Abstract
The attachment organelle of Mycoplasma pneumoniae is a polar, tapered cell extension containing an intracytoplasmic, electron-dense core. This terminal structure is the leading end in gliding motility, and its duplication is thought to precede cell division, raising the possibility that mutations affecting cytadherence also confer a defect in motility or cell development. Mycoplasma surface protein P30 is associated with the attachment organelle, and P30 mutants II-3 and II-7 do not cytadhere. In this study, the recombinant wild-type but not the mutant II-3 p30 allele restored cytadherence when transformed into P30 mutants by recombinant transposon delivery. The mutations associated with loss of P30 in mutant II-3 and reacquisition of P30 in cytadhering revertants thereof were identified by nucleotide sequencing of the p30 gene. Morphological abnormalities that included ovoid or multilobed cells having a poorly defined tip structure were associated with loss of P30. Digital image analysis confirmed quantitatively the morphological differences noted visually. Transformation of the P30 mutants with the wild-type p30 allele restored a normal morphology, as determined both visually and by digital image analysis, suggesting that P30 plays a role in mycoplasma cell development. Finally, the P30 mutants localized the adhesin protein P1 to the terminal organelle, indicating that P30 is not involved in P1 trafficking but may be required for its receptor-binding function.
The attachment organelle of the cell wall-less prokaryote Mycoplasma pneumoniae mediates colonization of respiratory epithelium, leading to bronchitis and atypical pneumonia in humans (32). This differentiated structure is defined ultrastructurally as a polar, tapered cell extension containing an intracytoplasmic electron-dense core (3) which is part of a cytoskeleton-like scaffolding in the mycoplasma cell (9, 28). The adhesin protein P1 is densely clustered on the mycoplasma surface at the attachment organelle (32). In addition, several mycoplasma proteins have been identified, the loss of which is accompanied by the inability to cytadhere, the failure to localize adhesin P1 to the attachment organelle, an altered morphology, and avirulence (1, 20, 22, 23). The terminal organelle is the leading end as mycoplasmas move by gliding motility (17), and its duplication is thought to precede cell division (4). The multifunctional complexity of the terminal organelle raises the possibility that mutations affecting cytadherence have pleiotropic consequences involving cell growth, development, and/or motility (21).
Like the adhesin P1, protein P30 is associated with the attachment organelle of M. pneumoniae (2), but its function is not clear. P30 is synthesized as a 29.7-kDa polypeptide having a positively charged amino terminus followed by a hydrophobic domain of 23 residues (6, 7) that may serve as a signal peptide (28). A second hydrophobic domain follows 40 residues after the first, but P30 is otherwise predicted to be highly hydrophilic. Antibody and protease accessibility studies indicate that P30 is oriented in the mycoplasma membrane with the C terminus on the cell exterior (7, 19, 26). The C-terminal domain of P30 shows substantial sequence homology with the C terminus of the adhesin P1 and exhibits immunological cross-reactivity with fibrinogen, keratin, and myosin, perhaps accounting for autoimmune sequelae associated with M. pneumoniae infections (7). Complete loss of P30 (mutant II-3) or a 144-bp deletion near the 3′ end of the p30 gene (mutant II-7) results in the inability to cytadhere (2, 7, 25, 26). Monoclonal antibodies directed against the extracellular domain of P30 block adherence (2), but this may reflect stearic interference with neighboring molecules on the mycoplasma surface.
In this study, we identified the mutations that are associated with loss of P30 in mutant II-3 and the reacquisition of P30 in cytadhering revertants thereof. Furthermore, transformation of this mutant with a recombinant transposon expressing the cloned wild-type p30 allele, but not the mutant allele, restored hemadsorption. Examination of P30 mutants by scanning and transmission electron microscopy (SEM and TEM, respectively) and digital image analysis revealed dramatic morphological abnormalities, which were corrected by introduction of recombinant wild-type P30. Finally, in contrast to other noncytadhering mutants examined to date, the P30 mutants appear to localize the adhesin P1 to the attachment organelle, suggesting that P30 is required for P1 function rather than its trafficking to the tip structure.
MATERIALS AND METHODS
Organisms and culture conditions.
The M. pneumoniae strains used in this study included wild-type strain M129, broth passage 17 (27), and the spontaneously arising class II mutants II-3 and II-7, derived from M129 (7, 23). Mycoplasmas were cultured in Hayflick medium (13) at 37°C to mid-log phase, when the phenol red pH indicator became orange-red. Gentamicin was included at 18 μg/ml for the growth of M. pneumoniae transformed with recombinant Tn4001mod (11, 12, 18). Mycoplasmas were washed and harvested by centrifugation as described previously (14).
Nucleotide sequence analysis.
DNA sequencing was carried out by the chain termination method, using custom synthesized primers, dye terminators, and an Applied Biosystems (Foster City, Calif.) model 373A automated sequencer according to the manufacturer’s instructions.
Recombinant transposon construction and transformation of M. pneumoniae.
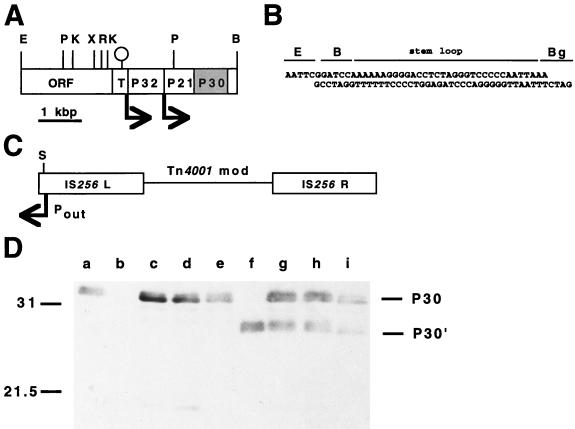
Plasmid pSP72 (Promega Corp., Madison, Wis.) was used for initial cloning steps. Previous studies suggested that an outward-reading promoter in IS256L of Tn4001mod (Fig. 1C) can affect the expression of genes cloned into this transposon (11). Therefore, a transcriptional terminator (31) was generated by annealing two long oligonucleotides (Fig. 1B) and ligated into the EcoRI and BglII sites in pSP72 to generate pKV91. The XbaI-BamHI fragment containing the gene for P30 (Fig. 1A) was cloned into the corresponding sites in pKV91 to generate pKV112. The p30 gene and adjacent transcriptional terminator were excised intact by digestion of pKV112 with EcoRV and BglII. After the single-stranded end at the BglII site was filled in with deoxynucleoside triphosphate and DNA polymerase, this fragment was cloned into the SmaI site of Tn4001mod in pISM2062 (Fig. 1C) (18) to generate pKV124. Plasmid DNA was purified in each case by using pZ523 columns (5′→3′, Inc., Boulder, Colo.). Mycoplasma cultures were transformed by electroporation and expanded as described previously (14, 15).
FIG. 1.
Construction of recombinant transposon Tn4001 for delivery of the cloned p30 gene into M. pneumoniae. (A) Restriction map of the region containing the genes for P32, P21, and P30. A predicted terminator is indicated by the stem-loop; arrows correspond to putative promoters. Restriction sites: B, BamHI; E, EcoRI; K, KpnI; P, PstI; R, EcoRV; X, XbaI. T indicates Ser-tRNA genes. The oligonucleotide containing a transcriptional terminator (B) was cloned into the EcoRI and BglII sites of pSP72 (Promega) to create pKV91. Restriction sites: B, BamHI; Bg, BglII; E, EcoRI. This stem-loop was introduced in order to minimize potential inhibition of P30 production due to an outward-reading promoter (Pout) in IS256L of Tn4001mod (C). The wild-type p30 allele was cloned into the XbaI and BamHI sites of pKV91 to yield pKV112, which was digested with EcoRV and BglII to release the p30 allele and downstream terminator for cloning into the SmaI site (S) of Tn4001mod (5) to generate pKV124. The relative location and orientation of the Pout promoter are indicated, as are the IS256 elements of this composite transposon. (D) Western immunoblotting analysis of II-3 and II-7 transformants for production of recombinant P30. The anti-P30 serum was specific for residues 103 to 181 (25). Lanes: a, wild-type M. pneumoniae; b, mutant II-3; c to e, transformants of II-3 with pKV124; f, mutant II-7; g to i, transformants of II-7 with pKV124. Omission of the transcriptional terminator from pKV124 had no effect on production of recombinant P30 by M. pneumoniae transformants (data not shown). Positions of protein size standards are given in kilodaltons on the left; positions of P30 and P30′ are indicated on the right.
Characterization of transformants.
Mycoplasma chromosomal DNA was extracted and probed by Southern blot hybridization (11). DNA probes specific for the gentamicin resistance gene, the gene encoding P30, and a region of pISM2062 outside Tn4001mod (18) were labeled by random-primed incorporation of digoxigenin-dUTP (Boehringer Mannheim Corp., Indianapolis, Ind.), using Klenow enzyme. Alkaline phosphatase-conjugated antidigoxigenin antibodies and 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (Boehringer Mannheim) were used as specified by the manufacturer to detect hybridization.
Mycoplasma protein profiles were analyzed by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 3% acrylamide stacking gel and a 12% separating gel (12). Mycoplasma cell pellets were prepared for electrophoresis, and proteins were visualized by silver staining or analyzed by Western immunoblotting (11) with anti-P30 antiserum (25). Antisera were also prepared against four-branch multiple antigenic peptides (MAPs) (37) corresponding to portions of P30 as described in more detail below. MAPs were synthesized by 9-fluorenylmethoxy carbonyl-based solid-phase technology, using an Advanced Chem Tech (Louisville, Ky.) model 350 synthesizer according to the manufacturer’s protocol. Immunoreactive protein bands were visualized by using horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse immunoglobulin G (1:1,000) and color development reagent (Bio-Rad Corp., Hercules, Calif.).
Mycoplasma colonies were screened for hemadsorption (HA) or evaluated by a quantitative HA assay as described in detail elsewhere (22, 23).
SEM and TEM.
Mycoplasma cell pellets were suspended in fresh Hayflick medium, passed through a 25-gauge needle three times to disperse cell aggregates, and filtered (0.45-μm pore size) to generate largely single-cell suspensions. These suspensions were used to seed individual wells of 24-well dishes containing either Formvar-coated, carbon-coated nickel grids (for TEM) or glass coverslips (for SEM), in either case precoated with poly-l-Lys as described previously (35). After 1 h at 37°C, grids and coverslips were either removed and processed for fixation (1-h time point) or transferred to fresh Hayflick medium and incubated further. Samples were fixed at the indicated time points in 1% paraformaldehyde–1% glutaraldehyde–0.1% picric acid–0.1 M sodium cacodylate (pH 7.2) for 45 min at 4°C and then rinsed twice with 0.1 M sodium cacodylate buffer (pH 7.2). For TEM, grids were rinsed with distilled water, allowed to air dry, and examined in a Philips 400 TEM. For SEM, coverslips were treated sequentially (10 min for each treatment) with 30, 50, 70, 85, 95, and 100% (three times) ethanol, critical-point dried, and sputter-coated with 20-nm-diameter gold for examination in a Philips 505 SEM or coated with chromium for examination in a LEO982 SEM.
Mycoplasmas cultured on electron microscopy grids were examined by immunoelectron microscopy as described in detail previously (12), using monoclonal anti-P1 antibodies generously provided by Steve Geary, University of Connecticut. For the analysis of thin sections of wild-type M. pneumoniae and mutant II-3, mycoplasma cells were rinsed in phosphate-buffered saline (pH 7.4) and fixed for 1 h at 5°C in 0.2 M cacodylate buffer (pH 7.2)–2% glutaraldehyde. Fixed mycoplasmas were rinsed three times with 0.1 M cacodylate (pH 7.2)–5% sucrose and then postfixed with 1% OsO4 for 1 h at 5°C. Samples were dehydrated in an ethanol series and rinsed twice for 15 min each time in propylene oxide and for 45 min each in 2:1, 1:1, and 1:2 propylene oxide-Epon 812 (Polysciences, Inc., Warrington, Pa.), followed by 100% Epon. Embedded samples were polymerized at 60°C for 24 h, and silver-gold thin sections were cut with a diamond knife on a Sorvall microtome and poststained with uranyl acetate and lead citrate.
Digital image analysis.
Mycoplasma morphology was analyzed quantitatively by digital capture of TEM images of individual cells. Cultures were examined in a double-blind manner, and images of at least 200 cells for each culture at each time point were analyzed by using the IM series morphometric and densitometric system program (Analytical Imaging Concepts, Irving, Calif.). A shape factor was calculated for each cell as the measurement of the greatest distance along an axis perpendicular to the length (breadth), divided by cell length. Comparative studies using hand-drawn images representative of mycoplasma morphologies were conducted. Shape factors were calculated with each image in four different orientations by this program and evaluated for consistency. A coccoid image yielded shape factors of 0.9 to 1.0, an extended oval image yielded shape factors of 0.4 to 0.5, and a filamentous image representative of typical wild-type M. pneumoniae cells yielded a shape factor of approximately 0.1. The only notable weakness in the program was the inability to distinguish a smooth round image from a round image with multiple lobes.
RESULTS
Transformation with recombinant p30 restored cytadherence.
Recombinant transposons were used to shuttle the wild-type p30 gene into M. pneumoniae, yielding transformants having both resident and recombinant p30 alleles. Several transformants were analyzed for each to control for possible positional effects of transposon insertion. Southern blot hybridization analysis confirmed the presence of both alleles and established that insertion was by transposition rather than whole-plasmid integration (data not shown), consistent with previous studies using recombinant transposons in M. pneumoniae (11, 12).
Transformants of mutants II-3 and II-7 with the recombinant wild-type p30 allele synthesized P30 at levels that were equal to or greater than that seen in wild-type M. pneumoniae, as demonstrated by SDS-PAGE and Western immunoblotting (Fig. 1D). As expected, the truncated P30 (P30′) was present in mutant II-7 and transformants thereof, but for reasons that are not clear, the level of P30′ in these transformants was generally lower than that in untransformed II-7. The same result was observed previously in comparable transformants of mutant M6 (12), which produces a truncated P30 due to a deletion nearly identical to that described for II-7 (25). All transformants of mutants II-3 and II-7 examined that synthesized recombinant P30 were HA+ by direct colony screening. Furthermore, these transformants exhibited comparable HA levels when examined quantitatively (data not shown). Thus, a direct correlation was observed between restoration of wild-type P30 in these transformants and the ability to cytadhere. Finally, the recombinant p30 gene in pKV124 was truncated by removing an internal NarI fragment within the repeat region at the 3′ end of the gene. When transformed into mutant II-3, this subclone failed to restore full-length P30 or HA (data not shown), indicating that P30 was produced from the recombinant rather than the resident p30 allele.
Dallo et al. (7) previously reported no difference in the nucleotide sequence between wild-type M. pneumoniae and mutant II-3 from approximately 1 kbp upstream of p30 through the p30 gene. However, transformation of mutant II-3 with the recombinant II-3 p30 allele cloned into the transposon vector failed to restore P30 production or confer an HA+ phenotype (data not shown). These observations raised the possibility of an upstream mutation affecting P30 synthesis in mutant II-3 or a sequencing error in the study by Dallo and coworkers (7).
Identification of the genetic defect in the P30-mutant II-3.
Nucleotide sequence analysis following PCR amplification revealed a frameshift due to the loss of a single adenine nucleotide at position 453 within the p30 gene (Fig. 2A). This mutation was confirmed by sequencing the p30 allele cloned directly from purified II-3 chromosomal DNA, and no other differences were found in the nucleotide sequence from the EcoRV site upstream of the gene for P32 through the BamHI site in hmw3 for wild-type and mutant II-3 M. pneumoniae. To verify further that this mutation was responsible for the mutant phenotype, we isolated three HA+ P30+ revertants of mutant II-3 by repeated enrichment for attachment to plastic. The p30 gene from these revertants was amplified by PCR and sequenced, confirming the original mutation in mutant II-3 while revealing that reversion had resulted from a second-site mutation within the p30 gene, restoring the proper open reading frame (ORF) (Fig. 2A). Taken together, these data clearly demonstrate that the loss of P30 and cytadherence in mutant II-3 is due to a frameshift in the p30 gene.
FIG. 2.
(A) Nucleotide sequence comparison of the indicated portion of the p30 gene of wild-type M. pneumoniae, the HA− mutant II-3, and an HA+ revertant of II-3 (II-3R1). The adenine nucleotide present in the wild-type sequence but absent from the II-3 sequence is shown in bold, as is the cytosine nucleotide absent from the wild-type and II-3 sequences but present in the revertant. Numbers correspond to nucleotide positions relative to the beginning of the p30 ORF. (B) Comparison of the deduced amino acid sequence of the indicated portions of P30 from wild-type, II-3, and II-3R1 M. pneumoniae. The sequence altered as a result of the second-site mutation is underlined. Numbers indicate amino acid positions from the deduced N terminus of P30. Accession no. AF090171 and AF090172 have been assigned to the mutant and revertant M. pneumoniae sequences shown in Fig. 2B.
Morphological defect correlates with loss of P30.
Cell morphology of wild-type M. pneumoniae and mutants II-3 and II-7 was characterized either directly by SEM or by TEM and digital image analysis. Mycoplasmas were cultured for the indicated times on glass coverslips or TEM grids that were precoated with poly-l-Lys to promote attachment of the mutants, which otherwise adhere poorly to inert surfaces. Wild-type cells frequently appeared elongated with a well-defined, tapered tip structure and long, filamentous tail (Fig. 3), consistent with previous descriptions (4). Ovoid or pleomorphic cells were present in the wild-type population in small numbers, but elongated mycoplasmas were clearly predominant.
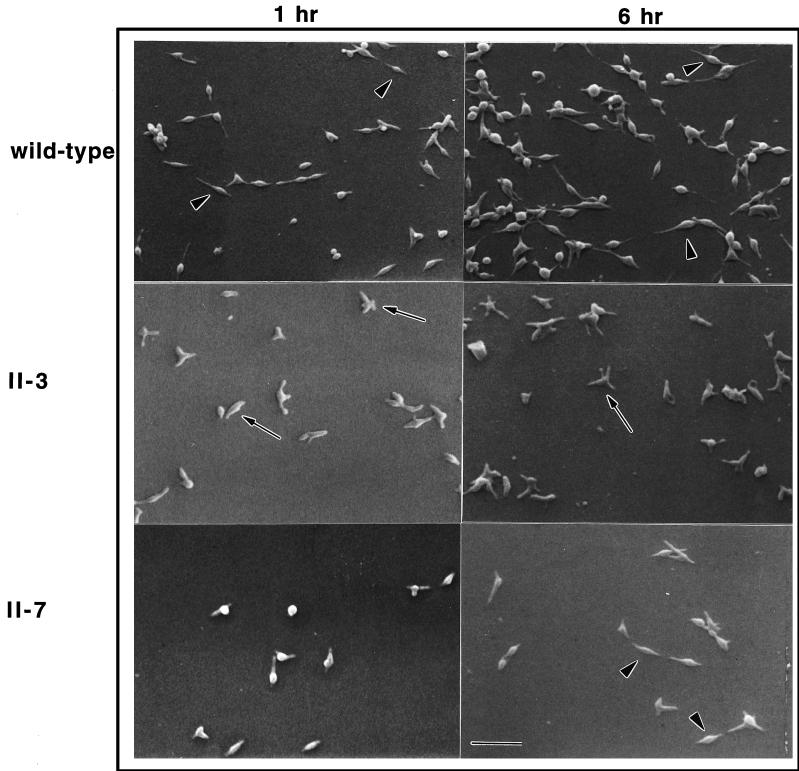
FIG. 3.
Morphological comparison of wild-type M. pneumoniae and noncytadhering mutants II-3 and II-7 by SEM after culture for the indicated times. Elongated cells having a tapered tip (arrowheads) were commonly seen in the wild-type cultures. Ovoid and multilobed cells (arrows) were seen with a much higher frequency in the mutant II-3 cultures. Cells in the mutant II-7 cultures were commonly ovoid initially but elongated by 6 h (arrowheads). Scale bar = 2.5 μm.
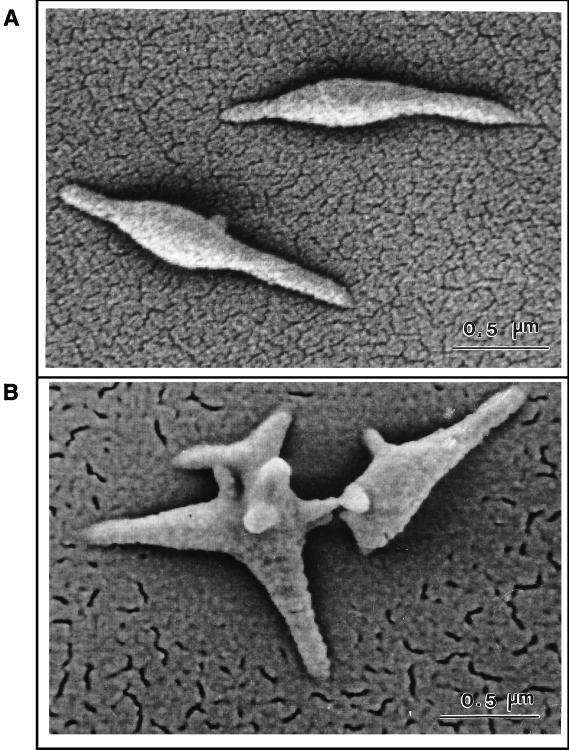
The cell morphology of cytadherence mutant II-3 was dramatically distinct from that of wild-type M. pneumoniae (Fig. 3). Mutant cells at 1 h were frequently ovoid, and cell extensions were short and thick when present, making it difficult at times to identify the tip structure. Cells exhibiting a wild-type morphology with an elongated, tapered tip were rare. By 6 h, a novel star-shaped morphology distinguished by multiple, short, broad lobes was common with this mutant. Examination at higher magnification (Fig. 4) confirmed that this star-shaped morphology represented individual cells and clearly established the contrasting appearance of wild-type M. pneumoniae and mutant II-3.
FIG. 4.
High-magnification SEM images of wild-type M. pneumoniae (A) and noncytadhering mutant II-3 (B). The bar in each panel represents 0.5 μm.
Cytadherence mutant II-7 produces a truncated P30 (P30′) lacking a portion of the extracellular, C-terminal domain that is dominated by Pro-rich repeats (7). This mutant exhibited features similar to those of both wild-type M. pneumoniae and mutant II-3 at the 1-h time point (Fig. 3). Specifically, some cells were beginning to elongate to form tapered filaments, but ovoid and multilobed cells were more common than with wild-type cultures. However, by 6 h mutant II-7 was generally more comparable morphologically to wild-type M. pneumoniae. Elongated cells predominated, with ovoid or pleomorphic cells present only in small numbers.
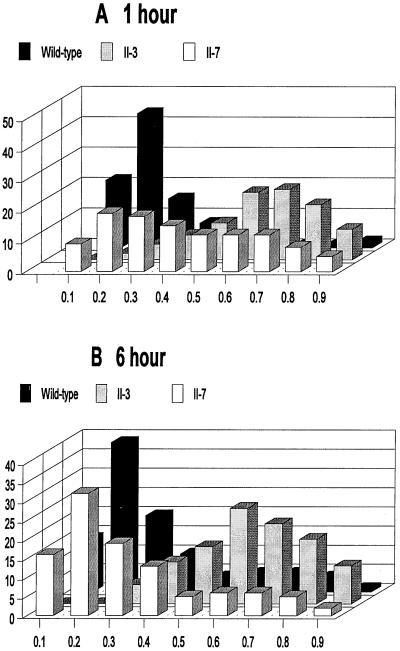
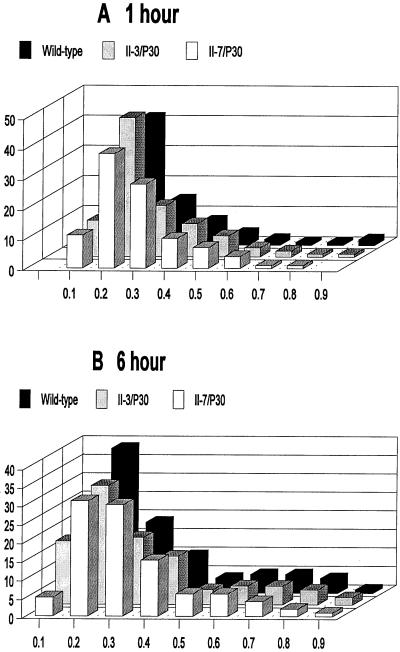
Morphological variability is typical of mycoplasma cultures (4); hence it was necessary to develop a means to compare populations quantitatively for morphological differences. Shape factors were calculated for wild-type and mutant mycoplasmas by digital image analysis of at least 200 individual cells at each time point. Shape factors were determined as the measurements of cell breadth divided by cell length, as defined in detail in Materials and Methods. Shape factor values approaching 1.0 and 0.0 reflect circular and linear, filamentous morphologies, respectively. Approximately 75% of the wild-type population exhibited a shape factor of 0.1 to 0.3 at both 1- and 6-h time points (Fig. 5), consistent with the filamentous forms typically exhibited by most wild-type mycoplasma cells at these time points. In contrast, over 60% of the population for mutant II-3 yielded a shape factor in the range 0.6 to 0.8 at both the 1- and 6-h time points, corresponding to a more ovoid morphology. This technique failed to distinguish smooth coccoid cells from the multilobed cells seen at 1 and 6 h, respectively, with mutant II-3. This limitation was confirmed in control experiments with hand-drawn images (see Materials and Methods). Nevertheless, digital image analysis of populations of wild-type and mutant cells underscored quantitatively the differences noted visually by SEM. Finally, >80% of the cells for mutant II-7 were evenly distributed across a shape factor range of 0.2 to 0.8 at 1 h. But by 6 h, approximately 70% of the population yielded shape factors in the range of 0.1 to 0.3, comparable to the shape factor distribution seen with wild-type M. pneumoniae and consistent with the morphological similarity noted by SEM.
FIG. 5.
Comparison of wild-type M. pneumoniae and mutants II-3 and II-7 cell morphology at 1-h (A) and 6-h (B) time points by TEM and digital image analysis. Shape factors were determined at each time point for a minimum of 200 cells for each culture. The frequencies for the indicated shape factor values were determined as percentages of the total, and the results from a representative experiment are shown.
The morphology of the mutant II-3 and II-7 transformants was also examined visually by SEM and quantitatively by TEM and digital image analysis. Visually the II-3 and II-7 transformants were indistinguishable from their wild-type counterparts by SEM (data not shown). The same was true for TEM and digital image analysis, where the shape factor distributions for II-3 and II-7 transformants were very similar to that of wild-type M. pneumoniae at both 1- and 6-h time points (Fig. 6).
FIG. 6.
Comparison by TEM and digital image analysis of the morphologies of wild-type M. pneumoniae and mutant II-3 and II-7 transformants having a recombinant wild-type p30 allele, determined as described in the legend to Fig. 5.
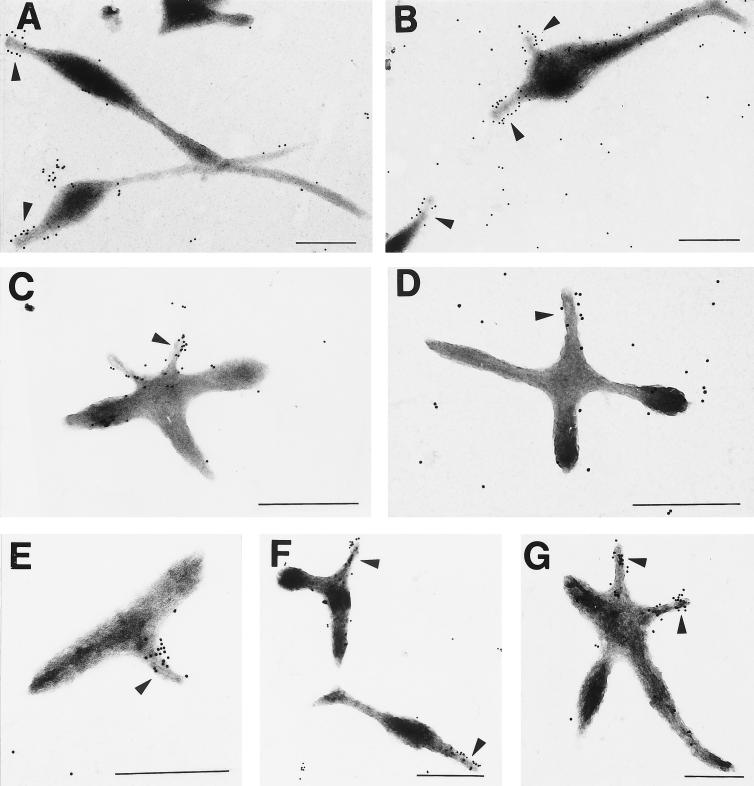
Analysis of thin sections of mutant II-3 by TEM confirmed the presence of the electron-dense core characteristic of the terminal organelle. Typically, the core structure was seen in this mutant in closer proximity to the cell body than in wild-type M. pneumoniae cells (data not shown). Usually one but occasionally two electron-dense cores were seen in a single cell, but examination of thin sections is limited by the ability to capture multiple tips in the same plane in a cell. For this reason we were unable to establish definitively that the multiple lobes associated with mutant II-3 at the 6-h time point did not each contain the core structure that defines the attachment organelle. However, anti-P1 antibodies generally labeled only one or occasionally two lobes on the star-shaped mutant II-3 cells (Fig. 7C and D), consistent with the single or occasional duplicate attachment organelle observed generally in wild-type cells. The same was true with mutant II-7 (Fig. 7E to G), for which P1 likewise localized primarily to the terminal organelle. Thus, unlike class I mutants (lacking HMW1-HMW3) and class III mutants (lacking A, B, and C), which fail to cluster P1 at the attachment organelle (1, 23), P1 trafficking appeared normal in the class II mutants.
FIG. 7.
Immunogold labeling of wild-type M. pneumoniae (A and B) and mutants II-3 (C and D) and II-7 (E to G) with monoclonal anti-P1 antibodies (1:75 dilution). Arrows indicate likely attachment organelles. Scale bar = 0.5 μm.
DISCUSSION
The results presented here clearly demonstrate that loss of P30 and the inability to cytadhere in M. pneumoniae mutant II-3 are due to a frameshift in the p30 gene. The recombinant wild-type p30 allele, but not that from the II-3 mutant, restored P30 production and cytadherence to P30 mutants when introduced by transposon delivery. Some variability in P30 levels was observed among the transformants and probably reflects a positional effect of transposon insertion on recombinant p30 expression. A second-site mutation in the p30 gene of HA+ revertants of II-3 restored the proper reading frame and wild-type levels of P30. These findings do not agree with the work of Dallo et al. (7), who reported no difference in the nucleotide sequences of the p30 gene in wild-type M. pneumoniae and mutant II-3. The cultures used in the two studies came from the same source (23) but may have differed in passage level; hence, the sequence discrepancy may be due to an additional mutation event with passage or may reflect a sequencing error in the Dallo study.
The mutation in II-3 yielded an alternate ORF ending only one nucleotide from the normal stop codon for the p30 gene and encoding an alternate P30 (Fig. 2B) predicted to have an antigenicity profile dramatically different from that of P30. This finding raised the possibility that a frameshift in the p30 gene represents a means for antigenic variation in M. pneumoniae. Antibodies directed against a central domain of P30 (Fig. 1), and antibodies specific for MAPs corresponding to two different regions of the deduced alternate P30 (data not shown), all failed to detect the predicted protein product in M. pneumoniae mutant II-3, suggesting that the mutant protein may be unstable and quickly turned over.
The adhesin P1 appeared to localize to the attachment organelle in mutants II-3 and II-7, consistent with recent findings for the M6 mutant transformed with recombinant hmw1 but lacking P30 (12). However, this was a departure from the immunogold labeling patterns observed previously with other cytadherence mutants, which fail to localize P1 to the terminal organelle (1). The inability of mutants II-3 and II-7 to cytadhere suggests that P1 is nonfunctional, despite localizing to what appears to be the attachment organelle in these mutants. This finding indicates that P1 trafficking to the terminal organelle is necessary but not sufficient for cytadherence. It is noteworthy that P1 lacks Cys residues (36), and given the proposed tripartite nature of its receptor-binding domains (8), P1 may require stabilizing in order to be functional. The final steps in protein folding often occur at the subcellular site where that protein functions, which for P1 is probably the tip structure. While antibody inhibition studies implicate P30 as an adhesin (2), data presented here raise the possibility that P30 is required for development of a functional attachment organelle.
The attachment organelle of M. pneumoniae is thought to function in cell development; hence, wild-type and mutant mycoplasma cells were compared to determine if loss or truncation of P30 also affected cell morphology. Wild-type cells typically formed tapered tip structures and filamentous tails within the first hour of incubation. With the exception of more coccoid forms, probably reflecting cells undergoing cell division, this morphology was maintained until cell density increased to the point of microcolony formation (Fig. 1 and data not shown). Mutant II-7, producing a truncated P30, exhibited an abnormal morphology early in culture but appeared more like wild-type cells with time. This is consistent with the appearance of the mutant M6 transformed with recombinant hmw1, as described previously (12). Poly-l-Lys coating permits mutant attachment to inert surfaces, but not necessarily by the same mechanism or with the same efficiency as that of wild-type M. pneumoniae cells. Thus, the slower morphological development of this mutant might be attributable to a potential difference in the capacity of this mutant to initiate adhesion and elongation on the glass surface.
The complete loss of P30 (mutant II-3) resulted in dramatic morphological changes, including predominantly ovoid or multilobed cells having an untapered tip structure. Cells having the typical, filamentous morphology characteristic of wild-type M. pneumoniae were rare in the mutant II-3 population. Conversely, cells exhibiting multiple filaments or otherwise atypical morphologies were seen only occasionally in wild-type cultures, as reported previously by Boatman (4). The reason for these exceptions within each population is not clear. Nevertheless, the major shift in the predominant morphology between wild-type and mutant II-3 M. pneumoniae is striking. Altered morphologies have been described previously for noncytadhering mycoplasma mutants (1, 12) but did not include the multiple filaments seen here. Furthermore, the present study provides a quantitative assessment of morphological differences seen visually within a mutant mycoplasma population and correlates those changes with a specific gene defect.
Mycoplasma cell morphology can be influenced by a variety of factors. For example, supplementation with unsaturated fatty acids induces filament formation in Mycoplasma gallisepticum (33), which, like M. pneumoniae, possesses a differentiated terminal organelle. Spiroplasmas are typically helical filaments in culture but are coccoid inside host cells, perhaps due to changes in ion concentrations (10). Binary fission is the characteristic mode of replication for M. pneumoniae, and formation of a second tip structure is thought to coincide with initiation of cell division (4). Multiple filaments might allow the cell to accommodate and redistribute increased cell mass in the same way that Rhizobium and Agrobacterium form multiple branches when cell division is blocked (24). Multiple branches were likewise common with Mycoplasma capricolum when DNA replication was inhibited by nucleoside starvation (34). In those cells the nucleoids commonly localized at the branch point of the cells. Multiple filaments might likewise result in M. pneumoniae if cell division or chromosome partitioning were impaired, for example, due to abnormal assembly of the tip organelle. The ovoid morphology and lack of tapering observed with mutant II-3 are consistent with a defect in organelle assembly, which may require P30 in order to yield a tip structure that can function normally in both cytadherence and cell division. Evaluation of nucleoid partitioning in mutant II-3 by phase-combined fluorescence microscopy may shed light on the nature of the developmental defect associated with loss of P30. Finally, the more normal morphology exhibited by mutant II-7 indicates that the C-terminal repeat domain of P30 is not required in cell development-related function. The N terminus of P30 probably lies in the cell interior, where it may interact with cytoskeletal elements during tip development or chromosome partitioning.
ACKNOWLEDGMENTS
We thank Cathy Kelloes for expert technical assistance, and we thank S. Geary and R. Herrmann for providing antibodies to P1 and P30, respectively.
This work was supported by Public Health Service research grants AI23362 and AI33396 from the National Institute for Allergy and Infectious Diseases to D.C.K.
REFERENCES
- 1.Baseman J B, Cole R M, Krause D C, Leith D K. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J Bacteriol. 1982;151:1514–1522. doi: 10.1128/jb.151.3.1514-1522.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baseman J B, Morrison-Plummer J, Drouillard D, Puelo-Scheppke B, Tryon V V, Holt S C. Identification of a 32-kilodalton protein of Mycoplasma pneumoniae associated with hemadsorption. Isr J Med Sci. 1987;23:474–479. [PubMed] [Google Scholar]
- 3.Biberfeld G, Biberfeld P. Ultrastructural features of Mycoplasma pneumoniae. J Bacteriol. 1970;102:855–861. doi: 10.1128/jb.102.3.855-861.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boatman E S. Morphology and ultrastructure of the mycoplasmatales. In: Barile M F, Razin S, editors. The mycoplasmas. 1. Cell biology. Washington, D.C: Academic Press; 1979. pp. 63–102. [Google Scholar]
- 5.Byrne M E, Rouch D A, Skurray R A. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene. 1989;81:361–367. doi: 10.1016/0378-1119(89)90197-2. [DOI] [PubMed] [Google Scholar]
- 6.Dallo S F, Chavoya A, Baseman J B. Characterization of the gene for a 30-kilodalton adhesin-related protein of Mycoplasma pneumoniae. Infect Immun. 1990;58:4163–4165. doi: 10.1128/iai.58.12.4163-4165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dallo S F, Lazzell A L, Chavoya A, Reddy S P, Baseman J B. Biofunctional domains of the Mycoplasma pneumoniae P30 adhesin. Infect Immun. 1996;64:2595–2601. doi: 10.1128/iai.64.7.2595-2601.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerstenecker B, Jacobs E. Topological mapping of the P1-adhesin of Mycoplasma pneumoniae with adherence-inhibiting monoclonal antibodies. J Gen Microbiol. 1990;136:471–476. doi: 10.1099/00221287-136-3-471. [DOI] [PubMed] [Google Scholar]
- 9.Göbel U, Speth V, Bredt W. Filamentous structures in adherent Mycoplasma pneumoniae cells treated with nonionic detergents. J Cell Biol. 1981;91:537–543. doi: 10.1083/jcb.91.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackett K J, Clark T B. Ecology of spiroplasmas. In: Whitcomb R F, Tully J G, editors. The mycoplasmas. Vol. 5. New York, N.Y: Academic Press; 1989. pp. 113–200. [Google Scholar]
- 11.Hahn T-W, Krebes K A, Krause D C. Expression in Mycoplasma pneumoniae of the recombinant gene encoding the cytadherence-associated protein HMW1 and identification of HMW4 as a product. Mol Microbiol. 1996;19:1085–1093. doi: 10.1046/j.1365-2958.1996.455985.x. [DOI] [PubMed] [Google Scholar]
- 12.Hahn T W, Wilby M J, Krause D C. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J Bacteriol. 1998;180:1270–1276. doi: 10.1128/jb.180.5.1270-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayflick L. Tissue cultures and mycoplasmas. Tex Rep Biol Med. 1965;23(Suppl. 1):285–303. [PubMed] [Google Scholar]
- 14.Hedreyda C T, Krause D C. Identification of a possible cytadherence regulatory locus in Mycoplasma pneumoniae. Infect Immun. 1995;63:3479–3483. doi: 10.1128/iai.63.9.3479-3483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedreyda C T, Lee K K, Krause D C. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid. 1993;30:170–175. doi: 10.1006/plas.1993.1047. [DOI] [PubMed] [Google Scholar]
- 16.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchoff H. Motility. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 289–306. [Google Scholar]
- 18.Knudtson K L, Minion F C. Construction of Tn4001lac derivatives to be used as promoter probe vectors in mycoplasmas. Gene. 1993;137:217–222. doi: 10.1016/0378-1119(93)90009-r. [DOI] [PubMed] [Google Scholar]
- 19.Krause, D. C. Unpublished data.
- 20.Krause D C. Mycoplasma pneumoniae cytadherence: unraveling the tie that binds. Mol Microbiol. 1996;20:247–253. doi: 10.1111/j.1365-2958.1996.tb02613.x. [DOI] [PubMed] [Google Scholar]
- 21.Krause D C. Mycoplasma pneumoniae cytadherence: organization and assembly of the attachment organelle. Trends Microbiol. 1998;6:15–18. doi: 10.1016/S0966-842X(97)01168-2. [DOI] [PubMed] [Google Scholar]
- 22.Krause D C, Baseman J B. Inhibition of Mycoplasma pneumoniae hemadsorption and attachment to respiratory epithelium by antibodies to a membrane protein. Infect Immun. 1983;39:1180–1186. doi: 10.1128/iai.39.3.1180-1186.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krause D C, Leith D K, Wilson R M, Baseman J B. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect Immun. 1982;35:809–817. doi: 10.1128/iai.35.3.809-817.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Latch J N, Margolin W. Generation of buds, swellings, and branches instead of filaments after blocking the cell cycle of Rhizobium meliloti. J Bacteriol. 1997;179:2373–2381. doi: 10.1128/jb.179.7.2373-2381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Layh-Schmitt G, Hilbert H, Pirkl E. A spontaneous hemadsorption-negative mutant of Mycoplasma pneumoniae exhibits a truncated adhesin-related 30-kilodalton protein and lacks the cytadherence-accessory protein HMW1. J Bacteriol. 1995;177:843–846. doi: 10.1128/jb.177.3.843-846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Layh-Schmitt G, Himmelreich R, Leibfried U. The adhesin related 30-kDa protein of Mycoplasma pneumoniae exhibits size and antigen variability. FEMS Microbiol Lett. 1997;152:101–108. doi: 10.1111/j.1574-6968.1997.tb10415.x. [DOI] [PubMed] [Google Scholar]
- 27.Lipman R P, Clyde W A., Jr The interrelationship of virulence, cytadsorption and peroxide formation in Mycoplasma pneumoniae. Proc Soc Exp Biol Med. 1969;131:1163–1167. doi: 10.3181/00379727-131-34061. [DOI] [PubMed] [Google Scholar]
- 28.Meng K E, Pfister R M. Intracellular structures of Mycoplasma pneumoniae revealed after membrane removal. J Bacteriol. 1980;144:390–399. doi: 10.1128/jb.144.1.390-399.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagarajan V. Protein secretion. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 713–726. [Google Scholar]
- 30.Popham P L, Hahn T-W, Krebes K A, Krause D C. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs post-translationally. Proc Natl Acad Sci USA. 1997;94:13979–13984. doi: 10.1073/pnas.94.25.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 32.Razin S, Jacobs E. Mycoplasma adhesion. J Gen Microbiol. 1992;138:407–422. doi: 10.1099/00221287-138-3-407. [DOI] [PubMed] [Google Scholar]
- 33.Rodman A W, Mitchell A. Nutrition, growth, and reproduction. In: Barile M F, Razin S, editors. The mycoplasmas. 1. Cell biology. Washington, D.C: Academic Press; 1979. pp. 103–139. [Google Scholar]
- 34.Seto S, Miyata M. Abstracts of the 12th International Congress of the International Organization for Mycoplasmology. 1998. Cell reproduction in Mycoplasma capricolum, abstr. D.54. [Google Scholar]
- 35.Stevens M K, Krause D C. Cytadherence-accessory protein HMW3 of Mycoplasma pneumoniae is a component of the attachment organelle. J Bacteriol. 1992;174:4265–4274. doi: 10.1128/jb.174.13.4265-4274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su C J, Tryon V V, Baseman J B. Cloning and sequence analysis of cytadhesin P1 gene from Mycoplasma pneumoniae. Infect Immun. 1987;55:3023–3029. doi: 10.1128/iai.55.12.3023-3029.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam J P. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc Natl Acad Sci USA. 1988;85:5409–5413. doi: 10.1073/pnas.85.15.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]