Abstract
STUDY QUESTION
Can an artificial intelligence (AI) model predict human embryo ploidy status using static images captured by optical light microscopy?
SUMMARY ANSWER
Results demonstrated predictive accuracy for embryo euploidy and showed a significant correlation between AI score and euploidy rate, based on assessment of images of blastocysts at Day 5 after IVF.
WHAT IS KNOWN ALREADY
Euploid embryos displaying the normal human chromosomal complement of 46 chromosomes are preferentially selected for transfer over aneuploid embryos (abnormal complement), as they are associated with improved clinical outcomes. Currently, evaluation of embryo genetic status is most commonly performed by preimplantation genetic testing for aneuploidy (PGT-A), which involves embryo biopsy and genetic testing. The potential for embryo damage during biopsy, and the non-uniform nature of aneuploid cells in mosaic embryos, has prompted investigation of additional, non-invasive, whole embryo methods for evaluation of embryo genetic status.
STUDY DESIGN, SIZE, DURATION
A total of 15 192 blastocyst-stage embryo images with associated clinical outcomes were provided by 10 different IVF clinics in the USA, India, Spain and Malaysia. The majority of data were retrospective, with two additional prospectively collected blind datasets provided by IVF clinics using the genetics AI model in clinical practice. Of these images, a total of 5050 images of embryos on Day 5 of in vitro culture were used for the development of the AI model. These Day 5 images were provided for 2438 consecutively treated women who had undergone IVF procedures in the USA between 2011 and 2020. The remaining images were used for evaluation of performance in different settings, or otherwise excluded for not matching the inclusion criteria.
PARTICIPANTS/MATERIALS, SETTING, METHODS
The genetics AI model was trained using static 2-dimensional optical light microscope images of Day 5 blastocysts with linked genetic metadata obtained from PGT-A. The endpoint was ploidy status (euploid or aneuploid) based on PGT-A results. Predictive accuracy was determined by evaluating sensitivity (correct prediction of euploid), specificity (correct prediction of aneuploid) and overall accuracy. The Matthew correlation coefficient and receiver-operating characteristic curves and precision-recall curves (including AUC values), were also determined. Performance was also evaluated using correlation analyses and simulated cohort studies to evaluate ranking ability for euploid enrichment.
MAIN RESULTS AND THE ROLE OF CHANCE
Overall accuracy for the prediction of euploidy on a blind test dataset was 65.3%, with a sensitivity of 74.6%. When the blind test dataset was cleansed of poor quality and mislabeled images, overall accuracy increased to 77.4%. This performance may be relevant to clinical situations where confounding factors, such as variability in PGT-A testing, have been accounted for. There was a significant positive correlation between AI score and the proportion of euploid embryos, with very high scoring embryos (9.0–10.0) twice as likely to be euploid than the lowest-scoring embryos (0.0–2.4). When using the genetics AI model to rank embryos in a cohort, the probability of the top-ranked embryo being euploid was 82.4%, which was 26.4% more effective than using random ranking, and ∼13–19% more effective than using the Gardner score. The probability increased to 97.0% when considering the likelihood of one of the top two ranked embryos being euploid, and the probability of both top two ranked embryos being euploid was 66.4%. Additional analyses showed that the AI model generalized well to different patient demographics and could also be used for the evaluation of Day 6 embryos and for images taken using multiple time-lapse systems. Results suggested that the AI model could potentially be used to differentiate mosaic embryos based on the level of mosaicism.
LIMITATIONS, REASONS FOR CAUTION
While the current investigation was performed using both retrospectively and prospectively collected data, it will be important to continue to evaluate real-world use of the genetics AI model. The endpoint described was euploidy based on the clinical outcome of PGT-A results only, so predictive accuracy for genetic status in utero or at birth was not evaluated. Rebiopsy studies of embryos using a range of PGT-A methods indicated a degree of variability in PGT-A results, which must be considered when interpreting the performance of the AI model.
WIDER IMPLICATIONS OF THE FINDINGS
These findings collectively support the use of this genetics AI model for the evaluation of embryo ploidy status in a clinical setting. Results can be used to aid in prioritizing and enriching for embryos that are likely to be euploid for multiple clinical purposes, including selection for transfer in the absence of alternative genetic testing methods, selection for cryopreservation for future use or selection for further confirmatory PGT-A testing, as required.
STUDY FUNDING/COMPETING INTEREST(S)
Life Whisperer Diagnostics is a wholly owned subsidiary of the parent company, Presagen Holdings Pty Ltd. Funding for the study was provided by Presagen with grant funding received from the South Australian Government: Research, Commercialisation, and Startup Fund (RCSF). ‘In kind’ support and embryology expertise to guide algorithm development were provided by Ovation Fertility. ‘In kind’ support in terms of computational resources provided through the Amazon Web Services (AWS) Activate Program. J.M.M.H., D.P. and M.P. are co-owners of Life Whisperer and Presagen. S.M.D., M.A.D. and T.V.N. are employees or former employees of Life Whisperer. S.M.D, J.M.M.H, M.A.D, T.V.N., D.P. and M.P. are listed as inventors of patents relating to this work, and also have stock options in the parent company Presagen. M.V. sits on the advisory board for the global distributor of the technology described in this study and also received support for attending meetings.
TRIAL REGISTRATION NUMBER
N/A.
Keywords: assisted reproduction, embryo quality, IVF, ICSI outcome, artificial intelligence, machine learning, genetics, PGT-A, preimplantation genetic testing for aneuploidy
Introduction
Embryo aneuploidy during conception is a leading cause of implantation failure, pregnancy loss and congenital defects in newborns (Scott et al., 2012; Schaeffer et al., 2018). During IVF procedures, preimplantation genetic testing for aneuploidy (PGT-A) is used to evaluate the genetic integrity of embryos and assist in identifying euploid embryos for transfer (Greco et al., 2020). Owing to the invasive nature of the biopsy procedure required for PGT-A assessment, only a small sample of cells, usually from the trophectoderm, can be taken for sequencing. Aside from the potential for damage during the embryo biopsy procedure (Rubino et al., 2020; Makhijani et al., 2021), one of the recent concerns raised with PGT-A testing is the reporting of chromosomal mosaicism in up to 40% of cases, which often results in embryos being deprioritized for transfer (Cram et al., 2019). Although meiotic errors originate in the oocyte and uniformly affect the embryo, recent evidence suggests that aneuploidies can also occur during the mitotic division process resulting in mosaic embryos that have a mixture of genetically normal and abnormal cells (Capalbo and Rienzi, 2017). Importantly, mosaic embryos can still result in normal live births, likely due to self-correction pathways that limit proliferation of abnormal cells throughout the embryo (Victor et al., 2019b). Embryos with low levels of mosaicism in particular have relatively good outcomes compared to those with high levels, including ongoing pregnancy rates, miscarriage rates and live birth outcomes, and in some cases result in similar clinical outcomes to that of euploid embryos (Abhari and Kawwass, 2021). The accuracy of PGT-A sequencing is high; however, the results are dependent on how representative the biopsy sample is of the whole embryo.
There is a strong incentive for the development of alternative, less invasive methods for evaluation of embryo genetic status, to complement current PGT-A assessment. The main methods currently under investigation include isolation of DNA from embryonic blastocoel fluid, or from spent culture medium (Kuznyetsov et al., 2020; Orvieto et al., 2021). The clinical benefit of these approaches remains largely unknown, but they are unlikely to be able to account for embryo DNA damage and self-correction mechanisms that occur with mitotic aneuploidy.
The aim of the current study was to develop and evaluate a robust artificial intelligence (AI)-based method for non-invasive analysis of embryo genetic (ploidy) status using static 2-dimensional images. This approach is designed to assess the embryo as a whole and use the phenotype or morphology of the embryo as a developmental readout of severe genetic damage. While one AI-based algorithm for analysis of embryo images to predict euploidy reports predictive ability on a test dataset of 84 embryo images from 19 patients (Chavez-Badiola et al., 2020), the study presented herein describes the development and testing of an AI for predicting embryo ploidy using datasets comprising thousands of images from multiple IVF clinics representing a range of demographics, including double-blind evaluation of prospectively collected data. The present work represents the first report of such an AI model that can be used with images taken on a range of imaging and microscope systems including both standard microscope-mounted camera types and time-lapse imaging systems.
Materials and methods
Experimental design
This study was designed to collect and analyze data for the development of an AI model for the evaluation of embryo genetic (ploidy) status during IVF. Subjects included female patients aged at least 18 years who underwent IVF procedures between 2011 and 2021. Most data were collected retrospectively, with additional data collected prospectively for double-blind evaluation of the final genetics AI model. Primary data for analysis included images of embryos taken using optical light microscopy systems, with matched PGT-A results as the ground truth outcome (Supplementary Table SI shows PGT-A testing details). Images were collected of embryos on Day 5, Day 6 and Day 7 of culture, with only Day 5 embryo images used for training and development of the AI model. The study was non-interventional and results were not used to influence treatment decisions in any way.
For inclusion in the study, images were required to be of in vitro cultured embryos taken using a standard optical light microscopy imaging system prior to biopsy or freezing. All images were required to have a minimum resolution of 480 × 480 pixels with the complete embryo in the field of view and the focus on the inner cell mass (ICM). This minimum resolution was chosen so that each image contained sufficient details of embryo morphology for machine learning applications, noting that imaging systems used in this study typically output images of at least this resolution. Details of imaging systems used in training and testing the AI model are provided in Supplementary Table SII.
Collection of retrospective data for this study was exempted from ethical review and approval, and from the requirement for patient informed consent, as confirmed by Sterling IRB #7751 for protocol ID LW-C-004A. Collection of prospective data for this study was performed in accordance with the Life Whisperer patient privacy policy, constituting informed consent for research purposes. This study was conducted according to the guidelines of the Declaration of Helsinki of 1975, as amended. This study was not registered as a clinical trial as it did not meet the definition of an applicable clinical trial as defined by the ICMJE, that is: ‘a clinical trial is any research project that prospectively assigns people or a group of people to an intervention, with or without concurrent comparison or control groups, to study the relationship between a health-related intervention and a health outcome’.
Statistical analyses
This study involved standard statistical methods used in performance evaluation of machine learning classifiers, including accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), Matthew correlation coefficient (MCC) and AUC value for both receiver-operating characteristic (ROC) curves and precision-recall curves (PRC; Florkowski, 2008). For binary classification of embryos, an AI score of at least 5.0/10.0 was considered a normal/positive prediction, and below 5.0/10.0 an abnormal/negative prediction as follows: normal/positive predictions included euploid embryos, mosaic-low embryos, embryos without the specific abnormality being evaluated and embryos predicted to lead to successful clinical pregnancy (determined by the presence of a fetal heartbeat at first ultrasound scan). Abnormal/negative predictions included aneuploid embryos, mosaic-high embryos, embryos with the specific abnormality being evaluated and embryos predicted not to lead to clinical pregnancy.
Statistical analyses comparing groups were performed using GraphPad Prism version 9.0.0 (GraphPad Software, Inc., San Diego, CA, USA). Average AI scores for multiple groups were compared using ordinary one-way ANOVA with Tukey’s multiple comparisons post-test, or Students t-test between pairs of groups, where indicated. Trends in the proportion of euploid embryos were evaluated using Chi-square test for trend. Error bars indicate SEM where presented, and P-values of <0.05 were considered significant.
To investigate the correlation of the genetics AI score with euploidy rate, images were assigned to one of four score brackets (euploid likelihood categories) as follows: low likelihood: scores of 0.0–2.4/10.0, medium likelihood: scores of 2.5–7.4/10.0, high likelihood: scores of 7.5–8.9/10.0 and very high likelihood: scores of 9.0–10.0/10.0. The percentage of euploid embryos was calculated in each category, and the correlation evaluated using Chi-square test for trend.
To investigate the ability of the genetics AI model to rank euploid embryos, a ranking analysis was performed using simulated embryo cohorts as follows: embryo images drawn from the 1001 blind test images with known PGT-A outcomes were randomized into ∼100 000 unique simulated cohorts for analysis, with an average of 10 embryos per cohort. Cohort sizes were drawn from a distribution of actual embryo cohort sizes obtained from a prior clinical dataset. Cohorts consisting of a single embryo, or cohorts with no euploid embryos, were excluded.
Embryos in the simulated cohorts were then ranked according to likelihood of euploidy separately by the genetics AI score, random ranking and Gardner score ranking. Two different methods were used for Gardner score ranking. Firstly, the commonly used 3BB threshold was applied to define good- versus poor-quality embryos (Kemper et al., 2021). The second method involved a four-group ranking system (Irani et al., 2017; Zhao et al., 2018), with rank groups defined as follows: excellent quality (3-6AA), good quality (3-6AB/BA, 1-2AA), average quality (3-6BB/AC/CA, 1-2AB/BA) and poor quality (1-6BC/CB/CC, 1-2BB). For cohorts where multiple embryos were of the same rank group, these embryos were ordered randomly within their shared position of the cohort. Note that Gardner scores were only available for a subset of 918 of the 1001 embryos in the Day 5 blind test dataset.
Ranking was evaluated using three methods: (i) whether the top-ranked embryo was euploid, (ii) whether at least one of the top two ranked embryos was euploid or (iii) whether the top two ranked embryos were both euploid. The result was expressed as a percentage of the total number of cohorts. Note that in cases (ii) and (iii), cohorts with only one euploid embryo were also excluded.
Computer vision image processing methods
Prior to analysis, the images underwent multiple preprocessing, as in VerMilyea et al. (2020), but were updated to include more advanced computer vision techniques as follows.
Step 1: Images were stripped of the alpha channel and encoded in a three-channel format.
Step 2: Each image was transformed to a tensor as the appropriate input for deep learning AI models.
Step 3: Images were padded to square dimensions to ensure consistent input for the models. The previous method used for padding in VerMilyea et al. (2020) was replaced with a Region Based Convolutional Neural Network (R-CNN) deep learning model called Faster R-CNN, which is known to be robust to background artifacts in the image (Ren et al., 2015). The training parameters used for this model included a ResNet50 Feature Pyramid Network backbone, prediction size = 800 pixels, minimum box size = 30 pixels, non-maximum suppression threshold = 0.25 and score threshold = 0.4. Small detected boxes (with at least one box edge smaller than 30 pixels) that lie completely within another box were removed to prevent the model from under-fitting the boundary. In cases where two over-lapping boxes were output, the larger box encompassing the two boxes was selected.
Step 4: Each image was cropped to the final detected bounding box to remove excess background and center the embryo. The previous elliptical Hough transformation method was replaced by the Faster R-CNN method described in Step 3.
Step 5: Segmentation was optionally applied to each image for the appropriate constituent models. Segmentation is used to divide each image into respective embryo sections to show either the zona pellucida region only, or the intra-zonal cavity (IZC) region only (i.e. the interior of the blastocyst including both the ICM and trophectoderm). The previous method of snake segmentation (active contour models) described in VerMilyea et al. (2020) was replaced with a faster and more robust semantic segmentation method called U-Net (Ronneberger et al., 2015).
Note that color normalization and resolution scaling were removed from the preprocessing procedure described in VerMilyea et al. (2020) to allow the AI model to become more robust to color offsets and resolution variation. However, during the training process, additional image augmentations were applied to anticipate changes to lighting conditions, rotation of the embryo, focal length, color bias and different resolutions, so that the final AI is robust to these conditions in new unseen datasets (Supplementary Materials and Methods).
Model training and selection process
The final genetics AI model is an ensemble model, consisting of several constituent CNN models combined according to method described in VerMilyea et al. (2020). Methods for selecting CNN architectures, AI model training, model validation and selection of the final ensemble model were generally performed as described in VerMilyea et al. (2020). In brief, the training process employed a wide range of model architectures, learning rates, momentum values and regularization methods. All models were evaluated on a holdback validation dataset primarily using confidence metrics designed to measure translatability. A curated list of models was considered candidates for inclusion in a distillation training process (Hinton et al., 2014). After distillation training, the resulting models were considered as candidates for ensembling, based on their performance on the validation dataset including prediction accuracy, model stability and confidence metrics. The final ensemble model was evaluated on a blind test dataset, independent of the validation dataset used for model selection, as well as additional double-blind test datasets. Throughout the training and selection process, the most effective CNN architectures for classifying embryo ploidy status were found to include ResNet (He et al., 2016) and DenseNet (Huang et al., 2017) architectures. Refer to Supplementary Materials and Methods for more information.
There were two key methods applied in training the current genetics AI model that extend beyond the methods employed in VerMilyea et al. (2020), namely untrainable data cleansing (UDC; Dakka et al., 2021) and distillation (Hinton et al., 2014). UDC is an AI method for identifying mislabeled data, removing (cleansing) images that multiple AI models are unable to correctly label (classify) during the AI training process, because the images are either: of such poor quality that there are no distinguishing features that correlate with any label (noisy data); or highly correlated to the opposite label of what would reasonably be expected, based on the classification of the majority of images in the dataset (incorrect or mislabeled data). Noisy or incorrect (mislabeled) images reduce the overall quality and usefulness of datasets when used in AI training and can present a misleading estimation of AI performance when included in testing datasets. Methods for handling such data have been shown to improve generalizability of AI models (Dakka et al., 2021). In this study, UDC was applied to the initial Day 5 dataset to produce cleansed datasets for AI training (n = 3174) and AI validation (n = 300). AI model performance was evaluated on both uncleansed (n = 1001) and cleansed (n = 786) versions of the blind test dataset (Table I). Note that the purpose of UDC is to arrive at a training and validation dataset with cleaner labels, allowing production of more stable constituent models, and direct comparisons of their performance when selecting those to incorporate into the final model. Therefore, all double-blind datasets obtained prospectively for this study did not have UDC applied and were representative of real-world clinical data. Additional details are provided in Supplementary Materials and Methods.
Table I.
Composition of datasets used for development of the Day 5 genetics artificial intelligence model.
| Datasets | Total Day 5 dataset (uncleansed) | Day 5 blind test dataset (uncleansed) | Day 5 blind test dataset (cleansed)a |
|---|---|---|---|
| Number of embryo images | 5050 | 1001 | 786 |
| Number of patients | 2438 | 788 | 658 |
| Dates treated | 2011–2020 | 2011–2020 | 2011–2020 |
| Number of cycles | 2485 | 798 | 664 |
| Average cycles per patient (range) | 1.0 (1–3) | 1.0 (1–2) | 1.0 (1–2) |
| Average embryo cohort size (range)b | 2.0 (1–17) | 1.3 (1–5) | 1.2 (1–4) |
| Average patient age in years (range) | 36.2 (19–53) | 35.6 (19–53) | 35.2 (19–51) |
| Number of donor gamete(s) used (%) | 1106 (22.0%) | 211 (21.1%) | 170 (21.6%) |
| Number of euploid embryos (%) | 3251 (64.4%) | 645 (64.4%) | 613 (78.0%) |
| Number of aneuploid embryos (%) | 1799 (35.6%) | 356 (35.6%) | 173 (22.0%) |
| Number of transferred embryos (%) | ND | 156 (15.6%) | 148 (18.8%) |
|
| |||
| Clinical pregnancy outcomes | |||
|
| |||
| Number of successful pregnancies (%) | ND | 92 (59.0%) | 87 (58.8%) |
| Number of unsuccessful pregnancies (%) | ND | 64 (41.0%) | 61 (41.2%) |
| Origin of images | |||
| Ovation—Austin (TX, USA) | 3328 (65.9%) | 671 (67.0%) | 522 (66.4%) |
| San Antonio IVF (TX, USA) | 538 (10.7%) | 103 (10.3%) | 78 (9.9%) |
| Midwest Fertility Specialists (IN, USA) | 236 (4.7%) | 45 (4.5%) | 37 (4.7%) |
| California Fertility Partners (CA, USA) | 943 (18.7%) | 182 (18.2%) | 149 (19.0%) |
| Ovation—Baton Rouge (LA, USA) | 5 (0.1%) | 0 (0.0%) | 0 (0.0%) |
|
| |||
| Chromosomal abnormalities involved c | |||
|
| |||
| Monosomy—n (%)d | 483 (26.8%) | 80 (22.5%) | 39 (22.5%) |
| Trisomy—n (%)d | 466 (25.9%) | 88 (24.7%) | 35 (20.2%) |
| Full gains or losses—n (%)e | 1217 (67.6%) | 237 (66.6%) | 119 (68.8%) |
| Segmental duplications or deletions—n (%)e | 382 (21.2%) | 86 (24.2%) | 33 (19.1%) |
| Single chromosomal abnormalities—n (%) | 1093 (60.8%) | 219 (60.6%) | 101 (58.4%) |
| Multiple abnormalities (complex)—n (%) | 657 (36.5%) | 137 (39.4%) | 72 (41.6%) |
| Chromosome 1—n (%) | 83 (4.6%) | 23 (6.6%) | 9 (5.2%) |
| Chromosome 2—n (%) | 120 (6.7%) | 19 (5.5%) | 12 (6.9%) |
| Chromosome 3—n (%) | 77 (4.3%) | 20 (5.8%) | 10 (5.8%) |
| Chromosome 4—n (%) | 111 (6.2%) | 27 (7.8%) | 8 (4.6%) |
| Chromosome 5—n (%) | 102 (5.7%) | 27 (7.8%) | 11 (6.4%) |
| Chromosome 6—n (%) | 86 (4.8%) | 17 (4.9%) | 10 (5.8%) |
| Chromosome 7—n (%) | 99 (5.5%) | 21 (6.1%) | 6 (3.5%) |
| Chromosome 8—n (%) | 110 (6.1%) | 24 (6.9%) | 11 (6.4%) |
| Chromosome 9—n (%) | 104 (5.8%) | 27 (7.8%) | 10 (5.8%) |
| Chromosome 10—n (%) | 86 (4.8%) | 11 (3.2%) | 5 (2.9%) |
| Chromosome 11—n (%) | 98 (5.4%) | 14 (4.0%) | 7 (4.0%) |
| Chromosome 12—n (%) | 71 (3.9%) | 16 (4.6%) | 7 (4.0%) |
| Chromosome 13—n (%) | 140 (7.8%) | 36 (10.4%) | 20 (11.6%) |
| Chromosome 14—n (%) | 121 (6.7%) | 25 (7.2%) | 12 (6.9%) |
| Chromosome 15—n (%) | 196 (10.9%) | 33 (9.5%) | 20 (11.6%) |
| Chromosome 16—n (%) | 255 (14.2%) | 49 (14.1%) | 26 (15%) |
| Chromosome 17—n (%) | 75 (4.2%) | 17 (4.9%) | 10 (5.8%) |
| Chromosome 18—n (%) | 127 (7.1%) | 31 (8.9%) | 17 (9.8%) |
| Chromosome 19—n (%) | 113 (6.3%) | 25 (7.2%) | 15 (8.7%) |
| Chromosome 20—n (%) | 78 (4.3%) | 21 (6.1%) | 12 (6.9%) |
| Chromosome 21—n (%) | 221 (12.3%) | 46 (13.3%) | 19 (11.0%) |
| Chromosome 22—n (%) | 323 (18.0%) | 52 (15.0%) | 28 (16.2%) |
| Sex chromosomes—n (%) | 161 (8.9%) | 34 (9.8%) | 15 (8.7%) |
The Day 5 blind test dataset of 1001 images was cleansed by the UDC method to remove poor quality and mislabeled images (remaining n = 786).
Some cohorts consisted of a combination of Day 5 and Day 6 embryos—these were separated according to dataset (see Supplementary Table SI).
Percentage calculated as proportion of aneuploid embryos in dataset. Embryos could have multiple chromosomes involved.
Number of embryos with monosomic/trisomic changes include those with single abnormalities only and those with a single full gain or loss accompanied by segmental changes.
Number of embryos with full/segmental changes include those with single or multiple abnormalities of the same type.
ND, not determined; UDC, untrainable data cleansing.
Development of the genetics AI model also included an emerging machine learning technique called knowledge distillation (Hinton et al., 2014). This technique allows for candidate constituent models to act as ‘teacher’ models during training of the constituent model, which is ultimately chosen to be part of the final ensemble model. Teacher models can encompass a diverse range of different architectures and machine learning parameters. Once specific teacher models have been selected, the parameters are fixed while they are used to train each constituent model.
In the current study, individual constituent models were trained and evaluated separately using a train-validate cycle process, as described in VerMilyea et al. (2020). A range of optimizers, learning rates, momentum values, regularization strategies and batch sizes were considered. In particular, batch-normalization and dropout methods were employed in order to stabilize the constituent models during training and prevent against overfitting. The training-validation cycle was carried out for 300 epochs each until a sufficiently stable model was developed with low loss function. At the conclusion of the series of training-validation cycles, the highest-performing models, selected using log loss and tangent score as metrics, were chosen as teacher models for distillation, and the training-validation cycle was repeated, but this time using distillation training. The highest-performing models were then selected as candidates for ensembling based on log loss, tangent score, AUC, total accuracy, sensitivity and specificity. Details regarding specific machine learning methods and evaluation metrics are provided in Supplementary Materials and Methods.
The final ensemble model developed in the current study includes three deep learning models selected using a majority-mean-based voting strategy. Every constituent model in the final ensemble model was a binary classification model, used cosine annealing as a learning rate scheduler, a stochastic gradient descent optimizer and uniform image normalization RGB = (0.5, 0.5, 0.5) for each input image. Note that the knowledge distillation method used a Kullback–Leibler divergence to modify the loss function while training, with a weighting, alpha, as reported below. The final model configuration used in this study was as follows:
Model 1: One full (no segmentation) DenseNet-161 model, trained on a dataset of images with UDC applied using full (no segmentation) training, learning rate = 1.0e−4, momentum = 0.95, dropout value = 0.25 and batch size = 16 images. This model was not a distilled model.
-
Model 2: One full (no segmentation) ResNet-50 model, trained on a dataset of images with UDC applied using full (no segmentation), learning rate = 3.0e−4, momentum = 0.90, dropout value = 0.10, batch size = 32 images and distillation alpha = 0.2. Two teacher models were used as follows:
One DenseNet-161 model, learning rate = 1.0e−4, momentum = 0.95, dropout value = 0.25 and batch size = 16 images.
One ResNet-50 model, learning rate = 1.0e−4, momentum = 0.90, dropout value = 0.10 and batch size = 32 images.
Model 3: One IZC DenseNet-121 model, trained on a dataset including images with UDC applied using IZC, learning rate = 1.0e−4, momentum = 0.90, dropout value = 0.15 and batch size = 16 images. This model was not a distilled model.
The ensemble method has recently been shown to sometimes overfit data, leading to poor performance of the ensemble AI model on test datasets (poor generalizability). Overfitting of the genetics AI model in the current study was tested for by comparing the performance of the final ensemble model to that of the individual constituent AI models making up the final model. Results demonstrated a similar or superior performance of the final ensemble model compared to constituent models, as described in Supplementary Materials and Methods.
Results
Datasets used in development of a genetics AI model for predicting the likelihood of embryo euploidy
A total of 15 192 embryo images with associated metadata were provided by 10 different clinics in the USA, India, Spain and Malaysia. Of these images, a total of 5050 images from 2438 patients treated at five clinics in the USA were used for development of the Day 5 genetics AI model. The following images were excluded from model training and validation:
PGT-A result was inconclusive or missing: 228 images.
Technical issues (duplicate images, unmatched images/metadata, etc.): 836 images.
Day 6 embryos: 5574 images (excluded from training but used to evaluate model performance, as described).
Days other than Day 5 or Day 6 (or day not recorded): 2584 images.
Mosaic embryos: 403 images (excluded from training but used to evaluate model performance, as described).
Double-blind test dataset images (datasets from independent clinics in India, Spain and Malaysia): 517 images (images were collected after development of the Day 5 genetics AI model and were used for double-blind testing and analysis of time-lapse images, as described).
Note that no images were excluded because of image quality (e.g. poor focus, low resolution, etc.). One thousand and one of the 5050 US-based images were held back for blind testing the genetics AI model. The composition of the total Day 5 dataset, as well as constituent blind test datasets used for evaluating performance (cleansed and uncleansed versions), is shown in Table I. Average age of women for the complete Day 5 dataset was 36.2 years, and the proportion of euploid and aneuploid embryos was 64.4% and 35.6%, respectively.
Over one-third of all aneuploid embryos displayed a complex karyotype, consisting of abnormalities in multiple chromosomes, and ∼9% of aneuploid embryos displayed an abnormality in one of the sex chromosomes. The frequency of individual autosomal abnormalities ranged from 3.9% (chromosome 12) to 18.0% (chromosome 22) for individual chromosomes.
Additional datasets used in evaluating performance of the genetics AI model are shown in Supplementary Table SIII, including the Day 6 dataset, the mosaic dataset and the prospectively collected double-blind test datasets to evaluate translatability and performance on time-lapse imaging systems.
A complete list of AI scores and relevant metadata is provided in Supplementary Table SIV.
The genetics AI model is predictive of embryo ploidy status for Day 5 embryos in a blind test dataset
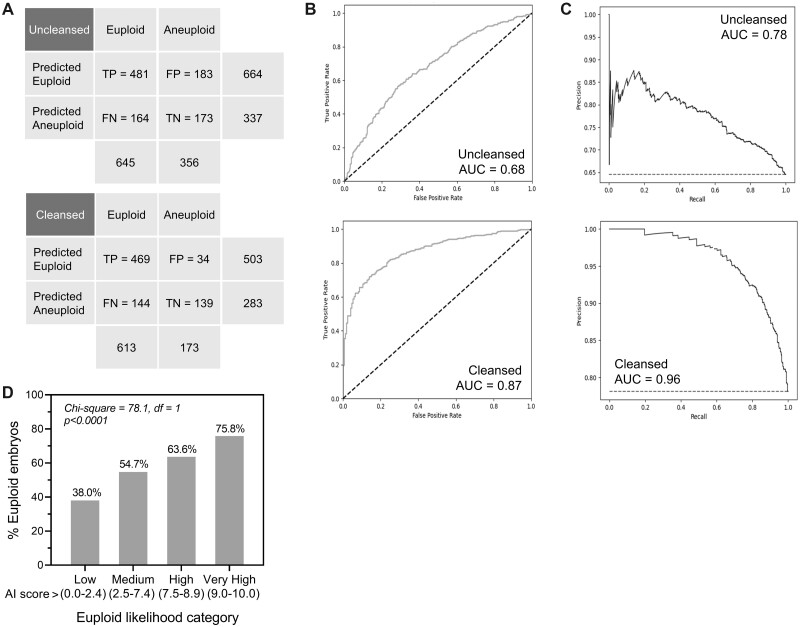
The overall accuracy of the genetics AI model for predicting euploid embryos on the uncleansed Day 5 blind test dataset was 65.3%, with a sensitivity of 74.6% and a specificity of 48.6%. Figure 1A shows the confusion matrix. PPV and NPV were 72.4% and 51.3%, respectively, and the MCC value was 0.235. Removal of poor quality and mislabeled images using UDC considerably improved overall accuracy on the cleansed dataset as expected, to 77.4%, with a corresponding MCC value of 0.491. This was largely due to improvement in specificity (80.3%). ROC are shown in Fig. 1B for both the uncleansed and cleansed test datasets, with AUC values of 0.68 and 0.87, respectively, and PRC are similarly presented in Fig. 1C, with AUC values of 0.78 and 0.96 for uncleansed and cleansed data. While evaluating AI performance on the cleansed blind test dataset is not strictly representative of real-world clinical performance, it is, however, informative for evaluating performance in the absence of poor-quality or potentially mislabeled images. In this way, it represents a cleaner measure of performance that is generalizable across clinics when accounting for the presence of potential confounding variables (see Dakka et al., 2021).
Figure 1.
Performance of the Day 5 artificial intelligence (AI) algorithm for predicting the likelihood of human embryo euploidy on uncleansed and cleansed blind test datasets. (A) Confusion matrices depicting true positives (TP), false positives (FP), false negatives (FN) and true negatives (TN) for the Day 5 AI model predicting embryo euploid status. Matrices are shown for uncleansed (top panel) and cleansed (bottom panel) blind test datasets. (B) Receiver-operating characteristic (ROC) curves for uncleansed (top panel) and untrainable data cleansing (UDC)-cleansed (bottom panel) Day 5 blind test datasets. The AUC values are depicted. (C) Precision-recall curves (PRC) for uncleansed (top panel) and UDC-cleansed (bottom panel) Day 5 blind test datasets. The AUC values are depicted. (D) The correlation between the genetics AI score and the proportion of euploid embryos was evaluated using four defined euploid likelihood categories as depicted. The statistical method used was Chi-square test for trend (df, degrees of freedom).
The genetics AI score showed a significant positive correlation with an increasing percentage of euploid embryos on the uncleansed dataset (Fig. 1D). For this evaluation, a random sampling of 80 mosaic embryos was taken from the Day 5 mosaic test dataset (Supplementary Table SIII) and added to the blind test dataset (overall ratio of ∼7% mosaic embryos, similar to the proportion of mosaic embryos in the full dataset of images originally provided). Results showed that the proportion of euploid embryos doubled from the lowest to the highest euploid likelihood category, with the proportion increasing from 30.8% to 75.8%. These results suggest that embryos of higher scores may be ranked over those of lower scores to aid in selection of euploid embryos during IVF procedures.
A significant positive correlation was also observed between euploid likelihood categories and the proportion of euploid embryos in subgroups based on patient age (<35 and ≥35 years, Supplementary Fig. S1A), and Gardner expansion Grades 3, 4 and 5 (non-significant trend for Grade 6, Supplementary Fig. S1B). Accuracy for binary prediction ranged from 60.0% to 75.4% based on age or expansion grade subgroup (Supplementary Fig. S1).
Enrichment for euploid embryos was investigated using ranking analyses with simulated embryo cohorts, as described in Materials and methods section. When embryo cohorts in the uncleansed dataset were ranked using the genetics AI model, the proportion of cohorts with a euploid embryo as the top-ranked embryo was 82.4% (Table II). This was a 26.4% improvement over randomly ranked cohorts (65.2% cohorts with euploid top-ranked embryo). On the subset of images with an associated Gardner score (n = 918), the AI model showed a 19.3% improvement in identification of a euploid embryo as the top-ranked embryo compared to the commonly used 3BB threshold for defining good- versus poor-quality embryos according to the Gardner method (81.1% and 68.0% of cohorts for the AI model and Gardner ranking, respectively). The AI model maintained an improvement of 12.8% over the Gardner score for identification of a top-ranked euploid embryo when further subdividing embryos according to four Gardner quality rank groups (81.1% and 71.9% of cohorts, respectively). At least one of the top two embryos was found to be euploid in 97.0% of cohorts when ranked by the AI model, and both of the top two ranked embryos were euploid in 66.4% of cohorts. In all cases, the genetics AI model showed an improvement in the ability to rank and enrich for euploid embryos within a patient cohort compared to random ranking and the Gardner ranking method (Table II).
Table II.
Results of simulated cohort ranking analyses to evaluate the ability of the genetics artificial intelligence model to enrich for euploid embryos over random ranking and the Gardner score.
| Measurement | Proportion of cohorts with top one ranked embryo euploid | Proportion of cohorts with one of top two ranked embryos euploid | Proportion of cohorts with both top two ranked embryos euploid |
|---|---|---|---|
| Compared to random ranking | |||
|
| |||
| Genetics AI model | 82.4% | 97.0% | 66.4% |
| Random | 65.2% | 88.9% | 43.2% |
| Improvement | 26.4% | 9.1% | 53.7% |
|
| |||
| Compared to Gardner ranking—3BB thresholda | |||
|
| |||
| Genetics AI model | 81.1% | 96.3% | 63.7% |
| Gardner | 68.0% | 90.6% | 46.6% |
| Improvement | 19.3% | 6.3% | 36.7% |
|
| |||
| Compared to Gardner ranking—four-group systemb | |||
|
| |||
| Genetics AI model | 81.1% | 96.3% | 63.7% |
| Gardner | 71.9% | 92.2% | 50.2% |
| Improvement | 12.8% | 4.4% | 27.4% |
A subset of 918 of the 1001 images in the Day 5 blind test dataset had associated Gardner grades. Gardner ranking was performed using a 3BB threshold as described in Materials and methods section.
A subset of 918 of the 1001 images in the Day 5 blind test dataset had associated Gardner grades. Gardner ranking was performed using a four-group system as described in Materials and methods section.
AI, artificial intelligence.
Studies have demonstrated improved clinical outcomes, such as pregnancy rates, for euploid embryos (Scott et al., 2012). As such, it might be expected that the genetics AI model, trained to evaluate the likelihood of euploidy, would also demonstrate predictive ability for clinical outcomes. A subset of 156 transferred embryos from the uncleansed Day 5 blind test dataset was utilized to evaluate the performance of the AI model for predicting clinical pregnancy outcome, as measured by the presence of a fetal heartbeat at first ultrasound scan. The accuracy of the AI model for predicting a successful clinical pregnancy on these embryos was 57.1%, indicating some predictive ability, although this was lower than the accuracy of 65.3% for predicting euploidy. This outcome is not unexpected, as a euploid result on PGT-A test is not a definitive marker of clinical success.
The genetics AI model is effective for use with images of mosaic embryos and Day 6 embryos, and can predict monosomic abnormalities
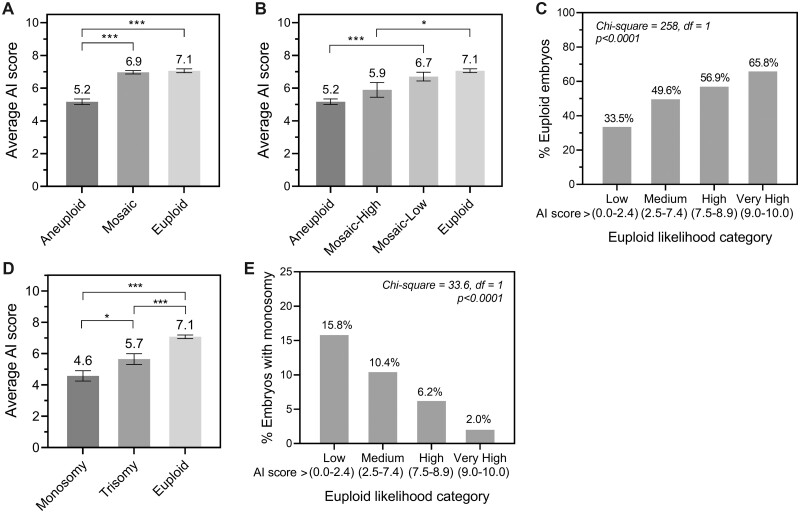
Mosaicism occurs when there are two or more cell populations with different karyotypes present within an individual embryo, including both euploid and aneuploid cell types. Consistent with this, the average AI score for mosaic embryos fell between the average scores for full aneuploid and euploid embryos (Fig. 2A; Day 5 mosaic test dataset described in Supplementary Table SIII). When subdividing mosaic embryos according to the level of mosaicism, mosaic-low embryos were found to have similar average scores to euploid cells, whereas mosaic-high embryos had similar average scores to aneuploid cells (Fig. 2B). Predictive accuracy of the genetics AI model on a dataset including mosaic embryos (uncleansed Day 5 blind test dataset + Day 5 mosaic dataset) was evaluated by treating mosaic-low embryos as a euploid classification and mosaic-high embryos as an aneuploid classification. Accuracy in this situation was 64.5%, which was similar to the predictive accuracy on the original Day 5 blind test dataset (65.3%) consisting of full euploid and aneuploid embryos only.
Figure 2.
Correlations between the Day 5 genetics artificial intelligence (AI) score and level of mosaicism, monosomic abnormalities, and performance on Day 6 human embryos. (A) Correlation between average genetics AI score and embryos based on ploidy status, including euploid, aneuploid, or mosaic status. (B) Correlation between average AI score and embryo ploidy status, separating mosaic embryos according to level of mosaicism. (C) The correlation between the AI score and the proportion of euploid embryos was evaluated using euploid likelihood categories on a dataset of images taken of blastocyst-stage embryos on Day 6 of in vitro culture. (D) Average genetics AI score in embryos with monosomic or trisomic changes compared to euploid embryos. (E) Correlation between AI score and the proportion of embryos with monosomic changes in different AI score categories. Average AI scores were compared using one-way ANOVA with Tukey’s multiple comparisons post-test (Student’s t-test was used to compare monosomic with trisomic changes), and Chi-square test for trend was used where indicated (df = degrees of freedom). P-values are represented as follows: *P <0.05, ***P <0.001.
The Day 5 genetics AI model was also evaluated on Day 6 blastocyst-stage embryo images for comparison (Day 6 dataset described in Supplementary Table SIII). Accuracy on Day 6 images was 59.6% with a sensitivity of 74.9% for prediction of euploid embryos, and a corresponding MCC of 0.184. Euploid likelihood category analysis demonstrated a significant positive correlation of AI score with percentage of euploid embryos, with twice as many euploid embryos in the very high category compared to the low category (Fig. 2C). Collectively, these results suggest that, while the accuracy is marginally reduced for Day 6 embryos, the Day 5 genetics AI model can be used to rank Day 6 embryos according to the likelihood of euploidy if desired.
The genetic complement of a human embryo is complex and can have myriad influences on morphology that are as yet unknown. While the genetics AI model was trained simply to detect euploidy in general, the ability of the AI model to detect different types of abnormalities was also of interest. Performance of the AI model was evaluated for predicting monosomic versus trisomic changes, full chromosomal gains or losses versus segmental duplications or deletions, and for single versus multiple (complex) chromosomal abnormalities on the original Day 5 blind test dataset of 1001 embryos (Supplementary Fig. S2). Of these analyses, it was found that there was a significant difference in average AI score between embryos with monosomic versus trisomic changes (Fig. 2D). Accuracy for predicting monosomic changes was slightly higher than predicting euploidy in general, at 67.3% compared with 65.3%, respectively. This accuracy increased to 72.6% when predicting embryos with monosomic changes compared to euploid embryos only. Analysis of correlation demonstrated that monosomic changes were almost 8-fold more likely to be found in the lowest AI score category than in the very high euploid likelihood category (Fig. 2E).
The genetics AI model generalizes well to different demographics and to images taken using time-lapse systems
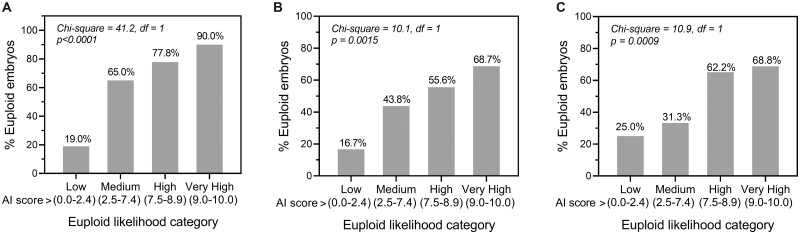
Three additional datasets were collected prospectively through the course of clinical use for evaluation of performance in different demographics. These data were collected from independent clinics that did not contribute data to model training, and therefore represent double-blind test datasets. The first of these was a dataset of 178 Day 5 embryo images taken using a standard microscope-mounted camera system for 31 patients treated between June and August 2021 at an IVF clinic in India. A total of 74% of embryos were euploid and 26% were aneuploid. Results showed a significant positive correlation between genetics AI score and euploidy rate on these images, with the proportion of euploid embryos increasing 4.7-fold from the lowest likelihood category (19.0%) to the highest likelihood category (90.0%, Fig. 3A). Overall accuracy was 81.5% and sensitivity for prediction of euploid embryos was 90.9%. Corresponding MCC was 0.488.
Figure 3.
Performance of the Day 5 genetics artificial intelligence (AI) model in different demographics and using time-lapse images. (A) The correlation between the AI score and the proportion of human euploid embryos was evaluated using euploid likelihood categories on a double-blind test dataset of images from a clinic in India. (B) The correlation between AI score and the proportion of euploid embryos evaluated on a double-blind test dataset of images taken using the GERI time-lapse imaging system by three clinics in Spain. (C) The correlation between AI score and the proportion of euploid embryos evaluated on a blind test dataset of images taken using the EmbryoScope time-lapse imaging system by a clinic in Malaysia. The statistical method used was Chi-square test for trend (df, degrees of freedom).
The second double-blind test dataset was of 182 embryo images taken using the GERI time-lapse imaging system (Supplementary Table SIII). Data were provided for 63 patients treated between May and September 2021 at three IVF clinics in Spain, with 62% of embryos being euploid and 38% aneuploid. Results again showed a significant positive correlation between genetics AI score and euploidy rate on these time-lapse images, with the proportion of euploid embryos increasing ∼4-fold from the lowest likelihood category (16.7%) to the highest likelihood category (68.7%, Fig. 3B). Overall accuracy was 65.4% and sensitivity 97.3% (MCC of 0.219).
To evaluate the generalizability of the genetics AI model to additional time-lapse imaging systems, a third double-blind test dataset of 141 embryo images taken using the EmbryoScope time-lapse system was studied (Supplementary Table SIII). The dataset consisted of images for 65 patients treated at a single clinic in Malaysia between November 2019 and October 2020. The ratio of euploid to aneuploid embryos was from 59% to 41%. The correlation of genetics AI score with proportion of euploid embryos was similar to that observed for the GERI system, with the likelihood of euploidy increasing just under 3-fold from lowest to highest score categories (25.0% and 68.8%, respectively, Fig. 3C). Accuracy was 61.0% and sensitivity 94.0% (MCC of 0.132). These results collectively support use of the genetics AI model in multiple ethnic demographics, and with images obtained using time-lapse systems.
Time-series data were available for a subset of 89 of the 141 EmbryoScope images for investigation of the optimal Day 5 time-point for evaluation (110, 115 and 120 h post-insemination). Note that the 120-h time-point included embryos at 117.5–122.5 h post-insemination, representing the latest time-point evaluated prior to biopsy. Performance of the genetics AI model was found to increase over time, with the highest overall accuracy and sensitivity observed at the latest time-point. Accuracy values were 56.2%, 58.4% and 62.9% for 110, 115 and 120 h, respectively, and sensitivity values for detection of euploid embryos were 70.0%, 86.0% and 96.0%, respectively. These results suggest that the optimal time-point for evaluation is the latest time-point possible on Day 5, with imaging performed immediately prior to biopsy for PGT-A.
A novel machine learning method may be identifying mislabeled aneuploid embryo images associated with variability in PGT-A testing
The UDC method can be used to identify images that may have been mislabeled, which in this case could be the result of inherent variability in the PGT-A method related to sampling bias of the trophectoderm (Victor et al., 2019b; Abhari and Kawwass, 2021). To evaluate PGT-A variability with regards to the performance of the Day 5 genetics AI model, a dataset of 16 embryo images classified as either mosaic or aneuploid at first PGT-A test was rebiopsied at a later date and PGT-A testing repeated. A list of these embryos and associated results is presented in Supplementary Table SV.
In this dataset, 13/16 (81%) embryos showed a similar outcome on PGT-A retest (euploid/mosaic-low or aneuploid/mosaic-high). Variability between tests was 19%, with 3/16 embryos switching classifications. Two of five embryos initially classified as aneuploid/mosaic-high switched to euploid/mosaic-low, demonstrating 40% variability in PGT-A for embryos initially labeled as aneuploid or mosaic-high. These preliminary data support the UDC results suggesting that a significant proportion of aneuploid embryos in the original dataset could have been mislabeled and might have been euploid or mosaic-low on retest.
Performance of the genetics AI model was also evaluated on both sets of test results. Accuracy for predicting whether embryos were euploid/mosaic-low or aneuploid/mosaic-high using the results of the first PGT-A test was 50.0% for Day 5/6 embryos and 50.0% for Day 5 embryos only. Accuracy improved considerably on the second set of PGT-A outcomes to 68.8% for Day 5/6 embryos and 75.0% for Day 5 only.
Discussion
An AI model trained on Day 5 embryo images and PGT-A outcomes were found to be predictive of ploidy status in both Day 5 and Day 6 blastocyst images. The genetics AI model generalized well to datasets from independent clinics representing different demographics, including prospectively collected data, and images taken using time-lapse systems. Overall accuracies ranged from ∼60% to 80% based on the dataset, with sensitivity for predicting euploid embryos ranging between ∼75% and 95%. In all cases, there was a significant positive correlation between the genetics AI score and the proportion of euploid embryos, providing support for the intended clinical application of ranking and selecting embryos that are more likely to be euploid within a patient cohort.
The ability of the genetics AI model to enrich for euploid embryos within a patient cohort was further evaluated using simulated cohort ranking studies, which showed that the probability of the top-ranked embryo being euploid was significantly higher (26.4%) than when ranked randomly. Random ranking is an appropriate comparison, as there is no recognized procedure for visually detecting genetic status. However, the ranking ability of the AI model was also compared to ranking using the Gardner score as a proxy for embryologist selection in the absence of any invasive genetic testing methods. Ranking using the AI model was also superior to ranking using two different Gardner ranking methods, although the improvement was somewhat lower than the improvement over random ranking (19.3% and 12.8% for the two Gardner methods). This is consistent with research demonstrating some limited correlation of the Gardner score with embryo euploidy (Capalbo et al., 2014; Minasi et al., 2016). Improvements compared to random ranking and Gardner ranking also held true when considering the proportion of cohorts with at least one of the top two embryos being euploid, and the proportion with both top two embryos being euploid.
The performance of the genetics AI model for selecting a euploid embryo in the top-ranked position was somewhat higher than that of the ERICA algorithm (Chavez-Badiola et al., 2020; 82.4% for the current study and 78.9% for ERICA), although the overall accuracy quoted for ERICA on a set of 84 images was somewhat higher than the value quoted for genetics AI on the blind test dataset of 1001 images (70% and 65.3%, respectively). It is noted, however, that the generalizability of ERICA has not yet been evaluated on a substantial number of patients, IVF clinics or double-blind datasets, and its performance on time-lapse images remains to be determined.
In the current study, genetics AI scores were also positively correlated with the proportion of euploid cells when evaluating embryo classifications of aneuploid, mosaic-high, mosaic-low or euploid. Historically, mosaic embryos were treated clinically as aneuploid embryos and generally not used for transfer. However, recent evidence suggests that mosaic embryos have improved clinical outcomes compared to full aneuploid embryos, and depending on the proportion of abnormal cells present, can even have outcomes similar to that of euploid embryos (Abhari and Kawwass, 2021). Mosaic embryos were therefore excluded from development of the genetics AI model, as they were expected to confound model training owing to the unclear biological classification of these embryos. Subsequent evaluation of mosaic embryos in this study supported a biological distinction between mosaic-low and mosaic-high embryos, as suggested by previous research groups (Spinella et al., 2018; Munné et al., 2020), and thus supported the hypothesis that the genetics AI model would correctly generalize to evaluating mosaic embryos. Predictive accuracy of the AI model was not affected when considering mosaic-low embryos as euploid, and mosaic-high embryos as aneuploid, suggesting that the AI model might be useful in differentiating mosaic embryos based on their level of mosaicism. This finding could be of marked clinical significance, as multiple educational societies have published recommendations for transfer of mosaic-low embryos over mosaic-high embryos in situations where there are no euploid embryos available (CoGEN, 2017; Cram et al., 2019).
While the genetics AI model was developed solely to detect the likelihood of overall embryo euploidy, subanalyses demonstrated a higher performance for identifying monosomic abnormalities than for predicting euploidy in general. Monosomic abnormalities are mostly non-viable, with only monosomy X carrying through to live birth (O’Conner, 2008). Studies have shown that autosomal monosomies are more detrimental to blastocyst development than other abnormalities, causing arrest earlier during blastocyst formation (Rubio et al., 2003). These observations are consistent with the genetics AI model being able to differentiate a more severe genetic phenotype, further validating its performance and potential clinical utility. It would be of interest to explore production of an AI algorithm specifically for detection of monosomic changes.
Development of the genetics AI model utilized a novel machine learning method, namely UDC (see Dakka et al., 2021), which identified a high proportion of embryos that appeared morphologically euploid to the AI model but were actually aneuploid based on PGT-A test results (false positives). There are a number of factors that could contribute to the occurrence of false positive predictions, including mosaicism, where the different proportions of normal and abnormal cells could conceivably interfere with evaluation of ploidy status (Viotti, 2020); embryo self-correction, where mosaic embryos containing a proportion of abnormal cells exclude these cells during further development to become euploid embryos (which could similarly interfere with evaluation of ploidy status; Orvieto et al., 2020); and lastly, there is variability in the PGT-A process itself. Studies have shown that the biopsied trophectoderm sample is not necessarily representative of the full trophectoderm, nor the ICM (Victor et al., 2019a). PGT-A testing results presented here demonstrated a relatively high variability for embryos initially classified as aneuploid or mosaic, which is consistent with previously published reports demonstrating that for mosaic embryos in particular there can be a very low concordance between trophectoderm biopsies (∼5–30% concordance, Navratil et al., 2020; Sachdev et al., 2020). These observations suggest that the UDC technique may be identifying euploid/mosaic-low embryos that were mislabeled as aneuploid/mosaic-high by PGT-A testing. However, further direct evaluation of the embryos identified by UDC would be necessary to confirm this finding.
The genetics AI model was also able to predict clinical pregnancy, to a degree, amongst transferred embryos all recorded as euploid by PGT-A. While this could simply represent a correlation between embryo ploidy status and overall embryo quality (Capalbo et al., 2014), on another level it might also support the hypothesis that some of these embryos were mislabeled as euploid owing to PGT-A sampling bias, and/or embryo self-correction. In this scenario, it is conceivable that the genetics AI model was in fact differentiating embryos that actually displayed a level of mosaicism, and therefore had a reduced likelihood of pregnancy, even though the PGT-A result was recorded as euploid. However, as with the previous observation on UDC label identification, additional direct evidence would be needed to support this conclusion. Nevertheless, these findings highlight the impact of PGT-A variability on performance evaluation of alternative, non-invasive procedures for predicting embryo ploidy status and, consistent with recent discussions, suggest that early embryonic ploidy status may be a dynamic process that could one day be better assessed using alternative methods to PGT-A (Bouba et al., 2021).
In conclusion, this study collectively supports the use of the genetics AI for non-invasive evaluation of embryo ploidy to aid in selection of euploid embryos for use in IVF procedures. It also poses broader considerations for the use of AI in comparison with PGT-A in embryo assessment for IVF, highlighting the challenges with current approaches and potential complementary solutions.
Data availability
The AI scores and relevant metadata used in this study are provided in Supplementary Table SIV. The embryo images and other patient data collected in this study are not publicly available owing to reasonable ethics and privacy concerns, and are not redistributable to researchers other than those engaged in the approved research collaborations with the named medical centers. For any interested collaborators, please contact the corresponding author. The AI model developed in this article is available for commercial use as part of the Life Whisperer Genetics embryo assessment software. The computer code developed is not publicly available owing to commercial restrictions.
Authors’ roles
S.M.D., J.M.M.H., M.D.V., D.P. and M.P. conceived the study and designed methodology. D.P. and M.P. were also responsible for project management and supervision of research activity. M.D.V., J.A. (Wings), J.A. (IVF-Spain), L.G., Y.G.B. and A.Y.X.L. provided significant resources. S.M.D, J.M.M.H., M.A.D. and T.V.N. were responsible for performing the research and formal analysis, and J.M.M.H., M.A.D. and T.V.N. were also responsible for data curation and AI development. S.M.D. and J.M.M.H wrote the article. All authors contributed to review and editing of the final manuscript.
Funding
Life Whisperer Diagnostics is a wholly owned subsidiary of the parent company, Presagen Holdings Pty Ltd. Funding for the study was provided by Presagen with grant funding received from the South Australian Government: Research, Commercialisation, and Startup Fund (RCSF). ‘In kind’ support and embryology expertise to guide algorithm development were provided by Ovation Fertility. 'In kind’ support in terms of computational resources provided through the Amazon Web Services (AWS) Activate Program.
Conflict of interest
J.M.M.H., D.P. and M.P. are co-owners of Life Whisperer and Presagen. S.M.D., M.A.D. and T.V.N. are employees or former employees of Life Whisperer. S.M.D., J.M.M.H., M.A.D., T.V.N., D.P. and M.P. are listed as inventors of patents relating to this work, and also have stock options in the parent company Presagen. M.V. sits on the advisory board for the global distributor of the technology described in this study and also received support for attending meetings.
Supplementary Material
Contributor Information
S M Diakiw, Life Whisperer Diagnostics (a subsidiary of Presagen), San Francisco, CA, USA, and Adelaide, SA, Australia.
J M M Hall, Life Whisperer Diagnostics (a subsidiary of Presagen), San Francisco, CA, USA, and Adelaide, SA, Australia; Australian Research Council Centre of Excellence for Nanoscale BioPhotonics, The University of Adelaide, Adelaide, SA, Australia; School of Physical Sciences, Faculty of Sciences, The University of Adelaide, Adelaide, SA, Australia.
M D VerMilyea, Ovation Fertility, Austin, TX, USA; Texas Fertility Center, Austin, TX, USA.
J Amin, Wings IVF Women’s Hospital, Ahmedabad, Gujarat, India.
J Aizpurua, IVF-Spain, Alicante, Spain.
L Giardini, IVF-Spain, Alicante, Spain.
Y G Briones, IVF-Spain, Alicante, Spain.
A Y X Lim, Alpha IVF & Women's Specialists, Petaling Jaya, Selangor, Malaysia.
M A Dakka, Life Whisperer Diagnostics (a subsidiary of Presagen), San Francisco, CA, USA, and Adelaide, SA, Australia.
T V Nguyen, Life Whisperer Diagnostics (a subsidiary of Presagen), San Francisco, CA, USA, and Adelaide, SA, Australia.
D Perugini, Life Whisperer Diagnostics (a subsidiary of Presagen), San Francisco, CA, USA, and Adelaide, SA, Australia.
M Perugini, Life Whisperer Diagnostics (a subsidiary of Presagen), San Francisco, CA, USA, and Adelaide, SA, Australia; Adelaide Medical School, Faculty of Health Sciences, The University of Adelaide, Adelaide, SA, Australia.
References
- Abhari S, Kawwass JF.. Pregnancy and neonatal outcomes after transfer of mosaic embryos: a review. J Clin Med 2021;10:1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouba I, Hatzi E, Ladias P, Sakaloglou P, Kostoulas C, Georgiou I.. Biological and Clinical Significance of Mosaicism in Human Preimplantation Embryos. J Dev Biol 2021;9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capalbo A, Rienzi L.. Mosaicism between trophectoderm and inner cell mass. Fertil Steril 2017;107:1098–1106. [DOI] [PubMed] [Google Scholar]
- Capalbo A, Rienzi L, Cimadomo D, Maggiulli R, Elliott T, Wright G, Nagy ZP, Ubaldi FM.. Correlation between standard blastocyst morphology, euploidy and implantation: an observational study in two centers involving 956 screened blastocysts. Hum Reprod 2014;29:1173–1181. [DOI] [PubMed] [Google Scholar]
- Chavez-Badiola A, Flores-Saiffe-Farías A, Mendizabal-Ruiz G, Drakeley AJ, Cohen J.. Embryo Ranking Intelligent Classification Algorithm (ERICA): artificial intelligence clinical assistant predicting embryo ploidy and implantation. Reprod Biomed Online 2020;41:585–593. [DOI] [PubMed] [Google Scholar]
- CoGEN. COGEN position statement on chromosomal mosaicism detected in preimplantation blastocyst biopsies. 2017. https://www.ivf-worldwide.com/index.php?option=com_content&view=article&id=733&Itemid=464 (15 January 2021, date last accessed).
- Cram DS, Leigh D, Handyside A, Rechitsky L, Xu K, Harton G, Grifo J, Rubio C, Fragouli E, Kahraman S. et al. PGDIS position statement on the transfer of mosaic embryos 2019. Reprod Biomed Online 2019;39(Suppl 1):e1–e4. [DOI] [PubMed] [Google Scholar]
- Dakka MA, Nguyen TV, Hall JMM, Diakiw SM, VerMilyea M, Linke R, Perugini M, Perugini D.. Automated detection of poor-quality data: case studies in healthcare. Sci Rep 2021;11:18005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florkowski CM. Sensitivity, specificity, receiver-operating characteristic (ROC) curves and likelihood ratios: communicating the performance of diagnostic tests. Clin Biochem Rev 2008;29(Suppl 1):S83–S87. [PMC free article] [PubMed] [Google Scholar]
- Greco E, Litwicka K, Minasi MG, Cursio E, Greco PF, Barillari P.. Preimplantation genetic testing: where we are today. Int J Mol Sci 2020;21:4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J.. Deep residual learning for image recognition. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 27–30 June 2016, pp. 770–778. [Google Scholar]
- Hinton G, Vinyals O, Dean J.. Distilling the knowledge in a neural network. In: Neural Information Systems (NIPS) Deep Learning Workshop. 2014. https://arxiv.org/abs/1503.02531 (12 December 2014, date last accessed). [Google Scholar]
- Huang G, Liu Z, Maaten LVD, Weinberger KQ.. Densely connected convolutional networks. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 21–26 July 2017, pp. 2261–2269. [Google Scholar]
- Irani M, Reichman D, Robles A, Melnick A, Davis O, Zaninovic N, Xu K, Rosenwaks Z.. Morphologic grading of euploid blastocysts influences implantation and ongoing pregnancy rates. Fertil Steril 2017;107:664–670. [DOI] [PubMed] [Google Scholar]
- Kemper JM, Liu Y, Afnan M, Hammond ER, Morbeck DE, Mol BWJ.. Should we look for a low-grade threshold for blastocyst transfer? A scoping review. Reprod Biomed Online 2021;42:709–716. [DOI] [PubMed] [Google Scholar]
- Kuznyetsov V, Madjunkova S, Abramov R, Antes R, Ibarrientos Z, Motamedi G, Zaman A, Kuznyetsova I, Librach CL.. Minimally invasive cell-free human embryo aneuploidy testing (miPGT-A) utilizing combined spent embryo culture medium and blastocoel fluid—towards development of a clinical assay. Sci Rep 2020;10:7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhijani R, Bartels CB, Godiwala P, Bartolucci A, DiLuigi A, Nulsen J, Grow D, Benadiva C, Engmann L.. Impact of trophectoderm biopsy on obstetric and perinatal outcomes following frozen–thawed embryo transfer cycles. Hum Reprod 2021;36:340–348. [DOI] [PubMed] [Google Scholar]
- Minasi MG, Colasante A, Riccio T, Ruberti A, Casciani V, Scarselli F, Spinella F, Fiorentino F, Varricchio MT, Greco E. et al. Correlation between aneuploidy, standard morphology evaluation and morphokinetic development in 1730 biopsied blastocysts: a consecutive case series study. Hum Reprod 2016;31:2245–2254. [DOI] [PubMed] [Google Scholar]
- Munné S, Spinella F, Grifo J, Zhang J, Beltran MP, Fragouli E, Fiorentino F.. Clinical outcomes after the transfer of blastocysts characterized as mosaic by high resolution Next Generation Sequencing—further insights. Eur J Med Genet 2020;63:103741. [DOI] [PubMed] [Google Scholar]
- Navratil R, Horak J, Hornak M, Kubicek D, Balcova M, Tauwinklova G, Travnik P, Vesela K.. Concordance of various chromosomal errors among different parts of the embryo and the value of re-biopsy in embryos with segmental aneuploidies. Mol Hum Reprod 2020;26:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Conner C. Chromosomal abnormalities: aneuploidies. Nat Educ 2008;1:172. [Google Scholar]
- Orvieto R, Aizer A, Gleicher N.. Is there still a rationale for non-invasive PGT-A by analysis of cell-free DNA released by human embryos into culture medium? Hum Reprod 2021;36:1186–1190. [DOI] [PubMed] [Google Scholar]
- Orvieto R, Shimon C, Rienstein S, Jonish-Grossman A, Shani H, Aizer A.. Do human embryos have the ability of self-correction? Reprod Biol Endocrinol 2020;18:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, He K, Girshick R, Sun J.. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell 2015;39:1137–1149. [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, Brox T.. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells W, Frangi A (eds). Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015. Cham: Springer, 2015. [Google Scholar]
- Rubino P, Tapia L, Ruiz de Assin Alonso R, Mazmanian K, Guan L, Dearden L, Thiel A, Moon C, Kolb B, Norian JM. et al. Trophectoderm biopsy protocols can affect clinical outcomes: time to focus on the blastocyst biopsy technique. Fertil Steril 2020;113:981–989. [DOI] [PubMed] [Google Scholar]
- Rubio C, Simon C, Vidal F, Rodrigo L, Pehlivan T, Remohi J, Pellicer A.. Chromosomal abnormalities and embryo development in recurrent miscarriage couples. Hum Reprod 2003;18:182–188. [DOI] [PubMed] [Google Scholar]
- Sachdev NM, McCulloh DH, Kramer Y, Keefe D, Grifo JA.. The reproducibility of trophectoderm biopsies in euploid, aneuploid, and mosaic embryos using independently verified next-generation sequencing (NGS): a pilot study. J Assist Reprod Genet 2020;37:559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer E, Porchia L, Lopez-Luna A, Hernandez-Melchor D, Lopez-Bayghen E.. Aneuploidy rates inversely correlate with implantation during in vitro fertilization procedures: in favor of PGT. In: Gomy I (ed). Modern Medical Genetics and Genomics. London, UK: IntechOpen, 2018,1–19. [Google Scholar]
- Scott RT, Ferry K, Su J, Tao X, Scott K, Treff NR.. Comprehensive chromosome screening is highly predictive of the reproductive potential of human embryos: a prospective, blinded, nonselection study. Fertil Steril 2012;97:870–875. [DOI] [PubMed] [Google Scholar]
- Spinella F, Fiorentino F, Biricik A, Bono S, Ruberti A, Cotroneo E, Baldi M, Cursio E, Minasi MG, Greco E. et al. Extent of chromosomal mosaicism influences the clinical outcome of in vitro fertilization treatments. Fertil Steril 2018;109:77–83. [DOI] [PubMed] [Google Scholar]
- VerMilyea M, Hall JMM, Diakiw SM, Johnston A, Nguyen T, Perugini D, Miller A, Picou A, Murphy AP, Perugini M. et al. Development of an artificial intelligence-based assessment model for prediction of embryo viability using static images captured by optical light microscopy during IVF. Hum Reprod 2020;35:770–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor AR, Griffin DK, Brake AJ, Tyndall JC, Murphy AE, Lepkowsky LT, Lal A, Zouves CG, Barnes FL, McCoy RC. et al. Assessment of aneuploidy concordance between clinical trophectoderm biopsy and blastocyst. Hum Reprod 2019a;34:181–192. [DOI] [PubMed] [Google Scholar]
- Victor AR, Tyndall JC, Brake AJ, Lepkowsky LT, Murphy AE, Griffin DK, McCoy RC, Barnes FL, Zouves CG, Viotti M. et al. One hundred mosaic embryos transferred prospectively in a single clinic: exploring when and why they result in healthy pregnancies. Fertil Steril 2019b;111:280–293. [DOI] [PubMed] [Google Scholar]
- Viotti M. Preimplantation genetic testing for chromosomal abnormalities: aneuploidy, mosaicism, and structural rearrangements. Genes (Basel) 2020;11:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YY, Yu Y, Zhang XW.. Overall blastocyst quality, trophectoderm grade, and inner cell mass grade predict pregnancy outcome in euploid blastocyst transfer cycles. Chin Med J (Engl) 2018;131:1261–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The AI scores and relevant metadata used in this study are provided in Supplementary Table SIV. The embryo images and other patient data collected in this study are not publicly available owing to reasonable ethics and privacy concerns, and are not redistributable to researchers other than those engaged in the approved research collaborations with the named medical centers. For any interested collaborators, please contact the corresponding author. The AI model developed in this article is available for commercial use as part of the Life Whisperer Genetics embryo assessment software. The computer code developed is not publicly available owing to commercial restrictions.