantitative trait locus (eQTL), Hi-C 3D chromatin interaction, and positional. Specifically, FUMA compiles chromosome conformation capture with high throughput sequencing (Hi-C) data from multiple databases. Hi-C is an assay that looks for enrichment of DNA sequences associated with chromatin loops at different locations in the genome. FUMA also looks to see if these enriched regions overlap with gene promotor or enhancer sequences. All types of mapping are performed in a tissue and a cell-type specific manner.34
Alzheimer’s disease risk loci analysis
We compiled Alzheimer’s disease risk variants from three well-known, published Alzheimer’s disease genome-wide meta-analyses.35–37 Leveraging our meta-analysis results, we looked at each risk variant’s association with residual cognitive resilience and with combined resilience in males, in females, and in the sex-interaction models.
Gene and pathway-based tests
Gene and pathway-level tests were performed with Multi-marker Analysis of GenoMic Annotation (MAGMA v1.09—the version of MAGMA with the known P-value inflation bug fixed)38 on all meta-analysis results. First, permutation-like gene tests were performed to determine if a higher number of significant variant-level P-values existed in a predefined gene window than expected by chance. This process was conducted across the entire genome. All gene-level results were then entered into permutation-like pathway tests to determine if there were more significant gene test P-values associated with known biological pathways than expected by chance. We leveraged two sets of curated pathway annotations from the Molecular Signatures Database (MSigDB) v.7.0 (downloaded on 5 February 2020), the curated gene set (C2) and the ontology gene set (C5).39 In total, we tested 18 243 genes and 12 173 biological pathways. All gene and pathway tests were adjusted for multiple comparisons using the false discovery rate (FDR) procedure, and an a priori significance threshold was set at PFDR < 0.05.
Genetic correlation analyses
Genetic correlation tests were performed between our resilience meta-analysis summary statistics and GWAS summary statistics of 65 complex traits using the Genetic Covariance Analyzer (GNOVA).40 To calculate genetic covariances with GNOVA, z-scores were quantified from each variant-level association in each set of GWAS summary statistics. Linkage disequilibrium scores were also quantified from an ancestry-matched reference panel (e.g. 1000 Genomes European reference panel). Then genetic covariances between trait pairs were calculated with the z-scores mentioned above. Inflation due to linkage disequilibrium structure was adjusted by implementing the ancestry-matched genome-wide linkage disequilibrium scores. Genetic covariances were also adjusted for sample overlap (between GWASs). For all genetic correlation analyses, we implemented GNOVA’s simplest, no annotation model. After conducting the genetic correlation analyses, genetic covariances were adjusted for multiple comparisons using the FDR procedure, and an a priori significance threshold was set at FDR < 0.05.
APOE × sex sensitivity analysis
A set of linear regressions were performed in R (v.4.0.3) with our resilience phenotypes as the outcomes, age and the first three genetic principal components as covariates, and inclusion of an APOE genotype × sex interaction term. APOE genotype was first coded with an APOE ε4 additive model and then in subsequent analyses with an APOE ε2 dominant model. An ε2 dominant model was implemented due to sample size constraints of homozygous ε2 individuals. All linear regressions were performed in the combined autopsy dataset and in the combined PET dataset and then meta-analysed across cohorts (R metafor package).41
Data availability
Data from the ADNI and A4 studies are shared through the LONI Image and Data Archive (https://ida.loni.usc.edu/). Data from ROS/MAP can be requested at www.radc.rush.edu. Data from ACT can be accessed through the Data Query Tool (http://act.kpwashingtonresearch.org/dqt/). GWAS summary statistics will be available through NIAGADS (https://www.niagads.org/datasets/).
Results
Cohort demographics stratified by sex are presented in Table 1. T-tests were performed between sexes for age, education, residual cognitive resilience score, and combined resilience score. Education and combined resilience score significantly differed between sexes based on P-values from the t-tests, whereby education and combined resilience were higher in males compared to females. Age and residual cognitive resilience score did not significantly differ between sexes. Chi-square tests were performed between sexes for amyloid status, Alzheimer’s disease diagnosis, and APOE ε4 carrier status. Amyloid status and Alzheimer’s disease diagnosis significantly differed between sexes based on P-values from chi-square tests, whereby amyloid status and Alzheimer’s disease diagnosis were both higher in females compared to males. APOE ε4 carrier status did not significantly differ between sexes.
SNP-heritability results
We calculated SNP-heritability estimates among the entire sample and among cognitively normal individuals, using the GCTA restricted maximum likelihood method.32 All results are presented in Table 2, but we will discuss the results among cognitively normal individuals which were statistically significant. Estimated among cognitively normal individuals of both sexes, resilience was 20–25% heritable. Estimates among male cognitively normal individuals were 26–44%, whereas among female cognitively normal individuals, estimates were 15–27%. We next tested to see if these heritability estimates significantly differed between sexes by calculating z-scores and generating P-values from the normal distribution from a one-tailed test. Heritability estimates did not significantly differ between sex for any of our phenotypes. Additionally, SNP-heritability estimates were attenuated when estimated among the whole sample but remained nominally significant when estimated among males and females combined and among males only.
Table 2.
SNP-heritability estimates by sex
| Both sexes | Males | Females | Sex differences test | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| h2 | h2(SE) | P-value | h2 | h2(SE) | P-value | h2 | h2(SE) | P-value | Z-score | P-value | |
| Residual Cognitive Resilience | 4.16% | 4.75% | 0.11 | 7.65% | 9.09% | 0.10 | 5.61% | 10.60% | 0.31 | −0.15 | 0.88 |
| Residual Cognitive Resilience (CN) | 20.90% | 5.48% | 6.80 × 10−11 | 25.82% | 11.60% | 5.07 × 10−5 | 27.17% | 9.86% | 1.24 × 10−4 | 0.09 | 0.93 |
| Combined Resilience | 19.71% | 8.89% | 0.01 | 29.42% | 19.48% | 0.03 | 0.0001% | 14.50% | 0.50 | −1.21 | 0.23 |
| Combined Resilience (CN) | 25.25% | 7.95% | 2.33 × 10−7 | 44.11% | 18.64% | 9.57 × 10−5 | 14.93% | 12.37% | 0.06 | −1.30 | 0.19 |
CN = cognitively normal. h2 values are the V(G)/Vp estimates calculated from the GCTA restricted maximum likelihood statistical method (with genetic relatedness matrices). Sex differences test P-values were generated from the normal distribution based on a one-tailed test.
Variant-level results
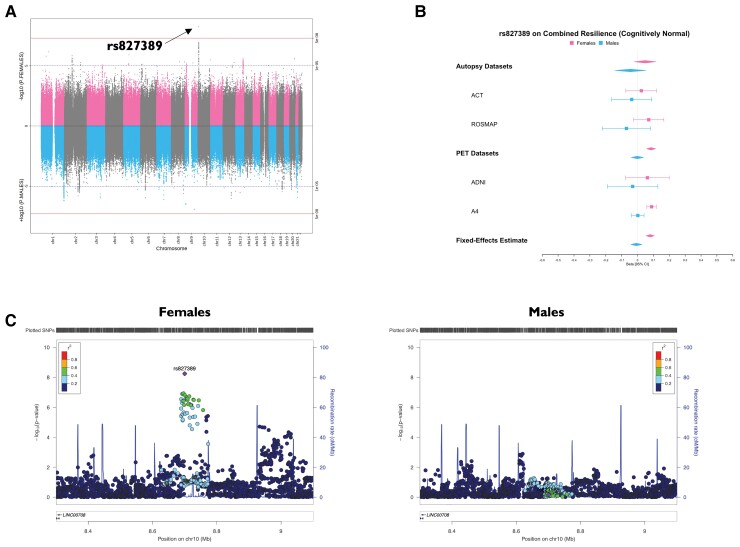
Variant-level results are presented in Supplementary Tables 1–12. QQ plots for all genome-wide meta-analyses with estimates of genomic lambda are presented in Supplementary Figs 1–3. For our residual cognitive resilience phenotype, we did not identify any sex-specific genome-wide significant loci in all participants or among cognitively normal individuals. For our combined resilience phenotype, we identified a genome-wide significant female-specific locus on chromosome 10 [rs827389, β(females) = 0.08, P(females) = 5.76 × 10−09, β(males) = −0.01, P(males) = 0.70, β(interaction) = 0.09, P(interaction) = 1.01 × 10−04] among cognitively normal individuals (Fig. 2) whereby the minor allele was associated with higher resilience scores. Although this locus was not genome-wide significant in residual cognitive resilience, the direction of effect was the same and the locus fell just below the genome-wide significance threshold in females [β(females) = 0.16, P(females) = 3.65 × 10−07]. No significant sex-specific associations were observed for combined resilience in all participants. We conducted APOE-by-sex sensitivity analyses on all resilience phenotypes. No associations were statistically significant (Supplementary Table 13).
Figure 2.
Minor allele of female-specific genome-wide significant locus on chromosome 10 (with rs827389) associated with higher combined resilience scores among cognitively normal females. (A) Miami plot with female variant associations on the top in pink and male variant associations on the bottom in blue. (B) Forest plot of rs827389 by cohort and by sex, including fixed-effects meta-analysis estimates. (C) Locus Zoom plots displaying the genomic region surrounding the chromosome 10 locus, by sex.
Functional annotation was performed on the genome-wide significant female chromosome 10 resilience locus (among cognitively normal individuals). All variants in linkage disequilibrium with rs827389 (top variant) were considered in annotation. The female locus was significantly enriched in Hi-C chromatin loops in multiple tissues, including foetal and adult cortex, aorta, left/right ventricle, liver, spleen, mesendoderm, mesenchymal stem cells, and trophoblast-like cells (Supplementary Table 14). Furthermore, enriched chromatin loops overlapped with promoter regions for multiple genes involved in RNA processing (Supplementary Table 15), including GATA3.
Alzheimer’s disease risk loci analysis
Sex-stratified and sex-interaction resilience associations with known Alzheimer’s disease genetic loci are presented in Supplementary Tables 16 and 17. In our residual cognitive resilience analysis, we observed eight nominally significant sex-interactions at three Alzheimer’s disease loci, whereby associations were male-specific for two loci, MS4A6A and PTK2B. Among males, the MS4A6A locus was positively associated with resilience and the PTK2B locus was negatively associated with resilience. Additionally, PICALM showed a flipped effect between sexes, whereby it was negatively associated in males and positively associated in females. Similarly, in our combined resilience analysis, we observed nine nominally significant sex-interactions at six Alzheimer’s disease loci, whereby associations were male-specific for four loci, MS4A6A, PTK2B, KAT8, and SORL1. Among males, the KAT8 and PTK2B loci were negatively associated with resilience, whereas the MS4A6A and SORL1 loci were positively associated with resilience.
Gene and pathway-level results
Gene test results are presented in Supplementary Tables 18–21 and pathway test results are presented in Supplementary Tables 22–25. We did not observe any genes or pathways that survived adjustment for multiple comparisons (FDR < 0.05) for any of the resilience phenotypes. However, we did observe one gene test for combined resilience that was close to surviving adjustment for multiple comparisons among females, and that gene was LEAP2 on chromosome 5 [PFDR(females) = 0.0998, PFDR(males) = 0.9971].
Genetic correlation results
We performed sex-stratified and sex-interaction genetic correlation analyses between our resilience phenotypes and 65 complex traits (Supplementary Tables 26–30). In this section, we will be presenting the male and female results, leveraging the sex-interaction results to aid in interpretation. For the entirety of the male, female and sex-interaction genetic correlation analysis results, see Supplementary Tables 26–30.
In our residual cognitive resilience phenotype, associations with 11 traits in males and six traits in females survived adjustment for multiple comparisons (FDR < 0.05) among all participants. For residual cognitive resilience among cognitively normal participants, 13 traits in males and nine traits in females survived adjustment for multiple comparisons (FDR < 0.05). In our combined resilience phenotype, associations with 22 traits in males and seven traits in females survived adjustment for multiple comparisons (FDR < 0.05) among all participants. For combined resilience among cognitively normal participants, 20 traits in males and 12 traits in females survived adjustment for multiple comparisons (FDR < 0.05).
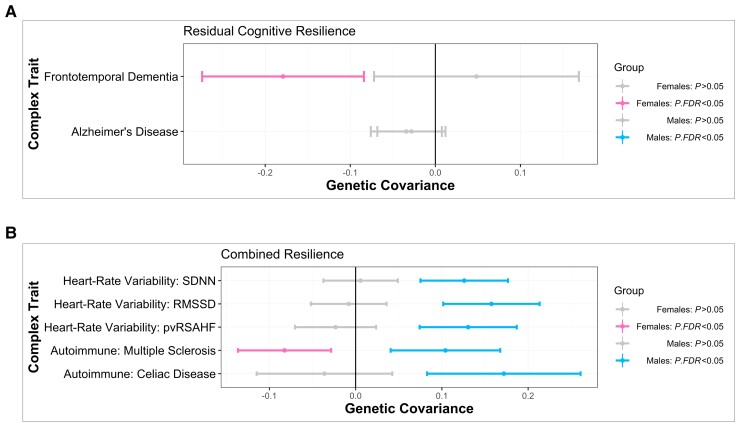
Of the traits that survived adjustment for multiple comparisons in at least one sex for residual cognitive resilience, one trait displayed a significant sex-interaction effect. This trait was frontotemporal dementia (FTD, Fig. 3A), and it was negatively associated in females, but was not associated in males. Of the traits that survived adjustment for multiple comparisons in at least one sex for residual cognitive resilience among cognitively normal individuals, three traits displayed a significant sex-interaction effect. These traits included asthma, cannabis dependence, and multiple sclerosis, whereby asthma and multiple sclerosis were negatively associated in females and cannabis dependence was positively associated in females, and none of the three traits were associated in males.
Figure 3.
Sex-specific shared genetic architecture between resilience and complex traits. Genetic covariance estimates with 95% confidence intervals are shown in the figure, with female estimates in pink and male estimates in blue. Grey confidence intervals denote a non-significant covariance estimate irrespective of sex. (A) Genetic covariance estimates with residual cognitive resilience, by sex, for Alzheimer’s disease and for FTD. (B) Genetic covariance estimates with combined resilience, by sex, for three HRV traits and for two autoimmune traits.
Of the traits that survived adjustment for multiple comparisons in at least one sex for combined resilience, six traits displayed a significant sex-interaction effect. These included Coeliac disease (Fig. 3B), SDNN (resting heart rate SD of NN intervals), resting heart rate pvRSA/HF (peak‐valley respiratory sinus arrhythmia or high frequency power) and resting heart rate RMSSD (root mean square of successive differences) (Fig. 3B), which were positively associated in males and not associated in females, as well as inflammatory bowel disease and sleep duration, which were negatively associated in males and were not associated in females. In addition, multiple sclerosis had opposing effects between sexes (Fig. 3B) such that it was negatively associated in females and positively associated in males. Of the traits that survived adjustment for multiple comparisons in at least one sex for combined resilience among cognitively normal individuals, seven traits displayed a significant sex-interaction effect. Internalizing problems was positively associated in females and not associated in males. Body mass index was negatively associated in males and not associated in females. Resting heart rate pvRSA/HF and resting heart rate RMSSD were positively associated in males and not associated in females. Multiple sclerosis and asthma had opposing effects between sexes such that both traits were positively associated in males and negatively associated in females.
Discussion
We performed a series of sex-aware genetic analyses on cognitive resilience phenotypes to characterize sex-specific variant, gene and pathway-level effects contributing to cognitive resilience to Alzheimer’s disease neuropathology. We identified a novel female-specific locus on chromosome 10, and we highlighted a number of high-quality female-specific candidate genes implicated in RNA processing linked to the top variant in the region. Finally, we characterized a number of novel sex-specific genetic covariances between cognitive resilience and relevant traits, including a female-specific association with frontotemporal dementia, male-specific associations with inflammatory bowel disease, sleep duration, body mass index and HRV traits, and opposing associations between sexes for asthma and multiple sclerosis. Overall, our results highlight the value of incorporating sex-stratified analyses into genetic studies of Alzheimer’s disease and suggest that female-specific genetic drivers of resilience may lie along immune-related pathways, while male-specific genetic drivers may fall along cardiovascular-related pathways.
Cognitive resilience is a highly heritable trait in both sexes
As we reported previously, the heritability estimates of resilience traits are slightly higher when restricting the sample to cognitively normal individuals,2 perhaps due to an increase in phenotypic heterogeneity when including individuals with dementia, and indeed we observed a similar pattern in the present heritability analyses when stratifying by sex. Specifically, in both male-stratified and female-stratified analyses, we observed higher heritability estimates that reached statistical significance among normal cognition participants (Table 2) and lower heritability estimates when including individuals with dementia (Table 2). While we did note slightly higher heritability estimates for males compared to females (Table 2), particularly for the combined resilience trait, the difference between sexes did not reach statistical significance. As sample sizes expand in future analyses, it will be interesting to see whether sex differences in heritability do emerge, but at present we can only conclude that these resilience traits appear to be heritable in a similar manner across sexes.
Sex-specific shared genetic architecture between cognitive resilience and autoimmune disorders
We observed sex-specific genetic covariances between resilience and autoimmune traits (Fig. 3B and Supplementary Tables 26–29), whereby genetic predisposition towards resilience was associated with reduced genetic risk for autoimmune traits among females (e.g. lupus, multiple sclerosis) and increased genetic risk for autoimmune traits among males (e.g. lupus, multiple sclerosis, Coeliac disease). It is notable that there are well-documented sex differences in trait prevalence for autoimmune disorders, with much higher trait prevalence in females compared to males.42,43 Alzheimer’s disease has known immune dysregulations and shares biology with autoimmune disorders, such as the imbalance of Th1 pro-inflammatory and Th2 anti-inflammatory cytokines in both Alzheimer’s disease and multiple sclerosis.42,43 Thus, it is perhaps intuitive that genetic factors that predispose lower susceptibility to autoimmune diseases are related to resilience among females. In contrast, it is unclear why males would show an inverse association with higher genetic susceptibility to autoimmune conditions relating to more genetic resilience to Alzheimer’s disease.
A sex difference in the genetic architecture of cognitive resilience could be due to differences in sex hormones, sex chromosomes or both. Sex hormones may modulate sex differences in the genetic aetiology of autoimmune disorders and resilience. Reproductive years account for the largest sex differences in autoimmune trait prevalence,42,43 and loss of oestrogen has a well established relationship with cognitive decline.44–46 In addition, males lose their sex hormones later in life than females, coinciding with when males tend to be diagnosed with autoimmune disorders.42,43 X-chromosome effects are another possible explanation, given the role of X-inactivation and X-chromosome instability in both autoimmunity and cognition.47–49 Recently, a second X chromosome was shown to promote survival in ageing50 and harbour resilience51 against Alzheimer’s disease in an ageing mouse model (irrespective of clinical Alzheimer’s disease risk), further supporting the possibility of X-chromosome effects as a mechanism underlying both autoimmunity and resilience. However, in contrast, mouse models of multiple sclerosis and lupus have shown that a second sex chromosome confers increased susceptibility.52,53 Additionally, having two X genes is aligned with more susceptibility to lupus in humans.54,55 Thus, it is unclear at this point whether X-chromosome effects could possibly be driving the resilience and autoimmunity sex difference we observed. A third possibility is that differences in metabolic processes between males and females explain genetic sex differences in autoimmunity and resilience. Age-related metabolic shifts tend to be coupled with increased neuroinflammation in females, whereas metabolic shifts do not show this same coupling in males.56,57 Regardless of the mechanism, our results suggest dramatic sex differences in the cognitive consequences of polygenic protection against autoimmune traits that deserve future attention.
Male-specific shared genetic architecture between cognitive resilience and cardiovascular traits
We observed male-specific positive genetic covariances with three heart rate variability traits, such that more resilient males had a higher genetic susceptibility to more favourable heart rate variability (Fig. 3B). It is well established that higher HRV is a marker of good heart health and that HRV decreases with ageing.58–60 The association between the genetic architecture of HRV and the genetic architecture of resilience could be due to (i) HRV driving more resilience in males; (ii) reverse causality with genetic factors that predispose towards cognitive resilience driving better HRV in males; or (iii) common genetic factors drive both HRV and resilience through independent pathways (i.e. pleiotropy). In support of a causal connection, evidence suggests that lower degrees of HRV are associated with cognitive impairment61,62 and that age-related HRV differences are sex dependent. Young, healthy females have a lower HRV compared to males, but with advanced age this sex difference is no longer apparent. In fact, elderly males tend to have lower HRV than elderly females,59 perhaps due to a survival bias4 from male susceptibility to midlife cardiovascular events.63 In support of reverse causality, resilience to both cognitive and HRV decline may work through similar circuitry, with prefrontal cortical brain circuitry as an example of this possible shared circuitry that could potentially drive better HRV.58,61 Multiple groups have shown a link between prefrontal cortical brain activity and HRV, with more than one group pointing towards HRV as a possible early marker of cognitive decline.58,61 While the possibility of better HRV driving resilience is exciting with some supporting evidence in the literature, future work must investigate each of these scenarios in great detail to determine causality.
Female-specific shared genetic architecture between cognitive resilience and FTD
We observed a significant negative genetic covariance in females between FTD and residual cognitive resilience (Fig. 3A), suggesting more resilient females are less genetically susceptible to FTD. Notably, the genetic covariance between resilience and Alzheimer’s disease was not significant in either sex (Fig. 3A). Illán-Gala and colleagues64 conducted a sex-aware analysis on behavioural variant FTD (bvFTD), the most common form of FTD, and observed that females had more cognitive reserve in FTD compared to males. Leveraging a residual approach similar to our study’s models, this group observed that females had better-than-expected executive function scores and less behavioural changes given pathology burden compared to males.64 Importantly, females had a higher amount of atrophy than males at FTD diagnosis, yet had similar disease progression.64 As mentioned previously, a recent study in ageing mouse models demonstrated that a second X chromosome promotes survival in ageing50 and resilience to Alzheimer’s disease51 irrespective of clinical Alzheimer’s disease risk. Taken together, this evidence from the literature and our FTD and Alzheimer’s disease genetic covariance findings all suggest that it may be the case that there is sex-specific shared genetic architecture of reserve/resilience across dementias subtypes, which contributes to disease protection agnostic to the underlying neuropathology.
However, Illán-Gala et al.64 also points out that Alzheimer’s disease is a posterior brain disease, with the anterior cingulum serving as a region of resilience in Alzheimer’s disease, whereas bvFTD is more of an anterior brain disease. Thus, possibly bvFTD sex-specific cognitive reserve/resilience brain regions are not the same as Alzheimer’s disease sex-specific cognitive reserve/resilience brain regions.64 Therefore, it may alternatively be the case that sex-specific reserve/resilience brain regions across dementia subtypes differ, but sex-specific genetic factors driving reserve/resilience are shared across subtypes and harbour some protection from the downstream consequences of neuropathology agnostic to dementia subtype. Perhaps, it could also be the case that we are observing an indirect effect between FTD and resilience, such that similar genetic architecture is independently contributing to both disorders. More sex-aware studies on the genetic architecture of reserve/resilience to different dementia subtypes will need to be conducted to determine causality.
Female-specific candidate genes implicated in RNA processing
Functional annotation of the chromosome 10 genome-wide significant locus (with rs827389) in cognitively normal females (Fig. 2) suggests its possible regulatory effects. This locus was significantly enriched in Hi-C chromatin loops at multiple gene promoters of genes implicated in RNA processing. These genes included: KIN, a DNA/RNA binding protein, TAF3, a TATA-box binding protein, and GATA3, a zinc-finger transcription factor (Supplementary Table 15). It is notable that a promising female-specific candidate gene we identified through functional annotation is GATA3, which encodes a ‘pioneer transcription factor’ that can bind heterochromatin and recruit factors to change chromatin state.65GATA3 is involved in sonic hedgehog signalling, a quintessential signalling pathway for pattern formation in neuronal development.66 It is also involved in embryonic development, influencing genes involved in extracellular matrix formation.67
In addition to involvement with neuronal development, GATA3 also controls immune T-cell fate.65 Specifically, GATA3 controls CD4+ effector cells: Th2 cells. CD4+ effector cells can produce autoantibodies against amyloid and therefore harbour protection against amyloid burden.68 It is thought that genetic drivers of immune-cell profiles may in part explain sex differences observed in response to Alzheimer’s disease pathology.69,70 Thus, it is noteworthy to see an immune-target arise at both the variant-level with GATA3, and at the whole genome level with our genetic correlation analyses. Taken together, this evidence points towards GATA3 as a female-specific candidate gene, and overall alludes to the idea that female resilience to Alzheimer’s disease pathology may involve regulation of RNA processing, although future studies need to replicate and to further elucidate this finding.
Alzheimer’s disease genetic loci associations with cognitive resilience trend towards male-specific effects
As shown in Supplementary Tables 16 and 17, we observed nominally significant sex-specific associations with resilience at well-known Alzheimer’s disease loci.35–37 These associations were male-specific at MS4A6A, PTK2B, KAT8 and SORL1, and a flipped effect between sexes was observed at the PICALM locus. Multiple groups have shown that SORL1 may exhibit sex-specificity, including evidence showing SORL1 variants to be detrimental in females.71–73 This aligns with what we observed in our study, as we saw a negative association with resilience at a SORL1 variant in females but a positive association in males. In addition, prior evidence suggests sex-specificity at the PICALM locus. A recent study of cognitively unimpaired individuals demonstrated a negative association in males as well as a sex-interaction association at a protective PICALM variant.74 This finding is consistent with what we observed, as we saw a negative association with resilience at a PICALM risk variant in males but a positive association in females. It is also noteworthy that PICALM contributes to multiple mechanisms involved in the Alzheimer’s disease neuropathological cascade, including neuroimmune processes.74 Taken together, this evidence may suggest that subtle sex differences at Alzheimer’s disease genetic loci may contribute to sex differences in the downstream response to Alzheimer’s disease pathology.
APOE effects not observed in resilient females
It was notable that we did not observe any statistically significant APOE × sex effects with resilience in our sensitivity analyses (Supplementary Table 13). Multiple groups have shown differential APOE effects by sex in females.12,75–78 As highlighted in our original manuscript, accounting for cognitive variance related to amyloid appears to massively reduce the APOE signal, and thus the reduced effect of APOE may partially explain the lack of APOE-by-sex effects here. Additionally, our cohort is on average older (mean age = 77) when the effects of APOE on cognition are attenuated,8,9 and the effects of circulating oestrogens (that have been hypothesized to modify APOE effects) are dramatically reduced.
Comparison of study findings across resilience phenotypes
Although significant variant associations and genetic covariances between residual cognitive resilience and combined resilience differed, the pattern of results was largely shared across resilience phenotypes. For example, the female chromosome 10 resilience locus that was genome-wide significant for combined resilience (with rs827389) fell just below genome-wide significant for residual cognitive resilience. In addition. the HRV genetic covariance findings in males and the multiple sclerosis genetic covariance finding in females in the combined resilience phenotype also held true in the residual cognitive resilience phenotype. However, it is notable that genetic covariances for more traits were significant in at least one sex for combined resilience compared to residual cognitive resilience. Overall, we are gaining additional power by leveraging a second-order latent variable, combined resilience, in genetic analyses due to the contributions of educational attainment in conferring resilience. We believe this framework should be integrated into future analyses.
Strengths and limitations
Our study had multiple strengths. We harmonized data across four deeply characterized cohorts of cognitive ageing, leveraged well-validated measures of amyloid, and implemented sex-aware statistical genetic analysis pipelines to identify sex-specific effects. Moreover, our variant, gene, pathway, and cross-trait analyses provide novel insight into the shared genetic architecture between cognitive resilience and other complex traits. However, our study also had limitations. We were underpowered to detect genome-wide sex-interaction effects. While we did not detect X-chromosome variant-level sex effects and we did not investigate imprinting, epigenetic or transcriptomic X-chromosome effects, we are excited for future projects to dive into X-chromosome biology in greater depth. Our study did not include measures of tau or other known age-related neuropathologies. In addition, we did not include measures of neurodegeneration. We were additionally limited in the cognitive domains included in our cognitive resilience models, including investigation of sub-domain effects. Finally, our study was limited to non-Hispanic white individuals, and there was limited heterogeneity in educational attainment, both attenuating the generalizability to other populations.
Conclusions
The findings of our sex-aware genetic study identified a locus, candidate genes, and molecular pathways that relate to resilience to the cognitive consequences of the Alzheimer’s disease neuropathological cascade in a sex-specific manner. Our findings suggest that the best target to enhance cognitive resilience to Alzheimer’s disease pathology may depend on both the biological sex and the genetic context of an individual.
Supplementary Material
Acknowledgements
Data used in preparation of this article were obtained from the ADNI database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf The results published here are in part based on data obtained from the AMP-AD Knowledge Portal (doi:10.7303/syn2580853). MSBB data were generated from post-mortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank and were provided by Dr Eric Schadt from Mount Sinai School of Medicine. MayoRNAseq data were provided by the following sources: The Mayo Clinic Alzheimer’s Disease Genetic Studies, led by Dr Nilufer Ertekin-Taner and Dr Steven G. Younkin, Mayo Clinic, Jacksonville, FL using samples from the Mayo Clinic Study of Aging, the Mayo Clinic Alzheimer’s Disease Research Center, and the Mayo Clinic Brain Bank. Study data includes samples collected through the Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories, and the NIMH Human Brain Collection Core. CMC Leadership: Panos Roussos, Joseph Buxbaum, Andrew Chess, Schahram Akbarian, Vahram Haroutunian (Icahn School of Medicine at Mount Sinai), Bernie Devlin, David Lewis (University of Pittsburgh), Raquel Gur, Chang-Gyu Hahn (University of Pennsylvania), Enrico Domenici (University of Trento), Mette A. Peters, Solveig Sieberts (Sage Bionetworks), Thomas Lehner, Stefano Marenco, Barbara K. Lipska (NIMH). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. The Alzheimer’s Disease Genetics Consortium supported genotyping, and data processing of samples through National Institute on Aging (NIA) grants U01-AG032984. Data for this study were prepared, archived, and distributed by the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania (U24-AG041689-01).
Abbreviations
- ACT
Adult Changes in Thought study
- ADNI
The Alzheimer’s Disease Neuroimaging Initiative
- APOE
apolipoprotein E
- FTD
frontotemporal dementia
- GWAS
genome-wide association study
- HRV
heart rate variability
- ROS/MAP
Religious Orders Study/Rush Memory and Aging Project
- SNP
single nucleotide polymorphism
- SUVR
standardized uptake value ratio
Contributor Information
Jaclyn M Eissman, Vanderbilt Memory and Alzheimer's Center, Vanderbilt University Medical Center, Nashville, TN, USA; Vanderbilt Genetics Institute, Vanderbilt University Medical Center, Nashville, TN, USA.
Logan Dumitrescu, Vanderbilt Memory and Alzheimer's Center, Vanderbilt University Medical Center, Nashville, TN, USA; Vanderbilt Genetics Institute, Vanderbilt University Medical Center, Nashville, TN, USA.
Emily R Mahoney, Vanderbilt Memory and Alzheimer's Center, Vanderbilt University Medical Center, Nashville, TN, USA; Vanderbilt Genetics Institute, Vanderbilt University Medical Center, Nashville, TN, USA.
Alexandra N Smith, Vanderbilt Memory and Alzheimer's Center, Vanderbilt University Medical Center, Nashville, TN, USA; Vanderbilt Genetics Institute, Vanderbilt University Medical Center, Nashville, TN, USA.
Shubhabrata Mukherjee, Department of Medicine, University of Washington, Seattle, WA, USA.
Michael L Lee, Department of Medicine, University of Washington, Seattle, WA, USA.
Phoebe Scollard, Department of Medicine, University of Washington, Seattle, WA, USA.
Seo Eun Choi, Department of Medicine, University of Washington, Seattle, WA, USA.
William S Bush, Cleveland Institute for Computational Biology, Department of Population and Quantitative Health Sciences, Case Western Reserve University, Cleveland, OH, USA.
Corinne D Engelman, Department of Population Health Sciences, School of Medicine and Public Health, University of Wisconsin-Madison, Madison, WI, USA.
Qiongshi Lu, Department of Statistics, University of Wisconsin-Madison, Madison, WI, USA; Department of Biostatistics and Medical Informatics, University of Wisconsin-Madison, Madison, WI, USA.
David W Fardo, Department of Biostatistics, College of Public Health, University of Kentucky, Lexington, KY, USA; Sanders-Brown Center on Aging, University of Kentucky, Lexington, KY, USA.
Emily H Trittschuh, Department of Psychiatry and Behavioral Sciences, University of Washington School of Medicine, Seattle, WA, USA; VA Puget Sound Health Care System, GRECC, Seattle, WA, USA.
Jesse Mez, Department of Neurology, Boston University School of Medicine, Boston, MA, USA.
Catherine C Kaczorowski, The Jackson Laboratory, Bar Harbor, ME, USA.
Hector Hernandez Saucedo, UC Davis Alzheimer's Disease Research Center, Department of Neurology, University of California Davis Medical Center, Sacramento, CA, USA.
Keith F Widaman, University of California at Riverside, Riverside, CA, USA.
Rachel F Buckley, Department of Neurology, Massachusetts General Hospital/Harvard Medical School, Boston, MA, USA; Center for Alzheimer's Research and Treatment, Department of Neurology, Brigham and Women’s Hospital/Harvard Medical School, Boston, MA, USA; Melbourne School of Psychological Sciences, University of Melbourne, Melbourne, Australia.
Michael J Properzi, Department of Neurology, Massachusetts General Hospital/Harvard Medical School, Boston, MA, USA.
Elizabeth C Mormino, Department of Neurology and Neurological Sciences, Stanford University, Stanford, CA, USA.
Hyun Sik Yang, Department of Neurology, Massachusetts General Hospital/Harvard Medical School, Boston, MA, USA; Center for Alzheimer's Research and Treatment, Department of Neurology, Brigham and Women’s Hospital/Harvard Medical School, Boston, MA, USA.
Theresa M Harrison, Helen Wills Neuroscience Institute, University of California Berkeley, Berkeley, CA, USA.
Trey Hedden, Icahn School of Medicine at Mount Sinai, New York City, NY, USA.
Kwangsik Nho, Department of Radiology and Imaging Sciences, Indiana Alzheimer Disease Center, Indiana University School of Medicine, Indianapolis, IN, USA; Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, IN, USA.
Shea J Andrews, Icahn School of Medicine at Mount Sinai, New York City, NY, USA.
Douglas Tommet, Department of Psychiatry and Human Behavior, Brown University School of Medicine, Providence, RI, USA.
Niran Hadad, The Jackson Laboratory, Bar Harbor, ME, USA.
R Elizabeth Sanders, Department of Medicine, University of Washington, Seattle, WA, USA.
Douglas M Ruderfer, Vanderbilt Genetics Institute, Vanderbilt University Medical Center, Nashville, TN, USA.
Katherine A Gifford, Vanderbilt Memory and Alzheimer's Center, Vanderbilt University Medical Center, Nashville, TN, USA.
Xiaoyuan Zhong, Department of Statistics, University of Wisconsin-Madison, Madison, WI, USA; Department of Biostatistics and Medical Informatics, University of Wisconsin-Madison, Madison, WI, USA.
Neha S Raghavan, Department of Neurology, Columbia University, New York, NY, USA; The Taub Institute for Research on Alzheimer's Disease and The Aging Brain, Columbia University, New York, NY, USA; The Institute for Genomic Medicine, Columbia University Medical Center and The New York Presbyterian Hospital, New York, NY, USA.
Badri N Vardarajan, Department of Neurology, Columbia University, New York, NY, USA; The Taub Institute for Research on Alzheimer's Disease and The Aging Brain, Columbia University, New York, NY, USA; The Institute for Genomic Medicine, Columbia University Medical Center and The New York Presbyterian Hospital, New York, NY, USA.
Margaret A Pericak-Vance, John P. Hussman Institute for Human Genomics, University of Miami School of Medicine, Miami, FL, USA.
Lindsay A Farrer, Department of Neurology, Boston University School of Medicine, Boston, MA, USA; Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA; Department of Medicine (Biomedical Genetics), Boston University School of Medicine, Boston, MA, USA.
Li San Wang, Penn Neurodegeneration Genomics Center, Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
Carlos Cruchaga, Department of Psychiatry, Washington University School of Medicine, St. Louis, MO, USA.
Gerard D Schellenberg, Penn Neurodegeneration Genomics Center, Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
Nancy J Cox, Vanderbilt Genetics Institute, Vanderbilt University Medical Center, Nashville, TN, USA.
Jonathan L Haines, Cleveland Institute for Computational Biology, Department of Population and Quantitative Health Sciences, Case Western Reserve University, Cleveland, OH, USA.
C Dirk Keene, Department of Pathology, University of Washington, Seattle, WA, USA.
Andrew J Saykin, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, USA.
Eric B Larson, Department of Medicine, University of Washington, Seattle, WA, USA; Kaiser Permanente Washington Health Research Institute, Seattle, WA, USA.
Reisa A Sperling, Department of Neurology, Massachusetts General Hospital/Harvard Medical School, Boston, MA, USA.
Richard Mayeux, Department of Neurology, Columbia University, New York, NY, USA; The Taub Institute for Research on Alzheimer's Disease and The Aging Brain, Columbia University, New York, NY, USA; The Institute for Genomic Medicine, Columbia University Medical Center and The New York Presbyterian Hospital, New York, NY, USA.
Michael L Cuccaro, John P. Hussman Institute for Human Genomics, University of Miami School of Medicine, Miami, FL, USA.
David A Bennett, Rush Alzheimer's Disease Center, Rush University Medical Center, Chicago, IL, USA.
Julie A Schneider, Rush Alzheimer's Disease Center, Rush University Medical Center, Chicago, IL, USA.
Paul K Crane, Department of Medicine, University of Washington, Seattle, WA, USA.
Angela L Jefferson, Vanderbilt Memory and Alzheimer's Center, Vanderbilt University Medical Center, Nashville, TN, USA.
Timothy J Hohman, Vanderbilt Memory and Alzheimer's Center, Vanderbilt University Medical Center, Nashville, TN, USA; Vanderbilt Genetics Institute, Vanderbilt University Medical Center, Nashville, TN, USA.
Funding
This research was supported in part by K01-AG049164, R01-AG059716, R21-AG05994, K12-HD043483, K24-AG046373, HHSN311201600276P, S10-OD023680, R01-AG034962, R01-NS100980, R01-AG056534, P30-AG010161, P30-AG072975, R01-AG057914, R01-AG015819, R01-AG017917, R13-AG030995, U01-AG061356, U01-AG006781, U19-AG066567, K99/R00-AG061238, U01-AG046152, U01-AG068057, UL1-TR000445, T32-GM080178, R01-AG073439, U24-AG074855, P20-AG068082 (Vanderbilt Alzheimer’s Disease Research Center), and the Vanderbilt Memory & Alzheimer’s Center. Data collection was supported through funding by NIA grants P50-AG016574, P50-AG005136, R01-AG032990, U01-AG046139, R01-AG018023, U01-AG006576, U01-AG006781, U01-AG006786, R01-AG025711, R01-AG017216, R01-AG003949, P30-AG019610, U01-AG024904, U01-AG032984, U24-AG041689, R01-AG046171, RF1-AG051550, 3U01-AG024904-09S4, NINDS grant R01-NS080820, CurePSP Foundation, and support from Mayo Foundation.
The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24-NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30-AG019610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson's Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research. Data were generated as part of the CommonMind Consortium supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and NIH grants R01-MH085542, R01-MH093725, R01-AG074012, P50-MH066392, P50-MH080405, R01-MH097276, R01-MH075916, P50-M096891, P50-MH084053S1, R37-MH057881, AG02219, AG05138, MH06692, R01-MH110921, R01-MH109677, R01-MH109897, U01-MH103392, and contract HHSN271201300031C through IRP NIMH. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01-AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc; Biogen Inc Cambridge, MA 02139, provided support for genotyping of the A4 Study cohort; Bristol-Myers Squibb Company; CereSpir, Inc; Cogstate; Eisai Inc; Elan Pharmaceuticals, Inc; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). Additional data collection and sharing for this project was funded by the Alzheimer's Disease Metabolomics Consortium (National Institute on Aging R01-AG046171, RF1-AG051550 and 3U01-AG024904-09S4).
Competing interests
T.J.H. sits on the scientific advisory board for Vivid Genomics. R.A.S. receives research funding from Eli Lilly and Janssen and has served as a paid consultant to AC Immune, Biogen, Janssen and Neurocentria. A.J.S. receives [F18]Flortaucipir (AV-1451) precursor support from Avid Radiopharmaceuticals. E.B.L. reports royalties from UpToDate. J.A.S. reports personal fees from Avid Radiopharmaceuticals and from Navidea Biopharmaceuticals. The remaining authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Driscoll I, Troncoso J. Asymptomatic Alzheimers disease: A prodrome or a state of resilience? Curr Alzheimer Res. 2011;8:330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dumitrescu L, Mahoney ER, Mukherjee S, et al. . Genetic variants and functional pathways associated with resilience to Alzheimer’s disease. Brain. 2020;143:2561–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barnes LL, Wilson RS, Bienias JL, Schneider JA, Evans DA, Bennett DA. Sex differences in the clinical manifestations of Alzheimer disease pathology. Arch Gen Psychiatry. 2005;62:685–691. [DOI] [PubMed] [Google Scholar]
- 4. Dumitrescu L, Mayeda ER, Sharman K, Moore AM, Hohman TJ. Sex differences in the genetic architecture of Alzheimer’s disease. Curr Genet Med Rep. 2019;7:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koran MI, Wagener MA, Hohman TJ. Sex differences in the association between AD biomarkers and cognitive decline. Brain Imaging Behav. 2017;11:205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buckley RF, Scott MR, Jacobs HIL, et al. . Sex mediates relationships between regional tau pathology and cognitive decline. Ann Neurol. 2020;88:921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buckley RF, Mormino EC, Chhatwal J, et al. . Associations between baseline amyloid, sex, and APOE on subsequent tau accumulation in cerebrospinal fluid. Neurobiol Aging. 2019;78:178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Farrer LA, Cupples LA, Haines JL, et al. . Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 9. Neu SC, Pa J, Kukull W, et al. . Apolipoprotein E genotype and sex risk factors for Alzheimer disease: A meta-analysis. JAMA Neurol. 2017;74:1178–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sampedro F, Vilaplana E, de Leon MJ, et al. . APOE-by-sex interactions on brain structure and metabolism in healthy elderly controls. Oncotarget. 2015;6:26663–26674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buckley RF, Mormino EC, Amariglio RE, et al. . Sex, amyloid, and APOE ε4 and risk of cognitive decline in preclinical Alzheimer’s disease: Findings from three well-characterized cohorts. Alzheimers Dement. 2018;14:1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hohman TJ, Dumitrescu L, Barnes LL, et al. . Sex-specific effects of Apolipoprotein E with cerebrospinal fluid levels of tau. JAMA Neurol. 2018;75:989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deming Y, Dumitrescu L, Barnes LL, et al. . Sex-specific genetic predictors of Alzheimer’s disease biomarkers. Acta Neuropathol. 2018;136:857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dumitrescu L, Barnes LL, Thambisetty M, et al. . Sex differences in the genetic predictors of Alzheimer’s pathology. Brain. 2019;142:2581–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu Y, Zhang H, Liu B, et al. . rs34331204 Regulates TSPAN13 expression and contributes to Alzheimer’s disease with sex differences. Brain. 2020;143:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hohman TJ, McLaren DG, Mormino EC, Gifford KA, Libon DJ, Jefferson AL. Asymptomatic Alzheimer disease. Neurology. 2016;87:2443–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sperling RA, Rentz DM, Johnson KA, et al. . The A4 study: stopping AD before symptoms begin? Sci Transl Med. 2014;6:228fs13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kukull WA, Higdon R, Bowen JD, et al. . Dementia and Alzheimer disease incidence: a prospective cohort study. Arch Neurol. 2002;59:1737–1746. [DOI] [PubMed] [Google Scholar]
- 19. Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA. Religious orders study and rush memory and aging project. J Alzheimers Dis. 2018;64:S161–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Properzi MJ, Buckley RF, Chhatwal JP, et al. . Nonlinear Distributional Mapping (NoDiM) for harmonization across amyloid-PET radiotracers. Neuroimage. 2019;186:446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Raghavan NS, Dumitrescu L, Mormino E, et al. . association between common variants in RBFOX1, an RNA-binding protein, and brain amyloidosis in early and preclinical Alzheimer disease. JAMA Neurol. 2020;77:1288–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mirra SS, Heyman A, McKeel D, et al. . The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology. 1991;41:479–486. [DOI] [PubMed] [Google Scholar]
- 23. Crane PK, Trittschuh E, Mukherjee S, et al. . Incidence of cognitively defined late-onset Alzheimer’s dementia subgroups from a prospective cohort study. Alzheimers Dement. 2017;13:1307–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muthén LK, Muthén BO. Mplus User’s Guide. 7th edn. Los Angeles, CA: Muthén & Muthén; 1998. p. 2015.
- 25. Das S, Forer L, Schonherr S, et al. . Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taliun D, Harris DN, Kessler MD, et al. . Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. Nature. 2021;590:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Demontis D, Walters RK, Martin J, et al. . Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin J, Khramtsova EA, Goleva SB, et al. . Examining sex-differentiated genetic effects across neuropsychiatric and behavioral traits. Biol Psychiatry. 2021;89:1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watanabe K, Umićević Mirkov M, de Leeuw CA, van den Heuvel MP, Posthuma D. Genetic mapping of cell type specificity for complex traits. Nat Commun. 2019;10:3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jansen IE, Savage JE, Watanabe K, et al. . Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51:404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kunkle BW, Grenier-Boley B, Sims R, et al. . Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. . Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu Q, Li B, Ou D, et al. . A powerful approach to estimating annotation-stratified genetic covariance via GWAS summary statistics. Am J Hum Genet. 2017;101:939–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Viechtbauer W. Conducting meta-analyses in R with the metafor Package. J Stat Softw. 2010;36:1–48. [Google Scholar]
- 42. Voskuhl R. Sex differences in autoimmune diseases. Biol Sex Differ. 2011;2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Voskuhl RR. The effect of sex on multiple sclerosis risk and disease progression. Mult Scler. 2020;26:554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McLay RN, Maki PM, Lyketsos CG. Nulliparity and late menopause are associated with decreased cognitive decline. J Neuropsychiatry Clin Neurosci. 2003;15:161–167. [DOI] [PubMed] [Google Scholar]
- 45. Ryan J, Carrière I, Scali J, Ritchie K, Ancelin ML. Life-time estrogen exposure and cognitive functioning in later life. Psychoneuroendocrinology. 2009;34:287–298. [DOI] [PubMed] [Google Scholar]
- 46. Ryan J, Scali J, Carriere I, et al. . Impact of a premature menopausezon cognitive function in later life. BJOG. 2014;121:1729–1739. [DOI] [PubMed] [Google Scholar]
- 47. Bianchi I, Lleo A, Gershwin ME, Invernizzi P. The X chromosome and immune associated genes. J Autoimmun. 2012;38:J187–J192. [DOI] [PubMed] [Google Scholar]
- 48. Brooks WH. X chromosome inactivation and autoimmunity. Clin Rev Allergy Immunol. 2010;39:20–29. [DOI] [PubMed] [Google Scholar]
- 49. Ozcelik T. X chromosome inactivation and female predisposition to autoimmunity. Clin Rev Allergy Immunol. 2008;34:348–351. [DOI] [PubMed] [Google Scholar]
- 50. Davis EJ, Lobach I, Dubal DB. Female XX sex chromosomes increase survival and extend lifespan in aging mice. Aging Cell. 2019;18:e12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davis EJ, Broestl L, Abdulai-Saiku S, et al. . A second X chromosome contributes to resilience in a mouse model of Alzheimer’s disease. Sci Transl Med. 2020;12:eaaz5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith-Bouvier DL, Divekar AA, Sasidhar M, et al. . A role for sex chromosome complement in the female bias in autoimmune disease. J Exp Med. 2008;205:1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sasidhar MV, Itoh N, Gold SM, Lawson GW, Voskuhl RR. The XX sex chromosome complement in mice is associated with increased spontaneous lupus compared with XY. Ann Rheum Dis. 2012;71:1418–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Scofield RH, Bruner GR, Namjou B, et al. . Klinefelter’s syndrome (47, XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008;58:2511–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu K, Kurien BT, Zimmerman SL, et al. . X chromosome dose and sex bias in autoimmune diseases: increased prevalence of 47. XXX in systemic lupus erythematosus and Sjögren’s syndrome. Arthritis Rheumatol. 2016;68:1290–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Klosinski LP, Yao J, Yin F, et al. . White matter lipids as a ketogenic fuel supply in aging female brain: implications for Alzheimer’s disease. EBioMedicine. 2015;2:1888–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shang Y, Mishra A, Wang T, et al. . Evidence in support of chromosomal sex influencing plasma based metabolome vs APOE genotype influencing brain metabolome profile in humanized APOE male and female mice. PLoS One. 2020;15:e0225392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Grässler B, Hökelmann A, Cabral RH. Resting heart rate variability as a possible marker of cognitive decline. Kinesiology. 2020;52:72–84. [Google Scholar]
- 59. Umetani K, Singer DH, McCraty R, Atkinson M. Twenty-four hour time domain heart rate variability and heart rate: relations to age and gender over nine decades. J Am Coll Cardiol. 1998;31:593–601. [DOI] [PubMed] [Google Scholar]
- 60. Umetani K, Duda CL, Singer DH. Aging effects on cycle length dependence of heart rate variability. IEEE; 1996:361–364. [Google Scholar]
- 61. Shah AJ, Su S, Veledar E, et al. . Is heart rate variability related to memory performance in middle-aged men? Psychosom Med. 2011;73:475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zulli R, Nicosia F, Borroni B, et al. . QT dispersion and heart rate variability abnormalities in Alzheimer’s disease and in mild cognitive impairment. J Am Geriatr Soc. 2005;53:2135–2139. [DOI] [PubMed] [Google Scholar]
- 63. Gao Z, Chen Z, Sun A, Deng X. Gender differences in cardiovascular disease. Med Nov Technol Devices. 2019;4:100025. [Google Scholar]
- 64. Illán-Gala I, Casaletto KB, Borrego-Écija S, et al. . Sex differences in the behavioral variant of frontotemporal dementia: A new window to executive and behavioral reserve. Alzheimers Dement. 2021;16:1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tremblay M, Sanchez-Ferras O, Bouchard M. GATA transcription factors in development and disease. Development. 2018;145:dev164384. [DOI] [PubMed] [Google Scholar]
- 66. Liu J, Wang X, Li J, Wang H, Wei G, Yan J. Reconstruction of the gene regulatory network involved in the sonic hedgehog pathway with a potential role in early development of the mouse brain. PLoS Comput Biol. 2014;10:e1003884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lee B, Kroener LL, Xu N, et al. . Function and hormonal regulation of GATA3 in human first trimester placentation. Biol Reprod. 2016;95:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kubick N, Flournoy PCH, Enciu AM, Manda G, Mickael ME. Drugs modulating CD4+ T cells blood-brain barrier interaction in Alzheimer’s disease. Pharmaceutics. 2020;12:880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lopez-Lee C, Kodama L, Gan L. Sex differences in neurodegeneration: the role of the immune system in humans. Biol Psychiatry. 2021;91:72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mathys H, Davila-Velderrain J, Peng Z, et al. . Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cellini E, Tedde A, Bagnoli S, et al. . Implication of sex and SORL1 variants in italian patients with Alzheimer disease. Arch Neurol. 2009;66:1260–1266. [DOI] [PubMed] [Google Scholar]
- 72. Liang Y, Li H, Lv C, et al. . Sex moderates the effects of the Sorl1 gene rs2070045 polymorphism on cognitive impairment and disruption of the cingulum integrity in healthy elderly. Neuropsychopharmacology. 2015;40:1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Reynolds CA, Zavala C, Gatz M, et al. . Sortilin receptor 1 predicts longitudinal cognitive change. Neurobiol Aging. 2013;34:1710.e11–1710.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Heath L, Earls JC, Magis AT, et al. . Manifestations of genetic risk for Alzheimer’s Disease in the blood: a cross-sectional multiomic analysis in healthy adults aged 18-90 + . bioRxiv. https://www.biorxiv.org/content/10.1101/2021.03.26, 2021 [Google Scholar]
- 75. Yaffe K, Haan M, Byers A, Tangen C, Kuller L. Estrogen use, APOE, and cognitive decline: evidence of gene-environment interaction. Neurology. 2000;54:1949–1954. [DOI] [PubMed] [Google Scholar]
- 76. Riedel BC, Thompson PM, Age BR. APOE and sex: Triad of risk of Alzheimer’s disease. J Steroid Biochem Mol Biol. 2016;160:134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stephen TL, Cacciottolo M, Balu D, et al. . APOE genotype and sex affect microglial interactions with plaques in Alzheimer’s disease mice. Acta Neuropathol Commun. 2019;7:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang Y, Hernandez G, Mack WJ, Schneider LS, Yin F, Brinton RD. Retrospective analysis of phytoSERM for management of menopause-associated vasomotor symptoms and cognitive decline: a pilot study on pharmacogenomic effects of mitochondrial haplogroup and APOE genotype on therapeutic efficacy. Menopause. 2020;27:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the ADNI and A4 studies are shared through the LONI Image and Data Archive (https://ida.loni.usc.edu/). Data from ROS/MAP can be requested at www.radc.rush.edu. Data from ACT can be accessed through the Data Query Tool (http://act.kpwashingtonresearch.org/dqt/). GWAS summary statistics will be available through NIAGADS (https://www.niagads.org/datasets/).