Abstract
Inositol pyrophosphates are signaling molecules containing at least one phosphoanhydride bond that regulate a wide range of cellular processes in eukaryotes. With a cyclic array of phosphate esters and diphosphate groups around myo-inositol, these molecular messengers possess the highest charge density found in nature. Recent work deciphering inositol pyrophosphate biosynthesis in Arabidopsis revealed important functions of these messengers in nutrient sensing, hormone signaling, and plant immunity. However, despite the rapid hydrolysis of these molecules in plant extracts, very little is known about the molecular identity of the phosphohydrolases that convert these messengers back to their inositol polyphosphate precursors. Here, we investigate whether Arabidopsis Plant and Fungi Atypical Dual Specificity Phosphatases (PFA-DSP1-5) catalyze inositol pyrophosphate phosphohydrolase activity. We find that recombinant proteins of all five Arabidopsis PFA-DSP homologues display phosphohydrolase activity with a high specificity for the 5-β-phosphate of inositol pyrophosphates and only minor activity against the β-phosphates of 4-InsP7 and 6-InsP7. We further show that heterologous expression of Arabidopsis PFA-DSP1-5 rescues wortmannin sensitivity and deranged inositol pyrophosphate homeostasis caused by the deficiency of the PFA-DSP-type inositol pyrophosphate phosphohydrolase Siw14 in yeast. Heterologous expression in Nicotiana benthamiana leaves provided evidence that Arabidopsis PFA-DSP1 also displays 5-β-phosphate-specific inositol pyrophosphate phosphohydrolase activity in planta. Our findings lay the biochemical basis and provide the genetic tools to uncover the roles of inositol pyrophosphates in plant physiology and plant development.
Introduction
Inositol pyrophosphates (PP-InsPs), such as InsP7 and InsP8, are molecules derived from myo-inositol (Ins) esterified with unique patterns of monophosphates (P) and diphosphates (PP) and have been described as versatile messengers in yeast, amoeba, and animal cells.1−4 With recent discoveries that PP-InsPs regulate nutrient sensing and immunity in plants, these molecules are a novel focus of research in plant physiology.5−13 The synthesis of PP-InsPs is partially conserved in eukaryotes, with some important distinctions in plants. In baker’s yeast and mammals, 5-InsP7 is synthesized by Kcs1/IP6K-type proteins, whereas Vip1/PPIP5K-type kinases phosphorylate the C1 position of both InsP6 (also termed phytic acid) and 5-InsP7, generating 1-InsP7 and 1,5-InsP8, respectively.14−17 In plants, detection, quantification, and characterization of PP-InsPs have been challenging due to the low abundance of these molecules and their susceptibility to hydrolytic activities during extraction.18,19 Employing [3H] myo-inositol labeling and subsequent analysis of plant extracts by strong-anion exchange high-performance liquid chromatography (SAX-HPLC) allowed the detection of PP-InsPs in different plant species.5,20−22 The recent development of capillary electrophoresis (CE) coupled to electrospray ionization mass spectrometry (ESI-MS), has enabled the detection and quantification of many InsP and PP-InsP isomers in various cell extracts including all InsP7 isomers, except enantiomers (labeled, e.g., as 1/3 or 4/6-InsP7).23 Similar to yeast and mammals, the Arabidopsis PPIP5K isoforms VIH1 and VIH2 catalyze the synthesis of InsP85,10 and are likely involved in the synthesis of 1/3-InsP7.11 However, Kcs1/IP6K-type proteins are absent in land plants. The question of how plants synthesize 5-InsP7 has been partially solved by work on Arabidopsis inositol (1,3,4) triphosphate 5/6 kinases ITPK1 and ITPK2. Notably, ITPK1 and ITPK2 were reported to catalyze the synthesis of 5-InsP7 from InsP6in vitro24−26 and consequently itpk1 mutant plants display reduced 5-InsP7 levels.11,13
In Arabidopsis, disturbances in the synthesis of InsP7 and/or InsP8 result in defective signaling of the plant hormones jasmonate5,6 and auxin,13 as well as defects in salicylic acid-dependent plant immunity12 and impaired phosphate (Pi) homeostasis.9−11,27 In the case of auxin and jasmonate perception, 5-InsP7 and InsP8, respectively, are proposed to function as co-ligands of the respective receptor complexes.5,6,13 The role of PP-InsPs in Pi signaling is related to their ability to bind to SPX proteins, which act as receptors for these messenger molecules.7,9,10,28−30 InsP8 has been found as the preferred ligand for stand-alone SPX proteins in vivo.9,10,28 InsP8-bound SPX receptors inactivate the MYB-type transcription factors PHR1 and PHL1, which control the expression of a majority of Pi starvation-induced (PSI) genes to regulate various metabolic and developmental adaptations induced by Pi deficiency.28,31−33 The tissue levels of various PP-InsPs, including 5-InsP7 and InsP8, respond sensitively to the plant’s Pi status,9,11 suggesting that their synthesis and degradation are tightly regulated. While the steps involved in the synthesis of PP-InsPs in plants are now better understood, still little is known about how these molecules are degraded.
Vip1/PPIP5Ks are bifunctional enzymes that harbor an N-terminal ATP-grasp kinase domain and a C-terminal phosphatase domain conserved in yeast, animals, and plants.5,10,15,17,34In vitro, the phosphatase domain of Arabidopsis PPIP5K VIH2 hydrolyzes PP-InsPs to InsP6,10 similar to the respective C-terminal domains of fission yeast and mammalian PPIP5Ks.35,36 Although Arabidopsis ITPK1 harbors no phosphatase domain, under conditions of low adenylate charge, it can shift its activity in vitro from kinase to ADP phosphotransferase activity using 5-InsP7 but no other InsP7 isomer.11,26 Apart from relying on the reversible activities of ITPK1 and Vip1/PPIP5Ks, the degradation of PP-InsPs may also be controlled by specialized phosphohydrolases.
In mammalian cells, diphosphoinositol polyphosphate phosphohydrolases (DIPPs), members of the nudix hydrolase family, have been shown to catalyze the hydrolysis of the diphosphate groups of InsP7 and InsP8 at the C1 and C5 position.4,37,38 The baker’s yeast genome encodes a single homologue of mammalian DIPP1, named diadenosine and diphosphoinositol polyphosphate phosphohydrolase (DDP1), which hydrolyzes various substrates including diadenosine polyphosphates, 5-InsP7 and InsP8, but has a preference for inorganic polyphosphates (poly-P) and for the β-phosphate of 1-InsP7.39−41 In addition, baker’s yeast has an unrelated PP-InsP phosphohydrolase, Siw14 (also named Oca3) with a high specificity for the β-phosphate at position C5 of 5-InsP7.42,43 This enzyme is a member of the Plant and Fungi Atypical Dual Specificity Phosphatases (PFA-DSPs) that belong to a large family of protein tyrosine phosphatases (PTPs).43−45
Blast search analyses revealed that the Arabidopsis thaliana genome encodes five PFA-DSPs, with AtPFA-DSP1 sharing 61% identity and 76% similarity with yeast Siw14.44,45 X-ray crystallography revealed that the protein adopts an α/β-fold typical for cysteine phosphatases, with the predicted catalytic cysteine (Cys150) residing at the bottom of a positively charged pocket.44,46 Of a number of putative phosphatase substrates tested, recombinant AtPFA-DSP1 displayed the highest activity against inorganic polyphosphate, as well as against deoxyribo- and ribonucleoside triphosphates, and less activity against phosphotyrosine-containing peptides and phosphoinositides.46 Here, we investigated whether Arabidopsis PFA-DSPs might function as PP-InsP phosphohydrolases.
Methods
Plant Materials and Growth Conditions
Seeds of A. thaliana T-DNA insertion lines pfa-dsp1-3 (WiscDsLox_473_B10, Col-0), pfa-dsp1-4 (CSHL_GT1415, Ler-0), pfa-dsp1-6 (SAIL_116_C12, Col-0) and mrp5 (GK-068B10) were obtained from The European Arabidopsis Stock Centre (http://arabidopsis.info/). To identify homozygous lines, F2 and F3 plants were genotyped by PCR using the primers indicated in Table S2.
For sterile cultures, Arabidopsis seeds were surface sterilized in 1.2% (v/v) NaHClO4 and 0.05% (v/v) Triton X-100 for 3 min, in 70% (v/v) ethanol and 0.05% (v/v) Triton X-100 for 3 min and in 100% (v/v) ethanol before transferring onto sterile filter paper. Sterilized seeds were sown onto half-strength Murashige and Skoog (MS) medium59 containing 1% sucrose, pH 5.7 and solidified with 0.7% (w/v) Phytagel (Sigma-Aldrich). After 2 days of stratification at 4 °C, the plates were transferred to a growth incubator and the seedlings were grown under short-day conditions with the following regime: 8/16 h light/dark; light intensity 120 μmol m–2 s–1; temperature 22 °C/20 °C.
Constructs
The following full-length ORFs were amplified by PCR from an Arabidopsis whole seedling cDNA preparation: PFA-DSP1 (At1g05000), PFA-DSP2 (At2g32960), PFA-DSP3 (At3g02800) PFA-DSP4 (At4g03960), and PFA-DSP5 (At5g16480). Likewise, the SIW14 ORF sequence was amplified from yeast genomic DNA. Primers used for amplification are listed in Table S2. The reverse primers contained a V5 sequence (underlined) allowing a translational fusion of the resulting gene products with a C-terminal V5 epitope tag. Amplification products were cloned into pDONR221 (Invitrogen) via BP clonase II (Invitrogen) reaction following the manufacturer’s instructions. The ORFs were then swapped into the episomal yeast expression vector pDRf1-GW60 by the LR clonase II (Invitrogen) reaction following the manufacturer’s instructions. For expression of SIW14 under control of the endogenous promoter from a CEN-based plasmid, the SIW14 gDNA was amplified from purified yeast gDNA using the primers listed in Table S2. The SIW14 gDNA was inserted into YCplac33 (ATCC #87586) using the restriction enzymes PstI and EcoRI.
For protein expression, PFA-DSP1–5 were amplified as described before but with a reverse primer containing a stop codon. Amplified products were cloned into pDONR221 (Invitrogen), then swapped by LR clonase II (Invitrogen) into the bacterial expression vector pDEST566 (Addgene plasmid # 11517), which contains a sequence encoding an N-terminal His6-maltose-binding protein (MBP) epitope tag. Free His-tagged MBP protein was expressed from a modified pET28 vector carrying an N-terminal sequence encoding a His8-maltose-binding protein (MBP) epitope tag.5
For transient expression in Nicotiana benthamiana, the ORF of PFA-DSP1 (wild-type sequence and with a mutated sequence encoding the C150S substitution) was swapped by LR clonase II (Invitrogen) from pDONR221 into the plant expression vector pGWB641,61 which harbors a viral CaMV 35S promoter to allow gene expression and a sequence encoding a C-terminal EYFP tag. Site-directed mutagenesis was performed on the respective plasmids with the primers listed in Table S2.
N. benthamiana Infiltration
A single colony of transformed Agrobacteria was inoculated in 2 mL of LB media containing the appropriate antibiotics and cultivated overnight at 26 °C in a spinning wheel. On the next morning, 1 mL of overnight culture was added to 5 mL of fresh LB with antibiotics and grown for another 4 h at 26 °C. Afterward, the cultures were harvested by centrifugation at 4 °C with 3000g for 20 min. The pellet was then resuspended in 3 mL of infiltration solution containing 10 mM MgCl2, 10 mM MES-KOH (pH 5.6), and 150 μM acetosyringone. OD600 was determined using a 1:10 dilution and adjusted to 0.8 in infiltration solution. Then, the working solution was prepared by pooling equal amounts of cultures (e.g., P19 + PFA-DSP1), which were then co-infiltrated in the abaxial surface of the leaf using a 1 mL syringe without a needle. Afterward, the plants were placed in a dark incubator at 26 °C for ∼1 day before keeping them for another 4 days on the workbench. The leaves were then harvested and frozen in liquid nitrogen before continuing with the extraction of inositol phosphates.
Yeast Strains
Different strains of the budding yeast Saccharomyces cerevisiae were used. The BY4741 wild-type (MATa his3Δ leu2Δ met15Δ ura3Δ), siw14Δ (YNL032w::kanMX4), vip1Δ (YLR410w::kanMX4),5kcs1Δ (YDR017c::kanMX4), and ipk2Δ (YDR173c::kanMX4) were obtained from Euroscarf. vip1Δ siw14Δ, kcs1Δ siw14Δ, ipk2Δ siw14Δ were generated using loxP/Cre gene disruption and the ble resistance marker, which confers phleomycine/Zeocin (Invitrogen) resistance62 using the primers listed in Table S2. In addition, the following mutants in the DDY1810 background (MATa; leu2Δ ura3-52 trp1Δ; prb1-1122 pep4-3 pre1-451)63 were used: kcs1Δ and kcs1Δ ddp1Δ. kcs1Δ siw14Δ, kcs1Δ ddp1Δ siw14Δ, and siw14Δ were generated in this background as described before. For all assays, the yeast cells were transformed by the Li-acetate method64 and cultured in either 2 × YPD + CSM medium or selective synthetic deficiency (SD) medium.
Yeast Growth Complementation Assay
Yeast transformants were inoculated in selective synthetic deficiency (SD) medium and grown overnight at 28 °C while shaking (200 rpm). Then, OD600 was measured, adjusted to 1.0, and an 8-fold dilution series was prepared in a 96-well plate. Subsequently, 10 μL of each dilution were spotted on selective solid media as described earlier65 and incubated at 26 °C for 2–4 days. To prepare selective solid media supplemented with wortmannin, autoclaved media was cooled down to 60 °C, wortmannin was added from a 10 mM stock in DMSO (Sigma-Aldrich) to a final concentration of 1–3 μM. Since the activity of wortmannin changed by age and by the number of freezing/thawing cycles, aliquots were kept at −20 °C and were not thawed more than five times. In addition, several concentrations were employed for the spotting assays to be able to identify the activity at which growth differences between siw14Δ, kcs1Δ, and their isogenic wild-type transformants became most obvious. Pictures were taken with a Bio-Rad ChemiDoc MP imager using white backlight.
Protein Preparation
His6-MBP-PFA-DSP protein fusions or free His8-MBP were expressed in Escherichia coli BL21 CodonPlus (DE3)-RIL cells (Stratagene). Overnight bacterial cultures were inoculated 1:1000 into fresh 2YT medium (1.6% tryptone, 1% yeast extract, 0.5% NaCl) with 100 mg/L ampicillin (pDEST566) or 50 mg/L kanamycin (pET28) and 25 mg/L chloramphenicol. Cells were grown at 37 °C while shaking (200 rpm) for 4 h (∼0.6 OD600), and protein expression was induced at 16 °C overnight with 0.1 mM isopropyl-d-1-thiogalactopyranoside. The cells were lysed as described66 using the following lysis buffer: 50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 25 mM imidazole, 10% (v/v) glycerol, 0.1% (v/v) Tween 20, 5 mM β-mercaptoethanol, and EDTA-free complete ULTRA protease inhibitor cocktail (Roche). Proteins were batch-purified using Ni-NTA agarose resin (Macherey-Nagel) and eluted using the above-mentioned lysis buffer with increased imidazole concentration (250 mM). Three elutions were combined and dialyzed using Slide-A-Lyzer Dialysis Cassettes (Thermo Scientific) following the manufacturer’s instructions and a buffer containing 50 mM Tris-Cl, pH 7.5 and 100 mM NaCl. The concentrated protein preparations were then stored at −20 °C. Purified proteins were analyzed using SDS-PAGE followed by Coomassie blue staining. Proteins were compared with PageRuler plus prestained protein ladder (Thermo Fisher) and with designated amounts of a BSA standard to estimate target protein concentrations.
In Vitro PP-InsP Phosphohydrolase Assay
The phosphohydrolase assay was carried out in a 15 μL reaction mixture containing 0.35–2 μM recombinant PFA-DSP or Siw14 protein, 50 mM HEPES (pH 7.0), 10 mM NaCl, 5% (v/v) glycerol, 0.1% (v/v) β-mercaptoethanol, and 0.33 mM of various InsP7 and InsP8 isomers as indicated, and was incubated for 1, 2, or 24 h at 22 °C. The PP-InsP isomers were synthesized as described previously.67,68 Reactions were separated by 33% PAGE and visualized by toluidine blue or DAPI staining.
Titanium Dioxide Bead Extraction and PAGE/CE-ESI-MS
Purification of inositol polyphosphates using TiO2 beads and analysis via PAGE was performed as described previously.11 CE-ESI-MS analyses of in vitro, yeast and plant samples were performed as described previously.11,23
Inositol Polyphosphate Extraction from Yeast Cells and Seedlings and HPLC Analyses
For inositol polyphosphate analyses from yeast, transformants were inoculated into a selective synthetic deficiency (SD) medium and grown overnight at 28 °C while shaking (200 rpm). They were then diluted 1:200 in 2 mL of fresh medium supplemented with 6 μCi mL–1 [3H]-myo-inositol (30–80 Ci mmol–1; Biotrend; ART-0261-5) and grown overnight at 28 °C in a spinning wheel. After centrifugation and washing of the cell pellet, inositol polyphosphates were extracted and analyzed as described before.5,69,70
Extraction of [3H]-myo-inositol polyphosphates from Arabidopsis seedlings and subsequent SAX-HPLC analyses were performed as described previously.70
RNA Isolation and Quantitative Real-Time PCR
Fifteen-day-old seedlings were transferred from solid half-strength MS plates to liquid half-strength MS media (supplemented with 1% sucrose) for 5 days before harvest and immediately frozen in liquid N2. Total RNA was extracted with NuceloSpin RNA Plant and Fungi kit (Macherey-Nagel). cDNA was synthesized using RevertAid RT reverse transcription kit (Thermo Fisher). Quantitative PCR reactions were conducted with the CFX384 real-time system (Bio-Rad) and the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) using the primers listed in Table S2. TIP41-like and PP2AA3 were used as reference genes to normalize relative expression levels of all tested genes. Relative expression was calculated using the CFX Maestro software (Bio-Rad).
Yeast Protein Extraction and Immunodetection
Multiple transformants were inoculated into 4 mL of YPD (with 3% glucose) or selective SD-media and grown for up to 24 h at 28 °C. On the following day, the yeast was reinoculated into 4 mL of fresh media and grown for another day. Afterward, the cells were harvested and resuspended in 500 μL of extraction buffer (300 mM sorbitol, 150 mM NaCl, 50 mM Na2HPO4, 1 mM EDTA, pH 7.5), supplemented with 100 mM β-mercaptoethanol and a 1:50 dilution of protease inhibitor cocktail for fungal extracts (Sigma-Aldrich). The cells were lysed with bead beating using 150–200 μL of glass beads (ø 0.5 mm). The lysate was spun down and the supernatant boiled for 10 min after the addition of sample buffer. The protein extracts were then resolved by SDS-PAGE. Target proteins were detected by immunoblot. As the primary antibody, a mouse anti-V5 (Invitrogen, R960-25, 1:2000 dilution) antibody was used, followed by either an Alexa fluor plus 800 goat anti-mouse antibody (Invitrogen, 1:20 000 dilution) or a goat anti-mouse HRP antibody (Bio-Rad, 1:10 000 dilution). As a loading control, Gal4 was detected using a rabbit polyclonal anti-Gal4 antibody (Santa Cruz, 1:1000 dilution), followed by a goat anti-rabbit StarBright Blue 700 antibody (Bio-Rad, 1:2500 dilution). The signal was detected using the multi-plex function of the ChemiDoc MP imager (Bio-Rad). Alternatively, for blots where a secondary antibody coupled to HRP was used, the chemiluminescence signal of the ECL reagent was detected, followed by Ponceau staining as loading control.
Results and Discussion
Arabidopsis PFA-DSP Proteins Display In Vitro PP-InsP Phosphohydrolase Activity with High Specificity for 5-InsP7
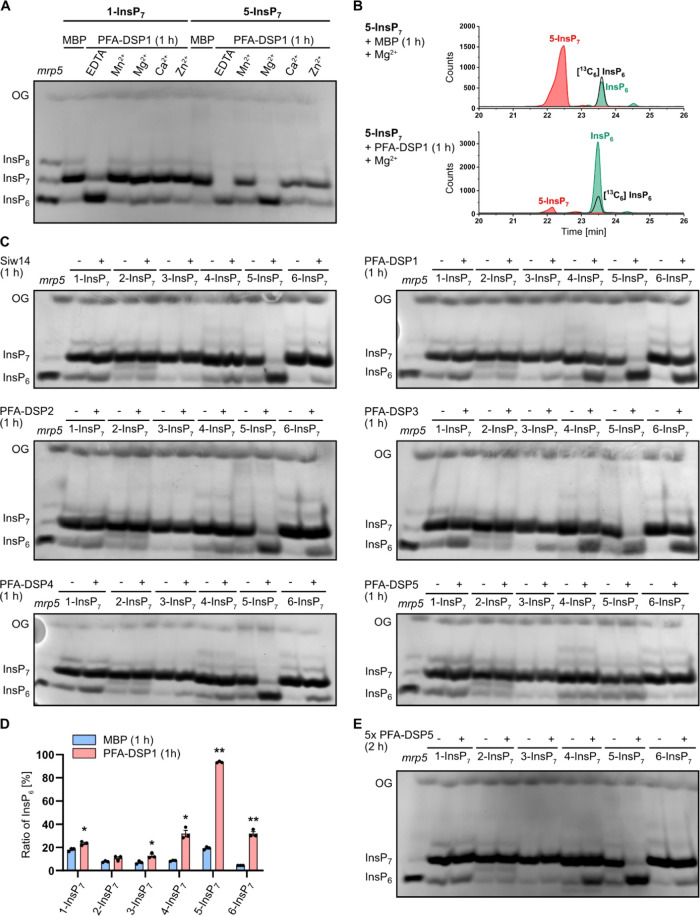
To explore the potential role of Arabidopsis PFA-DSP proteins in PP-InsP hydrolysis, we first generated translational fusions of PFA-DSPs with an N-terminal hexahistidine tag followed by a maltose-binding protein (MBP) and expressed recombinant proteins in bacteria. Corresponding His-MBP-Siw14 and free His-MBP constructs were generated as controls. All constructs allowed the purification of soluble recombinant proteins (Figure S1). We then tested potential PP-InsP phosphohydrolase activities of PFA-DSP1 with 1-InsP7 or 5-InsP7 in the presence of various divalent cations. Notably, PFA-DSP1 failed to catalyze the hydrolysis of 1-InsP7 or 5-InsP7 in the presence of Mn2+, Ca2+, or Zn2+. However, in the presence of the cytoplasmic prevalent cation Mg2+, PFA-DSP1 displayed a robust hydrolytic activity against 5-InsP7, likely resulting in the generation of InsP6, as deduced from the mobility of the reaction product compared to TiO2-purified mrp5 (multidrug resistance-associated protein 5) seed extract separated by polyacrylamide gel electrophoresis (PAGE) and visualized by toluidine blue staining (Figure 1A). Seeds of Arabidopsis mrp5 mutants that have a defective ABC-transporter involved in vacuolar loading of InsP647 display reduced InsP6 levels and simultaneously increased InsP7 and InsP8 levels.20,24 Therefore, TiO2-purified mrp5 seed extract serves as a marker to visualize InsP6, InsP7, and InsP8 on PAGE. CE-ESI-MS analysis of the reaction product spiked with a [13C6] InsP6 standard confirmed that the resulting product indeed had the migration behavior and the mass of phytic acid (Figure 1B). In contrast, 1-InsP7 was largely resistant to PFA-DSP1 also in the presence of Mg2+ (Figure 1A). In the absence of divalent cations (i.e., in buffer not supplemented with divalent cations but instead supplemented with EDTA, a condition unlikely to represent any cellular condition), both InsP7 isomers were hydrolyzed to InsP6, as deduced from the mobility of the reaction product by PAGE (Figure 1A).
Figure 1.
In vitro, Arabidopsis PFA-DSPs display Mg2+-dependent PP-InsP phosphohydrolase activity with high specificity for 5-InsP7. Recombinant His-MBP-PFA-DSPs and His-MBP-Siw14 (indicated with the plus symbol in (C) and (E)) were incubated with 0.33 mM InsP7 at 22 °C. His-MBP served as a negative control (as indicated with the minus symbol in (C) and (E)). (A) 0.4 μM His-MBP-PFA-DSP1 was incubated for 1 h with 1-InsP7 or 5-InsP7, and 1 mM EDTA, MnCl2, MgCl2, CaCl2, or ZnCl2 as indicated. The reaction products were then separated by 33% PAGE and visualized by toluidine blue. (B–D) The InsP7 phosphohydrolase activity of ∼0.4 μM His-MBP-PFA-DSPs and His-MBP-Siw14 was analyzed in the presence of 1 mM MgCl2. After 1 h, the reaction products were then (B, D) spiked with isotopic standards mixture ([13C6] 1,5-InsP8, [13C6] 5-InsP7, [13C6] 1-InsP7, [13C6] InsP6, [13C6] 2-OH InsP5) and subjected to CE-ESI-MS analyses or (C) separated by 33% PAGE and visualized by toluidine blue/DAPI staining. (D) Data represent mean ± SEM (n = 3). Representative extracted-ion electropherograms are shown in Figure S2. Asterisks indicate values that are significantly different from the MBP control reactions (according to Student’s t test, P < 0.05 (*); P < 0.01 (**)). (E) Recombinant His-MBP-PFA-DSP5 (2 μM) was incubated with 0.33 mM InsP7 isomers for 2 h. The reaction product was separated by 33% PAGE and visualized with toluidine blue. (A, C, E) Identity of bands was determined by migration compared to TiO2-purified mrp5 seed extract.
We then tested the hydrolytic activities of the Arabidopsis PFA-DSP homologues with all six “simple,” synthetic InsP7 isomers and with the two enantiomeric InsP8 isomers 1,5-InsP8 and 3,5-InsP8 in the presence of Mg2+. Of note, myo-inositol is a meso compound with a mirror plane dissecting the C2 and C5 positions. Derivatives differentially (pyro)phosphorylated at the C1 and C3 positions, as well as at the C4 and C6 positions are enantiomeric forms that can only be distinguished in the presence of appropriate chiral selectors.48,49 Yeast Siw14 and all Arabidopsis PFA-DSPs with the exception of PFA-DSP5, displayed robust activity with a high specificity toward 5-InsP7 (Figure 1C), confirming earlier reports that 5-InsP7 is a preferred substrate for yeast Siw14 compared to 1-InsP7.42,43 PFA-DSP1–4 and Siw14 also displayed partial hydrolytic activities against the enantiomers 4-InsP7 and 6-InsP7, as well as very weak hydrolytic activities against enantiomeric 1-InsP7 and 3-InsP7 (Figure 1C). The latter activities were more pronounced in PFA-DSP1 and PFA-DSP3 compared to Siw14 and PFA-DSP2. As for 5-InsP7, the reaction products with the other InsP7 isomers had the mass and the migration behavior of the InsP6 isomer phytic acid, as deduced from CE-ESI-MS analyses (Figures 1D and S2). Notably, PFA-DSP5 only showed very weak activities at the 0.4 μM concentration tested in our assay. However, when the reaction time was extended from 1 h to 2 h and the enzyme concentration was increased to 2 μM, PFA-DSP5 displayed robust activity with a substrate specificity similar to PFA-DSP1-4 and yeast Siw14, with a high selectivity for 5-InsP7 and only weak hydrolytic activities against 4-InsP7 and 6-InsP7 (Figure 1E).
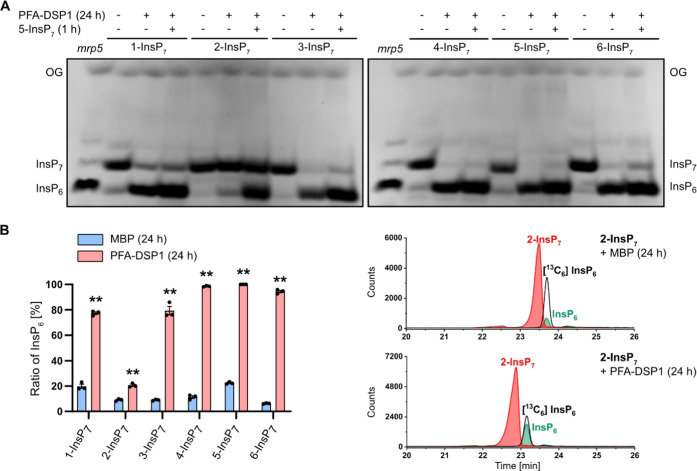
Notably, the meso InsP7 isomer 2-InsP7 was completely resistant to Siw14 or any of the Arabidopsis PFA-DSP proteins under the assay conditions. This was also the case in the absence of divalent cations (i.e., in buffer not supplemented with divalent cations but instead supplemented with EDTA), where Siw14 and Arabidopsis PFA-DSP1-4 failed to hydrolyze 2-InsP7 to a significant extent while all other InsP7 isomers were at least partially converted to an InsP isomer with the mobility of phytic acid (Figures S3 and S4). Even after a 24 h-long incubation with Arabidopsis PFA-DSP1, 2-InsP7 remained largely resistant to hydrolysis. In contrast, all other PP-InsP7 isomers were hydrolyzed to InsP6 under these conditions, as revealed by PAGE and CE-ESI-MS analyses (Figures 2A,B and S5). Corresponding control reactions that were supplemented with 5-InsP7 after 23 h validated the activity of Arabidopsis PFA-DSP1 after such long incubation times (Figures 2A and S6). These spiking experiments also rule out the possibility that 2-InsP7 contained a contaminant that inhibits PFA-DSP-dependent hydrolysis, as 5-InsP7 was still efficiently hydrolyzed in the presence of 2-InsP7.
Figure 2.
Under prolonged incubation time Arabidopsis PFA-DSP1 hydrolyzes various InsP7 isomers in vitro, except for 2-InsP7. Recombinant His-MBP-PFA-DSP1 (0.4 μM) (indicated with the plus symbol in the first line of (A)) was incubated with 0.33 mM InsP7 and 1 mM MgCl2 for 24 h at 22 °C. To ensure that PFA-DSP1 is active during the whole incubation time, after 23 h, 0.33 mM 5-InsP7 was added to a replicate and incubated for another 1 h (indicated with the plus symbol in the second line of (A)). His-MBP served as a negative control (as indicated with the minus symbol in (A)). (A) An aliquot of the reaction product was separated by 33% PAGE and visualized with toluidine blue. The identity of bands was determined by migration compared to TiO2-purified mrp5 seed extract. (B) Another reaction was spiked with an isotopic standards mixture ([13C6] 1,5-InsP8, [13C6] 5-InsP7, [13C6] 1-InsP7, [13C6] InsP6, [13C6] 2-OH InsP5) and subjected to CE-ESI-MS analyses. Data represent mean ± SEM (n = 3). Asterisks indicate values that are significantly different from the MBP control reactions (according to Student’s t test, P < 0.05 (*); P < 0.01 (**)). Representative extracted-ion electropherograms are shown in Figure S5.
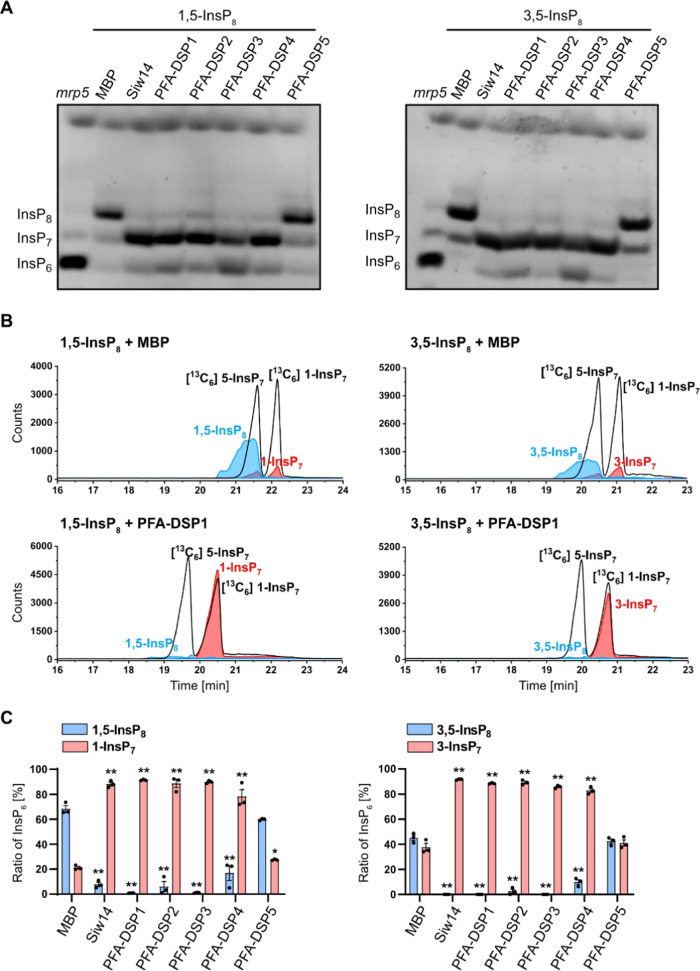
Finally, we tested whether the enantiomeric InsP8 isomers 1,5-InsP8 and 3,5-InsP8 serve as substrates for PFA-DSPs. As reported earlier for Siw14,43 PFA-DSP1–4 hydrolyzed 1,5-InsP8 to an InsP7 isomer based on the mobility of the reaction product in PAGE analyses (Figure 3A). Also the enantiomeric 3,5-InsP8 was efficiently hydrolyzed by Siw14 and PFA-DSP1–4 (Figure 3A), and CE-ESI-MS analysis of the reaction products showed the migration behavior and the mass of 1/3-InsP7 (Figures 3B,C and S7).
Figure 3.
Arabidopsis PFA-DSPs display robust 1/3,5-InsP8 phosphohydrolase activity in vitro. (A) Approximately 0.4 μM His-MBP-PFA-DSPs and His-MBP-Siw14 were incubated with 0.33 mM 1,5-InsP8 or 3,5-InsP8 for 1 h, in the presence of 1 mM MgCl2, and analyzed by PAGE and subsequent toluidine blue staining. The identity of bands was determined by migration compared to TiO2-purified mrp5 seed extract. (B, C) Second and third reactions were spiked with isotopic standards mixture ([13C6] 1,5-InsP8, [13C6] 5-InsP7, [13C6] 1-InsP7, [13C6] InsP6, [13C6] 2-OH InsP5) and subjected to CE-ESI-MS analyses. (C) Data represent mean ± SEM (n = 3). Asterisks indicate values that are significantly different from the MBP control reactions (according to Student’s t test, P < 0.05 (*); P < 0.01 (**)). Representative extracted-ion electropherograms are shown in Figure S7.
Altogether, these findings reveal that Arabidopsis PFA-DSP proteins and yeast Siw14 have a high specificity for the 5-β-phosphate of 5-InsP7, 1,5-InsP8, and 3,5-InsP8, and a weak activity against the β-phosphates of 4-InsP7 and 6-InsP7, respectively. In contrast, InsP6 and 2-InsP7 are resistant to PFA-DSP-catalyzed hydrolysis (summarized in Table S1).
Heterologous Expression of Arabidopsis PFA-DSP Homologues Complement Yeast siw14Δ Defects
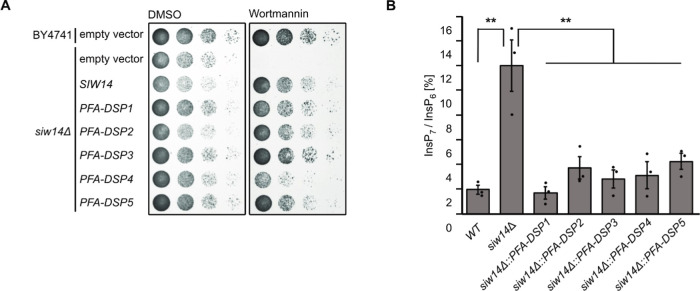
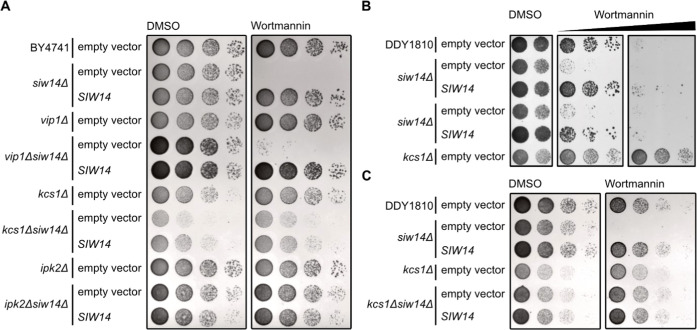
To investigate the physiological consequences of Arabidopsis PFA-DSP activities in vivo, we carried out heterologous expression experiments in baker’s yeast. To this end, we investigated the sensitivity of our siw14Δ yeast strain under conditions where phenotypes for siw14Δ strains and other yeast mutants defective in PP-InsP homeostasis have been reported previously5,24,50−52 and selected the fungal toxin wortmannin53 that caused a severe siw14Δ-associated growth defect. Previous observations that kcs1Δ yeast cells are resistant to wortmannin54 suggest that wortmannin sensitivity of siw14Δ yeast might be related to Kcs1-dependent PP-InsPs. The siw14Δ-associated growth defect was fully complemented by heterologous expression of either of the five Arabidopsis PFA-DSP homologues or of yeast SIW14 from episomal plasmids under control of a PMA1 promoter fragment (Figure 4A). Immunoblot analyses taking advantage of a C-terminal V5-tag revealed that all PFA-DSP homologues were expressed in yeast with PFA-DSP1 and PFA-DSP4 showing the highest protein abundance (Figure S8). Reduced growth of siw14Δ transformants expressing PFA-DSP4 on media supplemented with wortmannin is therefore likely not caused by inefficient expression of this homologue in yeast but might rather be a consequence of excess protein activity in this heterologous expression system. To investigate the contribution of PFA-DSPs in InsP metabolism, we monitored InsP profiles using SAX-HPLC analyses of various [3H]-myo-inositol labeled yeast transformants. Of note, conventional SAX-HPLC analyses as employed here do not allow the discrimination of different InsP7 or InsP8 isomers.11,49 Heterologous expression of PFA-DSPs in siw14Δ restored InsP7/InsP6 ratios to wild-type levels, indicating that Arabidopsis PFA-DSP proteins are functionally similar to Siw14 (Figure 4B).
Figure 4.
Heterologous expression of Arabidopsis PFA-DSPs complements siw14Δ-associated wortmannin sensitivity in yeast. (A) Growth complementation assays of an siw14Δ yeast strain. Wild-type yeast (BY4741) and an isogenic siw14Δ yeast mutant were transformed with either the empty episomal pDRf1-GW plasmid or different pDRf1-GW plasmids carrying the respective PFA-DSP gene or SIW14. Yeast transformants were then spotted in 8-fold serial dilutions (starting from OD600 1.0) onto selective media supplemented with either wortmannin or DMSO as control. Plates were incubated at 26 °C for 2 days before photographing. The yeast growth assay was repeated twice (n = 3) with similar results. (B) Relative amounts of InsP7 of wild-type yeast, siw14Δ and siw14Δ transformed with pDRf1-GW carrying the PFA-DSP genes are shown as InsP7/InsP6 ratios. InsP6 and InsP7 levels were determined by analysis of SAX-HPLC profiles using OriginPro 8. Data represent mean ± SEM (n = 3). Asterisks indicate values that are significantly different from siw14Δ (according to Student’s t test, P < 0.05 (*); P < 0.01 (**)).
Notably, the InsP7 signal was the only one consistently affected by the loss of SIW14 and heterologous expression of any PFA-DSP gene (Figure S9). We generated variants of Siw14 or PFA-DSP1, in which the catalytic cysteine was replaced by a serine resulting in a C214S and a C150S substitution in Siw14 and PFA-DSP1, respectively, and observed that complementation of siw14Δ-associated growth defects of respective transformants requires the catalytic activity of these proteins (Figure S10A,B). The inability of catalytic dead mutants to complement siw14Δ-associated growth defects was not caused by compromised expression or protein stability of these variants, as confirmed by immunoblot analyses (Figure S10C). In agreement with the growth complementation assays, the catalytically inactive versions of Siw14 and PFA-DSP1 also failed to restore wild-type InsP7 levels in siw14Δ transformants (Figure S10D,E). These experiments suggest that Arabidopsis PFA-DSPs can substitute for endogenous Siw14 in yeast with respect to wortmannin tolerance and InsP7 homeostasis and that complementation of the siw14Δ-associated defects depends on the catalytic activity of these proteins.
Growth Defects of siw14Δ Yeast on Wortmannin Require Kcs1-Dependent 5-InsP7 Synthesis
For a deeper understanding of the wortmannin phenotype of siw14Δ yeast, we investigated genetic interactions between Siw14 and different InsP kinases. We generated different double mutants with defects in Siw14 and the PP-InsP synthases Kcs1 and Vip1, and tested their performance on wortmannin-containing media (Figure 5A). Again, siw14Δ cells did not survive on media supplemented with 3 μM wortmannin, a defect that was fully complemented by the expression of SIW14 under control of the endogenous promoter from a CEN-based single-copy plasmid (Figure 5A). The growth of vip1Δ cells was comparable to wild-type yeast. In contrast, the vip1Δ siw14Δ double mutant showed a severe growth defect on media supplemented with wortmannin similar to single siw14Δ cells (Figure 5A). Like the vip1Δ yeast strain, a kcs1Δ strain did not show growth defects on media supplemented with wortmannin compared to control media. In contrast, at increased concentrations, we observed kcs1Δ-associated wortmannin resistance (Figure 5B), as reported earlier.54 Importantly, deletion of KCS1 in siw14Δ cells rescued siw14Δ-associated wortmannin sensitivity since the resulting kcs1Δ siw14Δ double-mutant yeast strain, despite growing overall weaker than the kcs1Δ single-mutant strain, showed no increased sensitivity to wortmannin (Figure 5A). These findings indicate that the presence of Kcs1 is critical for the growth defects displayed by siw14Δ single-mutant cells on wortmannin. We then investigated whether the presence of Kcs1 itself or of Kcs1-dependent PP-InsPs such as 5-InsP7 are relevant for siw14Δ-associated wortmannin sensitivity. To this end, we examined the phenotypes of ipk2Δ and of ipk2Δ siw14Δ yeast transformants. Both mutants lack IPK2, an inositol polyphosphate multikinase that sequentially phosphorylates Ins(1,4,5)P3 to Ins(1,3,4,5,6)P5 and is hence required for InsP6 and subsequent Kcs1-dependent 5-InsP7 or PP-InsP4 synthesis.54,55 Neither of the strains showed growth defects on media supplemented with wortmannin compared to the isogenic wild-type yeast strain, suggesting that also the loss of IPK2 rescues siw14Δ-associated wortmannin sensitivity (Figure 5A). We further tested wortmannin sensitivity of kcs1Δ and kcs1Δ siw14Δ yeast transformants in a different genetic background and observed similar results (Figure 5B,C). Taken together, these results provide a causal link between Kcs1 (but not Vip1)-dependent PP-InsPs and siw14Δ-associated wortmannin sensitivity with Kcs1 and Siw14/PFA-DSPs playing antagonistic roles in regulating this sensitivity.
Figure 5.
Yeast siw14Δ-associated wortmannin sensitivity requires Kcs1-dependent 5-InsP7. (A) Wild-type yeast (BY4741), siw14Δ, vip1Δ, kcs1Δ, ipk2Δ, vip1Δ siw14Δ, kcs1Δ siw14Δ, and ipk2Δ siw14Δ double-mutant yeast strains were transformed with either an empty (CEN-based) YCplac33 vector or a YCplac33 vector carrying a genomic fragment of SIW14 including a 653 bp promoter and 5′UTR and a 289 bp terminator region. Yeast transformants were then spotted in 8-fold serial dilutions (starting from OD600 1.0) onto selective media supplemented with either wortmannin or DMSO as control. Plates were incubated at 26 °C for 2 days before photographing. (B, C) Wild-type yeast (DDY1810), siw14Δ, kcs1Δ, and (C) kcs1Δ siw14Δ double-mutant yeast strains were transformed with either an empty YCplac33 vector or a YCplac33 vector carrying the genomic fragment of SIW14. The growth assay was performed as described for (A). All yeast growth assays (A–C) were repeated twice (n = 3) with similar results.
Increased PFA-DSP1 Expression Coincides with Decreased InsP7 Levels In Planta
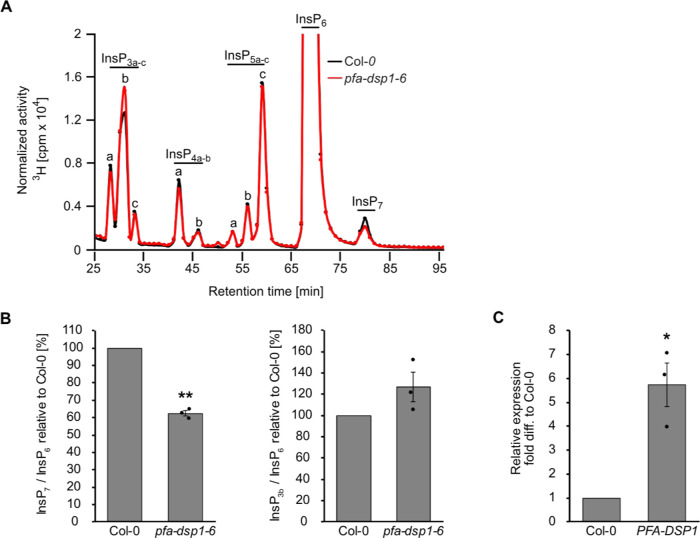
To gain insight into PFA-DSP functions in planta, we searched for Arabidopsis T-DNA insertion lines of PFA-DSP1 and were able to identify three lines, pfa-dsp1-3 and pfa-dsp1-6 in the Col-0 background and pfa-dsp1-4 in the Ler-0 background, for which homozygous progeny could be obtained. None of these lines displayed an obvious growth phenotype under our standard growth conditions. SAX-HPLC profiles of extracts of 20-day-old [3H]-myo-inositol labeled pfa-dsp1-3 and pfa-dsp1-4 seedlings did not reveal a significant difference compared to the respective wild-types (Figure S11A,B). However, SAX-HPLC analyses of the pfa-dsp1-6 line revealed a significant average reduction (around 36%) of the InsP7/InsP6 ratio compared to Col-0 (Figure 6B). The levels of other InsP species remained largely unaffected (Figure 6A,B). The available sequencing data for this line, as well as our analysis, indicated that the insertion of the T-DNA is 18 bp upstream of the start codon, suggesting that the full-length transcript and PFA-DSP1 protein might be expressed in this line. We therefore conducted qPCR analyses of pfa-dsp1-6 seedlings that were grown under identical conditions as the seedlings for SAX-HPLC analyses and detected ca. 6-fold increased expression of PFA-DSP1 in pfa-dsp1-6 in comparison to Col-0 seedlings (Figure 6C).
Figure 6.
Increased expression of PFA-DSP1 in Arabidopsis decreases InsP7 levels. (A) Representative SAX-HPLC profile of 20-day-old wild-type (Col-0) and pfa-dsp1-6 Arabidopsis seedlings radiolabeled with [3H]-myo-inositol. All visible peaks are highlighted and assigned to the corresponding InsP species. Based on published chromatographic mobilities,56,57 InsP4a likely represents Ins(1,4,5,6)P4 or Ins(3,4,5,6)P4, InsP5a likely represents InsP5 [2-OH], InsP5b likely represents InsP5 [4-OH] or its enantiomeric form InsP5 [6-OH], and InsP5c likely represents InsP5 [1-OH] or its enantiomeric form InsP5 [3-OH]. The isomeric natures of InsP3a-c, InsP4b, InsP7, and InsP8 are unknown. (B) InsP7/InsP6 and InsP3b/InsP6 ratio of pfa-dsp1-6 relative to Col-0 determined by the analysis of SAX-HPLC profiles using OriginPro 8. (C) Relative expression of PFA-DSP1 in plants grown under identical conditions as for SAX-HPLC analyses, presented as fold difference compared to Col-0. (B, C) Data represent mean ± SEM (n = 3). Asterisks indicate values that are significantly different from the Col-0 control (according to Student’s t test, P < 0.05 (*); P < 0.01 (**)).
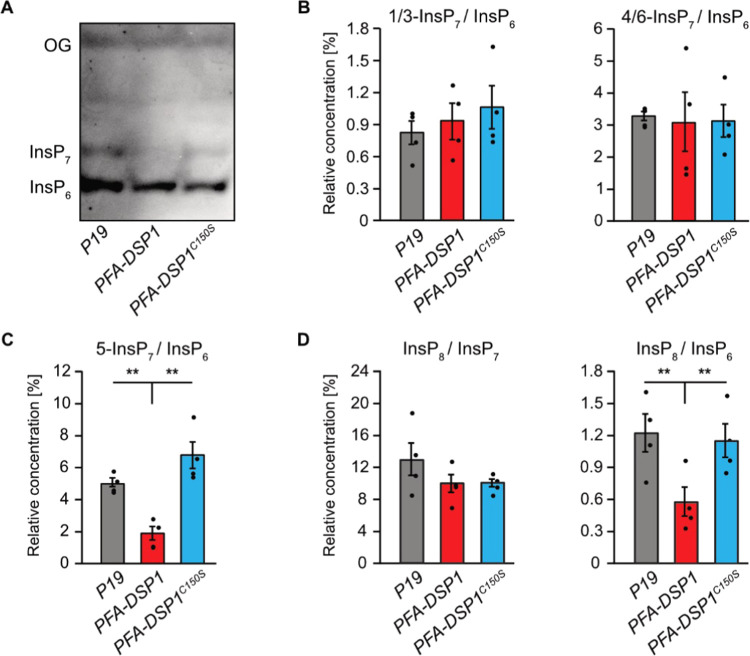
Since the analyses of the pfa-dsp1-6 line indicated that the T-DNA insertion causes an overexpression of PFA-DSP1, resulting in decreased InsP7 levels, we investigated whether PP-InsP phosphohydrolase activity is also observed in a heterologous plant expression system. To this end, we transiently expressed a translational fusion of PFA-DSP1 with a C-terminal EYFP under control of the strong viral CaMV 35S promoter in N. benthamiana using agrobacterium-mediated transfection. The respective catalytically inactive PFA-DSP1C150S-EYFP fusion protein was also expressed and InsPs were then extracted from N. benthamiana leaves and purified by TiO2 pulldown 5 days after infiltration. PAGE analyses showed that transient expression of PFA-DSP1 or expression of its catalytic inactive version did not alter InsP6 levels (Figure 7A). In contrast, InsP7 levels were reduced by the transient expression of PFA-DSP1 but not by the expression of its catalytic inactive version (Figure 7A). These findings were strengthened by subsequent CE-ESI-MS analyses that revealed no changes in the ratios of 1/3-InsP7/InsP6 or 4/6-InsP7/InsP6 compared to control leaves infiltrated with agrobacteria carrying the silencing inhibitor P19 alone (Figure 7B). In contrast, the 5-InsP7/InsP6 ratio was significantly reduced in plants expressing PFA-DSP1 compared to plants expressing the inactive version of PFA-DSP1 or P19 alone (Figure 7C). The InsP8/InsP6 ratio, in turn, was strongly reduced by the expression of PFA-DSP1 (Figure 7D) in agreement with a partial hydrolytic activity of PFA-DSP proteins against InsP8 isomers (Figures 3 and S7) and in agreement with the finding that 5-InsP7, a substrate hydrolyzed by PFA-DSP1, represents the major precursor for InsP8 synthesis.11 In summary, these results demonstrate that ectopic expression of Arabidopsis PFA-DSP1 results in a specific decrease of 5-InsP7 and InsP8in planta.
Figure 7.
Transient expression of PFA-DSP1 in N. benthamiana leaves specifically decreases 5-InsP7 and InsP8. InsPs from infiltrated N. benthamiana leaves, transiently expressing the silencing inhibitor P19 alone or together with PFA-DSP1-EYFP or PFA-DSP1C150S-EFYP, were purified with TiO2 pulldown and analyzed using CE-ESI-MS. Detected peaks were assigned to specific InsP isomers and quantified by comparing them with 13C InsP standards that were spiked into the purified samples. (A) Representative PAGE gel of a sample set used for CE-ESI-MS analysis. (B–D) Relative amounts of PP-InsPs compared to InsP6 or of InsP8 compared to all InsP7 isomers as indicated. Data represent mean ± SEM (n = 4). Asterisks indicate values that are significantly different from wild-type (according to Student’s t test, P < 0.05 (*); P < 0.01 (**)). Note that 4- or 6-InsP7, as well as 1- or 3-InsP7 represent enantiomeric forms that cannot be distinguished by CE-ESI-MS analyses.
Conclusions
Recent studies elucidating the identity and substrate specificity of InsP6/PP-InsP kinases have allowed us to establish important functions of PP-InsPs in nutrient sensing, hormone signaling, and plant immunity.5−13,20,28 In contrast, information on enzymatic activities removing PP-InsPs to switch off their signaling functions in plants is sparse. Intriguingly, the first robust detection of PP-InsP messengers in mammalian cells was made possible by blocking mammalian PP-InsP phosphohydrolases with fluoride.1 While substantial progress in elucidating the role of various PP-InsP phosphohydrolases in regulating these messengers in yeast and mammalian cells has been made,37,41,42,58 we are unaware of any study about PP-InsP degrading enzymes in plants at the onset of this study. Here, we provide evidence that the Arabidopsis PFA-DSP proteins are functional homologues of yeast Siw14 with high phosphohydrolase specificity for the 5-β-phosphate of various PP-InsPs.
The striking biochemical similarities between Arabidopsis PFA-DSPs as deduced from in vitro assays and heterologous expressions analyses in yeast might well explain redundancies of these enzymes and consequently a lack of obvious phenotypes in single pfa-dsp loss-of-function lines in Arabidopsis. A search in transcriptome studies revealed that PFA-DSP1, 2, and 4 are strongly induced by Pi deficiency (Figure S12). Such Pi-dependent regulation is in line with the disappearance of PP-InsPs in tissues of Pi-starved plants9,11 but future studies are required to establish the involvement of PFA-DSPs in the removal of messengers controlling Pi signaling. The high specificity of PFA-DSPs observed in this study establishes these enzymes as ideal tools to investigate the physiological roles of 5-β-phosphate containing PP-InsPs in plant development, plant immunity, nutrient perception, and abiotic stress tolerance. This is particularly important because of potentially confounding effects caused by the recently discovered plant 4/6-InsP711 and also because higher-order mutants involved in the synthesis of 5-β-phosphate containing PP-InsPs such as itpk1 itpk2 and vih1 vih2 display severe developmental defects or die at the young seedling stage.10,11 With the availability of a variety of promoters with tight spatial and temporal regulation, ectopic expression of PFA-DSPs in a tissue and developmentally controlled manner will provide helpful insights to unravel the roles of 5-InsP7 and 1/3, 5-InsP8 in plant development and plant physiology.
Acknowledgments
The authors thank Li Schlüter and Brigitte Ueberbach (Department of Plant Nutrition, Institute of Crop Science and Resource Conservation, University of Bonn) for excellent technical assistance and Marília Kamleitner (Department of Plant Nutrition, Institute of Crop Science and Resource Conservation, University of Bonn) for critically reading this manuscript. They also thank Priyanshi Rana (Department of Biochemistry, Indian Institute of Science) for her inputs during the revision of the manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.2c00145.
Purification of PFA-DSP proteins (Figure S1); in vitro, Arabidopsis PFA-DSP1 displays robust PP-InsP phosphohydrolase activity against 5-InsP7 and partial phosphohydrolase activity against 4-InsP7 and 6-InsP7, respectively (Figure S2); in the absence of divalent cations, all InsP7 isomers with the exception of 2-InsP7 become substrates for selected Arabidopsis PFA-DSPs in vitro (Figure S3); in the presence of Mg2+, PFA-DSP1 and PFA-DSP3 display robust in vitro InsP7 phosphohydrolase activity with high specificity for the 5-β-phosphate (Figure S4); under prolonged incubation time, Arabidopsis PFA-DSP1 efficiently hydrolyzes 5-InsP7, 4-InsP7, and 6-InsP7 but only displays partial activities against 1-InsP7 and 3-InsP7, and a very weak activity against 2-InsP7 (Figure S5); Arabidopsis PFA-DSP1 maintains 5-InsP7 phosphohydrolase activity during prolonged incubation times in vitro (Figure S6); in vitro, Arabidopsis PFA-DSPs display robust 1/3,5-InsP8 phosphohydrolase activity (Figure S7); all five PFA-DSP homologues are stably expressed in the siw14Δ yeast strain (Figure S8); heterologous expression of Arabidopsis PFA-DSPs complements siw14Δ-associated defects in InsP7/InsP6 ratios in yeast (Figure S9); complementation of siw14Δ-associated growth defects depends on catalytic activity (Figure S10); single-mutant Arabidopsispfa-dsp1 loss-of-function lines do not display InsP/PP-InsP defects (Figure S11); ArabidopsisPFA-DSP1, 2, and 4 are strongly induced by Pi deficiency (Figure S12); overview of Arabidopsis PFA-DSP substrate specificities in the presence of Mg2+ showing a robust PP-InsP phosphohydrolase activity against 5-InsP7, 1,5-InsP8, and 3,5-InsP8, in vitro (Table S1); and oligonucleotide sequences (Table S2) (PDF)
Accession Codes
DNA and Protein Sequences can be obtained from the Saccharomyces Genome database (https://www.yeastgenome.org/), TAIR (https://www.arabidopsis.org), and UniProt (https://www.uniprot.org/) under the following accession numbers: SIW14 (YNL032W, NC_001146.8), Arabidopsis PFA-DSP1 (At1g05000, NM_100379.3), Arabidopsis PFA-DSP2 (At2g32960, NM_128856.5), Arabidopsis PFA-DSP3 (At3g02800, NM_111148.3), Arabidopsis PFA-DSP4 (At4g03960, NM_116634.4), Arabidopsis PFA-DSP5 (At5g16480, NM_121653.4), Arabidopsis PP2AA3 (At1g13320, NM_101203), and Arabidopsis TIP41-like (At3g54000, NM_115260). PDB ID: 1XRI.
Author Present Address
¶ Department of Biomedicine, University of Basel, 4058 Basel, Switzerland
Author Contributions
◆ P.G. and R.S. contributed equally to this manuscript. D.L., G.S., and P.G. conceived the study. P.G., D.L., G.S., H.J.J., and R.F.H.G. designed experiments. P.G., R.S., D.L., G.L. D.Q., J.W., M.H., N.J., K.R., J.S., N.F.-R., and R.F.H.G. performed experiments. P.G. generated yeast mutants and performed all yeast experiments, generated constructs, isolated T-DNA insertion lines, performed HPLC analyses of plants, performed qPCR analyses, performed plant infiltration and TiO2 pulldowns, and analyzed most of the experiments. R.S. purified recombinant proteins and carried out and analyzed in vitro kinase assays. G.L. and D.Q. performed CE-ESI-MS/MS analysis and isomer identification. J.W. and J.S. generated constructs and established the expression and purification of recombinant proteins. N.J., M.H., and K.R. synthesized InsP7 and InsP8 isomers. N.F.-R. isolated T-DNA insertion lines, performed HPLC analyses of plants, generated constructs, and performed qPCR analyses. R.F.H.G. generated plant samples for CE-ESI-MS analysis and did transcriptome analysis. M.N.T. synthesized 13C-InsP standards. V.G. analyzed and quantified HPLC analyses. P.G., G.S., D.L., H.J.J., and D.F. supervised the experimental work. P.G., G.S., R.S., D.L., and R.Y. wrote the manuscript with input from all authors.
This work was funded by grants from the Deutsche Forschungsgemeinschaft (SCHA 1274/4-1, SCHA 1274/5-1, Research Training Group GRK 2064 and under Germany’s Excellence Strategy, EXC-2070-390732324, PhenoRob to G.S.; JE 572/4-1 and under Germany’s Excellence Strategy, CIBSS-EXC-2189–Project ID 390939984 to H.J.J.; and HE 8362/1-1, DFG Eigene Stelle, to R.F.H.G.). D.L. acknowledges the Department of Biotechnology (DBT) for HGK-IYBA award (BT/13/IYBA/2020/04) and a DBT Indian Institute of Science Partnership Program. H.J.J. acknowledges funding from the Volkswagen Foundation (Momentum Grant 2021).
The authors declare no competing financial interest.
Notes
During the revision of this manuscript, a study by Wang and colleagues71 reported high-resolution crystal structures of Arabidopsis PFA-DSP1 in complex with 5-InsP7, 6-InsP7, and 5-InsP7 analogues and provided evidence for efficient in vitro phosphatase activity of this enzyme against 5-InsP7 as well as weaker in vitro activities against 4-InsP7 and 6-InsP7 in agreement with our findings.
Supplementary Material
References
- Menniti F. S.; Miller R. N.; Putney J. W. Jr.; Shears S. B. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J. Biol. Chem. 1993, 268, 3850–3856. 10.1016/S0021-9258(18)53551-1. [DOI] [PubMed] [Google Scholar]
- Stephens L.; Radenberg T.; Thiel U.; Vogel G.; Khoo K. H.; Dell A.; Jackson T. R.; Hawkins P. T.; Mayr G. W. The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s). J. Biol. Chem. 1993, 268, 4009–4015. 10.1016/S0021-9258(18)53571-7. [DOI] [PubMed] [Google Scholar]
- Thota S. G.; Bhandari R. The emerging roles of inositol pyrophosphates in eukaryotic cell physiology. J Biosci. 2015, 40, 593–605. 10.1007/s12038-015-9549-x. [DOI] [PubMed] [Google Scholar]
- Shears S. B. Intimate connections: Inositol pyrophosphates at the interface of metabolic regulation and cell signaling. J. Cell. Physiol. 2018, 233, 1897–1912. 10.1002/jcp.26017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha D.; Johnen P.; Azevedo C.; Dynowski M.; Weiss M.; Capolicchio S.; Mao H.; Iven T.; Steenbergen M.; Freyer M.; Gaugler P.; de Campos M. K.; Zheng N.; Feussner I.; Jessen H. J.; Van Wees S. C.; Saiardi A.; Schaaf G. VIH2 Regulates the Synthesis of Inositol Pyrophosphate InsP8 and Jasmonate-Dependent Defenses in Arabidopsis. Plant Cell 2015, 27, 1082–1097. 10.1105/tpc.114.135160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha D.; Parvin N.; Dynowski M.; Johnen P.; Mao H.; Bitters S. T.; Zheng N.; Schaaf G. Inositol Polyphosphate Binding Specificity of the Jasmonate Receptor Complex. Plant Physiol. 2016, 171, 2364–2370. 10.1104/pp.16.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild R.; Gerasimaite R.; Jung J. Y.; Truffault V.; Pavlovic I.; Schmidt A.; Saiardi A.; Jessen H. J.; Poirier Y.; Hothorn M.; Mayer A. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 2016, 352, 986–990. 10.1126/science.aad9858. [DOI] [PubMed] [Google Scholar]
- Couso I.; Evans B.; Li J.; Liu Y.; Ma F.; Diamond S.; Allen D. K.; Umen J. G. Synergism between inositol polyphosphates and TOR kinase signaling in nutrient sensing, growth control and lipid metabolism in Chlamydomonas. Plant Cell 2016, 28, 2026–2042. 10.1105/tpc.16.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J.; Ma G.; Sui L.; Wei M.; Satheesh V.; Zhang R.; Ge S.; Li J.; Zhang T. E.; Wittwer C.; Jessen H. J.; Zhang H.; An G. Y.; Chao D. Y.; Liu D.; Lei M. Inositol Pyrophosphate InsP8 Acts as an Intracellular Phosphate Signal in Arabidopsis. Mol. Plant 2019, 12, 1463–1473. 10.1016/j.molp.2019.08.002. [DOI] [PubMed] [Google Scholar]
- Zhu J.; Lau K.; Puschmann R.; Harmel R. K.; Zhang Y.; Pries V.; Gaugler P.; Broger L.; Dutta A. K.; Jessen H. J.; Schaaf G.; Fernie A. R.; Hothorn L. A.; Fiedler D.; Hothorn M. Two bifunctional inositol pyrophosphate kinases/phosphatases control plant phosphate homeostasis. eLife 2019, 8, e43582 10.7554/eLife.43582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer E.; Qiu D.; Laha D.; Harmel R. K.; Gaugler P.; Gaugler V.; Frei M.; Hajirezaei M. R.; Laha N. P.; Krusenbaum L.; Schneider R.; Saiardi A.; Fiedler D.; Jessen H. J.; Schaaf G.; Giehl R. F. H. ITPK1 is an InsP6/ADP phosphotransferase that controls phosphate signaling in Arabidopsis. Mol. Plant 2021, 14, 1864–1880. 10.1016/j.molp.2021.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulabani H.; Goswami K.; Walia Y.; Roy A.; Noor J. J.; Ingole K. D.; Kasera M.; Laha D.; Giehl R. F. H.; Schaaf G.; Bhattacharjee S. Arabidopsis inositol polyphosphate kinases IPK1 and ITPK1 modulate crosstalk between SA-dependent immunity and phosphate-starvation responses. Plant Cell Rep. 2021, 41, 347–363. 10.1007/s00299-021-02812-3. [DOI] [PubMed] [Google Scholar]
- Laha N. P.; Dhir Y. W.; Giehl R. F. H.; Schäfer E. M.; Gaugler P.; Shishavan Z. H.; Gulabani H.; Mao H.; Zheng N.; von Wirén N.; Jessen H. J.; Saiardi A.; Bhattacharjee S.; Laha D.; Schaaf G.. ITPK1-Dependent Inositol Polyphosphates Regulate Auxin Responses in Arabidopsis thaliana, 2020. 10.1101/2020.04.23.058487. [DOI]
- Lin H.; Fridy P. C.; Ribeiro A. A.; Choi J. H.; Barma D. K.; Vogel G.; Falck J. R.; Shears S. B.; York J. D.; Mayr G. W. Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J. Biol. Chem. 2009, 284, 1863–1872. 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulugu S.; Bai W.; Fridy P. C.; Bastidas R. J.; Otto J. C.; Dollins D. E.; Haystead T. A.; Ribeiro A. A.; York J. D. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 2007, 316, 106–109. 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- Saiardi A.; Erdjument-Bromage H.; Snowman A. M.; Tempst P.; Snyder S. H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 1999, 9, 1323–1326. 10.1016/S0960-9822(00)80055-X. [DOI] [PubMed] [Google Scholar]
- Wang H.; Falck J. R.; Hall T. M.; Shears S. B. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 2012, 8, 111–116. 10.1038/nchembio.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha D.; Kamleitner M.; Johnen P.; Schaaf G. Analyses of Inositol Phosphates and Phosphoinositides by Strong Anion Exchange (SAX)-HPLC. Methods Mol. Biol. 2021, 2295, 365–378. 10.1007/978-1-0716-1362-7_20. [DOI] [PubMed] [Google Scholar]
- Qiu D. Y.; Eisenbeis V. B.; Saiardi A.; Jessen H. J. Absolute Quantitation of Inositol Pyrophosphates by Capillary Electrophoresis Electrospray Ionization Mass Spectrometry. J. Vis. Exp. 2021, 174, e62847 10.3791/62847. [DOI] [PubMed] [Google Scholar]
- Desai M.; Rangarajan P.; Donahue J. L.; Williams S. P.; Land E. S.; Mandal M. K.; Phillippy B. Q.; Perera I. Y.; Raboy V.; Gillaspy G. E. Two inositol hexakisphosphate kinases drive inositol pyrophosphate synthesis in plants. Plant J. 2014, 80, 642–653. 10.1111/tpj.12669. [DOI] [PubMed] [Google Scholar]
- Flores S.; Smart C. C. Abscisic acid-induced changes in inositol metabolism in Spirodela polyrrhiza. Planta 2000, 211, 823–832. 10.1007/s004250000348. [DOI] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F.; MacRobbie E. A.; Brearley C. A. Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 8687–8692. 10.1073/pnas.140217497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D. Y.; Wilson M. S.; Eisenbeis V. B.; Harmel R. K.; Riemer E.; Haas T. M.; Wittwer C.; Jork N.; Gu C. F.; Shears S. B.; Schaaf G.; Kammerer B.; Fiedler D.; Saiardi A.; Jessen H. J. Analysis of inositol phosphate metabolism by capillary electrophoresis electrospray ionization mass spectrometry. Nat. Commun. 2020, 11, 6035 10.1038/s41467-020-19928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha D.; Parvin N.; Hofer A.; Giehl R. F. H.; Fernandez-Rebollo N.; von Wiren N.; Saiardi A.; Jessen H. J.; Schaaf G. Arabidopsis ITPK1 and ITPK2 Have an Evolutionarily Conserved Phytic Acid Kinase Activity. ACS Chem. Biol. 2019, 14, 2127–2133. 10.1021/acschembio.9b00423. [DOI] [PubMed] [Google Scholar]
- Adepoju O.; Williams S. P.; Craige B.; Cridland C. A.; Sharpe A. K.; Brown A. M.; Land E.; Perera I. Y.; Mena D.; Sobrado P.; Gillaspy G. E.. Inositol Trisphosphate Kinase and Diphosphoinositol Pentakisphosphate Kinase Enzymes Constitute the Inositol Pyrophosphate Synthesis Pathway in Plants, 2019. 10.1101/724914. [DOI]
- Whitfield H.; White G.; Sprigg C.; Riley A. M.; Potter B. V. L.; Hemmings A. M.; Brearley C. A. An ATP-responsive metabolic cassette comprised of inositol tris/tetrakisphosphate kinase 1 (ITPK1) and inositol pentakisphosphate 2-kinase (IPK1) buffers diphosphosphoinositol phosphate levels. Biochem. J. 2020, 477, 2621–2638. 10.1042/BCJ20200423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. R.; Kuo H. F.; Chiou T. J. Intracellular phosphate sensing and regulation of phosphate transport systems in plants. Plant Physiol. 2021, 187, 2043–2055. 10.1093/plphys/kiab343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried M. K.; Wild R.; Zhu J. S.; Pipercevic J.; Sturm K.; Broger L.; Harmel R. K.; Abriata L. A.; Hothorn L. A.; Fiedler D.; Hiller S.; Hothorn M. Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. Nat. Commun. 2021, 12, 384 10.1038/s41467-020-20681-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimaite R.; Pavlovic I.; Capolicchio S.; Hofer A.; Schmidt A.; Jessen H. J.; Mayer A. Inositol Pyrophosphate Specificity of the SPX-Dependent Polyphosphate Polymerase VTC. ACS Chem. Biol. 2017, 12, 648–653. 10.1021/acschembio.7b00026. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Hu Q.; Xiao X.; Yao D.; Ge S.; Ye J.; Li H.; Cai R.; Liu R.; Meng F.; Wang C.; Zhu J. K.; Lei M.; Xing W. Mechanism of phosphate sensing and signaling revealed by rice SPX1-PHR2 complex structure. Nat. Commun. 2021, 12, 7040 10.1038/s41467-021-27391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V.; Linhares F.; Solano R.; Martin A. C.; Iglesias J.; Leyva A.; Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R.; Castrillo G.; Linhares F.; Puga M. I.; Rubio V.; Perez-Perez J.; Solano R.; Leyva A.; Paz-Ares J. A Central Regulatory System Largely Controls Transcriptional Activation and Repression Responses to Phosphate Starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga M. I.; Mateos I.; Charukesi R.; Wang Z.; Franco-Zorrilla J. M.; de Lorenzo L.; Irigoye M. L.; Masiero S.; Bustos R.; Rodriguez J.; Leyva A.; Rubio V.; Sommer H.; Paz-Ares J. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 14947–14952. 10.1073/pnas.1404654111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridy P. C.; Otto J. C.; Dollins D. E.; York J. D. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J. Biol. Chem. 2007, 282, 30754–30762. 10.1074/jbc.M704656200. [DOI] [PubMed] [Google Scholar]
- Wang H.; Nair V. S.; Holland A. A.; Capolicchio S.; Jessen H. J.; Johnson M. K.; Shears S. B. Asp1 from Schizosaccharomyces pombe binds a [2Fe-2S](2+) cluster which inhibits inositol pyrophosphate 1-phosphatase activity. Biochemistry 2015, 54, 6462–6474. 10.1021/acs.biochem.5b00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Ortiz M.; Saiardi A.; Walla E.; Jakopec V.; Kunzel N. A.; Span I.; Vangala A.; Fleig U. Asp1 Bifunctional Activity Modulates Spindle Function via Controlling Cellular Inositol Pyrophosphate Levels in Schizosaccharomyces pombe. Mol. Cell. Biol. 2018, 38, e00047-18 10.1128/MCB.00047-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilari R. S.; Weaver J. D.; Shears S. B.; Safrany S. T. Understanding inositol pyrophosphate metabolism and function: Kinetic characterization of the DIPPs. FEBS Lett. 2013, 587, 3464–3470. 10.1016/j.febslet.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey J. J.; Hidaka K.; Matsuda M.; Hirata M.; Shears S. B. The human and rat forms of multiple inositol polyphosphate phosphatase: functional homology with a histidine acid phosphatase up-regulated during endochondral ossification. FEBS Lett. 1999, 442, 99–104. 10.1016/S0014-5793(98)01636-6. [DOI] [PubMed] [Google Scholar]
- Safrany S. T.; Ingram S. W.; Cartwright J. L.; Falck J. R.; McLennan A. G.; Barnes L. D.; Shears S. B. The diadenosine hexaphosphate hydrolases from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human diphosphoinositol polyphosphate phosphohydrolase - Overlapping substrate specificities in a MutT-type protein. J. Biol. Chem. 1999, 274, 21735–21740. 10.1074/jbc.274.31.21735. [DOI] [PubMed] [Google Scholar]
- Andreeva N.; Ledova L.; Ryazanova L.; Tomashevsky A.; Kulakovskaya T.; Eldarov M. Ppn2 endopolyphosphatase overexpressed in Saccharomyces cerevisiae: Comparison with Ppn1, Ppx1, and Ddp1 polyphosphatases. Biochimie 2019, 163, 101–107. 10.1016/j.biochi.2019.06.001. [DOI] [PubMed] [Google Scholar]
- Lonetti A.; Szijgyarto Z.; Bosch D.; Loss O.; Azevedo C.; Saiardi A. Identification of an Evolutionarily Conserved Family of Inorganic Polyphosphate Endopolyphosphatases. J. Biol. Chem. 2011, 286, 31966–31974. 10.1074/jbc.M111.266320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidle E. A.; Chong L. S.; Wu M.; Crooke E.; Fiedler D.; Resnick A. C.; Rolfes R. J. A Novel Inositol Pyrophosphate Phosphatase in Saccharomyces cerevisiae: Siw14 PROTEIN SELECTIVELY CLEAVES THE beta-PHOSPHATE FROM 5-DIPHOSPHOINOSITOL PENTAKISPHOSPHATE (5PP-IP5). J. Biol. Chem. 2016, 291, 6772–6783. 10.1074/jbc.M116.714907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Gu C.; Rolfes R. J.; Jessen H. J.; Shears S. B. Structural and biochemical characterization of Siw14: A protein-tyrosine phosphatase fold that metabolizes inositol pyrophosphates. J. Biol. Chem. 2018, 293, 6905–6914. 10.1074/jbc.RA117.001670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romá-Mateo C.; Sacristan-Reviriego A.; Beresford N. J.; Caparros-Martin J. A.; Culianez-Macia F. A.; Martin H.; Molina M.; Tabernero L.; Pulido R. Phylogenetic and genetic linkage between novel atypical dual-specificity phosphatases from non-metazoan organisms. Mol. Genet. Genomics 2011, 285, 341–354. 10.1007/s00438-011-0611-6. [DOI] [PubMed] [Google Scholar]
- Romá-Mateo C.; Rios P.; Tabernero L.; Attwood T. K.; Pulido R. A novel phosphatase family, structurally related to dual-specificity phosphatases, that displays unique amino acid sequence and substrate specificity. J. Mol. Biol. 2007, 374, 899–909. 10.1016/j.jmb.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Aceti D. J.; Bitto E.; Yakunin A. F.; Proudfoot M.; Bingman C. A.; Frederick R. O.; Sreenath H. K.; Vojtik F. C.; Wrobel R. L.; Fox B. G.; Markley J. L.; Phillips G. N. Structural and functional characterization of a novel phosphatase from the Arabidopsis thaliana gene locus At1g05000. Proteins 2008, 73, 241–253. 10.1002/prot.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy R.; Grob H.; Weder B.; Green P.; Klein M.; Frelet-Barrand A.; Schjoerring J. K.; Brearley C.; Martinoia E. The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J. Biol. Chem. 2009, 284, 33614–33622. 10.1074/jbc.M109.030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine R. F.; Schell M. J. Back in the water: the return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2001, 2, 327–338. 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Blüher D.; Laha D.; Thieme S.; Hofer A.; Eschen-Lippold L.; Masch A.; Balcke G.; Pavlovic I.; Nagel O.; Schonsky A.; Hinkelmann R.; Wörner J.; Parvin N.; Greiner R.; Weber S.; Tissier A.; Schutkowski M.; Lee J.; Jessen H.; Schaaf G.; Bonas U. A 1-phytase type III effector interferes with plant hormone signaling. Nat. Commun. 2017, 8, 2159 10.1038/s41467-017-02195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada S.; Kageyama K.; Ohnishi Y.; Nishikawa J.; Nishihara T.; Imagawa M. Inositol phosphate kinase Vip1p interacts with histone chaperone Asf1p in Saccharomyces cerevisiae. Mol. Biol. Rep. 2012, 39, 4989–4996. 10.1007/s11033-011-1295-z. [DOI] [PubMed] [Google Scholar]
- Steidle E. A.; Morrissette V. A.; Fujimaki K.; Chong L.; Resnick A. C.; Capaldi A. P.; Rolfes R. J. The InsP7 phosphatase Siw14 regulates inositol pyrophosphate levels to control localization of the general stress response transcription factor Msn2. J. Biol. Chem. 2020, 295, 2043–2056. 10.1074/jbc.RA119.012148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. A.; Sherlock G.; Myers C. L.; Burrows N. M.; Deng C.; Wu H. I.; McCann K. E.; Troyanskaya O. G.; Brown J. M. Global analysis of gene function in yeast by quantitative phenotypic profiling. Mol. Syst. Biol. 2006, 2, 2006.0001 10.1038/msb4100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcaro A.; Wymann M. P. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem. J. 1993, 296, 297–301. 10.1042/bj2960297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A.; Resnick A. C.; Snowman A. M.; Wendland B.; Snyder S. H. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 1911–1914. 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez F.; Pulford D.; Stark M. J.; Carter A. N.; Downes C. P. Inositol trisphosphate metabolism in Saccharomyces cerevisiae: identification, purification and properties of inositol 1,4,5-trisphosphate 6-kinase. Biochem. J. 1994, 302, 709–716. 10.1042/bj3020709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H. F.; Hsu Y. Y.; Lin W. C.; Chen K. Y.; Munnik T.; Brearley C. A.; Chiou T. J. Arabidopsis inositol phosphate kinases IPK1 and ITPK1 constitute a metabolic pathway in maintaining phosphate homeostasis. Plant J. 2018, 95, 613–630. 10.1111/tpj.13974. [DOI] [PubMed] [Google Scholar]
- Stevenson-Paulik J.; Bastidas R. J.; Chiou S. T.; Frye R. A.; York J. D. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 12612–12617. 10.1073/pnas.0504172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safrany S. T.; Caffrey J. J.; Yang X.; Bembenek M. E.; Moyer M. B.; Burkhart W. A.; Shears S. B. A novel context for the ‘MutT’ module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 1998, 17, 6599–6607. 10.1093/emboj/17.22.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber B. D.; Giehl R. F.; Friedel S.; von Wiren N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179. 10.1104/pp.113.218453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D.; Lalonde S.; Looger L. L.; von Wiren N.; Frommer W. B. A cytosolic trans-activation domain essential for ammonium uptake. Nature 2007, 446, 195–198. 10.1038/nature05579. [DOI] [PubMed] [Google Scholar]
- Nakamura S.; Mano S.; Tanaka Y.; Ohnishi M.; Nakamori C.; Araki M.; Niwa T.; Nishimura M.; Kaminaka H.; Nakagawa T.; Sato Y.; Ishiguro S. Gateway Binary Vectors with the Bialaphos Resistance Gene, as a Selection Marker for Plant Transformation. Biosci., Biotechnol., Biochem. 2010, 74, 1315–1319. 10.1271/bbb.100184. [DOI] [PubMed] [Google Scholar]
- Gueldener U.; Heinisch J.; Koehler G. J.; Voss D.; Hegemann J. H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002, 30, e23 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onnebo S. M.; Saiardi A. Inositol pyrophosphates modulate hydrogen peroxide signalling. Biochem. J. 2009, 423, 109–118. 10.1042/BJ20090241. [DOI] [PubMed] [Google Scholar]
- Gietz R. D.; Schiestl R. H.; Willems A. R.; Woods R. A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 1995, 11, 355–360. 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Zonneveld B. J. M. B.J.M. Zonneveld. 1986. Cheap and simple yeast media. J. Microb. Methods 1986, 4, 287–291. 10.1016/0167-7012(86)90040-0. [DOI] [Google Scholar]
- Schaaf G.; Betts L.; Garrett T. A.; Raetz C. R.; Bankaitis V. A. Crystallization and preliminary X-ray diffraction analysis of phospholipid-bound Sfh1p, a member of the Saccharomyces cerevisiae Sec. 14p-like phosphatidylinositol transfer protein family. Acta Crystallogr., Sect. F 2006, 62, 1156–1160. 10.1107/S1744309106041728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capolicchio S.; Thakor D. T.; Linden A.; Jessen H. J. Synthesis of unsymmetric diphospho-inositol polyphosphates. Angew. Chem., Int. Ed. 2013, 52, 6912–6916. 10.1002/anie.201301092. [DOI] [PubMed] [Google Scholar]
- Capolicchio S.; Wang H.; Thakor D. T.; Shears S. B.; Jessen H. J. Synthesis of Densely Phosphorylated Bis-1,5-Diphospho-myo-Inositol Tetrakisphosphate and its Enantiomer by Bidirectional P-Anhydride Formation. Angew. Chem., Int. Ed. 2014, 53, 9508–9511. 10.1002/anie.201404398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C.; Saiardi A. Extraction and analysis of soluble inositol polyphosphates from yeast. Nat. Protoc. 2006, 1, 2416–2422. 10.1038/nprot.2006.337. [DOI] [PubMed] [Google Scholar]
- Gaugler P.; Gaugler V.; Kamleitner M.; Schaaf G. Extraction and Quantification of Soluble, Radiolabeled Inositol Polyphosphates from Different Plant Species using SAX-HPLC. J. Vis. Exp. 2020, 160, e61495 10.3791/61495. [DOI] [PubMed] [Google Scholar]
- Wang H.; Perera L.; Jork N.; Zong G.; Riley A. M.; Potter B. V. L.; Jessen H. J.; Shears S. B. A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop. Nat. Commun. 2022, 13, 2231 10.1038/s41467-022-29673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.