Abstract
Frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) is a complex heterogeneous neurodegenerative disorder for which mechanisms are poorly understood.
To explore transcriptional changes underlying FTLD-TDP, we performed RNA-sequencing on 66 genetically unexplained FTLD-TDP patients, 24 FTLD-TDP patients with GRN mutations and 24 control participants. Using principal component analysis, hierarchical clustering, differential expression and coexpression network analyses, we showed that GRN mutation carriers and FTLD-TDP-A patients without a known mutation shared a common transcriptional signature that is independent of GRN loss-of-function. After combining both groups, differential expression as compared to the control group and coexpression analyses revealed alteration of processes related to immune response, synaptic transmission, RNA metabolism, angiogenesis and vesicle-mediated transport. Deconvolution of the data highlighted strong cellular alterations that were similar in FTLD-TDP-A and GRN mutation carriers with NSF as a potentially important player in both groups. We propose several potentially druggable pathways such as the GABAergic, GDNF and sphingolipid pathways.
Our findings underline new disease mechanisms and strongly suggest that affected pathways in GRN mutation carriers extend beyond GRN and contribute to genetically unexplained forms of FTLD-TDP-A.
Keywords: frontotemporal lobar degeneration, transcriptome, GRN mutation, GABA, deconvolution
To better understand the disease processes underlying FTLD, Pottier et al. compare gene expression in patients versus healthy controls. Patients with GRN mutations have a similar transcriptional profile to genetically unexplained patients with similar brain pathology, with alterations in several potentially druggable pathways.
Introduction
Frontotemporal lobar degeneration (FTLD), the second most common early onset neurodegenerative disorder, is highly heterogeneous. A definite diagnosis of FTLD can only be obtained after brain autopsy. The main pathology observed in FTLD patients is FTLD-TDP, named after the TDP-43 protein aggregates found in the brain. According to the type and localization of TDP-43 aggregates in the cortical layers, FTLD-TDP is classified into five pathological subtypes A to E, with A to C being the most common.1,2 FTLD-TDP subtype A (FTLD-TDP-A) is characterized by abundant short dystrophic neurites, oval neuronal cytoplasmic inclusions in layer II of the neocortex and scattered lentiform intranuclear inclusions; FTLD-TDP subtype B (FTLD-TDP-B) patients on the other hand have few short dendritic neurites and moderate amount of neuronal cytoplasmic inclusions in deep cortical laminae; and in FTLD-TDP subtype C (FTLD-TDP-C) numerous long dystrophic neurites in upper cortical layers are reported, with low amount of neuronal cytoplasmic inclusion.
Several genes and genetic risk factors of FTLD have been described.3–10 Importantly, there is a correlation between monogenic causes and FTLD-TDP subtypes. For instance, patients with a repeat expansion in the C9orf72 gene present mostly with FTLD-TDP-B at autopsy, while even more strikingly, patients with mutations in the gene encoding progranulin (GRN) invariably present with FTLD-TDP-A pathology.11 Most FTLD-TDP-A patients, however, do not carry GRN mutations and are genetically unexplained. They represent ∼32% of the FTLD-TDP population.8 We reported that homozygous carriers of the rare allele of rs5848, located in a microRNA binding site within the GRN 3′UTR,12 are enriched in a genetically unexplained FTLD-TDP-A population as compared to controls with a 5-fold increased odds ratio.8 The rs5848 risk allele is associated with reduced GRN expression in cerebellum, plasma and CSF.12–14 Such correlations suggest a common aetiology between GRN mutation carriers and genetically unexplained FTLD-TDP-A patients. On the contrary, no genetic factors or specific pathways, have been associated with FTLD-TDP type C patients (FTLD-TDP-C).
Apart from genetic studies, important clues about the disease pathophysiology can be obtained from transcriptomic studies. Recently, a bulk RNA-sequencing study showed that Golgi vesicular transport, GABAergic signalling and cell death are affected in patients carrying C9orf72 repeat expansions as compared to non-expansion carriers and controls.15 Also, Chen-Plotkin et al.16 reported impairment of synaptic transmission and calcium signalling in GRN mutation carriers using a microarray approach.
To further dissect the transcriptional events underlying the different FTLD-TDP subtypes, with particular emphasis on the relationship between GRN-positive and GRN-negative FTLD-TDP-A, we performed bulk RNA-sequencing on the frontal cortex and cerebellum (a relatively less affected brain region) in a large cohort of FTLD-TDP patients and controls. We used multiple analytical methods including differential expression and coexpression analyses to identify genes and pathways associated with each FTLD-TDP subtype. We also aimed to identify targetable pathways relevant for FTLD-TDP patients.
Materials and methods
Participants
Participants with the frontal cortex and cerebellum available were selected from the Mayo Clinic Florida Brain Bank (n = 114; Table 1). Frontal-cortex tissue was collected from the middle frontal gyrus at the level of the nucleus accumbens. We included 24 FTLD-TDP-A, 20 FTLD-TDP-B and 22 FTLD-TDP-C, 24 GRN mutation carriers and 24 neuropathologically normal participants without any neurological disease (referred to as controls). FTLD-TDP-A patients had a median age at death of 83 [interquartile range (IQR): 79.0–86.8] and 12% were female. FTLD-TDP-B patients had a median age at death of 66.5 (IQR: 58.5–72.0) and 10% were female. FTLD-TDP-C patients had a median age at death of 74.5 (IQR: 67.8–78.0) and 8% were female. GRN mutation carriers had a median age at death of 66.5 (IQR: 64.0–75.8) and 14% were female. Controls had a median age at death of 86.5 (IQR: 79.3–89.8) and 16% were female. Patients with FTLD-TDP-A were all negative for GRN mutations as determined by Sanger sequencing. All patients were negative for C9orf72 repeat expansion using our previously reported protocol.6 Cerebellum tissue was collected for a subset of these samples including 22 FTLD-TDP-A, 18 FTLD-TDP-B, 19 FTLD-TDP-C, 20 GRN mutation carriers and 21 controls. The tissues sampled did not include primary motor cortex. All GRN mutation carriers had type A histology at autopsy. Using principal component analysis on available genomic data, we determined that all samples were non-Hispanic white, except for two controls from African–American ancestry (Supplementary Fig. 1). The pathological diagnosis of FTLD-TDP was considered sufficient for inclusion, irrespective of the clinical diagnosis of the patient. However, 50% of FTLD-TDP-A patients were referred to the brain bank as clinical Alzheimer's disease, 21% as frontotemporal dementia (FTD) and 29% with other diagnoses that are part of the FTLD spectrum. GRN mutation carriers had a variety of clinical diagnoses with 20% referred to the brain bank as Alzheimer's disease, 20% with nfvPPA, 20% with FTD and 40% with other diagnoses that are part of the FTLD spectrum. Twenty-five per cent of patients with FTLD-TDP-B were diagnosed with behavioural variant FTD (bvFTD) with motor neuron disease, 20% with motor neuron disease alone and 60% with other diagnoses that are part of the FTLD spectrum. Thirty-six per cent of FTLD-TDP-C patients were diagnosed with bvFTD, 22% with svPPA and 42% with other diagnoses that are part of the FTLD spectrum. Genotypes of the rs5848 polymorphism were obtained by either direct Sanger sequencing of the GRN gene, or by a standard protocol TaqMan reaction with assay ID C_7452046_20 run on a QuantStudio 7 PCR System.
Table 1.
Cohort characteristics
| Median age at death (IQR) | n female (%) | RIN FCX (IQR) | n total | |
|---|---|---|---|---|
| Controls | 86.5 (79.3–89.8) | 16 (66.7) | 9.1 (8.7–9.8) | 24 |
| FTLD-TDP-A | 83 (79–86.8) | 12 (50.0) | 9.5 (9.0–9.7) | 25 |
| FTLD-TDP-B | 66.5 (58.5–72) | 10 (50.0) | 9.8 (9.0–9.9) | 20 |
| FTLD-TDP-C | 74.5 (67.8–78) | 8 (36.4) | 9.3 (8.7–9.7) | 22 |
| GRN mutation carriers | 66.5 (64–75.8) | 14 (56.0) | 8.9 (8.2–9.5) | 24 |
Information is shown for FTLD-TDP patients without mutation in the known genes, control participants without neurological diseases (Controls) and patients carrying a pathogenic mutation in the GRN gene (GRN mutation carriers). Age at death, RNA integrity number (RIN), number (n) of female participants and pathological diagnosis (FTLD-TDP-A, FTLD-TDP-B, FTLD-TDP-C) are specified. Data presented are median age at death [interquartile range (IQR)] or n and percentages (%).
RNA-sequencing
RNA from frontal-cortex tissue of 90 FTLD-TDP patients and 24 controls as well as from cerebellum tissue of 79 FTLD-TDP patients and 21 controls were extracted using the RNeasy Plus mini kit (Qiagen, Venlo, Netherlands). RNA quality and quantity were assessed using an Agilent 2100 Bioanalyzer and the RNA Nano Chip (Agilent Technologies). Only samples with an RNA integrity number (RIN) above seven were included in the study, with a median of 9.3 for the frontal cortex (IQR: 8.70–9.73) and 9.15 for cerebellum (IQR: 8.43–9.70). Library preparation was performed using Illumina TruSeq mRNA v.2 prep and sequenced at 10 samples/lane as paired-end 101 base pair reads on the HiSeq4000 (Illumina). Raw RNA-sequencing reads were aligned to the human reference genome (GRCh38) using the spliced transcripts alignment to a reference (STAR, v.2.5.2b).17 Library quality assessment was performed using the RSeQC (v.3.0.0) package.18 Gene-level expression was quantified using the featureCounts command in the Subread package (v.1.5.1).19
Differential expression analyses
Statistical analyses and plots were generated using open source R software packages available from CRAN and Bioconductor. Counts per million were obtained using the edgeR package using default parameters.20 Samples were removed if their sex estimated from the RNA-sequencing data did not match the phenotypic information. Transcripts with low expression were removed using the filterByExpr function from edgeR package. Principal component and hierarchical cluster analysis using the Pearson correlation distance with average linkage were performed using the stats (v.3.6.2) and gplots packages. A negative binomial model was used to determine differentially expressed genes between groups. All analyses were adjusted for sex, RIN, batch and age. A Benjamini–Hochberg false discovery rate (FDR) correction was used for multiple testing. Genes with an FDR below 5% were considered statistically significant. Gene ontology (GO) analysis was performed using the anRichment package. GO terms with an FDR < 0.05 were considered significant. The up- and downregulated genes were used independently as input for STRING protein–protein interaction (PPI) network analysis. Only physical networks were considered based on text-mining, experiments and databases. Interacting proteins with a high confidence of 0.7 were used as input for Metascape PPI network analyses using default parameters.21 In addition to using the STRING database, Metascape also queries the Biogrid database. The resulting networks and subnetworks were obtained with the subset of proteins that form physical interactions with at least one other list member. Using the means of Molecular Complex Detection algorithm, we identified connected subnetwork components. Each subnetwork was further subjected to GO enrichment analyses and drug target finding using the Opentarget database.22 Venn diagrams were generated with the ggvenn package, volcano plots were created using the EnhancedVolcano package.
Coexpression analyses
We used the R package weighted gene coexpression network analysis (WGCNA) to identify sets of highly correlated transcripts (modules),23 using as input the residual expression values adjusted for the aforementioned covariates. Residuals were obtained after conditional quantile normalization using the cqn package and the use of a multivariable linear regression model. Signed hybrid networks were created using the bi-weight mid-correlation method. To achieve a scale-free topology, we selected a power appropriate for each comparison (between 9 and 16). A dynamic tree cutting method was used with a minimum module size of 30 and a merged height varying from 0.2 to 0.3, depending on the comparison. Modules generated using these settings were represented by their first principal component (module eigengene) and a unique colour. For every gene, correlations between expression levels and each module's eigengene value (module membership) were calculated.
Modules that correlated with disease status were visualized using the top 20 most connected genes (hubs) as input for Cytoscape. In these networks, the connectivity of each gene was represented by the size of its node, the strength of the correlation is reflected by the thickness of its edges and their differential expression status is represented by the colour of the outer circle. The entire modules were then subject to enrichment analyses, as described before. Additionally, GO terms were collapsed using default parameters of the REViGO algorithm and the CirGO python library.24,25 The top 500 hub genes from the green, purple and turquoise modules were used as input in the Metascape website for PPI analysis, as previously described. Networks were visualized with Cytoscape software,26 followed by GO enrichment and drug target finding using the Opentarget database.
To construct consensus networks, all frontal-cortex data or only transcripts in common between frontal-cortex and cerebellum datasets were included, depending on the analysis. Similar parameters as the ones described previously were used for scale-free topology, module size and merged height. Correlation with disease status was performed as previously described. Fisher's exact test was used to compare the number of modules correlating with disease status in the FTLD-TDP-A and GRN consensus as compared to FTLD-TDP-C and GRN consensus. Conservation between modules across both tissues was assessed using a hypergeometric test for each of the pairwise overlaps.
Cell proportion estimation
Cell proportions were estimated using the R DSA package.27 Marker genes were obtained using methods described elsewhere.28 Briefly, we used the top 500 human specific markers from BRETIGEA R package for each cell type (neurons, oligodendrocytes, astrocytes and endothelial cells).29 From this list, we randomly selected 100 genes for each cell type, and estimated cell proportions using the DSA algorithm. We repeated this 100 times and computed a Pearson correlation of estimated cell proportions between different runs. We then selected a representative subset of 100 marker genes per cell type as marker input for DSA analysis, which consists of computing the average marker gene expression in the dataset per individual. Differences in cell-type proportion per group of patients was assessed using a FDR corrected Kruskal–Wallis test.
We then applied the PSEA30 method that allows model selection of the cell type(s) that should be included in controls or FTLD patients for each gene. Differential expression was subsequently estimated in specific cell types. For this, we used the functions em_quantvg and lmfitst and FDR correction. Finally, dysregulated genes for each cell type with a P-valueFDR < 0.05 were used as input into the IPA software for canonical pathway analysis using default settings (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis).31
Data availability
The datasets generated herein are available in the Synapse platform under accession number ‘syn25991921’. All other relevant data supporting key findings of this study are available in the article and Supplementary material.
Results
Differential expression of FTLD-TDP subgroups compared to control individuals
We performed RNA-sequencing on frontal-cortex tissue of FTLD-TDP patients (n = 66) without mutations in known FTLD-TDP genes including 24 FTLD-TDP-A, 20 FTLD-TDP-B and 22 FTLD-TDP-C, 24 patients with GRN mutations and 24 controls (Table 1). When performing quality control, two participants were excluded from this dataset based on sex discrepancies between reported and estimated sex, for a total of 88 patients and 24 controls.
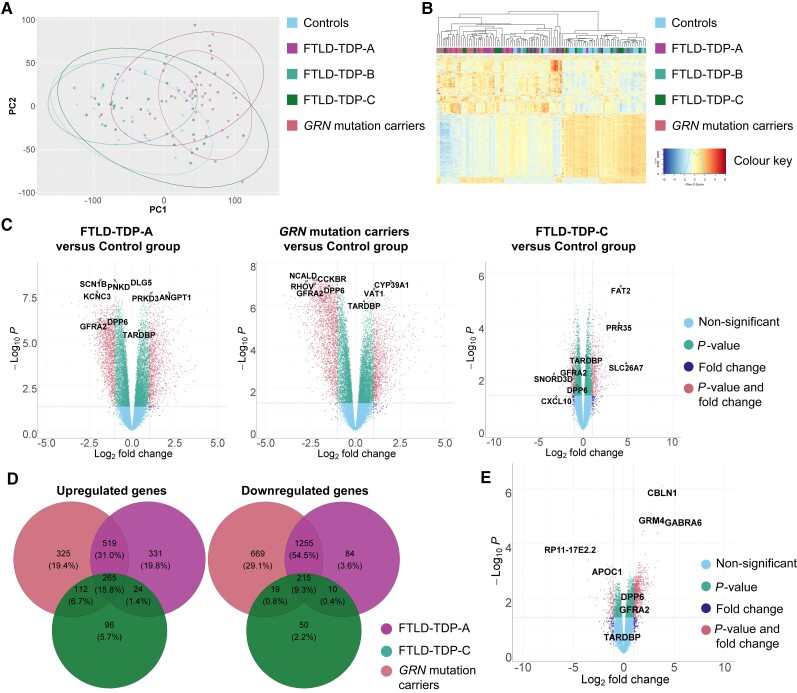
Performing a principal component analysis, we found that the FTLD-TDP-A and GRN groups cluster together (Fig. 1A). Principal components 1 and 2 explained 34% and 9% of the variance, respectively (eigenvalue = 3.944 × 103, eigenvalue = 1.04 × 102, respectively). Confirming this finding, hierarchical clustering showed clustering of the FTLD-TDP-A and GRN mutation carriers (Fig. 1B).
Figure 1.
Differential expression in FTLD-TDP groups versus controls. (A) Principal component (PC) 1 is represented on the x-axis and principal component 2 on the y-axis. Individuals with GRN mutation and pertaining to the FTLD-TDP-A group are coloured in pink and purple. Individuals pertaining to the FTLD-TDP-B and FTLD-TDP-C groups are coloured in light and dark green. Control individuals are coloured in blue. Ellipses delimit the different groups and are coloured accordingly. (B) Cluster analysis of all samples using the top 1000 most variable genes. Individuals with GRN mutation are coloured in pink, FTLD-TDP-A patients are coloured in purple. Individuals pertaining to the FTLD-TDP-B and FTLD-TDP-C groups are coloured in light and dark green, respectively. Control individuals are coloured in blue. Heat map rows show standardized expression levels of individual genes with red denoting high expression, yellow denoting medium expression and blue denoting low expression levels. (C) Volcano plots representing the differentially expressed genes in FTLD-TDP-A, GRN mutation carriers and FTLD-TDP-C groups versus control group. The fold change is presented in a log2 scale at the x-axis, while the FDR adjusted P-value is presented on the y-axis on a −log10 scale. (D) Venn diagram showing the overlap of up- and downregulated genes between groups (FTLD-TDP-A in purple, GRN mutation carriers in pink and FTLD-TDP-C in light green) as compared to controls. (E) Volcano plots representing the differentially expressed genes in GRN mutation carriers versus FTLD-TDP-C group.
When comparing FTLD-TDP-A to controls, and after adjustment for sex, RIN, age at death and plate, we detected 2704 differentially expressed genes (with P-valueFDR < 0.05 and at least two-fold change), from which 1139 were upregulated and 1565 downregulated (Fig. 1C and Supplementary Table 1). For the GRN group, 3380 genes were differentially expressed as compared to controls, from which 1221 were upregulated and 2159 downregulated (Fig. 1C and Supplementary Table 1). With these fold change and P-value thresholds, we detected only one differentially expressed gene in the FTLD-TDP-B group as compared to controls. We observed 791 differentially expressed genes in the FTLD-TDP-C group compared to controls, with 497 being upregulated and 294 downregulated (Fig. 1C and Supplementary Table 1).
Among the upregulated genes (found in the differential expression analyses of each patient group with controls), 31.0% were shared solely between GRN mutation carriers and FTLD-TDP-A compared to 6.7% shared between GRN carriers and FTLD-TDP-C (Fig. 1D): 15.8% of upregulated genes were shared between the three groups. Among the downregulated genes, 54.5% were shared solely between GRN mutation carriers and FTLD-TDP-A compared to 0.8% shared with FTLD-TDP-C (Fig. 1D): 9.3% of downregulated genes were shared between the three groups. AC104532.2 and AHNAK were the most upregulated and EFNA3 and SCN1B the most downregulated genes specifically in GRN mutation carriers and FTLD-TDP-A groups (Supplementary Table 1).
Since the presence of two risk alleles of the rs5848 variant, located in the 3′ UTR of GRN, is a known risk factor for FTLD-TDP-A, we next assessed whether the presence of homozygous rs5848 carriers was driving the similarities between GRN mutation carriers and FTLD-TDP-A. For this analysis, we removed all individuals carrying two copies of the rs5848 risk allele from all groups. This did not change substantially the principal component analysis and clustering results (Supplementary Fig. 2). In addition, when comparing groups of patients to controls, the strong similarity between GRN and FTLD-TDP-A was still obvious with 32.6% of the upregulated genes shared solely by GRN mutation carriers and FTLD-TDP-A, compared to 4.5% shared between GRN mutation carriers and FTLD-TDP-C (Supplementary Fig. 2). Among the downregulated genes, 40.8% were shared solely between GRN mutation carriers and FTLD-TDP-A, as compared to 1.7% shared with FTLD-TDP-C.
Even though we did not detect GRN differential expression in GRN mutation carriers as compared to controls in the frontal cortex, we detected an increased GRN expression in FTLD-TDP-A group (log2 foldchange = 0.48, P-valueFDR = 7.75 × 10−3). To assess differential expression in a brain region relatively unaffected by neuronal loss, we performed similar analyses in cerebellum tissue of patients and controls. We detected a reduced GRN expression in cerebellum in GRN mutation carriers although not significant (Supplementary Fig. 3, log2 foldchange = −0.22, P-valueFDR = 0.35).
Adding to our observations, when performing differential expression analysis on GRN mutation carriers versus FTLD-TDP-A patients, no significant genes were identified with P-valueFDR < 0.05 and log2 foldchange higher than 1 or smaller than −1. On the other hand, differential expression of GRN mutation carriers versus FTLD-TDP-C patients identified 1030 differentially expressed genes (Fig. 1E and Supplementary Table 1).
To further assess the similarities between FTLD-TDP-A and GRN mutation carriers, we performed consensus network analysis between GRN mutation carriers and FTLD-TDP-A or FTLD-TDP-C (Supplementary Fig. 4). Within the GRN and FTLD-TDP-A consensus analysis, we identified 21 modules, with eight correlating with disease status after Bonferroni correction (38.1%), while a consensus network between GRN mutation carriers and FTLD-TDP-C identified 23 modules, with only two correlating with disease status after Bonferroni correction (8.7%, P-valueFisher = 0.03). Altogether our results suggest that FTLD-TDP-A and GRN mutation carrier expression patterns are similar.
Differential expression analyses of combined FTLD-TDP-A with and without GRN mutations compared to control individuals
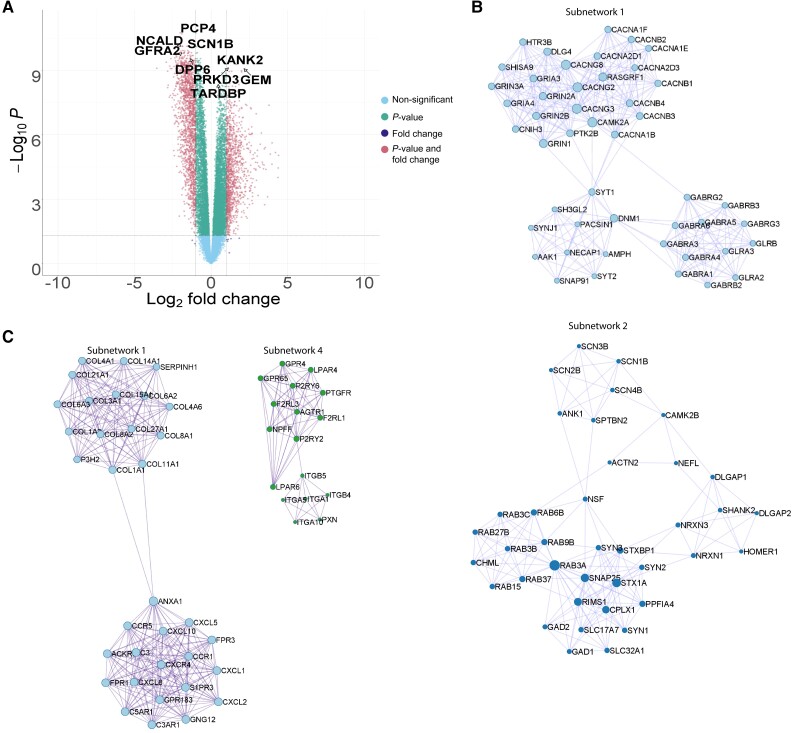
Given that our analyses suggested that FTLD-TDP-A and GRN mutation carriers share a common transcriptional signature, we combined the FTLD-TDP-A and GRN mutation carrier groups and compared them to controls. We identified 2902 differentially expressed genes (fold change < −2 or fold change > 2 and P-valueFDR < 0.05, Fig. 2A and Supplementary Table 2) from which 1235 were upregulated, including 766 genes that were part of the intersection between GRN mutation carriers and FTLD-TDP-A; and 1667 genes were downregulated including 1443 genes that were part of the intersection between the two groups. KANK2 was the most significant upregulated gene detected (log2 foldchange = 1.66, P-valueFDR = 5.34 × 10−10). PCP4 was the most significant downregulated gene detected (log2 foldchange = −2.02, P-valueFDR = 1.76 × 10−11). Interestingly, we detected the two risk genes GFRA2 and DPP6 in the top 50 downregulated genes (log2 foldchange = −1.98, P-valueFDR = 2.02 × 10−10; log2 foldchange = −1.34, P-valueFDR = 2.82 × 10−10). We further investigated the most significant genes using enrichment analyses (Supplementary Table 3). Among the upregulated genes, we detected an enrichment for the terms ‘immune system process’ and ‘defence response’ (P-valueFDR = 2.74 × 10−45, enrichmentRatio = 2.30; P-valueFDR = 6.45 × 10−43, enrichmentRatio = 2.93, respectively). On the other hand, we detected among the downregulated genes an enrichment in ‘synaptic signalling’ and ‘nervous system process’ terms (P-valueFDR = 4.13 × 10−78, enrichmentRatio = 4.35; P-valueFDR = 3.80 × 10−38, enrichmentRatio = 2.96, respectively).
Figure 2.
Differential expression analyses of FTLD-TDP-A with and without GRN mutation. (A) Volcano plot representing the differentially expressed genes in FTLD-TDP-A and GRN mutation carriers combined, as compared to the control group. The fold change is presented in a log2 scale at the x-axis, while the FDR adjusted P-value is presented on the y-axis on a −log10 scale. (B) PPI subnetworks of genes negatively differentially expressed. (C) PPI subnetworks of genes positively differentially expressed.
We then performed PPI network analysis on the differentially expressed genes (up- and downregulated). Among the upregulated genes, we identified 10 subnetworks of interacting proteins (Supplementary Tables 4–6 and Supplementary Fig. 5). Of the 140 genes, 29 are known therapeutic targets for unrelated disorders. Among the downregulated genes, we identified 10 subnetworks of interacting proteins (Supplementary Tables 4–6 and Supplementary Fig. 6). Subnetwork 1 was enriched in ‘ion transmembrane transport’ terms (P-valueFDR = 6.93 × 10−5) and particularly in GABAergic genes (Fig. 2B). Out of the 203 genes, 87 are known therapeutic targets.
Coexpression analysis of combined FTLD-TDP-A with and without GRN mutations compared to control individuals
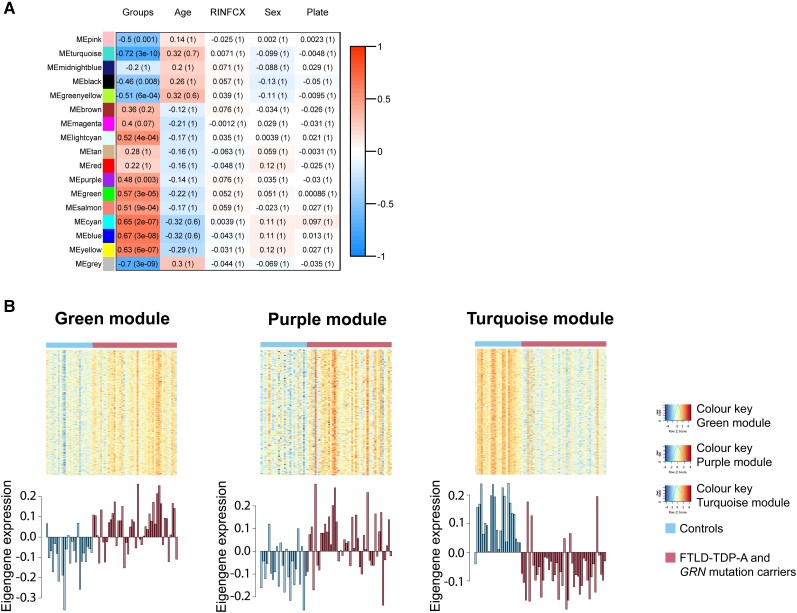
Next, we performed module-level analyses using WGCNA, adjusting for RIN, age at death, sex and plate. When comparing the combined group of FTLD-TDP-A and GRN carriers to controls, we identified 17 modules. Visualization of the module-trait relationships (Fig. 3A) revealed that the strongest relationships were dependent on the disease status with the identification of 11 modules of interest, after Bonferroni correction. As expected, no module correlated with potential confounders (RIN, age at death, sex or plate). GO enrichment analysis of these 11 modules (Supplementary Table 7) showed that they were involved in synaptic signalling (pink and turquoise, P-valueFDR = 4.90 × 10−3 and P-valueFDR = 8.18 × 10−58), Golgi vesicle transport (green-yellow, P-valueFDR = 2.67 × 10−2), RNA processing (lightcyan, P-valueFDR = 2.37 × 10−11), immune response (purple, P-valueFDR = 2.31 × 10−59), cardiovascular system development (green, P-valueFDR = 5.80 × 10−30), retrograde vesicle-mediated transport (salmon, P-valueFDR =1.87 × 10−3), RNA metabolic process (cyan, P-valueFDR = 1.30 × 10−7), SRP-dependent cotranslational protein targeting to membrane (blue, P-valueFDR = 2.23 × 10−31), and response to stimulus (yellow, P-valueFDR = 6.59 × 10−11). Among the genes in the green, purple and turquoise modules, a large number (43.85%, 45.55% and 44.56%, respectively, Supplementary Table 8) were differentially expressed and we therefore decided to focus the rest of the analyses on these modules. The eigengene of each module (Fig. 3B) showed that the green and purple modules were upregulated in patients compared to controls, while the turquoise module was downregulated in patients. More specifically, hierarchical clustering of GO terms revealed that the green module was enriched in terms associated with circulatory system development (41.8%) and immune response (21.4%, Fig. 4A). Although not a hub gene, GRN was upregulated (log2 foldchange = 0.33, P-valueFDR = 4.36 × 10−2) and belonged to this module. The purple module was enriched in GO terms associated exclusively with immune response (immune response 39%, positive regulation of immune system process 37%, Fig. 4B). Finally, the turquoise module was enriched in chemical synaptic transmission and regulation of ion transport GO terms (Fig. 4C). Interestingly, the hub gene for the turquoise module was NSF, which was identified in the PPI subnetwork 2 of downregulated genes (Fig. 2B). This PPI subnetwork was enriched in ‘vesicle-mediated transport’ terms (P-valueFDR = 6.23 × 10−3). To assess if the correlations observed in the frontal cortex were dependent on cellular composition, we performed consensus analysis on cerebellum tissue. Consensus network analysis between frontal-cortex data and cerebellum revealed that the green, purple and turquoise modules were conserved and correlated with disease status in cerebellum (Supplementary Figs 7–10).
Figure 3.
Coexpression analyses of FTLD-TDP-A with and without GRN mutation. (A) Module-trait relationships are presented for patients and controls. Modules with genes going up (red) or down (blue) together are shown, and a unique name has been attributed to each of these modules. Correlations and P-values are shown for variables of interest, including disease group (patients and controls), age at death, RNA integrity number (RIN), sex and plate. The strongest correlations (brightest colours) are observed for the disease group. (B) Heat maps are displayed for the green, purple and turquoise modules. High expression levels are shown in red and low levels in blue. Below every heat map, the first principal component of a given module (module eigengene) is displayed for each sample.
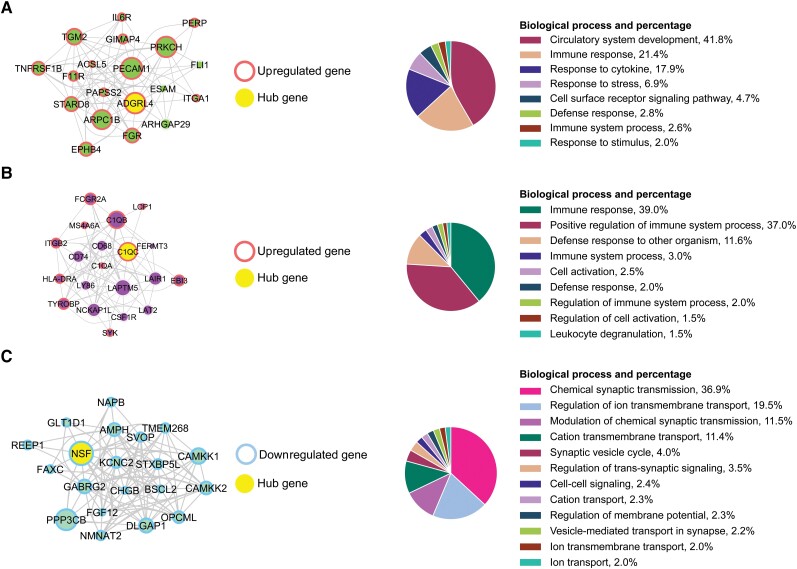
Figure 4.
Top hub genes from the coexpression analyses of FTLD-TDP-A with and without GRN mutation. (A) Top 20 hub genes for the green module are displayed. (B) Top 20 hub genes for the purple module are displayed. (C) Top 20 hub genes for the turquoise module are displayed. The size of the node is proportional to the connectivity of the gene. The strength of the correlation between two genes (nodes) is reflected by the thickness of the edge. Hierarchical GO analysis of biological process terms is presented on the right. Upregulated genes are circled in red and downregulated genes in blue. The main hub gene is coloured in yellow. Hierarchical GO analysis of biological process terms is presented on the right.
We then performed PPI network analysis on these modules of interest identified in the frontal-cortex WGCNA analysis. In the green module (Supplementary Fig. 11), we identified 13 subnetworks with the ITGA1 gene being part of subnetwork 4 and S1PR3 part of subnetwork 11 (Fig. 2C and Supplementary Tables 9 and 10). S1PR3 and ITGA1 were previously identified in subnetworks 1 and 4, respectively, from the differentially upregulated gene analysis. GO analysis revealed an enrichment of primary lysosome terms (P-valueFDR = 2.36 × 10−2) in subnetwork 8, which contains GRN (Supplementary Tables 9 and 10).
In the purple module (Supplementary Fig. 12 and Supplementary Tables 9 and 10), we identified nine subnetworks and more specifically, one enriched in major histocompatibility complex (MHC) protein complex (P-valueFDR = 4.59 × 10−2, subnetwork 1) and one enriched in neutrophil activation involved in immune response (P-valueFDR = 5.81 × 10−5, subnetwork 2). Interestingly, subnetwork 7 contains the C1q complex genes. Within the nine subnetworks, 14 out of 74 genes were reported drug targets (Supplementary Table 7).
In the turquoise module (Supplementary Fig. 13 and Supplementary Tables 9 and 10), we identified nine subnetworks and, similar to what we observed in the downregulated genes, we detected an enrichment in GO terms, such as regulation in calcium ion transmembrane transport (P-valueFDR = 1.48 × 10−3, subnetwork 2), and GABA receptor activity (P-valueFDR = 1.73 × 10−3, subnetwork 4). Subnetwork 4 contains the previously identified hub gene NSF. Within the nine subnetworks, 26 out of 125 genes were reported drug targets (Supplementary Tables 9 and 10).
Cell-type deconvolution of RNA-sequencing data
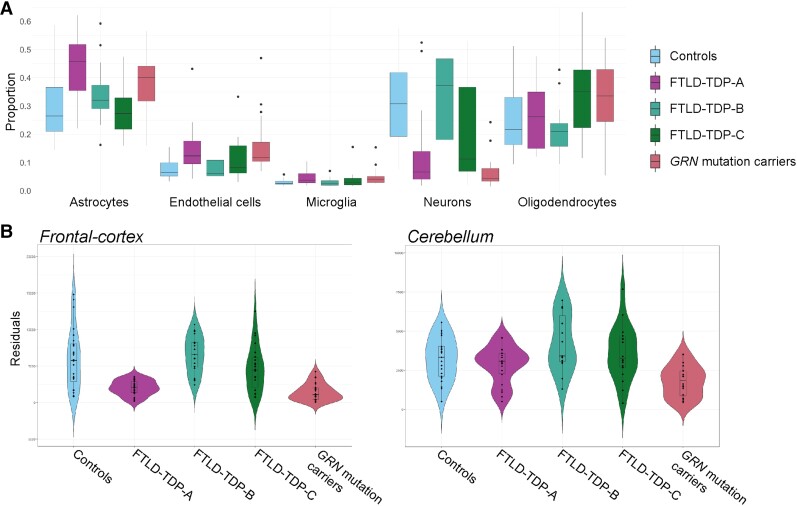
We further estimated the cell proportions in the frontal cortex of patients and controls. We observed that there was a strong variability between the different subtypes of FTLD patients, with a significant reduction of the relative proportion of neurons observed in GRN and FTLD-TDP-A patients, compared to controls (medianControls = 0.31, medianGRN = 0.04, P-valueGRN = 3.5 × 10−8; medianFTLD-TDP-A = 0.07, P-valueFTLD-TDP-A = 8.2 × 10−5; Fig. 5). We also observed a significant increase in the relative proportion of microglia in GRN compared to controls (medianControls = 0.03, medianGRN = 0.04, P-valueGRN = 2.7 × 10−2), and in the relative proportion of astrocytes in FTLD-TDP-A compared to controls (medianControls = 0.25, medianFTLD-TDP-A = 0.46, P-valueFTLD-TDP-A = 2.65 × 10−2). Finally, a significant increase in endothelial cell proportion was detected in GRN and FTLD-TDP-A compared to controls (medianControls = 0.06, medianGRN = 0.12, P-valueGRN = 5.10 × 10−4; medianFTLD-TDP-A = 0.12, P-valueFTLD-TDP-A = 4.34 × 10−3). These data indicate that the cellular composition in the frontal cortex of FTLD-TDP-A and GRN mutation carriers is similar, further highlighting the commonalities between the two groups.
Figure 5.
Cell-type deconvolution analysis of FTLD-TDP-A with and without GRN mutation. (A) Cell proportions estimations in the frontal cortex for five cell types (astrocytes, endothelial cells microglia, neurons and oligodendrocytes). (B) Residual expression in the frontal cortex and cerebellum of NSF per group.
Of the genes pertaining to the green module, 87% were predicted to be expressed by endothelial cells, of which 19% were specific for endothelial cells. Similarly, 97% of the genes pertaining to the purple module were predicted to be expressed by microglia, of which 18% were microglia-specific. Finally, 86% of the genes pertaining to the turquoise module were predicted to be expressed by neurons, of which 27% were neuron-specific.
We then performed differential expression in FTLD-TDP-A and GRN mutation carriers versus controls on the deconvoluted data. We detected 566 genes that were differentially expressed in neurons, 1144 genes in oligodendrocytes, 240 genes in microglia, 793 genes in astrocytes and 343 in endothelial cells (Supplementary Table 11).
Within neurons, PTGER3 and HECW1 were the most dysregulated genes (P-valueFDR = 1.59 × 10−9, beta = 0.08; P-valueFDR = 2.62 × 10−7, beta = −0.19, respectively) and the synaptogenesis signalling pathway was identified as the top perturbed canonical pathway (P-value = 2.05 × 10−14, Supplementary Table 11). Among other pathways, glutamate and GABA receptor signalling were also altered (P-value = 3.61 × 10−7 and P-value = 1.07 × 10−3, respectively). We detected two GABAergic genes in the top 10 differentially expressed genes (GABRB3 and GABRA5, P-valueFDR = 1.53 × 10−4, beta = 0.92; P-valueFDR = 1.53 × 10−4, beta = 0.33, respectively) confirming the implication of the GABAergic system in disease.
Within microglia, DDX46 and SLC22A9 were the most dysregulated genes (P-valueFDR = 4.15 × 10−4, beta = 0.86; P-valueFDR = 4.15 × 10−4, beta = −0.23, respectively). The GDNF family ligand-receptor interactions pathway was identified as the top perturbed canonical pathway perturbed (P-value = 1.58 × 10−3).
Within oligodendrocytes, GSTA1 and MAL were the most dysregulated genes (P-valueFDR = 1.73 × 10−5, beta = −0.03; P-valueFDR = 1.06 × 10−4, beta = −2.2, respectively). The most significant canonical pathway identified was the Ataxia-telangiectasia mutated (ATM) signalling pathway (P-value = 4.55 × 10−5).
Within astrocytes, WNT7B and TMCO4 were the most dysregulated genes (P-valueFDR = 1.65 × 10−4, beta = −0.17; P-valueFDR = 1.81 × 10−4, beta = 0.05, respectively). We detected an enrichment in mitochondrial dysfunction terms in differentially expressed genes (P-value = 5.24 × 10−6).
Within endothelial cells, HRCT1 and RNASEK-C17orf49 were the most dysregulated genes (P-valueFDR = 1.59 × 10−4, beta = −0.86; P-valueFDR = 4.71 × 10−4, beta = −2.15, respectively). The inhibition of matrix metalloproteases pathway was identified as the top perturbed canonical pathway (P-value = 1.72 × 10−4).
The NSF gene was identified in several of our analyses. It was significantly associated with disease status in the deconvoluted data in neurons (P-valueFDR = 2.15 × 10−2, beta = −0.79). Note that no significant difference in expression of NSF was observed in the deconvoluted data for FTLD-TDP-C and FTLD-TDP-B versus controls. We also highlighted NSF in the PPI analysis in the bulk differential expression analysis, as well as part of the top 20 hub genes from one of the three modules (green, turquoise or purple) and in their respective PPI analyses. The decreased expression of NSF in the bulk RNA-sequencing data appeared to be specific to the GRN mutation carriers and FTLD-TDP-A groups in the frontal cortex (Fig. 5B, log2 foldchange-FTLD-TDP-A = −1.66, P-valueFDR = 1.08 × 10−8; log2foldchange-GRN = −1.95, P-valueFDR = 2.78 × 10−7). We observed a trend in the same direction in cerebellum (log2 foldchange-FTLD-TDP-A = −0.31, P-valuenominal = 0.04; log2 foldchange-GRN = −0.38, P-valuenominal = 0.03).
Discussion
In this study, we used an unbiased whole transcriptomic approach to characterize expression patterns of FTLD-TDP patients in the affected frontal-cortex brain region. We investigated a total of 88 FTLD-TDP patients and controls. We found that GRN mutation carriers and FTLD-TDP-A patients have similar expression patterns using hierarchical clustering of the samples per gene expression and differential expression. We also reported that their frontal-cortex cellular composition is similar. Differential expression and coexpression network analyses identified an upregulation of inflammatory processes and downregulation of neuronal activity in patients compared to controls. Additionally, we highlighted vascular system, GABAergic and complement system alterations in FTLD-TDP-A patients. This study provides a comprehensive analysis of transcriptional changes in FTLD-TDP-A regardless of their underlying genetic aetiology and sheds light on potential new therapeutic targets and disease biomarkers.
GRN mutation carriers always present with FTLD-TDP-A at autopsy; however, it was not clear whether these patients shared common disease mechanisms with genetically unexplained FTLD-TDP-A patients. Previous genetic studies hinted towards a shared aetiology, reporting shared genetic associations such as TMEM106B and GFRA2 between both FTLD-TDP-A groups.72 In this study, we showed that FTLD-TDP-A patients with and without GRN mutations shared a common transcriptional signature using unbiased approaches with principal component analysis and hierarchical clustering. We further showed more commonalities between GRN mutation carriers and FTLD-TDP-A than with FTLD-TDP-C patients regarding differentially expressed genes and coexpression networks. We previously reported that rs5848, located in the 3′ UTR of GRN, is a risk factor for FTLD-TDP-A, linking GRN to genetically unexplained FTLD-TDP-A8; however, we show that the similar transcriptomic profile was independent of rs5848. Furthermore, genetically unexplained FTLD-TDP-A patients have increased levels of GRN compared to controls, similar to findings in FTLD-TDP-C and FTLD-TDP-B patients, whereas GRN mutation carriers have levels comparable to controls. These data indicate increased levels of GRN in the frontal cortex as a result of activated microglia in all FTLD-TDP patients; however, since GRN mutation carriers have less GRN expression to begin with, their levels appear similar to controls.32 These observations strongly suggest that the shared transcriptomic profile between FTLD-TDP-A and GRN mutation carriers is independent and likely downstream of loss of GRN. It is also noteworthy that we observed these highly similar transcriptomic signatures despite differences between FTLD-TDP-A and GRN mutation carriers with an average age at death 11 years later in the former as compared to the latter. Even though we observed overall similar coexpression patterns in cerebellum, a less affected brain region than the frontal cortex, the apparent similar transcriptomic signature is very likely to reflect similarities in cell-type composition. In fact, we observed a downregulation of PCP4 that encodes for PEP-19. PEP-19 binds to calmodulin, which activates several processes that are associated with neuronal death and play a role in the regulation of synaptic activity and maintenance of neuronal plasticity.33 A downregulation of PCP4 may reflect the neuronal loss we observed in the deconvoluted data. Furthermore, deconvolution of our bulk RNA-sequencing data indicated a comparable cellular structure in the frontal cortex. In addition, the three highlighted modules were enriched in cell-type-specific genes suggesting that each cell type plays a role in disease. This observation is evocative of the cellular phase observed in Alzheimer's disease and argues strongly in favour of single-cell approaches to gain more granular insights into the pathophysiology.34
However, bulk RNA-sequencing is still a valuable resource to understand system level alterations. We highlighted the NSF gene that is significantly dysregulated in the frontal cortex of FTLD-TDP-A and GRN mutation carriers, even after deconvolution. NSF, which encodes for the N-Ethylmaleimide Sensitive Factor protein, is essential for vesicle-mediated transport through its interaction with the soluble NSF attachment protein receptor (SNARE) complex35 and SNAPs.36 The SNARE complex mediates synaptic vesicle fusion that is orchestrated by the assembly of three proteins: VAMP2, SNAP25 and STX1A and can be regulated by SNARE binding proteins such as complexin 1 (CPLX1).37,38 In our data, we observed a downregulation of all these transcripts. Recently, an association between cognitive decline and dysregulation of the SNARE protein interactome, including CPLX1, was reported, with lower levels correlating with lower cognitive performance.39,40 As CPLX1 is mainly expressed by GABAergic neurons, this indicates that impairment of the inhibitory system may contribute to cognitive decline.41 To further elaborate, we also found a downregulation of GABAergic signalling pathways within the differential expression and the coexpression network analyses (turquoise module) and a dysregulation of GABA receptors in the deconvoluted neuronal population. In addition, the top dysregulated gene in the deconvoluted neurons, PTGER3, encoding for the Prostaglandin E Receptor 3, has been shown to act as a fever modulator through GABAergic signalling.42 The GABA receptors expression patterns are highly complex and have been shown to present cell and regional specificity, suggesting that single-cell approaches would be appropriate to further dissect the GABergic system in FTLD.43 A loss of GABAergic neurons has been reported in frontal and temporal cortices of FTLD patients, as well as reduced gamma oscillations and coherence in frontal lobes of bvFTD patients, reflecting lower GABAergic neuronal inhibition.44,45 A recent study reported that GABA levels are reduced in the right inferior frontal gyrus of bvFTD.46 While our observations are strongly associated with cellular composition, our data indicate that GABAergic synaptic vesicle fusion is impaired in FTLD-TDP-A patients. One could hypothesize that drugs targeting the GABAergic system might be beneficial.
ITGA1, that encodes the α1 subunit of integrin receptors, was identified in several of our analyses as upregulated in patients. Integrins are transmembrane heterodimeric receptors that are involved in cell adhesion and nerve regeneration.47–49 The α1β1 complex is involved in neurite outgrowth through activation of the CDK5 signalling pathway.50 Interestingly, the most upregulated gene identified in FTLD-TDP-A and GRN mutation carriers versus controls was KANK2. KANK2 has been shown to be the key protein involved in cell adhesion and migration through its interaction with integrins αβ.51 In our deconvoluted data, ITGA1 was expressed by endothelial cells and part of the green module that was enriched in vasculature development terms. We also report an increased proportion of endothelial cells in FTLD-TDP-A and GRN mutation carrier groups as compared to controls, similarly to what is observed in Alzheimer's disease brains52 that could explain the apparent upregulation of ITGA1. ITGA1 binds to type IV collagen53 and COL4A2, which encodes one of the six subunits of type IV collagen, was also upregulated in our patient cohort. Type IV collagen is the major structural component of basement membranes and used as a marker for vasculature components in brain tissue. Intriguingly, Olofsson and Englund54 recently reported, using this marker, a higher amount of a specific microvascular structure in the frontal cortex of FTLD patients. These raspberry-like structures are thought to be a sign of angiogenesis linked to hypoperfusion of the brain present in the bilateral frontal lobes of FTLD patients.55–59 The green module was defined by a mixture between inflammatory signal and angiogenesis, suggesting a coregulation between both processes supporting the effect of microglial activation enhancing angiogenesis through pro-inflammatory TNF-alpha signalling.60 Highlighting the importance of endothelial cells in FTLD, progranulin overexpression in endothelial cells impairs angiogenesis in vivo, increasing vessel growth and altering vessels’ integrity.61 On the other hand, Grn knock-out mice have a profound disruption of the blood brain barrier associated with structural defects of the endothelial junctional complexes.62 Among our deconvoluted data, we identified an enrichment in the inhibition of matrix metalloproteases pathway, with tissue inhibitors of metalloproteinases 4 and 2 (TIMP4 and TIMP2). Both play a critical role in extracellular matrix homeostasis, angiogenesis and inflammation.63–65 Altogether, this suggests that angiogenesis is a fundamental part of FTLD-TDP type A pathology and further studies are necessary to elucidate its contribution.
Additionally, within the purple module, we highlight the C1q recognition component of C1, the classical complement pathway. C1q is a hexamer of trimers composed of A, B and C chains, encoded by the C1QA, C1QB and C1QC genes.66 All three genes are upregulated in FTLD-TDP-A patients and GRN mutation carriers, probably reflecting increased numbers of microglia. Interestingly, C1QA has been shown to promote synaptic pruning by microglia in Grn−/− mice. In addition, knock-out of C1qa in Grn−/− mice protects against synapse loss, reduces behavioural deficits and improves survival, suggesting that complement activation and microglia-mediated synaptic pruning are drivers of neurodegeneration due to progranulin deficiency.67 The C1q complex genes appear to be overexpressed in Grn−/− mice, and treating cultured Grn+/+ (wild-type) and Grn−/− neurons with C1q induces cytoplasmic TDP-43 granule formation.68 Here we report that C1q upregulation is not only present in GRN mutation carriers but also in genetically unexplained FTLD-TDP-A patients.
Within the deconvoluted microglia, we highlighted the GDNF signalling pathway, which comprised the downregulated GFRA1 gene. GFRA1 is a RET coreceptor and part of the GDNF family ligands playing a neuroprotective role through modulation of microglial activation.69–71 We previously reported a common variant at the GFRA2 locus as a genetic modifier for GRN mutation carriers,72 expanding the number of GDNF family ligands involved in FTLD. In addition, GDNF implication appears to be common to several neurological disorders including Parkinson's disease where GDNF has been studied for its neuroprotective potential, even leading to clinical trials with GDNF brain delivery in patients.73–75
Within the deconvoluted oligodendrocyte data, we identified an alteration of the ATM signalling pathway. ATM, is a causal gene for Ataxia-telangiectasia (A-T) and encodes for a key protein involved in cell cycle checkpoint control during DNA damage repair.76 The ATM kinase phosphorylates a number of downstream targets involved in DNA damage repair, cell cycle arrest and apoptosis.77–80 Interestingly, A-T is characterized by cerebellar neurodegeneration and myelin abnormalities.81 Recently, ATM deficiencies were shown to cause DNA damage accumulation in oligodendrocytes leading to a reduction of the mature oligodendrocyte lineage.82 This is reminiscent of the TMEM106B−/− mouse model where a reduction of the number of differentiated oligodendrocytes was reported.83 Along this line, TDP-43 plays a pivotal role in proper functioning of mature oligodendrocytes. Indeed, TDP-43 deletion in mature oligodendrocytes leads to progressive disruption of myelin and selective cell death of mature oligodendrocytes.84 Interestingly, presence of TDP-43 inclusions in oligodendrocytes has been previously reported in FTLD-TDP, including GRN mutation carriers.85,86 Altogether, our data add to the growing body of evidence that oligodendrocytes play a role in FTLD-TDP pathology.
Finally, the gene S1PR3, identified within several PPI analyses, encodes a G protein-coupled receptor for sphingosine 1-phosphate (S1P). Sphingolipids are highly abundant in the brain and play an essential role in neuronal plasticity.87,88 We and others showed that GRN interacts with prosaposin,89–91 a critical protein for sphingolipid metabolism.92,93 In addition, GRN has been reported to bind β-hexosaminidase A and β-glucocerebrosidase, regulating their activities in sphingolipid metabolism and trafficking to lysosomes.94,95 Importantly, S1P3 receptors are involved in neurite retraction and deceleration of axonal growth,96 as well as cellular immunity function97–99 and S1PR3 promotes inflammatory macrophage activation.100 Knock-down of S1PR3 eliminates the anti-inflammatory effect of Fingolimod (FTY720), a neuroprotective drug used as immunosuppressant in multiple sclerosis.101 Since S1PR3 is predominantly expressed by astrocytes, it was shown to be a crucial player in astrocytic inflammatory response on treatment with Fingolimod. Therefore, an increased expression of S1PR3 might increase neuroinflammation.
We also detected an alteration of RNA metabolism in the coexpression network analysis. This finding is in agreement with the dysregulation of spliceosome processes observed in neuronal cells on treatment with PGRN.102 TDP-43 plays major roles in several aspects of RNA metabolism103 and in particular, transcriptome analyses highlighted aberrant splicing in neurons with altered TDP-43 levels.104,105 More recently alternative splicing was also reported in FTLD-TDP brains,106–108 and additional studies are required to further dissect the specificity of splicing patterns observed in the different FTLD-TDP subtypes.
Our findings may have implications for other common neurodegenerative disorders. Indeed, TDP-43 pathology is present in late-onset Alzheimer's disease brains and is associated with memory loss and hippocampal volume.109,110 Recently, a new pathological subtype of TDP-43 aggregates named TDP-43 type α has been described and consists of widespread distribution of dystrophic neurites and neuronal cytoplasmic inclusions in amygdala, hippocampus and frontotemporal cortex.111 These features, reported in a subset of Alzheimer's disease patients, are similar to the TDP-43 immunoreactivity observed in FTLD-TDP type A.111,112 Over half of these Alzheimer's disease patients were reported to have FTLD symptomatology. Further highlighting a potential similarity with FTLD-TDP-A subtype, the frequency of the homozygous TMEM106B protective allele (rs3173615) is reduced in non-FTLD brains with TDP-43 type α.111 Our work now suggests that they may also share transcriptional profiles however, further studies are necessary to address this question.
Our study has some limitations. Even though this is a large cohort of FTLD-TDP patients studied by RNA-sequencing, the actual number of samples is still limited. Our findings of loss of neuronal activity and increased inflammation signalling might reflect end-stage molecular events. In contrast to our findings, Chen-Plotkin et al.16 reported that GRN mutation carriers presented with a unique transcriptional signature. A possible explanation for this might lie in the higher number of brains studied here, and the use of more sensitive next-generation sequencing methods instead of microarrays. Additionally, we included only one affected brain region, whereas the aforementioned study used a mixture of two brain regions.
In sum, using an integrative approach from single gene differential expression to gene and protein network scale, we report for the first time a shared transcriptional signature in the frontal cortex of FTLD-TDP-A patients with and without GRN mutations. These transcriptional similarities are likely to be associated with comparable cell-type populations in both disease groups. We also describe an alteration in genes involved in vascularization through endothelial cells and importantly, we emphasize the dysregulation of the GABAergic signalling pathway, as well as the potential involvement of astrocytic inflammation. Overall, this study provides new avenues for research into FTLD-TDP type A and GRN and potentially new therapeutic targets.
Supplementary Material
Acknowledgements
We thank Xue Wang for her advice concerning the deconvolution analyses.
Abbreviations
- bvFTD
behavioural variant frontotemporal dementia
- DSA
digital sorting algorithm
- FTLD
frontotemporal lobar degeneration
- GO
gene ontology
- PPI
protein–protein interaction
- RIN
RNA integrity number
- TDP-43
Tar DNA binding protein 43
Contributor Information
Cyril Pottier, VIB Center for Molecular Neurology, VIB, Antwerp, Belgium; Department of Biomedical Sciences, University of Antwerp, Antwerp, Belgium.
Ligia Mateiu, VIB Center for Molecular Neurology, VIB, Antwerp, Belgium.
Matthew C Baker, Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA.
Mariely DeJesus-Hernandez, Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA.
Cristina Teixeira Vicente, VIB Center for Molecular Neurology, VIB, Antwerp, Belgium; Department of Biomedical Sciences, University of Antwerp, Antwerp, Belgium.
NiCole A Finch, Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA.
Shulan Tian, Department of Quantitative Health Sciences, Mayo Clinic, Rochester, MN, USA.
Marka van Blitterswijk, Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA.
Melissa E Murray, Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA.
Yingxue Ren, Department of Quantitative Health Sciences, Mayo Clinic, Jacksonville, FL, USA.
Leonard Petrucelli, Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA.
Björn Oskarsson, Department of Neurology, Mayo Clinic, Jacksonville, FL, USA.
Joanna M Biernacka, Department of Quantitative Health Sciences, Mayo Clinic, Rochester, MN, USA.
Neill R Graff-Radford, Department of Neurology, Mayo Clinic, Jacksonville, FL, USA.
Bradley F Boeve, Department of Neurology, Mayo Clinic, Rochester, MN, USA.
Ronald C Petersen, Department of Neurology, Mayo Clinic, Rochester, MN, USA.
Keith A Josephs, Department of Neurology, Mayo Clinic, Rochester, MN, USA.
Yan W Asmann, Department of Quantitative Health Sciences, Mayo Clinic, Jacksonville, FL, USA.
Dennis W Dickson, Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA.
Rosa Rademakers, VIB Center for Molecular Neurology, VIB, Antwerp, Belgium; Department of Biomedical Sciences, University of Antwerp, Antwerp, Belgium; Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA.
Funding
The work was supported by the University of Antwerp Research Funds (BOF) and Vlaams Instituut voor Biotechnologie (VIB), as well as the National Institutes of Health (NIH) grants R35 NS097261, UG3 NS103870, P50 AG016574, P30 AG062677, U01 AG045390, U19 AG063911 and U54 NS092089.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Mackenzie IR, Neumann M, Baborie A, et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122:111–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee EB, Porta S, Michael BG, et al. Expansion of the classification of FTLD-TDP: distinct pathology associated with rapidly progressive frontotemporal degeneration. Acta Neuropathol. 2017;134:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hutton M, Lendon CL, Rizzu P, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998;393:702–705. [DOI] [PubMed] [Google Scholar]
- 4. Baker M, Mackenzie IR, Pickering-Brown SM, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006;442:916–919. [DOI] [PubMed] [Google Scholar]
- 5. Cruts M, Gijselinck I, van der Zee J, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 2006;442:920–924. [DOI] [PubMed] [Google Scholar]
- 6. DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011;72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pottier C, Ren Y, Perkerson RB, et al. Genome-wide analyses as part of the international FTLD-TDP whole-genome sequencing consortium reveals novel disease risk factors and increases support for immune dysfunction in FTLD. Acta Neuropathol. 2019;137:879–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferrari R, Hernandez DG, Nalls MA, et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol. 2014;13:686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Deerlin VM, Sleiman PMA, Martinez-Lage M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mackenzie IR, Baker M, Pickering-Brown S, et al. The neuropathology of frontotemporal lobar degeneration caused by mutations in the progranulin gene. Brain. 2006;129:3081–3090. [DOI] [PubMed] [Google Scholar]
- 12. Rademakers R, Eriksen JL, Baker M, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17:3631–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hsiung GY, Fok A, Feldman HH, Rademakers R, Mackenzie IR. rs5848 polymorphism and serum progranulin level. J Neurol Sci. 2011;300:28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nicholson AM, Finch NA, Thomas CS, et al. Progranulin protein levels are differently regulated in plasma and CSF. Neurology. 2014;82:1871–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dickson DW, Baker MC, Jackson JL, et al. Extensive transcriptomic study emphasizes importance of vesicular transport in C9orf72 expansion carriers. Acta Neuropathol Commun. 2019;7:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen-Plotkin AS, Geser F, Plotkin JB, et al. Variations in the progranulin gene affect global gene expression in frontotemporal lobar degeneration. Hum Mol Genet. 2008;17:1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. [DOI] [PubMed] [Google Scholar]
- 19. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. [DOI] [PubMed] [Google Scholar]
- 20. McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ochoa D, Hercules A, Carmona M, et al. Open Targets Platform: supporting systematic drug-target identification and prioritisation. Nucleic Acids Res. 2021;49:D1302–D1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuznetsova I, Lugmayr A, Siira SJ, Rackham O, Filipovska A. CirGO: an alternative circular way of visualising gene ontology terms. BMC Bioinformatics. 2019;20:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhong Y, Wan YW, Pang K, Chow LM, Liu Z. Digital sorting of complex tissues for cell type-specific gene expression profiles. BMC Bioinformatics. 2013;14:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang X, Allen M, Li S, et al. Deciphering cellular transcriptional alterations in Alzheimer’s disease brains. Mol Neurodegener. 2020;15:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McKenzie AT, Wang M, Hauberg ME, et al. Brain cell type specific gene expression and co-expression network architectures. Sci Rep. 2018;8:8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuhn A, Thu D, Waldvogel HJ, Faull RL, Luthi-Carter R. Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nat Methods. 2011;8:945–947. [DOI] [PubMed] [Google Scholar]
- 31. Kramer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen-Plotkin AS, Xiao J, Geser F, et al. Brain progranulin expression in GRN-associated frontotemporal lobar degeneration. Acta Neuropathol. 2010;119:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharma RK, Parameswaran S. Calmodulin-binding proteins: A journey of 40 years. Cell Calcium. 2018;75:89–100. [DOI] [PubMed] [Google Scholar]
- 34. De Strooper B, Karran E. The cellular phase of Alzheimer’s disease. Cell. 2016;164:603–615. [DOI] [PubMed] [Google Scholar]
- 35. Sollner T, Whiteheart SW, Brunner M, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. [DOI] [PubMed] [Google Scholar]
- 36. Clary DO, Griff IC, Rothman JE. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell. 1990;61:709–721. [DOI] [PubMed] [Google Scholar]
- 37. Chen X, Tomchick DR, Kovrigin E, et al. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. [DOI] [PubMed] [Google Scholar]
- 38. McMahon HT, Missler M, Li C, Sudhof TC. Complexins: Cytosolic proteins that regulate SNAP receptor function. Cell. 1995;83:111–119. [DOI] [PubMed] [Google Scholar]
- 39. Ramos-Miguel A, Jones AA, Sawada K, et al. Frontotemporal dysregulation of the SNARE protein interactome is associated with faster cognitive decline in old age. Neurobiol Dis. 2018;114:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramos-Miguel A, Sawada K, Jones AA, et al. Presynaptic proteins complexin-I and complexin-II differentially influence cognitive function in early and late stages of Alzheimer’s disease. Acta Neuropathol. 2017;133:395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klichev BM. Conservative treatment of rectal prolapse in children. Pediatriia. 1989;99–100. [PubMed] [Google Scholar]
- 42. Tsuchiya H, Oka T, Nakamura K, Ichikawa A, Saper CB, Sugimoto Y. Prostaglandin E2 attenuates preoptic expression of GABAA receptors via EP3 receptors. J Biol Chem. 2008;283:11064–11071. [DOI] [PubMed] [Google Scholar]
- 43. Kwakowsky A, Calvo-Flores Guzman B, Govindpani K, Waldvogel HJ, Faull RL. Gamma-aminobutyric acid A receptors in Alzheimer’s disease: highly localized remodeling of a complex and diverse signaling pathway. Neural Regen Res. 2018;13:1362–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ferrer I. Neurons and their dendrites in frontotemporal dementia. Dement Geriatr Cogn Disord. 1999;10(Suppl. 1):55–60. [DOI] [PubMed] [Google Scholar]
- 45. Hughes LE, Ghosh BC, Rowe JB. Reorganisation of brain networks in frontotemporal dementia and progressive supranuclear palsy. Neuroimage Clin. 2013;2:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murley AG, Rouse MA, Jones PS, et al. GABA and glutamate deficits from frontotemporal lobar degeneration are associated with disinhibition. Brain. 2020;143:3449–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hemler ME. VLA proteins in the integrin family: structures, functions, and their role on leukocytes. Annu Rev Immunol. 1990;8:365–400. [DOI] [PubMed] [Google Scholar]
- 48. Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. [DOI] [PubMed] [Google Scholar]
- 49. Trigg DJ, O’Grady KM, Bhattacharyya T, Reinke M, Toriumi DM. Peripheral nerve regeneration: comparison of laminin and acidic fibroblast growth factor. Am J Otolaryngol. 1998;19:29–32. [DOI] [PubMed] [Google Scholar]
- 50. Li BS, Zhang L, Gu J, Amin ND, Pant HC. Integrin α1β1-mediated activation of cyclin-dependent kinase 5 activity is involved in neurite outgrowth and human neurofilament protein H Lys-Ser-Pro tail domain phosphorylation. J Neurosci. 2000;20:6055–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paradzik M, Humphries JD, Stojanović N, et al. KANK2 links αVβ5 focal adhesions to microtubules and regulates sensitivity to microtubule poisons and cell migration. Front Cell Dev Biol. 2020;8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lau SF, Cao H, Fu AKY, Ip NY. Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer’s disease. Proc Natl Acad Sci USA. 2020;117:25800–25809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Olofsson HE, Englund E. A cortical microvascular structure in vascular dementia, Alzheimer’s disease, frontotemporal lobar degeneration and nondemented controls: a sign of angiogenesis due to brain ischaemia? Neuropathol Appl Neurobiol. 2019;45:557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dopper EG, Chalos V, Ghariq E, et al. Cerebral blood flow in presymptomatic MAPT and GRN mutation carriers: A longitudinal arterial spin labeling study. Neuroimage Clin. 2016;12:460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Binnewijzend MA, Kuijer JPA, van der Flier WM, et al. Distinct perfusion patterns in Alzheimer’s disease, frontotemporal dementia and dementia with Lewy bodies. Eur Radiol. 2014;24:2326–2333. [DOI] [PubMed] [Google Scholar]
- 57. Du AT, Jahng GH, Hayasaka S, et al. Hypoperfusion in frontotemporal dementia and Alzheimer disease by arterial spin labeling MRI. Neurology. 2006;67:1215–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hu WT, Wang Z, Lee VM, Trojanowski JQ, Detre JA, Grossman M. Distinct cerebral perfusion patterns in FTLD and AD. Neurology. 2010;75:881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shimizu S, Zhang Y, Laxamana J, et al. Concordance and discordance between brain perfusion and atrophy in frontotemporal dementia. Brain Imaging Behav. 2010;4:46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Welser JV, Li L, Milner R. Microglial activation state exerts a biphasic influence on brain endothelial cell proliferation by regulating the balance of TNF and TGF-β1. J Neuroinflammation. 2010;7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Toh H, Cao M, Daniels E, Bateman A. Expression of the growth factor progranulin in endothelial cells influences growth and development of blood vessels: a novel mouse model. PLoS ONE. 2013;8:e64989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jackman K, Kahles T, Lane D, et al. Progranulin deficiency promotes post-ischemic blood-brain barrier disruption. J Neurosci. 2013;33:19579–19589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jiang Y, Goldberg ID, Shi YE. Complex roles of tissue inhibitors of metalloproteinases in cancer. Oncogene. 2002;21:2245–2252. [DOI] [PubMed] [Google Scholar]
- 64. Lee EJ, Kim HS. The anti-inflammatory role of tissue inhibitor of metalloproteinase-2 in lipopolysaccharide-stimulated microglia. J Neuroinflammation. 2014;11:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cabral-Pacheco GA, et al. The roles of matrix metalloproteinases and their inhibitors in human diseases. Int J Mol Sci. 2020;21:9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Reid KB, Porter RR. Subunit composition and structure of subcomponent C1q of the first component of human complement. Biochem J. 1976;155:19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lui H, Zhang J, Makinson SR, et al. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell. 2016;165:921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang J, Velmeshev D, Hashimoto K, Huang Y-H, et al. Neurotoxic microglia promote TDP-43 proteinopathy in progranulin deficiency. Nature. 2020;588:459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rickert U, Grampp S, Wilkms H, et al. Glial cell line-derived neurotrophic factor family members reduce microglial activation via inhibiting p38MAPKs-mediated inflammatory responses. J Neurodegener Dis. 2014;2014:369468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xing B, Xin T, Zhao L, Hunter RL, Chen Y, Bing G. Glial cell line-derived neurotrophic factor protects midbrain dopaminergic neurons against lipopolysaccharide neurotoxicity. J Neuroimmunol. 2010;225:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chang YP, Fang KM, Lee TI, Tzeng SF. Regulation of microglial activities by glial cell line derived neurotrophic factor. J Cell Biochem. 2006;97:501–511. [DOI] [PubMed] [Google Scholar]
- 72. Pottier C, Zhou X, Perkerson RB, et al. Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study. Lancet Neurol. 2018;17:548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bartus RT, Weinberg MS, Samulski RJ. Parkinson’s disease gene therapy: success by design meets failure by efficacy. Mol Ther. 2014;22:487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lindholm D, Mäkelä J, Di Liberto V, et al. Current disease modifying approaches to treat Parkinson’s disease. Cell Mol Life Sci. 2016;73:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kirik D, Cederfjall E, Halliday G, Petersen A. Gene therapy for Parkinson’s disease: Disease modification by GDNF family of ligands. Neurobiol Dis. 2017;97:179–188. [DOI] [PubMed] [Google Scholar]
- 76. Taylor AM, Lam Z, Last JI, Byrd PJ. Ataxia telangiectasia: More variation at clinical and cellular levels. Clin Genet. 2015;87:199–208. [DOI] [PubMed] [Google Scholar]
- 77. Shiloh Y, Rotman G. Ataxia-telangiectasia and the ATM gene: linking neurodegeneration, immunodeficiency, and cancer to cell cycle checkpoints. J Clin Immunol. 1996;16:254–260. [DOI] [PubMed] [Google Scholar]
- 78. Barlow C, Brown KD, Deng CX, Tagle DA, Wynshaw-Boris A. ATM selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nat Genet. 1997;17:453–456. [DOI] [PubMed] [Google Scholar]
- 79. Khanna KK. Cancer risk and the ATM gene: a continuing debate. J Natl Cancer Inst. 2000;92:795–802. [DOI] [PubMed] [Google Scholar]
- 80. Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle. 2007;6:931–942. [DOI] [PubMed] [Google Scholar]
- 81. Sahama I, Sinclair K, Fiori S, et al. Motor pathway degeneration in young ataxia telangiectasia patients: A diffusion tractography study. Neuroimage Clin. 2015;9:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tse K-H, Cheng A, Yeung SH-S, et al. Myelin pathology in ataxia-telangiectasia is the cell-intrinsic consequence of ATM deficiency in the oligodendrocytes. medRxiv [Preprint]. 2021. doi: 10.1101/2021.01.22.20245217 [DOI]
- 83. Zhou X, Nicholson AM, Ren Y, et al. Loss of TMEM106B leads to myelination deficits: Implications for frontotemporal dementia treatment strategies. Brain. 2020;143:1905–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang J, Ho WY, Lim K, et al. Cell-autonomous requirement of TDP-43, an ALS/FTD signature protein, for oligodendrocyte survival and myelination. Proc Natl Acad Sci USA. 2018;115:E10941–E10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hiji M, Takahashi T, Fukuba H, Yamashita H, Kohriyama T, Matsumoto M. White matter lesions in the brain with frontotemporal lobar degeneration with motor neuron disease: TDP-43-immunopositive inclusions co-localize with p62, but not ubiquitin. Acta Neuropathol. 2008;116:183–191. [DOI] [PubMed] [Google Scholar]
- 86. Neumann M, Kwong LK, Truax AC, et al. TDP-43-positive white matter pathology in frontotemporal lobar degeneration with ubiquitin-positive inclusions. J Neuropathol Exp Neurol. 2007;66:177–183. [DOI] [PubMed] [Google Scholar]
- 87. van Echten-Deckert G, Herget T. Sphingolipid metabolism in neural cells. Biochim Biophys Acta. 2006;1758:1978–1994. [DOI] [PubMed] [Google Scholar]
- 88. Choi JW, Chun J. Lysophospholipids and their receptors in the central nervous system. Biochim Biophys Acta. 2013;1831:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nicholson AM, Finch NA, Almeida M, et al. Prosaposin is a regulator of progranulin levels and oligomerization. Nat Commun. 2016;7:11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhou X, Sun L, Bastos de Oliveira F, et al. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J Cell Biol. 2015;210:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhou X, Sun L, Bracko O, et al. Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations. Nat Commun. 2017;8:15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sandhoff R, Sandhoff K. Emerging concepts of ganglioside metabolism. FEBS Lett. 2018;592:3835–3864. [DOI] [PubMed] [Google Scholar]
- 93. Schulze H, Sandhoff K. Sphingolipids and lysosomal pathologies. Biochim Biophys Acta. 2014;1841:799–810. [DOI] [PubMed] [Google Scholar]
- 94. Chen Y, Jian J, Hettinghouse A, et al. Progranulin associates with hexosaminidase A and ameliorates GM2 ganglioside accumulation and lysosomal storage in Tay-Sachs disease. J Mol Med. 2018;96:1359–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Jian J, Tian Q-Y, Hettinghouse A, et al. Progranulin recruits HSP70 to β-glucocerebrosidase and is therapeutic against Gaucher disease. EBioMedicine. 2016;13:212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Quarta S, Camprubí-Robles M, Schweigreiter R, et al. Sphingosine-1-phosphate and the S1P3 receptor initiate neuronal retraction via RhoA/ROCK associated with CRMP2 phosphorylation. Front Mol Neurosci. 2017;10:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Awojoodu AO, Ogle ME, Sefcik LS, et al. Sphingosine 1-phosphate receptor 3 regulates recruitment of anti-inflammatory monocytes to microvessels during implant arteriogenesis. Proc Natl Acad Sci USA. 2013;110:13785–13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Nussbaum C, Bannenberg S, Keul P, et al. Sphingosine-1-phosphate receptor 3 promotes leukocyte rolling by mobilizing endothelial P-selectin. Nat Commun. 2015;6:6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Murakami K, Kohno M, Kadoya M, et al. Knock out of S1P3 receptor signaling attenuates inflammation and fibrosis in bleomycin-induced lung injury mice model. PLoS ONE. 2014;9:e106792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Weigert A, Olesch C, Brune B. Sphingosine-1-phosphate and macrophage biology-how the sphinx tames the big eater. Front Immunol. 2019;10:1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dong YF, Guo R-B, Ji J, et al. S1PR3 is essential for phosphorylated fingolimod to protect astrocytes against oxygen-glucose deprivation-induced neuroinflammation via inhibiting TLR2/4-NFkappaB signalling. J Cell Mol Med. 2018;22:3159–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Rollinson S, Young K, Bennion-Callister J, Pickering-Brown SM. Identification of biological pathways regulated by PGRN and GRN peptide treatments using transcriptome analysis. Eur J Neurosci. 2016;44:2214–2225. [DOI] [PubMed] [Google Scholar]
- 103. Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Melamed Z, López-Erauskin J, Baughn MW, et al. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat Neurosci. 2019;22:180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Klim JR, Williams LA, Limone F, et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci. 2019;22:167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Prudencio M, Humphrey J, Pickles S, et al. Truncated stathmin-2 is a marker of TDP-43 pathology in frontotemporal dementia. J Clin Invest. 2020;130:6080–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ma XR, Prudencio M, Koike Y, et al. TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature. 2022;603(7899):124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Brown A-L, Wilkins OG, Keuss MJ, et al. Common ALS/FTD risk variants in UNC13A exacerbate its cryptic splicing and loss upon TDP-43 mislocalization. bioRxiv [Preprint]. 2021. doi: 10.1101/2021.04.02.438170 [DOI]
- 109. Josephs KA, Whitwell JL, Weigand SD, et al. TDP-43 is a key player in the clinical features associated with Alzheimer’s disease. Acta Neuropathol. 2014;127:811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Josephs KA, Whitwell JL, Tosakulwong N, et al. TAR DNA-binding protein 43 and pathological subtype of Alzheimer’s disease impact clinical features. Ann Neurol. 2015;78:697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Josephs KA, Murray ME, Tosakulwong N, et al. Pathological, imaging and genetic characteristics support the existence of distinct TDP-43 types in non-FTLD brains. Acta Neuropathol. 2019;137:227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Tome SO, Vandenberghe R, Ospitalieri S, et al. Distinct molecular patterns of TDP-43 pathology in Alzheimer’s disease: relationship with clinical phenotypes. Acta Neuropathol Commun. 2020;8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated herein are available in the Synapse platform under accession number ‘syn25991921’. All other relevant data supporting key findings of this study are available in the article and Supplementary material.