Abstract
Toxicological risk assessment is essential in the evaluation and authorization of different classes of chemical substances. Genotoxicity and mutagenicity testing are of highest priority and rely on established in vitro systems with bacterial and mammalian cells, sometimes followed by in vivo testing using rodent animal models. Transcriptomic approaches have recently also shown their value to determine transcript signatures specific for genotoxicity. Here, we studied how transcriptomic data, in combination with in vitro tests with human cells, can be used for the identification of genotoxic properties of test compounds. To this end, we used liver samples from a 28-day oral toxicity study in rats with the pesticidal active substances imazalil, thiacloprid, and clothianidin, a neonicotinoid-type insecticide with, amongst others, known hepatotoxic properties. Transcriptomic results were bioinformatically evaluated and pointed towards a genotoxic potential of clothianidin. In vitro Comet and γH2AX assays in human HepaRG hepatoma cells, complemented by in silico analyses of mutagenicity, were conducted as follow-up experiments to check if the genotoxicity alert from the transcriptomic study is in line with results from a battery of guideline genotoxicity studies. Our results illustrate the combined use of toxicogenomics, classic toxicological data and new approach methods in risk assessment. By means of a weight-of-evidence decision, we conclude that clothianidin does most likely not pose genotoxic risks to humans.
Keywords: Liver toxicity, Pesticide, Genotoxicity, Clastogenicity, HepaRG cells
Highlights
-
•
Analysis of clothianidin genotoxicity in silico, in vitro and in vivo.
-
•
Application of a toxicogenomics approach to analyze genotoxicity.
-
•
Weight-of-evidence decision supports classification as “non-genotoxic”.
1. Introduction
Risk assessment of chemical substances has two major goals: protection of the public from unacceptable risks to these substances and prediction of hazards from data e.g. for the purpose of classification and labelling. While traditionally batteries of in vivo tests are used to achieve these goals, there have been several advancements in the field of in vitro methods as well as in toxicogenomics over the past decade (Krewski et al., 2020; Sewell et al., 2021). These methods may allow achieving both goals – protection and prediction – by comprehensive tiered approaches that make use of such new approach methods (NAM) either as stand alone or in combination with traditional in vivo tests with the aim to refine, reduce or ad ultimo when possible replace animal testing.
In the present paper we use transcriptomic data from a previous 28-day in vivo study on the three pesticidal active substances clothianidin (CTD), imazalil (IMZ) and thiacloprid (THI) (Alarcan et al., 2021) as a starting point for our analyses. The choice of pesticides in the aforementioned study was based on the hepatotoxicity of the three compounds, and their presumed toxicological mode(s) of action relevant for possible combination effects; for further details please refer to (Alarcan et al., 2021). Pathway-focused transcriptomic analysis of these in vivo data revealed alerts on the endpoint genotoxicity, more specifically clastogenicity, for CTD only (Alarcan et al., 2021). CTD belongs to the family of neonicotinoids, a group of insecticides with chemical similarity to nicotine, acting through binding to the nicotinic acetylcholine receptor of insects. For a number of neonicotinoids including CTD, the European Food Safety Agency (EFSA) put restrictions into place for the risk they represent to wild bees and honey bees (EFSA, 2018). Besides these risks for non-target insects, neonicotinoids are also associated with hazards to other non-target organisms including mammals. Here, acute neurotoxic effects have been reported, while chronic exposure led to reduced body weight but also liver effects (cp. the report by the Joint FAO/WHO Meeting on Pesticide Residues; https://inchem.org/documents/jmpr/jmpmono/v2010pr01.pdf).
Results for three pesticidal active compounds at the transcriptomic level were compared to results from existing genotoxicity and mutagenicity guideline studies. Follow-up experiments were conducted to check if the presence or absence of alerts is in line with results from a battery of guideline studies. To this end, we chose HepaRG cells as an up-to-date in vitro model of human liver, in connection with Comet and γH2AX assays, as well as transcript marker pattern analyses to unravel potential genotoxic properties of the test compound CTD, complementing the aforementioned standard regulatory tests. Overall, the identification of a genotoxicity alert for CTD, but not IMZ or THI, in line with previous clastogenicity in vitro results for CTD, supports the usefulness of the toxicogenomics approach to detect alerts also detected by a traditional test battery, to achieve the goal of correctly predicting specific hazards.
This gives further evidence that transcriptomics as a new approach method may be implemented in next generation risk assessment as a first step screening method. Future directions of the use of transcriptomics in the analysis of genotoxicity has been discussed in detail (David, 2020): Besides traditional array-based testing also other tools like RNA sequencing or machine learning may be used, in order to utilize omics data to unravel characteristic fingerprint signatures of genotoxins (David, 2020). The use of transcriptomics for regulatory purposes such as read across and grouping has been successfully tested in the EU-ToxRisk project (Escher et al., 2019; Rovida et al., 2020) and implemented in a number of case studies.
Kohonen et al. could show the high predictive power of toxicogenomics for drug induced liver injury by building a ‘predictive toxicogenomics space’ using a ‘big data compacting and data fusion’ concept (Kohonen et al., 2017). Toxicogenomics based approaches have been successfully applied to predict other forms of liver toxicity within adverse outcome pathways (Heise et al., 2018; Knebel et al., 2019; Seeger et al., 2019).
Overall, the present study provides additional evidence that toxicogenomics can be used as a first line tool for hazard identification, and can be useful for risk assessment in combination with in vitro and/or in silico studies.
2. Materials and methods
2.1. Chemicals
CTD (CAS no. 210880-92-5, batch no. BCB53968V) was purchased from Sigma-Aldrich (Taufkirchen, Germany) in analytical grade with purity >99%. A 200 mM stock solution of CTD was prepared in dimethyl sulfoxide (DMSO; AppliChem, Darmstadt, Germany). Different positive controls were used depending on the individual assays and endpoints. All positive controls were dissolved in DMSO. For the Comet assay, the alkylating agent methyl methanesulfonate (MMS; Sigma-Aldrich) was used, with the 10 mM stock solution always freshly prepared. For the detection of double-strand breaks by γH2AX antibody staining, doxorubicin (Doxo; Cayman, Ann Arbor, MI, USA) and etoposide (Eto; Enzo Life Sciences, Farmingdale, NY, USA), as well as aflatoxin B1 (AB1; Sigma Aldrich) and benzo[a]pyrene (BaP; Supelco, Bellefonte, PA, USA) were used. All other chemicals were purchased from Merck (Darmstadt, Germany) or Sigma, at the highest purity available, if not stated otherwise below.
2.2. Cell culture
Undifferentiated HepaRG cells (Biopredic International, Saint Grégoire, France) were seeded in 96-well plates (9000 cells per well; for γH2AX staining), in 12-well plates (100,000 cells per well, for the Comet assay) or in 6-well plates (200,000 cells per well; for real-time RT-PCR) and cultured as previously described (Gripon et al., 2002; Luckert et al., 2018). Briefly, cells were cultivated and proliferated at 37 °C in a humidified atmosphere in William's E medium with 2 mM glutamine (Pan-Biotech, Aidenbach, Germany) supplemented with 10% fetal calf serum (FCS; Pan-Biotech), 100 U/mL penicillin, and 100 μg/mL streptomycin (both from Capricorn Scientific, Ebsdorfergrund, Germany), 0.05% human insulin (Pan-Biotech), and 50 μM hydrocortisone hemisuccinate (Sigma-Aldrich). After a 2-week proliferation phase, the cells were differentiated in the abovementioned culture medium additionally supplemented with 1.7% DMSO, for two further weeks. Differentiated HepaRG cells were treated with the test compounds in differentiation medium for 24 h.
2.3. Comet assay
The Comet assay allows for the detection of DNA damage in single cells. The test routine was conducted based on the Organisation for Economic Co-operation and Development (OECD) test guideline No. 489 (in vivo mammalian alkaline comet assay) (OECD, 2016), whereas the protocol was adapted from Singh et al. (1988). HepaRG cells were seeded in 12-well plates at a density of 100,000 cells per well, differentiated and incubated with CTD (0.5 mM and 1 mM for 24 h) as described in the cell culture section. MMS (100 μM) was used as a positive control for the assay. After incubation, cells were washed with 200 μl phosphate-buffered saline (PBS) and harvested with 200 μl trypsin per well by incubation at 37 °C for 5 min. Trypsinization was stopped with 500 μl culture medium. After a centrifugation step at 300×g for 5 min, the supernatant was removed and the pellet was washed twice with 200 μl PBS. The pellet was dissolved in 20 μl PBS and stored on ice. A total of 240 μl of heated low melting point agarose (Biozym, Vienna, Austria) was mixed with the cell suspension. The cell-agarose mixture was distributed equally on glass slides coated with 1% NEEO high quality agarose (Carl Roth, Karlsruhe, Germany). Slides were incubated in lysis buffer (2.25 M NaCl, 89 mM Na2EDTA, 8.9 mM Tris, 10% DMSO, 0.1% Triton X-100, pH 10) for 24 h at 4 °C. The next day, slides were briefly washed in electrophoresis buffer (300 mM NaOH, 1 mM Na2EDTA, pH > 13) and then randomly placed in the electrophoresis chamber. The run at 450 mA was performed for 30 min. Slides were incubated in neutralization buffer (0.4 M Tris, pH 7.5) for 10 min at room temperature. Next, the slides were dried in 100% ethanol for 10 min at room temperature. DNA staining was performed with SYBRGold (Invitrogen, Waltham, MA, USA). The high-content screening microscope Celldiscoverer 7 (Zeiss, Jena, Germany) including the software ZEN 3.1 was used for image acquisition. DNA tail intensity was assessed with the software Comet Assay IV (Instem, Staffordshire, United Kingdom). For each sample, two slides were prepared and analyzed. Per slide, 30 pictures were taken and at least 60 cells were analyzed. Statistical analysis was performed using one-way ANOVA and the Bonferroni post-hoc test: *p < 0.05, **p < 0.01, ***p < 0.001.
2.4. γH2AX assay
Differentiated HepaRG cells in 96-well format were treated with 0.1, 0.5 and 1 mM CTD and the positive controls Doxo (1 μM), Eto (25 μM), AB1 (1 μM), or BaP (5 μM). After 24 h, the cells were carefully washed once with cold PBS and fixed with 50 μl ice-cold methanol per well for 30 min. Cells were washed with PBS-T (0.1% Tween-20 in PBS) and blocked with 50 μl/well of 1% bovine serum albumin (BSA) in PBS-T for 1 h at room temperature. For antibody staining, blocking solution was aspirated and 40 μl/well (1:500 dilution in blocking solution) of the primary antibody anti-phospho histone H2A.X (S139) (EMD Milllipore, Burlington, MA, USA) was added. After 1 h, the antibody was removed and washed three times with PBS-T. The secondary antibody AlexaFluor 647 (Life Technologies, Carlsbad, CA, USA) was diluted 1:400 in blocking solution and 40 μl per well was incubated for 1 h. The cells were again washed thoroughly three times with PBS-T, then 50 μl DAPI (3 μM; TCI Deutschland, Eschborn, Germany) was added per well. After 30 min, the fluorescence intensity was measured with the Celldiscoverer 7 microscope; the software ZEN 3.1 was used for image acquisition and analysis. Statistical analysis was performed using one-way ANOVA and the Bonferroni post-hoc test: *p < 0.05, **p < 0.01, ***p < 0.001.
2.5. In vitro gene expression analysis
Quantitative real-time reverse transcriptase polymerase chain reaction (real-time RT-PCR) was used to investigate the accordance of gene expression by CTD treatment with a published gene signature for genotoxicity in HepaRG cells (Kreuzer et al., 2020). Cultivation and treatment were performed as described above. RNA isolation was performed with the RNeasy Mini Kit (Qiagen, Hilden, Germany) and transcription of RNA into cDNA was conducted with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), both according to the manufacturers’ protocol. Real-time RT-PCR was performed on ABI7900HT (Thermo Fisher, Waltham, MA, USA) with Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher). Thermal cycling started with an initial denaturation at 95 °C for 15 min, followed by 40 cycles of denaturation for 15 s at 95 °C and primer binding and elongation for 60 s at 60 °C. A final elongation step at 60 °C for 15 min and the addition of a dissociation curve step ended the procedure. ACTB, GAPDH and GUSB were used as housekeepers. For analysis of gene expression, their geometric mean was calculated. The 2−ΔΔCT method was used to calculate relative gene expression levels (Livak and Schmittgen, 2001). Primers were purchased from Eurofins Genomics (Ebersberg, Germany); corresponding sequences are shown in Supplemental Table 1.
2.6. In vivo gene expression data from rat liver
In vivo gene expression data from rat livers treated with different pesticidal active compounds were available from a previous study (Alarcan et al., 2021). The study had been designed as a repeated-dose toxicity study with different hepatotoxic pesticidal active compounds, reflecting different presumed modes of action. For detailed information about the choice of compounds, doses, and timing, please refer to the aforementioned paper. In brief, young adult female Wistar rats were gavaged with daily doses of 288 mg/kg body weight (bw) of CTD (dissolved in 0.5% aqueous carboxymethyl cellulose) for 28 days. Additional treatment groups received IMZ (93 mg/kg bw) or THI (108 mg/kg bw). After sacrifice, livers were removed and total RNA was isolated. Global gene expression was analyzed by RNA sequencing, using 100 ng of RNA on the Illumina NovaSeq6000 platform, and compared to vehicle controls. Transcriptome data have been published previously Alarcan et al. (2021), are available from GEO under accession number GSE153986 and can also be found in Supplemental Table 2. For details on statistical analysis and data processing, please refer to the aforementioned paper. An adjusted p-value < 0.05 was chosen as criterion to define statistically significant regulation of gene expression.
2.7. Gene ontology and gene network analysis
Gene ontology (GO) Biological Process term enrichment analysis was performed using a hypergeometric test by the R-package clusterProfiler version 3.14.3 (Yu et al., 2012) in R version 4.0.3 (R Core Team, 2020). False discovery rate (FDR) was applied to control for multiple testing (Benjamini and Hochberg, 1995) and an adjusted p-value < 0.05 was considered as significant (Supplementary Table 3). BH-corrected p-values served as input for REViGO (http://revigo.irb.hr/) to visualize GO terms according to semantic similarity and to reduce the redundancy of GO terms (Supek et al., 2011). The software Ingenuity Pathway Analysis (IPA) (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) was used to analyze data as described in Alarcan et al. (2021) and was used to generate a network between genes annotated with repair of DNA and related processes. Additional nodes were added to the network manually based on literature (Bonner et al., 2008; Hauer and Gasser, 2017; Hustedt and Durocher, 2016; Panier and Boulton, 2014; Polo and Jackson, 2011). Classification of genotoxicity was performed by Support Vector Machine ("svmRadial”) and Random Forest ("rf”) method from R-package caret version 6.0–88 (Kuhn, 2021). The training with 10-fold cross-validation was repeated three times and the final model was chosen according to accuracy.
2.8. In silico genotoxicity prediction
In silico tools are generally divided into expert systems and statistical systems. The combined use of both test systems is recommended in order to reduce the number of false-positive and false-negative predictions (Benigni et al., 2019; ICH Expert Working Group, 2014; OECD, 2014). To follow this, the expert system Derek Nexus for the endpoint genotoxicity and the statistical system Sarah Nexus for the endpoint bacterial mutagenicity were used in the present study. Both systems are distributed by the not-for-profit organization Lhasa Limited. For the prediction with Derek Nexus (version 6.1.0), the endpoint genotoxicity was selected for all species. Predictions with a likelihood level of at least equivocal were accepted. This means that there is a balanced number of arguments that support or do not support the hypothesis (Barber et al., 2017). All likelihood levels above equivocal (plausible, probable, certain) are generally accepted by regulatory authorities and included in a toxicological evaluation. Furthermore, tautomers were considered. For the prediction with Sarah Nexus (version 3.1.0), the reasoning type weighed was selected and the equivocal level and sensitivity level were each set to 8%. These settings are recommended by Lhasa Limited to correctly predict more mutagenic substances. In addition to the two commercially available software tools Derek and Sarah Nexus, the open source software Toxtree (version 3.1.0) was used as a further expert system. All in silico analyses were carried out in March 2021. The predictions of Toxtree are carried out using the rule base for mutagenicity and carcinogenicity (Benigni and Bossa, 2011; Benigni et al., 2013). Within the decision tree implemented in Toxtree, the following endpoints were selected: a) carcinogenicity (genotox and nongenotox) and mutagenicity rulebase by ISS, b), in vitro bacterial mutagenicity (Ames test) alerts by ISS, c) structure alerts for the in vivo micronucleus assay in rodents, and d) DNA binding alerts. The combined use of Derek Nexus, Sarah Nexus and Toxtree (being an alternative for the Derek Nexus expert system) has already proven successful for the endpoint bacterial mutagenicity (Herrmann et al., 2020). In addition to CTD, in silico analyses were also performed for the main described metabolites in the work from Yokota et al. (2003) and Khidkhan et al. (2021), i.e. N-methyl-N′-nitroguanidine (MNG), 2-(methylthio)-thiazole-5-carboxylic acid (MTCA) and N-(2-chlorothiazol-5-ylmethyl)-N′-nitroguanidine (TZNG).
3. Results
3.1. Transcriptome analysis indicates genotoxicity of CTD in rat liver
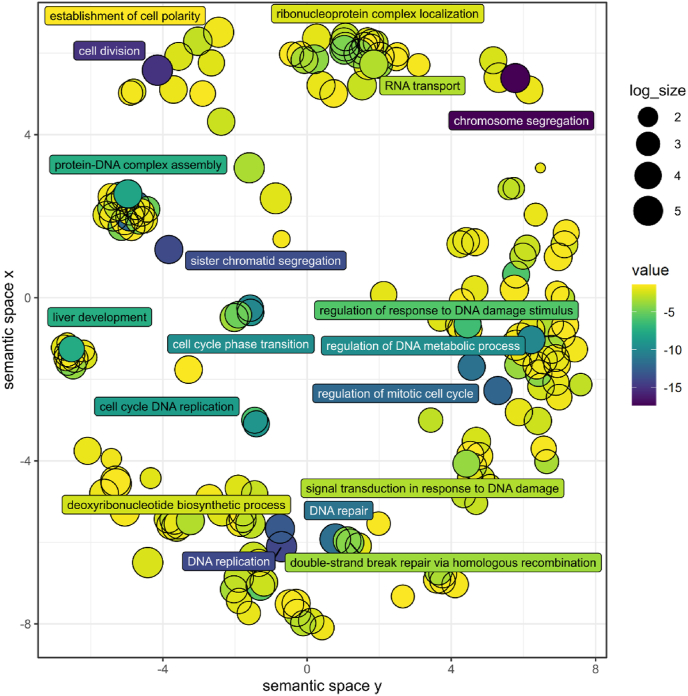
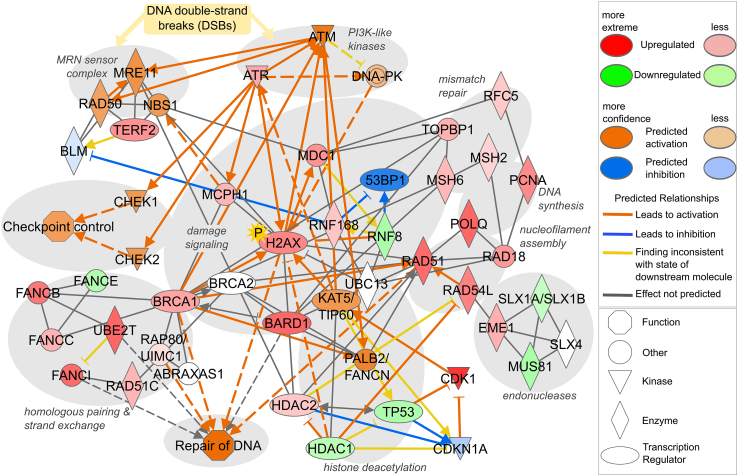
To investigate the possibilities for an implementation of transcriptomic analysis in risk assessment, global gene expression data sets of aberrant gene expression in rat livers from animals treated with the pesticidal active compounds CTD, IMZ or THI were subjected to bioinformatics analyses to predict potentially regulated biochemical functions and signaling pathways (data set contained in Supplemental Table 2; cp. Alarcan et al. (2021)). GO term enrichment analysis revealed that CTD, but not the other two compounds, affected various cellular functions related to DNA damage and DNA repair (Fig. 1, Supplemental Table 3). Similarly, IPA analysis predicted CTD-dependent regulation of, amongst others, H2ax, Atr, Mdc1, Mcph1, Brca1, Bard1 and Topbp1. Fig. 2 shows the network of CTD-regulated genes related to DNA damage, DNA repair and checkpoint control. In summary, these data indicate potential DNA damage and genotoxicity of CTD, but not of IMZ and THI in rat liver. In particular, the relation of altered transcripts with cellular processes indicative of DNA double-strand breaks, homologous recombination and sister chromatid exchange pointed towards a clastogenic potential of CTD.
Fig. 1.
GO term analysis of the specific gene expression effects of clothianidin (CTD) in rat liver following repeated-dose oral administration for 28 days. Transcriptomic data sets were taken from Alarcan et al., 2021. Enriched GO terms were visualized using REVIGO which groups GO terms according to semantic similarity after removing redundant terms. Bubble color indicates the BH-adjusted p-value on as -log10(p-value). Exposure to CTD, but not to the other two compounds, leads to changes in mRNA levels indicative of DNA damage and repair. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
Network of DNA damage-, DNA repair- and checkpoint control-related genes affected in rat liver by CTD, as obtained from previously published data (Alarcan et al., 2021) using IPA software. Continuous lines indicate direct functional relationships, dashed lines indicate indirect connections. Network nodes are labeled with the gene name and the functional classification of genes is denoted by their different shapes. Additional nodes were added to the network manually based on literature (Bonner et al., 2008; Hauer and Gasser, 2017; Hustedt and Durocher, 2016; Panier and Boulton, 2014; Polo and Jackson, 2011).
3.2. Outcome of regulatory genotoxicity studies
As active substances of pesticides, the three compounds CTD, IMZ and THI have been toxicologically characterized prior to their introduction to the European market, including genotoxicity studies. Available information from regulatory studies as presented in the respective reports by the European Food Safety Authority (EFSA) and the Joint FAO/WHO meeting on Pesticide Residues (JMPR) was compiled and is briefly reported in Table 1 (see also the respective links to the JMPR reports provided as footnotes to Table 1).
Table 1.
Outcome of regulatory studies of genotoxicity of CTD, IMZ and THI.
| Study type endpoint | Clothianidin | Imazalil | Thiacloprid |
|---|---|---|---|
| Ames test | Negative (5/6 studies) | Negative in all studies | Negative in all studies |
| Point mutation in mammalian cells | Negative in all studies | Negative in all studies | Negative in all studies |
| Clastogenicity in vitro | Positive (2/3 studies) | Negative in all studies | Negative in all studies |
| Genotoxicity in vivo | Negative 6/6 studies | Negative in all studies | Negative in all studies |
| Overall conclusion | WoE not genotoxic in vivo | Not genotoxic | Not genotoxic |
| Source | FAO/WHO JMPR 2011 a | FAO/WHO JMPR 2000 b | FAO/WHO JMPR 2006 c |
Footnotes:
, https://inchem.org/documents/jmpr/jmpmono/v2010pr01.pdf.
, https://inchem.org/documents/jmpr/jmpmono/v00pr08.htm#_00082240.
As summarized in the respective monographies by the JMPR and also in respective conclusions by EFSA, IMZ and THI were negative in all tests conducted according to OECD test protocols (see footnotes to Table 1). In contrast, CTD was positive in two out of three in vitro clastogenicity assays (EFSA, 2018; JMPR report as cited in Table 1). As it was, however, negative in a number of follow up in vivo tests, the overall conclusion “not genotoxic in vivo” was achieved.
Considering the fact that an obvious discrepancy was evident between the overall regulatory conclusion on CTD genotoxicity and the transcriptome-based prediction of genotoxicity, and based on the observation that some hints for clastogenicity were contained in regulatory in vitro studies, in line with the transcriptomic findings, we decided to follow up the genotoxicity by in silico and in vitro approaches.
3.3. In silico genotoxicity prediction of CTD and its metabolites
For CTD and its metabolites MNG and TZNG, prediction with the in silico tools Derek Nexus and Toxtree gave indications for a clastogenic potential in vitro and in vivo (Supplemental Tables 4–7). The functional group N-Nitro-/N-Nitroso or H-acceptor-path3-H-acceptor was indicated as alert by Derek Nexus and Toxtree, respectively. Regarding mutagenicity, the prediction with Derek Nexus, Sarah Nexus and Toxtree was different. While Derek Nexus predicted mutagenic potential in vitro and vivo, no alert for bacterial in vitro mutagenicity (Ames test) was identified with Toxtree. Since the output of the Sarah Nexus system was outside domain for CTD, MNG and TZNG, a prediction with this tool remained open. Overall, a clastogenic potential based on in silico analyses may be assumed for CTD and its metabolites MNG and TZNG, while only a weak suspicion exists for mutagenic activity. For the metabolite MTCA, a mutagenic potential was reliably excluded with the expert system Derek (Williams et al., 2016). The complementary statistical system Sarah Nexus also indicated that the compound is not mutagenic in vitro. This is consistent with the output by Toxtree. However, the prediction with Toxtree indicated also clastogenic activity in vivo which was not predicted by Derek Nexus. For this genotoxic endpoint, the H-acceptor-path3-H-acceptor was identified as alert. Taken together, the computational approaches supported the assumption of a clastogenic potential for CTD.
3.4. Transcript signature of CTD in human HepaRG liver cells
The human hepatocarcinoma cell line HepaRG was selected for in vitro studies due to its metabolic capacities closely resembling the features of primary hepatocytes. Cells were incubated for 24 h with different concentrations of CTD. For comparison, incubations were performed with a selection of food-relevant compounds primarily acting through genotoxic (acrylamide, aflatoxin B1, benzo[a]pyrene, lasiocarpine, methyleugenol, N-nitrosopyrrolidine) or non-genotoxic (ethanol, methapyrilene, 3-monochloropropanole, perfluorooctanesulfonic acid, perfluoroctanoic acid, Wy14,643) mechanisms. All chosen test compound concentrations did not induce overt cytotoxicity, as measured by the WST-1 assay (data not shown; see previously published data (Lichtenstein et al., 2020)). A high non-toxic concentration and two lower concentrations (50% and 10% of the high concentration) were chosen for incubation.
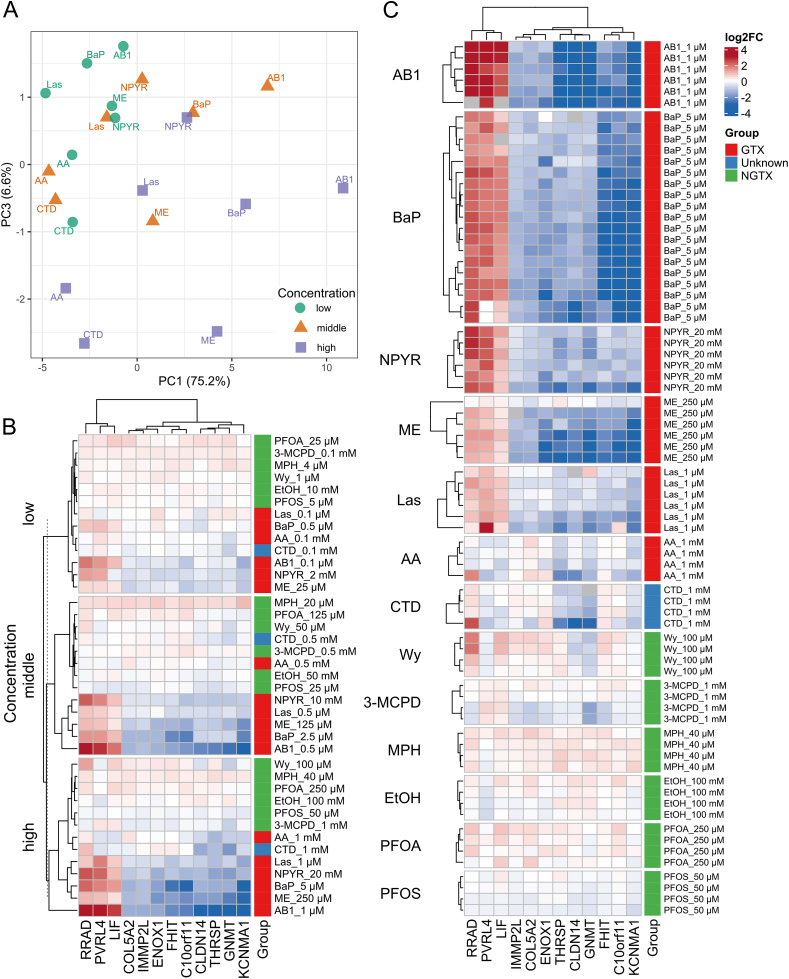
RNA was isolated and analyzed for the expression of a previously defined panel of marker genes aimed at the identification of genotoxic compounds in HepaRG cells and the discrimination between compounds primarily acting through genotoxic or non-genotoxic mechanisms (Kreuzer et al., 2020). Initially, an analysis of a panel of 33 marker genes for genotoxic carcinogens was performed. As evident from the principal component analysis scores plot depicted in Fig. 3A, the CTD samples showed similar behavior over all three concentrations, and the highest similarity was observed with data from acrylamide-treated cells. For detailed data on individual transcripts and replicates, please refer to Supplemental Table 8. CTD was not clearly separated from the compounds with known genotoxicity, but nonetheless appeared somewhat distinct from the genotoxic carcinogens. To further follow up on this observation, a set of 12 genes compiled to discriminate between genotoxic and non-genotoxic hepatocarcinogens was measured (Kreuzer et al., 2020). Data showed that, with the only exception of the middle concentration of acrylamide, all test compounds were correctly separated as genotoxic or non-genotoxic (Fig. 3B; see Supplemental Table 8 for detailed data). CTD clustered with the genotoxic compounds at the low and high concentration, but with the non-genotoxic compounds at the middle concentration. To elucidate the potential genotoxicity of CTD further, Random Forest (RF) and Support Vector Machine (SVM) models were applied for classification of CTD-dependent alterations in gene expression (Fig. 3C). For this purpose, data from all four individual replicates, obtained from independent experiments with the highest concentration of CTD, were analyzed separately using the two algorithms. Two replicates were classified as genotoxic by the SVM approach, whereas the other two replicates were classified as non-genotoxic (Fig. 4). A similarly inconclusive picture was obtained with the RF approach, yielding the genotoxic and one non-genotoxic prediction. In summary, no clear-cut classification of CTD as genotoxic or non-genotoxic was possible using the transcript-based approaches.
Fig. 3.
Gene expression signature for genotoxicity in human HepaRG liver cells. A PCA scores plot of genotoxic (GTX) compounds (benzo[a]pyrene (BaP), aflatoxin B1 (AB1), lasiocarpine (Las), N-nitrosopyrrolidine (NPYR), methyleugenol (ME), acrylamide (AA)) and CTD at three concentration levels (low, middle, high), indicated by color and symbols. Expression was measured for 33 genes and individual samples were averaged. B Heatmap showing log2 fold change (FC) of 12 selected genes for discriminating GTX and non-GTX substances, for GTX (red), non-GTX (green) and CTD (blue) averaged samples at three concentration levels. Non-GTX compounds: perfluorooctanoic acid (PFOA), 3-monochloropropane-diol (3-MCPD), ethanol (EtOH), perfluorooctanesulfonic acid (PFOS), methapyrilene hydrochloricde (MPH). C Heatmap showing log2 FC of 12 selected genes for individual samples of all compounds at high concentration. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
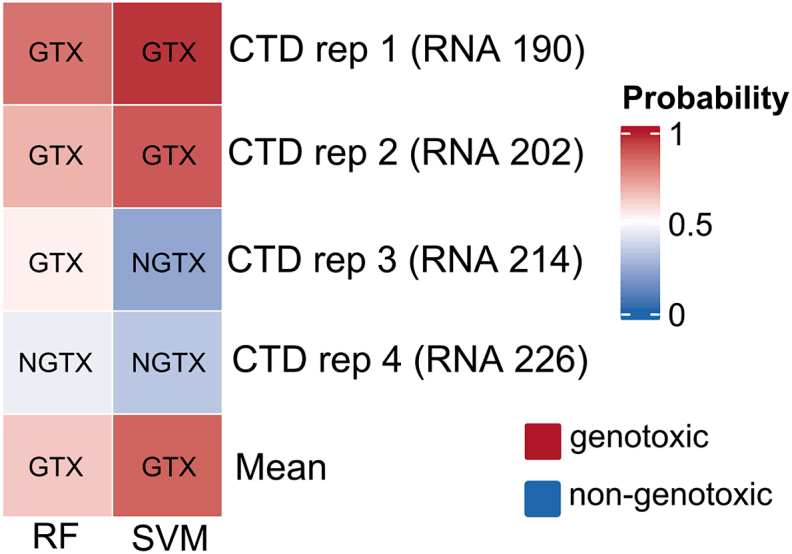
Results of classification by Random Forest (RF) and Support Vector Machine (SVM) for four individual CTD replicates, as well as on the mean of replicates, at high concentration levels.
3.5. Comet and γH2AX assays in human HepaRG liver cells
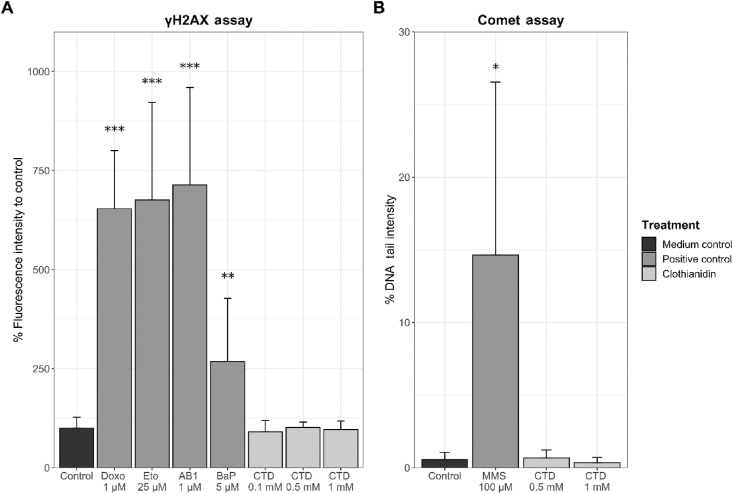
Additional assays were conducted aiming at the identification of potential genetic damage in HepaRG cells after incubation with CTD. Induction of DNA damage in HepaRG cells by 24 h of incubation with CTD was further checked by the γH2AX and Comet assays. In the γH2AX assay, the different genotoxic positive controls clearly led to statistically significant increases of the detected fluorescence signal, as expected (Fig. 5A; see Supplemental Table 9 for detailed data). CTD, however, did not provoke any changes (Fig. 5A). Similarly, a positive control for genotoxicity clearly induced DNA tail intensity in the Comet assays, whereas again no response was observed when incubating the cells with CTD (Fig. 5B; see Supplemental Table 10 for detailed data). Altogether, the two tests did not yield evidence for genotoxicity of CTD in HepaRG cells.
Fig. 5.
Analysis of DNA damage in HepaRG cells treated with different concentrations of CTD for 24 h. A Fluorescence intensity of phosphorylated histone H2AX (γH2AX) was captured with a Celldiscoverer 7 microscope after antibody staining with anti-phospho histone H2A.X (S139) and AlexaFluor647. Data were normalized to solvent control (1.7% DMSO). Mean + SD (n = 3 experiments) are provided. B Comet assay data were obtained by staining with SYBRGold, image recording with a Celldiscoverer 7 microscope and data acquisition with Comet Assay IV software. DNA damage was measured as DNA tail intensity (% of DNA in the tail). Mean + SD (n = 5 experiments, except for 0.5 mM CTD n = 2) are given. Statistical analysis was performed using one-way ANOVA and the Bonferroni post-hoc test: *p < 0.05, **p < 0.01, ***p < 0.001, treatment vs. control. Abbreviations: AB1, aflatoxin B1; BaP, benzo[a]pyrene; CTD, clothianidin; Doxo, doxorubicin; Eto, etoposide; MMS, methyl methanesulfonate.
4. Discussion
In this study, we investigated the genotoxic potential of CTD using a combination of different methodologies. Transcriptome analysis and subsequent GO term enrichment analysis identified pathways that were affected by CTD treatment in rats. Among them were DNA damage, DNA repair and checkpoint control. CTD treatment resulted specifically in up-regulation of genes that are playing an important role for these processes (Bonner et al., 2008; Hustedt and Durocher, 2016; Polo and Jackson, 2011). As these genes indicate potential genotoxicity/clastogenicity, we further applied a previously established gene expression pattern to compare CTD to known genotoxic compounds in human HepaRG liver cells. The classification model yielded ambiguous results for CTD, which was also the case for acrylamide, a known genotoxicant that had not been used to create the original transcriptomic signature. Possibly the selection of only five genotoxic compounds for establishing the marker gene signature (Kreuzer et al., 2020) might have been too small to unequivocally predict genotoxicity of CTD, which is chemically not related to the genotoxins used in the aforementioned study. Often, machine learning approaches for the identification of biomarkers and prediction of genotoxicity include more compounds, e.g. 13 genotoxic versus 15 non-genotoxic agents in TK6 cells (Li et al., 2015) followed up by validation of the classification with 45 test agents (Li et al., 2017). It is noteworthy that several studies have shown clear differences between the transcriptomic responses upon treatment comparing in vitro and in vivo systems, e.g. primary rat hepatocytes vs. rat liver (Grinberg et al., 2014; McMullen et al., 2019; Sutherland et al., 2018). For example, Luijten et al. (2021) revealed that the majority of matches were observed only in vitro or in vivo when comparing gene expression changes for 137 substances.
To complement the gene expression data, Comet assays as well as γH2AX stainings were performed in HepaRG cells. Both assays showed negative results, which would support the conclusion that CTD is not genotoxic. Overall, even though the predictions yielded from livers of rats repeatedly treated with high doses of CTD (Alarcan et al., 2021) can be considered clearly in favor of a genotoxic hazard posed by the insecticide, functional assays in human HepaRG liver cells (γH2AX, Comet) were negative, and also the transcriptional responses in HepaRG cells were rather equivocal. This illustrates possible challenges resulting from the use of batteries of new approach methods and omics techniques, in addition to classic toxicological assays: while model toxicants may yield clear answers in all or almost all test systems, real-life chemicals with less pronounced and specific toxicological properties might result in a diversity of test outcomes, including positive, negative and equivocal outcomes, making overall interpretation difficult. In the present case, as a weight-of-evidence decision, considering in addition to the regulatory studies especially the findings in human cells and the extraordinarily high doses of CTD administered in our animal study used for the transcriptomic approach, we conclude that CTD does most likely not pose a genotoxic risk to consumers exposed to residues of CTD via the diet.
Of note, several studies additional to the regulatory studies mentioned in the Results section of this paper have been published regarding the genotoxic potential of CTD. No significant effect towards the Comet assay was observed in HepG2 human hepatoma cells, while CTD induced increase in tail intensity in neuronal SHSY-5Y and bronchial epithelial BEAS-2B cells (Atli Sekeroglu et al., 2020; Senyildiz et al., 2018). Additionally, CTD induced the formation of DNA double strand breaks in BEAS-2B cells as evidenced by the increase in phosphorylated H2AX protein (Atli Sekeroglu et al., 2020). Another study conducted on human lymphocytes showed that CTD induced significant increases in chromosomal aberrations, aberrant cells, and micronuclei formation solely in the absence of an S9 external metabolic system (Atlı Şekeroğlu et al., 2018). It is noteworthy that cell systems featuring metabolic capacities (HepG2 cells, lymphocytes incubated with S9 mix, HepaRG cells in the present study) did not reveal genotoxicity of CTD, while other cell lines with lower metabolic capacities did. This supports the idea that detoxification processes may play an important role in the toxicity profile of CTD in rats and humans. Moreover, possible inter-species metabolic differences between rat and human could be relevant for the interpretation of our data. In that context, Khidkhan and colleagues showed, using liver microsomes, that both species produce the same metabolites although at different rates of formation (Khidkhan et al., 2021). Besides, a toxicokinetics study in rats showed that CTD was extensively distributed to all tissues and organs but was rapidly eliminated via urine, mainly in unchanged form (Yokota et al., 2003). Taken together, although the in silico analyses indicated clastogenic activity for both the parent compound CTD and some of the metabolites, this does not appear to manifest itself in vivo. One possible explanation could be the high efflux of CTD and/or detoxifying mechanisms of relevant metabolites that may counteract a possible expression of clastogenicity in vivo.
Transcriptomic studies have become more and more attractive for the toxicological characterization of chemical compounds, due to technological improvements, reduced costs and increasing efficiency (Harrill et al., 2019). However, despite their broad application in basic science, transcriptomics and more generally omics data are not yet routinely used in risk assessment for hazard identification and/or characterization. This is, at least in parts, based on difficulties with standardization and validation of such methods, as well as with the issue of how the classic toxicological concept of adversity can be adapted to complex omics endpoints such as transcriptome analyses (Marx-Stoelting et al., 2015). Nonetheless, omics data are frequently used for hypothesis generation regarding toxicological modes of action, based on predictions of affected pathways and functions using bioinformatics data mining tools. Thus omics data can be used to discover the most relevant pathways and identify potential molecular initiating events (MIE) and key events (KE) in the AOP framework (Brockmeier et al., 2017). Besides, there is more and more evidence accumulating that the use of transcriptomics data works well for regulatory tasks such as read across to help to fill data gaps (EU-ToxRisk) (Escher et al., 2019; Rovida et al., 2020). Furthermore, transcriptome data can serve as a tool for class comparison, prediction or discovery when testing chemical substances for hazardous effects (Sauer et al., 2017). Thus, our results provide further evidence that toxicogenomics may be used combination with other methods (cp. (Allemang et al., 2021)) to classify potentially genotoxic compounds.
CRediT authorship contribution statement
Heike Sprenger: bioinformatics, preparation of manuscript. Katrin Kreuzer: in vitro experimentation. Jimmy Alarcan: preparation of manuscript, literature search. Kristin Herrmann: in silico studies. Julia Buchmüller: in vitro experimentation. Philip Marx-Stoelting: preparation of manuscript. Albert Braeuning: study design, preparation of manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was partially funded by the European Union's Horizon 2020 research and innovation program under grant agreement No. 733032 HBM4EU (www.HBM4EU.eu). The work was also supported by the German Federal Institute for Risk Assessment (grant 1322-710).
Handling Editor: Dr. Jose Luis Domingo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fct.2022.113212.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alarcan J., Sprenger H., Waizenegger J., et al. Transcriptomics analysis of hepatotoxicity induced by the pesticides imazalil, thiacloprid and clothianidin alone or in binary mixtures in a 28-day study in female Wistar rats. Arch. Toxicol. 2021;95(3):1039–1053. doi: 10.1007/s00204-020-02969-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemang A., De Abrew K.N., Shan Y.K., Krailler J.M., Pfuhler S. A comparison of classical and 21st century genotoxicity tools: a proof of concept study of 18 chemicals comparing in vitro micronucleus, ToxTracker and genomics-based methods (TGx-DDI, whole genome clustering and connectivity mapping) Environ. Mol. Mutagen. 2021;62(2):92–107. doi: 10.1002/em.22418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlı Şekeroğlu Z., Şekeroğlu V., Uçgun E., Kontaş Yedier S., Aydın B. Cytotoxicity and genotoxicity of clothianidin in human lymphocytes with or without metabolic activation system. Drug Chem. Toxicol. 2018;42(4):364–370. doi: 10.1080/01480545.2018.1438458. [DOI] [PubMed] [Google Scholar]
- Atli Sekeroglu Z., Sekeroglu V., Aydin B., Kontas Yedier S., Ilkun E. Clothianidin induces DNA damage and oxidative stress in bronchial epithelial cells. Environ. Mol. Mutagen. 2020;61(6):647–655. doi: 10.1002/em.22376. [DOI] [PubMed] [Google Scholar]
- Barber C., Hanser T., Judson P., Williams R. Distinguishing between expert and statistical systems for application under ICH M7. Regul. Toxicol. Pharmacol. 2017;84:124–130. doi: 10.1016/j.yrtph.2016.12.012. [DOI] [PubMed] [Google Scholar]
- Benigni R., Bossa C. Mechanisms of chemical carcinogenicity and mutagenicity: a review with implications for predictive toxicology. Chem. Rev. 2011;111(4):2507–2536. doi: 10.1021/cr100222q. [DOI] [PubMed] [Google Scholar]
- Benigni R., Bossa C., Tcheremenskaia O. In vitro cell transformation assays for an integrated, alternative assessment of carcinogenicity: a data-based analysis. Mutagenesis. 2013;28(1):107–116. doi: 10.1093/mutage/ges059. [DOI] [PubMed] [Google Scholar]
- Benigni R., Laura Battistelli C., Bossa C., et al. Evaluation of the applicability of existing (Q)SAR models for predicting the genotoxicity of pesticides and similarity analysis related with genotoxicity of pesticides for facilitating of grouping and read across. EFSA Supporting Publications. 2019;16(3) doi: 10.2903/sp.efsa.2019.EN-1598. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Bonner W.M., Redon C.E., Dickey J.S., et al. GammaH2AX and cancer. Nat. Rev. Cancer. 2008;8(12):957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmeier E.K., Hodges G., Hutchinson T.H., et al. The role of omics in the application of adverse outcome pathways for chemical risk assessment. Toxicol. Sci. 2017;158(2):252–262. doi: 10.1093/toxsci/kfx097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team R. R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- David R. The promise of toxicogenomics for genetic toxicology: past, present and future. Mutagenesis. 2020;35(2):153–159. doi: 10.1093/mutage/geaa007. [DOI] [PubMed] [Google Scholar]
- EFSA Peer review of the pesticide risk assessment for bees for the active substance clothianidin considering the uses as seed treatments and granules. EFSA J. 2018;16(2) doi: 10.2903/j.efsa.2018.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher S.E., Kamp H., Bennekou S.H., et al. Towards grouping concepts based on new approach methodologies in chemical hazard assessment: the read-across approach of the EU-ToxRisk project. Arch. Toxicol. 2019;93(12):3643–3667. doi: 10.1007/s00204-019-02591-7. [DOI] [PubMed] [Google Scholar]
- Grinberg M., Stober R.M., Edlund K., et al. Toxicogenomics directory of chemically exposed human hepatocytes. Arch. Toxicol. 2014;88(12):2261–2287. doi: 10.1007/s00204-014-1400-x. [DOI] [PubMed] [Google Scholar]
- Gripon P., Rumin S., Urban S., et al. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. Unit. States Am. 2002;99(24):15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J., Shah I., Setzer R.W., et al. Considerations for strategic use of high-throughput transcriptomics chemical screening data in regulatory decisions. Curr Opin Toxicol. 2019;15:64–75. doi: 10.1016/j.cotox.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer M.H., Gasser S.M. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 2017;31(22):2204–2221. doi: 10.1101/gad.307702.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise T., Schmidt F., Knebel C., et al. Hepatotoxic combination effects of three azole fungicides in a broad dose range. Arch. Toxicol. 2018;92(2):859–872. doi: 10.1007/s00204-017-2087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann K., Holzwarth A., Rime S., Fischer B.C., Kneuer C. Q)SAR tools for the prediction of mutagenic properties: are they ready for application in pesticide regulation? Pest Manag. Sci. 2020;76(10):3316–3325. doi: 10.1002/ps.5828. [DOI] [PubMed] [Google Scholar]
- Hustedt N., Durocher D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2016;19(1):1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- ICH Expert Working Group . 2014. ICH Guideline M7 on Assessment and Control of DNA Reactive (Mutagenic) Impurities in Pharmaceuticals to Limit Potential Carcinogenic Risk EMA/CHMP/ ICH/83812/2013. [Google Scholar]
- Khidkhan K., Ikenaka Y., Ichise T., et al. Interspecies differences in cytochrome P450-mediated metabolism of neonicotinoids among cats, dogs, rats, and humans. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2021;239 doi: 10.1016/j.cbpc.2020.108898. [DOI] [PubMed] [Google Scholar]
- Knebel C., Buhrke T., Sussmuth R., Lampen A., Marx-Stoelting P., Braeuning A. Pregnane X receptor mediates steatotic effects of propiconazole and tebuconazole in human liver cell lines. Arch. Toxicol. 2019;93(5):1311–1322. doi: 10.1007/s00204-019-02445-2. [DOI] [PubMed] [Google Scholar]
- Kohonen P., Parkkinen J.A., Willighagen E.L., et al. A transcriptomics data-driven gene space accurately predicts liver cytopathology and drug-induced liver injury. Nat. Commun. 2017;8 doi: 10.1038/ncomms15932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer K., Bohmert L., Alhalabi D., Buhrke T., Lampen A., Braeuning A. Identification of a transcriptomic signature of food-relevant genotoxins in human HepaRG hepatocarcinoma cells. Food Chem. Toxicol. 2020;140 doi: 10.1016/j.fct.2020.111297. [DOI] [PubMed] [Google Scholar]
- Krewski D., Andersen M.E., Tyshenko M.G., et al. Toxicity testing in the 21st century: progress in the past decade and future perspectives. Arch. Toxicol. 2020;94(1):1–58. doi: 10.1007/s00204-019-02613-4. [DOI] [PubMed] [Google Scholar]
- Kuhn M. 2021. Caret: Classification and Regression Training.https://CRAN.R-project.org/package=caret [Google Scholar]
- Li H.H., Hyduke D.R., Chen R., et al. Development of a toxicogenomics signature for genotoxicity using a dose-optimization and informatics strategy in human cells. Environ. Mol. Mutagen. 2015;56(6):505–519. doi: 10.1002/em.21941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.H., Chen R., Hyduke D.R., et al. Development and validation of a high-throughput transcriptomic biomarker to address 21st century genetic toxicology needs. Proc. Natl. Acad. Sci. U. S. A. 2017;114(51):E10881–E10889. doi: 10.1073/pnas.1714109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein D., Luckert C., Alarcan J., et al. An adverse outcome pathway-based approach to assess steatotic mixture effects of hepatotoxic pesticides in vitro. Food Chem. Toxicol. Int. J. Publ. Br. Indus. Biol. Res. Assoc. 2020;139 doi: 10.1016/j.fct.2020.111283. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luckert C., Braeuning A., Lampen A., Hessel-Pras S. PXR: structure-specific activation by hepatotoxic pyrrolizidine alkaloids. Chem. Biol. Interact. 2018;288:38–48. doi: 10.1016/j.cbi.2018.04.017. [DOI] [PubMed] [Google Scholar]
- Luijten M., Wackers P.F.K., Rorije E., Pennings J.L.A., Heusinkveld H.J. Relevance of in vitro transcriptomics for in vivo mode of action assessment. Chem. Res. Toxicol. 2021;34(2):452–459. doi: 10.1021/acs.chemrestox.0c00313. [DOI] [PubMed] [Google Scholar]
- Marx-Stoelting P., Braeuning A., Buhrke T., et al. Application of omics data in regulatory toxicology: report of an international BfR expert workshop. Arch. Toxicol. 2015;89(11):2177–2184. doi: 10.1007/s00204-015-1602-x. [DOI] [PubMed] [Google Scholar]
- McMullen P.D., Pendse S.N., Black M.B., et al. Addressing systematic inconsistencies between in vitro and in vivo transcriptomic mode of action signatures. Toxicol. Vitro. 2019;58:1–12. doi: 10.1016/j.tiv.2019.02.014. [DOI] [PubMed] [Google Scholar]
- OECD . second ed. 2014. Guidance on Grouping of Chemicals. Series on Testing & Assessment No 194. Paris, France. [Google Scholar]
- OECD . Vivo Mammalian Alkaline Comet Assay. 2016. Test No. 489. [DOI] [Google Scholar]
- Panier S., Boulton S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014;15(1):7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Polo S.E., Jackson S.P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25(5):409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovida C., Barton-Maclaren T., Benfenati E., et al. Internationalization of read-across as a validated new approach method (NAM) for regulatory toxicology. ALTEX. 2020;37(4):579–606. doi: 10.14573/altex.1912181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U.G., Deferme L., Gribaldo L., et al. The challenge of the application of 'omics technologies in chemicals risk assessment: background and outlook. Regul. Toxicol. Pharmacol. 2017;91(Suppl. 1):S14–S26. doi: 10.1016/j.yrtph.2017.09.020. [DOI] [PubMed] [Google Scholar]
- Seeger B., Mentz A., Knebel C., et al. Assessment of mixture toxicity of (tri)azoles and their hepatotoxic effects in vitro by means of omics technologies. Arch. Toxicol. 2019;93(8):2321–2333. doi: 10.1007/s00204-019-02502-w. [DOI] [PubMed] [Google Scholar]
- Senyildiz M., Kilinc A., Ozden S. Investigation of the genotoxic and cytotoxic effects of widely used neonicotinoid insecticides in HepG2 and SH-SY5Y cells. Toxicol. Ind. Health. 2018;34(6):375–383. doi: 10.1177/0748233718762609. [DOI] [PubMed] [Google Scholar]
- Sewell F., Lewis D., Mehta J., Terry C., Kimber I. Rethinking agrochemical safety assessment: a perspective. Regul. Toxicol. Pharmacol. 2021;127 doi: 10.1016/j.yrtph.2021.105068. [DOI] [PubMed] [Google Scholar]
- Singh N.P., McCoy M.T., Tice R.R., Schneider E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175(1):184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland J.J., Webster Y.W., Willy J.A., et al. Toxicogenomic module associations with pathogenesis: a network-based approach to understanding drug toxicity. Pharmacogenomics J. 2018;18(3):377–390. doi: 10.1038/tpj.2017.17. [DOI] [PubMed] [Google Scholar]
- Williams R.V., Amberg A., Brigo A., et al. It's difficult, but important, to make negative predictions. Regul. Toxicol. Pharmacol. 2016;76:79–86. doi: 10.1016/j.yrtph.2016.01.008. [DOI] [PubMed] [Google Scholar]
- Yokota T., Mikata K., Nagasaki H., Ohta K. Absorption, tissue distribution, excretion, and metabolism of clothianidin in rats. J. Agric. Food Chem. 2003;51(24):7066–7072. doi: 10.1021/jf034760f. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.