Abstract
The study aims at detecting and characterizing haplotypes of red palm weevil (RPW) Rhynchophorus sp. in the Western region of Saudi Arabia based on the cytochrome oxidase subunit I (COI) gene sequence. The results indicated the occurrence of 17 nucleotide substitutions, of which three were nonsynonymous (NS). These three NS substitutions resulted in the variation in amino acid sequence in three positions, out of 133. These amino acids are isoleucine/valine, glycine/serine, and arginine/histidine. Based on the chemical properties of the cytochrome C oxidase (COX) enzyme, it is likely that the change at the first position has no effect, while changes at the other two positions can affect the chemical properties of the enzyme. At the three-dimensional (3D) level, the first two positions exist at the border or inside loop 3–4 of the enzyme, while the third position exists inside loop 4–5. These two loops influence the folding pattern of the enzyme, thus, likely affecting the function of the enzyme. However, it is unlikely that variations in the three positions will affect the binding ability of the heme group, which promotes the action of the COX enzyme in the electron transport chain. Variations in chemical properties and 3D structure of COX enzyme might be an evolutionary process (positive selection) that promotes in-time and in-site adaptation to the insect. In conclusion, this study can be helpful in pest management programs and in tracing RPW geographic spread and migration in Saudi Arabia.
Keywords: Date palm, Rhynchophorus, COI gene, Barcode, Makkah Al Mukarramah Region
1. Introduction
Date palm Phoenix dactylifera L. is one of the main pillars of the agriculture sector in MENA countries. It has been cultivated since the early civilizations as a major part of the local heritage with several religious and social symbolism. There are several cultivars of date palm producing different types of sweet drupes that can be consumed dry or soft, or can be pitted, stuffed, and transformed to be used in different culinary and confectionary preparations (Jain et al., 2011). Date palm fruits are considered a staple food with several nutritional and health benefits due to their high content in fiber, sugar, amino acids, oligo-elements. Moreover, the fruit is very rich in secondary metabolites like tocopherol, flavonoids, tannins, and terpenoids that have a wide range of medicinal and biological (e.g., bactericidal, fungicidal, antioxidant, anticancer, and anti-ulcer) properties (Vayalil, 2002, Al-Farsi et al., 2005, Baloch et al., 2006, El Hadrami and Al-Khayri, 2012, Oni et al., 2015). MENA region possesses around 84 million date palm trees out of 120 million trees worldwide contributing about 67% of the global date production (https://www.bioenergyconsult.com/date-palm-biomass).
Date palm pests can significantly reduce the yield by up to 75% causing irreversible damage to palm tree oasis (Haldhar et al., 2017). Red palm weevil Rhynchophorus ferrugineus in the Arabic peninsula and the parasitic fungus Bayoud in North Africa are becoming the most serious and common threats to date palm cropping in these regions. RPW has a large economic burden and is the most damaging pest of date palm species in Saudi Arabia. Since it was first reported in the Indian subcontinent (Ghosh, 1912), RPW was proven to attack 40 different palm species including date palms, coconut, areca palms, and many other ornamental palm species (Ojeu, 2008, Anonymous, 2013, Al-Saroj et al., 2017, EPPO, 2020). It is almost present everywhere in the globe and raises concerns about the future of palm tree cultivation in the MENA region (Murphy and Briscoe, 1999, Bozbuga and Hazir, 2008, EPPO, 2008, Melifronidou-Pantelidou, 2009, Pelikh, 2009, CABI, 2020). In Saudi Arabia, RPW was first recorded by the Ministry of Environment, Water, and Agriculture in date palm farms in the Eastern region (Alqatif province). Since then, it subsequently invaded new areas and it is almost found in all provinces in KSA including AL- Riyadh, AL-Madinah, Eastern region, Makkah Al-Mukarramah, Hail, Al-Bahah, Al-Qassim, Western region, Tabuk, and Najran (MOA, 2014).
Red palm weevil is about three cm long with a long-curved rostrum and reddish-brown color. They can fly without interruption for more than one km. Marked insects were found Seven km away after five days from the initial release site. Thus, RPW can be considered a very strong fly and highly invasive pest. A single female of RPW may lay more than 300 eggs and all biological stages (e.g., eggs, larvae, pupae, and adults) can be found inside one tree. Larvae excavate the trunk and feed until the destruction of the vascular system and collapse of the tree. It requires around four months to differentiate from egg to adult stage. The early infestation stages are difficult to be discovered, but when the symptoms appear, then the tree is no longer capable to survive and adult weevils start to leave it to infest new ones (Sallam et al., 2012). Thus, the management control approach and strategy are costly and very difficult to apply.
Documentation of insect diversity in Saudi Arabia has recently taken place (Sabir et al., 2019) based on the criteria of the International Union for Conservation of Nature that allows recognition and map threatened as well as economically damaging insect species. However, it is a challenge to discriminate insect species at the morphological level in addition to the huge number of cryptic species making the global species count uncertain (Scheffers et al., 2012). Several genetic markers have been used to investigate the diversity of RPW in the Middle East and Mediterranean Basin, including Saudi Arabia, of which is the cytochrome oxidase subunit I (COI) gene (Al-Ayied et al., 2006, El-Mergawy et al., 2011, Rugman-Jones et al., 2013, Sadder et al., 2015, Sabir et al., 2019, Sukirno et al., 2020). DNA barcoding, originally generated by Hebert et al. in 2003, is becoming a satisfactory alternative. This approach relies on the use of the COI gene encoding subunit I (658 nt) of the mitochondrial cytochrome C oxidase (COX) enzyme that became the most universal marker for the animal, especially insects, species identification (Hebert et al., 2003, Hebert et al., 2004, Pentinsaari et al., 2016). The originators developed a Barcode Index Number (BIN) system (Ratnasingham and Hebert, 2013) that acts as a powerful alternative to morphological species for easily distinguishing among species and instant scoring of diversity or occurrence of speciation (Hebert et al., 2013; 2016; Cristescu, 2014, Gwiazdowski et al., 2015, Mutanen et al., 2015, Borges et al., 2016, Ashfaq and Hebert, 2016, Ren et al., 2018). COX enzyme originally participates in the electron transport chain across the inner mitochondrial membrane. The encoded protein of the COI gene comprises 219 amino acids (AAs) to form a six-polypeptide chain structure bound to two heme groups (Tsukihara et al., 1995, Tsukihara et al., 1996, Tsukihara et al., 2003, Balsa et al., 2012).
The study aims to characterize RPW haplotypes in the Western region of Saudi Arabia based on COI gene at DNA and amino acids (AAs) levels compared with the published haplotypes either in Saudi Arabia or in the MENA region (El-Mergawy et al., 2011, Sukirno et al., 2020). This study also deciphers some evolutionary dynamics of the insect towards its smart decisions of in-time and in-site migration. In addition, changes in COI protein structure of the generated haplotypes at the three-dimensional (3D) level compared to that of cattle (Bos taurus) (Tsukihara et al., 1995) are projected to scope the light on the possible structural–functional architecture of COX enzyme that promotes electron transport and the ability to migrate.
2. Materials and methods
2.1. Collection of red palm weevil samples
Samples of red palm weevils were collected from three governorates in the Western region of Saudi Arabia, namely Taif, Makkah, and Jeddah (Fig. 1). Collection spots were selected based on the information received from the Ministry of Environment, Water, and Agriculture, KSA for areas with the most infested palm trees in this region. Collection of insects from the infested date palm trees was made using pesticide-free traps, following the method of Abraham et al. (2006), or otherwise, insects were collected manually during removal of damaged trees.
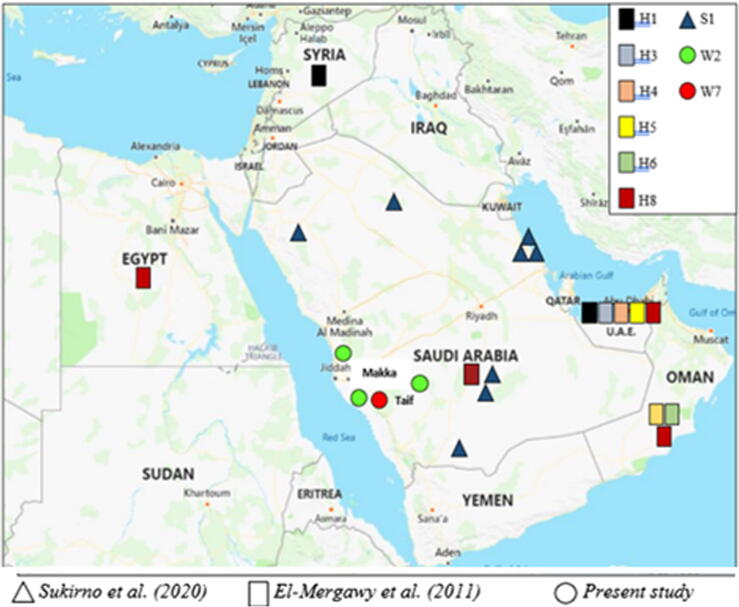
Fig. 1.
Collection spots of red palm weevils were collected from three governorates in the Western region of KSA, namely Jeddah, Makkah, and Taif. The figure shows collection spots in the MENA region of other studies of Sukirno et al. (2020) and El-Mergawy et al. (2020). Same haplotypes have the same color. All samples collected in the present study refer to two haplotypes where the first (green circle) includes seven samples collected from Jeddah governorate (e.g., W1, W2, and W3), Makkah (e.g., W5 and W9), and Taif governorate (e.g., W12, W13 and W14), while the second haplotype (red circle) includes one sample collected from Makkah governorate (e.g., W7).
2.2. DNA isolation and PCR amplification
Total genomic DNAs were extracted from different samples of Rhynchophorus sp. using QIAamp DNA Mini kit (Germany) following the manufacturer’s protocol. PCR was performed to recover cytochrome C oxidase I (COI(gene fragment of ∼650 nt as previously described (Folmer et al., 1994). The reaction was performed in a total volume of 50 µL containing 1x GoTaq®Green Master Mix (Promega, USA), 50 ng of genomic DNA, and 1 µM from each of the universal forward LCO1490 (5‘GGTCAACAAATCATAAAGATATTGG3‘) and reverse HCO2198 (5‘TAAACTTCAGGGTGACCAAAAAATCA3‘) primers (Vrijenhoek, 1994). PCR cycling conditions were as follows: 94 °C for 5 min, followed by 32 cycles of 94 °C for 30 sec, 50 °C for 30 sec and 68 °C for 1 min and a final extension step at 68 °C for 10 min (Sabir et al., 2019). Recovered amplicons were analyzed by agarose gel electrophoresis (1.2%). The molecular size of the amplified products was estimated using a 100-bp DNA ladder (Genedirex, South Korea).
2.3. Sanger sequencing and bioinformatics
Amplicons were purified using a QIAquick PCR Purification kit (Qiagen, Germany), then shipped for Sanger sequencing at Macrogen, South Korea using the two universal primers LCO1490 and HCO2198. Obtained sequences were visually cleaned and trimmed and clean sequences were aligned together using MEGA 6.0 software (Tamura et al., 2013).
Then, sequences were subjected to haplotyping using an Identification Engine publicly available (https://www.boldsystems.org/index.php/IDS_OpenIdEngine). For investigating haplotype barcodes, we recruited previous information of Rhynchophorus sp. haplotypes, based on cytochrome oxidase subunit I, in the MENA region as well as in other regions of the world. These haplotypes were published by El-Mergawy et al. (2011), e.g., H1, H3-H6 and H8, and Sukirno et al. (2020), e.g., S1. The haplotypes of El-Mergawy et al. (2011) were deposited in the National Center for Biotechnology Information (NCBI) as accession no. GU581582 for H1, GU581548 for H3, GU581545 for H4, GU581539 for H5, GU581526 for H6 and GU581519 for H8, while that of Sukirno et al. (2020) was deposited in the Barcode of Life Database System (BOLD Systems http://boldsystems.org/) under accession numbers RPWSA001–16 for S1, while had analogue sequence in the NCBI under accession no. MH016278. Then, genetic distances (the number of nucleotide substitutions per site) were estimated, and a phylogenetic tree, based on DNA COI gene sequence, was constructed using MEGA 6.0 software with maximum like hood (MLH) method (Tamura and Nei, 1993, Tamura et al., 2013).
Codon positions of the mRNA after trimming, which include 1st + 2nd + 3rd + Noncodings, were analyzed, and deduced AA sequences were generated and compared among haplotypes. Nucleotide substitution patterns, e.g., synonymous (S) and nonsynonymous (NS), were also generated using the joint Maximum Likelihood reconstructions of ancestral states (Muse and Gaut, 1994) of codon substitution and Felsenstein model (Felsenstein, 1981) of nucleotide substitution. Changes in AA sequences were scored referring to the generated consensus haplotype sequence.
The protein three-dimensional structure of the assigned consensus haplotype was predicted at I-TASSER Suite (Yang et al., 2015) and aligned to the AA sequence of cattle (Bos taurus) that is routinely used as a COI protein reference (Tsukihara et al., 1995). The bovine protein X-ray structure (Protein Data Bank ID 1OCC) was used as it is the recommended homology model of the COI barcode used in predicting 3D structures of other COI proteins (Pentinsaari et al., 2016).
3. Results and discussion
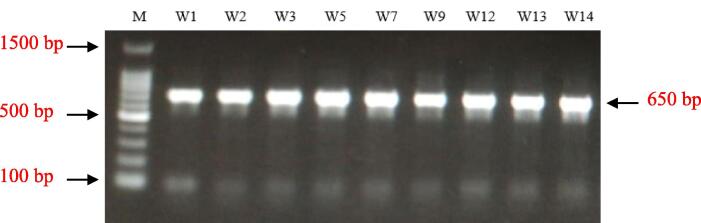
In the present study, 18 specimens of RPW were collected from three governorates in the Makkah region, namely Jeddah, Makkah, and Taif. Insects were morphologically characterized based on exoskeleton structure according to the Wattanapongsiri (1966). We claim that all collected samples phenotypically belong to Rhynchophorus ferrugineus as species identification using bold system showed 100% sequence similarity. Then, we approached to investigate RPW at a molecular level as recently described (Mujahid et al., 2018). Based on the quality of isolated DNA, the number of samples was narrowed to nine. These samples were given codes W1, W2, and W3 for those collected from Jeddah governorate, W5, W7, and W9 for those collected from Makkah governorate, while W12, W13, and W14 for samples collected from Taif governorate (Fig. 1 and Table 1). PCR to amplify the COI gene successfully generated the correct amplicon size (619 nt) (Fig. 2). Amplicons were shipped for sequencing as previously indicated and the generated molecular data (Fig. S1) was submitted to NCBI to receive accession numbers.
Table 1.
Codes of RPW samples were collected from spots in three governorates of the Western region of KSA (e.g., Jeddah, Makkah, and Taif).
| Code | Collection spot |
|---|---|
| W1 | Jeddah |
| W2 | |
| W3 | |
| W5 | Makkah |
| W7 | |
| W9 | |
| W12 | Taif |
| W13 | |
| W14 |
Fig. 2.
PCR products of the COI gene fragment (∼650 bp) amplified from DNAs of red palm weevil samples collected from Jeddah (e.g., W1, W2, and W3), Makkah (e.g., W5, W7, and W9), and Taif (e.g., W12, W13 and W14) governorates. M = 100-bp DNA ladder (Genedirex, South Korea, 100–1500 bp).
Results of the multiple DNA sequence alignment indicated that the nine RPW samples fall within two haplotypes (Fig. 3). One of these haplotypes (e.g., W7) exists in Makkah, while the other haplotype exists in the three governorates of Makkah region referring to the other eight samples (W1, W2, W3, W5, W9, W12, W13, and W14). Therefore, we decided to use the term W2 in further analysis referring to the second haplotype. Through the recent study conducted by Sukirno group, DNA barcodes of the RPW haplotypes in Saudi Arabia were generated (Sukirno et al., 2018a, Sukirno et al., 2018b, Sukirno et al., 2020), but the authors did not cover Makkah region. Bioinformatics analysis of Sukirno’s haplotypes indicated that the submitted sequencing data to the gene bank was not processed, which explains the existence of large variations in the generated haplotypes. Therefore, we decided to use a consensus COI gene sequence detected in the present study (namely S1, Fig. S2) for studying haplotypes growing in Saudi Arabia (Fig. 1). Then, we had to search COI gene sequences of other earlier studies in the region that complement those recovered from the present study. We recognized one successful study in generating RPW haplotypes in Saudi Arabia as well as in other African and Asian countries. This study was done by El-Mergawy’s group (El-Mergawy et al. 2011) who detected COI gene sequences for a number of several haplotypes (namely H1, H3-H6, and H8) existing in the MENA region (Fig. S2). Of which, the H8 haplotype was collected from Al-Ahsa (The eastern region of KSA) and described by El-Mergawy et al. (2011) as the international COI haplotype of RPW.
Fig. 3.
Multiple DNA sequence alignment of the COI gene fragment (619 nt) of red palm weevil samples collected from Jeddah (e.g., W1, W2 and W3), Makkah (e.g., W5 and W9) and Taif (e.g., W12, W13 and W14) governorates.
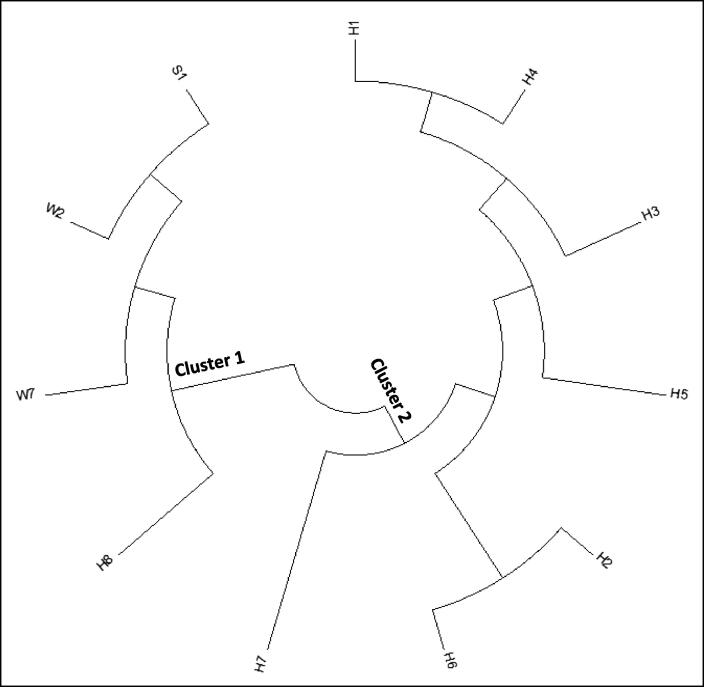
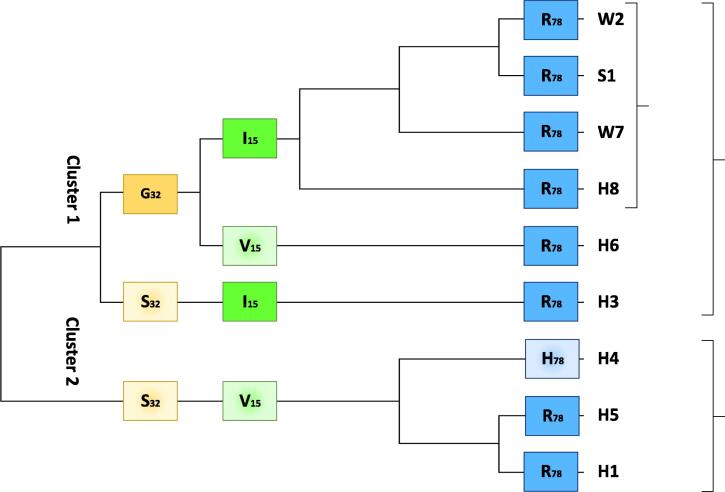
Barcodes (Fig. 4) and multiple DNA sequence alignment (Fig. 5) of the COI gene fragment of the RPW haplotypes collected from Makkah region (e.g., W2 and W7), as well as those of Sukirno et al. (2020) (e.g., S1) and El-Mergawy et al. (2011) (e.g., H1, H3-H6, and H8), indicated that fragment sizes are 619, 618 and 545 nt, respectively. Trimming of non-common DNA sequences of these haplotypes resulted in the recovery of a common DNA sequence of 399 nt. This common region of COI gene fragment in the nine haplotypes was used in generating the phylogenetic tree (Fig. 6). The results indicated the occurrence of two main clusters. The first cluster involves haplotypes W2, W7, S1, and H8, while the second main cluster involves the rest of the haplotypes. The first main cluster also indicates a close relationship between W2 and S1 haplotypes, on one hand, and W7 and H8, on the other hand. The second main cluster indicates that H7 is the most genetically distant among all the nine haplotypes. Also, haplotypes H2 and H6 fell in a separate subcluster (Fig. 6). Overall, we concluded that the international haplotype H8, as well as the S1 haplotype, is the closest to the two haplotypes of the Western region of Saudi Arabia.
Fig. 4.
Barcodes of COI haplotypes of RPW samples collected from governorates of the Western region of KSA, namely Jeddah, Makkah, and Taif (e.g., W2 and W7) (619 nt) in addition to the haplotype consensus sequence of Sukirno et al. (2020) (e.g., S1) (618 nt) and haplotypes of MENA region of El-Mergawy et al. (2011) (e.g., H1, H3-H6, and H8) (545 nt). Haplotype W2 represents samples collected from Jeddah (e.g., W1, W2, and W3), Makkah (e.g., W5 and W9), and Taif (e.g., W12, W13, and W14) governorates.
Fig. 5.
Multiple DNA sequence alignment of the COI gene fragment of RPW haplotypes of the present study (619 nt) (e.g., W2 and W7) in addition to the haplotype with consensus sequence of Sukirno et al. (2020) (618 nt) (e.g., S1) and haplotypes of MENA region of El-Mergawy et al. (2020) (545 nt) (e.g., H1, H3-H6 and H8). Haplotype W2 represents samples collected from Jeddah (e.g., W1, W2 and W3), Makkah (e.g., W5 and W9) and Taif (e.g., W12, W13 and W14) governorates.
Fig. 6.
Phylogenetic tree based on DNA sequence alignment of trimmed COI gene fragment of RPW haplotypes of the present study (e.g., W2 and W7) in addition to the haplotype consensus sequence of Sukirno et al. (2020) (e.g., S1) and haplotypes of different regions in the world of El-Mergawy et al. (2011) (e.g., H1-H8). Haplotype W2 represents samples collected from Jeddah (e.g., W1, W2, and W3), Makkah (e.g., W5 and W9), and Taif (e.g., W12, W13, and W14) governorates. The tree divided haplotypes based on COI gene sequence into two clusters. The first involves the international haplotype H8 that seems to be closest to the two haplotypes of the present study (e.g., W2 and W7) and the consensus haplotype of Sukirno et al. (2020) (e.g., S1).
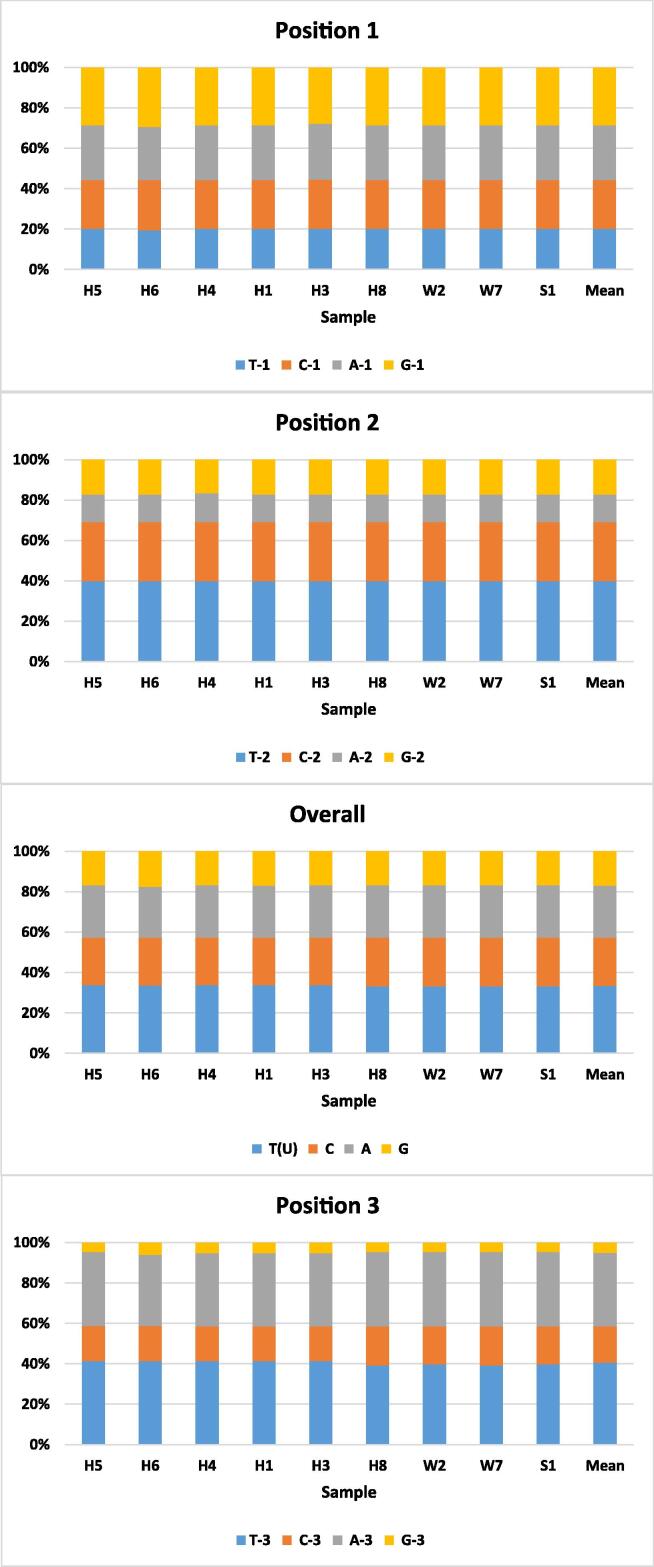
As indicated by Pentinsaari et al., 2016, Sabir et al., 2019, animal mitochondrial DNA is AT-rich (Fig. 7). The DNA sequences of COI gene fragment (399 nt) analyzed for the nine haplotypes have AT content of ∼60%. The AT content was lowest (∼47%) for the nucleotides in the first position across the 133 codons, while ∼53% in the second position and as high as ∼77% in the third position (Fig. 7). The G nucleotide was shown to be the least among the four nucleotides in the third position (∼5.0%) and across the three positions (∼17%). Nucleotide A in the second position was unexpectedly lowest (<14%). Overall results align with those of Sabir et al. (2019) in their study on different insect orders.
Fig. 7.
Nucleotide frequencies at each of the three codon positions in COI sequences of RPW haplotypes of the present study (e.g., W2 and W7) in addition to the haplotype with consensus sequence of Sukirno et al. (2020) (e.g., S1) and haplotypes of MENA region of El-Mergawy et al. (2011) (e.g., H1, H3-H6, and H8). Haplotype W2 represents samples collected from Jeddah (e.g., W1, W2, and W3), Makkah (e.g., W5 and W9), and Taif (e.g., W12, W13, and W14) governorates.
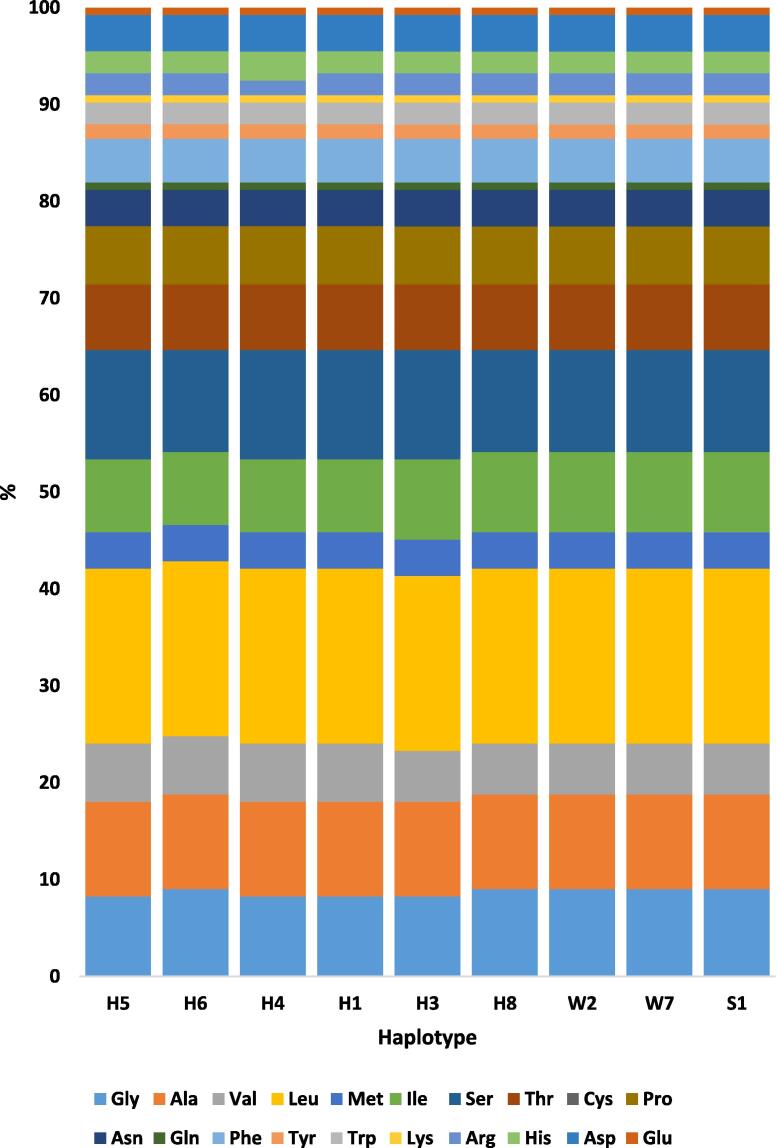
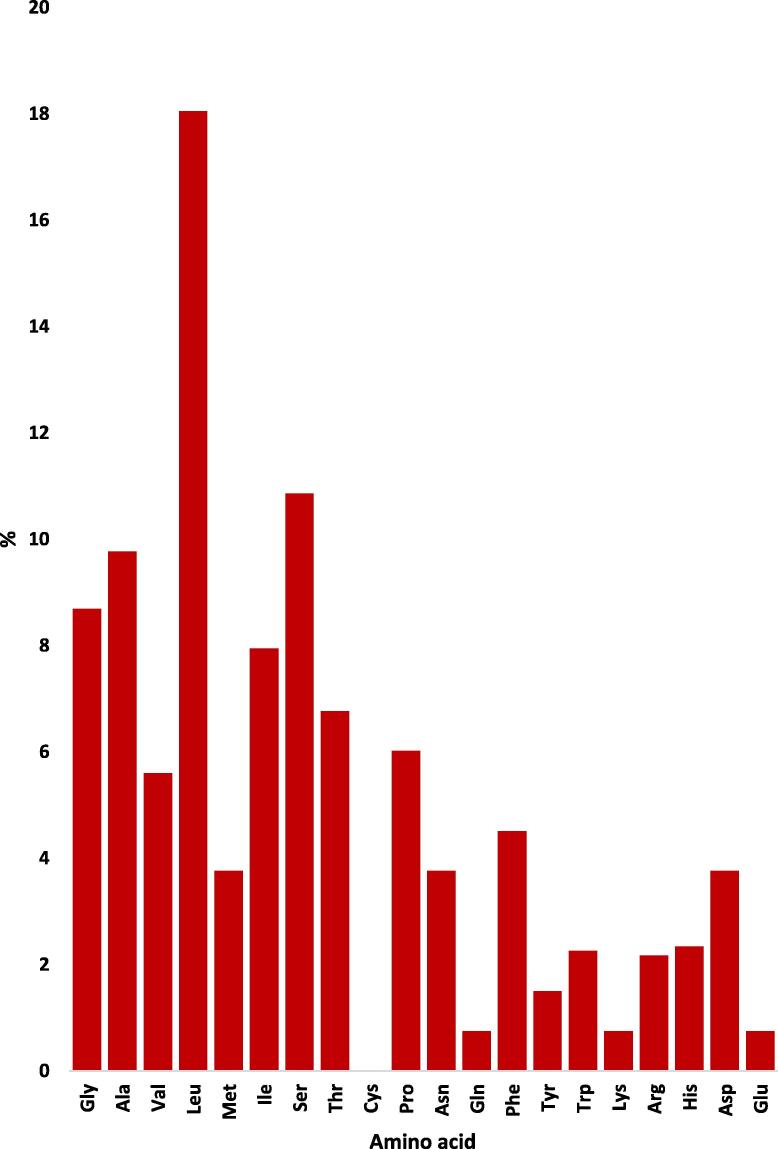
Amino acid sequences of the nine RPW haplotypes were arranged from left to right - based on their chemical properties - into the five standard groups: nonpolar aliphatic (G, A, V, L, M and I), polar uncharged (S, T, C, P, N, and Q), aromatic (F, Y, and W), positively charged (K, R, and H) and negatively charged (D and E) (Fig. 8). Percentages of AAs in COI indicate no clear differences among RPW haplotypes. The COI gene sequences of different haplotypes mostly encode four nonpolar aliphatic AAs namely glycine (G), alanine (A), leucine (L), and isoleucine (I), and two polar uncharged namely serine (S) and threonine (T) (Fig. 9). Interestingly, cysteine (C) was completely absent in all RPW haplotypes. This AA does not also exist in the COI protein sequence of cattle (Pentinsaari et al., 2016). The absence of cysteine in the COI sequence can affect the secondary structure as well as stabilization of tertiary and quaternary structures of the COI protein in RPW compared to this protein in other insects or animals. Three other AAs seem to be encoded in very small quantities. They are glutamine (Q), tyrosine (Y), and glutamic acid (E). Similar results were previously obtained by Pentinsaari et al. (2016) when studying Coleopteran and Lepidopteran species and Sabir et al. (2019) when studying almost all insect orders. Based on the criterion followed by Pentinsaari et al. (2016), the substitution of AAs with the same chemical property may not make a significant influence on enzyme function. Thus, the rare events of such AAs can possibly be compensated by AAs of the same chemical groups. For example, the polar uncharged serine and threonine existing at ∼11 and ∼7%, respectively, can compensate for the complete absence of cysteine (C) and the low abundance of glutamine (Q), while the positively charged arginine (R) and histidine (H), existing at ˃2%, can compensate the low abundance of lysine (K). In addition, aspartic acid, existing at ∼4%, can compensate glutamic acid (E) (Fig. 9). Interestingly, the four rare AAs (C, Q, K, and E) are encoded by twofold degenerate codons.
Fig. 8.
Percentages of amino acids within COI sequences of RPW haplotypes of the present study (e.g., W2 and W7) in addition to the haplotype with consensus sequence of Sukirno et al. (2020) (e.g., S1) and haplotypes of MENA region of El-Mergawy et al. (2011) (e.g., H1, H3-H6 and H8). Haplotype W2 represents samples collected from Jeddah (e.g., W1, W2 and W3), Makkah (e.g., W5 and W9) and Taif (e.g., W12, W13 and W14) governorates. The AA sequence (133 AAs) is located between positions 87 and 219 referring to the whole AA sequence of this protein (211 AAs) recently published by Sabir et al. (2019). AAs were arranged from left to right as the nonpolar aliphatic (G, A, V, L, M and I), polar uncharged (S, T, C, P, N and Q), aromatic (F, Y and W), positively charged (K, R and H) and negatively charged (D and E).
Fig. 9.
Abundances of amino acids across COI sequences of RPW haplotypes of the present study (e.g., W2 and W7) in addition to the haplotype with consensus sequence of Sukirno et al. (2020) (e.g., S1) and haplotypes of MENA region of El-Mergawy et al. (2011) (e.g., H1, H3-H6 and H8). Haplotype W2 represents samples collected from Jeddah (e.g., W1, W2 and W3), Makkah (e.g., W5 and W9) and Taif (e.g., W12, W13 and W14) governorates. The AA sequence (133 AAs) is located between positions 87 and 219 referring to the whole AA sequence of this protein (211 AAs) recently published by Sabir et al. (2019).
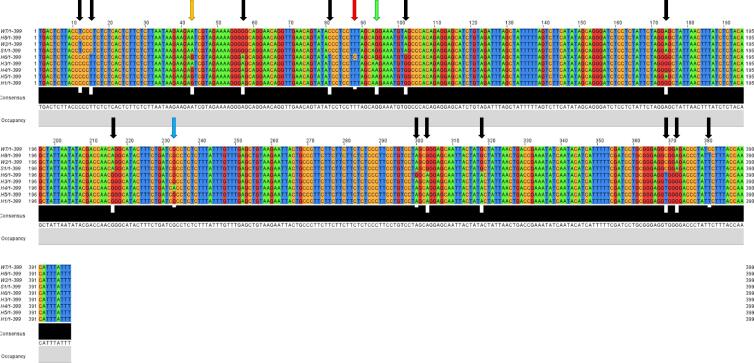
Multiple alignments of trimmed DNA sequences (399 nt) of the nine haplotypes indicated a number of 17 nucleotide substitutions among different haplotypes referring to consensus haplotype sequence (Fig. 10). Of which, only three nucleotides (at positions 43, 94 and 233, respectively) can contribute to nonsynonymous (NS) substitutions, while 14 nucleotides (at positions 12, 15, 57, 81, 88, 102, 174, 216, 300, 303, 318, 369, 372 and 381) contribute to synonymous (S) substitutions. Accordingly, multiple sequence alignment of the deduced amino acids (133 AAs) resulted in three NS substitutions, referring to the consensus haplotype sequence, at AA positions 15, 32, and 87 (Fig. 11). The first two NS substitutions occurred in the first nucleotide of the two codons ATC (position 43–45) coding for isoleucine (I), and GGA (position 94–96) coding for glycine (G). These two codons were changed to GTC coding for valine (V), and AGA coding for serine (S), respectively. The third NS substitution occurred in the second nucleotide of codon CGC (position 232–234) that codes for arginine (R). This codon was changed to CAC those codes for histidine (H) (Fig. 11). The 14 S substitutions of RPW haplotypes occurred generally in the third nucleotide of the codons except for one at nucleotide position 88. Interestingly, the substitution in this codon (position 88–90) occurred in the first nucleotide of codon TTA that codes for leucine (L). This codon was changed to CTA also codes for leucine.
Fig. 10.
Multiple sequence alignment of the trimmed COI gene fragment (399 nt) of RPW haplotypes of the present study (e.g., W2 and W7) in addition to the haplotype with consensus sequence of Sukirno et al. (2020) (e.g., S1) and haplotypes of MENA region of El-Mergawy et al. (2020) (e.g., H1, H3-H6 and H8). Haplotype W2 represents samples collected from Jeddah (e.g., W1, W2 and W3), Mecca (e.g., W5 and W9) and Taif (e.g., W12, W13 and W14) governorates. Alignment indicated a number of 17 nucleotide substitutions among different haplotypes. Orange, green and blue arrows refer to positions of nucleotide changes that resulted in non-synonymous (NS) substitutions (nt 43, 94 and 233, respectively), while black arrows refer to synonymous (S) substitutions. Red arrow refers to S substitution due to change of the second nucleotide of codon.
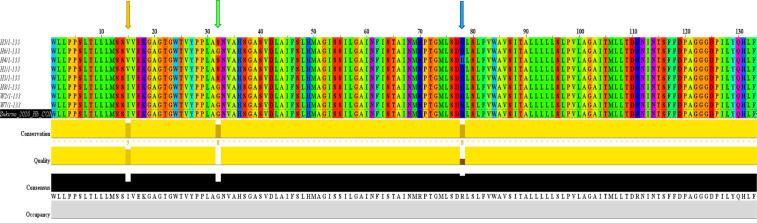
Fig. 11.
Multiple sequence alignment of the COI protein (133 AAs) for RPW haplotypes of the present study (e.g., W2 and W7) in addition to the haplotype with consensus sequence of Sukirno et al. (2020) (e.g., S1) and haplotypes of MENA region of El-Mergawy et al. (2020) (e.g., H1, H3-H6, and H8). Haplotype W2 represents samples collected from Jeddah (e.g., W1, W2, and W3), Makkah (e.g., W5 and W9), and Taif (e.g., W12, W13, and W14) governorates. Alignment indicated a number of three AA changes across COI proteins of different haplotypes at positions 15, 32, and 78 (at positions 15, 32, and 78) referring to amino acids isoleucine/valine, glycine/serine, and arginine/histidine, respectively. These three AAs are located at positions 88, 105, and 151, respectively, referring to the whole AA sequence of this protein (211 AAs) recently published by Sabir et al. (2019). The AA sequence (133 AAs) in this study expands between AA positions 87 and 219 referring to the whole AA sequence of this protein recently published by Sabir et al. (2019). Orange, green, and blue arrows refer to positions of the three amino acids (15, 32, and 78), respectively.
Fig. 12 describes the cladogram based on the three variable AAs in the nine haplotypes at position 15 for isoleucine/valine (I/V), position 32 for glycine/serine (G/S), and position 78 for arginine/histidine (R/H) (Fig. 11). Referring to the consensus AA sequence of this protein in insects recently published by Sabir et al. (2019) (211 AAs), the three AAs are located at positions 88, 105, and 151, respectively, while located at positions 101, 117, and 164, respectively, referring to the COI AA animal consensus sequence introduced by Pentinsaari et al. (2016). The cladogram almost showed pairwise relationships similar to those generated by COI gene sequences shown in Fig. 2. Two main clusters based on AA sequences of GOI were shown (Fig. 12). The first cluster involves a subcluster with AAs I, G, and R of haplotypes W2, S1, W7, and H8 that was used further as the consensus AA-based haplotype. Two other haplotypes exist in this cluster with AAs G, V, and R for H6 haplotypes, on one hand, and S, I, and R for H3 haplotype, on the other hand. The second cluster involves haplotype H4 with AAs S, V, and H at the three positions, while S, V, and R for the two haplotypes H5 and H1 (Fig. 12).
Fig. 12.
Cladogram based on multiple AA sequence alignment (133 AAs) of trimmed COI sequences for RPW haplotypes of the present study (e.g., W2 and W7) in addition to the haplotype with consensus sequence of Sukirno et al. (2020) (e.g., S1) and haplotypes of MENA region of El-Mergawy et al. (2011) (e.g., H1, H3-H6, and H8). The tree also indicates non-synonymous substitutions (NS) of three AAs at positions 15, 32, and 78 referring to amino acids isoleucine/valine, glycine/serine, and arginine/histidine, respectively. Glycine, isoleucine, and arginine (solid colors) refer to AAs in the consensus sequence, while serine, valine, and histidine (gradient colors) refer to substituted AAs due to NS DNA substitutions. Different colors are given to different AA positions, while the same color is given to the same AA position. Haplotype W2 represents samples collected from Jeddah (e.g., W1, W2, and W3), Makkah (e.g., W5 and W9), and Taif (e.g., W12, W13, and W14) governorates. The three AAs are located at positions 88, 105, and 151, respectively, referring to the whole AA sequence of this protein (211 AAs) recently published by Sabir et al. (2019).
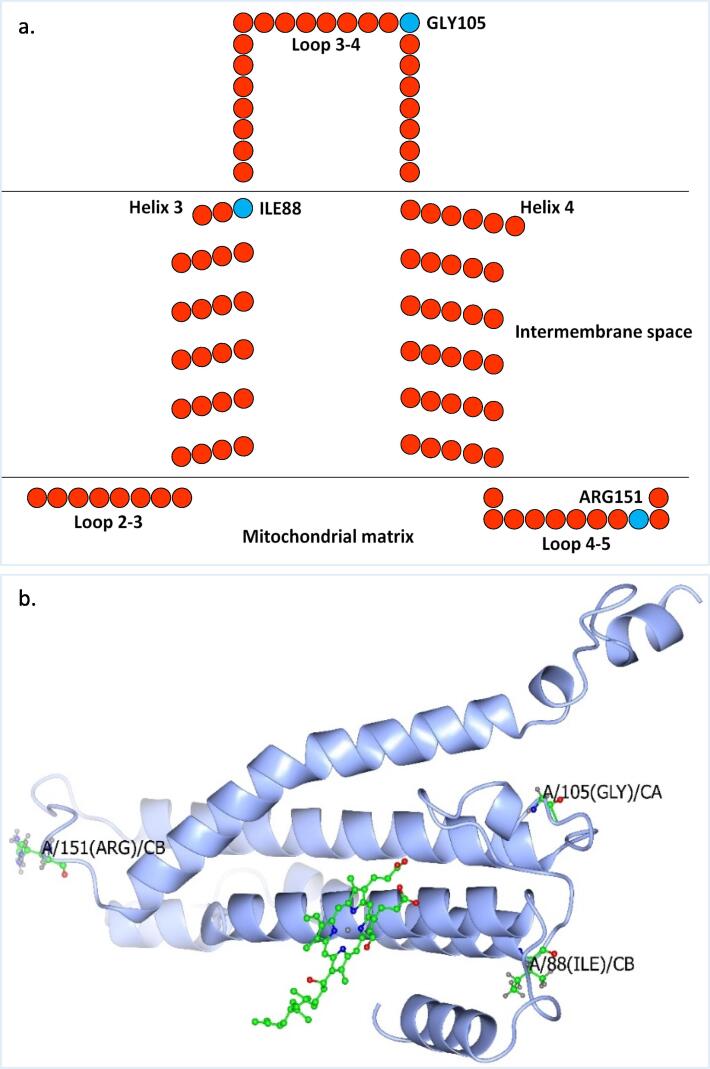
The AA sequences encoded by the COI gene fragment across the nine haplotypes covers 133 AAs starting from position 74 to 206 based on the AA insect consensus sequence published by Sabir et al. (2019), while from position 87 to 219 for AA animal consensus sequence introduced by Pentinsaari et al. (2016). Fortunately, this portion of the barcode includes the active site of COX enzyme mediating electron transfer from copper (Cu) to heme. Based on the criterion followed by Pentinsaari et al. (2016), variation of AAs with the same chemical property at any given position is not considered to significantly influence enzyme function. This is also true for variations of AAs from one chemical group to the other at a position far from the enzyme ligands. Schematic representation of the portion of the consensus COI haplotype of RPW indicates positions (88, 105, and 151) of the three AAs isoleucine, glycine, and arginine, respectively, referring to Sabir et al. (2019) (Fig. 5a). The figure also indicates the three-dimensional (3D) structure of the 133 AAs shown in Fig. 13. At position 88 for isoleucine/valine (I/V), Sabir et al. (2019) indicated that this position is for methionine/isoleucine (M/I), while methionine (M) in Pentinsaari et al. (2016). The three AAs I/V/M are nonpolar aliphatic, hence, it is likely that the presence of any of the three AAs will not change the function of the enzyme. At position 105 for glycine/serine (G/S), Sabir et al. (2019) indicated that this position is for serine/alanine (S/A) and glycine (G) only for order Odonata, while alanine (A) in Pentinsaari et al. (2016). Two of the three AAs (G/A) are nonpolar aliphatic, while the third AA (S) is polarly uncharged. Thus, it is possible that the change from G or A to S might affect the chemical properties of the enzyme and might also affect the folding pattern in the 3D structure. At position 151 for arginine/histidine (R/H), Sabir et al. (2019) indicated that this position is for arginine/glutamine (R/Q), while glutamine (A) in Pentinsaari et al. (2016). Two of the three AAs (R/H) are positively charged, while the third AA (Q) is polarly uncharged. Thus again, it is possible that the change from R or H to Q might affect the structure/function properties of the enzyme.
Fig. 13.
a. Schematic representation of the portion of the COI protein structure of haplotypes of the first cluster, the consensus AA sequence of RPW (e.g., isoleucine, glycine, and arginine) located at positions 88, 105, and 151, respectively, referring to the insect consensus AA sequence of this protein (211 AAs) recently published by Sabir et al. (2019). (b) Predicted three-dimensional COI protein structure of haplotype W7. b) Alignment of the predicted three-dimensional structure of COI proteins of consensus AA sequence of RPW versus documented structure of cattle (Bos taurus). The position of the heme molecule (in green) is shown.
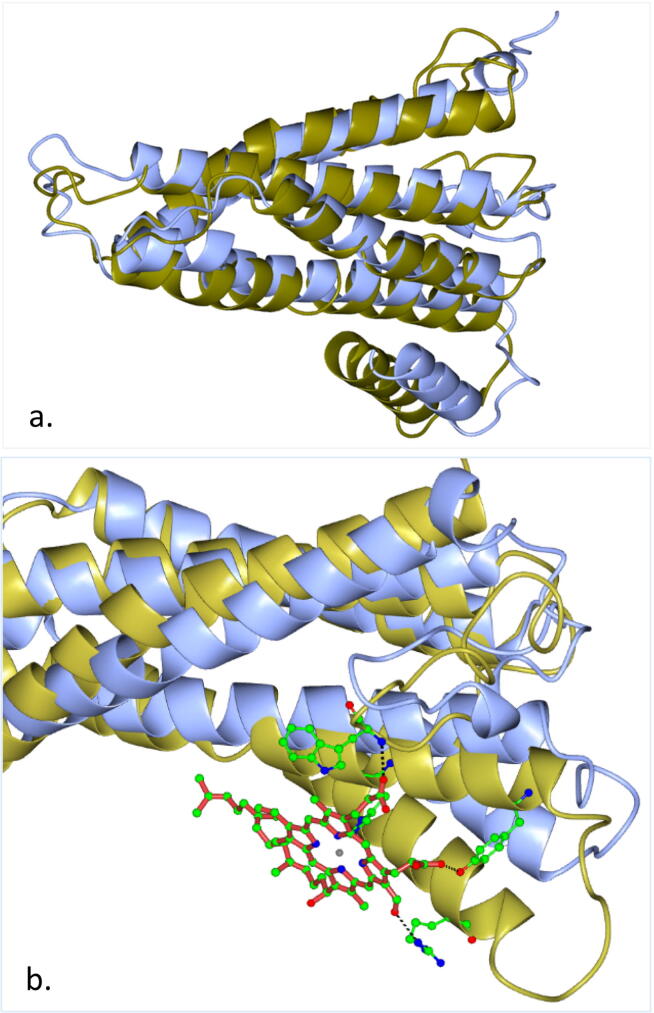
The predicted 3D structure of the consensus COI haplotype of RPW was aligned to the documented 3D structure of COI in cattle (Bos taurus) that was introduced by Pentinsaari et al. (2016) (Fig. 14). As indicated earlier, position 88 is I or V for the RPW consensus COI sequence, while M in Pentinsaari et al. (2016), position 105 is G or S for the RPW consensus COI sequence, while A in Pentinsaari et al. (2016) and position 151 is R or H for the RPW consensus COI sequence, while Q in Pentinsaari et al. (2016). As indicated above, a change of AA at position 88 might not affect the chemical properties of the COX enzyme, while changes at positions 105 and 151 can affect the chemical properties of the enzyme. In addition, the first two positions exist at the border or inside loop 3–4 of the enzyme, while the third position exists inside loop 4–5 (Fig. 13a). These two loops influence the folding pattern of the enzyme, thus, might affect the function of the enzyme. Sabir et al. (2019) indicated that AA variation was proven to be more pronounced in loops than in helices. Besides, loop 3–4 is important for pointing towards the heme group at the active site of the COI protein. These results explain the differential activity of the COX enzyme in different classes of the animal kingdom. Variation in chemical properties and 3D structure of COX enzyme might be an evolutionary process to adapt to different environment (in-time and in-site changes), which support the fact that RPW is a very strong fly and highly invasive pest that can cause severe damages whenever palm trees exist. The alignment pattern of the COI proteins of RPW versus that in cattle supports our previous statement (Fig. 6a). It is clear that the helices of the two proteins are not fully aligned in the 3D structure. Sabir et al. (2019) indicated that a number of 16 AAs are in close proximity with heme 515 to enforce the action of COX enzyme in the electron transport chain. Referring to AAs numbering of Sabir et al. (2019), these AAs exist at positions 15, 18, 21, 22, 38, 41, 42, 45, 46, 49, 50, 53, 54, 57, 109 and 110. Therefore, it is unlikely that variations in the three positions 88, 105, and 151 will affect the binding ability of the heme group to the COI protein (Fig. 14b). Overall, AA variations occurring at positions 105 and 151 might affect the chemical properties of COX enzyme, while variations in the three positions 88, 105, and 151 might affect the structure of the enzyme. However, none of these variations has any influence on the binding of the COX enzyme with the heme group.
Fig. 14.
a) Alignment of the predicted three-dimensional structure of COI proteins of consensus AA sequence of RPW (blue) versus documented structure (gold) of cattle (Bos taurus). b) Position of the heme molecule (in red) of the two aligned protein structures.
Previous results indicated scarcity of AAs variations in the COI gene (Pesole et al., 1999, Meiklejohn et al., 2007, Castoe et al., 2008, Mathews et al., 2013). However, evidence of nonsynonymous substitution of AAs in insects is a result of positive selection that could be caused by lifestyle changes towards rescuing the organism (Meiklejohn et al., 2007, Castoe et al., 2008, Galtier et al., 2009). Evolutionary, most insects of the RPW came to the Middle East from Pakistan that is sometimes called Pakistani weevil. In Saudi Arabia, the Ministry of Environment, Water, and Agriculture has implemented several programs to control RPW including spraying with pesticides, insect trapping, and management of the infested areas (Abraham et al., 2002). However, the weevil is still intensively spreading in the Western region of Saudi Arabia, especially in governorates in the Makkah region. It seems that the W2 haplotype is the strongest in the Makkah region and is expected to be migrating to other regions of Saudi Arabia and neighboring countries to cause more damage to existing palm trees.
Although the date production has quadrupled between 1996 and 2013, the record of FAO statistics demonstrated that this sector is under severe degradation (FAOSTAT, 2013). Since the past few decades, the date palm tree was subjected to several challenges including biotic and abiotic stresses due to human activities, intensive farming, climate change, etc. Thus, the rehabilitation of this valuable resource is becoming an emergency for all Arabian countries and requires more collaborative efforts, networks, and dedicated funds. In this respect, several recommendations, like genetic conservation, integrated bio-control approaches, sustainable farming procedures, the best way of processing technologies, valorization techniques, and better marketability of the fruit are recommended (El-Juhany, 2010, Johnson et al., 2015a, Johnson et al., 2015b). Of which, RPW, identified as the synonym Rhynchophorus vulneratus (Hallett et al. 2004), was confirmed as a dangerous fast-evolving species (Rugman-Jones et al., 2013, Sukirno et al., 2018a, Sukirno et al., 2018b).
In conclusion, we have utilized the COI subunit of RPW collected from Saudi Arabia to generate DNA barcodes of the insect and detect variations at the structural and functional levels among these and other specimens collected from Saudi Arabia by Sukirno et al. (2020) as well as from the MENA region by El-Mergawy et al. (2011). This data can help in detecting the evolutionary dynamics of the insect and the possible emergence of new dangerous versions of the insect. The data can also be helpful in quarantine and pest management programs and in tracing RPW geographic spread and migration in Saudi Arabia. Through this study, we gained new insights into the distribution of RPW haplotypes in the region and detected possible mechanisms by which COI protein evolves in Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2022.103388.
Contributor Information
Wafa Mohammed Al-Otaibi, Email: w.alotaibi@tu.edu.sa.
Khalid Mohammed Alghamdi, Email: Kalghamdy@kau.edu.sa.
Jazem A. Mahyoub, Email: jabdulrahman@kau.edu.sa.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Abraham, V.A., Faleiro, J.R., Radhakrishnan Nair, C.P., Nair, S.S., 2002. Present management technologies for red palm weevil, Rhynchophorus ferrugineus Oliver (Coleopteran: Curculionidae). In: Palms And Future Thrusts.
- Abraham V.A., Faleiro J.R., Al Shuaibi M.A., Abdan S.A. Status of pheromone trap captured female red palm weevils from date gardens in Saudi Arabia. J. Trop. Agric. 2006;39(2):197–199. [Google Scholar]
- Al-Ayied H.Y., Alswailem A.M., Shair O., Al Jabr A.M. Evaluation of phylogenetic relationship between three phenotypically different forms of Red date palm weevil Rhynchophorus ferrugineus Oliv. using PCR-based RAPD technique. Arch. Phytopathol. Plant Protect. 2006;39(4):303–309. [Google Scholar]
- Al-Farsi M., Alasalvar C., Morris A., Baron M., Shahidi F. Compositional and sensory characteristics of three native sun-dried date (Phoenix dactylifera L.) varieties grown in Oman. J. Agric. Food. Chem. 2005;53(19):7586–7591. doi: 10.1021/jf050578y. [DOI] [PubMed] [Google Scholar]
- Al-Saroj, S., Al-Abdallah, E., Al-Shawaf, A. M., Al-Dandan, A. M., Al-Abdullah, I., Al-Shagag, A., ALFehaid, Y., Abdallah, A.B., Faleiro, J.R., 2017. Efficacy of bait free pheromone trap (ElectrapTM) for management of red palm weevil, Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae). Pest Manage. Horticul. Ecosyst. 23(1), 55–59.
- Anonymous, 2013. Save Algarve palms. http://www.savealgarvepalms.com/en/weevilfacts/hostpalmtrees (accessed on 1/8/2020).
- Ashfaq M., Hebert P.D. DNA barcodes for bio-surveillance: regulated and economically important arthropod plant pests. Genome. 2016;59(11):933–945. doi: 10.1139/gen-2016-0024. [DOI] [PubMed] [Google Scholar]
- Baloch M.K., Saleem S.A., Baloch A.K., Baloch W.A. Impact of controlled atmosphere on the stability of Dhakki dates. LWT-food Sci. Technol. 2006;39(6):671–676. [Google Scholar]
- Balsa E., Marco R., Perales-Clemente E., Szklarczyk R., Calvo E., Landázuri M.O., Enríquez J.A. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16(3):378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Borges L.M., Hollatz C., Lobo J., Cunha A.M., Vilela A.P., Calado G., Coelho R., Costa A.C., Ferreira M.S., Costa M.H., Costa F.O. With a little help from DNA barcoding: investigating the diversity of Gastropoda from the Portuguese coast. Sci. Rep. 2016;6(1):1–11. doi: 10.1038/srep20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozbuga R., Hazir A. Pests of the palm (Palmae sp.) and date palm (Phoenix dactylifera) determined in Turkey and evaluation of red palm weevil (Rhynchophorus ferrugineus Olivier)(Coleoptera: Curculionidae) EPPO Bulle. 2008;38(1):127–130. [Google Scholar]
- CABI, 2020. Rhynchophorus ferrugineus (red palm weevil). In: Crop Protection Compendium. Wallingford, UK: CAB International. https://www.cabi.org/isc/datasheet/47472.
- Castoe T.A., Jiang Z.J., Gu W., Wang Z.O., Pollock D.D. Adaptive evolution and functional redesign of core metabolic proteins in snakes. PLoS ONE. 2008;3(5) doi: 10.1371/journal.pone.0002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristescu M.E. From barcoding single individuals to metabarcoding biological communities: towards an integrative approach to the study of global biodiversity. Trends Ecol. Evol. 2014;29(10):566–571. doi: 10.1016/j.tree.2014.08.001. [DOI] [PubMed] [Google Scholar]
- El Hadrami A., Al-Khayri J.M. Socioeconomic and traditional importance of date palm. Emirates J. Food Agric. 2012;24(5):371. [Google Scholar]
- El-Juhany L.I. Degradation of date palm trees and date production in Arab countries: causes and potential rehabilitation. Aust. J. Basic Appl. Sci. 2010;4(8):3998–4010. [Google Scholar]
- El-Mergawy R.A., Faure N., Nasr M.I., Avand-Faghih A., Rochat D., Silvain J.F. Mitochondrial genetic variation and invasion history of Red palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae), in middle-east and Mediterranean basin. Int. J. Agric. Biol. 2011;13:631–637. [Google Scholar]
- EPPO (European and Mediterranean Plant Protection Organization), 2008. Data sheets on quarantine pests—Rhynchophorus ferrugineus. EPPO Bull., 38, 55–59.
- EPPO, 2020. Rhynchophorus ferrugineus, EPPO datasheets on pests recommended for regulation. Available from: https://gd.eppo.int [Accessed: 15 May 2020].
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17(6):368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;5:294–299. [PubMed] [Google Scholar]
- Galtier N., Nabholz B., Glémin S., Hurst G.D.D. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 2009;18(22):4541–4550. doi: 10.1111/j.1365-294X.2009.04380.x. [DOI] [PubMed] [Google Scholar]
- Ghosh C.C. Life history of Indian insects–III. The Rhinoceros beetle (Oryctes rhinoceros) and the red palm weevil (Rhynchophorus ferrugineus) Memoirs Dept. Agric. India (Int. Ser.) II. 1912;(10):205–217. [Google Scholar]
- Gwiazdowski R.A., Foottit R.G., Maw H.E.L., Hebert P.D. The Hemiptera (Insecta) of Canada: constructing a reference library of DNA barcodes. PLoS ONE. 2015;10(4):e0125635. doi: 10.1371/journal.pone.0125635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldhar S.M., Maheshwari S.K., Muralidharan C.M. Pest status of date palm (Phoenix dactylifera) in arid regions of India. J. Agric. Ecol. 2017;3:1–11. [Google Scholar]
- Hallett R.H., Crespi B.J., Borden J.H. Synonymy of Rhynchophorus ferrugineus (Olivier), 1790 and R. vulneratus (Panzer), 1798 (Coleoptera, Curculionidae, Rhynchophorinae) J. Natural History. 2004;38(22):2863–-2882. [Google Scholar]
- Hebert P.D., Cywinska A., Ball S.L., Dewaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B Biol. Sci. 2003;270(1512):313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D., Penton E.H., Burns J.M., Janzen D.H., Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. 2004;101(41):14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D., DeWaard J.R., Zakharov E.V., Prosser S.W., Sones J.E., McKeown J.T., Mantle B., La Salle J. A DNA ‘Barcode Blitz’: Rapid digitization and sequencing of a natural history collection. PLoS ONE. 2013;8(7):e68535. doi: 10.1371/journal.pone.0068535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, S.M., Al-Khayri, J.M., Johnson, D.V. (Eds.), 2011. Date Palm Biotechnology. Springer Science & Business Media.
- Johnson D.V., Al-Khayri J.M., Jain S.M. Date palm genetic resources and utilization. Springer; Dordrecht: 2015. Introduction: Date production status and prospects in Africa and the Americas; pp. 3–18. [Google Scholar]
- Johnson D.V., Al-Khayri J.M., Jain S.M. Date palm genetic resources and utilization. Springer; Dordrecht: 2015. Introduction: Date production status and prospects in Asia and Europe; pp. 1–16. [Google Scholar]
- Mathews C.K., Van Holde K.E., Appling D.R., Anthony-Cahill S.J. 3 utg. Pearson; Toronto, Ont: 2013. Biochemistry. [Google Scholar]
- Meiklejohn C.D., Montooth K.L., Rand D.M. Positive and negative selection on the mitochondrial genome. Trends Genet. 2007;23(6):259–263. doi: 10.1016/j.tig.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Melifronidou-Pantelidou A. Eradication campaign for Rhynchophorus ferrugineus in Cyprus 1. EPPO Bull. 2009;39(2):155–160. [Google Scholar]
- MOA . Ministry of Agriculture; Riyadh: 2014. The current distribution of red palm weevil in the Kingdom of Saudi Arabia. [Google Scholar]
- Mujahid M., Ahmad J.N., Arif M.J., Nazir J., Giblin-Davis R.M. Molecular identification and phylogenetic analysis of distinct geographical populations of Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae) in Pakistan. Int. J. Agric. Biol. 2018;20(9):1997–2004. [Google Scholar]
- Murphy S.T., Briscoe B.R. The red palm weevil as an alien invasive: biology and the prospects for biological control as a component of IPM. Biocontrol News Informat. 1999;20:35N–46N. [Google Scholar]
- Muse S.V., Gaut B.S. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 1994;11(5):715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- Mutanen M., Kekkonen M., Prosser S.W., Hebert P.D., Kaila L. One species in eight: DNA barcodes from type specimens resolve a taxonomic quagmire. Mol. Ecol. Resour. 2015;15(4):967–984. doi: 10.1111/1755-0998.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeu Commission decision of 6 October 2008 amending decision of 2007/365/EC on emergency measures to prevent the introduction into and the spread within the community of Rhynchophorus ferrugineus (Olivier) 2008/5550/C. Off. J. Eur. Union L. 2008;26:6–14. [Google Scholar]
- Oni S.O., Adeosun A.M., Ladokun O.A., Ighodaro O.M., Oyedele M.O. Nutritional and phytochemical profile of Niger cultivated date palm (Phoenix dactilyfera L) J. Food Nutr. Sci. 2015;3:114–118. [Google Scholar]
- Pelikh, K., 2009. First report of RPW in Republic of Georgia. Red Palm Weevil.
- Pentinsaari M., Salmela H., Mutanen M., Roslin T. Molecular evolution of a widely-adopted taxonomic marker (COI) across the animal tree of life. Sci. Rep. 2016;6(1):1–12. doi: 10.1038/srep35275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G., Gissi C., De Chirico A., Saccone C. Nucleotide substitution rate of mammalian mitochondrial genomes. J. Mol. Evol. 1999;48(4):427–434. doi: 10.1007/pl00006487. [DOI] [PubMed] [Google Scholar]
- Ratnasingham S., Hebert P.D. A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS ONE. 2013;8(7):e66213. doi: 10.1371/journal.pone.0066213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J.M., Ashfaq M., Hu X.N., Ma J., Liang F., Hebert P.D., Lin L., Germain J.F., Ahmed M.Z. Barcode index numbers expedite quarantine inspections and aid the interception of nonindigenous mealybugs (Pseudococcidae) Biol. Invasions. 2018;20(2):449–460. [Google Scholar]
- resource FAOSTAT, E., 2013. Access mode: http://www.faostat3.fao.org/Q/QC.
- Rugman-Jones P.F., Hoddle C.D., Hoddle M.S., Stouthamer R. The lesser of two weevils: molecular-genetics of pest palm weevil populations confirm Rhynchophorus vulneratus (Panzer 1798) as a valid species distinct from R. ferrugineus (Olivier 1790), and reveal the global extent of both. PloS One. 2013;8(10):e78379. doi: 10.1371/journal.pone.0078379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir J.S., Rabah S., Yacoub H., Hajrah N.H., Atef A., Al-Matary M., Edris S., Alharbi M.G., Ganash M., Mahyoub J., Al-Hindi R.R. Molecular evolution of cytochrome C oxidase-I protein of insects living in Saudi Arabia. PLoS ONE. 2019;14(11):e0224336. doi: 10.1371/journal.pone.0224336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadder M.T., Vidyasagar P.S., Aldosari S.A., Abdel-Azim M.M., Al-Doss A.A. Phylogeny of red palm weevil (Rhynchophorus ferrugineus) based on ITS1 and ITS2. Oriental insects. 2015;49(3–4):198–211. [Google Scholar]
- Sallam A.A., El-Shafie H.A.F., Al-Abdan S. Influence of farming practices on infestation by red palm weevil Rhynchophorus ferrugineus (Olivier) in date palm: a case study. Int. Res. J. Agric. Sci. Soil Sci. 2012;2(8):370–376. [Google Scholar]
- Scheffers B.R., Joppa L.N., Pimm S.L., Laurance W.F. What we know and don’t know about Earth's missing biodiversity. Trends Ecol. Evol. 2012;27(9):501–510. doi: 10.1016/j.tree.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Sukirno S., Tufail M., Rasool K.G., Aldawood A.S. Palm weevil diversity in Indonesia: Description of phenotypic variability in Asiatic palm weevil, Rhynchophorus vulneratus (Coleoptera: Curculionidae) J. Entomol. Res. Soc. 2018;20(3):1–22. [Google Scholar]
- Sukirno S., Tufail M., Rasool K.G., Aldawood A.S. Undescribed color polymorphism of the Asiatic palm weevil, Rhynchophorus vulneratus Panzer (Coleoptera: Curculionidae) in Indonesia: biodiversity study based on COI gene. Florida Entomol. 2018:642–648. [Google Scholar]
- Sukirno S., Tufail M., Rasool K.G., Husain M., Aldawood A.S. Diversity of red palm weevil, Rhynchophorus ferrugineus Oliv. (Coleoptera: Curculionidae) in the Kingdom of Saudi Arabia: studies on the phenotypic and DNA barcodes. Int. J. Trop. Insect Sci. 2020 [Google Scholar]
- Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science. 1995;269(5227):1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272(5265):1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Tsukihara T., Shimokata K., Katayama Y., Shimada H., Muramoto K., Aoyama H., Mochizuki M., Shinzawa-Itoh K., Yamashita E., Yao M., Ishimura Y. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc. Natl. Acad. Sci. 2003;100(26):15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayalil P.K. Antioxidant and antimutagenic properties of aqueous extract of date fruit (Phoenix dactylifera L. Arecaceae) J. Agric. Food. Chem. 2002;50(3):610–617. doi: 10.1021/jf010716t. [DOI] [PubMed] [Google Scholar]
- Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3(5):294–299. [PubMed] [Google Scholar]
- Wattanapongsiri, A., 1966. A revision to the genera Rhynchophorus and Dynamis (Coleoptera: Curculionidae). Department of Agriculture Science Bulletin, Vol. 1, Bangkok.
- Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.