Abstract
Homologous enzymes often exhibit different catalytic rates despite a fully conserved active site. The canonical view is that an enzyme sequence defines its structure and function and, more recently, that intrinsic protein dynamics at different time scales enable and/or promote catalytic activity. Here, we show that, using the protein tyrosine phosphatase PTP1B, residues surrounding the PTP1B active site promote dynamically coordinated chemistry necessary for PTP1B function. However, residues distant to the active site also undergo distinct intermediate time scale dynamics and these dynamics are correlated with its catalytic activity and thus allow for different catalytic rates in this enzyme family. We identify these previously undetected motions using coevolutionary coupling analysis and nuclear magnetic resonance spectroscopy. Our findings strongly indicate that conserved dynamics drives the enzymatic activity of the PTP family. Characterization of these conserved dynamics allows for the identification of novel regulatory elements (therapeutic binding pockets) that can be leveraged for the control of enzymes.
Conserved motions allow the PTP enzyme family to exhibit different catalytic rates.
INTRODUCTION
Enzymes catalyze chemical reactions that direct essential metabolic and signaling processes. Landmark studies led to the canonical view that a protein amino acid sequence defines its structure and function (1). Accordingly, it is well established that the active site residues among homologs are exceptionally conserved (2). However, homologous enzymes do not have identical kinetic properties, with each member often exhibiting different rates or affinities for reaction intermediates, despite having the identical set of residues performing chemistry. Consistent with this, recent data suggest that the sequence-structure-function dogma is incomplete. Current data show that intrinsic enzyme dynamics are equally important for regulating enzymatic function (3–9).
The extent to which intrinsic enzyme dynamics, like structure, is encoded in sequence and the role(s) it plays in regulating enzyme activities remain open questions (4, 10–12). Computational results have highlighted that such correlations might exist (13, 14). However, experimental evidence is critically needed to confirm and advance these computational predictions. Understanding the molecular differences that give rise to divergence in activities will provide new routes for evolving enzymes with enhanced activities and novel strategies for the development of enzyme-specific therapeutics. Unfortunately, general methods for identifying residues outside the active site that function as catalytic modulators are not well established.
One approach, coevolutionary coupling analysis, uses patterns of pairwise correlations in sequence alignments coupled with spectral clustering to identify groups of residues that have evolved in a concerted manner [evolutionary domains (EDs)] (15). While the mathematical framework has been applied to canonical enzymes, i.e., adenylate kinase (15), experimental validation of EDs identified using coevolution analysis is currently scant. Nearly 20 years ago, a correlation between sequence conservation and side-chain rigidity using picosecond to nanosecond (ps-ns) dynamics in an SH3 domain was discovered (16). Furthermore, energetic pathways identified by evolutionary covariance analysis have been shown to coincide with networks of side chains connected by shared ps-ns dynamics in a PDZ domain (17). Last, it was shown that this ps-ns dynamics in multiple PDZ domains is conserved (18). However, little additional work has been reported to determine whether this finding is general, or whether evolutionary networks coincide with structural pathways or dynamics on other time scales in larger proteins or ones that have catalytic activity.
Here, we tested the ability of coevolutionary coupling analysis to identify evolutionarily conserved domains that modulate the enzyme activity of protein tyrosine phosphatase (PTP) 1B (PTP1B/PTPN1) and then used nuclear magnetic resonance (NMR) spectroscopy, x-ray crystallography, and biochemical assays to define their molecular origin. PTP1B is a key regulator of both insulin and leptin signaling pathways with additional roles in cancer and neuronal signaling (19–21). Similar to all members of the PTP family, PTP1B has a highly conserved active site defined by the following (Fig. 1A): (i) The PTP loop ([I/V]HC215xxGxxR221[S/T]G), which contains the invariant catalytic nucleophilic cysteine (C215); (ii) the E loop, which contains a conserved glutamate (E115) that coordinates the PTP-loop R221; (iii) the WPD loop, 179WPD181, which contains the catalytic aspartate (D181) that functions as the proton donor and acceptor during phosphoryl transfer; and (iv) the Q loop centered on Q262, which coordinates a nucleophilic water to mediate phosphocysteine hydrolysis (22). Although it was originally thought that PTP1B function depends only on the rate of WPD loop closure (23), recent work has shown that the active site loops are dynamically coordinated, exhibiting uniform dynamics throughout the PTP1B catalytic cycle (24). Last, many additional reports highlight the functional role of these active site loops in the chemistry of the catalytic reaction in PTP1B (25–27).
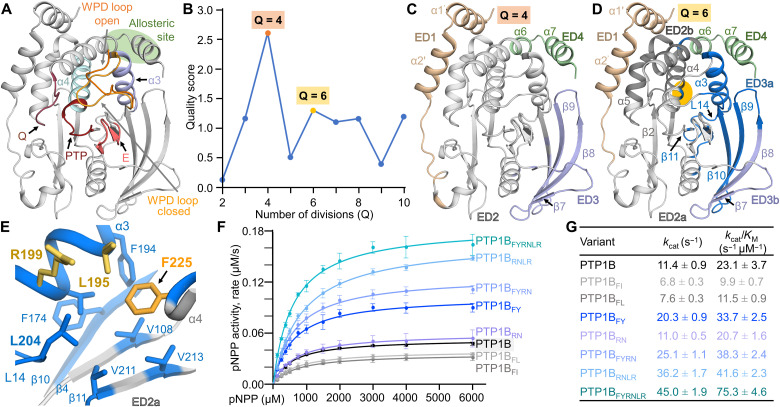
Fig. 1. Coevolutionary coupling analysis identifies a conserved domain that influences PTP1B activity.
(A) Overlay of the open [Protein Data Bank (PDB): 5K9V; WPD loop only) and closed (PDB: 5K9W; gray cartoon) states of PTP1B. Highlighted functional elements include the PTP and Q loops (red), the WPD loops (closed, light orange; open, orange), the E loop (pink), helix α3 (lavender) and helix α4 (light blue), and the allosteric pocket (green circle). (B) Evolutionary partitioning (quality score versus Q divisions) identifying groups of coevolving residues (ED) in PTP1B using multiple sequence aålignments (MSA). (C) PTP1B EDs identified for Q = 4: ED1, N terminus (beige); ED2, core PTP domain (gray); ED3, β strands 7 to 10 (lavender); ED4, allosteric pocket (green). (D) PTP1B EDs identified for Q = 6: ED1 and ED4 same as in (C); ED2a, key catalytic loops (light gray); ED2b, support helices α4/α5 and part of helix α6 (dark gray); ED3a, β strands β9/β10 (with parts of β4 and β8) that are now grouped with helix α3 and loop L14 (blue); ED3b, the remainder of β strands β8 and β7 (lavender). Orange circle highlights the position of F225 on helix α4, which is part of ED3b. (E) ED3a hydrophobic pocket centered on F225. (F) Catalytic activity of ED3a variants (n = 3 to 4). (G) Kinetic parameters for measurements in (F).
Here, we report that a group of residues, ~20 Å distant from the PTP1B active site, perform motions in the microsecond time scale that directly correlate with the enzymatic turnover of PTP1B. Furthermore, most of these residues are conserved in the PTP family of enzymes and thus the protein dynamics that drive PTP activity are also likely conserved. Our data explain how PTPs perform identical chemistry with different turnover rates, ultimately allowing diverse biological functions being controlled by the identical PTP fold.
RESULTS
Coevolutionary coupling analysis of PTP1B
To determine whether PTP1B contains EDs outside of the active site, we analyzed 4406 homologous PTP1B sequences using coevolutionary coupling analysis. The best scores were obtained when PTP1B was divided into four (Q4) and six (Q6) conserved domains (Fig. 1B). The four EDs in Q4 are the following (Fig. 1C): (i) The PTP1B N terminus, including helices α1 and α2, which, within the PTP family, is largely unique to PTP1B (28); (ii) the core PTP catalytic domain (29); (iii) the β strands β7/β8/β9 and part of β10 that comprise the outermost strands of the PTP1B central β sheet; and (iv) a part of helix α6 and all of helix α7, which define the PTP1B allosteric pocket (30). The function of each of these domains in PTP1B activity has been experimentally established, demonstrating that coevolutionary analysis successfully identifies clusters of residues critical for PTP1B function.
To identify subdomains that have the potential to influence PTP1B activity via as yet unknown mechanisms, we examined Q6 (Fig 1D), which resulted in the division of the core PTP catalytic domain and the β7/β8/β9/β10 strand domain into four distinct groups (the N-terminal and C-terminal EDs were unchanged between Q4 and Q6): (2a) the key catalytical loops; (2b) support helices α4/α5 and part of helix α6, which bridge the catalytic site to the N terminus; (3a) β strands β9/β10 (with parts of β4 and β8) that are now grouped with helix α3 and loop L14; and (3b) the remainder of β strands β8 and β7. While the initial four groups are largely contiguous in sequence, the groups in Q6 are not; rather, they are continuous in space. Particularly unique was ED3a, in which a single residue from helix α4, F225 (8.5 Å from C215; Cα-Cα distance; the remainder of α4 is part of group 2b), was identified to coevolve as part of the extended hydrophobic pocket defined by ED3a residues from β strands β9/β10, helix α3, and L14 (Fig. 1E). Q7/8/10 showed similar scores as Q6, but without understanding the function of the Q6 EDs, it is impossible to interpret the molecular basis of the EDs in Q7/8/10.
PTP1B helix α4 residue F225 defines a coevolved regulatory center
The multiple sequence alignment leveraged for the coevolutionary coupling analysis identified that the most frequently observed variants of ED3a residue F225 are F225I, F225L, and F225Y. To understand how the identity of the residue at position 225 affects PTP1B stability and activity, we generated each variant and then measured their thermal denaturation melting temperatures (Tm) and activities (Fig. 1, F and G, and table S1). The thermal denaturation experiments show that each variant is less thermally stable than wild-type (wt) PTP1B [ΔTm: −11.6°C (PTP1BF225I), −7.9°C (PTP1BF225L), and −8.0°C (PTP1BF225Y); all constructs are PTP1B1–301 unless otherwise noted; hereafter, PTP1B refers to wt-PTP1B1–301]. Furthermore, PTP1BF225A is completely insoluble, a result consistent with the observation that the F225 pocket is exclusively hydrophobic extending to loop L14 residue L204. These data demonstrate that not only is a bulky hydrophobic amino acid at this position required for proper folding but also replacing it with other hydrophobic amino acids negatively affects PTP1B stability. Despite this reduction in PTP1b stability, activity assays showed that the catalytic activities of the variants were either similar (PTP1BF225I and PTP1BF225L) or, unexpectedly, increased (PTP1BF225Y, increased by 1.8-fold) compared to wt-PTP1B.
To understand how an amino acid more than 8 Å from the active site affects PTP1B activity, we investigated the consequences of mutating residues within the F225 hydrophobic pocket to amino acids identified in our analysis to coevolve with PTP1BF225Y with the highest frequencies: helix α3 residues L195(R) and R199(N). We first tested the consequences of mutating these residues, both alone and in combination with F225Y. PTP1BL195R was not soluble, likely due to the steric and charge repulsion of two arginine residues present in the same pocket (R199 and L195R). In contrast, PTP1B variants PTP1BR199N, PTP1BR199N/L195R (PTP1BRNLR), PTP1BF225Y/R199N (PTP1BFYRN), and PTP1BF225Y/R199N/L195R (PTP1BFYRNLR; this was the most frequently observed variant in the coevolution analysis) expressed solubly. These variants also exhibit reduced thermal stabilities compared to PTP1B (table S1). Although L195 and R199 are, respectively, 14.5 and 18.9 Å away from C215, enzymatic assays showed that, like PTP1BF225Y, the catalytic activities of all variants except PTP1BR199N increased relative to PTP1B (PTP1BFYRN, 1.7-fold; PTP1BRNLR, 1.8-fold; PTP1BFYRNLR, 3.3-fold; Fig. 1, F and G). Furthermore, an analysis of the kinetic parameters showed that the increase depends solely on kcat, i.e., it is not achieved because substrates engage PTP1B differently but instead reflects increased turnover.
The increase in kcat of F225 variants is not due to a structural change or allostery
We then set out to define the molecular mechanism(s) by which these mutations alter PTP1B activity. It is well established that PTP1B is regulated by allostery, which is mediated by protein dynamics, and, further, that helix α7 is essential for this allosteric control (its deletion, PTP1BΔ7, frees PTP1B from allosteric regulation) (30). Although our previous work did not identify L195, R199, or F225 as part of the allosteric pathway (31), we tested whether the observed activity increases in these variants requires helix α7–mediated allostery. Deletion of α7, unlike these mutations, alters both kcat and KM (table S1). However, the activities of PTP1BF225Y, PTP1BFYRN, and PTP1BFYRNLR in PTP1B∆7 (Fig. 2A and table S1) show similar increases in kcat for all PTP1B∆7 and PTP1B coevolving variants [the crystal structures of PTP1BΔ7FYRN and PTP1BΔ7FYRNLR are unchanged, with root mean square deviations (RMSDs) of 0.12 and 0.17 Å versus PTP1BΔ7, respectively; fig. S1 and table S2]. These data demonstrate that the increases in activity in these variants do not depend on helix α7–mediated PTP1B allostery but instead are achieved by a distinct mechanism.
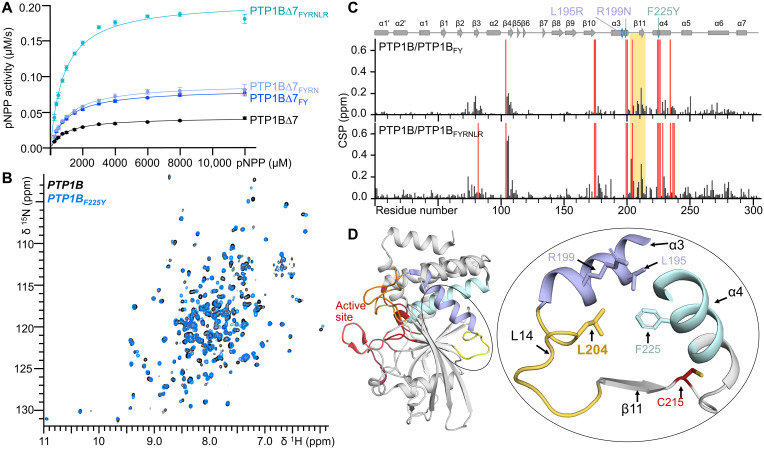
Fig. 2. Modulation of PTP1B activity by ED3a.
(A) Catalytic activity for PTP1B∆7 variants (n = 3 to 4). (B) Overlay of the 2D [1H,15N] TROSY spectrum of PTP1B (black) and PTP1BF225Y (blue). (C) CSPs of PTP1B versus PTP1BF225Y (top) or PTP1BF225Y-R199N-L195R (bottom). Red bars indicate peaks that broadened beyond detection; PTP1B secondary structural elements are shown, and the location of mutations is indicated by a diamond. (D) Loop L14 and L204 and its relationship to ED3a (residues F225, L195, and R199) and the active site PTP loop (C215). Structural elements are colored as in Fig. 1A. ppm, parts per million.
To identify this mechanism, we used biomolecular NMR spectroscopy. The two-dimensional (2D) [1H,15N] transverse relaxation optimized spectroscopy (TROSY) spectra of PTP1BF225Y (Fig. 2B) and PTP1BFYRNLR (fig. S2) are of exceptional quality, with nearly identical overall peak counts as the 2D [1H,15N] TROSY spectrum of PTP1B. This demonstrates that there are no major differences in backbone chemical exchange processes due to the mutation(s). It was previously shown (31) that the binding of a substrate mimicking inhibitor, TCS401, to PTP1B results in numerous chemical shift perturbations (CSPs) not only near the TCS401 binding site (i.e., the PTP1B active site) but also at the PTP1B allosteric site (30) and residues that connect the active and allosteric sites. The binding of TCS401 to both PTP1BF225Y and PTP1BFYRNLR leads to CSPs that are highly similar to those identified in TC401-bound PTP1B (fig. S3, A to C). Likewise, the CSPs that define the PTP1B allosteric pathway are also observed, confirming that the allosteric pathway is maintained among PTP1BF225Y, PTP1BFYRNLR, and PTP1B (31). When further comparing the 2D [1H,15N] TROSY spectra of PTP1BF225Y and PTP1BFYRNLR with that of PTP1B, additional novel CSPs were observed (Fig. 2C). Most new CSPs are readily attributable to the F225Y and FYRNLR mutations. However, new CSPs were also observed for residues in strand β4 and loop L14 (L14, residues 202 to 209, is part of the Q6 ED3a and connects helix α3 to strand β11; Fig. 2D), suggesting a role for these residues in the F255Y-mediated increase in PTP1B activity. The crystal structures of PTP1BF225Y and TCS401-bound PTP1BF225Y and PTP1BFYRNLR showed that the observed CSPs were not due to changes in conformation (free and TCS401-bound; RMSDs of 0.1, 0.13, and 0.18 Å, respectively) (fig. S4 and table S2), suggesting that neither conformational/structural changes nor allostery explains the altered activity.
PTP1B loop L14
PTP1B loop L14 is part of the Q6 ED3a. To test whether and how L14 hydrophobicity and/or flexibility affects PTP1B catalytic activity, we generated PTP1BL204A, PTP1BP206G, and PTP1BGGGGGG (L14, residues 203 to 208, each mutated to a glycine). While the catalytic activity of PTP1BP206G was unchanged from PTP1B, the activities of both PTP1BL204A and PTP1BGGGGGG increased (Fig. 3A and table S1). Michaelis-Menten kinetic analysis of PTP1BGGGGGG showed that this increase was largely attributable to an increase in KM. In contrast, the increased activity of PTP1BL204A (a residue of ~16 Å from the catalytic C215) was due to a change in kcat, i.e., reflecting the behavior observed for PTP1BF225Y and PTP1BFYRNLR.
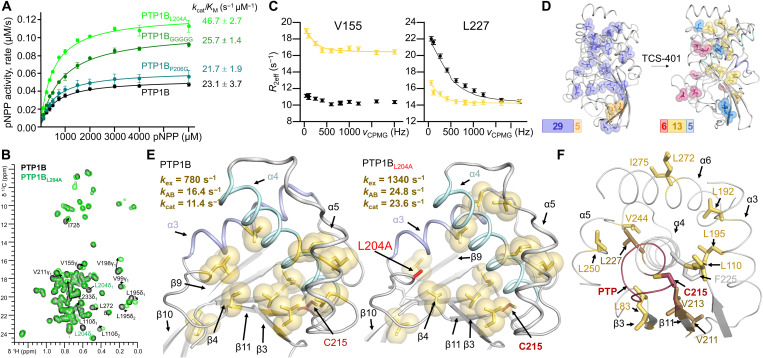
Fig. 3. Group 2 dynamics are related to enzyme catalysis.
(A) Catalytic activity for PTP1B L14 variants (n = 3 to 4). (B) Overlay of 13C ILV 2D [1H,13C] HSQC spectrum of PTP1B (black) and PTP1BL204A (green). Moving peaks are annotated and indicated with arrows. (C) Representative 13C-CPMG dispersion profiles for V155 [fast exchange in free PTP1BL204A (black) and intermediate exchange in TCS401-saturated PTP1BL204A (yellow)] and L227 [free PTP1BL204A (black) and intermediate exchange in TCS401-saturated PTP1BL204A (yellow)]. (D) Left: ct-CPMG relaxation dispersion grouping for PTP1BL204A (PDB 7MNC), group 1 (kex = 3000 ± 40 s−1; dark blue) and group 2 (kex = 5000 ± 210 s−1; orange). Right: ct-CPMG dispersion relaxation grouping for PTP1BL204A saturated with TCS401 (PDB 7MND), group 1 (kex = 3130 ± 280 s−1; blue), group 2 (kex = 1340 ± 120 s−1; yellow), and group 3 (kex = 680 ± 40 s−1; raspberry). (E) ct-CPMG dispersion relaxation group 2 in TCS401-saturated PTP1B (left) (24) and TCS401-saturated PTP1BL204A (right). The exchange frequency (kex) and kAB, along with the measured kcat, for TCS401-saturated PTP1B and PTP1BL204A are shown. (F) Group 2 residues (yellow sticks) cradle helix α4 (residues that anchor the PTP loop are shown in dark yellow). The PTP loop, including the catalytic C215, is shown in red.
To define the molecular basis for the increase, we first determined the crystal structures of free and TCS401-bound PTP1BL204A. The structures of PTP1BL204A are essentially identical to PTP1B, both in the absence of and bound to TCS401 (RMSDs of 0.15 and 0.14 Å, respectively; fig. S5, A and B, and table S2). Similarly, the 2D [1H,15N] TROSY spectra of PTP1BL204A (fig. S6A) and PTP1B are nearly identical to those observed for PTP1BF225Y (Fig. 2C) and PTP1BFYRNLR (fig. S2), except for CSPs in loop L4, a loop immediately adjacent to L14 (fig. S6B). Last, binding of TCS401 to PTP1BL204A leads to all expected CSPs (fig. S6, C and D). Together, no structural changes or unexpected CSPs are identified in PTP1B due to the L204A mutations, despite the change in enzymatic activity.
A novel role for microsecond/millisecond time scale dynamics in PTP1B activity
Because there were no notable changes in the conformation of PTP1B and its variants to explain the increased catalytic activity of PTP1BL204A, we reasoned that PTP1B dynamics may play a role. Because the increased catalytic efficiency is driven by a change in kcat, this correlates with changes in PTP1B dynamics in the microsecond/millisecond regime (23). To exclude the possibility that fast time scale dynamics (i.e., fluctuations in the ps-ns time regime) might play a role, we measured the 15N backbone (fig. S7) and 13C side-chain dynamics [Ile, Leu, and Val (ILV)] relaxation data (T1 and T2; all side-chain dynamics measurements were performed using 13CHD2 ILV-labeled PTP1B; Fig. 3B and figs. S8 and S9) of PTP1BL204A in both the absence and presence of TCS401. The data show that the PTP1BL204A fast time scale dynamics mirror the dynamics observed for PTP1B (31), demonstrating that the increase in kcat is not due to changes in fast time scale dynamics.
We then measured PTP1BL204A 13C ILV constant-time Carr-Purcell-Meiboom-Gill (ct-CPMG) side-chain dynamics, which directly report on dynamics on microsecond/millisecond regime (32). Here, residues undergoing microsecond/millisecond conformational exchange dynamics between two populations show changes in the effective rate of relaxation R2eff, which are measured as a function of repetition frequency (νcpmg) (Fig. 3C and fig. S10, A and B). The curve of the plots of R2eff versus νcpmg are fit using a two-state model (Carver-Richards) to extract the two populations (pA and pB) and the rate of exchange between them (kex). Our previous 13C ILV ct-CPMG side-chain relaxation measurements on PTP1B revealed that the chemically critical elements of PTP1B fluctuate coherently and distinctly from the rest of the protein when a substrate analog and/or inhibitor binds the PTP1B active site, demonstrating that all catalytic residues work in dynamic unity (24).
As observed for PTP1B (24), PTP1BL204A 13C ILV ct-CPMG side-chain dynamics experiments showed that many residues in free PTP1BL204A (34 residues) exhibited microsecond/millisecond exchange dynamics. These clustered into two groups with distinct motions (groups are collections of residues that experience similar exchange dynamics; Fig. 3D, left, and table S3). Group 1 contained 29 residues and experienced uniform fast exchange (kex = 3000 ± 40 s−1 and pB = 3.4 ± 0.2%; fig. S10A). Group 2 contains the remaining residues (five residues) and had distinct exchange parameters (kex = 5000 ± 210 s−1 and pB = 0.9 ± 0.1%; table S3). These residues are on the surface of the protein far from the active site and therefore likely reflect areas with increased flexibility and interactions with bulk solvent. The PTP1BL204A residues that comprise groups 1 and 2, and their associated exchange frequencies and populations, are similar to those previously observed for PTP1B (24). While these groups are statistically distinct, the reliability of populations extracted from the Carver-Richards equation for conformational dynamics in the fast exchange regime are low. However, the Carver-Richards equation is applicable when the kex >> R2 and when the one of the populations is significantly dominant. This is exactly the case for these measurements performed on free PTP1B (i.e., PTP1B is predominately in an open conformation).
As also previously observed for PTP1B (24), the formation of the TCS401-saturated PTP1BL204A complex (1:6 ratio) led to a large decrease in overall dynamics (fig. S10B and table S4). Specifically, 24 residues exhibited microsecond/millisecond exchange dynamics, which clustered into three groups with distinct motions (Fig. 3D, right). Group 1 (five residues, strand β8 or helix α7) had the fastest exchange dynamics (kex = 3130 ± 280 s−1 and pB = 0.3 ± 0.1%). Group 2 (13 residues, helices α3/α4/α6 or strands β4/β9/β10/β11) had significantly slower exchange dynamics (kex = 1340 ± 120 s−1 and pB = 1.9 ± 0.9%); these secondary structure elements are part of the hydrophobic cluster centered around F225 and loop L14 and include L195 (Fig. 3, C and D, and fig. S10B). Group 3 (six residues, active site loops) had the slowest exchange dynamics (kex = 680 ± 40 s−1 and pB = 11 ± 1%). We expect these changes in dynamics to mimic those that occur under conditions of catalysis because TCS401 is a substrate-like compound (33). Unlike substrates, TCS401 saturates PTP1B at experimentally accessible concentrations, permitting the careful and reproducible investigation of a substrate-bound–like PTP1B state.
As observed for free PTP1BL204A and PTP1B, the number of groups and the residues that comprise each group are nearly identical between TCS401-bound PTP1BL204A and TCS401-bound PTP1B (Fig. 3E). However, their associated exchange rates differ. Namely, for TCS401-bound PTP1BL204A, the kex of all three groups increased compared to the corresponding groups in PTP1B. The kex values for groups 1 and 3 are 1.5- and 1.2-fold faster, respectively, than those observed for PTP1B; while the kex for group 2, which includes F225 and L14, increases ~2-fold. This increase in kex mirrors the twofold increase in the PTP1BL204A kcat (Fig. 3E). Using the fitted populations, the unidirectional exchange rates can be calculated. Group 2 (kAB = 24.8 s−1) is remarkably similar to the independently measured unidirectional kcat = 23.6 s−1. Group 2 in PTP1B shows a similar relationship (kAB = 16.4 s−1 and kcat = 11.4 s−1).
Dynamics that controls PTP1B activity is distal to the active site and conserved
The high resemblance of kcat and kAB for group 2 residues in PTP1B, combined with the observed correlated change in kcat and kAB for group 2 in PTP1BL204A, clearly suggests that group 2 dynamics are related to enzyme catalysis. An examination of the residues that constitute group 2 reveal why this is the case (Fig. 3F). Namely, group 2 contains residues from both helix α4 (L227) and strand β11 (V211 and V213), the two secondary structural elements that anchor the PTP loop. Furthermore, additional group 2 residues L83 and L110 contact strand β11 to form a contiguous surface that packs against helix α4, while residues L192 and L195 from helix α3 bind directly to helix α4. Last, residue L250 packs against L227, while V244 contacts four residues at the helix α4 C terminus. Thus, changes in the dynamics of the side chains within group 2 residues are poised to alter the dynamics of the catalytic loop and/or its anchoring elements and thus C215. In this way, these motions may increase enzyme efficiency without altering the global structure or the mechanism of dephosphorylation. Last and most critically, the information flow through this pathway is bidirectional, with binding events at the active site altering the dynamics of loop L14 (L204) and mutations in loop L14 (L204A) altering dynamics in the active site.
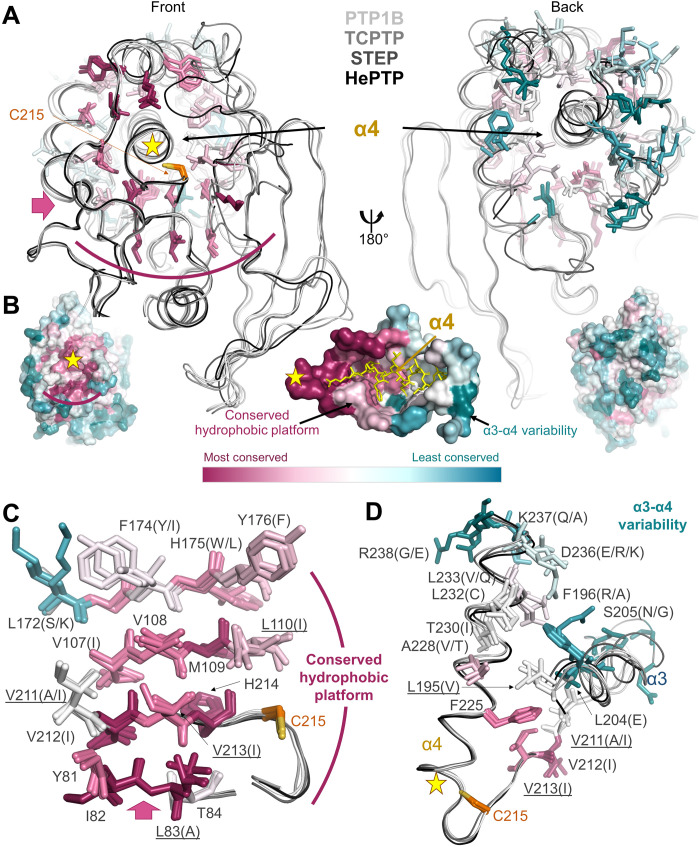
Notably, the majority of residues outside the active site that exhibit the highest sequence conservation in the PTP family are on the central β sheet (strands β2/β3/β4/β10/β11), where many of the group 2 residues are found (Fig. 4, A and B, left). This conservation extends even to bacterial tyrosine phosphatases such as YopH from Yersinia pestis (fig. S11). This platform of conserved residues supports the N-terminal portion of helix α4, which connects directly to the PTP loop containing the catalytic C215 (Fig. 4C). In contrast to the C-terminal portion of helix α4, all of helix α3 and the residues that support these secondary structural elements are much less conserved [Fig. 4, A (right), B (right), and D]. These sequence variations likely allow for different helix α4 motions resulting in distinct catalytic activities among PTP family members. It is therefore probable that this dynamic pathway is conserved across the PTP family, providing further support for the emerging view that conservation of dynamics in parts of enzymes that are distal from the active site is essential for enzyme function, i.e., the regulation of catalytic turnover.
Fig. 4. Group 2 residues are highly conserved.
(A) Overlay of PTP1B (white), TCPTP (light gray), STEP (dark gray), and HePTP (black). Residues surrounding helix α4 are shown as sticks and colored by conservation within the PTP family. Left: Front view looking down helix 4 from the active site (yellow star; catalytic Cys shown in orange sticks). Pink arrow indicates the view of the hydrophobic platform (curved magenta line) shown in (B). Right: Same overlay rotated by 180°. (B) PTP1B is shown as a surface and colored according to PTP family sequence conservation [most conserved (magenta) and least conserved (teal)]. Left: Same orientation as in (A), left. Right: Same orientation as in (A), right. Middle: View of helix α4 looking down illustrating the conserved hydrophobic platform and the increased variability at the α3-α4 junction. (C) Residues (shown as sticks) that comprise the conserved hydrophobic platform that support helix α4 are colored according to conservation and labeled (underline indicates a group 2 residue). (D) Residues (shown as sticks) illustrating the increased variability at the C-terminal portion of helix α4 adjacent to the N terminus of helix α3. Residues are colored and labeled as in (C).
DISCUSSION
Protein dynamics, at various time scales, are essential for the regulation, interaction, and function of prokaryotic and eukaryotic enzymes (3–9, 11). PTPs, especially PTP1B, are one of the best-studied groups of enzymes, as they have critical roles in the regulation of essential signaling pathways. As for many enzymes, crystallographic studies were leveraged to understand the molecular mechanism of the catalytic reaction (29, 34–37). This foundational work showed that the WPD loop adopts two distinct states, an open state and a closed state, and that these two states are critical for the enzymatic mechanism that drives the underlying chemistry of their dephosphorylation. Multiple studies since then have greatly augmented this view. For example, it was also shown that the rigidity of this loop is critical for the function of the loop itself (25, 31). Furthermore, multiple reports have provided key insights not only into the mechanism of allostery in PTP1B (30, 31, 38, 39) but also, more recently, to its closest homolog, TCPTP (28, 40). These and a plethora of other findings led to multiple in silico, high-throughput screening, structure-based drug design, and other efforts to identify specific inhibitors of PTPs.
Here, we used PTP1B, the best-studied member of the PTP family of enzymes, as a test bed to determine whether residues that coevolve are important for its catalytic function. Previous experiments investigating a small, noncatalytic domain demonstrated that networks of coevolving residues coincide with networks of residues connected by fast fluctuations of side chains (41). Here, we significantly extend that finding and show that coevolutionary coupling analysis identifies catalytically important residues more than 16 Å away from the active site of PTP1B. More significantly, we discovered that this distal network of PTP1B residues causes changes in side-chain fluctuations on the microsecond time scale that result in correlated changes in kcat. These results strongly support the view that the relationships discovered by coevolutionary coupling analysis reflect pathways of energetic distribution correlated with protein dynamics. They also reveal that, just like protein structure, the protein dynamics that drives protein activity is also likely conserved. Together, this work establishes that the dynamics of evolutionarily coevolving domains distal from the active site controls enzyme activity, explaining how PTPs can perform identical chemistry yet do so with different turnover rates and ultimately allowing diverse biological functions being controlled by the identical PTP fold. Together, this work advances protein design, disease mutation analysis, and therapeutic development of enzymes.
MATERIALS AND METHODS
ED analysis
To identify groups of coevolving residues in PTP1B, we obtained a multiple sequence alignment containing PTP1B homologs by building a hidden Markov model of the protein family, based on four iterations of jackhammer (42), and extracting the sequences from the UniProt UniRef100 database (43). We refined the alignment by requiring that all sequences covered at least 50% of the PTP1B sequence and excluding all sequences that contain more than 50% of gaps. We then used the asymmetric plmDCA algorithm (44), using default input parameters (including a 90% cutoff in sequence similarity resulting in 4408 sequences), to find pairs of residues with direct correlated mutations through evolution. We used the derived couplings to divide the PTP1B sequence into EDs (15), i.e., to find groups of residues that evolved together and almost independently of each other. The obtained quality score indicates the best partitioning for the underlying coupling graph based on the separability between the different clusters. For the analysis, we used the web server at spectrus.sissa.it/spectrus-evo_webserver with default parameters.
Protein expression
DNA coding the human PTP1B catalytic domains PTP1B (residues 1 to 301) and PTP1B∆7 (residues 1 to 284) was used to generate all PTP1B variants. PTP1B F225Y, F225Y/R199N, R199N, R199N/L195R, F225Y/R199N/L195R, L204A, P206G, and GGGGG (203-SLSPH-207) were generated using the QuikChange (Agilent) site-directed mutagenesis kit. DNA plasmids for each variant were transformed into Escherichia coli BL21 (DE3) RIL cells (Agilent) for protein expression. For activity assays and crystallization, cells were grown in LB in the presence of selective antibiotics at 37°C to an optical density at 600 nm (OD600) of ~0.8. Expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and proceeded for ~20 hours at 18°C. Cell pellets were harvested by centrifugation at 8000g and stored at −80°C until purification. For 15N-based NMR experiments (CSP and 15N-relaxation), PTP1B expression was facilitated by growing cells in D2O-based M9 minimal media containing 15NH4Cl (1 g/liter). For 13C ILV relaxation measurements, PTP1B was expressed with [2H,12C]-d-glucose (4 g/liter) in D2O-based M9 minimal media; upon reaching an OD600 of ~0.6, [3-13C, 3-methyl-2H2, 3,4,4,4-2H4] α-ketoisovaleric acid (120 mg/liter; CDLM 7354) and [4-13C, 4-2H2, 3-2H2] α-ketobutyric acid (60 mg/liter; CDLM 7353) in 100% D2O were added. After an OD600 of ~0.8 was reached, expression was induced by the addition of 1 mM IPTG (dissolved 100% D2O) and continued for ~20 hours at 18°C. Cells were harvested and stored at −80°C until purification. Typically, wt-PTP1B protein yields are ∼55 mg/liter, and variant yields are ~5 to 48 mg/liter in both LB and D2O-based M9 minimal media.
Protein purification
Cell pellets were resuspended in lysis buffer [25 mM tris (pH 8.0), 500 mM NaCl, 5 mM imidazole, and 0.1% Triton X-100] containing EDTA-free protease inhibitor cocktail (Roche). The cells were then lysed using high-pressure homogenization (Avestin). The lysate was clarified by centrifugation at 45,000g and filtered through a 0.22-μm filter. The clarified lysate was loaded onto a His-Trap HP column (GE Healthcare). The protein was eluted over a gradient of increasing amounts of buffer B [50 mM tris (pH 8.0), 500 mM NaCl, and 500 mM imidazole]. Fractions containing PTP1B were pooled and dialyzed overnight at 4°C in dialysis buffer [50 mM tris (pH 8.0) and 500 mM NaCl] with a 5:1 volume ratio of tobacco etch virus (TEV) protease. The next day, a “subtraction” His6-tag purification was performed to remove the TEV protease and the cleaved His6-tag. Cleaved PTP1B was further purified using size exclusion chromatography (Superdex 75 26/60, Cytiva) into storage buffer [20 mM Hepes (pH 7.4), 150 mM NaCl, and 0.5 mM tris(2-carboxyethyl)phosphine (TCEP)], crystallization buffer [20 mM tris (pH 7.4), 50 mM NaCl, and 0.5 mM TCEP], or NMR buffer [10 mM Hepes (pH 7.4), 150 mM NaCl, and 5 mM dithiothreitol]. Purified protein was either used immediately or flash-frozen in liquid nitrogen for storage at −80°C. Samples for the 15N-based/13C-based ILV relaxation measurements were aliquoted into 550 μl of aliquots and lyophilized; for measurements, lyophilized samples were resuspended in 550 μl of D2O.
pNPP activity assay
The activities of freshly prepared (50 nM) PTP1B/PTP1B∆7 and variants were measured in assay buffer (50 mM bis-tris pH 6.0, 150 mM NaCl, 0.5 mM TCEP) containing varying concentrations of p-nitrophenyl phosphate (pNPP; 0 to 6000 mM for PTP1B or 0 to 12000 mM for PTP1B∆7). PTP1B was incubated with pNPP at 30°C for 30 min. The reaction was stopped using 0.5 M NaOH, and the absorbance was measured at 405 nm using an EPOCH2 plate reader (BioTek). Measured absorbance from blanks contained substrate, but no protein was subtracted from all measurements. The rate of dephosphorylation of pNPP was analyzed using the molar extinction coefficient for pNPP of 18,000 M−1 cm−1 and an optical path length of 0.3 cm (Costar 96-well plates). Km and vmax were determined by fitting to the Michaelis-Menten equation, y = vmax*x/(Km + x); kcat was extracted using y = Et*kcat*x/(Km + x). The catalytic efficiency was obtained as kcat /Km. SigmaPlot 14.5 was used for data analysis. The experiments were carried out in duplicate and repeated three to six times.
Protein crystallization and data collection
Crystallization trials of PTP1B/PTP1B∆7 variants, including PTP1BF225Y, PTP1BFYRN, PTP1BFYRNLR, and PTP1BL204A, were performed at 6 mg/ml using vapor diffusion sitting drop experiments. For TCS401-bound structures, purified PTP1B (2 mg/ml) was incubated with a 10-M excess of TCS401 [25 mM stock in 100% dimethyl sulfoxide (DMSO)] for an hour on ice and then concentrated to 6 mg/ml for crystallization. PTP1B and TCS401-cocrystalized PTP1B crystals were obtained using a fine screen that ranged from pH 7.4 to 8.0 (0.1 M tris/Hepes) and in polyethylene glycol 8000 concentrations ranging from 13 to 24% (w/v). The MgCl2 concentration was held constant at 0.2 M. All crystals were grown at 4°C. Crystals were cryo-protected by a quick soak (few seconds) in mother liquor supplemented with 30% glycerol and immediately flash-frozen in liquid nitrogen. X-ray data were collected using the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 12-2 (PILATUS 6M detector), and the data were processed using autoXDS script [which runs X-ray Detector Software (XDS), pointless, aimless, and truncate].
X-ray structure determination
The crystal structures were phased using molecular replacement [search model Protein Data Bank (PDB) ID:5k9v] using Phaser as implemented in PHENIX (45). For TCS401-bound structures, clear density of the ligand was observed after PHENIX.AutoBuild and PHENIX.Refine. PHENIX.LigandFit was used to fit TCS401 into the electron density. All structures were subjected to iterative rounds of refinement in PHENIX and manually building using Coot (46). All data collection and refinement statistics are reported in table S2.
NMR spectroscopy
NMR data were collected on Bruker Advance Neo 600- and 800-MHz spectrometers equipped with TCI HCN Z-gradient cryoprobes at 298 K. NMR measurements of PTP1B were recorded using (2H,15N)-labeled protein at a final concentration of 0.2 mM in NMR buffer and 90% H2O/10% D2O. Data were processed using TopSpin 4.05 (Bruker) and analyzed using CcpNmr (47). The sequence-specific backbone assignments of PTP1BL204A were achieved using 3D triple resonance experiments including 2D [1H,15N] TROSY, 3D TROSY-HNCA, 3D TROSY-HN(CO)CA, 3D TROSY-HN(CO)CACB, and 3D TROSY-HNCACB. All NMR assignment data were processed using TopSpin 4.05 (Bruker) and analyzed using CARA (computer-aided resonance assignment).
NMR analysis of TCS401 inhibitor binding
TCS401 was titrated into 0.25 mM PTP1B at molar ratios of 0:1, 0.5:1, 1:1, 2:1, and 3:1 (TCS401:PTP1B). 2D [1H,15N] TROSY or 2D [1H,13C] HSQC spectra were recorded for each titration point. TCS401 was solubilized in d6-DMSO (25 mM). No significant chemical shift differences were identified in the PTP1B 2D [1H,15N] TROSY or 2D [1H,13C] HSQC spectrum upon the addition of d6-DMSO. CSPs (∆δ) between apo PTP1B and inhibitor-bound PTP1B spectra were calculated using .
15N relaxation measurements and analysis
Relaxation measurements were performed on (2H,15N)-labeled ligand-free and TCS401-bound PTP1B (at saturating 6:1 ratios of TCS401:PTP1B) at a final concentration of 0.2 mM in NMR buffer [10 mM Hepes (pH 7.4), 150 mM NaCl, and 0.5 mM TCEP] and 90% H2O/10% D2O at 298 K. 15N longitudinal (R1) and transverse (R2) relaxation rate measurements were acquired using sensitivity-enhanced experiments. TROSY versions of R1 and R2 experiments were acquired with a recycle delay of 2.5 s between experiments and the following relaxation delays for T1: 100, 1000, 2000, 2200, 2400, 2600, 3200, and 4800 ms (2000 and 2600 ms were repeated for measurement error assessment) and T2: 3.94, 15.78, 23.66, 27.61, 31.55, 35.50, 39.44, 47.33, and 63.10 ms (27.61 and 39.44 ms were repeated for measurement error assessment). T1 and T2 values were calculated using CcpNmr by fitting the intensity of peaks to exponential decay function, and errors were determined via relaxation curve fitting.
13C-methyl relaxation measurements
T1 and T1ρ experiments (32, 48) were recorded on (2H,12C,15N)-labeled PTP1BL204A with 13CHD2-labeled ILV methyl groups, either free or TCS401 saturated (1:6 ratio) at a final protein concentration of 0.25 mM in NMR buffer and 100% D2O. Sample concentration was tightly monitored to ensure no effect on τc and thus T1ρ measurements. TCS401 inhibitor was titrated to achieve full saturation. All relaxation data were recorded as a pseudo-3D in a fully interleaved manner at 298 K.
T1 relaxation delays are as follows: 20, 500, 1000, 1200, 1400, 1600, 2000, 4000, and 5500 ms [D1 (recycle delay) of 4.5 s; 800 MHz; 1200 and 4000 ms were repeated for measurement error assessment] and 20, 800, 1000, 1200, 1400, 1600, 2000, 2400, 3000, 3200, and 4000 ms (D1 of 4.2 s; 600 MHz; 1200 ms was repeated for measurement error assessment). T1ρ relaxation delays are as follows: 5, 30, 50, 60, 90, 100, 120, 150, and 180 ms (D1 of 2.5 s; 800 MHz; 50 and 150 ms were repeated for measurement error assessment) and 5, 30, 50, 70, 80, 90, 100, 160, 170, and 200 ms (D1 of 3.2 s; 600 MHz; 70 and 130 ms were repeated for measurement error assessment). The measurement errors between repeat measurements were 2.8% at 18.8 T and 2.6% at 14.1 T.
ct-CPMG (32) relaxation dispersion experiments on (2H,12C,15N)-labeled PTP1BL204A with 13CHD2-labeled ILV methyl groups were performed at 298 K at two magnetic field strengths (14.1 and 18.8 T). A constant time of 40 ms between 15N refocusing pulses and 10 different delay times corresponding to the following CPMG frequencies of 50, 100, 250, 400, 600, 800, 1000, 1200, 1600, and 2000 Hz were used (100 and 800 Hz were repeated for measurement error assessment, which was ~2.8%). D1 (relaxation delay) was set to 3.2 and 3.8 s for experiments performed on 14.1- and 18.8-T magnetic field strengths, respectively. Upon saturation (chemical shifts of interacting residues stopped changing position in spectrum; ~1:3 ratio), additional TCS401 (to 1:6 ratio) was added to ensure that all experiments were performed under fully inhibitor-saturated conditions and thus that the observed ct-CPMG dispersions are independent of ligand on/off exchange events.
13C-methyl (CHD2) relaxation analysis
T1 and T1ρ values were calculated using NMRviewJ (49) using the peak intensities (jitter function) and exponential decay fitting function. Errors were also determined via relaxation curve fitting. T2 was extracted from T1ρ by R2 = (R1ρ − R1cos2β)/sin2β, where β is the effective rotation angle for each 15N nucleus as determined by the strength of the spin-lock field and the chemical shift offset of the nucleus from the spin-lock frequency.
ct-CPMG relaxation dispersion intensity measurements were performed in NMRviewJ (jitter function) and converted to R2eff by R2eff(υCPMG) = (−1/Trelax) ln (ICPMG/I0). PTP1BL204A residues were fit individually to the Carver-Richards equation (50) for a system in two-state exchange using a Levenberg-Marquardt algorithm written by D. Korzhnev (University of Connecticut Health Center) (51). These residues where then fit into groups according to a procedure that we have previously described (24, 52, 53). The quality of the group fit was evaluated using the Bayesian information criterion (BIC) to compare the group fit to the results of the individual fits using ΔBIC = BICgroup − BICindividual. The more negative the ΔBIC, the better the fit. Groups were refined through several rounds. A residue was kept in a specific group if the BIC was <-1, and the residuals were randomly distributed. In-house scripts and spreadsheets were used to perform this analysis.
Quantification and statistical analysis
All activity measurements were repeated between three and four times.
Acknowledgments
Funding: This work was supported by the American Diabetes Association Pathway to Stop Diabetes Grant 1-14-ACN-31 to W.P., National Institute of General Medicine Science grant R01GM098482 to R.P., and the Novo Nordisk Foundation grant NNF15OC0016360 to K.L.-L. This research used beamline 12.2 at the SSRL. Use of the SSRL, SLAC National Accelerator Laboratory is supported by the U.S. Department of Energy, Office of Science and Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Author contributions: W.P., R.P., and K.L.-L. designed the research. D.G. performed the sequence alignment and ED calculation. K.R.T. carried out and analyzed the enzymatic, NMR, and crystallography experiments. M.W.C. performed the NMR experiments and assisted with the dynamics analysis. M.W.C., R.P., and W.P. wrote the paper. All authors discussed the data and manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All NMR chemical shifts have been deposited in the BioMagResBank (BMRB): 50913. Atomic coordinates and structure factors have been deposited in the PDB: 7MKZ, 7MN7, 7MN9, 7MNA, 7MNB, 7MNC, 7MND, 7MNE, 7MNF, 7MOU, 7MOV, and 7MOW. All alignment datasets are freely available at https://github.com/KULL-Centre/papers/tree/master/2021/ptp1b-torgeson-et-al or https://data.mendeley.com/datasets/64sn7vp7sj/1. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Tables S1 to S4
Figs. S1 to S11
REFERENCES AND NOTES
- 1.Anfinsen C. B., Principles that govern the folding of protein chains. Science 181, 223–230 (1973). [DOI] [PubMed] [Google Scholar]
- 2.Ribeiro A. J. M., Tyzack J. D., Borkakoti N., Holliday G. L., Thornton J. M., A global analysis of function and conservation of catalytic residues in enzymes. J. Biol. Chem. 295, 314–324 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oyen D., Fenwick R. B., Aoto P. C., Stanfield R. L., Wilson I. A., Dyson H. J., Wright P. E., Defining the structural basis for allosteric product release from E. coli dihydrofolate reductase using NMR relaxation dispersion. J. Am. Chem. Soc. 139, 11233–11240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehr D. D., McElheny D., Dyson H. J., Wright P. E., The dynamic energy landscape of dihydrofolate reductase catalysis. Science 313, 1638–1642 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Wolf-Watz M., Thai V., Henzler-Wildman K., Hadjipavlou G., Eisenmesser E. Z., Kern D., Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat. Struct. Mol. Biol. 11, 945–949 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Peng Y., Hansen A. L., Bruschweiler-Li L., Davulcu O., Skalicky J. J., Chapman M. S., Brüschweiler R., The Michaelis complex of arginine kinase samples the transition state at a frequency that matches the catalytic rate. J. Am. Chem. Soc. 139, 4846–4853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhabha G., Lee J., Ekiert D. C., Gam J., Wilson I. A., Dyson H. J., Benkovic S. J., Wright P. E., A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science 332, 234–238 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saleh T., Kalodimos C. G., Enzymes at work are enzymes in motion. Science 355, 247–248 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Villali J., Kern D., Choreographing an enzyme’s dance. Curr. Opin. Chem. Biol. 14, 636–643 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alderson T. R., Kay L. E., NMR spectroscopy captures the essential role of dynamics in regulating biomolecular function. Cell 184, 577–595 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Petrović D., Risso V. A., Kamerlin S. C. L., Sanchez-Ruiz J. M., Conformational dynamics and enzyme evolution. J. R. Soc. Interface 15, 20180330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenmesser E. Z., Millet O., Labeikovsky W., Korzhnev D. M., Wolf-Watz M., Bosco D. A., Skalicky J. J., Kay L. E., Kern D., Intrinsic dynamics of an enzyme underlies catalysis. Nature 438, 117–121 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Osuna S., The challenge of predicting distal active site mutations in computational enzyme design. WIREs Comput Mol Sci. 11, e1502 (2021). [Google Scholar]
- 14.Modi T., Risso V. A., Martinez-Rodriguez S., Gavira J. A., Mebrat M. D., Van Horn W. D., Sanchez-Ruiz J. M., Banu Ozkan S., Hinge-shift mechanism as a protein design principle for the evolution of β-lactamases from substrate promiscuity to specificity. Nat. Commun. 12, 1852 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granata D., Ponzoni L., Micheletti C., Carnevale V., Patterns of coevolving amino acids unveil structural and dynamical domains. Proc. Natl. Acad. Sci. U.S.A. 114, E10612–E10621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittermaier A., Davidson A. R., Kay L. E., Correlation between 2H NMR side-chain order parameters and sequence conservation in globular proteins. J. Am. Chem. Soc. 125, 9004–9005 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Fuentes E. J., Der C. J., Lee A. L., Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J. Mol. Biol. 335, 1105–1115 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Law A. B., Fuentes E. J., Lee A. L., Conservation of side-chain dynamics within a protein family. J. Am. Chem. Soc. 131, 6322–6323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tonks N. K., Diltz C. D., Fischer E. H., Purification of the major protein-tyrosine-phosphatases of human placenta. J. Biol. Chem. 263, 6722–6730 (1988). [PubMed] [Google Scholar]
- 20.Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T., Protein tyrosine phosphatases in the human genome. Cell 117, 699–711 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z.-Y., Dodd G. T., Tiganis T., Protein tyrosine phosphatases in hypothalamic insulin and leptin signaling. Trends Pharmacol. Sci. 36, 661–674 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Andersen J. N., Mortensen O. H., Peters G. H., Drake P. G., Iversen L. F., Olsen O. H., Jansen P. G., Andersen H. S., Tonks N. K., Møller N. P. H., Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 21, 7117–7136 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whittier S. K., Hengge A. C., Loria J. P., Conformational motions regulate phosphoryl transfer in related protein tyrosine phosphatases. Science 341, 899–903 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torgeson K. R., Clarkson M. W., Kumar G. S., Page R., Peti W., Cooperative dynamics across distinct structural elements regulate PTP1B activity. J. Biol. Chem. 295, 13829–13837 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui D. S., Lipchock J. M., Brookner D., Loria J. P., Uncovering the molecular interactions in the catalytic loop that modulate the conformational dynamics in protein tyrosine phosphatase 1B. J. Am. Chem. Soc. 141, 12634–12647 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keedy D. A., Hill Z. B., Biel J. T., Kang E., Rettenmaier T. J., Brandão-Neto J., Pearce N. M., von Delft F., Wells J. A., Fraser J. S., An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering. eLife 7, e36307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crean R. M., Biler M., van der Kamp M. W., Hengge A. C., Kamerlin S. C. L., Loop dynamics and enzyme catalysis in protein tyrosine phosphatases. J. Am. Chem. Soc. 143, 3830–3845 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh J. P., Li Y., Chen Y.-Y., Hsu S.-T. D., Page R., Peti W., Meng T.-C., The catalytic activity of TCPTP is auto-regulated by its intrinsically disordered tail and activated by integrin alpha-1. Nat. Commun. 13, 94 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barford D., Flint A. J., Tonks N. K., Crystal structure of human protein tyrosine phosphatase 1B. Science 263, 1397–1404 (1994). [PubMed] [Google Scholar]
- 30.Wiesmann C., Barr K. J., Kung J., Zhu J., Erlanson D. A., Shen W., Fahr B. J., Zhong M., Taylor L., Randal M., McDowell R. S., Hansen S. K., Allosteric inhibition of protein tyrosine phosphatase 1B. Nat. Struct. Mol. Biol. 11, 730–737 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Choy M. S., Li Y., Machado L. E. S. F., Kunze M. B. A., Connors C. R., Wei X., Lindorff-Larsen K., Page R., Peti W., Conformational rigidity and protein dynamics at distinct timescales regulate PTP1B activity and allostery. Mol. Cell. 65, 644–658.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rennella E., Schuetz A. K., Kay L. E., Quantitative measurement of exchange dynamics in proteins via 13C relaxation dispersion of 13CHD2-labeled samples. J. Biomol. NMR 65, 59–64 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Iversen L. F., Andersen H. S., Branner S., Mortensen S. B., Peters G. H., Norris K., Olsen O. H., Jeppesen C. B., Lundt B. F., Ripka W., Møller K. B., Møller N. P., Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B. J. Biol. Chem. 275, 10300–10307 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Pannifer A. D., Flint A. J., Tonks N. K., Barford D., Visualization of the cysteinyl-phosphate intermediate of a protein-tyrosine phosphatase by x-ray crystallography. J. Biol. Chem. 273, 10454–10462 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Groves M. R., Yao Z. J., Roller P. P., Burke T. R., Barford D., Structural basis for inhibition of the protein tyrosine phosphatase 1B by phosphotyrosine peptide mimetics. Biochemistry 37, 17773–17783 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Sarmiento M., Puius Y. A., Vetter S. W., Keng Y. F., Wu L., Zhao Y., Lawrence D. S., Almo S. C., Zhang Z. Y., Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition. Biochemistry 39, 8171–8179 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Salmeen A., Andersen J. N., Myers M. P., Tonks N. K., Barford D., Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol. Cell 6, 1401–1412 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Chio C. M., Yu X., Bishop A. C., Rational design of allosteric-inhibition sites in classical protein tyrosine phosphatases. Bioorg. Med. Chem. 23, 2828–2838 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hongdusit A., Fox J. M., Optogenetic analysis of allosteric control in protein tyrosine phosphatases. Biochemistry 60, 254–258 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Singh J. P., Lin M.-J., Hsu S.-F., Peti W., Lee C.-C., Meng T.-C., Crystal structure of TCPTP unravels an allosteric regulatory role of helix α7 in phosphatase activity. Biochemistry 60, 3856–3867 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Narayanan C., Bernard D. N., Bafna K., Gagné D., Chennubhotla C. S., Doucet N., Agarwal P. K., Conservation of dynamics associated with biological function in an enzyme superfamily. Structure 26, 426–436.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finn R. D., Clements J., Arndt W., Miller B. L., Wheeler T. J., Schreiber F., Bateman A., Eddy S. R., HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzek B. E., Wang Y., Huang H., McGarvey P. B., Wu C. H.; the UniProt Consortium , UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ekeberg M., Lövkvist C., Lan Y., Weigt M., Aurell E., Improved contact prediction in proteins: Using pseudolikelihoods to infer Potts models. Phys. Rev. E. 87, 012707 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst D. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D., The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Tugarinov V., Kay L. E., Quantitative 13C and 2H NMR relaxation studies of the 723-residue enzyme malate synthase G reveal a dynamic binding interface. Biochemistry 44, 15970–15977 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Johnson B. A., Blevins R. A., NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994). [DOI] [PubMed] [Google Scholar]
- 50.Carver J., Richards R. E., A general two-site solution for the chemical exchange produced dependence of T2 upon the carr-Purcell pulse separation. J. Magn. Reson. 6, 89–105 (1972). [Google Scholar]
- 51.Korzhnev D. M., Karlsson B. G., Orekhov V. Y., Billeter M., NMR detection of multiple transitions to low-populated states in azurin. Protein Sci. 12, 56–65 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar G. S., Clarkson M. W., Kunze M. B. A., Granata D., Wand A. J., Lindorff-Larsen K., Page R., Peti W., Dynamic activation and regulation of the mitogen-activated protein kinase p38. Proc. Natl. Acad. Sci. U.S.A. 115, 4655–4660 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lixa C., Clarkson M. W., Iqbal A., Moon T. M., Almeida F. C. L., Peti W., Pinheiro A. S., Retinoic acid binding leads to CRABP2 rigidification and dimerization. Biochemistry 58, 4183–4194 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S4
Figs. S1 to S11