Abstract
Burkholderia cepacia has emerged as an important pathogen in patients with cystic fibrosis. Many gram-negative pathogens regulate the production of extracellular virulence factors by a cell density-dependent mechanism termed quorum sensing, which involves production of diffusible N-acylated homoserine lactone signal molecules, called autoinducers. Transposon insertion mutants of B. cepacia K56-2 which hyperproduced siderophores on chrome azurol S agar were identified. One mutant, K56-R2, contained an insertion in a luxR homolog that was designated cepR. The flanking DNA region was used to clone the wild-type copy of cepR. Sequence analysis revealed the presence of cepI, a luxI homolog, located 727 bp upstream and divergently transcribed from cepR. A lux box-like sequence was identified upstream of cepI. CepR was 36% identical to Pseudomonas aeruginosa RhlR and 67% identical to SolR of Ralstonia solanacearum. CepI was 38% identical to RhlI and 64% identical to SolI. K56-R2 demonstrated a 67% increase in the production of the siderophore ornibactin, was protease negative on dialyzed brain heart infusion milk agar, and produced 45% less lipase activity in comparison to the parental strain. Complementation of a cepR mutation restored parental levels of ornibactin and protease but not lipase. An N-acylhomoserine lactone was purified from culture fluids and identified as N-octanoylhomoserine lactone. K56-I2, a cepI mutant, was created and shown not to produce N-octanoylhomoserine lactone. K56-I2 hyperproduced ornibactin and did not produce protease. These data suggest both a positive and negative role for cepIR in the regulation of extracellular virulence factor production by B. cepacia.
The phenomenon of quorum sensing is a regulatory mechanism that is involved in the control of cell density-dependent expression of many bacterial phenotypes (21, 55, 71). Quorum sensing, or autoinduction, is the process of producing and responding to high intracellular concentrations of N-acylhomoserine lactones (N-acyl-HSLs), which bind to specific proteins that regulate the transcription of selected genes. This process was first reported to control the bioluminescence (lux) phenotype in the marine organism Vibrio fischeri (42). In V. fischeri, the two components necessary for cell density-dependent lux expression are the LuxR and LuxI proteins (15, 16). The LuxI protein is required for the synthesis of the autoinducer N-(3-oxohexanoyl)-l-HSL (14). When present in sufficient amounts, the freely diffusible signaling molecule binds to LuxR, which activates the lux genes. The threshold concentration of autoinducer necessary for the induction of bioluminescence is attained when cultures achieve a sufficiently high cell density (for reviews, see references 21, 55, and 71).
Quorum sensing has since been shown to regulate the production of virulence factors in several gram-negative species (21, 55, 71), including the opportunistic pathogen Pseudomonas aeruginosa (47). Quorum sensing in P. aeruginosa involves two unique systems, lasRI and rhlRI (6, 31, 45, 47). The las system is composed of the transcriptional activator LasR and the autoinducer N-(3-oxododecanoyl)-l-HSL (22, 48). LasR activates the expression of elastase (lasB), alkaline protease (aprA), LasA protease (lasA), exotoxin A (toxA), the type II secretion apparatus (xcpP through xcpZ) and the autoinducer synthase lasI (7, 22, 47, 60, 74). The rhl system is composed of the RhlR transcriptional activator and the autoinducer N-butyryl-l-HSL (43, 49). RhlR activates the expression of rhamnolipids (rhlAB), elastase (lasB), lipase (lipA), the stationary-phase sigma factor gene rpoS, and other genes (6, 28, 31, 32, 44, 45, 50). These two systems form a hierarchical quorum-sensing cascade in which LasR regulates the expression of rhlR (32, 51). There is considerable overlap within this dual-level control system in the regulation of elastase and the alkaline protease (6, 22, 32).
Burkholderia cepacia (previously Pseudomonas cepacia) is an important pathogen in patients with cystic fibrosis (23). Twenty percent of cystic fibrosis patients infected with B. cepacia suffer from cepacia syndrome, a necrotizing pneumonia with fever and occasionally bacteremia (27). This condition leads to a rapid and fatal pulmonary decline and is a unique clinical outcome in comparison to respiratory infections with other pathogens. Most cystic fibrosis patients infected with B. cepacia are coinfected with P. aeruginosa (73). Due to the genetic conservation of quorum-sensing regulatory elements and similarities in the structure of N-acyl-HSLs, the potential for cell-to-cell communication between different species exists. McKenney et al. provided some evidence of cell-to-cell communication between B. cepacia and P. aeruginosa (36). Culture fluids from B. cepacia demonstrated autoinducer activity in several autoinducer bioassays. B. cepacia produces several extracellular virulence factors, including protease (37), lipase (33), and four types of siderophores: salicylic acid, ornibactin, pyochelin, and cepabactin (40, 65, 67, 69). The addition of concentrated culture fluids from P. aeruginosa stationary-phase cultures to B. cepacia cultures increased the production of siderophores, protease, and lipase, suggesting the presence of a quorum-sensing system (36). In the present study, we report the identification of the LuxRI homologs, CepRI, and an N-octanoyl-HSL autoinducer in B. cepacia. We also present evidence for the involvement of this quorum-sensing system in the regulation of siderophore and protease production.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. B. cepacia K56-2 was originally isolated from the sputum of a cystic fibrosis patient. K56-2 belongs to genomovar III (76) and contains the B. cepacia epidemic strain marker and the cable pilus gene (cblA) (35, 54, 66). This strain produces the siderophores ornibactin, salicylic acid, and negligible amounts of pyochelin and does not produce cepabactin (9).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 56 |
| SM10 | Mobilizing strain; RP4 tra genes integrated in chromosome; Kmr | 62 |
| MG4 | Δ(argF-lac)U169 zah-735::Tn10 recA56 srl::Tn10 | 53 |
| HB101 | supE44 hsdS20(rB mB) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 56 |
| VJS533 | recΔA56 ara Δ(lac-proAB)X111 rpsL φ80dΔ(lacZ)M15 | 70 |
| B. cepacia | ||
| K56-2 | Cystic fibrosis respiratory isolate | 9 |
| K56-R2 | cepR::Tn5-OT182 derivative of K56-2 | This study |
| K56-I2 | cepI::tmp derivative of K56-2 | This study |
| R. solanacearum AW1-A18 | solI8::SP | 19 |
| Plasmids | ||
| pOT182 | pSUP102(GM)::Tn5-OT182 Cmr Gmr Apr Tcr | 38 |
| pNOT19 | Modified pUC19 cloning vector; Apr | 57 |
| pUCP28T | Broad-host-range vector; IncP OriT pRO1600 ori Tpr | 58 |
| p34E-Tp | Source of tmp cassette; Tpr | 11 |
| pRK2013 | ColE1 Tra (RK2)+ Kmr | 18 |
| pEX18Tc | Suicide vector; sacB Tcr | 26 |
| pCRR2.1TOPO | Cloning vector for PCR products | Invitrogen |
| pSLR2-1 | 11.3-kb ClaI fragment from K56-R2 obtained by self-cloning; Tcr | This study |
| pSLR2-2 | 11.5-kb XhoI fragment from K56-R2 obtained by self-cloning; Tcr | This study |
| pSLA3.2 | pUCP28T with 3.2-kb SphI fragment from K56-2 containing cepIR genes; Tpr | This study |
| pSLR100 | pUCP28T with 1.65-kb KpnI-SphI fragment from pSLA3.2 containing cepR gene; Tpr | This study |
| pSLS201 | pNOT19 with 1.55-kb SphI-KpnI fragment from pSLA3.2 containing cepI gene; Apr | This study |
| pSLS201-T | pNOT19 with 2.2-kb fragment containing cepI gene inactivated with tmp cassette; Apr Tpr | This study |
| pEXCEPI | PCR product (inactivated cepI fragment from pSLS201-T) in pEX18Tc; Tcr Tpr | This study |
| pKDT17 | lasB::lacZ plac-lasR Apr | 48 |
| pHV2001− | luxR luxI′CDABE Apr | 49 |
| p395B | aidA::lacZ Tcr | 19 |
| pECP61.5 | rhlR rhlA::lacZ Apr | 51 |
For genetic manipulations, Escherichia coli DH5α and B. cepacia K56-2 were grown at 37°C in Luria-Bertani (LB) (Life Technologies, Burlington, Ontario, Canada) or Bacto-Terrific broth or agar plates (Difco, Detroit, Mich.). The following amounts of antibiotics (per milliliter) were used when necessary: 100 μg of ampicillin, 15 μg of tetracycline, 25 μg of kanamycin, 25 μg of chloramphenicol, and 1.5 mg of trimethoprim for E. coli and 300 μg of tetracycline, 100 μg of streptomycin, and 100 μg of trimethoprim for B. cepacia. A 100-mg/ml stock solution of trimethoprim was prepared in N,N-dimethyl-acetamide. For ornibactin production, protease, and chrome azurol S (CAS) assays, cultures were grown in succinate medium (39) at 37°C. For salicylic acid assays, cultures were grown in CAA medium (65) at 37°C. For lipase assays, cultures were grown in Anwar defined medium at 37°C (1). For β-galactosidase assays, cultures were grown in Trypticase soy broth medium (Difco) at 37°C. For all N-acyl-HSL bioassays and for partial purification of N-acyl-HSLs, B. cepacia cultures were grown for 24 h (stationary phase) in Trypticase soy broth adjusted to a pH of 7.0 at 30°C with shaking (200 rpm).
Tn5-OT182 mutagenesis and allelic exchange in B. cepacia.
For transposon mutagenesis, Tn5-OT182 (38) was transferred into K56-2 from SM10(pOT182) by conjugation. The cultures were mixed (100 μl of each), and cells were pelleted by centrifugation. The cells were resuspended in 0.1 ml of phosphate-buffered saline and spotted onto sterile 0.45 μm-pore-size nitrocellulose filters on LB agar plates containing 10 mM MgSO4 and incubated for 4 h at 37°C. The donor and recipient strains were also spotted individually as described above for controls. The filters were washed with 1 ml of sterile phosphate-buffered saline, and 100 μl was plated on LB containing 300-μg/ml tetracycline, 100-μg/ml streptomycin, and 50 μM FeCl3. Tetracycline- and streptomycin-resistant transconjugants were identified after incubation for 36 to 48 h at 37°C. Transconjugants were screened for siderophore hyperproduction on CAS plates (59). Mutants that produced zones larger than that of the parent after 2 to 3 days incubation were selected for further selection.
For allelic exchange, a K56-2 insertion mutant in cepI was constructed by using the suicide vector pEX18Tc containing the counterselectable marker sacB (26). The plasmid pSLS201-T (see Fig. 1B) contains a 2.25-kb fragment encoding cepI that was insertionally inactivated with a tmp cassette. The tmp cassette was isolated by AccI digestion of p34E-Tp (11), blunt ended by using DNA polymerase I Klenow fragment (Life Technologies), and cloned into a blunt-ended AccI site within the cepI reading frame. The inactivated cepI region was amplified by PCR from pSLS201-T and cloned into pEX18Tc (pEXCEPI). Triparental matings were performed to transfer pEXCEPI from E. coli DH5α to B. cepacia K56-2 by using pRK2013 as the mobilizing plasmid. Transconjugants were plated onto Pseudomonas isolation agar (Difco) plates containing 100-μg/ml trimethoprim to select for single crossover events in B. cepacia. Tpr transconjugants were streaked for isolated colonies on LB agar plates containing 100-μg/ml trimethoprim and 5% sucrose to select for double crossover events and excision of the plasmid. The insertional inactivation of cepI was confirmed by Southern hybridization.
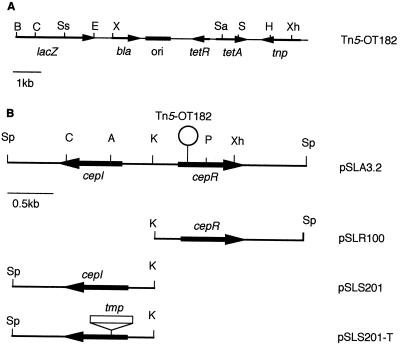
FIG. 1.
Physical and genetic map of Tn5-OT182 and various cepIR constructs. (A) Tn5-OT182 (38). The arrows represent the orientation and position of genes, and the black box represents the pBR325 origin of replication. Designations and abbreviations: lacZ, promoterless β-galactosidase reporter gene; bla, β-lactamase gene; ori, origin of replication; tetRA, gene encoding tetracycline resistance determinant; tnp, gene encoding transposase; B, BamHI; C, ClaI; Ss, SstI; E, EcoRI; X, XmnI; Sa, SalI; S, StuI; H, HindIII; Xh, XhoI. (B) The cepIR locus from B. cepacia (pSLA3.2). The arrows represent the location and orientation of genes, and the “lollipop” represents the site of transposon insertion. Designations and abbreviations: pSLR100, 1.65-kb cepR subclone in pUCP28T; pSLS201, 1.55-kb cepI subclone in pNOT19; pSLS201-T, trimethoprim cassette (tmp) introduced into pSLS201; cepI, gene encoding autoinducer synthase; cepR, gene encoding transcriptional activator; Sp, SphI; C, ClaI; A, AccI; K, KpnI; P, PstI; Xh, XhoI.
DNA manipulations.
Molecular biology techniques were performed as generally described by Sambrook et al. (56). Restriction enzymes, agarose, and molecular mass markers were purchased from Life Technologies. T4 DNA ligase was purchased from Promega Corp. (Madison, Wis.). Genomic DNA was isolated as described by Ausubel et al. (2). DNA fragments were separated on 0.7 to 1.5% agarose gels in Tris-borate or Tris-acetate buffer and purified with GeneClean II (Bio 101). For Southern hybridization analysis, restriction endonuclease digests of genomic DNA were transferred to GeneScreen Plus membranes (Dupont Canada, Mississauga, Ontario, Canada) and hybridization was performed at 65°C in 15 ml of 1% sodium dodecyl sulfate–10% dextran sulfate–salmon sperm DNA (0.1 mg/ml) according to the manufacturer’s recommendations. The blots were dried and subjected to autoradiography at −70°C, using Kodak X-Omat AR film. Colony hybridizations were performed as previously described (77). Recombinant plasmids were introduced into E. coli and B. cepacia by electroporation using a Gene Pulser (Bio-Rad, Richmond, Calif.) as previously described (10). PCR products were cloned by using the TOPO TA cloning system according to the manufacturer’s recommendations (Invitrogen, Carlsbad, Calif.).
For the self-cloning of flanking DNA from Tn5-OT182 mutants, approximately 5 μg of genomic DNA was digested with appropriate restriction enzymes, boiled for 5 min, ethanol precipitated, and resuspended in 60 μl of distilled H2O. Twenty μl of this suspension was ligated in a 25-μl reaction volume overnight at 12°C, and 2 μl was used to electroporate E. coli DH5α. For the cloning of the cepIR region, K56-2 subgenomic DNA libraries were created by cloning SphI-digested sucrose gradient fractions that reacted with probes consisting of self-cloned flanking DNA in Southern hybridization analysis into pUCP28T.
Nucleotide sequencing.
Nucleotide sequencing was performed with the ABI PRISM DyeDeoxy Termination Cycle Sequencing System with AmpliTaq DNA polymerase (Perkin-Elmer Corp.) and an ABI 1371A DNA sequencer by the University Core DNA Services (University of Calgary). The oligonucleotide OT182-LT (5′-GATCCTGGAAAACGGGAAAG-3′) was used to initiate DNA sequence reactions with plasmids obtained from Tn5-OT182 mutants by self-cloning. A primer walking strategy was employed for extended sequencing of recombinant plasmids. The nucleotide sequence of both DNA strands was determined. Custom oligonucleotides were synthesized by the University of Calgary Core DNA Services or Life Technologies. Analysis of the sequence was performed with PC/Gene software (Intelligenetics, Mountain View, Calif.). The BLASTX and BLASTN programs were used to search the nonredundant sequence database for homologous sequences (34).
Siderophore production assays.
Siderophore activity was measured by CAS assays (59). On CAS agar, siderophores remove iron from the CAS dye complex, resulting in a blue-to-orange color change in zones surrounding the colonies. The same dye complex was used to quantitate siderophore activity in culture supernatant fluid by measuring the increase in orange color at A630. CAS assays were performed on 100 μl of supernatant fluid. The A630 was measured and divided by the A600 to normalize for cell density, and this ratio was reported as CAS activity.
Ornibactin production was assayed as previously described (9). Briefly, the supernatant fluid from 100-ml cultures was lyophilized, extracted with methanol, and applied to a Sephadex LH-20 column (35 by 1.5 cm; Pharmacia) with methanol as the eluting solvent. Four-milliliter fractions were collected and assayed for iron-binding activity. Fractions containing CAS activity were pooled, and the total ornibactin amounts were estimated by the CAS assay.
For salicylic acid production, 50 ml of culture fluid was adjusted to pH 2.5 and extracted with 20 ml of ethyl acetate. The ethyl acetate layer was concentrated, and salicylic acid was isolated by thin-layer chromatography on Silica Gel G as previously described (67). All glassware for siderophore assays was washed with 2.4 M HCl and rinsed with deionized water to remove iron. All reagents were made with water purified by the Milli-Q System (Millipore, Missisauga, Ontario, Canada).
Protease and lipase assays.
For protease assays, cultures were grown overnight, normalized to an optical density at 600 nm (OD600) of 0.3, and spotted (3 μl) onto dialyzed brain heart infusion (D-BHI) agar–1.5% D-BHI milk (68). The plates were incubated for 2 days at 37°C and examined for clear zones surrounding the colonies.
Lipase activity was assayed as previously described by Lonon et al. (33). Cultures were assayed for lipase activity throughout growth. The reaction mixture consisted of 0.5 ml of concentrated supernatant, 0.15 ml of 10% Tween 20, 0.1 ml of 1 M CaCl2, and 2.3 ml of Tris buffer (pH 7.6). After 2 h of incubation at 37°C, the increase in turbidity (OD400) was measured. One unit of lipase activity is defined as an OD400 of 0.01.
Detection of N-acyl-HSLs from B. cepacia culture fluid.
N-acyl-HSLs were extracted from clarified culture fluid twice with equal volumes of acidified ethyl acetate as described elsewhere (47–49), and four different bioassays were employed to screen for N-acyl-HSLs. Each assay was selective for molecules with different acyl side chain lengths. The V. fischeri autoinducer assay (48), with E. coli VJS533 (pHV200I−), shows greatest sensitivity to C6-acyl-HSLs, particularly N-(3-oxohexanoyl)-HSL; the P. aeruginosa las assay (48), with E. coli MG4 (pKDT17), shows greatest sensitivity to N-(3-oxododecanoyl)-HSL; and the P. aeruginosa rhl assay (51), with E. coli DH5α (pECP61.5), shows greatest sensitivity to N-butyryl-HSL. The fourth bioassay used Ralstonia solanacearum containing p395B (19). This construct contains an N-acyl-HSL-dependent aidA-lacZ fusion. The R. solanacearum bioassay shows greatest sensitivity to N-octanoyl-HSL. None of the assays show absolute N-acyl-HSL specificity. They each respond to other autoinducers with greatly reduced sensitivity. For this assay, an overnight culture was grown in BG broth (19) plus tetracycline (10 μg/ml) and spectinomycin (10 μg/ml). The culture was diluted to an OD600 of 0.1 in fresh BG broth, and 0.5 ml of the diluted cell suspension was incubated with culture fluid extracts at 30°C with shaking. After a 5-h incubation, β-galactosidase activity was measured. Synthetic N-octanoyl-HSL (14) was used to construct a standard curve. We extracted 500 ml of culture fluid, concentrated the ethyl acetate extract to 1 ml, and tested an amount of ethyl acetate extract equivalent to 5 ml of culture fluid. Based on standard curves and the amount of extract tested, we should have been able to detect any of the following compounds at a culture fluid concentration of 0.5 to 1 nM or more: N-butyryl-HSL, N-hexanoyl-HSL, N-(3-oxohexanoyl)-HSL, N-octanoyl-HSL, N-(3-oxooctanoyl)-HSL, N-decanoyl-HSL, N-(3-oxodecanoyl)-HSL, N-dodecanoyl-HSL, and N-(3-oxododecanoyl-HSL).
Identification of B. cepacia N-acyl-HSL.
The procedure for characterizing the B. cepacia N-acyl-HSL is based on those previously described for identification of the P. aeruginosa autoinducers (48, 49). Cells were separated from the fluid of a 2-liter culture by centrifugation, and the culture fluid was extracted twice with equal volumes of acidified ethyl acetate. The extract was concentrated by rotary evaporation at 40 to 45°C and fractionated by C18 reverse-phase high-performance liquid chromatography (HPLC). The activity, as measured by the R. solanacearum bioassay (see above), was eluted as a sharp peak at 61 to 63% methanol in a linear 20-to-100% gradient of methanol and water. Fractions constituting this peak were pooled, concentrated by rotary evaporation, and subjected to a further separation by HPLC in 48% methanol in water. The active fractions were concentrated and analyzed by gas chromatography-mass spectrometry (GC-MS) as described previously (49).
Nucleotide sequence accession number.
The nucleotide sequences of the cepIR genes have been deposited in GenBank and assigned accession no. AF019654.
RESULTS
Isolation of B. cepacia siderophore hyperproduction mutants.
The objective of this study was to identify regulatory components involved in the control of siderophore production in B. cepacia. The suicide plasmid pOT182 containing the transposon Tn5-OT182 (Fig. 1A) (38) was introduced into B. cepacia K56-2 by conjugation. Tn5-OT182 contains an E. coli origin of replication which facilitates the cloning of DNA adjacent to the Tn. Sequencing of the cloned chromosomal DNA allows the identification of the interrupted gene without the construction of genomic libraries.
Approximately 1,350 Tcr Smr transconjugants from four independent mutagenesis experiments were screened on CAS agar for mutants altered in siderophore production. Orange zones are formed around colonies that produce siderophores on this medium due to the removal of iron from the blue CAS dye-iron complex. Mutants that produced zones larger than parental zones were selected for further characterization.
One mutant, K56-R2, which produced CAS zones approximately 50% larger than the parent (Table 2) is described in this study. Southern hybridization analysis was performed to confirm the presence of a unique Tn5-OT182 insertion in the chromosome (data not shown) and to map restriction endonuclease sites in the region of the chromosome flanking the Tn. Genomic DNA from K56-R2 was digested with ClaI or XhoI to produce fragments that contained the origin of replication and the Tcr determinant as well as chromosomal DNA flanking the Tn. Plasmids pSLR2-1 and pSLR2-2 were obtained from self-cloning of ClaI- and XhoI-digested DNA, respectively, from K56-R2. The OT182-LT primer is specific to the ends of the transposon and was used to perform cycle sequencing reactions on these plasmids. Approximately 300 to 400 bp of sequence was obtained per reaction and used to search the nonredundant protein sequence database by using the local alignment search tool BLASTX on the National Center for Biotechnology Information website. The sequences flanking the transposon showed sequence similarity to a number of members of the LuxR family of transcriptional regulators (21).
TABLE 2.
Effect of a cepR mutation on siderophore, protease, and lipase productiona
| Strain | Genotype | Supernatant CAS activity (A630/A600) | CAS agar zone diam (mm) | Ornibactinb (μg/ml/A600) | Salicylic acid (μg/ml/A600) | Protease zone radius (mm) | Lipase activity (U/ml/A600) |
|---|---|---|---|---|---|---|---|
| K56-2 | Wild type | 1.74 ± 0.30 | 9.5 ± 0.5 | 49.8 ± 4.2 | 0.54 ± 0.10 | 5.67 ± 0.29 | 0.53 ± 0.02 |
| K56-R2c | cepR::Tn5 | 2.47 ± 0.20c | 13.7 ± 0.3c | 83.1 ± 1.1c | 0.49 ± 0.04 | 0c | 0.29 ± 0.05c |
All values are means ± standard deviations of triplicate experiments unless indicated otherwise.
Mean ± standard deviation of duplicate experiments.
Significantly different from parent K56-2 in unpaired t test (P < 0.05).
Cloning of B. cepacia luxRI gene homologs cepRI.
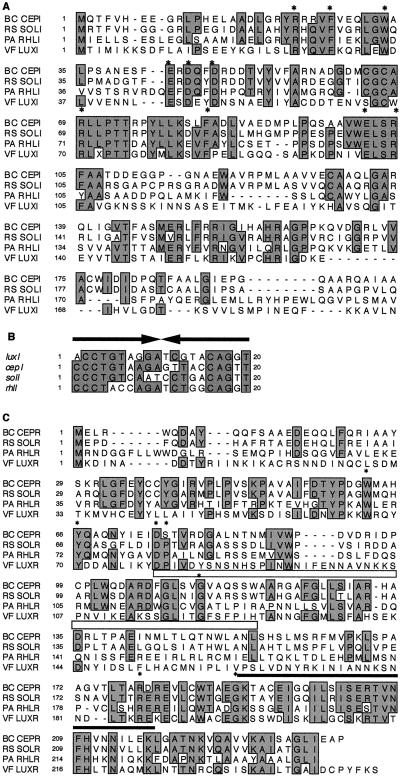
A subgenomic library consisting of K56-2 SphI fragments that were approximately 3 to 4 kb in size was constructed in E. coli. To detect the clone that contained the specific cepR fragment, a 2.0-kb XhoI-ClaI fragment derived from pSLR2-1 was used as a probe in colony hybridization analysis. Plasmids were isolated from those clones that reacted to the probe. The plasmid pSLA3.2 (Fig. 1B) contained a 3.2-kb SphI fragment that was sequenced and found to encode two complete open reading frames (ORFs), designated cepI and cepR. The cepI ORF encodes a protein with 202 amino acids and a predicted molecular weight of 22,263. CepI showed greatest homology with gene products from Ralstonia (formerly Burkholderia) and Pseudomonas spp. The putative cepI gene product has 64% identity and 70% similarity to R. solanacearum SolI (19), 38% identity and 52% similarity to P. aeruginosa RhlI (45), and 28% identity and 39% similarity to V. fischeri LuxI (12, 17, 24). The amino acid alignment of these amino acid sequences is shown in Fig. 2A. CepI contains each of the 10 amino acids that are conserved among all LuxI family members (46). A lux box-like sequence was identified upstream of cepI, matching the consensus lux box in 15 of 20 positions (25). The lux box-like sequence in the cepI promoter is aligned with the proposed lux box sequences from the promoters of luxI, solI, and rhlI in Fig. 2B.
FIG. 2.
Multiple alignments of amino acid sequences from LuxIR family members and of lux box promoter elements. (A) Amino acid alignment of various LuxI family members with B. cepacia CepI (BC CEPI) generated by using the programs PC/Gene CLUSTAL and SeqVu. Boxed, shaded regions highlight amino acids conserved in at least three of the proteins. The 10 invariant amino acids characteristic of LuxI homologs are denoted with asterisks (46). Additional sequences shown are those of proteins abbreviated as follows: RS SOLI, R. solanacearum SolI (accession no. AF021840 [19]); PA RHLI, P. aeruginosa RhlI (accession no. U11811 [45]); and VF LUXI, V. fischeri LuxI (accession no. 225903 [12]). (B) Comparison of lux box sequences in the promoter regions of LuxI homologs. The sequences shown are from luxI (V. fischeri [12]), cepI (B. cepacia), solI (R. solanacearum [19]), and rhlI (P. aeruginosa [31]). The black arrows represent the inverted repeats of the palindrome sequences. Boxed, shaded regions highlight nucleotides that are identical in at least three of the sequences. (C) Amino acid alignment of various LuxR family members with B. cepacia CepR (BC CEPR) generated by using the programs PC/Gene CLUSTAL and SeqVu. Boxed, shaded regions highlight amino acids that are identical in three of the four proteins. The open bar below LuxR residues 79 to 127 represents the autoinducer binding domain (61). The solid bar above CepR residues 190 to 217 represents the putative helix-turn-helix motif that was identified by PROSITE (3). The seven invariant amino acids of LuxR homologs are denoted with asterisks (19). Additional sequences shown are those of proteins abbreviated as follows: RS SOLR, R. solanacearum SolR (accession no. AF021840 [19]); PA RHLR, P. aeruginosa RhlR (accession no. L08962 [43]); and VF LUXR, V. fischeri LuxR (accession no. 225902 [12]).
The cepR ORF is divergently transcribed from cepI with an intergenic region of 727 bp. It encodes a protein with 239 amino acids and a predicted molecular weight of 26,592. The putative cepR gene product has 67% identity and 78% similarity to SolR (19), 36% identity and 51% similarity to RhlR (43), and 29% identity and 45% similarity to LuxR (V. fischeri) (12, 17, 24). The alignment of these amino acid sequences is shown in Fig. 2C. The location of the two most highly conserved regions, the DNA and autoinducer binding regions, are highlighted (61, 63). CepR contains only six of the seven amino acids which are identical in many of the luxR homologs studied to date (Fig. 2C) (19, 21, 52).
Characterization of a cepR mutant.
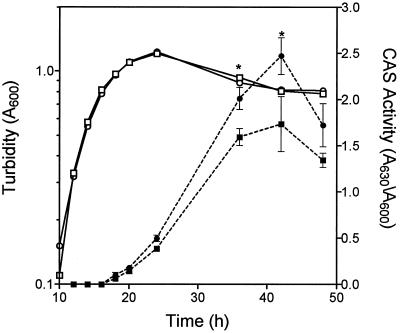
K56-R2 produced 44% larger zones than K56-2 on CAS agar (Table 2). The CAS activity in culture fluids was 42% greater in K56-R2 in comparison to the parent strain (Table 2). CAS activity was also measured in culture fluids throughout the growth of K56-2 and K56-R2 (Fig. 3). The growth of K56-R2 was similar to that of the parent. Although K56-R2’s siderophore production in log phase was similar to that of the parent, K56-R2 produced 26 to 42% more siderophore activity during stationary phase (Fig. 3).
FIG. 3.
Effect of growth on CAS activity in K56-2 and K56-R2. The CAS activity (solid symbols) and turbidity (open symbols) of K56-2 (squares) and K56-R2 (circles) were measured at selected intervals during batch culture in succinate medium supplemented with 10 mM ornithine. The values shown are the means ± standard deviations (error bars) from triplicate experiments. Asterisks denote a statistically significant difference from K56-2 as determined by the t test for unpaired observations (P < 0.05).
The CAS assay measures total siderophore activity. To determine if all siderophores were hyperproduced or if the effect was specific for individual siderophores, ornibactin and salicylic acid were individually isolated and quantitated. Ornibactin was purified by gel filtration chromatography and quantitated by CAS activity. The ornibactin yield was 67% greater in K56-R2 than in the parent strain (Table 2). This is greater than the difference in total CAS activity in culture fluids, possibly due to increased sensitivity in the CAS assay by purified ornibactins. The amount of salicylic acid produced in stationary phase cultures, however, was similar to that produced by the parent strain (Table 2). K56-2 produces barely detectable levels of pyochelin. There was no apparent increase in pyochelin production by the cepR mutant, as determined by thin-layer chromatography (data not shown), suggesting that the regulation of siderophores by cepR is specific for ornibactin.
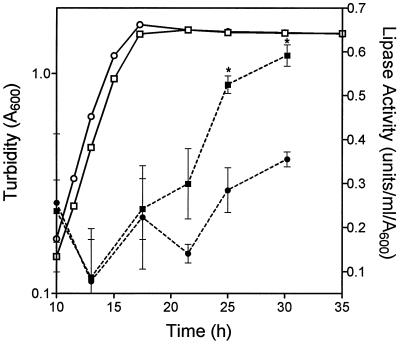
Both the las and rhl systems are involved in the regulation of secreted proteases, and the rhl system is implicated in the control of lipase production (28) in P. aeruginosa. B. cepacia produces two extracellular proteases, a 36-kDa zinc metalloprotease similar to elastase (encoded by lasB) and a 40-kDa protease which may be a precursor form (30, 37). K56-R2 did not produce detectable protease in the D-BHI milk agar plate assay (Table 2). Lipolytic activity has been detected in 60% of B. cepacia strains (33), and the lipase gene (lipA) in B. cepacia has been cloned and sequenced (29). Lipase activity was measured in concentrated culture fluids throughout the growth of K56-2 and K56-R2 (Fig. 4). Lipase production is growth phase-dependent, with maximal activity produced in the stationary phase. The cepR mutant produced 40 to 45% less lipase activity than the parent during the period of maximal lipase production.
FIG. 4.
Effect of growth on lipase activity in K56-2 and K56-R2. The lipase activity (solid symbols) and turbidity (open symbols) of K56-2 (squares) and K56-R2 (circles) were measured at selected intervals during batch culture in Anwar’s defined medium. The values shown are the means ± standard deviations (error bars) from duplicate experiments. Asterisks denote a statistically significant difference from K56-2 as determined by the t test for unpaired observations (P < 0.05).
To determine if the wild-type copy of cepR could restore the parental phenotype to K56-R2, pSLR100 (Fig. 1B) was introduced into the mutant strain by electroporation. Siderophore activity was measured on CAS agar to determine if ornibactin yields were reduced to parental levels. K56-R2(pSLR100) produced similar amounts of CAS activity to K56-2 (pUCP28T) (Table 3). Protease activity was also restored to parental levels in K56-R2(pSLR100) (Table 3). There was no difference in lipase activity between K56-R2(pUCP28T) and K56-R2(pSLR100), indicating that cepR was not able to complement the lipase phenotype of the cepR mutant (Table 3). Similar results were observed when pSLA3.2, which contains both cepI and cepR, was introduced into K56-R2 (data not shown).
TABLE 3.
Complementation of a B. cepacia cepR mutant with cepR in transa
| Strain | Genotype | CAS agar zone diam (mm) | Protease zone radius (mm) | Lipase activity (U/ml/A600) |
|---|---|---|---|---|
| K56-2(pUCP28T) | Wild type | 9.3 ± 0.1 | 5.33 ± 0.29 | 0.68 ± 0.11 |
| K56-R2(pUCP28T) | cepR::Tn5 | 12.5 ± 0.5 | 0 | 0.28 ± 0.00 |
| K56-R2(pSLR100) | cepR+ | 9.4 ± 0.1 | 6.00 ± 0.50 | 0.23 ± 0.07 |
All values are means ± standard deviations of triplicate experiments.
Characterization of a B. cepacia N-acyl-HSL.
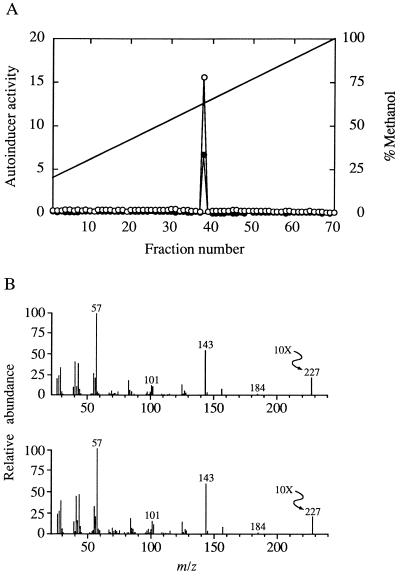
LuxI homologs are involved in the synthesis of N-acyl-HSL molecules. To determine if K56-2 produces an N-acyl-HSL molecule, we used the bioassays described in Materials and Methods to screen for N-acyl-HSLs. Each bioassay shows a specificity for N-acyl-HSLs with different acyl groups. Activity was detected with the R. solanacearum bioassay, and traces of activity were detected with the V. fischeri and P. aeruginosa las assays. The R. solanacearum assay shows greatest sensitivity towards N-octanoyl-HSL. An ethyl acetate extract was then subjected to HPLC and a single peak of activity, as measured with the R. solanacearum assay, was eluted at a position identical to that at which synthetic N-octanoyl-HSL was eluted (Fig. 5A). The amount of activity that was eluted in the single peak was equivalent to the amount of activity applied to the HPLC (recovery, 109% ± 23%), and none of the fractions contained materials detected by any of the other bioassays. A GC-MS analysis showed a molecule with a retention time and a mass spectrum that made it indistinguishable from synthetic N-octanoyl-HSL (Fig. 5B). From these data it appears that the only N-acyl-HSL we detected in cultures of B. cepacia was N-octanoyl-HSL. By comparing the response of the R. solanacearum reporter to culture fluid extracts with the reporter’s responses to different amounts of synthetic N-octanoyl-HSL, we estimate that the concentration of this signal molecule in the culture was approximately 25 nM. If present, the other N-acyl-HSLs listed in Materials and Methods were at concentrations below 1 nM. For comparison, the two P. aeruginosa N-acyl-HSL autoinducers are found at concentrations 1,000-fold higher in fully grown cultures (48, 49). Ethyl acetate extracts were prepared from K56-R2 culture fluids and examined for autoinducer activity in the R. solanacearum bioassay. There was very low autoinducer activity at approximately the limits of detection in extracts from K56-R2 culture fluids, suggesting that cepI expression requires the transcriptional regulator CepR.
FIG. 5.
Analysis of the N-acyl-HSL produced by B. cepacia K56-2. (A) HPLC analysis of a culture fluid extract (○) and synthetic N-octanoyl-HSL (•). HPLC conditions are described in Materials and Methods. Each fraction was 1 ml. Activity was measured by the R. solanacearum reporter system. The solid line indicates methanol concentration. (B) Analysis of purified B. cepacia N-acyl-HSL (top) and synthetic octanoyl-HSL (bottom) by GC-MS. The m/z of the molecular ion was 227 for both, as expected for N-octanoyl-HSL. The molecular ion at 227 is amplified 10-fold (10×).
Characterization of a cepI mutant.
To determine if the cepI gene directs the synthesis of N-octanoyl-HSL and is involved in the regulation of ornibactin, protease, and lipase production, we constructed a cepI mutant and characterized its phenotype. The cepI gene was inactivated with a trimethoprim cassette and introduced into the chromosome by allelic exchange techniques (57). Ethyl acetate extracts from this mutant, designated K56-I2, did not contain detectable levels of autoinducer as shown by the R. solanacearum bioassay (limit of detection, 25 pM), therefore confirming that cepI directs the synthesis of N-octanoyl-HSL.
K56-I2 produced 54% more CAS activity in supernatant fluids and 42% larger zones on CAS agar than K56-2 (Table 4). The ornibactin yield in culture supernatants of K56-I2 was 61% greater than that in K56-2 supernatants. CAS agar zones were also measured on CAS plates supplemented with 10 μM FeCl3. K56-2 produced zones with radii from the edge of the colony of 1.2 ± 0.3 mm, while K56-I2 and K56-R2 produced zones with radii of 4.0 ± 0.1 and 4.2 ± 0.3 mm, respectively, on high-iron CAS agar plates. Therefore, ornibactins are hyperproduced in high-iron medium in both cepI and cepR mutants. K56-I2 did not produce protease activity detectable by the D-BHI milk agar assay. Lipase activity, however, was not significantly less in the cepI mutant than in K56-2 (Table 4). Mutations in cepI and cepR, therefore, result in similar phenotypes with regard to N-octanoyl-HSL, ornibactin, and protease production but not lipase activity.
TABLE 4.
Effect of a cepI mutation on siderophore, protease, and lipase productiona
| Strain | Genotype | Supernatant CAS activity (A630/A600) | CAS agar zone diam (mm) | Ornibactinb (μg/ml/A600) | Protease zone radius (mm) | Lipase activity (U/ml/A600) |
|---|---|---|---|---|---|---|
| K56-2 | Wild type | 1.39 ± 0.00 | 9.5 ± 0.5 | 28.9 ± 7.6 | 5.5 ± 0.5 | 0.53 ± 0.02 |
| K56-I2 | cepI::tmp | 2.14 ± 0.44c | 13.5 ± 0.3c | 46.5 ± 10.3c | 0c | 0.46 ± 0.07 |
All values are the means ± standard deviations of triplicate experiments, unless indicated otherwise.
Mean ± standard deviation of quadruplicate experiments.
Significantly different from parent K56-2 in unpaired t test (P < 0.05).
To determine if the addition of exogenous autoinducer could restore protease production in K56-I2, the following assays were performed. Autoinducer extracts were prepared from K56-2 and added to sterile filter discs in amounts ranging from 12.5 to 125 pmol. The discs containing N-octanoyl-HSL were added to D-BHI skim milk agar plates inoculated with the protease-negative cepI mutant K56-I2. Protease production by K56-I2 detectable by zones around the colony was restored in this assay and was also restored when the parent K56-2 was streaked at right angles to K56-I2 in a cross-feeding assay (data not shown). However, neither supplementation with autoinducer extracts from K56-R2 nor cross-feeding experiments with this mutant restored protease activity in K56-I2. This suggests that the autoinducer N-octanoyl-HSL produced by K56-2 is required for protease production or secretion.
DISCUSSION
Many gram-negative pathogens regulate the expression of virulence genes through the cell density-dependent process known as quorum sensing. The spectrum of phenotypes regulated in this manner includes the production of exoenzymes in the opportunistic pathogen P. aeruginosa, the conjugal transfer of Ti plasmids in the plant pathogen Agrobacterium tumefaciens, and the production of antibiotics and degradative enzymes in the plant pathogen Erwinia carotovora (for reviews, see references 21, 55, and 71). The ability to coordinate the behavior of a population of bacterial pathogens may contribute to establishing a successful infection.
With the identification of the cepIR genes in B. cepacia, we extend the number of LuxIR homologs identified in gram-negative bacteria to date. The cepI and cepR genes are divergently transcribed and separated by an intergenic region of 727 bp. Divergent arrangements are also found in luxIR (17), solIR (19), ahyIR (72), and asaIR (72). Although the intergenic region between cepI and cepR is considerably larger than that in these other homologs, there are no ORFs with significant similarity to any known genes within this intergenic region. A noncoding region between solI and solR in R. solanacearum of 396 bp was also reported (19). In Rhodobacter sphaeroides, an ORF in the intergenic region, designated Orf2, has been suggested to play a role in the posttranslational regulation of cerI (52). Within the 3.2-kb SphI fragment containing cepI and cepR, the only ORF with similarity to a known gene was a partial ORF downstream of cepR that shows significant homology to a gene involved in Mg2+ transport (mgtC) in Salmonella typhimurium (64).
Four different bioassays that are sensitive to a range of N-acyl-HSLs were employed to detect autoinducer activity from K56-2 cultures. The R. solanacearum bioassay was the only system to detect significant amounts of activity. We present evidence that the activity is N-octanoyl-HSL (Fig. 5). N-octanoyl-HSL and N-hexanoyl-HSL are the two autoinducers produced by R. solanacearum (19). In addition to the high similarity between CepIR and SolIR of B. cepacia and R. solanacearum, these closely related species produce similar N-acyl-HSL structures. It was previously reported that cell culture fluids from B. cepacia contained at least three types of signaling molecules (36), whereas we detected a single autoinducer molecule in K56-2 and a cepI mutant did not produce detectable amounts of this autoinducer. There may be strain variation in the production of autoinducer molecules by B. cepacia, and therefore it would be interesting to examine the types of autoinducers produced by different genomovars as well as clinical and environmental isolates. In fact we have found that B. cepacia G4, an environmental isolate, produces much higher levels of N-octanoyl-HSL than does strain K56-2, and although N-octanoyl-HSL is the predominant signal produced by G4, several other N-acyl-HSLs can be detected at nanomolar concentrations in culture fluid extracts (8).
The concentration of N-octanoyl-HSL in K56-2 culture fluids was approximately 1,000-fold lower than N-acyl-HSL molecules in P. aeruginosa culture fluids (48, 49). McKenney et al. (36) reported that addition of concentrated culture fluids from B. cepacia stationary phase cultures prior to inoculation with B. cepacia caused a slight increase in production of siderophores, lipase, and protease during the mid-log phase of growth. Addition of concentrated P. aeruginosa culture fluids to B. cepacia cultures, however, promoted a much greater increase in the production of these exoproducts. In our study, extracts from K56-2 culture fluids were able to restore protease production by a cepI mutant. It is possible that B. cepacia produces low amounts of N-octanoyl-HSL or that the laboratory conditions we used were not optimal for production of this autoinducer molecule. Other factors or signals may be required to activate expression of the cepI gene. An example of this type of regulation is found in A. tumefaciens. The traR gene, which is involved in the transfer of the Ti plasmid, is expressed in the presence of compounds called opines, which are produced by plants within crown gall tumors. In the presence of opines traR is activated, which subsequently activates traI (20).
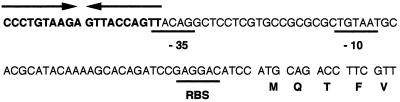
The cepR mutant produces low but detectable levels of N-octanoyl-HSL. The cepI promoter region contains putative −10 and −35 sites. The presence of a 20-bp lux box-like sequence that partially overlaps the putative −35 region (Fig. 6) suggests that CepR binds to the cepI promoter to activate cepI expression. The observation that K56-R2 produces low levels of N-octanoyl-HSL in the Ralstonia bioassay is consistent with the role of CepR in cepI regulation. In several related systems the LuxI homolog is under this type of autoregulatory control. For example, in P. aeruginosa LasR and low concentrations of autoinducer activate lasI expression (60). LuxR also activates luxI expression (15, 16), and SolR is required for expression of solI (19).
FIG. 6.
Nucleotide sequence of the cepI promoter region. The lux box-like sequence is shown in boldface type, with arrows indicating the imperfect inverted repeats. Putative promoter elements and the ribosome binding site (RBS) are underlined. The first five amino acids encoded by the cepI gene are shown in single-letter code below the nucleotide sequence.
CepR appears to function as both a positive and a negative regulator of extracellular virulence factor production in B. cepacia. The siderophore hyperproduction phenotype in K56-R2 was specific for ornibactin, suggesting that CepR normally functions to decrease the production of ornibactin at higher cell densities. Iron availability is an important signal involved in the regulation of siderophore production. We speculate that cell density serves as a second signal involved in limiting siderophore biosynthesis under high cell densities in stationary phase, since cells are no longer growing at a logarithmic rate and therefore would require less iron. CepR may either be a repressor of ornibactin synthesis or may activate a repressor of ornibactin biosynthetic genes.
Mutations in either cepI or cepR result in a protease-negative phenotype on D-BHI milk agar. The parental phenotype was restored in K56-R2 by complementation with cepR in trans and in K56-I2 by exogenous addition of ethyl acetate extracts of culture fluids, suggesting that CepR positively regulates protease production. In P. aeruginosa, both lasR and rhlR are involved in the regulation of the xcp secretion system (7) in addition to regulation of lasB transcription. This general secretory pathway mediates the transport of a variety of secreted virulence factors across the bacterial membrane (75). It is possible that cepIR regulates the production of protease at the transcriptional level or that cepIR regulates the production of the secretion apparatus necessary for the export of protease.
Other quorum-sensing systems have also been shown to negatively regulate expression of their target genes. For example, Erwinia stewartii EsaR represses its own expression (4) and acts as a repressor of cps genes required for capsular polysaccharide synthesis (5). It was reported that mutations in solI and solR do not affect the production of extracellular virulence determinants in R. solanacearum (19); however, mutations in either solR or solI result in an ∼1.7-fold increase in the cell wall-degrading enzyme polygalacturonase. This observation suggests that the SolIR system also plays a negative regulatory role in the control of polygalacturonase production.
K56-R2 produced significantly lower lipase activity than the parent strain. In contrast to protease and siderophore activity, lipase activity was not restored to parental levels when K56-R2 was complemented in trans with a plasmid containing either cepR or cepIR. The cepI mutant also produced parental levels of lipase. These data suggest that the transposon insertion in K56-R2 has a polar effect on a downstream gene required for lipase production or that K56-R2 has acquired a random second mutation responsible for decreased lipase production. McKenney et al. reported slight increases in lipase activity from B. cepacia cultures supplemented with concentrated B. cepacia culture fluids (36). One or more of the multiple autoinducers detected in the concentrated culture fluids may be involved in the regulation of lipase production although the results from our study indicate that the cepIR quorum-sensing system does not regulate lipase production in K56-2.
The role of quorum sensing and the control of virulence factor production in the pathogenesis of B. cepacia infections are not fully understood. Additional studies are needed to determine the target genes for CepR. We have recently cloned and sequenced pvdA, a gene involved in the biosynthesis of ornibactin in B. cepacia (66). The promoter region of pvdA contains a possible lux box-like sequence (data not shown). It will be interesting to examine the possible transcriptional regulation of pvdA by CepR. The sequence(s) of the B. cepacia protease gene(s) has not yet been reported. The lipase gene (lipA) does not contain a lux box-like sequence similar to the consensus sequence. Further studies are needed to determine the mechanisms by which cepIR regulate production of ornibactins, protease, and possibly other factors in B. cepacia.
ACKNOWLEDGMENTS
This study was supported by grants from the Canadian Cystic Fibrosis Foundation and the U.S. Cystic Fibrosis Foundation. S.L. is the recipient of an Alberta Heritage Foundation for Medical Research Studentship award.
REFERENCES
- 1.Anwar H, Brown M R W, Lambert P A. Effect of nutrient depletion on sensitivity of Pseudomonas cepacia to phagocytosis and serum bactericidal activity at different temperatures. J Gen Microbiol. 1983;129:2021–2027. doi: 10.1099/00221287-129-7-2021. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons; 1989. pp. 2.0.1–2.9.7. [Google Scholar]
- 3.Barioch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991;19:2241–2245. doi: 10.1093/nar/19.suppl.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck von Bodman S, Farrand S K. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J Bacteriol. 1995;177:5000–5008. doi: 10.1128/jb.177.17.5000-5008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck von Bodman S, Majerczak D R, Coplin D L. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc Natl Acad Sci USA. 1998;95:7687–7692. doi: 10.1073/pnas.95.13.7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapon-Herve V, Akrim M, Latifi A, Williams P, Lazdunski A, Bally M. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol Microbiol. 1997;24:1169–1178. doi: 10.1046/j.1365-2958.1997.4271794.x. [DOI] [PubMed] [Google Scholar]
- 8.Conway, B., and E. P. Greenberg. Unpublished observations.
- 9.Darling P, Chan M, Cox A D, Sokol P A. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect Immun. 1998;66:874–877. doi: 10.1128/iai.66.2.874-877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis J J, Sokol P A. Electrotransformation of Pseudomonas. In: Nickoloff J A, editor. Electroporation and electrofusion of microorganisms. Clifton, N.J: Humana Press; 1995. pp. 125–133. [DOI] [PubMed] [Google Scholar]
- 11.Deshazer D, Woods D E. Broad-host-range cloning and cassette vectors based on the R388 trimethoprim resistance gene. BioTechniques. 1996;20:762–764. doi: 10.2144/96205bm05. [DOI] [PubMed] [Google Scholar]
- 12.Devine J H, Countryman C, Baldwin T O. Nucleotide sequence of the luxR and luxI genes and structure of the primary regulatory region of the lux regulon of Vibrio fischeri. Biochemistry. 1988;27:837–842. [Google Scholar]
- 13.Eberhard A, Burlingame A L, Eberhard C, Kenyon G L, Nealson K H, Oppenheimer N J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 14.Eberhard A, Widrig C A, McBath P, Schineller J B. Analogs of the autoinducer of bioluminescence in Vibrio fischeri. Arch Microbiol. 1986;146:35–40. doi: 10.1007/BF00690155. [DOI] [PubMed] [Google Scholar]
- 15.Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 16.Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci USA. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engebrecht J, Silverman M. Nucleotide sequence of the regulatory locus controlling expression of bacterial genes for bioluminescence. Nucleic Acids Res. 1987;15:10455–10467. doi: 10.1093/nar/15.24.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flavier A B, Ganova-Raeva L M, Schell M A, Denny T P. Hierarchical autoinduction in Ralstonia solanacearum: control of N-acyl-homoserine lactone production by a novel autoregulatory system responsive to 3-hydroxypalmitic acid methyl ester. J Bacteriol. 1997;179:7089–7097. doi: 10.1128/jb.179.22.7089-7097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuqua W C, Winans S C. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 22.Gambello M J, Kaye S, Iglewski B H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Govan J R W, Hughes J E, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 24.Gray K M, Greenberg E P. Sequencing and analysis of luxR and luxI, the luminescence regulatory genes from the squid light organ symbiont Vibrio fischeri ES114. Mol Mar Biol Biotechnol. 1992;1:414–419. [Google Scholar]
- 25.Gray K M, Passador L, Iglewski B H, Greenberg E P. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J Bacteriol. 1994;176:3076–3080. doi: 10.1128/jb.176.10.3076-3080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H P. A broad-host-range Flp-FRT recombination system for sip-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 27.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levinson H. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 28.Jaeger K-E, Schneidinger B, Liebeton K, Haas D, Reetz M T, Philippou S, Gerritse G, Ransac S, Dijkstra B W. Lipase of Pseudomonas aeruginosa: molecular biology and biotechnological application. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of psuedomonads. Washington, D.C: ASM Press; 1996. pp. 319–330. [Google Scholar]
- 29.Jorgensen S, Skov K W, Diderichsen B. Cloning, sequence, and expression of a lipase gene from Pseudomonas cepacia: lipase production in heterologous hosts requires two Pseudomonas genes. J Bacteriol. 1991;173:559–567. doi: 10.1128/jb.173.2.559-567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kooi C, Cox A, Darling P, Sokol P A. Neutralizing monoclonal antibodies to an extracellular Pseudomonas cepacia protease. Infect Immun. 1994;62:2811–2817. doi: 10.1128/iai.62.7.2811-2817.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 32.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR to expression of the stationary phase sigma factor rpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 33.Lonon M K, Woods D E, Straus D C. Production of lipase by clinical isolates of Pseudomonas cepacia. J Clin Microbiol. 1988;26:979–984. doi: 10.1128/jcm.26.5.979-984.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madden T L, Tatusov R L, Zhang J. Applications of network BLAST server. Methods Enzymol. 1996;266:131–141. doi: 10.1016/s0076-6879(96)66011-x. [DOI] [PubMed] [Google Scholar]
- 35.Mahenthiralingam E, Simpson D A, Speert D P. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKenney D, Brown K E, Allison D G. Influence of Pseudomonas aeruginosa exoproducts on virulence factor production in Burkholderia cepacia: evidence of interspecies communication. J Bacteriol. 1995;177:6989–6992. doi: 10.1128/jb.177.23.6989-6992.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKevitt A I, Bajaksouzian S, Klinger J D, Woods D E. Purification and characterization of an extracellular protease from Pseudomonas cepacia. Infect Immun. 1989;57:771–778. doi: 10.1128/iai.57.3.771-778.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merriman T R, Lamont I L. Construction and use of a self-cloning promoter probe vector for gram-negative bacteria. Gene. 1993;126:17–23. doi: 10.1016/0378-1119(93)90585-q. [DOI] [PubMed] [Google Scholar]
- 39.Meyer J-M, Abdallah M A. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physico-chemical properties. J Gen Microbiol. 1978;107:319–328. [Google Scholar]
- 40.Meyer J M, Hohnadel D, Halle F. Cepabactin from Pseudomonas cepacia, a new type of siderophore. J Gen Microbiol. 1989;135:1479–1487. doi: 10.1099/00221287-135-6-1479. [DOI] [PubMed] [Google Scholar]
- 41.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 42.Nealson K H, Platt T, Hastings J W. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochsner U A, Koch A K, Fiechter A, Reiser J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochsner U A, Fiecher A, Reiser J. Isolation, characterization and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem. 1994;269:19787–19795. [PubMed] [Google Scholar]
- 45.Ochsner U A, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parsek M R, Schaefer A L, Greenberg E P. Analysis of random and site-directed mutations in rhlI, a Pseudomonas aeruginosa gene encoding an acylhomoserine lactone synthase. Mol Microbiol. 1997;26:301–310. doi: 10.1046/j.1365-2958.1997.5741935.x. [DOI] [PubMed] [Google Scholar]
- 47.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 48.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pearson J P, Pesci E C, Iglewski B H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puskas A, Greenberg E P, Kaplan S, Schaeffer A L. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J Bacteriol. 1997;179:7530–7537. doi: 10.1128/jb.179.23.7530-7537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ralling G, Bodrug S, Linn T. Growth rate-dependent regulation of RNA polymerase synthesis in Escherichia coli. Mol Gen Genet. 1985;201:379–386. doi: 10.1007/BF00331327. [DOI] [PubMed] [Google Scholar]
- 54.Sajjan U S, Sun L, Goldstein R, Forstner J F. Cable (Cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J Bacteriol. 1995;177:1030–1038. doi: 10.1128/jb.177.4.1030-1038.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. The bacterial ’enigma’: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 57.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 58.Schweizer H P. Improved methods for gene analysis and expression in Pseudomonas spp. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: ASM Press; 1996. pp. 229–236. [Google Scholar]
- 59.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 60.Seed P C, Passador L, Iglewski B H. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J Bacteriol. 1995;177:654–659. doi: 10.1128/jb.177.3.654-659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shadel G S, Young R, Baldwin T O. Use of regulated cell lysis in a lethal genetic selection in Escherichia coli: identification of the autoinducer-binding region of the LuxR protein from Vibrio fischeri ATCC 7744. J Bacteriol. 1990;172:3980–3987. doi: 10.1128/jb.172.7.3980-3987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. BioTechnology. 1983;1:784–791. [Google Scholar]
- 63.Slock J, Kolibachuk D, Greenberg E P. Critical regions of the Vibrio fischeri LuxR protein defined by mutational analysis. J Bacteriol. 1990;172:3974–3979. doi: 10.1128/jb.172.7.3974-3979.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith R L, Maquire M E. Microbial magnesium transport: unusual transporters searching for identity. Mol Microbiol. 1998;28:217–226. doi: 10.1046/j.1365-2958.1998.00810.x. [DOI] [PubMed] [Google Scholar]
- 65.Sokol P A. Production and utilization of pyochelin by clinical isolates of Pseudomonas cepacia. J Clin Microbiol. 1986;23:560–562. doi: 10.1128/jcm.23.3.560-562.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sokol, P. A. Unpublished observations.
- 67.Sokol P A, Lewis C J, Dennis J J. Isolation of a novel siderophore from Pseudomonas cepacia. J Med Microbiol. 1992;36:184–189. doi: 10.1099/00222615-36-3-184. [DOI] [PubMed] [Google Scholar]
- 68.Sokol P A, Ohman D E, Iglewski B H. A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. J Clin Microbiol. 1979;9:538–540. doi: 10.1128/jcm.9.4.538-540.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stephan H, Freund S, Beck W, Jung G, Meyer J-M, Winkelmann G. Ornibactins-a new family of siderophores from Pseudomonas. Biometals. 1993;6:93–100. doi: 10.1007/BF00140109. [DOI] [PubMed] [Google Scholar]
- 70.Stewart V J, Parales J V., Jr Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol. 1988;170:1589–1587. doi: 10.1128/jb.170.4.1589-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Swift S, Throup J P, Williams P, Salmond G P C, Stewart G S A B. Quorum sensing a population-density component in the determination of bacterial phenotype. Trends Biochem Sci. 1996;21:214–219. [PubMed] [Google Scholar]
- 72.Swift S, Karlyshev A V, Fish L, Durant E L, Winson M K, Chhabra S R, Williams P, Macintyre S, Stewart G S A B. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taylor R F H, Gaya H, Hodson M E. Pseudomonas cepacia: pulmonary infection in patients with cystic fibrosis. Respir Med. 1993;87:187–192. doi: 10.1016/0954-6111(93)90090-m. [DOI] [PubMed] [Google Scholar]
- 74.Toder D S, Gambello M J, Iglewski B H. Pseudomonas aeruginosa LasA: a second elastase gene under transcriptional control of lasR. Mol Microbiol. 1991;5:2003–2010. doi: 10.1111/j.1365-2958.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 75.Tommassen J, Filloux A, Bally M, Murgier M, Lazdunski A. Protein secretion in Pseudomonas aeruginosa. FEMS Microbiol Rev. 1992;103:73–90. doi: 10.1016/0378-1097(92)90336-m. [DOI] [PubMed] [Google Scholar]
- 76.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;45:600–603. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 77.Woods D. Oligonucleotide screening of cDNA libraries. Focus. 1984;6:1–2. [Google Scholar]