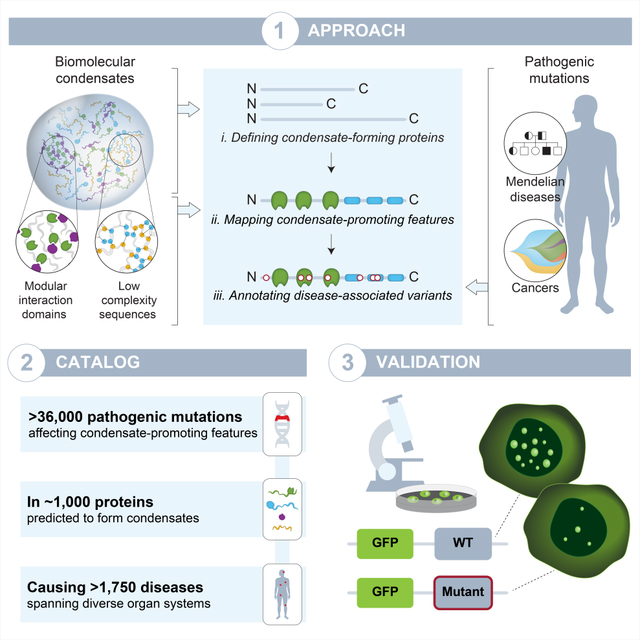
SUMMARY
A multitude of cellular processes involve biomolecular condensates, which has led to the suggestion that diverse pathogenic mutations may dysregulate condensates. While proof-of-concept studies have identified specific mutations that cause condensate dysregulation, the full scope of pathological genetic variation that affects condensates is not yet known. Here we comprehensively map pathogenic mutations to condensate-promoting protein features in putative condensate-forming proteins and find over 36,000 pathogenic mutations that plausibly contribute to condensate dysregulation in over 1,200 Mendelian diseases and 550 cancers. This resource captures mutations presently known to dysregulate condensates and experimental tests confirm that additional pathological mutations do indeed affect condensate properties in cells. These findings suggest that condensate dysregulation may be a pervasive pathogenic mechanism underlying a broad spectrum of human diseases, provide a strategy to identify proteins and mutations involved in pathologically altered condensates, and serve as a foundation for mechanistic insights into disease and therapeutic hypotheses.
Graphical Abstract
eTOC Blurb
Banani et al. map 36,000 pathogenic mutations to condensate-promoting features in proteins. The data lead to a prediction that more than 1,000 proteins are involved in condensate dysregulation in disease, providing a foundation for investigating the roles of condensates in disease and their potential therapeutic applications.
INTRODUCTION
How genetic variation gives rise to human disease is understood largely from the effects of mutations on the structure and function of individual protein molecules. Genetic and biochemical studies have revealed how mutations in protein coding sequences affect molecular-scale properties, such as conformation, stability, and catalytic activity, providing mechanistic hypotheses of disease causality that have led to valuable therapeutics (Stefl et al., 2013; Wan et al., 2004). However, underlying pathogenic mechanisms for many genetic diseases remain elusive, despite extensive cataloging of associated mutations. Recent studies have shown that disease-causing mutations may also affect properties related to mesoscale cellular organization (Kasza et al., 2019; Lewis et al., 2019). Many cellular proteins are compartmentalized within biomolecular condensates (Banani et al., 2017; Shin and Brangwynne, 2017), which are membraneless organelles that concentrate functionally related proteins and nucleic acids and organize many vital cellular processes, such as DNA replication, DNA repair, transcription, chromatin organization, RNA biosynthesis and homeostasis, ribosome biosynthesis, protein quality control, innate immunity, cell division, cell-cell adhesions, signaling, and synaptic transmission (Alberti, 2017; Beutel et al., 2019; Boija et al., 2018; Cai et al., 2019; Case et al., 2019; Cho et al., 2018; Du and Chen, 2018; Frottin et al., 2019; Gibson et al., 2019; Guo et al., 2019; Huang et al., 2019; Jiang et al., 2015; Kilic et al., 2019; King and Petry, 2020; Larson et al., 2017; Lu et al., 2020; Lyon et al., 2020; Milovanovic et al., 2018; Parker et al., 2019; Riback et al., 2020; Schwayer et al., 2019; Sheu-Gruttadauria and MacRae, 2018; Strom et al., 2017; Su et al., 2016; Woodruff et al., 2017; Zamudio et al., 2019; Zeng et al., 2016). A subset of condensate components directly governs the formation, maintenance, organization, composition, and physicochemical and material properties of the condensate (Banani et al., 2016; Feric et al., 2016; Jain et al., 2016; Li et al., 2012; Lin et al., 2015; Wang et al., 2018). Thus, a protein-coding mutation in a condensate-forming protein may affect not only the individual protein, but also the biomolecular condensate in which the protein is found. Specifically, mutations that affect regions of proteins that promote condensate formation can significantly alter the properties of condensates, including their formation (Ahn et al., 2021; Chandra et al., 2021; Li et al., 2020; Wan et al., 2019), material properties (Patel et al., 2015), localization (Boulay et al., 2017), or composition (Basu et al., 2020). These condensate-promoting features include modular interaction domains (MIDs) and stretches of low complexity sequences (LCSs) found within intrinsically disordered regions (IDRs) (Figure 1A). These observations have led us and others to postulate that condensate dysregulation may play a role across a broad spectrum of diseases (Alberti and Dormann, 2019; Alberti and Hyman, 2021; Boija et al., 2021; Kim et al., 2013; Molliex et al., 2015; Tsang et al., 2020).
Figure 1. A proteome-wide map of pathogenic mutations in condensate-promoting features. See also Figures S1–S2 and Table S1.
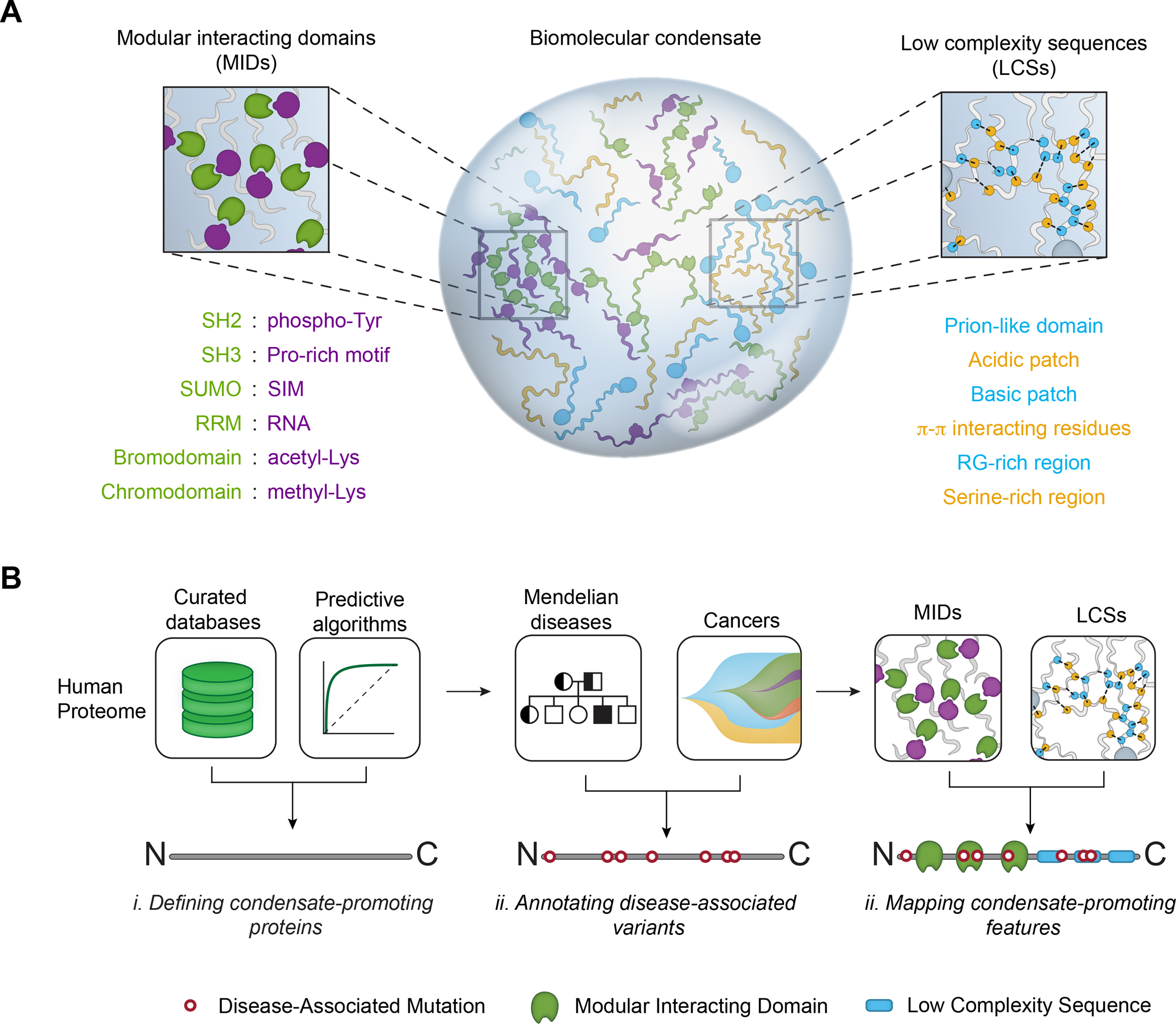
A. Multivalent interacting features in proteins that promote biomolecular condensate formation, including modular interacting domains (MIDs, left, green and purple) and low complexity sequences (LCSs, right, blue and green).
B. Approach to generate a map of pathogenic mutations that affect condensate-promoting features across the proteome (see also Figure S1A). MIDs and LCSs were mapped across the proteome (left, top) and used to define multivalent proteins (middle). Mendelian and cancer variants were mapped across the proteome (left, bottom), in particular on across the set of multivalent proteins (middle), to identify pathogenic mutations that affect MIDs and LCSs (Methods). The approach allows analysis of diseases, condensates, and mutational signatures associated with pathogenic mutations that affect condensate-promoting features in multivalent proteins (right).
A resource that links data on pathogenic genetic variation to condensate-promoting protein features could promote further study of diseases likely to involve dysregulated condensates. To this end, we collected putative condensate-forming proteins, annotated condensate-promoting sequence features (MIDs and LCSs) onto these proteins, and mapped a broad spectrum of human disease variants associated with Mendelian diseases and cancers to these features. This approach produced a catalog of over 36,000 pathogenic mutations associated with 1,790 diseases that may involve condensate dysregulation as an underlying pathogenic mechanism. To demonstrate the utility of this approach and estimate its predictive accuracy, we performed experimental tests across 12 proteins from the catalog and found most tested mutations do indeed cause condensate dysregulation phenotypes in cells. This resource and its associated analyses provide a foundation for the study of condensate-associated disease mechanisms by facilitating the generation of novel mechanistic and therapeutic hypotheses.
RESULTS
Generating a resource for the study of condensate dysregulation in disease
A set of putative condensate-forming proteins was defined by integrating existing databases of proteome-wide subcellular immunofluorescence (Yu et al., 2020), sequence-based predictions (Mierlo et al., 2021), and curation of phase-separating proteins from the literature (Li et al., 2019; Mészáros et al., 2019; You et al., 2019) (Figure 1B, Figure S1A, Table S4A–B, Methods). This approach defined 3,941 putative condensate-forming proteins.
Condensate-promoting features, consisting of MIDs and LCSs (Figure 1A), within these 3,941 putative condensate-forming proteins were then identified. MIDs, such as SH2, SH3, RRM, and Bromodomains, were defined by integrating annotations of the subset of conserved protein domains (Blum et al., 2020; Letunic et al., 2020; Lu et al., 2019; Mistry et al., 2020) known to participate in binding interactions (Bienz, 2020; Hentze et al., 2018; Lambert et al., 2018; Lunde et al., 2007; Pawson and Nash, 2003; Seet et al., 2006; Vaquerizas et al., 2009; Yun et al., 2011) (Table S4F). LCSs, such as prion-like domains (Alberti et al., 2009; Martin et al., 2020; Wang et al., 2018), low-complexity aromatic-rich kinked segments (LARKS) (Hughes et al., 2018), regions enriched with pi-interacting residues (Vernon et al., 2018), and acidic/basic charge blocks, were mapped using existing approaches where available or by scanning human protein sequences for statistically identified regions of low complexity (Figure S1A–C, Table S1). This analysis produced a map of condensate-promoting features across the set of putative condensate-forming proteins and recovered the MIDs and LCSs of known condensate-forming proteins with high fidelity (Figure S1D, Table S4B, Table S4F–G).
We then identified the pathogenic mutations that affect condensate-promoting features (Figure 1B), hypothesizing that such mutations would have a high likelihood of affecting condensate properties. We extracted pathological human disease variants from existing datasets of variants associated with Mendelian diseases and cancers (Methods). Variants were defined as pathogenic based on clinical assessments of pathogenicity provided in the source datasets for Mendelian variants, or integrated from independent knowledgebases for cancer variants (Chakravarty et al., 2017; Griffith et al., 2017; Landrum et al., 2017; Stenson et al., 2020; Tamborero et al., 2018) (Figure S1A, Table S4C, Methods). Such assessments of pathogenicity are largely based on established guidelines that integrate various sources of evidence, including associations with clinical phenotypes, population or tumor frequencies, and computational predictions, as well as knowledge of functional or molecular properties of the mutation and the affected protein (Li et al., 2017; Richards et al., 2015). Within pathogenic variants, we focused on the types of variants where we could reasonably predict the effect of mutations on condensate-promoting features (Methods). These variant types consisted of missense variants, in-frame insertions and deletions (indels), as well as nonsense and frameshift variants (hereafter, referred to together as truncating variants). Together, these variant types comprised over 98% of the observed pathogenic mutations (Figure S2A). Truncating variants may lead to nonsense-mediated decay (NMD), confounding whether a truncating mutation imparts its effects primarily through the loss of a condensate-promoting feature versus the loss of the protein. To minimize this confounding effect, we chose to eliminate all truncations predicted to elicit NMD (Lindeboom et al., 2016) from the analyses (Table S4D, Methods). In total, we extracted 322,825 pathogenic variants associated with 5,342 Mendelian diseases and 659 cancer types for further study (Cerami et al., 2012; Consortium, 2017; Hoadley et al., 2018; Landrum et al., 2017; Stenson et al., 2020) (Figure 1B, Figure S1A).
We mapped these pathogenic variants to the condensate-promoting features annotated within condensate-forming proteins. Mutations were defined as affecting condensate-promoting features if they were missense mutations or in-frame insertions within the bounds of an MID or LCS, or if they were in-frame deletions and truncating mutations removing part of an MID or LCS (Methods). This resulted in a catalog of 36,777 pathogenic mutations found to affect condensate-promoting features in 1,745 of the putative condensate-forming proteins, spanning 1,233 distinct Mendelian phenotypes and 557 cancer types (Figure S1A, Figure S2B–D, Table S4H). This catalog recovered pathogenic mutations shown in the literature to cause condensate dysregulation with a sensitivity of 76%, including mutations in proteins such as UBQLN2, FUS, MECP2, TIA1, HNRNPA1, and SPOP (Bouchard et al., 2018; Conicella et al., 2016; Dao et al., 2019; Li et al., 2020; Mackenzie et al., 2017; Molliex et al., 2015; Patel et al., 2015; Quiroz et al., 2020) (Figure S2E, Figure S1D, Table S2). Thus, this catalog of pathogenic mutations, with annotations of associated diseases, disrupted condensate-promoting features, and affected condensate-forming proteins for each mutation (Table S4H), provides a foundation for further studies of condensate dysregulation in disease.
The spectrum of diseases predicted to involve dysregulated condensates
Thus far, a small fraction of known diseases has been shown to arise from condensate dysfunction, so most diseases have not been directly linked to pathogenic mechanisms involving condensates. Condensates that have been linked to specific diseases thus far have provided important new insights into the biological regulation of the condensate as well as the pathogenic mechanisms underlying the disease (Ahn et al., 2021; Boija et al., 2021; Cai et al., 2021; Chandra et al., 2021; Kim et al., 2013; Li et al., 2020; Min et al., 2019; Molliex et al., 2015; Nedelsky and Taylor, 2019; Patel et al., 2015; Quiroz et al., 2020; Ramaswami et al., 2013; Spannl et al., 2019; Zhang et al., 2020). Thus, we next asked what types of diseases were most associated with the mutations predicted to dysregulate condensates. We categorized Mendelian diseases and cancers by the organ systems or tissue types they involved (Methods). Mutations affecting condensate-promoting features were involved in nearly all types of Mendelian diseases and cancers (Figure 2A–B). The proportion of such mutations affecting a particular organ system was more or less comparable across all organ systems, and these mutations accounted for 5–10% of pathogenic mutations across Mendelian diseases and 15–25% of mutations across cancer types.
Figure 2. Condensate dysregulation across the spectrum of disease. See also Table S4I–K.
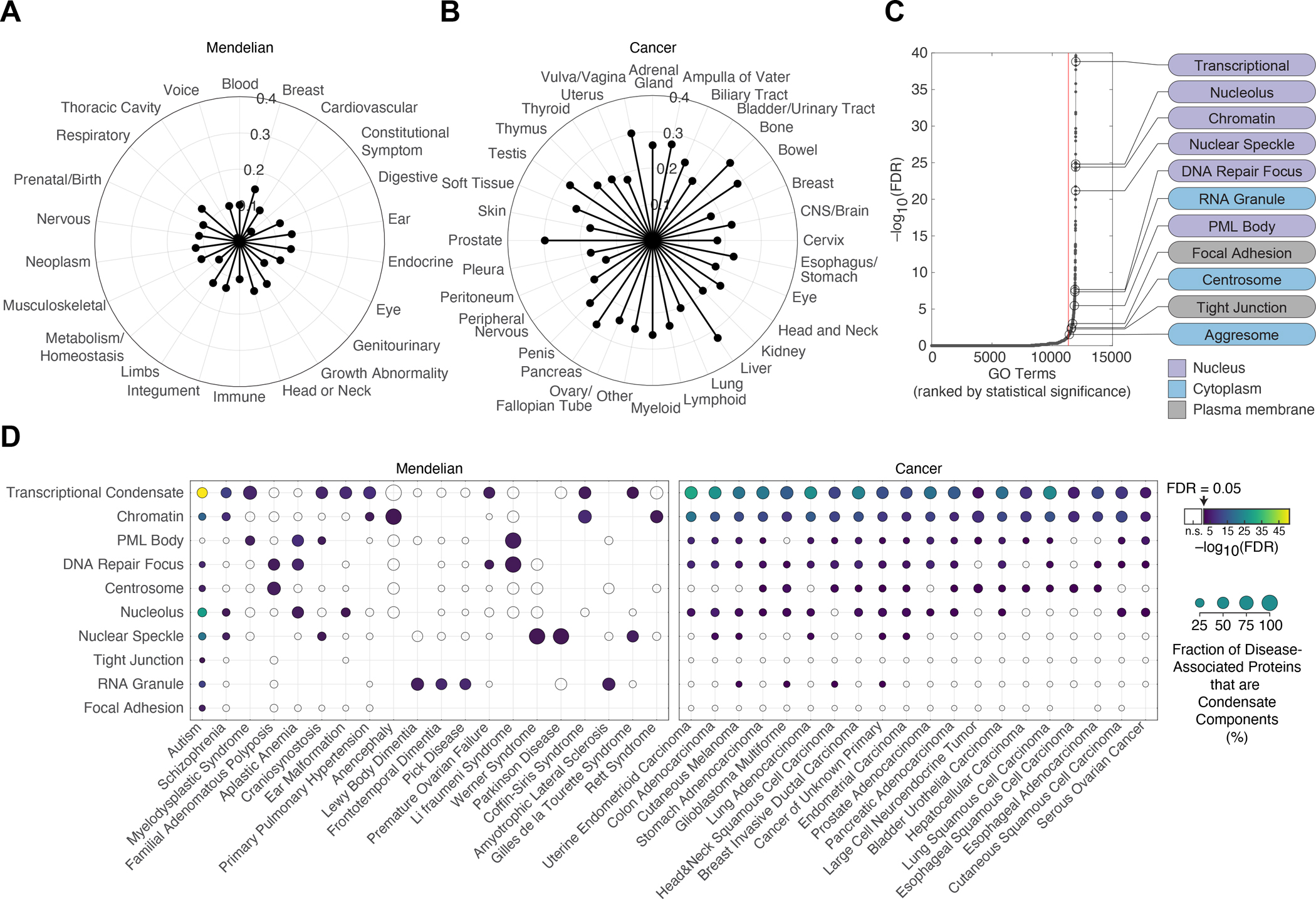
A. Proportion of pathogenic mutations (depicted as distance from center of radar plot) affecting condensate-promoting features in multivalent proteins across Mendelian diseases. Mendelian diseases are stratified by organ systems in which the diseases had a phenotypic effect (Methods).
B. Proportion of pathogenic mutations (depicted as distance from center of radar plot) affecting condensate-promoting features in multivalent proteins across cancers. Cancers are stratified by tissues of origin (Methods).
C. Enrichment of GO terms among the set of condensate-forming proteins that have pathogenic mutations that affect condensate-promoting features. GO terms (black dots) are ranked (x-axis) by statistical significance (−log10(FDR), y-axis). Red line denotes GO term rank corresponding to threshold for statistical significance (FDR < 0.05). The subset of significantly enriched GO terms that correspond to biomolecular condensates (Table S4I) are highlighted (black open circles and labels). Nuclear, cytoplasmic, and plasma membrane-associated condensates are indicated by purple, blue, or gray labels, respectively.
D. Significant associations between specific diseases and specific condensates. The set of condensate-forming proteins with pathogenic mutations affecting condensate-promoting features were mapped to specific condensates using Gene Ontology (see Methods) as well as associated with specific diseases. Overlaps between subsets of proteins associated with specific condensates (y-axis) and those associated with specific diseases (x-axis) were tested for statistical significance. Selected examples of Mendelian diseases (left) and cancer types (right) are shown (see also Table S4J–K). Filled data points correspond to a statistically significant association between the indicated disease with the indicated condensate, with the data point color corresponding to the Benjamini-Hochberg adjusted p-value (FDR) for the enrichment of proteins defined as components of the indicated condensate based on GO (Methods) among the set of condensate-forming proteins that have pathogenic mutations involved in the indicated disease that affect condensate-promoting features. Unfilled datapoints correspond to a lack of a statistically significant enrichment. Size of data point is proportional to the fraction of the indicated disease-associated condensate-forming proteins that are components of the indicated condensates.
Specific mutations have been shown to cause dysregulation of a small subset of the biomolecular condensates described thus far (Basu et al., 2020; Kim et al., 2013; Li et al., 2020; Mackenzie et al., 2017; Molliex et al., 2015; Patel et al., 2015; Quiroz et al., 2020; Ramaswami et al., 2013), while the majority of known condensates have not directly been linked to human disease. To evaluate the breadth of known condensates that could be dysregulated in disease, we looked for associations with specific condensates among the set of disease-associated, condensate-forming proteins within our catalog (Methods). The mutations predicted to dysregulate condensates occurred in proteins associated with a broad range of functions and condensates, but were particularly evident among components of nuclear condensates, such as those involved in transcription, chromatin structure, RNA splicing and pre-ribosome biosynthesis (Figure 2C, Table S4I–K). Stratifying this analysis by disease type revealed known associations of condensates and diseases—including those of RNA granules with FTD, ALS, and other neurodegenerative phenotypes (Conicella et al., 2016; Kim et al., 2013; Mackenzie et al., 2017; Molliex et al., 2015; Ramaswami et al., 2013); of transcriptional condensates with polydactyly (Basu et al., 2020); of heterochromatin with Rett syndrome (Li et al., 2020); and of keratohyalin granules with atopic dermatitis (Quiroz et al., 2020)—and nominated numerous additional putative associations between known condensates and specific Mendelian diseases or cancers (Figure 2D, Table S4J–K). These results corroborate the hypothesis that condensate dysregulation may be an underlying pathogenic mechanism across a broad spectrum of human diseases.
It is important to note that some nominated mutations are also likely to contribute to pathogenesis via other known molecular-scale mechanisms, such as disruption of protein fold, catalysis, ligand binding, post-translational modifications, and subcellular localization signals (Figure S2G, Table S4B). Condensate dysregulation does not exclude these canonical models of protein dysfunction, but rather, provides an additional framework with which to better understand the pathogenic basis of disease and a foundation for mechanistic and therapeutic hypotheses.
Pathogenic mutations affecting condensate-promoting features alter condensate properties in cells
To confirm that a subset of pathogenic mutations identified in this catalog can affect condensate properties in cells, we selected 25 putative condensate-forming proteins spanning a range of biological functions and diseases for study (Table S3). Murine embryonic stem cells (mESCs) were selected for use in this study because cell lines can be engineered to provide a consistent cellular environment for comparisons of multiple pairs of wild-type (WT) and mutant proteins and mESCs also have proven utility in the study of condensate properties (Cho et al., 2018; Guo et al., 2019; Henninger et al., 2021; Li et al., 2020; Sabari et al., 2018). Cell lines were generated in which genes encoding the 25 wild-type proteins were stably integrated and expressed with an mEGFP tag, and these were subjected to live-cell imaging with Airyscan confocal laser-scanning microscopy (Figure 3A). As controls, we used MECP2, a validated condensate-forming protein in mESCs (Li et al., 2020), and mEGFP, which exhibits a non-punctate distribution throughout the nucleus and cytoplasm (Figure 3B). The use of any one cell type for condensate studies is naturally limiting because condensate formation can depend on cell type, environmental stress and external signals. Nonetheless, approximately half of the proteins studied in mESCs (13/25) were found concentrated within punctate structures that exhibited dynamics typically observed in condensates, and two additional proteins formed puncta in another cell line or with exposure to oxidative stress (Figure S3A–C, Table S3).
Figure 3. Pathogenic mutations in condensate-promoting features alter condensate properties in live cells. See also Figures S3–S4 and Table S3.
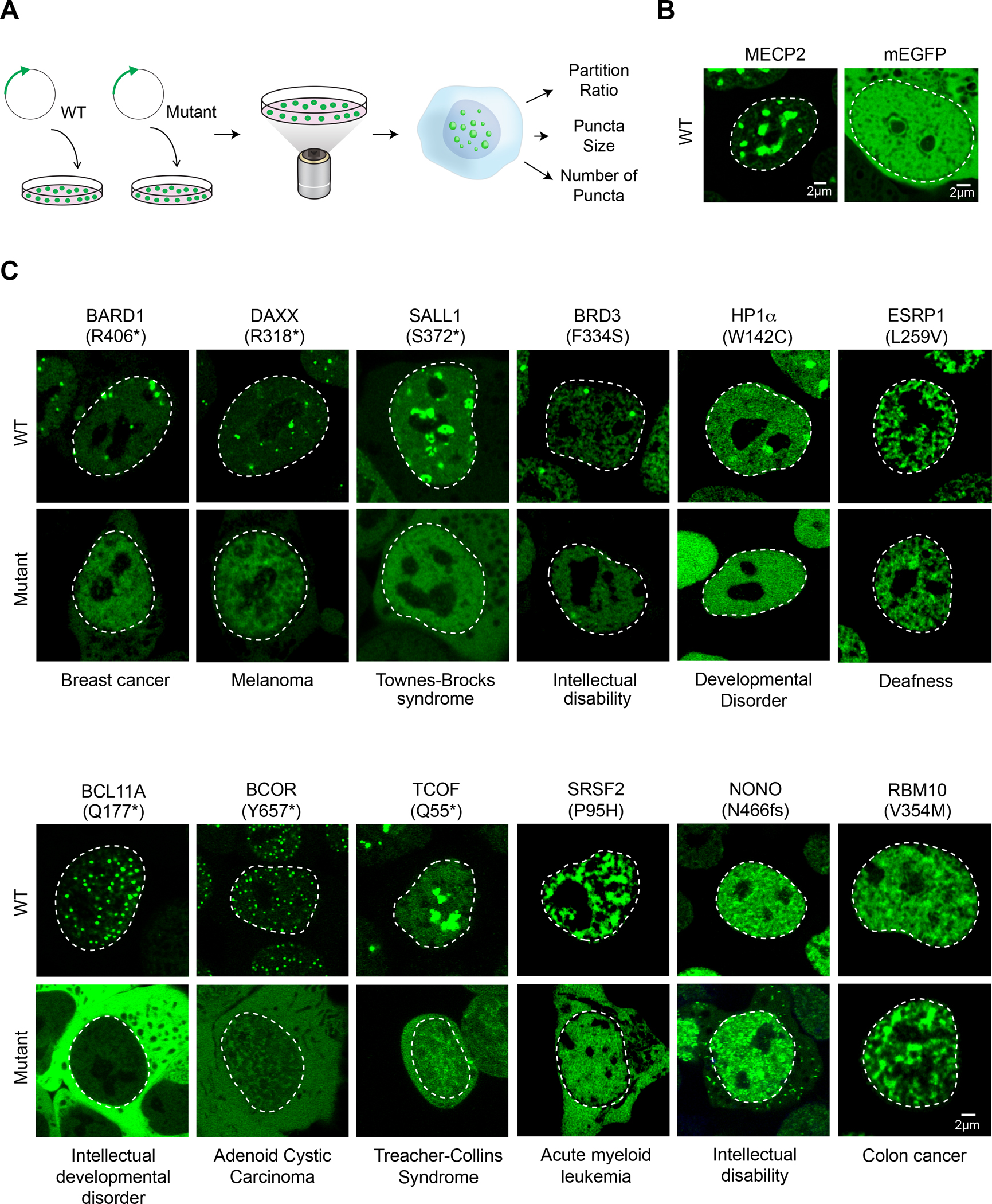
A. Experimental approach for testing the effect of a subset of identified mutations predicted to affect condensates. N-terminal mEGFP-tagged wild-type or mutant forms of candidate proteins were stably expressed in mESCs and condensate properties were assessed using live cell imaging and quantitative image analysis.
B. Representative images of wild-type MECP2 (positive control for condensate incorporation) mEGFP alone (negative control). Nuclei are outlined with white dashed lines.
C. Representative images of wild-type versus mutant mEGFP-tagged candidate proteins BARD1, DAXX, SALL1, BRD3, RBM10, BCL11A, NONO, BCOR, TCOF1, HP1A, SRSF2, ESRP1. Specific mutations that were tested along with their associated disease are indicated adjacent to the images.
We then asked whether pathological mutations that affect condensate-promoting features within the 13 proteins that formed condensates in mESCs affect measurable properties of the observed puncta. For each of the 13 proteins, we generated at least one analogous cell line expressing a representative missense or truncation mutation (Figure 3A, Table S3). Mutations were selected to represent the approximate proportion of mutation types and affected condensate-promoting features in the catalog (Figure S3D). For each WT and mutant pair of live cell lines, the area, number, and partitioning of the corresponding protein into condensates was measured (Figure 3A, Methods). We found that 87% of mutations tested (13/15) as well as a known condensate-disrupting mutation in MECP2 (Li et al., 2020) showed qualitative and quantitative differences in the properties of WT versus mutant puncta (Figure 3C, Figure S3E, Figure 4A–C). In 11 of 13 cases, the mutations caused significant reductions in partitioning of the protein into condensates. In the remaining two cases, one mutation enhanced the ability of the proteins to associate with condensates while the other caused the protein to form puncta in other cellular locations (Figure 3C, Figure S4B, Figure S4D). Two lines of evidence indicate these observations in mESCs were relevant in humans: all thirteen candidates have previously been observed to occur in condensate-like puncta in at least one human cell line (Thul et al., 2017), and at least five of these occur in condensate-like punctate structures in disease-relevant human tissues or human cell lines (Figure S4F–G). While our experimental tests represent a relatively small sampling of mutations compared to the full catalog, our results suggest a predictive accuracy of the catalog to be between 60–98% (95% confidence interval) (Figure S4E). These results suggest that a substantial fraction of pathogenic mutations that were mapped to condensate-promoting features of condensate-forming proteins do produce condensate dysregulation phenotypes in cells, and that these phenotypes include reduced condensate incorporation, enhanced condensate incorporation, and altered condensate localization (Figure 4A–B).
Figure 4. Mutations in condensate-promoting features cause diverse condensate dysregulation phenotypes.
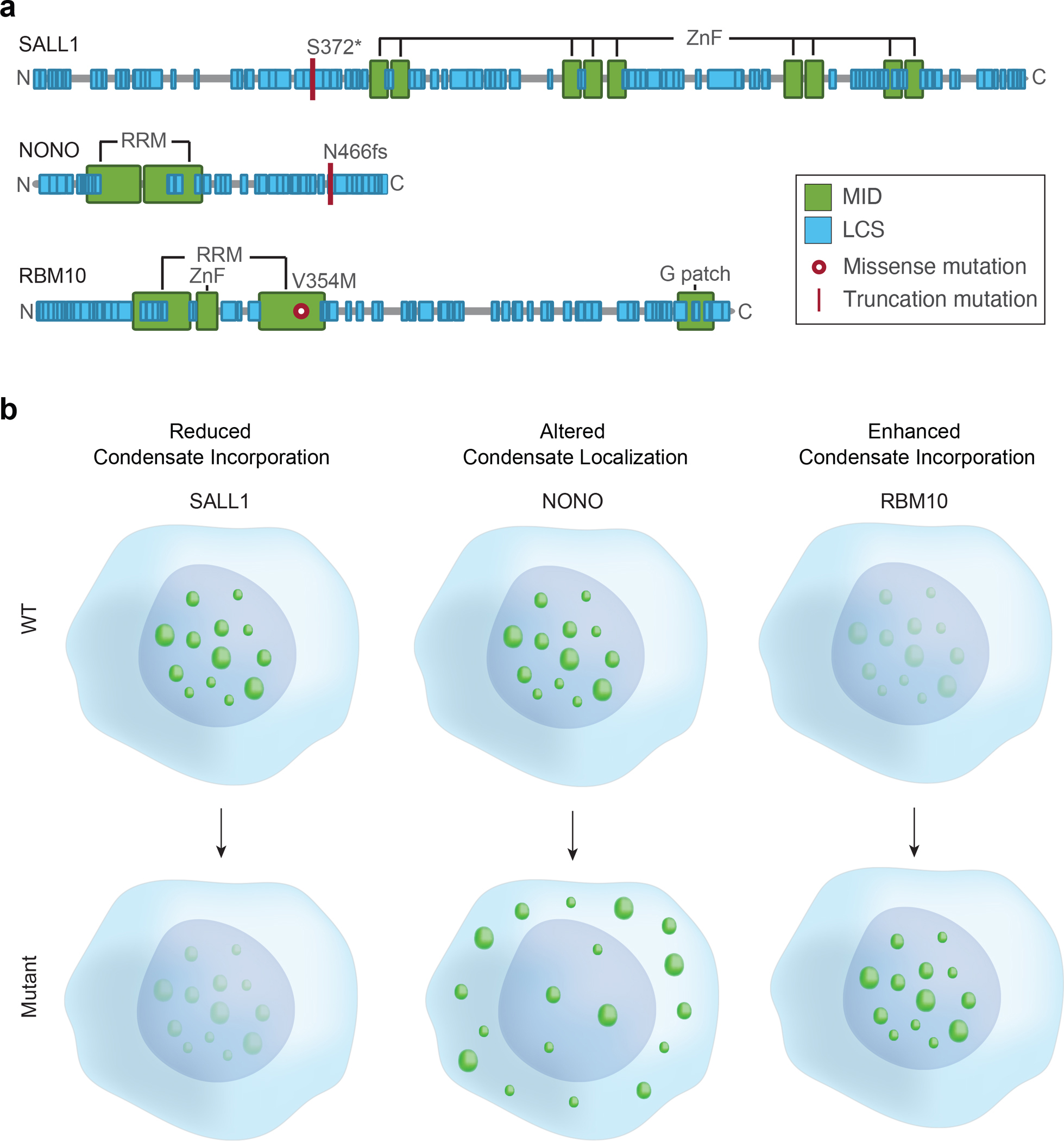
Models for observed types of condensate dysregulation resulting from pathogenic mutations that affect condensate-promoting features of condensate-forming proteins, including altered condensate incorporation (left), enhanced condensate formation (middle), and altered condensate localization (right). Candidate where these phenotypes were observed (Figure 3C) are listed.
DISCUSSION
Much of human disease is understood through the lens of mutations affecting proteins at the molecular length scale, due in part to advances afforded by structural biology and hypotheses of protein function and disease causality that emerge from 3D structural models. This understanding plays a considerable role in therapeutic advances, as it enables medicinal chemistry employing structure-based drug design. In contrast, far less is known about how mutations affect properties that organize cellular processes at the mesoscale, such as the propensity to form biomolecular condensates, although this propensity has recently been linked to a variety of protein features. Thus, a map of known condensate-promoting protein features and the pathological mutations that affect these features could be a powerful tool in the investigation of disease mechanisms derived from disruption of this mesoscale organization.
The approach we present identified over 36,000 pathogenic mutations that plausibly contribute to condensate dysregulation across over 1,200 Mendelian diseases and 500 cancers. The premise of the approach is supported by many studies that have identified various types of MIDs and LCSs as predominant determinants of the formation and macroscopic properties of condensates as well as evidence that pathological mutations in these condensate-promoting features can lead to altered condensate properties (Banani et al., 2017; Bouchard et al., 2018; Choi et al., 2020; Molliex et al., 2015; Patel et al., 2015; Wan et al., 2019). Several observations suggest that this resource will prove to be a useful predictive guide to studying condensate-associated diseases. The analytical approach used here captured nearly 80% of known disease-causing mutations that affect condensates. Our experimental validation results show that nearly all of the thirteen tested mutations alter condensate properties in cells, and that these span condensate dysregulation phenotypes such as dissolution, enhancement, and mislocalization. Despite a small experimental sample size compared to the full catalog, our estimates suggest the predictive accuracy of the catalog to be between 60–98% (95% confidence interval). We thus expect this catalog of mutations to be substantially enriched for those that directly affect condensate properties.
This resource suggests that a substantial fraction of pathogenic mutations impart their phenotypic effects by altering the physicochemical properties of condensates that compartmentalize the diverse regulatory functions of cells. It predicts that mutations affecting condensate-promoting features of condensate-forming proteins contribute to diseases spanning all human organ systems, suggesting that the potential for novel disease mechanism discovery, therapeutic hypotheses, and consequent impact on medicine, is considerably vast. The mechanistic evaluation of these mutations will likely require evolving paradigms that address phase-separating systems across disciplines, including polymer physics, nonequilibrium thermodynamics, pharmacodynamics and pharmacokinetics, and medicinal chemistry. The therapeutic opportunities for diseases caused by condensate dysregulation have yet to be fully explored, but evidence that therapeutic small molecules can selectively interact with specific condensates (Babinchak et al., 2020; Howard and Roberts, 2020; Klein et al., 2020; Lemos et al., 2020; Viny and Levine, 2020; Wheeler et al., 2019) suggests that such therapies can be devised.
LIMITATIONS OF THE STUDY
Validation of predicted condensate dysregulation is at present practically and technologically limited to relatively small experimental sample sizes. Our experimental studies do not establish a direct link between observed condensate dysregulation and the ultimate cellular and organismal defects that create the disease phenotype. Our analyses are primarily restricted to MIDs and LCSs, and while these are thought the be the predominant determinants of condensate properties, many additional protein features that we do not explicitly consider have been directly or indirectly associated with condensate properties (Bouchard et al., 2018; Gibson et al., 2019; Nott et al., 2015; Rai et al., 2018; Yoshizawa et al., 2018), suggesting that mutations not captured in our catalog may also affect condensates. We anticipate that technological and conceptual advances in condensate biology, as well as detailed molecular studies of specific proteins in disease-appropriate model systems may help to overcome these limitations in the future.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Richard A. Young (young@wi.mit.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact upon reasonable request with a completed Materials Transfer Agreement.
Data and code availability
Datasets used in this study are available from the sources cited in the Key Resources Table. Processed data used in the study is available in the Supplemental Tables and on the Young laboratory’s website.
Code generated during this study is available via https://github.com/bananisf/2022_Banani_Afeyan_Hawken_Condensate_Dysregulation.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-rabbit AlexaFluor-488 | ThermoFisher | A11008; RRID:AB_143165 |
| Rabbit polyclonal anti-TCOF1 | ThermoFisher | PA558309; RRID:AB_2648324 |
| Rabbit polyclonal anti-SALL1 | Invitrogen | PA562057; RRID:AB_2646913 |
| Rabbit polyclonal anti-BARD1 | abcam | ab226854 |
| Biological samples | ||
| Human adult large intestine adjacent normal tissue | BioIVT | HMLGINTAD J2-PPG7 |
| Human adult breast adjacent normal tissue | BioIVT | HMNBREASTN ADJ2 |
| Human adult kidney normal | BioIVT | HMKIDNEYNO RC2 |
| Chemicals, peptides, and recombinant proteins | ||
| Poly-L-ornithine | Sigma | P4957-50ML |
| Doxycycline | Sigma | D9891-5G |
| Hoechst 33258 | Life Technologies | H3569 |
| Vectashield | Vector Laboratories | 101098-042 |
| Laminin (Mouse) | ThermoFisher | CB40232-1MG |
| Critical commercial assays | ||
| Lipofectamine 3000 Transfection Agent | ThermoFisher | L3000001 |
| Experimental models: Cell lines | ||
| V6.5 murine embryonic stem cells | Laboratory of Rudolph Jaenisch | N/A |
| Human: HCT116 | ATCC | CCL-247 |
| Human: MCF7 | ATCC | HTB-22 |
| Human: HEK293T | ATCC | CRL-3216 |
| Recombinant DNA | ||
| pbfh_GFP | (Henninger et al., 2021) | Modified version of pJH135_pb_MCPx2_mCherry_rTTA |
| pbfh_GFP_MECP2 | This study | N/A |
| pbfh_GFP_MECP2_R186* | This study | N/A |
| pbfh_GFP_BARD1 | This study | N/A |
| pbfh_GFP_BARD1_R406* | This study | N/A |
| pbfh_GFP_BCL11A | This study | N/A |
| pbfh_GFP_BCL11A_Q177* | This study | N/A |
| pbfh_GFP_BCOR | This study | N/A |
| pbfh_GFP_BCOR_Y657* | This study | N/A |
| pbfh_GFP_BRD3 | This study | N/A |
| pbfh_GFP_BRD3_F334S | This study | N/A |
| pbfh_GFP_HP1A | This study | N/A |
| pbfh_GFP_HP1A_V21I | This study | N/A |
| pbfh_GFP_HP1A_W142C | This study | N/A |
| pbfh_GFP_DAXX | This study | N/A |
| pbfh_GFP_DAXX_R318* | This study | N/A |
| pbfh_GFP_EDC4 | This study | N/A |
| pbfh_GFP_EDC4_I371V | This study | N/A |
| pbfh_GFP_ESRP1 | This study | N/A |
| pbfh_GFP_ESPR1_L259V | This study | N/A |
| pbfh_GFP_NONO | This study | N/A |
| pbfh_GFP_NONO_446fs | This study | N/A |
| pbfh_GFP_RBM10 | This study | N/A |
| pbfh_RBM10_V354M | This study | N/A |
| pbfh_GFP_SALL1 | This study | N/A |
| pbfh_GFP_SALL1_S372* | This study | N/A |
| pbfh_GFP_SRSF2 | This study | N/A |
| pbfh_GFP_SRSF2_S54F | This study | N/A |
| pbfh_GFP_SRSF2_P95H | This study | N/A |
| pbfh_GFP_TCOF | This study | N/A |
| pbfh_GFP_TCOF_Q55* | This study | N/A |
| pbfh_GFP_ASXL1 | This study | N/A |
| pbfh_GFP_BCL6 | This study | N/A |
| pbfh_GFP_BRCA1 | This study | N/A |
| pbfh_GFP_DVL2 | This study | N/A |
| pbfh_GFP_DYR1A | This study | N/A |
| pbfh_GFP_ENC1 | This study | N/A |
| pbfh_GFP_G3BP1 | This study | N/A |
| pbfh_GFP_HMGA2 | This study | N/A |
| pbfh_GFP_NIPBL | This study | N/A |
| pbfh_GFP_NKX21 | This study | N/A |
| pbfh_GFP_SNCAP | This study | N/A |
| pbfh_GFP_TERT | This study | N/A |
| Software and algorithms | ||
| Fiji image processing package | (Schindelin et al., 2012) | https://fiji.sc/ |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Code generated by the study | This study | https://github.com/bananisf/ |
| Python v3.6.9 | www.python.org | N/A |
| CellPose v0.7.2 | (Stringer et al., 2021) | http://www.cellpose.org/ |
| MATLAB R2019b | Mathworks | N/A |
| metapredict | (Emenecker et al., 2021) | N/A |
| PLAAC | (Lancaster et al., 2014) | N/A |
| PSP | (Vernon et al., 2018) | N/A |
| Ensembl VEP v102 | (Yates et al., 2019) | N/A |
| Other | ||
| 35 mm glass-bottom imaging dishes | Mattek Corporation | P35G-1.5-20-C |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
V6.5 murine embryonic stem cells (mESCs) were a gift from R. Jaenisch. Human cell lines (HCT116, MCF7, and HEK293T) were obtained from ATCC.
Cell culture conditions
mESCs were cultured in 2i/LIF media on tissue culture treated plates coated with 0.2% gelatin (Sigma G1890) in a humidified incubator with 5% CO2 at 37°C. Cells were passaged every 2–3 days using TrypLE Express (Gibco) for dissociation quenched with serum/LIF media. 2i/LIF media: DMEM/F12 (Gibco) supplemented with 1x N2 and 1x B27 (Gibco), 1x MEM Non-essential amino acids (Gibco), 100 U/mL Penicillin-Streptomycin (Gibco), 1mM L-glutamine (Gibco), 0.25% BSA Fraction V (Gibco), 0.1 mM 2-mercaptoethanol (Sigma), 3 μM CHIR99021 (Stemgent), 1 μM PD0325901 (Stemgent), and 1000 U/mL leukemia inhibitor factor (LIF) (ESGRO). Serum/LIF media: KnockOut DMEM (Gibco) supplemented with 15% fetal bovine serum (Sigma), 2 mM L-glutamine (Gibco), 1x MEM non-essential amino acids (Gibco), 100 U/mL penicillin-streptomycin (Gibco), 0.1 mM 2-mercaptoethanol (Sigma), and 1000 u/mL leukemia inhibitor factor (LIF) (ESGRO).
Human cell lines HCT116 (ATCC), MCF7 (ATCC) and HEK293T (ATCC) were cultured in complete DMEM media (DMEM (Life Technologies 11995073), 10% Fetal Bovine Serum, FBS (Sigma Aldrich, F4135), 1% L-glutamine (GIBCO, 25030-081), 1% Penicillin Streptomycin (Life Technologies, 15140163) at 37°C with 5% CO2 in a humidified incubator. For passaging, cells were washed in PBS (Life Technologies, AM9625) and TrypLE Express Enzyme (Life Technologies, 12604021) was used to detach cells from plates by incubating them in TrypLE at 37°C with 5% CO2 in a humidified incubator for 5 minutes. TrypLE was quenched with complete DMEM, described above, and cells were plated in new tissue culture-grade plates.
Human tissue samples
Human tissue samples were purchased from bioIVT. Healthy human breast tissue was from a 27-year-old female with infiltrating lobular carcinoma of the breast and stage group T4N2M0. Health human kidney tissue was from a diabetic 28-year-old male.
Human tissue sample storage conditions
Human tissue samples were kept frozen, embedded in OCT, and stored at −80°C until use.
METHOD DETAILS
Definition of Condensate-Forming Proteins
The set of 20,394 Homo sapiens proteins and their sequences in the UniProt/SwissProt database v2020_06 (Consortium et al., 2020) were defined as the human proteome in this study (Figure S1A, Table S4A). The list of known and predicted condensate-forming proteins was defined by integrating: (i) proteins with a DeepPhase (Yu et al., 2020) score (based on analysis of proteome-wide immunofluorescence) of ≥ 0.9, a validated threshold provided by the developers of the algorithm; (ii) proteins scoring within the top 10% of PSAP scores (Mierlo et al., 2021) (based on proteome-wide sequence-based analysis); and (iii) known phase-separating proteins curated from LLPSDB (Li et al., 2019) (accessed January, 2021), PhaSePro (Mészáros et al., 2019) (accessed January, 2021), and PhaSepDB v1.3 (You et al., 2019). The resulting list of proteins is shown in Table S4E.
Mapping of Protein Features
Multiple classes of canonical and condensate-promoting protein features were mapped onto the proteome as follows, with the mapping results provided in Table S4B.
Domains.
Integrated domain annotations from CDD v3.18 (Lu et al., 2019), PFAM v33.1 (Mistry et al., 2020), SMART v7.1(Letunic et al., 2020) corresponding to the UniProt entries for the set of human proteins were obtained via InterPro v83.0 (Blum et al., 2020). We found that using integrated annotations provides a more comprehensive mapping of domains across the proteome. The integration in InterPro ensures that the same instance of a given domain within a protein from multiple domain databases is annotated as a single entry. Related domains (e.g. different subtypes of SH3 domains) were further grouped into a single parent entry using hierarchies provided in InterPro. Catalytic domains were defined as those having a molecular function annotation in InterPro including “enzyme activity” or ending with “-ase activity”. For the analyses in Figure S2G, protein regions with UniProt annotations of active site were also included within the catalytic domains category. Interaction Domains (or Modular Interaction Domains [MIDs], as we refer to them in the context of condensate formation) were defined by combining the following subsets of domains: (i) domain entries that had any of the following molecular function annotations in InterPro: protein complex, oligomerization, protein dimerization activity, protein tetramerization, protein homodimerization activity, protein heterodimerization activity, protein homooligomerization, DNA binding, RNA binding, protein binding, nucleic acid binding, actin binding, microtubule binding, actin filament binding, histone binding, integrin binding, clathrin binding, cellulose binding, telomeric DNA binding, cadherin binding, starch binding, protein kinase binding, ubiquitin binding, tubulin binding, cytoskeletal protein binding, collagen binding, chitin binding, mismatched DNA binding, chromatin binding, double-stranded DNA binding, or phospholipid binding; (ii) domain entries that matched (based on manual assessment) curated lists of domains from the literature corresponding to head-to-tail interacting domains (Bienz, 2020), RNA-binding domains (Hentze et al., 2018; Lunde et al., 2007), DNA-binding domains in transcription factors (Lambert et al., 2018; Vaquerizas et al., 2009), protein-protein interaction domains found in cell signaling (Pawson and Nash, 2003; Seet et al., 2006) (including those that recognize PTMs), or domains that recognize histone modifications (Yun et al., 2011); and (iii) manually curated list of domains not included in (i) or (ii) assembled from prior knowledge of or domain descriptions in InterPro documenting their known or suspected involvement in binding interactions. We note that the same list of MIDs were used for analyses involving canonical protein properties in Figure S2G as well as for the mapping of condensate-promoting features within condensate-forming proteins (Figure 1B, Figure S1A), given the known roles of MIDs in both canonical protein-protein interactions and in condensate formation. The final list of MIDs used in this study is provided in Table S4F.
It is important to note that the because the definition of a domain primarily relies on sequence conservation, it does not explicitly consider predictions of structure or disorder. While it turns out that most of the amino acid residues within most domains are often demonstrated or predicted to be folded/structured, this does not preclude occasional overlaps of domains with predicted intrinsically disordered regions and LCSs therein (see below).
Structural Elements.
Structural elements were defined by integrating the following protein annotations: (i) all protein regions annotated in UniProt (Consortium et al., 2020) as having structural elements helix, beta strand, turn, disulfide bond, and coiled coil; and (ii) all conserved domains (see above), which are often structured, that did not meet the definition of catalytic domain or interaction domain, filtered to remove any regions predicted to be intrinsically disordered (see below).
Interaction Motifs.
Interaction motifs were defined by integrating: (i) short linear interacting motifs (SLiM) annotations corresponding to UniProt entries from the ELM database (Kumar et al., 2019) (accessed November, 2020); (ii) all protein regions annotated in UniProt as region or motif (with the exception of those with a description including “Nuclear localization signal”); and (iii) molecular recognition features (MoRFs) that undergo coupled folding upon binding from MFIB database (Fichó et al., 2017) (version 26-06-2017). For (i), SLiMs were filtered to only include those annotated with the logic of true positive.
Protein Processing.
Regions involved in protein processing were defined by using regions annotated in UniProt as peptide, signal peptide, transit peptide, propeptide, and initiator methionine.
Post-translational Modifications.
Sites of PTMs were defined by integrating: (i) all protein regions annotated in UniProt as modified residue, lipidation, glycosylation, and cross-link; and (ii) all PTM sites corresponding to UniProt entries in PhosphoSitePlus (Hornbeck et al., 2015) (accessed November, 2020).
Nuclear Localization and Nuclear Export Signals.
NLSs and NESs were defined by integrating: (i) all protein regions annotated in UniProt as motif and with a description containing “Nuclear localization signal”; and (ii) NLS and NES sites corresponding to UniProt in NLSdb (Bernhofer et al., 2017) (accessed November, 2020). For (ii), NLSs and NESs were filtered to only include those annotated as Experimental or By Expert.
Other Functional Regions.
The miscellaneous category of other functional regions was defined by integrated all other protein regions annotated in UniProt presumed to be susceptible to disruption by mutation. The following annotations were integrated to define these regions: site, binding site, metal binding, calcium binding, DNA binding, nucleotide binding, and mutagenesis.
Intrinsically Disordered Regions.
IDRs were mapped using metapredict (Emenecker et al., 2021) using a threshold of 0.2, which was within the recommended range of cutoffs suggested by the developers of the algorithm.
Low Complexity Sequences.
The list of thirteen types of LCSs used in this study was assembled manually from literature evidence of their involvement in IDR-mediated phase separation (Table S1). Prion-like domains were mapped using PLAAC (Lancaster et al., 2014) with core length set to 60 and relative weighting of background probabilities set to 100, as done in prior work that globally mapped this LCS type across the proteome (Wang et al., 2018). pi-pi interacting residues were mapped using PSP (Vernon et al., 2018), and residues with a PScore of > 4, based on the confidence thresholds provided in the algorithm, were considered to be LCSs of this type. LARKS were obtained from a prior study (Hughes et al., 2018). For the remaining types of LCSs, established, validated approaches for mapping these LCSs to our knowledge do not exist to date, so an approach based on functions for analogous purposes in localCIDER (Holehouse et al., 2017) and on a previously applied procedure (Li et al., 2020) was used to map these LCS regions. Amino acid compositions in sliding 5-residue windows were computed for each protein. LCS regions were defined as stretches of ≥ 5 consecutive residues (at minimum 1 window length) that consisted of characteristic residues corresponding to the particular type of LCS occurring at a frequency above a predefined threshold, set as described below (Table S1). All identified regions were filtered for those that occurred within predicted IDRs, determined as described above. This approach performed well when benchmarked against a set of experimentally validated condensate-promoting LCSs, with a receiver operating characteristic (ROC) area under the curve (AUC) ranging from 0.8–1.0 across the mapped LCSs (Figure S1B, Table S1). Optimal cutoffs for frequencies of the characteristic amino acids within 5-residue window were determined for each LCS from the ROC curve as the point of minimum Euclidean distance from perfect performance (0% false positive rate, 100% true positive rate) (Figure S1B). LCS mapping results and the overlap between the different types of LCSs identified are shown in Figure S1C, Table S1, and Table S4G.
Annotation of Mendelian and Cancer Mutations
Variants associated with Mendelian diseases were obtained from ClinVar (Landrum et al., 2017) (accessed January 29, 2021) and HGMD v2020.4 (Stenson et al., 2020) in hg38. Cancer variants were obtained from AACR Project GENIE v8.1 (Consortium, 2017) and various TCGA and TARGET studies via cBioPortal (Cerami et al., 2012; Hoadley et al., 2018) (accessed January, 2021) (Figure S1A) (cBioPortal study identifiers: ucec_tcga_pan_can_atlas_2018, skcm_tcga_pan_can_atlas_2018, coadread_tcga_pan_can_atlas_2018, luad_tcga_pan_can_atlas_2018, stad_tcga_pan_can_atlas_2018, lusc_tcga_pan_can_atlas_2018, blca_tcga_pan_can_atlas_2018, brca_tcga_pan_can_atlas_2018, hnsc_tcga_pan_can_atlas_2018, cesc_tcga_pan_can_atlas_2018, gbm_tcga_pan_can_atlas_2018, lihc_tcga_pan_can_atlas_2018, ov_tcga_pan_can_atlas_2018, lgg_tcga_pan_can_atlas_2018, esca_tcga_pan_can_atlas_2018, prad_tcga_pan_can_atlas_2018, paad_tcga_pan_can_atlas_2018, kirp_tcga_pan_can_atlas_2018, kirc_tcga_pan_can_atlas_2018, sarc_tcga_pan_can_atlas_2018, thca_tcga_pan_can_atlas_2018, acc_tcga_pan_can_atlas_2018, ucs_tcga_pan_can_atlas_2018, laml_tcga_pan_can_atlas_2018, dlbc_tcga_pan_can_atlas_2018, thym_tcga_pan_can_atlas_2018, meso_tcga_pan_can_atlas_2018, kich_tcga_pan_can_atlas_2018, tgct_tcga_pan_can_atlas_2018, chol_tcga_pan_can_atlas_2018, pcpg_tcga_pan_can_atlas_2018, uvm_tcga_pan_can_atlas_2018, wt_target_2018_pub, all_phase2_target_2018_pub, aml_target_2018_pub, nbl_target_2018_pub, and rt_target_2018_pub). For cancer variants, genomic coordinates were converted from hg19 to hg38 using liftover (Kent et al., 2002). Deletions larger than 100kb were omitted from analysis. Variants were mapped to protein-coding sequence changes within our set of 20,394 human proteins in SwissProt/UniProt using Ensembl VEP v102 (Yates et al., 2019) and ID mappings between Ensembl and UniProt. Given that the biological relevance of alternate isoforms is not comprehensively understood across the proteome, we chose to focus on protein isoforms considered to be the canonical isoforms (Consortium et al., 2020) which represent the best annotated and understood isoforms across all proteins (although we acknowledge that alternative splicing can affect IDRs (Buljan et al., 2012; Romero et al., 2006; Smith et al., 2020) and condensate-forming properties (Batlle et al., 2020; Gueroussov et al., 2017; Tsang et al., 2020), and therefore mutations in alternative isoforms can in principle affect condensate properties as well). Canonical isoforms are selected based on criteria such as prevalence, similarity to homologs, and in the absence of other information, sequence length (Consortium et al., 2020). A total of n = 2,644,688 DNA variants (62% of all variants in the source datasets) mapped to the 20,394 canonical protein isoforms in UniProt. Beyond this point, variants were counted as protein variants—i.e., DNA variants resulting in the same protein-coding alteration, regardless of their similarity or differences at the DNA level, were counted as the same variant. All synonymous variants were removed from further analysis. For non-synonymous variants, only the primary (often the most severe) protein-coding change of the variant was considered for classification of mutation types (e.g. missense, nonsense, frameshift, etc.) based on the established hierarchy of mutation effect severity within Ensembl variant annotations.
Pathogenicity of Mendelian variants was classified based on designations of clinical significance for ClinVar variants (pathogenic or likely pathogenic) or of variant class for HGMD variants (DM or DM?), and of cancer variants was determined by assessing the variants for their inclusion in CIViC (August 1, 2020 release) (Griffith et al., 2017), their inclusion the list of CGI’s (Tamborero et al., 2018) Validated Oncogenic Mutations, or their designations of oncogenicity in OncoKB v2.10 (oncogenic, likely oncogenic, or predicted oncogenic) (Chakravarty et al., 2017) (Figure S1A). We note that these definitions of pathogenicity do rely on computational predictions of pathogenicity, but not to the same extent as clinical, biological/functional, or evolutionary evidence of pathogenicity (Li et al., 2017; Richards et al., 2015). The resulting 322,825 pathogenic mutations analyzed are shown in Table S4C.
Among pathogenic mutations, we chose to evaluate mutation types that were prevalent pathogenic mutations and where the effect of the mutation on the condensate-promoting feature at the protein level could reasonably be predicted. These mutation types included missense, frameshift, nonsense, in-frame deletion, and in-frame insertion mutations. Together these mutations accounted for 98.9% of pathogenic mutations (not shown), capturing the vast majority of pathogenic mutations. We did not evaluate mutations that affect splicing (0.5% of pathogenic mutations; splice region, splice donor, and splice acceptor mutations) or the start codon (0.3% of pathogenic mutations), or mutations that represented complex changes to the protein sequence (e.g. deletion-insertions, 0.2% of pathogenic mutations).
Nonsense and frameshift variants were considered together to be truncating variants and assessed for their predicted propensity to elicit NMD. Predictive rules for NMD were obtained from prior work (Lindeboom et al., 2016). A truncating variant was considered to elicit NMD if the corresponding premature stop codon it introduced occurred (i) >200 residues C-terminal to the start codon; (ii) >50 residues N-terminal to the final exon-exon junction; and (iii) in an exon ≤400 base pairs in length. Pathogenic variants classified as NMD-eliciting in this manner are shown in Table S4D and were omitted from further analyses of truncating variants.
Mutations were defined as affecting condensate-promoting features if they were missense mutations or in-frame insertions within the bounds of an MID or LCS, or if they were in-frame deletions and truncating mutations removing part of an MID or LCS. Truncation mutations can affect the valency of condensate-promoting features to varying degrees depending on the position of the truncation, and thus not all truncations are expected to lead to a substantial effect on condensates. We defined MID valency as total number of MIDs of any type and LCS content (a proxy for LCS valency) as the total number of LSC residues of any type and implemented a filter requiring that a truncation mutation remove at least 25% of the protein’s total MID or LCS valency. This cutoff was set based on studies of known condensate forming proteins that suggest that a fractional valency loss of 0.2–0.4 was necessary in these cases to observe substantial effects on condensate formation (Li et al., 2020; Quiroz et al., 2020). The catalog of 36,777 pathogenic mutations that affect condensate-promoting features within the set of putative condensate-forming proteins is shown in Table S4H.
Disease Terminology and Stratification of Diseases by Organ System
Different datasets use distinct terminologies for the same diseases, and we found that in some cases even within the same dataset, terminologies for the same diseases had semantic differences. For this reason, variants were mapped to a common disease nomenclature for analysis (Figure S1A). Mendelian variants were mapped to the list of 7,507 Mendelian phenotypes in OMIM (Amberger et al., 2015) (accessed January, 2021) (only phenotypes with the prefixes # or % were included) using links to OMIM provided in ClinVar or HGMD. 65% of pathogenic Mendelian mutations (n = 176,976 mutations) mapped to a Mendelian phenotype and were used for disease-related analyses. The Mendelian phenotypes were mapped to organ systems using HPO annotations (Köhler et al., 2018) (December 9, 2020 release) of OMIM phenotypes. Cancer variants were mapped to the list of 836 tumor types in OncoTree (Kundra et al., 2021) (accessed January, 2021; only terms at level 2 or greater were included, as level 1 indicated tissues of origin) using links to OncoTree provided in the cancer datasets. Nearly all of the pathogenic cancer variants (99.8%; n = 58,437 mutations) mapped to a tumor type and were used for disease-related analyses. Tumor types were mapped to tissues of origin using the hierarchy provided in OncoTree by mapping each tumor type to the corresponding term at level 1 of the hierarchy.
Gene Ontology Analysis
GO annotations associated with UniProt entries for human proteins were obtained from the Gene Ontology Resource (January 1, 2021 release) (Carbon et al., 2018). Annotations with the NOT qualifier were removed. Only annotations with the evidence codes EXP, IDA, IPI, IMP, IGI, IEP, HTP, HAD, HMP, HGI, or HEP were included in order restrict the analysis to annotations with supporting experimental evidence and to exclude computationally or phylogenetically derived annotations. A subset of GO terms that correspond to biomolecular condensates were manually curated from GO, and components of those condensates were defined as all proteins with GO annotations corresponding to those GO terms or any descendent terms thereof in the GO hierarchy. For known condensates without exact GO terms, a set of GO terms thought to best represent known properties of the condensate were used as the definitions for the condensate components. The correspondence between GO terms, known biomolecular condensates, and resultant proteins mapped to those condensates as used in this study is shown in Table S4I. For all analyses, the set of proteins associated with a particular GO term included all proteins annotated with the GO term or with all terms associated with the GO term at lower levels of the GO hierarchy.
For the analyses in Figure 2, all GO terms associated with the set of condensate-promoting proteins that had pathogenic mutations affecting condensate-promoting features were tested for statistical enrichment within the set. An analogous analysis performed by stratifying the set of condensate-promoting proteins that had pathogenic mutations affecting condensate-promoting features by the specific disease types associated with those mutations.
Selection of Candidates and Mutations for Experimental Validation
Candidate proteins for experimental validation (Table S3) were selected in a manner constrained only by the practical limitation of availability of DNA reagents by manually searching Addgene or commercially available cDNA repositories from the MGC (Team et al., 2009) for availability of DNA material encoding full length proteins. This practical limitation is likely to bias the candidate selection toward proteins that are well-characterized (and therefore deposited by investigators in plasmid repositories), but we are not presently aware of any variables in this ad hoc selection process that would confound the selection of candidates toward those that that are more or less likely to harbor pathogenic mutations that impact condensates. 12 of these proteins (48%) did not show punctate localization in mESCs when ectopically expressed with a GFP (Figure S3A, Table S3). The false discovery rate for the set of condensate-forming proteins from which these candidates were selected is expected to be much less than 48%, based on the validation reported in the source databases or algorithms (Li et al., 2019; Mészáros et al., 2019; Mierlo et al., 2021; You et al., 2019; Yu et al., 2020). This suggests that the failure to detect punctate localization is unlikely to be due to a false designation of a protein as condensate-forming. Rather, as condensate formation can often to be cell-type and cell-state specific, this suggests that mESCs in resting state may not provide a conducive environment to observe condensation for all proteins (Figure S3B). These 12 proteins were not analyzed further. For the remaining 13 proteins that did show evidence of punctate localization in mESCs, pathogenic mutations in these protein candidates were selected ad hoc such that the relative distribution of selected mutations was similar to mutations in the full catalog with respect to the types of condensate-promoting features (MIDs or LCSs) they affected (Figure S3D). Given the relatively small sample size of mutations selected for experimental testing compared to the full catalog, a strictly random selection did not guarantee that the distribution of selected mutations would meet this criterion. While we do not expect our selection process for mutations to be biased toward mutations more or less likely to exhibit condensate effects, we cannot strictly rule this out this possibility. We initially selected 1–2 mutations per protein candidate, with the intention of testing additional mutations if the initial set of mutations did not show effects on condensate properties. However, we found that the majority of mutants (87%) selected in this initial cycle had effects measurable effects on condensate properties in cells (Figure 3C, Figure S4B–C)
mESC Cell Line Generation
Stable cell lines were generated by cloning WT and mutant gene sequences using NEBuilder HiFi DNA Assembly (NEB) into a doxycycline-inducible, N-terminal mEGFP-tagged expression construct with a hygromycin-resistance gene, which was integrated into mESCs using the PiggyBac transposon system (Systems Biosciences). 0.5 × 106 wildtype mESCs were plated in 6-well format and simultaneously transfected with 1 μg of the expression vector and 1 μg of the PiggyBac transposase using Lipofectamine 3000 (ThermoFisher), according to manufacturer instructions in serum/LIF media. The next day, media was changed to 2i, and cells were split into 100 mm gelatin-coated plates with 2i-media supplemented with 500 μg/mL hygromycin (ThermoFisher) for selection. Selection media was exchanged every day and un-transfected control cells were monitored to assess selection.
Imaging and Image Analysis in mESCs
Cells were grown on 35 mm glass plates (MatTek) coated with poly-L-ornithine (Sigma) for 30 minutes at 37°C followed by coating with laminin (Corning) for 2 hours at 37°C. Expression of the mEGFP-tagged protein was induced by adding doxycycline to the media at 1μg/mL for 24 hours. Cells were imaged live in a heated chamber at 37°C with humidified air and 5% CO2 in 2i media supplemented with 5μM Draq5 dye (ThermoFisher) for nuclear staining. Images were acquired with a Zeiss LSM880 Confocal Microscope with Airyscan processing with a 63x Objective and 2x Zoom using ZenBlack acquisition software (W.M. Keck Microscopy Facility, MIT). Images were processed using Fiji is Just ImageJ (Fiji) (Schindelin et al., 2012). Image analysis was conducted on z-stacks with 10–20 slices per cell at 0.2–0.36 μm per slice. Condensate partition ratio, cross-sectional area, number per cell were calculated using a custom script written in Python v3.6.9. A Python package, cellpose (Stringer et al., 2021), was used on the Draq5-DNA signal to segment nuclei in each cell. For each z-stack image, the maximum intensity projection was determined and puncta were identified as objects within the nuclear boundary (nucleoplasm) in which signal within the condensate was above a particular threshold cutoff. A threshold cutoff of 3 standard deviations above the mean of the image intensity was used for all candidates except for MECP2, in which a threshold cutoff of 2 standard deviations above the mean was used. Once identified, the area and number of each condensate was quantified. Partition ratios were calculated as the ratio of the mean pixel intensity within puncta relative to the mean pixel intensity of the nucleoplasmic signal, excluding signal within other segmented puncta.
FRAP analysis was performed on LSM880 Airyscan microscope with 488 nm laser. Photobleaching was performed by defining and exposing a region of interest in or around a punctum to 100% laser power. Images were collected every 0.5–2 seconds for up to a minute or until the majority of the fluorescence intensity was recovered. Fiji was used to calculate intensity values within the bleached region of interest during the timelapse before, during, and after bleaching. Fluorescence intensities in the region were normalized to pre-bleached values and the recovery profile was fit to a single exponential model.
Stress Condition Experiments
Cells were treated with 0.5mM NaAsO2 (Sigma) solution in cell culture media for 60 minutes and imaged. Images were acquired using Zeiss LSM880 Confocal Microscope with Airyscan processing with a 63x Objective and 2x Zoom using ZenBlack acquisition software. Images were postprocessed using Fiji.
Immunofluorescence with Human Tissue Samples
Fresh frozen kidney and breast tissues were purchased from BioIVT. Tissue was embedded in OCT and frozen. Fresh frozen colon tissue was embedded in OCT and frozen at −80°C. Tissue was sectioned into 10 μm sections using the cryostat with temperature set at −15°C or −20°C, respectively. Sections were stored at −20°C until use.
For the immunofluorescence sections were brought to room temperature, they were fixed in 4% PFA in PBS for 10 minutes. Following three washes in PBS, tissues were permeabilized using 0.5% TX100 in PBS, washed three times in PBS and blocked with 4% BSA in PBS for 1 hours. Primary antibodies were diluted into 4% BSA in PBS and added to the tissue sample for overnight incubation at RT. Following three washes in PBS, samples were incubated with secondary antibodies diluted 1:500 in 4% BSA in PBS. Samples was washed in PBS, DNA was stained using 20μm/mL Hoechst 33258 (Life Technologies, H3569) for 5 minutes and mounted using Vectashield (VWR, 101098-042). Images were acquired using Zeiss LSM880 Confocal Microscope with Airyscan processing with a 63x Objective and 2x Zoom using ZenBlack acquisition software. Images were postprocessed using Fiji.
Primary antibodies used: TCOF1 (PA558309, Thermofisher); SALL1 (PA562057, Invitrogen); and BARD1 (ab226854, Abcam). Secondary antibody used: Goat anti-rabbit AlexaFluor-488 (A11008, Thermofisher).
Human Cell Line Experiments
Cells were transiently transfected with the 1μg of WT or mutant DNA constructs (same as those used for mESC experiments, see above) into 0.5 × 106 cells that were plated onto 35 mm glass plates (MatTek). Transfections were performed using Lipofectamine 3000 (ThermoFisher) according to manufacturer instructions in complete DMEM media, described above. On the day following transfection, the media was changed and expression of the mEGFP-tagged protein was induced by adding doxycycline to the media at 1μg/mL for 24 hours, followed by imaging, processing, and analysis as described for mESCs above.
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise noted, all statistical analyses involving overlaps between sets of proteins and were done using one-tailed Fisher’s exact tests, and p-values were adjusted for multiple comparisons using the Benjamini-Hoechberg procedure. Unless otherwise noted, all statistical analyses involving comparisons between distributions were done using two-sided Wilcoxon rank sum / Mann Whitney U tests, and p-values were adjusted for multiple comparisons using the Bonferroni correction.
Supplementary Material
Table S4. Results from analyses reported in this study. Related to Figures 1–3. This file contains several tables containing additional information on the mapping of condensate-promoting features, canonical protein features, and pathogenic mutations across the proteome, as well as on the analyses reported in this study.
HIGHLIGHTS.
Disease mutations mapped to condensate-promoting features in proteins
36,000 mutations predicted to cause condensate dysregulation in 1,700 diseases
Condensate dysregulation in cells confirmed for subset of mutations
Resource provides foundation for novel disease models and therapeutic hypotheses
ACKNOWLEDGEMENTS
We thank Philip A. Sharp, Arup Chakraborty, Krishna Shrinivas, Brennan Decker, Fei Dong, Neal Lindeman, Isaac Klein, Charles Li, Eliot Coffey, Jie Wang, and members of the Young Lab for helpful discussions. We thank Caitlin Rausch for assistance with the graphical abstract. The work was supported by National Institutes of Health (NIH) grant R01 GM123511 (R.A.Y.), National Science Foundation (NSF) grant PHY1743900 (R.A.Y.), NIH grant 2 R01 MH104610-20 (R.A.Y.), the St. Jude Children’s Research Hospital Collaborative on Cohesin, CTCF and the 3D Regulatory Nuclear Landscape of Pediatric Cancer Cells, funds from Novo Nordisk, Brigham and Women’s Hospital Clinical Pathology Residency Program (S.F.B), NIH National Cancer Institute (NCI) T32 CA251062-02 (S.F.B.), NIH National Cancer Institute Award F31 CA250171 (L.K.A.), NCI F32 1F32CA254216 (J.E.H.), Hope Funds for Cancer Research (A.D.), DOD PRCRP Horizon Award W81XWH-20-10716 (V.E.C.), NREF Andrew T. Parsa Research Fellowship Grant (V.E.C.), NIH T32 5T32DK007191-45 (J.M.P.), and the Gruss-Lipper, Rothschild, and Zuckerman postdoctoral fellowships (I.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
DECLARATION OF INTERESTS
R.A.Y. is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics, and Dewpoint Therapeutics. S.F.B. and A.D. are consultants to Dewpoint Therapeutics. J.E.H. and O.O. are consultants to Camp4 Therapeutics. T.I.L. is a shareholder of Syros Pharmaceuticals and a consultant to Camp4 Therapeutics. All other authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahn JH, Davis ES, Daugird TA, Zhao S, Quiroga IY, Uryu H, Li J, Storey AJ, Tsai Y-H, Keeley DP, et al. (2021). Phase separation drives aberrant chromatin looping and cancer development. Nature 1–5. 10.1038/s41586-021-03662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S (2017). The wisdom of crowds: regulating cell function through condensed states of living matter. J Cell Sci 130, jcs.200295. 10.1242/jcs.200295. [DOI] [PubMed] [Google Scholar]
- Alberti S, and Dormann D (2019). Liquid–Liquid Phase Separation in Disease. Annu Rev Genet 53, 171–194. 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- Alberti S, and Hyman AA (2021). Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Bio 22, 196–213. 10.1038/s41580-020-00326-6. [DOI] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, and Lindquist S (2009). A Systematic Survey Identifies Prions and Illuminates Sequence Features of Prionogenic Proteins. Cell 137, 146–158. 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, and Hamosh A (2015). OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 43, D789–D798. 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babinchak WM, Dumm BK, Venus S, Boyko S, Putnam AA, Jankowsky E, and Surewicz WK (2020). Small molecules as potent biphasic modulators of protein liquid-liquid phase separation. Nat Commun 11, 5574. 10.1038/s41467-020-19211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, and Rosen MK (2016). Compositional Control of Phase-Separated Cellular Bodies. Cell 166, 651–663. 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Bio 18, 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Mackowiak SD, Niskanen H, Knezevic D, Asimi V, Grosswendt S, Geertsema H, Ali S, Jerković I, Ewers H, et al. (2020). Unblending of Transcriptional Condensates in Human Repeat Expansion Disease. Cell 181, 1062–1079.e30. 10.1016/j.cell.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle C, Yang P, Coughlin M, Messing J, Pesarrodona M, Szulc E, Salvatella X, Kim HJ, Taylor JP, and Ventura S (2020). hnRNPDL Phase Separation Is Regulated by Alternative Splicing and Disease-Causing Mutations Accelerate Its Aggregation. Cell Reports 30, 1117–1128.e5. 10.1016/j.celrep.2019.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhofer M, Goldberg T, Wolf S, Ahmed M, Zaugg J, Boden M, and Rost B (2017). NLSdb—major update for database of nuclear localization signals and nuclear export signals. Nucleic Acids Res 46, gkx1021-. 10.1093/nar/gkx1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutel O, Maraspini R, Pombo-García K, Martin-Lemaitre C, and Honigmann A (2019). Phase Separation of Zonula Occludens Proteins Drives Formation of Tight Junctions. Cell 179, 923–936.e11. 10.1016/j.cell.2019.10.011. [DOI] [PubMed] [Google Scholar]
- Bienz M (2020). Head-to-Tail Polymerization in the Assembly of Biomolecular Condensates. Cell 182, 799–811. 10.1016/j.cell.2020.07.037. [DOI] [PubMed] [Google Scholar]
- Blum M, Chang H-Y, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, et al. (2020). The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49, gkaa977-. 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. (2018). Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175, 1842–1855.e16. 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, and Young RA (2021). Biomolecular condensates and cancer. Cancer Cell 39, 174–192. 10.1016/j.ccell.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N, Granata D, Marzahn MR, Lindorff-Larsen K, Salvatella X, et al. (2018). Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol Cell 72, 19–36.e8. 10.1016/j.molcel.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, Awad ME, Rengarajan S, Volorio A, McBride MJ, et al. (2017). Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 171, 163–178.e19. 10.1016/j.cell.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, and Babu MM (2012). Tissue-Specific Splicing of Disordered Segments that Embed Binding Motifs Rewires Protein Interaction Networks. Mol Cell 46, 871–883. 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, and Fawzi NL (2015). Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell 60, 231–241. 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Feliciano D, Dong P, Flores E, Gruebele M, Porat-Shliom N, Sukenik S, Liu Z, and Lippincott-Schwartz J (2019). Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat Cell Biol 21, 1578–1589. 10.1038/s41556-019-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Liu Z, and Lippincott-Schwartz J (2021). Biomolecular Condensates and Their Links to Cancer Progression. Trends Biochem Sci 10.1016/j.tibs.2021.01.002. [DOI] [PubMed] [Google Scholar]
- Carbon S, Douglass E, Dunn N, Good B, Harris NL, Lewis SE, Mungall CJ, Basu S, Chisholm RL, Dodson RJ, et al. (2018). The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47, D330–D338. 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Ditlev JA, and Rosen MK (2019). Regulation of Transmembrane Signaling by Phase Separation. Annu Rev Biophys 48, 1–30. 10.1146/annurev-biophys-052118-115534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov 2, 401–404. 10.1158/2159-8290.cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty D, Gao J, Phillips S, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, et al. (2017). OncoKB: A Precision Oncology Knowledge Base. Jco Precis Oncol 2017, 1–16. 10.1200/po.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra B, Michmerhuizen NL, Shirnekhi HK, Tripathi S, Pioso BJ, Baggett DW, Mitrea DM, Iacobucci I, White MR, Chen J, et al. (2021). Phase Separation Mediates NUP98 Fusion Oncoprotein Leukemic TransformationPhase Separation Drives Oncogenesis by NUP98 Fusion Proteins. Cancer Discov 12, 1152–1169. 10.1158/2159-8290.cd-21-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, and Cisse II (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, eaar4199. 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J-M, Holehouse AS, and Pappu RV (2020). Physical Principles Underlying the Complex Biology of Intracellular Phase Transitions. Annu Rev Biophys 49, 1–27. 10.1146/annurev-biophys-121219-081629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloer EW, Siesser PF, Cousins EM, Goldfarb D, Mowrey DD, Harrison JS, Weir SJ, Dokholyan NV, and Major MB (2018). p62-Dependent Phase Separation of Patient-Derived KEAP1 Mutations and NRF2. Mol Cell Biol 38, e00644–17. 10.1128/mcb.00644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella AE, Zerze GH, Mittal J, and Fawzi NL (2016). ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure 24, 1537–1549. 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TAPG (2017). AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 7, 818–831. 10.1158/2159-8290.cd-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TU, Bateman A, Martin M-J, Orchard S, Magrane M, Agivetova R, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R, et al. (2020). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49, D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao TP, Kolaitis R-M, Kim HJ, O’Donovan K, Martyniak B, Colicino E, Hehnly H, Taylor JP, and Castañeda CA (2018). Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol Cell 69, 965–978.e6. 10.1016/j.molcel.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao TP, Martyniak B, Canning AJ, Lei Y, Colicino EG, Cosgrove MS, Hehnly H, and Castañeda CA (2019). ALS-Linked Mutations Affect UBQLN2 Oligomerization and Phase Separation in a Position- and Amino Acid-Dependent Manner. Structure 27, 937–951.e5. 10.1016/j.str.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, and Chen Z (2018). DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, eaat1022. 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emenecker RJ, Griffith D, and Holehouse AS (2021). metapredict: a fast, accurate, and easy-to-use predictor of consensus disorder and structure. Biophys J 120, 4312–4319. 10.1016/j.bpj.2021.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasciani A, D’Annunzio S, Poli V, Fagnocchi L, Beyes S, Michelatti D, Corazza F, Antonelli L, Gregoretti F, Oliva G, et al. (2020). MLL4-associated condensates counterbalance Polycomb-mediated nuclear mechanical stress in Kabuki syndrome. Nat Genet 52, 1397–1411. 10.1038/s41588-020-00724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, and Brangwynne CP (2016). Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 165, 1686–1697. 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichó E, Reményi I, Simon I, and Mészáros B (2017). MFIB: a repository of protein complexes with mutual folding induced by binding. Bioinformatics 33, 3682–3684. 10.1093/bioinformatics/btx486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca M. de C., Oliveira J.F. de, Araujo BHS, Canateli C, Prado P.F.V. do, Neto DPA, Bosque BP, Rodrigues PV, Godoy J.V.P. de, Tostes K, et al. (2021). Molecular and cellular basis of hyperassembly and protein aggregation driven by a rare pathogenic mutation in DDX3X. Iscience 24, 102841. 10.1016/j.isci.2021.102841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottin F, Schueder F, Tiwary S, Gupta R, Körner R, Schlichthaerle T, Cox J, Jungmann R, Hartl FU, and Hipp MS (2019). The nucleolus functions as a phase-separated protein quality control compartment. Sci New York N Y 365, 342–347. 10.1126/science.aaw9157. [DOI] [PubMed] [Google Scholar]
- Gibson B, Doolittle L, Schneider M, Jensen L, Gamarra N, Henry L, Gerlich D, Redding S, and Rosen M (2019). Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 179, 470–484.e21. 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, Danos AM, Ainscough BJ, Ramirez CA, Rieke DT, Kujan L, et al. (2017). CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet 49, 170–174. 10.1038/ng.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueroussov S, Weatheritt RJ, O’Hanlon D, Lin Z-Y, Narula A, Gingras A-C, and Blencowe BJ (2017). Regulatory Expansion in Mammals of Multivalent hnRNP Assemblies that Globally Control Alternative Splicing. Cell 170, 324–339.e23. 10.1016/j.cell.2017.06.037. [DOI] [PubMed] [Google Scholar]
- Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall’Agnese A, Hannett NM, Spille J-H, Afeyan LK, Zamudio AV, Shrinivas K, et al. (2019). Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548. 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henninger JE, Oksuz O, Shrinivas K, Sagi I, LeRoy G, Zheng MM, Andrews JO, Zamudio AV, Lazaris C, Hannett NM, et al. (2021). RNA-Mediated Feedback Control of Transcriptional Condensates. Cell 184, 207–225.e24. 10.1016/j.cell.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Castello A, Schwarzl T, and Preiss T (2018). A brave new world of RNA-binding proteins. Nat Rev Mol Cell Bio 19, 327–341. 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et al. (2018). Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 173, 291–304.e6. 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holehouse AS, Das RK, Ahad JN, Richardson MOG, and Pappu RV (2017). CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophys J 112, 16–21. 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, and Skrzypek E (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43, D512–D520. 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard TP, and Roberts CWM (2020). Partitioning of Chemotherapeutics into Nuclear Condensates—Opening the Door to New Approaches for Drug Development. Mol Cell 79, 544–545. 10.1016/j.molcel.2020.07.029. [DOI] [PubMed] [Google Scholar]
- Huang WYC, Alvarez S, Kondo Y, Lee YK, Chung JK, Lam HYM, Biswas KH, Kuriyan J, and Groves JT (2019). A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103. 10.1126/science.aau5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MP, Sawaya MR, Boyer DR, Goldschmidt L, Rodriguez JA, Cascio D, Chong L, Gonen T, and Eisenberg DS (2018). Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 359, 698–701. 10.1126/science.aan6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, and Parker R (2016). ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 164, 487–498. 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang S, Huang Y, He X, Cui H, Zhu X, and Zheng Y (2015). Phase Transition of Spindle-Associated Protein Regulate Spindle Apparatus Assembly. Cell 163, 108–122. 10.1016/j.cell.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaan NM, Hamel C, Grabinski T, and Combs B (2020). Liquid-liquid phase separation induces pathogenic tau conformations in vitro. Nat Commun 11, 2809. 10.1038/s41467-020-16580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasza KE, Supriyatno S, and Zallen JA (2019). Cellular defects resulting from disease-related myosin II mutations in Drosophila. Proc National Acad Sci 116, 22205–22211. 10.1073/pnas.1909227116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler, and D. (2002). The Human Genome Browser at UCSC. Genome Res 12, 996–1006. 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, and Altmeyer M (2019). Phase separation of 53 BP 1 determines liquid-like behavior of DNA repair compartments. Embo J 38, e101379. 10.15252/embj.2018101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang Y-D, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. (2013). Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473. 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MR, and Petry S (2020). Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat Commun 11, 270. 10.1038/s41467-019-14087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall’Agnese A, Oksuz O, Henninger JE, Shrinivas K, Sabari BR, et al. (2020). Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392. 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Carmody L, Vasilevsky N, Jacobsen JOB, Danis D, Gourdine J-P, Gargano M, Harris NL, Matentzoglu N, McMurry JA, et al. (2018). Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res 47, D1018–D1027. 10.1093/nar/gky1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Gouw M, Michael S, Sámano-Sánchez H, Pancsa R, Glavina J, Diakogianni A, Valverde JA, Bukirova D, Čalyševa J, et al. (2019). ELM—the eukaryotic linear motif resource in 2020. Nucleic Acids Res 48, D296–D306. 10.1093/nar/gkz1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundra R, Zhang H, Sheridan R, Sirintrapun SJ, Wang A, Ochoa A, Wilson M, Gross B, Sun Y, Madupuri R, et al. (2021). OncoTree: A Cancer Classification System for Precision Oncology. Jco Clin Cancer Informatics 5, 221–230. 10.1200/cci.20.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, and Weirauch MT (2018). The Human Transcription Factors. Cell 172, 650–665. 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- Lancaster AK, Nutter-Upham A, Lindquist S, and King OD (2014). PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30, 2501–2502. 10.1093/bioinformatics/btu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al. (2017). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46, D1062–D1067. 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, and Narlikar GJ (2017). Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240. 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos C, Schulze L, Weiske J, Meyer H, Braeuer N, Barak N, Eberspächer U, Werbeck N, Stresemann C, Lange M, et al. (2020). Identification of Small Molecules that Modulate Mutant p53 Condensation. Iscience 23, 101517. 10.1016/j.isci.2020.101517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Khedkar S, and Bork P (2020). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49, gkaa937-. 10.1093/nar/gkaa937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Caldara AL, Zimmer SE, Stahley SN, Seybold A, Strong NL, Frangakis AS, Levental I, Wahl JK, Mattheyses AL, et al. (2019). The desmosome is a mesoscale lipid raft-like membrane domain. Mol Biol Cell 30, 1390–1405. 10.1091/mbc.e18-10-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Coffey EL, Dall’Agnese A, Hannett NM, Tang X, Henninger JE, Platt JM, Oksuz O, Zamudio AV, Afeyan LK, et al. (2020). MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature 1–8. 10.1038/s41586-020-2574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ, Younes A, et al. (2017). Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer. J Mol Diagnostics 19, 4–23. 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Peng X, Li Y, Tang W, Zhu J, Huang J, Qi Y, and Zhang Z (2019). LLPSDB: a database of proteins undergoing liquid–liquid phase separation in vitro. Nucleic Acids Res 48, D320–D327. 10.1093/nar/gkz778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, and Parker R (2015). Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 60, 208–219. 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom RGH, Supek F, and Lehner B (2016). The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet 48, 1112–1118. 10.1038/ng.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, Darzacq X, and Zhou Q (2018). Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323. 10.1038/s41586-018-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, et al. (2019). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48, D265–D268. 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wu T, Gutman O, Lu H, Zhou Q, Henis YI, and Luo K (2020). Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat Cell Biol 22, 453–464. 10.1038/s41556-020-0485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, and Varani G (2007). RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Bio 8, 479–490. 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon AS, Peeples WB, and Rosen MK (2020). A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Bio 1–21. 10.1038/s41580-020-00303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, Annu K, Baker M, Perkerson RB, Kurti A, et al. (2017). TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron 95, 808–816.e9. 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, and Mittag T (2020). Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699. 10.1126/science.aaw8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Yu Z, Zhang D, Chen S, Guan H, Zhou R, Wu Q, Zhang Q, Liu S, Ramani MKV, et al. (2021). Induced phase separation of mutant NF2 imprisons the cGAS-STING machinery to abrogate antitumor immunity. Mol Cell 10.1016/j.molcel.2021.07.040. [DOI] [PubMed] [Google Scholar]
- Mészáros B, Erdős G, Szabó B, Schád É, Tantos Á, Abukhairan R, Horváth T, Murvai N, Kovács OP, Kovács M, et al. (2019). PhaSePro: the database of proteins driving liquid–liquid phase separation. Nucleic Acids Res 48, D360–D367. 10.1093/nar/gkz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierlo G. van, Jansen JRG, Wang J, Poser I, Heeringen S.J. van, and Vermeulen M (2021). Predicting protein condensate formation using machine learning. Cell Reports 34, 108705. 10.1016/j.celrep.2021.108705. [DOI] [PubMed] [Google Scholar]
- Milovanovic D, Wu Y, Bian X, and Camilli PD (2018). A liquid phase of synapsin and lipid vesicles. Science 361, eaat5671. 10.1126/science.aat5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Wright WE, and Shay JW (2019). Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Gene Dev 33, 814–827. 10.1101/gad.324905.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. (2020). Pfam: The protein families database in 2021. Nucleic Acids Res 49, gkaa913-. 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, and Kriwacki RW (2016). Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 5, e13571. 10.7554/elife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea DM, Cika JA, Stanley CB, Nourse A, Onuchic PL, Banerjee PR, Phillips AH, Park C-G, Deniz AA, and Kriwacki RW (2018). Self-interaction of NPM1 modulates multiple mechanisms of liquid–liquid phase separation. Nat Commun 9, 842. 10.1038/s41467-018-03255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahm M, Lim SM, Kim Y-E, Park J, Noh M-Y, Lee S, Roh JE, Hwang S-M, Park C-K, Kim YH, et al. (2020). ANXA11 mutations in ALS cause dysregulation of calcium homeostasis and stress granule dynamics. Sci Transl Med 12, eaax3993. 10.1126/scitranslmed.aax3993. [DOI] [PubMed] [Google Scholar]
- Nedelsky NB, and Taylor JP (2019). Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat Rev Neurol 15, 272–286. 10.1038/s41582-019-0157-5. [DOI] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. (2015). Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57, 936–947. 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MW, Bell M, Mir M, Kao JA, Darzacq X, Botchan MR, and Berger JM (2019). A new class of disordered elements controls DNA replication through initiator self-assembly. Elife 8, e48562. 10.7554/elife.48562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077. 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Pawson T, and Nash P (2003). Assembly of Cell Regulatory Systems Through Protein Interaction Domains. Science 300, 445–452. 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Peskett T, Rau F, O’Driscoll J, Patani R, Lowe A, and Saibil H (2018). A Liquid to Solid Phase Transition Underlying Pathological Huntingtin Exon1 Aggregation. Mol Cell 70, 588–601.e6. 10.1016/j.molcel.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz FG, Fiore VF, Levorse J, Polak L, Wong E, Pasolli HA, and Fuchs E (2020). Liquid-liquid phase separation drives skin barrier formation. Sci New York N Y 367, eaax9554. 10.1126/science.aax9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai AK, Chen J-X, Selbach M, and Pelkmans L (2018). Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216. 10.1038/s41586-018-0279-8. [DOI] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, and Parker R (2013). Altered Ribostasis: RNA-Protein Granules in Degenerative Disorders. Cell 154, 727–736. 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Singh N, Kumar R, Patel K, Pandey S, Datta D, Mahato J, Panigrahi R, Navalkar A, Mehra S, et al. (2020). α-Synuclein aggregation nucleates through liquid–liquid phase separation. Nat Chem 1–12. 10.1038/s41557-020-0465-9. [DOI] [PubMed] [Google Scholar]
- Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei M-T, Kriwacki RW, and Brangwynne CP (2020). Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214. 10.1038/s41586-020-2256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17, 405–423. 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, et al. (2006). Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc National Acad Sci 103, 8390–8395. 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958. 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, and Görlich D (2015). Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. Elife 4, e04251. 10.7554/elife.04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JW, Oommen S, Qureshi MY, Goetsch SC, Pease DR, Sundsbak RS, Guo W, Sun M, Sun H, Kuroyanagi H, et al. (2020). Dysregulated ribonucleoprotein granules promote cardiomyopathy in RBM20 gene-edited pigs. Nat Med 26, 1788–1800. 10.1038/s41591-020-1087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwayer C, Shamipour S, Pranjic-Ferscha K, Schauer A, Balda M, Tada M, Matter K, and Heisenberg C-P (2019). Mechanosensation of Tight Junctions Depends on ZO-1 Phase Separation and Flow. Cell 179, 937–952.e18. 10.1016/j.cell.2019.10.006. [DOI] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou M-M, and Pawson T (2006). Reading protein modifications with interaction domains. Nat Rev Mol Cell Bio 7, 473–483. 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- Sharkey LM, Safren N, Pithadia AS, Gerson JE, Dulchavsky M, Fischer S, Patel R, Lantis G, Ashraf N, Kim JH, et al. (2018). Mutant UBQLN2 promotes toxicity by modulating intrinsic self-assembly. Proc National Acad Sci 115, 201810522. 10.1073/pnas.1810522115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J, and MacRae IJ (2018). Phase Transitions in the Assembly and Function of Human miRISC. Cell 173, 946–957.e16. 10.1016/j.cell.2018.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382. 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Smith JA, Curry EG, Blue RE, Roden C, Dundon SER, Rodríguez-Vargas A, Jordan DC, Chen X, Lyons SM, Crutchley J, et al. (2020). FXR1 splicing is important for muscle development and biomolecular condensates in muscle cells. J Cell Biol 219. 10.1083/jcb.201911129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannl S, Tereshchenko M, Mastromarco GJ, Ihn SJ, and Lee HO (2019). Biomolecular condensates in neurodegeneration and cancer. Traffic 20, 890–911. 10.1111/tra.12704. [DOI] [PubMed] [Google Scholar]
- Stefl S, Nishi H, Petukh M, Panchenko AR, and Alexov E (2013). Molecular mechanisms of disease-causing missense mutations. J Mol Biol 425, 3919–3936. 10.1016/j.jmb.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Chapman M, Evans K, Azevedo L, Hayden M, Heywood S, Millar DS, Phillips AD, et al. (2020). The Human Gene Mutation Database (HGMD®): optimizing its use in a clinical diagnostic or research setting. Hum Genet 139, 1197–1207. 10.1007/s00439-020-02199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C, Wang T, Michaelos M, and Pachitariu M (2021). Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18, 100–106. 10.1038/s41592-020-01018-x. [DOI] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, and Karpen GH (2017). Phase separation drives heterochromatin domain formation. Nature 547, 241–245. 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, and Vale RD (2016). Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599. 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborero D, Rubio-Perez C, Deu-Pons J, Schroeder MP, Vivancos A, Rovira A, Tusquets I, Albanell J, Rodon J, Tabernero J, et al. (2018). Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med 10, 25. 10.1186/s13073-018-0531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team TMP, Temple G, Gerhard DS, Rasooly R, Feingold EA, Good PJ, Robinson C, Mandich A, Derge JG, Lewis J, et al. (2009). The completion of the Mammalian Gene Collection (MGC). Genome Res 19, 2324–2333. 10.1101/gr.095976.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Blal HA, Alm T, Asplund A, Björk L, Breckels LM, et al. (2017). A subcellular map of the human proteome. Science 356. 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- Tsang B, Pritišanac I, Scherer SW, Moses AM, and Forman-Kay JD (2020). Phase Separation as a Missing Mechanism for Interpretation of Disease Mutations. Cell 183, 1742–1756. 10.1016/j.cell.2020.11.050. [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Wang Y-D, Parker M, Patmore DM, Kanagaraj A, Moore J, Rusch M, Finkelstein D, Ellison DW, Gilbertson RJ, et al. (2016). Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci Rep-Uk 6, 25996. 10.1038/srep25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, and Luscombe NM (2009). A census of human transcription factors: function, expression and evolution. Nat Rev Genet 10, 252–263. 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, and Forman-Kay JD (2018). Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife 7, e31486. 10.7554/elife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viny AD, and Levine RL (2020). Drug modulation by nuclear condensates. Sci New York N Y 368, 1314–1315. 10.1126/science.abc5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Chong S, Xuan F, Liang A, Cui X, Gates L, Carroll TS, Li Y, Feng L, Chen G, et al. (2019). Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 577, 121–126. 10.1038/s41586-019-1842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan PTC, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, et al. (2004). Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855–867. 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Choi J-M, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. (2018). A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 174, 688–699.e16. 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RJ, Lee HO, Poser I, Pal A, Doeleman T, Kishigami S, Kour S, Anderson EN, Marrone L, Murthy AC, et al. (2019). Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. Biorxiv 721001. 10.1101/721001. [DOI] [Google Scholar]
- Woodruff JB, Gomes BF, Widlund PO, Mahamid J, Honigmann A, and Hyman AA (2017). The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 169, 1066–1077.e10. 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]
- Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, et al. (2019). Ensembl 2020. Nucleic Acids Res 48, D682–D688. 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Ali R, Jiou J, Fung HYJ, Burke KA, Kim SJ, Lin Y, Peeples WB, Saltzberg D, Soniat M, et al. (2018). Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell 173, 693–705.e22. 10.1016/j.cell.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You K, Huang Q, Yu C, Shen B, Sevilla C, Shi M, Hermjakob H, Chen Y, and Li T (2019). PhaSepDB: a database of liquid–liquid phase separation related proteins. Nucleic Acids Res 48, D354–D359. 10.1093/nar/gkz847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Shen B, You K, Huang Q, Shi M, Wu C, Chen Y, Zhang C, and Li T (2020). Proteome-scale analysis of phase-separated proteins in immunofluorescence images. Brief Bioinform 22. 10.1093/bib/bbaa187. [DOI] [PubMed] [Google Scholar]
- Yun M, Wu J, Workman JL, and Li B (2011). Readers of histone modifications. Cell Res 21, 564–578. 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio AV, Dall’Agnese A, Henninger JE, Manteiga JC, Afeyan LK, Hannett NM, Coffey EL, Li CH, Oksuz O, Sabari BR, et al. (2019). Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Mol Cell 76, 753–766.e6. 10.1016/j.molcel.2019.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Shang Y, Araki Y, Guo T, Huganir RL, and Zhang M (2016). Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity. Cell 166, 1163–1175.e12. 10.1016/j.cell.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhao R, Tones J, Liu M, Dilley RL, Chenoweth DM, Greenberg RA, and Lampson MA (2020). Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol Biol Cell 31, 2048–2056. 10.1091/mbc.e19-10-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Xie J, Kong W, Xie J, Li Y, Du L, Zheng Q, Sun L, Guan M, Li H, et al. (2020). Phase Separation of Disease-Associated SHP2 Mutants Underlies MAPK Hyperactivation. Cell 183, 490–502.e18. 10.1016/j.cell.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. Results from analyses reported in this study. Related to Figures 1–3. This file contains several tables containing additional information on the mapping of condensate-promoting features, canonical protein features, and pathogenic mutations across the proteome, as well as on the analyses reported in this study.
Data Availability Statement
Datasets used in this study are available from the sources cited in the Key Resources Table. Processed data used in the study is available in the Supplemental Tables and on the Young laboratory’s website.
Code generated during this study is available via https://github.com/bananisf/2022_Banani_Afeyan_Hawken_Condensate_Dysregulation.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-rabbit AlexaFluor-488 | ThermoFisher | A11008; RRID:AB_143165 |
| Rabbit polyclonal anti-TCOF1 | ThermoFisher | PA558309; RRID:AB_2648324 |
| Rabbit polyclonal anti-SALL1 | Invitrogen | PA562057; RRID:AB_2646913 |
| Rabbit polyclonal anti-BARD1 | abcam | ab226854 |
| Biological samples | ||
| Human adult large intestine adjacent normal tissue | BioIVT | HMLGINTAD J2-PPG7 |
| Human adult breast adjacent normal tissue | BioIVT | HMNBREASTN ADJ2 |
| Human adult kidney normal | BioIVT | HMKIDNEYNO RC2 |
| Chemicals, peptides, and recombinant proteins | ||
| Poly-L-ornithine | Sigma | P4957-50ML |
| Doxycycline | Sigma | D9891-5G |
| Hoechst 33258 | Life Technologies | H3569 |
| Vectashield | Vector Laboratories | 101098-042 |
| Laminin (Mouse) | ThermoFisher | CB40232-1MG |
| Critical commercial assays | ||
| Lipofectamine 3000 Transfection Agent | ThermoFisher | L3000001 |
| Experimental models: Cell lines | ||
| V6.5 murine embryonic stem cells | Laboratory of Rudolph Jaenisch | N/A |
| Human: HCT116 | ATCC | CCL-247 |
| Human: MCF7 | ATCC | HTB-22 |
| Human: HEK293T | ATCC | CRL-3216 |
| Recombinant DNA | ||
| pbfh_GFP | (Henninger et al., 2021) | Modified version of pJH135_pb_MCPx2_mCherry_rTTA |
| pbfh_GFP_MECP2 | This study | N/A |
| pbfh_GFP_MECP2_R186* | This study | N/A |
| pbfh_GFP_BARD1 | This study | N/A |
| pbfh_GFP_BARD1_R406* | This study | N/A |
| pbfh_GFP_BCL11A | This study | N/A |
| pbfh_GFP_BCL11A_Q177* | This study | N/A |
| pbfh_GFP_BCOR | This study | N/A |
| pbfh_GFP_BCOR_Y657* | This study | N/A |
| pbfh_GFP_BRD3 | This study | N/A |
| pbfh_GFP_BRD3_F334S | This study | N/A |
| pbfh_GFP_HP1A | This study | N/A |
| pbfh_GFP_HP1A_V21I | This study | N/A |
| pbfh_GFP_HP1A_W142C | This study | N/A |
| pbfh_GFP_DAXX | This study | N/A |
| pbfh_GFP_DAXX_R318* | This study | N/A |
| pbfh_GFP_EDC4 | This study | N/A |
| pbfh_GFP_EDC4_I371V | This study | N/A |
| pbfh_GFP_ESRP1 | This study | N/A |
| pbfh_GFP_ESPR1_L259V | This study | N/A |
| pbfh_GFP_NONO | This study | N/A |
| pbfh_GFP_NONO_446fs | This study | N/A |
| pbfh_GFP_RBM10 | This study | N/A |
| pbfh_RBM10_V354M | This study | N/A |
| pbfh_GFP_SALL1 | This study | N/A |
| pbfh_GFP_SALL1_S372* | This study | N/A |
| pbfh_GFP_SRSF2 | This study | N/A |
| pbfh_GFP_SRSF2_S54F | This study | N/A |
| pbfh_GFP_SRSF2_P95H | This study | N/A |
| pbfh_GFP_TCOF | This study | N/A |
| pbfh_GFP_TCOF_Q55* | This study | N/A |
| pbfh_GFP_ASXL1 | This study | N/A |
| pbfh_GFP_BCL6 | This study | N/A |
| pbfh_GFP_BRCA1 | This study | N/A |
| pbfh_GFP_DVL2 | This study | N/A |
| pbfh_GFP_DYR1A | This study | N/A |
| pbfh_GFP_ENC1 | This study | N/A |
| pbfh_GFP_G3BP1 | This study | N/A |
| pbfh_GFP_HMGA2 | This study | N/A |
| pbfh_GFP_NIPBL | This study | N/A |
| pbfh_GFP_NKX21 | This study | N/A |
| pbfh_GFP_SNCAP | This study | N/A |
| pbfh_GFP_TERT | This study | N/A |
| Software and algorithms | ||
| Fiji image processing package | (Schindelin et al., 2012) | https://fiji.sc/ |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Code generated by the study | This study | https://github.com/bananisf/ |
| Python v3.6.9 | www.python.org | N/A |
| CellPose v0.7.2 | (Stringer et al., 2021) | http://www.cellpose.org/ |
| MATLAB R2019b | Mathworks | N/A |
| metapredict | (Emenecker et al., 2021) | N/A |
| PLAAC | (Lancaster et al., 2014) | N/A |
| PSP | (Vernon et al., 2018) | N/A |
| Ensembl VEP v102 | (Yates et al., 2019) | N/A |
| Other | ||
| 35 mm glass-bottom imaging dishes | Mattek Corporation | P35G-1.5-20-C |


