Abstract
The prenyltransferase undecaprenyl pyrophosphate synthetase (di-trans,poly-cis-decaprenylcistransferase; EC 2.5.1.31) was purified from the soluble fraction of Escherichia coli by TSK-DEAE, ceramic hydroxyapatite, TSK-ether, Superdex 200, and heparin-Actigel chromatography. The protein was labeled with the photolabile analogue of the farnesyl pyrophosphate analogue (E,E)-[1-3H]-(2-diazo-3-trifluoropropionyloxy)geranyl diphos-phate and was detected on a sodium dodecyl sulfate-polyacrylamide gel as a protein with an apparent molecular mass of 29 kDa. This protein band was cut out from the gel, trypsin digested, and subjected to matrix-assisted laser desorption ionization mass spectrometric analysis. Comparison of the experimental data with computer-simulated trypsin digest data for all E. coli proteins yielded a single match with a protein of unassigned function (SWISS-PROT Q47675; YAES_ECOLI). Sequences with strong similarity indicative of homology to this protein were identified in 25 bacterial species, in Saccharomyces cerevisiae, and in Caenorhabditis elegans. The homologous genes (uppS) were cloned from E. coli, Haemophilus influenzae, and Streptococcus pneumoniae, expressed in E. coli as amino-terminal His-tagged fusion proteins, and purified over a Ni2+ affinity column. An untagged version of the E. coli uppS gene was also cloned and expressed, and the protein purified in two chromatographic steps. We were able to detect Upp synthetase activity for all purified enzymes. Further, biochemical characterization revealed no differences between the recombinant untagged E. coli Upp synthetase and the three His-tagged fusion proteins. All enzymes were absolutely Triton X-100 and MgCl2 dependent. With the use of a regulatable gene disruption system, we demonstrated that uppS is essential for growth in S. pneumoniae R6.
Isoprenoids are among the most structurally and functionally diverse compounds ubiquitously occurring in nature. The enzymes for isoprenoid biosynthesis, termed prenyltransferases, catalyze the head-to-tail condensation between isopentenyl pyrophosphate (IPP), the fundamental five-carbon building block in the pathway, and allylic pyrophosphate to generate a variety of prenyl pyrophosphates. These participate in the biosynthesis of a variety of isoprenoid compounds such as terpens, steroids, dolichols, carotinoids, glycosyl carrier lipids, the side chains of respiratory quinones and natural rubber (28).
Four biosynthetic enzymes that utilize IPP as a substrate are found in extracts of Escherichia coli (17): an IPP isomerase (idi; SWISS-PROT Q46822 [putative assignment]) and the three prenyltransferases farnesyl pyrophosphate synthetase (Fpp synthetase, ispA; EC 2.5.1.10) (18), octaprenyl pyrophosphate synthetase (Opp synthetase, ispB) (5), and undecaprenyl pyrophosphate synthetase (Upp synthetase; EC 2.5.1.31). Fpp synthetase catalyzes the condensation of dimethylallyl pyrophosphate (DMAPP) and IPP to yield geranyl pyrophosphate (GPP), which the enzyme uses in a second condensation reaction with IPP to yield the ultimate product, farnesyl pyrophosphate (FPP). The other two prenyltransferases use FPP as the starting molecule in further rounds of sequential condensation with IPP. Opp synthetase generates the long-chain polyprenyl pyrophosphate-like isoprenoid quinones (ubiquinone-8, menaquinone-8, and dimethylmenaquinone-8), which have an all-trans-octaprenyl side chain (14). Upp synthetase generates undecaprenyl pyrophosphate (UPP, C55-PP), which contains a trans,cis-mixed isoprenoid chain (38). UPP is required as a lipid carrier of glycosyl transfer in the biosynthesis of a variety of cell wall polysaccharide components in bacteria.
Upp synthetase has been characterized after partial purification from several bacteria, including Salmonella newington (13), Bacillus subtilis (35), E. coli (10, 17), and Micrococcus luteus (9, 22), but it has been studied most extensively in Lactobacillus plantarum (2–4, 8, 20). Despite the interest in the biochemistry of this enzyme, bacterial genes encoding it have not been identified.
By bringing together classical biochemical and new genomics-proteomics approaches, we identified the gene encoding Upp synthetases in E. coli. Specific photolabeling of partially purified E. coli Upp synthetase with the FPP analogue (E,E)-[1-3H]-(2-diazo-3-trifluoropropionyloxy)geranyl diphosphate ([3H]DAFTP-GDP) allowed the identification of the corresponding Upp synthetase-containing band in a sodium dodecyl sulfate (SDS)-polyacrylamide gel of an unlabeled preparation. This unlabeled band was cut out and digested with trypsin. The molecular masses of the peptides thus produced were determined by matrix-assisted laser desorption ionization mass spectrometric (MALDI-MS) analysis, and the data obtained were used to search a database containing the molecular masses of peptides generated by computer-simulated trypsin digestion of all E. coli proteins. A single entry was found to match the experimental data (SWISS-PROT Q47675, YAES_ECOLI). This open reading frame and the corresponding open reading frames from Haemophilus influenzae and Streptococcus pneumoniae were cloned and expressed, and all were demonstrated to encode prenyltransferase activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, enzymes, and chemicals.
The E. coli strains used in this study were XL2-Blue and BL21(DE3)(pLysS) (Stratagene, Basel, Switzerland). The strains were grown in Luria-Bertani (LB) medium (Difco Laboratories, Detroit, Mich.) with aeration at 37°C. The S. pneumoniae strain used, R6 (7), was routinely grown on sheep blood (3%) agar plates (liquid cultures were propagated in Todd-Hewitt broth [Difco]) and incubated with 10% CO2 at 37°C. Plasmids pET-15b, pET-16b, and pET-28b were from Novagen (Abington, England), and pJDC9 (12) was provided by R. Hakenbeck. Restriction enzymes were from New England Biolabs (Beverly, Mass.) or Amersham Pharmacia Biotech (Dübendorf, Switzerland) and used as specified by the manufacturer. All other chemicals were from Sigma (St. Louis, Mo.).
Purification of native Upp synthetase from E. coli.
A total of 185 g (wet weight) of E. coliBL21(DE3) paste was subjected to chromatographic fractionation in four batches. Typically 45 g (wet weight) of E. coli paste was suspended in 100 ml of breaking buffer (20 mM HEPES-OH [pH 8.0], 10% glycerol, 1 mM MgSO4, 150 mM NaCl, 0.1% Triton X-100). Benzonase (Merck, Darmstadt, Germany) was added to 250 U/ml to hydrolyze DNA and RNA. The protease inhibitors Trasylol (Bayer, Leverkusen, Germany) and ɛ-aminocaproic acid (Serva, Heidelberg, Germany) were added to 100 U/ml and 5 mM, respectively. Cells were broken in a precooled French pressure cell (SLM Instruments, Urbana, Ill.) at 1.33 × 108 Pa. Na2 EDTA was added to 5 mM (final concentration), and the lysed cell suspension was subjected to two consecutive centrifugation steps of 20,000 × g for 30 min and 150,000 × g for 2 h. Solid (NH4)2SO4 was added to the supernatant to 30% saturation, and the suspension was stirred on ice for 30 min. The precipitate formed was removed by centrifugation; the supernatant fraction was brought to 50% ammonium sulfate saturation, and the precipitated proteins were pelleted as described above. The 50% ammonium sulfate pellet was dissolved in an appropriate buffer and subjected to five consecutive chromatographic steps: TSK-DEAE 650M (TosoHaas, Stuttgart, Germany), ceramic hydroxyapatite (Bio-Rad, Glattbrugg, Switzerland), TSK-Ether 5PW (TosoHaas), HiLoad Superdex 200 prep grade (Amersham Pharmacia Biotech), and heparin-Actigel ALD (Sterogene, Carlsbad, Calif.). Active fractions were pooled after each chromatographic step and dialyzed against the appropriate buffer prior to application onto the next column. Active fractions from the last chromatographic step were pooled and concentrated to about 0.5 ml by centrifugation at 2,000 × g in a Millipore Ultrafree-15 device with Biomax-10 membrane. Concentrated proteins were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) analysis. Gels were stained with colloidal Coomassie blue (GelCode blue stain; Pierce, Rockford, Ill.) and destained with deionized water.
Purification of recombinant Upp synthetase from an overproducing E. coli strain.
E. coli paste (25 g [wet weight]), expressing wild-type (wt) E. coli Upp synthetase (Fig. 1A), was suspended in 50 ml of breaking buffer and treated as described above for the native enzyme. Proteins precipitating between 35 and 50% ammonium sulfate saturation were collected by centrifugation, dissolved in buffer A (20 mM Tris-HCl [pH 8.0], 10% glycerol, 1 mM Na2 EDTA), and dialyzed against the same buffer overnight at 4°C. The dialysate was centrifuged at 30,000 × g for 15 min, and the supernatant was filtered through a 0.22-μm-pore-size membrane (Millex-GV; Millipore, Molsheim, France). The filtrate was applied onto a 50-ml Phospho-Ultrogel A6R (Biosepra, Idstein, Germany) column in buffer A. The column was washed with buffer A, and bound proteins were eluted with a 10-column-volume linear gradient of buffer B (buffer A, 1 M NaCl). Fractions containing Upp synthetase, as determined by SDS-PAGE analysis, were pooled and concentrated by dialysis against 60% saturated ammonium sulfate. Precipitated proteins were pelleted by centrifugation and dissolved in 5 ml of GF buffer (25 mM HEPES-OH [pH 8.0], 150 mM NaCl, 1 mM Na2 EDTA). The solution was filtered through a 0.22-μm-pore-size membrane and subjected to gel filtration chromatography on a HiLoad 26/60 Superdex 200 prep grade column (Pharmacia) in GF buffer. The His-tag fusion proteins (Fig. 1A) were purified with HisTrap kit (Amersham Pharmacia Biotech) as specified by the manufacturer.
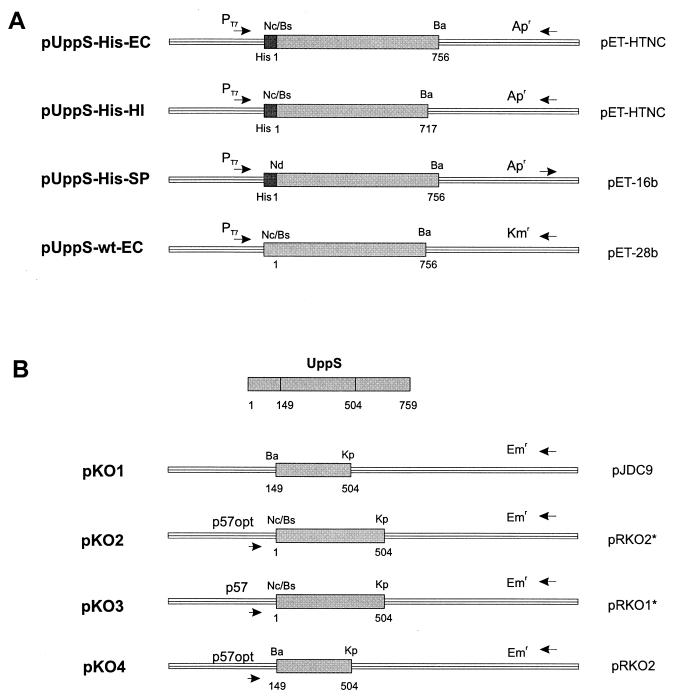
FIG. 1.
Plasmids used in this study. (A) Schematic representation of plasmids for expression of uppS genes from E. coli, H. influenzae, or S. pneumoniae as His-tag fusion protein and from E. coli uppS as untagged protein under the control of the T7 promoter. (B) Schematic representation of plasmids for generating pneumococcal mutants. For disrupting uppS and terminating expression of the downstream genes in the operon, a small internal fragment of the uppS gene was cloned into pJDC9, resulting in pKO1. To place the uppS operon under the control of the two tetracycline-regulatable promoters, p57opt and p57, an amino-terminal fragment of the uppS gene was cloned in the promoter vectors pRKO2* and pRKO1*, resulting in plasmids pKO2 and pKO3, respectively. To generate a disruption of the uppS gene while placing downstream genes under the control of the tetracycline-regulatable promoter p57opt, an internal fragment of uppS was cloned into the promoter vector pRKO2, resulting in plasmid pKO4. For details, see Materials and Methods. Numbers indicate nucleotides, staring with 1 at the putative initiation codon of each gene. Abbreviations for restriction enzymes: Ba, BamHI; Bs, BsaI; Kp, KpnI; Nc, NcoI; Nd, NdeI.
Radiolabeling of native Upp synthetase.
[3H]DAFTP-GDP, a photolabile analogue of the allylic pyrophosphate substrate t,t-farnesyl pyrophosphate, was prepared as described by Liu et al. (26). Baba et al. (10) have shown that E. coli and Lactobacillus plantarum Upp synthetase can be labeled with [3H]DAFTP-GDP upon UV irradiation in the presence of IPP. Partially purified native Upp synthetase was photolabeled with [3H]DAFTP-GDP in an attempt to isolate a radiolabeled polypeptide. The UV irradiation was performed by the method of Baba et al. (10). Briefly, purified native enzyme was UV irradiated (15 min at 254 nm) in the presence of 2.4 μM [3H]DAFTP-GDP (10 Ci/mmol) in 100 mM Tris-HCl (pH 7.5)–20 μM IPP. Multiple samples of the mixture were loaded onto an SDS-polyacrylamide gel and electrophoresed, and the gel was stained with Coomassie blue and destained. One half of the gel was incubated in the fluorographic reagent Amplify (Amersham Pharmacia Biotech) as instructed by the manufacturer. The gel was dried onto filter paper sheets and exposed to Biomax MR film (Kodak, Rochester, N.Y.). From the other half of the gel, the zone of each lane that corresponded to proteins of 20 to 40 kDa was cut into 1-mm slices and extracted with 1 ml of Soluene-350 (Packard, Zürich, Switzerland) at 55°C for 3 h, and radioactivity was counted after the addition of 10 ml of Hionic-Fluor (Packard) scintillation fluid.
MALDI-MS.
The MALDI-MS analysis was performed as reported earlier (16). Briefly, the protein band of interest was excised from an SDS-polyacrylamide gel and digested in situ. The gel piece was destained with 50% acetonitrile in 0.1 M ammonium bicarbonate and dried in a vacuum speed evaporator. The dried gel fragment was reswollen in 3 μl of 3 mM Tris-HCl (pH 8.0) containing 0.2 μg of trypsin (Wako, Neuss, Germany) and incubated at 37°C for 12 h. Three microliters of 30% acetonitrile–0.1% trifluoroacetic acid was added. After sonication for 3 min, 1 μl of the peptide extract was applied onto 0.5 μl of air-dried matrix. The matrix solution was prepared by dissolving 15 mg of nitrocellulose (Bio-Rad, Hercules, Calif.) and 20 mg of α-cyano-4-hydroxycinnamic acid (Sigma) in 1 ml of 1:2 (vol/vol) acetone-isopropanol. The sample was analyzed on a time-of-flight mass spectrometer (PerSeptive Biosystems, Cambridge, Mass.) equipped with a reflectron. An accelerating voltage of 20 kV was used. Calibration was internal to the samples.
Assay for Upp synthetase.
The standard incubation mixture for the Upp synthetase assay contained, in a final volume of 0.1 ml, 100 mM Tris-HCl buffer (pH 7.5), 0.2 mM MgCl2, 0.05% Triton X-100, 10 μM FPP, and 10 μM [1-14C]IPP (0.5 μCi/μmol, 30,000 dpm; Amersham Pharmacia Biotech). The reaction was initiated by addition of a suitable amount of enzyme. After incubation for 30 min at 35°C, the reaction was terminated by the addition of 0.1 ml of 50% (wt/vol) trichloroacetic acid, and the mixture was kept at 100°C for 30 min to complete the hydrolysis of the product. The mixture was cooled and neutralized by the addition of 0.1 ml of 5 M NaOH, followed by the addition of 0.5 ml of 2 M KCl. The products were extracted twice with 2 ml of n-hexane and backwashed with 1 ml of water. A fixed volume of the extract was evaporated to dryness, and scintillation fluid was added for subsequent determination of the radioactivity (with modifications as specified in references 10 and 35).
Coupled assay for Upp synthetase.
The assay used is a modification of the method of Kodama et al. (21) for inorganic phosphate determination. Upp synthetase was incubated at 35°C with 0.003 U of inorganic pyrophosphatase from Saccharomyces cerevisiae in a total reaction volume of 100 μl containing 100 mM Tris-HCl (pH 7.5), 0.2 mM MgCl2, 10 μM IPP, 10 μM FPP, and 0.05% (wt/vol) Triton X-100. The reaction was terminated after 30 min by addition of 100 μl of acidic malachite green solution (0.03% [wt/vol] malachite green–0.2% ammonium molybdate–0.05% Triton X-100 in 0.7 N KCl, filtered through Whatman no. 1 paper). Increase in the optical density at 660 nm (OD660) was measured spectrophotometrically, and the amount of phosphate released from IPP was calculated from a standard curve prepared with KH2PO4.
Electrophoresis.
Proteins were subjected to electrophoresis in a discontinuous slab gel system using the buffers described by Laemmli (24). An aliquot of cell lysate or purified protein was diluted with at least 1/4 volume of loading buffer (0.04% bromphenol blue–1.6% SDS–27% glycerol in 50 mM Tris-HCl [pH 6.8]) and was applied to an SDS–10% polyacrylamide gel. Electrophoresis was carried out in a Bio-Rad mini-Protean II cell unit at room temperature.
General DNA techniques and transformation.
Chromosomal DNA preparation was performed with the Qiagen (Hilden, Germany) genomic DNA purification system. To prepare plasmid DNA, the Promega (Zürich, Switzerland) Wizard mini or maxi purification system was used. Plasmids, PCR products, and chromosomal DNA were cleaved with the appropriate restriction enzymes, ligated, and transformed into E. coli XL1-Blue or XL2-Blue cells (6, 30). Transformants were selected on LB agar plates containing the appropriate antibiotic: ampicillin (100 μg/ml) for pET-13b and pET-16b, kanamycin (20 μg/ml) for pET-28a, or erythromycin (500 μg/ml) for pJDC9 and its derivatives. Transformation of S. pneumoniae was performed as described by Havarstein et al. (19), with modification. Briefly, frozen aliquots of competent cells were thawed, diluted to 1/10 with prewarmed Streptococcus medium (23), and incubated for 20 min at 37°C in an atmosphere of 10% CO2. Then 1 μl of plasmid DNA (1 μg/μl) was added to 500 μl of the mixture, and incubation continued for an additional 3 h. Transformants were selected on sheep blood (3%) agar plates containing erythromycin (0.5 μg/ml). Competent cells were obtained by growing strain R6 in Todd-Hewitt medium, supplemented with 5% calf serum, to an OD660 of 0.3 to 0.5. The culture was diluted 1/10, 10% glycerol was added, and aliquots were flash-frozen at −80°C.
PCR and sequencing.
PCR was performed with the Expand high-fidelity DNA system (Boehringer, Mannheim, Germany), using the manufacturer’s recommended protocol, in a Perkin-Elmer (Foster City, Calif.) thermocycler. Sequencing was performed by the dideoxy-chain termination method (31), using a modified DNA sequencing kit (dye terminator cycle sequencing; PE Applied Biosystems, Foster City, Calif.), and analyzed with an automated DNA sequencing system (ABI Prism 310 genetic analyzer; PE Applied Biosystems). Nucleotide and amino acid sequences were analyzed with the help of the University of Wisconsin Genetics Computer Group sequence analysis package (15) and with the program Lasergene (DNASTAR, Madison, Wis.).
Construction of the expression plasmids for uppS.
uppS genes from E. coli, H. influenzae, and S. pneumoniae were expressed as His-tag fusion proteins under the control of the T7 promoter (Fig. 1A). In all cases, full-length genes were amplified by using the appropriate forward and reverse primers. The forward primer used introduced a BsaI site (5′-GGTCTCTCATG-3′, for E. coli and H. influenzae) or an NdeI site (5′-GCCATATG-3′, for S. pneumoniae) overlapping the potential methionine starting codon (for E. coli uppS, the GTG immediately upstream of the ATG) and the reverse primer a BamHI site downstream of the stop codon. The PCR products were restricted with BsaI or NdeI and BamHI and then separated on an agarose gel; corresponding bands were eluted and cloned in pET-HTNC restricted with NcoI and BamHI (for E. coli and H. influenzae) and in pET-16b restricted with NdeI and BamHI (for S. pneumoniae), resulting in pUppS-His-EC, pUppS-His-HI, and pUppS-His-SP, respectively (Fig. 1A). pET-HTNC is a derivative of pET-15b which was restricted with NcoI and BamHI and in which the small 72-bp fragment was replaced by a linker which destroys the original NcoI site, regenerates the 6-His tag and a thrombin recognition site, and generates new singular sites for NcoI and BamHI downstream of the His tag (5′-CATGAGCAGCCATCATCACATCATCATAGCAGCGGCCTGGTTCCGCTGGGTTCCATGGTTCG-3′). To express the uppS gene from E. coli as untagged protein, the corresponding PCR product (see above) was cloned in pET-28b restricted with NcoI and BamHI.
Construction of plasmids for generating uppS gene disruption.
For disrupting the uppS gene and terminating expression of the downstream genes in the operon, a small internal fragment of the uppS gene (from bp 149 to 504) was amplified by PCR. The forward primer used introduced a BamHI site upstream of the first nucleotide, and the reverse primer introduced a KpnI site downstream of the last nucleotide. The PCR product was cloned as BamHI-KpnI fragment into pJDC9 (BamHI-KpnI), resulting in pKO1. To have the uppS operon under the control of the two different tetracycline-regulatable promoters, an amino-terminal fragment of the uppS gene (from bp 1 to 504) was amplified by PCR. The forward primer introduced a BsaI site (5′-GGTCTCTCATG…-3′) upstream of the starting methionine codon, and the reverse primer introduced a KpnI site downstream of the last nucleotide. The PCR product was cloned as a BsaI-KpnI fragment in the promoter vectors pRKO2* and pRKO1* (NcoI-KpnI), resulting in plasmids pKO2 and pKO3, respectively (asterisks indicate derivatives of pRKO1 and pRKO2 [34] where the BamHI site is replaced by an NcoI site). To generate a disruption of the uppS gene while placing downstream genes under the control of the tetracycline-regulatable promoter p57opt, an internal fragment of uppS from bp 149 to 504 was amplified by PCR. The forward primer used introduced a BamHI site upstream of the first nucleotide, and the reverse primer introduced a KpnI site downstream of the last nucleotide. The PCR product was cloned as a BamHI-KpnI fragment into the promoter vector pRKO2 (BamHI-KpnI), resulting in plasmid KO4. Recombinants were propagated in E. coli XL2. Mutants were generated in S. pneumoniae R6 by transformation using the protocol described above. The authenticity of each clone was verified by sequencing.
RESULTS
Purification of native Upp synthetase.
In earlier studies, different groups partially purified Upp synthetase from several bacteria (4, 9, 10, 13, 35) by using two or three chromatographic steps, but bacterial genes for Upp synthetase were not identified. As part of our strategy for isolating the gene from E. coli, we purified the Upp synthetase by using a different purification scheme with an additional chromatographic purification step to attain sufficient purity to identify clearly the corresponding band by SDS-PAGE.
For the purification, we started with a total of 185 g (wet weight) of E. coli BL21(DE3) paste collected in the logarithmic growth phase. The Upp synthetase of E. coli was purified as described in Materials and Methods. To monitor Upp synthetase activity during the purification, we used the radioactive Upp synthetase assay for the initial steps of the procedure. Later, when interfering endogenous phosphate had been removed, the coupled assay was used. For the first chromatographic step, we used a TSK-DEAE 650M column. Assaying the fractions revealed two peaks of prenyltransferase activity. The activity in the first peak was absolutely Triton X-100 dependent, in contrast to the second peak; this result was in accordance with observations published earlier (10). The assay used did not distinguish between Upp synthetase and Opp synthetase activities, but we were able to show that recombinant Opp synthetase, which is not Triton X-100 dependent, coeluted from the column with the second peak of prenyltransferase activity (data not shown). Therefore, the activity in the first peak was subjected to four additional chromatographic steps using ceramic hydroxyapatite, TSK-Ether 5PW, HiLoad Superdex 200, and heparin-Actigel ALD, respectively. On the HiLoad Superdex 200 column, Upp synthetase activity eluted at a molecular mass corresponding to the dimer. This is in agreement with earlier findings (10) that Upp synthetase is a dimer. Despite these multiple purification steps, Upp synthetase was not purified to homogeneity (Fig. 2A), possibly because of a very low copy number of Upp synthetase per cell.
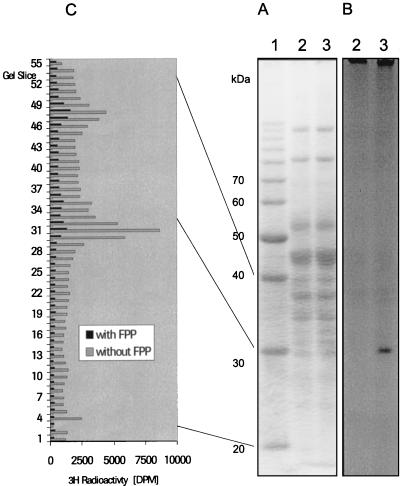
FIG. 2.
SDS-PAGE of heparin column-purified Upp synthetase and radiolabeling with [3H]DAFTP-GDP. (A) Aliquots of the pool from the heparin column-purified Upp synthetase were radiolabeled with [3H]DAFTP-GDP and applied to an SDS-polyacrylamide gel, which then was stained with Coomassie blue. Lanes: 1, protein molecular size markers as indicated on the left; 2, radiolabeled pool in the presence of 20 μM FPP; 3, radiolabeled pool without FPP. (B) Autoradiograph of the same gel. (C) On a duplicate gel, the region between 20 and 40 kDa of lanes 2 and 3 was cut into 55 gel slices each, and the radioactivity in each slice was measured.
Identification of the gene encoding Upp synthetase.
Because we could not purify the Upp synthetase to homogeneity and could not assign the protein to a specific band on an SDS-polyacrylamide gel, it was necessary to further decrease the number of possible candidates. We labeled the native Upp synthetase by cross-linking it specifically with a radiolabeled substrate analogue, [3H]DAFTP-GDP, and subjected the complex to SDS-PAGE (Fig. 2A, lane 3; Coomassie blue staining). Specificity was shown in a parallel sample in which the UV irradiation was carried out in the presence of 20 μM unlabeled FPP (Fig. 2A, lane 2). The entire gel was analyzed for radioactivity in two ways: (i) by direct autoradiography of the dried gel (Fig. 2B; exposure time, >45 days) and (ii) by cutting the lanes from a parallel gel in small 1-mm slices and counting the radioactivity in each slice (Fig. 2C). In both cases, an unambiguous signal corresponding to a band of 29 kDa was detected. These results are in agreement to those of Baba et al. (10), who found, using similar methods, a protein with a size of about 30 kDa. However, with their purification scheme it was not possible to clearly assign an activity to a band on an SDS-polyacrylamide gel. The band of 29 kDa was cut out and subjected to MALDI-MS analysis (see Materials and Methods). The monoisotopic masses found were matched to the theoretical peptide masses of the proteins of E. coli. Three peptides out of 14 matched peptides derived from the protein described in SWISS-PROT entry Q47675, YAES_ECOLI. The sequence of the matching peptides covered 16% of the amino acid sequence of the protein. The sequence coverage is an indication of confidence of protein identification. The selected protein showed the highest sequence coverage.
Deduced amino acid sequence and homology comparison.
The deduced amino acid sequence of SWISS-PROT Q47675 (YAES_ECOLI; designated here uppS [Upp synthetase]), contains 253 residues, which could encode a protein with a deduced total molecular mass of 28,444 kDa. The genomic sequence record indicates GTG as the initiation codon which lies immediately upstream of the alternative start codon. To clarify whether either one or both are used, we determined the N-terminal sequence of the purified Upp synthetase. We identified both forms of Upp synthetase with one or two methionines (ratio of around 3:2). In the subsequent experiments (expression of Upp synthetase), we took the GTG as the initiation codon. Computer-assisted analysis and comparison of DNA sequences were performed with the BLAST program. Comparison of the deduced Upp synthetase amino acid sequence from E. coli with entries in publicly available and in-house databases revealed a total of 28 sequences, including 25 from bacteria (7 of which were only partial sequences), 2 from S. cerevisiae, and 1 from Caenorhabditis elegans. Alignment of these proteins (Fig. 3) revealed strongly conserved regions where the amino acids were identical or nearly identical in all 28 sequences. No function has been experimentally determined for any of the proteins listed.
FIG. 3.
Multiple amino acid sequence alignment of the 28 potential Upp synthetases which show homology to Upp synthetase from E. coli. Shaded areas represent residues that are identical in at least 21 of the 29 sequences (black) and similar amino acids (gray). Amino acid positions are indicated on the right. Sequences: E. coli (ECOLI; SWISS-PROT Q47675) and its homologues from H. influenzae (HAEIN; SWISS-PROT P44938), Helicobacter pylori (HELPY; SWISS-PROT P55984), S. pneumoniae (STRPN; ftp://ftp.tigr.org/pub/data/s_pneumoniae); Bacillus subtilis (BACSU; SWISS-PROT O31751), Aquifex aeolicus (AQUAE; TrEMBL O67291), Archaeoglobus fulgidus (ARCFU; TrEMBL O29049), Borrelia burgdorferi (BORBU; TrEMBL O51146), Mycobacterium tuberculosis (MYCTU; TrEMBL O53434), Mycobacterium leprae (MYCLE; SWISS-PROT P38119), Methanococcus jannaschii (METJA; SWISS-PROT Q58767), Streptomyces fradiae (STRFR; SWISS-PROT P20182), Pseudomonas aeruginosa (PSEAE; unannotated translation from EMBL entry D50811 for the cds gene), Pyrococcus horikoshii (PYRHO; TrEMBL O59258), Synechocystis (SYNY; SWISS-PROT Q55482), Chlamydia trachomatis (CHLTR; TrEMBL G3328883); Methanobacterium thermoauotrophicum (METTH; TrEMBL O26334), Neisseria gonorrhoeae (NEIGO; http://dna1.chem.ou.edu/gono.html), Saccharomyces cerevisiae (YEAST1; SWISS-PROT P35196), S. cerevisiae (YEAST2; SWISS-PROT Q03175), Caenorhabditis elegans (C.ELE; TrEMBL O18007) (amino acids 1 to 275 of 1893 amino acids), Enterococcus faecalis (ENTFA; ftp://ftp.tigr.org/pub/data/e_faecalis) (for this sequence, only the amino-terminal part is available), Corynebacterium glutamicum (CORGL; SWISS-PROT P38118), Treponema pallidum (TREPA; EMBL AE001235), Campylobacter jejuni (CAMJU; http://www.sanger.ac.uk/Projects/C_jejuni), Streptococcus pyogenes (STRPY; http://dna1.chem.ou.edu/strep.html), Brucella abortus (BRUAB; TrEMBL Q44626), Staphylococcus aureus (STAAU; (unreleased Hoffmann-La Roche data). The sequence information for the carboxy-terminal part is available for the last six sequences only. The highly conserved regions of Upp synthetase are indicated by bars and numbered I to V. The following consensus patterns can be derived for each region: I, H-x-x-x-x-M-D-G-N-(RG)-R-(WYF)-A; II, G-H-x-x-G; III, (TS)-x-x-A-F-S-(ST)-E-N-x-x-R-x-x-x-E-V-x-x-L-M-x-L; IV, A-x-x-Y-G-G-R-x-(DE)-(LIVM)-x-x-A; V, (DE)-L-x-I-R-T-(SAG)-G-E-x-R-x-S-N-F-(ML)-(LMP)-W-Q-x-x-Y-(SAT)-E-x-x-F-x-x-x-x-W-P-(DE)-F.
Cloning and overexpression in E. coli of the gene encoding Upp synthetase from different bacteria and purification of recombinant proteins.
To verify the functionality of the Upp synthetase, the genes from E. coli, H. influenzae, and S. pneumoniae were cloned by PCR as N-terminal His-tag fusion proteins under the control of the T7 promoter (pET system) (Fig. 1A). In addition to the fusion proteins, we also expressed the E. coli wt Upp synthetase to compare the activity with those of the His-tag fusion proteins. To generate the expression plasmid, we cloned the DNA fragment encoding the E. coli uppS gene in pET-28b (Fig. 1A). The crude lysates of all four BL21(DE3)(pLysS) strains transformed with the corresponding expression plasmids (induced by 2 mM isopropyl-β-d-thiogalactopyranoside [IPTG] for 2 h) contained more than 1,000-fold-higher Upp synthetase activity than BL21(DE3)(pLysS) transformed with the expression vector without insert (data not shown).
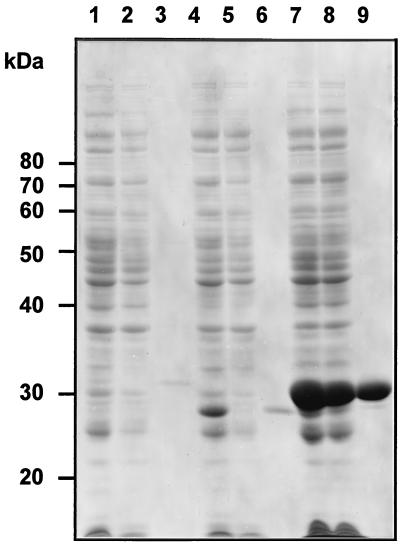
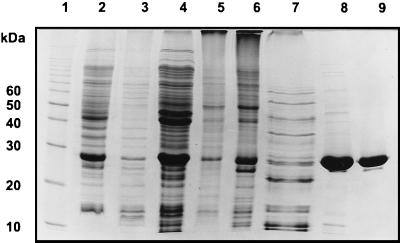
After purifying the fusion proteins from E. coli, H. influenzae, and S. pneumoniae Upp synthetase over a Ni2+ column as described in Materials and Methods, we obtained 0.8, 1.5, 10 mg, respectively, of >95% pure protein (Fig. 4) from 250 ml of induced E. coli culture. In the case of the S. pneumoniae Upp synthetase, the expression was so strong that we overloaded the column, because at least half of the His-tag fusion protein was found in the flowthrough (Fig. 4, lane 8). The apparent molecular masses of the three His-tag fusion proteins were 31, 29, and 30 kDa, respectively, in good agreement to the expected molecular masses of 30.4, 29.3, and 30.7 kDa (including 2 kDa for the His tag). To purify recombinant E. coli wt Upp synthetase, we started with 25 g (wet weight) of paste; after ammonium sulfate precipitation cuts (35 to 50%), Phospho-Ultrogel A6R chromatography, and HiLoad 26/60 Superdex gel filtration chromatography, we obtained about 12 mg of protein at a purity of over 98% (Fig. 5).
FIG. 4.
Purification of the His-tagged Upp synthetase fusion proteins from overproducing E. coli strains. The protein was purified from E. coli BL21(DE3)(pLysS) carrying pUppS-His-EC, pUppS-His-HI, or pUppS-His-SP as indicated below. Samples from various stages of the purification procedure were analyzed by SDS-PAGE, and proteins were visualized after staining with Coomassie blue. Lanes: 1 to 3, pUppS-His-EC; 4 to 6, pUppS-His-HC; 7 and 8, pUppS-His-SP; 1, 4, and 7, soluble cell extract after IPTG induction; 2, 5, and 8, flowthrough from the Ni2+ column; 3, 6, and 9, specific elution from the Ni2+ column with 500 mM imidazole buffer.
FIG. 5.
Purification of the E. coli Upp synthetase protein from the overproducing strain. The protein was purified from E. coli BL21(DE3)(pLysS)(pUppS-wt-EC) as described in the text. Samples from various stages of the purification procedure were analyzed on an SDS-polyacrylamide gel, and proteins were visualized after staining with Coomassie blue. Lanes: 1, protein standards as indicated on the left; 2, soluble cell extract after IPTG induction; 3, 150,000 × g pellet; 4; 150,000 × g supernatant; 5, 0 to 30% saturated (NH4)2SO4 fraction; 6, 30 to 50% saturated (NH4)2SO4 fraction; 7, 50 to 80% saturated (NH4)2SO4 fraction; 8, pool after purification on Phospho-Ultrogel column; 9, pool after purification on Superdex 200 gel filtration column.
Structural and functional analysis of the overproduced proteins.
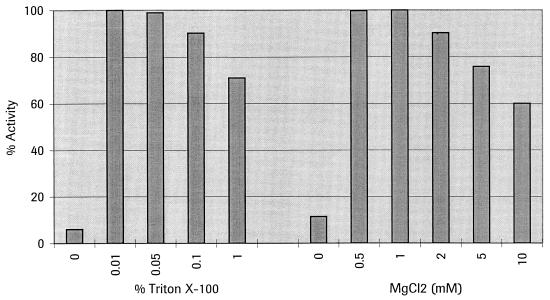
To determine whether we had correctly identified the cloned genes as coding for Upp synthetase, we characterized several aspects of the purified proteins. For the recombinant E. coli wt Upp synthetase, an amino-terminal sequence analysis of the first 10 residues (on an Applied Biosystems 494 protein sequencer) revealed a protein sequence completely consistent with that predicted from the DNA sequence. The overproduced wt E. coli protein comigrates with the band that was radiolabeled after purification of the native Upp synthetase (Fig. 2 and 5). In functional terms (Km values), all four proteins behave more or less identically in the direct and the coupled assays for Upp synthetase. They were absolutely Triton X-100 and MgCl2 dependent, as exemplified in Fig. 6 for the E. coli wt Upp synthetase in the coupled assay. The optimal concentration for Triton X-100 is between 0.01 and 0.1%, and that for MgCl2 is between 0.5 and 2 mM. Omission of either Triton X-100 or MgCl2 results in a residual activity of about 5 to 10%. Using reversed-phase thin-layer chromatography, we found that the major products of the Upp synthetase reaction after potato acid phosphatase treatment had the same mobility as authentic undecaprenol or decaprenol. We detected small amounts of intermediate-length (C30 to C45) products (data not shown).
FIG. 6.
Triton X-100 and MgCl2 dependency of purified E. coli wt Upp synthetase. The coupled assay for Upp synthetase was performed as described in Materials and Methods. The Triton X-100 and MgCl2 concentration was varied in the indicated range. The highest activity in each experiment represents 100%. Results of a representative experiment are shown. Triplicate values varied from 5 to 10%.
Construction and analysis of uppS deletion mutants.
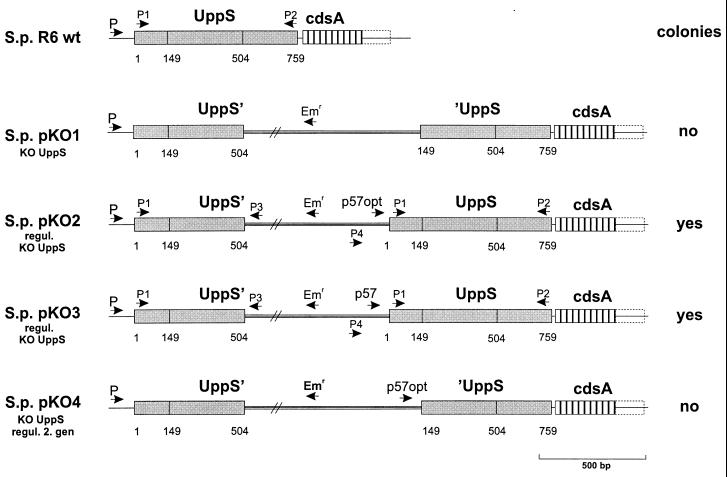
To investigate the function of the uppS gene, pneumococcal uppS gene mutants were obtained by site-directed insertional mutagenesis (29). A small internal gene fragment was cloned in plasmid pJDC9, which does not replicate in pneumococci (pKO1 [Fig. 1B]). The construct was transformed into S. pneumoniae R6, and plasmid-encoded erythromycin resistance (Emr) was used to select for single homologous recombination events. However, no Emr transformants were obtained. To further study the essentiality of the uppS gene for bacterial growth, we used a regulatable knockout system that allows the conditional expression of genes (34). The potential promoter region in front of the streptococcal uppS gene was replaced by the strong tetracycline-regulatable p57opt promoter (pKO2 [Fig. 1B]). Transformation of S. pneumoniae R6 with this plasmid resulted in many Emr transformants (S.p. pKO2 [Fig. 7]). Correct integration of the plasmid was confirmed by PCR analysis (data not shown). No difference in growth rate was detectable in the presence or absence of tetracycline (20 ng/ml), indicating that the basal expression from this promoter was sufficient to allow normal growth of the transformants. Even in the absence of induction, the basal expression level from p57opt is relatively high (34). Attempts to generate viable regulatable uppS knockout mutants by using the weaker and tighter promoter p57 (S.p. pKO3 [Fig. 7]) were successful only in the presence of tetracycline, and we could isolate six colonies. Replating these colonies on sheep blood (3%) agar plates with and without tetracycline resulted in growth on the tetracycline-containing plates, whereas in absence of tetracycline no colonies were detected. In growth experiments in liquid cultures in the presence of tetracycline, S.p. pKO3 characteristically reached a plateau after 5 h with an OD620 of 0.5, starting at an OD620 of 0.08 to 0.1. Without tetracycline in the medium, the regulatable S.p. pKO3 knockouts grow normally for about 2 h, but then they slow down and after 4 h stop further growth at an OD620 of 0.2 to 0.3 (data not shown). The correctness of integration was checked by PCR. Using the primer combination P1-P2 (Fig. 7), we found a band of the expected size (759 bp) for S.p. R6 wt and S.p. pKO3. Using the primer combination P1-P3 or P4-P2 (Fig. 7), we found a band of the expected size, 607 or 792 bp, respectively, only for the regulatable knockout strains and no bands for S.p. R6 wt (data not shown).
FIG. 7.
Schematic representation of the expected chromosomal uppS mutants of S. pneumoniae R6 with various gene disruption plasmids. Numbers indicate the nucleotides, starting with 1 at the initiating codon of each gene. P1 to P4 indicate the primers used in the PCR to confirm the correct integration of the plasmids.
From the S. pneumoniae sequence data, it is very likely that the uppS gene is the first gene in an operon. Therefore, it was necessary to exclude the possibility that the disruption of uppS causes a polar effect on the expression of downstream genes which themselves could be essential. Knowing that the transcription level of the constructs containing the p57opt promoter is sufficient for cell viability, we generated a construct which should disrupt the first gene, uppS, but place the downstream genes under the control of the tetracycline-regulatable promoter p57opt. No Emr transformants were obtained with this plasmid (S.p. pKO4 [Fig. 7]), indicating that the first gene of the operon, uppS, is essential.
DISCUSSION
To gain further insight into the structure and function of Upp synthetase, the only known bacterial enzyme which catalyzes the production of long-chain isoprenoids with cis-chain configuration, we attempted to identify and isolate the Upp synthetase gene from E. coli. Our strategy was to make use of the recently available E. coli genomic sequence by preparing a database containing the masses of peptides produced by computer-simulated trypsin digestion of all E. coli proteins predicted from the E. coli genomic sequence. The masses of peptides produced by experimental trypsin digestion of a purified protein can be used to search the databank for proteins with similar profiles and so identify a candidate gene. However, this approach requires a sample of the protein of interest that is largely free from contaminating proteins. For this reason, it was necessary to improve the purification scheme for the Upp synthetase of E. coli described earlier (10), as analysis of material produced this way resulted in a list of several possible candidate genes (data not shown).
Subjecting 185 g (wet weight) of E. coli paste, after cell breakage, to two differential centrifugation steps, ammonium sulfate precipitation, and fractionation by five different chromatographic steps yielded only approximately 50 to 150 μg of Upp synthetase protein (judged from the band visible after SDS-PAGE). Although this extensive purification scheme was not sufficient to purify the Upp synthetase to homogeneity, the remaining proteins in the mixture could be well resolved by SDS-PAGE. There are several possible explanations for the low yield of Upp synthetase: (i) instability of the protein, which seems to be unlikely since the recombinant protein is stable; (ii) losses during purification; and (iii) very low copy number per cell. Assuming 10−12 g (wet weight) per E. coli cell and an optimistic recovery of 10%, there would be less than 300 to 1,000 Upp synthetase molecules per cell. These numbers are certainly within a possible range, because two steps later in the pathway of cell wall biosynthesis (after dephosphorylation of UPP and transfer of the pentapeptide), the two membrane intermediates in peptidoglycan metabolism, pentapeptide lipid I and lipid II, are present in E. coli at cell copy numbers not higher than 700 and 2,000, respectively (39). In addition, it is interesting that the ribosomal binding site has an extra G relative to the AGGAG consensus and the spacing is rather long and nonoptimal (−11 for GTG and −14 for the ATG), which might cause a low level of transcription. Specific labeling of the purified Upp synthetase preparation with [3H]DAFTP-GDP allowed us to positively identify the protein band on an SDS-polyacrylamide gel that corresponds to Upp synthetase. MALDI-MS analysis of this band’s trypsin-digested profile led to the identification of a single candidate gene in E. coli (SWISS-PROT Q47675, YAES_ECOLI).
Comparison of the 28 predicted Upp synthetase sequences which we identified in various databases revealed several strongly conserved regions (Fig. 3). We detected a single homologous protein in all bacterial genomes for which the full sequence information was available at the time with the exceptions of Mycoplasma genitalium and Mycoplasma pneumoniae, for which no homologue was found. This is consistence with the absence of other cell wall biosynthetic enzymes in these bacteria. In eukaryotic genomes, two genes were identified in S. cerevisiae and one was found in C. elegans. There are two entries in GenBank, AA086931 and W61940, that could encode N-terminal and C-terminal portions of a mouse UppS homolog. In addition, a partial sequence which showed very good homology especially to the strongly conserved regions at the carboxy-terminal end (boxes IV and V [see below]) was found in an in-house human database. Experiments to clone and express the full-length human homologue and to characterize its enzymatic activity are ongoing. It remains to be determined whether that enzyme is identical to the dolichol synthetase in eukaryotes (1, 11, 27), which synthesizes long-chain (up to C100) prenyl pyrophosphates (also in a cis conformation).
Sequence comparison reveals at least five strongly conserved regions as well as some single conserved amino acids (outside of the defined boxes), which very likely represent the active site of the protein (Fig. 3). Box V is analogous to the Prosite consensus sequence described in Prosite document PS01066, which is annotated as “uncharacterized protein family UPF0015 signature” and lists 13 sequences, all of which were also identified in our homology search.
It is interesting that in E. coli the gene immediately preceding uppS is dxr, a 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in the terpenoid biosynthesis pathway (36). This clustering is not found in H. influenzae and S. pneumoniae.
The experiments for generating uppS gene knockouts provide very strong evidence that the gene is essential for growth of S. pneumoniae. We cannot exclude the possibility that some other downstream gene(s) of the operon is essential as well. The tetracycline-regulatable knockouts illustrate the importance of selecting a regulatable promoter of appropriate strength for replacing the natural promoter of a gene and interpreting results cautiously. If basal expression level of the promoter is too high, no difference between induced and uninduced state is detectable. This is the most likely explanation for the recovery of tetracycline-unresponsive transformants using pKO2. Where the promoter is weaker, only in the induced situation is the expression level high enough for cell growth, and in the noninduced situation the cells eventually stop growing. The results obtained with pKO4 and pKO3 provide very strong evidence that the uppS gene is essential for growth of S. pneumoniae. It remains to be shown whether this is also true for the uppS genes of other bacteria.
Upp synthetase shows at least three differences from other known prenyltransferases. (i) All other synthetases, like Fpp synthetase, Opp synthetase, geranylgeranyl pyrophosphate synthetase, and hexaprenyl pyrophosphate synthetase, condense the new isopentenyl unit in a trans configuration on the growing chain. In contrast, Upp synthetase uses trans,trans-FPP as an initiating building block, whereas subsequent condensations of isopentenyl units are in cis conformation. (ii) The primary structure of Upp synthetase is completely different from those of the other prenyltransferases. (iii) All other known prenyltransferases are believed to use the highly conserved aspartate-rich motif (DDxxD) for binding each pyrophosphate. This was demonstrated for avian Fpp synthetase by X-ray crystallography (37). Even isoprenoid cyclases (e.g., pentalenene synthase, epi-aristolochene synthase, and hopene synthase), which also use FPP as substrate but have little overall similarity to chain elongation prenyltransferases, use the DDxxD motif as a binding site for the pyrophosphate, as demonstrated by X-ray crystallography (25, 33, 40). Upp synthetase is a new class of isoprenoid synthetase because it has no such motif. We postulate that there is another motif responsible for the binding of both pyrophosphates. The only related and duplicated sequence motif identified is a single aspartate (boldface) followed by an arginine (underlined) with a spacing of three amino acids (DE)GNxR and DLxLR (positions 25 to 29 and 189 to 193, respectively, in E. coli Upp synthetase) which could potentially serve as the pyrophosphate binding site. From a structural point of view, it would be interesting to examine the three-dimensional structure of Upp synthetase to see how nature solved the problem of pyrophosphate binding in two different ways.
ACKNOWLEDGMENTS
We thank T. Hartung for preparing (E,E)-[1-3H]-(2-diazo-3-trifluoropropionyloxy)geranyl diphosphate, Marie-Françoise Takács for helping in purification of the proteins, and Elizabeth Gillick and Richard Moon for critically reading the manuscript. We also acknowledge Olivier Partouche, Bernard Rutten, and Christian Lacoste (all from F. Hoffmann-La Roche Ltd., Basel, Switzerland) for expert technical assistance.
We acknowledge the Streptococcal Genome Sequencing Project, funded by USPHS/NIH grant AI38406, and B. A. Roe, S. P. Linn, L. Song, X. Yuan, S. Clifton, M. McShan, and J. Ferretti; the Gonococcal Genome Sequencing Project, funded by USPHS/NIH grant AI38399, and B. A. Roe, S. P. Lin, L. Song, X. Yuan, S. Clifton, Tom Ducey, Lisa Lewis, and D. W. Dyer; and the C. jejuni Sequencing Group at the Sanger Center and the E. faecalis early releases from The Institute for Genomic Research.
ADDENDUM
During the review process, a paper describing the Micrococus luteus UppS protein and gene was published (32).
REFERENCES
- 1.Adair W L, Cafmeyer N, Keller R K. Solubilization and characterization of the long chain prenyltransferases involved in dolichyl phosphate biosynthesis. J Biol Chem. 1984;259:4441–4446. [PubMed] [Google Scholar]
- 2.Allen C M, Keenan M V, Sack J. Lactobacillus plantarum undecaprenyl pyrophosphate synthetase: purification and reaction requirements. Arch Biochem Biophys. 1976;175:236–248. doi: 10.1016/0003-9861(76)90504-x. [DOI] [PubMed] [Google Scholar]
- 3.Allen C M, Muth J D. Lipid activation of undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum. Biochemistry. 1977;16:2908–2915. doi: 10.1021/bi00632a017. [DOI] [PubMed] [Google Scholar]
- 4.Allen C M. Purification and characterization of undecaprenyl-pyrophosphate synthetase. Methods Enzymol. 1985;110:281–299. doi: 10.1016/s0076-6879(85)10085-6. [DOI] [PubMed] [Google Scholar]
- 5.Asai K, Fujisaki S, Nishimura Y, Nishino T, Okada K, Nakagawa T, Kawamukai M, Matsuda H. The identification of E. coli ispB (cel) gene encoding the octaprenyl diphosphate synthase. Biochem Biophys Res Commun. 1994;202:340–345. doi: 10.1006/bbrc.1994.1933. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1997. [Google Scholar]
- 7.Avery O T, MacLeod C M, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baba T, Allen C M. Substrate specificity of undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum. Biochemistry. 1978;17:5598–5604. doi: 10.1021/bi00619a003. [DOI] [PubMed] [Google Scholar]
- 9.Baba T, Allen C M. Prenyl transferases from Micrococcus luteus: characterization of undecaprenyl pyrophosphate synthetase. Arch Biochem Biophys. 1980;200:474–484. doi: 10.1016/0003-9861(80)90379-3. [DOI] [PubMed] [Google Scholar]
- 10.Baba T, Muth J, Allen C M. Photoaffinity labeling of undecaprenyl pyrophosphate synthetase with a farnesyl pyrophosphate analogue. J Biol Chem. 1985;260:10467–10473. [PubMed] [Google Scholar]
- 11.Bukhtiyarov Y E, Shabalin Y A, Kulaev I S. Solubilization and characterization of dehydrodolichyl diphosphate synthase from the yeast Saccharomyces carlbergensis. J Biochem. 1993;113:721–728. doi: 10.1093/oxfordjournals.jbchem.a124110. [DOI] [PubMed] [Google Scholar]
- 12.Chen J D, Morrison D A. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA of Streptococcus pneumoniae. Gene. 1988;64:155–164. doi: 10.1016/0378-1119(88)90489-1. [DOI] [PubMed] [Google Scholar]
- 13.Christenson J G, Gross S K, Robbins P W. Enzymatic synthesis of the antigen carrier lipid. J Biol Chem. 1969;244:5436–5439. [PubMed] [Google Scholar]
- 14.Collins M D, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implementations. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fountoulakis M, Langen H. Identification of proteins by matrix-assisted laser desorption ionization-mass spectrometry following in-gel digestion in low-salt, nonvolatile buffer and simplified peptide recovery. Anal Biochem. 1997;250:153–156. doi: 10.1006/abio.1997.2213. [DOI] [PubMed] [Google Scholar]
- 17.Fujisaki S, Nishino T, Katsuki H. Isoprenoid synthesis in Escherichia coli. Separation and partial purification of four enzymes involved in the synthesis. J Biochem. 1986;99:1327–1337. doi: 10.1093/oxfordjournals.jbchem.a135600. [DOI] [PubMed] [Google Scholar]
- 18.Fujisaki S, Hara H, Nishimura Y, Horiuchi K, Nishino T. Cloning and nucleotide sequence of the ispA gene responsible for farnesyl diphosphate synthase activity in E. coli. J Biochem. 1990;108:995–1000. doi: 10.1093/oxfordjournals.jbchem.a123327. [DOI] [PubMed] [Google Scholar]
- 19.Havarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keenan M V, Allen C M. Characterization of undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum. Arch Biochem Biophys. 1974;161:375–383. doi: 10.1016/0003-9861(74)90318-x. [DOI] [PubMed] [Google Scholar]
- 21.Kodama T, Fukui K, Kometani K. The initial phosphate burst in ATP hydrolysis by myosin and subfragment-1 as studied by a modified malachite green method for determination of inorganic phosphate. J Biochem. 1986;99:1465–1472. doi: 10.1093/oxfordjournals.jbchem.a135616. [DOI] [PubMed] [Google Scholar]
- 22.Kurokawa T, Ogura K, Seto S. Formation of polyprenyl phosphates by a cell-free enzyme of Micrococcus lysodeikticus. Biochem Biophys Res Commun. 1971;45:251–257. doi: 10.1016/0006-291x(71)90077-5. [DOI] [PubMed] [Google Scholar]
- 23.Lacks S. Integration efficiency and genetic recombination in pneumococcal transformation. Genetics. 1966;53:207–235. doi: 10.1093/genetics/53.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lesburg C A, Zhai G, Cane D E, Christianson D W. Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Stipanovic R D, Benedict C R. Synthesis of a tritium labeled photolabile analogue of farnesyl-diphosphate: (E,E)-[1-3H]-(2-diazo-3-trifluoropropionyloxy)geranyl diphosphate. J Labelled Compd Radiopharm. 1996;38:139–148. [Google Scholar]
- 27.Matsuoka S, Sagami H, Kurisaki A, Ogura K. Variable product specificity of microsomal dehydrodolichyl diphosphate synthase from rat liver. J Biol Chem. 1991;266:3464–3668. [PubMed] [Google Scholar]
- 28.Ogura K, Koyama T, Sagami H. Polyprenyl diphosphate synthases. Subcell Biochem. 1997;28:57–87. doi: 10.1007/978-1-4615-5901-6_3. [DOI] [PubMed] [Google Scholar]
- 29.Pozzi G, Oggioni M R, Manganelli R, Plevani P. Genetic manipulation of streptococci by chromosomal integration of recombinant DNA. In: Dunny G M, Cleary P P, McKay L L, editors. Genetics and molecular biology of streptococci, lactococci, and enterococci. 1991. pp. 59–61. Washington, D.C. [Google Scholar]
- 30.Sambrook R, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;71:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu N, Koyama T, Ogura K. Molecular cloning, expression, and purification of undecaprenyl diphosphate synthase: no sequence similarity between E- and Z-prenyl diphosphate synthases. J Biol Chem. 1998;273:19476–19481. doi: 10.1074/jbc.273.31.19476. [DOI] [PubMed] [Google Scholar]
- 33.Starks C M, Back K, Chappell J, Noel J P. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 34.Stieger, M., B. Wohlgensinger, M. Kamber, R. Lutz, and W. Keck. Integrational plasmids for the tetracycline regulated expression of genes in Streptococcus pneumoniae. Gene, in press. [DOI] [PubMed]
- 35.Takahashi I, Ogura K. Prenyltransferase of Bacillus subtilis: undecaprenyl pyrophosphate synthetase and geranylgeranyl pyrophosphate synthetase. J Biochem. 1982;92:1527–1537. doi: 10.1093/oxfordjournals.jbchem.a134077. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi S, Kuzuyama T, Watanabe H, Seto H. A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc Natl Acad Sci USA. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarshis L C, Proteau P J, Kellogg B A, Sacchettini J C, Poulter C D. Regulation of product chain length by isoprenyl diphosphate synthases. Proc Natl Acad Sci USA. 1996;93:15018–150223. doi: 10.1073/pnas.93.26.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umbreit J N, Strominger J L. Isolation of the lipid intermediate in peptidoglycan biosynthesis from Escherichia coli. J Bacteriol. 1972;122:1306–1309. doi: 10.1128/jb.112.3.1306-1309.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Heijenoort Y, Gómez M, Derrien M, Ayala J, van Heijenoort J. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J Bacteriol. 1992;174:3549–3557. doi: 10.1128/jb.174.11.3549-3557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wendt K U, Poralla K, Schulz G E. Structure and function of a squalene cyclase. Science. 1997;277:1811–1815. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]