Abstract
Cell types are the basic functional units of an organism. They exhibit diverse phenotypic properties at multiple levels, making them challenging to define, categorize and understand. This Review provides an overview of the basic principles of cell types rooted in evolution and development, and discusses approaches to characterize and classify cell types and investigate how they contribute to the organism’s function, using the mammalian brain as a primary example. I propose a roadmap towards a conceptual framework and knowledge base of cell types that will enable a deeper understanding of the dynamic changes of cellular function under healthy and diseased conditions.
In this Review, Zeng discusses how insights learned from the mammalian brain have begun to reveal generalizable organizing principles of cell types and proposes a roadmap based on these principles for taking a multilevel, iterative approach to define cell types and for generating a knowledge base of cell types across lifespan, species and the brain and body.
A cell is the basic unit of all living organisms (except for viruses) (Mazzarello, 1999). The evolution from unicellular to increasingly complex multicellular organisms involves multiplication of individual cells as well as groups of cells and diversification of the function of the cells. As such, billions of years of evolutionary process leads to the vast array of species whose diverse biological attributes are built upon their cellular compositions that exhibit similarities and differences both between species and among different organs within an individual organism (e.g., an animal or a plant). Thus, understanding the organization and function of cells within an organism lays the essential foundation for understanding how an organism works. Similarly, comparing the organization and function of cells between species allows understanding of functional diversity across species.
Studies over the past century have revealed that cells within an organism can be grouped into types – cells within a type exhibit similar structure and function that are distinct from cells in other types (Arendt, 2008). Categorizing cells into types greatly reduces the complexity of investigating the organization and function of cells, especially for large organisms with billions to trillions of cells in the body, e.g., mammals. Researchers have measured a wide range of cellular properties and used these measurements to classify cell types (Petilla Interneuron Nomenclature Group et al., 2008; Regev et al., 2017; Zeng and Sanes, 2017). However, there has not been a consistent and standard definition of cell types even though it is critical for reproducible investigation. It is often unclear if cell types defined by different phenotypic features agree with each other, nor which feature is the “right” one to define cell types. Furthermore, lacking a systematic approach and effort, we do not know if all the cell types in an organism have been identified and where the gaps are.
Recent advent in single cell transcriptomics is revolutionizing the way we understand cell types, with its unprecedented depth and scalability. It has been used to define cell types in a variety of species, tissue organs and brain regions (Armand et al., 2021; Svensson et al., 2020; Tanay and Sebe-Pedros, 2021). However, despite many illuminating studies it remains an open question to what extent transcriptomic clusters represent true cell types and what level of granularity is appropriate for defining cell types. Nonetheless, over the past few years, tremendous progress has been made and many new insights have been generated around these questions. In this review, I will mainly use the mammalian brain as an example (but also refer to other organs or species) to address key questions pertaining to the conceptual and operational definition of cell types.
Approaches to characterize cell types
Cell types in the brain and the body exhibit diverse properties in many modalities – molecular, morphological, physiological, and functional. Numerous studies in these different modalities in the brain over the past century, dating back to Ramón y Cajal and his contemporaries, have converged on a consistent high-level picture of cell type organization across brain regions (Fishell and Heintz, 2013; Markram et al., 2004; Masland, 2012; Mukamel and Ngai, 2019; Nelson et al., 2006; Petilla Interneuron Nomenclature Group et al., 2008; Sanes and Masland, 2015; Seung and Sumbul, 2014; Somogyi and Klausberger, 2005; Yuste et al., 2020; Zeng and Sanes, 2017). At the same time, cellular properties at individual cell level are highly heterogeneous, variations in different modalities do not necessarily exhibit high degrees of concordance, making it often impossible to define exactly what is a cell type and draw clear boundaries between “types”. In many cases, lacking a way to reproducibly label a cell type (typically using a molecular genetic approach) presents a major hurdle to relate different studies and findings to each other.
To untangle this complexity, it is necessary to adopt approaches that provide comprehensive, unbiased, quantitative and standardizable measurements and are scalable to densely sample a sufficient number of cells within a brain region or tissue organ as well as across the entire brain and body to eventually reach completeness, and then perform data-driven computational clustering and analysis to obtain cell type classification. The Petilla convention to define criteria for defining cortical interneuron types represents a major community effort to specify such approaches (Petilla Interneuron Nomenclature Group et al., 2008). Given that physiological properties can take many different forms under different conditions, and functional properties are unknown or poorly defined for many types of cells, as well as the fact that these two modalities are better examined in vivo, it is challenging to scale up the physiological and functional approaches, such as in vivo electrode recording or functional imaging, in a comprehensive and unbiased manner as a primary way to define cell types. On the other hand, molecular and anatomical approaches are more suited for this purpose (Fig. 1A). Molecular approaches include the profiling of chromatin modifications (epigenomics), RNA transcripts (transcriptomics), and proteins (proteomics). Anatomical approaches include the characterization of the spatial distribution, morphology and connectivity of individual cells. Currently single-cell transcriptomics and connectomics (i.e., delineating the patterns of interconnections between individual neurons) are the two primary approaches that have the potential to meet the completeness requirement. Both approaches are now being realized in simpler model organisms including C. elegans (Taylor et al., 2021; White et al., 1986; Witvliet et al., 2021) and Drosophila (Hulse et al., 2021; Li et al., 2022; Scheffer et al., 2020), whereas in mammals transcriptomics is currently feasible and connectomics is still in development (Abbott et al., 2020).
Figure 1. Approaches to characterize cell types.
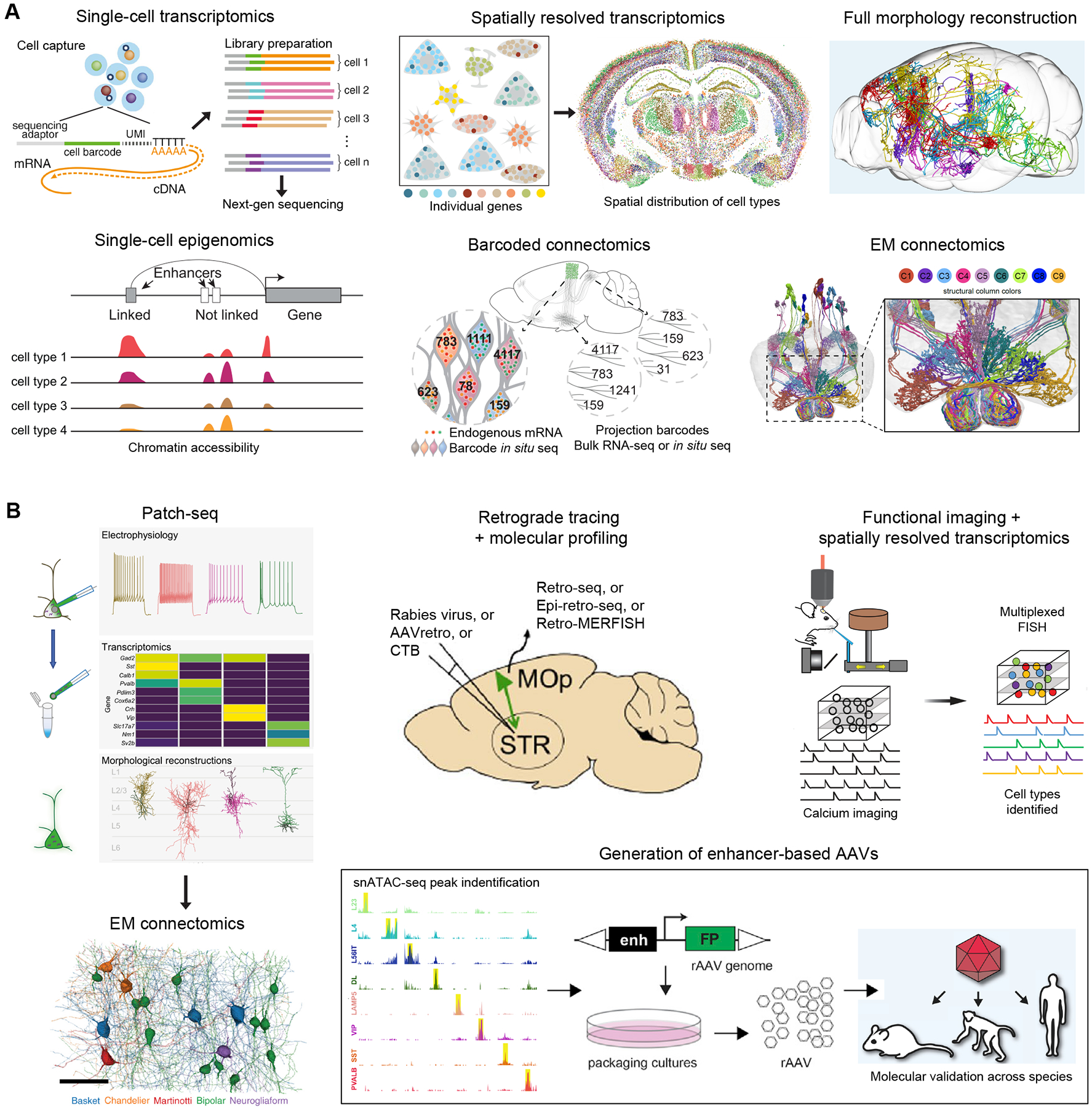
(A) Molecular and anatomical approaches as primary ways of single-cell characterization include single-cell transcriptomics by sc/snRNA-seq, single-cell epigenomics exampled by snATAC-seq, spatially resolved transcriptomics exampled by MERFISH, full morphology reconstruction exampled by MouseLight (image adopted from Winnubst et al., 2019), EM connectomics (image adopted from Hulse et al., 2021), and barcoded connectomics exampled by BARseq (image adopted from Chen et al., 2019). (B) Cross-modality integrated approaches include Patch-seq (image adopted from Lee et al., 2021), retrograde tracing followed by molecular profiling, functional imaging followed by spatially resolved transcriptomics, using Patch-seq data as a Rosetta stone to assign molecular identities to neurons reconstructed from EM dataset (image adopted from Turner et al., 2022), and generation of enhancer based viral vectors (image adopted from Mich et al., 2021).
Transcriptomics by single-cell or single-nucleus RNA-sequencing (scRNA-seq or snRNA-seq) is now the most widely used approach to generate cell type taxonomies or atlases from many species, tissue organs and brain regions, due to its comprehensiveness and high dimensionality (i.e., profiling thousands of expressed genes per cell in a largely unbiased manner) as well as its high scalability (to hundreds of thousands or millions of cells). Transcriptomic cell atlases at the whole organism level have been generated for Drosophila (Li et al., 2022), Ciona (Cao et al., 2019a), and the nervous system of C. elegans (Taylor et al., 2021). The Human Cell Atlas community effort aims to create cell atlases for all organs in the human body (Lindeboom et al., 2021; Regev et al., 2017). The BRAIN Initiative cell census effort has the goal of creating high-resolution whole-brain cell type atlases for mouse, human and non-human primates (Brain Initiative Cell Census Network, 2021; Ecker et al., 2017; Ngai, 2022). A variety of transcriptomic cell atlases have been generated in mouse from many different regions of the nervous system, such as cortex, hippocampus, striatum, thalamus, hypothalamus, cerebellum, spinal cord, retina, etc. (Cembrowski et al., 2018; Hashikawa et al., 2020; Kozareva et al., 2021; Macosko et al., 2015; Marques et al., 2016; Phillips et al., 2019; Pool et al., 2020; Poulin et al., 2014; Ren et al., 2019; Romanov et al., 2017; Russ et al., 2021; Sathyamurthy et al., 2018; Saunders et al., 2018; Shekhar et al., 2016; Stanley et al., 2020; Tasic et al., 2018; Van Hove et al., 2019; Yao et al., 2021a; Yao et al., 2021b; Zeisel et al., 2018; Zeisel et al., 2015), and body organs (Han et al., 2018a; Jaitin et al., 2014; Tabula Muris et al., 2018), and increasingly more in human and non-human primates (Bakken et al., 2021; Darmanis et al., 2015; Drokhlyansky et al., 2020; Garcia et al., 2022; Han et al., 2022; Hodge et al., 2019; Kamath et al., 2022; Lake et al., 2016; Masuda et al., 2019; Tabula Sapiens et al., 2022; Winkler et al., 2022; Yang et al., 2022).
Single-cell epigenomics, such as single-nucleus ATAC-seq (to characterize chromatin accessibility) or DNA methylation-sequencing, has also been used to generate cell type atlases for different brain regions that are consistent with transcriptomic cell atlases and further reveal cell type-specific gene and chromatin regulatory landscapes (Cusanovich et al., 2018; Lake et al., 2018; Li et al., 2021; Liu et al., 2021; Luo et al., 2017; Preissl et al., 2018; Yao et al., 2021a). Spatially resolved transcriptomics, including a variety of techniques based on in situ imaging, in situ capture or in situ sequencing (Close et al., 2021; Larsson et al., 2021; Lein et al., 2017; Moses and Pachter, 2022; Rao et al., 2021; Zhuang, 2021), is a powerful approach combining molecular and spatial characterization at single cell or near-single cell level, revealing spatial relationships between cell types in both local environment and global architecture (Chen et al., 2021; Moffitt et al., 2018; Ortiz et al., 2020; Rao et al., 2021; Wang et al., 2021; Zhang et al., 2021a). Other attributes in the transcriptomes, such as alternatively spliced variants, can provide further information and help to refine cell types (Booeshaghi et al., 2021). An area awaiting critical technology development is single-cell proteomics (Cho et al., 2022; Slavov, 2021), as the expression and subcellular distribution of proteins provides a crucial link between gene expression and cellular structure and function, and it may not have lock-step correlation with the transcriptome of the same cell type.
A cell’s morphology (i.e., shape) and connectivity (especially for neurons) has been regarded as the most defining feature of brain cell types ever since Cajal, though its place may be overtaken by transcriptome. A cell’s morphology can be reconstructed from high-resolution light microscopy (LM) (coupled with colorimetric or fluorescent sparse labeling) (Gao et al., 2022; Jenett et al., 2012; Peng et al., 2021; Winnubst et al., 2019; Wolff and Rubin, 2018) or electron microscopy (EM) datasets (Hulse et al., 2021; Scheffer et al., 2020; Seung and Sumbul, 2014). Connections among individual neurons can be identified using approaches such as EM (Gour et al., 2021; Helmstaedter et al., 2013; Hildebrand et al., 2017; Hulse et al., 2021; Morgan et al., 2016; Scheffer et al., 2020; Schneider-Mizell et al., 2021; Turner et al., 2022; Witvliet et al., 2021), single-neuron trans-synaptic tracing (Schwarz and Remy, 2019) and barcoded connectomics (Chen et al., 2019; Clark et al., 2021; Gergues et al., 2020; Han et al., 2018b; Kebschull et al., 2016; Sun et al., 2021). Again, for definitive cell type classification, one needs to use a fully representative, rather than partial and biased, set of morphological and connectional features. In this regard, with the acquisition of whole brain EM connectomic and LM morphological datasets in Drosophila, refined cell type classification in the brain of this species has been primarily driven by morphology and connectivity (Hulse et al., 2021; Jenett et al., 2012; Scheffer et al., 2020; Wolff and Rubin, 2018).
Most critically, these various approaches need to be integrated to achieve a coherent understanding of cell types and their function and to resolve issues such as which approach(es) (e.g., between transcriptomics and connectomics) can define cell types more clearly. The most common type of integration is to relate transcriptomic profiles with other modalities. Technically there are three ways to achieve such integration (Fig. 1B).
First, conduct multimodal characterization from the same cell using approaches such as single-cell multi-omics (Zhu et al., 2020), Patch-seq which collects electrophysiological, morphological and transcriptomic data from a single patched cell (Berg et al., 2021; Cadwell et al., 2016; Fuzik et al., 2016; Gouwens et al., 2020; Lee et al., 2021; Munoz-Manchado et al., 2018; Scala et al., 2021), retrograde connectivity tracing coupled with single-cell molecular profiling (e.g., Retro-seq, Epi-retro-seq, or Retro-MERFISH) (Kim et al., 2020; Tasic et al., 2018; Zhang et al., 2021a; Zhang et al., 2021b), or in vivo calcium imaging followed by multiplexed FISH (Bugeon et al., 2021; Condylis et al., 2022; Lovett-Barron et al., 2020; von Buchholtz et al., 2021; Xu et al., 2020).
Second, perform label transfer between independently collected datasets through “Rosetta stone” features, e.g., integration between single-cell transcriptomic and epigenomic datasets through marker genes and nearby chromatin modification sites (Armand et al., 2021; Yao et al., 2021a), or assigning molecular identities to neurons in EM and LM datasets using morphologies obtained from Patch-seq data. Integration between transcriptomics and epigenomics is now further empowered by various single-cell multi-omic techniques (Armand et al., 2021; Zhu et al., 2020).
Third, create cell type-targeting genetic tools (e.g., transgenic lines or recombinant viral vectors) using marker genes, promoters and enhancer elements identified from transcriptomic and epigenomic cell atlases (Chan et al., 2017; Daigle et al., 2018; Dimidschstein et al., 2016; Graybuck et al., 2021; Hrvatin et al., 2019; Matho et al., 2021; Mich et al., 2021; Vormstein-Schneider et al., 2020), and use these tools for structural and functional studies. Currently available genetic tools are mostly targeting more coarse-level cell classes or subclasses, or a mixture of cell types. These tools have nonetheless proven to be tremendously powerful as the vast majority of our current knowledge of cell types in the brain and body and their functions has been derived from studies utilizing these tools. The emergence of comprehensive transcriptomic and epigenomic cell atlases now makes it possible to create highly specific tools targeting nearly all identified cell types, and even extended to non-genetic-model organisms and species (Ngai, 2022). This will have a paradigm-shifting effect to the study of function and dysfunction of broad biological systems.
Overall, application of these approaches to characterize cell types in different brain regions and tissue organs as well as across species has begun to reveal generalizable organizing principles of cell types. Below I will discuss the large body of studies supporting these principles, and then conclude with a proposed roadmap based on these principles for taking a multilevel, iterative approach to define cell types and build an overarching knowledge base of cell types across the brain and body, across lifespan and across species.
Cell types are the product of evolution
The concept of cell types needs to be established based on where cell types originated and how they have diversified. Cell type classification has been compared to species classification (Stadler et al., 2021; Tanay and Sebe-Pedros, 2021; Zeng and Sanes, 2017). Indeed, species specialization is an overall culmination of the function of all the cell types within that species, thus they may follow similar evolutionary principles. There have been several ways proposed to classify species. One is based on the notion of reproductive isolation. However, this approach is not universally implementable, and many exceptions have also been found. A more fruitful approach is phylogenetic analysis, that is, comparing the relatedness between species using a wide range of structural and functional phenotypic features. Such analysis led to the foundational “tree of life” as we understand it today. Nonetheless, many issues remain unresolvable in the phylogenetic classification of species, often due to the highly specialized phenotypic features acquired by some species as they adapt to their ecological niches, as well as convergent phenotypic evolution in other cases, both of which could skew comparative analysis. Over the past decade, evolutionary approach based on comparative genomics (i.e., phylogenomics) has brought an entirely new paradigm to species classification, providing a systematic, rational, unbiased, universally applicable and extensible classification scheme (Murphy et al., 2021; Preuss and Wise, 2022; Stephan et al., 2022).
Similarly, cell types are inherited through the genome. Relatedness between cell types reflects their evolutionary distance as they were created through cell type duplication and segregation events. It has been proposed that the formation of a new cell type identity requires the evolution of a unique cell type regulatory signature that includes a cell type-specific core regulatory complex (CoRC) of transcription factors, which defines the identity and coordinated gene expression pattern of the new cell type (Arendt et al., 2016). This set of master regulatory transcription factors, sometimes called terminal selectors, have been identified in a number of neuron types in worms, flies and mice (Hobert and Kratsios, 2019; Reilly et al., 2020). The master transcription factors should be identifiable when the transcriptomes of evolutionarily related cell types are compared. A large body of studies (see above) have now shown that clustering of single-cell transcriptomes can systematically categorize cells into putative types, many of which are consistent with existing knowledge and thus can be considered as bona fide cell types. Evolutionarily conserved cell types can be systematically identified by cross-species comparison of single-cell transcriptomic types in the brain (Bakken et al., 2021; Colquitt et al., 2021; Hodge et al., 2019; Kebschull et al., 2020; Krienen et al., 2020; Tosches et al., 2018; Yamagata et al., 2021). Thus, this approach appears to “make sense”; it is not coincidental, but strongly supports the notion that transcriptomes harbor the molecular genetic code for cell type identity.
However, there are several challenges that must be surmounted to arrive at a complete and accurate evolutionary definition of cell types through cross-species comparisons. Accurate cross-species comparison of cell types at transcriptomic level requires well-annotated genomes, comparative gene ontologies and consistently high-quality transcriptomic data generation from many species (Tanay and Sebe-Pedros, 2021). Furthermore, species mostly diverged millions of years ago, as did cellular identities. Cell type homologies between related species are often discernible only at a relatively coarse level which do not fully capture the biological complexity. Many gaps also exist due to the extinction of intermediate species. These challenges could limit a deeper understanding of cell types (see below) and how they contribute to the body or brain function. On the other hand, one does not need to characterize cells from a large number of evolutionarily related species in order to define cell types. It is possible to gain a deep understanding of cell types from even a single species, since each species has evolved from its simpler ancestors through many rounds of cellular and regional duplications in which the newly created cell types and regions adopt new functions, and thus comparing between cell types and between regions within the species (in the same way as comparing between species) can reveal the evolutionary relationships between cell types as well. Then, we can expand the investigation into as many other species as possible, which will further clarify the description of cell types, their origins and how their functions manifest.
Therefore, the first and foremost principle is that cell types are the product of evolution and cell type identities are encoded in the genome. Like phylogenomics for species classification, transcriptomic (and epigenomic) classification is a good proxy of cell type classification as the gene regulatory mechanisms that encode and maintain cell type identities are embedded in the transcriptomes and epigenomes. This core concept has led to a systematic delineation of the relationship between cell types both within a species and, increasingly, across species. At the same time, cell type conservation may be imposed more by function than natural selection directly as in organismal evolution. As such, evolution of individual cell types may be more complicated than organismal evolution as a whole, and it will be interesting to see if different cell types evolve in similar or different ways as the whole organism. Finally, a transcriptome also contains gene expression profiles that underlie arguably all phenotypic features of the cell at the time or state when the cell is characterized. What else are transcriptomes and transcriptomic clusters telling us?
Hierarchical organization of transcriptomically-defined cell types
Transcriptomically derived cell type taxonomies in the adult mammalian brain, with majority of the studies conducted in mouse, have consistently revealed a hierarchical organization of cell types (Fig. 2) (Brain Initiative Cell Census Network, 2021; Macosko et al., 2015; Romanov et al., 2017; Russ et al., 2021; Saunders et al., 2018; Shekhar et al., 2016; Tasic et al., 2016; Tasic et al., 2018; Yao et al., 2021b; Zeisel et al., 2018; Zeisel et al., 2015; Zeng and Sanes, 2017). The first (highest) level of branches is the separation of neuronal and various non-neuronal cell classes (Fig. 2A). For neurons, the second level of branches is driven by major brain structures/regions, and the third level comprises various cell subclasses and types within each major brain structure, although there may be cell types crossing or shared between brain structures due to cell migration during development.
Figure 2. Hierarchical organization of cell types.
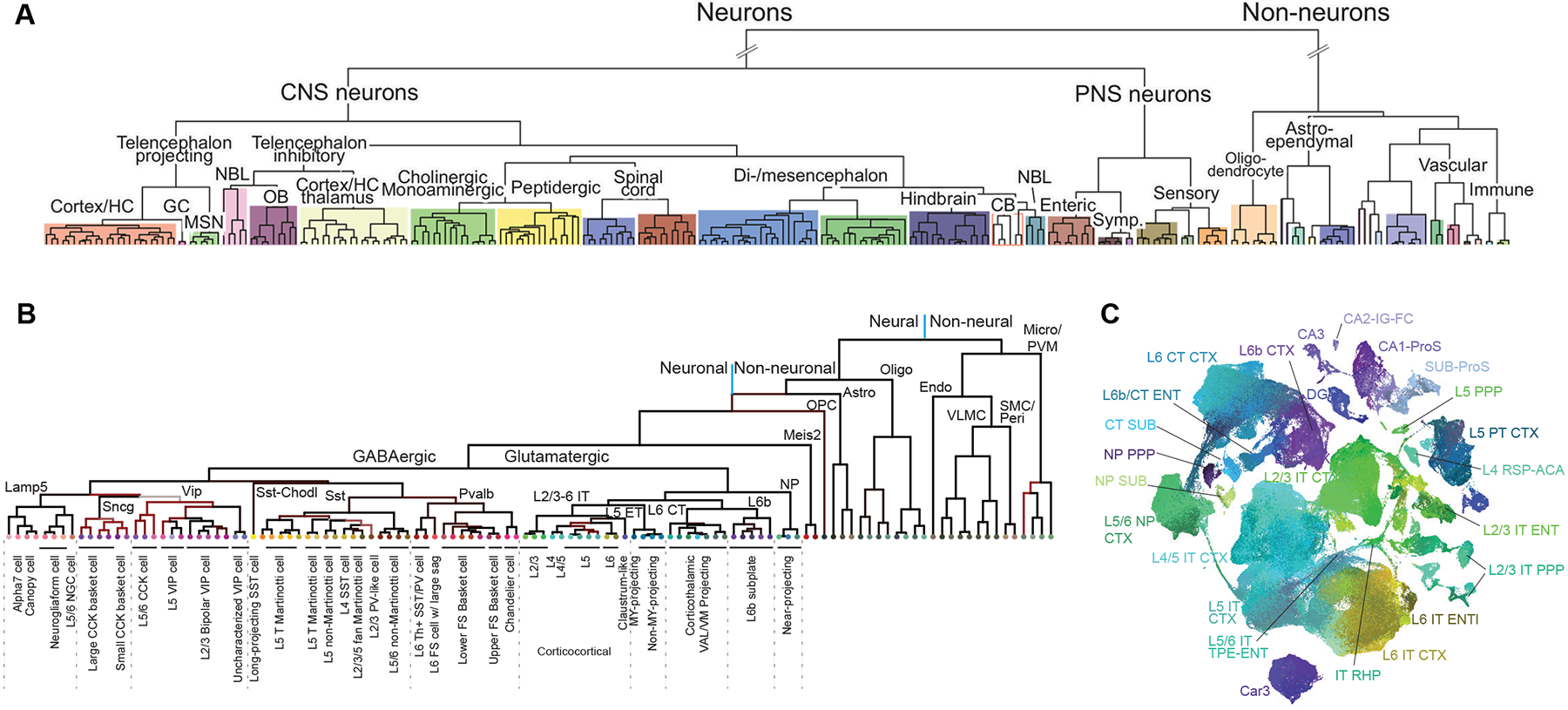
(A) A transcriptomic cell atlas for the mouse nervous system (image adopted from Zeisel et al., 2018). (B) A transcriptomic cell type taxonomy for the mouse primary motor cortex, with annotation (image adopted from Brain Initiative Cell Census Network, 2021). (C) UMAP representation of a transcriptomic cell type taxonomy for the glutamatergic neuron types in mouse isocortex and hippocampal formation, revealing discrete and continuous variations (image adopted from Yao et al., 2021b).
The basic architecture of the mammalian brain (Swanson, 2000, 2012) is composed of telencephalon, diencephalon, mesencephalon (midbrain) and rhombencephalon (hindbrain). Telencephalon (consisting of five major brain structures – isocortex, hippocampal formation, olfactory area, cortical subplate and cerebral nuclei) and diencephalon (including thalamus and hypothalamus) are collectively called forebrain. Midbrain is divided into tectum and tegmentum. And hindbrain is divided into pons, medulla and cerebellum. Within each of these major brain structures there are multiple regions and subregions, each with many cell types. A cell type can be specific to a subregion, a region or a major brain structure.
Here I use isocortex (or simply called cortex) as an example to illustrate the organization of cell types within a major brain structure. Isocortex is composed of multiple cortical areas, each mediating sensory, motor or associational function. Transcriptomic cell type taxonomies from visual cortex and motor cortex display a similar organization (Fig. 2B) (Brain Initiative Cell Census Network, 2021; Tasic et al., 2018; Yao et al., 2021a). In each of these areas, there are two neuronal classes based on the dominant neurotransmitters they release, glutamatergic and GABAergic, as well as several non-neuronal classes. The glutamatergic excitatory neurons mostly have long-range axon projections to other cortical and/or subcortical regions. They are divided into nine subclasses based on their layer specificity and long-range projection patterns: L2/3 IT, L4/5 IT, L5 IT, L6 IT, Car3 IT, L5 ET, L5/6 NP, L6 CT, and L6b (IT: intratelencephalic projecting; ET: extratelencephalic projecting; NP: near-projecting; CT: corticothalamic projecting). The GABAergic inhibitory neurons mostly have their axon projections confined within the local area. They are divided into six subclasses named after canonical marker genes: Lamp5, Sncg, Vip, Sst, Sst-Chodl and Pvalb. Within each of the glutamatergic or GABAergic subclasses, as well as each non-neuronal class, there are several transcriptomic clusters or types, resulting in a total of ~110 transcriptomic cell types in each cortical area (Brain Initiative Cell Census Network, 2021; Tasic et al., 2018). This organization is highly consistent with the existing knowledge about cortical cell types which have been extensively studied in a variety of phenotypic modalities over the past 50 years (Harris and Shepherd, 2015; Tremblay et al., 2016; Yuste et al., 2020; Zeng and Sanes, 2017), suggesting that single-cell transcriptomics alone can faithfully capture the overall cell type organization at class and subclass levels, although the validity of transcriptomic clusters at the lowest branch level has yet to be fully tested.
Comparison of transcriptomic cell types across different cortical areas reveals new insights. Glutamatergic neuron types are distinct between visual and motor cortical areas whereas GABAergic neuron types are shared between the two areas (Tasic et al., 2018). This dichotomy may be rooted in the developmental origins of these two cell classes. During development, glutamatergic neurons are generated within the cortex in which different areas are laid out by gradient expression of morphogens (Cadwell et al., 2019; O’Leary et al., 2007), whereas GABAergic neurons are generated in the subcortical ganglionic eminence and migrate into cortex (Hu et al., 2017; Lim et al., 2018). A larger transcriptomic study covering all areas from both isocortex and hippocampal formation (HPF) further identifies hundreds of transcriptomic types and a high degree of diversity in the glutamatergic neuron class across both brain structures (Fig. 2C) (Yao et al., 2021b). Within isocortex, cell types that are specific to a cortical area or shared among areas are both identified, and the shared cell types often exhibit gradient distribution or gradient gene expression across areas. This highly complex transcriptomic cell type landscape along the cortical sheet likely results from the series of cortical developmental events from “Protomap” to “Protocortex” (Cadwell et al., 2019; O’Leary et al., 2007). Compared between the two brain structures, the glutamatergic cell types in isocortex and HPF are highly distinct from each other, yet they also display one-to-one homology at subclass level suggesting a similar evolutionary origin (Yao et al., 2021b). This homologous relationship suggests that parallel neural networks can be formed by homologous sets of cell types.
Overall, based on these findings, we may hypothesize that the adult-stage transcriptomic landscape can reveal the organization of cell types within and between brain regions that reflect their evolutionary and developmental histories. The hierarchical organization of transcriptomic cell types likely represents evolutionary origins of and distinctions between cell types; major branches represent earlier division of cell classes, and minor branches represent more recent segregation events. This hierarchical organization is laid out via an elaborate developmental program involving a series of highly coordinated processes and events. This hypothesis can be tested by studying the evolution and development of cell types (see below).
Another prominent feature of the relationship between cell types revealed by transcriptomic studies is the coexistence of discrete and continuous variations between types. Continuous variations have been observed in a variety of forms in cortical excitatory and inhibitory neurons, medium spiny neurons in the striatum and excitatory projection neurons across different nuclei of the thalamus (Phillips et al., 2019; Stanley et al., 2020; Tasic et al., 2018; Yao et al., 2021b). Discrete variations exist among cell subclasses and major types that are usually at the higher branches of the hierarchy. Continuous variations are usually found among closely related transcriptomic clusters or subtypes at lower branches, such as the many IT neuron types across the cortical depth from L2/3 to L6 (Fig. 2C). Cells at opposite ends of the continuum have clearly distinct transcriptomic profiles, but the transition from one end to the other is gradual among the cells composing the continuum. This makes it difficult to subdivide the cells into types using statistical criteria and name an exact number of cell types. But the continuous variation itself is nonetheless biologically meaningful and needs to be properly represented in cell type descriptions. One way to better understand the significance of the continuous variation is to examine how it correlates with other modalities of cell type properties (see below).
Regarding non-neuronal cells in the mammalian brain, there are multiple classes which can be divided into neural and non-neural groups (Fig. 2A–B) (Brain Initiative Cell Census Network, 2021; Zeisel et al., 2018). The non-neural group contains cell classes of the immune origin, i.e., microglia and border-associated macrophages (BAMs) (Butovsky and Weiner, 2018; Masuda et al., 2019; Munro et al., 2022; Prinz et al., 2019; Thion and Garel, 2020; Van Hove et al., 2019), and of the vascular origin, i.e., endothelial cells, smooth muscle cells (SMCs), pericytes, and vascular leptomeningeal cells (VLMCs) (Schaeffer and Iadecola, 2021; Sweeney et al., 2019; Vanlandewijck et al., 2018). The neural group contains cell classes of the neuroectoderm origin (same as the origin of neurons), including oligodendrocytes, oligodendrocyte progenitor cells (OPCs), astrocytes and ependymal cells (Ben Haim and Rowitch, 2017; Dimou and Simons, 2017; Escartin et al., 2021; Khakh and Deneen, 2019; Kuhn et al., 2019; Ortiz-Alvarez et al., 2019; Redmond et al., 2019). In brain transcriptomic cell type taxonomies, non-neuronal cells generally display less diversity than neurons, with little regional specificity except for astrocytes. There are two major subclasses of astrocytes, one specific to the telencephalon and the other to non-telencephalon regions, in addition to several other highly specialized astrocyte-like cell types such as Müller glia of the retina and Bergmann glia of the cerebellum (Zeisel et al., 2018). Immature and mature oligodendrocytes form a long continuous trajectory originating from OPCs, indicating coexistence of multiple states of gradually maturing oligodendrocytes (Marques et al., 2016; Zeisel et al., 2018).
Like comparative genomics for species classification, single-cell transcriptomics is highly effective for cross-species comparison of cell types to reveal their evolutionary relationships. Comparative studies of cortical cell types among mouse, human and non-human primates (Bakken et al., 2021; Brain Initiative Cell Census Network, 2021; Hodge et al., 2019; Krienen et al., 2020) show that the hierarchical organization described above along with all the neuronal and non-neuronal cell classes and subclasses (major branches of the hierarchy) is well conserved across these mammalian species. Main differences across species lie in the heterogeneity of individual gene expression within each subclass and type, as well as the ambiguity of cross-species correspondence of the leaf-node transcriptomic types which likely will require multimodal characterization to clarify. Similarly, comparative transcriptomic studies reveal homologous cortical glutamatergic and GABAergic cell classes between mammal and reptile (Tosches et al., 2018), as well as homologies and variations of neuron subclasses and types in the cerebellar nuclei or the retina of mouse, chicken and primates (Kebschull et al., 2020; Yamagata et al., 2021). These studies further suggest that the hierarchical organization of brain cell types is a framework extensible to describing cell type evolution.
Correspondence between transcriptomic cell types and other cellular properties
Cell types are also considered to be the basic functional units of an organism. For the categorization of cell types based on their transcriptomes to be meaningful and to understand their relevance to the structure and function of the tissue organ where the cells reside, it is necessary to characterize other modalities of cellular properties. Multimodal correspondence of cell types in the mouse retina is a classic example where independent anatomical, functional and transcriptomic studies uncover similar numbers of neuron types (~130) and find that molecular profiles and anatomical distribution patterns (laminar specificity and mosaicism) are well correlated with visual response properties (Baden et al., 2020; Masland, 2012; Sanes and Masland, 2015; Seung and Sumbul, 2014; Shekhar and Sanes, 2021). Recent work from the BRAIN Initiative Cell Census Network (BICCN) in creating a multimodal cell census and atlas of the mammalian primary motor cortex represents the most comprehensive multimodal study to date, integrating transcriptomics with epigenomics, spatially resolved transcriptomics, morpho-electrical properties, and connectivity (Brain Initiative Cell Census Network, 2021).
Integration of transcriptomic and epigenomic datasets using computational approaches allows consolidation of robust molecular cell types and identification of hundreds of thousands of cis-regulatory elements (CREs) associated with specific cell types (Yao et al., 2021a). Some of these CREs are associated with specific marker genes whereas others may represent past events.
Integration of transcriptomics and MERFISH, a spatially resolved transcriptomic method, reveals the spatial organization of mouse motor cortex cell types (Zhang et al., 2021a). A major finding of the study is that, in addition to the laminar distribution of glutamatergic neuron subclasses as expected, even the GABAergic types within each subclass exhibit layer-selective localization, and the continuous variations among individual glutamatergic types or GABAergic types correlate well with their continuous distribution along cortical layers/depth (with a prominent example being that all the excitatory L2/3-L6 IT types line up along the cortical depth from L2/3 to L6) (Fig. 3A). Thus, a strong correspondence is demonstrated here between the continuous variations among cortical neuron types in the transcriptomic space and their continuous and directed spatial distribution patterns. Other MERFISH studies of neuron types in hypothalamus or nucleus accumbens also reveal strong correlation between transcriptomic specificity and anatomical/subregional specificity (Chen et al., 2021; Moffitt et al., 2018).
Figure 3. Multimodal correspondence of cell type phenotypic properties.
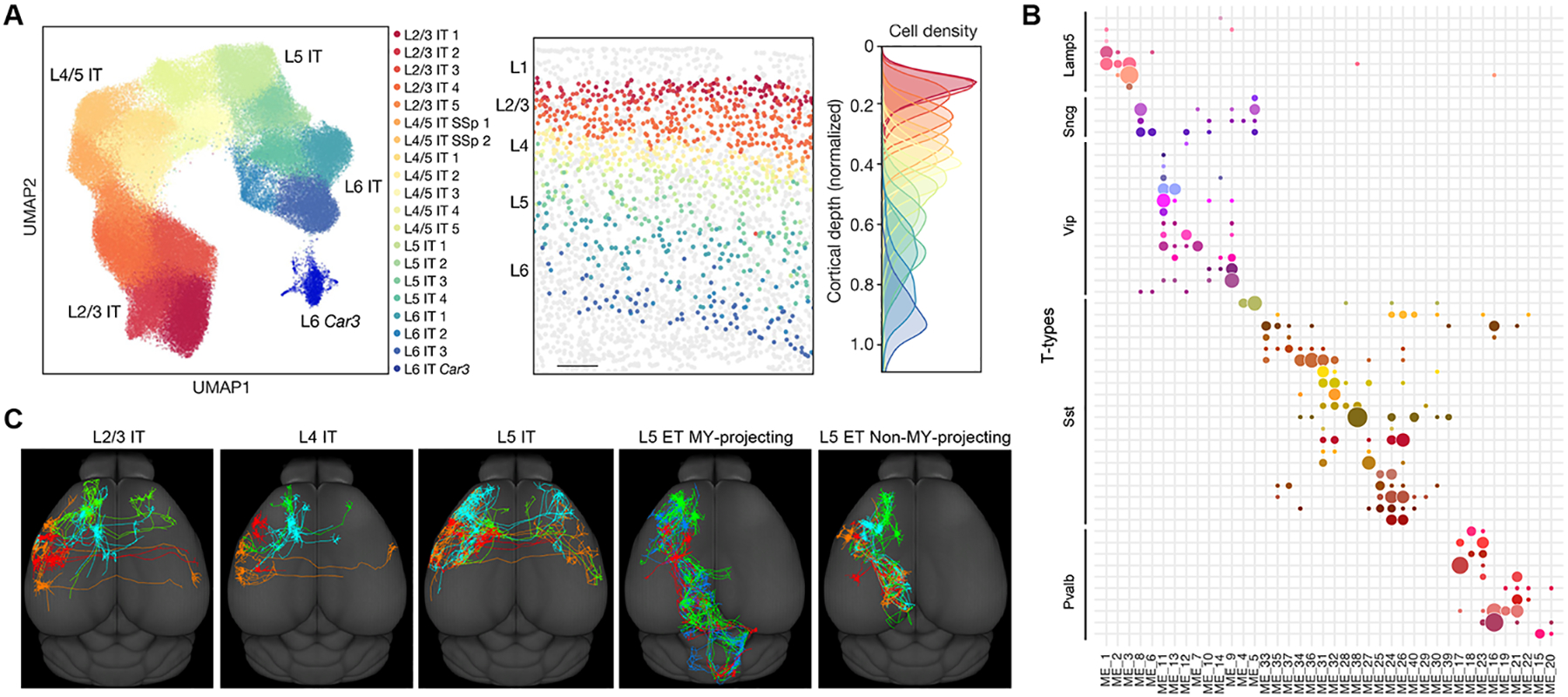
(A) MERFISH data from mouse motor cortex shows that continuous variation of glutamatergic IT transcriptomic types is correlated with their continuous spatial distribution along the cortical depth from L2/3 to L6 (image adopted from Zhang et al., 2021a). (B) Patch-seq data on GABAergic interneurons from mouse visual cortex shows correspondence between transcriptomic (T) types and morpho-electrical (ME) types (image adopted from Gouwens et al., 2020). (C) Brain-wide complete morphology reconstruction of cortical glutamatergic neurons shows distinct axon projection patterns between major transcriptomic types and further heterogeneity within each type (image adopted from Peng et al., 2021). Each color outlines the projection of one neuron within the type in each panel.
Integration of transcriptomic, electrophysiological and morphological properties by Patch-seq reveals multimodal corresponding distinctions of mouse motor cortex cell types at subclass level (Scala et al., 2021). Within each subclass the morpho-electrical properties vary continuously along with the transcriptomic types. There is also additional heterogeneity of morpho-electrical properties within some transcriptomic types, indicating a more complex picture. Another Patch-seq study of mouse visual cortical GABAergic neurons also reveals a relatively high degree of corresponding continuous variations of transcriptomic types with their anatomical distribution along the cortical depth and the variations of their morpho-electrical properties (Fig. 3B) (Gouwens et al., 2020). To overcome heterogeneity at individual type level, a set of triple-modality MET types are defined at an intermediate level of granularity (between transcriptomic subclasses and types). These visual cortex GABAergic MET types show robust cross-modality concordance and mutual predictability.
The vast majority of cortical and subcortical neuron types have long-range axon projections to form circuit networks throughout the brain. To examine the long-range axon projection specificity of transcriptomic cell types, Retro-seq and related methods (e.g., Epi-retro-seq, Retro-MERFISH) have been used (Kim et al., 2020; Tasic et al., 2018; Zhang et al., 2021a; Zhang et al., 2021b). Since a neuron type usually has multiple projection targets and a neuron within that type can choose a subset of those targets either specifically or randomly, a single-target-site Retro-seq assay is often insufficient to resolve the target specificity of a transcriptomic type except in special cases. Brain-wide complete reconstruction of single neuron morphology is currently the only approach to capture the full extent of a neuron’s axon projection pattern and define projection neuron types (Gao et al., 2022; Peng et al., 2021; Winnubst et al., 2019). A study using this approach in cortical excitatory neuron subclass-specific Cre driver lines (Peng et al., 2021) reveals distinct projection patterns between subclasses, e.g., not only between IT and ET but also between L2/3 IT and L5 IT neurons, confirming subclass level projection specificity (Fig. 3C). Within each subclass, the study also finds extensive heterogeneity among individual neurons; this heterogeneity reflects three axes of variations: regional specificity, topographic specificity and individual (potentially stochastic) variation, which do not readily correlate with transcriptomic types within the subclass. Thus, it remains an open question how axon projection patterns correlate with transcriptomic types, which needs to be addressed in future studies using approaches such as coupling complete morphology reconstruction with multiplexed FISH, or performing MAPseq/BARseq with sequencing of both starter cells and axon targets. It may also be necessary to extend such studies into developmental periods, to identify potentially clearer molecular correlates when the projection specificity is established (Klingler et al., 2021).
Systematic investigation of connectivity among transcriptomic types at synaptic level and relating them to conventional studies where morphology and individual molecular markers were used to identify cell types is much needed to better understand the connectional specificity between transcriptomic types. It has been suggested that neuron types may be defined by their unique communication properties implemented as synaptic input-output patterns (Huang and Paul, 2019; Paul et al., 2017). The emerging large-scale EM datasets (e.g., from the MICrONS project, https://www.iarpa.gov/research-programs/microns) hold great promise to tackle synaptic-level connectivity between cell types and individual cells in the mammalian brain (Abbott et al., 2020). Perhaps a greater opportunity lies in the Drosophila field where comprehensive catalogs of both transcriptomic cell types and connectional cell types have been obtained independently (Hulse et al., 2021; Li et al., 2022; Scheffer et al., 2020) and a systematic comparison and cross-correlation between them may be realized soon.
To compare functional properties among transcriptomic cell types, two general approaches have been taken – coupling in vivo calcium imaging with post hoc multiplexed FISH to decode the molecular identities of the imaged cells (Bugeon et al., 2021; Condylis et al., 2022; Lovett-Barron et al., 2020; von Buchholtz et al., 2021; Xu et al., 2020), or mapping immediate early gene (IEG) activation during sensory response or behavior using scRNA-seq or MERFISH (Hrvatin et al., 2018; Kim et al., 2019; Moffitt et al., 2018; Sathyamurthy et al., 2018; Wu et al., 2017). Using the former approach, it has been shown that GABAergic transcriptomic types in mouse visual cortex differ in their response to behavioral states (e.g., running versus resting), whereas visual response properties (e.g., orientation or direction selectivity) only differ at subclass level (Bugeon et al., 2021); in somatosensory cortex, higher sensory response is seen in a specific L2/3 IT excitatory transcriptomic type (Condylis et al., 2022). In hypothalamus, several studies using either of the two approaches in mice demonstrate that activated neurons during a specific behavioral state are often distributed across a range of transcriptomic cell types (Kim et al., 2019; Moffitt et al., 2018; Xu et al., 2020). Understanding the functional roles of different transcriptomic cell types is a huge undertaking. Obviously, studies mentioned here are just the beginning; many more will come in the future and will allow us to gain a much deeper understanding.
In summary, for a definition of cell types to be meaningful, it must be associated with what cell types do. A transcriptomic cell type taxonomy must be linked to anatomical and functional information to evaluate the validity of the transcriptomic taxonomy and determine the appropriate level of granularity (since in theory transcriptomic clusters can be infinitely subdivided and the more cells profiled the more clusters can be obtained). So far it has been shown that transcriptomic types have excellent correspondence with their spatial distribution patterns. Since the spatial distribution pattern is defined during development, this suggests that transcriptomes may retain the developmental plan. At the same time, whether specific transcriptomic types (beyond the subclass level) have specific connectional or functional attributes or not is still unclear in many cases. Since transcriptomes are rich in containing the molecular correlates of all sorts of cellular properties, specific molecular signatures responsible for certain essential anatomical or functional features may be hidden below noise and will need to be brought out through supervised approaches and used to refine the classification of cell types towards more functional relevance. It is also necessary to trace back into development to identify potentially clearer molecular correlates as different connectional or functional properties may be established at different developmental time points. On the other hand, it is also reasonable to propose that some connectional or functional properties should not be used to define cell types, because they may be emerging properties arising from the interaction of a network of cell types, or from experience and/or activity dependent processes that represent a cell “state” rather than a defining feature for a cell “type”.
Cell types versus cell states
A key question arising when evaluating a transcriptomic taxonomy is whether some clusters actually represent a particular cell state – i.e., a transient or dynamically responsive property of a cell to a context – rather than a cell type, as a cell type can exist in different states. This is a difficult question since most of the phenotypic measurements including the single-cell transcriptome are only a one-time snapshot of the cell. However, one can compare transcriptomes collected from different time points or different behavioral, physiological or pathological states and see which clusters appear, disappear or shift under different conditions. Cell type-specific gene expression changes associated with different cell states may be seen during circadian cycles, variable metabolic states, development, aging, or under behavioral, pharmacological or diseased conditions (Fig. 4) (Mayr et al., 2019; Morris, 2019). Furthermore, individual variations within a species (e.g., within the human population) that are driven by genetic or environmental factors may be manifested as a variety of cell type or cell state variations. Studying the various states of cell types will enhance our ability to distinguish core gene sets (e.g., master transcription factors) maintaining cell type identities versus genes associated with specific functional states, and further our understanding of the diverse function of cell types as well as the biological basis of individual variability.
Figure 4. Dynamic changes of cell types and states during development, aging and various physiological or pathological contexts.
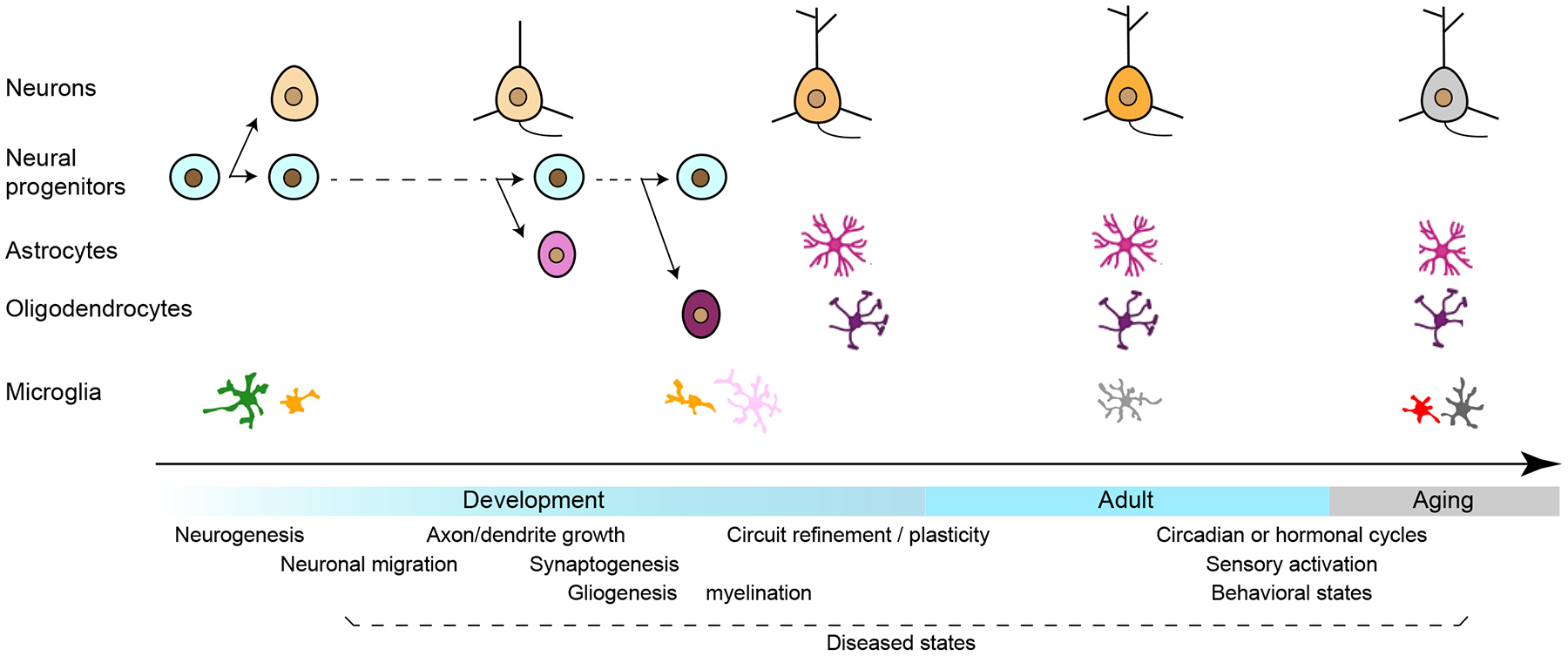
Major neuronal and non-neuronal classes are shown along the life stages of development, adulthood and aging. Neural progenitors generate different neuronal types, astrocytes and oligodendrocytes at different developmental timepoints, whereas microglia have a separate developmental origin. Major developmental events, various physiological states in adulthood, and different diseased states throughout the lifespan are shown below the timeline.
The distinction between cell types and cell states is particularly challenging during development, as cells continually change their states and, at certain key time points, they may switch their cell type identities. Can single cell phenotypic properties such as transcriptomes distinguish types versus states? Although not absolute, it is reasonable to assume that transcriptomic changes tend to be more continuous during cell state transitions, and more abrupt or discrete when cells switch their types. More often, emergence of a new cell type during development is the consequence of cell division from which a daughter cell takes up a new cell type identity (Fig. 4). Trajectory analysis or lineage tracing coupled with single-cell transcriptomics across developmental time points has now often been used to identify the time course of emergence and maturation of each cell type, as well as the ancestor-descendant relationship across cell types that are present at different developmental stages (e.g., progenitors versus differentiated cells) (McKenna and Gagnon, 2019; Saelens et al., 2019; Tritschler et al., 2019; Wagner and Klein, 2020).
Coordinated neuronal activities in brain circuits generate sensory perception and behavior. Specific neuronal populations activated during a particular perceptual or behavioral episode can be identified by screening for the activation of IEGs in them, via immunostaining, transgenic reporter lines, or single-cell or spatial transcriptomics in more recent years (DeNardo and Luo, 2017; Hrvatin et al., 2018; Moffitt et al., 2018; Wu et al., 2017). IEG activation leads to expression of downstream effectors, such as ion channels or synaptic proteins, that shape the cell states and remodel neuronal connections. Cell state changes in the brain are closely related to neural plasticity. Similarly, various diseased conditions can induce pathological changes in cell states in different brain regions or tissue organs. Numerous studies have revealed selective vulnerability of specific cell types for specific diseases. Pharmacological or genetic (e.g., CRISPR-based) perturbations in normal or diseased conditions, in combination with single-cell profiling (e.g., Perturb-seq), are powerful approaches to gain a mechanistic understanding of how disruptive or restorative cell state changes can affect cell type function or dysfunction (Adamson et al., 2016; Dixit et al., 2016; Jaitin et al., 2016; Replogle et al., 2022).
Here I highlight a prominent feature of the non-neuronal cell types in the brain, which is that despite having lower diversity than neurons in baseline adult state, many non-neuronal cell types undergo pronounced changes, i.e., they exhibit many different cell states, under different physiological or diseased conditions. Astrocytes exhibit diverse morphological and physiological properties in different brain regions and contribute to essential functions in blood-brain barrier, synaptogenesis, neurotransmitter buffering, ion homeostasis, and secretion of neuroactive agents (Ben Haim and Rowitch, 2017; Khakh and Deneen, 2019). Astrocytes become reactive under pathological conditions. Reactive astrocytes undergo morphological, molecular, and functional changes in response to injury or CNS diseases; they may adopt multiple, heterogeneous states depending on context (Escartin et al., 2021). Oligodendrocytes are the myelinating cells of the central nervous system (CNS) that are generated from OPCs throughout life. Myelination process also exhibits activity-dependent plasticity (Monje, 2018). Oligodendrocyte pathology is evident in a range of disorders including multiple sclerosis, schizophrenia and Alzheimer’s disease (Kuhn et al., 2019). Regarding cerebrovascular cell types, recent single-cell transcriptomic studies in the human brain reveal gene expression changes in them that can impact blood-brain barrier integrity in Huntington’s and Alzheimer’s diseases (Garcia et al., 2022; Yang et al., 2022). Finally, microglia are the primary innate immune cells in the CNS and have a distinct developmental origin from peripheral immune cells. They are generated from mesodermal progenitors that arise from the yolk sac and are among the earliest residential cell types in the brain. Microglia display diverse and dynamic phenotypic states and play a plethora of roles in development, adulthood (homeostasis), aging and diseases (Butovsky and Weiner, 2018; Prinz et al., 2019; Thion and Garel, 2020). Single-cell transcriptomic studies reveal a relatively homogeneous adult microglia population, and greater heterogeneity of microglia states at different developmental stages, during aging and in pathological conditions (Hammond et al., 2019; Li et al., 2019; Masuda et al., 2019). In particular, microglia can be both responders to and inducers of various neurodegenerative, neuroinflammatory and neurodevelopmental diseases. Taken together, these studies paint a collective picture on how the variety of non-neuronal cell types actively respond to and contribute to different physiological and pathological changes in the brain.
Cell type development
A deep understanding of a subject often comes from understanding how it is built. The entire repertoire of cell types in the brain and the body is built through a sequential and parallel series of spatially and temporally coordinated developmental events starting from a single fertilized egg, the zygote. This developmental program carries out a remarkable implementation plan that unravels the identities of all cell types which are encoded in the genome through evolution. Transcriptional and epigenetic regulatory programs are unfolded from the genome sequences and drive a cascading series of cell proliferation and differentiation processes leading to the manifestation of diverse cellular phenotypes. In the developmental ontogeny of cell types, earlier-stage ancestral cell types are fewer and are more multipotent, they give rise to a larger number of descendant cell types with increasingly restricted fates. The developmental program rolls out not only a temporal but also an elaborate spatial plan, specifying the location of each tissue organ and the spatial organization of all the cell types within each. This is a highly dynamic spatiotemporal process involving specific cell-cell interactions, cell migration streams, and formation of niches and microenvironments that allow functional specialization.
For the brain, the main series of events of brain development leading to the mature adult-stage cell types and circuits include: patterning and regionalization (laying out the master plan of brain architecture), neurogenesis and neuronal migration, gliogenesis and glia cell differentiation, neuronal differentiation and circuit formation (axonogenesis, dendritic arborization, synaptogenesis, myelination), and circuit refinement and plasticity (Fig. 4). Systematic single-cell transcriptomic, epigenomic and spatially resolved transcriptomic profiling with high temporal resolution, coupled with lineage tracing and other phenotypic characterization, holds tremendous potential to capture key sets of genes and genomic regulatory networks involved in these series of events and begin to resolve the extremely complex spatial and temporal transition of cell types and states leading to the adult-stage repertoire of cell types (Allaway et al., 2021; Bandler et al., 2022; Bhaduri et al., 2021; Cao et al., 2019b; Chen et al., 2022; Delgado et al., 2022; Di Bella et al., 2021; Klingler et al., 2021; La Manno et al., 2021; Romanov et al., 2020; Schmitz et al., 2022; Sharma et al., 2020; Shekhar et al., 2022; Tiklova et al., 2019; Zhu et al., 2018). Studying brain cell type development using these approaches will allow us to establish the developmental trajectory for each cell type from progenitors to transitional cell types and states to adult mature cells, discover the set of master transcription factors that define and maintain the identity of each cell type, and identify key events and molecular correlates that lead to the acquisition of a cell type’s specific connectional or functional properties.
The generation and patterning of mouse cortex and spinal cord cell types are two example systems where extensive historical studies have uncovered several common principles (Cadwell et al., 2019; Catela et al., 2015; Jessell, 2000; O’Leary et al., 2007; Osseward and Pfaff, 2019; Sagner and Briscoe, 2019). First, opposing morphogen gradients establish the basic plan for cortex (anterior-posterior) or spinal cord (dorsal-ventral) patterning and provide instructive signals for the expression of complementary sets of transcription factor activators and repressors, which in turn define distinct neural progenitor domains within cortex or spinal cord. Second, driven by the transcription factor network, each type of neural progenitors generates a series of neuronal cell types. The set of cell type-defining transcription factors in a progenitor or a daughter cell can change with time, such that different neuronal types emanate from the same progenitor in a precisely timed “birth order”. Later in development, the same neural progenitors also generate non-neuronal cell types such as astrocytes and oligodendrocytes. Third, specific sets of cell adhesion molecules provide guidance cues for axon path finding and synapse formation, leading to the assembly of region- and cell type-specific local and global circuit networks. Fourth, patterned neural activities spontaneously emerged from the circuits and/or influenced by external inputs further sculpt synaptic connections and circuit organization to refine functional specificity of the circuit networks.
In addition to these general principles, it is worth noting the many kinds of complexity already encountered. The development of a cell type may not follow a simple trajectory but involves multiple steps of divergent or convergent differentiation (Shekhar et al., 2022), the former due to the multipotency of progenitors or transitional cell types and the latter due to convergence of different transitional types. Developmental trajectory also is not the same as developmental lineage, as a lineage is defined as all the cells descended from a single precursor/progenitor and it has been shown in multiple systems that a progenitor can produce cells belonging to several neuronal and non-neuronal types, ordered by developmental timing (Agathocleous and Harris, 2009; Sagner and Briscoe, 2019; Sulston et al., 1983; Zeng and Sanes, 2017). Finally, there are transient cell types and circuits that mainly exist during development and have developmental stage-specific functions (Cossart and Garel, 2022; Molnar et al., 2020). All these observations, and more to be discovered, contribute to a nuanced understanding that cell type development is not a simple linear process, but a highly multifaceted process leading to the complex cell type landscape described in the above sections, which underlies the richness of cell type function.
A comprehensive atlas of mammalian cell type development, likely first generated in mouse and then extended to other species including human (Haniffa et al., 2021), will provide the foundation for matching developmental events and their timelines across species, better understanding the evolutionary relationships between cell types, evaluating and guiding human iPSC and organoid in vitro development, and ultimately, transforming our investigation and treatment of developmental disorders.
How to define cell types?
In conclusion, cell types are the product of evolution, and they are the basic functional units of an organism. To unify these two concepts and define cell types properly, we need to take a multilevel approach, progressing from simple and singular to complex and multifaceted. In such a roadmap, with each iteration, the definition of cell types will become more mature and unified, and the repertoire of cell types defined will better align with the functional architecture of the organism.
The logical first step is to use single-cell transcriptomics-based cell type taxonomy as the initial framework and anchor for defining cell types. The transcriptomic taxonomy contains evolutionarily rooted molecular signatures and allows effective label transfer and linking with all other modalities. Conversely, relating other cellular properties will help to refine transcriptomic types. The transcriptomic taxonomy organizes cell types in a hierarchical manner, laying out different levels of descriptions from major divisions at class and subclass levels to more granular and fuzzier divisions at type and subtype levels (due to the more prevailing continuous variations at the latter levels). To account for the biological reality, a hierarchical presentation of cell types is more meaningful than ascertaining an exact number of types. The transcriptomic taxonomy should be supported by comprehensive epigenomic and spatially resolved transcriptomic characterizations (Fig. 1A), to associate chromatin modification and gene regulatory elements to transcriptomic profiles and assign precise spatial distribution patterns to transcriptomic cell types.
Second, we need to conduct comprehensive anatomical, physiological and functional studies of transcriptomic types using approaches that allow molecular identification of the cells under study (Fig. 1A–B). Such studies will help to resolve differing opinions in lumping or splitting cell types and provide rationales for determining the appropriate level of granularity in defining cell types. They will also provide a context for understanding cell type function and associated cell state changes. In particular, generating complete neuronal morphology reconstructions and comprehensive brain-wide connectomics datasets and relating them to transcriptomic types (Fig. 1A) will be extremely informative in understanding the ultimate synaptic-level brain architecture and its underlying organizing principles, which will lay the foundation for understanding circuit-based brain function.
Third, we need to systematically study the entire developmental process of cell types, at least in mouse. Extending the above-mentioned approaches into development will reveal causal relationships and molecular mechanisms underlying the unique identities, connectivity or other forms of cell-cell interactions, and functions of the vast array of cell types. We should also extend such cell type studies into other species as much as possible, to further uncover evolutionary principles of cell type diversity and how it supports the common or species-specific biological functions including those of the human itself. The studies of cell types and evolution-development (Evo-Devo) are truly interdependent; to achieve meaningful progress one cannot just do one without the other.
Finally, to put all these together, we need a conceptual framework and knowledge base to organize all the knowledge gained from these studies. A tantalizing idea of a “periodic table of cell types” has been proposed (Xia and Yanai, 2019). Considering the Evo-Devo root and the consequential hierarchical organization of cell types, here I suggest that a “tree of cell types” might be more appropriate for an overarching classification of cell types and delineation of their origins and relationships (Stadler et al., 2021; Tanay and Sebe-Pedros, 2021). One can define a tree of cell types for each species, covering its entire life span, and compare such trees across species. Obviously, the “tree” will be a very complex, multi-dimensional graph, and there will likely be multiple branches connecting each node to account for convergence, divergence and other multipronged interrelatedness. To make the tree of cell types widely applicable, it will be critical to adopt explicitly definable and standardized criteria, develop a common cell type ontology and nomenclature, and create computational tools to allow mapping and comparison across datasets as well as genetic tools to enable consistent access of cell types (Osumi-Sutherland, 2017; Yuste et al., 2020). To extract knowledge and insights from the vast amount of data, a list of associated rules, logics and principles will need to be established and articulated, and this will be greatly facilitated by computational modeling. Ultimately, this knowledge base of cell types, interweaving cell type function, development and evolution, will provide the blueprint of life to enable a deeper understanding of the dynamic changes of cellular function under a wide range of healthy and diseased conditions, and lead to innovations that improve human health in many ways.
Acknowledgments
I thank my colleagues in the NIH BRAIN Initiative Cell Census Network (BICCN) and at the Allen Institute for many insightful discussions that deepened and broadened my understanding of many topics covered in this review. This work is supported by Allen Institute for Brain Science and by NIH grant U19MH114830.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The author declares no competing interests.
References
- Abbott LF, Bock DD, Callaway EM, Denk W, Dulac C, Fairhall AL, Fiete I, Harris KM, Helmstaedter M, Jain V, et al. (2020). The Mind of a Mouse. Cell 182, 1372–1376. [DOI] [PubMed] [Google Scholar]
- Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. (2016). A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dis of the Unfolded Protein Response. Cell 167, 1867–1882 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agathocleous M, and Harris WA (2009). From progenitors to differentiated cells in the vertebrate retina. Annu Rev Cell Dev Biol 25, 45–69. [DOI] [PubMed] [Google Scholar]
- Allaway KC, Gabitto MI, Wapinski O, Saldi G, Wang CY, Bandler RC, Wu SJ, Bonneau R, and Fishell G (2021). Genetic and epigenetic coordination of cortical interneuron development. Nature 597, 693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D (2008). The evolution of cell types in animals: emerging principles from molecular studies. Nature reviews Genetics 9, 868–882. [DOI] [PubMed] [Google Scholar]
- Arendt D, Musser JM, Baker CVH, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD, et al. (2016). The origin and evolution of cell types. Nature reviews Genetics 17, 744–757. [DOI] [PubMed] [Google Scholar]
- Armand EJ, Li J, Xie F, Luo C, and Mukamel EA (2021). Single-Cell Sequencing of Brain Cell Transcriptomes and Epigenomes. Neuron 109, 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden T, Euler T, and Berens P (2020). Understanding the retinal basis of vision across species. Nature reviews Neuroscience 21, 5–20. [DOI] [PubMed] [Google Scholar]
- Bakken TE, Jorstad NL, Hu Q, Lake BB, Tian W, Kalmbach BE, Crow M, Hodge RD, Krienen FM, Sorensen SA, et al. (2021). Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandler RC, Vitali I, Delgado RN, Ho MC, Dvoretskova E, Ibarra Molinas JS, Frazel PW, Mohammadkhani M, Machold R, Maedler S, et al. (2022). Single-cell delineation of lineage and genetic identity in the mouse brain. Nature 601, 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Haim L, and Rowitch DH (2017). Functional diversity of astrocytes in neural circuit regulation. Nature reviews Neuroscience 18, 31–41. [DOI] [PubMed] [Google Scholar]
- Berg J, Sorensen SA, Ting JT, Miller JA, Chartrand T, Buchin A, Bakken TE, Budzillo A, Dee N, Ding SL, et al. (2021). Human neocortical expansion involves glutamatergic neuron diversification. Nature 598, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri A, Sandoval-Espinosa C, Otero-Garcia M, Oh I, Yin R, Eze UC, Nowakowski TJ, and Kriegstein AR (2021). An atlas of cortical arealization identifies dynamic molecular signatures. Nature 598, 200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booeshaghi AS, Yao Z, van Velthoven C, Smith K, Tasic B, Zeng H, and Pachter L (2021). Isoform cell-type specificity in the mouse primary motor cortex. Nature 598, 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brain Initiative Cell Census Network (2021). A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugeon S, Duffield J, Dipoppa M, Ritoux A, Prankerd I, Nicolout-sopoulos D, Orme D, Shinn M, Peng H, Forrest H, et al. (2021). A transcriptomic axis predicts state modulation of cortical interneurons. bioRxiv, 2021.2010.2024.465600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, and Weiner HL (2018). Microglial signatures and their role in health and disease. Nature reviews Neuroscience 19, 622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell CR, Bhaduri A, Mostajo-Radji MA, Keefe MG, and Nowakowski TJ (2019). Development and Arealization of the Cerebral Cortex. Neuron 103, 980–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, et al. (2016). Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nature biotechnology 34, 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Lemaire LA, Wang W, Yoon PH, Choi YA, Parsons LR, Matese JC, Wang W, Levine M, and Chen K (2019a). Comprehensive single-cell transcriptome lineages of a proto-vertebrate. Nature 571, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019b). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catela C, Shin MM, and Dasen JS (2015). Assembly and function of spinal circuits for motor control. Annu Rev Cell Dev Biol 31, 669–698. [DOI] [PubMed] [Google Scholar]
- Cembrowski MS, Wang L, Lemire AL, Copeland M, DiLisio SF, Clements J, and Spruston N (2018). The subiculum is a patchwork of discrete subregions. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu WL, Sanchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, et al. (2017). Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nature neuroscience 20, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Liao S, Cheng M, Ma K, Wu L, Lai Y, Qiu X, Yang J, Xu J, Hao S, et al. (2022). Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 e21. [DOI] [PubMed] [Google Scholar]
- Chen R, Blosser TR, Djekidel MN, Hao J, Bhattacherjee A, Chen W, Tuesta LM, Zhuang X, and Zhang Y (2021). Decoding molecular and cellular heterogeneity of mouse nucleus accumbens. Nature neuroscience 24, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Sun YC, Zhan H, Kebschull JM, Fischer S, Matho K, Huang ZJ, Gillis J, and Zador AM (2019). High-Throughput Mapping of Long-Range Neuronal Projection Using In Situ Sequencing. Cell 179, 772–786 e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NH, Cheveralls KC, Brunner AD, Kim K, Michaelis AC, Raghavan P, Kobayashi H, Savy L, Li JY, Canaj H, et al. (2022). OpenCell: Endogenous tagging for the cartography of human cellular organization. Science 375, eabi6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IC, Gutierrez-Vazquez C, Wheeler MA, Li Z, Rothhammer V, Linnerbauer M, Sanmarco LM, Guo L, Blain M, Zandee SEJ, et al. (2021). Barcoded viral tracing of single-cell interactions in central nervous system inflammation. Science 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close JL, Long BR, and Zeng H (2021). Spatially resolved transcriptomics in neuroscience. Nature methods 18, 23–25. [DOI] [PubMed] [Google Scholar]
- Colquitt BM, Merullo DP, Konopka G, Roberts TF, and Brainard MS (2021). Cellular transcriptomics reveals evolutionary identities of songbird vocal circuits. Science 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condylis C, Ghanbari A, Manjrekar N, Bistrong K, Yao S, Yao Z, Nguyen TN, Zeng H, Tasic B, and Chen JL (2022). Dense functional and molecular readout of a circuit hub in sensory cortex. Science 375, eabl5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart R, and Garel S (2022). Step by step: cells with multiple functions in cortical circuit assembly. Nature reviews Neuroscience. [DOI] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. (2018). A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell 174, 1309–1324 e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, Takeno MM, Huang L, Gu H, Larsen R, et al. (2018). A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced BrainCell-Type Targeting and Functionality. Cell 174, 465–480 e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, and Quake SR (2015). A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112, 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado RN, Allen DE, Keefe MG, Mancia Leon WR, Ziffra RS, Crouch EE, Alvarez-Buylla A, and Nowakowski TJ (2022). Individual human cortical progenitors can produce excitatory and inhibitory neurons. Nature 601, 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo L, and Luo L (2017). Genetic strategies to access activated neurons. Current opinion in neurobiology 45, 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bella DJ, Habibi E, Stickels RR, Scalia G, Brown J, Yadollahpour P, Yang SM, Abbate C, Biancalani T, Macosko EZ, et al. (2021). Molecular logic of cellular diversification in the mouse cerebral cortex. Nature 595, 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi GA, Guo L, Xu Q, Liu R, Lu C, Chu J, et al. (2016). A viral strategy for targeting and manipulating interneurons across vertebrate species. Nature neuroscience 19, 1743–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimou L, and Simons M (2017). Diversity of oligodendrocytes and their progenitors. Current opinion in neurobiology 47, 73–79. [DOI] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. (2016). Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866 e1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drokhlyansky E, Smillie CS, Van Wittenberghe N, Ericsson M, Griffin GK, Eraslan G, Dionne D, Cuoco MS, Goder-Reiser MN, Sharova T, et al. (2020). The Human and Mouse Enteric Nervous System at Single-Cell Resolution. Cell 182, 1606–1622 e1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR, Geschwind DH, Kriegstein AR, Ngai J, Osten P, Polioudakis D, Regev A, Sestan N, Wickersham IR, and Zeng H (2017). The BRAIN Initiative Cell Census Consortium: Lessons Learned toward Generating a Comprehensive Brain Cell Atlas. Neuron 96, 542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escartin C, Galea E, Lakatos A, O’Callaghan JP, Petzold GC, Serrano-Pozo A, Steinhauser C, Volterra A, Carmignoto G, Agarwal A, et al. (2021). Reactive astrocyte nomenclature, definitions, and future directions. Nature neuroscience 24, 312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G, and Heintz N (2013). The neuron identity problem: form meets function. Neuron 80, 602–612. [DOI] [PubMed] [Google Scholar]
- Fuzik J, Zeisel A, Mate Z, Calvigioni D, Yanagawa Y, Szabo G, Linnarsson S, and Harkany T (2016). Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nature biotechnology 34, 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Liu S, Gou L, Hu Y, Liu Y, Deng L, Ma D, Wang H, Yang Q, Chen Z, et al. (2022). Single-neuron projectome of mouse prefrontal cortex. Nature neuroscience 25, 515–529. [DOI] [PubMed] [Google Scholar]
- Garcia FJ, Sun N, Lee H, Godlewski B, Mathys H, Galani K, Zhou B, Jiang X, Ng AP, Mantero J, et al. (2022). Single-cell dissection of the human brain vasculature. Nature 603, 893–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergues MM, Han KJ, Choi HS, Brown B, Clausing KJ, Turner VS, Vainchtein ID, Molofsky AV, and Kheirbek MA (2020). Circuit and molecular architecture of a ventral hippocampal network. Nature neuroscience 23, 1444–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gour A, Boergens KM, Heike N, Hua Y, Laserstein P, Song K, and Helmstaedter M (2021). Postnatal connectomic development of inhibition in mouse barrel cortex. Science 371. [DOI] [PubMed] [Google Scholar]
- Gouwens NW, Sorensen SA, Baftizadeh F, Budzillo A, Lee BR, Jarsky T, Alfiler L, Baker K, Barkan E, Berry K, et al. (2020). Integrated Morphoelectric and Transcriptomic Classification of Cortical GABAergic Cells. Cell 183, 935–953 e919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybuck LT, Daigle TL, Sedeno-Cortes AE, Walker M, Kalmbach B, Lenz GH, Morin E, Nguyen TN, Garren E, Bendrick JL, et al. (2021). Enhancer viruses for combinatorial cell-subclass-specific labeling. Neuron 109, 1449–1464 e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, et al. (2019). Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253–271 e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Wei X, Liu C, Volpe G, Zhuang Z, Zou X, Wang Z, Pan T, Yuan Y, Zhang X, et al. (2022). Cell transcriptomic atlas of the non-human primate Macaca fascicularis. Nature 604, 723–731. [DOI] [PubMed] [Google Scholar]
- Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, et al. (2018a). Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 172, 1091–1107 e1017. [DOI] [PubMed] [Google Scholar]
- Han Y, Kebschull JM, Campbell RAA, Cowan D, Imhof F, Zador AM, and Mrsic-Flogel TD (2018b). The logic of single-cell projections from visual cortex. Nature 556, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M, Taylor D, Linnarsson S, Aronow BJ, Bader GD, Barker RA, Camara PG, Camp JG, Chedotal A, Copp A, et al. (2021). A roadmap for the Human Developmental Cell Atlas. Nature 597, 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, and Shepherd GM (2015). The neocortical circuit: themes and variations. Nature neuroscience 18, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashikawa Y, Hashikawa K, Rossi MA, Basiri ML, Liu Y, Johnston NL, Ahmad OR, and Stuber GD (2020). Transcriptional and Spatial Resolution of Cell Types in the Mammalian Habenula. Neuron 106, 743–758 e745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, and Denk W (2013). Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500, 168–174. [DOI] [PubMed] [Google Scholar]
- Hildebrand DGC, Cicconet M, Torres RM, Choi W, Quan TM, Moon J, Wetzel AW, Scott Champion A, Graham BJ, Randlett O, et al. (2017). Whole-brain serial-section electron microscopy in larval zebrafish. Nature 545, 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, and Kratsios P (2019). Neuronal identity control by terminal selectors in worms, flies, and chordates. Current opinion in neurobiology 56, 97–105. [DOI] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, Zilionis R, Ratner A, Borges-Monroy R, Klein AM, et al. (2018). Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nature neuroscience 21, 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, Tzeng CP, Nagy MA, Stroud H, Koutsioumpa C, Wilcox OF, Assad EG, Green J, Harvey CD, Griffith EC, et al. (2019). A scalable platform for the development of cell-type-specific viral drivers. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JS, Vogt D, Sandberg M, and Rubenstein JL (2017). Cortical interneuron development: a tale of time and space. Development 144, 3867–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, and Paul A (2019). The diversity of GABAergic neurons and neural communication elements. Nature Reviews Neuroscience 20, 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse BK, Haberkern H, Franconville R, Turner-Evans DB, Takemura SY, Wolff T, Noorman M, Dreher M, Dan C, Parekh R, et al. (2021). A connectome of the Drosophila central complex reveals network motifs suitable for flexible navigation and context-dependent action selection. eLife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. (2014). Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, and Amit I (2016). Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 167, 1883–1896 e1815. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell reports 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell TM (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nature reviews Genetics 1, 20–29. [DOI] [PubMed] [Google Scholar]
- Kamath T, Abdulraouf A, Burris SJ, Langlieb J, Gazestani V, Nadaf NM, Balderrama K, Vanderburg C, and Macosko EZ (2022). Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease. Nature neuroscience 25, 588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebschull JM, Garcia da Silva P, Reid AP, Peikon ID, Albeanu DF, and Zador AM (2016). High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron 91, 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebschull JM, Richman EB, Ringach N, Friedmann D, Albarran E, Kolluru SS, Jones RC, Allen WE, Wang Y, Cho SW, et al. (2020). Cerebellar nuclei evolved by repeatedly duplicating a conserved cell-type set. Science 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, and Deneen B (2019). The Emerging Nature of Astrocyte Diversity. Annual review of neuroscience 42, 187–207. [DOI] [PubMed] [Google Scholar]
- Kim DW, Yao Z, Graybuck LT, Kim TK, Nguyen TN, Smith KA, Fong O, Yi L, Koulena N, Pierson N, et al. (2019). Multimodal Analysis of Cell Types in a Hypothalamic Node Controlling Social Behavior. Cell 179, 713–728 e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Zhang Z, Huang L, Ito-Cole T, Jacobs MW, Juavinett AL, Senturk G, Hu M, Ku M, Ecker JR, et al. (2020). Extraction of Distinct Neuronal Cell Types from within a Genetically Continuous Population. Neuron 107, 274–282 e276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler E, Tomasello U, Prados J, Kebschull JM, Contestabile A, Galinanes GL, Fievre S, Santinha A, Platt R, Huber D, et al. (2021). Temporal controls over inter-areal cortical projection neuron fate diversity. Nature 599, 453–457. [DOI] [PubMed] [Google Scholar]
- Kozareva V, Martin C, Osorno T, Rudolph S, Guo C, Vanderburg C, Nadaf N, Regev A, Regehr WG, and Macosko E (2021). A transcriptomic atlas of mouse cerebellar cortex comprehensively defines cell types. Nature 598, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krienen FM, Goldman M, Zhang Q, R CHDR, Florio M, Machold R, Saunders A, Levandowski K, Zaniewski H, Schuman B, et al. (2020). Innovations present in the primate interneuron repertoire. Nature 586, 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn S, Gritti L, Crooks D, and Dombrowski Y (2019). Oligodendrocytes in Development, Myelin Generation and Beyond. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Siletti K, Furlan A, Gyllborg D, Vinsland E, Mossi Albiach A, Mattsson Langseth C, Khven I, Lederer AR, Dratva LM, et al. (2021). Molecular architecture of the developing mouse brain. Nature 596, 92–96. [DOI] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. (2016). Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, Duong TE, Gao D, Chun J, Kharchenko PV, et al. (2018). Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nature biotechnology 36, 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson L, Frisen J, and Lundeberg J (2021). Spatially resolved transcriptomics adds a new dimension to genomics. Nature methods 18, 15–18. [DOI] [PubMed] [Google Scholar]
- Lee BR, Budzillo A, Hadley K, Miller JA, Jarsky T, Baker K, Hill D, Kim L, Mann R, Ng L, et al. (2021). Scaled, high fidelity electrophysiological, morphological, and transcriptomic cell characterization. eLife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E, Borm LE, and Linnarsson S (2017). The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358, 64–69. [DOI] [PubMed] [Google Scholar]
- Li H, Janssens J, De Waegeneer M, Kolluru SS, Davie K, Gardeux V, Saelens W, David FPA, Brbic M, Spanier K, et al. (2022). Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science 375, eabk2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, Gulati G, Bennett ML, Sun LO, Clarke LE, et al. (2019). Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 101, 207–223 e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YE, Preissl S, Hou X, Zhang Z, Zhang K, Qiu Y, Poirion OB, Li B, Chiou J, Liu H, et al. (2021). An atlas of gene regulatory elements in adult mouse cerebrum. Nature 598, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Mi D, Llorca A, and Marin O (2018). Development and Functional Diversification of Cortical Interneurons. Neuron 100, 294–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom RGH, Regev A, and Teichmann SA (2021). Towards a Human Cell Atlas: Taking Notes from the Past. Trends Genet 37, 625–630. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhou J, Tian W, Luo C, Bartlett A, Aldridge A, Lucero J, Osteen JK, Nery JR, Chen H, et al. (2021). DNA methylation atlas of the mouse brain at single-cell resolution. Nature 598, 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Barron M, Chen R, Bradbury S, Andalman AS, Wagle M, Guo S, and Deisseroth K (2020). Multiple convergent hypothalamus-brainstem circuits drive defensive behavior. Nature neuroscience 23, 959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Keown CL, Kurihara L, Zhou J, He Y, Li J, Castanon R, Lucero J, Nery JR, Sandoval JP, et al. (2017). Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, and Wu C (2004). Interneurons of the neocortical inhibitory system. Nature reviews Neuroscience 5, 793–807. [DOI] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcao A, Xiao L, Li H, Haring M, Hochgerner H, Romanov RA, et al. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH (2012). The neuronal organization of the retina. Neuron 76, 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Sankowski R, Staszewski O, Bottcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, et al. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392. [DOI] [PubMed] [Google Scholar]
- Matho KS, Huilgol D, Galbavy W, He M, Kim G, An X, Lu J, Wu P, Di Bella DJ, Shetty AS, et al. (2021). Genetic dissection of the glutamatergic neuron system in cerebral cortex. Nature 598, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr U, Serra D, and Liberali P (2019). Exploring single cells in space and time during tissue development, homeostasis and regeneration. Development 146. [DOI] [PubMed] [Google Scholar]
- Mazzarello P (1999). A unifying concept: the history of cell theory. Nat Cell Biol 1, E13–15. [DOI] [PubMed] [Google Scholar]
- McKenna A, and Gagnon JA (2019). Recording development with single cell dynamic lineage tracing. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mich JK, Graybuck LT, Hess EE, Mahoney JT, Kojima Y, Ding Y, Somasundaram S, Miller JA, Kalmbach BE, Radaelli C, et al. (2021). Functional enhancer elements drive subclass-selective expression from mouse to primate neocortex. Cell reports 34, 108754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, et al. (2018). Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar Z, Luhmann HJ, and Kanold PO (2020). Transient cortical circuits match spontaneous and sensory-driven activity during development. Science 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje M (2018). Myelin Plasticity and Nervous System Function. Annual review of neuroscience 41, 61–76. [DOI] [PubMed] [Google Scholar]
- Morgan JL, Berger DR, Wetzel AW, and Lichtman JW (2016). The Fuzzy Logic of Network Connectivity in Mouse Visual Thalamus. Cell 165, 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA (2019). The evolving concept of cell identity in the single cell era. Development 146. [DOI] [PubMed] [Google Scholar]
- Moses L, and Pachter L (2022). Museum of spatial transcriptomics. Nature methods 19, 534–546. [DOI] [PubMed] [Google Scholar]
- Mukamel EA, and Ngai J (2019). Perspectives on defining cell types in the brain. Current opinion in neurobiology 56, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Manchado AB, Bengtsson Gonzales C, Zeisel A, Munguba H, Bekkouche B, Skene NG, Lonnerberg P, Ryge J, Harris KD, Linnarsson S, et al. (2018). Diversity of Interneurons in the Dorsal Striatum Revealed by Single-Cell RNA Sequencing and PatchSeq. Cell reports 24, 2179–2190 e2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro DAD, Movahedi K, and Priller J (2022). Macrophage compartmentalization in the brain and cerebrospinal fluid system. Sci Immunol 7, eabk0391. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Foley NM, Bredemeyer KR, Gatesy J, and Springer MS (2021). Phylogenomics and the Genetic Architecture of the Placental Mammal Radiation. Annu Rev Anim Biosci 9, 29–53. [DOI] [PubMed] [Google Scholar]
- Nelson SB, Sugino K, and Hempel CM (2006). The problem of neuronal cell types: a physiological genomics approach. Trends in neurosciences 29, 339–345. [DOI] [PubMed] [Google Scholar]
- Ngai J (2022). BRAIN 2.0: Transforming neuroscience. Cell 185, 4–8. [DOI] [PubMed] [Google Scholar]
- O’Leary DD, Chou SJ, and Sahara S (2007). Area patterning of the mammalian cortex. Neuron 56, 252–269. [DOI] [PubMed] [Google Scholar]
- Ortiz-Alvarez G, Daclin M, Shihavuddin A, Lansade P, Fortoul A, Faucourt M, Clavreul S, Lalioti ME, Taraviras S, Hippenmeyer S, et al. (2019). Adult Neural Stem Cells and Multiciliated Ependymal Cells Share a Common Lineage Regulated by the Geminin Family Members. Neuron 102, 159–172 e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz C, Navarro JF, Jurek A, Martin A, Lundeberg J, and Meletis K (2020). Molecular atlas of the adult mouse brain. Sci Adv 6, eabb3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osseward PJ 2nd, and Pfaff SL (2019). Cell type and circuit modules in the spinal cord. Current opinion in neurobiology 56, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi-Sutherland D (2017). Cell ontology in an age of data-driven cell classification. BMC bioinformatics 18, 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A, Crow M, Raudales R, He M, Gillis J, and Huang ZJ (2017). Transcriptional Architecture of Synaptic Communication Delineates GABAergic Neuron Identity. Cell 171, 522–539 e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Xie P, Liu L, Kuang X, Wang Y, Qu L, Gong H, Jiang S, Li A, Ruan Z, et al. (2021). Morphological diversity of single neurons in molecularly defined cell types. Nature 598, 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petilla Interneuron Nomenclature Group, Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsaki G, Cauli B, Defelipe J, et al. (2008). Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nature reviews Neuroscience 9, 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JW, Schulmann A, Hara E, Winnubst J, Liu C, Valakh V, Wang L, Shields BC, Korff W, Chandrashekar J, et al. (2019). A repeated molecular architecture across thalamic pathways. Nature neuroscience 22, 1925–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool AH, Wang T, Stafford DA, Chance RK, Lee S, Ngai J, and Oka Y (2020). The cellular basis of distinct thirst modalities. Nature 588, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin JF, Zou J, Drouin-Ouellet J, Kim KY, Cicchetti F, and Awatramani RB (2014). Defining midbrain dopaminergic neuron diversity by single-cell gene expression profiling. Cell reports 9, 930–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, Zhang Y, Sos BC, Afzal V, Dickel DE, et al. (2018). Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nature neuroscience 21, 432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss TM, and Wise SP (2022). Evolution of prefrontal cortex. Neuropsychopharmacology 47, 3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz M, Jung S, and Priller J (2019). Microglia Biology: One Century of Evolving Concepts. Cell 179, 292–311. [DOI] [PubMed] [Google Scholar]
- Rao A, Barkley D, Franca GS, and Yanai I (2021). Exploring tissue architecture using spatial transcriptomics. Nature 596, 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond SA, Figueres-Onate M, Obernier K, Nascimento MA, Parraguez JI, Lopez-Mascaraque L, Fuentealba LC, and Alvarez-Buylla A (2019). Development of Ependymal and Postnatal Neural Stem Cells and Their Origin from a Common Embryonic Progenitor. Cell reports 27, 429–441 e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. (2017). The Human Cell Atlas. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MB, Cros C, Varol E, Yemini E, and Hobert O (2020). Unique homeobox codes delineate all the neuron classes of C. elegans. Nature 584, 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Isakova A, Friedmann D, Zeng J, Grutzner SM, Pun A, Zhao GQ, Kolluru SS, Wang R, Lin R, et al. (2019). Single-cell transcriptomes and whole-brain projections of serotonin neurons in the mouse dorsal and median raphe nuclei. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle JM, Saunders RA, Pogson AN, Hussmann JA, Lenail A, Guna A, Mascibroda L, Wagner EJ, Adelman K, Lithwick-Yanai G, et al. (2022). Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov RA, Tretiakov EO, Kastriti ME, Zupancic M, Haring M, Korchynska S, Popadin K, Benevento M, Rebernik P, Lallemend F, et al. (2020). Molecular design of hypothalamus development. Nature 582, 246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov RA, Zeisel A, Bakker J, Girach F, Hellysaz A, Tomer R, Alpar A, Mulder J, Clotman F, Keimpema E, et al. (2017). Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nature neuroscience 20, 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ DE, Cross RBP, Li L, Koch SC, Matson KJE, Yadav A, Alkaslasi MR, Lee DI, Le Pichon CE, Menon V, et al. (2021). A harmonized atlas of mouse spinal cord cell types and their spatial organization. Nature communications 12, 5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelens W, Cannoodt R, Todorov H, and Saeys Y (2019). A comparison of single-cell trajectory inference methods. Nature biotechnology 37, 547–554. [DOI] [PubMed] [Google Scholar]
- Sagner A, and Briscoe J (2019). Establishing neuronal diversity in the spinal cord: a time and a place. Development 146. [DOI] [PubMed] [Google Scholar]
- Sanes JR, and Masland RH (2015). The types of retinal ganglion cells: current status and implications for neuronal classification. Annual review of neuroscience 38, 221–246. [DOI] [PubMed] [Google Scholar]
- Sathyamurthy A, Johnson KR, Matson KJE, Dobrott CI, Li L, Ryba AR, Bergman TB, Kelly MC, Kelley MW, and Levine AJ (2018). Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity during Behavior. Cell reports 22, 2216–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030 e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala F, Kobak D, Bernabucci M, Bernaerts Y, Cadwell CR, Castro JR, Hartmanis L, Jiang X, Laturnus S, Miranda E, et al. (2021). Phenotypic variation of transcriptomic cell types in mouse motor cortex. Nature 598, 144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer S, and Iadecola C (2021). Revisiting the neurovascular unit. Nature neuroscience 24, 1198–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura SY, Hayworth KJ, Huang GB, Shinomiya K, Maitlin-Shepard J, Berg S, et al. (2020). A connectome and analysis of the adult Drosophila central brain. eLife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz MT, Sandoval K, Chen CP, Mostajo-Radji MA, Seeley WW, Nowakowski TJ, Ye CJ, Paredes MF, and Pollen AA (2022). The development and evolution of inhibitory neurons in primate cerebrum. Nature 603, 871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Mizell CM, Bodor AL, Collman F, Brittain D, Bleckert A, Dorkenwald S, Turner NL, Macrina T, Lee K, Lu R, et al. (2021). Structure and function of axo-axonic inhibition. eLife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz MK, and Remy S (2019). Rabies virus-mediated connectivity tracing from single neurons. Journal of neuroscience methods 325, 108365. [DOI] [PubMed] [Google Scholar]
- Seung HS, and Sumbul U (2014). Neuronal cell types and connectivity: lessons from the retina. Neuron 83, 1262–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Flaherty K, Lezgiyeva K, Wagner DE, Klein AM, and Ginty DD (2020). The emergence of transcriptional identity in somatosensory neurons. Nature 577, 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323 e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, and Sanes JR (2021). Generating and Using Transcriptomically Based Retinal Cell Atlases. Annu Rev Vis Sci 7, 43–72. [DOI] [PubMed] [Google Scholar]
- Shekhar K, Whitney IE, Butrus S, Peng YR, and Sanes JR (2022). Diversification of multipotential postmitotic mouse retinal ganglion cell precursors into discrete types. eLife 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N (2021). Single-cell protein analysis by mass spectrometry. Curr Opin Chem Biol 60, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, and Klausberger T (2005). Defined types of cortical interneurone structure space and spike timing in the hippocampus. The Journal of physiology 562, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler T, Pybus OG, and Stumpf MPH (2021). Phylodynamics for cell biologists. Science 371. [DOI] [PubMed] [Google Scholar]
- Stanley G, Gokce O, Malenka RC, Sudhof TC, and Quake SR (2020). Continuous and Discrete Neuron Types of the Adult Murine Striatum. Neuron 105, 688–699 e688. [DOI] [PubMed] [Google Scholar]
- Stephan T, Burgess SM, Cheng H, Danko CG, Gill CA, Jarvis ED, Koepfli KP, Koltes JE, Lyons E, Ronald P, et al. (2022). Darwinian genomics and diversity in the tree of life. Proc Natl Acad Sci U S A 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, and Thomson JN (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental biology 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Sun YC, Chen X, Fischer S, Lu S, Zhan H, Gillis J, and Zador AM (2021). Integrating barcoded neuroanatomy with spatial transcriptional profiling enables identification of gene correlates of projections. Nature neuroscience 24, 873–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson V, da Veiga Beltrame E, and Pachter L (2020). A curated database reveals trends in single-cell transcriptomics. Database (Oxford) 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW (2000). What is the brain? Trends in neurosciences 23, 519–527. [DOI] [PubMed] [Google Scholar]
- Swanson LW (2012). Brain architecture: understanding the basic plan, 2nd edn (New York, NY: Oxford University Press; ). [Google Scholar]
- Sweeney MD, Zhao Z, Montagne A, Nelson AR, and Zlokovic BV (2019). Blood-Brain Barrier: From Physiology to Disease and Back. Physiological reviews 99, 21–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris, C., Overall, c., Logistical, c., Organ, c., processing, Library, p., sequencing, Computational data, a., Cell type, a., Writing, g., et al. (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Sapiens C, Jones RC, Karkanias J, Krasnow MA, Pisco AO, Quake SR, Salzman J, Yosef N, Bulthaup B, Brown P, et al. (2022). The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, and Sebe-Pedros A (2021). Evolutionary cell type mapping with single-cell genomics. Trends Genet 37, 919–932. [DOI] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nature neuroscience 19, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SR, Santpere G, Weinreb A, Barrett A, Reilly MB, Xu C, Varol E, Oikonomou P, Glenwinkel L, McWhirter R, et al. (2021). Molecular topography of an entire nervous system. Cell 184, 4329–4347 e4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thion MS, and Garel S (2020). Microglial ontogeny, diversity and neurodevelopmental functions. Current opinion in genetics & development 65, 186–194. [DOI] [PubMed] [Google Scholar]
- Tiklova K, Bjorklund AK, Lahti L, Fiorenzano A, Nolbrant S, Gillberg L, Volakakis N, Yokota C, Hilscher MM, Hauling T, et al. (2019). Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nature communications 10, 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, and Laurent G (2018). Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360, 881–888. [DOI] [PubMed] [Google Scholar]
- Tremblay R, Lee S, and Rudy B (2016). GABAergic Interneurons in the Neocortex: From Cellular Properties to Circuits. Neuron 91, 260–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler S, Buttner M, Fischer DS, Lange M, Bergen V, Lickert H, and Theis FJ (2019). Concepts and limitations for learning developmental trajectories from single cell genomics. Development 146. [DOI] [PubMed] [Google Scholar]
- Turner NL, Macrina T, Bae JA, Yang R, Wilson AM, Schneider-Mizell C, Lee K, Lu R, Wu J, Bodor AL, et al. (2022). Reconstruction of neocortex: Organelles, compartments, cells, circuits, and activity. Cell 185, 1082–1100 e1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hove H, Martens L, Scheyltjens I, De Vlaminck K, Pombo Antunes AR, De Prijck S, Vandamme N, De Schepper S, Van Isterdael G, Scott CL, et al. (2019). A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nature neuroscience 22, 1021–1035. [DOI] [PubMed] [Google Scholar]
- Vanlandewijck M, He L, Mae MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Lavina B, Gouveia L, et al. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480. [DOI] [PubMed] [Google Scholar]
- von Buchholtz LJ, Ghitani N, Lam RM, Licholai JA, Chesler AT, and Ryba NJP (2021). Decoding Cellular Mechanisms for Mechanosensory Discrimination. Neuron 109, 285–298 e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vormstein-Schneider D, Lin JD, Pelkey KA, Chittajallu R, Guo B, Arias-Garcia MA, Allaway K, Sakopoulos S, Schneider G, Stevenson O, et al. (2020). Viral manipulation of functionally distinct interneurons in mice, non-human primates and humans. Nature neuroscience 23, 1629–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, and Klein AM (2020). Lineage tracing meets single-cell omics: opportunities and challenges. Nature reviews Genetics 21, 410–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Eddison M, Fleishman G, Weigert M, Xu S, Wang T, Rokicki K, Goina C, Henry FE, Lemire AL, et al. (2021). EASI-FISH for thick tissue defines lateral hypothalamus spatio-molecular organization. Cell 184, 6361–6377 e6324. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, and Brenner S (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical transactions of the Royal Society of London Series B, Biological sciences 314, 1–340. [DOI] [PubMed] [Google Scholar]
- Winkler EA, Kim CN, Ross JM, Garcia JH, Gil E, Oh I, Chen LQ, Wu D, Catapano JS, Raygor K, et al. (2022). A single-cell atlas of the normal and malformed human brain vasculature. Science 375, eabi7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnubst J, Bas E, Ferreira TA, Wu Z, Economo MN, Edson P, Arthur BJ, Bruns C, Rokicki K, Schauder D, et al. (2019). Reconstruction of 1,000 Projection Neurons Reveals New Cell Types and Organization of Long-Range Connectivity in the Mouse Brain. Cell 179, 268–281 e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witvliet D, Mulcahy B, Mitchell JK, Meirovitch Y, Berger DR, Wu Y, Liu Y, Koh WX, Parvathala R, Holmyard D, et al. (2021). Connectomes across development reveal principles of brain maturation. Nature 596, 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, and Rubin GM (2018). Neuroarchitecture of the Drosophila central complex: A catalog of nodulus and asymmetrical body neurons and a revision of the protocerebral bridge catalog. The Journal of comparative neurology 526, 2585–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YE, Pan L, Zuo Y, Li X, and Hong W (2017). Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron 96, 313–329 e316. [DOI] [PubMed] [Google Scholar]
- Xia B, and Yanai I (2019). A periodic table of cell types. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Yang H, Menon V, Lemire AL, Wang L, Henry FE, Turaga SC, and Sternson SM (2020). Behavioral state coding by molecularly defined paraventricular hypothalamic cell type ensembles. Science 370. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Yan W, and Sanes JR (2021). A cell atlas of the chick retina based on single-cell transcriptomics. eLife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AC, Vest RT, Kern F, Lee DP, Agam M, Maat CA, Losada PM, Chen MB, Schaum N, Khoury N, et al. (2022). A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature 603, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Liu H, Xie F, Fischer S, Adkins RS, Aldridge AI, Ament SA, Bartlett A, Behrens MM, Van den Berge K, et al. (2021a). A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex. Nature 598, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, van Velthoven CTJ, Nguyen TN, Goldy J, Sedeno-Cortes AE, Baftizadeh F, Bertagnolli D, Casper T, Chiang M, Crichton K, et al. (2021b). A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241 e3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Hawrylycz M, Aalling N, Aguilar-Valles A, Arendt D, Armananzas R, Ascoli GA, Bielza C, Bokharaie V, Bergmann TB, et al. (2020). A community-based transcriptomics classification and nomenclature of neocortical cell types. Nature neuroscience 23, 1456–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, Haring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014 e1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. (2015). Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. [DOI] [PubMed] [Google Scholar]
- Zeng H, and Sanes JR (2017). Neuronal cell-type classification: challenges, opportunities and the path forward. Nature reviews Neuroscience 18, 530–546. [DOI] [PubMed] [Google Scholar]
- Zhang M, Eichhorn SW, Zingg B, Yao Z, Cotter K, Zeng H, Dong H, and Zhuang X (2021a). Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature 598, 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhou J, Tan P, Pang Y, Rivkin AC, Kirchgessner MA, Williams E, Lee CT, Liu H, Franklin AD, et al. (2021b). Epigenomic diversity of cortical projection neurons in the mouse brain. Nature 598, 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Preissl S, and Ren B (2020). Single-cell multimodal omics: the power of many. Nature Methods 17, 11–14. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Sousa AMM, Gao T, Skarica M, Li M, Santpere G, Esteller-Cucala P, Juan D, Ferrandez-Peral L, Gulden FO, et al. (2018). Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X (2021). Spatially resolved single-cell genomics and transcriptomics by imaging. Nature methods 18, 18–22. [DOI] [PMC free article] [PubMed] [Google Scholar]


