Abstract
Antibodies against epitopes in S1 give the most accurate CoP against infection by the SARS‐CoV‐2 coronavirus. Measurement of those antibodies by neutralization or binding assays both have predictive value, with binding antibody titers giving the highest statistical correlation. However, the protective functions of antibodies are multiple. Antibodies with multiple functions other than neutralization influence efficacy. The role of cellular responses can be discerned with respect to CD4+ T cells and their augmentation of antibodies, and with respect to CD8+ cells with regard to control of viral replication, particularly in the presence of insufficient antibody. More information is needed on mucosal responses.
Keywords: correlates, antibodies, fc effector, neutralization, T cells
1. INTRODUCTION
The worldwide pandemic of COVID‐19 has evoked more vaccine development than the world has ever seen, justified by the deaths, disease, economic effects, and social disruption resulting from the disease. All old and many new strategies for vaccine development have been invoked to produce COVID‐19 vaccines. Many of the vaccines have been proven effective in classical phase 3 studies, but more vaccines are needed to cover the entire population of the world. Soon it will be no longer be ethical or practical to perform phase 3 placebo‐controlled studies, and therefore, licensure through demonstration of protective immune responses will be critical. Thus, identification of correlates of protection against SARS‐CoV‐2 coronavirus infection is needed for practical and theoretical reasons.
The identification of a correlate of protection is often an onerous task owing to the complexity of immune responses. It must be recognized that the immune system has evolved to be redundant, so that protection can be multifactorial. This fact leads to arguments among scientists who are more focused on one or another part of immunity. However, while we agree that immunity is usually a composite of different responses, the identification of a measure that best correlates with protection is a necessary practical exercise. Even if multiple immune responses contribute to protection, it is often the case that one response is most important for protection. As the subsequent article will attest, it is likely that protection against coronaviruses results from a composite of antibody and cellular responses, including Fc effector antibodies, mucosal antibodies, memory B cells, and T cells. However, in the interests of clarity, the subsequent text will separate immune responses into four parts: neutralizing antibodies, memory B cells, Fc effector antibodies, and T cell functions. Conclusions will be drawn as to their relative importance and predictive ability.
Certain principles relating to correlates should be kept in mind when considering the CoP for vaccines against SARS‐CoV‐2 coronavirus, the agent of COVID‐19 disease. Among the most important principles are that protection against infection is different from protection against disease, that more than one immune factor may correlate with protection, and that protection against mucosal infection may depend on different factors than protection against systemic disease. Moreover, memory may be a mechanism of protection if it results in rapid induction of immune functions after exposure, which in the case of COVID‐19 memory must likely act within the first week of infection to contribute to protection.
The need for rapid protection of populations against COVID‐19 has evoked the use of multiple vaccine platforms. However, thus far the common feature of those platforms is that protection correlates with the induction of neutralizing antibodies. Although evidence for the primacy of those antibodies for protection will be presented below, we will also summarize the evidence that memory B cells, antibody Fc effector functions, and T cell responses contribute to protection, with the relative importance of each being dependent on the specific vaccine and the population in which it is used. Primary contributors to sections were David Goldblatt (neutralizing antibodies), Shane Crotty (memory B cells and T cells), and Galit Alter (Fc functions).
2. HUMORAL IMMUNITY/NEUTRALIZING ANTIBODIES
SARS‐CoV‐2 is a single‐stranded positive‐sense RNA virus of approximately 29.9kb and its genome codes for four structural proteins and sixteen non‐structural proteins (nsp1−16). The structural nucleocapsid protein (N) forms the capsid outside the genome and the genome is further packed by an envelope which is associated with three structural proteins: membrane protein (M), transmembrane spike protein (S), and envelope protein (E). The heavily glycosylated S protein is post‐translationally cleaved by mammalian furin into two subunits, S1 and S2; the S1 subunit contains an amino N terminal domain (NTD) and a receptor‐binding domain (RBD) that binds to the host cell surface angiotensin‐converting enzyme 2 receptor (ACE2) while the S2 subunit is responsible for the host‐virus membrane fusion. The spike protein transiently undergoes conformational changes under the influence of furin which results in a hinge like lifting of RBD (so‐called “open” conformation) which facilitates ACE2 binding (reviewed in 1 ). SARS‐CoV‐2 infects host cells through this attachment of S1 to ACE2 followed by fusion of the viral envelope and host cell mediated by S2.
Antibodies to structural proteins of SARS‐CoV‐2 are induced following natural infection. 2 Serum IgM and IgA appear earlier than IgG, peak between 2‐ and 5‐week post‐infection and then decline with IgA persisting longer than IgM. IgG peaks slightly later (3‐7 weeks post‐symptom onset) and then persists. 3 Concentrations of all isotypes correlate with severity of disease; the highest concentrations are seen in those with severe disease and the lowest concentrations in those with asymptomatic infection. 4 , 5 Neutralizing antibodies are detectable within seven to 15 days following disease onset, with levels increasing until days 14–22 before leveling off and then decreasing, but titers are lower in those with asymptomatic or clinically mild disease. Ninety percent of antibodies derived from serum or plasma of individuals infected with SARS‐CoV‐2 which have neutralizing activity are targeted at the spike RBD. 6 Analysis of the crystal structure of RBD‐bound antibody revealed that steric hindrance inhibited viral engagement with ACE2, thereby blocking viral entry. 7 The most potent neutralizing antibodies were the most competitive with ACE2, indicating that blocking the interaction between RBD and ACE2 is a useful surrogate for neutralization. Detailed analysis of neutralizing antibody interaction with the RBD has revealed that ACE2 blocking antibodies can bind spike and RBD in both open and closed conformations while some antibodies bind RBD but do not block ACE2 and a 4th class of neutralizing antibody bind outside of the ACE2 blocking site, but only in the open confirmation. 8
Although RBD is immunodominant, there is evidence for a substantial role of other spike regions in antigenicity, most notably the N terminal domain (NTD). NTD antibodies may also have neutralizing activity; McCallum and colleagues identified a “supersite” on the NTD that was recognized by all NTD‐specific neutralizing monoclonal antibodies derived from memory B cells isolated from 3 survivors of SARS‐CoV‐2 infection. 9 The mechanism of neutralization by which NTD‐specific antibodies act remains to be fully determined, although it may involve the inhibition of conformational changes 10 or interaction with C type lectins such as DC‐SIGN, L‐SIGN, and SIGLEC1. 11 Recent reports also suggest that antibodies directed at the NTD may enhance the infectivity of the virus by inducing the open conformation of RBD, thus enhancing the binding capacity of the spike protein to ACE2 and infectivity of SARS‐CoV‐2 12 although such in vitro enhancement of infection does not necessarily translate into enhanced infection in vivo. 13
The potency of neutralizing antibodies has been shown to be a predictor of survival in patients with COVID‐19. 14 The presence of neutralizing antibodies induced by a previous infection has also been shown to provide robust protection to subsequent reinfection with the same strain. 15 ACE2 receptor inhibition assays as a surrogate for neutralization and pseudo‐virus neutralization assays utilizing pseudo‐typed viruses transfected with SARS‐CoV‐2 spike protein, that do not require BSL3 laboratories for the handling of live virus have contributed to the description of the role of neutralizing antibodies and disease. 16
Mutations can occur in any region of the SARS‐CoV‐2 genome although most do not modify the primary amino acid sequence and hence the function of the translated proteins or viral infectivity. However, a single mutation, or a combination of mutations, can yield variants with selective and survival advantages and improved viral fitness and several variants of concern (VOC) have spread worldwide. The first VOC, designated Alpha by the WHO (B.1.1.7 lineage) demonstrated increased transmissibility and had several mutations in the spike protein including D614G, N501Y and deletions DH69/DV70. The RBD N501Y mutation was shown to increase the binding affinity for the ACE2 receptor 17 although antibody binding and neutralizing activity induced by previous infection or vaccine was generally preserved. 18 The Beta VOC (B.1.351) emerged in South Africa in October 2020 with several structural and non‐structural mutations, including three critical mutations in the RBD of the S protein (K417N, E484K, and N501Y). These seemed to play a crucial role in the improved “viral fitness” and survival adaptations compared to the other strains and reduced binding of neutralizing antibodies to spike. 19
In late 2020, the Delta variant (B.1.617) was detected in India and spread rapidly worldwide displacing other variants. Notable mutations in the B.1.617.2 variant included L452R, T478K, and E484Q in the S RBD and P681R in the cleavage site between S1 and S2. The combination of mutations in the Delta variant seems to impart the virus a selective advantage compared to the original virus and other variants, as evidenced by high transmissibility and infectivity, and immune evasion. 20
In late 2021, the B.1.1.529 variant emerged in Southern Africa and was designated Omicron by the WHO. Omicron contains several mutations present in other variants, such as N501Y (alpha), E484A~E484K (beta and gamma), and T478K; P681H~P681R (delta) although in total has more than 50 mutations with more than 30 identified in the S gene alone. 21 These mutations are associated with enhanced infectivity and transmissibility, and Omicron has also been shown to escape neutralization by monoclonal antibodies, convalescent serum, and post‐vaccine antibody. 22 Overall, with the exception of the Alpha VOC, the emerging VOCs have been associated with reductions in neutralizing activity of antibodies derived from previously infected or individuals who have undergone primary vaccination 23 , 24 , 25 , 26 while Omicron VOC also appears to escape the neutralizing activity of most, but not all of the therapeutic antibodies currently available. 27 Interestingly, a booster dose of mRNA vaccines has been shown to largely restore neutralization activity against wild type, Delta, and Omicron. 28 The mechanism behind this enhanced functionality has been shown to be due to the third dose expanding memory B clone present after the second dose as well as stimulating new clones both of which showed increased potency and breadth due to targeting more conserved areas of the RBD. 29 , 30
The study of the persistence of antibodies post‐infection has been complicated by the lack of standardization of antibody assays, differences in sensitivity and specificity of commercially available assays and the characteristics of patients studied. Nevertheless, consensus has emerged that nucleocapsid antibodies decline faster than those specific for spike or RBD, with the latter persisting for up to 13 months following infection 26 , 31 , 32 and models suggesting years of persistence. 33 Key to long‐term protection is the persistence of neutralizing rather than just binding antibody and in general while titers decline in the months following infection, neutralizing and binding antibodies correlate well with each other. 26 , 32 Gallias and colleagues demonstrated that healthcare workers who were infected with the original Wuhan strain retained neutralizing activity against the D614G and alpha variants but reduced titers to the beta variant of concern. Moriyama and colleagues 34 showed that despite a decline in IgG to RBD following infection, the ability of convalescent serum to neutralize variants of concern (beta and gamma) improved in the months after infection suggesting a temporal maturation of neutralizing antibody that was attributed to affinity maturation of anti‐RBD antibody. It is unclear if this improvement extends to newer variants, and specifically Omicron which has infected individuals with previous natural or vaccine‐induced immunity. Infection, however, has in general been relatively mild with Omicron reinforcing the notion that immunity other than that mediated by antibody is required for modulating disease.
Antibody to SARS‐CoV‐2 has been identified in urine, feces, upper and lower respiratory trach secretions and in sputum although the role of mucosal immunity has not been as extensively studied as that of serum‐based immunity but is likely to be important for rationally designing vaccines that provide maximal protection against mucosal pathogens. Chan and colleagues studied pediatric and adult COVID‐19 patients and were able to show spike‐specific IgA in the nasal epithelial lining fluid which appeared to inversely correlate with severity of disease; those with mild disease having higher titers of neutralizing antibody within the first week of illness. 35 A protective role for IgA has also been postulated by Hennings and colleagues who found healthcare workers who did not contract COVID‐19 had higher serum IgA specific for spike protein although they did not study mucosal antibody. 36 Relatively, few studies have focussed on mucosal responses post‐vaccination. Nickel and colleagues 37 studied serum and salivary responses following natural infection and vaccination and were unable to detect spike and RBD specific IgG following BNT162b2 vaccination in saliva, in contrast to patients with COVID‐19 who all developed salivary IgG 15‐30 days following the onset of symptoms. Tang and colleagues compared the spike‐specific total and neutralizing antibody (Ab) responses in bronchoalveolar lavage fluid (BAL) and blood of COVID‐19 vaccinated individuals and hospitalized patients. Vaccinated individuals (BNT162b2 or mRNA‐1273) had significantly lower levels of neutralizing Ab against D614G, Delta, and Omicron in the BAL compared to COVID‐19 convalescents, despite robust S‐specific Ab responses in the blood. Furthermore, vaccination induced significant circulating S‐specific B and T cell immunity, but in contrast to COVID‐19 convalescents, these responses were absent in the BAL of vaccinated individuals. 38 SARS‐CoV‐2‐specific IgG may also appear in oral and nasal secretions after vaccination with mRNA‐1273 but it is not clear if this is locally produced or reflects antibodies passively transferred from serum. 39 Sheikh‐Mohamed 40 demonstrated that a single dose of mRNA vaccine (BNT162b2 or mRNA‐1273) induced salivary antibodies of both IgG and IgA but a second dose only induced a further increase in salivary IgG, with IgA persisting in only 30% of vaccine recipients positive after the first dose. At 6 months post‐vaccination, concentrations had declined as had neutralizing activity of saliva. Also in this study, higher spike and RBD‐specific IgA (but not IgG) measured at 2‐4 weeks post‐dose 2 was associated with protection from subsequent infection. In this study too, it is not clear if salivary antibody was simply transduced from serum or locally produced. Mucosal responses do seem to be more robust after natural infection compared to vaccines delivered via the intramuscular route which may be a consequence of initial SARS‐CoV‐2 entry into nasopharyngeal and oral mucosal cells thus stimulating local immunity. Whether mucosally delivered vaccines could replicate the qualitative nature of immunity induced by natural infection remains to be seen.
The central role of immunity to the spike antigen in preventing coronavirus infections has driven vaccine development for SARS and MERS and thus was a focus of early vaccine development for SARS‐CoV‐2. All 4 of the vaccines initially authorized for emergency use were based on spike antigen, and thus, most has been learnt about immunity to spike and RBD in the context of vaccines. Once vaccine efficacy estimates from clinical trials had been published, it became possible to relate vaccine efficacy to antibody responses measured in the serum of subjects post‐vaccination. The absence of standardization of neutralizing assays, however, meant that researchers had to compare post‐vaccination titers to those seen following convalescence. Khoury et al 41 analyzed the relationship between in vitro neutralization levels and the observed protection from COVID‐19 infection using data from vaccinated and convalescent cohorts. They were able to demonstrate that neutralization levels are highly predictive of immune protection although this relation would likely be slightly reduced to variants. They were also able to predict a decline in vaccine‐induced immunity over time. Earle and colleagues 42 using similar modeling confirmed the relationship between neutralizing titers and vaccine efficacy and demonstrated in addition that binding antibodies to spike were highly predictive of protection although, due to the lack of standardization of assays, the ratio of vaccine‐induced antibody to convalescent antibody was used as a readout (Figure 1). Lustig et al. studying a large cohort of vaccinated adults in Israel confirmed the tight correlation between vaccine‐induced anti‐RBD IgG, neutralizing titers, and protection. 43 These studies raised the possibility that a threshold of protection could be identified that would aid in the licensure of future vaccines.
FIGURE 1.
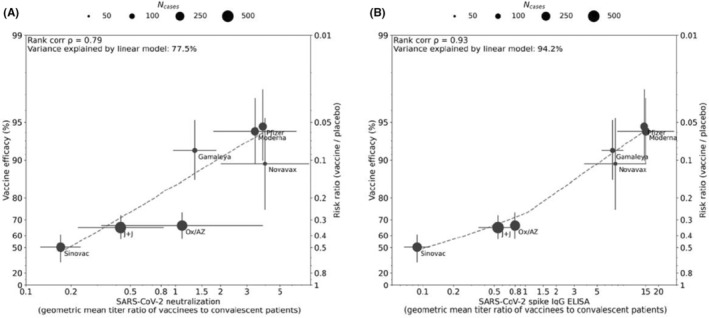
Correlation between antibody responses and efficacy rate for 7 COVID‐19 vaccines. Panels A and B display correlations of antibody responses for neutralization and ELISA assay ratios, respectively, normalized to HCS panel titers from the same assay. Dot size corresponds to the number of cases reported for Phase III efficacy analyses. The y‐axis is estimated log risk ratio reported on the vaccine efficacy scale. The x‐axis is ratio of the peak geometric mean neutralization titer or ELISA titer at 7‐28 days post‐vaccination, relative to HCS. Error bars indicate 95% confidence Intervals (except for Oxford/AZ antibody responses, which represent ratios of median titers with interquartile ranges) with dashed line showing non‐parametric LOESS fit. A rank correlation value was calculated with R2 in a linear model utilized for variance explanation. Reprinted from Vaccine Volume 39, Earle KA, Ambrosino DM, Fiore‐Gartland A, et al. Evidence for Antibody as a protective correlate for COVID‐19 vaccines, 4423‐4428, 2021
Goldblatt and colleagues 44 measured immune responses to four COVID‐19 vaccines of proven efficacy using a single serological platform. IgG anti‐spike antibodies were highly correlated with ID50 neutralization in a validated pseudoviral assay and correlated significantly with efficacies for protection against infection with wild‐type, alpha and delta variant SARS‐CoV‐2 virus (Figure 2). The protective threshold for each vaccine was calculated for IgG anti‐spike antibody and then combined to propose a population‐based correlate of protection for anti‐spike IgG.
FIGURE 2.
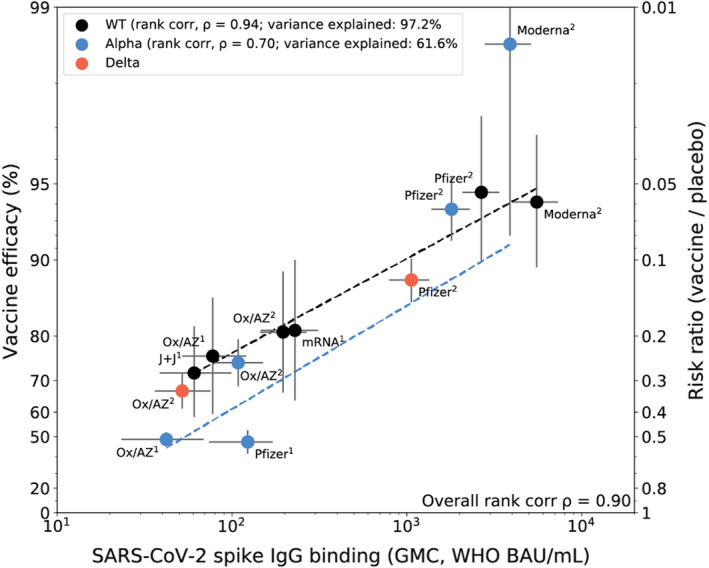
Correlation of spike IgG binding antibody measured on the same platform with vaccine efficacy for wild‐type, alpha and delta variants. Vaccine efficacy/effectiveness (VE) and SARS‐CoV‐2 spike binding IgG GMC, against original (WT), alpha and delta variants. Superscript 1 or 2 indicates the number of doses for the vaccine regimen. The y‐axis is estimated log risk‐ratio reported on the vaccine efficacy scale. The x‐axis is the geometric mean concentration (GMC) of spike‐specific IgG antibody binding measured by MSD and calibrated to the WHO standard (binding antibody units per mL). Error bars indicate 95% confidence intervals for either the GMC IgG level (x‐axis) or VE (y‐axis). Weighted least‐squares linear regression fit using inverse variance weighting on VE estimates (dashed line black for WT, dashed line blue for alpha variant). Rank correlation coefficient, variance explained by the model, and mean squared error (MSE) are indicated for the WT, and alpha variant models. Reprinted from Vaccine Volume 40, Goldblatt D, Fiore‐Gartland A, Johnson M, et al., Towards A Population‐Based Threshold of Protection for COVID‐19 Vaccines, 306‐315, 2022
Data from breakthrough infection in participants in clinical trials have also been interrogated in an attempt to derive a meaningful correlate of protection. Gilbert and colleagues studied subjects who participated in an efficacy trial of Moderna's SARS‐CoV‐2 mRNA‐1273 vaccine. 45 Through case‐cohort sampling participants were selected for measurement of IgG to S and RBD as well as neutralizing titers. Day 57 concentrations and titers were each inversely correlated with the risk of COVID‐19 infection although no threshold of protection was defined. A similar analysis was embedded in an efficacy trial of the ChadOx1 NCoV19 vaccine (Astra Zeneca). Binding spike and RBD IgG as well as neutralizing antibodies at 28 days after the second dose were measured in infected and noninfected vaccine recipients. 46 Higher levels of all immune markers were correlated with a reduced risk of symptomatic but not asymptomatic infection. Levels of binding IgG and neutralizing activity that correlated with a vaccine efficacy of 80% against symptomatic infection were defined although because of the overlap of antibody levels between the infected and uninfected subjects no absolute threshold of efficacy could be defined. However, neutralizing titer was directly related to efficacy, indicating that antibody levels are directly related to efficacy as shown in Figure 1. The hope with the studies cited above is that data can be used to bridge to new populations using validated assays, and allow extrapolation of efficacy estimates to new COVID‐19 vaccines.
The waning of vaccine‐induced antibody and the observed reduction in vaccine‐induced protection against infection (as distinct from disease) has raised the question of the utility of a protective threshold of anti‐spike or RBD antibody when measured immediately following the completion of a priming course of vaccine although with suitably designed longitudinal serological studies a threshold of antibody that would trigger revaccination could be defined.
3. MEMORY B CELLS IN PROTECTIVE IMMUNITY AGAINST COVID‐19
Memory B cells do not actively secrete antibodies; they are quiescent. Memory B cell frequencies and antibody titers exhibit different kinetics in response to SARS‐CoV‐2 infection. 47 Since the memory B cells and plasma cells are separate immunological compartments, conditions can occur like that seen after 2‐dose mRNA COVID‐19 vaccination, when spike IgG titers decline substantially over 6 months but memory B cells are stable or even increasing. 48 Memory B cells are re‐activated upon an infection or vaccination and are the source of classic anamnestic antibody responses. Memory B cells actually serve two important purposes. The first is a cellular source for the anamnestic antibody response if a pathogen gets past the circulating antibody titers and tissue‐resident memory T cells. The second important value of memory B cells is to serve as a library of “guesses” by the immune system regarding possible future viral variants. This was suggested in an animal model with protection data. 49 Extensive data from the influenza memory B cell literature are consistent with this concept, 50 , 51 though it is generally not possible to disentangle from the long history of previous diverse influenza exposures. 52
Memory B cells likely play a role in protective immunity against SARS‐CoV‐2 infection by both of the mechanisms above. 47 , 53 , 54 , 55 Antibodies are more effective at the time of exposure compared to post‐exposure; nevertheless, memory B cells can plausibly reactive and begin an anamnestic antibody response within 3‐5 days, 50 which is well within a time frame that would likely be valuable for limiting SARS‐CoV‐2 viral spread within the body and reducing the likelihood of hospitalization‐level COVID‐19 (Figure 3). While memory B cells have been associated with COVID‐19 protective immunity in a non‐human primate model, 56 direct evidence of SARS‐CoV‐2‐specific memory B cell activation, proliferation, or differentiation within a timeframe relevant for COVID‐19 protective immunity remains absent and is an important knowledge gap. Regarding the second mechanism—diversity of memory B cell specificities for variants—the COVID‐19 pandemic has dramatically demonstrated the importance of memory B cell diversity in the recognition of a pathogen and variants, also highlighting the brilliance of the immune system at predicting viral mutations, embedding those predictions in the memory B cell repertoire. Memory B cell biology in response to SARS‐CoV‐2 infection and COVID‐19 vaccines is discussed further below.
FIGURE 3.
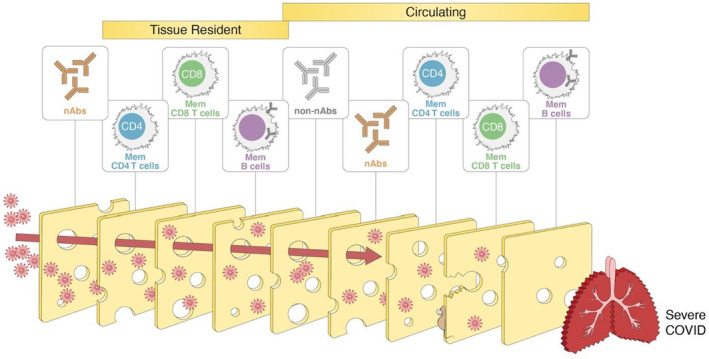
Layered defenses against SARS‐CoV‐2, or the “Swiss cheese” model of immunity. Multiple types of adaptive immunity with diverse mechanisms and locations likely provide layers of defense against COVID‐19. Conceptually, layered defenses are like a “Swiss cheese model”: even though each layer is imperfect, all together they make it highly unlikely that the pathogen breaches all of the layers of defense. Graphic inspired by the masking and public health layered defenses Swiss cheese model of Ian M. Mackay
Local tissue immunity can be an important component of protective immunity in addition to circulating memory T cells, memory B cells, and antibodies. Memory B cells have now been demonstrated in lungs of individuals after SARS‐CoV‐2 infection 57 and thus may play a role in protection from reinfection. Data are lacking on memory B cells in the upper respiratory tract or oral mucosa. The COVID‐19 vaccines elicit robust serum antibody titers for a period of months that manage to be transudated at relatively low levels into the nasal passages. In contrast, much higher amounts of antibody are produced in the nose after infection. 58 , 59 This suggests that local plasma cells may develop after infection, in addition to tissue‐resident memory B cells, both of which may contribute to protective immunity. Overall, between circulating memory B cells and tissue memory B cells, B cell memory likely contributes to protection against severe COVID‐19 (Figures 3 and 4).
FIGURE 4.
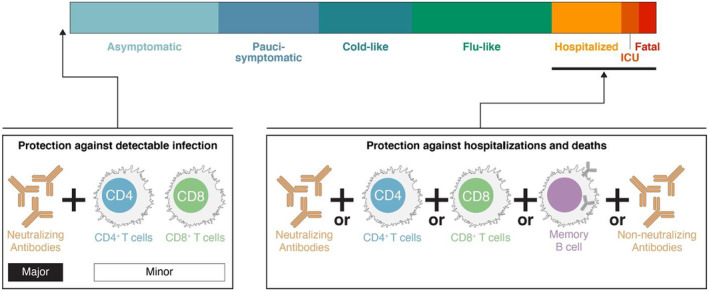
Gradations of protective immunity. “Protection” can be defined many ways and can be categorized based on COVID‐19 disease severity. Sterilizing immunity can only be provided by antibodies at the portal of entry. Prevention of detectable infection (e.g., a positive test) can be accomplishing by neutralizing antibodies and possibly tissue‐resident T cells. Prevention of hospitalization‐level COVID‐19 or fatal COVID‐19 can likely be accomplished by multiple branches of adaptive immunity acting together over time
4. Fc‐EFFECTOR FUNCTION
Antibodies are bi‐functional molecules, comprised of (1) two antigen‐binding domains (2× Fabs) that provide specificity and can block infection and (2) a constant domain (Fc) involved in directing immune clearing effects via the recruitment of the immune system (Figure 5). In the context of vaccine development, antibody binding and neutralizing activity are primarily evaluated to predict protection, however, for many pathogens, the Fab and Fc antibodies collaborate to achieve maximal protection against disease. 60 For example, the elimination of Fc‐effector function from neutralizing antibodies to HIV, influenza, and RSV reduces the clinical potency of those neutralizing antibodies. 61 , 62 , 63 Fc‐effector function can compensate for incomplete neutralization in Ebola virus infection 64 rescuing the protective activity of antibodies that only partially neutralize the virus. Moreover, several non‐neutralizing antibodies confer robust and cross‐strain protection against Influenza 65 and Ebola virus 66 via Fc‐effector functions highlighting the importance of the Fc‐effector function, rather than the Fab‐activity alone, in protection from some infections. Likewise, Fc‐effector function is key to the therapeutic activity of neutralizing bacterial‐toxin‐specific antibodies, that require both blockade of toxin action but also clearance of the toxins from the immune system. 67 However, whether both ends of the antibody are required for protection against SARS‐CoV‐2 has been poorly studied.
FIGURE 5.
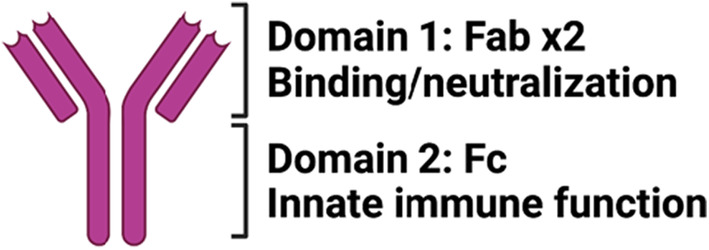
Antibody anatomy. Antibody molecules can be divided into 2 functional domains: Domain #1—composed of 2 antigen‐binding domains that contribute to antigen specificity and drive neutralization and Domain #2—consisting of a single constant domain that provides instructions to the immune system for elimination of antibody‐opsonized material
Antibodies have the ability to deploy a wide array of immune effector functions via their ability to bind to complement or Fc‐receptors, which are present on all immune cells. 60 Owing to the low‐affinity binding of Fc antibody—for complement and Fc‐receptors, aggregates of antibodies, found in immune complexes, are required to bind and activate cells. Moreover, distinct antibody subclasses/isotypes possess differing affinities for Fc‐receptors and complement. For example, IgG subclasses demonstrate distinct affinities for FcRs and human complement (IgG3>IgG1>IgG2=IgG4), IgM exhibits robust affinity for complement C1q, and IgA interacts largely with its own Fc‐receptor, called the Fcα‐receptor. Additional post‐translational modifications of the Fc‐domain of antibodies, during inflammatory immune responses, via altered Fc‐glycosylation, further tune the binding affinity of antibodies for FcRs and complement, providing another mechanism to augment antibody effector function. Thus, depending on the combination of glycosylated‐ isotype/subclasses generated during an immune response, distinct swarms of antibodies can trigger FcRs bound on a variety of cell types to induce cytokine secretion, degranulation, phagocytosis, cytotoxicity, etc.
While neutralizing titers are highest in severely ill individuals who ultimately succumb to COVID‐19, 68 , 69 antibodies do clearly contribute to protection against SARS‐CoV‐2 infection. 70 Specifically, passive transfer studies in non‐human primates (NHP) using convalescent serum have demonstrated the protective activity of serum‐antibodies in limiting infection and attenuating viral replication. 70 Most interestingly, while administration of high titers of convalescent neutralizing antibody‐containing plasma to naive NHP prior to SARS‐CoV‐2 challenge resulted in complete protection from infection, the transfer of sub‐neutralizing antibody titers did not completely block infection, but were able to attenuate the magnitude and duration of viremia, demonstrating that antibodies alone in the naive animals were able to both provide protection against infection at high titers (transmission blockade) but also to reduce disease following infection at lower titers (disease attenuation) (Figure 6). While transmission blockade can be clearly explained via the simple binding of virus at high titers, disease attenuation is likely critically dependent on the ability of transferred antibodies to drive enhanced clearance of the pathogen or attenuation of inflammation. 70 Along these lines, immune profiling of convalescent serum samples has demonstrated the importance of antibody Fc‐effector function, in addition to neutralization, in the disease attenuating function of convalescent serum therapy. 71 , 72 However, because selection of convalescent serum for therapy did not take antibody function into account, mixed results were observed across most serum‐transfer protection studies. 73 However, taking Fc effector functions into account could be a critical means to strategically improve convalescent plasma therapy. The protective activity of convalescent plasma treatment has been linked to SARS‐CoV‐2‐specific antibody‐dependent cellular cytotoxicity, 71 Fc‐receptor binding, 74 or antibody‐dependent opsonophagocytic functions, 75 pointing to the critical role of Fc‐effector functions in the therapeutic activity of antibodies.
FIGURE 6.
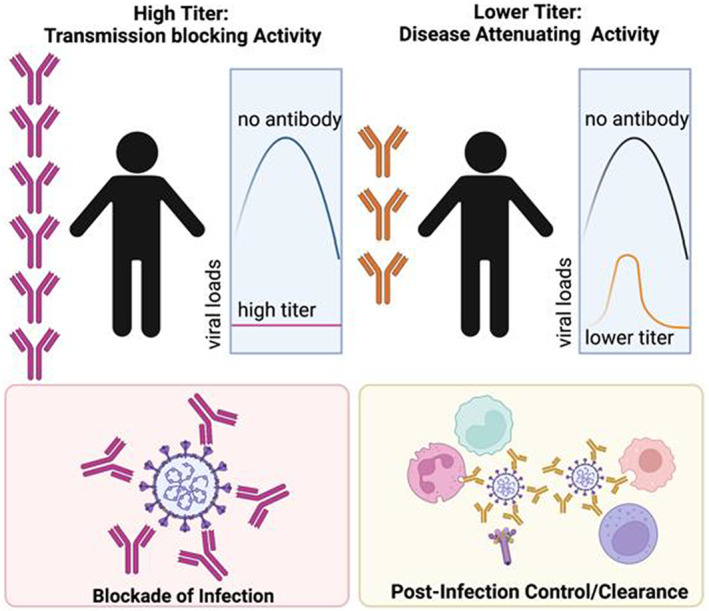
Antibody mechanisms of action. The cartoon depicts that potential contribution of Fab versus Fc mediated antibody functions at different antibody titers. Where neutralization alone may be sufficient to block transmission at peak titers (left). However, as titers wane, or variants evade large fractions of antibodies, the ability of antibodies to leverage immune effector functions may be vital to protection from disease
While nearly all approved SARS‐CoV‐2 vaccines induce robust spike‐specific binding titers and neutralization, their ability to drive Fc‐effector function remains less well defined. Previous studies on adenoviral 26 (Ad26) based vaccination against the human immunodeficiency virus (HIV) had demonstrated robust antibody Fc‐effector functions induced by this vector in both humans and NHPs, that were linked to protective immunity in an NHP challenge study. 76 , 77 Likewise, antibody Fc effector functions were observed following SARS‐CoV‐2 Ad26 vaccination in humans, 78 NHP, 79 and hamsters, 80 that co‐evolved with neutralizing antibodies and T cell immune responses, all of which were correlated with protection upon animal challenge. Similarly, robust Fc‐effector functions were observed following the Novavax adjuvanted SARS‐CoV‐2 spike‐protein immunization in macaques and humans, 81 induced most robustly with the use of an adjuvant. Moreover, correlate analyses of antibody profiles associated with viral restriction in the upper and lower respiratory tract revealed enhanced neutralization in animals protected from infection in the lower respiratory tract. However, in animals with complete protection in both the upper and lower respiratory tract, enhanced Fc effector functions were observed compared to animals with breakthrough upper respiratory viral loads. These data point to a critical role for Fc‐function in supplementing neutralization to achieve complete protection against the virus across the entire respiratory tract. Similar results were observed with an adjuvanted SARS‐CoV‐2 protein from Sanofi/GSK. 82 Analysis of mRNA vaccine‐induced immune responses also highlighted moderate levels of Fc‐effector functions after a single dose of the vaccine, 83 that were significantly augmented by the second dose of the vaccine. Moreover, mRNA vaccine‐induced antibodies demonstrated enhanced cross‐reactive Fc‐effector function across variants of concern whereas antibodies induced via natural infection, 84 , 85 adjuvanted‐protein immunization, 86 or adenoviral vaccination did not, 87 marking potentially distinct cross‐reactive Fc‐effector induction across vaccine platforms. Thus, while Fc‐effector function co‐evolves with binding titers across all vaccine platforms tested to date, the flexibility of the cross‐variant Fc‐effector response may vary across vaccine platform. These differences may be related to distinct epitope‐specific immunodominance profiles elicited by the platforms (more RBD versus NTD targeting) or perhaps related to differences in affinity maturation of the humoral immune response. Along these lines, recent comparison of the BNT162b2 and mRNA1273 vaccines pointed to significant differences in the functional quality of the response even across the two mRNA vaccines that have been deployed globally, with higher spike‐specific opsinophagocytic and NK cell activating, and NTD‐specific Fc‐receptor binding antibodies induced by the mRNA1273 vaccine. 88 Whether this is related to differences in mRNA dose, the extended interval between doses, or differences in formulation remains unclear, but tracks with enhanced real‐world vaccine effectiveness noted for the mRNA1273 vaccine, 89 highlighting that even within a platform, antibody effector function may be tuned to enhance the generation of more functional antibodies. Collectively, the data clearly point to a critical need to define epitope‐specific Fc‐effector specificities to fully dissect and define the minimal functional footprints that may play a vital role in protective immunity.
Opsonophagocytic, complement activating, and NK cell recruiting functions have been detected after most SARS‐CoV‐2 vaccines, 78 , 83 , 84 yet, whether a precise Fc‐function may collaborate with neutralizing antibodies or T cells to provide complete protection remains incompletely understood. Given the striking differences in Fc‐effector function that were elicited using distinct clinical adjuvants (SWE, alum, CpG‐alum, AS37, and AS03 86 with a SARS‐CoV‐2 spike‐receptor binding domain (RBD)‐nanoparticle vaccination in non‐human primates, it was possible to define the specific Fc‐effector function that tracked with neutralization to confer complete protection from challenge. Oil‐in water emulsion, SWE, and alum induced moderate Fc‐functions following immunization. Nucleic acid TLR sensors, TLR7 and TLR7 triggering agonists induced robust NK cell activating antibodies. Conversely, the GSK AS03 adjuvant elicited a robust opsonophagocytic response. While neutralizing antibodies were directly correlated with protection following challenge, several animals with robust neutralization still experienced breakthrough infection. Closer analysis of the Fc‐profiles among animals with matched neutralizing antibody titers that experienced or resisted breakthrough demonstrated the presence of robust neutrophil phagocytic activity and IgA levels in monkeys completely protected from challenge. These data point to a critical collaboration between Fab and Fc mediated functions, whereby neutralizing antibodies may be supplemented by opsonophagocytic or isotype‐specific functional activity to fully control and clear the virus upon exposure.
Similarly, early immune correlates studies of immune profiles associated with survival of severe disease pointed to two major differences between survivors and non‐survivors of severe COVID‐19: (1) Survivors of severe COVID‐19 generated neutralizing antibodies more rapidly 90 and at proportionately higher levels than binding antibodies, 14 pointing to a higher quality Fab‐evolutionary response linked to survival from infection, and (2) survivors of severe COVID‐19 evolved Fc‐effector functions more rapidly and to a higher level than individuals that ultimately died from COVID‐19. 69 However, interestingly, not all Fc‐effector functions were induced equally, NK cell activation was not differentially induced in survivors, but neutrophil and monocyte opsonophagocytic activating antibodies were induced most rapidly and to a higher magnitude in survivors compared to non‐survivors.
While the ability of antibodies to drive antibody‐dependent cellular cytotoxicity (ADCC) via the recruitment of NK cells is well established in another respiratory viral infection, influenza, 91 the precise role for ADCC versus opsonophagocytic functions, known to be key to respiratory bacterial infections 92 remains unclear in COVID‐19 disease. SARS‐CoV‐2 infection begins with viral attaching to the surface of the epithelium, via lectin‐like interactions between the spike and cell surface glycosylation 93 , 94 (Figure 7). However, upon interaction with the human angiotensin‐converting enzyme‐2 (ACE‐2), the virus is taken up endosomally where it is able to fuse with the cell membrane and cause infection. 95 Moreover, following intracellular replication, viral assembly occurs via the Golgi apparatus, resulting in endosomal release of viruses. Thus, unlike influenza virus which fuses and exits via the plasma membrane where it can leave remnants of its surface proteins, SARS‐CoV‐2 fuses and exits via endosomal compartments, leaving limited viral proteins on the surface of cells. Thus, killing of SARS‐CoV‐2‐infected cells via ADCC may be difficult. Conversely, antibodies able to rapidly clear viruses prior to infection or following release, via opsonophagocytic functions, may lead to rapid viral clearance. Attenuation of inflammation may provide an opportunity for other immune mechanisms, such as T cells, to ultimately eliminate all infected cells. Thus, while antibodies may exist that can elicit both ADCC and opsonophagocytic functions, one of those functions may be more tightly linked to protective immunity, due to the life‐cycle of the virus.
FIGURE 7.
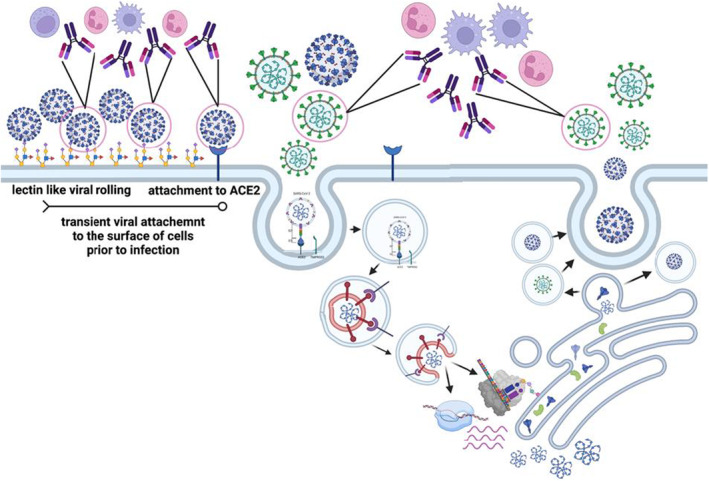
Relevance of antibody effector functions throughout the SARS‐CoV‐2 viral life cycle. The cartoon depicts the interactions of the virus with the host cell, and the moments when the spike antigen may be visible to circulating antibodies. As the virus roles across the cell surface and may be targetable by many effector mechanisms including those driven by phagocytic cells and natural killer (NK) cells (left). However, once binding to ACE2 has occurred, the virus is rapidly endocytosed, leaving limited to no spike on the surface of cells. Moreover, new viruses assemble and release from the Golgi, leaving little to no spike on the surface at the time of egress (right). Thus, functional spike‐specific antibodies likely confer the bulk of their protective functions via the recognition and elimination of free particles prior to infection or soon after egress, providing a critical bottleneck for the virus
Both neutralizing antibody levels and binding titers were robust correlates of protection in the phase 3 immune correlates analyses, largely conducted at peak immunogenicity against the wild‐type SARS‐CoV‐2 spike. 96 However, waning neutralizing antibody titers and the emergence of more neutralization resistant SARS‐CoV‐2 variants have resulted in enhanced numbers of breakthrough infections globally. 20 , 97 Although there is a rise in detectable cases, a concomitant rise in severity of disease has not been observed, 98 suggesting that non‐neutralizing vaccine‐induced immunity affords persistent protection against severe COVID‐19, despite the decline in neutralization. Importantly, emerging vaccine profile analyses suggest that not all antibody subpopulations decay with equal kinetics, with a steeper decline in neutralizing antibodies compared to binding antibodies that retain Fc effector functional activity. 99 While only a fraction of total antibodies contributes to neutralization (Figure 8), a larger fraction of antibodies targets the entire surface of the SARS‐CoV‐2 virus and can contribute to additional antibody effector functions, potentially continuing to confer protective immunity against the virus even in the setting of decaying neutralizing antibody titers. Likewise, variant mutations that evade the limited sub‐population of antibodies involved in strict neutralization do not affect all spike‐specific binding antibodies that may continue to drive antibody effector functions and contribute to antiviral immunity. Thus, while a loss of neutralization may result in loss of transmission blockade, the persistence of Fc‐effector functional antibodies may continue to drive rapid control and clearance of the virus following transmission, reducing disease severity and death.
FIGURE 8.
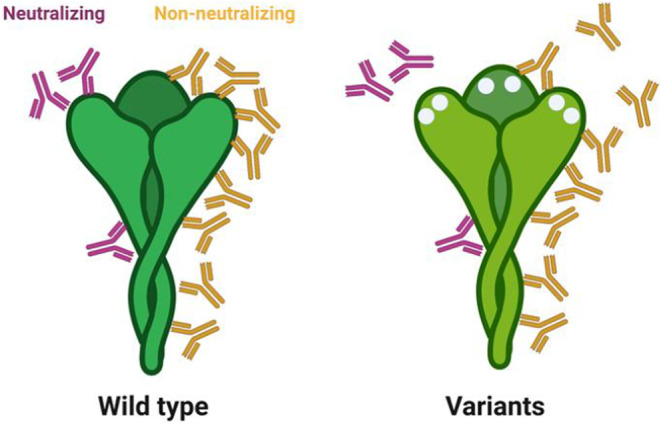
Impact of viral mutation on antibody recognition. The cartoon on the left depicts the restricted binding sites for neutralizing antibodies (purple), that either interfere directly with binding or fusion machinery, or may allosterically interfere with binding/fusion. Conversely, non‐neutralizing Fc‐functional antibodies (yellow) may bind to the entire surface of the spike. Yet, with the incorporation of mutations, in Variations of Concern, changes that may impede neutralizing antibody binding may disrupt a few, but only a fraction of non‐neutralizing antibodies
The definitive role of Fc‐effector function in immunity to SARS‐CoV‐2 was most clearly demonstrated in monoclonal antibody passive transfer studies. 100 , 101 , 102 While high titers of potent neutralizing antibodies do not require Fc‐effector functions to confer protection from infection in small animal models, potent neutralizing antibodies required Fc‐effector function to control and clear the virus after infection, 101b and less potent neutralizing antibodies have demonstrated a strong Fc‐dependency, conferring protection only in the setting of functional Fc‐domains. 101 , 104 Furthermore, these protective functions of weakly neutralizing antibodies, that target the more conserved S2‐domain of the spike antigen, can be further functionally enhanced via the addition of Fc‐point mutations that improve antibody interactions with Fc‐receptors on innate immune cells, resulting in more potent protection against viral challenge and disease. These data clearly highlight the critical role of Fc‐effector function in antibody‐mediated protection from disease, particularly in the setting of diminished neutralizing antibody activity. Moreover, the data argue that as neutralizing titers wane or more neutralization resistant variants appear, vaccine able to elicit antibody responses to conserved antigenic sites on the spike antigen, with robust Fc‐effector functionality, may drive longer lived protection against disease.
5. T CELLS IN PROTECTIVE IMMUNITY AGAINST COVID‐19
While there is robust evidence for important roles of nAbs in protective immunity against COVID‐19, various lines of evidence also point to contributions of T cells against COVID‐19, particularly severe COVID‐19 disease. There are two general ways to consider the roles of T cells in protective immunity. The first is that T cells and antibodies have different kinds of functionality that are often valuable in controlling viruses. Antibodies tend to be most effective when present prior to the start of an infection, with less efficacy after an infection has already started, because antibodies are more effective against extracellular virus than infected cells. Reciprocally, T cells cannot recognize a virus until after cells are infected, but they are the branch of adaptive immunity that specifically evolved to recognize and eliminate infected cells.
Some of the strongest indirect evidence for an important role of T cells in controlling SARS‐CoV‐2 infections comes from monoclonal antibody (mAb) clinical trials for COVID‐19. In clinical trials of people treated with mAbs within a few days of symptomatic COVID‐19, in an outpatient setting, mAb treatment clearly provided clinical benefit to those individuals, reducing the likelihood of hospitalization. 105 However, mAb treatment only reduced viral loads by fourfold in treated seronegative subjects. 105 In contrast, placebo group individuals who seroconverted on their own exhibited 1000‐fold to 10,000‐fold lower viral loads. 105 Given that the mAb infusions provide >100‐fold higher nAb titers, there is a large discordance between the modest drop in viral load after mAb infusion compared to the massive drop in viral load in individuals who develop their own immune response. Overall, the results are consistent with an important role for T cells in reducing viral loads by control and elimination of infected cells. 106 Multiple clinical trials have shown no beneficial effect of high‐dose neutralizing mAb treatment in hospitalized individuals. In contrast, mAbs provided in advance of infection have proven to be highly effective at preventing infections both in humans and non‐human primates, 107 , 108 consistent with high efficacy of mAbs before infection instead of during infection. To be absolutely clear on this point: early mAb treatment has a clinical benefit—and should be given immediately to high‐risk individuals—but the actual impact of mAbs on viral loads in those subjects is surprisingly modest, suggesting that antibodies are not big contributors in viral clearance and that the mAb treatment may be largely “buying time” for a patient's own T cell responses to amplify and clear the virus.
A second conceptual framework for considering protective benefits of T cells is a “layered defenses” model, sometimes colloquially referred to as a Swiss cheese model of defenses (Figure 3). This type of model can be applied to many scenarios, wherein by having a series of layers of defenses, even if the first layer of defense is incomplete, there are additional secondary layers that also provide defense, which in sum provide sufficient immunity to be highly effective, even if the first layer fails. In the case of adaptive immunity to SARS‐CoV‐2, the first layer is neutralizing antibodies (nAbs), with the adaptive immune system having CD4 T cells, CD8 T cells, memory B cells, and non‐nAbs each providing an additional layer of defense (Figure 9), providing a diversity of mechanisms of protective immunity, some of which are more relevant in particular tissues or time windows. This conceptual framework highlights the challenges of quantifying the contributions of different aspects of adaptive immunity to COVID‐19 if multiple components of immunity are present simultaneously.
FIGURE 9.
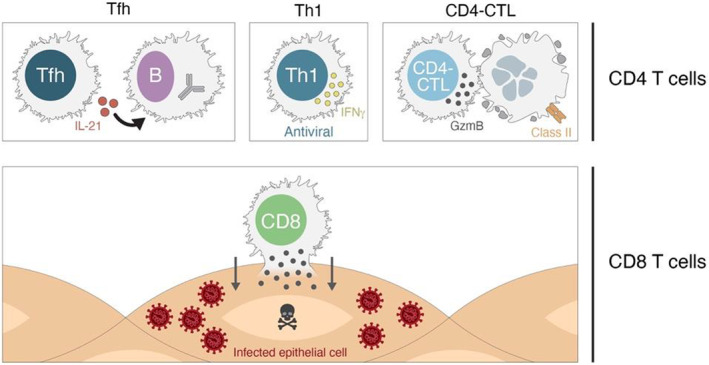
T cell mechanisms of action in protection against disease. CD4 T cells and CD8 T cells possess multiple mechanisms of action that are valuable in protection against viral infections
Overall, as discussed below, a reasonable working model is that nAbs are important for protection against SARS‐CoV‐2 infection, but once infection occurs T cells are significant contributors to control and clearance of SARS‐CoV‐2 infection, limiting symptomatic COVID‐19 and preventing hospitalization‐level disease and death (Figure 9), with the T cell functions provided by circulating and/or local tissue‐resident T cells depending on the circumstances.
5.1. T cell mechanisms of protection
There are multiple potential mechanisms by which T cells can contribute to protective immunity against infectious diseases. These are worth briefly reviewing and then addressing which mechanisms may be active in SARS‐CoV‐2 infections (Figure 9).
CD8 T cells recognize and kill infected cells via direct contact and are important in many viral infections 109 (Figure 9). In those viral infections, it is not possible to fully eliminate the virus without the activity of CD8 T cells.
For CD4 T cells, there are at least three distinct mechanisms of action involved in protective immunity against viruses: T‐follicular helper cells (TFH), TH1 cells, and CD4‐CTL cells (Figure 9). The first mechanism is T cell help to B cells, which is mediated by TFH cells. 110 This functionality of CD4 T cells is critical for the generation of nAbs against most viral infections, the generation of affinity matured memory B cells, and the generation of durable antibody responses. 110 , 111 In most contexts, there are weak or no nAbs in the absence of TFH cells and no long‐term antibody production (which comes from long‐lived plasma cells). Since most antibody responses to vaccines are dependent on TFH cells, antibody titers also serve as surrogate markers of CD4 T cell responses.
TH1 cells are classically associated with facilitating antiviral immunity. These cells produce interferon‐γ (IFNγ) and other cytokines that can act on infected cells to enhance a cell‐intrinsic antiviral state, as well as act in a local tissue environment to enhance the antiviral state of the tissue and recruit other effector cells to that site of infection. TH1 cells have been associated with protective immunity against influenza in human challenge studies. 112 TH1 cells have also been associated with protective immunity against SARS‐CoV in a mouse model. 113 CD4‐CTL cells are CD4 T cells related to TH1 cells, with cytotoxic activity similar to that of CD8 T cells. CD4‐CTL cells express granzymes. CD4‐CTL have been observed in multiple viral infections, including CMV, yellow fever virus, and Dengue virus. 114 , 115 , 116 , 117 , 118 CD4‐CTL cells have been specifically associated with protection in human Dengue, 114 and possibly influenza. 112
Location of T cells is also an important attribute of their biology. Unlike antibodies which can function at a distance from the B cells that produce them, T cells must directly contact an infected cell (or antigen‐presenting cell) to exert their antiviral functions. Thus, tissue‐resident memory T cells (a.k.a. TRM) are an important aspect of T cell biology. 119
5.2. T cell protection in SARS‐CoV‐2 infection
Available mechanisms of immunity relate to the kinetics of clinical illness. The longer the time window before clinical disease onset, the more possibilities there are for different components of adaptive immunity to contribute to protective immunity against an acute infection. 106 , 120 A disease that evolves slowly increases the likelihood that memory T cells could contribute to protective immunity. Importantly, COVID‐19 in humans is a relatively slow disease, with symptoms most often first reported 5 days after infection, hospitalization‐level disease often occurring 5 days later (~10 days post‐infection), and an average hospital stay lasting 5 days. 121 , 122 As a result, there is a large window of time for multiple branches of the immune system to control the infection before it progresses to hospitalization‐level of COVID‐19 illness. 106
It is intrinsically challenging to demonstrate roles for T cells in protection in humans. The simple passive transfer burden‐of‐proof for antibodies is not available for T cells in humans. Additionally, antigen‐specific T cells are more expensive and more technically challenging to measure than antibody responses. As noted above, mAb therapy studies during SARS‐CoV‐2 infections provide substantial indirect evidence of protective roles of T cells. Even with non‐physiologically high levels of nAbs in such patients, viral loads were reduced by only fourfold, while subjects making their own immune response had 1,000‐fold greater reductions in viral loads in the same period of time. 105 However, none of those clinical trials directly measured T cells.
In response to SARS‐CoV‐2 infection, humans make CD4 T cell responses in nearly 100% of cases, recognizing spike as well as many other SARS‐CoV‐2 proteins. 47 , 123 , 124 The SARS‐CoV‐2 CD4 T cell response is composed of TH1 cells, TFH cells, and CD4‐CTL cells. 123 , 125 , 126 Memory TH1 and TFH cells develop in the vast majority of infected individuals. 47 CD8 T cell responses are detectable in ~70% of individuals, 123 , 127 recognizing multiple SARS‐CoV‐2 proteins, 123 , 124 and CD8 T cell memory is detectable in the majority of individuals. 47 , 128 In sum, CD4 and CD8 T cell responses do develop in the majority of human SARS‐CoV‐2 infections.
One study aimed to address the relationship between T cells and control of SARS‐CoV‐2 infection by longitudinally tracking T cell responses and viral loads after symptom onset. 129 In that study, the presence of strong early T cell responses was correlated with mild disease and rapid viral clearance. 129 Antibodies did not exhibit the same pattern. Individuals with very few virus‐specific T cells early on were associated with sustained high viral loads and the subsequent development of severe COVID‐19. 129 While those are the clearest data available indicating viral control by T cells, the study did have limitations. A relatively small number of individuals were enrolled, and the study did not distinguish between CD4 T cells and CD8 T cells.
Moderbacher et al. measured SARS‐CoV‐2‐specific CD8 T cells, CD4 T cells, and nAbs in 52 individuals followed longitudinally for disease severity. 127 The study observed positive statistical associations between the presence of SARS‐CoV‐2‐specific CD4 T cells or CD8 T cells and reduced disease severity, while no association was seen between nAbs and reduction of COVID‐19 severity. 127 Furthermore, age was correlated with low frequencies of naive T cells and weak SARS‐CoV‐2‐specific CD4 and CD8 T cell responses, 127 providing a potential causal link between age and COVID‐19 severity. The data suggest that one reason age is such a major risk factor for COVID‐19 is that older individuals often have significantly smaller naive T cell repertoires, and thus have more difficulty making T cell responses to new viral infections. Limitations of this study were that it did not measure viral loads, or longitudinally track T cell responses in all individuals, and the cohort was somewhat limited in size. Three independent studies also found reduced SARS‐CoV‐2‐specific CD8 T cell responses in hospitalized patients by intracellular cytokine staining. 130 , 131 , 132 Lastly, a study assessing nucleoprotein‐specific CD8 T cells found significant associations between stronger CD8 T cell responses and mild disease. 133 In contrast, a study of only hospitalized patients did not find an association, which may be due to the lack of a non‐hospitalized COVID‐19 comparator group. 134 Higher levels of activated CD8 T cells in blood and poor disease outcomes were observed in a subset of individuals in a different study, but virus‐specific CD8 T cells were not measured. 135 Overall, in 6 studies that measured SARS‐CoV‐2‐specific CD4 T cells and CD8 T cells, early and/or larger T cell responses were associated with faster viral clearance and/or better clinical outcomes, indicating important roles for T cells in control and clearance of SARS‐CoV‐2.
Autopsies of fatal COVID‐19 cases and bronchoalveolar lavage (BAL) samples from hospitalized COVID‐19 cases are valuable sources of information about immune responses to SARS‐CoV‐2 in lungs. Severe COVID‐19 is associated with elevated neutrophil counts in blood, and most COVID‐19 autopsy studies observed massive infiltrates of neutrophils and other myeloid cells with limited numbers of T cells present, 136 , 137 , 138 , 139 consistent with an association between reduced T cells and severe COVID‐19. Studies of BAL samples also found that a paucity of T cells was associated with severe COVID‐19. 140 , 141 , 142 One extensive single‐cell transcriptomics study of BAL samples observed that viral RNA in monocytes was associated with CD8 T cell activation. 143 This was largely the expected outcome for an acute viral infection, wherein the presence of viral material should drive CD8 T cell responses to produce cytokines and kill virally infected cells in the lung. SARS‐CoV‐2‐specific CD4 T cells may assist in controlling infected cells both by production of TH1 cytokines that inhibit viral replication and recruit additional effector cells, and by CD4‐CTL that can direct kill virally infected cells. Class II expression is widely expressed on inflamed lung epithelial and endothelial cells, 126 making the SARS‐CoV‐2‐infected cell targets for CD4‐CTL killing. Tissue‐resident memory CD4 T cells and CD8 T cells specific for SARS‐CoV‐2 have been found in the lungs of humans after non‐hospitalization mild cases of COVID‐19, 57 indicating T cells do migrate to the lungs during successful resolution of SARS‐CoV‐2 infection. Overall, data from lungs and BAL have been consistent with important roles of T cells in control and clearance of SARS‐CoV‐2.
Separate lines of evidence regarding potential roles of T cells in prevention of severe COVID‐19 come from epidemiological findings in particular patient populations, including agammaglobulinemic individuals and patients on B cell depleting therapies. Such individuals have been found to have only a moderate increase in risk of hospitalization‐level COVID‐19, 144 , 145 , 146 , 147 , 148 , 149 suggesting that T cells can control and clear SARS‐CoV‐2 in the absence of a significant antibody response. Caveats to those data are that T cell responses were not directly studied in those individuals, and relative risks of those patient populations for COVID‐19 could be skewed by behavioral differences.
Roles for T cells in animal models of COVID‐19 are challenging to define because the major animal models currently in use have much faster disease progression than in humans, making any efficacy of T cells less likely to be measured. For example, in the majority of mouse and hamster models, death occurs in 6 days; whereas in humans, it is quite common to not even have the first symptoms of mild COVID‐19 reported until Day 6.
5.3. Cross‐reactive memory T cells
Pre‐existing cross‐reactive memory CD4 T cells recognizing SARS‐CoV‐2 provide insights into potential T cell mechanisms of protective immunity against COVID‐19. Cross‐reactive memory CD4 T cells against SARS‐CoV‐2 have been detected in approximately 50% of uninfected individuals. 123 , 150 , 151 Many of these memory CD4 T cells were generated in response to common cold coronavirus infections earlier in life. 152 Two studies have now reported that under the controlled conditions of vaccination, subjects having preexisting cross‐reactive SARS‐CoV‐2‐spike‐specific memory CD4 T cells have more robust CD4 T cell and antibody responses to COVID‐19 mRNA vaccines. 153 , 154 These studies give direct evidence that cross‐reactive T cells are biologically functional in vivo and enhance immunity. It was speculated that these cross‐reactive memory CD4 T cells may provide some degree of protective immunity against COVID‐19, independently of vaccination. 123 , 155 Epidemiological data support a reduction in severe COVID‐19 among individuals with a history of common cold coronavirus infection within the previous five years. 156 The most compelling evidence for a protective role of cross‐react memory T cells against COVID‐19 comes from a UK healthcare worker (HCW) study. 157 During the first COVID‐19 wave, HCWs were highly exposed to the virus. Many of those HCWs remained seronegative. Seronegative HCWs were found to have significantly higher frequencies of SARS‐CoV‐2 reactive T cells than pre‐pandemic samples from other HCWs. 157 Notably, seronegative HCWs with evidence of a potential SARS‐CoV‐2 infection based on the IFI27 gene biomarker were individuals with significantly higher frequencies of cross‐reactive memory T cells. These cross‐reactive T cells were enriched in targeting the SARS‐CoV‐2 replication machinery, which is highly conserved between coronaviruses. Altogether, those data suggest that individuals with high levels of cross‐reactive memory T cells had protective immunity that confined SARS‐CoV‐2 to a brief abortive infection, without seroconversion. 157 Similar HCW T cell patterns were observed in another study. 158 Altogether, the findings suggest a role for tissue‐resident memory T cells in protective immunity against COVID‐19. Tissue‐resident memory T cells are theoretically present in the nasal passages and mouth and could potentially locally restrict the viral infection. In a mouse model of SARS‐CoV, CD4 T cells were protective when present at the site of infection. 113 Overall, cross‐reactive memory CD4 T cell results indicate the potential of memory CD4 T cells in protection against SARS‐CoV‐2. Previously infected individuals are expected to have local upper respiratory tract tissue‐resident memory T cells. Thus, protective mechanisms by cross‐reactive memory T cells would also likely occur due to SARS‐CoV‐2‐specific tissue‐resident memory T cells in individuals with previous SARS‐CoV‐2 infection.
5.4. T cell protection against reinfection
Rhesus monkeys previously infected with SARS‐CoV‐2 are protected against reinfection. 159 , 160 Notably, depletion of CD8 T cells after SARS‐CoV‐2 infection resulted in significantly higher viral loads upon rechallenge of the animals with SARS‐CoV‐2, 70 demonstrating a role for CD8 T cells in protection against reinfection. While tissue‐resident memory CD8 T cells were not directly assessed, it may be that CD8 tissue‐resident memory T cells were the primary mechanism of protection in that model, given the speed of the CD8 T cell impact on viral loads within two days of rechallenge. 70
Humans infected with SARS‐CoV‐2 do develop immune memory and have a high level of protection against reinfection. Protection against infection with the same variant or a similar viral variant was greater than 90% against symptomatic disease in multiple studies for a period of at least 8‐12 months. 161 , 162 , 163 , 164 However, Omicron has a high degree of antibody escape. Most individuals infected with previous variants have undetectable nAb titers against Omicron, 165 and Omicron infection of people with natural immunity to previous variants is more common. 166 While there is limited protection against detectable infection with Omicron, there is still a high level of protection against hospitalization or fatality, 166 indicating a possible role for T cells in protection against Omicron. T cell recognition of Omicron is highly retained, 167 , 168 , 169 just as it has been for recognition of all variants of concern prior to Omicron. 170 Thus, whatever protective immunity is being provided by T cells against previous variants is still being provided against Omicron.
In the context of protection against SARS‐CoV‐2, tissue‐resident memory T cells need to be present in the epithelial layers of the nasal passages or oral cavity, or present in the epithelium of lung tissue, including bronchi and alveoli. An additional function of tissue‐resident memory T cells can be an alarm function, whereby memory T cells can recognize a new infection and rapidly alert other immune system branches. 171 , 172 , 173 This is potentially relevant in the context of COVID‐19, as one of the defining features of SARS‐CoV‐2 is an unusually efficient evasion of detection by early innate immunity by SARS‐CoV‐2, resulting in a lengthy delay before recognition of SARS‐CoV‐2 infection in humans and subsequent onset of symptoms. 106 An alternative early warning system by tissue‐resident memory T cells may overcome that innate immune silence.
There is a relatively long time window between SARS‐CoV‐2 infection and hospitalization‐level disease. Thus, even if the virus gets past the nAbs and tissue‐resident memory T cells at the portal of entry, there is time for additional mechanisms of protective adaptive immunity to activate and provide layers of defenses against severe COVID‐19 (Figure 3). Disease kinetics affect the likelihood that circulating memory cells contribute to viral control. Given that memory T cells can proliferate rapidly (their numbers can increase 10‐fold within 24 hours), every day is a substantial increase in the possibility that a circulating memory T cell response contributes to protective immunity. This same principle of the race between memory recall kinetics and disease progression also applies to memory B cell protection.
5.5. COVID‐19 vaccine T cell mediated protection
TH1 and TFH cells are generated in response to the Pfizer BNT162b2 or Moderna mRNA‐1273 mRNA COVID‐19 vaccines, and memory CD4 T cells are present in the vast majority of individuals six months post‐vaccination. Regarding CD8 T cells, there was initial confusion regarding CD8 T cell responses to the mRNA‐1273 vaccine. Multiple groups have now consistently reported that a majority of individuals develop spike‐specific CD8 T cells in response to the BNT162b2 or mRNA‐1273 COVID‐19 vaccines, but those cells are less well detected by certain short stimulation assays. T cells are intrinsically an essential component of protective immunity generated by the mRNA COVID‐19 vaccines, as provision of TFH cell help to B cells is central for affinity matured nAb responses to mRNA vaccines and affinity matured memory B cells. 174 , 175 , 176 , 177 , 178
No T cell correlates‐of‐protection COVID‐19 vaccine studies have been done in humans. No resources were set aside for such studies for any of the major COVID‐19 vaccines, even though such trials are practical. Intracellular cytokine (ICS) CD4 T cell and CD8 T cell assays can be implemented in vaccine clinical trials. 179 , 180 Alternatively, minimal handling quantiferon‐type whole blood IFNγ release assays (IGRA) can be implemented as higher throughput T cell assays, without distinguishing between CD4 and CD8 T cells. 181 , 182 , 183 Antibody titers can frequently be used as correlates of vaccine‐specific CD4 T cells, due to the dependence of nAbs on TFH cells. TH1 cell responses broadly correlate with TFH cells stimulated by the Pfizer BNT162b2, Moderna mRNA‐1273, Janssen/J&J Ad26.COV2.S, and Novavax NVX‐CoV2373 COVID‐19 vaccines. 153 , 169 , 176 , 184 , 185 Peak antibody titers thus may serve as a proxy indicator of an individual's T cell response to these COVID‐19 vaccines.
Significant protective efficacy against the ancestral strain, or Alpha, was observed after a single dose of BNT162b2mRNA vaccine, even though nAbs were low or undetectable. This was reported by Pfizer as evidence of a potential role of T cells in protection from COVID‐19. 186 Similar 1‐dose protection data have been observed for the mRNA‐1273 vaccine in humans. 187 CD4 T cells were found to be a correlate of protection against SARS‐CoV‐2 for the mRNA‐1273 vaccine in non‐human primates. 188 Spike‐specific CD4 T cells expressing CD40L, IL21, or any TH1 cytokine were all found to be significantly associated with lower viral loads in BAL and/or nasal swabs (e.g., reported P = 0.000, 0.000, and 0.001). 188 All three of these spike‐specific CD4 T cell populations were still associated with protective immunity when also considering spike‐specific IgG titers in multivariate analysis. The CD4 T cell and antibody responses showed evidence of linkage, which was expected as the CD4 TFH cell response was required for the antibody responses. A separate consideration, as noted above, is that the rhesus monkey model is a challenging model to observe T cell protective immunity, as the kinetics of the infection are faster and shorter than human clinical disease. Additionally, the monkeys were challenged with 800,000 PFU of the WA1 strain, 188 whereas the 50% infectious dose in humans is 10 PFU. 189 Thus, it could be considered impressive that any impact was observed between vaccine CD4 T cells and lower viral loads. Limitations of the study were that the T cell and antibody responses are linked and the role of the T cells could not be independently demonstrated. 188 Additionally, CD8 T cells were measured using an assay that largely did not detect spike‐specific CD8 T cells after mRNA‐1273 immunization, 185 whereas other CD8 T cell assays have found that a majority of immunized humans do make CD8 T cell responses. 153 Thus, any potential association between CD8 T cell responses and protective immunity in the rhesus monkey vaccination model may have gone undetected. In a separate vaccine study, CD4 T cell responses again correlated with protection, and CD8 T cell responses exhibited even strong correlation with protection. 190 Lastly, a T cell‐only SARS‐CoV‐2 intranasal vaccine also demonstrated protection in rhesus monkeys with no neutralizing antibodies. 191
T cell responses vary depending on the type of COVID‐19 vaccine. In response to the adenoviral COVID‐19 vaccines (AstraZeneca ChAdOx1 or J&J Ad26.COV2.S), TH1 cell, TFH cell, and CD8 T cell responses are detected in many individuals, and T cell memory does develop (see accompanying memory review). The primary COVID‐19 protein vaccine is the Novavax vaccine, NVX‐CoV2373. NVX‐CoV2373 elicits substantial TH1 and TFH cells responses, as well as detectable CD8 cell responses in a minority of individuals (unpublished data). There are two categories of inactivated virus COVID‐19 vaccines. CoronaVac, adjuvanted with alum, likely generates relatively weak CD4 T cell responses with a mixture of TH1 and TH2 cells, and no CD8 T cells. Covaxin (BBV152), adjuvanted with a TLR7/8 agonist, elicits substantial TH1 and TFH cell responses, 192 likely explaining the greater efficacy of Covaxin compared to CoronaVac.
There is waning mRNA vaccine protection against detectable infection over the course of 6 months after two‐dose vaccination. 164 , 193 , 194 Importantly, protection against hospitalizations and deaths was relatively stable over the same period of time. 164 , 192 The uncoupling of infection rates from hospitalization and fatality rates is consistent with a role of vaccine‐elicited T cells in protective immunity. It has been widely suggested that this is also seen for Omicron, where after two doses of mRNA vaccine most individuals have no detectable Omicron nAbs, and yet there is still significant immunity from hospitalizations or fatalities. 166 , 195 This again is consistent with a meaningful role for T cells in protective immunity. However, these analyses are currently limited in terms of temporal follow‐up and comparator groups.
5.6. Long COVID
It is unknown if T cells play a role in protecting against long COVID. There is reasonable evidence of viral RNA and protein persisting for at least 90 days in the intestines of greater than 50% of unvaccinated SARS‐CoV‐2‐infected individuals. 196 Thus, persistent SARS‐CoV‐2 infection as a cause of some cases of long COVID is a reasonable hypothesis. As a corollary, it is then plausible that weak CD8 T cell responses in some individuals could be associated with persistent SARS‐CoV‐2 in some tissues. In one case study, an 80‐year‐old man had substantial viral shedding for over 90 days that was associated with an impaired CD8 T cell response but an intact CD4 T cell and nAb response. 197 The potential involvement of insufficient T cell responses and multiple other immunological concepts of long COVID need to be tested and may shed light on protective immunity against COVID‐19 more broadly.
5.7. T cell protection summary
Numerous lines of evidence point to roles of T cells in protective immunity against COVID‐19. One key aspect is that the kinetics of severe COVID‐19 are slow enough that a T cell recall response is likely to have sufficient time to contribute to protection before the onset of hospitalization‐level disease. T cell recall can occur in 3‐5 days for other infections, and T cells have evidence of protection against symptomatic influenza. Given that COVID‐19 is often symptomatic after 5 days, it is quite plausible that circulating T cells could prevent or moderate symptoms of COVID‐19. 106 The biggest immunological difference between protective immunity generated by SARS‐CoV‐2 infection compared to vaccination is mostly likely the presence of local immunity in the upper respiratory tract and lungs. There are reasonable data that local tissue‐resident memory T cells are present and can limit viral replication sufficiently to moderate or prevent symptomatic disease. The most striking data are that these T cells can possibly fully prevent even seroconversion to infection. Infection generates tissue‐resident T cells, but vaccination presumably does not. The durability of immunity in previously infected individuals is also consistent with roles of T cells in immunity, given the low nAb titers in many individuals. Current vaccines are not designed to elicit tissue‐resident memory T cells at those tissue sites. Thus, this local mechanism of T cell protection by previous SARS‐CoV‐2 infection (or cross‐reactive T cells) would not be expected for current COVID‐19 vaccines. A mucosal vaccine would be required to generate local T cell immunity after immunization.
6. DISCUSSION
The identification of correlates of protection by vaccines requires understanding of the utility of correlates for enabling predictions but also the limitation that the immune system is complex and that CoPs are simplifications of a biological situation in which different arms of the immune system may all contribute to protection. Mucosal infections are often contrasted with viremic or bacteremic infections but in the case of SARS‐CoV‐2 one is dealing with an intermediate pathogenesis that has aspects of both local and systemic pathogenesis that does not involve viremia. Our analysis turned up multiple correlates of protection. However, although the most useful and easily applied is the quantity of antibodies that inhibit viral infection, it is manifest that opsonophagocytic functions and T cell responses impact on the ability of an infected person to control viral replication. Thus, there is a correlate of protection against SARS‐CoV‐2 infection and other correlates against severity of disease limiting spread from mucosal sites. There is value in identifying separate CoPs for protection against mucosal infection, protection against serious disease, and protection against transmission to others.
In this review, we have focused on neutralizing antibodies, memory B cells, Fc effector functions, and T cell responses, each of which appear to have roles in protection. For the vaccines currently in use, antibodies against the S1 protein of the spike are most important in preventing infection. Those antibodies have multiple functions including neutralization but also helping natural killer cells to function. Of course, CD4+ T cells must be available to promote B cell function and CD8+ T cells appear to be important in killing infected cells. Inapparent infection by SARS‐CoV‐2 is common in vaccines, but functional T cells operate to destroy infected cells.
We know that antibody titers fall rapidly after vaccination, which allows vaccines to again become susceptible to infection and symptomatic disease, suggesting that long‐lived plasma cells are not well produced by current vaccination. The result is that reinfection is common, although cellular immunity usually prevents serious disease. Thus, the ideal vaccine would generate neutralizing antibodies as well as both CD4+ and CD8+ T cells. Ideally, B cell memory and resident plasma cells would be generated to guarantee long‐term immunity.
The functionality of antibodies is also important. While antibodies are generally tested for neutralization against a fixed quantity of virus in the laboratory, protection against larger quantities of virus may be needed if a virus variant generates a higher challenge dose and therefore is more likely to overcome antibody on the mucosal surface. Human challenge studies suggest that only small amounts of viable virus are needed for infection. 198 Knowledge concerning SARS‐CoV‐2 mucosal antibody of either IgG or IgA class is insufficient.
The ideal vaccine against COVID‐19 disease would generate high levels of neutralizing antibodies, Fc Effector antibodies, and T cells of both the CD4+ and CD8+ type, all broadly active against virus variants and maintained for long periods. At the moment, we lack such a vaccine. mRNA vaccines do elicit high levels of neutralizing and Fc effector antibodies after two doses, but those responses fade with time, and although third dose gives powerful increases of antibody height and breadth, those boosts do not necessarily persist. It should be noted that such a defect is common with other mucosal pathogens, and it may be that repeated doses of vaccines against COVID‐19 will be necessary to maintain protection against infection. That being said, epidemiological evidence to date is that protection against serious and fatal disease is much easier to achieve even if vaccines do not induce long‐lasting plasma cells and thus durable antibody production.
AUTHOR CONTRIBUTIONS
Stanley Plotkin (stanley.plotkin@vaxconsult.com) organized and led the overall review. The primary authors of the neutralizing antibody, memory B cell, Fc effect functions, and T cell functions sections were David Goldblatt (d.goldblatt@acl.ac.uk), Shane Crotty (shane@lji.org), Galit Alter (galter@mgh.harvard.edu), and Shane Crotty, respectively. All authors contributed to editing and discussions. Each of the authors is a co‐correspondent.
CONFLICT OF INTEREST
David Goldblatt has no conflicts of interest. Galit Alter is employed by Seromyx, Inc. an Leyden Labs, and consults for Sanofi. Shane Crotty consults for Citi, Adagio, Morgan Stanley, Roche, JP Morgan, and Guggenheim. Stanley Plotkin consults for Merck, Moderna, New Link, Novavax, Omvax, Takeda, X Vax, Ntx, Valneva, Vasart, and Sanofi.
ACKNOWLEDGEMENTS
Thanks to Parham Ramezani‐Rad and Camila H. Coelho for helpful input. Thanks to Christina Corbaci and Eleanor G. Crotty for graphical design assistance. This work was funded by the NIH NIAID under award AI142742 (Cooperative Centers for Human Immunology, CCHI) and CHAVD UM1 AI144462, and was additionally supported in part by La Jolla Institute for Immunology Institutional Funds and an anonymous donor. Shane Crotty has consulted for GSK, JP Morgan, Citi, Morgan Stanley, Avalia NZ, Nutcracker Therapeutics, University of California, California State Universities, United Airlines, Adagio, and Roche. DG is part supported by the NIHR Great Ormond Street Biomedical Research Centre. We thank Wendy D'Arcy for her hard work putting this manuscript together.
Goldblatt D, Alter G, Crotty S, Plotkin SA. Correlates of protection against SARS‐CoV‐2 infection and COVID‐19 disease. Immunol Rev. 2022;00:1‐21. doi: 10.1111/imr.13091
Funding information
No funds were used to produce this manuscript
REFERENCES
- 1. Harvey WT, Carabelli AM, Jackson B, et al. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Post N, Eddy D, Huntley C, et al. Antibody response to SARS‐CoV‐2 infection in humans: a systematic review. PLoS One. 2020;15(12):e0244126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Y, Zhang L, Sang L, et al. Kinetics of viral load and antibody response in relation to COVID‐19 severity. J Clin Invest. 2020;130(10):5235‐5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carsetti R, Zaffina S, Piano Mortari E, et al. Different innate and adaptive immune responses to SARS‐CoV‐2 infection of asymptomatic, mild, and severe cases. Front Immunol. 2020;11:610300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Piccoli L, Park YJ, Tortorici MA, et al. Mapping neutralizing and immunodominant sites on the SARS‐CoV‐2 spike receptor‐binding domain by structure‐guided high‐resolution serology. Cell. 2020;183(4):1024‐1042.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS‐CoV‐2 infection. Nature. 2020;584(7819):115‐119. [DOI] [PubMed] [Google Scholar]
- 8. Barnes CO, Jette CA, Abernathy ME, et al. SARS‐CoV‐2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCallum M, de Marco A, Lempp FA, et al. N‐terminal domain antigenic mapping reveals a site of vulnerability for SARS‐CoV‐2. Cell. 2021;184(9):2332‐2347.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N‐terminal domain of the Spike protein of SARS‐CoV‐2. Science. 2020;369(6504):650‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lempp FA, Soriaga LB, Montiel‐Ruiz M, et al. Lectins enhance SARS‐CoV‐2 infection and influence neutralizing antibodies. Nature. 2021;598(7880):342‐347. [DOI] [PubMed] [Google Scholar]
- 12. Liu Y, Soh WT, Kishikawa JI, et al. An infectivity‐enhancing site on the SARS‐CoV‐2 spike protein targeted by antibodies. Cell. 2021;184(13):3452‐3466.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li D, Edwards RJ, Manne K, et al. In vitro and in vivo functions of SARS‐CoV‐2 infection‐enhancing and neutralizing antibodies. Cell. 2021;184(16):4203‐4219.e32. doi: 10.1016/j.cell.2021.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia‐Beltran WF, Lam EC, Astudillo MG, et al. COVID‐19‐neutralizing antibodies predict disease severity and survival. Cell. 2021;184(2):476‐488 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Addetia A, Crawford KH, Dingens A, et al. Neutralizing antibodies correlate with protection from SARS‐CoV‐2 in humans during a fishery vessel outbreak with a high attack rate. J Clin Microbiol. 2020;58(11):e02107‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nie J, Li Q, Wu J, et al. Quantification of SARS‐CoV‐2 neutralizing antibody by a pseudotyped virus‐based assay. Nat Protoc. 2020;15(11):3699‐3715. [DOI] [PubMed] [Google Scholar]
- 17. Starr TN, Greaney AJ, Hilton SK, et al. Deep mutational scanning of SARS‐CoV‐2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295‐1310 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muik A, Wallisch AK, Sänger B, et al. Neutralization of SARS‐CoV‐2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine‐elicited human sera. Science. 2021;371(6534):1152‐1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Focosi D, Maggi F. Neutralising antibody escape of SARS‐CoV‐2 spike protein: risk assessment for antibody‐based Covid‐19 therapeutics and vaccines. Rev Med Virol. 2021;31(6):e2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS‐CoV‐2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276‐280. [DOI] [PubMed] [Google Scholar]
- 21. Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600(7887):21. [DOI] [PubMed] [Google Scholar]
- 22. Planas D, Saunders N, Maes P, et al. Considerable escape of SARS‐CoV‐2 Omicron to antibody neutralization. Nature. 2022;602(7898):671‐675. [DOI] [PubMed] [Google Scholar]
- 23. Chen X, Chen Z, Azman AS, et al. Neutralizing antibodies against SARS‐CoV‐2 variants induced by natural infection or vaccination: a systematic review and pooled meta‐analysis. Clin Infect Dis. 2021;73(9):1722‐1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021;602(7898):654‐656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmidt F, Muecksch F, Weisblum Y, et al. Plasma neutralization of the SARS‐CoV‐2 Omicron variant. N Engl J Med. 2022;386(6):599‐601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gallais F, Gantner P, Bruel T, et al. Evolution of antibody responses up to 13 months after SARS‐CoV‐2 infection and risk of reinfection. EBioMedicine. 2021;71:103561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. VanBlargan LA, Errico JM, Halfmann PJ, et al. An infectious SARS‐CoV‐2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2022;28(3):490‐495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garcia‐Beltran WF, Denis KJ, Hoelzemer A, et al. mRNA‐based COVID‐19 vaccine boosters induce neutralizing immunity against SARS‐CoV‐2 Omicron variant. Cell. 2022;185(3):457‐466 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Muecksch F, Wang Z, Cho A, et al. Increased potency and breadth of SARS‐CoV‐2 neutralizing antibodies after a third mRNA vaccine dose. bioRxiv. 2022. [Google Scholar]
- 30. Kotaki R, Adachi Y, Moriyama S, et al. SARS‐CoV‐2 Omicron‐neutralizing memory B‐cells are elicited by two doses of BNT162b2 mRNA vaccine. Sci Immunol. 2022;7(70):eabn8590. doi: 10.1126/sciimmunol.abn8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. L'Huillier AG, Meyer B, Andrey DO, et al. Antibody persistence in the first 6 months following SARS‐CoV‐2 infection among hospital workers: a prospective longitudinal study. Clin Microbiol Infect. 2021;27(5):784.e1‐784.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wajnberg A, Amanat F, Firpo A, et al. Robust neutralizing antibodies to SARS‐CoV‐2 infection persist for months. Science. 2020;370(6521):1227‐1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grandjean L, Saso A, Torres Ortiz A, et al. Long‐term persistence of spike antibody and predictive modeling of antibody dynamics following infection with SARS‐CoV‐2. Clin Infect Dis. 2022;74(7):1220‐1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moriyama S, Adachi Y, Sato T, et al. Temporal maturation of neutralizing antibodies in COVID‐19 convalescent individuals improves potency and breadth to circulating SARS‐CoV‐2 variants. Immunity. 2021;54(8):1841‐1852 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chan RW, Chan KC, Lui GC, et al. Mucosal antibody response to SARS‐CoV‐2 in paediatric and adult patients: a longitudinal study. medRxiv. 2022;11(4):397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hennings V, Thörn K, Albinsson S, et al. The presence of serum anti‐SARS‐CoV‐2 IgA appears to protect primary health care workers from COVID‐19. Eur J Immunol. 2022;52(5):800‐809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nickel, O. , Rockstroh A, Wolf J, et al., Evaluation of the systemic and mucosal immune response induced by COVID‐19 and the BNT162b2 mRNA vaccine for SARS‐CoV‐2. medRxiv, 2022: p. 2022.01.29.22270066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang J, Zeng C, Cox TM, et al. Mucosal immunity against SARS‐CoV‐2 variants of concern including Omicron following vaccination. medRxiv. 2022. [Google Scholar]
- 39. Mades A, Chellamathu P, Kojima N, et al. Detection of persistent SARS‐CoV‐2 IgG antibodies in oral mucosal fluid and upper respiratory tract specimens following COVID‐19 mRNA vaccination. Sci Rep. 2021;11(1):24448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sheikh‐Mohamed S, Isho B, Chao GYC, et al. Systemic and mucosal IgA responses are variably induced in response to SARS‐CoV‐2 mRNA vaccination and are associated with protection against subsequent infection. medRxiv. 2022. doi: 10.1101/2021.08.01.21261297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khoury DS, Cromer D, Reynaldi A, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS‐CoV‐2 infection. Nat Med. 2021;27(7):1205‐1211. [DOI] [PubMed] [Google Scholar]
- 42. Earle KA, Ambrosino DM, Fiore‐Gartland A, et al. Evidence for antibody as a protective correlate for COVID‐19 vaccines. Vaccine. 2021;39(32):4423‐4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lustig Y, Sapir E, Regev‐Yochay G, et al. BNT162b2 COVID‐19 vaccine and correlates of humoral immune responses and dynamics: a prospective, single‐centre, longitudinal cohort study in health‐care workers. Lancet Respir Med. 2021;9(9):999‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goldblatt D, Fiore‐Gartland A, Johnson M, et al. Towards a population‐based threshold of protection for COVID‐19 vaccines. Vaccine. 2022;40(2):306‐315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gilbert PB, Montefiori DC, AB MD, et al. Immune correlates analysis of the mRNA‐1273 COVID‐19 vaccine efficacy clinical trial. Science. 2022;375(6576):43‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Feng S, Phillips DJ, White T, et al. Correlates of protection against symptomatic and asymptomatic SARS‐CoV‐2 infection. Nat Med. 2021;27(11):2032‐2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS‐CoV‐2 assessed for up to 8 months after infection. Science 2021;371(6529):eabf4063. doi: 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goel RR, Painter MM, Apostolidis SA, et al. mRNA vaccines induce durable immune memory to SARS‐CoV‐2 and variants of concern. Science. 2021;374(6572):abm0829. doi: 10.1126/science.abm0829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Purtha WE, Tedder TF, Johnson S, Bhattacharya D, Diamond MS. Memory B cells, but not long‐lived plasma cells, possess antigen specificities for viral escape mutants. The J Exp Med. 2011;208(13):2599‐2606. doi: 10.1084/jem.20110740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li GM, Chiu C, Wrammert J, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross‐reactive memory B cells. Proc Natl Acad Sci USA. 2012;109(23):9047‐9052. doi: 10.1073/pnas.1118979109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Andrews SF, Chambers MJ, Schramm CA, et al. Activation dynamics and immunoglobulin evolution of pre‐existing and newly generated human memory B cell responses to influenza hemagglutinin. Immunity. 2019;51(2):398‐410.e5. doi: 10.1016/j.immuni.2019.06.024 [DOI] [PubMed] [Google Scholar]
- 52. Dugan HL, Guthmiller JJ, Arevalo P, et al. Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci Transl Med. 2020;12(573):eabd3601. doi: 10.1126/scitranslmed.abd3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Z, Muecksch F, Schaefer‐Babajew D, et al. Naturally enhanced neutralizing breadth against SARS‐CoV‐2 one year after infection. Nature. 2021;595(7867):426‐431. doi: 10.1038/s41586-021-03696-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cho A, Muecksch F, Schaefer‐Babajew D, et al. Anti‐SARS‐CoV‐2 receptor binding domain antibody evolution after mRNA vaccination. Nature. 2021;600(7889):517‐522. doi: 10.1038/s41586-021-04060-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Crotty S. Hybrid immunity. Science. 2021;372(6549):1392‐1393. doi: 10.1126/science.abj2258 [DOI] [Google Scholar]
- 56. Gagne M, Corbett KS, Flynn BJ, et al. Protection from SARS‐CoV‐2 Delta one year after mRNA‐1273 vaccination in rhesus macaques coincides with anamnestic antibody response in the lung. Cell. 2022;185(1):113‐130.e15. doi: 10.1016/j.cell.2021.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Poon MML, Rybkina K, Kato Y, et al. SARS‐CoV‐2 infection generates tissue‐localized immunological memory in humans. Sci Immunol. 2021;6(65):eabl9105. doi: 10.1126/sciimmunol.abl9105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Collier A, Ris Y, Brown CM, Mcmahan K, et al. Immune responses in fully vaccinated individuals following breakthrough infection with the SARS‐CoV‐2 Delta variant in Provincetown, Massachusetts. Medrxiv. 2021. doi: 10.1101/2021.10.18.21265113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sajadi MM, Myers A, Logue J, et al. Mucosal and systemic responses to SARS‐CoV‐2 vaccination in infection naïve and experienced individuals. Biorxiv. 2021. doi: 10.1101/2021.12.13.472159 [DOI] [Google Scholar]
- 60. Lu LL, Suscovich TJ, Fortune SM, Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol. 2018;18:46‐61. doi: 10.1038/nri.2017.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sutton TC, Lamirande EW, Bock KW, et al. In vitro neutralization is not predictive of prophylactic efficacy of broadly neutralizing monoclonal antibodies CR6261 and CR9114 against lethal H2 influenza virus challenge in mice. J Virol. 2017;91:e01603‐17. doi: 10.1128/JVI.01603-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. DiLillo DJ, Palese P, Wilson PC, Ravetch JV. Broadly neutralizing anti‐influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest. 2016;126:605‐610. doi: 10.1172/JCI84428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bournazos S, Klein F, Pietzsch J, et al. Broadly neutralizing anti‐HIV‐1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158:1243‐1253. doi: 10.1016/j.cell.2014.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gunn BM, Yu WH, Karim MM, et al. A role for Fc function in therapeutic monoclonal antibody‐mediated protection against Ebola virus. Cell Host Microbe. 2018;24:221‐233 e225. doi: 10.1016/j.chom.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dreyfus C, Laursen NS, Kwaks T, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343‐1348. doi: 10.1126/science.1222908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Saphire EO, Schendel SL, Gunn BM, Milligan JC, Alter G. Antibody‐mediated protection against Ebola virus. Nat Immunol. 2018;19:1169‐1178. doi: 10.1038/s41590-018-0233-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Abboud N, Chow SK, Saylor C, et al. A requirement for FcgammaR in antibody‐mediated bacterial toxin neutralization. J Exp Med. 2010;207:2395‐2405. doi: 10.1084/jem.20100995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581:465‐469. doi: 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 69. Zohar T, Loos C, Fischinger S, Atyeo C, et al. Compromised humoral functional evolution tracks with SARS‐CoV‐2 mortality. Cell. 2020;183:1508‐1519 e1512. doi: 10.1016/j.cell.2020.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McMahan K, Yu J, Mercado NB, et al. Correlates of protection against SARS‐CoV‐2 in rhesus macaques. Nature. 2021;590:630‐634. doi: 10.1038/s41586-020-03041-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Begin P, Callum J, Jamula E, et al. Convalescent plasma for hospitalized patients with COVID‐19: an open‐label, randomized controlled trial. Nat Med. 2021;27:2012‐2024. doi: 10.1038/s41591-021-01488-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Natarajan H, Crowley AR, Butler SE, et al. SARS‐CoV‐2 antibody signatures robustly predict diverse antiviral functions relevant for convalescent plasma therapy. medRxiv. 2020. doi: 10.1101/2020.09.16.20196154 [DOI] [Google Scholar]
- 73. Klassen SA, Senefeld JW, Johnson PW, et al. The effect of convalescent plasma therapy on COVID‐19 patient mortality: systematic review and meta‐analysis. medRxiv. 2021. doi: 10.1101/2020.07.29.20162917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Natarajan H, Crowley AR, Butler SE, et al. Markers of polyfunctional SARS‐CoV‐2 antibodies in convalescent plasma. mBio. 2021;12(2):e00765‐21. doi: 10.1128/mBio.00765-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Herman JD, Wang C, Loos C, et al. Functional antibodies in COVID‐19 convalescent plasma. medRxiv. 2021. doi: 10.1101/2021.03.08.21253157 [DOI] [Google Scholar]
- 76. Barouch DH, Tomaka FL, Wegmann F, et al. Evaluation of a mosaic HIV‐1 vaccine in a multicentre, randomised, double‐blind, placebo‐controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13‐19). Lancet. 2018;392:232‐243. doi: 10.1016/S0140-6736(18)31364-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Alter G, Yu WH, Chandrashekar A, et al. Passive transfer of vaccine‐elicited antibodies protects against SIV in rhesus macaques. Cell. 2020;183:185‐196 e114. doi: 10.1016/j.cell.2020.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stephenson KE, Le Gars M, Sadoff J, et al. Immunogenicity of the Ad26.COV2.S vaccine for COVID‐19. JAMA. 2021;325:1535‐1544. doi: 10.1001/jama.2021.3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mercado NB, Zahn R, Wegmann F, et al. Single‐shot Ad26 vaccine protects against SARS‐CoV‐2 in rhesus macaques. Nature. 2020;586:583‐588. doi: 10.1038/s41586-020-2607-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tostanoski LH, Wegmann F, Martinot AJ, et al. Ad26 vaccine protects against SARS‐CoV‐2 severe clinical disease in hamsters. Nat Med. 2020;26:1694‐1700. doi: 10.1038/s41591-020-1070-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gorman MJ, Patel N, Guebre‐Xabier M, et al. Collaboration between the Fab and Fc contribute to maximal protection against SARS‐CoV‐2 in nonhuman primates following NVX‐CoV2373 subunit vaccine with Matrix‐M vaccination. bioRxiv. 2021. doi: 10.1101/2021.02.05.429759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Francica JR, Flynn BJ, Foulds KE, et al. Protective antibodies elicited by SARS‐CoV‐2 spike protein vaccination are boosted in the lung after challenge in nonhuman primates. Sci Transl Med. 2021;13(607):1‐17. doi: 10.1126/scitranslmed.abi4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tauzin A, Nayrac M, Benlarbi M, et al. A single dose of the SARS‐CoV‐2 vaccine BNT162b2 elicits Fc‐mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29:1137‐1150 e1136. doi: 10.1016/j.chom.2021.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kaplonek P, Cizmeci D, Fischinger S, et al. Subtle immunological differences in mRNA‐1273 and BNT162b2 COVID‐19 vaccine induced Fc‐functional profiles. bioRxiv. 2021. doi: 10.1101/2021.08.31.458247 [DOI] [Google Scholar]
- 85. Kaplonek P, Fischinger S, Cizmeci D, et al. mRNA‐1273 vaccine‐induced antibodies maintain Fc effector functions across SARS‐CoV‐2 variants of concern. Immunity. 2022;55(2):355‐365.e4. doi: 10.1016/j.immuni.2022.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Arunachalam PS, Walls AC, Golden N, et al. Adjuvanting a subunit COVID‐19 vaccine to induce protective immunity. Nature. 2021;594:253‐258. doi: 10.1038/s41586-021-03530-2 [DOI] [PubMed] [Google Scholar]
- 87. Alter G, Yu J, Liu J, et al. Immunogenicity of Ad26.COV2.S vaccine against SARS‐CoV‐2 variants in humans. Nature. 2021;596:268‐272. doi: 10.1038/s41586-021-03681-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kaplonek P, Cizmeci D, Fischinger S, et al. mRNA‐1273 and BNT162b2 COVID‐19 vaccines elicit antibodies with differences in Fc‐mediated effector functions. Sci Transl Med. 2022;14(645):eabm2311. doi: 10.1126/scitranslmed.abm2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pawlowski C, Lenehan P, Puranik A, Agarwal V, Venkatakrishnan AJ, Niesen MJM, O'Horo JC, Virk A, Swift MD, Badley AD, Halamka J, Soundararajan V. FDA‐authorized mRNA COVID‐19 vaccines are effective per real‐world evidence synthesized across a multi‐state health system Med (N Y). 2021. Aug 13;2(8):979‐992.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lucas C, Klein J, Sundaram M, et al. Kinetics of antibody responses dictate COVID‐19 outcome. medRxiv. 2020. doi: 10.1101/2020.12.18.20248331 [DOI] [Google Scholar]
- 91. Von Holle TA, Moody MA. Influenza and antibody‐dependent cellular cytotoxicity. Front Immunol. 2019;10:1457. doi: 10.3389/fimmu.2019.01457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Paschall AV, Middleton DR, Avci FY. Opsonophagocytic killing assay to assess immunological responses against bacterial pathogens. J Vis Exp. 2019:(146). doi: 10.3791/59400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sun XL. The role of cell surface sialic acids for SARS‐CoV‐2 infection. Glycobiology. 2021;31:1245‐1253. doi: 10.1093/glycob/cwab032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nguyen L, KA MC, Bui DT, et al. Sialic acid‐containing glycolipids mediate binding and viral entry of SARS‐CoV‐2. Nat Chem Biol. 2022;18:81‐90. doi: 10.1038/s41589-021-00924-1 [DOI] [PubMed] [Google Scholar]
- 95. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS‐CoV‐2. Nat Rev Microbiol. 2021;19:155‐170. doi: 10.1038/s41579-020-00468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cromer D, Steain M, Reynaldi A, et al. Neutralising antibody titres as predictors of protection against SARS‐CoV‐2 variants and the impact of boosting: a meta‐analysis. Lancet Microbe. 2022;3:e52‐e61. doi: 10.1016/S2666-5247(21)00267-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Puranik A, Lenehan PJ, Silvert E, et al. Comparison of two highly‐effective mRNA vaccines for COVID‐19 during periods of Alpha and Delta variant prevalence. medRxiv. 2021. doi: 10.1101/2021.08.06.21261707 [DOI] [Google Scholar]
- 98. Bergwerk M, Gonen T, Lustig Y, et al. Covid‐19 breakthrough infections in vaccinated health care workers. N Engl J Med. 2021;385:1474‐1484. doi: 10.1056/NEJMoa2109072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Collier AY, Yu J, McMahan K, et al. Differential kinetics of immune responses elicited by Covid‐19 vaccines. N Engl J Med. 2021;385:2010‐2012. doi: 10.1056/NEJMc2115596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wec AZ, Wrapp D, Herbert AS, et al. Broad neutralization of SARS‐related viruses by human monoclonal antibodies. Science. 2020;369:731‐736. doi: 10.1126/science.abc7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Pinto D, Sauer MM, Czudnochowski N, et al. Broad betacoronavirus neutralization by a stem helix‐specific human antibody. Science. 2021;373:1109‐1116. doi: 10.1126/science.abj3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kreer C, Zehner M, Weber T, et al. Longitudinal isolation of potent near‐germline SARS‐CoV‐2‐neutralizing antibodies from COVID‐19 patients. Cell. 2020;182:843‐854 e812. doi: 10.1016/j.cell.2020.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Winkler ES, Gilchuk P, Yu J, et al. Human neutralizing antibodies against SARS‐CoV‐2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021. Apr 1;184(7):1804‐1820.e16. doi: 10.1016/j.cell.2021.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhou P, Yuan M, Song G, et al. A human antibody reveals a conserved site on beta‐coronavirus spike proteins and confers protection against SARS‐CoV‐2 infection. bioRxiv. 2022. doi: 10.1101/2021.03.30.437769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Weinreich DM, Sivapalasingam S, Norton T, et al. REGN‐COV2, a neutralizing antibody cocktail, in outpatients with Covid‐19. New Engl J Med. 2020;384(3):238‐251. doi: 10.1056/nejmoa2035002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sette A, Crotty S. Adaptive immunity to SARS‐CoV‐2 and COVID‐19. Cell. 2021;184(4):861‐880. doi: 10.1016/j.cell.2021.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. O'Brien MP, Forleo‐Neto E, Musser BJ, et al. Subcutaneous REGEN‐COV antibody combination to prevent Covid‐19. New Engl J Med. 2021;385(13):1184‐1195. doi: 10.1056/nejmoa2109682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zost SJ, Gilchuk P, Case JB, et al. Potently neutralizing and protective human antibodies against SARS‐CoV‐2. Nature. 2020;584(7821):443‐449. doi: 10.1038/s41586-020-2548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol. 2014;15(12):1104‐1115. doi: 10.1038/ni.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50(5):1132‐1148. doi: 10.1016/j.immuni.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. 2015;15(3):185‐189. doi: 10.1038/nri3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wilkinson TM, Li CKF, Chui CSC, et al. Preexisting influenza‐specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18(2):274‐280. doi: 10.1038/nm.2612 [DOI] [PubMed] [Google Scholar]
- 113. Zhao J, Zhao J, Mangalam AK, et al. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44(6):1379‐1391. doi: 10.1016/j.immuni.2016.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Weiskopf D, Bangs DJ, Sidney J, et al. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc Natl Acad Sci USA. 2015;112(31):E4256‐E4263. doi: 10.1073/pnas.1505956112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Patil VS, Madrigal A, Schmiedel BJ, et al. Precursors of human CD4+ cytotoxic T lymphocytes identified by single‐cell transcriptome analysis. Sci Immunol. 2018;3(19):eaan8664. doi: 10.1126/sciimmunol.aan8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Verma S, Weiskopf D, Gupta A, et al. Cytomegalovirus‐specific CD4 T cells are cytolytic and mediate vaccine protection. Longnecker RM, ed. J Virol. 2016;90(2):650‐658. doi: 10.1128/jvi.02123-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Watson AM, Lam LKM, Klimstra WB, Ryman KD. The 17D‐204 vaccine strain‐induced protection against virulent yellow fever virus is mediated by humoral immunity and CD4+ but not CD8+ T cells. Pierson TC, ed. PloS Pathogens. 2016;12(7):e1005786. doi: 10.1371/journal.ppat.1005786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. McKinstry KK, Strutt TM, Kuang Y, et al. Memory CD4+ T cells protect against influenza through multiple synergizing mechanisms. J Clin Invest. 2012;122(8):2847‐2856. doi: 10.1172/jci63689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Masopust D, Soerens AG. Tissue‐resident T cells and other resident leukocytes. Annu Rev Immunol. 2019;37(1):1‐26. doi: 10.1146/annurev-immunol-042617-053214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kaslow DC. Certainty of success: three critical parameters in coronavirus vaccine development. npj Vaccines. 2020;5(1):42. doi: 10.1038/s41541-020-0193-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Garg S, Patel K, Pham H, et al. Clinical trends among U.S. adults hospitalized with COVID‐19, March to December 2020. Ann Intern Med. 2021;174(10):M21‐M1991. doi: 10.7326/m21-1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID‐19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172(9):577‐582. doi: 10.7326/m20-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS‐CoV‐2 coronavirus in humans with COVID‐19 disease and unexposed individuals. Cell. 2020;181(7):1489‐1501.e15. doi: 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tarke A, Sidney J, Kidd CK, et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS‐CoV‐2 epitopes in COVID‐19 cases. Cell Rep Med. 2021;2(2):100204. doi: 10.1016/j.xcrm.2021.100204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Meckiff BJ, Ramírez‐Suástegui C, Fajardo V, et al. Imbalance of regulatory and cytotoxic SARS‐CoV‐2‐reactive CD4+ T cells in COVID‐19. Cell. 2020;183(5):1340‐1353. doi: 10.1016/j.cell.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Kaneko N, Boucau J, Kuo HH, et al. Expansion of cytotoxic CD4+ T cells in the lungs in severe COVID‐19. Medrxiv. 2021. doi: 10.1101/2021.03.23.21253885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Moderbacher CR, Ramirez SI, Dan JM, et al. Antigen‐specific adaptive immunity to SARS‐CoV‐2 in acute COVID‐19 and associations with age and disease severity. Cell. 2020;183(4):996‐1012.e19. doi: 10.1016/j.cell.2020.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zuo J, Dowell AC, Pearce H, et al. Robust SARS‐CoV‐2‐specific T‐cell immunity is maintained at 6 months following primary infection. Nat Immunol. 2021;22(5):620‐626. doi: 10.1038/s41590-021-00902-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Tan AT, Linster M, Tan CW, et al. Early induction of functional SARS‐CoV‐2 specific T cells associates with rapid viral clearance and mild disease in COVID‐19 patients. Cell Rep. 2021;34(6):108728. doi: 10.1016/j.celrep.2021.108728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Cohen KW, Linderman SL, Moodie Z, et al. Longitudinal analysis shows durable and broad immune memory after SARS‐CoV‐2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med. 2021;2(7):100354. doi: 10.1016/j.xcrm.2021.100354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Peng Y, Mentzer AJ, Liu G, et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS‐CoV‐2 in UK convalescent individuals following COVID‐19. Nat Immunol. 2020;21(11):1336‐1345. doi: 10.1038/s41590-020-0782-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sekine T, Perez‐Potti A, Rivera‐Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID‐19. Cell. 2020;183(1):158‐168.e14. doi: 10.1016/j.cell.2020.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Peng Y, Felce SL, Dong D, et al. An immunodominant NP105–113‐B*07:02 cytotoxic T cell response controls viral replication and is associated with less severe COVID‐19 disease. Nat Immunol. 2022;23(1):50‐61. doi: 10.1038/s41590-021-01084-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Thieme CJ, Anft M, Paniskaki K, et al. Robust T cell response toward spike, membrane, and nucleocapsid SARS‐CoV‐2 proteins is not associated with recovery in critical COVID‐19 patients. Cell Rep Med. 2020;1(6):100092. doi: 10.1016/j.xcrm.2020.100092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Mathew D, Giles JR, Baxter AE, et al. Deep immune profiling of COVID‐19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508):eabc8511. doi: 10.1126/science.abc8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Li S, Jiang L, Li X, et al. Clinical and pathological investigation of patients with severe COVID‐19. JCI Insight. 2020;5(12). doi: 10.1172/jci.insight.138070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Radermecker C, Detrembleur N, Guiot J, et al. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID‐19. J Exp Med. 2020;217(12). doi: 10.1084/jem.20201012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Schurink B, Roos E, Radonic T, et al. Viral presence and immunopathology in patients with lethal COVID‐19: a prospective autopsy cohort study. Lancet Microbe. 2020;1(7):e290‐e299. doi: 10.1016/s2666-5247(20)30144-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Masso‐Silva JA, Moshensky A, Lam MTY, et al. Increased peripheral blood neutrophil activation phenotypes and NETosis in critically ill COVID‐19 patients: a case series and review of the literature. Clin Infect Dis. 2021;21(10):ciab437. doi: 10.1093/cid/ciab437 [DOI] [Google Scholar]
- 140. Liao M, Liu Y, Yuan J, et al. Single‐cell landscape of bronchoalveolar immune cells in patients with COVID‐19. Nat Med. 2020;26(6):842‐844. doi: 10.1038/s41591-020-0901-9 [DOI] [PubMed] [Google Scholar]
- 141. Oja AE, Saris A, Ghandour CA, et al. Divergent SARS‐CoV‐2‐specific T‐ and B‐cell responses in severe but not mild COVID‐19 patients. Eur J Immunol. 2020;50(12):1998‐2012. doi: 10.1002/eji.202048908 [DOI] [PubMed] [Google Scholar]
- 142. Szabo PA, Dogra P, Gray JI, et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell‐driven lung inflammation in severe COVID‐19. Immunity. 2021;54(4):797‐814. doi: 10.1016/j.immuni.2021.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Grant RA, Morales‐Nebreda L, Markov NS, et al. Circuits between infected macrophages and T cells in SARS‐CoV‐2 pneumonia. Nature. 2021;590(7847):635‐641. doi: 10.1038/s41586-020-03148-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Soresina A, Moratto D, Chiarini M, et al. Two X‐linked agammaglobulinemia patients develop pneumonia as COVID‐19 manifestation but recover. Pediatr Allergy Immunol. 2020;31(5):565‐569. doi: 10.1111/pai.13263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Hughes R, Whitley L, Fitovski K, et al. COVID‐19 in ocrelizumab‐treated people with multiple sclerosis. Mult Scler Relat Dis. 2021;49:102725. doi: 10.1016/j.msard.2020.102725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Coyle PK, Gocke A, Vignos M, Newsome SD. Vaccine considerations for multiple sclerosis in the COVID‐19 era. Adv Ther. 2021;38(7):3550‐3588. doi: 10.1007/s12325-021-01761-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Yek C, Warner S, Wiltz JL, et al. Risk factors for severe COVID‐19 outcomes among persons aged ≥18 years who completed a primary COVID‐19 vaccination series—465 health care facilities, United States, December 2020–October 2021. Mmwr Morbidity Mortal Wkly Rep. 2022;71(1):19‐25. doi: 10.15585/mmwr.mm7101a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Apostolidis SA, Kakara M, Painter MM, et al. Cellular and humoral immune responses following SARS‐CoV‐2 mRNA vaccination in patients with multiple sclerosis on anti‐CD20 therapy. Nat Med. 2021;27(4):2001‐11. doi: 10.1038/s41591-021-01507-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Sormani MP, Salvetti M, Labauge P, et al. DMTs and Covid‐19 severity in MS: a pooled analysis from Italy and France. Ann Clin Transl Neur. 2021;8(8):1738‐1744. doi: 10.1002/acn3.51408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Braun J, Loyal L, Frentsch M, et al. SARS‐CoV‐2‐reactive T cells in healthy donors and patients with COVID‐19. Nature. 2020;587(7833):270‐274. doi: 10.1038/s41586-020-2598-9 [DOI] [PubMed] [Google Scholar]
- 151. Bert NL, Tan AT, Kunasegaran K, et al. SARS‐CoV‐2‐specific T cell immunity in cases of COVID‐19 and SARS, and uninfected controls. Nature. 2020;584(7821):457‐462. doi: 10.1038/s41586-020-2550-z [DOI] [PubMed] [Google Scholar]
- 152. Mateus J, Grifoni A, Tarke A, et al. Selective and cross‐reactive SARS‐CoV‐2 T cell epitopes in unexposed humans. Sci New York NY. 2020;370(6512):89‐94. doi: 10.1126/science.abd3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Mateus J, Dan JM, Zhang Z, et al. Low‐dose mRNA‐1273 COVID‐19 vaccine generates durable memory enhanced by cross‐reactive T cells. Sci New York NY. 2021;374(6566):eabj9853. doi: 10.1126/science.abj9853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Loyal L, Braun J, Henze L, et al. Cross‐reactive CD4+ T cells enhance SARS‐CoV‐2 immune responses upon infection and vaccination. Science;374(6564). 2021;eabh1823. doi: 10.1126/science.abh1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Sette A, Crotty S. Pre‐existing immunity to SARS‐CoV‐2: the knowns and unknowns. Nat Rev Immunol. 2020;20(8):457‐458. doi: 10.1038/s41577-020-0389-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Sagar M, Reifler K, Rossi M, et al. Recent endemic coronavirus infection is associated with less severe COVID‐19. J Clin Invest. 2020;131(1):e143380. doi: 10.1172/jci143380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Swadling L, Diniz MO, Schmidt NM, et al. Pre‐existing polymerase‐specific T cells expand in abortive seronegative SARS‐CoV‐2. Nature. 2021;601:1‐8. doi: 10.1038/s41586-021-04186-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Antunes R da S, Pallikkuth S, Williams E, et al. Differential T cell reactivity to endemic coronaviruses and SARS‐CoV‐2 in community and health care workers. J Infect Dis 2021;224(1):jiab176. doi: 10.1093/infdis/jiab176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Chandrashekar A, Liu J, Martinot AJ, et al. SARS‐CoV‐2 infection protects against rechallenge in rhesus macaques. Science. 2020;369(6505):812‐817. doi: 10.1126/science.abc4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Deng W, Bao L, Liu J, et al. Primary exposure to SARS‐CoV‐2 protects against reinfection in rhesus macaques. Science. 2020;369:818‐823. doi: 10.1126/science.abc5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Lumley SF, O'Donnell D, Stoesser NE, et al. Antibody status and incidence of SARS‐CoV‐2 infection in health care workers. New Engl J Med. 2020;384(6):533‐540. doi: 10.1056/nejmoa2034545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Abu‐Raddad LJ, Chemaitelly H, Coyle P, et al. SARS‐CoV‐2 antibody‐positivity protects against reinfection for at least seven months with 95% efficacy. Eclinicalmedicine. 2021;35:100861. doi: 10.1016/j.eclinm.2021.100861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Hall VJ, Foulkes S, Charlett A, et al. SARS‐CoV‐2 infection rates of antibody‐positive compared with antibody‐negative health‐care workers in England: a large, multicentre, prospective cohort study (SIREN). Lancet. 2021;397(10283):1459‐1469. doi: 10.1016/s0140-6736(21)00675-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. León TM, Dorabawila V, Nelson L, et al. COVID‐19 cases and hospitalizations by COVID‐19 vaccination status and previous COVID‐19 diagnosis—California and New York, May–November 2021. MMWR Morbidity Mortal Wkly Rep. 2022;71(4):125‐131. doi: 10.15585/mmwr.mm7104e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Walls AC, Sprouse KR, Joshi A, et al. Delta breakthrough infections elicit potent, broad and durable neutralizing antibody responses. bioRxiv. Published online 2022. doi: 10.1101/2021.12.08.471707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Altarawneh HN, Chemaitelly H, Hasan MR, et al. Protection against the Omicron variant from previous SARS‐CoV‐2 infection. New Engl J Med. 2022;386(13):1288‐1290. doi: 10.1056/nejmc2200133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Tarke A, Coelho CH, Zhang Z, et al. SARS‐CoV‐2 vaccination induces immunological T cell memory able to cross‐recognize variants from Alpha to Omicron. Cell. 2022;185(5):847‐859. doi: 10.1016/j.cell.2022.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Keeton R, Tincho MB, Ngomti A, et al. T cell responses to SARS‐CoV‐2 spike cross‐recognize Omicron. Nature. 2022;603(7901):488‐492. doi: 10.1038/s41586-022-04460-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Gao Y, Cai C, Grifoni A, et al. Ancestral SARS‐CoV‐2‐specific T cells cross‐recognize the Omicron variant. Nat Med. 2022;28(3):472‐476. doi: 10.1038/s41591-022-01700-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Tarke A, Sidney J, Methot N, et al. Impact of SARS‐CoV‐2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med. 2021;2(7):100355. doi: 10.1016/j.xcrm.2021.100355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory CD8+ T cells. Nat Immunol. 2013;14(5):509‐513. doi: 10.1038/ni.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Schenkel JM, Fraser KA, Beura LK, et al. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science (New York, NY). 2014;346(6205):98‐101. doi: 10.1126/science.1254536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Strutt TM, McKinstry KK, Dibble JP, et al. Memory CD4+ T cells induce innate responses independently of pathogen. Nat Med. 2010;16(5):558‐564‐1p following 564. doi: 10.1038/nm.2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Pardi N, Hogan MJ, Naradikian MS, et al. Nucleoside‐modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med. 2018;215(6):1571‐1588. doi: 10.1084/jem.20171450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Alameh MG, Tombácz I, Bettini E, et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity. 2021;54(12):2877‐2892.e7. doi: 10.1016/j.immuni.2021.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Lederer K, Bettini E, Parvathaneni K, et al. Germinal center responses to SARS‐CoV‐2 mRNA vaccines in healthy and immunocompromised individuals. medRxiv. 2021. doi: 10.1101/2021.09.16.21263686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Kim W, Zhou JQ, Horvath SC, et al. Germinal centre‐driven maturation of B cell response to mRNA vaccination. Nature. 2022;604(7904):141‐145. doi: 10.1038/s41586-022-04527-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Mudd PA, Minervina AA, Pogorelyy MV, et al. SARS‐CoV‐2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell. 2022;185(4):603‐613. doi: 10.1016/j.cell.2021.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Sadoff J, Gars ML, Shukarev G, et al. Interim results of a phase 1–2a trial of Ad26.COV2.S Covid‐19 vaccine. New Engl J Med. 2021;384(19):1824‐1835. doi: 10.1056/nejmoa2034201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Atmar RL, Lyke KE, Deming ME, et al. Homologous and heterologous Covid‐19 booster vaccinations. medRxiv. Published online 2021. doi: 10.1101/2021.10.10.21264827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Murugesan K, Jagannathan P, Pham TD, et al. Interferon‐gamma release assay for accurate detection of SARS‐CoV‐2 T cell response. Clin Infect Dis. 2021;73(9):e3130‐e3132. doi: 10.1093/cid/ciaa1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Goletti D, Petrone L, Manissero D, et al. The potential clinical utility of measuring SARS‐CoV‐2‐specific T‐cell responses. Clin Microbiol Infec. 2021;27(12):1784‐1789. doi: 10.1016/j.cmi.2021.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Tan AT, Lim JME, Bert NL, et al. Rapid measurement of SARS‐CoV‐2 spike T cells in whole blood from vaccinated and naturally infected individuals. J Clin Invest. 2021;131(17):e152379. doi: 10.1172/jci152379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Painter MM, Mathew D, Goel RR, et al. Rapid induction of antigen‐specific CD4+ T cells is associated with coordinated humoral and cellular immunity to SARS‐CoV‐2 mRNA vaccination. Immunity. 2021;54(9):2133‐2142.e3. doi: 10.1016/j.immuni.2021.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Corbett KS, Flynn B, Foulds KE, et al. Evaluation of the mRNA‐1273 vaccine against SARS‐CoV‐2 in nonhuman primates. New Engl J Med. 2020;383(16):1544‐1555. doi: 10.1056/nejmoa2024671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Thomas SJ, Moreira ED, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine through 6 months. New Engl J Med. 2021;385(19):1761‐1773. doi: 10.1056/nejmoa2110345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Sahly HME, Baden LR, Essink B, et al. Efficacy of the mRNA‐1273 SARS‐CoV‐2 vaccine at completion of blinded phase. New Engl J Med. 2021;385(19):1774‐1785. doi: 10.1056/nejmoa2113017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Corbett KS, Nason MC, Flach B, et al. Immune correlates of protection by mRNA‐1273 vaccine against SARS‐CoV‐2 in nonhuman primates. Science. 2021;373(6561):eabj0299. doi: 10.1126/science.abj0299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Chiu C, Killingley B, Mann A, et al. Safety, tolerability and viral kinetics during SARS‐CoV‐2 human challenge. 2022. doi: 10.21203/rs.3.rs-1121993/v1 [DOI] [PubMed]
- 190. Chandrashekar A, Yu J, McMahan K, et al. Vaccine protection against the SARS‐CoV‐2 Omicron variant in macaques. Cell. 2022;185(9):1549‐1555.e11. doi: 10.1016/j.cell.2022.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Ishii H, Nomura T, Yamamoto H, et al. Neutralizing‐antibody‐independent SARS‐CoV‐2 control correlated with intranasal‐vaccine‐induced CD8+ T cell responses. Cell Rep Med. 2022;3(2):100520. doi: 10.1016/j.xcrm.2022.100520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Vikkurthi R, Ansari A, Pai AR, et al. Inactivated virus vaccine BBV152/Covaxin elicits robust cellular immune memory to SARS‐CoV‐2 and variants of concern. Medrxiv. 2021. doi: 10.1101/2021.11.14.21266294 [DOI] [PubMed] [Google Scholar]
- 193. Goldberg Y, Mandel M, Bar‐On YM, et al. Waning immunity after the BNT162b2 vaccine in Israel. New Engl J Medicine. 2021;385(24):2114228. doi: 10.1056/nejmoa2114228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Tartof SY, Slezak JM, Fischer H, et al. Effectiveness of mRNA BNT162b2 COVID‐19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet Lond Engl. 2021;398(10309):1407‐1416. doi: 10.1016/s0140-6736(21)02183-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Danza P, Koo TH, Haddix M, et al. SARS‐CoV‐2 infection and hospitalization among adults aged ≥18 years, by vaccination status, before and during SARS‐CoV‐2 B.1.1.529 (Omicron) variant predominance—Los Angeles County, California, November 7, 2021–January 8, 2022. MMWR Morbidity Mortal Wkly Rep. 2022;71(5):177‐181. doi: 10.15585/mmwr.mm7105e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Gaebler C, Wang Z, Lorenzi JCC, et al. Evolution of antibody immunity to SARS‐CoV‐2. Nature. 2021;591(7851):639‐644. doi: 10.1038/s41586-021-03207-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Turner JS, Day A, Alsoussi WB, et al. SARS‐CoV‐2 viral RNA shedding For More Than 87 days in an individual with an impaired CD8+ T cell response. Front Immunol. 2021;11:618402. doi: 10.3389/fimmu.2020.618402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Rapeport G, Smith E, Gilbert A, Catchpole A, McShane H, Chiu C. SARS‐CoV‐2 human challenge studies—establishing the model during an evolving pandemic. N Engl J Med. 2021. Sept.;385(11):961‐964. doi: 10.1056/NEJMp2106970 [DOI] [PubMed] [Google Scholar]


