Abstract
UV-B radiation acts as a developmental cue and a stress factor for plants, depending on dose. Activation of the transcription factor ELONGATED HYPOCOTYL 5 (HY5) in a UV RESISTANCE LOCUS 8 (UVR8)-dependent manner leads to the induction of a broad set of genes under UV-B. However, the underlying molecular mechanisms regulating this process are less understood. Here, we use molecular, biochemical, genetic, and metabolomic tools to identify the B-BOX transcription factor B-BOX PROTEIN 11 (BBX11) as a component of the molecular response to UV-B in Arabidopsis (Arabidopsis thaliana). BBX11 expression is induced by UV-B in a dose-dependent manner. Under low UV-B, BBX11 regulates hypocotyl growth suppression, whereas it protects plants exposed to high UV-B radiation by promoting the accumulation of photo-protective phenolics and antioxidants, and inducing DNA repair genes. Our genetic studies indicate that BBX11 regulates hypocotyl elongation under UV-B partially dependent on HY5. Overexpression of BBX11 can partially rescue the high UV-B sensitivity of hy5, suggesting that HY5-mediated UV-B stress tolerance is partially dependent on BBX11. HY5 regulates the UV-B-mediated induction of BBX11 by directly binding to its promoter. BBX11 reciprocally regulates the mRNA and protein levels of HY5. We report here the role of a BBX11-HY5 feedback loop in regulating photomorphogenesis and stress tolerance under UV-B.
The B-box protein BBX11 regulates photomorphogenesis and stress tolerance under UV-B.
Introduction
Plants are often exposed to the solar UV-B radiation. While a low dose of UV-B acts as a morphogenetic cue and promotes the accumulation of beneficial metabolites, high UV-B irradiation induces stress and hampers plant growth (Galvão and Fankhauser, 2015; Jenkins, 2017; Shi and Liu, 2021; Podolec et al., 2021a, 2021b, 2021c). In plants, the photoreceptor UV RESISTANCE LOCUS 8 (UVR8) is known to perceive both UV-B and short-wave UV-A radiation (Kliebenstein et al., 2002; Brown et al., 2005; Favory et al., 2009; Rizzini et al., 2011; Christie et al., 2012; Tilbrook et al., 2013; Jenkins, 2014; Rai et al., 2020, 2021; Podolec et al., 2021a, 2021b, 2021c). UVR8 exists as a dimer in the ground state which upon absorption of UV-B photons, monomerizes to activate UV-B signaling (Kaiserli and Jenkins, 2007; Heijde et al., 2013; Qian et al., 2016). The UVR8 monomer binds competitively to the E3 ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) via its VP motif to inhibit targeting its substrates for degradation (Oravecz et al., 2006; Favory et al., 2009; Huang et al., 2013; Yin et al., 2015, 2016; Hayes et al., 2017; Lau et al., 2019; Ren et al., 2019; Ponnu and Hoecker, 2021). The UVR8–COP1 complex formation also stabilizes the transcription factor ELONGATED HYPOCOTYL 5 (HY5) that regulates the expression of several downstream genes involved in UV-B protection (Ulm et al., 2004; Oravecz et al., 2006; Favory et al., 2009; Huang et al., 2012; Binkert et al., 2014). UVR8 is reverted back to its inactive dimer form by the action of REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) and RUP2 (Gruber et al., 2010; Heijde and Ulm, 2013; Podolec et al., 2021a, 2021b, 2021c). UVR8 also directly binds to some transcription factors like WRKY36, BES1-INTERACTING MYC-LIKE1 (BIM1)/BRASSINAZOLE-RESISTANT 2 (BES1), and MYB73/77 to inhibit their DNA binding and transcriptional regulation (Liang et al., 2018; Yang et al., 2018; Liang et al., 2020; Yang et al., 2020). PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and PIF5 are other transcription factors that are inactivated under UV-B (Tavridou et al., 2020). In the absence of UV-B, WRKY36 inhibits the transcription of HY5 (Yang et al., 2018). However, under UV the binding of UVR8 to WRKY36 leads to the release of this inhibition leading to HY5-mediated signaling and stress tolerance (Yang et al., 2018).
A high dose of UV-B leads to the activation of a UV-B acclimation response leading to UV-B tolerance. Protection against UV-B in plants involves the accumulation of several metabolites which act as sunscreen compounds and shield plants from UV-B radiation (Yoshiyama et al., 2013). These include several flavonoids, pigments, and other phenolics that absorb UV radiation to enhance cellular protection (Hectors et al., 2014; Liang et al., 2020). UV-B induces the expression of genes like CHALCONE ISOMERASE (CHI) and CHALCONE SYNTHASE (CHS) that are involved in the synthesis of the photoprotective pigments anthocyanins (Shi and Liu, 2021). In addition to the protective metabolites, plants also mitigate UV-B stress by activating their DNA repair mechanisms to amend UV-B DNA damage (Podolec et al., 2021a, 2021b, 2021c). Protection of the photosynthetic apparatus is an integral part of UV-B acclimation and tolerance across several photosynthetic organisms, including algae (Allorent et al., 2016). UVR8 plays a crucial role in activating the phenylpropanoid pathway and synthesizing reactive oxygen species (ROS) scavenging metabolites under UV-B (Brown and Jenkins, 2008; Hideg et al., 2013). The transcription factor HY5 plays an important role in the activation of several genes that are involved in the biosynthesis of phenolic acids, anthocyanins, and induction of the DNA repair pathway (Ulm et al., 2004; Brown and Jenkins, 2008; Binkert et al., 2014).
In addition to HY5, several other transcription factors are regulated by UV-B. Notable among them are members of the B-BOX PROTEIN (BBX) family of transcriptional regulators. Recently, BBX20, BBX21, and BBX22 were identified as cofactors of HY5 that render transcriptional activation potential to HY5, which lacks an activation domain (Bursch et al., 2020). BBX proteins are zinc-finger transcription factors that play a myriad of roles in plant developmental processes including photomorphogenesis, flowering, thermomorphogenesis, and shade avoidance (Crocco and Botto, 2013; Gangappa and Botto, 2014; Vaishak et al., 2019; Xu, 2019; Yadav et al., 2020). A small subset of BBX proteins have been found to play roles in UV-B signaling and stress tolerance (Jiang et al., 2012; Bai et al., 2014; Yadav et al., 2019a, 2019b). BBX24 is a negative regulator of UV-B photomorphogenesis (Jiang et al., 2012). UV-B induces both BBX24 mRNA and protein, and the accumulated protein binds to HY5 to inhibit its transcriptional activity (Jiang et al., 2012). BBX31, which codes for a microprotein acts as a positive regulator of UV-B signaling and protection (Yadav et al., 2019a). Under UV-B, BBX31 positively regulates photomorphogenesis independent of HY5, whereas it promotes UV-B stress tolerance in an HY5-dependent manner (Yadav et al., 2019a). RUP2 delays flowering under UV-B by physically binding with CONSTANS, which happens to be BBX1, and inhibiting its association with the promoter of FLOWERING LOCUS T (Arongaus et al., 2018). A number of other UV-B-induced BBX proteins have been identified and are predicted to function in UV-B signaling (Lyu et al., 2020).
Here, we identify the role of BBX11 in the regulation of UV-B signaling and stress tolerance. UV-B induces BBX11 expression in a dose-dependent manner. BBX11 regulates hypocotyl elongation under UV-B partially dependent on HY5. Under high fluence UV-B, BBX11 promotes the accumulation of phenolic compounds and induces photoprotection genes to enhance UV-B stress tolerance. Our data indicate that HY5 directly binds to the promoter of BBX11 and positively regulates its expression under UV-B. We propose here a BBX11-HY5 feedback loop regulating UV-B photomorphogenesis and stress tolerance.
Results
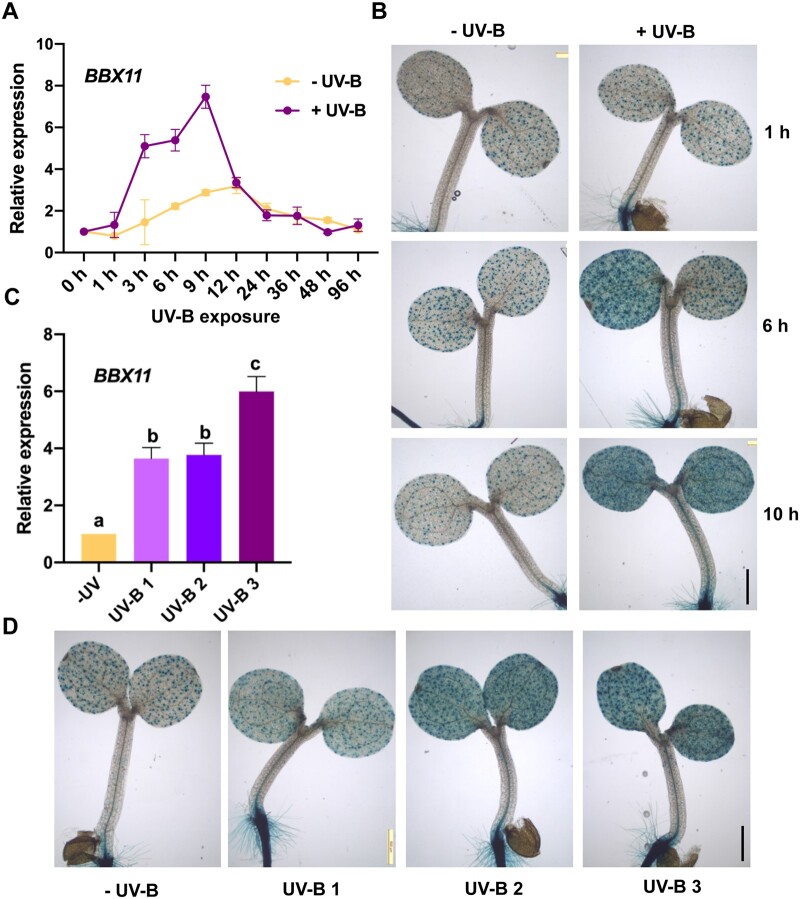
BBX11 expression is induced by UV-B
Microarray data suggest that the Arabidopsis BBX coding gene, BBX11, is induced by various fluence rates (7 W/m2 and 3 μmol m−2 s−1) of UV-B (Ulm et al., 2004; Brown and Jenkins, 2008). These reports prompted us to investigate if BBX11 plays any role in UV-B signaling. To validate the UV-B-mediated induction of BBX11, we performed RT-qPCR analysis of BBX11 expression in 5-day-old Columbia-0 (Col-0) seedlings grown in constant white light and exposed to supplemental UV-B (+UV-B) (2.2 W/m2) or not (−UV-B) for 0–96 h (Figure 1A). Under the control conditions, BBX11 transcript level marginally increased over this time period; however, UV-B treatment strongly upregulated the expression level of BBX11. BBX11 mRNA steadily increased after 3 h of UV-B treatment and peaked to about eight-fold upregulation around 9 h (Figure 1A). In order to determine if the UV-B-mediated induction of BBX11 is UVR8 dependent, we checked the expression of BBX11 in uvr8-6. Under UV-B 1 (0.9 W/m2), the mRNA levels of BBX11 in Col-0 seedlings are upregulated three- to four-fold after 3 and 8 h of exposure to UV-B (Supplemental Figure S1A). Under UV-B, BBX11 transcript levels were reduced in uvr8-6, suggesting that the UV-B-mediated induction of BBX11 is at least partially dependent on UVR8 (Supplemental Figure S1A). Under 2.2 W/m2 of UV-B, BBX11 expression in uvr8-6 was reduced at 3 h but comparable to Col-0 at 8 h (Supplemental Figure S1B). This suggests that at higher fluence and longer exposure, the UV-B-mediated induction of BBX11 might involve factors other than UVR8. We further examined if BBX12 and BBX13, which are structurally similar to BBX11 and belong to the same clade, are also induced by UV-B. BBX12 and BBX13 mRNA levels, unlike BBX11, did not show a strong upregulation when irradiated with supplemental UV-B 2 (2.2 W/m2) as compared to their levels in (−UV-B) conditions (Supplemental Figure S1C).
Figure 1.
BBX11 expression is induced by UV-B. A, RT-qPCR analysis of BBX11 expression in 5-day-old Col-0 seedlings grown in constant white light and exposed to supplemental UV-B 2 (2.2 W/m2) (+UV-B) or not (−UV-B) for the indicated time period. GAPDH was used as internal control. Error bars represent sem, n = 3 (B) Histochemical staining of 5-day-old pBBX11:GUS seedlings grown in constant white light and exposed to supplemental UV-B (+UV-B) or not (−UV-B) for the indicated durations, Scale bar- 500 μm. In (A and B) fluence of UV-B used was 2.2 W/m2 (C) RT-qPCR analysis of BBX11 expression in 5-day-old Col-0 seedlings grown in constant white light and exposed to supplemental UV-B of different doses UV-B 1 (0.91 W/m2), UV-B 2 (2.2 W/m2), UV-B 3 (3.9 W/m2), or not (−UV) for 6 h. GAPDH was used as internal control. Error bar represents sem, n = 3, Statistical groups indicated by letters were determined by one-way ANOVA, with Tukey’s test P ≤ 0.05. D, Histochemical staining of 5-day-old pBBX11:GUS seedlings grown in constant white light and exposed to supplemental UV-B 1, UV-B 2, UV-B 3, or not (−UV) for 10 h before staining. Scale bar: 500 μm.
In order to determine the spatio-temporal expression pattern of BBX11 in response to UV-B, we performed histochemical staining of 5-day-old pBBX11:GUS seedlings grown in constant white light and exposed to supplemental UV-B (+UV-B) or not (−UV-B) for 1, 6, and 10 h (Figure 1B). Under UV-B, speckled GUS staining was detected in the cotyledons as reported earlier for cWL-grown pBBX11:GUS seedlings (Figure 1B; Job and Datta, 2021). Low staining was also detected in the hypocotyl vasculature whereas high GUS staining was seen in the hypocotyl root junction (Figure 1B). Consistent with our RT-qPCR data, staining in the cotyledons was substantially higher in plants treated with UV-B for 6 and 10 h (Figure 1B). Next, we quantified the relative expression of BBX11 under various fluence levels of UV-B (0.9, 2.2, and 3.9 W/m2) (Figure 1C). For the convenience of the readers, we have denoted various fluence levels of UV-B used in different experiments throughout the manuscript as follows: UV-B 1 (near 1 W/m2), UV-B 2 (2–2.5 W/m2), UV-B 3 (3.1–4.8 W/m2). The hypocotyl elongation experiments were performed under UV-B 1, whereas in the stress experiments plants were exposed to UV-B 3. The biochemical and gene expression analysis were performed using seedlings treated with the intermediate UV-B 2. Our results indicate a gradual increase in the expression levels as the dose of UV-B is elevated (Figure 1C). We further checked the spatial expression of BBX11 under the different fluences. We found enhanced GUS staining in the cotyledons with the increase in the UV-B irradiation, indicating that UV-B induces BBX11 expression in a dose-dependent manner (Figure 1D).
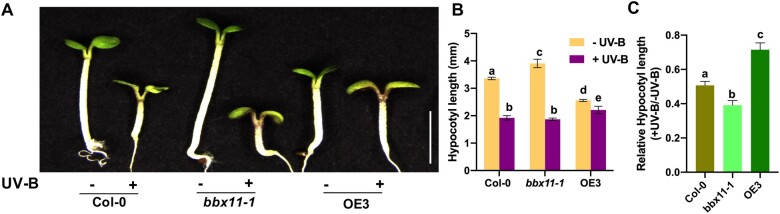
BBX11 regulates hypocotyl growth suppression under UV-B
The low dose of UV-B inhibits hypocotyl elongation in Arabidopsis (Jenkins, 2017). In order to investigate if BBX11 has any role to play in UV-B-mediated photomorphogenesis, we characterized the loss and gain of function mutants of BBX11. For loss of function mutants, we used the T-DNA insertion mutant bbx11-1 and the clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 mutants bbx11-2 and bbx11-3 generated by us and described previously (Job and Datta, 2021). We also characterized OE1, OE2, and OE3, three independent lines overexpressing BBX11, also reported in our previous study (Job and Datta, 2021). Seedlings were grown for 5 days under low fluence cWL (9 μmol m−2 s−1) without UV-B (−UV-B) and their hypocotyl lengths compared with seedlings grown under cWL with UV-B 1 supplementation (+UV-B) (Figure 2, A–C). As previously reported, under cWL bbx11 showed elongated hypocotyl length and BBX11-OE showed short hypocotyl length compared to Col-0 (Figure 2B). Under UV-B, hypocotyl growth was reduced in seedlings of all three genotypes. However, bbx11-1 showed enhanced suppression of hypocotyl length, whereas OE3 showed less inhibition in hypocotyl growth under UV-B relative to Col-0 (Figure 2, A–C). This suggests that BBX11 regulates hypocotyl growth suppression under UV-B.
Figure 2.
BBX11 regulates UV-B mediated hypocotyl growth inhibition. A–C, Representative images (A), quantification of hypocotyl lengths (B) and relative hypocotyl lengths (C) of 5-day-old seedlings of the indicated genotypes grown in constant white light supplemented with UV-B 1 (+) or not (−). Scale bar in (A) indicates 2 mm. In (B) and (C), error bars represent sem, n = 3. Statistical groups indicated by letters were determined by two-way ANOVA, with Sidaks’s multiple comparison test, P ≤ 0.05.
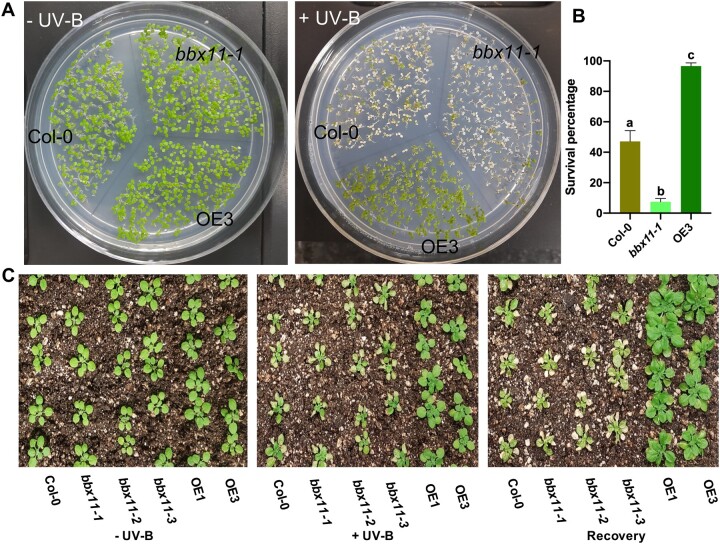
BBX11 provides protection against UV-B stress
Since higher UV-B irradiation elevates BBX11 expression, we asked if BBX11 plays any role in high UV-B stress tolerance in plants. Four-day-old Col-0, bbx11-1, and OE3 seedlings grown on plates under constant white light were irradiated with supplemental UV-B 3 (+UV-B) or not (−UV-B) for 30 h and allowed to recover under white light for 1 day before imaging (Figure 3A). We estimated the survival percentage of seedlings by counting seedlings in which the cotyledons remained green and did not bleach under UV-B radiation (Figure 3B). The survival percentage in Col-0 was close to 50%, while in bbx11-1 it was ˂10% and in OE3 almost 95% of seedlings survived (Figure 3B). This suggests that BBX11 protects plants from the damage caused by high dose of UV-B. Similar results were obtained in another independent experiment where seedlings on plates were exposed to high UV-B radiation for 20 h (Supplemental Figure S3A). In another experiment, Col-0, bbx11-1, and OE3 seedlings were initially acclimatized by growing them under constant white light supplemented with UV-B 1 for 5 days. These seedlings were then irradiated with UV-B 3 for another 24 h followed by recovery under white light for 1 day before imaging. Here, we did not find substantial difference in the stress tolerance phenotype between the three genotypes (Supplemental Figure S3B). In order to investigate the effect in older plants, we treated plants grown in the soil under white light for 18 days with supplemental UV-B 3 (+UV-B) or not (−UV-B) for 3 days and allowed them to recover under white light for 5 days. After the recovery period, the Col-0 and bbx11-1 plants showed signs of bleaching, with slightly more chlorotic patches in the mutant compared to the wild-type (Figure 3C). However, the OE3 plants showed enhanced tolerance to bleaching (Figure 3C). Together, these data suggest that BBX11 promotes tolerance to high doses of UV-B in plants.
Figure 3.
BBX11 protects plants from UV-B radiation. A, 4-day-old Col-0, bbx11-1, and OE3 seedlings grown on plates under constant white light were treated with supplemental UV-B 3 (+UV-B) or not (−UV-B) for 30 h and allowed to recover under white light for 1 day before imaging. B, quantification of seedling survival percentage, calculated as mentioned in methods. Error bars represent sem, n = 4, statistical groups indicated by letters were determined by one-way ANOVA, with Tukey’s test P ≤ 0.05. C, Plants of the indicated genotypes were grown in soil under long-day conditions (16/8 h) in white light for 18 days and exposed to supplemental UV-B 3 (+UV-B) or not (−UV-B) for 3 days and allowed to recover under white light for 5 days before being photographed.
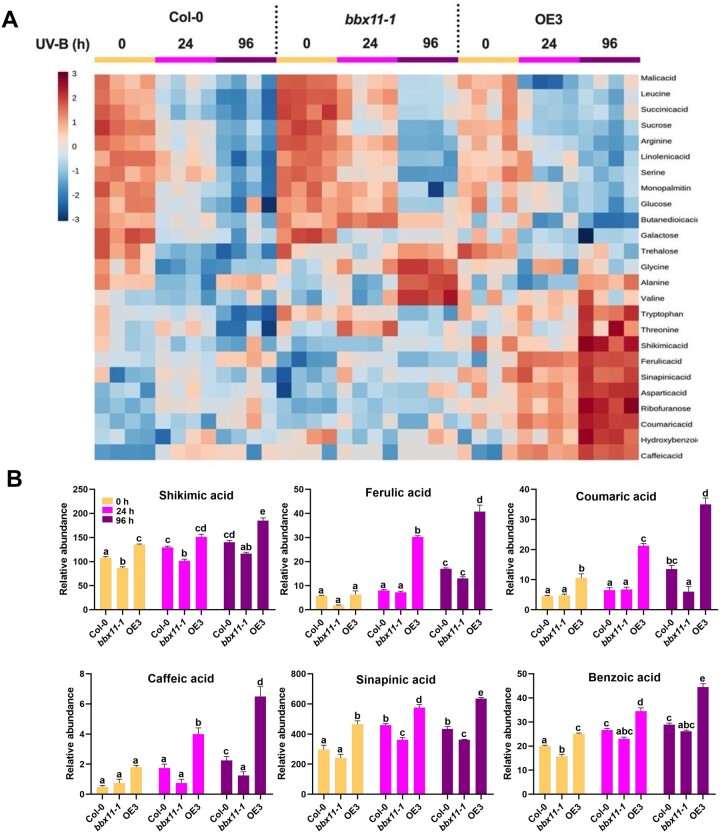
BBX11 promotes accumulation of photo-protective phenolic compounds
To determine if the enhanced tolerance of plants overexpressing BBX11 could be due to elevated levels of known or unknown protective metabolites we compared the metabolome of UV-B treated and untreated Col-0, bbx11-1, and OE3 plants. Untargeted metabolic profiling of 18-day-old plants grown under constant white light and exposed to 0, 24, and 96 h of supplemental UV-B radiation captured 379 metabolic features showing differential accumulation in the three genotypes. Partial least squares-discriminant analysis revealed that the metabolomes of Col-0, bbx11-1, and OE3 plants were altered differently after exposure to UV-B radiation for 24 and 96 h (Supplemental Figure S4A). We used comprehensive cluster analysis to visualize the response of metabolites under white light (0 h) and the two durations of UV-B treatments (24 and 96 h) in the three genotypes as a heat map (Figure 4A). The top 25 metabolites showing maximum variability among the genotypes in the absence (0 h) or presence of UV-B for 24 and 96 h were identified by this approach (Figure 4A). These include several amino acids, sugars, organic acids, carboxylic, fatty acids, and phenolic acids. OE3 substantially accumulated the intermediate metabolites of the phenylpropanoid biosynthesis pathway under UV-B treatment (Figure 4A). The accumulation of phenolics in Col-0, bbx11-1, and OE3 genotypes showed much variation after 96 h of UV-B treatment (Figure 4A). The relative accumulation of phenolic acids, for example, shikimic acid, sinapinic acid, cinnamic acid, vanillic acid, coumaric acid, ferulic acid, caffeic acid, hydroxybenzoic acid, and benzoic acid, was significantly higher in OE3 (Figure 4B; Supplemental Figure S4B). These phenolics play a crucial role in conferring UV-B stress tolerance. In our pBBX11:GUS line, we could also detect enhanced staining in the trichomes which are sites of secondary metabolite synthesis and storage (Supplemental Figure 4C). NitroBlue Tetrazolium (NBT) staining suggested reduced accumulation of superoxide radicals in OE3 seedlings irradiated with high fluence UV-B (Supplemental Figure S4D). All these data together suggest that BBX11 promotes the accumulation of photoprotective phenolic compounds and antioxidants to enhance protection against UV-B radiation.
Figure 4.
BBX11 promotes accumulation of photo-protective metabolites. A, Heat map showing the relative accumulation of central and secondary metabolites and (B) relative accumulation of phenolic acids, in 18-day-old seedlings of the indicated genotypes grown in constant white light and exposed to 0, 24, and 96 h of supplemental UV-B 3 before harvesting. In (A) heat map scale (+3 to −3) indicates abundance of metabolite. Four biological replicates were taken for the analyses from each of the treatments. In (B) error bars represent sem, n = 4. Statistical groups indicated by letters were determined by one-way ANOVA, with Tukey’s test P ≤ 0.05.
HY5 regulates the UV-B-mediated induction of BBX11 by binding to its promoter
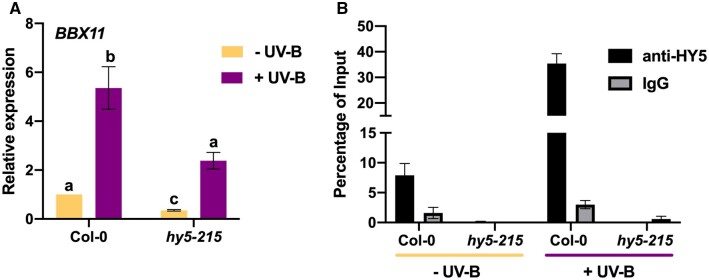
We further went on to determine the role of BBX11 in the UV-B signaling pathway. In a previous study BBX11 was identified as one of the genes that is induced by UV-B in WT seedlings but not in the hy5 mutant (Brown and Jenkins, 2008). In order to verify the HY5-dependent induction of BBX11 under UV-B, we checked the transcript levels of BBX11 in Col-0 and hy5-215 (Figure 5A). While there was five-fold upregulation in the mRNA levels of BBX11 in the WT seedlings after UV-B treatment, we could find just over two-fold upregulation in hy5 seedlings (Figure 5A). This suggests that HY5 positively regulates the expression of BBX11 under UV-B. HY5 has been previously shown to physically bind to the BBX11 promoter fragment containing the G-box variant, in seedlings grown under white light (Zhao et al., 2020; Job and Datta, 2021). In order to verify whether similar binding also happens under UV-B, we performed Chromatin Immunoprecipitation (ChIP) followed by qPCR on seedlings grown under WL +/− UV-B and compared the enrichment of HY5 on the BBX11 promoter (Figure 5B). Our results indicate that HY5 binding on the BBX11 promoter in Col-0 seedlings is almost five-fold enriched under +UV-B conditions (Figure 5B). Such enrichment was not seen when chromatin from hy5 seedlings (negative control) was used for a similar experiment (Figure 5B). We also did not find any enrichment of HY5 on a fragment from the 3′-untranslated region of BBX11 (Supplemental Figure S5A). All these data together suggest that under UV-B, the binding of HY5 to the promoter of BBX11 is increased, which leads to the enhanced expression of BBX11.
Figure 5.
HY5 regulates the UV-B-mediated induction of BBX11 by directly binding to its promoter. A, RT-qPCR analysis of BBX11 expression in 5-day-old Col-0 and hy5-215 seedlings grown in constant white light and exposed to supplemental UV-B 2 (+) or not (−) for 6 h. GAPDH was used as internal control. Error bars represent sem, n = 3. Statistical groups indicated by letters were determined by one-way ANOVA, with Tukey’s test, P ≤ 0.05 (B) ChIP-qPCR analyses of HY5 binding to the BBX11 promoter in vivo. Fourteen-day-old Col-0 and hy5-215 seedlings grown in constant white light and exposed to supplemental UV-B 3 (+) or not (−) for 3 h before crosslinking. DNA–protein complexes were immunoprecipitated using antibodies against HY5 (anti-HY5) and rabbit IgG as negative control (IgG). ChIP DNA was quantified by RT-qPCR with primers specific to the previously known HY5-binding site. Error bars represent sd, n = 2.
BBX11 and HY5 regulate UV-B photomorphogenesis and stress tolerance in a partially interdependent manner
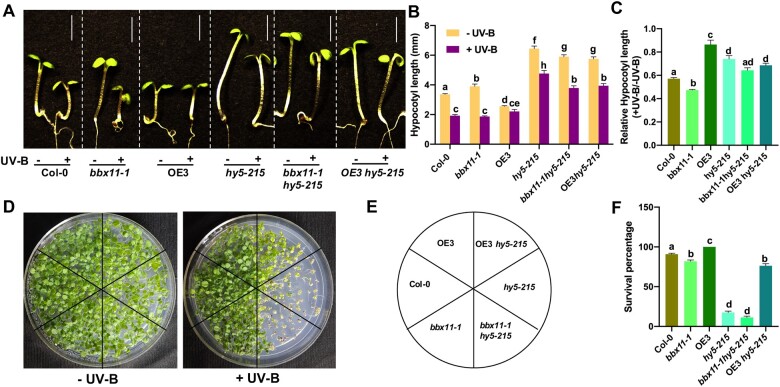
HY5 positively regulates photomorphogenesis and stress tolerance under UV-B (Brown and Jenkins, 2008; Binkert et al., 2014). Our results have indicated that BBX11 also regulates UV-B photomorphogenesis and stress tolerance. In order to understand the genetic interaction between BBX11 and HY5, we characterized bbx11-1hy5-215 and OE3hy5-215 grown under UV-B. In our experimental conditions, the average hypocotyl length of bbx11 seedlings under UV-B was around 1.9 mm, whereas it was around 4.8 mm for hy5 (Figures 2 and 6, A–C). The hypocotyl length of bbx11-1hy5215 seedlings under UV-B was around 3.8 mm (Figure 6B). The relative hypocotyl length in +UV-B/−UV-B for bbx11-1hy5215 was almost similar to hy5-215 (Figure 6C). The OE3hy5-215 seedlings grown in +UV-B conditions, exhibited hypocotyl length intermediate between OE3 and hy5-215 (Figure 6B), however, the relative hypocotyl length of OE3hy5-215 was similar to hy5-215 (Figure 6C). This suggests that BBX11 regulates hypocotyl elongation under UV-B partially dependent on HY5.
Figure 6.
BBX11 acts partially independent of HY5 to regulate photomorphogenesis and stress tolerance under UV-B. A–C, Representative images (A) quantification of hypocotyl lengths (B) and relative hypocotyl lengths (C) of 5-day-old seedlings of the indicated genotypes grown in constant white light supplemented with UV-B 1 (+) or not (−). Scale bars in (A) indicate 2 mm. In (B) and (C), error bars represent sem, n = 3. Statistical groups indicated by letters were determined by one-way ANOVA, with Tukey’s test (multiple comparison for (B), P ≤ 0.05 (D, E) 5-day-old seedlings of the indicated genotypes as shown in (E) grown in constant white light were exposed to supplemental UV-B 3 (+UV-B) or not (−UV-B) for 20 h and allowed to recover under white light for 10 days before imaging. F, quantification of seedling survival percentage, calculated as mentioned in methods. Error bars represent sem, n = 3, Statistical groups indicated by letters were determined by one-way ANOVA, with Tukey’s test, P ≤ 0.05.
To investigate the genetic interaction between BBX11 and HY5 to regulate UV-B stress tolerance we irradiated 5-day-old white light (WL)-grown seedlings of all genotypes indicated in Figure 6E with UV-B-3 for 20 h following which seedlings were allowed to recover for 10 days before imaging (Figure 6, D–F). Under these conditions, the survival % was about 90% in Col-0. bbx11-1 showed reduced survival, whereas OE3 exhibited enhanced survival compared to WT (Figure 6F). hy5-215 and bbx11-1hy5-215 seedlings showed ˂10% survival under the UV-B stress. However, under these conditions the survival percentage of OE3hy5-215 seedlings enhanced to around 75%. This suggests that BBX11 might be partially dependent on HY5 to provide protection against UV-B stress. The incomplete rescue of the hy5 phenotype by BBX11 OE also suggests that there might be other targets of HY5 regulating the UV-B acclimation and stress tolerance response.
Overexpression of BBX11 induces photoprotection and DNA damage repair genes
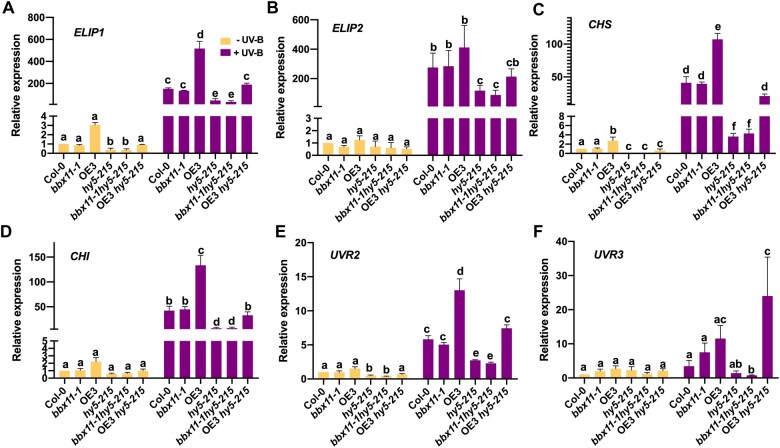
Since overexpression of BBX11 could rescue the bleaching phenotype of hy5 under moderate UV-B stress, we hypothesized that BBX11 might act downstream of HY5 in the same pathway and regulate similar photoprotective genes. HY5 is known to regulate the EARLY LIGHT-INDUCABLE PROTEIN (ELIP) genes which code for the early light-induced proteins that prevent photooxidative damage in plants. HY5 also induces the expression of genes involved in the synthesis of anthocyanins such as CHI, CHS, and FLAVANONE 3-HYDROXYLASE (F3H). Additionally, the UVR2 and UVR3 gene coding for DNA damage repair protein is activated by HY5 under UV-B. The mRNA expression levels of all these genes under UV-B were downregulated in hy5 as reported earlier (Figure 7, A–F). The transcript level of the six genes were upregulated in the BBX11 overexpressing line OE3, while it was similar to WT in bbx11-1 and almost equal to hy5-215 in the bbx11hy5-215 double mutant (Figure 7, A–F). Interestingly, in plants overexpressing BBX11 in the hy5-215 background the mRNA levels of ELIP1, CHS, CHI, UVR2, and UVR3 were enhanced compared to the levels detected in hy5-215. The transcript level of F3H was upregulated in OE3 compared to Col-0 but mRNA levels in OE3hy5-215 were similar to that in hy5-215 (Supplemental Figure S6). The upregulation of these photoprotective genes can contribute to the enhanced tolerance to UV-B seen in OE3hy5-215 seedlings. This suggests that the overexpression of BBX11 can induce photoprotection and DNA damage repair genes to enhance UV-B stress tolerance.
Figure 7.
BBX11 induces photoprotection and DNA damage repair genes under UV-B. A–F, RT-qPCR analysis of ELIP1 (A) ELIP2 (B), CHS (C), CHI (D), UVR2 (E), and UVR3 (F) in 5-day-old seedlings grown in constant white light and exposed to supplemental UV-B 2 (+) or not (−) for 4 h. GAPDH was used as internal control. Error bars represent sem, n = 3. Statistical groups indicated by letters were determined by one-way ANOVA, with Tukey’s test, P ≤ 0.05.
The HY5-BBX11 feedback loop regulates UV-B photomorphogenesis and stress tolerance
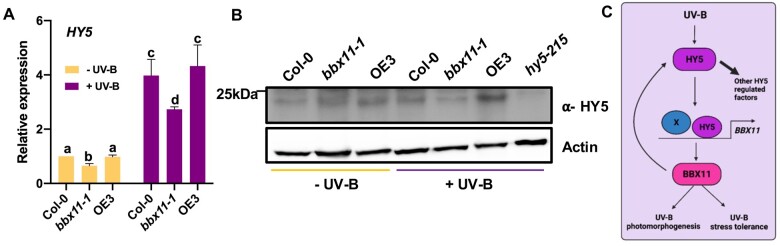
Previously, we found that HY5 activates BBX11 transcription by binding to its promoter under UV-B (Figure 5). Under UV-B stress conditions (UV-B 3), BBX11 acts downstream of HY5 to promote stress tolerance (Figure 6, D–F). However, OE3hy5-215 seedlings exhibit photomorphogenesis phenotypes similar to hy5 (Figure 6, A and B). This suggests that HY5 can also act downstream of BBX11. Together these indicate that a feedback loop between HY5 and BBX11 might be operating under UV-B to modulate responses. Since several BBX proteins are known to regulate transcription, we asked if BBX11 might regulate HY5 transcription under UV-B. We quantified the relative expression of HY5 in Col-0, bbx11-1 and OE3 exposed to supplemental UV-B 2 (+UV-B) or not (−UV-B). The HY5 transcript level in bbx11-1 was reduced compared to WT under both conditions, suggesting that BBX11 might modulate HY5 transcription (Figure 8A). The HY5 mRNA levels were not significantly different between the WT and OE3 in −UV-B as well as +UV-B conditions (Figure 8A). We further went on to check if BBX11 modulates HY5 protein accumulation under UV-B. Our Western blot data indicate that HY5 protein accumulation is suppressed in bbx11-1 under UV-B (Figure 8B). On the other hand there is enhanced accumulation of HY5 in OE3 in presence of UV-B (Figure 8B). Such a difference in the accumulation of HY5 protein between the loss and gain of function mutants of BBX11 was not evident in −UV-B conditions. Overall, our data suggest that under UV-B, BBX11 might be regulating the transcriptional and protein levels of HY5, thereby forming a feedback loop to regulate plant responses under UV-B (Figure 8C).
Figure 8.
HY5-BBX11 feedback loop regulates photomorphogenesis and stress tolerance under UV-B. A, qRT-PCR analysis of HY5 expression in 5-day-old seedlings of the indicated genotypes grown in constant white light and exposed to supplemental UV-B 2 (+) or not (−) for the 4 h. GAPDH was used as internal control. Error bars represent sem, n = 3. Statistical groups indicated by letters were determined by one-way ANOVA, with Tukey’s test, P ≤ 0.05. B, Immunoblot analysis of HY5 and actin (loading control) levels in Col-0, bbx11-1, and OE3 seedlings exposed to 6 h of supplemental UV-B 2 (+UV-B) or not (−UV-B). C, Working model indicating the role of an HY5-BBX11 feedback loop in regulating UV-B photomorphogenesis and stress tolerance. “X” indicates an unknown factor that might regulate BBX11 transcription in addition to HY5 under UV-B.
Discussion
BBX11 regulates UV-B photomorphogenesis
BBX11 has been previously characterized as a positive regulator of photomorphogenesis under red light (Zhao et al., 2020; Job and Datta, 2021; Liu et al., 2021). BBX11 directly interacts with phytochrome B (phyB) and PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and promotes phyB-mediated degradation of PIF4 under red light to inhibit hypocotyl elongation (Song et al., 2021). The inhibition of PIF4 activity might suppress hypocotyl elongation in the BBX11 overexpressing line OE3 in the absence of UV-B (Figure 2, A and B). Here we report the role of BBX11 in regulating UV-B mediated hypocotyl growth inhibition. bbx11-1 showed enhanced suppression of hypocotyl length while the over-expressor exhibited less inhibition of hypocotyl length under UV-B (Figure 2C). This might suggest that BBX11 negatively regulates UV-B mediated hypocotyl growth inhibition. However, it can be argued that the “enhanced suppression of hypocotyl length” in the mutant can be a result of the elongated hypocotyl of the mutant in −UV-B conditions (Figure 2). Moreover, the enhanced accumulation of flavanols in OE3 could reduce the effective dose of UV-B exposure, which could be responsible for the slightly elongated hypocotyl of this genotype under UV-B (Figure 2, A and B). Based on these arguments it may be concluded that BBX11 promotes UV-B photomorphogenesis and stress tolerance. Previously, another BBX protein BBX31 has been reported to positively regulate photomorphogenesis and stress tolerance under UV-B (Yadav et al., 2019a). BBX24, the third BBX protein known to play a role in UV-B signaling, negatively regulate photomorphogenesis under white light as well as under UV-B (Jiang et al., 2012). Our results also indicate that BBX11 regulates UV-B-mediated photomorphogenesis partially dependent on HY5. Although the induction of BBX11 by UV-B is compromised in hy5, it is not completely reduced to the −UV-B levels (Figure 5A). This indicates that there can be HY5-independent mechanisms functioning through BBX11. Our pBBX11:GUS lines indicated that BBX11 is strongly expressed in the guard cells of the cotyledons. However upon UV-B treatment for longer durations (6 and 10 h) or high intensity (UV-B 2 and UV-B 3), the blue staining becomes more diffused throughout the cotyledon as indicated in Figure 1, B and D. In the future it would be interesting to study the physiological role of BBX11 expression in the guard cells both under visible light and UV-B.
BBX11 protects plants from UV-B radiation
High UV-B irradiation causes stress and leads to UV-B acclimation and tolerance response in plants. The molecular and metabolic response of plants to protect themselves from damaging UV-B radiation involves the induction of genes and accumulation of metabolites offering photoprotection. Plants exposed to high UV-B irradiation often resort to the following strategies: (1) Accumulate photoprotective sunscreen compounds to shield the radiation exposure of cellular DNA and other macromolecules; (2) Detoxify the radiation-induced ROS that causes lipid and protein peroxidation; and (3) Repair damaged DNA. In our study, we found that plants overexpressing BBX11 exhibit enhanced UV-B tolerance. Our metabolomic analysis indicated differential accumulation of several primary and secondary metabolites in OE3 and bbx11-1 compared to Col-0. These include amino acids, sugars, and several phenolic acids. Glycine, Alanine, and Valine accumulated at high levels in bbx11-1, whereas the levels of Tryptophan and Tyrosine were higher in OE3 after 96 h of UV-B exposure. The two aromatic amino acids might play roles in the synthesis of pigments, hormones, and polyphenolic compounds that help shielding the plants from UV-B radiation. Trehalose showed an increased accumulation in the less tolerant bbx11-1 mutant. The role of sugars in UV-B acclimation response needs further investigation. Several phenolic acids like shikimic acid, sinapinic acid, coumaric acid, ferulic acid, caffeic acid, and benzoic acid showed higher accumulation in the UV-tolerant OE3 plants. Most of these phenolics play critical roles in the synthesis of cell wall components and act as intermediates in the phenylpropanoid pathway leading to the synthesis of UV-B absorbing flavonoids and pigments. In the future, it might be interesting to study the tissue-specific localization of these sunscreen metabolites. Previously we reported the role of BBX11 in the regulation of protochlorophyllide levels and chlorophyll synthesis during deetiolation. Our gene expression analysis also indicated that the transcript levels of the anthocyanin biosynthesis genes CHS and CHI are upregulated in plants overexpressing BBX11. We detected reduced NBT staining in OE3 plants indicating the reduced accumulation of superoxides in these lines. The mRNA levels of ELIP1 and ELIP2 were also upregulated in OE3 suggesting enhanced photoprotection activities. The increased expression of UVR2 and UVR3 might indicate that DNA repair pathways are also activated in plants overexpressing BBX11 contributing to UV-B acclimation and tolerance.
HY5-BBX11 feedback loop regulates UV-B responses
BBX proteins function as critical cofactors of HY5 regulating HY5 activity (Bursch et al., 2020). Some BBX factors physically interact with HY5 to modulate its transcriptional potential whereas some others form a BBX-HY5 feedback loop (Job et al., 2018; Zhao et al., 2020). A BBX11-BBX21-HY5-positive feedback loop operates under white light to regulate photomorphogenesis (Zhao et al., 2020). We found that HY5 directly binds to the promoter of BBX11 and activates its expression under UV-B (Figure 5). Furthermore, we detected enhanced HY5 accumulation in seedlings overexpressing BBX11 under UV-B (Figure 8B). All our results together indicate the operation of a BBX11-HY5 feedback loop to regulate photomorphogenesis and stress tolerance under UV-B (Figure 8). The loop adds partial interdependence to the partially independent functions of the two transcription factors to regulate UV-B photomorphogenesis and stress tolerance. Under UV-B stress conditions, overexpression of BBX11 in hy5 could partially rescue the acute bleaching phenotype of hy5 (Figure 6, D–F). This suggests that BBX11 can at least partially activate the tolerance response even in the absence of HY5. BBX11 might regulate a subset of HY5-modulated photoprotective genes/metabolites that can provide partial tolerance. However, as HY5 acts as a master regulator with several targets, the total repertoire of HY5-regulated genes and metabolites might be required to enhance tolerance. While the accumulation of the metabolites is required for the long-term acclimation response the activation of the genes is generally an early response. The mRNA levels of HY5 peak early (1.5–3 h) after exposure to UV-B radiation (Supplemental Figure S7). During these early hours, BBX11 transcript levels also increase constantly and reach a peak at around 9 h after initial exposure (Figure 1A). Our results indicate that BBX11 activates genes like CHS, CHI, ELIPs, UVR2, UVR3 within 4 h after UV-B exposure. This suggests that BBX11 might be acting as a coactivator of HY5 for activating these genes in response to UV-B exposure. It is interesting to note that under UV-B, the mRNA levels of BBX11 exhibit only a partial reduction in hy5. This suggests the role of some additional factor (X) in regulating BBX11 transcription under UV-B (Figure 8C). In white light, HY5 and BBX21 are known to bind directly to the promoter of BBX11 and positively regulate its transcription. A recent study indicates that BBX20, BBX21, and BBX22 might act as functional partners of HY5 during prolonged UV-B treatment (Podolec et al., 2021a, 2021b, 2021c). In the future, it might be interesting to investigate how different BBX proteins partner with HY5 and enable it to multitask under different conditions including UV-B exposure. The 5 times enhanced binding of HY5 on the BBX11 promoter under UV-B is interesting but lacks molecular explanation at the moment. Any possible role of UVR8, COP1, and RUPs in BBX11 mediated UV-signaling is also left to be explored. Nonetheless, this work introduces BBX11 as a key component of plant’s molecular armory to respond to UV-B radiation.
Materials and methods
Plant material
All overexpression lines and mutants used in this study are in the Arabidopsis Col-0 ecotype. The mutants bbx11-1, bbx11-2, bbx11-3, OE1, OE3, bbx11-1hy5-215, and OE3hy5-215 are described previously (Job and Datta, 2021). hy5-215 and uvr8-6 have been described in previous studies (Datta et al., 2006; Favory et al., 2009).
Growth condition and light treatment
Seeds were surface sterilized with 4% w/v bleach for 3 min and washed with sterilized water before sowing on 0.5× Murashige and Skoog (MS) medium containing 0.8% agar and 1% (w/v) sucrose. Plates were stratified at 4°C for 3 days under dark and illuminated with white light (80 μmol m−2 s−1) for 8 h to induce uniform germination before putting them out under experimental conditions. Plants were grown at 21°C in Percival growth chambers under low fluence constant white light (9 μmol m−2 s−1) unless otherwise stated. UV-B supplementation was provided through the UV-B narrowband tube (TL 20W/01 RS SLV; waveband between 305 and 315 nm and λmax 311 nm). Intensity of UV-B radiation was measured using a UV light meter (UVA/UVB 850009; Sper Scientific, Scottsdale, AZ, USA). The fluence and duration of treatment used in various experiments have been mentioned in the respective experiments.
Histochemical GUS staining
The pBBX11:GUS line has been reported previously (Job and Datta, 2021). Histochemical GUS staining was carried out as described previously (Weigel and Glazebrook, 2002). Briefly, 5-day-old seedlings were kept in 90% acetone on ice and rinsed with GUS staining solution (0.5-M sodium phosphate buffer, 10% Triton X-100, 100-mM potassium Ferrocyanide, 100-mM potassium Feriicyanide, and 2-mM X-Gluc). Vacuum infiltration of samples for 30 min under dark on ice was performed following which the seedlings were incubated at 37°C overnight. Seedlings were washed with 20%, 35%, and 50% ethanol, fixed using FAA solution, and kept in 70% ethanol for imaging. The seedlings were documented using Leica (DM2500) microscope.
ChIP
ChIP assay was done as described previously (Yadav et al., 2019a). Briefly, 14-day old seedlings were treated with the indicated light conditions for 3 h before cross-linking with 1% formaldehyde, and the chromatin was isolated. Purified chromatin was sonicated and precipitated with anti-HY5 (Agresera, Vannas, Sweden; 1:100) or IgG antibody (negative control; Invitrogen, Waltham, MA, USA), washed, and reversed cross-linked. The eluted DNA fragments were used in qPCR with the primers listed in Supplemental Table S1.
Hypocotyl length measurement
Seeds were sterilized, stratified, and uniformly germinated as indicated above. For hypocotyl length measurements seeds were grown for 5 days under constant white light (9 μmol m−2 s−1) or together with UV-B 1 (0.94 W/m2) supplementation. Approximately 30 seedlings of each genotype were photographed with Leica (M165 FC), and hypocotyl length was measured with ImageJ software. Three independent biological replicates (n = 3) with at least 30 seedlings each were analyzed for all hypocotyl length assays.
UV-B stress tolerance
For UV-B stress tolerance analysis, 4- to 5-day-old seedlings grown on 0.5 MS without sucrose plates under constant white light (20 μmol m−2 s−1) were treated with supplemental UV-B 3 (4.2 or 4.8 W/m2) or not (−UV-B) for the time indicated and allowed to recover under white light for durations as mentioned in respective figure legends before imaging. Seedling survival rate was measured by scoring green, expanded, and healthy seedlings as positive and pale green/bleached seedlings as negative. An average of 50 seedlings per genotype were analyzed per condition/replicate. For UV-B stress analysis in adult plants, plants were grown in soil under long-day conditions (16/8 h) in white light for 18 days and exposed to supplemental UV-B 3 (3.9 W/m2) or not (−UV-B) for 3 days and allowed to recover under white light for 5 days before being photographed.
Metabolite extraction and derivatization for metabolomics using GC–MS (Gas Chromatography-Mass Spectrometry)
For comprehensive metabolomics, 20 mg/mL of lyophilized tissue was used for metabolite extraction as described previously (Yadav et al., 2019a; Lingwan and Masakapalli, 2021). The extraction buffer was prepared in a ratio of 3:1:1 by mixing up prechilled methanol, chloroform, and water, respectively; 0.01-mg/mL ribitol was added to the extraction buffer as an internal standard. The extracted metabolites were subjected to drying using a speed vac. Samples were TMS (TriMethylSilyl) derivatized using methoxamine hydrochloride, Pyridine, and N-Methyl-N-trimethylsilyl trifluoroacetamide (Lisec et al., 2006). The derivatized samples were subjected to data acquisition using Agilent GC–MS 5977B equipped with 5% phenyl methyl siloxane column. Mass Hunter software was used for all data analysis. The metabolite was identified based on their fragments, retention time, and probability match against the NIST 17 library. Metaboanalyst version 5.0 was used for multivariate statistical analysis (Pang et al., 2021).
NBT staining
For superoxide anion detection, seedlings were stained in freshly prepared NBT solution (0.2% solution in 50-mM sodium phosphate buffer, pH 7.5) overnight with gentle shaking. The seedlings were washed and decolorized with bleaching solution (3 (ethanol):1 (acetic acid):1 (glycerol)) and imaged using Leica (M165 FC) Microscope.
RNA isolation and RT-qPCR analysis
For BBX11, BBX12, BBX13, and HY5 mRNA expression analysis, 5-day-old seedlings grown under white light were treated with supplemental UV-B (2.2 W/m2) for the indicated time. For dose-dependent BBX11 expression, analysis seedlings were grown under white light for 5 days and treated with supplemental UV-B 1 (0.91 W/m2), UV-B 2 (2.2 W/m2), and UV-B 3 (3.9 W/m2) for 6 h before harvesting the samples for RNA isolation. Total RNA was isolated using the TRIzol (Invitrogen, Waltham, MA, USA) method. The samples were immediately frozen in liquid nitrogen after the UV-B treatment. The tissue was grounded into a fine powder, and 1 mL of TRIzol reagent was added immediately and mixed by vortexing. Extraction of total RNA was performed as manufactures protocol. Purified RNA was quantified, and the cDNA library was made using Takara Primescipt first-strand cDNA synthesis Kit according to manufactures instructions. Three-fold diluted cDNA was used for RT-qPCR using SYBR Green (Takara Shiga, Japan) in Roche LC 96 qPCR machine. GAPDH was used as the internal control.
Total protein isolation and western blot
Five-day-old seedlings grown under continuous white light (60 μmol m−2 s−1) were treated with supplemental UV-B (3.1 W/m2) before freezing in liquid N2. The tissue was ground into a fine powder, and total protein was extracted in extraction Buffer (50-mM Tris–HCl pH 6.8, 0.5% SDS (Sodium Dodecyl Sulfate) w/v, 10% glycerol, 150-mM NaCl, 2 M Urea) with 50 μM MG132 (Sigma, St Louis, MO, USA) and 1× proteinase inhibitor cocktail (Sigma). The protein extract was cleared by centrifugation at 12,000 g for 20 min, and equal amounts of proteins were separated in 12% SDS–PAGE. Anti HY5 (Agresera, Vännäs, Sweden; 1:500) and anti-ACTIN (Sigma; 1:1,000) were used to perform western blot.
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 8.0 software. Details of the analysis are given in the respective figure legends.
Accession numbers
BBX11 (AT2G47890), HY5 (AT5G11260), CHS (AT5G13930), CHI (AT3G55120), ELIP1 (AT3G22840), ELIP2 (AT4G14690), UVR2 (AT1G12370), and UVR3 (AT3G15620).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. UV-B mediated induction of BBX11 is partially dependent on UVR8 whereas BBX12 and BBX13 mRNA expression is not induced by UV-B.
Supplemental Figure S2. Relative hypocotyl length of loss- and gain-of-function mutants of BBX11 under UV-B.
Supplemental Figure S3. Overexpression of BBX11 enhances protection against UV-B stress.
Supplemental Figure S4. BBX11 modulates accumulation of photoprotective secondary metabolites.
Supplemental Figure S5. HY5 directly binds to the promoter of BBX11.
Supplemental Figure S6. Expression of F3H is upregulated in OE3 seedlings overexpressing BBX11.
Supplemental Figure S7. HY5 is an early UV-B inducible gene.
Supplemental Table S1. List of primers used in this study.
Supplementary Material
Acknowledgments
The authors acknowledge the assistance provided by Shubhi Dwivedi and Arundhati Khenwar for some experiments. The authors thank Yadukrishnan Prem and Henrik Johansson for their critical reading of the manuscript and providing valuable comments and the PCDB lab members especially Arpan Mukherjee and Debojyoti Kar for valuable comments and helping to revise the final manuscript.
Funding
N.J. acknowledges DST-INSPIRE for PhD fellowship. S.D. acknowledges DBT (BT/HRD/NBA-NWB/39/2020-21 (8) and CSIR 38(1510)/21/EMR-II for funding.
Conflict of interest statement. The authors declare that there is no conflict of interest. Some of the results reported here have been incorporated as part of a patent application where S.D. and N.J. are the inventors.
Contributor Information
Nikhil Job, Department of Biological Sciences, Indian Institute of Science Education and Research-Bhopal, Bhopal 462066, Madhya Pradesh, India.
Maneesh Lingwan, BioX School of Basic Sciences, Indian Institute of Technology-Mandi, Mandi 175005, Himachal Pradesh, India.
Shyam Kumar Masakapalli, BioX School of Basic Sciences, Indian Institute of Technology-Mandi, Mandi 175005, Himachal Pradesh, India.
Sourav Datta, Department of Biological Sciences, Indian Institute of Science Education and Research-Bhopal, Bhopal 462066, Madhya Pradesh, India.
S.D. and N.J. conceived the project. N.J. planned and performed all the experiments except for the metabolomics experiments, which were performed by M.L. under the supervision of S.K.M. S.D. wrote the manuscript with help from N.J. and M.L. All the authors analyzed the data and helped prepare the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Sourav Datta (sdatta@iiserb.ac.in).
References
- Allorent G, Lefebvre-Legendre L, Chappuis R, Kuntz M, Truong TB, Niyogi KK, Ulm R, Goldschmidt-Clermont M (2016) UV-B photoreceptor-mediated protection of the photosynthetic machinery in Chlamydomonas reinhardtii. Proce Natl Acad Sci USA 113: 14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arongaus AB, Chen S, Pireyre M, Glöckner N, Galvão VC, Albert A, Winkler JB, Fankhauser C, Harter K, Ulm R (2018) Arabidopsis RUP2 represses UVR8-mediated flowering in noninductive photoperiods. Genes Dev 32: 1332–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai SL, Saito T, Honda C, Hatsuyama Y, Ito A, Moriguchi T (2014) An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 240: 1051–1062 [DOI] [PubMed] [Google Scholar]
- Binkert M, Kozma-Bognár L, Terecskei K, De Veylder L, Nagy F, Ulm R (2014) UV-B-Responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26: 4200–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102: 18225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Jenkins GI (2008) UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 146: 576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch K, Toledo-Ortiz G, Pireyre M, Lohr M, Braatz C, Johansson H (2020) Identification of BBX proteins as rate-limiting cofactors of HY5. Nat Plants 6: 921–928 [DOI] [PubMed] [Google Scholar]
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocco CD, Botto JF (2013) BBX proteins in green plants: insights into their evolution, structure, feature and functional diversification. Gene 531: 44–52 [DOI] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi G, Deng XW, Holm M (2006) Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C (2015) Sensing the light environment in plants: photoreceptors and early signaling steps. Curr Opin Neurobiol 34: 46–53 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19: 460–470 [DOI] [PubMed] [Google Scholar]
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R (2010) Negative feedback regulation of UV-B–induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA 107: 20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Sharma A, Fraser DP, Trevisan M, Cragg-Barber CK, Tavridou E, Fankhauser C, Jenkins GI, Franklin KA (2017) UV-B perceived by the UVR8 photoreceptor inhibits plant thermomorphogenesis. Curr Biol 27: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hectors K, Van Oevelen S, Geuns J, Guisez Y, Jansen MAK, Prinsen E (2014) Dynamic changes in plant secondary metabolites during UV acclimation in Arabidopsis thaliana. Physiol Plant 152: 219–230 [DOI] [PubMed] [Google Scholar]
- Heijde M, Binkert M, Yin R, Ares-Orpel F, Rizzini L, Van De Slijke E, Persiau G, Nolf J, Gevaert K, De Jaeger G, et al. (2013) Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc Natl Acad Sci USA 110: 20326–20331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M, Ulm R (2013) Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci USA 110: 1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideg É, Jansen MAK, Strid Å (2013) UV-B exposure, ROS, and stress: inseparable companions or loosely linked associates? Trends Plant Sci 18: 107–115 [DOI] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Yang P, Lau OS, Chen L, Wei N, Deng XW (2013) Conversion from CUL4-based COP1–SPA E3 apparatus to UVR8–COP1–SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci USA 110: 16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Yang P, Lau OS, Li G, Li J, Chen H, Deng XW (2012) Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24: 4590–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI (2014) The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI (2017) Photomorphogenic responses to ultraviolet-B light. Plant Cell Environ 40: 2544–2557 [DOI] [PubMed] [Google Scholar]
- Jiang L, Wang Y, Li QF, Bjorn LO, He JX, Li SS (2012) Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res 22: 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job N, Datta S (2021) PIF3/HY5 module regulates BBX11 to suppress protochlorophyllide levels in dark and promote photomorphogenesis in light. New Phytologist 230: 190–204 [DOI] [PubMed] [Google Scholar]
- Job N, Yadukrishnan P, Bursch K, Datta S, Johansson H (2018) Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol 176: 2963–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserli E, Jenkins GI (2007) UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B–specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lim JE, Landry LG, Last RL (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol 130: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K, Podolec R, Chappuis R, Ulm R, Hothorn M (2019) Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. EMBO J 38: e102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Mei S, Shi C, Yang Y, Peng Y, Ma L, Wang F, Li X, Huang X, Yin Y, Liu H (2018) UVR8 Interacts with BES1 and BIM1 to Regulate Transcription and Photomorphogenesis in Arabidopsis. Dev Cell 44: 512–523.e515 [DOI] [PubMed] [Google Scholar]
- Liang T, Shi C, Peng Y, Tan H, Xin P, Yang Y, Wang F, Li X, Chu J, Huang J, Yin Y, Liu H (2020) Brassinosteroid-activated BRI1-EMS-SUPPRESSOR 1 inhibits flavonoid biosynthesis and coordinates growth and UV-B stress responses in plants. Plant Cell 32: 3224–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwan M, Masakapalli SK (2021) A robust method of extraction and GC-MS analysis of Monophenols exhibited UV-B mediated accumulation in Arabidopsis. Physiol Mol Biol Plants 28: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat Protocol 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Liu B, Long H, Yan J, Ye L, Zhang Q, Chen H, Gao S, Wang Y, Wang X, Sun S (2021) A HY5-COL3-COL13 regulatory chain for controlling hypocotyl elongation in Arabidopsis. Plant Cell Environ 44: 130–142 [DOI] [PubMed] [Google Scholar]
- Lyu G, Li D, Li S (2020) Bioinformatics analysis of BBX family genes and its response to UV-B in Arabidopsis thaliana. Plant Signal Behav 15: 1782647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám E, Schäfer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques P-É, Li S, Xia J (2021) MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 49: W388–W396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolec R, Demarsy E, Ulm R (2021a) Perception and signaling of ultraviolet-B radiation in plants. Ann Rev Plant Biol 72: 793–822 [DOI] [PubMed] [Google Scholar]
- Podolec R, Lau K, Wagnon TB, Hothorn M, Ulm R (2021b) A constitutively monomeric UVR8 photoreceptor confers enhanced UV-B photomorphogenesis. Proc Natl Acad Sci 118: e2017284118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolec R, Wagnon TB, Leonardelli M, Johansson H, Ulm R (2021c) BBX proteins promote HY5-mediated UVR8 signaling in Arabidopsis. bioRxiv: 2021.2010.2014.464399 10.1101/2021.10.14.464399 [DOI] [Google Scholar]
- Ponnu J, Hoecker U (2021) Illuminating the COP1/SPA ubiquitin ligase: fresh insights into its structure and functions during plant photomorphogenesis. Front Plant Sci 12: 662793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Mao W, Liu Y, Ren H, Lau On S, Ouyang X, Huang X (2016) Dual-source nuclear monomers of UV-B light receptor direct photomorphogenesis in Arabidopsis. Mol Plant 9: 1671–1674 [DOI] [PubMed] [Google Scholar]
- Rai N, Morales LO, Aphalo PJ (2021) Perception of solar UV radiation by plants: photoreceptors and mechanisms. Plant Physiol 186: 1382–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai N, O'Hara A, Farkas D, Safronov O, Ratanasopa K, Wang F, Lindfors AV, Jenkins GI, Lehto T, Salojärvi J, et al. (2020) The photoreceptor UVR8 mediates the perception of both UV-B and UV-A wavelengths up to 350 nm of sunlight with responsivity moderated by cryptochromes. Plant Cell Enviro 43: 1513–1527 [DOI] [PubMed] [Google Scholar]
- Ren H, Han J, Yang P, Mao W, Liu X, Qiu L, Qian C, Liu Y, Chen Z, Ouyang X, et al. (2019) Two E3 ligases antagonistically regulate the UV-B response in Arabidopsis. Proc Natl Acad Sci USA 116: 4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 Protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Shi C, Liu H (2021) How plants protect themselves from ultraviolet-B radiation stress. Plant Physiol 187: 1096–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Heng Y, Bian Y, Xiao Y, Liu J, Zhao X, Jiang Y, Deng XW, Xu D (2021) BBX11 promotes red light-mediated photomorphogenic development by modulating phyB-PIF4 signaling. aBIOTECH 2: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavridou E, Pireyre M, Ulm R (2020) Degradation of the transcription factors PIF4 and PIF5 under UV-B promotes UVR8-mediated inhibition of hypocotyl growth in Arabidopsis. Plant J 101: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K, Arongaus AB, Binkert M, Heijde M, Yin R, Ulm R (2013) The UVR8 UV-B photoreceptor: perception, signaling and response. Arabidopsis Book 11: e0164–e0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Ádám É, Oakeley EJ, Schäfer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishak KP, Yadukrishnan P, Bakshi S, Kushwaha AK, Ramachandran H, Job N, Babu D, Datta S (2019) The B-box bridge between light and hormones in plants. J Photochem Photobiol B-Biol 191: 164–174 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Xu D (2020) COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol 228: 1748–1753 [DOI] [PubMed] [Google Scholar]
- Yadav A, Bakshi S, Yadukrishnan P, Lingwan M, Dolde U, Wenkel S, Masakapalli SK, Datta S (2019a) The B-box-containing microprotein miP1a/BBX31 regulates photomorphogenesis and UV-B protection. Plant Physiol 179: 1876–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A, Lingwan M, Yadukrishnan P, Masakapalli SK, Datta S (2019b) BBX31 promotes hypocotyl growth, primary root elongation and UV-B tolerance in Arabidopsis. Plant Signal Behav 14: e1588672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A, Ravindran N, Singh D, Rahul PV, Datta S (2020) Role of Arabidopsis BBX proteins in light signaling. J Plant Biochem Biotechnol 29: 623–635 [Google Scholar]
- Yang Y, Liang T, Zhang L, Shao K, Gu X, Shang R, Shi N, Li X, Zhang P, Liu H (2018) UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat Plants 4: 98–107 [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang L, Chen P, Liang T, Li X, Liu H (2020) UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J 39: e101928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Arongaus AB, Binkert M, Ulm R (2015) Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV-B signaling in Arabidopsis. Plant Cell 27: 202–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Skvortsova MY, Loubéry S, Ulm R (2016) COP1 is required for UV-B–induced nuclear accumulation of the UVR8 photoreceptor. Proc Natl Acad Sci USA 113: E4415–E4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama KO, Sakaguchi K, Kimura S (2013) DNA damage response in plants: conserved and variable response compared to animals. Biology 2: 1338–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Heng Y, Wang X, Deng XW, Xu D (2020) A positive feedback loop of BBX11–BBX21–HY5 promotes photomorphogenic development in Arabidopsis. Plant Commun 1: 100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.