Abstract
Broad domains of H3K4 methylation have been associated with consistent expression of tissue-specific, cell identity, and tumor suppressor genes. Here, we identified broad domain–associated genes in healthy human thymic T cell populations and a collection of T cell acute lymphoblastic leukemia (T-ALL) primary samples and cell lines. We found that broad domains are highly dynamic throughout T cell differentiation, and their varying breadth allows the distinction between normal and neoplastic cells. Although broad domains preferentially associate with cell identity and tumor suppressor genes in normal thymocytes, they flag key oncogenes in T-ALL samples. Moreover, the expression of broad domain–associated genes, both coding and noncoding, is frequently deregulated in T-ALL. Using two distinct leukemic models, we showed that the ectopic expression of T-ALL oncogenic transcription factor preferentially impacts the expression of broad domain–associated genes in preleukemic cells. Finally, an H3K4me3 demethylase inhibitor differentially targets T-ALL cell lines depending on the extent and number of broad domains. Our results show that the regulation of broad H3K4me3 domains is associated with leukemogenesis, and suggest that the presence of these structures might be used for epigenetic prioritization of cancer-relevant genes, including long noncoding RNAs.
Chromatin states play an essential role in the establishment of gene expression programs (Flavahan et al. 2017). Combinatorial patterns of histone modifications are expected to convey specific information potential that, in the end, impinges on gene expression. Important information regarding specific transcriptional outputs could be contained in the spread of epigenetic modifications over a genomic locus. Large megabase-sized chromatin domains have been described for the repressive chromatin mark H3K9me3 in senescence cells spanning large chromosomal regions (Soufi et al. 2012) or activating H3K4me3 in mammalian oocytes (Dahl et al. 2016; Liu et al. 2016; Zhang et al. 2016a). Large epigenetic domains along the genome are also emerging as a new concept in defining tissue-specific gene regulation and in the control of normal development and cancer (Hnisz et al. 2017). Active chromatin marks usually restricted to cis-regulatory elements, such as enhancers and promoters, have also been observed in broader deposits (more than several kilobases). For instance, in contrast to typical enhancers that are usually < 1 kb long, super-enhancers are large clusters of enhancers spanning several tens of kilobytes and are associated with large domains of H3K27ac (Hnisz et al. 2017). Super-enhancers are specifically bound by cell type–specific master transcription factors to specifically drive the expression of cell identity genes. Moreover, they have been found at key oncogenes in many cancer types, and disruption of super-enhancers by inhibition of chromatin regulators leads to selective inhibition of key oncogenes (Zhao and Shilatifard 2019).
We and others have uncovered, reminiscent to super-enhancers, that tissue-specific and cell identity gene promoters display a unique pattern of histone modifications featuring broad enrichment of H3K4me2/me3 that extends from the transcription start site (TSS) into the gene body (Pekowska et al. 2010; Koch et al. 2011; Benayoun et al. 2014; Chen et al. 2015; Zacarías-Cabeza et al. 2015; Suzuki et al. 2017). H3K4 methylation is usually confined to nucleosomes surrounding transcriptionally active gene promoters (Bernstein et al. 2005; Barski et al. 2007) and some highly active enhancers (Pekowska et al. 2011). We have previously shown that tissue-specific genes expressed in T cells generally display extended H3K4 methylation within the gene body (Pekowska et al. 2010; Zacarías-Cabeza et al. 2015). Along the same line, it has been shown that H3K4me3 domains that spread more broadly over genes in a given cell type (hereafter, broad H3K4me3 domains) preferentially mark genes that are essential for the identity and function of that cell type (Benayoun et al. 2014). Besides, several complex loci have been shown to display very large H3K4me3 domains including the HOX gene clusters (Bernstein et al. 2005) and the T cell receptor loci (Zacarías-Cabeza et al. 2015). In addition, a subset of tumor suppressors is associated with broad H3K4me3 domains in nonmalignant cells and is lost in cancer cells (Chen et al. 2015). Broad H3K4me3 domains are associated with high levels of RNA Pol II recruitment and pausing (Pekowska et al. 2010; Koch et al. 2011; Benayoun et al. 2014; Chen et al. 2015), leading to transcriptional consistency (Benayoun et al. 2014) and limited perturbation to external factors (Chen et al. 2015). Although the presence of a broad H3K4me3 domain is associated with a relatively high level of gene expression (Benayoun et al. 2015), it rather defines a specialized chromatin structure related to tissue- and cell-specific gene regulation. Although the functional and biological relevance of broad H3K4me3 domains is clearly established, their dynamics during normal development and precise deregulation in cancer are largely unexplored. To gain insight into this issue, here we aimed to analyze the dynamics of broad H3K4me3 domains during early human T cell differentiation in comparison with those found in a collection of primary T cell acute lymphoblastic leukemia (T-ALL) and cell lines.
T-ALL corresponds to a heterogeneous group of acute leukemia that is arrested at various stages of early T cell development with respect to normal thymic cell differentiation. Over the last decade, great progress has been made toward the identification of molecular and genetic abnormalities in T-ALL (Aifantis et al. 2008; Belver and Ferrando 2016; Girardi et al. 2017). A number of these genetic events, which generally involve transcription factors or epigenetic regulators, act mainly to block T cell differentiation, delineating T-ALL subgroups with specific gene expression profiles (Aifantis et al. 2008). It is therefore hypothesized that direct or indirect alterations of cis-regulatory elements could play a central role in the process of leukemogenesis; however, the precise mechanisms and the regulatory elements involved remain poorly characterized (Ntziachristos et al. 2016). The availability of relatively homogenous normal precursors matching the main T-ALL blockade makes it a suitable model to explore the epigenetic changes between normal and neoplastic cells. In this study, we identify broad H3K4me3 domains in human T cell precursors and primary T-ALL samples and cell lines in order to shed light on the dynamics of broad H3K4me3 domains between normal and neoplastic cells.
Results
Identification of broad H3K4me3 domains in human thymic subpopulations
We first compared two different approaches to isolate genes associated with broad domains based on H3K4me3 ChIP-seq data (Supplemental Fig. S1). The first approach was based on density coverage of H3K4me3 on the 5′ side of the gene bodies (Supplemental Fig. S1A; Pekowska et al. 2010). The second approach was based on the association of broad H3K4me3 peaks (MACS tool) with the TSS of genes (Supplemental Fig. S1B; Benayoun et al. 2014; Chen et al. 2015). The later studies defined broad domains from size-ranked H3K4me3 peaks based on 5% of larger peaks (Benayoun et al. 2014) or by selecting peaks >4 kb (Chen et al. 2015). To provide a more unbiased approach to set a threshold for broad H3K4me3 domains, we also tested a selection method based on the inflection point of ranked H3K4me3 peaks (Supplemental Figs. S1B, S2). Using two cut-offs based on the high and low inflection points of ranked H3K4me3 peaks allows the determination of broad H3K4me3 domains, intermediates, and sharp peaks (Supplemental Fig. S1C). Broad domains identified based on H3K4me3 coverage or size-ranked peaks were enriched in similar biological processes (Supplemental Fig. S1D). However, broad H3K4me3 domains identified with size-ranked approaches were preferentially enriched in tissue-specific gene functions. We subsequently used the inflection point–based method to identify broad H3K4me3 domains.
Previous studies have shown that broad H3K4me3 domains are associated with a specific chromatin signature linked to transcriptional elongation (Pekowska et al. 2010; Benayoun et al. 2014; Chen et al. 2015; Zacarías-Cabeza et al. 2015). We confirmed these findings by analyzing broad H3K4me3 domains in the Jurkat cell line (Supplemental Fig. S3). We first selected three subsets of sharp peak–associated genes with the closest expression to broad H3K4me3 domain–associated genes in Jurkat cells (Supplemental Fig. S3A). As expected, the average H3K4me3 profiles at broad domain–associated genes were larger than the sharp-associated controls and enriched in the 5′ part of the gene body (Supplemental Fig. S3B). Compared with sharp-associated genes, broad domains were associated with higher levels of POLR2A, CDK7, and H3K79me2 in the gene body and, to a lesser extent, of H3K36me3. Moreover, broad H3K4me3 domain–associated genes displayed a significant reduction of POLR2A pausing (Supplemental Fig. S3C). To assess whether broad H3K4me3 domains loci can be a preferential target of transcription elongation perturbation, we analyzed published experiments in which the effect of THZ1 (an inhibitor of the cyclin kinase CDK7) and EPZ-5676 (an inhibitor of the H3K79 histone methyltransferase DOT1L) has been assayed on gene expression and H3K4me3 levels, respectively, in Jurkat cells (Kwiatkowski et al. 2014; Orlando et al. 2014). Both inhibitors had a significantly higher impact on broad domain–associated loci compared with sharp-associated genes (Supplemental Fig. S3D). Thus, consistent with previous results, we found that broad H3K4me3 domains are associated with an early transcriptional elongation signature and are shown to be more sensitive to perturbation of transcriptional elongation.
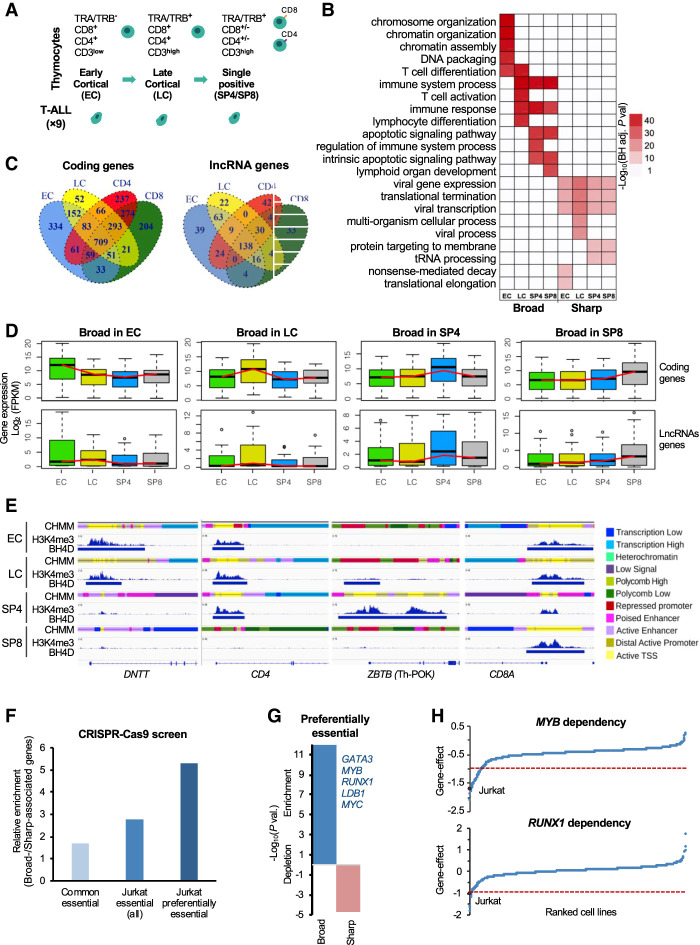
Subsequently, we identify H3K4me3 sharp peaks and broad H3K4me3 domain in human thymic subpopulations using previously generated H3K4me3 ChIP-seq data from sorted human thymocytes encompassing the main thymic T cell differentiation stages and analyzed within the frame of the BLUEPRINT Epigenome Project (Fig. 1A; Supplemental Fig. S2; Supplemental Tables S1–S3; Stunnenberg and Hirst 2016; Cieslak et al. 2020). This included early cortical (EC; TRA−/TRB−/CD3−/CD4+/CD8+), late cortical (LC; TRA+/TRB+/CD3+low/CD4+/CD8+), single positive CD4 (SP4; TRA+/TRB+/CD3+/CD4+/CD8−), and single positive CD8 (SP8; TRA+/TRB+/CD3+/CD4−/CD8+) thymocytes. Sharp peaks and broad H3K4me3 domain displayed similar H3K4me3 signals in the different T cell subpopulations (Supplemental Fig. S2B,C). To assess whether broad H3K4me3 domain–associated genes were specifically enriched in cell identity genes, as previously suggested (Pekowska et al. 2010; Benayoun et al. 2014; Chen et al. 2015; Zacarías-Cabeza et al. 2015), we compared the enriched Gene Ontology (GO) biological process between the broad and sharp H3K4me3 peak-associated genes for each developmental stage (Fig. 1B). Broad H3K4me3 domains in EC thymocytes were associated with chromatin regulation and T cell differentiation, whereas the broad H3K4me3 domains in the more committed LC thymocytes were exclusively associated with T cell and immune functions. The most mature thymocytes, SP4 and SP8, were enriched in immune response and apoptosis, reflecting the positive and negative selection processes. In contrast, genes associated with sharp H3K4me3 peaks were associated with unrelated (e.g., viral process) or metabolic (tRNA processing) terms, consistent with previous results (Chen et al. 2015).
Figure 1.
Dynamics of broad H3K4me3 domains during early T cell differentiation. (A) Schematic representation of the major stages of human thymopoiesis. (B) Top enriched pathways associated with broad H3K4me3 domain genes. (C) Overlap of broad H3K4me3 domains associated with coding or lncRNA genes between the thymic T cell subpopulations. (D) Box plot displaying the distribution of gene expression in each T cell subpopulation of genes associated with broad H3K4me3 domains in the indicated subpopulation. (E) Examples of dynamic broad H3K4me3 domain loci. H3K4me3 ChIP-seq signals, broad domains, and chromatin state (CHMM) profiles are shown for the indicated thymocyte subpopulations. The color-code is indicated at the right of the panel. (F) Ratio between the percentage of broad- and sharp-associated genes that were defined as commonly essential, essential in Jurkat cells, or preferentially essential in Jurkat cells based on the DepMap data set of loss-of-function CRISPR-Cas9-based screen for essential genes (for details, see Methods). (G) Significant overlap between broad- and sharp-associated genes in Jurkat cells and preferential essential genes was assessed by the hypergeometric test. (H) Example of two preferentially essential genes associated with broad H3K4me3 domains in Jurkat cells. The gene effect from the loss-of-function CRISPR-Cas9-based assays in all the cell lines is shown, and the Jurkat cell line is highlighted.
Next, we compared the presence of broad H3K4me3 domains between the thymic subpopulations and observed this to be highly dynamic throughout T cell differentiation (Fig. 1C). We next assessed the expression of the broad H3K4me3 domain–associated genes using our previously published RNA-seq data set (Fig. 1D; Cieslak et al. 2020). We observed that average gene expression was higher in the thymic subpopulation, where the broad H3K4me3 domain was present over the gene body, suggesting that the dynamics of broad domains correlates with changes in gene expression. This is highlighted by the epigenomic profiles of several key T cell–specific genes, such as DNTT, CD4, CD8A, ZBTB, CD1C, CD44, and RUNX3 (Fig. 1E; Supplemental Fig. S4A), where the presence of the broad H3K4me3 domain generally correlates with a transcriptionally-active chromatin, based on chromatin states defined in the same thymocyte populations. In contrast, the absence of broad H3K4me3 domain at these loci is often associated with repressive chromatin enriched with Polycomb-associated states. As described before for cell lines and primary tissues, here we showed that broad H3K4me3 domains dynamically associate with cell identity genes throughout T cell differentiation, highlighting that the presence of broad H3K4me3 domains is a hallmark of key stage-specific genes.
Broad H3K4me3 domains are associated with preferentially essential genes
Previous studies have suggested that broad H3K4me3 domains in a given cell type preferentially mark genes that are essential for the identity and function of that cell type (Benayoun et al. 2014; Chen et al. 2015). To directly assess the functional relevance of broad H3K4me3 domain–associated genes, we analyzed the DepMap data set, a large-scale loss-of-function CRISPR-Cas9-based screen for essential genes, which was performed in 808 human cancer cell lines, including the T-ALL Jurkat cell line (Behan et al. 2019; https://depmap.org). We assessed the relative enrichment of broad H3K4me3 domain–associated versus sharp-associated genes in the Jurkat cell line that were found to be essential in the majority of the cell lines (common essential), essential in Jurkat (all), or preferentially essential in Jurkat (Fig. 1F; Supplemental Table S4). Preferentially essential genes were defined as the top 25% of essential genes in Jurkat cells for which the gene effect is higher in Jurkat cells compared with the average of all the other cell lines (see Methods section). We observed that broad H3K4me3 domains in Jurkat cells have the strongest relative enrichment for genes that are preferentially essential in this cell line, which functionally define cell identity genes (Fig. 1F,G). Moreover, broad H3K4me3 domains are significantly associated with preferentially essential genes in Jurkat, whereas the sharp-associated genes are significantly depleted (Chi-square test, P-value < 0.0001) (Fig. 1G). Among the broad H3K4me3 domain genes that are preferentially essential in Jurkat cells, we found several transcriptional regulators highly relevant for T-ALL leukemogenesis (Fig. 1F, inset; Supplemental Table S4A), including MYC and the GATA3, MYB, RUNX1, and LDB1 transcription factors (Fig. 1H), which are parts of the TAL1-associated complex, a critical oncogenic complex active in Jurkat cells (Porcher et al. 2017). Thus, broad H3K4me3 domains mark genes that are preferentially essential in the given cell type.
Broad H3K4me3 domains flag driver oncogenes in T-ALL
Previous work has shown that broad H3K4me3 domains are associated with tumor suppressor genes in normal tissues (Chen et al. 2015), whereas the above results suggested that they can also mark oncogenic factors in cancer cells. We first analyzed the association of broad H3K4me3 domains present in normal thymocytes with a comprehensive list of 50 tumor suppressor gene in T-ALL (Supplemental Table S5), including frequently mutated genes (Liu et al. 2017). We found that broad domains present in at least one thymocyte sample were significantly associated with tumor suppressor genes (P-value = 1.3 × 10−23, hypergeometric test) (Supplemental Fig. S4B). This is illustrated by the lymphoid-specific tumor suppressor gene IKZF1 and the pan-cancer tumor suppressor gene PTEN, which are both associated with broad domains in all thymic subpopulations (Supplemental Fig. S4C) and are previously associated with broad domains in mature CD4+ T cells (Chen et al. 2015). Broad domains present in thymocytes and are conserved across other normal tissues (i.e., present in at least 50% of ENCODE normal tissues as defined by Chen et al. 2015) and are more frequently overlapped with tumor suppressor genes (P-value = 0.002; two-sided Fisher's exact test) (Supplemental Fig. S4B), supporting the previous finding that conserved broad domains are associated with pan-cancer tumor suppressor genes (Chen et al. 2015).
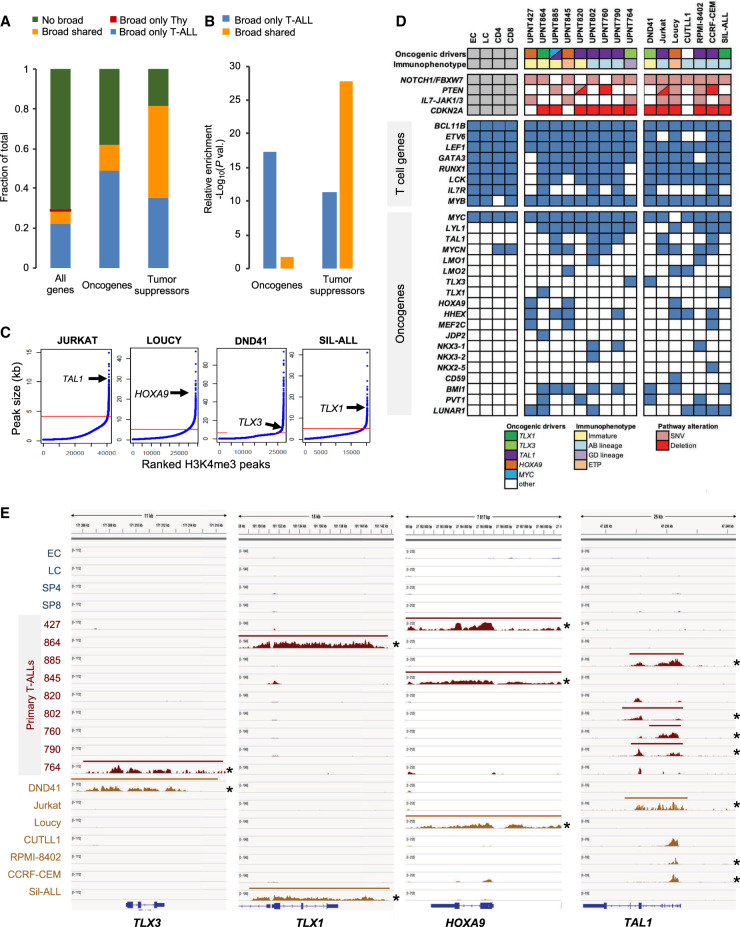
To assess the impact of broad H3K4me3 domain changes between normal T cell precursors and T-ALL neoplasia, we analyzed H3K4me3 ChIP-seq data in a series of nine primary T-ALLs and seven T-ALL-derived cell lines, using published and newly generated experiments (Supplemental Fig. S2; Supplemental Tables S1–S3). We analyzed the association of the broad H3K4me3 domain with a comprehensive list of T-ALL oncogenes and tumor suppressor genes (Supplemental Table S5), including lncRNAs, as well as, frequently mutated genes (Liu et al. 2017). Tumor suppressor genes were preferentially associated with broad H3K4me3 domains shared between T-ALL and thymocytes (Fig. 2A,B). However, we also found that T-ALL oncogenes were significantly associated with a gain of broad H3K4me3 domains in T-ALL (Fig. 2B), suggesting that the increased breadth of H3K4 trimethylation is linked to oncogenic up-regulation in cancer cells. Indeed, major driver oncogenes frequently up-regulated in T-ALL (Aifantis et al. 2008; Belver and Ferrando 2016; Girardi et al. 2017), such as TLX1, TLX3, TAL1, or HOXA9, were found to be associated with broad H3K4me3 domains in the T-ALL cell lines where they are overexpressed (Fig. 2C).
Figure 2.
Characterization of broad H3K4me3 domains in T-ALL samples. (A) Distribution of the indicated category of genes associated with broad H3K4me3 domains only in thymocytes (Tc), only in T-ALL, in both thymocytes and T-ALL (shared), or without broad H3K4me3 domain (no broad). Oncogenes and tumor suppressors are listed in Supplemental Table S5. Note that no oncogene or tumor suppressor gene was associated with a broad H3K4me3 domain only in thymocytes. (B) Relative enrichment for oncogenes and tumor suppressors associated with the broad H3K4me3 domain only in T-ALL or shared between T-ALL and thymocytes was assessed by hypergeometric tests. (C) The H3K4me3 peaks were ranked according to their size for the indicated T-ALL cell lines. The inflection point allowing the selection of broad H3K4me3 domains is indicated by a red line. The position of the broad H3K4me3 domains associated with the representative oncogene is shown for each cell line. (D) Table showing the association of broad H3K4me3 domains with oncogenes frequently activated in T-ALL. The maturation states and main oncogenetic alterations of T-ALL samples are shown. (E) Examples of key oncogenes associated with broad H3K4me3 domains in T-ALL cells. H3K4me3 ChIP-seq signal and broad H3K4me3 domain tracks are shown for each thymocyte population and T-ALL sample. Asterisk indicates ectopic expression of the oncogene.
To have a more precise view of the dynamics of broad H3K4me3 domains between normal and leukemic cells, we compared their presence for a subset of genes involved in T cell differentiation or driver oncogenes frequently up-regulated in T-ALL (Aifantis et al. 2008; Belver and Ferrando 2016; Girardi et al. 2017). Although T cell genes were associated with broad H3K4me3 domains in normal and malignant cells, we found that T-ALL oncogenes were generally associated with broad H3K4me3 domains in primary and cell line T-ALLs, in agreement with their oncogenic alterations (Fig. 2D). A marked exception was the MYC oncogene, which harbors a broad H3K4me3 domain in normal and malignant cells, consistent with its known role in T cell differentiation and the multiple mechanisms leading to MYC deregulation, not necessarily involving epigenetic deregulation (Sanchez-Martin and Ferrando 2017). Several examples provided clear evidence of broad H3K4me3 domain specifically flagging oncogenes in the T-ALL samples where they are overexpressed (Fig. 2E; Supplemental Fig. S5, asterisk indicates overexpression), also including oncogenic lncRNAs such as PVT1, LUNAR1, and NEAT1 (Supplemental Fig. S5). Thus, driver oncogenes overexpressed in T-ALL appear to acquire a broad H3K4me3 domain, likely linked to their genetic abnormalities.
Leukemic broad H3K4me3 domains switch from cell identity to cancer-related genes
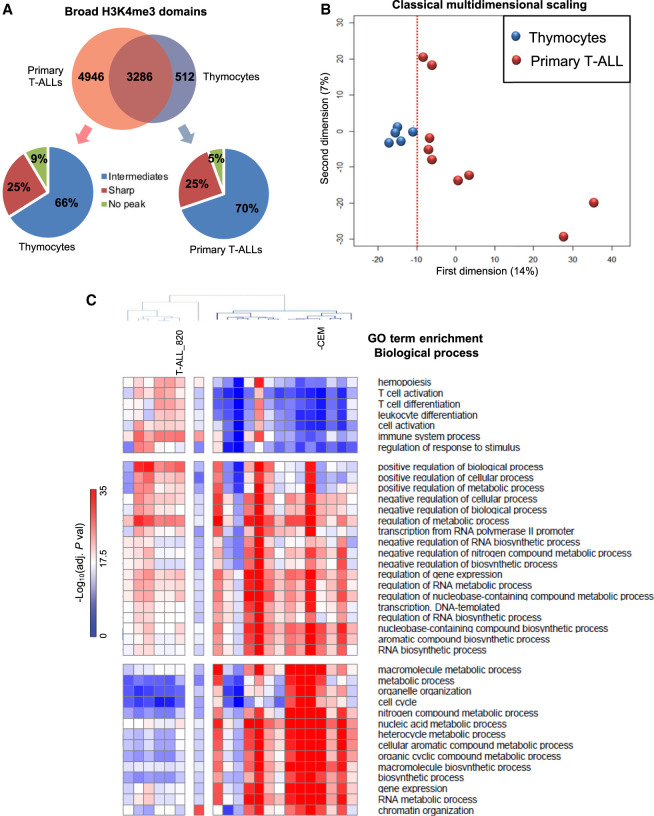
We next compared the number of gains and losses of broad H3K4me3 domains between normal and T-ALL samples. We found a strong bias for a gain of broad H3K4me3 domains in T-ALL and cell lines samples (Fig. 3A; Supplemental Fig. S6), including both coding genes and intergenic lncRNAs (2.2-fold in primary T-ALL and 3.4-fold in T-ALL cells). For instance, although 4946 broad H3K4me3 domains were specific to primary T-ALL samples, only 512 were thymocyte-specific. Note, however, that a substantial percentage of specific broad H3K4me3 domains were found as intermediate H3K4me3 peaks in the leukemic or normal counterpart, suggesting that many cancer-related changes involve a dynamic breadth of H3K4me3 peaks. Multidimensional scaling (MDS) analysis of T-ALL and thymocytes samples based solely on broad H3K4me3 domains showed that thymocyte subpopulations were grouped together, whereas the primary T-ALL samples were more sparsely distributed, suggesting a greater degree of heterogeneity among the leukemic samples (Fig. 3B). Finally, we compared the enrichment in GO terms for biological processes for the list of thymic-specific and T-ALL-specific broad H3K4me3 domain–associated genes (Fig. 3C). We found that the enrichment scores for biological processes associated with T cell identity (e.g., T cell activation, T cell differentiation) and hematopoiesis were higher for broad H3K4me3 domains found in normal T cell precursors, whereas the enrichment scores for biological processes associated with metabolism and cell proliferation were higher for broad H3K4me3 domains found in leukemic cells. Overall, these results suggest that the dynamics of broad H3K4me3 domains in T-ALL is associated with a switch between T cell–identity and cancer-related genes.
Figure 3.
Comparison of broad H3K4me3 domains between normal T cell populations and T-ALL. (A) Overlap between the broad-associated genes in primary T-ALL samples and thymocytes. The distribution of T-ALL- and thymocyte-specific broad domains in thymocytes and T-ALL samples, respectively, are indicated in the bottom panels. (B) Multidimensional scaling classification of T cell populations and T-ALL samples according to the presence or absence of broad H3K4me3 domains. The dotted red line indicated separation between the normal and leukemic samples. (C) Clustering of normal and T-ALL samples based on the top 10 enriched biological process GO terms of each sample.
The expression of broad H3K4me3 domain–associated genes is preferentially deregulated in leukemia
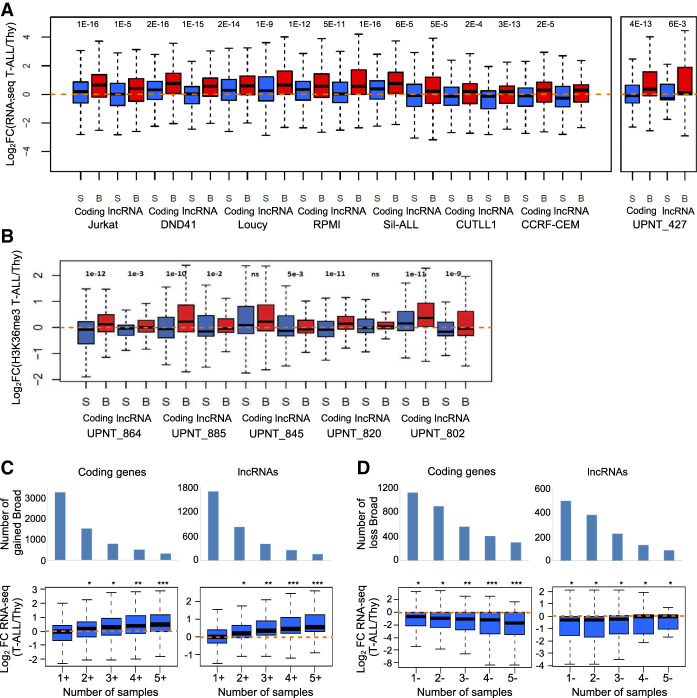
To gain more insight into the regulation of broad H3K4me3 domain–associated genes between normal and leukemic cells, we analyzed the changes in gene expression between normal thymocytes and T-ALL samples for both coding genes and lncRNAs (Fig. 4). In the case of T-ALL cell lines, the comparison was performed with total thymus using a consistent RNA-seq data set (Fig. 4A; Atak et al. 2013). Primary T-ALLs were compared with the averaged thymocyte subpopulations using, depending on the availability, either RNA-seq (Fig. 4A) or gene-body enrichment in H3K36me3 (Fig. 4B), a histone modification directly correlated with gene expression (Barski et al. 2007). We selected broad H3K4me3 domain–associated genes in the T-ALL samples, as well as a set of genes with equal expression levels in T-ALL samples but associated with sharp H3K4me3 profiles around the TSS. We observed that broad H3K4me3 domain–associated genes in each of the T-ALL samples were significantly up-regulated compared with the control gene set (Fig. 4A,B). On the contrary, broad H3K4me3 domain–associated genes in thymocytes were significantly down-regulated in the T-ALL samples compared with the control set (Supplemental Fig. S7). Similar analyses comparing normal B cells and primary chronic lymphocytic leukemia (CLL) lead to consistent results (Supplemental Fig. S8). Therefore, broad H3K4me3 domain–associated genes are more significantly deregulated in leukemia, independent of their relative expression level.
Figure 4.
Dynamic expression of broad H3K4me3 domain–associated genes. (A,B) Two sets of equally expressed genes associated with broad (B) or sharp H3K4me3 peak (S) in the indicated T-ALL sample were defined based on RNA-seq data (A) or H3K4me3 gene-body enrichment (B). The fold change in expression between the T-ALL samples and the normal T cell populations was determined. (C,D) Gene expression in the function of the frequency of association with broad H3K4me3 domains in primary T-ALL samples. Genes specifically associated with broad H3K4me3 domains in T-ALL samples were separated according to the cumulative number of samples with gain (C) or loss (D). The fold change expression of coding genes and lncRNAs between the thymus and a series of independent 41 T-ALLs was analyzed. (***) P < 0.001, (**) P < 0.01, (*) P < 0.05, Wilcoxon rank-sum test; (ns) not significant.
To explore the relevance of the deregulation of gene expression in the function of broad H3K4me3 domain dynamics, we asked whether the frequency of gain or loss of these domains could be generally associated with gene deregulation in T-ALL. To answer this question, we analyzed the fold change expression between normal thymocytes and an independent series of 41 T-ALLs (Bond et al. 2017). We grouped T-ALL broad H3K4me3 domain–associated genes according to the number of T-ALL samples with cancer-specific gain (Fig. 4C) or loss (Fig. 4D) of broad H3K4me3 domains at the associated genes (Fig. 4C,D). The recurrence of gain and loss of broad H3K4me3 domains was significantly associated with the level of, respectively, up- and down-regulation of the associated coding genes and, to a less extent, of the associated lncRNAs, thus suggesting that the recurrence of broad H3K4me3 changes is highly predictive of gene deregulation in an independent T-ALL series.
Overall, these results show that broad H3K4me3 domain–associated genes are more frequently deregulated in leukemia samples, indicating that mechanisms affecting the general expression of broad H3K4me3 domain–associated genes might be highly relevant for the oncogenic transformation. This phenomenon was true for both coding and noncoding genes, suggesting that regulation of lncRNAs associated with broad H3K4me3 domains might represent relevant candidate markers of leukemogenesis.
Sensitivity to epigenetic perturbation in the function of broad H3K4me3 domain spread
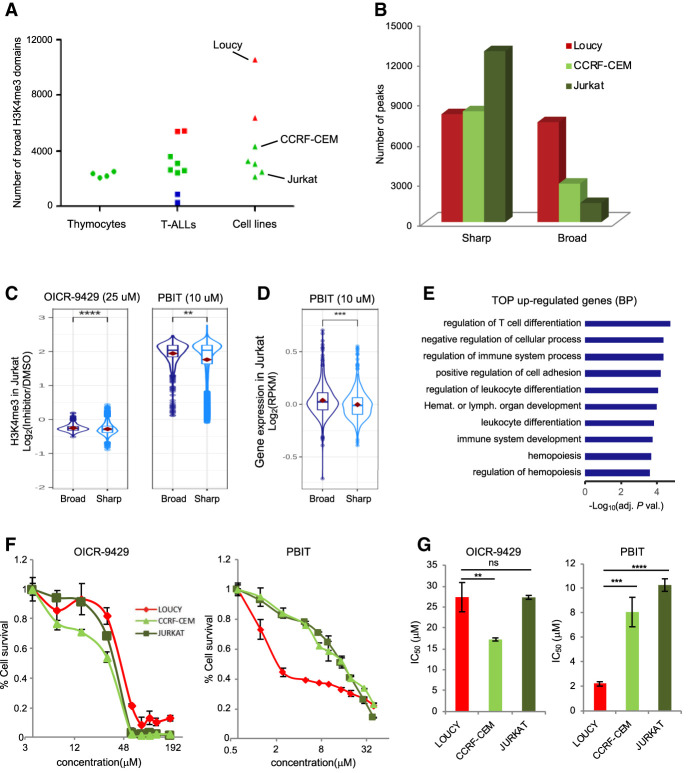
Epigenetic drugs are attractive approaches for cancer therapy (Zhao and Shilatifard 2019). Thus, we aimed to explore whether addiction to broad H3K4me3 domains in cancer cells could provide a rationale for epidrug targeting. We first compared the number of broad H3K4me3 domains identified in normal thymic subpopulations and T-ALL primary and cell line samples. We observed that the number of broad H3K4me3 domains was more variable among the T-ALL samples compared with normal cells (Fig. 5A; Supplemental Table S2). Indeed, some leukemic cells displayed a very low number of broad H3K4me3 domains (primary T-ALL samples 764, 885, and 427), whereas others showed higher numbers (cell lines Loucy and DND41 and primary T-ALL samples 864 and 845) of broad H3K4me3 domains, pointing out to potential global deregulation of epigenetic complexes affecting H3K4me3 spreading. Moreover, the Loucy and DND41 cell lines have broad H3K4me3 domains of a larger size compared with thymocytes and other leukemic cells (P-value < 10−313, Wilcoxon rank-sum test) (Supplemental Fig. S9). For instance, the Loucy cell line harbors a SET-NUP214 fusion oncogene that has been shown to recruit DOT1L methyltransferase, leading to aberrant H3 methylation patterns and transcriptional activation of the HOXA gene cluster (Okada et al. 2005; Van Vlierberghe et al. 2008). Consistently, an extremely large broad H3K4me3 domain at the HOXA gene cluster is observed in the Loucy cell line (Fig. 2E).
Figure 5.
Extent of broad H3K4me3 domains and differential drug sensitivity. (A) Number of broad H3K4me3 domains in the thymic T cell populations, the primary T-ALL, and the T-ALL cell lines. (B) Number of sharp and broad-H3K4me3 peaks in the indicated cell lines. (C) Box plots showing the distribution of the observed fold change of H3K4me3 ChIP-seq signals at broad and sharp promoters in Jurkat cells treated with the indicated inhibitor or DMSO. (D) Box plots showing the distribution of the observed fold change of RNA-seq FPKM at broad- and sharp-associated genes in Jurkat cells treated with PBIT or DMSO. (E) Top 10 GO terms for biological process enriched at genes associated with broad H3K4me3 domains that were up-regulated after PBIT treatment of Jurkat cells. (F) Proliferation assay of Loucy, CCRF-CEM, and Jurkat T-ALL cell lines in the presence of the indicated concentrations of OICR-9429 or PBIT inhibitor for 72 h. Results are the mean of three independent experiments. (G) Bar chart shows average ± SD IC50 values (n = 3) obtained in the proliferation assays shown in F. Significance in C and D was assessed by the Wilcoxon rank-sum test. Significance in G was assessed by a two-sided Student t-test. Error bars, SD. (****) P < 0.0001, (***) P < 0.001, (**) P < 0.01.
To assess whether broad H3K4me3 domains are more or less sensitive than sharp H3K4me3 peaks to perturbation of H3K4me3 levels, we performed H3K4me3 ChIP-seq experiments from Jurkat cells treated with DMSO or with either 2-(4-methylphenyl)-1,2-benzisothiazol-3(2H)-one (PBIT), a specific inhibitor of the KDM5 family of H3K4me3 demethylases (Blair et al. 2011; Sayegh et al. 2013), or OICR-9429, an inhibitor of the MLL-WDR5 complex, previously shown to decrease cellular levels of H3K4me3 (Grebien et al. 2015). As expected, OICR-9429 treatment decreased the overall level of H3K4me3 at broad- and sharp-associated promoters, whereas treatment with PBIT resulted in increased levels of H3K4me3 (note that ChIP-seq experiments were performed with the presence of a spike-in control to account for global changes in H3K4me3 levels; see Methods) (Fig. 5C). However, we found that broad H3K4me3 domains were more sensitive to inhibition of H3K4me3 demethylases. This finding was confirmed by the analyses of gene expression after PBIT treatment (Fig. 5D). Broad H3K4me3 domain–associated genes up-regulated by the PBIT treatment were linked to hematopoiesis and T cell differentiation (Fig. 5E), suggesting that inhibition of H3K4 demethylation can induce the expression of genes involved in normal cell differentiation. Examples of broad H3K4me3 domain–associated genes that were up-regulated by PBIT included genes important for T cell differentiation and function, such as TCF7, ZAP70, CD1A, IKZF1, and BCL11A (Supplemental Table S6).
Next, we explored the sensitivity to the OICR-9429 and PBIT inhibitors for three cell lines—Loucy, CCRF-CEM, and Jurkat—that displayed relatively large, intermediate, and low numbers of broad H3K4me3 domain, respectively (Fig. 5B). We reasoned that perturbing the global amount of H3K4me3 in the cells could reveal the discriminating sensitivity of the cell lines in the function of their relative number of broad H3K4me3 domains. As shown in Figure 5F, treatment with PBIT, but not with OICR-9429, inhibits proliferation of Loucy cells at lower concentrations than CCRF-CEM and Jurkat cells. Indeed, the IC50 of PBIT for the Loucy cell line (IC50 = 2.2 μM) was significantly lower than for CCRF-CEM (IC50 = 8.1 μM) and Jurkat (IC50 = 10.3 μM; P-value < 0.001, two-side t-test) (Fig. 5G) cell lines, suggesting that Loucy cells are more sensitive to increased levels of H3K4me3 than the other cell lines. These suggest that targeting broad H3K4me3 domain–addicted samples with H3K4 demethylating inhibitors might represent a rational anticancer strategy.
Oncogene overexpression preferentially impacts broad H3K4me3 domain–associated genes
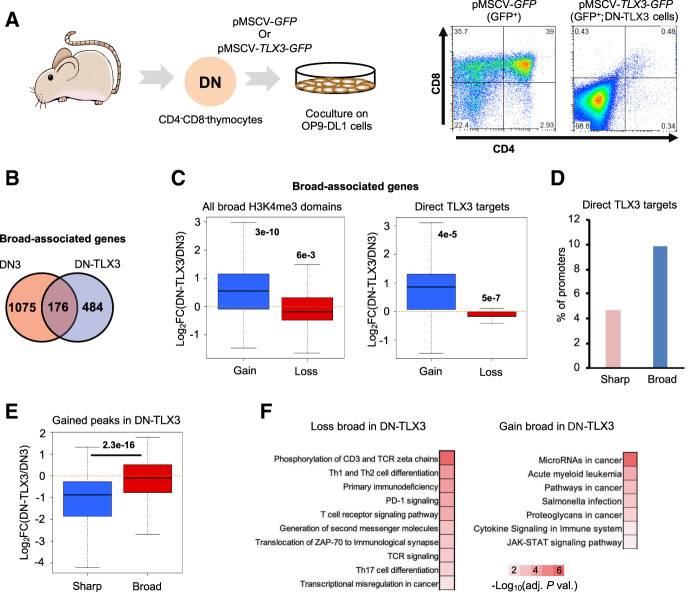
To further explore the link between oncogenic transformation and deregulation of broad H3K4me3 domains, we developed an experimental model of T-ALL by overexpressing TLX3, a major transcription factor oncogene in T-ALL, in mouse developing thymocytes (Fig. 6A). Sorted CD4−/CD8− (double negative [DN]) thymocytes from wild-type mice were infected with either GFP- or TLX3-expressing retrovirus and cocultured on OP9-DL1 stroma cells. TLX3 overexpression blocked T cell differentiation and led to a homogenous DN population of TLX3-expressing cells. These cells (hereafter, DN-TLX3 cells) proliferate indefinitely on OP9-DL1 coculture but were dependent on IL7R (see Methods) and thus resembled a preleukemic cell state. DN-TLX3 cells were subjected to gene expression and H3K4me3 ChIP-seq analyses and compared with DN thymocytes sorted from wild-type thymus (Oravecz et al. 2015). Broad H3K4me3 domains were highly dynamic between normal and transformed cells (Fig. 6B), with their gain and loss being significantly associated with up- and down-regulation of the associated genes, respectively (Fig. 6C, left). The same trend was observed when restricting the analysis to direct TLX3 targets, defined by TLX3 ChIP-seq peaks from DN-TLX3 (Fig. 6C, right). Indeed, the percentage of broad-associated promoters that were bound by TLX3 was significantly higher than for sharp-associated promoters (P-value < 0.00001, Chi-square test) (Fig. 3D). Moreover, the expression of genes associated with broad domains in DN-TLX3 cells was significantly up-regulated compared with a set of expression-matched genes with sharp H3K4me3 peaks (Fig. 6E). Finally, broad H3K4me3 domains that were lost in the leukemic DN-TLX3 cells were significantly associated with genes related to normal immunological and T cell functions, whereas gained broad H3K4me3 domains were associated with genes related to cancer and leukemogenesis (Fig. 6F). Similar results were obtained when using DN thymocytes derived from Rag2−/− mice (Rag2-DN) as control samples (Supplemental Fig. S10). To extend our findings to another T-ALL model, we used a recently published data set from transgenic mice overexpressing NUP214-ABL1 oncogenic fusion along with the TLX1 transcription factor, two cooperative oncogenic factors in T-ALL (Supplemental Table S1; Vanden Bempt et al. 2018). Analysis of broad and sharp H3K4me3 peaks in the wild-type and NUP214-ABL1+/TLX1+ thymus, along with analysis of gene expression and TLX1 binding sites, provided consistent results with those obtained with the TLX3 model (Supplemental Fig. S11). In particular, we found that the expression of genes associated with broad H3K4me3 domains in NUP214-ABL1+/TLX1+ thymus was significantly up-regulated compared with a set of expression-matched genes with sharp H3K4me3 peaks (Supplemental Fig. S11E). Thus, ectopic expression of oncogenic factors in preleukemic models of T-ALL selectively impacts on the expression of broad-associated genes, likely reproducing the situation in naturally occurring leukemias and suggesting that the epigenetic switch of broad H3K4me3 domain association from cell identity to cancer-related genes is an early process in the leukemic ontogeny.
Figure 6.
Dynamics of broad H3K4me3 domains in a mouse model of T-ALL. (A) Experimental workflow. Double-negative thymocytes were infected with a retroviral vector expressing TLX3 oncogene and/or the GFP. After 1 mo of coculture with OP9-DL1 stromal cells, the GFP-positive cells were purified and analyzed by FACS. TLX3 expression was confirmed by western blot using a custom-made antibody (Supplemental Figure S10A; Dadi et al. 2012). (B) Overlap between the broad H3K4me3 domains found in DN thymocytes from wild-type mice and TLX3+ leukemic cells (DN-TLX3). (C) Boxplot showing the fold change of expression of genes associated with broad H3K4me3 domains only in DN-TLX3 (gain) or DN (loss; left panel). (Right) Only the direct targets of TLX3 were analyzed. (D) Percentage of broad- and sharp-associated genes in DN-TLX3 cells that are direct targets of TLX3 based on TLX3 ChIP-seq. (E) Two sets of equally expressed genes associated with broad or sharp H3K4me3 peaks in DN-TLX3 cells were defined. The fold change of the expression between DN-TLX3 and DN was determined. (F) KEGG pathways significantly enriched for the gene sets associated with broad H3K4me3 domains only in DN-TLX3 (gain) or DN (loss; adjusted P-value < 0.01). The inverted log10 of the Benjamin-corrected P-value is shown. (C) Significance was assessed by a Wilcoxon signed-rank test. (E) Significance was assessed by a Wilcoxon rank-sum test.
Discussion
In this study, we took advantage of our comprehensive epigenomic resource encompassing thymic populations of human T cell precursors and primary T-ALL samples to shed light on the dynamics of broad H3K4me3 domains between normal and neoplastic cells. Previous studies have associated broad H3K4me2/3 domains with cell identity and tumor suppressor genes (Pekowska et al. 2010; Koch et al. 2011; Benayoun et al. 2014; Chen et al. 2015; Zacarías-Cabeza et al. 2015; Suzuki et al. 2017). Consistent with these results, in normal T cell precursors, the broad domains were found associated with key developmentally regulated genes marking T cell identity. Although tumor suppressor genes were significantly associated with broad domains in normal and leukemic cells, oncogenes were significantly associated with a gain of a broad domain in T-ALL samples. Indeed, major T-ALL oncogenes were flagged by broad domain in malignant samples where they are ectopically expressed. This indicates that the acquisition of broad domains, rather than regular sharp H3K4me3 peaks, is linked to the oncogenic events leading to ectopic expression of these oncogenes. Previous work from Chen et al. (2015) described a significant association of broad domains with tumor suppressor genes but not oncogenes. However, the apparent discrepancy with our current results is not likely owing to the different approaches used in defining broad H3K4me3 domains in the two studies (also see below) but rather the focus on normal tissues in the former study. More generally, we observed that coding and noncoding broad domain–associated genes are more frequently deregulated in leukemia compared with genes associated with sharp H3K4me3 peaks. Leukemic cells lose broad domains at T cell–related genes and gain broad domains at cancer-related genes. Using two mouse models of preleukemic cells, we show that selective deregulation of broad domain–associated genes can be an early oncogenic event driven by the ectopic expression of an oncogenic transcription factor. Overall, the dynamics of H3K4me3-broad domains between normal T cell precursors and leukemic blasts entail an epigenetic switch between cell identity and cancer-related gene regulation.
Extended H3K4 methylation and, in particular, the so-called broad H3K4me3 domains have been shown to be enriched at tissue-specific and cell identity genes in many tissues and species, from Drosophila to human (Pekowska et al. 2010; Benayoun et al. 2014; Chen et al. 2015; Dincer et al. 2015; Zacarías-Cabeza et al. 2015; Lv and Chen 2016; Park et al. 2020). Accordingly, the presence of a broad domain might be an important criteria to identify genes involved in key cellular processes (Benayoun et al. 2014; Kurum et al. 2016; Suzuki et al. 2017; Cildir et al. 2019; Collins et al. 2019; Park et al. 2020) and diseases (Dincer et al. 2015; Zhang et al. 2016b). Our study confirms and extends these observations by showing that broad domains are dynamically associated with genes playing key T cell functions and different stages of human T cell differentiation. In addition, we showed in a genome-wide manner, that for a given cell line, the presence of broad domains marks genes that are specifically essential for cell survival and proliferation in that cell line.
Extended H3K4 methylation is associated with unique epigenetic and transcriptional properties compared with sharp or TSS-centered peaks, including increased Pol II–mediated transcriptional elongation (Pekowska et al. 2010; Benayoun et al. 2014; Chen et al. 2015; Zacarías-Cabeza et al. 2015). Ours and previous studies have used different criteria to define broad H3K4me3 domains (Benayoun et al. 2014; Chen et al. 2015). For instance, whereas the signal of broad H3K4me3 defined by Chen et al. (2015) is broader but lower than sharp peaks, those defined in this paper appeared to be broader but as high as sharp peaks (Supplemental Fig. S3B). Thus, technical differences in defining broad domains might result in different broad domain subsets. Despite the use of different criteria to define broad H3K4me3 domains, we confirmed the transcriptional characteristics of broad-associated genes and further showed that broad domains are more sensitive than H3K4me3-sharp peaks to inhibitors of transcriptional elongation. These domains appear to work as “buffer” chromatin structures, providing a better transcriptional consistency and increased transcriptional resistance to cellular perturbation (Benayoun et al. 2014; Chen et al. 2015). However, whether the formation of a broad domain is the consequence or the cause of transcriptional consistency will need to be explored in the future (Soares et al. 2017). Although the intrinsic genomic features allowing the establishment of broad domains still need elucidation, recent studies have provided evidence that specific chromatin complexes are required to establish and maintain these domains (Dhar et al. 2018; Quevedo et al. 2019). Our observation of large broad H3K4me3 domains in the Loucy cell line, which harbor the SET-NUP214 fusion oncogene, provides an additional link between epigenetic deregulation in cancer cells and the establishment of the broad H3K4me3 domains.
Epigenetic dysregulation plays profound and ubiquitous roles in cancer progression and has appeared as an important contributor to hematological malignancies (Ntziachristos et al. 2016; Flavahan et al. 2017). In particular, the broad H3K4me3 domains have been shown to be associated with the regulation of tumor suppressors and oncogenes in several cancer types (Chen et al. 2015; Cao et al. 2017; Thibodeau et al. 2017; Dhar et al. 2018; Gerrard et al. 2019; Mikulasova et al. 2022) and could regulate leukemia stem cell maintenance (Wong et al. 2015). In acute lymphoblastic leukemia, promoters with high DNase I hypersensitivity and broad H3K4 methylation have a higher level of RAG-mediated cleavages and were found to be preferential targets for genomic alterations (Heinäniemi et al. 2016). Here, we found that broad domains in T-ALL flag key leukemia-associated oncogenes and entail an epigenetic switch from cell identity to cancer-related genes. Several mechanisms might contribute to the establishment and maintenance of broad domain in cancer cells (Supplemental Fig. S12). One specific mechanism leading to ectopic oncogene expression, such as in the cases of TAL1, TLX1, or TLX3 oncogenes, is a direct consequence of genomic alterations. In these cases, the presence of the broad H3K4me3 domain is associated with the hijacking of a T cell–specific enhancer or super-enhancer (Fig. 3), leading to an epigenetic translocation. Several studies have linked the broad H3K4me3 domains with the presence of super-enhancers (Chen et al. 2015; Cao et al. 2017; Suzuki et al. 2017; Thibodeau et al. 2017; Mikulasova et al. 2022) and suggested that similar mechanisms might be involved in the establishment and maintenance of both chromatin structures (Dhar et al. 2018; Collins et al. 2019; Quevedo et al. 2019). In particular, the most gene-proximal super-enhancers colocalize with the broad H3K4me3 domains in the tumor suppressor genes and oncogenes (Cao et al. 2017; Thibodeau et al. 2017). It is therefore plausible that the proximity of the proto-oncogene with a tissue-specific (super)-enhancer impacts on the chromatin structure of the locus by establishing a broad H3K4me3 domain.
Our results also showed an overall gain of broad H3K4me3 domain at cancer-related genes, which is more compatible with an indirect role, not involving a locus-specific genetic abnormality. Supporting this idea, we observed that ectopic expression of T-ALL oncogenes directly leads to a biased gain of broad domains at cancer-related genes and up-regulation of associated genes in preleukemic cells. In addition, specific epigenetic alterations such as mutations of chromatin remodeling complexes might particularly impact the formation of the broad domains. This is illustrated by the Loucy T-ALL cell line expressing the SET-NUP214 fusion oncogene, which is associated with a higher number and larger size of broad domains. Independently of the mechanisms behind the establishment of the broad domains in cancer cells, these large chromatin structures might provide novel characteristics to the transcription process, including transcriptional fidelity, increased robustness of gene expression, and resistance to environmental signals, ultimately rendering the cells addicted to broad domain–associated genes. The presence of broad H3K4me3 domains could be used for epigenetic prioritization in the identification of cancer-relevant genes, including lncRNAs.
Recent successes with epigenetic therapies suggest that cancer epigenetics can have a major clinical impact (Flavahan et al. 2017). Chromatin-related proteins and modifications are considered as favorable drug targets (epidrugs), and several agents have been designed and used in different stages of clinical trials combined with currently available chemotherapies (Bradner et al. 2017; Zhao and Shilatifard 2019). In particular, super-enhancers are exceptionally vulnerable to perturbation of components commonly associated with most enhancers, such as the BET-bromodomain inhibitor JQ1 (Lovén et al. 2013; Knoechel et al. 2014) or the CDK7 inhibitor THZ1 (Kwiatkowski et al. 2014). Super-enhancers are highly sensitive to perturbation by epigenetic inhibitors (Bradner et al. 2017; Hnisz et al. 2017). This is mainly explained by the increased cooperative and synergistic binding of multiple transcription factors and coactivators at super-enhancers. Reminiscent of super-enhancers, the broad H3K4me3 domains are also large chromatin structures predicted to recruit a large number of transcription factors and chromatin remodelers and therefore are likely to be highly sensitive to epigenetic perturbations. Indeed, treatment of cancer cells with JQ1 or HAT inhibitors preferentially affects the regulation of broad H3K4me3 promoters relative to typical promoters (Suzuki et al. 2017; Gerrard et al. 2019). Here we found that inhibition of transcriptional elongation by targeting CDK7 (THZ1) or DOT1L (EPZ-5676) or inhibition of H3K4me3 demethylation with the PBIT inhibitor also has a preferential impact on broad domains. Moreover, we show that cell lines with a high number of broad domains are more sensitive to perturbation of H3K4me3 levels. Reminiscent of this finding, the LOUCY and DND41 cell lines (the two cell lines with the highest numbers of broad domains) are about 100 times more sensitive to the CDK7 inhibitor THZ1 than Jurkat cells (Kwiatkowski et al. 2014). Altogether, these results suggest that inhibition of transcriptional elongation or H3K4 demethylation might represent promising approaches to target broad H3K4me3 domain–addicted leukemic cells. As previously proposed, resistance to BRD4 inhibitors may be prevented through combined treatment with additional inhibitors targeting other chromatin-related enzymatic activities, such as H3K4 methylation (Hnisz et al. 2017). Several epidrugs have been recently proposed as attractive therapeutic targets in T-ALL (Kwiatkowski et al. 2014; Ntziachristos et al. 2014; Yashiro-Ohtani et al. 2014; Benyoucef et al. 2016). Here, we suggest that targeting broad H3K4me3 domain–addicted cells could be an additional therapeutic strategy in T-ALL and, likely, other cancer types.
Methods
Cell lines
Loucy (ACC-394), Jurkat (ACC-282), CEM-CCRF (ATCC CRM-CLL-119) RPMI-8402 (ATCC CRL-1994), and Sil-ALL (ACC-511) cell lines were cultured in RPMI-1640 medium (Thermo Fisher Scientific 21875091) supplemented with 10% FBS. Platinum-E (Plat-E) cells were used for packaging retroviral vectors, as previously described (Morita et al. 2000). Plat-E cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FCS, 50 µg/mL streptomycin, 50 IU penicillin, 10 µg/mL blasticidin, and 1 µg/mL puromycin. OP9-DL1 stromal cell lines were maintained in α-MEM, GlutaMAX supplement medium (Thermo Fisher Scientific 32561094) supplemented with 20% FBS hyclone SH30070.03HI (Thermo Fisher Scientific 10772634) and 1% penicillin/streptomycin.
Immunophenotypic and molecular characterization of T-ALL samples
Diagnostic T-ALL samples were analyzed for immunophenotype, fusion transcripts (SIL-TAL1, CALM-AF10, NUP214-ABL, MLL), oncogenic transcripts (TLX1 and TLX3), T cell receptor rearrangements, NOTCH1/FBXW7/RAS/PTEN mutations, and IL7-JAK1/3 alterations, as previously described (Asnafi et al. 2003; Bergeron et al. 2007; van Dongen et al. 2012; Trinquand et al. 2013; Kim et al. 2020). Multiplex ligation-dependent probe amplification (MLPA) analysis (CDKN2A and PTEN status) was performed using the MRC Holland SALSA MLPA probe mix P383-A1 TALL according to the manufacturer's recommendations. Polymerase chain reaction products were separated by capillary electrophoresis on an ABI-3130 device. Coffalyser software (https://www.mrcholland.com) was used for the analysis. Quantification of HOXA9 by qRT-PCR and definition of HOXA9-positive T-ALL samples were performed as previously described (Bond et al. 2016). Quantification of TAL1 by qRT-PCR and definition of TAL1-positive T-ALL samples were performed as previously described (Asnafi et al. 2004). Immunophenotypic and molecular characterization of T-ALL samples is depicted in Figure 2D.
ChIP-seq experiments
ChIP-seq of H3K4me3 from human samples was performed following the BLUEPRINT protocol (Cieslak et al. 2020; http://dcc.blueprint-epigenome.eu/#/md/methods). ChIP-seq libraries were generated with the MicroPlex library preparation kit (Diagenode), according to the manufacturer's instructions. ChIP samples were sequenced in-house in single-end 75-nt mode using the NextSeq 500/550 (Illumina) according to the manufacturer's instructions and processed following the BLUEPRINT protocol.
Processing of ChIP-seq data
Publicly available data sets are listed in Supplemental Table S1. The reads were trimmed for low-quality reads with sickle (https://github.com/najoshi/sickle) and mapped to the hg19 reference genome using Bowtie 0.12.7 (Langmead et al. 2009). The peaks were called using MACS2.08 (Feng et al. 2012) with a P-value cut-off less than 0.1 and using the “‐‐ broad” and “-bdg” options. For each cell population, the respective ChIP-seq inputs were used as control data during peak calling. In addition to the peak files, MACS2 generated bedGraph files, which are transformed to bigWig files with bedGraphToBigWig. When spike-in from Drosophila S2 cells was available, bigWig files were produced using deepTools bamCoverage and scaling the signal by 100,000 divided by the number of reads mapping to the Drosophila exogenous BDGP6 genome. Peak-size distribution between two replicates of LC thymocytes was highly reproducible (Supplemental Fig. S1D). ChIP-seq of H3K4me3 and TLX3 from DN-TLX3 cells was performed as previously described (Pekowska et al. 2011; Renou et al. 2017), sequenced on an AB SOLiD v3.0 (Life Technologies) according to the manufacturer's protocol, and mapped onto the NCBI37/mm9 reference genome using the SOLiD preprocessing pipeline. TLX3 direct targets were identified with the GREAT tool using default parameters (McLean et al. 2010). Note that realigning the reads to GRCh38 (hg38) or GRCm38 (mm10) would not significantly affect the conclusions of this study as our study focus on TSS regions that inherently show low variability relative to most of the changes introduced by the hg38 or mm10 builds (Guo et al. 2017). ChIP-seq data were visualized using the Integrative Genome Viewer (IGV) (Thorvaldsdóttir et al. 2013). Metaprofiles and heatmaps were produced with deepTools v3.4.3 (Ramírez et al. 2014).
Peak annotation and identification of broad H3K4me3 domains
H3K4me3 broad, intermediate, or sharp domains were determined by identifying high (IpH) and low (IpL) inflection points of the H3K4me3 peak length (PL) versus gene rank (Supplemental Fig. S1E). The inflection point was computed using the R package “inflection” (Christopoulos 2014). The peaks are classified as broad if PL > IpH, as sharp if PL < IpL, and as intermediate if IpL < PL < IpH. The inflection points and the number of peaks per sample are listed in the Supplemental Table S2. Peaks were assigned to the transcripts with the overlapped TSS (between TSS and TSS + 1.5 kb). Concerning the assignment of broad H3K4me3 domains to gene transcripts, we considered that the two methods previously described (Benayoun et al. 2014; Chen et al. 2015) might select peaks that are not necessarily associated with the gene promoters. In the study from Chen et al. (2015), the overlapped region consisted of a window spanning from −10 kb to +4 kb from the TSS. In the study by Benayoun et al. (2014), peaks were assigned to the gene with the closest TSS, with no mention of a distance cut-off. We, therefore, choose to use a more conservative definition of gene association to retrieve broad H3K4me3 domains that overlapped the 5′ region of the genes and might therefore be considered as being part of their promoters. Coding genes were based on RefSeq from hg19 assembly. Intergenic lncRNAs were defined using a comprehensive catalog of lncRNAs, including de novo lncRNAs expressed in T-ALL (Kermezli et al. 2019). The total number of histone modification peaks and the associated transcripts are provided in Supplemental Table S3. For comparison, broad H3K4me3 domains were also defined using previously published criteria (Benayoun et al. 2014; Chen et al. 2015), as described in Supplemental Figure S1B.
Average tag density profile of histone marks
The average tag density profiles around the TSS for the histone mark H3K4me3, in a given cell line, were generated by calculating the mean tag density normalized as coverage per million (CPM) within −5 kb of the TSSs of reference and control lists genes. The TSSs were grouped into different classes based on their H3K4me3 breadth at each T cell developmental stage and different cell lines. The tag density profiles were computed individually for each class, and average density over 50-bp windows was used to generate the density profiles. The average densities were computed using R script, and the profiles were plotted using the package ggplot2 under R (R Code Team 2020).
POLR2A pausing index calculation
We defined the pausing region as the region from 250 bp upstream of to 250 bp downstream from the TSS and defined the elongation region as the region from 200 bp downstream from the TSS to 200 bp upstream of the TTS. The pausing index is the pausing-to-elongation ratio of POLR2A ChIP-seq read density and was computed using R package BRGenomics getPausingIndices function (R Code Team 2020).
Functional analyses
Functional enrichment analyses of broad H3K4me3 domain–associated genes were produced using a custom pipeline to automate multisample queries to the GREAT web service (McLean et al. 2010). In summary, broad and sharp peaks were queried with the rGREAT R package (https://github.com/jokergoo/rGREAT) against GO biological process. Genomic association rules with genes were performed using default GREAT “basal plus extension.” Significant terms were filtered as those having binomial fold enrichment higher than two and both binomial and hypergeometric test Benjamini–Hochberg (BH)-adjusted P-values lower than 0.05. The GOSemSim R package (Yu 2020) was used to compute Wang similarity distance between all terms. Terms with a similarity distance higher than 0.8 were grouped. Terms were further filtered for heatmap display to keep only the best five terms for each sample according to the binomial P-value. The color scale on the heatmaps displays binomial BH-adjusted P-values. Functional comparison between thymocytes and T-ALL samples was performed using g:Profiler (Reimand et al. 2016) and the default set of all human genes as the standard background set. Overrepresented KEGG pathways for the gene sets were obtained using g:Profiler, similar to GO term enrichment analysis. The top 20 overrepresented KEGG pathways or biological GO terms with the best P-values were identified for each of the samples.
Preferentially essential genes
Gene effects from CRISPR-Cas9 screens for essential genes in 808 human cell lines and common essential genes were retrieved from the DepMap website (DepMap Public 20Q4 release; https://depmap.org). Essential genes in Jurkat cells were defined as genes for which the gene effect is lower than −1, as suggested by DepMap (Behan et al. 2019). Preferentially essential genes in Jurkat were defined as the 25% of essential genes with the lowest mean-subtracted score across all cell lines, as suggested by DepMap (Behan et al. 2019). The list of Jurkat essential and preferential essential genes is provided in Supplemental Table S4.
Definition of cancer-related genes
Oncogenes and tumor suppressors involved in T-ALL were manually curated following authoritative reviews and original publications as indicated in Supplemental Table S5. Tumor suppressors included also frequently mutated genes (driver mutations) in T-ALL as previously published (Liu et al. 2017).
RNA-seq
The extraction of total RNA was performed using the RNeasy plus mini kit (Qiagen) according to the protocol recommended by the supplier. Total RNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific) and stored at −80°C until needed. Extracted RNA was used for the RNA-seq library preparation, using the TruSeq RNA library prep kit v2 (Illumina). Libraries were paired-end sequenced on the Illumina NextSeq 500 sequencer.
RNA-seq data analysis
Processed RNA-seq data for a series of 41 adult T-ALLs and thymic subpopulations have been previously published (Bond et al. 2017; Cieslak et al. 2020). Normalization and differential expression analyses were performed using DESeq2 (Anders and Huber 2010). Control lists were determined based on a reference gene list computed as optimally matched for similar signal level of expression using R script. The statistical comparisons were performed using the Wilcoxon test unless mentioned otherwise.
Inhibitor treatment
PBIT (14111) was purchased from Active Motif and diluted in DMSO. Cells were seeded at the concentration of 2 × 105/mL (in triplicate) in media containing the indicated amount of PBIT. Cell concentrations were measured after 72 h of culture using the Scepter 2 cell counter (Merck Millipore) or the disposable hematocyte C chip (393012) from Dutscher. The results were normalized with respect to the vehicle control, DMSO. OICR-9429 was purchased from Sigma-Aldrich and diluted in DMSO. The 25 × 103 Jurkat, CCRF-CEM, and Loucy cells were plated in each well of a 96-well plate per triplicate with media containing a serial dilution of OICR-9429. Plates were incubated for 72 h at 37°C. Ten microliters of cell proliferation reagent WST-1 (Sigma-Aldrich) was added to each well. Plates were then incubated for further 4 h and read using a multimode plate reader VICTOR Nivo (PerkinElmer). The values of IC50 for PBIT and OICR-9429 were determined using GraphPad Prism 5.0 software. For ChIP-seq experiments, Jurkat cells were treated with either DMSO or 10 μM PBIT or 25 μM OICR-9429 for 5 h. The immunoprecipitation reaction was complemented with 10% of Drosophila chromatin from S2 cells as a spike control (Orlando et al. 2014). For RNA-seq experiments, Jurkat cells were treated with either DMSO or 10 μM PBIT for 5 or 24 h.
Production of retroviral particles
For the production of ecotropic retroviral vector supernatants, Plat-E packaging cells (8 × 106 cells/dish) were cultured on 100-mm dishes and were transiently transfected with pMSCV-IRES-GFP or pMSCV-IRES-TLX3-GFP vectors (24 µg/dish) using the Lipofectamine 2000 transfection reagent (Invitrogen). The cells were washed 6 h later, and vector supernatants were collected 48 and 72 h after transfection and were frozen (−80°C) for further transduction of thymocytes.
Isolation and transduction of mouse DN thymocytes
Thymuses from 4- to 6-wk-old C57Bl/6 mice were extracted and mechanistically disrupted on a 75-µm nylon cell strainer (BD Biosciences Pharmingen). CD4−CD8− DN thymocytes were purified by the depletion of CD4+CD8+ double-positive (DP) cells, as well as CD4+CD8− and CD4−CD8+ single-positive (SP) cells, using the AutoMACS system (Miltenyi Biotec). Single-cell suspensions of thymocytes were stained with magnetic bead–conjugated anti-CD4 and anti-CD8 antibodies (Miltenyi Biotec 130-117-043 and 130-104-075) for 15 min at 4°C and then sorted in AutoMACS. For maximum purity, the sorting of DN cells was performed using a FACS Aria sorter (BD Biosciences). Sorted DN cells to be transduced with pMSCV-GFP or pMSCV-GFP-TLX3 retroviral vectors were resuspended in vector supernatants in the presence of 10 µg/mL polybrene and spinoculated for 2 h at 3500 rpm. Transduced thymocytes were seeded in six-well plates in coculture with OP9-DL1 γ-irradiated cells for further T cell differentiation in vitro over 5 wk. The differentiation medium (referred to as IL7/Flt3L medium) consisted of α-MEM medium supplemented with 10% FCS, 50 µM 2-ME, 2 mM glutamine, 10 mM HEPES (pH 7.5), 1 mM sodium pyruvate, 100 U/mL penicillin, 0.1 mg/mL streptomycin, 10 µg/mL gentamicin, 1% supernatant of rIL7-secreting J558L cells, and 2.5% supernatant of rFlt3L-producing SP2.0 cells. Note that DN-TLX3 cells required IL7 addition to the medium for continuous growth. All cells were maintained at 37°C in a humidified incubator with 5% CO2.
Flow cytometry
Transduced thymocytes were analyzed for GFP expression and immunophenotyped by flow cytometry using the following antibodies: Pacific Blue Rat Anti-Mouse CD4 (558107), PE-Cy7 Rat Anti-Mouse CD8a (561097; both from BD Biosciences Pharmingen) at days 4, 15, 22, and 30 after transduction. All data were acquired on a BD CantoII instrument, and seven-color analysis was performed using FlowJo software (Tree Star).
Microarray gene expression analyses
RNA from DN-TLX3+ cells and Rag2−/− thymocytes (Rag2-DN) were analyzed in triplicate using the Affymetrix mouse gene 1.0 ST. Microarray data from DN thymocytes were obtained from Oravecz et al. (2015). Expression data were MAS normalized using the Affy library from R-Bioconductor (https://www.bioconductor.org). Gene expression levels were determined by using the RMA-Level Gene Expression software (Affymetrix).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the metaseries GSE164228 and are listed in Supplemental Table S1. All scripts used in this study are available at GitHub (https://github.com/guillaumecharbonnier/mw-belhocine2021) and as Supplemental Code S1.
Supplementary Material
Acknowledgments
We thank the Cell Imaging Platform of Institut des Maladies Génétiques Imagine, the Transcriptomics and Genomics Marseille-Luminy (TGML) sequencing platform, the Marseille-Luminy cell biology platform for the management of cell culture, and the CRISPR Screen Action platform from the Canceropôle PACA. We thank students from the master “Biologie Structurale et Génomique” (class 2020–2021, Aix-Marseille University) who provided technical help during a practical session. Work in the laboratory of S.S. was supported by recurrent funding from INSERM and Aix-Marseille University and by specific grants from the European Union's FP7 Programme (agreement no. 282510-BLUEPRINT), the Foundation for Cancer Research ARC (ARC PJA 20151203149), A*MIDEX (ANR-11-IDEX-0001-02), Plan Cancer 2015 (C15076AS), Institut National du Cancer (INCA)-PLBIO, and Ligue Contre le Cancer (Equipe Labellisée). Work in the V.A. laboratory was supported by ITMO_C15075KS “Domaine de l’épigénétique et Cancer”, Association pour la Recherche contre le Cancer (Equipe labellisée 2018), and grants from INCA-PLBIO 2018-00253 and PRT-K 18-071.
Author contributions: M.B., V.A., and S.S. designed the study and wrote the manuscript. J.D.A.F., A.C., I.M., L.P., E.-L.M., J.H.A.M., H.G.S., and S.S. generated data and performed experiments. M.S., C.S., and V.A. characterized T-ALL samples. M.B., G.C., D.P., and S.S. performed bioinformatics analysis. I.M. and M.A.M. contributed to the analyses of the CRISPR-Cas9 screens data. P.F. contributed to the TLX3 model. A.M., L.J.R., and D.R. provided conceptual insights. V.A. and S.S. oversaw the conceptual development of the project. A.C., M.S., E.-L.M., C.S., and A.T. performed research and data analysis. All authors revised and validated the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.266924.120.
Competing interest statement
The authors declare no competing interests.
References
- Aifantis I, Raetz E, Buonamici S. 2008. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol 8: 380–390. 10.1038/nri2304 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnafi V, Beldjord K, Boulanger E, Comba B, Le Tutour P, Estienne MH, Davi F, Landman-Parker J, Quartier P, Buzyn A, et al. 2003. Analysis of TCR, pTα, and RAG-1 in T-acute lymphoblastic leukemias improves understanding of early human T-lymphoid lineage commitment. Blood 101: 2693–2703. 10.1182/blood-2002-08-2438 [DOI] [PubMed] [Google Scholar]
- Asnafi V, Beldjord K, Libura M, Villarese P, Millien C, Ballerini P, Kuhlein E, Lafage-Pochitaloff M, Delabesse E, Bernard O, et al. 2004. Age-related phenotypic and oncogenic differences in T-cell acute lymphoblastic leukemias may reflect thymic atrophy. Blood 104: 4173–4180. 10.1182/blood-2003-11-3944 [DOI] [PubMed] [Google Scholar]
- Atak ZK, Gianfelici V, Hulselmans G, De Keersmaecker K, Devasia AG, Geerdens E, Mentens N, Chiaretti S, Durinck K, Uyttebroeck A, et al. 2013. Comprehensive analysis of transcriptome variation uncovers known and novel driver events in T-cell acute lymphoblastic leukemia. PLoS Genet 9: e1003997. 10.1371/journal.pgen.1003997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, Santos R, Rao Y, Sassi F, Pinnelli M, et al. 2019. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 568: 511–516. 10.1038/s41586-019-1103-9 [DOI] [PubMed] [Google Scholar]
- Belver L, Ferrando A. 2016. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer 16: 494–507. 10.1038/nrc.2016.63 [DOI] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, et al. 2014. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158: 673–688. 10.1016/j.cell.2014.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, et al. 2015. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 163: 1281–1286. 10.1016/j.cell.2015.10.051 [DOI] [PubMed] [Google Scholar]
- Benyoucef A, Palii CG, Wang C, Porter CJ, Chu A, Dai F, Tremblay V, Rakopoulos P, Singh K, Huang S, et al. 2016. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev 30: 508–521. 10.1101/gad.276790.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron J, Clappier E, Radford I, Buzyn A, Millien C, Soler G, Ballerini P, Thomas X, Soulier J, Dombret H, et al. 2007. Prognostic and oncogenic relevance of TLX1/HOX11 expression level in T-ALLs. Blood 110: 2324–2330. 10.1182/blood-2007-04-079988 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181. 10.1016/j.cell.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Blair LP, Cao J, Zou MR, Sayegh J, Yan Q. 2011. Epigenetic regulation by lysine demethylase 5 (KDM5) enzymes in cancer. Cancers (Basel) 3: 1383–1404. 10.3390/cancers3011383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Marchand T, Touzart A, Cieslak A, Trinquand A, Sutton L, Radford-Weiss I, Lhermitte L, Spicuglia S, Dombret H, et al. 2016. An early thymic precursor phenotype predicts outcome exclusively in HOXA-overexpressing adult T-cell acute lymphoblastic leukemia: a group for research in adult acute lymphoblastic leukemia study. Haematologica 101: 732–740. 10.3324/haematol.2015.141218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Graux C, Lhermitte L, Lara D, Cluzeau T, Leguay T, Cieslak A, Trinquand A, Pastoret C, Belhocine M, et al. 2017. Early response-based therapy stratification improves survival in adult early thymic precursor acute lymphoblastic leukemia: a group for research on adult acute lymphoblastic leukemia study. J Clin Oncol 35: 2683–2691. 10.1200/JCO.2016.71.8585:JCO2016718585 [DOI] [PubMed] [Google Scholar]
- Bradner JE, Hnisz D, Young RA. 2017. Transcriptional addiction in cancer. Cell 168: 629–643. 10.1016/j.cell.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Fang Y, Tan HK, Goh Y, Choy JYH, Koh BTH, Hao Tan J, Bertin N, Ramadass A, Hunter E, et al. 2017. Super-enhancers and broad H3K4me3 domains form complex gene regulatory circuits involving chromatin interactions. Sci Rep 7: 2186. 10.1038/s41598-017-02257-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Chen Z, Wu D, Zhang L, Lin X, Su J, Rodriguez B, Xi Y, Xia Z, Chen X, et al. 2015. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat Genet 47: 1149–1157. 10.1038/ng.3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos DT. 2014. Developing methods for identifying the inflection point of a convex/concave curve. arXiv:1206.5478v2 [math.NA]. [Google Scholar]
- Cieslak A, Charbonnier G, Tesio M, Mathieu EL, Belhocine M, Touzart A, Smith C, Hypolite G, Andrieu GP, Martens JHA, et al. 2020. Blueprint of human thymopoiesis reveals molecular mechanisms of stage-specific TCR enhancer activation. J Exp Med 217: e20192360. 10.1084/jem.20192360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cildir G, Toubia J, Yip KH, Zhou M, Pant H, Hissaria P, Zhang J, Hong W, Robinson N, Grimbaldeston MA, et al. 2019. Genome-wide analyses of chromatin state in human mast cells reveal molecular drivers and mediators of allergic and inflammatory diseases. Immunity 51: 949–965.e6. 10.1016/j.immuni.2019.09.021 [DOI] [PubMed] [Google Scholar]
- Collins BE, Sweatt JD, Greer CB. 2019. Broad domains of histone 3 lysine 4 trimethylation are associated with transcriptional activation in CA1 neurons of the hippocampus during memory formation. Neurobiol Learn Mem 161: 149–157. 10.1016/j.nlm.2019.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadi S, Le Noir S, Payet-Bornet D, Lhermitte L, Zacarias-Cabeza J, Bergeron J, Villarèse P, Vachez E, Dik WA, Millien C, et al. 2012. TLX homeodomain oncogenes mediate T cell maturation arrest in T-ALL via interaction with ETS1 and suppression of TCRα gene expression. Cancer Cell 21: 563–576. 10.1016/j.ccr.2012.02.013 [DOI] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, et al. 2016. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537: 548–552. 10.1038/nature19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Zhao D, Lin T, Gu B, Pal K, Wu SJ, Alam H, Lv J, Yun K, Gopalakrishnan V, et al. 2018. MLL4 is required to maintain broad H3K4me3 peaks and super-enhancers at tumor suppressor genes. Mol Cell 70: 825–841.e6. 10.1016/j.molcel.2018.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dincer A, Gavin DP, Xu K, Zhang B, Dudley JT, Schadt EE, Akbarian S. 2015. Deciphering H3K4me3 broad domains associated with gene-regulatory networks and conserved epigenomic landscapes in the human brain. Transl Psychiatry 5: e679. 10.1038/tp.2015.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740. 10.1038/nprot.2012.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Gaskell E, Bernstein BE. 2017. Epigenetic plasticity and the hallmarks of cancer. Science 357: eaal2380. 10.1126/science.aal2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard DL, Boyd JR, Stein GS, Jin VX, Frietze S. 2019. Disruption of broad epigenetic domains in PDAC cells by HAT inhibitors. Epigenomes 3: 11. 10.3390/epigenomes3020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi T, Vicente C, Cools J, De Keersmaecker K. 2017. The genetics and molecular biology of T-ALL. Blood 129: 1113–1123. 10.1182/blood-2016-10-706465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebien F, Vedadi M, Getlik M, Giambruno R, Grover A, Avellino R, Skucha A, Vittori S, Kuznetsova E, Smil D, et al. 2015. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPα N-terminal leukemia. Nat Chem Biol 11: 571–578. 10.1038/nchembio.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Dai Y, Yu H, Zhao S, Samuels DC, Shyr Y. 2017. Improvements and impacts of GRCh38 human reference on high throughput sequencing data analysis. Genomics 109: 83–90. 10.1016/j.ygeno.2017.01.005 [DOI] [PubMed] [Google Scholar]
- Heinäniemi M, Vuorenmaa T, Teppo S, Kaikkonen MU, Bouvy-Liivrand M, Mehtonen J, Niskanen H, Zachariadis V, Laukkanen S, Liuksiala T, et al. 2016. Transcription-coupled genetic instability marks acute lymphoblastic leukemia structural variation hotspots. eLife 5: e13087. 10.7554/eLife.13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. 2017. A phase separation model for transcriptional control. Cell 169: 13–23. 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermezli Y, Saadi W, Belhocine M, Mathieu EL, Garibal MA, Asnafi V, Aribi M, Spicuglia S, Puthier D. 2019. A comprehensive catalog of LncRNAs expressed in T-cell acute lymphoblastic leukemia. Leuk Lymphoma 60: 2002–2014. 10.1080/10428194.2018.1551534 [DOI] [PubMed] [Google Scholar]
- Kim R, Boissel N, Touzart A, Leguay T, Thonier F, Thomas X, Raffoux E, Huguet F, Villarese P, Fourrage C, et al. 2020. Adult T-cell acute lymphoblastic leukemias with IL7R pathway mutations are slow-responders who do not benefit from allogeneic stem-cell transplantation. Leukemia 34: 1730–1740. 10.1038/s41375-019-0685-4 [DOI] [PubMed] [Google Scholar]
- Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, Gillespie SM, Fernandez D, Ku M, Wang H, et al. 2014. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet 46: 364–370. 10.1038/ng.2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C, et al. 2011. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol 18: 956–963. 10.1038/nsmb.2085 [DOI] [PubMed] [Google Scholar]
- Kurum E, Benayoun BA, Malhotra A, George J, Ucar D. 2016. Computational inference of a genomic pluripotency signature in human and mouse stem cells. Biol Direct 11: 47. 10.1186/s13062-016-0148-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, Dastur A, Amzallag A, Ramaswamy S, Tesar B, et al. 2014. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511: 616–620. 10.1038/nature13393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang C, Liu W, Li J, Li C, Kou X, Chen J, Zhao Y, Gao H, Wang H, et al. 2016. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537: 558–562. 10.1038/nature19362 [DOI] [PubMed] [Google Scholar]
- Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, McCastlain K, Edmonson M, Pounds SB, Shi L, et al. 2017. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 49: 1211–1218. 10.1038/ng.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. 10.1016/j.cell.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Chen K. 2016. Broad H3K4me3 as a novel epigenetic signature for normal development and disease. Genomics Proteomics Bioinformatics 14: 262–264. 10.1016/j.gpb.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulasova A, Kent D, Trevisan-Herraz M, Karataraki N, Fung KTM, Ashby C, Cieslak A, Yaccoby S, van Rhee F, Zangari M, et al. 2022. Epigenomic translocation of H3K4me3 broad domains over oncogenes following hijacking of super-enhancers. Genome Res (this issue) 32: 1343–1354. 10.1101/gr.276042.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 7: 1063–1066. 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, Loizou E, Holmfeldt L, Strikoudis A, King B, et al. 2014. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 514: 513–517. 10.1038/nature13605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Abdel-Wahab O, Aifantis I. 2016. Emerging concepts of epigenetic dysregulation in hematological malignancies. Nat Immunol 17: 1016–1024. 10.1038/ni.3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. 2005. hDOT1L links histone methylation to leukemogenesis. Cell 121: 167–178. 10.1016/j.cell.2005.02.020 [DOI] [PubMed] [Google Scholar]
- Oravecz A, Apostolov A, Polak K, Jost B, Le Gras S, Chan S, Kastner P. 2015. Ikaros mediates gene silencing in T cells through Polycomb repressive complex 2. Nat Commun 6: 8823. 10.1038/ncomms9823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, Fritz CC, Bradner JE, Guenther MG. 2014. Quantitative ChIP-seq normalization reveals global modulation of the epigenome. Cell Rep 9: 1163–1170. 10.1016/j.celrep.2014.10.018 [DOI] [PubMed] [Google Scholar]
- Park S, Kim GW, Kwon SH, Lee JS. 2020. Broad domains of histone H3 lysine 4 trimethylation in transcriptional regulation and disease. FEBS J 287: 2891–2902. 10.1111/febs.15219 [DOI] [PubMed] [Google Scholar]
- Pekowska A, Benoukraf T, Ferrier P, Spicuglia S. 2010. A unique H3K4me2 profile marks tissue-specific gene regulation. Genome Res 20: 1493–1502. 10.1101/gr.109389.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H, Imbert J, Andrau JC, Ferrier P, Spicuglia S. 2011. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J 30: 4198–4210. 10.1038/emboj.2011.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C, Chagraoui H, Kristiansen MS. 2017. SCL/TAL1: a multifaceted regulator from blood development to disease. Blood 129: 2051–2060. 10.1182/blood-2016-12-754051 [DOI] [PubMed] [Google Scholar]
- Quevedo M, Meert L, Dekker MR, Dekkers DHW, Brandsma JH, van den Berg DLC, Ozgür Z, van IWFJ, Demmers J, Fornerod M, et al. 2019. Mediator complex interaction partners organize the transcriptional network that defines neural stem cells. Nat Commun 10: 2669. 10.1038/s41467-019-10502-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J. 2016. g:Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res 44: W83–W89. 10.1093/nar/gkw199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renou L, Boelle PY, Deswarte C, Spicuglia S, Benyoucef A, Calvo J, Uzan B, Belhocine M, Cieslak A, Landman-Parker J, et al. 2017. Homeobox protein TLX3 activates miR-125b expression to promote T-cell acute lymphoblastic leukemia. Blood Adv 1: 733–747. 10.1182/bloodadvances.2017005538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Martin M, Ferrando A. 2017. The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia. Blood 129: 1124–1133. 10.1182/blood-2016-09-692582 [DOI] [PubMed] [Google Scholar]
- Sayegh J, Cao J, Zou MR, Morales A, Blair LP, Norcia M, Hoyer D, Tackett AJ, Merkel JS, Yan Q. 2013. Identification of small molecule inhibitors of Jumonji AT-rich interactive domain 1B (JARID1B) histone demethylase by a sensitive high throughput screen. J Biol Chem 288: 9408–9417. 10.1074/jbc.M112.419861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LM, He PC, Chun Y, Suh H, Kim T, Buratowski S. 2017. Determinants of histone H3K4 methylation patterns. Mol Cell 68: 773–785.e6. 10.1016/j.molcel.2017.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunnenberg HG, International Human Epigenome Consortium, Hirst M. 2016. The International Human Epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell 167: 1145–1149. 10.1016/j.cell.2016.11.007 [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Young RA, Sharp PA. 2017. Super-enhancer-mediated RNA processing revealed by integrative microRNA network analysis. Cell 168: 1000–1014.e15. 10.1016/j.cell.2017.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau A, Márquez EJ, Shin DG, Vera-Licona P, Ucar D. 2017. Chromatin interaction networks revealed unique connectivity patterns of broad H3K4me3 domains and super enhancers in 3D chromatin. Sci Rep 7: 14466. 10.1038/s41598-017-14389-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinquand A, Tanguy-Schmidt A, Ben Abdelali R, Lambert J, Beldjord K, Lengliné E, De Gunzburg N, Payet-Bornet D, Lhérmitte L, Mossafa H, et al. 2013. Toward a NOTCH1/FBXW7/RAS/PTEN–based oncogenetic risk classification of adult T-cell acute lymphoblastic leukemia: a group for research in adult acute lymphoblastic leukemia study. J Clin Oncol 31: 4333–4342. 10.1200/JCO.2012.48.5292 [DOI] [PubMed] [Google Scholar]
- Vanden Bempt M, Demeyer S, Broux M, De Bie J, Bornschein S, Mentens N, Vandepoel R, Geerdens E, Radaelli E, Bornhauser BC, et al. 2018. Cooperative enhancer activation by TLX1 and STAT5 drives development of NUP214-ABL1/TLX1-positive T cell acute lymphoblastic leukemia. Cancer Cell 34: 271–285.e7. 10.1016/j.ccell.2018.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JJ, Lhermitte L, Böttcher S, Almeida J, van der Velden VH, Flores-Montero J, Rawstron A, Asnafi V, Lécrevisse Q, Lucio P, et al. 2012. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia 26: 1908–1975. 10.1038/leu.2012.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, van Grotel M, Tchinda J, Lee C, Beverloo HB, van der Spek PJ, Stubbs A, Cools J, Nagata K, Fornerod M, et al. 2008. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood 111: 4668–4680. 10.1182/blood-2007-09-111872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Goode DL, Iwasaki M, Wei MC, Kuo HP, Zhu L, Schneidawind D, Duque-Afonso J, Weng Z, Cleary ML. 2015. The H3K4-methyl epigenome regulates leukemia stem cell oncogenic potential. Cancer Cell 28: 198–209. 10.1016/j.ccell.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro-Ohtani Y, Wang H, Zang C, Arnett KL, Bailis W, Ho Y, Knoechel B, Lanauze C, Louis L, Forsyth KS, et al. 2014. Long-range enhancer activity determines Myc sensitivity to notch inhibitors in T cell leukemia. Proc Natl Acad Sci 111: E4946–E4953. 10.1073/pnas.1407079111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. 2020. Gene ontology semantic similarity analysis using GOSemSim. Methods Mol Biol 2117: 207–215. 10.1007/978-1-0716-0301-7_11 [DOI] [PubMed] [Google Scholar]
- Zacarías-Cabeza J, Belhocine M, Vanhille L, Cauchy P, Koch F, Pekowska A, Fenouil R, Bergon A, Gut M, Gut I, et al. 2015. Transcription-dependent generation of a specialized chromatin structure at the TCRβ locus. J Immunol 194: 3432–3443. 10.4049/jimmunol.1400789 [DOI] [PubMed] [Google Scholar]
- Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, et al. 2016a. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537: 553–557. 10.1038/nature19361 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shi L, Dawany N, Kelsen J, Petri MA, Sullivan KE. 2016b. H3k4 tri-methylation breadth at transcription start sites impacts the transcriptome of systemic lupus erythematosus. Clin Epigenetics 8: 14. 10.1186/s13148-016-0179-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Shilatifard A. 2019. Epigenetic modifications of histones in cancer. Genome Biol 20: 245. 10.1186/s13059-019-1870-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.