Abstract
Clostridium histolyticum collagenase contains a number of different active components. Previously we have shown that colH encodes a 116-kDa collagenase (ColH) and a 98-kDa gelatinase. We purified a different 116-kDa collagenase (ColG) from the culture supernatant and sequenced its gene (colG). We also identified four other gelatinases (105, 82, 78, and 67 kDa) and determined their N-terminal amino acid sequences, all of which coincided with that of either ColG or ColH. Hybridization experiments showed that each gene is present in a single copy and each gene is transcribed into a single mRNA. These results suggest that all the gelatinases are produced from the respective full-length collagenase by the proteolytic removal of C-terminal fragments. The substrate specificities of the enzymes suggest that colG and colH encode class I and class II enzymes, respectively. Analysis of their DNA locations by pulsed-field gel electrophoresis and nucleotide sequencing of their surrounding regions revealed that the two genes are located in different sites on the chromosome. C. histolyticum colG is more similar to C. perfringens colA than to colH in terms of domain structure. Both colG and colA have a homologous gene, mscL, at their 3′ ends. These results suggest that gene duplication and segment duplication have occurred in an ancestor cell common to C. histolyticum and C. perfringens and that further divergence of the parent gene produced colG and colA.
Clostridium histolyticum, a pathogenic clostridium causing myonecrosis, produces collagenase, a zinc metalloproteinase that degrades various types of collagen and gelatin. This enzyme has a broad substrate specificity and potent collagenolytic activity compared to vertebrate collagenases (38). Furthermore, C. histolyticum can grow well in simple media, producing fairly large amounts of enzyme in the culture medium. Because of these characteristics, this enzyme is widely used for biochemical and physiological studies, e.g., collagen depletion from vertebrate tissues (40, 46), cleavage of collagen linkers (3, 15), and therapeutic purposes (14, 41).
The biochemical and physicochemical properties of this collagenase have been extensively studied (34). Several lines of evidence show that it has multiple forms (7, 23, 26, 27, 33, 38, 48), with molecular masses ranging from 68 to 130 kDa and isoelectric points between pH 5.35 and 6.20 (34). On the basis of the ratio of the activities toward collagen and synthetic peptide substrates, the forms are divided into two classes (6). Amino acid analysis (6), peptide mapping (7), and circular dichroism spectroscopy (7) have revealed that there is extensive similarity between the enzymes within the same class and that there are distinct differences between the two classes in both their primary and secondary structures. This led to the prediction that the two classes are encoded by different genes and that one class evolved from the other by gene duplication followed by divergent evolution (7, 34). For the origin of the multiple enzymes in each class, the following three possibilities were postulated: they are encoded by different genes having extensive similarity, they result from different transcripts from a single gene, or they are produced by proteolytic cleavage of a single precursor protein encoded by a single gene (34).
In a previous study, we have cloned and sequenced the colH gene, encoding a 116-kDa collagenase (ColH), and obtained evidence that a 98-kDa gelatinase is derived from the colH gene (49). Gelatin zymography showed the presence of two additional enzymes (78 and 67 kDa), which are highly gelatinolytic compared to the two ColH enzymes, in our ColH preparation (49). This observation led us to suspect that the two smaller gelatinases are encoded by a gene(s) other than colH. However, only one gene was detected when C. histolyticum DNA was analyzed by Southern hybridization with a colH probe. Furthermore, an 80-kDa recombinant ColH (rColH) protein lacking a C-terminal peptide retained activity toward water-soluble substrates (29). Thus, these gelatinases could be truncated forms of ColH. One direct approach to solving these questions is to identify the enzymes by their primary sequence and to clone and characterize their genes. This study was designed to show the relationships between all the gelatinolytic and collagenolytic enzymes present in the culture supernatant of C. histolyticum. We cloned a novel collagenase gene, determined its nucleotide sequence, and investigated locations of the collagenase genes. Based on these observations, we discuss a likely explanation for the multiplicity of C. histolyticum collagenases and discuss their molecular evolution and the structure-function relationship of these multidomain enzymes (29, 36).
MATERIALS AND METHODS
Bacterial strains, plasmids, and bacteriophage.
C. histolyticum JCM 1403 (ATCC 19401) was obtained from the Institute of Physical and Chemical Research (Saitama, Japan) and used throughout this study. pT7Blue T vector and the host strain, Escherichia coli NovaBlue (Novagen, Madison, Wis.), were used for cloning DNA fragments obtained by PCR. The plasmid vector pUC19 (32) and the host strain E. coli DH5α (4) were used for all other recombinant DNA experiments.
Media and culture conditions.
C. histolyticum was precultured in cooked meat medium (Nissui, Tokyo, Japan) at 37°C for 12 h and grown in Warren and Gray’s medium (44) at 37°C for the preparation of enzymes and total RNA. Transformed E. coli cells were selected on Luria-Bertani plates containing 20 g of LB broth base (Gibco, Paisley, United Kingdom) and 15 g of agar per liter, supplemented with 100 μg of ampicillin per ml, 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Wako Pure Chemical Industries, Ltd., Osaka, Japan), and 0.004% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Wako).
Assay for collagenase.
Collagenase activity was determined with azocoll as described previously (31), 4-phenylazobenzyloxycarbonyl-Pro-Leu-Gly-Pro-d-Arg (PZ-peptide; Sigma Chemical Co., St. Louis, Mo.) as described by Wünsch and Heidrich (47), or insoluble collagen as described previously (19). Protein concentrations were determined by using the bicinchoninic acid protein assay reagent (Pierce, Rockford, Ill.), with bovine serum albumin as a standard. All assays were carried out in triplicate.
Zymography.
Collagen zymography was performed by the method of Birkedal-Hansen and Taylor (5), using acid soluble type I collagen (Sigma) as described previously (31). Zymography with gelatin (Wako) was carried out by the method of Wilson et al. (45). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 7.5% polyacrylamide gel and staining with Coomassie brilliant blue R were performed as described by Laemmli (24).
Purification of the 78-kDa gelatinase and 116-kDa collagenase.
C. histolyticum was grown in 10 ml of cooked meat medium at 37°C for 12 h. The preculture was diluted 100-fold with 500 ml of Warren and Gray’s medium and incubated at 37°C for 16 h. Cells were removed by centrifugation at 12,000 × g for 10 min at 4°C. Ammonium sulfate was added to 80% saturation, and the precipitate was collected by centrifugation at 12,000 × g for 30 min at 4°C. The pellet was dissolved in 3.5 ml of 50 mM Tris-HCl buffer (pH 7.5), and then it (approximately 5 ml) was applied to a Sephacryl S-200 column (2.2 by 70 cm). Proteins were eluted with the same buffer at a flow rate of 20 ml/h. Enzyme activity was monitored by determination of azocoll-hydrolyzing activity. The fractions which were eluted in the first active peak were collected.
For purification of the 78-kDa gelatinase, a one-third volume (approximately 6.7 ml) of the pooled fraction was applied to an ion-exchange fast protein liquid chromatography column (MonoQ; bed volume, 1 ml; Pharmacia LKB Biotechnology, Uppsala, Sweden) which had been preequilibrated with 50 mM Tris-HCl (pH 7.5). Proteins were eluted with a 20-ml linear gradient of 0 to 0.5 M NaCl in 50 mM Tris-HCl (pH 7.5). The 78-kDa enzyme was eluted in a single peak at 192 mM NaCl. This step was repeated three times, and the active fractions were combined.
The 116-kDa ColG collagenase was purified in the same manner by ammonium sulfate precipitation, gel filtration, and ion-exchange chromatography. A one-fourth volume (5 ml) of the pooled collagenase fraction from the gel filtration column was applied to the ion-exchange fast protein liquid chromatography column. The collagenase was eluted in a single peak at 214 mM NaCl. This step was repeated four times, and the active fractions were combined.
Determination of the N-terminal amino acid sequence.
Enzyme fractions were applied to an SDS–7.5% polyacrylamide gel. Electrophoresis were carried out at 100 V for 3.5 h so that the band moved to the middle of the gel. Proteins were electrophoretically transferred to a sheet of polyvinylidene difluoride membrane (Trans-Blot transfer medium; Bio-Rad Laboratories, Hercules, Calif.) and visualized by staining with Coomassie brilliant blue R as described by the supplier. The area corresponding to the band was cut out and subjected to N-terminal amino acid sequence analysis on a protein sequencer (model 492; Perkin-Elmer, Foster City, Calif.).
Amino acid sequence similarity search.
A search for similar protein sequences was carried out by using the Blastp World Wide Web server (2) in the DNA Data Bank of Japan at the Center for Information Biology, National Institute of Genetics (Mishima, Japan). The N-terminal amino acid sequence determined for the 78-kDa ColG gelatinase was used as the query sequence, and this was searched against the nonredundant protein database, including SWISS-PROT, PIR, GenPept, and GenPept updates of the databases. Default parameters of the program were used for this search. A similar search using the amino acid sequences deduced from colG and the open reading frames (ORFs) found around the two collagenase genes was carried out in the same way.
DNA manipulations.
Restriction endonucleases were purchased from Takara Shuzo Co. (Kyoto, Japan), Toyobo (Osaka, Japan), and New England Biolabs (Beverly, Mass.). The DNA ligation kit was a product of Takara Shuzo. All recombinant DNA procedures were carried out as described by Maniatis et al. (28).
Cloning of a portion of colG.
A pair of degenerate oligonucleotide primers were designed to amplify a portion of colG by PCR. Their sequences, 5′-GA(A/G)AA(A/G)TA(C/T)GA(C/T)TT(C/T)GA(A/G)TA-3′ and 5′-TG(A/G)TTCCA(C/T)TT(A/G/T)AT(A/G)TT(C/T)TT-3′, correspond to the N-terminal amino acid sequence (Glu7 to Gln33) of the ColG enzymes. PCR was performed in a 100-μl mixture containing a 1 μM concentration of each primer, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.01% gelatin, 2.5 mM MgCl2, a 0.2 mM concentration of each deoxynucleotide triphosphate, and 2.5 U of Taq DNA polymerase (Takara) by using a thermal cycler (model TC1; Perkin-Elmer). C. histolyticum DNA was prepared as described previously (49) and used as the template. After a denaturation step at 94°C for 5 min, PCR (30 cycles) was carried out as follows: 94°C for 30 s, 45°C for 30 s, and 72°C for 30 s. The PCR product was purified from an acrylamide gel and cloned into pT7Blue T vector (Novagen). The resulting plasmid was designated pCHG1.
Construction of partial genomic libraries and their screening by PCR.
The insert DNA in pCHG1 was amplified by PCR and isolated by PAGE. The fragment was labelled with digoxigenin-11-dUTP (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) by PCR as described by Lanzillo (25) and used as a probe. Southern hybridization was carried out at 60°C. DNA fragments around the positive signal were recovered from an agarose gel and ligated into the HindIII site of pUC19. E. coli DH5α was transformed with the ligation mixture, and colonies were subjected to PCR screening as follows. Fifteen colonies were suspended in one tube containing 20 μl of distilled water, and 1 μl of the mixed suspension was added to 19 μl of the PCR mixture described above. After PCR under the above-described conditions, the products were examined by PAGE. To isolate a positive clone, the 15 clones in a positive group were examined separately by the same PCR screen.
The downstream colG fragments were obtained in the same way. A partial DraI library was screened by PCR with a pair of synthetic oligonucleotides, 5′-TGCCTTGGTATGGAAAAATTGA-3′ and 5′-TTGGCAGATAATGTTTTTCAGC-3′, to clone a middle portion of colG (pCHG3). Another pair of synthetic oligonucleotides, 5′-GGATATTTGGCTAAGGATAA-3′ and 5′-GTGTTTGTAAGAGAAGCAGC-3′, were used to screen a partial EcoRI library to clone a 3′-terminal portion of colG (pCHG5).
Nucleotide sequencing of colG.
The nucleotide sequence was determined by the dideoxy chain termination method (39), using an automated nucleotide sequencer (model ABI PRISM 377; Perkin-Elmer). Plasmid template DNA was prepared with the Wizard plus miniprep DNA purification system (Promega, Madison, Wis.). A Thermo Sequenase fluorescently labelled primer cycle sequencing kit with 7-deaza-dGTP (Amersham International plc, Little Chalfont, Buckinghamshire, England) and M13 dye primers (Perkin-Elmer) was used for sequencing. The ABI PRISM dye terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase, FS (Perkin-Elmer), and various synthetic primers were also used to determine ambiguous nucleotides and to fill in sequence gaps.
Southern hybridization.
Nucleotide fragments corresponding to the N-terminal amino acid sequences of the two collagenases (Ile1 to Tyr40 for the ColG enzymes and Val1 to Tyr39 for the ColH enzymes [49]) were prepared by PCR with two pairs of primers (5′-ATAGCGAATACTAATTCTGA-3′ plus 5′-ATAATTAAATAAACCATTAA-3′ and 5′-GTACAAAATGAAAGTAAGAG-3′ plus 5′-ATACTGAAAAAGGTCTGGTA-3′) and cloned into pT7Blue T vector (Novagen). After their nucleotide sequences were confirmed, they were amplified with the same primers, isolated by PAGE, and labelled with digoxigenin by PCR as described above. C. histolyticum DNA (0.5 μg) was digested with the appropriate restriction enzymes, applied to a 0.8% agarose minigel, and electrophoresed at 100 V for 1 h. Treatment of the gel and transfer of DNA onto a sheet of nylon membrane (Hybond-N; Amersham) were performed as described previously (31). After hybridization at 50°C, the hybridized probes were detected with anti-digoxigenin-alkaline phosphatase Fab fragments (Boehringer) and a chemiluminescent dye (Lumi-Phos 530; Lumigen, Detroit, Mich.).
Northern hybridization.
C. histolyticum cells were grown in Warren and Gray’s medium at 37°C until the culture reached an optical density at 600 nm of 1.8 or 2.7. Total RNA was prepared by the SDS-phenol method (1), and the sample (3 μg) was separated on a denaturing agarose gel (1%). Hybridization was carried out at 50°C with the same probes as described above.
Pulsed-field gel electrophoresis and Southern hybridization.
Genomic DNA of C. histolyticum was prepared and digested by the method of Canard and Cole (9). Endonuclease I-CeuI was purchased from New England Biolabs. After the plugs were digested for 6 h at the appropriate temperature, the fragments were separated by contour-clamped homogeneous electric field electrophoresis with a CHEF-DR III apparatus (Bio-Rad). Electrophoresis was carried out with a 1% agarose gel at 5 V/cm with a ramping pulse from 5 to 100 s for 22 h. Gels were calibrated with Saccharomyces cerevisiae chromosomes (size range, 225 to 1,900 kb) and a mixture of λ concatemers and HindIII fragments (New England Biolabs). To determine the sizes of the larger fragments (above 1 Mb), electrophoresis was carried out under the following conditions: agarose gel, 0.8%; electric field, 3.7 V/cm; ramping pulse, 120 to 500 s; duration, 36 h; and size marker, Hansenula wingei chromosomes (size range, 1.05 to 3.13 Mb; Bio-Rad).
Ribosomal DNA probes were used to show the general arrangement of the C. histolyticum chromosome. Three C. histolyticum rrlB fragments (a 1.6-kb HindIII–I-CeuI fragment containing the upstream rrlB region, a 1.1-kb I-CeuI–HindIII fragment containing the downstream rrlB region, and a 2.7-kb HindIII fragment containing both regions) were isolated by agarose gel electrophoresis, labelled with digoxigenin, and used for hybridization. The rrs and rrf probes were prepared from the clones isolated from Borrelia burgdorferi (12). The colG and colH probes were also used for hybridization.
Nucleotide sequencing of regions adjacent to colG and colH.
The nucleotide sequences of the regions adjacent to colG were determined as described above. The following plasmids, derived from a phage clone, λcol18 (49), were used for nucleotide sequencing of the regions adjacent to colH: pCHC16 carrying a 2.5-kb Sau3AI-SacI fragment, pCHC111 carrying a 1.4-kb SacI-XbaI fragment, pCHC113 carrying a 1.6-kb XbaI fragment, pCHC1145 carrying a 0.5-kb XbaI-HindIII fragment, pCHC1146 carrying a 2.0-kb HindIII fragment, pCHC1147 carrying a 2.3-kb HindIII fragment, and pCHC116 carrying a 2.9-kb PstI-Sau3AI fragment (49). Sequencing templates were prepared by nested deletion of these plasmids, and their nucleotide sequences were determined with an ABI PRISM BigDye terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase, FS (Perkin-Elmer).
Statistical analyses of the deduced amino acid sequence.
The presence and location of signal peptide cleavage sites in possible prepropeptides of ColA and ColG were predicted by using the SignalP server (35) at the Center of Biological Sequence Analysis, Department of Biotechnology, The Technical University of Denmark (Lyngby, Denmark). Sequence alignment was carried out with the Blast server or the Clustal W program (43). The statistical significance of the sequence similarity was examined by a Monte Carlo test by using the Lipman-Pearson alignment algorithm with the Dayhoff similarity scoring (37). The best alignment of two original sequences was compared with a control obtained from the best alignments between 1,000 different randomized sequences by using the rdf2 program at the National Institute of Genetics. Default parameters were used except for the ktup value, which was set at 1.
Nucleotide sequence accession numbers.
The DNA sequences of C. histolyticum colG and colH are available from the GenBank/EMBL/DDBJ databases (accession no. D87215 and AB014075, respectively). The sequences of a part of C. histolyticum 23S rRNA, i.e., rrlA, rrB, and rrlC, are available from the databases (accession no. AB013089, AB013090, and AB013091, respectively).
RESULTS
N-terminal sequence of a 78-kDa gelatinase.
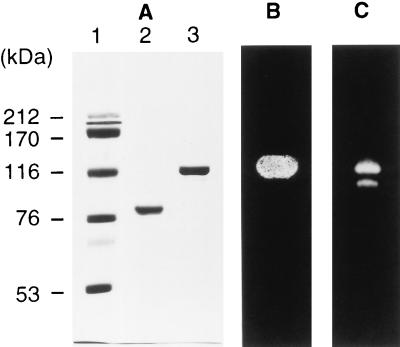
ColH collagenase (49) contained two minor gelatinases besides the 116-kDa ColH collagenase and the 98-kDa ColH gelatinase. One of these gelatinases (78 kDa) was purified to homogeneity (Fig. 1A), and its N-terminal amino acid sequence was determined. The sequence, Ile-Ala-Asn-Thr-Asn-Ser-Glu-Lys-Tyr-Asp-Phe-Glu-Tyr-Leu-Asn-Gly-Leu-Ser-Tyr-Thr-Glu-Leu-Thr-Asn-Leu-Ile-Lys-Asn-Ile-Lys-Trp-Asn-Gln-Ile-Asn-Gly-Leu-Phe-Asn-Tyr, was different from that of the ColH enzymes. A Blastp similarity search showed that it is similar (Poisson possibility value, 0.00018) to the N-terminal sequence of the Clostridium perfringens collagenase (ColA) (31). This enzyme was designated the 78-kDa ColG gelatinase.
FIG. 1.
Analysis of purified ColG enzymes by SDS-PAGE and zymography. (A) SDS-PAGE. Lanes: 1, molecular mass markers (sizes are indicated on the left); 2, 78-kDa ColG gelatinase (2 μg); 3, 116-kDa ColG collagenase (2 μg). (B) Collagen zymography. Five micrograms of a 116-kDa ColG collagenase sample was electrophoresed on an SDS-polyacrylamide gel, followed by zymography with collagen fibrils (type I). (C) Gelatin zymography. Ten nanograms of the same sample as in panel B was electrophoresed on a gelatin-impregnated SDS-polyacrylamide gel. The gel was stained with Coomassie brilliant blue R to detect gelatinolytic activity as an unstained area.
Purification of the C. histolyticum collagenase.
The ColG collagenase was purified by ammonium sulfate precipitation, gel filtration, and ion-exchange chromatography. The azocoll-hydrolyzing activity was eluted in two peaks near the void volume of the size exclusion column. Fractions in the first peak were combined, and the proteins were separated by ion-exchange chromatography. The activity which was eluted at 214 mM NaCl differed from that of ColH, which was eluted at 100 mM NaCl. The enzyme was purified to near homogeneity (Fig. 1A), with a recovery of 0.53 mg of protein from a 500-ml culture. Its apparent molecular mass was estimated to be 116 kDa by SDS-PAGE, and its hydrolytic activity against insoluble collagen fibrils was shown by collagen zymography (Fig. 1B). Gelatin zymography also showed that it possesses gelatinolytic activity (Fig. 1C). The specific activities of the enzyme against insoluble collagen and PZ-peptide were (means ± standard deviations) 826 ± 42 and 26.0 ± 2.3 U/mg of protein, respectively. The sequence of its N-terminal 40 amino acid residues was determined to be identical to that of the 78-kDa gelatinase. This enzyme was designated the 116-kDa ColG collagenase.
N-terminal sequencing of various gelatinases.
To find other enzymes, gelatin zymography was carried out with the enzyme fractions obtained as described previously (49). There were three bands exhibiting gelatinolytic activity besides the ColG and ColH enzymes. These proteins were separated by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane, and their N-terminal amino acid sequences were determined. Two enzymes (82 and 67 kDa) had the same sequence as ColG, while one (105 kDa) had the same sequence as ColH.
Cloning of the colG gene.
To amplify a portion of the colG gene, a pair of degenerate PCR primers were designed based on the N-terminal amino acid sequence of colG. They yielded a single amplification product of the expected size (approximately 80 bp). The fragment was cloned into a T vector, and the nucleotide sequence was determined by using 12 independent clones. The sequences of the 80-bp insert coincided with the N-terminal amino acid sequence of the ColG enzymes except for some variation due to the degeneracy of the primers. One of the plasmids was designated pCHG1. The insert fragment was labelled with digoxigenin and used as a probe for Southern hybridization to detect a single 2.4-kb HindIII fragment (data not shown). A partial genomic library was constructed by using 2.2- to 2.7-kb HindIII fragments, and 600 recombinant colonies were subjected to screening by PCR with the same degenerate primers. Five clones showed a positive signal, and all but one contained the same 2.4-kb HindIII insert. One of these plasmids was chosen and designated pCHG2. Nucleotide sequencing of this plasmid showed that it carries the 5′ portion of colG (nucleotide positions 1 to 2376 [Fig. 2]).
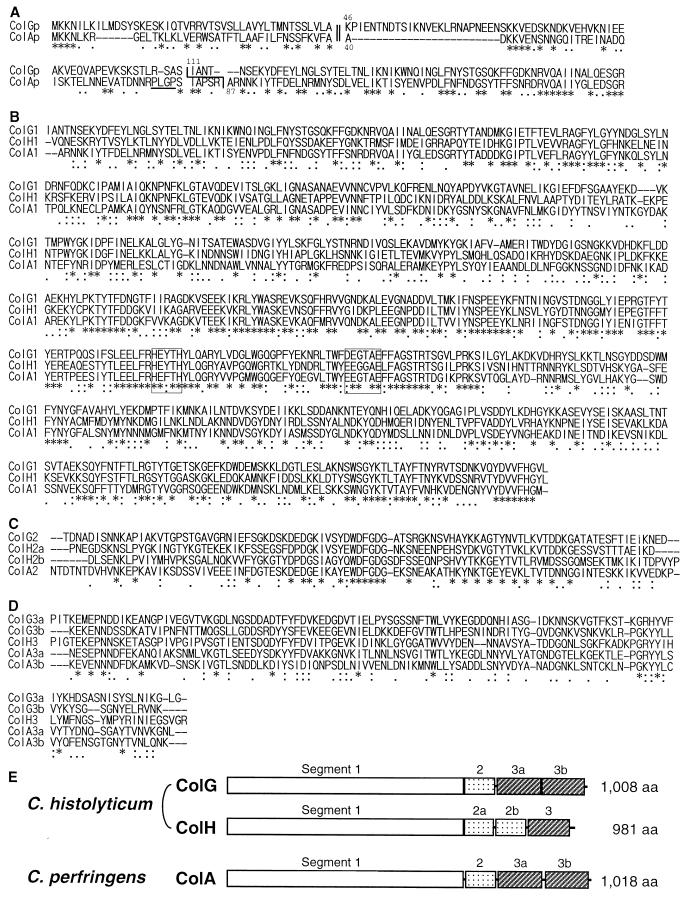
FIG. 2.
Nucleotide sequence of the colG gene and deduced amino acid sequence. The amino acid sequence determined for the N terminus of the 116-kDa ColG collagenase and the 78-kDa gelatinase is indicated by a thick underline. The potential ribosome binding site is indicated by a thin underline. The putative transcriptional terminator sequence is indicated by arrows. Adjacent regions are also shown. Restriction sites are indicated as follows: Dra, DraI; RI, EcoRI; and Hd, HindIII.
A 1.6-kb DraI fragment was detected by another round of Southern hybridization with a probe made from a 0.49-kb EcoRI-HindIII fragment (nucleotide positions 1882 to 2371) (data not shown). A partial genomic library carrying 1.2- to 2.0-kb DraI fragments was constructed and screened by PCR. One positive clone (pCHG3) was obtained from 250 clones; it carries a 1.6-kb DraI fragment (nucleotide positions 1625 to 3203). A fragment of colG further downstream was cloned in the same way by using a partial library carrying 3.0- to 4.0-kb EcoRI fragments. Four positive clones were obtained from 250 clones by PCR screening, all having the same 3.2-kb EcoRI fragment (nucleotide positions 2676 to 5914), designated pCHG5.
Nucleotide sequence of the colG gene.
The nucleotide sequences of the cloned inserts in pCHG2, pCHG3, and pCHG5 were determined by using various subclones constructed from them. The sequences were aligned by their overlaps to form a single contig (Fig. 2). The sequence which coincides with the N-terminal amino acid sequence of the ColG enzymes was found within an ORF starting at nucleotide position 1002 and ending at 4358. This ORF encodes a protein with a possible prepropeptide of 110 amino acid residues which when removed would give a mature protein of 113,897 Da. This is in good agreement with the molecular mass determined by SDS-PAGE (116 kDa) for the purified ColG collagenase. There exist three possible ATG initiation codons beginning at nucleotide positions 1002, 1029, and 1110. Since only the first ATG codon is preceded by a possible ribosome binding sequence (AGGGGG) with a 7-bp gap, this could be the translational initiation site. A stem-loop sequence (nucleotide positions 4370 to 4405) with a short run of T’s is present downstream of the termination codon; this could be a transcription terminator.
Deduced amino acid sequence of ColG.
A database search with the Blastp server revealed that the deduced amino acid sequence of mature ColG aligns well with those of C. histolyticum ColH and C. perfringens ColA. The latter showed higher similarity (Poisson P value, 8.0e–249) than the former (Poisson P value, 1.8e–215). When the possible prepropeptide of ColG (110 amino acid residues) was included in the query sequence, the P value became 1.2e–255 due to the significant alignment of the relevant sequences (Fig. 3A).
FIG. 3.
Amino acid sequence alignment and domain structure of clostridial collagenases. (A) The amino acid sequences of the putative prepropeptides were aligned with the Blast2 server. The amino acid positions are numbered from the putative translation initiation site. The predicted signal cleavage sites are indicated by a double line. The observed N-terminal sequences are indicated by a single line. The PLGP sequence is underlined. (B to D) The amino acid sequences of the mature collagenases are divided into three segments, and each segment was aligned separately with the Clustal W program. (B) Segment 1; (C) segment 2; (D) segment 3. The duplicated segments are indicated by the letters a and b. The sequences forming the catalytic center are boxed. Identical and similar residues among the three sequences are indicated by asterisks and colons or dots, respectively. (E) Schematic representation of the segmental structure of clostridial collagenases. aa, amino acids.
The signal peptides of ColA and ColG were predicted by the SignalP server to be a peptide extending from the N terminus to Ala39 (maximum C score, 0.734 at Ala40) and a peptide extending from the N terminus to Ala45 (maximum C score, 0.371 at Lys46), respectively. Although the score of the latter was low compared to that of the former, these predicted sites aligned well (Fig. 3A). This reinforces the proposed translation initiation site and the presence of a long prepropeptide in the ColG precursor. In a previous study we suggested that the PLGP sequence located near the C terminus of the ColA prepropeptide would play a role in postsignal cleavage maturation (31). It might be confined to ColA if it does, since such a sequence is not present in ColG.
A segmental structure of the mature enzyme was observed in the amino acid sequence deduced from colG, and segment 3 is duplicated (Fig. 3B to E). ColG is different from ColH and similar to C. perfringens ColA in its domain structure. The statistical significance of the sequence similarity was confirmed by a Monte Carlo test (Table 1), as the alignment between any pair of the sequences gave a score that was at least 14.9 standard deviations higher than the control. All of the clostridial enzymes showed significant similarity to Vibrio alginolyticus collagenase (42) in segments 1 and 2, but not in segment 3 (alignment data not shown). A consensus motif for metalloproteinases, HEXXH, was conserved in segment 1. We proposed that the sequence EEXXXE, which is located at the C-terminal side of the HEXXH sequence with a gap of 26 amino acid residues (20), participates in forming the catalytic center of ColH. This second motif is also conserved in ColG, although the first glutamate residue is replaced with an aspartate residue. Each of the segment 3s of ColG produced in E. coli bound to insoluble collagen (unpublished data), suggesting that the region forms a collagen binding domain in ColG like that of ColH (29).
TABLE 1.
Similarities among segments of collagenasesa
| Segment | Degree of sequence similarity with:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ColH1 | ColA1 | Val1 | ColH2a | ColH2b | ColA2 | Val2 | ColG3b | ColH3 | ColA3a | ColA3b | Val3 | |
| ColG1 | +163.1 | +160.1 | +18.3 | |||||||||
| ColH1 | +150.8 | +9.4 | ||||||||||
| ColA1 | +10.0 | |||||||||||
| ColG2 | +23.5 | +24.6 | +23.5 | +21.2 | ||||||||
| ColH2a | +23.0 | +17.1 | +21.7 | |||||||||
| ColH2b | +20.7 | +18.2 | ||||||||||
| ColA2 | +21.0 | |||||||||||
| ColG3a | +17.0 | +24.1 | +23.9 | +14.9 | +0.37 | |||||||
| ColG3b | +20.4 | +24.5 | +26.4 | −1.37 | ||||||||
| ColH3 | +22.8 | +18.0 | +1.67 | |||||||||
| ColA2a | +29.9 | +0.44 | ||||||||||
| ColA2b | +0.89 | |||||||||||
The degree of sequence similarity is expressed as the standard deviation of the alignment score obtained for an original pair of sequences against the scores obtained for 1,000 randomized pairs. Segment designations are composed of the enzyme designation plus the segment number; for example, ColH1 is segment 1 of ColH. Val1, Val2, and Val3 are segments 1, 2, and 3 of the V. alginolyticus collagenase, respectively (31, 42, 49).
Southern hybridization.
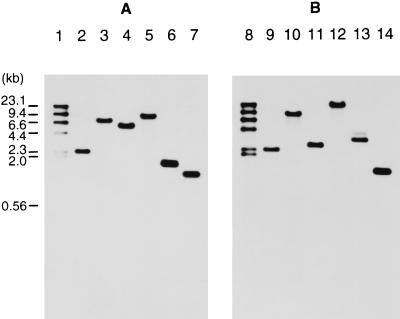
C. histolyticum DNA was digested with HindIII, XbaI, EcoRI, PstI, Sau3AI, or SspI and separated on a 0.8% agarose gel. colG- and colH-specific probes were prepared by PCR. Only one band was detected for a given digestion in each hybridization profile (Fig. 4). The sizes of the positive bands were in good agreement with the size of the fragment predicted from the nucleotide sequence (Fig. 4A). The differences between the two hybridization profiles were clearly observed for the EcoRI, PstI, and Sau3AI digests.
FIG. 4.
Southern hybridization with colG-specific (A) and colH-specific (B) probes (corresponding to the N termini). Lanes: 1 and 8, size markers (sizes are indicated on the left [in kilobase pairs] [kb]); 2 and 9, HindIII digests; 3 and 10, XbaI digests; 4 and 11, EcoRI digests; 5 and 12, PstI digests; 6 and 13, Sau3AI digests; 7 and 14, SspI digests.
Northern hybridization with the colG and colH gene probes.
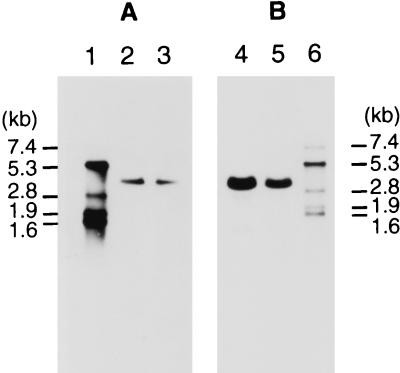
Total RNA from C. histolyticum was prepared in late logarithmic growth phase, when production of the collagenases continued at a high level. The RNAs were separated on a denaturing agarose gel and subjected to Northern hybridization with the colG and colH probes. Only one band was detected in each hybridization experiment (Fig. 5), and the sizes of the colG and colH transcripts were estimated to be 3.7 and 3.25 kb, respectively.
FIG. 5.
Northern hybridization with colG-specific (A) and colH-specific (B) probes (corresponding to the N termini). Lanes: 1 and 6, RNA markers (sizes are indicated on the left and right); 2 and 4, total RNA isolated at an optical density at 600 nm of 1.8 (3 μg); 3 and 5, total RNA isolated at an optical density at 600 nm of 2.7 (3 μg).
Loci of the collagenase genes.
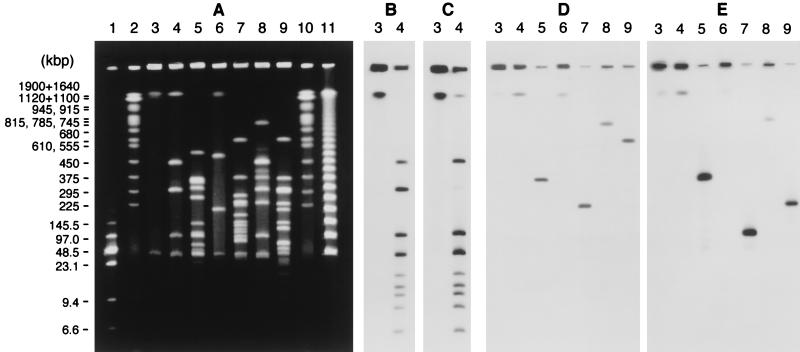
Genomic DNA of C. histolyticum was analyzed by pulsed-field gel electrophoresis (Fig. 6A). A 45-kb band was observed in the undigested sample. Endonuclease I-CeuI cleaved the chromosome DNA into 10 fragments (fragment A, 2,030 kb; B, 430 kb; C, 280 kb; D, 100 kb; E, 42 kb; F, 15 kb; G, 11 kb; H, 9.8 kb; I, 8.3 kb, and J, 6.2 kb). The size of fragment A was determined by electrophoresis as described in Materials and Methods. I-CeuI fragment E (42 kb) was broader than the other bands in the digest, indicating that it overlapped the 45-kb band in the undigested DNA. The C. histolyticum rrl probe hybridized to all the I-CeuI fragments (data not shown). The upstream and downstream rrl probes hybridized to all the I-CeuI fragments except A and C, respectively (Fig. 6B and C). Similar results were obtained with the rrs and rrf probes (data not shown). The rrl probes did not hybridize to the 45-kb band in the undigested sample. Each of the collagenase gene probes hybridized to a single fragment in a given digest (Fig. 6D and E). Although the 2.03-Mb I-CeuI fragment, the 2.23-Mb MluI fragment, and the 760-kb SacII fragment showed positive signals with both probes, different NruI and SmaI fragments were detected by the two probes.
FIG. 6.
(A) Plugs were subjected to endonuclease digestion, and the DNA fragments were separated by pulsed-field gel electrophoresis. Lanes: 1, size markers consisting of a mixture of λ concatemers and HindIII fragments (size range, 6.6 to 145.5 kb) (sizes are indicated on the left); 2 and 10, size markers consisting of Saccharomyces cerevisiae chromosomes (size range, 225 to 1,900 kb); 3, undigested DNA; 4, I-CeuI digest; 5, ApaI digest; 6, MluI digest; 7, NruI digest; 8, SacII digest; 9, SmaI digest; 11, size markers consisting of λ concatemers. (B through E) Southern hybridization with an upstream rrlB probe (B), a downstream rrlB probe (C), a colG probe (D), and a colH probe (E).
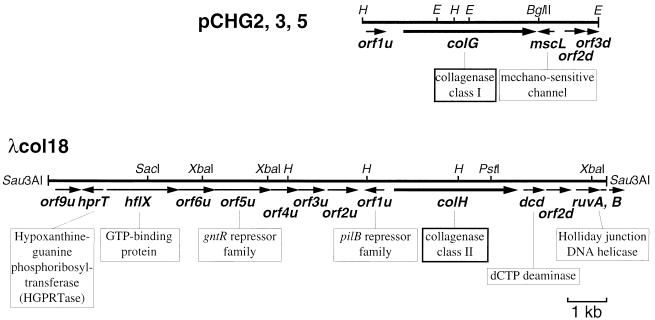
Nucleotide sequences of the adjacent regions of the two collagenase genes.
In order to see if the two collagenase genes were adjacent, the nucleotide sequences adjacent to colG and colH were determined (5.9 kb for colG and 14.0 kb for colH [Fig. 7]). The two fragments did not form a single contig. Around colG, one ORF (orf1uG) and three ORFs (mscL, orf2dG, and orf3dG) were found in the upstream (∼1.0 kb) and downstream (∼1.6 kb) regions, respectively. Around colH, nine ORFs (orf9uH, hprT, hflX, orf6uH, orf5uH, orf4uH, orf3uH, orf2uH, and orf1uH) and four ORFs (dcd, orf2dH, ruvA, and ruvB) were found in the upstream (∼8.7 kb) and downstream (∼2.3 kb) regions, respectively. Results of Blastp similarity searches with the amino acid sequences deduced from these ORFs as the query sequences are shown in Table 2.
FIG. 7.
Genetic organization of the DNA fragments carrying colG and colH. The amino acid sequence deduced from the ORF was significantly similar to that of the protein shown below the respective arrow. Some restriction sites are also shown. H, HindIII; E, EcoRI.
TABLE 2.
Proteins deduced from the ORFs around the two collagenase genes
| ORF | Size (aaa) of deduced protein | Blast, P value | Accession no. | Description of protein similar to the deduced product |
|---|---|---|---|---|
| orf1uG | 164 | None (no sequences with significant similarity) | ||
| mscL | 133 | 8.9e–52 | E70065 | Large conductance-mechanosensitive channel from B. subtilis |
| orf2dG | 180 | 6.4e–61 | B65002 | Hypothetical protein b2300 from E. coli |
| orf3dG | 58 | 9.5e–15 | G69538 | Conserved hypothetical protein from Archaeglobus fulgidus |
| orf9uH | 215 | 1.5e–17 | E70015 | Hypothetical protein YunB from B. subtilis |
| hprT | 175 | 2.4e–53 | S30100 | Hypoxanthine phosphoribosyltransferase from Lactococcus lactis |
| hflX | 596 | 3.5e–105 | S75472 | GTP binding protein from a Synechocystis sp. |
| orf6uH | 298 | None (no sequences with significant similarity) | ||
| orf5uH | 466 | 2.8e–44 | I40492 | Hypothetical protein 8 (srfA operon) from B. subtilis |
| orf4uH | 224 | 5.8e–6 | S75919 | Hypothetical protein from a Synechocystis sp. |
| orf3uH | 216 | 4.7e–54 | A70049 | Conserved hypothetical protein YvyE from B. subtilis |
| orf2uH | 246 | 2.1e–90 | B69972 | Spore coat protein homolog YrbC from B. subtilis |
| orf1uH | 159 | 5.7e–63 | E64249 | Pilin repressor PilB homolog from Mycoplasma genitalium |
| dcd | 172 | 3.4e–15 | F64353 | dCTP deaminase from Methanococcus jannaschii |
| orf2dH | 204 | None (no sequences with significant similarity) | ||
| ruvA | 197 | 5.7e–44 | E69702 | Holliday junction DNA helicase RuvA from B. subtilis |
| ruvB | >54 | (5.3e–11) | C64652 | Holliday junction DNA helicase from Helicobacter pylori |
aa, amino acids.
DISCUSSION
The similarity between the N-terminal sequences of the 78-kDa gelatinase and the C. perfringens ColA collagenase (31) suggested the presence of an additional collagenase besides ColH (49) in C. histolyticum. We purified the 116-kDa collagenase (ColG) from the culture supernatant. The molecular mass of ColG is close to that of ColH. This explains why we detected only a single collagenolytic band in the culture supernatant by collagen zymography. The present study showed that C. histolyticum possesses the two distinct collagenase genes. We determined the N-terminal sequences of five enzymes in this study in addition to the two ColH enzymes already determined (49). All the N-terminal sequences of the seven enzymes correspond to either colG or colH. The results of Southern hybridization showed that both colG and colH are single-copy genes. As shown by northern hybridization, each gene is transcribed into a single message, eliminating the possibility that the smaller enzymes are produced by premature transcription termination. The size of each transcript indicated that each gene is monocistronic. Thus, it is likely that the smaller enzymes are produced from their respective precursor enzymes by proteolytic cleavage at the C terminus, leading to the multiplicity in each class.
Bond and Van Wart isolated six enzymes from a commercial collagenase preparation and divided them into two classes based on substrate specificity and amino acid analysis. Class I enzymes (α, β, and γ) are less active against various peptide substrates, including the PZ-peptide, than class II enzymes (δ, ɛ, and ζ) (6). In our study the PZ-peptidase activity of ColG was 2.96 ± 0.26 mU/pmol of protein, much lower than that of rColH (85.9 ± 3.6 mU/pmol of protein). On the other hand, their activities against insoluble collagen are similar; the activities of ColG and ColH are 94.1 ± 4.8 and 164 ± 6 mU/pmol of protein, respectively (19). The 78-kDa ColG gelatinase also showed low hydrolytic activity against the PZ-peptide (1.32 ± 0.10 mU/pmol of protein). Previously, we showed that two C-terminally truncated forms of rColH (rColH′; 87 and 80 kDa) possess high PZ-peptidase activities (62.9 ± 4.8 and 123 ± 6 mU/pmol of protein, respectively), similar to that of full-length ColH (29). The activity on the peptide substrates regardless of C-terminal truncation can be explained by the presence of the catalytic domains at the N termini of ColG and ColH, which determine their specificities on the water-soluble substrates. The amino acid compositions predicted from colG and colH were compared with the amino acid analysis data for the largest enzymes in each class, β (class I, 115 kDa) and ζ (class II, 125 kDa) (6). The combinations of ColG plus β and ColH plus ζ showed higher correlation values (r2 = 0.978 and 0.963) than the alternative combination (r2 = 0.937 and 0.922). Based on these observations, it is highly possible that β (class I) and ζ (class II) enzymes are encoded by colG and colH, respectively. We can assume that the smaller species of enzymes described by Bond and Van Wart (6) are also the C-terminal truncates of the respective full-length enzyme: α (68 kDa) and γ (79 kDa) from β and δ (100 kDa) and ɛ (110 kDa) from ζ.
We analyzed the loci of the two collagenase genes by pulsed-field gel electrophoresis. An intron-encoded endonuclease, I-CeuI, specifically cleaves DNA at a 26-bp recognition site present in the 23S rRNA genes of many species. We cloned three rrl fragments (rrlA, rrlB, and rrlC) from C. histolyticum to confirm the presence of an I-CeuI site in each gene (unpublished data). The number and sizes of the I-CeuI fragments of the C. histolyticum chromosome were similar to those from C. perfringens (fragment A, 2,280 kb; B, 400 kb; C, 250 kb; D, 250 kb; E, 150 kb; F, 95 kb; G, 60 kb; H, 25 kb; I, 9.5 kb, and J, 9.5 kb) (22). The total genome size of C. histolyticum is estimated to be 2.9 Mb from their sizes, which coincides well with the value calculated from the sizes of the MluI fragments. Hybridization with the rrl, rrs, and rrf probes indicated the presence of 10 copies of the rrn operon on the C. histolyticum chromosome, which is similar to findings for many gram-positive species (11). The results suggest that fragment C (280 kb) has two copies of the 5′ region of the rrn operon outbound at both ends and that fragment A (2,030 kb) has two copies of the 3′ region of the rrn operons inbound at both ends. All the other fragments have a 3′ region of the rrn operon at one end and a 5′ region at the other end. The number and orientation of rrn operons showed that the organization of the C. histolyticum chromosome is similar to that of the C. perfringens chromosome (13).
The presence of a plasmid (approximately 45 kb) was revealed by the comparison of the electrophoretic patterns of uncut and I-CeuI-cut DNA and the rrl hybridization experiment. In C. perfringens the collagenase (κ toxin) gene (colA) is chromosomal, while some of the other toxin genes (ɛ, ι, β, and λ) are plasmid borne (21). The enterotoxin gene (cpe) is either chromosomal or plasmid borne depending on the strain (10). However, Southern hybridization with the colG and colH probes showed that neither gene is plasmid borne. The locus of the two genes on the C. histolyticum chromosome differs from that of colA on the C. perfringens chromosome. The former is located on I-CeuI fragment A (2,030 kb), while the latter is on I-CeuI fragment E (150 kb) (22). The distance between colG and colH genes in C. histolyticum is less than 760 kb, as shown by the SacII digest, but they are expected to be well separated, since there is at least one NruI site and one SmaI site between them. The nucleotide sequences of the regions adjacent to the two collagenase genes revealed that the genes are not tandemly located and that they are separated from each other by at least 3.3 kb.
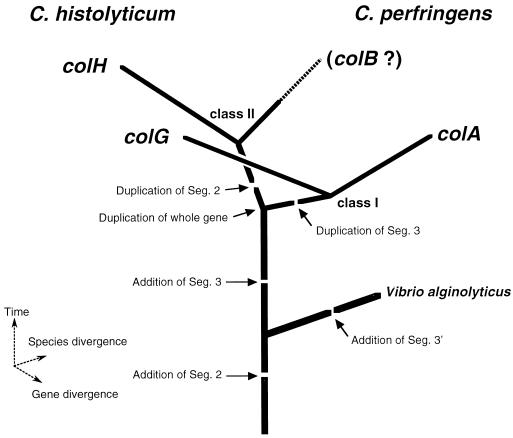
In pathogenic clostridia some of the toxin genes are located on lysogenized phages or transposable elements (8, 16, 17). However, we could not find any such genes around the collagenase genes by a Blastn similarity search. Downstream of colH are two ORFs which encode peptides homologous to a Holliday junction helicase, an enzyme essential for general recombination. The evolution of the clostridial collagenase genes can be explained by gene duplication and divergence as follows: (i) in an ancestral cell common to C. histolyticum and C. perfringens, an ancestral collagenase gene carrying a single copy of each segment (segments 1, 2, and 3) was duplicated; (ii) segments 3 and 2 were duplicated in each descendant to diverge into the subsequent ancestral genes encoding class I (segments 1, 2, 3a, and 3b) and class II (segments 1, 2a, 2b, and 3) enzymes, respectively; and (iii) species divergence occurred by an unknown mechanism and mutations were accumulated in each copy. colG and colA are considered to have derived from the former ancestor gene by divergence. This speculation is supported by the following observations. First, the segmental structure of C. histolyticum ColG is the same as that of C. perfringens ColA, and their sequences align with a significantly high similarity (Blastp P value, 1.2e–255). Second, both genes have a region encoding a long prepropeptide (110 amino acid residues for ColG and 86 residues for ColA), unlike colH. Finally, these genes are accompanied by the homologous mscL gene, encoding the mechanosensitive channel homolog (Blastp P value for similarity to C. perfringens MscL [29], 7.7e–44). We are now searching for the counterpart of colH in C. perfringens, tentatively named colB, although it might have been lost during evolution. Alternatively, the class divergence could have occurred after the species divergence. If this is the case, ColG should be more closely related to ColH than to ColA. In order to evaluate this hypothesis, the similarity between their segment 1s was examined with the Blastp server, so the effects of their different C-terminal structures could be ignored. Since ColG1 showed higher similarity to ColA1 (P value, 1.4e–182) than to ColH1 (P value, 3.6e–175), this hypothesis seems less likely. The International Polycystic Kidney Disease Consortium suggested that a segment 2-encoding fragment has been horizontally transferred from eukaryotic cells to prokaryotic cells (18). If this is the case, the event should have occurred before these genes diverged. Although the V. alginolyticus collagenase (42) has segments 1 and 2 that are significantly similar to those of the clostridial collagenases, its C-terminal segment 3 shows no significant similarity to that of the clostridial enzymes. This suggests that the addition of segment 2 had taken place before these genera diverged and that a different C-terminal segment was added to each. On the basis of this discussion, we propose a hypothesis for the evolution of the bacterial collagenases, as depicted schematically (Fig. 8). More systematic investigation of bacterial collagenase genes is necessary to draw a conclusive evolutionary picture.
FIG. 8.
Schematic representation of a plausible evolutionary pathway of the bacterial collagenase genes. Seg., segment.
ACKNOWLEDGMENTS
We thank David B. Wilson (Section of Biochemistry, Molecular and Cell Biology, Cornell University, Ithaca, N.Y.) for invaluable discussions and assistance in preparing the manuscript.
This work was partly supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture in Japan.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Becker A, Theuring F, Gottwald M, Kauser K, Schleuning W-D, Donner P. Purification of human big endothelin 1 derived through cleavage with collagenase and dipeptidylpeptidase IV from a fusion protein expressed in Escherichia coli. Protein Expr Purif. 1994;5:50–56. doi: 10.1006/prep.1994.1007. [DOI] [PubMed] [Google Scholar]
- 4.Bethesda Research Laboratories. BRL pUC host: E. coli DH5α™ competent cells. Focus. 1986;8:9. [Google Scholar]
- 5.Birkedal-Hansen H, Taylor R E. Detergent-activation of latent collagenase and resolution of its component molecules. Biochem Biophys Res Commun. 1982;107:1173–1178. doi: 10.1016/s0006-291x(82)80120-4. [DOI] [PubMed] [Google Scholar]
- 6.Bond M D, Van Wart H E. Characterization of the individual collagenases from Clostridium histolyticum. Biochemistry. 1984;23:3085–3091. doi: 10.1021/bi00308a036. [DOI] [PubMed] [Google Scholar]
- 7.Bond M D, Van Wart H E. Relationship between the individual collagenases of Clostridium histolyticum: evidence for evolution by gene duplication. Biochemistry. 1984;23:3092–3099. doi: 10.1021/bi00308a037. [DOI] [PubMed] [Google Scholar]
- 8.Brynestad S, Synstad B, Granum P E. The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning strains. Microbiology. 1997;143:2109–2115. doi: 10.1099/00221287-143-7-2109. [DOI] [PubMed] [Google Scholar]
- 9.Canard B, Cole S T. Genome organization of the anaerobic pathogen Clostridium perfringens. Proc Natl Acad Sci USA. 1989;86:6676–6680. doi: 10.1073/pnas.86.17.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornillot E, Saint-Joanis B, Daube G, Katayama S, Granum P E, Canard B, Cole S T. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol Microbiol. 1995;15:639–647. doi: 10.1111/j.1365-2958.1995.tb02373.x. [DOI] [PubMed] [Google Scholar]
- 11.de Bruijn F J, Lupski J R, Weinstock G M. Bacterial genomes: physical structure and analysis. New York, N.Y: Chapman & Hall; 1998. [Google Scholar]
- 12.Fukunaga M, Yanagihara Y, Sohnaka M. The 23S/5S ribosomal RNA genes (rrl/rrf) are separate from the 16S ribosomal RNA gene (rrs) in Borrelia burgdorferi, the aetiological agent of Lyme disease. J Gen Microbiol. 1992;138:871–877. doi: 10.1099/00221287-138-5-871. [DOI] [PubMed] [Google Scholar]
- 13.Garnier T, Canard B, Cole S T. Cloning, mapping, and molecular characterization of the rRNA operons of Clostridium perfringens. J Bacteriol. 1991;173:5431–5438. doi: 10.1128/jb.173.17.5431-5438.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelbard M K, James K, Riach P, Dorey F. Collagenase versus placebo in the treatment of Peyronie’s disease: a double-blind study. J Urol. 1993;149:56–58. doi: 10.1016/s0022-5347(17)35998-0. [DOI] [PubMed] [Google Scholar]
- 15.Hanada K, Yamato I, Anraku Y. Purification and reconstitution of Escherichia coli proline carrier using a site specifically cleavable fusion protein. J Biol Chem. 1988;263:7181–7185. [PubMed] [Google Scholar]
- 16.Inoue K, Iida H. Conversion of toxigenicity in Clostridium botulinum type C. Jpn J Microbiol. 1970;14:87–89. doi: 10.1111/j.1348-0421.1970.tb00495.x. [DOI] [PubMed] [Google Scholar]
- 17.Inoue K, Iida H. Phage-conversion of toxigenicity in Clostridium botulinum types C and D. Jpn J Med Sci Biol. 1971;24:53–56. [PubMed] [Google Scholar]
- 18.The International Polycystic Kidney Disease Consortium. Polycystic kidney disease: the complete structure of the PKD1 gene and its protein. Cell. 1995;81:289–298. doi: 10.1016/0092-8674(95)90339-9. [DOI] [PubMed] [Google Scholar]
- 19.Jung C-M, Matsushita O, Katayama S, Minami J, Ohhira I, Okabe A. Expression of the colH gene encoding Clostridium histolyticum collagenase in Bacillus subtilis and its application to enzyme purification. Microbiol Immunol. 1996;40:923–929. doi: 10.1111/j.1348-0421.1996.tb01161.x. [DOI] [PubMed] [Google Scholar]
- 20.Jung, C.-M., O. Matsushita, S. Katayama, J. Minami, J. Sakurai, and A. Okabe. Analysis of the catalytic center in the Clostridium histolyticum ColH collagenase. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 21.Katayama S, Dupuy B, Daube G, China B, Cole S T. Genome mapping of Clostridium perfringens strains with I-CeuI shows many virulence genes to be plasmid-borne. Mol Gen Genet. 1996;251:720–726. doi: 10.1007/BF02174122. [DOI] [PubMed] [Google Scholar]
- 22.Katayama S-I, Dupuy B, Garnier T, Cole S T. Rapid expansion of the physical and genetic map of the chromosome of Clostridium perfringens CPN50. J Bacteriol. 1995;177:5680–5685. doi: 10.1128/jb.177.19.5680-5685.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kono T. Purification and partial characterization of collagenolytic enzymes from Clostridium histolyticum. Biochemistry. 1968;7:1106–1114. doi: 10.1021/bi00843a031. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lanzillo J J. Chemiluminescent nucleic acid detection with digoxigenin-labeled probes: a model system with probes for angiotensin converting enzyme which detect less than one attomole of target DNA. Anal Biochem. 1991;194:45–53. doi: 10.1016/0003-2697(91)90149-n. [DOI] [PubMed] [Google Scholar]
- 26.Lwebuga-Mukasa J S, Harper E, Taylor P. Collagenase enzymes from Clostridium: characterization of individual enzymes. Biochemistry. 1976;15:4736–4741. doi: 10.1021/bi00666a031. [DOI] [PubMed] [Google Scholar]
- 27.Mandl I, Keller S, Manahan J. Multiplicity of Clostridium histolyticum collagenase. Biochemistry. 1964;3:1737–1741. doi: 10.1021/bi00899a026. [DOI] [PubMed] [Google Scholar]
- 28.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 29.Matsushita O, Jung C-M, Minami J, Katayama S, Nishi N, Okabe A. A study of the collagen-binding domain of a 116-kDa Clostridium histolyticum collagenase. J Biol Chem. 1998;273:3643–3648. doi: 10.1074/jbc.273.6.3643. [DOI] [PubMed] [Google Scholar]
- 30.Matsushita O, Jung C M, Okabe A. Identification of the gene encoding a mechanosensitive channel MscL homologue in Clostridium perfringens. Gene. 1995;165:147–148. doi: 10.1016/0378-1119(95)00490-w. [DOI] [PubMed] [Google Scholar]
- 31.Matsushita O, Yoshihara K, Katayama S-I, Minami J, Okabe A. Purification and characterization of a Clostridium perfringens 120-kilodalton collagenase and nucleotide sequence of the corresponding gene. J Bacteriol. 1994;176:149–156. doi: 10.1128/jb.176.1.149-156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 33.Miyoshi M, Rosenbloom J. General proteolytic activity of highly purified preparations of clostridial collagenase. Connect Tissue Res. 1974;2:77–84. doi: 10.3109/03008207409152091. [DOI] [PubMed] [Google Scholar]
- 34.Mookhtiar K A, Van Wart H E. Clostridium histolyticum collagenases: a new look at some old enzymes. Matrix Suppl. 1992;1:116–126. [PubMed] [Google Scholar]
- 35.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Nishi N, Matsushita O, Yuube K, Miyanaka H, Okabe A, Wada F. Collagen-binding growth factors: production and characterization of functional fusion proteins having a collagen-binding domain. Proc Natl Acad Sci USA. 1998;95:7018–7023. doi: 10.1073/pnas.95.12.7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterkofsky B. Bacterial collagenase. Methods Enzymol. 1982;82:453–471. [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seglen P O. Preparation of isolated rat liver cells. Methods Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- 41.Starkweather K D, Lattuga S, Hurst L C, Badalamente M A, Guilak F, Sampson S P, Dowd A, Wisch D. Collagenase in the treatment of Dupuytren’s disease: an in vitro study. J Hand Surg. 1996;21:490–495. doi: 10.1016/S0363-5023(96)80368-6. [DOI] [PubMed] [Google Scholar]
- 42.Takeuchi H, Shibano Y, Morihara K, Fukushima J, Inami S, Keil B, Gilles A-M, Kawamoto S, Okuda K. Structural gene and complete amino acid sequence of Vibrio alginolyticus collagenase. Biochem J. 1992;281:703–708. doi: 10.1042/bj2810703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warren G H, Gray J. A simplified culture medium for the production of collagenase. Nature (London) 1961;192:755–756. doi: 10.1038/192755a0. [DOI] [PubMed] [Google Scholar]
- 45.Wilson M J, Strasser M, Vogel M M, Sinha A A. Calcium-dependent and calcium-independent gelatinolytic proteinase activities of the rat ventral prostate and its secretion: characterization and effect of castration and testosterone treatment. Biol Reprod. 1991;44:776–785. doi: 10.1095/biolreprod44.5.776. [DOI] [PubMed] [Google Scholar]
- 46.Wolters G H J, Vos-Scheperkeuter G H, Lin H-C, Van Schilfgaarde R. Different roles of class I and class II Clostridium histolyticum collagenase in rat pancreatic islet isolation. Diabetes. 1995;44:227–233. doi: 10.2337/diab.44.2.227. [DOI] [PubMed] [Google Scholar]
- 47.Wünsch E, Heidrich H-G. Zur quantitativen Bestimmung der Kollagenase. Hoppe-Seyler’s Z Physiol Chem. 1963;333:149–151. doi: 10.1515/bchm2.1963.333.1.149. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida E, Noda H. Isolation and characterization of collagenases I and II from Clostridium histolyticum. Biochim Biophys Acta. 1965;105:562–574. doi: 10.1016/s0926-6593(65)80239-9. [DOI] [PubMed] [Google Scholar]
- 49.Yoshihara K, Matsushita O, Minami J, Okabe A. Cloning and nucleotide sequence analysis of the colH gene from Clostridium histolyticum encoding a collagenase and a gelatinase. J Bacteriol. 1994;176:6489–6496. doi: 10.1128/jb.176.21.6489-6496.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]