Abstract
The novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2 or COVID-19) has caused a global pandemic. The SARS-CoV-2 RNA genome is replicated by a conserved “core” replication-transcription complex (RTC) containing an error-prone RNA-dependent RNA polymerase holoenzyme (holo-RdRp, nsp12-nsp7-nsp8) and a RNA proofreading nuclease (nsp14-nsp10). Although structures and functions of SARS-CoV-2 holo-RdRp have been extensively studied and ribonucleotide-analog inhibitors, such as Remdesivir, have been treated for COVID-19 patients, the substrate and nucleotide specificity of SARS-CoV-2 holo-RdRp remain unknown. Here, our biochemical analysis of SARS-CoV-2 holo-RdRp reveals that it has a robust DNA-dependent RNA polymerase activity, in addition to its intrinsic RNA-dependent RNA polymerase activity. Strikingly, SARS-CoV-2 holo-RdRp fully extends RNAs with a low-fidelity even when only ATP and pyrimidine nucleotides, in particular CTP, are provided. This ATP-dependent error-prone ribonucleotide incorporation by SARS-CoV-2 holo-RdRp resists excision by the RNA proofreading nuclease in vitro. Our collective results suggest that a physiological concentration of ATP likely contributes to promoting the error-prone incorporation of ribonucleotides and ribonucleotide-analogs by SARS-CoV-2 holo-RdRp and provide a useful foundation to develop ribonucleotide analogs as an effective therapeutic strategy to combat coronavirus-mediated outbreak.
Keywords: SARS-CoV-2, COVID-19, RNA replication-Transcription complex, RNA-Dependent RNA polymerase, RNA proofreading
1. Introduction
The novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2 or COVID-19) is an enveloped positive-strand RNA virus that belong to the order Nidovirales [1,2]. Although several types of vaccines have been used for the prevention of COVID-19 and its variants [3], and nucleoside-analogs, such as Remdesivir [4,5], are being used for treatment of COVID-19, yet safe and effective therapeutic strategies for COVID-19 are limited.
The SARS-CoVs contain an unusually large genome of ∼30 kb, which is replicated by a replication and transcription complex (RTC). The “core” RTC includes an error-prone RNA-dependent RNA polymerase holoenzyme (holo-RdRp) that consists of nonstructural protein 12 (nsp12) containing the RdRp domain and two accessory proteins, nsp7 and nsp8 [[5], [6], [7]]. The two accessory subunits, nsp7 and nsp8 (nsp7:nsp8:nsp12 of 1:2:1 ratio), substantially extend the processivity of SARS-CoV RdRp activity [8]. In the structures of SARS-CoV-2 RTC [5,6,9,10], two nsp8 subunits are differently positioned to support RNA backbones interactions, consistent with the key role of nsp8 in the processivity of SARS-CoV RdRp. However, SARS-CoV-2 RTC itself has a relatively low-fidelity RdRp activity with a high mutation rate, which provides CoVs with genomic diversity that helps them to dynamically adapt to selective pressure [[11], [12], [13]].
On the other hand, CoVs encode a unique nsp14-nsp10 RNA proofreading complex that contains 3′-to-5′ exonuclease activity and mitigates the intrinsic error-prone RNA replication by SARS-CoV-2 holo-RdRp [14]. Notably, genetic deletion of nsp14 is lethal to SARS-CoV-2, suggesting that the balance between the low-fidelity RNA replication and proofreading function is critical for SARS-CoV-2 viability [13,15]. Furthermore, the nucleotide excision by nsp14-nsp10 has been proposed as an important barrier to developing nucleoside analogs, such as an adenine nucleoside analog Remdesivir, as anti-SARS-CoV-2 therapeutics [13,16,17].
Although structural and biochemical analyses of SARS-CoV-2 RTC and RNA proofreading complexes have been active research area [[4], [5], [6], [7],[18], [19], [20], [21], [22], [23]], limited information is available regarding the activities ad substrate specificity of the SARS-CoV-2 RdRp. Here, we characterized the activities and substrate specificity of the SARS-CoV-2 holo-RdRp complex in vitro to better understand the basis of the low-fidelity RNA replication by SARS-CoV-2. We show that SARS-CoV-2 holo-RdRp has robust DNA- and RNA-dependent RNA polymerase activities, and it requires at least one strand of RNA, either in template or primer, for efficient ribonucleotide incorporation. Importantly, SARS-CoV-2 holo-RdRp can fully extend RNA even when only ATP and any pyrimidine nucleotides, in particular with CTP, are present. Furthermore, unexpectedly, this ATP-dependent error-prone RNA replication by SARS-CoV-2 holo-RdRp resists excision by the nsp14-nsp10 RNA proofreading nuclease. Together, our results provide new insights into the mechanism of SARS-CoV-2 RNA replication and identify ATP as a key factor that triggers the high mutation-rate RNA replication by SARS-CoV-2.
2. Results
2.1. SARS-CoV-2 holo-RdRp has robust DNA- and RNA-dependent RNA polymerase activities
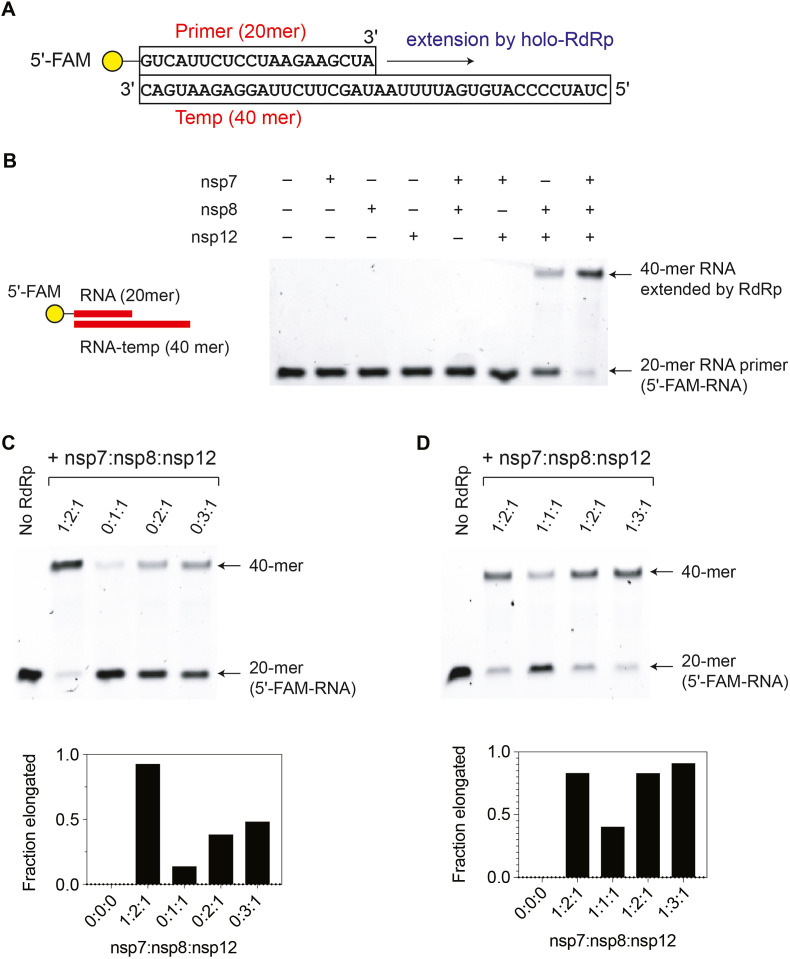
To study the activities and specificity of SARS-CoV-2 RTC, we reconstituted and characterized the primer extension functions of the recombinant nsp12-nsp7-nsp8 complex in vitro. SARS-CoV-2 nsp12, nsp7, and nsp8 were purified to near homogeneity (Fig. S1). As expected, nsp12, nsp7, and nsp8 itself did not show any detectable RNA primer extension activity (40-mer RNA template and 20-mer RNA primer). Unlike SARS-CoV-1 RdRp [8], we found that SARS-CoV-2 nsp8 alone, not with nsp7, substantially stimulates the RdRp activity of nsp12 (Fig. 1 B and C). However, both nsp7 and nsp8 subunits are required to form a fully functional SARS-CoV-2 holo-RdRp unit (Fig. 1B–D). Our data also support that the optimal ratio between nsp7, nsp8, and nsp12 is 1:2:1, given that the addition of excess nsp18 (nsp7:nsp8:nsp12 of 1:3:1) did not show further significant increase in RNA extention (Fig. 1D).
Fig. 1.
SARS-CoV-2 nsp8 stimulates the RNA primer extension by nsp12. (A) The RNA template and primer (5′-fluorescein labeled) used for the primer extension assay in this study. DNA template and primer (in Fig. 2) have identical sequences, except that DNAs have T instead U. (B) SARS-CoV-2 nsp8 stimulates the primer extension by nsp12. The RNA primer extension was assayed with different combinations of nsp12 (1 μM), nsp7 (1 μM), and nsp8 (2 μM). The RdRp activity of nsp12 was significantly increased in the presence of nsp8. (C) A nsp8-dependent stimulation of the RdRp activity of nsp12. SARS-CoV-2 nsp8 shows a dose-dependent enhancement of the RdRp activity of nsp12. (D) SARS-CoV-2 nsp7 completes the SARS-CoV-2 holo-RdRp. In the presence of nsp7 and nsp8, SARS-CoV-2 nsp12 forms a fully functional holo-RdRp (nsp7:nsp8:nsp12 of 1:2:1), efficiently extending the RNA primer (20 mer) to the full-length product (40 mer). Normalized fraction elongated [I40−mer/(I20−mer + I40−mer)] is shown in panels C and D. This is a representative assay of two independent experiments.
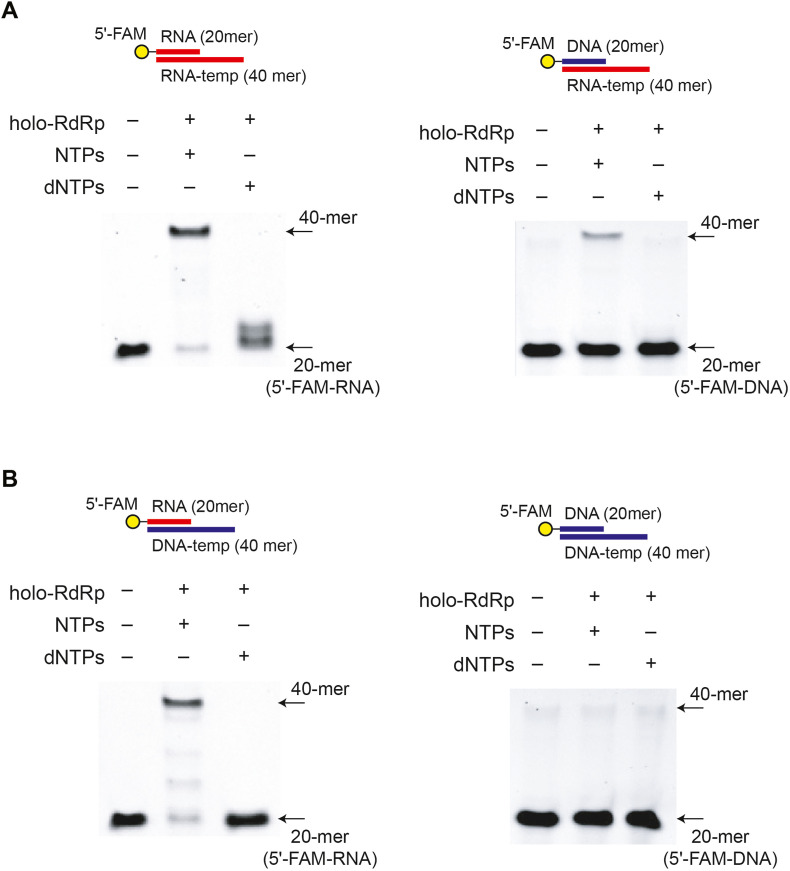
Next, we tested the primer extension by SARS-CoV-2 holo-RdRp against different combinations of RNA and DNA oligonucleotides in the template or primer. Surprisingly, SARS-CoV-2 holo-RdRp shows a robust DNA-dependent RNA polymerase activity, although this unexpected activity appears to be less efficient in RNA extension compared to its intrinsic RNA-dependent RNA polymerase activity (Fig. 2 ). Furthermore, SARS-CoV-2 holo-RdRp also incorporates ribonucleotides from a DNA primer (Fig. 2A), when RNA was used as a template. SARS-CoV-2 holo-RdRp did not show any detectable DNA-dependent DNA polymerase activity (Fig. 2B). Collectively, these data suggest that SARS-CoV-2 holo-RdRp can efficiently extend the RNA primer using either RNA- or DNA-template, and requires at least one RNA strand (either template or primer) for ribonucleotide incorporation.
Fig. 2.
SARS-CoV-2 holo-RdRp has robust RNA- and DNA-dependent RNA polymerase activities. (A) RNA-dependent RNA polymerase activity of SARS-CoV-2 holo-RdRp. SARS-CoV-2 holo-RdRp shows a RNA-dependent RNA polymerase activity with either RNA or DNA primer, although RNA primer is more efficient. (B) DNA-dependent RNA-polymerase activity of SARS-CoV-2 holo-RdRp. Unextectedly, SARS-CoV-2 holo-RdRp shows a robust DNA-dependent RNA polymerase activity with a RNA primer. Collectively, SARS-CoV-2 RdRp requires at least one strand of RNA, either in the template or primer, to extend RNA.
2.2. ATP substantially enhances the error-prone ribonucleotide incorporation by SARS-CoV-2 holo-RdRp
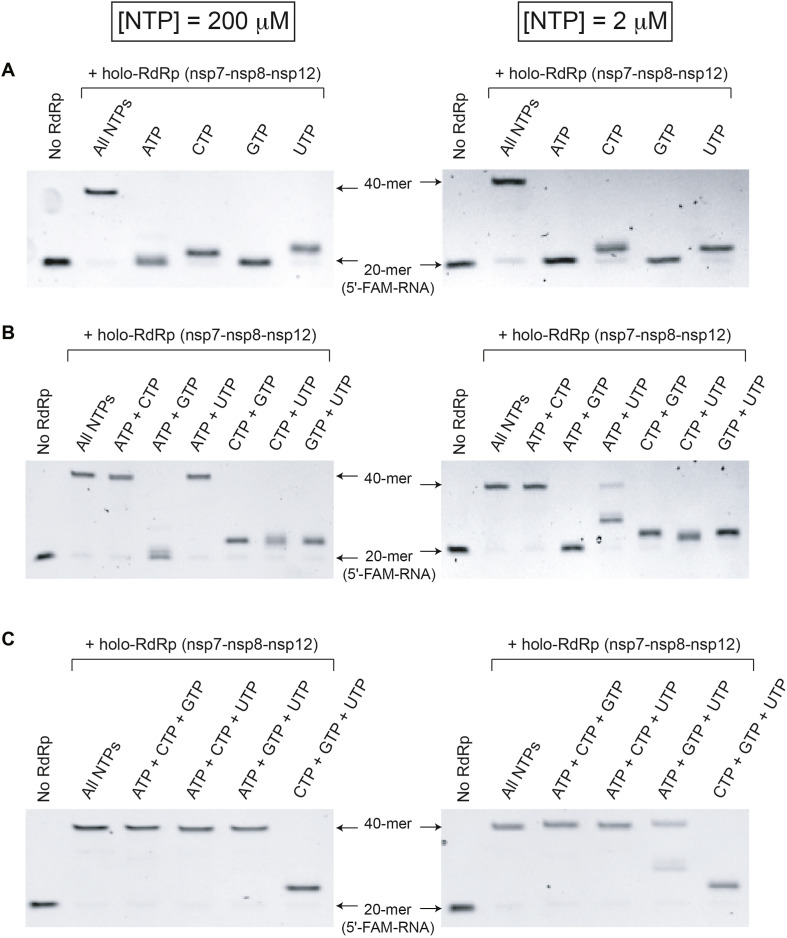
Although CoVs are known to show a high mutation rate [11], the basis for this error-prone RNA replication remains poorly understood and the nucleotide specificity of SARS-CoV-2 RdRp has not been determined. To address these issues, we compared the RNA-dependent RNA polymerase activity of SARS-CoV-2 holo-RdRp using different combinations of ribonucleotides (NTPs) at physiological (200 μM of each NTP) or low (2 μM of each NTP) nucleotide concentrations (Fig. 3 ). The 20 nucleotides of the RNA template that is extended by holo-RdRp include all four types of NTPs (five As, eight Us, five Cs, and two Gs) with the first two sites for incorporation of U (Fig. 1A).
Fig. 3.
ATP substantially enhances the error-prone ribonucleotide incorporation by SARS-CoV-2 holo-RdRp. The low-fidelity ribonucleotide incorporation by SARS-CoV-2 RdRp was monitored in the presence of different combinations of ribonucleotides, one NTP (A), two NTPs (B), and three NTPs (C). Two different concentrations of ribonucleotides (200 μM and 2 μM of each NTP) were used with the RNA substrate shown in Fig. 1A. These results suggest that ATP substantially enhances the low-fidelity RNA replication by SARS-CoV-2 RdRp. The 20 nucleotides of the RNA template that is extended by SARS-CoV-2 holo-RdRp include all four types of NTPs (five As, eight Us, five Cs, and two Gs) with the first two sites for incorporation of U.
Overall, SARS-CoV-2 showed a large-degree of a low-fidelity ribonucleotide incorporation both at high and low concentrations of NTPs (Fig. 3). In the presence of one ribonucleotides (ATP, UTP, CTP, and GTP), the addition of UTP showed a primer extension, which is likely corresponding to incorporation to the first two sites (complementary to two As in the template) (Fig. 1, Fig. 3A). Interestingly, the addition of CTP also showed a short RNA extension, presumably incorporation to the site complementary to A, suggesting a poor pyrimidine selectivity by SARS-CoV-2 holo-RdRp (Fig. 3A).
In contrast, a substantial amount of low-fidelity ribonucleotide incorporation products started to be detected from reactions with combinations of more than one type of NTP. Strikingly, even when only ATP and one type of pyrimidine nucleotides (CTP or UTP) were added, SARS-CoV-2 holo-RdRp fully extended the primer to the full-length product (40-mer) as efficiently as control reactions containing all four NTPs (Fig. 3B). Notably, a combination of ATP + CTP seems to be more efficient than ATP + UTP at 2 μM concentration (Fig. 3B). In contrast, the addition of GTP (ATP + GTP, CTP + GTP, and GTP + UTP) or two pyrimidines (CTP + UTP) showed only moderate or low-level of rinucleotide incorporation (Fig. 3B).
When three types of NTPs were added, all combinations of NTPs, except for the combination of GTP + CTP + UTP that lacks ATP, produced fully extended RNA products (Fig. 3C). Taken together, these findings suggest that ATP plays an important role in triggering the low-fidelidy ribonucleotide incorporation by SARS-CoV-2 holo-RdRp and that ATP and pyrimidines, in particular CTP, are sufficient for SARS-CoV-2 holo-RdRp to fully extend the RNA primer.
2.3. ATP-dependent error-prone ribonucleotide incorporation resists excision by the nsp14-nsp10 RNA proofreading nuclease
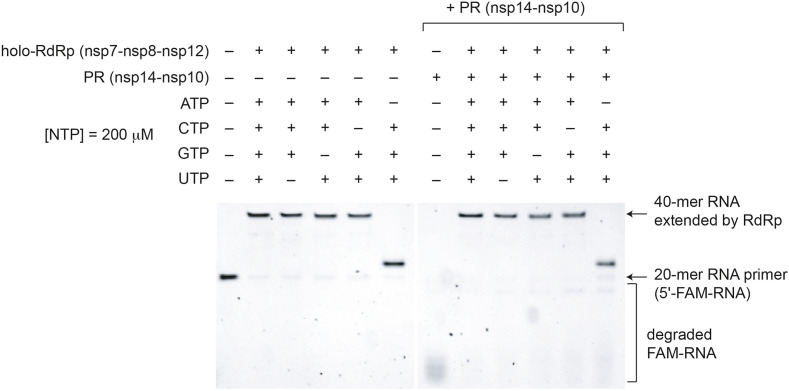
SARS-CoV-2 nsp14-nsp10 functions as an RNA proofreading nuclease that can remove the mismatched base pairs during RNA replication [[13], [14], [15],23]. We previously showed that SARS-CoV-2 nsp14-nsp10 is required to restart the RNA replication stalled by a mismatch [23]. Interestingly, RNA extension products that are terminated by Sofosbuvir, a UMP analog, was shown to be resistant to the excision by the SARS-CoV-2 proofreading nuclease [24]. Given the observed ATP-dependent low-fidelity ribonucleotide incorporation when ATP and pyrimidines are present, we wondered whether this observed mismatched product formation can be mitigated by the proofreading nuclease. To this end, we compared the RNA primer extension by SARS-CoV-2 holo-RdRp in the presence and absence of nsp14-nsp10, when two and three ribonucleotides are added (Figs. 4, S2, and S3).
In the control reaction, SARS-CoV-2 nsp14-nsp10 functions as an exonuclease, efficiently degrading the RNA primer (Fig. 4 ). However, the SARS-CoV-2 proofreading nuclease did not stop or attenuate the low-fidelity RNA replication by SARS-CoV-2 holo-RdRp. Similarly, the low-fidelity extension at 2 μM concentration of two or three types of NTPs was not corrected by SARS-CoV-2 proofreading nuclease (Fig. S3). Taken together, these results suggest the ATP-dependent low-fidelity ribonucleotide incorporation by SARS-CoV-2 holo-RdRp resists excision by the proofreading nuclease in the experimental conditions used in this study.
Fig. 4.
ATP-dependent error-prone ribonucleotide incorporation resists excision by SARS-CoV-2 proofreading nuclease. The low-fidelity ribonucleotide incorporation by SARS-CoV-2 RdRp was monitored in the presence and absence of SARS-CoV-2 proofreading nuclease (PR, nsp14-nsp10). A different combination of three NTPs were used at 200 μM concentration. Similarly, SARS-CoV-2 proofreading nuclease did not stop low-fidelity RNA extension when two NTPs were used (200 μM) (Fig. S2) or when two or three NTPs were used at 2 μM concentration (Fig. S3).
3. Discussion
In this study, we have established an ATP-dependent error-prone ribonucleotide incorporation by SARS-CoV-2 holo-RdRp (nsp12-nsp7-nsp8) (Fig. 3). Unexpectedly, this low-fidelity RNA extension product was resistant to excision by the SARS-CoV-2 RNA proofreader in our experimental conditions (Fig. 4). Notably, among four types of ribonucleotides (NTPs), ATP exists in a highest concentration, a millimolar concentration, in cells [25]. Therefore, our data suggest that ATP is likely the key factor that can potentially trigger the observed high mutation-rate RNA replication by SARS-CoV-2. And physiological ATP concentrations would likely be sufficient to trigger this error-prone ribonucleotide incorporation during SARS-CoV-2 RNA replication in vivo. Furthermore, our data revealed that SARS-CoV-2 holo-RdRp possesses a robust DNA-dependent RNA polymerase, in addition to its intrinsic RNA-dependent RNA polymerase function. In fact, SARS-CoV-2 holo-RdRp extends RNAs if there is at least one strand of RNA either in the template or primer, although it seems to most efficiently extend RNA from RNA duplex substrates (Fig. 2).
Ribonucleoside analogs have been widely tested to inhibit the activity of SARS-CoV-2 RdRp, including Remdesivir, Favipiravir, NHC EIDD-2801, and Sofobuvir [26,27]. Interestingly, EIDD-2801(β-D-N4-hydroxycytidine, a cytidine analog) were 3–10 times more potent than remdesivir (an adenosine analog) in inhibiting SARS-CoV-2 replication [27]. Consistently, CTP in combination with ATP resulted in more efficient RNA extension by SARS-CoV-2 holo-RdRp (even at 2 μM concentration) than UTP + ATP or ATP alone (Fig. 3). Although more work is needed to determine if there is nucleotide preference by SARS-CoV-2 holo-RdRp, this observation raises the possibility that ATP might preferentially trigger incorporation of specific pyrimidine-based nucleotides into RNA templates.
CoVs have a highly conserved nsp14-nsp10 proofreading exonuclease complex that can correct the mismatched base pairs during RNA replication, which thereby compensates for the high error-rate of RTC. Our group previously proved that SARS-CoV-2 nsp14-nsp10 functions as an actual proofreading nuclease for the SARS-CoV-2 RNA replication machinery in vitro, enabling SARS-CoV-2 holo-RdRp to resume the stalled replication [23]. In this study, however, SARS-CoV-2 nsp14-nsp10 was unable to stop the ATP-dependent error-prone RNA extension by SARS-CoV-2 RdRp when two or more NTPs are provided (Fig. 4). This suggests that SARS-CoV-2 holo-RdRp might protect the extended mismatched RNA duplexes or other subunits of SARS-CoV-2 RTC, such as a RNA helicase nsp13, is required for the efficient proofreading function of nsp14-nsp10. It is also possible that ATP might modulate activities of holo-RdRp and nsp14-nsp10, either alone or in combination with other ribonucleotides, by an unknown mechanism, so SARS-CoV-2 holo-RdRp can avoid the proofreading function to conduct the error-prone RNA synthesis. In this regard, it is notable that ATP has been shown to modulate the substrate specificity of SARS-CoV RNA helicase nsp13 [28]. Nonetheless, understanding the substrate specificity and processivity of the complete SARS-CoV-2 RTC machinery, in the context of a longer or intact coronaviral genomic RNA, will be required to determine the precise biochemical mechanism of SARS-CoV-2 replication.
In summary, our combined results provide previously unknown insights into the mechanism and function of SARS-CoV-2 RTC, in particular regarding its high mutation-rate RNA replication. Both holo-RdRp and ExoN of nsp14-nsp10 is structurally and functionally conserved across CoVs, which makes these complexes an attractive target for the development of broad-spectrum anti-CoV agents. Along with ongoing efforts to pharmacologically modulate the RNA replication by SARS-CoV-2 holo-RdRp, simultaneous blockade of both holo-RdRp and ExoN activity of nsp14-nsp10 would likely synergize to effectively combat and control current SARS-CoV-2 pandemic as well as future zoonotic CoV-mediated diseases.
4. Experimental procedures
Plasmids and protein purification: Recombinant SARS-CoV-2 nsp12, nsp7, nsp8, nsp14, and nsp10 proteins were overexpressed in E. coli BL21 (DE3) and purified as described previously [23]. Purified SARS-CoV-2 nsp12, nsp7, nsp8, nsp14, and nsp10 proteins were concentrated and then stored at − 80 °C as described previously [23].
RNA polymerase and proofreading assays: The RNA oligos (Table S1) were synthesized by Integrated DNA technologies. Inc. The RNA polymerase assay was performed as described previously [23]. Briefly, 20 μl reaction mixtures containing 1 μM SARS-CoV-2 nsp7, 2 μM SARS-CoV-2 nsp8, and 1 μM SARS-CoV-2 nsp12, and 100 nM annealed RNA oligos were preincubated in a buffer containing 10 mM Tris (pH 7.5), 10 mM KCl, 1 mM β-mercaptoethanol, 2 mM MgCl2 for 2 min in 30 °C. The reaction was initiated by adding different combinations of 200 μM or 2 μM of each NTP or dNTP (New England Biolabs). The reactions were incubated at 30 °C for 30 min and then quenched by adding an 80 μl loading buffer (formamide with 50 mM EDTA) followed by boiling at 95 °C for 5 min. RNA polymerization products were analyzed in 13% polyacrylamide gels containing 8 M urea, 89 mM Tris-borate (pH 8.0), and 2.0 mM EDTA, and visualized by fluorescence using iBrightFL1000 (ThermoFisher). Bands were quantified using ImageJ software (NIH).
The RNA proofreading assay was performed as described previously [23]. Briefly, mixtures (20 μl) containing 1 μM SARS-CoV-2 nsp7, 2 μM SARS-CoV-2 nsp8, 1 μM SARS-CoV-2 nsp12, and 100 nM annealed RNA oligos were preincubated in a buffer containing 10 mM Tris (pH 7.5), 10 mM KCl, 1 mM β-mercaptoethanol, 2 mM MgCl2 for 2 min in 30 °C. Then 500 nM of SARS-CoV-2 nsp14-nsp10 was added to reactions. The reaction was initiated by adding different combinations of 200 μM or 2 μM of each NTP. Reactions were quenched and RNA products were visualized as described above. Of note, in this assay condition, SARS-CoV-2 nsp14-nsp10 effectively excised the A:A mismatch at the 3′ end of the RNA primer, restoring the RNA replication by SARS-CoV-2 holo-RdRp [23].
Data availability
All data are contained within the article and in the supporting information.
Author contributions
Y.P., Z.M., and I.K.K conceptualized this study and wrote the original draft. Y.P. and Z.M. performed all data analysis, investigation, and methodology. I.K.K. supervised overall studies, edited the manuscript, and provided funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
I.K.K. was supported by National Institutes of Health (R01 GM141226 to I.K.K.) and American Cancer Society RSG Grant (133405-RSG-19-200-01-DMC to I.K.K.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2022.07.087.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gulyaeva A.A., Gorbalenya A.E. A nidovirus perspective on SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021;538:24–34. doi: 10.1016/j.bbrc.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tregoning J.S., Flight K.E., Higham S.L., Wang Z., Pierce B.F. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021;21:626–636. doi: 10.1038/s41577-021-00592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dangerfield T.L., Huang N.Z., Johnson K.A. Remdesivir is effective in combating COVID-19 because it is a better substrate than ATP for the viral RNA-dependent RNA polymerase. iScience. 2020;23 doi: 10.1016/j.isci.2020.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J., Sun Q., Yang X., Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428. doi: 10.1016/j.cell.2020.05.034. e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Hobartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3900–3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drake J.W., Holland J.J. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., Scherbakova S., Graham R.L., Baric R.S., Stockwell T.B., Spiro D.J., Denison M.R. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., Rocchi P., Ng W.L. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol. Cell. 2020;79:710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Sullivan J., Tedim Ferreira M., Gagne J.P., Sharma A.K., Hendzel M.J., Masson J.Y., Poirier G.G. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat. Commun. 2019;10:1182. doi: 10.1038/s41467-019-08859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogando N.S., Zevenhoven-Dobbe J.C., van der Meer Y., Bredenbeek P.J., Posthuma C.C., Snijder E.J. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020;94 doi: 10.1128/JVI.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chien M., Anderson T.K., Jockusch S., Tao C., Li X., Kumar S., Russo J.J., Kirchdoerfer R.N., Ju J. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. J. Proteome Res. 2020;19:4690–4697. doi: 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jockusch S., Tao C., Li X., Anderson T.K., Chien M., Kumar S., Russo J.J., Kirchdoerfer R.N., Ju J. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antivir. Res. 2020;180 doi: 10.1016/j.antiviral.2020.104857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M., Liu Z., Wang H., Li Y., Chen Y., Guddat L.W., Wang Q., Rao Z., Lou Z. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184:184–193. doi: 10.1016/j.cell.2020.11.016. e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573. doi: 10.1016/j.cell.2020.07.033. e1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C., Shi W., Becker S.T., Schatz D.G., Liu B., Yang Y. Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Science. 2021;373:1142–1146. doi: 10.1126/science.abi9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan L., Yang Y., Li M., Zhang Y., Zheng L., Ge J., Huang Y.C., Liu Z., Wang T., Gao S., Zhang R., Huang Y.Y., Guddat L.W., Gao Y., Rao Z., Lou Z. Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell. 2021;184:3474–3485. doi: 10.1016/j.cell.2021.05.033. e3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Z., Pourfarjam Y., Kim I.K. Reconstitution and functional characterization of SARS-CoV-2 proofreading complex. Protein Expr. Purif. 2021;185 doi: 10.1016/j.pep.2021.105894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jockusch S., Tao C., Li X., Chien M., Kumar S., Morozova I., Kalachikov S., Russo J.J., Ju J. Sofosbuvir terminated RNA is more resistant to SARS-CoV-2 proofreader than RNA terminated by Remdesivir. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-73641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 26.Buonaguro L., Tagliamonte M., Tornesello M.L., Buonaguro F.M. SARS-CoV-2 RNA polymerase as target for antiviral therapy. J. Transl. Med. 2020;18:185. doi: 10.1186/s12967-020-02355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., 3rd, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang K.J., Jeong S., Kang D.Y., Sp N., Yang Y.M., Kim D.E. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci. Rep. 2020;10:4481. doi: 10.1038/s41598-020-61432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and in the supporting information.