Abstract
Microorganisms often live in symbiosis with their hosts, and some are considered mutualists, where all species involved benefit from the interaction. How free-living microorganisms have evolved to become mutualists is unclear. Here we report an experimental system in which non-symbiotic Escherichia coli evolves into an insect mutualist. The stinkbug Plautia stali is typically associated with its essential gut symbiont, Pantoea sp., which colonizes a specialized symbiotic organ. When sterilized newborn nymphs were infected with E. coli rather than Pantoea sp., only a few insects survived, in which E. coli exhibited specific localization to the symbiotic organ and vertical transmission to the offspring. Through transgenerational maintenance with P. stali, several hypermutating E. coli lines independently evolved to support the host’s high adult emergence and improved body colour; these were called ‘mutualistic’ E. coli. These mutants exhibited slower bacterial growth, smaller size, loss of flagellar motility and lack of an extracellular matrix. Transcriptomic and genomic analyses of ‘mutualistic’ E. coli lines revealed independent mutations that disrupted the carbon catabolite repression global transcriptional regulator system. Each mutation reproduced the mutualistic phenotypes when introduced into wild-type E. coli, confirming that single carbon catabolite repression mutations can make E. coli an insect mutualist. These findings provide an experimental system for future work on host–microbe symbioses and may explain why microbial mutualisms are omnipresent in nature.
Subject terms: Experimental evolution, Symbiosis, Bacterial evolution
Experimental evolution of Escherichia coli in an insect host reveals that a single mutation in the carbon catabolite repression pathway can transform a non-symbiotic microbial species into an insect mutualist.
Main
Microbial symbioses are among the major evolutionary drivers underpinning biodiversity, wherein relationships range from parasitism through commensalism to mutualism1,2. Originally, however, such microbial symbionts must have been without association with their host organisms, deriving from environmental microbes at the beginning. How ordinary free-living microbes have become sophisticated mutualists is an important but unanswered question. To address this fundamental issue, experimental evolutionary approaches may provide valuable insights3–9. If a model microbe like Escherichia coli with elaborate molecular genetic tools and resources can establish a mutualistic association with a host organism via experimental evolution, such a ‘model experimental symbiotic system’ will be extremely useful for understanding the evolutionary processes of symbiosis towards mutualism. Recently, the stinkbug Plautia stali (Hemiptera: Pentatomidae) has emerged as an experimentally tractable model system for investigating the diversity, evolution and mechanism of gut symbiosis with bacterial mutualists10,11. In this study, we report an experimental system in which E. coli evolves into a bacterial mutualist that supports survival and reproduction of P. stali, thereby demonstrating that evolution of mutualism can proceed very easily and quickly via disruption of a global transcriptional regulator system.
Results
E. coli is potentially capable of symbiosis with P. stali
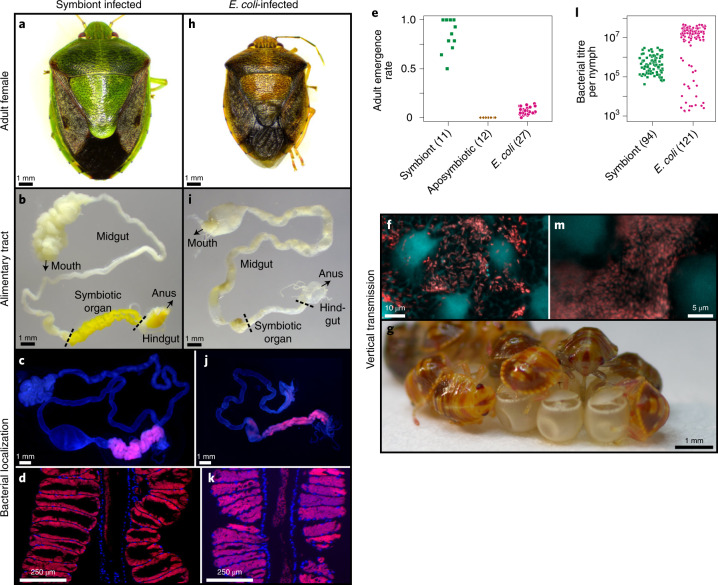
Plant-sucking heteropteran bugs generally possess specific symbiotic bacteria in the midgut, which contribute to their growth and survival via provisioning of essential amino acids and/or vitamins12,13. The brown-winged green stinkbug P. stali (Hemiptera: Pentatomidae) (Fig. 1a) developed a specialized symbiotic organ consisting of numerous crypts in a posterior region of the midgut (Fig. 1b). The crypt cavities are densely populated by a specific bacterial symbiont of the genus Pantoea (Fig. 1c,d). The symbiont is essential for growth and survival of the host insect. Normal insects infected with the uncultivable obligatory symbiont, Pantoea sp. A10,14, attained over 70% adult emergence rates (Fig. 1e), smeared the symbiont cells onto the eggs on oviposition (Fig. 1f) and transmitted the symbiont vertically to the offspring via nymphal probing of the eggshell (Fig. 1g and Supplementary Video 1). Aposymbiotic insects generated by egg surface sterilization died out with no adult emergence (Fig. 1e). Non-symbiotic bacteria, such as Bacillus subtilis and Burkholderia insecticola, cannot establish infection and symbiosis with P. stali10. Meanwhile, when E. coli was inoculated to sterilized newborn nymphs, the insects certainly exhibited retarded growth and high mortality; however, a small number of adult insects emerged, attaining 5–10% adult emergence rates (Fig. 1e and Extended Data Fig. 1)10. Such adult insects, which were dwarf in size and dark in colour (Fig. 1h), tended to die early but some insects managed to survive, mate and produce a small number of eggs. We dissected and inspected these insects, and found that, surprisingly, although the symbiotic organ was atrophied (Fig. 1i), E. coli localized to the midgut crypts just like the original symbiont, although the infection patterns were often patchy (Fig. 1j,k and Extended Data Fig. 2). Furthermore, E. coli cells were smeared on the eggshell and vertically transmitted to the offspring (Fig. 1l,m), although the transmission rates and the infection titres were unstable in comparison with those of the original symbiont (Fig. 1l). These results suggested that, although incipiently, E. coli is capable of localized infection, vertical transmission and supporting host survival in P. stali. Considering that E. coli belongs to the same Enterobacteriaceae as the original Pantoea symbiont, E. coli may be able to co-opt the mechanisms for infection and localization of the symbiont to establish the incipient symbiosis10. In this context, it seems relevant that, in the stinkbug family Pentatomidae, gut symbiotic bacteria have evolved repeatedly from the Enterobacteriaceae through recurrent acquisitions and replacements15,16.
Fig. 1. Infection, localization and vertical transmission of E. coli in the gut symbiotic system of P. stali.
a, Normal symbiotic adult female, large in size and green in colour. b, Dissected alimentary tract, in which the symbiotic organ is well developed and yellow in colour. c, FISH localization of symbiont cells to the symbiotic organ. d, Magnified FISH image showing symbiont localization to crypt cavities of the symbiotic organ. The image is reconstructed by merging three microscopic images. e, Adult emergence rates of newborn nymphs inoculated with normal symbiont (Pantoea sp. A), no bacteria (aposymbiotic) and E. coli. f, Symbiont cells smeared on egg surface. g, Newborn nymphs sucking symbiont cells from the eggshell (Supplementary Video 1). h, E. coli-infected adult female, dwarf in size and brown in colour. i, Dissected alimentary tract, in which the symbiotic organ is atrophied. j, FISH localization of E. coli to the symbiotic organ. k, Magnified FISH image visualizing E. coli localization to crypt cavities of the symbiotic organ. l, Bacterial titres in symbiont-inoculated and E. coli-inoculated nymphs 1 d after second instar moult in terms of groEL and nptII gene copies per insect, respectively. m, E. coli cells smeared on the egg surface. e,l, The numbers of biological replicates are shown after the strain names. Level adjustments without non-linear change were applied to c, d, f, j, k and m.
Extended Data Fig. 1. Phenotypes of P. stali adults infected with laboratory strains of E. coli.
a, Adult emergence rate. b, Body colour (greenish hue) of females (left) and males (right). (c) Body size (thorax width) of females (left) and males (right). Sym A is Pantoea sp. A, the original, uncultivable and essential gut symbiont of P. stali10. BW25113, EPI300, DH5a, JM109 and BL21 are commonly used laboratory strains of E. coli. The numbers of biological replicates are shown after the strain names.
Extended Data Fig. 2. FISH localization of E. coli and original symbiont Pantoea sp. A (=Sym A) in P. stali.
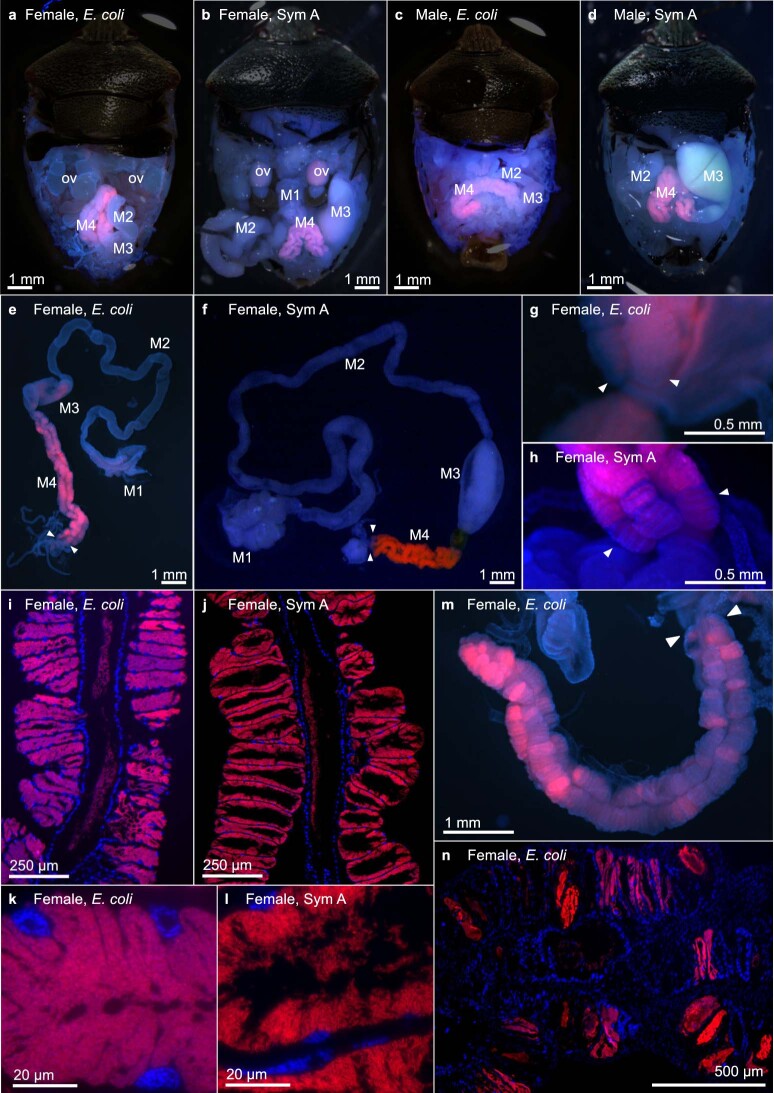
a–d, Localization in abdominal body cavity of adult insects: (a) E. coli in adult female, (b) Sym A in adult female, (c) E. coli in adult male, and (d) Sym A in adult male. FISH signals are localized to the midgut M4 region. Signals in oocytes are due to autofluorescence. Abbreviations: M1, M2, M3, and M4, midgut regions M1, M2, M3, and M4 (=symbiotic organ); ov, ovary. e,f, Localization of E. coli (e) and Sym A (f) in dissected alimentary tract of adult females. Arrowheads indicate female-specific enlarged end crypts at the posterior end of the symbiotic organ, which are presumably involved in vertical symbiont transmission by storing bacteria-containing secretion49. g,h, Magnified images of the end crypts infected with E. coli (g) and Sym A (h). Note that E. coli-infected end crypts are atrophied in comparison with Sym A-infected ones. i,j, Localization of E. coli (i) and Sym A (j) in the crypt cavities of the symbiotic organ. k,l, Magnified images of E. coli cells (k) and Sym A cells (l) packed in the crypt cavity. m,n, Patchy localization patterns of E. coli in the symbiotic organ, which are often found with E. coli but seldom observed with Sym A. Red signals represent the distribution of bacteria, except for red autofluorescence of ovaries in (a) and (b). j is reconstructed by merging five microscopic images. The level adjustment without non-linear change is applied to the images.
Experimental evolution using hypermutating E. coli
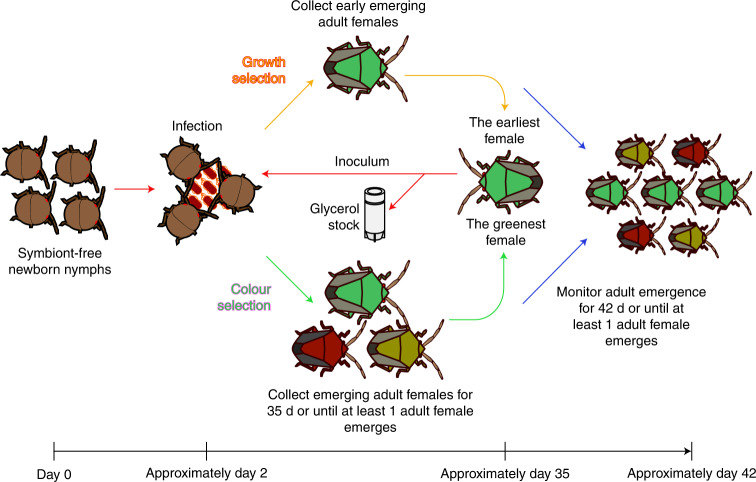
This finding prompted us to apply experimental evolutionary approaches to the P. stali-E. coli relationship. By continuous inoculation to and maintenance with P. stali, would E. coli improve the symbiosis-related traits and finally evolve into a symbiont-like entity? Considering the expected difficulty in observing the evolution of elaborate symbiosis in a realistic time frame, we adopted the hypermutating the E. coli strain, ΔmutS, in which the DNA mismatch repair enzyme gene mutS is disrupted and the molecular evolutionary rate is elevated by two orders of magnitude17. The E. coli strain of the same genetic background, ΔintS, in which the phage integrase gene is disrupted without phenotypic consequences, was used as control. Two selection schemes, growth and colour selection, were conducted (Fig. 2). In growth selection lines (GmL for hypermutating ΔmutS lines; GiL for non-mutating ΔintS lines), the first-emerged adult insect was subjected to dissection of the symbiotic organ for inoculation to the next generation as well as freeze-storing (for example, GmL07G12 is Growth-selected ΔmutS Line 07, Generation 12). In colour selection lines (CmL for ΔmutS lines; CiL for ΔintS lines), the most greenish adult insect was subjected to dissection of the symbiotic organ for inoculation to the next generation as well as freeze storing (for example, CiL05G02 is Colour-selected ΔintS Line 05, Generation 02). Throughout the evolutionary experiments, the host insects were supplied from a mass-reared inbred population of P. stali, thereby homogenizing the host genetic background and focusing on the evolutionary changes of the E. coli side. Since it takes around 1 month for newborn nymphs of P. stali to become adults under the rearing condition, it was expected that, ideally, we would be able to run 12 host generations per year. Actually, however, it took almost two years because (1) the E. coli-inoculated insects generally exhibited high mortality and retarded growth, (2) for keeping the insects under a good condition, frequent care without overcrowding was essential, which limited the manageable number of insects per evolutionary line ranging from 50 to 100 and (3) consequently, extended generation time and stochastic extinction of the evolutionary lines occurred frequently, which had to be restarted from the frozen E. coli stocks.
Fig. 2. Experimental scheme for the evolution of mutualistic E. coli with P. stali.
Evolutionary lines of E. coli were inoculated to sterilized newborn nymphs of P. stali and subjected to either host growth selection or host colour selection in this way.
Evolution of mutualistic E. coli
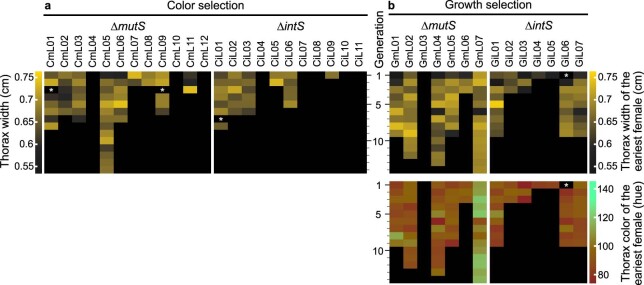
We established and maintained 12 CmL colour selection lines with 11 CiL control lines and 7 GmL growth selection lines with 7 GiL control lines (Fig. 3a,b). While the control ΔintS-infected lines almost constantly exhibited low adult emergence rates, some of the hypermutating ΔmutS-infected lines started to produce more adult insects. Notably, in the colour selection line CmL05, the adult emergence rate jumped up at generation seven and high emergence rates were maintained thereafter (Fig. 3a). In the growth selection line GmL07, the adult emergence rate improved as early as at generation two, which was maintained thereafter (Fig. 3b). In CmL05 and GmL07, coincident with the improvement of the adult emergence rate, the body colour of the adult insects improved from dark to greenish (Fig. 3a–c and Extended Data Fig. 3); furthermore, the colony morphology of E. coli changed from large and flat with rich extracellular matrix to small and convex with little extracellular matrix (Fig. 3c). When the frozen stocks of CmL05 and GmL07 were inoculated to P. stali, the improved adult emergence rate, the greenish body colour and the small and convex colony shape were reproducibly observed (Fig. 3d,e and Extended Data Fig. 4). These results indicated that some evolutionary lines of hypermutating E. coli have evolved mutualistic traits for the host insect and that the phenotypic effects are attributable to genetic changes in the evolutionary E. coli lines.
Fig. 3. Evolution of mutualistic traits for P. stali in hypermutating E. coli lines.
a, Evolutionary E. coli lines subjected to the host’s body colour selection. Data of adult emergence rate and body colour are displayed by heatmaps. The white asterisks indicate missing data of body colour measurement. b, Evolutionary E. coli lines subjected to the host’s growth speed selection. Data of adult emergence rate and days to the first adult emergence are displayed by heatmaps. a,b, When an evolutionary line produced no adult insect and recovery from the freeze stock failed twice consecutively, the evolutionary line was terminated due to shortage of inoculum. From generation 10 and onwards, selected evolutionary lines were maintained. c, Host’s body colour and colony morphology of evolutionary E. coli lines. Red colonies are due to rich extracellular matrix produced on the agar plates containing Congo red. d,e, Adult emergence patterns of P. stali infected with the representative E. coli lines, CmL05, GmL07, GmL02 and GmL04, in the original evolutionary experiments (d) and those in the confirmation experiments using frozen E. coli stocks (e). d, Pink lines represent the emergence rates of the original E. coli evolutionary lines, whereas the red lines in e represent the mean emergence rates (n = 3 biological replicates shown with brown dots) of the frozen E. coli stock infection experiments. c,e, The magenta and blue lines highlight ‘non-improved’ and ‘improved’ generations, respectively.
Extended Data Fig. 3. Effects of evolutionary E. coli lines on body size and colour of P. stali.
a, Evolutionary E. coli lines subjected to host’s body colour selection. Data of host’s body width are displayed by heat maps. Also see Fig. 3a. b, Evolutionary E. coli lines subjected to host’s growth speed selection. Data of host’s body width and colour are displayed by heat maps. Also see Fig. 3b.
Extended Data Fig. 4. Adult phenotypes of P. stali infected with the evolutionary E. coli lines CmL05, GmL07, GmL02 and GmL04.
a,b, Nymphal period. c,d, Nymphal period of the earliest adult females. e,f, Body colour. g,h, Thorax width. a,c,e,g, Phenotypes of the adult females used for inoculation to the next generation and preparation of glycerol stocks of the original evolutionary E. coli lines. b,d,f,h, Phenotypes of the adult insects inoculated with the frozen E. coli stocks. In b, f and h, line charts show mean values while dots indicate individual data points. Note that, corresponding to each original evolutionary E. coli line, three insect groups were inoculated with the frozen E. coli stock. In b, f and h, the yellow bands indicate the typical phenotypic ranges of the control insects infected with the original symbiont.
Microbial traits of mutualistic E. coli
In addition to colony size, shape and extracellular matrix on agar plates (Fig. 3c), the mutualistic E. coli lines CmL05 and GmL07 in culture exhibited distinct microbial traits in comparison with the original E. coli strains: slower growth rate; smaller cell size; loss of flagellar motility; and unstable cell shape (Fig. 4a–g and Supplementary Videos 2–4). Within the host insect, the evolutionary E. coli lines CmL05 and GmL07 showed significantly higher infection densities than the original E. coli strains (Fig. 4h and Extended Data Fig. 5). These observations revealed that mutualistic E. coli lines certainly have evolved a variety of ‘symbiont-like’ microbial traits.
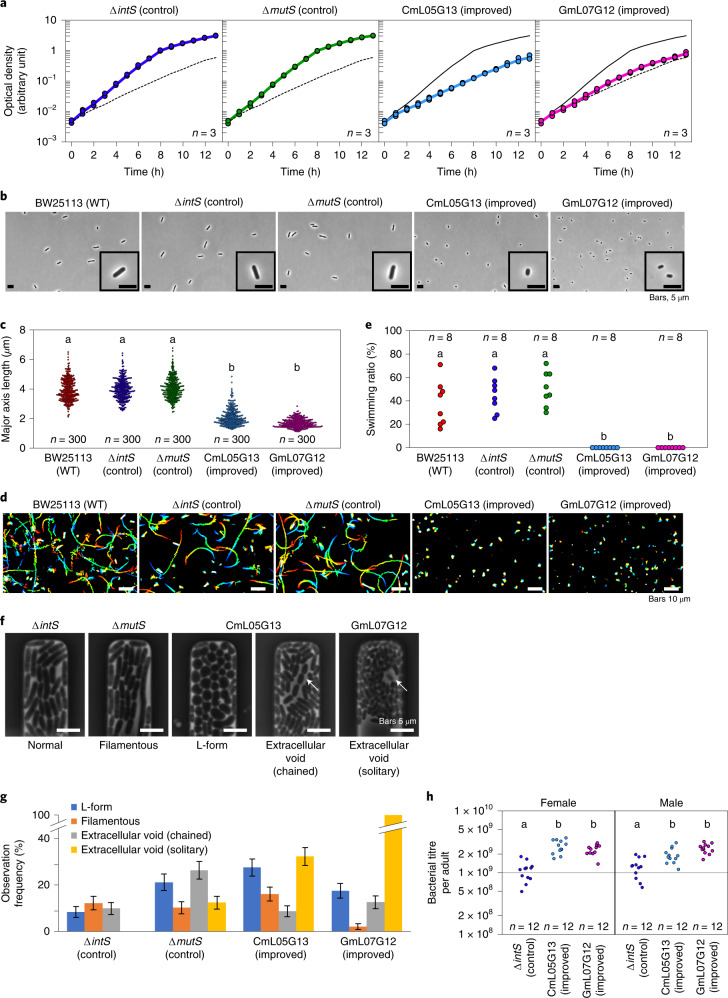
Fig. 4. Microbial traits of evolutionary E. coli lines CmL05 and GmL07 in comparison with the original E. coli strains BW25113, ΔintS and ΔmutS cultured in liquid medium.
a, Growth curves (three replicates each). The upper solid line is the trace of the ΔintS growth curve, whereas the lower dotted line is the trace of the CmL05 growth curve (Supplementary Video 2). b, Morphology of bacterial cells. c, Quantification of cell size in terms of major axis length. d, Motility of bacterial cells visualized by rainbow plot for 2 s (Supplementary Video 3). e, Quantification of bacterial motility in terms of the number of swimming cells per 100 cells observed. f, Characteristic cellular shape and growth mode in microfluidic channels. From left to right, the micrographs show the microchannels harbouring E. coli cells with normal rod-like shape (ΔintS), filamentation shape (ΔmutS), L form-like round shape (CmL05), extracellular void space and chained growth (CmL05) and extracellular void space and solitary growth (GmL07). The arrows indicate the cells showing the extracellular void space (Supplementary Video 4). g, Frequency of the microchannels in which E. coli cells exhibited characteristic cell shape and growth mode. The total numbers of microchannels observed in the time-lapse measurements (N) were 131 (ΔintS), 137 (ΔmutS), 149 (CmL05G13) and 143 (GmL07G12). The error bars represent the s.e. for the mean of binomially distributed samples, that is,, where P = C/N and C is the number of microchannels in which the cells with the indicated phenotype appeared. h, Bacterial titres in adult females 35 d after emergence in terms of ntpII gene copies per insect. a,c,e,h, The numbers of biological replicates are shown. c,e,h, Different alphabetical letters indicate statistically significant differences (two-sided pairwise Wilcoxon rank-sum test with Bonferroni correction: P < 0.05). The exact P values are provided with the source data.
Extended Data Fig. 5. FISH localization of the improved evolutionary E. coli lines CmL05G13 and GmL07G12 in P. stali.
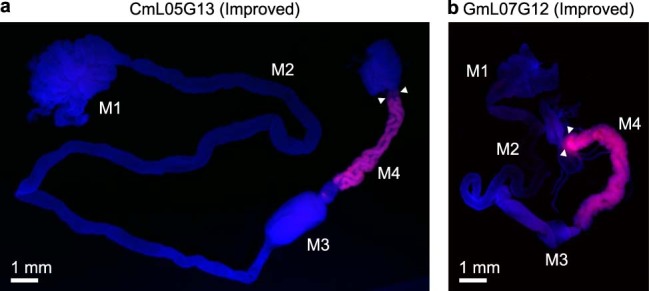
a, CmL05G17. b, GmLG12. FISH signals are localized to the midgut M4 region. Abbreviations: M1, M2, M3, and M4, midgut regions M1, M2, M3, and M4 (=symbiotic organ). Arrowheads indicate female-specific enlarged end crypts at the posterior end of the symbiotic organ, which are presumably involved in vertical symbiont transmission by storing bacteria-containing secretion49. The level adjustment without non-linear change is applied to the images.
Transcriptomics and genomics of mutualistic E. coli
An aliquot of the dissected symbiotic organ from each generation of the colour selection line CmL05 was subjected to RNA sequencing, from which E. coli-derived reads were extracted and analysed (Supplementary Table 1). Interestingly, the gene expression patterns of E. coli at generations 7–14 after the improvement of host phenotypes were separately clustered in contrast to those at generations 1–6 before the improvement (Fig. 5a). In the growth selection line GmL07, similarly, the gene expression patterns of E. coli at generations 2–12 after the improvement were distinct from that at generation 1 before the improvement and also from those of the other growth selection lines GmL02 and GmL04 in which the improvement of host phenotypes did not occur (Fig. 5b). These results suggested that the evolution of the mutualistic E. coli lines entails a specific and global change of gene expression patterns.
Fig. 5. Transcriptomics and genomics of evolutionary E. coli lines.
a,b, Clustering dendrograms and heatmaps based on gene expression levels across generations of evolutionary E. coli lines subjected to colour (left, 3,401 genes) (a) and growth selection (right, 3,360 genes) (b). The dendrograms represent the hierarchical clustering of the E. coli RNA-seq libraries. The grey and coloured areas depict non-improved and improved generations, respectively. c, Mutations identified in the genomes of CmL05 and GmL07 as coincident with the improvement of host phenotypes. SNV, single nucleotide variant. d, Candidate mutations disrupting the carbon catabolite repression: a frame shift mutation in cyaA of CmL05 (top) and a non-synonymous mutation causing change from leucine to proline at a functionally important cAMP binding domain in crp of GmL07 (bottom). e, Schematic presentation as to how the carbon catabolite repression pathway is disrupted by the cyaA and crp mutations.
In the growth selection line GmL07 and colour selection line CmL05, we surveyed differentially expressed genes before and after the improvement of host phenotypes, which identified 193 commonly downregulated genes and 95 commonly upregulated genes across GmL07 and CmL05 (Extended Data Fig. 6a,b). The commonly downregulated genes contained a number of metabolism-related genes, such as transporter genes for sugars and other nutrients like maltose, ribose, galactitol, trehalose, mannose and branched chain amino acids, glyoxylate bypass genes, fatty acid degradation genes and others. Notably, core genes involved in extracellular matrix (= Curli fimbriae) production were significantly downregulated after the improvement (Extended Data Fig. 6c), which likely accounted for the altered colony morphology of E. coli associated with the improvement of host phenotypes (Fig. 3c).
Extended Data Fig. 6. Gene expression changes of evolutionary E. coli lines GmL07 and CmL05 before and after improvement of host phenotypes.
a,b, Venn diagrams showing down-regulated genes (a) and up-regulated genes (b) after the improvement of host phenotypes. c, Expression levels of genes involved in extracellular matrix (Curli fimbriae) production before and after the improvement of host phenotypes. Asterisks indicate statistically significant differences (FDR q-value < 0.01). The numbers of the biological replicates and exact FDR q-values are provided with the source data.
The improved lines CmL05 and GmL07 and the non-improved lines GmL02 and GmL04 were subjected to genome sequencing throughout the evolutionary course (Supplementary Table 2), which identified many mutations accumulated in the hypermutating E. coli lines (Extended Data Fig. 7 and Supplementary Table 3). In an attempt to identify candidate mutations that are correlated with the improvement of the host’s phenotype, we surveyed the mutations that appeared at generation seven of CmL05 and then fixed, which yielded seven candidate genes, and also the mutations that appeared at generation two of GmL07 and then fixed, which yielded nine candidate genes (Fig. 5c).
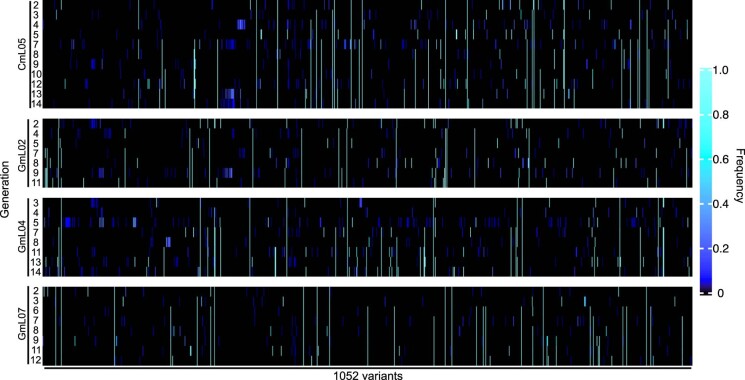
Extended Data Fig. 7. Mutations in the genomes of evolutionary E. coli lines CmL05, GmL02, GmL04 and GmL07 in the experimental evolutionary course.
Frequencies of 1,052 variants identified in the experimental evolution lines and generations are colour-coded. Vertical axis represents the generations of the experimental evolution lines whereas horizontal axis represents an array of 1,052 variants.
Disrupted carbon catabolite repression pathway in mutualistic E. coli
Of these candidates, we focused on a frameshift mutation that disrupted adenylate cyclase (CyaA) in CmL05 and a non-synonymous mutation that changed a functionally important cAMP binding site of the cAMP receptor protein (Crp) from leucine to proline in GmL07 (Fig. 5d). Despite their independent origins in distinct evolutionary lines, CyaA and Crp are pivotal components of the same global metabolic regulatory system, the carbon catabolite repression (CCR) pathway, operating in diverse bacteria including E. coli18,19 (Fig. 5e). With sufficient availability of glucose as the primary carbon source for E. coli, the CCR components are subjected to glucose-mediated suppression, being in an unphosphorylated form incapable of activating CyaA, by which the intracellular cAMP is maintained at a low level (Extended Data Fig. 8a). When glucose is used up, the glucose-mediated suppression is released, by which the CCR components are phosphorylated and activate CyaA, which results in an elevated intracellular cAMP level and promotes allosteric binding of cAMP to Crp. The resultant global transcriptional regulator Crp-cAMP activates and/or represses several hundreds of operons throughout the bacterial genome, referred to as the Crp-cAMP regulon, by which the bacterial metabolic pathways are switched to exploit other carbon sources for adaptation to nutrient-deficient and/or high bacterial density conditions (Extended Data Fig. 8b)20,21. According to RegulonDB22, the Crp-cAMP regulon of E. coli consists of some 390 upregulated genes and 80 downregulated genes (Extended Data Fig. 8c), which are involved in, for example, upregulation of transporters and catabolic enzymes for non-glucose sugars23, quorum sensing induction24 and production of extracellular matrix25.
Extended Data Fig. 8. Carbon catabolite repression (CCR) pathway and Crp-cAMP regulon of E. coli.

a, CCR pathway repressed in the presence of glucose. b, CCR pathway activated in the absence of glucose. c, Number of genes constituting the Crp-cAMP regulon of E. coli estimated by RegulonDB22.
Both the cyaA mutation in CmL05 and the crp mutation in GmL07 are disruptive of the CCR pathway. Considering that E. coli cells are packed in the host symbiotic organ very densely (Fig. 1k and Extended Data Fig. 2i,k), it seems likely that the symbiotic E. coli may be under a nutrient-limited condition in the host insect, at least locally. If so, it is expected that, in the evolutionary E. coli lines, while the Crp-cAMP transcriptional regulator was activated before the mutations occurred, the activation was disabled after the mutations occurred. Notably, of 193 genes commonly downregulated after the yaA mutation in CmL05 and the crp mutation in GmL07, 55 genes were reported as being activated by Crp-cAMP (Extended Data Fig. 9a). These genes, which are expected to be silenced on disruption of the CCR system, were significantly downregulated in CmL05 and GmL07, which represented many transporter genes for non-glucose sugars, carbohydrate metabolism genes, quorum sensing genes, extracellular matrix production genes, transcription factor genes and others (Extended Data Fig. 9b–i).
Extended Data Fig. 9. Genes commonly down-regulated in GmL07 and CmL05 after the improvement of host phenotypes, and also down-regulated by disruption of Crp-cAMP in E. coli.
a, Venn diagram showing the commonly down-regulated genes. b–i, Expression levels of the commonly down-regulated genes in GmL07 and CmL05 after the improvement of host phenotypes. (b) Transporter genes. (c) Carbohydrate metabolism genes. (d) Amino acid metabolism genes. (e) Lipid metabolism genes. (f) Quorum sensing genes. (g) Transcription factor genes. (h) Biofilm (= Curli fimbriae) formation genes. (i) Other genes. The inset figure at the bottom right represents the explanations of the elements in the plots. The numbers of the biological replicates and exact FDR q-values are provided with the source data.
Disrupted CCR genes make E. coli an insect mutualist
To test whether these mutations are involved in the mutualistic traits of the evolutionary E. coli lines, we prepared E.coli strains that carry the mutations under the wild-type (WT) genetic background: the strain ΔcyaA in which the cyaA gene is disrupted; and the strain crp221T>C whose crp gene was engineered to carry the leucine-proline replacement at the cAMP binding site. Both mutant E. coli strains exhibited small and convex colonies with little extracellular matrix, somewhat slower growth rate, smaller cell size and loss of flagellar motility (Fig. 6a and Extended Data Fig. 10a–e), which were generally reminiscent of the characteristic traits of the improved evolutionary E. coli lines CmL05 and GmL07 (Fig. 3c and Fig. 4a–e). When the mutant E. coli strains were inoculated to sterilized newborn nymphs of P. stali, both the ΔcyaA- and crp221T>C-infected insects exhibited remarkably high adult emergence rates, which were comparable to the insects infected with the improved evolutionary E. coli lines and were significantly higher than the insects infected with the control E. coli strains (Fig. 6b). Moreover, the ΔcyaA- and crp221T>C-infected insects were greenish in colour, which were comparable to the greenish insects infected with the improved evolutionary E. coli lines and distinct from the dwarf brown insects infected with the control E. coli strains (Fig. 6c). On the other hand, the infection densities of crp221T>C and ΔcyaA were not comparable to those of the improved evolutionary E. coli lines (Extended Data Fig. 10f). These results demonstrated that, strikingly, the single mutations that disrupt the CCR global regulator system make E. coli mutualistic to the host insect P. stali.
Fig. 6. Single mutations disrupting CCR make E. coli mutualistic to P. stali.

a, Small, convex and white colonies of ΔcyaA and crp221T>C. b, Adult emergence rates of P. stali infected with ΔcyaA and crp221T>C. The numbers of biological replicates are shown in the figure. Different alphabetical letters indicate statistically significant differences (two-sided pairwise Wilcoxon rank-sum test with Bonferroni correction: P < 0.05). The exact P values are provided with the source data. c, Adult insects infected with ΔcyaA and crp221T>C, which are larger in size and green in colour in comparison with those infected with control ΔintS. Note that the revertant of crp221T>C, crp-rev, exhibits ΔintS-like inferior phenotypes.
Extended Data Fig. 10. Phenotypic traits of ∆cyaA and crp221T>C mutants of E. coli.
a, Growth curves (3 replicates each). Upper solid line is the trace of ΔintS growth curve, whereas lower dotted line is the trace of CmL05 growth curve. b, Morphology of bacterial cells. c, Quantification of cell size in terms of major axis length. d, Motility of bacterial cells visualized by rainbow plot for 2 sec. e, Quantification of bacterial motility in terms of number of swimming cells per 100 cells observed. f, Bacterial titres in adult females 35 days after emergence in terms of ntpII gene copies per insect. In a, c, e and f, the numbers of biological replicates are shown in the plots. In c, e and f, different alphabetical letters indicate statistically significant differences (two-sided pairwise Wilcoxon rank sum test with Bonferroni correction: P < 0.05). The exact P-values are provided with the source data.
Discussion
We established an experimental insect-E. coli symbiotic system in which the model bacterium is localized to the host symbiotic organ, transmissible to the host offspring vertically and supportive of host survival, although not comparable to the original symbiont. By infecting and passaging a hypermutating E. coli strain with the host insect trans-generationally, several evolutionary lines rapidly developed improved adult emergence and body colour, realizing recurrent evolution of mutualism in the laboratory. Strikingly, the evolution of E. coli into insect mutualist was ascribed to single mutations that convergently disrupted the bacterial CCR pathway, uncovering unexpected involvement of the nutrient-responsive global transcriptional regulator in the establishment of symbiosis.
Our finding sheds light on the evolvability of symbiosis—elaborate mutualistic symbiosis can evolve much more easily and rapidly than conventionally envisaged. We suggest the possibility that the inactivation of the CCR global regulator may represent a pivotal evolutionary step at an early stage of symbiosis. Densely packed in the symbiotic organ, symbiotic bacteria are expected to constantly suffer nutritional shortage and activate the CCR pathway in vain, which may incur substantial metabolic cost and destabilize the symbiotic association. In this context, the disruption of the CCR pathway should benefit and stabilize symbiosis. Our finding may also be relevant to the general evolutionary trend of symbiont genomes towards size reduction26,27 and lack of transcription factors28,29. The disruption of the CCR pathway causes silencing of otherwise activated about 400 genes under the Crp-cAMP regulon22, which accounts for about 10% of the whole E. coli genome and provides potential targets for gene disruption, IS amplification and insertion, intragenomic recombination and reductive genome evolution. We propose that, although speculative, inactivation of transcriptional regulators and genome size reduction might have concurrently proceeded in this way during the symbiont genome evolution.
On the other hand, we found that the original symbiont of P. stali retained the complete CCR pathway genes, although the uncultivable symbiont accumulated hundreds of pseudogenes in the genome10. Plausibly, the CCR disruption observed in the evolutionary E. coli lines may represent one of multiple possible evolutionary trajectories towards symbiosis with P. stali and we expect that other mechanisms would be uncovered by larger scale and longer-span evolutionary experiments.
The P. stali-E. coli experimental symbiotic system will open a window to directly observe and analyse the evolutionary processes and mechanisms of mutualistic symbiosis in real time. E. coli is among the best understood cellular organisms30, whose 4.5–5.5 megabase genome encodes over 4,000 genes and around 70% of them carry functional information31,32. Laboratory evolution of mutualism using such a model bacterium with ample technological and genetic resources will lead to understanding of previously unapproachable evolutionary aspects of symbiosis. Considering that E. coli represents a universal component of the gut microbiome of human, mouse and other vertebrates33,34, the insect-E. coli system in combination with the germ-free mouse-E. coli experimental evolution systems35,36 would enable us to pursue not only the differences but also the commonality underpinning the mechanisms of gut symbiosis across vertebrates and invertebrates.
Methods
Insect and bacterial strains used in this study
An inbred laboratory strain of the brown-winged green stinkbug P. stali was established from several adult insects collected at Tsukuba, Ibaraki, Japan in September 2012 and has been maintained in the laboratory for years. This strain is associated with an essential and uncultivable gut symbiont Pantoea sp. A10 in the posterior midgut region specialized as the symbiotic organ (Fig. 1 and Extended Data Fig. 2). The insects were reared on raw peanuts, soya beans and water containing 0.05% ascorbic acid (Merck) at 25 ± 1 °C and 50 ± 5% relative humidity under a long-day regime of 16 h light and 8 h dark. The E. coli strains and mutants used in this study are listed in Supplementary Table 4. The mutants ∆intS, ∆mutS and crp221T>C were generated as described later.
Construction of E. coli mutants
The E. coli mutant ∆intS was established by replacing the intS gene of E. coli BW25113 with the nptII gene that confers kanamycin resistance by λ-Red homologous recombination using the pRed/ET plasmid (Gene Bridges GmbH). The E. coli mutant ∆mutS was established by replacing the mutS gene of ∆intS with the FRT-Cm-FRT cassette (Gene Bridges GmbH) by λ-Red homologous recombination; then CmR was eliminated by Flp-FRT recombination. The E. coli mutant crp221T>C was established from ∆intS by replacing the 221st nucleotide T of the WT crp gene with C, which changed the 74th amino acid leucine of the Crp protein to proline. This replacement was introduced by the MAGE method37,38 with a 90-mer DNA oligonucleotide (5′-taaagaaatg-atcctctcct-atctgaatca-gggtgatttt-attggcgaac-Cgggcctgtt-tgaagagggc-caggaacgta-gcgcatgggt-3′) whose 1st to 4th nucleotides were phosphorothioated. To establish the revertant of crp221T>C (crp221C>T), a 90-mer oligonucleotide (5′-taaagaaatg-atcctctcct-atctgaatca-gggtgatttt-attggTgaac-Tgggcctgtt-tgaagagggc-caggaacgta-gcgcatgggt-3′) was used. This oligonucleotide was designed not only to revert the 221st nucleotide but also replace the 216th nucleotide C to T to introduce a synonymous mutation, which allowed us to discriminate the resultant revertant clone from the unmodified WT.
Preparation of symbiont-free nymphs by surface sterilization of eggs
Egg clutches produced by the stock culture of P. stali were soaked in 4% formaldehyde for 10 min, rinsed with sterilized water several times and kept in sterilized plastic boxes until use. While this treatment does not affect hatchability and survival of the eggs, newborn nymphs fail to acquire the symbiotic bacteria and become symbiont-free11.
Experimental evolution of P. stali-E. coli artificial symbiotic system
Evolutionary experiments in this study consisted of, for each evolutionary P. stali line, (1) preparation of an inoculum either from the E. coli culture of ΔmutS or ΔintS (only G1) or from an adult female of the previous generation (from G2 and on), (2) oral administration of the inoculum to symbiont-free nymphs, (3) rearing of the nymphs either to their adulthood or death, (4) selection of an adult female for inoculation to the next generation, (5) contamination check of the selected adult female, (6) preparation of an inoculum and a glycerol stock from the symbiotic organ dissected from the selected female and (6) morphological measurements of all adult insects obtained.
Either diluted E. coli culture (2.5 ml adjusted to OD600 = 0.1) or homogenate of the symbiotic organ dissected from a selected female of the previous generation (2.5 ml containing 1/2 organ equivalent) was soaked in a cotton pad and orally administered to around 84 symbiont-free hatchlings derived from 6 surface-sterilized egg masses, by making use of the nymphal behaviour that, after egg surface probing for about 30 min and resting for around 1 d, they take water without feeding and moult to second instar in a few days10,11. These nymphs were reared on sterilized peanuts, soya beans and ascorbic acid water as described previously11. In the evolutionary experiments, two selection schemes, growth and colour selection, were conducted (Fig. 2). In the growth selection lines (GmL for hypermutating ΔmutS lines; GiL for non-mutating ΔintS lines), the first-emerged adult female was subjected to dissection of the symbiotic organ for inoculation to the next generation as well as freeze storing. In the colour selection lines (CmL for the ΔmutS lines; CiL for the ΔintS lines), adult females were collected for 35 d after hatching or until at least 1 adult female emerged. These adult females were anaesthetized on ice and photographed from the ventral side using a digital camera. Their body colour was measured using the image analysing software Natsumushi v.1.10 (ref. 39); the adult female that exhibited the highest hue angle (= greenness) was subjected to dissection of the symbiotic organ for inoculation to the next generation as well as freeze-storing.
The symbiotic organ of the selected female was dissected in PBS (0.8% NaCl, 0.02% KCl, 0.115% Na2HPO4, 0.02% KH2PO4, pH 7.4), rinsed with 70% ethanol and homogenized in 200 µl sterile water. Of the 200-µl homogenate, 5 µl was used for contamination checking by quantitative PCR (qPCR). The number of E. coli genome copies was evaluated in terms of kanamycin resistance gene copies, which are present in the ΔintS and ΔmutS mutants but absent in WT E. coli and other bacteria. The number of total bacterial genome copies was evaluated based on bacterial 16S ribosomal RNA gene copies. When the former E. coli genome copy number was approximately the same as the latter bacterial genome copy number, the specimen was diagnosed as free of contamination. When the specimen was diagnosed as contaminated, the next best female was used. For qPCR, the primers Tn5-1789F (5′-TGCTCGACGTTGTCACTGAA-3′) and Tn5-1879R (5′-GCAGGAGCAAGGTGAGATGA-3′) were used for the kanamycin resistance gene, while the primers 16S-967F (5′-CAACGCGAAGAACCTTACC-3′) and 16S-1046R (5′-CGACAGCCATGCANCACCT-3′) were used for the bacterial 16S rRNA gene. The PCR reaction was performed using the Brilliant PCR Mix (Agilent Technologies). The standard curve was drawn using serially diluted ∆intS genomic DNA, which contains one kanamycin gene copy and seven 16S rRNA gene copies per genome. The thermal profile was the initial denaturation at 95 °C for 3 min followed by 40 cycles of incubation at 95 °C for 5 s and at 60 °C for 10 s. To confirm specific amplification, melting curve analysis was also included. The reaction was conducted on Mx3000p (Agilent Technologies). While 100 µl of the homogenate of the female symbiotic organ diagnosed as free of contamination was used as the inoculum to the next generation, the remaining homogenate (approximately 95 µl) was mixed with an equal volume of 20% glycerol and stored at −80 °C.
Inoculation of E. coli frozen stocks to P. stali
The frozen glycerol stocks were thawed; 50 μl was taken and diluted with sterile water to 3 ml. Each of 3 replicates of around 84 symbiont-free hatchlings from 6 surface-sterilized egg masses was fed with 1 ml inoculum soaked in a cotton pad as described above. The symbionts A and ΔmutS were included in the evaluation as positive and negative controls, respectively. Adult emergence of the insects was monitored for 50 d after hatching. All the adult insects were photographed from the dorsal side with a digital camera; the hue angle (= greenness) of the scutellum and thorax width were measured with ImageJ v.1.5340. For the subsequent RNA sequencing (RNA-seq) analyses and resequencing of E. coli genomes, the symbiotic organs were isolated from the adult insects and homogenized in 100 µl PBS. Of the 100 µl homogenate, 50 µl was subjected to RNA-seq and the remaining 50 µl was used for genome resequencing.
RNA-seq analyses
The homogenate of the symbiotic organ was subjected to total RNA extraction using RNAiso (Takara Bio) and the RNeasy Mini Kit (QIAGEN). Then, rRNAs of both insect and bacterial origins were removed from the total RNA samples using the Ribo-Zero Gold rRNA Removal Kit (Epidemiology) (Illumina). The rRNA-depleted RNAs were converted to paired-end libraries using the Sure Select Strand Specific RNA Kit (Agilent Technologies) or TruSeq RNA Library Prep Kit v2 (Illumina) (Supplementary Table 1). The libraries were sequenced with HiSeq 3000 or HiSeq X (Illumina). The obtained sequences were trimmed, mapped to the E. coli BW25113 genome sequence (accession no. NZ_CP009273) and read-counted with CLC Genomics Workbench 10.0 (QIAGEN). Normalizations and differential expression analyses were conducted with EdgeR v.3.32.1 (ref. 41). Complex Heatmap v.2.10.0 (ref. 42) was used for the clustering analyses and to draw the heatmaps of the RNA-seq libraries.
Genome resequencing and detection of structural changes
DNA samples were extracted from the homogenates of the symbiotic organ using the QIAamp DNA Mini Kit (QIAGEN). The extracted DNAs were converted to paired-end libraries using the Nextera XT DNA Library Prep Kit (Illumina) and the libraries were sequenced with MiSeq system (Illumina). CLC Genomic Workbench v.10.0 was used to detect the E. coli genome variants that emerged during the evolutionary experiments. The heatmaps of the variant frequency data were drawn using Complex Heatmap v.2.10.0 (ref. 42).
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) analyses were performed essentially as described by Koga et al.43. The whole insect bodies or isolated digestive tracts were fixed with PBS containing 4% formaldehyde (Fujifilm). The fixed samples were embedded in Technovit 8100 (Kulzer) and processed into 2-µm tissue sections using a rotary microtome RM2255 (Leica Biosystems). The Alexa Fluor 555-labelled oligonucleotide probes Eco934 (5′-CATGCTCCACCGCTTGTG-3′) and SymAC89R (5′-GCAAGCTCTTCTGTGCTGCC-3′) were used to detect E. coli and symbiont A, respectively14. Host nuclei were counterstained with 4′,6-diamidino-2-phenylindole (Dojindo). The hybridized specimens were observed using a fluorescence dissection microscope M165FC with Leica Application Suite v.4.13.0 (Leica Microsystems), an epifluorescence microscope DM6B with Leica Application Suite X v.3.7.1.21655 (Leica Microsystems) and a laser confocal microscope LSM700 with ZEN 2011 v.7.0.7.0 (ZEISS). For Fig. 1d and Extended Data Fig. 2j, panoramic images were constructed by merging multiple images using Affinity Photo v.1.10.5 (Serif Ltd).
Infection of E. coli mutants and effects on host phenotypes
E. coli mutants were cultured, diluted and orally administrated to symbiont-free newborn nymphs of P. stali as described above. The insects were reared to monitor their adult emergence for 42 d after hatching. The dorsal images of the adults were taken with an image scanner GT-X830 (Epson) and the hue angle of the scutellum and thorax width was measured and analysed using the Natsumushi software v.1.1039. P. stali harbouring the original symbiont Pantoea sp. A was also included as a reference. As for the adult females infected with E. coli, bacterial titres in the symbiotic organs were measured by qPCR. The KAPA SYBR FAST qPCR Kit (Roche) and Tn5-1789F and Tn5-1879R primer sets were used for quantification. The standard curves were drawn using serially diluted pT7Blue (Takara Bio) plasmid carrying a kanamycin resistance gene fragment. The qPCR reactions were conducted on the Light Cycler 96 (Roche).
Measurement of E. coli phenotypes
To inspect colony morphology and extracellular matrix production, E. coli cultures were spread onto lysogeny broth agar plates containing 80 µg ml−1 Congo Red (Merck) and incubated at 25 °C for 3 d. Colonies formed on the plate were photographed by using a scanner GT-X850 and/or dissection microscope S9i (Leica Microsystems).
For growth curve measurements, each glycerol stock of E. coli was inoculated to 2 ml lysogeny broth (Becton Dickinson) and incubated at 25 °C for 16 h with shaking at 200 r.p.m. The cell culture was diluted to OD600 = 0.005 in 25 ml lysogeny broth and incubated at 25 °C with shaking at 200 r.p.m. From the bacterial culture, 120 µl of cell suspension was sampled every hour and samples were subjected to the measurement of OD600 using a spectrometer UV-1800 (Shimadzu).
For time-lapse analyses of growth and morphology of individual E. coli cells, two types of microfluidic devices were used. One type was a microfluidic device where bacterial cells were enclosed in microchambers etched on a glass coverslip. A cellulose membrane was attached to a coverslip via biotin-streptavidin binding, on which the microchambers were created as described elsewhere44,45. Another type was a microfluidic device made of polydimethylsiloxane with a channel structure similar to Mother Machine as described by Wang et al.46 (Fig. 4f). The width of the cell observation channels in this device was 9 µm, which was broader than that of the Mother Machine; thus, each cell observation channel could harbour 30–70 individual E. coli cells depending on cell size. E. coli cells in the exponential phase were introduced into both types of microfluidic devices and observed under a Nikon Ti-E microscope equipped with an ORCA-fusion camera (Hamamatsu Photonics). In the time-lapse measurements, phase-contrast images were acquired with a 100× oil immersion objective lens (plan apochromat, numerical aperture 1.45) at an interval of 3 min, in which 50–100 XY positions were simultaneously observed. The microscope was controlled from a computer using Micro-Manager v.4. In the microchamber device measurements, lysogeny broth was supplemented with 0.1% bovine serum albumin and 0.02% Tween-80 to suppress cell adhesion and was introduced into the devices at a flow rate of 2 ml h−1.
For measurements of size and flagellar motility, E. coli cells were grown in lysogeny broth medium with shaking at 25 °C to around OD600 = 2.0, observed under a phase-contrast microscope IX71 (Olympus), recorded by a charge-coupled device camera DMK33UP5000.WG (The Imaging Source) at 30 frames per second and analysed using ImageJ v.1.53 (ref. 40) and IGOR Pro 8.02J (WaveMetrics). The cell size data were measured for six individual cultures. The swimming ratio data were obtained as the number of swimming cells in 100 cells from 8 individual cultures.
Statistics and reproducibility
Statistical analyses were conducted with R v.4.1.2 (ref. 47) and RStudio48. R was also used to plot the data. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications10,11,14. All data points were plotted as long as applicable. Data distribution was assumed to be normal but this was not formally tested. For Figs. 1, 4 and 6, and Extended Data Figs. 1 and 10, the numbers of biological replicates are shown in the figures. Exact P values are provided with the source data. For Fig. 1a–d,f,h–k,m and Extended Data Figs. 2 and 5, at least two replicate analyses were conducted, and all replicates gave essentially the same results.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Supplementary Tables 1–4.
Newborn nymphs of P. stali probing the eggshell to acquire the symbiotic bacteria.
Slow growth of the evolutionary E. coli lines GmL07 and CmL05 at the single-cell level.
Motility of the WT E. coli strain BW25113, the control E. coli strains ΔιntS and ΔmutS and the improved evolutionary E. coli lines GmL07 and CmL05.
Characteristic cell shapes and growth modes of E. coli cells in microfluidic channels.
Acknowledgements
We thank U. Asaga, S. Kimura, J. Makino and T. Matsushita for insect rearing and technical assistance. This study was supported by the Japan Science and Technology Agency ERATO grant nos. JPMJER1803 and JPMJER1902 (T.F., C.F., Y.W. and R.K.) and the Japan Society for the Promotion of Science (JSPS) KAKENHI grant no. JP25221107 (T.F. and R.K.). Genome sequencing and analyses were supported by the JSPS KAKENHI grant no. JP16H06279.
Extended data
Source data
Numeric source data for Fig. 1e,l.
Numeric source data for Fig. 3a,b,d,e.
Numeric source data for Fig. 4a,c,e,g,h and statistical source data for Fig. 4.
Numeric source data for Fig. 5a,b,c.
Numeric source data for Fig. 6b and statistical source data for Fig. 6.
Numeric source data for Extended Data Fig. 1a,b,c.
Numeric source data for Extended Data Fig. 3a,b.
Numeric source data for Extended Data Fig. 4a,b,c,d,e,f,g,h.
Numeric source data for Extended Data Fig. 6a,b,c and statistical source data for Extended Data Fig. 6.
Numeric source data for Extended Data Fig. 7.
Numeric source data for Extended Data Fig. 8c.
Numeric source data for Extended Data Fig. 9a–i and statistical source data for Extended Data Fig. 9.
Numeric source data for Extended Data Fig. 10a,c,e,f and statistical source data for Extended Data Fig. 10.
Author contributions
R.K. and T.F. conceived the project and designed the experiments. R.K., M. Moriyama, N.O.-T. and Y.N. performed the insect-E. coli evolutionary experiments. R.K., M. Moriyama, N.O.-T., Y.I., H.T., Y.N. and T. Hosokawa analysed the insect phenotypes. R.K., M. Mizutani, K.O., R.O. and Y.W. analysed the E. coli phenotypes. R.K., M. Moriyama, N.O.-T., Y.G. and T. Hayashi performed the genome sequencing and analyses. M. Moriyama, R.K., N.O.-T., M.S. and Y.S. conducted the RNA sequencing and analyses. R.K., H.T., S.S. and C.F. designed and generated the hypermutating and other E. coli strains. T.F. wrote the article with input from all the other authors.
Peer review
Peer review information
Nature Microbiology thanks Martin Kaltenpoth, John McCutcheon and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
All RNA and DNA sequencing data produced in this study were deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (Supplementary Tables 1 and 2). The data have been deposited with links to BioProject accession no. PRJDB5544 in the DDBJ BioProject database. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ryuichi Koga, Email: r-koga@aist.go.jp.
Takema Fukatsu, Email: t-fukatsu@aist.go.jp.
Extended data
is available for this paper at 10.1038/s41564-022-01179-9.
Supplementary information
The online version contains supplementary material available at 10.1038/s41564-022-01179-9.
References
- 1.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert SF, Bosch TCG, Ledón-Rettig C. Eco-Evo-Devo: developmental symbiosis and developmental plasticity as evolutionary agents. Nat. Rev. Genet. 2015;16:611–622. doi: 10.1038/nrg3982. [DOI] [PubMed] [Google Scholar]
- 3.Hoang KL, Moran LT, Gerardo NM. Experimental evolution as an underutilized tool for studying beneficial animal–microbe interactions. Front. Microbiol. 2016;7:1444. doi: 10.3389/fmicb.2016.01444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King KC, et al. Rapid evolution of microbe-mediated protection against pathogens in a worm host. ISME J. 2016;10:1915–1924. doi: 10.1038/ismej.2015.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pankey MS, et al. Host-selected mutations converging on a global regulator drive an adaptive leap towards symbiosis in bacteria. eLife. 2017;6:e24414. doi: 10.7554/eLife.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tso GHW, et al. Experimental evolution of a fungal pathogen into a gut symbiont. Science. 2018;362:589–595. doi: 10.1126/science.aat0537. [DOI] [PubMed] [Google Scholar]
- 7.Robinson CD, et al. Experimental bacterial adaptation to the zebrafish gut reveals a primary role for immigration. PLoS Biol. 2018;16:e2006893. doi: 10.1371/journal.pbio.2006893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta AP, et al. Engineering yeast endosymbionts as a step toward the evolution of mitochondria. Proc. Natl Acad. Sci. USA. 2018;115:11769–11801. doi: 10.1073/pnas.1813143115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drew GC, Stevens EJ, King KC. Microbial evolution and transitions along the parasite–mutualist continuum. Nat. Rev. Microbiol. 2021;19:623–638. doi: 10.1038/s41579-021-00550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosokawa T, et al. Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations. Nat. Microbiol. 2016;1:15011. doi: 10.1038/nmicrobiol.2015.11. [DOI] [PubMed] [Google Scholar]
- 11.Nishide Y, et al. Aseptic rearing procedure for the stinkbug Plautia stali (Hemiptera: Pentatomidae) by sterilizing food-derived bacterial contaminants. Appl. Entomol. Zool. 2017;52:407–415. doi: 10.1007/s13355-017-0495-y. [DOI] [Google Scholar]
- 12.Nikoh N, Hosokawa T, Oshima K, Hattori M, Fukatsu T. Reductive evolution of bacterial genome in insect gut environment. Genome Biol. Evol. 2011;3:702–714. doi: 10.1093/gbe/evr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salem H, et al. Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc. Biol. Sci. 2014;281:20141838. doi: 10.1098/rspb.2014.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oishi S, Moriyama M, Koga R, Fukatsu T. Morphogenesis and development of midgut symbiotic organ of the stinkbug Plautia stali (Hemiptera: Pentatomidae) Zoological Lett. 2019;5:16. doi: 10.1186/s40851-019-0134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosokawa T, Matsuura Y, Kikuchi Y, Fukatsu T. Recurrent evolution of gut symbiotic bacteria in pentatomid stinkbugs. Zoological Lett. 2016;2:24. doi: 10.1186/s40851-016-0061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otero-Bravo A, Sabree ZL. Multiple concurrent and convergent stages of genome reduction in bacterial symbionts across a stink bug family. Sci. Rep. 2021;11:7731. doi: 10.1038/s41598-021-86574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giraud A, et al. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- 18.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 20.Martínez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos-Zavaleta A, et al. RegulonDB v 10.5: tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 2019;47:D212–D220. doi: 10.1093/nar/gky1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimada T, Fujita N, Yamamoto K, Ishihama A. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS ONE. 2011;6:e20081. doi: 10.1371/journal.pone.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 2005;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller CM, et al. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog. 2009;5:1000303. doi: 10.1371/journal.ppat.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 27.McCutcheon JP, Boyd BM, Dale C. The life of an insect endosymbiont from the cradle to the grave. Curr. Biol. 2019;29:R485–R495. doi: 10.1016/j.cub.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 28.Moran NA, Dunbar HE, Wilcox JL. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J. Bacteriol. 2005;187:4229–4237. doi: 10.1128/JB.187.12.4229-4237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Blount ZD. The unexhausted potential of E. coli. eLife. 2015;4:e05826. doi: 10.7554/eLife.05826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol. Biol. Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- 32.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walk ST, et al. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 2009;75:6534–6544. doi: 10.1128/AEM.01262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 35.Giraud A, et al. Dissecting the genetic components of adaptation of Escherichia coli to the mouse gut. PLoS Genet. 2008;4:e2. doi: 10.1371/journal.pgen.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barroso-Batista J, et al. Specific eco-evolutionary contexts in the mouse gut reveal Escherichia coli metabolic versatility. Curr. Biol. 2020;30:1049–1062. doi: 10.1016/j.cub.2020.01.050. [DOI] [PubMed] [Google Scholar]
- 37.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nyerges Á, et al. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proc. Natl Acad. Sci. USA. 2016;113:2502–2507. doi: 10.1073/pnas.1520040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanahashi M, Fukatsu T. Natsumushi: image measuring software for entomological studies. Entomol. Sci. 2018;21:347–360. doi: 10.1111/ens.12315. [DOI] [Google Scholar]
- 40.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 43.Koga R, Tsuchida T, Fukatsu T. Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl. Entomol. Zool. 2009;44:281–291. doi: 10.1303/aez.2009.281. [DOI] [Google Scholar]
- 44.Inoue I, Wakamoto Y, Moriguchi H, Okano K, Yasuda K. On-chip culture system for observation of isolated individual cells. Lab Chip. 2001;1:50–55. doi: 10.1039/b103931h. [DOI] [PubMed] [Google Scholar]
- 45.Hashimoto M, et al. Noise-driven growth rate gain in clonal cellular populations. Proc. Natl Acad. Sci. USA. 2016;113:3251–3256. doi: 10.1073/pnas.1519412113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang P, et al. Robust growth of Escherichia coli. Curr. Biol. 2010;20:1099–1103. doi: 10.1016/j.cub.2010.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2021).
- 48.RStudio Team. RStudio: Integrated Development Environment for R (RStudio, 2020).
- 49.Hayashi T, Hosokawa T, Meng X-Y, Koga R, Fukatsu T. Female-specific specialization of a posterior end region of the midgut symbiotic organ in Plautia splendens and allied stinkbugs. Appl. Environ. Microbiol. 2015;81:2603–2611. doi: 10.1128/AEM.04057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1–4.
Newborn nymphs of P. stali probing the eggshell to acquire the symbiotic bacteria.
Slow growth of the evolutionary E. coli lines GmL07 and CmL05 at the single-cell level.
Motility of the WT E. coli strain BW25113, the control E. coli strains ΔιntS and ΔmutS and the improved evolutionary E. coli lines GmL07 and CmL05.
Characteristic cell shapes and growth modes of E. coli cells in microfluidic channels.
Data Availability Statement
All RNA and DNA sequencing data produced in this study were deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (Supplementary Tables 1 and 2). The data have been deposited with links to BioProject accession no. PRJDB5544 in the DDBJ BioProject database. Source data are provided with this paper.