Abstract
The spread of coronavirus disease 2019 (COVID‐19) viral pneumonia caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has become a worldwide pandemic claiming several thousands of lives worldwide. During this pandemic, several studies reported the use of COVID‐19 convalescent plasma (CCP) from recovered patients to treat severely or critically ill patients. Although this historical and empirical treatment holds immense potential as a first line of response against eventual future unforeseen viral epidemics, there are several concerns regarding the efficacy and safety of this approach. This critical review aims to pinpoint the possible role of mass spectrometry‐based analysis in the identification of unique molecular component proteins, peptides, and metabolites of CCP that explains the therapeutic mechanism of action against COVID‐19. Additionally, the text critically reviews the potential application of mass spectrometry approaches in the search for novel plasma biomarkers that may enable a rapid and accurate assessment of the safety and efficacy of CCP. Considering the relative low‐cost value involved in the CCP therapy, this proposed line of research represents a tangible scientific challenge that will be translated into clinical practice and help save several thousand lives around the world, specifically in low‐ and middle‐income countries.
Keywords: antibodies, convalescent plasma, COVID‐19, mass spectrometry, metabolomics, proteomics, SARS‐CoV‐2
1. INTRODUCTION
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2) outbreak in 2019 has led to the human coronavirus disease‐19 (COVID‐19) pandemic (Guan et al., 2020), spreading across different continents and to almost every country. SARS‐CoV‐2 belongs to the family of Coronaviridae which are characterized as large, enveloped, positive‐sense, single‐stranded RNA (26‐32 kilobases in length) viruses (Masters, 2006). Members of the Coronaviruses are distributed largely among vertebrates, mainly mammals and birds, and can cause respiratory, enteric, hepatic, and neurological diseases in various animal species (Lu et al., 2020). Over the last two decades, there have been two other coronavirus disease outbreaks, namely Severe Acute Respiratory Syndrome (SARS) (Nie et al., 2003) and Middle East Respiratory Syndrome (MERS) (Zaki et al., 2012) in 2003 and 2012, respectively. SARS and MERS are characterized by the symptoms they cause, such as severe fever, non‐productive cough, myalgia, dyspnea and, ultimately, admission to intensive care units (ICUs) (Nie et al., 2003; Phua et al., 2020). In this regard, there are many similarities between SARS, MERS, and the clinical findings of COVID‐19. However, unlike SARS and MERS coronaviruses, the novel SARS‐CoV‐2, is highly transmissible and responsible for the unprecedented numbers of infections, claiming an alarming number of deaths worldwide. According to the World Health Organizations’ (WHO) last update on 7 February 2022, there have been 394,381,395 confirmed cases of COVID‐19 worldwide, including 5,735,179 reported deaths. Countries such as the United States of America, India, and Brazil, with more than 75,725,243, 42,272,014 and 26,473,273 confirmed cases, respectively, are now breeding ground for variants. On the same date, the WHO reported 857,657 confirmed cases and 2264 deaths in the United Arab Emirates (World Health Organization, 2021). As such, the COVID‐19 pandemic has had a tremendous impact in the health & socioeconomic condition of billions of people across the globe. All the while, the scientific community joined forces to find a secure solution for COVID‐19.
In the initial waves of the pandemic, the potential to use the classical and historical interventions of convalescent plasma (CP) to treat severely ill patients emerged as a viable and highly practical option, especially in the absence of alternative effective treatments (Duan et al., 2020; Kumar et al., 2010; Klassen et al., 2021). Subsequently, four antiviral drugs have demonstrated clinical benefit in randomized controlled trials (RCT), namely dexamethasone (RECOVERY Collaborative Group, 2021b), remdesivir (Beigel et al., 2020), molnupiravir (Jayk Bernal et al., 2022), and ritonavir‐boosted nirmatrelvir (Paxlovid) (Hammond et al., 2022). Anti‐SARS‐CoV‐2 monoclonal antibody (mAbs) treatments, such as bebtelovimab and sotrovimab, have also been developed and are recommended for non‐hospitalized patients with mild to moderate COVID‐19 who are at elevated risk of progressing to severe disease. Immunization to SARS‐CoV‐2 infection is made possible through multiple authorized, safe, and effective vaccines, however equitable vaccine access has yet to be realized. Similarly, the accessibility and cost of currently available antiviral and mAb treatments remain problematic in middle‐ and low‐income countries. The use of CP to treat COVID‐19 in countries with little access to vaccines or new generation treatments thus remains a relevant strategy to save lives.
2. CP TO TREAT VIRAL CONDITIONS, INCLUDING COVID‐19 DISEASE
CP administration for the treatment of severe acute respiratory viral infections is a longstanding strategy of passive immunization and was first used in the early twentieth century to control outbreaks of viral diseases (Marano et al., 2016). CP was used in the management of the H1N1 influenza virus pandemic where convalescent serum antibody preparations were used to remedy severely ill patients requiring intensive care (Hung et al., 2011). During the West African Ebola epidemic in 2013, convalescent blood was administered to treat patients suffering from Ebola virus disease (EVD) (Sahr et al., 2017). Importantly, CP was applied in the treatment of cases with SARS‐CoV and MERS‐CoV infections, diminishing viral load and improving clinical outcomes (Cheng et al., 2005; Ko et al., 2018).
Since the COVID‐19 outbreak, the FDA has provided guidelines for the collection and use of COVID‐19 convalescent plasma (CCP) (Figure 1) under emergency use authorization (EUA) or investigational new drug application (IND) regulations (FDA and HHS, 2021). A pilot study from China reported clinical findings CP from recovered patients (Shen et al., 2020). Within 3 days after CCP treatment, the body temperatures for four of the five patients normalized, and 12 days after the transfusion, their viral loads became undetectable. Within two weeks post‐treatment, three of the patients were dischar of five severely ill patients with COVID‐19 and acute respiratory distress syndrome (ARDS) that were treated with Cged, and two were in a stable condition (Shen et al., 2020). A separate study also reported a successful outcome where the administration of CCP resulted in the significant improvement of all 10 severely ill patients with no severe adverse effects (Duan et al., 2020). Here, within seven days, the viral load was undetectable in patients who had suffered viremia. A randomized controlled trial also revealed the immunomodulatory potential of CP in reducing the cytokine storm and disease severity indices in COVID‐19 patients, but also found CP to have no significant impact on mortality (Pouladzadeh et al., 2021).
FIGURE 1.
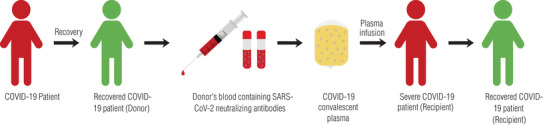
Diagram of the process of COVID‐19 convalescent plasma (CCP) collection and treatment. Fully recovered COVID‐19 patients donate blood containing anti‐SARS‐CoV‐2 antibodies. Neutralizing antibody‐rich plasma is separated and administered to severely or critically ill COVID‐19 patients via intravenous infusion
In contrast, results of a randomized clinical trial of CCP treatment in hospitalized COVID‐19 patients with moderate to severe disease showed no clinical improvement relative to the placebo, providing support against the use of CCP treatment for COVID‐19 pneumonia (van den Berg et al., 2022). The results from a randomized controlled, open‐label, platform trial also showed no improvement in survival or other clinical outcomes in hospitalized COVID‐19 patients treated with high‐titer CP (RECOVERY Collaborative Group, 2021a). Indeed, CCP can be used for either prophylaxis of infection or treatment of disease (Casadevall & Pirofski, 2020), however as highlighted above (Duan et al., 2020; Shen et al., 2020; van den Berg et al., 2022), and by other experts in the field (Dzik, 2020), it is important to conduct additional studies and randomized controlled investigations to confirm the effectiveness and safety of using CCP in all types of COVID‐19 hospitalized patients, particularly for those at risk of progressing to critical state. For example, Allahyari et al. (2021) evaluated the efficacy and safety of CCP in severe COVID‐19 patients with mild or moderate ARDS and found that early administration of CP assisted with symptom resolution (Allahyari et al., 2021).
While the United States Food and Drug Administrations (FDA) has very recently approved the use of CCP to treat critically ill COVID‐19 patients (Tanne, 2020), the potential use of CP treatments remains somehow controversial and, as already highlighted, a matter of safety concern (Arabi et al., 2015; Roback & Guarner, 2020). For example, non‐infectious risks such as transfusion‐related acute lung injury (TRALI) and transfusion‐associated circulatory overload (TACO) are well‐documented serious adverse effect of serum/plasma transfusion. Therefore, there is a justified concern that CCP administration to patients who are affected by severe pulmonary disease is associated with high risk of TACO or TRALI (Caccamo et al., 2020). Additionally, antibody‐dependent enhancement (ADE) is a risk that has already occurred in numerous viral diseases and entails the exacerbation of disease severity in the presence of certain antibodies from a prior infection. In the case of coronaviruses, various mechanisms of ADE have been reported, raising concern that one type of coronavirus could introduce antibodies that enhance infection of a different strain (Wan et al., 2020).
Neutralizing antibody (nAb) titers and the symptomatic state of the patient are key considerations in establishing CCP efficacy (Bloch et al., 2020). Evidence from recently published RCTs (Agarwal et al., 2020; Li et al., 2020; Rasheed et al., 2020) and large observational studies (Liu et al., 2020; Perotti et al., 2020; Salazar et al., 2021) suggest that CCP is most likely to be effective when high nAb titer units are administered early in the course of disease. However, the dynamic pandemic landscape has introduced confounding factors that present challenges when assessing the efficacy of CCP in observational studies and RCTs. A global review of experimental and clinical evidence concerning CP therapy for COVID‐19 was presented by Klassen et al. (2021). The authors also performed a meta‐analysis and qualitative inspection on all available daily Kaplan–Meier survival data from RCTs and matched‐control studies and observed a directionally consistent pattern, suggesting convalescent plasma efficacy in COVID‐19 patients.
3. SARS‐CoV‐2 VARIANTS AND CCP
Newly emerged SARS‐CoV‐2 variants of concern (VOCs) include Omicron (lineage B.1.1.529), Delta (lineage B.1.617.2), Beta (lineage B.1.351), Alpha (lineage B.1.1.7) and Gamma (lineage P.1). The Beta variant identified in South Africa harbours nine mutations in the spike gene, including three substitutions (K417N, E484K and N501Y) within the receptor‐binding domain (RBD) (Tegally et al., 2020). The Alpha variant first contains eight mutations in the spike gene, including one substitution (N501Y) in the RBD. The Gamma variant identified in Brazil contains three substitutions (K417T, E484K and N501Y) at the same RBD residues as Beta (Faria et al., 2021). Compared to wild‐type SARS‐CoV‐2, the Beta variant is significantly more resistant to neutralization by CCP and sera from vaccinated individuals (Hoffmann et al., 2021; Wang et al., 2021). On the other hand, the Alpha variant shows no increased resistance to CCP or sera from SARS‐CoV‐2 vaccinated individuals. The E484K substitution shared by Beta and Gamma appears to confer partial resistance against neutralization by multiple individual mAbs against the RBD receptor‐binding motif (Weisblum et al., 2020). Therefore, Beta and Gamma, but not Alpha, could potentially increase the risk of infection in convalescent and/or vaccinated individuals (Greaney et al., 2021; Planas et al., 2021; Wibmer et al., 2021).
First identified in India in late 2020, the Delta variant became the dominant strain worldwide prior to the Omicron variant. Similar to the other VOCs, the transmission of the Delta variant was found to be associated with an escape from neutralizing mAbs elicited by previous infection with SARS‐CoV‐2 or by COVID‐19 vaccination (Planas et al., 2021). Outcompeting Delta, the highly transmissible Omicron variant was first detected in Botswana. It is the most heavily mutated VOC with over 30 mutations in the spike protein, which is the primary neutralization target of CP, vaccines, and therapeutic mAbs (Hoffmann et al., 2020; Lan et al., 2020). Cao et al. (2022) demonstrated that 85% of mAbs of diverse epitopes were ineffective against Omicron (Cao et al., 2022). In addition, VanBlargan et al. (2022) found that several, but not all, clinically utilized anti‐receptor‐binding domain (RBD) mAbs lost neutralizing activity against the variant (VanBlargan et al., 2022).
The diverse mutations in the spike genes of these variants indicate antigenic drift and raise notable concerns regarding potential immune evasion. Moreover, these variants pose new challenges for mAb treatments and threaten the protective efficacy of current vaccines (Wang et al., 2021). Regarding CCP administration, the FDA also clearly stated that CP is a promising solution, although it has not been shown to be effective in every disease studied. So far, studies in CCP have not reported any adverse event associated with its administration (Chen & Xia, 2020; RECOVERY Collaborative Group, 2021a). Nevertheless, it remains crucial that we find molecular tools that allow us to measure and predict the safety of CCP before its administration. Thus, beside the need to further investigate the safety and efficacy of CCP in high‐quality trials as a consensus reached by a majority of experts in the field (Dzik, 2020), herein we advocate a technological approach to understand the performance of CP at molecular level.
4. SUGGESTED CP MECHANISMS OF ACTION
As previously mentioned, despite the historical and recurrent use of CP as a medical emergency response to unforeseen viral diseases, the exact molecular mechanism of action by which the plasma neutralizes the virus remains unclear (Casadevall & Pirofski, 2020; Cheng et al., 2005; Rojas et al., 2020). One obvious explanation for successful use of CP therapy is that the nAbs from the plasma mediate protection and viral neutralization. Although, the efficacy of the CP treatment has been often associated with the concentration of nAb in plasma from recovered donors (Rajendran et al., 2020; Tiberghien et al., 2020; van Griensven et al., 2016). Other probable mechanisms, including phagocytosis and/or antibody‐dependent cellular cytotoxicity, could also enhance the efficacy of CP (Casadevall & Pirofski, 2020; Rojas et al., 2020). Aside from antibodies, other mediators present in the plasma might protect against SARS‐CoV‐2 infection. For example, molecules such as CCL28 chemokines, which were reported to protect against HIV‐1 infection at mucosal sites (Rainone et al., 2011), might also be involved in protecting against coronavirus infection (Mohan et al., 2017).
Based on the concept that CP immunotherapy is linked with the concentration of nAbs, Arabi et al. (2015) proposed a protocol for MERS‐CoV infection where CP is evaluated through antibody based assay (ELISA) to inform whether the donor has sufficiently high antibody titers (Arabi et al., 2015, 2016). This method has been recently adapted to recovered patients with SARS‐CoV‐2 (Duan et al., 2020; Shen et al., 2020). Interestingly, in a study that utilized a sensitive SARS‐CoV‐2 pseudotyped‐lentiviral‐vector‐based neutralization assay to quantify specific nAbs in plasma from recovered COVID‐19 patients (Rojas et al., 2020), the data presented clearly indicated an unexpected variation in nAbs titers where nearly one‐third of patients did not develop high nAbs titers post‐infection (Wu et al., 2020). Different factors, such as age, lymphocyte count, and C‐reactive protein level in blood, have been linked to these disparities in nAbs titers, indicating that other plasma constituents contribute to patient recovery (Rojas et al., 2020; Smith et al., 2021; Wu et al., 2020). Finally, a more recent study evaluated multiple sero‐diagnostic assays for anti‐SARS‐CoV‐2 antibody detection. The head‐to‐head comparison of the serological tests revealed an agreement between ELISAs and latera flow assays up to 94.8% (Whitman et al., 2020). The authors concluded that studies aimed at understanding the serological mechanistic correlates of protective immunity to SARS‐CoV‐2 are necessary for guiding rational clinical and public health policies (Whitman et al., 2020). Therefore, despite the potential utility of this therapy, there is a need to take a more sophisticate approach to characterize CP and gain important molecular insights into its mechanism of action of CP.
5. MASS SPECTROMETRY ANALYSIS AS A WAY FORWARD TO COMPREHENSIVELY CHARACTERIZE CCP
Plasma proteomics can provide relevant and unbiased data relating to disease progression and potential therapeutic agents (Messner et al., 2020). Proteomic biomarkers are easily accessible and have proven useful for infectious diseases as they vary with different disease states of the patient (Whetton et al., 2020). Owing to the number of different analytes and their range of concentrations, plasma has one of the most complex peptidomes, proteomes and metabolomes in the human body. Because the dynamic range of compound concentration can vary >10 orders of magnitude, attaining in‐depth profiling of plasma peptidome/proteome/metabolome has proven difficult (Baker et al., 2012). In contrast to enzymatic and antibody‐based methods, mass spectrometry (MS) methods measure the highly accurate mass and fragmentation of small molecules such as peptide and metabolites (Baker et al., 2012; Geyer et al., 2017). In this regard, MS‐based analyses offer an attractive avenue to comprehensively profile human plasma both at proteomic (Geyer et al., 2017; Ignjatovic et al., 2019; Schwenk et al., 2017) and metabolomic‐levels (Anesi et al., 2019; Telu et al., 2016).
MS‐based techniques have been applied for plasma biomarker discovery and profiling for various human diseases (Bringans et al., 2017; Geyer et al., 2017, 2021; Huang et al., 2017,) and may be valuable for early COVID‐19 diagnosis, disease surveillance, and predictions of treatment response in patients (Ignjatovic et al., 2019). Patel et al. (2021) demonstrated that blood protein profiling of mild‐to‐critical COVID‐19 positive patients lead to the identification of six proteins associated with disease severity (Patel et al., 2021). Likewise, other studies have shown the successful application of proteomics in the detection of COVID‐19 biomarkers (Li et al., 2020; Messner et al., 2020; Shen et al., 2020; Whetton et al., 2020). Hence, identifying COVID‐19 biomarkers of clinical value, such as defining disease severity, predicting disease phases and patient outcomes may be useful in helping to manage the current pandemic (Park et al., 2020). COVID‐19 also comprises several phases and it is important to identify biomarkers to aid clinical decisions at each point (Whetton et al., 2020).
Plasma metabolic profiles provide unique information concerning disease severity as it reflects both endogenous and exogenous influences while supplying a functional readout of relevant cellular biochemistry (Telu et al., 2016). Correlating metabolic profiles with patient outcomes can therefore be used for the assessment of patient response to treatments and biomarker discovery (Figure 2) (Dunn et al., 2011). Gas chromatography‐mass spectrometry (GC–MS), liquid chromatography‐mass spectrometry (LC–MS/MS) and nuclear magnetic resonance spectroscopy (NMR) are commonly used analytical techniques for metabolite analysis. Properties of the metabolites of interest and factors such as the sample matrix, quantity, and concentration determine the optimal analysis technique. GC‐MS and LC‐MS/MS exceed the sensitivity, resolution and dynamic range afforded by NMR, making them more suitable for the characterization of blood disease biomarkers (Zeki et al., 2020). In addition, metabolomic profiling may represent a step toward personalized therapeutics for COVID‐19 patients as there is prospect for the stratification of COVID‐19 patients based on their metabolic phenotype to optimize drug efficacy (Migaud et al., 2020).
FIGURE 2.
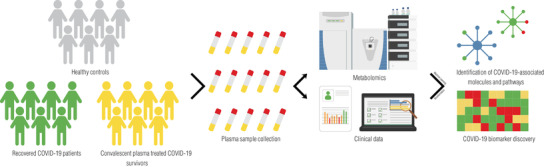
Experimental workflow for metabolomic characterization of COVID‐19 patient CCP. Plasma is harvested from a cohort of recovered COVID‐19 patients (CCP donors), CCP treated COVID‐19 survivors, and healthy controls. The plasma metabolomes are analysed using mass spectrometry. The metabolomic profiles coupled with clinical patient information can be used to identify COVID‐19‐associated molecules, pathways, and biomarkers
Metabolomics analyses are based on either targeted or untargeted approaches. With targeted metabolomic applications being most common, information on analysing different classes of metabolites is readily available. However, metabolomics studies are moving toward untargeted profiling to identify a broader range of novel metabolites in each measurement without bias. Untargeted metabolomics has been used to discover new plasma biomarkers for the early diagnosis of CKD in plasma from paediatric patients (Benito et al., 2018) and urea cycle disorders (UCDs) (Burrage et al., 2019). Table 1 summarizes the MS‐based proteomic and metabolic studies mentioned in this review.
TABLE 1.
Summary of MS‐based proteomic and metabolic studies analysing human plasma
| S/N | Sample size | Methodology | Results | References |
|---|---|---|---|---|
| 1 | The results of a primary exploratory cohort (31 COVID‐19 patients) were validated on a smaller cohort of 17 independent patients and 15 healthy volunteers | Ultra‐high‐throughput clinical proteomics using complementary LC systems coupled to a TripleTOF 6600. Data acquisition using sequential window acquisition of all theoretical fragment ion spectra (SWATH) or data‐independent acquisition (DIA) | Identification of 27 potential biomarkers that are differentially expressed depending on COVID‐19 disease severity | (Messner et al., 2020) |
| 2 | Standard Reference Material (SRM 1950) | Non‐targeted metabolomics using nano, conventional and UHPLC coupled to a Q‐TOF MS or Orbitrap MS | Highly reproducible results when using a single platform; but significantly different metabolite profiles when using different platforms | (Telu et al., 2016) |
| 3 | 42 participants | LC‐MS‐based clinical proteomics | Comparison of longitudinal proteomes of women and men in a weight loss study revealed that estrogen‐regulated proteins were significantly higher in women | (Geyer et al., 2021) |
| 4 | 572 chronic kidney disease patients | MS‐based proteomics platform using iTRAQ labelling and Matrix‐Assisted Laser Desorption/Ionization (MALDI) MS using a 4800 TOF/TOF system | Albuminuria, renal impairment (eGFR), and chronic kidney disease staging (CKD Stage 1, ROC curve of 0.77) were all found to be significantly associated with diabetic kidney disease | (Bringans et al., 2017) |
| 5 | 46 COVID‐19 patients and 53 control individuals | Stable isotope labeled proteomics and UPLC‐MS/MS untargeted metabolomic profiling using a Q‐Exactive HF‐X hybrid Quadrupole‐Orbitrap | The study observed that 105 proteins were differentially expressed in COVID‐19 patients' sera but not in control patients' sera. In severe patients, 93 proteins showed specific modulation | (Shen et al., 2020) |
| 6 | 40 urine specimens that passed quality check, including 32 healthy controls, 6 COVID‐19 patients and 2 corresponding recovery persons | LC‐MS/MS analysis on an Orbitrap Fusion Lumos coupled with EASY‐nLC 1200 | The correlation of samples within the health and recovery groups was shown to be greater than the correlation of samples between the healthy and patient groups. Overall, the study demonstrated that urine proteomics can reliably and sensitively differentiate COVID‐19 patients from healthy individuals | (Li et al., 2020) |
| 7 | Undepleted plasma samples from 8 COVID‐19 patients, including 3 non‐severe (mild) and 5 severe cases | LC–MS/MS analyses using Quadrupole Orbitrap MSs in data‐dependent acquisition (DDA) mode | The study discovered 91 differentially expressed plasma proteins between the mild and severe COVID‐19 groups, demonstrating the utility of plasma proteome signatures. Furthermore, the bioinformatics analysis revealed that several inflammatory modulators, particularly IL‐6, IL‐1B, and TNF, have a high specificity | (Park et al., 2020) |
| 8 | 32 patients diagnosed with CKD (aged 3–18 years) and 26 control patients (aged 6–19 years) | Untargeted metabolomics based on LC‐QTOF‐MS | Control and CKD paediatric patients' metabolic fingerprints were compared, and 5 metabolites that showed a significant change in both data analysis procedures were identified | (Benito et al., 2018) |
| 9 | 48 individuals with various urea cycle disorders | Untargeted mass spectrometry‐based metabolomics using a Waters ACQUITY UPLC and a Thermo Scientific Q‐Exactive MS | Plasma metabolomic analysis identified multiple potentially neurotoxic arginine metabolites in arginase deficiency, which may be useful in monitoring treatment efficacy in arginase deficiency. Furthermore, multiple biochemical perturbations in all UCDs were detected that likely reflect clinical management | (Burrage et al., 2019) |
| 10 | 76 subjects that included 26 healthy controls and 50 COVID‐19 patients of differing disease severity (mild, moderate, and severe) | Targeted lipidomics and untargeted lipidomics using high‐resolution TOF MS on a 5600 TripleTOF Plus | Untargeted metabolomics detected an initial pool of 1,552 metabolite peaks with coefficients of variation of 20% across quality control samples. The consolidated plasma metabolome contained 1,002 metabolites (598 lipids and 404 polar metabolites) quantified using 71 internal standards after structural confirmation using MS/MS spectra | (Song et al., 2020) |
| 11 | 9 patients with fatal outcome, 11 patients with severe symptoms, 14 patients with mild symptoms, and 10 healthy individuals | Sample extracts of hydrophilic compounds were analyzed using an LC‐ESI‐MS/MS system | In total, 431 metabolites and 698 lipids were identified and quantified, and both the metabolome and the lipidome revealed dramatic changes in the plasma of COVID‐19 patients. When comparing patients who died with healthy volunteers, malic acid and glycerol 3‐phosphate showed the greatest reduction, but they also showed dramatic reduction in groups with severe and mild symptoms | (Wu et al., 2020) |
| 12 | 5 patients with fatal outcome, 7 patients with severe symptoms, 10 patients with mild symptoms, and 8 healthy individuals | Proteomics profiling of plasma using LC‐MS/MS analysis on a Q Exactive HF‐X MS coupled with an Easy‐nLC 1200 system | Eleven biomarkers and a set of biomarker combinations were developed that can accurately differentiate or predict different COVID‐19 outcomes. For the cohort, 530 proteins were mutually quantified in >70% of the samples | (Shu et al., 2020) |
6. MULTI‐OMICS DATASETS MAY LEAD TO AN UNDERSTATING CCP MECHANISMS OF ACTION AND TO PREDICT ITS TREATMENT EFFICACY
A recent study by Song et al. (2020) examined the plasma lipidome and metabolome of COVID‐19 patients with varying disease severity against healthy controls. It was revealed that the plasma lipidome of COVID‐19 patients mirrors that of monosialodihexosyl ganglioside (GM3)‐enriched exosomes. They further demonstrated a positive association between GM3‐enriched exosomes and COVID‐19 disease severity. Lastly, the study identified a subset of 10 plasma metabolites that can distinguish COVID‐19 patients from healthy controls (Song et al., 2020). Similarly, plasma lipidomic and metabolomic profiling of a cohort of COVID‐19 patients revealed a correlation between lipid and metabolite alteration and clinical symptoms (Wu et al., 2020). Shu et al. (2020) performed a quantitative proteomic analysis on a cohort of COVID‐19 patients to profile host responses to SARS‐CoV‐2 infection. The study used plasma samples from survivors that recovered from mild or severe symptoms, as well as non‐survivors, and revealed COVID‐19‐related alterations for host proteins implicated with inflammation and coagulation. In addition, they developed and utilized a machine‐learning‐based pipeline for biomarker identification to accurately classify different samples. This pipeline demonstrated the potential of MS‐based proteomics to produce predictive biomarker signatures that support clinical decision making through the identification of 11 protein biomarkers and a set of biomarker combinations capable of accurately predicting different COVID‐19 outcomes or differentiating between them (Shu et al., 2020).
Acosta‐Ampudia et al. (2021) assessed untargeted metabolomic and lipidomic profiles of severe COVID‐19 patients, recovered COVID‐19 patients, and un‐exposed healthy controls to evaluate CP composition. They confirmed altered sera cytokine and metabolite composition in severe COVID‐19 patients when compared to recovered patients. Interestingly, recovered COVID‐19 patients exhibited sustained perturbed unsaturated fatty acid levels and did not revert to a pre‐pandemic phenotype. In addition, it was determined that total immunoglobulin A (IgA) and immunoglobulin G (IgG) anti‐S1‐SARS‐CoV‐2 antibodies correlate with nAbs and should be considered as important factors for CP selection (Acosta‐Ampudia et al., 2021).
7. CONCLUSION AND FUTURE DIRECTIONS
Before clinicians can select safe and efficient CP before administration, future multi‐omics translational studies are needed to determine the presence of CP biomarkers, other than nAbs, that correlate with treatment response and could be used to inform the selection of specific CP samples for therapeutic use. Employing multi‐omics studies to shed light on the mechanisms of CP will also lead to a robust MS‐based quality control protocol that supports the use of CP therapy as a sophisticated and reliable frontline resource for current and future viral epidemics based on a panel of biomarkers. Such a protocol will assist clinicians all over the world in optimizing CCP therapy for both safety and effectiveness, thereby providing confidence and quality assurance to this controversial treatment, and ultimately allowing it to be utilized to save millions of lives. Given that CCP therapy can be produced locally and affordably, we envisage that this projected approach will have enormous potential to improve the health care systems in low‐ and middle‐income countries, subsequently having a significant positive impact on their socioeconomic condition.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
Special thanks to the University of Sharjah for the financial support, competitive grant numbers 1901110133 and 2001110138; This study is part of the Human Disease Biomarkers Discovery Research Group's (SIMR‐UOS) strategy.
Baros‐Steyl, S. S. , Al Heialy, S. , Semreen, A. H. , Semreen, M. H. , Blackburn, J. M. , & Soares, N. C. (2022). A review of mass spectrometry‐based analyses to understand COVID‐19 convalescent plasma mechanisms of action. Proteomics, 22, e2200118. 10.1002/pmic.202200118
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Acosta‐Ampudia, Y. , Monsalve, D. M. , Rojas, M. , Rodríguez, Y. , Gallo, J. E. , Salazar‐Uribe, J. C. , Santander, M. J. , Cala, M. P. , Zapata, W. , Zapata, M. I. , Manrique, R. , Pardo‐Oviedo, J. M. , Camacho, B. , Ramírez‐Santana, C. , & Anaya, J. M. (2021). COVID‐19 convalescent plasma composition and immunological effects in severe patients. Journal of Autoimmunity, 118, 102598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal, A. , Mukherjee, A. , Kumar, G. , Chatterjee, P. , Bhatnagar, T. , & Malhotra, P. (2020). Convalescent plasma in the management of moderate covid‐19 in adults in India: Open label phase II multicentre randomised controlled trial (PLACID Trial). BMJ, 371, m3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahyari, A. , Seddigh‐Shamsi, M. , Mahmoudi, M. , Amel Jamehdar, S. , Amini, M. , Mozdourian, M. , Javidarabshahi, Z. , Eslami Hasan Abadi, S. , Amini, S. , Sedaghat, A. , Emadzadeh, M. , Moeini Nodeh, M. , Rahimi, H. , Bari, A. , Mozaheb, Z. , Kamandi, M. , Ataei Azimi, S. , Abrishami, M. , … Saeedian, N. (2021). Efficacy and safety of convalescent plasma therapy in severe COVID‐19 patients with acute respiratory distress syndrome. International Immunopharmacology, 93, 107239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anesi, A. , Rubert, J. , Oluwagbemigun, K. , Orozco‐Ruiz, X. , Nöthlings, U. , Breteler, M. M. B. , & Mattivi, F. (2019). Metabolic profiling of human plasma and urine, targeting tryptophan, tyrosine and branched chain amino acid pathways. Metabolites, 9, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi, Y. , Balkhy, H. , Hajeer, A. H. , Bouchama, A. , Hayden, F. G. , Al‐Omari, A. , Al‐Hameed, F. M. , Taha, Y. , Shindo, N. , Whitehead, J. , Merson, L. , AlJohani, S. , Al‐Khairy, K. , Carson, G. , Luke, T. C. , Hensley, L. , Al‐Dawood, A. , Al‐Qahtani, S. , Modjarrad, K. , … Fowler, R. (2015). Feasibility, safety, clinical, and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory syndrome coronavirus infection: A study protocol. Springerplus, 4, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi, Y. M. , Hajeer, A. H. , Luke, T. , Raviprakash, K. , Balkhy, H. , Johani, S. , Al‐Dawood, A. , Al‐Qahtani, S. , Al‐Omari, A. , Al‐Hameed, F. , Hayden, F. G. , Fowler, R. , Bouchama, A. , Shindo, N. , Al‐Khairy, K. , Carson, G. , Taha, Y. , Sadat, M. , & Alahmadi, M. (2016). Feasibility of using convalescent plasma immunotherapy for MERS‐CoV infection, Saudi Arabia. Emerging Infectious Diseases, 22, 1554–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, E. S. , Liu, T. , Petyuk, V. A. , Burnum‐Johnson, K. E. , Ibrahim, Y. M. , Anderson, G. A. , & Smith, R. D. (2012). Mass spectrometry for translational proteomics: progress and clinical implications. Genome Medicine, 4, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel, J. H. , Tomashek, K. M. , Dodd, L. E. , Mehta, A. K. , Zingman, B. S. , Kalil, A. C. , Hohmann, E. , Chu, H. Y. , Luetkemeyer, A. , Kline, S. , Lopez de Castilla, D. , Finberg, R. W. , Dierberg, K. , Tapson, V. , Hsieh, L. , Patterson, T. F. , Paredes, R. , Sweeney, D. A. , Short, W. R. , … Lane, H. C. (2020). Remdesivir for the treatment of Covid‐19 — Final report. New England Journal of Medicine, 383, 1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito, S. , Sánchez‐Ortega, A. , Unceta, N. , Andrade, F. , Aldámiz‐Echevarria, L. , Goicolea, M. A. , & Barrio, R. J. (2018). Untargeted metabolomics for plasma biomarker discovery for early chronic kidney disease diagnosis in pediatric patients using LC‐QTOF‐MS. Analyst, 143, 4448–4458. [DOI] [PubMed] [Google Scholar]
- Bloch, E. M. , Shoham, S. , Casadevall, A. , Sachais, B. S. , Shaz, B. , Winters, J. L. , Van Buskirk, C. , Grossman, B. J. , Joyner, M. , Henderson, J. P. , Pekosz, A. , Lau, B. , Wesolowski, A. , Katz, L. , Shan, H. , Auwaerter, P. G. , Thomas, D. , Sullivan, D. J. , Paneth, N. , … Tobian, A. A. R. (2020). Deployment of convalescent plasma for the prevention and treatment of COVID‐19. Journal of Clinical Investigation, 130, 2757–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringans, S. D. , Ito, J. , Stoll, T. , Winfield, K. , Phillips, M. , Peters, K. , Davis, W. A. , Davis, T. M. E. , & Lipscombe, R. J. (2017). Comprehensive mass spectrometry based biomarker discovery and validation platform as applied to diabetic kidney disease. EuPA Open Proteomics, 14, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrage, L. C. , Thistlethwaite, L. , Stroup, B. M. , Sun, Q. , Miller, M. J. , Nagamani, S. C. S. , Craigen, W. , Scaglia, F. , Sutton, V. R. , Graham, B. , Kennedy, A. D. , Milosavljevic, A. , Lee, B. H. , & Elsea, S. H. (2019). Untargeted metabolomic profiling reveals multiple pathway perturbations and new clinical biomarkers in urea cycle disorders. Genetics in Medicine, 21, 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo, N. , Sullivan, L. C. , Brooks, A. G. , & Dieli, F. (2020). Harnessing HLA‐E‐restricted CD8 T lymphocytes for adoptive cell therapy of patients with severe COVID‐19. British Journal of Haematology, 190, e185–e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y. , Wang, J. , Jian, F. , Xiao, T. , Song, W. , Yisimayi, A. , Huang, W. , Li, Q. , Wang, P. , An, R. , Wang, J. , Wang, Y. , Niu, X. , Yang, S. , Liang, H. , Sun, H. , Li, T. , Yu, Y. , Cui, Q. , … Xie, X. S. (2022). Omicron escapes the majority of existing SARS‐CoV‐2 neutralizing antibodies. Nature, 602, 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall, A. , & Pirofski, L. A. (2020). The convalescent sera option for containing COVID‐19. Journal of Clinical Investigation, 130, 1545–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B. , & Xia, R. (2020). Early experience with convalescent plasma as immunotherapy for COVID‐19 in China: Knowns and unknowns. Vox Sanguinis, 115, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Wong, R. , Soo, Y. O. Y. , Wong, W. S. , Lee, C. K. , Ng, M. H. L. , Chan, P. , Wong, K. C. , Leung, C. B. , & Cheng, G. (2005). Use of convalescent plasma therapy in SARS patients in Hong Kong. European Journal of Clinical Microbiology & Infectious Diseases, 24, 44–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, K. , Liu, B. , Li, C. , Zhang, H. , Yu, T. , Qu, J. , Zhou, M. , Chen, L. , Meng, S. , Hu, Y. , Peng, C. , Yuan, M. , Huang, J. , Wang, Z. , Yu, J. , Gao, X. , Wang, D. , Yu, X. , Li, L. , … Yang, X. (2020). Effectiveness of convalescent plasma therapy in severe COVID‐19 patients. Proceedings of the National Academy of Sciences of the United States of America, 117, 9490–9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, W. B. , Broadhurst, D. , Begley, P. , Zelena, E. , Francis‐Mcintyre, S. , Anderson, N. , Brown, M. , Knowles, J. D. , Halsall, A. , Haselden, J. N. , Nicholls, A. W. , Wilson, I. D. , Kell, D. B. , & Goodacre, R. (2011). Procedures for large‐scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols, 6, 1060–1083. [DOI] [PubMed] [Google Scholar]
- Dzik, S. (2020). COVID‐19 convalescent plasma: Now is the time for better science. Transfusion Medicine Reviews, 34, 141–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria, N. R. , Claro, I. M. , Candido, D. , Franco, L. A. M. , Andrade, P. S. , Thais, M. , Silva, C. A. M. , Sales, F. C. , Erika, R. , Aguiar, R. S. , Gaburo, N. , Cecília, C. , Fraiji, N. A. , Crispim, M. A. E. , Carvalho, P. S. S. , & Rambaut, A. (2021). Genomic characterisation of an emergent SARS‐CoV‐2 lineage in Manaus: preliminary findings. Virological.Org, 1–9. [Google Scholar]
- FDA and HHS (2021). Investigational COVID‐19 Convalescent Plasma | FDA.
- Geyer, P. E. , Holdt, L. M. , Teupser, D. , & Mann, M. (2017). Revisiting biomarker discovery by plasma proteomics. Molecular Systems Biology, 13, 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, P. E. , Mann, S. P. , Treit, P. V. , & Mann, M. (2021). Plasma proteomes can be reidentifiable and potentially contain personally sensitive and incidental findings. Molecular & Cellular Proteomics, 20, 100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney, A. J. , Loes, A. N. , Crawford, K. H. D. , Starr, T. N. , Malone, K. D. , Chu, H. Y. , & Bloom, J. D. (2021). Comprehensive mapping of mutations in the SARS‐CoV‐2 receptor‐binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host & Microbe, 29, 463–476.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, W. , Ni, Z. , Hu, Y. , Liang, W. , Ou, C. , He, J. , Liu, L. , Shan, H. , Lei, C. , Hui, D. S. C. , Du, B. , Li, L. , Zeng, G. , Yuen, K.‐Y. , Chen, R. , Tang, C. , Wang, T. , Chen, P. , Xiang, J. , … Zhong, N. (2020). Clinical characteristics of coronavirus disease 2019 in China. New England Journal of Medicine, 382, 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, J. , Leister‐Tebbe, H. , Gardner, A. , Abreu, P. , Bao, W. , Wisemandle, W. , Baniecki, M. , Hendrick, V. M. , Damle, B. , Simón‐Campos, A. , Pypstra, R. , & Rusnak, J. M. (2022). Oral nirmatrelvir for high‐risk, nonhospitalized adults with Covid‐19. New England Journal of Medicine, 386, 1397–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Arora, P. , Groß, R. , Seidel, A. , Hörnich, B. F. , Hahn, A. S. , Krüger, N. , Graichen, L. , Hofmann‐Winkler, H. , Kempf, A. , Winkler, M. S. , Schulz, S. , Jäck, H. M. , Jahrsdörfer, B. , Schrezenmeier, H. , Müller, M. , Kleger, A. , Münch, J. , & Pöhlmann, S. (2021). SARS‐CoV‐2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell, 184, 2384–2393.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Krüger, N. , Herrler, T. , Erichsen, S. , Schiergens, T. S. , Herrler, G. , Wu, N. H. , Nitsche, A. , Müller, M. A. , Drosten, C. , & Pöhlmann, S. (2020). SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181, 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Z. , Ma, L. , Huang, C. , Li, Q. , & Nice, E. C. (2017). Proteomic profiling of human plasma for cancer biomarker discovery. Proteomics, 17, 1600240. [DOI] [PubMed] [Google Scholar]
- Hung, I. F. N. , To, K. K. W. , Lee, C. K. , Lee, K. L. , Chan, K. , Yan, W. W. , Liu, R. , Watt, C. L. , Chan, W. M. , Lai, K. Y. , Koo, C. K. , Buckley, T. , Chow, F. L. , Wong, K. K. , Chan, H. S. , Ching, C. K. , Tang, B. S. F. , Lau, C. C. Y. , Li, I. W. S. , … Yuen, K. Y. (2011). Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clinical Infectious Diseases, 52, 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignjatovic, V. , Geyer, P. E. , Palaniappan, K. K. , Chaaban, J. E. , Omenn, G. S. , Baker, M. S. , Deutsch, E. W. , & Schwenk, J. M. (2019). Mass spectrometry‐based plasma proteomics: Considerations from sample collection to achieving translational data. Journal of Proteome Research, 18, 4085–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayk Bernal, A. , Gomes da Silva, M. M. , Musungaie, D. B. , Kovalchuk, E. , Gonzalez, A. , Delos Reyes, V. , Martín‐Quirós, A. , Caraco, Y. , Williams‐Diaz, A. , Brown, M. L. , Du, J. , Pedley, A. , Assaid, C. , Strizki, J. , Grobler, J. A. , Shamsuddin, H. H. , Tipping, R. , Wan, H. , Paschke, A. , … De Anda, C. (2022). Molnupiravir for oral treatment of covid‐19 in nonhospitalized patients. New England Journal of Medicine, 386, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen, S. A. , Senefeld, J. W. , Senese, K. A. , Johnson, P. W. , Wiggins, C. C. , Baker, S. E. , van Helmond, N. , Bruno, K. A. , Pirofski, L. A. , Shoham, S. , Grossman, B. J. , Henderson, J. P. , Wright, R. S. , Fairweather, D. L. , Paneth, N. S. , Carter, R. E. , Casadevall, A. , & Joyner, M. J. (2021). Convalescent plasma therapy for COVID‐19: A graphical mosaic of the worldwide evidence. Frontiers of Medicine, 8, 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, J. H. , Seok, H. , Cho, S. Y. , Ha, Y. E. , Baek, J. Y. , Kim, S. H. , Kim, Y. J. , Park, J. K. , Chung, C. R. , Kang, E. S. , Cho, D. , Müller, M. A. , Drosten, C. , Kang, C. I. , Chung, D. R. , Song, J. H. , & Peck, K. R. (2018). Challenges of convalescent plasma infusion therapy in Middle East respiratory coronavirus infection: A single centre experience. Antiviral Therapy, 23, 617–622. [DOI] [PubMed] [Google Scholar]
- Kumar, D. , Nath, L. , Kamal, M. A. , Varshney, A. , Jain, A. , Singh, S. , & Rao, K. V. S. (2010). Genome‐wide analysis of the host intracellular network that regulates survival of mycobacterium tuberculosis. Cell, 140, 731–743. [DOI] [PubMed] [Google Scholar]
- Lan, J. , Ge, J. , Yu, J. , Shan, S. , Zhou, H. , Fan, S. , Zhang, Q. , Shi, X. , Wang, Q. , Zhang, L. , & Wang, X. (2020). Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature, 581, 215–220. [DOI] [PubMed] [Google Scholar]
- Li, L. , Zhang, W. , Hu, Y. , Tong, X. , Zheng, S. , Yang, J. , Kong, Y. , Ren, L. , Wei, Q. , Mei, H. , Hu, C. , Tao, C. , Yang, R. , Wang, J. , Yu, Y. , Guo, Y. , Wu, X. , Xu, Z. , Zeng, L. , … Liu, Z. (2020). Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life‐threatening COVID‐19: A randomized clinical trial. JAMA ‐ Journal of the American Medical Association, 324, 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Wang, Y. , Liu, H. , Sun, W. , Ding, B. , Zhao, Y. , Chen, P. , Zhu, L. , Li, Z. , Li, N. , Chang, L. , Wang, H. , Bai, C. , & Xu, P. (2020). Urine proteome of COVID‐19 patients. URINE, 2, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. T. H. , Lin, H. M. , Baine, I. , Wajnberg, A. , Gumprecht, J. P. , Rahman, F. , Rodriguez, D. , Tandon, P. , Bassily‐Marcus, A. , Bander, J. , Sanky, C. , Dupper, A. , Zheng, A. , Nguyen, F. T. , Amanat, F. , Stadlbauer, D. , Altman, D. R. , Chen, B. K. , Krammer, F. , … Bouvier, N. M. (2020). Convalescent plasma treatment of severe COVID‐19: A propensity score–matched control study. Nature Medicine, 26, 1708–1713. [DOI] [PubMed] [Google Scholar]
- Lu, R. , Zhao, X. , Li, J. , Niu, P. , Yang, B. , Wu, H. , Wang, W. , Song, H. , Huang, B. , Zhu, N. , Bi, Y. , Ma, X. , Zhan, F. , Wang, L. , Hu, T. , Zhou, H. , Hu, Z. , Zhou, W. , Zhao, L. , … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet, 395, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marano, G. , Vaglio, S. , Pupella, S. , Facco, G. , Catalano, L. , Liumbruno, G. M. , & Grazzini, G. (2016). Convalescent plasma: New evidence for an old therapeutic tool? Blood Transfus, 14, 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters, P. S. (2006). The molecular biology of coronaviruses. Advances in Virus Research, 65, 193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner, C. B. , Demichev, V. , Wendisch, D. , Michalick, L. , White, M. , Freiwald, A. , Textoris‐Taube, K. , Vernardis, S. I. , Egger, A. S. , Kreidl, M. , Ludwig, D. , Kilian, C. , Agostini, F. , Zelezniak, A. , Thibeault, C. , Pfeiffer, M. , Hippenstiel, S. , Hocke, A. , von Kalle, C. , … Ralser, M. (2020). Ultra‐high‐throughput clinical proteomics reveals classifiers of COVID‐19 infection. Cell Systems, 11, 11–24.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migaud, M. , Gandotra, S. , Chand, H. S. , Gillespie, M. N. , Thannickal, V. J. , & Langley, R. J. (2020). Metabolomics to predict antiviral drug efficacy in COVID‐19. American Journal of Respiratory Cell and Molecular Biology, 63, 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan, T. , Deng, L. , & Wang, B. Z. (2017). CCL28 chemokine: An anchoring point bridging innate and adaptive immunity. International Immunopharmacology, 51, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, Q. H. , Luo, X. D. , & Hui, W. L. (2003). Advances in clinical diagnosis and treatment of severe acute respiratory syndrome. World Journal of Gastroenterology, 9, 1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Kim, H. , Kim, S. Y. , Kim, Y. , Lee, J. S. , Dan, K. , Seong, M. W. , & Han, D. (2020). In‐depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non‐severe COVID‐19 patients. Science Reports, 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, H. , Ashton, N. J. , Dobson, R. J. B. , Andersson, L. M. , Yilmaz, A. , Blennow, K. , Gisslen, M. , & Zetterberg, H. (2021). Proteomic blood profiling in mild, severe and critical COVID‐19 patients. Science Reports, 11, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotti, C. , Baldanti, F. , Bruno, R. , Del Fante, C. , Seminari, E. , Casari, S. , Percivalle, E. , Glingani, C. , Musella, V. , Belliato, M. , Garuti, M. , Meloni, F. , Frigato, M. , Di Sabatino, A. , Klersy, C. , & de Donno, G. (2020). Mortality reduction in 46 patients with severe COVID‐19 treated with hyperimmune plasma. A proof‐of‐concept, single‐arm, multicenter trial. Haematologica, 105, 2834–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua, J. , Weng, L. , Ling, L. , Egi, M. , Lim, C. M. , Divatia, J. V. , Shrestha, B. R. , Arabi, Y. M. , Ng, J. , Gomersall, C. D. , Nishimura, M. , Koh, Y. , & Du, B. (2020). Intensive care management of coronavirus disease 2019 (COVID‐19): Challenges and recommendations. The Lancet Respiratory Medicine, 8, 506–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas, D. , Bruel, T. , Grzelak, L. , Guivel‐Benhassine, F. , Staropoli, I. , Porrot, F. , Planchais, C. , Buchrieser, J. , Rajah, M. M. , Bishop, E. , Albert, M. , Donati, F. , Prot, M. , Behillil, S. , Enouf, V. , Maquart, M. , Smati‐Lafarge, M. , Varon, E. , Schortgen, F. , … Schwartz, O. (2021). Sensitivity of infectious SARS‐CoV‐2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nature Medicine, 27, 917–924. [DOI] [PubMed] [Google Scholar]
- Planas, D. , Veyer, D. , Baidaliuk, A. , Staropoli, I. , Guivel‐Benhassine, F. , Rajah, M. M. , Planchais, C. , Porrot, F. , Robillard, N. , Puech, J. , Prot, M. , Gallais, F. , Gantner, P. , Velay, A. , Le Guen, J. , Kassis‐Chikhani, N. , Edriss, D. , Belec, L. , Seve, A. , … Schwartz, O. (2021). Reduced sensitivity of SARS‐CoV‐2 variant Delta to antibody neutralization. Nature, 596, 276–280. [DOI] [PubMed] [Google Scholar]
- Pouladzadeh, M. , Safdarian, M. , Eshghi, P. , Abolghasemi, H. , bavani, A. G. , Sheibani, B. , Moradi Choghakabodi, P. , Feghhi, A. , Ghafourian Boroujerdnia, M. , Forouzan, A. , Jalali Far, M. A. , Kaydani, G. A. , Rajaei, E. , Amin, M. , Torabizadeh, M. , Yousefi, F. , & Hadaddezfuli, R. (2021). A randomized clinical trial evaluating the immunomodulatory effect of convalescent plasma on COVID‐19‐related cytokine storm. Internal and Emergency Medicine, 16, 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainone, V. , Dubois, G. , Temchura, V. , Überla, K. , Clivio, A. , Nebuloni, M. , Lauri, E. , Trabattoni, D. , Veas, F. , & Clerici, M. (2011). CCL28 induces mucosal homing of HIV‐1‐specific IgA‐secreting plasma cells in mice immunized with HIV‐1 virus‐like particles. Plos One, 6, e26979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran, K. , Krishnasamy, N. , Rangarajan, J. , Rathinam, J. , Natarajan, M. , & Ramachandran, A. (2020). Convalescent plasma transfusion for the treatment of COVID‐19: Systematic review. Journal of Medical Virology, 92, 1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed, A. M. , Fatak, D. F. , Hashim, H. A. , Maulood, M. F. , Kabah, K. K. , Almusawi, Y. A. , & Abdulamir, A. S. (2020). The therapeutic effectiveness of Convalescent plasma therapy on treating COVID‐19 patients residing in respiratory care units in hospitals in Baghdad, Iraq. medRxiv, 2020.06.24.20121905. [PubMed] [Google Scholar]
- RECOVERY Collaborative Group . (2021a). Convalescent plasma in patients admitted to hospital with COVID‐19 (RECOVERY): a randomised controlled, open‐label, platform trial. Lancet, 397, 2049–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RECOVERY Collaborative Group . (2021b). Dexamethasone in hospitalized patients with Covid‐19. New England Journal of Medicine, 384, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roback, J. D. , & Guarner, J. (2020). Convalescent plasma to treat COVID‐19: Possibilities and challenges. JAMA ‐ Journal of the American Medical Association, 323, 1561–1562. [DOI] [PubMed] [Google Scholar]
- Rojas, M. , Rodríguez, Y. , Monsalve, D. M. , Acosta‐Ampudia, Y. , Camacho, B. , Gallo, J. E. , Rojas‐Villarraga, A. , Ramírez‐Santana, C. , Díaz‐Coronado, J. C. , Manrique, R. , Mantilla, R. D. , Shoenfeld, Y. , & Anaya, J. M. (2020). Convalescent plasma in Covid‐19: Possible mechanisms of action. Autoimmunity Reviews, 19, 102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahr, F. , Ansumana, R. , Massaquoi, T. A. , Idriss, B. R. , Sesay, F. R. , Lamin, J. M. , Baker, S. , Nicol, S. , Conton, B. , Johnson, W. , Abiri, O. T. , Kargbo, O. , Kamara, P. , Goba, A. , Russell, J. B. W. , & Gevao, S. M. (2017). Evaluation of convalescent whole blood for treating Ebola Virus Disease in Freetown, Sierra Leone. Journal of Infection, 74, 302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar, E. , Christensen, P. A. , Graviss, E. A. , Nguyen, D. T. , Castillo, B. , Chen, J. , Lopez, B. V. , Eagar, T. N. , Yi, X. , Zhao, P. , Rogers, J. , Shehabeldin, A. , Joseph, D. , Masud, F. , Leveque, C. , Olsen, R. J. , Bernard, D. W. , Gollihar, J. , & Musser, J. M. (2021). Significantly Decreased Mortality in a Large Cohort of Coronavirus Disease 2019 (COVID‐19) Patients Transfused Early with Convalescent Plasma Containing High‐Titer Anti–Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2) Spike Protein IgG. American Journal of Pathology, 191, 90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk, J. M. , Omenn, G. S. , Sun, Z. , Campbell, D. S. , Baker, M. S. , Overall, C. M. , Aebersold, R. , Moritz, R. L. , & Deutsch, E. W. (2017). The human plasma proteome draft of 2017: Building on the human plasma peptideatlas from mass spectrometry and complementary assays. Journal of Proteome Research, 16, 4299–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, B. , Yi, X. , Sun, Y. , Bi, X. , Du, J. , Zhang, C. , Quan, S. , Zhang, F. , Sun, R. , Qian, L. , Ge, W. , Liu, W. , Liang, S. , Chen, H. , Zhang, Y. , Li, J. , Xu, J. , He, Z. , Chen, B. , … Guo, T. (2020). Proteomic and metabolomic characterization of COVID‐19 patient sera. Cell, 182, 59–72.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, C. , Wang, Z. , Zhao, F. , Yang, Y. , Li, J. , Yuan, J. , Wang, F. , Li, D. , Yang, M. , Xing, L. , Wei, J. , Xiao, H. , Yang, Y. , Qu, J. , Qing, L. , Chen, L. , Xu, Z. , Peng, L. , Li, Y. , … Liu, L. (2020). Treatment of 5 critically ill patients with COVID‐19 with convalescent plasma. JAMA ‐ Journal of the American Medical Association, 323, 1582–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, T. , Ning, W. , Wu, D. , Xu, J. , Han, Q. , Huang, M. , Zou, X. , Yang, Q. , Yuan, Y. , Bie, Y. , Pan, S. , Mu, J. , Han, Y. , Yang, X. , Zhou, H. , Li, R. , Ren, Y. , Chen, X. , Yao, S. , … Zhou, X. (2020). Plasma proteomics identify biomarkers and pathogenesis of COVID‐19. Immunity, 53, 1108–1122.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. , Abdesselem, H. B. , Mullins, M. , Tan, T. M. , Nel, A. J. M. , Al‐Nesf, M. A. Y. , Bensmail, I. , Majbour, N. K. , Vaikath, N. N. , Naik, A. , Ouararhni, K. , Mohamed‐Ali, V. , Al‐Maadheed, M. , Schell, D. T. , Baros‐Steyl, S. S. , Anuar, N. D. , Ismail, N. H. , Morris, P. E. , Mamat, R. N. R. , … Blackburn, J. M. (2021). Age, disease severity and ethnicity influence humoral responses in a multi‐ethnic covid‐19 cohort. Viruses, 13, 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J. W. , Lam, S. M. , Fan, X. , Cao, W. J. , Wang, S. Y. , Tian, H. , Chua, G. H. , Zhang, C. , Meng, F. P. , Xu, Z. , Fu, J. L. , Huang, L. , Xia, P. , Yang, T. , Zhang, S. , Li, B. , Jiang, T. J. , Wang, R. , Wang, Z. , … Shui, G. (2020). Omics‐driven systems interrogation of metabolic dysregulation in COVID‐19 pathogenesis. Cell Metabolism, 32, 188–202.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanne, J. H. (2020). Covid‐19: FDA approves use of convalescent plasma to treat critically ill patients. BMJ, 368, m1256. [DOI] [PubMed] [Google Scholar]
- Tegally, H. , Wilkinson, E. , Giovanetti, M. , Iranzadeh, A. , Fonseca, V. , Giandhari, J. , Doolabh, D. , Pillay, S. , San, E. J. , Msomi, N. , Mlisana, K. , von Gottberg, A. , Walaza, S. , Allam, M. , Ismail, A. , Mohale, T. , Glass, A. J. , Engelbrecht, S. , van Zyl, G. , … de Oliveira, T. (2020). Emergence and rapid spread of a new severe acute respiratory syndrome‐related coronavirus 2 (SARS‐CoV‐2) lineage with multiple spike mutations in South Africa. medRxiv, 10.1101/2020.12.21.20248640 [DOI] [Google Scholar]
- Telu, K. H. , Yan, X. , Wallace, W. E. , Stein, S. E. , & Simõn‐Manso, Y. (2016). Analysis of human plasma metabolites across different liquid chromatography/mass spectrometry platforms: Cross‐platform transferable chemical signatures. Rapid Communications in Mass Spectrometry, 30, 581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiberghien, P. , de Lamballerie, X. , Morel, P. , Gallian, P. , Lacombe, K. , & Yazdanpanah, Y. (2020). Collecting and evaluating convalescent plasma for COVID‐19 treatment: Why and how? Vox Sanguinis, 115, 488–494. [DOI] [PubMed] [Google Scholar]
- VanBlargan, L. A. , Errico, J. M. , Halfmann, P. J. , Zost, S. J. , Crowe, J. E. , Purcell, L. A. , Kawaoka, Y. , Corti, D. , Fremont, D. H. , & Diamond, M. S. (2022). An infectious SARS‐CoV‐2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nature Medicine, 28, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg, K. , Glatt, T. N. , Vermeulen, M. , Little, F. , Swanevelder, R. , Barrett, C. , Court, R. , Bremer, M. , Nyoni, C. , Swarts, A. , Mmenu, C. , Crede, T. , Kritzinger, G. , Naude, J. , Szymanski, P. , Cowley, J. , Moyo‐Gwete, T. , Moore, P. L. , Black, J. , … Wasserman, S. (2022). Convalescent plasma in the treatment of moderate to severe COVID‐19 pneumonia: A randomized controlled trial (PROTECT‐Patient Trial). Science Reports, 12, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Griensven, J. , Edwards, T. , de Lamballerie, X. , Semple, M. G. , Gallian, P. , Baize, S. , Horby, P. W. , Raoul, H. , Magassouba, N. , Antierens, A. , Lomas, C. , Faye, O. , Sall, A. A. , Fransen, K. , Buyze, J. , Ravinetto, R. , Tiberghien, P. , Claeys, Y. , De Crop, M. , … Haba, N. (2016). Evaluation of convalescent plasma for Ebola virus disease in Guinea. New England Journal of Medicine, 374, 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y. , Shang, J. , Sun, S. , Tai, W. , Chen, J. , Geng, Q. , He, L. , Chen, Y. , Wu, J. , Shi, Z. , Zhou, Y. , Du, L. , & Li, F. (2020). Molecular mechanism for antibody‐dependent enhancement of coronavirus entry. Journal of Virology, 94, 2015–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Nair, M. S. , Liu, L. , Iketani, S. , Luo, Y. , Guo, Y. , Wang, M. , Yu, J. , Zhang, B. , Kwong, P. D. , Graham, B. S. , Mascola, J. R. , Chang, J. Y. , Yin, M. T. , Sobieszczyk, M. , Kyratsous, C. A. , Shapiro, L. , Sheng, Z. , Huang, Y. , & Ho, D. D. (2021). Antibody resistance of SARS‐CoV‐2 variants B.1.351 and B.1.1.7. Nature, 593, 130–135. [DOI] [PubMed] [Google Scholar]
- Weisblum, Y. , Schmidt, F. , Zhang, F. , DaSilva, J. , Poston, D. , Lorenzi, J. C. C. , Muecksch, F. , Rutkowska, M. , Hoffmann, H. H. , Michailidis, E. , Gaebler, C. , Agudelo, M. , Cho, A. , Wang, Z. , Gazumyan, A. , Cipolla, M. , Luchsinger, L. , Hillyer, C. D. , Caskey, M. , … Bieniasz, P. D. (2020). Escape from neutralizing antibodies 1 by SARS‐CoV‐2 spike protein variants. Elife, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetton, A. D. , Preston, G. W. , Abubeker, S. , & Geifman, N. (2020). Proteomics and informatics for understanding phases and identifying biomarkers in COVID‐19 disease. Journal of Proteome Research, 19, 4219–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman, J. D. , Hiatt, J. , Mowery, C. T. , Shy, B. R. , Yu, R. , Yamamoto, T. N. , Rathore, U. , Goldgof, G. M. , Whitty, C. , Woo, J. M. , Gallman, A. E. , Miller, T. E. , Levine, A. G. , Nguyen, D. N. , Bapat, S. P. , Balcerek, J. , Bylsma, S. A. , Lyons, A. M. , Li, S. , … Marson, A. (2020). Evaluation of SARS‐CoV‐2 serology assays reveals a range of test performance. Nature Biotechnology, 38, 1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer, C. K. , Ayres, F. , Hermanus, T. , Madzivhandila, M. , Kgagudi, P. , Oosthuysen, B. , Lambson, B. E. , de Oliveira, T. , Vermeulen, M. , van der Berg, K. , Rossouw, T. , Boswell, M. , Ueckermann, V. , Meiring, S. , von Gottberg, A. , Cohen, C. , Morris, L. , Bhiman, J. N. , & Moore, P. L. (2021). SARS‐CoV‐2 501Y.V2 escapes neutralization by South African COVID‐19 donor plasma. Nature Medicine, 27, 622–625. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2021). WHO Coronavirus Disease (COVID‐19) Dashboard With Vaccination Data | WHO Coronavirus (COVID‐19) Dashboard With Vaccination Data. World Heal. Organ.
- Wu, D. , Shu, T. , Yang, X. , Song, J. X. , Zhang, M. , Yao, C. , Liu, W. , Huang, M. , Yu, Y. , Yang, Q. , Zhu, T. , Xu, J. , Mu, J. , Wang, Y. , Wang, H. , Tang, T. , Ren, Y. , Wu, Y. , Lin, S. H. , … Zhou, X. (2020). Plasma metabolomic and lipidomic alterations associated with COVID‐19. National Science Review, 7, 1157–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Wang, A. , Liu, M. , Wang, Q. , Chen, J. , Xia, S. , Ling, Y. , Zhang, Y. , Xun, J. , Lu, L. , Jiang, S. , Lu, H. , Wen, Y. , & Huang, J. (2020). Neutralizing antibody responses to SARS‐CoV‐2 in a COVID‐19 recovered patient cohort and their implications. SSRN Electronic Journal, 10.1101/2020.03.30.20047365 [DOI] [Google Scholar]
- Zaki, A. M. , van Boheemen, S. , Bestebroer, T. M. , Osterhaus, A. D. M. E. , & Fouchier, R. A. M. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine, 367, 1814–1820. [DOI] [PubMed] [Google Scholar]
- Zeki, Ö. C. , Eylem, C. C. , Reçber, T. , Kır, S. , & Nemutlu, E. (2020). Integration of GC–MS and LC–MS for untargeted metabolomics profiling. Journal of Pharmaceutical and Biomedical Analysis, 190, 113509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


