Abstract
The first step of eukaryotic gene expression is the assembly of RNA polymerase (Pol) II and general transcription factors on promoter DNA. This highly regulated process involves ~80 different proteins that together form the preinitiation complex (PIC). Decades of work have gone into understanding PIC assembly using biochemical and structural approaches. These efforts have yielded significant, but partial descriptions of PIC assembly. Over the past few years, cryo-electron microscopy has provided the first high-resolution structures of the near complete mammalian PIC assembly. These structures have revealed that PIC assembly is a highly dynamic process. This review will summarize recent structural findings and discuss their implications for understanding cell-type specific gene expression.
Keywords: Transcription initiation, cryo-EM, Mediator, TFIIH, TFIID
Introduction
Expression of eukaryotic protein-coding genes is dependent on their transcription by RNA polymerase (Pol) II. The first step of Pol II-mediated transcription is initiation. Initiation involves preinitiation complex (PIC) formation. PIC formation entails the recruitment of Pol II to gene promoters by general transcription factors. Appropriate PIC assembly is critical for defining cell-type specific gene expression.
A step-wise model for PIC assembly has emerged from decades of biochemical and structural work[1]. In the classical model of PIC assembly, the multi-subunit complex Transcription Factor (TF) IID first recognizes promoter DNA sequence elements and deposits TATA-binding protein (TBP) on promoter DNA[2]. This engagement of TBP with DNA is mediated by the TFIIA dimer. Next, DNA bound TBP-TFIIA recruits TFIIB, and TFIIB permits binding of the Pol II-TFIIF complex to the closed promoter DNA. The multi-subunit Mediator complex associates with the unphosphorylated Pol II C-terminal domain (CTD) of the largest Pol II subunit, RPB1[3]. Additionally, Mediator contacts the Pol II stalk subcomplex (RPB4/7), the Pol II foot (RPB8), and the TFIIH kinase module[4-8]. Finally, TFIIE and TFIIH engagement with Pol II induces architectural rearrangements within the PIC that result in promoter DNA melting and the formation of an open complex that is competent for synthesizing RNA.
Previous structural studies of PIC assembly have focused on TFIID with DNA, the core PIC with TBP, or the characterization of the Mediator and TFIIH bound PIC[6,8-15]. These efforts have produced partial pictures of PIC assembly, leaving important questions about the assembly process itself open. For example, it is unclear how DNA sequence influences initiation factor recruitment, how transcription initiation factors interact with each other in a PIC containing all factors (holo-PIC), and how enzymatic activities by kinases and ATPases support the transition into transcription elongation. This review will summarize recent structures of PIC assembly[4,5,7,16-18]. These structures have illuminated the dynamic landscapes of TFIID, Mediator, and TFIIH, further clarified the assembly process of the holo-PIC and described structural rearrangements that allow for the deposition of phosphorylations. Advances in cryo-electron microscopy (EM) technology, advent of direct electron detectors, and the development of cryo-EM processing procedures that accommodate structural flexibility have facilitated PIC structure determination[13]. Together, the latest structural efforts have brought a fresh understanding of PIC assembly to better explain how promoters are utilized in a cell type specific manner.
TFIID
TFIID is a 1.3 MDa complex consisting of TBP and 13 TFIID associated factors (TAFs)[20]. TFIID’s extreme flexibility and dynamic nature have made it a challenging structural target. The Nogales lab has pioneered structural studies of TFIID and has resolved structures of isolated TFIID and TFIID bound to DNA [10,11,21,22]. These structures and others have provided the overall architecture and assembly pathway for TFIID on promoters [10,11,16].
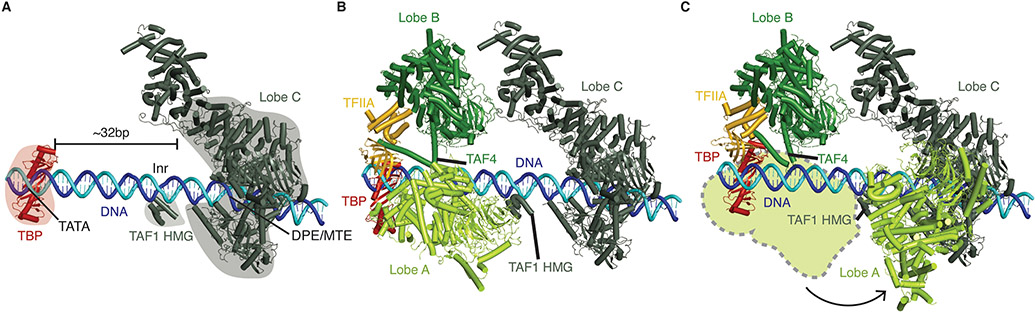
TFIID can be divided into three lobes termed A, B, and C [22] (Figure 1). Lobes B and C are relatively static whereas lobe A is highly flexible. Overall, TFIID dynamics are dictated by promoter DNA positioning sequences. These sequences include the motif ten element (MTE), the downstream promoter element (DPE), the Initiator (Inr), and the TATA sequence. Lobe C binds the MTE and DPE. Its subunit TAF1 additionally interacts with the Inr through its HMG domain [10,16]. TBP binds the TATA sequence (Figure 1A). TBP is a strong DNA binder and its interactions with lobe A subunits TAF11/13 and TAF1 prevent TBP from binding and bending DNA until it associates with TFIIA[10,11]. TFIIA supports deposition of TBP onto the upstream DNA (Figure 1B). Formation of the TBP-TFIIA complex results in TBP release from lobe A and stable TBP association with DNA. After TBP release, lobe A rotates towards lobe C (Figure 1C)[10,11,16].
Figure 1. TFIID organization and interactions with DNA.
A. Binding sites for TFIID lobes on promoter DNA. Lobe C binds the MTE/DPE elements. The Inr element is associated with the TAF1 HMG domain. TBP binds the upstream TATA element (PDB ID 7EGJ)[16].
B-C. TFIID lobe A is highly mobile. In panel B, lobe A is associated with TBP at the TATA site (PDB ID 7EGD) [16]. In panel C, lobe A has rotated and lies next to lobe B/C (PDB ID 7EGJ) [16]. The position of lobe A as seen in panel B is shown as a lime green surface.
Recent structures of TFIID-TFIIA complexes determined on various promoter sequences have shown that the ~32 base pair spacing between TBP and lobe C is maintained regardless of the sequence context (e.g., promoters lacking TATA or DPE elements)[16]. This suggests that lobes B and C serve to space lobe C and TBP on DNA in a largely sequence independent manner (Figure 1)[16].
The Mediator Complex
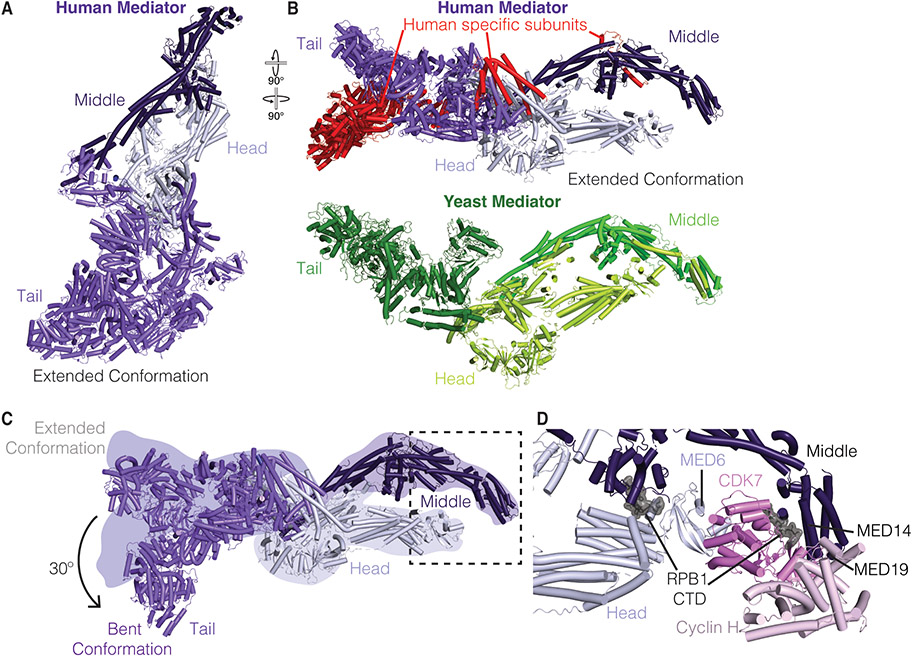
Mediator is a multi-subunit complex that associates with Pol II at promoters. Mammalian Mediator consists of 26 subunits and a dissociable 4-subunit kinase module that is not incorporated into the PIC[23]. Mediator complex lacking the kinase can be divided into three regions termed the head, middle, and tail modules (Figure 2A). Over the course of the last decade, structures of yeast head and middle modules were obtained by X-ray crystallography[23-25]. High resolution structures of the entire Mediator complex, however, have proven challenging to obtain due to size constraints (>1 MDa) and the dynamic and asymmetric nature of the complex. Excitingly, recent cryo-EM work has led to near complete structures of yeast, mouse, and human Mediator in isolation or bound to Pol II (Figure 2A,B) [4,5,7,26,27].
Figure 2. Structural of overview of Mediator and its interactions with TFIIH.
A. Human Mediator in the extended conformation (PDB ID 7EMF)[4]. Head, Middle, and Tail module colored in lilac, dark purple, and violet, respectively.
B. Top: Human Mediator in the extended conformation (PDB ID 7EMF)[4]. Model rotated relative to A. Human specific subunits are colored red. Bottom: Composite structure of yeast Mediator tail (Chaetomium thermophilum, PDB ID 7JMN, Med1, Med24, and unknown subunit Chaetomium thermophilum, PDB ID 6XP5, forest green), and Mediator middle (green) and head (limon) (S. cerevisiae, PDB ID 5OQM)[6,27].
C. Dynamics of the Mediator tail module in extended and bent conformations. Position of extended conformation shown as colored silhouettes. Bent conformation shown in cartoon (PDB ID 7ENJ) [4]. Dotted box indicates binding position of the CAK on Mediator.
D. Interactions of Mediator with TFIIH CAK module (pink). Figure corresponds to boxed region in Panel B. A portion of the RPB1 CTD (grey, surface) was observed in the structure (PDB ID 7LBM)[5].
The isolated structures of Mediator are remarkably similar despite low sequence identity (10-30% sequence identity between yeast and human Mediator subunits)[28] (Figure 2B). The most substantial differences both structurally and compositionally can be attributed to differences in the tail module (yeast/human subunits Med2/MED29, Med3/MED27, Med5/MED24, Med14/MED14, Med15/MED15, Med16/MED16 and additional metazoan specific subunits MED25 and MED23) and metazoan specific subunits (MED26 in the middle module, MED28 and MED30 connecting the head and tail modules)[26,27,29,30] (Figure 2B). The tail module is important for engaging transcription activators that link signal transduction to gene expression[31]. This review will focus on novel aspects of recent Mediator structures, including the tail module and Mediator interactions with Pol II and TFIIH. Readers are referred to other current reviews for in depth discussion of the head and middle modules[23,32].
Dynamics of mammalian Mediator
Recent cryo-EM structures have enabled near complete visualization of the Mediator tail module and have defined how the tail module is connected to both the head and middle modules [4,5,26]. Multiple interfaces connect the tail module to the rest of Mediator. Most notably, the MED14 subunit traverses all modules of Mediator and thereby connects the head, middle, and tail modules. Additionally, subunits MED27, MED28, MED29, MED30, and the N-terminus of MED15 together with MED14 coordinate the interaction between the tail and head modules. The Middle module forms a more dynamic interface with the tail module through subunits MED1, MED16, MED24, MED25, and the MED15 C-terminus.
Unexpectedly, the human Mediator tail module has been visualized in extended and bent states[4] (Figure 2A-C). In the extended state, the Mediator tail is elongated and MED23 interacts with MED24. In contrast, the bent state, which has a kinked tail conformation, lacks resolved density for MED25 and parts of MED16. Additionally, the contact between MED24 and MED23 is lost. To adopt the bent conformation, the tail module rotates by ~30° towards the inner face of the complex (Figure 2C). The bent conformation, but not the extended conformation, is observed in structures of Mediator bound to Pol II and TFIID[4]. In structures of initiation complexes lacking TFIID, however, Mediator adopts the extended conformation[5,7]. It is currently unclear why the tail module adopts these two different states and how this flexibility is linked to transcription initiation.
Despite the recent structural breakthroughs, significant portions of Mediator have not been resolved in the latest cryo-EM reconstructions[4,5,7]. This includes MED26, a metazoan specific subunit, where weak density for its C-terminal region is observed in the middle module of Mediator[4]. Other Mediator subunits are only partially resolved including MED1, MED4, MED15, MED19, and MED25[4,5,26]. The missing regions are associated with the Super Elongation Complex (complex formed between factors AFF1/4, AF9/ENL, CDK9, CyclinT1, ELL1/2, and EAF1/2), transcriptional activators, and condensate formation[31,33,34]. Future work is required to understand how inherent flexibility of Mediator subunits contributes to gene activation.
TFIIH-Mediator network enables CTD phosphorylation
The structures of human Mediator bound to the PIC have been resolved by three independent groups[4,5,7]. These structures have clarified how Mediator engages TFIIH and positions the TFIIH kinase subcomplex termed the CDK-activating kinase (CAK) module in context of the PIC (Figure 2D). The MAT1, CDK7, and Cyclin H containing CAK module signals the transition from initiation to elongation by phosphorylating the RPB1 CTD[35]. Mediator itself interacts with the unphosphorylated CTD through contacts in its middle module[4,5,7]. In structures of the yeast Mediator-PIC, the CAK was poorly resolved[6,15]. In contrast, the mammalian structures have enabled unambiguous docking of the CAK module adjacent to the Mediator head and middle modules. Specifically, the CAK binds Mediator subunits MED6, MED14, and MED19. The TFIIH subunit MAT1 connects the TFIIH core to the CAK. Densities for two CTD heptad repeats are resolved within the CAK active site (Figure 2D)[4,5]. Additional density for the CTD is observed in the Mediator middle module and has the same directionality as the CTD fragment bound within the CAK. From these observations, the He lab has proposed a model for sequential CTD heptad phosphorylation starting from the proximal repeats[4,5]. CTD phosphorylation eventually leads to the displacement of Mediator from Pol II, although the mechanism is not fully understood[36]. In addition to phosphorylating Pol II, TFIIH also contributes to promoter opening.
TFIIH and promoter opening
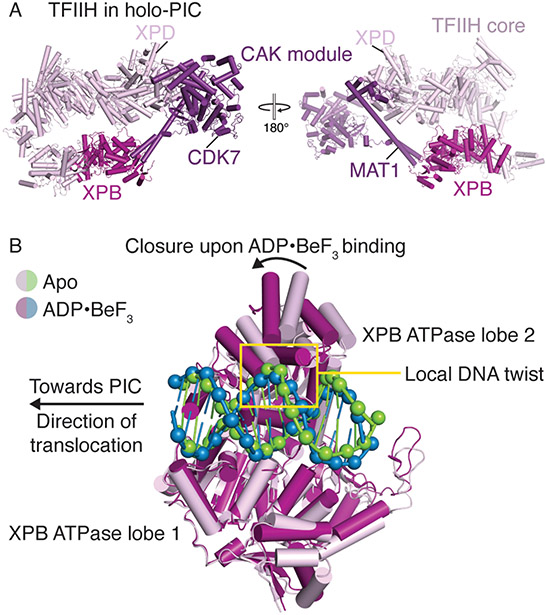
CAK containing TFIIH is a horseshoe-shaped 10-subunit complex with the XPB and XPD translocases flanking opposite ends (Figure 3A)[37]. Whereas the XPD translocase is in an inactive conformation during transcription initiation, the double-stranded DNA translocase XPB engages the downstream DNA and is believed to play an important role in the formation of an open promoter complex[38-41]. The process of promoter DNA opening is a critical checkpoint during transcription initiation and has been the focus of several studies. Structures of the yeast PIC revealed promoter DNA that was spontaneously opened[6,9,14]. In stark contrast, mammalian PIC structures determined over the past few years have captured only the closed promoter state[4,5,7,16]. This striking distinction in promoter opening is believed to stem from differences in yeast and mammalian TFIIE and TFIIH where opening of mammalian promoter DNA appears to be dependent on the ATPase activity of the XPB translocase[17].
Figure 3. TFIIH architecture and conformational changes in TFIIH XPB.
A. Structural overview of human TFIIH. XPB and the CAK module are colored in fuchsia and purple, respectively (PDB ID 7ENC)[4]. The positions of CDK7, MAT1, and XPD are indicated.
B. DNA translocation by XPB facilitates promoter opening. XPB ATPase lobe 1 in the apo (PDB ID 7NVW, pink and green)[17] and ADP·BeF3 bound states (PDB ID 7NVV, fuchsia and blue)[17] are aligned. Binding of ADP·BeF3 induces closure of the XPB ATPase and propagation of the local DNA twist upstream.
The Cramer group has recently captured snapshots of mammalian promoter DNA opening induced by conformational changes in the TFIIH XPB translocase [17,18]. Specifically, binding of XPB to the closed promoter DNA results in the introduction of local DNA twist (Figure 3B). ATP binding to XPB then induces closure of the XPB ATPase motor that results in upstream propagation of the DNA twist. Because the upstream DNA is fixed by TBP, XPB ATPase closure creates a torsional force that may facilitate opening of the promoter DNA by a single XPB translocation event. This finding is surprising because previous data suggested that multiple cycles of ATP hydrolysis and translocation by XPB were required for promoter DNA to open[38].
Structure of the Mammalian Holo-PIC
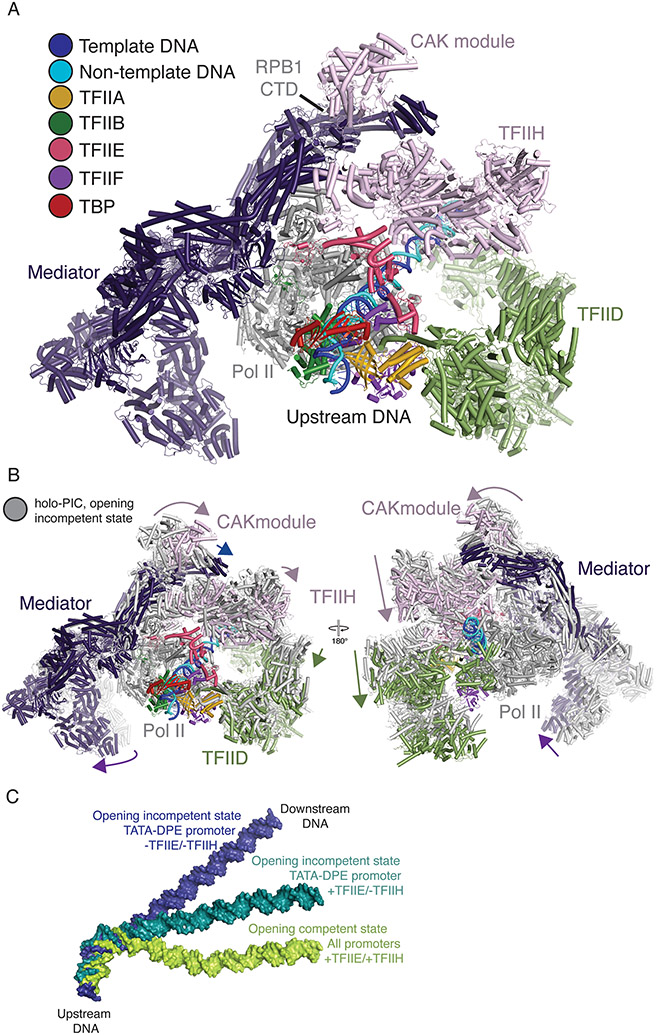
Advances made over the last ten years have broadened our understanding of PIC assembly, but the structure of the holo-PIC remained elusive. In a recent breakthrough, the Xu group has reconstituted and obtained reconstructions of the 76-subunit, 4.1 MDa mammalian holo-pre-initiation complex with Mediator, TFIID, and TFIIH (Figure 4A) [4]. The reconstructions reveal two distinct states of PIC assembly with an overall conserved architecture. Pol II, TFIIA, TBP, TFIIF, TFIIE, and TFIIB form the core of the complex. Mediator, TFIID, and TFIIH encircle the core PIC from three sides. Mediator contacts Pol II subunits RPB1, the polymerase stalk subunits RPB4/7, the RPB1 CTD as well as TFIIH and TFIIE via its head and middle modules. TFIID reaches from the upstream to the downstream DNA of Pol II and does not contact the polymerase directly but rather interacts with general transcription factors TFIIA, TFIIB, and TFIIF. TFIIH bridges Mediator and TFIID. TFIIH also contacts the core PIC via interactions of XPD and MAT1 with TFIIE and the Pol II stalk.
Figure 4. Structural overview of the holo-PIC and DNA dynamics.
A. Structure of the holo-PIC ready for promoter DNA opening (PDB ID 7ENC)[4]. Mediator (dark purple), TFIID (green), TFIIH (pink) and general transcription factors TFIIA, TFIIB, TFIIE, TFIIF, and TBP (gold, dark green, light pink, purple, and red) encircle Pol II from three sides. Template and non-template DNA are colored blue and cyan, respectively.
B. Conformational changes in the arrangement of Mediator, TFIID and TFIIH reposition the promoter DNA to form the promoter opening competent holo-PIC as shown in A (PDB ID 7ENC and 7ENA)[4]. Conformational changes are indicated by arrows.
C. DNA conformations of PIC-TFIID complexes in the presence or absence of TFIIE and/or TFIIH show transition of TATA-DPE promoter DNA from opening incompetent states (PDB ID 7EG7, blue and PDB ID 7EG9, teal)[16] to an opening competent state (PDB ID 7EGB, green)[16]. TATA-less promoter DNA immediately adopts the opening competent state.
The two observed PIC states exhibit closed promoter DNA. In the first state, the TFIIH translocase XPB does not fully engage with DNA, and TFIID lobe C and the TAF1 HMG contact the DPE and Inr elements, respectively (Figure 4B). The second state is competent for promoter opening. The DNA shifts by ~20 Å, and Mediator rotates by ~20° relative to the Pol II stalk. In this state, TFIIH subunits p52 and p8 associate with TFIID lobe C subunit TAF2 and prevent its reassociation with the promoter DNA. The promoter DNA that was formerly occupied by TFIID lobe C is now bound by the TFIIH core. TFIID and Mediator sandwich TFIIH and position XPB to bind promoter DNA and facilitate XPB driven promoter DNA opening. Structures of human PIC lacking TFIID but containing Mediator and TFIIH have only been observed in the second state, which is competent for promoter DNA opening[5,7]. The association of Mediator with the Pol II foot (RPB8) is only observed in the first state and is lost in the DNA opening competent state. Notably, the Mediator-Pol II foot interaction appears to be stable throughout initiation in the yeast system[6,14].
Together, these structures of the holo-PIC define how Mediator, TFIID, and TFIIH dynamically interact to regulate PIC assembly. TFIID together with Mediator helps stabilize TFIIH to facilitate promoter opening and CTD phosphorylation. Thus, PIC assembly is finely choreographed and is dependent on all PIC components to ensure appropriate gene expression.
DNA rearrangements in the holo-PIC
To address how promoter sequence context affects PIC assembly, PIC structures lacking Mediator were acquired on various promoter sequences and in the absence and presence of TFIIE and TFIIH[16]. These structures have revealed that sequence and factor context affect the conformational state of DNA within the PIC. Surprisingly, the TFIID-PIC structures determined on TATA-less promoters immediately adopt a conformation that supports promoter opening[16]. These structures bear striking resemblance to previously determined structures of the TBP-PIC complex and the promoter opening competent state of the holo-PIC[4,6,12,13]. Conversely, TATA-containing promoters that include the DPE transition through at least two additional DNA arrangements to reach the promoter opening competent state (Figure 4C). Initially, TATA-DPE promoters formed in the absence of TFIIE and TFIIH, adopt an opening incompetent state. Subsequent association of TFIIE then leads to an intermediate state, and finally, the inclusion of TFIIH facilitates the DNA transition into opening competency[16].
Overall, these structures suggest that promoter sequence can directly affect the conformational landscape of PIC formation and DNA positioning[16]. It is unclear, however, why PICs formed on ideal promoter sequences (e.g., TATA-DPE) go through several conformational intermediates, whereas PICs formed on non-ideal promoter sequences (e.g., TATA-less) directly adopt the state competent for promoter opening[16].
Outstanding questions
The past few years have yielded an array of structures that are required for understanding PIC assembly and promoter opening[4-6,9,16-18]. There are several outstanding questions in the field that require additional structural interrogation. Gene activation frequently requires activator proteins that interact with Mediator or TFIID[42]. The regions of Mediator that bind activators are not resolved in any of the recent structures[5]. Structures of the TFIID-PIC bound to the co-activator p53 have been determined, but it is still unclear how p53 mediates gene activation[16]. It also remains unknown if Mediator itself can bridge enhancer and promoter elements. A plethora of activator proteins exist and the catalog of structural interactions with the PIC remain to be characterized[5,16,42].
Once Pol II escapes from promoters, it enters the elongation phase of transcription. In vitro data suggests that initiation is coupled to elongation and that particularly pausing in promoter proximal regions can affect initiation frequency[43,44]. A preprint from the Murakami group has shown that two Pol II molecules can be resolved at high-resolution on a single DNA molecule after initiation from the same promoter[45]. It will now be important to build on this system and investigate how initiation factor bound Pol II interacts with an elongating or paused downstream Pol II molecule.
PIC formation is thought to require a nucleosome free or depleted region at gene promoters[46]. In addition to sequence elements that help place PICs on promoters in the correct orientation, the first gene body (+1) nucleosome affects where transcription initiates[47]. Structures of PIC complexes with the +1 nucleosome will help explain how PIC positioning on promoters is influenced by nucleosomes. Recently, a structure of TBP bound to a nucleosome was determined[48]. However, it remains unclear if this binding would allow for PIC formation.
The SAGA complex can place TBP at some promoters[49-52]. Currently it is unknown if SAGA can interact with the PIC, and how sequence context affects its conformation on DNA. Given the recent structural successes with TFIID, it should now be possible to probe SAGA association with a wide array of promoter sequences and assess whether SAGA can be incorporated into the PIC[16].
Recent studies have provided a wealth of structural information for transcriptional pre-initiation complexes. These studies have helped to illuminate how conformational flexibility can be used to engage specific DNA sequences, position elements for phosphorylation, and result in promoter DNA opening. It will be exciting to see how these structures are used to test hypotheses in cells and yield new insights into the process of transcription initiation.
Acknowledgements
We apologize to colleagues we could not cite due to space constraints. We thank members of our laboratories for discussions and comments. S.M.V acknowledges funding from the Smith Family Foundation, ALSF Crazy 8 Initiative, NIH DP2-GM146254, and the MIT Surdna Fund.
Footnotes
Declaration of Interests
The authors declare no conflict of interest.
References
•special interest
••outstanding interest
- 1.Sainsbury S, Bernecky C, Cramer P: Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol 2015, 16:129–143. [DOI] [PubMed] [Google Scholar]
- 2.Schier AC, Taatjes DJ: Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev 2020, 34:465–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myers LC, Gustafsson CM, Bushnell DA, Lui M, Erdjument-Bromage H, Tempst P, Kornberg RD: The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev 1998, 12:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. ••. Chen X, Yin X, Li J, Wu Z, Qi Y, Wang X, Liu W, Xu Y: Structures of the human Mediator and Mediator-bound preinitiation complex. Science 2021, 372. This paper describes the structure of the holo-PIC. Human Mediator is observed in the extended and bent conformations.
- 5. ••. Abdella R, Talyzina A, Chen S, Inouye CJ, Tjian R, He Y: Structure of the human Mediator-bound transcription preinitiation complex. Science 2021, 372:52–56. Structure of the human PIC-Mediator complex. This structure resolves the RPB1 CTD bound in the TFIIH CAK and provides a model for CTD phosphorylation.
- 6.Schilbach S, Hantsche M, Tegunov D, Dienemann C, Wigge C, Urlaub H, Cramer P: Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 2017, 551:204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. ••. Rengachari S, Schilbach S, Aibara S, Dienemann C, Cramer P: Structure of the human Mediator-RNA polymerase II pre-initiation complex. Nature 2021, 594:129–133. This paper resolves a partial structure of human Mediator bound to a mammalian PIC. The structure shows how TFIIH and Mediator interact.
- 8.Plaschka C, Larivière L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko EV, Borchers CH, Baumeister W, Herzog F, et al. : Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 2015, 518:376–380. [DOI] [PubMed] [Google Scholar]
- 9.Dienemann C, Schwalb B, Schilbach S, Cramer P: Promoter distortion and opening in the RNA polymerase II cleft. Mol Cell 2019, 73:97–106.e4. [DOI] [PubMed] [Google Scholar]
- 10.Patel AB, Louder RK, Greber BJ, Grünberg S, Luo J, Fang J, Liu Y, Ranish J, Hahn S, Nogales E: Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 2018, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louder RK, He Y, López-Blanco JR, Fang J, Chacón P, Nogales E: Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 2016, 531:604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y, Fang J, Taatjes DJ, Nogales E: Structural visualization of key steps in human transcription initiation. Nature 2013, 495:481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E: Near-atomic resolution visualization of human transcription promoter opening. Nature 2016, 533:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P: Transcription initiation complex structures elucidate DNA opening. Nature 2016, 533:353–358. [DOI] [PubMed] [Google Scholar]
- 15.Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei P-J, Burlingame AL, Kornberg RD: Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell 2016, 166:1411–1422.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. ••. Chen X, Qi Y, Wu Z, Wang X, Li J, Zhao D, Hou H, Li Y, Yu Z, Liu W, et al. : Structural insights into preinitiation complex assembly on core promoters. Science 2021, 372. Structure of TFIID bound PIC reveals dynamics of PIC assembly. These structures also show that DNA sequence affects the conformational state adopted by TFIID on DNA.
- 17. ••. Aibara S, Schilbach S, Cramer P: Structures of mammalian RNA polymerase II pre-initiation complexes. Nature 2021, 594:124–128. This paper describes promoter DNA opening in the mammalian system. The authors were able to capture partially open promoters in the presence of a nucleotide analog and propose a model for DNA opening by XPB.
- 18.•. Schilbach S, Aibara S, Dienemann C, Grabbe F, Cramer P: Structure of RNA polymerase II pre-initiation complex at 2.9 Ådefines initial DNA opening. Cell 2021, 184:4064–4072.e28. This paper describes how DNA opening is promoted by the Ssl2 translocase and provides high resolution structures of yeast promoter DNA opening.
- 19.Cheng Y, Glaeser RM, Nogales E: How Cryo-EM Became so Hot. Cell 2017, 171:1229–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burley SK, Roeder RG: Biochemistry and structural biology of transcription factor IID (TFIID). Annu Rev Biochem 1996, 65:769–799. [DOI] [PubMed] [Google Scholar]
- 21.Andel F, Ladurner AG, Inouye C, Tjian R, Nogales E: Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 1999, 286:2153–2156. [DOI] [PubMed] [Google Scholar]
- 22.Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E: Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 2013, 152:120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verger A, Monté D, Villeret V: Twenty years of Mediator complex structural studies. Biochem Soc Trans 2019, 47:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larivière L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P: Structure of the Mediator head module. Nature 2012, 492:448–451. [DOI] [PubMed] [Google Scholar]
- 25.Nozawa K, Schneider TR, Cramer P: Core Mediator structure at 3.4 Å extends model of transcription initiation complex. Nature 2017, 545:248–251. [DOI] [PubMed] [Google Scholar]
- 26. •. Zhao H, Young N, Kalchschmidt J, Lieberman J, El Khattabi L, Casellas R, Asturias FJ: Structure of mammalian Mediator complex reveals Tail module architecture and interaction with a conserved core. Nat Commun 2021, 12:1355. This paper describes the near complete mouse Mediator structure.
- 27.Zhang H, Chen D-H, Mattoo RUH, Bushnell DA, Wang Y, Yuan C, Wang L, Wang C, Davis RE, Nie Y, et al. : Mediator structure and conformation change. Mol Cell 2021, 81:1781–1788.e4. [DOI] [PubMed] [Google Scholar]
- 28.Poss ZC, Ebmeier CC, Taatjes DJ: The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol 2013, 48:575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai K-L, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ: Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 2014, 157:1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Khattabi L, Zhao H, Kalchschmidt J, Young N, Jung S, Van Blerkom P, Kieffer-Kwon P, Kieffer-Kwon K-R, Park S, Wang X, et al. : A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers. Cell 2019, 178:1145–1158.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeronimo C, Langelier M-F, Bataille AR, Pascal JM, Pugh BF, Robert F: Tail and kinase modules differently regulate core mediator recruitment and function in vivo. Mol Cell 2016, 64:455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plaschka C, Nozawa K, Cramer P: Mediator architecture and RNA polymerase II interaction. J Mol Biol 2016, 428:2569–2574. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CAS, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. : Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 2011, 146:92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. : Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiekhattar R, Mermelstein F, Fisher RP, Drapkin R, Dynlacht B, Wessling HC, Morgan DO, Reinberg D: Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature 1995, 374:283–287. [DOI] [PubMed] [Google Scholar]
- 36.Wong KH, Jin Y, Struhl K: TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol Cell 2014, 54:601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greber BJ, Nguyen THD, Fang J, Afonine PV, Adams PD, Nogales E: The cryo-electron microscopy structure of human transcription factor IIH. Nature 2017, 549:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fishburn J, Tomko E, Galburt E, Hahn S: Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc Natl Acad Sci U S A 2015, 112:3961–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alekseev S, Nagy Z, Sandoz J, Weiss A, Egly J-M, Le May N, Coin F: Transcription without XPB Establishes a Unified Helicase-Independent Mechanism of Promoter Opening in Eukaryotic Gene Expression. Mol Cell 2017, 65:504–514.e4. [DOI] [PubMed] [Google Scholar]
- 40.Kuper J, Braun C, Elias A, Michels G, Sauer F, Schmitt DR, Poterszman A, Egly J-M, Kisker C: In TFIIH, XPD helicase is exclusively devoted to DNA repair. PLoS Biol 2014, 12:e1001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peissert S, Sauer F, Grabarczyk DB, Braun C, Sander G, Poterszman A, Egly J-M, Kuper J, Kisker C: In TFIIH the Arch domain of XPD is mechanistically essential for transcription and DNA repair. Nat Commun 2020, 11:1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zabidi MA, Stark A: Regulatory Enhancer–Core-Promoter Communication via Transcription Factors and Cofactors. Trends Genet 2016, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gressel S, Schwalb B, Decker TM, Qin W, Leonhardt H, Eick D, Cramer P: CDK9-dependent RNA polymerase II pausing controls transcription initiation. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao W, Zeitlinger J: Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet 2017, 49:1045–1051. [DOI] [PubMed] [Google Scholar]
- 45.Yang C, Fujiwara R, Kim HJ, Colón JJG, Steimle S, Garcia BA, Murakami K: Structural visualization of de novo initiation of RNA polymerase II transcription. BioRxiv 2021, doi: 10.1101/2021.05.03.442346. [DOI] [Google Scholar]
- 46.Imbalzano AN, Kwon H, Green MR, Kingston RE: Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 1994, 370:481–485. [DOI] [PubMed] [Google Scholar]
- 47.Rhee HS, Pugh BF: Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 2012, 483:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Xiong L, Cramer P: Structures and implications of TBP-nucleosome complexes. Proc Natl Acad Sci U S A 2021, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donczew R, Warfield L, Pacheco D, Erijman A, Hahn S: Two roles for the yeast transcription coactivator SAGA and a set of genes redundantly regulated by TFIID and SAGA. Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papai G, Frechard A, Kolesnikova O, Crucifix C, Schultz P, Ben-Shem A: Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature 2020, 577:711–716. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Dienemann C, Stützer A, Urlaub H, Cheung ACM, Cramer P: Structure of the transcription coactivator SAGA. Nature 2020, 577:717–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herbst DA, Esbin MN, Louder RK, Dugast-Darzacq C, Dailey GM, Fang Q, Darzacq X, Tjian R, Nogales E: Structure of the human SAGA coactivator complex. Nat Struct Mol Biol 2021, 28:989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]