Abstract
Cancer is a predominant disease across animals. We applied a comparative genomics approach to systematically characterize genes whose conservation levels correlate positively (PC) or negatively (NC) with cancer resistance estimates across 193 vertebrates. Pathway analysis reveals that NC genes are enriched for metabolic functions and PC genes in cell cycle regulation, DNA repair, and immune response, pointing to their corresponding roles in mediating cancer risk. We find that PC genes are less tolerant to loss-of-function (LoF) mutations, are enriched in cancer driver genes, and are associated with germline mutations that increase human cancer risk. Their relevance to cancer risk is further supported via the analysis of mouse functional genomics and cancer mortality of zoo mammals’ data. In sum, our study describes a cross-species genomic analysis pointing to candidate genes that may mediate human cancer risk.
Genes and pathways associated with cancer resistance across species are relevant to human cancers.
INTRODUCTION
Animal species are known to have marked differences in their cancer rates and life spans, and several animals are considered cancer resistant, while others are considered to be cancer prone (1, 2). Studying the genomic underpinnings of these differences across various branches of life may provide insights into cancer development and cancer prevention/treatment options in humans (3).
The multistage carcinogenesis model states that “individual cells become cancerous after accumulating a specific number of mutational hits” (3, 4). On the basis of this model, larger (and longer-living) animals are expected to have higher cancer incidence as they have more stem cell divisions overall, resulting in a higher likelihood of producing and propagating carcinogenic mutations. For humans, it has been shown that the risks of cancer development across different tissue types are correlated with their corresponding estimated number of lifetime stem cell divisions (5, 6); consistent with that, human cancer risk is correlated with body height (7). However, cancer risk does not correlate with body size across species, a contradiction known as Peto’s paradox (3, 8, 9). For example, humans do not have a higher cancer risk than mice despite having thousands of times more cells (10–12). More drastically, the cancer-resistant bowhead whale (13) can weigh 100 metric tons, live for over 200 years (14), and have a million times more cells than mice. It follows that different species must have evolved different cancer resistance mechanisms to fit their lifestyles, modifying the “baseline” probability of malignant transformation determined by body size, life span, and tissue stem cell division (see note S1 for a short review of such mechanisms).
Numerous studies have adopted comparative genomics approaches to understand the evolution of cancer resistance mechanisms across mammals. Some have focused on known human cancer genes and their homologs. For example, Vicens and Posada (15) found that genes related to DNA repair and T cell proliferation have evolved under positive selection in mammals. Tollis et al. (16) found that the number of paralogs of human cancer genes across mammals is positively correlated with the species’ life span but not body size. Vazquez and Lynch (17) reported widespread tumor suppressor gene (TSG) duplications across both large and small Afrotherian species. Other studies focused on body size and longevity, yielding some insights into Peto’s paradox. Kowalczyk et al. (18) analyzed genes whose evolutionary rates across mammals correlate with body size and life span and discovered cancer resistance–related genes that are under increased evolutionary constraints in larger and longer-living mammals. Ferris et al. (19) identified regions with accelerated evolution in specific mammals, including several cancer-resistant species, which provided some insights on the cancer resistance mechanisms they have developed.
We base our current study on a similar hypothesis, i.e., that resistance to cancer across species evolved by increased selection (either positive or negative) of certain genes with functional relevance to cancer. However, unlike previous studies that focused exclusively on mammals, here, we perform a comprehensive genome-wide comparative study aimed at identifying genes related to cancer resistance across a wide range of vertebrate species. To this end, we estimated the protein conservation scores across species including mammals, birds, and fish, identifying genes whose conservation levels are associated with cancer resistance estimated based on the species’ life span and body size. We then use these cancer resistance–associated genes to build the first genomics-based predictor of cancer resistance for any species. We show that the biological processes associated with cancer resistance vary across taxonomic groups (classes and orders of species), pointing to the diversity in the evolutionary paths and mechanisms for resisting cancer. We see that the genes identified from this phylogenetic analysis are enriched for cancer driver genes and genes associated with cancer risk in humans. Some of these genes are also further shown to be associated with recent cancer mortality risk (CMR) data obtained by studying 110,148 adult zoo animals (20). These results show that a comparative genomics approach can help identify genes involved in human cancers.
RESULTS
Computing gene conservation and species cancer resistance estimates
We computed a matrix (21, 22) of gene conservation scores (phylogenetic profiles) for over 1600 species for which we got sequence information from UniProt (23), RefSeq (24), Keane et al. (13), and Ensembl (25) databases. These 240 species (237 of them belong to the Animalia kingdom) had some phenotypic information in the AnAge database (26). To do this, the protein sequence similarity between each gene in the genome of a reference species and its orthologs in each of the rest of the species (termed phylogenetic profiling) (27) was measured using the bit score computed with BLASTP (28). The BLASTP bit scores were normalized by their gene length (22, 29) and then rank-normalized across all genes within each species to control for the evolutionary distance between the reference and each species (Methods and note S9). These rank-normalized values range from 0 to 1, with higher values corresponding to higher conservation levels. This method is termed rank-based phylogenetic profiling. We primarily focused on the human as the reference species (30) as we are interested in making our findings relevant to human cancers. However, we demonstrated that our conclusions are robust to the choice of reference (Methods and note S4), largely because the normalization effectively removes dependency on phylogenetic distance.
Given that the strength of intrinsic cancer resistance mechanisms of a species is a “latent” property that is not directly observable, we used two proxy cancer resistance estimates that have been proposed in the literature—MLTAW and MLCAW. MLTAW is based on Peto’s paradox, i.e., cancer incidence within the normal life span of a species appears to have comparable orders of magnitudes across large or small, and long-lived or short-lived species. It follows that the intrinsic level of cancer resistance in a given species needs to roughly counteract its risk of cancer development due to cell division, which, according to a simple cancer development model, is proportional to ML6 × AW, where ML denotes the species maximum longevity and AW denotes its adult weight (Method and note S10) (8, 17, 31). MLCAW considers the well-established correlation between life span and body weight (AW) across many species (32) and thus regresses out the species AW from its ML (Methods). We computed MLTAW and MLCAW for 193 of the 240 species for which both ML and AW data were publicly available (table S1 and Methods). These 193 species are from multiple Vertebrata classes, including Mammalia (mammals, n = 108), Aves (birds, n = 55), Teleostei (teleost fishes, n = 18), and Reptilia (reptiles, n = 7).
Genes associated with cancer resistance are enriched in cell cycle, DNA repair, immune response, and different metabolic pathways
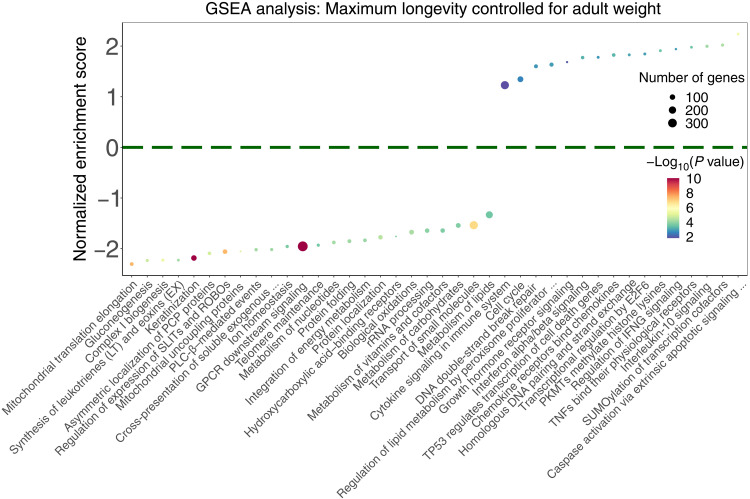
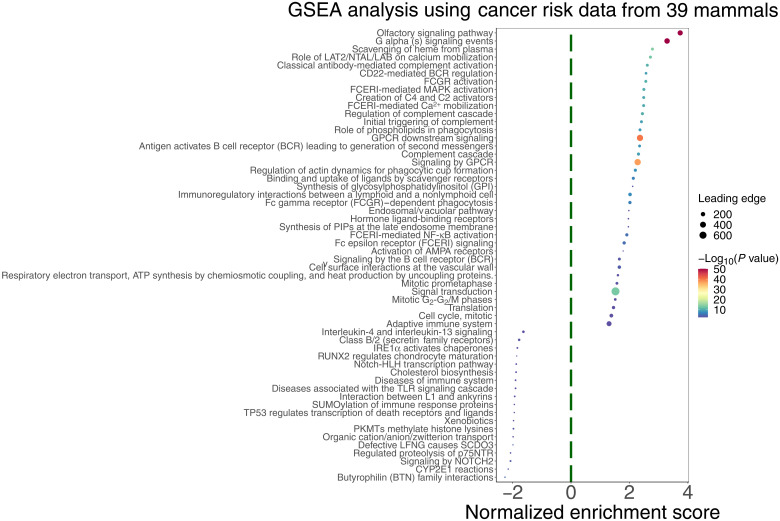
For each gene, we computed the Pearson correlation coefficient between its conservation scores and the cancer resistance estimates (MLTAW and MLCAW) across all species (table S2, A and B, and Methods). We then computed the pathway enrichment of the positively and the negatively correlated genes (termed PC or NC genes, respectively) with gene set enrichment analysis (GSEA; table S3, A and B; note S2; and Methods). Positively enriched pathways based on either the MLCAW (Fig. 1) or MLTAW measures (fig. S1) include cell cycle, immune response, DNA repair, and transcription regulation pathways [false discovery rate (FDR) < 0.1], indicating that many genes in these pathways are more conserved in the relatively long-lived cancer-resistant species. Negatively enriched pathways include a diverse range of metabolic pathways (FDR < 0.1; Fig. 1 and fig. S1). The positive enrichment in cell cycle and DNA repair–related pathways persists even after excluding genes that are associated with regulating life span or body size (Methods and tables S13 and S14), further indicating that these pathways are associated with cancer resistance. In addition, we obtained the top PC and NC genes with significant correlations with MLTAW or MLCAW (FDR < 0.1; note that GSEA does not require any fixed cutoff as such). Using a permutation test (Methods), we observe that the PC and NC genes exist in mutually exclusive pathways, compared to a random shuffled background model (P < 0.02).
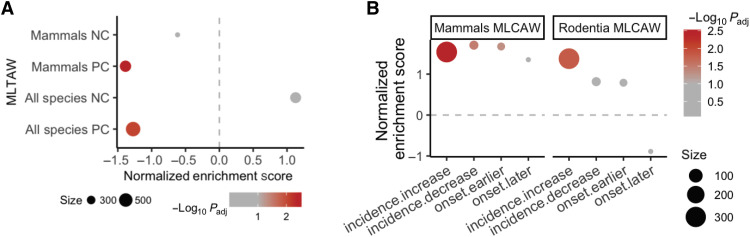
Fig. 1. Summary of the top significantly enriched pathways (adjusted P < 0.1) by the genes whose conservation scores are correlated with cancer resistance estimates (MLCAW), using GSEA with gene set annotations from the Reactome database.
The cancer resistance estimate used is “maximum longevity controlled for adult weight” (MLCAW). Normalized enrichment score is plotted on the y axis, where positive values correspond to enrichment by the PC genes and negative values correspond to enrichment by the NC genes. The dot color represents the significance of the enrichment (negative log10 GSEA P value), and the dot size represents the number of genes in the “leading edge,” i.e., the set of genes that are enriched in a pathway. For the sake of clarity, only a subset of the enriched pathways (FDR < 0.1) are shown, and long pathway names have been shortened (using “…”). The complete pathway enrichment results are given in table S3B.
PC and NC gene conservation scores are predictive of species cancer resistance
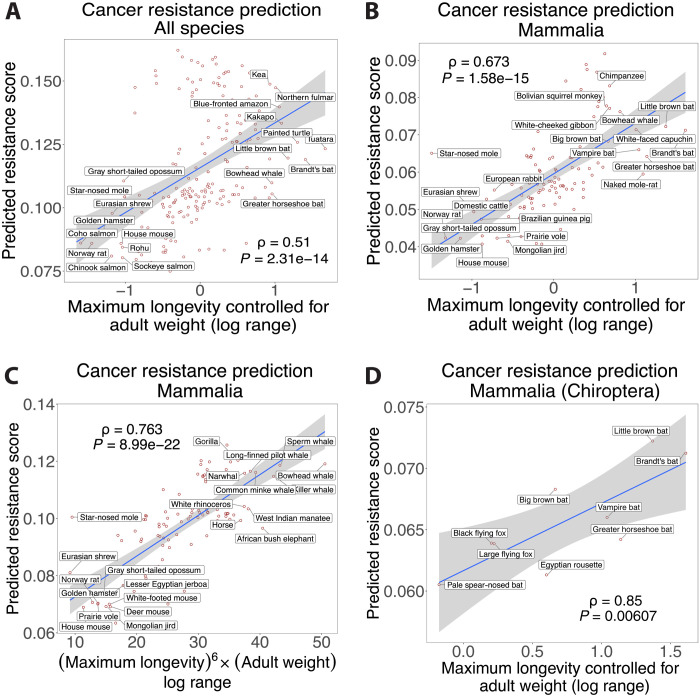
We next asked whether it is possible to accurately predict the cancer resistance estimates of individual species from their gene conservation scores. For a species, given the median conservation score (MCS) of all its genes, we defined a cancer resistance (CR) score that quantifies how many of the PC genes have conservation scores > MCS and how many NC genes have conservation scores < MCS (normalized by the total number of genes; Methods). Using a standard leave-one-out cross-validation (LOOCV) procedure, both across all species and then focusing on mammals or birds (as these groups contain a sufficient number of species), we find that the CR score is strongly predictive of the cancer resistance estimates of a left-out species using the PC/NC genes identified from the other species [all species: MLTAW Spearman’s ρ = 0.44, P = 1.32 × 10−10 (fig. S2) and MLCAW ρ = 0.51, P = 2.31 × 10−14 (Fig. 2A); mammals: MLCAW ρ = 0.67, P = 1.58 × 10−15 (Fig. 2B) and MLTAW ρ = 0.76, P = 8.99 × 10−22 (Fig. 2C); the results for birds are provided in note S3 and fig. S3]. Note S4 and figs. S4 to S9 present both technical controls (choosing random sets of PC and NC genes to predict cancer resistance estimates) and robustness analysis showing that these results hold when (i) using twofold cross-validation instead of LOOCV, (ii) under changes in the choice of reference species (using 12 different nonhuman species including mammals, birds, fish, and plants), (iii) under changes in threshold parameters, (iv) using alternate predictors showing the contributions of PC or NC genes separately, and finally (v) using Spearman’s instead of Pearson correlation to identify PC/NC genes (tables S1 and S2 and Methods). The predicted CR scores learnt from all mammals (LOOCV) also show significant correlation within different subgroups [as an example, MLCAW Spearman’s ρ = 0.85, P = 0.0061 for the order Chiroptera, i.e., bats (Fig. 2D); for others, see note S5 and fig. S10]. Similarly, the predicted CR scores learnt from all birds’ species (LOOCV) show significant correlation within the order Passeriformes for which we have the largest number of samples (Spearman’s ρ = 0.79, P = 0.0012; fig. S3B and note S3).
Fig. 2. Correlation between predicted cancer resistance (CR) scores and cancer resistance estimates.
Scatterplots showing the correlation between the predicted CR scores computed based on gene conservation (y axes) and either of the two cancer resistance estimates (x axes): MLCAW, i.e., maximum longevity controlled for adult weight, or MLTAW, i.e., (maximum longevity)6 × (adult weight), with LOOCV. Results for (A) MLCAW across all species; (B and C) MLCAW and MLTAW within mammalian species, respectively; (D) using the MLCAW mammalian-specific predictions only within a subgroup: order Chiroptera. Species with the top and bottom 5% MLCAW values in (A), the top and bottom 10% MLTAW or MLCAW values in (B) and (C), and all data points in (D) are labeled by their common names. In each panel, the Spearman’s ρ and P values (P) are shown.
Our results show that high CR scores are predicted for many long-living species that are considered to be cancer resistant, including the bowhead whale, the African elephant, the chimpanzee, the Brandt’s bat, the naked mole rat, etc. (Fig. 2, A to D; figs. S6 and S10; and note S5) (2, 3, 33, 34). Predictions of cancer resistance in additional species without documented body weight or life span are provided in table S1. The PC/NC genes derived from one clade do not, however, yield accurate predictions in another taxonomic group (across classes: fig. S11; across mammalian orders: fig. S12 and tables S2 and S4). This indicates that different taxonomic groups may have evolved to have some differences in their cancer resistance mechanisms, which we study next.
Cancer resistance–associated genes in mammals, birds, and teleost fishes
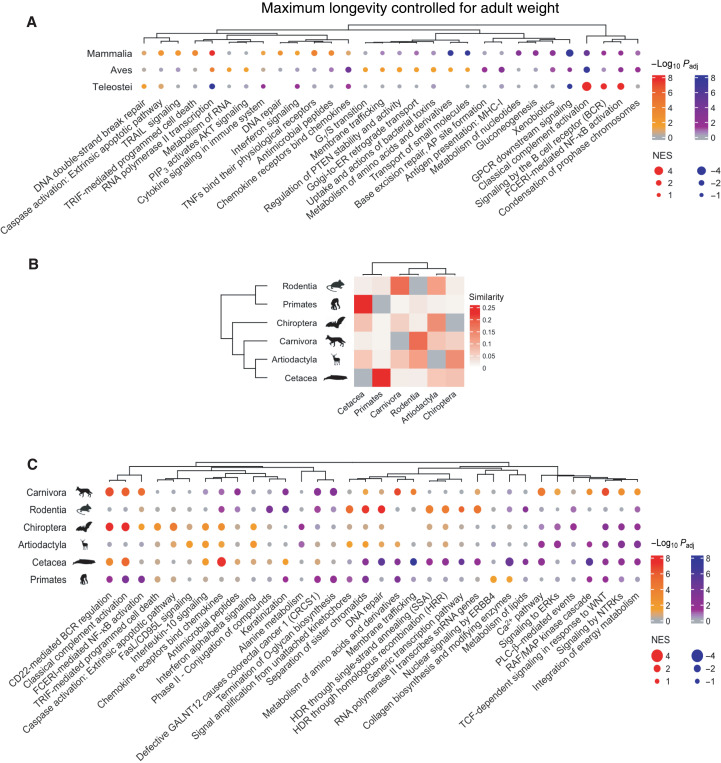
We next repeated the correlation analysis between gene conservation score and MLTAW/MLCAW scores separately for mammals, birds, and the teleost fish and computed the PC/NC gene–enriched pathways for each of the three groups (Methods). There are overall significant overlaps among the NC gene–enriched pathways of the three classes, especially based on MLCAW [odds ratio (OR), i.e., OR as large as 18.9, Fisher’s exact test adjusted P as small as 1.8 × 10−11; fig. S13, A and B, and table S3I], while the overlaps among the PC gene–enriched pathways are mostly insignificant (other than between mammals and birds using MLCAW: OR = 5.06, adjusted P = 0.037; fig. S13, A and B, and table S3I). Both common pathways [e.g., G protein–coupled receptor (GPCR) signaling] and pathways unique to specific classes [e.g., fatty acid and amino acid metabolism and phosphatidylinositol 3-kinase (PI3K)–AKT signaling pathway in birds] were observed (details in Fig. 3A, fig. S13C, and table S3).
Fig. 3. GSEA of gene conservation correlations with the cancer resistance estimate “maximum longevity controlled for adult weight” (MLCAW) specifically in different taxonomic groups.
(A) Summary visualization of the top enriched pathways (with GSEA) based on gene conservation correlations with MLCAW in Mammalia (mammals), Aves (birds), and Teleostei (teleost fishes). A selected subset of top gene sets are shown to save space, all with adjusted P < 0.1 in at least one of the classes (Methods). GSEA significance (negative log10 adjusted P values) is encoded by dot color, with two sets of colors (red-orange and blue-purple) representing positive or negative enrichment, respectively; gray color means adjusted P ≥ 0.1. Dot size represents the absolute value of normalized enrichment scores (NES) measuring the effect size of enrichment. The complete GSEA results are given in table S3. (B) Heatmap showing the similarity (Jaccard index) between the significantly enriched gene sets (FDR < 0.1) from each pair of mammalian orders, based on the MLCAW correlation. The dendrogram on the left is the phylogenetic tree of the mammalian orders, and the rows of the heatmap are arranged accordingly. The dendrogram on the top represents the hierarchical clustering of the orders based on their similarities in the GSEA results. (C) Summary visualization of the top enriched pathways (with GSEA) based on gene conservation correlations with MLCAW in different mammalian orders. This figure panel should be read as in (A).
The class Mammalia contains the largest number of species (n = 108) with available data, allowing us to further investigate the specificities in several orders, including Rodentia (rodents, n = 20), Primates (n = 18), Carnivora (carnivores, n = 18), Artiodactyla (even-toed hoofed mammals, n = 11), Cetacea (aquatic mammals like whales, n = 10), and Chiroptera (bats, n = 9). Figure 3B and fig. S13D visualize the similarities (using a Jaccard index–like measure) between the significant PC/NC gene–enriched pathways from pairs of orders (Methods). The different orders exhibit an overall similarity pattern that does not fully coincide with their phylogenetic relations (dendrograms in Fig. 3B and fig. S13D). Primates share the highest pathway-level similarity with Cetacea (Fisher’s exact test adjusted P < 2.2 × 10−16; table S5). Rodentia appears the most similar to Carnivora (Fisher’s exact test adjusted P < 2.2 × 10−16; table S5) and Artiodactyla. However, specific enriched pathways are shared across orders (table S5, Fig. 3C, and fig. S13E). This includes various cytokine signaling pathways and extrinsic apoptotic pathways that are mostly enriched by PC genes (Fig. 3C and fig. S13E), recapitulating the role of the innate immune system in the evolution of more cancer-resistant mammalian species. WNT and vascular endothelial growth factor (VEGF) signaling and lipid metabolism are among the pathways showing consistent NC gene enrichment across orders (especially based on MLTAW; Fig. 3C and fig. S13E). DNA repair–related pathways, showing PC enrichment in Rodentia and other orders, exhibit very strong NC enrichment in Cetacea (based on MLCAW; Fig. 3C). Complement cascade/activation also exhibits an order specificity (Fig. 3C and fig. S13E). These observations point to the diversity in pathways associated with cancer resistance in different mammalian orders.
Cancer resistance–associated genes are enriched for human cancer driver genes
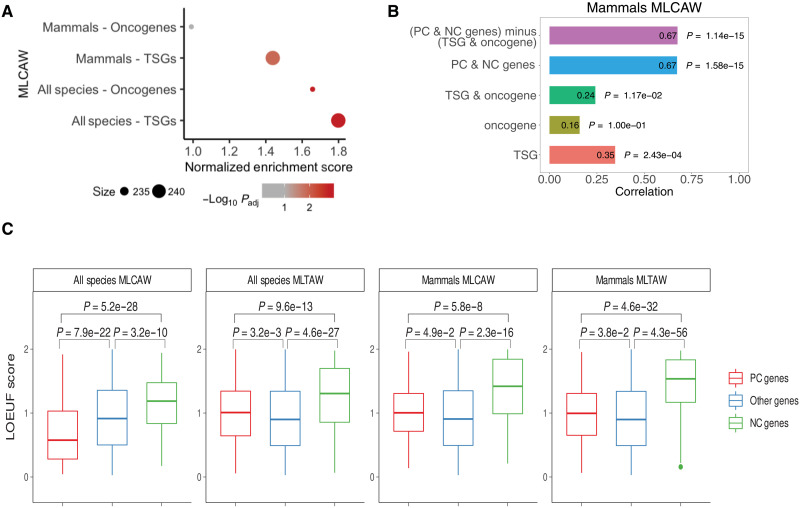
We turned to ask whether PC and NC genes are enriched for well-established human cancer driver genes [from the COSMIC database (35)]. PC genes (but not NC genes) inferred either across all species or mammals are highly enriched for human TSGs (GSEA adjusted P = 0.0011 and 0.013, respectively; Fig. 4A and table S6B) and oncogenes in the all-species analysis (GSEA adjusted P = 0.0011; Fig. 4A). These strong enrichments still hold with PC genes identified while excluding all primates (table S6B). These enriched cancer driver genes are mainly from DNA repair, RNA transcription, and PI3K-AKT signaling pathways, but not other signal transduction pathways such as tyrosine kinase receptors and estrogen receptors (table S7B and Methods). We note that excluding the human TSGs and oncogenes from the PC/NC genes when computing the CR score does not reduce the accuracy in predicting cancer resistance across species (Fig. 4B and fig. S14). Last, we find that the PC genes inferred across all species are enriched for the genes reported in various human cancer genome-wide association studies (GWAS) studies curated from the EBI GWAS Catalog [GSEA adjusted P = 0.02; enrichments still hold with PC genes identified after excluding all primates (table S6C); results were obtained using the MLCAW measure (Methods)].
Fig. 4. Enrichment analysis of PC/NC genes with human TSGs and oncogenes and comparing the LOEUF scores of PC and NC genes with other genes.
(A) Summary of enrichment via GSEA results for human TSGs or oncogenes whose conservation scores correlate with MLCAW measure in all species or mammals. Dot size corresponds to gene set size. Dot color denotes negative log10 adjusted P value from GSEA; gray corresponds to adjusted P ≥ 0.1. Positive normalized enrichment score (x axis) corresponds to enrichment by PC genes, and vice versa for NC genes. (B) Spearman’s correlation (ρ) in predicting cancer resistance (MLCAW) in mammals using only TSGs, only oncogenes, both TSGs and oncogenes, PC and NC genes in cross-validation, and PC and NC genes after removing TSGs and oncogenes in cross-validation is shown (Methods). (C) Box plots comparing the LOEUF scores of the genes whose conservation score positively (PC) or negatively correlates (NC) with a cancer resistance estimate, and the other genes in the genome, based on the two cancer resistance estimates (maximum longevity)6 × (adult weight) (MLTAW) and the residue of maximum longevity after regressing out the adult weight (MLCAW), either in all species or in mammalian species.
To study the nature of selection operating on the PC and NC genes in human evolution, we compared the LOEUF [loss-of-function (LoF) observed/expected upper bound fractions] scores of PC, NC, and the rest of the genes (background) in the human genome (with PC and NC genes defined with an adaptive cutoff based on the number of false discoveries, given in table S6A, Methods, and note S2); the higher the LOEUF score, the greater the tolerance to LoF mutations (36). We find that the NC genes have significantly higher LOEUF scores compared to PC genes and the rest of the genes in the genome (Fig. 4C and table S6D), indicating that they were subject to weaker purifying selection pressure than the PC and other genes, which is expected given that humans are considered a relatively cancer-resistant species (37).
The expression of PC genes in normal human tissues is associated with their lifetime cancer risk
As PC genes are enriched for human TSGs and oncogenes, they may also have roles in modulating human cancer risk. We hence examined whether their expression levels across different noncancerous human tissues are associated with lifetime cancer risks across these tissues, which are highly variable (6). Analyzing lifetime risk data [the SEER program (6, 38)] and the GTEx RNA sequencing (RNA-seq) data (39), we find that the MLTAW PC genes (but not MLCAW ones) are enriched for genes whose expression levels negatively correlate with cancer risk across tissues [adjusted P = 0.0088 in the all-species analysis and 0.003 in the mammal-specific analysis (Fig. 5A); results still hold after excluding primates when identifying the PC genes (table S6E)]. We do not see a similar pattern using NC genes (Fig. 5A).
Fig. 5. Enrichment analysis of PC/NC genes with genes whose expression levels correlate with the tissue-specific cancer incidence across human tissues and whose knockout causes cancer-related phenotypes in mice.
(A) Summary of the GSEA results of the top PC/NC genes from the MLTAW correlation in all species or mammals for genes whose expression levels correlate with the tissue-specific cancer incidence across human tissues (38). Dot size corresponds to gene set size. Dot color denotes negative log10 adjusted P value from GSEA; gray corresponds to adjusted P ≥ 0.1. Positive normalized enrichment score (x axis) corresponds to enrichment by genes whose higher expression is associated with higher cancer incidence across human tissues and vice versa. (B) Summary of enrichment (via GSEA) for mouse genes whose knockout causes cancer-related phenotypes in the genes whose conservation scores correlate with MLCAW in all mammals or specifically rodents. “incidence.increase” denotes the mouse genes whose knockout results in an increase in observed cancer incidence obtained from the MGI database, similarly for other gene sets listed on the x axis. Dot size and color are interpreted as in (A). Positive normalized enrichment score (y axis) corresponds to enrichment by PC genes, and vice versa for NC genes.
PC genes are associated with cancer incidence in mice and canine transmissible venereal tumors
We investigated the relevance of PC and NC genes to cancer risk in other mammalian species. We first focused on the mouse, which has been extensively studied genetically. Mining the MGI database (40), we assembled lists of genes whose knockout in the mouse results in cancer-related phenotypes including the increase/decrease of cancer incidence and cancer onset time (Methods). We find strong enrichment of the MLCAW PC genes (in all mammals and specifically rodents) in cancer incidence–increasing genes (P = 0.003; Fig. 5B and table S6F). In the all-mammal analysis, however, a weaker PC enrichment was observed for incidence-decreasing genes and “earlier onset” genes (adjusted P < 0.05; Fig. 5B).
To investigate the role of PC genes in tumorigenesis, we analyzed the expressed mutated genes in a single-cell phylogeny of a mouse melanoma model (41), in which five subclones (B1 to B5) were identified (42). The mutated genes are significantly enriched with the PC genes from the all-species MLTAW and MLCAW analysis (table S8), consistent with the putative function of PC genes as safeguards of cellular transformation. The mutated PC genes in each subclone are enriched in distinct pathways (table S8), implying that, following the initial common mutations, each subclone evolved independently by overcoming different cancer-resistant mechanisms. These results illustrated how PC genes are involved in the carcinogenic process.
In addition, we investigated canine transmissible venereal tumors (CTVTs), a naturally occurring transmissible cancer in dogs that first arose about 11,000 years ago (43). In CTVTs, more than 10,000 genes carry nonsynonymous mutations, and 646 genes have LoF via different mechanisms (43). Notably, there is a significant enrichment of the PC genes from the mammals MLTAW analysis for CTVT LoF genes (adjusted P = 0.017; fig. S15 and table S6G).
Genes associated with CMR in mammals
A recent publication by Vincze et al. (20) provided cancer-related mortality data for 191 mammalian species using data on 110,148 individual adult zoo mammals. Among the 191 mammalian species, the genomes of 39 mammals are available in publicly available datasets. We computed gene conservation scores for each of these 39 mammals and normalized them using the same procedure as before (Methods). We then identified genes whose conservation scores are significantly correlated with the CMR measure reported by Vincze et al. (20). We identified 93 and 95 genes whose conservation scores are significantly positively (termed PCMR genes) and negatively (NCMR genes) correlated with CMR (Pearson correlation, FDR < 0.2; table S15). As expected, the genes that are positively correlated (PC genes) with the cancer resistance estimate MLTAW are enriched with the genes that are negatively correlated with CMR (NCMR genes) [Fisher’s exact test; all-species analysis (for PC genes): OR = 1.79, P = 0.01, overlap = 26 genes; mammals-only analysis (for PC genes): OR = 1.64, P = 0.055, overlap = 16 genes]. Similarly, genes that are negatively correlated (NC genes) with cancer resistance are enriched with the genes that are positively correlated with CMR (PCMR genes) [Fisher’s exact test; all-species analysis (for NC genes): OR = 1.95, P = 0.0025, overlap = 31 genes; mammals-only analysis (for NC genes): OR = 3.03, P = 1.84 × 10−5, overlap = 24 genes]. There was, however, no significant overlap enrichment of PCMR/NCMR genes with NC/PC genes using the MLCAW measure.
We next computed the pathway enrichment of the genes whose conservation scores are strongly correlated with cancer risk (CMR) across the 39 species. We find 39 positively enriched pathways including complement cascade, GPCR downstream signaling, signaling by the B cell receptor, and mitotic cell cycle and 19 negatively enriched pathways including interleukin signaling, cholesterol biosynthesis, and signaling by NOTCH2 (FDR < 0.1; Fig. 6 and table S15). As expected, the positively enriched pathways with CMR significantly overlap with the negatively enriched pathways based on MLTAW/MLCAW cancer resistance estimates (Fisher’s exact test; all-species MLTAW analysis: OR = 26.93, P = 6.12 × 10−20, overlap = 26 pathways; all-species MLCAW analysis: OR = 1.64, P = 0.17, overlap = 7 pathways; mammals-only MLTAW analysis: OR = 51.2, P = 5.48 × 10−26, overlap = 26 pathways; mammals-only MLCAW analysis: OR = 29.78, P = 6.85, overlap = 26 pathways). Similarly, the negatively enriched pathways with CMR significantly overlap with the positively enriched pathways based on cancer resistance estimates (Fisher’s exact test; all-species MLTAW analysis: OR = 3.58, P = 0.068, overlap = 3 pathways; all-species MLCAW analysis: OR = 4.35, P = 0.043, overlap = 3 pathways; mammals-only MLTAW analysis: OR = 1.82, P = 0.43, overlap = 1 pathway; mammals-only MLCAW analysis: OR = 4.56, P = 0.084, overlap = 2 pathways). In sum, these results provide additional support to the association of many of the pathways pointed out earlier with cancer resistance.
Fig. 6. Summary of the top significantly pathways enriched in genes whose conservation scores are correlated with cancer risk (CMR value), using GSEA with gene set annotations from the Reactome database (adjusted P < 0.1).
Normalized enrichment scores are plotted on the y axis, where positive values correspond to enrichment by the positively correlated (PCMR) genes and negative values correspond to enrichment by the negatively correlated (NCMR) genes. The dot color represents the significance of the enrichment (negative log10 GSEA P value), and the dot size represents the number of genes in the leading edge.
Specific PC genes with strong evidence of cancer relevance across many different analyses
We manually curated the lists of PC genes, identifying a subset showing relevance to cancers based on multiple criteria according to the various analyses performed above (e.g., being human cancer drivers, genes whose knockout results in cancer-related phenotypes in mice, specific to cancer resistance estimates, and is an NCMR gene; Methods and table S9), and investigated their functions closely. Several of these curated genes have known or investigated associations with germline cancer risk syndromes. For instance, mutations in BRCA1 and BRCA2 are extremely well established in defining hereditary breast and ovarian cancer syndrome (44, 45). Risk syndromes have been defined more recently for moderate penetrance genes such as CHEK2 (breast and colon cancer) and BRIP1 (ovarian cancer) (46, 47). NBN is currently under investigation for contribution to germline breast and ovarian risk (48, 49). Some of the manually prioritized genes we identified are currently being studied for their association with cancer risk, and our results may support greater consideration of their contribution to human cancer development. For example, BUB1B, prioritized strongly in our list, is under investigation for association with early-onset colorectal cancer (50) but does not have clinically relevant screening or management recommendations at this time.
Other curated genes have known clinical associations with cancer. NPM1 and TET2 are currently used for prognostication with acute myeloid leukemias (51, 52). Bacillus Calmette-Guerin (BCG), a therapy used in early-stage bladder cancer, is a ligand for TLR2 (53, 54). Interferon-γ (IFNG) is currently being evaluated therapeutically with other immunotherapies across multiple trials (55), and mutations in DEK are currently being used as biomarkers in multiple hematologic trials (56). Numerous genes in our curated list (table S9), while linked to cancer as per our enrichment analysis, have not yet had their functional relevance clarified, such as RBM27, STAM2, SCAF4, SP140, RSBN1, SECISBP2L, THUMPD2, PIFO, and POLK. These genes may warrant higher prioritization to study their role across human cancers and potential therapeutic relevance based on our findings.
Furthermore, we ranked all the PC genes from the all-species analysis (identified using MLTAW and MLCAW estimates), based on the percentage of cancer patients from the pan-cancer TCGA cohort with nonsilent mutations (downloaded from Xena browser; table S10) (57). We find that 22 PC genes have nonsilent mutations in at least 5% of TCGA cancer patients (table S10). Some of the top-ranked genes like FAT3, KMT2C, and DNAH7 have been known to be associated with cancer (58–60).
DISCUSSION
We systematically analyzed the genomes of almost 200 species to identify genes whose conservation levels are correlated with cancer resistance estimates across different taxonomic groups and characterized their functional enrichment. We built the first genomics-based predictor of cancer resistance across species. We further studied the relevance of these phylogenetically derived cancer resistance–associated PC/NC genes to cancer development in humans.
Overall, we found that PC genes are highly relevant to carcinogenesis and enriched with cell cycle, DNA repair, immune response, and transcription regulation genes in the all-species analysis (Fig. 1 and fig. S1). These results echo those of a recent study showing that cell cycle, DNA repair, nuclear factor κB–related, and immunity pathways have higher evolutionary constraints in larger and longer-living mammals (18). Notably, this is also consistent with a long history of research establishing the association between DNA repair or genomic maintenance and longevity across species (61–63). MLTAW and MLCAW were used as two cancer resistance estimates of species, and per definition, they are correlated with each other (Spearman’s ρ = 0.45, P = 4.12 × 10−11). However, despite the overall similarity at a high level, the MLTAW and MLCAW analyses uncover different aspects of the cancer resistance mechanisms. The top PC-enriched pathways using the MLTAW measure, where both body size and life span are multiplication factors, are dominated by cell cycle regulation and transcription/RNA regulation (fig. S1), suggesting a stronger role of tissue stem cell division. The MLCAW measure, however, controls for body size, and its PC-enriched pathways include innate immunity or cell death for eradicating defective cells (64), highlighting the involvement of these factors after reaching adult size. NC genes computed with both MLTAW and MLCAW are notably enriched for processes related to cell metabolism, indicating either evolutionary metabolic constraints in the smaller/shorter-lived species or accelerated evolution of metabolism in the larger/longer-lived species (32).
Another notable pattern is the variability in the PC/NC gene functions across different taxonomic groups—it is frequently observed that genes of one pathway can be PC in one group but NC in another. Such variation may reflect a trade-off between individual life span and survival/reproductive function dependent on the different lifestyles in different groups of species. Some of the observed order-specific enrichments are consistent with known mechanisms of cancer resistance for the corresponding species. For example, the naked mole rat is known to have more efficient excision repair systems and be more resistant to bleomycin-induced somatic mutations than the mouse (63, 65), and an active complement system has been observed in bats (66, 67). In comparison, the enrichment results differ considerably across the taxonomic classes (mammals, birds, and teleost fishes), and it is perhaps not surprising that the PC/NC genes identified in birds or teleost fishes were not found to be strongly enriched for mammalian cancer-related genes with GSEA. Therefore, in the latter part of our study, we mainly relied on the PC/NC genes from the analyses involving mammals to identify and further test potential novel genes related to cancer resistance in humans. The PC genes are enriched for known cancer driver genes in several mammalian species (human, mouse, and dog), demonstrating the validity of our comparative genomics approach in identifying genes relevant to cancer development or resistance.
We outline several limitations of our study. First, the gene conservation computation is based on comparison to a reference species and rank normalization, which does not consider gene copies, paralogous genes, or the phylogenetic tree structure. Yet, notably, the rank normalization gene conservation scores used in our analysis effectively remove potentially confounding effects of phylogenetic distance. While alternative methods may be used to adjust for the inter-phylogeny distances, we showed that our results are robust to the choice of the reference species (e.g., with house mouse, which is a known cancer-prone species unlike humans, large species like sperm whales, cancer-resistant species like the naked mole rat, and evolutionarily distant species like birds, fish, and plants; details in note S4) and various other conservation scoring parameters. Yet, we should note that we have chosen to use a simple approach that does not use phylogeny-aware parametric models, and the possible use of the latter in follow-up studies may possibly alter some of the results. We also note that we do not account for the number of paralogs, which is another potential confounding factor. However, as many downstream analyses have been performed on the pathway level, this may mitigate the potential confounding effect of paralogs. The full-scale identification of gene copy numbers across all species is quite challenging and out of the scope of the current investigation. Performing this analysis just for top-ranked NC and PC genes, we find that most mammalian species harbor only a single copy of the top PC/NC genes, suggesting that copy number variation is unlikely to greatly modify our results. However, a few PC/NC genes do have an increased copy number in well-known cancer-resistant species (see note S6 and table S11). Future studies are warranted to comprehensively investigate the association between gene copy number and cancer resistance across species. Second, while the MLTAW and MLCAW analysis is based on established proxy cancer resistance estimates, some of the PC/NC genes we identified may be mainly or even solely involved in body size or life-span evolution. This may be due to the well-recognized close relation between body size, life span, and cancer development. Although we have tried to identify a subset of PC/NC genes that are also associated with CMR in 39 mammals (table S15), some of the PC/NC genes may be simply constrained or important for development and this may drive the signal independently of cancer. However, while further studies are deemed to test the causal roles of PC and NC genes (and see the curated gene list presented in table S9) in human carcinogenesis, the significant enrichment in known cancer genes and the knockout mouse data supports the causal role of many of the PC genes in cancer resistance (Fig. 5B). We also note though that the MLTAW and MLCAW estimates cannot capture variations in cancer resistance that are not reflected through body size and life span, e.g., those related to adaptation to different oxygen and oxidative stress levels (note S7) (68). Yet, given these cautionary notes, we think that this customized use of previously established cancer resistance measures does serve for identifying cancer resistance–related genes beyond associations with either body size or life span solely.
In summary, this study presents a systematic species comparison identifying key genes and pathways associated with cancer resistance across species. Many of the genes identified are implicated in human cancers, and their further study may increase our understanding of human cancer development, prevention, and treatment.
METHODS
Computation of gene conservation scores
We created a matrix of gene conservation scores for across over 1600 species with human genome as a reference [240 species out of them were a part of the AnAge phenotypic database (26)]. The amino acid sequence of the proteins in all of these species is available in UniProt (23), RefSeq (24), Keane et al. (13), and Ensembl (25) databases. The conservation scores (or ranked phylogenetic profiling) were calculated using the protein sequence similarity between each gene in the human genome, and its homologs in each of the species were measured by the bit score computed with BLASTP (28). For each human gene and each species, we only considered the matched gene with the highest bit score. While other good approaches are available like reciprocal blast, our method has been widely used and worked well using human and other reference genomes (69–71). To reduce the influence of random matches, the bit scores were set to 0 for matches with E value > 1 × 10−5. Bit score is known to be affected by the length of the reference protein. To eliminate the protein length effect, we normalize to protein length by dividing each bit score by the score of the reference protein against itself, resulting in values between 0 and 1 (21, 22). Last, the conservation scores were obtained by rank-normalizing the protein length normalized bit scores across genes within each species to control for the evolutionary distance between human and each species. These rank-normalized values range from 0 to 1, with higher values corresponding to higher levels of conservation (note S9). To examine whether the use of human (considered a relatively cancer-resistant species) as the reference affects the results, in a similar manner, we also repeated the above computation using a cancer-prone species like house mouse as reference.
Recently, Vincze et al. (20) provided cancer-related mortality of 191 mammalian species using data on 110,148 individual adult zoo mammals. We downloaded and analyzed the 39 genomes of these mammalian species that were available in public datasets (Ensembl, UniProt, and RefSeq). We computed gene conservation scores for these 39 mammals and normalized them using the same procedures as those described above.
Cancer resistance estimates
Since the cancer incidence in nonhuman species is unknown, we used two indirect methods to estimate the level of cancer resistance in a species. Let AW stand for adult weight and ML for maximum longevity of a species; we define the two cancer resistance estimates/measures as follows: MLTAW measure: log(ML6 × AW); MLCAW or “maximum longevity controlled for adult weight” measure: residue obtained by regressing out log(AW) from log(ML), using linear regression.
MLTAW and MLCAW were computed for 193 of the 240 species for which both ML and AW data are available in the AnAge database (26). These 193 species are from various classes or taxonomy groups: 108 Mammalia (mammals), 55 Aves (birds), 18 Teleostei (ray-finned fishes), 7 Reptilia (reptiles), 1 Amphibia (amphibians), 1 Cephalaspidomorphi (jawless fishes), 1 Chondrichthyes (cartilaginous fishes), 1 Coelacanthi (lobe-finned fishes), and 1 Holostei (bony fishes).
Identification of cancer resistance–associated genes
To identify cancer resistance–associated genes (PC or NC genes), we computed the Pearson correlation coefficient between the conservation scores of each gene and each of the two cancer resistance estimates (MLTAW and MLCAW) after proper transformation (described above). Pearson correlation was chosen (instead of Spearman’s correlation) to reduce the number of ties in further GSEA analysis for pathway enrichment. The robust identification of PC/NC genes is independent of the correlation measure used (see note S4 for details). Among the genes with Benjamini-Hochberg adjusted P values (FDR) less than 0.1 or 0.01, those with correlation estimates > 0 are defined as PC genes, while those with correlation estimates < 0 are NC genes. This analysis was done for all species or within certain groups of species. PC and NC genes were identified separately based on each of the two cancer resistance estimates (MLTAW/MLCAW).
Identification of PC/NC genes associated with cancer resistance, but not with longevity or biomass
We identify genes whose conservation scores are correlated with either maximum longevity or adult weight across species (Pearson’s correlation, FDR < 0.1; from the AnAge data resource, we had maximum longevity and adult weight data for 226 and 205 species, respectively). We remove these genes from our original lists of PC and NC genes (from all-species analysis), resulting in lists of genes that significantly correlate exclusively with the cancer resistance measures but not with either maximum longevity or adult weight alone. Pathway enrichment analysis was done on these genes in the same manner as reported above (we also repeated this analysis using only mammals instead of all-species).
Cancer resistance predictor
Since higher conservation scores of the PC genes correspond to a higher level of cancer resistance, and vice versa for the NC genes, we define a cancer resistance (CR) score for each species as follows: CR score = [(Number of PC genes with conservation scores > MCS) + (Number of NC genes with conservation scores < MCS)]/(Total number of genes), where MCS is the median conservation score of all genes in a species. PC and NC genes are chosen for FDR < 0.1. We also repeat this analysis for different thresholds (some other quantile other than median) or FDR thresholds for robustness studies. The total number of genes is 20,076 in our analysis when we used human as reference.
Cross-validation analysis was mainly done in a leave-one-out manner. For each test sample, we identify PC and NC genes on the training set and predict CR scores on the test set. For robustness tests, we also did a twofold cross-validation, i.e., identify PC and NC genes in the training group and test the accuracy of the CR predictions in the left-out group. We also do cross-validation by leaving out an entire group of species and identifying PC and NC genes from the remaining species, and testing on the left-out group. For the all-species analysis, we left one class out (for different classes), and for the mammalian analysis, we left one order out (for different orders).
Modifications of cancer resistance predictions
We explored the prediction of cancer resistance using only PC genes or NC genes as follows: CR score = (Number of PC genes with conservation scores > MCS)/(Total number of genes) or CR score = (Number of NC genes with conservation scores < MCS)/(Total number of genes). We also predicted cancer resistance using either human TSGs or oncogenes obtained from the Cancer Gene Census dataset from the COSMIC database (35). Specifically, we used either TSGs alone, oncogenes alone, or TSGs combined with oncogenes to compute the CR score: (Number of TSGs, or oncogenes, or combined > MCS)/(Total number of genes), where MCS is the median conservation score of all genes in a species.
Pathway, cancer driver gene, and other cancer-related GSEA
The biological pathway annotation data were downloaded from the Reactome database (72). The sets of curated human oncogenes and TSGs were obtained from the Cancer Gene Census dataset from the COSMIC database (35). Significant markers reported in various GWAS studies linked to human cancers were collected from the EBI GWAS Catalog database (73) using the keyword “cancer” as the phenotypes/traits. Variants in stronger linkage disequilibrium (LD) (with D′ ≥ 0.8 and r2 ≥ 0.3) with the GWAS-associated markers (within 500,000 base pairs in each side) in loci, replicated in more than one study, were selected using the R package, LDlinkR (74), and genes containing such variants were selected. The sets of genes whose knockout can result in various cancer-related phenotypes in mice were obtained from the MGI database (40). Specifically, we selected the genes for which the allele attributes are “Null/knockout,” and the corresponding phenotype terms are “increased cancer incidence,” “decreased cancer incidence,” “increased cancer latency,” and “decreased cancer latency.” The set of LoF genes identified in CTVTs was obtained from Murchison et al. (43). The enrichment for each of the biological pathways and cancer-related gene sets based on the MLTAW or MLCAW correlation results was tested with GSEA on a ranked list of all 20,076 genes [GSEA (75); ranking based on the correlation coefficients between conservation scores and MLTAW/MLCAW values].
Testing for the mutual exclusivity of the pathway membership for PC and NC genes
Permutation tests were used to determine whether PC and NC genes tend to coexist in the same biological pathways or exist in distinct nonoverlapping pathways. More specifically, given a preidentified set of m PC genes and n NC genes, in each permutation (shuffling), we randomly assigned m out of the same set of (m + n) genes to be PC genes, and the rest to be NC genes. For each set of PC and NC genes identified from the analysis across all species (with either MLCAW or MLTAW, using the fixed cutoff of FDR < 0.1), the unique pathways that they are part of were identified on the basis of the Reactome database (72). The total number of pathways shared between the PC genes and the NC genes was computed and compared to the null distribution formed by permuting the PC and NC category labels of the genes to obtain a permutation test P value. We provide the number of PC and NC genes in each pathway in table S12.
Detailed analysis of PC/NC gene enrichment in the known human cancer genes
To investigate whether our method may be specifically effective in recovering a subset of cancer genes acting via certain mechanisms, we identified the subset of known human cancer genes from the COSMIC database (35) that overlap with either significant PC or NC genes (FDR < 0.1), as well as the complementary subset that do not overlap with any significant PC/NC genes. Pathway enrichment of each of these cancer gene subsets was performed with Fisher’s exact test using the pathway annotation from the Reactome database (72).
Comparing the pathway enrichment results from different taxonomic groups
The pathway enrichment results from mammals, birds, and teleost fishes, as well as from different mammalian orders were compared. The mammalian orders we analyzed include Rodentia (rodents, n = 20), Primates (n = 18), Carnivora (carnivores, n = 18), Artiodactyla (even-toed hoofed mammals, n = 11), Cetacea (aquatic mammals like whales, n = 10), and Chiroptera (bats, n = 9). We used a Jaccard index–like metric to measure the similarity of the sets of enriched pathways between each pair of taxonomic groups. Specifically, let PA and NA be the sets of significant (FDR < 0.1) positively and negatively enriched pathways from a taxonomic group A, and similarly, let PB and NB be the sets of positively and negatively enriched pathways from a taxonomic group B, then the Jaccard-like similarity measure between group A and group B was computed as follows: (|PA∩PB| + |NA∩NB|)/(|PA∪PB| + |NA∪NB|). For visualization (Fig. 3, A and C), because of space limitation, only a subset of the top enriched pathways was displayed. Specifically, the pathways included in the visualizations were those among the top 10 most significantly positively enriched or the top 10 most significantly negatively enriched pathways in at least one of the taxonomic groups being compared.
LOEUF score analysis for PC and NC genes
The LOEUF scores of all human genes were obtained from Karczewski et al. (36). Lower LOEUF scores correspond to less tolerance to LoF genomic variations in humans. The LOEUF scores of the PC (or NC) genes we identified were compared with each other or to the rest of the genes in the human genome with Wilcoxon rank sum tests. Given that a homogeneous adjusted P value cutoff produced drastically different numbers of significant genes from different analyses (e.g., with adjusted P < 0.05, there are more than 2800 significant genes from the primate MLTAW correlation analysis, but only 78 from the rodent MLTAW correlation), here, the PC and NC genes were selected instead based on the criterion of “expected number of false discoveries (76) smaller than 1.” However, if this criterion results in fewer than 100 genes, then a less stringent criterion of “expected number of false discoveries smaller than 5” was used. These sets of PC and NC genes are given in table S6A.
Analysis of the genes associated with lifetime cancer risk across human tissues
The data on the lifetime cancer risk in each of the human tissue/organ sites were obtained from the SEER database (38), and RNA-seq data of normal human tissue samples across multiple tissue types were downloaded from the GTEx database (39). For each gene, we computed its median expression level in each tissue type and then computed the Spearman’s correlation coefficient between the median expression value and the lifetime cancer risk across tissue types. All the genes were ranked by the Spearman’s correlation coefficient, and the enrichment of the PC or NC genes for genes associated with lifetime cancer risk across human tissues was tested with GSEA. The criterion for identifying PC and NC genes is the same as that described in the previous section.
Selection of subsets of PC/NC genes with high relevance to human cancers based on multiple criteria
For each of the PC/NC genes from the various analyses (at FDR < 0.1), we look for supporting evidence from many of the different analyses described in the article. Evidence considered for cancer relevance includes those instances where a gene (i) is a PC or NC gene (at FDR < 0.1) for the all-species, mammals-only, birds-only analysis using both the estimates; (ii) is a human oncogene or tumor suppressor; (iii) whose knockout causes early cancer incidence or early cancer onset in mice; (iv) is a LoF gene in CTVT; (v) is a GWAS gene associated with human cancers; (vi) expressed mutated genes in single-cell phylogeny of a mouse melanoma model; (vii) is specifically associated with cancer resistance estimates but not associated with maximum longevity and adult weight in all-species and mammals-only analysis; and (viii) is an NCMR gene (negatively correlated with true CMR in 39 mammals). We then rank each gene by the number of times of support in table S9.
Acknowledgments
This work used the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). We thank X. Wei Wang for helpful comments on this work.
Funding: This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, and the Center for Cancer Research.
Author contributions: N.U.N., K.C., Y.T., and E.R. conceived the project. E.R. and Y.T. supervised the research work. N.U.N. and K.C. carried out most of the analysis with help from L.N., E.S., and L.R.P. The gene conservation scores were computed by E.S. The data were analyzed and interpreted by N.U.N., K.C., Y.T., E.R., L.N., P.S.R., I.U., K.A., S.H., and C.-P.D. The paper was written by N.U.N., K.C., Y.T., and E.R. with inputs from all authors. All authors have read and commented on the manuscript.
Competing interests: E.R. is a co-founder of Metabomed Ltd. and MedAware, and a (divested) co-founder and nonpaid scientific consultant for Pangea Biomed. The other authors declare no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The code and data used for cancer resistance prediction, and a few other key analyses are made available at (https://hpc.nih.gov/~Lab_ruppin/species_cancer_resistance_ScienceAdvances.zip) for the sake of reproducibility.
Supplementary Materials
This PDF file includes:
Figs. S1 to S17
Notes S1 to S10
References
Other Supplementary Material for this manuscript includes the following:
Table S1 to S15
REFERENCES AND NOTES
- 1.Albuquerque T. A. F., Drummond do Val L., Doherty A., de Magalhães J. P., From humans to hydra: Patterns of cancer across the tree of life. Biol. Rev. Camb. Philos. Soc. 93, 1715–1734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbunova V., Seluanov A., Zhang Z., Gladyshev V. N., Vijg J., Comparative genetics of longevity and cancer: Insights from long-lived rodents. Nat. Rev. Genet. 15, 531–540 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seluanov A., Gladyshev V. N., Vijg J., Gorbunova V., Mechanisms of cancer resistance in long-lived mammals. Nat. Rev. Cancer 18, 433–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordling C. O., A new theory on the cancer-inducing mechanism. Br. J. Cancer 7, 68–72 (1953). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomasetti C., Li L., Vogelstein B., Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355, 1330–1334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomasetti C., Vogelstein B., Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khankari N. K., Shu X.-O., Wen W., Kraft P., Lindström S., Peters U., Schildkraut J., Schumacher F., Bofetta P., Risch A., Bickeböller H., Amos C. I., Easton D., Eeles R. A., Gruber S. B., Haiman C. A., Hunter D. J., Chanock S. J., Pierce B. L., Zheng W.; Colorectal Transdisciplinary Study (CORECT); Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE); Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE); Transdisciplinary Research in Cancer of the Lung (TRICL) , Association between adult height and risk of colorectal, lung, and prostate cancer: Results from meta-analyses of prospective studies and mendelian randomization analyses. PLOS Med. 13, e1002118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.R. Peto, Origins of Human Cancer (Cold Spring Harbor Publications, 1977), pp. 1403–1428. [Google Scholar]
- 9.Tollis M., Boddy A. M., Maley C. C., Peto’s Paradox: How has evolution solved the problem of cancer prevention? BMC Biol. 15, 60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeno Y., Hubbard G. B., Lee S., Cortez L. A., Lew C. M., Webb C. R., Berryman D. E., List E. O., Kopchick J. J., Bartke A., Reduced incidence and delayed occurrence of fatal neoplastic diseases in growth hormone receptor/binding protein knockout mice. J. Gerontol. A Biol. Sci. Med. Sci. 64, 522–529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipman R., Galecki A., Burke D. T., Miller R. A., Genetic loci that influence cause of death in a heterogeneous mouse stock. J. Gerontol. A Biol. Sci. Med. Sci. 59, 977–983 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szymanska H., Lechowska-Piskorowska J., Krysiak E., Strzalkowska A., Unrug-Bielawska K., Grygalewicz B., Skurzak H. M., Pienkowska-Grela B., Gajewska M., Neoplastic and nonneoplastic lesions in aging mice of unique and common inbred strains contribution to modeling of human neoplastic diseases. Vet. Pathol. 51, 663–679 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Keane M., Semeiks J., Webb A. E., Li Y. I., Quesada V., Craig T., Madsen L. B., van Dam S., Brawand D., Marques P. I., Michalak P., Kang L., Bhak J., Yim H.-S., Grishin N. V., Nielsen N. H., Heide-Jørgensen M. P., Oziolor E. M., Matson C. W., Church G. M., Stuart G. W., Patton J. C., George J. C., Suydam R., Larsen K., López-Otín C., O’Connell M. J., Bickham J. W., Thomsen B., de Magalhães J. P., Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10, 112–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George J. C., Bada J., Zeh J., Scott L., Brown S. E., O’Hara T., Suydam R., Age and growth estimates of bowhead whales (Balaena mysticetus) via aspartic acid racemization. Can. J. Zool. 77, 571–580 (1999). [Google Scholar]
- 15.Vicens A., Posada D., Selective pressures on human cancer genes along the evolution of mammals. Genes 9, 582 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tollis M., Schneider-Utaka A. K., Maley C. C., The evolution of human cancer gene duplications across mammals. Mol. Biol. Evol. 37, 2875–2886 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vazquez J. M., Lynch V. J., Pervasive duplication of tumor suppressors in Afrotherians during the evolution of large bodies and reduced cancer risk. eLife 10, e65041 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowalczyk A., Partha R., Clark N. L., Chikina M., Pan-mammalian analysis of molecular constraints underlying extended lifespan. eLife 9, e51089 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferris E., Abegglen L. M., Schiffman J. D., Gregg C., Accelerated evolution in distinctive species reveals candidate elements for clinically relevant traits, including mutation and cancer resistance. Cell Rep. 22, 2742–2755 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vincze O., Colchero F., Lemaître J.-F., Conde D. A., Pavard S., Bieuville M., Urrutia A. O., Ujvari B., Boddy A. M., Maley C. C., Thomas F., Giraudeau M., Cancer risk across mammals. Nature 601, 263–267 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabach Y., Billi A. C., Hayes G. D., Newman M. A., Zuk O., Gabel H., Kamath R., Yacoby K., Chapman B., Garcia S. M., Borowsky M., Kim J. K., Ruvkun G., Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature 493, 694–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabach Y., Golan T., Hernández-Hernández A., Messer A. R., Fukuda T., Kouznetsova A., Liu J.-G., Lilienthal I., Levy C., Ruvkun G., Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling. Mol. Syst. Biol. 9, 692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.UniProt Consortium , UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Leary N. A., Wright M. W., Brister J. R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., Astashyn A., Badretdin A., Bao Y., Blinkova O., Brover V., Chetvernin V., Choi J., Cox E., Ermolaeva O., Farrell C. M., Goldfarb T., Gupta T., Haft D., Hatcher E., Hlavina W., Joardar V. S., Kodali V. K., Li W., Maglott D., Masterson P., McGarvey K. M., Murphy M. R., O’Neill K., Pujar S., Rangwala S. H., Rausch D., Riddick L. D., Schoch C., Shkeda A., Storz S. S., Sun H., Thibaud-Nissen F., Tolstoy I., Tully R. E., Vatsan A. R., Wallin C., Webb D., Wu W., Landrum M. J., Kimchi A., Tatusova T., DiCuccio M., Kitts P., Murphy T. D., Pruitt K. D., Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howe K. L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M. R., Armean I. M., Azov A. G., Bennett R., Bhai J., Billis K., Boddu S., Charkhchi M., Cummins C., Fioretto L. D. R., Davidson C., Dodiya K., Houdaigui B. E., Fatima R., Gall A., Giron C. G., Grego T., Guijarro-Clarke C., Haggerty L., Hemrom A., Hourlier T., Izuogu O. G., Juettemann T., Kaikala V., Kay M., Lavidas I., Le T., Lemos D., Martinez J. G., Marugán J. C., Maurel T., McMahon A. C., Mohanan S., Moore B., Muffato M., Oheh D. N., Paraschas D., Parker A., Parton A., Prosovetskaia I., Sakthivel M. P., Salam A. I. A., Schmitt B. M., Schuilenburg H., Sheppard D., Steed E., Szpak M., Szuba M., Taylor K., Thormann A., Threadgold G., Walts B., Winterbottom A., Chakiachvili M., Chaubal A., Silva N. D., Flint B., Frankish A., Hunt S. E., IIsley G. R., Langridge N., Loveland J. E., Martin F. J., Mudge J. M., Morales J., Perry E., Ruffier M., Tate J., Thybert D., Trevanion S. J., Cunningham F., Yates A. D., Zerbino D. R., Flicek P., Ensembl 2021. Nucleic Acids Res. 49, D884–D891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tacutu R., Thornton D., Johnson E., Budovsky A., Barardo D., Craig T., Diana E., Lehmann G., Toren D., Wang J., Fraifeld V. E., de Magalhães J. P., Human Ageing Genomic Resources: New and updated databases. Nucleic Acids Res. 46, D1083–D1090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellegrini M., Marcotte E. M., Thompson M. J., Eisenberg D., Yeates T. O., Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. U.S.A. 96, 4285–4288 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 29.Sherill-Rofe D., Rahat D., Findlay S., Mellul A., Guberman I., Braun M., Bloch I., Lalezari A., Samiei A., Sadreyev R., Goldberg M., Orthwein A., Zick A., Tabach Y., Mapping global and local coevolution across 600 species to identify novel homologous recombination repair genes. Genome Res. 29, 439–448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braun M., Sharon E., Unterman I., Miller M., Shtern A. M., Benenson S., Vainstein A., Tabach Y., ACE2 co-evolutionary pattern suggests targets for pharmaceutical intervention in the COVID-19 pandemic. iScience 23, 101384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peto R., Quantitative implications of the approximate irrelevance of mammalian body size and lifespan to lifelong cancer risk. Philos. Trans. R Soc. Lond. B Biol. Sci. 370, 20150198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speakman J. R., Body size, energy metabolism and lifespan. J. Exp. Biol. 208, 1717–1730 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Varki N. M., Varki A., On the apparent rarity of epithelial cancers in captive chimpanzees. Philos. Trans. R Soc. Lond. B Biol. Sci. 370, 20140225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson G. S., Adams D. M., Recurrent evolution of extreme longevity in bats. Biol. Lett. 15, 20180860 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forbes S. A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., Kok C. Y., Jia M., De T., Teague J. W., Stratton M. R., McDermott U., Campbell P. J., COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karczewski K. J., Francioli L. C., Tiao G., Cummings B. B., Alföldi J., Wang Q., Collins R. L., Laricchia K. M., Ganna A., Birnbaum D. P., Gauthier L. D., Brand H., Solomonson M., Watts N. A., Rhodes D., Singer-Berk M., England E. M., Seaby E. G., Kosmicki J. A., Walters R. K., Tashman K., Farjoun Y., Banks E., Poterba T., Wang A., Seed C., Whiffin N., Chong J. X., Samocha K. E., Pierce-Hoffman E., Zappala Z., O’Donnell-Luria A. H., Minikel E. V., Weisburd B., Lek M., Ware J. S., Vittal C., Armean I. M., Bergelson L., Cibulskis K., Connolly K. M., Covarrubias M., Donnelly S., Ferriera S., Gabriel S., Gentry J., Gupta N., Jeandet T., Kaplan D., Llanwarne C., Munshi R., Novod S., Petrillo N., Roazen D., Ruano-Rubio V., Saltzman A., Schleicher M., Soto J., Tibbetts K., Tolonen C., Wade G., Talkowski M. E.; Genome Aggregation Database Consortium, Neale B. M., Daly M. J., MacArthur D. G., The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balmain A., Harris C. C., Carcinogenesis in mouse and human cells: Parallels and paradoxes. Carcinogenesis 21, 371–377 (2000). [DOI] [PubMed] [Google Scholar]
- 38.E. SEER. Surveillance, and End Results (SEER) Program (www.seer.cancer.gov) SEER*Stat Database Incidence, SEER 9 RegsResearch Data, Nov 2017 Sub (1973–2015) - Linked To County Attributes - Total U.S., 1969–2016 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2018, based on the November 2017 submission. (2018).
- 39.GTEx Consortium , The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bult C. J., Blake J. A., Smith C. L., Kadin J. A., Richardson J. E.; Mouse Genome Database Group , Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 47, D801–D806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pérez-Guijarro E., Yang H. H., Araya R. E., Meskini R. E., Michael H. T., Vodnala S. K., Marie K. L., Smith C., Chin S., Lam K. C., Thorkelsson A., Iacovelli A. J., Kulaga A., Fon A., Michalowski A. M., Hugo W., Lo R. S., Restifo N. P., Sharan S. K., Dyke T. V., Goldszmid R. S., Ohler Z. W., Lee M. P., Day C.-P., Merlino G., Multimodel preclinical platform predicts clinical response of melanoma to immunotherapy. Nat. Med. 26, 781–791 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.K. L. Marie, E. Pérez-Guijarro, S. Malikić, E. S. Azer, H. H. Yang, C. Kızılkale, C. Gruen, W. Robinson, H. Liu, M. C. Kelly, C. Marcelus, S. Burkett, A. Buluç, F. Ergün, M. P. Lee, G. Merlino, C.-P. Day, S. C. Sahinalp, Profiles of expressed mutations in single cells reveal subclonal expansion patterns and therapeutic impact of intratumor heterogeneity. bioRxiv 2021.03.26.437185 [Preprint]. 23 May 2021. 10.1101/2021.03.26.437185. [DOI]
- 43.Murchison E. P., Wedge D. C., Alexandrov L. B., Fu B., Martincorena I., Ning Z., Tubio J. M. C., Werner E. I., Allen J., De Nardi A. B., Donelan E. M., Marino G., Fassati A., Campbell P. J., Yang F., Burt A., Weiss R. A., Stratton M. R., Transmissible dog cancer genome reveals the origin and history of an ancient cell lineage. Science 343, 437–440 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S., Parmigiani G., Meta-analysis of BRCA1 and BRCA2 penetrance. J. Clin. Oncol. 25, 1329–1333 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuchenbaecker K. B., Hopper J. L., Barnes D. R., Phillips K.-A., Mooij T. M., Roos-Blom M.-J., Jervis S., van Leeuwen F. E., Milne R. L., Andrieu N., Goldgar D. E., Terry M. B., Rookus M. A., Easton D. F., Antoniou A. C.; the BRCA1 and BRCA2 Cohort Consortium, Guffog L. M., Evans D. G., Barrowdale D., Frost D., Adlard J., Ong K.-R., Izatt L., Tischkowitz M., Eeles R., Davidson R., Hodgson S., Ellis S., Nogues C., Lasset C., Stoppa-Lyonnet D., Fricker J.-P., Faivre L., Berthet P., Hooning M. J., van der Kolk L. E., Kets C. M., Adank M. A., John E. M., Chung W. K., Andrulis I. L., Southey M., Daly M. B., Buys S. S., Osorio A., Engel C., Kast K., Schmutzler R. K., Caldes T., Jakubowska A., Simard J., Friedlander M. L., McLachlan S.-A., Machackova E., Foretova L., Tan Y. Y., Singer C. F., Olah E., Gerdes A.-M., Arver B., Olsson H., Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317, 2402–2416 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Cybulski C., Wokołorczyk D., Jakubowska A., Huzarski T., Byrski T., Masojć J. G., Dębniak T., Górski B., Blecharz P., Narod S. A., Lubiński J., Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer. J. Clin. Oncol. 29, 3747–3752 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Ramus S. J., Song H., Dicks E., Tyrer J. P., Rosenthal A. N., Intermaggio M. P., Fraser L., Gentry-Maharaj A., Hayward J., Philpott S., Anderson C., Edlund C. K., Conti D., Harrington P., Barrowdale D., Bowtell D. D., Alsop K., Mitchell G.; AOCS Study Group, Cicek M. S., Cunningham J. M., Fridley B. L., Alsop J., Jimenez-Linan M., Poblete S., Lele S., Sucheston-Campbell L., Moysich K. B., Sieh W., Guire V. M., Lester J., Bogdanova N., Dürst M., Hillemanns P.; Ovarian Cancer Association Consortium, Odunsi K., Whittemore A. S., Karlan B. Y., Dörk T., Goode E. L., Menon U., Jacobs I. J., Antoniou A. C., Pharoah P. D. P., Gayther S. A., Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J. Natl. Cancer Inst. 107, dvj214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurian A. W., Hughes E., Handorf E. A., Gutin A., Allen B., Hartman A.-R., Hall M. J., Breast and ovarian cancer penetrance estimates derived from germline multiple-gene sequencing results in women. JCO Precis. Oncol. 1, 1–12 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Zhang B., Beeghly-Fadiel A., Long J. R., Zheng W., Genetic variants associated with breast-cancer risk: Comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol. 12, 477–488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hahn M.-M., Vreede L., Bemelmans S. A. S. A., van der Looij E., van Kessel A. G., Schackert H. K., Ligtenberg M. J. L., Hoogerbrugge N., Kuiper R. P., de Voer R. M., Prevalence of germline mutations in the spindle assembly checkpoint gene BUB1B in individuals with early-onset colorectal cancer. Gene Chromosome Cancer 55, 855–863 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Chou W.-C., Chou S.-C., Liu C.-Y., Chen C.-Y., Hou H.-A., Kuo Y.-Y., Lee M.-C., Ko B.-S., Tang J.-L., Yao M., Tsay W., Wu S.-J., Huang S.-Y., Hsu S.-C., Chen Y.-C., Chang Y.-C., Kuo Y.-Y., Kuo K.-T., Lee F.-Y., Liu M.-C., Liu C.-W., Tseng M.-H., Huang C.-F., Tien H.-F., TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood 118, 3803–3810 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Verhaak R. G. W., Goudswaard C. S., van Putten W., Bijl M. A., Sanders M. A., Hugens W., Uitterlinden A. G., Erpelinck C. A. J., Delwel R., Löwenberg B., Valk P. J. M., Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): Association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood 106, 3747–3754 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Fuge O., Vasdev N., Allchorne P., Green J. S., Immunotherapy for bladder cancer. Res. Rep. Urol. 7, 65–79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urban-Wojciuk Z., Khan M. M., Oyler B. L., Fåhraeus R., Marek-Trzonkowska N., Nita-Lazar A., Hupp T. R., Goodlett D. R., The role of TLRs in anti-cancer immunity and tumor rejection. Front. Immunol. 10, 2388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ni L., Lu J., Interferon gamma in cancer immunotherapy. Cancer Med. 7, 4509–4516 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanden C., Gullberg U., The DEK oncoprotein and its emerging roles in gene regulation. Leukemia 29, 1632–1636 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Ellrott K., Bailey M. H., Saksena G., Covington K. R., Kandoth C., Stewart C., Hess J., Ma S., Chiotti K. E., McLellan M., Sofia H. J., Hutter C., Getz G., Wheeler D., Ding L.; MC3 Working Group; Cancer Genome Atlas Research Network , Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 6, 271–281.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gala K., Li Q., Sinha A., Razavi P., Dorso M., Sanchez-Vega F., Chung Y. R., Hendrickson R., Hsieh J. J., Berger M., Schultz N., Pastore A., Abdel-Wahab O., Chandarlapaty S., KMT2C mediates the estrogen dependence of breast cancer through regulation of ERα enhancer function. Oncogene 37, 4692–4710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo Z., Yan X., Song C., Wang Q., Wang Y., Liu X.-P., Huang J., Li S., Hu W., FAT3 mutation is associated with tumor mutation burden and poor prognosis in esophageal cancer. Front. Oncol. 11, 603660 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu C., Yang Q., Xu J., Zhao W., Zhang Z., Xu D., Zhang Y., Zhao E., Zhao G., Somatic mutation of DNAH genes implicated higher chemotherapy response rate in gastric adenocarcinoma patients. J. Transl. Med. 17, 109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cagan A., Baez-Ortega A., Brzozowska N., Abascal F., Coorens T. H. H., Sanders M. A., Lawson A. R. J., Harvey L. M. R., Bhosle S., Jones D., Alcantara R. E., Butler T. M., Hooks Y., Roberts K., Anderson E., Lunn S., Flach E., Spiro S., Januszczak I., Wrigglesworth E., Jenkins H., Dallas T., Masters N., Perkins M. W., Deaville R., Druce M., Bogeska R., Milsom M. D., Neumann B., Gorman F., Constantino-Casas F., Peachey L., Bochynska D., Smith E. S. J., Gerstung M., Campbell P. J., Murchison E. P., Stratton M. R., Martincorena I., Somatic mutation rates scale with lifespan across mammals. Nature 604, 517–524 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hart R. W., Setlow R. B., Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc. Natl. Acad. Sci. U.S.A. 71, 2169–2173 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L., Dong X., Tian X., Lee M., Ablaeva J., Firsanov D., Lee S.-G., Maslov A. Y., Gladyshev V. N., Seluanov A., Gorbunova V., Vijg J., Maintenance of genome sequence integrity in long- and short-lived rodent species. Sci. Adv. 7, eabj3284 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woo S. R., Corrales L., Gajewski T. F., Innate immune recognition of cancer. Annu. Rev. Immunol. 33, 445–474 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Evdokimov A., Kutuzov M., Petruseva I., Lukjanchikova N., Kashina E., Kolova E., Zemerova T., Romanenko S., Perelman P., Prokopov D., Seluanov A., Gorbunova V., Graphodatsky A., Trifonov V., Khodyreva S., Lavrik O., Naked mole rat cells display more efficient excision repair than mouse cells. Aging 10, 1454–1473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mellors J., Tipton T., Longet S., Carroll M., Viral evasion of the complement system and its importance for vaccines and therapeutics. Front. Immunol. 11, 1450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moore M. S., Reichard J. D., Murtha T. D., Zahedi B., Fallier R. M., Kunz T. H., Specific alterations in complement protein activity of little brown Myotis (Myotis lucifugus) Hibernating in white-nose syndrome affected sites. PLOS ONE 6, e27430 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hammarlund E. U., von Stedingk K., Pahlman S., Refined control of cell stemness allowed animal evolution in the oxic realm. Nat. Ecol. Evol. 2, 220–228 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Bloch I., Sherill-Rofe D., Stupp D., Unterman I., Beer H., Sharon E., Tabach Y., Optimization of co-evolution analysis through phylogenetic profiling reveals pathway-specific signals. Bioinformatics 36, 4116–4125 (2020). [DOI] [PubMed] [Google Scholar]
- 70.Omar I., Guterman-Ram G., Rahat D., Tabach Y., Berger M., Levaot N., Schlafen2 mutation in mice causes an osteopetrotic phenotype due to a decrease in the number of osteoclast progenitors. Sci. Rep. 8, 13005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsaban T., Stupp D., Sherill-Rofe D., Bloch I., Sharon E., Schueler-Furman O., Wiener R., Tabach Y., CladeOScope: Functional interactions through the prism of clade-wise co-evolution. NAR Genom. Bioinform. 3, lqab024 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R., Loney F., May B., Milacic M., Rothfels K., Sevilla C., Shamovsky V., Shorser S., Varusai T., Weiser J., Wu G., Stein L., Hermjakob H., D’Eustachio P., The reactome pathway knowledgebase. Nucleic Acids Res. 48, D498–D503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buniello A., MacArthur J. A. L., Cerezo M., Harris L. W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., Suveges D., Vrousgou O., Whetzel P. L., Amode R., Guillen J. A., Riat H. S., Trevanion S. J., Hall P., Junkins H., Flicek P., Burdett T., Hindorff L. A., Cunningham F., Parkinson H., The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Myers T. A., Chanock S. J., Machiela M. J., LDlinkR: An R package for rapidly calculating linkage disequilibrium statistics in diverse populations. Front. Genet. 11, 157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gordon A., Glazko G., Qiu X., Yakovlev A., Control of the mean number of false discoveries, Bonferroni and stability of multiple testing. Ann. Appl. Stat. 1, 179–190 (2007). [Google Scholar]
- 77.Tian X., Doerig K., Park R., Qin A. C. R., Hwang C., Neary A., Gilbert M., Seluanov A., Gorbunova V., Evolution of telomere maintenance and tumour suppressor mechanisms across mammals. Philos. Trans. R Soc. B Biol. Sci. 373, 20160443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abegglen L. M., Caulin A. F., Chan A., Lee K., Robinson R., Campbell M. S., Kiso W. K., Schmitt D. L., Waddell P. J., Bhaskara S., Jensen S. T., Maley C. C., Schiffman J. D., Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA 314, 1850–1860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sulak M., Fong L., Mika K., Chigurupati S., Yon L., Mongan N. P., Emes R. D., Lynch V. J., TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife 5, e11994 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buffenstein R., Negligible senescence in the longest living rodent, the naked mole-rat: Insights from a successfully aging species. J. Comp. Physiol. B 178, 439–445 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Tian X., Azpurua J., Hine C., Vaidya A., Myakishev-Rempel M., Ablaeva J., Mao Z., Nevo E., Gorbunova V., Seluanov A., High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499, 346–349 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hadi F., Kulaberoglu Y., Lazarus K. A., Bach K., Ugur R., Beattie P., Smith E. S. J., Khaled W. T., Transformation of naked mole-rat cells. Nature 583, E1–E7 (2020). [DOI] [PubMed] [Google Scholar]
- 83.Gorbunova V., Hine C., Tian X., Ablaeva J., Gudkov A. V., Nevo E., Seluanov A., Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proc. Natl. Acad. Sci. U.S.A. 109, 19392–19396 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S. K., Cook H., Mende D. R., Letunic I., Rattei T., Jensen L. J., von Mering C., Bork P., eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waris G., Ahsan H., Reactive oxygen species: Role in the development of cancer and various chronic conditions. J. Carcinog. 5, 14 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ilacqua A. N., Kirby A. M., Pamenter M. E., Behavioural responses of naked mole rats to acute hypoxia and anoxia. Biol. Lett. 13, 20170545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McIntyre I. W., Campbell K. L., MacArthur R. A., Body oxygen stores, aerobic dive limits and diving behaviour of the star-nosed mole (Condylura cristata) and comparisons with non-aquatic talpids. J. Exp. Biol. 205, 45–54 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S17
Notes S1 to S10
References
Table S1 to S15