Abstract
In Klebsiella pneumoniae, transcription of the nitrogen fixation (nif) genes is regulated in response to molecular oxygen or availability of fixed nitrogen by the coordinated activities of the nifA and nifL gene products. NifA is a nif-specific transcriptional activator, the activity of which is inhibited by interaction with NifL. Nitrogen control of NifL occurs at two levels: transcription of the nifLA operon is regulated by the global ntr system, and the inhibitory activity of NifL is controlled in response to fixed nitrogen by an unknown factor. K. pneumoniae synthesizes two PII-like signal transduction proteins, GlnB, which we have previously shown not to be involved in the response of NifL to fixed nitrogen, and the recently identified protein GlnK. We have now cloned the K. pneumoniae glnK gene, studied its expression, and shown that a null mutation in glnK prevents NifL from responding to the absence of fixed nitrogen, i.e., from relieving the inhibition of NifA activity. Hence, GlnK appears to be involved, directly or indirectly, in NifL-dependent regulation of nif gene expression in K. pneumoniae. Comparison of the GlnB and GlnK amino acid sequences from six species of proteobacteria identifies five residues (residues 3, 5, 52, 54, and 64) which serve to distinguish the GlnB and GlnK proteins.
In diazotrophic proteobacteria, transcription of the nitrogen fixation (nif) genes is dependent on the nif-specific activator protein NifA, which is a member of the ςN-dependent family of bacterial activators (18, 28, 36). In most organisms, the activity of NifA is controlled in response to two major environmental factors, namely, the availability of oxygen and of fixed nitrogen, but the mechanism of this control differs from one organism to another (20, 36). In Klebsiella pneumoniae and Azotobacter vinelandii, nifA is coordinately transcribed with a second gene, nifL, the product of which antagonizes NifA activity in response to both oxygen and fixed nitrogen (6, 23, 34, 46). In K. pneumoniae, nifLA expression is itself regulated in response to the cellular N status, whereas in A. vinelandii, it is constitutive (6, 16, 54). The response of NifL to oxygen is at least partly understood in that NifL in both organisms is a flavoprotein with flavin adenine dinucleotide as a prosthetic group and the oxidized form of NifL inhibits NifA activity (23, 48, 49). However, the mechanism by which NifL senses and responds to the cellular nitrogen status is unknown.
In enteric bacteria, global responses to changes in nitrogen status are mediated by the nitrogen regulation (ntr) system, which is considered to comprise four proteins. Two of these proteins, a uridylyltransferase (encoded by glnD) and PII (a small, trimeric protein encoded by glnB), comprise a sensory transduction system whereby GlnD uridylylates GlnB under N-limiting conditions and deuridylylates GlnB-UMP in N excess. Hence, the uridylylation status of GlnB signals the intracellular nitrogen status, which is then communicated to the NtrB-NtrC two-component regulatory system. In N limitation, NtrB is autophosphorylated on residue His139 and these phosphoryl groups are subsequently transferred to NtrC with the concomitant activation of NtrC-P-dependent genes. In N excess, GlnB stimulates the NtrB-dependent dephosphorylation of NtrC and inactivation of ntr-dependent operons (37, 40).
The potential roles of one or more of the ntr genes in NifL-dependent nif regulation has been investigated in both K. pneumoniae and A. vinelandii. In K. pneumoniae, NifL-dependent nitrogen control occurs in the absence of GlnB or GlnD, suggesting that neither protein is essential for the sensing of fixed nitrogen by NifL (19, 24). However, it should be noted that experiments with the glnD null mutant utilized a strain with a secondary ntrB mutation that allowed constitutive expression of NtrC-dependent genes, including nifLA. In A. vinelandii, a mutation in nfrX (a glnD homologue) produces a Nif− phenotype that can be suppressed by a secondary mutation in nifL, suggesting that in this organism, the absence of uridylyltransferase results in permanent inhibition of NifA activity by NifL (11).
Our understanding of the global Ntr system in enteric bacteria was recently complicated by the recognition of a second PII-like protein (encoded by glnK) in Escherichia coli (56). E. coli glnK appears to be cotranscribed with a downstream gene (amtB), the product of which has been proposed to be an ammonium transporter (31, 41, 56), and expression of the operon is NtrC dependent (51). The amino acid sequence of GlnK is 67% identical to that of GlnB, and GlnK can also be uridylylated in N limitation, presumably at the conserved Tyr51 residue (56). The presence of two PII-like proteins has since been reported in Rhizobium etli, Azospirillum brasilense, Azorhizobium caulinodans, Herbaspirillum seropedicae, Azoarcus sp. strain BH72, Rhodobacter sphaeroides, and Acetobacter diazotrophicus (5, 13, 14, 32, 38, 42, 43, 53).
The precise role of GlnK in nitrogen metabolism remains to be elucidated (2), but recent experiments which examined the activities of K. pneumoniae NifL and NifA in a heterologous E. coli or Salmonella typhimurium background found that the release of NifA from NifL-dependent inhibition does not occur in an ntrC mutant (22). It was concluded that NtrC activates transcription of a gene the product of which functions to relieve NifL inhibition of NifA under N-limiting conditions. One candidate for such a gene is glnK.
We have now cloned and sequenced the glnK gene of K. pneumoniae. We have studied the control of glnK expression and investigated the effect of a glnK null mutation. We found that in the absence of GlnK, NifA activity is inhibited in an NifL-dependent manner, even under N-limiting conditions, suggesting that GlnK is involved, directly or indirectly, in the sensing of cellular nitrogen status by NifL.
MATERIALS AND METHODS
Strains and media.
The strains and plasmids used in this work are listed in Table 1. All strains were grown on Luria medium (Luria broth or Luria agar [LA]). The nitrogen-free medium used (NFDM) was as described by Dixon et al. (15). When used for growth of K. pneumoniae strains, NFDM was supplemented with 25-μg/ml l-histidine, and for E. coli, it was supplemented with 1-μg/ml thiamine. Ammonium sulfate was added to NFDM as a nitrogen source at a final concentration of 1 mg/ml. The antibiotics used for E. coli were 100-μg/ml carbenicillin, 15-μg/ml kanamycin, 15-μg/ml chloramphenicol, and 30-μg/ml streptomycin; those used for K. pneumoniae were 200-μg/ml both carbenicillin and ampicillin together, 30-μg/ml kanamycin, 40-μg/ml chloramphenicol, and 200-μg/ml spectinomycin.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Relevant phenotype | Source or reference |
|---|---|---|---|
| K. pneumoniae strains | |||
| UNF122 | hisD2 Δlac-2002 | Wild type | 25 |
| UNF923 | Δ(his-nifH)2639 nifH::MudAplac Δlac-2002 sbl-300::Tn10 | Nif | 35 |
| UNF1537 | hisD2 Δlac-2002 glnB502 glnB21::Tn5 | GlnB | 25 |
| UNF2792 | hisD2 Δlac-2002 sbl-300::Tn10 recA56 rpoN71::kan | RpoN | 12 |
| UNF3440 | Δ(his-nifH)2639 nifH::MudAplac Δlac-2002 sbl-300::Tn10 glnB502 glnB21::Tn5 | Nif GlnB | This work |
| UNF3432 | hisD2 Δlac-2002 glnK1::KIXX | GlnK | This work |
| UNF3433 | Δ(his-nifH)2639 nifH::MudAplac Δlac-2002 sbl-300::Tn10 glnK1::KIXX | Nif GlnK | This work |
| E. coli strains | |||
| YMC10 | ΔlacU169 endA1 thi-1 hsdR17 supE44 | Wild type | 4 |
| YMC26 | ΔlacU169 endA1 thi-1 hsdR17 supE44 glnD99::Tn10 | GlnD | 44 |
| RB9060 | ΔlacU169 endA1 thi-1 hsdR17 supE44 ΔglnB2306 | GlnB | 7 |
| RB9066 | ΔlacU169 endA1 thi-1 hsdR17 supE44 ntrC10::Tn5 | NtrC | 7 |
| TH1 | ΔlacU169 endA1 thi-1 hsdR17 supE44 ΔrpoN2518 | RpoN | T. Hunt |
| Plasmids | |||
| pCC46 | K. pneumoniae nifLA expressed from placZ in pHSG575 | NifL+ NifA+ | 10 |
| pCC47 | K. pneumoniae nifA expressed from placZ in pHSG575 | NifA+ | 14 |
| pNM480 | pBR322-based lacZ fusion vector | Cbr | 39 |
| pRJ5 | PCR fragment from K. pneumoniae glnK cloned in pBluescript KS+ | ′GlnK′ Cbr | This work |
| pRJ16 | BamHI-HindIII fragment containing glnK expressed from pglnK in pTZ18 | GlnK+ Cbr | This work |
| pRJ32 | PvuII fragment from pRJ16 containing promoter region and start of glnK in pNM480 | pGlnK::LacZ | This work |
| pRJ44 | K. pneumoniae glnK expressed from pcat in Spr (Ω) derivative of pACYC184 | GlnK+ Spr | This work |
DNA methods.
Plasmids were isolated by using a plasmid miniprep kit (Qiagen) in accordance with the manufacturer’s instructions. Restriction enzyme digests, blunting of DNA, ligation, and Southern hybridization reactions were carried out as described by Sambrook et al. (45). Plasmid pRJ32 was constructed by cloning the PvuII fragment carrying the 3′ end of mdl, the glnK promoter region, and the 5′ end of glnK into lacZ translational fusion plasmid pNM480. Plasmid pRJ44 was constructed by cloning an EcoRI-DraIII fragment (which extends from 21 bp upstream of the glnK ATG to 15 bp downstream of the termination codon) in place of the DraI fragment internal to the cat gene of pACYC184. In order to change the selective marker on this plasmid from tetracycline resistance to streptomycin resistance, an Ω cassette was then cloned into the BamHI site.
Isolation of fragment containing glnK.
An internal fragment of the K. pneumoniae glnK gene was isolated from genomic DNA by PCR using primers based on conserved regions of GlnB (5′ CTGCGAATTCGATHATHAAACCHTTCAARCTGGA 3′ and 5′ ACGCGGATCCTCRCCGGTRCGRATRCGAATSACSCG 3′, where H is A, C, or T, R is A or G, and S is C or G). The resultant fragment was cloned into pBluescript KS+ by using BamHI and EcoRI sites included in the primers, giving pRJ5, and sequenced to confirm its likely identity. Chromosomal DNA was isolated from wild-type K. pneumoniae UNF122 by the method of Ausubel et al. (3). Isolated DNA (10 μg) was subjected to either single or double digestion with EcoRI, BamHI, HindIII, SspI, and SphI and separated on 0.8% agarose. Fragments containing glnK were identified by Southern hybridization using the BamHI-EcoRI fragment from pRJ5 as a probe labelled with [32P]dCTP using the Rediprime labelling kit (Amersham). DNA was redigested with BamHI and HindIII, and fragments of 1.5 kb were isolated from agarose and ligated into BamHI-HindIII-digested pTZ18 to form a minilibrary. Ligated DNA was used to transform E. coli 71-18. White, ampicillin-resistant colonies were patched onto nitrocellulose filters, laid onto LA-carbenicillin plates, and grown for 3 h. Filters were then transferred onto LA-chloramphenicol plates and incubated overnight. Clones containing glnK were identified by colony hybridization (45) using the [32P]dCTP-labelled fragment described above. Sequencing of one hybridizing plasmid (pRJ16) was performed by using the dideoxy sequencing kit and protocol from Pharmacia.
Primer extension analysis of total RNA.
Cells were grown overnight in the same media as used for β-galactosidase assays and subcultured into fresh media, and total RNA was isolated from exponentially growing cells by using the RNeasy kit (Qiagen) in accordance with the manufacturer’s instructions. Primer extension analysis was performed by using 10 μg of total RNA and 0.2 pmol of a primer end labelled with [γ-32P]dATP as described by Sawers and Böck (47). The oligonucleotide used (5′ GAATGGTTTGATTACCACGG 3′) hybridizes to bases 33 to 14 of glnK. Primer extension reactions were carried out as described by Sawers and Böck (47). Products were separated on a 6% polyacrylamide sequencing gel alongside a sequence produced by using the same 32P-labelled primer and pRJ16 as the template.
Construction of glnK::KIXX mutation.
The kanamycin resistance-encoding KIXX cassette from pUC4KIXX was cloned into the EagI site blunted 5′→3′ by using mung bean nuclease (New England Biolabs), creating pRJ17, in which the kan gene is transcribed in the direction opposite to that of glnK. The BamHI-PvuI fragment of pRJ17, containing all of the cloned region, was then blunted 3′→5′ by using large-fragment Klenow polymerase (Pharmacia), ligated into the SmaI site of sacB-containing vector pSG335 (21) to give pRJ25, and transformed into UNF122. Recombinant strains were identified by the ability to grow on LA supplemented with 5% sucrose and resistance to kanamycin. The glnK::KIXX mutation was then transduced, by using the P1 phage, into K. pneumoniae UNF923 to create UNF3433. The correct insertion of glnK::KIXX into both chromosomes was checked by PCR. Reactions were carried out in 100-μl volumes using Taq DNA polymerase (Boehringer) and primers homologous to the sequence of the glnK PCR fragment (sense, 5′ TTCAAGCTGGAAGATGTG 3′; antisense, 5′ CCTTTCTGACGGCGGAAC 3′) at a concentration of 20 pmol/reaction mixture.
β-Galactosidase assays.
For K. pneumoniae strains, β-galactosidase assays were carried out as described by Holtel and Merrick (24): for N-limitation, the nitrogen source was 100-μg/ml glutamine (see Table 2 experiments) or 100-μg/ml serine (see Table 3 experiments), and for N sufficiency, it was 100-μg/ml glutamine plus 1-mg/ml (NH4)2SO4 (see Table 2) or 1-mg/ml (NH4)2SO4 (see Table 3). For E. coli strains, cultures were grown for 24 h in Luria broth before subculture in NFDM supplemented with 0.02% Casamino Acids rather than glutamine. The data in Tables 2 and 3 are the means of at least three experiments in which the standard deviations were not greater than ±10%.
TABLE 2.
Regulation of pglnK-lacZ in response to nitrogen
| Strain | Relevant genotype | β-Galactosidase activitya
|
|
|---|---|---|---|
| −Nb | +Nc | ||
| Escherichia coli | |||
| YMC10(pRJ32) | Wild type | 4,395 | 1,104 |
| YMC26(pRJ32) | glnD | 129 | 132 |
| RB9060(pRJ32) | glnB | 3,427 | 4,734 |
| RB9060(pRJ32, pRJ44) | glnB (glnK+) | 3,807 | 535 |
| RB9066(pRJ32) | ntrC | 189 | 137 |
| TH1(pRJ32) | rpoN | 92 | 97 |
| Klebsiella pneumoniae | |||
| UNF122(pRJ32) | Wild type | 4,623 | 473 |
| UNF1537(pRJ32) | glnB | 5,367 | 4,921 |
| UNF2792(pRJ32) | rpoN | 248 | 167 |
| UNF3432(pRJ32) | glnK | 4,705 | 634 |
From pRJ32 (in Miller units).
−N, nitrogen-limiting medium.
+N, nitrogen-sufficient medium.
TABLE 3.
Effect of GlnK on the regulation of K. pneumoniae pnifH
| Strain | Relevant genotype
|
β-Galactosidase activitya
|
||
|---|---|---|---|---|
| Chromosome | Plasmid | −Nb | +Nc | |
| UNF923(pCC46) | Wild type | nifL+ nifA+ | 618 | 34 |
| UNF3440(pCC46) | glnB | nifL+ nifA+ | 498 | 31 |
| UNF3433(pCC46) | glnK | nifL+ nifA+ | 161 | 33 |
| UNF3433(pCC46, pRJ44) | glnK | nifL+ nifA+, glnK+ | 613 | 109 |
| UNF923(pCC46, pRJ44) | Wild type | nifL+ nifA+, glnK+ | 609 | 56 |
| UNF923(pCC47) | Wild type | nifA+ | 1,336 | 1,124 |
| UNF3440(pCC47) | glnB | nifA+ | 1,075 | 967 |
| UNF3433(pCC47) | glnK | nifA+ | 1,045 | 1,233 |
Expressed from the chromosomal nifH::MudAplac fusion (in Miller units).
−N, nitrogen-limiting medium.
+N, nitrogen-sufficient medium.
[14C]methylamine transport assays.
Cells were grown overnight in the same way as for β-galactosidase assays. Cultures were then washed two times in saline phosphate and resuspended in 4.8 ml of NFDM. The optical density at 650 nm was measured to determine the protein concentration. At zero time, 20 μl of 14CH3NH3+ (826 Ci/mol) was added to give a final concentration of 10 μM 14CH3NH3+. Samples of 500 μl were taken at 0, 2, 4, 6, 10, and 20 min, and uptake was terminated by filtration through nitrocellulose filters (Millipore type HA; 0.45 μm pore size) under a constant vacuum. Filters were then washed five times with NFDM and exposed for 1 h to a PhosphoImager plate from which counts were determined. Data were calibrated by using internal standards spotted on filters and counted in the same experiment.
Nucleotide sequence accession number.
The sequence of the BamHI-HindIII fragment carried on pRJ16 (Fig. 1) has been assigned EMBL accession no. AJ006531.
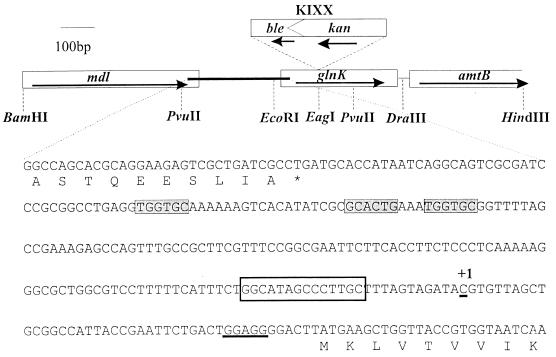
FIG. 1.
Map of the cloned BamHI-HindIII fragment (pRJ16) showing restriction sites used for genetic manipulations and the site of insertion of the KIXX cassette in glnK. The sequence of a region of the cloned fragment (300 bp, bp 481 to 781 of the sequence under accession no. AJ006531) comprising the 3′ end of mdl, the glnK promoter region, and the start of glnK is shown. The proposed NtrC binding sites (filled boxes), ς54-dependent promoter (clear box), and ribosome-binding site (underlined) are highlighted, and the transcriptional start site (+1) is marked.
RESULTS
Cloning of glnK.
A 300-bp fragment encoding part of K. pneumoniae glnK was amplified by PCR using degenerate primers based on the conserved regions of the E. coli and K. pneumoniae glnB sequences. This fragment was cloned into Bluescript KS+ by using EcoRI and BamHI restriction sites incorporated into the primers. The sequence of the cloned fragment confirmed that it encoded a polypeptide with 93% identity to the Ile8-to-Ala94 region of E. coli GlnK. This cloned fragment was then used as a probe to isolate a 1.5-kb hybridizing fragment from a minigene bank of BamHI-HindIII-digested chromosomal K. pneumoniae DNA cloned in pTZ18. The resultant plasmid was designated pRJ16.
Sequencing.
Sequencing of the fragment in pRJ16 identified three open reading frames, all of which were transcribed in the same direction, from BamHI to HindIII, and which, by homology to the equivalent E. coli genes, encode the 3′ end of mdl, all of glnK, and the 5′ end of amtB (Fig. 1). Within the 240-bp glnK promoter region (Fig. 1), a consensus ςN-dependent −24, −12 promoter sequence is located 60 bp upstream of the glnK translation start and two potential NtrC-binding sites are present; one complete site is 100 bp upstream of the proposed promoter, and a half site is 30 bp upstream of that. There are two potential initiating methionine codons for amtB, of which we consider the second the most likely to be correct, based on homology with E. coli amtB and the presence of an appropriate ribosome-binding site. In this case, the K. pneumoniae amtB gene is separated by 35 bp from the 3′ end of glnK, a spacing almost identical to that found in E. coli (56). No obvious termination or promoter sequences are apparent between glnK and amtB, suggesting that the two genes comprise a single operon, as proposed for the homologous genes in E. coli, R. etli, and A. vinelandii (33, 50, 53). The K. pneumoniae glnK gene encodes a polypeptide that is 94% identical to E. coli GlnK but only 69% identical to E. coli GlnB.
Comparison of GlnB and GlnK.
Database searching for homologues of K. pneumoniae GlnK identified 21 gene products in the α and γ subdivisions of the class Proteobacteria. These fall into two distinct groups, which are homologous to E. coli GlnB and GlnK, respectively. In six organisms, both GlnB and GlnK have been identified, and alignment of their amino acid sequences using ClustalW indicates that the two proteins are characterized by differences at just 5 of the 112 residues (residues 3, 5, 52, 54, and 64) (Fig. 2). In GlnB proteins, residue 3 is lysine, residue 5 is glutamate or aspartate, residue 52 is methionine or valine, residue 54 is aspartate, and residue 64 is valine. By contrast, in GlnK, residue 3 is leucine or isoleucine, residue 5 is threonine, methionine, or isoleucine, residue 52 is serine or alanine, residue 54 is serine or asparagine, and residue 64 is alanine (with the exception of R. sphaeroides GlnK, in which residue 64 is valine).
FIG. 2.
Alignment of GlnB and GlnK polypeptide sequences. Residues that distinguish GlnK and GlnB (residues 3, 5, 52, 54, and 64) are in boldface type.
This subdivision on the basis of polypeptide homology is consistent with the genetic organization of the PII structural genes within the α and γ subdivisions of the class Proteobacteria. The glnB genes are either monocistronic, as in E. coli and K. pneumoniae (25, 30), or linked to glnA, as in Rhizobium or Rhodospirillum spp. (9, 26), whereas genes encoding GlnK proteins are almost always linked to amtB homologues. One exception is the A. brasilense glnZ gene, which encodes a GlnK-like protein on the basis of the predicted amino acid sequence but which is not linked to amtB (55).
In vivo transcript analysis.
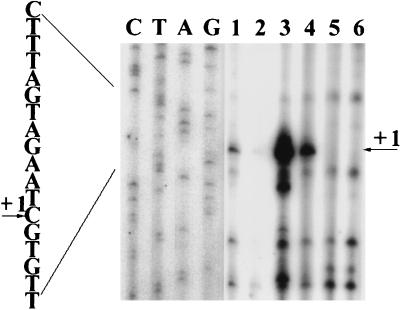
Primer extension analysis was carried out on RNA from wild-type K. pneumoniae (UNF122), an rpoN derivative (UNF2792), and the wild-type strain carrying a glnK-lacZ fusion plasmid (pRJ32). One specific signal was identified as the major transcript with a number of minor signals which appeared to be nonspecific in that they were present in all samples and did not correspond to any obvious motifs in the nontranslated sequence. Transcription was shown to begin 44 bp upstream of the translational start and at the appropriate distance from the proposed ςN-binding site (Fig. 3). Transcription from this site was greatly reduced when cells were grown under nitrogen-rich conditions and was undetectable in an rpoN mutant which does not contain ςN.
FIG. 3.
Transcriptional start site of the glnK amtB operon determined by primer extension analysis of RNA from wild-type K. pneumoniae UNF122 (lanes 1 and 2), UNF122 with glnK-lacZ fusion plasmid pRJ32 (lanes 3 and 4), and UNF2792 rpoN (lanes 5 and 6). The RNAs in lanes 1, 3, and 5 were from strains grown under nitrogen-limiting conditions [NFDM containing histidine (25 μg/ml)], and glutamine (100 μg/ml)], and those in lanes 2, 4, and 6 were from strains grown under nitrogen-sufficient conditions [NFDM containing histidine (25 μg/ml) and (NH4)2SO4 (1 mg/ml)]. Reaction mixtures run alongside the sequence were generated by using the same primer.
In vivo analysis of glnK expression.
To analyze the expression of the glnK amtB operon in vivo, a translational glnK-lacZ fusion (pRJ32) was constructed and β-galactosidase activity was assayed in a variety of genetic backgrounds under nitrogen-limiting and nitrogen-sufficient conditions (Table 2). The pattern of expression of glnK was largely as predicted from the promoter elements identified in the DNA sequence. Expression was elevated under nitrogen-limiting compared to nitrogen-sufficient conditions and was essentially eliminated in the absence of ςN (rpoN), NtrC, or GlnD. Expression from pglnK was constitutive in the absence of GlnB, whereas it was essentially wild type in the absence of GlnK. In both K. pneumoniae and E. coli, the level of expression in nitrogen sufficiency was considerably greater than in an rpoN or ntrC mutant strain.
Polarity of glnK::KIXX mutation glnK1.
The introduction of a glnK::KIXX mutation into the glnK amtB operon might be expected to have a polar effect on expression of amtB. This was analyzed by using [14C]methylamine transport assays to measure the activity of AmtB. [14C]methylamine transport was assessed in three strains: wild-type UNF122, glnK::KIXX mutant UNF3432, and UNF3432 carrying a plasmid-borne, constitutively expressed glnK gene [UNF3432(pRJ44)]. In N-limited wild-type cells, methylamine uptake was linear for 20 min at a rate of 140 pmol/mg of dry weight/min, and this uptake was reduced to less than 20 pmol/mg of dry weight/min in ammonia-grown cells. Methylamine transport was less than 5 pmol/mg of dry weight/min in glnK::KIXX mutant strain UNF3433 and was not complemented by the reintroduction of a plasmid-borne copy of glnK (pRJ44). Expression of glnK from pRJ44 was shown to be constitutive by introducing the plasmid into a glnB glnK double mutant of E. coli and analyzing GlnK levels in N limitation and N sufficiency by Western blotting with an antibody raised against an oligopeptide equivalent to conserved residues 31 to 50 of GlnB (14) (data not shown). These data confirm the polarity of the glnK1 mutation on amtB and support the proposal that the two genes constitute an operon.
Phenotypic characterization of a glnK::KIXX mutation.
The effect of the glnK::KIXX mutation on the activity of the nifLA gene products was assessed by using, as a genetic background, K. pneumoniae UNF923, which carries a chromosomal pnifH-lacZ fusion and has all of the nif genes, from nifD through to nifQ and into the his operon, deleted. The nifLA genes were cloned on low-copy-number plasmid pCC46 so that their expression from plac on this replicon was independent of the cellular nitrogen status. This independence of expression was confirmed by measuring lacZ expression from the same promoter on the same replicon in N limitation and N sufficiency (data not shown). When pCC46 is introduced into UNF923, expression from pnifH is normally regulated in response to the cellular nitrogen status (Table 3). Hence, in this strain, where nifLA expression is uncoupled from ntr-dependent regulation, the response of pnifH to nitrogen status reflects the activity of the NifA protein as regulated by its interaction with NifL. As a control plasmid, pCC47, which expresses NifA alone from the same promoter as that in pCC46, was used.
The absence of GlnB (UNF3440) had only a minor effect on pnifH expression in this assay system, resulting in a slight reduction in the fully derepressed level of activity. By comparison, the glnK::KIXX mutation reduced the derepressed level by 75%, indicating that in the absence of GlnK, NifA was unable to escape fully from the inhibitory effects of NifL, even under nitrogen-limiting growth conditions. Normal NifLA-dependent regulation was restored to the glnK mutant by introduction of a constitutively expressed glnK gene on pRJ44. Neither the glnB nor the glnK mutation had any effect on activation by NifA alone (pCC47).
DISCUSSION
Sequence comparison of GlnB and GlnK proteins.
The occurrence of duplicate copies of genes encoding PII-like proteins now appears to be common among members of the α and γ subdivisions of the class Proteobacteria. The tertiary structures of E. coli GlnB and GlnK are very similar, the major differences being in the conformations of the T loops and of C-terminal residues 109 to 112 (57). Nevertheless, under some conditions, GlnB and GlnK are simultaneously expressed in the cell, and this raises questions about their respective roles. The fact that in some organisms, specific phenotypes can be assigned to mutations in glnB demonstrates that, at least in some respects, the two proteins have discrete functions, but the precise function of GlnK is still unclear (2).
Three of the residues, 3, 5, and 64, which distinguish GlnB and GlnK, cluster closely together in their three-dimensional structures (8, 57). In GlnB, Lys3 and Glu5 form a ring of alternating charged residues in the central cavity of the protein, whereas in GlnK, residues 3 and 5 are uncharged. These differences may have a functional significance in that the charge differences could serve to prevent trimer formation between heterologous polypeptides, i.e., to ensure that only homogeneous GlnB or GlnK trimers are formed at times when both glnB and glnK are expressed in the same cell. The other two distinguishing residues, 52 and 54, are at the base of the T loop and could affect the structure or mobility of the T loop, which is that part of PII which interacts with other proteins.
Regulation of glnK and amtB.
Expression of K. pneumoniae glnK is dependent on ςN and NtrC, as in E. coli, R. etli, A. caulinodans, and A. brasilense (glnZ) (13, 38, 53, 56). The functionality of the proposed ςN promoter in K. pneumoniae glnK was supported by transcript mapping and by analysis of the expression of a pglnK-lacZ fusion which showed a significant level of expression under nitrogen-sufficient conditions that was still NtrC dependent. Very similar results were reported for the A. caulinodans glnK-amtB promoter (38) and for A. brasilense glnZ expression (13). These data suggest that pglnK could be activated by low levels of NtrC-P present under nitrogen-sufficient growth conditions. However, studies with single-copy chromosomal lacZ fusions to the E. coli glnK promoter did not show such expression (2, 51). Our observation of pglnK-lacZ expression in ammonia may be a consequence of NtrC titration by the fusion plasmid, and indeed, we recognize that regulation in this system may be very sensitive to gene expression levels and copy number. A mutation in glnK did not affect pglnK-lacZ expression, indicating that GlnK is not autoregulatory.
Data on the effects of a glnB deletion on glnK expression are contradictory. We observed constitutive expression of K. pneumoniae glnK in glnB deletion strain RB9060 and, using the same strain, Atkinson and Ninfa (2) also found very significant levels of E. coli glnK expression in ammonium. By contrast, van Heeswijk et al. (56) also used the same glnB strain in Western blots and detected no GlnK in the presence of ammonium.
Role of GlnK in nitrogen fixation.
The NifL protein of K. pneumoniae was first implicated in nif-specific nitrogen regulation by Merrick et al. (34), but the means by which NifL senses the nitrogen status of the cell has remained elusive. Our previous studies indicated that neither uridylyltransferase (GlnD) nor the PII protein (GlnB) was directly involved in the regulation of NifL activity in response to nitrogen status. We concluded that another nitrogen-sensing component, possibly an alternative PII-like protein, might be responsible (19, 24), and we have now shown that this is so.
The absence of GlnK severely impairs the ability of NifA to adopt an active form in the presence of NifL when cells are grown in nitrogen limitation. There is still some residual NifL-dependent regulation present in the glnK mutant (approximately 25% of that in the wild type), but given the similarities between GlnK and GlnB, we suggest that GlnB may be able to substitute partially for GlnK in this situation. Studies with E. coli have shown that in other situations, e.g., regulation of adenylyltransferase, either GlnB or GlnK can mediate regulation (2). Likewise, multicopy glnK restores regulation in a glnB mutant (Table 2), indicating that, when expressed at high levels in ammonium, GlnK can substitute for GlnB, presumably in promoting NtrB-dependent dephosphorylation of NtrC. Normal regulation was restored to a glnK mutant by the presence of a constitutively expressed plasmid-borne copy of glnK, establishing that GlnK is only effective in relieving the inhibitory effects of NifL on NifA in nitrogen-limiting medium. Our present results do not distinguish between a direct or an indirect effect of GlnK on NifL activity but are consistent with the observations of He et al. (22), who concluded that the product of an NtrC-dependent gene was required to relieve NifL inhibition of NifA activity.
GlnB or GlnK proteins have now been implicated in the regulation of NifA-dependent gene expression in a number of diazotrophs. In A. brasilense glnB mutants, NifA is inactive (29) but nif gene expression is restored by deletions in the N-terminal domain of NifA, suggesting that GlnB is required to activate NifA by preventing the inhibitory effect of its N-terminal domain (1). The N-terminal domains of H. seropedicae NifA and A. vinelandii VnfA and AnfA have also been implicated in nitrogen sensing (17, 52). Furthermore, an H. seropedicae glnB mutant is Nif− while nifA expression, which is NtrC dependent, is expected to be constitutive in this background (5). Finally the Nif− phenotype of a glnD (nfrX) mutant of A. vinelandii implicates a PII-like protein in the regulation of NifA activity in that organism, and a candidate glnK gene has recently been identified (11, 33).
Our data are consistent with a model in which the nitrogen-responsive inhibition of NifA activity by NifL in K. pneumoniae is regulated by an interaction between GlnK and the amino-terminal domain of NifA. Under nitrogen-limiting conditions, a particular form of GlnK could interact with NifA, thereby preventing the inhibitory effects of NifL. In a glnK mutant, this would not occur (although GlnB might partially substitute for GlnK) and NifL would inhibit NifA activity even in N limitation. In the wild type, a change to nitrogen sufficiency would alter the activity of GlnK and would also repress glnK transcription, thereby potentially allowing NifL to interact with NifA. With the present data, we cannot, however, exclude an alternative model in which the regulatory interaction is between GlnK and NifL.
The binding of 2-ketoglutarate and ATP to GlnB has been shown to activate the protein with respect to its control of adenylyltransferase and NtrB, and it has been proposed that an allosteric alteration of GlnB upon the binding of 2-ketoglutarate activates the uridylyltransferase reaction (27). Our previous data indicated that uridylylation is not required for control of NifL inhibition of NifA activity (19), and it is therefore possible that an allosteric change induced in GlnK by the binding of a low-molecular-weight ligand such as 2-ketoglutarate might be sufficient to control interaction with NifA and/or NifL.
ACKNOWLEDGMENTS
R.J. was supported by a BBSRC studentship. M.M. acknowledges support from the BBSRC through a grant-in-aid to the John Innes Centre.
We thank Wally van Heeswijk for supplying strains, Gary Sawers for technical advice, and Gavin Thomas for help with methylammonium transport assays. We also thank Tania Arcondeguy, Sara Austin, Ray Dixon, and Gary Sawers for helpful comments on the manuscript.
ADDENDUM IN PROOF
After this article was accepted for publication, He at al. published an article in the Journal of Bacteriology (L. He, E. Soupene, A. Ninfa, and S. Kustu, J. Bacteriol. 180:6661–6667, 1998) which also identifies a role for GlnK in relieving inhibition of NifA activity by NifL.
REFERENCES
- 1.Arsene F, Kaminski P A, Elmerich C. Modulation of NifA activity by PII in Azospirillum brasilense: evidence for a regulatory role of the NifA N-terminal domain. J Bacteriol. 1996;178:4830–4838. doi: 10.1128/jb.178.16.4830-4838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson M, Ninfa A J. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol Microbiol. 1998;29:431–447. doi: 10.1046/j.1365-2958.1998.00932.x. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1987. [Google Scholar]
- 4.Backman K, Chen Y M, Magasanik B. Physical and genetic characterisation of the glnA-glnG region of the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1981;78:3743–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benelli E M, Souza E M, Funayama S, Rigo I U, Pedrosa F O. Evidence for two possible glnB-type genes in Herbaspirillum seropedicae. J Bacteriol. 1997;179:4623–4626. doi: 10.1128/jb.179.14.4623-4626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco G, Drummond M, Woodley P, Kennedy C. Sequence and molecular analysis of the nifL gene of Azotobacter vinelandii. Mol Microbiol. 1993;9:869–880. doi: 10.1111/j.1365-2958.1993.tb01745.x. [DOI] [PubMed] [Google Scholar]
- 7.Bueno R, Pahel G, Magasanik B. Role of glnB and glnD gene products in regulation of the glnALG operon of Escherichia coli. J Bacteriol. 1985;164:816–822. doi: 10.1128/jb.164.2.816-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr P D, Cheah E, Suffolk P M, Vasudevan S G, Dixon N E, Ollis D L. X-ray structure of the signal transduction protein PII from Escherichia coli at 1.9Å. Acta Crystallogr. 1996;D52:93–104. doi: 10.1107/S0907444995007293. [DOI] [PubMed] [Google Scholar]
- 9.Colonna-Romano S, Riccio A, Guida M, Defez R, Lamberti A, Iaccarino M, Arnold W, Priefer U, Pühler A. Tight linkage of glnA and a putative regulatory gene in Rhizobium leguminosarum. Nucleic Acids Res. 1987;15:1951–1964. doi: 10.1093/nar/15.5.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contreras A, Drummond M. Cys184 and Cys187 of NifL protein of Klebsiella pneumoniae are not absolutely required for inhibition of NifA activity. Gene. 1991;103:83–86. doi: 10.1016/0378-1119(91)90395-r. [DOI] [PubMed] [Google Scholar]
- 11.Contreras A, Drummond M, Bali A, Blanco G, Garcia E, Bush G, Kennedy C, Merrick M. The product of the nitrogen fixation regulatory gene nfrX of Azotobacter vinelandii is functionally and structurally homologous to the uridylyltransferase encoded by glnD in enteric bacteria. J Bacteriol. 1991;173:7741–7749. doi: 10.1128/jb.173.24.7741-7749.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppard J R, Merrick M J. Cassette mutagenesis implicates a helix-turn-helix motif in promoter recognition by the novel RNA polymerase sigma factor ς54. Mol Microbiol. 1991;5:1309–1317. doi: 10.1111/j.1365-2958.1991.tb00777.x. [DOI] [PubMed] [Google Scholar]
- 13.de Zamaroczy M. Structural homologues PII and PZ of Azospirillum brasilense provide intracellular signalling for selective regulation of various nitrogen-dependent functions. Mol Microbiol. 1998;29:449–463. doi: 10.1046/j.1365-2958.1998.00938.x. [DOI] [PubMed] [Google Scholar]
- 14.de Zamaroczy M, Paquelin A, Peltre G, Forchhammer K, Elmerich C. Coexistence of two structurally similar but functionally different PII proteins in Azospirillum brasilense. J Bacteriol. 1996;178:4143–4149. doi: 10.1128/jb.178.14.4143-4149.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon R, Kennedy C, Kondorosi A, Krishnapillai V, Merrick M. Complementation analysis of Klebsiella pneumoniae mutants defective in nitrogen fixation. Mol Gen Genet. 1977;157:189–198. doi: 10.1007/BF00267397. [DOI] [PubMed] [Google Scholar]
- 16.Drummond M, Clements J, Merrick M, Dixon R. Positive control and autogenous regulation of the nifLA promoter in Klebsiella pneumoniae. Nature. 1983;301:302–307. doi: 10.1038/301302a0. [DOI] [PubMed] [Google Scholar]
- 17.Drummond M, Frise E, Green A, Jacob J. Nitrogen regulation of VnfA and AnfA function in Azotobacter vinelandii analysed by chimera formation. In: Tikonovich I A, Provorov N A, Romanov V I, Newton W E, editors. Proceedings of the 10th International Congress on Nitrogen Fixation. Dordrecht, The Netherlands: Kluwer Academic; 1995. p. 214. [Google Scholar]
- 18.Drummond M, Whitty P, Wootton J. Sequence and domain relationships of ntrC and nifA from Klebsiella pneumoniae: homologies to other regulatory proteins. EMBO J. 1986;5:441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards R, Merrick M. The role of uridylyltransferase in the control of Klebsiella pneumoniae nif gene regulation. Mol Gen Genet. 1995;247:189–198. doi: 10.1007/BF00705649. [DOI] [PubMed] [Google Scholar]
- 20.Fischer H-M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geissler S, Drummond M. A counterselectable pACYC184-based lacZα-complementing plasmid with novel multiple cloning sites: construction of chromosomal deletions in Klebsiella pneumoniae. Gene. 1993;136:253–255. doi: 10.1016/0378-1119(93)90474-h. [DOI] [PubMed] [Google Scholar]
- 22.He L, Soupene E, Kustu S. NtrC is required for control of Klebsiella pneumoniae NifL activity. J Bacteriol. 1997;179:7446–7455. doi: 10.1128/jb.179.23.7446-7455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill S, Austin S, Eydmann T, Jones T, Dixon R. Azotobacter vinelandii NIFL is a flavoprotein that modulates transcriptional activation of nitrogen-fixation genes via a redox-sensitive switch. Proc Natl Acad Sci USA. 1996;93:2143–2148. doi: 10.1073/pnas.93.5.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holtel A, Merrick M J. The Klebsiella pneumoniae PII protein (glnB gene product) is not absolutely required for nitrogen regulation and is not involved in NifL-mediated nif gene regulation. Mol Gen Genet. 1989;217:474–480. doi: 10.1007/BF02464920. [DOI] [PubMed] [Google Scholar]
- 25.Holtel A H, Merrick M. Identification of the Klebsiella pneumoniae glnB gene: nucleotide sequence of wild-type and mutant alleles. Mol Gen Genet. 1988;215:134–138. doi: 10.1007/BF00331314. [DOI] [PubMed] [Google Scholar]
- 26.Johannson M, Nordlund S. Transcription of the glnB and glnA genes in the photosynthetic bacterium Rhodospirillum rubrum. Microbiology. 1996;142:1265–1272. doi: 10.1099/13500872-142-5-1265. [DOI] [PubMed] [Google Scholar]
- 27.Kamberov E S, Atkinson M R, Ninfa A J. The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J Biol Chem. 1995;270:17797–17807. doi: 10.1074/jbc.270.30.17797. [DOI] [PubMed] [Google Scholar]
- 28.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of ς54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang Y Y, de Zamaroczy M, Arsene F, Paquelin A, Elmerich C. Regulation of nitrogen fixation in Azospirillum brasilense Sp7: involvement of nifA, glnA and glnB gene products. FEMS Microbiol Lett. 1992;79:113–119. doi: 10.1111/j.1574-6968.1992.tb14028.x. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Magasanik B. The glnB region of the Escherichia coli chromosome. J Bacteriol. 1993;175:7441–7449. doi: 10.1128/jb.175.22.7441-7449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marini A-M, Vissers S, Urrestarazu A, Andre B. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 1994;13:3456–3463. doi: 10.1002/j.1460-2075.1994.tb06651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin D, Reinhold-Hurek B. Identification and molecular analysis of multiple glnB homologues in Azoarcus sp. BH72. In: Elmerich C, Kondorosi A, Newton W E, editors. Proceedings of the 11th International Congress on Nitrogen Fixation. Dordrecht, The Netherlands: Kluwer Academic; 1998. p. 405. [Google Scholar]
- 33.Meletzus D, Rudnick P, Doetsch N, Green A, Kennedy C. Characterization of the glnK-amtB operon of Azotobacter vinelandii. J Bacteriol. 1998;180:3260–3264. doi: 10.1128/jb.180.12.3260-3264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merrick M, Hill S, Hennecke H, Hahn M, Dixon R, Kennedy C. Repressor properties of the nifL gene product of Klebsiella pneumoniae. Mol Gen Genet. 1982;185:75–81. [Google Scholar]
- 35.Merrick M J. Nitrogen control of the nif regulon in Klebsiella pneumoniae: involvement of the ntrA gene and analogies between ntrC and nifA. EMBO J. 1983;2:39–44. doi: 10.1002/j.1460-2075.1983.tb01377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merrick M J. Regulation of nitrogen fixation genes in free-living and symbiotic bacteria. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman & Hall; 1992. pp. 835–876. [Google Scholar]
- 37.Merrick M J, Edwards R A. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michel-Reydellet N, Desnoues N, de Zamaroczy M, Elmerich C, Kaminski P A. Characterisation of the glnK-amtB operon and involvement of AmtB in methylammonium uptake in Azorhizobium caulinodans. Mol Gen Genet. 1998;258:671–677. doi: 10.1007/s004380050781. [DOI] [PubMed] [Google Scholar]
- 39.Minton N P. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene. 1984;31:269–273. doi: 10.1016/0378-1119(84)90220-8. [DOI] [PubMed] [Google Scholar]
- 40.Ninfa A J, Atkinson M R, Kamberov E S, Feng J, Ninfa E G. Control of nitrogen assimilation by the NRI-NRII two-component system of enteric bacteria. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 67–88. [Google Scholar]
- 41.Ninnemann O, Jauniaux J-C, Frommer W B. Identification of a high affinity NH4+ transporter from plants. EMBO J. 1994;13:3464–3471. doi: 10.1002/j.1460-2075.1994.tb06652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perlova O, Nawroth R, Meletzus D. Identification and characterisation of genes involved in the ammonium sensing mechanism in Acetobacter diazotrophicus. In: Elmerich C, Kondorosi A, Newton W E, editors. Proceedings of the 11th International Congress on Nitrogen Fixation. Dordrecht, The Netherlands: Kluwer Academic; 1998. p. 166. [Google Scholar]
- 43.Qian Y, Tabita F R. Rhodobacter sphaeroides glnK. EMBL; 1997. accession no. AF023909. [Google Scholar]
- 44.Reitzer L J, Magasanik B. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc Natl Acad Sci USA. 1985;82:1979–1983. doi: 10.1073/pnas.82.7.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Santero E, Toukdarian A, Humphrey R, Kennedy C. Identification and characterisation of two nitrogen fixation regulatory regions nifA and nfrX in Azotobacter vinelandii and Azotobacter chroococcum. Mol Microbiol. 1988;2:303–314. doi: 10.1111/j.1365-2958.1988.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 47.Sawers G, Böck A. Novel transcriptional control of the pyruvate formate-lyase gene: upstream regulatory sequences and multiple promoters regulate anaerobic expression. J Bacteriol. 1989;171:2485–2498. doi: 10.1128/jb.171.5.2485-2498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitz R A. NifL of Klebsiella pneumoniae carries an N-terminally bound FAD cofactor, which is not directly required for the inhibitory function of NifL. FEMS Microbiol Lett. 1998;157:313–318. doi: 10.1111/j.1574-6968.1997.tb12791.x. [DOI] [PubMed] [Google Scholar]
- 49.Söderbäck E, Reyes-Ramirez F, Eydmann T, Austin S, Hill S, Dixon R. The redox- and fixed nitrogen-responsive regulatory protein NIFL from Azotobacter vinelandii comprises discrete flavin and nucleotide-binding domains. Mol Microbiol. 1998;28:179–192. doi: 10.1046/j.1365-2958.1998.00788.x. [DOI] [PubMed] [Google Scholar]
- 50.Son H S, Rhee S G. Cascade control of Escherichia coli glutamine synthetase. Purification and properties of PII protein and nucleotide sequence of its structural gene. J Biol Chem. 1987;262:8690–8695. [PubMed] [Google Scholar]
- 51.Soupene E, He L, Yan D, Kustu S. Ammonia acquisition in enteric bacteria: physiological role of the ammonium/methylammonium transport B (AmtB) protein. Proc Natl Acad Sci USA. 1998;95:7030–7034. doi: 10.1073/pnas.95.12.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Souza E M, Pedrosa F O, Drummond M, Rigo L U, Yates M G. Control of Herbaspirillum seropedicae NifA activity by ammonium ions and oxygen. J Bacteriol. 1999;181:681–684. doi: 10.1128/jb.181.2.681-684.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tate R, Riccio A, Merrick M, Patriarca E J. The Rhizobium etli amtB gene coding for an NH4+ transporter is down-regulated early during bacteroid differentiation. Mol Plant-Microbe Interact. 1998;11:188–198. doi: 10.1094/MPMI.1998.11.3.188. [DOI] [PubMed] [Google Scholar]
- 54.Toukdarian A, Kennedy C. Regulation of nitrogen metabolism in Azotobacter vinelandii: isolation of ntr and glnA genes and construction of ntr mutants. EMBO J. 1986;5:399–407. doi: 10.1002/j.1460-2075.1986.tb04225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Dommelen A, Keijers V, Vanderleyden J, de Zamaroczy M. (Methyl)ammonium transport in the nitrogen-fixing bacterium Azospirillum brasilense. J Bacteriol. 1998;180:2652–2659. doi: 10.1128/jb.180.10.2652-2659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Heeswijk W, Hoving S, Molenaar D, Stegeman B, Kahn D, Westerhoff H V. An alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol Microbiol. 1996;21:133–146. doi: 10.1046/j.1365-2958.1996.6281349.x. [DOI] [PubMed] [Google Scholar]
- 57.Xu Y, Cheah E, Carr P D, van Heeswijk W, Westerhof H V, Vasudevan S G, Ollis D. GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J Mol Biol. 1998;282:149–165. doi: 10.1006/jmbi.1998.1979. [DOI] [PubMed] [Google Scholar]