Summary
Naturally coloured cotton (NCC) fibres need little or no dyeing process in textile industry to low‐carbon emission and are environment‐friendly. Proanthocyanidins (PAs) and their derivatives were considered as the main components causing fibre coloration and made NCCs very popular and healthy, but the monotonous fibre colours greatly limit the wide application of NCCs. Here a G. hirsutum empurpled mutant (HS2) caused by T‐DNA insertion is found to enhance the anthocyanidins biosynthesis and accumulate anthocyanidins in the whole plant. HPLC and LC/MS‐ESI analysis confirmed the anthocyanidins methylation and peonidin, petunidin and malvidin formation are blocked. The deficiency of GhOMT1 in HS2 was associated with the activation of the anthocyanidin biosynthesis and the altered components of anthocyanidins. The transcripts of key genes in anthocyanidin biosynthesis pathway are significantly up‐regulated in HS2, while transcripts of the genes for transport and decoration were at similar levels as in WT. To investigate the potential mechanism of GhOMT1 deficiency in cotton fibre coloration, HS2 mutant was crossed with NCCs. Surprisingly, offsprings of HS2 and NCCs enhanced PAs biosynthesis and increased PAs levels in their fibres from the accumulated anthocyanidins through up‐regulated GhANR and GhLAR. As expected, multiple novel lines with improved fibre colours including orange red and navy blue were produced in their generations. Based on this work, a new strategy for breeding diversified NCCs was brought out by promoting PA biosynthesis. This work will help shed light on mechanisms of PA biosynthesis and bring out potential molecular breeding strategy to increase PA levels in NCCs.
Keywords: naturally coloured cotton, fibre colour, proanthocyanidin biosynthesis, flavonoid O‐methyltransferase gene, anthocyanidin methylation
A putative flavonoid O‐methyltransferase (GhOMT1) in Gossypium hirsutum is found to play a critical role in anthocyanidin methylation. Deficiency of GhOMT1 blocks the anthocyanidins methylation, increases anthocyanidins accumulation and enhances PAs biosynthesis in fibres of NCCs which broaden our understanding on mechanisms of plant colour formation and open up huge possibilities to achieve cotton fibres with new colours.

Introduction
Flavonoids are widely distributed plant secondary metabolites, which play important roles not only in the colour formation of most flowers, fruits and seeds, but also in protecting plants against various biotic and abiotic stresses (Chaves‐Silva et al., 2018; Falcone Ferreyra et al., 2012; Tanaka and Brugliera, 2013). Anthocyanins, a major form of flavonoids particularly responsible for the red, purple and blue colours of many flowers and fruits, are synthesized via the general phenylpropanoid pathway which has been well documented during the past decades (Falcone Ferreyra et al., 2012; Tanaka and Brugliera, 2013). In contrast, the biosynthesis of another subgroup of flavonoids, proanthocyanidins and their derivatives (PAs), is not well understood due to their complex structure and varied composition (Dixon et al., 2005; He et al., 2008). PAs are present in fruits, bark and seeds of many plants, conferring flavour and astringency on them and often acting as protectants against predator. Since PAs are increasingly recognized to be beneficial for human health, considerable attention has been drawn on the regulation of their biosynthesis (Dixon et al., 2005; He et al., 2008; Sun et al., 2021). PAs are oligomeric and polymeric products of flavan‐3‐ols (+)‐catechin and (−)‐epicatechin, and their biosynthesis shares the common anthocyanidin pathway until the step forming flavan‐3,4‐diol (i.e. leucopelargonidin) (Abrahams et al., 2002; He et al., 2008; Tanner et al., 2003; Xie and Dixon, 2005). Leucopelargonidin, including its hydroxylated form leucocyanidin and leucodelphinidin, are colourless, can be converted to anthocyanins with different colours by further oxidation, glycosylation and methylation (Koes et al., 2005). PA synthesis starts in some species with the reduction of leucoanthocyanidin by leucoanthocyanidin reductase (LAR), and others with the reduction of anthocyanidin by anthocyanidin reductase (ANR) as well (Bogs et al., 2005; Liu et al., 2016; Yu et al., 2019). The resulting catechin or epicatechin can then be transported into vacuole and further polymerized and oxidized to form brown proanthocyanidin derivatives, especially in testa (Debeaujon et al., 2003; Koes et al., 2005; Pang et al., 2007). Flavonoid pathway genes are known to be co‐ordinately induced and transcription factors that directly regulate the expression of the structural genes of the pathway have been identified in several species (Jaakola, 2013). A complex of three regulatory proteins consisting of MYB, bHLH and WD40 (MBW) regulates a set of genes involved in the biosynthesis of anthocyanins and PAs, participates in different types of controls ranging from fine‐tuned transcriptional regulation by environmental factors to the initiation of the flavonoid biosynthesis pathway by positive regulatory feedback (Naik et al., 2022; Wang et al., 2022; Xu et al., 2015).
Upland cotton (Gossypium hirsutum L.) is one of the most important resources of natural textile materials and has a great value in both agricultural and industrial economies. The fibres from most cultivated cottons are white, with some cultivars producing brown or light green fibres. The coloured fibres have attracted increasing attention since they need little or no dyeing processes and greatly reduce carbon emissions during the fabric manufacture, and become increasingly important especially in facing global warming (Gong et al., 2018; Kohel, 1985). Quinones, direct contributors to colour formation in brown fibres, are the oxidation products of PAs that mainly include the 2,3‐cis forms (epigallocatechin and epicatechin) of procyanidin (PC) and prodelphinidin (PD) units (Feng et al., 2013, 2014; Li et al., 2012; Liu et al., 2018a; Sun et al., 2022), which endow fibres with beautiful colours, antibacterial activity, mildew resistance, flame retardant and UV protection (Chen and Cluver, 2010; Emiliani et al., 2013; Hinchliffe et al., 2016; Hustvedt, 2005). The potential benefit to human health and environment‐friendly characteristics of NCC fibres make them very popular nature textile materials and have attracted increasing interests. However, the relative poorer fibre quality (such as short fibre length) and limited colour choices of NCCs have greatly restricted their use in textile manufacture (Dutt et al., 2004; Sun et al., 2021). In addition, the scarcity of germplasm resources, and relatively poor understanding on the mechanisms underlying the colour formation of NCCs have greatly hindered the progress on the breeding of NCCs with improved colours and fibre quality. Previous studies have confirmed that the central pathway for flavonoid biosynthesis is conserved in plants with minor modification depending on the species, for example, F3′5′H (flavonoid‐3′‐5′‐hydroxylase) pathway does not exist in Arabidopsis, maize and some flowering plants (Falcone Ferreyra et al., 2012; Katsumoto et al., 2007; Noda et al., 2013), and PAs biosynthesis sub‐pathway cannot be found in maize. In NCCs, main genes involved in anthocyanidin and PAs synthesis, including GhCHS, GhCHI, GhDFR, GhC4H, GhF3H, GhF3′H, GhF3′5′H, GhANS, GhLAR and GhANR, have been cloned and proved to contribute to the colour formation with PAs (procyanidins and prodelphinidins) and derivatives accumulated in brown fibres (Feng et al., 2013, 2014; Gao et al., 2019; Gong et al., 2014; Tang et al., 2020; Wang et al., 2014; Xiao et al., 2007), and the transcription factor GhTT2_A07 (Hinchliffe et al., 2016), GhTT2‐3A (Yan et al., 2018), GsTT8 (Sun et al., 2022) linked to fibre colour, indicating that the anthocyanidin and PA pathways are relatively intact and complicated in NCCs and wild cotton G. stocksii (Li et al., 2020; Sun et al., 2022). However, better elucidation of the mechanisms of PAs synthesis is still required for molecular breeding of cotton for novel colour trait, and uncoupling fibre colour with poor fibre quality by modifying the expression level of genes in anthocyanin and PAs biosynthetic pathway.
In the present work, an upland cotton mutant HS2, being purple in whole plant except fibre, was identified and found to accumulate a large amount of free anthocyanidins in the purple tissues, most likely caused by a function deficiency of a flavonoid O‐methyltransferase (GhOMT1) gene, resulting in a consequent accumulation of unmethylated anthocyanidin monomers and dramatical decrease of methylated anthocyanidin monomers. To take full advantage of the accumulated anthocyanidins and further clarify the relationship between anthocyanin and PAs synthesis in NCCs, HS2 was crossed with different NCC varieties and the fibre colours of the progenies were characterized. Interestingly, various new fibre colours could be observed in the offsprings, ranging from light brown to dark brown, light green to blackish green, and new colours of orange‐red and blue. The over‐accumulated unmethylated anthocyanidins of cyanidin and delphinidin could continue to form various types of PAs by enhanced expression of GhLAR and GhANR genes in the offsprings of HS2 and NCC varieties. Not only our results open up a new route to molecular breeding for more fibre colours other than brown and green, but also shed new light on the deep understanding of mechanism underlying the colour formation of cotton fibre.
Results
A cotton mutant HS2 arisen by T‐DNA insertion shows anthocyanidins over‐accumulation
An empurpled mutant (hereafter referred to as HS2) was identified from T‐DNA insertion lines of Coker 312 (G. hirsutum L.). The newly germinated seedlings of homozygous HS2 displayed yellow‐red cotyledons, red veins and purple stems. After several hours, the whole seedlings turned purple colour, which persisted in all visible tissues of the adult plants throughout the whole growth period, including leaves, stems, petals, bolls and even testa (Figures 1a–c and S1). Further investigation revealed that anthocyanidins were over‐accumulated in HS2 leaves (Figures 1a–c and S1), whereas chlorophyll content was slightly decreased (Figure 1e). The other main agricultural traits, such as the plant height and the number of bolls, were not significantly affected in HS2 (Figure S1). Southern blot analysis confirmed that only one copy of T‐DNA was present in HS2 (Figure S2). The results of TAIL‐PCR revealed that the T‐DNA insertion occurred in a sequence rich in transposons and repeat sequences in the Scaffold EF457753.1 (Figure S2).
Figure 1.
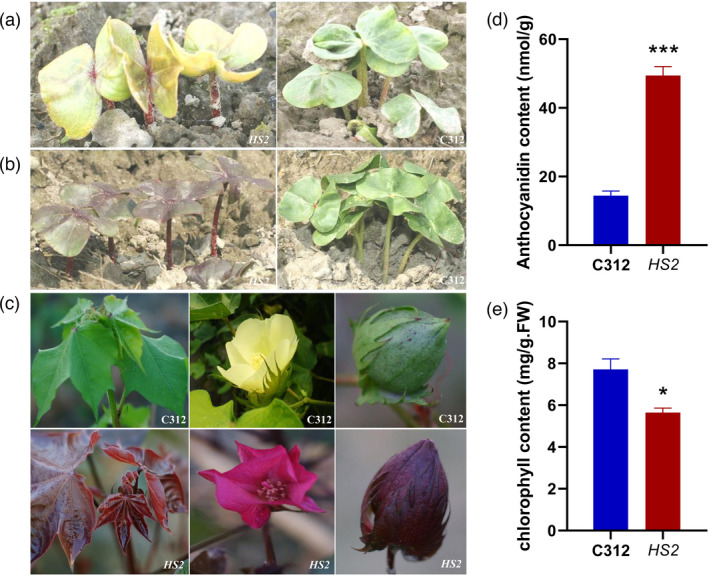
The phenotypes of empurpled mutant HS2 and the parental line C312. (a, b) Freshly germinated seedlings of HS2 (left) and C312 (right) at 8:00 a.m. (a) and 16:00 p.m. (b); (c) Coloured leaves, flowers and bolls of C312 (up) and HS2 (down) plants; (d) Anthocyanidins content in the leaves of C312 and HS2. (e) Chlorophyll contents in the leaves of C312 and HS2. (d, e: mean ± sd; n = 7; Student’s t‐test, *P < 0.05, ***P < 0.001).
To investigate the genetic nature of the mutation, the homozygous HS2 was crossed with its parental line C312 and another okra‐leaf cultivar YZ1 (G. hirsutum L.), respectively. All the F1 plants were purple‐red, while the F2 plants presented green, purple‐red and purple with the segregation ratio of 1:2:1, suggesting the purple trait was caused by a monogenous semi‐dominant mutation (Figures S3, S4; Table S1). PCR analysis with T‐DNA and flanking sequence‐specific primers confirmed the co‐segregation of T‐DNA insertion and the empurpled phenotype among F2 progenies and their derived lines from HS2 crossed with other cotton cultivars (Figure S3).
Methylation of anthocyanidins was blocked in HS2
To reveal the mechanism responsible for the empurpled phenotype, the profiles of anthocyanidins in HS2 and C312 were analysed using HPLC‐LC/MS chromatograms. Several peaks were found to be significantly different between HS2 and C312, such as the one at acquisition time of 28 min which increased from 48.7% to 78.4% (Figure 2a–d). Further analysis confirmed that in C312, all of the six common monomers, pelargonidin, cyanidin, delphinidin, peonidin, petunidin and malvidin, could be detected by ESI analysis, whereas in HS2 plants three methylated monomers, i.e. peonidin, petunidin and malvidin were almost undetectable (Figure 2e–j). The result also indicated that the accumulation of total anthocyanidins in HS2 tissues was arisen due to the increase of the unmethylated monomers, including pelargonidin, cyanidin and delphinidin.
Figure 2.
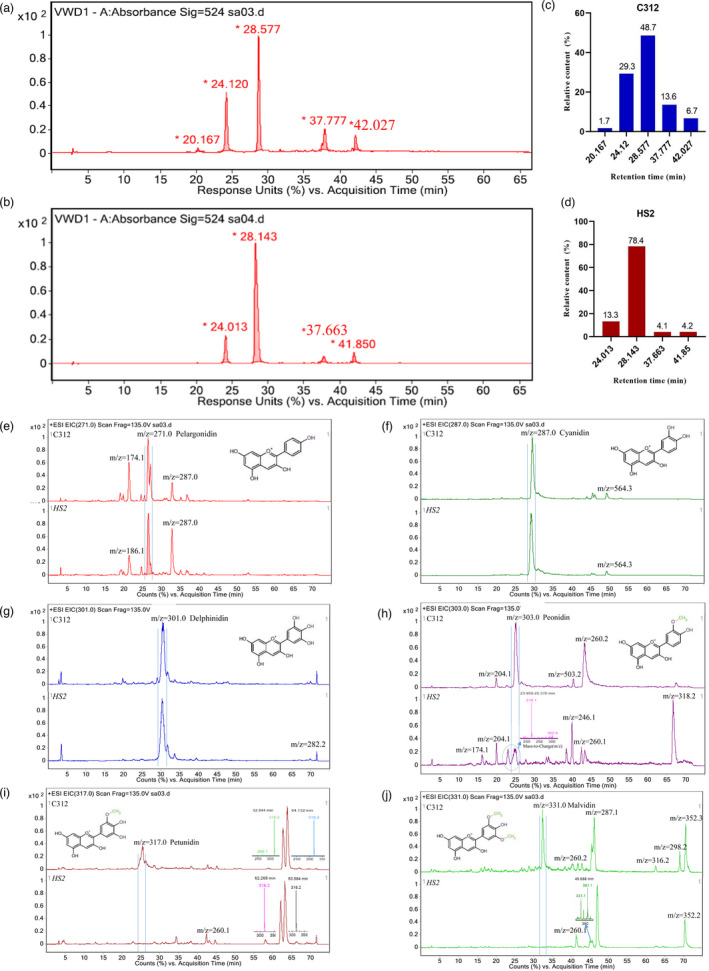
Analysis of anthocyanidin gradients in C312 and HS2. (a, b) HPLC‐LC/MS chromatograms of anthocyanins in C312 (a) and HS2 (b); c, d. Statistics of the relative proportion of different anthocyanin components in C312 (c) and HS2 (d) respectively. (e–j) Six main anthocyanidins profiles in cotton leaves determined by LC/MS ESI positive ion scanning in C312 and HS2. (e) Pelargonidin ([M + H]+ = 271); (f) Cyanidin ([M + H]+ = 287); (g) Delphinidin ([M + H]+ = 301); (h) Peonidin ([M + H]+ = 303); (i) Petunidin ([M + H]+ = 317); (j) Malvidin ([M + H]+ = 331). Peaks marked with vertical lines indicate the specific anthocyanidin monomer in each picture.
Down‐regulation of GhOMT1 altered anthocyanidin compositions in HS2
Since the T‐DNA was inserted in a putative gypsy retrotransposon region, it is possible that the structure or the expression of the surrounding genes is affected. To find out the affected gene, about 200‐kb region in genomic sequence around the insertion site was analysed. Many tandemly arranged transposable elements could be found upstream of the T‐DNA insertion site, including retrotransposon copia, transposon GORGE3‐like, retrotransposon gypsy, transposon MuDR, and some other unknown repetitive sequences and LTRs, with only several coding genes were found in this region (GenBank EF457753.1).
Interestingly, a flavonoid O‐methyltransferase gene (GhOMT1, GH_A01G2052), being reportedly responsible for the methylation in anthocyanidin, was found to be located downstream of the putative gypsy retrotransposon where the T‐DNA was inserted (Figure 3a). Therefore, quantitative real‐time PCR (RT‐qPCR) analysis was performed to compare the expression level of GhOMT genes in C312 and HS2. The transcript of GhOMT1 was hardly detectable in leaves of HS2 (Figure 3d), while that of other GhOMTs showed no significant difference between HS2 and C312 (Figure S5). G. hirsutum is a tetraploid cotton species with two subgenomes of At and Dt, with each containing a GhOMT1 gene, we would like then to investigate whether the transcripts of two GhOMT1 genes are differentially affected. Comparing with the GhOMT1 transcript from At subgenome, the one from Dt subgenome has a 65‐bp fragment deletion as revealed by RNA‐Sequence and 5′ RACE at the region close to the start codon. With gene‐specific primers based on the sequence differences, GhOMT1 transcript from the Dt subgenome (GH_D01G2164) was found to be expressed at a very low level in both HS2 and C312, whereas the one from the At subgenome was dramatically down‐regulated in HS2 (Figure S5).
Figure 3.
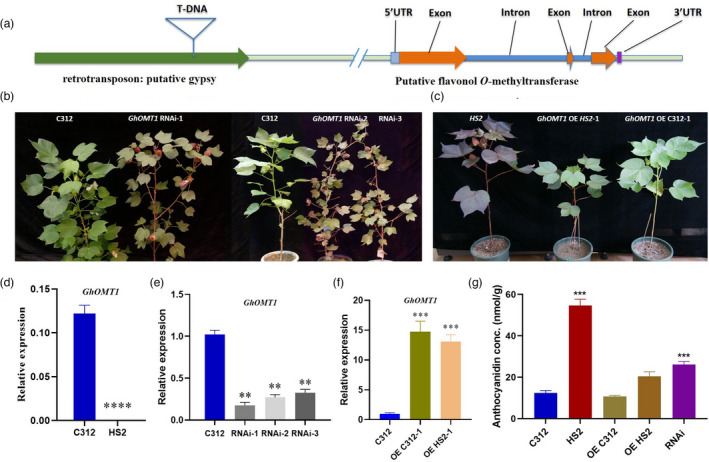
T‐DNA insertion caused GhOMT1 deficiency and anthocyanidins accumulation. (a) A T‐DNA inserted in a putative gypsy retrotransposon upstream of GhOMT1 gene; (b) Phenotypes of GhOMT1 RNA interfered lines (GhOMT1 RNAi) and C312; (c) Phenotypes of GhOMT1 overexpressed lines. Left: HS2, Middle: GhOMT1 overexpressed in HS2 (GhOMT1‐OE HS2), Right: GhOMT1 overexpressed in C312 (GhOMT1‐OE C312); (d) The transcript level of GhOMT1 in leaves of C312 and HS2. (e) The transcript level of GhOMT1 in leaves of C312 and GhOMT1 RNAi lines. (f) The transcript level of GhOMT1 in leaves of GhOMT1 overexpressed line in C312 and HS2. (g) Anthocyanidin content in C312, HS2, GhOMT1 overexpressed and GhOMT1 RNAi lines. (d, e, f, g: mean ± sd; n = 7 (d), n = 3 (e), n = 5 (f), n = 5 (g); Student’s t‐test, **P < 0.01,***P < 0.001).
To further investigate the function of GhOMT1 (GhOMT1‐At), overexpression lines of GhOMT1 driven by the 35S promoter (GhOMT1‐OE) and RNA interfered lines (GhOMT1‐RNAi) were generated on C312 background, in which the transcript level of GhOMT1 was changed by 22‐ and 0.25‐fold, respectively (Figures 3e, f and S6). The GhOMT1‐RNAi lines showed red leaves and deep red colour in stems, branches, petioles, buds, and sepals, similar to that in HS2 (Figure 3b), while no visible alternation in phenotype was found in the GhOMT1‐OE lines of C312 (Figure 3c). The phenotypes of homozygous lines of GhOMT1‐OE in HS2 (GhOMT1‐OE HS2) were similar to the wild type C312 (Figures 3c and S6); therefore, overexpression of GhOMT1 in HS2 could partially restore the purple phenotype of HS2 to normal green phenotype. Corresponding to the phenotype, the anthocyanidin content was found to increase significantly in GhOMT1‐RNAi lines, while not in GhOMT1‐OE lines (Figure 3g). Leaf anthocyanidins profiles further confirmed that the methylated anthocyanidins decreased in GhOMT1‐RNAi lines as that in HS2, while the other three unmethylated anthocyanidins showed obvious increase (Figure S7). These findings indicated that GhOMT1 played critical roles in anthocyanidin biosynthesis and the functional deficiency of GhOMT1 in HS2 was truly associated with the phenotypic change to purple colour and changes in content and composition of anthocyanidins in cotton.
Key genes in flavonoid core‐pathway were up‐regulated in HS2
To reveal the mechanism of the increased unmethylated anthocyanidins in HS2, the expressions of genes involved in anthocyanin and PA biosynthesis were analysed. No significant expression changes were found in the genes involved in phenylpropanoid pathway, such as GhPAL1, GhPAL2 and GhC4H, with the only exception that Gh4CL was significantly increased. Interestingly, however, the transcript level of most of the structural genes in the central pathway of anthocyanidin biosynthesis was notably up‐regulated in HS2 compared to that in C312, but not GhGST involved in the decoration of anthocyanidins, and GhLAR and GhANR in the biosynthesis of PAs (Figure 4).
Figure 4.
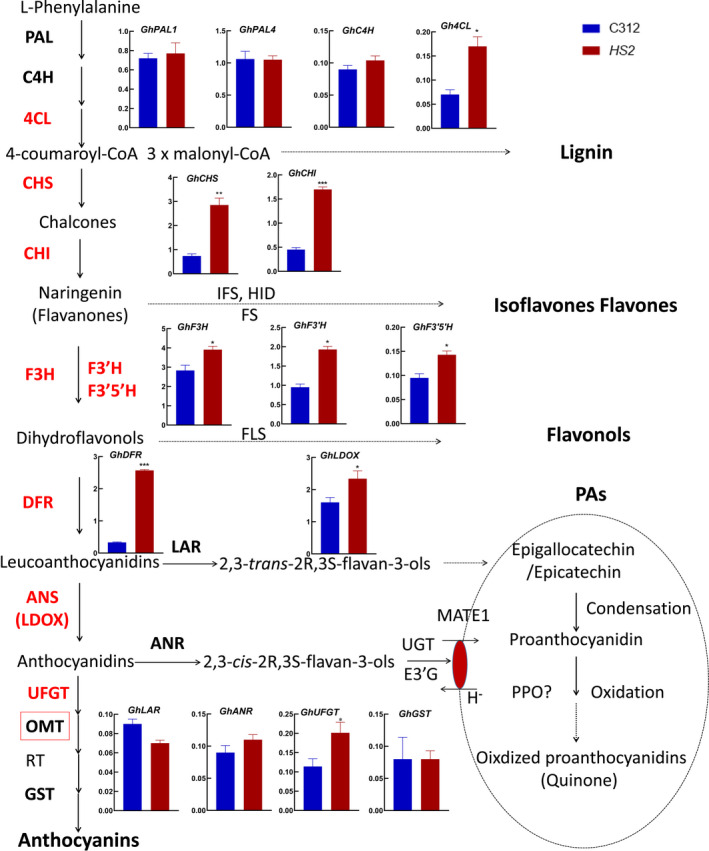
Expression analysis of genes involved in phenylpropanoid pathway, anthocyanins pathway and PA pathway in C312 and HS2. Main products and enzymes in phenylpropanoid pathway, anthocyanins pathway and PA pathway are shown in diagram. Genes that encode enzymes in bold were analysed by qRT‐PCR and the relative expression levels in C312 and HS2 were listed nearby. The relative genes of enzymes in red were significantly up‐regulated in HS2 while that in black did not show apparent changes. Significance analysis was performed by Student’s t‐test between C312 and HS2 (mean ± sd; n = 5; *P < 0.05, **P < 0.01).
In higher plants, the well‐known and conserved MBW (MYB‐bHLH‐WDR) complexes participate in and control different types of flavonoid biosynthesis pathway. Expression of potential regulators of cotton anthocyanin biosynthesis was also investigated through RNA sequencing for HS2 and C312. The genes encoding MBW (MYB‐bHLH‐WDR) complexes in HS2 were focused on. The differential expression of 14 MYBs was identified between C312 and HS2 (Table S2), including the up‐regulated genes like MYB16 (GH_A10G1686) and MYB113 (GH_D07G0852), and the down‐regulated genes like MYB3 (GH_A08G0383), MYB5 (GH_D12G1315 and GH_A13G1170) and MYB44 (GH_D05G2657, GH_D06G0201, GH_A11G2346 and GH_D11G2108). Five and six bHLHs were down‐ and up‐regulated in HS2, respectively, including GLABRAs (GL3, down‐regulated; GH_D11G0216 and GH_A11G0214). From the DEGs related to MBW complexes, in contrast to the down‐regulation of MYB5, two MYB regulators MYB305 and MYB113 were up‐regulated in HS2. TTG1 was not significantly altered in expression between C312 and HS2, whereas GL3 was unexpectedly down‐regulated. Therefore, MYB113‐GL3‐TTG1 complex, conserved in plants such as Arabidopsis, possibly works in HS2 and might be responsible for the enhanced anthocyanidin accumulation, although remains to be further revealed. Some other transcription factors showed differentially expressed at varying degrees (Figure S8), which might imply their participation in anthocyanin synthesis.
It is possible that knock‐down of GhOMT1 blocked the generation of methylated anthocyanidins (peonidin, petunidin and malvidin) and provided compensatory enhancement of the biosynthesis of unmethylated anthocyanidin monomers (pelargonidin, cyanidin and delphinidin), and as a result led to the empurpled phenotypes of HS2. PAs acted as the main pigment components of brown cotton fibre of NCCs, and were not accumulated in HS2 fibres. The HS2 fibres were still white colour like wild type C312, suggesting that the PAs biosynthesis was not significantly affected in HS2, which is unaltered fibre colour.
Therefore, we reasoned that NCCs fibres are generally rich in PAs most likely due to the highly active PAs biosynthesis during fibre development. What is more, this prompted us to further investigate the potential application of the accumulated anthocyanidins arising out of GhOMT1‐deficiency to increase PA levels in fibres.
PAs biosynthesis was increased in the progenies derived from NCCs crossed by HS2
Nine NCC varieties, including seven varieties with brown fibres and two varieties with green fibres, were crossed to HS2 in 18 cross combinations (Table S3). Interestingly, their offsprings displayed a wider variety of fibre colour ranging from relatively light colours as observed in the parental lines to more dark colours (Figure S9). For example, cross combinations of HS2 and NCC ZX1 or XC5 (brown fibres) resulted in the fibre colours of brown, yellow, dark yellow and dark brown (Figure S9A). The cross combinations between LX1 and XC7 (light green fibres) and HS2 resulted in the fibre colours of deep green, army green and dark green (Figure S9B). Meanwhile, for the combination of HS2 and T586 (brown fibres), a series of blue colour fibres were found in the offsprings (Figure S9C).
All stably inherited F4 coloured cotton lines derived from NCCs crossed by HS2 keep the extremely low transcript level of GhOMT1 as parental line of HS2 (Figure S10), and co‐segregated with the purple trait (Figures S3, S10 and S11). The biosynthesis of anthocyanidins and PAs in ZX1, LX1 and their F4 coloured cotton lines derived from ZX1 and LX1 crossed by HS2 were further investigated by checking the expression level of three key enzyme genes (GhCHS, GhLAR and GhANR) in the leaves and fibres at different developmental stages. In leaves, the expression of GhCHS was relatively higher in HS2 compared to that in the F4 lines from ZX1×HS2 and LX1×HS2 (P < 0.05), while the expression of GhLAR and GhANR were very low in all samples, with a slightly higher expression of GhLAR in both F4 lines from ZX1×HS2 and LX1×HS2 (Figure 5a,e). In the developing fibres from 3 to 15 days post anthesis (DPA), the expression levels of GhCHS, GhLAR and GhANR in the F4 lines derived from HS2×ZX1 were all significantly increased (P < 0.05) (Figure 5b–d). Particularly, GhLAR expression was notably up‐regulated in all stages (P < 0.01), and GhCHS and GhANR expression was especially up‐regulated from 9 DPA (P < 0.01). Similar results were found when HS2 was crossed with the green fibre cotton LX1, i.e. GhCHS, GhLAR and GhANR expression was significantly upregulated in the F4 lines from LX1×HS2 (P < 0.05) (Figure 5f–h). The fibre colour changes may result from the highly expressed genes in the anthocyanidins biosynthesis and especially in PAs biosynthesis through NCCs crossed by HS2.
Figure 5.
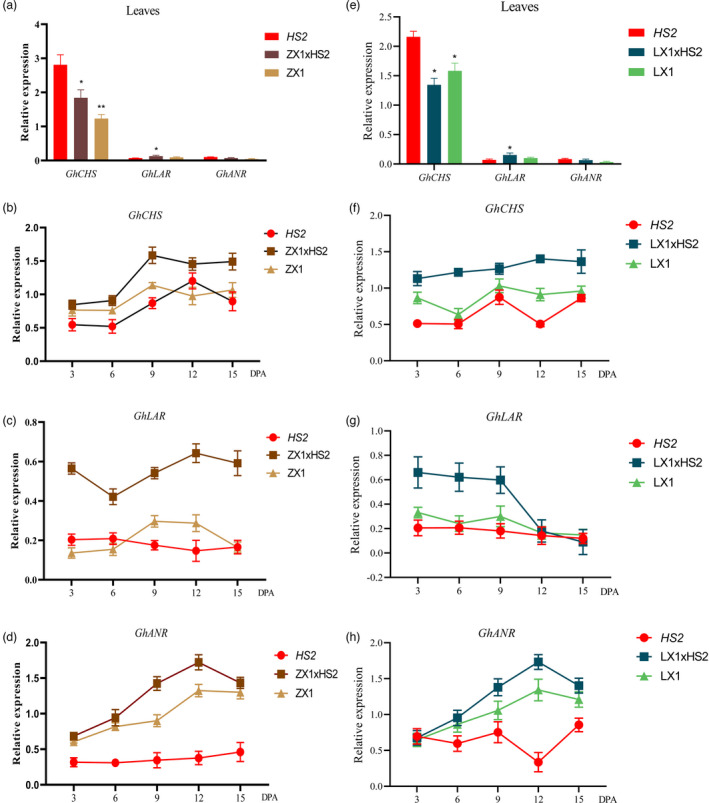
The transcript levels of GhCHS, GhLAR and GhANR in HS2, ZX1, LX1 and their F4 lines. (a) The transcript levels of GhCHS, GhLAR and GhANR in leaves in HS2, ZX1 and their F4 lines; (b–d). The transcript levels of GhCHS (b), GhLAR (c) and GhANR (d) in developing fibres of HS2, ZX1 and F4 lines from ZX1×HS2; (e). The transcript levels of GhCHS, GhLAR and GhANR in leaves in HS2, LX1 and their F4 lines; (f–h). The transcript levels of GhCHS (f), GhLAR (g) and GhANR (h) in developing fibres of HS2, LX1 and F4 lines from LX1×HS2. DPA: Days Post‐Anthesis.
Novel coloured fibre lines were raised by crossbreeding with HS2
To further investigate the application potential of the empurpled mutant with over‐accumulated anthocyanidins in the genetic improvement of cotton fibre colour, continuous field trials and selection of fibre colours in crossing offsprings were conducted in 2012. Lines with novel fibre colours, such as orange‐red, saddle brown, olive green, emerald green and navy blue were brought out in the homozygous progeny lines and proved inherited stably in consequent field experiment (Figure 6a,b). In lines with improved fibre colours, the leaf colour of adult plants became light purple or purple red and the amount of anthocyanidins in the leaves of F4 lines was less than that in HS2 but significantly higher than that in the NCC parents (Figures 6c and S10). In the 20 DPA fibres, however, anthocyanidin contents were dramatically increased in the offsprings (Figure 6d). These results indicated that the application of HS2 effectively enhanced the entire colour range of cotton fibres. Based on these results, a strategy was raised to improve and create new colour fibres, by increasing PA levels in fibres through enhancing activities of LAR and ANR to form flavan‐3‐ols and blocking anthocyanidins to form anthocyanins simultaneously, or regulating potential partners of MBW (MYB‐bHLH‐WDR) complexes to enhance the total anthocyanidins and PAs in the molecular breeding of naturally coloured cotton (Figure 7).
Figure 6.
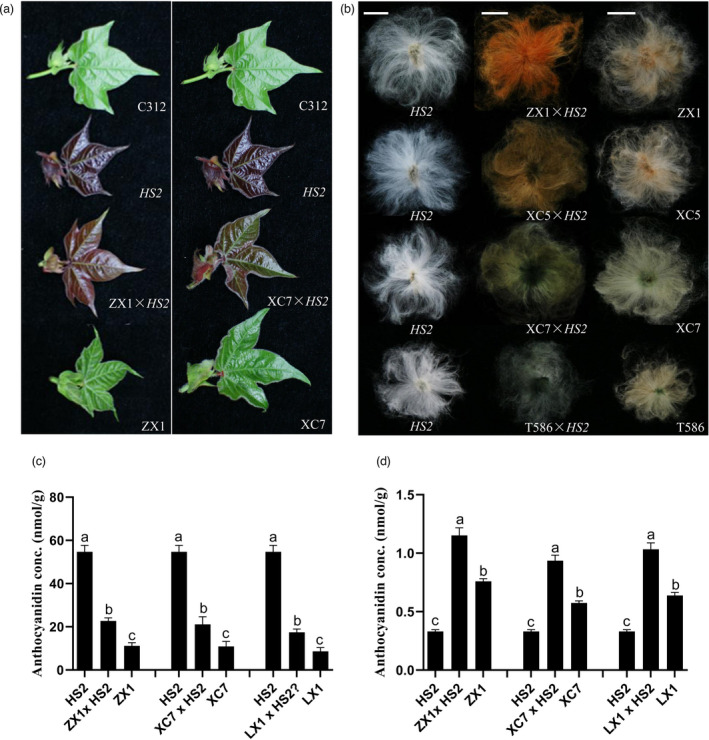
Leaf, fibre and anthocyanidin contents in HS2, NCCs and their F4 lines with fibre colour stably improved. (a) Leaves of C312, HS2, homozygous stable F4 lines derived from ZX1×HS2, XC7×HS2 and naturally coloured cotton (ZX1 and XC7); (b) Fibres of HS2, NCC and homozygous stable F4 lines (from left to right: HS2; F4 progenies from the crosses; NCCs ZX1, T586, XC5 or XC7). Hybrid crosses and their parent lines marked in the pictures. (c) Anthocyanidin content in leaves of HS2, NCC lines, and their stable fibre colour improved lines. (d) Anthocyanidin content in 20 DPA fibres of HS2, NCC lines, and their stable fibre colour improved lines. DPA: Days Post‐Anthesis. ‘a’ to ‘c’ indicate statistically significant differences between the anthocyanidin contents of the indicated seedlings, as determined by one‐way analysis of variance (ANOVA), followed by Tukey’s least significant difference (LSD) test (P < 0.05).
Figure 7.
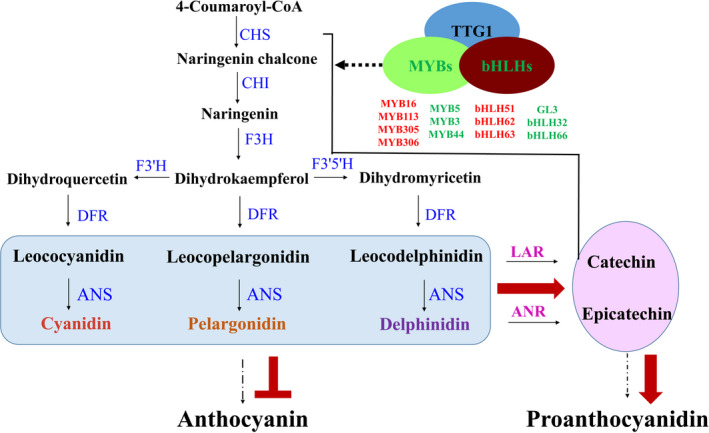
A comprehensive diagram to enhance PA levels. Flow chart of increasing proanthocyanidin levels in fibres by promoting biosynthesis of common substrates (leoanthocyanidins and anthocyanidins in blue box), enhancing activities of LAR and ANR to form flavan‐3‐ols and blocking anthocyanidin to form anthocyanins simultaneously, or regulating potential TFs to increase the total anthocyanidins and PAs. Red arrows indicated the direction to increase PA biosynthesis. The dotted arrows indicated some steps omitted. The up‐regulated TFs of MBW (MYB‐bHLH‐WDR) complexes in HS2 were labelled as red colour and down‐regulated TFs labelled as green colour (listed in Table S2).
Discussion
T‐DNA insertion in transposon regions caused functional loss of GhOMT1
In this work, we reported an empurpled cotton mutant HS2 where the T‐DNA was inserted between a transposon and the coding region of GhOMT1. To our interest, transcripts of GhOMT1 were sharply reduced in HS2 compared to C312. Transposable elements (TEs) are reported to play important roles in regulating neighbouring gene expression in many respects. Recently, for example, the red fruit colour of apple is thought to associate with the insertion of a gypsy‐like retrotransposon into transcription activator MdMYB1 (Zhang et al., 2019), and in Capsella rubella TE insertion at the 3′ UTR of FLOWERING LOCUS C (FLC) affects mRNA stability and promotes the onset of flowering (Niu et al., 2019). In G. hirsutum genome TEs account for 67.2% and the insertion of different TEs in the upstream region of DtMYB or DtERF has been proved to enhance its expression significantly, which confirmed the roles of TEs in rewiring gene regulatory networks (Wang et al., 2016). Herein, GhOMT1 transcripts in At subgenome were found greatly higher than that in Dt subgenome (GH_D01G2164) in C312, while it could hardly be detectable in HS2, which indicated T‐DNA insertion in HS2 resulted in its functional loss.
GhOMT1 played a critical role in anthocyanidin methylation and total anthocyanidins accumulation
O‐methyltransferases (OMTs), which catalyses the formation of O‐methylated compounds including chalcone, flavone, isoflavone, flavonol and anthocyanin families are divided into two major categories according to their molecular mass and substrate specificity (Akita et al., 2011; Joshi and Chiang, 1998; Lucker et al., 2010). GhOMT1 (GH_A01G2052), class II OMT, can recognize caffeic acid, coumarin, flavonoids and alkaloids as their substrates, and was previously predicted as a putative caffeic acid O‐methyltransferase by sequence analysis (Akita et al., 2011; Flagel et al., 2009; Hugueney et al., 2009). In this study, we found that functional loss of GhOMT1 in HS2 resulted in the loss of methylated anthocyanins including peonidin, petunidin and malvidin, thus indicating anthocyanidins are also substrates of GhOMT1. Therefore, GhOMT1 was proposed to function as a flavonoid O‐methyltransferase similar to that in other plants, such as Vitis vinifera, Cyclamen persicum, and Catharanthus roseus (Akita et al., 2011; Cacace et al., 2003; Hugueney et al., 2009; Lucker et al., 2010; Schroder et al., 2004). Strikingly, the GhOMT1 deficiency in HS2 not only hindered the formation of methylated anthocyanins and altered the composition of anthocyanidins, but also in turn significantly compensatorily promoted anthocyanidin biosynthesis. As we have observed, the expression of the genes encoding almost all the key enzymes involved in flavonoid biosynthesis was largely enhanced in HS2.
The MBW (MYB‐bHLH‐WDR) complexes regulating flavonoid genes at the transcriptional level were well conserved in plants (Chaves‐Silva et al., 2018; Xu et al., 2015). Comparing the gene expression between C312 and HS2, several potential MYB and bHLH regulators of cotton anthocyanin biosynthesis were identified. In contrast to the down‐regulation of MYB5 that mainly regulates the PAs biosynthesis, two MYB regulators that positively regulate the anthocyanin biosynthesis, MYB305 and MYB113 (Liu et al., 2009; Sablowski et al., 1994), were up‐regulated in HS2. MYB113 might activate the anthocyanin biosynthesis by forming a complex together with GL3 and TTG1, which are also activators of anthocyanin biosynthesis (Humphries et al., 2005; Wada et al., 2014). Derived from the RNA‐seq, TTG1 was not significantly altered in expression between C312 and HS2, whereas GL3 was unexpectedly down‐regulated. Regulatory roles of several of these TFs in anthocyanin biosynthesis have been characterized in other organisms. For instance, MYB5 in Medicago truncatula could activate the promoters of ANR and LAR, and overexpression of MYB5 would significantly induce PA accumulation (Liu et al., 2014). In addition, MYB305 and MYB113 positively regulate the anthocyanin biosynthesis, with MYB113 activating the anthocyanin biosynthesis by forming a complex together with GL3 and TTG1 (Humphries et al., 2005; Liu et al., 2009; Sablowski et al., 1994; Wada et al., 2014). These results emphasized that the precise roles of the three partners and the functioning of the MBW complexes are not yet fully understood even in Arabidopsis (Chaves‐Silva et al., 2018; Xu et al., 2015). Therefore, if MYB113‐GL3‐TTG1 works in HS2 and is responsible for the enhanced anthocyanin accumulation remains to be further demonstrated.
Flavonoids as developmental regulators play important roles in the biology of plants by affecting several developmental processes (Taylor and Grotewold, 2005). This might be ascribed to the requirement for methylated anthocyanins in many physiological processes of plants, such as responses to varied environmental stresses and development (Falcone Ferreyra et al., 2012; Hernández et al., 2009; Kovinich et al., 2014; Nakabayashi et al., 2014), so blocking the methylation step provided positive feedback to flavonoid biosynthesis. Due to the highly biochemical activity, over‐accumulation of anthocyanin in the cytosol, where it is synthesized, would be toxic to the cell (Grotewold et al, 1998; Hrazdina and Jensen, 1992; Klein et al., 1996; Marrs et al., 1995). Perhaps the over‐accumulated hydroxylated anthocyanidins and/or the lack of methylated anthocyanidins acted as signal or stimuli to feedback the expression of flavonoid genes. Therefore, the phenotype in HS2, i.e. white fibres and purple colour in the other tissues, is probably the result of a ‘dammed lake’ of anthocyanidins that was formed from the enhanced biosynthesis of unmethylated anthocyanidin monomers and the unaltered efficiency of subsequent decoration to form anthocyanins and polymerization to form PAs.
Up‐regulation of GhANR and GhLAR increased PA biosynthesis and improved fibre colour
Previous studies have shown that PAs are the major pigments in brown cotton fibres, and their main components are procyanidins, prodelphinidins and their derivatives (Feng et al., 2014; Li et al., 2012). Nevertheless, details of fibre colour formation are still largely unclear, and a wide gap remains between the unravelled mechanisms and creation of novel coloured cotton by genetic modification. LAR and ANR provide two separate pathways to form flavan‐3‐ols, i.e. primarily (−)‐epicatechin and (+)‐catechin, and were regarded as the key enzymes in PAs biosynthesis (Bassolino et al., 2013; Bogs et al., 2005; Kovinich et al., 2012; Tanner et al., 2003; Wang et al., 2018; Xie et al., 2003). Modulation of tissue colours by manipulation of ANR and LAR has been successful in several plants (Bogs et al., 2005; Gao et al., 2019; Liu et al., 2016; Yu et al., 2019). In G. hirsutum NCCs, GhANR and GhLAR were thought to act on anthocyanidins and leucoanthocyanidins respectively in the PAs biosynthesis pathway in brown fibres (Feng et al., 2014; Gao et al., 2019; Hinchliffe et al., 2016; Yan et al., 2018). In fibre of HS2, the expression of GhLAR and GhANR was not enhanced (Figure 5), which may have limited the further conversion of leucoanthocyanidins and anthocyanidin to generate enough PAs, despite the accumulation of the substrates of GhLAR and GhANR. This is probably the reason why the fibres of HS2 mutant remained white colour. When HS2 crossed with NCC varieties, a series of progeny lines with increased anthocyanidins, PAs and improved fibre colours were produced. We proposed that this is due to the upregulation of GhANR and GhLAR expression in developing fibres of the crossing progenies, GhANR and GhLAR could reduce the accumulated substrates of leucoanthocyanidins and anthocyanidins to form catechin and epicatechin, which would facilitate PAs biosynthesis in fibres and resulted in a wider range of fibre colours. As PAs accumulated in fibres, the leaf colour of these lines turned light purple or purple‐red, indicating that the anthocyanins synthesis reduced as the PAs biosynthesis increasing. This fact suggested that anthocyanidins were shared substrates for anthocyanin and PA biosynthesis, and the restriction of anthocyanin biosynthesis at the modification and oxidation stages might increase PAs accumulation in plants with both pathways. So it is reasonable to speculate that high content of anthocyanidins produced in HS2 combined with high expression of GhANR and GhLAR through crossbreeding or gene manipulation will be an efficient way to improve fibre colour.
Crossbreeding with HS2 also improved green fibre colour
Different from brown fibres, the pigment compositions and formation mechanisms of green fibres are more complex and less understood. Cinnamic acid and its derivatives, including caffeic acid, ferulic acid, caffeoyl quinic acid, 3,4‐dihydroxystyrene, coniferylaldehyde, 5‐hydroxy coniferaldehyde and sinapaldehyde, have been identified in green fibre (Feng et al., 2017; Ma et al., 2016; Sun et al., 2019; Yatsu et al., 1983). Recently, flavonoids, such as colourless anthocyanidin, flavone, flavanonol, and flavanol were also detected and proposed to contribute to green cotton pigmentation (Sun et al., 2019). In this study, dark green fibres were generated in the offsprings from the cross between XC7 and HS2 mutant, where the expressions of GhANR and GhLAR were also found to be upregulated, suggesting that anthocyanidins even PAs might also contribute to green fibre coloration. Blue fibre was produced from the hybrid of brown fibre cotton T586 and HS2, and also indicated PAs were one of the major contributors to blue pigment. In plants, both cinnamic acid and its derivatives and flavonoids are produced through the phenylpropanoid pathway (Berni et al., 2019). Gh4CL4, an enzyme in the phenylpropanoid pathway to catalyse the formation of CoA‐esters of cinnamic acids and their derivatives, is thought to be involved in the metabolism of caffeic and ferulic residues to affect pigmentation in green fibres (Feng et al., 2017; Sun et al., 2019). The deficiency of GhOMT1 in HS2 up‐regulated the expression of most genes in the flavonoid core pathway and Gh4CL in the phenylpropanoid pathway. The upregulation of Gh4CL might be another contributor to affect the pigmentation in their offsprings. This result helps shed light on the pigment compositions and their formation mechanisms in NCCs with green fibres.
Application of HS2 in NCC fibre colour improvement
Due to the apparent advantages in environmental protection, human healthy and facing increasingly serious global warming, NCC fibres are favoured by the textile industry with increasing demand in recent decades (Gong et al., 2018). However, the creation of cotton varieties with a wider range of fibre colours has been a major challenge until recently, with the main problems concerning limited germplasm resources and the rarity of knowledge about the mechanisms of colour formation in NCC fibres.
In the mutant HS2, the deficiency of GhOMT1 led to a remarkable increased content and altered composition of anthocyanidins, which provides a valuable resource for future utilization in cotton breeding of novel fibre colours. Based on this study, a strategy was proposed and confirmed to improve fibre colours by increasing total PAs in cotton fibres through regulating the structural genes involved in flavonoid biosynthesis, as well as enzymes involved in the decoration steps and transcription factors regulating anthocyanidin synthesis. Besides, on the basis of the ample supply of free anthocyanidins in the HS2 mutant, subsequent genetic manipulation of GhDFR, GhF3’H and GhF3’5’H to change the content and composition of anthocyanidins together with manipulation of GhANR and GhLAR to modify the content and composition of PAs are likely to become valuable strategies to create various fibre colours for future breeding work.
Above hypothesis on PA synthesis in cotton fibres revealed the great value of HS2 in crossbreeding of NCCs. In addition, after crossing with NCCs, the hybrid offsprings maintained purple and purple‐red traits in leaves from the germinated seedling to mature plant stages (Figures S1, S3, S4, S10, S11) which might be the dose effect of GhOMT1 enzyme in heterozygous and homozygous plants, and this phenotype can be used as a co‐dominant genetic marker for early selection. In mature individuals, lines with changed fibre colour in the progenies were selected for further study and breeding. In this way, novel lines could be selected through visible phenotypes that greatly simplified the selection and breeding progress.
Interestingly, apart from the alteration in fibre colours, some of the traits related to stress responses of insect‐resistant, freezing‐resistant and growth vigour were also improved in the hybrids and their derivatives of HS2 and the NCCs, such as enhanced photosynthesis, stomatal conductance, transpiration rate and water use efficiency (Figure S12), which also showed the vast value of using the empurpled mutant to generate novel cotton cultivars. It is largely reported previously that anthocyanidins contributed in many ways to the growth and survival of plants and serve as coping stress‐responsive mechanisms (Bhattacharya et al., 2010; Chaves‐Silva et al., 2018; Dixon et al., 2005; Emiliani et al., 2013; Falcone Ferreyra et al., 2012; Kovinich et al., 2014; Landi et al., 2015), and the accumulation of flavonoids/anthocyanins increases cotton resistance to bollworms and the fungal pathogen Verticillium dahlia (Fan et al., 2016; Long et al., 2019). Therefore, in HS2, the blocked methylation resulted in increased unmethylated anthocyanidins, which also indicated the importance of HS2 in future breeding to generate cultivars with enhanced stress resistance.
Materials and methods
Plant materials and growth conditions
The upland cotton varieties Coker 312 (C312), Yuzao1 (YZ1) (Gossypium hirsutum L.), the empurpled mutant HS2 with C312 as background, 7 brown‐fibre NCC cultivars including Zongxu1 (ZX1), Xincai1 (XC1), Xincai5 (XC5), Zongxu1‐61 (ZX1‐61), Zongxu1‐52 (ZX1‐52), Xincai20 (XC20) and T586 (T586), and 2 light‐green‐fibre NCC cultivars including Xincai7 (XC7) and Lvxu1 (LX1) were used in this study. All plants were grown in the greenhouse or experimental farm under standard conditions at Zhejiang Sci‐Tech University (Xiasha Campus), Hangzhou, China. Combinations of HS2, C312, YZ1 and NCC cultivars were reciprocally crossed (Table S2). At least 200 F2 plants of each cross combination were cultivated. The lines with fibre colour notably changed in F2/F3 populations were selected for self‐crossing at least two generations to get stable population (F4 lines with stably inherited purplish colour phenotype and coloured fibre); other agronomic characters were also investigated and determined.
Flowers on the day of flowering were tagged as 0 DPA (Days Post Anthesis), and cotton bolls were harvested at 0, 3, 6, 9, 12, 15, 20 and 25 DPA. On the harvesting day, cut the cotton bolls, took out the ovule and scraped the fibres from the ovules, and then put collected materials immediately in liquid nitrogen and stored at −80 °C before use.
PCR analysis and Southern blotting
Genomic DNAs were isolated from the leaves of transgenic plants according to a modified cetyl trimethyl ammonium bromide method (Paterson et al., 1993). Specific primers designed to amplify a coding region fragment of the npt II (kanamycin resistance) gene and T‐DNA sequence were used for PCR analysis to check the T‐DNA insertion (Table S4).
Southern blot was conducted to identify the copy number of inserted T‐DNA in different transgenic T1 plants by using a DIG‐High Prime DNA labelling and detection Starter Kit I as previously reported (Liu et al., 2018b). A specific PCR‐amplified fragment of T‐DNA sequence was used as a probe, and the hybridization of DIG‐labelled DNA probes was performed at 42 °C for 24 h. For Southern blot assays, total nucleic acids were fractionated by 1% (w/v) agarose gel electrophoresis and transferred to Hybond‐N+ membranes (Amersham Pharmacia Biotech, Buckinghamshire, UK). Probe labelling and hybridization were performed according to the instructions of the DIG High Prime DNA Labelling and Detection Starter I Kit (Roche, Mannheim, Germany). Radioactive signals were detected using the Typhoon 9200 imager (GE Healthcare, Piscataway, NJ).
Flanking sequence identification of T‐DNA insertion site
Tail‐PCR (Thermal asymmetric interlaced PCR) was performed and the arbitrary degenerate primers were designed in accordance with previous reports (Liu and Chen, 2007). Genes specific primers (GSP1L, GSP2L and GSP3L; GSP1R, GSP2R and GSP3R) were designed according to the inside adjacent sequences of the T‐DNA border in pBI121 (Table S4). Then the TAIL‐PCR products with single band or clearly dominated by a single band were cloned in pGEM‐T easy vector and sequenced.
5′ RACE of GhOMT1 were performed according to the instruction manual (5′ RACE System for Rapid Amplification of cDNA Ends, Version 2.0, Catalog no. 18374‐058; Invitrogen, Shanghai, China), primers were listed in Table S4.
Vectors construct and genetic transformation
The GhOMT1 full‐length complementary DNA and a fragment of GhOMT1 for interference were amplified by PCR from cDNA (here GhOMT1 was GhOMT1‐At; primer list in Table S5). The DNA fragment was cloned into pENTR/D TOPO vector (Invitrogen). Coding sequences were then transferred from the entry clones to gateway cloning vector pK7GWIWG2(II) with LR Clonase II to construct RNA interference vector for GhOMT1 silencing (Karimi et al., 2002). The amplified DNA fragments of GhOMT1 full‐length cDNA were digested with Xba I+Sac I and then ligated into pBI121 to overexpress the GhOMT1 gene. Transgenic cotton lines of GhOMT1‐RNAi and GhOMT1 over‐expression in C312 were generated by Agrobacterium‐mediated transformation according to the previous studies (Zhang, 2019). Homozygous transgenic T3 lines overexpressing GhOMT1 in C312 carrying a single insertion were used to cross with HS2 to obtain HS2‐overexpressing GhOMT1 to complete complementation experiments. Homozygous transgenic T3 lines of GhOMT1‐RNAi were used for further analysis.
RNA extraction, RNA sequencing and qRT‐PCR
Total RNA was extracted from different samples using an RNAsimple Total RNA Kit (Tiangen, Beijing, China). After removing residual DNA with a DNase Mini Kit (Qiagen, Hilden, Germany), total RNA was reverse transcribed using a ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan) following the manufacturer’s instructions. qRT‐PCR assays were performed with SYBR Premix Ex Taq (TaKaRa, Shenzhen, China) on a Mastercycler ep realplex system (Eppendorf, Hamburg, Germany). The primers are shown in Table S6. The cotton Ubiquitin7 gene (GhUBQ7, GenBank accession number: DQ116441, GH_D11G1140.1) was used as an internal standard for the assays. All experiments were performed in triplicate with three biological repeats.
Expression of potential regulators of cotton anthocyanin biosynthesis was also investigated in HS2 and C312, such as the well‐known R2R3‐MYB, bHLH and WD40 components of different MBW (MYB‐bHLH‐WD40) ternary complexes. The high‐quality RNA of C312 and HS2 (OD260/280: 2.0–2.2 and RIN ≥ 7.0) was used to construct the stranded library after depletion of rRNA by using ribo‐zero kit (Epicentre, Madison, WI). The strand‐specific libraries were sequenced on Illumina Hiseq2500 platform at Novogene (Beijing, China) with pair‐end strategy (2 × 150 bp). Clean data from RNA sequencing were aligned to the reference genome (ZJU_v2.1, downloaded from CottonGen (Yu et al., 2014) by TopHat (v2.1.0, ‐‐library‐type fr‐firststrand) (Kim et al., 2013). HTSeq was used to calculate the number of short reads aligned to the characterized gene loci and DESeq2 was then used to identify the differentially expressed genes (cut‐off fold change ≥ 2 and P‐value ≤ 0.05) (Anders et al., 2015; Love et al., 2014). Enrichment of GO terms and pathways in DEGs was estimated by chi‐square test. Only the GO terms referring to more than three genes were retained and the redundancy of GO terms was removed through the online tool REViGO with default settings (Supek et al., 2011).
Anthocyanidin extraction for content analysis
The samples of leaves or immature fibres (approx. 100 mg) were collected for anthocyanidin extraction with HCl (0.5% v/v) and methanol buffer, and measurements of anthocyanidin accumulation were performed as described by Wade et al. (2003). This process was repeated 3 times. The supernatant was assayed spectrophotometrically, and anthocyanidin absorbance units (A530‐A657) per gram fresh weight were calculated. The blank was 480 ml of methanol with 0.5% (v/v) HCl and 320 ml of Milli‐Q H2O for a total of 800 ml. A spectrophotometer (UV‐2600, Shimadzu, Japan) was used for absorbance measurements at 530, 620 and 650 nm. The optical density (OD) was determined based on the following equation: OD = (A530−A620)−[0.1 × (A650−A620)].
HPLC and LC/MS analysis
Phenylpropanoids and anthocyanidins were extracted for HPLC and LC/MS analysis. The samples (200 mg) were collected and extracted with three repeats according to the process in the previous paper (Berni et al., 2019; Sun et al., 2019). The purified powders were resuspended in 0.1% (v/v) methanol‐HCl solution for HPLC on an Agilent 1290 Infinity HPLC system and LC/MS analysis (Berni et al., 2019; Han et al., 2013; Sun et al., 2019).
Samples for MS analyses were injected into an Agilent 6460 triple quadruple mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) equipped with an electrospray ionization source (ESI) and an Agilent 1200 separation module operating in the positive ionization mode with the following parameters: nitrogen drying gas temperature 325 °C, nitrogen sheath gas temperature 350 °C, nitrogen drying gas flow 5 L/min, nitrogen sheath gas flow 11 L/min, nebulizer pressure 45 psi, and capillary voltage 3000 V. The levels of anthocyanidins in leaves or fibres were expressed as nanograms per gram fresh weight material.
For anthocyanidin component analysis, LC was performed with an Agilent ZORBAX SB‐C18 column (50 × 4.6 mm, 1.8 μm particle size) (Merck KGaA, Darmstadt, Germany) using aqueous formic acid (0.2% v/v) (mobile phase A) and acetonitrile plus formic acid (0.2% v/v) (mobile phase B). The elution profile was as follows: 0–0.5 min, 95% A; 0.5–5 min, 5%–20% B in A; 5–7 min 90% B in A; and 7.1–10 min 95% A. The flow rate was 0.8 mL/min at a column temperature of 50 °C. LC was coupled to an Agilent MSD Trap XCT‐Plus mass spectrometer equipped with an electrospray operated in negative ionization mode (capillary voltage, 4000 eV; temperature, 350 °C; nebulizing gas, 60 p.s.i.; dry gas 12 L/min) and an Agilent 1100 diode array detector (detection 200–700 nm; J&M Analytik, Jena, Germany). MS/MS was used to monitor daughter ion formation. LC/MSD Trap Software 5.2 (Bruker Daltonik) was used for data acquisition and processing. Metabolites were quantified using an MbA standard curve. Enzyme products were quantified using the UV spectra (350–370 nm) and external standard curves of pelargonidin ([M + H]+ = 271), cyanidin ([M + H]+ = 287), delphinidin ([M + H]+ = 301), peonidin ([M + H]+ = 303), petunidin ([M + H]+ = 317), and malvidin ([M + H]+ = 331), which were also used as internal standards (Sigma‐Aldrich, St. Louis, MO, USA). Compounds were tentatively identified using their molecular masses and specific fragmentation patterns. The raw data were extracted using MassHunter software (Agilent Technologies) and examined in Excel (Microsoft).
Statistical analysis
All data are presented as the mean ± SD from at least three independent experiments with at least three replicates each. The statistical significance of the differences was determined using Student’s t‐test through GraphPad 8.0. Differences between treatments were considered significant when P < 0.05 or 0.01 or 0.001 in a two‐tailed analysis.
Conflict of interest
No conflict of interest declared.
Author contributions
Y.S. and X.Z. conceived the study. L.K., D.Y., H.Z., Y.X., Y.W., J.J., X.W., J.M., F.C., Y.Z. and J.S. performed the experiments and data analyses. Y.S., L.K. and X.Z. wrote the manuscript.
Supporting information
Figure S1 The phenotype of HS2 and C312 in the field.
Figure S2 T‐DNA insertion and the flanking sequence analysis of HS2 mutant.
Figure S3 GhOMT1 mutation by T‐DNA insertion associated with empurpled phenotypic change.
Figure S4 The phenotypes of HS2, YZ1 and their F1 plant.
Figure S5 The transcript level of five GhOMT genes and GhOMT1‐At and GhOMT1‐Dt in C312 and HS2.
Figure S6 Phenotypes and GhOMT1 expression level in the GhOMT1 overexpressed C312 and HS2 lines.
Figure S7 The contents and profiles of anthocyanidins in the leaves of GhOMT1 RNAi plants.
Figure S8 Summary of differentially expressed transcription factors between C312 and HS2.
Figure S9 Fiber colors of HS2, NCCs and their progenies derived from NCCs crossed by HS2.
Figure S10 Leave color and GhOMT1 expression analysis of C312, HS2, natural colored cottons and their homozygous F4 hybrids.
Figure S11 The phenotypes of HS2, ZX1 and their hybrid offpring (F4) in the field.
Figure S12 The photosynthetic abilities significantly improved in hybrid lines including photosynthesis rate, stomatal conductivity, transpiration rate and water use efficiency.
Table S1 The genetic analysis of cross combinations of HS2 and 6 upland cotton cultivars (P < 0.95).
Table S2 The differentially expressed genes in HS2 and C312 from RNA‐sequence data.
Table S3 The combinations of crosses of HS2 and naturally coloured cotton cultivars.
Table S4 Primers for PCR detection and the flanking sequence used in this paper.
Table S5 Primers for vector constuction and flanking sequencing.
Table S6 Primers for Real time PCR used in this paper.
Acknowledgements
The authors greatly appreciate Dr. Xuejun Hua from College of Life Sciences and Medicine, Zhejiang Sci‐Tech University for his constructive comments during manuscript preparation. This work was supported by the National Natural Science Foundation of China (U1903204, 32170623) and the Natural Science Foundation of Zhejiang Province (LZ21C130004). The funding agencies had no role in research design, data collection and analysis, or manuscript writing.
Ke, L. , Yu, D. , Zheng, H. , Xu, Y. , Wu, Y. , Jiao, J. , Wang, X. , Mei, J. , Cai, F. , Zhao, Y. , Sun, J. , Zhang, X. and Sun, Y. (2022) Function deficiency of GhOMT1 causes anthocyanidins over‐accumulation and diversifies fibre colours in cotton (Gossypium hirsutum). Plant Biotechnol. J., 10.1111/pbi.13832
Contributor Information
Xianlong Zhang, Email: xlzhang@mail.hzau.edu.cn.
Yuqiang Sun, Email: sunyuqiang@zstu.edu.cn.
References
- Abrahams, S. , Tanner, G.J. , Larkin, P.J. and Ashton, A.R. (2002) Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis. Plant Physiol. 130, 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akita, Y. , Kitamura, S. , Hase, Y. , Narumi, I. , Ishizaka, H. , Kondo, E. , Kameari, N. et al. (2011) Isolation and characterization of the fragrant cyclamen O‐methyltransferase involved in flower coloration. Planta, 234, 1127–1136. [DOI] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P.T. and Huber, W. (2015) HTSeq–a Python framework to work with high‐throughput sequencing data. Bioinformatics, 31(2), 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassolino, L. , Zhang, Y. , Schoonbeek, H.J. , Kiferle, C. , Perata, P. and Martin, C. (2013) Accumulation of anthocyanins in tomato skin extends shelf life. New Phytol. 200, 650–655. [DOI] [PubMed] [Google Scholar]
- Berni, R. , Cai, G. , Hausman, J.‐F. and Guerriero, G. (2019) Plant fibers and phenolics: A review on their synthesis, analysis and combined use for biomaterials with new properties. Fibers, 7, 80. [Google Scholar]
- Bhattacharya, A. , Sood, P. and Citovsky, V. (2010) The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol. Plant Pathol., 11, 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs, J. , Downey, M.O. , Harvey, J.S. , Ashton, A.R. , Tanner, G.J. and Robinson, S.P. (2005) Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 139, 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacace, S. , Schroder, G. , Wehinger, E. , Strack, D. , Schmidt, J. and Schroder, J. (2003) A flavonol O‐methyltransferase from Catharanthus roseus performing two sequential methylations. Phytochemistry, 62, 127–137. [DOI] [PubMed] [Google Scholar]
- Chaves‐Silva, S. , Santos, A.L.D. , Chalfun‐Junior, A. , Zhao, J. , Peres, L.E.P. and Benedito, V.A. (2018) Understanding the genetic regulation of anthocyanin biosynthesis in plants ‐ Tools for breeding purple varieties of fruits and vegetables. Phytochemistry, 153, 11–27. [DOI] [PubMed] [Google Scholar]
- Chen, H.‐L. and Cluver, B. (2010) Biodegradation and mildew resistance of naturally colored cottons. Text. Res. J. 80, 2188–2194. [Google Scholar]
- Debeaujon, I. , Nesi, N. , Perez, P. , Devic, M. , Grandjean, O. , Caboche, M. and Lepiniec, L. (2003) Proanthocyanidin‐accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell, 15, 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R.A. , Xie, D.Y. and Sharma, S.B. (2005) Proanthocyanidins–a final frontier in flavonoid research? New Phytol. 165, 9–28. [DOI] [PubMed] [Google Scholar]
- Dutt, Y. , Wang, X.D. , Zhu, Y.G. and Li, Y.Y. (2004) Breeding for high yield and fibre quality in colored cotton. Plant Breed. 123, 145–151. [Google Scholar]
- Emiliani, J. , Grotewold, E. , Falcone Ferreyra, M.L. and Casati, P. (2013) Flavonols protect Arabidopsis plants against UV‐B deleterious effects. Mol. Plant, 6, 1376–1379. [DOI] [PubMed] [Google Scholar]
- Falcone Ferreyra, M.L. , Rius, S.P. and Casati, P. (2012) Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, X. , Fan, B. , Wang, Y. and Yang, W. (2016) Anthocyanin accumulation enhanced in Lc‐transgenic cotton under light and increased resistance to bollworm. Plant Biotechnol. Rep. 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, H. , Li, Y. , Wang, S. , Zhang, L. , Liu, Y. , Xue, F. , Sun, Y. et al. (2014) Molecular analysis of proanthocyanidins related to pigmentation in brown cotton fibre (Gossypium hirsutum L.). J. Exp. Bot. 65, 5759–5769. [DOI] [PubMed] [Google Scholar]
- Feng, H. , Tian, X. , Liu, Y. , Li, Y. , Zhang, X. , Jones, B.J. , Sun, Y. et al. (2013) Analysis of flavonoids and the flavonoid structural genes in brown fiber of upland cotton. PLoS One, 8, e58820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, H. , Yang, Y. , Sun, S. , Li, Y. , Zhang, L. , Tian, J. , Zhu, Q. et al. (2017) Molecular analysis of caffeoyl residues related to pigmentation in green cotton fibers. J. Exp. Bot. 68, 4559–4569. [DOI] [PubMed] [Google Scholar]
- Flagel, L.E. , Chen, L. , Chaudhary, B. and Wendel, J.F. (2009) Coordinated and fine‐scale control of homoeologous gene expression in allotetraploid cotton. J. Hered. 100, 487–490. [DOI] [PubMed] [Google Scholar]
- Gao, J. , Shen, L.I. , Yuan, J. , Zheng, H. , Su, Q. , Yang, W. , Zhang, L. et al. (2019) Functional analysis of GhCHS, GhANR and GhLAR in colored fiber formation of Gossypium hirsutum L. BMC Plant Biol. 19, 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, W. , Du, X. , Jia, Y. and Pan, Z. (2018) Color cotton and its utilization in China. In Cotton Fiber: Physics, Chemistry and Biology ( Fang, D. , ed), pp. 117–132. Cham: Springer. [Google Scholar]
- Gong, W. , He, S. , Tian, J. , Sun, J. , Pan, Z. , Jia, Y. , Sun, G. et al. (2014) Comparison of the transcriptome between two cotton lines of different fiber color and quality. PLoS One, 9, e112966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold, E. , Chamberlin, M. , Snook, M. , Siame, B. , Butler, L. , Swenson, J. , Maddock, S. et al. (1998) Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell, 10, 721–740. [PMC free article] [PubMed] [Google Scholar]
- Han, L.‐B. , Li, Y.‐B. , Wang, H.‐Y. , Wu, X.‐M. , Li, C.‐L. , Luo, M. , Wu, S.‐J. et al. (2013) The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell, 25, 4421–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F. , Pan, Q.H. , Shi, Y. and Duan, C.Q. (2008) Biosynthesis and genetic regulation of proanthocyanidins in plants. Molecules, 13, 2674–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández, I. , Alegre, L. , Van Breusegem, F. and Munné‐Bosch, S. (2009) How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 14, 125–132. [DOI] [PubMed] [Google Scholar]
- Hinchliffe, D.J. , Condon, B.D. , Thyssen, G. , Naoumkina, M. , Madison, C.A. , Reynolds, M. , Delhom, C.D. et al. (2016) The GhTT2_A07 gene is linked to the brown colour and natural flame retardancy phenotypes of Lc1 cotton (Gossypium hirsutum L.) fibres. J. Exp. Bot. 67, 5461–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrazdina, G. and Jensen, R.A. (1992) Spatial organization of enzymes in plant metabolic pathways. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 241–267. [Google Scholar]
- Hugueney, P. , Provenzano, S. , Verries, C. , Ferrandino, A. , Meudec, E. , Batelli, G. , Merdinoglu, D. et al. (2009) A novel cation‐dependent O‐methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiol. 150, 2057–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries, J.A. , Walker, A.R. , Timmis, J.N. and Orford, S.J. (2005) Two WD‐repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol. Biol. 57(1), 67–81. [DOI] [PubMed] [Google Scholar]
- Hustvedt, G.C. and Cox, P. (2005) The ultraviolet protection factor of naturally‐pigmented cotton. J Cotton Sci. 9, 47–55. [Google Scholar]
- Jaakola, L. (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477. [DOI] [PubMed] [Google Scholar]
- Joshi, C.P. and Chiang, V.L. (1998) Conserved sequence motifs in plant S‐adenosyl‐L‐methionine‐dependent methyltransferases. Plant Mol. Biol. 37, 663–674. [DOI] [PubMed] [Google Scholar]
- Karimi, M. , Inze, D. and Depicker, A. (2002) GATEWAY vectors for Agrobacterium‐mediated plant transformation. Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Katsumoto, Y. , Fukuchi‐Mizutani, M. , Fukui, Y. , Brugliera, F. , Holton, T.A. , Karan, M. , Nakamura, N. et al. (2007) Engineering of the rose flavonoid biosynthetic pathway successfully generated blue‐hued flowers accumulating delphinidin. Plant Cell Physiol. 48, 1589–1600. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. and Salzberg, S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, M. , Weissenböck, G. , Dufaud, A. , Gaillard, C. , Kreuz, K. and Martinoia, E. (1996) Different energization mechanisms drive the vacuolar uptake of a flavonoid glucoside and a herbicide glucoside. J. Biol. Chem. 271, 29666–29671. [DOI] [PubMed] [Google Scholar]
- Koes, R. , Verweij, W. and Quattrocchio, F. (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242. [DOI] [PubMed] [Google Scholar]
- Kohel, R.J. (1985) Genetic analysis of fiber color variant in cotton. Crop Sci. 25, 793–797. [Google Scholar]
- Kovinich, N. , Kayanja, G. , Chanoca, A. , Riedl, K. , Otegui, M.S. and Grotewold, E. (2014) Not all anthocyanins are born equal: distinct patterns induced by stress in Arabidopsis. Planta, 240, 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovinich, N. , Saleem, A. , Rintoul, T.L. , Brown, D.C. , Arnason, J.T. and Miki, B. (2012) Coloring genetically modified soybean grains with anthocyanins by suppression of the proanthocyanidin genes ANR1 and ANR2. Transgenic Res. 21, 757–771. [DOI] [PubMed] [Google Scholar]
- Landi, M. , Tattini, M. and Gould, K.S. (2015) Multiple functional roles of anthocyanins in plant‐environment interactions. Environ. Exp. Bot. 119, 4–17. [Google Scholar]
- Li, T. , Fan, H. , Li, Z. , Wei, J. , Lin, Y. and Cai, Y. (2012) The accumulation of pigment in fiber related to proanthocyanidins synthesis for brown cotton. Acta Physiol. Plant. 34, 813–818. [Google Scholar]
- Li, Z. , Su, Q. , Xu, M. , You, J. , Khan, A.Q. , Li, J. , Zhang, X. et al. (2020) Phenylpropanoid metabolism and pigmentation show divergent patterns between brown color and green color cottons as revealed by metabolic and gene expression analyses. J. Cotton Res. 3(4), 11. [Google Scholar]
- Liu, C. , Jun, J.H. , and Dixon, R.A. (2014) MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula . Plant Physiol. 165(4), 1424–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Wang, X. , Shulaev, V. and Dixon, R.A. (2016) A role for leucoanthocyanidin reductase in the extension of proanthocyanidins. Nat. Plants, 2, 16182. [DOI] [PubMed] [Google Scholar]
- Liu, G. , Ren, G. , Guirgis, A. and Thornburg, R. W. (2009) The MYB305 transcription factor regulates expression of nectarin genes in the ornamental tobacco floral nectary. Plant Cell, 21(9), 2672–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.‐F. , Luo, C. , Song, W.U. , Shen, H. , Li, G. , He, Z.‐G. , Chen, W.‐G. et al. (2018a) Flavonoid biosynthesis controls fiber color in naturally colored cotton. PeerJ, 6, e4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G. and Chen, Y. (2007) High‐efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques, 43, 649–656, 652, 654 passim. [DOI] [PubMed] [Google Scholar]
- Liu, Z.J. , Zhao, Y.P. , Liang, W. , Cui, Y.P. , Wang, Y.M. and Hua, J.P. (2018b) Over‐expression of transcription factor GhWRI1 in upland cotton. Biol. Plant. 62, 335–342. [Google Scholar]
- Long, L. , Liu, J. , Gao, Y. , Xu, F.C. , Zhao, J.R. , Li, B. and Gao, W. (2019) Flavonoid accumulation in spontaneous cotton mutant results in red coloration and enhanced disease resistance. Plant Physiol. Biochem. 143, 40–49. [DOI] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucker, J. , Martens, S. and Lund, S.T. (2010) Characterization of a Vitis vinifera cv. Cabernet Sauvignon 3',5'‐O‐methyltransferase showing strong preference for anthocyanins and glycosylated flavonols. Phytochemistry, 71, 1474–1484. [DOI] [PubMed] [Google Scholar]
- Ma, M. , Hussain, M. , Memon, H. and Zhou, W. (2016) Structure of pigment compositions and radical scavenging activity of naturally green‐colored cotton fiber. Cellulose, 23, 955–963. [Google Scholar]
- Marrs, K. , Alfenito, M.R. , Lloyd, A.M. and Walbot, V. (1995) A glutathione S‐transferase involved in vacuolar transfer encoded by the maize gene Bronze‐2. Nature, 375, 397–400. [DOI] [PubMed] [Google Scholar]
- Naik, J. , Misra, P. , Trivedi, P.K. and Pandey, A. (2022) Molecular components associated with the regulation of flavonoid biosynthesis. Plant Sci. 317, 111196. [DOI] [PubMed] [Google Scholar]
- Nakabayashi, R. , Yonekura‐Sakakibara, K. , Urano, K. , Suzuki, M. , Yamada, Y. , Nishizawa, T. , Matsuda, F. et al. (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, X.M. , Xu, Y.C. , Li, Z.W. , Bian, Y.T. , Hou, X.H. , Chen, J.F. , Zou, Y.P. et al. (2019) Transposable elements drive rapid phenotypic variation in Capsella rubella . Proc. Natl Acad. Sci. USA, 116, 6908–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, N. , Aida, R. , Kishimoto, S. , Ishiguro, K. , Fukuchi‐Mizutani, M. , Tanaka, Y. and Ohmiya, A. (2013) Genetic engineering of novel bluer‐colored chrysanthemums produced by accumulation of delphinidin‐based anthocyanins. Plant Cell Physiol. 54, 1684–1695. [DOI] [PubMed] [Google Scholar]
- Pang, Y. , Peel, G.J. , Wright, E. , Wang, Z. and Dixon, R.A. (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula . Plant Physiol. 145, 601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H. , Brubaker, C.L. and Wendel, J.F. (1993) A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Rep. 11, 122–127. [Google Scholar]
- Sablowski, R.W. , Moyano, E. , Culianez‐Macia, F.A. , Schuch, W. , Martin, C. and Bevan, M. (1994) A flower‐specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 13(1), 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder, G. , Wehinger, E. , Lukacin, R. , Wellmann, F. , Seefelder, W. , Schwab, W. and Schroder, J. (2004) Flavonoid methylation: a novel 4'‐O‐methyltransferase from Catharanthus roseus, and evidence that partially methylated flavanones are substrates of four different flavonoid dioxygenases. Phytochemistry, 65, 1085–1094. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Sun, Y. and Zhu, Q.H. (2021) Breeding next‐generation naturally colored cotton. Trends Plant Sci. 26, 539–542. [DOI] [PubMed] [Google Scholar]
- Sun, S. , Xiong, X.P. , Zhu, Q. , Li, Y.J. and Sun, J. (2019) Transcriptome sequencing and metabolome analysis reveal genes involved in pigmentation of green‐colored cotton fibers. Int. J. Mol. Sci. 20, 4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Zhang, D. , Zheng, H. , Wu, Y. , Mei, J. , Ke, L. , Yu, D. et al. (2022) Biochemical and expression analyses revealed the involvement of proanthocyanidins and/or their derivatives in fiber pigmentation of Gossypium stocksii . Int. J. Mol. Sci. 23, 1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek, F. , Bošnjak, M. , Škunca, N. and Šmuc, T. (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One, 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y. and Brugliera, F. (2013) Flower colour and cytochromes P450. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z. , Fan, Y. , Zhang, L. , Zheng, C. , Chen, A. , Sun, Y. , Guo, H. et al. (2020) Quantitative metabolome and transcriptome analysis reveals complex regulatory pathway underlying photoinduced fiber color formation in cotton. Gene, 767, 145180. [DOI] [PubMed] [Google Scholar]
- Tanner, G.J. , Francki, K.T. , Abrahams, S. , Watson, J.M. , Larkin, P.J. and Ashton, A.R. (2003) Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J. Biolog. Chem. 278, 31647–31656. [DOI] [PubMed] [Google Scholar]
- Taylor, L.P. and Grotewold, E. (2005) Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 8, 317–323. [DOI] [PubMed] [Google Scholar]
- Wada, T. , Kunihiro, A. and Tominaga‐Wada, R. (2014) Arabidopsis CAPRICE (MYB) and GLABRA3 (bHLH) control tomato (Solanum lycipersicum) anthocyanin biosynthesis. PLoS One, 9(9), e109093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, H.K. , Sohal, A.K. and Jenkins, G.I. (2003) Arabidopsis ICX1 is a negative regulator of several pathways regulating flavonoid biosynthesis genes. Plant Physiol. 131, 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Huang, G. and Zhu, Y. (2016) Transposable elements play an important role during cotton genome evolution and fiber cell development. Sci. China Life Sci. 59, 112–121. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Liu, H. , Li, X. , Xiao, X. , Ai, X. , Luo, C. , Zhu, L. et al. (2014) Genetic mapping of fiber color genes on two brown cotton cultivars in Xinjiang. SpringerPlus, 3, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Zhang, L. , Jiang, X. , Dai, X. , Xu, L. , Li, T. , Xing, D. et al. (2018) Evolutionary and functional characterization of leucoanthocyanidin reductases from Camellia sinensis . Planta, 247, 139–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Chen, L. , Yang, Q. , Hu, Z. , Guo, P. , Xie, Q. and Chen, G. (2022) New insight into the pigment composition and molecular mechanism of flower coloration in tulip (Tulipa gesneriana L.) cultivars with various petal colors. Plant Sci. 317, 111193. [DOI] [PubMed] [Google Scholar]
- Xiao, Y.H. , Zhang, Z.S. , Yin, M.H. , Luo, M. , Li, X.B. , Hou, L. and Pei, Y. (2007) Cotton flavonoid structural genes related to the pigmentation in brown fibers. Biochem. Biophys. Res. Comm. 358, 73–78. [DOI] [PubMed] [Google Scholar]
- Xie, D.Y. and Dixon, R.A. (2005) Proanthocyanidin biosynthesis–still more questions than answers? Phytochemistry, 66, 2127–2144. [DOI] [PubMed] [Google Scholar]
- Xie, D.Y. , Sharma, S.B. , Paiva, N.L. , Ferreira, D. and Dixon, R.A. (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science, 299, 396–399. [DOI] [PubMed] [Google Scholar]
- Xu, W. , Dubos, C. and Lepiniec, L. (2015) Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 20, 176–185. [DOI] [PubMed] [Google Scholar]
- Yan, Q. , Wang, Y. , Li, Q. , Zhang, Z. , Ding, H. , Zhang, Y. , Liu, H. et al. (2018) Up‐regulation of GhTT2‐3A in cotton fibres during secondary wall thickening results in brown fibres with improved quality. Plant Biotechnol. J. 16, 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsu, L.Y. , Espelie, K.E. and Kolattukudy, P.E. (1983) Ultrastructural and chemical evidence that the cell wall of green cotton fiber is suberized. Plant Physiol. 73, 521–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Jung, S. , Cheng, C.‐H. , Ficklin, S.P. , Lee, T. , Zheng, P. , Jones, D. et al. (2014) CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42, D1229–D1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, K. , Jun, J.H. , Duan, C. and Dixon, R.A. (2019) VvLAR1 and VvLAR2 are bifunctional enzymes for proanthocyanidin biosynthesis in Grapevine. Plant Physiol. 180, 1362–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. (2019) Agrobacterium‐mediated genetic transformation of cotton. Methods Mol. Biol. 1902, 19–33. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Hu, J. , Han, X. , Li, J. , Gao, Y. , Richards, C.M. , Zhang, C. et al. (2019) A high‐quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 10, 1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The phenotype of HS2 and C312 in the field.
Figure S2 T‐DNA insertion and the flanking sequence analysis of HS2 mutant.
Figure S3 GhOMT1 mutation by T‐DNA insertion associated with empurpled phenotypic change.
Figure S4 The phenotypes of HS2, YZ1 and their F1 plant.
Figure S5 The transcript level of five GhOMT genes and GhOMT1‐At and GhOMT1‐Dt in C312 and HS2.
Figure S6 Phenotypes and GhOMT1 expression level in the GhOMT1 overexpressed C312 and HS2 lines.
Figure S7 The contents and profiles of anthocyanidins in the leaves of GhOMT1 RNAi plants.
Figure S8 Summary of differentially expressed transcription factors between C312 and HS2.
Figure S9 Fiber colors of HS2, NCCs and their progenies derived from NCCs crossed by HS2.
Figure S10 Leave color and GhOMT1 expression analysis of C312, HS2, natural colored cottons and their homozygous F4 hybrids.
Figure S11 The phenotypes of HS2, ZX1 and their hybrid offpring (F4) in the field.
Figure S12 The photosynthetic abilities significantly improved in hybrid lines including photosynthesis rate, stomatal conductivity, transpiration rate and water use efficiency.
Table S1 The genetic analysis of cross combinations of HS2 and 6 upland cotton cultivars (P < 0.95).
Table S2 The differentially expressed genes in HS2 and C312 from RNA‐sequence data.
Table S3 The combinations of crosses of HS2 and naturally coloured cotton cultivars.
Table S4 Primers for PCR detection and the flanking sequence used in this paper.
Table S5 Primers for vector constuction and flanking sequencing.
Table S6 Primers for Real time PCR used in this paper.


