Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV), SARS-CoV-2, and human coronavirus (hCoV)-NL63 utilize ACE2 as the functional receptor for cell entry, which leads to zoonotic infection. Horses (Equus caballus) attracted our attention because the spike protein receptor-binding domains (RBDs) of SARS-CoV-2 and SARS-CoV-2-related coronaviruses bind equine ACE2 (eACE2) with high affinity. Here we show that eACE2 binds the RBDs of these three coronaviruses and also SARS-CoV-2 variants but with lower affinities compared with human ACE2 (hACE2). Structural analysis and mutation assays indicated that eACE2-H41 accounts for the lower binding affinity of eACE2 to the RBDs of SARS-CoV-2 variants (Alpha, Beta, and Gamma), SARS-CoV, and hCoV-NL63. Pseudovirus infection assays showed that the SARS-CoV-2 Delta strain (B.1.617.2) displayed a significantly increased infection efficiency in eACE2-expressing HeLa cells. Our results reveal the molecular basis of eACE2 binding to the RBDs of SARS-CoV, SARS-CoV-2, and hCoV-NL63, which provides insights into the potential animal transmission of these ACE2-dependent coronaviruses.
Keywords: SARS-CoV-2, SARS-CoV, hCoV-NL63, receptor binding domain, horse, ACE2, complex structure, pseudovirus infection
Graphical abstract
Binding of the spike protein to the cell surface receptor determines the host range of coronaviruses. Combining structural data and pseudovirus infection analysis, Lan et al. show that H41 of equine ACE2 confers a lower binding affinity to the RBDs of SARS-CoV-2 variants and SARS-CoV in comparison with human ACE2.
Introduction
Coronaviruses are enveloped, positive-sense, single-stranded RNA viruses that are commonly classified into the four genera of alphacoronavirus, betacoronavirus, gammacoronavirus, and deltacoronaviruse. Coronavirus infection causes respiratory and enteric diseases with varying levels of pathogenicity in humans and other animals. Zoonotic transmission of coronaviruses has been posing a tremendous threat to global health and economic development in the 21st century, as evidenced by the emergence of three epidemic betacoronaviruses, including severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002–2003 (Drosten et al., 2003) and Middle East respiratory syndrome (MERS)-CoV in 2012 (Zaki et al., 2012) as well as the pandemic of SARS-CoV-2 since the end of 2019 (Zhu et al., 2020). A growing body of evidence has revealed the important roles of zoonotic transmission during the evolution and diversification of coronaviruses. Horseshoe bats (Rhinolophus spp.) have been identified as the natural reservoir of a group of SARS-related coronaviruses. SARS-CoV has been suggested to originate in Chinese rufous horseshoe bats (Rhinolophus sinicus) (Lau et al., 2005; Ge et al., 2013; Yang et al., 2015; Hu et al., 2017) and is thought to have been zoonotically transmitted into palm civets and other animals in a live-animal market and, finally, into humans (Guan et al., 2003; Kan et al., 2005). Bats are also the likely origin of MERS-CoV (Ithete et al., 2013; Lau et al., 2013). MERS-CoV has been suggested to circulate in dromedary camels through zoonotic transmission, resulting in frequent infection of humans since 2012 (Alagaili et al., 2014; Hemida et al., 2013). Although the natural reservoirs and intermediate hosts of SARS-CoV-2 remain elusive, several lines of evidence support the idea that it may also originate in horseshoe bats. The full genome of SARS-CoV-2 exhibits 96.2% and 96.8% nucleotide sequence identity to that of RaTG13 and BANAL-52 coronaviruses from horseshoe bats, respectively (Boni et al., 2020; https://www.researchsquare.com/article/rs-871965/v1). Pangolin coronaviruses (PCoV) exhibiting 85.5%–92.4% nucleotide sequence identity to SARS-CoV-2 have also been identified in smuggled Malayan pangolins (Manis javanica) in China’s Guangxi (GX) and Guangdong (GD) provinces (Lam et al., 2020; Xiao et al., 2020). There is also evidence of an ancestral association of the alphacoronaviruses human CoV (hCoV)-229E and hCoV-NL63 with bats (Corman et al., 2015; Donaldson et al., 2010; Huynh et al., 2012).
The spike (S) glycoprotein of coronaviruses mediates viral entry by binding to the host cell receptor and inducing fusion of viral and cellular membranes. Therefore, the receptor binding specificity of the S glycoprotein plays a critical role in determining host tropism. Since the outbreak of coronavirus disease 2019 (COVID-19), several studies have evaluated the potential host range of SARS-CoV-2, SARS-CoV, RaTG13, and PCoVs based on different methods (Wu et al., 2020; Liu et al., 2021a, 2021c; Li et al., 2020, 2021; Niu et al., 2021; Alexander et al., 2020), illuminated the underlying binding mechanism (Zhang et al., 2021b; Wu et al., 2020; Liu et al., 2021a, 2021b). Using the receptor-binding domains (RBDs) of SARS-CoV-2, SARS-CoV, RaTG13, and PCoV-GD to perform surface staining with HEK 293T cells expressing 16 different ACE2 orthologs, Li et al. (2020) found that the SARS-CoV and SARS-CoV-2 RBDs have a nearly identical ACE2-binding profile spanning 11 ACE2 orthologs, 10 of which they have in common, including those from humans, camels, cattle, horses, goat, sheep, cats, rabbits, pangolins, and civets. The same authors also found that the PCoV-GD RBD has an ACE2-binding profile similar to that of the SARS-CoV-2 RBD, whereas the RaTG13 RBD has a significantly narrower ACE2-binding profile than the other three RBDs (Li et al., 2020). Of 16 examined ACE2 orthologs, horse and rabbit ACE2 were the only two receptors exhibiting a capacity to bind all four RBDs from SARS-CoV, SARS-CoV-2, PCoV-GD, and RaTG13 (Li et al., 2020). The binding affinity between equine ACE2 (eACE2) and the RBDs of SARS-CoV-2, SARS-CoV, or pangolin-CoV was comparable with that of hACE2, whereas the affinity of eACE2 to the RaTG13 RBD has been reported to be even higher than that of hACE2 (Li et al., 2020; Liu et al., 2021a; Wu et al., 2020). Although natural infection of horses by SARS-CoV-2 or SARS-CoV has not been observed to date, some other betacoronaviruses have been reported to cause enteric infection in adult horses (Haake et al., 2020; Pusterla et al., 2018; Bryan et al., 2019). Thus, there is accumulating evidence that horses might be a suitable host for coronaviruses to replicate, recombine, and finally obtain the ability to infect humans in the future. In addition to animal conservation, close surveillance of horses may also be important to reduce the risk of virus spillover from humans to horses and vice versa.
So far, there is no direct evidence of binding variance between humans and horses for the three ACE2-dependent coronaviruses, and the underlying structural differences are also still elusive. Here we found that eACE2 bound the three ACE2-dependent coronaviruses and a part of the tested SARS-CoV-2 variants with lower affinities than hACE2. We also determined the crystal structures of eACE2 in complex with the RBDs of SARS-CoV-2, SARS-CoV, and hCoV-NL63 at resolutions of 2.9, 2.65, and 3.2 Å, respectively. Based on structural comparison and mutational analysis, we found that the amino acid residue at position 41 of ACE2 was the critical determinant of differential binding between equine and human receptors to these three ACE2-dependent coronaviruses. Pseudovirus infection assays of emerging SARS-CoV-2 variants, including variants of concern (VOCs) and variants of interest (VOIs), suggested enhanced infection of eACE2-expressing HeLa cells by the Delta strain (B.1.617.2).
Results
Binding affinities of SARS-CoV-2, SARS-CoV, and hCoV-NL63 RBDs for eACE2
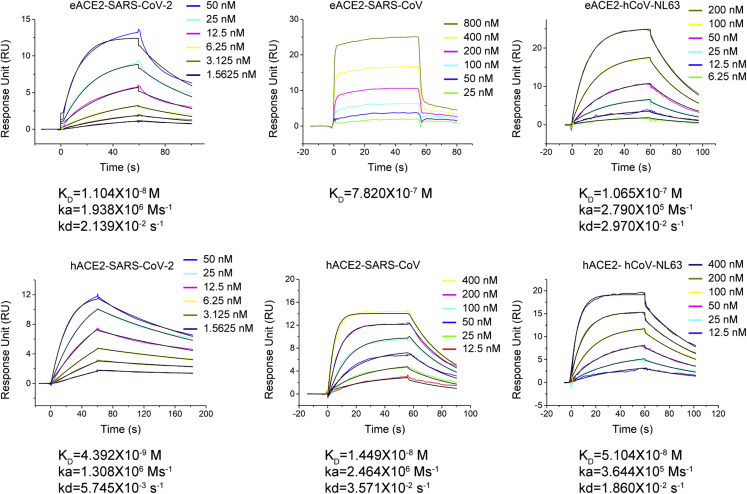
Previous studies have suggested that horses might be a susceptible host for SARS-CoV-2 and SARS-CoV (Li et al., 2020; Liu et al., 2021a; Wu et al., 2020). Here we utilized surface plasmon resonance (SPR) spectroscopy to measure and compare the binding affinities of these three ACE2-dependent coronaviruses with eACE2 and hACE2. The peptidase domain of eACE2 or hACE2 was immobilized on a CM5 sensor, and the RBD of SARS-CoV-2, SARS-CoV, or hCoV-NL63 was flowed through (Figure 1 ). The SARS-CoV-2 RBD bound to eACE2 with an affinity of 11.0 nM, approximately 2.5-fold weaker than the 4.4-nM binding affinity between the SARS-CoV-2 RBD and hACE2. The measured binding affinity of the SARS-CoV RBD for eACE2 was 782 nM, which was much weaker (∼54-fold) than its binding to hACE2 (KD = 14.5 nM). At the same time, we also examined binding of the hCoV-NL63 RBD to eACE2, and the measured affinity of 106.5 nM was approximately 2-fold weaker than the binding to hACE2 (KD = 51.0 nM). These results revealed that, although not as strong as hACE2, eACE2 also demonstrated a capacity to bind the RBDs of SARS-CoV-2, SARS-CoV, and hCoV-NL63. The SARS-CoV RBD exhibited the largest difference in binding affinity between eACE2 (KD = 782 nM) and hACE2 (KD = 14.5 nM).
Figure 1.
Binding curves of RBDs from ACE2-dependent coronaviruses with eACE2 or hACE2
Shown is SPR characterization of the binding between eACE2 or hACE2 and RBDs from SARS-CoV-2, SARS-CoV, and hCoV-NL63. eACE2 or hACE2 was immobilized on the CM5 chip, and RBDs were flowed through. The raw and fitted curves are displayed as colored and black lines, respectively.
Complex structures of SARS-CoV-2, SARS-CoV, and hCoV-NL63 RBDs bound to eACE2
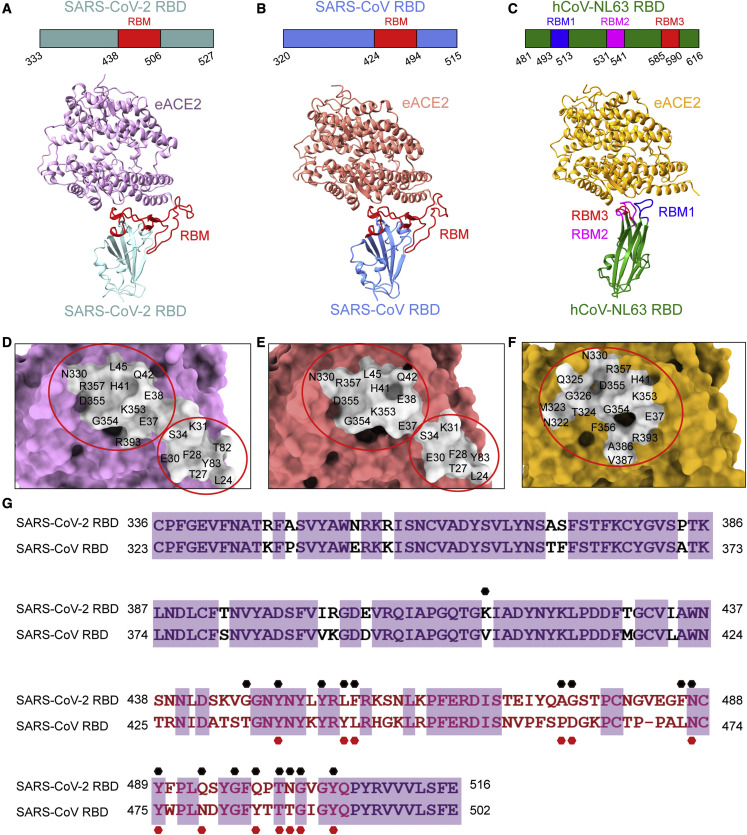
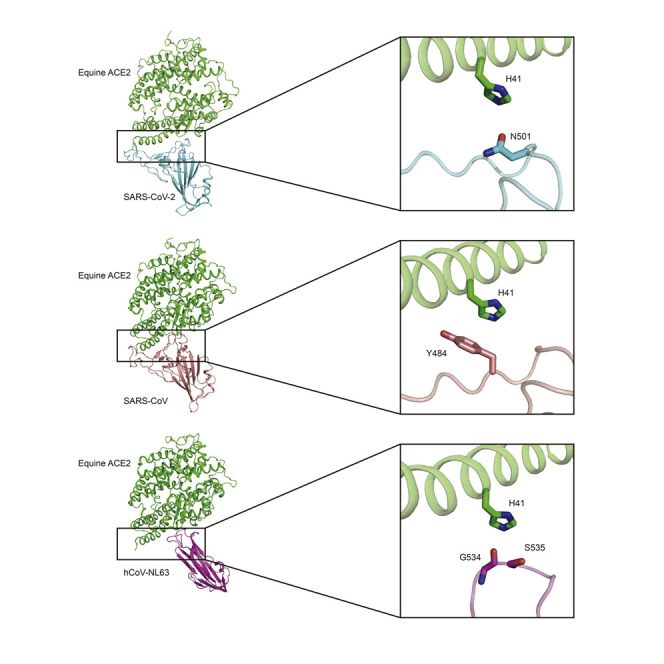
The crystal structures of SARS-CoV-2, SARS-CoV, or hCoV-NL63 RBDs bound to hACE2 have been reported previously (Li et al., 2005; Lan et al., 2020; Wang et al., 2020; Wu et al., 2009). To decipher the structural basis for the distinct binding between eACE2 and hACE2, we crystallized these three RBDs bound to the peptidase domain of eACE2 and obtained structures with a resolution of 2.9, 2.65, and 3.2 Å, respectively (Table S1; Figures 2A–2C). The overall structures of the SARS-CoV-2 RBD/eACE2 and SARS-CoV-2 RBD/hACE2 complexes were nearly identical, with a root-mean-square deviation (RMSD) of 0.67 Å for 727 aligned Cα atoms (Figure S1A). In the SARS-CoV RBD/eACE2 and SARS-CoV RBD/hACE2 complexes, the overall structures were also similar except for the varied angles between the two lobes of ACE2, which resulted in an RMSD of 3.35 Å for 756 aligned Cα atoms (Figure S1B). Similar to the binding mode of hACE2, eACE2 also bound to the continuous RBM (residues 438–506 for SARS-CoV-2 and residues 424–494 for SARS-CoV) of the SARS-CoV-2 and SARS-CoV RBDs (Figures 2A and 2B; Table S2). The footprint of SARS-CoV-2 and SARS-CoV RBD binding to eACE2 can be clustered into two patches (Figures 2D and 2E; Table S2). One patch mainly includes residues from the N-terminal half of the ACE2 N-terminal helix (residues 19–70), and the other patch is primarily composed of a hairpin loop (residues 350–357) and the C-terminal half of the ACE2 N-terminal helix (Figures 2D and 2E; Table S2). These two patches together form a buried surface area of ∼800 Å2 on the surface of eACE2 for binding to the SARS-CoV-2 and SARS-CoV RBDs. Similarly, the overall structures of hCoV-NL63 RBD/eACE2 and hCoV-NL63 RBD/hACE2 complexes were also highly similar, with a slightly varied angle between ACE2 and the RBD (Figure S1C). The hCoV-NL63 RBD/eACE2 interface consisted of three discontinuous RBMs (residues 493–513, 531–541, and 585–590) on the RBD that interact with one focused patch on eACE2, which is mainly composed of residues from three loops (residues 319–330, 350–357, and 386–393) (Figures 2C and 2F; Table S2). This patch has a relatively smaller buried surface area of ∼560 Å2 compared with ∼800 Å2 for binding with the SARS-CoV-2 and SARS-CoV RBDs.
Figure 2.
Complex structures of eACE2 with RBDs from ACE2-dependent coronaviruses
(A–C) Domain structures of RBDs and overall structures of the RBDs of SARS-CoV-2 (left), SARS-CoV (center), and hCoV-NL63 (right) in complex with eACE2. The boundaries of RBMs were identified from the crystal structures. The RBMs of SARS-CoV-2 and SARS-CoV are colored red, and that of hCoV-NL63 is colored blue, magenta, and red.
(D–F) Footprint of SARS-CoV-2 (left), SARS-CoV (center), and hCoV-NL63 (right) on the surface of eACE2. eACE2 residues binding the RBDs within a 4-Å cutoff are labeled as the footprint. The epitope is colored light gray. RBDs are colored powder blue, cornflower blue, and green for SARS-CoV-2, SARS-CoV, and hCoV-NL63, respectively; eACE2 is colored plum, light coral, and goldenrod for these three complexes, respectively. Red circles indicate the cluster of interacting residues.
(G) Sequence alignment of the SARS-CoV-2 and SARS-CoV RBDs. Residues contacting eACE2 in the SARS-CoV-2 RBD are indicated by black dots. Residues contacting eACE2 in the SARS-CoV RBD are indicated by red dots.
The binding epitopes of the SARS-CoV and SARS-CoV-2 RBDs on eACE2 are highly similar, although the binding affinities were vastly different, with values of 11 and 782 nM for SARS-CoV-2 and SARS-CoV, respectively. To illustrate the determinants of the differential interactions, we compared the eACE2-interacting residues on the SARS-CoV-2 and SARS-CoV RBDs (Figure 2G; Table S2). A total of 18 and 13 RBD residues were involved in eACE2 binding for SARS-CoV-2 and SARS-CoV, respectively. Among these interacting residues, only eight residues were conserved between SARS-CoV-2 and SARS-CoV (Figure 2G; Table S2). 15 hydrogen bonds and 2 salt bridges were formed between SARS-CoV-2 and eACE2. In the SARS-CoV RBD/eACE2 complex, there were 9 hydrogen bonds and 2 salt bridges. We propose that these differential interactions endow the two coronaviruses with distinct binding affinities.
Comparison of the binding interfaces between RBDs and eACE2 or hACE2
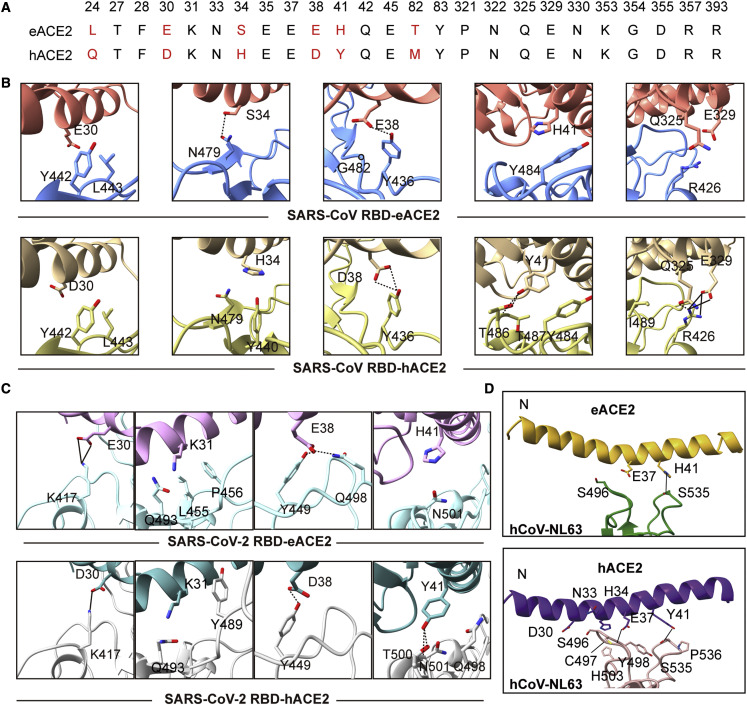
The N-terminal helix (residues 19–50) and three separated regions (residues 319–330, 350–357, and 386–393) of ACE2 are involved in binding the SARS-CoV, SARS-CoV-2, and hCoV-NL63 RBDs (Table S2; Figure S2). Sequence alignments showed that eACE2 and hACE2 are similar, with a sequence identity of 87%. The regions spanning residues 319–330, 350–357, and 386–393 are strictly conserved, whereas residues of the N-terminal helix display the largest variation between eACE2 and hACE2 (Figure S2). Among all ACE2 residues implicated in RBD binding, six positions with different residues were found at the N-terminal helix of human and horse ACE2, including 24 (L/Q), 30 (E/D), 34 (S/H), 38 (E/D), 41 (H/Y), and 82 (T/M), respectively (Figure 3A). All contacting residues at the binding interfaces of RBD and ACE2 orthologs with a distance cutoff of 4 Å are listed in Table S2.
Figure 3.
Comparison of the binding interface of three ACE2-dependent coronaviruses with eACE2 or hACE2
(A) Alignment of key residues for interaction with the RBDs of the ACE2-dependent coronaviruses.
(B) Comparison of interactions at the differential residues between horse and human ACE2 for interacting with the SARS-CoV RBD.
(C) Comparison of differential residues between horse and human ACE2 for interacting with the SARS-CoV-2 RBD.
(D) The observed bend around the N-terminal helix between the eACE2/hCoV-NL63 RBD and hACE2/hCoV-NL63 RBD. All complex structures are aligned according to the RBD in each complex. In the SARS-CoV-2 RBD/eACE2 complex, eACE2 is colored plum, and SARS-CoV-2 RBD is colored powder blue. In the SARS-CoV-2 RBD/hACE2 complex, hACE2 is colored dark cyan, and SARS-CoV-2 RBD is colored gray. In the SARS-CoV RBD/eACE2 complex, eACE2 is colored light coral, and SARS-CoV RBD is colored cornflower blue. In the SARS-CoV RBD/hACE2 complex, hACE2 is colored rose brown, and SARS-CoV RBD is colored tan. In the hCoV-NL63 RBD/eACE2 complex, eACE2 is colored goldenrod, and hCoV-NL63 RBD is colored green. In the hCoV-NL63 RBD/hACE2 complex, hACE2 is colored purple, and hCoV-NL63 RBD is colored pink. Hydrogen bonds and salt bridges are indicated by dashed and solid black lines, respectively.
We first analyzed the distinct interactions between horse and human ACE2 when bound to the SARS-CoV RBD. These differential interactions of the SARS-CoV RBD bound to eACE2 or hACE2 are located at variable (24, 30, 34, 38, 41, and 82) and conserved residues (79, 325, and 329) (Figure 3A; Table S2). Among these sites with differential interactions, L24, T82, and L79 are located near the interface of crystal packing of SARS-CoV RBD/hACE2. Thus, we mainly focused on the remaining sites for the detailed analysis and comparison. At position 30 of ACE2, E30 of eACE2 interacts with Y442 and L443 of the SARS-CoV RBD, whereas D30 of hACE2 has no contacts with the SARS-CoV RBD because of the shorter side chain. Similarly, E38 of eACE2 forms interactions with Y486 and G482, whereas D38 of hACE2 only interacts with Y486. On the other hand, hACE2 displayed much closer contact than eACE2 via the variable residues 34 and 41 as well as the conserved residues 325 and 329. H34 of hACE2 interacts with N479 and Y440, whereas only N479 is involved in binding with S34 of eACE2. Y41 of hACE2 forms hydrogen bonds with Y484, T486, and T487, whereas only Y484 is in contact with H41 of eACE2. The conserved residues Q325 and E329 in hACE2 form hydrogen bonds and salt bridges with R426 of the SARS-CoV RBD, which is absent in the SARS-CoV RBD/eACE2 complex (Figure 3B; Table S2).
When binding the RBD of SARS-CoV-2, varied interactions are found at four positions of the ACE2 orthologs, including 30, 31, 38, and 41. E30 of eACE2 forms two salt bridges with K417 of the SARS-CoV-2 RBD, whereas only one salt bridge was found between D30 of hACE2 and K417 of the SARS-CoV-2 RBD. E38 of eACE2 interacts with Y449 and Q498 by forming two hydrogen bonds, whereas D38 of hACE2 only forms one hydrogen bond with Y449.K31 of eACE2 interacts with L455, P456, and Q493, whereas K31 of hACE2 interacts with Y489 and Q493. H41 of eACE2 only forms interaction with N501, whereas Y41 of hACE2 interacts with Q498 and N501 and forms hydrogen bonds with T500 (Figure 3C; Table S2). We also compared the binding interfaces of the hCoV-NL63 RBD with eACE2 and hACE2. The two binding interfaces were highly similar except for the bent N-terminal helix in eACE2 (Figure 3D; Table S2). Because the crystals of hCoV-NL63 RBD/eACE2 and hCoV-NL63 RBD/hACE2 were in different space groups, we could not exclude the possibility that the observed bending of the N-terminal helix was a result of different crystal packing.
eACE2 displayed lower binding affinities for the RBDs of SARS-CoV-2 Alpha, Beta, and Gamma variants
A number of novel variants of SARS-CoV-2 have emerged since early 2020, some of which displayed increased transmissibility, virulence, antibody resistance, or broadened host range (Table S3; Cele et al., 2021; Planas et al., 2021; Hoffmann et al., 2021; Supasa et al., 2021; Liu et al., 2021d; Zhou et al., 2021; Bushman et al., 2021). To evaluate whether these circulating variants of SARS-CoV-2 might be more transmissible to horses, we first analyzed the binding of eACE2 or hACE2 to the 13 mutated S proteins expressed on the surface of HEK293T cells, including two SARS-CoV-2 VOCs, three previous VOCs, and seven previous VOIs (Figures 4A and S3). The cell-based surface staining assay suggested that eACE2 had extremely low affinity for the S of SARS-CoV-2 Alpha, Beta, and Gamma as well as significantly increased and slightly lower affinity for the Delta and Omicron variants, respectively. In contrast, eACE2 showed comparable or enhanced affinities for all tested VOIs and Delta plus (Figure 4A). With hACE2, almost all variants displayed comparable or even enhanced binding, especially the five tested VOCs (Figure 4A), which might be a result of the evolution of these SARS-CoV-2 variants in the human population, which made them more adaptable to hACE2. The effect of these five VOCs on eACE2 binding was further supported by the SPR assay (Figures 4B and S4). The KD values of eACE2 binding to the SARS-CoV-2 Alpha, Beta, and Gamma RBDs were 3.855 × 10−7 M, 7.381 × 10−6 M, and 6.937 × 10−6 M, which was 35-, 668-, and 628-fold lower, respectively, than that of the wild type (WT). These results were similar to the cell surface staining assay. The binding of eACE2 to the RBDs of Delta and Omicron had similar or slightly decreased affinity compared with the WT RBD. On other hand, the RBDs of all five SARS-CoV-2 VOCs bound hACE2 with comparable or enhanced affinity (Figure 4B), which is in agreement with previous reports (McCallum et al., 2021; Zhang et al., 2021a; Mannar et al., 2021; Cai et al., 2021; Wang et al., 2021b). Cell surface staining and SPR assays suggested that the mutations in SARS-CoV-2 Alpha, Beta, and Gamma variants have a dramatic effect on the binding to eACE2.
Figure 4.
Binding of eACE2 and hACE2 to SARS-CoV-2 variants
(A) The variant change ratio in binding activity, determined by measuring the mean fluorescence intensity (MFI) relative to that of WT D614G. Values over 100% indicate an increase, and those lower than 100% indicate a decrease. Results were calculated from two independent experiments performed in technical duplicates (n = 2) and represent mean ± SD. pVOC, previous variant of concern; pVOI, previous variant of interest. See also Figure S3.
(B) The binding affinities of horse or human ACE2 for the RBDs of SARS-CoV-2 variants are summarized. Horse or human ACE2 was immobilized on the CM5 chip, and RBDs were flowed through. See also Figure S4.
H41 accounts for the lower binding affinity of eACE2 for the SARS-CoV-2 Alpha, Beta, and Gamma variants
Next we tried to decipher the critical determinant of the lower binding affinity of eACE2. The decreased binding affinity (∼35-fold lower) of SARS-CoV-2 Alpha (N501Y) and the further decreased affinity (∼600-fold lower) of SARS-CoV-2 Beta and Gamma (K417 N/T, E484K, N501Y) indicates that the N501Y, K417N/T, and E484K mutations would disfavor binding of eACE2 with the SARS-CoV-2 RBD. In the complexes of the SARS-CoV-2 RBD bound to eACE2, K417 and N501 are located at the binding interface, whereas E484 is located at its outer edge. We proposed that the E484K mutation would not disrupt binding of the SARS-CoV-2 RBD to eACE2, which was supported by the higher binding affinity of the Iota (E484K) variant for eACE2 (Figure 4A). Thus, we mainly focused on K417 and N501 for the following analysis. As shown in Figure 3C, the residues in eACE2/hACE2 for interacting with SARS-CoV-2 RBD K417 and N501 are E30/D30 and H41/Y41. In previous studies, the K417N mutation has been reported to lower the binding affinity of hACE2 for the RBD of SARS-CoV-2 (Barton et al., 2021; Upadhyay et al., 2021; Zhang et al., 2021b), which is consistent with the mode of binding to eACE2. Thus, we considered E30/D30 as the critical binding site but not a differential site for eACE2/hACE2 binding to SARS-CoV-2 RBD. We hypothesized that the H41/Y41 residue, which interacts with N501, mainly accounts for the differential binding affinity of equine and human ACE2 for the RBDs of SARS-CoV-2 variants.
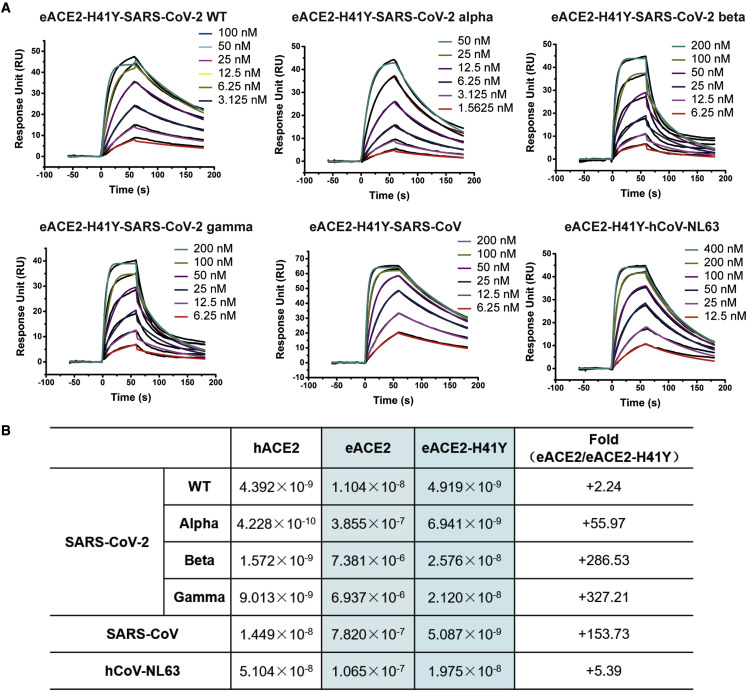
To confirm the role of H41/Y41 in RBD binding, we constructed a variant of eACE2 with the H41Y mutation (eACE2-H41Y) and evaluated its binding affinity (Figure 5A). The mutation of H41Y in eACE2 rescued the low affinity of eACE2 for the RBDs of the SARS-CoV-2 Alpha, Beta, and Gamma variants, with ∼55-, ∼280-, and ∼320-fold increases in affinity, respectively (Figure 5B). We also evaluated the influence of the H41Y mutation on binding of eACE2 to the RBDs of SARS-CoV and hCoV-NL63. The results suggested that the H41Y mutation also endowed eACE2 with dramatically increased affinity for SARS-CoV (∼153-fold) and somewhat increased affinity for hCoV-NL63 (∼5-fold), which confirmed the importance of Y41 for receptor binding of these ACE2-dependent coronaviruses (Figure 5B). Although the H41Y mutation increased binding of eACE2 to the RBDs of SARS-CoV-2 Alpha, Beta, and Gamma, there were disparities between the binding of these SARS-CoV-2 variants to eACE2-H41Y and hACE2. We hypothesized that other direct and indirect interacting residues might also account for the remaining gap in binding affinity.
Figure 5.
H41Y enhances the binding of eACE2 to SARS-CoV-2, SARS-CoV, and hCoV-NL63
(A) SPR curves of the binding between the eACE2-H41Y RBD and SARS-CoV-2 variants, SARS-CoV, or hCoV-NL63. Purified eACE2-H41Y was immobilized on the CM5 chip, and the indicated RBDs were flowed through.
(B) The binding affinities and fold changes are summarized.
Pseudovirus entry of SARS-CoV-2 variants into cells expressing eACE2 and hACE2
To further study the potential of SARS-CoV-2 variants to infect horses, we characterized the ability of eACE2 to support entry of 13 SARS-CoV-2 pseudoviruses bearing mutations from two VOCs, three previous VOCs, and seven previous VOIs. HeLa cells stably expressing eACE2 or hACE2 were subjected to a pseudovirus infection assay (Figure 6A), and the cell entry efficiency was presented as the changed ratio relative to the WT pseudovirus bearing the D614G mutation. As shown in Figure 6B, all of these SARS-CoV-2 variants displayed comparable or increased pseudovirus infection of HeLa-hACE2 cells, whereas only a part of the variants, including Alpha, Beta, Delta, Eta, Iota, Lambda, and Delta plus, exhibited enhanced infection of HeLa-eACE2 cells. The Delta variant exhibited the most significantly enhanced infectivity in HeLa-eACE2 cells, which was similar to that of HeLa-hACE2 cells.
Figure 6.
Entry efficiency of pseudoviruses based on SARS-CoV-2 variants in HeLa cells expressing eACE2 or hACE2
(A) The work flow of pseudovirus infection assay of SARS-CoV-2 variants.
(B) The values show the variant change ratio in luciferase activity for each indicated mutated pseudovirus variant compared with WT D614G. Values over 100% indicate an increase, and those lower than 100% indicate a decrease. Results were calculated from two independent experiments performed in technical duplicates (n = 2) and represent mean ± SD.
Discussion
A variety of animals have been reported as confirmed or potential hosts for SARS-CoV-2, SARS-CoV, and hCoV-NL63 infection. In this study, we confirmed that eACE2 is able to bind the RBDs of SARS-CoV-2, SARS-CoV, and hCoV-NL63, although with a lower affinity. We determined the complex structures of these three RBDs bound to eACE2. Compared with the previously reported structures of the RBD/hACE2 complex, we found that eACE2 engaged a binding mode almost identical to these three ACE2-dependent coronaviruses. The binding assay of eACE2-H41Y with SARS-CoV-2 variants suggested that H41 is the critical determinant of the lower binding affinity of eACE2 for the RBDs of the SARS-CoV-2 Alpha, Beta, and Gamma variants. We also evaluated the infection ability of SARS-CoV-2 variant pseudoviruses in HeLa-eACE2 cells, which indicated that some SARS-CoV-2 variants may have the potential to infect horses.
The importance of H41 for binding of ACE2 to SARS-CoV-2 was also supported by other studies (Bouricha et al., 2020; Liu et al., 2021b, 2021c). Substitution of tyrosine with histidine at position 41 of ACE2 in silico has been proposed to lower the binding affinity of SARS-CoV-2 for the ACE2 of horses (Bouricha et al., 2020), and a Y41H variant of ACE2 from bats (Rhinolophus macrotis) showed attenuated binding affinity for the RBD of SARS-CoV-2 (Liu et al., 2021b). New World monkeys with H41-E42 in their ACE2 are resistant to SARS-CoV-2 infection, and the H41Y-E42Q double mutation could dramatically increase the binding and cell entry of SARS-CoV-2 (Liu et al., 2021c). Sequence alignment of ACE2 orthologs from domestic and wild animals suggested that Y41 is highly conserved in most of these animals, except for donkeys, horses, and some species of bats, which harbor the H41 variation (Li et al., 2020). Because bats have extensive species diversity, H41 occurred frequently in their ACE2s, which is consistent with the relatively low binding of SARS-CoV-2 to ACE2s of these bats (Liu et al., 2021b).
In addition to H41, which disfavors binding, E30 of eACE2 with a longer acidic side chain forms more interactions than D30 of hACE2 when binding to the RBDs of SARS-CoV-2 and SARS-CoV. This was consistent with the results of a previous deep mutagenesis assay (Chan et al., 2020) and the higher neutralization potency of ACE2-immunoglobulin (Ig) with a D30E mutation (Li et al., 2020). E38 of eACE2 also forms more complex interactions than hACE2-D38, which, we hypothesized, might contribute to stronger binding. These affinity-enhancing mutations might be applied to optimize soluble ACE2 decoys for therapeutic applications.
In addition to the direct interaction between RBD and ACE2, indirect regions might also play an important role in receptor binding. The ACE2 of donkeys (Equus asinus, XM_014857647.1) is completely identical to eACE2 at these key interacting residues (Figure S2A), and donkeys have been predicted in silico to be potential hosts of SARS-CoV-2 (Damas et al., 2020). Experimental assays indicated that eACE2 could bind the RBD of SARS-CoV-2, and HEK293T cells expressing eACE2 could be infected effectively by SARS-CoV-2 live virus but those expressing the ACE2 of donkeys could not (Li et al., 2020). Sequence alignment of the ACE2 orthologs from horses and donkeys revealed a 20-amino-acid deletion in the latter. This deletion is positioned in the larger lobe of ACE2, which is far away from the direct RBD-binding region (Figures S2A and S2B). This special case reminds us that in addition to the direct interacting regions, attention should also be paid to other parts of ACE2.
Binding between S protein and host receptors is a prerequisite for coronaviruses to realize cell entry. However, low binding affinity does not always equal low infection efficiency because the infection process is rather complicated and driven by multiple factors, such as the possibility of direct contact, receptor binding and membrane fusion, replication of the genome and translation of structural proteins, as well as viral particle assembly and release. Mink and mouse are two examples of species with low receptor binding affinity but efficient cell entry. The binding of mink ACE2 to the SARS-CoV-2 RBD was undetectable at a concentration of 400 nM RBD (Ren et al., 2021), but SARS-CoV-2 successfully infected mink naturally. HeLa cells expressing mouse ACE2 have been reported to be permissive for infection by the SARS-CoV-2 Alpha, Beta, and Gamma strains (Wang et al., 2021a), although the binding affinity is as low as 353 μM (Sun et al., 2021). These results indicate that, if direct contact is achieved, then weak or intermittent binding is enough for SARS-CoV-2 to realize infection.
Bats are widely considered the most likely natural reservoir of SARS-CoV-2. However, the intermediate hosts of SARS-CoV-2 are still mysterious, which hinders our understanding of the interspecies transmission and evolution of SARS-CoV-2. Although there is no evidence that horses can be infected naturally by any of the three ACE2-dependent pathogenic coronaviruses tested in this study, the underlying mechanism of horse receptor binding indicates that caution is warranted to guard against future interspecies transmission of these ACE2-dependent coronaviruses.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-coronavirus (SARS-CoV-2) spike S2 mouse mAb | MP Biomedicals | Cat#08720401; RRID: AB_2920626 |
| PE-labeled anti-his antibody | Miltenyi | Cat#130120787; RRID: AB_2752196 |
| FITC-labeled anti-mouse IgG | ThermoFisher Scientific | Cat# A16073; RRID: AB_2534746 |
| Chemicals, peptides, and recombinant proteins | ||
| Fetal bovine serum | GIBCO | Cat#16000-044 |
| Trypsin | Macgene | Cat#CC017 |
| ACE2 recombinant protein | This paper | N/A |
| Recombinant coronavirus RBDs | This paper | N/A |
| Cellfectin II Reagents | GIBCO | Cat#10362100 |
| High Affinity Ni-Charged Resin FF | Genscript | Cat# L00666 |
| Critical commercial assays | ||
| Bright-Glo™ Luciferase Assay Buffer | Promega | Cat#E264B |
| Bright-Glo™ Luciferase Assay Substrate | Promega | Cat#E263B |
| Deposited data | ||
| Horse ACE2 with SARS-CoV-2 RBD | This paper | PDB: 7FC5 |
| Horse ACE2 with SARS-CoV RBD | This paper | PDB: 7FC6 |
| Horse ACE2 with hCoV-NL63 RBD | This paper | PDB: 7FC3 |
| Experimental models: Cell lines | ||
| Human: HEK293T, embryo | ATCC | CRL-3216 |
| Human: HeLa, Female | ATCC | CCL-2 |
| HeLa expressing ACE2 from diverse origin, Female | Dr. Qiang Ding’s lab | Liu et al., 2021c |
| Spodoptera frugiperda cells | ATCC | CRL-1711 |
| Trichoplusia ni cells | ATCC | PTA-9384 |
| Oligonucleotides | ||
| eACE2for: TCCCACCATCGGGCGCGGATC CATGAGCGGCAGCTCCTGGCTGCT |
RuiBiotech | N/A |
| eACE2rev: CCTCTAGTACTTCTCGACAAGC TTTCAATGGTGATGGTGATGGTGGTCGGC GTAGGGGGACC |
RuiBiotech | N/A |
| eACE2-H41Yfor: CTGAGCTACCAGTCTA GCCTGGCCTCT |
RuiBiotech | N/A |
| eACE2-H41Yrev: AGACTGGTAGCTCAGC TCCTCGGCCTCA |
RuiBiotech | N/A |
| SARS-CoV-2for: TCTGCCTTTGCGGCGGATCCCACCAAT CTGTGCCCTTTCGGCGA |
RuiBiotech | N/A |
| SARS-CoV-2rev: TCCTCTAGTACTTCTC GACAAGCTTTCAATGGTGATGGTGATG GTGAGGGCCGCACACGGTA |
RuiBiotech | N/A |
| hCoV-NL63for: TCTGCCTTTGCGGCGG ATCCCCAGCACACCGACATCAACT |
RuiBiotech | N/A |
| hCoV-NL63rev: TCCTCTAGTACTTCTCGACAAGCTTTCAA TGGTGATGGTGATGGTGGATACCGGA CACAGGGT |
RuiBiotech | N/A |
| SARS-CoVfor: TCTGCCTTTGCGGCGGATCCCCGGGT GGTGCCCAGCGGCGAC |
RuiBiotech | N/A |
| SARS-CoVrev: TCCTCTAGTACTTCTCGACAAGCTTTCA ATGGTGATGGTGATGGTGCAGCTTGGG GCCGCACAC |
RuiBiotech | N/A |
| Recombinant DNA | ||
| pcDNA3.1 | Thermo Fisher | Cat#V79020 |
| pFastBac-Dual vector | GIBCO | Cat#10712024 |
| pLVX-IRES-zsGreen1 vector | Clontech Laboratories, Inc | Cat#632187 |
| pNL4-3-R-E-luciferase | NIH-AIDS Reagent Program | Cat#3418 |
| Software and algorithms | ||
| FlowJo 10 software | FlowJo | https://www.flowjo.com/ |
| Graphpad Prism 7 | GraphPad | www.graphpad.com |
| Biacore Evaluation Software | GE Healthcare | https://www.cytivalifesciences.com/en/us/shop/protein-analysis/spr-label-freeanalysis/software/biacore-insightevaluation-software-p-23528 |
| HKL2000 | Otwinowski and Minor (1997) | https://www.hkl-xray.com/hkl-2000 |
| PHASER (CCP4 Program Suite) | McCoy et al. (2007) | http://www.phaser.cimr.cam.ac.uk/inind.php/Phaser_Crystallographic_Software |
| COOT | Emsley and Cowtan (2004) | http://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot/ |
| PHENIX | Adams et al. (2002) | http://www.phenix-online.org/ |
| UCSF Chimera X | Pettersen et al. (2021) | https://www.cgl.ucsf.edu/chimerax/ |
| Other | ||
| Superdex 200 High-Performance column | GE Healthcare | N/A |
| Nanodrop One Spectrophotometer | Thermo Scientific | N/A |
| CM5 sensor chip | GE Healthcare | Cat#BR100530 |
| Biacore T200 | GE Healthcare | N/A |
Resource availability
Lead contact
Further information and requests for resources and primary data should be directed to and will be fulfilled by the Lead Contact, Jiwan Ge (gejw@mail.tsinghua.edu.cn).
Materials availability
We will share these reagents without any restrictions.
Experimental model and subject details
HEK 293T cells (ATCC, CRL-3216), HeLa cells (ATCC, CCL-2) and HeLa cells expressing ACE2 orthologs (HeLa-hACE2, HeLa-eACE2) were maintained at 37°C in a humidified atmosphere comprising 5% CO2 in Dulbecco’s minimal essential medium (DMEM) containing 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 100 U/mL of penicillin–streptomycin. Escherichia coli DH5α used for vector amplification and DH10bac used for recombinant bacmid production were both cultured in LB media.
Method details
Crystallization and data collection
Residues 319 to 541 of SARS-CoV-2 S, 306 to 527 of SARS-CoV S, 481–616 of hCoV-NL63S, and 19–615 of eACE2 were individually cloned into the pFastBac dual vector with a gp67 signal peptide at the N-terminus and expressed using the Bac-to-bac baculovirus system. Proteins were purified as previously reported (Li et al., 2005; Lan et al., 2020; Wu et al., 2009). Complexes were obtained by mixing ACE2 and RBD at a molar ratio of 1:1.2 and further purified on a Superdex 200 size-exclusion column pre-equilibrated with HBS buffer. Fractions were collected and concentrated to 10 mg/mL for crystal screening. After about one week, the crystal of the hCoV-NL63 RBD/eACE2 complex was grown in 1.8 M ammonium sulfate, 0.1 M BIS-TRIS pH 6.5, 2% (v/v) polyethylene glycol monomethyl ether 550; the crystal of the SARS-CoV-2/eACE2 complex was grown in 0.2 M sodium formate, 20% (w/v) polyethylene glycol 3350; the crystal of the SARS-CoV/eACE2 complex was grown in 0.03 M citric acid, 0.07 M BIS-TRIS propane, pH 7.6, 20% (w/v) polyethylene glycol 3350. Diffraction data were collected at the SSRF BL18U1 beamline and processed using HKL 2000 (Otwinowski and Minor, 1997). The data-processing statistics are listed in Table S1.
Structure determination and refinement
Structures were solved by molecular replacement using Phaser in the CCP4 suite (McCoy et al., 2007). Structures with PDB codes 3KBH, 6M0J and 2AJF were used as search models for hCoV-NL63 RBD/eACE2, SARS-CoV-2/eACE2 and SARS-CoV/eACE2, respectively. The structure was later refined in PHENIX and COOT (Emsley and Cowtan, 2004; Adams et al., 2002). Final Ramachandran statistics were as follows: 94.66% favored, 5.08% allowed and 0.25% outliers for SARS-CoV-2 RBD/eACE2; 95.16% favored, 4.59% allowed and 0.25% outliers for SARS-CoV RBD/eACE2; 95.01% favored, 4.85% allowed and 0.14% outliers for hCoV-NL63 RBD/eACE2. The structure refinement statistics are listed in Table S1. All structure figures were generated using UCSF Chimera X (Pettersen et al., 2021).
Surface plasmon resonance spectroscopy
Purified hACE2 or eACE2 was immobilized on a CM5 sensor chip (GE Healthcare) to a level of ∼450 response units on a Biacore T200 instrument (GE Healthcare). Serial dilutions of the SARS-CoV RBD, SARS-CoV-2 RBD, hCoV-NL63 RBD, and mutants of SARS-CoV-2 RBD were flowed through in a running buffer composed of 10 mM HEPES, pH 7.2, 150 mM NaCl and 0.05% Tween 20. SPR was performed with RBD concentrations ranging from 200 to 1.5625 nM with double dilution. The resulting data were fitted to a 1:1 binding model using Biacore Evaluation Software (GE Healthcare).
Binding of ACE2 to cell surface-expressed wild-type and mutated S glycoprotein
The entire procedure was conducted as previously published (Starr et al., 2020). Briefly, HEK 293T cells were transfected with plasmids encoding either wild-type or mutated SARS-CoV-2 S glycoproteins, and incubated at 37°C for 36 h. Cells were digested from the plate with trypsin and distributed into 96-well plates. Cells were washed twice with 200 μL staining buffer (PBS with 2% heat-inactivated fetal bovine serum (FBS) between each of the following steps. First, cells were stained with the human and horse ACE2, S2-specific monoclonal antibody (MP Biomedicals, Singapore 08720401), at 4°C for 30 min in 100 μL staining buffer. Then, a PE-labeled anti-his antibody (Miltenyi 130120787) or FITC-labeled anti-mouse IgG (ThermoFisher Scientific A10673), was added in 40 μL staining buffer at 4°C for 30 min. After extensive washing, the cells were resuspended and analyzed using a BD LSRFortessa fluoresce-activated cell sorting (FACS) instrument (BD Biosciences, USA) with FlowJo 10 software (FlowJo, USA). Serial dilutions of ACE2 and S2-specific monoclonal antibodies were tested, and the lowest saturated concentrations were used in the assay (5 μg/mL for ACE2, 1 μg/mL for S2-specific monoclonal antibody). Mock-transfected HEK 293T cells were stained using the same procedure as background control. The ACE2 or antibody binding percentages were calculated based on the ratio between mutated and wild-type MFI normalized to that of the S2 specific antibody. All MFI values were weighted by multiplying the number of positive cells in the selected gates.
Production of SARS-CoV-2 wild-type and variant pseudoviruses
The wild-type pseudovirus used throughout the analysis was based on the prototype strain (GenBank: MN908947.3) with the D614G mutation (WT D614G). Thirteen mutated strains were constructed, including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), Omicron (B.1.1.529), Kappa (B.1.617.1), Eta (B.1.525), Epsilon (B.1.427/429), Iota (B.1.526), Lambda (C.37), Mu (B.1.621), A23.1 and Delta plus. The genes of variants were synthesized by Genewiz, Inc. The single mutations identified from the GISAID database were introduced into the pcDNA3.1 vector encoding WT D614G using a QuickChange site-directed mutagenesis kit (Agilent 210519). SARS-CoV-2 pseudoviruses were generated by co-transfecting HEK 293T cells (ATCC) with human immunodeficiency virus backbones expressing firefly luciferase (pNL4-3-R-E-luciferase) and a pcDNA3.1 vector encoding either wild-type or mutated S proteins. The virion-containing supernatant was collected 48h or 72h later, centrifuged to remove cell lysis, and stored at −80°C until use.
HeLa cell lines expressing hACE2 or eACE2
HeLa cells expressing ACE2 orthologs were kindly provided by Dr. Qiang Ding from Tsinghua University School of Medicine. The species names and accession numbers for the ACE2 orthologs used are as follows: Human, Homo sapiens, NP_001358344.1; Horse, Equus caballus, XP_001490241.1. The cDNA of ACE2 orthologs, each with a C-terminal FLAG-tag, were synthesized by GenScript and cloned into the pLVX-IRES-zsGreen1 vector (Catalog No. 632187, Clontech Laboratories, Inc). HeLa cells were transduced with lentiviruses expressing the ACE2 orthologs as previously described (Liu et al., 2021c).
Quantification and statistical analysis
Statistical analysis of the different experiments can be found in each methods section and figure legends. The cell surface binding and pseudovirus entry were performed as two independent experiments in technical duplicates. Graphpad Prism 7 was used for data plotting.
Acknowledgments
We thank the staff at the SSRF BL18U1 beamline for data collection and processing. We thank staff at the X-ray crystallography platform of the Tsinghua University Technology Center for Protein Research for providing facility support. This work was supported by funds from the National Key Research and Development Program of China (2021YFC2300104), the National Natural Science Foundation of China (32171202 and 32100973), the Tsinghua University Spring Breeze Fund (2020Z99CFY031), and the China Postdoctoral Science Foundation (2020M670295).
Author contributions
J.L. carried out protein expression, purification, crystallization, diffraction data collection, and structure determination, and J.G. and W.L. provided help. J.L. and J.G. carried out the SPR assays, analyzed the data, and drafted all figures. Q.Z., P.C., and L.Z. performed the cell-based binding assay and pseudovirus infection assay. W.R. and Q.D. constructed HeLa cell lines expressing hACE2 or eACE2. J.G., X.W., and Q.Z. conceived, designed, and directed the study and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: August 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.str.2022.07.005.
Supplemental information
Data and code availability
All the data reported in this paper will be shared by the lead contact upon request. The coordinates and structure factors files for horse ACE2 with hCoV-NL63, SARS-CoV-2 and SARS-CoV RBDs have been deposited to the Protein Data Bank (http://www.rcsb.org) with accession code 7FC3, 7FC5 and 7FC6 respectively. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., Mccoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Alagaili A.N., Briese T., Mishra N., Kapoor V., Sameroff S.C., Burbelo P.D., de Wit E., Munster V.J., Hensley L.E., Zalmout I.S., et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio. 2014;5:e00884-14. doi: 10.1128/mbio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M.R., Schoeder C.T., Brown J.A., Smart C.D., Moth C., Wikswo J.P., Capra J.A., Meiler J., Chen W., Madhur M.S. Predicting susceptibility to SARS-CoV-2 infection based on structural differences in ACE2 across species. FASEB J. 2020;34:15946–15960. doi: 10.1096/fj.202001808r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M.I., Macgowan S.A., Kutuzov M.A., Dushek O., Barton G.J., van der Merwe P.A. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife. 2021;10:e70658. doi: 10.7554/elife.70658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T.Y., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Bouricha E.M., Hakmi M., Akachar J., Belyamani L., Ibrahimi A. In silico analysis of ACE2 orthologues to predict animal host range with high susceptibility to SARS-CoV-2. 3 Biotech. 2020;10:483. doi: 10.1007/s13205-020-02471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan J., Marr C.M., Mackenzie C.J., Mair T.S., Fletcher A., Cash R., Phillips M., Pusterla N., Mapes S., Foote A.K. Detection of equine coronavirus in horses in the United Kingdom. Vet. Rec. 2019;184:123. doi: 10.1136/vr.105098. [DOI] [PubMed] [Google Scholar]
- Bushman M., Kahn R., Taylor B.P., Lipsitch M., Hanage W.P. Population impact of SARS-CoV-2 variants with enhanced transmissibility and/or partial immune escape. Cell. 2021;184:6229–6242.e18. doi: 10.1016/j.cell.2021.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Lavine C.L., Rawson S., Peng H., Walsh R.M., Jr., Zhu H., Anand K., Tong P., et al. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. Science. 2021;373:642–648. doi: 10.2210/pdb7n1w/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Gazy I., Jackson L., Hwa S.H., Lustig G., Giandhari J., Wilkinson E., Naidoo Y., Ganga Y., Khan K., et al. Network for genomic surveillance in South Africa. COMMIT-KZN Team Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Baldwin H.J., Tateno A.F., Zerbinati R.M., Annan A., Owusu M., Nkrumah E.E., Maganga G.D., Oppong S., Adu-Sarkodie Y., et al. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015;89:11858–11870. doi: 10.1128/jvi.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M., Hiller M., Koepfli K.P., Pfenning A.R., Zhao H., et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA. 2020;117:22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Haskew A.N., Gates J.E., Huynh J., Moore C.J., Frieman M.B. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 2010;84:13004–13018. doi: 10.1128/jvi.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/s0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Haake C., Cook S., Pusterla N., Murphy B. Coronavirus infections in companion animals: virology, epidemiology, clinical and pathologic features. Viruses. 2020;12:1023. doi: 10.3390/v12091023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida M.G., Perera R.A., Wang P., Alhammadi M.A., Siu L.Y., Li M., Poon L.L., Saif L., Alnaeem A., Peiris M. Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveill. 2013;18:20659. doi: 10.2807/1560-7917.es2013.18.50.20659. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hornich B.F., Hahn A.S., Kruger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Zeng L.P., Yang X.L., Ge X.Y., Zhang W., Li B., Xie J.Z., Shen X.R., Zhang Y.Z., Wang N., et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J., Li S., Yount B., Smith A., Sturges L., Olsen J.C., Nagel J., Johnson J.B., Agnihothram S., Gates J.E., et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86:12816–12825. doi: 10.1128/jvi.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithete N.L., Stoffberg S., Corman V.M., Cottontail V.M., Richards L.R., Schoeman M.C., Drosten C., Drexler J.F., Preiser W. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg. Infect. Dis. 2013;19:1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79:11892–11900. doi: 10.1128/jvi.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.Y., Jia N., Zhang Y.W., Shum M.H.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lau S.K.P., Li K.S.M., Tsang A.K., Lam C.S.F., Lam C.S., Ahmed S., Chen H., Chan K.H., Woo P.C., Yuen K.Y. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:8638–8650. doi: 10.1128/jvi.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.W., Wong B.H.L., Wong S.S.Y., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang H., Tang X., Fang S., Ma D., Du C., Wang Y., Pan H., Yao W., Zhang R., et al. SARS-CoV-2 and three related coronaviruses utilize multiple ACE2 orthologs and are potently blocked by an improved ACE2-ig. J. Virol. 2020;94 doi: 10.1128/jvi.01283-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Guo R., Liu Y., Zhang Y., Hu J., Ou X., Mi D., Chen T., Mu Z., Han Y., et al. The Rhinolophus affinis bat ACE2 and multiple animal orthologs are functional receptors for bat coronavirus RaTG13 and SARS-CoV-2. Sci. Bull. 2021;66:1215–1227. doi: 10.1016/j.scib.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Pan X., Li L., Yu F., Zheng A., Du P., Han P., Meng Y., Zhang Y., Wu L., et al. Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species. Cell. 2021;184:3438–3451.e10. doi: 10.1016/j.cell.2021.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Tan S., Niu S., Wang J., Wu L., Sun H., Zhang Y., Pan X., Qu X., Du P., et al. Cross-species recognition of SARS-CoV-2 to bat ACE2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2020216118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hu G., Wang Y., REN W., Zhao X., Ji F., Zhu Y., Feng F., Gong M., Ju X., et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Plante K.S., Plante J.A., Xie X., Zhang X., Ku Z., An Z., Scharton D., Schindewolf C., et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. 2021;602:294–299. doi: 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Zhou S., Tuttle K.S., Kim A., Li W., Dimitrov D.S., Subramaniam S. Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding. Cell Rep. 2021;37:110156. doi: 10.1016/j.celrep.2021.110156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., Walls A.C., Sprouse K.R., Bowen J.E., Rosen L.E., Dang H.V., de Marco A., Franko N., Tilles S.W., Logue J., et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science. 2021;374:1621–1626. doi: 10.1126/science.abl8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/s0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S., Wang J., Bai B., Wu L., Zheng A., Chen Q., Du P., Han P., Zhang Y., Jia Y., et al. Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin. EMBO J. 2021;40:e107786. doi: 10.15252/embj.2021107786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/s0076-6879(97)76066-x. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Pusterla N., Vin R., Leutenegger C.M., Mittel L.D., Divers T.J. Enteric coronavirus infection in adult horses. Vet. J. 2018;231:13–18. doi: 10.1016/j.tvjl.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Lan J., Ju X., Gong M., Long Q., Zhu Z., Yu Y., Wu J., Zhong J., Zhang R., et al. Mutation Y453F in the spike protein of SARS-CoV-2 enhances interaction with the mink ACE2 receptor for host adaption. PLoS Pathog. 2021;17:e1010053. doi: 10.1371/journal.ppat.1010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Gu H., Cao L., Chen Q., YE Q., Yang G., Li R.T., Fan H., Deng Y.Q., Song X., et al. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat. Commun. 2021;12:5654. doi: 10.1038/s41467-021-25903-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay V., Lucas A., Panja S., Miyauchi R., Mallela K.M.G. Receptor binding, immune escape, and protein stability direct the natural selection of SARS-CoV-2 variants. J. Biol. Chem. 2021;297:101208. doi: 10.1016/j.jbc.2021.101208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54:1611–1621.e5. doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu C., Wang Y., Hong Q., Zhang C., Li Z., Xu S., Zuo Q., Liu C., Huang Z., Cong Y. Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM. Nat. Commun. 2021;12:7345. doi: 10.1038/s41467-021-27350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Li W., Peng G., Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. USA. 2009;106:19970–19974. doi: 10.2210/pdb3kbh/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Chen Q., Liu K., Wang J., Han P., Zhang Y., Hu Y., Meng Y., Pan X., Qiao C., et al. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020;6:68. doi: 10.1038/s41421-020-00210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Yang X.L., Hu B., Wang B., Wang M.N., Zhang Q., Zhang W., Wu L.J., Ge X.Y., Zhang Y.Z., Daszak P., et al. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J. Virol. 2015;90:3253–3256. doi: 10.1128/jvi.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/nejmoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang J., Xiao T., Cai Y., Lavine C.L., Peng H., Zhu H., Anand K., Tong P., Gautam A., Mayer M.L., et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science. 2021;374:1353–1360. doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang Y., Liu K., Li Y., Lu Q., Wang Q., Zhang Y., Wang L., Liao H., Zheng A., et al. The molecular basis for SARS-CoV-2 binding to dog ACE2. Nat. Commun. 2021;12:4195. doi: 10.1038/s41467-021-24326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data reported in this paper will be shared by the lead contact upon request. The coordinates and structure factors files for horse ACE2 with hCoV-NL63, SARS-CoV-2 and SARS-CoV RBDs have been deposited to the Protein Data Bank (http://www.rcsb.org) with accession code 7FC3, 7FC5 and 7FC6 respectively. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.