Abstract
Malignant pleural mesothelioma is a rare and aggressive neoplasm, which has primarily been attributed to the exposure to asbestos fibers (83% of cases); yet, despite a ban of using asbestos in many countries, the incidence of malignant pleural mesothelioma failed to decline worldwide. While little progress has been made in malignant pleural mesothelioma diagnosis, bevacizumab at first, then followed by double immunotherapy (nivolumab plus ipilumumab), were all shown to improve survival in large phase III randomized trials. The morphological analysis of the histological subtyping remains the primary indicator for therapeutic decision making at an advanced disease stage, while a platinum-based chemotherapy regimen combined with pemetrexed, either with or without bevacizumab, is still the main treatment option. Consequently, malignant pleural mesothelioma still represents a significant health concern owing to poor median survival (12–18 months). Given this context, both diagnosis and therapy improvements require better knowledge of the molecular mechanisms underlying malignant pleural mesothelioma’s carcinogenesis and progression. Hence, the Hippo pathway in malignant pleural mesothelioma initiation and progression has recently received increasing attention, as the aberrant expression of its core components may be closely related to patient prognosis. The purpose of this review was to provide a critical analysis of our current knowledge on these topics, the main focus being on the available evidence concerning the role of each Hippo pathway’s member as a promising biomarker, enabling detection of the disease at earlier stages and thus improving prognosis.
Key Points
| This review highlights the common and multiple alterations of the Hippo pathway and its regulators, thus suggesting that this pathway is strongly inactivated in malignant pleural mesothelioma, and that YAP/TAZ would likely be major players in pleural carcinogenesis. |
| The Hippo pathway is a promising biomarker, possibly allowing for the detection of malignant pleural mesothelioma at earlier stages, thus improving patient prognosis. |
Epidemiology and Clinic of Malignant Pleural Mesothelioma (MPM)
Malignant pleural mesothelioma (MPM) is a rare (prevalence 8–30 cases/million/year) and aggressive neoplasm, originating from the mesothelium lining the pleural surface of the lungs [1], which has been largely attributed to environmental/occupational exposure to asbestos fibers (83% of cases) [2, 3]. However, despite the ban of using asbestos in many countries for over 20 years, the incidence of MPM has failed to decline worldwide, with a total of 250,000 deaths anticipated for the next 30 years [4, 5] because of the following reasons: (i) a significant lag between first-time exposure to asbestos and the onset of symptoms in an aging, genetically susceptible population (≥ 30–60 years) [6, 7]; (ii) on-going use of asbestos in middle-income and low-income countries [8]; (iii) presence of other risk factors of MPM’s pathogenesis, including refractory ceramic fibers [9], hormonal factors [10, 11], and some mineral (erionite, fluoro-edenite) or chemical exposures [12–15] such as ionizing radiation [16–18].
Although MPM’s etiology is rather well consolidated, only little progress has been made in MPM diagnosis and treatment. Over the past 20 years, the morphological analysis of the three histological subtypes, including epithelioid, biphasic also called mixed, and sarcomatoid forms, has been the primary indicator for therapeutic decision making at an advanced disease stage [5, 19, 20]. In daily practice, only a few effective biomarkers have been recommended for MPM [21], whereas an invasive biopsy followed by first-line systematic chemotherapy with cisplatin plus pemetrexed, with or without bevacizumab, are still the main treatment options [22–24]. Indeed, double immunotherapy combining nivolumab and ipilimumab has recently emerged for frontline [25] or second-line therapy [26], with an overall survival (OS) benefit that was mainly observed in patients with the non-epithelioid histological subtype, while median OS in patients with the epithelioid subtype was close to that recorded in patients who underwent chemotherapy plus bevacizumab (around 18 months) [21]. Consequently, MPM still represents a significant health concern, with poor median survival (12–18 months [21]). Given this context, both diagnosis and therapy improvements require better knowledge of the molecular mechanisms underlying the carcinogenesis and progression of MPM.
Cellular Processes and Molecular Alterations Leading to MPM
Damages Induced by Asbestos Fibers
The underlying mechanisms by which asbestos fibers induce malignant transformation of mesothelial cells remain enigmatic [27]. Fiber dimension and surface properties are critical determinants of asbestos bioactivity and toxicity [28]. Nevertheless, the three following theories have been proposed to explain how asbestos fibers could induce carcinogenesis:
The controversial “chromosome tangling theory” suggests that the direct action of asbestos by physical interactions with chromosomes of mesothelial cells during cell division causes DNA breaks and lesions, responsible for structural and numerical chromosomal abnormalities [29, 30].
The “oxidative stress theory” explains that an indirect action of asbestos, due to the inability of phagocytic cells to digest asbestos fibers, results in a massive release of reactive oxygen species and reactive nitrogen from macrophages (Fig. 1), thus causing chronic inflammation [31, 32]. Consequently, oxidative stress impairs DNA repair mechanisms [33, 34] and leads to genetic and epigenetic alterations responsible for uncontrolled growth, resistance to apoptosis, and ultimately the occurrence of MPM [33, 35, 36].
According to the “adsorption theory,” the presence of negative or positive charges on asbestos imprisons specific proteins or chemicals, including the components of cigarette smoke in vivo [37].
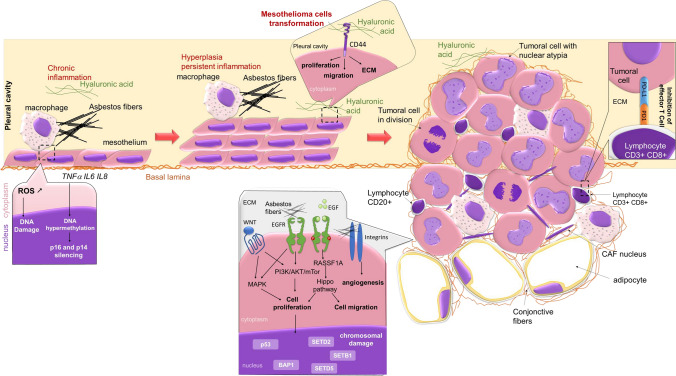
Fig. 1.
Cellular processes leading to malignant pleural mesothelioma (MPM) initiation and growth. The asbestos fibers induce MPM by causing chronic inflammation of the parietal pleura recruiting phagocytes [279]. Phagocytes fail to degrade these fibers, which (i) generates reactive oxygen species (ROS) and nitrogen species resulting in a chronic inflammatory reaction, (ii) alters the DNA molecule of the mesothelial cells, with the appearance of chromosomal aberrations and mutations, (iii) impairs DNA repair mechanisms [33, 34], and (iv) leads to epigenetic alterations responsible for uncontrolled growth, resistance to apoptosis, and hence the occurrence of MPM [33, 35, 46, 279]. In addition to their action on the DNA of mesothelial cells, asbestos fibers would activate several signaling pathways (MAPK, PI3K/AKT/mammalian target of rapamycin) involved in the survival of mesothelial cells downstream of tyrosine kinase receptors such as the epidermal growth factor (EGF), hepatocyte growth factor, or downstream integrin receptors. In parallel, signaling from growth factor receptors can be reinforced, these receptors often being overexpressed, thus accentuating the stimulation of the Ras association domain family 1 isoform A (RASSF1A)/Hippo signaling pathway, a pathway involved when it functions physiologically in cellular homeostasis. Malignant pleural mesothelioma are lesions rich in T lymphocytes and fibroblasts, but expression of PDL-1 by tumoral mesothelial cells could inhibit CD4+ and CD8+ T-cell activation or lead to T-cell apoptosis, allowing tumor growing [102]. Finally, the interaction of hyaluronic acid with its main receptor, CD44, regulates matrix assembly, cytoskeleton architecture, cell migration, proliferation and differentiation of cancer stem cells [155, 156], and activates matrix metalloproteinases (MMPs) involved in tumor progression [165]. CD cluster of differentiation, ECM extracellular cell matrix, EGFR EGF receptor, MAPK mitogen activated protein kinase, PD-L1 programmed death-ligand 1, PD1 programmed death-1, PI3K phosphoinositide 3-kinase, STAT Signal Transducers and Activators of Transcription
Furthermore, growing evidence suggests that asbestos-induced damage is supported by a genetic predisposition, mainly concerning tumor suppressor genes involved in DNA damage repair/checkpoint, including BRCA2 [34, 38–40], CHECK1, TOP2A [41], ERCC1, and XRCC1 [42]. In addition to their action on mesothelial cells’ DNA, asbestos fibers could activate several signaling pathways (STAT3, MAPK, or PI3K/AKT/mammalian target of rapamycin [mTOR]) involved in the survival of mesothelial cells downstream of tyrosine kinase receptors such as epidermal growth factor, hepatocyte growth factor, or downstream integrin receptors [43–45]. Other early epigenetic modifications have likewise been described such as histone modification [46, 47] (Fig. 1).
In response to chronic inflammation caused by asbestos deposition, the single flat layer of mesothelial cells often forms multicellular layers that start to release numerous cytokines and growth factors that stimulate neo-angiogenesis and autophagy, which then, in turn, promote cell survival and malignant transformation [46, 48, 49]. For instance, it has been shown that the release of tumor necrosis factor-α from surrounding immune cells activates the nuclear factor-kappa B pathway, which induces cell proliferation and resistance to apoptosis, thus increasing the number of mesothelial cells that survive following asbestos exposure [50, 51].
Recurrent (Epi)genetic Alterations of MPM Apart from Alteration of the Hippo Signaling Pathway
Several recurrent genetic or epigenetic alterations in MPM lesions are now well defined. These molecular considerations reveal the lack of many oncogenic events that are typical to other tumors (KRAS, BRAF, epidermal growth factor receptor [EGFR], and PIK3CA gene mutations) in MPM, while loss of tumor suppressor genes (TSGs) emerges as a molecular signature [23, 46, 52, 53]. The recurrent alterations of MPM are listed below.
Inactivation of Tumor Suppressor Genes
p16 and p14 (Cyclin-Dependent Kinase Inhibitor 2A/2B CDKN2A/2B Gene)
The cyclin-dependent kinase inhibitor 2A (CDKN2A) gene encodes, by alternative splicing, two cell-cycle repressors, p16INK4a and p14ARF, which are involved in the cell-cycle control by regulating retinoblastoma and p53 proteins [54]. The p16INK4a protein binds to CDK4/6 and inhibits the phosphorylation of retinoblastoma protein, thereby blocking the cell’s transition from G1 to S phase (control point) and, thus, arresting cell division [55]. The loss of p16 INK4a expression leads to the phosphorylation and inactivation of the retinoblastoma protein and, consequently, to unchecked cell-cycle progression into the S phase [56]. The alterations of the p16 tumor suppressor gene, commonly described in MPM, are point mutations (2%), hypermethylation of the promoter (10%), associated with either a loss of heterozygosity of the locus at 9p21 or homozygous deletions (70–80%) [46, 57, 58].
The loss or inactivation of p16INK4a is usually associated with the loss or inactivation of CDKN2A/p14. The p14 ARF protein also participates in cell-cycle control, interacting with human double minute 2 homolog, which is a p53 negative regulator. The p14 ARF loss of expression leads to the degradation of p53 by the proteasome, which is linked to its interaction with human double minute 2 homolog. The coding sequence of p14ARF is deleted in 62% of MPM cases (55% of epithelioid forms, 65% of sarcomatoids, and 71% of biphasics). Among the most common genetic alterations in TSGs, only those in CDKNA2A/2B are associated with poor survival of MPM-affected patients [23, 59]. Importantly, three CDK4/6 inhibitors, namely palbociclib, ribociclib, and abemaciclib, are currently under clinical investigation in MPM management [23].
BAP1
BRCA1-associated protein 1 (BAP1) is a nuclear deubiquitinase acting on lysine 119 of histone H2A, which is involved in regulating essential target genes that are implicated in transcription, DNA repair, cell-cycle control, cell differentiation, and apoptosis [60–62]. It also acts as a homologous recombination DNA repair component found in the BRCA1/BARD1 complex [63]. Somatic inactivating aberrations in BAP1, including point mutations, copy number loss, and rearrangements, are another common inactivation process in 20–64% of MPM cases [52, 59]. Additionally, the germline mutations (7%) are similarly responsible for a cancer susceptibility syndrome [38, 64, 65]. Indeed, some families display a high incidence of MPM after exposure to asbestos due to the loss of one of the asbestos-induced alleles, while the second allele is inactivated by a germline mutation [40, 66]. In a recent study, the alterations of BAP1 in MPM have been revealed to act as a negative response predictor for cisplatin-based chemotherapy via transcriptional downregulation of apoptotic genes [67].
p53
The p53 protein is a transcription factor, which has been described as the genome guardian, given that this agent is essential for maintaining cellular integrity by controlling the expression of many genes involved in cell proliferation, apoptosis, and DNA repair. In healthy cells, only little p53 is detected, owing to its rapid degradation by the proteasome after binding with MDM2, which is responsible for its short half-life (approximately 20 minutes). However, under stress conditions (DNA damage, cell-cycle abnormality, or cellular metabolism disturbance), the p53/MDM2 interaction is abolished leading to p53 stabilization and cell division (in G1/S and G2/M) arrest for DNA repair. If the damage is non-repairable, these cells undergo apoptosis following the activation of other apoptotic genes such as Bax [68].
While a Tp53 mutation is likely the most common genetic abnormality in human cancers, in MPM, p53 loss of function through post-translational regulation is more common than the somatic Tp53 gene mutations that account for about 10% of cases [59, 69–72]. The rarity of Tp53 mutations is related to the low mutational load of these tumors, as compared with bronchial cancers, which has been estimated at 0.79 mutations/megabase, on average [73]. More recently, Tp53 mutations have been associated with poorer OS [74, 75]. Yet, the nuclear accumulation of p53 protein in malignant mesothelial cells is detected in more than 50% of cases, following its stabilization by phosphorylation or acetylation in response to chronic inflammation and the release of reactive oxygen species under the effect of asbestos fibers. It must be emphasized that the first favorable results of the immunotherapy trials conducted in the mesothelioma indication were the results published by Bueno et al. These data underlined the potential immunogenicity of the mutations found on NF2, BAP1, and p53, resulting in the formation of neo-epitopes that are strongly binding (half-maximal inhibitory concentration <50 nM) to the HLA Class I molecules [71].
WIF1
The binding of the Wnt (Wingless Integrase) glycoprotein to the Frizzled receptor activates the Wnt/βcatenin intracellular signaling cascade, leading to the stabilization and accumulation of β-catenin. The latter then enters into the nucleus where it regulates cell proliferation and dedifferentiation through interaction with TCF/LET transcription factors. In MPM, common alterations of the Wnt inhibitory factor-1 and Wnt inhibitory factor-2 regulatory pathways have been well described, resulting in the overexpression of Wnt proteins in tumor cells [76]. For instance, WIF1 is an essential negative regulatory factor of Wnt, which is commonly inactivated through hypermethylation of its promoter in MPM, thereby promoting the activation of this signaling pathway, in addition to uncontrolled mesothelial proliferation [77–79].
SETD2, SETB1, and SETD5
These genes encode histone methyltransferase that regulates the conformation of histones and thus gene transcriptional activity regulation. A decrease in histone methylation leads to tumorigenesis, progression, chemotherapy resistance, and an unfavorable prognosis, suggesting that these genes possibly act as tumor suppressors [80]. Inactivating mutations of these genes, through recurrent mechanism of gene fusions, splice alterations, and nonsynonymous mutations, or their functional loss is a common molecular feature in MPM, as shown in two large studies of massively parallel genomic sequencing and RNA sequencing with a quite probable functional impact according to the prediction program structure/function [71, 80]. Although the functional consequences of these genes’ inactivation in MPM are not yet known, recent research has highlighted that the SETD2 expression level would possibly represent an influential factor for MPM prognosis [81].
Receptor Tyrosine Kinase (RTK) Overexpression
While the acquisition of mutations that activate proto-oncogenes has not been described so far in large genomic studies of MPM, RTK alterations essentially result from protein overexpression without true gene amplification [56, 82–85].
Epidermal Growth Factor Receptor (EGFR)
Overexpression of EGFR has been described in 50–95% of MPM cases, and the pathway is biologically functional [84, 86–88], but activating mutations of EGFR have not been reported in the Western population of patients with mesothelioma [87] (Fig. 1). Accordingly, first-generation tyrosine kinase inhibitors against EGFR, including erlotinib and gefitinib, have failed to reveal any significant efficacy in MPM [89]. Additionally, mutations of signaling pathways downstream of EGFR have been identified in the few publications dealing with this topic. The functional significance of these mutations is still uncertain, and some of them could rather be sequencing artifacts of the paraffin-embedded samples or just constitute a “passenger” event. The most commonly identified event is a PIK3CA mutation, the consequences of which on mesothelial cancer cells are still unknown [59, 90–92].
Hepatocyte Growth Factor Receptor (or Met)
Met activation may contribute to MPM’s pathogenesis. Indeed, mutations in Met receptor’s juxta-membrane domain have been described in 3–16% of MPM cases, the functional consequences of which are still unclear, with overexpression detected in 74–100% of cases upon immunohistochemistry [82, 93–95] (Fig. 1). A predominant location of Met at the mesothelial cancer cells’ membrane is of better prognosis compared with that of nuclear or cytoplasmic localization [94].
Vascular Endothelial Growth Factor Receptor (VEGFR)
A comparative analysis of mesothelioma tumor samples to non-neoplastic mesothelium revealed an increased expression of multiple proangiogenic cytokines, including vascular endothelial growth factor [VEGF] (81% vs 20%), fibrocyte growth factor-1 (67% vs 50%), and fibrocyte growth factor-2 (92% vs 40%) [35]. In addition, the expression of angiogenic cytokines was shown to correlate with increased intra-tumoral microvessel density and worse patient survival [35]. The expression of VEGF receptors has been confirmed in patients’ mesothelioma samples, and it was shown to vary between 20% and 70%, depending on the receptor subtype [96].
In the past 2 decades, different antiangiogenic agents have been trialed in patients with mesothelioma. However, only the Avastin Cisplatin Pemetrexed Study (MAPS) trial demonstrated the benefit of combining bevacizumab (a full-length recombinant humanized monoclonal antibody directed against VEGF) with cisplatin/pemetrexed doublet on OS and progression-free survival in 448 patients with MPM [21, 97]. For a more detailed discussion about the history and role of anti-angiogenic strategies in patients with mesothelioma, we direct the reader to a recent review [98].
Place of the Immune Microenvironment in MPM Development
Numerous studies, involving numerous cancers, have reported that surrounding stromal cells sustain tumor cells’ growth by enhancing genomic instability and epigenetic dysregulation [99–102]. Among them, immune cells have been shown to attempt retaining tumor development in the malignant transformation’s early stages, but they end up being inevitably overwhelmed and, thus, too ineffective to kill tumor cells. The host immune response against cancer cells was actually shown to be negatively regulated by the complex consisting of programmed death-1 (PD-1) and its main ligand PD-L1 (programmed death-ligand 1) [103], as cancer cells expressing PD-L1 inhibit CD4+ and CD8+ T-cell activation or lead to T-cell apoptosis, thereby enabling tumor growth [103].
As in carcinomas, in the early steps of MPM development, CD8+ tumor-infiltrating lymphocytes, which are activated by chronic inflammation due to irritation caused by asbestosis fiber deposits within the pleural space or deep lung, after either inhalation [104] or brought by via blood vessels [105, 106], predict a favorable prognosis for the patient with MPM following tumor resection [107]. Investigation on eight tumor-infiltrating immune cell types, as well as evaluation of the expression of five cytokine/chemokine receptors in 230 patients with MPM demonstrated that CD163/CD8 and CD163/CD20 were independent prognostic factors of survival [108]. However, evaluation of high PD-L1 expression by immunohistochemistry is a poor predictive response marker for PD-L1 inhibitors in patients with MPM [109, 110]. The analysis of PD-L1 expression in 214/448 patients from the phase II/III MAPS trial only revealed that PD-L1 expression was higher and more common in sarcomatoid and biphasic MPM cells than in epithelioid subtype cells, which negatively impacted patients’ outcome [111].
Interestingly, in the multicenter randomized, non-comparative, open-label, phase II clinical MAPS2 trial, in which the patients with MPM who relapsed after one or two lines of pemetrexed-platinum chemotherapy were randomly allocated to receive either the PD1-inhibitor nivolumab alone or in associated with the CTLA4 inhibitor ipilimumab, the authors found encouraging clinical activity of the combination therapy and survival benefits for patients [26]. Consistently, the data from another multicenter, randomized, open-label, phase III trial (CheckMate 743) have supported the beneficial and meaningful effects of first-line nivolumab plus ipilimumab on OS of the patients with unresectable MPM versus platinum plus pemetrexed chemotherapy (median OS: 18.1 months) [25]. It is of note that following these results, nivolumab plus ipilimumab has now been authorized in the USA and in Brazil as first-line treatment for unresectable MPM.
Hippo Signaling
Originally identified and characterized in Drosophila, the Hippo signaling pathway takes its name from one of its major members, the Hippo kinase (MST [Mammalian Ste20-like serine/threonine kinase] in mammals); mutations of this gene lead to tissue hyperplasia, generating a phenotype that evokes features of a hippopotamus. Highly conserved from flies to mammals, the Hippo signaling pathway not only regulates organ size through regulation of cell proliferation and apoptosis [112–114], but it also plays a key role in the self-renewal of stem cells and tissues [115, 116].
Whilst many of the core proteins that constitute the pathway are well established, the full components and their interaction remain to be fully elucidated. In fact, the regulation of this pathway has become increasingly complex in the course evolution, as Drosophila’s protein homologs exist at several isoforms in mammals; they are not only encoded by different genes but also through the alternative splicing of each gene, thereby enabling the production of multiple mature mRNAs from a pre-mRNA [117]. Although these isoforms are structurally very similar and often exhibit interchangeable roles, specific roles of each isoform are currently emerging in the literature. For the sake of clarity, only the mammalian Hippo signaling pathway will be described hereafter, with only a few exceptions.
In mammals, the main proteins of the Hippo pathway are subdivided into three groups (Fig. 2): (i) the upstream regulators (CD44, NF2, RASSF1A…); (ii) the core kinases (MST and NDR [nuclear Dbf2-related kinase]) and their respective adapters; and (iii) the downstream effectors, namely yes-associated protein (YAP) and its paralogous transcriptional co-activator (TAZ) with the PDZ-binding motif.
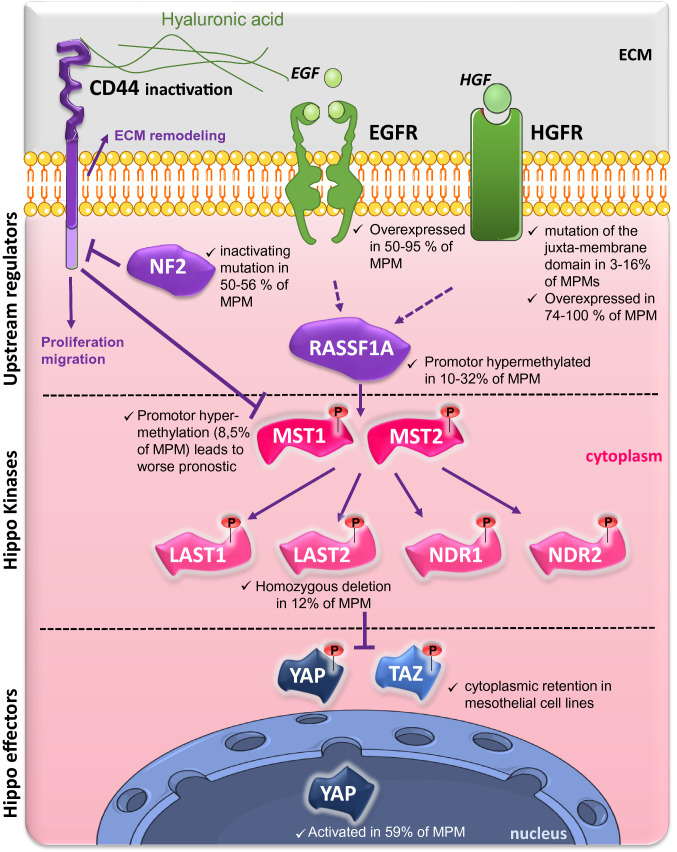
Fig. 2.
Indexed alterations of the expression of members of the RASSF1A/Hippo pathway in malignant pleural mesothelioma (MPM) at a glance. In mammals, the Hippo pathway is subdivided into three groups of proteins: upstream regulators (CD44, NF2, RASSF1A), the core kinases (MST and NDR [nuclear Dbf2-related kinase]) and their respective adapters (not shown) and the end effectors, namely YAP (Yes-associated protein) and its paralogous TAZ (transcriptional co-activator with PDZ-binding motif). CD44, linked to hyaluronic acid (HA), a glycosaminoglycan found abundantly in the pleural cavity leads to cell proliferation/invasion. In healthy mesothelial cells, NF2 negatively regulates the CD44-HA (HA) interaction and thus the pro-tumorigenic activity of CD44 [171], but in MPM, NF2 is inactive in near 50% of MPM [59, 71, 92]. The intracellular segment of CD44 also fixes MST and prevents its action, thus, YAP/TAZ activity cannot be inhibited by their phosphorylation by LATS kinases [172]. Next to this direct inhibition of the activity of Hippo kinases, there are losses of expression of MST1 (MST1 promoter is methylated in 8.5% of MPM cases and leads to worse prognostic of patient [252]) and LATS2 (homozygous deletion mutations are found in 12% of the MPM [250]). Each of these anomalies results in an aberrant activation of YAP, that why, YAP has been shown to be constitutively activated in 59% of patients with MPM [254] while TAZ is reported to be sequestered in cytoplasm from MPM cell lines [252]. CD cluster of differentiation, ECM extracellular cell matrix, EGF epidermal growth factor, EGFR EGF receptor, HGFR hepatocyte growth factor receptor, LATS large tumor suppressor kinase, MST mammalian Ste20-like serine/threonine kinase, NDR nuclear Dbf2-related kinase, TAZ transcriptional co-activator with PDZ-binding motif, YAP Yes-associated protein
During development, the Hippo pathway participates in the transition from a proliferative/undifferentiated state into a non-proliferative/differentiated state of epithelial cells. In undifferentiated and proliferating (non-confluent) cells, core kinases of the Hippo pathway remain inactive. As a result, non-phosphorylated transcriptional coactivator YAP/TAZ enter the nucleus and regulate the expression of several target genes that are mainly involved in cell proliferation and apoptosis. At confluence, the activation of upstream Hippo components leads to the subsequent phosphorylation of YAP/TAZ and their cytoplasmic sequestration blocking their activation of target genes in the nucleus [118, 119].
Importantly, with the exception of YAP and TAZ, all the other Hippo pathway members are considered to be the tumor-suppressor genes, which exhibit frequent alterations in multiple cancers through a variety of mechanisms including amplification, mutations in upstream signaling factors, and gene fusions (reviewed in [120, 121]). In this case, despite contact inhibition, YAP/TAZ continue to translocate into the nucleus and induce target gene expression. This aberrant nuclear localization has been correlated with poor prognosis [122–124].
Upstream Regulators of the Hippo Pathway
A large number of proteins, which can regulate Hippo kinases (MST and NDR [LATS1/2, NDR1/2) and their effectors (YAP and TAZ), are localized at intercellular junctions (tight and adherent). These regulators are involved in the maintenance of apical-basal polarity or epithelial cell shape and geometry [125]. These components are also known to sense, integrate, and respond to extracellular physical forces, such as attachment and mechanical forces, and chemical signals, such as hormones and growth factors, from neighboring cells, in addition to the extracellular matrix (ECM), and surrounding biological fluids [113, 119, 126].
Regulators of the Hippo pathway include:
components of apical-basal polarity complexes such as Scribble [127], Crumbs [128–130], PAR (protease-activated receptors) [131], Ex (Expanded), Kibra (kidney and brain expressed protein), NF2, α-catenin [132–134], AMOT (angiomotin) [115, 135–137], and PTPN14 (protein tyrosine phosphatase non-receptor type 14) [138, 139],
components of the FAT4 polarity complexes [140],
guardians of the epithelial phenotype such as RASSF1A [141],
G-protein coupled receptors [112],
cytoskeletal proteins [123],
ECM, including its composition, density and, therefore, rigidity [143, 144].
The Hippo pathway regulators that are best characterized and most frequently recognized as implicated in tumorigenic processes are as follows:
Neurofibromatosis Type 2 (NF2)
The Neurofibromatosis Type 2 (NF2) gene encodes a “Merlin” protein of 595 amino acids with strong sequence homology to the ezrin/radixin/moesin family proteins. This protein plays a role in the dynamics and structure of the cell surface by linking transmembrane proteins to the actin cytoskeleton. Consequently, NF2 is known to regulate the interaction of mesothelial cells with ECM cycle elements, as well as that of mesothelial cells with each other, as it stabilizes the intercellular junctions. By stabilizing these junctions, NF2 contributes to establishing and maintaining the apical-basal polarity of mesothelial cells, which is lost early during the cellular transformation process [145]. By its functional interaction with adhesion proteins such as CD44 (hyaluronic acid [HA] receptor) [146], FAK [147, 148], or Met receptor, NF2 represses several signaling cascades including the mTOR pathway and Hippo signaling pathway [56]. The inactivation of NF2-Hippo signaling induces the activation of YAP, which is responsible for the transcription of several genes that promote cell proliferation and anti-apoptotic activity. Last, NF2 participates in the regulation of several cellular processes including the invasion, proliferation, and survival of mesothelial cells [149, 150]. Thus, Merlin negative expression has been suggested to represent a biomarker of worse prognosis, as based on 344 patients with MPM treated with either the FAK inhibitor defactinib or placebo as maintenance therapy following induction chemotherapy [151]. Nonetheless, such subgroup analysis did not support this compound’s further clinical development, given that defactinib failed to significantly prolong survival as compared with placebo. However, YAP tyrosine 357 phosphorylation by FAK was shown to mediate nuclear localization of YAP and favor drug-persistent cancer cells in an EFGR-mutated lung cancer organoid model, whereas a FAK inhibitor was able to suppress such escape to treatment. This observation shed a new light on the potential role of FAK inhibition in YAP-driven cancer processes [152].
Finally, a role for NF2/Merlin in the nucleus has also been evidenced, as Merlin was shown to translocate into the nucleus in order to inhibit the culling-RING E3 ubiquitin ligase CRL4DCAF1 [153]. CRL4DCAF1 directly ubiquitinylates and destabilizes the Hippo pathway tumor suppressor kinases LATS1 and LATS2 within the nucleus. Such inhibition of LATS1/2 activates the Hippo pathway terminal effector and transcriptional co-activator YAP, resulting in the transcription of genes involved in tumorigenesis. More recently, blocking of CRL4DCAF1 ubiquitin ligase activity by targeting its upstream activator NEDD8-activating enzyme was shown to suppress LATS1/2 ubiquitinoylation in vitro, leading to inhibitory YAP phosphorylation at serine 127. Furthermore, activation of mTOR in NF2-mutated cells was shown to play a role, given that an in vitro synergy was found between a CRL4DCAF1 inhibitor and dual mTOR/PI3K inhibitor, though the interplay between the Hippo pathway and mTOR signaling is still imperfectly understood.
CD44
CD44, a non-kinase transmembrane glycoprotein, is widely expressed on the surface of multiple cell types including endothelial cells, epithelial cells, mesothelial cells, fibroblasts, and leukocytes [154, 155]. The interaction of the CD44 receptor with appropriate extracellular ligand was shown to be involved in cell-cell and cell-matrix interactions and, consequently, regulates cells proliferation, adhesion, migration, lymphocytes activation, and release of cytokines [156, 157]. All these biological properties are essential for the physiological activities of normal cells, but they can also influence the behavior of cancer cells.
CD44 was originally known to be the main receptor of HA, a glycosaminoglycan component of the ECM, expressed not only by stromal but also abundantly by cancerous cells [158, 159] (Fig. 1). Elevated HA levels in tumor stroma were shown to correlate with poorly differentiated tumors and short OS through stimulating cancer cell migration and cell invasion [160]. The interaction of HA and CD44 results in activation of various intracellular signaling pathways, including the Rho GTPases [161], MAP kinases, and PI3K/AKT [162] through heterodimerization with tyrosine kinase receptors such as EGFR or Met [163–165]. In addition, CD44 isoforms serve as substrates for matrix metalloproteinases and can modulate cellular signaling and cytoskeleton independently of the RTKs, through the release of physiologically active cleavage products [166–168] (Fig. 2).
Overall, through activation of these pathways, CD44 plays a role in tumor progression through promoting drug resistance, tumor invasiveness, and other oncogenic properties [156, 169, 170]. However, CD44 acts also as a tumor suppressor gene. This activity is related to the interaction of its cytoplasmic domain with NF2, thus preventing HA binding [146, 171], and through its relationship with MST kinase [172, 173].
RASSF1A
RASSF1A belongs to a protein superfamily with strong sequence homologies for a Ras (domain family association) protein-binding domain, which comprises ten members including RASSF1 to RASSF 10. The first identified member in humans is RASSF1, which is encoded by a gene located on the short arm of chromosome 3, measuring 11 Kb and consisting of eight exons. Alternative splicing and the use of both transcription promoters (distance of about 3.5 Kb) allow the synthesis of eight variants, including RASSF1A to RASSF1H [174]. The RASSF1A variant encodes a 39-kDa protein with multiple association domains (Fig. 3), as follows:
Ras association (RA) domain, which constitutes the main structural feature of the RASSF family and extends from amino acids 194–289. This domain allows for a specific interaction with activated members of the Ras family (GTP-bound form), most often after stimulation of RTK, among those most expressed by tumor mesothelial cells, such as EGFR, platelet-derived growth factor receptor, or c-Met hepatocyte growth factor receptor.
C-terminal Salvador/RASSF1/Hippo (SARAH) mediates the direct interaction of RASSF1A with the members of the Hippo signaling pathway, including the SAV/WW45 adapter and MST1/2 kinase.
ataxia telangiectasia mutant domain, which is potentially phosphorylatable by the kinase of the same name, in response to DNA damage in the form of single-strand and double-strand breaks.
The N-terminus CI/DAG domain, which is involved in the associations of RASSF1A with the death receptors complex (TNF-R1/MOAP-1 or TRAIL-R1/MOAP-1).
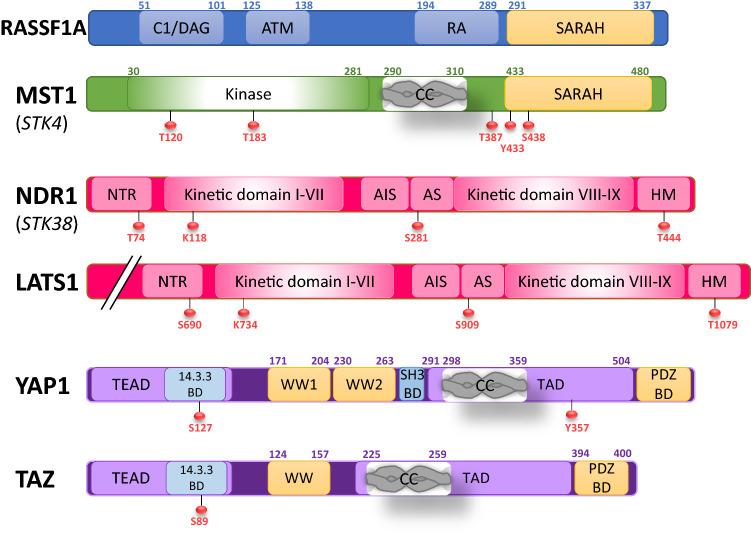
Fig. 3.
Protein structure of the different members of the Ras association domain family 1 isoform A (RASSF1A)/Hippo pathway. Schematic of the main domains of the members of the RASSF1A/Hippo pathway as the main sites of phosphorylation on tyrosine (Y), serine (S), and threonine (T) regulating their activity (https://www.uniprot.org/uniprot). AIS auto-inhibitory sequence, AS activation segment, ATM ataxia telangiectasia mutant, BD binding domain, CC coiled coil, DAG diacylglycerol, NTR N-terminal regulatory domain, RA Ras association, SARAH SAlvador, RASSF1, Hippo, TAD transactivator domain, TEAD TEA domain family member 1)
RASSF1A interacts with nearly 50 different partners, which enables its action as a nexus for the coordination of numerous signaling pathways that control cell fate, cell metabolism, cell communication, cell motility, cell growth and division, as well as cell death [174, 175]. Additionally, RASSF1A is one of the main regulators of the epithelial phenotype (reviewed in [176]). The studies conducted by our group have indeed demonstrated that RASSF1A allows for the maintenance of the epithelial phenotype of human bronchial cells; therefore, altering this pathway in this cellular model causes: (i) epithelial-mesenchymal transition; (ii) acquisition of a migratory phenotype (two-dimensional migration, three-dimensional migration, invasion, and trans-endothelial migration) [141]. Indeed, RASSF1A maintains this epithelial phenotype by regulating the activity of the Hippo pathway kinases and their YAP end effector [141, 177].
The RASSF1 gene encodes another variant named RASSF1C. Despite being encoded by the same gene, RASSF1C has opposite actions to those of RASSF1A, and unlike RASSF1A, the expression of RASS1C is conserved in most cancers thus enabling the expression of its oncogenic actions [178]. Deregulated, the RASSF/Hippo pathway is often involved in tumorigenesis processes including those leading to MPM [179, 180].
Kinases (MST and NDR) of the Hippo Pathway and their Respective Adapters
In mammals, the core of the Hippo pathway is largely characterized by kinase signaling cascade, formed by two types of serine/threonine kinases (MST1 and NDR) and their respective adapters (WW45 and MOB1), activating each other through phosphorylation.
MST Family
The five mammalian-related proteins of MST comprise MST1 (STK4), MST2 (STK3), MST3 (STK24), MST4 (STK26), and YSK1 (STK25 or SOK1). These kinases are members of the “STE20” superfamily [181]. The five MST kinases are divided into two subgroups: MST1/2 and MST3/4/YSK1.
MST1/2 kinases are closely related (76% homology) and widely expressed in human tissues [182]. They are structurally and functionally distinct from MST3/MST4/YSK1, giving them unique roles. In addition to a kinase domain, the protein sequence of the MST1/2 kinases comprises a coiled-coil domain, as well as a SARAH domain, by which they interact with both adaptor proteins, WW45 and RASSF1A (Fig. 3).
MST1/2 are involved in the control of cell growth, apoptosis, and migration, while MST3/MST4/YSK1 plays an essential role in the distribution of the cytoskeleton and subsequent control of cell movements [183]. In fact, MST3 and MST4 regulate the activities of ezrin/radixin/moesin and Paxiline proteins [184]; in addition, they control cell polarity and the organization of the Golgi apparatus through cavernous malformation-3 protein (also known as PDCD10) [185]. YSK1 also controls cell polarity and the organization of the Golgi apparatus, yet through its substrate 14.3.3 protein [186, 187].
Protein WW45 (Sav1)
The WW45 (Sav1) tumor suppressor protein contains two WW domains, a SARAH domain and a coiled-coil region. WW45 is ubiquitously expressed in adult tissues and plays an essential role in the regulation of a wide variety of cellular functions, including protein degradation, transcription, and RNA splicing. This protein was initially identified as an adaptor protein associated with the MST kinases, which promotes MST1-induced cell-cycle blockage and apoptosis [188]. However, WW45 is also required for the stabilization of MST1/2 and forms a complex that brings MST1/2 and LATS1/2 kinases together so that MST1/2 could phosphorylate Lats1/2 [189, 190].
NDR Kinases
The NDR kinases comprise four members in mammals, including the LATS1 and LATS2 kinases, as well as the NDR1 and NDR2 (aka STK38/STK38L) kinases. These kinases belong to the subclass of AGC serine/threonine kinases (protein kinase A, G, and C) that present in species from yeast to humans, and they regulate cellular proliferation and morphogenesis [191] (Fig. 3).
The NDRs are composed of a binding domain at their Mob-called NTR (N-terminal regulatory domain) that other AGC kinases do not have; two kinesics domains I–VII and VIII–IX separated by an auto-inhibitory sequence, and an activation segment located in the catalytic domain, where here is a serine residue, and a C-terminal hydrophobic motif, outside of the catalytic domain, containing a threonine phosphorylation site [192]. The NDR/LATS kinases must be phosphorylated on two residues to achieve full kinase activation. In mammalian cells, MST1/2 activates NDR kinases by phosphorylation, although other upstream regulators are similarly involved in their regulation [193].
NDR1/2
NDR1/2 kinases exhibit strong sequence homology (87%), which provides them with many common roles [194]. NDR1/2 kinases are fully activated by: (i) phosphorylation of their hydrophobic motifs on threonine 444/442 via MST1/2/3 kinases; (ii) their binding to Mob1 via their NTR domain on serine 281/282 in their T-loop activation loop, which is necessary to support the auto-phosphorylation of NDR1/2; and (iii) PP2A, which by dephosphorylating the two threonine 444/442 NDR1/2 inactivates these kinases [193, 195] (Fig. 3).
YAP stability is regulated through its phosphorylation at serine127 residue by NDR1/2 kinases, which results in YAP cytoplasmic sequestration and proteasomal degradation, and subsequently inhibition of the YAP transcriptional activity [196]. Additionally, through phosphorylation of other substrates, NDR1/2 kinases have likewise been implicated in the regulation of ciliogenesis, centrosome duplication, chromosome alignment during mitosis, cytokinesis, and apoptosis [197–201]. Recent studies additionally demonstrated critical functions of NDR kinases in autophagy [202], cell motility [177], innate immunity, and inflammation [203].
The LATS1 and LATS2 kinases share considerable sequence similarity within their kinase domain (85% similarity) that is located at the proteins’ C terminus, while the N terminus portion exhibits significantly lower conservation. Each kinase displays two main regulatory sites, including an activation loop where the serine residues 909 and 841 are harbored, in addition to a phosphorylation site by MST kinases on threonine 1079 and 1041 [204] (Fig. 3). The phosphorylation of these two residues promotes the activity of YAP/TAZ transcription cofactors through their phosphorylation on four to five serine residues (including S127 and S381 for YAP and S89 for TAZ). LATS1/2 can be dephosphorylated at both sites by PP2A phosphatase.
LATS1/2
However, YAP/TAZ are not the only substrates of LATS1/2 kinases, as the latter also control cellular homeostasis, cell-cycle regulation, cell mobility, and apoptosis through the regulation of the actin cytoskeleton and microtubules, particularly through modulation of the LIMK/Cofilin pathway [113, 141, 205]. Overall, by modulating the functions of both oncogenic and tumor suppressor effector, LATS kinases have emerged as central regulators of cell fate [206].
Mob1 Protein
The Mob adaptor proteins impact the Hippo pathway through interactions with both effector kinases and an inactivating phosphatase [207]. The human genome encodes seven MOB-related proteins (hMOB1A/B, hMOB2 A/C, and hMOB3A/B/C), whereas hMOB1A and hMOB1B share 95% sequence identity [208]. Interestingly, only hMOB1 protein directly interacts with all four human NDR kinases, whereas hMOB2 only forms a complex with NDR1/2 but not with LATS1/2 kinases [195]. The effectiveness of hMOB1 binding with NDR kinases has proven to be significantly increased upon phosphorylation on threonine residues (T12/35) by MST1/2 kinases [209–212]. This results in conformational changes [195]. In addition, the formation of the hMob1/NDR and hMob1/LATS complexes leads to an increased autophosphorylation of NDR and LATS kinases on their regulatory loops. The interaction of hMob1 with NDR is also essential for the phosphorylation of NDR1/2 hydrophobic motifs by MST kinases, whereas the phosphorylation of LATS/2 motifs is independent from this interaction [213]. Importantly, there is a competition between hMob1 and hMob2 for NDR kinase binding, whereas hMob1 interaction is associated with increased NDR1/2 activity, while hMob2 binding to NDR1/2 blocks kinase activation [214, 215].
Downstream Effectors, YAP and TAZ
Following the sequential phosphorylation/activation of MST and then NDR kinases, transcription cofactors YAP and TAZ are directly phosphorylated by NDR kinases, on Ser127 and Ser89 residues, respectively, which leads to their cytoplasmic sequestration by the protein 14.3.3 [216] or alpha-catenin [217, 218]. When the NDR kinases remain inactive, non-phosphorylated YAP/TAZ accumulate within the nucleus, and thus drive a pro-oncogenic transcriptional program. Interestingly, YAP/TAZ transcriptional activity can be promoted by phosphorylation on tyrosine 357 by the SRC/YES1 kinase, c-Abl, or FAK, following nuclear localization [137, 219–223].
The 46% homologous YAP and TAZ proteins are derived from the transcription of two distinct genes, YAP1 (11q22) and WWTR1 (3q25), which are probably derived from a duplication of the YAP1 gene during evolution [224]. Importantly, these two transcriptional cofactors do not harbor an intrinsic DNA-binding domain, whereas they share several functional domains enabling them to interact with more than 20 proteins, including transcription factors that allow them to activate different clusters of downstream target genes (Fig. 3) as follows:
TEAD-binding domain (TB), which recognizes the transcriptional enhancer factor domain (TEAD) family of transcription factors;
14.3.3 binding domain;
one or two WW interaction domains (isoform dependent), where two conserved tryptophan (W) residues enable the interaction with a number of PPxY motif-containing proteins (particularly LATS1/2 and Kibra);
coiled-coil domain;
transcriptional activation domain, which is rich in serine, threonine, and acidic amino acids, which governs the transcriptional activity of DNA-binding proteins, most notably TEADs, along with others, such as SMADs, RUNXs, p63/p73, and AP-1;
C-terminal PDZ-binding motif enabling YAP and TAZ to interact with all the proteins containing the same PDZ motif [225];
N-terminal proline-rich domain and an SH3-binding motif that is present on YAP but not on TAZ.
YAP has eight known alternately spliced isoforms (from YAP1-1α to YAP1-1δ and YAP1-2α to YAP1-2δ) separated into two groups, YAP1-1 and YAP1-2, which differ in the transcriptional activation domain regions and the second WW domain (four YAP-1 isoforms display a single WW domain, whereas the four YAP1-2 isoforms display only two) [226]. However, the functional importance of different YAP isoforms is still incompletely understood.
The gene WWTR1, which encodes the protein TAZ (400 amino acids), consists of seven exons. At present, no work has been published focused on the existence of the TAZ isoforms in humans. The TAZ protein structure is largely similar to that of YAP, yet with only one WW domain present on this YAP paralog (Fig. 3).
Following activation and interaction with partner transcription factors, the cofactors YAP and TAZ are then rendered able to regulate apoptosis, motility, growth, and cell proliferation. We should keep in mind that transcriptional activity of Yap and Taz can be modulated via other signaling pathways, which are also “non-canonical” pathways (for a review, please see [112, 227]).
Alteration of Hippo Pathway Members in MPM
Most members of the Hippo signaling pathway are potent tumor suppressors, as their inactivation promotes the migration, invasion, and malignancy of cancer cells through strong activation of downstream effectors YAP and TAZ, with the latter being the only oncogenes of this pathway. These events strongly correlate with poor patient prognosis (Fig. 2).
Here, below, is the current situation of Hippo signaling alteration in MPM:
CD44 Inactivation
The tumor suppressor role of CD44 is related, at least to some extent, to its interaction with NF2. Indeed, dephosphorylated NF2 binds to cytoplasmic domain of CD44 (first 50 amino acids of NF2), instead of ezrin/radixin/moesin proteins, thus blocking the signal transduction and consequent enhancement of cell proliferation and invasion [146]. By blocking the interaction of HA with CD44, NF2 prevents the pro-tumorigenic activity of CD44 [171]. In addition, NF2 overexpression inhibits CD44 ectodomain cleavage by the metalloprotease, thus preventing cellular migration as a crucial tumor property [228]. These results support not only the idea that proteolytic processing of CD44 promotes tumor growth, but also the hypothesis that cells exhibiting NF2 gene mutations have become unable to block CD44 cleavage. These cells are thus naturally predisposed to malignant degeneration.
The tumor suppressor activity of CD44 additionally depends on its relationship with MST kinase [172], namely, the first core kinase of the Hippo pathway cascade. In the presence of high standard CD44 levels, YAP/TAZ activity cannot be inhibited by their phosphorylation by LATS kinases that are localized downstream of the MST kinases. Indeed, the interaction of cytoplasmic CD44 domain with MST prevents its action occurring [173]. Following cleavage of CD44, the intracellular domain can be translocated into the nucleus and then modulates the expression of certain genes, including matrix metalloproteinase 9, which further increases the invasion capacity of cells.
In addition, high HA expression levels have been associated with drug resistance and progression in different tumor cells that initially displayed initial sensitivity to the drugs, particularly on account of the CD44/HA interaction’s capacity to stimulate the expression of MDR1 [229] or MRP2 [230] and their corresponding ATP-binding cassette (ABC) transporters, which can induce the selective expulsion of anti-cancer treatments out of cells.
Already many years ago, several studies had revealed the existence of high HA levels in patients’ pleural effusions [231, 232] and reported the interaction of HA with CD44 in the MPM setting [233]. Indeed, some MPM cell lines display HA binding sites on their cell surface, whereas these sites are absent in normal mesothelial cells [234]. In MPM, it is more specifically established that HA and its receptors facilitate neoplastic cell motility and invasion, and thus that CD44/HA interactions facilitate tumor progression. Malignant pleural mesothelioma cell lines expressing high CD44 amounts thus exhibit increased proliferation and invasion capacity following HA treatments. These abilities are suppressed following inhibition of CD44 function by gene knockdown or neutralizing antibodies [232, 235]. Remarkably, by using lipid nanoparticles equipped with a new HA derivative conjugated with an anti-cancer drug, in the form of an advanced drug-delivery system against CD44-expressing cells, a recent study reported an accumulative effect on cellular drug uptake, which subsequently permitted significant suppression of MPM progression [236].
NF2 (Merlin) Deletion
Loss-of function mutations in the NF2 gene (22q12 locus) were the first molecular alterations identified in MPM [237, 238]. These mutations have been frequently observed, accounting for 20–50% of MPM cases depending on the molecular assessment technique used [59, 71, 92]. These mutations are either deletions of the entire gene or only part of 22q, or insertions leading to truncated and inactive NF2 proteins, and thus the deregulation of all the cellular processes that NF2 helps control [239]. It is worth mentioning that NF2 deficiency contributes to tumorigenesis, independently of the canonical Hippo-Yap pathway [240]. In support of these data, a recent elegant transcriptomic and proteomic study revealed that dysregulation of NF2 and Hippo-Yap engage different protein interactions and exhibit different tumor-infiltrating immune cells in MPM [72].
RASSF1A Silencing
Hypermethylation of the RASSF1 gene promoter (3p21.3), which is known for its tumor suppressor activity, has been reported in 10–32% of MPMs, which was associated with shorter patient OS [241–244]. Surprisingly, our observation demonstrated that RASSF1A’s depletion enhances not only formation of long-membrane protrusion, which is also known as tunneling nanotubes, but it also increases tunneling nanotube-mediated intercellular propagation of different organelles such as mitochondria or lysosomes in mesothelioma cell lines [245]. By facilitating intercellular communication between cells, tunneling nanotubes play a critical role in cancer progression and metastasis [246, 247]. It is worth mentioning that the depletion of all or only part of the 3p21.1 locus has also been related to the function loss of another tumor suppressor gene, which is known as BRCA-associated protein 1 (BAP1) [247, 248].
MST1 and Large Tumor Suppressor 2 (LATS2) Inactivation
In MPM, the Hippo pathway may be inactivated by MST1 or LATS2 inactivation, which could play a crucial role in the deregulation of cell proliferation or survival in mesothelial cells [249, 250]. In a recent study, our group monitored the methylation status of the Hippo pathway components using methylation-specific polymerase chain reaction in 223 samples from patients with MPM who participated in a phase III trial (MAPS/ IFCT-GFPC-0701). This trial assessed the prognostic value of the methylation status with regard to OS and disease-free survival, using both univariate and multivariate analyses. The methylation of RASSF1A, RASSF2A, RASSF6, and RASSF10 were detected in 11.1%, 14.5%, 21.5%, and 4.4% of samples, respectively, whereas no sample exhibited any RASSF5, MST2, LATS1, or LATS2 methylation. Furthermore, none of these methylations influenced patient survival in a univariate analysis [251]. Interestingly, MST1 promoter was methylated in 19/223 samples (8.5%), the median OS of patients with the methylated MST1 promoter being 1.4-fold lower than that of patients with the unmethylated MST1 promoter [251].
The first observation of an inactivating homozygous deletion or point mutation of LATS2 gene (13q12) was first described in 12% of MPM tumor samples (35% of MPM cell lines) using comparative genomic hybridization and sequencing analyses [249]. These results were further confirmed by another study showing that LATS2 mutation results in an increased nuclear accumulation of YAP, where it can activate the transcription of target genes [252]. In complementary experiments, the authors showed that simultaneous mutations of LATS2 and NF2 were observed in 8% of patients with MPM with poor prognosis. Interestingly, this co-mutation appears to be specific to MPM’s carcinogenesis [252]. While the normal NF2 protein promotes YAP’s sequestration and thus inactivation at intercellular adhesion structures, the inactivating NF2 mutations result in an increased non-phosphorylated YAP pool within the cytoplasm (as LATS2 is inactivated), then followed by its nuclear translocation. Another study in this line showed the mutations of both LATS1 and SAV (Salvador or WW45), which represents a chaperone protein that negatively regulates Hippo kinases, were observed in a MPM tumor sample, reflecting again the relevance of this signaling pathway in this tumor [71].
Subsequent YAP Activation
All these alterations of the Hippo pathway and its regulators suggest that this pathway is commonly inactivated in MPM, and that YAP/TAZ could be major players in pleural carcinogenesis. YAP, but not TAZ, could indeed be the effector of the Hippo pathway involved in pleural carcinogenesis. Consistent with this hypothesis, YAP has been shown to be constitutively activated in 59% of patients with MPM tested [253]. Additionally, our group reported that MST1 inactivation reduced cellular basal apoptotic activity, thereby increasing proliferation, invasion, and soft agar or in suspension growth, resulting in nuclear YAP accumulation (yet TAZ cytoplasmic retention in mesothelial cell lines), and that YAP silencing decreased the invasion of MST1-depleted mesothelial cell lines [251]. Based on this observation, YAP could be a putative target for therapeutic interventions in human MPM.
What Therapeutic Targets Among Hippo Pathway Members Should be Considered in Patients with MPM?
The alteration of the Hippo/RASSF signaling pathway is, therefore, a key event in pleural carcinogenesis [250–252, 255]. Restoring the proper functioning of this signaling pathway and monitoring YAP’s nuclear activity could block the progression of various cancers including MPM. Nevertheless, we should bear in mind that, given the multiple number and functions of YAP targets, it would be unfortunate that by blocking one YAP/TAZ downstream effector, we would fully recapitulate the effects of direct YAP/TAZ inhibition. Several strategies could be considered:
-
Increase the activity of core Hippo pathway kinases (MST/NDR) in order to restore YAP cytoplasmic sequestration
Importantly, Hippo pathway agonists, such as C19 or GGTI-298, are under development; these agonists induce the phosphorylation of both MST1/2 and LATS1/2, increase YAP cytoplasmic levels, and enhance TAZ degradation [255, 256].
Using the DNA methyltransferase inhibitor can also activate Hippo pathway upstream regulators RASSF1 and RASSF5 by demethylating their promoter sequence, which ultimately enhances the activation of Hippo pathway, thus reducing tumor growth [257].
-
Inhibit YAP transcription activity
By preventing YAP interaction with TEAD: as mentioned, the YAP-TEAD complex regulates the expression of genes involved in oncogenic transformation [258]. Verteporfin is the first compound identified that directly inhibits YAP-TEAD’s binding and reduces organ overgrowth caused by increased YAP activity [259]. In mesothelial cells, verteporfin significantly reduces the expression of YAP through downregulating its transcription and orientation of the YAP protein to the proteasome for degradation [260]. Unfortunately, verteporfin exhibits many undesirable effects that prevent its use in clinical practice. Another study showed that using the extract from the top flower of a medicinal plant, Dropwort, restrains the oncogenic activity of both YAP and TAZ through promoting ubiquitination in both “in vitro” and “in vivo” MPM models [261]. Additionally, peptide inhibitors that disrupt the interaction between TEAD and YAP/TAZ could represent a potential therapeutic option, as recently shown in gastric cancer [262]. In another study, the authors validated the newly generated tri-functional cell-penetrating peptides with anti-tumoral effects, which block the interaction between YAP and TEAD in the nucleus in both in vitro and in vivo experiments [263]. In addition, the vestigial-like protein VGLL4 is a transcriptional repressor that directly competes with YAP for TEAD binding [196].
-
By inhibiting non-canonical signals and pathways participating in controlling YAP activity. For instance, statins, bisphosphonates, and nitrogen-containing bisphosphates inhibit the mevalonate cascade, which is necessary for the activation of Rho GTPase and thereby suppresses YAP/TAZ activity [264]. The Rho/ROCK inhibitor, GSK269962A, prevents YAP’s activation and inhibits the transcriptional activity of YAP-TEAD [260]. Moreover, ibudilast and rolipram block the phosphodiesterase, thereby activating cAMP-dependent protein kinase A signaling, which ultimately suppresses Yap’s activity [112]. Accordingly, forskolin, which is an adenylyl cyclase activator that leads to cAMP production, can significantly increase YAP phosphorylation and its cytoplasmic accumulation [265]. Likewise, strategies inhibiting both SRC family kinase and PI3K-AKT signaling could likewise complete inhibition of YAP/TAZ in tumors. Nevertheless, they are not yet available [266].
It should be noted that many targeted therapies are used, in addition to conventional therapies, at the second/third therapeutic line for patients, the tumors of whom do not meet the usual protocol inclusion criteria or for those patients who start to develop resistance. Currently, only YAP is being described to modulate the efficacy of therapeutic inhibitors or antibodies, in contrast to conventional chemotherapy for which both YAP and TAZ are likely to be implicated [267, 268]. Indeed, YAP would possibly increase the chemo-sensitivity of hepatocellular carcinoma cells by modulating p53, whereas p53 most probably exerts positive feedback for controlling YAP expression through promoter interaction [269].
Inhibiting YAP’s Anarchic Activity: Is it the Right Therapeutic Strategy to be Adopted for Patients with MPM?
The results obtained from our work on mesothelioma or bronchial epithelial cells raise the question of the relevance of inhibiting YAP activity as a therapeutic strategy in patients with MPM presenting an alteration of the RASSF/Hippo pathway. Indeed, we showed that the concomitant inactivation of YAP and RASSF1A in mesothelial (H28 and H2052) or bronchial (HBEC-3) cell lines failed to prevent all the cellular disorders caused by disrupting the RASSF1/Hippo signaling pathway, while these cells retained their ability to grow without adhesion (agar) [141, 251].
This result could be explained by the observation that there is not one single YAP but rather several YAP isoforms, and that the siRNAs used in our work, as well as in other studies, are not able to inactivate the expression of all these variants. Although only very few data exist regarding the specificities of the different YAP variants, especially because of a lack of proper tools, it has, however, been established that these isoforms do not exert the same function. For instance, as YAP1-1 does not bind to the p73 factor, this agent is unable to induce apoptosis when HEK293 cells are deprived of nutrients [270]. As another example, through interaction with YAP1-2, not YAP1-1, angiomotin inhibits YAP1-2’s proapoptotic function and nuclear shuttling [271]. Similarly, in the melanoma setting, only YAP2 would promote the invasion of tumor cells [272].
If YAP’s inactivation does not prevent all the disorders induced by the disruption of the RASSF1/Hippo pathway in in vitro experiments, this is explained by the observation that the siRNAs used in our work do not inactivate the expression of all of the variants. Given this scenario, it must be ensured that the YAP activity inhibitors that are currently under development actually inhibit all of these isoforms. There may indeed be a compensation phenomenon between these isoforms, when only one of the YAP’s isoforms is being targeted by anti-YAP/TEAD therapies. Additionally, it should be kept in mind that YAP does not contribute to controlling gene expression only by associating with the TEAD transcription factor. Therefore, YAP’s interactions with other transcription factors (Smad1/2/3, RUNX1/2/3, p73) should also be considered for explaining YAP’s oncogenic activity of YAP [79, 273, 274]. Furthermore, YAP is able to interact with p53 mutant proteins, modulating the transcriptional activity of these p53 mutants (cyclin A, cyclin B, and CDK1), which undoubtedly contributes to the oncogenic properties of mutants p53 “function gains” [275]. Finally, another possible scenario to be taken into account is that YAP can play contradictory roles in carcinogenesis. Indeed, if YAP is mostly described as being an oncogene, it can at times behave as having tumor suppressor gene properties [276–278].
Conclusions
This literature review highlights the common and multiple alterations of the Hippo signaling pathway in patients with MPM. It must, however, be noted that signaling pathways interfere in its regulation; therefore, the essential place that this pathway’s deregulations actually play in the natural MPM history is still unclear. Further experiments are required to better understand the manner in which we could correct these deregulations and, thus, halt the development of this devastating cancer. Although we still lack the necessary tools, it has already been established that the Hippo pathway turns out to be a promising biomarker, enabling us to detect MPM at earlier stages and thus improve overall patient prognosis. Given this context, systematic screening of either abnormal promoter methylation patterns of the Hippo pathway component in samples of human tissues or stable circulating microRNA with the capacity of regulating the Hippo pathway (ex: miR-122, miR-847-3p) is likely to be a relevant foundation for possible MPM monitoring and early detection in the post-professional follow-up of workers who were previously exposed to asbestos.
Declarations
Funding
No external funding was received for the preparation of this article.
Conflicts of interest/competing interests
Fatéméh Dubois, Céline Bazille, Jérôme Levallet, Elodie Maille, Solenn Brosseau, Jeannick Madelaine, Emmanuel Bergot, Gérard Zalcman, and Guénaëlle Levallet have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
No datasets were generated or analyzed during the current study.
Code availability
Not applicable.
Author contributions
FD, EM, and GL wrote the original draft of this article, which was then proofread, corrected, and validated by FD, CB, JL, EM, SB, JM, EB, GZ, and GL.
Footnotes
Gérard Zalcman and Guénaëlle Levallet contributed equally to the article.
References
- 1.Robinson BM. Malignant pleural mesothelioma: an epidemiological perspective. Ann Cardiothorac Surg. 2021;1(14):491–497. doi: 10.3978/j.issn.2225-319X.2012.11.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonald JC, McDonald AD. The epidemiology of mesothelioma in historical context. Eur Respir J. 1996;9(9):1932–1942. doi: 10.1183/09031936.96.09091932. [DOI] [PubMed] [Google Scholar]
- 3.Gilham C, Rake C, Burdett G, Nicholson AG, Davison L, Franchini A, et al. Pleural mesothelioma and lung cancer risks in relation to occupational history and asbestos lung burden. Occup Environ Med. 2016;73(5):290–299. doi: 10.1136/oemed-2015-103074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pira E, Donato F, Maida L, Discalzi G. Exposure to asbestos: past, present and future. J Thorac Dis. 2018;10(Suppl. 2):S237–S245. doi: 10.21037/jtd.2017.10.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brims F. Epidemiology and clinical aspects of malignant pleural mesothelioma. Cancers. 2021;13(16):4194–4209. doi: 10.3390/cancers13164194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen D. Working with asbestos and the possible health risks. Occup Med. 2015;65(1):6–14. doi: 10.1093/occmed/kqu175. [DOI] [PubMed] [Google Scholar]
- 7.Abbott DM, Bortolotto C, Benvenuti S, Lancia A, Filippi AR, Stella GM. Malignant pleural mesothelioma: genetic and microenviromental heterogeneity as an unexpected reading frame and therapeutic challenge. Cancers (Basel). 2020;12(5):1186–1209. doi: 10.3390/cancers12051186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen LP, Baez J, Stern MEC, Takahashi K, George F. Trends and the economic effect of asbestos bans and decline in asbestos consumption and production worldwide. Int J Environ Res Public Health. 2018;15(3):531–540. doi: 10.3390/ijerph15030531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andujar P, Lecomte C, Renier A, Fleury-Feith J, Kheuang L, Daubriac J, et al. Clinico-pathological features and somatic gene alterations in refractory ceramic fibre-induced murine mesothelioma reveal mineral fibre-induced mesothelioma identities. Carcinogenesis. 2007;28(7):1599–1605. doi: 10.1093/carcin/bgm023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodríguez-Cid JR, García-Acevedo O, Benjamin-Contreras J, Bonilla-Molina D, Flores-Mariñelarena RR, Martínez-Barrera L, et al. Expression of estrogen receptor beta (ERβ) and its prognostic value in pleural mesothelioma. J Thorac Dis. 2019;11(4):1456–1464. doi: 10.21037/jtd.2019.03.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinton G, Manzotti B, Balzano C, Moro L. Expression and clinical implications of estrogen receptors in thoracic malignancies: a narrative review. J Thorac Dis. 2021;3(3):1851–1863. doi: 10.21037/jtd-20-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metintas S, Metintas M, Ucgun I, Oner U. Malignant Mesothelioma due to Environmental Exposure to Asbestos: follow-up of a Turkish cohort living in a rural area. Chest. 2002;122(6):2224–2229. doi: 10.1378/chest.122.6.2224. [DOI] [PubMed] [Google Scholar]
- 13.Carbone M, Emri S, Dogan AU, Steele I, Tuncer M, Pass HI, et al. A mesothelioma epidemic in Cappadocia: scientific developments and unexpected social outcomes. Nat Rev Cancer. 2007;7(2):147–154. doi: 10.1038/nrc2068. [DOI] [PubMed] [Google Scholar]
- 14.Carbone M, Baris YI, Bertino P, Brass B, Comertpay S, Dogan AU, et al. Erionite exposure in North Dakota and Turkish villages with mesothelioma. Proc Nat Acad Sci USA. 2011;108(33):13618–13623. doi: 10.1073/pnas.1105887108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortega-Guerrero MA, Carrasco-Núñez G, Barragán-Campos H, Ortega MR. High incidence of lung cancer and malignant mesothelioma linked to erionite fibre exposure in a rural community in Central Mexico. Occup Environ Med. 2015;72(3):216–218. doi: 10.1136/oemed-2013-101957. [DOI] [PubMed] [Google Scholar]
- 16.Hodgson DC, Gilbert ES, Dores GM, Schonfeld SJ, Lynch CF, Storm H, et al. Long-term solid cancer risk among 5-year survivors of Hodgkin’s lymphoma. J Clin Oncol. 2007;25(12):1489–1497. doi: 10.1200/JCO.2006.09.0936. [DOI] [PubMed] [Google Scholar]
- 17.Chirieac LR, Barletta JA, Yeap BY, Richards WG, Tilleman T, Bueno R, et al. Clinicopathologic characteristics of malignant mesotheliomas arising in patients with a history of radiation for Hodgkin and non-Hodgkin lymphoma. J Clin Oncol. 2013;31(36):4544–4549. doi: 10.1200/JCO.2013.49.9616. [DOI] [PubMed] [Google Scholar]
- 18.Farioli A, Violante FS, Mattioli S, Curti S, Kriebel D. Risk of mesothelioma following external beam radiotherapy for prostate cancer: a cohort analysis of SEER database. Cancer Causes Control. 2013;24(8):1535–1545. doi: 10.1007/s10552-013-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taioli E, Wolf AS, Camacho-Rivera M, Kaufman A, Lee DS, Nicastri D, et al. Determinants of survival in malignant pleural mesothelioma: a Surveillance, Epidemiology, and End Results (SEER) study of 14,228 patients. PLoS ONE. 2015;10(12):e0145039. doi: 10.1371/journal.pone.0145039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Süveg K, Putora PM, Berghmans T, Glatzer M, Kovac V, Cihoric N. Current efforts in research of pleural mesothelioma: an analysis of the ClinicalTrials.gov registry. Lung Cancer. 2018;124:12–18. doi: 10.1016/j.lungcan.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Zalcman G, Mazieres J, Margery J, Greillier L, Audigier-Valette C, Moro-Sibilot D, et al. Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): a randomised, controlled, open-label, phase 3 trial. Lancet. 2016;387(10026):1405–1414. doi: 10.1016/S0140-6736(15)01238-6. [DOI] [PubMed] [Google Scholar]
- 22.Mutti L, Peikert T, Robinson BWS, Scherpereel A, Tsao AS, de Perrot M, et al. Scientific advances and new frontiers in mesothelioma therapeutics. J Thorac Oncol. 2018;13(9):1269–1283. doi: 10.1016/j.jtho.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Xu D, Schmid RA, Peng RW. Biomarker-guided targeted and immunotherapies in malignant pleural mesothelioma. Ther Adv Med Oncol. 2020;12:1758835920971421. doi: 10.1177/1758835920971421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson BW, Takahashi K, Cheng YY. Preclinical models and resources to facilitate basic science research on malignant mesothelioma: a review. Front Oncol. 2021;11:748444. doi: 10.3389/fonc.2021.748444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baas P, Scherpereel A, Nowak AK, Fujimoto N, Peters S, Tsao AS, et al. First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (CheckMate 743): a multicentre, randomised, open-label, phase 3 trial. Lancet. 2021;397(10272):375–386. doi: 10.1016/S0140-6736(20)32714-8. [DOI] [PubMed] [Google Scholar]
- 26.Scherpereel A, Mazieres J, Greillier L, Lantuejoul S, Dô P, Bylicki O, et al. Nivolumab or nivolumab plus ipilimumab in patients with relapsed malignant pleural mesothelioma (IFCT-1501 MAPS2): a multicentre, open-label, randomised, non-comparative, phase 2 trial. Lancet Oncol. 2019;20(2):239–253. doi: 10.1016/S1470-2045(18)30765-4. [DOI] [PubMed] [Google Scholar]
- 27.Gaudino G, Xue J, Yang H. How asbestos and other fibers cause mesothelioma. Transl Lung Cancer Res. 2020;9(Suppl. 1):S39–46. doi: 10.21037/tlcr.2020.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez VC, Pietruska JR, Miselis NR, Hurt RH, Kane AB. Biopersistence and potential adverse health impacts of fibrous nanomaterials: what have we learned from asbestos? Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2009;1(5):511–529. doi: 10.1002/wnan.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toyokuni S. Mechanisms of asbestos-induced carcinogenesis. Nagoya J Med Sci. 2009;71(1–2):1–10. [PMC free article] [PubMed] [Google Scholar]
- 30.Huang SXL, Jaurand MC, Kamp DW, Whysner J, Hei TK. Role of mutagenicity in asbestos fiber-induced carcinogenicity and other diseases. J Toxicol Environ Health B Crit Rev. 2011;14(1–4):179–245. doi: 10.1080/10937404.2011.556051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J, Thompson J, Shukla A. Asbestos-induced oxidative stress in lung pathogenesis. In: Laher I, editor. Systems biology of free radicals and antioxidants. Berlin: Springer; 2014. [Google Scholar]
- 32.Urso L, Cavallari I, Sharova E, Ciccarese F, Pasello G, Ciminale V. Metabolic rewiring and redox alterations in malignant pleural mesothelioma. Br J Cancer. 2020;122(1):52–56. doi: 10.1038/s41416-019-0661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betti M, Aspesi A, Ferrante D, Sculco M, Righi L, Mirabelli D, et al. Sensitivity to asbestos is increased in patients with mesothelioma and pathogenic germline variants in BAP1 or other DNA repair genes. Genes Chromosomes Cancer. 2018;57(11):573–583. doi: 10.1002/gcc.22670. [DOI] [PubMed] [Google Scholar]
- 34.Carbone M, Ly BH, Dodson RF, Pagano I, Morris PT, Dogan UA, et al. Malignant mesothelioma: facts, myths, and hypotheses. J Cell Physiol. 2012;227(1):44–58. doi: 10.1002/jcp.22724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar-Singh S, Weyler J, Martin MJ, Vermeulen PB, Van Marck E. Angiogenic cytokines in mesothelioma: a study of VEGF, FGF-1 and -2, and TGF beta expression. J Pathol. 1999;189(1):72–78. doi: 10.1002/(SICI)1096-9896(199909)189:1<72::AID-PATH401>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 36.Neri M, Ugolini D, Boccia S, Canessa PA, Cesario A, Leoncini G, et al. Chemoprevention of asbestos-linked cancers: a systematic review. Anticancer Res. 2012;32(3):1005–1013. [PubMed] [Google Scholar]
- 37.Jiang L, Toyokuni S. Elucidation of asbestos-induced mesothelial carcinogenesis toward its prevention. Genes Environ. 2011;33(1):4–9. doi: 10.3123/jemsge.33.4. [DOI] [Google Scholar]
- 38.Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43(10):1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohar JA, Cheung M, Talarchek J, Howard SE, Howard TD, Hesdorffer M, et al. Germline BAP1 mutational landscape of asbestos-exposed malignant mesothelioma patients with family history of cancer. Cancer Res. 2016;76(2):206–215. doi: 10.1158/0008-5472.CAN-15-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carbone M, Yang H. Mesothelioma: recent highlights. Ann Transl Med. 2017;5(11):238. doi: 10.21037/atm.2017.04.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arulananda S, Thapa B, Walkiewicz M, Zapparoli GV, Williams DS, Dobrovic A, et al. Mismatch repair protein defects and microsatellite instability in malignant pleural mesothelioma. J Thorac Oncol. 2018;13(10):1588–1594. doi: 10.1016/j.jtho.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 42.Levpuscek K, Goricar K, Kovac V, Dolzan V, Franko A. The influence of genetic variability of DNA repair mechanisms on the risk of malignant mesothelioma. Radiol Oncol. 2019;53(2):206–212. doi: 10.2478/raon-2019-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuzaki H, Maeda M, Lee S, Nishimura Y, Kumagai-Takei N, Hayashi H, et al. Asbestos-induced cellular and molecular alteration of immunocompetent cells and their relationship with chronic inflammation and carcinogenesis. J Biomed Biotechnol. 2012;2012:492608. doi: 10.1155/2012/492608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamminen JA, Myllärniemi M, Hyytiäinen M, Keski-Oja J, Koli K. Asbestos exposure induces alveolar epithelial cell plasticity through MAPK/Erk signaling. J Cell Biochem. 2012;113(7):2234–2247. doi: 10.1002/jcb.24094. [DOI] [PubMed] [Google Scholar]
- 45.Bonelli M, Terenziani R, Zoppi S, Fumarola C, La Monica S, Cretella D, et al. Dual inhibition of CDK4/6 and PI3K/AKT/mTOR signaling impairs energy metabolism in MPM cancer cells. Int J Mol Sci. 2020;21(14):5165–5181. doi: 10.3390/ijms21145165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chernova T, Murphy FA, Galavotti S, Sun XM, Powley IR, Grosso S, et al. Long-fiber carbon nanotubes replicate asbestos-induced mesothelioma with disruption of the tumor suppressor gene Cdkn2a (Ink4a/Arf) Curr Biol. 2017;27(21):3302–14.e6. doi: 10.1016/j.cub.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshikawa Y, Kuribayashi K, Minami T, Ohmuraya M, Kijima T. Epigenetic alterations and biomarkers for immune checkpoint inhibitors: current standards and future perspectives in malignant pleural mesothelioma treatment. Front Oncol. 2020;14(10):554570. doi: 10.3389/fonc.2020.554570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benedetti S, Nuvoli B, Catalani S, Galati R. Reactive oxygen species a double-edged sword for mesothelioma. Oncotarget. 2015;6(19):16848–16865. doi: 10.18632/oncotarget.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue J, Patergnani S, Giorgi C, Suarez J, Goto K, Bononi A, et al. Asbestos induces mesothelial cell transformation via HMGB1-driven autophagy. Proc Natl Acad Sci USA. 2020;117(41):25543–25552. doi: 10.1073/pnas.2007622117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang H, Bocchetta M, Kroczynska B, Elmishad AG, Chen Y, Liu Z, et al. TNF-alpha inhibits asbestos-induced cytotoxicity via a NF-kappaB-dependent pathway, a possible mechanism for asbestos-induced oncogenesis. Proc Natl Acad Sci USA. 2006;103(27):10397–10402. doi: 10.1073/pnas.0604008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bononi A, Napolitano A, Pass HI, Yang H, Carbone M. Latest developments in our understanding of the pathogenesis of mesothelioma and the design of targeted therapies. Expert Rev Respir Med. 2015;9(5):633–654. doi: 10.1586/17476348.2015.1081066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hylebos M, Van Camp G, van Meerbeeck JP, Op de Beeck K. The genetic landscape of malignant pleural mesothelioma: results from massively parallel sequencing. J Thorac Oncol. 2016;11(10):1615–1626. doi: 10.1016/j.jtho.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 53.Andujar P, Lacourt A, Brochard P, Pairon J-C, Jaurand M-C, Jean D. Five years’ update on relationships between malignant pleural mesothelioma and exposure to asbestos and other elongated mineral particles. J Toxicol Environ Health B Crit Rev. 2016;19(5–6):151–172. doi: 10.1080/10937404.2016.1193361. [DOI] [PubMed] [Google Scholar]
- 54.Potrony M, Puig-Butillé JA, Aguilera P, Badenas C, Carrera C, Malvehy J, et al. Increased prevalence of lung, breast, and pancreatic cancers in addition to melanoma risk in families bearing the cyclin-dependent kinase inhibitor 2A mutation: implications for genetic counseling. J Am Acad Dermatol. 2014;71(5):888–895. doi: 10.1016/j.jaad.2014.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao R, Choi BY, Lee MH, Bode AM, Dong Z. Implications of genetic and epigenetic alterations of CDKN2A (p16(INK4a)) in cancer. EBioMedicine. 2016;8:30–39. doi: 10.1016/j.ebiom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sekido Y. Molecular pathogenesis of malignant mesothelioma. Carcinogenesis. 2013;34(7):1413–1419. doi: 10.1093/carcin/bgt166. [DOI] [PubMed] [Google Scholar]
- 57.Campbell K, Brosseau S, Reviron-Rabec L, Bergot E, Lechapt E, Levallet G, Zalcman G. Mésothéliomes malins pleuraux: le point en 2013. Bull Cancer. 2013;100(12):1283–1293. doi: 10.1684/bdc.2013.1857. [DOI] [PubMed] [Google Scholar]
- 58.Kettunen E, Savukoski S, Salmenkivi K, Böhling T, Vanhala E, Kuosma E, Anttil S, Wolff H. CDKN2A copy number and p16 expression in malignant pleural mesothelioma in relation to asbestos exposure. BMC Cancer. 2019;19(1):507. doi: 10.1186/s12885-019-5652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Rienzo AD, Archer MA, Yeap BY, Dao N, Sciaranghella D, Sideris AC, et al. Gender-specific molecular and clinical features underlie malignant pleural mesothelioma. Cancer Res. 2016;76(2):319–328. doi: 10.1158/0008-5472.CAN-15-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hebert L, Bellanger D, Guillas C, Campagne A, Dingli F, Loew D, et al. Modulating BAP1 expression affects ROS homeostasis, cell motility and mitochondrial function. Oncotarget. 2017;8(42):72513–72527. doi: 10.18632/oncotarget.19872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.LaFave LM, Béguelin W, Koche R, Teater M, Spitzer B, Chramiec A, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21(11):1344–1349. doi: 10.1038/nm.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pillappa R, Maleszewski JJ, Sukov WR, Bedroske PP, Greipp PT, Boland JM, et al. Loss of BAP1 expression in atypical mesothelial proliferations helps to predict malignant mesothelioma. Am J Surg Pathol. 2017;42(2):256–263. doi: 10.1097/PAS.0000000000000976. [DOI] [PubMed] [Google Scholar]
- 63.Srinivasan G, Sidhu GS, Williamson EA, Jaiswal AS, Najmunnisa N, Wilcoxen K, et al. Synthetic lethality in malignant pleural mesothelioma with PARP1 inhibition. Cancer Chemother Pharmacol. 2017;80(4):861–867. doi: 10.1007/s00280-017-3401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Panou V, Gadiraju M, Wolin A, Weipert CM, Skarda E, Husain AN, et al. Frequency of germline mutations in cancer susceptibility genes in malignant mesothelioma. J Clin Oncol. 2018;36(28):2863–2871. doi: 10.1200/JCO.2018.78.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ladanyi M, Zauderer MG, Krug LM, Ito T, McMillan R, Bott M, et al. New strategies in pleural mesothelioma: BAP1 and NF2 as novel targets for therapeutic development and risk assessment. Clin Cancer Res. 2012;18(17):4485–4490. doi: 10.1158/1078-0432.CCR-11-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Betti M, Casalone E, Ferrante D, Romanelli A, Grosso F, Guarrera S, et al. Inference on germline BAP1 mutations and asbestos exposure from the analysis of familial and sporadic mesothelioma in a high-risk area. Genes Chromosomes Cancer. 2015;54(1):51–62. doi: 10.1002/gcc.22218. [DOI] [PubMed] [Google Scholar]
- 67.Oehl K, Vrugt B, Wagner U, Kirschner MB, Meerang M, Weder W, et al. Alterations in BAP1 are associated with cisplatin resistance through inhibition of apoptosis in malignant pleural mesothelioma. Clin Cancer Res. 2021;27(8):2277–2291. doi: 10.1158/1078-0432.CCR-20-4037. [DOI] [PubMed] [Google Scholar]
- 68.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303(5660):1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 69.Papp T, Schipper H, Pemsel H, Bastrop R, Muller KM, Wiethege T, et al. Mutational analysis of N-ras, p53, p16INK4a, p14ARF and CDK4 genes in primary human malignant mesotheliomas. Int J Oncol. 2001;18(2):425–433. doi: 10.3892/ijo.18.2.425. [DOI] [PubMed] [Google Scholar]
- 70.Daubriac JD, Le Pimpec-Barthes J, Galateau-Salle FF, Jaurand MC. Molecular changes in mesothelioma with an impact on prognosis and treatment. Arch Pathol Lab Med. 2012;136(3):277–293. doi: 10.5858/arpa.2011-0215-RA. [DOI] [PubMed] [Google Scholar]
- 71.Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48(4):407–416. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 72.Yang H, Hall SRR, Sun B, Zhao L, Gao Y, Schmid RA, et al. NF2 and canonical Hippo-YAP pathway define distinct tumor subsets characterized by different immune deficiency and treatment implications in human pleural mesothelioma. Cancers (Basel). 2021;13(7):1561–1584. doi: 10.3390/cancers13071561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo G, Chmielecki J, Goparaju C, Heguy A, Dolgalev I, Carbone M, et al. Whole-exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res. 2015;75(2):264–269. doi: 10.1158/0008-5472.CAN-14-1008. [DOI] [PubMed] [Google Scholar]
- 74.Quetel L, Meiller C, Assié JB, Blum Y, Imbeaud S, Montagne F, et al. (Genetic alterations of malignant pleural mesothelioma: association with tumor heterogeneity and overall survival. Mol Oncol. 2020;14(6):1207–1223. doi: 10.1002/1878-0261.12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanagiri T, Shinohara S, Takenaka M, Shigematsu Y, Yasuda M, Shimokawa H, et al. Effects of hyaluronic acid and CD44 interaction on the proliferation and invasiveness of malignant pleural mesothelioma. Tumour Biol. 2012;33(6):2135–2141. doi: 10.1007/s13277-012-0473-5. [DOI] [PubMed] [Google Scholar]
- 76.Fox SA, Richards AK, Kusumah I, Perumal V, Bolitho EM, Mutsaers SE, et al. Expression profile and function of Wnt signaling mechanisms in malignant mesothelioma cells. Biochem Biophys Res Commun. 2013;440(1):82–87. doi: 10.1016/j.bbrc.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 77.Batra S, Shi Y, Kuchenbecker KM, He B, Reguart N, Mikami I, et al. Wnt inhibitory factor-1, a Wnt antagonist, is silenced by promoter hypermethylation in malignant pleural mesothelioma. Biochem Biophys Res Commun. 2006;2(4):1228–1232. doi: 10.1016/j.bbrc.2006.02.084. [DOI] [PubMed] [Google Scholar]
- 78.Kohno H, Amatya VJ, Takeshima Y, Kushitani K, Hattori N, Kohno N, Inai K. Aberrant promoter methylation of WIF-1 and SFRP1, 2, 4 genes in mesothelioma. Oncol Rep. 2010;24(2):423–431. doi: 10.3892/or_00000875. [DOI] [PubMed] [Google Scholar]
- 79.de Assis LVM, Locatelli J, Isoldi MC. The role of key genes and pathways involved in the tumorigenesis of Malignant Mesothelioma. Biochim Biophys Acta. 2014;1845(2):232–247. doi: 10.1016/j.bbcan.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 80.Guo L, Teng L. YAP/TAZ for cancer therapy: opportunities and challenges (review) Int J Oncol. 2015;46(4):1444–1452. doi: 10.3892/ijo.2015.2877. [DOI] [PubMed] [Google Scholar]
- 81.Yu M, Yu M, Zhu LJ, Yuan XY, Zhang X. Expression and clinical significance of SETD2 in maligant pleural mesothelioma. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 2021;39(2):91–98. doi: 10.3760/cma.j.cn121094-20200831-00505. [DOI] [PubMed] [Google Scholar]
- 82.Kawaguchi K, Murakami H, Taniguchi T, Fujii M, Kawata S, Fukui T, et al. Combined inhibition of MET and EGFR suppresses proliferation of malignant mesothelioma cells. Carcinogenesis. 2009;30(7):1097–1105. doi: 10.1093/carcin/bgp097. [DOI] [PubMed] [Google Scholar]
- 83.Brevet M, Shimizu S, Bott MJ, Shukla N, Zhou Q, Olshen AB, et al. Coactivation of receptor tyrosine kinases in malignant mesothelioma as a rationale for combination targeted therapy. Thorac Oncol. 2011;6(5):864–874. doi: 10.1097/jto.0b013e318215a07d. [DOI] [PubMed] [Google Scholar]
- 84.Bonelli MA, Fumarola C, La Monica S, Alfieri R. New therapeutic strategies for malignant pleural mesothelioma. Biochem Pharmacol. 2017;123:8–18. doi: 10.1016/j.bcp.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 85.Grosso S, Marini A, Gyuraszova K, Voorde JV, Sfakianos A, Garland GD, et al. The pathogenesis of mesothelioma is driven by a dysregulated translatome. Nat Commun. 2021;12(1):4920–4937. doi: 10.1038/s41467-021-25173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Destro A, Ceresoli GL, Falleni M, Zucali PA, Morenghi E, Bianchi P, et al. EGFR overexpression in malignant pleural mesothelioma. Lung Cancer. 2006;51(2):207–215. doi: 10.1016/j.lungcan.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 87.Schildgen V, Pabst O, Tillmann RL, Lüsebrink J, Schildgen O, Ludwig C, et al. Low frequency of EGFR mutations in pleural mesothelioma patients, Cologne, Germany. Appl Immunohistochem Mol Morphol. 2015;23(2):118–125. doi: 10.1097/PDM.0b013e3182a3645e. [DOI] [PubMed] [Google Scholar]
- 88.Chia PL, Scott AM, John T. Epidermal growth factor receptor (EGFR)-targeted therapies in mesothelioma. Expert Opinion Drug Deliv. 2019;16(4):441–451. doi: 10.1080/17425247.2019.1598374. [DOI] [PubMed] [Google Scholar]
- 89.Broeckx G, Pauwels P. Malignant peritoneal mesothelioma: a review. Transl Lung Cancer Res. 2018;7(5):537–542. doi: 10.21037/tlcr.2018.10.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mezzapelle R, Miglio U, Rena O, Paganotti A, Allegrini S, Antona J, et al. Mutation analysis of the EGFR gene and downstream signalling pathway in histologic samples of malignant pleural mesothelioma. Br J Cancer. 2013;108(8):1743–1749. doi: 10.1038/bjc.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shukuya T, Serizawa M, Watanabe M, Akamatsu H, Abe M, Imai H, et al. Identification of actionable mutations in malignant pleural mesothelioma. Lung Cancer. 2014;86(1):35–40. doi: 10.1016/j.lungcan.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 92.Lo Iacono M, Monica V, Righi L, Grosso F, Libener R, Vatrano S, et al. Targeted next-generation sequencing of cancer genes in advanced stage malignant pleural mesothelioma: a retrospective study. J Thorac Oncol. 2015;10(3):492–499. doi: 10.1097/JTO.0000000000000436. [DOI] [PubMed] [Google Scholar]
- 93.Jagadeeswaran R, Ma PC, Seiwert T, Jagadeeswaran S, Zumba O, Nallasura V, et al. Functional analysis of c-Met/hepatocyte growth factor pathway in malignant pleural mesothelioma. Cancer Res. 2006;66(1):352–361. doi: 10.1158/0008-5472.CAN-04-4567. [DOI] [PubMed] [Google Scholar]
- 94.Levallet G, Vaisse-Lesteven M, Stang NL, Ilg AGS, Brochard P, Astoul P, et al. Plasma cell membrane localization of c-MET predicts longer survival in patients with malignantmMesothelioma: a series of 157 cases from the MESOPATH Group. J Thorac Oncol. 2012;7(3):599–606. doi: 10.1097/JTO.0b013e3182417da5. [DOI] [PubMed] [Google Scholar]
- 95.Bois MC, Mansfield AS, Sukov WR, Jenkins SM, Moser JC, Sattler CA, et al. c-Met expression and MET amplification in malignant pleural mesothelioma. Ann Diagn Pathol. 2016;23:1–7. doi: 10.1016/j.anndiagpath.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 96.Ohta Y, Shridhar V, Bright RK, Kalemkerian GP, Du W, Carbone M, et al. VEGF and VEGF type C play an important role in angiogenesis and lymphangiogenesis in human malignant mesothelioma tumours. Br J Cancer. 1999;81(1):54–61. doi: 10.1038/sj.bjc.6690650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brosseau S, Assoun S, Naltet C, Steinmetz C, Gounant V, Zalcman G. A review of bevacizumab in the treatment of malignant pleural mesothelioma. Future Oncol. 2017;13(28):2537–2546. doi: 10.2217/fon-2017-0307. [DOI] [PubMed] [Google Scholar]
- 98.Nowak AK, Brosseau S, Cook A, Zalcman G. Antiangiogeneic strategies in mesothelioma. Front Oncol. 2020;10:126. doi: 10.3389/fonc.2020.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pietras K, Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316(8):1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 100.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen DS, Mellman I. Oncology meets immunology: the cancer immunity cycle. Immunity. 2013;39(1):1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 102.Svokos KA, Salhia B, Toms SA. Molecular biology of brain metastasis. Int J Mol Sci. 2014;15(6):9519–9530. doi: 10.3390/ijms15069519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dong Y, Sun Q, Zhang X. PD-1 and its ligands are important immune checkpoints in cancer. Oncotarget. 2017;8(2):2171–2186. doi: 10.18632/oncotarget.13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yamada N, Oizumi S, Kikuchi E, Shinagawa N, Konishi-Sakakibara J, Ishimine A, et al. CD8+ tumor-infiltrating lymphocytes predict favorable prognosis in malignant pleural mesothelioma after resection. Cancer Immunol Immunother. 2010;59(10):1543–1549. doi: 10.1007/s00262-010-0881-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Campesato LF, Merghoub T. Antiangiogenic therapy and immune checkpoint blockade go hand in hand. Ann Transl Med. 2017;5(24):497–502. doi: 10.21037/atm.2017.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alcala N, Caux C, Girard N, McKay JD, Galateau-Salle F, Foll M, Fernandez-Cuesta L. Redefining mesothelioma types as a continuum uncovers the immune and vascular systems as key players in the diagnosis and prognosis of this disease. bioRxiv. 2018 doi: 10.1101/334326. [DOI] [Google Scholar]
- 107.Vogl M, Rosenmayr A, Bohanes T, Scheed A, Brndiar M, Stubenberger E, Ghanim B. Biomarkers for malignant pleural mesothelioma—a novel view on inflammation. Cancers (Basel) 2021;13(4):658. doi: 10.3390/cancers13040658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ujiie H, Kadota K, Nitadori JI, Aerts JG, Woo KM, Sima CS, et al. The tumoral and stromal immune microenvironment in malignant pleural mesothelioma: a comprehensive analysis reveals prognostic immune markers. Oncoimmunology. 2015;4(6):e1009285. doi: 10.1080/2162402X.2015.1009285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yap TA, Aerts JG, Popat S, Fennell DA. Novel insights into mesothelioma biology and implications for therapy. Nat Rev Cancer. 2017;17(8):475–488. doi: 10.1038/nrc.2017.42. [DOI] [PubMed] [Google Scholar]
- 110.Pasello G, Zago G, Lunardi F, Urso L, Kern I, Vlacic G, et al. Malignant pleural mesothelioma immune microenvironment and checkpoint expression: correlation with clinical-pathological features and intratumor heterogeneity over time. Ann Oncol. 2018;29(5):1258–1265. doi: 10.1093/annonc/mdy086. [DOI] [PubMed] [Google Scholar]
- 111.Brosseau S, Danel C, Scherpereel A, Mazières J, Lantuejoul S, Margery J, et al. Shorter survival in malignant pleural mesothelioma patients with high PD-L1 expression associated with sarcomatoid or biphasic histology subtype: a series of 214 cases from the Bio-MAPS cohort. Clin Lung Cancer. 2019;20(5):e564–e575. doi: 10.1016/j.cllc.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 112.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27(4):355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ma S, Meng Z, Chen R, Guan KL. The Hippo pathway: biology and pathophysiology. Annu Rev Biochem. 2019;88:577–604. doi: 10.1146/annurev-biochem-013118-111829. [DOI] [PubMed] [Google Scholar]
- 115.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13(8):877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hiemer SE, Varelas X. Stem cell regulation by the Hippo pathway. Biochem Biophys Acta. 2013;1830(2):2323–2334. doi: 10.1016/j.bbagen.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 117.Porazinski S, Ladomery M. Alternative splicing in the Hippo pathway-implications for disease and potential therapeutic targets. Genes (Basel) 2018;9(3):161–180. doi: 10.3390/genes9030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boggiano JC, Fehon RG. Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling. Dev Cell. 2012;22(4):695–702. doi: 10.1016/j.devcel.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rausch V, Hansen CG. The Hippo pathway, YAP/TAZ, and the plasma membrane. Trends Cell Biol. 2020;30(1):32–48. doi: 10.1016/j.tcb.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 120.Han Y. Analysis of the role of the Hippo pathway in cancer. J Transl Med. 2019;17(1):1–17. doi: 10.1186/s12967-019-1869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Calses PC, Crawford JJ, Lill JR, Dey A. Hippo Pathway in cancer: aberrant regulation and therapeutic opportunities. Trends Cancer. 2019;5(5):297–307. doi: 10.1016/j.trecan.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 122.Shreberk-Shaked M, Oren M. New insights into YAP/TAZ nucleo-cytoplasmic shuttling: new cancer therapeutic opportunities? Mol Oncol. 2019;13(6):1335–1341. doi: 10.1002/1878-0261.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Janse van Rensburg HJ, Yang X. The roles of the Hippo pathway in cancer metastasis. Cell Signaling. 2016;28(11):1761–1772. doi: 10.1016/j.cellsig.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 124.Zygulska AL, Krzemieniecki K, Pierzchalski P. Hippo pathway—brief overview of its relevance in cancer. J Physiol Pharmacol. 2017;68(3):311–335. [PubMed] [Google Scholar]
- 125.Elbediwy A, Vincent-Mistiaen ZI, Thompson BJ. YAP and TAZ in epithelial stem cells: a sensor for cell polarity, mechanical forces and tissue damage. BioEssays. 2016;38(7):644–653. doi: 10.1002/bies.201600037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guo X, Zhao B. Integration of mechanical and chemical signals by YAP and TAZ transcription coactivators. Cell Biosci. 2013;3(1):33–42. doi: 10.1186/2045-3701-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang CC, Graves HK, Moya IM, Tao C, Hamaratoglu F, et al. Differential regulation of the Hippo pathway by adherens junctions and apical–basal cell polarity modules. Proc Natl Acad Sci USA. 2015;112(6):1785–1790. doi: 10.1073/pnas.1420850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, et al. The crumbs complex couples cell density sensing to hippo-dependent control of the TGF-β-SMAD pathway. Dev Cell. 2010;19(6):831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 129.Mao X, Li P, Wang Y, Liang Z, Liu J, Li J, et al. CRB3 regulates contact inhibition by activating the Hippo pathway in mammary epithelial cells. Cell Death Dis. 2017;8(1):e2546. doi: 10.1038/cddis.2016.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martin E, Girardello R, Dittmar G, Ludwig A. New insights into the organization and regulation of the apical polarity network in mammalian epithelial cells. FEBS J. 2021;288(4):7073–7095. doi: 10.1111/febs.15710. [DOI] [PubMed] [Google Scholar]
- 131.Lv XB, Liu CY, Wang Z, Sun YP, Xiong Y, Lei QY, Guan KL. PARD3 induces TAZ activation and cell growth by promoting LATS1 and PP1 interaction. EMBO Rep. 2015;16(8):975–985. doi: 10.15252/embr.201439951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18(2):300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18(2):288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xiao L, Chen Y, Ji M, Dong J. KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J Biol Chem. 2011;286(10):7788–7796. doi: 10.1074/jbc.M110.173468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by Angiomotin. J Biol Chem. 2011;286(9):7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li CY, Lu H, Hu J, Lan J, Galicia N, Klein OD, Du W. αE-catenin inhibits YAP/TAZ activity to regulate signalling centre formation during tooth development. Nat Commun. 2016;7:12133. doi: 10.1038/ncomms12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li P, Silvis MR, Honaker Y, Lien W-H, Arron ST, Vasioukhin V. αE-catenin inhibits a Src–YAP1 oncogenic module that couples tyrosine kinases and the effector of Hippo signaling pathway. Genes Dev. 2016;30(7):798–811. doi: 10.1101/gad.274951.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Michaloglou C, Lehmann W, Martin T, Delaunay C, Hueber A, Barys L, et al. The tyrosine phosphatase PTPN14 is a negative regulator of YAP activity. PLoS ONE. 2013;8(4):e61916. doi: 10.1371/journal.pone.0061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang W, Huang J, Wang X, Yuan J, Li X, Feng L, et al. PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 2012;26(17):1959–1971. doi: 10.1101/gad.192955.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Deel MD, Li JJ, Crose LES, Linardic CM. A review: molecular aberrations within Hippo signaling in bone and soft-tissue sarcomas. Front Oncol. 2015;5:190. doi: 10.3389/fonc.2015.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dubois F, Keller M, Calvayrac O, Soncin F, Hoa L, Hergovich A, et al. RASSF1A suppresses the invasion and metastatic potential of human non-small cell lung cancer cells by inhibiting YAP activation through the GEF-H1/RhoB pathway. Can Res. 2016;76(6):1627–1640. doi: 10.1158/0008-5472.CAN-15-1008. [DOI] [PubMed] [Google Scholar]
- 142.Lee MG, Jeong SI, Ko KP, Park SK, Ryu BK, Kim IY, et al. RASSF1A directly antagonizes RhoA activity through the assembly of a Smurf1-mediated destruction complex to suppress tumorigenesis. Can Res. 2016;76(7):1847–1859. doi: 10.1158/0008-5472.CAN-15-1752. [DOI] [PubMed] [Google Scholar]
- 143.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 144.Chakraborty S, Njah K, Pobbati AV, Lim YB, Raju A, Lakshmanan M, et al. Agrin as a mechanotransduction signal regulating YAP through the Hippo Pathway. Cell Rep. 2017;18(10):2464–2479. doi: 10.1016/j.celrep.2017.02.041. [DOI] [PubMed] [Google Scholar]
- 145.Petrilli AM, Fernández-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 2016;35(5):537–548. doi: 10.1038/onc.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Morrison H, Sherman LS, Legg J, Banine F, Isacke C, Haipek CA, et al. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001;15(8):968–980. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Poulikakos PI, Xiao GH, Gallagher R, Jablonski S, Jhanwar SC, Testa JR. Re-expression of the tumor suppressor NF2/merlin inhibits invasiveness in mesothelioma cells and negatively regulates FAK. Oncogene. 2006;25(44):5960–5968. doi: 10.1038/sj.onc.1209587. [DOI] [PubMed] [Google Scholar]
- 148.Shapiro IM, Kolev VN, Vidal CM, Kadariya Y, Ring JE, Wright Q, et al. Merlin deficiency predicts FAK inhibitor sensitivity: a synthetic lethal relationship. Sci Transl Med. 2014;6(237):237ra68. doi: 10.1126/scitranslmed.3008639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mota M, Shevde LA. Merlin regulates signaling events at the nexus of development and cancer. Cell Commun Signal. 2020;18(1):63–71. doi: 10.1186/s12964-020-00544-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hong AW, Meng Z, Plouffe SW, Lin Z, Zhang M, Guan KL. Critical roles of phosphoinositides and NF2 in Hippo pathway regulation. Genes Dev. 2020;34(7–8):511–525. doi: 10.1101/gad.333435.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fennell DA, Baas P, Taylor P, Nowak AK, Gilligan D, Nakano T, Pachter JA, Weaver DT, Scherpereel A, Pavlakis N, van Meerbeeck JP, Cedrés S, Nolan L, Kindler H, Aerts JGJV. Maintenance defactinib versus placebo after first-line chemotherapy in patients with merlin-stratified pleural mesothelioma: COMMAND-a double-blind, randomized, phase II study. J Clin Oncol. 2019;37(10):790–798. doi: 10.1200/JCO.2018.79.0543. [DOI] [PubMed] [Google Scholar]
- 152.Haderk F, Fernández-Méndez C, Čech L, Yu J, Meraz IM, Olivas V, Barbosa Rabago D, Kerr DL, Gomez C, Allegakoen DV, Guan J, Shah KN, Herrington KA, Gbenedio OM, Nanjo S, Majidi M, Tamaki W, Rotow JK, McCoach CE, Riess JW, Gutkind JS, Tang TT, Post L, Huang B, Santisteban P, Goodarzi H, Bandyopadhyay S, Kuo CJ, Roose JP, Wu W, Blakely CM, Roth JA, Bivona TG. A focal adhesion kinase-YAP signaling axis drives drug tolerant persister cells and residual disease in lung cancer. BioRxiv. 2022 doi: 10.1101/2021.10.23.465573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cooper J, Xu Q, Zhou L, Pavlovic M, Ojeda V, Moulick K, de Stanchina E, Poirier JT, Zauderer M, Rudin CM, Karajannis MA, Hanemann CO, Giancotti FG. Combined inhibition of NEDD8-activating enzyme and mTOR suppresses NF2 loss-driven tumorigenesis. Mol Cancer Therap. 2017;16(8):1693–1704. doi: 10.1158/1535-7163.MCT-20-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Goodison S, Urquidi V, Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52(4):189–196. doi: 10.1136/mp.52.4.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Misra S, Ghatak S, Toole BP. Regulation of MDR1 expression and drug resistance by a positive feedback loop involving hyaluronan, phosphoinositide 3-kinase, and ErbB2. J Biol Chem. 2005;280(21):20310–20315. doi: 10.1074/jbc.M500737200. [DOI] [PubMed] [Google Scholar]
- 156.Naor D, Sionov RV, Ish-Shalom D. CD44: structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- 157.Williams K, Motiani K, Giridhar PV, Kasper S. CD44 integrates signaling in normal stem cell, cancer stem cell and (pre)metastatic niches. Exp Biol Med (Maywood, N.J.) 2013;238(3):324–338. doi: 10.1177/1535370213480714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Karihtala P, Soini Y, Auvinen P, Tammi R, Tammi M, Kosma VM. Hyaluronan and breast cancer: correlations with nitric oxide synthases and tyrosine nitrosylation. J Histochem Cytochem. 2007;55(12):1191–1198. doi: 10.1369/jhc.7A7270.2007. [DOI] [PubMed] [Google Scholar]
- 159.Chen C, Zhao S, Karnad A, Freeman JW. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol. 2018;11(1):64–87. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Stamenkovic I, Yu Q. Shedding light on proteolytic cleavage of CD44: the responsible sheddase and functional significance of shedding. J Investig Dermatol. 2009;129(6):1321–1324. doi: 10.1038/jid.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bourguignon LYW, Peyrollier K, Xia W, Gilad E. Hyaluronan-CD44 interaction activates stem cell marker nanog, stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J Biol Chem. 2008;283(25):17635–17651. doi: 10.1074/jbc.M800109200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Toole BP. Hyaluronan-CD44 interactions in cancer: paradoxes and possibilities. Clin Cancer Res. 2009;15(24):7462–7468. doi: 10.1158/1078-0432.CCR-09-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 2002;16(23):3074–3086. doi: 10.1101/gad.242602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yin J, Zhang H, Wu X, Zhang Y, Li J, Shen J, et al. CD44 inhibition attenuates EGFR signaling and enhances cisplatin sensitivity in human EGFR wild-type non-small-cell lung cancer cells. Int J Mol Med. 2020;45(6):1783–1792. doi: 10.3892/ijmm.2020.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Joosten SPJ, Spaargaren M, Clevers H, Pals ST. Hepatocyte growth factor/MET and CD44 in colorectal cancer: partners in tumorigenesis and therapy resistance. Biochimica et biophysica acta. Rev Cancer. 2020;1874(2):188437. doi: 10.1016/j.bbcan.2020.188437. [DOI] [PubMed] [Google Scholar]
- 166.Bourguignon LYW, Shiina M, Li JJ. Chapter ten—hyaluronan–CD44 interaction promotes oncogenic signaling, microRNA functions, chemoresistance, and radiation resistance in cancer stem cells leading to tumor progression. In: Simpson MA, Heldin P, editors. Advances in cancer research. Amsterdam: Elsevier; 2014. pp. 255–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Terawaki S, Kitano K, Aoyama M, Mori T, Hakoshima T. MT1-MMP recognition by ERM proteins and its implication in CD44 shedding. Genes Cells. 2015;20(10):847–859. doi: 10.1111/gtc.12276. [DOI] [PubMed] [Google Scholar]
- 168.Chen L, Fu C, Zhang Q, He C, Zhang F, Wei Q. The role of CD44 in pathological angiogenesis. FASEB J. 2020;34(10):13125–13139. doi: 10.1096/fj.202000380RR. [DOI] [PubMed] [Google Scholar]
- 169.Orian-Rousseau V. CD44 acts as a signaling platform controlling tumor progression and metastasis. Front Immunol. 2015;6:154. doi: 10.3389/fimmu.2015.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Senbanjo LT, Chellaiah MA. CD44: a multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front Cell Dev Biol. 2017;5:18–24. doi: 10.3389/fcell.2017.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bai Y, Liu YJ, Wang H, Xu Y, Stamenkovic I, Yu Q. Inhibition of the hyaluronan-CD44 interaction by merlin contributes to the tumor-suppressor activity of merlin. Oncogene. 2007;26(6):836–850. doi: 10.1038/sj.onc.1209849. [DOI] [PubMed] [Google Scholar]
- 172.Xu Y, Stamenkovic I, Yu Q. CD44 attenuates activation of the Hippo signaling pathway and is a prime therapeutic target for glioblastoma. Can Res. 2010;70(6):2455–2464. doi: 10.1158/0008-5472.CAN-09-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ooki T, Murata-Kamiya N, Takahashi-Kanemitsu A, Wu W, Hatakeyama M. High-molecular-weight hyaluronan is a hippo pathway ligand directing cell density-dependent growth inhibition via PAR1b. Dev Cell. 2019;49(4):590–604. doi: 10.1016/j.devcel.2019.04.018. [DOI] [PubMed] [Google Scholar]
- 174.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120(Pt 18):3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 175.Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Can Res. 2005;65(9):3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 176.Dubois F, Bergot E, Zalcman G, Levallet G. RASSF1A, puppeteer of cellular homeostasis, fights tumorigenesis, and metastasis—an updated review. Cell Death Dis. 2019;10(12):928–941. doi: 10.1038/s41419-019-2169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Keller M, Dubois F, Teulier S, Martin APJ, Levallet J, Maille E, et al. NDR2 kinase contributes to cell invasion and cytokinesis defects induced by the inactivation of RASSF1A tumor-suppressor gene in lung cancer cells. J Exp Clin Cancer Res. 2019;38(1):158–174. doi: 10.1186/s13046-019-1145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dubois F, Bergot E, Levallet G. Cancer and RASSF1A/RASSF1C, the Two Faces of Janus. Trends Cancer. 2019;5(11):662–665. doi: 10.1016/j.trecan.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 179.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283(9):5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 180.Gomez M, Gomez V, Hergovich A. The Hippo pathway in disease and therapy: cancer and beyond. Clin Transl Med. 2014;3:22–34. doi: 10.1186/2001-1326-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Creasy CL, Chernoff J. Cloning and characterization of a member of the MST subfamily of Ste20-like kinases. Gene. 1995;167(1–2):303–306. doi: 10.1016/0378-1119(95)00653-2. [DOI] [PubMed] [Google Scholar]
- 182.Xu CM, Liu WW, Liu CJ, Wen C, Lu HF, Wan FS. Mst1 overexpression inhibited the growth of human non-small cell lung cancer in vitro and in vivo. Cancer Gene Ther. 2013;20(8):453–460. doi: 10.1038/cgt.2013.40. [DOI] [PubMed] [Google Scholar]
- 183.Dempsey LA. Mst kinase roles. Nat Immunol. 2019;20(1):1. doi: 10.1038/s41590-018-0284-y. [DOI] [PubMed] [Google Scholar]
- 184.Gloerich M, Ten Klooster JP, Vliem MJ, Koorman T, Zwartkruis FJ, Clevers H, Bos JL. Rap2A links intestinal cell polarity to brush border formation. Nat Cell Biol. 2012;14(8):793–801. doi: 10.1038/ncb2537. [DOI] [PubMed] [Google Scholar]
- 185.Mardakheh FK, Self A, Marshall CJ. RHO binding to FAM65A regulates Golgi reorientation during cell migration. J Cell Sci. 2016;129(24):4466–4479. doi: 10.1242/jcs.198614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Preisinger C, Short B, De Corte V, Bruyneel E, Haas A, Kopajtich R, et al. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14–3-3ζ. J Cell Biol. 2004;164(7):1009–1020. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Thompson BJ, Sahai E. MST kinases in development and disease. J Cell Biol. 2015;210(6):871–882. doi: 10.1083/jcb.201507005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Luo X, Li Z, Yan Q, Li X, Tao D, Wang J, et al. The human WW45 protein enhances MST1-mediated apoptosis in vivo. Int J Mol Med. 2009;23(3):357–362. [PMC free article] [PubMed] [Google Scholar]
- 189.Graves JD, Gotoh Y, Draves KE, Ambrose D, Han DK, Wright M, et al. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase Mst1. EMBO J. 1998;17(8):2224–2234. doi: 10.1093/emboj/17.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Avruch J, Zhou D, Fitamant J, Bardeesy N, Mou F, Barrufet LR. Protein kinases of the Hippo pathway: regulation and substrates. Semin Cell Dev Biol. 2012;23(7):770–784. doi: 10.1016/j.semcdb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tamaskovic R, Bichsel SJ, Hemmings BA. NDR family of AGC kinases–essential regulators of the cell cycle and morphogenesis. FEBS Lett. 2003;546(1):73–80. doi: 10.1016/s0014-5793(03)00474-5. [DOI] [PubMed] [Google Scholar]
- 192.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11(1):9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 193.Sharif AAD, Hergovich A. The NDR/LATS protein kinases in immunology and cancer biology. Semin Cancer Biol. 2018;48:104–114. doi: 10.1016/j.semcancer.04.010. [DOI] [PubMed] [Google Scholar]
- 194.Stegert MR, Tamaskovic R, Bichsel SJ, Hergovich A, Hemmings BA. Regulation of NDR2 protein kinase by multi-site phosphorylation and the S100B calcium-binding protein. J Biol Chem. 2004;279(22):23806–23812. doi: 10.1074/jbc.M402472200. [DOI] [PubMed] [Google Scholar]
- 195.Hergovich A. Regulation and functions of mammalian LATS/NDR kinases: looking beyond canonical Hippo signalling. Cell Biosci. 2013;3(1):32–44. doi: 10.1186/2045-3701-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang W, Gao Y, Li P, Shi Z, Guo T, Li F, et al. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 2014;24(3):331–343. doi: 10.1038/cr.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Emoto K, Parrish JZ, Jan LY, Jan YN. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443(7108):210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- 198.Vichalkovski A, Gresko E, Cornils H, Hergovich A, Schmitz D, Hemmings BA. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr Biol. 2008;18(23):1889–1895. doi: 10.1016/j.cub.2008.10.060. [DOI] [PubMed] [Google Scholar]
- 199.Chiba S, Ikeda M, Katsunuma K, Ohashi K, Mizuno K. MST2- and Furry-mediated activation of NDR1 kinase is critical for precise alignment of mitotic chromosomes. Curr Biol. 2009;19(8):675–681. doi: 10.1016/j.cub.2009.02.054. [DOI] [PubMed] [Google Scholar]
- 200.Cornils H, Kohler RS, Hergovich A, Hemmings BA. (Downstream of human NDR kinases: impacting on c-myc and p21 protein stability to control cell cycle progression. Cell Cycle. 2011;10(12):1897–1904. doi: 10.4161/cc.10.12.15826. [DOI] [PubMed] [Google Scholar]
- 201.Chiba S, Amagai Y, Homma Y, Fukuda M, Mizuno K. NDR2-mediated Rabin8 phosphorylation is crucial for ciliogenesis by switching binding specificity from phosphatidylserine to Sec15. EMBO J. 2013;32(6):874–885. doi: 10.1038/emboj.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Joffre C, Dupont N, Hoa L, Gomez V, Pardo R, Gonçalves-Pimentel C, et al. The pro-apoptotic STK38 kinase is a new Beclin1 partner positively regulating autophagy. Curr Biol. 2005;25(19):2479–2492. doi: 10.1016/j.cub.2015.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ye X, Ong N, An H, Zheng Y. The emerging roles of NDR1/2 in infection and inflammation. Front Immunol. 2020;11:534. doi: 10.3389/fimmu.2020.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chan EHY, Nousiainen M, Chalamalasetty RB, Schäfer A, Nigg EA, Silljé HHW. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24(12):2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 205.Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, et al. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with H-warts/Lats1 tumor suppressor. J Cell Biol. 2000;149(5):1073–1086. doi: 10.1083/jcb.149.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Furth N, Aylon Y. The LATS1 and LATS2 tumor suppressors: beyond the Hippo pathway. Cell Death Differ. 2017;24(9):1488–1501. doi: 10.1038/cdd.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Duhart JC, Raftery LA. Mob family proteins: regulatory partners in Hippo and Hippo-like intracellular signalingpPathways. Front Cell Devel Biol. 2020;8:161–183. doi: 10.3389/fcell.2020.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hergovich A. MOB control: reviewing a conserved family of kinase regulators. Cell Signal. 2011;23(9):1433–1440. doi: 10.1016/j.cellsig.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Praskova M, Xia F, Avruch J. (MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol. 2008;18(5):311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ni L, Zheng Y, Hara M, Pan D, Luo X. Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling. Genes Dev. 2015;29(13):1416–1431. doi: 10.1101/gad.264929.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kim SY, Tachioka Y, Mori T, Hakoshima T. Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway. Sci Rep. 2016;6:28488. doi: 10.1038/srep28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Couzens AL, Xiong S, Knight JDR, Mao DY, Guettler S, Picaud S, et al. MOB1 mediated phospho-recognition in the core mammalian Hippo pathway. Mol Cell Proteomics. 2017;16(6):1098–1110. doi: 10.1074/mcp.M116.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hergovich A. Mammalian Hippo signalling: a kinase network regulated by protein–protein interactions. Biochem Soc Trans. 2012;40(1):124–128. doi: 10.1042/BST20110619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kohler RS, Schmitz D, Cornils H, Hemmings BA, Hergovich A. Differential NDR/LATS interactions with the human MOB family reveal a negative role for human MOB2 in the regulation of human NDR kinases. Mol Cell Biol. 2010;30(18):4507–4520. doi: 10.1128/MCB.00150-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gundogdu R, Hergovich A. The possible crosstalk of MOB2 with NDR1/2 kinases in cell cycle and DNA damage signaling. J Cell Signal. 2016;1:125–133. doi: 10.4172/2576-1471.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21(21):2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 2011;144(5):782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Silvis MR, Kreger BT, Lien WH, Klezovitch O, Rudakova GM, Camargo FD, et al. α-Catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal. 2011;4(174):ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell. 2008;29(3):350361. doi: 10.1016/j.molcel.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 220.Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, et al. β-catenin driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151(7):1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kim NG, Gumbiner BM. Adhesion to fibronectin regulates Hippo signaling via the FAK–Src–PI3K pathway. J Cell Biol. 2015;210(3):503–515. doi: 10.1083/jcb.201501025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Taniguchi K, Wu LW, Grivennikov SI, de Jong PR, Lian I, Yu F-X, et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519(7451):57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hsu PC, Yang CT, Jablons DM, You L. The crosstalk between Src and Hippo/YAP signaling pathways in non-small celllLung cancer (NSCLC) Cancers (Basel). 2020;12(6):1361–1385. doi: 10.3390/cancers12061361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hilman D, Gat U. The evolutionary history of YAP and the hippo/YAP pathway. Mol Biol Evol. 2011;28(8):2403–2417. doi: 10.1093/molbev/msr065. [DOI] [PubMed] [Google Scholar]
- 225.Hong W. Guan KL The YAP and TAZ transcription coactivators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012;23(7):785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gaffney CJ, Oka T, Mazack V, Hilman D, Gat U, Muramatsu T, et al. Identification, basic characterization and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1 gene. Gene. 2012;509(2):215–222. doi: 10.1016/j.gene.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Cho YS, Jiang J. Hippo-independent regulation of Yki/Yap/Taz: a non-canonical view. Front Cell Dev Biol. 2021;9:658481. doi: 10.3389/fcell.2021.658481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hartmann M, Parra LM, Ruschel A, Böhme S, Li Y, Morrison H, et al. Tumor suppressor NF2 blocks cellular migration by inhibiting ectodomain cleavage of CD44. Mol Cancer Res. 2015;13(5):879–890. doi: 10.1158/1541-7786.MCR-15-0020-T. [DOI] [PubMed] [Google Scholar]
- 229.Misra S, Hascall VC, Markwald RR, Ghatak S. Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Front Immunol. 2015;6:201. doi: 10.3389/fimmu.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ohashi R, Takahashi F, Cui R, Yoshioka M, Gu T, Sasaki S, et al. Interaction between CD44 and hyaluronate induces chemoresistance in non-small cell lung cancer cell. Cancer Lett. 2007;252(2):225–234. doi: 10.1016/j.canlet.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 231.Martensson G, Thylen A, Lindquist U, Hjerpe A. The sensitivity of hyaluronan analysis of pleural fluid from patients with malignant mesothelioma and a comparison of different methods. Cancer. 1994;73(5):1406–1410. doi: 10.1002/1097-0142(19940301)73:5<1406::aid-cncr2820730515>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 232.Fujimoto N, Gemba K, Asano M, Fuchimoto Y, Wada S, Ono K, et al. Hyaluronic acid in the pleural fluid of patients with malignant pleural mesothelioma. Respir Invest. 2013;51(2):92–97. doi: 10.1016/j.resinv.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 233.Cortes-Dericks L, Schmid RA. CD44 and its ligand hyaluronan as potential biomarkers in malignant pleural mesothelioma: evidence and perspectives. Respir Res. 2017;18(1):58–70. doi: 10.1186/s12931-017-0546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Asplund T, Heldin P. Hyaluronan receptors are expressed on human malignant mesothelioma cells but not on normal mesothelial cells. Cancer Res. 1994;54(16):4516–4523. [PubMed] [Google Scholar]
- 235.Ohno Y, Shingyoku S, Miyake S, Tanaka A, Fudesaka S, Shimizu Y, et al. Differential regulation of the sphere formation and maintenance of cancer-initiating cells of malignant mesothelioma via CD44 and ALK4 signaling pathways. Oncogene. 2018;37:6357–6367. doi: 10.1038/s41388-018-0405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sakurai Y, Kato A, Hida Y, Hamada J, Maishi N, Hida K, et al. Synergistic enhancement of cellular uptake with CD44-expressing malignant pleural mesothelioma by combining cationic liposome and hyaluronic acid-lipid conjugate. J Pharm Sci. 2019;108(10):3218–3224. doi: 10.1016/j.xphs.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 237.Bianchi AB, Mitsunaga SI, Cheng JQ, Klein WM, Jhanwar SC, Seizinger B, et al. High frequency of inactivating mutations in the neurofibromatosis type 2 gene (NF2) in primary malignant mesotheliomas. Proc Natl Acad Sci USA. 1995;92(24):10854–10858. doi: 10.1073/pnas.92.24.10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sekido Y, Pass HI, Bader S, Mew DJ, Christman MF, Gazdar AF, et al. Neurofibromatosis type 2 (NF2) gene is somatically mutated in mesothelioma but not in lung cancer. Cancer Res. 1995;55(6):1227–1231. [PubMed] [Google Scholar]
- 239.Thurneysen C, Opitz I, Kurtz S, Weder W, Stahel RA, Felley-Bosco E. Functional inactivation of NF2/merlin in human mesothelioma. Lung Cancer. 2009;64(2):140–147. doi: 10.1016/j.lungcan.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 240.Li Y, Zhou H, Li F, Chan SW, Lin Z, Wei Z, et al. Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway. Cell Res. 2015;25(7):801–817. doi: 10.1038/cr.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Fischer JR, Ohnmacht U, Rieger N, Zemaitis M, Stoffregen C, Kostrzewa M, et al. Promoter methylation of RASSF1A, RARbeta and DAPK predict poor prognosis of patients with malignant mesothelioma. Lung Cancer. 2006;54(1):109–116. doi: 10.1016/j.lungcan.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 242.Destro A, Ceresoli GL, Baryshnikov E, Garassino I, Zucali PA, De Vincenzo F, et al. Gene methylation in pleural mesothelioma: correlations with clinico-pathological features and patient’s follow-up. Lung Cancer. 2008;59(3):369–376. doi: 10.1016/j.lungcan.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 243.Fujii M, Fujimoto N, Hiraki A, Gemba K, Aoe K, Umemura S, et al. Aberrant DNA methylation profile in pleural fluid for differential diagnosis of malignant pleural mesothelioma. Cancer Sci. 2012;103(3):510–514. doi: 10.1111/j.1349-7006.2011.02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Dubois F, Jean-Jacques B, Roberge H, Bénard M, Galas L, Schapman D, et al. A role for RASSF1A in tunneling nanotube formation between cells through GEFH1/Rab11 pathway control. Cell Commun Signal. 2018;6(1):66–86. doi: 10.1186/s12964-018-0276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wang X, Gerdes HH. Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ. 2015;22(7):1181–1191. doi: 10.1038/cdd.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Vignais ML, Caicedo A, Brondello JM, Jorgensen C. Cell connections by tunneling nanotubes: effects of mitochondrial trafficking on target cell metabolism, homeostasis, and response to therapy. Stem Cells Int. 2017;2017:6917841. doi: 10.1155/2017/6917941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43(7):668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Pulford E, Huilgol K, Moffat D, Henderson DW, Klebe S. Malignant mesothelioma, BAP1 immunohistochemistry, and VEGFA: does BAP1 have potential for early diagnosis and assessment of prognosis? Dis Markers. 2017;2017:1310478. doi: 10.1155/2017/1310478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Murakami H, Mizuno T, Taniguchi T, Fujii M, Ishiguro F, Fukui T, et al. LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res. 2011;71(3):873–883. doi: 10.1158/0008-5472.CAN-10-2164. [DOI] [PubMed] [Google Scholar]
- 250.Miyanaga A, Masuda M, Tsuta K, Kawasaki K, Nakamura Y, Sakuma T, et al. Hippo pathway gene mutations in malignant mesothelioma: revealed by RNA and targeted exon sequencing. J Thorac Oncol. 2015;10(5):844–851. doi: 10.1097/JTO.0000000000000493. [DOI] [PubMed] [Google Scholar]
- 251.Maille E, Brosseau S, Hanoux V, Creveuil C, Danel C, Bergot E, et al. MST1/Hippo promoter gene methylation predicts poor survival in patients with malignant pleural mesothelioma in the IFCT-GFPC-0701 MAPS phase 3 trial. Br J Cancer. 2019;120(4):387–397. doi: 10.1038/s41416-019-0379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tranchant R, Quetel L, Tallet A, Meiller C, Renier A, Koning L, et al. Co-occurring mutations of tumor suppressor genes, LATS2 and NF2, in malignant pleural mesothelioma. Clin Cancer Res. 2017;23(12):3191–3202. doi: 10.1158/1078-0432.CCR-16-1971. [DOI] [PubMed] [Google Scholar]
- 253.Meerang M, Bérard K, Friess M, Bitanihirwe BKY, Soltermann A, Vrugt B, et al. Low Merlin expression and high Survivin labeling index are indicators for poor prognosis in patients with malignant pleural mesothelioma. Mol Oncol. 2016;10(8):1255–1265. doi: 10.1016/j.molonc.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sekido Y. Inactivation of Merlin in malignant mesothelioma cells and the Hippo signaling cascade dysregulation: Merlin inactivation in mesothelioma. Pathol Int. 2011;61(6):331–344. doi: 10.1111/j.1440-1827.2011.02666.x. [DOI] [PubMed] [Google Scholar]
- 255.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13(1):63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Mi W, Lin Q, Childress C, Sudol M, Robishaw J, Berlot CH, et al. Geranylgeranylation signals to the Hippo pathway for breast cancer cell proliferation and migration. Oncogene. 2015;34(24):3095–3106. doi: 10.1038/onc.2014.251. [DOI] [PubMed] [Google Scholar]
- 257.Slemmons KK, Yeung C, Baumgart JT, Juarez JO, McCalla A, Helman LJ. Targeting Hippo-dependent and Hippo-independent YAP1 signaling for the treatment of childhood rhabdomyosarcoma. Cancer Res. 2020;80(14):3046–3056. doi: 10.1158/0008-5472.CAN-19-3853. [DOI] [PubMed] [Google Scholar]
- 258.Felley-Bosco E, Stahel R. Hippo/YAP pathway for targeted therapy. Transl Lung Cancer Res. 2014;3(2):75–83. doi: 10.3978/j.issn.2218-6751.2014.02.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26(12):1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhang WQ, Dai YY, Hsu PC, Wang H, Cheng L, Yang YL, et al. Targeting YAP in malignant pleural mesothelioma. J Cell Mol Med. 2017;21(11):2663–2676. doi: 10.1111/jcmm.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Pulito C, Korita E, Sacconi A, Valerio M, Casadei L, Lo Sardo F, et al. Dropwort-induced metabolic reprogramming restrains YAP/TAZ/TEAD oncogenic axis in mesothelioma. J Exp Clin Cancer Res. 2019;38(1):349–371. doi: 10.1186/s13046-019-1352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wu D, Luo L, Yang Z, Chen Y, Quan Y, Min Z. Targeting human Hippo TEAD binding interface with YAP/TAZ-derived, flexibility-reduced peptides in gastric cancer. Int J Peptide Res Ther. 2021;27(1):119–128. doi: 10.1007/s10989-020-10069-9. [DOI] [Google Scholar]
- 263.Dominguez-Berrocal L, Cirri E, Zhang X, Marin GH, Lebel-Bina S, Rebollo A. New therapeutic approach for targeting Hippo signalling Pathway. Sci Rep. 2019;9(1):4771–4782. doi: 10.1038/s41598-019-41404-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16(4):357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- 265.Sapio L, Gallo M, Illiano M, Chiosi E, Naviglio D, Spina A, et al. The natural cAMP elevating compound forskolin in cancer therapy: is it time? J Cell Physiol. 2017;232(5):922–927. doi: 10.1002/jcp.25650. [DOI] [PubMed] [Google Scholar]
- 266.Thompson BJ. YAP/TAZ: drivers of tumor growth, metastasis, and resistance to therapy. BioEssays. 2020;42(5):e1900162. doi: 10.1002/bies.201900162. [DOI] [PubMed] [Google Scholar]
- 267.Mohajan S, Jaiswal PK, Vatanmakarian M, Yousefi H, Sankaralingam S, Alahar SK, et al. Hippo pathway: regulation, deregulation and potential therapeutic targets in cancer. Cancer Lett. 2021;507:112–123. doi: 10.1016/j.canlet.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Zeng R, Dong J. The Hippo signaling pathway in drug resistance in cancer. Cancers (Basel). 2021;13(2):318. doi: 10.3390/cancers13020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Bai N, Zhang C, Liang N, Zhang Z, Chang A, Yin J, et al. Yes-associated protein (YAP) increases chemosensitivity of hepatocellular carcinoma cells by modulation of p53. Cancer Biol Ther. 2013;14(6):511–520. doi: 10.4161/cbt.24345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J Biol Chem. 2008;283(41):27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 271.Oka T, Schmitt AP, Sudol M. Opposing roles of angiomotin-like-1 and zona occludens-2 on pro-apoptotic function of YAP. Oncogene. 2012;31(1):128–134. doi: 10.1038/onc.2011.216. [DOI] [PubMed] [Google Scholar]
- 272.Nallet-Staub F, Marsaud V, Li L, Gilbert C, Dodier S, Bataille V, et al. Pro-invasive activity of the Hippo pathway effectors YAP and TAZ in cutaneous melanoma. J Invest Dermatol. 2014;134(1):123–132. doi: 10.1038/jid.2013.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014;94(4):1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 274.Zhu C, Li L, Zhao B. The regulation and function of YAP transcription co-activator. Acta Biochim Biophys Sin. 2015;47:16–28. doi: 10.1093/abbs/gmu110. [DOI] [PubMed] [Google Scholar]
- 275.Di Agostino S, Sorrentino G, Ingallina E, Valenti F, Ferraiuolo M, Bicciato S, et al. YAP enhances the pro-proliferative transcriptional activity of mutant p53 proteins. EMBO Rep. 2016;17(2):188–201. doi: 10.15252/embr.201540488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Bertini E, Oka T, Sudol M, Strano S, Blandino G. YAP: At the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8(1):49–57. doi: 10.4161/cc.8.1.7259. [DOI] [PubMed] [Google Scholar]
- 277.Perra A, Kowalik MA, Ghiso E, Ledda-Columbano GM, Di Tommaso L, Angioni MM, et al. YAP activation is an early event and a potential therapeutic target in liver cancer development. J Hepatol. 2014;61(5):1088–1096. doi: 10.1016/j.jhep.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 278.Wang H, Du YC, Zhou X, Liu H. Tang SC (2014) The dual functions of YAP-1 to promote and inhibit cell growth in human malignancy. Cancer Metastasis Rev. 2014;33(1):173–181. doi: 10.1007/s10555-013-9463-3. [DOI] [PubMed] [Google Scholar]
- 279.Lee AY, Raz DJ, He B, Jablons DM. Update on the molecular biology of malignant mesothelioma. Cancer. 2007;109(8):1454–1461. doi: 10.1002/cncr.22552. [DOI] [PubMed] [Google Scholar]