Abstract
Linker histone H1 proteins contain many variants in mammalian and can stabilize the condensed state of chromatin by binding to nucleosomes and promoting a more inaccessible structure of DNA. However, it is poorly understood how the binding of histone H1s to chromatin DNA is regulated. Screened as one of a collection of epithelial cells-enriched long non-coding RNAs (lncRNAs), here we found that small nucleolar RNA host gene 8 (SNHG8) is a chromatin-localized lncRNA and presents strong interaction and phase separation with histone H1 variants. Moreover, SNHG8 presents stronger ability to bind H1s than linker DNA, and outcompetes linker DNA for H1 binding. Consequently, loss of SNHG8 increases the amount of H1s that bind to chromatin, promotes chromatin condensation, and induces an epithelial differentiation-associated gene expression pattern. Collectively, our results propose that the highly abundant SNHG8 in epithelial cells keeps histone H1 variants out of nucleosome and its loss contributes to epithelial cell differentiation.
Subject terms: Gene expression, Chromatin structure, Histone variants
Introduction
Long non-coding RNAs (lncRNAs), a class of non-coding RNAs longer than 200 nucleotides, can exert their functions via various mechanisms from regulating gene expression to impacting mRNA and protein stability [1, 2]. Although lncRNAs have been found to play roles in almost all biological processes including cell differentiation and organ development [3–6], only a few lncRNAs have been characterized to date. In order to find and characterize functionally important lncRNAs, the high-throughput screening technologies including genome-wide CRISPR interference and CRISPR activation were developed and have greatly advanced the field in the past years [7–10]. Compared to the large number of lncRNAs, however, these methods are time-consuming and laborious. On the other hand, there are enormous RNA-seq data deposited in public database. Especially, lncRNAs present striking tissue- and cell type-specific expression patterns, and a great part of those public data could be classified according to tissue and cell type origin [11, 12]. These features would make lncRNAs especially suitable for bioinformatic screening.
Eukaryotic genomic DNA is highly compacted into chromatin, with nucleosome as its basic unit [13]. The occupancy and topological organization of nucleosomes as well as other chromatin-binding factors that occlude access to DNA determines chromatin accessibility [14]. Linker histone H1 proteins are the most abundant chromatin-binding proteins. There are eleven H1 variants (H1s) in mammalian, including the somatic variants H1.0−H1.5, H1.x and four germ cell-specific H1s. All these variants have the same general structure consisting of a central globular domain (GD), a short N-terminal tail, and a longer C-terminal domain (CTD) [15]. H1s have been known to stabilize the condensed state of chromatin by binding to nucleosomes and promoting a more inaccessible structure of DNA [16, 17]. H1s-mediated chromatin compaction and transcriptional repression have been proposed to play a critical role in human stem cell differentiation [18, 19]. However, it is poorly understood how the binding ability of H1s to chromatin DNA is regulated. Especially, it remains unknown whether lncRNAs play a role in this process.
Herein, we identify the epithelial cells-enriched small nucleolar RNA host gene 8 (SNHG8) as a histone H1-binding lncRNA that regulates chromatin accessibility and contributes to epithelial differentiation.
Materials and methods
Cell-type specific score (CTSS)
According to the tree structure of cell lines based on Cell Ontology annotation, we supposed the cell line ontology tree has a number of nodes, and a score at each tree node for each gene was firstly calculated. This score is a measure of the degree to which a gene is up-regulated or down-regulated in a particular node of the tree, and was calculated according the formula as followed:
Then, the specificity scores of a gene in each cell type was summed up according to the tree structure and node scores. For any node in the tree and the subtree rooted at that node, these subtree p nodes in total. For a gene, its specificity scores the maximum value in the vector of p node scores.
Gene Set Enrichment Analysis (GSEA) [20] was applied to the rankings of epithelial signature genes and RNA-seq data defined by the t-statistic of the differential expression analysis. Genesets for analysis were derived from the MsigDB or previous publication [21]. GSEA was performed on each of these gene set collections separately.
The Gene Ontology analysis in the current study was performed using the PANTHER 15.0 and P value smaller than 5% indicated a significant difference.
Cell lines, transfection and lentiviral infection
Human cell lines including MCF10A, RWPE-1 and HEK-293T cells were purchased from the Cell Bank of the Chinese Academy of Sciences in Shanghai (http://www.cellbank.org.cn/). HaCaT was obtained from Zhigang Zhang’s Lab, Shanghai Cancer institute. MCF10A cells were maintained in a 1:1 mixture of Dulbecco’s modified Eagle’s medium and F12 medium (DMEM/F12, 11320033, Gibco, Grand Island, NY) supplemented with 5% horse serum (Gibco, 16050122), hydrocortisone (0.5 μg/mL, 803146, Sigma-Aldrich, Germany), insulin (10 μg/mL, I0305000, Sigma-Aldrich), epidermal growth factor (20 ng/mL, AF-100-15, PeproTech, USA), Cholera toxin (100 ng/mL, C8052, Sigma-Aldrich). HaCaT and HEK-293T cells were cultured in DMEM with 10% fetal bovine serum. RWPE-1 cells were maintained in Keratinocyte-SFM (Gibco). To induce cell differentiation, RWPE-1 cells were grown for 36 h in Keratinocyte-SFM supplemented with 1 mM calcium chloride. Lentivirus was produced by co-transfecting HEK-293T cells with the lentiviral construct pCMV-dR8.91 (Δ8.9) plasmid, containing the genes gag, pol and rev, and the pMDG envelope-expressing plasmid, using X-treme GENE 9 DNA Transfection Reagent (06366244001, Roche, Basel, Switzerland). Viral supernatant was harvested at 24–48 h post-transfection, passed through a 0.45 μm filter, diluted 2:3 with fresh medium containing 8 μg/mL polybrene, and used to infect the target cells at 40% confluence.
RNA isolation, quantitative real time PCR (qRT-PCR) and Northern blot
Total RNA from indicated cells was extracted with Trizol Reagent (15596-026 Thermo, USA) according to the manufacturer’s protocol. Indicated total RNAs were subjected to RNA-seq analysis. Details of qRT-PCR have been described previously [22]. The primers used for qRT-PCR are listed in the Supplementary Table S1A.
Northern blot was carried out according to the manufacturer’s protocol (Northern Max-Gly Kit, Invitrogen). RNA was loaded on denatured SDS-PAGE gels. Biotin-labeled antisense probes and positive control SNHG8 (T1-T5) viariants were generated using T7 RNA polymerase by in vitro transcription with the Ribo MAX Large Scale RNA Production System (Promega). Primers used for in vitro transcription are listed in the Supplementary Table S1B.
Plasmids
The oligonucleotides for shRNAs targeting SNHG8 were cloned into pLKO.1-puro vectors. The empty pLKO.1-puro vector was used as a negative control. Two pairs of shRNA against SNHG8 (shG8#1 and shG8#2) produced the best knockdown efficiency. The target sequences of shG8#1 and #2 were listed in the Supplementary Table S1C.
For overexpression of SNHG8, the full-length of SNHG8 were synthesized by Sagon Biotech, and inserted into pLVX-puro expression vector. For RNA-pulldown assays, the full-length of 4×S1m were synthesized by Shanghai Qihe Biotech and inserted into pcDNA3.1(+) vector using EcoR I and Xba I. For 4×S1m-SNHG8 and its truncations, the PCR products containing the full-length of SNHG8 or truncations were inserted into the pcDNA 3.1(+)-S1m vectors using Xba I and Apa I, and primers used for constructing these plasmids were listed in the Supplementary Table S1B. For Flag-tagged RIP assays, the Full-length of H1.0, H1.2, H1.3, H1.5 and HP1α were synthesized by Shanghai TsingKe Biotech and inserted into pQCXIN vector using BamH I and EcoR I with Flag tag on their N-terminal, primers used for constructing these plasmids were listed in the Supplementary Table S1D. All of the pLVX-puro vectors contain the H1 subtypes with C-terminal domain were purchased from Shanghai Qihe Biotech. For MS2-MCP (MS2 coated protein) living cell imaging assay, the full-length of SNHG8 and MCP were inserted into the pLVX-12×MS2 and pLVX-mCherry vectors respectively, and all these plasmids were purchased from Shanghai TsingKe Biotech.
For prokaryotic expression and purification of the His-tagged H1.3 protein, the coding sequence of H1.3 was amplified from pQCXIN-H1.3 plasmid, and inserted into pGEX-6p-1 vector using BamH I and Xho I with 6×His tag on its C-terminal. See the Supplementary Table S1E for all of these primers.
In vitro transcription/translation
In vitro translation assay was performed using the TnT Quick Coupled Transcription/Translation System (L1171, Promega, Wisconsin, USA), according to the manufacturer’s instructions. Reactions were carried out using 1 mM transcend biotin-lysyl-tRNA. The eventual labeled translation products were then separated on SDS-PAGE gels, transferred onto nitrocellulose membrane and visualized by binding of streptavidin–horseradish peroxidase, followed by chemiluminescent detection. pcDNA3.1(+) vector containing the full length of SNHG8 was used to examine the coding potential of SNHG8. pcDNA3.1(+)-MYC/HA-UHRF1/NKILA were used as positive control for the experiment.
Immunoblotting
Details of immunoblotting have been described previously [22]. The antibodies used in these experiments are listed in the Supplementary Table S2.
CRISPR/Cas9
A pair of complementary oligos sgRNA1 and sgRNA2, for each CRISPR targeting sgRNA was annealed with 5’ overhangs of ‘ACCG’ and ‘AAAC’. The annealed DNA is inserted into a Bsa I linearized pGL3 vector with the U6 promoter [23]. Individual clones were obtained by electroporation of the plasmid containing gRNA and Cas9 into MCF10A cells, followed by selection by puromycin (2 μg/mL) before plating. Genomic DNA of selected single clones were extracted either for the genotyping validation with appropriate sets of primer listed in the Supplementary Table S1C. For depletion of histone H1 subtypes, lentiCRISPRv2 plasmids were constructed to target corresponding H1 subtype, the guide RNA targeted sequences were listed in the Supplementary Table S1C.
Three-dimensional culture and immunofluorescence
Three-dimensional cell culture was performed as previously described [24]. Briefly, 5000 indicated cells were suspended in 400 μL of medium containing 2% GFR Matrigel and then seeded onto a GFR Matrigel (356230, BD, New Jersey, USA) pre-coated eight-well chambers (80826, IBIDI, Germany). Cells were maintained at 37 °C and 5% CO2 and the culture medium was changed every two days. Images of mammospheres were collected using a Zeiss LSM 880 microscope. Luminal filling was examined and scored as described previously [25]. Acini were scored as clear (90–100% of luminal space was clear), mostly clear (50–90% clear), mostly filled (10–50% clear), and filled (0–10% clear).
RNA pull-down assay
4×S1m RNA pull-down assays were carried out as described [26] with modifications. SNHG8 full-length or indicated truncations were cloned into pcDNA3.1(+) plasmid with the 4×S1m tag at their 5′ end. RNA products were in vitro transcribed using the T7 Ribo MAX Large-Scale RNA Production System (Promega). 10 μg per reaction of synthetic RNAs were denatured for 5 min at 65 °C in RNA Structure buffer (10 mM HEPES pH 7.0, 10 mM MgCl2) and slowly cooled down to room temperature. Then, folded RNAs were incubated with 50 μL of streptavidin Dynabeads (11205D, Invitrogen, USA) for 20 min at 4 °C in the presence of 2 U/mL RNase inhibitor (2313 A, TAKARA, China). MCF10A and HaCaT cells (1 × 108) were harvested and re-suspended in 10 mL of lysis buffer (10 mM HEPES pH 7.0, 200 mM NaCl, 1 mM DTT, 1% Triton X-100, protease inhibitor cocktail) followed by sonication for 6 × 20 s with an interval of 2 min on ice and then centrifuged at 13,000 rpm for 10 min at 4 °C. The supernatant was pre-cleared with 50 μL of streptavidin Dynabeads for 20 min at 4 °C followed by the addition of 20 μg/mL yeast tRNA for 20 min at 4 °C. Then the pre-cleared lysate was added to folded RNAs and incubated for 3.5 h at 4 °C followed by washing 5 × 10 min with wash buffer [10 mM HEPES pH 7.0, 400 mM NaCl, 1 mM DTT, 1% Triton X-100, protease inhibitor cocktail (Roche), 1 mM PMSF]. To harvest the protein complex, 50 μL of 1×SDS loading buffer was added and boiled for 15 min at 100 °C. Retrieved proteins were analyzed by immunoblotting.
Coomassie blue staining and mass spectrometry
After 4×S1m RNA pull-down, equal amounts of proteins were loaded on NuPAGE 4–12 % Bis-Tris Gel (Invitrogen).Then the gel was stained using Coomassie Brilliant Blue G250 (Thermo) for liquid chromatography−tandem mass spectrometry (LC − MS/MS) analysis according to the manufacturer’s instructions. After coomassie blue staining, specific bands were cut and sent to the Core Facility of Basic Medical Sciences (Shanghai Jiao Tong University College of Basic Medical Science, Shanghai) for mass spectrometry analysis.
RNA-protein complex immunoprecipitation
Indicated MCF10A or MCF10A stable cell lines and then 1×108 each stable cells were harvested, re-suspended in 3 mL lysis buffer [50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% NP-40, 1 mM PMSF, protease inhibitor cocktail (04693116001, Roche)] followed by sonication for 6 × 20 s with an interval of 2 min on ice. After centrifuging at 13,000 rpm for 10 min at 4 °C, the supernatant was pre-cleared with 25 μL Protein A/G agarose beads (26159, Thermo). 80 μL Protein A/G agarose beads were suspended with lysis buffer and washed twice. The beads were divided into two parts and incubated with 5 μg anti-mouse IgG or anti-Flag antibody (or anti-pan H1 antibody) for 45 min at room temperature and then washed twice with lysis buffer, and then blocked with 1% BSA and 20 μg/mL yeast tRNA (Thermo).The beads were then incubated with previously pre-cleared lysates for 3 h at 4 °C and followed by washing 5 × 10 min with wash buffer [50 mM Tris-HCl pH 7.4, 300 mM NaCl, 0.05% Sodium Deoxycholate, 0.5% NP-40, 1 mM PMSF, protease inhibitor cocktail (Roche)]. The RNA-protein complex was eluted with 100 μL elution buffer (100 mM Tris-HCl PH 6.8, 4% SDS) at room temperature for 15 min. For immunoblotting, 10 μL each eluted samples were added with 10 μL of 1×SDS loading buffer was and boiled for 15 min at 100 °C and analyzed by immunoblotting. For qRT-PCR, each eluted sample was treated with Trizol (Invitrogen) and RNA purified according to manufactory’s instruction then reverse transcription was performed with MMLV system (M1701, Promega) with random primers followed by qRT-PCR analysis. Primers are listed in the Supplementary Table S1A.
Chromatin isolation by RNA purification (ChIRP)
ChIRP was performed using MCF10A cells adapting the protocols described previously [27] with minor modifications. Briefly, a set of 20-mer antisense DNA probes targeting SNHG8 and Lac Z specific regions were designed using the online probe designer http://www.singlemoleculefish.com/designer.html as listed in the Supplementary Table S1F. MCF10A cells were crosslinked with 1% formaldehyde for 10 min at room temperature. Crosslinking was stopped by the addition of glycine to a final concentration of 0.25 M, followed by incubation at room temperature for 5 min. The cross-linked chromatin was incubated with the biotinylated DNA probes, subjected to streptavidin magnetic beads capturing and subsequent wash/elution steps essentially performed as described [27]. The eluted RNA fragments and chromatin were analyzed by qRT-PCR and immunoblotting. Primers and antibodies are listed in the Supplementary Table S1A and the Supplementary Table S2, respectively.
RNA fluorescence in situ hybridization (FISH)
To detect the subcellular localization of SNHG8, RNA FISH was carried out as previously described [28] with in vitro transcribed Dig-labeled antisense probe (Supplementary Table S1G). Briefly, cells were fixed with 3.6% PFA and 10% acetic acid for 15 min, followed by permeabilization with 0.5% Triton X-100 for 5 min on ice. Then cells were subjected to incubation with denatured Dig-labeled FISH probes in hybridization buffer (50% formamide in 2×SSC) at 50 °C for 16 h. After hybridization, anti-Dig primary antibody and fluorescent secondary antibody were sequentially added to visualize signal with Zeiss LSM 880 confocal microscope. The nuclei were counterstained with DAPI. For co-localization, RNA FISH assays were performed in MCF10A and HaCaT cells with stable expression of GFP-tagged H1.3.
Subcellular fractionation
Isolation of cell nucleus and cytoplasm in MCF10A cells was performed as previously described [29]. The RNA and protein were extracted from the cytoplasm, nucleoplasm and chromatin according to the number of cells. RNAs were extracted by TRIzol and proteins were lysed by 1×SDS loading buffer. The isolated RNAs were used for cDNA synthesis and qRT-PCR analysis. Primers are listed in the Supplementary Table S1A.
MS2-MCP living cell imaging and Fluorescence recovery after photobleaching assay (FRAP)
SNHG8-12×MS2 or 12×MS2 was overexpressed in MCF10A cells which stably expressed GFP-tagged H1.3 and mCherry-tagged MCP (MS2 coated protein). For live-cell imaging, cell density of 2 × 104 cells/mL were plated on a confocal imaging dish (JET BIOFIL, Guangzhou, China) and incubated for two days at 37 °C and under 5% CO2 before imaging. FRAP experiments were performed as previously described [30].
Micrococcal nuclease (MNase) digestion and salt extraction analysis
MNase digestion and salt extraction analyses were performed as described previously [30]. Briefly, 4 × 105 indicated cells for MNase digestion were suspended in 100 μL of buffer A [(0.32 M sucrose, 15 mM HEPES-NaOH (pH 7.9), 60 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5% bovine serum albumin (w/v), 0.5 mM spermidine, 0.15 mM spermine and 0.5 mM dithiothreitol)], layered on a cushion of 100 μL buffer A containing 30% sucrose (w/v) and centrifuged at 3000 × g for 5 min at 4 °C. For MNase digestion analysis, the nuclear pellet was suspended in 100 μL of buffer C [(20 mM Tris-HCl (pH 7.5), 70 mM NaCl, 20 mM MgCl2, 3 mM CaCl2, 0.5 mM spermidine, 0.15 mM spermine and 0.5 mM dithiothreitol)] and digested with MNase (Thermo) at 37 °C for indicated time. For salt extraction analysis, pelleted nuclei were resuspended in buffer B [(15 mM HEPES-NaOH (pH 7.9), 60 mM KCl, 15 mM NaCl, 0.34 mM sucrose, 10% glycerol (v/v)] and incubated at 4 °C for 30 min with indicated concentrations of NaCl. The mixtures were then centrifuged at 15,000 × g for 5 min at 4 °C, and the resulting supernatants were subjected to immunoblotting. Image J software was used for immunoblotting band quantification.
Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq)
ATAC-seq was performed as previously described [31]. Briefly, 7500 indicated cells were washed with cold PBS, collected by centrifugation then re-suspended in resuspension buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2). After collection, cells were lysed in lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.2% NP-40) and collected before incubating in transposition mix containing Tn5 transposase (Vazyme). Purified DNA was then ligated with adapters, amplified and size selected for sequencing. Library DNA was sequenced with paired end 42 bp reads.
RNA-seq and ATAC-seq data analysis
ATAC-seq and RNA-Seq was performed by Novogene. For RNA-seq analysis, reads were mapped to Human Genome Assembly GRC38 by STAR version 2.5. Gene and isoform expression quantification were called by RSEM version 1.2 with default parameters on the GENCODE human v28 gene annotation file. Differential expression analysis was performed using the Bioconductor package edgeR (version 3.18.1). Significant differentially expressed genes were chosen according to two criteria: (1) a significance level FDR < 0.05; and (2) expression-level average fragments per kilobase of transcript per million mapped reads values >1 in either the treatment or the control groups.
For ATAC-seq the sequencing reads were filtered by Trimmomatic (0.35) and Cutadapt (1.13), and then mapped to human reference sequence for hg38 using Bowtie2 with parameters “-X 2000 –local”. Mapped reads were then sorted and deduplicated using Samtools (1.8) with parameters, and Picard tools Mark Duplicates (1.90). We performed peak calling using the MACS2 (2.1.0) callpeak module with parameters “-p 0.01 –no model –extsize 150 -B –SPMR –keep-dup all –call-summits” on pooled replicates. Tracks of signal were computed using MACS2 bdgcmp module with parameter “-m ppois”. Signal density was computed using compute Matrix and Bedtools (2.25.0). ChIP peak Anno (3.16.1) was used for identifying nearby genes from the peaks obtained from MACS. MergePeaks software was used to identify the same peaks between different groups with peaks submits within 500 bp. Differential genes were computed according to signal density using DESeq2 (1.22.2) and then defined these closed and opened genes with fold changes ≥2 and P < 0.05.
Protein expression and purification
The pGEX-6p-1-H1.3-6×His plasmid was transformed into Escherichia coli Rosetta 2 (DE3). After induction for 18 h with 0.1 mM IPTG at 20 °C, the cells were harvested by centrifugation and the pellets were re-suspended in lysis buffer (25 mM Tris-HCl, pH 7.5, 1 M NaCl, 500 mM Urea, 10% glycerol, 1 mM PMSF, 5 mM benzamidine, 1 μg/mL leupeptin and 1 μg/mL pepstatin). The cells were then lysed by sonication and the cell debris was removed by ultracentrifugation. The supernatant was mixed with glutathione Sepharose-4B beads (GE healthcare) and rocked for 2 h at 4 °C before elution with 20 mM reduced glutathione (Sigma). Pre-Scission protease (GE healthcare) was then added to remove the N-terminal GST tags. The proteins were further mixed with Ni-NTA agarose beads (QIAGEN) and rocked for 2 h at 4 °C before elution with 300 mM imidazole. The proteins were further purified by gel-filtration chromatography equilibrated with 25 mM Tris-HCl, pH 7.5, 150 mM NaCl. The H1.3 were labeled by Alexa Fluor 647 protein labeling system (A20006, Invitrogen) according to manufactory’s instruction. The purified proteins were concentrated by centrifugal filtrations (Millipore), then stored in aliquots at −80 °C.
SNHG8 and p601 DNA array labeling
SNHG8 was in vitro transcribed using the T7 Ribo MAX Large-Scale RNA Production System (P1320, Promega) with or without fluorescein-12-UTP (134367-01-4, Roche) labeling. And in vitro transcribed SNHG8 was purified by chromatography and fractions were loaded on and a natural SDS-PAGE gel and staining by Gelred dye for quality control. 2×p601 arrays were prepared according to a previous description [32] and the purified 2×p601 was labeled by Alexa Fluor 594 Nucleic Acid Labeling Kit (Invitrogen) according to manufactory’s instruction. Labeled p601 fragments were purified by a NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel).
Droplet assembly
Phase-separated droplets of H1.3 or H1.3 truncations with SNHG8 or 4×p601 arrays were formed by a quick dilution of the purified H.3 and SNHG8 or 4×p601 arrays [(protein: RNA/DNA) molar ratio, 5:1] into a reaction buffer containing NacCl and Tris-HCl and adjusted the concentration to 20 mM Tris-HCl (pH 7.5), 150 mM NaCl. The phase-separated droplet solution was incubated for 30 min to 4 h at room temperature. Droplets were imaged immediately to 4 h using Zeiss LSM 880 confocal microscope with DIC or fluorescence mode.
Electrophoretic mobility shifts assay (EMSA)
The in vitro transcribed SNHG8 and 2×p601 was mixed with or without H1.3 at molecular ratio 1:1 in the buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2) at 25 °C for 10 min. For preparation of the competitive sample, 2×p601 mixed with H1.3 and then added SNHG8 into the p601−H1.3 mixture. The reactions were quenched by adding the loading dye. The samples were then loaded into a 6% de-natural polyacrylamide gel with 0.5×TBE buffer and stained with Gelred dye.
Quantification and statistical analysis
The statistical analyses are described in the figure legends. Multiple t-test is corrected by Bonferroni–Dumn method. Differences were considered significant at P < 0.05. The experiments were repeated independently for at least 3 times with similar results.
Results
Epithelial cells-enriched SNHG8 expression
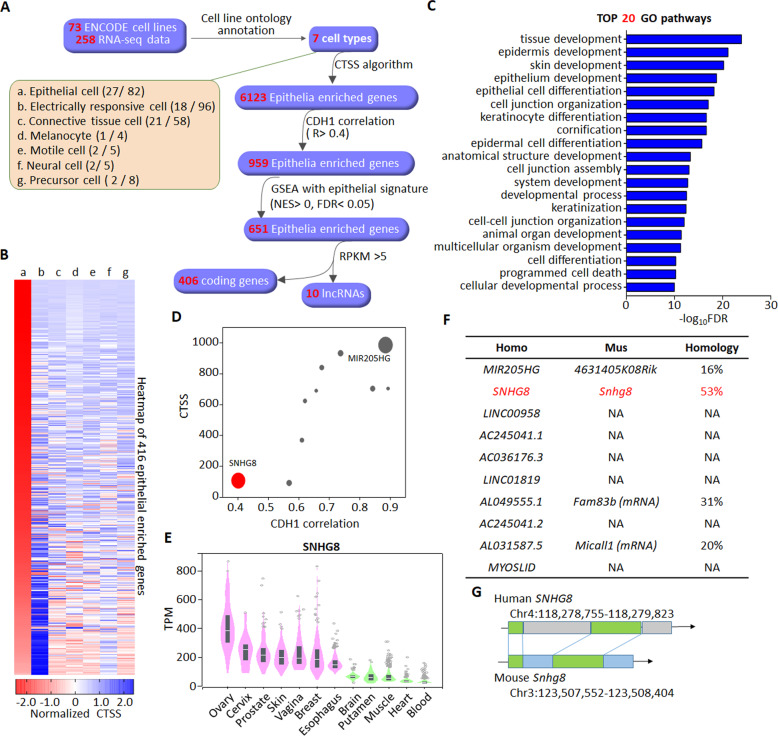
A total of 258 RNA-seq data from 73 cell lines of different origins were deposited in Encyclopedia of DNA Elements (ENCODE) project [33, 34], and these cell lines can be classified into 7 annotated cell types according to their Cell Ontology annotations [35] (Fig. 1A, Supplementary Data Set 1). To identify functional lncRNAs in epithelial cells from these resources, we designed a bioinformatic screening pipeline named the CTSS. A workflow illustrating the screening process is shown in Fig. 1A. The screening generated a collection of 6123 epithelial cells-enriched genes consisting of 4064 coding genes and 2059 non-coding genes including 536 long non-coding genes (Supplementary Data Set 2). Some known epithelia-associated lncRNAs, such as MALAT1 [36], NEAT1 [37] and TINCR [38], were also included. To obtain the most probably functional lncRNAs, the list of epithelial cells-enriched genes was further narrowed down according to their correlations with the prototypical epithelial marker cadherin 1 (CDH1), GSEA with an epithelial signature [39] and their abundances, referred to as reads per kilobase per million mapped reads (RPKM). A final list of 416 genes was yielded, including 406 coding genes and 10 lncRNAs (Fig. 1A, B, Supplementary Data Set 3). The CTSSs of these genes in each cell type from a to g were calculated and normalized by Z-score, followed by ranking based on the normalized CTSSs in epithelial cells (Fig. 1B). Of note, protein coding genes were also included throughout the whole screening process because most of them had defined functions and could be used as positive controls. As revealed by GO analysis, the identified 406 protein-coding genes were predominantly involved in epithelia-specific processes (Fig. 1C). Among the identified 10 lncRNAs (Fig. 1D), miR-205 host gene (MIR205HG) with high abundance was previously reported to be mainly expressed in the basal layer of prostate epithelium and capable of regulating basal-luminal differentiation [40]. Besides MIR205HG, all other 9 lncRNAs showed an epithelia-enriched feature, although their expressional patterns in individual epithelial tissue were different (Fig. 1E and Supplementary Fig. S1). Considering that the functions of these 9 lncRNAs in epithelial cells were unknown, we focused on SNHG8 for further evaluation because of its similar abundance/RPKM to MIR205HG (Fig. 1D) and its homology between human and mouse (Fig. 1F, G).
Fig. 1. CTSS-based screening of epithelial cell-enriched lncRNAs.
A Scheme of the screening process of epithelia-specific lncRNAs. The 73 cell lines recorded by ENCODE are classified into 7 cell types as indicated by a-g according to their cell ontology annotations. The numbers of cell lines and corresponding RNA-seq data in each cell type are shown in the parentheses. Following the CTSS screening of cell-type enriched genes, the resultant 6123 epithelia-enriched genes were further filtered by the correlation with CDH1 (Pearson R > 0.4), GSEA analysis with epithelial signature (NES, normalized enrichment score, NES > 0, false discovery rate, FDR < 0.05) and RPKM > 5, yielding a final list of 406 coding genes and 10 lncRNAs. B Heatmap depicting the CTSSs of the 416 epithelia-enriched genes across different cell types as indicated by group a-g like panel A. Genes are ranked based on the normalized CTSSs in epithelial cells. C Top 20 GO biological processes enriched by the 406 epithelia-enriched coding genes. D Scatter plot depicting the CTSSs and correlations with CDH1 of the 10 epithelia-enriched lncRNAs. The size of the dot represents the average RPKM. E The level of SNHG8 in indicated normal tissues from GTEx was analyzed by GTEx portal website (https://www.gtexportal.org). Expression values are shown in transcripts per million (TPM) calculated from a gene model with isoforms collapsed to a single gene. Box plots are shown as median and 25th and 75th percentiles; points are displayed as outliers if they are above or below 1.5 times the interquartile range. F Table summarizing the homology of indicated lncRNAs with corresponding mouse analogs. NA not applicable. G The homology sequence analysis depiction of SNHG8 and its conserved analog in mouse. The green regions are highly conserved.
Characterization of SNHG8 in epithelial cells
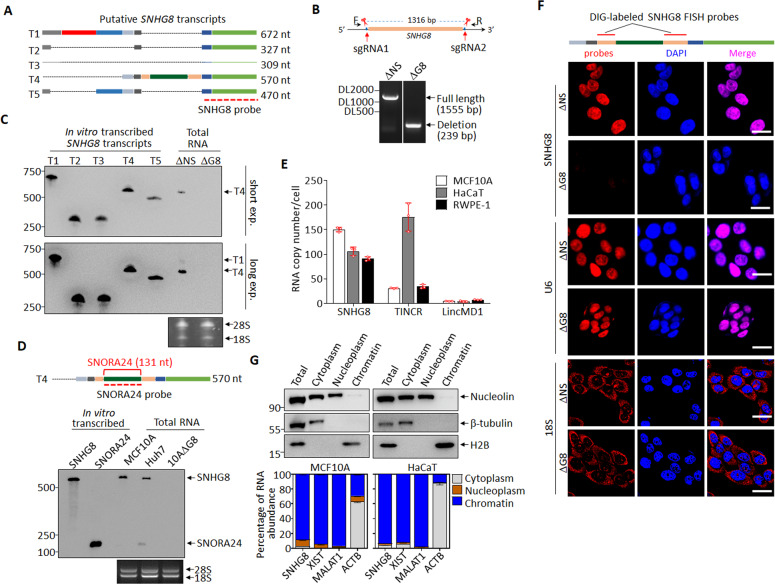
Although SNHG8 had been reported to play a role in carcinogenesis [41, 42], the molecular and cellular characterization of SNHG8 is absent to date. According to the Ensembl annotation, SNHG8 gene encodes five transcripts (Fig. 2A). To validate their expressions in epithelial cells, we performed Northern blot in CRISPR-Cas9 system-generated SNHG8-knockout (MCF10A-ΔG8) and non-specific knockout (MCF10A-ΔNS) clones from the human mammary epithelial cell line MCF10A (Fig. 2B) with a probe targeting all five transcripts (Fig. 2A), in which in vitro transcribed five putative SNHG8 transcripts were used as positive controls (Fig. 2C). The results showed that SNHG8 mainly encoded transcript T4 (ENST00000602573.1) in MCF10A-ΔNS cells, which could be completely knocked out in MCF10A-ΔG8 (Fig. 2C). Of note, a minor band was also detected upon long exposure, whose length was most close to that of transcript T1 (ENST00000602414.6) (Fig. 2A). According to Ensembl database, T4 is the only transcript conserved with the mouse homology Snhg8. Hereafter, T4 was used to represent SNHG8. Considering that SNHGs are a group of lncRNAs with introns and exons in their sequences and generate small nucleolar RNAs through alternative splicing [43], and the small nucleolar RNA SNORA24 is included in SNHG8 (Fig. 2D), we examined whether SNHG8 gene is also processed into SNORA24 in MCF10A cells. Northern blot assay using a probe to target both SNORA24 and SNHG8 (Fig. 2D) revealed that hepatocellular carcinoma Huh7 cells expressed SNORA24 [44] and SNHG8. However, MCF10A cells predominantly expressed SNHG8 with almost undetectable SNORA24 expression (Fig. 2D). By the way, prediction with the Coding Potential Assessment Tool (CPAT) (Supplementary Fig. S2A) and in vitro transcription/translation assay with MYC, UHRF1 and lncRNA NKILA respectively as coding (Supplementary Fig. S2B) and non-coding controls also confirmed the non-coding nature of SNHG8.
Fig. 2. Characterization of SNHG8 in epithelial cells.
A Schematic diagram of putative SNHG8 transcripts (data from Ensembl database), with the SNHG8 probe for Northern blot indicated. B Schematic diagram of guide RNAs (sgRNA1 and sgRNA2) targeting SNHG8 gene loci and electrophoresis of the SNHG8 genomic PCR products. C Northern blots with SNHG8 probe in total RNA extracts from MCF10A-ΔNS and MCF10A-ΔG8 cells, with 1 ng in vitro transcribed putative SNHG8 transcripts (T1-T5) used as positive controls, and 28S/18S used as total RNA loading controls. D Schematic diagram of SNHG8 (T4), with SNORA24 and the SNORA24 probe indicated (top). Northern blots with SNORA24 probe in total RNA extracts from MCF10A and Huh7 cells, with 0.3 ng in vitro transcribed putative SNHG8 (T4) and SNORA24 transcripts used as positive controls, and 28S/18S used as total RNA loading controls (bottom). E The average copy numbers of SNHG8, TINCR and LincMD1 per cell in MCF10A, RWPE-1 and HaCaT cells as quantified by absolute qRT-PCR. F Schematic diagram of SNHG8 with probes used for DIG-labeled RNA-FISH assay indicated (Top). Representative confocal microscopy images of SNHG8 detected by Digoxin-labeled RNA-FISH assay with U6 snRNA and 18S RNA respectively as nuclear and cytoplasm RNA controls in MCF10A-ΔNS and MCF10A-ΔG8 cells. Scale bar, 20 μm. G MCF10A and HaCaT cells were fractionated and subjected to immunoblotting (top) and qRT-PCR (bottom) for the indicated proteins and RNAs. For immunoblotting, β-tubulin, Nucleolin, and H2B were respectively used as cytoplasm, non-chromatin, and chromatin markers; for qRT-PCR, XIST and MALAT1 were used as chromatin markers, and β-actin mRNA (ACTB) as cytoplasm marker. Data in panels E and G are the mean ± SD (n = 3), and all experiments were repeated for three times with similar results.
qRT-PCR analyses showed that, like TINCR but not the muscle cell-enriched lncRNA LincMD1 [45], the copy of SNHG8 was abundant in MCF10A cells, prostate epithelial cell line RWPE-1 and human keratinocyte cell line HaCaT (Fig. 2E). To define the subcellular localization of SNHG8, we performed RNA FISH, and found that SNHG8 was almost exclusively localized in the nucleus like U6 snRNA rather than 18S RNA, the latter being in cytoplasm (Fig. 2F). Specifically, subcellular fractionation followed by qRT-PCR revealed that SNHG8 was mainly detected in the chromatin of MCF10A and HaCaT cells like non-coding RNA XIST [46] and MALAT1 [47] (Fig. 2G).
SNHG8 binds to linker histone H1 family members
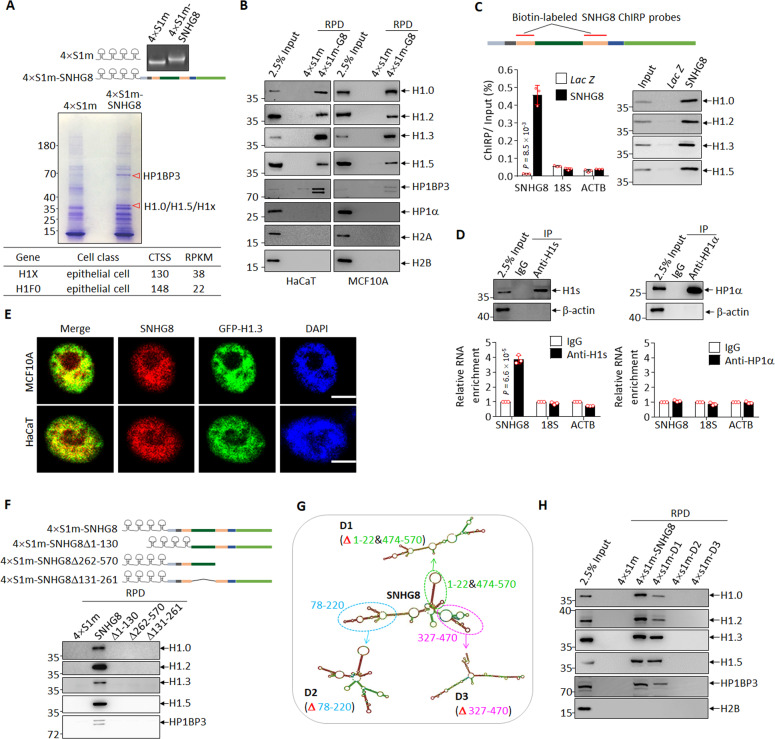
The chromatin localization of SNHG8 inspired us to ask whether SNHG8 binds to chromatin modulators to exert biological functions. To explore the binding partners of SNHG8 in epithelial cells, in vitro transcribed 4-fold repeat of a modified streptavidin-binding RNA aptamer termed S1m (4×S1m) and 4×S1m-fused full-length SNHG8 (4×S1m−SNHG8, Fig. 3A) were incubated with MCF10A cell lysates, followed by pulldown with streptavidin magnetic beads. The bound proteins were separated by SDS-PAGE and the differential bands were analyzed by LC − MS/MS. Two differential bands at 35 and 70 kD were identified as linker histone H1 family members H1.0/H1.5/H1x and heterochromatin protein 1 binding protein 3 (HP1BP3), respectively (Fig. 3A, Supplementary Data Set 4), the latter being evolutionarily and structurally related to the H1s [48]. Interestingly, H1F0 (coding H1.0) and H1X (coding H1x) were also present in the epithelia-enriched gene list after CTSS screening (Fig. 3A, Supplementary Data Set 2).
Fig. 3. SNHG8 binds to linker histone H1 family members.
A 4×S1m and 4×S1m-SNHG8 were constructed and transcribed in vitro, followed by electrophoresis on formaldehyde-denatured agarose gel (top). In vitro transcribed 4×S1m and 4×S1m-SNHG8 were incubated with MCF10A cell lysates and subjected to RNA pulldown assay, followed by SDS-PAGE separation and coomassie blue staining of bound proteins. The red triangles point to two differentially stained bands subjected to LC − MS/MS analysis (middle). CTSSs of H1FX and H1F0 from Fig.1A are shown in the bottom panel. B In vitro transcribed 4×S1m and 4×S1m-SNHG8 were incubated with HaCaT (left) or MCF10A (right) cell lysates, followed by RNA pulldown and immunoblotting for the indicated proteins. RPD, RNA-pulldown. C Schematic diagram of SNHG8 with probes used for ChIRP assay indicated (top). qRT-PCR analysis (bottom left) and immunoblotting (bottom right) for the indicated RNAs and proteins were performed. Lac Z probes were used as negative control. D Endogenous histone H1 or HP1α were respectively purified with anti-pan-H1 (anti-H1s) and anti-HP1α antibodies from MCF10A cells. Immunoblotting (top) and qRT-PCR analysis (bottom) for the indicated proteins and RNAs were performed. β-actin was used as a negative control for immunoblotting, and 18S RNA and β-actin mRNA (ACTB) were used as negative controls for qRT-PCR. E Representative confocal microscopy images showing SNHG8 visualized by DIG-labeled RNA-FISH assay and GFP-tagged H1.3 in MCF10A and HaCaT cells. Scale bar, 7.5 μm. F 4×S1m-SNHG8 and its linear sequence-based truncations (top) were incubated with MCF10A cell lysates and subjected to RNA pulldown, followed by immunoblotting for the indicated proteins (bottom). FL full length, RPD RNA-pulldown. SNHG8 and its secondary structure-based truncations (G) were incubated with MCF10A cell lysates and subjected to RNA pulldown, followed by immunoblotting for the indicated proteins (H). RNA structures were predicted by RNA fold web server (http://rna.tbi.univie.ac.at). Data in panels C and D are the mean ± SD (n = 3), and P values between SNHG8 and Lac Z (C) or IgG and Anti-H1s (D) were calculated by multiple T-test corrected by Bonferroni-Dumn method. All experiments except for A were repeated three times with similar results.
We continued to validate whether SNHG8 interacted with H1s. As shown in Fig. 3B, in vitro transcribed SNHG8 pulled down histone H1 variants detected in MCF10A and HaCaT cells, including H1.0, H1.2, H1.3 and H1.5, as well as HP1BP3, but not chromatin associated proteins heterochromatin protein 1α (HP1α) as well as core histones H2A and H2B. We also performed ChIRP assay to verify whether endogenous SNHG8 could pull down endogenous H1s, in which a set of probes to target the Lac Z mRNA, normally absent from human cells [49], were used as negative control. As depicted in Fig. 3C, SNHG8 probes specifically enriched endogenous SNHG8 but not 18S RNA or ACTB mRNA, and efficiently pulled down endogenous histone H1.0, H1.2, H1.3 and H1.5. Reciprocally, Flag-tagged histone H1s but not Flag-tagged HP1α abundantly immunoprecipitated endogenous SNHG8 rather than 18S RNA in MCF10A cells (Supplementary Fig. S3). Similarly, antibody against pan-H1s but not HP1α immunoprecipitated endogenous SNHG8 but not 18S RNA or ACTB mRNA (Fig. 3D). Additionally, RNA FISH assay also showed co-localization of SNHG8 with GFP-tagged H1.3 in MCF10A and HaCaT cells (Fig. 3E). Cumulatively, our results propose that histone H1s are endogenous binding partners for SNHG8.
To map the minimal sequence of SNHG8 required to interact with histone H1s, SNHG8 mutants deleted of the 5′, middle, and 3′ region were individually incubated with MCF10A cell lysates, followed by RNA pulldown. The results showed that the truncation of either region completely disrupted the interaction between SNHG8 and H1s (Fig. 3F). Considering that lncRNAs tend to carry out interactions through secondary structure rather than linear sequence [50], the secondary structure of SNHG8 was predicted by RNAfold web server [51] and three structure-based SNHG8 mutants D1, D2 and D3 were generated (Fig. 3G). Because deletion variants are folded into their own stable secondary structures, they are not necessarily complete superimposable to that of SNHG8 [51]. RNA pulldown assay showed that the D1 mutant, which preserves two stem loops of SNHG8 (Fig. 3G), maintained interaction with histone H1 to a degree (Fig. 3H). In contrast, the other two mutants which largely disrupted the conformation of SNHG8 abrogated the interaction (Fig. 3G, H).
SNHG8 competes with DNA for histone H1 binding
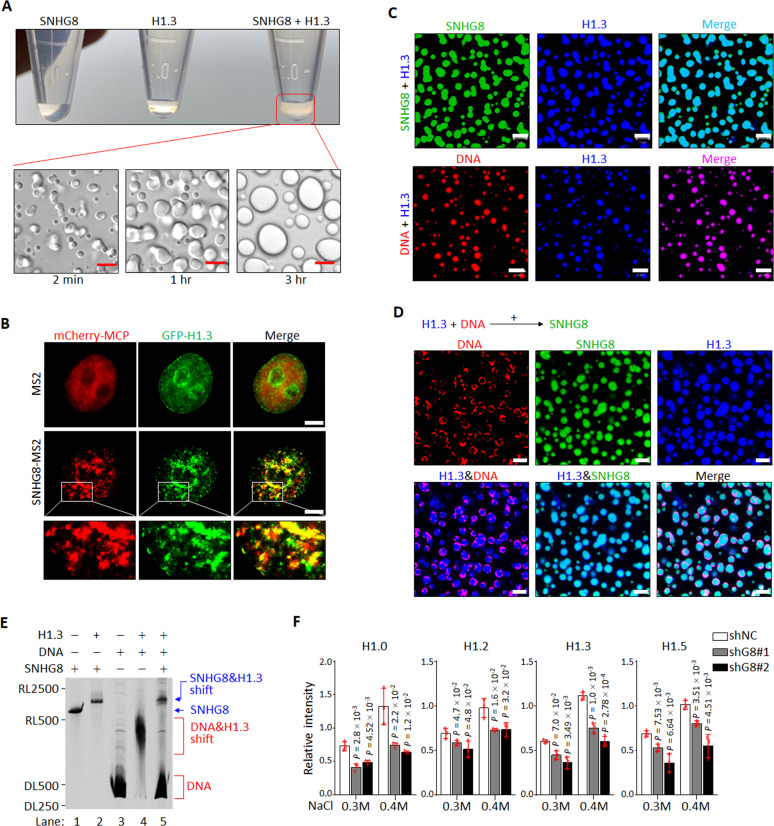
To examine whether SNHG8 and histone H1 are directly associated with each other, we purified SNHG8 and H1.3 in vitro (Supplementary Figs. S4A and S4B). Upon mixing them in physiological salt, a turbid solution was unexpectedly observed (Fig. 4A). Under a microscope, the turbid material revealed liquid droplets, which fused with each other into larger ones over time (Fig. 4A; Supplementary Videos 1, 2), indicative of liquid-liquid phase separation (LLPS), a characteristic of proteins with intrinsically disordered regions and the capacity for multivalency [52, 53]. Indeed, both the N- and C-terminal regions of H1s are intrinsically disordered [32, 54] (Supplementary Fig. S4C). SNHG8−H1.3 phase separation required the full-length of H1.3, as the CTD of H1.3 greatly compromised the formation of droplets, and the GD failed to form droplets with SNHG8 (Supplementary Fig. S4D).
Fig. 4. SNHG8 competes with DNA for histone H1 binding.
A Photographs of tubes containing in vitro-transcribed SNHG8, recombinant H1.3 or SNHG8−H1.3 mixture (top), and images of turbid SNHG8−H1.3 mixture observed by differential interference contrast confocal microscopy (bottom). Scale bar, 5 μm. B MCF10A cells stably expressing GFP-tagged H1.3 and mcherry-MCP were transfected with MS2 or MS2-SNHG8 and subjected to living cell imaging under a confocal microscopy. Scale bar, 5 μm. MCP, MS2 coated protein. C Representative confocal microscopy images of fluorescein-labeled SNHG8 (green) or AF594-labeled p601 DNA (red) mixed with AF647-labeled H1.3 (blue) in 150 mM NaCl by molecular ratio 1:1. Scale bar, 5 μm. D Fluorescein-labeled SNHG8 (green) was added to the pre-mixed AF647 labeled H1.3 (blue)−AF355-labeled p601 DNA (red) mixture, followed by incubation for 30 min. Representative confocal microscopy images were shown. Scale bar, 5 μm. E In vitro electrophoretic mobility shift assay showing the competitive relationship between SNHG8 and p601 DNA for H1.3. F Quantification of salt extraction assays showing the relative intensity of each H1 variant extracted by increasing salt concentrations from MCF10A cells with and without knockdown of SNHG8. Data are the mean ± SD (n = 3), and P values compared with shNC cells were calculated by multiple T-test corrected by Bonferroni–Dumn method. All experiments were repeated for three times with similar results.
To test whether droplets of H1-bound SNHG8 exist in epithelial cells, living cell imaging of SNHG8 was achieved by using the MS2-MCP system [55]. When MS2-SNHG8 and mcherry-MCP were co-expressed in MCF10A cells together with GFP-H1.3, the RNA and protein complex formed by MS2-SNHG8 and mcherry-MCP (MS2-SNHG8−mcherry-MCP), rather than MS2-mcherry−MCP complex, was strikingly colocalized with GFP-H1.3, and condensed into puncta with GFP-H1.3 under live cell imaging system (Fig. 4B). Notably, the puncta that came into contact with one another were coalesced and most of them were not perfectly circular. These nonspherical condensates were proposed to be likely due to the local rigidity of chromatin partitioned in the puncta [56]. To test whether these puncta/nonspherical condensates display rapid exchange kinetics with their surroundings, a characteristic of phase-separated condensates [57], FRAP was performed and showed that MS2-SNHG8−mcherry-MCP and GFP-H1.3 were able to recover (Supplementary Fig. S4E, Supplementary Video 3). These results suggest that SNHG8 and H1.3 form LLPS condensates in the cells.
In line with the previous reports [56, 58], histone H1s can phase separate when mixed with DNA (Fig. 4C). When SNHG8 and DNA were separately mixed with H1.3 at molecule ratio 1:1, the droplets formed by SNHG8−H1.3 were much larger than DNA−H1.3 droplets at the same time point (Fig. 4C). Moreover, the addition of SNHG8 to the H1.3−DNA mixture disrupted the H1.3−DNA droplets by excluding DNA to the droplet exterior (Fig. 4D), a phenomenon that has also been observed in other competitive phase separation models [59]. Further, EMSA demonstrated that addition of H1.3 to either SNHG8 or DNA reduced their mobility and shifted them to higher molecular weight (Fig. 4E). When SNHG8 was added to the pre-incubated DNA−H1.3 mixture, however, DNA was replaced from H1.3, and the shift of SNHG8−H1.3 but not DNA−H1.3 was formed (Fig. 4E). All these results from competitive phase separation and EMSA assays (Fig. 4D, E) suggested that SNHG8 has a higher binding ability to H1.3 than chromatin DNA. To verify this assumption in cells, SNHG8 was knocked down in MCF10A cells with two specific shRNAs (Supplementary Fig. S4F). By using chromatin extraction assay [60], we found that, under low salt concentration (0.3 M and 0.4 M), H1s were extracted ~2 fold less in SNHG8 knockdown cells compared to control cells (Fig. 4F, Supplementary Fig. S4G), suggesting that there is more H1s bound to chromatin to result in chromatin compaction. Collectively, SNHG8 phase separates H1s and competes with DNA for H1 binding.
SNHG8 knockdown leads to chromatin condensation and epithelial differentiation-associated gene expression
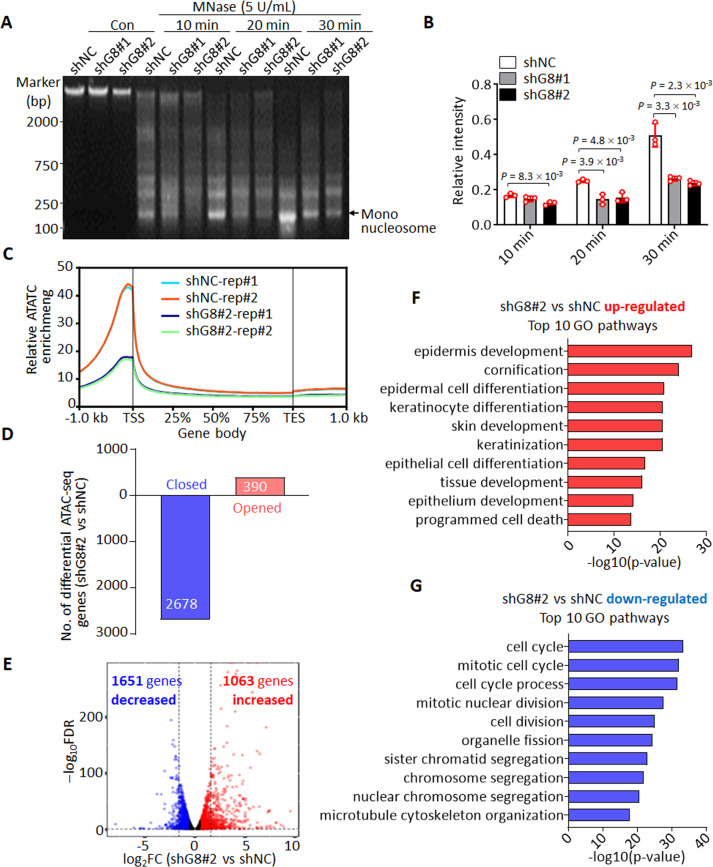
To investigate whether the increased binding of histone H1 to chromatin upon SNHG8 depletion affects chromatin accessibility, we performed chromatin MNase digestion assay. The results showed that the chromatin in MCF10A cells with SNHG8 knockdown was more inaccessible compared to control cells (Fig. 5A, B). The result could be further confirmed in MCF10A-ΔG8 cells compared with MCF10A-ΔNS cells (Supplementary Fig. S5A). We also performed ATAC-seq assay to comprehensively determine alterations of gene accessibility after SNHG8 knockdown. As for this, shNC and shG8#2-infected MCF10A cells were subjected to ATAC-seq. The ATAC-seq peaks of open chromatin were highly consistent between independent experiments in both groups (Supplementary Fig. S5B). Out of the 54975 ATAC-seq peaks detected, 33913 peaks were common between shNC and shG8#2 cells, while 18307 and 2755 peaks were respectively specific to shNC and shG8#2 cells (Fig. 5C, Supplementary Fig. S5C, Supplementary Data Set 5), indicating the chromatin of shG8#2 cells was in a more inaccessible state. By mapping those differential ATAC-seq peaks to the genomic locus, we found that 2678 genes tended to be closed and 390 genes tended to be opened under SNHG8 knockdown (Fig. 5D, Supplementary Fig. S5D). GO pathway analysis revealed that those opened genes could be significantly enriched in pathways such as regulations of cell migration and cell differentiation (Supplementary Fig. S5E), and those closed genes were remarkably enriched in cell cycle-associated pathways (Supplementary Fig. S5F). To find out whether the alteration of genomic accessibility could be reflected on gene expression level, we also performed RNA-seq analysis in shNC- and shG8#2-infected MCF10A cells (Supplementary Data Set 6). Generally, the results showed that down-regulated genes outnumbered up-regulated genes (Fig. 5E), consistent to the more inaccessible genomic status upon SNHG8 knockdown. More importantly, GO pathway analysis of genes deregulated after SNHG8 knockdown showed that the up-regulated genes were most significantly enriched in epithelial differentiation-related pathways (Fig. 5F), while the top pathways enriched by down-regulated genes are all cell cycle-associated pathways (Fig. 5G). Cell cycle arrest is one of the hallmarks of epithelial differentiation [61]. Thus, both the ATAC-seq and RNA-seq results suggested that SNHG8 knockdown resulted in an epithelial cell differentiation-associated genomic expression pattern.
Fig. 5. Knockdown of SNHG8 leads to chromatin condensation.
A, B Nuclei isolated from MCF10A cells with and without knockdown of SNHG8 were subjected to MNase digestion analysis (A). The relative intensities of mono-nucleosome were calculated by normalizing the intensity of mono-nucleosomes in each lane against the intensity of total DNA in corresponding lane (B). DNA intensities were measured by Image J. Data are the mean ± SD (n = 3), and P values between two line-linked groups were calculated by multiple T-test corrected by Bonferroni–Dumn method. C Metaplot ATAC-seq signals in MCF10A cells with and without knockdown of SNHG8 in two independent assays. TSS transcription start site, TES transcription end site. D Bar plot depicting the number of genes opened and closed after knockdown of SNHG8 (FC ≥ 2 and FDR < 0.05 peaks). FC fold change, FDR false discovery rate. E Comparative transcriptome analysis between MCF10A cells with and without knockdown of SNHG8. log2FC vs log10FDR shows gene changes between shG8#2 and shNC. Significantly regulated hits (according to FC ≥ 2 and FDR < 0.05) are shown as colored dots (red, higher in shG8#2 cells; blue, higher in shNC cells). RNA-seq was repeated twice. FC fold change, FDR false discovery rate. The top 10 GO biological processes respectively enriched by genes up-regulated (F) and down-regulated (G) after knockdown of SNHG8 in MCF10A cells.
SNHG8 loss contributes to epithelial differentiation
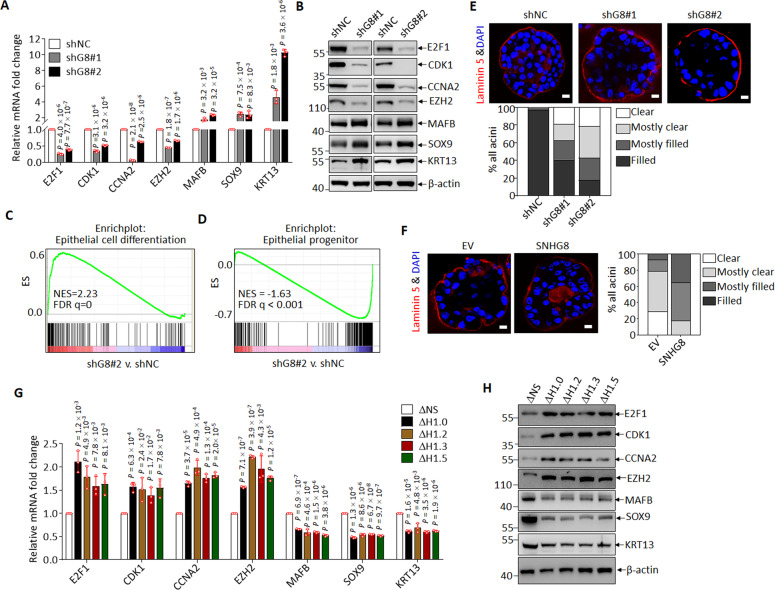
MCF10A cells had been shown to have stem/progenitor-like properties [62]. We thus asked whether knockdown of SNHG8 indeed induced MCF10A cells to undergo differentiation as the transcriptomic data indicated. Firstly, we validated the decreased expression of key cell cycle regulators such as E2F1, CDK1, CCNA2, EZH2, and the increased expression of epithelial cell differentiation associated transcription factors and markers, including MAFB, SOX9 and KRT13 [21], in SNHG8-knocked down MCF10A cells compared to control cells both at mRNA (Fig. 6A) and protein level (Fig. 6B), as well as in MCF10A-ΔG8 cells compared to parental MCF10A cells (Supplementary Fig. S6A). Secondly, GSEA revealed that epithelial cell differentiation-associated gene signatures could be enriched in the up-regulated genes (Fig. 6C), and the established gene sets representing epithelial progenitor genes could be enriched in the down-regulated genes (Fig. 6D). Thirdly, knockdown of SNHG8 impedes cell cycle at G0/G1 stage (Supplementary Fig. S6B). More importantly, 3D mammosphere formation assay, a classical model to evaluate epithelial cell differentiation as characterized by hollow lumen formation [63], showed that SNHG8 knockdown accelerated acini maturation of MCF10A cells (Fig. 6E). Finally, re-expression of SNHG8 in MCF10A-ΔG8 cells to the comparable level of endogenous SNHG8 rescued cell cycle arrest (Supplementary Fig. S6C), acini maturation delay (Fig. 6F) and gene expression deregulation (Supplementary Fig. S6A). In line with its negative regulating role in acinar formation, additionally, SNHG8 was significantly down-regulated during this process (Supplementary Fig. S6D). All these results supported that SNHG8 loss probably induces differentiation of MCF10A cells. This differentiation-inducing effect of SNHG8 knockdown could also be seen in RWPE-1 and HaCaT cells (Supplementary Figs. S6E and S6F). Similarly, the expression of SNHG8 was down-regulated during calcium-induced differentiation of RWPE-1 cells (Supplementary Fig. S6G).
Fig. 6. Loss of SNHG8 induces epithelial differentiation through histone H1.
A, B qRT-PCR analysis (A) and immunoblotting (B) for the indicated RNAs and proteins in MCF10A cells with and without knockdown of SNHG8. C, D GSEA of RNA-seq data from MCF10A cells with and without knockdown of SNHG8 with the indicated epithelial differentiation (C) and progenitor-associated genesets (D). E, F Representative confocal microscopy images and quantification of the acini formed in 3D culture by MCF10A cells with and without knockdown of SNHG8 for 8 days (E) and by MCF10A-ΔG8 cells with and without re-expression of SNHG8 for 10 days (F). Scale bar, 10 μm. G, H qRT-PCR analysis (G) and immunoblotting (H) for the indicated RNAs and proteins in MCF10A cells with and without CRISPR/Cas9-mediated depletion of H1 subtypes. Data in panels A and G are the mean ± SD (n = 3), and P values compared with shNC(A) or ΔNS (G) were calculated by multiple T-test corrected by Bonferroni–Dumn method. All experiments except for A were repeated three times with similar results.
Next, we sought to validate the role of histone H1 in SNHG8 knockdown-induced differentiation-associated gene expression. On the one hand, four somatic histone H1 subtypes (H1.0, H1.2, H1.3 and H1.5) were individually depleted by CRISPR/Cas9 in MCF10A-ΔG8 cells (Supplementary Fig. S6H), and the results showed that the depletion of all these subtypes reversed the repressed expression of cell cycle regulators and the activated expression of differentiation-associated genes caused by SNHG8 deficiency both at mRNA and protein levels (Fig. 6G, H). On the other hand, histone H1 with duplicate C-terminal domain (H1CC), which confer stronger binding of H1s to chromatin [30], was overexpressed in MCF10A cells to simulate H1s with increased binding to chromatin upon SNHG8 depletion. As a result, H1CC phenocopied SNHG8 knockdown as evidenced by the repressed expression of cell cycle regulators and the activated expression of differentiation-associated genes (Supplementary Figs. S6I and S6J). Collectively, SNHG8 loss induces differentiation-associated gene expression, probably through causing H1-mediated chromatin condensation.
Discussion
With the CTSS-based pipeline to identify epithelial cells-enriched lncRNAs, we identified 10 epithelial cells-enriched lncRNAs due to our strict evaluating standard, although the first level of screening by CTSS got 536 long non-coding genes including some known epithelia-associated lncRNAs, such as MALAT1, NEAT1 and TINCR. Therefore, the methodology might have missed a lot of information. However, we believe that these 10 lncRNAs possess the most typical epithelial features among the list of 536 epithelia-enriched lncRNAs screened by CTSS, even though other important lncRNAs could be filtered out. Herein, we chose to focus on epithelial cells-enriched SNHG8 to elucidate its potential biological functions. Indeed, SNHG8 was abundant in typical epithelial cell lines, such as MCF10A, RWPE-1 and HaCaT.
More intriguingly, SNHG8, which is mainly localized in the chromatin of epithelial cells, binds to almost all H1s but not core histone partners H2A and H2B. H1s have been reported to present intrinsically disordered regions and obtain the capacity for multivalency so as to undergo LLPS with many chromatin factors including linker DNA, H2A and HP1α during chromatin organization [32, 56]. Our in vitro and living cell images also showed the interaction between SNHG8 and H1s in way of LLPS. Compared to DNA, SNHG8 manifested stronger ability in phase separating H1s, raising the possibility that SNHG8 might have higher binding ability to H1s than DNA, which was supported by the fact that SNHG8 outcompetes DNA for H1s binding in EMSA. In line, SNHG8 loss increased the amount of H1s binding to chromatin DNA, leading to chromatin condensation and remodeled gene expression. Actually, unlike the core histones, the linker H1 binds to the nucleosome in a dynamic manner to form higher-order chromatin structures, thus stabilizing the nucleosomes and provides the structural and functional flexibility of chromatin [32]. Therefore, although SNHG8 was localized on the chromatin fraction, it could keep H1s out of nucleosome to regulate chromatin flexibility.
Due to the large number of H1 subtypes and the redundancy between them, both the overall and individual biological functions of H1s are poorly understood. It has been reported that single and double knockouts of H1 variants lacked general defects [64], whereas combined knockout of more than two major H1 variants leads to severe phenotypes from mild growth retardation to embryonic lethality in mice [65]. Therefore, functional redundancy among H1 family members and compensation of the lost H1 variants by the remaining H1s are thought to account for the lack of phenotypes in single knockouts of H1s. Interestingly, SNHG8 binds and regulates all H1s. Correspondingly, SNHG8 knockdown leads to chromatin condensation and has a profound effect on gene expression. Therefore, SNHG8 might be a useful tool to study the overall biological effects of H1s.
Finally, although that SNHG8 is associated with epithelial identity, SNHG8 was significantly down-regulated during differentiation of MCF10A and RWPE-1 cells, which present progenitor cell-like characteristics with differentiation potentials [40, 62]. In line, SNHG8 knockdown induced epithelial differentiation-associated gene expression pattern together with closing of cell cycle-associated genes. Also, 3D mammosphere formation assay supported that SNHG8 knockdown accelerated acini maturation of MCF10A cells. These data suggested that epithelial cells with differentiation potentials present higher SNHG8 expression than mature epithelial cells, and its down-regulation contributes to their differentiation, although it remained to be further confirmed in Snhg8-knocked out mice.
Supplementary information
Acknowledgements
We are grateful to Prof. Robert G. Roeder for providing p601 plasmid [66]. We thank Prof. Yongzhong Liu for providing pLKO.1-puro and pLVX-puro plasmids. This work was supported by National Natural Science Foundation (82103233, 82022053, 81972583) and its innovative group support (No. 81721004), National Key R&D Program of China (2020YFA0803403), CAMS Innovation Fund for Medical Sciences (CIFMS) (2019-I2M-5-051), and Shanghai Science and Technology Commission (20JC1410100).
Author contributions
PH and CZ performed most experiments. YJ performed bioinformatics analysis. M-KG, NZ, and SY conducted partial experiments. YY and J-XY provided constructive comments and discussion. S-MS and G-QC designed and supervised the entire project and prepared the manuscript.
Data availability
The RNA- and ATAC-seq data from this publication have been deposited to the GEO database (https://www.ncbi.nlm.nih.gov/geo) and assigned the identifiers GSE151626.
Competing interests
The authors declare no competing interests.
Ethical approval
This study did not include human participants, human data, or human tissue and animal experiments.
Footnotes
Edited by M. Sibilia
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ping He, Cheng Zhang, Yan Ji.
Contributor Information
Shao-Ming Shen, Email: smshen@shsmu.edu.cn.
Guo-Qiang Chen, Email: chengq@shsmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-00944-x.
References
- 1.Delás MJ, Hannon GJ. LncRNAs in development and disease: from functions to mechanisms. Open Biol. 2017;7:170121–30. doi: 10.1098/rsob.170121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saw PE, Xu X, Chen J, Song EW. Non-coding RNAs: the new central Dogma of cancer biology. Sci China Life Sci. 2021;64:22–50. doi: 10.1007/s11427-020-1700-9. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–92. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Li G, Wu Y, Xi J, Kang J. LincRNA1230 inhibits the differentiation of mouse ES cells towards neural progenitors. Sci China Life Sci. 2016;59:443–54. doi: 10.1007/s11427-016-5008-7. [DOI] [PubMed] [Google Scholar]
- 6.Andergassen D, Rinn JL. From genotype to phenotype: genetics of mammalian long non-coding RNAs in vivo. Nat Rev Genet. 2021, 10.1038/s41576-021-00427-8. [DOI] [PubMed]
- 7.Bester AC, Lee JD, Chavez A, Lee Y-R, Nachmani D, Vora S, et al. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173:649–64. doi: 10.1016/j.cell.2018.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:eaah7111. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, et al. Genome-scale deletion screening of human hong non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 2016;34:1279–86. doi: 10.1038/nbt.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Cao Z, Wang Y, Guo Y, Xu P, Yuan P, et al. Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites. Nat Biotechnol. 2018;36:1203–10. doi: 10.1038/nbt.4283. [DOI] [PubMed] [Google Scholar]
- 11.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarropoulos I, Marin R, Cardoso-Moreira M, Kaessmann H. Developmental dynamics of LncRNAs across mammalian organs and species. Nature. 2019;571:510–4. doi: 10.1038/s41586-019-1341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olins DE, Olins AL. Chromatin history: our view from the bridge. Nat Rev Mol Cell Biol. 2003;4:809–14. doi: 10.1038/nrm1225. [DOI] [PubMed] [Google Scholar]
- 14.Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet. 2019;20:207–20. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- 15.Crane-Robinson C. Linker histones: history and current perspectives. Biochim Biophys Acta. 2016;1859:431–5. doi: 10.1016/j.bbagrm.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 16.McBryant SJ, Lu X, Hansen JC. Multifunctionality of the linker histones: an emerging role for protein-protein interactions. Cell Res. 2010;20:519–28. doi: 10.1038/cr.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fyodorov DV, Zhou BR, Skoultchi AI, Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol. 2018;19:192–206. doi: 10.1038/nrm.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li JY, Patterson M, Mikkola HK, Lowry WE, Kurdistani SK. Dynamic distribution of linker histone H1.5 in cellular differentiation. PLoS Genet. 2012;8:e1002879. doi: 10.1371/journal.pgen.1002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Cooke M, Panjwani S, Cao K, Krauth B, Ho PY, et al. Histone H1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 2012;8:e1002691. doi: 10.1371/journal.pgen.1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment aanalysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopezpajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, et al. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell. 2015;32:693–706. doi: 10.1016/j.devcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen SM, Ji Y, Zhang C, Dong SS, Yang S, Xiong Z, et al. Nuclear PTEN safeguards Pre-mRNA splicing to link golgi apparatus for its tumor suppressive role. Nat Commun. 2018;9:2392. doi: 10.1038/s41467-018-04760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 24.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 25.Schafer ZT, Grassian AR, Song L, Jiang Z, Gerharthines Z, Irie HY, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–13. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leppek K, Stoecklin G. An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins. Nucleic Acids Res. 2014;42:e13. doi: 10.1093/nar/gkt956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP) J Vis Exp. 2012;25:3912. doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing YH, Yao RW, Zhang Y, Guo CJ, Jiang S, Xu G, et al. SLERT regulates DDX21 rings associated with Pol I transcription. Cell. 2017;169:664–78. doi: 10.1016/j.cell.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Conrad T, Ørom UA. Cellular fractionation and isolation of chromatin-associated RNA. Methods Mol Biol. 2017;1468:1–9. doi: 10.1007/978-1-4939-4035-6_1. [DOI] [PubMed] [Google Scholar]
- 30.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–16. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.21–21.29.29. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, et al. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179:470–84. doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarntivijai S, Lin Y, Xiang Z, Meehan TF, Diehl AD, Vempati UD, et al. CLO: The cell line ontology. J Biomed Semant. 2014;5:37–46. doi: 10.1186/2041-1480-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–14. doi: 10.1080/15476286.2017.1358347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong P, Xiong Y, Yue J, Hanley SJB, Kobayashi N, Todo Y, et al. Long non-coding RNA NEAT1: a novel target for diagnosis and therapy in human tumors. Front Genet. 2018;9:471. doi: 10.3389/fgene.2018.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–5. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107:15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Profumo V, Forte B, Percio S, Rotundo F, Doldi V, Ferrari E, et al. LEADeR role of miR-205 host gene as long noncoding RNA in prostate basal cell differentiation. Nat Commun. 2019;10:307. doi: 10.1038/s41467-018-08153-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan X, Yan Y, Xue M. Small nucleolar RNA host gene 8: A rising star in the targets for cancer therapy. Biomed Pharmacother. 2021;139:111622. doi: 10.1016/j.biopha.2021.111622. [DOI] [PubMed] [Google Scholar]
- 42.He P, Zhang C, Chen G, Shen S. Loss of lncRNA SNHG8 promotes epithelial-mesenchymal transition by destabilizing CDH1 mRNA. Sci China Life Sci. 2021;64:1858–67. doi: 10.1007/s11427-020-1895-2. [DOI] [PubMed] [Google Scholar]
- 43.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12:84–88. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- 44.McMahon M, Contreras A, Holm M, Uechi T, Forester CM, Pang X, et al. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. Elife. 2019;8:e48847. doi: 10.7554/eLife.48847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerase A, Pintacuda G, Tattermusch A, Avner P. Xist localization and function: new insights from multiple levels. Genome Biol. 2015;16:166. doi: 10.1186/s13059-015-0733-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sridhar B, Rivas-Astroza M, Nguyen TC, Chen W, Yan Z, Cao X, et al. Systematic mapping of RNA-chromatin interactions in vivo. Curr Biol. 2017;27:602–9. doi: 10.1016/j.cub.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garfinkel BP, Melamed-Book N, Anuka E, Bustin M, Orly J. HP1BP3 is a novel histone H1 related protein with essential roles in viability and growth. Nucleic Acids Res. 2015;43:2074–90. doi: 10.1093/nar/gkv089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McFadden EJ, Hargrove AE. Biochemical methods to investigate lncRNA and the influence of lncRNA: protein complexes on chromatin. Biochemistry. 2016;55:1615–30. doi: 10.1021/acs.biochem.5b01141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathews DH, Turner DH, Watson RM. RNA secondary structure prediction. Curr Protoc Nucleic Acid Chem. 2016;67:11. 12. 11–11. 12. 19. doi: 10.1002/cpnc.19. [DOI] [PubMed] [Google Scholar]
- 52.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–98. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Ji X, Li P, Liu C, Lou J, Wang Z, et al. Liquid-liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci China Life Sci. 2020;63:953–85. doi: 10.1007/s11427-020-1702-x. [DOI] [PubMed] [Google Scholar]
- 54.Gibbs EB, Kriwacki RW. Linker histones as liquid-like glue for chromatin. Proc Natl Acad Sci USA. 2018;115:11868–70. doi: 10.1073/pnas.1816936115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urbanek MO, Galka-Marciniak P, Olejniczak M, Krzyzosiak WJ. RNA imaging in living cells - methods and applications. RNA Biol. 2014;11:1083–95. doi: 10.4161/rna.35506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shakya A, Park S, Rana N, King JT. Liquid-liquid phase separation of histone proteins in cells: role in chromatin organization. Biophys J. 2020;118:753–64. doi: 10.1016/j.bpj.2019.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–40. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner AL, Watson M, Wilkins OG, Cato L, Travers A, Thomas JO, et al. Highly disordered histone H1-DNA model complexes and their condensates. Proc Natl Acad Sci USA. 2018;115:11964–9. doi: 10.1073/pnas.1805943115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou W, Mohr L, Maciejowski J, Kranzusch PJ. cGAS phase separation inhibits TREX1-mediated DNA degradation and enhances cytosolic DNA sensing. Mol Cell. 2021;81:739–55. doi: 10.1016/j.molcel.2021.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kishi Y, Fujii Y, Hirabayashi Y, Gotoh Y. HMGA regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nat Neurosci. 2012;15:1127–33. doi: 10.1038/nn.3165. [DOI] [PubMed] [Google Scholar]
- 61.Caldon CE, Sutherland RL, Musgrove E. Cell cycle proteins in epithelial cell differentiation: implications for breast cancer. Cell Cycle. 2010;9:1918–28. doi: 10.4161/cc.9.10.11474. [DOI] [PubMed] [Google Scholar]
- 62.Qu Y, Han B, Yu Y, Yao W, Bose S, Karlan BY, et al. Evaluation of MCF10A as a reliable model for normal human mammary epithelial cells. PLoS One. 2015;10:e0131285. doi: 10.1371/journal.pone.0131285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaiko-Shcherbak A, Fabris G, Dreissen G, Merkel R, Hoffmann B, Noetzel E. The acinar cage: basement membranes determine molecule exchange and mechanical stability of human breast cell acini. Plos One. 2015;10:e0145174. doi: 10.1371/journal.pone.0145174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan YH, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1 degrees replacement subtype. Mol Cell Biol. 2001;21:7933–43. doi: 10.1128/MCB.21.23.7933-7943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan YH, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 66.Shimada M, Chen WY, Nakadai T, Onikubo T, Guermah M, Rhodes D, et al. Gene-Specific H1 Eviction through a Transcriptional Activator→p300→NAP1→H1 Pathway. Mol Cell. 2019;74:268–83. doi: 10.1016/j.molcel.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA- and ATAC-seq data from this publication have been deposited to the GEO database (https://www.ncbi.nlm.nih.gov/geo) and assigned the identifiers GSE151626.